Abstract
Burn injury induces a systemic hyperinflammatory response with detrimental side effects. Studies have described the biochemical changes induced by severe burns, but the transcriptome response is not well characterized. The goal of this work is to characterize the blood transcriptome after burn injury. Burn patients presenting to a regional center between 2012 and 2017 were prospectively enrolled. Blood was collected on admission and at predetermined time points (hours 2, 4, 8, 12, and 24). RNA was isolated and transcript levels were measured with a gene expression microarray. To identify differentially regulated genes (false-discovery rate ≤0.1) by burn injury severity, patients were grouped by TBSA above or below 20% and statistically enriched pathways were identified. Sixty-eight patients were analyzed, most patients were male with a median age of 41 (interquartile range, 30.5–58.5) years, and TBSA of 20% (11%–34%). Thirty-five patients had % TBSA injury ≥20%, and this group experienced greater mortality (26% vs 3%, P = .008). Comparative analysis of genes from patients with </≥20% TBSA revealed 1505, 613, 380, 63, 1357, and 954 differentially expressed genes at hours 0, 2, 4, 8, 12, and 24, respectively. Pathway analysis revealed an initial up-regulation in several immune/inflammatory pathways within the ≥20% TBSA groups followed by shutdown. Severe burn injury is associated with an early proinflammatory immune response followed by shutdown of these pathways. Examination of the immunoinflammatory response to burn injury through differential gene regulation and associated immune pathways by injury severity may identify mechanistic targets for future intervention.
Burn injury is a global health concern, approximately 11 million people require medical care and 180,000 die as a result of burns annually.1 In the United States, burn injury results in approximately 486,000 care encounters and 40,000 hospitalizations annually.2 Despite recent advances in care, burn injury and the resultant immune and inflammatory responses continue to be associated with significant morbidity and mortality.3,4 Burns greater than 20% to 25% TBSA are associated with the development of burn shock and require carefully titrated fluid resuscitation.5 The majority of deaths in hospitalized burn patients occur in the acute resuscitative phase of care, following a rapid decline in clinical condition.4,6 Burn survivors experience significant morbidity related to their injuries, and growing evidence characterizes burns as chronic condition.7 In severe burns there is evidence that hypermetabolism persists at one year after injury.8 Increased infection-related morbidities among burn survivors suggests short- and long-term immune dysfunction.9 Severe burn injury puts patients at risk for negative short- and long-term clinical outcomes.
Given the link between a dysregulated systemic response to burns and detrimental outcomes, this hyperinflammatory and hypermetabolic state is a target of current research.10 Prior studies have elucidated key biochemical changes that occur after burn, but the underlying transcriptome response is not well characterized. Surviving a critical injury requires appropriate inflammatory and immune responses; signals which can be revealed through study of the whole blood transcriptome. However, transcriptomic research has revealed that maladaptive inflammatory and immune responses are associated with poor outcomes in a variety of shock and disease states.11 Transcriptome-level studies in burn injured patients have identified a “genomic storm” in patients sustaining burns >20% TBSA compared to healthy controls,11 but few differentially regulated genes when comparing >20% to >40% TBSA burns.12 This prior work established that transcriptome-level response can be appreciated early after injury. However, these studies did not investigate smaller burns and also lacked time point resolution within the first 24 hours. The goal of this work is to characterize the blood transcriptome in the acute phase following burn injury and investigate differences between patients with and without severe burns defined by a TBSA ≥20%.
METHODS
Study Population
This observational cohort study was approved by the Institutional Review Board of MedStar Health Research Institute and the Human Research Protection Offices of the Army. All patients over the age of 18 years who presented to a regional burn center within 5 hours of thermal injury due to flash, flame, or contact with anticipated need for hospital admission were screened for enrollment. Patients who arrived to the hospital >4 hours postinjury were not enrolled in this study. The admission blood draw for the study (hour 0) had to occur within 1 hour of admission to the hospital, or the patient was not enrolled. In addition, patients with a preexisting history of coagulopathy, those taking anticoagulant medications, pregnant women, chemically injured patients, children, and patients not fluent in English or Spanish were excluded. Sixty-eight individuals were included in the present analysis (Table 1). Patients were enrolled prospectively into this observational study between 2012 and 2017. Sampling and other procedures have been described in detail elsewhere.13 The current analysis is a retrospective study of these prospectively collected samples. In future work, additional patients from this cohort may be added to this analysis to allow for more strict statistical cutoffs that will hence report data with higher stringency.
Table 1.
