Abstract
To withstand the high intracellular pressure, the cell wall of most bacteria is stabilized by a unique cross-linked biopolymer called murein or peptidoglycan. It is made of glycan strands [poly-(GlcNAc-MurNAc)], which are linked by short peptides to form a covalently closed net. Completely surrounding the cell, the murein represents a kind of bacterial exoskeleton known as the murein sacculus. Not only does the sacculus endow bacteria with mechanical stability, but in addition it maintains the specific shape of the cell. Enlargement and division of the murein sacculus is a prerequisite for growth of the bacterium. Two groups of enzymes, hydrolases and synthases, have to cooperate to allow the insertion of new subunits into the murein net. The action of these enzymes must be well coordinated to guarantee growth of the stress-bearing sacculus without risking bacteriolysis. Protein-protein interaction studies suggest that this is accomplished by the formation of a multienzyme complex, a murein-synthesizing machinery combining murein hydrolases and synthases. Enlargement of both the multilayered murein of gram-positive and the thin, single-layered murein of gram-negative bacteria seems to follow an inside-to-outside growth strategy. New material is hooked in a relaxed state underneath the stress-bearing sacculus before it becomes inserted upon cleavage of covalent bonds in the layer(s) under tension. A model is presented that postulates that maintenance of bacterial shape is achieved by the enzyme complex copying the preexisting murein sacculus that plays the role of a template.
Reinforcement for the Wall
To enable the cell wall of bacteria to resist the intracellular pressure of several atmospheres and to maintain a specific cell shape, it needs to be reinforced by a strong scaffolding. Interestingly, the murein (peptidoglycan) of eubacteria (116, 126) and the pseudomurein of archaebacteria (60, 82) are quite analogous structures. In both cases, two chemically different threads, glycan strands and peptide chains, form a tight fabric. Unlike chitin, murein and pseudomurein are held together by covalent bonds in two or even three dimensions, resulting in a mono- or multilayered structure (Fig. 1). The meshwork character makes both compounds ideal structures to protect the cytoplasmic membrane against cell turgor.
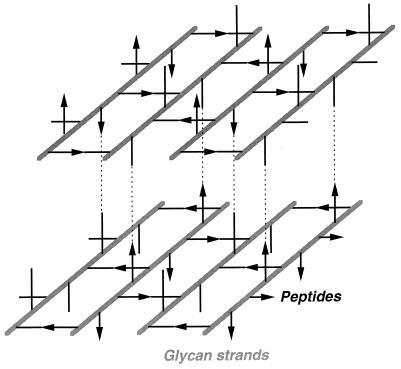
FIG. 1.
Architecture of the murein layer. The drawing shows two murein layers and indicates (by dotted lines) how they can be stacked on one another to form a multilayered murein. Glycan strands are represented by solid bars. The peptide cross bridges are indicated by black lines (acceptor stem peptides) and black arrows (donor stem peptides).
MUREIN, A CROSS-LINKED BIOPOLYMER
Structure
In murein (Fig. 2) the glycan strands are made of the aminosugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), which are linked together by β-1,4 glycosidic bonds. The presence of the lactyl group of the muramic acid (see Fig. 4A) allows the covalent attachment of peptides, which can be cross-linked to form the characteristic net structure of murein. Whereas the chemistry of the glycan strands shows only few variations among different bacteria, such as O or N acetylation, the peptides vary a lot (126, 134). Importantly, a dibasic amino acid has to be present to enable the cross-linking peptide bond to be formed. In Escherichia coli, this is meso-diaminopimelic acid, an intermediate in the biosynthetic pathway leading to lysine. The stem peptides linked to the glycan strands are arranged helically along the strand, protruding in all directions and forming angles to one another of about 90° (97) (Fig. 1). Therefore, in a monolayered murein, only every second peptide can be cross-linked by turns to the right and left. Determination of the degree of cross-linkage indicates that under certain growth conditions all peptides in the plane of the glycan strands can be cross-linked (50).
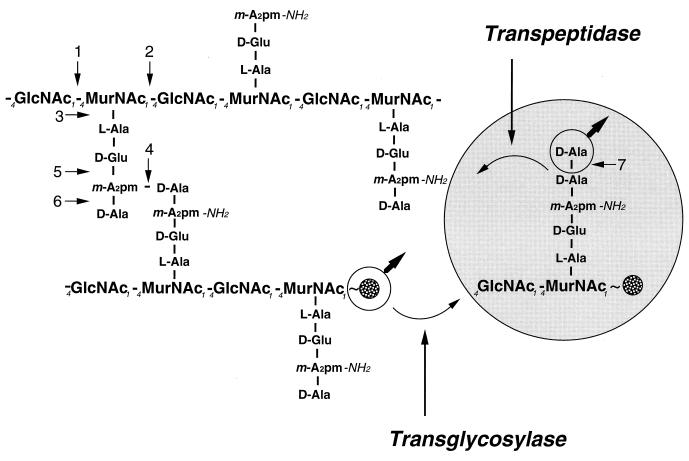
FIG. 2.
Chemistry of the murein of E. coli. A section of the murein of E. coli is shown. On the right is shown how a new murein precursor linked to the undecaprenyl pyrophosphate group (represented by a spotted circle) is linked to the preexisting murein by the formation of two bonds. Concomitant with cleavage of the d,d-peptide bond between the two d-Ala residues of the pentapeptide precursor, a transpeptidase forms a d,d-peptide bond between the carboxyl group of the penultimate d-Ala of the precursor and the epsilon amino group of a diaminopimelic acid residue present in a peptide moiety of the growing murein sacculus. A transglycosylase splits the pyrophosphate bond between the undecaprenyl group and the MurNAc of a nascent glycan strand in the sacculus and forms a glycosidic bond to the hydroxyl group at carbon 4 of the GlcNAc of the precursor molecule. The numbers point to the bonds cleaved by specific murein hydrolases present in E. coli: 1, N-acetylglucosaminidase; 2, lytic transglycosylase; 3, N-acetylmuramyl-l-alanine amidase; 4, d,d-endopeptidase; 5, γ-d-glutamyl-l-diaminopimelic acid endopeptidase; 6, l,d-carboxypeptidase; 7, d,d-carboxypeptidase; m-A2pm, meso-diaminopimelic acid.
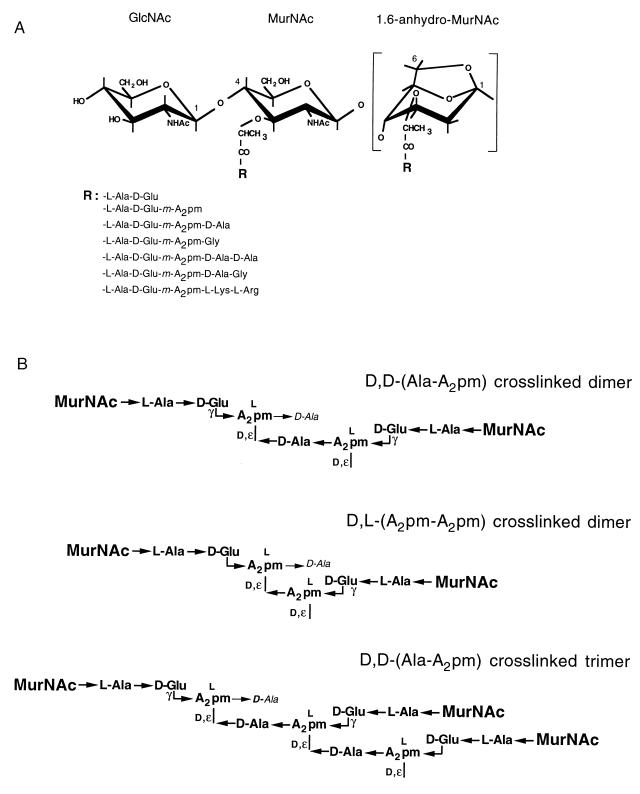
FIG. 4.
Muropeptide structures. (A) Monomeric muropeptide structures identified from the murein of E. coli. The different peptide moieties (R) that can substitute the lactyl group of MurNAc are listed. (B) Chemistry of the major cross-linked dimers and trimers found in the murein of E. coli. A2pm, diaminopimelic acid.
Detailed insight into the structure of murein has been made possible by establishing two powerful analysis methods (48, 62). The high-molecular-weight polymer murein is first completely degraded with specific enzymes, such as muramidases that degrade the glycan strands or amidases that cleave off the peptides from the glycans (Fig. 2). This is then followed by fractionation of the reaction products by reversed-phase high-pressure liquid chromatography (HPLC).
Muropeptide Analysis
The murein degradation products, called muropeptides, which are obtained by muramidase digestion can be separated by reversed-phase HPLC on ODS-Hypersil (48, 50) (Fig. 3). An unexpected complexity was found, which is explained by the unrestricted combination of only a few different monomers via two different cross-links [d-Ala–(d)-m-A2pm or (l)-m-A2pm–(d)-m-A2pm] to dimeric and also tri- and tetrameric structures (Fig. 4B). The structures of all identified monomeric muropeptides are depicted in Fig. 4A, and a summary of all the muropeptides separated by HPLC is given in Table 1. Nevertheless, the basic structure of the murein of E. coli is a simple meshwork of glycan strands cross-linked with short peptides.
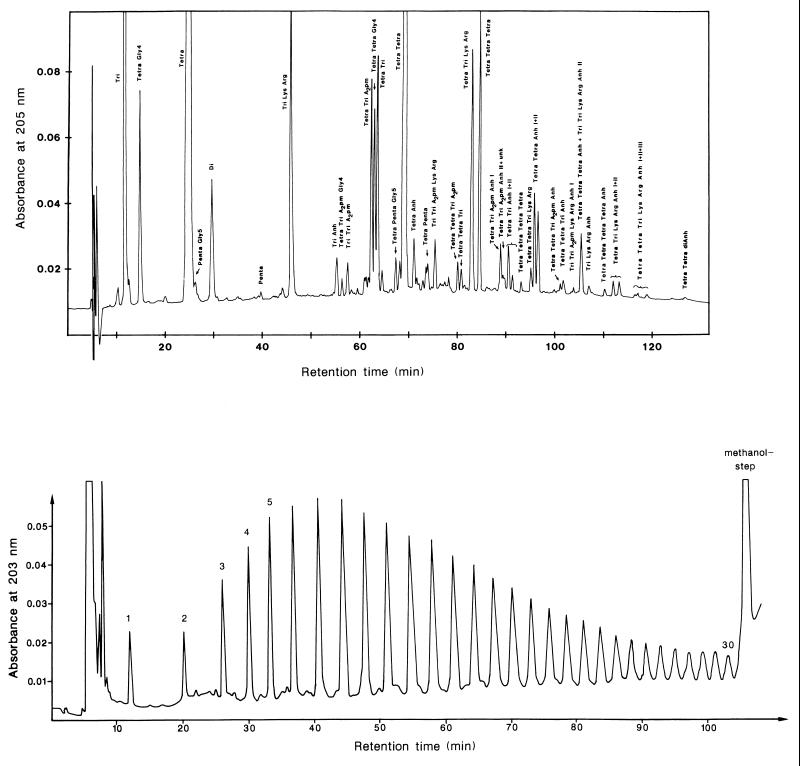
FIG. 3.
Fractionation of enzymatic degradation products of murein by HPLC. The upper panel shows the separation of sodium borohydride-reduced muropeptides obtained after complete digestion of isolated murein sacculi of E. coli with the muramidase Cellosyl. Chromatography was performed at 55°C on a 3-μm Hypersil ODS column with a linear gradient from 50 mM sodium phosphate (pH 4.32) to 15% methanol in 50 mM sodium phosphate (pH 4.95) (for details, see reference 48). The lower panel shows the separation of murein glycan strands obtained by digestion of isolated sacculi with human serum amidase. Chromatography was performed on a 5-μm Nucleosil ODS column. Elution was done at 50°C with a convex gradient of 100 mM sodium phosphate (pH 2.0) containing 5% acetonitrile to 100 mM sodium phosphate (pH 2.0) containing 11% acetonitrile (for details, see reference 62). This was followed by elution with 100% methanol.
TABLE 1.
Muropeptide composition of the murein of E. coli KN126a
| Muropeptide | Retention time (min) | Amount (%) (mean ± SD) |
|---|---|---|
| Tri | 11.9 | 7.41 ± 5 |
| Tetra(Gly4) | 15.0 | 1.71 ± 8 |
| Tetra | 25.2 | 35.90 ± 4 |
| Penta(Gly5) | 26.1 | 0.26 ± 18 |
| Di | 29.6 | 2.13 ± 11 |
| Penta | 39.7 | 0.07 ± 23 |
| Tri-Lys-Arg | 45.9 | 4.68 ± 6 |
| Tri(Anh) | 55.1 | 0.36 ± 8 |
| Tetra-tri(A2pm,Gly4) | 56.1 | 0.07 ± 22 |
| Tri-tri(A2pm) | 57.3 | 0.28 ± 13 |
| Tetra-tri(A2pm) | 62.4 | 1.55 ± 11 |
| Tetra-tetra(Gly4) | 63.0 | 1.47 ± 10 |
| Tetra-tri | 63.6 | 2.99 ± 9 |
| Tetra-penta(Gly5) | 67.3 | 0.32 ± 18 |
| Tetra-tetra | 69.2 | 27.30 ± 3 |
| Tetra(Anh) | 71.0 | 0.60 ± 11 |
| Tetra-penta | 73.8 | 0.17 ± 24 |
| Tri-tri-Lys-Arg(A2pm) | 75.4 | 0.43 ± 20 |
| Tetra-tetra-tri(A2pm) | 80.1 | 0.24 ± 14 |
| Tetra-tetra-tri | 80.9 | 0.28 ± 10 |
| Tetra-tri-Lys-Arg | 83.0 | 2.92 ± 7 |
| Tetra-tetra-tetra | 84.6 | 2.33 ± 5 |
| Tetra-tri(A2pm,Anh)I | 88.9 | 0.26 ± 15 |
| Tetra-tri(A2pm,Anh)II | 89.4 | 0.27 ± 16 |
| Tetra-tri(Anh)I | 90.6 | 0.42 ± 12 |
| Tetra-tri(Anh)II | 91.3 | 0.14 ± 15 |
| Tetra-tetra-tetra-tetra | 93.1 | 0.08 ± 17 |
| Tetra-tetra-tri-Lys-Arg | 95.1 | 0.27 ± 10 |
| Tetra-tetra(Anh)I | 95.9 | 0.67 ± 7 |
| Tetra-tetra(Anh)II | 96.6 | 0.67 ± 5 |
| Tetra-tetra-tri(A2pm,Anh) | 101.1 | 0.08 ± 13 |
| Tetra-tetra-tri(Anh) | 101.7 | 0.18 ± 12 |
| Tetra-tetra-tetra-(Anh) | 105.4 | 0.55 ± 10 |
| Tetra-tetra-tetra-tetra(Anh) | 110.2 | 0.05 ± 12 |
| Tetra-tri-Lys-Arg(Anh)I | 111.9 | 0.13 ± 13 |
| Tetra-tri-Lys-Arg(Anh)II | 113.2 | 0.16 ± 21 |
| Tetra-tetra-tri-Lys-Arg(Anh)I | 116.4 | 0.03 ± 20 |
| Tetra-tetra-tri-Lys-Arg(Anh)II | 116.9 | 0.05 ± 24 |
| Tetra-tetra-tri-Lys-Arg(Anh)III | 118.8 | 0.05 ± 17 |
Muropeptides were prepared from isolated murein sacculi by digestion with cellosyl and fractionated by reversed-phase HPLC on Hypersil ODS as described by Glauner et al. (50). Di, disaccharide dipeptide; Tri, disaccharide tripeptide; Tetra, disaccharide tetrapeptide; Penta, disaccharide pentapeptide (disaccharide, GlcNAc–β-1,4–MurNAc); Gly4/Gly5, glycine in position 4 or 5 of a peptide side chain; Lys-Arg, Lys-Arg residue from lipoprotein; Anh, 1,6-anhydro muramic acid; A2pm indicates a d,l-A2pm-A2pm cross bridge rather than a d,d A2pm-Ala cross bridge (present in all other oligomeric muropeptides).
