Abstract
The formation of mitotically derived spores, called conidia, is a common reproductive mode in filamentous fungi, particularly among the large fungal class Ascomycetes. Asexual sporulation strategies are nearly as varied as fungal species; however, the formation of conidiophores, specialized multicellular reproductive structures, by the filamentous fungus Aspergillus nidulans has emerged as the leading model for understanding the mechanisms that control fungal sporulation. Initiation of A. nidulans conidipohore formation can occur either as a programmed event in the life cycle in response to intrinsic signals or to environmental stresses such as nutrient deprivation. In either case, a development-specific set of transcription factors is activated and these control the expression of each other as well as genes required for conidiophore morphogenesis. Recent progress has identified many of the earliest-acting genes needed for initiating conidiophore development and shown that there are at least two antagonistic signaling pathways that control this process. One pathway is modulated by a heterotrimeric G protein that when activated stimulates growth and represses both asexual and sexual sporulation as well as production of the toxic secondary metabolite, sterigmatocystin. The second pathway apparently requires an extracellular signal to induce sporulation-specific events and to direct the inactivation of the first pathway, removing developmental repression. A working model is presented in which the regulatory interactions between these two pathways during the fungal life cycle determine whether cells grow or develop.
Asexual sporulation is a common reproductive mode for a diverse group of fungi that includes many medically, industrially, and agriculturally important species. Asexual spores of higher fungi are called conidia, and although there is great variety in conidial form and function, all conidia represent nonmotile asexual propagules that are usually made from the side or tip of specialized sporogenous cells and do not form through progressive cleavage of the cytoplasm. The process of conidiation involves many common developmental themes including temporal and spatial regulation of gene expression, cell specialization, and intercellular communication. The genetic mechanisms controlling these processes have been addressed in any detail for only two well-studied ascomycetes, Aspergillus nidulans and Neurospora crassa. Although the general mechanisms governing sporulation in these two fungi are similar, there are clearly important differences. This review will concentrate on the progress made over the last few years toward understanding the genetic regulation of development in A. nidulans. A recent review by Springer (144) describes the N. crassa conidiation pathway.
From a fairly simplistic point of view, the A. nidulans asexual reproductive cycle can be divided into at least three different stages, beginning with a growth phase that is required for cells to acquire the ability to respond to induction signals, proceeding through initiation of the developmental pathway, and culminating with execution of the developmentally regulated events leading to sporulation. It is possible to at least partially separate these stages in the laboratory because the vegetative or hyphal state is continuously maintained during submerged growth in nutritionally sufficient liquid medium (14). After cells have acquired the ability to respond to inducing signals, development can be induced synchronously by exposing hyphae to air (see below). The kinetics of conidiation under these conditions are highly reproducible (14, 84), and this approach can be used for obtaining large quantities of developmentally staged materials for biochemical analysis.
There are numerous other experimental features that have made A. nidulans a particularly useful organism with which to study sporulation and development in general (153). Conidiation can easily be observed macroscopically due to conidial pigmentation; because conidiophores and conidia are dispensable structures, it is simple to obtain mutants that are defective in their ability to produce normal conidiophores. The fact that A. nidulans can reproduce sexually as well as asexually means that, once generated, most conidiation-deficient mutants can be manipulated to construct strains with desired genotypes for epistasis studies or for mapping mutations to specific locations on one of the eight chromosomes (42, 74, 126). A. nidulans also has a well-defined parasexual cycle providing additional tools for genetic analysis (74, 126). Hundreds of genes have been mapped to specific locations by classical methods, and it is typically quite simple to map a newly identified mutation to a given chromosome by using parasexual genetics and then to determine its linkage to previously defined genes by meiotic analyses (39, 155). With the advent of electrophoretic karyotyping, it is also possible to map cloned genes to determine their chromosomal locations by probing blots of resolved chromosomes (22). In addition, a physical map of overlapping cosmids from the two commonly used cosmid libraries has been constructed and provides even simpler tools for gene mapping (23) (available at http://fungus.genetics.uga.edu:5080/Physical_Maps.html).
As with yeast and many other fungi, a significant advantage of genetic studies of A. nidulans comes from the ready availability of sophisticated molecular genetic tools for specific functional analysis. DNA-mediated transformation with genomic DNA libraries allows relatively straightforward isolation of genes through complementation of mutant phenotypes followed by plasmid or cosmid recovery (154). Because transformation often results in homologous integration, it is feasible to disrupt, delete, or otherwise modify specific genes in the genome (1, 104, 150). Transcriptional promoters can be analyzed in vivo by fusing the promoter of interest to a reporter gene like the Escherichia coli lacZ gene and introducing the construct into the fungus (158). β-Galactosidase activity can be determined at various times during development by isolating crude protein from cell extracts. In situ staining is also possible and is one way to determine the cellular location of a given protein (5, 7). Recently, several laboratories have shown that the Aequorea victoria gene encoding green fluorescent protein (GFP) (128) can be used in A. nidulans to examine protein expression and localization in living cells. However, to date this approach has been successful only when GFP is expressed at very high levels with strong promoters. Several strong inducible promoters that can drive the expression of a gene of interest are available and are useful for studying the developmental effects of misscheduled gene expression (1, 63, 90).
MORPHOGENESIS AND SPORULATION
Colony Formation
Vegetative growth in A. nidulans, as in other filamentous fungi, begins with the germination of a spore. This could be either a mitotically derived conidiospore or a meiotically produced ascospore. Spore germination leads to the formation of tubular hyphae that grow in a polar fashion by apical extension and branching to form a network of interconnected cells known as a mycelium. The mycelium forms a radially symmetric colony that expands indefinitely at a constant rate of about 0.5 mm h−1 at 37°C (87). Macroscopically, the fungal mycelium appears to be an amorphous collection of equivalent vegetative cells. Functionally, however, the various cells within the mycelium interact to form an ordered network with different hyphae or cells presumably playing distinct roles in the acquisition of nutrients from the environment and in determining the precisely timed development of specialized reproductive structures. While various parameters affecting mycelial development have been described for numerous fungi (129), the molecular mechanisms controlling hyphal growth and colony formation remain largely uncharacterized. Substantial research aimed toward understanding basic cell biological problems associated with hyphal growth, including cell cycle, cytokinesis, and polarity determinants, has been performed, but this is beyond the scope of this review (15, 102, 122, 167).
Approximately 16 h after spore germination, the first phenotypic evidence of hyphal specialization within the colony becomes readily apparent (35, 87). At this time, aerial hyphal branches are formed in the center of the colony and some of these branches subsequently differentiate into conidiophores, the term applied to the asexual spore bearing structures of A. nidulans. It takes between 6 and 8 h from the initiation of aerial growth to the formation of the first asexual spore or conidium, so that a new spore is formed and the asexual cycle can be reinitiated within about 24 h after the original spore germinates (35). Following formation of the first conidiophores within the center of the colony, developmental initiation moves out toward the edge of the colony, leaving the oldest conidiophores in the center and the newly forming conidiophores at the margin of the growing colony. While it takes approximately 16 h of growth for hyphae in the center of the colony to become competent to develop the first conidiophores, newly forming hyphal elements in the radially expanding colony are able to elaborate conidiophores very soon after their appearance. Given that the radial expansion rate of an A. nidulans colony growing on complete medium at 37°C is about 0.5 mm/h and conidiophores can be observed within 1 to 2 mm of the colony margin, it can be surmised that these structures are formed within 2 to 4 h of the appearance of the new hyphae. This is particularly interesting in that the cell cycle in A. nidulans is about 100 min (112) and conidiophores contain dozens of nuclei. Presumably, such rapid formation of conidiophores requires recruitment of nuclei from hyphae, but it could also involve uncharacterized alterations in the cell cycle. Although A. nidulans will also grow vegetatively in liquid submerged culture, with few exceptions (see below) such a culture is aconidial. Typically, hyphae must be exposed to an air interface to stimulate conidiophore production (33, 84).
Termination of asexual reproduction begins in the center of the colony and is followed by commencement of another clearly recognizable developmental process, meiotic (or sexual) spore formation. This stage involves the formation of multicellular fruiting bodies, called cleistothecia, that are surrounded by specialized cells termed Hülle cells (35). Maturation of the sexual fruiting body includes karyogamy and meiosis within the specialized ascogenous hyphae found inside the cleistothecium and concludes with formation of hundreds of asci, each containing eight binucleate ascospores (35, 169). The mechanisms controlling cleistothecial development have not been studied as thoroughly as for conidiation. However, there are many indications that important interactions occur between the asexual and sexual sporulation pathways in A. nidulans (described below). As with conidia, ascospore germination results in the regeneration of the vegetative hyphae and reinitiation of the lifecycle.
Conidiophore Formation
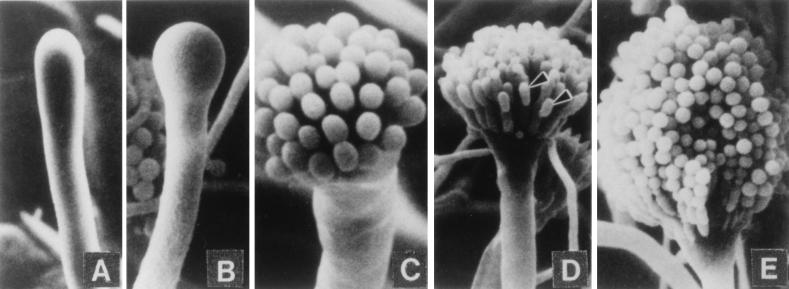
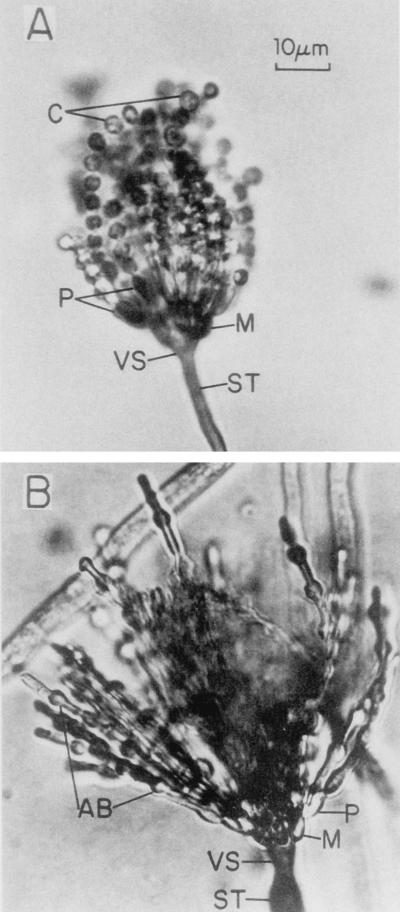
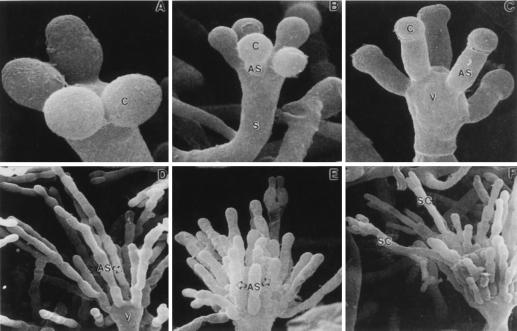
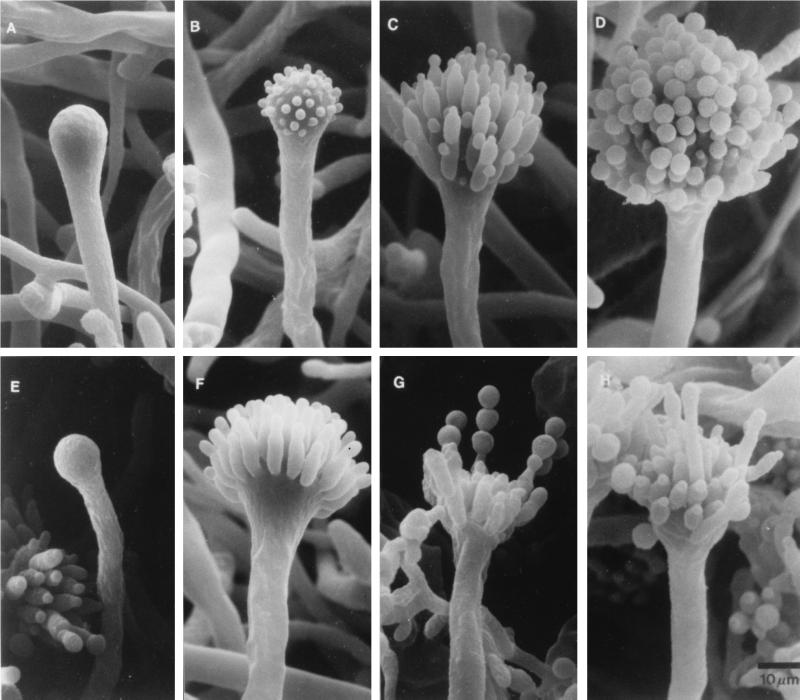
The formation of conidiophores in a colony is a complex process that can be divided into several morphologically distinct stages (Fig. 1) (153). This process begins with the growth of a conidiophore stalk that elongates by apical extension of an aerial branch, much like vegetative hyphal growth (Fig. 1A). The conidiophore stalk differs from a vegetative hyphae in at least three easily recognizable ways (109, 153). First, the stalk cell extends from a specialized thick-walled cell, termed a footcell, that anchors the stalk to the growth substratum. Because it is difficult to identify a footcell in the mycelium before a stalk is observed, it is not really known whether the formation of the thick-walled footcell actually precedes the formation of the stalk, as is typically stated. In maturing conidiophores, the footcell can be distinguished from other mycelial cells by the presence of a two-layered wall in which the outer layer is continuous with the rest of the mycelium while the inner layer is unique to the footcell and the emerging conidiophore stalk (109). The second distinguishing feature of the conidiophore stalk is that it has a 4- to 5-μm diameter as opposed to the 2- to 3-μm diameter more typical of vegetative aerial hyphae. Finally, unlike vegetative aerial hyphae that grow indefinitely and are capable of branching, conidiophore stalks rarely branch and their length is relatively determinate. For A. nidulans, the conidiophore stalk attains a height of about 100 μm, but this is a species-specific trait and varies among the different species of aspergilli.
FIG. 1.
Morphological changes during conidiophore formation. Shown are scanning electron micrographs of the stages of conidiation. (A) Early conidiophore stalk. (B) Vesicle formation from the tip of the stalk. (C) Developing metulae. (D) Developing phialides. (E) Mature conidiophores bearing chains of conidia. Reproduced from reference 109 with permission of the publisher.
