Abstract
Aims
Until recently, the pluripotency factor Octamer (ATGCAAAT)-binding transcriptional factor 4 (OCT4) was believed to be dispensable in adult somatic cells. However, our recent studies provided clear evidence that OCT4 has a critical atheroprotective role in smooth muscle cells. Here, we asked if OCT4 might play a functional role in regulating endothelial cell (EC) phenotypic modulations in atherosclerosis.
Methods and results
Specifically, we show that EC-specific Oct4 knockout resulted in increased lipid, LGALS3+ cell accumulation, and altered plaque characteristics consistent with decreased plaque stability. A combination of single-cell RNA sequencing and EC-lineage-tracing studies revealed increased EC activation, endothelial-to-mesenchymal transitions, plaque neovascularization, and mitochondrial dysfunction in the absence of OCT4. Furthermore, we show that the adenosine triphosphate (ATP) transporter, ATP-binding cassette (ABC) transporter G2 (ABCG2), is a direct target of OCT4 in EC and establish for the first time that the OCT4/ABCG2 axis maintains EC metabolic homeostasis by regulating intracellular heme accumulation and related reactive oxygen species production, which, in turn, contributes to atherogenesis.
Conclusions
These results provide the first direct evidence that OCT4 has a protective metabolic function in EC and identifies vascular OCT4 and its signalling axis as a potential target for novel therapeutics.
Keywords: Atherosclerosis, OCT4, ABCG2, ROS, Heme
Graphical Abstract
Translational Perspective.
Despite advances in lipid-lowering treatment, there is little understanding of the molecular and cellular mechanisms that can help to prevent or reverse atherosclerosis development. We have previously discovered that the pluripotency factor OCT4 plays an atheroprotective role in smooth muscle cells but does not play a critical role in myeloid cells. Here, we show that OCT4 has an atheroprotective role in endothelial cells at least in part via activation of the ABCG2 transporter, which in turn protects endothelial cells from the intracellular accumulation of excessive heme and ROS, indicating that OCT4-dependent signaling can be used for developing new therapeutic interventions.
1. Introduction
Atherosclerosis is a common progressive macrovascular disease that drives myocardial infarctions and strokes, making it the leading cause of death worldwide.1 A monolayer of endothelial cells (ECs) lines the inner wall of all blood vessels and plays a critical role in regulating vascular homeostasis. ECs form a selectively permeable barrier that controls the transport of macromolecules, such as low-density lipoprotein (LDL) and transmigration of white blood cells from the bloodstream into the vessel wall, processes critical for atherogenesis.2 EC activation by factors such as non-laminar flow and oxidized LDL leads to EC dysfunction, which is defined by the down-regulation of nitric oxide synthase (NOS), up-regulation of cell surface adhesion molecules (ICAM1 and VCAM1) and pro-inflammatory molecules, the production of reactive oxygen species (ROS),3 and is thought to be the inciting event in atherosclerosis development. The resulting endothelial dysfunction is often coupled with changes in cellular phenotype, including the process of endothelial-to-mesenchymal transition (EndoMT), which is correlated with atherosclerotic plaque vulnerability.4–7
Although previous studies have demonstrated the importance of EC activation and phenotypic modulation during atherosclerosis, little is known about key mechanisms regulating EC plasticity and how they influence lesion growth and/or stability. Using EC-specific knockout mouse models, several groups have reported on signalling pathways involved in EC-specific mechanisms of atherosclerosis, including fibroblast growth factor receptor 1 (FGFR1)/transforming growth factor β (TGFβ),8 Kruppel-like factor 2 (KLF2)/Forkhead Box P1 (FOXP1),9 and KLF4.10 Interestingly, it has been shown that similar signalling pathways can play opposite roles in different vascular cell types. For example, previous studies of KLF4 provided evidence that it has an opposite effect in smooth muscle cell (SMC) vs. EC and myeloid cells, in that KLF4 deficiency in SMC is atheroprotective,11 but the loss of Klf4 in EC10 or myeloid cells12 promotes atherosclerosis. Similarly, studies have found that TGFβ-signalling is pro-atherogenic in EC6 and atheroprotective in SMC.13 Therefore, there is a critical need to discover cell-specific pathways that can be targeted to induce beneficial effects in multiple cell types without causing significant detrimental off-target effects in other cells.
The Octamer (ATGCAAAT)-binding transcriptional factor 4, OCT4 [also known as POU5F1, OCT4A (in human), and OCT3/4 (in mouse)], regulates pluripotency in embryonic stem cells14 and is one of the critical factors in the induction of pluripotent stem cells.15 Until recently, OCT4 was believed to be completely silenced and therefore non-functional in somatic cells.16 Consistently with these reports, we did not observe OCT4 activation in heathy arteries.17 However, recently our group17,18 and others19–21 have demonstrated that OCT4 is reactivated in vascular SMC in pathological conditions, including atherosclerosis, vascular injury, aortic aneurism, and pulmonary hypertension. Using SMC and pericyte (SMC-P)-specific conditional Oct4 knockout mice, we have shown that OCT4 plays an atheroprotective role by regulating SMC phenotypic transitions.17 Furthermore, we found that SMC-P-specific loss of Oct4 markedly impaired angiogenesis following vascular injury, including corneal burn and hindlimb ischaemia, at least in part through defective migration of perivascular cells leading to pathological vascular permeability.18 These data were the first evidence that the pluripotency factor OCT4 can play functional roles in adult somatic cells. Interestingly, using a myeloid-selective LysM-Cre mouse model, we have shown that OCT4 in myeloid cells does not play a critical role in atherosclerosis.17 The recent observation that ketone bodies such as β-hydroxybutyrate protect cultured vascular EC from cellular aging in vitro by mediating nuclear ribonucleoprotein A1-induced stabilization of Oct4 mRNA21 indicates that OCT4 is functional in EC. Therefore, given our findings in SMC-P and the growing appreciation of EC plasticity in atherosclerosis, we sought to determine if OCT4 was also a key regulator of EC phenotype in atherosclerosis.
Here, we use mice with EC-specific tamoxifen-inducible knockout of Oct4 to demonstrate that OCT4 in EC is atheroprotective by controlling EC activation and improving indices of plaque stability. Mechanistically, we found that OCT4 acts at least in part via activation of adenosine triphosphate (ATP)-binding cassette (ABC) transporter G2 (ABCG2), which in turn protects EC from the intracellular accumulation of excessive heme and ROS.
Despite multiple reports showing reactivation of the pluripotent isoform OCT4 in somatic cells, they have mainly relied on detecting the mRNA or protein levels of OCT4 in these cells.22,23 These approaches are highly controversial due to several potential false-positives associated with OCT4 transcript and protein detection, including low OCT4 expression levels, possible antibody cross-reactivity with other non-pluripotent isoforms, and multiple pseudo-genes.24,25 To avoid ambiguities in detecting low OCT4 levels, we chose to use a genetic loss-of-function approach to conditionally and specifically inactivate the pluripotency isoform of OCT4 in EC. Of major importance, this approach allows us to use the antibody- and gene expression-independent strategies to test the role of OCT4 in different cell types and biological processes.
2. Methods
2.1. Mice
Protocols for experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) of the Cleveland Clinic and the University of Virginia. All animal procedures conformed to the National Institutes of Health (NIH) guidelines. Oct4Flox/Flox [Pou5f1tm1Scho] mice,26 Cdh5-CreERT2 [Tg(Cdh5-cre/ERT2)1Rha] mice,27 Rosa-Stop-eYFP [B6.129X1-GT(ROSA)26Sortm1(EYFP)Cos] mice (The Jackson Laboratory, #006148), and Apoe−/− (B6.129P2-Apoetm1Unc) mice (The Jackson Laboratory, #002052) were used in this study. We first bred Oct4Flox/Flox mice with Apoe−/−, Cdh5-CreERT2, or Rosa-Stop-eYFP mice to generate Oct4Flox/Flox; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; and Apoe−/− mice. We then bred Oct4Flox/WT; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; Apoe−/− (Cre+) males with Oct4Flox/WT; Rosa-Stop-eYFP+/+; Apoe−/− (Cre−) females to generate Oct4Flox/Flox; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; Apoe−/−; and Oct4WT/WT; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; Apoe−/− littermate experimental mice. Experimental mice were backcrossed nine generations to the C57BL/6J line (The Jackson Laboratory). We genotyped conditional Oct4 mice and Cdh5-CreERT2 mice, as described previously,17,27 and ROSA26-STOPfloxeYfP mice were genotyped as previously described.11
2.2. Tamoxifen administration and animal diet
We achieved activation of Cre-recombinase through 10 intraperitoneal injections of tamoxifen (Sigma–Aldrich, T-5648) [1 mg in 100 µL of peanut oil (Sigma–Aldrich)] over a 10-day period starting at 6–8 weeks of age. All experimental and control mice were treated with tamoxifen in an identical manner. Following the tamoxifen administration, experimental mice (both male and female) were placed on a high-fat Western-type diet (WD) containing 21% milk fat and 0.15% cholesterol (Harlan Teklad; TD.88137), for 4, 10, or 18 weeks starting at 8 weeks of age. Irradiated mouse standard chow diet was purchased from Harlan (TD.7012).
Mice were fasted for 4 h prior to euthanasia, and blood and blood plasma were collected. Glucose was measured using a handheld AimStrip Plus blood glucose measuring device (Cat# 37321, Germaine Lab, TX, USA) via tail tip cut prior to euthanasia. Mice were euthanized via CO2 asphyxiation, followed by cervical dislocation. Following WD feeding for target duration, the brachiocephalic artery (BCA) and lungs were harvested, fixed in 4% paraformaldehyde (Cat# 157-4-100, Electron Microscopy Sciences, PA, USA), and embedded to paraffin. BCAs were sectioned at 10 μm thickness, starting at the aortic arch and ending at the bifurcation to the right subclavian artery. Morphometric and immunohistochemical analyses were performed using 2–3 sections per artery. Left lung lobes were sectioned longitudinally at 5 μm thickness. Total plasma cholesterol, triglyceride levels, and complete blood counts were analysed by the University of Virginia Clinical Pathology Laboratory.
2.3. Lipid accumulation analysis by Sudan IV staining
Aortas from Oct4Flox/Flox; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; ApoE–/– and Oct4WT/WT; Cdh5-CreERT2; Rosa-Stop-eYFP+/+; ApoE–/– mice fed WD for either 10 or 18 weeks were dissected from the aortic arch to the iliac bifurcation and subjected to en face Sudan IV staining. In brief, after meticulously cleaning the peri-adventitial fat, aortas were dehydrated in 70% ethanol for 5 min and incubated in Sudan IV solution for 6 min, which was prepared as follows: 1 g Sudan IV (Sigma–Aldrich, S4261) was diluted in 100 mL 70% ethanol and 100 mL 100% acetone. Finally, aortas were differentiated in 80% ethanol for 3 min and stored in phosphate buffered saline (PBS) at 4°C.
2.4. Morphometric and immunohistochemical analyses
We performed modified Russell–Movat (Movat) staining or Masson’s Trichrome staining for morphometric analysis of the BCAs. The areas within the internal elastic lamina, lesion, and lumen were measured. Collagen content was observed by PicroSirius red staining and birefringence imaging using plane-polarized light with an Olympus IX83. Immunohistochemistry was performed with antibodies for LGALS3 (Cedarlane; CL8942AP; 1:500) and TER-119 (rat anti-mouse, Santa-Cruz Biotechnology Inc.; 1:200). Staining for immunohistochemistry was visualized by 3,3′-diaminobenzidine (DAB, Acros Organics). Images were acquired with an Olympus IX83 coupled with Cellsens Dimension software. Settings were fixed at the beginning of both acquisition and analysis steps and were unchanged. Vessel morphometry and areas of DAB-positive immunohistochemical or PircoSirius red staining were quantified using Image-Pro 10 software (Media Cybernetics Inc.), as previously described.11,17,28
Immunofluorescence was performed with antibodies for OCT4-biotin (Santa-Cruz Biotechnology Inc.; clone C10; 1:50) with tyramide signal amplification (Invitrogen; T-30955) or OCT4 antibody (Abcam; ab181557; 1:500), eYFP/eGFP (Abcam; ab6673; 1:250), ACTA2-Cy3 (Sigma–Aldrich; C6198, clone1A4; 1:500), LGALS3 (Cedarlane; CL8942AP; 1:500), VCAM1 (Abcam; ab115135; 1:250), vWF (Abcam; ab11713; 1:500), and ABCG2 (Santa-Cruz Biotechnology Inc.; SCBT-58224; BXP-53; 1:30). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI). The secondary antibodies were donkey anti-rabbit Alexa Fluor 647 (Invitrogen; A31573; 1:100), donkey anti-rat DyLight 650 (Invitrogen; SA5-10029; 1:100), and donkey anti-goat Alexa Fluor 488 (Invitrogen; A11055; 1:100). Incubations with isotype-matched IgG antibodies were used as a negative control for all immunostaining. Confocal images were acquired using a Zeiss LSM700 scanning confocal microscope and a Leica DM2500M confocal microscope. Brightness and contrast were adjusted uniformly across all images of the same set during analysis. Cells within the lesion area made up of 30 µm depth from the lumen were counted (designated as 30 µm fibrous cap area). The depth of 30 µm has been chosen based on our previous extensive investigation of the overall thickness of the contiguous subluminal ACTA2 staining, a commonly accepted way to define the fibrous cap area.29 The number of eYFP (YFP+), LGALS3 (LGALS3+), ACTA2 (ACTA2+), VCAM1 (VCAM1+), and ABCG2 (ABCG2+) cells were normalized to either the total number of cells, as assessed by DAPI staining, or to the number of YFP+ cells. High-resolution z-stack analysis was performed using Zen 3.0 Light Edition Software or LAS X Software (Leica) to ensure staining was limited to a single cell. For pixel analysis, z-stack slices were collapsed into maximum intensity projections and analysed using Image-Pro 10 software. Researchers were blinded to the genotype of the animals until the end of the analysis.
2.5. Image analysis
For morphometry analysis, we used the software program Image-Pro 10. All images were taken with an Olympus IX83 coupled with Cellsens Dimension software and uploaded as TIFF files. Each image was traced via the polygon tool within the Image-Pro program, with a total of three regions of interest (ROIs). The three regions include the external elastic lamina, the internal elastic lamina, and the lesion. Each specimen was calibrated and standardized via a 100 µm scale bar. The total area of each ROI was measured.
2.6. Flow cytometry
For flow cytometry analyses, mice were euthanized by CO2 asphyxiation and then perfused with 10 mL of PBS. Aortas from the iliac bifurcation to the aortic root or lung left lobes were cleaned from fat and fascia, cut longitudinally, dissected, and placed into an enzyme cocktail containing 0.75 mg/mL Liberase™ (Roche, 355374) and 57 IU/10 mL Elastase (Worthington Bio. Corp., LS002279) in RPMI-1640. Aortas were minced and placed in a 37°C incubator for 1.5 h. Cells were passed through a 70 µm strainer (Corning), spun down at 500 g for 5 min, and resuspended in red blood cell lysis buffer (BD PharmLyse, 555899) for 2 min and then inactivated in fluorescence-activated cell sorting (FACS) buffer [1% bovine serum albumin (BSA) in PBS]. The remaining cells were stained with Viability dye (Live/Dead Red, ThermoFisher Scientific, L23102). EC-derived cells were identified based on the endogenous yellow fluorescence protein (YFP). Samples were run on a Beckman Coulter LSR Fortessa flow cytometer and analysed by Flow Jo software. Cdh5-negative littermate mice were used as a negative control to set the YFP+ gate.