Patient demographics and injury characteristics
| <20 pct (N = 33) | >20 pct (N = 35) | Total (N = 68) | P | |
|---|---|---|---|---|
| Age | .151 | |||
| Mean (SD) | 41.333 (19.410) | 45.853 (14.569) | 43.627 (17.145) | |
| Median (Q1, Q3) | 34.000 (24.000, 58.000) | 47.000 (38.000, 58.500) | 41.000 (30.500, 58.500) | |
| Min–max | 18.000–85.000 | 19.000–77.000 | 18.000–85.000 | |
| Missing | 0 | 1 | 1 | |
| Gender | .662 | |||
| Female (%) | 7 (21.2%) | 9 (25.7%) | 16 (23.5%) | |
| Male (%) | 26 (78.8%) | 26 (74.3%) | 52 (76.5%) | |
| % TBSA | <.001 | |||
| Mean (SD) | 10.521 (5.151) | 42.671 (22.996) | 27.069 (23.304) | |
| Median (Q1, Q3) | 11.500 (6.700, 14.500) | 34.000 (24.500, 50.250) | 20.250 (11.688, 34.500) | |
| Min–max | 1.750–18.500 | 20.000–97.000 | 1.750–97.000 | |
| Race | .774 | |||
| African American (%) | 12 (36.4%) | 16 (45.7%) | 28 (41.2%) | |
| Asian (%) | 1 (3.0%) | 1 (2.9%) | 2 (2.9%) | |
| Caucasian (%) | 16 (48.5%) | 14 (40.0%) | 30 (44.1%) | |
| Other (%) | 4 (12.1%) | 3 (8.6%) | 7 (10.3%) | |
| Unknown (%) | 0 (0.0%) | 1 (2.9%) | 1 (1.5%) | |
| ICU days | <.001 | |||
| Mean (SD) | 4.242 (6.270) | 32.000 (46.863) | 18.529 (36.449) | |
| Median (Q1, Q3) | 1.000 (0.000, 6.000) | 17.000 (5.000, 34.500) | 5.500 (1.000, 19.000) | |
| Min–max | 0.000–20.000 | 0.000–248.000 | 0.000–248.000 | |
| Ventilator days | <.001 | |||
| Mean (SD) | 1.818 (4.538) | 16.382 (27.520 | 9.209 (21.035) | |
| Median (Q1, Q3) | 0.000 (0.000, 0.000 | 3.000 (1.000, 19.500 | 1.000 (0.000, 10.000) | |
| Min–max | 0.000–20.000 | 0.000–114.000 | 0.000–114.000 | |
| Missing | 0 | 1 | 1 | |
| Mortality | .008 | |||
| Alive (%) | 32 (97.0%) | 26 (74.3%) | 58 (85.3%) | |
| Death (%) | 1 (3.0%) | 9 (25.7%) | 10 (14.7%) |
Clinical Data and Sample Collection
Patient demographic information, laboratory data, and treatment information were collected prospectively from the medical record. Blood samples were collected after admission within 5 hours of injury (hour 0) and at predetermined time points (hours 2, 4, 8, 12, and 24) during the first 24 hours in hospital. Samples were collected into PAXgene Blood RNA tubes (BD Biosciences, San Jose, CA).
Transcriptome Analysis
Total RNA was isolated using the PAXgene Blood RNA kit (Qiagen, Valencia, CA) following manufacturer directions, and global transcript levels were measured over time using the Agilent SurePrint G3 Human GE v2 8x60K Microarray Design ID: 039494 (Agilent, Santa Clara, CA) as described elsewhere.14,15
Bioinformatics Analysis
Microarray data were between- and within-array normalized using the limma package in R.16,17 The combat batch effect removal tool from the swamp package in R was used to remove an evident batch effect based on the year the arrays were processed.18,19 Limma was used to fit a linear model to each probe and calculate differential expression between patient groups using a moderated t-test. A false-discovery rate (FDR) of ≤0.1 as calculated by the Benjamini–Hochberg method was used as a significance cutoff. An FDR <0.1 resulted in too few genes to run meaningful pathway enrichment analysis. Different % TBSA cutoffs were chosen (such as 50%, 30%, and 20%), and 20% TBSA was ultimately chosen because this cutoff led to two groups which were well matched in the numbers of patients and in their demographics. In addition, 20% TBSA was chosen due to the clinical correlate for the definition of burn shock requiring formal resuscitation.
Patients were grouped by burn size (TBSA ≥20%) and mortality for differential expression analysis. This study did not include a healthy, noninjured cohort. Therefore, each patient’s data was normalized to the hour 0 time point to obtain differentially expressed genes. Therefore, for each patient, while the differentially expressed genes (DEGs) identified at the “hour 2” time point may not be 2 hours from the onset of their burn injury, it was always 2 hours from the hour 0 time point which the hour 2 sample was normalized to.
Differentially expressed probes were mapped to their respective pathways using Ingenuity Pathway Analysis (IPA).20 IPA’s Canonical Pathways and Upstream Regulator features were used to predict the differential activity of cellular pathways and regulatory networks, respectively. Volcano plots and pathway heatmaps were generated with the ggplot2 package in R. Principal components analysis was calculated using the prcomp command in base R. The prince plot was generated using the swamp package in R.18,19
Statistical Analysis
Descriptive statistics characterized the demographics and injuries of the patients by burn severity. Categorical variables were presented as frequencies and percentages and tested using the χ 2 or Fisher’s exact test. Continuous variables were expressed as means and SDs or medians and interquartile ranges (IQRs) and tested for differences between defined groups using the t-test or Mann–Whitney test as appropriate. Statistical significance was determined at the P < .05 level (two sided).
Data Availability
The raw gene expression data have been uploaded to GEO: GSE182616.
