Abstract
BirA*, a mutant form of the biotinylating enzyme BirA, can nonspecifically biotinylate ε-amino groups on lysines of proteins. Based on the promiscuous labeling nature of BirA*, plasmids expressing fusion constructs of BirA* to a given ligand have been used to transfect eukaryotic cells, leading to the biotinylation of intracellular proteins interacting or in close proximity to such Ligand.BirA* constructs. Mass spectrometry performed on the recovered biotinylated partners allows one to map intracellular protein interactors, a technique known as BioID. In contrast, the expression and purification of recombinant Ligand.BirA* constructs could serve as a powerful tool for labeling and detecting cell surface receptors. Here, we report the design and expression of recombinant Affibody.BirA* constructs, ZEGFR:1907.BirA* and ZHER2:243.BirA*, as protein bispecifics able to biotinylate their respective receptors EGFR and HER2 on the surface of MDA-MB-231 (EGFR+, EpCaM+, and CD44+) and SK-OV-3 (HER2++, EGFR+, EpCaM+, and CD44+) cancer cells. These Affibody.BirA* constructs retain both their BirA* enzymatic activity as well as their receptor-binding function. Importantly, MDA-MB-231 and SK-OV-3 cells biotinylated with Affibody.BirA* constructs did label their receptors EGFR and HER2 but did not biotinylate irrelevant antigens such as EpCaM or CD44 present on the surface of both cell lines. Ligand.BirA* bispecifics may represent a promising class of agents to identify unknown receptors on cell surfaces.
Keywords: BirA*, cell surface receptor discovery, bispecific, affibody, enzyme ligand fusion
The detection and identification of a cell surface interaction can provide valuable information, leading to intracellular signaling cascades. Importantly, labeling strategies for assessing a membrane-bound, surface–protein interaction must be performed under physiological conditions and the labeled proteins being stable enough to be captured using pull-down or immunoprecipitation approaches under denaturing conditions.
Biotinylation methods have been particularly favored for identifying intracellular protein interactions based on the near-covalent binding and the recovery of biotinylated proteins using immobilized avidin or streptavidin matrices. Two biotinylation approaches, namely, BioID and APEX, have been developed for tagging and mapping intracellular protein interactions.1,2 These techniques rely on the intracellular expression of gene-encoded, ligand–enzyme fusion constructs responsible for biotinylating proximal proteins.3 However, these techniques are limited to the detection of protein–protein interactions (PPIs) inside cells.
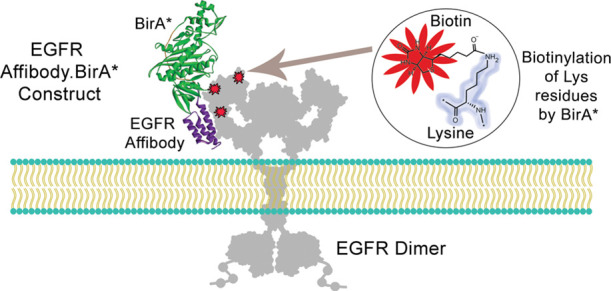
Here, we designed and expressed protein Ligand.BirA* constructs as targeted biotinylation agents to label receptors on a cell surface. Specifically, BirA is a 35 kDa bacterial protein that is a member of a family of enzymes called biotin protein ligases (BPL).4 BPLs are responsible for post-translationally attaching biotin to a specific lysine ε-amino group within a defined short peptide substrate. This peptide is usually present at the C-terminal end of carboxylases, and the resulting biotin modification allows these enzymes to carry out their functions.4 Mechanistically, BirA generates a mixed anhydride amino-reactive intermediate bio-5′-AMP in the presence of adenosine triphosphate (ATP) and biotin, resulting in the catalytic ligation of biotin to the ε-amino group of Lys-122 on the biotin carboxyl carrier protein (BCCP) subunit of acetyl-CoA carboxylase.5 This post-translational modification of acetyl-CoA carboxylase enables this enzyme to catalyze the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA, which is an important step in the regulation pathway of fatty acid synthesis and degradation inside cells.6 Site-specific mutations introduced within BirA has led to the characterization of its structural domains and their roles in its two functions.7,8 One particular mutation of BirA at arginine 118 [R118G] located in the biotin-binding motif causes the mutated BirA to display a reduced affinity for the amino-reactive bio-5′-AMP intermediate, causing its premature release from the active site of R118G-mutated BirA (termed BirA*), resulting in the random biotinylation of any protein lysine ε-amino groups located within its proximity (Figure 1).8−11 The nonspecific biotinylation of proximal protein lysine residues by BirA* has been used for the purpose of identifying intracellular PPIs in a technique called BioID.1 As a proof-of-concept, we report the expression and use of recombinant BirA* fusion proteins, ZEGFR:1907.BirA* and ZHER2:243.BirA*, as labeling agents for the targeted biotinylation of their cell surface receptors, namely, the epidermal growth factor receptor (EGFR) and HER2.
Figure 1.
