Abstract
The development of checkpoint blockade immunotherapy has transformed the medical oncology armamentarium. But despite its favorable impact on clinical outcomes, immunotherapy benefits only a subset of patients, and a substantial proportion of these individuals eventually manifest resistance. Serious immune-related adverse events and hyperprogression have also been reported. It is therefore essential to understand the molecular mechanisms and identify the drivers of therapeutic response and resistance. In this review, we provide an overview of the current and emerging clinically relevant genomic biomarkers implicated in checkpoint blockade outcome. US Food and Drug Administration–approved molecular biomarkers of immunotherapy response include mismatch repair deficiency and/or microsatelliteinstability and tumor mutational burden of at least 10 mutations/megabase. Investigational genomic-associated biomarkers for immunotherapy response include alterations of the following genes/associated pathways: chromatin remodeling (ARID1A, PBRM1, SMARCA4, SMARCB1, BAP1), major histocompatibility complex, specific (eg, ultraviolet, APOBEC) mutational signatures, T-cell receptor repertoire, PDL1, POLE/POLD1, and neo-antigens produced by the mutanome, those potentially associated with resistance include β2-microglobulin, EGFR, Keap1, JAK1/JAK2/interferon-gamma signaling, MDM2, PTEN, STK11, and Wnt/Beta-catenin pathway alterations. Prospective clinical trials are needed to assess the role of a composite of these biomarkers to optimize the implementation of precision immunotherapy in patient care.
Immunotherapies such as checkpoint inhibitors have demonstrated durable responses in selected patients with diverse tumor types. However, only about 20% of patients respond to immune checkpoint blockade, and a clinically significant proportion of patients who derive benefit from this treatment eventually develop resistance (1). Additionally, serious immune-related adverse events have been reported in a clinically significant proportion of patients who receive checkpoint inhibitors, and others have experienced accelerated disease progression (known as hyperprogression) (2-4). It is therefore essential to identify robust biomarkers of immune checkpoint blockade efficacy to select the optimal treatment for each patient.
Programmed cell death-ligand 1 (PD-L1) expression on tumor or immune cells was the first US Food and Drug Administration (FDA)–approved immune-related biomarker, predicting response to checkpoint blockade in diverse tumor types, including non-small cell lung cancer (NSCLC), melanoma, urothelial cancer, renal cell cancer, and triple-negative breast cancer (5). However, patients with PD-L1–negative tumors often respond to immunotherapy, and therefore PD-L1 expression as a solitary biomarker for checkpoint blockade benefit is suboptimal.
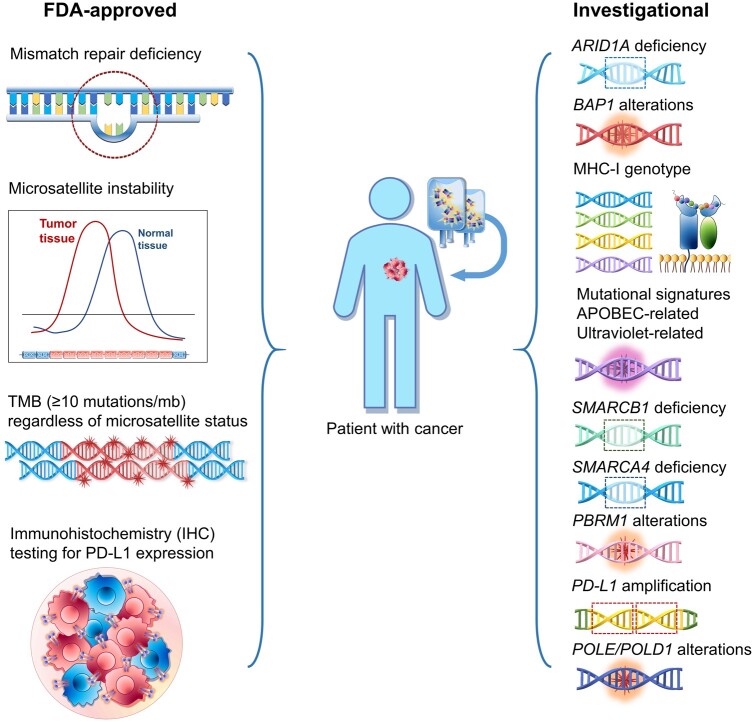
Deficient mismatch repair (dMMR)/microsatellite instability (MSI) (6,7) and intermediate-to-high tumor mutational burden (TMB) (>10 mutations/megabase [mut/mb]) (8,9) predict salutary effects from checkpoint inhibitors, regardless of tumor of origin. Recently, the FDA approved these genomic alterations as pan-solid cancer immunotherapy response predictors. Current efforts and this review (Table 1) (6,8,10,12-38,40-46,48-52) are focused on the rapidly evolving field evaluating novel genomic immunotherapy biomarkers in diverse tumor types.
Table 1.
Genomic biomarkers for response or resistance to checkpoint blockadea
| Genomic biomarker | Cancer type | Role | Predictive value and comments | Selected references |
|---|---|---|---|---|
| FDA-approved biomarkers associated with response | ||||
| Deficient mismatch repair/microsatellite instability | Across solid tumor types, adult and pediatric |
|
|
|
| TMB (>10 mutations/mb) regardless of microsatellite status | Across solid tumor types, adult and pediatric | Increased mutations/neo-antigens |
|
|
| Reported/Investigational response alterations | ||||
| Chromatin remodeling (SWI/SNF complex) | ||||
| ARID1A | Across solid tumors (eg, ovarian clear cell, endometrial, and gastric) | SWI/SNF chromatin remodeling | ARID1A deficiency leads to impaired MMR function | |
| PBRM1 | Clear cell renal cancer | SWI/SNF chromatin remodeling | Contradictory data; some papers suggest that PBRM1 alterations predict response to immunotherapy, others do not | |
| SMARCA4 | Driver in small cell ovarian cancer with hypercalcemia (found in uterine and thoracic sarcomas [undifferentiated], NSCLC, bladder, colorectal) | SWI/SNF chromatin remodeling |
|
|
| SMARCB1 | Rhabdoid tumors | SWI/SNF chromatin remodeling | Preliminary: SMARCB1 loss in rhabdoid tumors may correlate with immunotherapy response | Bakouny, et al. 2020 (24) |
| Other alterations | ||||
| BAP1 alterations | Mesothelioma | Promotes immune inflammatory environment in mesothelioma |
|
|
| Major histocompatibility complex class-I (MHC-I) genotype | Across solid tumors | Efficient presentation of driver neoantigens to CD8+ T cells |
|
Goodman, et al. 2020 (28) |
| Mutational signatures APOBEC-related ultraviolet-related | Across solid tumors | Associated with high immunogenicity |
|
|
| Mutational signatures ultraviolet-related | Across solid tumors | Associated with high immunogenicity |
Predicts response in patients with low TMB
Requires validation |
Pham, et al. 2019 (30) |
| PD-L1 amplification | Across solid tumors (and in Hodgkin lymphoma) | PD-L1 ligand is important in the immune checkpoint machinery |
|
Goodman, et al. 2018 (31) |
| POLE/POLD1 | Across solid tumors | High tumor mutational rates, high TIL rates, and increased expression of cytotoxic T-cell markers | Predicts response | |
| Biomarkers associated with resistance/Hyperprogression | ||||
| Beta-2 microglobulin mutations | Melanoma | Defects in antigen presentation, escape of immune recognition |
|
|
| EGFR alterations | Across tumor types | Unclear |
|
|
| KEAP1 mutations | NSCLC | Associated with “cold” tumor microenvironment | Not clear if KEAP1 alterations are predictive or prognostic | |
| JAK1/2 loss | Across tumor types (melanoma, colorectal) | Defects in interferon-receptor signaling pathways |
|
|
| MDM2 amplification | Melanoma | Unclear |
|
|
| PTEN loss | Melanoma | Upregulation of immunosuppressive cytokines; may decrease CD8+ T cell infiltration |
|
|
| STK11 mutations with KRAS alterations | Lung | Altered cytokines/chemokines, metabolic restriction of T cells, impaired antigenicity | Not clear if STK11 alterations are predictive or prognostic | |
| Wnt/Beta-catenin pathway alterations | Melanoma, colon cancer | Decreases T-cell infiltration |
|
BAP1 = BRCA1-associated protein 1; EGFR = epidermal growth factor receptor; FDA = US Food and Drug Administration; JAK = Janus kinase; KEAP1 = Kelch-like ECH associated protein 1; mb = megabase; MDM2 = murine double minute 2; MHC-I = major histocompatibility complex class-I; MMR = mismatch repair; NSCLC = non-small cell lung cancer; PBRM1 = polybromo-1; PD-1 = programmed cell death-1; PD-L1 = programmed cell death-ligand 1; POLE = DNA polymerase epsilon; PTEN = phosphatase and TENsin homolog deleted on chromosome 10; SMARCB = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B; STK11 = serine/threonine kinase 11; TIL = tumor infiltrative lymphocyte; TMB = tumor mutational burden.
