Abstract
The maltose system of Escherichia coli offers an unusually rich set of enzymes, transporters, and regulators as objects of study. This system is responsible for the uptake and metabolism of glucose polymers (maltodextrins), which must be a preferred class of nutrients for E. coli in both mammalian hosts and in the environment. Because the metabolism of glucose polymers must be coordinated with both the anabolic and catabolic uses of glucose and glycogen, an intricate set of regulatory mechanisms controls the expression of mal genes, the activity of the maltose transporter, and the activities of the maltose/maltodextrin catabolic enzymes. The ease of isolating many of the mal gene products has contributed greatly to the understanding of the structures and functions of several classes of proteins. Not only was the outer membrane maltoporin, LamB, or the phage lambda receptor, the first virus receptor to be isolated, but also its three-dimensional structure, together with extensive knowledge of functional sites for ligand binding as well as for phage λ binding, has led to a relatively complete description of this sugar-specific aqueous channel. The periplasmic maltose binding protein (MBP) has been studied with respect to its role in both maltose transport and maltose taxis. Again, the combination of structural and functional information has led to a significant understanding of how this soluble receptor participates in signaling the presence of sugar to the chemosensory apparatus as well as how it participates in sugar transport. The maltose transporter belongs to the ATP binding cassette family, and although its structure is not yet known at atomic resolution, there is some insight into the structures of several functional sites, including those that are involved in interactions with MBP and recognition of substrates and ATP. A particularly astonishing discovery is the direct participation of the transporter in transcriptional control of the mal regulon. The MalT protein activates transcription at all mal promoters. A subset also requires the cyclic AMP receptor protein for transcription. The MalT protein requires maltotriose and ATP as ligands for binding to a dodecanucleotide MalT box that appears in multiple copies upstream of all mal promoters. Recent data indicate that the ATP binding cassette transporter subunit MalK can directly inhibit MalT when the transporter is inactive due to the absence of substrate. Despite this wealth of knowledge, there are still basic issues that require clarification concerning the mechanism of MalT-mediated activation, repression by the transporter, biosynthesis and assembly of the outer membrane and inner membrane transporter proteins, and interrelationships between the mal enzymes and those of glucose and glycogen metabolism.
INTRODUCTION AND SCOPE
Historically, some of the earliest work on Escherichia coli genetics and physiology in the Jacob and Monod groups concentrated on the maltose system, but it was rapidly overshadowed by the spectacular success in understanding the lactose operon. Fortunately, interest in the maltose system persisted as a result of its being an example of a “positively regulated” group of genes and the mysterious connection between maltose metabolism and the susceptibility of E. coli to phage λ infection.
The maltose system is responsible for the uptake and efficient catabolism of α(1→4)-linked glucose polymers (maltodextrins) up to 7 to 8 glucose units. This system has turned out to be a far richer source of interesting molecules and regulatory phenomena than anyone might have anticipated 30 years ago when the malA and malB regions were mapped on the E. coli chromosome (239). For example, in the course of studying the receptor activity of E. coli responsible for λ attachment, Randall and Schwartz discovered the λ receptor (LamB) (207) and, in collaboration with Hofnung, Szmelcman, and one of us (W.B.), showed that this outer membrane protein constituted the channel for the passage of sugars, especially maltodextrins, across the outer membrane (263, 265). This information focused attention on the possibility of porins as specific channels and culminated in the solution of the three-dimensional structure of the maltoporin channel associated with malto-oligosaccharide ligands (231). Together with a large body of information about the functional sites within LamB, the structure represents one of the most complete descriptions of a membrane transport protein.
From the earliest time that the maltose system was studied, transcriptional control was a major focus because of the interest in a positively controlled system. The MalT protein, which is the activator at all mal promoters (112, 206, 214), and the sites at which it binds (MalT boxes) have provided an important alternative view to the lac operon about how transcription is activated. Although the precise mechanism of how MalT interacts with and stimulates RNA polymerase is still unknown, there is considerable information about the structural requirements for MalT binding to DNA as well as its ability to form a nucleoprotein complex with the catabolite activator protein (CAP).
The periplasmic binding protein-dependent ATP binding cassette (ABC) transporter of the maltose system has been studied to understand the mechanism of transport and the mechanism of protein localization to different cellular compartments (e.g., inner membrane, outer membrane, and periplasm) (21, 156, 173, 250), as well as the determinants of membrane protein topology. The periplasmic maltose binding protein (MBP) has been characterized structurally (257), and a variety of functional sites within its structure for interacting with the chemosensory apparatus and the ABC transporter have been identified. The actual membrane transporter (MalFGK2), made up of two integral membrane proteins (MalF and MalG) and two copies of the ATP-hydrolyzing subunit (MalK), has been characterized biochemically, and there is some information about functional sites that are important for substrate recognition, ATP binding, and subunit contacts and interactions with MBP. The ability to manipulate the transporter genetically offers a rare opportunity to dissect the mechanism of transport by a combined approach of biochemistry and genetics. The direct participation of the transporter in transcriptional regulation indicates that gene regulation in bacteria may depend on the rates of substrate entry in addition to the physical presence of the substrate itself.
Reviews on the maltose system of E. coli have appeared (22, 241). In this review, we attempt to summarize what is currently known about this system. We have focused on three areas: (i) transport of maltodextrins into the cell via an outer membrane porin and a periplasmic binding protein-dependent ABC transporter, (ii) metabolism of the internalized sugars, and (iii) transcriptional regulation of the mal genes. We attempt to clarify what we believe are significant functional relationships between transport activity and transcriptional control. Also, we discuss the interconnections between the maltose-degrading enzymes and those of glucose catabolism and gluconeogenesis from the perspective of how endogenous inducers of the mal system are produced. Because the initial breakdown of maltodextrins results in both glucose and glucose-1-phosphate, these enzyme activities must be coordinated with those for glycogen synthesis and breakdown. Finally, other less obvious connections with phosphate metabolism and trehalose metabolism are also discussed. We will not consider aspects of secretion (169), folding (272), or assembly (151, 223) of the components of the maltose system.
MALTOSE GENES AND THEIR PRODUCTS
Table 1 summarizes all known mal genes in E. coli. Their definition is based on the function of MalT. Thus, all genes regulated by MalT belong to the maltose regulon, while the expression of malT itself is independent of MalT. malP and malQ encode essential enzymes for maltose and maltodextrin metabolism, whereas malS and malZ encode nonessential maltodextrin-metabolizing enzymes. malEFG and malK lamB encode the binding protein-dependent ABC transporter. Some mal genes are organized in clusters. The malA region at 76.5 min contains two divergently orientated operons, with malT transcribed clockwise and malPQ transcribed counterclockwise (65, 126, 200). Likewise, all maltose transport genes are clustered at 91.4 min in the malB region with two divergently organized operons: malEFG in the counterclockwise orientation and malK lamB malM in the clockwise orientation (203, 254).
TABLE 1.
mal genes and their products, genes controlling mal gene expression, and genes related to maltodextrin metabolism
| Gene | Position on chromosome (min) | Gene product and function | Reference(s) |
|---|---|---|---|
| mal genes and their main regulator | |||
| malT | 76.5 | Transcriptional activator, essential for transcription of all mal genes except the malI/X/Y gene cluster. Binds ATP and maltotriose as inducer. | 34, 40, 66, 206, 214, 215, 280 |
| malE | 91.4 | Periplasmic MBP; binds maltose/maltodextrins with micromolar affinity. | 37, 76, 107–109, 128, 138, 244, 248, 257, 264, 265 |
| malF | 91.4 | Intrinsic membrane protein of the transport system. In association with MalG and MalK, it forms the MalFGK2 translocation complex. | 41, 80, 82, 93, 252, 273 |
| malG | 91.4 | Intrinsic membrane protein of the transport system. In association with MalF and MalK, it forms the MalFGK2 translocation complex. | 25, 50–52, 254, 273 |
| malK | 91.5 | Transport ATPase, responsible for energization of transport. In association with MalF and MalG, it forms the MalFGK2 translocation complex. Target of inducer exclusion by unphosphorylated EIIAGlc of the PTS. In the absence of inducer, it interacts with MalT to cause repression. | 7, 44, 53, 55, 60, 103, 115, 147, 151, 166, 167, 184, 210, 251 |
| lamB | 91.5 | Receptor for phage λ and specific pore for maltodextrins (maltoporin, glycoporin). | 16, 39, 85, 92, 124, 136, 207, 231, 263, 265 |
| malM | 91.5 | Periplasmic protein of unknown function, partially associated with the outer membrane. Contains an Ala-Pro linker also found in OmpA. | 104, 222, 234 |
| malP | 76.5 | Maltodextrin phosphorylase. Substrates are maltopentaose and larger maltooligosaccharides. malP mutants still grow on maltose but accumulate large amount of maltodextrins under these conditions. | 177, 181, 229, 242, 290 |
| malQ | 76.4 | Amylomaltase. Maltodextrinyltransferase with maltotriose as the smallest substrate. malQ mutants cannot grow on maltose, are sensitive to maltose, and are constitutive for mal gene expression. | 69, 165, 182, 194, 294, 295 |
| malS | 80.5 | Periplasmic α-amylase, cleaves preferentially maltohexaose from the nonreducing end of meltotextrins. | 91, 92, 235, 256 |
| malZ | 9.1 | Maltodextrin glucosidase and γ-cyclodextrinase, cleaves glucose sequentially from the reducing end of maltodextrins. Maltotriose is the smallest substrate. It linearizes γ-cyclodextrin but not α- and β-cyclodextrin. | 185, 211, 267 |
| Genes whose products control mal gene expression | |||
| cya | 85.9 | Adenylate cyclase. Production of cAMP, involvement in catabolite repression. | 75, 134, 187 |
| crp | 75.1 | cAMP-binding protein, needed for the transcription of malT and the transport gene cluster. | 35, 135, 143, 206 |
| malI | 36.6 | Repressor for malXY, not dependent on MalT, inducer unknown. | 209 |
| malX | 36.6 | Enzyme II of the PTS, transports and phosphorylates glucose, can transport maltose by diffusion. | 208 |
| malY | 36.6 | βC-S lyase (cystathionase). Overproduction reduces mal gene expression by interaction with MalT and its inactivation. | 208, 301 |
| aes (ybaC, orf203) | 10.8 | Esterase. Overproduction reduces mal gene expression, presumably by interaction with MalT and its inactivation. | 164, 186 |
| mlc | 35.9 | Gene regulator, represses the expression of malT and manXYZ. | 70, 131 |
| maa (mac, F183a) | 10.32 | Glucose/maltose transacetylase, not MalT dependent, responsible for exit of maltose and glucose in their acetylated forms. | 20, 26 |
| Genes whose products affect endogenous synthesis of inducer | |||
| glgA | 75.4 | Glycogen synthase. ADP-dependent synthesis of glycogen. Degradation of glycogen yields maltotriose, which, in malQ mutants, leads to constitutivity of the maltose system. glgA mutants have a lower level on uninduced mal gene expression than glgA+ strains. | 148, 176 |
| glgC | 75.4 | ADP-glucose-pyrophosphorylase. Synthesis of ADP-glucose, needed for constitutive mal gene expression in malQ mutants. glgC mutants have a lower uninduced mal gene expression than glgC+ strains. | 4, 176 |
| glgP | 75.4 | Glycogen phosphorylase. Glycogen degradation and formation of glucose-1-phosphate. Possibly involved in synthesis of endogenous inducer. | 300 |
| glgB | 75.4 | Branching enzyme | 5, 176 |
| glgX | 75.4 | Amylase-like enzyme, role in glycogen degradation unclear. | 221 |
| amyA | 43.2 | Cytoplasmic α-amylase, not MalT dependent, no apparent role in glycogen degradation. | 198 |
| galU | 27.8 | UDP-glucose pyrophosphorylase. Possible origin of cytoplasmic unphosphorylated glucose. | 292 |
| glgS | 68.7 | Short polypeptide, involved in RpoS-dependent glycogen synthesis. | 11, 118 |
| glk | 54.0 | Glucokinase. Reduces level of internal glucose which can form endogenous inducer, responsible for mal gene repression at high osmolarity. | 43, 95, 161 |
| treR | 96.2 | Repressor for treB and treC. treR mutants allow transport of maltose via the treB-encoded transport system and induce treC, whose product is involved in inducer synthesis. | 129 |
| treB | 96.1 | Enzyme II for trehalose of the PTS, allows transport of maltose. | 142 |
| treC | 96.1 | Trehalose-6-phosphate hydrolase, involved in inducer synthesis. | 21 |
| pgm | 15.4 | Phosphoglucomutase. Needed for the synthesis of endogenous inducer. A pgm mutant can still grow on maltose but only in the presence of MalZ. | 1, 69, 153, 220 |
| Genes that affect mal gene expression by an unknown mechanism when mutated | |||
| asuE | 25.6 | tRNA-modifying enzyme. An asuE mutant increases mal gene expression at high osmolarity. | 262 |
| yjeA (genX) | 94.4 | Homolog to lysyl-tRNA synthases LysS and LysU. A genX mutant interferes with the ability of a malQ malZ292 pgm strain to grow on maltose. | 146, 170 |
| envZ | 76.1 | Sensor kinase of the two-component osmoregulatory system. Certain envZ mutants that lead to the overphosphorylation of OmpR show reduced malT expression. | 33 |
| phoP phoQ | 25.7 | Two-component system responding to Mg2+ starvation. Overexpression of the response regulator leads to mal gene repression. | 278, 283 |
Other gram-negative enteric bacteria have additional genes, not present in E. coli, that are under the control of MalT. For instance, Klebsiella pneumoniae harbors a battery of malT-controlled pul genes that are necessary for the biosynthesis and the secretion of pullulanase (71), an extracellular enzyme that degrades pullulan to maltotriose (14, 285). Pullulan, first isolated from Pullularia pullulans (13), consists of α(1→6)-glucosidically linked maltotriose units. The MalT-dependent pulA gene, encoding the lipoprotein pullulanase (163), is positioned divergently from a series of genes, also dependent on MalT, that are necessary for the export of pullulanase through the outer membrane (193, 195). Likewise, Klebsiella oxytoca harbors a series of cym genes encoding proteins for the uptake and metabolism of cyclic dextrins (89). The expression of these genes may also directly or indirectly depend on malT. The expression of the maltose regulon in Vibrio cholerae affects the virulence of this organism, and mutations in malQ and malF render it less virulent (149). The products of all the MalT-dependent E. coli genes have been identified, but the function of one, the periplasmic MalM protein (104, 222), is still unclear. The observation that MalM is also present in Salmonella typhimurium may suggest that this periplasmic protein has an important function (234).
Table 1 contains many more genes that do not belong to the maltose regulon but whose encoded proteins affect the metabolism of maltodextrins or the regulation of mal gene expression at different levels. Dextrin-metabolizing or synthesizing enzymes affect the level of internal maltotriose, the inducer of the system which is recognized by MalT; cyclic AMP (cAMP)/CAP, as well as the product of the mlc gene, controls the expression of malT itself; and the levels of MalK, the ATP-hydrolyzing subunit of the transport system, as well as of MalY (a βC-S lyase) and of Aes (an esterase with homology to lipases) affect the expression of the maltose transport genes. There are some more genes which, when mutated, have a subtle effect on mal gene expression. Among them is asuE, encoding a tRNA-modifying enzyme, and treR, encoding a repressor for the trehalose-utilyzing system in E. coli. These aspects will be discussed in detail below.
POSITIVE TRANSCRIPTIONAL ACTIVATOR, MALT
malT, Its Product, and Regulation of Its Expression
An early genetic approach identified malT as a gene that appeared essential for the expression of all maltose-inducible functions (112, 113, 126). Since all maltose-inducible operons require malT and since malT fails to repress the operons in the absence of inducer (67, 126), it was clear that the protein encoded by malT is a purely positive regulator that is activated by an inducer and stimulates transcription by activating RNA polymerase (48). The malT gene was cloned (204) and sequenced (40), and its product, MalT, was purified and characterized (214). The protein appears monomeric in dilute solution, contains 901 amino acids, and exhibits a molecular weight of 103,000. It binds ATP (KD, 0.4 μM) (215) and maltotriose (KD, about 20 μM) (49), both of which are necessary for transcriptional activation, even though ATP hydrolysis is not required, since the binding of ADP or nonhydrolyzable ATP analogs also stimulates transcription (215).
Mutations in malT [malT(Con)], resulting in the constitutive expression of all mal genes, have been isolated (49, 67). These mutations center around amino acids 243 and 358 of the polypeptide chain. Whereas in an in vitro assay the wild-type MalT protein is inactive in the absence of maltotriose, the MalT(Con) proteins are active in the absence of maltotriose but still bind maltotriose with an increased affinity. All mutant proteins can still be stimulated further in their transcriptional activity by maltotriose (49). The last C-terminal 95 amino acids of MalT constitute the DNA-binding domain. In this area, MalT exhibits homology to a number of prokaryotic transcriptional activators (280) of the UhpA-LuxR family of regulatory proteins (260). Truncated forms of MalT lacking its C-terminal DNA binding domain are negatively dominant over the function of the wild-type protein, indicating that the protein interacts with itself when binding to DNA (40).
The expression of malT is not autoregulated by MalT but is subject to catabolite repression and therefore requires the presence of the cAMP/CAP complex (35, 36, 65). According to the current view, the mechanism of catabolite repression is based mainly on the regulation of intracellular cAMP levels. This, in turn, is a function of adenylate cyclase, controlled by the enzyme IIAGlc of the phosphoenolpyruvate (PEP)-dependent sugar phosphotransferase system (PTS) in its phosphorylated (activating) and dephosphorylated (inactivating) states (191). Indeed, mutants lacking adenylate cyclase or CAP are unable to grow on maltose. It is rather informative to monitor mal gene expression in a wild-type strain during growth in rich media such as Luria-Bertani broth or tryptone broth, in the absence of external inducer. After inoculation (1:20 dilution) from an overnight culture, expression of a malK-lacZ fusion or maltose transport activity is high. It dramatically decreases (indicating a lack of new synthesis) as growth commences and reaches its lowest level during mid-log-phase growth. Since this phenomenon coincides with a cessation of cell division, it was once erroneously concluded that the synthesis of maltose binding protein (74) as well as galactose binding protein (246) might be connected to cell division. However, this phenomenon most probably reflects the concentration of internal cAMP (15), pointing once more to the importance of cAMP in catabolite repression.
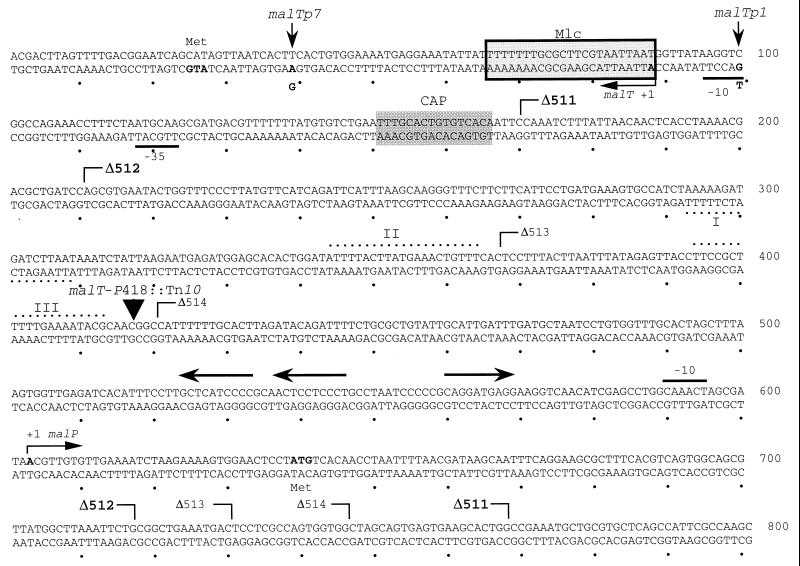
Mutations in the control region of the malT gene (Fig. 1) have shown that its expression in the wild type is limited at both the transcriptional and translational levels. Mutations with increased malT expression have been isolated (malTp1, malTp7) (Fig. 1) (34). The use of deletions introduced upstream of the malT promoter revealed that the 120 bp upstream of the transcriptional start site, encompassing the binding sites for the polymerase and the cAMP/CAP complex, are sufficient for malT expression. However, deletion of DNA further upstream increased the expression of malT, indicating that a binding site for a protein reducing malT expression had been removed (200, 205). In addition, a Tn10 insertion (malT-P418::Tn10) (Fig. 1) that increases malT expression has been isolated. The position of this insertion is still upstream of the DNA region which, when deleted, increases expression (Fig. 1). Whether or not these sites are involved in the dominant negative effect of certain mutations in envZ (105, 286, 289), resulting in an overphosphorylated OmpR protein (2, 224, 291), is still unclear. Interestingly, the DNA upstream of the malT promoter whose deletion results in an increase in malT expression contains sequences that are similar to sequences identified as binding sites for phosphorylated OmpR (133, 155) (Fig. 1). Although ompR null mutants do not exhibit an increase in malT expression, mutations in envZ leading to uncontrolled phosphorylation of OmpR reduce the transcription of malT (33).
FIG. 1.