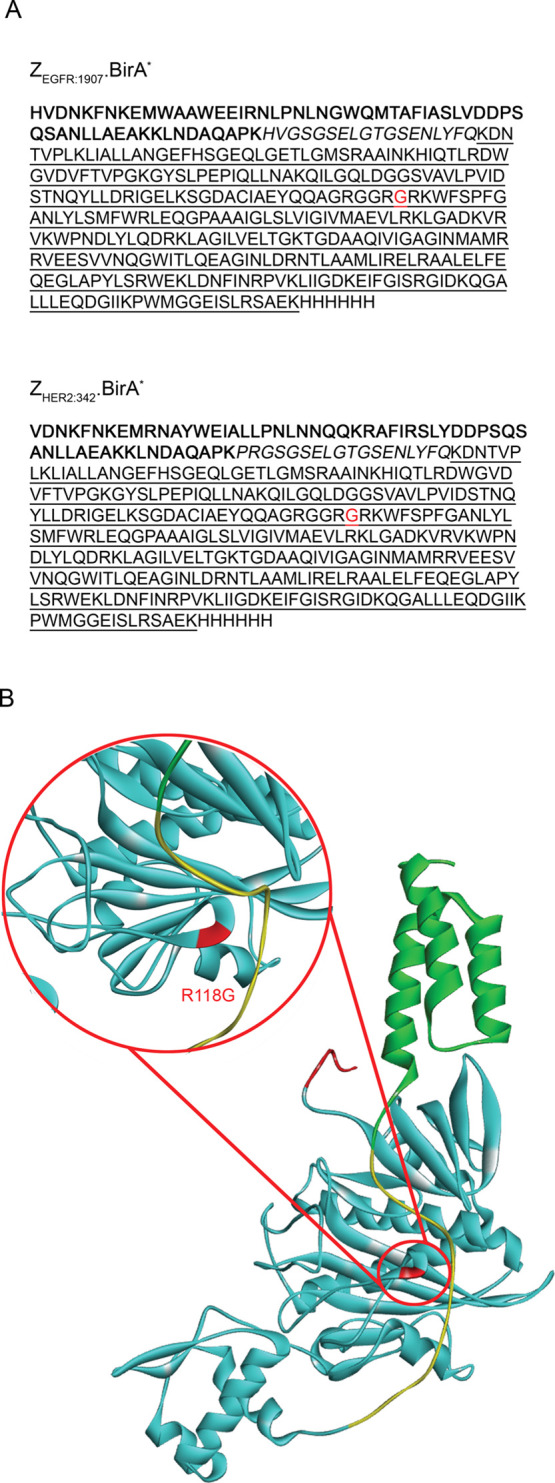
Design of the Affibody.BirA* fusion constructs, ZEGFR:1907.BirA* and ZHER2:342.BirA*. (A) Amino-acid sequences of ZEGFR:1907.BirA* (top) or ZHER2:342.BirA* (bottom). The affibody ligand sequence (in bold characters) is followed by a linker region (italics), a C-terminal BirA* catalytic domain (underlined), and ending with a hexahistidine tag. (B) Ribbon representation of the Affibody.BirA* fusion construct, ZEGFR:1907.BirA* generated from the I-TASSER server.17 Color pattern: affibody ligand (in green), linker containing a TEV cleavage site (in yellow), C-terminal BirA* catalytic domain (in cyan), and a histidine tag (in red). The location of the R118G mutation in BirA* is highlighted in red (magnified image).
Results and Discussion
BirA* is a mutated form of the enzyme BirA (R118G) that is able to nonspecifically biotinylate itself as well as proteins within its proximity. This feature of BirA* forms the basis of a technique called BioID, where mammalian cells are transfected with a plasmid coding for a Ligand.BirA* fusion construct to identify cellular proteins interacting or in close proximity to a specific ligand.1 The use of this technique is limited to the detection of intracellular interactions and assumes that each domain of a given Ligand.BirA* bispecific is functionally active. However, the most accessible targets for therapeutic intervention are receptors expressed on the surface of specific cell types for which there is an understanding of their mechanism of action in modulating cell functions upon binding a given ligand.12 Current techniques used to identify unknown cell surface receptors include regular pull-down approaches such as affinity purification mass spectrometry (AP-MS) and ligand–receptor capture-TRICEPS (LRC-TRICEPS).13,14 The main drawback with these techniques is that they detect only interacting proteins that remain bound to themselves and the beads, withstanding the lysis buffer conditions and wash steps. Weaker and transient interactions between protein pairs may not be detected using these approaches.
The present study explored the design, expression, and use of protein Ligand.BirA* bispecifics as tools to label cell surface determinants specifically bound or in spatial proximity to a ligand. Affibodies are small (6 kDa) engineered antibody mimics tailored to recognize specific targets.15,16 Two Affibody.BirA* bispecifics were expressed in Escherichia coli. These fusion proteins were composed of an N-terminal affibody ligand linked to a C-terminal BirA* domain using a short hinge region. The resulting expressed protein sequences were termed ZEGFR:1907.BirA* and ZHER2:342.BirA* (Figure 1). A histidine tag was inserted at the C-terminus of BirA* for purification purposes (Figure 1A). A tobacco etch virus (TEV) protease cleavage site was also included in the flexible hinge region between the affibody and BirA* domains for the purpose of separating the two domains. The predicted ribbon structure of ZEGFR:1907.BirA* is shown in Figure 1B, highlighting the R118G mutation.
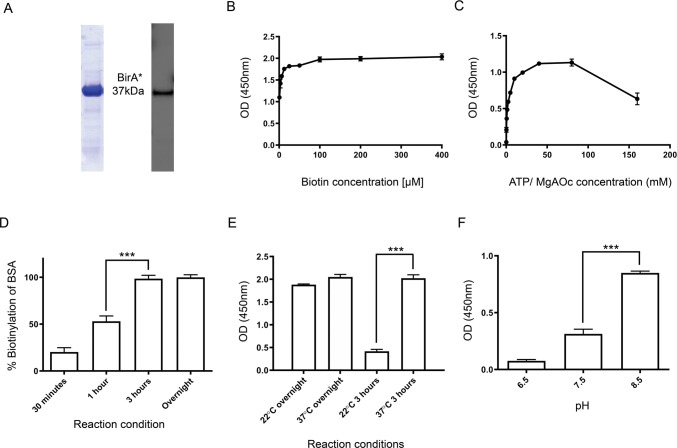
BirA* alone was also produced to establish the optimal conditions for the biotinylation activity of BirA* to use for the Affibody.BirA* constructs. Recombinant BirA* was expressed in E. coli and purified using Ni-nitrilotriacetic acid (NTA) affinity chromatography. Its identity and purity were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining and Western blot analysis, migrating as a 37 kDa band, the predicted molecular weight of BirA* (Figure 2A). Optimal reaction conditions to achieve maximal biotinylation using BirA* were then established by enzyme-linked immunosorbent assay (ELISA) using immobilized bovine serum albumin (BSA) as a lysine-rich substrate and streptavidin-horseradish peroxidase (HRP) as the detection agent. Specifically, parameters such as the concentrations of biotin and ATP/MgOAc were empirically defined at 37 °C [for subsequent cell-labeling conditions]. The levels of the biotinylation activity of BirA* reached a plateau at 100 μM biotin (Figure 2B) and at 40 mM ATP/MgOAc (Figure 2C). It was observed that BirA* maximally biotinylated immobilized BSA after 3 h of incubation (Figure 2D) and that reducing the reaction temperature to 22 °C (room temperature) significantly impacted the extent of biotinylation occurring over a 3 h period (Figure 2E). Finally, it was empirically determined that BirA* displayed maximal biotinylation activity at pH 8.5 (Figure 2F). Importantly, the solubility of Affibody.BirA* constructs is significantly reduced (precipitation), and ATP hydrolyzes at pH values above 8.5.
