Abstract
Intermediate CAG (polyQ) expansions in the gene ataxin-2 (ATXN2) are now recognized as a risk factor for amyotrophic lateral sclerosis. The threshold for increased risk is not yet firmly established, with reports ranging from 27 to 31 repeats.
We investigated the presence of ATXN2 polyQ expansions in 9268 DNA samples collected from people with amyotrophic lateral sclerosis, amyotrophic lateral sclerosis with frontotemporal dementia, frontotemporal dementia alone, Lewy body dementia and age matched controls.
This analysis confirmed ATXN2 intermediate polyQ expansions of ≥31 as a risk factor for amyotrophic lateral sclerosis with an odds ratio of 6.31. Expansions were an even greater risk for amyotrophic lateral sclerosis with frontotemporal dementia (odds ratio 27.59) and a somewhat lesser risk for frontotemporal dementia alone (odds ratio 3.14). There was no increased risk for Lewy body dementia. In a subset of 1362 patients with amyotrophic lateral sclerosis with complete clinical data, we could not confirm previous reports of earlier onset of amyotrophic lateral sclerosis or shorter survival in 25 patients with expansions.
These new data confirm ≥31 polyQ repeats in ATXN2 increase the risk for amyotrophic lateral sclerosis, and also for the first time show an even greater risk for amyotrophic lateral sclerosis with frontotemporal dementia. The lack of a more aggressive phenotype in amyotrophic lateral sclerosis patients with expansions has implications for ongoing gene-silencing trials for amyotrophic lateral sclerosis.
Keywords: amyotrophic lateral sclerosis, frontotemporal dementia, ataxin-2, polyQ expansion
Glass et al. examine ataxin-2 intermediate polyQ expansions in 9268 DNA samples from patients with ALS and related disorders. They confirm that expansions of ≥31 repeats are a risk factor for ALS, and an even greater risk for ALS with FTD, but did not find expansions to be associated with earlier onset or more aggressive disease.
Introduction
Ataxin-2 (ATXN2) is an RNA binding protein and regulator of stress granule assembly that is increasingly implicated in the pathogenesis of neurodegenerative diseases. Polyglutamine (CAG) expansions of >34 repeats within ATXN2 are causative of the disease spinocerebellar ataxia type 2 (SCA2).1 Intermediate expansions that do not reach the threshold for SCA2 are now recognized as a risk factor for amyotrophic lateral sclerosis (ALS). ATXN2 is shown to be a modifier of TDP43 toxicity in yeast, flies and mouse models of ALS.2–4 Indeed, in TDP-43 transgenic mice, an animal model of ALS, lifespan was significantly extended either by crossing these animals with ATXN2 knockout mice, or by treatment with antisense oligonucleotides directed at silencing the ATXN2 gene.3 The importance and clinical implications of these findings lead to the current ongoing trial of ATXN2 antisense oligonucleotides for the treatment of people with ALS (Clinical Trial NCT04494256).
The initial publication, which compared 915 ALS patients to 980 controls, identified polyglutamine (polyQ) expansions of 27–33 repeats to be enriched in the ALS population compared to controls.4 Subsequently, several groups have replicated the increased risk of intermediate repeat length expansions, although the appropriate threshold cut-off has varied from ≥29 to ≥31.5–10 Further, it has been reported that patients with intermediate expansions may experience a more aggressive ALS phenotype, including earlier age of onset4 and shorter survival.5,6 Last, no studies have directly investigated the association of ATXN2 intermediate expansions with ALS combined with frontotemporal dementia (ALSFTD) or with Lewy body dementia (LBD). We chose LBD as a comparison because it is a late life neurodegenerative disorder with clinical features and pathology distinct from FTD and where a large genomic dataset was available.
To address these issues, we investigated the prevalence of ATXN2 intermediate expansions in DNA collected from 9268 people, comparing age-appropriate controls to cohorts with diagnoses of ALS, ALSFTD, FTD without ALS and LBD. Further, we attempted to establish an association between ATXN2 intermediate repeat expansions and disease phenotypes, including age of onset and survival.