FDA-Approved Biomarkers
On May 23, 2017, the FDA granted accelerated approval to pembrolizumab for dMMR/MSI diverse solid cancers (Table 1). This was a hallmark approval because, for the first time, a regulatory agency approved an immunotherapy drug based on a genomic marker and, importantly, that approval was tissue agnostic. On June 16, 2020, the FDA gave a second similar (histology-agnostic) approval to pembrolizumab. This time, the approval was for solid tumors harboring TMB of at least 10 mut/mb. It is important to note that the vast majority of tumors with dMMR/MSI have a high TMB, but most tumors with a high TMB are not MSI. Therefore, it is probable that high TMB mediates the responsiveness to checkpoint blockade, perhaps by resulting in a high number of mutanome-derived neo-antigens, hence increasing the chances that one or more of these neo-antigens will be immunogenic and trigger T-cell activation, leading to eradication of the cancer cells bearing the immunogenic neo-antigens.
dMMR and Microsatellite Instability
MMR proteins control the excision of DNA mismatches introduced by DNA polymerase during cell division; these mismatches commonly occur in repetitive DNA sequences known as microsatellites. Impairment of the MMR system leads to MSI, which is characterized by the accumulation of mismatches in repeated sequences. Defects in the MMR system can be assessed using polymerase chain reaction–based assays that test for MSI or immunohistochemical (IHC) analyses of the expression of MMR proteins, including MLH1, MSH2, MSH6, and PMS2. Both tests are approved for selecting patients for treatment. MSI-high (MSI-H) or dMMR tumors can arise because of somatic or epigenetic MMR gene alterations (53,54) or to Lynch syndrome-associated germline MMR gene mutations (55-57).
The genomic analysis of more than 15 000 cancers (>50 types of malignancy) revealed MSI-H in 2.2% of specimens of which 16% had a germline mutation in an MMR gene, consistent with Lynch syndrome (58-60). MSI-H has been identified in 28% of patients with endometrial cancer and in 19% of patients with colon cancer (58,59). It is also reported in diverse tumor types (not classically considered in Lynch syndrome), including gastric (22%), adrenal cortical (5%), and esophageal (3%) carcinomas (58).
Several trials have shown statistically significant responses to anti-programmed cell death-1 (PD-1) agents in heavily pretreated patients with MSI-H/dMMR in diverse tumor types, leading to FDA approval of MSI/dMMR as a predictive biomarker for immune checkpoint blockade. Notably, in May 2017, pembrolizumab received the first tissue and site-agnosticFDA approval for the treatment of adult and pediatric patients with unresectable or metastatic MSI-H or dMMR solid tumors that had progressed after prior treatment (7). Five single-arm trials evaluated the use of pembrolizumab in patients with dMMR or MSI-H tumors; the overall response rate (ORR) was 39.6% (complete response [CR] = 7%), with a large subgroup of responses being durable (6,10,61,62). Additionally, pembrolizumab was approved for the treatment of unresectable or metastatic MSI-H or dMMR colorectal cancer (CRC) that had progressed after treatment with a fluoropyrimidine, oxaliplatin, and irinotecan (63). In June 2020, pembrolizumab was further approved for the first-line treatment of patients with advanced MSI-H or dMMR CRC, based on the results of a multicenter, open-label, randomized trial (KEYNOTE-177) (64). Patients who received treatment with pembrolizumab had longer median progression-free survival (PFS) compared with patients who received standard chemotherapy (16.5 vs 8.2 months; hazard ratio [HR] = 0.60, 95% confidence interval [CI] = 0.45 to 0.80, P = .0002). The ORR was 43.8% with pembrolizumab vs 33.1% with chemotherapy, whereas median duration of response was not reached with pembrolizumab vs 10.6 months with chemotherapy (64).
Nivolumab, another anti-PD-1 agent, was also granted FDA approval as a monotherapy and in combination with ipilimumab for the treatment of patients 12 years of age or older with MSI-H or dMMR metastatic CRC after prior treatment with a fluoropyrimidine, oxaliplatin, and irinotecan (65,66). Both approvals were based on the results of CHECKMATE-142, a nonrandomized phase II trial evaluating nivolumab with or without other anti-cancer drugs. Patients with MSI-H/dMMR CRC after 1 or more lines of treatment received nivolumab until disease progression, death, or unacceptable toxicity (50). Among the 74 patients studied, the ORR was 31%, and the median depth of response was not reached. In the combination arm, patients received nivolumab every 2 weeks in combination with low-dose ipilimumab (1 mg/kg) every 6 weeks. The ORR was 55%, and the median duration of response was not reached; the 12-month PFS rate was 71% (11). In an updated analysis, the ORR was 60% (95% CI = 44.3 to 74.3); median PFS and overall survival (OS) were not reached (12). These data demonstrate that MSI-H/dMMR is a powerful marker for response to pembrolizumab and nivolumab in diverse tumor types.
TMB of at Least 10 Mut/mb
TMB is defined as the number of somatic synonymous and nonsynonymous mutations (base substitutions and indels) per megabase of analyzed DNA. Nonsynonymous molecular alterations lead to transcription of abnormal proteins, which are recognized by the immune system as neoantigens and render the tumor highly immunogenic. In June 2020, the FDA granted accelerated approval to pembrolizumab for the treatment of patients with advanced solid tumors with intermediate to high TMB (ie, ≥10 mut/mb) whose disease had progressed on prior treatment and who had no satisfactory alternative treatment options (9). Additionally, the FoundationOneCDx assay was also approved by the FDA as a companion diagnostic test for pembrolizumab. This approval was based on the results of KEYNOTE-158, a nonrandomized clinical trial of patients with diverse solid tumor types who received treatment with pembrolizumab until disease progression or unacceptable toxicity (67). The trial aimed to evaluate predictive biomarkers through a prospectively planned retrospective analysis of 10 cohorts of patients. Among 102 patients with tumors with TMB of at least 10 mut/mb, the ORR (primary endpoint) was 29% (CR = 4%), and about half of the responses were durable at 2 years (67).
An analysis of 104 814 tumor tissue samples from the Foundation Medicine database showed that TMB of at least 10 mut/mb was identified in about 13% of all solid tumors (68-70). Other investigators retrospectively reviewed data on 1638 patients with diverse tumor types who had undergone comprehensive genomic profiling and reported that TMB-high (TMB-H) tumor status, which was defined as at least 20 mut/mb, was more commonly noted in patients with melanoma (34%) and NSCLC (24%) (8). In that study, among patients who received immunotherapy, TMB-H was independently associated with a higher ORR (58% vs 20%; P = .0001) and longer median PFS (12.8 vs 3.3 months; P < .0001) and OS (not reached vs 16.3 months; P = .0036). Several additional studies have shown a statistically significant clinical benefit of checkpoint inhibitor treatment in patients with higher TMB (13,14). In patients with NSCLC and TMB of at least 10 mut/mb, frontline nivolumab plus ipilimumab was associated with longer PFS compared with chemotherapy, irrespective of the PD-L1 expression level (13). Next-generation sequencing (NGS) of 148 803 samples from the University of California San Diego and Foundation Medicine databases further revealed that 9762 (6.6%) were TMB of at least 20 mut/mb, and 2179 (1.5%) were MSI-H (69). Among 60 patients who received immunotherapy, the median PFS was longer in patients with low MSI and TMB of at least 20 mut/mb compared with those with low MSI and TMB of less than 20 mut/mb (26.8 vs 4.3 months; P = .0173). Additionally, higher TMB was linearly correlated with better outcomes after immune checkpoint blockade (8), and the impact of TMB was independent of microsatellite status (69).
Despite robust data demonstrating the association of higher TMB with response to immunotherapy, several challenges remain. First, harmonization of TMB assays is critical. Some investigators suggest that TMB response thresholds might differ across tumor types (70). Additionally, neoantigen immunostimulation differs on the basis of the corresponding molecular alteration, with certain mutations likely inducing a strong immune response (71). Further, not all studies show a survival benefit for higher TMB, even when there are robust and durable responses. Importantly, current TMB evaluation algorithms do not take into consideration neoantigens that might be generated from posttranslational modifications or gene fusions. Furthermore, TMB calculation using circulating tumor DNA analysis is being assessed. In patients with advanced NSCLC treated with pembrolizumab, plasma analysis using a 500-gene panel demonstrated that TMB of at least 16 mut/mb was associated with longer PFS (72). Other biologic considerations such as major histocompatibility complex (MHC) presentation and T-cell receptor (TCR) repertoire may also be important.
PD-L1 Immunohistochemistry
PD-L1 expression is being evaluated in numerous clinical trials as a biomarker predicting response to immunotherapy agents. High PD-L1 membrane expression is assessed by immunohistochemistry and defined using various scoring methods and cutoff points ranging from more than 1% to more than 50%. PD-L1 expression has been approved by the FDA as a companion diagnostic for the administration of immune checkpoint inhibitors in various indications, including first-line treatment for advanced NSCLC, head and neck squamous cell carcinoma, urothelial carcinoma, and triple-negative breast cancer and second-line therapy and beyond for advanced NSCLC, esophageal squamous cell carcinoma, cervical cancer, and gastric-esophagogastric junction adenocarcinoma. High PD-L1 expression has been associated with clinical benefit in patients with selected tumor types (73-76). This variability in the predictive value of PD-L1 expression and in the cutoff used to determine PD-L1 expression positivity is attributed to differences in tumor type, disease stage, types of checkpoint inhibition (PD-1 vs PD-L1), and methodology. Methodological differences are associated with technical issues regarding assessment of PD-L1 expression in tumor cells and immune cells; the use of scoring systems combining expression in tumor cells, lymphocytes, and macrophages; and the antibodies used for biomarker evaluation. Examples of monoclonal antibodies used to assess PD-L1 expression include SP142 and SP263 (Ventana Medical Systems, Oro Valley, Arizona, USA) and 22C3 pharmDx (Agilent, Santa Clara, California, USA). Scoring methods include the combined positive score, defined as the number of PD-L1–positive cells (tumor cells, lymphocytes, and macrophages) divided by the total number of cells (tumor cells, lymphocytes, and macrophages) and multiplied by 100, and the tumor proportion score (TPS), defined as the number of PD-L1-positive tumor cells divided by the total number of tumor cells and multiplied by 100. To overcome the variability in PD-L1 assessment, standardization of methodology is warranted.
