Abstract
The purpose of this study was to investigate the effect of chemoresistant cancer-associated fibroblasts (R-CAFs) against cisplatin (DDP) on colorectal cancer (CRC) progression. First, clinical tissue samples of chemoresistant or chemosensitive CRC patients were collected to isolate R-CAFs or chemosensitive CAFs (S-CAFs), respectively. HT29 cells or HUVECs were co-cultured with R-CAFs by transwell device. Then the proliferation and apoptosis of HT29 cells were detected with Cell Counting Kit-8 (CCK-8) and flow cytometry. Transwell assay and tube formation assay was used to detect the migration and angiogenesis of HUVECs. In addition, a colorectal cancer transplantation model was established subcutaneously in nude mice by injecting stably transfected HT29 cells and exosomes from different CAF groups, and then the tumor volume and weight were measured and recorded. Hematoxylin and eosin staining, immunohistochemistry, and terminal deoxynucleotidyl transferase dUTP Nick-End Labeling (TUNEL) staining were performed to characterize the histopathological characteristics and apoptosis level of tumor tissues, respectively. S-CAFs and R-CAFs were isolated successfully. HT29 cell co-culture with R-CAFs significantly affected the proliferation and apoptosis of HT29 cells. Exosomes derived from R-CAFs (R-CAFs-Exo) were delivered to HT29 cells, which could induce viability, suppress apoptosis and accelerate the angiogenesis of CRC. In addition, VEGFA was highly expressed in R-CAFs-Exo, which might indicate that R-CAFs could transmit VEGFA through exosomes. Overexpressed VEGFA in R-CAFs apparently regulates the viability, apoptosis, DDP resistance, and angiogenesis of CRC. In-vivo experiments confirmed that R-CAFs-Exo promoted the progression of CRC and DDP resistance by delivering VEGFA. R-CAFs-derived exosomes promote the viability, apoptosis, DDP resistance, and angiogenesis of CRC by delivering VEGFA.
Keywords: colorectal cancer, cancer-associated fibroblast, cisplatin resistance, exosome, VEGFA
Introduction
Colorectal cancer (CRC), a kind of cancer that spreads widely worldwide, ranks as the third most prevalent malignancy [1]. Advanced diagnosis and metastasis are the main causes of high mortality in patients with CRC [2]. The underlying mechanisms of CRC are multifactorial, and the factors promoting its pathogenesis include genetics, age, chronic inflammation, and lifestyle [3]. Despite advances in awareness and early screening, a quarter of cases are still diagnosed with colorectal cancer at an advanced stage [4]. At present, the treatment of CRC covers surgical resection, chemotherapy, or adjuvant treatment, which will inevitably bring some serious side effects, such as vomiting, weight loss, and infection complications [5,6]. In addition, the majority of CRC patients develop drug resistance after a period of treatment, which eventually results in CRC recurrence [7,8]. Therefore, exploring novel and reliable biomarkers for CRC treatment is urgently needed.
To identify new non-invasive biomarkers for early diagnosis of CRC, we note that tumor microenvironment (TME) cell-related markers have special significance [9]. At the beginning of the tumor, the phenotype of surrounding cells changes in response to malignant transformation. One of the most significant transformations is to activate normal fibroblasts into tumor-associated fibroblasts (CAFs), showing specific and differential protein expression and producing regulatory signals to promote tumor development [10,11]. Studies have confirmed that fibroblasts in the TME achieve cross-talk with tumor cells by secreting exosomes [12]. Exosomes are small vesicles ranging from 30 to 100 nm in size, released upon exocytosis of multivesicular bodies, which contain and deliver mRNAs, proteins, and DNA [13]. They may mediate signal transduction in specific recipient cells by diffusing into neighboring cells or distant anatomical locations [14]. CAFs-derived exosomes remodel TME by regulating several processes, such as angiogenesis, immunosuppression, and drug acquisition, which is also thought to be the reason for promoting tumor progression and chemotherapy resistance [15,16]. LncRNA MEG3 released from CAF-derived exosomes enhances cisplatin chemoresistance in small-cell lung cancer via regulation of a miR-15a-5p/CCNE1 axis [17]. The latest research showed that CAF-derived exosomes delivered LINC01410 to promote epithelial–mesenchymal transition of esophageal squamous cell carcinoma [18]. Additionally, exosomes contain various substances that can accelerate angiogenesis and thus play an important role in cancer invasion [19]. Research demonstrated that CAF-derived exosomes upregulated miR-135b-5p to promote CRC cell growth and angiogenesis by inhibiting TXNIP [20]. However, the effect of CAFs-derived exosomes on CRC angiogenesis and chemoresistance is still largely unknown.
In our study, we first isolated CAFs from chemoresistant and chemosensitive patients, and then CAFs-derived exosomes were isolated and identified. Subsequently, a series of cell experiments and animal experiments were carried out to identify the role of CAFs-derived exosomes in the proliferation, apoptosis, angiogenesis, and DDP resistance of CRC, as well as the molecular mechanisms involved.
Methods
Collection of clinical tissue samples and separation of cancer-associated fibroblasts
Tumor tissues of CRC patients were obtained from the Third People’s Hospital of Yancheng City. The criteria of chemoresistance and chemosensitivity are divided according to the 2021 second edition of the National Comprehensive Cancer Network colon cancer guidelines. The primary CAFs were isolated from the collected tumor tissues of chemoresistant and chemosensitive patients, namely chemoresistant CAFs (R-CAFs) and chemosensitive CAFs (S-CAFs) [21]. And the CAFs were identified by the related marker in immunofluorescence assay. Ethical approval was obtained from the institutional ethics committee of Third People’s Hospital of Yancheng City.