Materials and methods
The sources of DNA for sequencing are as described previously,11,12 from Wellderly,13 and from the Baltimore longitudinal study of ageing (https://www.blsa.nih.gov).14 All samples were subject to whole-genome sequencing and the clinical phenotype of cases were provided by the source clinicians. As described previously,11,12 samples were subject to several quality control assessments, including call rate and heterozygosity/homozygosity rates. Further, samples were analysed to assess population stratification via principal component analysis, and divergent samples were removed from the analysis (Supplementary Fig. 1). Genomes were analysed with Expansion Hunter, v.3.015 to identify copy numbers in the genes for ATXN2 and C9orf72. A subset of the samples carrying ATXN2 expansions longer than 34, the established repeat threshold underlying SCA2, was re-examined manually for proper graphical realignment. This led to the exclusion of five genomes that did not align correctly at this locus. C9orf72 expansions were identified in 90 people with ≤30 ATXN2 repeats and one individual with an intermediate ATXN2 polyQ expansion. A subset of 1362 patients cared for by J.D.G. (Emory cohort) was used to investigate the influence of ATXN2 intermediate expansions on ALS phenotypes. Within this cohort, 350 patients who did not undergo whole-genome sequencing were genotyped for ATXN2 repeat length and the C9orf72 expansion using standard and repeat-primed PCR.16 These ATXN2 data are included in the Emory clinical phenotyping (see next).
Statistical analysis
The binary case-control status was regressed against the presence or absence of ATXN2 repeats ≥31. Analysis was performed using R (v.3.5.2) using the glm() function applying the binary and logit transformation to the model. Estimates from the model for the contribution of ATXN2 repeats ≥31 were converted to odds ratio by exponentiating the estimates. The lower and upper confidence intervals (CIs) were calculated as follows: exp(estimate +/1.96× standard error of estimate). In the per repeat length analysis, to accommodate very small numbers of cases and controls in each repeat length, the binary case-control status was regressed using the Firth logistics regression method17 using the logisdf package (v.1.23). The logistic and Firth regression models were also corrected for sex and principal components 1–10 to account for any subtle cohort and population stratification.
Data availability
The majority of genomes, including repeat expansion data, have been previously published11,12 and are publicly available on dbGaP (phs001369, n = 6907), the AMP-PD portal (n = 4579 including overlapping samples) and the New York Genome Center web portal.