After apical extension of the conidiophore stalk ceases, the tip begins to swell and reaches a diameter of ∼10 μm (Fig. 1B). This structure, known as the conidiophore vesicle, is not separated by a septum from the stalk, so that the stalk, vesicle, and footcell essentially comprise a single unit. Mims et al. (109) observed multiple nuclei aligned around the outside of the vesicle that undergo a more or less synchronous division associated with the synchronous production of buds from the vesicle surface to form a layer of primary sterigmata termed metulae (109). However, Fischer and Timberlake (58) reported that the nuclei underwent several synchronized mitoses while the conidiophore stalk was elongating and the vesicle was expanding whereas nuclear movement into the buds was a more gradual process. Microscopic analysis of conidiophore heads indicates that each contains about 60 metulae and that each metular bud forms a cigar-shaped cell about 6 μm long and 2 μm wide that contains a single nucleus (40, 109, 119) (Fig. 1C). Metulae in turn bud twice to produce a layer of about 120 uninucleate sterigmata termed phialides. This metular budding is polar so that both phialides are produced from the end of the metula that is farthest from the vesicle (Fig. 1D). The production of primary and secondary sterigmata in this way has been compared to pseudohyphal growth of Saccharomyces cerevisiae, and some similarities in the genes involved in these processes have been observed (62; see below). Although phenotypically like metulae in size and shape, phialides differ from metulae in that they give rise to chains of uninucleate spores called conidia (Fig. 1E). In this respect, phialides have been likened to stem cells because they undergo repeated asymmetric division to produce chains of spores, but maintain their own identity (153). Phialides also play a supportive role during conidiation producing products that function only in the spore (100, 118, 146, 153). Because each phialide can make upward of 100 spores, the total number of conidia from one conidiophore probably exceeds 10,000. Like stalk height, the formation of sterigmata differs among Aspergillus species (48). Some species lack metula and produce phialides directly from vesicles while others produce more than one layer of metula before making phialides. How and why this diversity in conidiophore structure among closely related fungi developed is an interesting evolutionary problem but has not been addressed experimentally.
PHYSIOLOGICAL REQUIREMENTS FOR CONIDIATION
Acquisition of Developmental Competence
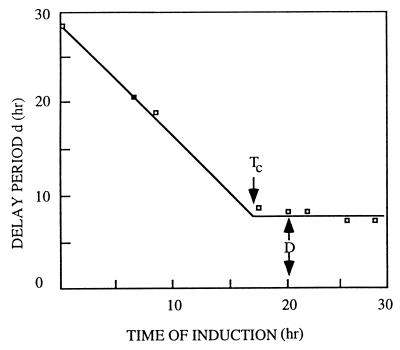
Conidiation does not usually occur in A. nidulans until cells have gone through a defined period of vegetative growth (14, 33). This was demonstrated by taking advantage of the fact that under normal media conditions, A. nidulans can be maintained indefinitely in the vegetative stage of its life cycle by growing hyphae submerged in liquid medium. Asexual sporulation only rarely takes place and sexual sporulation never occurs unless vegetative hyphae are exposed to air. This trait provides a means for synchronous developmental induction by harvesting hyphae grown in submerged culture and then exposing them to air (13, 84). By varying the times for which hyphal cultures are maintained in submerged culture, it has been shown that there is a minimum growth period before cells can respond to induction (14, 170). If development is induced following at least 18 h of vegetative growth, the time required to observe the initial conidiophore-specific structures remains constant at about 5 h (Fig. 2). If the submerged growth period is shorter than 18 h, the time required for development increases correspondingly. These results have been interpreted to mean that cells require approximately 18 h of growth before they are competent to respond to the inductive signal provided by exposure to air.
FIG. 2.
Experimental determination of the timing of developmental competence (Tc). Spores were inoculated into liquid medium at time zero and allowed to germinate in submerged culture. At the times indicated on the horizontal axis, mycelia were transferred to a solid substrate and observed to determine the earliest time of conidiophore formation. The delay time (d) is the interval between transfer to solid medium and the first appearance of conidiophores. The time Tc is the time after which d = D = constant. Reproduced from reference 35 with permission of the publisher.
Although the acquisition of competence is the earliest described conidiation-specific event in the A. nidulans life cycle, it is also the least understood. Neither the concentration of a limiting nutrient such as glucose nor continuous transfer to fresh medium alters the timing with which cells become competent (33, 123). However, precocious conidiation mutants with a decreased time requirement for the acquisition of competence (14, 51) have been isolated, suggesting that there is a genetic component to competence. Physiological changes ranging from differences in the inducibility of enzyme systems to altered glucose uptake have been correlated with cultures obtaining developmental competence, but how these changes are regulated and which events occur first are not known (33, 123). In this regard, it is interesting that these physiological changes take place more quickly in precocious conidiation mutants than in wild-type strains. Taken together, these results have been taken to support the hypothesis that this early aspect of Aspergillus conidiophore development occurs as an integral part of the life cycle rather than as a response to unfavorable environmental conditions.
Developmental Induction
As mentioned above, developmentally competent liquid-grown hyphae from A. nidulans typically do not conidiate until after they are exposed to air (14). The mechanism by which exposure of competent hyphae to air serves to induce conidiation is not understood. The signal is probably not associated with changes in O2 or CO2 levels but has been proposed to involve cell surface changes induced by abrupt formation of an air/water interface at the hyphal surface (113). It is important to note that the conidiation block in submerged culture is not absolute and is dependent on the strain as well as the medium composition (96, 135, 141). Experiments aimed at understanding the genetic events leading to the initiation of conidiation are described later in this review.
Conidiation in A. nidulans strains that have the wild-type allele of the velvet gene (veA+) is red light dependent in that veA+ A. nidulans strains do not conidiate as effectively in the dark as veA1 mutant strains do, even if exposed to air (111). However, 15 to 30 min of continuous exposure to red light at any time after induction and before the formation of differentiated structures apparently activates the developmental program. Exposure to far-red light immediately after exposure to red light at least partially suppresses the activation of conidiation. This photoreversibility of red light induction is reminiscent of phytochrome-mediated responses observed in higher plants, leading to the suggestion that a phytochrome-like molecule in A. nidulans may regulate development. A. nidulans strains carrying the veA1 mutation conidiate in the presence or absence of light, suggesting that the veA product may play a negative regulatory role in the absence of light. Thus, VeA may function similarly to numerous plant genes that are required to prevent photomorphogenesis in dark-grown plants and in roots (111).
Starvation Stress and Sporulation
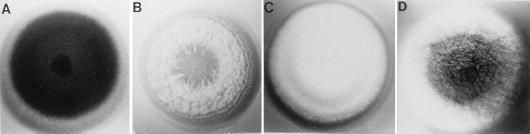
While sporulation by air-exposed A. nidulans colonies apparently occurs in a medium-independent fashion, there have been many reports describing A. nidulans conidiation in submerged culture under conditions where nutrients are limited or in response to other stresses (96, 113, 135). Most recently, Skromne et al. (141) showed that A. nidulans will sporulate following transfer from nutritionally complete medium to medium lacking either a nitrogen or a carbon source for growth (Fig. 3). Although these conditions are apparently sufficient to cause development in submerged culture, it is not yet clear whether such nutritional stress plays a direct role in developmental activation for hyphae grown on the surface of a plate. In fact, nutrient limitation reduces the overall conidiation of surface colonies and poor supplementation of many auxotrophic mutants makes them nearly asporogenous (136; see below). Moreover, continual replacement of the growth medium beneath a surface-grown colony does not inhibit sporulation (123). Thus, while nutritional stress may be sufficient to cause sporulation, it is apparently not required for development to take place. These observations might be explained if the development of conidiophores in wild-type colonies exposed to air occurs in response to an internal signal that activates the sporulation pathway in a genetically programmed way (see below). This signal either is absent or fails to function in submerged culture. However, in the absence of this signal, other environmental cues such as carbon and nitrogen starvation can cause developmental induction. In any case, it is interesting that as with development on the surface of a plate, development of submerged cultures induced by metabolic stress apparently requires that cells go through a period of vegetative growth to become competent before transfer to nitrogen or carbon-free medium will cause development (86, 141).
FIG. 3.
Starvation can induce conidiation in submerged culture. A wild-type strain was grown in glucose minimal medium for 18 h and then shifted to fresh minimal medium (A) or minimal medium lacking a carbon source (B). Conidiophores formed within 12 h after a shift to nitrogen starvation medium (development also occurred upon shifting to medium lacking a nitrogen source), but no conidiophores were seen in the culture shifted to minimal medium containing a usable carbon or nitrogen source. Reproduced from reference 86 with permission of the publisher.
GENETIC CONTROL OF SPORULATION
Differentiation of the multiple cell types making an A. nidulans conidiophore involves the controlled activation of several hundred genes (151–153). However, to date the functions of only a small number of these genes has been determined. Timberlake showed that there are approximately 1,200 diverse mRNAs that accumulate to varying concentrations specifically during conidiation (151). By contrast, Martinelli and Clutterbuck used mutational analysis to estimate that only 45 to 100 genes are uniquely required for asexual sporulation (98). This large discrepancy in predicted gene number could be explained if some genes detected based only on expression patterns encode redundant or incremental functions that would not be detectable by simple visual examination of mutants. In keeping with this idea, deletion from A. nidulans of a 38-kbp region containing numerous spore-specific genes did not result in detectable changes in development (10). Furthermore, subtle defects in sporulation, like the spore wall defect resulting from deletion of the rodA gene (see below), might be difficult to detect in broad screens based on tedious visual examination of colonies (146). Finally, Martinelli and Clutterbuck (98) limited their analysis to mutations that did not alter vegetative growth rates. mRNAs present at high levels during sporulation may also be present at low levels in vegetative cells and may be required for some aspect of normal growth and metabolism. In fact, many cDNAs isolated as conidiation-specific genes have turned out to contain genes involved in carbon metabolism or other cellular processes (115). Mutations in these genes would have been excluded by Martinelli and Clutterbuck. Regardless of the actual number of loci involved, it is certain that many genes are specifically required for sporulation and that, at least in some cases, their transcription is developmentally controlled.
Central Regulatory Pathway
Genetic and biochemical studies of A. nidulans asexual sporulation have led to the identification of only two genes, brlA and abaA, that are specifically required for conidiation (without affecting vegetative growth) and are absolutely necessary for spore formation (45). Together with a third gene called wetA, brlA and abaA have been proposed to define a central regulatory pathway that acts in concert with other genes to control conidiation-specific gene expression and determine the order of gene activation during conidiophore development and spore maturation (21, 110). Mutations in any one of these three genes blocks asexual sporulation at a specific stage in conidiophore morphogenesis and prevents expression of a broad class of developmentally regulated mRNAs (21, 110).
brlA encodes an early regulator of development.
The phenotype of brlA null mutants has been described as “bristle” because the early developmental block in these mutants prevents the transition from polar growth of the conidiophore stalk to swelling of the conidiophore vesicle (45). Instead, brlA mutants differentiate conidiophore stalks that grow somewhat indeterminately, reaching heights 20 to 30 times taller than wild-type conidiophores and giving the colony a “bristly” appearance (Fig. 4). These brlA mutants also fail to accumulate developmentally regulated transcripts including abaA and wetA mRNAs (21). Thus, brlA is proposed to become necessary for activating development-specific gene expression beginning at the time of conidiophore vesicle formation. Two lines of evidence indicate that brlA activity continues to be required up through and including spore formation. First, numerous hypomorphic mutant alleles of brlA that support more extensive conidiophore development than brlA null mutants, differentiating complex structures with different degrees of vesicle and sterigmata development but never spores, have been described (42, 47). These hypomorphic mutants presumably maintain partial BrlA function and result in altered expression patterns for numerous genes encoding development-specific activities. Second, developmental induction experiments with a temperature-sensitive brlA allele, brlA42ts, showed that development was arrested following a shift to the restrictive temperature for BrlA42 activity, regardless of the developmental stage (110).
FIG. 4.
brlA mutants form indeterminate conidiophore stalks. Conidiophores from a wild-type strain (A) and a brlA mutant strain (B) are shown. In the wild-type strain, the stalks grow to a fairly uniform height and bear additional conidiophore-specific structures, while the stalks of a brlA mutant grow somewhat indeterminately and fail to elaborate other conidiophore-specific cells. Arrows indicate stalks. Reproduced from reference 1 with permission of the publisher.
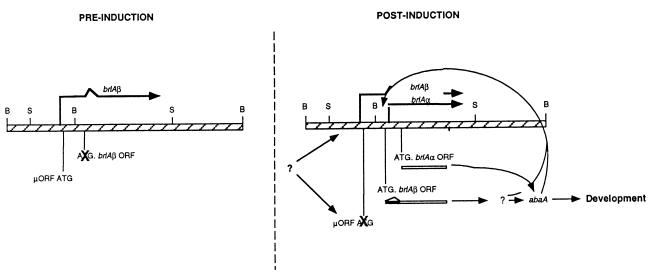
The wild-type brlA gene was isolated by complementation of the mutant (21, 72) and shown to result in two overlapping transcription units, designated brlAα and brlAβ, that each accumulate to detectable levels early in development at about the time conidiophore vesicles first appear (127). brlAβ transcription initiates about 1 kb upstream of brlAα transcription, which begins within brlAβ intronic sequences (127). The brlAβ transcript encodes two open reading frames (ORFs) that begin with AUGs, a short upstream ORF (μORF) and a downstream ORF that encodes the same polypeptide as brlAα except that it includes an additional 23 amino acids at the NH terminus (see Fig. 10). Mutations that block the expression of either transcript alone cause abnormal development (127). However, multiple copies of either brlAα or brlAβ can compensate for loss of the other gene. These results are consistent with the hypothesis that the brlAα and brlAβ transcription units are individually essential for normal development but the products of each gene have redundant functions. Han et al. (68) have shown that brlAα and brlAβ are controlled by different mechanisms and suggested that this complex locus has evolved to provide a mechanism to separate responses to the multiple regulatory inputs activating and maintaining brlA expression throughout development (see below).
FIG. 10.
Model for differential control of brlAα and brlAβ during conidiophore development. The brlAβ mRNA is transcribed in vegetative cells before developmental induction, but translation of the μORF represses translation of BrlA. Following induction, unknown regulatory factors activate BrlA translation from brlAβ by removing the translational block imposed by the μORF, increasing the transcription of brlAβ, or both. Activation of BrlA translation leads to transcription of abaA and other downstream regulatory proteins. This in turn activates a positive-feedback loop that leads to high levels of brlAα expression to cause further developmental changes. Reproduced from reference 68 with permission of the publisher.
