Abstract
Purpose:
While numerous biology-driven subtypes have been described in metastatic castration-resistant prostate cancer (mCRPC), unsupervised molecular subtyping based on gene expression has been less studied, especially using large cohorts. Thus, we sought to identify the intrinsic molecular subtypes of mCRPC and assess molecular and clinical correlates in the largest combined cohort of mCRPC samples with gene expression data available to date.
Methods:
We combined and batch effect corrected gene expression data from four mCRPC cohorts from the Fred Hutchinson Cancer Research Center (FHCRC, N=157), a small-cell neuroendocrine prostate cancer (SCNC)-enriched cohort from Weill Cornell Medicine (WCM, N=49), and cohorts from the Stand Up 2 Cancer/Prostate Cancer Foundation East Coast Dream Team (ECDT, N=266) and the West Coast Dream Team (WCDT, N=162).
Results:
Hierarchical clustering of RNA-seq data from these 634 mCRPC samples identified two distinct adenocarcinoma subtypes, one of which (adeno-immune) was characterized by higher gene expression of immune pathways, higher CIBERSORTx immune scores, diminished ASI benefit, and non-lymph node metastasis tropism compared to an adeno-classic subtype. We also identified two distinct subtypes with enrichment for a neuroendocrine (NE) phenotype, including an NE-liver subgroup characterized by liver metastasis tropism, PTEN loss, and APC and SPOP mutations compared to an NE-classic subgroup.
Conclusion:
Our results emphasize the heterogeneity of mCRPC beyond currently accepted molecular phenotypes, and suggests that future studies should consider incorporating transcriptome-wide profiling in order to better understand how these differences impact treatment responses and outcomes.
INTRODUCTION
Prostate cancer is a clinically and molecularly heterogenous disease. Outcomes for patients can vary substantially, as the presentation of prostate cancer can range from indolent tumors to highly aggressive and lethal disease. The molecular heterogeneity of the disease is reflected in the panoply of DNA alterations in key driver genes initially identified in localized prostate cancer(1–3). RNA-based gene expression profiling approaches further demonstrated a division between luminal-like or basal-like subtypes in prostate tumors(4–6) that have different prognoses and importantly may respond differently to anti-androgen therapy, the backbone of systemic therapy in prostate cancer. These clinically important subtypes are now being incorporated into national clinical trials to improve patient selection for various standard of care and experimental therapies.
Metastatic prostate cancer is a very different disease than localized prostate cancer; this is particularly true for metastatic castration-resistant prostate cancer (mCRPC), which represents end stage disease with poor outcomes. The genomic landscape of mCRPC is characterized by a higher frequency of DNA alterations across many oncogenic drivers, especially in the androgen receptor (AR) due to the selective pressure of therapy(7–10). However, these DNA alterations have failed to capture the full heterogeneity of the disease and, with the exception of DNA-repair deficiencies and microsatellite instability in a minority of cases, cannot be used to guide clinical decision-making. In addition to DNA alterations, important transcriptional changes also take place during disease progression to mCRPC. Transcriptional changes have been identified on the basis of histologic differences between subsets of mCRPC; a number of studies have identified lineage plasticity, most commonly towards a neuroendocrine (NE) lineage, in response to prolonged AR-targeted therapy through which prostate cancer become independent of AR signaling for proliferation(10–14). Indeed, more recent studies focusing on differences in the neuroendocrine (NE) and AR signaling axes between mCRPC samples have defined five different subgroups: AR-high prostate cancer (ARPC), AR-low prostate cancer (ARLPC), amphicrine tumors composed of cells co-expressing AR and NE genes (AMPC), double-negative tumors (i.e., AR–/NE–; DNPC), and tumors with small cell or NE gene expression without AR activity (SCNC)(15). Lineage plasticity without NE differentiation may also be present towards a more stem cell phenotype through epithelial to mesenchymal transition (EMT) or towards a gastrointestinal lineage(16). Additional RNA profiling studies have demonstrated that transcriptional subtypes that exist in localized prostate cancer may exist in mCRPC; a recent study demonstrated that luminal and basal subtypes in mCRPC predict response to AR signaling inhibitors (ASIs), analogous to localized prostate cancer(17).
While previous subtyping studies have revealed the biological heterogeneity of mCRPC, comprehensive unsupervised transcriptome-wide clustering approaches have seen limited use in metastatic samples to date, likely due to the lack of large cohorts of metastatic disease with molecular profiling, which requires metastatic tissue biopsies that are logistically difficult to obtain. Compared to localized prostate cancer, in which cohorts with thousands of patients samples have been published(18), the largest mCRPC cohorts with next-generation sequencing and clinical outcomes are comprised of <150 (7,19,20). To allow for unsupervised data-driven approaches that may reveal key information which have not yet been hypothesized and tested, we recently compiled publicly available RNA-seq from mCRPC samples to form the largest dataset of its kind(17) which represents a unique opportunity for identifying the intrinsic molecular subtypes of mCRPC and assessing for molecular and clinical correlates.
METHODS
mCRPC Clinical Cohorts
We combined and batch effect corrected data from four mCRPC cohorts to assess intrinsic subtypes, and analyzed RNA, DNA, and overall survival as previously published(17). The mCRPC cohorts used were from the Fred Hutchinson Cancer Research Center (FHCRC, N=157)(21), a small-cell/neuroendocrine prostate cancer (SCNC)-enriched cohort from Weill Cornell Medicine (WCM, N=49)(14), an cohort from the Stand Up 2 Cancer/Prostate Cancer Foundation (SU2C/PCF) East Coast Dream Team (ECDT, N=266)(8,19) and the West Coast Dream Team (WCDT, N=162)(7,10,22).