Figure 2.
Optimization of biotinylation reaction conditions using recombinantly expressed BirA*. (A) BirA* was expressed as described in the Methods section. Its purity and apparent molecular mass were confirmed by SDS-PAGE (Coomassie blue-stained band) and by Western blot (right) using an anti-histidine tag antibody. BirA* migrates as a single 37 kDa band in the presence of 50 mM dithiothreitol (DTT) (reducing conditions). Establishing the optimal biotin (B) and ATP/MgOAc(C) concentrations required for BirA* to biotinylate immobilized BSA at 37 °C overnight. (D) BirA* biotinylation of plate-bound BSA reaches a plateau when exposed to BirA* for 3 h at 37 °C. BSA is maximally biotinylated when exposed to BirA* at 37 °C for 3 h (E) at pH 8.5 (F). Each point represents the average and standard deviation of experiments performed in triplicate (n = 3). For panels D, E, and F, the statistical difference between the two means was significant (***P < 0.0001) as calculated using a Student’s t-test.
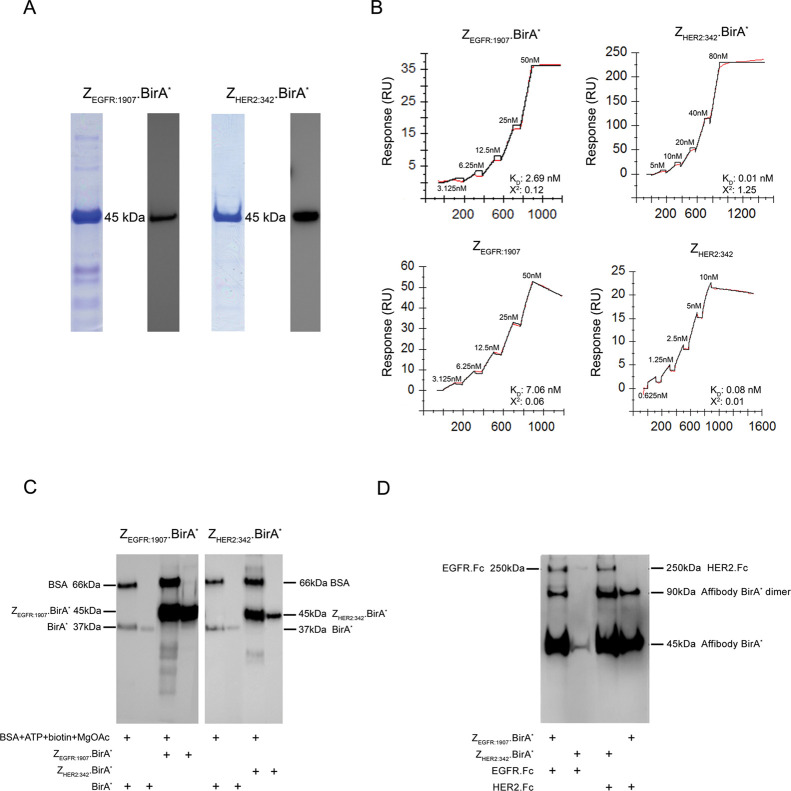
Recombinant ZEGFR:1907.BirA* and ZHER2:342.BirA* constructs were expressed in E. coli and purified by Ni-NTA affinity chromatography. Their apparent molecular weight (45 kDa) and purity were confirmed by SDS-PAGE, followed by Coomassie blue staining and Western blot analyses (Figure 3A). The functional activity of both domains composing these Affibody.BirA* constructs was assessed using surface plasmon resonance (SPR) by measuring the binding of each affibody ligand to its respective target as well as the biotinylation activity of BirA* within each construct toward immobilized BSA. ZEGFR:1907.BirA* was shown to bind strongly to immobilized EGFR with a dissociation constant (KD) of 2.69 nM, a value comparable to the affinity measured for ZEGFR:1907 alone to EGFR (KD, 7.06 nM; Figure 3B and Table 1). ZHER2:342.BirA* bound more strongly to its target receptor HER2 with a KD of 0.01 nM. This dissociation constant was similar to the one derived for ZHER2:342 alone interacting with HER2 (KD, 0.08 nM; Table 1). The slightly higher kon observed by both Affibody.BirA* constructs toward their receptors in comparison to the respective affibody alone could be explained by the presence of Affibody.BirA* construct dimers, increasing their avidity toward their respective targets. BirA* constructs can form dimers because of the presence of a single free cysteine (C107) thiol group located in the BirA* catalytic site, resulting in the formation of a disulfide bridge between two BirA* constructs. ZEGFR:1907 and ZEGFR:1907.BirA* did not bind to HER2, and reciprocally, ZHER2:342 and ZHER2:342.BirA* did not interact with EGFR, confirming their specificity (Figure S1).
Figure 3.
ZEGFR:1907.BirA* and ZHER2:34.BirA* are able to retain their binding and biotinylation activities. (A) ZEGFR:1907.BirA* and ZHER2:342.BirA* were expressed as described in the Methods section, and their purity and size confirmed by SDS-PAGE and by Western blot using an anti-Histidine tag antibody. Both ZEGFR:1907.BirA* and ZHER2:342.BirA* migrate as a single 45 kDa band in the presence of 50 mM DTT (reducing conditions). (B) Single-cycle kinetics sensorgrams derived from increasing concentrations of ZEGFR:1907.BirA* and ZHER2:342.BirA* interacting with their respective immobilized targets [EGFR and HER2], as monitored using SPR. The binding affinity of each Affibody.BirA* construct was compared to that of its corresponding affibody alone. The binding of ZEGFR:1907.BirA* and ZEGFR:1907 to HER2 and ZHER2:342.BirA* and ZHER2:342 to EGFR served as negative controls to confirm the specificity of each Affibody.BirA* construct for their target (Supplementary Figure 1). (C) Monitoring the biotinylation activity of ZEGFR:1907.BirA*, ZHER2:342.BirA*, and BirA* alone toward BSA by Western blot analysis in the presence of ATP, biotin, and MgOAc.BirA*, ZEGFR:1907.BirA*, and ZHER2:342.BirA* are also biotinylated during the reaction. The Western blot samples were prepared in 50 mM DTT (reducing conditions). (D) Monitoring the biotinylation activity of ZEGFR:1907.BirA* and ZHER2:342.BirA* toward their recombinant target human EGFR.Fc and HER2.Fc, respectively, by Western blot analysis. ZEGFR:1907.BirA* and ZEGFR:1907 were allowed to bind and biotinylate each immobilized target, serving as both negative and positive controls to confirm the specificity of each Affibody.BirA* construct for their respective target. ZEGFR:1907.BirA* and ZHER2:342.BirA* were also biotinylated during this reaction. No reducing agent was added to the samples in panel D to maintain EGFR.Fc and HER2.Fc as dimers. As such, Affibody.BirA* constructs can exist as monomers and as disulfide bridge-containing dimers.
