SUMMARY
Reciprocal interactions between host T helper cells and gut microbiota enforce local immunological tolerance and modulate extra-intestinal immunity. However, our understanding of antigen-specific tolerance to the microbiome is limited. Here, we developed a systematic approach to predict HLA class-II-specific epitopes using the humanized bacteria-originated T cell antigen (hBOTA) algorithm. We identified a diverse set of microbiome epitopes spanning all major taxa that are compatible with presentation by multiple HLA-II alleles. In particular, we uncovered an immunodominant epitope from the TonB-dependent receptor SusC that was universally recognized and ubiquitous among Bacteroidales. In healthy human subjects, SusC-reactive T cell responses were characterized by IL-10-dominant cytokine profiles, whereas in patients with active Crohn’s disease, responses were associated with elevated IL-17A. Our results highlight the potential of targeted antigen discovery within the microbiome to reveal principles of tolerance and functional transitions during inflammation.
In brief
The functional basis of tolerance to the gut microbiome is incompletely understood. Pedersen et al. develop an antigen-discovery approach to assess microbiota-directed T cell immunity in health and inflammation. They reveal the dynamic nature of T cell-mediated responses toward commensal Bacteroidales, highlighting inflammation-associated changes in Crohn’s disease.
Graphical Abstract

INTRODUCTION
The adaptive immune system actively maintains tolerance to commensals inhabiting the intestines (Honda and Littman, 2016; Zhao and Elson, 2018), with a continuous interaction between T cells and microbiome antigens (Honda and Littman, 2016; Belkaid and Harrison, 2017; Knoop et al., 2017; Zhao and Elson, 2018; Zegarra-Ruiz et al., 2021). Seminal efforts indicate wide diversity in the TCR repertoire of microbiome-specific T cells in humans (Hegazy and West, 2017), indicative of widespread recognition of diverse microbiome antigens. In this context, T cell tolerance to the microbiome is linked to the state of the local tissue microenvironment and exhibits considerable plasticity between T cell phenotypes (Hand et al., 2012; Brucklacher-Waldert et al., 2014; Chai et al., 2017). However, it remains to be determined how T cell fate is temporally regulated by the microbiome and how microbiome-specific T cells participate in local and peripheral protective immunities. Addressing these questions will require direct knowledge of T cell antigen specificity to track T cells in health and disease and connect TCR-specificity to diverse functional responses.
A limited number of MHCII-restricted microbiome antigens have been identified to date. In mouse models, T cell antigens are identified in segmented filamentous bacteria (Yang et al., 2014), Helicobacter hepaticus (Kullberg et al., 2003; Chai et al., 2017; Xu et al., 2018), Bacteroides thetaiotaomicron (Wegorzewska et al., 2019), and Akkermansia muciniphila (Ansaldo et al., 2019). Additionally, a T cell epitope derived from Cbir1 flagellin was initially discovered in mouse models of colitis and has been validated as a relevant antigen in Crohn’s disease (CD) (Lodes et al., 2004; Cong et al., 2009). Similarly, an Escherichia coli OmpC antigen recognized by peripheral T cells has been reported in humans (Uchida et al., 2020). These examples provide a proof of concept that microbiome-specific T cell responses occur in humans and highlight the need to thoroughly define the functional basis of adaptive immunity to the microbiome.
Identifying microbiome-derived antigens has been a major challenge. With the increased quality and availability of metagenomic datasets, there is an emerging opportunity to use systematic in silico strategies to predict and prioritize microbiome-encoded epitopes for experimental validation. Significant strides have been taken to improve our understanding of MHCII presentation and our ability to predict MHCII-restricted epitopes (Graham et al., 2018; Abelin et al., 2019); however, in silico epitope discovery efforts have been primarily focused on MHCI-restricted epitopes with relevance to immune oncology and infectious diseases (Bonomo and Deem, 2018; Doytchinova and Flower, 2018). As such, the commensal MHCII-restricted antigen landscape remains largely unexplored.
Here, we present a systematic effort for epitope identification within the human microbiome. We developed a humanized version of the bacteria-originated T cell antigen (BOTA) algorithm (Graham et al., 2018) to perform predictions of MHCII-restricted epitopes from human metagenomic data. In vitro validation of predicted epitopes using peripheral blood mononuclear cells (PBMCs) from healthy individuals revealed dozens of previously uncharacterized immunoreactive T cell epitopes from numerous unique commensal bacteria. In particular, we identified a Bacteroidetes SusC antigen and characterized functional T cell responses to this epitope in mice, healthy human subjects, and patients with inflammatory bowel disease (IBD) to define functional heterogeneity of T cell responses to the microbiome. Our results highlight the value of targeted antigen discovery within the microbiome and provide a suite of microbiome epitopes to facilitate ex vivo immunoprofiling of the microbiome-reactive T cell pool.
RESULTS
Development of hBOTA to discover microbiome epitopes recognized by human T cells
To address the challenge of antigen discovery within the human microbiome, we leveraged our previous work using the BOTA algorithm (Graham et al., 2018). Extending upon this approach, we developed a humanized BOTA (hBOTA) pipeline that (1) prioritized commensal peptides by extracting regions likely to have topological accessibility in their native state in phagosomes and (2) identified the corresponding commensal immunopeptidome by performing MHCII binding predictions using a predefined set of HLA class II alleles (Figure 1A). hBOTA accepts assembled metagenomic data, reference genomes, or microbiome gene catalog, and similarly to the original BOTA algorithm, it annotates the proteins using PSORT (Yu et al., 2010), HMMTOP (Tusnády and Simon, 2001), and Pfam domains (El-Gebali et al., 2019). The result is a list of protein regions that are predicted to show higher propensity for processing during antigen presentation. The corresponding immunopeptidome is then defined by parsing predicted candidate peptide sequences in a 15-mer sliding window through NetMHCIIpan (Jensen et al., 2018) using a predetermined set of HLA class II alleles. The hBOTA pipeline therefore enables the identification of commensal antigen candidates and facilitates predictions that account for interindividual variation in both microbiome and HLA locus composition.
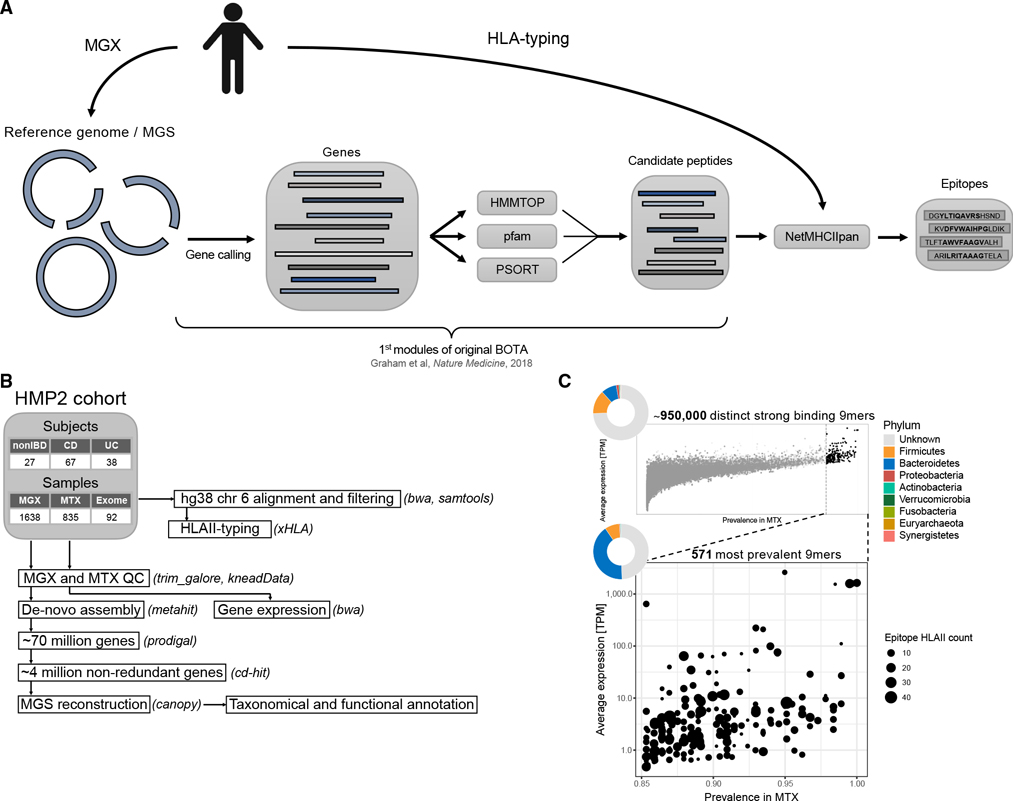
Figure 1. hBOTA predicts candidate commensal epitopes from the human microbiome.
(A) An overview of the hBOTA pipeline. Starting from an annotated human metagenome or reference genome, candidate peptides are extracted from predicted genes by integrating various features defined by HMMTOP, Pfam domain search using HMMER and PSORTb. Candidate peptides are then parsed through NetMHCIIpan to identify candidate epitopes.
(B) Processing pipeline for the HMP2 metagenomics (MGX), metatranscriptomics (MTX), and exome data. This pipeline includes quality control (QC), de novo assembly, gene prediction, metagenomic species (MGS) reconstruction, annotation, read mapping, and HLA typing. Software tools used at each step are indicated in parentheses.
(C) Prevalence and average expression in transcript per million (TPM) of encoding genes for unique 9-mer epitopes in the dataset. Dot color indicates epitope mean binding to HLA class II alleles; dot size indicates number of compatible HLA class II alleles (%rank < 2%). Pie charts indicate the relative fraction of each phylum that contributes epitope encoding genes plotted in each figure. Top: all unique 9-mer epitopes. Bottom: unique 9-mer epitopes expressed in greater than 85% of metatranscriptomic samples. See also Figure S1.
BOTA was originally designed based on MHCII peptidomics capturing Listeria epitopes (Graham et al., 2018). To provide additional evidence to support the utility of topological prioritization, a set of 2,842 validated bacterial MHCII epitopes in 629 proteins was curated from the Immune Epitope Database and Analysis Resource (IEDB). Using this dataset, we compared recovery of validated epitopes from full length protein between hBOTA and NetMHCIIpan against HLA class II alleles from a representative US Caucasian population. The immediate benefit of applying hBOTA was an 8-fold reduction of the search space due to topological prioritization (from 25,599 to 3,167 predicted amino acids) (Figure S1A). This in turn resulted in a 1.6-fold increase in the rate of true positives among the predicted strong binders in hBOTA compared with NetMHCIIpan (13.5% versus 8.2%). This improvement remained at a more stringent %rank of 1% or less, where we similarly observed an increase in the rate of true positives among the predicted strong binders in hBOTA compared with NetMHCIIpan (1.3-fold, 11.3% versus 8.9%) (Figure S1A). Thus, hBOTA improved the recovery of true bacterial epitopes compared with NetMHCIIpan alone.
We sought to apply hBOTA in defining candidate commensal epitopes within population-level microbiomes. Using publicly available data from The Integrative Human Microbiome Project (HMP2) cohort (Lloyd-Price et al., 2019) (Figure 1B), we created a reference-independent microbiome representation by performing de novo metagenomic assembly of 1,638 metagenomic samples (132 subjects) that resulted in a non-redundant gene catalog containing 3,914,507 genes. hBOTA prioritized protein regions in just 1.6% of these genes, which represented a significant contraction of the search space. We then used a set of 67 HLA class II alleles imputed from the HMP2 exome data (92 subjects, Figure S1B) to predict strong binding 15-mer epitopes (% rank < 2%) using NetMHCIIpan, which collectively represented 948,241 unique 9-mer binding cores (referred to epitope cores below). In searching the output for previously published microbiome epitopes, we found two of 9-mer cores (YIGSGAILS and GSGAILSG) that spanned a previously published H2-I-Ab restricted A. muciniphila epitope (TLYIGSGAILS) (Ansaldo et al., 2019). Our application of hBOTA on population-level microbiomes thus significantly deconvolved the search space to a smaller subset of MHCII-restricted candidate epitopes.