Bioinformatics
Normalized gene expression, mutation calls, and copy number for FHCRC, WCM, and the ECDT were obtained directly from cBioPortal(23). The same data from the WCDT were obtained from prior publications(7,10,17). RNA-seq batch correction was performed as previously described(17). In our primary analysis of the RNA sequencing data, we included genes that had expression data in all samples. Hierarchical clustering was performed on the top 1000 genes with the highest variance across all the samples. Spearman’s correlation was used as the distance function, and default “ward.D” agglomeration method was used. Using this method, we identified four distinct subtypes based on the gene expression patterns. Clinical and pathologic variables were obtained from the original publications of these cohorts. Pathway scores using the Hallmark Pathways from MSigDb(24) were calculated using gene set variation analysis (GSVA)(25). Oncogene activating alterations (amplification and/or mutation) and tumor suppressor bi-allelic inactivating alterations (copy number loss and/or mutation) were defined as previously published(17). CIBERSORTx was used to calculate an absolute immune score(26).
Statistics
Statistical testing between the subtypes and continuous variables was performed using a Wilcoxon rank-sum test. Statistical testing between the subtypes and categorical variables was performed using Fisher’s exact test. Survival analyses were performed using Cox regression and visualized using the Kaplan-Meier method. All statistical testing was performed in R version 4.0.4. All statistical testing was two-sided, and a p-value < 0.05 was considered statistically significant.
Data Availability Statement
Only previously published data were used for this study. FHCRC, WCM, and the ECDT were downloaded from www.cbioportal.org. WCDT genomics data are available from dbGaP (phs001648) and EGA (EGAD00001009065).
RESULTS
Distinct Gene Expression Clusters
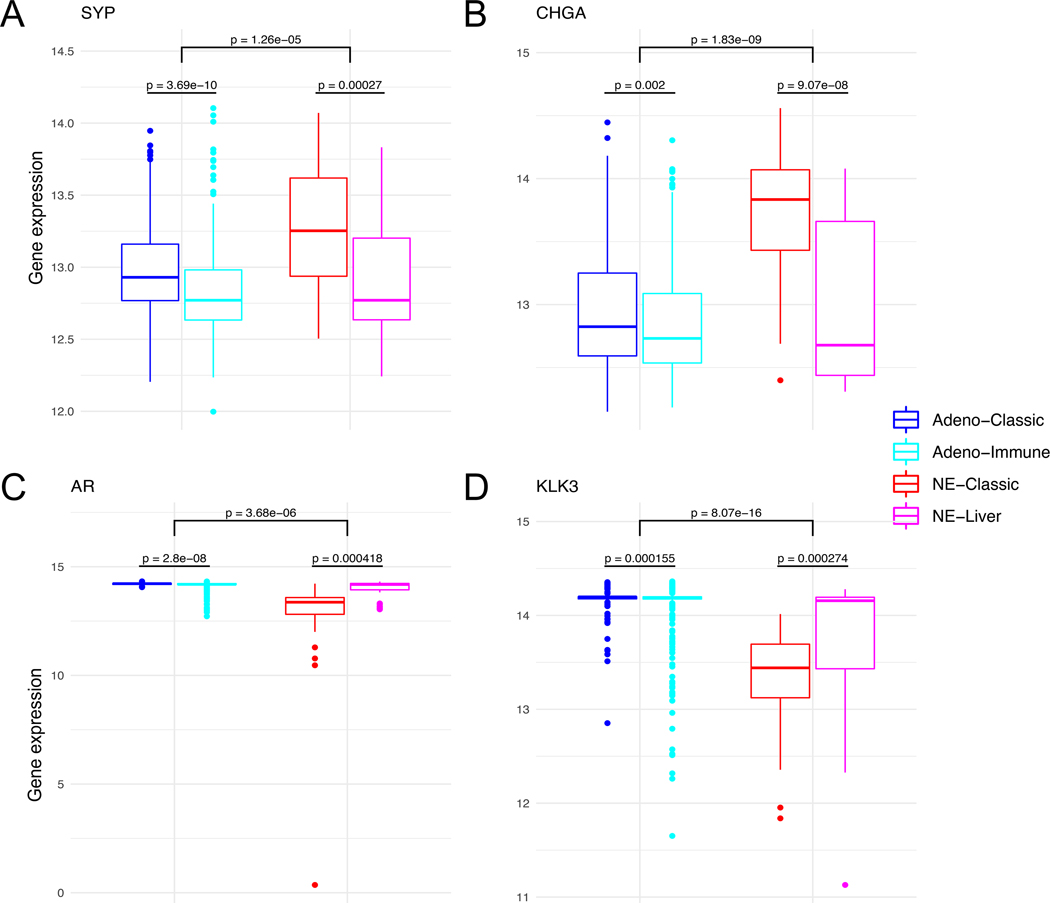
In order to identify intrinsic RNA subtypes within mCRPC, we performed hierarchical clustering on 634 total mCRPC samples with RNA sequencing, using the top 1000 genes with the highest variance (Figure 1). This resulted in four distinct subtypes based on RNA expression patterns. These subtypes were not associated with the cohort of origin, supporting the effectiveness of the batch correction. However, there visually did appear to be associations with both biopsy site and small-cell neuroendocrine cancer (SCNC, hereafter referring to the phenotypic definition as originally identified in each cohort). In the two subtypes associated with SCNC, the rate of SCNC was higher in one (78% SCNC, this RNA subtype hereafter referred to as NE-classic) vs. the other (29% SCNC, with all of the samples originating from a liver biopsy, this RNA subtype hereafter referred to as NE-liver). This was reflected in gene expression patterns of the neuroendocrine markers SYP and CHGA which were higher in the NE subtypes, more so in NE-classic (Figure 2A-B, Supplemental Table 1). This was also reflected in AR and KLK3 (encoding PSA) gene expression, with the lowest expression in NE-classical, and intermediate expression in NE-liver subtype (Figure 2C-D, Supplemental Table 1).