2.7. Single-cell RNA sequencing
2.7.1. FACS
Aortas, including the arch, thoracic, and abdominal portions, or lung left lobes from Oct4Δ/Δ (n = 3) and Oct4+/+ (n = 2) female mice after 5 weeks of WD feeding were cleaned, cut, and digested as described in Section 2.6 except that all buffers and solutions contained 1 µg/mL Actinomycin D (Gibco, 11805017) to prevent transcriptional changes induced by digestion. LoBind pipet tips (Xtip Biotix, R1000-9FC) were used to transfer samples. The cell suspension was transferred to 0.04% ultra-pure non-acetylated BSA (Thermo Fisher, AM2616) in PBS, filtered through 35 µm filters (Falcon, 352235) on ice, and sorted on a BD Influx sorter with the 100 µm nozzle into LoBind tubes (Eppendorf, 022431048) containing PBS with 0.04% ultra-pure non-acetylated BSA. Cdh5-negative littermate mice after 5 weeks of WD feeding were used as a control to set the YFP+ gate. The cells in each group were captured in Chromium 10X genomics libraries, which after barcoding, were pooled and sequenced on the Illumina MiSeq, 150 cycle high-output.
2.7.2. Cell Ranger processing of Oct4Δ/Δ and Oct4+/+ mouse samples
Samples were run through the standard Cell Ranger (version 3.0.0) pipeline. Samples were demultiplexed using cellranger mkfastq command with –qc option. Count matrices for the samples were then obtained with the cellranger count command using mm10 assembly of the mouse genome provided by 10X Genomics.
2.7.3. Differential gene expression of Oct4Δ/Δ and Oct4+/+
Oct4 Δ/Δ and Oct4+/+ samples were subjected to quality control (QC) filters, which removed cells with fewer than 800 (aortas) or 450 (lungs) UMI, fewer than 500 (aortas) or 330 (lungs) detected genes, and >20% (aortas) or 13% (lungs) of UMI coming from mitochondrial genes. In addition, genes not expressed by any of the cells or having >1 transcript detected in <3 cells were removed. Following these QC filters, we were left with 8783 genes and 1876 cells (of 1984) in aortas and with 7476 genes and 1288 cells (of 1335) in lungs. The data were then normalized using a scran30 package, and differential expression analyses between Oct4Δ/Δ and Oct4+/+ conditions were performed using the MAST31 package.
2.7.4. Pseudotime for Oct4Δ/Δ and Oct4+/+
Pseudotime cell trajectory analysis was performed using the Monocle 2 package:32 Samples were QC filtered as described above, cell data set was created using negbinomial size distribution and trajectory was created using differentially expressed genes found by the MAST package (see above) with q values <0.1. Gene expression as a function of pseudotime was plotted using the plot_genes_in_pseudotime monocle command.
2.7.5. Cell Ranger processing of human data from Wirka et al.33
Sra files were obtained using the prefetch command from sratoolkit (version 2.10.8), and fastq files were extracted using the fastq-dump command with split-files option (from the same toolkit). Count matrices were extracted from the fastq files using the ‘cellranger count’ command (cellranger version 3.0.0) with the human genome, assembly GRCh38. Then the correlation of expression of ABCG2 and all other genes was found using the base R cor function.
2.8. EC culture
The murine aortas were collected from Oct4Δ/Δ and Oct4+/+ mice and processed into aortic rings for further expansion, as described previously.34 Following the expansion of EC from the aortic rings, cells were plated on a gelatin-coated (Cat# 6950, Cell Biologics, IL, USA) T25 flask in endothelial growth media (EGM) consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% foetal bovine serum (FBS), 90 μg/mL heparin sulphate, and 50 ng/mL EC growth supplement. ECs and SMCs were purchased from Thermo Fisher {C0065C [human aortic endothelial cell (HAEC)]; C0035C [human umbilical vein endothelial cell (HUVEC)]; C0075C [human aortic smooth muscle cell (HASMC)] or Millipore Sigma [B354-05 bovine aortic smooth muscle cell (BAoSMC)]. We routinely used LookOut® Mycoplasma Erase (Sigma, Germany) to ensure a sterile cell culture platform, and mycoplasma testing to assure our cell culture quality.
2.9. CRISPR editing of exon1 of OCT4 in HUVEC
OCT4ex1_KO HUVEC were generated from Synthego Corporation (CA, USA) using a Cas9-guide RNA (5′-GAAGCTCACTTGCCTCCTCC-3′, PAM-GGG) complex targeting exon1 of the human OCT4 gene, whereas the control HUVEC cells (OCT4WT) were treated with Cas9 enzyme. We used these cells within passages 4–7 and cultured them in EGM as mentioned before. The same mycoplasma precautions were taken as above.
2.10. Oxygen consumption rate and extracellular acidification rate
Oxygen consumption rate (OCR) and extracellular acidification rate were measured using the Seahorse Extracellular Flux (XF24) Analyzer (Seahorse Bioscience Inc., North Billerica, MA, USA) according to the manufacturer’s protocol, and Seahorse assay medium was prepared according to the previous study.35 In brief, Seahorse assay media for ‘Mitostress Assay’ was prepared with DMEM without glucose, l-glutamine, phenol red, sodium pyruvate, and sodium bicarbonate (Sigma–Aldrich) supplemented with 1.08 g/L glucose, 1.85 g/L sodium chloride, 1 mM sodium pyruvate, and 15 mg/mL phenol red, followed by supplementation with 2 mM l-glutamine and pH adjustment to 7.35 with sodium hydroxide. HUVEC were plated at a density of 50 000 cells/well in EC growth media overnight in a gelatin-coated plate, with three wells/plate left empty for background correction. The growth medium was taken out, and cells were washed with the appropriate Seahorse assay medium three times. After the final wash, the assay medium was put on to each well at a final volume of 500 μL/well. Next, the plate was incubated in a 37°C non-CO2 incubator for 1 h and then transferred to the Seahorse XF24 Analyzer for analysis. Inhibitors for OCR measurements were added to the Mitostress assay media at the following concentrations: oligomycin A (1.5 µM, Sigma–Aldrich), FCCP (1.5 µM, Sigma–Aldrich), antimycin A (2.5 µM, Sigma–Aldrich), and rotenone (1.25 µM, Sigma–Aldrich).36 It allowed for OCR estimation coupled with ATP production, maximal respiratory capacity, and spare respiratory capacity. Basal respiration was estimated before injecting oligomycin A.
2.11. RNA interference
To knock down endogenous Oct4 in Oct4+/+ mouse aortic EC, custom made Dharmacon™ Accell™ specially modified siRNAs targeting Oct4 (herein siOct4, 0.5 µM) or control and non-targeting siRNA (herein siNT, 0.5 µM) were used according to the manufacturer’s protocol and without any use of transfection reagent. The level of suppression and specificity of Oct4 siRNA was evaluated by quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR).
2.12. Heme estimation
To estimate the heme concentration in the cell culture supernatant, we used supernatant from either siNT- and siOct4-treated mouse Oct4+/+ cells or OCT4ex1_KO and OCT4WT HUVEC and directly quantified them using the QuantiChrom Heme Assay (BioAssay Systems, CA, USA) kit and its manufacturer’s protocol. To measure the resultant colour product’s absorbance, we used the SpectraMax iD5 multimode plate reader (Molecular Devices LLC, CA, USA).
2.13. ROS estimation by 2′,7′-dichlorofluorescin diacetate
2′,7′-Dichlorofluorescin diacetate (H2DCFDA) was used to measure intracellular ROS using the H2DCFDA cellular ROS detection reagent (D399, Invitrogen™). Cells (either siNT- and siOct4-treated mouse Oct4+/+ cells or OCT4ex1_KO and OCT4WT HUVECs) were seeded at 1.5–2.5 × 104 cells/well on a gelatin-coated (Cat# 6950, Cell Biologics, IL, USA) 96-well plate for 24 h in the indicated conditions prior to assay in EGM [haem-depleted FBS37 in case of HUVEC experiment], and indicated drugs [Succinylacetone (Cat# 25501, Cayman Chemical Company,MI, USA), Hemin Chloride (Cat# 198820, MP Biomedicals), Fumitremorgin C (FTC; Cat# 11030, Cayman Chemical Company)]/DMSO/vehicle with no phenol red. Following 24 h of cell culture, cells were stained with 25 µM H2DCFDA in 1× PBS buffer for 45 min at 37°C, and then the signal was read by the SpectraMax iD5 multimode plate reader (Molecular Devices LLC, CA, USA) with an excitation/emission wavelength filter of 485/535 nm used to detect fluorescence of 2′,7′-dichlorofluorescein (DCF). The signal was background corrected and normalized to the vehicle.
2.14. Induction of EndoMT by recombinant TGFβ
Oct4 +/+ EC (1 × 105 cells/well) were plated in a 12-well gelatin-coated (Cat# 6950, Cell Biologics, IL, USA) culture plate in EGM. A day later, the culture media was changed to clear unbound or dead cells. Two days after initial seeding (60–70% confluency), cells were serum-starved and then treated with siNT (0.5 µM) and siOct4 (0.5 µM) for 24 h in Accell serum-free siRNA delivery media (Dharmacon™, Horizon Discovery, UK). Following siRNA treatment, each well was gently washed with serum-free EGM before treatment with recombinant mouse TGFβ1 (10 ng/mL) (Cat# 763102, Bio Legend, CA, USA) with/without 100 µM N-acetyl-l-cysteine (Cat# 20261, Cayman Chemical Company) for every other day, for a total period of 5 days. At the end of the 5th day, cells were harvested for gene expression analysis of endothelial marker (Pecam1), mesenchymal markers (Act2a, Tagln, Cnn1, Col1a1, and Col15a1), along with Oct4 and Nfkb (pro-inflammatory marker) by qRT–PCR.
2.15. Western blot
For the total protein extraction, cells were lysed by Pierce™ RIPA buffer (Cat# 89900, Thermo Scientific) in the presence of Halt™ protease inhibitor single-use cocktail (Cat# 78430, Thermo Scientific). The nuclear and cytoplasmic extracts were isolated by NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher, #78833). For western blot analyses, 1 µg (to detect GAPDH) or 15–50 µg (to detect OCT4 and ABCG2) of total protein were fractionated by electrophoresis under denaturing conditions on a gradient mini-PROTEAN TGX polyacrylamide precast gel (Bio-Rad) and transferred onto a transfer membrane (Millipore). Proteins were detected by probing western blots with antibodies specific to OCT4 (Abcam; ab181557; 1:200), ABCG2 (Santa-Cruz Biotechnology Inc.; SCBT-58224; BXP-53; 1:200), ACTA2 (Abcam; ab7817; 1:1000), vWF (Santa-Cruz Biotechnology Inc.; sc-53466; 1:500), eNOS (Cell Signaling; 32027S; 1:500), TAGLN (Santa-Cruz Biotechnology Inc.; sc-53466; 1:500), GAPDH (Cat# MAB374, Millipore, 1:1000; we did not detect any GAPDH band in BAoSMC), or beta-Actin (Santa-Cruz Biotechnology Inc.; sc-4778; 1:1000).
2.16. Chromatin immunoprecipitation assays
For in vitro chromatin immunoprecipitation (ChIP) assays, we follow previously described methods.38 Cells treated with or without water-soluble cholesterol (Sigma, C4951; 20 µg/mL for 24 h) were fixed with 1.42% paraformaldehyde for 15 min at room temperature. Crosslinked chromatin was sheared into fragments of 200–600 base pairs by temperature-controlled sonication (Qsonica, CT, USA). The sheared chromatin was immunoprecipitated with 2 µg of anti-OCT4 antibody (ab19857, Abcam), while negative control samples were incubated with non-immune rabbit IgG (Jackson ImmunoResearch Laboratories). Immune complexes were captured following the previously described protocol. After elution and purification of the genomic DNA (gDNA), RT–PCR was performed on immunoprecipitated (IP) and non-immunoprecipitated (INPUT) gDNA. Primer set used for the Abcg2 binding site and is mentioned in Supplementary material online, Table S12. The Human Negative Control Primer Set (Active Motif, 71001) amplifying a 78 base-pair fragment from a gene desert on human chromosome 12 was used as a negative control. Results were quantified as a percent of INPUT and normalized to non-immune IgG.
2.17. RNA isolation, cDNA preparation, and qRT–PCR
Total RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Isolated RNA was treated with DNaseI, and then RNA was reverse transcribed with the iScript™ gDNA Clear cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed on a C1000 Thermal Cycler CFX96 (Bio-Rad) using an IQ™ SYBR Green Supermix (Bio-Rad) for Oct4 and a Radiant™ Green 2×-qPCR Mix Lo-ROX (Alkali Scientific, FL, USA) for the rest of the genes. Primers used were specific for mouse Pou5f1 (exon 1), Abcg2, Nfkβ, Pecam1, Act2a, Tagln, Cnn1, Col1a1, Col15a1, and 18srRNA, and human ABCG2 (see Supplementary material online, Table S12). The expression of the genes was normalized to 18srRNA.
2.18. Mitochondrial membrane potential measurement
The HUVECs were seeded on a 96-well black culture plate. The cells were washed with PBS, and 5 µL of JC-1 staining solution (Cayman, #10009172) were added with culture media in each well. Cells were then incubated in a CO2 incubator for 30 min. After incubation, the cells were washed twice with assay buffer (Cayman, #10009322). The fluorescence intensity was measured by SpectraMax iD5 multimode plate reader (Molecular Devices LLC, CA, USA). The mitochondrial membrane potential was determined by the ratio of intensity J-aggregates (excitation and emission at 535 and 595 nm) to intensity of J-monomers (excitation and emission at 485 and 535 nm).
2.19. Mitochondrial fragmentation analysis
To obtain the mitochondria morphology images, ECs were stained with Mitospy (Biolegend, #424801) staining solution (500 nM) for 30 min at 37°C. Images were then processed using ImageJ (NIH) to subtract backgrounds and subjected to binary conversion. After the binary conversion, the total mitochondrial area (μm2) and mitochondria number were quantified in each imaged cell. Additionally, mitochondrial fragmentation counts were calculated using the mitochondrial particle number normalized by total mitochondria area as previously described.39
2.20. Adenovirus transduction
The enhanced Ad-CMV-GFP (CV10001) and the Ad-CMV-ABCG2 (VH806202) overexpressing adenoviruses were both purchased from Vigene Biosciences. The media containing adenovirus (500 MOI) was added to the cells and incubated at 37°C with 5% CO2 overnight. The next day, the media were replaced by fresh culture media.