Considering the enormous size and heterogeneity of the commensal proteomic landscape (Human Microbiome Project Consortium, 2012), we hypothesized that gut microbiome gene expression and prevalence, in addition to epitope accessibility and MHCII binding, is an essential feature governing in vivo exposure of commensal epitopes to host immune cells. To identify highly expressed epitopes, considered at the level of epitope cores defined above, expression levels for the epitope encoding genes across 741 metatranscriptomic samples from the HMP2 cohort were summarized to infer expression level and prevalence for each epitope core. We considered that the same epitope core could be encoded by homologous or even unrelated genes in the same or different species and represented their expression signal in each sample as a summed signal of all encoding genes. Highly abundant and prevalent epitopes were identified by selecting epitopes cores with at least 85% expression prevalence in at least one of the nonIBD, ulcerative colitis (UC), and CD sample groups in HMP2, thereby further reducing the epitope landscape from 948,241 to 571 epitope cores (Figure 1C). Within this reduced set of highly prevalent epitopes cores, the majority of the corresponding 15-mer epitopes displayed binding specificity to multiple HLA class II alleles (Figure 1C). Based on the HLA class II allele distribution in the HMP2 population and general European ancestry population (Figure S1B), this translates to a recognition potential across a large fraction of a population. In total, 2,871 low prevalent epitope genes encoded the 571 highly prevalent 9-mer cores (Figure S1C). Although almost half of the genes encoding highly prevalent epitope cores (49.5%) did not map to known species, 41% derived from Bacteroidetes and 8.5% were from Firmicutes, whereas the remaining epitopes were encoded by genes from Proteobacteria, Verrucomicrobia, and Actinobacteria (0.5%, 0.3%, and 0.2%, respectively) (Figure 1C). Considering epitopes mapping to at least one known species, 42.4% of epitopes were shared between more than one genus within the same phylum, whereas 4.4% were shared across multiple phyla (Figures S1D and S1E). In summary, with hBOTA, we prioritized a small set of predicted strong binding, universally recognized epitopes from the gut microbiome for experimental validation.
T cell responses to microbiome epitopes were widespread in healthy subjects and elicited diverse cytokine profiles
To validate immune recognition of the epitopes predicted with hBOTA, we curated a panel of 48 15-mer peptides that encode highly prevalent epitopes cores (Figure 2A), referred to as microbiome associated peptides (MAPs). We selected the top 22 peptides with the highest predicted frequency of recognition by HMP2 HLA class II alleles to ensure that the selected peptides would span multiple HLA class II allele specificities. The remaining tested peptides were selected manually to facilitate testing of epitopes from (1) disease relevant taxa, (2) a diverse set of bacteria reflecting the most abundant phyla in the human gut, and (3) taxa likely to be actively recognized by the host IgA and IgG in humans (Palm et al., 2014; Armstrong et al., 2019). In doing so, we also included peptides with lower predicted frequency of recognition within HMP2 with a taxonomically diverse origin or from a disease relevant strain. We note that some peptides failed synthesis, e.g., MAP16-MAP18, and were therefore replaced. The resulting panel consisted of MAPs with high abundance within the HMP2 population, binding specificities compatible with multiple different HLA class II alleles, and collectively included human microbiome epitopes predicted from every major phylum, covering multiple genera and species (Figures 2A and 2B; Table S1).
Figure 2. T cell responses to microbiome epitopes are widespread in healthy subjects and elicit diverse cytokine profiles.
(A) Number of total HMP2 HLA class II alleles with binding predicted by NetMHCIIpan (%rank < 2%) and the percentage of recognition in the HMP2 population for the 48 15-mer MAPs included in the screen. Percentage of population recognition was defined as the percentage of HMP2 subjects with at least one HLA class II allele with predicted binding.
(B) Peptide sequences for MAPs along with the taxonomic distribution of encoding genes. Tile color indicates the phyla of encoding genes, whereas the total number of genera and species of known identity are indicated by segment plots. See also Table S1.
(C) Left: barplot summarizing immune reactivity of MAPs and positive controls (CEFT pool, INFLA). Immune reactivity is reported as observed responses across 40 subjects, defined by at least one significant cytokine response (Z score ≥ 2) relative to negative controls (H2-DMA, OVA, and Scrambled). Right: ternary plots depicting cytokine response profile and Upset intersection plots of cytokine responses for the top 10 responding MAPs and CEFT pool positive control. Only subjects that responded with at least one significant cytokine response were included. For the ternary plots, each dot represents the response profile for one subject. Axes indicate percent of the total response for each cytokine after normalizing to negative controls for each cytokine individually. Coloring indicates the level of point density. See also Figure S2 and Data S1.
Given that microbiota-specific memory Th cells circulate in the periphery at steady state (Hegazy and West, 2017), we isolated PBMCs from 40 healthy human subjects and stimulated them in vitro with synthesized MAPs to validate immune reactivity. The intestinal Th cell pool is largely dominated by T helper 1 (Th1), Th17, and regulatory T (Treg) cell phenotypes (Maynard and Weaver, 2009), and we therefore chose IFNγ, IL-17A, and IL-10 responses classically associated with these three T cell phenotypes, respectively, as a measure of immune reactivity. All MAPs induced a significant cytokine response (Z score ≥ 2 compared with mean of negative controls) in at least one of the subjects tested, whereas 34/48 MAPs (70.8%) induced significant immune responses in more than five subjects (Figure 2C), indicative of antigen-specific immune reactivity to most hBOTA predicted MAPs. Although ranking responsiveness in this manner facilitates prioritization for deeper characterization, we note that MAPs eliciting weak cytokine responses in a few individuals are better characterized as inconclusive epitopes that would require testing in a larger cohort to conclusively validate.
Cytokine responses showed inter-subject heterogeneity based on cytokines observed and corresponding magnitude of response (Figure S2A), and cytokine responses to MAPs tittered to peptide concentration (Figure S2C). Among MAPs that elicited responses in 10 or more individuals (10/48 MAPs, 20.8%), significant responses were generally characterized by IFNγ and IL-17A, accompanied by varying levels of IL-10, and both singular cytokine responses and mixed cytokine responses were observed (Figure 2C). Few subject-specific responses were associated with all three cytokines but instead manifested as a singular or dual cytokine response (Figure 2C). Most of the MAPs were derived from multiple individual species, spanning genera and, in some cases, phyla (Figure 2B). These results therefore indicate host-dependent variations in microbiome-specific immune responses that are possibly linked to the dynamic and highly context-dependent tuning of T cell activation and differentiation resulting from host-microbiome interactions.
Identification of an immunodominant epitope in SusC-associated TonB-dependent receptor plug domain in Bacteroidales
Treg cells are central to intestinal homeostasis (Russler-Germain et al., 2017), and Th17 cells support tissue integrity and IgA responses during homeostasis (Hirota et al., 2013; Honda and Littman, 2016; Zhao and Elson, 2018). We therefore hypothesized that the continuous interaction between commensal antigens and host intestinal T cells would likely appear as predominantly IL-10 and/or IL-17A responses upon MHCII-restricted antigen recall. We adopted a dominance score to emphasize which MAPs consistently induced strong responses for each cytokine tested. The dominance score was created by counting the number of strongly (Z ≥ 6), moderately (4 ≤ Z <6), and weakly (2 ≥ Z < 4) significant responses for each cytokine with weights of 1.8, 1.6, and 1, respectively. Among 5 MAPs with the highest combined IL-10 and IL-17A dominance scores, 4 were predicted from TonB-dependent receptors (TBDRs) (Figure S2B). Of note, two of the five, MAP22 (AAIYGARAAFGVVLV) and MAP36 (AASAAVYGARAANGV), shared 66.7% amino acid sequence identity and originated from the same conserved domain within the TBDR plug as the murine H2-I-Ab restricted SusC-like TonB epitope (VLKDASAAAIYGSR) (Graham et al., 2018).
TBDRs are present in the outer membrane of gram-negative bacteria and mediate transport of a wide variety of substrates, including iron, nickel, vitamin B12, and carbohydrates. They share a common domain architecture composed of a 22-stranded transmembrane β-barrel and a highly solvated and flexible plug domain enclosed within the β-barrel (Schauer et al., 2008; Noinaj et al., 2010). TBDRs are present in multiple copies (>30 per genome) in most gram-negative bacteria, including species from the dominant gut microbiome phyla Bacteroidetes and α- or γ-Proteobacteria (Blanvillain et al., 2007; Schauer et al., 2008). 27.8% (159/571) of highly abundant 9-mer epitopes predicted by hBOTA were found in genes annotated as TBDRs (Figure S1C).
To investigate antigenicity of TBDRs encoding epitopes with similarity to experimentally validated epitopes, we performed an InterProScan Pfam domain search for TBDR homologs from Bacteroidetes annotated with plug (PF07715) and TBDR (PF00593) annotations, filtering for genes having at least one 9-mer epitope predicted by NetMHCIIpan 3.2 (Jensen et al., 2018) for HLA class II alleles found in HMP2. This resulted in 29,702 unique TBDR homologs. Next, we searched among these for sequence homology to MAP22 and MAP36 and detected 16,208 such TBDRs (55%). Half of these TBDR homologs were isolated from Bacteroidetes found in the human gut microbiome (feces or intestinal source), spanning 10 genera and with multiple TBDR homologs per species (54.3 ± 39.7) (Figures 3A and 3B). TBDR homologs matching our experimentally validated TBDR epitopes were more frequent among Bacteroidetes isolated from human feces or intestine, compared with Bacteroidetes from the human oral cavity or from environmental sources (Figure 3C). Together, this indicates that species within the Bacteroidetes phylum carry multiple TBDRs and that the antigenic epitope signature is enriched in the intestinal microbiome.
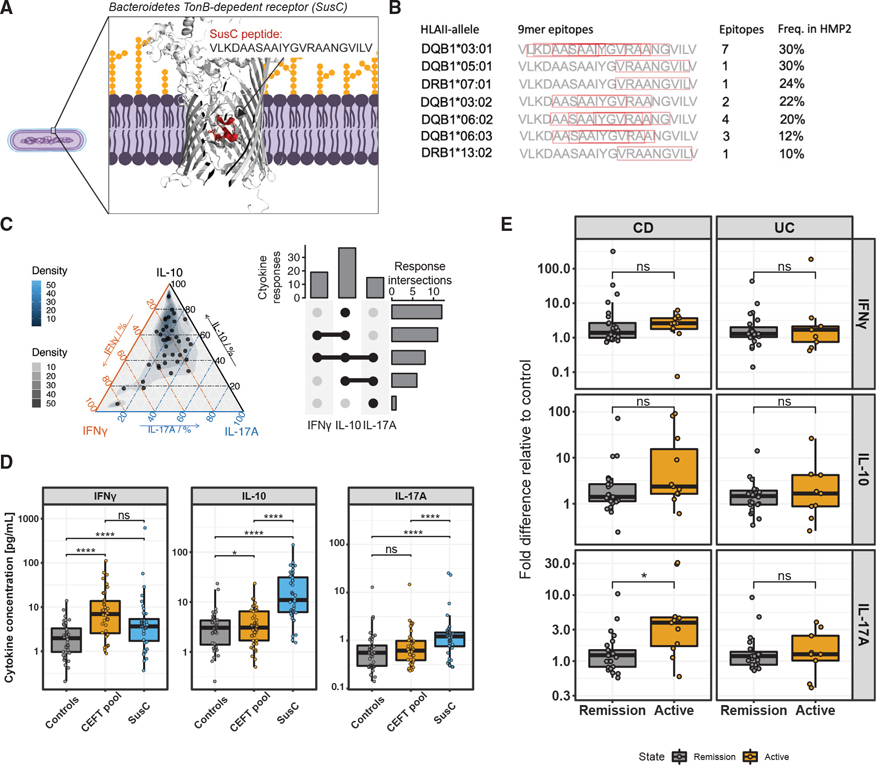
Figure 3. Bacteroidetes TonB-dependent receptors (TBDRs) contain a conserved epitope hotspot within the human microbiome.