Figure 1.
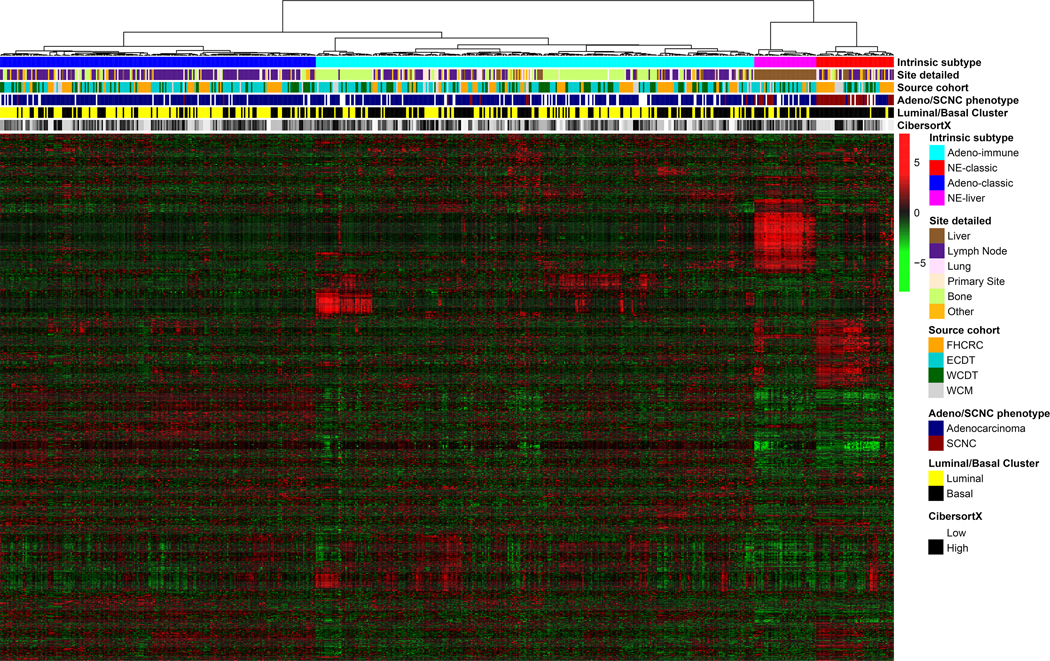
Heatmap of FHCRC, ECDT, WCDT, and WCM data showing gene expression profiles of the top 1000 differentially expressed genes (rows) in 634 patients (columns). Both patients and genes are clustered using unsupervised hierarchical clustering. Red represents high expression, and green represents low expression.
Figure 2.
Boxplots show that SYP (A), CHGA (B), AR (C) and KLK3 (D) expression across RNA subtypes. Gene expression shows the log transformed batch corrected rank.
Biological Pathways
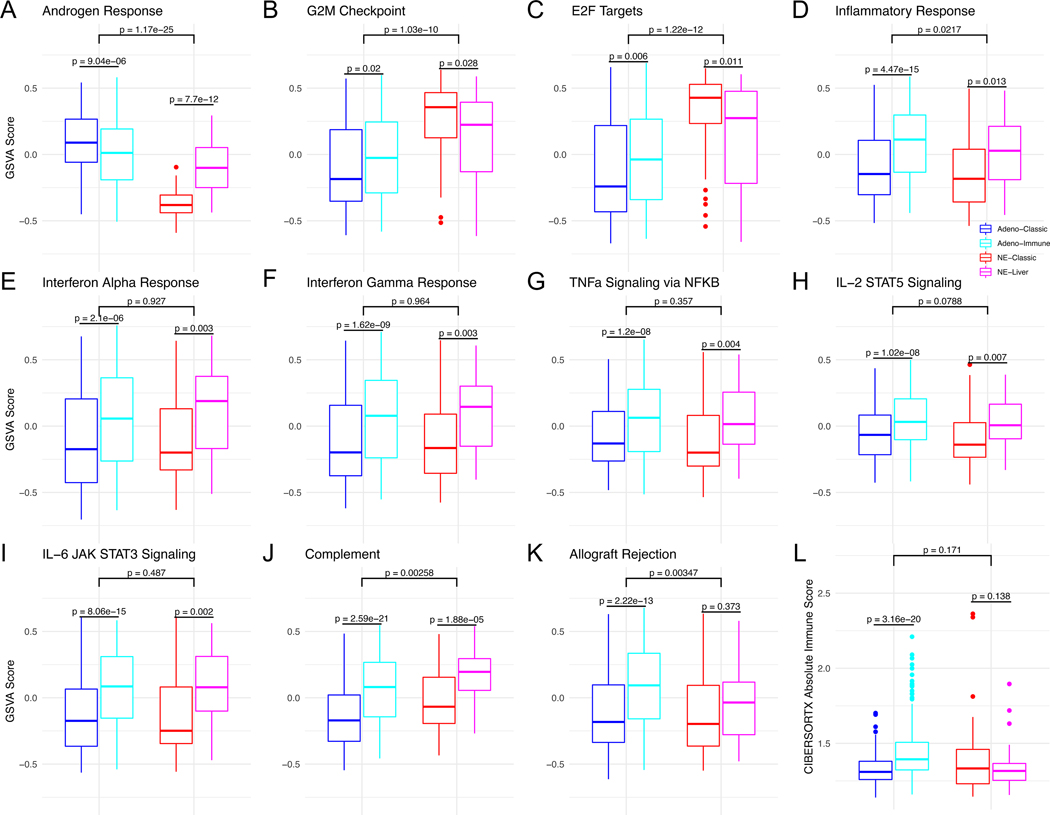
Next, we sought to understand the pathway level differences between the novel RNA subtypes using the MSigDb Hallmark pathways. We first examined the AR signaling pathway, where, as expected, the two adenocarcinoma subtypes had highest expression, followed by NE-liver, and then NE-classic, consistent with the AR and KLK3 expression above (Figure 3A, Supplemental Table 2). SCNC is also known to be highly proliferative, and this characteristic was reflected in NE-classic having higher expression of proliferative pathways such as the G2M checkpoint and E2F targets compared to the adenocarcinoma subtypes, with NE-liver between the two (Figures 3B and 3C, Supplemental Table 2).
