Abstract
Background
Diagnosis of invasive candidiasis (IC) relies on insensitive cultures; the relative utility of fungal biomarkers in children is unclear.
Methods
This multinational observational cohort study enrolled patients aged >120 days and <18 years with concern for IC from 1 January 2015 to 26 September 2019 at 25 centers. Blood collected at onset of symptoms was tested using T2Candida, Fungitell (1→3)-β-D-glucan, Platelia Candida Antigen (Ag) Plus, and Platelia Candida Antibody (Ab) Plus assays. Operating characteristics were determined for each biomarker, and assays meeting a defined threshold considered in combination. Sterile site cultures were the reference standard.
Results
Five hundred participants were enrolled at 22 centers in 3 countries, and IC was diagnosed in 13 (2.6%). Thirteen additional blood specimens were collected and successfully spiked with Candida species, to achieve a 5.0% event rate. Valid T2Candida, Fungitell, Platelia Candida Ag Plus, and Platelia Candida Ab Plus assay results were available for 438, 467, 473, and 473 specimens, respectively. Operating characteristics for T2Candida were most optimal for detecting IC due to any Candida species, with results as follows: sensitivity, 80.0% (95% confidence interval, 59.3%–93.2%), specificity 97.1% (95.0%–98.5%), positive predictive value, 62.5% (43.7%–78.9%), and negative predictive value, 98.8% (97.2%–99.6%). Only T2Candida and Platelia Candida Ag Plus assays met the threshold for combination testing. Positive result for either yielded the following results: sensitivity, 86.4% (95% confidence interval, 65.1%– 97.1%); specificity, 94.7% (92.0%–96.7%); positive predictive value, 47.5% (31.5%–63.9%); and negative predictive value, 99.2% (97.7%–99.8%).
Conclusions
T2Candida alone or in combination with Platelia Candida Ag Plus may be beneficial for rapid detection of Candida species in children with concern for IC.
Clinical Trials Registration
Keywords: Pediatrics, invasive candidiasis, biomarkers
In this multinational observational cohort study of children and adolescents with concern for invasive candidiasis, operating characteristics of the T2Candida assay alone or combined with the Platelia CandidaAg Plus assay support the potential clinical utility of these diagnostic tools.
Invasive candidiasis (IC) is a common cause of bloodstream infections in hospitalized patients [1] and associated with increased hospital lengths of stay, charges, and all-cause mortality rates [2, 3]. These infections can be particularly severe in children with cancer [4, 5] and after organ or hematopoietic cell transplantation [6–8].
Diagnosing IC in pediatric patients is challenging. Cultures are insensitive [9] and often take >24 hours for a preliminary positive result, delaying initial therapy [10] and leading to increased mortality rate [11, 12]. A single-center prediction rule to identify candidemia in pediatric intensive care unit patients found that multiple factors had a predictive probability of 46% [13]. However, an independent multicenter study failed to validate the model [14].
Fungal biomarkers are potential novel tools to identify Candida spp. in children with increased concern for IC. T2Candida, Fungitell, Platelia Candida Antigen (Ag) Plus, and Platelia Candida Antibody (Ab) Plus assays are approved for diagnosis of candidemia or IC in adults [15–23]. Furthermore, T2Candida and Fungitell are endorsed by an international consensus guideline for the diagnosis of IC in adults [24]. However, there are limited data on these biomarkers to diagnose IC in children [24–27].
The International Pediatric Fungal Network performed a prospective, multinational observational cohort study, BIOmarkers in Pediatric Invasive Candidiasis (BIOPIC), to define the operating characteristics of T2Candida, Fungitell (1→3)- β-D-glucan, Platelia Candida Ag Plus, and Platelia Candida Ab Plus assays individually and in combination among at-risk children and adolescents with signs concerning for IC.
METHODS
Study Design
BIOPIC is a prospective observational cohort study conducted by the International Pediatric Fungal Network [28]. Twenty-five sites (23 in the United States and 2 international) opened for enrollment (Supplementary Table 1). Each site obtained institutional review board approval locally.
Study Cohort
Eligible participants were aged >120 days and <18 years with clinical characteristics concerning for IC (Table 1). Participants were ineligible if they (1) had possible, probable, or proven invasive fungal disease according to European Organization for Research and Treatment of Cancer/Mycoses Study Group criteria within the past 30 days; (2) had been previously included in the study; (3) weighed <4 kg, disallowing sufficient phlebotomy; or (4) were receiving empirical or targeted antifungal therapy.
Table 1.
Criteria for Increased Clinical Concern for Invasive Candidiasis
| At least one of the following underlying diagnoses/clinical conditions: |
| Intestinal failure (eg, short-gut syndrome) |
| Solid tumor or hematologic malignancy |
| Aplastic anemia |
| Solid organ transplant recipient |
| Hematopoietic stem cell transplant recipient |
| Any patient currently in or imminently transferred to a non-neonatal ICU |
| All of the following current clinical management: |
| Presence of at least one central catheter (arterial or venous) |
| Blood culture drawn for clinical concern of infection |
| Initiation or change in systemic nonantifungal antimicrobial therapy at time of blood culture attainment |
Abbreviation: ICU, intensive care unit.
Blood Sample Collection and Processing
A single blood sample (≤10.5 mL) was obtained within 24 hours of blood culture collection and aliquoted into ethylenediaminetetraacetic acid (EDTA) and serum separator tubes in a specified order (Supplementary Table 2). Sites shipped frozen specimens to a central repository; after enrollment completion, specimens were shipped to 2 testing laboratories for performance of the 4 different biomarker assays (Appendix B in the Supplementary Materials). Laboratory personnel completing these assays were blinded to the clinical outcomes of the enrolled participants.
Preparation of Spiked Specimens
The estimated 5% event rate (pretest probability) of proven or probable IC was not achieved. An institutional review board amendment was approved to prospectively enroll 15 additional pediatric patients without concern for IC at 1 site. These patients’ blood specimens were spiked using a similar technique, detailed in Appendix A (Supplementary Materials), that led to Food and Drug Administration approval for the T2Candida assay [23]. Distribution of species spiked was determined by epidemiology in a large pediatric cohort of IC [3]. All biomarker assay testing for these specimens was done directly on the spiked blood specimen.
Fungal Biomarker Assays
T2Candida Assay
The T2Candida assay (T2Biosystems) was performed using frozen whole-blood samples thawed at room temperature. Samples with volumes ≥3mL were loaded directly to the sample inlet snorkel. Samples with volumes of 2–3 mL were pipetted by hand into the sample inlet; the opening of each sample inlet was pierced with a sterile 1-mL pipette tip, and a new sterile 1-mL pipette tip was used to load each sample. Samples <2.0 mL were excluded from testing. After a run time of approximately 4–8 hours, depending on number of samples, results of either “detected” or “not detected” were displayed. The lower limit of detection is 1 colony-forming unit/mL.