Results
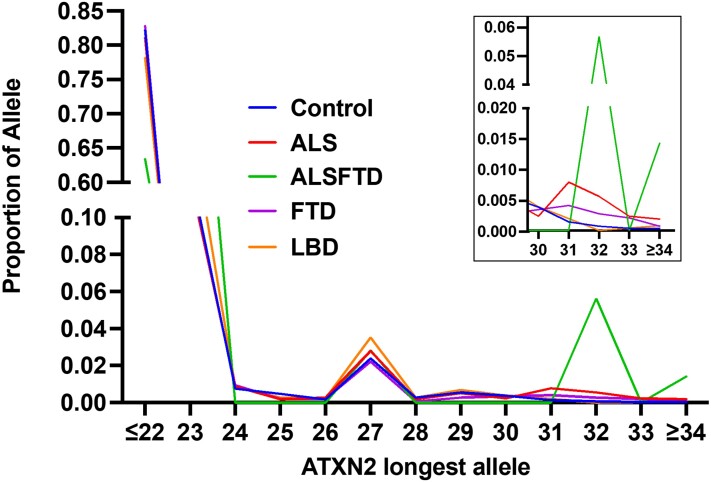
A total of 9268 genomes: 2181 ALS, 71 ALSFTD, 1485 FTD, 2610 LBD and 2921 controls, were evaluated for repeat length of ATXN2. Diagnosis was provided by the submitting physician, and age and site of onset were available for a subset of patients. The designation of ALSFTD was based on the combined clinical diagnoses of FTD and motor neuron disease. The control group was without neurodegenerative disease with a mean age of 73.6 years (median 81 years). Table 1 shows the numbers of patients with polyQ repeat numbers ranging from ≤22 to ≥34, with the frequencies for each repeat length shown graphically in Fig. 1. Comparisons across the four groups showed no differences in ATXN2 repeat length between any case group versus controls at a repeat length up to 30 repeats (Supplementary Table 2 and Supplementary Fig. 2). However, an increased frequency of ALS patients was observed at ≥31 repeats, establishing this as the cut-off for increased risk (Table 2 and Fig. 1). Comparing all ALS cases with ATXN2 repeat length ≥31 resulted in an odds ratio of 6.93 (P-value 9.50 × 10−7, 95% CI = 3.19–15.02; Table 2). Interestingly, comparing ALSFTD patients versus controls with ≥31 repeats displayed an even higher risk (odds ratio = 29.45, P-value 9.50 × 10−8, 95% CI = 8.5 to 102.02). FTD alone showed a lower risk, although there was still a significant odds ratio (odds ratio = 2.70, P-value 0.046, 95% CI = 1.02 to 7.13). The risk for a related neurodegenerative disease, LBD was not associated with ATXN2 repeat length (odds ratio = 1.19, P-value 0.735, 95% CI = 0.43 to 3.29).
Table 1.
Numbers of patients with ATXN2 repeats in each diagnosis group
| Longest repeat | ALS | ALSFTD | Control | FTD | LBD |
|---|---|---|---|---|---|
| ≤22 | 1767 | 45 | 2400 | 1230 | 2039 |
| 23 | 265 | 19 | 367 | 179 | 400 |
| 24 | 19 | 0 | 22 | 14 | 23 |
| 25 | 4 | 0 | 14 | 3 | 7 |
| 26 | 6 | 0 | 5 | 3 | 6 |
| 27 | 61 | 2 | 70 | 33 | 92 |
| 28 | 4 | 0 | 8 | 1 | 7 |
| 29 | 11 | 0 | 16 | 4 | 18 |
| 30 | 5 | 0 | 11 | 5 | 10 |
| 31 | 17 | 0 | 4 | 6 | 5 |
| 32 | 12 | 4 | 2 | 4 | 0 |
| 33 | 5 | 0 | 1 | 3 | 1 |
| ≥34 | 5 | 1 | 1 | 0 | 2 |
| Totals | 2181 | 71 | 2921 | 1485 | 2610 |
Figure 1.
Proportion of genomes with ATXN2 polyQ repeat number of the longest allele. Note that the majority of alleles register 22 and 27 repeats. The inset is a magnification of the proportion at ≥31 repeats, showing the increased proportion of people with ALS, FTD and ALSFTD.
Table 2.