Regulatory region upstream of malT. The unboxed shaded area represents the cAMP/CAP binding site for the malT promoter. The boxed shaded area represents the binding site of Mlc, the regulator of malT expression, as determined by footprint analysis. The arrows at malT +1 and +1 malP indicate the transcriptional startpoints of the malT and malP transcripts, respectively. The arrows upstream of the malP promoter show the single and tandem MalT boxes needed for malP expression. −10 and −35 regions are indicated for the malT promoter only. The changes of AT to GC at position 39 (malTp7) and of GC to TA at position 100 (malTp1) are mutations that increase malT expression at the translational and transcriptional levels, respectively. malTp1 renders malT expression independent of the cAMP/CAP complex (34). The extent of the deletions upstream of the malT promoter, Δ511, Δ512, Δ513, and Δ514, are indicated. Their malT expression relative to the wild type is increased by a factor of 2.8 (Δ511), 2.2 (Δ512), 1.1 (Δ513), and 1.0 (Δ514) (200). The Tn10(Cam) insertion at nucleotide 418 leads to a 2.5-fold increase in malT expression. Dotted lines indicate homology to sites that have been identified as OmpR binding sites. In particular, I and III indicate homology to site FII and II indicates site FI as defined in reference 155. Modified from reference 200 with permission of the publisher.
Expression of malT is controlled by a repressor called Mlc. This was detected by isolating a chromosomal insertion that increased malT expression. The insertion was found to be in mlc, a known gene whose product, when overproduced, impairs the utilization of glucose (131). The increase in malT expression could still be observed in mutants carrying the deletions as well as the insertion upstream of the malT promoter, excluding these regions as binding sites for Mlc. Mlc shows homology to NagC, a gene regulator functioning as a repressor for the divergent nag operons (encoding proteins for the uptake and degradation of N-acetyl-d-glucosamine) and as an activator as well as a repressor for the glmUS operon encoding enzymes for the biosynthesis of N-acetyl-d-glucosamine (188). By footprint analysis, it was shown that Mlc binds to malT DNA at a position 1 to 23 bp from the start of the malT transcript (Fig. 1). In mutants lacking Mlc, the expression of malT is increased by a factor of 2 to 3 when grown in glycerol. On the other hand, overexpression of Mlc from a multicopy plasmid strongly reduces malT expression (68). The identity of the effector for Mlc is still unclear. Internal free glucose or glucose derived from the metabolism of disaccharides such as trehalose (141) or even maltose (28) slightly induces malT expression in a Mlc-dependent fashion. However, free glucose, even 100 mM, does not prevent DNA binding of Mlc during footprint or band shift assays. Therefore, it is likely that a metabolic product of glucose could be the controlling compound for Mlc. Mlc not only regulates malT expression but also represses the man operon (259) when overproduced (189). Mlc may thus represent a new global regulator for sugar-metabolizing systems.
MalT Box
The first feature recognized for at least three MalT-dependent promoters was the lack of the usual −35 region, whereas the −10 region corresponds to those of constitutive promoters (47). Instead, the asymmetric hexanucleotide sequence 5′-GGA(G/T)GA-3′, the so-called MalT box, was identified centered at bp −37.5 or at −38.5 upstream of the transcriptional start point (9, 10, 106, 201). In addition, DNA upstream of the −38 region was found to be essential for mal gene expression (106, 201), whereas the 30-bp sequence preceding the transcriptional start point contains only a few positions that are essential for promoter activity (64).
The second structural feature found to be essential for MalT-dependent mal gene expression was two additional MalT boxes in a direct repeat upstream of the −38 region (206, 279, 281). The analysis of these MalT boxes also led to an extension in the consensus sequence, which is now defined as 5′-GGGGA(T/G)GAGG-3′ (281). In contrast to the orientation of the MalT box, which is proximal to the transcription initiation site and always oriented 5′ to 3′ in the direction of transcription, the orientation of the two repeats can be in either direction. This motif of MalT-dependent promoter structure with three MalT boxes has also been found by sequence analysis upstream of malS (235) and malZ (267), two MalT-dependent genes encoding maltodextrin-hydrolyzing enzymes in E. coli.
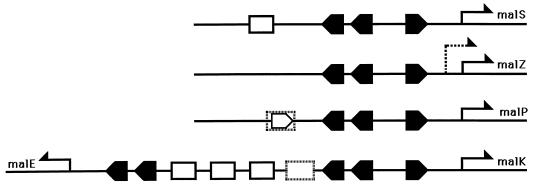
Figure 2 shows a schematic view of the arrangement of the MalT and cAMP/CAP-binding sites of the different E. coli mal gene promoters.
FIG. 2.
Scheme for the promoter structures of the different mal operons. Arrows indicate transcriptional start points. Solid large arrow-shaped bullets represent MalT binding sites (MalT boxes), and open rectangles represent cAMP/CAP binding sites. The open arrow-shaped bullet indicates a MalT box that has been identified by footprinting but seems dispensable for Mal-dependent transcriptional activation of malP (46). This site overlaps a CAP binding site (dashed cAMP/CAP binding site) as determined by footprint analysis. Again, mutation of this site does not affect the transcription of malP (46). Similarly, the cAMP/CAP binding site most proximal to the malK promoter (dashed rectangle) has been identified by footprint analysis but is not essential for transcription of malK (279). Having two MalT binding sites in a direct repeat is a recurrent feature of mal promoters and plays a crucial role in their activation (281). The dashed arrow in the malZ promoter indicates a second transcriptional start site that is independent of MalT and more frequently used at high osmolarity. The cAMP/CAP binding site in the malS promoter has not been tested for its functional importance. The three cAMP/CAP binding sites between malK and malE are essential for the transcription of both genes (279).
By using the decanucleotide 5′-GGGGAGGAGG-3′, the MalT-binding consensus box, and nucleotide sequences present in the polylinker of a vector plasmid as connecting sequences, Danot and Raibaud constructed several functional semisynthetic promoters and tested them for MalT-dependent gene expression (45). These studies confirmed the importance of the structural motif formed by two MalT-binding sites in a direct repeat. This motif is involved in promoter activation either alone or in conjunction with a third MalT-binding site proximal to the transcription start site. In this configuration, the promoters are active irrespective of the orientation of the repeat. Provided that the alignment along the axis of the helix is retained, the distance of the repeat to the proximal MalT-binding site can be varied to some extent. This analysis defines the inducible MalT-dependent promoter that does not require the cAMP/CAP complex (45). The role of these sites in the contact and activation of RNA polymerase by MalT has been assessed by mutating each site and measuring the contribution of the remaining sites (46).
Of the five known MalT-dependent operons in E. coli, two (malPQ and malZ) are independent of cAMP/CAP and two, malK lamB malM and malEFG, depend on it. The upstream region of the last mal gene, malS, does contain a sequence resembling a cAMP/CAP binding site, but whether it is functional is unclear. malK lamB malM and malEFG are oriented divergently from each other. Their transcription start sites are 271 bp apart. Located between these start sites is a 210-bp regulatory region which comprises two series of MalT boxes (three MalT boxes, including the repeat, in front of malK, and two MalT boxes, the repeat, in front of malE) separated by three cAMP/CAP binding sites. The entire control region is required for the full expression of both operons. Sequential removal of the two distal MalT boxes reduces but does not abolish the expression of malE, whereas the removal of any MalT box abolishes the expression of the malK operon (206). This suggested that multiple copies of MalT and cAMP/CAP form a unique nucleoprotein structure at this regulatory region that is required for maximal activity of both promoters. The observation that the function of these sites is sensitive to the phase of the DNA helix has led to a model in which the DNA is wrapped around the complex composed of MalT and cAMP/CAP (199, 206). The state of supercoiling also appears to be important for the effectiveness by which the two operons are transcribed. Not only is relaxed DNA less efficient in transcription initiation, but also the sites occupied by the activator complex are shifted in supercoiled relative to relaxed DNA (216).
By a series of elegant experiments, it was found that binding of cAMP/CAP in the intergenic regulatory region between malE and malK results in a repositioning of MalT binding. In the absence of cAMP/CAP, MalT binds with high affinity to the three MalT boxes upstream of the malK transcriptional start site (identified previously). In the presence of cAMP/CAP, MalT binding is shifted by three nucleotides toward the Pribnow box of the malK promoter. The cAMP/CAP effect requires the malKp-distal MalT binding sites (218). The repositioning is caused by DNA bending, which can be mimicked by replacing cAMP/CAP with the integration host factor (217). The role of the CAP and MalT binding sites in transcriptional regulation of the two divergently oriented promoters continues to be the subject of extensive studies (213).
Recently, it has been observed that Lrp, the leucine-responsive protein (32), also affects the transcription of malT as well as of some malT-dependent genes (268). By using operon fusions to malT, malE, and malK in the genetic background of strain MC4100, this Lrp dependence could not be observed in the laboratory of W.B. It has also been reported that the expression of the malK lamB malM and malEFG operons, but not of the malT gene, is controlled by the pH of the medium, being elevated at high pH (121). The mediator of this pH-dependent control is unknown.
MALTOSE/MALTODEXTRIN TRANSPORT SYSTEM
Periplasmic Substrate Recognition Site and Its Interaction with Membrane Components
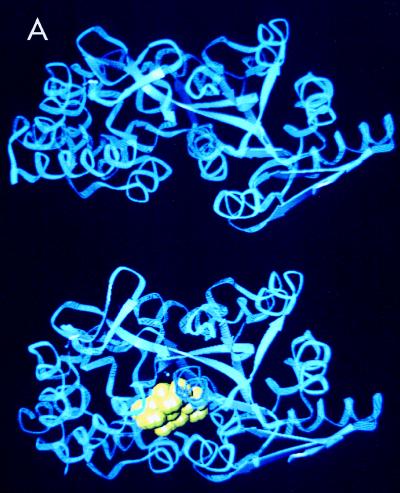
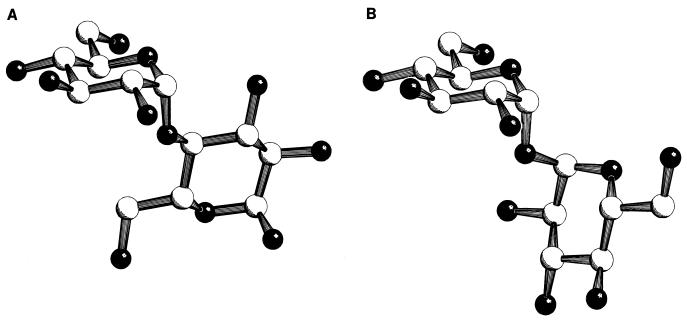
The maltose/maltodextrin transport system is a member of the family of multicomponent and periplasmic binding protein-dependent ABC high-affinity transport systems of gram-negative enteric bacteria (21, 57, 116, 173, 249, 250). The substrate recognition site of the system is determined primarily by the soluble binding protein with a high affinity for maltose and maltodextrins (KD around 1 μM) (138, 265) that is located in the periplasm in high concentration (around 1 mM and in 30- to 50-fold molar excess over the intrinsically membrane-bound proteins of the system) (74). This maltose binding protein (MalE protein or MBP) consists of two nearly symmetrical lobes between which the binding site is formed (197, 257). Substrate-loaded and substrate-free forms of MBP, as well as of other substrate-binding proteins, differ dramatically in their conformations, so that substrate bound to the protein no longer has access to bulk solvent. In the substrate-free form, the lobes are open and the substrate-binding site becomes accessible to the bulk solvent. Several physical, spectroscopic, and thermodynamic properties of MBP change when ligand is bound to it (100, 101, 109, 175, 244, 247, 265). Structural data on the open (244) and closed substrate-loaded (243, 244, 257) forms of the protein are available (Fig. 3A). The two lobes not only perform a bending movement relative to each other but also perform a twisting movement (Fig. 3B). In addition, the structures of a MBP with a dominant negative mutation (248) as well as of MBPs harboring a foreign epitope (226) or deletions and insertions (245) have been solved. Both the substrate-loaded and the substrate-free forms of MBP have access to the membrane components MalF and MalG (17, 158), even though it is unclear whether the interacting substrate-free form is of the open or the closed configuration.
FIG. 3.
Crystal structures of the open and closed forms of MBP. (A) Structural representation of MBP in the open (top) and closed (bottom) forms. The yellow structure represents maltose bound within the binding site. Courtesy of Sherry Mowbray (247); reprinted with permission of the publisher. (B) Perspective view of the superimposed backbone structure of unliganded (mauve) and maltose-bound (blue) MBP (looking into the binding cleft from the side) with bound maltose (ball-and-stick model). The direction and magnitude of the conformational change when going from the unliganded to the ligand-bound form are shown. Reprinted from reference 244 with permission of the publisher.
MBP has been subjected to a detailed mutational analysis, and regions affecting transport (78, 264) or chemotaxis (77) have been defined. Also, intensive studies on the renaturation process of denaturated MBP (37, 96) and on the biosynthesis and secretion of the protein (37, 73, 90, 261, 272) have been published. A recent report even indicates that periplasmic binding proteins, MBP among them, exhibit properties of molecular chaperonins in the periplasm (212).
The interaction of MBP with MalF and MalG has been studied by the genetic approach of mutant and suppressor analysis (274). This study, in combination with the knowledge of the crystal structure of MBP, has led to the conclusion that one lobe of MBP interacts with MalF and the other lobe interacts with MalG (128). The starting point for this genetic analysis was the isolation of mutants (actually, two point mutations were always required in either malF or malG) that transported maltose in the absence of MBP. Surprisingly, some of these MBP-independent mutations became maltose negative in the presence of wild-type MBP, a situation that allowed isolation of mutants with suppressor mutations in malE (274). Most of these suppressor mutations in MBP were dominant negative in combination with a wild-type MalFGK2 complex, indicating a higher affinity towards the membrane components when in the non-ATP hydrolysis-triggering mode (128, 248). Similarly, after the introduction of two cysteines (G69C and S337C) by site-directed mutagenesis into each domain of MBP, the formation of an interdomain disulfide cross-link that holds the protein in a closed conformation could be observed. This mutant MBP confers a dominant negative phenotype for growth on maltose, for maltose transport, and for maltose chemotaxis (303). The observation that wild-type MBP interferes with the transport activity of the MalFGK2 complex of the MBP-independent mutant permitted further studies on the interaction of MBP with the MalFGK2 complex. Transport studies with vesicles revealed that the wild-type binding protein inhibited transport only at high concentrations while it actually stimulated transport at low concentration (59). This analysis, together with the properties of dominant negative mutations in MBP, has led to the proposal that the MalFGK2 complex must be able to attain at least two different conformations in its interaction with MBP, only one of which is able to trigger ATP hydrolysis by the MalK subunit (21, 248).
Even though the major recognition entity of the maltose transport system is the MBP, it is clear that not all substrates that are bound by the binding protein are transported. This is particularly obvious with cyclodextrins and the p-nitrophenyl derivatives of maltooligosaccharides that are bound very well by MBP but are not transported by the intact system (87, 150, 269). This is due to the modes of substrate recognition by MBP. Substrates that can be attached to MBP at the reducing end are transported; those which are bound within the dextrinyl chain (such as cyclodextrins) are not (107, 108, 173).
MBP also functions as the substrate recognition site for maltose chemotaxis (114). The sites in MBP that are essential for the interaction with the maltose transport machinery and the chemotaxis machinery are distinct but partially overlapping (77, 97, 98, 302, 303). The α helix 7 of MBP appears to be exclusively involved in the interaction with the membrane components of the transport system. Mutations in this α helix belonging to the C-lobe of MBP affect transport without affecting the binding of substrate or the interaction with the chemotactic machinery (264).
Membrane-Spanning Subunits MalF and MalG of the Transport System
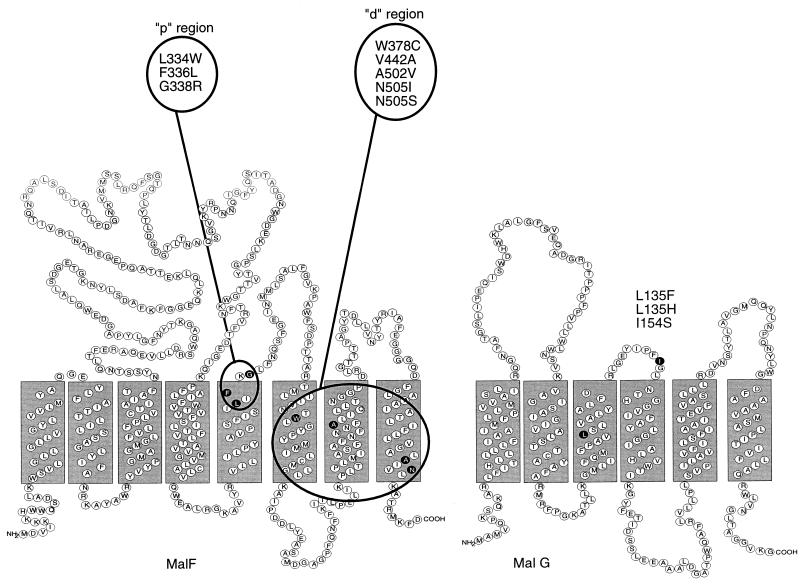
The application of the phoA fusion technique allowed determination of the two-dimensional topology of both MalF (24, 82, 94) and MalG (25, 52). Accordingly, MalF consists of eight membrane-spanning α-helical segments (MSS) with both termini of the polypeptide chain protruding into the cytoplasm. MalG, as judged by the same criteria, consists of six MSS, and again both termini extend into the cytoplasm (Fig. 4). Near their C termini, both proteins carry a sequence between MSS 6 and 7 (MalF) and MSS 4 and 5 (MalG), respectively, that is very similar to the consensus sequence EAA-X3-G-X9-I-X-LP conserved in all intrinsically membrane-bound subunits of binding protein-dependent ABC systems (227, 228). Substitutions at the same positions in MalF and MalG cause different phenotypes, and mutations in malG or malF that slightly affect or do not affect transport by themselves cause a completely defective phenotype when present together. This indicates that MalF and MalG are acting together asymmetrically in the translocation step. Suppressor mutations to transport negative mutations in malF or malG were found in malK, indicating that the EAA-harboring domain interacts with MalK (167).
FIG. 4.
Two-dimensional structure of MalF and MalG. A topological model of MalF and MalG based on the analysis of malF::phoA and malG::phoA fusions is shown. MBP-independent mutants always require two mutations, one in the p (proximal) region and one in the d (distal) region. The G338R mutation in MalF alone causes MBP-dependent lactose transport. The EAAXXLG consensus motif is located in the cytoplasmic loop between MSS 6 and 7 of MalF and MSS 4 and 5 of MalG. Reprinted from reference 41 with permission of the publisher.
Studies with mutations in malF and their intragenic suppressors represent the first attempts to determine the positions of the different MSS relative to each other. Thus, mutations in MSS 7 were suppressed by mutations in MSS 6 or 8, perhaps an indication that these helices are next neighbors. In addition, mutations in MSS 6 leading to altered substrate specificity occur only on one side of the helical wheel, pointing to a participation of this surface in the substrate-specific transport channel (80).
The MalF subunit of the maltose transport system of E. coli and other gram-negative enteric bacteria (44) represents something of an exception among the membrane-bound components of binding protein-dependent ABC transporters, since these MalF proteins contain a large periplasmic loop between MSS 3 and 4. This loop may not mediate the interaction with the binding protein, since all pairs of mutations in malF leading to an MBP-independent phenotype (and which can be suppressed by mutations in MBP for the dominant negative phenotype caused by wild-type MBP) are positioned outside the loop. It may be involved in the docking of MBP to the membrane components. Consistent with this view is the observation that in none of the MBP-independent mutants is docking of MBP prevented. Also, no periplasmic loop is found in the MalF homologs of binding protein-dependent maltose transport systems in gram-positive bacteria (196, 225, 276) or even in the Archaea (299), whose members contain lipid-anchored high-affinity binding proteins for maltose on the outside of the cytoplasmic membrane. In the latter case, docking is no problem because close vicinity to the membrane components is based on the lipid anchorage.
Even though it represents the major substrate recognition site, MBP cannot be the only substrate binding site of the transport system. MBP-independent mutants with mutations in MalF or MalG that retain specificity for maltose have been isolated, indicating the presence of a latent substrate binding site in the MalFGK2 membrane complex (273). The Vmax of these mutants for maltose transport can sometimes be similar to the wild-type value, while the apparent Km of uptake is always increased by a factor of about 103. Transport in these mutants is still active, i.e., against the concentration gradient, and dependent on the MalK-mediated hydrolysis of ATP. Characteristically, these MBP-independent mutants with mutations in MalF always carry two mutations, one in MSS 5 near the periplasmic surface, in the p (proximal) region, and the other one deep within the cytoplasmic membrane, in either MSS 6, 7, or 8, in the d (distal) region (41). It is interesting that substrate specificity mutations have been isolated in MSS 6 and that nearest-neighbor analysis predicts that MSS 6, 7, and 8 are close to each other (80) and at the same time form the d region. The attractive concept that mutations in the p region affect binding-protein recognition and mutations in the d region affect the coupling to the ATPase activity mediated by the associated MalK subunit apparently does not hold true, however. Both mutations are required for the uncoupled MalK-ATPase phenotype.