Fungitell Assay
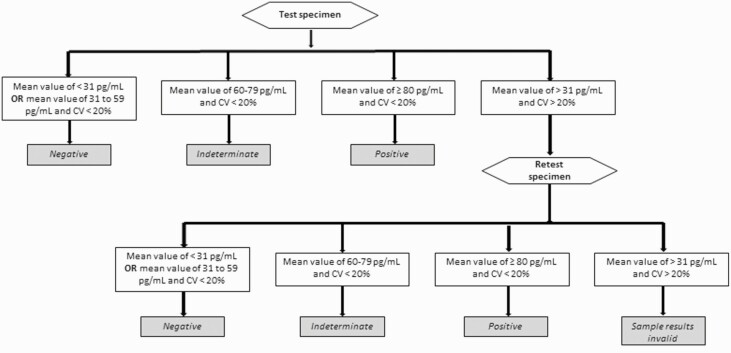
Fungitell assay (Associates of Cape Cod) was performed according to manufacturer using frozen serum. Each specimen was tested in duplicate. Specimens required a percentage coefficient of variation <20% to be resulted; excessive percentage coefficient of variation values caused specimens to be retested or resulted as invalid. Following manufacturer guidance, (1→3)- β-D-glucan values <60 pg/mL were negative, values of 60–79 pg/mL were indeterminate, and values ≥80 pg/mL were positive. Figure 1 shows the testing algorithm for interpretation based on manufacturer guidance and prior publications [29, 30].
Figure 1.
Testing algorithm for the Fungitell (1→3)-β-D-glucan assay. Abbreviation: CV, coefficient of variation.
Platelia Candida Ag Plus Assay
Platelia Candida Ag Plus (Bio-Rad) was performed using frozen serum, according to the manufacturer’s instructions. Specimens with concentrations <62.5 pg/mL were negative; concentrations from 62.5 to <125 pg/mL were indeterminate; and concentrations ≥125 pg/mL were positive.
Platelia Candida Ab Plus Assay
Platelia Candida Ab Plus (Bio-Rad) was performed using frozen serum, according to manufacturer instructions, to detect antibodies to Candida mannan antigen. Specimens with concentrations <5 AU/mL were negative, concentrations from 5 to <10 AU/mL were indeterminate, and concentrations ≥10 AU/mL were positive.
Outcome
The primary end point of proven or probable IC was defined by the 2008 European Organization for Research and Treatment of Cancer/Mycoses Study Group criteria for invasive fungal disease [31]. For a patient to meet the primary end point, diagnosis of proven or probable IC must have been made on or between day 0 (day of enrollment) through day 14.
Data Collection
Data were prospectively collected using Research Electronic Data Capture (REDCap) [32]. The coordinating center was Duke University, and the analysis center was Children’s Hospital of Philadelphia. Patient data were entered by sites and reviewed, with quarterly automated data checks and queries to sites. Baseline data elements collected included patient demographics, underlying disease, and clinical and immunologic risk factors. In addition, exposures to a range of products that may lead to a false result for ≥1 assay were collected.
Statistical Analysis
Sample Size and Power
Based on a study sample size of 500 and a pretest probability of 5%, power calculations estimated an assay with sensitivity and specificity of 90% would provide a positive predictive value (PPV) of 32% (95% confidence interval [CI], 26%–39%) and a negative predictive value (NPV) of 99% (98%–100%). These predictive probabilities were deemed a priori to be clinically useful, as a positive test result would substantially increase concern for IC (ie, increase from a 5% pretest probability of disease to a 32% posttest probability) and a negative test would substantially decrease a clinician’s concern for IC (ie, decrease from a 5% pretest probability of disease to a 1% posttest probability).
Primary Analysis
For each assay, sensitivity, specificity, PPV, and NPV with 95% CIs were calculated using manufacturer-recommended positivity thresholds; calculated sensitivity and specificity then informed likelihood ratios and posttest probability for the presence or absence of IC for a range of hypothetical IC prevalence rates (1%, 2%, 5%, and 10%). Each assay sensitivity, specificity, PPV, and NPV were recalculated with 95% CIs, excluding from the analysis spiked specimens (Appendix B [Supplementary Materials]).
Sensitivity Analysis
To consider diagnostic ability in a more contemporaneous time frame, sensitivity analyses considered shorter outcome windows (0–7 and 0 –2 days). Because the T2Candida assay is designed to target 5 Candida spp. (C. albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis), we repeated analyses for this assay limiting outcome to these 5 species. A final sensitivity analysis excluded specimens exposed within relevant exposure windows to products reported as possible sources of false-positive Fungitell (1→3)- β-D-glucan, Platelia Candida Ag Plus, or Platelia Candida Ab Plus results and false-negative T2Candida results (Appendix B [Supplementary Materials]).
Assay Results Considered in Combination
We hypothesized that multiple biomarker assays considered in combination could potentially optimize operating characteristics. We decided ad hoc that only quantitative assays with an area under the receiver operating characteristic (ROC) curve (AUC) >0.65 would be considered in subsequent combination algorithms. Potential combination testing considered assays performed in parallel or series. Further details are in the statistical analysis plan (Appendix B [Supplementary Materials]). Instead of using the manufacturer-recommended positivity threshold when evaluating the combinations incorporating quantitative assays, the positivity threshold was determined using an optimal operating slope (OOS) approach to minimize expected cost of mistakes from diagnosis decisions based on the test result [33–35]. The OOS was defined as (1 − p)CFP/pCFN, where p was the pretest probability; CFP, the cost of false-positive mistakes; and CFN, the cost of false-negative mistakes. The value at which the tangency on the ROC curve equals the OOS was considered the optimal threshold. If none of the candidate points’ tangencies on the ROC equaled the OOS, then we used the value whose tangency most closely approximated the OOS.
RESULTS
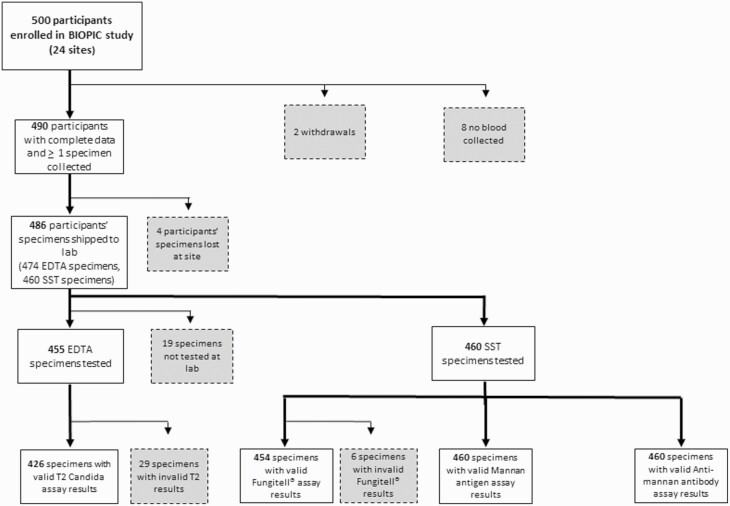
Between 1 January 2015 and 26 September 2019, a total of 500 patients with concern for IC were enrolled at 22 centers (Table 2). The cohort’s median age (interquartile range) was 6.0 (2.5–11.8) years, and patients were predominantly white (74.9%) and not Hispanic or Latino (84.9%). Accounting for withdrawals and inadequate or lost specimens, 486 participants provided ≥1 specimen for testing (Figure 2). Only 1 specimen was collected >24 hours after the blood culture was obtained. The 474 EDTA specimens yielded 426 valid T2Candida results (89.9%), and the 460 serum separator tube specimens yielded valid Fungitell, Platelia Candida Ag Plus, and Platelia Candida Ab Plus results for 454 (98.7%), 460 (100%), and 460 (100%) of the specimens, respectively. Thirteen IC events were identified in the 14-day follow-up period. All but 1 of the clinical infections were candidemia events (Table 3).