Cell culture
CRC cell line (HT29) and the human umbilical vein endothelial cells (HUVECs) were purchased from American Type Culture Collection (American Type Culture Collection, Manassas, VA, USA). HT29 cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS (Gibco, Carlsbad, CA, USA) under standard conditions (37°C, 5% CO2). And HUVEC were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Carlsbad, CA, USA) medium under the same conditions.
Extraction and identification of exosomes
According to the previous description [22], R-CAFs-derived exosomes (R-CAFs-exo) or S-CAFs-exo were extracted by ultra-centrifugation. Then, transmission electron microscopy (TEM) was performed: exosome suspension (10 μL) was absorbed onto carbon-coated copper grids (200 mesh) for 1 min. After washing with H2O, the samples were negatively stained with 2% uranyl acetate solution and dried naturally. Lastly, the samples were visualized with a Phillips Tecnai transmission electron microscope. The particle size range of exosomes was determined by a nanoparticle tracking analysis (NTA) system. To identify the exosomes, the expressions of exosomal markers CD9, CD81, and TSG101 were monitored by western blot.
Exosome tracking
Exosomes were incubated with PKH67 (Beyotime, Shanghai, China) with light avoidance for 30 min, and co-cultured with HT29 cells. The cells were stained with 4’,6-diamidino-2-phenylindole for 2 h and observed under a fluorescent microscope (Olympus, Tokyo, Japan).
Detection of DDP resistance in colorectal cancer cells
The resistance of CRC cells to DDP was evaluated at half-maximal inhibitory concentration (IC50). R-CAFs were transfected with OE-VEGFA or OE-NC and then the exosomes were isolated, respectively. After that, HT29 cells were cultured with R-CAFs-derived exosomes (OE-NC-Exo, OE-VEGFA-Exo) from the different transfected groups and treated with the DDP of different concentrations. Cell viability was measured by CCK-8 (Sigma, St. Louis, MO, USA). At last, the IC50 of HCT116 cells to DDP was obtained with relative survival curves.
Cell viability analysis
CCK-8 (DOJINDO, Tokyo, Japan) assay was performed for cell viability detection. Briefly, HT29 cells or HUVECs in each group were harvested and seeded in 96-well plates. 10 μL of CCK-8 reagent was added to each well and incubated for 2 h at 37°C. Then, the absorbance value (at 450 nm) was obtained by a microplate reader.
Flow cytometry analysis
Flow cytometry analysis was performed with an Annexin V/FITC kit (Thermo Fisher Scientific, China) to evaluate cell apoptosis. In brief, HT29 cells treated with the different groups were seeded in a six-well plate and digested with trypsin. The apoptotic cells were dual-stained with PI and Annexin V-FITC. Afterward, the stained cells were measured via the BDTM LSRII flow cytometer (BD Biosciences, San Jose, CA, USA).
Migration assay
Two-four-transwell chambers (pore sizes: 8 µm; Costar, Cambridge, MA, USA) were used for the cell migration detection. HUVECs of 5 × 104 with serum-free medium (200 µL) were added to the upper chamber, and the lower chamber contained a blank medium. After culturing for 24 h, cells on the upper membrane surface were scraped off and the invasive cells on the membrane surface were fixed with methanol, then stained with 4% paraformaldehyde and crystal violet (Sigma-Aldrich, USA). At last, HUVECs were counted and photographed under an inverted light microscope (Olympus).
Tube formation assay
Tube formation assay was performed to evaluate the effects of different treatments on angiogenesis of HUVECs. HUVECs of 3 × 104 were plated in 96-well plates coated with 50 µL Matrigel (Corning, Corning, NY, USA) and cultured in DMEM culture medium for 4 h. Tubules were photographed under a microscope and evaluated by Image-Pro Plus software.
qRT-PCR
Total RNA was extracted from cells using the Trizol reagent (Thermo Fisher Scientific). A total of 1 μg mRNA was used to synthesize cDNA using PrimeScript RT Master Mix (TaKaRa, Dalian, China). The quantitative real-time polymerase chain reaction (qRT-PCR) was performed using SYBR Premix ExTaq (TaKaRa, Shiga, Japan). The relative mRNA expression was normalized by internal control (β-actin). Sequences of primers are listed in Table 1.
Table 1.