The BrlA polypeptide contains a directly repeated sequence that closely resembles the CC/HH Zn(II) coordination sites that are typical of zinc finger DNA binding motifs, indicating that brlA probably encodes a nucleic acid binding protein (1). This possibility is supported by the fact that mutations altering the BrlA CC/HH motif eliminate activity in vivo (2). In addition, A. nidulans brlA expression in Saccharomyces cerevisiae results in brlA-dependent activation of Aspergillus genes in S. cerevisiae, provided that they contain the proposed consensus sites for BrlA protein interaction [BrlA response elements (BREs); (C/A)(G/A)AGGG(G/A)] (36). Although attempts to demonstrate BrlA binding to the BRE element in vitro have been unsuccessful to date, several developmentally regulated genes including abaA, wetA, rodA, and yA have multiple BREs closely associated with their transcription start sites. This observation is consistent with the idea that brlA encodes a primary transcriptional regulator of the central regulatory pathway for development.
The pivotal role for brlA in controlling development is most dramatically demonstrated by results of experiments involving misscheduled activation of brlA in inappropriate cell types. Adams et al. (1) constructed an A. nidulans strain that allowed the controlled transcription of brlA in vegetative cells by fusing the brlA coding region to the promoter for the A. nidulans catabolic alcohol dehydrogenase gene, alcA(p) (63, 124). Because alcA transcription is induced by threonine or ethanol and repressed in the presence of glucose, brlA could be induced or suppressed in vegetative cells by simply changing the medium. When the alcA(p)::brlA fusion strain was transferred from medium with glucose as a carbon source to medium with threonine, hyphal tips immediately stopped growing and differentiated into reduced conidiophores that produced viable conidia (1, 4) (Fig. 5). In addition, this forced activation of brlA resulted in activation of the abaA and wetA regulatory genes, as well as additional developmentally specific transcripts with known and unknown functions.
FIG. 5.
Overexpression of brlA activates sporulation in submerged culture. The brlA gene was placed under the control of the alcohol-inducible promoter alcA. The alcA(p)::brlA strain and a wild-type strain were grown for 12 h in liquid minimal medium containing glucose to repress brlA expression from the alcA promoter and then shifted to alcA(p)-inducing medium. (B) By 3 h after the medium shift, the alcA(p)::brlA strain produced spores from the tips of hyphae. (A) No conidiation was observed in the wild-type strain even 24 h after the medium shift.
Activation of abaA initiates a positive-feedback loop.
abaA encodes a developmental regulator that is activated by brlA during the middle stages of conidiophore development after sterigmata differentiation (9). The phenotype of abaA null mutants is described as “abacus” because these mutants produce aconidial conidiophores that differentiate sterigmata but do not form sporogenous phialides (45, 137) (Fig. 6). Instead, these mutants form branching sterigmata, leading to long chains of cells that appear like beads on a string as in an abacus. Although the brlA transcript accumulates to relatively normal levels in abaA mutants, many other developmentally regulated mRNAs (including the wetA mRNA) do not.
FIG. 6.
Conidiophores from a wild-type strain (A) and an abaA mutant strain (B). The abaA mutant produces normal conidiophore stalks (ST) and vesicles (VS), but metulae (M) and phialides are abnormal and form abacus (AB) structures instead of conidia (C). Magnifications for panels A and B are equivalent. Reproduced from reference 110 with permission of the publisher.
The predicted AbaA polypeptide contains a domain that is very closely related to the DNA binding domain of the simian virus 40 enhancer factor TEF-1 (9, 26). This domain is also closely related to the yeast Ty1 enhancer binding protein TEC1 and has been called the TEA (TEF-1 and TEC1, AbaA) or ATTS (AbaA, TEC1p, TEF-1 sequence) domain. AbaA also contains a potential leucine zipper for dimerization, and, like brlA, abaA is required for transcriptional activation of numerous sporulation-specific genes (21, 110). Results from in vitro biochemical analyses showed that AbaA binds to the consensus sequence 5′-CATTCY-3′ (ARE), where Y is a pyrimidine (8). Multiple AREs are present in the regulatory regions for developmentally regulated genes, including brlA, wetA, yA, rodA, and abaA itself. As with BREs, these cis-acting sequences are able to confer transcriptional activity in a heterologous S. cerevisiae gene expression system that is dependent on the expression of the Aspergillus activator, abaA.
Forced activation of abaA from the alcA(p) in vegetative hyphae resulted in growth cessation and accentuated cellular vacuolization but not in conidial differentiation (4, 110). abaA induction also led to the activation of several developmentally specific genes with known and unknown functions including wetA and, perhaps surprisingly, brlA. Thus, brlA and abaA are reciprocal inducers but brlA expression must occur before abaA expression for productive conidiophore development. The role of abaA in feedback regulation of brlA expression is somewhat complicated by the observation that brlA expression actually increases in abaA null mutants (6). Thus, while overexpression of abaA in hyphae causes brlA activation, brlA is apparently overexpressed in abaA null mutants, indicating that AbaA has both repressive and stimulatory activities toward brlA. Aguirre (6) attempted to explain this paradox by proposing that AbaA functions as a transcriptional repressor of brlA when present at low concentrations but acts as an activator when present at high concentrations, as would occur following alcA promoter-induced expression.
wetA is required late in development.
The wetA gene is required late in development for the synthesis of crucial cell wall components (95). The phenotype of a wetA null mutant is described as “wet-white.” These mutants differentiate normal conidiophores, but the conidia produced are defective in that they never become pigmented but instead autolyse (138).
wetA is predicted to encode a polypeptide that is rich in serine (14%), threonine (7%), and proline (10%) (95). Although analysis of the WetA sequence has given no clear indication of WetA function due to a lack of sequence similarities with other genes in databases, the wetA gene has been proposed to encode a regulator of spore-specific gene expression (95). This hypothesis is based on the finding that wetA mutants fail to accumulate many sporulation-specific mRNAs (21). In addition, forced activation of wetA in vegetative cells caused growth inhibition and excessive branching and resulted in the accumulation of transcripts from several genes that are normally expressed only during spore formation and whose mRNAs are found in mature spores (95). wetA activation in hyphae did not result in brlA or abaA activation and never led to premature conidiation.
stuA and medA modify development.
Two other developmental regulatory genes, stuA and medA, have been shown to be necessary for the precise spatial pattern seen in the multicellular conidiophore (6, 28, 107, 108). stuA and medA have been termed developmental modifiers and are required for a restricted series of cell divisions that establish the spatial organization of the conidiophore. Mutations in either gene give rise to spatially deranged conidiophores, but both stuA and medA mutants are able to produce some viable conidia and are therefore termed oligosporogenous mutants (45).
Mutations in the stuA gene result in the production of extremely shortened conidiophores that lack metulae and phialides and instead produce conidia directly from buds formed on the conidiophore vesicle (45) (Fig. 7A to C). Temporal expression of the central regulatory genes is normal in stuA mutants. However, mutations in stuA lead to the delocalized expression of abaA(p)::lacZ and brlA(p)::lacZ fusions, indicating that stuA may act to establish the proper cellular distribution of these regulatory proteins (108).
FIG. 7.
Electron micrographs of stuA and medA mutants. In the stuA mutant, abnormal conidia are formed either directly from the conidiophore vesicle (A) or from abnormal sterigmata (B and C). Normal metula and phialides are not produced. medA mutant conidiophores are initially nearly normal, but multiple layers of metulae are produced before phialides differentiate and begin to form conidia (D and E). Sometimes sterigmata redifferentiate to produce secondary conidiophores (F). Reproduced from reference 6 with permission of the publisher.
stuA is predicted to encode a transcription factor with significant similarity in its putative DNA binding domain to the binding domains present in several fungal transcriptional regulators (61). These include Swi4, Mpb1, and Phd1 of S. cerevisiae and Res1 and Cdc10 from Schizosaccharomyces pombe. Of these genes, stuA is most similar to the PHD1 gene, whose overexpression leads to pseudohyphal growth, which can be thought of as morphogenetically and perhaps functionally analogous to conidiophore production (61). During pseudohyphal growth, which occurs in response to nitrogen starvation, elongated mother cells bud in a polar fashion to produce elongated daughter cells that in turn bud in a polar fashion to begin the production of a “filament.” In a similar way during conidiophore morphogenesis, elongated metula and phialide cells result from polar budding off the vesicle. These polar divisions are lacking in stuA mutants. Furthermore, overexpression of stuA blocks the ability of BrlA to drive terminal differentiation and promotes a pseudohyphal type of growth pattern, indicating a role for stuA in this process (28). Functionally, it can be argued that both pseudohyphal growth and conidiophore development aid in the dispersal of asexually produced cells beyond growth media that is nutritionally spent.
Like the brlA locus, stuA is complex, and two transcripts, stuAα and stuAβ, are produced from different transcription start sites (108). stuAα initiates within the first intron found in stuAβ, and both transcripts have long 5′ leaders and mini-ORFs similar to those seen at the brlA locus. There is some indication the mini-ORFs play a role in the translational regulation of stuA. At the transcriptional level, there is a ∼50-fold increase in expression following the acquisition of developmental competence and an additional 15-fold increase in expression following developmental induction. A wild-type copy of the brlA gene is in some way required for the postinduction increase (108).
medA mutations result in the production of multiple layers of sterigmata before conidia form (40, 42, 97, 127, 137) (Fig. 7D to F). Occasionally, medA mutant conidiophores bear secondary conidiophores that sometimes produce conidia. The medA mutant phenotype thus indicates that medA is required to maintain commitment to or to pass through the program for conidial differentiation (45, 97). Like stuA and brlA, the medA locus also has two transcription start sites that give rise to two mRNAs (103). In addition, both mRNAs have long leader sequences that contain multiple mini ORFs. This again suggests the possibility of translational regulation. medA transcript levels are high during vegetative growth and decline following developmental induction (103).
While stuA is apparently important for proper spatial distribution of brlA, medA appears to be required for correct temporal expression of brlA (6, 28, 103). Both brlAα and brlAβ transcripts are detected earlier in medA mutant strains than in the wild type. In addition, brlAβ mRNA continues to increase throughout development, which is not the case in a wild-type strain (see below). Therefore, in a medA mutant strain, the ratio of brlAα to brlAβ is lower than in a wild-type strain. Given that abaA is essential for phialide differentiation, it was predicted that medA mutant strains would have abnormal abaA expression. In fact, expression of abaA is greatly reduced or entirely absent, depending on the medA mutant strain (28). Interestingly, the medA mutant phenotype of certain medA mutant strains is suppressed by an extra copy of brlA, but this suppression is cold sensitive (28). This result has been proposed to indicate a physical interaction between MedA and BrlA, and it is thought that MedA might act to stabilize transcription complexes that include BrlA (105).
Coordinated activation of gene expression.
(i) Gene classes.
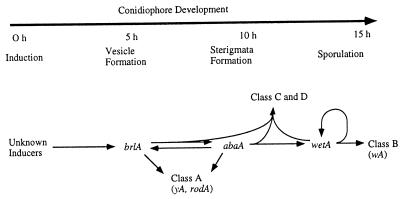
By examining patterns of RNA accumulation in a series of brlA, abaA, and wetA mutant strains during normally induced development or following alcA(p)::brlA, alcA(p)::abaA, or alcA(p)::wetA induction, the nonregulatory developmentally activated genes were divided into four categories (95, 110, 153). Class A genes are activated by either brlA or abaA or both, independent of wetA (Fig. 8). These class A genes are expected to be involved in early development events. abaA alone activates wetA, which in turn activates Class B genes, independent of brlA and abaA. Thus, class B genes are expected to encode spore-specific functions. Class C and D genes require the combined activities of brlA, abaA, and wetA for their expression and have been proposed to encode phialide-specific functions. Class C and D genes are distinguished from one another by their expression patterns during normally induced development in wild-type and mutant strains. Accumulation of wetA mRNA requires wetA+ activity both during normal conidiophore development and in forced-expression experiments, bringing up the interesting possibility that wetA is autogenously regulated (21, 95).
FIG. 8.
Timeline of conidiation and the central regulatory pathway. The genes in the central regulatory pathway for conidiation are predicted to activate other genes responsible for the production of conidiophores. The activation of the class A, B, C, and D genes give rise to the formation of a conidiophore with the timing indicated in the upper part of the figure.
These results have been incorporated into the model describing the genetic processes underlying the temporal and spatial control of gene expression during differentiation of conidiophores shown in Fig. 8. In this model, activation of brlA expression initiates a cascade of events that are coordinated by the interactions of brlA, abaA, and wetA. The timing and extent of expression of the regulatory and nonregulatory genes are determined, as development continues, by intrinsic changes in the relative concentrations of the regulatory gene products in the various conidiophore cell types. There remains a great deal to learn regarding the molecular genetic processes that lead to brlA expression as well as the precise mechanisms through which brlA, abaA, and wetA interact with each other and with stuA and medA in controlling their own expression and the expression of other developmentally regulated genes.
(ii) Pigmentation genes.
The characteristic dark green pigment of wild-type A. nidulans spores results from the production of a spore-specific pigment that gives colonies a green appearance and provides a straightforward means of identifying mutants with abnormal pigmentation (38, 41, 45, 126). These mutants have allowed the identification and characterization of genes involved in pigment synthesis that serve as excellent reporters of developmentally regulated gene expression (155). Although many loci contribute to the nuances of spore color (see below), two genes, wA and yA, are central to the process of spore pigmentation. The wA gene encodes a polyketide synthase, and mutations in wA result in white spored strains that lack melanin and an electron-dense outer spore wall layer that contains α-1,3-glucan (99, 100). yA is predicted to encode a p-diphenol oxidase known as conidial laccase, and yA mutants have yellow spores (12, 38, 84, 171). The fact that wA mutations are epistatic to mutations in yA and that conidial laccase accumulates in the walls of wA mutants has led to the hypothesis that wA catalyzes the synthesis of a yellow pigmented polyketide that is converted to the green form by the product of the yA gene (100). Although these products have not been characterized in A. nidulans, Brown et al. isolated and characterized a compound from a laccase-deficient strain of Aspergillus parasiticus and showed the compound to be an octaketide (24). The transcripts of both yA and wA are developmentally regulated by genes from the central regulatory pathway. wA is apparently regulated as a class B gene (above) in that it is directly controlled by the developmental regulatory gene wetA (99). Both brlA and abaA can direct the activation of yA, making this a class A gene (11, 110).
Many other genes are involved in the production of green pigmented wild-type spores. ygA and yB define two loci that when mutated give rise to yellow spored conidiophores (38, 82). yB was identified by a dominant mutation that could be remediated by addition of high levels of copper, a required cofactor for YA laccase activity. Because ygA mutant strains can also be remediated by the addition of copper, it has been proposed that YB is involved in copper uptake while YgA may function in intracellular copper distribution. wB (132) and wC (99) represent two additional loci that when mutated give rise to white-spored strains, although the relationship of these mutations to the activity of the WA polyketide synthase is not known. Other genes that function in determining spore color include drkA and drkB, which are apparently involved in production of the sac that encloses individual conidia, because mutations in these genes result in conidial chains enclosed in a single sac (46). Mutations in chA (chartreuse), fwA (fawn), bwA (brown), or dilA and dilB (dilute conidia color) result in modified spore color without affecting the levels of conidial laccase. These mutants are probably altered in the quality or quantity of a pigment precursor or in some uncharacterized aspect of spore wall structure (41, 73). Whether or not these genes are controlled by the central regulatory pathway, as in the case of yA and wA, is not known.