Table 2.
Demographic and Clinical Characteristics of Participants Contributing a Blood Specimen
| Any Specimen (N = 486) | EDTA Testinga (n = 474) | SST Testingb (n = 460) | |
|---|---|---|---|
| Age | Median, (IQR) | Median, (IQR) | Median, (IQR) |
| 6.0, (2.5–11.8) | 6.1, (2.6–11.8) | 6.2, (2.8–11.7) | |
| Race | n, (%) | n, (%) | n, (%) |
| Asian | 18, 3.7 | 18, 3.8 | 18, 3.9 |
| Black | 55, 11.4 | 55, 11.6 | 51, 11.1 |
| Other | 24, 4.9 | 24, 5.1 | 22, 4.8 |
| Unknown/not reported | 25, 5.1 | 24, 5.1 | 21, 4.6 |
| White | 364, 74.9 | 353, 74.5 | 348, 75.7 |
| Ethnicity | |||
| Hispanic/Latino | 65, 13.4 | 64, 13.5 | 59, 12.8 |
| Not Hispanic/Latino | 413, 85.0 | 402, 84.8 | 393, 85.4 |
| Unknown/not reported | 8, 1.6 | 8, 1.7 | 8, 1.7 |
| Gender | |||
| Female | 239, 49.2 | 231, 48.7 | 224, 48.7 |
| Male | 247, 50.8 | 243, 51.3 | 236, 51.3 |
| Underlying Condition c | |||
| Cardiac disorders | 31, 6.4 | 28, 5.9 | 28, 6.1 |
| Genetic/metabolic/congenital disorders | 42, 8.6 | 40, 8.4 | 38, 8.3 |
| Hematologic malignancy | 158, 32.5 | 156, 32.9 | 154, 33.5 |
| Intestinal failured | 93, 19.1 | 92, 19.4 | 86, 18.7 |
| Neurologic disorders | 36, 7.4 | 34, 7.2 | 32, 7.0 |
| Respiratory disorders | 42, 8.6 | 39, 8.2 | 37, 8.0 |
| Solid organ transplant | 31, 6.4 | 30, 6.3 | 29, 6.3 |
| Solid tumor malignancy | 114, 23.5 | 113, 23.8 | 112, 24.3 |
| Stem cell transplant | 46, 9.5 | 42, 9.2 | 42, 9.1 |
| Surgery/Trauma in the last 2 weeks | 45, 9.3 | 42, 8.9 | 43, 9.3 |
| Othere | 23, 4.7 | 23, 4.9 | 20, 4.3 |
| Previously healthy | 1, 0.2 | 1, 0.2 | 1, 0.2 |
Abbreviations: EDTA, ethylenediaminetetraacetic acid; IQR, interquartile range; SST, serum separator tube.
Used by T2Candida assay.
Used by Fungitell, Platelia Candida Antigen Plus and Platelia Candida Antibody Plus assays.
Sum of categories may be greater than the overall number of subjects because some subjects may have multiple underlying conditions.
Intestinal failure includes conditions such as short gut, bowel dismotility syndrome, etc.
Other includes: autoimmune disease, cutaneous disorders, endocrine disorders, inherited immunodeficiency, invasive infection, lymphatic disorders, prematurity, and renal disorders.
Figure 2.
Application of exclusion criteria to determine evaluable specimens for analysis. Gray boxes represent participants excluded from analysis. Results of 29 T2Candida assays were reported as invalid, included 5 specimens with instrument errors during assay performance and 24 whose results were invalidated owing to failure of the internal positive control. Abbreviations: BIOPIC, BIOmarkers in Pediatric Invasive Candidiasis; EDTA, ethylenediaminetetraacetic acid; SST, serum separation tube.
Table 3.
Listing of Candida Species Identified From Enrolled Patients and Used in Spiked Specimens
| Species | Day of first detectiona | Site of First Detectiona | Spiking Concentrationb (CFU/ mL) | |
|---|---|---|---|---|
| Specimens | ||||
| Clinical infections | ||||
| C. albicans | 0 | Blood | … | |
| C. albicans | 4 | Peritoneal | … | |
| C. guillermondii | 7 | Blood | … | |
| C. parapsilosis | 0 | Blood | … | |
| C. parapsilosis | 0 | Blood | … | |
| C. parapsilosis | 0 | Blood | … | |
| C. parapsilosis | 0 | Blood | … | |
| C. parapsilosis | 0 | Blood | … | |
| C. parapsilosis | 11 | Blood | … | |
| C. tropicalis | 0 | Blood | … | |
| C. tropicalis | 0 | Blood | … | |
| C. tropicalis | 0 | Blood | … | |
| C. tropicalis | 0 | Blood | … | |
| Spiked Specimens | ||||
| C. albicans | … | … | 8.7 | |
| C. albicans | … | … | 8.7 | |
| C. albicans | … | … | 8.7 | |
| C. albicans | … | … | 13.2 | |
| C. glabrata | … | … | 7.5 | |
| C. krusei | … | … | 10.0 | |
| C. lusitaniae | … | … | 11.1 | |
| C. parapsilosis | … | … | 6.5 | |
| C. parapsilosis | … | … | 8.6 | |
| C. parapsilosis | … | … | 10.0 | |
| C. parapsilosis | … | … | 15.0 | |
| C. tropicalis | … | … | 7.4 | |
| C. tropicalis | … | … | 7.4 |
Abbreviation: CFU, colony-forming unit.
Day and site of first detection applies only to natural infections.
Spiking concentration applies only to spiked specimens.
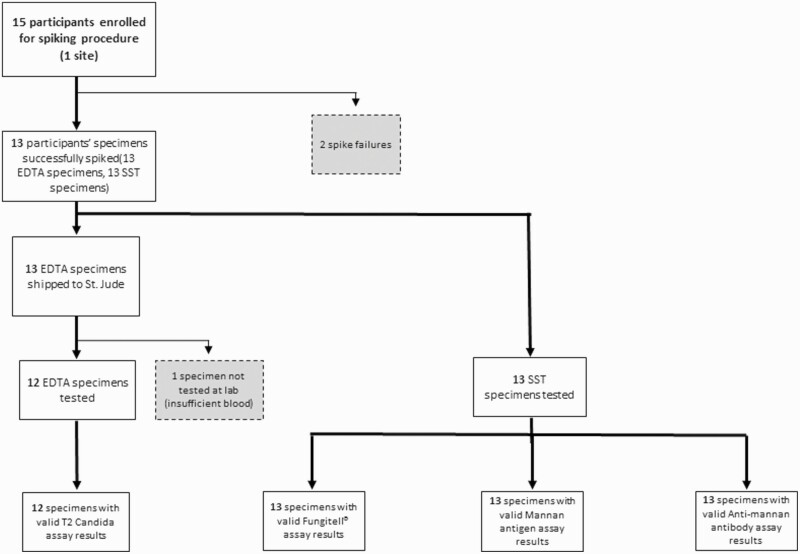
Separately, 15 patients without concern for IC were enrolled at 1 center and had a blood specimen collected, 13 of which were successfully spiked with Candida spp. (Table 3). All successfully spiked specimens had a valid result for the Fungitell, Platelia Candida Ag Plus, and Platelia Candida Ab Plus assays; 12 spiked specimens had a valid result for the T2Candida assay (Figure 3).