Comparison of patient groups: ATXN2 repeats ≤30 versus ≥31
| Adjustment | Group pair | P-value | Odds ratio | Confidence interval |
|---|---|---|---|---|
| with covariates | ALS versus controls | 9.50 × 10−7 | 6.93 | 3.19 to 15.02 |
| with covariates | ALSFTD versus controls | 9.50 × 10−8 | 29.45 | 8.5 to 102.02 |
| with covariates | FTD versus controls | 0.046 | 2.70 | 1.02 to 7.13 |
| with covariates | LBD versus controls | 0.735 | 1.19 | 0.43 to 3.29 |
| with covariates | ALS versus FTD | 9.18 × 10−3 | 2.40 | 1.24 to 4.64 |
| with covariates | ALSFTD versus FTD | 5.54 × 10−5 | 9.85 | 3.24 to 29.97 |
| with covariates | ALSFTD versus ALS | 3.91 × 10−3 | 4.21 | 1.59 to 11.18 |
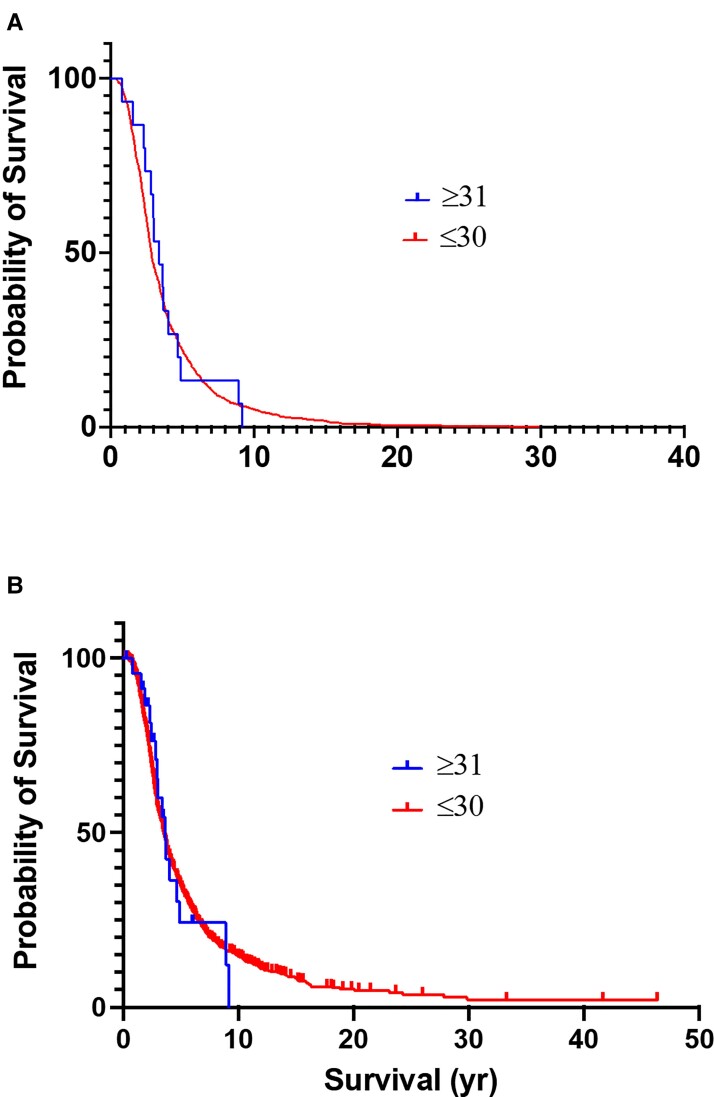
A subset of 1362 ALS patients with complete clinical information (Emory cohort) was examined for associations between ATXN2 repeat length and sub-phenotypes. Within this cohort, 25 patients (1.8%) harboured ≥31 ATXN2 repeats. Comparing ALS patients with and without expansions (≤30 repeats) did not reveal any difference in the mean age of onset of disease (58.7 years in those ≤30 versus 60.5 years in those ≥31, P = 0.46). Information on survival was available 900 people who were followed to the time of death. No differences in survival (P = 0.89) were found comparing patients with ≤30 repeats (mean = 2.86 years, range 1.4–29.8), and those with ≥31 repeats (3.39 years, range 2.4–8.9; Fig. 2A). Similarly, no difference (P = 0.44) was observed when comparing the survival including all patients and censoring those still alive: 3.5 years for ≤30 and 3.6 years for ≥31 repeats (Fig. 2B). Evaluating the cohorts separated by site of onset (limb versus bulbar) also showed no differences in age of onset or survival, with the caveat that there were only four bulbar onset patients with expansions ≥31 (Supplementary Table 1). Fifty-two patients in this clinical cohort had ALSFTD. Only one had an expansion of 31 repeats, and all others had repeat sizes of 28 or fewer.
Figure 2.
Comparison of survival for ALS patients with <30 or ≥31 ATXN2 polyQ repeats. (A) All deceased patients. (B) All patients with censoring of those still alive.