Figure 3.
Boxplots showing GSVA scores across RNA subtypes and Hallmark pathways (A-K) and the CibersortX immune score (L).
There was a small but statistically significant difference between the two RNA intrinsic adenocarcinoma subtypes for AR signaling (Figure 3A, Supplemental Table 2), consistent with differences noted with previous hypothesis-driven approaches(15,17). However, the most prominent difference was in immune pathways, with consistently higher expression of all eight immune-related hallmark pathways (Figure 3D–3K, Supplemental Table 2) in one RNA subtype (hereafter referred to adeno-immune) compared to the other (hereafter referred to adeno-classic). The results using the CIBERSORTx overall immune score were consistent with the pathway analyses, with overall immune scores higher in adeno-immune vs. adeno-classic (Figure 3L, Supplemental Table 2). These data suggest that there may be two immunogenically distinct subgroups of mCRPC adenocarcinoma. Comparison between the two RNA NE subtypes demonstrated that the NE-liver subtype exhibited higher expression of 7 out of the 8 immune-related hallmark pathways (all except for the allograft rejection signature) (Figures 3D-K, Supplemental Table 2). While there was no significant difference in the CIBERSORTx overall immune score between the two NE subtypes (Figure 3L, Supplemental Table 2), the pathway analysis suggests that there are qualitative differences in the tumor immunologic response between the two NE subtypes.
DNA Alterations
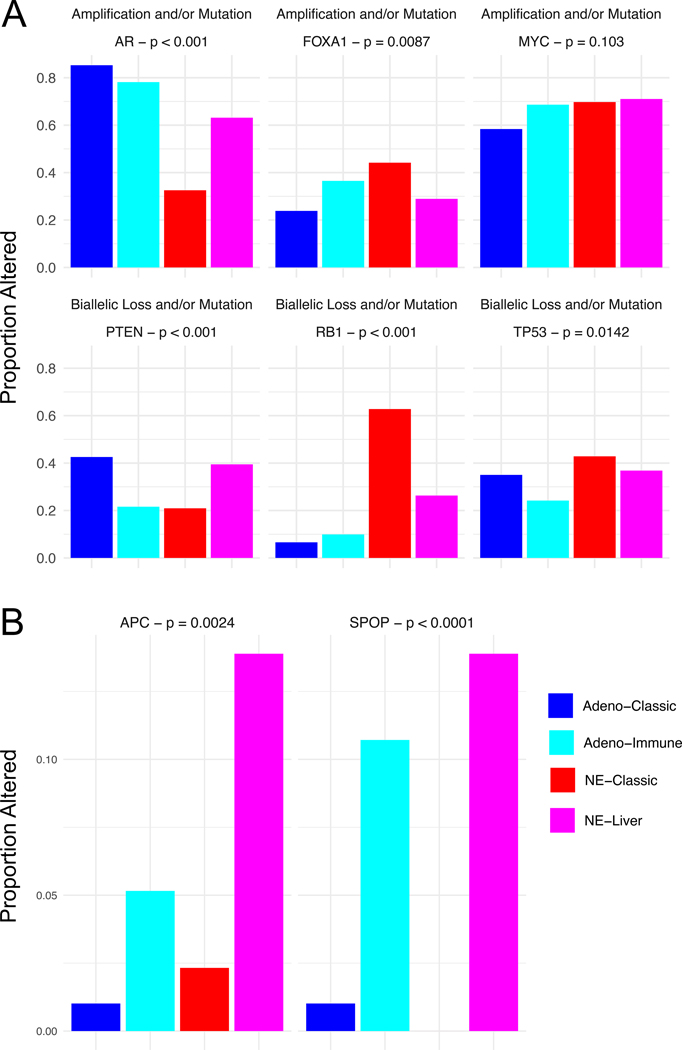
We next sought to investigate DNA alterations in key prostate cancer oncogenic driver genes. Alterations in the tumor suppressors RB1, PTEN, and TP53 are all known to be associated with aggressive prostate cancer and SCNC(27). Of the two SCNC-enriched RNA subtypes, the NE-classic was more enriched for RB1 alterations (63% in NE-classic vs. 26% in NE-liver) and the NE-liver for PTEN alterations (21% in NE-classic vs. 39% in NE-liver), with similar TP53 alteration rates (43% in NE-classic vs. 37% in NE-liver) (Figure 4A). While the rate of AR alterations was lower in both NE subtypes than in the adenocarcinoma subtypes, there were differences between the NE subtypes (33% in NE-classic vs. 63% in NE-liver) consistent with the AR and KLK3 expression data. There was also a difference in FOXA1 alteration rates (44% in NE-classic vs. 29% in NE-liver), but not MYC (70% in NE-classic vs. 71% in NE-liver) (Figure 4A, Supplemental Table 3). With regards to the adenocarcinoma subtypes, there were differences in PTEN alterations (43% in adeno-classic vs. 22% in adeno-immune). Mutation rates for APC and SPOP differed between both NE subtypes (APC 2.3% in NE-classic vs. 14% in NE-liver; SPOP 0% in NE-classic vs. 14% in NE-liver) and adenocarcinoma subtypes (APC 1% in adeno-classic vs. 5.2% in adeno-immune; SPOP 1% in adeno-classic vs. 11% in adeno-immune; Figure 4B, Supplemental Table 3). We found no differences between the subtypes in tumor mutational burden (as a surrogate for MSI/dMMR) or BRCA2 deep deletion or mutations, which are the two genomic markers currently most used for treatment selection in mCRPC (for immunotherapy or PARP-inhibitor therapy, respectively; Supplemental Figure 1).