(A) Number of TBDR homologs in Bacteroidetes species isolated from human feces or intestinal samples grouped at genus level. TBDR homologs were defined as genes containing both a plug (PF07715) and a TBDR (PF00593) Pfam annotation.
(B) Cumulative numbers of TBDR homologs in genomes of Bacteroidetes isolated from human feces or intestinal samples.
(C) Percentage of TBDR homologs with a plug domain matching experimentally validated TBDR epitopes (MAP22 and MAP36) in Bacteroidetes isolated from human oral, feces, or intestinal samples, or from environmental origin.
(D) Predicted HLA class II binding for Bacteroidetes TBDR genes isolated from human feces or intestinal samples. The cumulative number of binding HMP2 HLA class II alleles predicted using NetMHCIIpan are reported for each amino acid (aa) position and summarized (mean ± SEM). Vertical dotted lines indicate the 22 amino acid position of MAP22 and MAP36 22-mers. TBDR genes were aligned according to the aa position of sequence homology to MAP22 and MAP36.
(E) Amino acid sequences of synthesized TonB-dependent receptor peptides (TBDRPs).
(F) Cytokine concentrations and significance levels (paired t test) for stimulations of PBMCs from 3 healthy individuals with TBDRPs. p values ≤ 0.05 were regarded as significant. ns, not significant (p > 0.05), * p ≤ 0.05, ** p ≤ 0.01. Bars indicate mean ± SEM.
Based on the prevalence of TBDR epitopes in our predictions and the apparent antigenicity of plug domain epitopes, we hypothesized the TBDR plug domain to be an epitope hotspot. To investigate this, we aligned Bacteroidetes plug domains according to BLAST hit locations and summarized the average predicted positional antigenicity along the plug domains using a 9-mer search space for binding of HMP2 HLA class II alleles. The average number of HLA class II alleles predicted to bind 9-mer cores was considerably higher at the regions matching to MAP22 and MAP36, suggesting this region to be especially antigenic with multiple nested epitopes (Figure 3D). To validate predicted antigenicity, we synthesized four 22-mer TBDR peptides (TBDRP1–4, Figure 3E) covering multiple nested epitopes of the plug region and stimulated PBMCs from 3 healthy donors. Of these, one 22-mer peptide (TBDRP3) gave a potent IL-10 response in all 3 subjects (Figure 3F). Thus, we identified a conserved region within the plug domain of TBDRs that is highly antigenic and recognized by host T cells.
T cell responses to the immunodominant SusC epitope were associated with IL-10 production in healthy subjects but dynamically shifted toward IL-17 production in active Crohn’s disease
TBDRP3 was identified with 95.45% identity to multiple different TBDR-like SusC proteins originating from Bacteroidetes, when searching against non-redundant protein sequences from bacteria on NCBI/BLASTP suite (Figure 4A). We henceforth refer to TBDRP3 as “SusC peptide.” In Bacteroidetes, the encoding SusC protein is part of a multi-protein system referred to as the starch-utilization system (Sus) that is encoded by polysaccharide utilization loci (PUL) and is responsible for the sensing, binding, depolymerization, and transport of glycans (Martens et al., 2011; Bolam and Koropatkin, 2012). Like the conserved TBDR plug domain, the SusC peptide contained multiple nested strong binding registers to some of the most frequent HLA class II alleles in HMP2 (Figure 4B). To investigate population-wide immunodominance of the SusC peptide, we stimulated PBMCs from 40 healthy subjects and quantitatively assessed immune reactivity by cytokine bead arrays. We found that the SusC peptide induced a robust T cell response in most individuals and was largely dominated by IL-10 (Figures 4C and 4D). The peptide induced IL-10 responses in 37/40 subjects, with a marked elevated IL-10 response across subjects compared with negative controls (p < 0.0001), although also inducing IFNγ and IL-17A responses in 19/40 and 16/40 subjects, respectively (p < 0.0001 and p < 0.0001, respectively) (Figures 4C and 4D). Subject-specific recall responses skewed toward an anti-inflammatory profile composed mainly of IL-10 or a pro-inflammatory profile with predominantly IFNγ (Figure 4C). We expect subject-specific variations in the functional recall response to be directly linked to the functional output of intestinal T cells at the time of blood draw and therefore reflect the underlying intestinal health state.
Figure 4. The SusC epitope is widely recognized and reveals dynamic shifts in commensal-specific T cell responses.
(A) Schematic of the SusC peptide within the highly conserved plug domain of the TBDR SusC found in members of the Bacteroidales order. SusC structure was predicted using the online Robetta server (Kim et al., 2004) by using B. dorei TBDR (NCBI Reference Sequence: WP_007845382.1) to search for PDB homologs. The SusC peptide was aligned to the predicted SusC structure in PyMOL (Schrödinger LLC, 2021).
(B) Position and number of strong binding (%rank < 2%) 9-mer epitopes within the SusC peptide for the most frequent HLA class II alleles among HMP2 participants.
(C) Response profiles presented as a ternary plot (left) and Upset intersection plot with individual cytokine response and response intersections (Z score ≥ 2) for IFNγ, IL-10, and IL-17A (right) after stimulation of PBMCs from 40 subjects with the SusC peptide. Each dot in the ternary plot represents the response profile for one subject. Only subjects that responded with at least one significant cytokine response were plotted. Axes indicate percent share in the total response for each cytokine after normalizing to negative controls for each cytokine individually. Coloring indicates the level of density.
(D) Cytokine concentrations and FDR-corrected significance levels (paired Wilcoxon test) for peptide stimulations in 40 subjects. The mean of negative controls (H2-DMA, OVA, Scrambled) are plotted as “controls.” Each dot indicates the mean value for one subject. p values ≤ 0.05 were regarded as significant. ns, not significant (p > 0.05), * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, **** p ≤ 0.0001.
(E) Cytokine responses to SusC in patients with IBD (see Table 1 for patient demographics). Mean cytokine concentrations after peptide stimulation of PBMCs were normalized relative to negative control (DMSO). p values were generated using individual residualized models for cytokines and CD/UC accounting for significant covariates (see Table S2) using log10-transformed fold difference to negative control, whereas FDR correction was used to correct for multiple testing. Each dot indicates the mean value for one patient. ns, not significant (p > 0.05), * p ≤ 0.05.
Considering the continuous interaction between the adaptive immune system and commensal antigens, we therefore reasoned that the quantitative response magnitude and qualitative nature of SusC-specific T cell cytokine responses may be altered in the context of intestinal inflammation. In IBD, immune pathology is typified by uncontrolled CD4+ effector T cell responses toward commensals, which is dominated by Th1 and Th17 responses (Belkaid and Hand, 2014; Calderón-Gómez et al., 2016; Hegazy and West, 2017). To identify possible alterations in SusC reactivity in IBD, we recruited patients with CD and UC (Table 1) and compared SusC reactivity in PBMCs from remittive and active disease. To identify differences between cytokine responses in remittive and active disease for CD and UC individually, we used residualized linear regression models for each cytokine separately, by accounting for significant covariates (Table S2) and modeling the log10-transformed fold cytokine response over the negative control as a function of disease state and any significant covariates. We found an increased IL-17A response in CD with active disease compared with patients with CD in remission (p = 0.013) (Figure 4E). We observed a similar trend in UC, although not statistically significant. Together, this revealed an alteration of the SusC-specific T cell response with a shift toward IL-17A in patients with CD experiencing a flare in disease. Thus, T cell responses to commensal microorganisms are dynamic, reflecting cytokine skewing and phenotypic transitions associated with loss of tolerance.
Table 1.
IBD patient demographics
| CD | UC | |||
|---|---|---|---|---|
|
|
|
|||
| Remission (n = 25) | Active (n = 11) | Remission (n = 23) | Active (n = 9) | |
|
| ||||
| Sex (% males) | n = 11 (44%) | n = 3 (27%) | n = 13(57%) | n = 3 (33%) |
| Age at blood draw (mean ± SD) | 38.76 ± 14.18 | 36.18 ± 9.08 | 46.57 ± 18.41 | 40.56 ± 16.22 |
| Previous resection (% yes) | n = 5 (20%) | n = 4 (36%) | N/A | N/A |
| Antidiarrheal (% yes) | n = 1 (4%) | N/A | N/A | N/A |
| Antibiotic (% yes) | n = 1 (4%) | N/A | N/A | N/A |
| Antimetabolite (% yes) | n = 8 (32%) | n = 4 (36%) | n = 3 (13%) | N/A |
| Biologic (% yes) | n = 23 (92%) | n = 9 (82%) | n = 19(83%) | n = 5 (56%) |
| Steroid (% yes) | n = 3 (12%) | n = 2 (18%) | n = 1 (4%) | n = 3 (33%) |
Murine intestinal SusC-specific CD4+ T cells show active immunoregulatory and effector phenotypes
Consistent with our observations in healthy human donors, we previously described a homeostatic T cell response in healthy mice to the conserved VLKDASAAAIYGSR epitope in a SusC-like protein found in members of the Bacteroidales genus (Graham et al., 2018). In vivo mouse models provide a unique opportunity to dissect tissue-specific T cell phenotypes and have highlighted the importance of tissue-adaptation for localized immunosurveillance by T cells (Szabo et al., 2019). With the homeostatic SusC response conserved between human and mouse, we therefore leveraged mouse models to investigate tissue-specific phenotypes and TCR repertoires associated with TonB-specific T cell responses.
We utilized SusC VLKDASAAAIYGSR-loaded H2-I-Ab tetramers to isolate CD4+ T cells from mice to characterize the functional characteristics of ileal intraepithelial T cells and their lymphoid counterparts in the corresponding draining lymph nodes (mesenteric lymph node [mLN]) (Figure 5A). We confirmed that these mice indeed elicited IL-10 production upon peptide restimulation of splenocytes compared with controls (Figure S4A). Binding of SusC tetramer to CD4+ T cells from intraepithelial lymphocytes (IELs) and mLN indicated specific interaction with MHCII-restricted TCRs (Figure 5B). In these experiments, non-CD4+ T cells (B220+, CD11b+, CD11c+, F4/80+, CD8a+) were excluded before FACS-sorting of CD45+CD3+CD4+ SusC-specific T cells discriminated by double-positive staining with dual-labeled tetramer fluorophores (Figure S4B). We recovered a total of 2,782 SusC-specific CD4+ T cells (985 and 1,797 tetramer+ IELs and mLN T cells, respectively) by single-cell sequencing after quality filtering of sequencing reads (see STAR Methods). In parallel, we sorted and sequenced 24,992 SusC tetramer-negative CD4+ T cells (13,125 and 11,867 tetramer− IELs and mLN CD4+ T cells, respectively).