Length Distribution of Glycan Strands
The glycan strands, which are released by digestion with an amidase that cleaves the amide bond linking the peptides to the glycan strands, can be fractionated according to their length, i.e., their degree of polymerization, by reversed-phase HPLC on ODS Nucleosil (62) (Fig. 3). The method can separate all lengths from 1 to 30 disaccharide units. The average length of the unfractionated material that can be eluted from the column by a methanol step is about 80 disaccharide units. Thus, the length distribution of the glycans is extremely broad.
Particular Muropeptides
Several aspects of the structure of murein seem to be of particular importance (49, 50). The prevailing cross-linkage is via d,d-peptide bonds between m-A2pm and d-Ala, which are known to be formed by a penicillin-sensitive transpeptidation reaction (see below). By contrast, the cross-linkages via l,d-peptide bonds between two m-A2pm residues are uncommon (about 2% of the linkages) in exponentially growing cells. However, the relative amount of l,d-cross-linkage can more than double during the stationary phase of growth. The murein is cross-linked not only by dimeric peptide bridges connecting two glycan strands but also to a significant extent (4.6%) by trimeric peptide bridges connecting three glycan strands. Tetrameric cross-links are extremely rare and represent only about 0.16% of the linkages. Structural considerations indicate that the three (four) strands cross-linked by a trimeric (tetrameric) peptide cannot be arranged in one plane. One (two) of the three (four) strands must be positioned either above or below the plane defined by the other two strands. This raises the question about the function of such a structural element in a monolayered murein.
The covalent linkage of a lipoprotein to the murein, which can be cleaved off by pronase treatment during the preparation of pure murein sacculi gives rise to muropeptides substituted at their A2pm residues by the two carboxyl-terminal amino acids of the lipoprotein, Lys and Arg (14, 15, 74). Therefore, a muropeptide analysis also contains some information about the amount and specific linkage of the murein lipoprotein to the sacculus.
A peculiar structure of the murein of E. coli is a 1.6-anhydromuramic acid present at one end of the murein strands (67) (Fig. 4A). All of the normally reducing ends seem to be blocked thereby, since free reducing ends have not been found (64). The 1,6-anhydro ring is formed by an endogenous lysozyme-like enzyme (see below).
The 1,6-anhydromuramic acid that is a natural tag of one of the ends of the glycans can be used to calculate the average length of all glycan strands in the murein by determining the ratio of all muropeptides to all 1,6-anhydromuropeptides (48). Depending on the growth conditions, an average length of about 29 disaccharide units was found. A surprising result was obtained when analyzing the length distribution of the glycan strands by HPLC (62). It turned out that most of the strands are rather short, with lengths of around 5 to 10 disaccharide units, which equals about 1/300 of the total circumference of the cell. The distribution showed an amazingly broad variation from 1 to more than 100 disaccharide units. Whether the extremely long strands have a specific function and topological localization in the sacculus is not known yet.
SYNTHESIS OF THE MUREIN SACCULUS
Membrane Translocation of Precursors
The biosynthetic pathway leading to the polymer murein comprises three cellular compartments: cytoplasm, cytoplasmic membrane, and periplasm (Fig. 5). The subunits of the biopolymer are synthesized in the cytoplasm as UDP-activated precursors (for a review, see reference 160), which are then translocated as lipid-linked compounds across the membrane and finally inserted into the preexisting murein sacculus by murein synthases (Table 2). The translocation of the hydrophilic murein subunits is a crucial step in murein synthesis, but surprisingly little is known.
FIG. 5.
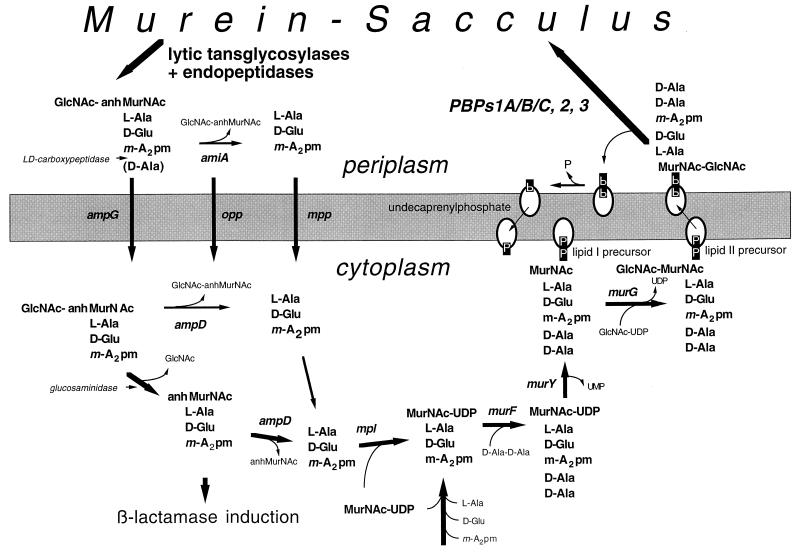
Recycling pathway of murein turnover products in E. coli. During growth, lytic transglycosylases together with d,d-endopeptidases release monomeric 1,6-anhydromuropeptides in the periplasm, which are mostly tripeptide derivatives due to the presence of a l,d-carboxypeptidase. The turnover products can enter two different metabolic pathways. In one case, they are first degraded by an amidase (AmiA) (161) in the periplasm to the disaccharide and the peptide, which can be transported into the cell by the general oligopeptide transport system (Opp) (53) or the specific murein peptide permease (Mpp) (118). The fate of the sugars is not well established. Alternatively, the turnover products can be taken up by the cell through the AmpG transporter, which accepts intact muropeptides (99, 114). In the cytoplasm, the muropeptides are further degraded by an amidase (AmpD) with a specificity for 1,6-anhydromuropeptides (72, 81) and an N-acetylglucosaminidase (176), yielding free tripeptides, anhydro-MurNAc, and GlcNAc. The intermediate 1,6-anhydro-N-acetylmuramyltripeptide functions as an inducer of AmpC β-lactamases by interacting with the AmpR regulator (79). The tripeptide can be reused directly for the synthesis of murein precursor molecules. A specific tripeptide ligase (Mpl) (103) forms UDPMurNAc tripeptide, which is a normal intermediate of the de novo murein biosynthetic pathway. Accordingly, a d-Ala–d-Ala dipeptide is linked to the tripeptide intermediate. The resulting UDPMurNAc-pentapeptide is then translocated to undecaprenyl phosphate, yielding the lipid I precursor that is supplemented by the addition of GlcNAc from UDPGlcNAc to the final lipid II precursor, the bactoprenol-linked disaccharide-pentapeptide. After its translocation to the periplasmic side of the membrane, the precursor is inserted into the murein sacculus by transpeptidation and transglycosylation reactions catalyzed by the high-molecular-weight PBP1A, PBP1B, PBP1C, PBP2, and PBP3. The released undecaprenyl pyrophosphate is hydrolyzed to undecaprenyl phosphate before it can enter a new cycle.
TABLE 2.
Murein synthases of E. coli
| Map position
|
Gene | Enzyme | Size (kDa) | Localization | Reference(s) | |
|---|---|---|---|---|---|---|
| kb | min | |||||
| 3,520 | 75.9 | ponA (mrcA) | Transglycosylase/transpeptidase (PBP1A) | 94.5 | Inner membrane | 17, 78 |
| 165 | 3.6 | ponB (mrcB) | Transglycosylase/transpeptidase (PBP1B) | 94.3 | Inner membrane | 17, 109 |
| 2,645 | 57.0 | pbpC | Transglycosylase/transpeptidase (PBP1C) | 85.1 | Inner membrane | 133 |
| 667 | 14.4 | pbpA (mrdA) | Transpeptidase (PBP2) | 70.8 | Inner membrane | 1 |
| 92 | 2.0 | ftsI (pbpB) | Transpeptidase (PBP3) | 63.9 | Inner membrane | 110 |
| 3,347 | 72.1 | mgt | Monofunctional glycosyltransferase | 27.3 | Inner membrane | 33 |
For export into the periplasm, first the P-MurNAc-pentapeptide moiety of the cytoplasmic UDPMurNAc-pentapeptide is hooked via a pyrophosphate onto the C55 isoprenoid undecaprenylphosphate, also called bactoprenol. This lipid I precursor is completed to the final lipid II murein precursor molecule by the addition of GlcNAc from UDPGlcNAc (Fig. 5). The transport of the lipid-bound disaccharide-pentapeptide precursors at a sufficient high rate that matches the rate of murein synthesis by all likelihood is catalyzed by additional proteins, not identified yet (flippases?).
Inhibition of phospholipid synthesis, either directly or indirectly by inducing the stringent response (23), results in cessation of murein synthesis (40, 77, 124). However, none of the enzymes involved in murein synthesis could be shown to be inhibited under these conditions. It has therefore been proposed that the translocation of the lipid II precursor depends on ongoing phospholipid synthesis (40). Likewise, the synthesis of the O-specific side chain of lipopolysaccharides, which also depends on the translocation of bactoprenol-linked subunits across the cytoplasmic membrane, is blocked when phospholipid synthesis is inhibited (40).
One of the missing links for an understanding of the translocation of the lipid-linked murein precursors may be the integral membrane proteins RodA (73) and FtsW (86), which show a high degree of similarity to each other and are predicted to have 10 membrane-spanning regions. An interaction of these two proteins with specific murein synthases, penicillin-binding proteins 2 and 3 (PBP2 and PBP3), respectively, which both are transpeptidases, has been shown (102). Thus, RodA and FtsW may be part of a tunnelling device directing the flow of the murein precursors directly to the membrane-anchored enzymes, which then insert the precursors into the preexisting murein sacculus.
Bifunctional Enzymes
Being transpeptidases, PBP2 and PBP3 are unable to synthesize or enlarge the murein net on their own (46, 51). The synthesis of cross-linked murein calls for the formation of two types of covalent bonds: glycosidic and peptide bonds (Fig. 2). All known synthases that are involved in the final steps of the synthesis of the murein sacculus are listed in Table 2. Transpeptidases have to cooperate with murein transglycosylases, which catalyze the polymerization of the precursors to glycan strands (102). Interestingly, besides monofunctional murein transglycosylases (33, 58), bifunctional enzymes that have transpeptidase as well as transglycosylase activity and thus catalyze both reactions also exist (78, 109, 133). These are PBP1A, PBP1B, and PBP1C. They have sequence homologies, and all three have the conserved sequence motifs characteristic of transglycosylases and transpeptidases. Despite this homology, PBP1C has a peculiar β-lactam specificity different from that of PBP1A and PBP1B (133). At least one of either PBP1A or PBP1B is essential for growth, since a deletion of both genes is lethal (143). As explained below, these bifunctional enzymes are probably the main murein-synthesizing enzymes.
Transglycosylation
The transglycosylation reaction is likely to catalyze the transfer of a nascent lipid-linked murein strand onto the C-4 carbon of the glucosamine residue in the lipid-linked murein precursor (Fig. 2). Undecaprenyldiphosphate is released during this process. This mechanism has experimentally been proven for murein transglycosylation in Bacillus licheniformis (169). Transglycosylation is inhibited by the glycolipid antibiotic moenomycin, which binds to the active site of the enzyme due to a structural analogy to the lipid-linked murein precursor (123). It is not clear what determines the length of the murein strands and how the growing strand is released from the enzyme. In any case, the strand is terminated by a 1.6-anhydro-MurNAc. Either it is accomplished by a kind of idling reaction that results in an intramolecular glycosyl transfer of the growing strand onto the C-6 hydroxyl group of the terminal muramic acid rather than onto the lipid II precursor, or a continuously polymerized strand is broken down by the action of endolytic transglycosylases. Such an enzymatic specificity has recently been identified in E. coli (94).
d,d-Transpeptidation: the Target of Penicillin
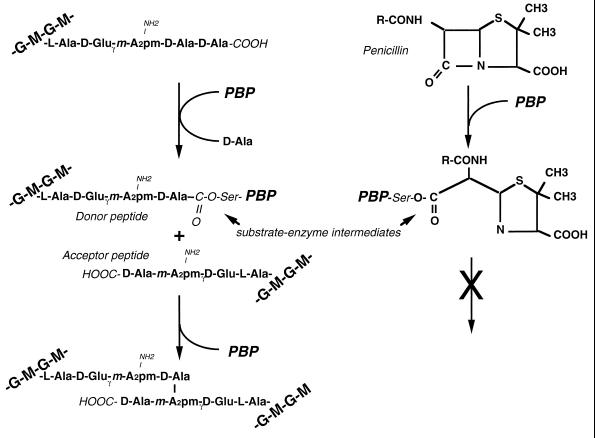
The transpeptidation reaction has been analyzed in quite some detail, because it is the target of β-lactam antibiotics (45, 46, 142, 150, 174) (Fig. 6). The reaction is a two-step mechanism. First, the d-Ala–d-Ala moiety of the pentapeptide precursor is cleaved and an enzyme-substrate intermediate is formed, with concomitant release of the terminal d-Ala. Due to a structural analogy of penicillin to d-alanyl–d-alanine, the enzyme can also react with penicillin and cleave the β-lactam bond, forming a penicilloyl-enzyme intermediate. In a second step, the mureinyl-tetrapeptidyl moiety but not the penicilloyl group is transferred to an acceptor, normally the non-alpha amino group of the dibasic amino acid in a second murein stem peptide. Penicillin functions as a suicide substrate in this reaction by freezing the reaction at the penicilloyl-enzyme intermediate.
FIG. 6.
d,d-Transpeptidation reaction. The general scheme of the two-step transpeptidation reaction, resulting in the formation of a cross-link between two glycan strands, is shown on the left. In the first step, the d-Ala–d-Ala peptide bond of a donor peptide is cleaved by a PBP enzyme, and a covalent bond between the carboxyl group of the penultimate d-Ala and the hydroxyl group of a serine in the active center of the enzyme is formed, giving rise to a mureinyl enzyme intermediate. In a second reaction step, the mureinyl moiety is transferred to a free epsilon amino group of a diaminopimelic acid residue in another peptide moiety (acceptor peptide). Concomitant with the formation of the cross-link, the PBP enzyme is released. The interaction of the PBP enzyme with penicillin is indicated on the right. By analogy to the cleavage of the d-Ala–d-Ala bond during the transpeptidation, the β-lactam ring is cleaved and a penicilloyl enzyme intermediate is formed. In contrast to the mureinyl enzyme intermediate, the penicilloyl enzyme intermediate is rather inert, and thus the enzyme is blocked by a covalent bond to the antibiotic, a kind of suicide substrate for PBPs. G, GlcNAc; M, MurNAc; A2pm, diaminopimelic acid; Ser is from the catalytic center of the PBP.