Figure 4.
(A) Proportion of samples with an amplification and/or mutation (for oncogenes) or bi-allelic loss and/or mutation (for tumor suppressor genes) for each RNA subtype. (B) Proportion of samples with a mutation for each subtype.
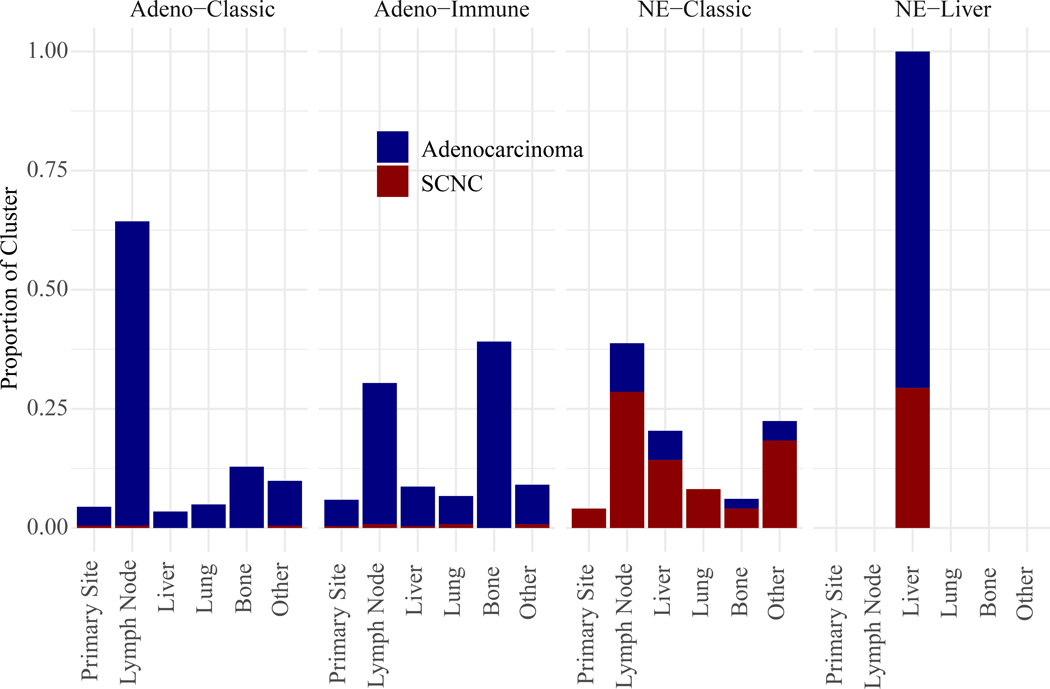
Tumor Site
The site of metastasis was different between the NE subtypes, with all of the NE-liver tumors coming from liver metastases, whereas the distribution was more evenly split in the NE-classic tumors between the different metastatic locations (Figure 5, Supplemental Table 4). Interestingly, a difference between the two adenocarcinoma RNA subtypes was also observed, with adeno-immune being enriched for distant metastases while adeno-classic was enriched for lymph node (LN) metastases. The gene expression from bulk RNA-sequencing of tumor biopsies is a mix of the tumor cells and the surrounding non-tumor tissue which will influence comparison across tumor sites. However, immune-related measures should favor LNs preferentially, as the non-tumor background is mostly immune cells, which is in contrast to what we observe for adeno-immune vs. adeno-classic, suggesting that there may indeed be a difference in immunogenicity between the two adenocarcinoma RNA intrinsic subtypes.
Figure 5.
Proportion of each RNA subtype originating from different biopsy sites, as well as the proportion of each subtype from adenocarcinoma vs. SCNC.