Table 1. Binding Kinetic Parameters of ZEGFR:1907.BirA* and ZHER2:342.BirA* to Immobilized EGFR and HER2, as Derived from SPR Sensorgrams.
| construct | kon (M–1 s–1) | koff (s–1) | KD (M) | Chi2 |
|---|---|---|---|---|
| ZEGFR:1907.BirA* | 3.82 × 104 | 1.03 × 10–4 | 2.69 × 10–9 | 0.12 |
| ZEGFR:1907 | 9.43 × 104 | 6.66 × 10–4 | 7.06 × 10–9 | 0.06 |
| ZHER2:342.BirA* | 1.04 × 104 | 1.14 × 10–7 | 1.09 × 10–11 | 1.25 |
| ZHER2:342 | 1.45 × 106 | 1.11 × 10–4 | 7.6 × 10–11 | 0.01 |
The biotinylation activities of both ZEGFR:1907.BirA* and ZHER2:342.BirA* were tested using their ability to nonspecifically biotinylate themselves and proximal lysine amino groups present in BSA as detected by Western blot probed with streptavidin-HRP. As expected, the observed bands represent biotinylated forms of each Affibody.BirA* construct as well as biotinylated BSA (Figure 3C). An equimolar amount of BirA* and Affibody.BirA* was used in this assay. Because ZEGFR:1907.BirA* and ZHER2:342.BirA* include 5 and 6 more lysine residues, respectively, than BirA* alone (Figure 1A), it was expected that these additional lysine ε-amino groups could be labeled by BirA* in these constructs during their expression in E. coli and their in vitro labeling. In addition, the ZHER2:342.BirA* band appears more pronounced in the BSA-containing sample because of the in vitro labeling occurring in the presence of ATP, biotin, and MgOAc (Figure 3C).
After confirming the binding ability and biotinylation activity of the Affibody.BirA* constructs, both ZEGFR:1907.BirA* and ZHER2:342.BirA* were used to biotinylate their target proteins EGFR and HER2, respectively, first as recombinant proteins and then on the surface of cancer cells expressing these markers. ZEGFR:1907.BirA* was shown to biotinylate recombinant EGFR.Fc, while ZHER2:342.BirA* was shown to modify HER2.Fc, when ATP, biotin, and MgOAc were added, as detected by Western blot probed with streptavidin-HRP. No biotinylation of EGFR.Fc and HER2.Fc was observed when ZHER2:342.BirA* was allowed to biotinylate EGFR.Fc or when ZEGFR:1907.BirA* was used to biotinylate HER2.Fc (Figure 3D), further confirming the specificity of binding of the Affibody.BirA* constructs. In addition to biotinylating their respective receptors immobilized on the surface of protein G beads, accessible proximal lysine ε-amino side chains on both Affibody.BirA* constructs are further biotinylated in vitro in the presence of BSA, biotin, ATP, and MgOAc, as evident by their increased band intensity from lane 2 compared to lane 1, and lane 4 compared to lane 3. In contrast, the relatively strong biotinylated signal observed for the ZEGFR:1907.BirA* band in lane 4 could be explained by the fact that endogenously biotinylated ZEGFR:1907.BirA* sticks nonspecifically to protein G beads. This nonspecific band pattern representing the monomer and dimer forms of biotinylated ZEGFR:1907.BirA was consistently observed by Western blot. These Affibody.BirA* constructs were then used to biotinylate their receptors on the surface of cells. ZEGFR:1907.BirA* was incubated with MDA-MB-231 cells, a triple negative human breast cancer cell line overexpressing EGFR,18 in the presence of 400 μM biotin, 40 mM ATP/MgOAc, and in tris buffer KCl (TBK) buffer (50 mM Tris, 100 mM KCl) pH 8.5 for 3 h at 37 °C. The labeled cells were then lysed and EGFR recovered by immunoprecipitation using an anti-EGFR antibody. The biotinylation of EGFR by ZEGFR:1907.BirA* on the surface of MDA-MB-231 cells was confirmed by Western blot analysis using streptavidin-HRP. ZEGFR:1907.BirA* biotinylated EGFR in a time-dependent manner, showing a gradual increase in EGFR labeling after 1 h and 3 h of exposure to ZEGFR:1907.BirA*. ZHER2:342.BirA* did not biotinylate EGFR on the surface of MDA-MB-231 cells (Figure 4A). The same experiment was carried out on the surface of the ovarian cancer SK-OV-3 cells known to overexpress HER2 and to modestly express EGFR.19 ZHER2:342.BirA* and ZEGFR:1907.BirA* were allowed to biotinylate HER2 and EGFR on the surface of these cells, over a 3 h time period. These two proteins were then immunoprecipitated using anti-HER2 or anti-EGFR antibodies. As in the case of MDA-MB-231, ZHER2:342.BirA* and ZEGFR:1907.BirA* were able to biotinylate HER2 and EGFR, respectively, on the surface of SK-OV-3 cells in a time-dependent manner over a 3 h time period (Figure 4B). Two other proteins expressed on the surface of MDA-MB-231 and SK-OV-3 cells, namely, the epithelial cellular adhesion molecule (EpCaM) and CD44, were also immunoprecipitated from these labeled cells to confirm that ZEGFR:1907.BirA* and ZHER2:342.BirA* only biotinylated their respective targets. No biotinylated EpCaM or CD44 was observed for MDA-MB-231 cells labeled using ZEGFR:1907.BirA* nor for SK-OV-3 cells labeled using ZHER2:342.BirA* and ZEGFR:1907.BirA*, after 1 h or 3 h of labeling. Biotinylated proteins were also pulled down using streptavidin beads from the lysates of MDA-MB-231 and SK-OV-3 cells after the intact cells were biotinylated with ZEGFR:1907.BirA* and ZHER2:342.BirA* for 3 h. Biotinylated EGFR was recovered among the biotinylated protein pool when ZEGFR:1907.BirA* was used to label both MDA-MB-231 and SK-OV-3 cells. Similarly, biotinylated HER2 was pulled down when the ZHER2:342.BirA* construct was used to label SK-OV-3 cells, but not MDA-MB-231 cells (Figure 4C). Equal loading of samples was confirmed by Western blot analyses, where in panels 4A and 4B, immunoprecipitation elution samples served as loading controls, probed with mAbs specific for EGFR, HER2, EpCaM, and CD44. In panel C, lysates of biotinylated MDA-MB-231 and SK-OV-3 cells were probed with either anti-HER2 or anti-EGFR mAbs to serve as gel loading controls for these markers. Eluted proteins recovered from biotinylated proteins bound to streptavidin dynabeads (Figure 4C) were also analyzed by Western blot probed using streptavidin-HRP, highlighting the overall range of biotinylated proteins pulled down by these beads (Figure S2). Flow cytometry was also used to confirm the level of expression of EGFR, HER2, EpCaM, and CD44 on the surface of MD-MB-231 and SK-OV-3 (Figure S3).
