Abstract
Summary
Spatially resolved transcriptomics promises to increase our understanding of the tumor microenvironment and improve cancer prognosis and therapies. Nonetheless, analytical methods to explore associations between the spatial heterogeneity of the tumor and clinical data are not available. Hence, we have developed spatialGE, a software that provides visualizations and quantification of the tumor microenvironment heterogeneity through gene expression surfaces, spatial heterogeneity statistics that can be compared against clinical information, spot-level cell deconvolution and spatially informed clustering, all using a new data object to store data and resulting analyses simultaneously.
Availability and implementation
The R package and tutorial/vignette are available at https://github.com/FridleyLab/spatialGE. A script to reproduce the analyses in this manuscript is available in Supplementary information. The Thrane study data included in spatialGE was made available from the public available from the website https://www.spatialresearch.org/resources-published-datasets/doi-10-1158-0008-5472-can-18-0747/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Spatially resolved transcriptomics (ST) has allowed a better understanding of the tumor microenvironment (TME), immune infiltration and its relationship with immunotherapy response (Tang et al., 2016), as well as promising to improve the development of cancer prognoses and therapies (Nederlof et al., 2021). Many ST methods involve mRNA probe hybridization in immobilized tissues on surfaces (Zhang et al., 2021). Hence, gene expression localization within the tissue architecture is preserved as opposed to other transcriptomic approaches, such as bulk RNA (RNA-seq) or single-cell RNA sequencing (scRNAseq) (Maniatis et al., 2021; Yu et al., 2021). Given the high-dimensional nature of ST experiments, data structures are needed to accommodate the gene expression abundance, spatial locations and clinical information for samples taken from multiple subjects. Similarly, methods for ST visualization and analysis are needed, with existing tools varying in flexibility and approaches (Bergenstrahle et al., 2020; Dries et al., 2021; Hao et al., 2021; Hu et al., 2021; Navarro et al., 2017; Sun et al., 2020; Tan et al., 2020; Zhao et al., 2021).
To meet these unmet computational needs, we have developed a pipeline for the processing and analysis of probe-based sequencing ST experiments and exploration of the TME, which we call spatialGE. Novel features of the software include the generation of transcriptomic surfaces; quantification of spatial heterogeneity and the association with clinical information; gene expression deconvolution; spatially informed clustering (STclust); and a new data structure to store multi-sample spatial data, subject-level metadata and analytical results.
2 Implementation
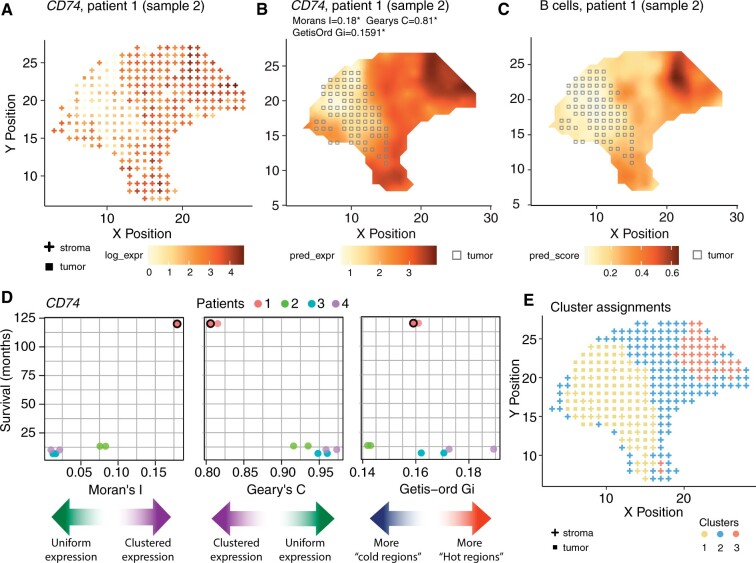
Our new software takes gene expression and spatial location data as input, and optionally, associated metadata (i.e. clinical information), and stores it in a new R object class referred to as ‘STList’. Other existing data structures can store metadata; however, the STList takes a single table with just one row of information for each sample. This approach makes inputting data easier for users and allows spatialGE to complete downstream analyses with a few lines of code. Users also have the possibility to provide Visium outputs directly from space ranger or Seurat objects, as well as .dcc/.pkc files from GeoMx experiments. In addition, results from spatialGE analyses are saved in the STList making downstream statistical analysis and visualization of results seamless. The software allows users to perform gene- or spot-wise filtering and generate quality control visualizations, followed by library size normalization and logarithmic or voom transformation (Law et al., 2014). The relative gene expression at each spot can be visualized using ‘quilt plots’ (Fig. 1A). If the STList is created from space ranger outputs or Seurat objects, histological images are imported automatically, allowing to plot gene expression and these images side by side for comparative analysis. Higher resolution plots are achieved with transcriptomic surfaces produced by spatial interpolation (‘kriging’; Fig. 1B, Supplementary Fig. S1) (Diggle and Ribeiro, 2007). Surfaces can also be generated for deconvoluted cell type scores (Fig. 1C), with cell type compositions at each spot inferred via gene expression deconvolution in xCell (Aran et al., 2017). Lastly, tumor purity scores are generated using ESTIMATE (Yoshihara et al., 2013), followed by tumor/stroma classification using model-based clustering (Fig 1A–C) (Scrucca et al., 2016). See Supplementary Methods for additional details.
