Abstract
The cell wall is essential to nearly every aspect of the biology and pathogenicity of Candida albicans. Although it was intially considered an almost inert cellular structure that protected the protoplast against osmotic offense, more recent studies have demonstrated that it is a dynamic organelle. The major components of the cell wall are glucan and chitin, which are associated with structural rigidity, and mannoproteins. The protein component, including both mannoprotein and nonmannoproteins, comprises some 40 or more moieties. Wall proteins may differ in their expression, secretion, or topological location within the wall structure. Proteins may be modified by glycosylation (primarily addition of mannose residues), phosphorylation, and ubiquitination. Among the secreted enzymes are those that are postulated to have substrates within the cell wall and those that find substrates in the extracellular environment. Cell wall proteins have been implicated in adhesion to host tissues and ligands. Fibrinogen, complement fragments, and several extracellular matrix components are among the host proteins bound by cell wall proteins. Proteins related to the hsp70 and hsp90 families of conserved stress proteins and some glycolytic enzyme proteins are also found in the cell wall, apparently as bona fide components. In addition, the expression of some proteins is associated with the morphological growth form of the fungus and may play a role in morphogenesis. Finally, surface mannoproteins are strong immunogens that trigger and modulate the host immune response during candidiasis.
Candida albicans is a serious agent of infection, particularly in immunocompromised patients. The delicate balance between the host and this otherwise normal commensal fungus may turn into a parasitic relationship, resulting in the development of infection, called candidiasis. The nature and extent of the impairment of normal host defense influence the manifestation and severity of infection. In general, superficial mucocutaneous candidiasis is frequent in patients with T-cell deficiencies, such as AIDS patients. The more serious, life-threatening, deep-seated or disseminated candidiasis is normally found in a spectrum of severely immunocompromised patients (29, 390). The fungus is not a mere passive participant in the infectious process, and a hypothetical set of virulence factors for C. albicans has been proposed and supported by various studies. These fungal attributes include the production of secreted hydrolytic enzymes, dimorphic transition (morphogenetic conversion from budding yeast to the filamentous growth form or hypha), antigenic variability, the ability to switch between different cell phenotypes, adhesion to inert and biological substrates, and immunomodulation of host defense mechanisms (for a review of these topics, see reference 96).
Initially, the cell wall was considered an almost inert structure that supplies rigidity and protection to the protoplast. Today, the cell wall is well established as being essential to almost every aspect of the biology and pathogenicity of C. albicans (64). The cell wall acts as a permeability barrier and is the structure that maintains the characteristic shape of the fungus. Also, as the most external part of the cell, the wall mediates the initial physical interaction between the microorganism and the environment, including the host. For these reasons, the cell wall of C. albicans is the focus of study by numerous research groups. Their objectives are the elucidation of both basic biological processes and functional mechanisms regulating the synthesis, organization, and environmental interactions of this complex macromolecular structure. Proteins have been implicated in most of the cell wall functions. Extensive reviews exist on different aspects of the cell wall of C. albicans (64, 478–482, 491, 493); however, recent reviews that focus on proteins have been limited primarily to the function of proteins as adhesins (49, 50, 110, 111, 151, 209, 406). This review focuses on both general characteristics of the cell wall and secreted proteins and specific aspects of individual proteins.
Although hydrolytic enzymes such as acid phosphatase were examined previously, studies on the identity and function of protein components began in the early to mid 1980s with studies in 1983 by Chaffin and Stocco (71), in 1985 by Elorza et al. (125) and Sundstrom and Kenny (524), and in 1986 by Pontón and Jones (414). In the first part of this decade, there has been an explosive growth in the number of studies of the cell wall proteins. These studies have been driven by presumed virulence functions of specific proteins and fueled by the realization that this is a complex, dynamic “organelle.” Out of such impetus has come the identification of specific proteins not yet associated with specific pathogenic function and observations with more general import for the cell and cell wall proteins. This increasing knowledge of the protein component of the cell wall may result in a better understanding of the pathogenic mechanisms of the fungus and also may contribute to the design of innovative therapeutic regimens and diagnostic procedures (175, 333). At times, these studies have revealed several surprises and unexpected findings, which only add more attraction to the study of this fascinating microorganism. In writing this review, we have focused on the protein component with three objectives: (i) to summarize general aspects of proteins; (ii) to summarize studies on specific proteins or protein families; and (iii) to consider the implications, unanswered questions, and future research directions suggested by these studies.
Cell Wall and Morphology
Although the terms “dimorphism” and “dimorphic fungus”, i.e., existing in two morphological forms, are well established and commonly accepted when referring to C. albicans, strictly speaking this fungus has the ability to adopt a spectrum of morphologies, and thus C. albicans could be considered a “polymorphic” or “pleomorphic” organism (244, 390). Since changes in the cell wall determine the shape of the whole fungal cell, the cell wall is the structure ultimately responsible for a given morphology. C. albicans can reproduce by budding, giving rise to the formation of yeast cells (also designated blastospores or blastoconidia). The production of germ tubes results in the conversion to a filamentous growth phase or hypha, also called the mycelial form. The formation of pseudohyphae occurs by polarized cell division when yeast cells growing by budding have elongated without detaching from adjacent cells. Under certain nonoptimal growing conditions, C. albicans can undergo the formation of chlamydospores, which are round, refractile spores with a thick cell wall. These morphological transitions often represent a response of the fungus to changing environmental conditions and may permit the fungus to adapt to different biological niches. The transition from a commensal to a pathogenic lifestyle may also involve changes in environmental conditions and dispersion within the human host. The ultrastructure, composition, and biological properties of the cell wall are affected by these morphological changes (64). Although progress has been achieved in the recent years, the molecular mechanisms governing these morphogenetic conversions are still not fully understood, partly due to the difficulty of genetic manipulations in this fungus (274, 275, 474). Recent reports that may herald rapid advances in this area have identified transcriptional regulatory genes, a general transcriptional repressor TUP1 (38), a putative transcriptional factor RBF1 (220), and a myc-like transcriptional factor EFG1 (516) that affect cellular morphology when their expression is altered. Most of the observations from these studies have been incorporated by Magee (310) into a model for the regulation of pseudohyphal growth.
Cell Wall and Interactions with the Host
Two major aspects of the host-parasite interactions are the adhesion of C. albicans cells to host cells and tissues and the immunomodulation of the host immune response.
Adhesion is a prerequisite for colonization and an essential step in the establishment of infection. C. albicans adheres to epithelial cells, endothelial cells, soluble factors, extracellular matrix, and inert materials implanted in the body of the host. Multiple adherence mechanisms appear to be used by C. albicans cells (49, 50, 110, 111, 151, 209, 252, 406). Physical interactions of this fungus with the host are mediated at the cell surface, and cell wall constituents implicated in binding have been designated adhesins (49). The large repertoire of adhesins displayed by this fungus may reflect the variety of host sites that it can invade (49, 50, 110, 111, 209). Specific characteristics of individual cell wall moieties participating in adhesion events are discussed later in this review.
Another important aspect of interactions with the host, with direct implications for pathogenesis, is the potential of this fungus to modulate the immune response mounted by the host (64, 96, 107). The capacity of cell wall constituents, including glucan, chitin, and mannoproteins, to modulate (activate or depress) the immune response is well documented (11, 64). Mannans and mannoproteins display the most potent immunomodulatory activity, being able to regulate the action of virtually all arms of the immune system (natural killer cells, phagocytic cells, cell-mediated immunity, and humoral mechanisms) (64, 107, 402, 412). Although individual cell wall moieties with immunomodulatory properties are described below, readers are referred to excellent reviews on this topic (11, 64, 107).
CELL WALL COMPOSITION AND ORGANIZATION
Composition
Approximately 80 to 90% of the cell wall of C. albicans is carbohydrate. Three basic constituents represent the major polysaccharides of the cell wall: (i) branched polymers of glucose containing β-1,3 and β-1,6 linkages (β-glucans); (ii) unbranched polymers of N-acetyl-d-glucosamine (GlcNAc) containing β-1,4 bonds (chitin); and (iii) polymers of mannose (mannan) covalently associated with proteins (glyco[manno]proteins). In addition, cell walls contain proteins (6 to 25%) and minor amounts of lipid (1 to 7%) (50, 64, 490, 493).
The microfibrillar polymers (β-glucans and chitin) represent the structural components of the wall. They form a rigid skeleton that provides strong physical properties to the cell. From a quantitative point of view, β-glucans are the main constituent, accounting for 47 to 60% by weight of the cell wall. Chitin is a minor (0.6 to 9%) but important component of the C. albicans wall, particularly of the septa between independent cell compartments, budding scars, and the ring around the constriction between mother cell and bud (126, 360).
On the other hand, mannose polymers (mannan), which do not exist as such but are found in covalent association with proteins (mannoproteins), represent about 40% of the total cell wall polysaccharide and are the main material of the cell wall matrix (50, 64, 480, 490, 493). The term “mannan” has been used also to refer to the main soluble immunodominant component present in the outer cell wall layer of C. albicans, called phosphomannoprotein or phosphopeptidomannan complex. This cell wall fraction contains homopolymers of d-mannose (as the main component), 3 to 5% protein, and 1 to 2% phosphate (436). The general features of cell wall mannoproteins in C. albicans are basically identical to those found for Saccharomyces cerevisiae, one of the most thoroughly investigated yeasts in this regard. Several studies have resulted in a detailed knowledge of the structure of this cell wall constituent in C. albicans (12, 262–265, 494–498). Thus, mannose polymers are linked to the protein moiety through asparagine (by N-glycosidic bonds through two GlcNAc [di-N-acetylchitobiose] residues) and threonine or serine (by O-glycosidic, alkali-labile linkages) residues. The N-glycosidically linked carbohydrate is composed of backbone chains of α-1,6-linked mannopyranosyl residues to which oligosaccharide side chains are attached. The side chain mannopyranosyl residues contain α-1,2, α-1,3, β-1,2, β-1,6, and phosphodiester linkages as well as branches (α-1,6) that are oversynthesized under acidic growth conditions (150, 152, 261–267, 494–498). The O-glycosidically-linked sugar component consists of single mannose residues and short, unbranched mannose oligosaccharides (412). Several studies raise the question of additional sugars present in cell wall constituents. These observations include the following: (i) not all proteinaceous moieties present in cell wall extracts from this fungus react with concanavalin A, a lectin recognizing α-mannosylpyranose, or with polyclonal and monoclonal antibodies that recognize other mannan epitopes, such as factor 6, a mannooligosaccharide that confers serotype A specificity (57); (ii) differences in glycosylation and in sensitivity to neuraminidase have been detected in candidal receptors for complement (4, 563); and (iii) treatment with neuraminidase affects the electrostatic surface properties of C. albicans as detected with a fluorescent probe (227). As suggested in these studies, the observations raise the possibility that additional sugars are cell wall constituents. However, the observations could reflect the existence of contaminating proteases in the glycosidase preparation. Sugar residues other than mannose may define either additional functional or antigenic motifs or both in cell wall glycoproteins.
The percent composition of walls from yeast cells and filamentous forms are similar, although the relative amounts of β-glucans, chitin, and mannan vary according to the C. albicans growth form considered (50, 480, 491). Hyphal cells contain at least three times as much chitin as yeast cells do (77, 127, 518). Chitin is the first polymer to appear in regenerating protoplasts (124, 375). Although the ratio of β-1,3- to β-1,6-glucan in the insoluble fraction is similar in yeast and hyphal cells, the insoluble glucan in the initial period of germ tube formation contains considerably more β-1,3 linkages than that found in yeast and mature hyphal cells (518). The literature contains several reports on the identification of morphology-specific proteins and mannoproteins that are discussed later in this review.
Organization
The different cell wall components interact with each other to give rise to the overall architecture of the cell wall. Besides hydrogen and hydrophobic bonds, there is also experimental evidence for the presence of covalent linkages between different components (453, 482). Surarit et al. (527) reported the presence of glycosidic linkages between glucan and chitin in the nascent wall of C. albicans. Recent evidence indicates that mannoproteins may also establish covalent associations with β-glucans (237, 238, 462, 463). It is suggested that β-1,3- and β-1,6-glucans are linked to proteins by phosphodiester linkages, a process that may involve the participation of a GPI (glycosyl phosphatidylinositol) anchor (238) (see below). Protein and mannoprotein species that are released only after digestion of the glucan cell wall network with β-glucanases may play a key role in configuring the final cell wall structure characteristic of each growth form (yeast and mycelium) of C. albicans (453, 479–482). Interactions between glyco(manno)proteins and chitin also appear to exist in the wall of C. albicans cells as deduced from two lines of evidence: (i) chitinase treatment of isolated cell walls solubilizes protein moieties, and (ii) the kinetics of incorporation of protein and mannoprotein constituents into the walls of regenerating protoplasts is altered in the presence of nikkomycin, an antibiotic that blocks chitin synthesis (124, 319).
Cell wall architecture has been studied most extensively in S. cerevisiae and is likely to be a model for C. albicans since there are some similar observations, in particular sensitivity to enzymatic digestion, glucan-mannoprotein linkages, and candidate proteins, that fit the same model (237, 238, 246, 247, 268, 462, 463). In a very recent study, Kollár et al. (268) detected the presence of material containing all four major cell wall components, β-1,3-glucan, β-1,6-glucan, chitin, and mannoprotein. Their analysis indicated that β-1,6-glucan has some β-1,3-glucan branches that may be linked to the reducing end of chitin. The β-1,6-glucan and mannoprotein are attached through a remnant of the mannoprotein GPI anchor. Reducing ends of β-1,6-glucan may also be attached to the nonreducing end of β-1,3-glucan. The proportion of cell wall polysaccharide involved in this type of structure is not clear. The following cell wall building block, where the linkages are indicated by the long dashes, is proposed (247, 268): Mannoprotein—GPI remnant—β-1,6-glucan—β-1,3-glucan—chitin. The authors point out that these linkages are likely to be formed in the periplasmic space as a common end of the individual biosynthetic pathways. Chitin and β-1,3-glucan are synthesized at the plasma membrane and extruded into the periplasm, mannoprotein is synthesized in the cytoplasm and transported through the secretory pathway, and β-1,6-glucan synthesis may occur partially in the endoplasmic reticulum or Golgi complex (268). Not all components are necessarily present in a complex; therefore, the authors suggest that more chitin may be present in inner cell wall layers and more mannoprotein may be present in the outer layers.
Layering.
Since polysaccharides are poorly reactive to the ordinary fixatives and stains used for transmission electron microscopy, only a few well-defined ultrastructural details are obtained by conventional protocols (Fig. 1A and B). However, transmission electron microscopy studies performed with more special techniques or with cytochemical stains and contrasting agents show several layers in the cell wall of C. albicans (Fig. 1C to F). The appearance of these layers is variable and seems to be related to the strain examined, growth conditions, morphology, and preparation of the specimens (50, 64, 422). Thus, there is no consensus about the number of layers present in the cell wall. Different authors have reported the presence of three to eight different layers (28, 64, 186, 421, 438). The outer cell wall layer appears as a dense network with a fibrillar or flocculent aspect (64, 480), whereas the inner wall layer appears contiguous with the plasmalemma with extensive membrane invaginations involved in anchoring of the cell wall to the membrane (192, 276). The microfibrillar polysaccharides glucan and chitin, the components that supply rigidity to the overall wall structure, appear to be more concentrated in the inner cell wall layer, adjacent to the plasma membrane. In contrast, proteins and mannoproteins appear to be dominant in the outermost cell wall layer (Fig. 1B), although they are also present through the entire wall and at the inner regions of the cell wall. Some of the latter proteins may be covalently associated with glucans. Evidence from several cytochemical and cytological studies indicate that the cell wall layering may be due to the distribution of mannoproteins at various levels within the wall structure (64). In any case, it seems clear that layering may be the result of quantitative differences in the proportions of the individual wall components (β-glucans, chitin, and mannoproteins) in each layer rather than of qualitative differences (389).
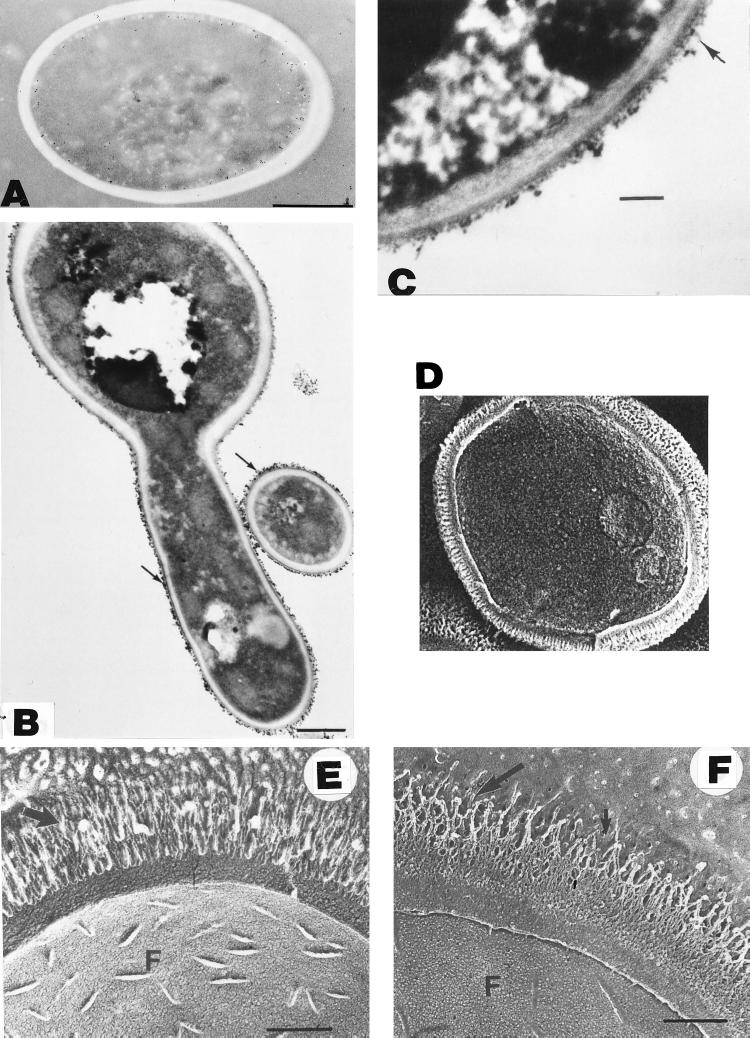
FIG. 1.
Cell wall structure. (A) Transmission electron micrograph of a section of a C. albicans cell prepared by freeze-substitution, showing the cell wall as a thick, electron-dense, homogeneous structure. The presence of distinct layers was not evident in this preparation. Bar, 1 μm. Reprinted from reference 3 with permission of the publisher. (B) Thin sections of cells treated with gold-conjugated concanavalin A, showing an intense labeling with gold particles of the external wall surface. The surface exhibits a fibrillar appearance (arrows), suggesting that concanavalin A-reactive cell wall components, i.e., mannoproteins, are particularly abundant at the most external wall layers. The remaining wall structure also appeared as a homogenous structure in this transmission electron micrograph. Bar, 0.5 μm. Reprinted from reference 549 with permission of the American Society for Microbiology. (C) Other procedures for transmission electron microscopy examination of thin sections of C. albicans cells revealed more clearly the presence of an outer floccular layer (arrow) and showed that the remaining cell wall structure is not homogeneous and that some layering exists. Bar, 200 nm. Reprinted from reference 240 with permission of the publisher. (D to F) Complexity of the wall ultrastructure and presence of distinct layers in the cell wall of C. albicans as revealed by different scanning electron microscopy-based procedures such as cryo-scanning electron microscopy (D) and freeze-fracture, freeze-etch analysis (E and F). The presence of well-ordered, regularly arranged, radiating fibrils in the outer layer is particularly evident in the micrographs shown in panels E (hydrophilic cells) and F (hydrophobic cells). Bar, 0.3 μm in both panels. Panel D reprinted from reference 246 with permission of the publisher. Panels E and F reprinted from reference 191 with permission of the publisher.
Fimbriae.
The outer cell wall layer that is composed mainly of mannoproteins appears as a dense network of radially projecting fibrils (28, 64), designated fimbriae (154, 583). These fibrils extend for 100 to 300 nm (190, 276) and are approximately 5 nm in diameter (28). Both filamentous forms and blastospores exhibit this characteristic feature (28). C. albicans fimbriae consist of many subunits assembled through noncovalent hydrophobic interactions (583). The major structural subunit of fimbriae is a glycoprotein with an apparent molecular mass of 66 kDa, while the unglycosylated protein has an approximate molecular mass of 8.64 kDa (583). In crude extracts, in addition to the 66-kDa moiety, components migrating with an electrophoretic mobility equivalent to proteins of 54, 47, and 39 kDa reacted with monoclonal antibodies (MAbs) raised against purified fimbriae, suggesting the presence of species with differing degrees of glycosylation (585). The hydrophobic status of the cells profoundly affects fimbrial structure. Hydrophilic cells have long, compact, evenly distributed fibrils, while hydrophobic cells have short, blunt fibrils (190) (Fig. 1E and F). The overall hydrophilic status may be due to masking of hydrophobic components by hydrophilic surface fibrils (190). Fimbrial components mediate the adherence of C. albicans to glycosphingolipid receptors on human epithelial cells (279, 583, 584, 586) as discussed later.
Celerin et al. (67, 68) reported that in the fungus Mycrobotryum violaceum, fimbriae are composed of a protein with strong similarity to collagen. The fimbrial protein from C. albicans discussed above does not appear to be related to this collagenous fimbrial protein (68). However, this type of collagenous fimbria appears to be conserved among fungal species since antiserum to the protein domain reacts with surface proteins of many fungi (67). The antiserum also reacts with 81- and 84-kDa surface moieties that may represent a second putative candidal fimbria (67). Although some fungi contain more than one type of fimbria (577), there is no additional evidence for multiple types of fimbriae in C. albicans. If such collagenous fimbriae are found in C. albicans, additional fimbria-mediated interactions of the microorganism with the host cells and tissues may be possible (see below).
CELL WALL PROTEINS
Cell Wall versus Secreted Proteins
Should location or function determine the classification of a cell wall protein or secreted protein? Are proteins either one or the other? Proteins that are found in the in vitro growth medium are often called secreted or extracellular proteins. To reach this location, these proteins travel through the cell wall, where they coexist with cell wall-bound moieties and by location are proteins that contribute the total cell wall proteinaceous component. However, it seems reasonable to consider transiently associated proteins with an environmental destination as being secreted. On the other hand, how do we classify a protein when the association does not appear to be transient or when the postulated function of the protein is within the cell wall? There are proteins that are cell associated under one growth condition and secreted under another (519). Cytochemical detection of phospholipase activity shows a localized cell wall location in cells grown on yeast extract medium and a development of a more generalized cell wall, cell surface, and secreted localization in cells in contact with the chorioallantoic membrane (425, 427). There are enzymes recovered from culture supernatants whose functions are thought to be cell wall biosynthesis and remodeling (75). For most proteins, extracellular locations have not all been examined, so that some proteins that are reported as cell wall associated might also be found in culture fluids if examined. Several cell wall components that are not thought to be secreted have been detected in supernatants of C. albicans cell cultures (2, 178, 299, 536, 543). The relationship of some of these moieties with the cell wall structure is unclear. They may come from the outer wall layers. Alternatively, they may be released by lysed cells or as a consequence of the controlled degradation of the cell wall structure, required for wall expansion during growth. One of the criteria that has been used to demonstrate a cell surface location is binding of a ligand or antibody. When similar observations are made with extracellular proteins such as secreted acid proteinase (397) or phospholipase (425, 427), should this finding be differently interpreted, particularly when there may be a function for the cell associated protein (147, 433)? This discussion makes clear that for some proteins, classification as cell wall or secreted may be dependent upon the growth conditions of the organism, the conditions under which localization has been examined, and our view of the function of the protein. As the mechanisms of targeting proteins to subcellular locations are elucidated and protein functions more completely examined, these issues should be resolved. In this review, we have included both proteins whose function is thought to be in the cell wall and those whose role is thought be primarily extracellular.
In the next four sections, we consider general questions of cell wall protein extraction, protein composition of the extracts, protein modification, and distribution of proteins within the cell wall. In these studies, the emphasis has been on definition of cell wall protein by location, since the principal concern has been the removal of a cytoplasmic contribution to the extract. In the following sections, we review specific proteins that have been grouped by various functional and identity relationships.
Extraction
Different techniques have been used to extract cell wall components of C. albicans. These include physical, chemical, and enzymatic methods and a combination of them. The choice of extracting reagents and techniques, the sequence of extraction methods, and the use of either intact cells or purified cell walls as the starting material may affect both qualitative and quantitative solubilization of cell wall components. In general, and due to the insolubility of both chitin and glucans, sequential alkali and acid treatments are required to effect their extraction (140, 141). In early studies, mannans were extracted from whole cells or isolated cell walls by alkali treatment and further precipitated with Fehling’s solution as a copper complex (408). A milder procedure involved mannan extraction from cells resuspended in citrate buffer (pH 7) by autoclaving and further purification by precipitation with Fehling’s solution (405) or with Cetavlon (66, 376, 392). This topic is covered more extensively in other reviews (141).
Proteinaceous components have been extracted or solubilized from the cell wall of C. albicans by a variety of techniques. Most of the studies have involved either detergents (such as sodium dodecyl sulfate or n-octylglucoside), reducing agents (such as dithiothreitol [DTT] and β-mercaptoethanol [βME]), or hydrolases (such as proteases, Zymolyase, or other β-glucanases, and chitinases) to release proteins from both isolated cell walls and intact cells (57, 59, 61, 71, 78, 122, 125, 319, 365, 414, 508, 524, 525). These reagents have been used alone or in combination. Although sulfhydryl compounds such as βME appear to be less efficient that hydrolases such as Zymolyase in releasing cell wall-bound proteins and glycoproteins (295), these chemical agents solubilize a complex array of proteinaceous components from the walls of intact C. albicans cells (57, 58). On the other hand, some β-glucanases used to solubilize cell wall moieties actually are enzymatic complexes that may contain other unidentified or uncontrolled hydrolytic activities, which may alter the native characteristics of the released molecules.
Other extraction procedures have been less frequently reported and include chemical, enzymatic, and physical methods alone or in combination. β-Elimination with NaOH has been used to release putative structural proteins (369). Ethylenediamine has also been used in structural studies to extract proteins (455). Salt (NaCl) was used to extract the surface determinant of a MAb (40) and a surface adhesin (225). Homogenization has been used to shear fimbriae (583), and α-mannosidase treatment followed by sonication has been used to release wall antigens (195).
There has been little comparison of the various methods used to extract the wall proteins. Casanova and Chaffin (57) compared five extracts for yeast cells and germ tubes: (i) βME extract of intact cells at alkaline pH; (ii) the Zymolyase extract of the treated cells; (iii) βME extract of isolated cell walls at alkaline pH; (iv) the Zymolyase extract of the treated cell walls; and (v) a sodium dodecyl sulfate (SDS)-βME extract of isolated cell walls. The extracts were examined by blotting with concanavalin A, two MAbs (MAb 4C12 to a high-molecular-weight component of germ tubes [59] and MAb 24.17 to a mannan epitope of a high-molecular-weight component [72]) and antiserum for factor 6. The authors concluded that the two sequential extracts obtained from intact cells were most satisfactory. In addition, although extraction with reducing agents is frequently thought to release medium to small components from the cell wall, this study showed that βME also released the high-molecular-weight components. These appeared to be larger than the same component present in Zymolyase extracts. βME and other reducing reagents are believed to solubilize mainly components associated with the outermost layers of the cell wall (50, 61, 295). These reagents also increase cell wall porosity and facilitate subsequent action of cell wall degrading enzymes (103, 589). The hydrolysis of glucan by Zymolyase or glucanases may release proteins enmeshed or covalently attached to the glucan. Proteins covalently attached to glucan are postulated to represent species contributing to cell wall structure (59, 122, 123, 125, 482). Covalent attachment of mannoprotein to glucan, perhaps through phosphodiester linkages, has been suggested, as noted below (237, 238, 462, 463).
A valid question is whether the proteins found in these extracts are genuine cell wall components. It has been suggested that treatment of intact C. albicans cells with reducing agents (DTT or βME) may release some intracellular macromolecular components (50). This question has been examined most thoroughly for extracts obtained with βME. As discussed later in this review, receptors or binding proteins for ligands that bind to the intact cell are found in such extracts. On the other hand, several proteins previously associated with a cytoplasmic function have also been found in the wall extracts (6, 8, 161, 303, 520). These observations led to additional experiments using different approaches to demonstrate that the proteins found in the extract were genuine wall components. Transmission electron microscopy demonstrated that each of the moieties was indeed present in the cell wall, including the cell wall interior (6, 8, 303). Chaffin and colleagues (6, 303) also used a more general method to identify genuine cell wall proteins. Intact cells were treated with a derivative of biotin that does not permeate the membrane and therefore does not label cytoplasmic proteins. The extracted biotinylated proteins included those previously thought to be confined to the cytoplasm. This demonstrated that the proteins were present in the cell wall prior to extraction and that their presence in the extract was not due to cytoplasmic contamination. Support for the validity of the extraction procedure was also obtained with parental and mutant strains of S. cerevisiae (294). Two members of the Ssa family of proteins (Ssa1p and Ssa2p) were detected in the cell wall and cytoplasm of the parental strain, whereas in the mutant strain missing these two proteins the remaining members of the family were detected in the cytoplasm but not in the cell wall. The failure to find Ssa proteins in the cell wall of the mutant strain demonstrated that the cell wall extract was not contaminated with the Ssa proteins of the cytoplasm. Hence, current evidence indicates that treatment with sulfhydryl compounds is a suitable method to release autochthonous cell wall protein and glycoprotein components without substantially altering their biological characteristics (6, 60, 61, 293, 295, 296, 300, 302, 377).
Composition
Analysis of the protein and glycoprotein constituents solubilized from isolated cell wall preparations and from intact cells of both candidal growth forms by different treatments has revealed both a complex array of protein-containing components and quantitative and qualitative differences in the protein composition of yeast and mycelial cell walls (57–59, 71, 125, 295, 319, 320, 324, 363, 414–416, 524) (Fig. 2). Some components have been characterized as high-molecular-weight mannoproteins (HMWM). The identity of these proteins may vary with the morphology of the organism. Several HMWM are released by treatment with β-glucanases and may be covalently attached to structural polysaccharides. These HMWM may play an important role in modulating the organization of the different cell wall constituents to obtain the final supramolecular structure of the wall specific for each C. albicans morphology. In addition, HMWM contain large amounts of carbohydrate and consequently could be major elicitors of anticandidal host immunity (59, 62, 123, 159, 160, 297, 301, 326, 327, 415, 479, 523). In the medium- to low-molecular-weight range, from 20 to more than 40 polypeptide species (depending on the study considered) have been identified (61, 71, 125) (Fig. 2). As discussed above, the evidence suggests that these proteins are bona fide cell wall constituents.
FIG. 2.
Polypeptide composition of cell wall extracts and whole protoplast homogenates from C. albicans as revealed by different electrophoretic techniques (A and B). C. albicans cells from which samples were obtained were incubated in the presence of 14C-labelled protein hydrolysate and subsequently tagged with biotin. Double-labelled cell wall proteins and glycoproteins were extracted from intact blastoconidia (lanes 1 and 3) and germinated blastoconidia (lanes 2 and 4) by sequential treatment with βME (lanes 1 and 2) and digestion with Zymolyase 20T (lanes 3 and 4). Samples of protoplast homogenates from blastoconidia and germinated blastoconidia were run in lanes 5 and 6, respectively. Polypeptides were separated by SDS-PAGE and detected by fluorography (A) or transferred to nitrocellulose and detected with an avidin-peroxidase conjugate (B; lane S in this panel shows a mixture of prestained molecular weight standards run in parallel). Numbers and letters are used to identify and compare bands detected in the cell wall extracts by the different experimental procedures used. Although qualitative differences were observed (i.e., some polypeptides exhibited a strong radioactive label but were weakly biotinylated [band 6; star]), surface labelling of cells with biotin appeared to be a suitable technique to detect proteins in the wall of C. albicans. Thus, from the complex polypeptide pattern found in the protoplast homogenate samples as revealed by fluorography (A, lanes 5 and 6), only few species were labelled with biotin, indicating that most proteins released by βME and Zymolyase from intact cells are bona fide cell wall components. Brackets indicate a cluster of bands within a molecular weight range where many candidal moieties that represent receptors for host ligands have been identified (see the text). Reprinted from reference 61 with permission of the American Society for Microbiology. (C) The complexity of the polypeptide pattern of the cell wall extracts was clearly evidenced when analysis was performed by two-dimensional PAGE and silver staining (the polypeptide pattern shown corresponds to the βME extract from blastoconidia). Reprinted from reference 486 with permission of the American Society for Microbiology.
There is a growing body of experimental evidence indicating that the properties—expression, distribution, and chemical characteristics—of cell wall proteins and glycoproteins observed in vitro and in vivo are dependent on multiple factors. These include growth conditions, organism-related factors (such as growth state, morphology of the cells, strain and serotype, phenotypic switching, cell surface hydrophobic or hydrophilic status), and the nature of the biological specimens (intact cells or isolated wall preparations) that are subjected to analysis (5, 7, 24, 44, 57, 63, 106, 159, 190, 191, 195, 210, 284, 297, 301, 326, 416, 422, 477, 513, 529). Iron availability, which has been shown to be important for pathogens in establishing infection (46, 400, 566), affects the cell surface (529). There are quantitative but not qualitative changes in the profile of surface proteins associated with growth at different iron concentrations. Yeast cells of most strains grown in limiting or excess iron do not adhere as well to human buccal epithelial cells as do organisms grown at intermediate concentrations that support optimum growth. The effects of growth conditions on the expression of specific proteins are discussed for each protein in later sections. The cell wall may be thus envisaged as a highly dynamic “organelle.” The fungus is capable of expressing differentially variable wall constituents that may be useful for switching between commensal and pathogenic lifestyles and for modulating and/or evading the immune host defense.
Modification
Posttranslational modifications of proteins include glycosylation, acetylation, prenylation, phosphorylation, ubiquitination and addition of a GPI moiety. Organisms use these modifications to confer structural options for proteins, to provide regulatory control of their functions and to target proteins to specific cellular locations. While not all of these modifications have been described in C. albicans, it is likely that the organism possesses the ability to modify its proteins by most, if not all, of the posttranslational modifications.
Glycosylation.
Without any doubt, glycosylation is the most important modification of the proteins in the fungal cell wall. Attachment of sugar moieties to proteins results in the formation of the glycoproteins, which in the case of C. albicans are mainly mannoproteins. The general features of mannoproteins have been discussed above. However, the presence of nonglycosylated proteins has also been found in the cell wall of C. albicans (6, 8, 58, 61, 161). Mannosylated proteins can be broadly divided into two classes. The HMWM, most of which are larger than 200 kDa, are postulated to have structural functions within the cell wall. Some medium- to low-molecular-weight proteins also react with concanavalin A, indicating their mannan content. Within this broad division, the amount of carbohydrate attached to the same polypeptide may vary (59, 122, 364). Within the high-molecular-weight class, there is a difference in the size of the side chains associated with morphology. Oligosaccharides obtained from mannoproteins from yeast cells average 600 residues, and those from germ tubes average 300 residues (122). Cell wall proteins may also be O glycosylated with unbranched mannose chains containing one to a few residues (412). Elorza et al. (123) have suggested that some proteins are initially secreted as O-glycosylated proteins and become cross-linked with glucan and/or other N-glycosylated proteins only after incorporation into the cell wall structure.
Phosphorylation.
Phosphorus is a minor component of the cell wall of C. albicans (77). It has been assumed that it is present in cell wall mannoproteins in phosphodiester linkages between mannose residues (17, 454). Bulk mannan from C. albicans can be fractionated into five fractions that differ in the amount of phosphate (393). Phosphomannoprotein complexes from cells of both C. albicans A and B serotypes have been characterized (261–263, 494–498). This material contained 0.9 to 1.6% phosphate depending on the morphology and strain considered, and the authors concluded that β-(1,2) oligomannosaccharides were attached by phosphodiester linkage to other branching moieties. β-(1,2) oligomannosidic epitopes were further observed on a C. albicans 14- to 18-kDa phospholipomannan moiety (545), a glycolipid with important immunologic properties (133–135, 228, 229, 424), whose mannose residues may be added differently from mannan (546). Not all the glycoproteins in the cell wall of C. albicans contain phosphate, and some proteins may contain phosphate but not carbohydrate (58).
Finally, there are results suggesting that β-1,6- and β-1,3-glucan moieties present in C. albicans cell wall mannoproteins may be connected to a GPI anchor, which is known to be phosphodiester linked to the C-terminal amino acid of the mature protein (237, 238, 247, 268). This is in agreement with the hypothesis that proteins destined to be incorporated into the cell wall are linked to β-glucan through the glycan part of their GPI anchors (104), as has been demonstrated for the S. cerevisiae α-agglutinin (306, 573). The GPI anchor targets α-agglutinin to the cell wall (573). Pulse-chase experiments indicate that a plasma membrane-bound form is released to periplasmic space as an intermediate form that is then incorporated into the cell wall (305, 306). Recent studies of the linkage between mannoprotein and glucan suggest that the GPI remnant consists of ethanolamine-phosphate-mannose5, with the terminal mannose attached to the nonreducing end of β-1,6-glucan (268). A transglycosylation reaction is proposed to effect the linkage.
Ubiquitination.
Ubiquitin is a small (approximately 8,500-Da) polypeptide first isolated from bovine thymus (167). Sequence analysis of various ubiquitin genes has revealed striking evolutionary conservation among species (138). Ubiquitin plays important roles in protein modification, protein degradation, gene transcription, organization of chromatin structure, and stress resistance in higher eukaryotes (138, 139, 197). It is also associated with some cell surface protein and signaling functions (69, 368, 404, 504, 553). In yeast, a role for ubiquitination in endocytosis and/or turnover of plasma membrane protein receptors including α-receptor has been demonstrated (198, 269, 442). The C terminus of ubiquitin is covalently attached to ɛ-amino groups of lysine in protein substrates by an enzymatic conjugation system. A large number of enzymes responsible for the formation and processing of ubiquitin-protein conjugates have been described (138), including in C. albicans (98). We have cloned a polyubiquitin gene (UBI1) of C. albicans that contains three tandem copies, head-to-tail spacerless repeats, of the sequence coding for the 76 amino acids of the ubiquitin protein (485). The gene has also been cloned from a different strain (15). Northern blot analysis revealed a single mRNA population of about 1 kb present in similar amounts in both yeast and mycelial cells (485). Indirect immunofluorescence demonstrated that ubiquitin determinants were located on the cell surface, and Western blot analysis of a βME extract demonstrated that several cell wall proteins contained ubiquitin-like epitopes. The cell wall species that are ubiquitinated are discussed below with the individual proteins. The role of ubiquitin in the cell wall whether in protein degradation, stress protection, or perhaps even modulation of activity of receptor-like molecules remains to be assessed.
Distribution and Expression
As described above, transmission electron microscopy studies of the C. albicans cell wall show the existence of several layers. The structural appearance of these layers is variable and seems to be related to the strain examined, the growth conditions, the morphology (yeast cells or germ tubes) exhibited by the microorganism, and the sample preparation protocol (50, 64, 422) (Fig. 1). After total removal of proteins and mannoproteins by treatment with strong alkali or heating, the cell wall appeared to be significantly thinner with loss of any appreciable layering. These structural changes were paralleled by the absence of electron-dense components detectable with ordinary electron microscopy dyes, concanavalin A binding sites, and positive staining with reagents specific for mannoproteins (periodate-silver). Thus, the cell wall layering appears attributable to the distribution of mannoproteins at various levels within the wall structure (64). Treatment of cells with sulfhydryl agents and hydrolytic enzymes, coupled with specific cytochemical staining, has consistently shown that mannoprotein constituents are preferentially located at the outermost layer of the wall of C. albicans cells. The surface has a fibrillar or flocculent aspect with thin, delicate filaments or fimbriae (Fig. 1; see below). This material is present mostly in virulent strains, and it is also more abundant in isolates exhibiting increased adherence to host tissues (64). Proteins whose biological function is at the cell surface or the extracellular environment may nevertheless be found at the innermost layer of the wall and through the wall as they travel from the plasma membrane and periplasmic space to their destination (478). Thus, some proteins destined for the extracellular environment may also be obtained in cell wall extracts. Evidence for this mannoprotein traffic in C. albicans has been reported (423). Immunoelectron microscopy shows that proteins on the cell surface visualized by indirect immunofluorescence are also detected at the cell surface as well as near the plasma membrane with some protein distributed through the wall (6, 303). On the other hand, a protein that is not detected at the cell surface by indirect immunofluorescence but is present in cell wall extracts is located only in the interior of the wall (8). Using a panel of MAbs to localize proteins, Pontón et al. (416) demonstrated various distribution classes of cell wall proteins: (i) expressed only on the germ tube surface, (ii) expressed on the germ tube surface and within the yeast cell wall, (iii) expressed on both yeast cell and germ tube surfaces, and (iv) expressed within the wall of both germ tubes and yeast cells. A fifth category, expressed only on yeast surfaces, is also reported (296). Proteins that are associated with β-glucans should be concentrated near the plasma membrane with the structural polysaccharide. In any case, since asymmetry in mannoprotein distribution is evident in C. albicans walls (64), layering is more likely to be the result of quantitative differences in the proportions of the individual wall components (β-glucans, chitin, and mannoproteins) in each layer rather than qualitative differences (389).
Differences in the distribution of proteins and glycoproteins at the cell surface are also noted. This asymmetry may be related to the physiologic role played by each particular moiety. High-molecular-weight mannoproteins that may play an important and active morphogenetic role in modulating the organization of the cell wall (59, 62, 123, 160, 326, 327, 480) are homogeneously distributed on the cell surface (59, 62, 159). Some proteins and mannoprotein moieties that are receptors for different host ligands exhibit clustering or asymmetric cell surface distribution (60, 296, 328). The distribution of common or morphology-associated, homogeneously or heterogeneously distributed cell surface antigens of C. albicans as revealed by immunofluorescence microscopy is shown in Fig. 3.
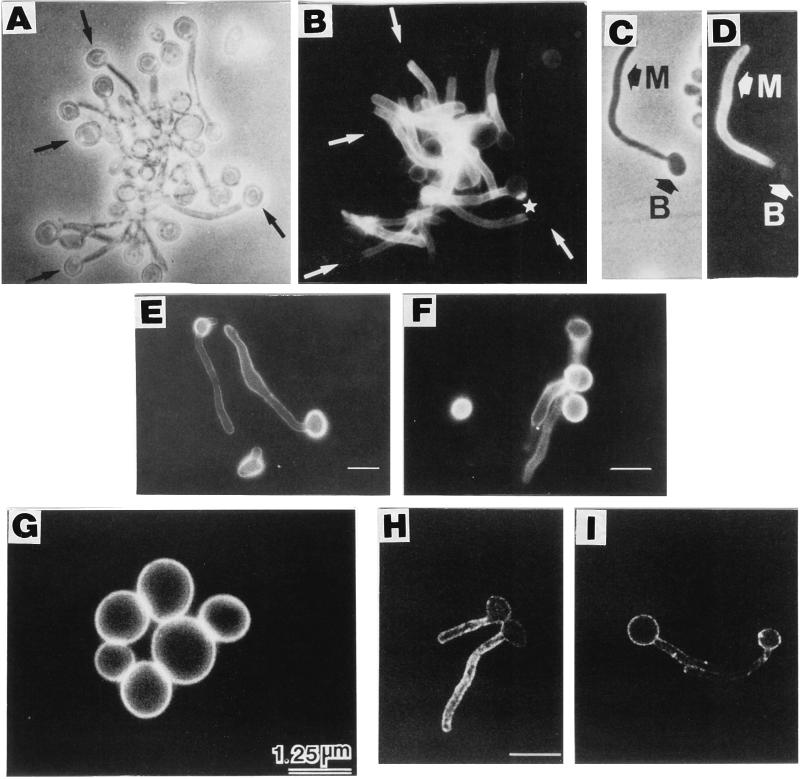
FIG. 3.
Surface expression of cell wall proteins. Phase-contrast (A and C) and immunofluorescence (B, D, and E to I) of C. albicans blastoconidia (B) and mycelial filaments (M) reacted with different polyclonal and monoclonal antibody preparations raised to protein and glycoprotein cell wall constituents. Some antibodies recognized antigens that appeared to be specific for or preferentially expressed in germ tubes (A and D) or blastoconidia (E and F). Arrows in panels A to D point to the location of mother blastoconidia (A and C) that exhibited no fluorescence (B and D). Some antigens appeared to be homogenously distributed on the surface of mycelial filaments (B and D) or blastoconidia (G). However, patches of greater fluorescence intensity were observed with other antisera preparations (H and I), suggesting that antigens recognized by such antisera were heterogenously and randomly distributed within the cell wall structure. The pictures in panels B and D to G are from standard immunofluorescence microscopy observations. Panels H and I show single-focal-plane sections of different cells obtained by confocal fluorescence microscopy and associated software. Bar, 10 μm (except for panel G, which is 1.25 μm). Panels A to D reprinted from reference 59 with permission of the American Society for Microbiology. Panels E and F reprinted from reference 24 with permission of the American Society for Microbiology. Panel G reprinted from reference 72 with permission of the American Society for Microbiology. Panels H and I reprinted from reference 328 with permission of the American Society for Microbiology.
At least three cell moieties appear to have posttranslational regulation of their localization in the cell wall. A MAb that recognized a hyphal surface protein detected a smaller protein in the plasma membrane of yeast cells (394). The C3d binding protein was detected as a 60-kDa moiety in germ tube cell walls, while a 50-kDa component was found in yeast cell membranes (563). The third example is a 30-kDa protein found in the cell wall of germ tubes but not yeast cells (5). However, the gene is expressed in yeast cells and presumably translated, although the cellular location of the protein is not known. Another group of proteins to be discussed below, i.e., enolase, hsp70, 3-phosphoglycerate kinase, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), well-known cytoplasmic proteins, have recently been described as also present in the cell wall. They may represent proteins with regulation of partition between two cellular compartments—cytoplasm and cell wall.
Other proteins, such as hydrolytic enzymes [discussed below], are altered in their expression by environmental substrates. Another example of an apparently environmentally regulated protein is the 58-kDa fibrinogen binding protein that is expressed by cells growing on Lee medium but not by cells growing on yeast extract-peptone-glucose medium (5). The foregoing discussion makes clear that the expression and localization of proteins in the cell wall are complex and dynamic processes. The presence and location of proteins and glycoproteins in the cell wall are likely to be affected by several mechanisms including gene expression, posttranslational regulation, subcellular partitioning, and targeting of destination within the cell wall.