Clinical Outcomes
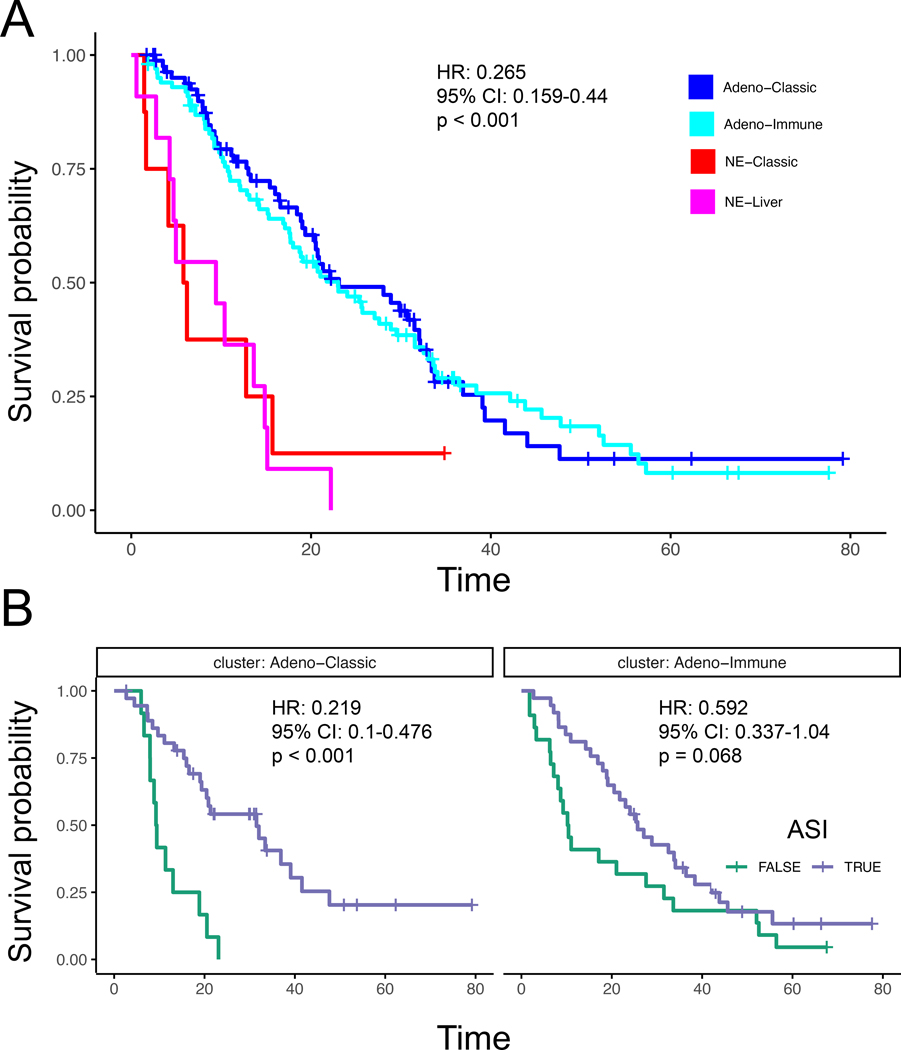
The association between different RNA subtypes and clinical outcomes was also investigated. As expected, the two adenocarcinoma subtypes had significantly improved overall survival compared to the NE subtypes (Hazard Ratio (HR) 0.265 (95% CI 0.159−0.44); p < 0.001; Figure 6A), without a difference between them. However, when we account for ASI therapy, we observed that the adeno-classic subtype showed a significant benefit from ASI therapy (HR 0.219 (95% CI 0.1−0.476); p < 0.001) after the biopsy, whereas the adeno-immune subtype had a weaker trend in the same direction that was not significant (HR 0.592 (95% CI 0.337−1.04); p = 0.068; Figure 6B). While the NE-classic and NE-liver RNA intrinsic subtypes also showed similar worse survival, there were different proportions of patients that were not originally identified as SCNC in these subtypes.
Figure 6.
(A) Survival compared between subtypes Adeno-Classic, Adeno-Immune, NE-Classic, and NE-Liver. (B) Survival compared between patients with and without ASI for Adeno-Classic and Adeno-Immune.
DISCUSSION
Data-driven and unsupervised approaches to derive intrinsic subtypes can reveal differences not immediately apparent based on hypothesis-driven approaches based on prior biological knowledge. Herein, we present the largest intrinsic RNA-based molecular subtyping of mCRPC in 634 samples with RNA-seq. We find two distinct adenocarcinoma subtypes, one of which (adeno-immune) is characterized by higher expression of immune pathways, higher CIBERSORTx immune scores, a lower hazard ratio for ASI benefit, and non-LN metastasis tropism compared to an adeno-classic subtype. We also identify two distinct NE subtypes, including an NE-liver subgroup characterized by liver metastasis tropism, PTEN loss, and APC and SPOP mutations compared to an NE-classic subgroup.
SCNC is an aggressive, androgen-independent subtype of mCRPC that can either be present de novo, or more commonly emerge during the course of treatment via lineage plasticity(12–14,27,28). SCNC has been shown to exhibit distinct transcriptomic and epigenomic patterns, and is genomically characterized by loss of the tumor suppressors PTEN, RB1, and TP53(12–14,27–31). When we compare our intrinsic RNA subtypes to the five subtypes previously described by AR and NE markers(15), theNE-classic subtype likely corresponds well to the SCNC (AR-, NE+) subgroup. The NE-liver subgroup shares similarities to some other subgroups described previously, such as AMPC (AR+,NE+) or intermediate atypical carcinoma (IAC)(32), a histologic subgroup between adenocarcinoma and SCNC. It is also possible that it represents a transition state between adenocarcinoma and emerging SCNC, which on bulk sequencing would have gene expression between the two, or specific biology driving the aggressive tumors that metastasize to the liver. Indeed, both intrinsic RNA subtypes enriched for SCNC tumors identify tumors not originally diagnosed as SCNC, especially in the NE-liver group. However, the prognosis of both subtypes is equally poor, and the transcriptional similarities would suggest that similar therapeutic approaches could be tried. Interestingly, the difference in RB1/PTEN alterations between the RNA NE subtypes also suggest that these alterations are not equivalent. As PTEN-altered tumors begin to have new treatment options(33), the management of NE-liver tumors may diverge from NE-classic. We did not observe a distinct double negative cluster in this unsupervised intrinsic clustering resulting in four main clusters(15), and such tumors may be grouped in with the other subtypes.