Figure 3.
Processing of specimens for subjects enrolled under specimen spiking protocol. Gray boxes represent specimens excluded from analysis. Abbreviations: EDTA, ethylenediaminetetraacetic acid; SST, serum separation tube.
Primary Analysis
Table 4 presents the sensitivity, specificity, PPV, and NPV individually for all 4 biomarkers across the outcome window of 0–14 days after specimen collection. The T2Candida assay had the best operating characteristics (sensitivity, 79.2%; specificity, 97.1%; PPV, 61.3%; NPV, 98.8%) for the 14-day outcome window, followed by the Platelia Candida Ag Plus (sensitivity, 39.1%; specificity, 96.7%; PPV, 37.5%; NPV 96.9%), Platelia Candida Ab Plus (sensitivity, 21.7%; specificity, 94.9%; PPV, 17.9%; NPV, 96.0%), and Fungitell (sensitivity, 9.1%; specificity, 85.4%; PPV, 3.0%; NPV, 95.0%). Supplementary Table 3 compares the operating characteristics for each biomarker and each outcome window when including and excluding spiked specimens. Positive and negative likelihood ratios for each assay are presented in Supplementary Table 4.
Table 4.
Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value for Each Biomarker by Different Outcome Assessment Windows
| Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | |||||
|---|---|---|---|---|---|---|---|---|
| T2Candidaa | n, (%) | 95% CI | n, (%) | 95% CI | n, (%) | 95% CI | n, (%) | 95% CI |
| Any Candida spp. | ||||||||
| 0–14 d | 19/24, (79.2) | 57.9–92.9 | 402/414, (97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 402/407, (98.8) | 97.2–99.6 |
| 0–7 d | 19/23, (82.6) | 61.2–95.1 | 403/415, (97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 403/407, (99.0) | 97.5–99.7 |
| 0–2 d | 19/21, (90.5) | 69.6–98.8 | 405/417, (97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 405/407, (99.5) | 98.2–99.9 |
| Targeted Candida spp.b | ||||||||
| 0–14 d | 19/22, (86.4) | 65.1–97.1 | 404/416, (97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 404/407, (99.3) | 97.9–99.9 |
| 0–7 d | 19/21, (90.5) | 69.6–98.8 | 405/417, (97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 405/407, (99.5) | 98.2–99.9 |
| 0–2 d | 19/20, (95.0) | 75.1–99.9 | 406/418,(97.1) | 95.0–98.5 | 19/31, (61.3) | 42.2–78.2 | 406/407, (99.8) | 98.6–100.0 |
| Fungitell c | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 2/22, (9.1) | 1.1–29.2 | 380/445, (85.4) | 81.8–88.5 | 2/67, (3.0) | 0.4–10.4 | 380/400, (95.0) | 92.4–96.9 |
| 0–7 d | 2/21, (9.1) | 1.2–30.4 | 381/446, (85.4) | 81.8–88.6 | 2/67, (3.0) | 0.4–10.4 | 381/400, (95.3) | 92.7–97.1 |
| 0–2 d | 2/19, (10.5) | 1.3–33.1 | 383/448, (85.5) | 81.9–88.6 | 2/67, (3.0) | 0.4–10.4 | 383/400, (95.8) | 93.3–97.5 |
| Platelia Candida Antigen Plus d | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 9/23, (39.1) | 19.7–61.5 | 435/450, (96.7) | 94.6–98.1 | 9/24, (37.5) | 15.4–30.8 | 435/449, (96.9) | 94.8–98.3 |
| 0–7 d | 9/22, (40.9) | 20.7–63.7 | 436/451, (96.7) | 94.6–98.1 | 9/24, (37.5) | 15.4–30.8 | 436/449, (97.1) | 95.1–98.5 |
| 0–2 d | 8/20, (40.0) | 19.1–64.0 | 437/453, (96.5) | 94.3–98.0 | 8/24, (33.3) | 15.6–55.3 | 437/449, (97.3) | 95.4–98.6 |
| Platelia Candida Antibody Plus e | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 5/23, (21.7) | 7.5–43.7 | 427/450, (94.9) | 92.4–96.7 | 5/28, (17.9) | 6.1–36.9 | 427/445, (96.0) | 93.7–97.6 |
| 0–7 d | 5/22, (22.7) | 7.8–45.4 | 428/451, (94.9) | 92.5–96.7 | 5/28, (17.9) | 6.1–36.9 | 428/445, (96.2) | 94.0–97.8 |
| 0–2 d | 4/20, (20.0) | 5.7–43.7 | 429/453, (94.7) | 92.2–96.6 | 4/28, (14.3) | 4.0–32.7 | 429/445, (96.4) | 94.2–97.9 |
Abbreviations: CI, confidence interval.
467 total specimens (455 specimens, 12 spikes) tested at lab and 438 specimens with valid results (93.8%); 24 specimens with valid result ever had event detected (5.5%).
One enrolled subject’s specimen and one spiked specimen had a Candida species which is not detectable by the T2Candida assay and was not considered an event for this analysis. Effective event rate was 5.0%.
473 specimens (460 specimens, 13 spikes) received at lab, 467 specimens with valid result (98.7%), 22 specimens with valid result ever had event detected (4.7%); manufacturer’s recommended positivity cutoff: ≥80 pg/mL.
473 specimens (460 specimens, 13 spikes) received at lab, all had valid result; 23 specimens ever had event detected (4.9%); manufacturer’s recommended positivity cutoff: ≥125 pg/mL.
473 specimens (460 specimens, 13 spikes) received at lab, all had valid result; 23 specimens ever had event detected (4.9%); manufacturer’s recommended positivity cutoff: ≥10 AU/mL.
Using sensitivity and specificity metrics for each biomarker, the positive and negative posttest probabilities were calculated for disease prevalence rates of 1%, 2%, 5%, and 10% (Table 5). The probabilities of IC after a positive assay result across the range of prevalence rates were most optimal for the T2Candida assay, ranging from 21.6% in a 1% prevalence scenario to 75.2% in a 10% prevalence scenario. Of the remaining assays, Platelia Candida Ag had the highest posttest probability positive results, ranging from 10.6% in a 1% prevalence scenario to 56.6% in a 10% prevalence scenario.
Table 5.