Primer sequences used for RT-qPCR
| Genes | Primer sequences |
|---|---|
| VEGFA | F: 5′-TGGCTCACTGGCTTGCTCTA-3′ R: 5′-ATCCAACTGCACCGTCACAG-3′ |
| bFGF | F: 5′-GGCACTGAAATGTGCAACAG-3′ R: 5′-TCCAGGTCCAGTTTTTGGTC-3′ |
| TGF-β | F: 5′-CACCGGAGAGCCCTGGATA-3′ R: 5′-TGCCGCACACAGCAGTTC-3′ |
| FIGF | F: 5′-GTATGGACTCTCGCTCAGCAT-3′ R: 5′-AGGCTCTCTTCATTGCAACAG-3′ |
| GAPDH | F: 5′-GGTGGTCTCCTCTGACTTCAA-3′ R: 5′-GTTGCTGTAGCCAAATTCGTTGT-3′ |
Western blotting
The cells were lysed in radio immunoprecipitation assay (RIPA) lysis buffer to produce homogenate. The total protein was obtained by centrifugation (15 000 × g, 15 min) at 4°C. And the concentration was tested by the BCA protein assay kit (Thermo Fisher Scientific, USA). The proteins were then separated using 10% SDS-PAGE and electrophoretically transferred to the polyvinylidene difluoride membranes (Millipore, USA). Then, the membranes were blocked, washed, and incubated with primary antibody: CD63 (1:5000, ab134045; Abcam), CD81 (1:5000, ab109201; Abcam), CD9 (1:1000, ab236630; Abcam, Cambridge, MA, USA), vascular endothelial growth factor A (VEGFA) (1:1000, ab214424; Abcam), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000, ab8245; Abcam), overnight at 4°C. The following day, the membrane was incubated with the goat anti-rabbit horseradish peroxidase (HRP) -conjugated IgG H&L (HRP) secondary antibodies (1:5000, ab6721; Abcam). The relative protein expression of CD163, CD206, Agr1, NOS2, and IL-1β was statistically evaluated by ImageJ software.
Immunofluorescence staining
R-CAFs, S-CAFs, or HT29 cells were assessed by immunofluorescence as indicated [23] using the following antibodies: primary antibody of α-SMA (19245S, 1:200; Cell Signaling Technology, USA), Vimentin (5741S, 1:100; Cell Signaling Technology), or VEGFA (1:500, ab52917; Abcam) at 4°C overnight, and then with goat anti-rabbit IgG (ab150077, 1:2000; Abcam) away from light (45 min, room temperature). Next, the cells were washed and then cultured with DAPI (5 min, room temperature) in the dark. Finally, the immunofluorescence was visualized under a fluorescence microscope (Olympus).
In-vivo studies
A total of 5 × 106 HT29 cells were subcutaneously injected into the flank of nude mice to establish the subcutaneous transplanted tumor model of CRC. Then, we established R-CAFs stably transfected with OE-VEGFA or Sh-VEGFA by lentivirus infection, and then the exosomes from each group were isolated. Two weeks later, the exosomes (5 μg) from different transfected CAFs groups (OE-VEGFA or Sh-VEGFA) were then injected intratumorally twice weekly. The mice were randomly divided into three groups: NC-Exo, OE-VEGFA-Exo, and Sh-VEGFA-Exo (n = 5). And the tumor volume was calculated over the next 5 weeks. The mice were sacrificed on day 35 to obtain xenograft. All animal work was performed according to the Health guidelines, and protocols were approved by the Institutional Animal Care and Use Committee of the Third People’s Hospital.
Immunohistochemistry
Immunohistochemistry staining was performed as previously described [24]. After conventional paraffin embedding and sectioning (5 μM), the tumor tissues were subjected to xylene dewaxing, gradient alcohol hydration, and treated with 3% H2O2. After blocking with 5% BSA, the primary antibodies of Ki67 (1:200, ab16667; Abcam) were added and incubated overnight at 4°C. Then, the sections were incubated with the goat and anti-rabbit IgG for 30 min, and DAB was employed for coloration. After hematoxylin counterstaining, the tissues were dehydrated, transparentized, and mounted for microscopic examination.
Hematoxylin and eosin staining and TUNEL assay
Tumor tissue was fixed in 4% formaldehyde, and then the samples were dehydrated in gradient concentrations of ethanol and embedded in paraffin. After that, the tissues were cut into 5-µm slices and dyed with HE staining. Finally, the stained sections were observed with a light microscope for morphologic changes. In addition, The apoptosis level in tumor tissues was examined with the TUNEL assay (Beyotime) in accordance with the manufacturer’s instruction.
Statistical analyses
All data from at least three independent experiments were presented as the mean ± SD. Statistical analyses were performed by GraphPad Prism 7.0 statistical software. Significant differences between groups were analyzed by the Student’s t-test and assigned as *P < 0.05; **P < 0.01; ***P < 0.001, respectively.
Results
Cancer-associated fibroblasts regulate the proliferation and apoptosis of colorectal cancer cells
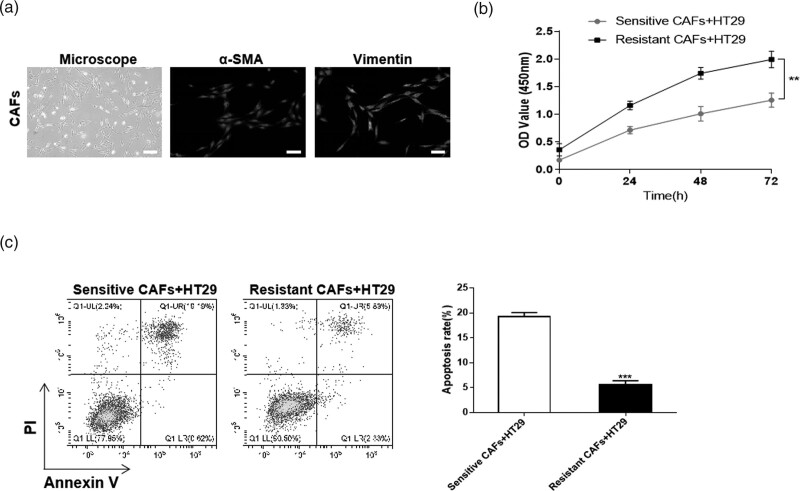
First, CAFs were isolated from CRC tumor tissues of 6 patients with chemoresistance (Resistant CAFs) or chemosensitivity (Sensitive CAFs). Then the markers of CAFs were identified by immunofluorescence, and the results showed a high expression of α-SMA and vimentin in CAFs, which confirmed the successful isolation (Fig. 1a). To prove the effect of CAFs from different groups on CRC cells, Resistant CAFs and Sensitive CAFs were co-cultured with HCT116. According to the CCK-8 assay, compared with the Sensitive CAFs group, the cell viability of Resistant CAFs co-cultured group was obviously increased (Fig. 1c). The results of flow cytometry revealed that the apoptosis level of HT29 cells was apparently decreased in Resistant CAFs co-cultured group (Fig. 1d). These results revealed that CAFs from the chemoresistance patients could promote proliferation and inhibit apoptosis of CRC cells.