Investigational Biomarkers Conferring Sensitivity to Immunotherapy
Beyond MSI-H (which results in high TMB) and TMB of at least 10 mut/mb, there are several biomarkers that may confer sensitivity to immune checkpoint blockade. These genomic biomarkers include, but are not limited to, alterations in chromatin remodeling genes, PD-L1 amplification, specific MHC types that permit neo-antigen presentation, and specific mutational signatures that are associated with increased TMB and/or increased immunogenicity of neo-antigens (Table 1 and Figure 1).
Figure 1.
US Food and Drug Administration–approved and investigational biomarkers associated with response to immuno-oncology therapy. APOBEC = apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like; ARID1A = AT-rich interaction domain 1A; BAP1 = BRCA1-associated protein 1; MHC-I = major histocompatibility complex class-I; PBRM1 = polybromo-1; PD-L1 = programmed cell death-ligand 1; POLD1 = DNA polymerase delta 1; POLE = DNA polymerase epsilon; SMARCA4 = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A, member 4; SMARCB1 = SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B, member 1; TMB = tumor mutational burden.
Chromatin Remodeling (SWI/SNF Complex)
Cells have developed several mechanisms to manipulate DNA and package it into chromatin. The building block of chromatin is the nucleosome. ATP-dependent chromatin remodeling complexes such as switch/sucrose non-fermentable (SWI/SNF) complexes are specialized protein machinery complexes able to restructure the nucleosome to make its DNA accessible during transcription, DNA repair, and replication. Several genes that make up these complexes, including those in the ARID and SMARC families and PBRM1, have been associated with responsiveness to immune checkpoint blockade (15,16,19,24,77). Mechanistically, reexpression of endogenous retroviruses resulting from deficiency of the SWI/SNF complex chromatin remodeling complex may contribute to activation of the immune response in these malignancies (78).
ARID1A alterations . The AT-rich interaction domain 1 A (ARID1A) gene encodes a subunit of the ATP-dependent chromatin remodeling SWI/SNF complex (79,80). Inactivating mutations in ARID1A are frequently identified in many tumor types: ovarian clear cell (45%), endometrial (45%), gastric (19%), and hepatocellular (14%) carcinomas (81-85). ARID1A functional loss has been clinically and experimentally correlated with immunotherapy response (15,16,86).
ARID1A deficiency interferes with MMR system regulation, leading to increased tumor mutational load and higher PD-L1 expression and more immune infiltrates (15,17,86,87). During DNA replication, ARID1A binds and recruits MSH2, a key MMR protein, to chromatin (15). ARID1A-deficient ovarian tumors in syngeneic mice were characterized by increased mutation load, tumor-infiltrating lymphocytes, and PD-L1 expression (15). Anti–PD-L1 antibody administration led to improved survival rates of mice with ARID1A-deficient vs ARID1A–wild-type ovarian tumors. Further, in 3304 patients with diverse tumor types, ARID1A molecular alterations correlated with MSI-H and TMB-H, but immunotherapy response in ARID1A-altered tumors was independent of these factors (16). In patients treated with immunotherapeutic agents, those with ARID1A-altered vs wild-type tumors had improved PFS (11 vs 4 months; P = .006) (16). These data indicate that ARID1A alterations should be further validated as a pan-cancer predictive biomarker for immune checkpoint blockade response.
PBRM1 molecular alterations . Polybromo-1 (PBRM1) is a tumor suppressor gene that encodes a subunit of the SWI/SNF complex and participates in regulation of the cell cycle, apoptosis, and centromeric cohesion. The data regarding PBRM1 alterations and response to checkpoint blockade are mixed, with some studies suggesting that aberrant PBRM1 is predictive and others indicating that it is not.
Molecular alterations in PBRM1 are commonly identified in clear cell renal cell carcinoma (RCC) (approximately 40%) (77,88) and are considered a biomarker predicting response to immunotherapy (18,19). Additionally, PBRM1 mutations have been identified in cholangiocarcinoma (17%) (89), mesothelioma (15%) (90), pancreatic cancer (8%) (91), and lung cancer (3%) (77). In patients with clear cell RCC treated with nivolumab, those harboring PBRM1 mutations had higher rates of clinical benefit (odds ratio [OR] = 2.14, 95% CI = 1.00 to ∞, P = .0497) and longer PFS (HR = 0.67, 95% CI = 0.47 to 0.96, P = .03) and OS (HR = 0.65, 95% CI = 0.44 to 0.96, P = .03) compared with patients without such mutations (19). Other investigators, using whole-exome sequencing in 35 patients with clear cell RCC, also demonstrated that loss-of-function mutations in PBRM1 were associated with improved clinical benefit from immune checkpoint inhibitors (OR = 12.93, 95% CI = 1.54 to 190.8, P = .012) (18). Their findings were independently validated in 63 patients with clear cell RCC who had been treated with PD-1 or PD-L1 inhibitors (OR for clinical benefit = 6.10, 95% CI = 1.42 to 32.64, P = .0071).
Contrary to these data, investigators recently showed that PBRM1 knockout (assessed by mRNA and protein levels) resulted in reduced interferon-gamma (IFN-γ)–STAT1 signaling in murine and human RCC cell lines (20). A retrospective analysis of approximately 700 patients with renal cancer from 3 independent clinical cohorts (IMmotion150 dataset, MSK-IMPACT cohort, and The Cancer Genome Atlas cohort) demonstrated that PBRM1 mutations were associated with fewer immune infiltrates and lower response rates to immunotherapy (20). Finally, in a retrospective analysis of 441 patients with NSCLC from 2 independent cohorts (385 patients from Memorial Sloan Kettering Cancer Center and 56 from Dana Farber Cancer Institute) who received immunotherapy, the presence of PBRM1 mutations correlated with shorter OS (6 months in PBRM1-mutated vs 13 months in PBRM1–wild-type patients; P = .03), including in multivariate analysis (77). These data collectively indicate that the predictive role of altered PBRM1 for immunotherapy is inconclusive.
SMARCA4 . SMARCA4 encodes another subunit of the chromatin remodeling SWI/SNF complex. SMARCA4 molecular alterations have been identified in 4% of cancers, including lung (10%), bladder (7%), colorectal (5%), and breast (2%) cancers (92,93). SMARCA4 alterations seem to be the “driver” molecular change in almost all small cell carcinomas of the ovary, hypercalcemic type (94,95); loss of SMARCA4 per IHC has been reported in up to 10% of patient with NSCLC (96).
Recently, SMARCA4 loss was used to define groups of undifferentiated carcinomas, including SMARCA4-deficient undifferentiated uterine sarcomas that share morphologic, IHC, and genetic similarities to small cell carcinoma of the ovary, hypercalcemic type (97), and undifferentiated thoracic carcinomas (98). Durable responses have been noted in anecdotal reports of patients with small cell carcinoma of the ovary, hypercalcemic type (21), NSCLC (22), and thoracic sarcoma (23,99). Preliminary data suggest that, although SMARC4-deficient tumors have low TMB, they have high PD-L1 expression and T-cell infiltration, suggestive of an immunogenic microenvironment (21).
SMARCB1 . SMARCB1, another component of the chromatin remodeling SWI/SNF complex, functions as a tumor suppressor gene. Complete SMARCB1 and/or SMARCA4 inactivation has been associated with aggressive tumor behavior in malignant rhabdoid and atypical teratoid rhabdoid tumors (100), and it has been suggested that SMARCB1-altered rhabdoid tumors respond to immunotherapy (101). In a recent study, the analysis of genomic, transcriptomic, and immune microenvironment data from sarcomatoid and rhabdoid RCC demonstrated the upregulation of immune pathways and greater CD8+ T-cell infiltration and PD-L1 expression on tumor cells in these subtypes compared with other RCC subtypes (24). Patients with sarcomatoid and rhabdoid tumors who received immunotherapy had improved outcomes compared with patients who did not (24).
Other Alterations Possibly Associated With Immunotherapy Response
BAP1 . BRCA1-associated protein 1 (BAP1) is a tumor suppressor gene involved in the regulation of chromatin remodeling and DNA damage repair (102,103). BAP1 copy number loss or inactivating mutations are frequently noted in mesothelioma (40%-64%) and result in the accumulation of DNA-damaged cells (104-106). BAP1 loss promotes an immune inflammatory environment in mesothelioma (27). Integrative genomic, proteomic, and transcriptomic analyses of 19 peritoneal mesotheliomas showed that, in tumors with BAP1 loss (vs wild type), there were higher rates of infiltrated immune cells (27). Clinical trials demonstrated that PD-1 inhibitors are active in patients with malignant pleural mesothelioma, and these mesotheliomas frequently harbor BAP1 alterations (25,26,106).