2.21. SORE6-GFP lentivirus transduction
SORE6-dscopGFP lentiviral particles were generated by transfection of 293T cells. 293T cells were transfected (Fugene transfection reagent) with pPACKH1 vectors and SORE6-dscopGFP plasmid DNA according to manufacturer’s protocols (System Biosciences). Viral supernatant was collected at 48 h, and the virus was concentrated with PEG-it Virus Precipitation Solution (System Biosciences). SORE6-GFP virus was then added to wild-type and CRISPR-Cas9-edited OCT4 knockout HUVECs in a six-well plate. Next, 48 h later, 2 µL/mL puromycin was added to cells. After puro selection, cells were collected and plated in a 96-well plate in EGM complete media (2500 cells/well). Fluorescence images were taken with Incucyte Live Cell Analysis System (Sartorius).
2.22. Statistics
The normality of the data was determined via either the Kolmogorov–Smirnov test or the Shapiro–Wilk test. For comparison of two groups of continuous variables with normal distribution, two-tailed Student’s t-tests (for equal variances) were used. Linear mixed-model analysis of variance (ANOVA) with Tukey’s or Sidak’s post hoc tests was used for multiple group comparisons. Two-group comparisons with non-normal distributions were analysed using the non-parametric ANOVA based on the Wilcoxon rank-sum test. Fisher’s exact test was used for categorical data. P < 0.05 was considered to be significant. Statistical outliers were identified as values beyond 3 s.d. of the mean level and were excluded from analyses. The sample size (number of mice) was chosen on the basis of our previous studies.11,17 Graph Pad Prism 8.1.1 software was used to analyse the data. All in vitro experiments were done in duplicates or triplicates for each experimental group (technical replicates) and performed in three independent experiments (biological replicates). Due to the limited availability of OCT4ex1_KO and OCT4WT HUVEC, western blots presented in Figures 6A, 7B, and Supplementary material online, Figure S1C were performed in two independent experiments. In experiments with viruses, an independent experiment corresponds to a separate virus infection. The number of mice (biological replicates) used for each in vivo analysis is indicated in the figure legends.
Figure 6.
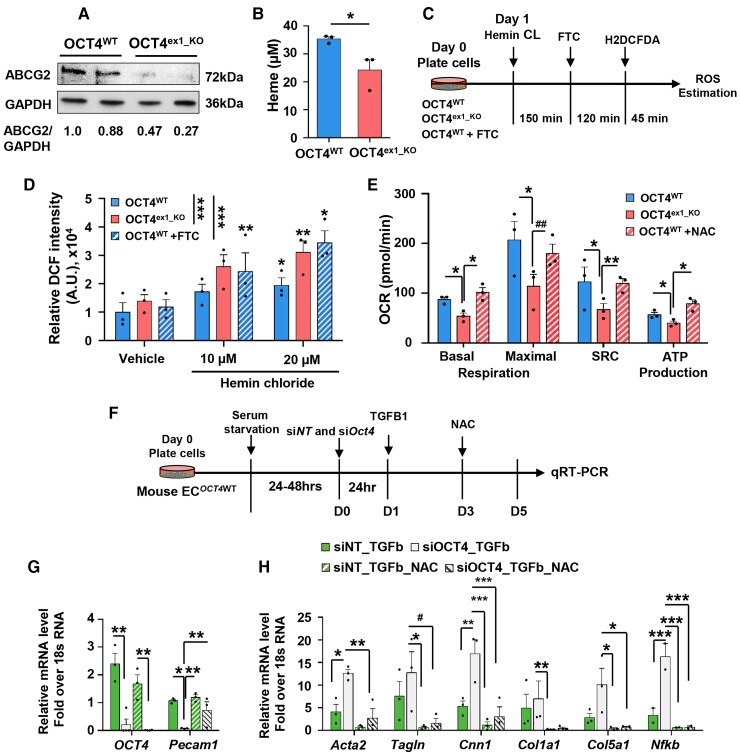
Inhibition of intracellular ROS rescues mitochondrial respiration and EndoMT in the OCT4-deficient ECs. (A) Representative western blot showing protein levels of ABCG2 and GAPDH in OCT4WT and OCT4ex1_KO HUVECs. Replicates correspond to independent experiments. The relative ABCG2/GAPDH ratio based on the densitometry analysis is indicated beneath the blots. (B) Quantification of extracellular haem concentrations in OCT4WT and OCT4ex1_KO HUVEC cultured media. Values = mean ± S.E.M.; *P < 0.05 by unpaired t-test; n = 3 independent experiments. (C and D) Loss of OCT4 increased ROS generation within HUVEC in response to hemin chloride treatment. (C) Schematic showing the experimental design for the loss-of-function assay of ABCG2 after treatment with succinylacetone (endogenous inhibitor of haem synthesis) followed by FTC (specific ABCG2 inhibitor) to inhibit ABCG2 in OCT4WT HUVECs and subsequent intracellular ROS estimation in OCT4WT, OCT4ex1_KO, and OCT4WT+FTC HUVECs. (D) Hemin chloride-induced ROS generation was then measured by DCF intensity and showed dose–response with increasing hemin chloride concentration. OCT4WT+FTC group recapitulated the OCT4ex1_KO phenotype in the presence of increasing hemin chloride. Values = mean ± S.E.M; Data were analysed by linear mixed-model ANOVA followed by Tukey’s post hoc test, *P < 0.05, **P < 0.01, and ***P < 0.001; n = 3 independent experiments. (E) Seahorse mitochondrial stress test measuring the oxygen consumption rates (OCR) in OCT4WT, OCT4ex1_KO HUVECs, and OCT4ex1_KO HUVECs cultured in the presence of the ROS inhibitor, N-acetyl-l-cysteine (NAC). NAC significantly improved basal mitochondrial respiration, spare respiratory capacity, and ATP production in OCT4ex1_KO HUVECs. Error bars represent mean ± S.E.M.*P < 0.05, **P < 0.01 by linear mixed-model ANOVA followed by Tukey’s post hoc test, n = 3 independent experiments. Results for OCT4WTand OCT4ex1_KO HUVECs are the same as in Figure 4H. (F) Schematic showing experimental design for in vitro EndoMT experiments. (G and H) qRT–PCR analyses demonstrated that EC transfected with blocking siOct4 have lower Oct4 and Pecam1 levels (G) but higher levels of the EndoMT markers and Nfkb mRNA gene expression (H) in response to TGFβ1 treatment as compared with siNT control cells. The presence of NAC significantly improved TGFb1-induced gene expression alterations. Data were analysed by one-way ordinary ANOVA (mixed-model), followed by Tukey’s multiple comparisons test (*P < 0.05, **P < 0.01, ***P < 0.001, and #P = 0.07). Values = mean ± S.E.M; n = 3 independent experiments.
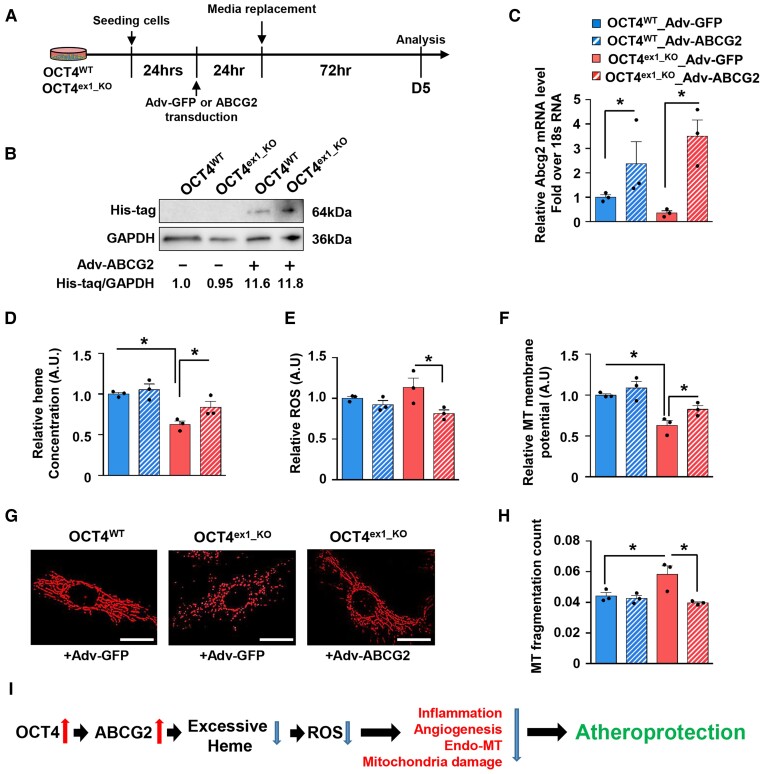
Figure 7.
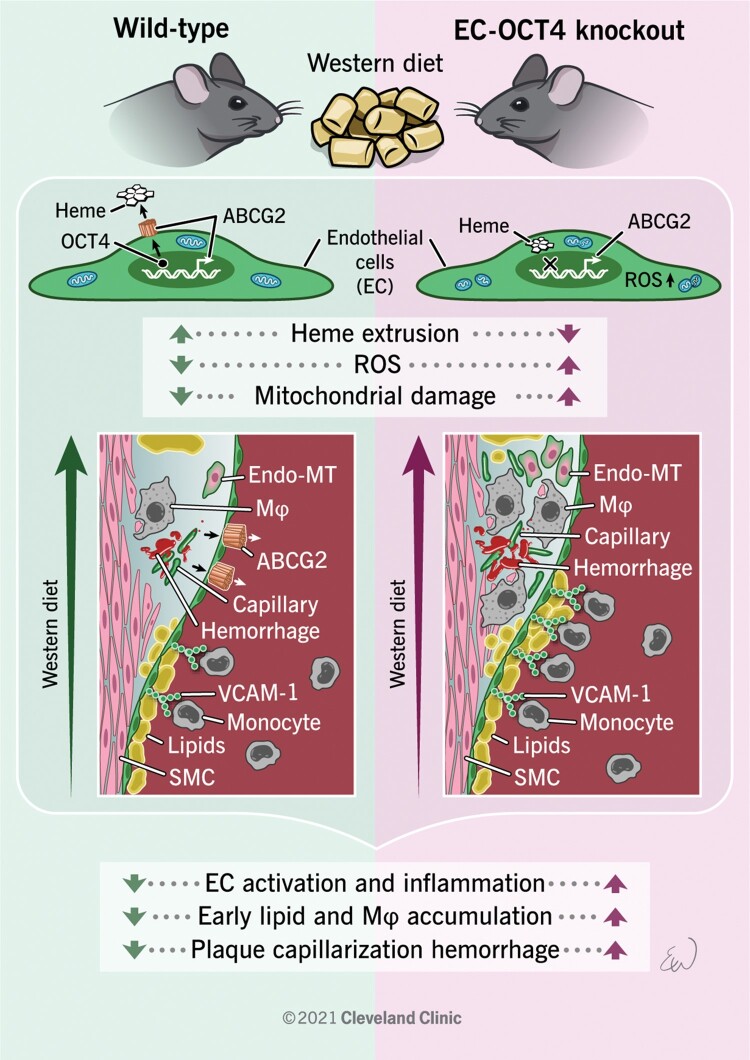
Overexpression of ABCG2 improves ROS accumulation and mitochondrial functions in OCT4-deficient ECs. (A) Schematic showing experimental design for the overexpression of ABCG2 in OCT4WT and OCT4ex1_KO HUVECs. Cells were infected with human ABCG2 overexpressing adenovirus containing His-Tag (Adv-ABCG2) or control adenovirus (Adv-GFP) for 24 h, followed by functional assays. (B) Representative western blot showing protein levels of His-tag and GAPDH in Adv-ABCG2-infected OCT4WT and OCT4ex1_KO HUVECs. The relative His-tag/GAPDH ratio based on the densitometry analysis is indicated beneath the blots. (C) qRT–PCR analysis showed significant increases in mRNA levels for ABCG2 in OCT4WT and OCT4ex1_KO HUVECs infected with Adv-ABCG2 as compared with Adv-GFP. *P < 0.05 by unpaired Student t-test; n = 3 independent experiments. (D–F) Quantification of extracellular haem concentrations in cultured media (D), intracellular ROS accumulation (E), and mitochondria membrane potential (F) in OCT4WT and OCT4ex1_KO HUVECs with and without ABCG2 overexpression. (G) Representative immunofluorescence staining using MitoSpy™ to visualize mitochondria morphology. Scale bar = 20 µm. (H) Quantification of the mitochondria fragmentation count based on the fragmentation score system. (D–F, H) Values = mean ± S.E.M.; *P < 0.05 by one-way ANOVA followed by Tukey’s post hoc test; n = 3 independent experiments. (I) Proposed model of the atheroprotective role of OCT4 in ECs: Activation of the pluripotency factor OCT4 in ECs at the early stages of atherosclerosis development results in up-regulating ABCG2 that in turn plays a role in regulating haem and ROS levels inside of cells and leads to atheroprotection. Loss of OCT4 results in haem and ROS accumulation leading to inflammation, EndoMT, pathological angiogenesis, and mitochondrial dysfunction and promoting atherosclerosis progression.
3. Results
3.1. Endothelial OCT4 deficiency exacerbates atherosclerosis in Apoe−/− mice
Given the ambiguities in detecting the pluripotency OCT4 isoform in adult cells,24,25 we took several antibody-dependent and antibody-independent approaches to unambiguously investigate whether EC express the pluripotency isoform of OCT4. We generated CRISPR-Cas9-edited OCT4 exon1 human umbilical vein endothelial cells (OCT4ex1_KO HUVECs) (see Supplementary material online, Figure S1A). The HUVEC line was chosen because previous studies by two independent groups have demonstrated that HUVECs express the pluripotency isoform of OCT4.21,40 Our CRISPR-Cas9 approach resulted in a 54% editing efficiency based on the Sanger sequencing (see Supplementary material online, Figure S1B). All subsequent experiments were carried out on a mixed cell population. The same lot of HUVECs treated with Cas9, but without the guide RNA were used as a wild-type control (OCT4WT). Western blotting with an anti-OCT4 antibody (Abcam, ab181557) demonstrated a ∼50% decrease in OCT4 protein levels in OCT4ex1_KO HUVECs correlating with editing efficiency based on the sequencing results (see Supplementary material online, Figure S1C). In addition, as an alternative antibody-independent approach, we used the SORE6-GFP reporter that was previously developed to detect and isolate human cancer stem cells.41 This reporter responds to the pluripotency factors OCT4 and SOX2 by expressing green fluorescence protein (GFP). OCT4WT and OCT4ex1_KO HUVECs were infected with the SORE6-GFP lentivirus and checked for GFP expression. Importantly, we observed GFP expression in OCT4WT HUVECs. OCT4ex1_KO HUVECs demonstrated a ∼40% decrease in GFP expression as compared with OCT4WT (see Supplementary material online, Figure S1D and E), indicating that GFP activation is OCT4-dependent.