Calculated Post-Test Probabilities for Each Biomarker by Fixed Prevalence Rates and by Different Outcome Assessment Windows
| Posttest Probability With 0–14-d Window, % | Posttest Probability With 0–7-d Window, % | Posttest Probability With 0–2-d Window, % | ||||
|---|---|---|---|---|---|---|
| Candidiasis (%) | No Candidiasis (%) | Candidiasis (%) | No Candidiasis (%) | Candidiasis (%) | No Candidiasis (%) | |
| T2Candida Any Candida spp. | (79.2% Sens, 97.1% Spec) | (82.6% Sens, 97.1% Spec) | (90.5% Sens, 97.12% Spec) | |||
| Prevalence (%) | ||||||
| 1 | 21.6 | 99.8 | 22.4 | 99.8 | 21.4 | 99.9 |
| 2 | 35.8 | 99.6 | 36.8 | 99.6 | 39.1 | 99.8 |
| 5 | 59.0 | 98.9 | 60.1 | 99.1 | 62.3 | 99.5 |
| 10 | 75.2 | 97.7 | 76.0 | 98.0 | 77.7 | 98.9 |
| T2 Candida Targeted Candida spp. | (86.4% Sens, 97.1% Spec) | (90.5% Sens, 97.1% Spec) | (95.0% Sens, 97.1% Spec) | |||
| Prevalence (%) | ||||||
| 1 | 23.2 | 99.9 | 24.1 | 99.9 | 25.1 | 99.9 |
| 2 | 37.9 | 99.7 | 39.1 | 99.8 | 40.3 | 99.9 |
| 5 | 61.2 | 99.3 | 62.3 | 99.5 | 63.5 | 99.7 |
| 10 | 76.9 | 98.5 | 77.7 | 98.9 | 78.6 | 99.4 |
| Fungitell | (91 % Sens, 85.4% Spec) | (9.1% Sens, 85.4% Spec) | (10.5% Sens, 85.5% Spec) | |||
| Prevalence (%) | ||||||
| 1 | 0.6 | 98.9 | 0.7 | 98.9 | 0.7 | 99.0 |
| 2 | 1.3 | 97.9 | 1.3 | 97.9 | 1.5 | 97.9 |
| 5 | 3.2 | 94.5 | 3.3 | 94.7 | 3.7 | 94.8 |
| 10 | 6.5 | 89.4 | 6.8 | 89.5 | 7.5 | 89.6 |
| Platelia Candida Antigen Plus | (39.1% Sens, 96.7% Spec) | (40.9% Sens, 96.7% Spec) | (40.0% Sens, 96.5% Spec) | |||
| Prevalence (%) | ||||||
| 1 | 10.6 | 99.4 | 11.1 | 99.4 | 10.3 | 99.4 |
| 2 | 19.3 | 98.7 | 20.1 | 98.8 | 18.8 | 98.7 |
| 5 | 38.2 | 96.8 | 39.3 | 96.9 | 37.3 | 96.8 |
| 10 | 56.6 | 93.5 | 57.7 | 93.6 | 55.7 | 93.5 |
| Platelia Candida Antibody Plus | (21.7% Sens, 94.9% Spec) | (22.7% Sens, 94.9% Spec) | (20.0% Sens, 94.7% Spec) | |||
| Prevalence (%) | ||||||
| 1 | 4.1 | 99.2 | 4.3 | 99.2 | 3.7 | 99.2 |
| 2 | 8.0 | 98.3 | 8.3 | 98.4 | 7.2 | 98.3 |
| 5 | 18.3 | 95.8 | 19.0 | 95.9 | 16.6 | 95.7 |
| 10 | 32.1 | 91.6 | 33.1 | 91.7 | 30.0 | 91.4 |
Abbreviations: Sens, sensitivity; Spec, specificity
Sensitivity Analysis
When limiting events to those Candida spp. targeted by T2Candida, the sensitivity, specificity, PPV, and NPV of T2Candida were 86.4%, 97.1%, 61.3%, and 99.3%, respectively (Table 4). Compared with the 0–14-day outcome window, windows of 0–7 and 0–2 days improved T2Candida sensitivity. Sensitivity was highest (90.5%) when restricting the outcome window to 0–2 days, while retaining similar specificity. Adjustment of the outcome window did not demonstrably affect the other 3 biomarkers (Table 4). The Candida spp. identified by traditional cultures for each patient with a false-negative biomarker result are displayed by biomarker in Supplementary Table 5.
Removing specimens that tested positive but in the presence of a condition associated with false-positive results decreased sensitivity and increased specificity and PPV for Fungitell and decreased the sensitivity and increased PPV for Platelia Candida Ag Plus. Removing specimens that tested negative but in the presence of a condition associated with false-negative results slightly improved T2Candida sensitivity (Table 6).
Table 6.
Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value for Each Biomarker By Different Outcome Assessment Windows, Excluding Specimens Exposed to Conditions Increasing Likelihood of False Positive or False Negative Results
| Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | |||||
|---|---|---|---|---|---|---|---|---|
| T2Candidaa | n, (%) | 95% CI | n, (%) | 95% CI | n, (%) | 95% CI | n, (%) | 95% CI |
| Any Candida spp. | ||||||||
| 0–14 d | 19/23, (82.6) | 61.2–95.1 | 311/323, (96.3) | 93.6–98.1 | 19/31, (61.3) | 42.2–78.2 | 311/315, (98.7) | 96.8–99.7 |
| 0–7 d | 19/22, (86.4) | 65.1–97.1 | 312/324, (96.3) | 93.6–98.1 | 19/31, (61.3) | 42.2–78.2 | 312/315, (99.1) | 97.2–99.8 |
| 0–2 d | 19/21, (90.5) | 69.6–98.8 | 313/325, (93.6) | 93.6–98.1 | 19/31, (61.3) | 42.2–78.2 | 313/315, (99.4) | 97.7–100.0 |
| Targeted Candida spp.b | ||||||||
| 0–14 d | 19/22, (86.4) | 65.1–97.1 | 312/324, (96.3) | 93.6–98.1 | 19/31, (61.3) | 42.2–78.2 | 312/315, (99.1) | 97.2–99.8 |
| 0–7 d | 19/21, (90.5) | 69.6–98.8 | 313/325, (96.3) | 93.6–98.1 | 19/31, (61.3) | 42.2–78.2 | 313/315, (99.4) | 97.7–99.9 |
| 0–2 d | 19/20, (95.0) | 75.1–99.9 | 314/326, (96.3) | 93.7–98.1 | 19/31, (61.3) | 42.2–78.2 | 314/315, (99.7) | 98.2–100.0 |
| Fungitell c | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 1/21, (4.8) | 0.1–23.8 | 380/387, (98.2) | 96.3–99.3 | 1/8, (12.5) | 0.3–52.7 | 380/400, (95.0) | 92.4–96.9 |
| 0–7 d | 1/20, (5.0) | 0.1–24.9 | 381/388, (98.2) | 96.3–99.3 | 1/8, (12.5) | 0.3–52.7 | 381/400, (95.3) | 92.7–97.1 |
| 0–2 d | 1/18, (5.6) | 0.1–27.3 | 383/390, (98.2) | 96.3–99.3 | 1/8, (12.5) | 0.3–52.7 | 383/400, (95.8) | 93.3–97.5 |
| Platelia Candida Antigen Plus d | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 6/20, (30.0) | 11.9–54.3 | 435/441, (98.6) | 97.1–99.5 | 6/12, (50.0) | 21.1–78.9 | 435/449, (96.9) | 94.8–98.3 |
| 0–7 d | 6/19, (31.6) | 12.6–56.6 | 436/442, (98.6) | 97.1–99.5 | 6/12, (50.0) | 21.1–78.9 | 436/449, (97.1) | 95.1–98.5 |
| 0–2 d | 6/18, (33.3) | 13.3–59.0 | 437/443, (98.7) | 97.1–99.5 | 6/12, (50.0) | 21.1–78.9 | 437/449, (97.3) | 95.4–98.6 |
| Platelia Candida Antibody Plus e | ||||||||
| Any Candida spp. | ||||||||
| 0–14 d | 5/23, (21.7) | 7.46–43.7 | 427/450, (94.9) | 92.4–96.7 | 5/28, (17.9) | 6.1–36.9 | 427/445, (96.0) | 94.0–97.6 |
| 0–7 d | 5/22, (22.7) | 7.82–45.4 | 428/451, (94.9) | 92.5–96.7 | 5/28, (17.9) | 6.1–36.9 | 428/445, (96.2) | 94.0–97.8 |
| 0–2 d | 4/20, (20.0) | 5.73–43.7 | 429/453, (94.7) | 92.2–96.6 | 4/28, (14.3) | 4.0–32.7 | 429/445, (96.4) | 94.2–97.9 |
See Table 2 for list of conditions and exposures considered to cause false positives for each assay.
