Abstract
Background:
Increasing evidence connects the gut microbiome to Parkinson’s disease (PD) etiology, but little is known about microbial contributions to PD progression and its clinical features.
Objective:
We aim to explore the association between the gut microbiome with PD, and the microbial association with PD-specific clinical features.
Methods:
In a community-based case-control study of 96 PD patients and 74 controls, microbiome data were obtained from 16S rRNA gene sequencing of fecal samples, and analyzed for microbial diversity, taxa abundance, and predicted functional pathways that differed in PD patients and controls, and their association with PD-specific features (disease duration, motor subtypes, L-DOPA daily dose, and motor function).
Results:
PD patients’ gut microbiome showed lower species diversity (p = 0.04) and were compositionally different (p = 0.002) compared to controls but had a higher abundance of three phyla (Proteobacteria, Verrucomicrobiota, Actinobacteria) and five genera (Akkermansia, Enterococcus, Hungatella, and two Ruminococcaceae) controlling for sex, race, age, and sequencing platform. Also, 35 Metacyc pathways were predicted to be differentially expressed in PD patients including biosynthesis, compound degradation/utilization/assimilation, generation of metabolites and energy, and glycan pathways. Additionally, the postural instability gait dysfunction subtype was associated with three phyla and the NAD biosynthesis pathway. PD duration was associated with the Synergistota phylum, six genera, and the aromatic compound degradation pathways. Two genera were associated with motor function.
Conclusion:
PD patients differed from controls in gut microbiome composition and its predicted metagenome. Clinical features were also associated with bacterial taxa and altered metabolic pathways of interest for PD progression.
Keywords: Parkinson’s disease, gut microbiome, brain-gut axis, Unified Parkinson’s Disease Rating Scale
INTRODUCTION
Parkinson’s disease (PD) is a complex neurodegenerative disease characterized by progressive motor impairment and non-motor features such as cognitive, mood, and peripheral autonomic nervous system disorders, including gastrointestinal dysfunction [1]. The gut microbiome contributes not only to well-known digestive tract disorders common in PD, but may also exert an influence on PD pathogenesis [2]. Gut symptoms, especially constipation, often occur decades before PD diagnosis, and pathologic hall-marks of PD, namely Lewy bodies and α-synuclein aggregation in the gut and enteric nervous system have been found to precede brain pathology [3]. Thus, processes key in PD may be initiated in the gut possibly followed by a prion-like spread of pathological α-synuclein to the brain [4]. Such spread can be stopped when the vagus nerve is severed [5]. Further-more, α-synuclein has biophysical characteristics of antimicrobial peptides and may be trafficked from the gut to the central nervous system to confer immunity in advance of an infection [6].
Investigating the gut-brain connection for PD onset and progression is especially important as during its long prodromal phase, preventative actions could stop or slow neurodegeneration. PD specific gut microbes or their metabolites might present avenues for finding early disease biomarkers and intervention strategies prior to motor symptom onset.
Here, we explore the gut microbiome diversity, bacterial abundance, and its predicted metagenome in a community-based PD study in rural California comparing both community and household-based controls to PD patients. In addition, we assessed gut microbiome associations with phenotypic diversity of clinical features in PD cross-sectionally.
METHODS
Study population
The Parkinson’s, Environment and Gene (PEG) study is a population-based, case-control study of PD in Kern, Tulare, and Fresno counties, California. Participants were recruited in two waves: 2001–2007 (PEG1) and 2012–2017 (PEG2). At baseline, eligible PD cases were 1) newly diagnosed (within 3–5 years); 2) residing in California for at least 5 years; 3) confirmed by UCLA movement disorder specialist as “probable” or “possible” PD; 4) without other neurological conditions or terminal illnesses; 5) consented to participation (for details, see [7-9]). We recruited community controls in the same counties from randomly selected residential addresses (tax assessor and Medicare lists). Since 2017, we asked participants from both waves who could be re-contacted to participate in fecal sample collection. We recruited two-types of controls: 1) Household members of the PD cases to control for potential bias from shared environmental factors, and 2) Community members to avoid overmatching on environmental exposures within households. All controls were required not to have PD or any terminal illness and all participants were required to not be immunocompromised or have taken antibiotics within the past 3 months. In total, 96 PD patients with 53 household controls, and 21 community controls were enrolled. This study was approved by the UCLA Institutional Review Board. Informed written consent was obtained from all participants.
Data and sample collection
Trained research staff collected data using standardized interviews including: 1) demographic information such as sex, race/ethnicity, education; 2) medical histories including family history of PD, other diseases and medications; 3) other standardized instruments including Wexner Constipation Scoring System (Wexner), Geriatric Depression Scale (GDS) and Diet History Questionnaire II (DHQ II). Participants collected a fecal sample at their homes using a Para-Pak® collection kit preserved in 96% ethanol and mailed these to UCLA within 14 days of collection where samples were stored at −80°C in a freezer until DNA extraction.
16S rRNA gene sequencing, rarefaction, and feature filtering
Bacterial DNA was extracted from fecal samples using the ZymoBIOMICS DNA kit with bead beating. The V4 region of the 16S gene was amplified and underwent pair-ended 250 × 2 sequencing on Illumina HiSeq 2500 or MiSeq platforms. Raw data were processed using the DADA2 pipeline (v1.22.0) where sequencing reads were quality-filtered, processed into amplicon sequence variants (ASVs)—a classification method that corresponds to species level, and assigned taxonomy by closed-reference picking against the Silva database [10]. The sequencing depths ranged from 6,054 to 135,162 with a mean depth of 53,235 ± 20,454 per sample. ASVs were filtered in two steps, first by total abundance—ASVs were removed if abundance was less than 50, then by prevalence—ASVs were removed if prevalence was less than 10% in all samples (7,975,543/9,954,854 sequences remained after filtering). ASVs were also rarefied to even depth without abundance and prevalence filtering to assess alpha diversity, because the filtering step may exclude rare species and thus affects the alpha diversity measure. Data processing steps were performed with the phyloseq package (v1.34.0) and the workflow is shown in Supplementary Figure 1.
Metagenomic prediction
The metagenomic profile of the gut microbiome, i.e., the functional potential of the bacterial community based on 16S rRNA marker sequencing data [11], was predicted with PICRUST2 (v2.4.1). In conjunction with ASV abundance, these profiles reflect predicted gene content, i.e., metagenes, classified by enzyme commission (EC) number or KEGG (Kyoto Encyclopedia of Genes and Genomes) Orthology (KO), and predicted functional pathways, i.e., Metacyc or KEGG pathway profiles. Metagenes and pathways were removed if the abundance was less than 100 in total, or the prevalence was less than 10% in all samples (1,842/2,100 EC, 6,121/7,045 KO, 368/399 Metacyc, and 153/172 KEGG pathways remained after filtering).