Next, using western blotting with the antibody characterized in experiments with OCT4ex1_KO HUVECs, we demonstrated that several EC and SMC primary lines, including HUVECs, HAEC, HASMC, BAoSMC, and mouse aortic SMC express OCT4 (see Supplementary material online, Figure S2A). Immunofluorescence staining in atherosclerotic lesions from Apoe−/− mice revealed intra-nuclear OCT4 staining in EC (see Supplementary material online, Figure S2B and C).
To test whether endothelial OCT4 expression has a role during atherogenesis, we generated a tamoxifen-inducible EC eYFP-lineage-tracing EC-specific Oct4 knockout Apoe−/− mouse model, Cdh5-CreERT2; Oct4Flox/Flox; Rosa-Stop-eYFP+/+Apoe−/− (designated as Oct4Flox/Flox) (Figure 1A). This model allows for highly efficient and permanent labelling of mature EC with enhanced YFP at the time of tamoxifen injections with subsequent determination of the fate of these cells and their progeny independent of detectable levels of EC endogenous lineage markers and simultaneous knockout of the OCT4 exon 1, specifically responsible for pluripotency, in EC (designated as Oct4Δ/Δ). Littermate Cdh5-CreERT2; Oct4WT/WT; Rosa-Stop-eYFP+/+Apoe−/− mice (designated as Oct4+/+) were used as wild-type controls. Both Oct4Flox/Flox and Oct4+/+ mice were injected with tamoxifen between 6 and 8 weeks of age, followed by high-fat Western diet (WD) feeding for 4 weeks (early EC injury stage of atherosclerosis), 10 weeks (fatty streak stage), or 18 weeks (advanced lesion stage) (Figure 1B).
Figure 1.
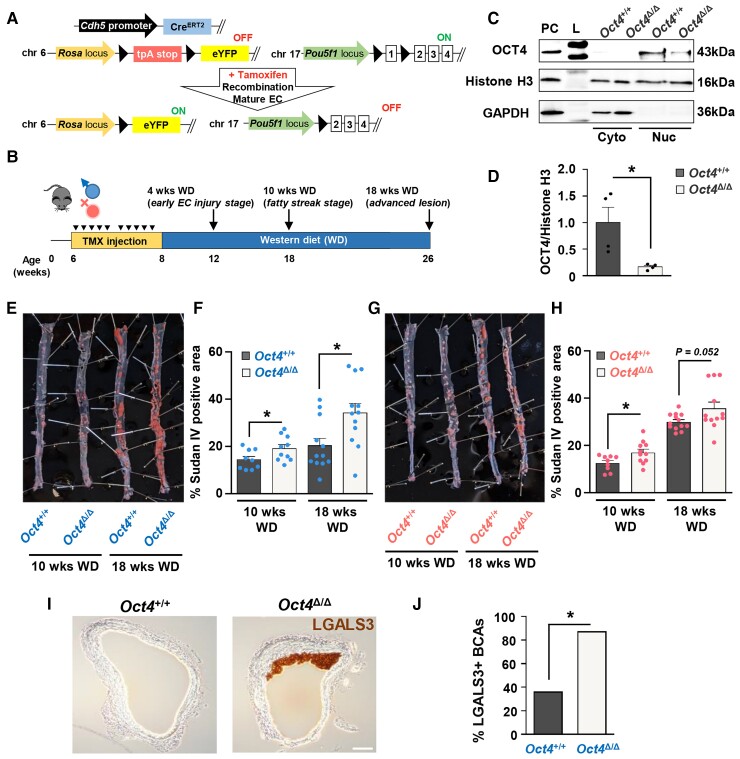
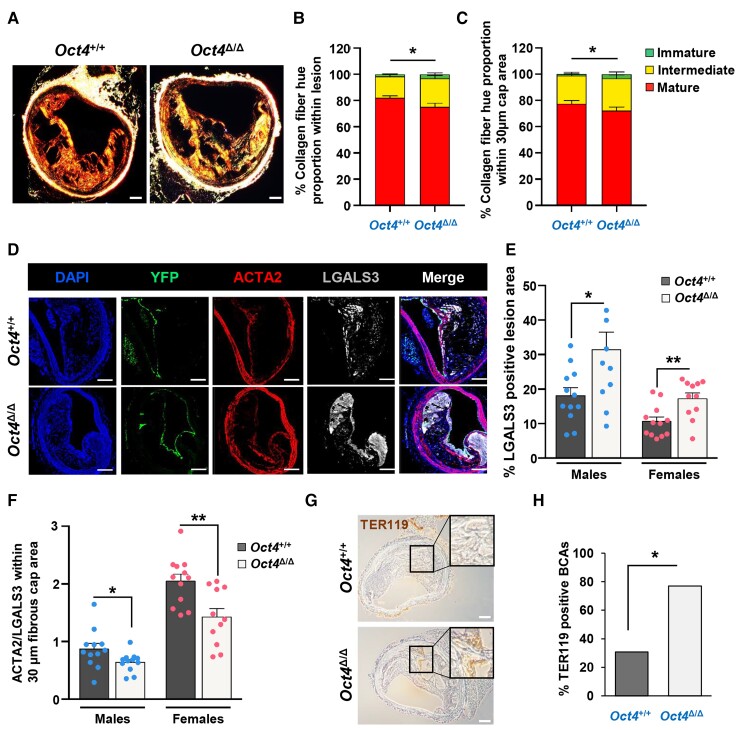
EC-specific conditional knockout of Oct4 exacerbates atherosclerosis in Apoe−/− mice. (A) EC-specific-lineage-tracing and conditional Oct4 knockout mouse model. Schematic of Cdh5-CreERT2; Oct4Flox/Flox; Rosa-Stop-eYFP+/+Apoe−/− mouse (designated as Oct4Flox/Flox). Mice received 10 injections of tamoxifen (TMX) between 6 and 8 weeks of age. In response to tamoxifen, Cre-recombinase under the control of the Cdh5 gene promoter excises LoxP sites (black triangles), removing Oct4 exon 1 (responsible for pluripotency) and STOP codon upstream of eYFP specifically in EC. Finally, it provides simultaneous EC-lineage-tracing and EC-specific Oct4 knockout. (B) Schematic of animal experiments. (C) Representative western blot showing protein levels of OCT4, Histone H3, and GAPDH in nuclear (Nuc) and cytoplasmic (Cyto) extracts from EC isolated from aortas of EC-specific OCT4 knockout (Oct4Δ/Δ) and wild-type (Oct4+/+) mice. L, ladder; PC, positive control, A404 precursor cells. (D) Densitometry quantification of Western blots. Values = mean ± S.E.M.. *P < 0.05 by unpaired t-test; n = 3 independent experiments. (E–H) Sudan IV en face staining and quantification of lesion area in aortas of Oct4Δ/Δ and Oct4+/+ male (E and F) and female (G and H) mice after 10 or 18 weeks of WD feeding. Values = mean ± S.E.M.. *P < 0.05, #P = 0.052 by unpaired t-test; Oct4+/+: n = 9 for 10 weeks and n = 12 for 18 weeks of WD; Oct4Δ/Δ: n = 11 (females) and n = 10 (males) for 10 weeks and n = 12 for 18 weeks of WD. (I) Representative immunostaining of LGALS3 in BCAs collected from Oct4Δ/Δ and Oct4+/+ male mice after 4 weeks of WD feeding. Scale bar = 100 µm. (J) The percentage of male mice with LGALS3-positive staining in Oct4+/+ (n = 4/11 positive BCAs) and Oct4Δ/Δ (n = 7/8 positive BCAs) BCAs. BCAs were analysed at 60 µm distance from aortic arch. *P < 0.05 by Fisher’s exact test.
To ensure Oct4 knockout in EC, we tested genetic recombination at the Oct4 exon 1 locus in lung EC. Specifically, YFP+ (EC-derived) and YFP− (non-EC-derived) cells were FACS-sorted from lungs of the Oct4Δ/Δ mice (see Supplementary material online, Figure S3), and recombination efficiency was detected using a genotyping reaction with specific primers.17,26 Of major importance, YFP+ EC demonstrated >96% recombination efficiency in contrast to YFP− cells (see Supplementary material online, Figure S2D and E). As further validation, aortic ECs were isolated from Oct4+/+ and Oct4Δ/Δ mice, and OCT4 protein levels were estimated by western blotting (Figure 1C and D). Protein extracts from mouse A404 cells, which express high levels of OCT4,17,42 were used as a positive control. Importantly, OCT4 protein was detected only in nuclear extracts from wild-type EC, whereas Oct4 knockout EC showed a marked significant decrease in OCT4 protein levels (Figure 1C and D).
Using these Oct4Δ/Δ mice, we observed significant increases in total lipid burden, based on the en face Sudan IV staining, after 10 and 18 weeks of WD feeding within aortas of Oct4Δ/Δ male mice as compared with Oct4+/+ male mice (Figure 1E and F). In Oct4Δ/Δ female mice, the percentage of Sudan IV+ area was significantly higher after 10 weeks of the diet and showed a trend toward an increase (P = 0.052) after 18 weeks of WD feeding (Figure 1G and H). Interestingly, there was an increase in the occurrence of LGALS3+ cells in atherosclerotic plaques within the pulmonary arteries of Oct4Δ/Δ male mice fed WD for 18 weeks compared with wild-type control mice (see Supplementary material online, Figure S2F and G), indicating that OCT4 plays a role in plaque development in EC from different vascular beds. Notably, after 10 and 18 weeks of WD, Oct4Δ/Δ mice had no significant differences in body weight, tissue weight, fasting cholesterol, triglyceride, and glucose levels as compared with Oct4+/+ mice (see Supplementary material online, Tables S1 and S2). In addition, the complete blood cell counts after 18 weeks of WD demonstrated no difference between Oct4Δ/Δ and Oct4+/+ male and female mouse profiles. However, a significant decrease in neutrophil counts in the Oct4Δ/Δ male group was found (see Supplementary material online, Table S3), likely resulting from the total increase in atherosclerosis in this group.
Interestingly, a transient reduction in body weight was documented in the male Oct4Δ/Δ mice after 4 weeks of WD. At the same time, we observed significant decreases in blood glucose levels in both male and female Oct4Δ/Δ mice compared with wild-type mice (see Supplementary material online, Tables S1 and S2). Nevertheless, counterintuitive to the improved glucose and body weight levels, we observed a significant increase in the early LGALS3+ cells accumulation (presumably macrophages) in the atheroprone areas of the BCA of Oct4Δ/Δ mice as compared with Oct4+/+ mice fed WD for 4 weeks (Figure 1I and J).
After 10 weeks of WD feeding, both male and female Oct4Δ/Δ mice demonstrated increases in BCA lesion size (see Supplementary material online, Figure S4A–C, and F). In addition, male mice had decreases in BCA lumen size and higher content of the elastic/collagen-enriched positive areas based on the Masson Trichrome staining as compared with wild-type littermate mice (see Supplementary material online, Figure S4D and E). Despite the increased lesion burden seen by Sudan VI, there was no significant difference in BCA lesion area, lumen size, or outward vessel remodelling between Oct4Δ/Δ and Oct4+/+ mice after 18 weeks of WD feeding (see Supplementary material online, Figure S5). However, 18-week WD Oct4Δ/Δ BCA lesions were found to have impaired indices of plaque stability, including reduced collagen maturation based on PicroSirius Red staining (males, Figure 2A–C), increased numbers of LGALS3+ cells (Figure 2D and E), reduced ratio of ACTA2+ cells to LGALS3+ cells within the area encompassing the inner 30 µm layer of the fibrous cap overlying lesions (designated as the 30 µm fibrous cap area) (Figure 2F), increased intraplaque haemorrhage (males, Figure 2G and H), and increased necrotic core size (males, Supplementary material online, Figure S5E). The relative ratio of SMC and macrophages has been shown to correlate with the fibrous cap stability.43 It is important to note that recent studies have shown that LGALS3 (MAC2) is expressed in both macrophages and SMC undergoing phenotypic transitions.33,44 Unfortunately, it is currently not possible to perform simultaneous dual-lineage-tracing for several cell types (e.g. EC and SMC). Nevertheless, based on our results and the fact that LGALS3 is considered as a potential cardiovascular inflammatory biomarker,45 we can conclude that EC-specific knockout of OCT4 leads to the early accumulation of macrophages (Figure 1I and J) and increased LGALS3+ cell content in the advanced lesions (Figure 2D and E), and also results in the decreased ratio of ACTA2+ to LGALS3+ cells (Figure 2F) that is associated with a less stable lesion. These changes are important since studies in humans show that changes in lesion composition, not size, are the primary determinant of clinical events.46
Figure 2.
EC-specific conditional knockout of Oct4 destabilizes atherosclerotic plaque. (A–C) Assessment of collagen fibre hue variations based on PicroSirius Red staining followed by polarized microscopy. (A) PicroSirius Red staining of representative BCA cross-sections of Oct4Δ/Δ and Oct4+/+ male mice fed WD for 18 weeks. Scale bar = 100 µm. The ratio between the amount of immature (green), intermediate (yellow), and mature (red) collagen fibrils showed a significant decrease in mature fibrils within the lesion (B) and the 30 µm fibrous cap area (C) of Oct4Δ/Δ mice (n = 13) as compared with control mice (n = 14), ±S.E.M.. *P < 0.05 (for mature fibrils) by unpaired Student t-test. (D) Immunofluorescence staining of DAPI, YFP, ACTA2, and LGALS3 on representative BCA sections of Oct4+/+ and Oct4Δ/Δ mice fed WD for 18 weeks, showing a noticeable increase in LGALS3+ cells in the lesion. Scale bar = 100 µm. (E and F) Quantification of the percentages of LGALS3+ lesion area (E) and the ratio of ACTA2+ cells over LGALS3+ cells within the 30 µm protective fibrous cap area (F) of BCAs collected from Oct4Δ/Δ and Oct4+/+ male and female mice after 18 weeks of WD feeding. Values = mean ± S.E.M., *P < 0.05 determined by either unpaired t-test with Welch correction (E) or unpaired t-test (F) for Oct4+/+ (n = 12 males; 12 females) vs. Oct4Δ/Δ (n = 12 males; 11 females) mice. (G) Immunostaining for the red blood cell marker TER119. Scale bar = 100 µm. Inserts indicate magnified areas. (H) The percentage of BCAs exhibiting intraplaque haemorrhage based on TER119 staining in Oct4+/+ (n = 4/13 positive BCAs) vs. Oct4Δ/Δ (n = 10/13 positive BCAs) male mice. BCAs were analysed at 120, 420, and 720 µm distance from aortic arch. If any of these three locations had any TER119-positive staining, that animal was marked as positive for haemorrhage. *P < 0.05 by Fisher’s exact test.
Taken together, these results indicate that EC-specific loss of OCT4 leads to increased lipid burden and atheroma formation in different vascular beds paired with detrimental changes to multiple indices of plaque stability within BCAs, suggesting an atheroprotective role for OCT4 in EC.