The screening of mutations in malF and malG for the ability to transport other sugars revealed that one mutation, malF515, which changes Leu334 to Trp (L334W) on the periplasmic side of MSS 5, permitted the transport of lactose in addition to maltose. MalK as well as MalG is required for lactose transport, as is MBP. The requirement for MBP is particularly intriguing because MBP clearly does not bind lactose. Therefore, these results are consistent with the idea that the unliganded form of MBP interacts with the membrane complex (160). The position for such a “specificity” mutation is surprising since it is on the periplasmic side of MalF, which is not expected to form the substrate recognition site. This may indicate that substrate specificity for lactose is already present in MalF or MalG but needs the proper opening of the gate for the substrate to enter. In a different construct, large portions of the MalF protein have been deleted (beginning from the C terminus up to MSS 1). These deletion mutants are able to transport lactose in a MalG-, MalK-, and MBP-dependent manner without an additional mutation (159). This may indicate that the substrate recognition site of the membrane complex for lactose resides in MalG.
MalK, the Energy-Coupling Protein of the Transport System
The MalK subunit contains a classical consensus sequence found in all ATP-hydrolyzing proteins (284). It consists of two subsites, the A and B domains. These two motifs, as well as the sequence surrounding these sites, are conserved in the equivalent subunits of all binding protein-dependent transport systems (123). In addition, they are found in many proteins of prokaryotic and eukaryotic origin whose function is in the transport of molecules, including polysaccharides, peptides, and proteins (83, 122). MalK and equivalent proteins of other binding protein-dependent transport systems have been characterized not only as ATP-binding proteins but also as enzymes that hydrolyze ATP. When MalK is embedded in the membrane as a complex with MalF and MalG, maximal rates of ATP hydrolysis are achieved only in the presence of substrate-loaded MBP (54, 57–59, 184). The ATPase activity of the detergent-solubilized MalFGK2 complex of the MBP-independent mutant F500 has also been measured in solution (53). The active complex consists of two MalK subunits connected by one molecule each of MalF and MalG (55). When this complex is reconstituted in liposomes, ATP-dependent active transport of maltose into the liposomes can be measured, demonstrating the function of the MalK dimer as an energy module driving the active transport of maltose. In agreement with the dimeric structure of MalK in the translocation complex is the observation that the ATP binding sites of both subunits are essential (56) and that the kinetics of ATP hydrolysis show cooperativity (53). Using the technique of Hu (132) to probe proteins for their ability to form dimers by fusing them to a truncated λ repressor (lacking the dimerization domain but retaining the operator binding), we were able to show that wild-type MalK has the ability to form dimers in vivo (68).
Little is known about the mechanism of energy coupling of ATP hydrolysis to the accumulation of maltose. Since neither the substrate nor any of the proteins involved have been seen to become phosphorylated during transport, it has become commonplace to interpret energy coupling as the transfer of the energy gained through ATP hydrolysis to protein conformational energy in MalF and MalG, followed by its subsequent binding protein-triggered release, resulting in the unidirectional translocation of the substrate through the membrane. Mutants with mutations in malF or malG that no longer require the triggering by substrate-loaded MBP have been isolated. It is noteworthy that the MalFGK2 complex of these mutants no longer requires the presence of substrate-loaded MBP to stimulate the ATPase activity of MalK (57). ATPase activity has become uncoupled in these mutants, similar to the uncoupled ATPase activity of the purified wild-type MalK subunit (166). This has led to the notion that the transport system is a signal transduction pathway that begins in the periplasm with the recognition of maltose by the binding protein and ends with the control of MalK-ATPase activity in the cytoplasm (41, 57). The helical domain of the ATPase in MalK is likely to be involved in the signal transduction between MalF and MalG (167).
No quantitative studies on the synthesis of MalK in comparison to its partners in the membrane, MalF and MalG, are available. Even though the stoichiometric composition of the biochemically active complex has been determined as MalFGK2 (55), it is unclear whether MalK is synthesized in excess or, depending on the regulatory conditions, in varying amounts. The latter possibility is not unlikely since malF and malG are located in a different operon from malK and since substantial amounts of MalK have been found, in contrast to MalF and MalG (7, 251). As discussed below, MalK plays an important role in the MalT-dependent control of mal gene expression. It is only natural to see MalK not only as an energy module to drive transport but also as a control unit of transport activity. Obviously, the availability of ATP should influence transport activity. However, since the Km of MalK for ATP is in the micromolar range (58) and the physiological concentration of ATP is in the millimolar range, the cells would presumably have to become quite exhausted of ATP before they stopped transporting maltose.
We would like to postulate an additional level of control for the function of MalK in transport, which works by regulating the affinity of MalK for its partners, MalF and MalG. We observed that transport of glycerol-3-phosphate by the binding protein-dependent Ugp transport system is inhibited by internal Pi (29). The Ugp system exhibits a surprising degree of sequence similarity to the maltose system in all subunits in spite of the differences in substrate specificity and regulation (178). In an attempt to find whether UgpC, the MalK analog of the Ugp system, was the target of Pi inhibition, we exchanged MalK with UgpC (115) and tested a possible inhibition of maltose transport by Pi. Although Pi did not inhibit the UgpC-mediated uptake of maltose, it also failed to inhibit the uptake of glycerol-3-phosphate via the Ugp system when UgpC was overproduced. Apparently, the reduced affinity of UgpC for its cognate membrane partners in the presence of Pi can be compensated for by increasing the UgpC concentration. It suggests the possibility that the degree of association of the ATP-hydrolyzing subunit with the membrane components may control transport activity. An equivalent finding in the histidine transport system in Salmonella typhimurium has been interpreted in the same way. High concentrations of internal histidine were able to inhibit transport of histidine in trans (152). These observations may indicate that the transport activity of ABC transporters is controlled from the inside via the ATP-hydrolyzing subunit by binding the accumulated substrate.
The MalK subunit is usually pictured as being peripherally membrane associated through binding to the intrinsic membrane proteins MalF and MalG. This membrane association of MalK may in fact be more intimate (296). Studies with the functionally related binding protein-dependent histidine transport system of Salmonella typhimurium have shown that HisP, the MalK analog of this system, might actually be accessible from the periplasm (6), even though the interaction of the cognate periplasmic binding protein, HisJ, takes place with the intrinsic membrane proteins of the system and not with HisP (192). Similar findings of accessibility of MalK on the periplasmic side of the membrane have been reported for MalK in S. typhimurium (236). Models of the transport mechanism mediated by binding protein-dependent ABC systems picture the input of energy by ATP hydrolysis as conservation of a strained protein structural conformation in MalF and MalG that is released by triggering with substrate-loaded binding protein (21, 59, 250). The analogy of the repetitive movement of SecA through the membrane that has been postulated as crucial for Sec-dependent protein secretion (140, 293) to a possible repetitive movement of a subdomain of MalK through the membrane (236) is less convincing. The transported molecule would be far smaller than the moving machinery, and the moving part of MalK should then exhibit substrate specificity for the transported molecule, which has never been observed. In contrast, UgpC, the ATP-hydrolyzing subunit of the glycerol phosphate transport system, can complement malK mutants to transport maltose (115), indicating that the ATPase subunit does not carry substrate specificity for the transported molecule.
Role of the Lambda Receptor (Maltoporin) in the Diffusion of Maltose and Maltodextrins through the Outer Membrane
Efficient uptake of maltose and, particularly, longer maltodextrins at low concentrations requires the presence of λ-receptor (maltoporin), the specific diffusion pore for maltodextrins and other carbohydrates, in the outer membrane (62, 63, 92, 141, 263, 265). It is interesting that lamB, the gene for the λ-receptor, is, together with malK, located in a different operon from the remaining genes of the transport system (malE, malF, and malG). Since MalK is involved in the regulation of the system, and since the λ-receptor is also required for the diffusion of carbohydrates other than maltodextrins (63, 141), a differential regulation of these two genes (in comparison to the malEFG operon) should be considered. In addition, translational control of lamB (72, 110) may be evoked when considering the need of carbohydrate uptake under starvation conditions (62, 174).
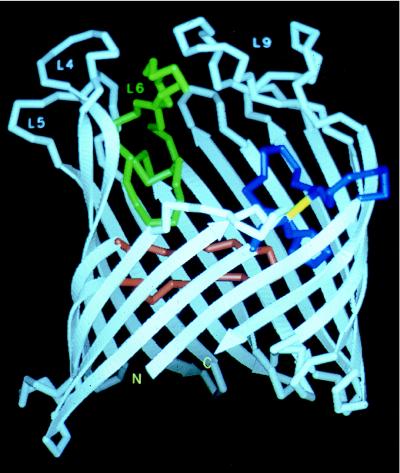
A number of studies have analyzed the function of LamB (maltoporin) in the diffusion of maltodextrins (16, 88, 92, 136, 154, 171). All these studies demonstrate the presence of a maltodextrin binding site that is essential for the facilitated diffusion process of maltodextrins. A major contribution to understanding the molecular basis of transport through porin channels has come from determination of the three-dimensional structure of LamB, which was done in the presence of a series of maltooligosaccharides. These studies showed that each subunit of the trimeric protein contains a wide channel formed by an 18-stranded (230), antiparallel β-barrel. Three inwardly folded loops contribute to a constriction about halfway through the channel (231) (Fig. 5). The crystal structures of maltoporin complexed with maltose, maltotriose, or maltohexaose reveal an extended binding site within the channel. The maltooligosaccharides are in apolar van der Waals contact with the “greasy slide,” a hydrophobic path that is composed of aromatic residues and is located at the channel lining (79, 287). Interestingly, the LamB protein contains two of four sequences (235) that are conserved in amylases and form the maltodextrin binding sites there (282). One of them, FYQRHD, at positions 106 to 111 of LamB, is actually part of the sugar binding site as determined by X-ray crystallography (79). A similar structure for the maltoporin from S. typhimurium, including the prominent greasy slide for maltodextrins, has been reported (162).
FIG. 5.
Crystal structure of LamB. A schematic drawing of the λ-receptor (maltoporin) monomer is shown. The cell exterior is at the top, and the periplasmic space at the bottom. The area of the subunit in trimer contacts is facing the viewer. The 18 antiparallel β strands of the barrel are represented by arrows. Strands are connected to their nearest neighbors by loops or regular turns. Loops L1 (blue), L3 (red), and L6 (green) fold inward toward the barrel. L3 is the major determinant of the constriction site. The yellow bond symbolizes the disulfide bridge Cys22-Cys38 within loop 1. Loop 2, facing the viewer, latches onto an adjacent subunit in the trimer. Loops L4 to L6 and L9 form a large protrusion. The horizontal lines delineate the boundaries of the hydrophobic core of the membrane as inferred from the hydrophobic area found on the molecular surface. Reprinted from reference 231 with permission of the publisher.
Calculations show that the diffusion of maltose through LamB and the following uptake by the ABC transporter are closely matched at maltose concentrations in the micromolar range. The Vmax for the disaccharide maltose in a fully induced strain has been measured to be 20 nmol per min per 109 cells (265). The diffusion of maltose through the outer membrane, mediated by about 10,000 trimeric LamB porin molecules per cell, is apparently not yet limiting at micromolar maltose concentrations, the Km of the transport system (27). We calculated the diffusion of maltose for different external concentrations through these 30,000 pores with an approximate diameter of 1 nm, using a diffusion constant of 10−5 cm2/sec and an outer membrane thickness of 6 nm. We found that a rate of diffusion of 20 nmol per min per 109 cells, the Vmax of the fully induced ABC transporter, is reached at 0.1 μM. Considering that not all maltose molecules that reach the external face of the lambda receptor will actually enter the pore, this concentration is most probably a lower limit, and in reality the concentration might be closer to 1 μM. Therefore, one can conclude that the overall uptake of maltose at its Km is close to the maximal rate of diffusion through the outer membrane. The validity of this conclusion has been experimentally tested with mutants containing reduced amounts of lambda receptor (27). In other words, a transport system that requires a Vmax of about 20 nmol per min per 109 cells and is equipped with at least equal amounts of a specific diffusion pore to those in the fully induced lambda receptor cannot exhibit an apparent Km considerably lower than 1 μM, in spite of a periplasmic binding protein with a considerably lower KD of binding. In contrast, transport systems with an inherently low Vmax, such as binding protein-dependent amino acid transport systems, may very well approach in their Km of transport the KD of the corresponding binding protein.
ENZYMES OF THE MALTOSE SYSTEM
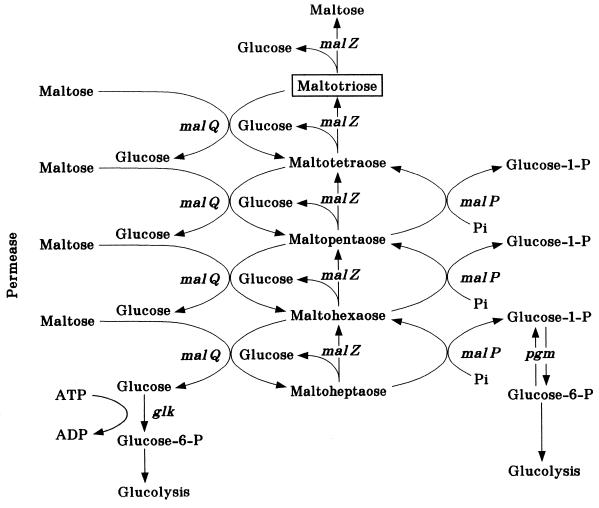
Incoming maltose and maltodextrins of up to seven glucose moieties are metabolized to glucose and glucose-1-phosphate by the combined action of three cytoplasmic enzymes, amylomaltase (MalQ), maltodextrin phosphorylase (MalP), and maltodextrin glucosidase (MalZ) (Fig. 6). Since maltodextrins larger than six glucose moieties are not very well transported by the ABC transporter, they are reduced in size by a periplasmic amylase, the MalS protein.
FIG. 6.
Maltose degradation by the maltose enzymes. The enzymes amylomaltase (malQ), maltodextrin phosphorylase (malP), and maltodextrin glucosidase (malZ) are indicated by their genes. After transport of maltose by the binding protein-dependent ABC transporter, a maltosyl, maltotriosyl, or maltotetraosyl (and so on) residue is transferred from maltotriose, maltotetraose, or maltopentaose (and so on) onto the incoming maltose releasing glucose in the process. Maltopentaose and longer maltodextrins are recognized by the maltodextrin phosphorylase, forming glucose-1-phosphate and a maltodextrin that is smaller by one glucosyl residue. Maltodextrin glucosidase recognizes maltotriose and longer maltodextrins (up to maltoheptaose), releasing glucose consecutively from the reducing end of the maltodextrin. This scheme demonstrates that maltose degradation by the maltose enzymes requires the endogenous presence of a maltodextrin primer with the minimal size of maltotriose. The activity of the last maltose enzyme, the periplasmic α-amylase, is not considered in this scheme. Adapted from reference 69 with permission of the American Society for Microbiology.
Amylomaltase
Amylomaltase (165, 194, 294, 295), encoded by malQ, is a dextrinyl transferase that can transfer maltosyl and longer dextrinyl residues onto glucose, maltose, and longer maltodextrins. The smallest substrate which amylomaltase recognizes is maltotriose (182). Acting on maltotriose, it releases glucose from the reducing end, forms a maltosyl-enzyme complex, and transfers the maltosyl residue onto the nonreducing end of an acceptor, be it glucose, maltose, or any larger maltodextrin. The paradoxical consequence of this scheme is that maltose itself should not be a primary substrate, but pure maltose should be a substrate only in the presence of small amounts of maltodextrins acting as a primer in the reaction. Amylomaltase, in contrast to the MalZ enzyme discussed below, is able to release not only glucose but also longer dextrins from the reducing end of its maltodextrin substrates (182). Since MalQ is not a hydrolase but a transferase, the sum of glycosidic linkages of all maltodextrins involved in the reaction must remain constant. Only when glucose is removed from this equilibrium of dextrins, for instance by glucokinase-mediated phosphorylation to glucose-6-phosphate, does the degradation of maltose continue while forming longer maltodextrins.
It is important to stress that maltose, strictly speaking, is not a substrate of amylomaltase but only an acceptor in the transfer reaction catalyzed by the enzyme. Thus, it follows that for maltose degradation, the cell has to be able to internally produce small amounts of maltodextrins as primers with the minimum size of maltotriose. Amylomaltase is essential for maltose degradation, and malQ mutants are unable to grow on maltose. Not only are malQ mutants Mal−, but also their growth is inhibited by maltose (127). Under these conditions, they accumulate large amounts of free maltose inside the cell (265). In the presence of an alternative carbon source, mutations arise that are found preferentially in malT or in malK. Why no mutations in other genes arise whose products are essential in transport remains a mystery (241). Even in the absence of external maltose, malQ mutants contain significant concentrations of free glucose, maltose, maltotriose, and larger maltodextrins, provided that the strain contains the glycogen-synthesizing enzymes (69, 81). Therefore, within the cell, glycogen must continually be degraded to maltodextrins, which cannot be repolymerized (under production of glucose) in the absence of amylomaltase.
The enzymatic reaction of amylomaltase can be exploited for the convenient synthesis of uniformly labeled maltodextrins (maltotriose and larger) of high specific activity starting from uniformly labeled maltose. When amylomaltase is incubated, even after extensive dialysis with radiochemically pure [14C]maltose as the only substrate, [14C]glucose and 14C-maltodextrins are formed immediately. They can be separated and purified by chromatographic procedures and exhibit the same specific radioactivity (with respect to the glucosyl moiety) as the initial maltose. The primers used for this reaction are present in minute amounts, bound to the enzyme (180).
Maltodextrin Phosphorylase
Maltodextrin phosphorylase, encoded by malP, forms glucose-1-phosphate by sequential phosphorolysis of the nonreducing end glucose moieties of larger dextrins. As discussed above, amylomaltase does not split the glycosidic bond of maltose, but the net products of the action of MalP on maltose are glucose and maltodextrins. Since glucose will be removed in vivo by glucokinase to form glucose-6-phosphate, maltodextrins would accumulate. Indeed, malP malQ+ mutants can grow on maltose. Under these conditions, they become very large, are filled with long, linear dextrins (238, 239), and stain blue with iodine (1). Surprisingly, they do not transform these dextrins into glycogen, even though the glgB-encoded branching enzyme is present. Maltodextrins do not accumulate in the malP+ wild-type strain. Maltodextrin phosphorylase (8, 181, 242) recognizes maltopentaose and longer linear maltodextrins and forms α-glucose-1-phosphate by phosphorolysis from the nonreducing end of the maltodextrin. The enzyme consists of a dimer with 796 amino acids per polypeptide. The three-dimensional structure of the enzyme has been solved recently (177, 290).
Obviously, it is important that maltodextrin phosphorylase does not attack maltotetraose and maltotriose, since dextrins of a minimum size are required for full activity of amylomaltase. Notably, glycogen phosphorylase, the glgP-encoded enzyme, supposedly involved in the degradation of glycogen and linear dextrins (300), cannot replace maltodextrin phosphorylase in the utilization of maltose and maltodextrins as a carbon source.
The reaction catalyzed by maltodextrin phosphorylase is, of course, reversible. The rate in the phosphorolysis direction will be stimulated by increasing concentrations of cytoplasmic Pi. In turn, the concentration of internal Pi will vary depending on the availability of external Pi and phosphorus-containing organic compounds, as well as the state of induction of the phoB-dependent pho regulon (298). There are other connections between the mal and the pho regulon. Aside from the sequence similarity between the proteins of the maltose- and phoB-dependent Ugp transport systems for glycerol-3-phosphate (178) and the functional exchangeability between their ATP binding subunits, MalK and UgpC (115), there is a common response (i.e., repression) of the two systems to dominant mutations in envZ, resulting in overphosphorylation of OmpR, the response regulator of the two-component regulatory EnvZ-OmpR system involved in osmoregulation (33).
In addition, the utilization of a fermentable carbon source such as glucose, trehalose, or maltose at Pi concentrations below 1 mM requires the derepression of the pho regulon. phoB mutants do not turn deep red on MacConkey indicator plates (0.3 mM Pi) unless 5 mM Pi is added. Also, whereas strains that turn red on these plates derepress the pho regulon, nonfermenters that appear pale remain repressed for the pho regulon. Apparently, the utilization of a fermentable carbon source requires higher cellular Pi concentrations than does that of a noncarbohydrate carbon source present in MacConkey plates (111).