PD clinical features
PD patients were examined by UCLA movement disorder specialists and symptoms assessed with the Unified Parkinson’s Disease Rating Scale (UPDRS) I-IV. Motor exams were conducted preferably during a functional “off” medication status (i.e., ≥12 hours since last PD medication); a correction factor, i.e., the mean difference of UPDRS III score between “on” and “off” scores in all patients, was added if a patient was “on” medication (N = 17). Missing items due to disability unrelated to PD (e.g., “arise from chair”) were imputed using the mean score of this item from all participants. We calculated the summary score for UPDRS III as an indicator of motor function, and further classified the patients into predominant motor subtypes including Postural Instability and Gait Dysfunction (PIGD), Tremor Dominant (TD), or Indeterminate (IND), as previously described [12]. Daily L-DOPA dose and other PD-related medications were collected on the exam day.
Statistical analysis
The microbiome was assessed for alpha diversity (Shannon index), beta diversity (Bray-Curtis dissimilarity), and taxa abundance comparing PD patients with community or household controls. The mean difference of alpha diversity between these groups was assessed using the Wilcoxon test statistic, while group-based beta diversity differences were tested with permutation multivariate analysis of variance (PERMANOVA). Differences in taxa abundance associated with PD status were assessed using DESeq2, an empirical Bayesian approach that shrinks dispersion and fits non-rarified count data to a negative binomial model [13]. We excluded taxa with less than 20% prevalence in either group (PD cases or controls) and adjusted all regression models for race, sex, age, and sequencing platform at a minimum, adding covariates during sensitivity analyses (see below). Additional factors we explored initially but did not enter into final models include smoking status, education, Wexner, GDS, and dietary factors from the DHQII. These factors did not change reported results more than minimally and this approach avoids sparse data issues. We used Benjamini-Hochberg (BH) corrections to control for false discovery rate (FDR).
Similarly, we explored associations between predicted metagenomic data and PD using the Wilcoxon test to assess differences in gene richness (i.e., alpha diversity of predicted bacterial functional genes), the PERMANOVA test to assess differences in beta diversity (Bray-Curtis dissimilarity of the gene count), and regression modeling to assess the differential abundance of Metacyc and KEGG pathways by PD status, controlling for the minimum covariate set. The analyses described above were performed with SAS 9.4 and R (v4.0.0).
Restricting to PD patients, we compared alpha and beta diversity, abundance of taxa and predicted metagenome by disease duration (years since diagnosis), predominant motor subtype (PIGD vs. others), motor scores (UPDRS III), and L-Dopa daily dosage at the time of fecal sampling adjusting for our minimum confounder set and using BH corrections to control for FDR.
Sensitivity analyses
First, we conducted sensitivity analyses to assess additional confounding by repeating analyses after adding constipation data into regression models. Second, we restricted analyses to PD cases and paired household controls (as matched sets) only. For differential taxa abundance, we modelled associations with mixed effects regression for case-control pairs, including pair indicators as random effects and confounders (race, sex, age, and sequencing platform) as fixed effects. Finally, as there is no one definitive method/package for microbiome analyses, and different methods may produce differences in results, it is recommended to use more than one differential abundance assessment method and check findings for consistency[14]. Therefore, we repeated the analysis using the R package MaAsLin2 (v1.7.3) to test for robustness of our results for differential taxa abundance.
RESULTS
Study population and microbiome profiles
The demographics of 170 participants (56% PD patients) who completed the study interview and provided a fecal sample are shown in Table 1. Participants were on average 72 years old, 52% males and 80% white. More PD patients than controls were men (67% vs. 34%), and patients were on average slightly older (73 years vs. 70 years).
Table 1.
Demographics of the study population
| PD (N = 96) |
Control (N = 74) |
Total (N = 170) |
|
|---|---|---|---|
| Race | |||
| White | 78 (81.3%) | 58 (78.4%) | 136 (80.0%) |
| Latino | 18 (18.8%) | 16 (21.6%) | 34 (20.0%) |
| Sex | |||
| Male | 64 (66.7%) | 25 (33.8%) | 89 (52.4%) |
| Female | 32 (33.3%) | 49 (66.2%) | 81 (47.6%) |
| Age | |||
| Mean (SD) | 72.9 (9.17) | 69.7 (8.61) | 71.5 (9.04) |
| Median [Min, Max] | 73.5 [43.0, 95.0] | 69.5 [44.0, 88.0] | 71.5 [43.0, 95.0] |
| Platform | |||
| HiSeq | 81 (84.4%) | 56 (75.7%) | 137 (80.6%) |
| MiSeq | 15 (15.6%) | 18 (24.3%) | 33 (19.4%) |
| Duration of PD Diagnosis | |||
| Mean (SD) | 9.6 (4.3) | ||
| Median [Min, Max] | 9 [3, 20] | ||
| Missing | 3 | ||
| Motor subtype | |||
| PIGD | 44 (47.3%) | ||
| Tremor Dominant and Indeterminate | 49 (52.7%) | ||
| Missing | 3 | ||
| LEDD | |||
| Mean (SD) | 563 (495) | ||
| Median [Min, Max] | 450 [0, 2190] | ||
| Missing | 3 | ||
| UPDRS III Score | |||
| Mean (SD) | 28.7 (13.4) | ||
| Median [Min, Max] | 29.0 [5.00, 66.2] | ||
| Missing | 3 |
PD, Parkinson’s disease; PIGD, postural instability and gait dysfunction; LEDD, levodopa equivalent doses; UPDRS, Unified Parkinson’s Disease Rating Scale.
Microbiome profiles
We identified 252 ASVs (corresponding to species), 105 genera, and 8 phyla from low abundance-filtered sequences based on 16S rRNA gene sequencing. The predicted metagenome included 1,842 ECs (corresponding to 368 Metacyc pathways) and 6,121 KOs (corresponding to 153 KEGG pathways) after low abundance/prevalence filtering. Microbial composition is shown in Supplementary Figures 2 and 3.