Discussion
Here we provide new data on the influence of ATXN2 intermediate polyQ expansions for the risk of ALS. These data are among the most extensive set of ALS genomes to address this issue, and among the first to examine ATXN2 expansions in large populations with ALS-related disorders (ALSFTD, FTD), and in an unrelated neurodegenerative disorder, LBD. We confirm the increased risk for ALS and establish a threshold cut-off ≥31 repeats. Further, we identified an even greater risk for the combination of ALS and FTD. Interestingly, we also found an increased risk for FTD without ALS, albeit associated with a lower level of risk. This observation, along with the lack of increased risk of polyQ expansions for LBD, provides some evidence for the specificity of ATXN2 expansions for ALS and ALSFTD.
Contrary to previous reports in Italian populations,5,6 but consistent with previous data from the UK and the Netherlands,9 we found no association of intermediate ATXN2 polyQ expansions with clinical features suggestive of a more aggressive disease, including younger age of onset and shorter survival. It is unclear why our findings differ; however, these new data may be important for the ongoing clinical trial using antisense technology to knock down ATXN2 protein. Although all sporadic ALS patients are eligible to participate, the later experimental cohorts are recruiting ALS patients only with ATXN2 polyQ expansions (Clinical Trial NCT04494256), on the basis of the supposition that ATXN2 knockdown will be more effective than in ALS patients without expansions. The lack of increased clinical severity in these patients may argue against this hypothesis since there are no data that we are aware of that those with polyQ expansions generate more ATXN2 protein than those without expansions.
Although ALS and FTD are clinically distinct, these disorders are inescapably linked by their common pathological and genetic underpinnings. These commonalities include the neuropathological finding of phosphorylated cytoplasmic TDP inclusions, and the overlap of ALS and FTD in individual patients and within families. Indeed, the most common disease associated mutation in familial ALS, the C9orf72 hexanucleotide expansion, is also the most common mutation found in familial FTD. Although it might be assumed that FTD seen with or without ALS is the ‘same’ disease, there are clinical18 and pathological/proteomic19 indications that they may have distinct underlying biology. Our finding that ATXN2 intermediate polyQ expansions impart a higher risk for ALSFTD than ALS alone, but a much lower increased risk for FTD, provides additional data supporting the possibility that FTD with and without ALS may represent distinct disease/phenotypes that share overlapping pathways contributing to pathogenesis. We note that a previous analysis of a smaller cohort of 368 FTD patients20 and another cohort consisting of a combination of 479 clinically diagnosed FTD and 162 pathologically diagnosed FTLD-TDP21 did not identify increased risk associated with intermediate ATXN2 repeat expansions.
The mechanisms underlying the relationship between ATX2 and ALS pathogenesis are unknown. ATX2 is a ubiquitously expressed protein with various cellular functions.22 Perhaps most germane to ALS is that ATXN2 is an RNA binding protein that regulates post-transcriptional control of expression, with direct effects on translation of subsets of RNA.23 It is not surprising, therefore, that ATXN2 interacts with other ALS genes and their protein products, including C9orf72, TDP43 and FUS.24 During stress, ATX2 is localized to stress granules where it is involved in stress granule assembly.25 These functions have been implicated in experimental ALS models, although it is unclear whether intermediate expansions interfere with these pathways.
Conclusion
The discovery of ATXN2 intermediate polyQ expansions in ALS is undoubtedly an important, yet unresolved, clue into the pathobiology ALS. The ongoing therapeutic trial targeting ATXN2 in ALS is a direct and positive outcome of this discovery. The lack of phenotypic and survival differences between ALS patients with and without expansions supports the inclusion of both groups in this trial. These new data demonstrating an even higher risk for ALSFTD in those with intermediate expansions adds to the genetic and pathological connections between these two neurodegenerative disorders.