Fig. 1.
Results from spatialGE analysis of a melanoma stage IIIc using spatial transcriptomics [Patient 1, Sample 2 (Thrane et al., 2018)]. (A) Quilt plot showing log-expression at each spot for immune gene CD74, with squares and crosses indicating tumor and stromal spot classifications respectively. (B) Transcriptomic surface of CD74 expression allows for a clearer visualization of the tissue heterogeneity, with squares indicating tumor compartment. (C) The inferred abundance of B cells in the tissue can be visualized by the predicted surface. (D) Scatter plots showing the relationships between spatial statistics calculated for CD74 and the survival of patients in months from Thrane et al. (2018). The colored arrows provide a guide for the interpretation of the spatial statistics. The dot with a border indicates the tissue section from Patient 1 featured in the other panels in Figure 1. (E) Tumor/stroma assignments and spatially informed clusters with STclust (using a spatial weight of 0.025) are shown. Cluster 1 and cluster 2 represent tumor and stroma regions of the tissue, respectively. Cluster 3 likely correspond to spots showing immune activity given the topological match with CD74 (B) and B cell scores (C)
spatialGE also provides users with spatial statistics to quantify TME heterogeneity (Supplementary Table S1 and Fig. S2). The Moran’s I (Moran, 1950) and Geary’s C (Geary, 1954) allow users to ascertain whether gene expression is uniform throughout the tissue. The Getis-Ord Gi statistic measures the tendency of a gene to produce expression hot- or cold-spots (Getis and Ord, 2010). Users can associate per-sample spatial heterogeneity statistics with other measurable clinical outcomes present in the sample metadata [spatial heterogeneity statistics (SThet); Fig. 1D]. Finally, spatialGE includes a computationally efficient spatially informed unsupervised clustering method, referred to as STclust, to detect TME compartments or ‘niches’ (Fig. 1E, Supplementary Figs S3–S5). In this approach, we begin by detecting genes with the highest spot-spot variation as calculated from standardized expression values (see Supplementary Methods for additional details). Then, a distance matrix is computed based on two scaled distance matrices: (i) transcriptomic autocorrelation between spots using the top variable genes (D_1) and (ii) spatial distances between spots (D_2). Next, the autocorrelation matrix (D1) is ‘shrunk’ toward the spatial distances matrix (D2) by calculating its weighted average as D=[(1-w)*D_1]+(w*D_2). The user specifies the weight (w) to apply. Based on our experience, a weight smaller weights seem to best capture tissue heterogeneity (Supplementary Figs S3–S5).
3 Discussion
The spatialGE package is a comprehensive analytical R package for the simultaneous analysis of multiple tissues assayed with probe-based ST technologies (i.e. Visium, GeoMx) using the new STList R object class. spatialGE is unique in implementing kriging to generate gene expression surfaces at high resolution for multiple ST tissue sections simultaneously (for an alternative method see (Zhao et al., 2021)). Several ST clustering approaches are already available (Supplementary Table S2) (Bergenstrahle et al., 2020; Dries et al., 2021; Hu et al., 2021; Tan et al., 2020; Zhao et al., 2021); however, our spatially informed clustering approach, STclust, is computationally efficient (Supplementary Fig. S6) and resembles tissue features by only weighting transcriptomic similarities by their distances between spots. A comparison between STclust and Louvain clustering (as implemented in the widely used software Seurat), showed a Rand similarity index of 0.91 for k = 4 (Patient 1, Sample 2; Supplementary Fig. S5), indicating good agreement of spot assignments between the two methods. Nonetheless, STclust yielded clusters that appeared more spatially contiguous than Louvain. Another innovation of spatialGE is the estimation of quantifiable metrics (SThet) to capture transcriptomic complexity and the ability to compare them against clinical outcomes (Fig. 1D).