Enzymes with Cell Wall Function
As discussed above, a number of hydrolytic enzymes have been recovered from both cell-associated locations (cell wall and periplasm) and culture medium whose function is postulated to be within the cell wall (Table 1). These enzymes are thought to be involved in cell wall biosynthesis or the remodeling that accompanies growth and division of cells.
TABLE 1.
Hydrolytic enzymes and proteins with cell wall and extracellular targets
| Enzyme | Gene | Location | Comments | Reference(s) |
|---|---|---|---|---|
| Cell wall substrates | ||||
| Exo-β-(1,3)-glucanase | EXG (XOG1) | Cell wall, extracellular | Cell wall morphogenesis | 75, 307, 308, 361, 387, 428, 429 |
| β-1,3-glucan transferase | BGL2 | Cell wall | Cell wall metabolism | 122, 178, 196, 466, 471 |
| Chitinase | CHT1-3 | Periplasm, cell wall, extracellular | Hydrolytic enzyme, cell wall morphogenesis | 141, 170, 346, 347 |
| β-N-acetylglucosaminidase | HEX1 | Periplasmic, extracellular | Hydrolytic enzyme, virulence factor? | 55, 221, 365, 426, 519 |
| Transglutaminase | Cell wall | Covalent cross-links? | 456 | |
| Extracellular substrates | ||||
| Secreted aspartyl proteinase | SAP1-9 | Extracellular, cell surface | Putative virulence factor, gene expression condition dependent | 1, 3, 18, 31, 84, 99–101, 109, 147, 157, 205, 206, 214, 215, 232, 233, 277, 348–350, 353, 359, 370, 372, 378, 379, 381, 397, 433–435, 441, 445–448, 450, 451, 509–511, 514, 515, 552, 570, 574 |
| Phospholipase | ||||
| Phospholipase A | Cell wall, surface, extracellular | Hydrolytic enzyme | 23, 174, 425, 427 | |
| Phospholipase B | PLB1 | Extracellular | Hydrolytic enzyme, putative virulence factor | 19, 217, 473, 533 |
| Phospholipase C | Extracellular | Hydrolytic enzyme | 425, 427 | |
| Lysophospholipase | Cell wall, surface, extracellular | Hydrolytic enzyme | 19, 23, 174, 355, 425, 427, 533 | |
| Lysophospholipase-transacylase | Extracellular | Hydrolytic enzyme, putative virulence factor | 19, 217, 533, 534 | |
| Esterase | Extracellular | Hydrolytic enzyme | 56, 419, 449, 551 | |
| Glucoamylase | Extracellular | Hydrolytic enzyme | 82 | |
| Hemolytic factor | Cell wall, extracellular | Hydrolytic enzyme | 317 | |
| Acid phosphatase | Periplasmic, surface | Hydrolytic enzyme | 78, 105, 391, 547 | |
| Lipase | LIP1 | Extracellular | Hydrolytic enzyme | 148 |
| Hyaluronidase | Extracellular | Hydrolytic enzyme, virulence factor? | 499, 500 | |
| Chondroitan sulfatase | Extracellular | Hydrolytic enzyme, virulence factor? | 499, 500 | |
| Metallopeptidase | Cell wall, extracellular? | Hydrolytic enzyme | 120, 121 | |
| Trehalase | Cell wall, extracellular | Hydrolytic enzyme | 362, 454 |
Exo-β-(1,3)-glucanase.
Secretory exo-β-glucan hydrolases (β-glucanases or β-glucosidases) are widely occurring enzymes in many yeast and fungal species. Although the exact physiological roles of these enzymes are unknown (141, 386), they participate in the metabolism of β-glucan, which is the main structural microfibrillar polymer of the cell wall in C. albicans (64). The most widely accepted biological role of glucanases is limited hydrolysis of cell wall glucan during morphogenetic events (141, 386). β-Glucanases have been described to be associated with the C. albicans cell wall (387, 428).
Detailed information on the chemical nature of secretory β-(1,3)-glucanases has been reported mainly for S. cerevisiae, where at least two isoenzymes, arising by differential glycosylation of a primary gene product, are secreted into the medium (430). In C. albicans, exo-β-(1,3)-glucanase activity was found to be secreted and exported mainly during germ tube formation. Negligible enzyme was released into the medium when yeast cells were grown (429). As with β-N-acetylglucosaminidase, this may be related to the more porous nature of the germ tube cell wall (519). In contrast to S. cerevisiae, only one exoglucanase has been detected in C. albicans, and it accounts for most of the total glucanase activity present in the growth medium and cell extracts (307, 361, 428). However, there are some discrepancies between results reported from different groups. The enzyme purified from cell extracts of C. albicans 1001 was reported to be a heterodimer of subunits with molecular masses of 63 and 44 kDa (362). Subsequently, the major exoglucanases secreted into the medium by strains 1001 and 3153A were found to be identical, single nonglycosylated polypeptides, with a molecular mass of about 38 kDa (307). The peptides had significant chemical and immunological similarity to the major exoglucanase secreted by S. cerevisiae. Cloning and sequencing of the gene EXG (previously XOG1) coding for the exo-β-(1,3)-glucanase of C. albicans (75, 308) revealed high identity to the β-(1,3)-exoglucanase EXG1 gene cloned from S. cerevisiae (561). A single transcript was detected in both yeast and hyphal forms, and the levels of expression appeared proportional to the growth rate (74). Sequence analysis indicated a signal peptide for secretion and a recognition by a KEX2-like protease (75). A mature enzyme of 400 amino acid residues with no sites for N-linked glycosylation was predicted. These results are consistent with the characteristics (carbohydrate content and molecular mass) of the secreted enzyme previously reported (307). Recombinant exo-β-(1,3)-glucanase of C. albicans purified from S. cerevisiae has been found to contain a number of short blocks of sequence homology to several genes for cellulases of the family A glucanases, including the conserved sequence site NEP, which has previously been shown to be important in the catalytic function of several cellulases (76). Glu-330 has been identified as the catalytic nucleophile in the enzyme (308).
β-1,3-Glucan transferase.
Many yeasts, including C. albicans and S. cerevisiae, contain a highly conserved protein with a size ranging from 31.5 to 34 kDa depending on the species (196). It is a major cell wall mannoprotein in S. cerevisiae (466). In C. albicans, a protein of 34 kDa is secreted by protoplasts and observed as an aggregate in gel filtration (122). When released by Zymolyase, it eluted as a low-molecular-weight species. The protein appeared to have a single N-linked oligosaccharide of 2.5 kDa and a residual protein moiety of 31.5 kDa. A cell wall protein with an approximate molecular mass of 34 kDa was isolated as a by-product during purification of an endo-(1,3)-β-glucanase from the material secreted to the medium by C. albicans (178). The enzyme displayed a unique glucanosyl transferase activity and did not contain any exo- or endo-β-glucanase activity. The authors suggested that this 34-kDa protein is a glucan-branching enzyme responsible for the transformation of the initial linear β-(1,3)-glucan into the branched β-(1,3)-β-(1,6)-glucan that is found in the cell wall of the fungus. The C. albicans BGL2 gene encoding the β-1,3 glucan transferase has been cloned (471) and is similar to the S. cerevisiae BGL2 gene (245). The S. cerevisiae enzyme has been described as an exoglucanase (245) and as an endoglucanase (373). However, more recently, the S. cerevisiae enzyme has been shown to be homologous to the sequence of the C. albicans enzyme and corresponds to Bgl2p (165). More sensitive assays revealed that at low concentrations of glucose oligosaccharides, glucanase activity was observed, while at higher concentrations, glucosyltransferase activity predominated. The enzyme transfers β-1,3 glucan oligosaccharides from a donor β-1,3 glucan to an acceptor β-1,3 glucan, forming a linear polymer joined by a β-1,6 linkage (587). This activity would permit the enzyme to participate in cross-linking or repair of glucan within the cell wall.
The C. albicans and S. cerevisiae genes have N-terminal signal sequences but not C-terminal sequences for the addition of a GPI anchor (245, 471). A mutant C. albicans strain was constructed by sequential disruption of alleles and was missing the protein in extracts (468). However, residual non-Bgl2p activity was detected, suggesting the presence of another transferase in C. albicans. In vivo, the mutant strain was less virulent for mice and fewer organisms were recovered from the kidneys of infected animals than for the parental strain. The reduction in virulence of C. albicans suggested that there were differences in the cell wall that affected the survival of the mutant strain in the animal model. In S. cerevisiae, the disruptant strain missing the protein had no obvious phenotype (245) and no residual glucosyl transferase activity (165). A strain in which Bgl2p was overproduced had a reduced growth rate (373). These observations suggest that the role of BGL2 in cell wall metabolism has yet to be fully elucidated.
Chitinases.
Chitinases are produced by many organisms including chitin-containing organisms that produce both chitin synthases and chitinases. In C. albicans, the most likely role for these enzymes, like glucanases, is limited hydrolysis of cell wall chitin during morphogenetic events (141). C. albicans contains three chitinase genes, CHT1, CHT2, and CHT3 (346). More than half the chitinase activity detectable in whole-cell extracts is associated with secreted enzyme (periplasmic and cell wall) (170). Reactivity is detected in supernatants of washed cells and with intact cells by using a substrate that does not enter the cell (346). Chitinase production increases during exponential growth and is greater in cells grown on yeast extract-peptone-dextrose (YEPD) medium than on a minimal medium. One of the roles of chitinase in S. cerevisiae, where chitin is confined primarily to the septum, is mother-daughter separation. Disruption of S. cerevisiae CTS1 results in clumps of cells due to failure of the cells to separate (273). Treatment of C. albicans yeast cells with a chitinase inhibitor also leads to inhibition of cell separation and clumps of cells (170).
The C. albicans chitinase gene(s) was identified by use of degenerate PCR primers based on conserved fungal chitinase sequences (346). The deduced amino acid sequence encoded by the CHT2 open reading frame (ORF) predicted a polypeptide of 583 amino acids, and that encoded by CHT3 predicted a polypeptide of 567 amino acids. The deduced sequence from CHT1 consisted of 416 amino acids (347). The C. albicans chitinases were similar (36 to 38%) to that of S. cerevisiae (346). However, the similarity was 55 to 65% in the N-terminal region containing the putative catalytic domain. There are potential N-glycosylation sites in the sequences. Near the 3′ end of the sequences of CHT2 and CHT3 is a sequence encoding a region that is rich in serine and threonine, which may be potential sites for O glycosylation. This region is not present in CHT1, which encodes a smaller putative protein (347). Expression of CHT2 and CHT3 was detected in both yeast cells and hyphae, although greater expression was associated with yeast growth (346). Expression of CHT1 was not detected under the various growth conditions examined. Preliminary results using a disruption of CHT2 suggested that the chitinase participated in cell separation as cells tended to form clumps or clusters.
β-N-Acetylglucosaminidase.
Production of β-N-acetylglucosaminidase by C. albicans cells is induced by the presence of GlcNAc in the medium (519). GlcNAc, the β-N-acetylglucosaminidase reaction product, also induces the synthesis of other enzymes required for the metabolism of this amino sugar (171, 492, 519). The enzyme may function as a chitobiase that, in conjunction with chitinase, completes the hydrolysis of chitin to provide both nitrogen and carbon sources from chitin. The enzyme, for which two molecular forms with different glycosylation levels appear to exist (364), is secreted and deposited into different regions of the cell envelope of both the yeast and mycelial forms (365, 426, 519). The enzyme is also released to the culture medium of both growth forms but to a greater extent (at least fourfold greater) during hyphal formation (519). It is suggested that the germ tube wall is more porous than that of yeast cells and, consequently, that release of extracellular enzymes to the medium is facilitated by the hyphal formation process. β-N-Acetylglucosaminidase acts on a number of natural and synthetic substrates including diacetylchitobiose and triacetylchitotriose. In its native form, the enzyme appears to be a monomer with a molecular mass of 66 kDa (519).
The metabolism of GlcNAc by C. albicans has attracted interest, since this amino sugar induces the yeast-to-mycelium transition in this fungal species (493). β-N-Acetylglucosaminidase from C. albicans is specifically a chitobiase (519). It may act coordinately with chitinase, which is also present in this fungus (22), in the hydrolysis of chitin to form GlcNAc. However, the role of these enzymes in cell metabolism is not clear (223), but there is no direct role in morphogenesis (383). In vivo, cleaving of GlcNAc from complex carbohydrates by C. albicans β-N-acetylglucosaminidase may provide a suitable carbon source for the fungus. Alternatively, removal of GlcNAc residues from glycoproteins of the fungal cell surface may cause conformational changes that modify adhesion of C. albicans cells to host tissues (55). Jenkinson and Shepherd (223) reported that a C. albicans mutant defective in the production of β-N-acetylglucosaminidase was less virulent in an experimentally induced candidiasis mouse model than was the wild-type strain, thus suggesting a possible role for this enzyme as a candidal virulence factor. On the other hand, the phenotypic properties of the mutant suggest that the enzyme is not essential for the growth of C. albicans cells (223). Hence, further work is required to determine the role of this enzyme in pathogenicity. The entire β-N-acetylglucosaminidase gene (HEX1) from C. albicans has been cloned (55). The organism appeared to use alternative transcription termination sites depending upon growth conditions, since the HEX1 mRNA from cells grown on GlcNAc was 200 nucleotides longer than the transcript from cells grown on glucose. Plasmid-borne HEX1 also responded to GlcNAc induction.
Glutaminyl-peptide-γ-glutamylyl-transferase.
The activity of the enzyme glutaminyl-peptide-γ-glutamylyl-transferase (transglutaminase) has been detected in cell extracts obtained from both growth phases of C. albicans. The activity was associated mostly with the cell wall fraction, whereas the cytosol contained almost negligible amounts of enzyme activity (456). This distribution suggested an extracellular (cell wall-bound) location for transglutaminase in intact cells. Although the wall component displaying this enzymatic activity has not been characterized, the postulated transglutaminase activity could be involved in the formation of covalent bonds between different wall proteinaceous moieties (456). In this context, it has been suggested that formation of such covalent linkages could play a role in maintaining the spatial organization of the cell wall during the biogenesis and assembly of this structure.
Hydrolytic Enzymes and Proteins with Extracellular Targets
The enzymes in the previous section are postulated to find substrates and to have their primary function within the cell wall. This section considers enzymes whose substrates are not associated with the cell wall but are found in the environment (Table 1). The action of these enzymes may provide access to nutrients for the organism. When hydrolysis of these substrates or action of extracellular proteins affects the function and viability of the host, the enzymes may be considered virulence factors that contribute to the establishment of infection. These proteins are at least transiently associated with the cell wall during their translocation across the cell wall to the external environment. However, in some cases, the association is less transient. The distribution of some of these enzymes is variable and, under some growth conditions, may be primarily cell associated in the periplasm and cell wall. Some of these proteins have also been localized to the cell surface by the same criteria that have established a cell surface location for adhesins whose function is at the cell surface. One of these extracellular proteins, Sap, also found at the cell surface, may contribute to adherence (209, 406). We have also included in this section a discussion of a secreted protein with immunomodulation properties whose relationship with other secreted moieties has not been established. Additional studies of the function, localization, and molecular mechanisms of targeting proteins to specific locations will help resolve questions about the cell-associated and extracellular distribution of these proteins.
Acid proteinase.
The extracellular proteolytic activity is one of the several hydrolytic enzyme activities described for C. albicans (390) and is due to aspartyl proteinase enzymes. The candidal secreted aspartyl proteinase was first identified by Staib (514, 515), and over the last 25 years its biological characteristics, including its role as a potential virulence factor of C. albicans, have been studied by a number of laboratories. The proteolytic activity is associated with a 42- to 45-kDa acid carboxyl proteinase. The enzyme has broad substrate specificity and is active in the range of pH 2.0 to 7.0, with the pH optimum varying from 2.5 to 4.5 depending on the substrate (109, 451). As discussed below, there is a family of aspartyl proteinases whose expression appears to be regulated by the strain, cell type, and environment. As a protease, the enzyme may have a variety of substrates, and these substrates may vary depending upon the host organ, e.g., skin or blood, that is colonized or infected. This potential spectrum of substrates and expression of isoenzymes may account for the different roles that have been postulated for the enzyme as a virulence factor. Purification of Sap1, Sap2, and Sap3 showed isoenzyme differences in antigenic similarity, thermal stability, and activity at low pH (509). The crystal structure of the enzyme showed details of the binding site, suggesting the possibility of structural differences among isoenzymes that might affect substrate specificities (1, 94). The techniques and strains that are being developed and constructed and understanding isoenzyme differences should contribute to elucidation of the role of the enzyme in colonization and infection.
Aspartyl proteinases are secreted by pathogenic species of Candida in vivo during infection (99, 100, 349, 450). The enzymes are secreted in vitro when the organism is cultured in the presence of exogenous protein (usually bovine serum albumin) as the nitrogen source (18, 109, 206, 434, 441, 447, 448, 451). However, exogenous protein in the culture medium was found not to be essential for induction of enzyme synthesis (205). Instead, the pH of the medium seems to act directly upon secretory aspartyl proteinase synthesis and not as a secondary effect of the nitrogen supply from the proteinase-mediated protein digestion, as initially thought. In any case, induction of C. albicans extracellular aspartyl proteinase appears to involve stimulation of a signal transduction event at the plasma membrane level (281). This stimulation appears to be induced by large proteins and/or polypeptides (containing eight or more amino acid residues), since internalization of small peptides with less than seven residues by peptide transport was not the inducing signal for proteinase production.
Initially designated Cap (Candida aspartyl proteinase), this enzymatic activity has been given a variety of labels. Thus, the first cloned gene in C. albicans was referred to as PEP1, because of its similarity to the pepsinogen gene (372). However, the Candida proteinase is similar to many different aspartyl proteinase genes and is not homologous to pepsinogen. The study reporting the cloning of the second gene referred to both secreted aspartyl proteinase genes as PRA genes (574), because of their similarity to the Pra gene product of S. cerevisiae. However, Pra proteinase is a vacuolar enzyme that is not secreted. A related Candida aspartyl proteinase that also is not secreted is more closely homologous to the Pra proteinase (304), while a Saccharomyces homolog of the Candida secreted aspartyl proteinase is not Pra (571). For these reasons, it was proposed that the Candida secreted aspartyl proteinase be referred to as Sap (SAP gene; Sap protein) (311, 571).
The biochemical properties of Sap (i.e., molecular weight, pI, sensitivity to inhibitors, substrate specificity, N-terminal amino acid sequence of the protein) vary depending on the strain and laboratory (109, 451). The recent cloning and sequencing of seven distinct SAP genes (from SAP1 to SAP7) (214, 215, 359, 366, 571, 574) may contribute to an explanation of such discrepancies. The seven cloned genes each encode a mature protein that is highly conserved and a precursor peptide that contains the most evolutionarily divergent region of the SAP genes (570). Comparison of the N-terminal protein sequences suggested the existence of at least one more SAP gene (SAP8) (371). SAP8 was subsequently cloned by Hube and colleagues (511). Also, a ninth gene (SAP9) has been cloned (461). Hence, a family consisting of at least nine SAP genes can be drawn upon to produce Sap enzymatic activity in C. albicans. Recently, the levels of the Sap1, Sap2, and Sap3 isoenzymes were monitored under a variety of growth conditions for several C. albicans strains (570), including strain WO-1, which alternates between two switch phenotypes, white (W) and opaque (O) (510). These studies revealed that the specific Sap isoenzyme produced is determined by the cell type (strain) whereas the level of Sap production is affected by environmental factors, and they showed that both the yeast-to-mycelium transition and phenotypic switching can determine which of the Sap isoenzymes is produced (570). A study of the expression of seven members of the SAP gene family in different strains and phenotypes and under different conditions was performed by another group (214). SAP1 and SAP3 levels were regulated during the phenotypic transition between W and O forms. SAP2 was the dominant transcript in the yeast form, and its expression was autoinduced by peptide products of its own enzymatic activity and repressed by amino acids. SAP4 and SAP6 expression was observed only at neutral pH during morphogenetic conversion from yeast to hypha induced by serum. Expression of SAP7 was not detected under any of the experimental conditions used throughout the study. The authors concluded that the different members of the SAP gene family may play distinct roles in colonization and invasion of the host (214). SAP8 is the third gene of the family to be expressed in the opaque phenotype (511).
Regardless of the question of how many C. albicans secreted proteinases exist and what role each may play, evidence suggests that aspartyl proteinase either is not glycosylated or is glycosylated at a very low level. One of the reported enzymes does not appear to contain any putative N-glycosylation sites in the deduced amino acid sequence (215). The addition of tunicamycin (an inhibitor of N glycosylation [112]) had no effect on the secretion, molecular weight, and activity of aspartyl proteinase (370). In addition, the purified enzyme does not react with concanavalin A (453) or stain with periodic acid-silver reagent (370). Therefore, if the functional protein does contain any oligosaccharide residues, they are believed to be O linked with the peptide chain. The Asn-Ala-Thr consensus glycosylation sequence has been found in the prepropeptide region, but there is no convincing evidence that this site is actually glycosylated (215). An immunocytochemistry analysis with affinity-purified antibodies to the enzyme detected reactivity in granules, and this reactivity was inhibited by glycogen, suggesting that the antibodies cross-reacted with glycogen-like polysaccharides (3). A 45-kDa intracellular form of the secretory aspartyl proteinase that seems to be the precursor of the 43-kDa mature enzyme has been reported (205, 206). It has been suggested that although the mature form is not glycosylated, that glycosylation may occur to some extent in the 45-kDa precursor (353), which may partly explain the results reported by Akashi et al. (3).
Proteinase production is believed to enhance the ability of the organism to colonize and penetrate host tissues and to evade the host immune system (31, 96, 109, 209, 232, 433, 435, 451, 552). A correlation between virulence and levels of proteinase production in both C. albicans clinical isolates (99, 157) and laboratory strains with altered proteinase levels (277, 350, 441) supports a potential role in virulence. More virulent strains of C. albicans can be isolated from patients with AIDS than from normal subjects, and this characteristic may be associated with an elevated production of aspartyl proteinase (101, 397). In this context, the enzyme has been reported to be rare or absent in nonpathogenic strains or species of Candida (348). Mutant strains of C. albicans that do not secrete this enzyme show significant reductions in lethality for mice (277, 350). It has also been suggested that the protease could promote the release of cell wall mannan by cleaving the peptide moiety of candidal cell wall mannoproteins (381). The released mannan may cause stimulation or suppression of cell-mediated (oligosaccharide fragments of mannan appear to be potent inhibitors of cell-mediated immunity) and humoral immune functions (381). The enzyme is able to degrade a number of important defensive host proteins such as immunoglobulins and complement (232, 446, 451). Infection with high-proteinase-generating C. albicans strains (378) and injection of purified candidal proteinase alone (445) can cause a decrease in the levels of host defense molecules. The decrease presumably renders the host more susceptible to candidiasis. In this context, it has also been reported that C. albicans proteinase can trigger the kininogen-kinin proteolytic cascade in the bloodstream (233). Activation of this and of other interrelated cascades that are triggered by this system (complement, fibrinolysis, and arachidonic cascades) can have a variety of negative immunologic consequences for the host. Proteolytic activity may also play a key role in providing a suitable environment that strongly affects the final outcome of Candida infections in burn patients (379), who are more susceptible to fatal candidiasis than are normal individuals. In vitro, at least Sap2p hydrolyzes mucin, and it may contribute in vivo to penetration of the gastrointestinal mucin barrier and provide access to underlying cells (84). The protease may also mediate adhesion to endothelial and epithelial cells, since enzyme inhibitors inhibit binding to endothelial cells and reduce cavitation of yeast cells binding to murine corneocytes (147, 433).
More direct evidence of the implication of Sap proteins in virulence has come from recent studies by Hube, Sanglard and colleagues (214a, 460a). The authors constructed strains harboring disruptions in a number of SAP genes, including SAP1, SAP2, and SAP3 (214a) and a triple-knockout of SAP4, SAP5, and SAP6 (460a). In all cases, mutants showed decreased virulence in an animal model of disseminated candidiasis.
Phospholipase.
Extracellular (secreted) phospholipase activities that have been reported for C. albicans include phospholipases A, B, and C (19, 23, 174, 217, 425, 427, 533). C. albicans also secretes lysophospholipase and lysophospholipase-transacylase (19, 23, 174, 217, 355, 425, 427, 533, 534); however, these activities appear to be associated with the same enzyme (355, 533, 534). Phospholipase A and lysophospholipase activities have been found in the cell wall of yeast cells and hyphae by cytochemical techniques (425, 427). Enzyme activity was associated more closely with the walls of older yeast cells than of younger cells and was more prominent at the tip of growing hyphae than in the lateral walls (425, 427). When yeast cells and germ tubes were grown in the same medium but at a different pH, the specific activity of extracellular phospholipase A was similar for yeast cells and germ tubes while that of lysophospholipase was higher for the yeast form (174).
Two forms of lysophospholipase-transacetylase have been purified from culture supernatants of yeast cells of one strain (533, 534). The enzymes appeared to be monomers of approximately 81 and 41 kDa. Both enzymes had hydrolase and transacetylase activities, with the hydrolase activity being more prominent at low substrate concentrations and the transacetylase activity being more prominent at higher concentrations. The larger species contained proline and higher specific activities for the two reactions than the smaller species. In addition, rabbit polyclonal antiserum against the smaller species did not react with the larger species in Western blot assays. A single enzyme was purified from another strain (355). This enzyme was a 84-kDa glycoprotein that had both hydrolase and transacetylase activities. The enzyme was biochemically distinct from the two enzymes purified in the first studies and did not react with the antibody prepared to the smaller species in the previous study.
Since these enzymes are associated with membrane damage of the host cells, adherence, and penetration, they are putative virulence factors of C. albicans. A correlation between phospholipase activity and adhesion to buccal epithelial cells and virulence in a mouse model has been reported (23). C. albicans strains that adhered and were virulent were also higher producers of extracellular phospholipase activity than were other C. albicans, C. parapsilosis, and S. cerevisiae strains that did not adhere. More recently, the role of phospholipases in the pathogenesis of C. albicans was explored by Ibrahim et al. (217). These authors used three different approaches to demonstrate the contribution and role of these enzymes as virulence factors. First, blood isolates of C. albicans produced higher extracellular phospholipase activities than did commensal isolates. Second, an isolate with high extracellular phospholipase activity was invasive in an infant mouse model of candidiasis whereas an isolate with low extracellular phospholipase activity was not. Third, among several selected putative virulence factors examined in an animal model of invasive candidiasis with different isolates, only the levels of phospholipase activity were found to be predictive of mortality. Characterization of phospholipases produced by three isolates showed the secretion mainly of phospholipase B and lysophospholipase-transacylase. These results implicated extracellular phospholipases in the pathogenesis of hematogenous infections caused by C. albicans. A partial PLB1 sequence is available (473).
Esterase.
Esterase activity has been found in a variety of pathogenic Candida species (56, 419, 452). Recently, induction of an extracellular esterase from C. albicans in culture media containing different Tweens (polyoxyethylenesorbitan compounds) as the sole carbon source was reported (551). Levels of enzyme activity correlated with fungal growth and substrate concentration. Activity was stimulated by activators of lipase activity. The induced esterase was heat labile and had maximum activity at pH 5.5. Thin-layer chromatography of the reaction products suggested that the enzyme is a monoester hydrolase and thus not a lipase in strict sense. Esterase activity was present in a variety of Candida clinical isolates, with no apparent correlation between enzymatic activity and relative pathogenicity. Its biological significance remains to be established.
Glucoamylase.
The C. albicans gene encoding secreted glucoamylase [α-(1,4)-d-glucan glucohydrolase] has been cloned by conferring on S. cerevisiae cells the ability to grow on media containing starch as the sole carbon source. The enzyme is efficiently secreted in the heterologous host (82). Coding and promoter regions in the sequence were identified. The gene, which contains no introns, codes for a protein of 946 amino acids with a deduced molecular mass of 105 kDa. However, there are 17 potential sites for N glycosylation, which may explain the fact that the partially purified enzyme has an apparent molecular mass of 200 kDa. Importantly, the first 20 amino acid residues are characteristic of a typical signal sequence found in secreted proteins. Because of the highly efficient translation and secretion of the gene product, this gene promoter and its signal sequence have been used for the expression of heterologous proteins in S. cerevisiae (292). The sequence was also used in vaccine development for the expression of glycoprotein antigens (83).
Hemolytic factor.
The ability of pathogenic organisms to acquire iron in the mammalian host has been shown to be critical in establishing infection (46, 400, 566). Iron availability is a limiting growth factor for C. albicans in human serum, where iron may be sequestered by transferrin (118, 367). In this context, it has been recently reported that C. albicans exhibits hemolytic activity when grown on glucose-enriched blood agar (317). The activity is present on intact organisms, and hyphae display greater activity than yeast cells. The factor is also secreted into the culture medium. However, the identity of the cell surface moiety responsible for this activity remains to be characterized. In any case, this hemolytic activity may be a putative virulence factor for C. albicans, allowing iron acquisition from hemoglobin released from lysed host erythrocytes and restoring the ability to grow in human serum.
Acid phosphatase.
Among the catalytic mannoproteins detected outside the plasma membrane barrier in C. albicans, acid phosphatase was one of the first to be characterized. It is a candidal hydrolase, for which some role in the pathogenesis of candidiasis has been suggested but not confirmed (390, 493). C. albicans acid phosphatase is an inducible enzyme that has been purified to homogeneity; the purified enzyme is a 125- to 130-kDa mannoprotein with a pH optimum of 3.6 to 4.5 (78, 391). The location (or distribution) within the cell wall structure of acid phosphatase in C. albicans (78, 105, 547) is similar to that of acid phosphatase in S. cerevisiae, where the enzyme was found to be located in the outermost and innermost cell wall layers (137, 290).
Miscellaneous.
Lipase activity is secreted by C. albicans cells growing in medium containing Tweens 80, 60, 40, and 20 as the carbon source (148). A candidal gene, LIP1, conferring lipase activity on S. cerevisiae has been isolated. The deduced sequence contained 351 amino acids with a predicted molecular mass of 38 kDa and five potential N-glycosylation sites. Southern blot analysis at low stringency suggested the presence of a lipase family.
Hyaluronidase and chondroitin sulfatase are produced by most isolates of C. albicans and secreted to the surrounding environment (499). The enzymes are produced by C. tropicalis, C. guilliermondii, C. parapsilosis, and C. krusei but by a smaller proportion of isolates. There was no apparent difference in the frequency of enzyme production by isolates from individuals with or without oral infection. These enzymes are considered to be important virulence factors for oral bacterial pathogens and may also contribute to oral candidal infection. A recent report by Shimizu et al. (500) supported the importance of the combined action of these two enzymes along with acid proteinase and phospholipase in the virulence of C. albicans.
Recently, El Moudni et al. showed that C. albicans cells contain high levels of metallopeptidase (121). The enzyme displays an electrophoretic mobility equivalent to about 52 kDa. By using a metallopeptidase-specific antiserum in indirect immunofluorescence and immunoelectron microscopy studies, the enzyme was localized in the cell wall and along the plasmalemma (120). Identification of such type of enzymatic activity could have important implications for the ability of C. albicans to invade host tissues if the enzyme is indeed proved to be a matrix proteinase and to degrade extracellular matrix.
Finally, a mannoprotein exhibiting trehalase activity is secreted into the cell envelope of C. albicans, from which it can be extracted by DTT treatment of intact cells (365, 429). About half the activity was associated with the cell envelope (429). Trehalase activity increased throughout yeast cell growth and remained elevated during the first hour after induction of germ tubes. In starved cells, all of the activity was extracellular.
An immunosuppressive, B-cell mitogenic (ISM) protein with an apparent molecular mass of 43 kDa (p43) has been purified from the culture filtrates of C. albicans (536). The immunosuppressive and B-cell mitogenic properties of this protein were associated with the host susceptibility to C. albicans infection. Treatment of BALB/c and C57BL/6 mice with p43 facilitated fungal growth, and loss of the capacity to produce p43 correlated with the loss of fungal virulence. Immunization of BALB/c mice with p43 fully protected the mice against the fungal infection (537). In addition, passive administration of specific anti-p43 antibodies significantly protected the animals against challenge with living microorganisms. In any case, immunomodulation may be not the primary biological function of this candidal moiety. The N-terminal sequence of the purified peptide did not correspond to known sequences.
Morphology-Associated Proteins
C. albicans is generally considered a dimorphic organism, capable of reproducing by budding, leading to the formation of yeast cells, or by production of germ tubes, resulting in filamentous growth (mycelium). Since the cell wall is the structure ultimately responsible for shape, and since the final steps of cell wall assemblage occur externally to the plasma membrane, the research efforts of a number of investigators in different laboratories have concentrated in the study of this morphogenetic event, as well as on the characterization of cell wall components (especially proteins and mannoproteins) that are growth phase specific. These studies have been fueled by four important considerations: (i) the interest in morphogenesis itself as an important biological phenomenon, (ii) the commonly postulated relationship between growth in the mycelial form and pathogenicity, (iii) the coordination between expression of adhesion factors and germ tube formation, and (iv) the idea that characterization of mycelium-specific antigens would provide the basis for the development of improved serodiagnostic test for the detection of invasive candidiasis (50, 64, 79, 96, 209, 352, 416, 418, 453, 480).
However, characterization of morphology-specific cell wall components is complicated by the fact that expression of cell wall components is a dynamic process. This process is influenced by environmental and nutritional factors and varies among strains (5, 42–44, 422, 550). Thus, for most phase-specific antigens, it is not clear whether they represent de novo synthesis of proteins associated with morphogenesis or, rather, constitute spatial and/or temporal rearrangements of preexisting components that were in a cryptic state before the morphogenetic event. There is evidence indicating the presence of specific antigens associated with yeast or mycelial cells, as reported by several groups who investigated the antigenic determinants of C. albicans during cellular proliferation (41–43, 50, 59, 62, 63, 72, 159, 207, 283, 394, 414–416, 422, 508, 525, 526, 534). On the other hand, several apparently morphology-specific proteins are also present in the other form in a cryptic state. These include the molecule recognized by a MAb, which is a surface protein in hyphae but a plasma membrane protein in yeast cells (394); a 30-kDa cell wall protein of germ tubes but not yeast cells, although mRNA was detected in both yeast cells and germ tubes (5); and differences in size and location reported for the receptor of the complement fragment C3d (563). In a final example, Gozalbo et al. (175) reported isolation of a gene that directed the synthesis of a 1.5 kb transcript in both yeast cells and germ tubes. Affinity-purified antibody to the gene product was used to determine cellular location and size. Two proteins of 25 and 48 kDa were detected in the membrane fraction of both yeast cells and germ tubes and the material secreted by protoplasts. In cell wall extracts, reactivity with two proteins of approximately 40 and 60 kDa was detected only in extracts from yeast cells obtained with Zymolyase. All of the observations indicate that the same gene products may be expressed in yeast and mycelial cells but located differentially within the cell. Thus, dimorphism may involve, in addition to differential gene expression and cell wall organization, a distinct pattern of both posttranscriptional and posttranslational processing of some gene products leading to differential location of the processed proteins.
Cell wall proteins associated with morphology are summarized in Table 2 and the following sections.
TABLE 2.
Morphology-associated and other cell wall proteins
| Protein | Gene | Location | Commentsa | Reference(s) |
|---|---|---|---|---|
| Morphology associated | ||||
| 4C12 antigen (180 kDa, 260 kDa) | Hyphae | Structural protein? | 59, 62, 122, 128, 160, 327 | |
| 3D9 antigen (110–170 kDa) | Hyphae | Structural protein? | 324, 325 | |
| DC3:H10 antigen | Hyphae | Present in complex with other proteins | 87, 300 | |
| Hwp1p (34 kDa) | HWP1 | Hyphae | N-terminal signal sequence, tandem repeats, C terminus ST rich | 513 |
| Hyr1p (deduced 94 kDa) | HYR1 | Hyphae | N-terminal signal sequence, C terminus ST rich, GPI anchor sequence | 16 |
| Cell wall protein (30 kDa) | Hyphal cell wall, yeast? | Probable posttranslation regulation of location | 5 | |
| MAb determinants | 106 | |||
| Yeast cell (68, 82, 96, 104 kDa) | Yeast cells and early germ tubes | Proteins have one or two common epitopes | ||
| Hyphae (104, 117 kDa) | Germ tubes and hyphal tip | Proteins have one or two common epitopes | ||
| Other | ||||
| Hsp70 (70 kDa) | SSA1 (HSP70), SSA2 | Yeast cells, hyphae | Chaperone?, translocation?; immunogenic, Ab protective? | 278, 294, 303 |
| Hsp90 | ||||
| 47-kDa fragment | HSP90 | Yeast cells, hyphae? | hsp90 degradation product, immunogenic, Ab protective | 331, 334, 335, 337–340, 342, 532 |
| Enolase (48 kDa) | ENO1-2 | Yeast cells, hyphae | Binds to glucan, internal cell wall, lacks N-terminal signal sequence, N terminus blocked, immunogenic, antigen and Abs may be diagnostic, Ab protective? | 8, 143, 177, 219, 221, 330, 356, 420, 520–522, 556, 558, 564 |
| Phosphoglycerate kinase (40 kDa) | PGK1 | Yeast cells, hyphae | Lacks N-terminal signal sequence, enzymatic activity? immunogenic | 6 |
| GAPDH (33 kDa) | TDH1 | Yeast cells, hyphae | Enzymatic activity, immunogenic | 161 |
| ADH | ADH1? | Cell wall? | Fibronectin binding? immunogenic | 27, 406, 489 |
| Als1p | ALS1 | Cell wall? | N-terminal signal sequence, variable tandem repeat, GPI-anchor sequence, member family | 212, 473 |
Abbreviations: S, serine; T, threonine; Ab, antibody.
Epitope recognized by MAb 4C12.
We have identified two major HMWM components solubilized from isolated mycelial-phase walls after degradation of the glucan network with Zymolyase (59). The proteins have an electrophoretic mobility corresponding to apparent molecular masses of 260 and 180 kDa (HMWM-260 and HMWM-180). These moieties were not detected in the wall of blastoconidia. These components behaved as a highly disperse population of mannoproteins with molecular masses ranging from about 150 kDa to more than 600 kDa on gel chromatography (122).
As shown by indirect immunofluorescence with MAb 4C12, which recognizes the peptide moiety of HMWM-260, expression of this species was dependent on the morphology of the cells of strain ATCC 26555 (59). No fluorescence was observed on blastoconidia or on short germ-tube-like structures. The HMWM-260 species appeared to be homogeneously distributed on the surface of long mycelial filaments. Western blot analysis of the Zymolyase extract of hyphal cells showed that MAb 4C12 reacted with its homologous antigen and with another high-molecular-weight species, HMWM-180. The reactive bands were polydisperse. In another strain (3153A), the reactive component could also be released, by treatment with βME, in a form that was larger than that released by Zymolyase (57). The reactive material released by Zymolyase was more polydisperse in this strain than in ATCC 26555 and had two major components of 260 and 220 kDa. Three observations suggested that the reactive species may represent different degrees of glycosylation of the same protein: (i) MAb 4C12 recognized only a single 180-kDa band in the material secreted by protoplasts in the presence of tunicamycin, and it was insensitive to endo-β-acetylglucosaminidase H (Endo H) treatment, suggesting that it was devoid of N-linked sugar chains (123); (ii) β-elimination and concanavalin A reactivity of the 180 kDa species indicated that this protein contains O-glycosidically linked sugars (59, 123); and (iii) treatment of the HMWM-260 species with Endo H resulted in the detection of the HMWM-180 as the only species recognized by the MAb 4C12 from ATCC 26555 (59). In strain 3153A, treatment of the material released by βME and Zymolyase with Endo H yielded two species of 260 and 220 kDa.
The involvement of the species recognized by MAb 4C12 in the mycelial cell wall construction and morphogenesis arises from several complementary observations.
(i) 1,4-Diaminobutanone, an ornithine decarboxylase inhibitor (327), and the chelating agent EDTA (160) both block normal germ tube formation in C. albicans without affecting the overall cell growth and protein synthesis. Although there may be other effects, these compounds appear to inhibit the synthesis and export, respectively, of the wall mannoprotein recognized by MAb 4C12.
(ii) A C. albicans yeast monomorphic mutant lacks this mycelium-specific epitope (128). Treatment of cells with cystamine, an inhibitor of glutamyltransferases postulated to be involved in the formation of covalent bonds between cell wall proteins, almost completely blocked the incorporation of the species containing the epitope recognized by MAb 4C12 into the nascent walls of regenerating protoplasts but did not inhibit their release into the medium (456).
(iii) Fab fragments prepared from MAb 4C12, an immunoglobulin G1 (IgG1) subclass antibody, inhibited germ tube formation (62). Binding of Fab fragments may alter the native configuration of the 260-kDa mannoprotein, causing its abnormal and irregular deposition in the cell wall as revealed by indirect immunofluorescence. Intact MAb 4C12 did not inhibit germ tube formation, and indirect immunofluorescence showed confluent and homogeneous binding of the antibody to the surface of mycelial cells. In this context, Fab fragments from a MAb directed against the peptide portion of a cell surface specific glycoprotein of Dictyostelium discoideum inhibited the initial stages of the cell differentiation process in this slime mold (506). Similarly, the yeast-to-mycelium transition in C. albicans can also be envisaged as a cell differentiation process. These observations support the contention that HMWM are not mere passive components of the cell wall but may play an important and active morphogenetic role in modulating the organization of the remainder of the cell wall constituents to obtain the final supramolecular structure specific for each morphology (327, 479–482).
Epitope recognized by MAb 3D9.
Protein from a germ tube culture purified by affinity chromatography on fibrinogen was used as immunogen in the production of MAbs. One of these antibodies, MAb 3D9 of the IgM class, reacted only with germ tube surfaces as judged by indirect immunofluorescence (324). The fluorescence was homogeneous along the germ tube. However, after 48 h of growth, the fluorescence was less intense and heterogeneous, with its greatest reactivity at the tip. This observation suggested that expression was greater at the point of active growth. Whether the diminution of reactivity behind the growing tip was a result of shedding of the determinant or alterations and rearrangements in the mature hyphal wall is unclear. While the presumed presence of the determinant in the culture medium points to the first explanation, the possibility that excess antigen is secreted to the exocellular environment during growth can not be eliminated. Western blot analysis of EDTA-βME and Zymolyase extracts from yeast cells and germ tubes confirmed reactivity with germ tube moieties. The reactive moiety was polydisperse. The determinant was larger than 210 kDa in the chemical extract and ranged from 110 to 220 kDa in the Zymolyase extract. The larger size in extract containing reducing agent parallels that observed with the determinant of MAb 4C12, as discussed above. The sensitivity of the determinant to proteases and insensitivity to periodate indicated that the epitope was a peptide. Analysis by SDS-polyacrylamide gel electrophoresis (PAGE) of the protein purified from Zymolyase extracts of germ tubes revealed a single disperse band of 110 to 170 kDa that was reactive with concanavalin A (325). The final product was enriched 126-fold for the protein and 16-fold for carbohydrate. MAb 3D9 did not react with Rhodotorula or Saccharomyces or any of 40 isolates distributed among six other Candida species (324). Reactivity was observed with all 58 isolates of C. albicans examined. The determinant appears to be specific to C. albicans and the hyphal form.
Hwp1p.
HWP1 (hyphal wall protein) was cloned by screening a cDNA library with adsorbed antiserum that reacted only with hyphal surfaces (513). Further analysis showed that the ORF encoded a 234-amino-acid sequence with a calculated molecular mass of 22,750 Da containing a typical signal sequence and signal site for yeast KEX2-encoded endoprotease. More interestingly, the sequence revealed a series of tandem repeats containing a 10-amino-acid motif rich in proline and glutamine that covered much of the remaining sequence. No consensus N-glycosylation sites were found. The C terminus was rich in serine and threonine, providing potential O-glycosylation sites. A 2.3-kb mRNA was detected only under growth conditions where hyphae were produced. Antiserum raised to the recombinant protein reacted only with hyphal surfaces. Similar expression of Hwp1p was detected on the surface of 20 other C. albicans strains. The antiserum also reacted with fungi in tissue sections of colonized mice. Again, only hyphal surfaces were reactive. Western blot analysis of Zymolyase extracts of hyphal forms showed a pattern of polydisperse reactivity at approximately 34 kDa. No reactivity was detected with extracts of yeast cells. A recombinant protein produced in Pichia pastoris was about twice the calculated size. The authors suggest that the differences may be attributable to anomalous migration of proline-rich proteins, glycosylation, and protease activity of Zymolyase. As a proline-rich protein, Hwp1p may share properties with similar proteins in which proline residues are proposed function to maintain polypeptide chains in extended conformations and to mediate noncovalent interactions between chains. As noted by the authors, salivary proline-rich proteins are found in this group. These proteins are a substrate for transglutaminase of buccal epithelial cells to which C. albicans can adhere. This protein on candidal surfaces may also be a substrate. The repeats also contain cysteine that may be involved in covalent linkages.
Hyr1p.
A hyphally regulated gene, HYR1, was isolated and characterized (16). Northern analysis confirmed that the 3-kb transcript of the gene was expressed under several conditions favoring formation of germ tubes but not under yeast growth conditions. The ORF encoded a predicted 937-amino-acid polypeptide of 94.1 kDa. The sequence contained an N-terminal signal sequence and C-terminal sequence for attachment of a GPI anchor. The sequence also contained 17 N-glycosylation sites and a domain rich in serine and threonine. Another domain was rich in serine, glycine, and asparagine and contained seven copies of a 4-amino-acid motif, NEGS, of unknown function. The presence of an N-terminal signal sequence, a C-terminal GPI signal sequence, and a serine- and threonine-rich region is shared with several other sequences that are associated with the S. cerevisiae cell wall (Cwp1p, Cwp2p, Tip1p, Aga1p, Agα1p, and Flo1p) (291, 444, 538, 555) or membrane sequences (388). Other C. albicans sequences have similar structural characteristics: the putative cell wall protein Als1p (discussed in a subsequent section) (212), the protein encoded by clone 8M (477), membrane protein Phr1p (467), and possibly the putative Hyr1-like protein associated with HYR99 (473).
A mutant strain containing disruptions of both alleles of HYR1 showed no obvious phenotype in yeast growth, germ tube formation, sensitivity to caffeine or calcofluor white, or aberrant forms microscopically. Although the gene is differentially expressed in germ tubes, expression of the gene as a fusion protein under the alcohol dehydrogenase promoter in yeast cells did not result in an apparent change in the yeast phenotype or in formation of germ tubes. Although the function of this protein is not known, the authors suggest that it may play a structural role. The lack of phenotype of the mutant is not inconsistent with this possibility, since the protein may be nonessential or have a function that overlaps with another protein.