Fig. 1.
CAFs regulate the proliferation and apoptosis of CRC cells. The CAFs isolated from CRC tumor tissues with chemoresistance (Resistant CAFs) or chemosensitivity (Sensitive CAFs) were identified and then co-cultured with CRC cell line, HT29 cells. (a) Immunofluorescence was used for the expression of α-SMA and vimentin in CAFs. (b) CCK-8 assay was performed for the cell viability of HT29 cells. (c) The level of apoptosis was detected by flow cytometry. **P < 0.01; ***P < 0.001. CAFs, cancer-associated fibroblasts; CCK-8, Cell Counting Kit-8; CRC, colorectal cancer.
Resistant cancer-associated fibroblasts-derived exosomes affect proliferation, apoptosis, and angiogenesis of colorectal cancer
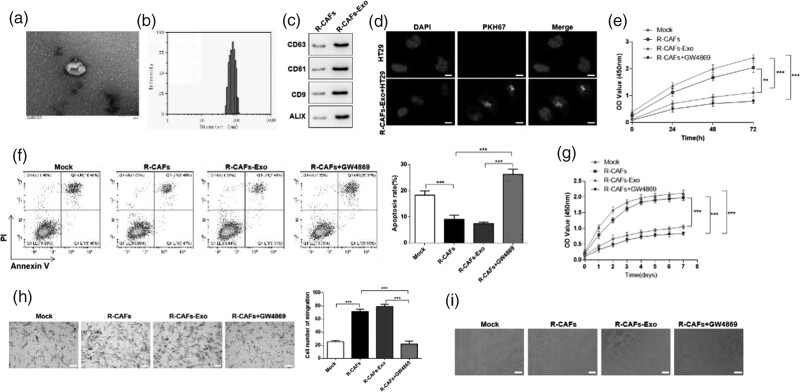
The exosomes secreted by Resistant CAFs (R-CAFs-Exo) were isolated and observed by TEM (Fig. 2a). NTA revealed that the particle size of R-CAFs-Exo ranged from 40 to 120 nm (Fig. 2b). Western blot was applied to measure the expressions of the exosomal marker CD63, CD81, CD9, and ALIX. The results manifested that exosomal markers were positively expressed in R-CAFs-Exo (Fig. 2c). After that, HT29 cells were incubated with PKH67-labeled R-CAFs-Exo, and we found that green fluorescence was localized in the cytoplasm of CRC cells, while no green fluorescence can be observed in PBS treated HT29 cells, indicating exosomes were successfully extracted by HT29 cells (Fig. 2d). From CCK-8 assay and flow cytometry, co-cultured with R-CAFs or R-CAFs-Exo markedly increased viability and suppress apoptosis of HT29 cells, while R-CAFs + GW4869 notably reversed the effects above, restraining the viability and inducing apoptosis of HT29 cells (Fig. 2e and f). There was evidence that CAFs regulate cancer progression by influencing tumor angiogenesis [25]. To further confirm the impact of R-CAFs-Exo on the angiogenesis of CRC, HUVEC were used to co-cultured with R-CAFs, R-CAFs-Exo, or R-CAFs + GW4869. CCK-8 and transwell assay confirmed that R-CAFs or R-CAFs-Exo promoted proliferation and migration of HUVEC, R-CAFs + GW4869 had the opposite effects (Fig. 2g and h). According to the tube formation experiment, co-cultured with R-CAFs or R-CAFs-Exo could notably increase the tube formation ability of HUVEC (Fig. 2i). The above results indicated that the exosomes secreted by Resistant CAFs affected the viability and apoptosis of HT29 cells, as well as angiogenesis.
Fig. 2.
Resistant CAFs-derived exosomes affect proliferation, apoptosis, and angiogenesis of CRC. The exosomes from Resistant CAFs (R-CAFs) was isolated and identified, then R-CAFs, R-CAFs-derived exosomes (R-CAFs-Exo) and R-CAFs + GW4968 (exosome inhibitor) were co-cultured with HT29 cells (e and f) or HUVEC (g–i) for further detection. (a) The isolated exosomes were observed under TEM. (b) The particle size range of exosomes was analyzed by NTA. (c) The expressions of the exosomal markers were measured by western blot. (d) The observation of exosome uptake by HT29 cells after co-culturing. (e) CCK-8 assay and (f) flow cytometry was used to detect the viability and apoptosis of HT29 cells. (g) CCK-8 and (h) transwell assay was used for the proliferation and migration detection. (i) Tube formation experiment was performed in HUVEC. **P < 0.01; ***P < 0.001. CAFs, cancer-associated fibroblasts; CCK-8, Cell Counting Kit-8; CRC, colorectal cancer; HUVECs, human umbilical vein endothelial cells; NTA, nanoparticle tracking analysis.