While conidial pigmentation is quite obvious from macroscopic examination of colonies, a less easily observed pigmentation of the conidiophore stalk, vesicle, metulae, and phialides also occurs. The normal grayish brown color of these structures becomes visible in mutants that fail to make pigmented spores such as wA mutants or aconidial strains like abaA mutants that are normally grayish (43). Conidiophore pigmentation requires the activity of two genes, ivoA and ivoB, that when mutated give rise to colorless conidiophores. The products of these genes are responsible for synthesis of a melanin-like pigment from N-acetyl-6-hydroxytryptophan (19). The developmentally regulated expression of ivoA and ivoB is most probably controlled by the activity of brlA (19, 43).
(iii) Spore wall protein genes.
Another class of genes with auxiliary functions that are regulated during conidiophore development encodes proteins that make up specific components of the spore wall. Two A. nidulans genes, rodA and dewA, have been shown to encode hydrophobic proteins that contribute to the hydrophobicity of conidia, presumably facilitating their dispersal by air (146, 147). Both of these genes were isolated from a group of cDNA clones called CAN (conidiation A. nidulans) that were isolated based on their developmentally regulated expression (21, 174). RodA and DewA belong to a general class of proteins found in fungi, termed hydrophobins, that have been found in several species including members of the Basidiomycetes, Ascomycetes, and Deuteromycetes (161). Like dewA and rodA, other fungal hydrophobins were identified principally because they are abundantly produced during various cellular differentiation processes. The predicted proteins have little sequence identity at the amino acid level but have certain structural features in common. Most importantly, they all contain eight cysteine residues arranged in a conserved pattern and they have a similar arrangement of hydrophobic domains and thus exhibit similar hydrophobicity plots (54). Hydrophobin-encoding genes in other fungi function in a wide range of processes including spore (16, 83) and hyphal (114, 134, 160) wall hydrophobicity and infection structure formation in plant (148) and insect (145) pathogens.
Disruption of the rodA gene resulted in conidia that were very easily wettable compared to the wild type and gave colonies with a dark center due to the abnormal accumulation of liquid on the conidiophores. This phenotype was shown to arise from loss of the conidial rodlet layer. Disruption of dewA also resulted in spore wall defects, but the phenotype was more subtle. dewA mutant conidia were not wettable with water alone, but addition of a small amount of detergent to the water made them more wettable than wild-type spores. dewA mutants still produced the hydrophobic rodlet layer that is lacking in rodA mutants, and the dewA protein was found to be located in the spore wall. rodA and dewA double mutants are less hydrophobic than either single mutant, indicating that dewA increases hydrophobicity independent of the rodlet layer. It is important to note that the phenotypes of dewA and rodA mutant colonies are sufficiently subtle that these mutants might have been overlooked in the classical screens used to identify genes that function during conidiation. This observation helps to reconcile the disparate estimates of the number of genes that contribute specifically to conidiophore development made by Martinelli and Clutterbuck (98) and by Timberlake (151).
(iv) Other sporulation-related gene functions.
Many of the mutants isolated from early screens for conidiation-specific genes were not only defective in aspects of conidiophore morphogenesis but were also at least partially defective in their vegetative growth properties (42, 45, 98). This implies that some cellular or metabolic functions are more critical during conidiation than during vegetative growth. For example, Champe’s group isolated a mutant that was unable to form asexual spores on complete medium (medium containing the chemically undefined supplement yeast extract), but this conidiation defect could be remediated by addition of arginine to the medium (136). On defined minimal medium (medium without yeast extract), arginine was required for growth but at a much lower concentration than that required for conidiation. Genetic analysis of this mutant showed it to be an allele of the argB locus, which encodes the enzyme ornithine transcarbamylase. Thus, what initially appeared to be a conidiation-specific defect turned out to be a defect in a metabolic process whose function is in greater demand during conidiation (136). In their survey of conidiation mutants, Martinelli and Clutterbuck (98) also isolated auxotrophic mutants for which partial starvation has a more deleterious effect on vegetative growth. These mutants may identify metabolic processes that are critical for asexual sporulation but do not identify genes that are specific to development.
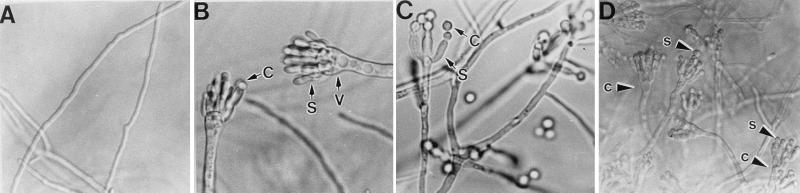
Other potentially interesting nonmetabolic developmental mutants have been neglected because they also had defects in vegetative growth that could not be remediated by medium supplementation. An example of two such genes which have only recently been characterized are the apsA and apsB genes (44, 58). Mutations in either gene result in conidiophores that are apparently normal up to the stage at which primary sterigmata (metulae) are formed. However, nuclei fail to migrate from the vesicle into the metulae, and development stops at the stage of secondary sterigmata bud initiation (Fig. 9). Even in apsA null mutants, this block is not complete, and any metula which by chance acquires a nucleus is able to complete development (58). This suggests that entry of a nucleus into the metula could be a developmental check point that ensures that two separate morphogenetic events, bud formation and nucleation, are complete before the initiation of other downstream events.
FIG. 9.
apsA mutants produce anucleate sterigmata. Electron micrographs showing conidiophore formation in a wild-type strain (A to D) are contrasted with micrographs of an apsA mutant (E to H). In apsA mutants, development of the conidiophore is normal up to the metula stage (E and F). Nuclei typically fail to enter the metulae, and development arrests with an anucleate metular bud (F). Occasionally, nuclei enter metulae, leading to production of functional phialides and normal chains of spores (G and H). Reproduced from reference 58 with permission of the publisher.
The machinery for nuclear migration and division appears to be normal in apsA and apsB mutants, but there is a defect in nuclear positioning. This nuclear positioning defect is not limited to development but can also be seen in the irregular distribution of nuclei in vegetative hyphae (58). The apsA gene has been isolated and sequenced, and it encodes a protein with a putative coiled-coil domain similar to one found in Num1 from S. cerevisiae, a gene product needed for nuclear migration in emerging buds (79). Kruger and Fischer (80) recently described the identification of two extragenic suppressor mutations (samA and samB) that partially bypass the need for apsA in conidium formation. These mutations had additional effects on growth, colony morphology, and sexual spore formation, suggesting that the functions of these genes are also not developmentally specific. There are many other examples of A. nidulans genes that are necessary during vegetative growth but apparently are even more critical during development. Understanding the many nuances of conidiophore morphogenesis and spore formation will certainly require a careful study of genes falling into this category.
Activation of the Central Regulatory Pathway
As described above, Prade and Timberlake (127) demonstrated that mutations separately eliminating brlAα or brlAβ activity caused developmental abnormalities but that additional doses of brlAα in a brlAβ mutant strain or of brlAβ in a brlAα mutant strain remediated these defects. These results have been interpreted to mean that the two brlA-encoded polypeptides have redundant functions and led to the hypothesis that brlAα and brlAβ have evolved to provide separate means of responding to the multiple regulatory inputs controlling brlA expression (68, 127). Han et al. (68) constructed translational fusions between each of the brlAα and brlAβ ORFs and the E. coli lacZ gene to begin to test this hypothesis. They found that brlAα and brlAβ are regulated in different ways (Fig. 10). The brlAβ mRNA is transcribed in vegetative cells before developmental induction but does not accumulate to appreciable levels, presumably because translation of the brlAβ μORF represses BrlA translation to block development. The importance of this translational block was demonstrated by eliminating the μORF AUG, which led to brlA expression in hyphae and inappropriate sporulation. This result implies that one way in which brlA expression could be initiated following induction is through a controlled change in the choice of translational initiation sites from the μORF AUG to the BrlAβ AUG. However, it is also possible that increased transcription of brlAβ provides sufficient template to allow inefficient BrlA translation from the internal AUG. In support of the latter hypothesis, it has been demonstrated that forced activation of brlAβ transcription in hyphae is sufficient to cause sporulation despite the presence of the μORF. In addition, there is a significant increase in brlAβ mRNA levels following developmental induction. Although this has not been shown to result from increased transcription, specific fragments from the brlAβ regulatory region have been shown to confer developmentally specific expression to an otherwise inactive promoter-lacZ fusion (69). In any case, the ultimate result of brlA activation is activation of other developmentally specific genes. Following BrlAβ translation, brlAα transcription is activated primarily through the brlA-dependent positive feedback loop by a mechanism that probably involves AbaA and may also involve BrlA directly. This prediction is supported by the observation that 5 AbaA and 12 BrlA binding sites are found upstream of brlAα (8, 36). We also found that forced overexpression of brlAβ resulted in the accumulation of brlAα mRNA whereas forced activation of brlAα did not result in brlAβ accumulation (68, 69).
Fluffy mutations define early developmental regulators.
In estimating the number of genes required for various stages in development, Martinelli and Clutterbuck proposed that 83% of all sporulation mutants were altered in their ability to initiate development before formation of the conidiophore vesicle and presumably before activation of brlA expression (98). This result indicates that regulation of the decision to initiate conidiophore development is highly complex, requiring the activities of numerous genes (98). The most common phenotype observed among these proposed early conidiation mutants has been described as fluffy. The term “fluffy” includes a diverse group of phenotypes, but in general all fluffy mutants grow as undifferentiated masses of vegetative hyphae forming large cotton-like colonies (56, 98, 149, 165). To begin to understand the role that fluffy genes have in controlling brlA activation, Wieser et al. undertook an extensive genetic analysis of both dominant and recessive mutations that cause a fluffy phenotype (165). The analysis of recessive fluffy mutations led to the characterization of six genes, designated fluG, flbA, flbB, flbC, flbD, and flbE. Loss-of-function mutations in any one of these six genes result in a fluffy colony morphology and greatly reduced brlA expression (Fig. 11). All of these genes have been physically isolated and studied in some detail (3, 85, 87, 162–164). Dominant mutations causing a fluffy phenotype have also been characterized with the thought that this approach could identify some important regulatory genes not found among the recessive fluffy mutants (166). Although these dominant mutants have proven difficult to study by classical genetic approaches, analysis of the mutants resulting from dominant activating mutations in fadA has proven useful for understanding the relationship between growth control and sporulation (173; see below).
FIG. 11.
Phenotypic classes of fluffy mutants. Colonies were grown for 3 days on solid minimal medium. (A) Developmentally wild-type strain of A. nidulans. (B) fluG mutant. (C) flbA mutant. (D) Delayed conidiation mutant typical of flbB, flbC, flbD, and flbE mutants.
(i) fluG is required for production of an extracellular factor and is related to prokaryotic glutamine synthetase I.
Although fluG mutants produce fluffy colonies (3, 85, 165) (Fig. 11B) and fail to activate brlA under normal growth conditions, there are two ways in which they can be induced to conidiate. First, when fluG mutants (even a fluG deletion mutant) are grown on a solid surface under suboptimal nutritional conditions they produce some conidiophores (3). This result has been interpreted to indicate that fluG is required for a genetically programmed aspect of conidiophore development. In the absence of fluG, it is possible to detect sporulation in response to environmental stress. This environmentally regulated response presumably also occurs in the wild type but is overwhelmed by the more prolific fluG-dependent programmed development response. As predicted in this model, fluG mutants can respond to starvation stress in submerged culture by producing conidiophores (86). However, the response differs from that observed for the wild type in two respects. First, although elimination of the carbon source causes fluG mutants to sporulate in submerged culture, the number of conidiophores observed is smaller than in the wild type. Second, although elimination of the nitrogen source does cause morphological changes in the fluG mutant in submerged culture, it does not cause sporulation. Taken together, these results might indicate that there are two aspects to stress-induced sporulation in submerged culture—one that is fluG dependent and a second that is fluG independent. Because overexpression of fluG in submerged culture can cause development (see below), one aspect of stress-induced development could be increased activity of fluG.
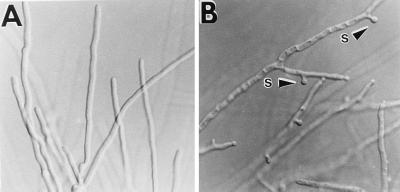
The second way that fluG mutants can be induced to conidiate is by growing next to wild-type strains or strains with mutations in different fluffy genes (85). In this case, a strong band of conidiation can be observed at the interface between two colonies, suggesting that the wild type provides to the mutant strain a missing factor that can stimulate conidiation. This phenotype rescue of the fluG mutant strain occurs even if the two strains are separated by a dialysis membrane with a pore size of 6 to 8 kDa. These results have led to the proposal that fluG is required for synthesis of a low-molecular-weight factor that signals the activation of the major, programmed pathway for conidiophore development independent of the environmental conditions. Without fluG, development can be activated by a mechanism that involves sensing the growth rate or nutrient status directly, bypassing the need for fluG.
The wild-type fluG gene was isolated by complementation of one mutant strain and characterized further (3). The FluG polypeptide was shown to have a molecular mass of about 96 kDa and is present in the cytoplasm of cells grown vegetatively and following developmental induction (85). The carboxy-terminal 436 amino acids of FluG have about 30% identity to prokaryotic glutamine synthetase I (GSI), and there is also limited similarity to eukaryotic glutamine synthetases—particularly in regions thought to compose the active site of the enzyme. While this was the first example of a eukaryotic gene with high similarity to prokaryotic GSI, we have recently found a related gene in the plant Arabadopsis thaliana EST database (e.g., F14033). There are also sequences in the plant EST databases that have high similarity to the amino-terminal half of FluG (59) which otherwise has no significant similarity to any proteins found in the current databases.
The importance of fluG in activating development is supported by the observation that overexpression of fluG in vegetative hyphae is sufficient to cause activation of brlA and sporulation in liquid medium, where conidial development is normally suppressed (86) (Fig. 12B). This result implies a direct role for fluG in controlling development and supports the idea that the concentration of the FluG-supplied conidiation signal normally limits wild-type development in liquid medium. We have shown that this ability to activate sporulation resides in the C-terminal domain that has similarity to prokaryotic GSI (50). However, an actual prokaryotic GSI is unable to substitute for this domain of FluG, indicating that FluG probably has other activities that are distinct from synthesis of glutamine. Taken together, these results have led to the speculation that fluG is involved in the constitutive synthesis of an extracellular sporulation-inducing factor that is related to glutamine or glutamate, but we have not yet been able to isolate this compound (85).