The differences between the intrinsic adenocarcinoma subtypes of mCRPC are also potentially clinically important. Interestingly, the two intrinsic adenocarcinoma subtypes do not differ via an AR high vs. low dichotomy described in these previous subtyping efforts(15,17). mCRPC has traditionally been thought of as not particularly immunogenic, with poor response rates to immune checkpoint blockade(34,35). Our adeno-immune subtype suggests that not all tumors may be equally quiescent, and suggest potential biomarker-driven approaches for immunotherapies in mCRPC. Despite analyzing the largest harmonized cohort of mCRPC tumors with gene expression data, a limitation to this study is that that limited clinical treatment annotation and follow-up data were only available for a subset of the patients, and did not allow for analysis of association between subtypes and treatments other than AR targeted therapy.
Ultimately, our results emphasize the molecular heterogeneity of mCRPC. Our unbiased clustering approach confirms key findings identified through hypothesis-driven approaches in the field, such as the existence of distinct subgroups driven by AR signaling versus neuroendocrine (NE) features; however, we also discover that AR-driven adenocarcinoma subgroups can be differentiated by signaling along immune pathways, and that NE subgroups may be grouped biologically, with distinctions in the site of metastases and DNA alterations. In total, our study suggests that future studies and clinical trials should consider transcriptome-wide profiling in order to better understand how these differences impact treatment and outcomes.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE
Data-driven unsupervised molecular subtyping of metastatic castration-resistant prostate cancer (mCRPC) can reveal insights into the disease heterogeneity observed clinically. Thus, we sought to leverage large sample cohorts with harmonized gene expression data to identify the intrinsic molecular subtypes of mCRPC and assess molecular and clinical correlates. Analysis of 634 mCRPC samples identified four distinct subtypes, two adenocarcinoma-enriched and two enriched for a neuroendocrine (NE) phenotype. One of the adenocarcinoma subtypes (adeno-immune) was characterized by higher gene expression of immune pathways, diminished benefit of androgen-signaling inhibitors, and non-lymph node metastasis tropism compared to an adeno-classic subtype. An NE-liver subtype was characterized by liver metastasis tropism, PTEN loss, and APC and SPOP mutations compared to an NE-classic subtype. Both NE-subtypes had worse survival than the adenocarcinoma subtypes. Our results emphasize the molecular heterogeneity of mCRPC and supports incorporating transcriptome-wide profiling in future studies investigating how these subtypes may influence treatment outcomes.
ACKNOWLEDGEMENTS
This research was supported by a Stand Up To Cancer-Prostate Cancer Foundation Prostate Cancer Dream Team Award (SU2C-AACR-DT0812 to E. J. Small) and by the Movember Foundation. Stand Up To Cancer is a division of the Entertainment Industry Foundation. This research grant was administered by the American Association for Cancer Research, the scientific partner of SU2C. M. Sjöström was supported by the Swedish Research Council (Vetenskapsrådet) with grant number 2018-00382, the Swedish Society of Medicine (Svenska Läkaresällskapet) and a Prostate Cancer Foundation Young Investigator Award. D. A. Quigley was funded by a Prostate Cancer Foundation Young Investigator Award and a BRCA Foundation Young Investigator Award. Additional funding was provided by a UCSF Benioff Initiative for Prostate Cancer Research award. S. G. Zhao was funded by the Department of Defense (PC190039) and the National Institutes of Health (1DP2CA271832). J. M. Lang and S. G. Zhao are supported by Department of Defense (PC200334) and the University of Wisconsin Carbone Cancer Center Support Grant (P30 CA014520).
Footnotes
CONFLICT OF INTERESTS
S.G.Z. has patent applications with Decipher Biosciences on molecular signatures in prostate cancer unrelated to this work, and a spouse who is an employee of Exact Sciences. J.M.L. has consulted or advised for Sanofi, Immunomedics, Janssen, Pfizer/Astellas, 4D Pharma, has received research funding from Medivation, Agensys, GlaxoSmithKline, Immunomedics, Bristol Myers Squibb, Janssen, and has a patent licensed by Salus Discovery LLC. M.B. has a spouse who is an employee of Luminex Corporation. K.T.H. has a spouse who is an employee of Epic Systems. E.J.S. reports other support from Fortis, Harpoon, Teon, Janssen, Johnson and Johnson, Ultragenyx outside the submitted work. The remaining authors declare no potential conflicts of interest.