Resistant cancer-associated fibroblasts-derived exosomes deliver VEGFA
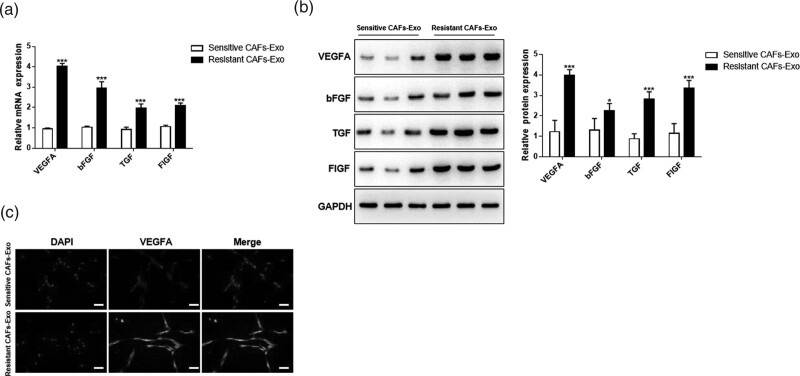
Previous studies have shown that VEGFA, bFGF, TGF-β, and FIGF are closely related to angiogenesis [26–28]. Therefore, in the next study, we detected the expression of these factors in CAF-Exo from the resistant or sensitive group. QRT-PCR and western blot results showed that compared with the Sensitive CAFs-Exo group, all the factors detected including VEGFA, bFGF, TGF-β, and FIGF were up-regulated, and among them, VEGFA existed a most markedly increased expression (Fig. 3a and b). Besides, the high expression of VEGFA in Resistant CAFs-Exo was verified by immunofluorescence assay (Fig. 3c). These results indicated that exosomes derived by Resistant CAFs could secret more VEGFA than Sensitive CAFs-derived exosomes.
Fig. 3.
Resistant CAFs-derived exosomes deliver VEGFA. Resistant CAFs-derived exosomes (Resistant CAFs-Exo) and Sensitive CAFs-derived exosomes (Sensitive CAFs-Exo) were isolated, then the angiogenesis-related factors were detected. (a) qRT-PCR was used to detect the mRNA expression of related factors. (b) Western blot was performed for the detection of related factors. (c) Immunofluorescence assay was used to detect the expression of VEGFA. *P < 0.05; **P < 0.01; ***P < 0.001. CAFs, cancer-associated fibroblasts.
Resistant cancer-associated fibroblasts-derived exosomes deliver VEGFA to promote the progression of colorectal cancer
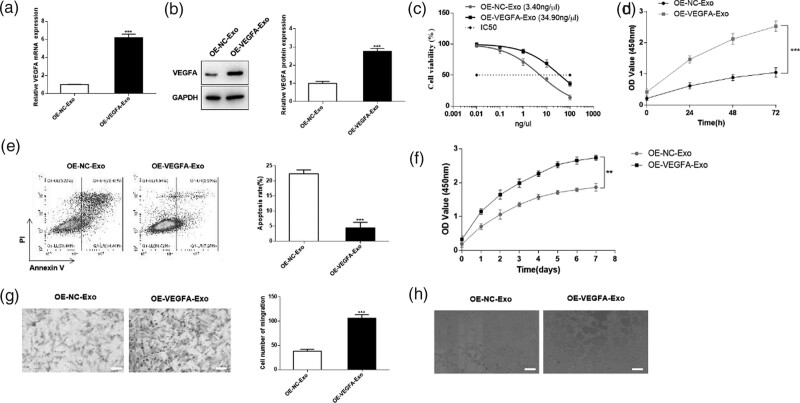
Subsequently, we overexpressed VEGFA in R-CAFs, qRT-PCR, and western blot was performed for the transfection efficiency (Fig. 4a and b). Then, exosomes derived from R-CAFs with different transfected group was co-cultured with CRC cells or HUVEC by transwell device. According to CCK-8 assay, overexpression of VEGFA significantly increased cell viability and IC50 value, indicating that overexpressed VEGFA could improve the resistance of HT29 cells to DDP (Fig. 4c and d). Flow cytometry analysis showed that the cell apoptosis was repressed by OE-VEGFA-Exo group (Fig. 4e). Moreover, overexpression of VEGFA also apparently promoted proliferation, migration, and tube formation of HUVEC (Fig. 4f–h). These results demonstrated that VEGFA secreted from R-CAFs-Exo could regulate the viability, apoptosis, DDP resistance, and angiogenesis of CRC.
Fig. 4.
Resistant CAFs-derived exosomes deliver VEGFA to promote the progression of CRC. R-CAFs were transfected with OE-NC or OE-VEGFA, and then the exosomes were isolated and co-cultured with HT29 cells (c–e) or HUVEC (f–h). (a) QRT-PCR was used to detect the transfection efficiency of CAFs. (b) Western blot was used for the protein expression of VEGFA in CAFs. (c–d) CCK-8 assay was used to detect cell viability and IC50 value of HT29 cells. (e) Flow cytometry was used to detect the apoptosis level of HT29 cells. (f) The proliferation, (g) migration, and (h) tube formation were detected in HUVEC. **P < 0.01; ***P < 0.001. CAFs, cancer-associated fibroblasts; CCK-8, Cell Counting Kit-8; CRC, colorectal cancer; HUVECs, human umbilical vein endothelial cells; NTA, nanoparticle tracking analysis.