Abbreviation: CI, confidence interval.
467 total specimens (455 specimens, 12 spikes) tested at lab and 438 specimens with valid results (93.8%); 92 negative specimens (92 specimens, 0 spikes) excluded as potential false negatives, 23/346 retained specimens had event detected (6.7%).
One retained spiked specimen had a Candida species cultured which is not detectable by the T2Candida assay and was not considered an event for this analysis. Effective event rate was 6.4%.
473 specimens (460 specimens, 13 spikes) received at lab, 467 specimens with valid result; 59 positive specimens (58 specimens, 1 spike) excluded as potential false positives, 21/408 retained specimens had event detected (5.2%); manufacturer’s recommended positivity cutoff: ≥80 pg/mL.
473 specimens (460 specimens, 13 spikes) received at lab; 12 positive specimens (11 specimens, 1 spike) excluded as potential false positives, 20/461 retained specimens had event detected (4.3%); manufacturer’s recommended positivity cutoff: ≥125 pg/mL.
473 specimens (460 specimens, 13 spikes) received at lab; no potential false positives identified; 23 specimens ever had event detected (4.9%); manufacturer’s recommended positivity cutoff: ≥10 AU/mL.
Assay Results Considered in Combination
The AUCs from the Fungitell (0.5265) and Platelia Candida Ab (0.5959) assays did not reach the ad hoc threshold of 0.65 necessary to be included in combination testing algorithms. The AUC for the Platelia Candida Ag (0.6535) exceeded the threshold, and this assay was thus considered in combination with T2Candida.
The prevalence in the subset of 418 specimens, including 12 spiked specimens, with both a valid T2Candida and Platelia Candida Ag Plus assay result available, was 5.3%; to approximate this, a prevalence of 5% was used to calculate the Platelia Candida Ag Plus OOS. The Platelia Candida Ag Plus cutoff point of 168.96 pg/mL performed best in scenarios where the cost of a false-negative was up to approximately 8 times the cost of a false-positive; this cutoff point was used as the positivity threshold for combination testing. The sensitivity was 86.4%, the specificity 94.7%, the PPV 47.5%, and the NPV 99.2% when either the T2Candida assay or Platelia Candida Ag Plus assay result was positive. Requiring both assay results to be positive resulted in a sensitivity of 31.8%, a specificity of 99.8%, a PPV of 87.5%, and an NPV of 96.3% (Table 7).
Table 7.
Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value for the T2Candida and Platelia Candida Antigen Plus Results Used in Combination
| Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | Specimens Exceeding Cutoff | |
|---|---|---|---|---|---|
| Test | % (95% CI) | % (95% CI) | % (95% CI) | % (95% CI) | n |
| T2Candida only | 77.3 (54.6, 92.2) | 97.2 (95.1, 98.6) | 60.7 (40.6, 78.5) | 98.7 (97.0, 99.6) | 28 |
| Platelia Candida Antigen Plus only | 40.9 (20.7, 63.7) | 97.2 (95.1, 98.6) | 45.0 (23.1, 68.5) | 96.7 (94.5, 98.3) | 20 |
| At least one test positive | 86.4 (65.1, 97.1) | 94.7 (92.0, 96.7) | 47.5 (31.5, 63.9) | 99.2 (97.7, 99.8) | 40 |
| Both tests positive | 31.8 (13.9, 54.9) | 99.8 (98.6, 100.0) | 87.5 (47.4, 99.7) | 96.3 (94.0, 97.9) | 8 |
418 specimen pairs (406 enrolled, 12 spikes) included in analysis, 22 specimens with event (5.3%).
DISCUSSION
This is the largest prospective study of fungal biomarkers for the diagnosis of IC in any age group. In this cohort of hospitalized children at increased risk for IC, the T2Candida assay had the highest sensitivity (79.2%) and specificity (97.1%), followed by the Platelia Candida Ag Plus assay (sensitivity, 39.1%; specificity, 96.7%). The T2Candida assay sensitivity improved when the outcome window was narrowed to 0–2 days (90.5% [95% CI, 69.6%–98.8%]), but narrowing the outcome window did not demonstrably affect operating characteristics of the Platelia Candida Ag Plus assay. While the specificities of Platelia Candida Ab Plus and the Fungitell assays were adequate, neither exceeded 25% sensitivity for any outcome window considered.
Based on these results, the T2Candida assay is the only assay of the 4 investigated with sufficient sensitivity and specificity to be considered individually as a tool for diagnosis of IC in at-risk children and adolescents. Notably, when IC prevalence (ie, the pretest probability) was 5% or 10%, a positive test result increased posttest probability to almost 60%, and a negative test result decreased posttest probability to just above 2%. These operating characteristics are similar to those reported in a cohort of adult patients for a similar clinical indication [23]. In contrast, the operating characteristics of the Fungitell assay in our cohort were inconsistent with prior adult studies [36, 37]. Sensitivity among adult patients is reportedly much higher, resulting in guideline endorsement of the assay for diagnosis of IC in adults [24].
When evaluating posttest probabilities of the T2Candida assay, one must consider similar metrics for blood cultures, the current diagnostic standard. In a review of autopsy-informed investigations, the sensitivity of blood cultures was as low as 50% [9]. Assuming the same specificity as the T2Candida assay of 97.1% and disease prevalence of 5.0%, the associated positive and negative posttest probabilities would be 51% and 3%, respectively, worse than for the T2Candida assay. In addition, using sterile culture results as the reference standard for T2Candida assay may have limited the estimated operating characteristics of this assay. Limited clinical details for patients with a false-positive T2Candida assay result were available and provided in Supplementary Table 6. Most patients had either gastrointestinal insufficiency, cancer, or both. It is possible that the assay was detecting Candida spp. transiently present in the bloodstream that were not detected by routine culture. The clinical relevance of this possibility cannot be determined from these data.
We assessed whether combining results of the Platelia Candida Ag with T2Candida could improve the diagnostic potential of either test alone. A testing approach of ≥1 positive result improved sensitivity to 86.4% and negative posttest probability to <1% compared with either assay alone, but it reduced specificity (94.7%) and positive posttest probability (47.5%). Requiring that both test results be positive optimized specificity (99.8%) and positive posttest probability (87.5%) but compromised sensitivity (31.8%) and negative posttest probability (3.7%).