Maltodextrin Glucosidase, an Enzyme of Unclear Function
MalZ was discovered in malF or malG mutants that transport maltose independently of MBP. In contrast to the wild type, these mutants also transport p-nitrophenyl α-maltoside (NPG2) and are able to hydrolyze this compound in the cytoplasm (211). Since amylomaltase is unable to hydrolyze NPG2 and since the observed NPG2-hydrolyzing activity was maltose inducible and MalT dependent, it was clear from the start that the novel enzyme must be a member of the maltose regulon. The cloning and sequencing of the malZ gene and the isolation and biochemical characterization of the encoded protein revealed an enzyme that hydrolyzed maltoheptaose and smaller maltodextrins to glucose and maltose. The smallest substrate is maltotriose; maltose is not a substrate. In contrast to other glucosidases, the MalZ enzyme preferentially removes glucose (and to some extent maltose) consecutively from the reducing end of the maltodextrin chain (267). The deduced amino acid sequence of MalZ reveals homology to cyclodextrinyl transferases (190). We found that the enzyme does not hydrolyze α-cyclodextrin or pullulan. However, γ-cyclodextrin (and, to a much smaller extent, β-cyclodextrin) is an excellent substrate, forming maltose and glucose. The significance of this finding is unclear (185). γ-Cyclodextrin is not a carbon source for E. coli. The compound cannot diffuse through the outer membrane or be taken up by the maltose transport system. malZ mutants grow normally on maltose and maltodextrins. Only in combination with a pgm mutation do malZ mutants have a Mal− phenotype.
malQ mutants cannot grow on maltose or maltotriose. This is somewhat surprising since MalZ should release glucose (which could be used as a carbon source) from maltotriose. Most probably, the simultaneous accumulation of maltose resulting from the MalZ-dependent hydrolysis of maltotriose in the malQ mutant is toxic. By selecting malQ mutants to grow on maltose, a mutation in malZ (malZ292) was obtained. Surprisingly, the purified mutant MalZ enzyme is not able to hydrolyze purified maltose and shows the same maltodextrin-hydrolyzing activity as the wild-type enzyme. However, by using [14C]glucose in the presence of unlabeled maltotriose or maltotetraose, the mutant enzyme, unlike the wild-type enzyme, is able to transfer maltodextrins onto the labeled glucose. The mutant MalZ enzyme has acquired a quality which is similar to that of amylomaltase. However, amylomaltase is strictly a transferase, which is not true for the mutant MalZ enzyme. The latter also remains a hydrolase, allowing the net hydrolysis of maltose to glucose in the presence of maltodextrins (185). As in the case of amylomaltase, the utilization of maltose by the mutant MalZ enzyme in vivo requires the endogenous formation of maltodextrins that can be used as primers. The first step in the catalysis performed by amylomaltase and the MalZ enzyme is identical, namely, release of glucose. However, whereas in the case of amylomaltase the maltodextrinyl residue can be transferred only onto the C-4 carbon hydroxyl group of the nonreducing glucose residue of the acceptor molecule, in the case of MalZ it can also be transferred to water.
The malZ gene has been found and characterized as a typical mal gene that is dependent on MalT (235, 267). Recently, we found that malZ expression can also occur in the absence of MalT, in particular under conditions of high medium osmolarity. Using the method of primer extension with reverse transcriptase, we found that besides the MalT-dependent transcript described first (235), malZ is also transcribed into a second mRNA, originating upstream of the start site of the main promoter (145). The significance of this phenomenon is unclear.
Role of Glucokinase and Phosphoglucomutase in Maltose/Maltodextrin Metabolism
The final products of the combined action of amylomaltase, maltodextrin phosphorylase, and maltodextrin glucosidase are glucose and α-glucose-1-phosphate. Therefore, to funnel these end products of the specific maltose enzymes into general metabolism, the cells rely on glucokinase (encoded by glk) for the phosphorylation of glucose to glucose-6-phosphate and on phosphoglucomutase (encoded by pgm) for the transformation of α-glucose-1-phosphate to glucose-6-phosphate, which enters into glycolysis. Mutants unable to phosphorylate glucose due to the lack of glucokinase, enzyme IIGlc (encoded by ptsG) and enzyme IIMan (ptsM) of the PTS-mediated phosphorylation, are unable to grow on maltose (30). Similarly, they are unable to grow on trehalose, which is degraded by an enzyme that produces internal glucose (219). Therefore, the utilization of internally produced glucose appears to be essential for growth. It is unclear whether the inability to grow on these sugars in the absence of glucose phosphorylation is due to a possible toxic effect of accumulating internal glucose (requiring extrusion) or the insufficient flow of carbon and energy via the phosphoglucomutase pathway alone. The drain of glucose-1-phosphate for the biosynthesis of polysaccharides might be another limiting factor.
pgm mutants lacking phosphoglucomutase activity are able to grow on maltose, even though seemingly only one half of the maltose molecule can be used as a carbon and energy source. These strains exhibit a maltose blue phenotype (1, 220) caused by the massive production of dextrins due to the accumulation of glucose-1-phosphate and the reversal of the maltodextrin phosphorylase reaction. The problem created by the accumulation of maltodextrins is eased by the action of maltodextrin glucosidase, the MalZ enzyme, which produces glucose from the maltodextrins, followed by glucokinase-dependent phosphorylation to glucose-6-phosphate. Indeed, pgm malZ double mutants are unable to grow on maltose. Similarly, pgm mutants can grow on galactose (1) but only when all the enzymes of the maltose system, including MalZ, are present (69). pgm mutants, even null mutations created by insertion elements (153), still show residual Pgm-like activity when the enzyme activity is assayed by coupling the formation of glucose-6-phosphate from glucose-1-phosphate to the NADP+-dependent oxidation by glucose-6-phosphate dehydrogenase. This activity is caused by a sugar-phosphate transferase which is able not only to hydrolyze sugar phosphates but also to transfer the phosphate moiety to another sugar molecule (258). Thus, in the presence of free glucose, glucose-1-phosphate can be transformed to glucose-6-phosphate, mimicking phosphoglucomutase activity.
Periplasmic α-Amylase
The malS gene (235) was discovered as a maltose-inducible and MalT-dependent gene by the lacZ fusion technique (91). MalS is a periplasmic α-amylase with weak homology to the cytoplasmic MalZ enzyme but not to amylomaltase. MalS cleaves maltodextrins except maltose. Its preferred product released from larger dextrins is maltohexaose. The enzyme needs Ca2+ for activity and DsbA (157) for proper folding in the periplasm. The four cysteine residues of MalS form intramolecular disulfide bonds. The disulfide bond at Cys40-Cys58 is located in an N-terminal extension of about 160 amino acids which has no homology to other amylases but to the proposed peptide-binding domain of GroEL, the Hsp60 of E. coli. The N-terminal extension is linked to the C terminal amylase domain via disulfide bond Cys104-Cys520. Reduction of the disulfide bonds by dithiothreitol treatment led to aggregation, suggesting that the N terminus of MalS may represent an internal chaperone domain (256). malS mutants have no recognizable maltose phenotype. The function of the enzyme is most probably the degradation of longer dextrins that enter the periplasm to shorter dextrins that can be transported by the binding protein-dependent maltose/maltodextrin transport system (92). Even though MBP, the recognition site of the transport system, binds all maltodextrins from maltose to amylose, only dextrins up to the size of maltohexaose can be transported across the membrane (84, 86).
Figure 6 summarizes the pathway by which, according to our view, maltose is degraded into glucose and α-glucose-1-phosphate by the maltose-specific enzymes.
Maltose Utilization in Other Bacteria
Maltose uptake and utilization in other bacteria, notably in gram-positive bacteria, usually proceeds differently. In Bacillus subtilis, maltose is probably taken up by a proton motive force-dependent transport system and split internally into glucose and glucose-1-phosphate by a maltose-specific phosphorylase (266). The observation that Lactobacillus sanfrancisco takes up maltose and excretes half of it as glucose points to the same mechanism (172). In addition, maltose may also be taken up in lactobacilli by a PEP-dependent sugar PTS system and is internally hydrolyzed to glucose and glucose-6-phosphate (270). The isolation of a maltose-6-phosphate hydrolase and the vicinity of its gene, malH, to malB, encoding a putative IIB-like enzyme of the PTS, points to the same maltose degradative pathway in Fusobacterium mortiferum (23). This is quite in analogy to the uptake and degradation of trehalose in E. coli (142, 219).
NONCLASSICAL REGULATORY PHENOMENA
MalK, the ATP-Hydrolyzing Subunit, as a Sensor for mal Expression
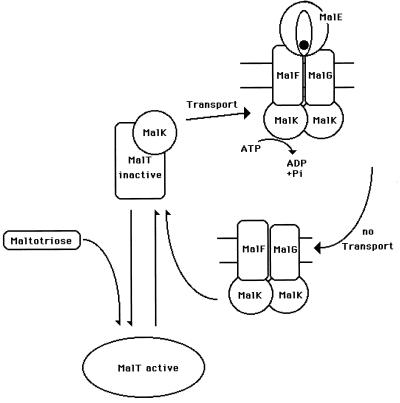
It has been known for a long time that mutations in the transport system, later identified as malK mutations, lead to elevated expression of the remaining mal genes (31, 125). This can be conveniently observed by using malK-lacZ fusions with a nonfunctional MalK, which exhibit high and constitutive β-galactosidase activity. An intact MalT activator is required for this constitutivity. The same malK-lacZ fusion in a malK+ merodiploid genetic background exhibits low β-galactosidase activity that can be induced by maltose, reflecting the induction observed in the wild type (31). These findings indicate that MalK acts as a repressor. Indeed, when plasmid-borne malK is overexpressed in the original malK-lacZ fusion strain, β-galactosidase activity is abolished. This effect can also be observed in a wild-type strain where the overexpression of malK renders the strain unable to grow on maltose, even though the strain can grow normally on glycerol (210). The mutational analysis of MalK has shown that the regulatory function resides in the C-terminal domain of the protein and that its functions in transport and regulation are independent of each other (147, 151, 237). Surprisingly, a malT(Con) mutation leading to expression of the mal genes in the absence of inducer can largely counteract the repressing effects of overproduced MalK (210).
To explain how MalK acts as a repressor, it has been argued that the protein could be an enzyme that degrades the inducer (31). This possibility was attractive, since malT(Con) mutants, which require less or no inducer (49), are resistant to the repression mediated by overproduced MalK. However, the synthesis of maltotriose, the inducer of the maltose system (202), is not disturbed by overproduction of MalK and does not lead to induction under these conditions (69). Thus, MalK does not seem to act on the level of inducer degradation. A second possibility is that MalK, as long as it is not engaged in transport, interacts with MalT and inactivates it. MalT has been proposed to exist in an equilibrium of two forms, only one of which is acting as a transcriptional activator. Thus, when stabilized by inducer, MalT would be able to act as a transcriptional activator (66), whereas when stabilized by MalK, it would be inactive. Hence, MalK and the inducer would compete for MalT, shifting the equilibrium either with maltotriose to the inducing conformation or with MalK to the inactive form, causing repression. The first evidence for such a scheme comes from the laboratory of Shuman (183). A biotinylation site was introduced at the C terminus of MalT, leading to a biotinylated MalT (MalT-BCCP). Avidin-coated beads were then used to attach MalT-BCCP on their surface. These beads were able to bind MalK, demonstrating an interaction between MalK and MalT. Also, the phenotype of strains harboring MalT-BCCP is in line with the consequences of a MalK-MalT interaction. The presence of MalT-BCCP is negatively dominant over wild-type MalT. Overproduction of MalK abolished the dominant negative phenotype, indicating the titration of MalT-BCCP by MalK (183).
As discussed above, MalK forms dimers in vivo, as indicated by fusions to a truncated λ repressor (which is unable to dimerize). MalK mutants that are unable to repress mal gene expression no longer show a tendency to form dimers in vivo (68). This may indicate that the dimeric form of MalK is the one that interacts with MalT.
Figure 7 shows a model of how MalK could mediate repression. MalK is pictured as a protein that is able to shift between a state of association with MalFG and a state of association with MalT. In the presence of substrate to be transported, MalK would shift to the transport conformation allowing MalT to function as mal gene activator. In the absence of substrate to be transported, MalK would be free to interact with MalT and cause repression. The model is reminiscent of findings with the proline utilization system, where PutA, the proline dehydrogenase, functions as a membrane-bound enzyme in the presence of proline and as a cytoplasmic repressor in its absence (168).
FIG. 7.
MalK cycle of transport and repression. A proposed model for the function of MalK in transport and regulation is shown. When maltose (solid circle) is present in the medium, it is recognized by MBP (MalE) and then transported through the membrane via MalFGK2. In the process, the signal of substrate recognition is related to MalK on the cytoplasmic side of the membrane and ATP is hydrolyzed. Under these conditions, MalK is tightly associated with the MalFG complex and has no affinity for MalT. In the absence of maltose, the affinity of MalK for MalFG is lowered and it becomes available to interact with MalT, keeping MalT in its inactive state as a mal gene transcriptional activator. The key point in controlling the function of MalK is its association with ATP. When ATP is bound to MalK (but not hydrolyzed due to the absence of triggering substrate-loaded MBP), the protein is a repressor and interacts with MalT. This interaction is counteracted by the inducer (maltotriose). Thus, MalK cycles between an exclusively transporting and an exclusively MalT-inhibiting state, the predominance of transport or inhibition being determined by exogenous substrate and endogenous inducer. It should be stressed that there is no compelling evidence that MalK actually dissociates from MalFG during MalT inhibition. However, it is known that overproduced, i.e., unbound, MalK can exert repression.
Two observations support such a MalK cycle. First, there is a particular mutation in malK (malK941) (147) which leads to a superrepressor MalK protein. This mutation is near the ATP binding site. Labelling of the mutant protein with radioactive 8-azido-ATP is still possible; therefore, binding of ATP, but no ATP hydrolysis, probably also takes place. This would mean that the absence or presence of ATP hydrolysis determines the quality of MalK as a repressor and that ATP bound to MalK is required for repression. Second, mutations in malF and malG that result in high constitutive levels of MalK ATPase activity (41, 57) show a high level of mal gene expression in the absence of inducer, presumably because the ATP-hydrolyzing form of MalK is mimicking transport activity and is unable to repress MalT activity (183).
The MalK protein is unusually long in comparison to other ATP-hydrolyzing subunits of binding protein-dependent ABC transport systems; the difference is the presence of a C-terminal extension. On the other hand, UgpC, the corresponding subunit of the Ugp system, exhibits the same C-terminal extension as MalK and shows sequence similarity to MalK except at its C-terminal extension. MalK and UgpC can be exchanged between the two systems and can complement the transport defect of the heterologous system, but only in the absence of the cognate subunit (115), indicating a lower affinity in forming the heterologous complex. A similar exchange of MalK from S. typhimurium by LacK, the ATP-hydrolyzing subunit of a binding protein-dependent ABC transporter for lactose in Agrobacterium radiobacter, has been reported (296). Similarly, CymD, the ATP-hydrolyzing subunit of the cyclodextrin transport system of K. oxytoca, can substitute for MalK in E. coli (179).
Considering the additional segment at the C terminus of MalK, where all the regulation-negative mutations have been found (147), it was of interest to find whether UgpC, which carries the same C-terminal extension as MalK, would also be active as a repressor. While the overproduction of UgpC had no repressing effect on mal gene expression, it did reduce the expression of the remaining ugp genes (271). Thus, repression exerted by an ATP-hydrolyzing subunit of a binding protein-dependent transport system is not a peculiarity of MalK but may represent a novel form of regulation occurring in this type of transport system.
The MalK subunit is also the target of PTS-mediated inducer exclusion that is brought about by the interaction of nonphosphorylated EIIAGlc with MalK (60, 147, 275). This interaction leads to a reduction in Vmax without changing the Km of the transport system (60). This is reminiscent of the effect of the UgpC-mediated inhibition of the Ugp transport system by internal Pi (29, 298), which we explain by the dissociation of UgpC from its protein partners in the membrane. Thus, it appears that the state of MalK (membrane bound or cytosolic) is important for its function in transport and regulation.
MBP-dependent ABC transport systems have also been found in gram-positive bacteria (102, 196), in thermophilic bacteria (120, 225), and in a hyperthermophilic archaeon (130, 299). The canonical composition of these transport systems is the same as in gram-negative bacteria, even though the respective binding proteins are anchored in the membrane by a diacylated glycerol linked via a thioether to the N-terminal cysteine of the binding protein. Surprisingly, in many of these systems that have been analyzed by sequencing, the genes encoding the MalEFG analogs are clustered in the same orientation as in the E. coli maltose system whereas the gene encoding the corresponding ATP-hydrolyzing enzyme is localized elsewhere on the chromosome or has not been found yet (225). In Streptomyces, the same ATP-hydrolyzing subunit appears to serve at least two different transport systems (233). These observations, together with the finding that ATP-hydrolyzing subunits can (under certain conditions) be functionally exchanged among heterologous systems, indicate that this component may have been the first one to appear during evolution, with additional proteins determining altered substrate specificity arriving later in evolution. Also, it would not be surprising if, in the future, several “ancient” systems that share their ATP-hydrolyzing subunits were found.
MalY as a mal Repressor
The finding that malK-lacZ fusions lacking MalK function are expressed at high levels has been used in a genetic screen (looking for white colonies on β-galactosidase indicator plates) with the intention of selecting mutations that prohibit the synthesis of an internal inducer. A chromosomal Tn10 insertion which abolished the constitutivity of the malK-lacZ fusion was isolated. The gene was mapped at 36 min on the chromosome, outside any known mal operon. It was named malI to indicate the expected function of the gene product in the synthesis of the endogenous mal gene inducer (81). malI was cloned and sequenced. The deduced amino acid sequence of the malI gene product revealed that it is highly similar to the classical repressor proteins, such as LacI or GalR (209) and must represent a repressor itself. An operon containing two genes, malX and malY, is located next to and transcribed divergently from malI. This operon, as well as malI itself, is repressed by the MalI protein (208). Therefore, MalI is not involved in the synthesis of the endogenous inducer of the mal system, as had been expected, but mutations in malI lead to the constitutive expression of the malX malY operon, which in turn represses the mal system. Sequencing revealed that malX encodes a protein exhibiting homology to the enzyme IICBGlc of the PTS, encoded by ptsG and catalyzing the transport of glucose. Indeed, the expression of malX could complement ptsG mutants for growth on glucose. However, the expression of malX does not cause repression of the mal genes. It is malY, the distal gene in the operon, that causes repression when overexpressed, either in a malI mutant or when cloned under tac promoter control (208). The sequence of malY indicates weak homology to aminotransferases, including the consensus sequence of a pyridoxal phosphate binding site, but the purified enzyme had no detectable aminotransferase activity with glutamate as a substrate. Instead, we found βC-S lyase activity (cleavage of a βC-S bond in amino acids) connected to the protein, similar to cystathionase activity (301). The latter enzyme, encoded by metC, is essential in methionine biosynthesis and cleaves cystathionine to homocysteine, ammonia, and pyruvate (12). Indeed, metC mutations can be complemented for growth in the absence of methionine by malI mutants or by multicopy plasmids harboring malY, even though MalY and MetC do not show significant sequence homology, and the quaternary structures (38) of the two proteins are different; MetC is a tetramer and MalY is a monomer.
The βC-S lyase activity of MalY is not the cause of the repression of the mal genes. A mutation in the pyridoxal phosphate binding site of MalY was constructed that resulted in the loss of βC-S lyase activity, even though the protein was still active as a mal gene repressor. In addition, overproduction of MetC had no effect on the regulation of the mal genes (301). As in the case of MalK, mutants with mutations in MalY that still exhibit nearly unchanged enzymatic activity but have lost their ability to repress the maltose system can be isolated. The clustering of these mutations favors a mechanism of protein-protein interaction with MalT as causing repression. There are several indications to suggest that MalY acts by the same mechanism as MalK: malT(Con) mutants are largely insensitive to the effect of either MalK or MalY. Also, when maltose enters the cell via a constitutive transport system for trehalose (see below), an inducer that renders MalT insensitive to overproduction of MalK as well as MalY can be formed. On the other hand, the inhibiting effect of MalY can still be observed in mlc mutants that have elevated levels of MalT. Therefore, the effect of MalY and MalK on the activity of MalT depends on the amount of MalT and on the presence of inducer. A high inducer concentration (or the malT(Con) mutation) is more effective than an increase in the concentration of MalT.
Aes as a mal Repressor
Another enzyme with similar regulatory functions on mal gene expression as MalY and MalK is Aes, an enzyme with acetyl esterase activity. aes, the gene encoding Aes, is identical to a previously described open reading frame (orf203) of unknown function whose deduced amino acid sequence showed homology to lipases (164). Overproduction of Aes strongly and specifically represses mal gene expression. Again, the effect of Aes on mal gene expression is similar to the effect of MalK and MalY. Thus, malT(Con) strains, which are resistant to the overproduction of MalK and MalY, are also resistant to the overproduction of Aes. Also, the insertion in mlc resulting in elevated malT expression reduces the effect of Aes (186). It is our working hypothesis that both MalY and Aes act in the same way as MalK by interacting with MalT to shift the equilibrium to its inactive state. Even though they do not exhibit sequence similarity, all three proteins must contain a similar structural motif by which they recognize MalT.
Effect of Phosphorylated PhoP on mal Expression
PhoQ-PhoP represents a two-component regulatory system that responds to low Mg2+ concentrations in the medium (277, 278, 283). By using a mutant with the sensor kinase PhoQ deleted and with pyruvate as the carbon source (to increase the phosphorylation capacity by introducing a high internal concentration of acetyl phosphate), malK as well as malE but not malT expression was reduced by a factor of 10. The same reduction in malEK expression was obtained by introducing phoP on a multicopy plasmid and after growth on glycerol. At present, it is not clear whether the effect of PhoP on mal gene expression is a direct one or is mediated by one of the many PhoQ- and PhoP-regulated proteins (18).