Mechanism of Resistant cancer-associated fibroblasts accelerating colorectal cancer progression in vivo
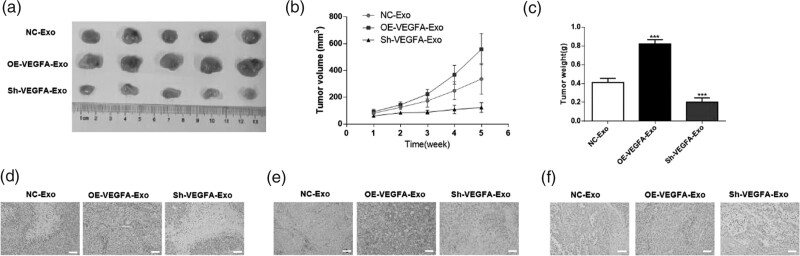
To observe the effect of VEGFA delivered by R-CAFs-Exo on the progression of CRC in vivo, HT29 cells were injected into Nude mice to construct CRC subcutaneous transplanted tumor model. One week later, the exosomes were then injected intratumorally and were isolated from stably transfected R-CAFs (OE-VEGFA or Sh-VEGFA). From the result of subcutaneous tumorigenesis, compared with the NC-Exo group, overexpressing VEGFA could apparently accelerate the tumor growth, including the tumor weight and volume, while it has the opposite effect on tumor growth in Sh-VEGFA-Exo (Fig. 5a–c). Then, H&E staining was then performed to examine the histopathological characteristics of the tumors (Fig. 5d). Furthermore, immunohistochemistry was performed using an antibody against Ki67, which is indicative of cell proliferation, in solid tumors. We observed that Sh-VEGFA-Exo exerts a suppressed effect on cell proliferation (Fig. 5e). Also, TUNEL staining showed VEGFA depletion resulted in elevated apoptosis rate (Fig. 5f). And not surprisingly, overexpression of VEGFA has the opposite effect on the above (Fig. 5d–f). In summary, VEGFA derived by Resistant CAFs aggravated CRC progression in vivo.
Fig. 5.
Mechanism of Resistant CAFs accelerating CRC progression in vivo. To establish the subcutaneous transplanted tumor model of CRC, HT29 cells were subcutaneously injected into the flank of nude mice. Then R-CAFs were stably transfected with OE-VEGFA or Sh-VEGFA by lentivirus infection and the exosomes were isolated from each group. (a) The tumor tissues of nude mice were dissected in each group. (b) Tumor volume and (c) weight were measured in each group. (d) H&E staining was performed in tumor tissues. (e) Immunohistochemistry was used to measure the proliferation level in tumor tissues. (f) Flow cytometry was carried out to detect the apoptosis level in tumor tissues. **P < 0.01; ***P < 0.001. CAFs, cancer-associated fibroblasts; CCK-8, Cell Counting Kit-8; CRC, colorectal cancer; HUVECs, human umbilical vein endothelial cells; H&E, hematoxylin and eosin.
Discussion
CRC is considered to be one of the most frequently occurring cancers with high metastatic potential [29]. Thanks to the increased sensitivity of colonoscopy screening [30] and the improvement of chemotherapy treatment in the past decades [31,32], there is a decline in the incidence and mortality rate of CRC. Unfortunately, most CRC cases present in an advanced metastatic stage at the time of diagnosis, and the 5-year survival rate is only 14% [33]. The basis of the current clinical treatment of CRC includes surgery, targeted therapy, and adjuvant radiotherapy [8]. However, drug resistance during treatment is still one of the deadlocks of low survival rates in CRC patients. Therefore, elucidating the resistance mechanism of chemotherapy is an important goal to improve the survival rate of CRC.
Accumulating evidence indicates that CAFs display the capacity to influence cancer progression, which highlights the potential of CAFs as therapeutic targets [34, 35]. CAFs, as an important component of TME, modulate cancer development through multiple aspects including inducing cancer cell invasion and metastasis, expediting angiogenesis, and promoting immune evasion and chemotherapy resistance [36–38]. Su et al. identified a specific CAF subgroup that promotes tumor formation and chemoresistance in breast and lung cancer through continuous normal fibroblast-kB activation [39]. Recently, CAFs derived exosomes have been identified to play a role in tumor growth and metastasis. For instance, it has been revealed that CAFs-Exo contributed to oral cancer cell proliferation and metastasis [40] and is connected with CRC development [41]. In another study, CAFs-derived exosomes activated colorectal cancer stem cells to promote chemoresistance [42]. Studies indicated that CAFs-Exo confers cisplatin resistance in head and neck cancer through exosomal miR-196a [43].
In this study, we first isolated CAFs from chemoresistant or chemosensitive patients, and they were co-cultured with CRC cells. Cell function experiments showed that CAFs in the chemoresistant group were significantly different from that in the chemosensitive group in regulating the proliferation and apoptosis of CRC. Next, we determined that R-CAF-derived exosomes could support CRC cell growth, suppress apoptosis, induce chemoresistance of DDP, and promote HUVEC angiogenesis.