Immunomodulatory 65-kDa Mannoprotein (MP65)
Cassone and colleagues (45, 65, 169, 542, 543) have studied the main mannoprotein components of C. albicans implicated in immunomodulation of host defense. Among the mannoproteins present in an acidic extract (MP-F2), a 65-kDa mannoprotein (MP65) is a main target of human T-cell response. The proliferative response stimulated by this component was of an antigenic nature rather than a mitogenic one, and the response was targeted mainly to polypeptide epitopes (65, 542). A similar constituent was present in the material released to the culture medium by C. albicans (45, 543). MP65 was further purified by immunoaffinity chromatography with MAbs generated against putative protein epitopes of the molecule (169). MP65 has a pI of 4.1 and a protein-to-polysaccharide ratio of 1.8:1. The moiety appears to contain only O-linked sugar residues, as deduced from its sensitivity to dilute alkali but not to several endoglycosidases. Nanogram doses of purified MP65 induced extensive T-cell proliferation of human peripheral blood mononuclear cells.
Heat Shock Proteins
One of the most highly evolutionarily conserved features of living organisms is the ability to respond to a sudden change of temperature (heat shock response) and to other adverse environmental conditions. This response involves the increased production of a set of proteins collectively referred to as heat shock proteins (hsps), whose production presumably contributes to the protection and damage repair of cells following stress (312). Initially identified as proteins whose synthesis was enhanced by an increase in temperature, several of the major hsps have been described since then to play important roles in all major growth-related processes, such as cell division, DNA synthesis, transcription, translation, protein folding and transport, and membrane translocation (312). Several hsps are synthesized constitutively, reflecting the important cellular functions performed by these proteins under nonstress conditions. hsps are also immunodominant antigens and major targets of host immune response during different types of infection (240, 241, 323, 332, 581). The response to hsps may also be involved in autoimmunity (241, 332, 582). Several families of hsps exist which are designated according to their average apparent molecular mass (e.g., hsp90, hsp70, and hsp60). Some of these families are divided into different subfamilies, and each subfamily may have different members.
The response of C. albicans to heat shock includes the synthesis of a set of proteins with an ample array of molecular masses (588). Genes coding for proteins belonging to the hsp90 and hsp70 families have been found in C. albicans (131, 278, 303, 316, 335, 532). Interestingly, the cellular location of some of these gene products is not confined to the cytosolic compartment, since they are also present in the cell wall and at the cell surface (303, 340). This extracellular localization could be a common feature of several fungal hsps, since it has been described also for Histoplasma capsulatum (168) and S. cerevisiae (294). As components of fungal cell walls, hsps may play roles similar to those described in the cytoplasm.
hsp90.
The structure of hsp90 is highly conserved in organisms ranging from bacteria to humans. Members of the hsp90 family of proteins play important roles in protein biogenesis and have been referred to as molecular chaperones. hsp90s interact with a variety of cellular proteins, such as steroid hormone receptors (410) and tyrosine kinases (95), and have been associated with morphological changes in pathogenic fungi and protozoa (322, 488, 554). S. cerevisiae contains two genes encoding hsp90, HSC82 and HSP82. HSC82 is a constitutively expressed gene only weakly induced upon stress exposure, while HSP82 is expressed at a much lower basal level and is strongly activated after exposure to heat shock (32).
In C. albicans, an immunodominant 47-kDa antigen was identified as a heat-stable breakdown product of hsp90 (335). This 47-kDa component was first identified as one of the major targets of the immune response mounted by patients with systemic candidiasis (334, 335) and is different from enolase (143, 329, 517). C. albicans hsp90 or its fragments circulate in the body fluids of patients with disseminated candidiasis. The 47-kDa antigen was isolated from patient sera by affinity chromatography (338), and detection of antigenemia has potential diagnostic value (334). Circulating fungal hsp90 may also play a role in pathogenesis (332, 336). Antibodies to the 47-kDa antigen correlated with survival to infection both in humans and in animal models (339, 342). Antibody to the 47-kDa antigen was present in the majority of patients with chronic mucocutaneous candidiasis and AIDS (47, 339), and it has been suggested that it mediates the protective effect of humoral immunity in systemic candidiasis (339, 342). Epitope mapping showed that patients recovering from systemic candidiasis produce antibodies against both fungus-specific and conserved epitopes of hsp90 (342). In particular, a highly conserved epitope, LKVIRK, was recognized by all patients with antibody to the 47-kDa antigen (341), and both patient sera and a murine MAb raised to this epitope when given prophylactically reduced mortality in a mouse model of invasive candidiasis (342). Treatment with a human recombinant antibody fragment to the same epitope also reduced the mortality of infected mice and improved renal function in sublethally challenged mice (331). The authors proposed that the fungal hsp90 may bind to host proteins, thereby interfering with host protein structure or function, and that antibody may neutralize the ability of the fungal hsp90 to bind to host proteins.
Matthews et al. used an affinity-purified monospecific antibody against the 47-kDa antigen and immunoelectron microscopy techniques to study the subcellular localization of this molecule (340). Results showed that it was present in the cytoplasm and the cell wall of both yeast and mycelial cells of C. albicans. This extracellular location implies that the 47-kDa antigen is naturally exposed and readily available to antibodies produced by the host, without the necessity for cellular breakage. This extracellular location may also be responsible for its high antigenicity and some of the other properties (mainly immunoprotection) associated with the immune response to this molecule.
More recently, a full-length C. albicans HSP90 gene was isolated by screening cDNA and genomic libraries with a probe derived from the S. cerevisiae homolog HSP82 (532). The predicted mass of C. albicans hsp90 was 81 kDa, and the C-terminal portion was identical to the previously described 47-kDa fragment (334). Expression of HSP90 was regulated during growth and morphological transition and after heat shock. Southern blotting analysis revealed only one HSP90 locus (532). The inability to isolate an homozygous null mutant is consistent with the notion that a single HSP90 locus exists in C. albicans and that hsp90 is essential for viability.
hsp70.
hsp70s are highly conserved in organisms ranging from bacteria to humans. The DnaK chaperones are the prokaryotic homologues of hsp70s (437), having about 50% identity to all eukaryotic hsp70s. The eukaryotic proteins are between 50 and 98% identical. Sequence similarity between hsp70 proteins extends over the entire protein, but particularly highly conserved regions are present in the N-terminal domain (30, 89). The hsp70s are not restricted to heat shock protection; they also play a role in protein folding, translocation of proteins across membranes, and gene regulation (90, 91, 155, 156, 431). In S. cerevisiae, at least 10 HSP70-like genes have been described and classified into five families from SSA to SSE (288, 374, 440, 502). Most of the proteins encoded by these genes are present at considerable levels under all growth conditions, although their synthesis is increased after exposure to stress. The SSA family includes four members, SSA1 to SSA4, with 80 to 97% DNA sequence similarity (218). Cell viability requires moderate to high levels of at least one of the proteins encoded by this family (567). Under normal growth conditions, both SSA1 and SSA2 genes were expressed at moderate levels, while heat shock resulted in increased expression of SSA1 and strong induction of both SSA3 and SSA4 expression (218). Inactivation of either SSA1 or SSA2 did not result in an obvious phenotype. However, ssa1 ssa2 double mutants were unable to form colonies at 37°C (88). They were viable and grew more slowly than the parental strain at temperatures lower than 37°C as a result of a high expression of SSA4 (567). Until recently, the gene products of the SSA subfamily of hsp70 have been considered to be located exclusively in the cytosol of S. cerevisiae (91, 312). However, we have described the presence of Ssa1p and Ssa2p in the cell wall of this budding yeast, where they could play a similar role to that in the cytoplasm (294).
In C. albicans, three different genes encoding members of the hsp70 protein family have been described (131, 278, 303, 316). By screening a C. albicans cDNA library with antibodies to whole fungal cells, La Valle et al. (278) isolated a clone encoding a stress protein of C. albicans whose deduced amino acid sequence was 84% similar to the product of S. cerevisiae SSA1. As expected, Southern blots of digested DNA probed with the cloned cDNA under low-stringency conditions indicated the presence of other members of the HSP70 gene family in C. albicans. Confirming this observation, antiserum raised to the recombinant protein recognized two bands in C. albicans whole cell extracts with apparent molecular masses of about 70 kDa. Northern blot experiments with the cloned cDNA as a probe showed the presence of 2.4- and 2.2-kb hybridization bands. Expression of the 2.4-kb constituent was heat inducible.
A second member of the hsp70 family of proteins was identified by our group (303). The nucleotide and deduced amino acid sequences of a cDNA clone coding for a constitutively expressed 70-kDa cell wall protein (5) showed a high degree of homology and identity to the SSA1 and SSA2 sequences of S. cerevisiae. The relationship with the SSA family was further supported by the reactivity of the 70-kDa candidal component with antibody recognizing the Ssa proteins of S. cerevisiae. Thus, to reflect the existence of multiple members of this family in C. albicans, and in accordance with the nomenclature of S. cerevisiae, we proposed that the original HSP70 designation of La Valle et al. (278) be replaced with SSA1 and that the gene corresponding to the second member of this family be named SSA2.
Several lines of evidence demonstrated that a member(s) of the hsp70 family of proteins is present in the cell wall of C. albicans (303). Affinity-purified antibody to the Ssa2 fusion protein produced by the cloned cDNA fragment identified a 70-kDa moiety in the βME extracts containing cell wall components from both morphologies of the organism. Indirect immunofluorescence demonstrated its presence at the cell surface. Immunoelectron microscopy showed that it was present in both the cytoplasm and the cell wall. Biotinylation of intact cells with a derivative that did not permeate the cell membrane demonstrated that the 70-kDa protein was present outside the cell membrane before extraction. Furthermore, and confirming the results obtained with C. albicans, members of the hsp70 family of proteins have been found in the cell wall of S. cerevisiae (294).
Members of the hsp70 family are highly immunogenic proteins and are major targets of the host immune response to different pathogenic microorganisms, including pathogenic fungi (240, 241, 323). La Valle et al. (278) reported that sera from three healthy individuals contained antibodies to C. albicans Ssa1p and suggested that presence of such antibodies could contribute to protection against infection. It was also pointed out that the variability in this generally conserved protein was predominantly in the C-terminal region, which is also the immunodominant region (241). Moreover, preliminary experiments suggested that recombinant Ssa1p induces T-lymphocyte proliferation. In the case of the product of C. albicans SSA2, serum samples from both normal individuals and patients with candidiasis contained antibodies against the C-terminal portion of Ssa2p (303). hsp70 is also recognized by sera of patients with oral and esophageal infection (524). It has also been demonstrated that the antibody response in a murine model of systemic candidiasis includes reactivity against a 75-kDa hsp (85). Due to the high levels of homology between the different hsp70s, a combination of antibodies to the different members of this family in C. albicans as well as to hsp70s from other microorganisms may be present in the host serum. These reactivity patterns are also likely to be the result of combination of antibodies against conserved epitopes and epitopes unique to individual C. albicans hsps.
In any case, the ubiquitous nature of hsps, along with their high degree of homology, poses interesting challenges to the immune system of the host. First, the presence of epitopes shared by a number of infectious agents may provide the immune system with a universal signal for infection, and antibodies to these conserved regions could provide some natural resistance to infection, somewhat in between innate and acquired immunity. Second, epitopes shared by the parasite and the host may trigger deleterious autoimmune responses (240). This could also be true for immune responses against highly conserved immunodominant glycolytic enzymes (see below).
Heat shock or stress mannoproteins.
Heat shock mannoproteins with approximate molecular masses of 180 to 200, 130 to 150, 90 to 110, and 60 to 70 kDa have been identified as being involved in the secretory immune response during mucosal candidiasis (413, 417). These molecules were expressed after a temperature shift from 25 to 37°C and were detected by indirect immunofluorescence. The antigenic determinants recognized by secretory IgAs in saliva and vaginal washings were polysaccharide in nature (413). Subculturing on agar at 25°C switched off expression of the antigens from mucosal isolates, while several transfers were required to reduce expression by isolates from deep infections (417). Since factors other than temperature can influence the in vitro and in vivo expression, these cell wall components should be considered “stress” proteins. The 200-kDa component appeared to be the main component recognized by vaginal secretory IgA. The 180- to 200-kDa component also enhanced tumor necrosis factor secretion by a murine macrophage cell line (411).
Glycolytic Enzymes
Glycolytic enzymes, which are highly conserved through evolution, appear to be important during C. albicans pathogenesis, since they act as major inducers of host immune responses and are major allergens during candidiasis (161, 219, 221, 489, 517, 530). Glycolytic enzymes can constitute up to 30% of total soluble proteins in S. cerevisiae (201). The presence of glycolytic enzymes in the cell wall of C. albicans has been reported recently. Enolase was found to be associated with glucan in the inner layers of the cell wall (8). Another report localized this enzyme on the surface of C. albicans cells, probably due to adventitious binding of enolase released from lysed cells (132). Phosphoglycerate kinase (PGK) was found in the cell wall and at the surface (6). We have also recently reported the presence of a cell wall-bound form of GAPDH (161). The presence of alcohol dehydrogenase (ADH) in the cell wall also has been suggested (406) as the fibronectin receptor. Together, localization of presumably “intracellular” glycolytic enzymes in the cell wall of C. albicans is rather intriguing, as are the possible roles that these molecules could play as part of the cell wall (e.g., energetic role, antigenicity, surface receptors), as well as the mechanisms involved in their secretion. However, the presence of glycolytic enzymes on microbial surfaces is not unprecedented. GAPDH has been reported as a major surface protein on group A streptococci (Streptococcus pyogenes), where it not only is enzymatically active but also serves as a binding protein for fibronectin, lysozyme, myosin, actin, and plasmin (403, 572). In Kluyveromyces marxianus, the same enzyme protein has been identified as a constitutive protein of the cell wall, and its level increases substantially upon induction of flocculation (136). ADH is also a surface protein of Entamoeba histolytica, where it has been reported to display receptor activities for a number of extracellular matrix (ECM) components (579). PGK, GAPDH, and triose-phosphate isomerase have been found on the surface of Schistosoma mansoni (173, 280, 503) and have been suggested as potential candidates for vaccine development.
Enolase.
Enolase (2-phospho-d-glycerate hydrolyase) is an enzyme of the glycolytic pathway, where it catalyzes the dehydration of 2-phosphoglycerate to phosphoenolpyruvate. It also catalyzes the reverse reaction during gluconeogenesis. It is among the most abundant proteins in C. albicans and S. cerevisiae (201). In S. cerevisiae, enolase is encoded by two genes whose expression is differentially regulated depending on the carbon source and growth phase (343) and is coordinately regulated with the expression of other glycolytic enzymes (314). Different groups have reported the cloning and characterization of the C. albicans enolase gene (143, 330, 521). Although only one gene was initially detected by Southern analysis (143, 330), further experiments demonstrated the presence of two enolase gene loci (420). However, it is not known whether both loci provide functional genes.
C. albicans enolase was first described as a cytoplasmic antigen, with an approximate molecular mass of 48 kDa, that elicited strong humoral responses in patients with disseminated candidiasis (517). Subsequently, this molecule was identified as enolase (144, 329). It is abundantly produced both in vivo and in vitro, where it is found in the liquid culture supernatant (520, 564). It circulates in the blood of patients with disseminated candidiasis, and detection of antigenemia has been shown to be a marker for invasive infection (564). In addition, it is an immunodominant antigen (517), and anti-enolase antibodies may have diagnostic value (177, 356, 557, 558). It has also been described as an allergen (219, 221). In a murine model of systemic candidiasis, it was demonstrated that C. albicans enolase stimulated both cellular and humoral responses (520). More recently, an immunoprotective effect of anti-enolase antibodies was suggested (556).
Recently, Angiolella et al. (8) very elegantly identified enolase as a glucan-associated integral component of the cell wall of C. albicans. These authors provided several lines of evidence suggesting that enolase was a bona fide component of the cell wall, including demonstration of its incorporation into the cell wall (which was inhibited by cilofungin, a lipopeptide inhibitor of glucan synthesis in fungi) and detection by immunoelectron microscopy in inner wall layers of intact cells and isolated cell walls. In accordance with previous reports (521), these authors reported the absence of enolase on the surface of fungal cells.
Phosphoglycerate kinase.
PGK catalyzes the hydrolysis of 1,3-bisphosphoglycerate to 3-phosphoglycerate with the production of ATP. Alloush et al. (6) reported cloning PGK1 by screening a cDNA expression library with antiserum to cell wall proteins. Antibody affinity purified to product of the clone detected a 40-kDa constitutively expressed cell wall protein and bound to the surface of intact cells. The presence of PGK in the cell wall was confirmed by two methods. Cell wall proteins of whole cells were biotinylated with a derivative which does not permeate the membrane, and PGK was found among the biotinylated proteins. Immunoelectron microscopy revealed that the protein was present at the outer surface of the cell membrane and in the cell wall as well as in the cytoplasm.
Glyceraldehyde-3-phosphate dehydrogenase.
GAPDH catalyzes the conversion of glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate and the concomitant reduction of NAD+. We have recently isolated a cDNA clone by immunoscreening an expression library with pooled sera from two patients with systemic candidiasis and five neutropenic patients with a high level of anti-C. albicans IgM antibodies (161). The encoded polypeptide was reactive only with sera from patients with systemic candidiasis. The nucleotide sequence of the coding region of the genomic clone showed homology (78 to 79%) to S. cerevisiae TDH1 to TDH3 genes coding for GAPDH, and their amino acid sequences showed 76% identity; thus, the gene has been designated TDH1. Western blot analysis with a polyclonal antiserum against the purified cytosolic C. albicans GAPDH (PAb anti-GAPDH) revealed reactivity of a single 33-kDa band in the βME cell wall extracts. Indirect immunofluorescence demonstrated the presence of GAPDH at the candidal cell surface. Semiquantitative flow cytometry analysis showed sensitivity of the GAPDH form to trypsin and its resistance to removal with 2 M NaCl or 2% SDS. The specificity of the reaction was indicated by decreased fluorescence in the presence of soluble GAPDH. A cell concentration-dependent GAPDH activity was detected in intact blastoconidia and germ tubes. Activity was reduced by pretreatment with trypsin, formaldehyde, and PAb anti-GAPDH. These observations indicate that an immunogenic, enzymatically active cell wall-associated form of the glycolytic enzyme GAPDH is found at the cell surface of C. albicans (161).
Alcohol dehydrogenase.
ADH catalyzes the reduction of acetaldehyde to ethanol with the formation of NAD+. A possible relationship between C. albicans ADH and receptors for vitronectin and fibronectin has been suggested (406). However, its putative surface location, deduced from its role as an adhesin, has not been further studied. The nucleotide sequence of the gene ADH1 encodes a 350-amino-acid polypeptide with high homology to other yeast ADHs (27, 489).
Binding Proteins (Receptors) for Host Ligands
Since the cell wall is the outer surface of the organism, it is inescapable that physical interactions between the fungal cell and the host will be mediated by cell wall moieties. The potential interactions involve contact with phagocytic cells, host cells of every organ infectable by the fungus, extracellular matrix, and soluble proteins. In the balance of the relationship between the fungus and the host, these interactions may serve to promote the “interest” of the fungus in maintaining itself as a human commensal or establishing metastatic disease or the “interest” of the host in protecting itself against invasion of sterile sites and overgrowth in areas of normal colonization. It is not clear that all of these interactions have been identified, and it is even less clear that we know the roles played by these interactions in the development of infection. The task of deciphering the interactions and their role in pathogenesis is complicated, since there are multiple virulence factors and their importance may be influenced by the strain of the fungus, by the host tissue that is infected, and by underlying factors associated with host status. The concept of a virulence set hypothesis has been discussed by Cutler (96) in his review of putative virulence factors.
Although other cell wall components such as chitin, β-glucan, and lipid play roles in the adhesion of C. albicans (158, 398, 476), proteins and mannoproteins are unquestionably the major mediators of adhesion. Early studies in the mid-1980s that sparked interest in the host ligands bound by C. albicans and the proteins that mediated such interactions were reports of the binding of human fibrinogen (34) and of complement fragments iC3b and C3d (194). Since then, additional host ligands and some of the fungal binding proteins or receptors for these ligands have been identified. The most relevant features of candidal cell wall adhesins and receptors for host ligands are summarized in Table 3. Most of these proteins appear to be among the medium-size polypeptide components present in the cell wall of C. albicans. For some ligands, several candidal proteins are able to bind the host protein, and it appears that some fungal proteins may be able to bind more than one ligand. The relationships between these multiple binding entities that have been detected in vitro and among isolated components and their functional role in the intact organism remain to be elucidated.
TABLE 3.
Adhesins and binding proteins
| Adhesin-ligand interaction | Gene | Location | Comments | Reference(s) |
|---|---|---|---|---|
| Protein-protein | ||||
| CR2 (C3d binding proteins) | 194, 395, 563 | |||
| 60, 50 kDa | Hyphal surface (60 kDa), yeast cell membrane? (50 kDa) | Isolated whole cell hyphal extracts; binding species differ in glycosylation | 52, 60, 235, 289, 563 | |
| 55–60 kDa (minor 67–68, 20 kDa) | Hyphae | Hyphal extract, Western blot with antiserum to CR2 | 235 | |
| 66, 40, 20 kDa | Yeast cells, hyphae | Western blot of cell wall extract with antiserum to CR2; 40 kDa ubiquitinated | 300, 486 | |
| 94, 68, 60, 50, 31 kDa | Hyphae | Western blot of cell wall extract with antiserum to CR2 | 145 | |
| iC3b binding proteins | Yeast cells, hyphae | Epithelial, endothelial cell adhesin | 25, 26, 115, 146, 162, 176, 194 | |
| 130 kDa (also 100, 50 kDa) | Hyphae | Immunoprecipitation of surface proteins by MAb OKM-1 | 117 | |
| 165 kDa | INT1? | Yeast cells | Western blot of yeast cell cytosol, membrane, but not cell wall extracts with MAb OKM-1 | 153, 210 |
| 66, 55, 42 kDa | Hyphae | C3 affinity chromatography of soluble hyphal cell extract; 42 kDa binds C3, C3b, iC3b; 55 kDa binds C3 >C3b, iC3b; 66 kDa no binding | 4 | |
| Fibrinogen binding proteins | 34, 358, 548 | |||
| mp58 (58 kDa) | FBP1 | Some yeast cells, hyphae | Fibrinogen binding; ubiquitinated, collagen-like domains | 5, 60, 484, 486 |
| 68, 60–62 kDa | Hyphae | Multifunctional binding; fibrinogen, plastic, laminin, C3d | 33–35, 550 | |
| Laminin binding proteins | ||||
| p37 (37 kDa), 67 kDa | Yeast cell, hyphal cell wall | 67 kDa only yeast cell wall; p37 of yeast but not hyphal cells binds laminin; reacts with human high-affinity receptor Ab; p37 ubiquitinated, collagen-like domains | 296, 484, 486 | |
| 68, 62, 60 kDa | Hyphae > yeast cells | Multifunctional binding: fibrinogen, plastic, laminin, C3d | 33–35 | |
| Multiple species | Ligand affinity blot of yeast cell wall extract | 164 | ||
| Fibronectin binding protein | Differences reported for fibronectin binding site | 222, 248, 253, 257, 380, 407, 466, 507 | ||
| 62, 72 kDa (60, 105 kDa unreduced) | Yeast cells, hyphae | Affinity chromatography gelatin or fibronectin; 60-kDa species reacts with MAb OKM-1; 60-kDa species highly glycosylated; more species identified Ab to human fibronectin receptor | 256, 259 | |
| Multiple species | Yeast cells | Yeast cell wall extract; anti-human fibronectin receptor antibody three proteins 40–50 kDa, polydisperse >143; ligand affinity blot additional proteins | 164 | |
| Collagen binding proteins | 249, 255, 259 | |||
| 62, 72 kDa (60, 105 kDa unreduced) | Yeast cells, hyphae | Affinity chromatography gelatin or fibronectin; 60-kDa species reacts with MAb OKM-1 | 259 | |
| Entactin binding proteins | ||||
| 65, 44, 25 kDa | Yeast cells, hyphae | Ligand affinity blot; RGD dependent and independent | 293 | |
| Vitronectin binding protein | ||||
| 30 kDa | Yeast cells | Ligand affinity blot with cell wall extract | 222, 286, 505 | |
| Ala1p | ALA1 | ? | Confers collagen IV, laminin and fibronectin binding on S. cerevisiae, member agglutinin-like gene family | 154a |
| Protein-sugar | ||||
| Fucose binding protein | Yeast cells, hyphae? | Buccal, vaginal epithelial cell adhesin; strain variation | 37, 53, 92, 93, 345, 544 | |
| >15.7 kDa | 53, 544 | |||
| GlcNAc binding protein | Yeast cells, hyphae? | Buccal, vaginal epithelial cell adhesin; strain variation | 37, 53, 92, 93, 345, 544 | |
| 190 kDa | Yeast cells | Cell wall extracts of galactose- but not glucose-grown cells; not known if is the same protein in reference 345 | 129 | |
| Protein-glycosphingolipid | ||||
| Fimbrial protein 66 kDa? | Yeast cells, hyphae? | Epithelial cell adhesin; adhesintope shared with P. aeruginosa pili | 279, 583–586 | |
| Protein-presumed protein | ||||
| Secreted aspartyl proteinase, 42–45 kDa |
SAP1-9 | Yeast cells, hyphae | Binding endothelial cells, corneocytes | 147, 433; Table 1 |
| Protein-plastic | ||||
| Plastic binding proteins | Binding to many plastics, environmental influences | 181, 243, 344, 458 | ||
| 60, 68, 200, >200 kDa | Hyphae | 60, 68 kDa may be multifunctional binding fibrinogen, laminin, C3d | 33, 34, 549 | |
| 30 kDa | AAF1 | Yeast cells, hyphae | Bind polystyrene and epithelial cells | 20, 21 |
| Carbohydrate-protein | ||||
| Acid labile mannan | ||||
| β-1,2 mannotetraose | Yeast cells, hyphae | Mannose receptor, splenic, lymph node macrophage | 73, 285 | |
| Carbohydrate-unknown | ||||
| Acid-stable mannan | ||||
| Factor 6 | Yeast cells, hyphae | BEC adhesion | 357 | |
| Unknown | Yeast cells, hyphae | Splenic, lymph node macrophage | 234 |
Serum proteins.
The fact that C. albicans can interact with and respond to serum proteins has been recognized for many years. The ability of serum to stimulate the in vitro formation of germ tubes is well established and has been used as an element of species identification (535). The ability of the organism to stimulate the alternative complement pathway was also described 20 years ago (432). An effect on coagulation proteins was also noted during infection (409) and in vitro a decade later by Maisch and Calderone (313). To a large extent, interest in the interactions of C. albicans with serum proteins initiated the cascade of the studies of interactions between the fungus and host ligands at the molecular level.
(i) Serum albumin and transferrin.
The ability of intact C. albicans cells to bind different serum proteins in vitro (including albumin, transferrin, and fibrinogen) was initially described by several authors on the basis of optical (immunofluorescence) and electron microscopy observations (33, 34, 401, 548). While the binding of fibrinogen has been examined more extensively, as discussed below, the binding of serum albumin and transferrin has not been investigated further.
(ii) Fibrinogen.
Interaction of serum proteins with C. albicans has been particularly well characterized for fibrinogen. In vitro binding of human fibrinogen was reported initially to occur exclusively on short germ tubes and mature hyphae of the organism and with higher avidity than other serum proteins such as albumin and transferrin (34, 401, 548). Binding was not detected to yeast cells grown on Sabouraud medium. Among different cell surface mannoproteins that appeared to be responsible for adhesion of C. albicans cells to plastic surfaces, two species with molecular masses of 68 and 60 to 62 kDa were shown to interact with fibrinogen. They also interacted with laminin and C3d, the terminal degradation product of the complement C3 component (35, 550). This multiple interaction lead to the suggestion that candidal fibrinogen-binding proteins may be related to mammalian integrins of the β2 or β3 subset (33, 34, 548) (see below). Using iodinated whole fibrinogen, and D and E fibrinogen fragments, Annaix et al. (9) showed that fibrinogen and the D fragment bound to C. albicans germ tubes. The D fragment is an 85-kDa moiety containing the carboxy-terminal portions of fibrinogen. The binding was time dependent, saturable, and reversible. An average of 6,000 binding sites were determined per germ tube, with a dissociation constant (Kd) of 5.2 × 10−8 M. Binding was not inhibited by several sugars, suggesting that the interaction was not a lectin type of binding. Ligand affinity blotting of a DTT-iodoacetamide extract with the D fragment identified a single 68-kDa candidal moiety with binding activity.
Using another C. albicans strain, we reported the binding of fibrinogen to both yeast cells and mycelial filaments of organisms grown on Lee medium (60). Ligand affinity blotting with fibrinogen and anti-fibrinogen antibody allowed the detection of a mannoprotein in a broad band with an apparent molecular mass of 58 kDa (mp58). The component was detected in the βME extracts of both yeast cells and germ tubes. The mp58 species may represent a specific candidal receptor for the ligand, since other mammalian proteins, such as laminin, fibronectin, type IV collagen, and C3d, did not bind in ligand affinity blotting experiments (60, 300). The protein is likely to be present in the cell wall as part of a multisubunit complex, since it migrates on native polyacrylamide gels at a much larger size (300). A cDNA clone for mp58 was isolated by immunoscreening a C. albicans expression library (5). The gene, FBP1 (fibrinogen binding protein), showed condition-dependent transcription, since the mRNA transcript was found only when both yeast and germ tubes were grown in a minimal medium and was not detected when the cells were incubated in rich medium (5). Since the expression of FBP1 appears to be regulated by nutrition, fibrinogen binding may not be the primary function of mp58. In addition, since expression is environment dependent, this may account, at least partly, for the differences in the pattern of fibrinogen binding to cells observed by the two research groups as mentioned above (9, 34, 60, 548). It may also account for differences in the fibrinogen-binding molecular species identified in these studies. However, since the experimental conditions were different in the various studies, more specific comparisons are required to determine whether there is a single fibrinogen binding protein.
The mp58 species contained N- and O-glycosidically linked sugar residues that represent 18 to 20% and 3 to 4%, respectively, of its molecular mass. These carbohydrate residues appear to be involved (particularly, the O-linked carbohydrate moiety) in the interaction with the ligand (60). mp58 has exposed hydrophobic domains, as determined by its ability to bind to polystyrene-latex microbeads (295, 299).
The mp58 species was found to possess two other interesting structural features. It contained epitopes that mimic collagenous domains or sequences of the type IV collagen molecule (484) as well as ubiquitin-like epitopes (486). The collageneous domains may define additional adhesive or binding motifs. Detection of collagenous domains in cell surface moieties seems not to be a unique feature of C. albicans. Celerin et al. (68) recently reported that amino acid sequences in surface fimbriae of the fungus M. violaceum showed strong similarity to sequences that are characteristic of the collagen gene family. Antiserum recognizing the fimbrial protein epitopes reacted with surface moieties from other fungal species including C. albicans (67).
The mp58 species is also ubiquitinated and shares this property with at least two other cell wall moieties (486). Two-dimensional electrophoretic analysis and immunoblot experiments with antibodies to ubiquitin and the purified mp58 showed colocalization of reactivity, suggesting that mp58 was ubiquitinated. In an enzyme-linked immunosorbent assay, antibody to mp58 reacted with ubiquitin. In the context of receptors, ubiquitination has intriguing implications. Ubiquitination of cell surface receptors, e.g., a lymphocyte homing receptor, has been described for higher eukaryotes (282, 504, 580). Reversible multiubiquitination and deubiquitination may turn the cell surface receptor signaling function on or off (368, 404). Thus, it is tempting to speculate that some C. albicans ubiquitinated receptors, such as mp58, are somehow involved in signaling the fungal cell about its external environment and triggering a response to that signal.
The surface distribution of the fibrinogen binding moieties has been determined by several methods. The initial studies involved indirect immunofluorescence. These studies suggested that the binding was generally heterogeneously dispersed on the surface of germ tubes (34, 60, 328) and on the small proportion of yeast cells (11 to 20%) that bound fibrinogen. This heterogeneous distribution was confirmed by other methods. Scanning electron microscopy showed that fibrinogen-coated latex beads would also decorate the surface of germ tubes with some areas of clustering (548). Transmission electron microscopy showed areas of increased concentration of bound gold particle-fibrinogen conjugates. On thin sections, the clusters extended from the surface into the wall, although the inner wall was only weakly labeled. Pretreatment with formaldehyde or βME reduced or abolished binding. We analyzed fibrinogen binding in serial sections of yeast cells and germ tubes by confocal fluorescence microscopy (328). These sections supported the heterogeneous dispersion of fibrinogen binding on the surface and indicated that the heterogeneous distribution observed by regular microscopy was not due to focal-plane artifacts. A similar heterogeneous surface distribution of mp58 was observed when a polyclonal antibody (PAb anti-mp58) raised to the purified molecule was used as a probe. All collection strains and fresh clinical isolates of C. albicans we have examined so far express mp58-like molecules on their surface (487). These strains exhibit both the ability to bind fibrinogen and react with PAb anti-mp58, although differences in expression that depended on growth conditions and strain were also found.
The mp58 fibrinogen binding protein is expressed in vivo (302). Immunohistochemical techniques with the PAb anti-mp58 were applied to tissue specimens from patients with confirmed systemic and mucocutaneous candidiasis. Most yeast cells and hyphal filaments were strongly positive. Although the proportion of positive yeast cells was not determined, there is a sense that this proportion may be higher than that observed in vitro (60, 328). The in vivo expression of mp58 provides support for the contention that fibrinogen binding may be important in host interactions. In this context, the ability of C. albicans cells to bind fibrinogen may be a virulence-related property, since coagulation proteins such as fibrin appear to be the receptor for C. albicans cells that adhere to blood clots in vivo (34). Disseminated intravascular coagulation was reported to occur in parallel, in some instances, with severe Candida septicemia (409). Moreover, binding to fibronectin that is believed to be one of the host ligands for Candida adhesion (51, 443) and binding of platelets to C. albicans cells appears to be mediated by fibrinogen (313, 439, 547).
(iii) Complement fragment C3d.
The ability of C. albicans cells to rosette antibody-sensitized erythrocytes coated with C3d as well as iC3b complement fragments was first described by Heidenreich and Dierich (194) and latter reported by other investigators (for a review of this topic, see reference 50).
A C3d-binding protein was purified from extracts of whole hyphal organisms by affinity chromatography. The purified receptor consisted of a doublet of 60 and 66 to 68 kDa. However, the biological activity, i.e., binding of C3d, appeared to be associated with the 60-kDa component that is expressed at the cell surface of hyphal elements (52, 289, 470, 563). Although it was initially thought that C. albicans yeast cells did not express C3d binding proteins, immunoelectron microscopy with a MAb to the purified C3d receptor revealed otherwise (235). An epitope cross-reacting with this MAb was detected on the plasma membrane of blastoconidia. During germination, it became associated with the cell wall, mostly at the cell surface of the organism. This component was a 50-kDa protein that, in addition to the hyphal 60-kDa moiety, has been reported to exhibit major C3d binding activity (563). Both the purified 50- and 60-kDa moieties were able to inhibit rosetting of erythrocytes coated with iC3b (90%) and C3d (50 to 64%) with hyphae. Other authors have identified additional components with different molecular masses that exhibited antigenic cross-reactivity toward antibodies (PAb anti-C3d) to the major cell wall C3d binding protein previously characterized (52, 289). In one study, a major component of 55 to 60 kDa and minor species of 67 to 68 and 20 kDa were detected in extracts from hyphae (235). In another study, moieties of 66, 40, and 21 kDa were detected in βME extracts from blastoconidia and hyphae (300). In a study of parental and clotrimazole-tolerant clones, reactive species of 94, 68, 60, 50 and 31 kDa were detected in the DTT extract of the parental strain (145). The differences observed in these studies could be due to one or more sources, including extraction procedures, strain and growth conditions, differences in glycosylation, and lack of specificity of the probe used for immunodetection. On the other hand PAb anti-C3d appeared to be a highly specific probe, since it did not cross-react with other well-characterized candidal cell surface receptors such as the mp58 fibrinogen binding moiety. This supports the contention of a separate identity for the mp58 moiety and the C3d receptor (300). C3d binding protein is presumably present in the cell wall in association with itself or other proteins, since it is found in cell wall extracts in a form with an apparent molecular weight larger than that detected by SDS-PAGE (300, 470).
Both direct and indirect evidence indicate that the 50- and 60-kDa species are mannoproteins that differ in the glycosylation of a unique polypeptide (52, 289, 470, 563). Mannoproteins obtained by concanavalin A affinity chromatography inhibited rosetting. The two components present in DTT extracts reacted in blotting experiments with concanavalin A and were sensitive to periodate treatment. Differences in the carbohydrate moiety were shown by the demonstration of differences in reactivity with sugar-specific reagents (563). The 60-kDa, but not the 50-kDa, mannoprotein reacted with wheat germ agglutinin. Endoglycosidase F and N-glycanase altered the 60-kDa species, with the generation of 40- to 45-kDa products (470, 563). Treatment with neuraminidase resulted in loss of immunoreactivity of the 60-kDa moiety, presumably due to loss of carbohydrate (563) or possibly contaminating protease activity (as noted above). Treatments that targeted protein but not those that targeted carbohydrate reduced rosetting. Thus, the carbohydrate moiety is unlikely to be involved in ligand binding. This is in contrast to mp58 fibrinogen binding species, where O-linked mannose appear to be involved in ligand binding (60).
One of the moieties that was recognized by the PAb anti-C3d receptor (300) also reacted with anti-ubiquitin antibody (486). This 40-kDa moiety, along with the mp58 molecule and the candidal laminin receptor characterized by our group (296), is a major ubiquitinated protein in the cell wall (see the discussion of mp58, above).
The relationship, if any, of the candidal and mammalian receptors for C3d is unclear. Significant differences in the isoelectric point and amino acid composition have been reported between the Candida 60-kDa C3d-binding protein and the mammalian C3d receptor (CR2) (470). Both MAbs and PAbs directed to CR2 failed to react directly with C. albicans cells (115, 117, 162). However, in one report, an anti-human CR2 antibody was able to inhibit rosetting of C3d-coated erythrocytes with C. albicans (115). In any case, due to the functional similarities, the term “CR2” or “CR2-like” has also been used for the candidal C3d receptor (149, 563).
The candidal receptor for C3d is expressed in vivo (235). Immunoelectron microscopy was used to examine expression by fungal cells in kidney tissue and peritoneal lavage fluid from infected mice. Fungal cells in peritoneal lavage fluid showed intense staining of mother cells of germ tubes, germ tubes, and pseudohyphae. Staining of the parental yeast cells was greatest at the innermost layers of the cell wall and minimal at the surface, which was consistent with the in vitro distribution. In contrast, germ tubes and pseudohyphae exhibited staining throughout the cell wall. In kidney tissue, the C3d receptor was expressed primarily on the cell wall of hyphae and pseudohyphae. The CR2 protein is also immunogenic in vivo (149). In a lymphoblastogeneic assay, lymphocytes from lymph nodes and spleens from infected mice were stimulated to a greater extent than were lymphocytes from control mice. In an intravenous murine model of infection, several avirulent clones had reduced expression of C3d binding ability but not iC3b binding ability (145). These clones were isolated as tolerant to clotrimazole, and, while all clones grew under conditions favoring hyphal formation, some differences in morphology of the organisms were noted. The clones differed in lipase activity but not acid proteinase. An analysis of the cell wall proteins of one clone demonstrated that the components associated with C3d binding were quantitatively and qualitatively altered. While this correlation with C3d binding but not iC3b binding and virulence supports a role for C3d binding in infection, it is not sufficient to determine its contribution to or role in pathogenesis.
The C3d receptor is one of the best-characterized glyco(manno)proteins present on the surface of C. albicans, yet its role in the infectious process is mostly unknown (50, 406). The fact that the C3d-binding protein is also expressed in vivo (149, 235) and the isolation of avirulent C. albicans strains with a reduced ability to recognize C3d (145) support a possible role for this candidal adhesin as virulence factor. However, the mechanism(s) by which it may contribute to pathogenesis remains speculative. Clumping between opsonized and nonopsonized C. albicans cells by using complement receptors may lead to aggregate formation that would protect the fungus from phagocytosis. Another possibility is participation in iron acquisition from complement-opsonized erythrocytes, as has been suggested for iC3b binding proteins discussed in the subsequent section. In this context, the deposition of complement fragments on any surface, e.g., endothelial cells, would provide a potential bridge for adhesion of C. albicans to that surface.
(iv) Complement fragment iC3b.
The presence of iC3b-binding components on the surface of hyphal forms of C. albicans was reported by Heidenreich and Dierich (194) and subsequently confirmed for both yeast and hyphal forms (115, 162). Very early in the study of the iC3b binding protein, various laboratories hypothesized that the functional similarity between the fungus and host cells in binding iC3b might extend to a structural similarity of the receptors involved. Antibodies to the human receptors were used as probes of candidal surfaces. Human complement receptors 3 and 4 (CR3 and CR4) bind iC3b and are two members of the human β2 subset of the integrin family of receptors (86, 460). These heterodimeric proteins differ in their α subunits. CR3 contains the αM subunit, and CR4 contains the αX subunit. αM and αX are polypeptides of 165 and 150 kDa, respectively, and, in mammals, they each combine with a common β2 chain of 95 kDa. At least eight MAbs that recognize αM exhibit high to moderate reactivity with C. albicans, and one MAb recognizing αX shows high reactivity with the fungus (25, 115, 117, 146, 162, 210; for a review, see reference 209).
The expression of iC3b binding capacity is altered by several environmental parameters. Expression is temperature dependent, since hyphae formed at 30°C strongly bound coated erythrocytes while those produced at 38.5°C did not (117). Initially, it was reported that binding was not Ca2+ dependent (117). More recently, this issue has been revisited, and these studies indicate that Ca2+ is required to stabilize the binding (512). When flow cytometry methods were used to monitor the binding of anti-αM antibodies as a measure of iC3b binding capacity, reactivity was also detected on yeast cells, although at a low level (162). Yeast cells grown in 50 mM glucose bound more antibody than did cells grown in 5 mM glucose, and the extent of the response was strain dependent. Binding of radiolabeled ligand to yeast cells demonstrated that binding was saturable and reversible, with an association constant of 2.45 × 106 liters/mol. The authors noted that this constant was very similar to that for human CR3. Carbon source also affected iC3b binding capacity as yeast cells grown in glucose had a four to six fold higher expression of the receptor compared to those grown in glutamate (210).
Identification of the candidal receptors for iC3b was initially attempted by Eigentler et al. (117). Using a monoclonal antibody to the αM chain (MAb OKM-1), these authors immunoprecipitated a 130-kDa protein along with minor components of 50 and 100 kDa from extracts of C. albicans cells previously labeled with 125I. Later, a 165-kDa moiety was detected in cytosolic and membrane fractions but not in cell wall extracts of the yeast cells by Western immunoblotting, again with MAb OKM-1 (210). More recently, a number of polypeptides with molecular masses of 70, 66, 55, and 42 kDa were purified by affinity chromatography from whole homogenates of hyphal forms of C. albicans (4). The 42-kDa moiety was affinity purified on C3 and reacted with MAb OKM-1. The three other components were isolated by affinity chromatography with antiserum prepared to the 42 kDa species. However, the high-molecular-weight components, i.e., the 130- and 165-kDa species, detected in previous studies were not identified in this work. The 66-, 55-, and 42-kDa proteins cross-reacted with MAb OKM-1 in Western immunoblotting experiments. However, differences were observed in the ability of these moieties to bind C3, C3b, and iC3b. Ligand affinity blots revealed binding of these three complement components to the 42-kDa species, whereas the 55-kDa protein reacted with C3 but only weakly with C3b and iC3b, and the 66-kDa component failed to bind any C3 ligand. Neuraminidase treatment of the 66-kDa component resulted in the appearance of a 42-kDa moiety and of two other proteins between 55 and 66 kDa, presumably due to loss of carbohydrate or possibly contaminating protease activity (as noted above). The initial 55-kDa species almost completely disappeared following enzyme treatment, with some slight evidence of aggregation products. The 42-kDa entity was essentially unaffected by the treatment. None of the components was affected by treatment with endoglycosidase F. The 55- and 66-kDa moieties were suggested to be glycosylated forms of the 42-kDa entity. The antiserum against the 42-kDa species inhibited the binding of iC3b-coated erythrocytes to the fungus and bound to a human cell line expressing CR3. The authors concluded that although candidal CR3-like and human CR3 receptors have antigenic similarity and bind identical ligands, they may differ in structure.
The binding of iC3b, the cross-reactivity of intact Candida cells and isolated candidal macromolecular components towards a number of MAbs recognizing epitopes on the αX and chains, and the similarity in molecular mass between the αM and αX chains and the 130- and 165-kDa candidal proteins reactive toward MAb OKM-1 indicate functional antigenic and structural relationships between the iC3b receptor of C. albicans and human CR3 and CR4. Because of these similarities, the C. albicans iC3b receptor is frequently termed the integrin analog (for reviews, see references 49, 50, 208, 209, and 406). Recently, these similarities have been used to construct probes for a candidal gene. Gale et al. (153) used as a probe a fragment of the transmembrane region of αM to screen a partial C. albicans genomic library. Isolated clones were then probed with a degenerate oligonucleotide for a conserved cytoplasmic sequence of αX. The isolated sequence, αINT1, revealed an ORF encoding 1,164 amino acids (ca. 185 kDa) with no extensive database homology. The sequence contains a putative membrane-spanning region and cytoplasmic tail. Within the deduced amino acid sequence there is an I region, found in five α-integrin subunits, that has 18% identity to the I domain of αM, and the region contains three potential partial motifs for binding divalent cations. PAbs prepared to the peptide corresponding to the second cation binding site and to an RGD-containing peptide were prepared. These antisera reacted with 82 and 64% of yeast cells, respectively. The gene was expressed in S. cerevisiae under the GAL1-10 promoter, and 19% of transformed cells reacted with MAb OKM-1, compared to 6% of control cells. However, the binding of iC3b to S. cerevisiae cells expressing the gene was not examined. When the gene was induced, the S. cerevisiae cells formed structures that resembled germ tubes. This is clearly an intriguing gene, and features of the deduced protein proposed to be membrane anchored and exposed at the cell wall surface differ from features of others identified or proposed as cell wall proteins.