Microbiome associated with PD
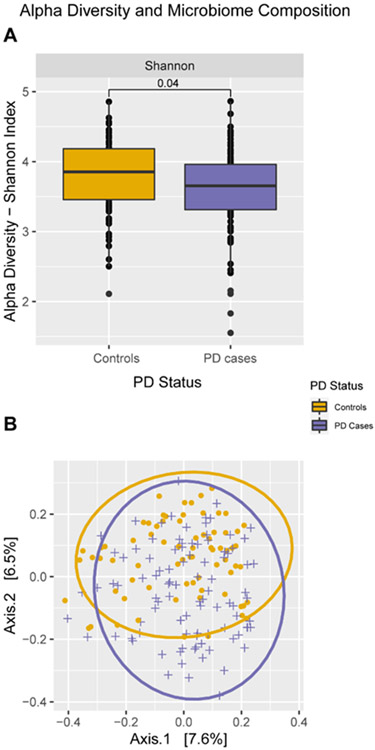
Compared to controls, PD patients had a lower mean Shannon index (p = 0.04, Fig. 1A), and a different microbial profile based on the Bray-Curtis dissimilarity (p = 0.002, Fig. 1B). These differences in alpha and beta diversity remained when we restricted to PD cases and paired household controls (Shannon index: p = 0.0036, Bray-Curtis dissimilarity: p = 0.01, Supplementary Figure 4). At the phylum level, PD patients exhibited higher abundances of Proteobacteria, Verrucomicrobiota, and Actinobacteriota (Table 2). At the genus level, PD patients showed increased abundance of UBA1819 (Ruminococcaceae), DTU089 (Ruminococcaceae), Akkermansia, Enterococcus, and Hungatella. Controlling for constipation removed associations with the Actinobacteriota phylum and the DTU089 genus (Ruminococcaceae). In PD-household control pair only analyses, higher abundance of the Akkermansia genus and Verrucomicrobiota phylum remained statistically significantly for PD patients, whereas other taxa differences were attenuated and no longer statistically significant. Additional evaluation of the change in estimates by removing the community controls (N = 21) yielded similar results with Verrucomicrobiota, Proteobacteria, Akkermansia, and UBA1819 remaining statistically significantly different (Supplementary Table 1). Analyses based on the MaAsLin2 package were very similar except that Actinobacteriota and Enterococcus were no longer identified as differentially abundant (see also Table 2).
Fig. 1.
Comparison of microbiome profile between PD patients and controls. A) Alpha diversity: Shannon index (p = 0.036). B) Beta diversity: Bray-Curtis Dissimilarity (PERMANOVA test: p = 0.002). PD, Parkinson’s disease; PERMANOVA, Permutation multivariate analysis of variance.
Table 2.
Differential taxa abundance associated with PD compared to two different control groups (total and household only)
| Taxa | Main Analys isa (N=170) |
Sensitivity Analys is 1b (N=153) |
Sensitivity Analys is 2c (N=100) |
Sensitivity Analys is 3d (N=170) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log2FCe | SE | Adj p | Log2FCe | SE | Adj p | Log2FCe | SE | Adj p | Log2FCe | SE | Adj p | |
| Proteobacteria (phylum) | 1.37 | 0.35 | 0.0007 | 1.40 | 0.41 | 0.0055 | 0.17 | 0.31 | 0.8341 | 1.20 | 0.26 | 0.0001 |
| Verrucomicrobiota (phylum) | 1.23 | 0.40 | 0.0078 | 1.34 | 0.47 | 0.0163 | 1.31 | 0.42 | 0.0270 | 1.05 | 0.30 | 0.0033 |
| Actinobacteriota (phylum) | 1.02 | 0.37 | 0.0135 | −0.07 | 0.44 | 0.8730 | 0.82 | 0.34 | 0.0918 | 0.57 | 0.26 | 0.0884 |
| UBA1819 (Ruminococcaceae family genus) | 2.27 | 0.39 | 0.0000 | 1.98 | 0.46 | 0.0018 | 0.40 | 0.35 | 0.8345 | 1.51 | 0.27 | 0.0000 |
| DTU089 (Ruminococcaceae family genus) | 3.08 | 0.81 | 0.0078 | NAg | NA | NA | −0.90 | 0.78 | 0.8345 | 2.06 | 0.63 | 0.0073 |
| Akkermansia (genus) | 1.43 | 0.42 | 0.0213 | 1.96 | 0.48 | 0.0021 | 1.50 | 0.41 | 0.0197 | 0.97 | 0.29 | 0.0066 |
| Enterococcus (genus) | 3.65 | 1.11 | 0.0252 | 2.79 | 1.25 | 0.1925 | 2.61 | 1.86 | 0.7327 | NAf | NA | NA |
| Hungatella (genus) | 2.54 | 0.80 | 0.0312 | 3.56 | 0.96 | 0.0070 | 1.86 | 0.67 | 0.1306 | 2.11 | 0.56 | 0.0014 |
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform. DESeq2 was used to perform the analysis.
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform and constipation status. DESeq2 was used to perform the analysis.
Included household controls only: PD patient and household control pair was treated as random effect. Adjusted for sex, race, age, and sequencing platform. MaAsLin2 was used to perform the analysis.
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform. MaAsLin2 was used to perform the analysis.
Coefficient indicates the difference in the log-transformed relative abundances between PD and Control.
This taxon was removed from the sensitivity analyses due to small cell count. Log2FC, Log2 fold change; SE, standard error; Adj, adjusted.
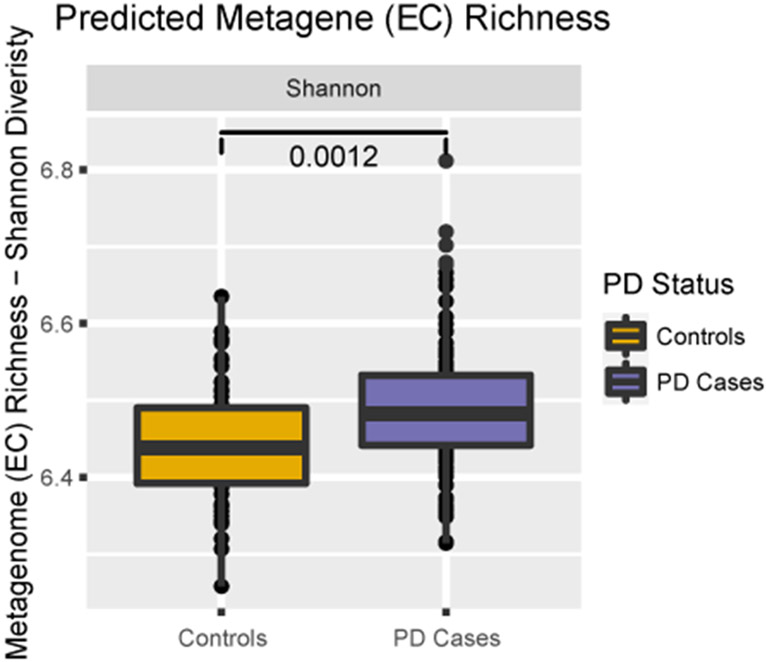
Predicted functional pathways associated with PD
PD patients exhibited higher EC bacterial gene diversity according to the Shannon index (p = 0.0012, Fig. 2), but no significant difference in the Bray-Curtis dissimilarity (p = 0.391, data not shown). We found 26 Metacyc pathways to be more abundant in PD patients and 9 pathways in controls belonging to 4 top-level superclasses: Biosynthesis, Degradation/Utilization/Assimilation, Generation of Precursor Metabolite and Energy, and Glycan Pathways (see Table 3 and Supplementary Figure 5). When controlling for constipation, most predicted pathways (26 Metacyc, 4 KEGG) remained statistically significant while in PD-household control pair only analyses, only 13 Metacyc pathways remained. Additional evaluation of the change in estimates by removing the community controls (N = 21) yielded similar results with the majority of pathways remaining statistically different (Supplementary Table 2). Using the MaAsLin2 package, results were consistent with those presented, albeit slightly attenuated. Results related to KOs and KEGG pathways are shown in Supplementary Tables 3 and 4 and Supplementary Figure 6.
