Abstract
Introduction:
CYP2D6 contributes to the metabolism of approximately 20–25% of drugs. However, CYP2D6 is highly polymorphic and different alleles can lead to impacts ranging from null to dramatic increases through gene duplication. Moreover, there are commonly used drugs that potently inhibit the CYP2D6 enzyme, thus causing “phenoconversion” which has potential to convert the genotypic normal metabolizer into phenotypic poor metabolizer. Despite growing literature on the clinical implications of non-normal CYP2D6 genotype and phenoconversion on patient-related outcomes, implementation of CYP2D6 pharmacogenetics and phenoconversion to guide prescribing in clinical care is rare. This review focuses on providing the clinical importance of CYP2D6 pharmacogenetics and phenoconversion in precision medicine and summarizes the challenges and approaches to implement CYP2D6 pharmacogenetics and phenoconversion into clinical practice.
Areas covered:
A literature search was performed using PubMed and clinical studies documenting effects of CYP2D6 genotypes and/or CYP2D6 inhibitor medications on pharmacokinetics, pharmacodynamics or treatment outcomes of CYP2D6-metabolized drugs, and studies on implementation challenges and approaches.
Expert opinion:
Considering the extent and impact of genetic polymorphisms of CYP2D6, phenoconversion by the comedications, and contribution of CYP2D6 in drug metabolism, CYP2D6 pharmacogenetics is essential to ensure drug safety and efficacy. Utilization of proper guidelines incorporating both CYP2D6 pharmacogenetics and phenoconversion in clinical care assists in optimizing drug therapy.
Keywords: CYP2D6, phenoconversion, enzyme inhibition, pharmacogenetics, opioids, antidepressants, tamoxifen, personalized medicine, precision medicine, drug-drug interaction, drug-drug-gene interaction
1. Introduction
Recent developments in pharmacogenetics research and clinical implementation have advanced pharmacogenetics-based personalized therapy, with the goal of improving drug safety and efficacy[1,2]. The drug metabolizing enzyme, cytochrome P450 2D6 (CYP2D6) is estimated to contribute to the metabolism of approximately 20–25% of drugs. The gene encoding CYP2D6 is also highly polymorphic and given its importance in drug metabolism and genetic variability, CYP2D6 pharmacogenetics is essential for designing and implementing personalized drug therapy. As a result, pharmacogenetic-based drug therapy guidelines have been developed for at least 50 CYP2D6-metabolized drugs based on the activity of CYP2D6 (Table 1)[3].
Table 1.
Drugs metabolized by CYP2D6 to their inactive or active metabolites
| Type of metabolism | Drug class | Drug names |
|---|---|---|
| Active drugs metabolized to less active or inactive drug by CYP2D6 # | Antidepressants | amitriptyline |
| citalopram | ||
| clomipramine | ||
| desipramine | ||
| doxepin | ||
| duloxetine | ||
| escitalopram | ||
| fluoxetine | ||
| fluvoxamine | ||
| imipramine | ||
| mirtazapine | ||
| nortriptyline | ||
| paroxetine | ||
| sertraline | ||
| trimipramine | ||
| venlafaxine | ||
| Antiphychotics | aripiprazole | |
| brexpiprazole | ||
| clozapine | ||
| flupenthixol | ||
| fluphenazine | ||
| haloperidol | ||
| olanzapine | ||
| pimozide | ||
| quetiapine | ||
| risperidone | ||
| zuclopenthixol | ||
| Beta blocker | atenolol | |
| bisoprolol | ||
| carvedilol | ||
| metoprolol | ||
| Antiarrhythmic | amiodarone | |
| disopyramide | ||
| flecainide | ||
| propafenone | ||
| quinidine | ||
| sotalol | ||
| Opioid | methadone | |
| oxycodone | ||
| ADHD treatment | atomoxetine | |
| methylphenidate | ||
| Gaucher’s disease treatment | eliglustat | |
| Antiemetic | odansetron | |
| tropisetron | ||
| Antihypertensive | clonidine | |
| Inactive drugs metabolized to active drug by CYP2D6 | Opioids | codeine |
| tramadol | ||
| hydrocodone | ||
| Antiestrogens | tamoxifen | |
| Anticancer | gefitinib |
Some metabolites might be similarly active as the active parent drug.