MHC genotype . In addition to the quality of the neo-antigens, the host immune system’s ability to efficiently present driver neo-antigens to T cells plays a critical role in immune response. The MHC class-I (MHC-I) genotype is predictive of response to immunotherapy in combination with TMB (28). The ability to present neo-antigens, defined by the Patient Harmonic-mean Best Rank (PHBR) score, was calculated in 83 patients with diverse malignancies who received immune checkpoint inhibitors. The PHBR score represents the ability of a specific human leukocyte antigen (HLA) class I genotype to bind and present a missense mutation; a lower PHBR score is suggestive of more efficient antigen presentation (107). Among patients with higher TMB (>20 mut/mb), those whose tumors had low PHBR scores had higher ORRs (78% vs 43%; P = .049) and longer PFS (26.8 vs 5.8 months; P = .03) compared with patients with PHBR-high tumors (28). The PHBR score did not predict response in patients with TMB-low tumors. Other investigators have also demonstrated that HLA-corrected TMB can reconcile the observed disparity in relationships between TMB and checkpoint blockade responses (108). Furthermore, MHC class II–restricted neo-antigens also play a crucial role in the antitumor response that is nonoverlapping with that of MHC class I–restricted neo-antigens and, therefore, needs to be considered when identifying patients who will most benefit from immunotherapy (109,110).
Mutational signatures . Neo-antigens are mutated peptides that enable the immune cells to recognize the tumor cell as foreign. The specific types of neo-antigens that stimulate a strong immune reaction remain unclear, but certain mutational signatures appear to be predictive of high immunogenicity and immunotherapy response. For instance, APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like, a family of evolutionarily conserved cytidine deaminases involved in viral infections) hyperactivity has been implicated in localized hypermutagenesis (also designated as kataegis) in patients with breast cancer (111). In a study of 99 patients with diverse tumor types who received immunotherapy, APOBEC-related mutagenesis was associated with higher response rates (OR = 9.69, P = .0106) and longer PFS (3.1 vs 2.1 months; P = .0239), independent of TMB (29). APOBEC-related neo-antigens tend to be more hydrophobic, and therefore more immunogenic, because they are better presented by the MHC and more easily recognized by T cells. Similarly, neo-antigens produced from an ultraviolet (UV)–mutated genome had increased hydrophobicity and immunogenicity (30). A high UV signature correlated with longer PFS and OS in patients with low or intermediateTMB tumors after checkpoint inhibitor treatment, but there was no association in patients with TMB-H (>20 mut/mb) tumors.
PD-L1 amplification . PD-L1 expression, assessed by IHC, is currently approved as a biomarker for treatment with anti–PD-1 or anti–PD-L1 agents, albeit one with technical limitations (5). PD-L1 gene amplification, as assessed by next-generation sequencing, is the hallmark of Hodgkin disease, which is highly responsive to checkpoint blockade (112), and also correlates with solid tumor immunotherapy responsiveness, although studied in only a small number of patients (31). In a retrospective analysis of comprehensive tumor molecular profiling data from 118 187 patients with diverse cancers, the prevalence of PD-L1 amplification was 0.7% (843 of 118 187 patients) (31). Solid tumors with the highest proportions of PD-L1 amplification (PD-L1 amplified/cases analyzed) were undifferentiated soft-tissue sarcoma (3.9%), head and neck squamous cell carcinoma (3.1%), breast carcinoma (1.9%), and lung squamous cell carcinoma (1.7%). Of 13 patients with solid tumors with PD-L1 amplification, 9 had received treatment with checkpoint blockade; the ORR was 66.7%, and the median PFS was 15.2 months (31). Interestingly, PD-L1 amplification did not always correlate with PD-L1 overexpression by IHC, even in responders, perhaps because of the technical limitations of IHC.
POLE/POLD1 mutations . DNA polymerase epsilon (POLE) and DNA polymerase delta 1 (POLD1) alterations correlate with immunotherapy response. These proteins play a critical role in DNA replication and repair regulation. In 47 721 patients (pan-cancer), POLE and POLD1 mutations were identified in 2.79% and 1.37%, respectively (32). Patients with POLE- or POLD1-mutated tumors had longer OS compared with patients with wild-type tumors after immunotherapy (32). Alterations in POLE or POLD1 have been associated with statistically significantly higher tumor mutational burden (33), higher CD8+ lymphocyte infiltration levels, and increased expression of cytotoxic T-cell markers compared with POLE or POLD1 wild-type tumors (113). In ongoing clinical trials (NCT02693535, NCT02912572, NCT03810339), patients with POLE or POLD1 mutations are selected for treatment with immunotherapy. For instance, in the Targeted Agent and Profiling Utilization Registry study (NCT02693535), a non-randomized clinical trial, patients with POLE or POLD1 mutations (or high mutational load) are selected for treatment with pembrolizumab or nivolumab and ipilimumab.
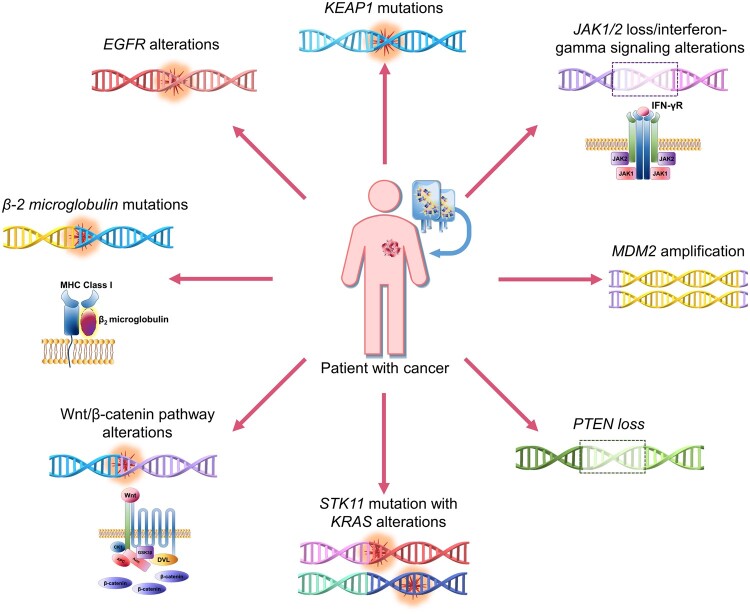
Investigational Biomarkers Conferring Resistance to Immunotherapy
Several genomic markers, such as loss of beta-2 (β2)-microglobulin and JAK1/2, interfere with the immune response and hence correlate with resistance to immune checkpoint blockade. In addition, loss of Phosphatase and TENsin homolog deleted on chromosome 10 (PTEN) and alterations in EGFR, KEAP1, STK11/KRAS, and the B-catenin pathway have been associated with immunotherapy resistance, although the mechanisms are unclear. EGFR alterations and murine double minute 2 (MDM2) amplification have been correlated with accelerated progression (also known as hyperprogression) after checkpoint blockade, but the underlying biology has not been elucidated (Table 1, Figure 2) (44,114).
Figure 2.
Investigational biomarkers associated with resistance to immuno-oncology therapy. It is not yet clear if some of these markers, such as KEAP1 and STK11 alterations, are prognostic or predictive. EGFR = epidermal growth factor receptor; JAK = Janus kinase; KEAP1 = Kelch-like ECH associated protein 1; KRAS = v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog; MDM2 = murine double minute 2; PTEN = phosphatase and TENsin homolog deleted on chromosome 10; STK11 = serine/threonine kinase 11.
β2-Microglobulin Mutations
β2-microglobulin is a low-molecular-weight protein that forms MHC-I molecules in combination with the heavy chain. Proper functioning of β2-microglobulin is critical for antigen presentation and assembly of HLA class I complexes. Loss of β2-microglobulin facilitates tumor escape from immune recognition (115-117) and is implicated in resistance to immunotherapy. In one study, a truncated mutation in β2-microglobulin was identified in tissue obtained from a patient with melanoma who progressed on anti–PD-1 therapy (34). IHC revealed loss of surface expression of MHC-I. In another study, analysis of longitudinal tumor biopsies demonstrated molecular alterations in β2-microglobulin in 5 (29.4%) of 17 patients with advanced melanoma who progressed on immunotherapy (35). Loss of heterozygosity was noted only in nonresponders, whereas there was no molecular alteration in β2-microglobulin detected in responders.
EGFR Alterations
Immunotherapy provides limited clinical benefit in patients with NSCLC harboring EGFR mutations (118). EGFR alterations are also associated with hyperprogression after treatment with immunotherapy in some studies, but not in others (3,37,114,119). In vivo investigation of tumor growth in T-cell–deficient mice, which were injected with EGFR-mutated patient-derived xenografts, showed that nivolumab triggered the accrual of macrophages and led to increased tumor growth and lung dissemination (120). Investigators have proposed upregulation of PD-1 and PD-L1 through EGFR activation as a mechanism of resistance (121).