RESULTS
Demographics
Sixty-eight patients were included in the present analysis. Patient demographics are shown in Table 1. Seventy-six percent of patients were male with a median age of 41 (IQR, 30.5–58.5) years, and TBSA of 20% (IQR, 11%–34%). Thirty-five patients had % TBSA injury ≥20%, and this group experienced greater mortality (26% vs 3%, P = .008). There were no significant differences in age, race, or gender. The time from injury to the first blood draw hour 0 is less than 5 hours (mean = 2.41 hours, SD = 1.09 hours, IQR 1.48–3.19 hours, min 0.67 hours, max 4.73). The time from injury to hour 0 blood draw did not correlate with % TBSA, and therefore is not a confounding factor when assessing the two groups (Pearson’s correlation r = 0.0038).
Principal Component Analysis
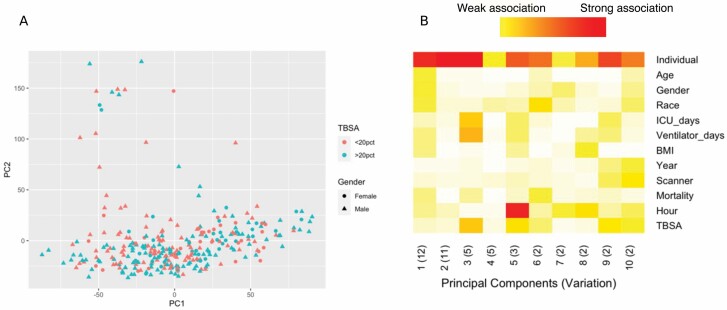
Principal component analysis indicated that samples largely cluster together without diverging based on TBSA or gender (Figure 1A). A prince plot of various clinical and technical features identified a substantial relationship between the individual and the expression of principal components but identified little else as potential confounding influencers of gene expression (Figure 1B).
Figure 1.
Unsupervised analysis of gene expression after burn injury. Principal components analysis does not segregate individuals with either similar burn severity or gender based on gene expression profiles (A). A prince plot showing the association of principal components with clinical and technical features shows little impact on gene expression by most variables but identifies variation among individuals. Red (high variance), white (low variance) (B).
Differential Expression by Burn Size
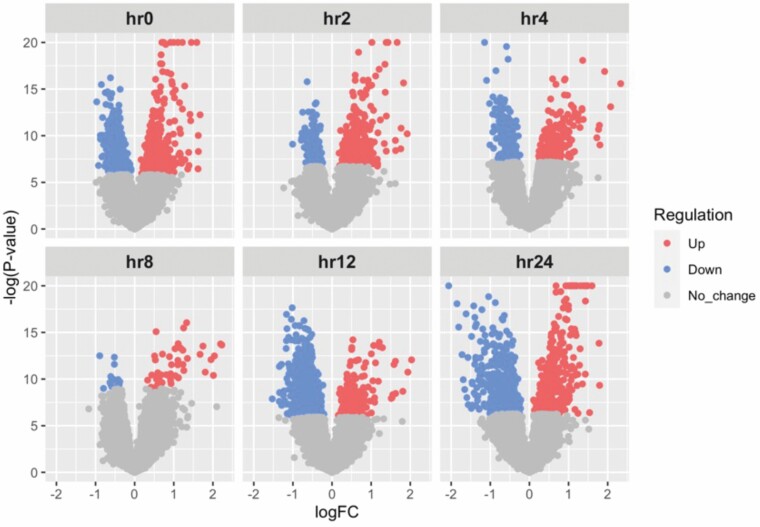
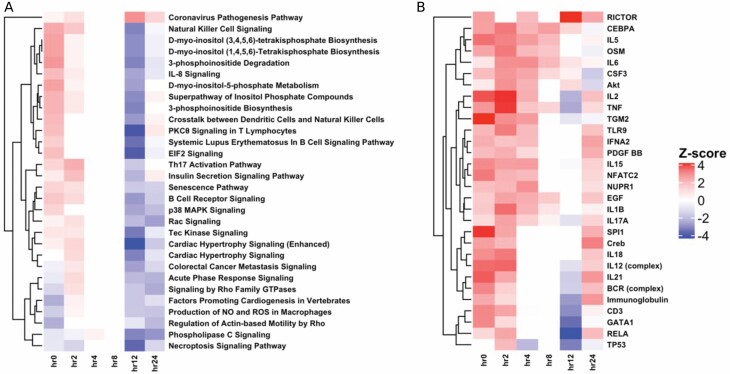
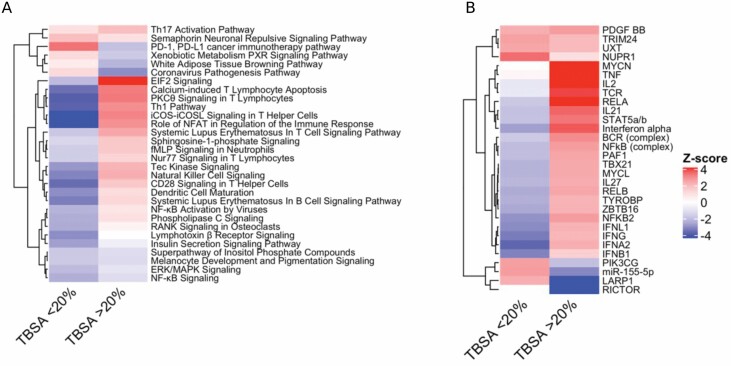
Differential expression analysis that compared patients with ≥20% TBSA to patients with <20% TBSA at each time point up to 24 hours was conducted. This analysis identified 1505, 613, 380, 63, 1357, and 954 differentially expressed genes at hours 0, 2, 4, 8, 12, and 24 respectively using an FDR threshold of significance of 0.1 (Figure 2). Using a P value significance threshold of .01, we identified 3052, 2143, 1695, 932, 2623, and 2210 differentially expressed genes at hours 0, 2, 4, 8, 12, and 24, respectively. All subsequent analysis was done using the genes that passed the FDR significance threshold (≤0.1). Pathway enrichment analysis at each time point was conducted using IPA and identified pathway trends across time points using IPA’s Comparison Analysis feature. This approach identified a modest activation (increased Z score) of several important immune pathways at early time points within the ≥20% TBSA individuals relative to the <20% TBSA individuals (Figure 3A). This activation is followed by a sharp deactivation of these same pathways starting at 12 hours. By 24 hours, differential regulation of the pathways between the TBSA groups have subsided for many pathways, but remain deactivated in several key immune regulation pathways, such as B-cell receptor signaling and IL-8 signaling pathways. In addition, several key processes of widespread cellular functions such as p38 MAPK signaling, Tec kinase signaling, and senescence pathways remained deactivated. A similar enrichment analysis was applied to common regulatory networks using IPA and identified a set of upstream regulators that are predicted to be differentially activated/suppressed at each time point within the ≥20% TBSA individuals relative to the <20% TBSA individuals (Figure 3B).