There are a number of questions to be resolved before an unambiguous identity can be assigned to the iC3b receptor and determination to what extent it is indeed an integrin analog. It should be noted that no evidence has been found for the presence of a β2 protein in C. albicans. In any case, the relationship of the medium-molecular-weight components reacting with MAb OKM-1 detected by Alaei et al. (4) and the other high-molecular-weight species, previously detected, that also cross-react with this antibody (117, 210) is unknown. The predicted unglycosylated size of αInt1p (153) is greater than any species that is detected by MAb OKM-1 in extracts or that is reported to bind ligand (4, 117, 208). There is also some question associated with the unambiguous subcellular location of the iC3b-binding species in intact cells. MAbs to αM and αX are detected by immunofluorescence as binding at the surface of C. albicans yeast cells and hyphae (25, 115, 117, 146, 162, 176, 210). This observation is consistent with the suspected role of epitopes recognized by these MAbs as cell surface-bound receptors for C3b and iC3b. However, immunoblotting and ligand affinity-blotting moieties that cross-react with MAb OKM-1 and bind C3 derivatives were detected mostly in cytosolic extracts or whole homogenates of C. albicans cells (4, 117, 210). While the functional and antigenic similarities between the candidal binding protein and the human CR3 and CR4 seem to rest on firm ground, the structural relationships between these molecules remain in question and require further work to be established clearly.
As for the C3d binding proteins, several functions have been suggested for the iC3b receptors of C. albicans in the pathogenesis of candidiasis, although in this case a more clear relationship of these candidal components with virulence and pathogenicity appears to exist. A spontaneous mutant strain selected as being resistant to cerelenin was also reduced in its ability to adhere to fibrin clots and to cause endocarditis or vaginitis in animal models. This strain was examined for the presence of C3d and iC3b binding proteins (395). Rosetting of iC3b-coated but not C3d-coated erythrocytes was impaired in the mutant strain, which showed a 53% reduction. Extracts from the wild-type strain but not from the mutant strain were able to inhibit rosetting. This further supported the loss of functional iC3b receptors in the mutant strain. The iC3b-binding moieties may participate in the process for acquiring iron from opsonized erythrocytes, as suggested by Moors et al. (367). C. albicans activates the alternate pathway of complement, and, in vivo, “bystander” deposition of complement fragments on erythrocytes may supply the ligands to mediate binding of the fungus. By rosetting such erythrocytes, C. albicans may gain access to their iron through the candidal cell surface hemolysin-mediated lysis (317). Gilmore et al. (162) proposed a protective mechanism in which iC3b that was noncovalently attached to the receptor on the surface of C. albicans may cause masking of the recognition site on iC3b for CR3 receptors of neutrophils, thus decreasing phagocytosis of iC3b-coated candidal cells. They observed that yeast cells of one strain coated with MAb Mo1 (an anti-human CR3 antibody) that blocked the receptor showed approximately 65% phagocytosis compared to 55% for uncoated yeast cells when incubated with serum and neutrophils.
The ability of C. albicans to bind to iC3b via a specific receptor may play an important role in the adherence of the fungus to the host epithelial and endothelial surfaces (25, 26, 146, 176). Yeast cells grown in glucose express more iC3b receptor than cells grown in glutamate and have increased binding to human umbilical vein cells (176). Blocking of the receptor with iC3b resulted in decreased binding. Among other Candida spp., C. stellatoidea but not C. tropicalis, C. parapsilosis, C. krusei, or C. lusitaniae rosetted iC3b-coated erythrocytes (115, 194). Bendel et al. (26) examined the correlation between expression of the iC3b receptor and adhesion to HeLa cells. The proportion of cells binding MAb OKM-1 was greatest for C. albicans (68%) followed by C. tropicalis (32%), with C. parapsilosis, C. glabrata, C. lusitaniae, C. krusei, and S. cerevisiae ranging from 18 to 1%. A similar hierarchy was obtained for adhesion, since C. albicans and C. tropicalis adhered similarly with 45 to 50% of yeast cells bound and C. krusei and S. cerevisiae adhered with 10 to 12% of yeast cells bound. In another study, it was noted that C. albicans showed greater surface fluorescence with MAb OKM-1 than did C. tropicalis but that adhesion of the two species to HeLa cells was similar (25). Incubation of C. albicans with anti-αM MAbs OKM-1 and Mo1, iC3b, or several RGD-containing peptides from iC3b significantly inhibited the adhesion of C. albicans to HeLa cells while C. tropicalis binding was not inhibited by iC3b or its peptides. On the other hand, purified fibronectin and fibronectin-RGD peptides inhibited the adhesion of C. tropicalis but not C. albicans. After growth in serum-free medium, the presence of iC3b and fibronectin was observed on the surface of HeLa cells. Treatment of HeLa cells with anti-C3 antibody but not anti-fibronectin antibody inhibited the adhesion of C. albicans, and the reverse was true for C. tropicalis. These observations support different mechanisms for adherence of C. albicans and C. tropicalis to epithelial cells. Overall, these studies, while implicating iC3b receptors in candidal adhesion, also show that other adhesins are involved, since inhibition of the iC3b receptor does not completely abolish the interaction with host cells.
Extracellular matrix proteins.
The initiation of studies on the ability of C. albicans to bind and adhere to components of ECM is attributable largely to Klotz (248, 249, 251). Klotz showed that C. albicans yeast cells bound at the junctions of endothelial cells in confluent layers and that binding was enhanced when endothelial cells were removed from the substrate or contracted, resulting in ECM exposure. ECM proteins, collagen type IV, laminin, and fibronectin immobilized on plastic plates mediated the binding of yeast cells to the coated wells. ECM components form a network by interactions with each other. For example, entactin (nidogen) has the ability to interact with other ECM components such as fibronectin, fibrinogen, laminin, and type IV collagen (142, 575, 576). Thus, multiple binding targets are simultaneously present and adjacent in the ECM. Another potential contributor to the interaction are the collageneous domains present in candidal proteins, e.g., p37 laminin binding protein of yeast cells and mp58 fibrinogen binding protein (484). Some ECM proteins have collagen binding sites, and these may bind to candidal proteins containing collagenous domains. Therefore, in vivo binding to ECM is likely to involve multiple receptors binding to different ligands. These may be candidal receptors that bind to similar binding sites on ECM components as do integrins (see below), as well as ECM components that have collagen binding sites that bind to candidal proteins via collagenous domains. In general, the receptors are expressed extensively on hyphae, and this has been postulated as one of the ways by which germ tubes contribute to the development of infection. Although yeast cells are the most likely form to be hematogenously disseminated and extravasate, only a few yeast cells express the various binding capacities in vitro. An obvious question, then, is whether interactions between extravasating yeast cells and ECM contribute to establishing and maintaining infection. Three possibilities may be considered. First, not all extravasating yeast cells may need to bind for an infection to be initiated. Second, more cells may express the various receptors in vivo. Third, with the multiplicity of binding interactions that are available to the fungus, most yeast cells may express at least one of these, and that may be sufficient for binding.
In mammalian systems, the majority of molecules involved in cell adhesion to ECM components are members of the supergene family of integrins. These binding proteins are highly conserved receptors that mediate cell-matrix and cell-cell interactions in a wide variety of biological processes including infectious diseases (216, 457). The parallel between candidal complement fragment receptors and host integrins and candidal ECM receptors and host integrins was not unappreciated. The presence of candidal moieties exhibiting cross-reactivity with antibodies raised against the α and β chains of integrins has been reported (25, 102, 115, 162, 176, 210, 318, 464, 465). In this case the β1 subunit found in some mammalian integrins mediating binding to ECM was also detected in C. albicans membranes or cell surface (318, 464, 465). It was suggested that binding of C. albicans to fluid-phase and immobilized human basement membrane glycoproteins and endothelium, e.g., laminin, fibronectin, entactin, and vitronectin, is mediated, at least partly, by integrin analogs (or perhaps homologs) present at the surface of fungal cells (49, 50, 102, 116, 176, 209, 250, 259, 406, 464, 465). The possible existence of integrin analogs for complement fragment receptors in C. albicans cells has been discussed above.
(i) Laminin.
Laminin, a major component of the basement membrane (539), is a large multidomain glycoprotein that seems to play a critical role not only in normal cell adhesion but also during tissue invasion and metastasis by tumor cells and pathogenic microorganisms. Contemporaneous with the study by Klotz (249) on the binding of the fungus to immobilized ECM components, Bouchara et al. (35) reported the binding of laminin to germ tubes grown in medium 199. As determined by indirect immunofluorescence, no binding to yeast cells grown in Lee medium was detected. Immunoelectron microscopy showed that this binding was in the outermost fibrillar layer. Radiolabeled laminin was used to show that binding was saturable and specific. Further analysis indicated about 8,000 binding sites per organism with a Kd of 1.3 × 10−9 M. Binding was reduced by fibrinogen but not fibronectin, serum albumin, or several sugars, suggesting that binding was not lectin-like.
The presence of receptors in a DTT-iodoacetamide extract of germ tubes was examined by ligand affinity blotting. Reactivity was detected with a 68 kDa moiety that was a major component of the extract and with a doublet of 60 and 62 kDa. The components of the doublet were suggested to be possible degradation products. These proteins seem to be the same candidal moieties that were also found to interact with fibronectin, fibrinogen, and C3d and that appeared to be responsible for an enhanced adhesion of C. albicans cells to plastic surfaces (33, 34, 253, 548), suggesting the existence of a single receptor-like molecule at the surface of C. albicans cells that may recognize several ligands.
We have reported the characterization of two polypeptides that bind laminin (296) and that appear to be different from those described by Bouchara et al. (35). A ligand affinity blot with laminin reacted with a 37-kDa (p37) and 67-kDa moiety only in extracts from intact yeast cells. The 67-kDa but not the 37-kDa component reacted with concanavalin A (61). The size of the components was similar to the human high-affinity laminin receptor, and antibody to that receptor was used for analysis of the candidal proteins (296). A rabbit polyclonal antibody (PAb 4160) directed toward the carboxyl terminus of the ligand binding domain of the human 67-kDa high-affinity laminin receptor and its 37-kDa precursor was used in a Western blot analysis. PAb 4160 reacted with the 37- and 67-kDa components in the extract from yeast cells. A second antibody, PAb 4056, directed toward the internal domain of the human receptors, reacted with a 37-kDa moiety in extracts from both yeast cells and germ tubes. The 37-kDa moiety from germ tubes did not bind laminin. Under the conditions used, neither the 37- nor the 67-kDa species bound fibrinogen, fibronectin, or collagen type IV. Thus, the p37 moiety did not appear to be a multifunctional protein in yeast cells and appeared to be nonfunctional in germ tubes.
The localization of the protein on the surface was examined by indirect immunofluorescence with PAb 4160. A patchy binding pattern was only detected on about 10% of yeast cells. The reactive yeast cells also appeared to be the larger cells in the population. These observations were confirmed using a polyclonal antibody raised against the purified p37 component (302). Since a heterogeneous surface dispersion was also observed for mp58 (328) and since mp58 is present in the cell wall in a multisubunit state (300), we considered the possibility that both receptors were present in a multireceptor complex. Using a dual fluorescence labeling technique, we have found that although both the mp58 and p37 receptors were heterogeneously distributed on the cell surface, they were not colocalized, since the areas of concentration of each receptor were different (302).
The p37 polypeptide was found to have some features in common with the mp58 species. Thus, p37 also possesses collagen-like domains (484) and ubiquitin-like epitopes (486) that may define additional structural, regulatory, or binding motifs (see the discussion of mp58, above). The 67-kDa moiety does not appear to be ubiquitinated. Thus, although these proteins appeared to be functionally and antigenically related, only one of them was ubiquitinated. Since the roles of ubiquitination include targeted protein turnover and receptor regulation, this suggests that there may be stability or regulatory differences between these proteins.
More recently, Glee et al. (164) have detected by ligand affinity blotting an even larger number of cell wall proteins with the ability to bind laminin. The number of reactive moieties in that study may be more than 10. The determination was made with extracts that were analyzed without dialysis or other treatment. The authors showed that dialysis altered the profile of proteins recovered from extracts. Also, the profile of fibronectin binding proteins differed in dialyzed and undialyzed extracts. In addition to treatment of the extract used for analysis, the rationale for the differences between results reported by different groups with respect to the binding of fibrinogen to C. albicans cells (see above) may also apply here, as may differences in the conditions used for analysis.
The p37 is expressed in vivo. Immunohistochemical analysis of tissue sections from patients with disseminated and superficial candidiasis with PAb anti-p37 revealed that the pattern of morphological expression was similar to the in vitro pattern. Expression was essentially restricted to yeast cells, with less than 50% of the organisms being reactive (302).
Laminin binding proteins are postulated to contribute to the pathogenesis of infection by mediating adhesion to host ligands and retention of the fungus in host tissue. Support for such a role is provided by several reports from in vitro studies. C. albicans appears to use multiple mechanisms to adhere to keratinocytes (396). Adhesion was inhibited by several sugars and peptides, including a synthetic peptide from the laminin B chain, although laminin itself was not very effective. Models for binding to ECM have utilized substrates immobilized in wells of microtiter plates. Yeast cells adhere to immobilized laminin, and binding is inhibited by anti-laminin antibody, RGD-containing peptides, and fibronectin (249, 253, 260, 462). Yeast cells also adhere to corneal endothelial cell ECM that contains multiple ligands (253). The ability of laminin to inhibit this binding was not examined, although fibronectin was a significant inhibitor. As noted previously, yeast cells have enhanced binding to junctions of endothelial cells in confluent monolayers and to ECM when cells are contracted (248, 251). Clearly, the role of adhesion to laminin in pathogenesis has yet to be established. This is likely to prove a challenge, since it appears that not all binding proteins may have been identified and characterized and the number of such proteins will make the construction or isolation of mutant strains deficient in laminin binding difficult.
(ii) Fibronectin.
C. albicans and other Candida species adhere to fibronectin. Fibronectin is a large (440-kDa) dimeric glycoprotein that circulates in plasma and is found as part of ECM. Various functional domains of the molecule have been identified; they include binding domains denoted as fibrin, collagen, DNA, cell (containing an RGD sequence), and heparin. Initial studies of the adhesion of Candida cells to endothelium suggest a role for fibronectin as the most prominent host ligand for candidal adhesion (248, 257, 472, 507), and binding of C. albicans to immobilized fibronectin was demonstrated by Skerl et al. (507). The binding was inhibited by mannan and was reduced by proteolytic treatment of fungal cell surfaces. The fibronectin receptor appeared to be distinct from C3d and iC3b binding activities when fibronectin binding in strains deficient in the other activities was examined (380). Several studies in different laboratories have examined the candidal receptor and its ligand binding characteristics. However, there are differences between these studies that may reflect the presence of multiple adhesins. A contribution by strains, experimental conditions, or assay methods is also a possible explanation for the differences in results reported. Comparisons between studies are also difficult, since the same parameters were not determined in each study. While a number of observations are consistent with homology between the candidal fibronectin receptor(s) and a mammalian integrin, some are not.
The first receptor study by Klotz and Smith (253) showed that binding of soluble fibronectin to C. albicans yeast cells was saturable, with approximately 8,000 binding sites per cells and a Kd of 1.1 × 10−8 M. Binding occurred rapidly and was not reversible in a 2-h period. In a later study, Nègre et al. (380) found two classes of binding sites: a small number (approximately 5,000 per cell) of high-affinity binding sites (Kd, 1.3 × 10−9 M) and a large number (approximately 30,000 per cell) of low-affinity sites (Kd, 1.2 × 10−7 M). The binding in this study occurred more slowly and was reversible. This study used a broader concentration range that facilitated the detection of two binding classes. Differences were also reported for the effect of cations, in particular calcium, on binding. Initially, binding was found to be enhanced by two- to more than threefold by the presence of Ca2+ (222, 259). The Ca2+ effect was more pronounced at pH 7, near the optimal binding pH 6, than at pH 4 (222). In a subsequent study, fibronectin binding decreased with increasing concentrations of Ca2+ and was enhanced by the presence of EDTA, since in the presence of the chelator the number of high-affinity binding sites doubled (380). Nègre et al. (380) pointed out that the lack of calcium dependence differed from that expected for an integrin-like receptor and suggested that previous investigations may have been looking at a calcium-dependent effect of binding to plastic, the substratum to which fibronectin is bound in assays with insoluble ligand. However, the Ca2+ effect observed by Jakab et al. (222) was with soluble fibronectin.
Some disagreements have been reported both in the identity of fibronectin fragments that interact with C. albicans and in the inhibitors of binding. Some results support the collagen binding domain as the site of soluble fibronectin binding, while others support the cell binding domain as the major recognition site. The cell binding domain is also suggested to be the major site of binding to immobilized fibronectin on the basis of some observations, while other results support multiple binding sites. Penn and Klotz (407) reported that among proteolytic fibronectin fragments, the 120-kDa soluble cell binding domain but not the gelatin or heparin binding domain bound to fungal cells. Santoni et al. (465) reported that the fungus bound to the immobilized 120-kDa fragment. On the other hand, Nègre et al. (380) found that the 120-kDa moiety was less active in inhibiting soluble fibronectin binding than the gelatin/collagen domain but more active than the fibrin and least active heparin binding domains. Adhesion was less specific with fragments from each of the four binding domains promoting adhesion of the fungus to acrylic disks coated with any of the fragments. Jakab et al. (222) reported that the collagen binding fragment of fibronectin was a less effective inhibitor (25%) of soluble fibronectin binding than was a fragment containing the cell binding domain (85%).
There is also disagreement concerning inhibitors of interactions of the fungus with soluble and immobilized fibronectin. Klotz and Smith (253) and Jakab et al. (222) reported that RGD and related peptides inhibited binding. RGD was the most effective peptide inhibitor (15 to 28%) of soluble fibronectin binding (222, 253). The GRGESP and GRGDTP peptides were less effective inhibitors, and GRGDSP was ineffective (253). However, GRGESP was an effective inhibitor (88%) of binding to immobilized ligand. In contrast, Santoni et al. (465) found that GRGDSP but not GRGESP could inhibit binding to insoluble fibronectin. The difference with respect to GRGESP and GRGDSP inhibition is of particular interest, since it affects the possibility that the fungal receptor is an integrin homolog (465). GRGDSP but not GRGESP is an effective inhibitor with mammalian integrins. In opposition to these observations, Nègre et al. (380) reported that a recombinant 120-kDa fibronectin fragment with the RGDS sequence deleted was as good a promoter of C. albicans binding as was the native sequence. They also found that RGD peptides did not block soluble fibronectin binding. The inhibitory effect of heparin has also differed among studies. Heparin has been reported to inhibit soluble fibronectin binding (222) but not the binding of the 120-kDa fragment containing the cell binding domain (407). Heparin and heparan sulfate were found to inhibit binding to immobilized fibronectin, apparently by binding to fibronectin and interfering with recognition or access to the binding site(s) by C. albicans (254).
Klotz et al. (259) used affinity chromatography to isolate a fibronectin binding component(s) from a detergent extract of yeast cells. Two bound proteins with unreduced molecular masses of 60 and 105 kDa were detected by SDS-PAGE, and two species of 62 and 72 kDa were detected under reducing conditions. The proteins were present in the cell membrane as well as in the cell wall of yeast cells and were also found in germ tubes. The proteins present in the yeast cell extract were eluted from columns of fibronectin or gelatin coupled to agarose with EDTA, α-methylmannopyranoside, or peptides containing the RGD sequence. Both moieties appeared to be mannoproteins on the basis of their ability to bind concanavalin A. The 60-kDa component in extracts was found to exhibit cross-reactivity with antibodies raised to the human integrin receptor for fibronectin, vitronectin, and complement (MAbs OKM-1 and Mo1 for the αM subunit). The anti-fibronectin receptor antiserum also identified two other major reactive moieties of 50 and 90 kDa and several less reactive species in the same size range. This reactivity further indicated that candidal cell wall glycoproteins that are probably related to integrins may be receptors responsible for yeast cell adherence to ECM (259). On the basis of this assumption, a method similar to that used to purify animal integrins was used to isolate the fungal binding protein (256). In SDS-PAGE, this procedure yielded a highly glycosylated protein of approximately 60 kDa, whose N terminus was blocked to Edman degradation. It eluted from a molecular mass sizing chromatography column with an apparent molecular mass of 42 kDa. A similar phenomenon was observed when the C3d binding protein was subjected to gel permeation chromatography, and these authors suggested that the apparently anomalous migration might be due to some interactions with the column matrix retarding elution (470). These experiences suggest that size determinations for cell wall proteins obtained by gel permeation chromatography must be considered carefully if they are not supported by other procedures.
On the other hand, the presence of a fibronectin receptor antigenically related to the α5β1 mammalian integrin has been reported for cells of C. albicans and other Candida spp. (465). The binding to C. albicans of antibodies to human integrin subunits and the human fibronectin receptor was examined by fluorescence flow cytometry. Reactivity was detected with yeast cells with anti-α5 antibodies (MAb SAM1 and marginally with MAb P1D6) and with two anti-fibronectin receptor antiserum. The percentage of positive organisms, approximately 97%, was greater when germ tubes were analyzed with one anti-fibronectin receptor antiserum. In addition, two MAbs (A1A5 and 4B4) for the β1 subunit that were negative with yeast cells stained 41 to 59% of the germ tubes. Anti-fibronectin antibody completely inhibited yeast cell and germ tube adhesion to immobilized fibronectin, while an RGD-containing peptide partially inhibited binding. MAb SAM1 (anti-α5) also inhibited binding, while MAb 4B4 (anti-β1) partially inhibited binding. These results suggest that the C. albicans receptor is antigenically and functionally related to α5β1 integrin. With the same commercial anti-human fibronectin receptor antiserum, Western blot analysis of undialyzed extract revealed three major reactive species between approximately 40 and 50 kDa and a polydisperse reactivity above 143 kDa (164). However, these species did not represent all the moieties in the extract that were able to bind ligand, since additional species larger than 50 kDa and smaller than 35 kDa were detected in a ligand affinity blot.
MAb SAM1 and the two anti-fibronectin receptor antisera were used to examine for a similar receptor on other species (464). Reactivity was observed with C. tropicalis, C. stellatoidea and C. glabrata, with C. tropicalis exhibiting the greatest activity with MAb SAM1. C. krusei showed marginal reactivity with the antibodies. C. tropicalis and C. stellatoidea bound to immobilized fibronectin to a greater extent than did C. glabrata, while C. krusei bound only marginally. Thus, a correlation appears to exist between the expression and function of the α5β1 integrin-like fibronectin receptor and the hierarchy of relative virulence among the pathogenic Candida spp. Quite recently, DeMuri and Hostetter (102) have examined a β1 fibronectin receptor in C. tropicalis. Binding of fibronectin was saturable and reversible, with 854 receptors per cell and a Kd of 2.3 × 10−9 M. The receptor density is about 6- to 10-fold lower than estimated for C. albicans (253, 380). An extract of a cell membrane preparation inhibited fibronectin binding (102). A ligand affinity blot of the extract identified a band of approximately 125 kDa. Immunoblotting with an antiserum to a human fibronectin receptor (α5β1) also identified a similar-sized band, although reactivity with smaller bands was detected with both receptor and control antisera. MAb P1D6 (anti-α5) did not react with the extract, while an antiserum to β1 reacted with the 125-kDa moiety. A fibronectin binding protein of 105 kDa was immunoprecipitated from a cell extract. These observations suggested that C. tropicalis contained the β1 subunit but not a corresponding α subunit, and the authors suggested that one subunit might be sufficient.
Although the regulation of fibronectin receptor expression is still poorly understood, expression of the fibronectin receptor(s) appears to be regulated in part by environmental conditions. The nutrient composition of the growth medium is one of the most commonly observed parameters affecting binding capacity. The binding of fibronectin is influenced by the batch of medium used for fungal growth (380). Differences in binding between organisms grown on Sabouraud dextrose medium in suspension and on agar were noted (222). Organisms grown in suspension at 37°C bound 60% more fibronectin than when grown at 21°C. When grown under conditions that resulted in differences in cell surface hydrophobicity (CSH), the more hydrophobic cells showed increased binding to fibronectin. Silva et al. (505) also demonstrated that hydrophobic cells bound in greater numbers than hydrophilic cells to immobilized fibronectin. Yeast cells grown on Sabouraud medium bound some 2.5 times more fibronectin than did organisms grown on Lee medium and more than 5 times as much as those grown on yeast nitrogen base (578). Metabolically active yeast cells (26 and 37°C) bound more ligand than did cells maintained at 4°C (253). Recently, hemoglobin has been identified as an inducer of fibronectin receptors in C. albicans cells (578). In the presence of hemoglobin, the binding of soluble fibronectin was enhanced 20- to 80-fold. The induction was specific for hemoglobin, since hemin, Fe, and protoporphyrin IX could not substitute. Thus, the effect was not just an increase in the Fe concentration. A small enhancement of expression was observed with globulin, globin, and myoglobin. Cells were required to grow in the presence of hemoglobin to manifest the increased expression, and the induction was reversible when hemoglobin was removed. Induced cells also showed increased binding to immobilized fibronectin and confluent endothelial cell layers. However, removal of endothelial cells from the wells did not result in an increased binding to the exposed matrix material. The effect of hemoglobin on adhesive potential is interesting, since, as discussed above, the organism has hemolytic activity (317). Exposure of C. albicans to the vascular compartment may induce receptors that enhance further interactions of the fungus with the host.
Considering all the experimental evidence reported, it seems likely that there is more than one mechanism mediating the binding of fibronectin to candidal surfaces. There may be multiple fibronectin receptors. As noted in the discussion of laminin binding (see above), the preparation of the extract used to identify reactive species may profoundly affect the outcome (164). In fresh, undialyzed extracts, multiple fibronectin binding species were identified. In dialyzed extracts, the number and relative contribution of the species were altered. In addition, only some of the reactive species identified by ligand affinity blots were detected with antiserum to the human receptor. Antigenic differences between human receptors and candidal receptors may also contribute to differences, as may the conditions used for analysis. Santoni et al. (465) found a difference in reactivity with C. albicans and other species among anti-α-subunit MAbs. In a later study that failed to identify a reactive moiety, the less reactive MAb was used (102). Alternatively, a single receptor could recognize multiple domains with different affinities. There is precedence for the latter among proteins such as the staphylococcal fibronectin binding protein (36). A third possibility is that there is a single receptor protein that in different forms or states of modification has affinity for different domains of fibronectin.
The expression of fibronectin binding proteins has not been examined specifically in vivo. However, both in vitro and in vivo observations support the hypothesis that binding to fibronectin is a contributor to host interactions. Since the preferential adhesion to ECM as opposed to endothelial cells was reported (248), adhesion studies have focused on soluble and immobilized fibronectin as discussed above. These provide models of exposure to serum fibronectin and ECM. However, several studies have suggested that fibronectin can mediate cellular adhesion (230, 248, 472, 507). Binding to both buccal and vaginal epithelial cells in suspension was reduced by the addition of fibronectin (230, 507). These epithelial cells were demonstrated by immunofluorescence techniques to have fibronectin on their surfaces. Vaginal epithelial cells could be demonstrated to be composed of two populations that differed in surface fibronectin (230). More C. albicans yeast cells bound to the high-fibronectin epithelial cells than to the low-fibronectin epithelial cells, and adhesion to the high-fibronectin cells was inhibited by the addition of exogenous fibronectin. The high-fibronectin epithelial cells were more abundant in epithelial-cell populations recovered during the first or fourth week of the menstrual cycle, during pregnancy, and from diabetic women (475). In a model of nonbacterial thrombotic endocarditis, it was noted that fibronectin was present on treated but not normal rabbit cardiac valvular endothelium and that there was a correlation between organisms that bound fibronectin and those that caused infection (472). In vivo evidence of a role of adhesion to fibronectin or any other ECM ligand in pathogenesis is at best scant. Binding of C. albicans to substrates with RGD recognition motifs contributed to studies to examine the effect of peptides containing the motif in model systems. Klotz et al. (260) reported that an RGD-containing peptide reduced the census of yeast cells in several tissues in a rabbit model of infection 4 h after intravenous inoculation and proposed that the effect could be attributable to blocking the adherence to host substrates. In a perfused murine liver model, treatment of yeast cells with RGD-containing peptides increased the trapping and killing of yeast cells when they were infused into the liver (469). The authors suggested that binding of the peptide to the yeast surface might serve as an opsonin, promoting killing by Kupffer cells. Another possibility would be that inhibition of adherence to other liver cells increased the proportion of yeast cells that were available for phagocytosis and killing. Whether the multiple fibronectin receptors indicated by several studies result from processing of the product or a single gene or from the expression of multiple genes, a genetic approach to examine the contribution interactions with soluble and insoluble fibronectin and role of the various binding moieties will be very challenging.
(iii) Entactin.
López-Ribot and Chaffin (293) examined the interaction between C. albicans and entactin (nidogen), a novel adhesion glycoprotein molecule present in the ECM, especially in the basement membrane (239). Entactin is a sulfated glycoprotein that contains an RGD sequence and that forms a tight complex with laminin and interacts with collagen IV, fibronectin, fibrinogen, and itself. The interaction of entactin with intact cells and the binding moieties present in cell wall extracts was examined. Cells of both the yeast and the hyphal morphologies of the fungus were found to bind entactin, as detected by an indirect immunofluorescence assay. Most germ tubes bound entactin, and binding was heterogenously distributed on the hyphal surface with no reactivity on the parent yeast cell. A small percentage of nongerminated yeast cells in a germ tube culture bound entactin, and about 10% of cells in a yeast culture expressed the entactin binding capacity.
Cell wall material in a βME extract from both morphologies bound to immobilized entactin. The specificity of the binding was demonstrated by the ability of either an anti-entactin antibody or an antiserum to cell wall components to block the binding. No morphological differences were associated with the binding components, since extracts from yeast cells were able to completely inhibit the binding of germ tube components to entactin and vice versa. Binding utilized both RGD-dependent and -independent binding sites, since an RGDS peptide inhibited binding by approximately 50%. Thus, there are at least two sites on the entactin molecule that are recognized by candidal binding proteins. A single RGD sequence is present in the central domain of entactin. Preincubation of the extracts with laminin or fibronectin depleted the extracts of some entactin binding capacity. This may be due to a promiscuous receptor that binds multiple ligands, e.g., RGD motif, or to a multisubunit receptor that contains binding proteins for several ECM components.
The entactin binding components of the extract were detected by ligand affinity blotting with entactin. Moieties with molecular masses of approximately 25, 44, and 65 kDa present in βME cell wall extracts from both growth forms reacted with entactin. These bands, which appear to be identical in the two growth forms, may represent different stages of modification of the same entactin binding protein or different binding proteins that may recognize the same or different adhesive domains on entactin. Since entactin has the ability to bind to collagen, as noted above, one or more of these proteins may contain collagenous domains that are interacting with the collagen binding site of entactin. The possible relationship (if any) of these molecules to some other receptor-like molecules described for C. albicans within the same molecular weight range remains undetermined.
(iv) Vitronectin.
C. albicans cells can bind vitronectin (serum-spreading factor) (222). Vitronectin is a constituent of vascular walls and dermis. It is present in serum and is involved in the regulation of blood coagulation (97). It contains a number of domains capable of interacting with microorganisms and mammalian cells, including an RGD sequence that binds eukaryotic integrin receptors and a glycosaminoglycan-binding domain that interacts with complex carbohydrates and glycoconjugates (182, 528). The characteristics of vitronectin binding to Candida were found to be the same as and to differ significantly from those of other ECM components such as fibronectin. The binding of vitronectin increased during late exponential growth of the organism (222). Two studies that have examined the effect of growth temperature and cell surface hydrophobicity (CSH) on the interaction have reached somewhat different conclusions. Jakab et al. (222) found that cells grown at 37°C bound 35% more soluble vitronectin than did cells grown 21°C and that cells with increased CSH bound somewhat more vitronectin than did cells with more hydrophilic surfaces. On the other hand, Silva et al. (505) found that hydrophobic cells grown at 23°C did not differ significantly in their adhesion to immobilized vitronectin from cells grown at 37°C. The interaction with soluble vitronectin was optimal at pH 4 and was enhanced substantially by calcium at pH 7 (222). A reduction in the binding capacity of cells following heating or treatment with several proteases suggested the presence of a protein receptor. Among potential binding inhibitors, a 64% reduction was observed with fibronectin and a minimal reduction of 10% with fibrinogen and collagen type I, while no effect was noted with type IV collagen and gelatin. Unlike other ECM ligands, no clear role for the RGD motifs present in the vitronectin molecule was established, since RGD and RGDS were ineffective inhibitors while GRGDS was somewhat effective (20% inhibition). Heparin was the most effective inhibitor (50 to 85% inhibition) (222, 286). This suggests that binding may be through the glycosaminoglycan binding region of vitronectin. Analysis of binding of plasma vitronectin revealed the presence of both high- and low-affinity receptors (286). For the high-affinity receptor, analysis yielded a determination of 98,000 binding sites per cell with a Kd of 3.5 × 10−7 M. This dissociation constant is similar to the low-affinity binding constant reported in one study for fibronectin (380), although the number of binding sites is larger than reported for any other ligand.
Both carbohydrate and protein receptors for vitronectin have been reported from the same laboratory. In agreement with the prediction of a protein receptor (222), ligand affinity blotting with radiolabeled vitronectin identified a 30-kDa species extracted from cells with SDS and βME (286). In the blotting analysis, heparin completely inhibited vitronectin binding, although it inhibited the binding to intact cells by only 50%. More recently, Olson et al. (398) have reported that fungi interact with vitronectin through cell wall β-glucan, similarly to the mechanism described for Pneumocystis carinii (287). The authors hypothesized that the glycosaminoglycan binding region of vitronectin bound cell wall β-glucan. Using β-glucan from S. cerevisiae, they demonstrated by suspension binding analysis that there was a concentration-dependent specific binding between vitronectin and β-glucan. Assuming that the candidal vitronectin receptor would be antigenically related to the human integrin vitronectin receptor, Klotz et al. (259) analyzed an octylglucoside extract of isolated cell walls with antiserum to the human receptor. Moieties of 50, 60, and 90 kDa were most reactive, although several other components in the same molecular weight range were also detected.
Since it has been reported that vitronectin is adsorbed to polymeric biomaterials to a much higher degree than other ECM and serum proteins, attachment of C. albicans to indwelling plastic catheters and prostheses (contaminated medical material is a major cause of nosocomial Candida infections [see below]) could take place through interactions with vitronectin. In conclusion, vitronectin binding may represent a novel adherence mechanism of C. albicans cells to host tissues and biomaterials (222). Binding to vitronectin may also be a protective mechanism for the host. Interaction with vitronectin appears to increase the binding of C. albicans to a macrophage cell line and phagocytosis (286). Vitronectin-coated β-glucan particles stimulate tumor necrosis factor alpha release from macrophages, although at high concentrations the β-glucan suppressed cytokine release (398). Since β-glucan is common to fungi, this may be a general fungal recognition system. Binding of C. albicans to macrophage may involve multiple mechanisms or may be specific to macrophages from different tissues. As noted below, moieties of both acid-labile and acid-stable mannan appear to mediate the binding to splenic macrophages (234, 285).
(v) Collagens.
Klotz (249) showed that C. albicans bound to immobilized type IV collagen, a component of basement membrane, and also to type I collagen. Binding to gelatin (denatured type I collagen) was about one-third lower than to the native form in this study, although in a more recent study the binding to native and denatured collagen was similar (255). Binding to type I collagen was reduced by about half to five fold by the removal of calcium and magnesium ions (249, 259). Calcium mediated most of the ion enhancement, since removal of calcium effected nearly a threefold reduction (259).
Adhesion of C. albicans to type I and type IV collagen is inhibited almost completely by fibronectin and substantially by a GRGRSP peptide (253). Adhesion to type I but not type IV collagen is inhibited about 40% by a GRGDTP peptide. On the other hand, adhesion to type IV but not type I collagen is inhibited by an RGD peptide. Heparin inhibited the binding to type I and type IV collagen (254). When tested with type I collagen, heparan sulfate and dextran sulfate were also inhibitors. These compounds appeared to mediate the inhibition by binding to the ECM ligand rather than to the fungus. An unfractionated tryptic digest of gelatin almost completely abolished the binding of C. albicans to immobilized type IV and type I collagen, gelatin, fibronectin, and laminin (255). Fragments of gelatin produced by CnBr digestion were also examined. Several peptides isolated by high-pressure liquid chromatography were very effective in inhibiting binding to immobilized gelatin. The only identifiable peptide was a 47-amino-acid fragment with residues 40 to 86 of the α-1 chain of type I collagen that contains three RGX sequences but not RGE or RGD sequences. Several peptides from the α-1 chain were synthesized and examined. Only a 10-amino-acid peptide (residues 778 to 787), GQRGVVGLPG-NH2, was active, causing 68% inhibition. The authors suggest that inhibitory fragments from gelatin are biocompatible and are potential candidates for reducing adherence to host proteins in vivo.
Klotz et al. (259) used affinity chromatography to isolate receptors for gelatin present in an detergent extract of yeast cells. Two proteins of 60 and 105 kDa were obtained. The same two proteins were obtained from extracts of germ tubes and by affinity isolation with a fibronectin matrix. These proteins were discussed in the section on fibronectin binding proteins, above.
Mannan adhesins and other binding proteins.
In addition to the cell wall receptors that interact with host proteins found in serum and ECM, C. albicans has mannoproteins and proteins that bind other host ligands or that contribute to hydrophobic interactions. Among these additional interactions are recognition between carbohydrates and proteins. The carbohydrate partner may be either the candidal adhesin, as in the mannan moiety of mannoproteins, or the host ligand recognized by candidal lectin-like proteins. Furthermore, C. albicans can bind to plastic materials. Adhesion to plastic is not an esoteric interaction, since binding of the fungus to plastic materials found in catheters and protheses may contribute to the development of infection and deterioration of these devices and since adherent organisms may be more resistant to antifungal drugs.
(i) Mannan adhesins.
In addition to the ability of protein moieties to interact with the host, the mannan portion of mannoprotein may be involved in the fungus-host interaction. Two different oligosaccharides of mannan have been implicated in the adhesion of hydrophilic yeast cells to the marginal zone of murine spleen. A mutant strain deficient in the acid-labile portion of mannan was reduced in its ability to bind to marginal zone areas and displayed less specificity in binding, since cells also adhered to white pulp (73). Cutler and colleagues demonstrated that yeast cells bound to macrophages of the marginal zone and that mannan inhibited the binding to spleen and lymph node tissue (236). They identified a β-1,2-linked mannotetraose as the entity possessing adhesive activity in the acid-labile mannan (285). An additional adhesin that contributes significantly to the binding of yeast cells to the marginal zone has been found in the acid-stable portion of mannan (234). Factor 6, a mannooligosaccharide that confers serotype A specificity, also contributes to epithelial adhesion (357). A factor 6-deficient strain or serotype B strains showed reduced adhesion to exfoliated human buccal epithelial cells compared to serotype A cells. Mannan from serotype A parental cells but not from the deficient strain, as well as antiserum to factor 6, inhibited the binding of C. albicans A cells. O-linked mannosyl moieties of cell wall proteins may also play a role. The O-linked oligosaccharides of the 58-kDa fibrinogen binding protein have been implicated in the binding of ligand (60).
(ii) Hydrophobic proteins.
CSH in C. albicans appears to be related to a plethora of host interactions and fungal functions. These interactions and functions include the ability of the microorganism to adhere to epithelial and endothelial cell surfaces, ECM proteins, and plastic indwelling devices; to evade the action of phagocytic cells; and to enhance the uptake of substances from the medium (10, 39, 130, 185–187, 252, 258, 298, 309, 500, 549, 562).
Preliminary attempts to characterize cell wall components that may confer a hydrophobic character to the surface of C. albicans suggested that several surface proteins were potentially involved in CSH of the organism. Tronchin et al. (549) reported the existence of germ tube-specific proteins with molecular masses of >200,000, 200,000, 68, and 60 kDa involved in the adherence of C. albicans to plastic as discussed below. Adherence to a plastic substrate suggests that these moieties have hydrophobic character. Later, Hazen et al. (193) reported that treatment of C. albicans cells with lyticase, a β-1,3-glucanase preparation, released at least four medium- to low-molecular-mass (<65-kDa) surface proteins that were unique or more abundant in hydrophobic yeast cells. Another experimental approach to detecting this class of proteins was developed based on the preferential binding of hydrophobic proteins to latex spheres (295). Material released from the surface of intact yeast cells and germ tubes by mild treatment with βME or Zymolyase was adsorbed onto latex-polystyrene microspheres. SDS-PAGE analysis of the adsorbed proteins showed that different sets of low- to medium-molecular-weight (from 20 to 67 kDa) proteins and mannoproteins were associated with each of the four fractions. Thus, different proteins, including species released both by the reducing agent and by hydrolysis of glucan linkages, appeared to contribute to hydrophobic properties of yeast cells and germ tubes. Hydrophobic components appeared to be more abundant in the extracts from germ tubes than from yeast cells. This is in agreement with observations indicating that mycelial filaments of C. albicans are invariably hydrophobic regardless of the CSH displayed by the mother blastoconidia from which the germ tubes emanate (183, 184, 186, 189, 295, 297). Hydrophobic interaction chromatography–high-pressure liquid chromatography analysis of C. albicans surface proteins indicated that hydrophobic proteins are usually smaller than 50 kDa while hydrophilic proteins are predominantly larger than 90 kDa (190, 191). This difference is most probably due to the distinct glycosylation levels between hydrophobic and hydrophilic cell wall proteins (188). On the other hand, freeze-fracture examination of the most external cell surface layers revealed structural differences between hydrophobic and hydrophilic cells. Thus, hydrophilic cells exhibit a dense layer of fibrils, composed mostly of high-molecular-weight mannoproteins, that is absent or scant in hydrophobic cells (190). Hazen and Hazen (191) suggested that the amount of cell wall-bound hydrophobic proteins remains constant but the amount of hydrophilic masking proteins varies during growth. Hence, conversion from surface hydrophilicity to hydrophobicity by C. albicans cells could be due to changes in the length and concentration of fibrils present in the external wall layer (190, 191) or to changes in the glycosylation levels of cell wall mannoproteins (188, 501). Such changes, in turn, may account for the shifts in CSH within a population observed as a consequence of the morphologic transition or of different physiologic and environmental conditions.
Overall, the observations in the studies mentioned above indicate that CSH of C. albicans cells could be ascribed to protein and mannoprotein species that appear to be quantitatively minor wall components. However, detection of a relatively large assortment of hydrophobic molecules suggests that they may simultaneously play other essential physiological roles rather than exclusively confer the hydrophobic character to the cell surface. In this context, the germ tube proteins with molecular masses of 68 and 60 kDa (sometimes a doublet of 60 and 62 kDa) described by Tronchin et al. (549), which appear to be involved in the adherence of C. albicans cells to plastic, may also act as receptors for laminin, fibrinogen, and C3d (35, 549). Evidence suggesting that the mp58 fibrinogen binding mannoprotein (60) and possibly the p37 laminin receptor (294) contain exposed hydrophobic domains has been reported (295, 299, 483). Whether these receptors are hydrophobic or whether hydrophobic forces facilitate the initial contact and enhanced stability for receptor interactions between fungal and host ligands is not known (185, 187). In the case of the mp58 moiety, its postulated hydrophobic character would facilitate self-association of individual molecules within the highly glycosylated hydrophilic environment of the whole-cell-wall structure, leading to areas of increased density. Self-association may thus explain the clustering or asymmetric distribution of the mp58 within the cell wall structure (328). Clustering of receptors is a phenomenon that, as stated above, could increase the security of interaction between the fungal cell and the host.
Using a polyclonal antiserum against yeast hydrophobic proteins, Glee et al. (163) demonstrated that hydrophobic species are also exposed on fungal cells present in host tissues. In vivo expression supports the contention that CSH may play a role in candidal virulence and pathogenesis, since hydrophobic cells are more virulent in an animal model (10). In agreement with previous reports, this antiserum recognized hydrophobic proteins predominantly in the low-molecular-mass range (163). Several C. albicans secreted enzymes and receptor-like molecules have been characterized in this size range. However, distinct antibodies to the candidal secreted aspartyl proteinase and exo-β-(1,3)-glucanase were negative for moieties recognized by the polyclonal antiserum anti-hydrophobic proteins (163). These two enzyme proteins appear not to be among the hydrophobic moieties or to share determinants with them. Hydrophobic cells showed increased binding to several ECM components, i.e., fibronectin, laminin, fibrinogen, and type IV collagen, compared to hydrophilic cells (505). Among the proteins recognized by antiserum to hydrophobic proteins are some proteins that also react with anti-human fibronectin receptor antiserum (164). Enhanced binding to the ECM by hydrophobic proteins may be a contributor to the enhanced virulence of hydrophobic cells. On the other hand, the rate of accumulation and the amount of bound complement factor C3 were similar for hydrophilic and hydrophobic yeast cells (270). Thus, there may be a difference in glycosylation patterns between proteins that bind complement and those that bind ECM.
(iii) Fimbriae.
The presence of long, thin filamentous protein cell surface appendages termed fimbriae in C. albicans strains was initially reported by Gardiner et al. (154). Fimbriae may mediate adhesive interactions between the fungus and the host. As discussed above, further characterization of purified fimbrial preparations revealed that the major structural subunit of the fimbriae is a glycoprotein with a molecular mass of approximately 66 kDa (583). The glycoprotein consists of 80 to 85% carbohydrate (primarily d-mannose) and 10 to 15% protein composed of 50% hydrophobic amino acid residues. It is unclear whether there are additional minor components that contribute to the fimbrial structure and function. The fimbriae bind directly to buccal epithelial cells (BECs), and purified fimbrial preparations inhibit C. albicans binding to BECs (583). The ability of C. albicans yeast cells to bind specifically to the glycosphingolipid lactosylceramide has been reported (226). In this regard, C. albicans fimbriae also bind to asialo-GM1 (gangliotetraosylceramide) in a saturable and concentration-dependent manner (584). Therefore, the fimbrial protein appears to be the moiety mediating binding to glycosphingolipids displayed on the surface of human BECs and could represent an adhesion motif for fungal cells (226). Bacterial pili may also bind to the glycosphingolipid asialo-GM1, and antibodies to Pseudomonas aeruginosa pili cross-reacted with fimbriae (585). Recent studies showed that the epitope that confers receptor-binding properties (adhesintope) to C. albicans fimbriae appears to be identical to that present in P. aeruginosa pili (279, 586). Antibodies prepared to the bacterial adhesintope inhibited binding (43 to 50%) of the fungus to BECs. Two synthetic peptides from the P. aeruginosa adhesintope were competitive inhibitors of binding to asialo-GM1 (58 to 69%) and BECs (48 to 59%). The smaller peptide, DEQGIPK, that was the central region of the larger peptide was somewhat less effective than the larger peptide. These conserved adhesive motifs present in adhesins from evolutionarily distant microorganisms may account for the recognition of host ligands bearing βGalNAc(1-4)βGal groups. It has been demonstrated that a number of pulmonary pathogens are able to utilize the βGalNAc(1-4)βGal sequence as the ligand or receptor to attach to host tissues (271). Hence, this carbohydrate motif may also play a role in the genesis of Candida infections.