KEAP1 Mutations
KEAP1 regulates cytoprotective responses to oxidative and electrophilic stress by binding the transcription factor NRF2 (122,123). Loss-of-function mutations in KEAP1 result in dissociation and constitutive activation of NRF2, cellular resistance to oxidative stress, and increased tumor cell growth (124,125). Additionally, NRF2 activates drug efflux pump genes that confer resistance to cytotoxic drugs (126). KEAP1 mutations occur in 2.7% of patients with cancer, most commonly in patients with NSCLC (15.8%) (38). Studies suggest that KEAP1 mutations confer resistance to immunotherapy because they are associated with a “cold” tumor microenvironment (38,39). In 1 study, among 1661 patients who received immunotherapy, those with KEAP1 mutations had shorter OS compared with those with wild-type tumors (10 vs 20 months; P = .0029) (38). Other investigators showed that co-mutation of KRAS and KEAP1 (27% of KRAS-mutated tumors) in patients with NSCLC is an independent prognostic factor associated with shorter OS and duration of response to first-line platinum-based chemotherapy in patients who received immunotherapy (39). In the phase III MYSTIC NSCLC study, KEAP1 mutations correlated with poor outcome across arms (durvalumab alone, durvalumab plus tremelimumab or chemotherapy). Other studies have, however, reported that KEAP1 mutations may be associated with poor prognosis regardless of therapy and are not specifically predictive for checkpoint blockade outcome (40,41).
Janus Kinase 1 and 2 Loss
Janus kinase (JAK) is a family of intracellular, nonreceptor tyrosine kinases that transduce cytokine signals via the JAK-STAT pathway. Importantly, IFN-γ, critical to immune response, binds to its receptor and recruits and activates JAK1 and JAK2, and subsequently STAT1, thus resulting in immune cell activation (127-129). Loss of IFN-γ signaling pathway genes has been implicated in immunotherapy resistance (34,42,43,130,131).
Whole-exome sequencing of paired melanoma tumors (primary tumor at diagnosis and metastatic tumor at the time of recurrence) after disease progression on anti–PD-1 therapy revealed acquired loss-of-function mutations in JAK1 or JAK2, with deletion of the wild-type allele (34). Functionally, these mutations resulted in lack of response to IFN-γ. In another study, investigators showed that patients with JAK1/2 inactivating mutations did not respond to immunotherapy (42). Additionally, in vitro exposure to IFN-γ failed to mediate downstream signaling. Finally, in another report, patients with JAK2-mutated advanced melanoma did not respond to anti-CTLA4 treatment, and melanoma cell lines with molecular alterations in IFN-γ pathway genes were refractory to immunotherapy (130). Together, these data indicate that loss-of-function JAK1/2 mutations can mediate primary or acquired resistance to immunotherapy.
MDM2 Amplification
MDM2 encodes a nuclear-localized E3 ubiquitin ligase, which functions as a negative regulator of the TP53 gene. MDM2 amplification occurred in approximately 4% of more than 100 000 cancers analyzed (44,93,132) and was most commonly noted in patients with liposarcoma (64% of cases), gallbladder adenocarcinoma (11%), sarcoma (11%), and urothelial carcinoma (10%) (44). Most patients (99%) had additional molecular alterations (44). MDM2 amplification has been linked to hyperprogression after treatment with immunotherapeutic agents (3,44,133,134). The biologic mechanism by which MDM2 amplification mediates hyperprogression after treatment with immunotherapy is unknown, and it is possible that another gene on the MDM2 amplicon is culpable. One study reported that MDM2 mediates resistance to immunotherapy through degradation of transcription factor NFATc2, leading to reduced T-cell activation (135).
PTEN Loss
PTEN is the second most commonly mutated tumor suppressor gene. PTEN loss has been described in diverse tumor types, including hepatocellular (57% of patients), prostate (52%), endometrial (49%), and colorectal (48%) cancers (136). PTEN inhibits PI-3 kinase and AKTsignaling by converting phosphoinositol-(3-5)-trisphosphate to phosphoinositol-(4, 5)-bisphosphate. PTEN loss, through mutations, epigenetic mechanisms, and gene silencing, disrupts its regulatory control of cell proliferation, energy metabolism, angiogenesis, and survival.
PTEN loss has been implicated in immunotherapy resistance in preclinical models, in an immune-suppressive microenvironment in prostate cancer and glioblastoma, and in acquired resistance in the clinic in a patient with metastatic uterine leiomyosarcoma, as well as to poor outcome from checkpoint blockade in melanoma (45,137-139).
STK11 Mutations With KRAS Alterations
Serine/threonine kinase 11 (STK11) is a tumor suppressor gene that regulates cell proliferation and energy metabolism. Patients with STK11/KRAS-mutated (compared with KRAS-only mutated) tumors have statistically significantly worse clinical outcomes after treatment with immunotherapy (47). In another study, the presence of STK11 alterations was associated with shorter PFS and OS and lower ORR in patients with NSCLC who received treatment with an anti–PD-1 agent combined with doublet chemotherapy (52). However, recently published real-world data (Flatiron Health Network) suggest that STK11 mutations confer poor prognosis to patients with NSCLC regardless of treatment and show no specific prediction for immunotherapy outcomes (41).
TCR Repertoire
The TCR repertoire refers to the multiple possible combinations of TCR sequences and represents a “footprint” for T cells’ recognition of tumor neo-antigens. Higher TCR clonality has been associated with response to anti–PD-1 therapy in patients with melanoma (140,141). In patients with pancreatic cancer, low baseline clonality and higher rates of expanded clones after treatment with ipilimumab were associated with longer OS (142). Finally, neo-adjuvant treatment with nivolumab in patients with NSCLC induced expansion of neo-antigen-specific T-cell clones (143). Patients who achieved a major pathological response had higher rates of T-cell clonality.
Wnt/β-Catenin Pathway Alterations
Wnt/β-catenin signaling plays a critical role in proliferation, migration, and division (144,145). Deregulation of this pathway is a strong “driver” in diverse tumors, mainly CRC, where it occurs in 93% of cases (146). Many investigators suggest that the Wnt/β-catenin pathway—as a candidate immune-related modulator—correlates with impaired immune cell recruitment in melanoma and other tumor types (48,137,147-149). An integrative The Cancer Genome Atlas data analysis, based on genomic, transcriptomic, or proteomic approaches, showed that non–T-cell–inflamed tumors (3137 of 9244 tumors, 33.9%) were enriched for WNT/β-catenin pathway activation (150). Preclinical and clinical models demonstrate that WNT/β-catenin activation leads to suppression of CD8+ T-cell tumor infiltration and evasion of immune elimination (48,147-149). T-cell suppression through activation of the Wnt/β-catenin pathway has been linked to decreased chemokine levels (151). In preclinical trials, RNAi-mediated inhibition of the Wnt/β-catenin pathway combined with anti–PD-1/CTLA-4 agents led to T-cell infiltration and tumor growth inhibition (152).
Other Immune-Related Biomarkers
Biomarkers beyond genomics predicting response and resistance to immune checkpoint inhibitors are included in Table 2. High PD-L1 expression, defined using various cutoff points ranging from more than 1% to more than 50%, is associated with clinical benefit in patients with various tumor types, but not in all clinical settings. Other emerging predictive biomarkers include gene expression signatures (153,154), oral and gut microbiome (155,156), neo-antigen load (157,158), PD-1 expression on immune cells (159), and TCR repertoire (140,142). Biomarkers predicting susceptibility to immune-related adverse events include circulating pro-inflammatory cytokines (160) and gut microbiota (155).
Table 2.
Biomarkers beyond genomics for response and resistance to immune checkpoint inhibitorsa
| Biomarker | Cancer type | Role | Predictive value and comments | Selected references |
|---|---|---|---|---|
| FDA-approved biomarkers | ||||
| PD-L1 expression | Bladder cancer, NSCLC, TNBC, cervical cancer, and gastric/GEJ cancer | Inhibits peripheral T-cell activation and suppresses immune surveillance | Predicts response to immune checkpoint inhibitors | Patel, et al. 2015 (5) |
| Reported/Investigational biomarkers | ||||
| Gene expression signatures | Melanoma, NSCLC | Immune-related signatures |
|
|
| Gut microbiome | Melanoma | Modulate immune-mediated colitis |
|
|
| Neoantigen load | Across tumor types (melanoma, NSCLC) | Increased mutations/neoantigens | Predicts response to adoptive T-cell therapy and checkpoint inhibitors | |
| PD-1 expression | Across tumor types (melanoma, gastric cancer, NSCLC) | PD-1 expression on CD8+ T cells negatively influences effector functions | Predicts response | Kumagai, et al. 2020 (159) |
| TCR diversity | Melanoma, NSCLC | Associated with immune expansion of T-cell clone that recognizes a specific tumor neoantigen | Predicts response | |
| Proinflammatory cytokines | Melanoma | Increase proinflammatory activities | Predicts toxicity | |
FDA = US Food and Drug Administration; GEJ = gastro-esophageal junction; NSCLC = non-small cell lung cancer; PD-1 = programmed cell death-1; PD-L1 = programmed cell death-ligand 1; TCR = T-cell receptor; TNBC = triple-negative breast cancer.
Tumor microenvironment typically comprises infiltrating immune cells, such as cytotoxic T cells, helper T-cell subsets, regulatory T cells, tumor-associated macrophages, and dendritic cells. The clinical relevance of the density and location of tumor-infiltrating lymphocytes (TILs) in patients with cancer is currently being investigated. Previous studies have demonstrated that increased TIL concentration is associated with improved prognosis in patients with various tumor types (162). Whether high TIL concentration is predictive of response to immune checkpoint inhibitors is currently under extensive evaluation (163). Investigators have shown that higher TIL concentrations are associated with higher response rates and improved clinical outcomes in patients who receive immunotherapy in different clinical settings (140,164). The presence of a high number of TILs has also been associated with high PD-L1 expression (165,166) and/or MSI-H and dMMR (167,168). Ongoing and future investigations will address the limitations involving the methodology, interpretation (invasive margin or central infiltration), and cutoff values for TILs and will optimize the standardization of TIL assessment to enable comparisons of various clinical trials (169).