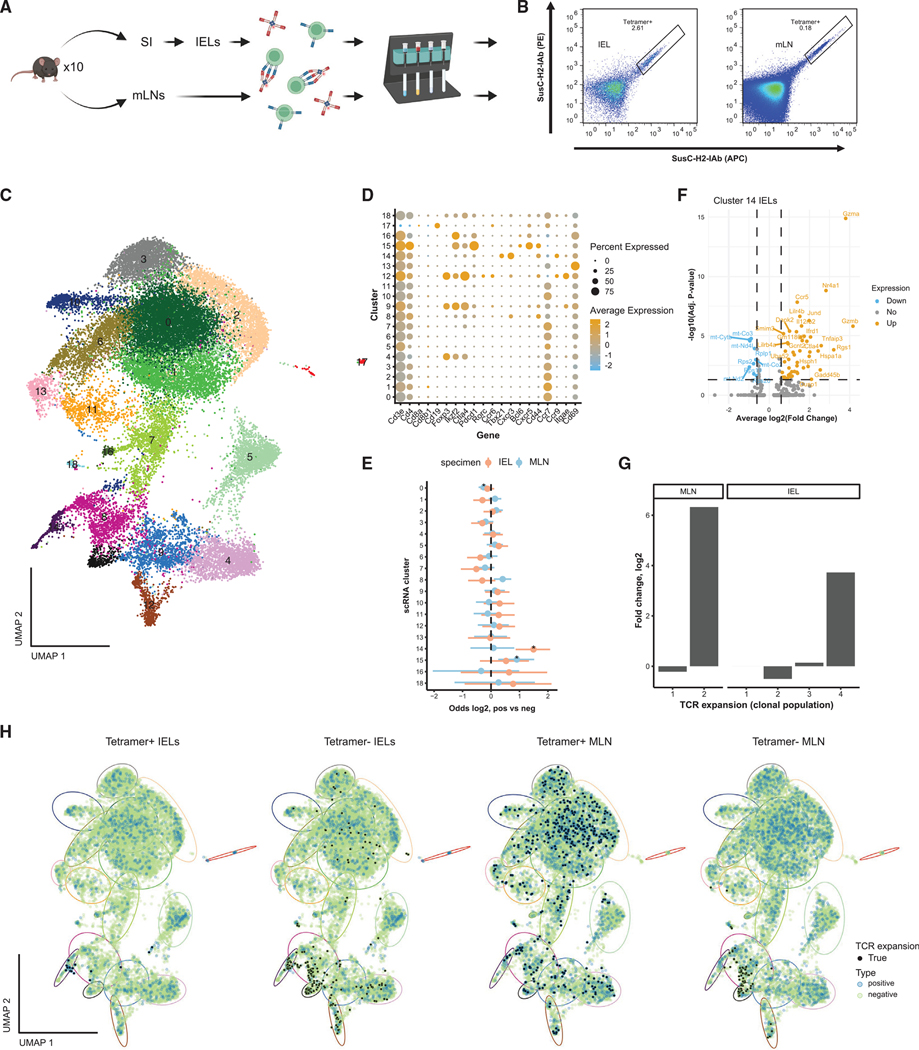
Figure 5. Clonally expanded small intestinal SusC-specific T cells take on multiple phenotypes during homeostasis.
(A) Experimental design. Intraepithelial lymphocytes (IELs) from the small intestine (SI) and mesenteric lymph nodes (mLNs) were isolated from ten female C57BL/6J mice and stained with SusC-H2-IAb tetramer before magnetic enrichment of tetramer bound cells. Created with BioRender.com.
(B) Representative tetramer staining of CD4 single-positive IELs and mLN T cells. See also Figure S4B.
(C) Integrated UMAP of tetramer+ and tetramer− IELs and mLN T cells.
(D) Dotplot of average gene expression (Z score) of canonical gene markers in UMAP clusters and percent of cells expressing these markers.
(E) Enrichment (odds ratio) between tetramer+ and tetramer− cells within UMAP clusters (* indicates significant enrichment, Fisher’s test, FDR < 0.05).
(F) Volcano plot for genes significantly upregulated or downregulated in cluster 14 tetramer+ IELs versus tetramer− IELs. Significance thresholds were set at average log2(fold change) > 0.6 and adjusted p value < 0.05 (Bonferroni correction). Colors indicate genes with significant down (blue) or up (orange) regulation or with no change in regulation (gray) for tetramer+ fraction.
(G) Log2-fold change in TCR clonal population sizes (number of cells per clonal population) between tetramer+ versus tetramer− cells in the mLN and IELs.
(H) Integration of TCR expansion (true indicates >1 TCR clone) with UMAP for individual samples (IELs and mLN samples). Dot color indicates sample type as tetramer+ or tetramer−. Ellipses indicate approximate regions for cellular clusters depicted in (C), with matched colors.
To identify cellular phenotypes enriched among tetramer+ T cells, we performed a differential expression (DE) analysis of genes recovered by scRNA-seq and clustering of cells by Uniform Manifold Approximation and Projection (UMAP). Together, tetramer+-and tetramer−-sorted cells clustered into 19 total clusters (0–18), in which the expression of CD3 (Cd3e) and CD4 markers was confirmed in all but one small cluster (cluster 17) of contaminating B cells that was excluded from further analysis (Figures 5C and 5D). Compared with tetramer− cells, per sample determination of cluster frequencies revealed enrichment in cluster 14 of tetramer+ IELs and in cluster 15 of tetramer+ mLN T cells (Figure 5E). Cluster 14 IELs express Th1-associated marker genes, and cluster 15 mLN T cells express Tfh-associated markers (Figure 5D).
To gain further insight into the characteristics of tetramer+ IELs in cluster 14 and tetramer+ mLN T cells in cluster 15, we compared their transcriptional signatures against their tetramer− counterparts. Although this revealed little difference for the mLN T cells (Figure S4C), the tetramer+ IELs in cluster 14 were characterized by the overexpression of cytotoxic programs (Gzma, Gzmb) (Figures 5F and S5A). Reclustering of only tetramer+ IELs revealed a transcriptionally distinct subset of cells (cluster 4, Figure S5A) expressing genes associated with the cytotoxic program (Tbx21, Ifng, Nkg7, and Fasl), early response to TCR engagement (Nr4a1 and Dusp1), activation (Tnfrsf4 and Ccl5), the NFkB pathway (Tnfaip3 and Nfkbia), immune suppression (Ctla4), and costimulation (Icos), together with the expression of markers for tissue residency (Itgae, Cd69, Ccr9, and Cxcr6) (Figure S5A). Furthermore, cells in this cluster exhibited expression of the transcription factor Runx3, but not ThPOK (Zbtb7b) (Figure S5A), which has been proposed to reflect recommitment of CD4+ T cells to the expression of CD8 lineage and cytotoxicity genes in transition to becoming cytotoxic IELs (Mucida et al., 2013; Reis et al., 2013; Sujino et al., 2016). However, as expected due to the FACS gating strategy, cells in this cluster did not express CD8a (Cd8a) or CD8b (Cd8b1) (Figure S5A), confirming that these cells are single-positive cytotoxic CD4+ IELs. Neither mLN T cells nor tetramer− IELs showed a similar T cell lineage. Thus, we identified SusC-specific CD4+ T cells in the ileal epithelial layer specialized in cytotoxic responses.
To investigate lymph-node-resident T cells, we defined SusC tetramer+ clonotypes and their corresponding transcriptional signatures in mLN T cells. Clonotypes were defined as those sharing the same TCR gene segments and nucleotide sequences of CDR3 regions. Overall, productive V-J spanning was found in 88.8% (±0.013%) of tetramer+ T cells. Although clonal expansion among tetramer+ IELs was largely restricted to the cytotoxic cells (Figures 5H and S5A), clonal expansion of tetramer+ T cells in the mLN was evident across multiple celltype clusters (Figure 5H). By reclustering tetramer+ mLN T cells, we identified expanded clones in a cluster (cluster 2) of activated (Tnfrsf4, Icos) Treg cells (Foxp3, Ikzf2, Il2ra, Il2rb, Ctla4, Capg, Nrp1, and Tnfrsf18), a cluster (cluster 4) with a lower expression of Ikzf2 and Il2rb representing Tfh cells (Bcl6, Tox2) expressing suppressive genes (Ctla4, Pdcd1, and Tigit) and genes associated with IL-10 production (Maf, Izumo1r), and a cluster (cluster 6) of cells expressing genes related to the early response to TCR engagement (Nr4a1, Erg1, and Tnfrsf9) and cell growth (Ncl) (Figure S5B). Furthermore, clonal expansion was particularly evident in a cluster of cells representing resting T cells (cluster 1). Comparing clonal expansion between tetramer-positive- and negative-sorted T cells from the mLN, we identified an 80-fold increased expansion among SusC-specific T cells (Figure 5G). These TCR clonal expansions were observed among Treg cells and resting T cells, thus indicating remarkable functional heterogeneity in SusC-specific T cell clones (Figure 5G).
Taken together, SusC-specific CD4+ T cells adopt multiple phenotypes based on their anatomic location in the intestinal epithelium versus lymphoid tissues. Specifically, SusC-specific T cells exhibit functional features associated with cytotoxic CD4+ IELs, Treg cells, Tfh cells, and resting T cells. These findings indicate active and dynamic functional regulation of T cell states associated with maintenance of intestinal tolerance to members of Bacteroidales.
DISCUSSION
Despite readily available metagenomic datasets, the commensal MHCII-restricted antigen landscape is largely unexplored. Here, we present a systematic effort of antigen prediction and validation of commensal antigens presented by human MHCII and report multiple immunoreactive T cell epitopes within the human microbiome from a range of commensal microbial phyla and species. Our findings of steady-state reactivity to commensal epitopes supports that tolerance to the microbiome is an active process, rather than a product of immunological ignorance. Moreover, our results also indicate that T cell responses to the microbiome are dynamic and change depending on local intestinal inflammation. This concept is exemplified by our observation that SusC is an immunodominant epitope in the microbiome and that T cell responses toward this epitope vary with respect to cytokine output in health and disease.
The SusC-derived T cell epitope associated with TBDRs (TBDRs) exhibited several unique features that impact its immunogenicity. We postulate that the immunodominance of the TBDR plug domain is shaped at several levels and that these mechanisms provide important insights into features influencing immunodominance in the intestine. First, the high gene abundance and prevalence of TBDRs (Blanvillain et al., 2007; Schauer et al., 2008) should increase the prevalence of MHCII-associated ligands. TBDRs are present in the genomes of several unique species in the microbiome and are duplicated within individual microbial genomes, particularly in Bacteroidales. Metatranscriptomics confirms the high expression of TBDRs in the human microbiome, and nutrient-dependent transcriptional regulation of SusC-like TBDRs (Sonnenburg et al., 2010) may further influence temporal changes in antigen availability and T cell recognition (Wegorzewska et al., 2019). Second, TBDRs can be packaged in Bacteroides outer membrane vesicles (OMVs) (Elhenawy et al., 2014), thus serving as a mechanism for long distance microbiota-host communication and transepithelial delivery of microbial antigens (Shen et al., 2012; Kaparakis-Liaskos and Ferrero, 2015; Jones et al., 2020). Third, upon phagocytosis of OMVs or whole bacteria by APCs, certain regions of TBDRs may be preferentially accessible to protease cleavage by lysosomal processing. The TBDR plug domain is tethered by a flexible linker, allowing it to occupy and occlude the transporter pore region or dissociate to open the pore. In the open conformation, the plug domain is solvent exposed and likely susceptible to protease cleavage and subsequent processing for loading onto MHCII. Fourth, the plug domain is an epitope hotspot containing multiple nested epitopes with predicted binding to prevalent HLAII-alleles. This hotspot within the TBDR plug domain is highly conserved across Bacteroidales and may diversify the total number of T cell clones specific for TBDRs, ensuring a diverse TCR repertoire that is refractory to species-level fluctuations in abundance. Considering the functional redundancy of genes and proteins within bacterial families in the microbiome (Tian et al., 2020), conservation across members of the microbiota may be an important feature dictating immunodominance in the gut. This notion is supported by the immunodominance of flagellin antigens that are conserved across members of the Lachnospiraceae family (Lodes et al., 2004; Targan et al., 2005; Duck et al., 2007). Additionally, evidence of TCR reactivity to bacteria of similar taxonomy (Lathrop et al., 2011) suggests the presence of pan-species TCR specificities. Collectively, our findings suggest that MHCII-restricted T cell responses are governed by features of immunogenicity conferred by antigen abundance and protein structure with additional contributing features such as microbial lifestyle.