FIG. 12.
Overexpression of fluG, flbA, and flbD causes conidiophore production in submerged culture. The alcA promoter was used to drive the expression of fluG, flbA, and flbD. The alcA(p)::fluG, alcA(p)::flbA, and alcA(p)::flbD strains and a wild-type strain were grown for 14 h in liquid minimal medium containing glucose to repress alcA and then shifted to alcA-inducing medium. Overexpression of fluG (B), flbA (C), and flbD (D) led to production of conidiophores in ∼18 h, 9 h, and 9 h after the shift, respectively. The wild-type strain (A) never produced conidiophores or spores. Panel D is reproduced from reference 162 with permission of the publisher.
(ii) flbB, flbC, and flbD encode nucleic acid binding proteins.
Another group of fluffy mutants can be distinguished by the observation that beginning 2 to 3 days after inoculation, conidiophores were produced in the center of the colony while the margins remained fluffy (Fig. 11D). All of the delayed-conidiation fluffy mutants characterized to date define four distinct complementation groups, designated flbB, flbC, flbD, and flbE (165). The wild-type genes were each isolated by complementation of the respective mutants (162–164). As described for fluG, the cloned flbB, flbC, flbD, and flbE genes recognize RNAs that are present in vegetative cells and continue to be expressed at relatively constant levels during development. flbD is predicted to encode a 314-codon polypeptide that includes a region with significant identity to the DNA binding domain in the human proto-oncogene c-myb (91, 162). flbC is predicted to encode a 354-amino-acid polypeptide (164) that includes two CC/HH zinc finger domains typical of a large group of DNA binding proteins including A. nidulans BrlA (1, 106). flbB is predicted to encode a 389-amino-acid polypeptide (163) with similarity to b-Zip DNA binding proteins (159). Thus, flbB, flbC, and flbD are likely to function as DNA binding proteins and could control the transcriptional activation of other developmental regulators, like brlA, in response to sporulation signals. To date, no similarities between flbE and other proteins in the available databases have been observed (163). As with fluG, overexpression of either flbC or flbD in submerged hyphae results in the activation of brlA expression and sporulation (Fig. 12D) (analyses of flbB and flbE overexpression are in progress), further supporting the hypothesis that these proteins play a direct role in initiation of development.
(iii) flbA antagonizes a G-protein-mediated signaling pathway that inhibits sporulation.
The final group (Fig. 11C) of fluffy mutants analyzed is distinguished by the fact that the center of the colony begins to disintegrate by 3 days after inoculation and the entire colony has autolysed so that only a few hyphal strands remain by 5 days postinoculation (87, 165). All of the recessive mutant strains that have been identified as having this fluffy autolytic phenotype have been shown to result from mutations in a single locus designated flbA. The flbA gene produces a 3-kb mRNA that, like fluG, flbB, flbC, flbD, and flbE, was present at low levels in vegetative cells and whose level remained relatively constant throughout the life cycle, although minor increases were observed following developmental induction (87). As with each of the other genes discussed, overexpression of flbA in submerged hyphae activated simple conidiophore development (Fig. 12C).
The DNA sequence of the flbA gene contains one long ORF predicting a 719-amino-acid polypeptide that is most significantly similar to the S. cerevisiae Sst2 polypeptide (55, 87). Recent work has shown that FlbA and Sst2 are members of a large protein family that includes examples from a wide range of organisms including yeast and humans (53, 57, 78). All of the proteins in this family share a conserved C-terminal domain of ∼120 amino acids that has been termed the RGS domain (regulator of G-protein signaling). Both genetic and biochemical experiments indicate that this region is important in negatively controlling heterotrimeric G-protein-mediated signal transduction pathways by facilitating intrinsic GTPase activity of the Gα subunit (18, 55, 78, 81, 173). Thus, one likely function for flbA is in similarly regulating an intracellular signaling pathway for conidiation.
The role of FlbA in controlling conidiation was clarified through characterization of a dominant mutation in a different gene called fadA (fluffy autolytic dominant A), which resulted in a fluffy autolysis phenotype similar to that of flbA loss-of-function mutants (173). The fadA gene encodes a heterotrimeric G protein α-subunit that has >90% identity to the Neurospora crassa Gna1 and the Cryphonectaria parasitica CPG1 proteins, and all three fungal proteins have ∼55% identity to mammalian Giα proteins (37, 156). Comparison of the sequence from fadA dominant mutant alleles that cause a fluffy autolytic phenotype with the wild-type fadA sequence and with other genes encoding Gα proteins revealed that the dominant fluffy phenotype results from mutations that cause a decrease in the intrinsic GTPase activity (i.e., Gly42 to Arg, Arg178 to Cys or Leu, or Gln204 to Leu) (49, 143, 166, 173). Such loss of GTPase activity mutations should cause constitutive signaling, indicating that activation of the FadA signaling pathway leads to proliferation and blocks sporulation (173). Thus, activation of FadA signaling causes the same phenotype as does loss of FlbA function. This is the expected result if the normal role of the FlbA RGS domain protein is to interfere with FadA activity. This hypothesis was further supported by the finding that loss-of-function fadA mutations suppressed the developmental defect of flbA deletion mutants. Moreover, a dominant interfering allele of fadA (fadAG203R) caused hyperactive asexual sporulation and, like flbA overexpression, caused sporulation in submerged culture. This particular mutation is known to affect the ability of GTP-bound Gα protein to dissociate from the Gβγ subunits (131), indicating that Gβγ may be partially involved in signaling growth and preventing sporulation.
(iv) Complex interactions among “fluffy” genes.
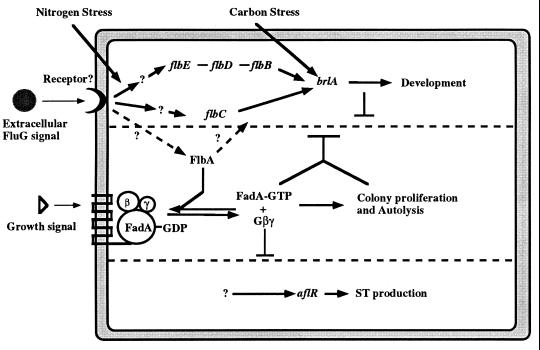
We have examined the possible interactions among various “fluffy” genes by assessing the phenotypes of different combinations of mutants (165, 173) and by testing the genetic requirements for sporulation induced by overexpression of fluG, flbA, and flbD (86, 162) (experiments with other genes are in progress). The results of these experiments have led to the proposal that there are two antagonistic signaling pathways regulating A. nidulans growth and development, as shown in Fig. 13 (173). In this model, growth signaling is mediated by the FadA G protein α-subunit (possibly in conjunction with Gβγ). Activation of FadA by exchange of GDP for GTP results in a proliferative phenotype and blocks sporulation. This presumably involves interaction of a ligand with a G-protein-coupled receptor (117), but no such molecules have been identified to date. Developmental activation requires that the FadA signaling pathway be at least partially inactivated, and this requires FlbA, which, by analogy to other RGS domain proteins (18), presumably activates the FadA GTPase to inactivate signaling. Thus, the major function of FlbA is not to activate development specifically but instead is to antagonize the FadA-GTP growth signal and thereby allow sporulation. However, it is also apparent that FlbA has additional functions, because overexpression is able to cause inappropriate activation of asexual development even in fadA deletion mutant strains (173).
FIG. 13.
Model describing fluffy gene interactions in controlling initiation of development. As described in the text, we have proposed that the activities of two antagonistic signaling pathways determine whether development and secondary metabolism occur (70, 86, 173). One pathway requires the product of FluG activity, which is proposed to work as an extracellular signal to activate a sporulation-specific pathway that requires flbB, flbC, flbD, and flbE. When the FadA Gα protein is GTP bound, it regulates downstream effectors to enhance proliferation and repress both sporulation and ST production. The FluG signal causes inactivation of FadA by activating FlbA, which functions as a GTPase-activating protein to turn off the FadA-dependent signaling pathway. This inactivation of FadA then allows both sporulation and ST biosynthesis to occur.
Development-specific signaling requires the fluG gene product, which is apparently necessary for the synthesis of a small diffusible factor that functions extracellularly to signal developmental initiation (85, 86). Because the wild type and other fluffy mutants are able to rescue the fluG mutant conidiation defect in an extracellular fashion, we have proposed that FluG is needed for the production of an extracellular developmental signal and that other gene products are needed for responding to this signal. This is also supported by the finding that fluG overexpression-induced sporulation requires the wild-type activities of other early-acting developmental regulatory genes including flbA (86). Because flbA-activated sporulation also requires wild-type fluG activity, we have proposed that there are two primary consequences of responding to FluG factor and that these are presumably mediated through interactions between the factor and an as yet unidentified membrane associated receptor. The first function is activation of FlbA, which then interferes with FadA signaling of proliferation. The second function involves activation of a development-specific pathway that requires the products of other genes including flbB, flbC, flbD, flbE, and brlA (1, 86, 165). Although the asexual sporulation defects in flbA null mutants are suppressed by either dominant or recessive loss-of-function mutations in fadA, the defects in fluG mutants are not (173). This is presumably because the development-specific role of FluG is FadA independent and both FluG-dependent processes must occur if development is to proceed.
flbB, flbC, and flbD all encode putative transcription factors that represent excellent candidates for direct activators of brlA expression. Mutations in each of these genes cause a similar delayed conidiation phenotype, but FlbC is proposed to function independently of FlbB and FlbD (and FlbE) because flbC mutations, in combination with flbB, flbD, or flbE mutations, result in additive developmental defects (165). Given that each of these genes is transcribed constitutively but apparently functions specifically in sporulation one possible consequence of FluG signaling is some posttranslational modification of one or all of the products of these genes leading to their activation. Whatever the mechanism, we expect that additional activities must be needed for developmental activation. It is not yet clear if the genes required for such early developmental events as sensing and transmitting the FluG signal or mediating FadA-dependent repression of sporulation will be found among the as yet uncharacterized fluffy mutations or if other approaches will be required to identify them.
(v) Early developmental regulators affect control of secondary metabolism.
A. nidulans contains a cluster of 25 genes that encode enzymes required to synthesize a toxic and carcinogenic secondary metabolite called sterigmatocystin (ST), a precursor of the better-known fungal toxin aflatoxin (AF) (25). One ST cluster gene (stc gene), aflR, functions as a pathway-specific transcriptional activator of other genes in the ST pathway, and aflR expression is regulated during the life cycle such that aflR mRNA begins to accumulate early in the stationary phase and activation of other ST biosynthesis genes quickly follows (168, 172). Numerous observations have supported the hypothesis that microbial secondary metabolite production and sporulation are intimately associated (17, 52). However, the general mechanisms controlling the activation of aflR and synthesis of ST and AF in conjunction with sporulation have not been understood (75, 76).
The genetic basis of the relationship between asexual development and secondary metabolism and possible common elements in the regulation of these two processes were studied by examining the early-acting developmental mutations on ST production in A. nidulans (70). We found that loss-of-function mutations in flbA or fluG or dominant activating mutations in fadA result in a block in both stc gene activation and asexual sporulation. By contrast, overexpression of flbA (but not fluG) or dominant interfering fadA mutations (fadAG203R) caused precocious stc gene activation and ST accumulation. Finally, fadA loss-of-function mutations suppressed the need for either flbA or fluG in activating ST production but only suppressed the flbA requirement for sporulation (70, 173). These results are consistent with a model presented above (Fig. 13) in which the proposed FluG factor stimulates both development-specific events and activation of FlbA, which in turn inactivates FadA-dependent proliferation signaling. Because FadA signaling antagonizes both sporulation and ST production, FadA inactivation by FlbA is a prerequisite for both processes. However, as described for sporulation, this model must have further complications in that FlbA must have some FadA-independent function. This follows from the observation that flbA overexpression in a fadA-deletion mutant activated stc gene expression, ST production, and sporulation just as well as it did in a wild-type strain.
Although ST and AF production usually occurs during sporulation in many Aspergillus spp., the opposite is not true, in that ST and AF production can take place even when no sporulation is observed (e.g., in submerged cultures). This finding presumably indicates an important difference in the control of development versus ST biosynthesis. This difference is reflected by the complex requirements for FluG in both processes. While ΔfluG mutants fail to make ST and do not sporulate, mutations that inactivate FadA suppress the requirement for FluG in ST production but not sporulation. This implies that the main role of FluG in ST biosynthesis is activation of FlbA, which then inactivates FadA. Although FluG-dependent activation of FlbA is also needed for sporulation, there is an additional pathway-specific role for FluG that results in the activation of genes directing conidiation (Fig. 13). It is not yet known if similar pathway-specific controls are also required for activating aflR and stc gene expression.
Other genes involved in developmental activation.
(i) Isolation of developmental regulators by overexpression.
An interesting consequence of forced overexpression of developmental regulators like brlA and flbA in vegetative hyphae is that in addition to activating development, their expression inhibits vegetative growth (4, 85, 86, 162). In the case of forced brlA expression, growth inhibition has been shown to be accompanied by generalized metabolic shutdown including decreases in protein and RNA accumulation, suggesting that activating development has broad consequences on metabolism (4). Marhoul and Adams (93) took advantage of this observation in designing a screen for developmental regulators among genes that negatively affect growth when overexpressed. This was accomplished by constructing a genomic DNA library in which random fragments from partially restricted wild-type genomic DNA were inserted next to the alcA(p) in a plasmid that also contained the argB+ gene (157) for use as a selectable marker in transformation experiments. This genomic DNA expression library was used to transform an argB A. nidulans mutant, selecting for arginine prototrophy on medium that causes repression of the alcA promoter (e.g., glucose-containing medium). Transformants were then replicated to medium with alcA(p)-repressing or -inducing (e.g., alcohol) carbon sources and screened for colonies that grew on repressing but not on inducing medium. In this way, a total of 66 different transformants with unique plasmid integration patterns were recovered.