Figure 4.
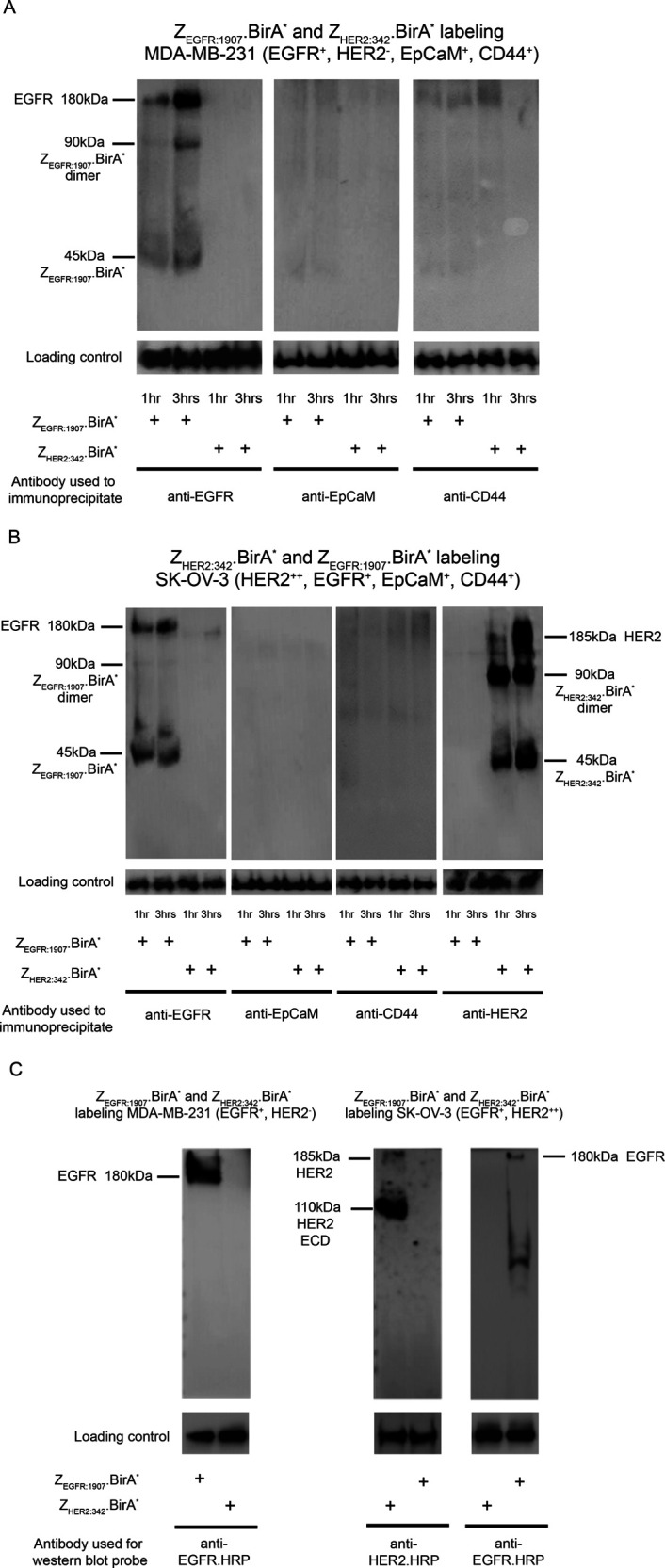
ZEGFR:1907.BirA* and ZHER2:34.BirA* are able to biotinylate EGFR and HER2, respectively, on the surface of human cancer cells expressing EGFR or HER2. (A) EGFR-expressing cells (MDA-MB-231) were only biotinylated with ZEGFR:1907.BirA* or ZHER2:342.BirA*. Labeled cells, at 1 h and 3 h time points, were lysed, and EGFR itself was pulled down (IP) using Cetuximab (human EGFR mAb). The captured biotinylated EGFR was detected by Western blot using streptavidin-HRP. (B) The same experiment was performed for ZHER2:342.BirA* and ZEGFR:1907.BirA* using the HER2-overexpressing and EGFR-expressing cells (SK-OV-3). Specifically, cells were biotinylated with either ZHER2:342.BirA* or ZEGFR:1907.BirA*. Cells were collected at 1 h and 3 h time points, lysed, and HER2 and EGFR were pulled down (IP) using Trastuzamab (human HER2 mAb) or Cetuximab (human EGFR mAb), respectively. The captured biotinylated HER2 or EGFR were detected by Western blot using streptavidin-HRP. For both cell lines, MDA-MB-231 (A) and SK-OV-3 (B), EpCaM, and CD44 were also pulled down, and their lack of biotinylation was confirmed in order to establish the specificity of the biotinylation event. For panels A and B, immunoprecipitation elution samples served as loading controls, probed with mAbs specific for EGFR, HER2, EpCaM, and CD44 to confirm that an equal amount of each marker was loaded on the beads. (C) MDA-MB-231 and SK-OV-3 cells were biotinylated with either ZEGFR:1907.BirA* or ZHER2:342.BirA*, as previously described for 3 h. Cells were then lysed, and biotinylated protein species were pulled down using streptavidin dynabeads. The captured biotinylated proteins were analyzed by Western blot. In the case of the EGFR-expressing MDA-MB-231 and SK-OV-3 cells, biotinylated EGFR was detected using an anti-EGFR-HRP conjugate. An anti-HER2.HRP conjugate was used to detect biotinylated HER2 recovered from HER2-expressing SK-OV-3 cells. Western blots of cell lysates probed with either anti-HER2 or anti-EGFR mAbs served as the loading controls of these markers. SDS-PAGE and Western blots were performed on samples prepared under nonreducing conditions, resulting in the observation of Affibody.BirA* monomers and dimers.