Figure 2.
Volcano plots of differential expression analysis between ≥20% TBSA and <20% TBSA patients. Using an FDR cutoff of 0.1, differential expression analysis identified 1505, 613, 380, 63, 1357, and 954 differentially regulated genes at hours 0, 2, 4, 8, 12, and 24 respectively. FDR, false-discovery rate.
Figure 3.
Pathways and regulators differentially regulated among </≥20% TBSA patients. Enriched canonical biological pathways identify a mild activation of immune pathways at hours 0 and 2 leading to a shutdown of many immune pathways by hour 12 (A). Top upstream regulators of many immune pathways show similar activation patterns at hour 0 leading to suppression by hour 12 (B).
Gene Expression Network
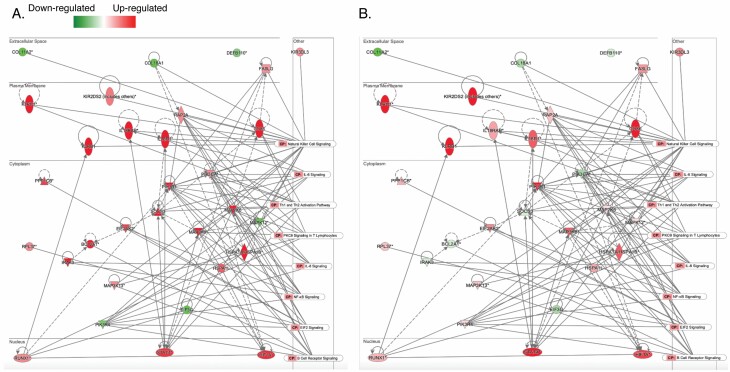
To better visualize the relationships between the genes driving the early immune activation observed in Figure 3B, we constructed a network from the significant genes in natural killer cell signaling, IL-6 signaling, Th1 and Th2 activation pathway, protein kinase C (PKC) θ signaling in T lymphocytes, IL-8 signaling, NF-kB signaling, EIF2 signaling, and B-cell receptor signaling and overlaid the network with the gene expression at hour 0 (Figure 4A) and hour 24 (Figure 4B). Many of the genes that are highly up-regulated at hour 0 are either up-regulated at a lower magnitude (including IL18RAP, IL18R1, HSPAL, MAP2K6, and PPP1CB) or are down-regulated at hour 24 (including IRAK3, BCL2A1, SOCS3, PIK3CA, and MAPK12).
Figure 4.
Network of selected immune pathways at 0 hours after injury (A) and 24 hours after injury (B). Differential expression analysis between ≥20% TBSA and <20% TBSA individuals shows reduced activation and/or suppression of key immune regulating molecules from 0 to 24 hours. Down-regulation and up-regulation are colored in red and green, respectively. Darker color means more difference between ≥20% and <20% TBSA individuals. * represents multiple probes of an individual gene were identified.
Differential Expression by Mortality
Pathway enrichment analysis for all samples was conducted using IPA and identified pathway trends between patients based on mortality using IPA’s Comparison Analysis feature. This approach identified a pattern of relative activation (increased Z score) in several immune pathways in the acute phase of injury within the ≥20% TBSA individuals who died relative to those who lived (Figure 5A). There was a different pattern observed in patients with <20% TBSA, where many of these immune pathways were inactivated (decreased Z score) in the individual who died (Figure 5B).
Figure 5.
Pathways and regulators differentially regulated among patients grouped by mortality. In patients with ≥20% TBSA who died, there was activation of most pathways, while the opposite trend was observed in the <20% group (A). Top upstream regulators showed the same pattern of expression (B).