The transpeptidation reaction is energized by the hydrolysis of the d-Ala–d-Ala bond of the pentapeptide. Therefore, the pentapeptide is called the donor. Whereas the donor for the transpeptidation must be a pentapeptide, the acceptor that presents a free amino group could be a penta-, tetra-, or tripeptide. In the final product, the peptide bridge, the presence of a free amino group at the A2pm residue marks the peptide that played the role of the donor during transpeptidation. Because of the free amino group in the donor peptidyl moiety, higher oligomers can be formed. Theoretically, there is no limit; however, only dimeric, trimeric, and tetrameric cross bridges have been detected in the murein of E. coli (50).
l,d-Transpeptidation
The overwhelming majority of cross bridges (about 93%) are formed by d,d-peptide bonds between d-Ala and m-A2pm. In addition, a small number of l,d-cross-links between two m-A2pm residues is present (50) (Fig. 4B). It is still unknown how the cross-links between two diaminopimelic acid residues are formed. The enzyme is not known, and we have no information about the energy donor for this reaction. However, it is likely that a tetrapeptide functions as a donor in a transpeptidation reaction catalyzed by an l,d-transpeptidase. The same enzyme may also be involved in the exchange of d-Ala with other d-amino acids that takes place when E. coli is grown in the presence of various d-amino acids (22). l,d-Transpeptidation seems important for the cell under conditions where no pentapeptides are present in sufficient amounts. A dramatic increase in the relative number of A2pm-A2pm cross-links has been observed at the onset of bacteriolysis caused by various agents and in the stationary phase of growth under conditions where pentapeptide donors that could drive the normal cross-linking reaction are in short supply (13, 39, 49, 92, 113, 120, 166). An l,d-transpeptidation reaction is expected to be insensitive to β-lactam antibiotics. Hence, this reaction may support the survival of bacteria in the presence of low concentrations of penicillins.
A BAG-SHAPED EXOSKELETON CALLED THE SACCULUS
Isolated Murein Sacculi
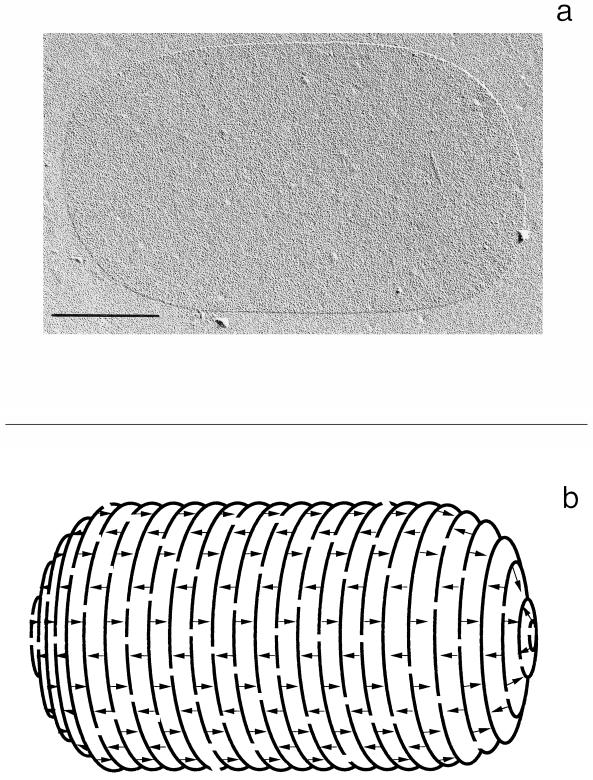
Murein is formed into a spectacular three-dimensional structure, a hollow body that completely surrounds the bacterial cell (71, 116, 126, 170). It has the shape and size of the bacterium; indeed, it is a gigantic molecule (Fig. 7). Sacculi can be isolated in their intact structure but never in their native three-dimensional shape, which inevitably is lost when the cellular pressure is released. Because of the high tension in the murein layer as a result of the cellular turgor, it has been argued that the meshes in the murein net must adopt a hexagonal form (89). However, due to the different elasticities of the materials from which the net is made, stiff glycans and highly flexible peptides (96, 97), a more rectangular shape of the meshes is expected to be formed, but we do not know the true native form. Nevertheless, isolated sacculi reflect the specific shape of the cell from which they have been prepared. This illustrates that the murein sacculus maintains the morphology of the bacterium. Unfortunately, until now it has not been possible to obtain a detailed image of the structure by electron microscopy or by any other modern technique (16, 87, 97). Thus, it is still not known how the threads in the net are arranged with respect to the overall shape of the sacculus. Moreover, it is still uncertain whether some kind of order exists at all. As discussed below, an ordered structure would greatly facilitate persistence of the shape during growth and division. Furthermore, the murein sacculus is likely to have a specific cutting pattern and therefore may not only maintain but also determine the specific shape of a bacterium.
FIG. 7.
Murein sacculus of E. coli. (a) Electron micrograph (agar filtration) (prepared by H. Frank) of an isolated murein sacculus. Bar, 0.5 μm. (b) Idealized schematic drawing of the architecture of the murein sacculus. The parallel lines represent a few of the vast number of glycan strands, and the arrows indicate the peptide bridges.
Arrangement of the Glycan Strands
For physical reasons, an arrangement of the glycan strands, which are rather stiff and have only restricted flexibility (97), perpendicular to the long axis of the rod-shaped sacculus would be most appropriate, since the stress in the wall is twice as high in the hoop direction as in the longitudinal direction. Indirect experimental evidence for an arrangement of the glycan strands perpendicular to the long axis was obtained when isolated murein sacculi were partially digested by an endopeptidase (162) or disrupted by sonication (163). This resulted in electron micrographs showing a preferential striping of the sacculus in the hoop direction. This conclusion has recently been questioned, since digestion with a muramidase resulted in a similar appearance of the sacculi (32), suggesting that the observed striping probably does not reflect the arrangement of the glycan strands. Hence, experimental proof is still missing. Furthermore, it is not known whether the strands are oriented parallel or antiparallel to one another.
As pointed out above, cross-linkage is the result of a transpeptidation from a donor peptide to an acceptor peptide (Fig. 6). The formation of cross-links between the glycan strands could therefore be either uniform, with all the peptides of one strand playing the role of either donors or acceptors, or random. It is more likely that during synthesis a given bifunctional transglycosylase/transpeptidase enzyme polymerizes the disaccharide precursors and catalyzes cross-linkage, always in the same direction, to a neighboring acceptor strand. As a consequence, strands with exclusively donor peptides, characterized by the presence of a free epsilon amino group at the A2pm residue, would alternate with strands substituted exclusively with acceptor peptides having their epsilon amino group of the A2pm in the cross-linking peptide bond (Fig. 7b; also see Fig. 12).
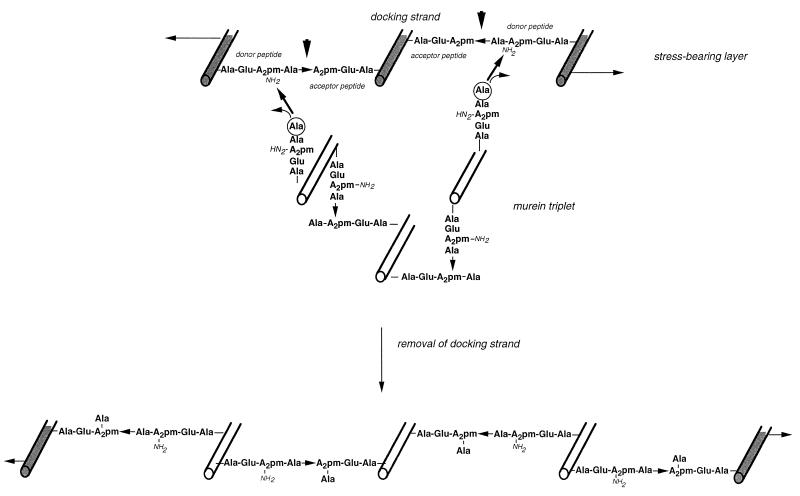
FIG. 12.
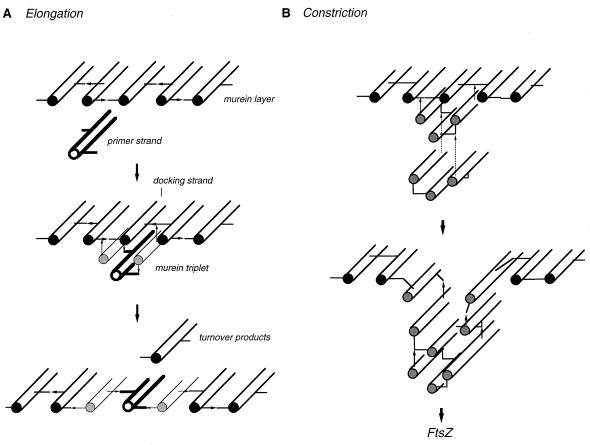
Model for the growth of murein following a make-before-break strategy (three-for-one growth model). Three cross-linked glycan strands from the monolayered, stress-bearing murein are shown in grey. A triple pack of newly synthesized, cross-linked glycans still in a relaxed state is shown in outline. The murein triplet is covalently attached to the free amino groups present in the donor peptides of the cross-links on both sides of a strand, called the docking strand, that is substituted by acceptor peptides. Specific cleavage of the preexisting cross-links (arrowheads) results in the replacement of the docking strand by the murein triplet. The rods represent the glycan strands. A2pm, diaminopimelic acid.
The analysis of the lengths of the glycan strands revealed that most of the strands are extremely short relative to the circumference of the cell (62; see above). For an ordered structure of the murein sacculus, the glycans are likely to be aligned with ring structures around the cell (Fig. 7b). During growth, the lengths are altered by the action of exo- and endolytic transglycosylases, as has been shown by pulse-chase experiments (see below). It is also possible that strands are linked by enzymes that utilize the energy stored in the 1,6-anhydromuramic acid residues.
Murein Surface Density
The murein sacculus of E. coli is probably made of only one continuous stress-bearing layer with a maximal thickness of about 7 nm (116, 173). A neutron small-angle scattering study suggested that about 75 to 80% of the murein sacculus is single layered, with the rest being triple layered (98). Despite this thin structure, the amount of murein per unit of cell surface area can be reduced by 50% without affecting the morphology and growth of the cells (121). This indicates that even an extremely loosely woven murein net is able to maintain mechanical stability. The permeability characteristics of the murein net have been studied by determining the effective pore sizes of isolated sacculi with fluorescein-labelled dextran probes with different molecular weights (30). The results of these studies suggest that the murein net is an effective hindrance for globular proteins of more than 50 kDa in their attempts to pass through the sacculus. Recent findings indicate that specialized murein hydrolases seem to be involved in the transenvelope transport of bulky proteins (35).
Elasticity of Murein
The rod-shaped murein sacculus can be extended in the direction of both axes because the threads of the murein net do not form a completely closed structure in either direction. In particular, the peptides are rather flexible and can be stretched to fourfold the length of their most compact conformation. Experiments with isolated murein sacculi have shown that they can reversibly increase in surface by up to 300% above the relaxed conformation (75, 89).
MUREIN METABOLISM DURING GROWTH
Cell Elongation and Cell Division
The metabolism of the murein sacculus during bacterial growth has been studied by continuous, pulse, and pulse-chase labelling experiments (21, 25, 29, 39, 50, 108, 112, 120, 171, 172, 175). It became clear that murein is by no means a static, rigid layer but a highly dynamic structure, as one would expect for a structure that grows and divides. Studies with synchronously growing cultures clearly showed that the mode of murein growth is different during elongation and cell division (26, 29, 49, 175). Whereas multiple murein strands are inserted side by side during constriction, single strands are inserted between preexisting old murein strands during elongation. It has been proposed on the basis of studies with morphology mutants of Klebsiella pneumoniae and E. coli, as well as of the specific effects of certain β-lactam antibiotics, that rod-shaped bacteria have at least two systems, one specifically responsible for cell elongation and one specifically involved in the formation of new poles by cell constriction (132). These two systems can be nicely correlated with the specific activity of two distinct proteins: PBP2, which is specific for elongation, and PBP3, which is specific for cell division (142). According to the “two-competing-site” model, the final shape (rod versus sphere) is determined by the balance of these two enzyme systems (100).
Maturation of Murein
Quite a number of changes in the muropeptide composition take place as the murein ages (13, 19, 31, 49, 120, 156). The most important ones are shown in Fig. 8. Some processes, such as the decrease in pentapeptide side chains, are rapid. Some are slow, taking almost one generation; these include the increase in tripeptide side chains, the increase in cross-linkage and the decrease in the average length of the glycan strands. Also, the addition of the murein lipoprotein is a slow process, as indicated by the kinetics of the increase in the number of LysArg-substituted muropeptides that represent the attachment points (14).
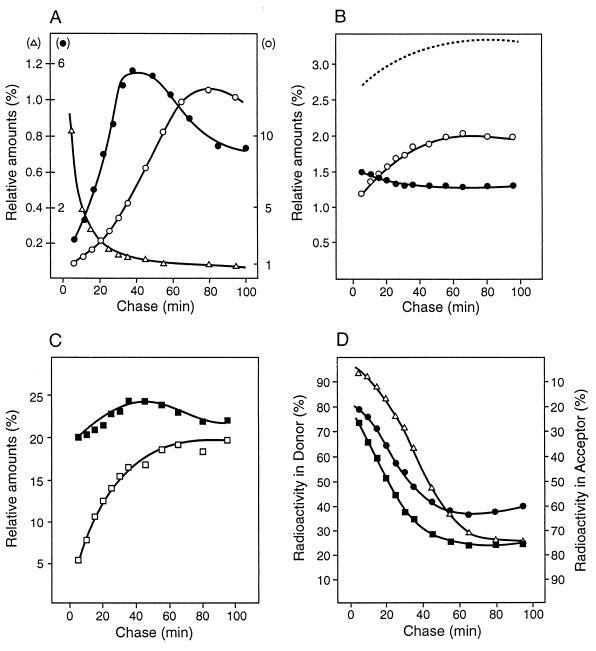
FIG. 8.
Maturation of murein. Changes in the relative amounts of defined muropeptide structures in the murein of E. coli were analyzed by pulse-chase labelling experiments. Exponentially growing A2pm auxotrophic E. coli W7 (with a generation time of 30 min) was pulse-labelled with [3H]diaminopimelic acid for 2 min and chased for different times as indicated. Experimental details are given in reference 49. (A) Changes of the major monomeric muropeptides. Solid circles, LysArg-modified disaccharide tripeptides; open circles, disaccharide tripeptides; triangles, disaccharide pentapeptides. (B) Alteration in time of the anhydromuropeptides representing the chain ends. Solid circles, monomers; open circles, cross-linked muropeptides; dotted line, total amount of anhydromuropeptides. (C) Changes in the cross-linkages. Solid squares, cross-linkage by d,d-A2pm-Ala; open squares, cross-linkage by l,d-A2pm-A2pm. (D) Distribution of labelled diaminopimelic acid in donor and acceptor peptides of cross-linked muropeptides. Solid circles, muropeptides cross-linked by two tetra peptides; open triangles, muropeptides cross-linked by one tri- and one tetrapeptide; solid squares, trimeric muropeptides cross-linked by three tetra-peptides.