Supplementary Material
Acknowledgements
We thank the Laboratory of Neurogenetics (NIH) staff for their collegial support and technical assistance. We thank the Biobanc-Hospital Clinic-Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), integrated in the Spanish National Biobank Network, for samples and data procurement. Data sources also funded by the Scripps Research Translational Institute, an NIH-NCATS Clinical and Translational Science Award (CTSA; 5 UL1 RR025774). We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona, USA, for the provision of human brain tissue (PI: Thomas G. Beach, MD) supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. We also thank the ROSMAP brain bank (David Bennett) for contribution of samples [P30AG72975 (ROS), R01AG15819 (Ros), R01AG17917 (Map) and U01AG61356 (Ros and Map)]. Sources of genomes and clinical data: The Accessible Genetics Consortium, Prospect Consortium and PSP Association, International FTD Consortium, International LBD Consortium, International ALS Consortium, New York Genome Center, Answer ALS. We also thank Doctors Robert H. Brown, Orla Hardiman, Vincenzo Silani and Nicola Ticozzi for their contributions of patient DNA.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- FTD
frontotemporal dementia
- LBD
Lewy body dementia
Contributor Information
Jonathan D Glass, Department of Neurology, Emory University School of Medicine, Atlanta, GA 30322, USA.
Ramita Dewan, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, NIH, Bethesda, MD 20892, USA.
Jinhui Ding, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, NIH, Bethesda, MD 20892, USA; Computational Biology Group, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20892, USA.
J Raphael Gibbs, Computational Biology Group, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD 20892, USA.
Clifton Dalgard, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Pamela J Keagle, Department of Neurology, University of Massachusetts Medical School, Worcester, MA 01605, USA.
Shankaracharya, Department of Neurology, University of Massachusetts Medical School, Worcester, MA 01605, USA.
Alberto García-Redondo, Instituto de Investigaciaon Hospital 12 de Octubre, Madrid, Spain.
Bryan J Traynor, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, NIH, Bethesda, MD 20892, USA; National Institute of Neurological Disorders and Stroke (NINDS), NIH, Bethesda, MD, USA; Therapeutic Development Branch, National Center for Advancing Translational Sciences (NCATS), NIH, Rockville, MD 20850, USA; Department of Neurology, Johns Hopkins University, Baltimore, MD 21287, USA; Reta Lila Weston Institute, UCL Queen Square Institute of Neurology, University College London, London, UK.
Ruth Chia, Neuromuscular Diseases Research Section, Laboratory of Neurogenetics, National Institute on Aging, NIH, Bethesda, MD 20892, USA.
John E Landers, Department of Neurology, University of Massachusetts Medical School, Worcester, MA 01605, USA.
Funding
This work was supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02), and grants from the ALS Association (17-SI-386 to J.D.G., J.E.L.) and the Muscular Dystrophy Association (827015 to J.D.G.).
Competing interests
B.J.T. holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 (patent numbers EP2751284A1, CA2846307A and 20180187262) and has received research grants from The Myasthenia Gravis Foundation, the Robert Packard Center for ALS Research, the ALS Association (ALSA), the Italian Football Federation (FIGC), the Center for Disease Control and Prevention (CDC), the United States Department of Veterans Affairs, the Muscular Dystrophy Association, Merck and Microsoft Research. B.J.T. receives funding through the Intramural Research Program at the National Institutes of Health. B.J.T. sits on the scientific advisory committee of the American Neurological Association, is an associate editor of Brain, and sits on the editorial boards of the Journal of Neurology, Neurosurgery, and Psychiatry, Neurobiology of Aging and eClinical Medicine. J.D.G. receives research support from the ALS Association, the Muscular Dystrophy Association, the National Institute of Ageing (P30AG066511), the National Institutes of Neurological Disorders and Stroke (P01NS08974), Biogen, Mitsubishi Tanabe, The Healy Center. J.E.L. is a member of the scientific advisory board for Cerevel Therapeutics, a consultant for ACI Clinical LLC sponsored by Biogen, Inc. or Ionis Pharmaceuticals, Inc. J.E.L. is also a consultant for Perkins Coie LLP and may provide expert testimony. J.E.L. was supported by funding from NIH/NINDS (R01NS073873 and R56NS073873).