Fig. 2.
Comparison of alpha diversity of predicted metagenome (EC): Shannon index (p = 0.0012). PD, Parkinson’s disease; EC, enzyme commission; PERMANOVA, Permutation multivariate analysis of variance.
Table 3.
Differential predicted Metacyc pathways abundance associated with PD compared to two different control groups (total and household only)
| Metacyc pathways | Main Analysisa (N=170) |
Sensitivity Analysis 1b (N=153) |
Sensitivity Analysis 2c (N=100) |
Sensitivity Analysis 3d (N=170) |
||||
|---|---|---|---|---|---|---|---|---|
| Log2FCe | Adj P | Log2FCe | Adj P | Log2FCe | Adj p | Log2FCe | Adj p | |
| Allantoin degradation to glyoxylate III | 1.06 | 0.0000 | 1.14 | 0.0005 | 0.39 | 0.0500 | 0.75 | 0.0000 |
| Superpathway of (R,R)-butanediol biosynthesis | 1.09 | 0.0064 | 1.37 | 0.0011 | 0.37 | 0.2696 | 0.78 | 0.0002 |
| Superpathway of UDP-glucose-derived O-antigen building blocks biosynthesis | 0.70 | 0.0064 | 0.68 | 0.0189 | 0.57 | 0.0031 | 0.55 | 0.0001 |
| Glucose and glucose-1-phosphate degradation | 1.00 | 0.0139 | 1.02 | 0.0189 | 0.75 | 0.0096 | 0.75 | 0.0005 |
| 1,4-dihydroxy-2-naphthoate biosynthesis I | 0.90 | 0.0155 | 0.91 | 0.0266 | 0.81 | 0.0048 | 0.68 | 0.0008 |
| Superpathway of phylloquinol biosynthesis | 0.88 | 0.0157 | 0.88 | 0.0271 | 0.80 | 0.0049 | 0.66 | 0.0009 |
| NAD salvage pathway II | 0.76 | 0.0250 | 0.59 | 0.0717 | 0.48 | 0.0819 | 0.55 | 0.0022 |
| 4-hydroxyphenylacetate degradation | 1.98 | 0.0265 | 3.04 | 0.0016 | 0.75 | 0.3603 | 1.72 | 0.0007 |
| L-arginine degradation II (AST pathway) | 1.41 | 0.0265 | 1.41 | 0.0408 | 0.32 | 0.6855 | 1.03 | 0.0043 |
| D-glucarate degradation I | 0.68 | 0.0265 | 0.71 | 0.0345 | 0.34 | 0.1477 | 0.50 | 0.0030 |
| Superpathway of demethylmenaquinol-8 biosynthesis | 0.72 | 0.0265 | 0.74 | 0.0345 | 0.70 | 0.0065 | 0.56 | 0.0023 |
| Superpathway of menaquinol-11 biosynthesis | 0.68 | 0.0265 | 0.71 | 0.0345 | 0.65 | 0.0085 | 0.53 | 0.0025 |
| Superpathway of menaquinol-12 biosynthesis | 0.68 | 0.0265 | 0.71 | 0.0345 | 0.65 | 0.0085 | 0.53 | 0.0025 |
| Superpathway of menaquinol-13 biosynthesis | 0.68 | 0.0265 | 0.71 | 0.0345 | 0.65 | 0.0085 | 0.53 | 0.0025 |
| Gondoate biosynthesis (anaerobic) | −0.15 | 0.0265 | −0.17 | 0.0317 | −0.07 | 0.1253 | −0.09 | 0.0076 |
| Superpathway of L-aspartate and L-asparagine biosynthesis | −0.16 | 0.0282 | −0.19 | 0.0281 | −0.07 | 0.1338 | −0.09 | 0.0134 |
| Fatty acid β-oxidation I | 0.95 | 0.0282 | 1.04 | 0.0345 | 0.10 | 0.8613 | 0.70 | 0.0060 |
| Superpathway of menaquinol-8 biosynthesis I | 0.65 | 0.0282 | 0.68 | 0.0345 | 0.63 | 0.0092 | 0.51 | 0.0031 |
| Superpathway of menaquinol-7 biosynthesis | 0.65 | 0.0282 | 0.68 | 0.0345 | 0.63 | 0.0092 | 0.51 | 0.0031 |
| Norspermidine biosynthesis | 2.10 | 0.0282 | 2.27 | 0.0345 | 1.18 | 0.1868 | 1.52 | 0.0079 |
| Polymyxin resistance | 1.27 | 0.0282 | 1.13 | 0.0601 | 0.65 | 0.2349 | 0.93 | 0.0060 |
| Cis-vaccenate biosynthesis | −0.14 | 0.0310 | −0.18 | 0.0283 | −0.07 | 0.1220 | −0.08 | 0.0196 |
| Allantoin degradation IV (anaerobic) | 1.60 | 0.0311 | 1.64 | 0.0556 | 0.82 | 0.2761 | 1.16 | 0.0077 |
| Adenosylcobalamin salvage from cobinamide I | −0.20 | 0.0340 | −0.24 | 0.0345 | −0.13 | 0.0561 | −0.14 | 0.0153 |
| Glycogen biosynthesis I (from ADP-D-Glucose) | −0.20 | 0.0340 | −0.24 | 0.0336 | −0.13 | 0.0229 | −0.12 | 0.0175 |
| Adenosylcobalamin biosynthesis from cobyrinate a,c-diamide I | −0.21 | 0.0340 | −0.24 | 0.0345 | −0.13 | 0.0498 | −0.14 | 0.0157 |
| Nitrate reduction VI (assimilatory) | −0.46 | 0.0340 | −0.57 | 0.0271 | −0.36 | 0.0277 | −0.34 | 0.0086 |
| Adenosylcobalamin salvage from cobinamide II | −0.21 | 0.0340 | −0.24 | 0.0345 | −0.13 | 0.0525 | −0.13 | 0.0162 |
| Arginine, ornithine and proline interconversion | 0.47 | 0.0354 | 0.35 | 0.1510 | 0.12 | 0.5778 | 0.34 | 0.0114 |
| Superpathway of 2,3-butanediol biosynthesis | 0.77 | 0.0368 | 1.20 | 0.0021 | 0.30 | 0.3243 | 0.58 | 0.0068 |
| Formaldehyde assimilation II (RuMP Cycle) | 0.63 | 0.0381 | 0.46 | 0.1757 | 0.14 | 0.6759 | 0.46 | 0.0134 |
| Mevalonate pathway I | 0.93 | 0.0381 | 0.98 | 0.0556 | 0.79 | 0.0691 | 0.68 | 0.0125 |
| NAD salvage pathway I | −0.16 | 0.0381 | −0.17 | 0.0556 | −0.08 | 0.1062 | −0.09 | 0.0252 |
| Superpathway of N-acetylneuraminate degradation | 0.25 | 0.0433 | 0.27 | 0.0561 | 0.19 | 0.1157 | 0.22 | 0.0047 |
| Superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 0.88 | 0.0450 | 0.93 | 0.0556 | 0.77 | 0.0706 | 0.65 | 0.0154 |
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform. DESeq2 was used to perform the analysis.