REFERENCES
- 1.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015;163(4):1011–25 doi 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerhauser C, Favero F, Risch T, Simon R, Feuerbach L, Assenov Y, et al. Molecular Evolution of Early-Onset Prostate Cancer Identifies Molecular Risk Markers and Clinical Trajectories. Cancer Cell 2018;34(6):996–1011 e8 doi 10.1016/j.ccell.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541(7637):359–64 doi 10.1038/nature20788. [DOI] [PubMed] [Google Scholar]
- 4.Zhao SG, Chen WS, Das R, Chang SL, Tomlins SA, Chou J, et al. Clinical and Genomic Implications of Luminal and Basal Subtypes Across Carcinomas. Clin Cancer Res 2019;25(8):2450–7 doi 10.1158/1078-0432.CCR-18-3121. [DOI] [PubMed] [Google Scholar]
- 5.Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol 2017;3(12):1663–72 doi 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, et al. Integrated Classification of Prostate Cancer Reveals a Novel Luminal Subtype with Poor Outcome. Cancer Res 2016;76(17):4948–58 doi 10.1158/0008-5472.CAN-16-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018;174(3):758–69 e9 doi 10.1016/j.cell.2018.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28 doi 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viswanathan SR, Ha G, Hoff AM, Wala JA, Carrot-Zhang J, Whelan CW, et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell 2018;174(2):433–47 e19 doi 10.1016/j.cell.2018.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjostrom M, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet 2020. doi 10.1038/s41588-020-0648-8. [DOI] [PMC free article] [PubMed]
- 11.Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25(23):6916–24 doi 10.1158/1078-0432.CCR-19-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J Clin Oncol 2018;36(24):2492–503 doi 10.1200/JCO.2017.77.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal RR, Quigley DA, Huang J, Zhang L, Beer TM, Rettig MB, et al. Whole Genome and Transcriptional Analysis of Treatment-Emergent Small Cell Neuroendocrine Prostate Cancer Demonstrates Intra-Class Heterogeneity. Mol Cancer Res 2019. doi 10.1158/1541-7786.MCR-18-1101. [DOI] [PMC free article] [PubMed]
- 14.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22(3):298–305 doi 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labrecque MP, Coleman IM, Brown LG, True LD, Kollath L, Lakely B, et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest 2019;129(10):4492–505 doi 10.1172/JCI128212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla S, Cyrta J, Murphy DA, Walczak EG, Ran L, Agrawal P, et al. Aberrant Activation of a Gastrointestinal Transcriptional Circuit in Prostate Cancer Mediates Castration Resistance. Cancer Cell 2017;32(6):792–806 e7 doi 10.1016/j.ccell.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aggarwal R, Rydzewski NR, Zhang L, Foye A, Kim W, Helzer KT, et al. Prognosis Associated With Luminal and Basal Subtypes of Metastatic Prostate Cancer. JAMA Oncol 2021. doi 10.1001/jamaoncol.2021.3987. [DOI] [PMC free article] [PubMed]
- 18.Zhao SG, Lehrer J, Chang SL, Das R, Erho N, Liu Y, et al. The Immune Landscape of Prostate Cancer and Nomination of PD-L2 as a Potential Therapeutic Target. J Natl Cancer Inst 2019;111(3):301–10 doi 10.1093/jnci/djy141. [DOI] [PubMed] [Google Scholar]
- 19.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116(23):11428–36 doi 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rydzewski NR, Peterson E, Lang JM, Yu M, Laura Chang S, Sjostrom M, et al. Predicting cancer drug TARGETS - TreAtment Response Generalized Elastic-neT Signatures. NPJ Genom Med 2021;6(1):76 doi 10.1038/s41525-021-00239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22(4):369–78 doi 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen WS, Aggarwal R, Zhang L, Zhao SG, Thomas GV, Beer TM, et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2019. doi 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7 doi 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37(7):773–82 doi 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wang Y, Ci X, Choi SYC, Crea F, Lin D, et al. Molecular events in neuroendocrine prostate cancer development. Nat Rev Urol 2021;18(10):581–96 doi 10.1038/s41585-021-00490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clin Cancer Res 2019. doi 10.1158/1078-0432.CCR-19-1423. [DOI] [PMC free article] [PubMed]
- 29.Aparicio AM, Shen L, Tapia EL, Lu JF, Chen HC, Zhang J, et al. Combined Tumor Suppressor Defects Characterize Clinically Defined Aggressive Variant Prostate Cancers. Clin Cancer Res 2016;22(6):1520–30 doi 10.1158/1078-0432.CCR-15-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beltran H, Romanel A, Conteduca V, Casiraghi N, Sigouros M, Franceschini GM, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020;130(4):1653–68 doi 10.1172/JCI131041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berchuck JE, Baca SC, McClure HM, Korthauer K, Tsai HK, Nuzzo PV, et al. Detecting Neuroendocrine Prostate Cancer Through Tissue-Informed Cell-Free DNA Methylation Analysis. Clin Cancer Res 2021. doi 10.1158/1078-0432.CCR-21-3762. [DOI] [PMC free article] [PubMed]
- 32.Small EJ, Aggarwal RR, Friedl V, Weinstein A, Thomas GV, True LD, et al. Intermediate atypical carcinoma (IAC): A discrete subtype of metastatic castration-resistant prostate cancer (mCRPC) suggesting that treatment-associated small cell/neuroendocrine prostate cancer (t-SCNC) may evolve from mCRPC adenocarcinoma (adeno)—Results from the SU2C/PCF/AACR West Coast Prostate Cancer Dream Team (WCDT). Journal of Clinical Oncology 2020;38(6_suppl):158- doi 10.1200/JCO.2020.38.6_suppl.158. [DOI] [Google Scholar]
- 33.Sweeney C, Bracarda S, Sternberg CN, Chi KN, Olmos D, Sandhu S, et al. Ipatasertib plus abiraterone and prednisolone in metastatic castration-resistant prostate cancer (IPATential150): a multicentre, randomised, double-blind, phase 3 trial. Lancet 2021;398(10295):131–42 doi 10.1016/S0140-6736(21)00580-8. [DOI] [PubMed] [Google Scholar]
- 34.Beer TM, Kwon ED, Drake CG, Fizazi K, Logothetis C, Gravis G, et al. Randomized, Double-Blind, Phase III Trial of Ipilimumab Versus Placebo in Asymptomatic or Minimally Symptomatic Patients With Metastatic Chemotherapy-Naive Castration-Resistant Prostate Cancer. J Clin Oncol 2017;35(1):40–7 doi 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- 35.Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJ, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184–043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol 2014;15(7):700–12 doi 10.1016/S1470-2045(14)70189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Only previously published data were used for this study. FHCRC, WCM, and the ECDT were downloaded from www.cbioportal.org. WCDT genomics data are available from dbGaP (phs001648) and EGA (EGAD00001009065).