ENDOGENOUS INDUCER OF THE MALTOSE SYSTEM
The relatively high “uninduced level” of mal expression in E. coli when the cells are grown without maltodextrins in the medium (i.e., with glycerol) points to the endogenous synthesis of the inducer. Several lines of evidence support this conclusion: first, malK-lacZ fusions (lacking MalK function) display a much higher β-galactosidase activity than does the corresponding malK+ merodiploid strain. This shows not only that MalK acts as a repressor but also that MalT is activated by internal sources. Second, as outlined above, maltose metabolism initiated by amylomaltase needs a maltodextrin primer with the minimal chain length of maltotriose. Thus, in the absence of endogenous maltodextrins, E. coli could not grow on maltose. Third, mutants with mutations in enzyme IIGlc (ptsG), enzyme IIMan (ptsM), or glucokinase (glk) can be induced by glucose entering the cell in its unphosphorylated form via the galactose permease. This indicates that the inducer is formed from glucose. The induction by glucose does not occur in a mutant also lacking phosphoglucomutase (pgm). Such a strain exhibits a very low level of mal gene expression (in comparison to its pgm+ parent) as measured by transport activity (69). Since the induction-preventing mutation is in pgm, it is clear that the synthesis of inducer can arise from gluconeogenesis. The only inducer established so far to be able to activate MalT in vitro is maltotriose (202). Therefore, the cell must have the ability to synthesize maltotriose or another sugar with inducer capabilities. There are at least two ways in which maltotriose can be synthesized endogenously.
Glycogen as a Source of Maltotriose
malQ mutants are constitutive for the maltose transport system and other malT-dependent mal gene products. This constitutivity is dependent on glycogen or, more correctly, on the presence of the glycogen-synthesizing enzymes (69). malQ mutants that carry an additional mutation in glgA (encoding glycogen synthetase) or glgC (encoding ADP-glucose pyrophosphorylase) are no longer constitutive but normally inducible by maltose. Thus, the constitutivity in malQ mutants must be caused by the production of endogenous inducer from glycogen, and the function of amylomaltase is to remove the inducer. With the knowledge of the activity of amylomaltase, it is clear that maltotriose is removed and kept at low concentrations by the formation of larger maltodextrins and glucose. Indeed, extracts of malQ mutants contain glucose, maltose, maltotriose, and larger maltodextrins that are absent or found in much lower concentration in the corresponding malQ+ strain (81). Whereas the removal of glycogen-derived endogenous inducer is clearly a function of amylomaltase, it is less clear how maltotriose is derived from glycogen. malQ malZ or malQ amyA double mutants are still constitutive in the expression of the mal genes, indicating that maltodextrin glucosidase (267) or a cytoplasmic amylase (198) is not involved in producing maltotriose from glycogen (69). A likely candidate for maltotriose production is glgX, a gene, found in the glycogen gene cluster, whose deduced amino acid sequence shows homology to amylases (221).
Second Pathway for Maltotriose Formation
Even in the absence of glycogen, i.e., in glgA and glgC mutants defective in the synthesis of glycogen, the mal genes can be induced in the absence of external maltodextrins but under conditions that generate glucose and α-glucose-1-phosphate (or glucose-6-phosphate) inside the cell. The best example is the metabolism of trehalose, which induces the mal genes (in the absence of glycogen) to about 30% of the fully induced or constitutive level of expression (141). The immediate products of trehalose metabolism are glucose and glucose-6-phosphate (219). Similarly, when glucose is transported into the cell without phosphorylation (via galactose permeases in a ptsG ptsM glk mutant), the maltose genes become induced. This glucose-dependent induction needs glucose-1-phosphate, since pgm mutants which lack phosphoglucomutase and hence are unable to deliver glucose-1-phosphate by gluconeogenesis are not induced by glucose. By using exogenous [14C]glucose, the glucose-1-phosphate-dependent formation of internal maltose and maltotriose can be observed in such a mutant (69). One has to stress, however, that mal gene induction by the external addition of glucose and the formation of maltotriose from exogenous glucose may simply be coincidental events. In addition, it is clear that ptsG ptsM glk and even ptsG ptsM glk pgm mutants do transform [14C]glucose into other, still unidentified, products (69).
It appears that even in the absence of trehalose metabolism or when the cells are growing on glycerol and no glucose is taken up from the medium, the mal genes are endogenously induced by the formation of small amounts of maltotriose (or another unidentified inducer). In wild-type strains, the basal expression of the mal genes is low, curbed by the action of the MalK protein, but it becomes significant in malK-lacZ fusion strains lacking MalK function. Since this constitutivity is also seen in strains defective in glycogen synthesis, it is obvious that the endogenous inducer can also be derived from gluconeogenesis without bringing in glucose from the medium. These observations have led us to formulate a scheme for the synthesis of the internal inducer that is based on the function of an as yet unidentified maltose/maltotriose phosphorylase. This enzyme should reversibly form maltose (and maltotriose) from glucose (or maltose) and glucose-1-phosphate (22). The formation of these maltodextrins and therefore the postulated enzyme(s) for their synthesis are independent of any maltose enzyme, since maltotriose is created in malT mutants as well (69). The gluconeogenetic origin of glucose-1-phosphate seems clear; less clear is the origin of free internal glucose. There are several possibilities. The first is the hydrolytic function of a phosphoglucotransferase, an enzyme that was purified some time ago but whose gene has not yet been identified. This enzyme transfers the phosphate moiety from glucose-phosphate not only to another sugar but also to water, thus forming free glucose (258). The other possibility is the release of free glucose from UDP-glucose by analogy to the release of galactose from UDP-galactose (297). Indeed, galU mutants lacking UDP-glucose pyrophosphorylase are somewhat impaired in the constitutivity of a malK-lacZ fusion.
If the model of inducer synthesis is correct, pgm mutants should be strongly reduced in their basal expression of the maltose genes and uninducible by trehalose. This is exactly what is observed when measuring maltose transport of pgm mutants grown in glycerol or, even more dramatically, grown on trehalose. However, malK-lacZ (Mal−) fusions are only weakly (two- to threefold) reduced in their constitutive expression when tested in pgm mutants. Similarly, the induction of a malK-lacZ fusion (be it Mal+ or Mal−) by trehalose is only weakly reduced in pgm mutants. At present, there is no reasonable explanation for the difference in these two tests for mal gene expression.
Elevated Expression of the mal Genes during Glucose Starvation
When E. coli is grown in the chemostat with glucose as the limiting carbon source and at micromolar concentrations, the maltose system is expressed at elevated levels (62, 63, 174). The authors concluded that this was due to the increased synthesis of endogenous maltotriose as inducer of the system. Most probably, glucose enters the cell via the Mgl transport system (61) that also recognizes this sugar with high affinity. The Mgl system may really be more of a glucose transporter than a galactose transporter since it not only transports glucose with high affinity but also is induced by internal glucose (99). Here again, the strong correlation between the appearance of endogenous free glucose and the induction of the maltose transport genes can be seen.
This is in sharp contrast to the situation for cells growing logarithmically in batch cultures at high glucose concentrations. Under these conditions, transport of glucose via the PTS exerts strong catabolite repression, blocking the transcription of malT and the malT-dependent genes of the malK lamB malM and malE malF malG operons. In addition, transport of maltose is diminished by inducer exclusion. These effects are caused by the interaction of unphosphorylated enzyme IIAGlc of the PTS with MalK (60, 147, 275).
Role of Glucose in Internal Induction of the Maltose System
There are several facts that emphasize the importance of free internal glucose in the induction of the maltose system: (i) trehalose whose metabolism yields internal glucose induces the maltose system; (ii) the transport of free glucose into the cell induces the system; (iii) maltose and maltodextrin metabolism itself results in the formation of free internal glucose; and (iv) maltose and [14C]glucose can endogenously form maltotriose whose reducing glucose moiety is 14C labelled. This obvious association of unphosphorylated internal glucose with the induction process is further stressed when the concentration of internal glucose is manipulated by phosphorylation via overproduced glucokinase. Raising the amount of glucokinase 10-fold over the chromosomally encoded level by plasmid-carried glk reduces mal gene expression. This repression occurs only when mal gene expression is due to endogenous induction either during growth under “uninduced conditions” (with glycerol as the carbon source) in a wild-type strain or in a constitutive malK-lacZ fusion lacking MalK function. The effect of glucokinase overproduction cannot be observed when constitutive mal gene expression is due to the formation of maltotriose from glycogen in malQ mutants (where free glucose is not involved) or in malT(Con) mutants, which are largely independent of inducer (161).
Osmoregulation of the Maltose System
A malK-lacZ fusion that lacks any MalK activity is expressed constitutively due to the presence of endogenous inducer. With increasing osmolarity of the growth medium, the expression of the fusion is abolished (31). Using a malK-lacZ fusion that is merodiploid for malK (and can therefore grow on maltose), the fusion is expressed at low levels when grown on glycerol but can be induced by maltose, thus reflecting the normal induction pattern by maltose in a wild-type strain. High osmolarity still leads to repression of the uninduced levels but not when induced with maltose (31), consistent with the observation that high osmolarity does not affect growth on maltose. This osmodependence of mal gene regulation does not affect malT expression (31), and it is not dependent on the envZ/ompR (255) regulatory circuit or the ςS-dependent (117) control system. However, during the attempt to isolate mutations that counteract the effects of high osmolarity, an insertion was isolated upstream of the promoter of malT (malT-P418::Tn10 [Fig. 1]). This insertion does not abolish osmoregulation but simply elevates expression of malT. Another mutation found in this selection (144) was an insertion (null mutation) in asuE, a gene known to encode a lysine tRNA-modifying enzyme (262). This mutation does not affect malT expression and only dampens the effect of high osmolarity on malK and malE expression. The mechanism by which AsuE is involved in the osmoregulation of mal gene expression is unclear.
Since only the uninduced level of mal gene expression is affected by osmolarity, one may argue that it is the synthesis of endogenous inducer that is inhibited under conditions of high osmolarity. Most recently, we found that the presence of glucokinase is necessary for the effective repression of uninduced or malK-dependent constitutive mal gene expression. Since the expression of glk at high osmolarity remains unchanged (as measured by a translational glk-lacZ fusion), the most likely explanation is a stimulation of glucokinase activity at high osmolarity. In strains carrying ptsM and ptsG mutations in addition to the glk mutation, no osmoregulation of mal gene expression is observed. This demonstrates that osmoregulation (i.e., repression) of mal gene expression is caused by the removal of internal free glucose.
INTERCONNECTION BETWEEN THE MALTOSE AND TREHALOSE SYSTEMS
Maltose and trehalose are disaccharides composed of two α-glycosidically linked glucose residues, but their structures are quite different. Whereas maltose contains an α(1→4)-glycosidic linkage and exhibits a reducing end glucose, trehalose contains two α(1→4)-glycosidic linkages and no reducing sugar residue (Fig. 8). Due to the particular symmetrical and backfolded structure of this nonreducing sugar, trehalose, in contrast to maltose, has properties that make it an excellent agent to preserve protein and membrane structures (42). It has been observed that trehalose is synthesized as a protecting agent or osmolyte in many cell types including E. coli under conditions of dehydration as well as high osmolarity (137). Thus, it is not surprising that E. coli has developed two entirely different systems for the metabolism of maltose and trehalose, both of which are excellent carbon sources for this organism. Unlike maltose, trehalose uses a PTS-dependent enzyme IITre (encoded by treB) for its uptake as trehalose-6-phosphate (142) followed by its internal hydrolysis by trehalose-6-phosphate hydrolase (encoded by treC) to glucose and glucose-6-phosphate (219). treB and treC form an operon that is controlled by the adjacent treR gene, encoding a repressor (129). The internally acting inducer of the system is trehalose-6-phosphate, which inactivates TreR. There is one complicating fact: in addition to its degradative abilities, E. coli is able to synthesize internal trehalose at high osmolarity. The two necessary enzymes are trehalose-6-phosphate synthase (encoded by otsA), catalyzing the transfer of glucose from UDP-glucose to glucose-6-phosphate, thus forming trehalose-6-phosphate, and trehalose-6-phosphate phosphatase (encoded by otsB), catalyzing the hydrolysis of trehalose-6-phosphate to free trehalose (137). otsB and otsA form an operon that is induced at high osmolarity in an RpoS-dependent manner (119). The expression of ostB otsA and of treB treC is mutually exclusive in that high osmolarity prevents the induction of the treB treC operon. Despite these entirely different pathways of maltose and trehalose uptake and metabolism, there are curious links between the two systems.
FIG. 8.
Structures of maltose and trehalose. (A) Maltose (α-d-glucopyranosyl-1→4-glucopyranose) in its predominant α enantiomeric form. (B) Trehalose, shown as the naturally occurring α-d-glucopyranosyl-1→1-α-d-glucopyranoside. The structures were calculated by minimizing the free energy of the axial and equatorial hydroxyl positions. The structures are shown in their chair conformation, with all substituents (except the anomeric oxygens) in the equatorial configuration.
Trehalose as an Inducer of the Maltose System
It has been observed that the presence of trehalose in the growth medium induces the maltose system (240, 253). Trehalose is not transported by the maltose system, nor does trehalose or trehalose-6-phosphate stimulate MalT; also, MalT does not regulate any of the genes involved in the degradation of trehalose, since malT mutants grow well on trehalose. Nonetheless, there is a subtle dependence of trehalose utilization on MalT function. When malT mutants are grown on trehalose, they exhibit an apparent Km of trehalose uptake that is in the order of millimolar, while malT+ strains exhibit a Km of 16 μM (19). The explanation for this difference is that the MalT-dependent λ receptor is needed for the diffusion of trehalose at concentrations below mM (141). This is not only true for the utilization of trehalose. Apparently, the λ receptor also functions as a “glycoporin” for the uptake of other sugars, even monosaccharides, at low external concentrations. mal gene expression at limiting glucose concentration may provide this increase in glycoporin activity (63). The ability of trehalose to induce the maltose system depends on the presence of TreC, the enzyme that degrades trehalose-6-phosphate to glucose and glucose-6-phosphate (141). We have interpreted this finding as an indication that glucose and glucose-6-phosphate (and, subsequently, glucose-1-phosphate via Pgm) are the precursors of the synthesis of maltose and maltotriose. However, in treR mutants, even in the absence of trehalose, the mal genes are expressed at an elevated level, indicating that TreC not only provides the precursors for maltotriose synthesis from trehalose-6-phosphate hydrolysis but also is in itself involved in inducer synthesis (68). Whether the product of TreC activity in the absence of trehalose-6-phosphate is maltotriose or another inducing compound is unclear.
Transport of Maltose and Trehalose in the Archaeon Thermococcus litoralis
Recently, we observed that the hyperthermophilic archaeon Thermococcus litoralis has high transport activity at 85°C for maltose and trehalose, with Km values of 15 to 20 nM (299). Both substrates are transported by the same system, since they mutually inhibit each other’s uptake. In addition, a membrane-bound binding protein, which shows the same specificity for maltose and trehalose as the intact system, was isolated from these cells. A gene cluster could be cloned from this organism harboring three genes with homology to malE, malF, and malG from E. coli, with the N-terminal protein sequence of the isolated MalE protein being identical to the deduced amino acid sequence of its malE gene (130). The transport system, as well as the binding protein, is induced by trehalose (299). This demonstrates that the transport system for maltose and trehalose in T. litoralis consists of a binding protein-dependent ABC transporter and suggests a common origin for trehalose and maltose transport. The development of a trehalose-specific and PTS-dependent transport system (which to some extent still remembers maltose) in bacteria might therefore have been a later event.
FINAL CONSIDERATIONS
One may ask what purpose these different regulatory circuits have that govern the expression of the maltose system. There are two main branches of regulation: one is stimulatory, represented by endogenous induction and cAMP/CAP dependency, and the other is repressive, represented by MalK and other proteins that inactivate the central gene activator. From the blueprint of the high-affinity and binding protein-dependent transport system that is also connected to the cell’s chemotactic machinery, it appears that the maltose system is geared for the scavenging of low levels of maltose and maltodextrins. Even before the arrival of the true substrate, the system is becoming prepared by endogenous induction, particularly when fasting on glucose or other carbohydrate carbon sources. However, only in the presence of the true substrate is full-fledged induction developed. It makes perfect sense that the function of holding back full induction should be connected to a component of the transport system itself. In this way, the system responds not only to the presence of substrate by induction but also to loss of substrate by repression. This could in principle also be achieved by a gene regulator that is an activator in the presence of the inducer and a repressor in the absence of the inducer (the ara system is the classical example [232]), but apparently, a finer tuning results from the regulation seen in the maltose system. That the transport step itself is connected to regulation is not unprecedented. We find this in the regulation of the pho regulon by the Pst transport system (288); in the Bgl system, where an antiterminator is controlled by its cognate phosphotransferase system (3); and in the regulation of the Fec system by the transport of ferric citrate through its cognate outer membrane receptor FecA (139).
Aside from these two main streams (endogenous induction and MalK-dependent repression), there appear to be several additional regulatory interconnections such as the existence of a repressor for the activator or the inhibition of the activator by seemingly unrelated enzymes (MalY or Aes). The characteristic feature of these effectors is that they have hardly any recognizable effect under normal physiological conditions but may become important under extreme growth conditions. It is possible that the maltose system is nothing special in this respect, and several more “classical” systems that show this kind of network regulation may be discovered.
ACKNOWLEDGMENTS
We are indebted to Armin Geyer (Department of Chemistry, University of Konstanz), who produced Fig. 8. We thank Sherry Mowbray, Tilman Schirmer, and Florante Quiocho for their help in providing Fig. 3 and 5. We thank Erika Oberer-Bley for her help in preparing the manuscript.
Work done in the laboratory of W. Boos was supported by grants from the Deutsche Forschungsgemeinschaft (SFB156 and Schwerpunkt Netzwerkregulation in Bakterien). Work in the Shuman laboratory was supported by NIH grant GM51118. The collaboration of the two laboratories was supported by the Max-Planck-Forschungspreis 1991.
REFERENCES
- 1.Adhya S, Schwartz M. Phosphoglucomutase mutants of Escherichia coli K12. J Bacteriol. 1971;108:621–626. doi: 10.1128/jb.108.2.621-626.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiba H, Nakasa F, Mizushima S, Mizuno T. Evidence for the importance of the phosphotransfer between the two regulatory components EnvZ and OmpR in osmoregulation in Escherichia coli. J Biol Chem. 1989;264:14090–14094. [PubMed] [Google Scholar]
- 3.Amster-Choder O, Wright A. Transcriptional regulation of the bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator. J Cell Biochem. 1993;51:83–90. doi: 10.1002/jcb.240510115. [DOI] [PubMed] [Google Scholar]
- 4.Baecker P A, Furlong C E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose synthase as deduced from the nucleotide sequence of the glgC gene. J Biol Chem. 1983;258:5084–5088. [PubMed] [Google Scholar]
- 5.Baecker P A, Greenberg E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli 1,4-α-d-glucan:1,4-α-d-glucan 6-α-d-(1,4-α-d-glucano)-transferase as deduced from the nucleotide sequence of the glgB gene. J Biol Chem. 1986;261:8738–8743. [PubMed] [Google Scholar]
- 6.Baichwal V, Liu D X, Ames G F L. The ATP-binding component of a prokaryotic traffic ATPase is exposed to the periplasmic (external) surface. Proc Natl Acad Sci USA. 1993;90:620–624. doi: 10.1073/pnas.90.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bavoil P, Hofnung M, Nikaido H. Identification of a cytoplasmic membrane-associated component of the maltose transport system of Escherichia coli. J Biol Chem. 1980;255:8366–8369. [PubMed] [Google Scholar]
- 8.Becker S, Palm D, Schinzel R. Dissecting differential binding in the forward and reverse reaction of Escherichia coli maltodextrin phosphorylase using 2-deoxyglucosyl substrates. J Biol Chem. 1994;269:2485–2490. [PubMed] [Google Scholar]
- 9.Bedouelle H. Mutations in the promoter regions of the malEFG and malK-lamB operons of Escherichia coli K12. J Mol Biol. 1983;170:861–882. doi: 10.1016/s0022-2836(83)80192-2. [DOI] [PubMed] [Google Scholar]
- 10.Bedouelle H, Schmeissner U, Hofnung M, Rosenberg M. Promoters of the malEFG and malK-lamB operons in Escherichia coli K12. J Mol Biol. 1982;161:519–531. doi: 10.1016/0022-2836(82)90405-3. [DOI] [PubMed] [Google Scholar]
- 11.Beglova N, Fischer D, Hengge-Aronis R, Gehring K. H-1, N-15 and C-13 NMR assignments, secondary structure and overall topology of the Escherichia coli GlgS protein. Eur J Biochem. 1997;246:301–310. doi: 10.1111/j.1432-1033.1997.t01-1-00301.x. [DOI] [PubMed] [Google Scholar]
- 12.Belfaiza J, Parsot C, Martel A, Bouthier de la Tour C, Margarita D, Cohen G N, Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender H, Lehmann J, Wallenfels K. Pullulan, ein extracelluläres Glucan von Pullularia pullulans. Biochim Biophys Acta. 1959;36:309–316. doi: 10.1016/0006-3002(59)90172-6. [DOI] [PubMed] [Google Scholar]
- 14.Bender H, Wallenfels K. Untersuchungen an Pullulan. II. Spezifischer Abbau durch ein bakterielles Enzyme. Biochem Z. 1961;334:79–95. [Google Scholar]
- 15.Benner D, Müller N, Boos W. Temperature-sensitive catabolite activator protein in Escherichia coli BUG6. J Bacteriol. 1985;161:347–352. doi: 10.1128/jb.161.1.347-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benz R, Francis G, Nakae T, Ferenci T. Investigation of the selectivity of maltoporin channels using mutant LamB proteins—mutations changing the maltodextrin binding site. Biochim Biophys Acta. 1992;1104:299–307. doi: 10.1016/0005-2736(92)90044-m. [DOI] [PubMed] [Google Scholar]
- 17.Bohl E, Shuman H A, Boos W. Mathematical treatment of the kinetics of binding protein dependent transport systems reveals that both the substrate loaded and unloaded binding proteins interact with the membrane components. J Theor Biol. 1995;172:83–94. doi: 10.1006/jtbi.1995.0006. [DOI] [PubMed] [Google Scholar]
- 18.Böhm, A., and W. Boos. 1997. Unpublished observations.