In accordance with the expected immunogenicity of SusC, immunoreactivity across several healthy subjects revealed a dynamic and highly subject-specific T cell reactivity profile with respect to cytokine production. The SusC antigen induced potent IL-10 responses, accompanied by varying levels of IFNγ and IL-17A, suggesting an overall tolerogenic response composed of underlying heterogeneous SusC-specific T cell phenotypes and reflecting differences in microbiome-specific immunity between healthy subjects at steady state.
To extend these observations, we characterized the transcriptional phenotypes and clonality of intestinal SusC-specific CD4+ T cells in mice. At steady state, we found SusC-specific CD4+ T cells across anatomic sites, with these cells adopting phenotypes of intestinal tissue-resident cytotoxic CD4+ IELs and lymphoid tissue Treg cells, Tfh cells, and resting T cells. These distinct transcriptional phenotypes are likely shaped by local environmental cues, such as cytokines, that balance compartmentalized protective and regulatory functions. We suggest a model in which priming and tuning of intestinal SusC-specific T cells during homeostasis lead to tolerance-inducing phenotypes acting to suppress aberrant immune responses. These findings highlight the potential of divergent functional differentiation trajectories of microbiome-specific T cells.
The T cell response to any given antigen is dynamic and can alternate between tolerogenic, inflammatory, or pathogenic depending on the encoding microbe and immune context (Hand et al., 2012). Historically, the main barrier to characterizing transitions from tolerance has been the difficulty in identifying immunodominant T cell epitopes. hBOTA makes it possible to perform accurate HLA-customized predictions of immunodominant T cell epitopes, and tracking T cell responses over time in the same individual would reflect changes in that individual’s underlying immunological state.
Consistent with previous reports of increased infiltration of Th17 cells and expression of IL-17A in inflamed mucosa of Crohn’s patients (Fujino et al., 2003; Hegazy and West, 2017), SusC-specific peripheral T cells from CD patients with disease flares exhibited increased IL-17A production upon restimulation, supporting a dynamic shift in T cell reactivity during intestinal inflammation. These findings suggest that circulating commensal-specific T cells in the periphery can serve as a proxy for monitoring mucosal immunity. Similarly, flagellin-specific CD4+ T cells in patients with CD have been reported to display Th1 and Th17 phenotypes producing either IFNγ or IL-17A, or both, upon recall encounter (Calderón-Gómez et al., 2016; Morgan and Mannon, 2021). Genetic studies of IBD have clearly identified risk genes in Th1 and Th17 pathways (Liu et al., 2015; de Lange et al., 2017; Huang et al., 2017). In particular, the IL-17 cytokine pathway has pleiotropic effects in mucosal inflammation both driving pathology and providing protective tissue immunity (O’Connor et al., 2009; Maxwell et al., 2015; Hall et al., 2018). Consistent with the notion that IL-17 plays pathogenic and protective roles in colitis, neutralizing antibodies directed against IL-17A or IL-17 receptor A (IL17RA) were not efficacious in treating CD and in some cases exacerbated symptoms (Hueber et al., 2012; Targan et al., 2012; Maxwell et al., 2015; Emond et al., 2019). Furthermore, pathogenic and homeostatic Th17 cells show differential transcriptional trajectories (Lee et al., 2012). Accordingly, Th17 cell states are plastic and shaped by local cues leading to transitions between T regulatory type 1 (Tr1) cells and Th1-like states (Hirota et al., 2011; Gagliani et al., 2015). In this context, our work suggests that microbiome-specific Th immunity actively maintains tolerance and can transition to pathogenic states during inflammation.
Taken together, our efforts in antigen discovery provide a suite of commensal epitopes to facilitate mechanistic dissection of T cell tolerance to the gut microbiota. Identifying immunodominant antigens shared across members of the microbiota, such as the SusC epitope, will help facilitate mechanistic studies of microbiota-directed T cell immunity in humans and guide future developments of immuno-monitoring technologies that have direct clinical utility.
Limitations of the study
Our prioritization strategy filtered out low abundance epitopes that might be immunodominant despite low abundance. We further prioritized epitopes that are predicted to bind multiple common HLAII heterodimers, thus excluding potentially immunodominant epitopes that are highly individualized or bind specifically to certain rare HLAII heterodimers. The SusC epitope is unique in its abundance and ability to be presented by multiple HLAII heterodimers. We show it is immunogenic in humans and mice. In humans, the T cell response to SusC is dynamic, changing in cytokine profile based on disease status in patients with CD. Larger cohorts will be needed for this analysis to be statistically powered in patients with UC. Also, longitudinal profiling over time in patients with IBD will add significant insight into the nature of dynamic shifts in T cell responses to SusC and to determine if these transitions can predict flares before their onset. Some of these questions can be addressed in mouse models. We characterized SusC-specific intestinal T cells at baseline homeostasis in mice. The tetramer-sorting strategy enriches for SusC-specific T cells but should not be considered a purification strategy, and further work will be required to validate TCR reactivity to SusC. Nevertheless, tetramer enrichment identified several distinct T cell phenotypes associated with tetramer+ populations. Future work holds great potential for identifying mechanisms underlying dynamic T cell shifts to microbiome epitopes associated with inflammation and breakdown of tolerance.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests may be directed to and will be fulfilled by the lead contact, Ramnik J. Xavier (xavier@molbio.mgh.harvard.edu).
Materials availability
Detailed protocols are available to generate tetramers. There are restrictions to the availability of VLKDASAAAIYGSR:H2-I-Ab tetramer, due to limited production capacity.
Data and code availability
Raw single-cell RNA and TCR-seq files have been deposited in the NCBI Gene Expression Omnibus under GEO accession GSE196426 and are publicly available upon publication.
Source data for Figure S2A are available as supplemental files:
Source data for Figure S2C are available as supplemental files:
Code for hBOTA has been deposited in GitLab: https://gitlab.com/xavier-lab-computation/public/hbota.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human samples
Blood samples (unpurified buffy coats, 25–50mL) from healthy donors (ages within 18–65 years, unknown distribution of male and female) were collected at Research Blood Components, LCC, MA, USA after obtaining a signed consent form. Exact age or sex was not provided by vendor. Standard testing for blood-borne pathogens was performed. IRB approval was obtained from WCG IRB (Puyallup, WA, USA), study number 1278235, IRB tracking number 120160613.
Whole blood samples (10mL) from IBD patients diagnosed with either CD or UC based on colonoscopy and clinical symptoms were collected at the Massachusetts General Hospital, Boston, MA, USA (Table 1). Active or inactive disease was assessed by colonoscopy and with the use of the Harvey Bradshaw Index for CD (HBI) and Simple Clinical Activity Index for UC (SCCAI). IBD patients were enrolled with signed consent in the Prospective Registry in IBD study at Massachusetts General Hospital (PRISM) before blood draw. IRB approval was obtained from Mass General Brigham IRB (Somerville, MA, USA), IRB protocol number 2004P001067.
For both healthy subjects and IBD patients sample sizes are indicated in the result section and figure legends where described.
Mice
Eight to ten week old female C57BL/6J mice were utilized for this study from the colony of mice originally purchased from Jackson Laboratories (n=10). Mice were maintained at the animal facility at Massachusetts General Hospital (MGH) and housed in cages with 3–5 mice per cage, with sterilized water and food given ad libitum. All experimental procedures were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at MGH, IACUC protocol number 2003N000158.
METHOD DETAILS
Processing and analysis of multi-omics data
Sequencing reads from 1638 metagenomic (MGX) and 835 metatranscriptomic (MTX) were processed using the quality control pipeline KneadData (0.7.0) (http://huttenhower.sph.harvard.edu/kneaddata) including, Trimmomatic (Bolger et al., 2014) and BMTagger to remove short reads (<50 bp), low quality reads (Phred score < 20), reads aligning to hg38 or hg38 mRNA, and Nextera and Truseq adaptors.
Metagenomic assembly was performed independently for each sample. Contigs were assembled from quality control reads using Megahit (v.1.1.2, default settings) (Li et al., 2015), followed by gene calling using Prodigal (-meta, default settings) (Hyatt et al., 2010) from contigs of ≥500 nucleotides. A non-redundant gene catalog was created using CD-hit (b-aS 0.9, -c 0.95, -r 0, -d 0 and -B 0, version 4.6.5) (Fu et al., 2012). Gene abundance and expression was calculated by mapping MGX and MTX samples with a minimum identity of 95% using BWA (mem, default settings) and normalized to transcripts per million (TPM).
Co-abundant genes (CAGs) were identified using co-abundance binning (Nielsen et al., 2014), and metagenomic species (MGS) containing at least 400 CAGs were taxonomically annotated at the phylum, genus and species level, based on at least 40% mapping of genes to NCBI RefSeq (v. July 2017), as previously described (Li et al., 2014). Functional annotation was performed using EggNOG-mapper (Huerta-Cepas et al., 2017).
hBOTA pipeline
The hBOTA pipeline is based on the original BOTA algorithm (Graham et al., 2018), with minor changes allowing metagenomic input data and accommodation of human HLA class II alleles for epitope predictions. To start, hBOTA takes in reference-based or de-novo assembled metagenomes, annotates protein-coding genes as gram-positive or gram-negative, extracts amino acid sequences and then informs (1) protein domain using HMMScan of HMMER version 3.1b2 (Eddy, 2011) against Pfam (El-Gebali et al., 2019) (2) cellular location using PSORTb version 3.0.2 (Yu et al., 2010) and (3) membrane topology predictions using HMMTOP (Tusnády and Simon, 1998). hBOTA then generates a list of candidate peptides that (1) are derived from cell wall, outer membrane or extracellular proteins, (2) are located in the outward facing part of the protein (cell wall and outer membrane proteins) and (3) meet the following criteria to ensure sufficient structural accessibility; (a) the peptide should be 8 or more amino acids away from an anchoring domain (cell wall and outer membrane proteins), (b) it cannot be located in a small domain (>20 amino acids) and (c) it cannot be flanked by two domains less than 20 amino acids apart. As described in the original BOTA article (Graham et al., 2018), the selection criteria for candidate peptides are based on observations from MHCII peptidomics capturing Listeria epitopes. Candidate peptides of varying lengths are then analyzed using a 15 amino acid long sliding window for MHCII binding using NetMHCIIpan 3.2 (Jensen et al., 2018), which returns a predicted binding rank, level (strong, weak) and a 9mer binding core according to a user provided HLA class II alleles.
hBOTA benchmarking
A list of validated MHCII epitopes was curated from the IEDB database (https://www.iedb.org/), by filtering for published linear bacterial MHCII epitopes (minimum length 9 amino acids) validated for human T cell reactivity or MHCII binding. The UniProt identifier associated with each epitope was used to download the corresponding sequence of a full protein from the UniProt website (www.uniprot.org) and these served as input for MHCII epitope prediction with hBOTA as described above or with NetMHCIIpan 3.2 only, using frequent HLA class II alleles (>1% population frequency) from a representative US Caucasian population (Allele Frequency Net Database - USA Caucasian pop 5, 268 samples, http://www.allelefrequencies.net/). Epitopes predicted as strong binders (%rank < 2% or %rank < 1% as indicated) that matched with the epitopes downloaded from IEDB were defined as true positives.