3.2. Single-cell RNA-sequencing demonstrates OCT4-driven changes in the gene expression in early-stage atherosclerosis
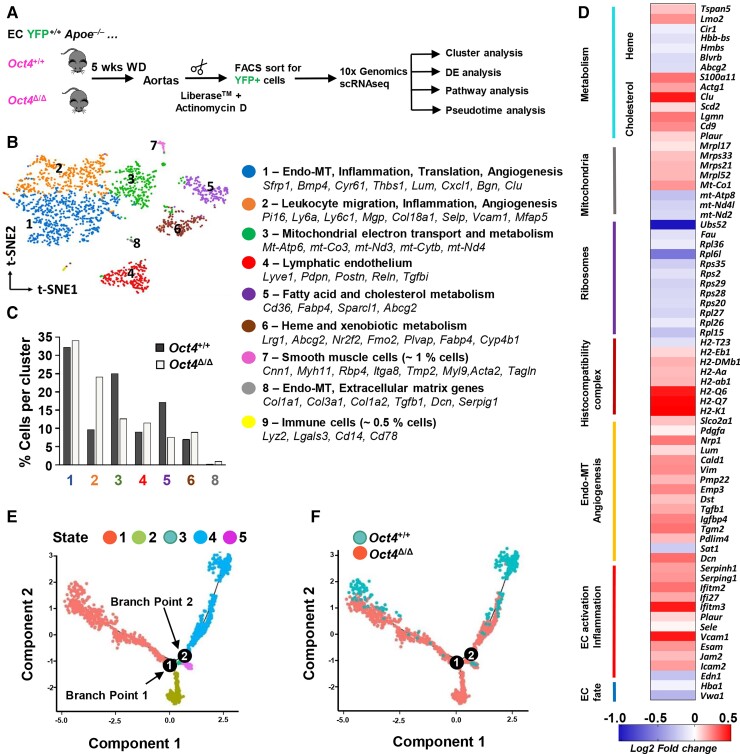
To systematically determine transcriptomic variation among EC states and the overall cell heterogeneity after conditional knockout of Oct4 during atherogenesis, we performed single-cell RNA-sequencing (scRNAseq) on FACS-sorted aortic YFP+ EC from Oct4Δ/Δ and Oct4+/+ female mice after 5 weeks of WD feeding (early stage of atherosclerosis) (Figure 3A). Following scRNAseq quality control, we obtained 449 wild-type EC (Oct4EC-WT) from the aortas of EC-Oct4+/+ mice and 1623 Oct4 knockout EC (Oct4EC-Δ) from the aortas of Oct4Δ/Δ mice for downstream analyses. Unsupervised clustering of the aortic EC revealed nine unique transcriptomic states (Figure 3B; Supplementary material online, Table S4). Integrated wild-type and Oct4 knockout datasets displayed satisfactory clustering alignment. Clusters 7 and 9 represented <1% contamination from SMC (Cluster 7; Cdh5−Myh11+) and myeloid cells (Cluster 9; Cdh5−Ptprc+). Although previous reports demonstrated that Cdh5-CreERT2 could initiate flox locus recombination in myeloid cells,47 our scRNAseq results indicate that tamoxifen treatment between 6 and 8 weeks of age leads to very minimal labelling of myeloid cells in Apoe−/− aortas after 5 weeks of WD feeding. EC Clusters 1–6 and 8 contained both Oct4EC-Δ and Oct4EC-WT cells. Nevertheless, the representation (percentage) of Oct4 knockout and wild-type cells within clusters was different (Figure 3C). Clusters 1 and 2 were enriched in multiple genes associated with activated endothelium (Vcam1, Icam2, and Fn1) and EndoMT markers (Vim, Tagln2, Cnn2, and Bmp4). In addition, Cluster 8 had fewer endothelial markers but was highly enriched in EndoMT markers, including Tgfb1, genes previously associated with osteogenic markers (Ogn, Dcn, Lum, and Postn) and collagens (Col3a1, Col1a1, and Col1a2). We found that the Oct4EC-Δ dataset had a higher percentage of cells in Clusters 2 and 8. Cluster 4 was enriched in markers of lymphatic endothelium (Lyve1and Pdpn), and was represented at a similar percentage for both Oct4EC-Δ and Oct4EC-WT datasets. Cluster 3, which was reduced in Oct4EC-Δ, was highly enriched in genes associated with mitochondrial electron transport and metabolism, suggesting that Oct4 knockout in EC may lead to mitochondrial dysfunction. Further metabolic changes were suggested by Cluster 5, which was enriched in genes associated with fatty acid and cholesterol metabolism and also reduced in the Oct4EC-Δ dataset. Haem and xenobiotic metabolism characterized Cluster 6, which was equivalently represented in both datasets. These results indicate that (i) aortic EC assume multiple heterogeneous transcriptomic states in early atherogenesis and (ii) OCT4 expression plays a role in regulating EndoMT and endothelial metabolism.
Figure 3.
Single-cell RNA sequencing reveals marked transcriptomic alterations in Oct4 knockout EC from mouse atherosclerotic aortas. (A) Schematic for 10× Genomic scRNA-seq experiment. ECs were isolated from aortas of Oct4+/+ (n = 2) and Oct4Δ/Δ (n = 3) female mice after 5 weeks of WD feeding. Aortas were digested in Liberase™ in the presence of the RNA polymerase inhibitor, actinomycin D, to prevent transcriptional changes during cell preparation. Aortic cells were FACS-sorted based on the EC lineage tracer, eYFP, followed by 10× scRNA-seq protocol. Following quality control, we obtained 449 ECs from Oct4+/+ mice and 1623 cells from Oct4Δ/Δ mice. (B) An aggregated t-distributed stochastic neighbour embedding (t-SNE) plot of YFP+ EC from Oct4+/+ and Oct4Δ/Δ mice coloured by cluster. Clusters were characterized based on the specific markers. (C) Percentage of Oct4+/+ and Oct4Δ/Δ cells in each cluster. (D) DE analysis revealed 405 up- and down-regulated genes in Oct4Δ/Δ as compared with Oct4+/+ EC, including up-regulation of adhesion and pro-inflammatory genes, as well as genes involved in EndoMT and angiogenesis, and down-regulation of EC fate genes and genes involved in mitochondria and heme metabolism. (E) Pseudotime analysis discovered five distinct states of the integrated Oct4+/+ and Oct4Δ/Δ EC with two cell fate decision points. (F) Ordering of Oct4+/+ and Oct4Δ/Δ EC along the pseudotime trajectory revealed two unique states (States 2 and 3) of EC from Oct4Δ/Δ mice as compared with Oct4+/+ mice.
Differential gene expression (DE) analysis identified 405 significantly up- or down-regulated genes in Oct4EC-Δ as compared with Oct4EC-WT cells (Figure 3D, Supplementary material online, Tables S5 and S6). Oct4EC-Δ cells demonstrated decreases in EC fate genes, ribosomal and mitochondrial genes, decreases in genes responsible for haem and xenobiotic metabolism, and increases in pro-inflammatory, pro-coagulant, and adhesion molecules, as well as increases in markers of EndoMT and angiogenesis as compared with Oct4EC-WT cells.
To better understand the cell’s transition dynamics, we performed pseudotime analysis. With pseudotime analysis, Oct4EC-Δ and Oct4EC-WT cells were ordered along a trajectory using genes that were differentially expressed between Oct4EC-Δ and Oct4EC-WT datasets (see Supplementary material online, Table S5) as a reference set. Interestingly, these analyses identified two branch points representing possible cell fate decisions (Figure 3E and F). Consistent with our cluster analysis, we found two unique states for Oct4EC-Δ cells that were completely absent in the wild-type dataset. These included States 2 and 3, which were enriched in markers of activated endothelium, including Vcam1 and Fn1, and markers of EndoMT (Lum) (see Supplementary material online, Figure S6).
Taken together, our scRNAseq results indicate that the loss of Oct4 shifted EC toward an activated endothelial state characterized by increased EndoMT, angiogenesis, and mitochondrial dysfunction.
3.3. EC-specific loss of Oct4 promotes EndoMT and intraplaque angiogenesis in atherosclerotic mice
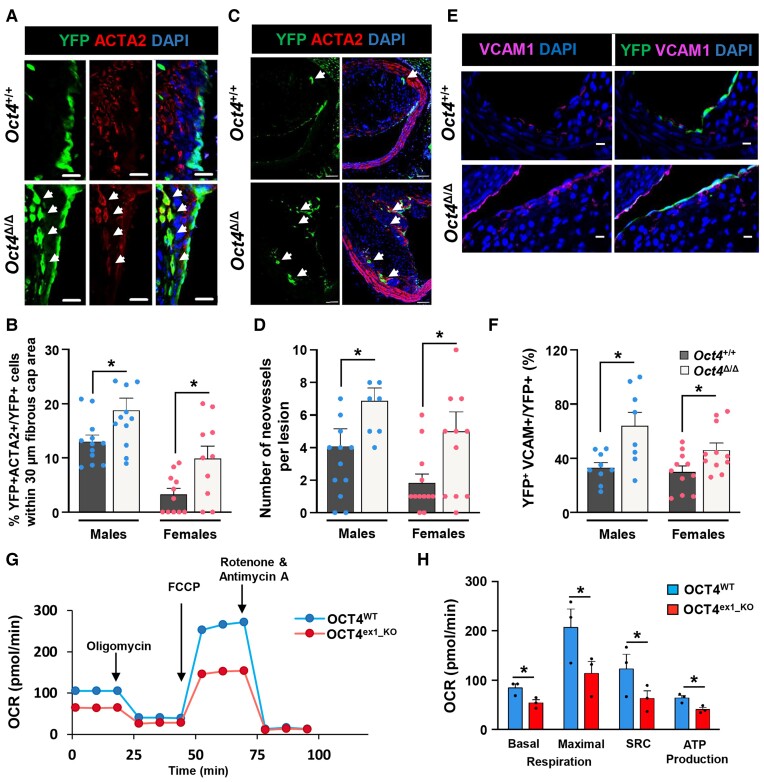
To further explore the role of OCT4 during the progression of atherosclerosis and to validate our scRNAseq outcomes, BCAs from 18-week WD-fed (advanced stage of atherosclerosis) Oct4Δ/Δ and Oct4+/+ male and female mice were evaluated for the cellular co-expression of YFP and the SMC/mesenchymal cell marker, ACTA2. Immunofluorescence staining followed by single-cell counting of z-stack confocal microscopy images from BCA plaques demonstrated a significant increase in YFP+ cells co-expressing ACTA2 (counted as YFP+ACTA2+ cells over YFP+ cells) within a 30 µm fibrous cap area in Oct4Δ/Δ as compared with Oct4+/+ BCAs (Figure 4A and B), indicating a higher incidence of EndoMT in the arteries of EC-specific Oct4 knockout mice. High-resolution confocal imaging and z-stack analysis addressed YFP+ACTA2+ double-positive cells at the single-cell level and removed experimental artefacts due to cell–cell superimposition.
Figure 4.
Loss of OCT4 in EC results in increased EndoMT, neovascularization, and the number of VCAM1+ cells in plaques and decreased mitochondrial respiration in vitro. (A and C) Representative immunostaining on serially sectioned BCAs collected from Oct4+/+ or Oct4Δ/Δ mice fed WD for 18 weeks; fibrous cap area (A) or lesion area (C). (A) White arrows indicating YFP+ACTA2+ EC undergoing EndoMT. Scale bar = 20 µm. (B) Quantification of the percentage of YFP+ACTA2+ cells within the total YFP+DAPI+ cell population in the 30 µm protective fibrous cap area of the male and female lesions. Data were analysed by unpaired t-test (males) or non-parametric Mann–Whitney test (females); *P < 0.05 (n = 12 males; 12 females) vs. Oct4Δ/Δ (n = 12 males; 11 females) mice. (C) White arrows indicating capillary-like YFP+ neovessels. Scale bar = 50 µm. (D) Quantification of the number of YFP+ intraplaque capillary-like neovessels. Values = mean ± S.E.M.; data were analysed by non-parametric Mann–Whitney test; *P < 0.05 Oct4Δ/Δ (n = 9 males; 12 females) vs. Oct4+/+ (n = 9 males; 11 females) mice. (E) Representative immunostaining serially sectioned BCAs collected from Oct4+/+ or Oct4Δ/Δ male fed WD for 10 weeks. Scale bar = 10 µm. (F) Quantification of the percentage of YFP+VCAM1 + DAPI+ cells within the total YFP+ cell population at the luminal surface of the male and female vessels. Values = mean ± S.E.M.; *P < 0.05 by unpaired t-test (females) or unpaired t-test with Welch’s correction (males). Oct4+/+ (n = 8 males; 11 females) and Oct4+/+ (n = 8 males; 11 females) mice after 10 weeks of WD feeding. (G) Graphical representation of the Seahorse XF24 Cell Mito Stress Test assays measuring the oxygen consumption rates (OCR) in OCT4WT and OCT4ex1_KO HUVECs with arrows indicating treatments with specific stressors: oligomycin, carbonyl cyanite-4 (trifluoromethoxy) phenylhydrazone (FCCP), and Rotenone/Antimycin A. (H) Quantification of OCR in OCT4WT and OCT4ex1_KO HUVECs revealed a significant difference in basal mitochondrial respiration, maximum respiration, spare respiratory capacity (SRC), and ATP production. Values = mean ± S.E.M.; *P < 0.05 by two-way ANOVA, n = 3 independent experiments.
Capillary-like neovessels have been previously observed and described within advanced atherosclerotic lesions of Apoe−/− mice.48,49 We found evidence of intraplaque angiogenesis based on the frequency of the YFP+ capillary-like neovessels (Figure 4C). Our data showed that the YFP+ capillary-like neovessels frequency was significantly higher in Oct4Δ/Δ mice than in Oct4+/+ mice (Figure 4D).
Furthermore, we checked whether EC-specific loss of Oct4 caused EC activation by up-regulating cell adhesion molecules, as evidenced by our scRNAseq. VCAM1 immunofluorescence staining of BCA 10-week WD lesions revealed significantly higher numbers of YFP+VCAM1+ EC in the Oct4Δ/Δ than in wild-type control mice (Figure 4E and F).
In addition, our scRNAseq analyses revealed that several genes related to mitochondrial electron transport were dysregulated following EC-specific Oct4 knockout. Indeed, several critical mitochondrial genes (Mt-Co1, Mt-Atp8, Mt-Nd4l, and Mt-Nd2) were down-regulated, and multiple mitochondrial ribosomal genes were up-regulated (Figure 3D), indicating that the loss of Oct4 may induce mitochondrial dysfunction. To explore this possibility, we used our CRISPR-Cas9-edited, OCT4ex1_KO HUVECs (see Supplementary material online, Figure S1). We evaluated mitochondrial function by real-time OCR measurements of OCT4ex1_KO and OCT4WT cells using a Seahorse XF Extracellular Flux Analyzer. Interestingly, OCT4ex1_KO HUVECs were significantly deficient in basal, maximal, and spare respiratory capacity as compared with OCT4WT HUVECs (Figure 4G and H). Consistent with these observations, OCT4ex1_KO cells produced fewer ATP molecules. There was no difference in non-mitochondrial respiration (Figure 4G, comparing the last three time points after Rotenone/Antimycin A treatment). In summary, loss of OCT4 significantly reduced the mitochondrial respiratory capacity of EC, which is highly consistent with our scRNAseq observations.