Angiogenesis has a central role in the development of cancer, from the initiation of carcinogenesis to the advanced stages of cancer [44]. Excessive abnormal angiogenesis is predominantly induced by an imbalance between pro- and anti-angiogenic factors, such as the overproduction of vascular endothelial growth factor (VEGF) [45]. VEGFA, a member of the VEGF family, could induce physiological and pathological angiogenesis by promoting endothelial cell growth, migration, differentiation, and vascular permeability [46,47]. Wang et al. noted that B7-H3 promoted the migration, invasion, and tube formation of HUVECs by up-regulating the expression of VEGFA [48]. In addition, miR-125a-5p and miR-150-5p have been reported to participate in CRC by targeting VEGFA [49,50]. It is worth noting that tumor angiogenesis is mediated not only by tumor cells but also by tumor matrix cells, such as CAF and immune cells [51–53]. However, there are no previous reports on the role of CAF in the transmission of VEGFA in tumor angiogenesis. Our study demonstrated for the first time that CAFs-derived exosomes regulate CRC angiogenesis and progression by transmitting VEGFA.
More importantly, we demonstrated that the mechanism by which R-CAF affects the function of CRC or HUVEC might be achieved by secreting exosomes containing VEGFA. Our results showed that VEGFA content was apparently overexpressed in the exosome. Subsequent rescue experiments confirmed that VEGFA can regulate the viability, apoptosis, DDP resistance, and angiogenesis of colorectal cancer. Finally, combined with animal experiments, we proved that CAF, from chemoresistant patients, derived exosomes delivery VEGFA accelerating CRC progression in vitro and in vivo.
Acknowledgements
We gratefully acknowledge all the researchers and study participants for their contributions.
All protocols of animal experiments were approved by the Medical Ethics Committee of Yancheng Third People’s Hospital, Jiangsu.
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.Y.S. and H.Z. performed experiments; H.J. and H.Z. designed and supervised the project; H.Y. and Y.S. performed experiments; F.Y. analyzed the data; Y.S. and F.W. designed the figures and wrote the article.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Yuanyuan Shi is the first author.
Hua Zhu is the co-first author.
References
- 1.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet 2019; 394:1467–1480. [DOI] [PubMed] [Google Scholar]
- 2.Shi Q, He Y, Zhang X, Li J, Cui G, Zhang X, et al. Two novel long noncoding RNAs – RP11-296E3.2 and LEF1-AS1can – separately serve as diagnostic and prognostic bio-markers of metastasis in colorectal cancer. Med Sci Monit 2019; 25:7042–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang P, Liu Y. A reasonable diet promotes balance of intestinal microbiota: prevention of precolorectal cancer. Biomed Res Int 2019; 2019:3405278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afrăsânie VA, Marinca MV, Alexa-Stratulat T, Gafton B, Păduraru M, Adavidoaiei AM, et al. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer – practical implications for the clinician. Radiol Oncol 2019; 53:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price TJ, Segelov E, Burge M, Haller DG, Ackland SP, Tebbutt NC, et al. Current opinion on optimal treatment for colorectal cancer. Expert Rev Anticancer Ther 2013; 13:597–611. [DOI] [PubMed] [Google Scholar]
- 6.Kamarudin MNA, Sarker MMR, Zhou J-R, Parhar I. Metformin in colorectal cancer: molecular mechanism, preclinical and clinical aspects. J Exp Clin Cancer Res 2019; 38:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Emburgh BO, et al. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol 2014; 8:1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Jeught K, Xu H-C, Li Y-J, Lu X-B, Ji G. Drug resistance and new therapies in colorectal cancer. World J Gastroenterol 2018; 24:3834–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petitprez F, Meylan M, de Reyniès A, Sautès-Fridman C, Fridman WH. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol 2020; 11:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saforo D, Omer L, Smolenkov A, Barve A, Casson L, Boyd N, et al. Primary lung cancer samples cultured under microenvironment-mimetic conditions enrich for mesenchymal stem-like cells that promote metastasis. Sci Rep 2019; 9:4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K, Ehata S, Koinuma D, Morishita Y, Soda M, Mano H, et al. Pancreatic tumor microenvironment confers highly malignant properties on pancreatic cancer cells. Oncogene 2018; 37:2757–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang F, Ning Z, Ma L, Liu W, Shao C, Shu Y, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer 2017; 16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol 2009; 19:43–51. [DOI] [PubMed] [Google Scholar]
- 14.Syn N, Wang L, Sethi G, Thiery J-P, Goh B-C. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol Sci 2016; 37:606–617. [DOI] [PubMed] [Google Scholar]
- 15.Giusti I, Di Francesco M, Poppa G, Esposito L, D'Ascenzo S, Dolo V. Tumor-derived extracellular vesicles activate normal human fibroblasts to a cancer-associated fibroblast-like phenotype, sustaining a pro-tumorigenic microenvironment. Front Oncol 2022; 12:839880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Li Y, Zou L, Zhu Z. Role of exosomes in crosstalk between cancer-associated fibroblasts and cancer cells. Front Oncol 2019; 9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Hao G, Zhuang M, Lv H, Liu C, Su K. MEG3 LncRNA from exosomes released from cancer-associated fibroblasts enhances cisplatin chemoresistance in SCLC via a MiR-15a-5p/CCNE1 axis. Yonsei Med J 2022; 63:229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Z, Jiang T, Cao B, Sun X, Liu JF, CAF-derived exosomes deliver LINC01410 to promote epithelial-mesenchymal transition of esophageal squamous cell carcinoma. 2022; 412:113033. [DOI] [PubMed] [Google Scholar]