We attempted to chemically conjugate BirA* to an antibody, as such proteins represent a large class of available ligands targeting a diverse range of cell surface markers. However, our preliminary attempts at modifying antibodies with BirA* generated complexes that retain both functions, but could only label the antibody structure itself (results not shown). The average length of a human or mouse IgG antibody is 15 nm, while the long axis of an affibody molecule has a span of only 4.4 nm (Figures S4 and 1).25 The labeling radius of BirA* has been reported to be ∼10 nm, based on the diffusion rate and half-life of the amino-reactive intermediate bio-5′-AMP released from the catalytic site of BirA*.20 BioID studies have mainly been performed with smaller ligands (∼20–100 kDa) than antibodies (∼150 kDa) that would have favored the biotinylation of interacting partners.1,21−23 This distance limit suggests that Ligand.BirA* bispecifics constructed using an antibody as a ligand would not lead to the efficient biotinylation of amino groups on a protein target upon binding of such Ligand.BirA* constructs. Consequently, we searched for alternative ligands smaller than antibodies, namely, affibodies, scFvs, and nanobodies. As BirA* is a protein domain that is minimally soluble in physiological buffers (this study),2 our initial scFv ligand.BirA* fusion constructs either aggregated, displayed limited solubility (results not shown and is given in a previous study2), or were poorly expressed. Affibodies were also used to create BirA* bispecifics as they are smaller (∼6 kDa) and bind to their targets with high affinity.15,16 Importantly, affibodies are sufficiently small to position the enzyme BirA* within a distance range for its released bio-5′-AMP intermediates to react with lysine side chains on proximal and adjacent proteins. Interestingly, the BirA* component of our affibody constructs could be replaced with an ascorbic acid peroxidase enzyme (as in the case of APEX), which upon the addition of H2O2 and a biotin-phenol substrate would create biotin-phenoxyl radicals that have been shown to covalently react with electron-rich amino acids such as Tyr, Trp, His, and Cys on proximal proteins located up to a radial distance of 20 nm from the enzyme catalytic site.15,24
In conclusion, these results suggest that Affibody.BirA* fusion proteins can serve as agents for selectively labeling cell surface receptors. This approach can also be used to identify unknown receptors or off-target interactions of clinically relevant scFvs, and nanobodies and related small protein ligands may yield ligand.BirA* bispecifics that will bind to their targets and locate their BirA* catalytic site within its ∼10 nm labeling range.
Methods
Recombinant Protein Design, Expression, and Purification
Recombinant constructs were designed from the published affibody sequences ZEGFR:1907 and ZHER2:243 recognizing EGFR and HER2, respectively.15,16 A gene-encoding BirA* was fused to each affibody gene, yielding constructs ZEGFR:1907.BirA* and ZHER2:243.BirA*, where BirA* is positioned at the C-terminus of affibodies in each expressed construct (Figure 1).8−11 The sequence HVGSGSELGTGSENLYFQ served as both a spacer and a TEV cleavage site in linking the affibody and BirA* domains. Each construct incorporates a C-terminal 6 histidine tag for purification purposes.
The gene constructs were cloned into a Champion pET301/CT-DEST Gateway Vector expression plasmid, and nucleotide sequences were optimized for the expression in E. coli. The resulting plasmids were transformed into the BL21 DE3 star strain (Invitrogen) and protein expression induced using 1 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG) over a period of 5 h shaking at room temperature. Each bacterial pellet was subsequently lysed in 50 mM Tris (pH 8), 300 mM NaCl, 1% Triton X-100, 10 mM 2-mercaptoethanol, 10 mM MgCl2, 0.1 mg/mL lysozyme (Sigma-Aldrich), and 25 units/mL benzonase nuclease (Sigma-Aldrich) lysis buffer using an ultrasonicator (Branson) at a 30% amplitude with 10-second pulses on and 15-s pulses off. The protein constructs were purified from lysates using IMAC NTA affinity chromatography. Protein fractions were eluted with 500 mM imidazole in TBK buffer, pH 8.5 and 50% glycerol. The recovered protein fractions were concentrated using a Centricon (10 kDa cutoff, Millipore) and desalted against 40 mM Tris (pH 8.0), 100 mM KCl, and 10% glycerol. Protein purity was confirmed by SDS-PAGE by running the samples in the presence of 50 mM DTT (reducing conditions), and the protein concentration was calculated from absorbance readings recorded at 280 nm.
Cell Lines
The human breast adenocarcinoma cell lines MDA-MB-231 and SK-OV-3 were used as representative cell lines expressing EGFR and both HER2 and EGFR. MDA-MB-231 was cultured at 37 °C, 5.0% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and dihydrostreptomycin (100 μg/mL). SK-OV-3 cells were cultured in RPMI Medium 1640 supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and dihydrostreptomycin (100 μg/mL).
BirA* Biotinylation Activity Assay
The catalytic activity of BirA* was measured using BSA as a lysine-rich protein substrate. The level of biotinylation of BSA arising from exposure to each of the BirA* construct was monitored either by ELISA or by Western blot analysis. For the ELISA-based assay, wells of high protein-binding, clear, flat-bottom, polystyrene 96-well plate (Corning) were coated with 1 μg/well BSA in 100 μL of PBS pH 7.4 overnight, then washed three times with PBS-T (PBS with 0.05% Tween 20). BirA* reaction components (2ug BirA*, ATP, Biotin, and MgOAc) diluted in TBK buffer were dispensed into each well. After washing each plate, the extent of biotinylation of BSA was monitored by treating each well with 1:2500 streptavidin-HRP (BioLegend) [1 h; room temperature], followed by washing with PBS-T and by adding the substrate 3,3′,5,5′-tetramethylbenzidine (TMB). For Western blot analysis, the biotinylation assay was carried out in solution, and the resulting mixtures of proteins were resolved by SDS-PAGE in the presence of 50 mM DTT (reducing conditions), transferred to a nitrocellulose membrane, and finally probed using 1:2500 Streptavidin-HRP.