Overall, loss of OCT4 results in significant changes in endothelial function associated with increased VCAM1 expression, increased rate of EndoMT and intraplaque angiogenesis, and profoundly impaired endothelial mitochondrial oxidation. Next, we sought to determine the mechanism by which OCT4 reactivation impacts EC function.
3.4. Bioinformatics analyses identified ABCG2 as a direct target of OCT4 in EC
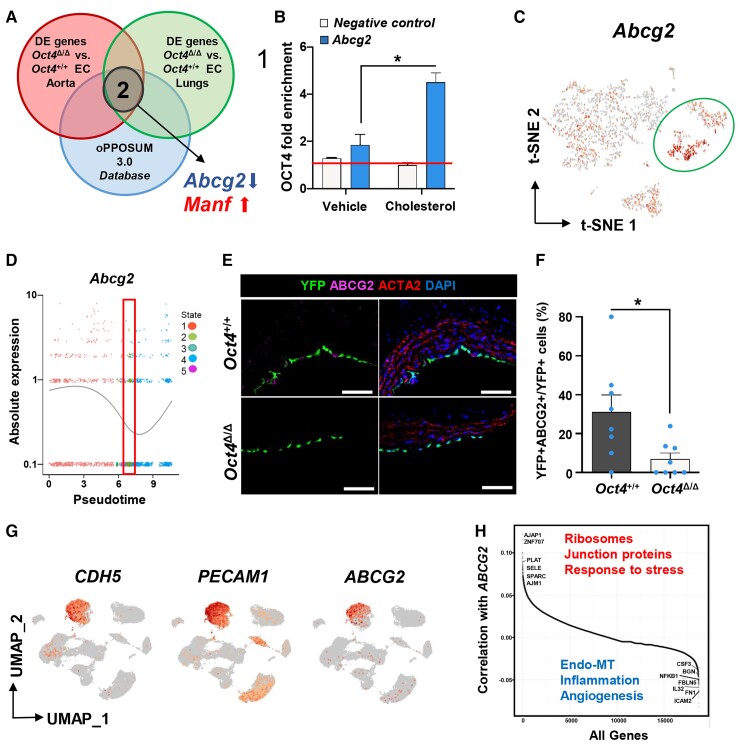
To further elucidate the molecular mechanisms responsible for OCT4-dependent atheroprotective changes in EC, we performed an additional scRNA-seq analysis on the FACS-sorted lung YFP+ EC from Oct4Δ/Δ and Oct4+/+ female mice fed WD for 5 weeks. Our rationale for using lung EC was to see if OCT4 plays a similar biological role in EC of different origins. DE analysis identified 250 significantly up- or down-regulated genes in lung knockout EC compared with wild-type cells (see Supplementary material online, Table S7). Surprisingly, we found that about a quarter of lung differentially expressed genes were similar to the differentially expressed genes from the aortic scRNAseq DE analysis (see Supplementary material online, Table S8), including decreases in genes responsible for haem and xenobiotic metabolism and increases in pro-inflammatory, pro-coagulant, and adhesion molecules, as well as increases in markers of EndoMT, and angiogenesis (see Supplementary material online, Table S8). Next, we compared these two scRNAseq data sets with a web-based system for the detection of over-represented conserved transcription factor binding sites and binding site combinations in groups of genes or publicly available ChIP-seq datasets, oPOSSUM 3.0 database50 (see Supplementary material online, Tables S9 and S10). Interestingly, our analysis revealed only two putative OCT4 target genes: Abcg2 (ABC superfamily G member 2) that was also significantly down-regulated in Oct4EC-Δ cells compared with Oct4EC-WT cells, and Manf (mesencephalic astrocyte-derived neurotrophic factor) that was up-regulated in Oct4EC-Δ cells compared with Oct4EC-WT cells (Figure 5A, Supplementary material online, Table S10).
Figure 5.
Abcg2 is a direct target of OCT4 and is associated with EC phenotypic transitions in human and mouse atherosclerotic arteries. (A) Comparing in vivo aortic and lung scRNA-seq data sets with the oPPOSUM 3.0 database revealed Abcg2 and Manf1 as putative targets of OCT4. (B) ChIP analysis on cultured mouse aortic wild-type EC with or without cholesterol loading using antibodies specific for OCT4 compared with non-immune IgG (red line). Values = mean ± S.E.M.; data were analysed by unpaired t-test, n = 2 representative experiments; *P < 0.05. NC—results for the negative control loci primer set. (C) A feature plot of Abcg2 gene distribution within the scRNA-seq t-SNE map. (D) The expression level of Abcg2 in different cellular states, which has been identified by the pseudotime analysis. The red rectangle indicates State 2, unique for Oct4Δ/Δ cells, with low Abcg2 expression levels. Dots represent individual cells at each time point. The curve line represents the average of Abcg2 expression between all cells at each time point. (E) Immunostaining of YFP, ABCG2, ACTA2, and DAPI on representative BCA sections collected from 4 weeks WD-fed Oct4Δ/Δ and Oct4+/+ male mice. Scale bar = 10 µm. (F) Quantification of the percentage of YFP+ABCG2+ cells within total YFP+ cells. Values = mean ± S.E.M.. Data were analysed by non-parametric Mann–Whitney test, *P < 0.05 Oct4+/+ (n = 7) vs. Oct4Δ/Δ (n = 8) mice. (G and H) Publicly available scRNAseq data on unsorted cells from atherosclerotic human coronary arteries (Wirka et al., 2019)33 were reanalysed to trace ABCG2 expression in EC. (G) Uniform manifold approximation and projection (UMAP) visualization of cells present in human atherosclerotic arteries, showing enrichment of ABCG2 expression specifically in EC based on EC genes CDH5 and PECAM1. (H) Pairwise Pearson correlation of ABCG2 with all other genes in cells extracted from the CDH5+PECAM1+ EC cluster. Examples of the top highly correlated and anti-correlated genes and molecular pathways are indicated on the graph.
Our rationale for focusing on ABCG2 was threefold: (i) pathway enrichment analysis revealed that both Oct4 knockout lung and aortic EC demonstrated down-regulated haem and xenobiotic pathways (see Supplementary material online, Table S8), indicating that ABCG2 plays a functional role downstream of OCT4; (ii) a role of ABCG2 in microvascular EC has been previously reported;51,52 (iii) numerous studies have identified ABCG2 as a marker of cancer stem cells and demonstrated that ABCG2 is a direct target of OCT4.53 To validate the bioinformatics prediction, we performed a ChIP assay with an OCT4 antibody on chromatin isolated from cultured mouse aortic EC with or without cholesterol loading for 24 h. Using the putative OCT4 binding site and ChIP primers generated based on the results from the oPPOSUM 3.0 analyses (see Supplementary material online, Figure S7), we confirmed that OCT4 was enriched at the Abcg2 promoter compared with IgG antibody or negative binding site controls and cholesterol loading significantly increased OCT4 enrichment at the Abcg2 promoter (Figure 5B).
Interestingly, although Abcg2 was present in all seven EC scRNAseq clusters, we found that Clusters 5 and 6, the two EC clusters enriched with genes involved in metabolic processes, were especially enriched with Abcg2 expression (Figure 5C). Moreover, tracing Abcg2 along the pseudotime trajectory demonstrated that States 2 and 3, two unique Oct4EC-Δ states showing EndoMT and activated EC signature, have the lowest Abcg2 expression levels compared with the other three states (Figure 5D). Next, we performed immunofluorescence staining for ABCG2 on atherosclerotic BCA cross-sections. We found that YFP+ABCG2+ ECs were significantly reduced in Oct4Δ/Δ mice as compared with wild-type mice (Figure 5E and F).
There are marked differences in atherosclerosis development between mouse models and human plaques.54 Nevertheless, our previous studies demonstrated that OCT4 is reactivated in human coronary artery atherosclerotic lesions.17 In parallel to our mouse studies, we examined whether ABCG2 is expressed in human EC within atherosclerotic arteries. We reanalysed the publicly available scRNAseq data on unsorted cells isolated from atherosclerotic human coronary arteries.33 Intriguingly, we found that ABCG2 expression was strictly localized in the CDH5+PECAM1+ EC cluster (Figure 5G). Furthermore, across the CDH5+PECAM1+ EC cluster, we performed a correlation analysis between ABCG2 expression and expression of all other genes. We found that ABCG2 expression was highly correlated with genes responsible for the normal function of EC (ribosome, junction-associated proteins, and response to stress) and negatively correlated with genes involved in EndoMT, inflammation, and angiogenesis (Figure 5H, Supplementary material online, Table S11).
Taken together, ABCG2 is an OCT4 target gene, present in normally functioning EC in both humans and mice, and down-regulated in cells expressing higher levels of EndoMT and pro-inflammatory genes.
3.5. OCT4/ABCG2 axis maintains intracellular haem levels to prevent oxidative stress in EC
ABCG2 is an ABC transporter responsible for extruding toxic xenobiotics, including porphyrin and haem, from cells.55 Although nothing is known about the potential role of ABCG2 in atherosclerosis, previous studies demonstrated that ABCG2 plays a protective role in brain microvascular EC by reducing haem and ROS-mediated toxicity and inflammation by inhibiting NFkB-signalling.51 In addition, it has been reported that EC-derived ABCG2 plays a pivotal role in cardiac repair after myocardial infarction by improving microvascular EC survival under oxidative stress.52,56 Intriguingly, we did not observe any other ATP-binding transporters dysregulated in the Oct4 knockout EC. We next sought to determine whether the OCT4/ABCG2 axis protects aortic EC against haem and ROS accumulation and whether its disruption leads to increases in EndoMT, angiogenesis, and mitochondria dysfunction.
Mouse aortic ECs were isolated from Oct4+/+ mice and transfected with an Oct4 small interfering (si)RNA (siOct4) or a non-target siRNA control (siNT). Cells transfected with siOct4 demonstrated a significant decrease in Oct4 and Abcg2 and an increase in Nfkb1 mRNA levels (see Supplementary material online, Figure S8A). Concomitantly, we found that siOct4 knockdown led to significantly reduced haem extrusion capacity and increased intracellular ROS accumulation compared with siNT transfection (see Supplementary material online, Figure S8B and C). To extend this to human EC, we performed similar experiments in HUVECs. We observed reduced ABCG2 protein levels in cultured OCT4ex1_KO HUVEC compared with OCT4WT HUVEC by western blotting (Figure 6A) and decreased haem extrusion capacity in OCT4ex1_KO compared with OCT4WT cells (Figure 6B). Next, to explore the role of OCT4, ABCG2, and intracellular haem and ROS levels on EC functions, we performed a series of experiments investigating different stages of their interactions in wild-type and OCT4-deficient EC.
First, to investigate whether failure in the ABCG2-dependent haem extrusion in OCT4-deficient EC is responsible for the intracellular ROS accumulation, we challenged OCT4WT and OCT4ex1_KO HUVEC with excess haem (in the form of the exogenous hemin chloride) to determine the importance of ABCG2 in protecting against overabundant haem levels as previously described.51 In parallel, OCT4WT cells were treated with succinylacetone, which inhibits endogenous haem synthesis, followed by treatment with an ABCG2-specific inhibitor, FTC,57 to prevent intracellular haem extrusion and to verify whether ABCG2 inhibition leads to increased intracellular ROS in wild-type cells (Figure 6C). Both OCT4WT cells treated with FTC and OCT4ex1_KO demonstrated significantly higher intracellular ROS than the OCT4WT untreated control group (Figure 6D). These results suggest that reduced levels of ABCG2 may be one of the reasons for the increased ROS accumulation in OCT4-deficient EC.
Second, to further explore whether ROS accumulation in OCT4-deficient EC is responsible for the reduced mitochondrial function of EC and the increased EndoMT, we used an antioxidant N-acetyl-l-cysteine (NAC)58 to test if inhibition of ROS could rescue the adverse phenotype caused by OCT4 deficiency. Of major importance, we found that the treatment of OCT4ex1_KO HUVEC with NAC improved the oxygen consumption rates to the levels found in OCT4WT cells (Figure 6E). In addition, NAC partially reduced TGFβ1-induced EndoMT in vitro. Mouse aortic ECs were transfected with siOct4 or siNT and treated with TGFβ1 with or without NAC for 5 days to induce EndoMT in vitro. At the end of the experiment, gene expression for Oct4, Pecam1, Nfkb, and EndoMT markers, including Acta2, Tagln, Cnn1, Col15a, and Col1a1, were assessed by qRT–PCR. Results showed that transfection with siOct4 markedly increased the TGFβ1-induced expression of the EndoMT markers and Nfkb compared with the siNT group, and the treatment of cells with NAC reduced the transition (Figure 6F–H).
Third, to get better insight into the connection between OCT4/ABCG2 and haem, ROS, and mitochondrial functions, we used a human ABCG2 overexpressing adenovirus (Adv-ABCG2) (Figure 7A–C) to test whether exogenous ABCG2 overexpression can rescue impaired phenotypes in OCT4-deficient EC. Results showed that Adv-ABCG2 significantly increased the secretion of haem into extracellular space (Figure 7D) and significantly decreased intracellular ROS accumulation in OCT4ex1_KO cells in response to hemin chloride treatment (Figure 7E) compared with a control Adenovirus (Adv-GFP). Furthermore, we found improved mitochondrial functions, including mitochondrial membrane potential (Figure 7F) and mitochondrial fragmentation (Figure 7G and H), providing evidence that ABCG2 is an important OCT4 target at least partially responsible for the OCT4-dependent protective properties.
In summary, our data provide compelling evidence that OCT4-dependent regulation of ABCG2 in EC plays an important role in protecting ECs from oxidative stress that in turn leads to atheroprotection by preventing mitochondrial dysfunction, inflammation, and pathological angiogenesis and EndoMT (Figure 7I).
4. Discussion
This study demonstrates an important functional role for OCT4 in EC in atherosclerosis. Loss of Oct4 in EC of Apoe−/− WD-fed mice resulted in accelerated atherosclerosis development, including increased lipid and macrophage accumulation in multiple vascular beds. Furthermore, the lesions were found to have multiple impairments in indices of plaque stability, including reduced collagen content, reduced ACTA2/LGALS3 fibrous cap ratio, and increased intraplaque haemorrhage. These findings provide the first evidence of a functional role for the pluripotency factor OCT4 in vascular EC and reinforce the critical atheroprotective role of OCT4 in atherosclerosis. Our results demonstrate that genetic inactivation of OCT4 leads to multiple alterations in EC phenotype, including increased EC activation, EndoMT, and angiogenesis within lesions.