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform and constipation status. DESeq2 was used to perform the analysis.
Included household controls only: PD patient and household control pair was treated as random effect. Adjusted for sex, race, age, and sequencing platform. MaAsLin2 was used to perform the analysis.
Included both community controls and population controls; adjusted for sex, race, age, and sequencing platform. MaAsLin2 was used to perform the analysis.
Coefficient indicates the difference in the log-transformed relative abundances between PD and Control. Log2FC, Log2 fold change; Adj, adjusted.
Microbiome profile and predicted metagenome associated with clinical PD-characteristics
Clinical characteristics of the 93 PD patients are shown in Table 1. Alpha and beta diversity were not associated with PD duration, L-DOPA dose, PD subtypes, or UPDRS III score (data not shown). No specific taxa were found to be associated with patients’ L-DOPA doses. The Verrucomicrobiota phylum was more abundant among PD patients with a PIGD motor subtype, while the Synergistota and Proteobacteria phyla were characteristic of PD patients exhibiting other motor subtypes. Longer PD duration was associated with decreased levels of the Synergistota phylum and significant shifts in six genera: increased levels of Fournierella, DTU089, and Haemophilus, and decreased levels of Pseudomonas, Lactobacillus, and Roseburia. Several Metacyc pathways involved with degradation of gallate, methygallate, catechol and toluene were also decreased in patient with longer PD duration. Additionally, two genera were associated with higher UPDRS III scores: increased level of Lachnospiraceae_NK4B4_group and decreased level of Senegalimassilia (See Table 4).
Table 4.
PD patient only analyses (N = 93)
| PD specific factors | Differentially expressed bacterial taxa/predicted pathways | log2 FoldChange |
Adjusted p |
|
|---|---|---|---|---|
| L-Dopa daily dosage | Phylum | - - - - | - - - - | - - - - |
| Genus | - - - - | - - - - | - - - - | |
| Metacyc pathway | - - - - | - - - - | - - - - | |
| PD Subtypes (PIGD vs. Tremor Dominate + Indeterminate) | Phylum | Proteobacteria | −1.664 | 0.0045 |
| Synergistota | −3.436 | 0.0129 | ||
| Verrucomicrobiota | 1.327 | 0.0129 | ||
| Genus | - - - - | - - - - | - - - - | |
| Metacyc pathway | NAD biosynthesis II (from tryptophan) | −10.991 | 0.0000 | |
| PD Duration | Phylum | Synergistota | −0.417 | 0.0437 |
| Genus | Pseudomonas | −1.799 | <0.0001 | |
| Haemophilus | 0.497 | 0.0005 | ||
| Fournierella | 1.073 | 0.0009 | ||
| DTU089 | 0.540 | 0.0096 | ||
| Roseburia | −0.161 | 0.0155 | ||
| Lactobacillus | −0.269 | 0.0155 | ||
| Metacyc pathway | Gallate degradation I | −1.842 | <0.0001 | |
| Catechol degradation to 2-oxopent-4-enoate II | −1.643 | <0.0001 | ||
| Catechol degradation II (meta-cleavage pathway) | −1.715 | 0.0001 | ||
| Methylgallate degradation | −1.184 | 0.0017 | ||
| Gallate degradation II | −1.167 | 0.0023 | ||
| Toluene degradation IV (aerobic) (via catechol) | −0.769 | 0.0157 | ||
| UDPRS III | Phylum | - - - - | - - - - | - - - - |
| Genus | Senegalimassilia | −0.435 | 0.0002 | |
| Lachnospiraceae_NK4B4_group | 0.491 | 0.0021 | ||
| Metacyc pathway | - - - - | - - - - | - - - - | |
PD, Parkinson’s disease; TD, tremor dominant; PIGD, postural instability and gait dysfunction; UPDRS, Unified Parkinson’s Disease Rating Scale.
DISCUSSION
Over the past decade, almost two dozen studies reported changes in gut microbiota with PD, but few investigated more than 100 patients [15-18]; only six studies recruited household members as controls. Also, findings vary considerably, likely due to small samples sizes, differences in sample collection and storage method, sequencing platforms and analytical protocols employed. The results we present, thus, improve our understanding of the bacterial diversity and taxonomic composition of the gut microbiome in PD and, importantly, we compare patients to both household and community controls. Our study also addresses a knowledge gap on bacterial genera and phyla associated with PD-specific characteristics. Finally, we predicted functional pathways that suggest the involvement of important processes such as the glycan pathways and the generation of cell energy in PD.
Our PD patients generally exhibited a reduced alpha diversity, i.e., less richness and evenness of their gut microbiome profiles compared to household and community controls combined. This is in agreement with two previous studies [19, 20], but counter to studies reporting increased alpha diversity in PD patients [16, 21], or no differences [17, 22, 23]. We also detected a beta-diversity difference (Bray-Curtis dissimilarities) in the microbiome composition of PD patients, which is consistent with most published studies [15-18].