Microbiome associated peptides
Microbiome Associated Peptides (MAPs) were predicted using the hBOTA pipeline with metagenomic data from the HMP2 cohort (1638 samples, 132 individuals) (Lloyd-Price et al., 2019), using annotated HLA class II alleles from 92 individuals in the HMP2 cohort (Figure S1B) as input for NetMHCIIpan 3.2 (Jensen et al., 2018). Strong binding 9mer epitopes (%rank < 2%) were further prioritized by using a minimum expression prevalence cutoff of 85% across corresponding HMP2 metatranscriptomics (835 samples). Only genes with expression in transcripts per million (TPM) greater than zero were considered. Final 15mer peptide sequences were then created by finding the top 9mer epitope from each annotated protein with the highest frequency of MHCII recognition within HMP2 and then selecting the corresponding 15mer sequences spanning the highest number of strong binding 9mer epitope registers. Resulting microbiome associated peptides (MAPs) were synthesized at GenScript, NJ, USA. Lyophilized peptides were reconstituted in DMSO at 2 mg/mL and saved at −20°C.
A final screening panel consisted of prioritized MAPs, along with negative and positive controls. Negative controls included mouse H2-DMA (LVCFVSNLFPPMLTV), Scrambled (GGGYSAPSANVAGGG) and OT-II ovalbumin (OVA; ISQAVHAAHAEINEAGR) peptides. Scrambled peptide binds mouse MHCII H2 strongly, but is not presented by human MHCII molecules and has no homology to known murine or microbiome proteins. Positive controls, the CEFT pool (JPT Technologies, cat. no. PM-CEFT-MHC-II) containing 14 MHCII-restricted T cell epitopes originating from Clostridium tetani, Epstein-Barr virus (HHV-4), Human cytomegalovirus (HHV-5) or Influenza, and an Influenza A (INFLA; PKYVKQNTLKLAT) peptide, were included to resemble antigen-specific positive controls, albeit to non-commensal pathogens.
Restimulation of human PBMCs
Cytometric bead arrays (CBAs, Flex Set; BD Biosciences) for IFNγ (cat no. 558274), IL-10 (cat no. 560111) and IL-17A (cat no. 560383) cytokines were used to quantitatively assess cytokine responses in PBMCs from healthy subjects and IBD patients after in vitro stimulation in triplicates with peptides or controls as indicated.
PBMCs were isolated by density gradient separation using Ficoll-Paque PLUS (VWR- GE Life Sciences, cat. no. 17144002). Isolated PBMCs were cultured for at least 1h at 37°C, 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, 2.5% sodium bicarbonate and 1% penicillin-streptomycin at 7.5×106 cells/mL before a 16h peptide stimulation at 37°C, 5% CO2 with 1μL of 2mg/mL peptide, unless otherwise indicated. For non-peptide controls, a volume of 1mL was used.
CBAs were performed on culture supernatants using the recommended protocol and analyzed by flow cytometry. Standard curves made from 2-fold dilutions of kit standards were used to interpolate pg/mL concentrations of cytokines in GraphPad Prism 8.1.2 from median PE fluorescence intensities. Median PE fluorescence intensities below the standard curve were set to the theoretical pg/mL detection limit determined by the vendor.
Ternary diagrams showing peptide response profiles were generated with R package ggtern version 3.1.0 (Hamilton and Ferry, 2018), using the fold difference of mean cytokine concentrations relative to negative controls. Only subject responses with at least one significant cytokine response (Z≥2) were plotted. Percentage on ternary diagram axis designates the share of the respective cytokine in the total cytokine response after normalizing to controls for each cytokine individually. Upset intersection plots of cytokine responses (Z≥2) were generated with R package UpSetR 1.4.0 (Conway et al., 2017) implemented though R package ComplexHeatmap 2.5.1 (Gu et al., 2016).
Duplication, conservation and antigenicity of TonB
A catalog of Bacteroidetes TonB homologs were generated by performing an InterProScan search for genes with plug (PF07715) and TonB_dep_Rec (PF00593) pfam annotations. The amino acid sequences of annotated plug domains were extracted based on annotated start and stop positions, plus an additional 80 amino acids extending C-terminally from the stop position or to the end of the protein.
We then parsed the extracted plug domain sequences through NetMHCIIpan 3.2 (Jensen et al., 2018) with HMP2 HLA class II alleles to predict 9mer epitopes and discarded plug domains not containing at least one epitope. From this set of plug domains, we extracted sequences homologous to the MAP22 and MAP36 22-mers by BLAST and filtering for additional amino acid patterns. For MAP22 and MAP36, the corresponding 22-mer peptides were generated to cover 22 amino acids towards the N-terminus with respect to MAP22. To BLAST, we used blastp (-max_target_seqs 100000 -evalue 10) to determine the plausible homologous sub-sequences in the plug regions and further accepted only the subsequences with the following amino acid composition rules: ^[TFIVLM][MLI][RK][DG] or ^[MLI][RK][DG] or ŜA[AS] or ÂSA[AS], where ’^’ indicates start of the sequence and characters in the square brackets indicate alternative amino acids at a given position.
To define regions of increased antigenicity within the plug domain, we aligned homologous plug domains according to the starting position of blast hit, summarized the cumulative number of binding HMP2 HLA class II alleles per amino acid using a 9mer sliding window for each plug domain.
Absolute counting of lymphocytes in IBD patients
Absolute counting of CD4+ T cells, CD8+ T cells, CD19+ B cells and CD14+ monocytes was done for each IBD patient. 500μL whole blood was incubated with 10mL pre-warmed 1x lyse/fix buffer (BD Biosciences cat. no. 558049) for 10min at 37°C for removal of red blood cells and fixation of remaining leukocytes. Leukocytes were then washed once with 10mL PBS containing 2% BSA and resuspended in 150μL PBS 2% BSA. Cells were saved at 4°C and analyzed by flow cytometry within 3 days. For flow cytometry, cells were stained with anti-CD3-PE-Cy7 (ThermoFischer Scientific cat.no 25-0038-42), anti-CD4-eFluor450 (eBioscienceTM cat. no. 48-0049-42), anti-CD8-PE (Biolegend cat. no. 344706), anti-CD19-FITC (Biolegend cat. no. 392508) and anti-CD14-APC (Biolegend cat. no. 367117), and 25μL CountBright™ Absolute Counting Beads (ThermoFischer Scientific cat. no. C36950) containing 0.255×105 beads was added. CD4+, CD8+, CD19+ and CD14+ populations were gated (Figure S3) and absolute cell numbers were calculated using following formula:
Tetramer production
Peptide:MHCII tetramers were generated as previously described (Moon et al., 2011). Briefly, soluble heterodimeric I-Ab molecules covalently linked to the SusC peptide epitope (VLKDASAAAIYGSR) were expressed and biotinylated in stably transfected Drosophila S2 cells. Following immunoaffinity purification, these biotinylated peptide:MHCII complexes were titrated and tetramerized to PE or APC fluorochrome-conjugated streptavidin (Prozyme).
Restimulation of mouse splenocytes
Splenocytes from 8–12-week-old female C57BL/6J mice (n=7) were isolated and cultured in complete media (DMEM with 10% FBS, 1% L-glutamine, 2.5% sodium bicarbonate, and 1% penicillin-streptomycin) at a concentration of 5 × 106 cells per mL. Cells from each well were pulsed with a final concentration of 10μg/mL of the SusC 14-mer peptide (VLKDASAAAIYGSR), Scrambled peptide (GGGYSAPSANVAGGG) or a vehicle control (dimethylsulfoxide) for 24 h at 37 °C in triplicates. Supernatants were collected and cytokine production was measured by cytometric bead array (Flex Set; BD Biosciences) for IFNγ (cat no. 558296), IL-17A (cat no. 560283), and IL-10 (cat no. 558300).
Tetramer staining of murine IELs and mLN T cells
Using MHCII tetramers, SusC-positive CD4+ T cells were isolated and stained from the intraepithelial lymphocytes (IELs) and mLN of mice. For murine IELs, the distal 5 cm of the small intestine of mice was excised, attached fat and Peyer’s patches were removed, and tissues were cut longitudinally to further remove luminal contents by washing with ice-cold PBS. Epithelial cells were isolated using a PBS buffer containing 2 mM EDTA, 1 mM dithiothreitol and 2% fetal bovine serum (FBS), shaking at 37°C for 30 min. Lymphocytes were further purified using a 40% Percoll gradient and were resuspended in RPMI 1640 with 5% FBS. For murine mLN lymphocytes, 2–5 lymph nodes per mouse were mashed with a sterile syringe through a 70um filter and washed with a buffer containing PBS and 2% FBS. Isolated lymphocytes from each tissue site were then incubated with 10uM of the SusC tetramer for 1 hour at room temperature. Tetramer positive cells were enriched using anti-APC and anti-PE microbeads (Miltenyi) on a MACS LS column using positive selection as previously described (Moon et al., 2011). Resulting cells eluted from the LS column were stained with a panel of antibodies specific to the following surface markers: CD45, CD3e, CD4, CD8a, F4/80, CD11b, CD11c and B220. Stained cells were run on a cell sorter (Sony SH700) and both tetramer positive and tetramer negative CD4+ T cells were sorted into a tube containing FACS buffer (PBS + 2% FBS), counted and sent for sequencing.
Single-cell RNA and TCR sequencing
Sorted single cells were separated into droplet emulsions using the Chromium Next GEM Single Cell 5′ Solution (v1). Approximately 10,000 cells for tetramer negative samples, 3,000 cells for tetramer positive mLN cells and 1200 tetramer positive IELs were loaded per channel. Gene expression and V(D)J TCR libraries were created according to manufacturer’s instructions (10x Genomics). Gene expression libraries were sequenced on the HiSeq X (Illumina) with the following read configuration: Read 1: 28 cycles, Read 2: 96 cycles, Index read 1: 8 cycles. TCR libraries were sequenced on the Nextseq (Illumina) with the following read configuration: Read 1: 150 cycles, Read 2: 150 cycles, Index read 1: 8 cycles.
Gene expression analysis
Single-cell RNA sequencing data was processed using the default settings of Cell Ranger (v3.1.0) as implemented in cellranger_workflow in the Terra platform (https://app.terra.bio/) (Li et al., 2020). Raw outputs from Cell Ranger were processed in the Seurat R package (4.0.5). We retained cells that had unique feature (gene) counts within range from 200 to 3000 and mitochondria counts of less than 6%, log-normalized the counts and scaled each gene. 2000 highly variable features were determined using the “vst” method (FindVariableFeatures function), and served as input to the principal component analysis for dimensionality reduction. We used the first 30 principal components to cluster cells using the Louvain algorithm. To integrate all samples (3 mLN tetramer−, 1 mLN tetramer+, 3 IEL tetramer− and 1 IEL tetramer+) we used the “anchoring” approach (FindIntegrationAnchors function) (Stuart et al., 2019). To determine cluster specific transcriptional programs, differential gene expression analysis was performed with Wilcoxon Rank Sum Test.