EndoMT represents an important link between inflammatory stress and endothelial dysfunction.59 Herein, we demonstrate that the loss of OCT4 results in marked increases in EndoMT both in vivo and in vitro. However, we cannot tell whether EndoMT directly leads to the adverse atherosclerotic phenotype in EC-Oct4 knockout mice. Recent studies from several groups have used EC-lineage-tracing mouse models to understand EC transitions in atherosclerosis. The Simons group has found that inhibition of FGFR signalling in all ECs in atherosclerotic Apoe−/− mice activated TGFβ signalling resulting in a significant increase in EndoMT, increased atherosclerotic lesion size, and changes consistent with decreased plaque stability, including larger necrotic core size and reduced size of the fibrous cap.4 In a follow-up study, this group performed an EC-specific knockout of both Tgfbr1 and Tgfbr2 and found reduced atherosclerotic plaque growth and vascular wall inflammation accompanied by decreased EndoMT.6 Moreover, the Kovacic group, using an EC-lineage-tracing Apoe−/− mouse model, demonstrated that a large fraction of EC acquires the mesenchymal marker, fibroblast activation protein, and that these cells make up a sizable fraction of lesion cells.7 Human studies from both groups showed a positive correlation between CD31+ACTA2+ cell number (presumably EC undergoing EndoMT) and the severity of atherosclerosis.4,7 Both groups concluded that their findings support the detrimental role of EndoMT in the pathology of atherosclerosis. However, although these results show a link between the frequency of EndoMT and the severity of atherosclerosis, they do not answer whether these transitions are the causes or consequences of atherosclerosis. Indeed, a recent scRNAseq study on EC sorted selectively from advanced Apoe−/− BCA lesions indicated that ECs give rise to cells transcriptomically similar to ACTA2+ SMC-derived cells likely via EndoMT.44 Given that increased ACTA2+ SMC and collagen production in atherosclerotic lesions are thought to be beneficial for lesion stability, these scRNAseq results suggest that EndoMT does not necessarily play a detrimental role in the lesion. Moreover, our recent study of the origins of fibrous cap cells demonstrated that approximately 20% of ACTA2+ cells in the fibrous cap of advanced BCA lesions are derived from EC that have undergone EndoMT and that this increases to 60–70% if SMC investment is impaired.29 In addition, we presented evidence based on identifying SMC within advanced human coronary artery lesions based on detection of H3K4diMe of the MYH11 promoter, an SMC-specific epigenetic signature that persists even in phenotypically modulated SMC,11,60 that 25% of fibrous cap cells in humans come from a non-SMC source, likely predominately EC based on detection of CD31+ACTA2+ cells.29 Results suggest that EndoMT may have a beneficial role in certain contexts, which is consistent with it being a known source of perivascular cells during vascular development and angiogenesis.61 Therefore, although our data demonstrate that loss of OCT4 increases EndoMT and is associated with impaired indices of lesion stability, it remains unclear whether the natural rate of EndoMT is beneficial or detrimental. Future studies are necessary to better understand the contribution of EndoMT and its role in modulating lesion stability and progression.
Although rare in mouse models of atherosclerosis, intraplaque neovascularization has been previously reported in Apoe−/− mouse models.48,49 Furthermore, angiogenesis inhibitors, endostatin, TNP-470,62 VEGFR1 inhibitor (axitinib),63 and mTOR inhibitor (Everolimus)64 have been shown to reduce plaque growth and neovascularization in Apoe−/− mice. Here, we demonstrate that EC-specific loss of OCT4 increases intraplaque neovascularization (Figure 4C and D) and is associated with increased intraplaque haemorrhage (Figure 2G and H), suggesting that this may be one of the causes of exacerbated atherosclerosis.
Interestingly, the increased EndoMT and neovascularization following EC-specific loss of OCT4 occurred in the context of impaired processing of haem/ROS and mitochondrial function. We have shown that ABCG2, an essential ATP-driven drug transporter55 is an OCT4 target gene and is significantly down-regulated upon EC-specific conditional loss of Oct4. Several studies have reported a critical role for ABCG2 in angiogenesis and endothelial intracellular redox regulation in health and diseases, including Alzheimer’s disease,51 myocardial infarction56 and pressure overload-induced cardiac hypertrophy and heart failure.52 The redox regulation mechanism of ABCG2 in EC has been linked to the extrusion of excessive intracellular haem and protoporphyrin IX (precursor of haem).51,52 Thus, tight regulation and detoxification of free haem and iron are essential to limiting ROS formation. On the other hand, haem is a part of haemoglobin and it has been shown that the EC expression of haemoglobin α plays a critical role in the regulation of NOS-mediated vascular reactivity.65 Inhibiting ABCG2 with FTC resulted in significant ROS accumulation in our wild-type EC, which looked similar to our OCT4 knockout EC. Moreover, ABCG2 overexpression in OCT4 knockout EC at least partially rescued ROS accumulation in response to hemin chloride treatment, confirming the preceding reports regarding the detrimental effects of intracellular haem accumulation on ROS burden and inflammatory response. Interestingly, there is prevalent human single nucleotide polymorphism in the ABCG2 gene, which has been shown to reduce the transport of statins and increase the risk of statin-induced adverse drug reactions like myopathies, rhabdomyolysis, and hepatotoxicity.66 Therefore, understanding key regulators of ABCG2 is not only critical for maintaining EC integrity but may also have implications in statin intolerance in patients.
In addition to the inefficient extrusion of haem, the OCT4-deficient EC demonstrated significant dysregulation of mitochondrial functions, including a reduction in mitochondrial respiration, ATP production, and membrane potential, as well as an increase in mitochondrial fragmentation. This agrees with a previous report on mitochondrial dysfunction in bovine aortic EC treated with a physiologically high dose of hemin.67 ECs have been shown to rely primarily on glycolysis instead of mitochondrial oxidative phosphorylation for ATP production.68 Consistent with this, the mitochondrial content of EC is relatively low compared with other cell types. Nevertheless, it is well known that EC mitochondria have an important role in EC response to cellular stress, including hyperlipidaemia,69 by regulating EC calcium homeostasis, ROS production, apoptosis, lipid and fatty acid metabolism, and autophagy.70 Importantly, using the antioxidant NAC and ABCG2 overexpression, we were able to rescue the dysregulated mitochondrial functions of OCT4-deficient EC. These findings are consistent with previous reports demonstrating that α-tocopherol (vitamin E) restored mitochondrial respiration in hemin overloaded bovine aortic EC.67 Moreover, NAC at least partially reversed in vitro EndoMT, implicating ROS as a causative factor in the phenotypic transitions of EC. Hence, antioxidants may have therapeutic potential in preventing EC dysfunction like that seen following OCT4 knockout, including increased intracellular haem overabundance and mitochondrial dysfunction during atherosclerosis. It is interesting to speculate that OCT4-signalling might act as a sensor in EC to compensate during times of high cellular stress (e.g. atherosclerosis or post-MI) by regulating ABCG2-dependent haem levels allowing for proper mitochondrial functioning.
Intriguingly, although we observed an OCT4-dependent phenotype in both male and female animals, females manifested a blunted phenotype suggesting they were less susceptible to OCT4 deficiency. One possibility to account for these findings is that estrogens play an important role in mitochondrial function and biogenesis by binding to mtDNA and regulating the expression of key mitochondrial genes, including the master regulator of energy metabolism and mitochondrial biogenesis, PGC-1α.71 Interestingly, the ABCG2 promoter region is also known to have oestrogen-responsive elements.72 Future studies are required to test these possibilities to better understand sex differences in atherosclerosis.
Given our previous studies in SMC and myeloid cells, we can conclude that OCT4 plays an atheroprotective role in both SMC and EC without any adverse effects in myeloid cells, which means that OCT4-dependent mechanisms can be potentially used to develop improved therapies for treating and preventing atherosclerosis. Pivotal issues for future studies will be: (i) to understand the metabolic pathways intertwined with OCT4 reactivation upon vascular injury and atherosclerosis development; (ii) investigating the impact of OCT4 activity on the mitochondrial structure, heterogeneity, and metabolomics during atherosclerosis, and pathological angiogenesis; (iii) defining the overlap of OCT4 deficiency in vascular SMC and EC and its impact on vascular health and pathology; (iv) identifying endogenous stimuli reactivating OCT4 in response to stress; and (v) determining how these OCT4-dependent mechanisms might be targeted for novel cardiovascular diseases therapeutics. Notably, using bioinformatics analysis, we found numerous putative OCT4 target genes in aortic EC, including genes involved in EndoMT, xenobiotic and haem metabolism, shear stress, and gap junctions (see Supplementary material online, Table S9). While we believe decreased ABCG2 expression is related to our detrimental atherosclerotic phenotype, we cannot rule out the possibility that this phenotype is likely the cumulation of numerous genes altered in response to a loss of Oct4. Future studies are needed to examine the role of other EC-specific OCT4 targets in atherosclerosis.
Correspondence and requests for materials should be addressed to O.A.C.
Supplementary Material
Acknowledgements
We thank R. Tripathi, M. Bevard, C. Casden, A. Saenz, and A. Washington from the University of Virginia and I. Zhevlakova, D. Szpak, and T. Masannat from the Cleveland Clinic Lerner Research Institute for technical assistance; A. Janocha and A. Mulya from the Cleveland Clinic Lerner Research Institute for help with Seahorse machine handling and training. M. Camhi from the Cleveland Clinic Lerner Research Institute Biomedical Engineering Histochemistry Core. G. Falcao Alencar from the University of Virginia for the help with scRNAseq analyses; A. Tomilin, O.F. Sarmento, K. Asosingh, and S. Erzurum for discussion; H.R. Schöler from the Max Plank Institute for Oct4 Flox mice; R. Adams from Max Plank Institute for Cdh5-CreERT2 mice; L.M. Wakefield from the National Institute of Health for the SORE6-dscopGFP plasmid. We thank the University of Virginia Advanced Microscopy Core, Flow Cytometry Core, and Genome Technologies Core for their technical assistance.
Contributor Information
Junchul Shin, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Svyatoslav Tkachenko, Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Malay Chaklader, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Connor Pletz, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Kanwardeep Singh, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Gamze B Bulut, Robert M. Berne Cardiovascular Research Center, University of Virginia, Charlottesville, VA, USA.
Young min Han, Center for Molecular and Translational Medicine, Georgia State University, Atlanta, GA, USA.
Kelly Mitchell, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Richard A Baylis, Robert M. Berne Cardiovascular Research Center, University of Virginia, Charlottesville, VA, USA.
Andrey A Kuzmin, Russian Academy of Sciences, Institute of Cytology, St Petersburg, Russian Federation.
Bo Hu, Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Justin D Lathia, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Olga Stenina-Adognravi, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Eugene Podrez, Department of Inflammation and Immunity, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Tatiana V Byzova, Department of Neuroscience, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Gary K Owens, Robert M. Berne Cardiovascular Research Center, University of Virginia, Charlottesville, VA, USA; Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA, USA.
Olga A Cherepanova, Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
J.S. and M.C. conducted most of the in vitro experiments, immunofluorescence staining, confocal microscopy, ChIP assay, data interpretation, analysis, and contributed to manuscript writing. S.T. performed scRNAseq and bioinformatics analyses and contributed to data interpretation and manuscript writing. C.P. and K.S. assisted in mouse colony maintenance, performed image analyses, and contributed to manuscript revisions. G.B.B. performed flow cytometry experiments and data analysis. Y.H. was involved in OCT4 antibody optimization and performed western blotting and immunofluorescence staining. K.M. performed SORE6-GFP experiments. R.A.B. was involved in experimental design and optimization at the early stage of the project and contributed to discussion and manuscript revisions. A.A.K. designed CRISPR-Cas9-guide RNA for OCT4 knockout HUVEC and helped with in vitro ChIP experiment design. B.H. assisted in bioinformatics analyses. J.D.L., O.S.-A., E.P., T.B., and G.K.O. provided reagents, resources, and advice throughout the project. All authors participated in making the final manuscript revision. O.A.C. supervised the entire project and had a major role in experimental design, performed scRNAseq, data analysis, and interpretation, generated all experimental mice, and wrote the manuscript.
Funding
This work was supported by the American Heart Association Innovative Research Grant 17IRG33370017 and NIH R56HL148025 grant (to O.A.C.), NIH R01 HL071625 and R01 HL145536 (to T.V.B.), and NIH R01 HL136314 (to G.K.O.).
Data Availability
The data and biological materials that support the findings of this study are available from the corresponding author upon reasonable request. High-throughput sequencing data (FASTQ) files for scRNAseq, as well as cell-gene count matrices for scRNAseq have been deposited at Gene Expression Omnibus (GEO) with SuperSeries reference number GSE162119. These data were used to generate the images in Figures 3 and 5, and Supplementary material online, Figure S6. Uncropped western blots are available as Supplementary Source files.