An interesting observation was made when the stability of different peptide bridges was monitored during a chase (49). It was found that they are cleaved with different kinetics (Fig. 8D) and reach different final plateaus even though murein recycling in particular affects the results of longer chase periods by levelling them to 50% cleavage, thereby obscuring the real degree of breakage of the cross bridges. Whereas most of the dimeric peptide bridges consisting of a tetrapeptide and a tripeptide (tetra-tri) and the trimeric cross-links were cleaved in one generation, only about half of the dimeric peptide bridges consisting of two tetrapeptides (tetra-tetra) were split in the same time. This was surprising because the two dimeric cross bridges, either tetra-tetra or tetra-tri, are cleaved by endopeptidases with similar Km and Vmax values when present in isolated muropeptides (69, 83, 153). Therefore, they must have different functions in the growing sacculus.
A most important finding was the demonstration not only that covalent bonds are cleaved during growth but also that muropeptides are released from the murein meshwork (56, 57). Although the sacculus is a monolayer, dramatic turnover is going on during growth.
Murein Turnover
The phenomenon of murein turnover in E. coli was detected rather late because turnover is coupled to murein recycling (52, 114). The turnover material that is released at a rate of 40 to 50% per generation is quite efficiently recycled (Fig. 5). About 90% of the turnover material is reinserted into the murein sacculus. The turnover products are 1.6-anhydro-MurNAc–β-1,4–GlcNAc–tetra- and tripeptides, indicating that they are released by the action of lytic transglycosylases (see below). The absence of dimers or trimers suggests that turnover is due to the concerted action of both lytic transglycosylases and endopeptidases. The turnover products accumulate in the periplasm, from where they are reimported into the cytoplasm. Several recycling pathways exist (Fig. 5).
Murein Recycling
The major route for the uptake of the turnover products is via the transporter protein AmpG (99, 114), a transmembrane protein proposed to act as a specific permease for intact muropeptides, which are mostly tripeptide-substituted muropeptides due to the presence of an l,d-carboxypeptidase in the periplasm (7, 159). In the cytoplasm, the muropeptides have to be trimmed to the size that allows reutilization for the synthesis of murein precursors. A β-N-acetylglucosaminidase splits the disaccharide (117, 176), and an amidase (AmpD) cleaves off the peptide (72, 81). Interestingly, AmpD has a specificity for 1,6-anhydro muropeptides and therefore does not hydrolyze the murein precursor molecules such as UDPMurNAc-pentapeptide also present in the cytoplasm.
Besides intact muropeptides, degradation products of the turnover material are also taken up by the cell. The presence of an amidase (AmiA) in the periplasm causes degradation of the turnover products to their peptide and sugar parts (161). The peptides are taken up by the cell via two peptide transport systems, by a general oligopeptide permease (Opp) (53), and by a specific murein peptide permease (Mpp) (118). To avoid the need for a complete breakdown of the peptides to the amino acid level, a specific ligase (mpl) exists that catalyzes the attachment of the tripeptide to UDPMurNAc (103).
β-Lactamase Induction
A recent finding established that the intracellular concentration of the murein turnover products can be measured by the cell (79, 80, 115). The induction of the inducible AmpC β-lactamase of enterobacteria responds to a signalling muropeptide that accumulates in the cytoplasm when growth is affected by the presence of penicillin (Fig. 5). Under these conditions, the rate of murein breakdown is increased compared to the rate of murein synthesis, resulting in an increase in murein turnover products. The signal seems to be the 1,6-anhydromuramyltripeptide, which, by interacting with the AmpR regulator protein, induces AmpC expression (79). It has been proposed that the cell uses murein turnover as a general sensing device to obtain information on the actual state of the stress-bearing sacculus to enable the cell to respond properly to changes that may affect the mechanical stability of the cell (115).
GROWTH AND DIVISION OF THE SACCULUS
Murein Hydrolytic Activity
To enlarge a completely closed string bag, meshes have to be cleaved to permit the insertion of new material between the existing netting (66, 69, 71, 137, 139, 170). For this reason, in addition to murein synthases, enzymes capable of cleaving bonds in the murein sacculus are expected to participate in the growth process. Moreover, it is predicted that murein hydrolytic activity is an essential function. However, since murein hydrolases are present in a surprisingly large number and broad variety of different specificities (44), final proof of this claim is difficult (Table 3).
TABLE 3.
Murein hydrolases of E. coli
| Hydrolase | Map position
|
Gene | Enzyme | Size (kDa) | Localization | Reference(s) | |
|---|---|---|---|---|---|---|---|
| kb | min | ||||||
| Lytic transglycosylase | |||||||
| 4628 | 99.7 | sltY | Soluble lt 70 (Slt70) | 70 | Periplasm | 42, 61 | |
| 2944 | 63.4 | mltA | Membrane-bound lt A (MltA) | 38 | Outer membrane | 101, 158 | |
| 2824 | 60.9 | mltB | Membrane-bound lt B (MltB) | 39 | Outer membrane | 41 | |
| Soluble lt 35 (Slt35) (proteolytic product of MltB) | 35 | Periplasm | 36, 43 | ||||
| 3102 | 66.9 | mltC | Membrane-bound lt C (MltC) | 40 | Outer membrane | 34 | |
| 1243 | 26.8 | emtA | Endospecific membrane-bound ltA (endo-Mlt) | 22 | Outer membrane | 94 | |
| β-N-Acetylglucosaminidase | |||||||
| 1163 | 25.1 | nagZ | β-N-Acetylglucosaminidase | 37 | Cytoplasm | 117, 145 | |
| N-Acetylmuramyl-l-alanine amidases | |||||||
| 2550 | 55.0 | amiA | Muramyl-l-alanine amidase A | 28 | Periplasm | 154 | |
| 4394 | 94.7 | amiB | Muramyl-l-alanine amidase B | 48 | Periplasm | 155 | |
| 2946 | 63.5 | amiC | Muramyl-l-alanine amidase C | 43 | Periplasm | 147 | |
| 119 | 2.6 | ampD | N-Acetylanhydromuramyl-l-alanine amidase | 20.5 | Cytoplasm | 72, 81 | |
| dd-Endopeptidases | |||||||
| 3326 | 71.7 | dacB | PBP4 (additional d,d-carboxypeptidase IB activity) | 48 | Membrane associated | 93, 106 | |
| 2232 | 48.1 | pbpG | PBP7, PBP8 (proteolytic product of PBP7) | 31 | Membrane associated | 63, 127, 157 | |
| 2444 | 52.7 | mepA | MepA | 28 | Periplasm | 84 | |
| dd-Carboxypeptidases | |||||||
| 663 | 14.3 | dacA | d,d-Carboxypeptidase (PBP5) | 40 | Inner membrane | 18 | |
| 880 | 19.0 | dacC | d,d-Carboxypeptidase (PBP6) | 40 | Inner membrane | 18 | |
| 2080 | 44.8 | dacD | d,d-Carboxypeptidase (PBP6B) | 43.5 | Inner membrane | 2 | |
As indicated in Fig. 2, specific hydrolases exist for almost each covalent bond in the murein in E. coli (139). Some of the enzymes accept only the intact high-molecular-weight murein sacculus as a substrate, whereas others are active only on soluble degradation products. The diversity of murein hydrolases (Table 3) reflects the manifold functions that these enzymes fulfill. Besides their essential pacemaker role for growth of the sacculus, they are involved in turnover and recycling of murein, as discussed above. Importantly, some of the murein hydrolases are potentially autolytic enzymes (151, 170). They can cause bacteriolysis when acting in an uncontrolled manner. The potentially autolytic system of E. coli consists of the lytic transglycosylases, the endopeptidases, and one of the amidases (AmiC).
An interesting reaction is catalyzed by the so-called lytic transglycosylases, which are peculiar lysozyme-like enzymes that cleave the β-1,4 glycosidic bond between GlcNAc and MurNAc (Fig. 9) (67, 144, 146). However, unlike lysozyme, they do not transfer the glycosyl moiety onto water but onto the C-6 hydroxyl group of the muramic acid. Hence, they are catalyzing an intramolecular glycosyl transferase reaction (149). The X-ray structure of one of these enzymes, Slt70, revealed that the active site resembles the fold of goose-type lysozyme (148). The product, the 1.6-anhydro muramic acid structure (Fig. 4A and 9), blocks the otherwise reducing end of the glycan strands. The meaning of this ring structure, which contains part of the chemical energy of the glycosidic bond that has been cleaved, is not known. It has been speculated that the bond energy may be utilized for some rearrangements of the glycan strands, such as the joining of strands to produce longer ones or the resealing of strands that had been cleaved. Another suggestion is that the 1,6-anhydro ring is a specific marker for murein turnover products as opposed to murein precursor molecules.
FIG. 9.
Mode of action of exoglycosylases. The processive degradation of murein strands by a lytic transglycosylase is shown. Structures shown in light grey depict released (soluble) muropeptides. The circle represents the enzyme molecule, with the thin circle marking the site from where the exoenzyme started its action. G, GlcNAc; M, MurNAc; bars and arrows indicate acceptor and donor peptides, respectively.
Recently, it has been shown that some of the lytic transglycosylases are lipoproteins residing in the outer membrane of the cell envelope (34, 41, 94, 101). Exo- and endolytic transglycosylases exist in E. coli (Table 3). The exolytic transglycosylases are processive enzymes that, starting at one end of the glycan strands, split off one disaccharide unit at a time. Consequently, these exoglycosylases cleave the murein sacculus in a zipperlike fashion (Fig. 9). Biochemical evidence indicates that the enzymes start at the glucosamine end (6, 129), whereas theoretical considerations based on the three-dimensional structure of the Slt70 suggest that the 1,6-anhydromuramic acid is the starting point (149).
The Problem of Enlarging a Stress-Bearing Structure
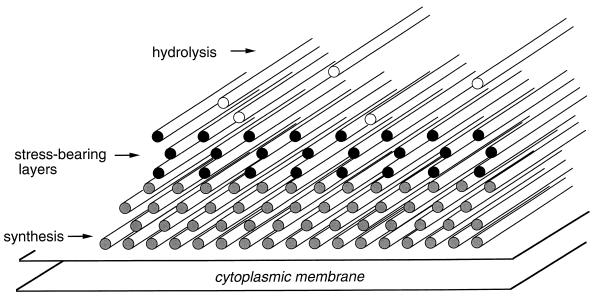
The concerted action of murein hydrolases and synthases that is needed to enlarge the covalently closed murein sacculus must be well coordinated to avoid the risk of lysis as a result of rupture of the stress-bearing sacculus and to maintain the rod shape (87, 88). There are two predominant ways in which murein hydrolases and synthases could cooperate during growth. One possibility is that the hydrolases act first and introduce cuts into the meshwork to create insertion points for fitting in new material by synthases (116). The other is that the new material is hooked to the sacculus by synthases before covalent bonds within the attachment sites of the added murein are cleaved by hydrolases (89, 90). The new material will then automatically be pulled into the layer due to the existing tension in the surface. The latter strategy, which has most appropriately been named by Arthur Koch the “make-before-break” strategy, is realized in the inside-to-outside growth mechanism of the multilayered peptidoglycan of gram-positive bacteria (Fig. 10) (90).
FIG. 10.
Inside-to-outside growth of the peptidoglycan of gram-positive bacteria. The addition of newly synthesized murein layers in a relaxed state underneath the stress-bearing layers with concomitant degradation of the outermost layers is schematically illustrated. The rods represent the murein strands. For the sake of simplicity, the cross-links between the glycans have been omitted.
It is obvious that enlargement of the monolayered sacculus of gram-negative bacteria is an even more difficult task and must follow a particular well-suited strategy. Any growth model that attempts to explain the enlargement and constriction of the monolayered murein of E. coli must find an answer to the important findings of murein turnover, the presence of trimeric cross-bridges, and the short half-life of trimers and tetra-tri dimers but not of tetra-tetra dimers (49, 50).
Speculative Views on the Growth Mechanism of the Murein Sacculus of Gram-Negative Bacteria
On the basis of the results of pulse-labelling experiments, which clearly show that single strands are inserted into the sacculus during cell elongation (26, 29, 119), it has been proposed that a bifunctional transglycosylase/transpeptidase in cooperation with an endopeptidase moves around the sacculus and inserts a single new strand between the preexisting ones (20, 116, 119). The endopeptidase, by cleaving the peptide cross bridges, would pave the way for the insertion and cross-linking to the neighboring strands of the newly synthesized murein strand. This model (Fig. 11), which has been presented by Park (116, 119), not only is in accordance with the pulse-labelling results, which show that during the pulse cross-links are exclusively formed between the newly synthesized glycan strands and the preexisting murein sacculus, but also could explain the finding that after a defined time span of about 8 min a steadily increasing number of cross-links is made between the newly synthesized and therefore labelled material. This has been interpreted to reflect a processive insertion of single glycan strands by a membrane-linked enzyme that moves unidirectionally around the circumference of the cytoplasmic membrane. After a certain period (8 min in the experiment referred to), one round is completed and a new ring of glycan strands is inserted next to and cross-linked to the previous ring of glycans. This mechanism, which does not follow the make-before-break strategy, would depend on a tight coordination of the synthase with the hydrolytic endopeptidase. According to this model, murein turnover would not be a result of the growth strategy but would be a process of its own. As mentioned above, it has been argued that murein turnover represents a sensing device for the structure of the murein sacculus (115). In addition, following this insertion mechanism of single glycan strands, the purpose of trimeric muropeptides remains unclear.
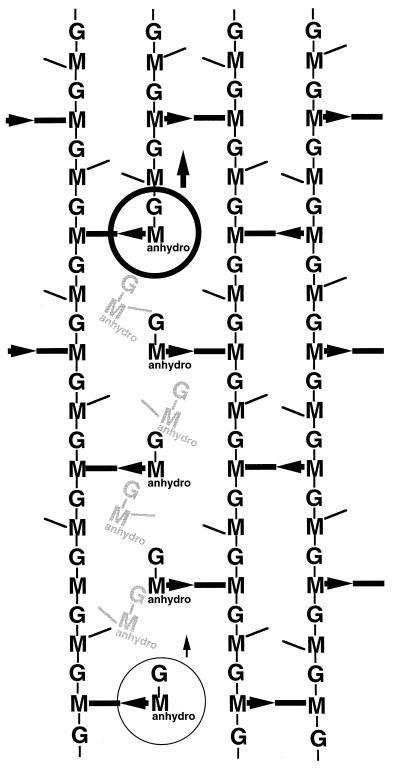
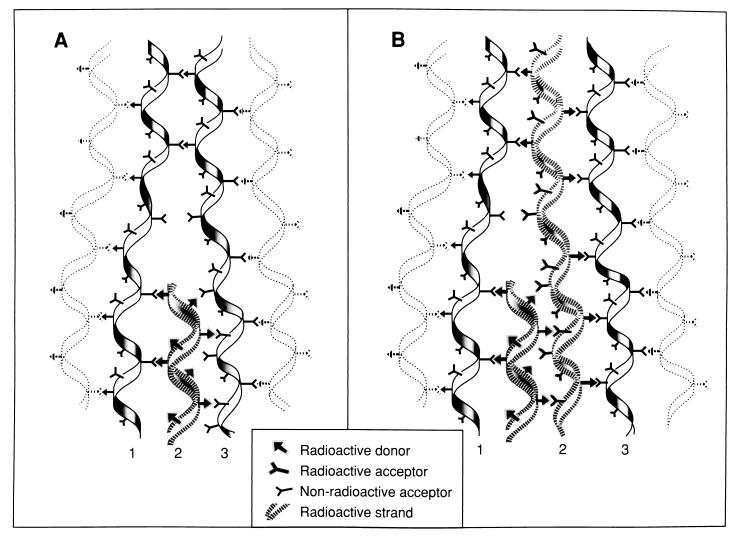
FIG. 11.