Ultimately, choosing a diagnostic approach that considers T2Candida assay alone or in combination with the Platelia Candida Ag Plus assay depends on the goals of care for a clinical situation. Posttest probabilities across differing prevalence rates of T2Candida assay performed alone may sufficiently inform initiation or cessation of antifungal therapy in many clinical circumstances. However, in clinical situations where greater certainty is desired before starting or stopping antifungal therapy, a 2-test approach may be more optimal. Additional considerations for testing approaches include assay availability, result turnaround time, and assay costs. Each of these factors will differ by institution; it is recommended that each center develop its own systematic diagnostic approach.
These results must be interpreted in the context of limitations. First, the prevalence rate for the assembled cohort was less than anticipated. The study inclusion criteria required patients to have an underlying medical condition and clinical presentation consistent with patients enrolled to a previous cohort of pediatric patients with IC [3]. Even with these inclusion criteria, it is possible that there was selection bias for less severely ill patients who were less likely to have IC. Future investigation of nonculture diagnostic tools should consider refined inclusion criteria that enrich for a higher pretest probability of IC. The lower rate required inclusion of spiked specimens, which could have altered the calculated operating characteristics of each biomarker. While inclusion of spiked specimens did not significantly alter the calculated sensitivity for each biomarker (Appendix B, section 1.7.1.5 [Supplementary Materials]) it did result in more favorable estimates of sensitivity (Supplementary Table 3).
Second, specimens were tested after enrollment was completed; certain assays could have performed better in real time. Third, the reference standard was reliant on detection of Candida spp. with conventional, imperfect diagnostic studies, potentially increasing the possibility of mislabeling a positive assay result as false-positive. Fourth, while the T2Candida assay tests 2 mL of sample, the automated loading feature for the testing system requires a 3-mL sample. We hand-pipetted samples of <3 mL into the system, obviating the automatic loading mechanism. Manual pipetting should not adversely affect test results. Moreover, this study’s findings suggest manually loading specimens is possible and potentially beneficial for pediatric patients in whom access to blood volume is limited. Fifth, although the outcome was inclusive of any form of IC, only 1 event in this cohort met the definition of IC in the absence of candidemia. As such, the findings are limited to candidemia and may not be generalizable to all forms of IC. Sixth, the results reported only provide the operating characteristics of the biomarkers studied and do not assess the impact of results on clinical outcomes. Finally, there are other available nonculture diagnostic tools that we did not assess, such as Candida-specific multiplex polymerase chain reaction [38].
The estimated operating characteristics of T2Candida alone or in combination with the Platelia Candida Ag Plus assay in children and adolescents with clinical characteristics associated with increased risk for candidemia were reasonable. The decision to order T2Candida alone or in combination with the Platelia Candida Ag Plus assay is reliant on the clinician’s assessed applicability of these data to the patient and clinical scenario under consideration (ie, starting or stopping antifungal therapy) in conjunction with awareness of the cost and turnaround time for test results.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments . The authors thank Yuan-Shung Wang, MS, from the Children’s Hospital of Philadelphia for her assistance with data organization and cleaning. They also thank Jhoanna Zaida Aquino, BS, and Allison Fullenkamp, MS, from Duke University for their study coordination efforts, and Jeff Hawley, BS, from Duke University for his efforts in database creation and management. Finally, they thank the clinical research assistants at all participating institutions for their commitment to patient screening and chart abstraction.
Disclaimer . The National Institute of Allergy and Infectious Diseases was the primary study sponsor. Performance of the T2Candida assay was supported in part by American Lebanese Syrian Associated Charities, using reagents and instrumentation provided by T2 Biosystems. None of the funding sponsors had a role in the conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. The opinions expressed in this article are the authors’ own and do not reflect the view of the National Institutes of Health (NIH), the Department of Health and Human Services, or the US government.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, NIH (grant R01HD081044 to B. T. F. and W. J. S.).
Contributor Information
Brian T Fisher, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA; Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Craig L K Boge, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Rui Xiao, Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Sydney Shuster, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Dawn Chin-Quee, Duke University, Durham, North Carolina, USA.
John Allen, IV, Duke University, Durham, North Carolina, USA.
Shareef Shaheen, Duke University, Durham, North Carolina, USA.
Randall Hayden, Department of Pathology, St Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Sri Suganda, Department of Pathology, St Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Theoklis E Zaoutis, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA; Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Yeh Chung Chang, Duke University, Durham, North Carolina, USA.
Dwight E Yin, Children’s Mercy and University of Missouri–Kansas City School of Medicine, Kansas City, Missouri, USA.
Anna R Huppler, Medical College of Wisconsin and Children’s Wisconsin, Milwaukee, Wisconsin, USA.
Lara Danziger-Isakov, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
William J Muller, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Emmanuel Roilides, Infectious Disease Unit, 3rd Department of Pediatrics, School of Medicine, Aristotle University and Hippokration Hospital, Thessaloniki, Greece.
José Romero, Arkansas Children’s Hospital Research Institute, Little Rock, Arkansas, USA.
Paul K Sue, University of Texas Southwestern Medical Center, Dallas, Texas, USA.
David Berman, John Hopkins All Children’s Hospital, St Petersburg, Florida, USA.
Rachel L Wattier, University of California–San Francisco, San Francisco, California, USA.
Natasha Halasa, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Alice Pong, University of California San Diego, San Diego, California, USA.
Gabriela Maron, St Jude Children’s Research Hospital, Memphis, Tennessee, USA.
Pere Soler-Palacin, Hospital Universitari Vall d’Hebron, Barcelona, Catalonia, Spain.
Susan C Hutto, University of Alabama, Birmingham, Birmingham, Alabama, USA.
Blanca E Gonzalez, Cleveland Clinic Foundation, Cleveland, Ohio, USA.
Christine M Salvatore, Weill Cornell Medicine, New York, New York, USA.
Sujatha Rajan, Cohen Children’s Medical Center of New York, New Hyde Park, New York, USA.
Michael Green, UPMC Children’s Hospital of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Elizabeth Doby Knackstedt, University of Utah, Salt Lake City, Utah, USAand.
Sarmistha B Hauger, Dell Children’s Medical Center, Austin, Texas, USA.
William J Steinbach, Duke University, Durham, North Carolina, USA.