T cell repertoire analysis
Single-cell TCR sequencing data was assembled and the clonotypes were determined using the default settings of Cell Ranger (v3.1.0) VDJ pipeline as implemented in cellranger_workflow in the Terra platform (https://app.terra.bio/) (Li et al., 2020). TCR alpha and beta chains were consolidated and integrated with scRNA gene expression using R package scRepertoire (1.3.2). Classification of TCR clones as expanded across cells required matching of genes that comprise the TCR chains and matching of nucleotide sequence in the CDR3 regions.
QUANTIFICATION AND STATISTICAL ANALYSIS
To define significant cytokine responses to predicted MAPs a Z-score threshold of Z≥2 (2 standard deviations) compared to mean cytokine concentrations for negative controls (H2-DMA, Scrambled and OVA) was used. Z-scores were calculated for each replicate individually and results are reported as the mean Z-score across triplicates for each peptide.
For analysis of significant cytokine responses in 3 healthy individuals to TonB-dependent receptor protein peptides 1–4 (TBDRP1–4) compared to Scrambled negative control a paired t-test comparing mean cytokine concentrations of donors was performed using R package ggpubr 0.4.0. P-values ≤ 0.05 were regarded as significant.
For analysis of significance levels in cytokine concentrations between negative controls, CEFT pool and SusC peptide stimulations in 40 healthy individuals, a paired Wilcoxon test was performed in R package rstatix 0.7.0 comparing mean cytokine concentrations of donors. P-values ≤ 0.05 were regarded as significant. P-values were adjusted using FDR correction.
In determining significance of SusC reactivity in CD and UC patients (see Table 1 for patient demographics), confounding variables were residualized for each cytokine in CD and UC separately before performing linear regression using the log10 transformed fold difference to negative control as dependent variable and disease state together with any significant covariates as independent variables (Table S2). Confounding variables considered include age, sex, previous resection (CD only), patient medications grouped as antidiarrheals, aminosalicylates, antibiotics, steroids, antimetabolites, probiotics, biologics or immunosuppressants, and levels of CD4+ T cells, CD8+ T cells, CD19+ B cells and CD14+ monocytes as determined by absolute counting. The p.adjust function in R was used to correct for multiple testing by controlling the FDR.
For analysis of significance levels in cytokine concentrations between DMSO, Scrambled and SusC 14mer stimulations in mouse splenocytes (7 mice), a paired Wilcoxon test comparing mean cytokine concentrations was performed in R package rstatix 0.7.0. FDR corrected P-values ≤ 0.05 were regarded as significant.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| FITC anti-human CD19 | Biolegend | Cat#392508; RRID: AB_2750099 |
| APC anti-human CD14 | Biolegend | Cat#367117; RRID: AB_2566791 |
| PE anti-human CD8 | Biolegend | Cat#344706; RRID: AB_1953244 |
| PE-Cy7 anti-human CD3 | Thermo Fisher Scientific | Cat#25-0038-42; RRID: AB_1582253 |
| eFluor450 anti-human CD4 | Thermo Fisher Scientific | Cat#48-0049-42; RRID: AB_1272057 |
| Anti-mouse CD45 (30-F11) | Thermo Fisher Scientific | Cat# 11-0451-82 RRID: AB_465050 |
| Anti-mouse CD3e (145-2C11) | Thermo Fisher Scientific | Cat#11-0031-82; RRID:AB_464882 |
| Anti-mouse CD4 (SK-3) | Thermo Fisher Scientific | Cat# 46-0047-42; RRID:AB_1834401 |
| PB anti-mouse CD8a (53-6.7) | Thermo Fisher Scientific | Cat# 14-0081-82; RRID:AB_467087 |
| PB anti-mouse F4/80 (BM8) | Thermo Fisher Scientific | Cat# 48-4801-80 RRID:AB_1548756 |
| PB anti-mouse CD11b (M1/70) | Thermo Fisher Scientific | Cat# 48-0112-80; RRID:AB_1582237 |
| PB anti-mouse CD11c (N418) | Thermo Fisher Scientific | Cat# 48-0114-80; RRID:AB_1548665 |
| PB anti-mouse B220 (RA3-6B2) | Thermo Fisher Scientific | Cat# 48-0452-80; RRID:AB_1548763 |
|
| ||
| Biological samples | ||
|
| ||
| Unpurified buffy coats | Research Blood Components, LCC, MA, USA | Cat#002 |
| Whole blood from IBD patients | Massachusetts General Hospital, Boston, MA, USA | NA |
| Animals | ||
| Female C57BL/6J mice | Jackson Laboratories |
Strain#000664 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Microbiome associated peptides (MAPs) | This paper | This paper |
| CEFT pool | JPT Technologies | Cat#PM-CEFT-MHC-II |
| INFLA peptide | This paper | This paper |
| Scrambled peptide | This paper | This paper |
| H2-DMA peptide | This paper | This paper |
| OVA peptide | This paper | This paper |
| SusC 22mer (human) | This paper | This paper |
| SusC 14mer (mouse) | Graham et al., 2018 | N/A |
| VLKDASAAAIYGSR:H2-I-Ab tetramer | This paper | This paper |
| Ficoll-Paque PLUS | VWR-GE Life Sciences | Cat#17144002 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#F4135 |
| Penicillin/streptomycin | Corning | Cat#45000-652 |
| L-glutamine | Thermo Fisher Scientific | Cat#35050-061 |
| Lyse/Fix Buffer 5X | BD Biosciences | Cat#558049 |
| EDTA | Thermo Fisher Scientific | Cat#15575020 |
| Dithiothreitol | Sigma-Aldrich | Cat#10197777001 |
| Percoll | VWR | Cat#17-0891-01 |
|
| ||
| Critical commercial assays | ||
|
| ||
| CBA Flex Set (IFNg) | BD Biosciences | Cat#558274 |
| CBA Flex Set (IL-10) | BD Biosciences | Cat#560111 |
| CBA Flex Set (IL-17A) | BD Biosciences | Cat#560383 |
| Mouse IFN-γ Flex Set | BD Biosciences | Cat#558296 |
| Mouse IL-10 Flex Set | BD Biosciences | Cat#558300 |
| Mouse IL-17A Flex Set | BD Biosciences | Cat#560283 |
| Anti-APC MicroBeads | Miltenyi Biotec | Cat#130-090-855 |
| Anti-PE MicroBeads | Miltenyi Biotec | Cat#130-048-801 |
| MACS LS columns | Miltenyi Biotec | Cat#130-042-401 |
| CountBright™ Absolute Counting Beads | Thermo Fischer Scientific | Cat#C36950 |
| Chromium Single Cell 5’ Library & Gel Bead Kit, 16 rxns | 10× Genomics | Cat#PN-1000006 |
| Chromium Single Cell 5’ Library Construction Kit, 16 rxns | 10× Genomics | Cat#PN-1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Mouse T Cell, 96 rxns | 10× Genomics | Cat#PN-1000071 |
| Culture media | ||
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat#11960-044 |
| RPMI 1640 | Gibco | Cat#11875-093 |
|
| ||
| Deposited data | ||
|
| ||
| scRNA-seq and TCR-seq data | NCBI Gene Expression Omnibus | NCBI GEO: GSE196426 |
| hBOTA algorithm | This paper | https://gitlab.com/xavier-lab-computation/public/hbota |
|
| ||
| Software and algorithms | ||
|
| ||
| KneadData 0.7.0 | N/A | http://huttenhower.sph.harvard.edu/kneaddata |
| Trimmomatic | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| Megahit 1.1.2 | Li et al., 2015 | https://github.com/voutcn/megahit |
| Prodigal | Hyatt et al., 2010 | https://github.com/hyattpd/Prodigal |
| CD-hit version 4.6.5 | Fu et al., 2012 | http://weizhong-lab.ucsd.edu/cd-hit/ |
| EggNOG-mapper | Huerta-Cepas et al., 2017 | http://eggnog-mapper.embl.de/ |
| HMMER version 3.1b2 | Eddy, 2011 | http://hmmer.org/ |
| PSORTb version 3.0.2 | Yu et al., 2010 | https://www.psort.org/psortb/ |
| HMMTOP | Tusnády and Simon, 1998 | http://www.enzim.hu/hmmtop/html/document.html |
| NetMHCIIpan 3.2 | Jensen et al., 2018 | https://services.healthtech.dtu.dk/service.php?NetMHCIIpan-3.2 |
| GraphPad Prism 8.1.2 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| RStudio | RStudio Team, 2021 | http://www.rstudio.com/ |
| R package ggtern version 3.1.0 | Hamilton and Ferry, 2018 | https://CRAN.R-project.org/package=ggtern |
| R package UpSetR 1.4.0 | Conway et al., 2017 | https://cran.r-project.org/web/packages/UpSetR/index.html |
| R package ComplexHeatmap 2.5.1 | Gu et al., 2016 | https://www.bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| 10× Genomics Cell Ranger 3.1.0 | Zheng et al., 2017 | N/A |
| Terra | Li et al., 2020 | https://app.terra.bio/ |
| R package Seurat 4.0.5 | Hao et al., 2021 | https://satijalab.org/seurat/index.html |
| R package scRepertoire 1.3.2 | Borcherding et al., 2020 | https://github.com/ncborcherding/scRepertoire |
| FlowJo v10 | BD Biosciences | https://www.flowjo.com/solutions/flowjo/downloads |
|
| ||
| Other | ||
|
| ||
| The Immune Epitope Database and Analysis Resource | Vita et al., 2019 | https://www.iedb.org/ |
| UniProt | The UniProt Consortium, 2021 | www.uniprot.org |
| Allele Frequency Net Database | Gonzalez-Galarza et al., 2020 | http://www.allelefrequencies.net/ |
Highlights.
Discovery of microbiome epitopes across all major taxa of the human gut microbiome
TonB-dependent receptor SusC contains a conserved and immunodominant epitope hotspot
SusC-specific T cell response shifts from IL-10 to IL-17A in active Crohn’s disease
SusC-specific intestinal T cells assume regulatory and effector roles in mice
ACKNOWLEDGMENTS
We greatly appreciate all the study participants without whom this research would not be possible. We thank all the involved clinical staff, including Sara M. Gregory and Cara McConaughey, for establishment and coordination of the patient cohort. We are grateful to Elizabeth Heppenheimer for editorial assistance with the manuscript and figures. We thank Spencer Vaughan and Olivia Venezia for assistance with tetramer production, and Natan Pirete and Patricia Rogers for support with cell sorting. We thank members of the Xavier laboratories for helpful feedback. We thank Kawther Abu Elneel, Yanhua Zhao, Daphney Chin, and Luke Besse for their help in coordinating this project. The graphical abstract was created with BioRender.com. This work was supported by grants from the Helmsley Foundation, the Center for Microbiome Informatics and Therapeutics (CMIT) at MIT grant #6938971 to D.B.G., and the National Institutes of Health (NIH) U19 AI110495, DK127171, HL157717, and P30 DK043351 to R.J.X.
Footnotes
DECLARATION OF INTERESTS
R.J.X. is a co-founder of Celsius Therapeutics and Jnana Therapeutics, and a member of the Scientific Advisory Board of Nestle, as well as a member of the Board of Directors at Moonlake Immunotherapeutics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.08.016.