Model for the growth of murein following a cut-and-insertion strategy as proposed by Park (116). The structure of the murein at a growth site for elongation of the murein sacculus as it may exist during a pulse-labelling experiment is shown. The stippled, helical strand represents a new radioactive strand of murein inserted between preexisting strands initially (A) and later when, as a result of continued insertion along a helical path, a radioactive strand contacts the radioactive strand formed a few minutes earlier (B). The arrows between strands indicate the direction of cross-links in dimers from donor to acceptor strands. Reprinted from reference 116 with permission.
With reference to the inside-to-outside growth mechanism of gram-positive bacteria and in consideration of the results of the studies on the murein metabolism, another model has been put forward to describe the growth of the murein sacculus of E. coli (65, 68–70). It is postulated that new material is attached to the existing murein before covalent bonds are cleaved to promote the insertion of the new material into the layer under stress (Fig. 12).
One idea suggested itself, namely, that the trimeric cross bridges may function as attachment structures for new material. At this specific structure in the sacculus, the three linked glycan strands are arranged in different planes: one strand is positioned above or below the plane defined by the other two strands. In an otherwise monolayered murein, the existence of such a structure suggests a specific function. Furthermore, the trimeric cross bridges were found to have short half-lives (Fig. 8), as would be expected for attachment structures, since they would represent functional intermediates. In addition, the total number could be quite small, depending on the number of growing points, which has been calculated to be around 200 (116). Hence, the low percentage of trimers is in accordance with the idea that they may function as attachment sites for new murein.
To allow for smooth growth of the murein layer, the amount of material first added and then inserted should be small (89). The smallest patch of murein that can be added to the smallest attachment structure, one glycan strand, for structural reasons is a patch of three (not two) cross-linked glycan strands. As depicted in Fig. 12, a murein triplet in a relaxed state can be hooked underneath a single strand, a kind of docking strand, in the murein layer under stress. The triplet can be covalently linked to the sacculus by transpeptidation of the peptide moieties of the two outer strands of the triplet to the free epsilon amino groups of A2pm residues in the peptide cross bridges on both sides of the docking strand. The attachment sites are trimeric cross bridges. Specific removal of the old strand without rupture of the murein netting is possible only if such murein strands function as docking strands that are substituted by peptides with no free amino groups and are cross-linked to strands substituted by peptides that have a free epsilon amino group at the A2pm residue. With such an arrangement, the attachment sites of the murein triplet will enclose the d-Ala–(d)-m-A2pm peptide bonds that have to be cleaved for the specific removal of the docking strand (Fig. 12). Upon release of the old strand, the newly added triplet is automatically pulled into the layer under tension. As a result of this operation, one preexisting strand is replaced by three new ones. Hence, the mechanism has been called the three-for-one growth strategy. Accordingly, the observed murein turnover would be a direct outcome of the growth mechanism used by the bacterium. The difference in the half-lives of the two types of dimeric cross bridges will be addressed below.
A stepwise synthesis of the triplet during growth of the cylindrical middle part of the sacculus is suggested by the findings of pulse-labelling studies, which show that during cell elongation newly synthesized (i.e., labelled) strands are attached one at a time to preexisting, nonlabelled murein (26, 29, 119). This result excludes the possibility that the murein triplet is synthesized in one piece. Rather, it may be formed by synthesis and cross-linkage of two newly synthesized strands to both sides of an already existing murein strand (Fig. 13A). The preexisting single murein strand, which may be called a primer, is assumed to be polymerized by monofunctional transglycosylases known to be present (Table 2). Although it must be pointed out that there is no convincing experimental evidence for the existence of unbound glycan strands that could function as primers (19, 54, 104), considering the small number of growth sites (100 to 200), the number may indeed be quite small and difficult to detect.
FIG. 13.
Cell elongation and cell constriction according to the three-for-one growth mechanism. (A) Murein synthesis during cell elongation is proposed to use preexisting free glycan strands (primer strands) that are supplemented to form murein triplets by the synthesis and cross-linkage of two strands on both sides of the primer strand. The triplet is then attached to the peptide bridges on the right and left of the docking strand in the stress-bearing murein layer. Removal of the docking strand by murein hydrolases results in the insertion of the murein triplet into the murein sacculus. (B) During cell constriction, three glycan strands are simultaneously synthesized and cross-linked to one another to form a murein triplet that is attached underneath a docking strand located at the site of cell division. Formation of a constriction is the outcome of the repeated addition (dotted lines) of murein triplets, each followed by the release of the middle strand and an inward pull of the membrane-anchored enzymes resulting from the contracting FtsZ ring (12, 107, 122, 130). The glycan strands are represented by rods, and the peptide bridges are shown by lines (acceptor peptides) and arrows (donor peptides).
Constriction of the Murein Sacculus
An appealing feature of the three-for-one model is the fact that cell division can be envisaged by the very same mechanism (Fig. 13B). During cell division, the addition of triplets of glycan strands is directed exclusively to the murein docking strands at the site of cell division. To keep pace with the rate of murein synthesis during cell division, which takes place in a sharp zone (32, 108, 112, 138, 171, 175), the triplets may have to be synthesized in one step by a complex of three murein synthases, as indicated in Fig. 14. Indeed, unlike cell elongation, cell division occurs by the simultaneous insertion of several newly synthesized strands side by side (29, 112). The three-for-one mechanism of murein synthesis that guarantees a safe enlargement of the sacculus during cell elongation would also make cell division a safe process. At no time point during growth and division is the integrity of the stress-bearing murein sacculus affected.
FIG. 14.
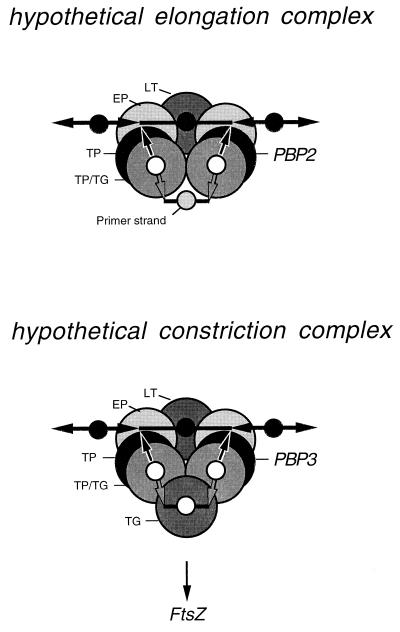
Hypothetical murein-synthesizing enzyme complexes. Two similar multienzyme complexes may exist that would differ in their specificities mainly as a result of the presence of either PBP2 (elongation complex) or PBP3 (constriction complex). The large circles represent the enzymes (LT, lytic transglycosylase; EP, endopeptidase; TP, transpeptidase; TP/TG, bifunctional transpeptidase-transglycosylase; TG, transglycosylase). The small circles indicate the glycan strands running perpendicular to the plane of the drawing (solid circles represent preexisting strands; open circles represent the newly synthesized murein triplet). Bars and arrows indicate the cross-linking donor and acceptor peptides.
Electron microscopic autoradiography studies have shown that murein synthesis during cell division takes place primarily in a sharp growth zone whereas the diffuse incorporation that occurs all over the cylindrical part of the sacculus during cell elongation is turned down (32, 175). Constriction is accomplished by a mechanical device that pulls the membrane-bound murein-synthesizing enzymes, probably assembled in a multienzyme complex (see below), inward and therefore prohibits the insertion of the newly added triplets into the surface of the cylindrical part of the sacculus. According to the three-for-one mechanism, a triple-layered murein diaphragm would be formed, if this structure was immediately cleaved by the hydrolytic removal of the middle layer (Fig. 13B). As a result, a cleavage furrow is formed. Constriction proceeds by the addition of further triplets to the leading edge of the furrow and the continuous pull by the closing contractile ring structure (130). The contractile organelle is not well understood, but an important component is the tubulin-like FtsZ protein, which polymerizes into a ring at the site of division (12, 107, 122). FtsZ was found to interact with a number of other proteins involved in cell division, including FtsA, FtsW, and ZipA (27, 86, 164).
The final separation of the daughter cells is mechanistically a special event, where the two fused dome-shaped poles must be closed at their tips and must be detached from one another (47). After cell separation, the two new hemispherical polar caps of the daughter cells stop further insertion of murein precursors and do not grow any more (32).
INDICATION FOR A MUREIN-SYNTHESIZING MACHINERY
Hypothetical Multienzyme Complex
During growth and division of the murein sacculus, the functions of several different enzymes, hydrolases and synthases, must be coordinated in both time and space. It has therefore been speculated that by analogy to the holoenzyme of DNA replication (85), murein growth may likewise be performed by a multienzyme complex, a holoenzyme of murein replication (65).
Protein-protein interaction studies by affinity chromatography indeed demonstrated that murein hydrolases and synthases do interact with each other (128, 165). Lytic transglycosylases immobilized to a matrix (activated Sepharose) specifically retained the bifunctional transglycosylase/transpeptidases PBP1A, PBP1B, and PBP1C, the transpeptidases PBP2 and PBP3, and the endopeptidases PBP4 and PBP7. The same interactions could be shown when some of the PBPs were used as specific ligands. Surprisingly, it is this very group of enzyme specificities that is needed to enlarge murein according to the three-for-one growth mechanism. As illustrated in Fig. 14, a dimer of a bifunctional PBP (PBP1A or PBP1B) could supplement a preexisting primer murein strand to form a murein triplet, which in turn is then attached to the sacculus by transpeptidation catalyzed by a dimer of a transpeptidase such as PBP2 or PBP3. The docking strand could be removed specifically by the concerted action of a dimer of an endopeptidase (PBP4 or PBP7) splitting the cross bridges to the left and right of the old strand and a lytic transglycosylase, either Slt70, MltA, or MltB, depolymerizing the polysaccharide strand.
Since cell division takes place with the simultaneous insertion of multiple strands, the murein-synthesizing machinery responsible for cell constriction may synthesize a murein triplet in one step (Fig. 14). Therefore, it is postulated that the division machinery contains an additional monofunctional transglycosylase that synthesizes the middle strand of the triplet. The cell division machine also differs from the elongation machinery by the presence of the transpeptidase PBP3. By contrast, PBP2, the transpeptidase that is known to be responsible for the rod shape, would be a specific component of the elongation complex (Fig. 14).
Control of Potentially Autolytic Enzymes
Formation of a multienzyme complex, a murein-synthesizing machinery, would be advantageous not only for the coordination of the action of the different enzymes involved in the multistep process of growth and division of the murein sacculus but also for control of the potentially autolytic murein hydrolases. It seems that insertion into the complex may be the only way for the murein hydrolases to attack their substrate, the murein. This kind of topological control of the murein hydrolases is indicated by the observation that overproduction of cloned lytic transglycosylases, with one exception, does not result in rapid lysis as one would expect (10, 11, 101). The overproduced enzymes seem to be hindered from attacking the murein sacculus. If indeed the membrane-bound hydrolases could reach their substrate only when inserted into the complex, it would be an ideal control: in the complex, the murein hydrolases can be positioned behind the synthetic enzymes, which guarantees that they can cleave covalent bonds in the stress-bearing structure only after new material has been attached (Fig. 15).
FIG. 15.
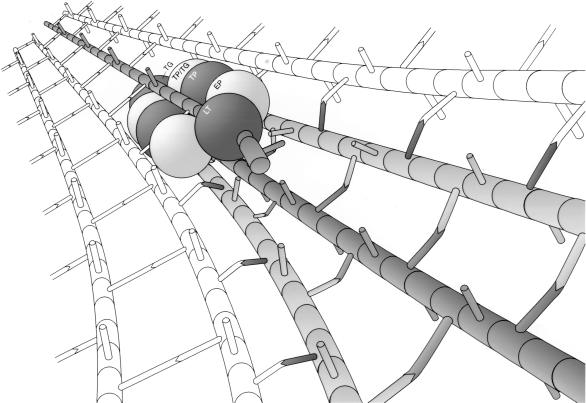
Proposed murein replicase holoenzyme in action. It is proposed that the multienzyme complex (Fig. 14) synthesizes three new murein strands (shown in grey), attaches these strands to the cross bridges on both sides of the docking strand (single grey strand), and at the same time degrades this strand by the concerted action of a processive lytic transglycosylase and a dimer of an endopeptidase. The complex slides along the docking strand with the murein synthases in front of the hydrolases, thereby acting according to the make-before-break strategy. LT, lytic transglycosylase; EP, endopeptidase; TP, transpeptidase; TP/TG, bifunctional transpeptidase-transglycosylase; TG, transglycosylase.
For the soluble Slt70, it turned out that the enzyme is rather labile except when it is being inserted into the complex. This was concluded from experiments showing that the addition of the endopeptidase PBP7 to purified enzyme preparations stabilized and greatly stimulated the enzymatic activity of Slt70 (128).
The lipoprotein lytic transglycosylases may be controlled by their anchorage to the outer membrane. Overproduction of the cloned lipoprotein MltA does not induce rapid lysis; only spheroplasts are formed (101). The membrane-bound enzymes may not be able to reach the murein layer, which is kept at a certain distance by the presence of the murein-bound lipoprotein (15, 74). Only by an interaction with the other proteins of the multienzyme complex may the membrane-anchored enzyme bind to the murein sacculus. By contrast to an overproduction of MltA and Slt70, rapid lysis is observed during overproduction of MltB (41). In this case, it was found that the enzyme can proteolytically be cleaved to a soluble, active form, called Slt35 (36), which can easily bind to the murein sacculus and cause autolysis.
A murein-synthesizing machinery containing one of the Mlts would consist of proteins anchored to both membranes, since the PBP1A, PBP1B, PBP1C, PBP2, and PBP3 are bound to the cytoplasmic membrane whereas the Mlts are linked to the outer membrane (101). Interestingly, even the soluble Slt70 was found to bind specifically to the outer membrane-facing side of the sacculus (167). Therefore, one would expect that the assembly of the machinery would give rise to the formation of membrane adhesion sites. Contacts between cytoplasmic membrane and murein at the sites of murein synthesis have been postulated before, since the murein precursors are still linked to the cytoplasmic membrane via the bactoprenol moiety when being incorporated into the sacculus (119). Indeed, adhesion sites, also called Bayer junctions, have been shown by electron microscopy (4), and increased murein-synthesizing activity has been observed in membrane fractions enriched in such sites (3, 76). More specifically, the presence of PBP1B in membrane contact sites has been demonstrated by immunogold labelling (5).