As discussed previously, there is a class of fimbriae composed of a protein with strong similarity to collagen that is widely distributed among fungi (67). Antiserum to protein epitopes of this collagen-type fungal fimbria reacts with surface moieties of C. albicans (67), although it is unclear whether this represents a putative second class of candidal fimbria. Celerin et al. (68) reported that native fungal fimbriae can function like a mammalian ECM component, acting as a substratum that allows the adherence, spreading, and proliferation of cells of a human melanoma cell line. They predicted that collagen-like fungal fimbriae have a role in fungus-host interactions. Along with other candidal cell surface moieties, including the mp58 and p37 receptors (see above), that contain collagenous domains, such a putative collagenous fimbrial protein may define additional binding motifs for the fungal cells.
(iv) Plastic binding proteins.
The ability of C. albicans cells to adhere to plastic medical devices was established some time ago. After adherence the organism may propagate and establish biofilms. The release of microorganisms from biofilms formed on different types of medical implants, prostheses, or catheters may contribute to or initiate acute disseminated nosocomial infections, whose frequency has increased dramatically in the last years due precisely to this reason (119, 166). In this context, a large number of nosocomial infections caused by Candida species derive from the use of different types of medical catheters and prostheses (390). Organisms may also colonize denture materials and adhere to saliva-coated plastic, as noted below. Organisms adhering to plastic may also be less susceptible to antifungal drugs (180, 231), and colonization may contribute to deterioration of the devices (48, 172, 321, 559).
It was suggested in early studies on plastic adherence that CSH plays a major role in this process (258, 354, 358). However, the nature of candidal cell surface moieties potentially involved in the attachment of C. albicans to plastic remained undetermined in these studies. McCourtie and Douglas (344) demonstrated that yeast cells grown in galactose were most strongly adherent to acrylic. Galactose-grown cells have an additional surface layer that may mediate the adherence to plastic. In later studies from their laboratory (discussed in the following section), similar conditions were shown to promote binding to epithelial cells, and material accumulating in the medium inhibited epithelial adhesion (92, 93, 345, 544). The relationship between material promoting epithelial adhesion and adherence to plastic is not known. Subsequently, Tronchin et al. (549) reported that germ tubes of C. albicans had a fibrillar surface layer that was responsible for enhanced adherence of the microorganism to polystyrene plastic. Analysis by SDS-PAGE of the material retained on plastic surfaces after removal of attached fungal cells allowed further characterization of the adhesins present in the fibrillar layer that could mediate binding to plastic. Two components of 68 and 60 kDa and two high-molecular-mass components (a 200-kDa moiety and another species with a molecular mass higher than 200 kDa) were solubilized from the plastic surface. The 68-kDa component appeared to be one of the major constituents of the germ tube surface layers. Further evidence for the role of cell wall surface moieties in binding to plastic has recently been reported (459). The authors described differential effects of several MAbs on germination and adhesion. MAb 3D9, discussed previously as identifying a germ tube protein, had no effect on either germination or adhesion. Incubation of yeast cells in the presence of MAb B9E decreased both germination and binding to polystyrene, whereas MAb 21E6 decreased germination but enhanced adhesion. Among Candida spp., C. albicans is less adherent than C. krusei and more adherent than C. glabrata (179).
Adhesion to plastic is likely to involve additional proteins. First, adhesion of C. albicans to plastic is regulated by environmental conditions and the phenotypic state of the organism (243, 344, 458). Second, as discussed in previous sections, the expression of cell wall proteins appears to be a very dynamic process. One may expect that the pattern of surface components with the ability to interact with plastic will vary depending on the strain, the morphologic transition process, or the nutritional and environmental shifts (e.g., temperature, composition of the culture medium, or phenotypic switching), factors that are known to affect the protein and mannoprotein composition of the cell wall of C. albicans (6, 59, 63, 72, 159, 195, 297, 301, 326, 422, 525). Hence, adhesion of C. albicans to plastic could be mediated by cell wall moieties that play other physiological roles (as suggested for protein and mannoprotein species conferring CSH) (295) rather than by specific surface components.
Because of their molecular mass and their topological location at the most external layers of the cell wall structure, the two high-molecular-weight species with affinity for plastic, detected by Tronchin et al. (549), could be equivalent to the HMWM that form the dense layer of fibrils present on the surface of hydrophilic cells (190). However, one may expect that these fibrillar proteins will display a hydrophilic character, while hydrophobic interactions appear to be essential for the attachment of C. albicans cells to plastic materials (258, 354, 358). These large plastic-adhering proteins were minor components and were not found in the culture medium (549). However, we have detected the presence of highly glycosylated HMWM in cell wall extracts from C. albicans (295) and in the material secreted to the culture medium by cells of both growth phases that were specifically retained on the surface on polystyrene-latex microspheres (299). These observations suggest that these species have exposed hydrophobic domains, most probably present in the peptide moiety. It is the peptide portion of the mannoprotein molecules that predominantly exhibited the ability to bind to plastic through hydrophobic bonds (540).
C. albicans is able to form biofilms (181). Biofilms consist of a monolayer or multilayer of cells embedded within a matrix of extracellular polymeric material (351). Three observations may account for the ability of this fungus to form such biofilms. (i) C. albicans cells can release into the medium protein and mannoprotein species chemically and antigenically related to the components bound to the cell wall that have exposed hydrophobic domains (2, 299, 543). (ii) Some wall components are not tightly bound to the wall structure since they are easily retained on the substratum where the cells attach (549). (iii) As noted in the following section, there may be complementary molecules in the released and surface-bound material that mediate adhesion. Hawser and Douglas (181) showed that the extent of biofilm formation depended on the type of catheter material. Biofilm formation was greatest on latex, which is frequently used in urinary catheters, followed by silicone elastomer and polyvinyl chloride, which is frequently found in central venous catheters. It was substantially lower on polyurethane and 100% silicone. In addition, strains differed in the extent of biofilm formation. Scanning electron microscopy showed that biofilm consists of a dense network of the various candidal morphological forms as well as extracellular material. The fungus can bind to silicone found in denture lining and voice prostheses (48, 565). Adhesion to denture soft lining materials was strain variable, greater to an acrylic base than to the liner, and reduced after coating with saliva (565). Changes in the material may offer potentiation of adhesion, since adherence to experimental silicone materials was lower than to a commercial lining material. Binding to silicone voice prostheses differed between two strains, with the strain having the more negative zeta potential binding more slowly to the negatively charged silicone rubber (48). Coating the silicone with saliva reduced binding, and bound organisms detached when an air bubble was passed through the flow chamber. Microorganisms may exist in mixed biofilms containing both bacteria and yeasts, as observed on silicone voice prostheses, where C. albicans strains were reported to have hydrophilic surfaces while the bacterial hydrophobicities were variable (382).
(v) Epithelial binding lectin-like protein.
The adhesion of C. albicans to BECs or vaginal epithelial cells facilitates colonization and can be regarded as the first step in the pathogenesis of Candida infections. Several categories of candidal adhesins appear to be involved in this process: (i) integrin analogs, (ii) fimbrial adhesins, (iii) lectin-like adhesins, and (iv) factor 6 (49, 50, 96, 110, 111, 209, 406). Of these, lectin-like interactions between the protein moiety of a candidal lectin-like mannoprotein adhesin and a carbohydrate receptor on the surface of the host cell seem to be a major mechanism of adhesion to epithelial cells. The presence of candidal lectin-like adhesins with specificity for l-fucose or GlcNAc has been reported (37, 53, 92, 93, 544). Fucose was the major inhibitor of binding of one strain of C. albicans to human BECs, while glucosamine and GlcNAc were major inhibitors of the binding of another strain (93). Synthesis of the lectin-like proteins was increased when yeasts were grown in galactose (345). While two different lectin-like proteins were produced, each strain appeared to produce only a single such protein with affinity for either l-fucose or GlcNAc (53, 93). None of these adhesins have been fully characterized, and there is no information on the molecular mass of the native moiety bearing the adhesion motif. However, a fucoside-binding protein fragment of an adhesin was purified from culture supernatants of C. albicans yeast cells by affinity adsorption chromatography on the trisaccharide determinant of the H blood group antigen that terminates in an l-fucose residue (53, 544). Before chromatography, the protein was treated with papain, N-glycanase, and dilute alkali. The purified adhesin fragment was devoid of carbohydrate, although no further biochemical characteristics of this molecule were reported. Analysis of the purified biotinylated adhesin by SDS-PAGE and Western blotting with 125I-streptavidin and subsequent autoradiography revealed a band corresponding to a protein with a molecular mass of 15.7 kDa (53). However, the intact mannoprotein moiety containing the fucoside-binding fragment is likely to be considerably larger.
These lectin-like proteins can mediate epithelial adhesion. Binding to human BECs and vaginal epithelial cells was examined with strains with different sugar specificities (93). The extracellular material recognizing l-fucose led to a more than 50% inhibition of binding of the homologous strain(s) producing the l-fucose lectin-binding material. Strains producing proteins recognizing GlcNAc were inhibited less than 30%. The material recognizing GlcNAc had no effect on the strains producing the l-fucose binding material. The purified fragment of the l-fucose binding protein inhibited the binding of yeast cells to BECs by up to 80% (53, 544). The failure to obtain complete inhibition suggests that other adhesins contributed to binding. A chromatography overlay assay was used to examine the binding of yeast cells, culture supernatant material, and purified fragment to glycosphingolipids extracted from human BECs and sheep erythrocytes (53). All bound to the same components that contained fucose. However, yeast cells but not the adhesin preparation bound to a component containing GlcNAc, possibly in the form of an A blood group antigen, suggesting that all adhesins may not be recovered in the culture supernatant. Binding to other extract components was not detected. This study also indicated that blood group antigens can act as epithelial cell receptors for C. albicans.
More recently, Enache et al. (129) have examined the binding of C. albicans to an esophageal cell line (HET1-A) that is mediated in part by a lectin-like interaction. Yeast cells grown in galactose bound more extensively than did cells grown in glucose. With this strain of C. albicans, GlcNAc and glucosamine inhibited binding by about 40%. A 190-kDa species was detected in the βME extracts of galactose-grown but not glucose-grown yeast cells by Aurodyne-stained blots of SDS-PAGE-separated proteins. This species was postulated to be associated with the increased adherence of galactose-grown cells. Whether this species is a cell wall-bound form of the material found by McCourtie and Douglas (345) is not known.
Such lectin-like interactions may contribute to other host or self interactions. Although binding to intact erythrocytes mediated by the lectin-like moiety was not examined by Cameron and Douglas (53), the recognition of sugar ligands on erythrocyte surfaces may contribute to a fungal system to obtain iron, as discussed above. The presence on the cell surface of ligands that are recognized by this lectin-like protein offers the possibility that fungal cell association is mediated, at least in part, by these complementary sugar-lectin moieties. In any case, further work is required to fully characterize these lectin-like candidal adhesins and to establish their relationship, if any, to other cell surface-bound components (e.g., the fimbrial adhesin) of the fungus, which are known to recognize other carbohydrate-containing molecules displayed on the surface of human epithelial cells such as glycosphingolipids (see above).
(vi) Agglutinin-like proteins.
Several genes have been identified that have homology to fungal agglutinin genes, in particular S. cerevisiae AGα1 encoding α-agglutinin (291). The proteins encoded by these genes are candidates for surface binding interactions. In C. albicans, the ALS (agglutinin-like sequence) family consists of at least four genes that may encode cell wall proteins (212, 473). In S. cerevisiae, α-agglutinin mediates cell-cell interaction during the mating of haploid yeast. It is possible that the ALS genes are remnants of such a cycle in C. albicans or encode proteins that have been subverted to another function. Like Agα1p, the Als proteins had hydrophobic N and C termini, suggestive of processing through the secretory pathway and addition of a GPI anchor. Recently, the GPI anchor of Agα1p was shown to be lost during covalent attachment of the protein to cell wall glucan (306). One unusual property of the Als proteins was a central domain of a tandemly repeated 36-amino-acid motif. These tandem repeats were not found in Agα1p. The number of copies of this repeated motif was highly variable, depending on which C. albicans strain was examined. In addition, ALS alleles in the same strain may exhibit different numbers of copies of the repeat motif. The repeat sequences were rich in serine, threonine, and proline and contained a consensus N-glycosylation site, suggesting that the repeat region would be highly N and O glycosylated. If indeed Als1p is in the cell wall, along with Hwp1p (513) (discussed above), it will be the second cell wall protein to contain a repeat motif. The repeat motifs of the Als proteins and Hwp1p are not the same. While ALS genes were originally isolated due to their expression in RPMI 1640-grown hyphae but not in YEPD-grown yeast forms, the genes do not appear to be regulated by the yeast-hyphal transition. Instead, regulation is dependent on components of the culture media.
Recently, Gaur and Klotz described the isolation of a gene, ALA1 (for agglutinin-like adhesin) encoding a C. albicans adhesin for extracellular matrix components and BECs that appears to be another member of the family (154a). A C. albicans genomic library was transformed into S. cerevisiae, and adherent clones were selected by using ECM protein-coated magnetic beads. Transformed S. cerevisiae bound to collagen IV, fibronectin, or laminin-coated beads. Plasmids conferring adherent properties were rescued, and sequencing revealed an ORF encoding a protein of 1,419 amino acid residues. The sequence predicted surface localization. Ala1p shows homology to C. albicans Als1p and S. cerevisiae Agα1, especially at the N terminus, where it shows characteristics reminiscent of Ig domains that may be implicated in binding. Its relationship to other ECM binding moieties described above remains to be established.
(vii) Adherence to Streptococcus spp. and other bacteria.
As a commensal of the oral cavity, C. albicans must adhere to surfaces and cells of the oral cavity to prevent clearance by the flushing action of saliva. In addition to adhesins that mediate the binding to BECs discussed in other sections, the organism can bind salivary proteins and oral and nonoral bacteria. These interactions appear to be effected through multiple mechanisms. Macroscopic and microscopic aggregation or agglutination of isolates with C. albicans has been observed with strains of Streptococcus sanguis, S. salivarius, S. mutans, S. mitis, Fusobacterium nucleatum, Actinomyces viscosus, Lactobacillus amylovorus, and Bacteroides gingivalis (13, 213). In general, agglutination with bacteria other than streptococcal species was sensitive to heat treatment of the bacteria and to inhibition by a sugar. For these species, the interaction suggested a lectin-like interaction with candidal surface carbohydrate. The streptococcal coagglutination appeared to involve a different mechanism, which has been examined more extensively in subsequent studies by Jenkinson and colleagues.
C. albicans and C. tropicalis but not C. kefyr and C. krusei were able to bind or coaggregate with S. gordonii (202). This interaction appeared to be a general property of the fungus, since numerous strains exhibited the ability to bind to the bacterium. In addition to S. gordonii, C. albicans yeast cells bound to S. oralis and S. sanguis but not to S. mutans and S. salivarius when the bacteria were immobilized in microtiter plate wells or cosedimented (203, 224). These results differ to some extent from the previous study in that binding to S. mutans and S. salivarius was not observed. Binding was greater for yeast cells grown at 37°C than at 28°C (224). The greatest coaggregation was observed when the cells were starved for glucose. The increase appeared to be due to synthesis of a new protein, since the response was inhibited by the presence of trichodermin or amphotericin B. The change was not related to hydrophobicity, since no change in this property was noted. The binding was inhibited by the addition of EDTA and heat or proteolytic treatment of yeast cells. Addition of calcium ions or treatment of yeast cells with βME did not alter coaggregation. Thus, candidate adhesins are among the proteins that can be released by β-glucanase treatment, although many proteins are present in both chemical and enzymatic extracts. Cell wall polysaccharide from S. gordonii inhibited the reaction (203). Antibody to the polysaccharide also reduced binding. This polysaccharide contained rhamnose, glucose, N-acetylgalactosamine, glucose, and GlcNAc. The binding was not inhibited by sugars found in the polysaccharide or fucose or mannose (203, 224), suggesting that the candidal recognition motif is a more complex oligosaccharide structure. The failure of sugars to inhibit the reaction suggests that the fungal adhesin is not the epithelial binding lectin-like adhesin discussed elsewhere in this review; on the other hand, the bacterial polysaccharide contains some of the same sugars recognized by this lectin-like adhesin.
However, the interactions are not restricted to recognition of bacterial carbohydrates (204). Treatment of the bacteria with alkali only partially reduced the interaction with the fungus and suggested the participation of alkali-insensitive moieties. Candidates for this interaction were found among surface proteins that were already known to contribute to bacterial interactions. Inactivation in S. gordonii of cshA that specified a high-molecular-mass protein resulted in about a 50% reduction in the binding of C. albicans cells. Antibody to the amino-terminal nonrepetitive domain of the protein but not the C-terminal portion containing repetitive amino acid blocks partially inhibited the binding of C. albicans. When the nonrepetitive and repetitive domains were immobilized in plate wells, they both supported candidal adhesion. Inactivation of the bacterial sspA or sspA and sspB genes, encoding antigen I/II adhesins, resulted in a greater decrease in binding (about 80%). When a recombinant Enterococcus faecalis strain expressed the sspB gene, binding of C. albicans was increased threefold compared to that for the nontransformed strain. The mutant strains of S. gordonii retained expression of the polysaccharide, supporting the contribution of the proteins to binding. Similar proteins are also expressed by S. oralis and S. sanguis but not by S. mutans and E. faecalis, providing a basis for differences in coadherence among bacterial species. These various studies support multiple interactions between streptococci and C. albicans.
(viii) Adherence to salivary proteins.
C. albicans can bind several salivary proteins, although very little is known about the cell wall components mediating the binding. C. albicans adheres to human salivary proteins when these proteins are use to coat dental acrylic or lining material or hydroxyapatite beads, a model for pellicle formation on tooth surfaces (54, 113, 385, 560). Several bacterial species, e.g., S. sanguis and A. viscosus, can bind to pellicle. Several C. albicans strains adhered to the beads, although differences in the extent of binding were noted (54). Glucose starvation of yeast cells grown at 28°C enhanced adhesion. Treatment of the bound salivary proteins but not the fungus with neuraminidase increased binding. Deglycosylation of salivary proteins bound to glass slides reduced the binding of the fungus (384). These observations suggest that carbohydrates may be involved in binding. Neuraminidase secreted by oral bacteria could increase the availability or identity of ligands participating in binding by exposing sugar residues that are recognized by candidal adhesins, e.g., the fucose-binding protein discussed above.
More than one salivary component may be involved. Fractionation of saliva showed that proline-rich proteins (PRPs) and statherin promoted binding to beads (54). In addition, an unknown component of another fraction of parotid saliva supported binding. Parotid saliva that is rich in PRPs significantly increases the binding to denture acrylic compared to that for submandibular/sublingual saliva (560). A number of PRPs have been described and implicated in the binding of some bacteria to pellicle (108). Binding of radiolabeled yeast cells to membranes containing electrophoretically separated saliva proteins identified four major reactive species (17, 20, 24, and 27 kDa) (399). Purification and partial-sequence analysis indicated that these were basic PRPs. PRPs may also become bound to oral epithelial cells and may contribute to the binding to those cells.
Mucins in human submandibular and sublingual saliva bind to both yeast cells and germ tubes, as detected by their presence in an extract of candidal and bound saliva proteins (113). Little binding was detected with components of parotid saliva. Similarly, the submandibular and sublingual saliva promoted yeast binding when bound to polymethylmethacrylate beads. On the other hand, in the same study, purified mucin failed to bind yeast cells to coated beads or to replace mucin-depleted saliva, suggesting that purification may alter the recognition structure. Mucins do not bind to the hydroxyapatite beads, and mucin fractions were not active in the study by Cannon et al. (54). In a solid-phase overlay assay with electrophoretically separated salivary components from either human or rat saliva transferred to nitrocellulose, a single reactive component was detected (199). The reactive component of human saliva was identified as a low-molecular-weight mucin, MG2 in human saliva and rat submandibular gland (RSMG) mucin in rat saliva. The purified RSMG component had characteristics of both mucin and proteoglycan (200). The binding moiety contained glycosaminoglycans and human blood group A oligosaccharide, which are generally associated with proteoglycans and mucins, respectively. Enzyme sensitivity assays suggested the presence of both chondroitin sulfate and heparan sulfate. The binding capacity was associated primarily with the heparan sulfate side chains. This finding contrasts with a previous report that while heparan sulfate inhibited fungal binding to ECM, it did so by binding to ECM and not directly to the fungus (254). In contrast to the observation that mucin was the only reactive species in an overlay assay of separated salivary components (199), as noted above, multiple PRPs were identified as reactive species in a recent study (399). These differences have yet to be resolved.
Binding of salivary components to silicone surfaces of denture soft lining material or voice prostheses may reduce binding compared to the uncoated material, as discussed previously (48, 565).
(ix) Miscellaneous.
A DNA sequence of C. albicans, AAFI, that confers enhanced adhesion to both polystyrene and epithelial cells as well as autoaggregation properties to S. cerevisiae transformants has been isolated (20, 114). A 30-kDa protein that may be responsible for these phenomena in S. cerevisiae transformants was identified on the surface of both yeast and hyphal C. albicans cells (21). The authors suggested that this moiety may be related to the cysteine-rich 30-kDa hydrophobins that have been isolated from the cell surface of Schizophyllum and Aspergillus species (568, 569). The ability of the 30-kDa protein to promote autoaggregation in S. cerevisiae transformants indicates that the cell surface molecule may also be involved in the interaction among the fungal cells, acting as an aggregation factor. In fact, the formation of macroscopic aggregates is commonly observed in C. albicans mycelial cultures. Further work is obviously required to determine the role of this protein in candidal biology and host interactions.
A 49-kDa component that was able to inhibit the binding of C. albicans to human BECs by about 40% was obtained from two strains of C. albicans yeast cells grown on galactose (225). The component that was present in culture supernatants or extracted from cells by salt and heat was about 96% proteinaceous with 4% carbohydrate and minor amounts of phosphate and sulfate. Although the similarity in size to enolase and the 47-kDa hsp90 fragment found in the cell wall was noted, the study did not examine the effect of sugars or other ligands or antibodies on the binding to epithelial cells. The relationship of this species to the lectin-like binding protein, iC3b receptor, or other epithelial cell adhesin is unknown.
WHERE ARE WE GOING? THE MYSTERIES AND CHALLENGES
A term that has been used in studies about cell wall proteins and in this review is “dynamic.” This term has been used to remind us that the cell wall is not a static, invariant structure but, rather, a structure that can change during the life of a single cell in response to the growth state and a variety of environmental stimuli. This plasticity of an organelle that is required to maintain the integrity of the cell in its normal milieu is perhaps surprising. However, the budding cycle, cell growth, and hyphal formation require that this protective envelope be modulated in even its structural role. As demonstrated by the studies considered in this review, the regulation of expression of components in the cell wall occurs at several levels. Even in the glucan and chitin component, the fraction that one may be tempted to consider most invariant, there is temporal regulation of synthesis during the cell cycle involving multiple enzymes (for recent reviews, see references 80 and 246).
In the previous sections of this review, we have focused primarily on individual components of the cell wall. These observations suggest the direction of studies in the near future. Thus, we can anticipate additional studies to identify cell wall proteins and their functions in vitro, particularly if they are postulated as being contributors to pathogenesis, and we can expect the extension of studies of identified proteins to demonstrate the role of specific components in host interactions. These latter studies are likely to take advantage of the increasing experience with molecular genetics with this organism and emerging views of host immune response. In these concluding remarks, we would like to summarize many of the observations from the standpoint of the questions they have raised about the mechanisms required to produce and regulate the structure and function of the proteins and mannoproteins in the cell wall. Some of the recent observations are quite surprising and raise questions that we would not have considered a few years ago.
The mannoprotein and protein fraction contains some 40 or more species that, in itself, introduces a measure of complexity to the cell wall. Redundancy appears to be common among binding proteins or receptors, with several moieties binding each ligand or plastic. Identification of proteins in cell wall or cell extracts is not synonymous with surface localization, which is a requirement to be the mediator of the binding observed in intact cells. Thus, with the exception of the plastic binding proteins that were isolated by a method that implies exposure of the fungal surface to the substratum, it is not clear that each of these components is present at the cell surface. For most of these proteins that bind the same ligand, we do not know whether they represent products of different genes, are related by processing, or both. Another complicating factor is the possibility that processing alters the ligand specificity or binding site used by the binding protein.
Another related issue is the suggestion made several years ago that candidal binding proteins are multifunctional. Supporting this contention is the similarity of apparent molecular size for some components, the competition among ligands for isolated components, and the analogy to mammalian integrins that may bind more than one ligand. On the other hand, some ligands bind to more than one integrin. In contrast to these observations, there are some proteins that seem to bind a single ligand, e.g., the mp58 fibrinogen binding protein and the p37 laminin receptor moieties. Two studies that directly addressed this point determined that the C3d binding and mp58 fibrinogen binding proteins were different and that laminin and fibrinogen binding were not colocalized in yeast cells (300, 302). The fungus also appears to use multiple interactions to bind to host cells and substrates. Two mannan adhesins have been identified in mediating the binding to splenic macrophage. C. albicans can bind to multiple ligands present in ECM and do so via more than one binding site, i.e., RGD-peptide dependent and independent. Multiple binding interactions have also been suggested to mediate the binding to epithelial cells, e.g., mannan factor 6, lectin-like protein, and iC3b binding protein. Unraveling the individual contribution of the various binding proteins, those that are singular and those that are promiscuous in their ligand specificity, to the complex substrates presented by the host tissue compared to the use of purified ligands in vitro will be challenging.
Multiple mechanisms affect the expression of various cell wall mannoproteins and proteins. First, expression may regulated at the level of gene transcription as judged from Northern blot analysis. Among the genes whose expression is regulated by nutrient conditions are the genes for mp58 (FBP1) and ALS1 (5, 212). At least two genes, HWP1 and HYR1, appear to be expressed only in hyphae (16, 513). There are also proteins whose level of expression responds to in vitro growth conditions. Thus, more iC3b binding protein is detected at elevated glucose levels and more lectin-like protein is detected when cells are grown on galactose rather than glucose (162, 210, 345). There are also proteins and protein modifications that are expressed on some cells but not others in the population (60, 72, 296). Some determinants are also expressed by a variable number of cells in the population, apparently depending on the stage of culture growth (41, 42, 72).
Expression of the SAP gene family clearly is subject to a variety of controls of environment, strain, and morphology. Determination of the conditions affecting locus-specific expression is under way, and elucidation of the regulatory circuits will surely follow (214, 359). Miyasaki et al. (359) have suggested that there may be different trans-acting elements present in different strains or whose expression is controlled by morphology that contribute to differences in locus expression. For other proteins, variable expression within a population exposed to the same environmental conditions poses some interesting questions. The phenomenon is similar to penetrance, in which not all organisms with an appropriate genotype express the phenotype. Nonexpressing yeast cells can give rise to expressing germ tubes, and subsequent yeast cultures have a population of variable expression. For the mp58 and p37 fibrinogen and laminin binding proteins, the expressing yeast cells were among the largest cells in the population. Since older cells are generally larger than young ones (70), an intriguing possibility is that yeast cell expression is associated either with age or with some alteration in control of cell size.
Multiple posttranslational mechanisms determine cell wall protein expression. Proteins whose cellular location is morphology dependent are examples of one type of regulation (5, 394, 563). Since at least two of these proteins differ in size in the different locations, a posttranslational processing-dependent mechanism of targeting may be hypothesized. Thus, when processed in one way, the protein bears a signal that targets one destination, and when processed in another way, it bears a different destination signal. Such a change could also be effected by a regulation of mRNA splicing that removed signals for processing and targeting under one condition but not the other. In the case of the one protein where gene expression has been examined (5), no obvious difference was noted in the size of the transcript in a Northern blot.
Another type of regulation is represented by proteins that are differentially distributed within the cell wall. First, there are proteins that are secreted through the cell wall to the exocellular environment, and then there are proteins that are retained within the cell wall. However, retained proteins can differ in their location. Some proteins are homogeneously distributed at the surface, and others are heterogeneously distributed (59, 62, 293, 296, 328). At least one protein, enolase, is located internally and is not expressed at the surface (8). Some proteins are thought to be covalently linked to glucan (237, 238, 247, 268, 462, 463). However, proteins that are postulated to be covalently linked, i.e., releasable by β-glucanases, may also be released by treatment with reducing agents (57). Among the proteins that have signal sequences are proteins such as Sap, whose presumed major destination is the exocellular environment, as well as proteins that remain predominantly wall associated. Therefore a signal sequence is not sufficient to determine the extracellular location. The retention or release mechanisms have yet to be explored fully.
These examples suggest the likely existence of multiple methods for targeting cell wall proteins to specific locations and retention of proteins in the cell wall. Gene sequences clearly indicate that some proteins are secreted by the classical endoplasmic reticulum-Golgi pathway. Proteins such as Hwp1, Hyr1p, Sap proteins, β-N-acetylglucosaminidase, and putative Als1p contain a signal peptide sequence associated with routing through this pathway (55, 215, 292, 359, 571). On the other hand, other cell wall proteins do not appear to contain such a signal peptide. These proteins include hsp70, enolase, PGK, and GAPDH, which that have been demonstrated to be bona fide cell wall components. A few exceptions of secreted proteins without such a signal sequence have been noted in plants, animals, and microbes (272). These proteins in prokaryotic organisms and the model yeast S. cerevisiae appear to be secreted by substrate-specific transport systems (ATP binding cassette [ABC] transporters), while the mechanisms for such proteins in mammalian cells are unknown. The secretion of a-factor in S. cerevisiae involves the ABC transporter Ste6p. Other cell wall proteins appear to be secreted by as yet unknown pathways (81). If this is a valid analogy, the existence of several such systems for this group of C. albicans proteins would be predicted. The presence of an hsp70 among this group raises another possibility. Among functions associated with cytoplasmic hsp70 is translocation across intracellular membranes. hsp70 may assist in its own translocation across the plasma membrane or may contribute to the translocation of other proteins. If members of this group of proteins provide important or essential functions for C. albicans, substrate-specific transporters may constitute a new target for antifungal agents.
One area in which we have minimal information, even from the model system of S. cerevisiae, is how proteins are targeted to specific cell wall locations. As noted above, there are several aspects to localization of a protein within the cell or cell wall. In S. cerevisiae, α-agglutinin is modified by addition of a GPI anchor that is lost during covalent attachment to cell wall glucan (306) and a truncated protein missing the GPI anchor is secreted to the medium (573). To date, no C. albicans cell wall proteins have been demonstrated to have a GPI anchor, although Hyr1p and the putative cell wall protein Als1p contain a likely sequence. Two proteins, determinants of which are recognized by MAb 4C12 or 3D9, are both released by β-glucanases, but both may also be found in βME extracts. If the enzymatically released moieties are covalently attached to glucan, there are several possibilities to explain this partitioning. First, the moieties could be secreted in both a GPI-tailed form (enzymatically released) and an unmodified form (chemically released). A second possibility would be that some of the bound form is subsequently released endogenously within the wall. A third possibility is that excess moiety is secreted and only a portion is attached.
Another subcellular partitioning problem is found with the hsp70, PGK, GAPDH, and enolase proteins that are found both in the cytoplasm and the cell wall. There must be some mechanism to apportion the protein between the two sites. For enolase, two genes have been identified, and the function of the two loci is unknown (420). One possibility is that the loci specify intra- and extracellular enolase. The cell wall enolase protein is blocked at its N terminus, and this type of modification could be used as a partitioning mechanism. Unlike enolase, only one locus is proposed for PGK (309). Subcellular partitioning may influence the regulatory circuit(s) affecting the regulation of the gene. The promoter region of C. albicans PGK1 did not contain some of the regulatory sequences found in S. cerevisiae (6), and the noncoding regions of other glycolytic enzymes are less highly conserved than the coding regions (530). These differences support the potential for different regulation between the two species that could be related in part to the subcellular partitioning in C. albicans.
A homogeneous and heterogeneous distribution of moieties on the cell surface has been demonstrated. However, mechanisms to effect such differences in distribution have not been addressed. Since there is thought to be minimal mobility within the cell wall, such an asymmetric distribution is likely to occur during cell wall synthesis. Indeed, surfaces of formaldehyde-fixed cells show the same distribution pattern of fibrinogen binding as do viable cells (60). Thus, this asymmetry is not induced by ligand binding. Furthermore, the mp58 and p37 binding proteins, which are both asymmetrically distributed, are not codistributed in cells that express both activities (302). Such a subcellular localization could be introduced by an asymmetric delivery of the proteins to the growing cell wall or by self-aggregation following delivery. However, the last possibility would suggest that different proteins are separately delivered or have different aggregation mechanisms.
Although none of the glycolytic enzyme proteins present in the cell wall have been demonstrated to also bind host ligands, there is a precedent among other microbial systems for this activity. With several proteins of the glycolytic pathway present, at least two of which appear to be enzymatically active, the possibility that the function of these proteins is indeed enzymatic will have to be considered. This is particularly interesting, since the reactions are able to generate ATP and NADH, which could be used for extracellular reactions, such as covalent linkages between cell wall components. However, this would also require that the substrates for the reaction be available.
Lastly, there is tantalizing evidence that cell wall adhesins/binding proteins are environmental sensors capable of initiating an alteration in fungal gene expression. Bailey et al. (14) showed that the profile of abundant proteins was altered by adherence to HeLa cells and that after adhesion, at least one protein appeared to become phosphorylated. Alterations of gene expression are mediated by some integrins (216). If the functional analogy of some surface proteins with integrins extends beyond the identity of the ligand, binding of ligands such as ECM may also initiate a cascade of signals that alter gene expression. If this suggestion is substantiated, a whole new avenue of investigation will open to determine the genes and functions altered by adhesion and the adhesive interactions that are able to initiate such events. In this context, it is provocative that the major ubiquitinated proteins of the cell wall also are binding proteins. There are suggestions that ubiquitination may be involved in the regulation of receptor signaling for some mammalian receptors (368, 404).
FINAL COMMENT AND OUTLOOK
Our investigation into the proteins and mannoproteins of the cell wall of C. albicans is little more than a decade old. From these studies, along with those of the glucan and chitin component, has emerged a picture of a complex organelle. The number of proteins and mannoproteins found in wall in itself provides some measure of complexity. To this is added the evidence that proteins and mannoproteins collectively and individually do not have the same patterns of distribution within the wall, at the cell surface, or secreted to the extracellular environment. Furthermore, proteins are not retained in the wall by the same mechanisms, i.e., covalent or noncovalent attachment, and this again applies to some individual proteins that appear to have both mechanisms. A schematic model showing some of these aspects of distribution of cell wall components and the distribution of various protein and glycoprotein species in the wall structure is presented in Fig. 4. The identification and function of some these extracellular proteins as hydrolyases, adhesins, and putative structural proteins were not unexpected. However, the identity of some proteins to intracellular enzymes, the antigenic, functional, or structural relationships that some proteins appear to have with mammalian integrins, and the redundancy of proteins with apparently similar functions were not anticipated. The cell wall proteins are subject to regulation at the transcriptional and posttranslational levels by mechanisms that are responsive to environmental conditions and growth state. In turn, some cell wall proteins may be sensors of environmental contacts that initiate a signal for alterations in fungal gene expression. The localization of proteins within the cell wall is apparently subject to regulatory mechanisms that we are only now recognizing and have not yet begun to explore. This organelle is integral to fungal biology and pathogenesis of candidal infection, and unraveling its mysteries will provide challenges for the next decade.
FIG. 4.
Schematic diagram of the cell wall (CW) structure of C. albicans, showing the presence of different layers enriched in particular components. The microfibrillar polymers of β-glucans (β-g, ) and chitin (c,  ) appear to be more heavily concentrated in the inner cell wall domains; β-glucan–chitin complexes that appear to be formed by glycosidic linkages between both polymers will be located adjacent to the plasma membrane (PM) and the periplasmic space (ps). Proteins and glyco(manno)proteins (gp) appear to be dominant in the outermost cell wall layer, although they are also distributed through the entire wall structure. Once secreted through the plasma membrane, some protein and glycoproteins species will remain at the periplasmic space, possibly playing enzymatic roles (▵); some others will establish functional (i.e., β-glucanases [▾]) or structural covalent associations with β-glucans and possibly also with chitin (•—•) adjacent to the plasma membrane; and, finally, other moieties will constitute the most external layer, where the different molecular entities may be homogeneously (□) or heterogeneously (fimbriae, cluster of receptor-like molecules, etc. [○]) distributed or specifically released (i.e., extracellular enzymes) to the extracellular medium (EM) (•, ▪). Proteins and glycoprotein species in the outermost wall layer (□, ○) may establish different types of covalent (disulfide linkages) and noncovalent (hydrophobic and hydrogen ionic bonds) interactions. During their passage through the wall from the plasma membrane and periplasmic space to the outermost cell wall layers (□, ○) and possibly the extracellular environment (•, ▪), proteins and glycoproteins are most likely to be in equilibrium with other proteinaceous constituents, thus contributing, at least from a functional point of view, to the cell wall layering. In any case, protein and glycoprotein species other than those specifically secreted to the exocellular medium may also be released to such locations by dying (lysed) cells or as a consequence of unbalanced processes of synthesis and degradation of the cell wall structure, required for wall expansion during cell growth. To simplify the scheme, some aspects such as possible interactions of cell wall components with the plasma membrane and proteins retained in the cell wall, apparently by either covalent or noncovalent linkages, are not depicted.
) appear to be more heavily concentrated in the inner cell wall domains; β-glucan–chitin complexes that appear to be formed by glycosidic linkages between both polymers will be located adjacent to the plasma membrane (PM) and the periplasmic space (ps). Proteins and glyco(manno)proteins (gp) appear to be dominant in the outermost cell wall layer, although they are also distributed through the entire wall structure. Once secreted through the plasma membrane, some protein and glycoproteins species will remain at the periplasmic space, possibly playing enzymatic roles (▵); some others will establish functional (i.e., β-glucanases [▾]) or structural covalent associations with β-glucans and possibly also with chitin (•—•) adjacent to the plasma membrane; and, finally, other moieties will constitute the most external layer, where the different molecular entities may be homogeneously (□) or heterogeneously (fimbriae, cluster of receptor-like molecules, etc. [○]) distributed or specifically released (i.e., extracellular enzymes) to the extracellular medium (EM) (•, ▪). Proteins and glycoprotein species in the outermost wall layer (□, ○) may establish different types of covalent (disulfide linkages) and noncovalent (hydrophobic and hydrogen ionic bonds) interactions. During their passage through the wall from the plasma membrane and periplasmic space to the outermost cell wall layers (□, ○) and possibly the extracellular environment (•, ▪), proteins and glycoproteins are most likely to be in equilibrium with other proteinaceous constituents, thus contributing, at least from a functional point of view, to the cell wall layering. In any case, protein and glycoprotein species other than those specifically secreted to the exocellular medium may also be released to such locations by dying (lysed) cells or as a consequence of unbalanced processes of synthesis and degradation of the cell wall structure, required for wall expansion during cell growth. To simplify the scheme, some aspects such as possible interactions of cell wall components with the plasma membrane and proteins retained in the cell wall, apparently by either covalent or noncovalent linkages, are not depicted.
ACKNOWLEDGMENTS
Work in the laboratory of W.L.C. was supported by Public Health Service grant AI23416 from the National Institutes of Health. Work in the laboratory of J.P.M. was supported by grants from DIGICyT (PM92-0246) and CICyT (SAF95-0595), Ministerio de Educación y Ciencia, and FISSS (93/0801), Ministerio de Sanidad y Consumo, Spain. The support of grant CRG 931457 (NATO Collaborative Research Grants Programme) to W.L.C. and J.P.M. is also acknowledged. J.L.L.R. acknowledges support from Public Health Service grant 1 R29 AI42401-01 and a Pfizer Medical Education Grant.
We thank Frans Klis for providing manuscripts prior to publication.
REFERENCES
- 1.Abad-Zapatero C, Goldman R, Muchmore S W, Hutchins C, Steart K, Navaza J, Payne C D, Ray T L. Structure of a secreted aspartic protease from C. albicans complexed with a potent inhibitor: implications for the design of antifungal agents. Protein Sci. 1996;5:640–652. doi: 10.1002/pro.5560050408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguiar J M, Baquero F, Jones J M. Candida albicans exocellular antigens released into a synthetic culture medium: characterization and serological response in rabbits. J Gen Microbiol. 1993;139:3005–3010. doi: 10.1099/00221287-139-12-3005. [DOI] [PubMed] [Google Scholar]
- 3.Akashi T, Homma M, Kanbe T, Tanaka K. Ultrastructure of proteinase-secreting cells of Candida albicans studied by alkaline bismuth staining and immunocytochemistry. J Gen Microbiol. 1993;139:2185–2195. doi: 10.1099/00221287-139-9-2185. [DOI] [PubMed] [Google Scholar]
- 4.Alaei S, Larcher C, Ebenbichler C, Prodinger W M, Janatova J, Dierich M P. Isolation and biochemical characterization of the iC3b receptor of Candida albicans. Infect Immun. 1993;61:1395–1399. doi: 10.1128/iai.61.4.1395-1399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alloush H M, López-Ribot J L, Chaffin W L. Dynamic expression of cell wall proteins of Candida albicans revealed by probes from cDNA clones. J Med Vet Mycol. 1996;34:91–97. [PubMed] [Google Scholar]
- 6.Alloush H M, López-Ribot J L, Masten B J, Chaffin W L. 3-Phosphoglycerate kinase: a glycolytic enzyme present in the cell wall of Candida albicans. Microbiology. 1997;143:321–330. doi: 10.1099/00221287-143-2-321. [DOI] [PubMed] [Google Scholar]
- 7.Anderson J, Mihalik R, Soll D R. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J Infect Dis. 1996;173:684–690. doi: 10.1093/infdis/173.3.684. [DOI] [PubMed] [Google Scholar]
- 9.Annaix Y, Bouchara J R, Tronchin G, Senet J M, Robert R. Structures involved in binding human fibrinogen to Candida albicans germ tubes. FEMS Microbiol Immunol. 1990;64:147–154. doi: 10.1111/j.1574-6968.1990.tb03514.x. [DOI] [PubMed] [Google Scholar]
- 10.Antley P P, Hazen K C. Role of yeast cell growth temperature on Candida albicans virulence in mice. Infect Immun. 1988;56:2884–2890. doi: 10.1128/iai.56.11.2884-2890.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashman R B, Papadimitriou J M. Production and function of cytokines in natural and acquired immunity to Candida albicans infection. Microbiol Rev. 1995;59:646–672. doi: 10.1128/mr.59.4.646-672.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ataoglu H, Zueco J, Sentandreu R. Characterization of epitopes recognized by Candida factor 1 and 9 antisera by use of Saccharomyces cerevisiae mnn mutants. Infect Immun. 1993;61:3313–3317. doi: 10.1128/iai.61.8.3313-3317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagg J, Silverwood R W. Coagglutination reactions between Candida albicans and oral bacteria. J Med Microbiol. 1986;22:165–169. doi: 10.1099/00222615-22-2-165. [DOI] [PubMed] [Google Scholar]
- 14.Bailey A, Wadsworth E, Calderone R A. Adherence of Candida albicans to human buccal epithelial cells: host-induced protein synthesis and signalling events. Infect Immun. 1995;63:569–572. doi: 10.1128/iai.63.2.569-572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey, D. A., N. A. R. Gow, and A. J. P. Brown. 1995. GenBank accession no. Z544197.