Beyond these measures of global diversity differences in the microbiome, we also found taxa level differences in bacterial abundance generally confirming previous findings. Specifically, we identified three phyla and five genera as more abundant in our PD patients.
Increases in the Verrucomicrobia phylum we identified were driven by the Akkermansia genus, consistent with previous findings [15, 24, 25]. Akkermansia, specifically Akkermansia muciniphila, has gotten attention as a species beneficial to gastrointestinal health and, possibly, a marker of healthy aging [26]. A. muciniphila are mucin-degrading bacteria that can produce short-chain fatty acids (SCFAs), including acetate and propionate, playing a role in maintaining epithelial integrity and regulating immune system and anti-inflammatory responses [27]. Decreased levels of Akkermansia and SCFAs in the gut have been associated with chronic disease conditions (e.g., ulcerative colitis) by affecting the integrity or thickness of the mucus layer and thereby the abundance of A. muciniphila [28].
Thus, finding Akkermansia in PD patients to be more abundant seems paradoxical and requires further investigation, especially as this result has now been replicated multiple times. Enrichment of Akkermansia may result from constipation, a common symptom in PD. Animal studies found proliferation of Akkermansia in unbalanced microbiota, where its mucin-degrading feature depletes the intestinal mucus layer, decreases the number of goblet cells, and causes drier stool [29]. It is also possibly a host response specific to PD, i.e., the gut microbiome reacting to an evolving gut and brain pathology. This bacterium not only degrades mucin but also stimulates mucin production and closely interacts with the host immune system, i.e., Akkermansia induces adaptive immune responses in a homeostatic environment [30]. On the other hand, animal models suggest that the microbiota transplanted from human PD patients into susceptible mouse strains induce PD-like motor dysfunction such as deficits in beam traversal and pole descent tasks, which would support a more causative role for these bacteria [31]. This is in line with our observation of higher Verrucomicrobia abundance in PD patients with PIGD motor subtype. PIGD is a more aggressive phenotype of PD with a highly disabling gait disorder and is believed to be indicative of rapid motor and cognitive deterioration [32]. With the supporting evidence from animal models, it is possible that the clinical presentation of PIGD can be explained by the contribution of gut microbes such as Verrucomicrobia Akkermansia. It is worth noting that several A. muciniphila strains with distinct metabolic and functional features may colonize the same environment [33]. In our study, the majority (83%) of Akkermansia belongs to the A. muciniphila species, yet due to the limitations of short-read 16S rRNA gene sequencing, we cannot distinguish between specific strains of A. muciniphila.
We also found higher abundance of the phyla Proteobacteria and Actinobacteriota in PD patients compared with all controls but not in analyses restricted to household members. Increased level of Proteobacteria in the gut has been associated with dysbiosis. Its role is widely studied in various diseases, including PD, because of its potential immunoregulation ability via the production of lipopolysaccharides (LPS). Gram-negative bacteria are the main source of LPS in humans and are well tolerated in the gut of healthy individuals. In conditions of inflammation, the integrity of the epithelial cells is compromised (also known as the “leaky gut”) and LPS enter the intestinal wall and interact with immune cells triggering the local innate and adaptive immune system [34]. Furthermore, an LPS-triggered immune process can affect the central nervous system via the gut-brain axis, activating microglia and leading to death of dopaminergic neurons [35, 36]. It is worth noting that LPS produced by different bacteria can vary in molecular structure, and not all are considered harmful. Proteobacteria came into focus in PD because there are several highly toxic opportunistic pathogens amongst them such as Escherichia coli, Salmonella, and Vibrio [37, 38]. Only one other study reported higher abundances of Actinobacteria in PD [39]. We speculate that gut inflammation in PD patients creates an oxidative state and is likely to promote colonization of aerotolerant taxa such as Actinobacteria compare to other strictly anaerobic taxa [40]. In our study, however, the differences were sensitive to adjustment for constipation, suggesting that future studies should also take this factor into account.
At a higher taxonomic resolution, we identified four other genera as more abundant in PD patients: UBA1819, DTU089, Hungatella, and Enterococcus. UBA1819 and DTU089, although less often reported in the PD literature, belong to the Ruminococcaceae family that is responsible for the production of the SCFA butyrate considered beneficial to gut epithelial integrity and immunoregulation [41]. However, it is important to note that these differences disappeared in household control pair-matched analyses suggesting that they may reflect characteristics of the PD household rather than being disease influencing features. Associations between Hungatella and PD have been previously reported, but results are not entirely consistent [42, 43]. Enterococcus, as well as the Enterococcaceae family, have been positively associated with PD [22, 44]. However, whether and how these specific genera are related to PD pathogenesis remains elusive.
Among PD patients with longer disease duration, an increase in the genera Fournierella and DTU089 (Ruminococcaceae family) and decrease in Roseburia (Lachnospiraceae family) is consistent with previous reports [15, 16]. We also observed a negative association between the abundance of Lactobacillus and PD duration. Lactobacillus are gut bacteria for which abundance has been associated with several human diseases [45]; however, no consensus about its influence on human health has been reached possibly due to strain and species-specific functional variation. Pseudomonas was decreased in patients with longer disease duration possibly related to reduced aromatic compound degradation pathways attributed to strains/species of Pseudomonas. In addition, we found two genera differentially abundant dependent on UPDRS III scores; however, the increase of Lachnospiraceae genera in patients with higher UPDRS III score is counter to previous reports [15, 22], and the decreased abundance of Senegalimassilia was novel and needs further investigation.
Medication has been suggested to alter the gut microbiome and gut microbes may affect the efficacy of medications [46]. Two studies reported a differential abundance of certain taxa associated with PD medication [21, 47]. However, we did not observe any association between microbial profile and L-DOPA medication doses in PD patients.
Based on predicted metagenomic data, we identified several pathways distinguishing PD patients from controls, and most remained statistically significant in sensitivity analyses adjusting for constipation, but not when we used a pair-matched approach that restricted comparisons to PD-household control sets (N = 100, Table 3). While this might partially reflect the reduction in statistical power, it may also suggest that it is important to distinguish disease related from household-related influences on the microbiome in PD. Several predicted functional shifts are consistent with the idea that the microbiota are a source of metabolites influencing PD pathogenesis or are showing metabolic shifts consistent with a response to host metabolic shifts in PD. For example, a higher abundance of the norspermidine biosynthesis pathway was predicted for the microbiome of our PD patients. Norspermidine is a polyamine (PA) and alterations of its metabolism have been implicated in neuronal degeneration, specifically, acceleration of the aggregation of pathologic α-synuclein [48]. We previously reported that N8-Acetylspermidine in PD patient serum was positively associated with faster progression [49]. Whether this or any other PA of microbial-origin interact with α-synuclein in the gut requires further investigation. We also predicted increased allantoin degradation in PD patients; this is consistent with the detection of lower levels of blood uric acid/urate in PD patients [50]. As allantoin is a major oxidative product of uric acid, this may indicate higher oxidative stress in PD [51]. Higher abundances of several manaquinol biosynthesis super-pathways in PD patients were predicted, which is consistent with a gut microbiome meta-analysis in PD [42]. Menaquinol is used as an electron donor by nitric oxide (NO) reductases to reduce two NO molecules to nitrous oxide (N2O). This may lead to a reduction of nitrative/oxidative stress that damages neurons. We and others have previously shown that nitric oxide synthase gene variants that possibly affect NO balance increase PD risk [50].