The control of the murein hydrolases brought about by their insertion into a complex together with murein synthases, although quite effective, is nevertheless not an absolute one, and autolysis can be triggered by several routes. The MS2 phage lysis protein, which by itself has no murein hydrolase activity, induces lysis by inducing the formation of contact sites between the inner and outer membranes (166, 168). It is tempting to speculate that this allows the membrane-bound lipoprotein lytic transglycosylases to interact with the murein without being inserted into the multienzyme complex and thus to cause uncontrolled lysis.
Surprisingly, all murein synthesis inhibitors, irrespective of their specific site of action, cause bacteriolysis rather than bacteriostasis (152). The general principle of all these murein synthesis inhibitors may be to cause the subcomplex of murein synthases to idle (152) while allowing the hydrolases to continue to degrade the docking strand (Fig. 16). The idling could be due to either an interruption of the flow of murein precursors or a direct inhibition of the murein-synthesizing bifunctional enzymes in the complex. Penicillin tolerance of nongrowing, starved cells that induced the stringent control (95, 125) is probably due to a dependence of the assembly process of the complex on ongoing phospholipid synthesis (40). Therefore, the complex is not formed in starved cells, with the result that the murein hydrolases have no access to the murein. This may be the molecular basis for the classical penicillin enrichment technique (105).
FIG. 16.
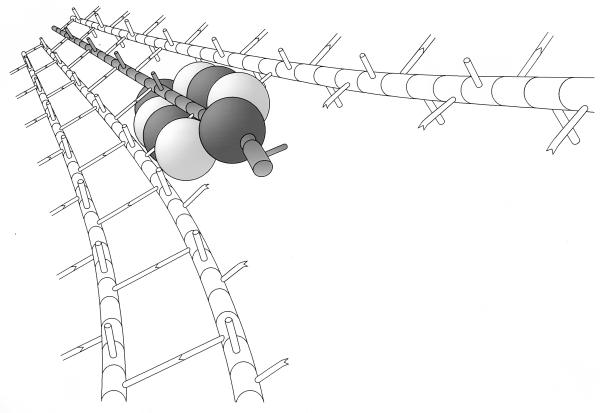
Idling of the murein replicase in the presence of murein synthesis inhibitors. It is assumed that in the presence of murein synthesis inhibitors, the multienzyme complex is still pushed along the murein strands by the action of the murein hydrolases processively degrading the docking strand while the synthases idle due to a block in the supply or utilization of the murein precursors (see the legend to Fig. 15).
MORPHOGENESIS OF E. COLI
Cell Cycle of a Rod
The specific, genetically determined shape of bacteria is precisely transmitted from generation to generation (24, 111, 135, 141). Therefore, growth and division of the shape-maintaining murein sacculus must result in the formation of two identical daughter sacculi. It has been proposed that this is accomplished by the parent sacculus playing the role of a template.
E. coli grows with a constant shape even when growing at different sizes (8, 24, 37). The specific rod shape of E. coli is defined by the ratio of the length to the width of the cylindrical middle part of the murein sacculus, assuming that the tube is closed at both ends by hemispherical caps with the diameter of the cylinder. The increase in cell size that takes place at higher growth rates (8, 38) is brought about by a proportional increase in length and width; hence, the ratio of the two parameters that defines shape is kept constant.
Only three conditions have to be met during growth for a rod-shaped bacterium to maintain its given form. First, the cylindrical middle part of the cell has to double in length; second, the diameter of the cylinder must be kept constant; and third, constriction has to take place exactly at the midpoint of the cell. How the cell can define its equator is still an intriguing and unsolved question (28, 91, 131). However, rather simple mechanisms could fulfill the other two requirements, as will be discussed below.
Maintenance of the Diameter
If the murein sacculus had a certain ordered arrangement of its glycan strands and peptide bridges as described above (Fig. 7), the diameter of the sacculus could be maintained by replacing the preexisting docking strands with strands of exactly the same length. This could be done by a copying mechanism with the old strands functioning as a template. It is conceivable that this is done by the murein-synthesizing machinery travelling along the docking strand (Fig. 15). By moving along the strand to be replicated, the enzyme complex synthesizes three copies and cross-links these along both sides of the template strand until it reaches the end of the template, where it dissociates off. Thus, the length of the newly synthesized strands would be determined by the lengths of the preexisting ones. Importantly, the total circumference of each ring structure, from whatever number and length of individual strands it is formed, will be maintained by such a copying mechanism. Growth at numerous sites, randomly distributed all over the sacculus, would result in a smooth elongation of the cell with the diameter being kept constant. Since, during maturation, the glycan strands in each cell cycle are altered in their length by ligation as well as cleavage reactions, the copying mechanism will not result in an accumulation of strands all with the same length, which was a criticism of the process (89).
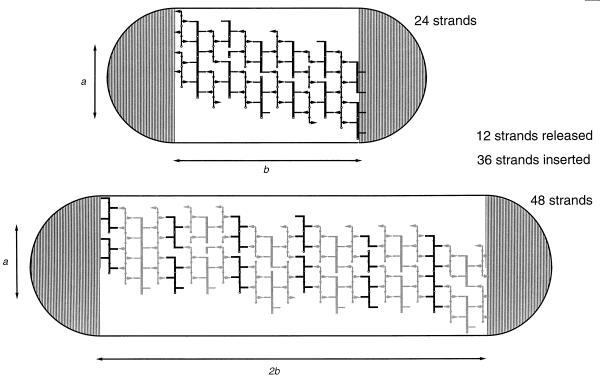
Doubling in Length
According to the three-for-one growth model, the murein sacculus grows and divides by inserting three new strands at the expense of one old strand. Replacement of all docking strands, i.e., each second strand, in the cylindrical part of the murein sacculus by a murein triplet will precisely double the length and thus the amount of murein (Fig. 17). However, this holds true only if the replacement of the docking strands in the newly inserted murein triplets is blocked until cell division. It is therefore proposed that the cross-links in newly inserted murein triplets differ from those present in the preexisting murein in such a way that only the preexisting murein but not the newly inserted one can accept new murein triplets.
FIG. 17.
Theoretical doubling of the cylindrical middle part of the murein sacculus by a three-for-one growth modus. The murein of the cylindrical middle part of a rod-shaped sacculus is schematically spread out inside the outlines of a cell, with the metabolically inert polar caps indicated by striped semicircles. Lines running perpendicular to the long axis of the rod-shaped cell indicate glycan strands. The docking strands are shown as fat lines. Arrows and lines arranged parallel to the long axis of the cell represent acceptor and donor peptides.
Possible Regulatory Function of Carboxypeptidases
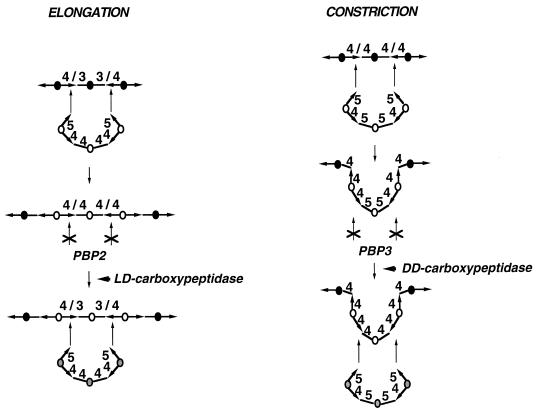
Indeed, there is a structural difference. As mentioned above, newly synthesized murein is cross-linked by tetra-tetra cross bridges whereas mature murein is characterized by the presence of tetra-tri cross-links (49) (Fig. 8D), which are the products of the action of l,d-carboxypeptidases. It is proposed that new murein triplets can be hooked only to preexisting tetra-tri cross bridges in the sacculus of the newborn cell due to the specificity of PBP2, the transpeptidase involved in cell elongation (142). It follows that further growth automatically stops when all docking strands of the preexisting murein have been replaced, since this results in murein exclusively cross-linked by tetra-tetra peptides. This coincides with a doubling in the amount of murein and the length of the cylindrical part of the sacculus. Hence, the cell can recognize when to divide. To reinitiate growth, the tetra-tetra bridges have to be converted into tetra-tri cross-links by the action of an l,d-carboxypeptidase (Fig. 18). A pulse in l,d-carboxypeptidase activity at the end of a cell cycle could therefore function as a signal for cell division and would start a new cycle.
FIG. 18.
Proposed changes in the cross-linkages during elongation and constriction of the murein sacculus. It is proposed that during cell elongation, murein triplets are exclusively hooked to tetra-tri peptide bridges due to the specificity of the PBP2-containing elongation machinery (Fig. 14). The newly inserted murein triplet is assumed to be cross-linked by tetra-tetra cross-links. Only after the action of an l,d-carboxypeptidase are the peptide bridges converted to tetra-tri cross-links that can function as acceptors for the addition of new murein triplets by the elongation complex. During cell constriction, the newly synthesized murein triplet is assumed to be hooked to tetra-tetra cross-links due to the specificity of the PBP3-containing cell division machinery (Fig. 14). The high rate of localized murein synthesis during cell division is likely to result in the presence of tetra-penta cross-links in the constriction site that may not be accepted by PBP3. Thus, the action of a d,d-carboxypeptidase converting the tetra-penta cross-links to tetra-tetra bridges may be needed for cell constriction. The circles represent the glycan strands, and the arrows and lines represent the donor and acceptor peptides of the cross-links. The numbers above the cross-links indicate whether a tri-, tetra-, or pentapeptide is present.
For cell division itself, d,d-carboxypeptidase rather than l,d-carboxypeptidase may be needed. Whereas PBP2 may depend on tetra-tri cross-links, PBP3, the transpeptidase specific for cell division, seems to depend on tetra-tetra cross-links (Fig. 18), as indicated by the finding that mostly tetra-tetra cross-links are present in septal murein (29, 113). Since tetra-penta cross bridges are formed first and then modified to tetra-tetra cross bridges by the action of d,d-carboxypeptidases, the activity of d,d-carboxypeptidase could be a limiting factor in the supply of sufficient amounts of tetra-tetra cross-links at the site of cell division. Consistent with this argument, it has been shown that certain PBP3 mutants (ftsI23) that do not divide at the restrictive temperature could be suppressed by an increase in the level of the d,d-carboxypeptidases PBP5 and/or PBP6 (9). However, in conflict with the model presented here, it was further concluded that the effect of the increase in d,d-carboxypeptidase activity was an indirect one due to a final stimulation of the formation for tripeptidyl residues by the action of l,d-carboxypeptidases on the tetrapeptidyl groups produced by the d,d-enzyme. This conclusion was drawn from the finding that d-cycloserine, which increases the number of tripeptidyl residues in the murein due to an inhibition of the d-Ala–d-Ala adding enzyme, could also suppress the ftsI23 phenotype. However, the effect could be observed only under certain conditions and only on agar plates, not in liquid cultures. Furthermore, as mentioned above, septal murein shows no increase in tetra-tri cross-links, and thus the claim that tripeptide side chains are the preferential acceptors for PBP3 in transpeptidation remains speculative.
After cell division, the tetra-tri murein skin of the newborn cells will grow old during cell elongation by getting tetra-tetra cross bridges, and the cell must await another l,d-carboxypeptidase “rejuvenation cure” to continue growth. Interestingly, an oscillation in l,d-carboxypeptidase activity during the cell cycle, with a peak at the time point of cell division, has indeed been demonstrated (7).
HEREDITY OF STRUCTURAL INFORMATION
Is the Murein Sacculus Playing the Role of a Template?
The morphogenetic changes that take place during the cell cycle of bacteria are minor compared to the complex developmental processes during the propagation of eukaryotes (59). This raises the question whether an elaborate morphogenetic program is needed to generate and maintain the specific shape of the bacterial cell as well. The existence of a simple structure, the murein sacculus, that is responsible for the specific form may facilitate bacterial morphogenesis by presenting a matrix for growth and division of the murein sacculus (135). Other examples of structural inheritance by nongenetic mechanisms are known in biology, in particular with ciliates (140).
Several experiments suggest that the existing murein sacculus indeed serves as a template. In one series of experiments, the matrix, i.e., the sacculus, was enzymatically removed during growth under osmotic protection, resulting in the formation of osmotically labile spheroplasts (64). When the bacteria were then allowed to regain a murein sacculus, the cells were unable to reshape themselves into rods but, rather, formed a spherical sacculus (136). By contrast, when osmotically stable spherical cells, surrounded by a spherical sacculus, were produced by growth of E. coli in the presence of the β-lactam mecillinam, which specifically inhibits cell elongation, rod formation started as soon as the antibiotic was removed (55). The bacteria were able to regrow the spherical sacculus into a rod-shaped sacculus with reestablishment of the original polarity of the cell. It seems that sufficient structural information was retained in the spherical sacculus to allow reversion to a rod. For the osmotically labile spheroplasts, this was not the case. Eventually, however, after a long trial-and-error process, even these cells became rod-shaped again. This has been interpreted as an accidental formation of the right primer structure, a rod-shaped outgrowth that can govern the controlled synthesis of a proper rod-shaped sacculus. Thus, there is no convincing evidence that a morphogenetic apparatus exists in bacteria that is able to create a specific shape de novo. Copying the existing sacculus may indeed be the mechanism by which bacteria transmit their shape from generation to generation.
ACKNOWLEDGMENTS
I thank Uli Schwarz for many stimulating discussions throughout my studies in his department. In addition, I acknowledge the critical comments by Miguel DePedro, Angelika Busch, David Edwards, Moritz von Rechenberg Guido Schiffer, Markus Templin, and Waldemar Vollmer. I also thank Markus Templin for his help in preparing the list of murein synthases and hydrolases of Tables 2 and 3.