- 19.Boos W, Ehmann U, Forkl H, Klein W, Rimmele M, Postma P. Trehalose transport and metabolism in Escherichia coli. J Bacteriol. 1990;172:3450–3461. doi: 10.1128/jb.172.6.3450-3461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boos W, Ferenci T, Shuman H A. Formation and excretion of acetylmaltose after accumulation of maltose in Escherichia coli. J Bacteriol. 1981;146:725–732. doi: 10.1128/jb.146.2.725-732.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boos W, Lucht J M. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1175–1209. [Google Scholar]
- 22.Boos W, Peist R, Decker K, Zdych E. The maltose system of Escherichia coli. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 201–229. [Google Scholar]
- 23.Bouma C L, Reizer J, Reizer A, Robrish S A, Thomson J. 6-Phospho-α-glucosidase from Fusobacterium mortiferum: cloning, expression, and assignment to family 4 of the glucosylhydrolases. J Bacteriol. 1997;179:4129–4137. doi: 10.1128/jb.179.13.4129-4137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd D, Manoil C, Beckwith J. Determinants of membrane protein topology. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd D, Traxler B, Beckwith J. Analysis of the topology of a membrane protein by using a minimum number of alkaline phosphatase fusions. J Bacteriol. 1993;175:553–556. doi: 10.1128/jb.175.2.553-556.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand B, Boos W. Maltose transacetylase of Escherichia coli. Mapping and cloning of its structural gene, mac, and characterization of the enzyme as a dimer of identical polypeptides with a molecular weight of 20.000. J Biol Chem. 1991;266:14113–14118. [PubMed] [Google Scholar]
- 27.Brass J M, Bauer K, Ehmann U, Boos W. Maltose-binding protein does not modulate the activity of maltoporin as a general porin in Escherichia coli. J Bacteriol. 1985;161:720–726. doi: 10.1128/jb.161.2.720-726.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bremer E, Silhavy T J, Weinstock J M. Transposable λplac Mu bacteriophages for creating lacZ operon fusions and kanamycin resistance insertions in Escherichia coli. J Bacteriol. 1985;162:1092–1099. doi: 10.1128/jb.162.3.1092-1099.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzoska P, Rimmele M, Brzostek K, Boos W. The pho regulon-dependent ugp uptake system for glycerol-3-phosphate in Escherichia coli is trans inhibited by Pi. J Bacteriol. 1994;176:15–20. doi: 10.1128/jb.176.1.15-20.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buhr A, Daniels G A, Erni B. The glucose transporter of Escherichia coli. Mutants with impaired translocation activity that retain phosphorylation activity. J Biol Chem. 1992;267:3847–3851. [PubMed] [Google Scholar]
- 31.Bukau B, Ehrmann M, Boos W. Osmoregulation of the maltose regulon in Escherichia coli. J Bacteriol. 1986;166:884–891. doi: 10.1128/jb.166.3.884-891.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case C C, Bukau B, Granett S, Villarejo M R, Boos W. Contrasting mechanisms of envZ control of mal and pho regulon genes in Escherichia coli. J Bacteriol. 1986;166:706–712. doi: 10.1128/jb.166.3.706-712.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapon C. Expression of malT, the regulator gene of the maltose region in Escherichia coli, is limited both at transcription and translation. EMBO J. 1982;1:369–374. doi: 10.1002/j.1460-2075.1982.tb01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapon C. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J Bacteriol. 1982;150:722–729. doi: 10.1128/jb.150.2.722-729.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chapon C, Kolb A. Action of CAP on the malT promoter in vitro. J Bacteriol. 1983;156:1135–1143. doi: 10.1128/jb.156.3.1135-1143.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun S Y, Strobel S, Bassford P, Randall L L. Folding of maltose-binding protein—evidence for the identity of the rate-determining step in vivo and in vitro. J Biol Chem. 1993;268:20855–20862. [PubMed] [Google Scholar]
- 38.Clausen T, Huber R, Laber B, Pohlenz H-D, Messerschmidt A. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Escherichia coli at 1.83 Å. J Mol Biol. 1996;262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- 39.Clement J M, Hofnung M. Gene sequence of the lambda receptor, an outer membrane protein of Escherichia coli K-12. Cell. 1981;27:507–514. doi: 10.1016/0092-8674(81)90392-5. [DOI] [PubMed] [Google Scholar]
- 40.Cole S T, Raibaud O. The nucleotide sequence of the malT gene encoding the positive regulator of the Escherichia coli maltose regulon. Gene. 1986;42:201–208. doi: 10.1016/0378-1119(86)90297-0. [DOI] [PubMed] [Google Scholar]
- 41.Covitz K M Y, Panagiotidis C H, Hor L I, Reyes M, Treptow N A, Shuman H A. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J. 1994;13:1752–1759. doi: 10.1002/j.1460-2075.1994.tb06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crowe L M, Reid D S, Crowe J H. Is trehalose special for preserving dry biomaterials? Biophys J. 1996;71:2087–2093. doi: 10.1016/S0006-3495(96)79407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis S J, Epstein W. Phosphorylation of d-glucose in Escherichia coli mutants defective in glucophosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol. 1975;122:1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahl M K, Francoz E, Saurin W, Boos W, Manson M D, Hofnung M. Comparison of sequences from the malB regions of Salmonella typhimurium and Enterobacter aerogenes with Escherichia coli K12: a potential new regulatory site in the intergenic region. Mol Gen Genet. 1989;218:199–207. doi: 10.1007/BF00331269. [DOI] [PubMed] [Google Scholar]
- 45.Danot O, Raibaud O. On the puzzling arrangement of the asymmetric MalT-binding sites in the MalT-dependent promoters. Proc Natl Acad Sci USA. 1993;90:10999–11003. doi: 10.1073/pnas.90.23.10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danot O, Raibaud O. Multiple protein-DNA and protein-protein interactions are involved in transcriptional activation by MalT. Mol Microbiol. 1994;14:335–346. doi: 10.1111/j.1365-2958.1994.tb01294.x. [DOI] [PubMed] [Google Scholar]
- 47.Danot O, Raibaud O. Which nucleotides in the “−10” region are crucial to obtain a fully active MalT-dependent promoter? J Mol Biol. 1994;238:643–648. doi: 10.1006/jmbi.1994.1324. [DOI] [PubMed] [Google Scholar]
- 48.Danot O, Vidal-Ingigliardi D, Raibaud O. Two amino acid residues from the DNA-binding domain of MalT play a crucial role in transcriptional activation. J Mol Biol. 1996;262:1–11. doi: 10.1006/jmbi.1996.0493. [DOI] [PubMed] [Google Scholar]
- 49.Dardonville B, Raibaud O. Characterization of malT mutants that constitutively activate the maltose regulon of Escherichia coli. J Bacteriol. 1990;172:1846–1852. doi: 10.1128/jb.172.4.1846-1852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dassa E. Sequence-function relationships in MalG, an inner membrane protein from the maltose transport system in Escherichia coli. Mol Microbiol. 1993;7:39–47. doi: 10.1111/j.1365-2958.1993.tb01095.x. [DOI] [PubMed] [Google Scholar]
- 51.Dassa E, Hofnung M. Sequence of gene malG in Escherichia coli K12: homologies between integral membrane components from binding protein-dependent transport systems. EMBO J. 1985;4:2287–2293. doi: 10.1002/j.1460-2075.1985.tb03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dassa E, Muir S. Membrane topology of MalG, an inner membrane protein from the maltose transport system of Escherichia coli. Mol Microbiol. 1993;7:29–38. doi: 10.1111/j.1365-2958.1993.tb01094.x. [DOI] [PubMed] [Google Scholar]
- 53.Davidson A L, Laghaeian S S, Mannering D E. The maltose transport system of Escherichia coli displays positive cooperativity in ATP hydrolysis. J Biol Chem. 1996;271:4858–4863. [PubMed] [Google Scholar]
- 54.Davidson A L, Nikaido H. Overproduction, solubilization, and reconstitution of the maltose transport system from Escherichia coli. J Biol Chem. 1990;265:4254–4260. [PubMed] [Google Scholar]
- 55.Davidson A L, Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 56.Davidson A L, Sharma S. Mutation of a single MalK subunit severely impairs maltose transport activity in Escherichia coli. J Bacteriol. 1997;179:5458–5464. doi: 10.1128/jb.179.17.5458-5464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson A L, Shuman H A, Nikaido H. Mechanism of maltose transport in Escherichia coli. Transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA. 1992;89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dean D A, Davidson A L, Nikaido H. The role of ATP as the energy source for maltose transport in Escherichia coli. Res Microbiol. 1990;141:348–352. doi: 10.1016/0923-2508(90)90010-n. [DOI] [PubMed] [Google Scholar]
- 59.Dean D A, Hor L I, Shuman H A, Nikaido H. Interaction between maltose-binding protein and the membrane-associated maltose transporter complex in Escherichia coli. Mol Microbiol. 1992;6:2033–2040. doi: 10.1111/j.1365-2958.1992.tb01376.x. [DOI] [PubMed] [Google Scholar]
- 60.Dean D A, Reizer J, Nikaido H, Saier M. Regulation of the maltose transport system of Escherichia coli by the glucose-specific enzyme III of the phosphoenolpyruvate-sugar phosphotransferase system. Characterization of inducer exclusion-resistant mutants and reconstitution of inducer exclusion in proteoliposomes. J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- 61.Death A, Ferenci T. The importance of the binding-protein-dependent Mgl system to the transport of glucose in Escherichia coli growing on low sugar concentrations. Res Microbiol. 1993;144:529–537. doi: 10.1016/0923-2508(93)90002-j. [DOI] [PubMed] [Google Scholar]
- 62.Death A, Ferenci T. Between feast and famine: endogenous inducer synthesis in the adaptation of Escherichia coli to growth with limiting carbohydrates. J Bacteriol. 1994;176:5101–5107. doi: 10.1128/jb.176.16.5101-5107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Death A, Notley L, Ferenci T. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J Bacteriol. 1993;175:1475–1483. doi: 10.1128/jb.175.5.1475-1483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Débarbouillé M, Raibaud O. Expression of the Escherichia coli malPQ operon remains unaffected after drastic alteration of its promoter. J Bacteriol. 1983;153:1221–1227. doi: 10.1128/jb.153.3.1221-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Débarbouillé M, Schwartz M. The use of gene fusions to study the expression of malT, the positive regulator gene of the maltose regulon. J Mol Biol. 1979;132:521–534. doi: 10.1016/0022-2836(79)90273-0. [DOI] [PubMed] [Google Scholar]
- 66.Débarbouillé M, Schwartz M. Mutants which make more malT product, the activator of the maltose regulon in Escherichia coli. Mol Gen Genet. 1980;178:589–595. doi: 10.1007/BF00337865. [DOI] [PubMed] [Google Scholar]
- 67.Débarbouillé M, Shuman H A, Silhavy T J, Schwartz M. Dominant constitutive mutations in malT, the positive regulator of the maltose regulon in Escherichia coli. J Mol Biol. 1978;124:359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- 68.Decker K. Ph.D. thesis. Konstanz, Germany: University of Konstanz; 1997. [Google Scholar]
- 69.Decker K, Peist R, Reidl J, Kossmann M, Brand B, Boos W. Maltose and maltotriose can be formed endogenously in Escherichia coli from glucose and glucose-1-phosphate independently of enzymes of the maltose system. J Bacteriol. 1993;175:5655–5665. doi: 10.1128/jb.175.17.5655-5665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Decker K, Plumbridge J, Boos W. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol. 1998;27:381–390. doi: 10.1046/j.1365-2958.1998.00694.x. [DOI] [PubMed] [Google Scholar]
- 71.d’Enfert C, Chapon C, Pugsley A P. Export and secretion of the lipoprotein pullulanase by Klebsiella pneumoniae. Mol Microbiol. 1987;1:107–116. doi: 10.1111/j.1365-2958.1987.tb00534.x. [DOI] [PubMed] [Google Scholar]
- 72.De Smit M H, Van Duin J. Control of translation by mRNA secondary structure in Escherichia coli. J Mol Biol. 1994;244:144–150. doi: 10.1006/jmbi.1994.1714. [DOI] [PubMed] [Google Scholar]
- 73.Diamond D L, Strobel S, Chun S-Y, Randall L L. Interaction of SecB with intermediates along the folding pathway of maltose-binding protein. Protein Sci. 1995;4:1118–1123. doi: 10.1002/pro.5560040610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dietzel I, Kolb V, Boos W. Pole cap formation in Escherichia coli following induction of the maltose-binding protein. Arch Microbiol. 1978;118:207–218. doi: 10.1007/BF00415731. [DOI] [PubMed] [Google Scholar]
- 75.Dumay V, Danchin A, Crasnier M. Regulation of Escherichia coli adenylate cyclase activity during hexose phosphate transport. Microbiology. 1996;142:575–583. doi: 10.1099/13500872-142-3-575. [DOI] [PubMed] [Google Scholar]
- 76.Duplay P, Bedouelle H, Fowler A, Zabin I, Saurin W, Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984;259:10606–10613. [PubMed] [Google Scholar]
- 77.Duplay P, Szmelcman S. Silent and functional changes in the periplasmic maltose binding protein of Escherichia coli K12. II. Chemotaxis towards maltose. J Mol Biol. 1987;194:675–678. doi: 10.1016/0022-2836(87)90244-0. [DOI] [PubMed] [Google Scholar]
- 78.Duplay P, Szmelcman S, Bedouelle H, Hofnung M. Silent and functional changes in the periplasmic maltose binding protein of Escherichia coli K12. I. Transport of maltose. J Mol Biol. 1987;194:663–673. doi: 10.1016/0022-2836(87)90243-9. [DOI] [PubMed] [Google Scholar]
- 79.Dutzler R, Wang Y-F, Rizkallah P J, Rosenbusch J P, Schirmer T. Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway. Structure. 1996;4:127–134. doi: 10.1016/s0969-2126(96)00016-0. [DOI] [PubMed] [Google Scholar]
- 80.Ehrle R, Pick C, Ulrich R, Hofmann E, Ehrmann M. Characterization of transmembrane domains 6, 7, and 8 of MalF by mutational analysis. J Bacteriol. 1996;178:2255–2262. doi: 10.1128/jb.178.8.2255-2262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ehrmann M, Boos W. Identification of endogenous inducers of the mal system in Escherichia coli. J Bacteriol. 1987;169:3539–3545. doi: 10.1128/jb.169.8.3539-3545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ehrmann M, Boyd D, Beckwith J. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferenci T. The recognition of maltodextrins by Escherichia coli. Eur J Biochem. 1980;108:631–636. doi: 10.1111/j.1432-1033.1980.tb04758.x. [DOI] [PubMed] [Google Scholar]
- 85.Ferenci T, Boos W. The role of the Escherichia coli lambda receptor in the transport of maltose and maltodextrins. J Supramol Struct. 1980;13:101–116. doi: 10.1002/jss.400130110. [DOI] [PubMed] [Google Scholar]
- 86.Ferenci T, Brass J, Boos W. The role of the periplasmic maltose-binding protein and the outer-membrane phage lambda receptor in maltodextrin transport of Escherichia coli. Biochem Soc Trans. 1980;8:680–681. doi: 10.1042/bst0080680. [DOI] [PubMed] [Google Scholar]
- 87.Ferenci T, Muir M, Lee K-S, Maris D. Substrate specificity of the Escherichia coli maltodextrin transport system and its component proteins. Biochim Biophys Acta. 1986;860:44–50. doi: 10.1016/0005-2736(86)90496-7. [DOI] [PubMed] [Google Scholar]
- 88.Ferenci T, Schwentorat M, Ullrich S, Vilmart J. Lambda receptor in the outer membrane of Escherichia coli as a binding protein for maltodextrins and starch polysaccharides. J Bacteriol. 1980;142:521–526. doi: 10.1128/jb.142.2.521-526.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fiedler G, Pajatsch M, Böck A. Genetics of a novel starch utilisation pathway present in Klebsiella oxytoca. J Mol Biol. 1996;256:279–291. doi: 10.1006/jmbi.1996.0085. [DOI] [PubMed] [Google Scholar]
- 90.Francetic O, Hanson M P, Kumamoto C A. prlA suppression of defective export of maltose-binding protein in secB mutants of Escherichia coli. J Bacteriol. 1993;175:4036–4044. doi: 10.1128/jb.175.13.4036-4044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Freundlieb S, Boos W. α-Amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986;261:2946–2953. [PubMed] [Google Scholar]
- 92.Freundlieb S, Ehmann U, Boos W. Facilitated diffusion of p-nitrophenyl-α-d-maltohexaoside through the outer membrane of Escherichia coli. J Biol Chem. 1988;263:314–320. [PubMed] [Google Scholar]
- 93.Froshauer S, Beckwith J. The nucleotide sequence of the gene for MalF protein, an inner membrane component of the maltose transport system of Escherichia coli. Repeated DNA sequences are found in the malE-malF intercistronic region. J Biol Chem. 1984;259:10896–10903. [PubMed] [Google Scholar]
- 94.Froshauer S, Green G N, Boyd D, McGovern K, Beckwith J. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol. 1988;200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 95.Fukuda Y, Yamaguchi S, Shimosaka M, Murata K, Kimura A. Cloning of the glucokinase gene in Escherichia coli B. J Bacteriol. 1983;156:922–925. doi: 10.1128/jb.156.2.922-925.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ganesh C, Shah A N, Swaminathan C P, Surolia A, Varadarajan R. Thermodynamic characterization of the reversible, two-state unfolding of maltose binding protein, a large two-domain protein. Biochemistry. 1997;36:5020–5028. doi: 10.1021/bi961967b. [DOI] [PubMed] [Google Scholar]
- 97.Gardina P, Conway C, Kossman M, Manson M. Aspartate and maltose-binding protein interact with adjacent sites in the tar chemotactic signal transducer of Escherichia coli. J Bacteriol. 1992;174:1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gardina P J, Bormans A F, Hawkins M A, Meeker J W, Manson M D. Maltose-binding protein interacts simultaneously and asymmetrically with both subunits of the Tar chemoreceptor. Mol Microbiol. 1997;23:1181–1191. doi: 10.1046/j.1365-2958.1997.3001661.x. [DOI] [PubMed] [Google Scholar]
- 99.Gerhardt, F., and W. Boos. 1997. Unpublished observations.