Discussion
Tumor microenvironment complexity, intratumoral heterogeneity, and distinct host immunity complicate the identification of immune checkpoint blockade predictive biomarkers. Tumors and their surrounding ecosystem have individually distinct and complex immune profiles. Biomarkers such as PD-L1, assessed by IHC, may be imprecise predictors of immune checkpoint blockade response for technical reasons or because of the complexity of the immune response (5). In addition, overexpression of specific biomarkers, such as alternate checkpoints TIM-3 and VISTA, was also statistically significantly associated with shorter PFS after anti–PD-1- and PD-L1-based therapies in patients with diverse malignancies (170). Therefore, patients may need customized combinations of immunotherapy. Ultimately, composite biomarkers that incorporate genomic and immune profiles, host pharmacogenome as reflected by MHC and the TCR repertoire, and other factors may be needed. Based on current data, it seems unlikely that a single biomarker will be predictive for all patients or all types of immunotherapeutic agents. Machine learning algorithms that can combine genomic, transcriptomic, epigenomic, and immune-related markers and consider their interactions may be exploitable to overcome the complexity of the immune system. Ideally, all patients with cancer should be screened for all available immuno-oncology markers and receive the corresponding best treatment. Prospective trials should focus on the systematic analysis of genomic and immune profiles of patients who are treated on immuno-oncology trials and the correlation of these profiles with response or resistance and toxicity. Given the plethora of biomarkers that have been correlated with resistance to immunotherapy, particularly hyperprogressive disease, caution is warranted when patients with these biomarkers are considered for immuno-oncology treatments. The identification of novel biomarkers and consensus regarding “molecular resistance” to immunotherapy would help optimize the deployment of these potentially curative treatments.
Funding
National Institutes of Health/National Cancer Institute, award number P30 CA016672 (The University of Texas MD Anderson Cancer Center). This work is also supported by donor funds from Jamie’s Hope, Mr and Mrs Zane W. Arrott, and Mr and Mrs Steven McKenzie for Dr Tsimberidou’s Personalized Medicine Program.
Notes
Role of the funder: The funders had no role in the writing of this review or the decision to submit it for publication.
Disclosures: Dr Elena Fountzilas has the following financial relationships to disclose: Travel grants: Merck, Pfizer, and K.A.M Oncology/Hematology; Speaker fees: from Roche, Leo, Pfizer; Stock ownership: Deciphera Pharmaceuticals, Inc. Dr Razelle Kurzrock has the following financial relationships to disclose: Research Funding (Institution): Incyte, Genentech, Merck Serono, Pfizer, Sequenom, Foundation Medicine, Konica Minolta, Grifols, Biologic Dynamics, Boehringer Ingelheim, Medimmune, and Guardant. Consulting role: X-Biotech, Loxo, Biologic Dynamics, Turning Point, TD2, Bicara, and Actuate Therapeutics. Speaker fees: Roche. Ownership interest: IDbyDNA and CureMatch, Inc. Board member: CureMatch and CureMetrix. Dr Apostolia-Maria Tsimberidou has the following financial relationships to disclose: Research Funding (Institution): Immatics, Parker Institute for Cancer Immunotherapy, Tempus, OBI Pharma, EMD Serono, Baxalta, ONYX, Bayer, Boston Biomedical, Placon Therapeutics, Karus Therapeutics, and Tvardi Therapeutics. Consulting or Advisory Role: Covance, Genentech and Tempus. Dr Henry Hiep Vo reports no relevant conflicts of interest.
Author contributions: RK and AT conceived the manuscript. All authors drafted the manuscript and/or figures, reviewed and modified drafts of the manuscript, and approved the final manuscript.
Data Availability
No new data were generated or analyzed in support of this research.
Contributor Information
Elena Fountzilas, Department of Medical Oncology, Euromedica General Clinic, Thessaloniki, Greece; European University Cyprus, Limassol, Cyprus.
Razelle Kurzrock, Center for Personalized Cancer Therapy and Division of Hematology and Oncology, UC San Diego Moores Cancer Center, San Diego, CA, USA.
Henry Hiep Vo, The University of Texas MD Anderson Cancer Center, Department of Investigational Cancer Therapeutics, Houston, TX, USA.
Apostolia-Maria Tsimberidou, The University of Texas MD Anderson Cancer Center, Department of Investigational Cancer Therapeutics, Houston, TX, USA.
References
- 1. Emens LA, Ascierto PA, Darcy PK, et al. Cancer immunotherapy: opportunities and challenges in the rapidly evolving clinical landscape. European Journal of Cancer. 2017;81:116–129. [DOI] [PubMed] [Google Scholar]
- 2. Champiat S, Dercle L, Ammari S, et al. Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin Cancer Res. 2017;23(8):1920–1928. [DOI] [PubMed] [Google Scholar]
- 3. Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23(15):4242–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adashek JJ, Kato S, Ferrara R, et al. Hyperprogression and immune checkpoint inhibitors: hype or progress? Oncologist. 2020;25(2):94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. [DOI] [PubMed] [Google Scholar]
- 6. Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: Results from the phase II KEYNOTE-158 study. J Clin Oncol. 2019;38(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lemery S, Keegan P, Pazdur R. First FDA approval agnostic of cancer site–when a biomarker defines the indication. N Engl J Med. 2017;377(15):1409–1412. [DOI] [PubMed] [Google Scholar]
- 8. Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subbiah V, Solit DB, Chan TA, et al. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥10: a decision centered on empowering patients and their physicians. Ann Oncol. 2020;31(9):1115–1118. [DOI] [PubMed] [Google Scholar]
- 10. Le DT, Kim TW, Van Cutsem E, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J Clin Oncol. 2020;38(1):11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. [DOI] [PubMed] [Google Scholar]
- 12. Lenz H-J, Lonardi S, Zagonel V, et al. Nivolumab plus low-dose ipilimumab as first-line therapy in microsatellite instability-high/DNA mismatch repair deficient metastatic colorectal cancer: Clinical update. J Clin Oncol. 2020;38(suppl 4):11–11.31725351 [Google Scholar]
- 13. Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. New Engl J Med. 2018;378(22):2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–1448. [DOI] [PubMed] [Google Scholar]
- 15. Shen J, Ju Z, Zhao W, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24(5):556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okamura R, Kato S, Lee S, et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J Immunother Cancer. 2020;8(1):e000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim YB, Ahn JM, Bae WJ, et al. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int J Cancer. 2019;145(4):916–926. [DOI] [PubMed] [Google Scholar]
- 18. Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Braun DA, Ishii Y, Walsh AM, et al. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol. 2019;5(11):1631–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X-D, Kong W, Peterson CB, et al. PBRM1 loss defines a nonimmunogenic tumor phenotype associated with checkpoint inhibitor resistance in renal carcinoma. Nat Commun. 2020;11(1):2135–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jelinic P, Ricca J, Van Oudenhove E, et al. Immune-active microenvironment in small cell carcinoma of the ovary, hypercalcemic type: rationale for immune checkpoint blockade. J Natl Cancer Inst. 2018;110(7):787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naito T, Umemura S, Nakamura H, et al. Successful treatment with nivolumab for SMARCA4-deficient non-small cell lung carcinoma with a high tumor mutation burden: a case report. Thoracic Canc. 2019;10(5):1285–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iijima Y, Sakakibara R, Ishizuka M, et al. Notable response to nivolumab during the treatment of SMARCA4-deficient thoracic sarcoma: a case report. Immunotherapy. 2020;12(8):563–569. [DOI] [PubMed] [Google Scholar]
- 24. Bakouny Z, Braun DA, Shukla SA, et al. Integrative molecular characterization of sarcomatoid and rhabdoid renal cell carcinoma (S/R RCC) to reveal potential determinants of poor prognosis and response to immune checkpoint inhibitors (ICI). J Clin Oncol. 2020;38(suppl 6):715.31922920 [Google Scholar]
- 25. Alley EW, Lopez J, Santoro A, et al. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2017;18(5):623–630. [DOI] [PubMed] [Google Scholar]
- 26. Scherpereel A, Mazieres J, Greillier L, et al. Nivolumab or nivolumab plus ipilimumab in patients with relapsed malignant pleural mesothelioma (IFCT-1501 MAPS2): a multicentre, open-label, randomised, non-comparative, phase 2 trial. Lancet Oncol. 2019;20(2):239–253. [DOI] [PubMed] [Google Scholar]
- 27. Shrestha R, Nabavi N, Lin YY, et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med. 2019;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goodman AM, Castro A, Pyke RM, et al. MHC-I genotype and tumor mutational burden predict response to immunotherapy. Genome Med. 2020;12(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boichard A, Pham TV, Yeerna H, et al. APOBEC-related mutagenesis and neo-peptide hydrophobicity: implications for response to immunotherapy. Oncoimmunology. 2019;8(3):1550341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pham TV, Boichard A, Goodman A, et al. Role of ultraviolet mutational signature versus tumor mutation burden in predicting response to immunotherapy. Mol Oncol. 2020;14(8):1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol. 2018;4(9):1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang F, Zhao Q, Wang Y-N, et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 2019;5(10):1504–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yao J, Gong Y, Zhao W, et al. Comprehensive analysis of POLE and POLD1 gene variations identifies cancer patients potentially benefit from immunotherapy in Chinese population. Sci Rep. 2019;9(1):15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. New Engl J Med. 2016;375(9):819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sade-Feldman M, Jiao YJ, Chen JH, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017;8(1):1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodig SJ, Gusenleitner D, Jackson DG, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. 2018;10(450):eaar3342. [DOI] [PubMed] [Google Scholar]