References
- 1. Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Das SR, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Jordan LC, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, O’Flaherty M, Pandey A, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Spartano NL, Stokes A, Tirschwell DL, Tsao CW, Turakhia MP, VanWagner LB, Wilkins JT, Wong SS, Virani SS. Heart disease and stroke Statistics—2019 update: a report from the American Heart Association. Circulation 2019:139:e56–e528. [DOI] [PubMed] [Google Scholar]
- 2. Busse R, Fleming I. Vascular endothelium and blood flow. Handb Exp Pharmacol 2006:43–78. [DOI] [PubMed] [Google Scholar]
- 3. Gimbrone MA, Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res 2016;118:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG, Kotelianski V, Wang F, Tellides G, Simons M. FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep 2012;2:1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen PY, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, Simons M. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest 2015;125:4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen PY, Qin L, Li G, Wang Z, Dahlman JE, Malagon-Lopez J, Gujja S, Cilfone NA, Kauffman KJ, Sun L, Sun H, Zhang X, Aryal B, Canfran-Duque A, Liu R, Kusters P, Sehgal A, Jiao Y, Anderson DG, Gulcher J, Fernandez-Hernando C, Lutgens E, Schwartz MA, Pober JS, Chittenden TW, Tellides G, Simons M. Endothelial TGF-β signalling drives vascular inflammation and atherosclerosis. Nat Metab 2019;1:912–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evrard SM, Lecce L, Michelis KC, Nomura-Kitabayashi A, Pandey G, Purushothaman KR, d’Escamard V, Li JR, Hadri L, Fujitani K, Moreno PR, Benard L, Rimmele P, Cohain A, Mecham B, Randolph GJ, Nabel EG, Hajjar R, Fuster V, Boehm M, Kovacic JC. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat Commun 2016;7:11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen PY, Qin L, Tellides G, Simons M. Fibroblast growth factor receptor 1 is a key inhibitor of TGF signaling in the endothelium. Sci Signal 2014;7:ra90. [DOI] [PubMed] [Google Scholar]
- 9. Zhuang T, Liu J, Chen X, Zhang L, Pi J, Sun H, Li L, Bauer R, Wang H, Yu Z, Zhang Q, Tomlinson B, Chan P, Zheng X, Morrisey E, Liu Z, Reilly M, Zhang Y. Endothelial Foxp1 suppresses atherosclerosis via modulation of Nlrp3 inflammasome activation. Circ Res 2019;125:590–605. [DOI] [PubMed] [Google Scholar]
- 10. Zhou G, Hamik A, Nayak L, Tian H, Shi H, Lu Y, Sharma N, Liao X, Hale A, Boerboom L, Feaver RE, Gao H, Desai A, Schmaier A, Gerson SL, Wang Y, Atkins GB, Blackman BR, Simon DI, Jain MK. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest 2012;122:4727–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharma N, Lu Y, Zhou G, Liao X, Kapil P, Anand P, Mahabeleshwar GH, Stamler JS, Jain MK. Myeloid Kruppel-like factor 4 deficiency augments atherogenesis in ApoE–/– mice–brief report. Arterioscler Thromb Vasc Biol 2012;32:2836–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen PY, Qin L, Li G, Tellides G, Simons M. Smooth muscle FGF/TGFβ cross talk regulates atherosclerosis progression. EMBO Mol Med 2016;8:712–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature 1990;344:435–439. [DOI] [PubMed] [Google Scholar]
- 15. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 16. Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 2006;8:188–194. [DOI] [PubMed] [Google Scholar]
- 17. Cherepanova OA, Gomez D, Shankman LS, Swiatlowska P, Williams J, Sarmento OF, Alencar GF, Hess DL, Bevard MH, Greene ES, Murgai M, Turner SD, Geng YJ, Bekiranov S, Connelly JJ, Tomilin A, Owens GK. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat Med 2016;22:657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hess DL, Kelly-Goss MR, Cherepanova OA, Nguyen AT, Baylis RA, Tkachenko S, Annex BH, Peirce SM, Owens GK. Perivascular cell-specific knockout of the stem cell pluripotency gene Oct4 inhibits angiogenesis. Nat Commun 2019;10:967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Firth AL, Yao W, Remillard C V, Ogawa A, Yuan JXJ. Upregulation of Oct-4 isoforms in pulmonary artery smooth muscle cells from patients with pulmonary arterial hypertension. Am J Physiol Cell Mol Physiol 2010;298:L548–L557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan Y, Tan MW, Xue X, Ding XY, Wang G-K, Xu ZY. Involvement of Oct4 in the pathogenesis of thoracic aortic dissection via inducing the dedifferentiated phenotype of human aortic smooth muscle cells by directly upregulating KLF5. J Thorac Cardiovasc Surg 2016;152:820–829.e4. [DOI] [PubMed] [Google Scholar]
- 21. Han YM, Bedarida T, Ding Y, Somba BK, Lu Q, Wang Q, Song P, Zou MH. β-Hydroxybutyrate prevents vascular senescence through hnRNP A1-mediated upregulation of Oct4. Mol Cell 2018;71:1064–1078.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell 2007;1:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lengner CJ, Welstead GG, Jaenisch R. The pluripotency regulator Oct4: a role in somatic stem cells? Cell Cycle 2008;7:725–728. [DOI] [PubMed] [Google Scholar]
- 24. Atlasi Y, Mowla SJ, Ziaee SAM, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 2008;26:3068–3074. [DOI] [PubMed] [Google Scholar]
- 25. Liedtke S, Stephan M, Kögler G. Oct4 expression revisited: potential pitfalls for data misinterpretation in stem cell research. Biol Chem 2008;389:845–850. [DOI] [PubMed] [Google Scholar]
- 26. Kehler J, Tolkunova E, Koschorz B, Pesce M, Gentile L, Boiani M, Lomelí H, Nagy A, McLaughlin KJ, Schöler HR, Tomilin A. Oct4 is required for primordial germ cell survival. EMBO Rep 2004;5:1078–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Lüthi U, Barberis A, Benjamin LE, Mäkinen T, Nobes CD, Adams RH. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010;465:483–486. [DOI] [PubMed] [Google Scholar]
- 28. Durgin BG, Cherepanova OA, Gomez D, Karaoli T, Alencar GF, Butcher JT, Zhou YQ, Bendeck MP, Isakson BE, Owens GK, Connelly JJ. Smooth muscle cell-specific deletion of col15α1 unexpectedly leads to impaired development of advanced atherosclerotic lesions. Am J Physiol Hear Circ Physiol 2017;312:H943–H958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newman AAC, Serbulea V, Baylis RA, Shankman LS, Bradley X, Alencar GF, Owsiany K, Deaton RA, Karnewar S, Shamsuzzaman S, Salamon A, Reddy MS, Guo L, Finn A, Virmani R, Cherepanova OA, Owens GK. Multiple cell types contribute to the atherosclerotic lesion fibrous cap by PDGFRβ and bioenergetic mechanisms. Nat Metab 2021;3:166–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 2016;5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahabeleshwar GH, Feng W, Phillips DR, Byzova T V. Integrin signaling is critical for pathological angiogenesis. J Exp Med 2006;203:2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu W, Comhair SAA, Chen R, Hu B, Hou Y, Zhou Y, Mavrakis LA, Janocha AJ, Li L, Zhang D, Willard BB, Asosingh K, Cheng F, Erzurum SC. Integrative proteomics and phosphoproteomics in pulmonary arterial hypertension. Sci Rep 2019;9:18623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yetkin-Arik B, Vogels IMC, Neyazi N, van Duinen V, Houtkooper RH, van Noorden CJF, Klaassen I, Schlingemann RO. Endothelial tip cells in vitro are less glycolytic and have a more flexible response to metabolic stress than non-tip cells. Sci Rep 2019;9:10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Hon T, Ye W, Zhang L. Heme deficiency interferes with the Ras-mitogen-activated protein kinase signaling pathway and expression of a subset of neuronal genes. Cell Growth Differ 2002;13:431–439. [PubMed] [Google Scholar]
- 38. Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 2006;1:179–185. [DOI] [PubMed] [Google Scholar]
- 39. Hong Z, Kutty S, Toth PT, Marsboom G, Hammel JM, Chamberlain C, Ryan JJ, Zhang HJ, Sharp WW, Morrow E, Trivedi K, Weir EK, Archer SL. Role of dynamin-related protein 1 (Drp1)-mediated mitochondrial fission in oxygen sensing and constriction of the ductus arteriosus. Circ Res 2013;112:802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Y, Chen X, Kang B, She S, Zhang X, Chen C, Li W, Chen W, Dan S, Pan X, Liu X, He J, Zhao Q, Zhu C, Peng L, Wang H, Yao H, Cao H, Li L, Herlyn M, Wang YJ. Endogenous authentic OCT4A proteins directly regulate FOS/AP-1 transcription in somatic cancer cells. Cell Death Dis 2018;9:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang B, Raviv A, Esposito D, Flanders KC, Daniel C, Nghiem BT, Garfield S, Lim L, Mannan P, Robles AI, Smith WI, Zimmerberg J, Ravin R, Wakefield LM. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Rep 2015;4:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle–specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ Res 2001;88:1127–1134. [DOI] [PubMed] [Google Scholar]
- 43. Finn A V, Nakano M, Narula J, Kolodgie FD, Virmani R. Concept of vulnerable/unstable plaque. Arterioscler Thromb Vasc Biol 2010;30:1282–1292. [DOI] [PubMed] [Google Scholar]
- 44. Alencar GF, Owsiany KM, Sukhavasi K, Sukhavasi K, Mocci G, Nguyen AT, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA, Haskins R, Baylis RA, Finn A V, McNamara CA, Zunder ER, Venkata V, Pasterkamp G, Björkegren J, Bekiranov S, Owens GK. The stem cell pluripotency genes Klf4 and Oct4 regulate complex SMC phenotypic changes critical in late-stage atherosclerotic lesion pathogenesis. Circulation 2020;142:2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao Z, Liu Z, Wang R, Zheng Y, Li H, Yang L. Galectin-3 is a potential mediator for atherosclerosis. J Immunol Res 2020;2020: 5284728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death. Arterioscler Thromb Vasc Biol 2000;20:1262–1275. [DOI] [PubMed] [Google Scholar]
- 47. He Q, Li X, Singh K, Luo Z, Meija-Cordova M, Jamalpour M, Lindahl B, Kriz V, Vuolteenaho R, Ulvmar M, Welsh M. The Cdh5-CreERT2 transgene causes conditional Shb gene deletion in hematopoietic cells with consequences for immune cell responses to tumors. Sci Rep 2019;9:7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Perrotta P, Van der Veken B, Van Der Veken P, Pintelon I, Roosens L, Adriaenssens E, Timmerman V, Guns PJ, De Meyer GRY, Martinet W. Partial inhibition of glycolysis reduces atherogenesis independent of intraplaque neovascularization in mice. Arterioscler Thromb Vasc Biol 2020;40:1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perrotta P, Pintelon I, de Vries MR, Quax PHA, Timmermans JP, De Meyer GRY, Martinet W. Three-dimensional imaging of intraplaque neovascularization in a mouse model of advanced atherosclerosis. J Vasc Res 2020;57:348–354. [DOI] [PubMed] [Google Scholar]
- 50. Kwon AT, Arenillas DJ, Hunt WR, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2012;2:987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen S, Callaghan D, Juzwik C, Xiong H, Huang P, Zhang W. ABCG2 reduces ROS-mediated toxicity and inflammation: a potential role in Alzheimer’s disease. J Neurochem 2010;114:1590–1604. [DOI] [PubMed] [Google Scholar]
- 52. Higashikuni Y, Sainz J, Nakamura K, Takaoka M, Enomoto S, Iwata H, Tanaka K, Sahara M, Hirata Y, Nagai R, Sata M. The ATP-binding cassette transporter ABCG2 protects against pressure overload–induced cardiac hypertrophy and heart failure by promoting angiogenesis and antioxidant response. Arterioscler Thromb Vasc Biol 2012;32:654–661. [DOI] [PubMed] [Google Scholar]
- 53. Fatima S, Zhou S, Sorrentino BP. Abcg2 expression marks tissue-specific stem cells in multiple organs in a mouse progeny tracking model. Stem Cells 2012;30:210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vedder VL, Aherrahrou Z, Erdmann J. Dare to compare. Development of atherosclerotic lesions in human, mouse, and Zebrafish. Front Cardiovasc Med 2020;7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krishnamurthy P, Schuetz JD. Role of Abcg2/Bcrp in biology and medicine. Annu Rev Pharmacol Toxicol 2006;46:381–410. [DOI] [PubMed] [Google Scholar]
- 56. Higashikuni Y, Sainz J, Nakamura K, Takaoka M, Enomoto S, Iwata H, Sahara M, Tanaka K, Koibuchi N, Ito S, Kusuhara H, Sugiyama Y, Hirata Y, Nagai R, Sata M. The ATP-binding cassette transporter BCRP1/ABCG2 plays a pivotal role in cardiac repair after myocardial infarction via modulation of microvascular endothelial cell survival and function. Arterioscler Thromb Vasc Biol 2010;30:2128–2135. [DOI] [PubMed] [Google Scholar]
- 57. Bakhsheshian J, Hall MD, Robey RW, Herrmann MA, Chen JQ, Bates SE, Gottesman MM. Overlapping substrate and inhibitor specificity of human and murine ABCG2. Drug Metab Dispos 2013;41:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vandekeere S, Dubois C, Kalucka J, Sullivan MR, García-Caballero M, Goveia J, Chen R, Diehl FF, Bar-Lev L, Souffreau J, Pircher A, Kumar S, Vinckier S, Hirabayashi Y, Furuya S, Schoonjans L, Eelen G, Ghesquière B, Keshet E, Li X, Vander Heiden MG, Dewerchin M, Carmeliet P. Serine synthesis via PHGDH is essential for heme production in endothelial cells. Cell Metab 2018;28:573–587.e13. [DOI] [PubMed] [Google Scholar]
- 59. Cho JG, Lee A, Chang W, Lee MS, Kim J. Endothelial to mesenchymal transition represents a key link in the interaction between inflammation and endothelial dysfunction. Front Immunol 2018;9:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods 2013;10:171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commun 2016;7:12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moulton KS, Heller E, Konerding MA, Flynn E, Palinski W, Folkman J. Angiogenesis inhibitors endostatin or TNP-470 reduce intimal neovascularization and plaque growth in apolipoprotein E–Deficient mice. Circulation 1999;99:1726–1732. [DOI] [PubMed] [Google Scholar]
- 63. Van der Veken B, De Meyer GRY, Martinet W. Axitinib attenuates intraplaque angiogenesis, haemorrhages and plaque destabilization in mice. Vascul Pharmacol 2018;100:34–40. [DOI] [PubMed] [Google Scholar]
- 64. Kurdi A, Roth L, Van der Veken B, Van Dam D, De Deyn PP, De Doncker M, Neels H, De Meyer GRY, Martinet W. Everolimus depletes plaque macrophages, abolishes intraplaque neovascularization and improves survival in mice with advanced atherosclerosis. Vascul Pharmacol 2019;113:70–76. [DOI] [PubMed] [Google Scholar]
- 65. Straub AC, Lohman AW, Billaud M, Johnstone SR, Dwyer ST, Lee MY, Bortz PS, Best AK, Columbus L, Gaston B, Isakson BE. Endothelial cell expression of haemoglobin α regulates nitric oxide signalling. Nature 2012;491:473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kellick KA, Bottorff M, Toth PP. A clinician’s guide to statin drug-drug interactions. J Clin Lipidol 2014;8:S30–S46. [DOI] [PubMed] [Google Scholar]
- 67. Higdon AN, Benavides GA, Chacko BK, Ouyang X, Johnson MS, Landar A, Zhang J, Darley-Usmar VM. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Circ Physiol 2012;302:H1394–H1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kluge MA, Fetterman JL, Vita JA. Mitochondria and endothelial function. Circ Res 2013;112:1171–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu E, Calvert PA, Mercer JR, Harrison J, Baker L, Figg NL, Kumar S, Wang JC, Hurst LA, Obaid DR, Logan A, West NEJ, Clarke MCH, Vidal-Puig A, Murphy MP, Bennett MR. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation 2013;128:702–712. [DOI] [PubMed] [Google Scholar]
- 70. Groschner LN, Waldeck-Weiermair M, Malli R, Graier WF. Endothelial mitochondria—less respiration, more integration. Pflügers Arch Eur J Physiol 2012;464:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ventura-Clapier R, Moulin M, Piquereau J, Lemaire C, Mericskay M, Veksler V, Garnier A. Mitochondria: a central target for sex differences in pathologies. Clin Sci 2017;131:803–822. [DOI] [PubMed] [Google Scholar]
- 72. Ee PLR, Kamalakaran S, Tonetti D, He X, Ross DD, Beck WT. Identification of a novel estrogen response element in the breast cancer resistance protein (ABCG2) gene. Cancer Res 2004;64:1247–1251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and biological materials that support the findings of this study are available from the corresponding author upon reasonable request. High-throughput sequencing data (FASTQ) files for scRNAseq, as well as cell-gene count matrices for scRNAseq have been deposited at Gene Expression Omnibus (GEO) with SuperSeries reference number GSE162119. These data were used to generate the images in Figures 3 and 5, and Supplementary material online, Figure S6. Uncropped western blots are available as Supplementary Source files.