- 19.Olejarz W, Kubiak-Tomaszewska C, Chrzanowska A, Lorenc T. Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int J Mol Sci 2020; 21: 5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin H, Yu S, Xie Y, Dai X, Dong M, Sheng C, Hu J. Cancer-associated fibroblasts-derived exosomes upregulate microRNA-135b-5p to promote colorectal cancer cell growth and angiogenesis by inhibiting thioredoxin-interacting protein. Cell Signal 2021; 84:110029. [DOI] [PubMed] [Google Scholar]
- 21.Sharon Y, Alon L, Glanz S, Servais C, Erez N. Isolation of normal and cancer-associated fibroblasts from fresh tissues by Fluorescence Activated Cell Sorting (FACS). J Vis Exp 2013:e4425. doi: 10.3791/4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, Möller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 2015; 4:27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzè E, Dinallo V, Laudisi F, Di Grazia A, Di Fusco D, Colantoni A, et al. Interleukin-34 stimulates gut fibroblasts to produce collagen synthesis. J Crohns Colitis 2020; 14:1436–1445. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, Sang Y, Song X, Zhang D, Wang L, Zhao W, et al. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 2021; 11:3932–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, McAndrews KM, Kalluri R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 2021; 18:792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui H, Zhao J, Zhou L, Wen H, Deng W, Li C, et al. Tanshinone IIA inhibits β-catenin/VEGF-mediated angiogenesis by targeting TGF-β1 in normoxic and HIF-1α in hypoxic microenvironments in human colorectal cancer. Cancer Lett 2017; 403:86–97. [DOI] [PubMed] [Google Scholar]
- 27.Mar AC, Chu C-H, Lee H-J, Chien C-W, Cheng J-J, Yang S-H, et al. Interleukin-1 receptor type 2 acts with c-Fos to enhance the expression of interleukin-6 and vascular endothelial growth factor A in colon cancer cells and induce angiogenesis. J Biol Chem 2015; 290:22212–22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marconcini L, Marchio S, Morbidelli L, Cartocci E, Albini A, Ziche M, et al. c-fos-induced growth factor/vascular endothelial growth factor D induces angiogenesis in vivo and in vitro. Proc Natl Acad Sci USA 1999; 96:9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan C, Li Y, Yu Y, Yu X, Liu H, Zhang Q, et al. Natural product pectolinarigenin exhibits potent anti-metastatic activity in colorectal carcinoma cells in vitro and in vivo. Bioorg Med Chem 2019; 27:115089. [DOI] [PubMed] [Google Scholar]
- 30.Kahi CJ, Rex DK. Current and future trends in colorectal cancer screening. Cancer Metastasis Rev 2004; 23:137–144. [DOI] [PubMed] [Google Scholar]
- 31.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet 2005; 365:153–165. [DOI] [PubMed] [Google Scholar]
- 32.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 2020; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel R, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020; 70:145-–1164. [DOI] [PubMed] [Google Scholar]
- 34.Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol 2015; 25:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017; 171:934–949.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev 2021; 101:147–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bu L, Baba H, Yoshida N, Miyake K, Yasuda T, Uchihara T, et al. Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene 2019; 38:4887–4901. [DOI] [PubMed] [Google Scholar]
- 38.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int J Cancer 2020; 146:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 2018; 172:841–856.e16. [DOI] [PubMed] [Google Scholar]
- 40.Li YY, Tao Y-W, Gao S, Li P, Zheng J-M, Zhang S-E, et al. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine 2018; 36:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera M, Llorens C, Rodríguez M, Herrera A, Ramos R, Gil B, et al. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and cancer-associated fibroblasts in colorectal cancer. Mol Cancer 2018; 17:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One 2015; 10:e0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol 2019; 20:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguilar-Cazares D, Chavez-Dominguez R, Carlos-Reyes A, Lopez-Camarillo C, Hernadez de la Cruz ON, Lopez-Gonzalez JS. Contribution of angiogenesis to inflammation and cancer. Front Oncol 2019; 9:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jászai J, Schmidt MHH. Trends and challenges in tumor anti-angiogenic therapies. Cells 2019; 8:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 1989; 246:1306–1309. [DOI] [PubMed] [Google Scholar]
- 47.Tischer E, Gospodarowicz D, Mitchell R, Silva M, Schilling J, Lau K, et al. Vascular endothelial growth factor: a new member of the platelet-derived growth factor gene family. Biochem Biophys Res Commun 1989; 165:1198–1206. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, Ma Y, Zhan S, Zhang G, Cao L, Zhang X, et al. B7-H3 promotes colorectal cancer angiogenesis through. Cell Death Dis 2020; 11:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Qiu Jianwei, Kang Haifeng, Wang Yaming, Qian Junbo. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res 2018; 10:5839–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X, Xu X, Pan B, Zeng K, Xu M, Liu X, et al. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging (Albany NY) 2018; 10:3421–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nyberg P, Salo T, Kalluri R. Tumor microenvironment and angiogenesis. Front Biosci 2008; 13:6537–6553. [DOI] [PubMed] [Google Scholar]
- 52.Mongiat M, Andreuzzi E, Tarticchio G, Paulitti A. Extracellular matrix, a hard player in angiogenesis. Int J Mol Sci 2016; 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neve A, Cantatore FP, Maruotti N, Corrado A, Ribatti D. Extracellular matrix modulates angiogenesis in physiological and pathological conditions. Biomed Res Int 2014; 2014:756078. [DOI] [PMC free article] [PubMed] [Google Scholar]