- 37. Ferrara R, Mezquita L, Texier M, et al. Hyperprogressive disease in patients with advanced non-small cell lung cancer treated with PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol. 2018;4(11):1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen X, Su C, Ren S, et al. Pan-cancer analysis of KEAP1 mutations as biomarkers for immunotherapy outcomes. Ann Transl Med. 2020;8(4):141–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24(2):334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rizvi N, Cho BC, Reinmuth N, et al. Mutations associated with sensitivity or resistance to immunotherapy in mNSCLC: analysis from the MYSTIC trial. J Thorac Oncol. 2019;14(10):S217. [Google Scholar]
- 41. Papillon-Cavanagh S, Doshi P, Dobrin R, et al. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5(2):e000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discovery. 2017;7(2):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horn S, Leonardelli S, Sucker A, et al. Tumor CDKN2A-associated JAK2 loss and susceptibility to immunotherapy resistance. J Natl Cancer Inst. 2018;110(6):677–681. [DOI] [PubMed] [Google Scholar]
- 44. Kato S, Ross JS, Gay L, et al. Analysis of MDM2 Amplification: next-generation sequencing of patients with diverse malignancies. J Clin Oncol Precision Oncology. 2018;2018;10–1200. /PO.1217.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nature Medicine. 2019;25(3):462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8(7):822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. [DOI] [PubMed] [Google Scholar]
- 49. Abril-Rodriguez G, Torrejon DY, Liu W, et al. PAK4 inhibition improves PD-1 blockade immunotherapy. Nat Cancer. 2020;1(1):46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim JY, Lee KH, Kang J, et al. Hyperprogressive disease during anti-PD-1 (PDCD1) / PD-L1 (CD274) therapy: a systematic review and meta-analysis. Cancers. 2019;11(11):1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skoulidis F, Arbour KC, Hellmann MD, et al. Association of STK11/LKB1 genomic alterations with lack of benefit from the addition of pembrolizumab to platinum doublet chemotherapy in non-squamous non-small cell lung cancer. J Clin Oncol. 2019;37(suppl 15):102–102. [Google Scholar]
- 53. Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95(15):8698–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95(12):6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lynch HT, Lynch J. Lynch syndrome: genetics, natural history, genetic counseling, and prevention. J Clin Oncol. 2000;18(suppl 21):19s–31s. [PubMed] [Google Scholar]
- 56. Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–261. [DOI] [PubMed] [Google Scholar]
- 57. Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75(6):1215–1225. [DOI] [PubMed] [Google Scholar]
- 58. Cortes-Ciriano I, Lee S, Park W-Y, et al. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun. 2017;8(1):15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22(11):1342–1350. [DOI] [PubMed] [Google Scholar]
- 60. Schwark LA, Srinivasan P, Kemel Y, et al. Pan-cancer microsatellite instability to predict for presence of Lynch syndrome. J Clin Oncol. 2018;36(suppl 18):LBA1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O’Neil BH, Wallmark JM, Lorente D, et al. Safety and antitumor activity of the anti-PD-1 antibody pembrolizumab in patients with advanced colorectal carcinoma. PLoS One. 2017;12(12):e0189848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mehra R, Seiwert TY, Gupta S, et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long-term follow-up in KEYNOTE-012. Br J Cancer. 2018;119(2):153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.US Food and Drug Administration. FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication. Accessed August 31, 2020.
- 64. Andre T, Shiu K-K, Kim TW, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: the phase 3 KEYNOTE-177 Study. Journal of Clinical Oncology. 2020;38(suppl 18):LBA4. [Google Scholar]
- 65.US Food and Drug Administration. FDA grants nivolumab accelerated approval for MSI-H or dMMR colorectal cancer. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-nivolumab-accelerated-approval-msi-h-or-dmmr-colorectal-cancer. Accessed August 31, 2020.
- 66.US Food and Drug Administration. FDA grants accelerated approval to ipilimumab for MSI-H or dMMR metastatic colorectal cancer. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-ipilimumab-msi-h-or-dmmr-metastatic-colorectal-cancer. Accessed August 31, 2020.
- 67.US Food and Drug Administration. FDA approves pembrolizumab for adults and children with TMB-H solid tumors. https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors. Accessed July 23, 2020.
- 68. Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goodman AM, Sokol ES, Frampton GM, et al. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res. 2019;7(10):1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen P, Zhang C, Meng Z, et al. Comparison of tumor mutational burden across eight types of human cancer. J Clin Oncol. 2020;38(suppl 15):e15170. [Google Scholar]
- 71. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aggarwal C, Thompson JC, Chien AL, et al. Baseline plasma tumor mutation burden predicts response to pembrolizumab-based therapy in patients with metastatic non-small cell lung cancer [published online ahead of print February 26, 2020]. Clin Cancer Res. 2020;10.1158/1078-0432.Ccr-19-3663:clincanres.3663.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. New Engl J Med. 2018;379(22):2108–2121. [DOI] [PubMed] [Google Scholar]
- 74. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. New Engl J Med. 2016;375(19):1823–1833. [DOI] [PubMed] [Google Scholar]
- 75. Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394(10212):1915–1928. [DOI] [PubMed] [Google Scholar]
- 76. Chung HC, Ros W, Delord JP, et al. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2019;37(17):1470–1478. [DOI] [PubMed] [Google Scholar]
- 77. Zhou H, Liu J, Zhang Y, et al. PBRM1 mutation and preliminary response to immune checkpoint blockade treatment in non-small cell lung cancer. NPJ Precis Oncol. 2020;4(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Leruste A, Tosello J, Ramos RN, et al. Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell. 2019;36(6):597–612.e598. [DOI] [PubMed] [Google Scholar]
- 79.CancerGenome. My cancer genome: Biomarkers: ARID1A. https://www.mycancergenome.org/content/gene/arid1a/#ref-3. Accessed June 28, 2020.
- 80. Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. [DOI] [PubMed] [Google Scholar]
- 81. Jones S, Wang T-L, Shih I-M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. [DOI] [PubMed] [Google Scholar]
- 83. Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li L, Li M, Jiang Z, et al. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells. 2019;8(7):678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jiang T, Chen X, Su C, et al. Pan-cancer analysis of ARID1A alterations as biomarkers for immunotherapy outcomes. J Cancer. 2020;11(4):776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci USA. 2016;113(47):13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dagogo-Jack I, Schrock AB, Kem M, et al. Clinicopathologic characteristics of BRG1-deficient NSCLC. J Thorac Oncol. 2020;15(5):766–776. [DOI] [PubMed] [Google Scholar]
- 93. The AACR Project GENIE Consortium. AACR Project GENIE: Powering precision medicine through an international consortium. Cancer Discov. 2017;7(8):818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Conlon N, Silva A, Guerra E, et al. Loss of SMARCA4 expression is both sensitive and specific for the diagnosis of small cell carcinoma of ovary, hypercalcemic type. Am J Surg Pathol. 2016;40(3):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moes-Sosnowska J, Szafron L, Nowakowska D, et al. Germline SMARCA4 mutations in patients with ovarian small cell carcinoma of hypercalcemic type. Orphanet Journal of Rare Diseases. 2015;10:32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Herpel E, Rieker RJ, Dienemann H, et al. SMARCA4 and SMARCA2 deficiency in non-small cell lung cancer: immunohistochemical survey of 316 consecutive specimens. Ann Diagn Pathol. 2017;26:47–51. [DOI] [PubMed] [Google Scholar]
- 97. Kolin DL, Dong F, Baltay M, et al. SMARCA4-deficient undifferentiated uterine sarcoma (malignant rhabdoid tumor of the uterus): a clinicopathologic entity distinct from undifferentiated carcinoma. Mod Pathol. 2018;31(9):1442–1456. [DOI] [PubMed] [Google Scholar]
- 98. Le Loarer F, Watson S, Pierron G, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47(10):1200–1205. [DOI] [PubMed] [Google Scholar]
- 99. Takada K, Sugita S, Murase K, et al. Exceptionally rapid response to pembrolizumab in a SMARCA4-deficient thoracic sarcoma overexpressing PD-L1: a case report. Thorac Cancer. 2019;10(12):2312–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jackson EM, Sievert AJ, Gai X, et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15(6):1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Leruste A, Tosello J, Ramos RN, et al. Clonally expanded T cells reveal immunogenicity of rhabdoid tumors. Cancer Cell. 2019;36:597-612.e8. [DOI] [PubMed] [Google Scholar]
- 102. Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci USA. 2014;111(1):285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Louie BH, Kurzrock R. BAP1: not just a BRCA1-associated protein. Cancer Treat Rev. 2020;90:102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. [DOI] [PubMed] [Google Scholar]
- 105. Nasu M, Emi M, Pastorino S, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10(4):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kato S, Tomson BN, Buys TPH, et al. Genomic landscape of malignant mesotheliomas. Mol Cancer Ther. 2016;15(10):2498–2507. [DOI] [PubMed] [Google Scholar]
- 107. Marty R, Kaabinejadian S, Rossell D, et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell. 2017;171(6):1272–1283. e1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shim JH, Kim HS, Cha H, et al. HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann Oncol. 2020;31(7):902–911. [DOI] [PubMed] [Google Scholar]