PKC θ in T Lymphocytes Signaling Pathway
The PKC θ in T lymphocytes pathway was originally activated in the patient group with ≥20% TBSA compared to the <20% TBSA group with a Z score = 1.89 at hour 0. At hours 2, 4, and 8, the activation of this pathway evened out between the groups. At hour 12, there was significant deactivation of this pathway with a Z score = −3.873. This pathway had the largest negative activation Z score of any of the identified pathways at any time point, suggesting that it was the most severely suppressed. Differentially regulated genes within this pathway include calcium voltage-gated channel subunit alpha1 E (CACNA1E), caspase recruitment domain family member 11 (CARD11), CD86 molecule (CD86), Fos proto-oncogene, AP-1 transcription factor subunit (FOS), FYN proto-oncogene, Src family tyrosine kinase (FYN), mitogen-activated protein kinase kinase kinase 6 (MAP3K6), nuclear factor kappa B subunit 1 (NFKB1), phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), and RAS-like proto-oncogene B (RALB).
Necroptosis Signaling Pathway
The necroptosis signaling pathway was shown to be deactivated at hour 0 and remained deactivated throughout the 24-hour time course with a deactivation peak at hour 12 (Z score = −3.464). Differentially expressed genes within this pathway include Cytochrome B-245 Beta Chain (CYBB), Peptidyl-prolyl cis-trans isomerase (FKBP1A), Glutamate-Ammonia Ligase (GLUL), Interferon Alpha and Beta Receptor Subunit 1 (IFNAR1), Pellino E3 Ubiquitin Protein Ligase 1 (PELI1), Peptidylprolyl Isomerase D (PPID), Toll Like Receptor Adaptor Molecule 1 (TICAM1), tumor necrosis factor (TNF), Translocator Protein (TSPO), and Z-DNA Binding Protein 1 (ZBP1).
DISCUSSION
This study aims to characterize longitudinal changes in the blood transcriptome during the first 24 hours after burn injury and compare the transcriptome response between patients with and without severe burns. These data show that severe burn injury is associated with a high number of differentially expressed genes as early as hospital admission (within 4 hours of injury), and that this gene expression changes dynamically during the first 24 hours. The greatest number of differentially regulated genes among patients with TBSA ≥20% were appreciated on admission and at hour 12. Pathway analysis reveals an initial up-regulation in several immune–inflammatory pathways immediately after injury and subsequent shutdown, or relative down-regulation by hours 12 to 24. This pattern suggests an initial systemic inflammatory response followed by a compensatory anti-inflammatory response that occurs rapidly in the setting of severe burns. Furthermore, among patients with severe burns, those who die show a pattern of relative activation in immune–inflammatory pathways, suggesting a more intense systemic response to injury.
The immune system is generally categorized into innate and adaptive immunity. The innate, or humoral immune response is the body’s first line of defense, while the adaptive, or cellular immune response is slower.21 The host immune response to burns, trauma, and myriad disease states is a topic of ongoing research. Despite different inciting factors, common patterns have been observed.11 The general pattern is initial systemic inflammation mediated by the innate immune system, followed by a compensatory anti-inflammatory response evidenced by suppression of the adaptive immune system.22,23 Burn injury is associated with an intense and sustained cytokine response.24 Late phase care complications often include infection.25,26 Burn injury is increasingly characterized as a chronic condition and patients are known to have long-term immune suppression.7,27,28 The data presented here suggest that dysregulated immune function in patients with severe burns may begin as early as the first 24 hours after injury.
Among the enriched pathways, the PKC θ signaling pathway in activation of IL-2 is particularly important. T cells play an essential role in the adaptive immune response and failure of T-cell-regulated immune responses can lead to wound infection and ultimate septic complications.29 The main receptor on T cells, T-cell receptor gets activated through binding of a ligand from antigen presenting cells, such as macrophages. Subsequent phosphorylation of PKC θ leads to nuclear translocation of NF-kB which acts as a transcription factor for IL-2. Hence, the PKC θ signaling pathway plays an important role in the body’s response to infection through activation of IL-2, an inflammatory cytokine.30 The data presented here show that T cells in patients with severe burns have deactivation of this important immune pathway suggesting their ability to combat infection is altered compared to those patients with smaller burns. If this activation pathway can be reactivated through a pharmacotherapy or genetic alteration, burn patient response to infection may be able to be altered.
Indeed, Hur et al have demonstrated decreased levels of IL-2 in serum of patients with severe burns (average TBSA = 50.4%) at day 1 postinjury compared to health controls (13.2 vs 24.08 pg/ml burn vs control). They also showed that IL-2 cytokine expression in the serum had a strong correlation to many other interleukins that are secreted by T helper cells (IFN-g, IL-4, IL-7, IL-12p70, and IL-17) suggesting the important pleiotropic effects of this signaling pathway. This analysis aids in providing mechanistic insight into the derangements to cytokine levels that are known to be apparent in burn patients. There are also conflicting reports that show increases to serum IL-2 levels postburn in mouse models31 and in patients,32–34 however, these studies report variable % TBSA, and time postburn. In addition, few studies compared burn patient serums levels to healthy cohorts. Our analysis contributes to the greater picture in that we provide granular time-point-specific data from which timely interventions can be made.