- 100.Gilardi G, Mei G, Rosato N, Agrò A F, Cass A E G. Spectroscopic properties of an engineered maltose binding protein. Protein Eng. 1997;10:479–486. doi: 10.1093/protein/10.5.479. [DOI] [PubMed] [Google Scholar]
- 101.Gilardi G, Zhou L Q, Hibbert L, Cass A E G. Engineering the maltose binding protein for reagentless fluorescence sensing. Anal Chem. 1994;66:3840–3847. doi: 10.1021/ac00093a047. [DOI] [PubMed] [Google Scholar]
- 102.Gilson E, Alloing G, Schmidt T, Claverys J P, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 1988;7:3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gilson E, Nikaido H, Hofnung M. Sequence of the malK gene in Escherichia coli K12. Nucleic Acids Res. 1982;10:7449–7458. doi: 10.1093/nar/10.22.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gilson E, Rousset J-P, Charbit A, Perrin D, Hofnung M. malM, a new gene of the maltose regulon in Escherichia coli K12. I. malM is the last gene of the malK-lamB operon and encodes a periplasmic protein. J Mol Biol. 1986;191:303–311. doi: 10.1016/0022-2836(86)90127-0. [DOI] [PubMed] [Google Scholar]
- 105.Granett S, Villarejo M. Regulation of gene expression in Escherichia coli by the local anesthetic procaine. J Mol Biol. 1982;160:363–367. doi: 10.1016/0022-2836(82)90181-4. [DOI] [PubMed] [Google Scholar]
- 106.Gutierrez C, Raibaud O. Point mutations that reduce the expression of malPQ, a positively controlled operon of Escherichia coli. J Mol Biol. 1984;177:69–86. doi: 10.1016/0022-2836(84)90058-5. [DOI] [PubMed] [Google Scholar]
- 107.Hall J A, Ganesan A K, Chen J, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Functional significance in active transport. J Biol Chem. 1997;272:17615–17622. doi: 10.1074/jbc.272.28.17615. [DOI] [PubMed] [Google Scholar]
- 108.Hall J A, Gehring K, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Correlation with the structure of ligands and the structure of binding protein. J Biol Chem. 1997;272:17605–17609. doi: 10.1074/jbc.272.28.17605. [DOI] [PubMed] [Google Scholar]
- 109.Hall J A, Thorgeirsson T E, Liu J, Shin Y-K, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli. Electron paramagnetic resonance study of ligand-induced global conformational changes by site-directed spin labeling. J Biol Chem. 1997;272:17610–17614. doi: 10.1074/jbc.272.28.17610. [DOI] [PubMed] [Google Scholar]
- 110.Hall M N, Gabay J, Débarbouillé M, Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982;295:616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- 111.Hartmann A, Boos W. Mutations in phoB, the positive gene activator of the pho regulon in Escherichia coli, affect the carbohydrate phenotype on MacConkey indicator plates. Res Microbiol. 1993;144:285–293. doi: 10.1016/0923-2508(93)90013-r. [DOI] [PubMed] [Google Scholar]
- 112.Hatfield D, Hofnung M, Schwartz M. Genetic analysis of the maltose A region in Escherichia coli. J Bacteriol. 1969;98:559–567. doi: 10.1128/jb.98.2.559-567.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hatfield D, Hofnung M, Schwartz M. Nonsense mutations in the maltose A region of the genetic map of Escherichia coli. J Bacteriol. 1969;100:1311–1315. doi: 10.1128/jb.100.3.1311-1315.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hazelbauer G L. The maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975;122:206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hekstra D, Tommassen J. Functional exchangeability of the ABC proteins of the periplasmic binding protein-dependent transport systems Ugp and Mal. J Bacteriol. 1993;175:6546–6552. doi: 10.1128/jb.175.20.6546-6552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hengge R, Boos W. Maltose and lactose transport in Escherichia coli. Examples of two different types of concentrative transport systems. Biochim Biophys Acta. 1983;737:443–478. doi: 10.1016/0304-4157(83)90009-6. [DOI] [PubMed] [Google Scholar]
- 117.Hengge-Aronis R. Back to log phase: ςS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 118.Hengge-Aronis R, Fischer D. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol. 1992;6:1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 119.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma-factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herrmann A, Schlösser A, Schmid R, Schneider E. Biochemical identification of a lipoprotein with maltose-binding activity in the thermoacidophilic Gram-positive bacterium Alicyclobacillus acidocaldarius. Res Microbiol. 1996;147:733–737. doi: 10.1016/s0923-2508(97)85120-0. [DOI] [PubMed] [Google Scholar]
- 121.Heyde M, Coll J L, Portalier R. Identification of Escherichia coli genes whose expression increases as a function of external pH. Mol Gen Genet. 1991;229:197–205. doi: 10.1007/BF00272156. [DOI] [PubMed] [Google Scholar]
- 122.Higgins C F. ABC transporters—from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 123.Higgins C F, Hiles I D, Salmond G P C, Gill D R, Downie J A, Evans I J, Holland I B, Gray L, Buckel S D, Bell A W, Hermodson M A. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986;323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 124.Hofnung M. An intelligent channel (and more) Science. 1995;267:473–474. doi: 10.1126/science.7824946. [DOI] [PubMed] [Google Scholar]
- 125.Hofnung M, Hatfield D, Schwartz M. malB region in Escherichia coli K-12: characterization of new mutations. J Bacteriol. 1974;117:40–47. doi: 10.1128/jb.117.1.40-47.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hofnung M, Schwartz M. Mutations allowing growth on maltose of Escherichia coli K 12 strains with a deleted malT gene. Mol Gen Genet. 1971;112:117–132. doi: 10.1007/BF00267490. [DOI] [PubMed] [Google Scholar]
- 127.Hofnung M, Schwartz M, Hatfield D. Complementation studies in the maltose-A region of Escherichia coli K12 genetic map. J Mol Biol. 1971;61:681–694. doi: 10.1016/0022-2836(71)90072-6. [DOI] [PubMed] [Google Scholar]
- 128.Hor L I, Shuman H A. Genetic analysis of periplasmic binding protein dependent transport in Escherichia coli. Each lobe of maltose-binding protein interacts with a different subunit of the MalFGK2 membrane transport complex. J Mol Biol. 1993;233:659–670. doi: 10.1006/jmbi.1993.1543. [DOI] [PubMed] [Google Scholar]
- 129.Horlacher R, Boos W. Characterization of TreR, the major regulator of the Escherichia coli trehalose system. J Biol Chem. 1997;272:13026–13032. doi: 10.1074/jbc.272.20.13026. [DOI] [PubMed] [Google Scholar]
- 130.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hosono K, Kakuda H, Ichihara S. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci Biotechnol Biochem. 1995;59:256–261. doi: 10.1271/bbb.59.256. [DOI] [PubMed] [Google Scholar]
- 132.Hu J C. Repressor fusions as a tool to study protein-protein interactions. Structure. 1995;3:431–433. doi: 10.1016/s0969-2126(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 133.Huang K-J, Igo M M. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 134.Inada T, Takahashi H, Mizuno T, Aiba H. Down regulation of cAMP production by cAMP receptor protein in Escherichia coli: an assessment of the contributions of transcriptional and posttranscriptional control of adenylate cyclase. Mol Gen Genet. 1996;253:198–204. doi: 10.1007/s004380050313. [DOI] [PubMed] [Google Scholar]
- 135.Ishizuka H, Hanamura A, Inada T, Aiba H. Mechanism of the down-regulation of cAMP receptor protein by glucose in Escherichia coli: role of autoregulation of the crp gene. EMBO J. 1994;13:3077–3082. doi: 10.1002/j.1460-2075.1994.tb06606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jordy M, Andersen C, Schülein K, Ferenci T, Benz R. Rate constants of sugar transport through two LamB mutants of Escherichia coli: comparison with wild-type maltoporin and LamB of Salmonella typhimurium. J Mol Biol. 1996;259:666–678. doi: 10.1006/jmbi.1996.0348. [DOI] [PubMed] [Google Scholar]
- 137.Kaasen I, Falkenberg P, Styrvold O B, Strøm A R. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli. Evidence that transcription is activated by KatF (AppR) J Bacteriol. 1992;174:889–898. doi: 10.1128/jb.174.3.889-898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kellerman O, Szmelcman S. Active transport of maltose in Escherichia coli K-12. Involvement of a periplasmic maltose-binding protein. Eur J Biochem. 1974;47:139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- 139.Kim I, Stiefel A, Plantör S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 140.Kim Y J, Rajapandi T, Oliver D. SecA protein is exposed to the periplasmic surface of the E. coli inner membrane in its active state. Cell. 1994;78:845–853. doi: 10.1016/s0092-8674(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 141.Klein W, Boos W. Induction of the λ receptor is essential for the effective uptake of trehalose in Escherichia coli. J Bacteriol. 1993;175:1682–1686. doi: 10.1128/jb.175.6.1682-1686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Klein W, Horlacher R, Boos W. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J Bacteriol. 1995;177:4043–4052. doi: 10.1128/jb.177.14.4043-4052.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 144.Kolbus, T., and W. Boos. 1997. Unpublished observations.
- 145.Kolbus, T., C. Braun, R. Peist, and W. Boos. 1997. Unpublished observations.
- 146.Kong L, Fromant M, Blanquet S, Plateau P. Evidence for a new Escherichia coli protein resembling a lysyl-tRNA synthetase. Gene. 1991;108:163–164. doi: 10.1016/0378-1119(91)90503-4. [DOI] [PubMed] [Google Scholar]
- 147.Kühnau S, Reyes M, Sievertsen A, Shuman H A, Boos W. The activities of the Escherichia coli MalK protein in maltose transport, regulation and inducer exclusion can be separated by mutations. J Bacteriol. 1991;173:2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kumar A, Larsen C E, Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose:α-1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J Biol Chem. 1986;261:16256–16259. [PubMed] [Google Scholar]
- 149.Lang H, Jonson G, Holmgren J, Palva E T. The maltose regulon of Vibrio cholerae affects production and secretion of virulence factors. Infect Immun. 1994;62:4781–4788. doi: 10.1128/iai.62.11.4781-4788.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lehmann J, Schiltz E, Steck J. Studies of the interaction of the maltose-binding protein of Escherichia coli, a closed-groove binder, with 4,6-O-ethylidenemalto-oligosaccharides (dp 2–5) and its regioselective labelling with 3-azibutyl 1-thio-alpha-(6-H-3) maltoside. Carbohydr Res. 1992;232:77–87. doi: 10.1016/s0008-6215(00)90995-9. [DOI] [PubMed] [Google Scholar]
- 151.Lippincott J, Traxler B. MalFGK complex assembly and transport and regulatory characteristics of MalK insertion mutants. J Bacteriol. 1997;179:1337–1343. doi: 10.1128/jb.179.4.1337-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu C E, Ames G F L. Characterization of transport through the periplasmic histidine permease using proteoliposomes reconstituted by dialysis. J Biol Chem. 1997;272:859–866. doi: 10.1074/jbc.272.2.859. [DOI] [PubMed] [Google Scholar]
- 153.Lu M, Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Luckey M, Nikaido H. Bacteriophage lambda receptor protein in Escherichia coli K-12: lowered affinity of some mutant proteins for maltose-binding protein in vitro. J Bacteriol. 1983;153:1056–1059. doi: 10.1128/jb.153.2.1056-1059.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Maeda S, Takayanagi K, Nishimura Y, Maruyama T, Sato K, Mizuno T. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: a study involving synthetic OmpR-binding sequences. J Biochem (Tokyo) 1991;110:324–327. doi: 10.1093/oxfordjournals.jbchem.a123579. [DOI] [PubMed] [Google Scholar]
- 156.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 157.Martin J L, Waksman G, Bardwell J C A, Beckwith J, Kuriyan J. Crystallization of DsbA, an Escherichia coli protein required for disulphide bond formation in vivo. J Mol Biol. 1993;230:1097–1100. doi: 10.1006/jmbi.1993.1226. [DOI] [PubMed] [Google Scholar]
- 158.Merino G, Boos W, Shuman H A, Bohl E. The inhibition of maltose transport by the unliganded form of the maltose-binding protein of Escherichia coli: experimental findings and mathematical treatment. J Theor Biol. 1995;177:171–179. doi: 10.1006/jtbi.1995.0236. [DOI] [PubMed] [Google Scholar]
- 159.Merino, G., and H. A. Shuman. Truncation of MalF results in lactose transport via the maltose transport system of Escherichia coli. J. Biol. Chem., in press. [DOI] [PubMed]
- 160.Merino G, Shuman H A. Unliganded maltose-binding protein triggers lactose transport in an Escherichia coli mutant with an alteration in the maltose transport system. J Bacteriol. 1997;179:7687–7694. doi: 10.1128/jb.179.24.7687-7694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Meyer D, Schneider-Fresenius C, Horlacher R, Peist R, Boos W. Molecular characterization of glucokinase from Escherichia coli K-12. J Bacteriol. 1997;179:1298–1306. doi: 10.1128/jb.179.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meyer J E W, Hofnung M, Schulz G E. Structure of maltoporin from Salmonella typhimurium ligated with a nitrophenyl-maltotrioside. J Mol Biol. 1997;266:761–775. doi: 10.1006/jmbi.1996.0823. [DOI] [PubMed] [Google Scholar]
- 163.Michaelis S, Chapon C, D’Enfert C, Pugsley A P, Schwartz M. Characterization and expression of the structural gene for pullulanase, a maltose-inducible secreted protein of Klebsiella pneumoniae. J Bacteriol. 1985;164:633–638. doi: 10.1128/jb.164.2.633-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miyamoto K, Nakahigashi K, Nishimura K, Inokuchi H. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol. 1991;219:393–398. doi: 10.1016/0022-2836(91)90180-e. [DOI] [PubMed] [Google Scholar]
- 165.Monod J, Torriani A M. De l’amylomaltase d’Escherichia coli. Ann Inst Pasteur (Paris) 1950;78:65–77. [PubMed] [Google Scholar]
- 166.Morbach S, Tebbe S, Schneider E. The ATP-binding cassette (ABC) transporter for maltose/maltodextrins of Salmonella typhimurium. Characterization of the ATPase activity associated with the purified MalK subunit. J Biol Chem. 1993;268:18617–18621. [PubMed] [Google Scholar]
- 167.Mourez M, Hofnung N, Dassa E. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Muro-Pastor A M, Ostrovsky P, Maloy S. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J Bacteriol. 1997;179:2788–2791. doi: 10.1128/jb.179.8.2788-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Murphy C K, Beckwith J. Export of proteins to the cell envelope in Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 967–978. [Google Scholar]
- 170.Nakamura Y, Ito K. Control and function of lysyl-tRNA synthase: diversity and coordination. Mol Microbiol. 1993;10:225–231. doi: 10.1111/j.1365-2958.1993.tb01948.x. [DOI] [PubMed] [Google Scholar]
- 171.Nekolla S, Andersen C, Benz R. Noise analysis of ion current through the open and the sugar-induced closed state of the LamB channel of Escherichia coli outer membrane: evaluation of the sugar binding kinetics to the channel interior. Biophys J. 1994;66:1388–1397. doi: 10.1016/S0006-3495(94)80929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Neubauer H, Glaasker E, Hammes W P, Poolman B, Konings W N. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. J Bacteriol. 1994;176:3007–3012. doi: 10.1128/jb.176.10.3007-3012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nikaido H, Pokrovskaya I D, Reyes L, Ganesan A K, Hall J A. Maltose transport system of Escherichia coli as a member of ABC transporters. In: Torriani A, Yagil E, Silver S, editors. Phosphate in microorganisms: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1994. pp. 91–96. [Google Scholar]
- 174.Notley L, Ferenci T. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient-stressed Escherichia coli. Mol Microbiol. 1995;16:121–129. doi: 10.1111/j.1365-2958.1995.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 175.Novokhatny V, Ingham K. Thermodynamics of maltose binding protein unfolding. Protein Sci. 1997;6:141–146. doi: 10.1002/pro.5560060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Okita T W, Rodriguez R L, Preiss J. Biosynthesis of bacterial glycogen: cloning of the glycogen enzyme structural genes of Escherichia coli. J Biol Chem. 1981;256:6944–6952. [PubMed] [Google Scholar]
- 177.O’Reilly M, Watson K A, Schinzel R, Palm D, Johnson L N. Oligosaccharide substrate binding in Escherichia coli maltodextrin phosphorylase. Nat Struct Biol. 1997;4:405–412. doi: 10.1038/nsb0597-405. [DOI] [PubMed] [Google Scholar]
- 178.Overduin P, Boos W, Tommassen J. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988;2:767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 179.Pajatsch, M., and A. Böck. 1997. Unpublished observations.
- 180.Pajatsch, M., W. Boos, and A. Böck. Preparation of radioactively labeled linear maltodextrins and cyclodextrins of high specific activity from [14C]-maltose. Carbohydr. Res., in press. [DOI] [PubMed]
- 181.Palm D, Goerl R, Burger K J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985;313:500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- 182.Palmer N T, Ryman B E, Whelan W J. The action pattern of amylomaltase from Escherichia coli. Eur J Biochem. 1976;69:105–115. doi: 10.1111/j.1432-1033.1976.tb10863.x. [DOI] [PubMed] [Google Scholar]
- 183.Panagiotidis, C. H., W. Boos, and H. A. Shuman. Control of transcription by transport monitoring of an ABC transporter. Submitted for publication.