Surface Plasmon Resonance
The ability of ZEGFR:1907.BirA* and ZHER2:243.BirA* to bind to their targets (the extracellular domains of EGFR and HER2, respectively) was determined by SPR (Biacore T200; GE Healthcare Mississauga, ON, Canada). The extracellular domain of EGFR (human EGFR.Fc; R&D) or HER2 (human HER2.Fc; R&D) was immobilized in separate flow cells of a protein G CM-5 chip. Binding analyses were performed using single-cycle kinetics by flowing ZEGFR:1907.BirA*, ZEGFR:1907, ZHER2:243.BirA*, or ZHER2:243 over either immobilized human EGFR.Fc or human HER2.Fc for 120 s at a flow rate of 30 μL/minute. Protein G reference flow cells served as control wells. Proteins were dissolved in HBS-P running buffer (10 mmoL/L HEPES pH 7.4, 150 mmoL/L NaCl, 0.05% v/v Tween 20), and the chip was regenerated using 10 mM glycine pH 1.7 buffer.
Biotinylation of Immobilized Targets
Human EGFR.Fc or HER2.Fc was immobilized on the surface of protein G dynabeads. The beads were then exposed to either ZEGFR:1907.BirA or ZHER2:243.BirA*. Unbound Affibody.BirA* was washed away, and BirA* reaction components [40 mM ATP, 400uM Biotin, 40 mM MgOAc] diluted in TBK buffer were added. The level of the biotinylation of EGFR.Fc and HER2.Fc arising from exposing them to Affibody.BirA* constructs was monitored by eluting the biotinylated proteins bound to the protein G beads by boiling the beads in lithium dodecyl sulfate (LDS) sample buffer and performing Western blot on the recovered samples under nonreducing conditions using 1:2500 streptavidin-HRP.
Biotinylation of Cell Surface Antigens
ZEGFR:1907.BirA* (or ZHER2:243.BirA* negative control) was incubated with 2 × 106 MDA-MB-231 cells in the presence of 400 μM biotin and 40 mM ATP/MgOAc in TBK buffer, pH 8.5 for 3 h at 37 °C. Cells were subsequently lysed using a radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (30mM HEPES, pH 7.4,150mM NaCl, 1% Nonidet P-40,0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 5mM EDTA, 1mM NaV04, 50mM NaF). Anti-EGFR antibody (10 μg) (Cetuximab) bound to protein G dynabeads (Invitrogen) was used to pull-down EGFR from the labeled cell lysate. The resulting immune complexes were loaded on an SDS-PAGE gel and transferred to a nitrocellulose membrane. The presence of biotinylated EGFR was confirmed by Western blot using 1:2500 streptavidin-HRP (BioLegend). The same procedure was performed with the lysates of SK-OV-3 cells previously biotinylated with either ZHER2:243.BirA* or ZEGFR:1907.BirA* to label EGFR and HER2. The antibodies used for the pull-down experiments were 10 μg of anti-HER2 (Trastuzumab) and anti-EGFR (Cetuximab) bound to protein G dynabeads. Anti-EpCaM (10 μg) (Aarigo biolaboratories) and anti-CD44 (SinoBiological) antibodies were used to capture EpCaM and CD44, respectively, from the labeled lysates derived from both MDA-MB-231 and SK-OV-3 cells. Streptavidin dynabeads (Invitrogen) were also used to capture biotinylated proteins from the lysates of MDA-MB-231 and SK-OV-3 cells labeled with either ZHER2:243.BirA* and ZEGFR:1907.BirA*. The presence of EGFR and HER2 among the biotinylated proteins pulled down was confirmed by Western blot using 1:5000 anti-EGFR.HRP (Abcam) and anti-HER2.HRP (Novus Biologicals) conjugates. Immunoprecipitation eluted samples were probed with mAbs specific for EGFR, HER2, EpCaM (R&D Systems), and CD44 (R&D Systems) on Western blots to serve as loading controls (Figure 4A, B). For the loading controls for these Western blot analyses shown in Figure 4C, the direct lysate was loaded to ensure that a relatively equal amount of receptor was present on the beads. Proteins bound to streptavidin dynabeads from samples shown in Figure 4C were also eluted and probed with 1:2500 streptavidin-HRP to show biotinylated proteins pulled down from each of these treatments. Samples analyzed in these Western blot analyses were run under nonreducing conditions.
Flow Cytometry
MDA-MB-231 or SK-OV-3 cells were incubated with 500 nM of either the primary antibody or the Affibody.BirA* at 4 °C for 1 h. Cells were subsequently washed, and anti-human IgG Fc-PE, anti-rat IgG Fc-PE, or streptavidin-PE (BioLegend) were added at a 1:1000 dilution. Cells were incubated at 4 °C for 30 min and washed, and the fluorescence signal was recorded using a BD LSR II (BD Biosciences).
Acknowledgments
This work was financially supported by project grants 148556 and 156138 from the Canadian Institutes of Health Research to J.G. The authors would like to thank Dr. Marzena Cydzik for her part in the data analysis of the BirA* kinetics, Dr. Peter Lee for helping generate predicted ribbon model structures, the abstract graphic, and the figure layouts, as well as Dr. Amanda Sparkes for her continuous guidance.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.1c00192.
Single-cycle kinetics sensorgrams derived from increasing concentrations of ZEGFR:1907.BirA* and ZEGFR:1907 interacting with immobilized HER2, and ZHER2:243.BirA* and ZHER2:243 or with immobilized EGFR, as monitored using surface plasmon resonance; Western blot analysis of immune complexes recovered from streptavidin dynabeads pull-downs of biotinylated proteins from cell lysate samples biotinylated using ZEGFR:1907.BirA* and ZEGFR:1907 analyzed, probed using a streptavidin-HRP Western blot; flow cytometry analyses of MDA-MB-231 and SK-OV-3 cells, showing the expression profiles of EGFR, HER2, EpCaM, and CD44 as well as the binding of Affibody.BirA* bispecifics; and ribbon representation of a human IgG antibody alongside that of an affibody (PDF)
Accession Codes
BirA: P06709.
Author Contributions
M.A. and J.G. conceptualized the project and wrote the manuscript. M.A. performed and designed experiments as well as analyzed the data.
The authors declare no competing financial interest.