Furthermore, the necroptosis signaling pathway is deactivated across all time points. Conventionally, cellular necrosis is termed “un-programmed cell death,” while apoptosis is termed “programmed cell death.” Necroptosis is a more newly defined type of cellular necrosis that goes through a programmed cell death that is distinct from apoptosis and is caspase independent. Necroptosis has a connection to the immune system and is often the preferred type of cell death for evading pathogens.35 It also has connections to chronic inflammatory diseases such as Crohn’s disease and pancreatitis. Necroptosis is classically activated through TNF-α ligand binding to TNF receptor. Under caspase-8-independent conditions, this ligand/receptor binding leads to the recruitment of receptor-interacting protein kinase 3 (RIPK3) to RIPK1 (receptor-interacting protein kinase 1) which forms a heterodimer complex that phosphorylates mixed lineage kinase domain-like (MLKL). MLKL is then oligomerized and translocated to cell membranes where it forms pores that cause cell plasma membrane destruction and ultimate cell death and secretion of intracellular waste.36 The role of necroptosis in burn injury has not been thoroughly examined, despite the potential connection between burn injury mechanisms and cellular death. It has been studied in a rat comb burn model where it was hypothesized that burn progression proceeds down a necroptosis signaling pathway and that treatment with necrostatin-1 (an inhibitor of RIPK1 kinase) may rescue this cell death.37 However, there was no evidence of necrostatin-1’s ability to change burn progression outcomes. In addition, RIPK3 was shown to be present in normal and burned skin. Although relevant to the necroptosis signaling pathway, this prior study evaluated necroptosis in the local skin burn wound environment, as opposed to our study examining the systemic response. In another study by Idrovo et al, a 15% TBSA mouse burn model was conducted and an increased expression of RIPK1 and 3 was observed in livers of burned mice, suggesting that hepatic inflammation is associated with necroptosis signaling.38 The inactivation of necroptosis signaling pathways throughout the first 24 hours postburn in the severely injured patients could suggest that cellular death occurs through apoptotic or necrotic pathways, or that the ability of cells to undergo programmed cell death is altered. These altered cellular responses may lead to downstream issues with response to pathogens that causes the severe immune dysregulation in patients. If these signaling pathways can be altered, perhaps these downstream effects can be mitigated such as has been suggested in ischemic brain injury, immune system disorders, and cancer.35
Prior work has established that severe burns are associated with differential gene expression when compared to healthy controls, and this “genomic storm” is similar in both blunt trauma and endotoxemia and can be appreciated early.11 Surviving severe injury requires an appropriate immunoinflammatory response but in many cases the host response can be maladaptive. From these data, a pattern of persistent or relatively increased activation in immunoinflammatory pathways is identified in patients with severe burn who die. The development of burn shock after severe burn injury leads to significant morbidity and mortality. Disordered systemic inflammation and immunomodulation likely contributes to the severity of burn shock and the poor short- and long-term clinical outcomes experienced by these patients. Further characterization of the differential transcriptome response induced by severe burns may help identify potential treatment targets.
Potential limitations of this study include the time from the injury to the hour 0 blood collection and blood cell composition. Although the first blood draw occurred within five hours of injury, there is a small difference between patients limited by the time required for transport, which can be large in our regional catchment area. Future studies will attempt to coordinate EMS blood draws in the field to obtain true “hour 0” samples, or will normalize to un-injured healthy control patients allowing us to shift each time point to the actual time postburn, which will improve the time point resolution of future studies and will eliminate potential confounding patients who are actually further along in their postburn period than others.
Prior research has highlighted the differential physiologic, biochemical, and underlying cellular responses in the elderly burn population compared to their younger counterparts. It is hypothesized that significant down-regulation of the immune response and a dysregulated inflammatory response contributes to the increased morbidity and mortality observed clinically in the elderly burn population.39,40 In our study, we did not appreciate clustering in gene expression associated with age. However, this is likely due to sample size, considering that in the present study, there were only four patients above the age of 70, three of which suffered a % TBSA burn >20%.
The RNA used to run microarrays was extracted using whole blood. Different cell composition may be a confounder to the gene expression data. For the limited samples having complete blood count hospital labs drawn within a similar timeframe (±30 minutes) of the study-related blood draw (43 samples), there is no significant in any of the cell compositions tested. The difference between % TBSA ≥20 and <20 subjects for lymphocyte counts as an example was not significant (t-test, P = .682). Comparing with first 10 principal components of the gene expression identified in Figure 1, there was no significant association with lymphocyte count was identified. During the sample collection phase of this experiment, peripheral blood mononuclear cells were isolated using a Ficoll separation and stored in Trizol for future analysis as described in detail elsewhere.13 In future studies, these samples may be used to further elucidate the effect of cell composition on transcriptomics postburn. In addition, blood plasma was stored for future analysis and should be used for further validation of our findings beyond the transcript level. No confirmatory analyses were performed at the mRNA level or protein level in the current analysis.
CONCLUSION
Severe burn injury is associated with an early proinflammatory immune response followed by shutdown of these pathways. Burn patients who die show relative activation of genes in the first 24 hours after injury in several proinflammatory pathways compared to those who live. Examination of the inflammatory response to burn injury and differentially regulated genes and immune pathways by injury severity and mortality may identify mechanistic targets for future intervention.