REFERENCES
- Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, Creech AL, Serrano LR, Nasir G, Nasrullah Y, et al. (2019). Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity 51, 766–779.e17. 10.1016/j.immuni.2019.08.012. [DOI] [PubMed] [Google Scholar]
- Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, and Barton GM (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong H, Alipour M, Valcheva R, Bording-Jorgensen M, Jovel J, Zaidi D, Shah P, Lou Y, Ebeling C, Mason AL, et al. (2019). Host immunoglobulin G selectively identifies pathobionts in pediatric inflammatory bowel diseases. Microbiome 7, 1. 10.1186/s40168-018-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Harrison OJ (2017). Homeostatic immunity and the microbiota. Immunity 46, 562–576. 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, and Arlat M (2007). Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2, e224. 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolam DN, and Koropatkin NM (2012). Glycan recognition by the Bacteroidetes Sus-like systems. Curr. Opin. Struct. Biol. 22, 563–569. 10.1016/j.sbi.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo ME, and Deem MW (2018). Predicting influenza H3N2 vaccine efficacy from evolution of the dominant epitope. Clin. Infect. Dis. 67, 1129–1131. 10.1093/cid/ciy323. [DOI] [PubMed] [Google Scholar]
- Borcherding N, Bormann NL, and Kraus G (2020). scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47. 10.12688/f1000research.22139.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucklacher-Waldert V, Carr EJ, Linterman MA, and Veldhoen M (2014). Cellular plasticity of CD4+ T cells in the intestine. Front. Immunol. 5, 488. 10.3389/fimmu.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Gómez E, Bassolas-Molina H, Mora-Buch R, Dotti I, Planell N, Esteller M, Gallego M, Martí M, Garcia-Martín C, Martínez-Torró C, et al. (2016). Commensal-specific CD4+ cells from patients with Crohn’s disease have a T-helper 17 inflammatory profile. Gastroenterology 151, 489–500.e3. 10.1053/j.gastro.2016.05.050. [DOI] [PubMed] [Google Scholar]
- Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z, Perry JSA, Knoop KA, Tanoue T, Narushima S, et al. (2017). Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2, eaal5068. 10.1126/sciimmunol.aal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, and Elson CO (2009). A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA 106, 19256–19261. 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, and Gehlenborg N (2017). UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji S-G, et al. (2017). Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261. 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, and Flower DR (2018). In silico prediction of cancer immunogens: current state of the art. BMC Immunol 19, 11. 10.1186/s12865-018-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, and Elson CO (2007). Isolation of flagellated bacteria implicated in Crohn’s disease. Inflamm. Bowel Dis. 13, 1191–1201. 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- Eddy SR (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res 47, D427–D432. 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, and Feldman MF (2014). Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5, e00909–e00914. 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond B, Ellis LA, Chakravarty SD, Ladouceur M, and Lefebvre P (2019). Real-world incidence of inflammatory bowel disease among patients with other chronic inflammatory diseases treated with interleukin-17a or phosphodiesterase 4 inhibitors. Curr. Med. Res. Opin. 35, 1751–1759. 10.1080/03007995.2019.1620713. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, and Fujiyama Y (2003). Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70. 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N, Amezcua Vesely MCA, Iseppon A, Brockmann L, Xu H, Palm NW, Zoete M.R. de, Licona-Limón P, Paiva RS, Ching T, et al. (2015). TH17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225. 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza FF, McCabe A, Santos E.J.M.D. dos, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, et al. (2020). Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 48, D783–D788. 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DB, Luo C, O’Connell DJ, Lefkovith A, Brown EM, Yassour M, Varma M, Abelin JG, Conway KL, Jasso GJ, et al. (2018). Antigen discovery and specification of immunodominance hierarchies for MHCII-restricted epitopes. Nat. Med. 24, 1762–1772. 10.1038/s41591-018-0203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hall AO, Towne JE, and Plevy SE (2018). Get the IL-17F outta here. Nat. Immunol. 19, 648–650. 10.1038/s41590-018-0141-z. [DOI] [PubMed] [Google Scholar]
- Hamilton NE, and Ferry M (2018). ggtern: ternary diagrams using ggplot2. J. Stat. Softw. 87, 1–17. 10.18637/jss.v087.c03. [DOI] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagán AJ, Pepper M, Maynard CL, Elson CO, and Belkaid Y (2012). Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 337, 1553–1556. 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. (2017). Circulating and tissue-resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 153, 1320–1337.e16. 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. (2011). Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263. 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner J-E, Villa M, Duarte JH, Demengeot J, Steinmetz OM, and Stockinger B (2013). Plasticity of TH17 cells in Peyer’s patches is responsible for the induction of T cell–dependent IgA responses. Nat. Immunol. 14, 372–379. 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Huang H, Fang M, Jostins L, Umićević Mirkov MU, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, et al. (2017). Fine-mapping inflammatory bowel disease loci to single variant resolution. Nature 547, 173–178. 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PDR, Wehkamp J, Feagan BG, Yao MD, Karczewski M, et al. (2012). Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700. 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Mering C. von, and Bork P (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol. Biol. Evol. 34, 2115–2122. 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, and Hauser LJ (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, and Nielsen M (2018). Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406. 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EJ, Booth C, Fonseca S, Parker A, Cross K, Miquel-Clopé s A, Hautefort I, Mayer U, Wileman T, Stentz R, and Carding SR (2020). The uptake, trafficking, and biodistribution of Bacteroides thetaiotaomicron generated outer membrane vesicles. Front. Microbiol. 11, 57. 10.3389/fmicb.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis-Liaskos M, and Ferrero RL (2015). Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 15, 375–387. 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- Kim DE, Chivian D, and Baker D (2004). Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32, W526–W531. 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, Kim D, Hsieh C-S, Hogan SP, Elson CO, et al. (2017). Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci. Immunol. 2, eaao1314. 10.1126/sciimmunol.aao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg MC, Andersen JF, Gorelick PL, Caspar P, Suerbaum S, Fox JG, Cheever AW, Jankovic D, and Sher A (2003). Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc. Natl. Acad. Sci. USA 100, 15830–15835. 10.1073/pnas.2534546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio C-W, Santacruz N, Peterson DA, Stappenbeck TS, and Hsieh C-S (2011). Peripheral education of the immune system by colonic commensal microbiota. Nature 478, 250–254. 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999. 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gould J, Yang Y, Sarkizova S, Tabaka M, Ashenberg O, Rosen Y, Slyper M, Kowalczyk MS, Villani A-C, et al. (2020). Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods 17, 793–798. 10.1038/s41592-020-0905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, and Lam T-W (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841. 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986. 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, and Hershberg RM (2004). Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 113, 1296–1306. 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, and Gordon JI (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9, e1001221. 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, Stevens E, Bigler J, Davis JA, Rottman JB, et al. (2015). Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity 43, 739–750. 10.1016/j.immuni.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Maynard CL, and Weaver CT (2009). Intestinal effector T cells in health and disease. Immunity 31, 389–400. 10.1016/j.immuni.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Dash P, Oguin TH, McClaren JL, Chu HH, Thomas PG, and Jenkins MK (2011). Quantitative impact of thymic selection on Foxp3+ and Foxp3− subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc. Natl. Acad. Sci. USA 108, 14602–14607. 10.1073/pnas.1109806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan NN, and Mannon PJ (2021). Flagellin-specific CD4 cytokine production in Crohn disease and controls is limited to a small subset of antigen-induced CD40L + T cells. J. Immunol. 206, 345–354. 10.4049/jimmunol.2000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, Wijk F. van, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. (2013). Transcriptional reprogramming of mature CD4+ T helper cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 14, 281–289. 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier EL, et al. (2014). Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828. 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- Noinaj N, Guillier M, Barnard TJ, and Buchanan SK (2010). TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor W, Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, Kolls JK, and Flavell RA (2009). A protective function for interleukin 17A in T cell–mediated intestinal inflammation. Nat. Immunol. 10, 603–609. 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, and Mucida D (2013). Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat. Immunol. 14, 271–280. 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team (2021). RStudio: integrated development environment for R. (RStudio, PBC; ). http://www.rstudio.com/. [Google Scholar]
- Russler-Germain EV, Rengarajan S, and Hsieh C-S (2017). Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol 10, 1375–1386. 10.1038/mi.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Rodionov DA, and Reuse H de. (2008). New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33, 330–338. 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. (2021). The PyMOL Molecular Graphics System, Version 2.5.0.
- Shen Y, Giardino Torchia MLG, Lawson GW, Karp CL, Ashwell JD, and Mazmanian SK (2012). Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520. 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, and Sonnenburg JL (2010). Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino T, London M, Hoytema van Konijnenburg D.P.H. van, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, and Mucida D (2016). Tissue adaptation of regulatory and intraepithelial CD4+ T cells controls gut inflammation. Science 352, 1581–1586. 10.1126/science.aaf3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PA, Miron M, and Farber DL (2019). Location, location, location: tissue resident memory T cells in mice and humans. Sci. Immunol 4, eaas9673. 10.1126/sciimmunol.aas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targan SR, Feagan BG, Vermeire S, Panaccione R, Melmed GY, Blosch C, Newmark R, Zhang N, Chon Y, Lin S-L, et al. (2012). Mo2083 a randomized, double-blind, placebo-controlled study to evaluate the safety, tolerability, and efficacy of amg 827 in subjects with moderate to severe Crohn’s disease. Gastroenterology 143, e26. 10.1053/j.gastro.2012.07.084. [DOI] [Google Scholar]
- Targan SR, Landers CJ, Yang H, Lodes MJ, Cong Y, Papadakis KA, Vasiliauskas E, Elson CO, and Hershberg RM (2005). Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn’s disease. Gastroenterology 128, 2020–2028. 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2020). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49, D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wang X-W, Wu A-K, Fan Y, Friedman J, Dahlin A, Waldor MK, Weinstock GM, Weiss ST, and Liu Y-Y (2020). Deciphering functional redundancy in the human microbiome. Nat. Commun. 11, 6217. 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády GE, and Simon I (1998). Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283, 489–506. 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- Tusnády GE, and Simon I (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Uchida AM, Boden EK, James EA, Shows DM, Konecny AJ, and Lord JD (2020). Escherichia coli–Specific CD4+ T cells have public T-cell Receptors and low interleukin 10 production in Crohn’s disease. Cell. Mol. Gastroenterol. Hepatol. 10, 507–526. 10.1016/j.jcmgh.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B (2019). The immune epitope database (IEDB): 2018 update. Nucleic Acids Res 47, D339–D343. 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegorzewska MM, Glowacki RWP, Hsieh SA, Donermeyer DL, Hickey CA, Horvath SC, Martens EC, Stappenbeck TS, and Allen PM (2019). Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol. 4, eaau9079. 10.1126/sciimmunol.aau9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. 10.1038/nature25500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal Th17 cells towards commensal bacterial antigens. Nature 510, 152–156. 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, and Brinkman FSL (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu W-JH, Saldana-Morales FB, Hill AA, Majumdar S, Orozco S, Bell R, et al. (2021). Thymic development of gut-microbiota-specific T cells. Nature 594, 413–417. 10.1038/s41586-021-03531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, and Elson CO (2018). Adaptive immune education by gut microbiota antigens. Immunology 154, 28–37. 10.1111/imm.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw single-cell RNA and TCR-seq files have been deposited in the NCBI Gene Expression Omnibus under GEO accession GSE196426 and are publicly available upon publication.
Source data for Figure S2A are available as supplemental files:
Source data for Figure S2C are available as supplemental files:
Code for hBOTA has been deposited in GitLab: https://gitlab.com/xavier-lab-computation/public/hbota.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.