REFERENCES
- 1.Asoh S, Matsuzawa H, Ishino F, Strominger J L, Matsuhashi M, Ohta T. Nucleotide sequence of the pbpA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur J Biochem. 1986;160:231–238. doi: 10.1111/j.1432-1033.1986.tb09961.x. [DOI] [PubMed] [Google Scholar]
- 2.Baquero M-R, Bouzon M, Quintela J C, Ayala J A, Moreno F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with d,d-carboxypeptidase activity. J Bacteriol. 1996;178:7106–7111. doi: 10.1128/jb.178.24.7106-7111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas J A, Diaz J, Rodriguez-Tebar A, Vazquez D. Specific location of penicillin-binding proteins within the cell envelope of Escherichia coli. J Bacteriol. 1986;165:269–275. doi: 10.1128/jb.165.1.269-275.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayer M E. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968;53:395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- 5.Bayer M H, Keck W, Bayer M E. Localization of penicillin-binding protein 1b in Escherichia coli: immunoelectron microscopy and immunotransfer studies. J Bacteriol. 1990;172:125–135. doi: 10.1128/jb.172.1.125-135.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beachey E H, Keck W, de Pedro M A, Schwarz U. Exoenzymatic activity of transglycosylase isolated from Escherichia coli. Eur J Biochem. 1981;116:355–358. doi: 10.1111/j.1432-1033.1981.tb05342.x. [DOI] [PubMed] [Google Scholar]
- 7.Beck B D, Park J T. Activity of three murein hydrolases during the cell division cycle of Escherichia coli K-12 as measured in toluene-treated cells. J Bacteriol. 1976;126:1250–1260. doi: 10.1128/jb.126.3.1250-1260.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begg K J, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg K J, Takasuga A, Edwards D H, Dewar S J, Spratt B G, Adachi H, Ohta T, Matsuzawa H, Donachie W D. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol. 1990;172:6697–6703. doi: 10.1128/jb.172.12.6697-6703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betzner A S, Keck W. Molecular cloning, overexpression and mapping of the slt gene encoding the soluble lytic transglycosylase of Escherichia coli. Mol Gen Genet. 1989;219:489–491. doi: 10.1007/BF00259625. [DOI] [PubMed] [Google Scholar]
- 11.Betzner A S, Ferreira L C, Höltje J-V, Keck W. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett. 1990;55:161–164. doi: 10.1016/0378-1097(90)90187-u. [DOI] [PubMed] [Google Scholar]
- 12.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 13.Blasco B, Pisabarro A G, de Pedro M A. Peptidoglycan biosynthesis in stationary-phase cells of Escherichia coli. J Bacteriol. 1988;170:5224–5228. doi: 10.1128/jb.170.11.5224-5228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun V, Wolff H. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem. 1970;14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- 15.Braun V, Hantke K. Biochemistry of bacterial cell envelopes. Annu Rev Biochem. 1974;43:89–121. doi: 10.1146/annurev.bi.43.070174.000513. [DOI] [PubMed] [Google Scholar]
- 16.Braun V, Gnirke H, Henning U, Rehn K. Model for the structure of the shape-maintaining layer of the Escherichia coli cell envelope. J Bacteriol. 1973;114:1264–1270. doi: 10.1128/jb.114.3.1264-1270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broome-Smith J K, Edelman A, Yousif S, Spratt B G. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding proteins 1A and 1B of Escherichia coli K12. Eur J Biochem. 1985;147:437–446. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- 18.Broome-Smith J K, Ioannidis I, Edelman A, Spratt B G. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 1988;16:1617. doi: 10.1093/nar/16.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burman L G, Park J T. Changes in the composition of Escherichia coli murein as it ages during exponential growth. J Bacteriol. 1983;155:447–453. doi: 10.1128/jb.155.2.447-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burman L G, Park J T. Molecular model for elongation of the murein sacculus of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:1844–1848. doi: 10.1073/pnas.81.6.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burman L G, Raichler J, Park J T. Evidence for diffuse growth of the cylindrical portion of the Escherichia coli murein sacculus. J Bacteriol. 1983;155:983–988. doi: 10.1128/jb.155.3.983-988.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caparros M, Pisabarro A G, de Pedro M A. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cashel M, Rudd K E. The stringent response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium. Washington, D.C: American Society for Microbiology; 1987. pp. 1410–1438. [Google Scholar]
- 24.Cooper S. Bacterial growth and division. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 25.Cooper S, Hsieh M L. The rate and topography of cell wall synthesis during the division cycle of Escherichia coli using N-acetylglucosamine as a peptidoglycan label. J Gen Microbiol. 1988;134:1717–1721. doi: 10.1099/00221287-134-6-1717. [DOI] [PubMed] [Google Scholar]
- 26.Cooper S, Hsieh M-L, Guenther B. Mode of peptidoglycan synthesis in Salmonella typhimurium: single-strand insertion. J Bacteriol. 1988;170:3509–3512. doi: 10.1128/jb.170.8.3509-3512.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Boer P A, Cook W R, Rothfield L I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- 28.de Boer P A, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jonge B L, Wientjes F B, Jurida I, Driehuis F, Wouters J T, Nanninga N. Peptidoglycan synthesis during the cell cycle of Escherichia coli: composition and mode of insertion. J Bacteriol. 1989;171:5783–5794. doi: 10.1128/jb.171.11.5783-5794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demchick P, Koch A L. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Pedro M A, Schwarz U. Heterogeneity of newly inserted and preexisting murein in the sacculus of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5856–5860. doi: 10.1073/pnas.78.9.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Pedro M A, Quintela J C, Höltje J-V, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Berardino M, Dijkstra A, Stüber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 34.Dijkstra A J, Keck W. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb Drug Resist. 1996;2:141–145. doi: 10.1089/mdr.1996.2.141. [DOI] [PubMed] [Google Scholar]
- 35.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dijkstra A J, Hermann F, Keck W. Cloning and controlled overexpression of the gene encoding the 35 kDa soluble lytic transglycosylase from Escherichia coli. FEBS Lett. 1995;366:115–118. doi: 10.1016/0014-5793(95)00505-4. [DOI] [PubMed] [Google Scholar]
- 37.Donachie W D. The cell cycle of Escherichia coli. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 38.Donachie W D, Begg K J, Vicente M. Changes in cell diameter during the division cycle of Escherichia coli. J Bacteriol. 1980;142:869–877. doi: 10.1128/jb.142.3.869-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driehuis F, Wouters J. Effect of growth rate and cell shape on the peptidoglycan composition in Escherichia coli. J Bacteriol. 1987;169:97–101. doi: 10.1128/jb.169.1.97-101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehlert K, Höltje J-V. Role of precursor translocation in coordination of murein and phospholipid synthesis in Escherichia coli. J Bacteriol. 1996;178:6766–6771. doi: 10.1128/jb.178.23.6766-6771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehlert K, Höltje J-V, Templin M F. Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16:761–768. doi: 10.1111/j.1365-2958.1995.tb02437.x. [DOI] [PubMed] [Google Scholar]
- 42.Engel H, Kazemier B, Keck W. Murein-metabolizing enzymes from Escherichia coli: sequence analysis and controlled overexpression of the slt gene, which encodes the soluble lytic transglycosylase. J Bacteriol. 1991;173:6773–6782. doi: 10.1128/jb.173.21.6773-6782.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel H, Smink A J, van Wijngaarden L, Keck W. Murein-metabolizing enzymes from Escherichia coli: existence of a second lytic transglycosylase. J Bacteriol. 1992;174:6394–6403. doi: 10.1128/jb.174.20.6394-6403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghuysen J M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 45.Ghuysen J-M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 46.Ghuysen J-M. Penicillin-binding proteins—wall peptidoglycan assembly and resistance to penicillin—facts, doubts and hopes. Int J Antimicrob Agents. 1997;8:45–60. doi: 10.1016/s0924-8579(96)00358-5. [DOI] [PubMed] [Google Scholar]
- 47.Giesbrecht P, Wecke J, Reinicke B. On the morphogenesis of the cell wall of Staphylococci. Int Rev Cytol. 1976;44:225–318. doi: 10.1016/s0074-7696(08)61651-4. [DOI] [PubMed] [Google Scholar]
- 48.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 49.Glauner B, Höltje J-V. Growth pattern of the murein sacculus of Escherichia coli. J Biol Chem. 1990;265:18988–18996. [PubMed] [Google Scholar]
- 50.Glauner B, Höltje J-V, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 51.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Disteche M, Ghuysen J-M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodell E W. Recycling of murein by Escherichia coli. J Bacteriol. 1985;163:305–310. doi: 10.1128/jb.163.1.305-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodell E W, Higgins C F. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol. 1987;169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodell E W, Markiewicz Z, Schwarz U. Absence of oligomeric murein intermediates in Escherichia coli. J Bacteriol. 1983;156:130–135. doi: 10.1128/jb.156.1.130-135.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodell E W, Schwarz U. Sphere-rod morphogenesis of Escherichia coli. J Gen Microbiol. 1975;86:201–209. doi: 10.1099/00221287-86-2-201. [DOI] [PubMed] [Google Scholar]
- 56.Goodell E W, Schwarz U. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J Bacteriol. 1985;162:391–397. doi: 10.1128/jb.162.1.391-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenway D L, Perkins H R. Turnover of the cell wall peptidoglycan during growth of Neisseria gonorrhoeae and Escherichia coli. Relative stability of newly synthesized material. J Gen Microbiol. 1985;131:253–263. doi: 10.1099/00221287-131-2-253. [DOI] [PubMed] [Google Scholar]
- 58.Hara H, Suzuki H. A novel glycan polymerase that synthesizes uncross-linked peptidoglycan in Escherichia coli. FEBS Lett. 1984;168:155–160. doi: 10.1016/0014-5793(84)80226-4. [DOI] [PubMed] [Google Scholar]
- 59.Harold F M. From morphogenes to morphogenesis. Microbiology. 1995;141:2765–2778. doi: 10.1099/13500872-141-11-2765. [DOI] [PubMed] [Google Scholar]
- 60.Hartmann E, König H. Comparison of the biosynthesis of the methanobacterial pseudomurein and the eubacterial murein. Naturwissenschaften. 1990;77:472–475. doi: 10.1007/BF01135923. [DOI] [PubMed] [Google Scholar]
- 61.Hartmann R, Bock-Hennig S B, Schwarz U. Murein hydrolases in the envelope of Escherichia coli. Properties in situ and solubilization from the envelope. Eur J Biochem. 1974;41:203–208. doi: 10.1111/j.1432-1033.1974.tb03261.x. [DOI] [PubMed] [Google Scholar]
- 62.Harz H, Burgdorf K, Höltje J-V. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal Biochem. 1990;190:120–128. doi: 10.1016/0003-2697(90)90144-x. [DOI] [PubMed] [Google Scholar]
- 63.Henderson T A, Templin M, Young K D. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J Bacteriol. 1995;177:2074–2079. doi: 10.1128/jb.177.8.2074-2079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henning U, Rehn K, Braun V, Höhn B, Schwarz U. Cell envelope and shape of Escherichia coli K12: properties of a temperature-sensitive rod mutant. Eur J Biochem. 1972;26:570–586. doi: 10.1111/j.1432-1033.1972.tb01800.x. [DOI] [PubMed] [Google Scholar]
- 65.Höltje J. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology. 1996;142:1911–1918. doi: 10.1099/13500872-142-8-1911. [DOI] [PubMed] [Google Scholar]
- 66.Höltje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 67.Höltje J-V, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Höltje J-V. “Three for one”—a simple growth mechanism that guarantees a precise copy of the thin, rod-shaped murein sacculus of Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 419–426. [Google Scholar]
- 69.Höltje J-V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 70.Höltje J-V. Molecular interplay of murein synthases and murein hydrolases in Escherichia coli. Microb Drug Resist. 1996;2:99–103. doi: 10.1089/mdr.1996.2.99. [DOI] [PubMed] [Google Scholar]
- 71.Höltje J-V, Schwarz U. Biosynthesis and growth of the murein sacculus. In: Nanninga N, editor. Molecular cytology of Escherichia coli. New York, N.Y: Academic Press, Inc.; 1985. pp. 77–119. [Google Scholar]
- 72.Höltje J-V, Kopp U, Ursinus A, Wiedemann B. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-amidase. FEMS Microbiol Lett. 1994;122:159–164. doi: 10.1111/j.1574-6968.1994.tb07159.x. [DOI] [PubMed] [Google Scholar]
- 73.Ikeda M, Sato T, Wachi M, Jung H K, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inouye M. Lipoprotein of the outer membrane of Escherichia coli. In: Manson L A, editor. Biomembranes. New York, N.Y: Plenum Press; 1979. pp. 141–207. [DOI] [PubMed] [Google Scholar]
- 75.Isaac L, Ware G C. The flexibility of bacterial cell walls. J Appl Bacteriol. 1974;37:335–339. doi: 10.1111/j.1365-2672.1974.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 76.Ishidate K, Creeger E S, Zrike J, Deb S, Glauner B, MacAlister T J, Rothfield L I. Isolation of differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J Biol Chem. 1986;261:428–443. [PubMed] [Google Scholar]
- 77.Ishiguro E E. Apparent obligatory dependence of peptidoglycan synthesis on phospholipid synthesis studied in ether-treated Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 177–181. [Google Scholar]
- 78.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97:287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 79.Jacobs C, Frere J-M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 80.Jacobs C, Huang L J, Bartowsky E, Normark S, Park J T. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobs C, Joris B, Jamin M, Klarsov K, Van Beeumen J, Mengin-Lecreulx D, van Heijenoort J, Park J T, Normark S, Frere J-M. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 82.Kandler O, König H. Cell envelopes of archaebacteria. In: Woese C R, Wolfe R S, editors. The bacteria. New York, N.Y: Academic Press, Inc.; 1985. pp. 413–457. [Google Scholar]
- 83.Keck W, Schwarz U. Escherichia coli murein-dd-endopeptidase insensitive to β-lactam antibiotics. J Bacteriol. 1979;139:770–774. doi: 10.1128/jb.139.3.770-774.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keck W, van Leeuwen A M, Huber M, Goodell E W. Cloning and characterisation of mepA, the structural gene of the penicillin-insensitive murein hydrolase from Escherichia coli. Mol Microbiol. 1990;4:209–219. doi: 10.1111/j.1365-2958.1990.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 85.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 86.Khattar M M, Addinall S G, Stedul K H, Boyle D S, Lutkenhaus J, Donachie W D. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J Bacteriol. 1997;179:784–793. doi: 10.1128/jb.179.3.784-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koch A L. On the growth and form of Escherichia coli. J Gen Microbiol. 1982;128:2527–2539. doi: 10.1099/00221287-128-11-2527. [DOI] [PubMed] [Google Scholar]
- 88.Koch A L. Additional arguments for the key role of “smart” autolysins in the enlargement of the wall of Gram-negative bacteria. Res Microbiol. 1990;141:529–541. doi: 10.1016/0923-2508(90)90017-k. [DOI] [PubMed] [Google Scholar]
- 89.Koch A L. Bacterial growth and form. New York, N.Y: Chapman & Hall; 1995. [Google Scholar]
- 90.Koch A L, Doyle R J. Inside-to-outside growth and turnover of the wall of gram-positive rods. J Theor Biol. 1985;117:137–157. doi: 10.1016/s0022-5193(85)80169-7. [DOI] [PubMed] [Google Scholar]
- 91.Koch A L, Höltje J-V. A physical basis for the precise location of the division site of rod-shaped bacteria: the central stress model. Microbiology. 1995;141:3171–3180. [Google Scholar]
- 92.Kohlrausch U, Höltje J-V. Analysis of murein and murein precursors during antibiotic-induced lysis of Escherichia coli. J Bacteriol. 1991;173:3425–3431. doi: 10.1128/jb.173.11.3425-3431.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korat B, Mottl H, Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol Microbiol. 1991;5:675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 94.Kraft, A. R., M. F. Templin, and J.-V. Höltje. Unpublished data.