SYSCOT STUDY GROUP AUTHORSHIP
Melissa M. McLawhorn, RN BSN, Rachael A. Callcut, MD, MSPH, Mitchell J. Cohen, MD, Linda R. Petzold, PhD, Jeffrey D. Varner, PhD, Maria Cristina Bravo, PhD, Kathleen E. Brummel-Ziedins, PhD, Kalev Freeman, MD, PhD, Kenneth G. Mann, PhD, Thomas Orfeo, PhD, and Anthony E. Pusateri, PhD.
ACKNOWLEDGMENTS
Seshamalini Srinivasan for her contribution to sample processing and initial data analysis. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
Funding: This work was conducted under the Systems Biology Coagulopathy of Trauma (SYSCOT) Research Program of the US Army Medical Research and Development Command and the Defense Health Program. Funding was provided under contracts, W911QY-15-C-0025 and W911NF-10-1-0459.
Conflict of interest statement. None declared.
Contributor Information
John W Keyloun, Department of Surgery, MedStar Georgetown University Hospital, Washington, District of Columbia, USA; Firefighters’ Burn and Surgical Research Laboratory, MedStar Health Research Institute, Washington, District of Columbia, USA.
Ross Campbell, The Geneva Foundation, Silver Spring, Maryland, USA; Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Bonnie C Carney, Firefighters’ Burn and Surgical Research Laboratory, MedStar Health Research Institute, Washington, District of Columbia, USA; Department of Surgery, Georgetown University School of Medicine, Washington, District of Columbia, USA; Department of Biochemistry, Georgetown University School of Medicine, Washington, District of Columbia, USA.
Ruoting Yang, Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Stacy-Ann Miller, Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA; Oak Ridge Institute for Science and Education, Silver Spring, Maryland, USA.
Leanne Detwiler, The Geneva Foundation, Silver Spring, Maryland, USA; Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Aarti Gautam, Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Lauren T Moffatt, Firefighters’ Burn and Surgical Research Laboratory, MedStar Health Research Institute, Washington, District of Columbia, USA; Department of Surgery, Georgetown University School of Medicine, Washington, District of Columbia, USA; Department of Biochemistry, Georgetown University School of Medicine, Washington, District of Columbia, USA.
Rasha Hammamieh, Medical Readiness Systems Biology, Center for Military Psychiatry and Neuroscience, Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Marti Jett, Headquarters Walter Reed Army Institute of Research, Silver Spring, Maryland, USA.
Jeffrey W Shupp, Firefighters’ Burn and Surgical Research Laboratory, MedStar Health Research Institute, Washington, District of Columbia, USA; Department of Surgery, Georgetown University School of Medicine, Washington, District of Columbia, USA; Department of Biochemistry, Georgetown University School of Medicine, Washington, District of Columbia, USA.
SYSCOT study group:
Melissa M McLawhorn, Rachael A Callcut, Mitchell J Cohen, Linda R Petzold, Jeffrey D Varner, Maria Cristina Bravo, Kathleen E Brummel-Ziedins, Kalev Freeman, Kenneth G Mann, Thomas Orfeo, and Anthony E Pusateri
REFERENCES
- 1. Burns. World Health Organization [17 Dec. 2020]; available from https://www.who.int/news-room/fact-sheets/detail/burns. Date accessed 17 December 2020.
- 2. Burn Incidence Fact Sheet. American Burn Association [updated 8 May 2017; 17 Dec. 2020]; available from https://ameriburn.org/who-we-are/media/burn-incidence-fact-sheet/. Date accessed 17 December 2020.
- 3. Jeschke MG, van Baar ME, Choudhry MA, Chung KK, Gibran NS, Logsetty S. Burn injury. Nat Rev Dis Primers 2020;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rae L, Fidler P, Gibran N. The physiologic basis of burn shock and the need for aggressive fluid resuscitation. Crit Care Clin 2016;32:491–505. [DOI] [PubMed] [Google Scholar]
- 5. Pham TN, Cancio LC, Gibran NS. American Burn Association practice guidelines burn shock resuscitation. J Burn Care Res 2008;29:257–66. [DOI] [PubMed] [Google Scholar]
- 6. Swanson JW, Otto AM, Gibran NSet al. Trajectories to death in patients with burn injury. J Trauma Acute Care Surg 2013;74:282–8. [DOI] [PubMed] [Google Scholar]
- 7. Barrett LW, Fear VS, Waithman JC, Wood FM, Fear MW. Understanding acute burn injury as a chronic disease. Burns Trauma 2019;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herndon DN, Tompkins RG. Support of the metabolic response to burn injury. Lancet 2004;363:1895–902. [DOI] [PubMed] [Google Scholar]
- 9. Duke JM, Randall SM, Wood FM, Boyd JH, Fear MW. Burns and long-term infectious disease morbidity: a population-based study. Burns 2017;43:273–81. [DOI] [PubMed] [Google Scholar]
- 10. Jeschke MG, Gauglitz GG, Kulp GAet al. Long-term persistance of the pathophysiologic response to severe burn injury. PLoS One 2011;6:e21245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiao W, Mindrinos MN, Seok Jet al. ; Inflammation and Host Response to Injury Large-Scale Collaborative Research Program . A genomic storm in critically injured humans. J Exp Med 2011;208:2581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sood RF, Gibran NS, Arnoldo BD, Gamelli RL, Herndon DN, Tompkins RG; Inflammation the Host Response to Injury Investigators . Early leukocyte gene expression associated with age, burn size, and inhalation injury in severely burned adults. J Trauma Acute Care Surg 2016;80:250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shupp JW, Brummel-Ziedins KE, Cohen MJet al. Assessment of coagulation homeostasis in blunt, penetrating, and thermal trauma: guidance for a multicenter systems biology approach. Shock 2019;52(1S Suppl 1):84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donohue DE, Gautam A, Miller SAet al. Gene expression profiling of whole blood: a comparative assessment of RNA-stabilizing collection methods. PLoS One 2019;14:e0223065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gautam A, Donohue D, Hoke Aet al. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS One 2019;14:e0225137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ritchie ME, Phipson B, Wu Det al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ritchie ME, Silver J, Oshlack Aet al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007;23:2700–7. [DOI] [PubMed] [Google Scholar]
- 18. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 19. Lauss M. swamp: visualization, analysis and adjustment of high-dimensional data in respect to sample annotations. R package version 1.5.1. https://CRAN.R-project.org/package=swamp2019. Date accessed 17 December 2020.