A number of different genes with known and unknown functions have been shown to block growth in yeast or Aspergillus when expressed at high levels (1, 27, 71, 89, 110, 120, 121, 133). Along with developmental activators, these include genes encoding cytoskeletal components like actin and tubulin, both kinases and phosphatases, and proteins involved in the movement of other proteins into the nucleus. To identify mutants that were growth inhibited due to overexpression of developmental activators from those that were inhibited for other reasons, each of the mutants was grown under alcA-inducing conditions and examined microscopically for inappropriate sporulation, and RNA was extracted and examined for brlA activation. High levels of brlA mRNA expression were detected in 16 of the 66 growth inhibited mutants, and in at least 7 of these, hyphal tips differentiated to form reduced conidiophores (93). The phenotype of these 16 mutants was described as FAB (for forced activators of brlA expression). The developmental roles of most of the genes identified in this screen have not yet been determined, but two genes, fabM and fabP, were recovered and characterized in some detail. fabM is predicted to encode an essential poly(A) binding protein (92). While it is not surprising that a gene encoding a poly(A) binding protein is essential for viability, it is not clear why overexpression of fabM caused sporulation. However, this activity did require other developmental regulators including brlAβ, flbA, flbB, flbC, and fluG. We have proposed that fabM is an example of an essential gene that also plays some specific developmental role, presumably related to allowing the translation of developmentally important mRNAs like brlA (92). We have found that fabP, in contrast to fabM, is not required for growth or development, although its overexpression can cause developmental activation in a brlA-dependent manner (94). Because the predicted FabP protein contains a domain with high similarity to serine/threonine kinase domains, we predict that FabP has a redundant function in a kinase cascade associated with developmental induction.
(ii) A role for ras?
Much of the data discussed above points to the idea that there is a signal transduction pathway(s) that leads to the initiation of conidiophore development in A. nidulans. The genes isolated thus far share motifs of genes commonly found in signal transduction pathways. For instance, the data on fluG indicates that it may produce a diffusible substance that could act as a signal, while flbA is similar to genes that modulate G-protein-mediated signals. In addition, putative transcription factors have been isolated that may function in response to the activities of the fluG or flbA to activate genes involved in conidiophore formation (85, 86, 162, 165). Although some of the standard components of signal transduction pathways have been isolated by complementation of a particular developmental mutant phenotype, genes that might have been expected such as kinases or receptors have not been isolated to date.
A more direct approach to studying signal transduction and development was taken by Som and Kolaparthi, who isolated a ras homolog (A-ras) from A. nidulans by probing an A. nidulans library with the S. cerevisiae RAS gene (142). In eukaryotes from yeast to humans, ras has been implicated in the transduction of signals involved in growth and differentiation (60, 67, 77, 125, 130, 140). The Ras protein has been shown to exist in two different forms in the cell membrane, an inactive GDP-bound form and an active GTP-bound form. In response to various signals, GDP is exchanged for GTP and Ras becomes active to transduce external signals across the cell membrane. Data from the Som laboratory has shown that the A. nidulans ras homolog is an essential gene (142). Two lines of experiments were conducted to determine a possible role for A-ras during development. A mutation known to be activating in other Ras proteins was made and placed under the control of the inducible promoter alcA, so that its levels could be altered during growth and development. Likewise, a mutation that inactivates other Ras proteins was made and placed under alcA(p) control. Altering the levels of active and inactive Ras during development indicated that the level of active Ras could play a role in determining progression through the stages of growth and conidiophore development. While a high level of active Ras is apparently required for germination, the level of active Ras must decrease for conidiophore development to proceed. Som has speculated that there are several concentration thresholds for active A-Ras that allow development to proceed to a certain point, giving rise to a particular cell type while inhibiting further development (142). That A-ras is an essential gene in A. nidulans made its isolation by traditional screens impossible; it is possible that other genes in the pathway were also missed because they are essential or alternatively because mutations in such genes cause no detectable phenotype.
(iii) Acquisition of developmental competence.
Another interesting developmental event occurring prior to brlA activation is the acquisition of developmental competence. Previously, Champe’s group used conditional mutations to show that three different aco (aconidial) genes, acoA, acoB, and acoC, are blocked in conidiation before the acquisition of developmental competence (29, 30, 33, 170). These precompetence mutants were isolated based on the idea that genes required for competence may no longer be essential once a strain has already become competent. It was reasoned that temperature-sensitive precompetence mutants could be distinguished from aconidial mutants affected in postcompetence events by growth at the permissive temperature in submerged culture past the time needed by the wild type for competence followed by a simultaneous shift to the restrictive temperature and inducing conditions. Precompetence mutants conidiate because the functions they are missing at the restrictive temperature are no longer needed. All three preinduction mutants identified by this screen are nonallelic, and all are blocked in both asexual and sexual sporulation.
acoA, acoB, and acoC mutants grown at the restrictive temperature all overproduce abundant quantities of a complex set of metabolites that absorb UV light (29, 30). At least some of these compounds are diphenyl ethers, which probably arise from acetate through the polyketide pathway (29, 35). Although acoA, acoB, and acoC mutants are sexually sterile at the restrictive temperature, they also produce large quantities of a hormone-like fatty acid metabolite called psi (precocious sexual inducer) factor that stimulates sexual sporulation while inhibiting asexual sporulation in wild-type strains (32, 34, 101). It has been suggested that the mutational defect in these preinduction mutants deregulates an enzymatic activity that is common to both fatty acid and polyketide biosynthetic pathways (35). The acoB gene was recently isolated; however, comparison of its sequence to the sequences of proteins in the databases yielded no information about its putative function (88).
Relationships between Asexual and Sexual Sporulation
As described above, A. nidulans is able to produce spores sexually as well as asexually. Sexual reproduction typically occurs after asexual sporulation has stopped and involves the formation of macroscopic (125- to 200-μm) fruiting bodies called cleistothecia that are filled with asci, each containing eight binucleate ascospores. While cleistothecium and ascospore formation is clearly a distinct process from conidiophore and conidia production, these two important events in the life cycle also have certain features in common. For instance, many mutations that were originally identified as causing defects in conidiophore development also block sexual development (116, 169). stuA mutants are unable to differentiate cleistothecia or the specialized Hülle cells that normally surround them, while medA mutants are unable to produce cleistothecia but do form Hülle cells (45). Therefore, these genes may play a regulatory role in both developmental processes.
The veA gene also plays an important role in determining sexual sporulation; however, the effect is directly opposite to what is observed for conidiation (169). veA1 mutant strains are generally reduced in cleistothecial production but conidiate more effectively than veA+ strains. Moreover, incubating veA+ strains in the dark reduces conidiation but cleistothecial formation initiates earlier and is more prolific than when the same strains are incubated in the light. Cleistotheciation and conidiation by veA1 mutants are not affected by light. Han et al. (64, 65) found that conidiation and cleistotheciation in veA+ and veA1 strains could also be affected by medium composition. When veA+ strains were grown in the light with either 2% glycerol, 1% lactose, or 1% galactose as the carbon source, there was a greater-than-100-fold reduction in sporulation in comparison to that seen for the same strain grown on medium with 1% glucose (Table 1). Conidiation by the veA1 mutant was the same on all media, and sexual development was not significantly affected in veA+ or veA1 strains. In contrast, medium containing 2% acetate or 6% glucose blocked cleistothecial development in both veA+ and veA1 strains. How veA affects and is affected by these diverse conditions remains to be understood.
TABLE 1.
Effect of carbon sources on sexual and asexual developmenta
| Carbon source | Sporulation in strain:
|
|||||
|---|---|---|---|---|---|---|
|
veA+
|
veA1
|
nsdD19 veA+
|
||||
| ASb | Sc | AS | S | AS | S | |
| 0.5% glucose | ++ | − | ++ | − | ++ | − |
| 1% glucose | ++ | ++ | +++ | + | +++ | − |
| 2% glucose | ++ | +++ | +++ | ++ | +++ | − |
| 6% glucose | +++ | − | +++ | − | +++ | − |
| 2% glycerol | − | ++ | ++ | ++ | +++ | − |
| 1% lactose | − | ++ | ++ | ++ | +++ | − |
| 1% galactose | − | +++ | ++ | ++ | +++ | − |
| 2% acetate | +++ | − | +++ | − | +++ | − |
Asexual sporulation. Approximately 106 conidia were inoculated onto 30 ml of solid medium containing various C sources and incubated at 37°C for 72 h. The number of conidia was determined per 0.8-mm2 area: −, <5 × 104; +, 5 × 104 to 1 × 106; ++, 1 × 106 to 1 × 107; +++, >1 × 107.
Sexual fruiting-body formation. Samples were grown as for the determination of conidial yield, except that cultures were incubated for 108 h before harvest. The number of cleistothecial initials was determined by staining for laccase II activity and is expressed per 0.8-mm2 area: −, 0; +, <10; ++, 10 to 50; and +++, >50.
Finally, although technical difficulties have limited efforts to study the genetics of sexual fruiting-body formation directly, some recent progress has been made. Han et al. (65, 66) have described an extensive screen for mutants blocked in cleistothecial development and defined three mutant classes. The first class includes 20 mutants called NSD (never in sexual development) because these strains fail to produce any cleistothecia or Hülle cells. The second class includes more than 100 mutants called BSD (blocked in sexual development) because these mutants initiate cleistothecial development but are blocked at some step before ascospore formation. The third class includes more than 100 mutants called ASD (abnormal sexual development) because these mutants produce cleistothecia that are small or appear with some delay compared to the situation in the wild type. The 20 NSD mutants all result from recessive mutations and define four separate complementation groups designated nsdA, nsdB, nsdC, and nsdD. The nsdD gene was recently cloned (31) and shown to encode a GATA-type zinc finger DNA binding protein (U70044). Interestingly, when nsdD was inserted into the A. nidulans genome in high copy number, it caused a nearly complete block in asexual sporulation. Understanding the complex regulatory interactions between genes like nsdD that function primarily in determining sexual sporulation and those that function in activating asexual sporulation will probably provide important insights into the mechanisms controlling both processes and how these important events are coordinated in the A. nidulans life cycle.
CONCLUSIONS
The ease with which A. nidulans can be manipulated through both classical and molecular genetics has made it possible to learn much about the genetic controls for asexual sporulation in this fungus; however, much remains to be learned. Work must be continued to determine the biochemical activities of the developmental regulatory genes isolated to date. New screens and selection methods will have to be implemented to identify genes that subtly modify the activities of the regulatory genes and formation of conidiophores. In addition, little is known about the internal and external signals that lead to the acquisition of developmental competence or, for that matter, the mechanisms that control spore dormancy and germination. Although genes that may function to transduce a signal for conidiophore development have been identified, the nature of the signals themselves has not been determined and many components of the signal transduction pathways remain to be elucidated. Finally, most work has concentrated on understanding the mechanisms involved in the formation of a single conidiophore and has neglected the colony as a developmental unit. Future work should therefore address the coordinate regulation of colony development.
It will also be interesting to learn if any of the signals and genes found to be important for asexual sporulation in A. nidulans are used for the same process in other filamentous fungi. While variation in conidiophore size and structure has been noted among the various aspergilli, it is not known how this variation is determined. Homologs of brlA, fluG, and flbA have been found in other species of Aspergillus (20), but nothing has been done to address potential differences in the activities of these genes. We have recently described the identification of a N. crassa homolog of flbD and shown that this gene can complement A. nidulans flbD mutants (139). However, deletion of this gene from N. crassa had no measurable effect on conidiation or growth. To date, no other putative homologs of A. nidulans developmentally regulatory genes have been identified in such distantly related members of the Ascomycetes, though very little effort has been made in this direction. It is possible that the mechanisms and genes that control conidiation in other filamentous fungi will prove to be as diverse as the types of conidiophores produced by these organisms.
ACKNOWLEDGMENTS
We thank Cletus D’Souza, Bee Na Lee, Sangtae Han, John Marhoul, and John Fondon for communicating results prior to publication, and we thank our other colleagues in the laboratory for their comments and assistance in assembling this review.
Work from the T.H.A. laboratory was partially supported by grants from USDA and NIH. J.W. was partially supported by an NSF training grant to the Program for Biology of Filamentous Fungi at Texas A&M University.