Our study, while relatively small, is one of the largest microbiome PD studies in the US. We conducted the study in a rural setting and enrolled patients from the community. The comprehensive data on demographics, medical history, and lifestyle factors allowed for comprehensive confounder assessment, foremost constipation. A strength is our investigation of gut microbiome composition according to clinical features and having two types of controls helped us control for shared household environments that may shape the gut microbiome. Our study has some limitations related to size as it limits confounder control and statistical power for subgroup analyses or identifying less prevalent microbial taxa. Second, the fecal samples were collected at a single time point, which does not allow us to establish temporality or causality, similar to the majority of previous studies. Third, the limited species-level resolution of our sequencing and annotation pipeline may have affected the interpretation of our results. However, this limitation of 16S rRNA-based microbiome studies will only be overcome as the resolution and size of the databases increase, and as sequencing technologies (e.g., full-length sequencing, shotgun sequencing) improve and become more affordable. Lastly, the metagenomic pathways were based on predicted metagenomic data using existing reference genomes and may not reflect the actual metagenome.
In conclusion, we found that PD patients have lower microbiota diversity and that microbial composition differed in three phyla and five genera resulting in some interesting pathways predicted to be different. Additionally, these differences extended to disease duration, motor subtypes and motor function scores. We are confirming some previous findings and added novel insights into what may drive some differences (constipation) and potential microbiome differences by PD subtype and duration.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Environmental Health Sciences of the National Institutes of Health (grants numbers: R01 ES031106, R01 ES010544, U54-ES012078, P01-ES016732, P50-NS038367, and initial pilot funding P30-ES07048), the American Parkinson’s disease Association (grant number 20161386), Parkinson Alliance (2018 and 2019 Grant), Burroughs Wellcome Fund Interschool Training Program in Chronic Diseases, and Toffler Scholar Award.
Footnotes
CONFLICT OF INTEREST
The authors have no competing interests to declare that are relevant to the content of this article.
SUPPLEMENTARY MATERIAL
The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JPD-223500.
REFERENCES
- [1].Khoo TK, Yarnall AJ, Duncan GW, Coleman S, O’Brien JT, Brooks DJ, Barker RA, Burn DJ (2013) The spectrum of nonmotor symptoms in early Parkinson disease. Neurology 80, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pellegrini C, Antonioli L, Colucci R, Blandizzi C, Fornai M (2018) Interplay among gut microbiota, intestinal mucosal barrier and enteric neuro-immune system: A common path to neurodegenerative diseases? Acta Neuropathol 136, 345–361. [DOI] [PubMed] [Google Scholar]
- [3].Shannon KM, Keshavarzian A, Mutlu E, Dodiya HB, Daian D, Jaglin JA, Kordower JH (2012) Alpha-synuclein in colonic submucosa in early untreated Parkinson’s disease. Mov Disord 27, 709–715. [DOI] [PubMed] [Google Scholar]
- [4].Braak H, Rüb U, Gai WP, Del Tredici K (2003) Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm (Vienna) 110, 517–536. [DOI] [PubMed] [Google Scholar]
- [5].Svensson E, Horvath-Puho E, Thomsen RW, Djurhuus JC, Pedersen L, Borghammer P, Sorensen HT (2015) Vagotomy and subsequent risk of Parkinson’s disease. Ann Neurol 78, 522–529. [DOI] [PubMed] [Google Scholar]
- [6].Barbut D, Stolzenberg E, Zasloff M (2019) Gastrointestinal immunity and alpha-synuclein. J Parkinsons Dis 9, S313–S322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Duarte Folle A, Paul KC, Bronstein JM, Keener AM, Ritz B (2019) Clinical progression in Parkinson’s disease with features of REM sleep behavior disorder: A population-based longitudinal study. Parkinsonism Relat Disord 62, 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liew Z, Wang A, Bronstein J, Ritz B (2014) Job exposure matrix (JEM)-derived estimates of lifetime occupational pesticide exposure and the risk of Parkinson’s disease. Arch Environ Occup Health 69, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ritz BR, Paul KC, Bronstein JM (2016) Of pesticides and men: A California story of genes and environment in Parkinson’s disease. Curr Environ Health Rep 3, 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, Langille MGI (2020) PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38, 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stebbins GT, Goetz CG, Burn DJ, Jankovic J, Khoo TK, Tilley BC (2013) How to identify tremor dominant and postural instability/gait difficulty groups with the movement disorder society unified Parkinson’s disease rating scale: Comparison with the unified Parkinson’s disease rating scale. Mov Disord 28, 668–670. [DOI] [PubMed] [Google Scholar]
- [13].Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nearing JT, Douglas GM, Hayes MG, MacDonald J, Desai DK, Allward N, Jones CMA, Wright RJ, Dhanani AS, Comeau AM, Langille MGI (2022) Microbiome differential abundance methods produce different results across 38 datasets. Nat Commun 13, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hill-Burns EM, Debelius JW, Morton JT, Wissemann WT, Lewis MR, Wallen ZD, Peddada SD, Factor SA, Molho E, Zabetian CP, Knight R, Payami H (2017) Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov Disord 32, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Barichella M, Severgnini M, Cilia R, Cassani E, Bolliri C, Caronni S, Ferri V, Cancello R, Ceccarani C, Faierman S, Pinelli G, De Bellis G, Zecca L, Cereda E, Consolandi C, Pezzoli G (2019) Unraveling gut microbiota in Parkinson’s disease and atypical parkinsonism. Mov Disord 34, 396–405. [DOI] [PubMed] [Google Scholar]
- [17].Cirstea MS, Yu AC, Golz E, Sundvick K, Kliger D, Radisavljevic N, Foulger LH, Mackenzie M, Huan T, Finlay BB, Appel-Cresswell S (2020) Microbiota composition and metabolism are associated with gut function in Parkinson’s disease. Mov Disord 35, 1208–1217. [DOI] [PubMed] [Google Scholar]
- [18].Wallen ZD, Appah M, Dean MN, Sesler CL, Factor SA, Molho E, Zabetian CP, Standaert DG, Payami H (2020) Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Petrov VA, Saltykova IV, Zhukova IA, Alifirova VM, Zhukova NG, Dorofeeva YB, Tyakht AV, Kovarsky BA, Alekseev DG, Kostryukova ES, Mironova YS, Izhboldina OP, Nikitina MA, Perevozchikova TV, Fait EA, Babenko VV, Vakhitova MT, Govorun VM, Sazonov AE (2017) Analysis of gut microbiota in patients with Parkinson’s disease. Bull Exp Biol Med 162, 734–737. [DOI] [PubMed] [Google Scholar]