- 16.Bailey D A, Feldman P J F, Bovey M, Gow N A R, Brown A J P. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballou C E. Structure and biosynthesis of the mannan component of the yeast cell envelope. Adv Microb Physiol. 1976;14:93–158. doi: 10.1016/s0065-2911(08)60227-1. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee A, Ganesan K, Datta A. Induction of secretory acid proteinase in Candida albicans. J Gen Microbiol. 1991;137:2455–2461. doi: 10.1099/00221287-137-10-2455. [DOI] [PubMed] [Google Scholar]
- 19.Banno Y, Yamada T, Nozawa Y. Secreted phospholipases of the dimorphic fungus Candida albicans: separation of three enzymes and some biological properties. Sabouraudia. 1985;23:47–54. doi: 10.1080/00362178585380081. [DOI] [PubMed] [Google Scholar]
- 20.Barki M, Koltin Y, Yanko M, Tamarkin A, Rosenberg M. Isolation of a Candida albicans DNA sequence conferring adhesion and aggregation on Saccharomyces cerevisiae. J Bacteriol. 1993;175:5683–5689. doi: 10.1128/jb.175.17.5683-5689.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barki M, Koltin Y, van Wetter M, Rosenberg M. A Candida albicans surface antigen mediating adhesion and autoaggregation in Saccharomyces cerevisiae. Infect Immun. 1994;62:4107–4111. doi: 10.1128/iai.62.10.4107-4111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett-Bee K, Hamilton M. The detection and analysis of chitinase activity from the yeast form of Candida albicans. J Gen Microbiol. 1984;130:1857–1861. doi: 10.1099/00221287-130-7-1857. [DOI] [PubMed] [Google Scholar]
- 23.Barret-Bee K, Hayes Y, Wilson R G, Riley J F. A comparison of phospholipase activity, cellular adherence and pathogenicity of yeasts. J Gen Microbiol. 1985;131:1217–1221. doi: 10.1099/00221287-131-5-1217. [DOI] [PubMed] [Google Scholar]
- 24.Barturen B, Bikandi J, San Millán R, Moragues M D, Regúlez P, Quindós G, Pontón J. Variability in expression of antigens responsible for serotype specificity in Candida albicans. Microbiology. 1995;141:1535–1543. doi: 10.1099/13500872-141-7-1535. [DOI] [PubMed] [Google Scholar]
- 25.Bendel C M, Hostetter M K. Distinct mechanisms of epithelial adhesion for Candida albicans and Candida tropicalis. J Clin Invest. 1993;92:1840–1849. doi: 10.1172/JCI116775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendel C M, St. Sauver J, Carlson S, Hostetter M K. Epithelial adhesion in yeast species: correlation with surface expression of the integrin analog. J Infect Dis. 1995;171:1660–1663. doi: 10.1093/infdis/171.6.1660. [DOI] [PubMed] [Google Scholar]
- 27.Bertram G, Swoboda R K, Gooday G W, Gow N A R, Brown A J P. Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast. 1996;12:115–127. doi: 10.1002/(sici)1097-0061(199602)12:2<115::aid-yea889>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 28.Bobichon H, Gache D, Bouchet P. Ultrarapid cryofixation of Candida albicans: evidence for a fibrillar reticulated external layer and mannan channels within the cell wall. Cryo-Lett. 1994;15:161–172. [Google Scholar]
- 29.Bodey G P. Candidiasis: pathogenesis, diagnosis, and treatment. 2nd ed. New York, N.Y: Raven Press; 1993. [Google Scholar]
- 30.Boorstein W, Ziegelhoffer T, Craig E A. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 31.Borg M, Rüchel R. Expression of extracellular acid protease by proteolytic Candida spp. during experimental infection of oral mucosa. Infect Immun. 1988;56:626–631. doi: 10.1128/iai.56.3.626-631.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borkovich K, Farrelly F, Finkelstein D, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperature. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouali A, Robert R, Tronchin G, Senet J-M. Binding of human fibrinogen to Candida albicans in vitro: a preliminary study. J Med Vet Mycol. 1986;24:345–348. doi: 10.1080/02681218680000511. [DOI] [PubMed] [Google Scholar]
- 34.Bouali A, Robert R, Tronchin G, Senet J M. Characterization of binding of human fibrinogen to the surface of germ-tubes and mycelium of Candida albicans. J Gen Microbiol. 1987;133:545–551. doi: 10.1099/00221287-133-3-545. [DOI] [PubMed] [Google Scholar]
- 35.Bouchara J P, Tronchin G, Annaix V, Robert R, Senet J M. Laminin receptors on Candida albicans germ tubes. Infect Immun. 1990;58:48–54. doi: 10.1128/iai.58.1.48-54.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bozzini M, Visai L, Pignatti P, Petersen T E, Spezial P. Multiple binding sites in fibronectin and the staphylococcal fibronectin receptor. Eur J Biochem. 1992;207:327–333. doi: 10.1111/j.1432-1033.1992.tb17054.x. [DOI] [PubMed] [Google Scholar]
- 37.Brassart D, Woltz A, Golliard M, Neeser J R. In vitro inhibition of adhesion of Candida albicans clinical isolates to human buccal epithelial cells by Fucα1→2Galβ-bearing complex carbohydrates. Infect Immun. 1991;59:1605–1613. doi: 10.1128/iai.59.5.1605-1613.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 39.Braun P C. Surface hydrophobicity enhances cortiscosterone incorporation in Candida albicans. Infect Immun. 1994;62:4087–4090. doi: 10.1128/iai.62.9.4087-4090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brawner D L, Cutler J E. Variability in expression of a cell surface determinant on Candida albicans as evidenced by an agglutinating monoclonal antibody. Infect Immun. 1984;43:966–972. doi: 10.1128/iai.43.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brawner D L, Cutler J E. Ultrastructural and biochemical studies of two dynamically expressed cell surface determinants on Candida albicans. Infect Immun. 1986;51:327–336. doi: 10.1128/iai.51.1.327-336.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brawner D L, Cutler J E. Variability in expression of cell surface antigens of Candida albicans during morphogenesis. Infect Immun. 1986;51:337–343. doi: 10.1128/iai.51.1.337-343.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brawner D L, Cutler J E. Cell surface and intracellular expression of two Candida albicans antigens during in vitro and in vivo growth. Microb Pathog. 1987;2:249–257. doi: 10.1016/0882-4010(87)90123-9. [DOI] [PubMed] [Google Scholar]
- 44.Brawner D L, Cutler J E, Beaty W L. Caveats in the investigation of form-specific molecules of Candida albicans. Infect Immun. 1990;58:378–383. doi: 10.1128/iai.58.2.378-383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bromuro C, Torosantucci A, Gómez M J, Urbani F, Cassone A. Differential release of an immunodominant 65 kDa mannoprotein antigen from yeast and mycelial forms of Candida albicans. J Med Vet Mycol. 1994;32:447–459. doi: 10.1080/02681219480000601. [DOI] [PubMed] [Google Scholar]
- 46.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 47.Burford-Mason A P, Matthews R C, Williams J R B. Transient abrogation of immunosuppression in a patient with chronic mucocutaneous candidiasis following vaccination with Candida albicans. J Infect. 1987;14:147–157. doi: 10.1016/s0163-4453(87)91977-3. [DOI] [PubMed] [Google Scholar]
- 48.Busscher H J, Geertsema-Doornbusch G I, van der Mei H C. Adhesion to silicone rubber of yeast and bacteria isolated from voice prostheses: influence of salivary conditioning films. J Biomed Mater Res. 1997;34:201–209. doi: 10.1002/(sici)1097-4636(199702)34:2<201::aid-jbm9>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 49.Calderone R A. Molecular interactions at the interface of Candida albicans and host cells. Arch Med Res. 1993;24:275–279. [PubMed] [Google Scholar]
- 50.Calderone R A, Braun P C. Adherence and receptor relationships of Candida albicans. Microbiol Rev. 1991;55:1–20. doi: 10.1128/mr.55.1.1-20.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calderone, R. A., and W. M. Scheld. 1987. Role of fibronectin in the pathogenesis of candidal infections. Rev. Infect. Dis. 9(Suppl. 4):400–403. [DOI] [PubMed]
- 52.Calderone R A, Linehan L, Wadsworth E, Sandberg A L. Identification of C3d receptors on Candida albicans. Infect Immun. 1988;56:252–258. doi: 10.1128/iai.56.1.252-258.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cameron B J, Douglas L J. Blood group glycolipids as epithelial cell receptors for Candida albicans. Infect Immun. 1996;64:891–896. doi: 10.1128/iai.64.3.891-896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cannon R D, Nand A K, Jenkinson H F. Adherence of Candida albicans to human salivary components adsorbed to hyroxyapatite. Microbiology. 1995;141:213–219. doi: 10.1099/00221287-141-1-213. [DOI] [PubMed] [Google Scholar]
- 55.Cannon R D, Niimi K, Jenkinson H F, Shepherd M G. Molecular cloning and expression of the Candida albicans β-N-acetylglucosaminidase (HEX1) gene. J Bacteriol. 1994;176:2640–2647. doi: 10.1128/jb.176.9.2640-2647.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casal M, Linares M J. Contribution to the study of the enzymatic profiles of yeast organisms with medical interest. Mycopathologia. 1983;81:155–159. doi: 10.1007/BF00436820. [DOI] [PubMed] [Google Scholar]
- 57.Casanova M, Chaffin W L. Cell wall glycoproteins of Candida albicans as released by different methods. J Gen Microbiol. 1991;137:1045–1051. doi: 10.1099/00221287-137-5-1045. [DOI] [PubMed] [Google Scholar]
- 58.Casanova M, Chaffin W L. Phosphate-containing proteins and glycoproteins of the cell wall of Candida albicans. Infect Immun. 1991;59:808–813. doi: 10.1128/iai.59.3.808-813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casanova M, Gil M L, Cardeñoso L, Martínez J P, Sentandreu R. Identification of wall-specific antigens synthesized during germ tube formation by Candida albicans. Infect Immun. 1989;57:262–271. doi: 10.1128/iai.57.1.262-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casanova M, López-Ribot J L, Monteagudo C, Llombart-Bosch A, Sentandreu R, Martínez J P. Identification of a 58-kilodalton cell surface fibrinogen-binding mannoprotein from Candida albicans. Infect Immun. 1992;60:4221–4229. doi: 10.1128/iai.60.10.4221-4229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casanova M, López-Ribot J L, Martínez J P, Sentandreu R. Characterization of cell wall proteins from yeast and mycelial cells of Candida albicans by labelling with biotin: comparison with other techniques. Infect Immun. 1992;60:4898–4906. doi: 10.1128/iai.60.11.4898-4906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casanova M, Martínez J P, Chaffin W L. Fab fragments from a monoclonal antibody against a germ tube mannoprotein block the yeast-to-mycelium transition in Candida albicans. Infect Immun. 1990;58:3810–3812. doi: 10.1128/iai.58.11.3810-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casanova M, Martínez J P, Chaffin W L. Identification of germ tube cell wall antigens of Candida albicans. J Med Vet Mycol. 1991;29:269–272. doi: 10.1080/02681219180000391. [DOI] [PubMed] [Google Scholar]
- 64.Cassone A. Cell wall of Candida albicans: its functions and its impact on the host. Curr Top Med Mycol. 1989;3:248–314. doi: 10.1007/978-1-4612-3624-5_10. [DOI] [PubMed] [Google Scholar]
- 65.Cassone A, Torosantucci A, Palma C, Gómez M J, Ausiello C M, Djeu J Y. Cell wall constituents of Candida albicans as biological response modifiers. In: Goldstein A L, Garaci E, editors. Combination therapies. New York, N.Y: Plenum Press; 1992. pp. 159–166. [Google Scholar]
- 66.Cawley T N, Harrington M G, Letters R. A study of the phosphate linkages in phosphomannan in cell walls of Saccharomyces cerevisiae. Biochem J. 1972;129:711–720. doi: 10.1042/bj1290711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Celerin M, Day A W, Castle A J, Laudenbach D E. A glycosylation pattern that is unique to fimbriae from the taxon Microbotryales. Can J Microbiol. 1995;41:452–460. [Google Scholar]
- 68.Celerin M, Ray J M, Schisler N J, Day A W, Stetler-Stevenson W G, Laudenbach D E. Fungal fimbriae are composed of collagen. EMBO J. 1996;15:4445–4453. [PMC free article] [PubMed] [Google Scholar]
- 69.Cenciarelli C, Hou D, Hsu K C, Rellahan B L, Wiest D L, Smith H T, Fried V A, Weissman A M. Activation-induced ubiquitination of the T cell antigen receptor. Science. 1992;257:795–797. doi: 10.1126/science.1323144. [DOI] [PubMed] [Google Scholar]
- 70.Chaffin W L. The relationship between yeast cell size and cell division in Candida albicans. Can J Microbiol. 1984;30:192–203. doi: 10.1139/m84-030. [DOI] [PubMed] [Google Scholar]
- 71.Chaffin W L, Stocco D M. Cell wall proteins of Candida albicans. Can J Microbiol. 1983;29:1438–1444. doi: 10.1139/m83-220. [DOI] [PubMed] [Google Scholar]
- 72.Chaffin W L, Skudlarek J, Morrow K J. Variable expression of a surface determinant during proliferation of Candida albicans. Infect Immun. 1988;56:302–309. doi: 10.1128/iai.56.2.302-309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chaffin W L, Collins B, Marx J N, Cole G T, Morrow K J., Jr Characterization of mutant strains of Candida albicans deficient in expression of a surface determinant. Infect Immun. 1993;61:3449–3458. doi: 10.1128/iai.61.8.3449-3458.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chambers R S, Sullivan P A. Expression of the exoglucanase gene in yeast and hyphal forms of Candida albicans. FEMS Microbiol Lett. 1993;111:63–67. doi: 10.1111/j.1574-6968.1993.tb06362.x. [DOI] [PubMed] [Google Scholar]
- 75.Chambers R S, Broughton M J, Cannon R D, Carne A, Emerson G W, Sullivan P A. An exo-β-(1,3)-glucanase of Candida albicans: purification of the enzyme and molecular cloning of the gene. J Gen Microbiol. 1993;139:325–334. doi: 10.1099/00221287-139-2-325. [DOI] [PubMed] [Google Scholar]
- 76.Chambers R S, Walden A R, Brooke G S, Cutfield J F, Sullivan P A. Identification of a putative active site residue in the exo-β-(1,3)-glucanase of Candida albicans. FEBS Lett. 1993;327:366–369. doi: 10.1016/0014-5793(93)81022-r. [DOI] [PubMed] [Google Scholar]
- 77.Chattaway F W, Holmes M R, Barlow A J E. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J Gen Microbiol. 1968;51:367–376. doi: 10.1099/00221287-51-3-367. [DOI] [PubMed] [Google Scholar]
- 78.Chattaway F W, Shenolikar S, Barlow A J E. Release of acid phosphatase and polysaccharide- and protein-containing components from surface of dimorphic forms of Candida albicans by treatment with dithiothreitol. J Gen Microbiol. 1974;83:423–426. doi: 10.1099/00221287-83-2-423. [DOI] [PubMed] [Google Scholar]
- 79.Chaturvedi V P, Vanegas R, Chaffin W L. Coordination of germ tube formation and surface antigen expression in Candida albicans. FEMS Microbiol Lett. 1994;134:99–106. doi: 10.1111/j.1574-6968.1994.tb07268.x. [DOI] [PubMed] [Google Scholar]
- 80.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cleves A E, Cooper D N W, Barondes S H, Kelly R B. A new pathway for protein export in Saccharomyces cerevisiae. J Cell Biol. 1996;133:1017–1026. doi: 10.1083/jcb.133.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen J, Groyne F, Merchez M. Nucleotide sequence and transcriptional analysis in Candida albicans and Saccharomyces cerevisiae of the C. albicans gene coding for a secreted glucoamylase. Abstracts of the 13th International Conference on Yeast Genetics and Molecular Biology. Yeast. 1986;2:S67. [Google Scholar]
- 83.Cohen J, Francotte M, Thiriart C, van Wijnendaele F, Bruck C, de Wilde M. Cold Spring Harbor Symposium on Modern Approaches to New Vaccines Including Prevention of AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. Expression of the HIV-1 ENV (gp160) in the yeast Saccharomyces cerevisiae via expression/secretion vectors and partial characterization of gene product; p. 64. [Google Scholar]
- 84.Colina A, Aumont F, Deslaureirs N, Belhemeru P, de Repentigny L. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996;64:4514–4519. doi: 10.1128/iai.64.11.4514-4519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Constantino P J, Franklyn K M, Gare N F, Warmington J R. Production of antibodies to antigens of Candida albicans in CBA/H mice. Infect Immun. 1994;62:1400–1405. doi: 10.1128/iai.62.4.1400-1405.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Corbi A L, Kishimoto T K, Miller L J, Springer T A. The human leukocyte adhesion glycoprotein Mac-1 (complement receptor type 3, CD11b) α subunit. Cloning, primary structure, and relation to the integrins, von Willebrand factor and factor B. J Biol Chem. 1988;263:12403–12411. [PubMed] [Google Scholar]
- 87.Cortlandt D A, López-Ribot J L, Morrow K J, Straus D C, Chaffin W L. Dynamic expression of a cell surface determinant of Candida albicans. Mycopathologia. 1995;132:1–8. doi: 10.1007/BF01103780. [DOI] [PubMed] [Google Scholar]
- 88.Craig E A, Jacobsen K. Mutations of the heat-inducible 70 kilodalton genes of yeast confer temperature-sensitive growth. Cell. 1984;38:841–849. doi: 10.1016/0092-8674(84)90279-4. [DOI] [PubMed] [Google Scholar]
- 89.Craig E A, Jacobsen K. Mutations in cognate gene of Saccharomyces cerevisiae HSP70 result in reduced growth at low temperatures. Mol Cell Biol. 1985;5:3517–3524. doi: 10.1128/mcb.5.12.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Craig E, Kang P J, Boorstein W. A review of the role of 70 kDa heat shock proteins in protein translocation across membranes. Antonie Leeuwenhoek. 1990;58:137–146. doi: 10.1007/BF00548924. [DOI] [PubMed] [Google Scholar]
- 91.Craig E A, Gambill B D, Nelson R J. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol Rev. 1993;57:402–414. doi: 10.1128/mr.57.2.402-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Critchley I A, Douglas L J. Isolation and partial characterization of an adhesin from Candida albicans. J Gen Microbiol. 1987;133:629–636. doi: 10.1099/00221287-133-3-629. [DOI] [PubMed] [Google Scholar]
- 93.Critchley I A, Douglas L J. Role of glycosides as epithelial cell receptors for Candida albicans. J Gen Microbiol. 1987;133:637–643. doi: 10.1099/00221287-133-3-637. [DOI] [PubMed] [Google Scholar]
- 94.Cutfield S M, Dodson E J, Anderson B F, Moody P C, Marshall C J, Sullivan P A, Cutfield J F. The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Structure. 1995;3:1261–1271. doi: 10.1016/s0969-2126(01)00261-1. [DOI] [PubMed] [Google Scholar]
- 95.Cutforth T, Rubin G M. Mutations in hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 96.Cutler J E. Putative virulence factors of Candida albicans. Annu Rev Microbiol. 1991;45:187–218. doi: 10.1146/annurev.mi.45.100191.001155. [DOI] [PubMed] [Google Scholar]
- 97.Dahlbäck K, Löfberg H, Dahlbäck B. Immunohistochemical demonstration of vitronectin in association with elastin and amyloid deposits in human kidney. Histochemistry. 1987;87:511–515. doi: 10.1007/BF00492465. [DOI] [PubMed] [Google Scholar]
- 98.Damagnez V, Rolfe M, Cottarel G. Schizosaccharomyces pombe and Candida albicans cDNA homologues of the Saccharomyces cerevisiae UBC4 gene. Gene. 1995;155:137–138. doi: 10.1016/0378-1119(94)00926-j. [DOI] [PubMed] [Google Scholar]
- 99.De Bernardis F, Agatensi L, Ross I K, Emerson G W, Lorenzini R, Sullivan P A, Cassone A. Evidence for a role for secreted aspartate proteinase of Candida albicans in vulvovaginal candidiasis. J Infect Dis. 1990;161:1276–1283. doi: 10.1093/infdis/161.6.1276. [DOI] [PubMed] [Google Scholar]
- 100.De Bernardis F, Cassone A, Sturtevant J, Calderone R A. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect Immun. 1995;63:1887–1892. doi: 10.1128/iai.63.5.1887-1892.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Bernardis F, Chiani P, Ciccozzi M, Pellegrini G, Ceddia T, DíOffizzi G, Quinti I, Sullivan P A, Cassone A. Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect Immun. 1996;64:466–471. doi: 10.1128/iai.64.2.466-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DeMuri G P, Hostetter M K. Evidence for a β1 integrin fibronectin receptor in Candida tropicalis. J Infect Dis. 1996;174:127–132. doi: 10.1093/infdis/174.1.127. [DOI] [PubMed] [Google Scholar]
- 103.de Nobel J G, Dijkers C, Hooijberg E, Klis F M. Increased cell wall porosity in Saccharomyces cerevisiae after treatment with dithiothreitol or EDTA. J Gen Microbiol. 1989;135:2077–2084. [Google Scholar]
- 104.de Nobel H, Lipke P N. Is there a role for GPIs in yeast cell wall assembly? Trends Cell Biol. 1994;4:41–45. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 105.De Nollin S, Thoné F, Borgers M. Enzyme cytochemistry of Candida albicans. J Histochem Cytochem. 1975;23:758–765. doi: 10.1177/23.10.172554. [DOI] [PubMed] [Google Scholar]
- 106.Deslauriers N, Michaud J, Carré B, Léveillée C. Dynamic expression of cell-surface antigens probed with Candida albicans-specific monoclonal antibodies. Microbiology. 1996;142:1239–1248. doi: 10.1099/13500872-142-5-1239. [DOI] [PubMed] [Google Scholar]
- 107.Domer J E. Candida cell wall mannan: a polysaccharide with diverse immunologic properties. Crit Rev Microbiol. 1989;17:33–51. doi: 10.3109/10408418909105721. [DOI] [PubMed] [Google Scholar]
- 108.Douglas C W. Bacterial-protein interactions in the oral cavity. Adv Dent Res. 1994;8:254–262. doi: 10.1177/08959374940080021901. [DOI] [PubMed] [Google Scholar]
- 109.Douglas L J. Candida proteinases and candidosis. Crit Rev Biotechnol. 1988;8:121–129. doi: 10.3109/07388558809150541. [DOI] [PubMed] [Google Scholar]
- 110.Douglas L J. Mannoprotein adhesins of Candida albicans. In: Bennet J E, Hay R J, Peterson P K, editors. New strategies in fungal disease. New York, N.Y: Churchill Livingstone, Inc.; 1992. pp. 34–50. [Google Scholar]
- 111.Douglas, L. J. 1995. Adhesin-receptor interactions in the attachment of Candida albicans to host epithelial cells. Can. J. Bot. 73(Suppl. 1):S1147–S1153.
- 112.Duksin D, Mahoney W C. Relationship of the structure and biological activity of the natural homologues of tunicamycin. J Biol Chem. 1982;257:3105–3109. [PubMed] [Google Scholar]
- 113.Edgerton M, Scannapieco F A, Reddy M S, Levine M J. Human submandibular-sublingual saliva promotes adhesion of Candida albicans to polymethylmethacrylate. Infect Immun. 1993;61:2644–2452. doi: 10.1128/iai.61.6.2644-2652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Edwards, J. E. 1994. GenBank accession no. U18983.
- 115.Edwards J E, Gaither T A, O’Shea J J, Rotrosen D, Lawley T J, Wright S A, Frank M M. Expression of specific binding sites on Candida with functional and antigenic characteristics of human complement receptors. J Immunol. 1986;137:3577–3583. [PubMed] [Google Scholar]
- 116.Edwards J E, Mayer C L, Filler S G, Wadsworth E, Calderone R A. Cell extracts of Candida albicans block adherence of the organisms to endothelial cells. Infect Immun. 1992;60:3087–3091. doi: 10.1128/iai.60.8.3087-3091.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eigentler A, Schulz T F, Larcher C, Breitwieser E M, Myones B L, Petzer A L, Dierich M P. C3bi-binding protein on Candida albicans: temperature-dependent expression and relationship to human complement receptor type 3. Infect Immun. 1989;60:3087–3091. doi: 10.1128/iai.57.2.616-622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ellin R J, Wolff S M. Effect of pH and iron concentration on growth of Candida albicans in human serum. J Infect Dis. 1973;127:705–708. doi: 10.1093/infdis/127.6.705. [DOI] [PubMed] [Google Scholar]
- 119.Elliot T S J. Intravascular-device infections. J Med Microbiol. 1988;27:161–167. doi: 10.1099/00222615-27-3-161. [DOI] [PubMed] [Google Scholar]
- 120.El Moudni B, Rodier M H, Babin P, Jacquemin J L. Metallopeptidase in Candida albicans: immunolocalization in yeast. J Mycol Med. 1997;7:5–9. [Google Scholar]
- 121.El Moudni B, Rodier M H, Barrault C, Ghazali M, Jacquemin J L. Purification and characterisation of a metallopeptidase of Candida albicans. J Med Microbiol. 1995;43:282–288. doi: 10.1099/00222615-43-4-282. [DOI] [PubMed] [Google Scholar]
- 122.Elorza M V, Marcilla A, Sentandreu R. Wall mannoproteins of the yeast and mycelial cells of Candida albicans: nature of the glycosidic bonds and polydispersity of their mannan moieties. J Gen Microbiol. 1988;134:2393–2404. doi: 10.1099/00221287-134-8-2393. [DOI] [PubMed] [Google Scholar]
- 123.Elorza M V, Mormeneo S, García de la Cruz F, Gimeno C, Sentandreu R. Evidence for the formation of covalent bonds between macromolecules in the domain of the wall of Candida albicans mycelial cells. Biochem Biophys Res Commun. 1989;162:1118–1125. doi: 10.1016/0006-291x(89)90789-4. [DOI] [PubMed] [Google Scholar]
- 124.Elorza M V, Murgui A, Rico H, Sentandreu R. Formation of a new cell wall by protoplasts of Candida albicans: effect of papulacandin B, tunicamycin and nikkomycin. J Gen Microbiol. 1987;133:2315–2325. doi: 10.1099/00221287-133-8-2315. [DOI] [PubMed] [Google Scholar]
- 125.Elorza M V, Murgui A, Sentandreu R. Dimorphism in Candida albicans: contribution of mannoproteins to the architecture of yeast and mycelial cells. J Gen Microbiol. 1985;131:2209–2216. doi: 10.1099/00221287-131-9-2209. [DOI] [PubMed] [Google Scholar]
- 126.Elorza M V, Rico H, Sentandreu R. Calcofluor white alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- 127.Elorza M V, Rico H, Gozalbo D, Sentandreu R. Cell wall composition and protoplast regeneration in Candida albicans. Antonie Leeuwenhoek J Microbiol Serol. 1983;49:457–469. doi: 10.1007/BF00399324. [DOI] [PubMed] [Google Scholar]
- 128.Elorza M V, Sentandreu R, Ruiz-Herrera J. Isolation and characterization of yeast monomorphic mutants of Candida albicans. J Bacteriol. 1994;176:2318–2325. doi: 10.1128/jb.176.8.2318-2325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Enache E, Eskandari T, Borja L, Wadsworth E, Hoxter B, Calderone R. Candida albicans adherence to a human oesophageal cell line. Microbiology. 1996;142:2741–2746. doi: 10.1099/13500872-142-10-2741. [DOI] [PubMed] [Google Scholar]
- 130.Ener B, Douglas L J. Correlation between cell-surface hydrophobicity of Candida albicans and adhesion to buccal epithelial cells. FEMS Microbiol Lett. 1992;78:37–42. doi: 10.1016/0378-1097(92)90284-u. [DOI] [PubMed] [Google Scholar]
- 131.Eroles P, Sentandreu M, Elorza M V, Sentandreu R. Cloning of a DNA fragment encoding part of a 70 kDa heat shock protein of Candida albicans. FEMS Microbiol Lett. 1995;128:95–100. doi: 10.1111/j.1574-6968.1995.tb07506.x. [DOI] [PubMed] [Google Scholar]
- 132.Eroles P, Sentandreu M, Elorza M V, Sentandreu R. The highly immunogenic enolase and Hsp70p are adventitious Candida albicans cell wall proteins. Microbiology. 1997;143:313–320. doi: 10.1099/00221287-143-2-313. [DOI] [PubMed] [Google Scholar]
- 133.Faille C, Mackenzie D W R, Michalski J C, Poulain D. The evaluation of an enzyme immunoassay using neoglycolipids constructed from Candida albicans oligomannosides to define the specificity of anti-mannan antibodies. Eur J Clin Microbiol Infect Dis. 1992;11:438–446. doi: 10.1007/BF01961859. [DOI] [PubMed] [Google Scholar]
- 134.Faille C, Michalski J C, Strecker G, Mackenzie D W R, Camus D, Poulain D. Immunoreactivity of neoglycolipids constructed from oligomannosidic residues of Candida albicans cell wall. Infect Immun. 1990;58:3537–3544. doi: 10.1128/iai.58.11.3537-3544.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Faille C, Wieruszeski J M, Lepage G, Michalski J C, Poulain D, Strecker G. 1H-NMR spectroscopy of manno-oligosaccharides of the β-1,2-linked series released from the phosphopeptidomannan of Candida albicans VW32 (serotype A) Biochem Biophys Res Commun. 1991;181:1251–1258. doi: 10.1016/0006-291x(91)92073-s. [DOI] [PubMed] [Google Scholar]
- 136.Fernandes P A, Keen J N, Findlay J B C, Moradas-Ferreira P. A protein homologous to glyceraldehyde-3-phosphate dehydrogenase is induced in the cell wall of a flocculent Kluyveromyces marxianus. Biochim Biophys Acta. 1992;1159:67–73. doi: 10.1016/0167-4838(92)90076-p. [DOI] [PubMed] [Google Scholar]
- 137.Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Finley D, Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 139.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 140.Fleet G H. Composition and structure of yeast cell walls. Curr Top Med Mycol. 1985;1:24–56. doi: 10.1007/978-1-4613-9547-8_2. [DOI] [PubMed] [Google Scholar]
- 141.Fleet G H. Cell walls. In: Rose A H, Harrison J S, editors. The yeast. 2nd ed. Vol. 4. London, United Kingdom: Academic Press, Ltd.; 1991. pp. 199–277. [Google Scholar]
- 142.Fox J F, Mayer U, Nischt R, Aumailley M, Reinhardt R, Wiedemann H, Maun K, Timpl R, Krieg T, Engel J, Chu M L. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Franklyn K M, Warmington J R. Cloning and nucleotide sequence analysis of the Candida albicans enolase gene. FEMS Microbiol Lett. 1993;111:101–108. doi: 10.1111/j.1574-6968.1993.tb06368.x. [DOI] [PubMed] [Google Scholar]
- 144.Franklyn K M, Warmington J R, Ott A K, Ashman R B. An immunodominant antigen of Candida albicans shows homology to the enzyme enolase. Immunol Cell Biol. 1990;68:173–178. doi: 10.1038/icb.1990.24. [DOI] [PubMed] [Google Scholar]
- 145.Franzke S, Calderone R A, Schaller K. Isolation of avirulent clones of Candida albicans with reduced ability to recognize the CR2 ligand C3d. Infect Immun. 1993;61:2662–2669. doi: 10.1128/iai.61.6.2662-2669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frey C L, Barone J M, Drutz D J. Abstracts of the 90th General Meeting of the American Society for Microbiology 1990. Washington, D.C: American Society for Microbiology; 1990. The role of the Candida albicans iC3b receptor in fungal adherence to endothelial cells, abstr. F-101; p. 425. [Google Scholar]
- 147.Frey C L, Barone J M, Dreyer G, Koltin Y, Petteway S R, Jr, Drutz D J. Abstracts of the 90th General Meeting of the American Society for Microbiology 1990. Washington, D.C: American Society for Microbiology; 1990. Synthetic protease inhibitors inhibit Candida albicans extracellular acid protease activity and adherence to endothelial cells, abstr. F-102; p. 425. [Google Scholar]
- 148.Fu Y, Ibrahim A S, Fonzi W, Zhoud X, Ramos C F, Ghannoum M A. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology. 1997;143:331–340. doi: 10.1099/00221287-143-2-331. [DOI] [PubMed] [Google Scholar]
- 149.Fukayama M, Wadsworth E, Calderone R A. Expression of the C3d-binding protein (CR2) from Candida albicans during experimental candidiasis as measured by lymphoblastogenesis. Infect Immun. 1992;60:8–12. doi: 10.1128/iai.60.1.8-12.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fukazawa Y. Antigenic structure of Candida albicans. In: Kurstak E, Marquis G, Auger P, de Repentigny L, Montplaisir S, editors. Immunology of fungal diseases. New York: Marcel Dekker, Inc.; 1989. pp. 37–62. [Google Scholar]
- 151.Fukazawa Y, Kagaya K. Molecular basis of adhesion of Candida albicans. J Med Vet Mycol. 1997;35:87–99. doi: 10.1080/02681219780000971. [DOI] [PubMed] [Google Scholar]
- 152.Fukazawa Y, Nishikawa A, Suzuki M, Shinoda T. Immunochemical basis of the specificity of the yeast: immunochemical determinants of several antigenic factors of yeast. In: Preusser H J, editor. Medical mycology. Stuttgart, Germany: Gustav Fischer Verlag; 1980. pp. 126–136. [Google Scholar]
- 153.Gale G, Finkel D, Meinke M, McClellan M, Olson J, Kendrick K, Hostetter M. Cloning and expression of a gene encoding an integrin-like protein in Candida albicans. Proc Natl Acad Sci USA. 1996;93:357–361. doi: 10.1073/pnas.93.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gardiner R B, Canton M, Day A W. Serological studies on the fimbriae of yeast and yeast-like species. Bot Gaz. 1982;143:534–541. [Google Scholar]
- 154a.Gaur N K, Klotz S A. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun. 1997;65:5289–5294. doi: 10.1128/iai.65.12.5289-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Georgopoulus C, Welch W. Roles of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- 156.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 157.Ghannoum M, Abu-Elteen K. Correlative relationship between proteinase production, adherence, and pathogenicity of various strains of Candida albicans. J Med Vet Mycol. 1986;24:407–413. doi: 10.1080/02681218680000621. [DOI] [PubMed] [Google Scholar]
- 158.Ghannoum M A, Burns G R, Elteen K A, Radwan S S. Experimental evidence for the role of lipids in adherence of Candida spp. to human buccal epithelial cells. Infect Immun. 1986;54:189–193. doi: 10.1128/iai.54.1.189-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gil M L, Casanova M, Martínez J P, Sentandreu R. Antigenic cell wall mannoproteins in Candida albicans isolates and in other Candida species. J Gen Microbiol. 1991;137:1053–1061. doi: 10.1099/00221287-137-5-1053. [DOI] [PubMed] [Google Scholar]
- 160.Gil M L, Casanova M, Martínez J P. Changes in the cell wall glycoprotein composition of Candida albicans associated to the inhibition of germ tube formation by EDTA. Arch Microbiol. 1994;161:489–494. doi: 10.1007/BF00307769. [DOI] [PubMed] [Google Scholar]
- 161.Gil-Navarro I, Gil M L, Casanova M, Martínez J P, Gozalbo D. The glycolytic enzyme glyceraldehyde-3-phosphate-dehydrogenase of Candida albicans is a surface antigen. J Bacteriol. 1997;179:4992–4999. doi: 10.1128/jb.179.16.4992-4999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gilmore B J, Retsinas E M, Lorenz J S, Hostetter M K. An iC3b receptor on Candida albicans. J Infect Dis. 1988;157:38–46. doi: 10.1093/infdis/157.1.38. [DOI] [PubMed] [Google Scholar]
- 163.Glee P M, Sundstrom P, Hazen K C. Expression of surface hydrophobic proteins by Candida albicans in vivo. Infect Immun. 1995;63:1373–1379. doi: 10.1128/iai.63.4.1373-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Glee P M, Masouka J, Ozier W T, Hazen K C. Presence of multiple laminin- and fibronectin-binding protein in cell wall extract of Candida albicans: influence of dialysis. J Med Vet Mycol. 1996;34:57–61. doi: 10.1080/02681219680000091. [DOI] [PubMed] [Google Scholar]
- 165.Goldman R C, Sullivan P A, Zakula D, Capobianco J O. Kinetics of β-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur J Biochem. 1995;227:372–378. doi: 10.1111/j.1432-1033.1995.tb20399.x. [DOI] [PubMed] [Google Scholar]
- 166.Goldmann D A, Pier G B. Pathogenesis of infections related to intravascular catheterization. Clin Microbiol Rev. 1993;6:176–192. doi: 10.1128/cmr.6.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Goldstein G, Scheid M, Hammerling U, Schlesinger D H, Niall H D, Boyse W A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gómez F J, Gómez A M, Deepe G S., Jr An 80-kilodalton antigen from Histoplasma capsulatum that has homology to heat shock protein 70 induces cell mediated immune responses and protection in mice. Infect Immun. 1992;60:2565–2571. doi: 10.1128/iai.60.7.2565-2571.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gómez M J, Torosantucci A, Arancia S, Maras B, Parisi L, Cassone A. Purification and biochemical characterization of a 65-kilodalton mannoprotein (MP65), a main target of anti-Candida cell-mediated immune responses in humans. Infect Immun. 1996;64:2577–2584. doi: 10.1128/iai.64.7.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gooday G W, Zhu W-Y, O’Donnell R W. What are the roles of chitinases in the growing fungus? FEMS Microbiol Lett. 1992;100:387–392. [Google Scholar]
- 171.Gopal P, Sullivan P A, Shepherd M G. Enzymes of N-acetylglucosamine metabolism during germ-tube formation in Candida albicans. J Gen Microbiol. 1982;128:2319–2326. doi: 10.1099/00221287-128-10-2319. [DOI] [PubMed] [Google Scholar]
- 172.Gottlieb K, Mobarhan S. Review: microbiology of the gastrostomy tube. J Am Coll Nutr. 1994;13:311–313. doi: 10.1080/07315724.1994.10718415. [DOI] [PubMed] [Google Scholar]
- 173.Goudot-Crozel V, Caillol D, Djabali M, Dessein A J. The major parasite surface antigen associated with human resistance to schistosomiasis is a 37 kDa glyceraldehyde-3P-dehydrogenase. J Exp Med. 1989;170:2065–2080. doi: 10.1084/jem.170.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Goyal S, Khuller G K. Phospholipid composition and subcellular distribution in yeast and mycelial forms of Candida albicans. J Med Vet Mycol. 1992;30:355–362. doi: 10.1080/02681219280000461. [DOI] [PubMed] [Google Scholar]
- 175.Gozalbo D, Elorza M V, Sanjuán R, Marcilla A, Valentín E, Sentandreu R. Critical steps in fungal cell wall synthesis: strategies for their inhibition. Pharmacol Ther. 1993;60:337–345. doi: 10.1016/0163-7258(93)90015-6. [DOI] [PubMed] [Google Scholar]
- 176.Gustafson K S, Vercellotti G M, Bendel C M, Hostetter M K. Molecular mimicry in Candida albicans. Role of an integrin analogue in adhesion of the yeast to human endothelium. J Clin Invest. 1991;87:1896–1902. doi: 10.1172/JCI115214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gutiérrez J, Maroto C, Piédrola E, Martín E, Pérez J A. Circulating Candida antigens and antibodies: useful markers of candidemia. J Clin Microbiol. 1993;31:2550–2552. doi: 10.1128/jcm.31.9.2550-2552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hartland R P, Emeson G W, Sullivan P A. A secreted β-glucan-branching enzyme from Candida albicans. Proc R Soc London Ser B. 1991;246:155–160. doi: 10.1098/rspb.1991.0138. [DOI] [PubMed] [Google Scholar]
- 179.Hawser S. Adhesion of different Candida spp. to plastic: XTT formazan determinations. J Med Vet Mycol. 1996;34:407–410. [PubMed] [Google Scholar]
- 180.Hawser S. Comparisons of the susceptibilities of planktonic and adherent Candida albicans to antifungal agents: a modified XTT tetrazolium assay using synchronized C. albicans cells. J Med Vet Mycol. 1996;34:149–152. [PubMed] [Google Scholar]
- 181.Hawser S P, Douglas L J. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 1994;62:915–921. doi: 10.1128/iai.62.3.915-921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hayman E G, Pierschbacher M D, Suzuki S, Ruoslahti E. Vitronectin: a major cell attachment-promoting protein in fetal bovine serum. Exp Cell Res. 1985;160:245–258. doi: 10.1016/0014-4827(85)90173-9. [DOI] [PubMed] [Google Scholar]
- 183.Hazen B W, Hazen K C. Dynamic expression of cell surface hydrophobicity during initial yeast cell growth and before germ tube formation of Candida albicans. Infect Immun. 1988;56:2521–2525. doi: 10.1128/iai.56.9.2521-2525.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hazen B W, Liebert R E, Hazen K C. Relationship of cell surface hydrophobicity to morphology of monomorphic and dimorphic fungi. Mycologia. 1988;80:348–355. [Google Scholar]
- 185.Hazen K C. Participation of yeast cell surface hydrophobicity in adherence of Candida albicans to human epithelial cells. Infect Immun. 1989;57:1894–1900. doi: 10.1128/iai.57.7.1894-1900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hazen K C. Cell surface hydrophobicity of medically important fungi, specially Candida species. In: Doyle R J, Rosenberg M, editors. Microbial cell surface hydrophobicity. Washington, D.C: American Society for Microbiology; 1990. pp. 249–295. [Google Scholar]
- 187.Hazen K C, Brawner D L, Riesselman M H, Cutler J E, Jutila M A. Differential adherence of hydrophobic and hydrophilic Candida albicans yeast cells to mouse tissues. Infect Immun. 1991;59:907–912. doi: 10.1128/iai.59.3.907-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hazen K C, Glee P M. Hydrophobic cell wall glycosylation by the pathogenic fungus Candida albicans. Can J Microbiol. 1994;40:266–272. doi: 10.1139/m94-043. [DOI] [PubMed] [Google Scholar]
- 189.Hazen K C, Hazen B W. A polystyrene microsphere assay for detecting surface hydrophobicity variations within Candida albicans populations. J Microbiol Methods. 1987;6:289–299. [Google Scholar]
- 190.Hazen K C, Hazen B W. Hydrophobic surface protein masking by the opportunistic fungal pathogen Candida albicans. Infect Immun. 1992;60:1499–1508. doi: 10.1128/iai.60.4.1499-1508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hazen K C, Hazen B W. Surface hydrophobic and hydrophilic protein alterations in Candida albicans. FEMS Microbiol Lett. 1993;107:83–88. doi: 10.1111/j.1574-6968.1993.tb06008.x. [DOI] [PubMed] [Google Scholar]
- 192.Hazen K C, Hazen B W, Allietta M M. Cell wall anchoring to cytoplasmic membrane of Candida albicans. FEMS Microbiol Lett. 1995;125:143–148. doi: 10.1111/j.1574-6968.1995.tb07350.x. [DOI] [PubMed] [Google Scholar]
- 193.Hazen K C, Lay J G, Hazen B W, Fu R C G, Murthy S. Partial biochemical characterization of cell surface hydrophobicity and hydrophilicity of Candida albicans. Infect Immun. 1990;58:3469–3476. doi: 10.1128/iai.58.11.3469-3476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Heidenreich F, Dierich M P. Candida albicans and Candida stellatoidea, in contrast to other Candida species, bind iC3b and C3d but not C3b. Infect Immun. 1985;50:598–600. doi: 10.1128/iai.50.2.598-600.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hernando F L, Estévez J J, Cebrián M, Poulain D, Pontón J. Identification of Candida albicans cell wall antigens lost during subculture in synthetic media. J Med Vet Mycol. 1993;31:227–237. [PubMed] [Google Scholar]
- 196.Herrero E, Sanz P, Sentandreu R. Cell wall proteins liberated by Zymolyase from several ascomycetous and imperfect yeasts. J Gen Microbiol. 1987;133:2895–2903. [Google Scholar]
- 197.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Microbiol. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 198.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 199.Hoffman M P, Haidaris C G. Analysis of Candida albicans adhesion to salivary mucin. Infect Immun. 1993;61:1940–1949. doi: 10.1128/iai.61.5.1940-1949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hoffman M P, Haidaris C G. Identification and characterization of a Candida albicans-binding proteoglycan secreted from rat submandibular salivary glands. Infect Immun. 1994;62:828–836. doi: 10.1128/iai.62.3.828-836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Holland M J, Holland J P. Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry. 1978;17:4900–4907. doi: 10.1021/bi00616a007. [DOI] [PubMed] [Google Scholar]
- 202.Holmes A R, Cannon R D, Jenkinson H R. Interactions of Candida albicans with bacterial and salivary molecules in oral biofilms. J Ind Microbiol. 1995;15:208–213. doi: 10.1007/BF01569827. [DOI] [PubMed] [Google Scholar]
- 203.Holmes A R, Gopal P K, Jenkinson H F. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infect Immun. 1995;63:1827–1834. doi: 10.1128/iai.63.5.1827-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Holmes A R, McNab R, Jenkinson H F. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun. 1996;64:4680–4685. doi: 10.1128/iai.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Homma M, Chibana H, Tanaka K. Induction of extracellular proteinase in Candida albicans. J Gen Microbiol. 1993;139:1187–1193. doi: 10.1099/00221287-139-6-1187. [DOI] [PubMed] [Google Scholar]
- 206.Homma M, Kanbe T, Chibana H, Tanaka K. Detection of intracellular forms of secretory aspartic proteinase in Candida albicans. J Gen Microbiol. 1992;138:627–633. doi: 10.1099/00221287-138-3-627. [DOI] [PubMed] [Google Scholar]
- 207.Hopwood V D, Poulain D, Fortier B, Evans G, Vernes A. A monoclonal antibody to a cell wall component of Candida albicans. Infect Immun. 1986;54:222–227. doi: 10.1128/iai.54.1.222-227.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hostetter M K. Complement receptors in Candida albicans. Clin Immunol Newsl. 1991;11:1–3. [Google Scholar]
- 209.Hostetter M K. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial cells. Clin Microbiol Rev. 1994;7:29–42. doi: 10.1128/cmr.7.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hostetter M K, Lorenz J S, Preus L, Kendrick K E. The iC3b receptor on Candida albicans: subcellular localization and modulation of receptor expression by glucose. J Infect Dis. 1990;161:761–768. doi: 10.1093/infdis/161.4.761. [DOI] [PubMed] [Google Scholar]
- 211.Howlett J A, Squier C. Candida albicans ultrastructure: colonization and invasion of oral epithelium. Infect Immun. 1980;29:252–260. doi: 10.1128/iai.29.1.252-260.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hoyer L L, Scherer S, Shatzman A R, Livi G P. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol Microbiol. 1995;15:39–54. doi: 10.1111/j.1365-2958.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 213.Hsu L Y, Minah G E, Peterson D E, Wingard J R, Merz W G, Altomonte V, Tylenda C A. Coaggregation of oral Candida isolates with bacterial from bone marrow transplant recipients. J Clin Microbiol. 1990;28:2621–2626. doi: 10.1128/jcm.28.12.2621-2626.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hube B, Monod M, Schofield D A, Brown A J P, Gow N A R. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 214a.Hube B, Sanglard D, Odds F C, Hess D, Monod M, Schafer W, Brown A J P, Gow N A R. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hube B, Turver C J, Odds F C, Eiffert H, Boulnois G J, Kochel H, Rüchel R. Sequence of the Candida albicans gene encoding the secretory aspartate proteinase. J Med Vet Mycol. 1991;29:129–132. [PubMed] [Google Scholar]
- 216.Hynes R O. Integrins: versatility, modulation and signalling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 217.Ibrahim A S, Mirbod F, Filler S G, Banno Y, Cole G T, Kitajima Y, Edwards J E, Jr, Nozawa Y, Ghannoum M A. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infect Immun. 1995;63:1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ingolia T D, Slater M R, Craig E A. Saccharomyces cerevisiae contains a complex multigene family related to the major heat-shock-inducible gene of Drosophila. Mol Cell Biol. 1982;2:1388–1398. doi: 10.1128/mcb.2.11.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ishiguro A, Homma M, Torii S, Tanaka K. Identification of Candida albicans antigens reactive with immunoglobulin E antibody of human sera. Infect Immun. 1992;60:1550–1557. doi: 10.1128/iai.60.4.1550-1557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ishii N, Yamamoto M, Yoshihara F, Arisawa M, Aoki Y. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology. 1997;143:429–435. doi: 10.1099/00221287-143-2-429. [DOI] [PubMed] [Google Scholar]
- 221.Ito K, Ishiguro A, Kanbe T, Tanaka K, Torii S. Detection of IgE antibody against Candida albicans enolase and its crossreactivity to Saccharomyces cerevisiae enolase. Clin Exp Allergy. 1995;25:522–528. doi: 10.1111/j.1365-2222.1995.tb01089.x. [DOI] [PubMed] [Google Scholar]
- 222.Jakab E, Paulsson M, Ascencio F, Ljungh A. Expression of vitronectin and fibronectin binding by Candida albicans yeast cells. APMIS. 1993;101:187–193. [PubMed] [Google Scholar]
- 223.Jenkinson H F, Shepherd M G. A mutant of Candida albicans deficient in β-N-acetylglucosaminidase (Chitobiase) J Gen Microbiol. 1987;133:2097–2106. doi: 10.1099/00221287-133-8-2097. [DOI] [PubMed] [Google Scholar]
- 224.Jenkinson H F, Lala H C, Shepherd M G. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 1990;58:1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jenq W, Chen C L, Chang C C, Crang R F E. Characterization of a major cell wall antigen and potential adhesin in three strains of Candida albicans. Arch Microbiol. 1994;162:33–40. doi: 10.1007/BF00264370. [DOI] [PubMed] [Google Scholar]
- 226.Jiménez-Lucho V, Ginsburg V, Krivan H C. Cryptococcus neoformans, Candida albicans, and other fungi bind specifically to the glycosphingolipid lactosylceramide (Galβ1-4Glcβ1-1Cer), a possible adhesion receptor for yeast. Infect Immun. 1990;58:2085–2090. doi: 10.1128/iai.58.7.2085-2090.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jones L, Hobden C, O’Shea P. Use of a real time fluorescent probe to study the electrostatic properties of the cell surface of Candida albicans. Mycol Res. 1995;99:969–976. [Google Scholar]
- 228.Jouault T, Bernigaud A, Lepage G, Trinel P A, Poulain D. The Candida albicans phospholipomannan induces in vitro production of tumor necrosis factor-α from human and murine macrophages. Immunology. 1994;83:268–273. [PMC free article] [PubMed] [Google Scholar]
- 229.Jouault T, Lepage G, Bernigaud A, Trinel P A, Fradin C, Wieruszeski M, Strecker G, Poulain D. β-1,2-linked oligomannosides from Candida albicans act as signals for tumor necrosis factor alpha production. Infect Immun. 1995;63:2378–2381. doi: 10.1128/iai.63.6.2378-2381.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kalo A, Segal E, Sahar E, Dayan D. Interaction of Candida albicans with genital mucosal surfaces: involvement of fibronectin in adherence. J Infect Dis. 1988;157:1253–1256. doi: 10.1093/infdis/157.6.1253. [DOI] [PubMed] [Google Scholar]
- 231.Kalya A V, Ahearn D G. Increased resistance to antifungal antibiotics of Candida spp. adhered to silicone. J Ind Microbiol. 1995;14:451–455. doi: 10.1007/BF01573956. [DOI] [PubMed] [Google Scholar]
- 232.Kaminishi H, Miyaguchi H, Tamaki T, Suenaga N, Hisamatsu M, Mihashi I, Matsumoto H, Maeda H, Hagihara Y. Degradation of humoral host defense by Candida albicans proteinase. Infect Immun. 1995;63:984–988. doi: 10.1128/iai.63.3.984-988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kaminishi H, Tanaka M, Cho T, Maeda H, Hagihara Y. Activation of the plasma kallikrein-kinin system by Candida albicans proteinase. Infect Immun. 1990;58:2139–2143. doi: 10.1128/iai.58.7.2139-2143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kanbe T, Cutler J E. Evidence for adhesin activity in the acid-stable moiety of the phosphomannoprotein cell wall complex of Candida albicans. Infect Immun. 1994;62:1662–1668. doi: 10.1128/iai.62.5.1662-1668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kanbe T, Li R K, Wadsworth E, Calderone R A, Cutler J E. Evidence for the expression of the C3d receptor of Candida albicans in vitro and in vivo by immunofluorescence and immunoelectron microscopy. Infect Immun. 1991;59:1832–1838. doi: 10.1128/iai.59.5.1832-1838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kanbe T, Han Y, Redgrave B, Riesselman M H, Cutler J E. Evidence that mannans of Candida albicans are responsible for adherence of yeast forms to spleen and lymph nodes tissue. Infect Immun. 1993;61:2578–2584. doi: 10.1128/iai.61.6.2578-2584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kapteyn J C, Montijn R C, Dijkgraaf G J, Klis F M. Identification of β-1,6-glucosylated cell wall proteins in yeast and hyphal forms of Candida albicans. Eur J Cell Biol. 1994;65:402–407. [PubMed] [Google Scholar]
- 238.Kapteyn J C, Montijn R C, Dijkgraaf G J, van den Ende H, Klis F M. Covalent association of β-1,3-glucan with β-1,6-glucosylated mannoproteins in cell walls of Candida albicans. J Bacteriol. 1995;177:3788–3792. doi: 10.1128/jb.177.13.3788-3792.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Katz A, Fish A J, Kleppel M M, Hagen S G, Michael A F, Butkwoski R J. Renal entactin (nidogen): isolation, characterization and tissue distribution. Kidney Int. 1991;40:643–652. doi: 10.1038/ki.1991.256. [DOI] [PubMed] [Google Scholar]
- 240.Kauffmann S H E. Heat shock proteins and the immune response. Immunol Today. 1990;11:129–136. doi: 10.1016/0167-5699(90)90050-j. [DOI] [PubMed] [Google Scholar]
- 241.Kauffmann S H E, Schoel B. Heat shock proteins as antigen in immunity against infection and self. In: Morimoto R I, Tissieres A, Georgopoulos G, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 495–531. [Google Scholar]