- 20. Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller AC, Rashid RM, Elamin EM. The “T” in trauma: the helper T-cell response and the role of immunomodulation in trauma and burn patients. J Trauma 2007;631407–17. [DOI] [PubMed] [Google Scholar]
- 22. Shen H, de Almeida PE, Kang KH, Yao P, Chan CW. Burn injury triggered dysfunction in dendritic cell response to TLR9 activation and resulted in skewed T cell functions. PLoS One 2012;7:e50238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valvis SM, Waithman J, Wood FM, Fear MW, Fear VS. The immune response to skin trauma is dependent on the etiology of injury in a mouse model of burn and excision. J Invest Dermatol 2015;135:2119–28. [DOI] [PubMed] [Google Scholar]
- 24. Matsuura H, Matsumoto H, Osuka Aet al. Clinical importance of a cytokine network in major burns. Shock 2019;51:185–93. [DOI] [PubMed] [Google Scholar]
- 25. Church D, Elsayed S, Reid O, Winston B, Lindsay R. Burn wound infections. Clin Microbiol Rev 2006;19:403–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lachiewicz AM, Hauck CG, Weber DJ, Cairns BA, van Duin D. Bacterial infections after burn injuries: impact of multidrug resistance. Clin Infect Dis 2017;65:2130–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson BZ, McAlister S, McGuire HMet al. Corrigendum: Pediatric burn survivors have long-term immune dysfunction with diminished vaccine response. Front Immunol 2020;11:598646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parment K, Zetterberg A, Ernerudh J, Bakteman K, Steinwall I, Sjoberg F. Long-term immunosuppression in burned patients assessed by in vitro neutrophil oxidative burst (Phagoburst). Burns 2007;33:865–71. [DOI] [PubMed] [Google Scholar]
- 29. Buchanan IB, Maile R, Frelinger JA, Fair JH, Meyer AA, Cairns BA. The effect of burn injury on CD8+ and CD4+ T cells in an irradiation model of homeostatic proliferation. J Trauma 2006;61:1062–8. [DOI] [PubMed] [Google Scholar]
- 30. Hayashi K, Altman A. Protein kinase C theta (PKCtheta): a key player in T cell life and death. Pharmacol Res 2007;55:537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Finnerty CC, Przkora R, Herndon DN, Jeschke MG. Cytokine expression profile over time in burned mice. Cytokine 2009;45:20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finnerty CC, Herndon DN, Przkora Ret al. Cytokine expression profile over time in severely burned pediatric patients. Shock 2006;26:13–9. [DOI] [PubMed] [Google Scholar]
- 33. Teodorczyk-Injeyan JA, Sparkes BG, Mills GB, Peters WJ. Immunosuppression follows systemic T lymphocyte activation in the burn patient. Clin Exp Immunol 1991;85:515–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teodorczyk-Injeyan JA, Sparkes BG, Lalani S, Peters WJ, Mills GB. IL-2 regulation of soluble IL-2 receptor levels following thermal injury. Clin Exp Immunol 1992;90:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Place DE, Kanneganti TD. The innate immune system and cell death in autoinflammatory and autoimmune disease. Curr Opin Immunol 2020;67:95–105. [DOI] [PubMed] [Google Scholar]
- 36. Dhuriya YK, Sharma D. Necroptosis: a regulated inflammatory mode of cell death. J Neuroinflammation 2018;15:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reddy AS, Abraham A, McClain SAet al. The role of necroptosis in burn injury progression in a rat comb burn model. Acad Emerg Med 2015;22:1181–6. [DOI] [PubMed] [Google Scholar]
- 38. Idrovo JP, Boe DM, Kaahui S, Yang WL, Kovacs EJ. Hepatic inflammation after burn injury is associated with necroptotic cell death signaling. J Trauma Acute Care Surg 2020;89:768–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dreckmann SC, Amini-Nik S, Tompkins RG, Vojvodic M, Jeschke MG. Genome-wide comparisons of gene expression in adult versus elderly burn patients. PLoS One 2019;14:e0226425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jeschke MG, Patsouris D, Stanojcic Met al. Pathophysiologic response to burns in the elderly. EBioMedicine 2015;2:1536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw gene expression data have been uploaded to GEO: GSE182616.