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Scoles DR, Pulst SM. Spinocerebellar ataxia type 2. Adv Exp Med Biol. 2018;1049:175–195. [DOI] [PubMed] [Google Scholar]
- 2. Becker LA, Gitler AD. Ataxin-2 is droppin’ some knowledge. Neuron. May. 2018;98:673–675. [DOI] [PubMed] [Google Scholar]
- 3. Becker LA, Huang B, Bieri G, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borghero G, Pugliatti M, Marrosu F, et al. ATXN2 is a modifier of phenotype in ALS patients of Sardinian ancestry. Neurobiol Aging. 2015;36:2906.e1–2906.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chio A, Calvo A, Moglia C, et al. ATXN2 polyQ intermediate repeats are a modifier of ALS survival. Neurology. 2015;84:251–258. [DOI] [PubMed] [Google Scholar]
- 7. Van Damme P, Veldink JH, van Blitterswijk M, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. [DOI] [PubMed] [Google Scholar]
- 8. Wang MD, Gomes J, Cashman NR, Little J, Krewski D. Intermediate CAG repeat expansion in the ATXN2 gene is a unique genetic risk factor for ALS–A systematic review and meta-analysis of observational studies. PLoS ONE. 2014;9:e105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sproviero W, Shatunov A, Stahl D, et al. ATXN2 trinucleotide repeat length correlates with risk of ALS. Neurobiol Aging. 2017;51:178.e1–178.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neuenschwander AG, Thai KK, Figueroa KP, Pulst SM. Amyotrophic lateral sclerosis risk for spinocerebellar ataxia type 2 ATXN2 CAG repeat alleles: A meta-analysis. JAMA Neurol. 2014;71:1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chia R, Sabir MS, Bandres-Ciga S, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dewan R, Chia R, Ding J, et al. Pathogenic huntingtin repeat expansions in patients with frontotemporal dementia and amyotrophic lateral sclerosis. Neuron. 2021;109:448–460.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erikson GA, Bodian DL, Rueda M, et al. Whole-genome sequencing of a healthy aging cohort. Cell. 2016;165:1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrucci L. The Baltimore Longitudinal Study of Aging (BLSA): A 50-year-long journey and plans for the future. J Gerontol A Biol Sci Med Sci. 2008;63:1416–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolzhenko E, Deshpande V, Schlesinger F, et al. Expansion Hunter: A sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35:4754–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 18. Saxon JA, Thompson JC, Harris JM, et al. Cognition and behaviour in frontotemporal dementia with and without amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2020;91:1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Umoh ME, Dammer EB, Dai J, et al. A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol Med. 2018;10:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rubino E, Mancini C, Boschi S, et al. ATXN2 intermediate repeat expansions influence the clinical phenotype in frontotemporal dementia. Neurobiol Aging. 2019;73:231.e7–231.e9. [DOI] [PubMed] [Google Scholar]
- 21. Ross OA, Rutherford NJ, Baker M, et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet. 2011;20:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ostrowski LA, Hall AC, Mekhail K. Ataxin-2: From RNA control to human health and disease. Genes (Basel). 2017;8:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dansithong W, Paul S, Figueroa KP, et al. Ataxin-2 regulates RGS8 translation in a new BAC-SCA2 transgenic mouse model. PLoS Genet. 2015;11:e1005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y, Halliday GM, Kiernan MC, Tan RH. TDP-43 levels in the brain tissue of ALS cases with and without C9ORF72 or ATXN2 gene expansions. Neurology. 2019;93:e1748–e1755. [DOI] [PubMed] [Google Scholar]
- 25. Zhang K, Daigle JG, Cunningham KM, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173:958–971.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of genomes, including repeat expansion data, have been previously published11,12 and are publicly available on dbGaP (phs001369, n = 6907), the AMP-PD portal (n = 4579 including overlapping samples) and the New York Genome Center web portal.