- 109. Alspach E, Lussier DM, Miceli AP, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019;574(7780):696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Marty Pyke R, Thompson WK, Salem RM, et al. Evolutionary pressure against MHC class II binding cancer mutations. Cell. 2018;175(2):416–428.e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. New Engl J Med. 2014;372(4):311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Domingo E, Freeman-Mills L, Rayner E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1(3):207–216. [DOI] [PubMed] [Google Scholar]
- 114. Kim S, Kim J, Hong S, et al. Hyperprogression and pseudoprogression in patients with non-small cell lung cancer on checkpoint blocking immunotherapy. J Thorac Oncol. 2018;13(10):S888–S889. [Google Scholar]
- 115. Restifo NP, Marincola FM, Kawakami Y, et al. Loss of functional beta 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst. 1996;88(2):100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hicklin DJ, Wang Z, Arienti F, et al. beta2-microglobulin mutations, HLA class I antigen loss, and tumor progression in melanoma. J Clin Invest. 1998;101(12):2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. del Campo AB, Kyte JA, Carretero J, et al. Immune escape of cancer cells with beta2-microglobulin loss over the course of metastatic melanoma. Int J Cancer. 2014;134(1):102–113. [DOI] [PubMed] [Google Scholar]
- 118. Lee CK, Man J, Lord S, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol. 2018;4(2):210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Huang X, Xia L, Lan F, et al. Treatment of nivolumab results in hyperprogressive disease in a patient harboring EGFR Exon 20 insertion and MYC amplification. J Thorac Oncol. 2019;14(9):e189–e191. [DOI] [PubMed] [Google Scholar]
- 120. Lo Russo G, Moro M, Sommariva M, et al. Antibody-Fc/FcR interaction on macrophages as a mechanism for hyperprogressive disease in non-small cell lung cancer subsequent to PD-1/PD-L1 blockade. Clin Cancer Res. 2019;25(3):989–999. [DOI] [PubMed] [Google Scholar]
- 121. Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol. 2015;10(6):910–923. [DOI] [PubMed] [Google Scholar]
- 122. Li QK, Singh A, Biswal S, et al. KEAP1 gene mutations and NRF2 activation are common in pulmonary papillary adenocarcinoma. J Hum Genet. 2011;56(3):230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10):e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ohta T, Iijima K, Miyamoto M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68(5):1303–1309. [DOI] [PubMed] [Google Scholar]
- 125. Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236(2):313–322. [DOI] [PubMed] [Google Scholar]
- 126. Bai X, Chen Y, Hou X, et al. Emerging role of NRF2 in chemoresistance by regulating drug-metabolizing enzymes and efflux transporters. Drug Metab Rev. 2016;48(4):541–567. [DOI] [PubMed] [Google Scholar]
- 127. Akada H, Akada S, Hutchison RE, et al. Critical role of Jak2 in the maintenance and function of adult hematopoietic stem cells. Stem Cells. 2014;32(7):1878–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Igarashi K, Garotta G, Ozmen L, et al. Interferon-gamma induces tyrosine phosphorylation of interferon-gamma receptor and regulated association of protein tyrosine kinases, Jak1 and Jak2, with its receptor. J Biol Chem. 1994;269(20):14333–14336. [PubMed] [Google Scholar]
- 129. Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. [DOI] [PubMed] [Google Scholar]
- 130. Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404.e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Benci JL, Xu B, Qiu Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554.e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Dembla V, Somaiah N, Barata P, et al. Prevalence of MDM2 amplification and coalterations in 523 advanced cancer patients in the MD Anderson phase 1 clinic. Oncotarget. 2018;9(69):33232–33243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mao S, Zhang J, Guo Y, et al. Hyperprogression after anti-programmed cell death ligand-1 therapy in a patient with recurrent metastatic urothelial bladder carcinoma following first-line cisplatin-based chemotherapy: a case report. Drug Des Devel Ther. 2019;13:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ju W, Chen S, Wang G, et al. Association between MDM2/MDM4 amplification and PD-1/PD-L1 inhibitors-related hyperprogressive disease: a pan-cancer analysis. J Clin Oncol. 2019;37(suppl 15):2557–2557. [Google Scholar]
- 135. Zou Q, Jin J, Hu H, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat Immunol. 2014;15(6):562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Millis SZ, Ikeda S, Reddy S, et al. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016;2(12):1565–1573. [DOI] [PubMed] [Google Scholar]
- 137. Trujillo JA, Luke JJ, Zha Y, et al. Secondary resistance to immunotherapy associated with β-catenin pathway activation or PTEN loss in metastatic melanoma. J Immuno Ther Cancer. 2019;7(1):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Vidotto T, Saggioro FP, Jamaspishvili T, et al. PTEN-deficient prostate cancer is associated with an immunosuppressive tumor microenvironment mediated by increased expression of IDO1 and infiltrating FoxP3+ T regulatory cells. Prostate. 2019;79(9):969–979. [DOI] [PubMed] [Google Scholar]
- 139. George S, Miao D, Demetri GD, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46(2):197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Roh W, Chen PL, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hopkins AC, Yarchoan M, Durham JN, et al. T cell receptor repertoire features associated with survival in immunotherapy-treated pancreatic ductal adenocarcinoma. JCI Insight. 2018;3(13):e122092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD-1 blockade in resectable lung cancer. New Engl J Med. 2018;378(21):1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. [DOI] [PubMed] [Google Scholar]
- 145. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xue J, Yu X, Xue L, et al. Intrinsic β-catenin signaling suppresses CD8+ T-cell infiltration in colorectal cancer. Biomed Pharmacother. 2019;115:108921. [DOI] [PubMed] [Google Scholar]
- 148. Berraondo P, Ochoa MC, Olivera I, et al. Immune desertic landscapes in hepatocellular carcinoma shaped by β-catenin activation. Cancer Discov. 2019;9(8):1003–1005. [DOI] [PubMed] [Google Scholar]
- 149. Grasso CS, Giannakis M, Wells DK, et al. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8(6):730–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Luke JJ, Bao R, Sweis RF, et al. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res. 2019;25(10):3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Spranger S, Dai D, Horton B, et al. Tumor-residing batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711–723.e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ganesh S, Shui X, Craig KP, et al. RNAi-mediated β-catenin inhibition promotes T cell infiltration and antitumor activity in combination with immune checkpoint blockade. Mol Ther. 2018;26(11):2567–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387(10030):1837–1846. [DOI] [PubMed] [Google Scholar]
- 155. Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. [DOI] [PubMed] [Google Scholar]
- 156. Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lauss M, Donia M, Harbst K, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nature Communications. 2017;8(1):1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kumagai S, Togashi Y, Kamada T, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nature Immunol. 2020;21(11):1346–1358. [DOI] [PubMed] [Google Scholar]
- 160. Lim SY, Lee JH, Gide TN, et al. Circulating cytokines predict immune-related toxicity in melanoma patients receiving anti-PD-1-based immunotherapy. Clin Cancer Res. 2019;25(5):1557–1563. [DOI] [PubMed] [Google Scholar]
- 161. Fujimura T, Sato Y, Tanita K, et al. Serum levels of soluble CD163 and CXCL5 may be predictive markers for immune-related adverse events in patients with advanced melanoma treated with nivolumab: a pilot study. Oncotarget. 2018;9(21):15542–15551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gooden MJM, de Bock GH, Leffers N, et al. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105(1):93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Badalamenti G, Fanale D, Incorvaia L, et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell Immunol. 2019;343:103753. [DOI] [PubMed] [Google Scholar]
- 164. Loi S, Schmid P, Aktan G, et al. Relationship between tumor infiltrating lymphocytes (TILs) and response to pembrolizumab (pembro)+chemotherapy (CT) as neoadjuvant treatment (NAT) for triple-negative breast cancer (TNBC): phase Ib KEYNOTE-173 trial. Ann Oncol. 2019;30:iii2. [Google Scholar]
- 165. Kitano A, Ono M, Yoshida M, et al. Tumour-infiltrating lymphocytes are correlated with higher expression levels of PD-1 and PD-L1 in early breast cancer. ESMO Open. 2017;2(2):e000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sanchez-Canteli M, Granda-Díaz R, del Rio-Ibisate N, et al. PD-L1 expression correlates with tumor-infiltrating lymphocytes and better prognosis in patients with HPV-negative head and neck squamous cell carcinomas. Cancer Immunol Immunother. 2020;69(10):2089–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Howitt BE, Shukla SA, Sholl LM, et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1(9):1319–1323. [DOI] [PubMed] [Google Scholar]
- 168. Howitt BE, Strickland KC, Sholl LM, et al. Clear cell ovarian cancers with microsatellite instability: a unique subset of ovarian cancers with increased tumor-infiltrating lymphocytes and PD-1/PD-L1 expression. Oncoimmunology. 2017;6(2):e1277308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Hendry S, Salgado R, Gevaert T, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: Part 2: TILs in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol. 2017;24(6):311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kato S, Okamura R, Kumaki Y, et al. Expression of TIM3/VISTA checkpoints and the CD68 macrophage-associated marker correlates with anti-PD1/PDL1 resistance: implications of immunogram heterogeneity. Oncoimmunology. 2020;9(1):1708065–1708065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.