REFERENCES
- 1.Adams T H, Boylan M T, Timberlake W E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 2.Adams T H, Deising H, Timberlake W E. brlA requires both zinc fingers to induce development. Mol Cell Biol. 1990;10:1815–1817. doi: 10.1128/mcb.10.4.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams T H, Hide W A, Yager L N, Lee B N. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol Cell Biol. 1992;12:3827–3833. doi: 10.1128/mcb.12.9.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams T H, Timberlake W E. Developmental repression of growth and gene expression in Aspergillus. Proc Natl Acad Sci USA. 1990;87:5405–5409. doi: 10.1073/pnas.87.14.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams T H, Timberlake W E. Upstream elements repress premature expression of an Aspergillus developmental regulatory gene. Mol Cell Biol. 1990;10:4912–4919. doi: 10.1128/mcb.10.9.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguirre J. Spatial and temporal controls of the Aspergillus brlA developmental regulatory gene. Mol Microbiol. 1993;8:211–218. doi: 10.1111/j.1365-2958.1993.tb01565.x. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre J, Adams T H, Timberlake W E. Spatial control of developmental regulatory genes in Aspergillus nidulans. Exp Mycol. 1990;14:290–293. [Google Scholar]
- 8.Andrianopoulos A, Timberlake W E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 1994;14:2503–2515. doi: 10.1128/mcb.14.4.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrianopoulos A, Timberlake W E. ATTS, a new and conserved DNA binding domain. Plant Cell. 1991;3:747–748. doi: 10.1105/tpc.3.8.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aramayo R, Adams T H, Timberlake W E. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics. 1989;122:65–71. doi: 10.1093/genetics/122.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aramayo R, Timberlake W E. The Aspergillus nidulans yA gene is regulated by abaA. EMBO J. 1993;12:2039–2048. doi: 10.1002/j.1460-2075.1993.tb05853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aramayo R, Timberlake W E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 1990;18:3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Axelrod D E. Kinetics of differentiation of conidiophores and conidia by colonies of Aspergillus nidulans. J Gen Microbiol. 1972;73:181–184. doi: 10.1099/00221287-73-1-181. [DOI] [PubMed] [Google Scholar]
- 14.Axelrod D E, Gealt M, Pastushok M. Gene control of developmental competence in Aspergillus nidulans. Dev Biol. 1973;34:9–15. doi: 10.1016/0012-1606(73)90335-7. [DOI] [PubMed] [Google Scholar]
- 15.Beckwith S M, Roghi C H, Morris N R. The genetics of nuclear migration in fungi. Genet Eng. 1995;17:165–180. [PubMed] [Google Scholar]
- 16.Bell-Pederson D, Dunlap J C, Loros J J. The Neurospora circadian clock-controlled gene, ccg-2 is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 1992;6:2382–2394. doi: 10.1101/gad.6.12a.2382. [DOI] [PubMed] [Google Scholar]
- 17.Bennett J W. Secondary metabolism and differentiation in fungi. In: Bennett J W, Ciegler A, editors. Secondary metabolism and differentiation in fungi. New York, N.Y: Marcel Dekker, Inc.; 1983. pp. 1–32. [Google Scholar]
- 18.Berman D M, Wilkie T M, Gilman A G. GAIP and RGS4 are GTPase activating proteins (GAPs) for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 19.Birse C E, Clutterbuck A J. Isolation and developmentally regulated expression of an Aspergillus nidulans phenol oxidase-encoding gene, ivoB. Gene. 1991;98:69–76. doi: 10.1016/0378-1119(91)90105-k. [DOI] [PubMed] [Google Scholar]
- 20.Borgia P T, Dodge C L, Eagleton L E, Adams T H. Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans. FEMS Microbiol Lett. 1994;122:227–232. doi: 10.1111/j.1574-6968.1994.tb07172.x. [DOI] [PubMed] [Google Scholar]
- 21.Boylan M T, Mirabito P M, Willett C E, Zimmerman C R, Timberlake W E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol. 1987;7:3113–3118. doi: 10.1128/mcb.7.9.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brody H, Carbon J. Electrophoretic karyotype of Aspergillus nidulans. Proc Natl Acad Sci USA. 1989;86:6260–6263. doi: 10.1073/pnas.86.16.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brody H, Griffith J, Cuticchia A J, Arnold J, Timberlake W E. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 1991;19:3105–3109. doi: 10.1093/nar/19.11.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown D, Hauser F, Salvo J. Structural elucidation of a putative conidial pigment intermediate in Aspergillus parasiticus. Tetrahedron Lett. 1993;34:419–422. [Google Scholar]
- 25.Brown D W, Yu J-H, Kelkar H S, Fernandes M, Nesbitt T C, Keller N P, Adams T H, Leonard T J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bürglin T R. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66:11–12. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- 27.Burke D, Gasdaska P, Hartwell L. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busby T, Miller K Y, Miller B L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of brlA and stuA genes. Genetics. 1996;143:155–163. doi: 10.1093/genetics/143.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butnick N Z, Yager L N, Hermann T E, Kurtz M B, Champe S P. Mutants of Aspergillus nidulans blocked at an early stage of sporulation secrete an unusual metabolite. J Bacteriol. 1984;160:533–540. doi: 10.1128/jb.160.2.533-540.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butnick N Z, Yager L N, Kurtz M B, Champe S P. Genetic analysis of mutants of Aspergillus nidulans blocked at an early stage of sporulation. J Bacteriol. 1984;160:541–545. doi: 10.1128/jb.160.2.541-545.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chae K S, Kim J H, Choi Y, Han D M, Jahng K Y. Isolation and characterization of a genomic DNA fragment complementing an nsdD mutation of Aspergillus nidulans. Mol Cells. 1995;5:146–150. [Google Scholar]
- 32.Champe S P, el Zayat A. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J Bacteriol. 1989;171:3982–3988. doi: 10.1128/jb.171.7.3982-3988.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champe S P, Kurtz M B, Yager L N, Butnick N J, Axelrod D E. Spore formation in Aspergillus nidulans: competence and other developmental processes. In: Turian G, Hohl H R, editors. The fungal spore: morphogenetic controls. New York, N.Y: Academic Press, Inc.; 1981. pp. 63–91. [Google Scholar]
- 34.Champe S P, Rao P, Chang A. An endogenous inducer of sexual development in Aspergillus nidulans. J Gen Microbiol. 1987;133:1383–1387. doi: 10.1099/00221287-133-5-1383. [DOI] [PubMed] [Google Scholar]
- 35.Champe S P, Simon L D. Cellular differentiation and tissue formation in the fungus Aspergillus nidulans. In: Rossomando E F, Alexander S, editors. Morphogenesis: an analysis of the development of biological form. New York, N.Y: Marcel Dekker, Inc.; 1992. pp. 63–91. [Google Scholar]
- 36.Chang Y C, Timberlake W E. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1992;133:29–38. doi: 10.1093/genetics/133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi G H, Chen B, Nuss D L. Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc Natl Acad Sci USA. 1995;92:305–309. doi: 10.1073/pnas.92.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clutterbuck A J. Absence of laccase from yellow-spored mutants of Aspergillus nidulans. J Gen Microbiol. 1972;70:423–435. doi: 10.1099/00221287-70-3-423. [DOI] [PubMed] [Google Scholar]
- 39.Clutterbuck A J. Aspergillus nidulans. In: King R C, editor. Handbook of Genetics. New York, N.Y: Plenum Publishing Corp.; 1974. pp. 447–510. [Google Scholar]
- 40.Clutterbuck A J. Cell volume per nucleus in haploid and diploid strains of Aspergillus nidulans. J Gen Microbiol. 1969;55:291–299. doi: 10.1099/00221287-55-2-291. [DOI] [PubMed] [Google Scholar]
- 41.Clutterbuck A J. A fawn conidia mutant in Aspergillus nidulans. Aspergillus Newsl. 1965;6:12. [Google Scholar]
- 42.Clutterbuck A J. The genetics of conidiation in Aspergillus nidulans. In: Pateman J A, Smith J E, editors. Genetics and physiology of Aspergillus. New York, N.Y: Academic Press, Inc.; 1977. pp. 305–317. [Google Scholar]
- 43.Clutterbuck A J. The genetics of conidiophore pigmentation in Aspergillus nidulans. J Gen Microbiol. 1990;135:1731–1738. doi: 10.1099/00221287-136-9-1731. [DOI] [PubMed] [Google Scholar]
- 44.Clutterbuck A J. Mutants of Aspergillus nidulans deficient in nuclear migration during hyphal growth and conidiation. Microbiology. 1994;140:1169–1174. doi: 10.1099/13500872-140-5-1169. [DOI] [PubMed] [Google Scholar]
- 45.Clutterbuck A J. A mutational analysis of conidial development in Aspergillus nidulans. Genetics. 1969;63:317–327. doi: 10.1093/genetics/63.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clutterbuck A J. New conidial color mutants in Aspergillus nidulans. Aspergillus Newsl. 1968;9:14. [Google Scholar]
- 47.Clutterbuck A J. A variegated position effect in Aspergillus nidulans. Genet Res Camb. 1970;16:303–316. doi: 10.1017/s0016672300002561. [DOI] [PubMed] [Google Scholar]
- 48.Cole G. Models of cell differentiation in conidial fungi. Microbiol Rev. 1986;50:95–132. doi: 10.1128/mr.50.2.95-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coleman D E, Berghuis A M, Lee E, Linder M E, Gilman A G, Sprang S R. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;2265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 50.D’Souza, C., B. N. Lee, and T. H. Adams. Unpublished results.
- 51.D’Souza, C., J.-H. Yu, and T. H. Adams. Unpublished results.
- 52.Demain A L. Regulation of secondary metabolism. In: Finkelstein D B, Ball C, editors. Biotechnology of filamentous fungi—technology and products. Boston, Mass: Butterworth-Heinemann; 1991. pp. 89–112. [Google Scholar]
- 53.deVries L, Mousli M, Wurmser A, Farquar M G. GAIP, a protein that specifically interacts with the trimeric G protein Gαi3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci USA. 1995;92:11926. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.deVries O, Fekkes M, Wosten H, Wessels J. Insoluble complexes in the walls of Schizophyllum communae and other filamentous fungi. Arch Microbiol. 1993;159:330–335. [Google Scholar]
- 55.Dietzel C, Kurjan J. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorn G L. Genetic and morphological properties of undifferentiated and invasive variants of Aspergillus nidulans. Genetics. 1970;66:267–279. doi: 10.1093/genetics/66.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Druey K M, Blumer K J, Kang V H, Kehrl J H. Inhibition of G-protein mediated MAP kinase activation by a new mammalian gene family. Nature. 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 58.Fischer R, Timberlake W E. Aspergillus nidulans apsA (anucleate primary sterigmata) encodes a coiled-coil protein required for nuclear positioning and completion of asexual development. J Cell Biol. 1995;128:485–498. doi: 10.1083/jcb.128.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamas P, Niebel F D C, Lescure N, Cullimore J. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant-Microbe Int. 1996;9:233–242. doi: 10.1094/mpmi-9-0233. [DOI] [PubMed] [Google Scholar]
- 60.Gibbs J B, Marshall M S, Scolnik E M, Dixon R A, Vogel U S. Modulation of guanine nucleotides bound to ras in NIH3T3 cells by oncogene growth factors and GTPase activating protein (GAP) J Biol Chem. 1990;265:20437–20442. [PubMed] [Google Scholar]
- 61.Gimeno C J, Fink G R. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol Cell Biol. 1994;14:2100–2112. doi: 10.1128/mcb.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gimeno C J, Fink G R. The logic of cell division in the life cycle of yeast. Science. 1992;257:626. doi: 10.1126/science.1496375. [DOI] [PubMed] [Google Scholar]
- 63.Gwynne D I, Buxton F P, Sibley S, Davies R W, Lockington R A, Scazzocchio C, Sealy-Lewis H. Comparison of the cis-acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene. 1987;51:205–216. doi: 10.1016/0378-1119(87)90309-x. [DOI] [PubMed] [Google Scholar]
- 64.Han D M, Han Y J, Chae K S, Jahng K Y, Lee Y H. Effects of various carbon sources on the development of Aspergillus nidulans with velA+ or velA1 allele. Kor J Mycol. 1994;22:332–337. [Google Scholar]
- 65.Han D M, Han Y J, Lee Y H, Jahng K Y, Jahng S H, Chae K S. Inhibitory conditions of asexual development and their application for the screening of mutants defective in sexual development. Kor J Mycol. 1990;18:225–232. [Google Scholar]
- 66.Han D M, Han Y J, Kim J H, Jahng K Y, Chung Y S, Chung J H, Chae K S. Isolation and characterization of NSD mutants in Aspergillus nidulans. Kor J Mycol. 1994;22:1–7. [Google Scholar]
- 67.Han M, Sternberg D. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- 68.Han S, Navarro J, Greve R A, Adams T H. Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J. 1993;12:2449–2457. doi: 10.1002/j.1460-2075.1993.tb05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han, S. T., and T. H. Adams. Personal communication.
- 70.Hicks J K, Yu J-H, Keller N P, Adams T H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein-dependent signaling pathway. EMBO J. 1997;16:4916–4923. doi: 10.1093/emboj/16.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hurt E C. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnstone I L, Hughes S G, Clutterbuck A J. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 1985;4:1307–1311. doi: 10.1002/j.1460-2075.1985.tb03777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kafer E. The process of spontaneous recombination in vegetative nuclei of Aspergillus nidulans. Genetics. 1961;46:1581–1609. doi: 10.1093/genetics/46.12.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 75.Kale S P, Bhatnagar D, Bennett J W. Isolation and characterization of morphological variants of Aspergillus parasiticus deficient in secondary metabolite production. Mycol Res. 1994;98:645–652. doi: 10.1017/s0953756203007998. [DOI] [PubMed] [Google Scholar]
- 76.Kale S P, Cary J W, Bhatnagar D, Bennett J W. Characterization of experimentally induced, nonaflatoxigenic variant strains of Aspergillus parasiticus. Appl Environ Microbiol. 1996;62:3399–3404. doi: 10.1128/aem.62.9.3399-3404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoka T, Poweres G, McGill O, Fasano J, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 78.Koelle M R, Horvitz R H. Eg110 Regulates G-protein signalling in C. elegans nervous system and shares a conserved domain with many mammalian genes. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 79.Kormanec J, Schaaff-Gerstenschlager I, Zimmermann F, Perecko D, Kuntzel H. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol Gen Genet. 1991;230:277–287. doi: 10.1007/BF00290678. [DOI] [PubMed] [Google Scholar]
- 80.Kruger M, Fischer R. Isolation of two apsA suppressor strains in Aspergillus nidulans. Genetics. 1996;144:533–540. doi: 10.1093/genetics/144.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurjan J, Dietzel C. Analysis of the role of SCG1, a G alpha homolog, and SST2 in pheromone response and desensitization in yeast. Cold Spring Harbor Symp Quant Biol. 1988;2:577–584. doi: 10.1101/sqb.1988.053.01.066. [DOI] [PubMed] [Google Scholar]
- 82.Kurtz M B, Champe S P. Dominant spore color mutants of Aspergillus nidulans defective in germination and sexual development. J Bacteriol. 1981;148:629–638. doi: 10.1128/jb.148.2.629-638.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lauter F-R, Russo V E A, Yanofsky C. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 1992;6:2373–2381. doi: 10.1101/gad.6.12a.2373. [DOI] [PubMed] [Google Scholar]
- 84.Law D J, Timberlake W E. Developmental regulation of laccase levels in Aspergillus nidulans. J Bacteriol. 1980;144:509–517. doi: 10.1128/jb.144.2.509-517.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee B N, Adams T H. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal. Genes Dev. 1994;8:641–651. doi: 10.1101/gad.8.6.641. [DOI] [PubMed] [Google Scholar]
- 86.Lee B N, Adams T H. fluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlAβ activation. EMBO J. 1996;15:299–309. [PMC free article] [PubMed] [Google Scholar]
- 87.Lee B N, Adams T H. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation leads to activation of brlA and premature initiation of development. Mol Microbiol. 1994;14:323–334. doi: 10.1111/j.1365-2958.1994.tb01293.x. [DOI] [PubMed] [Google Scholar]
- 88.Lewis C, Champe S P. A preinduction sporulation gene from Aspergillus nidulans. Microbiology. 1995;141:1821–1828. doi: 10.1099/13500872-141-8-1821. [DOI] [PubMed] [Google Scholar]
- 89.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lockington R A, Sealy-Lewis H M, Scazzocchio C, Davies R W. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene. 1985;33:137–149. doi: 10.1016/0378-1119(85)90088-5. [DOI] [PubMed] [Google Scholar]
- 91.Luscher B, Eisenman R N. New light on Myc and Myb. II. Myb. Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- 92.Marhoul J F, Adams T H. Aspergillus fabM encodes an essential product that is related to poly(A)-binding proteins and activates development when overexpressed. Genetics. 1996;144:1463–1470. doi: 10.1093/genetics/144.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marhoul J F, Adams T H. Identification of developmental regulatory genes in Aspergillus nidulans by overexpression. Genetics. 1995;139:537–547. doi: 10.1093/genetics/139.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marhoul, J. F., and T. H. Adams. Personal communication.