- 242.Kennedy M J, Sandin R E. Influence of growth conditions on Candida albicans adhesion, hydrophobicity and cell wall ultrastructure. J Med Vet Mycol. 1988;26:79–92. [PubMed] [Google Scholar]
- 243.Kennedy M J, Rogers A L, Yancey R J., Jr Environmental alterations and phenotypic regulation of Candida albicans adhesion to plastic. Infect Immun. 1989;57:3876–3881. doi: 10.1128/iai.57.12.3876-3881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kerridge D. Fungal dimorphism: a sideways look. In: Vanden Bossche H, Odds F C, Kerridge D, editors. Dimorphic fungi in biology and medicine. New York, N.Y: Plenum Press; 1993. pp. 3–10. [Google Scholar]
- 245.Klebl F, Tanner W. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J Bacteriol. 1989;171:6259–6264. doi: 10.1128/jb.171.11.6259-6264.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Klis F M. Review: cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 247.Klis F M, Caro L H P, Vossen J H, Kapteyn J C, Ram A RF J, Montijn R C, Van Berkel M A A, Van Den Ende H. Identification and characterization of a major building block in the cell wall of Saccharomyces cerevisiae. Biochem Soc Trans. 1997;25:856–860. doi: 10.1042/bst0250856. [DOI] [PubMed] [Google Scholar]
- 248.Klotz S A. The adherence of Candida yeast to human and bovine vascular endothelium and subendothelial extracellular matrix. FEMS Microbiol Lett. 1987;48:201–205. [Google Scholar]
- 249.Klotz S A. Adherence of Candida albicans to components of the subendothelial extracellular matrix. FEMS Microbiol Lett. 1990;68:249–254. doi: 10.1016/s0378-1097(05)80049-7. [DOI] [PubMed] [Google Scholar]
- 250.Klotz S A. Plasma and extracellular matrix proteins mediate in the fate of Candida albicans in the human host. Med Hypotheses. 1994;42:328–334. doi: 10.1016/0306-9877(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 251.Klotz S A, Maca R D. Endothelial cell contraction increases Candida adherence to exposed extracellular matrix. Infect Immun. 1988;56:2495–2498. doi: 10.1128/iai.56.9.2495-2498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Klotz S A, Penn R L. Multiple mechanisms may contribute to the adherence of Candida yeasts to living cells. Curr Microbiol. 1987;16:119–122. [Google Scholar]
- 253.Klotz S A, Smith R L. A fibronectin receptor on Candida albicans mediates adherence of the fungus to extracellular matrix. J Infect Dis. 1991;163:604–610. doi: 10.1093/infdis/163.3.604. [DOI] [PubMed] [Google Scholar]
- 254.Klotz S A, Smith R L. Glycosaminoglycans inhibit Candida albicans adherence to extracellular matrix proteins. FEMS Microbiol Lett. 1992;99:205–208. doi: 10.1016/0378-1097(92)90026-k. [DOI] [PubMed] [Google Scholar]
- 255.Klotz S A, Smith R L. Gelatin fragments block adherence of Candida albicans to extracellular matrix proteins. Microbiology. 1995;141:2681–2684. doi: 10.1099/13500872-141-10-2681. [DOI] [PubMed] [Google Scholar]
- 256.Klotz S A, Chen R C, Smith R L, Rouse J B. The fibronectin adhesin of Candida albicans. Infect Immun. 1994;62:4679–4681. doi: 10.1128/iai.62.10.4679-4681.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Klotz S A, Drutz D J, Harrison J L, Huppert M. Adherence and penetration of vascular endothelium by Candida yeasts. Infect Immun. 1983;42:374–384. doi: 10.1128/iai.42.1.374-384.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Klotz S A, Drutz D J, Zajic J E. Factors governing adherence of Candida species to plastic surfaces. Infect Immun. 1985;50:97–101. doi: 10.1128/iai.50.1.97-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Klotz S A, Rutten M J, Smith R L, Babcock S R, Cunningham M D. Adherence of Candida albicans to immobilized extracellular matrix proteins is mediated by calcium-dependent surface glycoproteins. Microb Pathog. 1993;14:133–147. doi: 10.1006/mpat.1993.1014. [DOI] [PubMed] [Google Scholar]
- 260.Klotz S A, Smith R L, Stewart B W. Effect of an arginine-glycine-aspartic acid-containing peptide on hematogenous candidal infections. Antimicrob Agents Chemother. 1992;36:132–136. doi: 10.1128/aac.36.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kobayashi H, Giummelly P, Takahashi S, Ishida M, Sato J, Takaku M, Nishidate Y, Shibata N, Okawa Y, Suzuki S. Candida albicans serotype A strains grow in yeast extract added Sabouraud liquid medium at pH 2.0, elaborating mannans without β-1,2 linkage and phosphate group. Biochem Biophys Res Commun. 1991;175:1003–1009. doi: 10.1016/0006-291x(91)91664-x. [DOI] [PubMed] [Google Scholar]
- 262.Kobayashi H, Shibata N, Mitobe H, Ohkubo Y, Suzuki S. Structural study of phosphomannan of yeast-form cells of Candida albicans J-1012 strain with special reference to application of mild acetolysis. Arch Biochem Biophys. 1989;272:364–375. doi: 10.1016/0003-9861(89)90230-0. [DOI] [PubMed] [Google Scholar]
- 263.Kobayashi H, Shibata N, Nakada M, Chaki S, Mizugami K, Ohkubo Y, Suzuki S. Structural analysis of cell wall phosphomannan of Candida albicans NIH B-792 (serotype B) strain, with special reference to 1H and 13C NMR analyses of acid-labile oligomannosyl residues. Arch Biochem Biophys. 1990;278:195–204. doi: 10.1016/0003-9861(90)90248-w. [DOI] [PubMed] [Google Scholar]
- 264.Kobayashi H, Takaku M, Nishidate Y, Takahashi S, Takikawa M, Shibata N, Okawa Y, Suzuki S. Structure of the d-mannan of the pathogenic yeasts, Candida stellatoidea ATCC 20408 (type II) strain, in comparison with that of C. stellatoidea ATCC 36232 (type I) strain. Carbohydr Res. 1992;231:105–116. doi: 10.1016/0008-6215(92)84012-h. [DOI] [PubMed] [Google Scholar]
- 265.Kobayashi H, Shibata N, Suzuki S. Evidences for oligomannosyl residues containing both β-1,2 and α-1,2 linkages as a serotype A-specific epitope(s) in mannans of Candida albicans. Infect Immun. 1992;60:2106–2109. doi: 10.1128/iai.60.5.2106-2109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kobayashi H, Tanaka S, Susuki J, Kiuchi Y, Shibata N, Suzuki S, Okawa Y. Amended structure of side chains in a cell wall mannan from Candida albicans serotype A strain grown in yeast extract-Sabouraud liquid medium under acidic conditions: detection of the branched side chains corresponding to antigenic factor 4. FEMS Microbiol Lett. 1997;152:235–242. doi: 10.1016/s0378-1097(97)00203-6. [DOI] [PubMed] [Google Scholar]
- 267.Kogan G, Pavliak V, Masler L. Structural studies of mannan of the cell wall of pathogenic yeast Candida albicans serotypes A and B and Candida parapsilosis. Carbohydr Res. 1988;172:243–253. doi: 10.1016/s0008-6215(00)90858-9. [DOI] [PubMed] [Google Scholar]
- 268.Kollár, R., B. B. Reinhold, E. Petráková, H. J. C. Yeh II, G. Ashwell, J. Drgonová, J. C. Kapteyn, F. M. Klis, and E. Cabib. Architecture of the yeast cell wall. J. Biol. Chem., in press. [DOI] [PubMed]
- 269.Kolling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kozel T R, Weinhold L C, Lupan D M. Distinct characteristics of initiation of the classical and alternative complement pathways by Candida albicans. Infect Immun. 1996;64:3360–3368. doi: 10.1128/iai.64.8.3360-3368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Krivan H C, Roberts D D, Ginsburg V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence βGalNAc(1-4)βGal found in some glycolipids. Proc Natl Acad Sci USA. 1988;85:6157–6161. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kuchler K. Unusual routes of protein secretion: the easy way out. Trends Cell Biol. 1993;3:421–426. doi: 10.1016/0962-8924(93)90030-5. [DOI] [PubMed] [Google Scholar]
- 273.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 274.Kurtz M B, Kelly R, Kirsch D R. Molecular genetics of Candida albicans. In: Kirsch D R, Kelly R, Kurtz M B, editors. The genetics of Candida. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 21–74. [Google Scholar]
- 275.Kurtz M B, Kirsch D R, Kelly R. The molecular genetics of Candida albicans. Microbiol Sci. 1988;5:58–63. [PubMed] [Google Scholar]
- 276.Kusamichi M, Monodane T, Tokunaga M, Koike H. Influence of surrounding media on preservation of cell wall structure of Candida albicans revealed by low temperature scanning electron microscopy. J Electron Microsc. 1990;39:477–486. [PubMed] [Google Scholar]
- 277.Kwon-Chung K J, Lehman D, Good C, Magee P T. Genetic evidence for role of extracellular proteinase in virulence of Candida albicans. Sabouraudia. 1985;23:81–83. doi: 10.1128/iai.49.3.571-575.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.La Valle R, Bromuro C, Ranucci L, Muller H M, Crisanti A, Cassone A. Molecular cloning and expression of a 70-kilodalton heat shock protein of Candida albicans. Infect Immun. 1995;63:4039–4045. doi: 10.1128/iai.63.10.4039-4045.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lee K K, Yu L, Macdonald D L, Paranchych W, Hodges R S, Irvin R T. Anti-adhesin antibodies that recognize a receptor-binding motif (adhesintope) inhibit pilus/fimbrial-mediated adherence of Pseudomonas aeruginosa and Candida albicans to asialo-GM1 receptors and human buccal epithelial cell surface receptors. Can J Microbiol. 1996;42:479–486. doi: 10.1139/m96-065. [DOI] [PubMed] [Google Scholar]
- 280.Lee K W, Shalaby K A, Thakur A, Medhat A, Karim A M, LoVerde P T. Cloning of the gene for phosphoglycerate kinase from Schistosoma mansoni and characterization of its gene product. Mol Biochem Parasitol. 1995;71:221–231. doi: 10.1016/0166-6851(95)91598-o. [DOI] [PubMed] [Google Scholar]
- 281.Lerner C G, Goldman R C. Stimuli that induce production of Candida albicans extracellular aspartyl proteinase. J Gen Microbiol. 1993;139:1643–1651. doi: 10.1099/00221287-139-7-1643. [DOI] [PubMed] [Google Scholar]
- 282.Leung D W, Spencer S A, Cachianes G, Hammonds R G, Collins C, Henzel W J, Barnard R, Waters M J, Wood W I. Growth hormone receptor and serum binding protein: purification, cloning and expression. Nature. 1987;330:537–543. doi: 10.1038/330537a0. [DOI] [PubMed] [Google Scholar]
- 283.Leusch H G. Detection and characterization of two antigens specific for cell walls of Candida albicans mycelial growth phase. Curr Microbiol. 1989;19:193–198. [Google Scholar]
- 284.Li R K, Cutler J E. A cell surface/plasma membrane antigen of Candida albicans. J Gen Microbiol. 1991;137:455–464. doi: 10.1099/00221287-137-3-455. [DOI] [PubMed] [Google Scholar]
- 285.Li R-K, Cutler J E. Chemical definition of an epitope/adhesin molecule on Candida albicans. J Biol Chem. 1993;268:18293–18299. [PubMed] [Google Scholar]
- 286.Limper A H, Standing J E. Vitronectin interacts with Candida albicans and augments organism attachment to the NR8383 macrophage cell line. Immunol Lett. 1994;42:139–144. doi: 10.1016/0165-2478(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 287.Limper A H, Standing J E, Hoffman O A, Castro M, Neese L W. Vitronectin binds to Pneumocystis carinii and mediates organism attachment to cultured epithelial cells. Infect Immun. 1993;61:4302–4309. doi: 10.1128/iai.61.10.4302-4309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lindquist S, Craig E A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 289.Linehan L, Wadsworth E, Calderone R A. Candida albicans C3d receptor, isolated by using a monoclonal antibody. Infect Immun. 1988;56:1981–1986. doi: 10.1128/iai.56.8.1981-1986.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Linnemans W A M, Boer P, Elbers P F. Localization of acid phosphatase in Saccharomyces cerevisiae: a clue to cell wall formation. J Bacteriol. 1977;131:638–644. doi: 10.1128/jb.131.2.638-644.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lipke P N, Wojciechowicz D, Kurjan J. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Livi G P, Lillquist J S, Miles L M, Ferrara A, Sathe G M, Simon P L, Meyers C A, Gorman J A, Young P R. Secretion of N-glycosylated interleukin-1β in Saccharomyces cerevisiae using a leader peptide from Candida albicans. Effect of N-linked glycosylation on biological activity. J Biol Chem. 1991;266:15348–15355. [PubMed] [Google Scholar]
- 293.López-Ribot J L, Chaffin W L. Binding of the extracellular matrix component entactin to Candida albicans. Infect Immun. 1994;62:4564–4571. doi: 10.1128/iai.62.10.4564-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.López-Ribot J L, Chaffin W L. Members of the hsp70 family of proteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1996;178:4724–4726. doi: 10.1128/jb.178.15.4724-4726.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.López-Ribot J L, Casanova M, Martínez J P, Sentandreu R. Characterization of cell wall proteins of yeast and hydrophobic mycelial cells of Candida albicans. Infect Immun. 1991;59:2324–2332. doi: 10.1128/iai.59.7.2324-2332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.López-Ribot J L, Casanova M, Monteagudo C, Sepúlveda P, Martínez J P. Evidence for the presence of a high-affinity laminin receptor-like molecule on the surface of Candida albicans yeast cells. Infect Immun. 1994;62:742–746. doi: 10.1128/iai.62.2.742-746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.López-Ribot J L, Navarro D, Sepúlveda P, Nogueira J M, Casanova M, Martínez J P. A comparative study on cell wall antigens and cell surface hydrophobicity in clinical isolates of Candida albicans. Mycopathologia. 1994;127:1–13. doi: 10.1007/BF01104005. [DOI] [PubMed] [Google Scholar]
- 298.López-Ribot J L, Vespa M N, Chaffin W L. Adherence of Candida albicans germ tubes to murine tissues in an ex vivo assay. Can J Microbiol. 1994;40:77–81. doi: 10.1139/m94-013. [DOI] [PubMed] [Google Scholar]
- 299.López-Ribot J L, Gozalbo D, Sepúlveda P, Casanova M, Martínez J P. Preliminary characterization of the material released to the culture medium by Candida albicans yeast and mycelial cells. Antonie Leeuwenhoek. 1995;68:195–201. doi: 10.1007/BF00871815. [DOI] [PubMed] [Google Scholar]
- 300.López-Ribot J L, Martínez J P, Chaffin W L. Comparative study of the C3d receptor and 58-kilodalton fibrinogen-binding mannoproteins of Candida albicans. Infect Immun. 1995;63:2126–2132. doi: 10.1128/iai.63.6.2126-2132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.López-Ribot J L, Casanova M, Gil M L, Martínez J P. Common and form-specific cell wall antigens of Candida albicans as released by chemical and enzymatic treatments. Mycopathologia. 1996;134:13–20. doi: 10.1007/BF00437047. [DOI] [PubMed] [Google Scholar]
- 302.López-Ribot J L, Monteagudo C, Sepúlveda P, Casanova M, Martínez J P, Chaffin W L. Expression of the fibrinogen binding mannoprotein and the laminin receptor of Candida albicans in vitro and in infected tissues. FEMS Microbiol Lett. 1996;142:117–122. doi: 10.1111/j.1574-6968.1996.tb08417.x. [DOI] [PubMed] [Google Scholar]
- 303.López-Ribot J L, Alloush H M, Masten B, Chaffin W L. Evidence for the presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect Immun. 1996;64:3333–3340. doi: 10.1128/iai.64.8.3333-3340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Lott T J, Page L S, Boiron P, Benson J, Reiss E. Nucleotide sequence of the Candida albicans aspartyl proteinase gene. Nucleic Acids Res. 1989;17:1779. doi: 10.1093/nar/17.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Lu C F, Kurjan J, Lipke P N. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol Cell Biol. 1994;14:4825–4833. doi: 10.1128/mcb.14.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lu C F, Montijn R C, Brown J L, Klis F M, Kurjan J, Bussey H, Lipke P N. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β-1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Luna-Arias J P, Andaluz E, Ridruejo J C, Olivero I, Larriba G. The major exoglucanase from Candida albicans: a non-glycosylated secretory monomer related to its counterpart from Saccharomyces cerevisiae. Yeast. 1991;7:833–841. doi: 10.1002/yea.320070808. [DOI] [PubMed] [Google Scholar]
- 308.Mackenzie L F, Brooke G S, Cutfield J F, Sullivan P A, Withers S G. Identification of glu-330 as the catalytic nucleophile of Candida albicans exo-β-(1,3)-glucanase. J Biol Chem. 1997;272:3161–3167. doi: 10.1074/jbc.272.6.3161. [DOI] [PubMed] [Google Scholar]
- 309.Macura A B. Hydrophobicity of Candida albicans related to their adherence to mucosal epithelial cells. Zentralbl Bakteriol Mikrobiol Hyg Ser A. 1987;266:491–496. doi: 10.1016/s0176-6724(87)80231-6. [DOI] [PubMed] [Google Scholar]
- 310.Magee P T. Which came first, the hypha or the yeast? Science. 1997;277:52–53. doi: 10.1126/science.277.5322.52. [DOI] [PubMed] [Google Scholar]
- 311.Magee B B, Hube B, Wright R J, Sullivan P A, Magee P T. The genes encoding the secreted aspartyl proteinases of Candida albicans constitute a family with at least three members. Infect Immun. 1993;61:3240–3243. doi: 10.1128/iai.61.8.3240-3243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Mager W H, Moradas-Ferreira P. Stress response of yeast. Biochem J. 1993;290:1–13. doi: 10.1042/bj2900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Maisch P A, Calderone R A. Adherence of Candida albicans to a fibrin-platelet matrix formed in vitro. Infect Immun. 1980;27:650–656. doi: 10.1128/iai.27.2.650-656.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Maitra P K, Lobo Z. A kinetic study of glycolytic enzyme synthesis in yeast. J Biol Chem. 1971;246:475–488. [PubMed] [Google Scholar]
- 315.Manavathu E K. Chromosomal localization of phosphoglycerate kinase (PGK) gene in Candida albicans using a heterologous probe. Indian J Exp Biol. 1993;31:206–209. [PubMed] [Google Scholar]
- 316.Maneu V E, Cervera A M, Martínez J P, Gozalbo D. Molecular cloning of a Candida albicans gene (SSB1) coding for a protein related to the hsp70 family. Yeast. 1997;13:677–681. doi: 10.1002/(SICI)1097-0061(19970615)13:7<677::AID-YEA131>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 317.Manns J M, Mosser D M, Buckley H R. Production of a hemolytic factor by Candida albicans. Infect Immun. 1994;62:5154–5156. doi: 10.1128/iai.62.11.5154-5156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Marcantonio E E, Hynes R O. Antibodies to the conserved cytoplasmic domain of the integrin β1 subunit react with proteins in vertebrates, invertebrates, and fungi. J Cell Biol. 1988;106:1765–1772. doi: 10.1083/jcb.106.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Marcilla A, Elorza M V, Mormeneo S, Rico H, Sentandreu R. Candida albicans mycelial cell wall structure: supramolecular complexes released by Zymolyase, chitinase and β-mercaptoethanol. Arch Microbiol. 1991;155:312–319. doi: 10.1007/BF00243448. [DOI] [PubMed] [Google Scholar]
- 320.Marcilla A, Mormeneo S, Elorza M V, Manclús J J, Sentandreu R. Wall formation by Candida albicans yeast cells: synthesis, secretion and incorporation of two types of mannoproteins. J Gen Microbiol. 1993;139:2985–2993. doi: 10.1099/00221287-139-12-2985. [DOI] [PubMed] [Google Scholar]
- 321.Marcuard S P, Finley J L, MacDonald K C. Large-bore feeding tube occulsion by yeast colonies. J Parenter Enteral Nutr. 1993;17:187–190. doi: 10.1177/0148607193017002187. [DOI] [PubMed] [Google Scholar]
- 322.Maresca B, Kobayashi G S. Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol Rev. 1989;53:186–209. doi: 10.1128/mr.53.2.186-209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Maresca B, Kobayashi G S. Hsp70 in parasites as an inducible protective protein and as an antigen. Experientia. 1994;50:1067–1074. doi: 10.1007/BF01923463. [DOI] [PubMed] [Google Scholar]
- 324.Marot-Leblond A, Robert R, Aubry J, Ezcurra P, Senet J-M. Identification and immunochemical characterization of a germ tube specific antigen of Candida albicans. FEMS Immunol Med Microbiol. 1993;7:175–186. doi: 10.1111/j.1574-695X.1993.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 325.Marot-Leblond A, Robert R, Senet J-M, Palmer T. Purification of a Candida albicans germ tube specific antigen. FEMS Immunol Med Microbiol. 1995;12:127–136. doi: 10.1111/j.1574-695X.1995.tb00184.x. [DOI] [PubMed] [Google Scholar]
- 326.Martínez J P, Gil M L, Casanova M, López-Ribot J L, García de Lomas J, Sentandreu R. Wall mannoproteins in cells from colonial phenotypic variants of Candida albicans. J Gen Microbiol. 1990;136:2421–2432. doi: 10.1099/00221287-136-12-2421. [DOI] [PubMed] [Google Scholar]
- 327.Martínez J P, López-Ribot J L, Gil M L, Sentandreu R, Ruiz-Herrera J R. Inhibition of the dimorphic transition of Candida albicans by the ornithine decarboxylase inhibitor 1,4-diaminobutanone: alterations in the glycoprotein composition of the cell wall. J Gen Microbiol. 1990;136:1937–1943. doi: 10.1099/00221287-136-10-1937. [DOI] [PubMed] [Google Scholar]
- 328.Martínez J P, López-Ribot J L, Chaffin W L. Heterogeneous surface distribution of the fibrinogen-binding protein on Candida albicans. Infect Immun. 1994;62:709–712. doi: 10.1128/iai.62.2.709-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Mason, A. B., M. E. Brandt, and H. R. Buckley. 1989. Enolase activity associated with a cytoplasmic antigen of Candida albicans. Yeast 5(Suppl.):S231–S239. [PubMed]
- 330.Mason A B, Buckley H R, Gorman J A. Molecular cloning and characterization of the Candida albicans enolase gene. J Bacteriol. 1993;175:2632–2639. doi: 10.1128/jb.175.9.2632-2639.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Matthews R, Hodgetts S, Burnie J. Preliminary assessment of a human recombinant antibody fragment to hsp90 in murine invasive candidiasis. J Infect Dis. 1995;171:1668–1671. doi: 10.1093/infdis/171.6.1668. [DOI] [PubMed] [Google Scholar]
- 332.Matthews R C. Candida albicans hsp90: link between protection and autoimmunity. J Med Microbiol. 1992;36:367–370. doi: 10.1099/00222615-36-6-367. [DOI] [PubMed] [Google Scholar]
- 333.Matthews R C. Pathogenicity determinants of Candida albicans: potential targets for immunotherapy? Microbiology. 1994;140:1505–1511. doi: 10.1099/13500872-140-7-1505. [DOI] [PubMed] [Google Scholar]
- 334.Matthews R C, Burnie J P. Diagnosis of systemic candidiasis by an enzyme-linked dot immunobinding assay for a circulating immunodominant 47-kilodalton antigen. J Clin Microbiol. 1988;26:459–63. doi: 10.1128/jcm.26.3.459-463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Matthews R C, Burnie J P. Cloning of a DNA sequence encoding a major fragment of the 47 kilodalton stress protein homologue of Candida albicans. FEMS Microbiol Lett. 1989;60:25–30. doi: 10.1016/0378-1097(89)90071-2. [DOI] [PubMed] [Google Scholar]
- 336.Matthews R C, Burnie J P. The role of hsp90 in fungal infection. Immunol Today. 1992;13:345–348. doi: 10.1016/0167-5699(92)90169-8. [DOI] [PubMed] [Google Scholar]
- 337.Matthews R C, Burnie J P, Tabaqchali S. Immunoblot analysis of the serological response in systemic candidosis. Lancet. 1984;ii:1415–1418. doi: 10.1016/s0140-6736(84)91618-0. [DOI] [PubMed] [Google Scholar]
- 338.Matthews R C, Burnie J P, Tabaqchali S. Isolation of immunodominant antigens from the sera of patients with systemic candidosis and characterization of the serological response to Candida albicans. J Clin Microbiol. 1987;25:230–237. doi: 10.1128/jcm.25.2.230-237.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Matthews R C, Burnie J P, Smith D, Clark I, Midgley J, Conolly M, Gazzard B. Candida and AIDS: evidence for protective antibody. Lancet. 1988;ii:263–266. doi: 10.1016/s0140-6736(88)92547-0. [DOI] [PubMed] [Google Scholar]
- 340.Matthews R C, Wells C, Burnie J P. Characterisation and cellular localisation of the immunodominant 47-kDa antigen of Candida albicans. J Med Microbiol. 1988;27:227–232. doi: 10.1099/00222615-27-4-227. [DOI] [PubMed] [Google Scholar]
- 341.Matthews R C, Burnie J P, Lee W. The application of epitope mapping to the development of a new serological test for systemic candidosis. J Immunol Methods. 1991;143:73–79. doi: 10.1016/0022-1759(91)90274-j. [DOI] [PubMed] [Google Scholar]
- 342.Matthews R C, Burnie J P, Howat D, Rowland T, Walton F. Autoantibody to hsp90 can mediate protection against systemic candidosis. Immunology. 1991;74:20–24. [PMC free article] [PubMed] [Google Scholar]
- 343.McAlister L, Holland M J. Targeted deletion of a yeast enolase structural gene: identification and isolation of yeast enolase isozymes. J Biol Chem. 1982;257:7181–7188. [PubMed] [Google Scholar]
- 344.McCourtie J, Douglas L J. Relationship between cell surface composition of Candida albicans and adherence to acrylic after growth on different carbon sources. Infect Immun. 1981;32:1234–1241. doi: 10.1128/iai.32.3.1234-1241.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.McCourtie J, Douglas L J. Extracellular polymer of Candida albicans: isolation, analysis and role in adhesion. J Gen Microbiol. 1985;131:495–503. doi: 10.1099/00221287-131-3-495. [DOI] [PubMed] [Google Scholar]
- 346.McCreath K J, Specht C A, Robbins P W. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc Natl Acad Sci USA. 1995;92:2544–2548. doi: 10.1073/pnas.92.7.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.McCreath K J, Specht C A, Liu Y, Robbins P W. Molecular cloning of a third chitinase gene (CHT1) from Candida albicans. Yeast. 1996;12:501–504. doi: 10.1002/(SICI)1097-0061(199604)12:5%3C501::AID-YEA931%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 348.McDonald F. Secretion of inducible proteinase by pathogenic Candida species. Sabouraudia. 1984;22:79–82. [PubMed] [Google Scholar]
- 349.McDonald F, Odds F C. Inducible proteinase of Candida albicans in diagnostic serology and in the pathogenesis of systemic candidosis. J Med Microbiol. 1980;13:423–435. doi: 10.1099/00222615-13-3-423. [DOI] [PubMed] [Google Scholar]
- 350.McDonald F, Odds F C. Virulence for mice of a proteinase-secreting strain of Candida albicans and a proteinase-deficient mutant. J Gen Microbiol. 1983;129:431–438. doi: 10.1099/00221287-129-2-431. [DOI] [PubMed] [Google Scholar]
- 351.Melo L F, Bott T R, Fletcher M, Capdeville B. Biofilms—science and technology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. [Google Scholar]
- 352.Merson-Davies L A, Hopwood V, Robert R, Marot-Leblond A, Senet J M, Odds F C. Reaction of Candida albicans cells of different morphology index with monoclonal antibodies specific for the hyphal form. J Med Mycol. 1991;35:321–324. doi: 10.1099/00222615-35-6-321. [DOI] [PubMed] [Google Scholar]
- 353.Milewski S, Mignini F, Covelli I, Borowski E. Specific inhibition of acid proteinase secretion in Candida albicans by Lys-Nva-FMDP. J Med Vet Mycol. 1994;32:1–11. [PubMed] [Google Scholar]
- 354.Minagi S, Miyake Y, Inagagi K, Tsuru H, Suginaka H. Hydrophobic interaction in Candida albicans and Candida tropicalis adherence to various denture base resin materials. Infect Immun. 1985;47:11–14. doi: 10.1128/iai.47.1.11-14.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Mirbod F, Banno Y, Ghannoum M A, Ibrahim A S, Nakashima S, Katajima Y, Cole G T, Nozawa Y. Purification and characterization of lysophospholipase-transacylase (h-LPTA) from a highly virulent strain of Candida albicans. Biochim Biophys Acta. 1995;1257:181–188. doi: 10.1016/0005-2760(95)00072-k. [DOI] [PubMed] [Google Scholar]
- 356.Mitsutake K, Kohno S, Miyazaki T, Miyazaki H, Maesaki S, Koga H. Detection of Candida enolase antibody in patients with candidiasis. J Clin Lab Anal. 1994;8:207–210. doi: 10.1002/jcla.1860080405. [DOI] [PubMed] [Google Scholar]
- 357.Miyakawa Y, Kuribayashi T, Kagaya K, Suzuki M, Nakase T, Fukazawa Y. Role of specific determinant in mannan of Candida albicans serotype A in adherence to human buccal epithelial cells. Infect Immun. 1992;60:2493–2499. doi: 10.1128/iai.60.6.2493-2499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Miyake Y, Fujita Y, Minagi S, Suginaka H. Surface hydrophobicity and adherence of Candida to acrylic surfaces. Microbios. 1986;46:7–14. [PubMed] [Google Scholar]
- 359.Miyasaki S H, White T C, Agabian N. A fourth secreted aspartyl proteinase gene (SAP4) and a CARE2 repetitive element are located upstream of the SAP1 gene in Candida albicans. J Bacteriol. 1994;176:1702–1710. doi: 10.1128/jb.176.6.1702-1710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Molano J, Bowers B, Cabib E. Distribution of chitin in the yeast cell wall. An ultrastructural and chemical study. J Cell Biol. 1980;85:199–212. doi: 10.1083/jcb.85.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Molina M, Cenamor R, Nombela C. Exo-1,3-β-glucanase activity in Candida albicans: effect of the yeast to mycelium transition. J Gen Microbiol. 1987;133:609–617. doi: 10.1099/00221287-133-3-609. [DOI] [PubMed] [Google Scholar]
- 362.Molina M, Cenamor R, Sánchez M, Nombela C. Purification and some properties of Candida albicans exo-1,3-β-glucanase. J Gen Microbiol. 1989;135:309–314. doi: 10.1099/00221287-135-2-309. [DOI] [PubMed] [Google Scholar]
- 363.Molloy C, Shepherd M G, Sullivan P A. Identification of envelope proteins of Candida albicans by vectorial iodination. Microbios. 1989;57:73–83. [PubMed] [Google Scholar]
- 364.Molloy C, Cannon R D, Sullivan P A, Shepherd M G. Purification and characterization of two forms of N-acetylglucosaminidase from Candida albicans showing widely different outer chain glycosylation. Microbiology. 1994;140:1543–1553. doi: 10.1099/13500872-140-7-1543. [DOI] [PubMed] [Google Scholar]
- 365.Molloy C, Shepherd M G, Sullivan P A. Differential extraction of N-acetylglucosaminidase and trehalase from the cell envelope of Candida albicans. Exp Mycol. 1995;19:178–185. doi: 10.1006/emyc.1995.1022. [DOI] [PubMed] [Google Scholar]
- 366.Monod M, Togni G, Hube B, Sanglard D. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 1994;13:357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- 367.Moors M A, Stull T L, Blank K J, Buckley H R, Mosser D M. A role for complement receptor-like molecules in iron acquisition by Candida albicans. J Exp Med. 1992;175:1643–1651. doi: 10.1084/jem.175.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Mori S, Heldin C H, Claesson-Welsh L. Ligand-induced polyubiquitination of the platelet-derived growth factor β-receptor. J Biol Chem. 1992;267:6429–6434. [PubMed] [Google Scholar]
- 369.Mormeneo S, Marcilla A, Iranzo M, Sentandreu R. Structural mannoproteins released by β-elimination from Candida albicans cell walls. FEMS Microbiol Lett. 1994;123:131–136. doi: 10.1111/j.1574-6968.1994.tb07212.x. [DOI] [PubMed] [Google Scholar]
- 370.Morrison C J, Hurst S F, Bragg S L, Kuykendall R J, Diaz H, McLaughlin D W, Reiss E. Purification and characterization of the extracellular aspartyl proteinase of Candida albicans: removal of extraneous proteins and cell wall mannoprotein and evidence for lack of glycosylation. J Gen Microbiol. 1993;139:1177–1186. doi: 10.1099/00221287-139-6-1177. [DOI] [PubMed] [Google Scholar]
- 371.Morrison C J, Hurst S F, Bragg S L, Kuykendall R J, Diaz H, Pohl J, Reiss E. Heterogeneity of the purified extracellular aspartyl proteinase from Candida albicans: characterization with monoclonal antibodies and N-terminal amino acid sequence analysis. Infect Immun. 1993;61:2030–2036. doi: 10.1128/iai.61.5.2030-2036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Mrša V, Klebl F, Tanner W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-1,3-glucanase. J Bacteriol. 1993;175:2102–2106. doi: 10.1128/jb.175.7.2102-2106.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the HSP70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- 375.Murgui A, Elorza M V, Sentandreu R. Tunicamycin and papulacandin B inhibit incorporation of specific mannoproteins into the wall of Candida albicans regenerating protoplasts. Biochim Biophys Acta. 1986;884:550–558. doi: 10.1016/0304-4165(86)90207-2. [DOI] [PubMed] [Google Scholar]
- 376.Nakajima T, Ballou C E. Characterization of the carbohydrate fragments obtained from Saccharomyces cerevisiae mannan by alkaline degradation. J Biol Chem. 1974;249:7679–7684. [PubMed] [Google Scholar]
- 377.Navarro D, Monzonis E, López-Ribot J L, Sepúlveda P, Casanova M, Nogueira J M, Martínez J P. Diagnosis of systemic candidiasis by enzyme immunoassay detection of specific antibodies to mycelial phase cell wall and cytoplasmic candidal antigens. Eur J Clin Microbiol Infect Dis. 1993;12:839–846. doi: 10.1007/BF02000404. [DOI] [PubMed] [Google Scholar]
- 378.Neely A N, Childress C M, Holder I A. Effect of challenge with Candida albicans of different virulence on plasma proteins in burned mice. Infect Immun. 1991;59:1576–1578. doi: 10.1128/iai.59.4.1576-1578.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Neely A N, Orloff M M, Holder I A. Candida albicans growth studies: a hypothesis for the pathogenesis of Candida infections in burns. J Burn Care Rehabil. 1992;13:323–329. doi: 10.1097/00004630-199205000-00004. [DOI] [PubMed] [Google Scholar]
- 380.Nègre E, Vogel T, Levanon A, Guy R, Walsh T J, Roberts D D. The collagen binding domain of fibronectin contains a high affinity binding site for Candida albicans. J Biol Chem. 1994;269:22039–22045. [PubMed] [Google Scholar]
- 381.Nelson R D, Shibata N, Podzorski R P, Herron M J. Candida mannan: chemistry, suppression of cell-mediated immunity, and possible mechanisms of action. Clin Microbiol Rev. 1991;4:1–19. doi: 10.1128/cmr.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Neu T R, Verkerke G J, Herrmann I F, Schutte H K, Van der Mei H V, Busscher H J. Microflora on explanted silicone rubber voice prostheses: taxonomy, hydrophobicity and electrophoretic mobility. J Appl Bacteriol. 1994;76:521–528. doi: 10.1111/j.1365-2672.1994.tb01111.x. [DOI] [PubMed] [Google Scholar]
- 383.Niimi M, Niimi K, Cannon R D. Temperature related expression of the vacuolar aspartic proteinase (APR1) gene and β-N-acetylglucosaminidase (HEX1) gene during Candida albicans morphogenesis. FEMS Microbiol Lett. 1997;148:247–254. doi: 10.1111/j.1574-6968.1997.tb10296.x. [DOI] [PubMed] [Google Scholar]
- 384.Nikawa H, Sadamori S, Hama T, Okuda K. Factors involved in the adherence of Candida albicans and Candida tropicalis to protein-adsorbed surfaces. An in vitro study using immobilized protein. Mycopathalogia. 1992;118:139–145. doi: 10.1007/BF00437146. [DOI] [PubMed] [Google Scholar]
- 385.Nikawa H, Hayashi S, Nikawa Y, Hamada T, Samaranayake L P. Interactions between denture lining material, protein pellicles and Candida albicans. Arch Oral Biol. 1993;38:631–634. doi: 10.1016/0003-9969(93)90132-6. [DOI] [PubMed] [Google Scholar]
- 386.Nombela C, Molina M, Cenamor R, Sánchez M. Yeast β-glucanases: a complex system of secreted enzymes. Microbiol Sci. 1988;5:328–332. [PubMed] [Google Scholar]
- 387.Notario V. β-Glucanases from Candida albicans: purification, characterization, and the nature of their attachment to cell wall components. J Gen Microbiol. 1982;128:747–759. doi: 10.1099/00221287-128-4-747. [DOI] [PubMed] [Google Scholar]
- 388.Nouffer C, Jenö P, Cozelman A, Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol Cell Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Odds F C. Morphogenesis in Candida albicans. Crit Rev Microbiol. 1985;12:45–93. doi: 10.3109/10408418509104425. [DOI] [PubMed] [Google Scholar]
- 390.Odds F C. Candida and candidosis. 2nd ed. London, United Kingdom: Bailliere Tindall; 1988. [Google Scholar]
- 391.Odds F C, Hierholzer J C. Purification and properties of a glycoprotein acid phosphatase from Candida albicans. J Bacteriol. 1973;114:257–266. doi: 10.1128/jb.114.1.257-266.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Okubo Y, Shibata N, Ichikawa T, Chaki S, Suzuki S. Immunochemical study on bakers’ yeast mannan prepared by fractional precipitation with cetyltrimethylammonium bromide. Arch Biochem Biophys. 1981;212:204–215. doi: 10.1016/0003-9861(81)90360-x. [DOI] [PubMed] [Google Scholar]
- 393.Okubo Y, Homma Y, Suzuki S. Relationship between phosphate content and serological activities of the mannans of Candida albicans strains NIH A-207, NIH B-792, and J-1012. J Bacteriol. 1979;137:677–680. doi: 10.1128/jb.137.1.677-680.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Ollert M W, Calderone R A. A monoclonal antibody that defines a surface antigen on Candida albicans hyphae cross-reacts with yeast cell protoplasts. Infect Immun. 1990;58:625–631. doi: 10.1128/iai.58.3.625-631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Ollert M W, Wadsworth E, Calderone R A. Reduced expression of the functionally active complement receptor for iC3b but not C3d on an avirulent mutant of Candida albicans. Infect Immun. 1990;58:909–913. doi: 10.1128/iai.58.4.909-913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Ollert M W, Sonchen R, Korting H C, Ollert U, Brautigam S, Brautigam W. Mechanisms of adherence of Candida albicans to cultured human epidermal keratinocytes. Infect Immun. 1993;61:4560–4568. doi: 10.1128/iai.61.11.4560-4568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Ollert M W, Wende C, Gorlich M, McMullan-Vogel C G, Borg-von Zepelin M, Vogel C W, Korting H C. Increased expression of Candida albicans secretory proteinase, a putative virulence factor, in isolates from human immunodeficiency virus-positive patients. J Clin Microbiol. 1995;33:2583–2589. doi: 10.1128/jcm.33.10.2543-2549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Olson E J, Standing J E, Griego-Harper N, Hoffman O A, Limper A H. Fungal β-glucan interacts with vitronectin and stimulates tumor necrosis factor alpha release from macrophages. Infect Immun. 1996;64:3548–3554. doi: 10.1128/iai.64.9.3548-3554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.O’Sullivan J M, Cannon R D, Sullivan P A, Jenkinson H F. Identification of salivary basic proline-rich proteins as receptors for Candida albicans adhesion. Microbiology. 1997;143:341–348. doi: 10.1099/00221287-143-2-341. [DOI] [PubMed] [Google Scholar]
- 400.Otto B R, Verweij-van Vught A M J J, MacLaren D M. Transferrins and heme compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–273. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 401.Page S, Odds F C. Binding of plasma membrane proteins to Candida species in vitro. J Gen Microbiol. 1988;134:2693–2702. doi: 10.1099/00221287-134-10-2693. [DOI] [PubMed] [Google Scholar]
- 402.Palma C, Serbousek D, Torosantucci A, Cassone A, Djeu J Y. Identification of a mannoprotein fraction from Candida albicans that enhances human polymorphonuclear leukocyte (PMNL) functions and stimulates lactoferrin in PMNL inhibition of candidal growth. J Infect Dis. 1992;166:1103–1112. doi: 10.1093/infdis/166.5.1103. [DOI] [PubMed] [Google Scholar]
- 403.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Paolini R, Kinet J P. Cell surface control of the multiubiquitination and deubiquitination of high-affinity immunoglobulin E receptors. EMBO J. 1993;12:779–786. doi: 10.1002/j.1460-2075.1993.tb05712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Peat S, Whelan W J, Edwards T E. Polysaccharides of baker’s yeast. IV. Mannan. 1961;1:29–34. [Google Scholar]
- 406.Pendrak M L, Klotz S A. Adherence of Candida albicans to host cells. FEMS Microbiol Lett. 1995;129:103–114. doi: 10.1111/j.1574-6968.1995.tb07566.x. [DOI] [PubMed] [Google Scholar]
- 407.Penn C, Klotz S A. Binding of plasma fibronectin to Candida albicans occurs through the cell binding domain. Microb Pathog. 1994;17:387–393. doi: 10.1006/mpat.1994.1084. [DOI] [PubMed] [Google Scholar]
- 408.Phaff H J. Cell wall of yeasts. Annu Rev Microbiol. 1963;17:15–30. doi: 10.1146/annurev.mi.17.100163.000311. [DOI] [PubMed] [Google Scholar]
- 409.Philippidis P, Naiman J L, Sibinga M S, Valdesdapnea M A. Disseminated intravascular coagulation in Candida albicans septicemia. J Pediatr. 1971;78:683–686. doi: 10.1016/s0022-3476(71)80476-6. [DOI] [PubMed] [Google Scholar]
- 410.Picard D, Khurshed B, Garabedian M J, Fortin M G, Linquist S, Yamamoto K R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature (London) 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 411.Pitzurra L, Polonelli L, Cantelli C, Gerloni M, Pontón J, Bikandi J, Blasi E. Candida albicans stress mannoprotein, SMP200, enhances tumour necrosis factor secretion in the murine macrophage cell line ANA-I. J Med Vet Mycol. 1996;34:219–222. doi: 10.1080/02681219680000371. [DOI] [PubMed] [Google Scholar]
- 412.Podzorski R P, Gray G R, Nelson R D. Different effects of Candida albicans mannan and mannan-derived oligosaccharides on antigen-stimulated lymphoproliferation in vitro. J Immunol. 1990;144:707–716. [PubMed] [Google Scholar]
- 413.Polonelli L, Gerloni M, Conti S, Fisicaro P, Cantelli C, Portincasa P, Almondo F, Barea P L, Hernando F L, Pontón J. Heat-shock mannoproteins as targets of secretory IgA in Candida albicans. J Infect Dis. 1994;169:1401–1405. doi: 10.1093/infdis/169.6.1401. [DOI] [PubMed] [Google Scholar]
- 414.Pontón J, Jones J M. Analysis of cell wall extracts of Candida albicans by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot techniques. Infect Immun. 1986;53:565–572. doi: 10.1128/iai.53.3.565-572.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Pontón J, Jones J M. Identification of two germ-tube-specific cell wall antigens of Candida albicans. Infect Immun. 1986;54:864–868. doi: 10.1128/iai.54.3.864-868.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Pontón J, Marot-Leblond A, Ezkurra P A, Barturen B, Robert R, Senet J M. Characterization of Candida albicans cell wall antigens with monoclonal antibodies. Infect Immun. 1993;61:4842–4847. doi: 10.1128/iai.61.11.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Pontón J, Hernando F L, Moragues M D, Barea P L, Gerloni M, Conti S, Fisicaro P, Cantelli C, Polonelli L. Candida albicans stress mannoproteins expression in superficial and systemic candidiasis. Mycopathologia. 1996;133:89–94. doi: 10.1007/BF00439119. [DOI] [PubMed] [Google Scholar]
- 418.Pontón J, Quindós G, Arilla M C, Mackenzie D W. Simplified adsorption method for detection of antibodies to Candida albicans germ tubes. J Clin Microbiol. 1994;32:217–219. doi: 10.1128/jcm.32.1.217-219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Pospisil L, Kabatova A. Lipolytic activity in some Candida strains. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1976;131:692–696. [PubMed] [Google Scholar]
- 420.Postlethwait P, Sundstrom P. Genetic organization and mRNA expression of enolase genes of Candida albicans. J Bacteriol. 1995;177:1772–1779. doi: 10.1128/jb.177.7.1772-1779.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Poulain D, Tronchin G, Dubremetz J F, Biguet J. Ultrastructure of the cell wall of Candida albicans blastospores: study of its constitutive layers by the use of a cytochemical technique revealing polysaccharides. Ann Microbiol (Inst Pasteur) 1978;129:140–153. [PubMed] [Google Scholar]
- 422.Poulain D, Hopwood V, Vernes A. Antigenic variability of Candida albicans. Crit Rev Microbiol. 1985;12:223–270. doi: 10.3109/10408418509104430. [DOI] [PubMed] [Google Scholar]
- 423.Poulain D, Cailliez J C, Dubremetz J F. Secretion of glycoproteins through the cell wall of Candida albicans. Eur J Cell Biol. 1989;50:94–99. [PubMed] [Google Scholar]
- 424.Poulain D, Faille C, Delaunoy C, Jaquinot P, Trinel P A, Camus D. Probable presence of β(1-2)-linked oligomannosides that act as human immunoglobulin G3 epitopes and are distributed over Candida albicans 14- to 18-kDa antigen. Infect Immun. 1993;61:1164–1166. doi: 10.1128/iai.61.3.1164-1166.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Pugh D, Cawson R A. The cytochemical localization of phospholipase A and lysophospholipase in Candida albicans. Sabouraudia. 1975;13:110–115. doi: 10.1080/00362177585190181. [DOI] [PubMed] [Google Scholar]
- 426.Pugh D, Cawson R A. The cytochemical localization of acid hydrolases in four common fungi. Cell Mol Biol (Oxford) 1977;22:125–132. [PubMed] [Google Scholar]
- 427.Pugh D, Cawson R A. The cytochemical characterization of phospholipase in Candida albicans infecting the chick chorioallantoic membrane. Sabouraudia. 1977;15:29–35. [PubMed] [Google Scholar]
- 428.Ram S P, Hynes K H, Romana L K, Shepherd M G, Sullivan P A. The β-glucanases and β-glucosidase of Candida albicans. Life Sci Adv Biochem. 1988;7:379–383. [Google Scholar]
- 429.Ram S P, Romana L K, Shepherd M G, Sullivan P A. Exo-(1-3)-β-glucanase, autolysin and trehalase activities during yeast growth and germ-tube formation in Candida albicans. J Gen Microbiol. 1984;130:1227–1236. doi: 10.1099/00221287-130-5-1227. [DOI] [PubMed] [Google Scholar]
- 430.Ramírez M, Muñoz M D, Basco R D, Giménez-Gallego G, Hernández L M, Larriba G. Two glycosylation patterns for a single protein (exoglucanase) in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1990;71:43–48. doi: 10.1111/j.1574-6968.1990.tb03796.x. [DOI] [PubMed] [Google Scholar]
- 431.Rassow J, Voos W, Pfanner N. Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 1995;5:207–212. doi: 10.1016/s0962-8924(00)89001-7. [DOI] [PubMed] [Google Scholar]
- 432.Ray T L, Wuepper D. Activation of the alternative (properdin) pathway of complement by Candida albicans and related species. J Invest Dermatol. 1976;67:700–703. doi: 10.1111/1523-1747.ep12598581. [DOI] [PubMed] [Google Scholar]
- 433.Ray T L, Payne C D. Scanning electron microscopy of epidermal adherence and cavitation in murine candidiasis: a role for Candida acid proteinase. Infect Immun. 1988;56:1942–1949. doi: 10.1128/iai.56.8.1942-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Ray T L, Payne C D. Comparative production and rapid purification of Candida acid proteinase from protein-supplemented cultures. Infect Immun. 1990;58:508–514. doi: 10.1128/iai.58.2.508-514.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Ray T L, Payne C D, Morrow B J. Candida albicans acid proteinase: characterization and role in candidiasis. Adv Exp Med Biol. 1991;306:173–183. doi: 10.1007/978-1-4684-6012-4_22. [DOI] [PubMed] [Google Scholar]
- 436.Reiss E. Immunochemistry of fungal antigens (part B): opportunistic pathogens. In: Murphy J W, Friedman H, Bendinelli M, editors. Fungal infections and immune responses. New York, N.Y: Plenum Press; 1993. pp. 439–468. [Google Scholar]
- 437.Rensing S A, Maier U G. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1994;39:80–86. doi: 10.1007/BF00178252. [DOI] [PubMed] [Google Scholar]
- 438.Rico H, Miragall F, Sentandreu R. Abnormal formation of Candida albicans walls produced by calcofluor white: an ultrastructural and stereologic study. Exp Mycol. 1985;9:241–253. [Google Scholar]
- 439.Robert R, Mahaza C, Miegeville M, Pontón J, Marot-Leblond A, Senet J M. Binding of resting platelets to Candida albicans germ tubes. Infect Immun. 1996;64:3752–3757. doi: 10.1128/iai.64.9.3752-3757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Rose M D, Misra L M, Vogel J P. KAR2, a karyogamy gene, is the yeast homologue of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 441.Ross I K, De Bernardis F, Emerson G W, Cassone A, Sullivan P A. The secreted aspartate proteinase of Candida albicans: physiology of secretion and virulence of a proteinase deficient mutant. J Gen Microbiol. 1990;136:687–694. doi: 10.1099/00221287-136-4-687. [DOI] [PubMed] [Google Scholar]
- 442.Roth A F, Davis N G. Ubiquitination of the yeast α-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Rotrosen D, Calderone R A, Edwards J E. Adherence of Candida species to host tissues and plastic surfaces. Rev Infect Dis. 1986;8:73–85. doi: 10.1093/clinids/8.1.73. [DOI] [PubMed] [Google Scholar]
- 444.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Rüchel R. A variety of Candida proteinases and their possible targets of proteolytic attack in the host. Zentralbl Bakteriol Mikrobiol Hyg Ser A. 1984;257:266–274. [PubMed] [Google Scholar]
- 446.Rüchel R. Cleavage of immunoglobulins by pathogenic yeasts of the genus Candida. Microbiol Sci. 1986;3:316–319. [PubMed] [Google Scholar]
- 447.Rüchel R, Tegeler R, Trost M. A comparison of secretory proteinases from different strains of Candida albicans. Sabouraudia. 1982;20:233–244. doi: 10.1080/00362178285380341. [DOI] [PubMed] [Google Scholar]
- 448.Rüchel R, Uhlemann K, Böning B. Secretion of acid proteinases by different species of the genus Candida. Zentralbl Bakteriol Mikrobiol Hyg Ser A. 1983;255:537–548. [PubMed] [Google Scholar]
- 449.Rüchel R, Böning B, Borg M. Characterization of a secretory proteinase of Candida parapsilosis and evidence for the absence of enzyme during infection in vitro. Infect Immun. 1986;53:411–419. doi: 10.1128/iai.53.2.411-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Rüchel R, Zimmermann F, Böning-Stutzer B, Helmchen U. Candidiasis visualized by proteinase-directed immunofluorescence. Virchows Arch A. 1991;419:199–202. doi: 10.1007/BF01626348. [DOI] [PubMed] [Google Scholar]
- 451.Rüchel, R., F. de Bernardis, T. L. Ray, P. A. Sullivan, and G. T. Cole. 1992. Candida acid proteinases. J. Med. Vet. Mycol. 30(Suppl. 1):123–132. [PubMed]
- 452.Rudek W. Esterase activity in Candida species. J Clin Microbiol. 1978;8:756–759. doi: 10.1128/jcm.8.6.756-759.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Ruiz-Herrera J. Fungal cell wall: structure, synthesis and assembly. Boca Raton, Fla: CRC Press, Inc.; 1992. [Google Scholar]
- 454.Ruiz-Herrera J, Sentandreu R. Fungal cell wall synthesis and assembly. Curr Top Med Mycol. 1989;3:168–217. doi: 10.1007/978-1-4612-3624-5_8. [DOI] [PubMed] [Google Scholar]
- 455.Ruiz-Herrera J, Mormeneo S, Vanaclocha P, Font de Mora J, Iranzo M, Puertes I, Sentandreu R. Structural organization of the components of the cell wall from Candida albicans. Microbiology. 1994;140:1513–1523. doi: 10.1099/13500872-140-7-1513. [DOI] [PubMed] [Google Scholar]
- 456.Ruiz-Herrera J, Iranzo M, Elorza M V, Sentandreu R, Mormeneo S. Involvement of transglutaminase in the formation of covalent cross-links in the cell wall of Candida albicans. Arch Microbiol. 1995;164:186–193. doi: 10.1007/BF02529970. [DOI] [PubMed] [Google Scholar]
- 457.Ruoslahti E. Integrins. J Clin Invest. 1991;87:1–5. doi: 10.1172/JCI114957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Samaranayake L P, MacFarlane T W. An in-vitro study of the adherence of Candida albicans to acrylic surfaces. Arch Oral Biol. 1980;25:603–609. doi: 10.1016/0003-9969(80)90075-8. [DOI] [PubMed] [Google Scholar]
- 459.San Millan R, Ezkurra P A, Quindos G, Robert R, Senet J M, Pontón J. Effect of monoclonal antibodies directed against Candida albicans cell wall antigens on the adhesion of the fungus to polystyrene. Microbiology. 1996;142:2271–2277. doi: 10.1099/13500872-142-8-2271. [DOI] [PubMed] [Google Scholar]
- 460.Sánchez-Madrid F, Nagy J A, Robbins E, Simon P, Springer T A. A human leukocyte differentiation antigen family with distinct α-subunits and a common β-subunit. J Exp Med. 1983;158:1785–1803. doi: 10.1084/jem.158.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460a.Sanglard D, Hube B, Monod M, Odds F C, Gow N A R. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infect Immun. 1997;65:3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Sanglard, D., and M. Monod. 1997. Personal communication.