- 95.Kusser W, Ishiguro E E. Involvement of the relA gene in the autolysis of Escherichia coli induced by inhibitors of peptidoglycan biosynthesis. J Bacteriol. 1985;164:861–865. doi: 10.1128/jb.164.2.861-865.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Labischinski H, Barnickel G, Bradaczek H, Giesbrecht P. On the secondary and tertiary structure of murein. Eur J Biochem. 1979;95:147–155. doi: 10.1111/j.1432-1033.1979.tb12949.x. [DOI] [PubMed] [Google Scholar]
- 97.Labischinski H, Barnickel G, Naumann D, Keller P. Conformational and topological aspects of the three-dimensional architecture of bacterial peptidoglycan. Ann Inst Pasteur/Microbiol. 1985;136A:45–50. doi: 10.1016/s0769-2609(85)80020-x. [DOI] [PubMed] [Google Scholar]
- 98.Labischinski H, Goodell E W, Goodell A, Hochberg M L. Direct proof of a “more-than-single-layered” peptidoglycan architecture of Escherichia coli W7: a neutron small-angle scattering study. J Bacteriol. 1991;173:751–756. doi: 10.1128/jb.173.2.751-756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lindquist S, Weston-Hafer K, Schmidt H, Pul C, Korfmann G, Erikson J, Sanders C, Martin H H, Normark S. AmpG, a signal transducer in chromosomal β-lactamase induction. Mol Microbiol. 1993;9:703–715. doi: 10.1111/j.1365-2958.1993.tb01731.x. [DOI] [PubMed] [Google Scholar]
- 100.Lleo M M, Canepari L P, Satta G. Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J Bacteriol. 1990;172:3758–3771. doi: 10.1128/jb.172.7.3758-3771.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lommatzsch J, Templin M, Kraft A R, Vollmer W, Höltje J-V. Outer membrane localization of murein hydrolases: MltA, a third lipoprotein lytic transglycosylase in Escherichia coli. J Bacteriol. 1997;179:5465–5470. doi: 10.1128/jb.179.17.5465-5470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matsuhashi M, Wachi M, Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990;141:89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- 103.Mengin-Lecreulx D, van Heijenoort J, Park J T. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mett H, Bracha R, Mirelman D. Soluble nascent peptidoglycan in growing E. coli cells. J Biol Chem. 1980;255:9884–9890. [PubMed] [Google Scholar]
- 105.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 230–234. [Google Scholar]
- 106.Mottl H, Keck W. Purification of penicillin-binding protein 4 of Escherichia coli as a soluble protein by dye-affinity chromatography. Eur J Biochem. 1991;200:767–773. doi: 10.1111/j.1432-1033.1991.tb16243.x. [DOI] [PubMed] [Google Scholar]
- 107.Mukherjee A K, Dai K, Lutkenhaus J. Escherichia coli cell division protein ftsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mulder E, Woldringh C L. Autoradiographic analysis of diaminopimelic acid incorporation in filamentous cells of Escherichia coli: repression of peptidoglycan synthesis around the nucleoid. J Bacteriol. 1991;173:4751–4756. doi: 10.1128/jb.173.15.4751-4756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakagawa J, Tamaki S, Matsuhashi M. Purified penicillin binding proteins 1Bs from Escherichia coli membrane showing activities of both peptidoglycan polymerase and peptidoglycan crosslinking enzyme. Agric Biol Chem. 1979;43:1379–1380. [Google Scholar]
- 110.Nakamura M, Maruyama I N, Soma M, Kato J I, Suzuki H, Hirota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 111.Nanninga N. Growth and form in microorganisms: morphogenesis of Escherichia coli. Can J Microbiol. 1988;34:381–389. doi: 10.1139/m88-069. [DOI] [PubMed] [Google Scholar]
- 112.Nanninga N, Wientjes F B, de Jonge B L, Woldringh C L. Polar cap formation during cell division in Escherichia coli. Res Microbiol. 1990;141:103–118. doi: 10.1016/0923-2508(90)90102-v. [DOI] [PubMed] [Google Scholar]
- 113.Obermann W, Höltje J-V. Alterations of murein structure and of penicillin-binding proteins in minicells from Escherichia coli. Microbiology. 1994;140:79–87. doi: 10.1099/13500872-140-1-79. [DOI] [PubMed] [Google Scholar]
- 114.Park J T. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J Bacteriol. 1993;175:7–11. doi: 10.1128/jb.175.1.7-11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park J T. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 116.Park J T. The murein sacculus. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 117.Park J T. The convergence of murein recycling research with β-lactamase research. Microb Drug Resist. 1996;2:105–112. doi: 10.1089/mdr.1996.2.105. [DOI] [PubMed] [Google Scholar]
- 118.Park, J. T. Personal communication.
- 119.Park J T, Burman L G. Elongation of the murein sacculus of Escherichia coli. Ann Inst Pasteur/Microbiol. 1985;136A:51–58. doi: 10.1016/s0769-2609(85)80021-1. [DOI] [PubMed] [Google Scholar]
- 120.Pisabarro A G, de Pedro M A, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prats R, de Pedro M A. Normal growth and division of Escherichia coli with a reduced amount of murein. J Bacteriol. 1989;171:3740–3745. doi: 10.1128/jb.171.7.3740-3745.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.RayChaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 123.Ritzeler O, Hennig L, Findeisen M, Welzel P, Muller D, Markus A, Lemoine G, Lampilas M, van Heijenoort J. Synthesis of a trisaccharide analogue of moenomycin A(12)—implications of new moenomycin structure-activity relationships. Tetrahedron. 1997;53:1675–1694. [Google Scholar]
- 124.Rodionov D G, Ishiguro E E. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol. 1995;177:4224–4229. doi: 10.1128/jb.177.15.4224-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rodionov D G, Pisabarro A G, de Pedro M A, Kusser W, Ishiguro E E. Beta-lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J Bacteriol. 1995;177:992–997. doi: 10.1128/jb.177.4.992-997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, United Kingdom: Chapman & Hall, Ltd.; 1980. [Google Scholar]
- 127.Romeis T, Höltje J-V. Penicillin-binding protein 7/8 of Escherichia coli is a d,d-endopeptidase. Eur J Biochem. 1994;224:597–604. doi: 10.1111/j.1432-1033.1994.00597.x. [DOI] [PubMed] [Google Scholar]
- 128.Romeis T, Höltje J-V. Specific interaction of penicillin-binding proteins 3 and 7/8 with the soluble lytic transglycosylase in Escherichia coli. J Biol Chem. 1994;269:21603–21607. [PubMed] [Google Scholar]
- 129.Romeis T, Vollmer W, Höltje J-V. Characterization of three different lytic transglycosylases in Escherichia coli. FEMS Microbiol Lett. 1993;111:141–146. doi: 10.1111/j.1574-6968.1993.tb06376.x. [DOI] [PubMed] [Google Scholar]
- 130.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 131.Rothfield L I, Zhao C R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 132.Satta G, Fontana R, Canepari P. The two-competing site (TCS) model for cell shape regulation in bacteria. The envelope as an integration point for the regulatory circuits of essential physiological events. Adv Microb Physiol. 1994;36:181–245. doi: 10.1016/s0065-2911(08)60180-0. [DOI] [PubMed] [Google Scholar]
- 133.Schiffer, G., and J.-V. Höltje. Unpublished data.
- 134.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schwarz U. The sacculus after four decades—seen from some distance. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 1–7. [Google Scholar]
- 136.Schwarz U, Leutgeb W. Morphogenetic aspects of murein structure and biosynthesis. J Bacteriol. 1971;106:588–595. doi: 10.1128/jb.106.2.588-595.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schwarz U, Asmus A, Frank H. Autolytic enzymes and cell division of Escherichia coli. J Mol Biol. 1969;41:419–429. doi: 10.1016/0022-2836(69)90285-x. [DOI] [PubMed] [Google Scholar]
- 138.Schwarz U, Ryter A, Rambach A, Hellio R, Hirota Y. Process of cellular division in Escherichia coli: differentiation of growth zones in the sacculus. J Mol Biol. 1975;98:749–759. doi: 10.1016/s0022-2836(75)80008-8. [DOI] [PubMed] [Google Scholar]
- 139.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishing; 1994. pp. 131–166. [Google Scholar]
- 140.Sonneborn T M. Does preformed cell structure play an essential role in cell heredity? In: Allen J M, editor. The nature of biological diversity. New York, N.Y: McGraw-Hill Inc.; 1963. pp. 165–221. [Google Scholar]
- 141.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suzuki H, Nishimura Y, Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA. 1978;75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Taylor A, Das B C, van Heijenoort J. Bacterial-cell wall peptidoglycan fragments produced by phage lambda or Vi II endolysin and containing 1,6-anhydro-N-acetylmuramic acid. J Biochem. 1975;53:47–54. [Google Scholar]
- 145.Templin, M. F. Unpublished data.
- 146.Templin M F, Edwards D H, Höltje J-V. A murein hydrolase is the specific target of Bulgecin in Escherichia coli. J Biol Chem. 1992;267:20039–20043. [PubMed] [Google Scholar]
- 147.Templin, M. F., A. Ursinus, and J.-V. Höltje. Unpublished data.
- 148.Thunnissen A M W H, Dijkstra A J, Kalk K H, Rozeboom H J, Engel H, Keck W, Dijkstra B W. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature. 1994;367:750–753. doi: 10.1038/367750a0. [DOI] [PubMed] [Google Scholar]
- 149.Thunnissen A-M W H, Rozeboom H J, Kalk K H, Dijkstra B W. Structure of the 70-kDa soluble lytic transglycosylase complexed with Bulgecin A. Implications for the enzymatic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]
- 150.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tomasz A. Murein hydrolases—enzymes in search of a physiological function. In: Hakenbeck R, Höltje J-V, Labischinski H, editors. The target of penicillin. Berlin, Germany: Walter de Gruyter; 1983. pp. 155–172. [Google Scholar]
- 152.Tomasz, A. 1986. Penicillin-binding proteins and the antibacterial effectiveness of beta-lactam antibiotics. Rev. Infect. Dis. 8(Suppl. 3):260–378. [DOI] [PubMed]
- 153.Tomioka S, Matsuhashi M. Purification of penicillin-insensitive d,d-endopeptidase, a new cell wall peptidoglycan-hydrolyzing enzyme in Escherichia coli, and its inhibition by deoxyribonucleic acids. Biochem Biophys Res Commun. 1978;84:978–984. doi: 10.1016/0006-291x(78)91679-0. [DOI] [PubMed] [Google Scholar]
- 154.Tomioka S, Nikaido T, Miyakawa T, Matsuhashi M. Mutation of the N-acetylmuramyl-l-alanine amidase gene of Escherichia coli. J Bacteriol. 1983;156:463–465. doi: 10.1128/jb.156.1.463-465.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsui H C, Zhao G, Feng G, Leung H-C E, Winkler M E. The mutL repair gene of Escherichia coli K-12 forms a superoperon with a gene encoding a new cell-wall amidase. Mol Microbiol. 1994;11:189–202. doi: 10.1111/j.1365-2958.1994.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 156.Tuomanen E, Cozens R. Changes in peptidoglycan composition and penicillin-binding proteins in slow-growing Escherichia coli. J Bacteriol. 1987;169:5308–5310. doi: 10.1128/jb.169.11.5308-5310.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tuomanen E, Schwartz J. Penicillin-binding protein 7 and its relationship to lysis of nongrowing Escherichia coli. J Bacteriol. 1987;169:4912–4915. doi: 10.1128/jb.169.11.4912-4915.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ursinus A, Höltje J-V. Purification and properties of a membrane-bound lytic transglycosylase from Escherichia coli. J Bacteriol. 1994;176:338–343. doi: 10.1128/jb.176.2.338-343.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ursinus A, Steinhaus H, Höltje J-V. Purification of a nocardicin A-sensitive ld-carboxypeptidase from Escherichia coli by affinity chromatography. J Bacteriol. 1992;174:441–446. doi: 10.1128/jb.174.2.441-446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Heijenoort J. Biosynthesis of the bacterial peptidoglycan unit. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science Publishing; 1994. pp. 39–54. [Google Scholar]
- 161.van Heijenoort J, Parquet C, Flouret B, van Heijenoort Y. Envelope-bound N-acetylmuramyl-l-alanine amidase of Escherichia coli K 12. Purification and properties of the enzyme. Eur J Biochem. 1975;58:611–619. doi: 10.1111/j.1432-1033.1975.tb02412.x. [DOI] [PubMed] [Google Scholar]
- 162.Verwer R W, Nanninga N, Keck W, Schwarz U. Arrangement of glycan chains in the sacculus of Escherichia coli. J Bacteriol. 1978;136:723–729. doi: 10.1128/jb.136.2.723-729.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Verwer R W, Beachey E H, Keck W, Stoub A M, Poldermans J E. Oriented fragmentation of Escherichia coli sacculi by sonication. J Bacteriol. 1980;141:327–332. doi: 10.1128/jb.141.1.327-332.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vicente M, Errington J. Structure, function and controls in microbial division. Mol Microbiol. 1996;20:1–7. doi: 10.1111/j.1365-2958.1996.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 165.von Rechenberg M, Ursinus A, Höltje J-V. Affinity chromatography as a means to study multi-enzyme-complexes involved in murein synthesis. Microb Drug Resist. 1996;2:155–157. doi: 10.1089/mdr.1996.2.155. [DOI] [PubMed] [Google Scholar]
- 166.Walderich B, Höltje J-V. Specific localization of the lysis protein of bacteriophage MS2 in membrane adhesion sites of Escherichia coli. J Bacteriol. 1989;171:3331–3336. doi: 10.1128/jb.171.6.3331-3336.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Walderich B, Höltje J-V. Subcellular distribution of the soluble lytic transglycosylase in Escherichia coli. J Bacteriol. 1991;173:5668–5676. doi: 10.1128/jb.173.18.5668-5676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Walderich B, Ursinus-Wössner A, van Duin J, Höltje J-V. Induction of the autolytic system of Escherichia coli by specific insertion of bacteriophage MS2 lysis protein into the bacterial cell envelope. J Bacteriol. 1988;170:5027–5033. doi: 10.1128/jb.170.11.5027-5033.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ward J B, Perkins H P. The direction of glycan synthesis in a bacterial peptidoglycan. Biochem J. 1973;135:721–728. doi: 10.1042/bj1350721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weidel W, Pelzer H. Bagshaped macromolecules—a new outlook on bacterial cell walls. Adv Enzymol. 1964;26:193–232. doi: 10.1002/9780470122716.ch5. [DOI] [PubMed] [Google Scholar]
- 171.Wientjes F B, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wientjes F B, Nanninga N. On the role of the high molecular weight penicillin-binding proteins in the cell cycle of Escherichia coli. Res Microbiol. 1991;142:333–344. doi: 10.1016/0923-2508(91)90049-g. [DOI] [PubMed] [Google Scholar]
- 173.Wientjes F B, Woldringh C L, Nanninga N. Amount of peptidoglycan in cell walls of gram-negative bacteria. J Bacteriol. 1991;173:7684–7691. doi: 10.1128/jb.173.23.7684-7691.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wise E M, Jr, Park J T. Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc Natl Acad Sci USA. 1965;54:75–81. doi: 10.1073/pnas.54.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Woldringh C L, Huls P, Pas E, Brakenhoff G J, Nanninga N. Topography of peptidoglycan synthesis during elongation and polar cap formation in a cell division mutant of Escherichia coli MC4100. J Gen Microbiol. 1987;133:575–586. [Google Scholar]
- 176.Yem D W, Wu H C. Purification and properties of β-N-acetylglucosaminidase from Escherichia coli. J Bacteriol. 1976;125:324–331. doi: 10.1128/jb.125.1.324-331.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]