- [20].Weis S, Schwiertz A, Unger MM, Becker A, Fassbender K, Ratering S, Kohl M, Schnell S, Schafer KH, Egert M (2019) Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Parkinsons Dis 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qian Y, Yang X, Xu S, Wu C, Song Y, Qin N, Chen SD, Xiao Q (2018) Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav Immun 70, 194–202. [DOI] [PubMed] [Google Scholar]
- [22].Pietrucci D, Cerroni R, Unida V, Farcomeni A, Pierantozzi M, Mercuri NB, Biocca S, Stefani A, Desideri A (2019) Dysbiosis of gut microbiota in a selected population of Parkinson’s patients. Parkinsonism Relat Disord 65, 124–130. [DOI] [PubMed] [Google Scholar]
- [23].Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P (2015) Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord 30, 350–358. [DOI] [PubMed] [Google Scholar]
- [24].Bedarf JR, Hildebrand F, Coelho LP, Sunagawa S, Bahram M, Goeser F, Bork P, Wullner U (2017) Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson’s disease patients. Genome Med 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Heintz-Buschart A, Pandey U, Wicke T, Sixel-Doring F, Janzen A, Sittig-Wiegand E, Trenkwalder C, Oertel WH, Mollenhauer B, Wilmes P (2018) The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov Disord 33, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W (2010) Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5, e10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Derrien M, Belzer C, de Vos WM (2017) Akkermansia muciniphila and its role in regulating host functions. Microb Pathog 106, 171–181. [DOI] [PubMed] [Google Scholar]
- [28].Geerlings SY, Kostopoulos I, de Vos WM, Belzer C (2018) Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC (2016) A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR, Brown EM, Graham DB, Xavier RJ, Moon JJ, Barton GM (2019) Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK (2016) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480 e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Skidmore FM, Monroe WS, Hurt CP, Nicholas AP, Gerstenecker A, Anthony T, Jololian L, Cutter G, Bashir A, Denny T, Standaert D, Disbrow EA (2022) The emerging postural instability phenotype in idiopathic Parkinson disease. NPJ Parkinsons Dis 8, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guo X, Li S, Zhang J, Wu F, Li X, Wu D, Zhang M, Ou Z, Jie Z, Yan Q, Li P, Yi J, Peng Y (2017) Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genomics 18, 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Candelli M, Franza L, Pignataro G, Ojetti V, Covino M, Piccioni A, Gasbarrini A, Franceschi F (2021) Interaction between lipopolysaccharide and gut microbiota in inflammatory bowel diseases. Int J Mol Sci 22, 6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shannon KM (2022) Gut-derived sterile inflammation and Parkinson’s disease. Front Neurol 13, 831090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Deng I, Corrigan F, Zhai G, Zhou XF, Bobrovskaya L (2020) Lipopolysaccharide animal models of Parkinson’s disease: Recent progress and relevance to clinical disease. Brain Behav Immun Health 4, 100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A (2017) Proteobacteria: A common factor in human diseases. Biomed Res Int 2017, 9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, Group DS, Xavier RJ (2016) Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 1551. [DOI] [PubMed] [Google Scholar]
- [39].Vascellari S, Palmas V, Melis M, Pisanu S, Cusano R, Uva P, Perra D, Madau V, Sarchioto M, Oppo V, Simola N, Morelli M, Santoru ML, Atzori L, Melis M, Cossu G, Manzin A (2020) Gut microbiota and metabolome alterations associated with Parkinson’s disease. mSystems 5, e00561–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ni J, Wu GD, Albenberg L, Tomov VT (2017) Gut microbiota and IBD: Causation or correlation? Nat Rev Gastroenterol Hepatol 14, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Morrison DJ, Preston T (2016) Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Romano S, Savva GM, Bedarf JR, Charles IG, Hildebrand F, Narbad A (2021) Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jin M, Li J, Liu F, Lyu N, Wang K, Wang L, Liang S, Tao H, Zhu B, Alkasir R (2019) Analysis of the gut microflora in patients with Parkinson’s disease. Front Neurosci 13, 1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hopfner F, Kunstner A, Muller SH, Kunzel S, Zeuner KE, Margraf NG, Deuschl G, Baines JF, Kuhlenbaumer G (2017) Gut microbiota in Parkinson disease in a northern German cohort. Brain Res 1667, 41–45. [DOI] [PubMed] [Google Scholar]
- [45].Heeney DD, Gareau MG, Marco ML (2018) Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr Opin Biotechnol 49, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A (2018) Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Palacios N, Hannoun A, Flahive J, Ward D, Goostrey K, Deb A, Smith KM (2021) Effect of levodopa initiation on the gut microbiota in Parkinson’s disease. Front Neurol 12, 574529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krasnoslobodtsev AV, Peng J, Asiago JM, Hindupur J, Rochet JC, Lyubchenko YL (2012) Effect of spermidine on misfolding and interactions of alpha-synuclein. PLoS One 7, e38099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Roede JR, Uppal K, Park Y, Lee K, Tran V, Walker D, Strobel FH, Rhodes SL, Ritz B, Jones DP (2013) Serum metabolomics of slow vs. rapid motor progression Parkinson’s disease: A pilot study. PLoS One 8, e77629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cipriani S, Chen X, Schwarzschild MA (2010) Urate: A novel biomarker of Parkinson’s disease risk, diagnosis and prognosis. Biomark Med 4, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Martinez-Moral M-P, Kannan K (2019) Allantoin as a marker of oxidative stress: Inter- and intraindividual variability in urinary concentrations in healthy individuals. Environ Sci Technol Lett 6, 283–288. [Google Scholar]
- [52].Paul KC, Sinsheimer JS, Rhodes SL, Cockburn M, Bronstein J, Ritz B (2016) Organophosphate pesticide exposures, nitric oxide synthase gene variants, and gene-pesticide interactions in a case-control study of Parkinson’s Disease, California (USA). Environ Health Perspect 124, 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.