References
- 1. Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol 2013; 34:1–14. [DOI] [PubMed] [Google Scholar]
- 2. Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 2005; 41:1232–9. [DOI] [PubMed] [Google Scholar]
- 3. Fisher BT, Zaoutis TE, Xiao R, et al. Comparative efficacy of echinocandins versus triazoles or amphotericin B formulations as initial directed therapy for invasive candidiasis in children and adolescents. J Pediatr Infect Dis Soc 2021; piab024. doi: 10.1093/jpids/piab024 [DOI] [PubMed] [Google Scholar]
- 4. Creutzig U, Zimmermann M, Reinhardt D, Dworzak M, Stary J, Lehrnbecher T. Early deaths and treatment-related mortality in children undergoing therapy for acute myeloid leukemia: analysis of the multicenter clinical trials AML-BFM 93 and AML-BFM 98. J Clin Oncol 2004; 22:4384–93. [DOI] [PubMed] [Google Scholar]
- 5. Pagano L, Caira M, Candoni A, et al. The epidemiology of fungal infections in patients with hematologic malignancies: the SEIFEM-2004 study. Haematologica 2006; 91:1068–75. [PubMed] [Google Scholar]
- 6. Profumo RJ. Pediatric liver transplant recipients: mortality analysis over 20 years. J Insur Med 2006; 38:3–8. [PubMed] [Google Scholar]
- 7. Martin SR, Atkison P, Anand R, Lindblad AS, Group SR. Studies of Pediatric Liver Transplantation 2002: patient and graft survival and rejection in pediatric recipients of a first liver transplant in the United States and Canada. Pediatr Transplant 2004; 8:273–83. [DOI] [PubMed] [Google Scholar]
- 8. Groll AH, Pana D, Lanternier F, et al. 8th European Conference on Infections in Leukaemia: 2020 guidelines for the diagnosis, prevention, and treatment of invasive fungal diseases in paediatric patients with cancer or post-haematopoietic cell transplantation. Lancet Oncol 2021; 22:e254–e269. [DOI] [PubMed] [Google Scholar]
- 9. Clancy CJ, Nguyen MH. Finding the “missing 50%” of invasive candidiasis: how nonculture diagnostics will improve understanding of disease spectrum and transform patient care. Clin Infect Dis 2013; 56:1284–92. [DOI] [PubMed] [Google Scholar]
- 10. Lai CC, Wang CY, Liu WL, Huang YT, Hsueh PR. Time to positivity of blood cultures of different Candida species causing fungaemia. J Med Microbiol 2012; 61:701–4. [DOI] [PubMed] [Google Scholar]
- 11. Garey KW, Rege M, Pai MP, et al. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis 2006; 43:25–31. [DOI] [PubMed] [Google Scholar]
- 12. Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother 2005; 49:3640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zaoutis TE, Prasad PA, Localio AR, et al. Risk factors and predictors for candidemia in pediatric intensive care unit patients: implications for prevention. Clin Infect Dis 2010; 51:e38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher BT, Ross RK, Roilides E, et al. Failure to validate a multivariable clinical prediction model to identify pediatric intensive care unit patients at high risk for candidemia. J Pediatric Infect Dis Soc 2016; 5:458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ostrosky-Zeichner L, Alexander BD, Kett DH, et al. Multicenter clinical evaluation of the (1–>3) beta-D-glucan assay as an aid to diagnosis of fungal infections in humans. Clin Infect Dis 2005; 41:654–9. [DOI] [PubMed] [Google Scholar]
- 16. Held J, Kohlberger I, Rappold E, Busse Grawitz A, Häcker G. Comparison of (1->3)-β-D-glucan, mannan/anti-mannan antibodies, and Cand-Tec Candida antigen as serum biomarkers for candidemia. J Clin Microbiol 2013; 51:1158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lunel FM, Donnelly JP, van der Lee HA, Blijlevens NM, Verweij PE. Performance of the new Platelia Candida Plus assays for the diagnosis of invasive Candida infection in patients undergoing myeloablative therapy. Med Mycol 2011; 49:848–55. [DOI] [PubMed] [Google Scholar]
- 18. Odabasi Z, Mattiuzzi G, Estey E, et al. Beta-D-glucan as a diagnostic adjunct for invasive fungal infections: validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin Infect Dis 2004; 39:199–205. [DOI] [PubMed] [Google Scholar]
- 19. León C, Ruiz-Santana S, Saavedra P, et al. Cava Trem Study Group. Contribution of Candida biomarkers and DNA detection for the diagnosis of invasive candidiasis in ICU patients with severe abdominal conditions. Crit Care 2016; 20:149. doi: 10.1186/s13054-016-1324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hartl B, Zeller I, Manhart A, Selitsch B, Lass-Flörl C, Willinger B. A retrospective assessment of four antigen assays for the detection of invasive candidiasis among high-risk hospitalized patients. Mycopathologia 2018; 183:513–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pini P, Bettua C, Orsi CF, et al. Evaluation of serum (1 → 3)-β-D-glucan clinical performance: kinetic assessment, comparison with galactomannan and evaluation of confounding factors. Infection 2016; 44:223–33. [DOI] [PubMed] [Google Scholar]
- 22. Clancy CJ, Nguyen MH. T2 magnetic resonance for the diagnosis of bloodstream infections: charting a path forward. J Antimicrob Chemother 2018; 73(suppl 4):iv2–iv5. [DOI] [PubMed] [Google Scholar]
- 23. Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 2015; 60:892–9. [DOI] [PubMed] [Google Scholar]
- 24. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 2020; 71:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao DS, Ghosh A, Singhi S, Chakrabarti A. Mannan antigen detection in the diagnosis of patients with invasive candidiasis. Indian J Med Res 2002; 116:13–20. [PubMed] [Google Scholar]
- 26. Hamula CL, Hughes K, Fisher BT, Zaoutis TE, Singh IR, Velegraki A. T2Candida provides rapid and accurate species identification in pediatric cases of candidemia. Am J Clin Pathol 2016; 145:858–61. [DOI] [PubMed] [Google Scholar]
- 27. Montagna MT, Coretti C, Lovero G, et al. Diagnostic performance of 1→3-β-d-glucan in neonatal and pediatric patients with candidemia. Int J Mol Sci 2011; 12:5871–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Steinbach WJ, Roilides E, Berman D, et al. International Pediatric Fungal Network. Results from a prospective, international, epidemiologic study of invasive candidiasis in children and neonates. Pediatr Infect Dis J 2012; 31:1252–7. [DOI] [PubMed] [Google Scholar]
- 29. Alexander BD, Smith PB, Davis RD, Perfect JR, Reller LB. The (1,3){beta}-D-glucan test as an aid to early diagnosis of invasive fungal infections following lung transplantation. J Clin Microbiol 2010; 48:4083–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Angebault C, Lanternier F, Dalle F, et al. Prospective evaluation of serum beta-glucan testing in patients with probable or proven fungal diseases. Open Forum Infect Dis 2016; 3:ofw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Pauw B, Walsh TJ, Donnelly JP, et al. European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group; National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 2008; 46:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schäfer H. Constructing a cut-off point for a quantitative diagnostic test. Stat Med 1989; 8:1381–91. [DOI] [PubMed] [Google Scholar]
- 34. Somoza E, Mossman D. Comparing and optimizing diagnostic tests: an information-theoretical approach. Med Decis Making 1992; 12:179–88. [DOI] [PubMed] [Google Scholar]
- 35. Halpern EJ, Albert M, Krieger AM, Metz CE, Maidment AD. Comparison of receiver operating characteristic curves on the basis of optimal operating points. Acad Radiol 1996; 3:245–53. [DOI] [PubMed] [Google Scholar]
- 36. Posteraro B, Tumbarello M, De Pascale G, et al. (1,3)-β-d-Glucan-based antifungal treatment in critically ill adults at high risk of candidaemia: an observational study. J Antimicrob Chemother 2016; 71:2262–9. [DOI] [PubMed] [Google Scholar]
- 37. Hanson KE, Pfeiffer CD, Lease ED, et al. β-D-glucan surveillance with preemptive anidulafungin for invasive candidiasis in intensive care unit patients: a randomized pilot study. PLoS One 2012; 7:e42282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fuchs S, Lass-Florl C, Posch W. Diagnostic performance of a novel multiplex PCR assay for candidemia among ICU patients. J Fungi 2019; 5:86. doi: 10.3390/jof5030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.