- 184.Panagiotidis C H, Reyes M, Sievertsen A, Boos W, Shuman H A. Characterization of the structural requirements for assembly and nucleotide binding of an ATP-binding cassette transporter—the maltose transport system of Escherichia coli. J Biol Chem. 1993;268:23685–23696. [PubMed] [Google Scholar]
- 185.Peist R, Schneider-Fresenius C, Boos W. The MalT-dependent and malZ-encoded maltodextrin glucosidase of Escherichia coli can be converted into a dextrinyltransferase by a single mutation. J Biol Chem. 1996;271:10681–10689. doi: 10.1074/jbc.271.18.10681. [DOI] [PubMed] [Google Scholar]
- 186.Peist R, Koch A, Bolek P, Kolbus T, Boos W. Characterization of the aes gene of Escherichia coli encoding an enzyme with esterase activity. J Bacteriol. 1997;179:7679–7686. doi: 10.1128/jb.179.24.7679-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Peterkofsky A, Svenson I, Amin N. Regulation of Escherichia coli adenylate cyclase activity by the phosphoenolpyruvate:sugar phosphotransferase system. FEMS Microbiol Rev. 1989;63:103–108. doi: 10.1016/0168-6445(89)90013-2. [DOI] [PubMed] [Google Scholar]
- 188.Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–3965. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- 190.Podkovyrov S M, Zeikus J G. Structure of the gene encoding cyclomaltodextrinase from Clostridium thermohydrosulfuricum 39E and characterization of the enzyme purified from Escherichia coli. J Bacteriol. 1992;174:5400–5405. doi: 10.1128/jb.174.16.5400-5405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase system. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1149–1174. [Google Scholar]
- 192.Prossnitz E. Determination of a region of the HisJ binding protein involved in the recognition of the membrane complex of the histidine transport system of Salmonella typhimurium. J Biol Chem. 1991;266:9673–9677. [PubMed] [Google Scholar]
- 193.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pugsley A P, Dubreuil C. Molecular characterization of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol Microbiol. 1988;2:473–479. doi: 10.1111/j.1365-2958.1988.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 195.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 196.Puyet A, Espinosa M. Structure of the maltodextrin-uptake locus of Streptococcus pneumoniae. Correlation to the Escherichia coli maltose regulon. J Mol Biol. 1993;230:800–811. doi: 10.1006/jmbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 197.Quiocho F A, Spurlino J C, Rodseth L E. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 198.Raha M, Kawagishi I, Müller V, Kihara M, Macnab R M. Escherichia coli produces a cytoplasmic α-amylase, AmyA. J Bacteriol. 1992;174:6644–6652. doi: 10.1128/jb.174.20.6644-6652.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Raibaud O. Nucleoprotein structures at positively regulated bacterial promoters: homology with replication origins and some hypotheses on the quaternary structure of the activator proteins in these complexes. Mol Microbiol. 1989;3:455–458. doi: 10.1111/j.1365-2958.1989.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 200.Raibaud O, Débarbouillé M, Schwartz M. Use of deletions created in vitro to map transcriptional regulatory signals in the malA region of Escherichia coli. J Mol Biol. 1983;163:395–408. doi: 10.1016/0022-2836(83)90065-7. [DOI] [PubMed] [Google Scholar]
- 201.Raibaud O, Gutierrez C, Schwartz M. Essential and nonessential sequences in malPp, a positively controlled promoter in Escherichia coli. J Bacteriol. 1985;161:1201–1208. doi: 10.1128/jb.161.3.1201-1208.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Raibaud O, Richet E. Maltotriose is the inducer of the maltose regulon. J Bacteriol. 1987;169:3059–3061. doi: 10.1128/jb.169.7.3059-3061.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Raibaud O, Roa M, Braun-Breton C, Schwartz M. Structure of the malB region in Escherichia coli K12. I. Genetic map of the malK-lamB operon. Mol Gen Genet. 1979;174:241–248. doi: 10.1007/BF00267796. [DOI] [PubMed] [Google Scholar]
- 204.Raibaud O, Schwartz M. Restriction map of the Escherichia coli malA region and identification of the malT product. J Bacteriol. 1980;143:761–771. doi: 10.1128/jb.143.2.761-771.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Raibaud O, Vidal-Ingigliardi D, Kolb A. Genetic studies on the promoter of malT, the gene that encodes the activator of the Escherichia coli maltose regulon. Res Microbiol. 1991;142:937–942. doi: 10.1016/0923-2508(91)90003-s. [DOI] [PubMed] [Google Scholar]
- 206.Raibaud O, Vidal-Ingigliardi D, Richet E. A complex nucleoprotein structure involved in activation of transcription of two divergent Escherichia coli promoters. J Mol Biol. 1989;205:471–485. doi: 10.1016/0022-2836(89)90218-0. [DOI] [PubMed] [Google Scholar]
- 207.Randall-Hazelbauer L, Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973;116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Reidl J, Boos W. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J Bacteriol. 1991;173:4862–4876. doi: 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Reidl J, Römisch K, Ehrmann M, Boos W. MalI, a novel protein involved in regulation of the maltose system of Escherichia coli, is highly homologous to the repressor proteins GalR, CytR, and LacI. J Bacteriol. 1989;171:4888–4899. doi: 10.1128/jb.171.9.4888-4899.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Reyes M, Shuman H A. Overproduction of MalK protein prevents expression of the Escherichia coli mal regulon. J Bacteriol. 1988;170:4598–4602. doi: 10.1128/jb.170.10.4598-4602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Reyes M, Treptow N A, Shuman H A. Transport of p-nitrophenyl-α-maltoside by the maltose transport system of Escherichia coli and its subsequent hydrolysis by a cytoplasmic maltosidase. J Bacteriol. 1986;165:918–922. doi: 10.1128/jb.165.3.918-922.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Richarme G, Caldas T D. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J Biol Chem. 1997;272:15607–15612. doi: 10.1074/jbc.272.25.15607. [DOI] [PubMed] [Google Scholar]
- 213.Richet E. On the role of the multiple regulatory elements involved in the activation of the Escherichia coli malEp promoter. J Mol Biol. 1996;264:852–862. doi: 10.1006/jmbi.1996.0682. [DOI] [PubMed] [Google Scholar]
- 214.Richet E, Raibaud O. Purification and properties of the MalT protein, the transcription activator of the Escherichia coli maltose regulon. J Biol Chem. 1987;262:12647–12653. [PubMed] [Google Scholar]
- 215.Richet E, Raibaud O. MalT, the regulatory protein of the Escherichia coli maltose system, is an ATP-dependent transcriptional activator. EMBO J. 1989;8:981–987. doi: 10.1002/j.1460-2075.1989.tb03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Richet E, Raibaud O. Supercoiling is essential for the formation and stability of the initiation complex at the divergent malEp and malKp promoters. J Mol Biol. 1991;218:529–542. doi: 10.1016/0022-2836(91)90699-7. [DOI] [PubMed] [Google Scholar]
- 217.Richet E, Søgaard-Andersen L. CRP induces the repositioning of MalT at the Escherichia coli malKp promoter primarily through DNA bending. EMBO J. 1994;13:4558–4567. doi: 10.1002/j.1460-2075.1994.tb06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Richet E, Vidal-Ingigliardi D, Raibaud O. A new mechanism for coactivation of transcription initiation: repositioning of an activator triggered by the binding of a second activator. Cell. 1991;66:1185–1195. doi: 10.1016/0092-8674(91)90041-v. [DOI] [PubMed] [Google Scholar]
- 219.Rimmele M, Boos W. Trehalose-6-phosphate hydrolase of Escherichia coli. J Bacteriol. 1994;176:5654–5664. doi: 10.1128/jb.176.18.5654-5664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Roehl R A, Vinopal R T. New maltose blue mutations in Escherichia coli K-12. J Bacteriol. 1979;139:683–685. doi: 10.1128/jb.139.2.683-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Romeo T, Kumar A, Preiss J. Analysis of the Escherichia coli glycogen gene cluster suggests that catabolic enzymes are encoded among the biosynthetic genes. Gene. 1988;70:363–376. doi: 10.1016/0378-1119(88)90208-9. [DOI] [PubMed] [Google Scholar]
- 222.Rousset J-P, Gilson E, Hofnung M. malM, a new gene of the maltose regulon in Escherichia coli K12. II. Mutations affecting the signal peptide of the MalM protein. J Mol Biol. 1986;191:313–320. doi: 10.1016/0022-2836(86)90128-2. [DOI] [PubMed] [Google Scholar]
- 223.Rouvière P E, Gross C A. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 224.Russo F D, Silhavy T J. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 225.Sahm K, Matuschek M, Müller H, Mitchell W J, Bahl H. Molecular analysis of the amy gene locus of Thermoanaerobacterium thermosulfurigenes EM1 encoding starch-degrading enzymes and a binding protein-dependent maltose transport system. J Bacteriol. 1996;178:1039–1046. doi: 10.1128/jb.178.4.1039-1046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Saul F A, Vulliez-le Normand B, Lema F, Bentley G A. Crystal structure of a recombinant form of the maltodextrin-binding protein carrying an inserted sequence of a B-cell epitope from the preS2 region of hepatitis B virus. Protein Struct Funct Genet. 1997;27:1–8. doi: 10.1002/(sici)1097-0134(199701)27:1<1::aid-prot2>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 227.Saurin W, Dassa E. Sequence relationships between integral inner membrane proteins of binding protein-dependent transport systems—evolution by recurrent gene duplications. Protein Sci. 1994;3:325–344. doi: 10.1002/pro.5560030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Saurin W, Köster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 229.Schinzel R, Palm D. Escherichia coli maltodextrin phosphorylase: contribution of active site residues glutamate-637 and tyrosine-538 to the phosphorolytic cleavage of α-glucans. Biochemistry. 1990;29:9956–9962. doi: 10.1021/bi00494a028. [DOI] [PubMed] [Google Scholar]
- 230.Schirmer T, Cowan S W. Prediction of membrane spanning β-strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schirmer T, Keller T A, Wang Y F, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 232.Schleif R. Two positively regulated systems, ara and mal. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1300–1309. [Google Scholar]
- 233.Schlösser A, Kampers T, Schrempf H. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J Bacteriol. 1997;179:2092–2095. doi: 10.1128/jb.179.6.2092-2095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schneider E, Francoz E, Dassa E. Completion of the nucleotide sequence of the ‘maltose B’ region in Salmonella typhimurium: the high conservation of the malM gene suggests a selected physiological role for its product. Biochim Biophys Acta. 1992;1129:223–227. doi: 10.1016/0167-4781(92)90492-i. [DOI] [PubMed] [Google Scholar]
- 235.Schneider E, Freundlieb S, Tapio S, Boos W. Molecular characterization of the MalT-dependent periplasmic α-amylase of Escherichia coli encoded by malS. J Biol Chem. 1992;267:5148–5154. [PubMed] [Google Scholar]
- 236.Schneider E, Hunke S, Tebbe S. The MalK protein of the ATP-binding cassette transporter for maltose of Escherichia coli is accessible to protease digestion from the periplasmic side of the membrane. J Bacteriol. 1995;177:5364–5367. doi: 10.1128/jb.177.18.5364-5367.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schneider E, Walter C. A chimeric nucleotide-binding protein, encoded by a hisP-malK hybrid gene, is functional in maltose transport in Salmonella typhimurium. Mol Microbiol. 1991;5:1375–1383. doi: 10.1111/j.1365-2958.1991.tb00784.x. [DOI] [PubMed] [Google Scholar]
- 238.Schwartz M. Aspects biochimiques et génétiques du metabolisme du maltose chez Escherichia coli K12. C R Acad Sci. 1965;260:2613–2616. [PubMed] [Google Scholar]
- 239.Schwartz M. Phenotypic expression and genetic localization of mutations affecting maltose metabolism in Escherichia coli K 12. Ann Inst Pasteur (Paris) 1967;112:673–698. [PubMed] [Google Scholar]
- 240.Schwartz M. Sur l’existence chez Escherichia coli K12 d’une régulation commune à la biosynthèse des récepteurs du bacteriophage λ et au métabolisme du maltose. Ann Inst Pasteur (Paris) 1967;113:687–704. [PubMed] [Google Scholar]
- 241.Schwartz M. The maltose regulon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1482–1502. [Google Scholar]
- 242.Schwartz M, Hofnung M. La maltodextrin phosphorylase d’Escherichia coli. Eur J Biochem. 1967;2:132–145. doi: 10.1111/j.1432-1033.1967.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 243.Sharff A J, Rodseth L F, Quiocho F A. Refined 1.8-Å structure reveals the mode of binding of β-cyclodextrin to the maltodextrin binding protein. Biochemistry. 1993;32:10553–10559. doi: 10.1021/bi00091a004. [DOI] [PubMed] [Google Scholar]
- 244.Sharff A J, Rodseth L E, Spurlino J C, Quiocho F A. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 245.Sharff A J, Rodseth L E, Szmelcman S, Hofnung M, Quiocho F A. Refined structures of two insertion/deletion mutants probe function of the maltodextrin binding protein. J Mol Biol. 1995;246:8–13. doi: 10.1006/jmbi.1994.0059. [DOI] [PubMed] [Google Scholar]
- 246.Shen B H P, Boos W. Regulation of the β-methylgalactoside transport system and the galactose-binding protein by the cell cycle of Escherichia coli. Proc Natl Acad Sci USA. 1973;70:1481–1485. doi: 10.1073/pnas.70.5.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shilton B H, Flocco M M, Nilsson M, Mowbray S L. Conformational changes of three periplasmic receptors for bacterial chemotaxis and transport: the maltose-, glucose/galactose- and ribose-binding proteins. J Mol Biol. 1996;264:350–363. doi: 10.1006/jmbi.1996.0645. [DOI] [PubMed] [Google Scholar]
- 248.Shilton B H, Shuman H A, Mowbray S L. Crystal structures and solution conformations of a dominant-negative mutant of Escherichia coli maltose-binding protein. J Mol Biol. 1996;264:364–376. doi: 10.1006/jmbi.1996.0646. [DOI] [PubMed] [Google Scholar]
- 249.Shuman H A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982;257:5455–5461. [PubMed] [Google Scholar]
- 250.Shuman H A, Panagiotidis C H. Tinkering with transporters: periplasmic binding protein-dependent maltose transport in Escherichia coli. J Bioenerg Biomembr. 1993;25:613–620. doi: 10.1007/BF00770248. [DOI] [PubMed] [Google Scholar]
- 251.Shuman H A, Silhavy T J. Identification of the malK gene product. A peripheral membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1981;256:560–562. [PubMed] [Google Scholar]
- 252.Shuman H A, Silhavy T J, Beckwith J R. Labeling of proteins with β-galactosidase by gene fusion. Identification of a cytoplasmic membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1980;255:168–174. [PubMed] [Google Scholar]
- 253.Silhavy T J, Hartig-Beecken I, Boos W. Periplasmic protein related to the sn-glycerol-3-phosphate transport system of Escherichia coli. J Bacteriol. 1976;126:951–958. doi: 10.1128/jb.126.2.951-958.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Silhavy T J, Brickman E, Bassford P J, Casadaban M J, Shuman H A, Schwartz V, Guarente L, Schwartz M, Beckwith J R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979;174:249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- 255.Slauch J M, Silhavy T J. The porin regulon: a paradigm for the two-component regulatory system. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 383–417. [Google Scholar]
- 256.Spiess S, Happersberger H P, Glocker M O, Spiess E, Rippe K, Ehrmann M. Biochemical characterization and mass spectrometric disulfide bond mapping of periplasmic α-amylase of Escherichia coli. J Biol Chem. 1997;272:22125–22133. doi: 10.1074/jbc.272.35.22125. [DOI] [PubMed] [Google Scholar]
- 257.Spurlino J C, Lu G-Y, Quiocho F A. The 2.3-Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 258.Stevens-Clark J S, Theisen M C, Conklin K A, Smith R A. Phosphoramidates. VI. Purification and characterization of a phosphoryl transfer enzyme from Escherichia coli. J Biol Chem. 1968;243:4468–4473. [PubMed] [Google Scholar]
- 259.Stolz B, Huber M, Markovic-Housley Z, Erni B. The mannose transporter of Escherichia coli. Structure and function of the IIABMan subunit. J Biol Chem. 1993;268:27094–27099. [PubMed] [Google Scholar]
- 260.Stout V, Torres-Cabassa A, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Strobel S M, Cannon J G, Bassford P J J. Regions of maltose-binding protein that influence SecB-dependent and SecA-dependent export in Escherichia coli. J Bacteriol. 1993;175:6988–6995. doi: 10.1128/jb.175.21.6988-6995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sullivan M A, Cannon J F, Webb F H, Bock R M. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J Bacteriol. 1985;161:368–376. doi: 10.1128/jb.161.1.368-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Szmelcman S, Hofnung M. Maltose transport in Escherichia coli K12. Involvement of the bacteriophage lambda receptor. J Bacteriol. 1975;124:112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Szmelcman S, Sassoon N, Hofnung M. Residues in the α helix 7 of the bacterial maltose binding protein which are important in interactions with the Mal FGK2 complex. Protein Sci. 1997;6:628–636. doi: 10.1002/pro.5560060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Szmelcman S, Schwartz M, Silhavy T J, Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and λ-resistant mutants with the dissociation constants of the maltose binding protein as measured by fluorescence quenching. Eur J Biochem. 1976;65:13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- 266.Tangney M, Buchanan C J, Priest F G, Mitchell W J. Maltose uptake and its regulation in Bacillus subtilis. FEMS Microbiol Lett. 1992;97:191–196. doi: 10.1016/0378-1097(92)90385-2. [DOI] [PubMed] [Google Scholar]
- 267.Tapio S, Yeh F, Shuman H A, Boos W. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. J Biol Chem. 1991;266:19450–19458. [PubMed] [Google Scholar]
- 268.Tchetina E, Newman E B. Identification of lrp-regulated genes by inverse PCR and sequencing: regulation of two mal operons of Escherichia coli by leucine-responsive regulatory protein. J Bacteriol. 1995;177:2679–2683. doi: 10.1128/jb.177.10.2679-2683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Thieme R, Lay H, Oser A, Lehmann J, Wrissenberg S, Boos W. 3-Azi-1-methoxybutyl d-maltooligosaccharides specifically bind to the maltose/maltooligosaccharide-binding protein of Escherichia coli and can be used as photoaffinity labels. Eur J Biochem. 1986;160:83–91. doi: 10.1111/j.1432-1033.1986.tb09943.x. [DOI] [PubMed] [Google Scholar]
- 270.Thompson J. Regulation of sugar transport and metabolism in lactic acid bacteria. FEMS Microbiol Rev. 1987;46:221–231. [Google Scholar]
- 271.Tommassen, J., and W. Boos. 1995. Unpublished observations.
- 272.Topping T B, Randall L L. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligands. J Biol Chem. 1997;272:19314–19318. doi: 10.1074/jbc.272.31.19314. [DOI] [PubMed] [Google Scholar]
- 273.Treptow N, Shuman H. Genetic evidence for substrate and periplasmic binding-protein recognition by MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J Bacteriol. 1985;163:654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Treptow N A, Shuman H A. Allele-specific malE mutations that restore interactions between maltose-binding protein and the inner-membrane components of the maltose transport system. J Mol Biol. 1988;202:809–822. doi: 10.1016/0022-2836(88)90560-8. [DOI] [PubMed] [Google Scholar]
- 275.van der Vlag J, van Dam K, Postma P W. Quantification of the regulation of glycerol and maltose metabolism by IIAGlc of the phosphoenolpyruvate-dependent glucose phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1994;176:3518–3526. doi: 10.1128/jb.176.12.3518-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.van Wezel G P, White J, Young P, Postma P W, Bibb M J. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacI-galR family of regulatory genes. Mol Microbiol. 1997;23:537–549. doi: 10.1046/j.1365-2958.1997.d01-1878.x. [DOI] [PubMed] [Google Scholar]
- 277.Véscovi E G, Ayala Y M, Di Cera E, Groisman E A. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+ J Biol Chem. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- 278.Véscovi E G, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 279.Vidal-Ingigliardi D, Raibaud O. Three adjacent binding sites for cAMP receptor protein are involved in the activation of the divergent malEp-malKp promoters. Proc Natl Acad Sci USA. 1991;88:229–233. doi: 10.1073/pnas.88.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Vidal-Ingigliardi D, Richet E, Danot O, Raibaud O. A small C-terminal region of the Escherichia coli MalT protein contains the DNA-binding domain. J Biol Chem. 1993;268:24527–24530. [PubMed] [Google Scholar]
- 281.Vidal-Ingigliardi D, Richet E, Raibaud O. Two MalT binding sites in direct repeat. A structural motif involved in the activation of all the promoters of the maltose regulons in Escherichia coli and Klebsiella pneumoniae. J Mol Biol. 1991;218:323–334. doi: 10.1016/0022-2836(91)90715-i. [DOI] [PubMed] [Google Scholar]
- 282.Vihinen M, Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol. 1989;24:329–418. doi: 10.3109/10409238909082556. [DOI] [PubMed] [Google Scholar]
- 283.Waldburger C D, Sauer R T. Signal detection by the PhoQ sensor-transmitter. Characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J Biol Chem. 1996;271:26630–26636. doi: 10.1074/jbc.271.43.26630. [DOI] [PubMed] [Google Scholar]
- 284.Walker J E, Saraste M, Runswik J M, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wallenfels K, Bender H, Rached I. Pullulanase from Aerobacter aerogenes; production in a cell-bound state. Purification and properties of the enzyme. Biochem Biophys Res Commun. 1966;22:254–261. doi: 10.1016/0006-291x(66)90474-8. [DOI] [PubMed] [Google Scholar]
- 286.Wandersman C, Moreno F, Schwartz M. Pleiotropic mutations rendering Escherichia coli K-12 resistant to bacteriophage TP1. J Bacteriol. 1980;143:1374–1383. doi: 10.1128/jb.143.3.1374-1383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wang Y F, Dutzler R, Rizkallah P J, Rosenbusch J P, Schirmer T. Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J Mol Biol. 1997;272:56–63. doi: 10.1006/jmbi.1997.1224. [DOI] [PubMed] [Google Scholar]
- 288.Wanner B L, Jiang W, Kim S-K, Yamagata S, Haldimann A, Daniels L L. Are the multiple signals transduction pathways of the Pho regulon due to cross talk or cross regulation? In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 287–315. [Google Scholar]
- 289.Wanner B L, Sarthy A, Beckwith J. Escherichia coli mutant that reduces amounts of several periplasmic and outer membrane proteins. J Bacteriol. 1979;140:229–239. doi: 10.1128/jb.140.1.229-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Watson K A, Schinzel R, Palm D, Johnson L N. The crystal structure of Escherichia coli maltodextrin phosphorylase provides an explanation for the activity without control in this basic archetype of a phosphorylase. EMBO J. 1997;16:1–14. doi: 10.1093/emboj/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Waukau J, Forst S. Molecular analysis of the signaling pathway between EnvZ and OmpR in Escherichia coli. J Bacteriol. 1992;174:1522–1527. doi: 10.1128/jb.174.5.1522-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Weissborn A C, Liu Q, Rumley M K, Kennedy E P. UTP:α-d-glucose-1-phosphate uridylyltransferase of Escherichia coli: isolation and DNA sequence of the galU gene and purification of the enzyme. J Bacteriol. 1994;176:2611–2618. doi: 10.1128/jb.176.9.2611-2618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wickner W, Leonard M R. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 294.Wiesmeyer H, Cohn M. The characterization of the pathway of maltose utilization by Escherichia coli. II. General properties and mechanism of action of amylomaltase. Biochim Biophys Acta. 1960;39:427–439. doi: 10.1016/0006-3002(60)90195-5. [DOI] [PubMed] [Google Scholar]
- 295.Wiesmeyer H, Cohn M. The characterization of the pathway of maltose utilization by Escherichia coli. I. Purification and physical chemical properties of the enzyme amylomaltase. Biochim Biophys Acta. 1960;39:417–426. doi: 10.1016/0006-3002(60)90194-3. [DOI] [PubMed] [Google Scholar]
- 296.Wilken S, Schmees G, Schneider E. A putative helical domain in the MalK subunit of the ATP-binding-cassette transport system for maltose of Salmonella typhimurium (MalFGK2) is crucial for interaction with MalF and MalG. A study using the LacK protein of Agrobacterium radiobacter as a tool. Mol Microbiol. 1996;22:655–666. doi: 10.1046/j.1365-2958.1996.d01-1724.x. [DOI] [PubMed] [Google Scholar]
- 297.Wu H C P, Kalckar H M. Endogenous induction of the galactose operon in Escherichia coli K12. Proc Natl Acad Sci USA. 1966;55:622–629. doi: 10.1073/pnas.55.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Xavier K B, Kossmann M, Santos H, Boos W. Kinetic analysis by in vivo 31P nuclear magnetic resonance of internal Pi during the uptake of sn-glycerol-3-phosphate by the pho regulon-dependent Ugp system and the glp regulon-dependent GlpT system. J Bacteriol. 1995;177:699–704. doi: 10.1128/jb.177.3.699-704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Xavier K B, Martins L O, Peist R, Kossmann M, Boos W, Santos H. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1996;178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Yu F, Jen J, Takeuchi E, Inouye M, Nakayama H, Tagaya M, Fukui T. α-Glucan phosphorylase from Escherichia coli. Cloning of the gene and purification and characterization of the protein. J Biol Chem. 1988;263:13706–13711. [PubMed] [Google Scholar]
- 301.Zdych E, Peist R, Reidl J, Boos W. MalY of Escherichia coli is an enzyme with the activity of a βC-S lyase (cystathionase) J Bacteriol. 1995;177:5035–5039. doi: 10.1128/jb.177.17.5035-5039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhang Y H, Conway C, Rosato M, Suh Y, Manson M D. Maltose chemotaxis involves residues in the N-terminal and C-terminal domains on the same face of maltose-binding protein. J Biol Chem. 1992;267:22813–22820. [PubMed] [Google Scholar]
- 303.Zhang Y H, Mannering D E, Davidson A L, Yao N H, Manson M D. Maltose-binding protein containing an interdomain disulfide bridge confers a dominant-negative phenotype for transport and chemotaxis. J Biol Chem. 1996;271:17881–17889. doi: 10.1074/jbc.271.30.17881. [DOI] [PubMed] [Google Scholar]