- 462.Sanjuán R, Zueco J, Pérez J, Peñarroja C, Sentandreu R. A comparative study of the incorporation of a 1,6-β-glucan and an O-glycosylated protein epitope into the cell wall of Candida albicans. Microbiology. 1996;142:2255–2262. doi: 10.1099/13500872-142-8-2255. [DOI] [PubMed] [Google Scholar]
- 463.Sanjuán R, Zueco J, Stock R, Font de Mora J, Sentandreu R. Identification of glucan-mannoprotein complexes in the cell wall of Candida albicans using a monoclonal antibody that reacts with a (1,6)-β-glucan epitope. Microbiology. 1995;141:1545–1551. doi: 10.1099/13500872-141-7-1545. [DOI] [PubMed] [Google Scholar]
- 464.Santoni G, Birarelli P, Jin-Hong L, Gamero A, Djeu J Y, Piccoli M. An α5β1-like integrin receptor mediates the binding of less pathogenic Candida species to fibronectin. J Med Microbiol. 1995;43:360–367. doi: 10.1099/00222615-43-5-360. [DOI] [PubMed] [Google Scholar]
- 465.Santoni G, Gismondi A, Liu J H, Punturieri A, Santoni A, Frati L, Piccoli M, Djeu J Y. Candida albicans expresses a fibronectin receptor antigenically related to α5β1 integrin. Microbiology. 1994;140:2971–2979. doi: 10.1099/13500872-140-11-2971. [DOI] [PubMed] [Google Scholar]
- 466.Sanz P, Herrero E, Sentandreu R. Secretory pattern of a major integral mannoprotein of the yeast cell wall. Biochim Biophys Acta. 1987;924:193–203. [Google Scholar]
- 467.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Sarthy A V, McGonigal T, Coen M, Frost D J, Muelbroek J, Goldman R C. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-β-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 469.Sawyer R T, Garner R E, Hudson J A. Arg-Gly-Asp (RGD) peptides alter hepatic killing of Candida albicans in the isolated perfused mouse liver model. Infect Immun. 1992;60:213–218. doi: 10.1128/iai.60.1.213-218.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Saxena A, Calderone R A. Purification and characterization of the extracellular C3d-binding protein of Candida albicans. Infect Immun. 1990;58:309–314. doi: 10.1128/iai.58.2.309-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Scadden, A. D., and P. A. Sullivan. 1994. GenBank accession no. U12975.
- 472.Scheld W M, Strunk R W, Balian G, Calderone R A. Microbial adhesion to fibronectin in vitro correlates with production of endocarditis in rabbits. Proc Soc Exp Biol Med. 1985;180:474–482. doi: 10.3181/00379727-180-42205. [DOI] [PubMed] [Google Scholar]
- 473.Scherer, S. Candida albicans information. http://alces.med.umn.edu/Candida.html.
- 474.Scherer S, Magee P T. Genetics of Candida albicans. Microbiol Rev. 1990;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Segal E, Soroka A, Schecter A. Correlative relationship between adherence of Candida albicans to human vaginal epithelial cells in vitro and candidal vaginitis. Sabouraudia. 1984;22:191–200. [PubMed] [Google Scholar]
- 476.Segal E, Kremer I, Dayan D. Inhibition of adherence of Candida albicans to acrylic by a chitin derivative. Eur J Epidemiol. 1992;8:350–355. doi: 10.1007/BF00158567. [DOI] [PubMed] [Google Scholar]
- 477.Sentandreu M, Elorza M V, Sentandreu R, Fonzi W A. Abstracts of the ASM Conference on Candida and Candidiasis: biology, pathogenesis, and management. Washington, D.C: American Society for Microbiology; 1996. A novel pH-regulated cell surface protein of C. albicans, abstr. A13. [Google Scholar]
- 478.Sentandreu R, Herrero E, Martínez J P, Larriba G. Biogenesis of the yeast cell wall. Subcell Biochem. 1984;10:193–235. doi: 10.1007/978-1-4613-2709-7_3. . Plenum Press, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 479.Sentandreu R, Herrero E, Martínez J P, Elorza M V. Yeast cell wall glycoproteins. NATO ASI Ser. 1991;H53:229–239. [Google Scholar]
- 480.Sentandreu R, Martínez J P, Elorza M V, Mormeneo S. Relationships between dimorphism, cell wall structure, and surface activities in Candida albicans. In: Prasad R, editor. Candida albicans: cellular and molecular biology. Berlin, Germany: Springer-Verlag KG; 1991. pp. 72–88. [Google Scholar]
- 481.Sentandreu R, Elorza M V, Mormeneo S, Sanjuán R, Iranzo M. Possible roles of mannoproteins in the construction of Candida albicans cell wall. In: Vanden Bosche H, Odds F C, Kerridge D, editors. Dimorphic fungi in biology and medicine. New York, N.Y: Plenum Press; 1993. pp. 169–175. [Google Scholar]
- 482.Sentandreu R, Mormeneo S, Ruiz-Herrera J. Biogenesis of the fungal cell wall. In: Wessels J G H, Meinhardt F, editors. The Mycota I: growth, differentiation and sexuality. Berlin, Germany: Springer-Verlag KG; 1994. pp. 111–124. [Google Scholar]
- 483.Sepúlveda P. Caracterización de adhesinas y marcadores antigénicos de la superficie celular de Candida albicans. Ph.D. thesis dissertation. Valencia, Spain: University of Valencia; 1995. [Google Scholar]
- 484.Sepúlveda P, Murgui A, López-Ribot J L, Casanova M, Timoneda J, Martínez J P. Evidence for the presence of collagenous domains in Candida albicans cell surface proteins. Infect Immun. 1995;63:2173–2179. doi: 10.1128/iai.63.6.2173-2179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Sepúlveda P, Cervera A M, López-Ribot J L, Chaffin W L, Martínez J P, Gozalbo D. Cloning and characterization of a cDNA coding for Candida albicans polyubiquitin. J Med Vet Mycol. 1996;34:315–322. [PubMed] [Google Scholar]
- 486.Sepúlveda P, López-Ribot J L, Gozalbo D, Cervera A M, Martínez J P, Chaffin W L. Ubiquitin-like epitopes associated with Candida albicans cell surface receptors. Infect Immun. 1996;64:4406–4408. doi: 10.1128/iai.64.10.4406-4408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Sepúlveda, P., J. L. López-Ribot, M. Casanova, D. Navarro, E. Cantón, and J. P. Martínez. Candida albicans fibrinogen binding mannoprotein: expression in clinical strains and immunogenicity in patients with systemic candidiasis. Microbiologia, in press. [PubMed]
- 488.Shapira M, McEwen J G, Jaffe C L. Temperature effects on molecular processes which lead to stage differentiation in Leishmania. EMBO J. 1988;7:2895–2901. doi: 10.1002/j.1460-2075.1988.tb03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Shen H D, Choo K B, Lee H H, Hsieh J C, Lin W L, Lee W R, Han S H. The 40-kilodalton allergen of Candida albicans is an alcohol dehydrogenase: molecular cloning and immunological analysis using monoclonal antibodies. Clin Exp Allergy. 1991;21:675–681. doi: 10.1111/j.1365-2222.1991.tb03195.x. [DOI] [PubMed] [Google Scholar]
- 490.Shepherd M G. Cell envelope of Candida albicans. Crit Rev Microbiol. 1987;15:7–25. doi: 10.3109/10408418709104445. [DOI] [PubMed] [Google Scholar]
- 491.Shepherd M G, Gopal P K. Nature and control of cell wall biosynthesis. In: Vanden Bossche H, Odds F C, Kerridge D, editors. Dimorphic fungi in biology and medicine. New York, N.Y: Plenum Press; 1993. pp. 153–167. [Google Scholar]
- 492.Shepherd M G, Ghazali H M, Sullivan P A. N-acetyl-d-glucosamine kinase and germ-tube formation in Candida albicans. Exp Mycol. 1980;4:147–159. [Google Scholar]
- 493.Shepherd M G, Poulter R T M, Sullivan P A. Candida albicans: biology, genetics, and pathogenicity. Annu Rev Microbiol. 1985;39:579–614. doi: 10.1146/annurev.mi.39.100185.003051. [DOI] [PubMed] [Google Scholar]
- 494.Shibata N, Kobayashi H, Tojo M, Suzuki S. Characterization of phosphomannan-protein complexes isolated from viable cells of yeast and mycelial forms of Candida albicans NIH B-792 strain by action of Zymolyase 100T. Arch Biochem Biophys. 1986;251:697–701. doi: 10.1016/0003-9861(86)90379-6. [DOI] [PubMed] [Google Scholar]
- 495.Shibata N, Fukasawa S, Kobayashi H, Tojo M, Yonezu T, Ambo A, Ohkubo Y, Suzuki S. Structural analysis of phospho-d-mannan-protein complexes isolated from yeast and mold form cells of Candida albicans NIH A-270 serotype A strain. Carbohydr Res. 1989;187:239–253. doi: 10.1016/0008-6215(89)80006-0. [DOI] [PubMed] [Google Scholar]
- 496.Shibata N, Kobayashi H, Takahashi S, Okawa Y, Hisamichi K, Suzuki S, Suzuki S. Structural study on a phosphorylated mannotetraose obtained from the phosphomannan of Candida albicans NIH B-792 strain by acetolysis. Arch Biochem Biophys. 1991;290:535–542. doi: 10.1016/0003-9861(91)90578-7. [DOI] [PubMed] [Google Scholar]
- 497.Shibata N, Arai M, Haga E, Kikuchi T, Najima M, Satoh T, Kobayashi H, Suzuki S. Structural identification of an epitope of antigenic factor 5 in mannans of Candida albicans NIH B-792 (serotype B) and J-1012 (serotype A) as β-1,2-linked oligomannosyl residues. Infect Immun. 1992;60:4100–4110. doi: 10.1128/iai.60.10.4100-4110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Shibata N, Hisamichi K, Kikuchi T, Kobayashi H, Okawa Y, Suzuki S. Sequential nuclear magnetic resonance assignment of β-1,2-linked mannooligosaccharides isolated from the phosphomannan of the pathogenic yeast Candida albicans NIH B-792 strain. Biochemistry. 1992;31:5680–5686. doi: 10.1021/bi00139a036. [DOI] [PubMed] [Google Scholar]
- 499.Shimizu M T, Jorge A O, Unterkircher C S, Fantinato V, Paulo C R. Hyaluronidase and chondroitin sulphatase production by different species of Candida. J Med Vet Mycol. 1995;33:27–31. [PubMed] [Google Scholar]
- 500.Shimizu M T, Almeida N Q, Fantinato V, Unterkircher C S. Studies on hyaluronidase, chondroitin sulphatase, proteinase and phospholipase secreted by Candida species. Mycoses. 1996;39:161–167. doi: 10.1111/j.1439-0507.1996.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 501.Shimokawa O, Nakayama H. A Candida albicans rough-type mutant with increased cell surface hydrophobicity and a structural defect in the cell wall mannan. J Med Vet Mycol. 1986;24:165–168. doi: 10.1080/02681218680000241. [DOI] [PubMed] [Google Scholar]
- 502.Shirayama M, Kawakami K, Matsui Y, Tanaka K, Tohe A. MSI3, a multicopy suppressor of mutants hyperactivated in the RAS-cAMP pathway, encodes a novel HSP70 protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993;240:323–332. doi: 10.1007/BF00280382. [DOI] [PubMed] [Google Scholar]
- 503.Shoemaker C, Gross A, Gebremichael A, Harn D. cDNA cloning and functional expression of the Schistosoma mansoni protective antigen triose-phosphate isomerase. Proc Natl Acad Sci USA. 1992;89:1842–1846. doi: 10.1073/pnas.89.5.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Siegelman M, Bond M W, Gallatin M W, John T S, Smith H T, Fried V A, Weissman I L. Cell surface molecule associated with lymphocyte homing is a ubiquitinated branched-chain glycoprotein. Science. 1986;231:823–829. doi: 10.1126/science.3003913. [DOI] [PubMed] [Google Scholar]
- 505.Silva T M, Glee P M, Hazen K C. Influence of cell surface hydrophobicity on attachment of Candida albicans to extracellular matrix proteins. J Med Vet Mycol. 1995;33:117–122. [PubMed] [Google Scholar]
- 506.Siu C H, Lam T Y, Choi A H C. Inhibition of cell-cell binding at the aggregation stage of Dictyostelium discoideum development by monoclonal antibodies directed against an 80,000-dalton surface glycoprotein. J Biol Chem. 1985;260:16030–16036. [PubMed] [Google Scholar]
- 507.Skerl K G, Calderone R A, Segal E, Sreevalsan T, Scheld W M. In vitro binding of Candida albicans yeast cells to human fibronectin. Can J Microbiol. 1984;30:221–227. doi: 10.1139/m84-033. [DOI] [PubMed] [Google Scholar]
- 508.Smail E H, Jones J M. Demonstration and solubilization of antigens expressed primarily on the surfaces of Candida albicans germ tubes. Infect Immun. 1984;45:74–81. doi: 10.1128/iai.45.1.74-81.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Smolenski G, Sullivan P A, Cutfield S M. Analysis of secreted aspartic proteinases for Candida albicans: purification and characterization of individual Sap1, Sap2, and Sap3 isoenzymes. Microbiology. 1997;143:349–356. doi: 10.1099/00221287-143-2-349. [DOI] [PubMed] [Google Scholar]
- 510.Soll D R. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Soll D R. The emerging molecular biology of switching in Candida albicans. ASM News. 1996;62:415–420. [Google Scholar]
- 512.Spötl L, Möst J, Dierich M P. Ca ions stabilize the binding of complement factor iC3b to the pseudohyphal form of Candida albicans. Infect Immun. 1994;62:1125–1127. doi: 10.1128/iai.62.3.1125-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Staab J F, Ferrer C A, Sundstrom P. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J Biol Chem. 1996;271:6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 514.Staib F. Serum-proteins as nitrogen source for yeast-like fungi. Sabouraudia. 1965;4:187–193. doi: 10.1080/00362176685190421. [DOI] [PubMed] [Google Scholar]
- 515.Staib F. Proteolysis and pathogenicity of Candida albicans strains. Mycopathol Mycol Appl. 1969;37:345–348. doi: 10.1007/BF02129881. [DOI] [PubMed] [Google Scholar]
- 516.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Strockbine N A, Largen M T, Zweibel S M, Buckley H R. Identification and molecular weight characterization of antigens from Candida albicans that are recognized by human sera. Infect Immun. 1984;43:715–721. doi: 10.1128/iai.43.2.715-721.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Sullivan P A, Chiew Y Y, Molloy C, Templeton M D, Shepherd M G. An analysis of the metabolism and cell wall composition of Candida albicans during germ-tube formation. Can J Microbiol. 1983;29:1514–1525. doi: 10.1139/m83-233. [DOI] [PubMed] [Google Scholar]
- 519.Sullivan P A, McHugh N J, Romana L K, Shepherd M G. The secretion of N-acetylglucosaminidase during germ-tube formation in Candida albicans. J Gen Microbiol. 1984;130:2213–2218. doi: 10.1099/00221287-130-9-2213. [DOI] [PubMed] [Google Scholar]
- 520.Sundstrom P, Jensen J, Balish E. Humoral and cellular immune responses to enolase after alimentary tract colonization or intravenous immunization with Candida albicans. J Infect Dis. 1994;170:390–395. doi: 10.1093/infdis/170.2.390. [DOI] [PubMed] [Google Scholar]
- 521.Sundstrom P M, Aliaga G R. Molecular cloning of cDNA and analysis of protein secondary structure of Candida albicans enolase, an abundant, immunodominant glycolytic enzyme. J Bacteriol. 1992;174:6789–6799. doi: 10.1128/jb.174.21.6789-6799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Sundstrom P M, Aliaga G R. A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J Infect Dis. 1994;169:452–456. doi: 10.1093/infdis/169.2.452. [DOI] [PubMed] [Google Scholar]
- 523.Sundstrom P M, Kenny G E. Characterization of antigens specific to the surface of germ tubes of Candida albicans by immunofluorescence. Infect Immun. 1984;43:850–855. doi: 10.1128/iai.43.3.850-855.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Sundstrom P M, Kenny G E. Enzymatic release of germ tube-specific antigens from cell walls of Candida albicans. Infect Immun. 1985;49:609–614. doi: 10.1128/iai.49.3.609-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Sundstrom P M, Nichols E J, Kenny G E. Antigenic differences between mannoproteins of germ tubes and blastospores of Candida albicans. Infect Immun. 1987;55:616–620. doi: 10.1128/iai.55.3.616-620.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Sundstrom P M, Tam M R, Nichols E J, Kenny G E. Antigenic differences in the surface mannoproteins of Candida albicans as revealed by monoclonal antibodies. Infect Immun. 1988;56:601–606. doi: 10.1128/iai.56.3.601-606.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Surarit R, Gopal P K, Shepherd M G. Evidence for a glycosidic linkage between chitin and glucan in the cell wall of Candida albicans. J Gen Microbiol. 1988;134:1723–1730. doi: 10.1099/00221287-134-6-1723. [DOI] [PubMed] [Google Scholar]
- 528.Suzuki S, Pierschbacher M D, Hayman E G, Nguyen K, Ohgren Y, Ruoslahti E. Domain structure of vitronectin. J Biol Chem. 1984;259:15307–15314. [PubMed] [Google Scholar]
- 529.Sweet S P, Douglas L J. Effect of iron deprivation on surface composition and virulence determinants of Candida albicans. J Gen Microbiol. 1991;137:859–865. doi: 10.1099/00221287-137-4-859. [DOI] [PubMed] [Google Scholar]
- 530.Swoboda R K, Bertram G, Hollander H, Greenspan D, Greenspan J S, Gow N A R, Gooday G W, Brown A J P. Glycolytic enzymes of Candida albicans are nonubiquitous immunogens during candidiasis. Infect Immun. 1993;61:4263–4271. doi: 10.1128/iai.61.10.4263-4271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Swoboda R K, Miyasaki S, Greenspan D, Greenspan J S. Heat-inducible ATP-binding proteins of Candida albicans are recognized by sera of infected patients. J Gen Microbiol. 1993;139:2995–3003. doi: 10.1099/00221287-139-12-2995. [DOI] [PubMed] [Google Scholar]
- 532.Swoboda R K, Bertram G, Budge S, Gooday G W, Gow N A R, Brown A J P. Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect Immun. 1995;63:4506–4514. doi: 10.1128/iai.63.11.4506-4514.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Takahashi M, Banno Y, Nozawa Y. Secreted Candida albicans phospholipases: purification and characterization of two forms of lysophospholipase-transacylase. J Med Vet Mycol. 1991;29:193–204. [PubMed] [Google Scholar]
- 534.Takahashi M, Banno Y, Shikano Y, Mori S, Nozawa Y. Purification and characterization of lysophospholipase-transacylase of pathogenic fungus Candida albicans. Biochim Biophys Acta. 1991;1082:161–169. doi: 10.1016/0005-2760(91)90190-s. [DOI] [PubMed] [Google Scholar]
- 535.Taschdijian C, Burchall J, Kozinn P. Rapid identification of Candida albicans by filamentation on serum and serum substitutes. Am J Dis Child. 1960;99:212. doi: 10.1001/archpedi.1960.02070030214011. [DOI] [PubMed] [Google Scholar]
- 536.Tavares D, Salvador A, Ferreira P, Arala-Chaves M. Immunological activities of a Candida albicans protein which plays an important role in the survival of the microorganism in the host. Infect Immun. 1993;61:1881–1888. doi: 10.1128/iai.61.5.1881-1888.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Tavares D, Ferreira P, Vilanova M, Videira A, Arala-Chaves M. Immunoprotection against systemic candidiasis in mice. Int Immunol. 1995;7:785–796. doi: 10.1093/intimm/7.5.785. [DOI] [PubMed] [Google Scholar]
- 538.Teunissen A W R H, Holub E, Van Der Hucht J, Van den Berg J S, Steensma H Y. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast. 1993;9:423–437. doi: 10.1002/yea.320090413. [DOI] [PubMed] [Google Scholar]
- 539.Timpl R, Rohde H, Gehron-Robey P, Rennard S I, Foidart J M, Martin G. Laminin—a glycoprotein from basement membranes. J Biol Chem. 1979;254:9933–9937. [PubMed] [Google Scholar]
- 540.Tojo M, Shibata N, Mikami T, Suzuki M, Suzuki S. Participation of peptide moieties in adhesive behavior of antigenic mannans of Candida albicans to the plastic microtiter plate in enzyme-linked immunosorbent assay. Clin Chem. 1987;33:1925–1928. [PubMed] [Google Scholar]
- 541.Torosantucci A, Boccanera M, Casalinuovo I, Pellegrini G, Cassone A. Differences in the antigenic expression of immunomodulatory mannoprotein constituents on yeasts and mycelial forms of Candida albicans. J Gen Microbiol. 1990;136:1421–1428. doi: 10.1099/00221287-136-7-1421. [DOI] [PubMed] [Google Scholar]
- 542.Torosantucci A, Bromuro C, Gómez M J, Ausiello C M, Urbani F, Cassone A. Identification of a 65-kDa mannoprotein as a main target of human cell-mediated immune response to Candida albicans. J Infect Dis. 1993;168:427–435. doi: 10.1093/infdis/168.2.427. [DOI] [PubMed] [Google Scholar]
- 543.Torosantucci A, Gómez M J, Bromuro C, Casalinuovo I, Cassone A. Biochemical and antigenic characterization of mannoprotein constituents released from yeast and mycelial forms of Candida albicans. J Med Vet Mycol. 1991;29:361–372. doi: 10.1080/02681219180000591. [DOI] [PubMed] [Google Scholar]
- 544.Tosh F D, Douglas L J. Characterization of a fucoside-binding adhesin of Candida albicans. Infect Immun. 1992;60:4734–4739. doi: 10.1128/iai.60.11.4734-4739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Trinel P A, Borg-von-Zepelin M, Lepage G, Jouault T, Mackenzie D W R, Poulain D. Isolation and preliminary characterization of the 14- to 18-kDa Candida albicans antigen as phospholipomannan containing β-1,2 linked oligomannosides. Infect Immun. 1993;61:4398–4405. doi: 10.1128/iai.61.10.4398-4405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Trinel P A, Cantelli C, Bernigaud A, Jouault T, Poulain D. Evidence for different mannosylation processes involved in the association of β-1,2-linked oligomannosidic epitopes in Candida albicans mannan and phospholipomannan. Microbiology. 1996;142:2263–2270. doi: 10.1099/13500872-142-8-2263. [DOI] [PubMed] [Google Scholar]
- 547.Tronchin G, Poulain D, Biguet J. Localisation ultrastructurale de l’activité phosphatasique acide chez Candida albicans. Biol Cell (Ivry sur Seine) 1980;38:147–152. [Google Scholar]
- 548.Tronchin G, Robert R, Bouali A, Senet J M. Immunocytochemical localization of in vitro binding of human fibrinogen to Candida albicans germ tube and mycelium. Ann Inst Pasteur Microbiol. 1987;138:177–187. doi: 10.1016/0769-2609(87)90194-3. [DOI] [PubMed] [Google Scholar]
- 549.Tronchin G, Bouchara J P, Robert R, Senet J M. Adherence of Candida albicans germ tubes to plastic: ultrastructural and molecular studies of fibrillar adhesins. Infect Immun. 1988;56:1987–1993. doi: 10.1128/iai.56.8.1987-1993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Tronchin G, Bouchara J P, Robert R. Dynamic changes of the cell wall surface of Candida albicans associated with germination and adherence. Eur J Cell Biol. 1989;50:285–290. [PubMed] [Google Scholar]
- 551.Tsuboi R, Komatsuzaki H, Ogawa H. Induction of an extracellular esterase from Candida albicans and some of its properties. Infect Immun. 1996;64:2936–2940. doi: 10.1128/iai.64.8.2936-2940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Tsushima H, Mine H, Kawakami Y, Hyodoh F, Ueki A. Candida albicans aspartic proteinase cleaves and inactivates human epidermal cysteine proteinase inhibitor, cystatin a. Microbiology. 1994;140:167–171. doi: 10.1099/13500872-140-1-167. [DOI] [PubMed] [Google Scholar]
- 553.van de Rijn M, Weissman I L, Siegelman M. Biosynthesis pathway of gp90MEL-14, the mouse lymph node-specific homing receptor. J Immunol. 1990;145:1477–1482. [PubMed] [Google Scholar]
- 554.Van der Ploeg L H T, Giannini S H, Cantor C R. Heat-shock genes: regulatory role for differentiation in parasitic protozoa. Science. 1985;228:1443–1445. doi: 10.1126/science.4012301. [DOI] [PubMed] [Google Scholar]
- 555.van der Vaart J M, Caro L H P, Chapman J W, Klis R M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:2953–2955. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.van Deventer A J M, Goessens W H F, van Vliet H J A, Verbrugh H A. Abstracts of the ASM Conference on Candida and Candidiasis: biology, pathogenesis, and management. Washington, D.C: American Society for Microbiology; 1996. Anti-enolase antibodies protective against systemic candidiasis in mice, abstr. B50. [Google Scholar]
- 557.van Deventer A J M, van Vliet H J A, Hop W C J, Goessens W H F. Diagnostic value of anti-Candida enolase antibodies. J Clin Microbiol. 1994;32:17–23. doi: 10.1128/jcm.32.1.17-23.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.van Deventer A J M, van Vliet H J A, Voogd L, Hop W C J, Goessens W H F. Increased specificity of antibody detection in surgical patients with invasive candidiasis with cytoplasmic antigens depleted of mannan residues. J Clin Microbiol. 1993;31:994–997. doi: 10.1128/jcm.31.4.994-997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.van Weissenbruch R, Bouckaert S, Remon J P, Nelis H J, Aerts R, Albers F W. Chemoprophylaxis of fungal deterioration of the Provox silicone tracheoesophageal prosthesis in postlaryngectomy patients. Ann Otol Rhinol Laryngol. 1997;106:329–337. doi: 10.1177/000348949710600413. [DOI] [PubMed] [Google Scholar]
- 560.Vasilas A, Molina L, Hoffman M, Haidaris C G. The influence of morphological variation on Candida albicans adhesion to denture acrylic in vitro. Arch Oral Biol. 1992;37:613–622. doi: 10.1016/0003-9969(92)90123-p. [DOI] [PubMed] [Google Scholar]
- 561.Vásquez de Aldana C R, Correa J, San Segundo P, Bueno A, Nebreda A R, Méndez E, Del Rey F. Nucleotide sequence of the exo-1,3-β-glucanase-encoding gene, EXG1, of the yeast Saccharomyces cerevisiae. Gene. 1991;97:173–182. doi: 10.1016/0378-1119(91)90049-h. [DOI] [PubMed] [Google Scholar]
- 562.Vespa M N, López-Ribot J L, Chaffin W L. Adherence of germ tubes of Candida albicans to tissues from immunocompromised mice. FEMS Immunol Med Microbiol. 1995;11:57–64. doi: 10.1111/j.1574-695X.1995.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 563.Wadsworth E, Prasad S C, Calderone R A. Analysis of mannoprotein from blastoconidia and hyphae of Candida albicans with a common epitope recognized by anti-complement receptor type 2 antibodies. Infect Immun. 1993;61:4675–4681. doi: 10.1128/iai.61.11.4675-4681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Walsh T J, Hathorn J W, Sobel J D, Merz W G, Sánchez V, Maret S M, Buckley H R, Pfaller M A, Schaufele R, Sliva C, Navarro E, Lecciones P, Chandrasekar P, Lee J, Pizzo P A. Detection of circulating Candida enolase by immunoassay in patients with cancer and invasive candidiasis. N Engl J Med. 1991;324:1026–1031. doi: 10.1056/NEJM199104113241504. [DOI] [PubMed] [Google Scholar]
- 565.Waters, M. G., D. W. Williams, R. G. Jagger, and M. A. Lewis. Adherence of Candida albicans to experimental denture soft lining materials. J. Prosthet. Dent. 77:306–312. [DOI] [PubMed]
- 566.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Werner-Washburne M, Stone D E, Craig E A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Wessels J G H, de Vries O M H, Ásgeirdóttir S A, Schuren F H J. Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in Schizophyllum. Plant Cell. 1991;3:793–799. doi: 10.1105/tpc.3.8.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Wessels J G H, de Vries O M H, Ásgeirdóttir S A, Springer J. The thn mutation of Schizophyllum commune, which suppresses formation of aerial hyphae, affects expression of the Sc3 hydrophobin gene. J Gen Microbiol. 1991;137:2439–2445. doi: 10.1099/00221287-137-10-2439. [DOI] [PubMed] [Google Scholar]
- 570.White T C, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.White T C, Miyasaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Winram S B, Lottenberg R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;142:2311–2320. doi: 10.1099/13500872-142-8-2311. [DOI] [PubMed] [Google Scholar]
- 573.Wojciechowicz D, Lu C-F, Kurjan J, Lipke P N. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol Cell Biol. 1993;13:2554–2563. doi: 10.1128/mcb.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Wright R J, Carne A, Hieber A D, Lamont I L, Emerson G W, Sullivan P A. A second gene for a secreted aspartate proteinase in Candida albicans. J Bacteriol. 1992;174:7848–7853. doi: 10.1128/jb.174.23.7848-7853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Wu C, Chung A E. Potential role of entactin in hemostasis. J Biol Chem. 1991;266:18802–18807. [PubMed] [Google Scholar]
- 576.Wu C, Reing J, Chung A E. Entactin forms a complex with fibronectin and co-localizes in the extracellular matrix of the embryonal carcinoma-derived 4CQ cell line. Biochem Biophys Res Commun. 1991;178:1219–1225. doi: 10.1016/0006-291x(91)91023-6. [DOI] [PubMed] [Google Scholar]
- 577.Xu J, Day A W. Multiple forms of fimbriae on the sporidia of corn smut, Ustilago maydis. Int J Plant Sci. 1992;153:531–540. [Google Scholar]
- 578.Yan S, Nègre E, Cashel J A, Guo N, Lyman C A, Walsh T J, Roberts D D. Specific induction of fibronectin binding activity by hemoglobin in Candida albicans grown in defined media. Infect Immun. 1996;64:2930–2935. doi: 10.1128/iai.64.8.2930-2935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Yang W, Li E, Kairong T, Stanley S L., Jr Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhE gene product of Escherichia coli. Mol Biochem Parasitol. 1994;64:253–260. doi: 10.1016/0166-6851(93)00020-a. [DOI] [PubMed] [Google Scholar]
- 580.Yarden Y, Escobedo J A, Kuang W J, Yang F T, Daniel T O, Tremble P M, Chen E Y, Ando M E, Harkins R N, Francke U, Fried V A, Ullrich A, Williams L T. Structure of the receptor for platelet-derived growth factor helps define a family of closely related growth factor receptors. Nature. 1986;232:226–232. doi: 10.1038/323226a0. [DOI] [PubMed] [Google Scholar]
- 581.Young D, Lathigra R, Hendrix R, Sweetser D, Young R A. Stress proteins are immune targets in leprosy and tuberculosis. Proc Natl Acad Sci USA. 1988;85:4267–4270. doi: 10.1073/pnas.85.12.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Young D B. Heat-shock proteins-immunity and autoimmunity. Curr Opin Immunol. 1992;4:396–400. doi: 10.1016/s0952-7915(06)80029-4. [DOI] [PubMed] [Google Scholar]
- 583.Yu L, Lee K K, Ens K, Doig P C, Carpenter M R, Staddon W, Hodges R S, Paranchych W, Irvin R T. Partial characterization of a Candida albicans fimbrial adhesin. Infect Immun. 1994;62:2834–2842. doi: 10.1128/iai.62.7.2834-2842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Yu L, Lee K K, Sheth H B, Lane-Bell P, Srivastava G, Hindsgaul O, Paranchych W, Hodges R S, Irvin R T. Fimbria-mediated adherence of Candida albicans to glycosphingolipid receptors on human buccal epithelial cells. Infect Immun. 1994;62:2843–2848. doi: 10.1128/iai.62.7.2843-2848.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Yu L, Lee K K, Hodges R S, Panachych W, Irvin R T. Adherence of Pseudomonas aeruginosa and Candida albicans to glycosphingolipis (asialo-GM1) receptors is achieved by a conserved receptor-binding domain present on their adhesins. Infect Immun. 1994;62:5213–5219. doi: 10.1128/iai.62.12.5213-5219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Yu L, Lee K K, Paranchych W, Hodges R S, Irvin R T. Use of synthetic peptides to confirm that the Pseudomonas aeruginosa PAK pilus adhesin and the Candida albicans fimbrial adhesin possess a homologous receptor-binding domain. Mol Microbiol. 1996;19:1107–1116. doi: 10.1046/j.1365-2958.1996.454982.x. [DOI] [PubMed] [Google Scholar]
- 587.Yu L, Goldman R, Sullivan P, Walker G F, Fesik S W. Heteronuclear NMR studies of 13C-labeled yeast cell wall β-glucan oligosaccharides. J Biomol NMR. 1993;3:429–441. doi: 10.1007/BF00176009. [DOI] [PubMed] [Google Scholar]
- 588.Zeuthen M L, Howard D H. Thermotolerance and the heat-shock response in Candida albicans. J Gen Microbiol. 1989;135:2509–2518. doi: 10.1099/00221287-135-9-2509. [DOI] [PubMed] [Google Scholar]
- 589.Zlotnik H, Fernandez M P, Bowers B, Cabib E. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol. 1984;159:1018–1026. doi: 10.1128/jb.159.3.1018-1026.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]