Applying spatialGE to the Thrane et al. (2018) melanoma ST data, we observed agreement between the transcriptomic surfaces (Supplementary Fig. S1) and tumor and stroma regions observed in pathology images. STclust provided TME compartments (i.e. clusters; Fig. 1E; Supplementary Figs S3 and S4) that resembled the features annotated in pathology images (Thrane et al., 2018). Finally, we observed an association between spatial heterogeneity and patient survival (Fig. 1D), providing support to the hypothesis that heterogeneity is a predictor of patient outcomes (Nederlof et al., 2021).
In future versions, spatialGE will include analysis tools for different ST technologies (e.g. Slide-seq). We are currently developing additional analytical methods and visualizations that use histological annotation tools for non-gridded technologies (e.g. GeoMx), as well as a web-based implementation of spatialGE. Another developing area is the integration of scRNA-seq data with ST for spot-level cell.
Supplementary Material
Acknowledgements
The authors thank the spatial research group at the KTH Royal Institute of Technology—SciLifeLab (Stockholm, Sweden) for making their melanoma data available.
Funding
This work was supported by the National Institutes of Health (NIH) [T32-CA233399, P30-CA076292, R00-CA226679]. This content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
Conflict of Interest: none declared.
Contributor Information
Oscar E Ospina, Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL 33612, USA.
Christopher M Wilson, Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL 33612, USA.
Alex C Soupir, Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL 33612, USA.
Anders Berglund, Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL 33612, USA.
Inna Smalley, Department of Tumor Biology, Moffitt Cancer Center, Tampa, FL 33612, USA.
Kenneth Y Tsai, Department of Anatomic Pathology, Moffitt Cancer Center, Tampa, FL 33612, USA.
Brooke L Fridley, Department of Biostatistics and Bioinformatics, Moffitt Cancer Center, Tampa, FL 33612, USA.
References
- Aran D. et al. (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol., 18, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergenstrahle J. et al. (2020) Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics, 21, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P.J., Ribeiro P.J. (2007) Spatial prediction. In: Model-Based Geostatistics. Springer, New York, NY. pp. 134–156. https://doi.org/10.1007/978-0-387-48536-2. [Google Scholar]
- Dries R. et al. (2021) Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol., 22, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary R.C. (1954) The contiguity ratio and statistical mapping. Incorp. Stat., 5, 115–146. [Google Scholar]
- Getis A., Ord J.K. (2010) The analysis of spatial association by use of distance statistics. In: Anselin,L. and Rey,S. (eds.) Perspectives on Spatial Data Analysis. Advances in Spatial Science (The Regional Science Series). Springer, Berlin, Heidelberg, pp. 127–145. [Google Scholar]
- Hao Y. et al. (2021) Integrated analysis of multimodal single-cell data. Cell, 184, 3573–3587. e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. et al. (2021) SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods, 18, 1342–1351. [DOI] [PubMed] [Google Scholar]
- Law C.W. et al. (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol., 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis S. et al. (2021) Spatially resolved transcriptomics and its applications in cancer. Curr. Opin. Genet. Dev., 66, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P.A. (1950) Notes on continuous stochastic phenomena. Biometrika, 37, 17–23. [PubMed] [Google Scholar]
- Navarro J.F. et al. (2017) ST Pipeline: an automated pipeline for spatial mapping of unique transcripts. Bioinformatics, 33, 2591–2593. [DOI] [PubMed] [Google Scholar]
- Nederlof I. et al. (2021) A high-dimensional window into the micro-environment of triple negative breast cancer. Cancers (Basel), 13, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L. et al. (2016) mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R. J., 8, 289–317. [PMC free article] [PubMed] [Google Scholar]
- Sun S. et al. (2020) Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods, 17, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X. et al. (2020) SpaCell: integrating tissue morphology and spatial gene expression to predict disease cells. Bioinformatics, 36, 2293–2294. [DOI] [PubMed] [Google Scholar]
- Tang H. et al. (2016) Immunotherapy and tumor microenvironment. Cancer Lett., 370, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane K. et al. (2018) Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res., 78, 5970–5979. [DOI] [PubMed] [Google Scholar]
- Yoshihara K. et al. (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun., 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. et al. (2021) Statistical and bioinformatics analysis of data from bulk and single-cell RNA sequencing experiments. Methods Mol. Biol., 2194, 143–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. et al. (2021) Spatial molecular profiling: platforms, applications and analysis tools. Brief. Bioinf., 22, bbaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E. et al. (2021) Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol., 39, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.