Supplementary Material
References
- Roux K. J.; Kim D. I.; Raida M.; Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. 2012, 196, 801–810, 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed]
- Rhee H.-W.; Zou P.; Udeshi N. D.; Martell J. D.; Mootha V. K.; Carr S. A.; Ting A. Y. Proteomic mapping of mitochondria in living cells Via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. Recent advances in proximity-based labeling methods for interactome mapping. F1000Research 2019, 8, 135. 10.12688/f1000research.16903.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-Smith A.; Cronan J. Jr. Molecular biology of biotin attachment to proteins. J. Nutr. 1999, 129, 477S–484S. 10.1093/jn/129.2.477s. [DOI] [PubMed] [Google Scholar]
- Prakash O.; Eisenberg M. A. Biotinyl 5′-adenylate: Corepressor role in the regulation of the Biotin genes of Escherichia coli K-12. Proc. Nat. Acad. Sci. U. S. A. 1979, 76, 5592–5595. 10.1073/pnas.76.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM; Tymoczko JL; Stryer L.. Acetyl Coenzyme A Carboxylase Plays a Key Role in Controlling Fatty Acid Metabolism. In Biochemistry, 5th ed.; Section 22.5, W.H. Freeman and Co.: New York, 2002. [Google Scholar]
- Wilson K. P.; Shewchuk L. M.; Matthews B. W. The E. coli biotin holoenzyme synthetase(slash)bio repressor crystal structure delineates the biotin and dna-binding domains. Proc. Natl. Acad. Sci. U. S. A. 1993, 89, 9257–9261. 10.2210/pdb1bib/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K.; Beckett D. Function of a conserved sequence motif in biotin holoenzyme synthetases. Protein Sci. 2000, 9, 1530–1539. 10.1110/ps.9.8.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streaker E. D.; Beckett D. Nonenzymatic biotinylation of a biotin carboxyl carrier protein: Unusual reactivity of the physiological target Lysine. Protein Sci. 2006, 15, 1928–1935. 10.1110/ps.062187306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E.; Schulman H.; Cronan J. E. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2008, 13, 3043–3050. 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J. E. Targeted and proximity-dependent promiscuous protein biotinylation by a mutant escherichia coli biotin protein ligase. J. Nutr. Biochem. 2005, 16, 416–418. 10.1016/j.jnutbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Yao S.; Zhu Y.; Chen L. Advances in targeting cell surface signalling molecules for immune modulation. Nat. Rev. Drug Discovery 2013, 12, 130–146. 10.1038/nrd3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Ozment T.; Wright G. L.; Peterson J. M. Identification of Putative Receptors for the Novel Adipokine CTRP3 Using Ligand-Receptor Capture Technology. PLoS One 2016, 11, e0164593 10.1371/journal.pone.0164593. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID: 27727322
- Bender J.; Schmidt C. Mass spectrometry of membrane protein complexes. Biol. Chem. 2019, 400, 813–829. 10.1515/hsz-2018-0443. [DOI] [PubMed] [Google Scholar]
- Friedman M.; Orlova A.; Johansson E.; Eriksson T. L. J.; Höidén-Guthenberg I.; Tolmachev V.; Nilsson F. Y.; Ståhl S. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding affibody molecule. J. Mol. Biol. 2008, 376, 1388–1402. 10.1016/j.jmb.2007.12.060. [DOI] [PubMed] [Google Scholar]
- Orlova A.; Magnusson M.; Eriksson T. L. J.; Nilsson M.; Larsson B.; Höidén-Guthenberg I.; Widström C.; Carlsson J.; Tolmachev V.; Ståhl S.; Nilsson F. Y. Tumor imaging using a picomolar affinity her2 binding affibody molecule. Cancer Res. 2006, 66, 4339–4348. 10.1158/0008-5472.can-05-3521. [DOI] [PubMed] [Google Scholar]
- Yang J.; Zhang Y. I-TASSER Server: New development for protein structure and Function predictions. Nucleic Acids Res. 2015, 43, W174. 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subik K.; Lee J.-F.; Baxter L.; Strzepek T.; Costello D.; Crowley P.; Xing L.; Hung M.-C.; Bonfiglio T.; Hicks D. G.; Tang P. The expression patterns of Er, pr, her2, ck5/6, EGFR, Ki-67 and AR by Immunohistochemical analysis in breast cancer cell lines. Breast Cancer: Basic Clin. Res. 2010, 4, 117822341000400 10.1177/117822341000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFazio-Eli L.; Strommen K.; Dao-Pick T.; Parry G.; Goodman L.; Winslow J. Quantitative assays for the measurement of her1-her2 heterodimerization and phosphorylation in cell lines and breast tumors: Applications for diagnostics and targeted drug mechanism of action. Breast Cancer Res. 2011, 13, R44. 10.1186/bcr2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. I.; Birendra K. C.; Zhu W.; Motamedchaboki K.; Doye V.; Roux K. J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Nat. Acad. Sci. U. S. A. 2014, 111, E2453. 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner S.; Cheng F.; Hogan A.; Grima N.; Yang S.; Ke Y.; Au C.; Morsch M.; De Luca A.; Davidson J.; Molloy M.; Shi B.; Ittner L.; Blair I.; Chung R.; Lee A. ALS/FTD-causing mutation In CYCLIN F causes THE dysregulation of SFPQ. Hum. Mol. Genet. 2021, 30, 971–984. 10.1093/hmg/ddab073. [DOI] [PubMed] [Google Scholar]
- Cabukusta B.; Berlin I.; van Elsland D. M.; Forkink I.; Spits M.; de Jong A. W. M.; Akkermans J. J. L. L.; Wijdeven R. H. M.; Janssen G. M. C.; van Veelen P. A.; Neefjes J. Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-related FFNT motifs. Cell Rep. 2020, 33, 108475 10.1016/j.celrep.2020.108475. [DOI] [PubMed] [Google Scholar]
- Li Y.; Sousa R. Novel system for in vivo biotinylation and its application to crab antimicrobial protein scygonadin. Biotechnol. Lett. 2012, 34, 1629–1635. 10.1007/s10529-012-0942-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V.; Zou P.; Rhee H.-W.; Udeshi N. D.; Cracan V.; Svinkina T.; Carr S. A.; Mootha V. K.; Ting A. Y. Proteomic mapping of the human MITOCHONDRIAL intermembrane space in live cells via RATIOMETRIC Apex tagging. Mol. Cell 2014, 55, 332–341. 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire E. O. Crystal structure of a Neutralizing Human Igg Against HIV-1: A template for Vaccine Design. Science 2001, 293, 1155–1159. 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.