- 95.Marshall M A, Timberlake W E. Aspergillus nidulans wetA activates spore-specific gene expression. Mol Cell Biol. 1991;11:55–62. doi: 10.1128/mcb.11.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martinelli S. Conidiation of Aspergillus nidulans in submerged culture. Trans Br Mycol Soc. 1976;67:121–128. [Google Scholar]
- 97.Martinelli S D. Phenotypes of double conidiation mutants of Aspergillus nidulans. J Gen Microbiol. 1979;114:277–287. doi: 10.1099/00221287-114-2-277. [DOI] [PubMed] [Google Scholar]
- 98.Martinelli S D, Clutterbuck A J. A quantitative survey of conidiation mutants in Aspergillus nidulans. J Gen Microbiol. 1971;69:261–268. doi: 10.1099/00221287-69-2-261. [DOI] [PubMed] [Google Scholar]
- 99.Mayorga M E, Timberlake W E. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthase. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 100.Mayorga M E, Timberlake W E. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics. 1990;126:73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mazur P, Meyers H V, Nakanishi K, El-Zayat A E, Champe S P. Structural elucidation of sporogenic fatty acid metabolites in Aspergillus nidulans. Tetrahedron Lett. 1990;31:3837. [Google Scholar]
- 102.McGoldrick C A, Gruver C, May G S. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J Cell Biol. 1995;128:577–587. doi: 10.1083/jcb.128.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miller B. The medA gene of Aspergillus nidulans. J Cell Biochem. 1993;17C:44. [Google Scholar]
- 104.Miller B, Miller K, Timberlake W. Direct and indirect gene replacements in Aspergillus nidulans. Mol Cell Biol. 1985;5:1714–1721. doi: 10.1128/mcb.5.7.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller, B. L., K. Y. Miller, and J. R. Dutton. 1995. Regulatory interactions that control multicellular development during Aspergillus sporulation. Fungal Genet. Newsl. 42A(Suppl.):7.
- 106.Miller J, McLachlan A D, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;41:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller K Y, Toennis T M, Adams T H, Miller B L. Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol Gen Genet. 1991;227:285–292. doi: 10.1007/BF00259682. [DOI] [PubMed] [Google Scholar]
- 108.Miller K Y, Wu J, Miller B L. stuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- 109.Mims C W, Richardson E A, Timberlake W E. Ultrastructural analysis of conidiophore development in the fungus Aspergillus nidulans using freeze-substitution. Protoplasma. 1988;44:132–141. [Google Scholar]
- 110.Mirabito P M, Adams T H, Timberlake W E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell. 1989;57:859–868. doi: 10.1016/0092-8674(89)90800-3. [DOI] [PubMed] [Google Scholar]
- 111.Mooney J L, Yager L N. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 1990;4:1473–1482. doi: 10.1101/gad.4.9.1473. [DOI] [PubMed] [Google Scholar]
- 112.Morris N R. Lower eukaryotic cell cycle: perspectives on mitosis from the fungi. Curr Opin Cell Biol. 1990;2:252–257. doi: 10.1016/0955-0674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 113.Morton A G. The induction of sporulation in mould fungi. Proc R Soc London Ser B. 1961;153:548–569. [Google Scholar]
- 114.Mulder G, Wessels J. Molecular cloning of RNA’s differentially expressed in monokaryons and dikaryons of Schizophyllum communae in relation to fruiting. Exp Mycol. 1986;10:214–227. [Google Scholar]
- 115.Navarro R E, Stringer M A, Hansburg W, Timberlake W E, Aguirre J. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr Genet. 1996;29:352–359. [PubMed] [Google Scholar]
- 116.Navarro-Bordonaba J, Adams T H. Development of conidia and fruiting bodies in ascomycetes. In: Wessels J G H, Meinhardt F, editors. Growth, differentiation, and sexuality. Vol. 1. Berlin, Germany: Springer-Verlag KG; 1994. pp. 333–349. [Google Scholar]
- 117.Neer E J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 118.O’Hara E B, Timberlake W E. Molecular characterization of the Aspergillus nidulans yA locus. Genetics. 1989;121:249–254. doi: 10.1093/genetics/121.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oliver P T P. Conidiophore and spore development in Aspergillus nidulans. J Gen Microbiol. 1972;73:45–54. doi: 10.1099/00221287-73-1-45. [DOI] [PubMed] [Google Scholar]
- 120.Osmani S A, Engle D B, Doonan J H, Morris N R. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell. 1988;52:241–251. doi: 10.1016/0092-8674(88)90513-2. [DOI] [PubMed] [Google Scholar]
- 121.Osmani S A, Pu R T, Morris N R. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- 122.Osmani S A, Ye X S. Cell cycle regulation in Aspergillus by two protein kinases. Biochem J. 1996;317:633–641. doi: 10.1042/bj3170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pastushok M, Axelrod D E. Effect of glucose, ammonium and media maintenance on the time of conidiophore initiation by surface colonies of Aspergillus nidulans. J Gen Microbiol. 1976;94:221–224. doi: 10.1099/00221287-94-1-221. [DOI] [PubMed] [Google Scholar]
- 124.Patemen J A, Doy C H, Olsen J E, Norris U, Creaser E H, Hynes M. Regulation of alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (AldDH) in Aspergillus nidulans. Proc R Soc London Ser B. 1983;217:243–264. doi: 10.1098/rspb.1983.0009. [DOI] [PubMed] [Google Scholar]
- 125.Perrimon M. The torso receptor protein tyrosine kinase signalling pathway: an endless story. Cell. 1993;74:219–222. doi: 10.1016/0092-8674(93)90412-j. [DOI] [PubMed] [Google Scholar]
- 126.Pontecorvo G, Roper J A, Hemmons L M, MacDonald K D, Bufton A W J. The Genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 127.Prade R A, Timberlake W E. The Aspergillus nidulans brlA regulatory locus encodes two functionally redundant polypeptides that are individually required for conidiophore development. EMBO J. 1993;12:2439–2447. doi: 10.1002/j.1460-2075.1993.tb05898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Prasher D C. Using GFP to see the light. Trends Genet. 1995;11:320–329. doi: 10.1016/s0168-9525(00)89090-3. [DOI] [PubMed] [Google Scholar]
- 129.Prosser J I. Growth kinetics of mycelial colonies and aggregates of ascomycetes. Mycol Res. 1992;97:513–528. [Google Scholar]
- 130.Qui M, Green S. NGF and EGF rapidly activate p21 ras in PC12 cells by distinct convergent pathways involving tyrosine phosphorylation. Neuron. 1991;7:937–946. doi: 10.1016/0896-6273(91)90339-2. [DOI] [PubMed] [Google Scholar]
- 131.Rens-Domiano S, Hamm H E. Structural and functional relationships of heterotrimeric G-proteins. FASEB J. 1995;9:1059–1066. doi: 10.1096/fasebj.9.11.7649405. [DOI] [PubMed] [Google Scholar]
- 132.Roper J, Walker M. wB, a new white mutant of Aspergillus nidulans on linkage group VII. Fungal Genet Newsl. 1990;37:35. [Google Scholar]
- 133.Rose M D, Fink G R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- 134.Ruiters M, Wessels J. In situ localization of specific mRNA’s in developing fruit bodies of the basidiomycete Schizophyllum communae. Exp Mycol. 1989;13:212–222. [Google Scholar]
- 135.Saxena R K, Sinha U. Conidiation of Aspergillus nidulans in submerged liquid culture. Gen Appl Microbiol. 1973;19:141–146. [Google Scholar]
- 136.Serlupi-Crescenzi O, Kurtz M B, Champe S P. Developmental defects resulting from arginine auxotrophy in Aspergillus nidulans. J Gen Microbiol. 1983;129:3535–3544. doi: 10.1099/00221287-129-11-3535. [DOI] [PubMed] [Google Scholar]
- 137.Sewall T C, Mims C W, Timberlake W E. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell. 1990;2:731–739. doi: 10.1105/tpc.2.8.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sewall T C, Mims C W, Timberlake W E. Conidium differentiation in Aspergillus nidulans wild-type and wet-white (wetA) mutant strains. Dev Biol. 1990;138:499–508. doi: 10.1016/0012-1606(90)90215-5. [DOI] [PubMed] [Google Scholar]
- 139.Shen, W.-C., J. Wieser, T. H. Adams, and D. J. Ebbole. The Neurospora myb-1 gene complements an Aspergillus flbD sporulation mutant but has no identifiable role in Neurospora growth or development. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 140.Simon M, Bowtell D, Dodson G, Laverty T, Rubin G. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signalling by sevenless protein Tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 141.Skromne I, Sanchez O, Aguirre J. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology. 1995;141:21–28. doi: 10.1099/00221287-141-1-21. [DOI] [PubMed] [Google Scholar]
- 142.Som T, Kolaparthi V S R. Developmental decisions in Aspergillus nidulans are regulated by Ras activity. Mol Cell Biol. 1994;14:5333–5348. doi: 10.1128/mcb.14.8.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sondek J, Lambright D G, Noel J P, Hamm H E, Sigler P B. GTPase mechanisms of G proteins from the 1.7 A crystal structure of transducin α center dot GDP center dot A1F4. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 144.Springer M L. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- 145.St Leger R, Staples R, Roberts D. Cloning and regulatory analysis of starvation-stress gene, ssgA encoding a hydrophobin-like protein from the entomopathogenic fungus Metarhizium anisophiae. Gene. 1992;120:119–124. doi: 10.1016/0378-1119(92)90019-l. [DOI] [PubMed] [Google Scholar]
- 146.Stringer M A, Dean R A, Sewall T C, Timberlake W E. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 1991;5:1161–1171. doi: 10.1101/gad.5.7.1161. [DOI] [PubMed] [Google Scholar]
- 147.Stringer M A, Timberlake W E. dewA encodes a fungal hydrophobin component of the Aspergillus nidulans spore wall. Mol Microbiol. 1995;16:33–44. doi: 10.1111/j.1365-2958.1995.tb02389.x. [DOI] [PubMed] [Google Scholar]
- 148.Talbot N, Ebbole D, Hamer J. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnoporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tamame M, Antequera F, Villanueva J R, Santos T. High-frequency conversion of a “fluffy” developmental phenotype in Aspergillus spp. by 5-azacytidine treatment: Evidence for involvement of a single nuclear gene. Mol Cell Biol. 1983;3:2287–2297. doi: 10.1128/mcb.3.12.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Timberlake W E. Cloning and analysis of fungal genes. In: Alic M, Bennett J W, Lasure L L, editors. More gene manipulations in fungi. San Diego, Calif: Academic Press, Inc.; 1991. pp. 51–83. [Google Scholar]
- 151.Timberlake W E. Developmental gene regulation in Aspergillus nidulans. Dev Biol. 1980;78:497–510. doi: 10.1016/0012-1606(80)90349-8. [DOI] [PubMed] [Google Scholar]
- 152.Timberlake W E. Isolation of stage- and cell-specific genes from fungi. NATO ASI Ser Ser H. 1986;1:343–357. [Google Scholar]
- 153.Timberlake W E. Molecular genetics of Aspergillus development. Annu Rev Genet. 1990;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- 154.Timberlake W E, Marshall M A. Genetic engineering of filamentous fungi. Science. 1989;244:1313–1317. doi: 10.1126/science.2525275. [DOI] [PubMed] [Google Scholar]
- 155.Timberlake W E, Marshall M A. Genetic regulation of development in Aspergillus nidulans. Trends Genet. 1988;4:162–169. doi: 10.1016/0168-9525(88)90022-4. [DOI] [PubMed] [Google Scholar]
- 156.Turner G E, Borkovich K A. Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J Biol Chem. 1993;268:14805–14811. [PubMed] [Google Scholar]
- 157.Upshall A, Gilbert T, Saari G, OHara P J, Weglenski P, Berse B, Miller K, Timberlake W E. Molecular analysis of the argB gene of Aspergillus nidulans. Mol Gen Genet. 1986;204:349–354. doi: 10.1007/BF00425521. [DOI] [PubMed] [Google Scholar]
- 158.van Gorcom R F M, Pouwels P H, Goosen T, Visser J, van der Broek H W J, Hamer J E, Timberlake W E, van den Hondel C A M J J. Expression of the E. coli β-galactosidase gene in Aspergillus nidulans. Gene. 1985;40:437–443. doi: 10.1016/0378-1119(85)90028-9. [DOI] [PubMed] [Google Scholar]
- 159.Vinson C R, Sigler P B, McKnight S L. Scissor-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 160.Wessels G H, deVries O M H, Asgeirsdottir S A, Springer J. The thn mutation of Schizophyllum commune, which suppresses formation aerial hyphae, affects expression of the Sc3 hydrophobin gene. J Gen Microbiol. 1991;137:2439–2445. doi: 10.1099/00221287-137-10-2439. [DOI] [PubMed] [Google Scholar]
- 161.Wessels J G H. Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol. 1994;32:413–437. [Google Scholar]
- 162.Wieser J, Adams T H. flbD encodes a myb-like DNA binding protein that controls initiation of Aspergillus nidulans conidiophore development. Genes Dev. 1995;9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- 163.Wieser, J., and T. H. Adams. Unpublished results.
- 164.Wieser, J., J. Fondon, and T. H. Adams. Unpublished results.
- 165.Wieser J, Lee B N, Fondon J W, Adams T H. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr Genet. 1994;27:62–69. doi: 10.1007/BF00326580. [DOI] [PubMed] [Google Scholar]
- 166.Wieser J, Yu J-H, Adams T H. Dominant mutations affecting both sporulation and sterigmatocystin biosynthesis in Aspergillus nidulans. Curr Genet. 1997;32:218–224. doi: 10.1007/s002940050269. [DOI] [PubMed] [Google Scholar]
- 167.Wolkow T D, Harris S D, Hamer J E. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J Cell Sci. 1996;109:2179–2188. doi: 10.1242/jcs.109.8.2179. [DOI] [PubMed] [Google Scholar]
- 168.Woloshuk C P, Foutz K R, Brewer J F, Bhatnagar D, Cleveland T E, Payne G A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yager L. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans. In: Bennet J W, Klich M A, editors. Aspergillus biology and industrial applications. Boston, Mass: Butterworth-Heinemann; 1992. pp. 19–41. [PubMed] [Google Scholar]
- 170.Yager L N, Kurtz M B, Champe S P. Temperature-shift analysis of conidial development in Aspergillus nidulans. Dev Biol. 1982;93:92–103. doi: 10.1016/0012-1606(82)90242-1. [DOI] [PubMed] [Google Scholar]
- 171.Yelton M M, Timberlake W E, van den Hondel C A M J J. A cosmid for selecting genes by complementation in Aspergillus nidulans: selection of the developmentally regulated yA locus. Proc Natl Acad Sci USA. 1985;82:834–838. doi: 10.1073/pnas.82.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yu J-H, Butchko R A E, Fernandes M, Keller N P, Leonard T J, Adams T H. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
- 173.Yu J-H, Wieser J, Adams T H. The Aspergillus FlbA RGS domain protein antagonizes G-protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- 174.Zimmermann C, Orr W, Leclerc R, Barnard E, Timberlake W. Molecular cloning and selection of genes regulated in Aspergillus development. Cell. 1980;21:709–715. doi: 10.1016/0092-8674(80)90434-1. [DOI] [PubMed] [Google Scholar]