Around 100 alleles of CYP2D6 with different impact on function of the encoded protein have been identified[4]. Variant alleles range from a loss of function and no enzyme activity to gene duplication that can lead to greater activity than normal[5,6]. Based on the activity level of the different alleles, an activity score is given to the CYP2D6 diplotype. The higher the activity score, the higher the enzyme activity of the CYP2D6. Based on the activity score of CYP2D6, a genotype-based phenotype is assigned. These include poor metabolizer (PM), intermediate metabolizer (IM), normal metabolizer (NM), and ultra-rapid metabolizer (UM) [5,6]. However, CYP2D6 is highly susceptible to inhibition by several commonly used medications, and the expected phenotype based on genotype alone could be erroneous if concomitant use of CYP2D6 inhibitors is not considered while predicting the CYP2D6 phenotype[7,8]. Those inhibitor medications can convert CYP2D6 NMs or UMs to IMs or PMs. This phenomenon is known as phenoconversion, which occurs when the individual’s genotype-based prediction of phenotype for drug metabolism does not match with the actual capacity for metabolizing drugs, due to the presence of the CYP2D6-inhibiting comedication[8].
The US Food and Drug Administration (FDA) lists and classifies CYP2D6 inhibitors as strong, moderate, and weak [9]. Strong (e.g., paroxetine, fluoxetine, bupropion) and moderate (e.g., duloxetine, sertraline) inhibitors can decrease the CYP2D6 activity such that genotypically NM individuals are converted phenotypically to PMs and IMs, respectively. Those inhibitor medications are highly prescribed and three of them (fluoxetine, bupropion, and duloxetine ) are in the list of top 50 most commonly prescribed drugs in the United States [10]. Moreover, concomitant use of those drugs cannot be overlooked while predicting the phenotype as they are very common among the patients prescribed CYP2D6 metabolized medications [6,11,12]. As personalized medicine is primarily dependent on the prediction of the actual metabolizing ability of the metabolizing enzyme, failure to consider the comedications while predicting the phenotype can be the Achilles’ heel of personalized medicine as it relates to CYP2D6 [8]. Considering the importance of phenoconversion, Clinical Pharmacogenetics Implementation Consortium (CPIC)[13] guidelines suggest incorporating the effects of CYP2D6 inhibitors while calculating the activity score of CYP2D6[7,14–16].
Integration of CYP2D6 pharmacogenetics and phenoconversion in clinical practice to benefit patients is challenging. CYP2D6 laboratory testing results often include the genotypes and activity scores without proper interpretation and guidelines for prescribing[17]. In addition, information on phenoconversion and interpretation of phenotypes after considering phenoconversion are challenging yet essential for proper implementation. Current electronic health record (EHR) systems also lack support tools to assist health providers with integrating drug interactions with pharmacogenetics to implement personalized medicine in practice.
This review aims to provide information about the clinical importance of CYP2D6 pharmacogenetics and phenoconversion in precision medicine for CYP2D6 metabolized drugs. Challenges and approaches to integrating CYP2D6 pharmacogenetics and phenoconversion into clinical practice are also reviewed.
2. CYP2D6 pharmacogenetics
The CYP2D6 enzyme is encoded by nine exons of the CYP2D6 gene located on chromosome 22(22q13.1). Being involved in metabolizing 20–25% of all the drugs, CYP2D6 is one of the essential enzymes for metabolism of drugs in humans[18–20]. Furthermore, CYP2D6 is involved not only in metabolizing active drug into its inactive metabolite, including antidepressants (e.g., amitriptyline, nortriptyline, venlafaxine, paroxetine), antipsychotics (e.g., aripiprazole, risperidone), cardiovascular drugs (e.g., metoprolol, carvedilol) but also in metabolizing the inactive drug into its active metabolite including opioid analgesics (e.g., codeine, tramadol, hydrocodone), and the anticancer anti-estrogenic agent (tamoxifen)[21–25]. Therefore, the metabolic activity of CYP2D6 can be associated with adverse drug reactions[7,21] or drug ineffectiveness[26–28]. Table 1 lists the 50 drugs that are metabolized by CYP2D6 either to inactive metabolites or active metabolites and have pharmacogenetics-based drug therapy guidelines. In addition to liver, CYP2D6 is also present in the brain. In the brain, CYP2D6 is involved in local drug biotransformation and metabolism of endogenous substrates[29].
CYP2D6 is extremely polymorphic, and more than 100 alleles have been identified[4]. Variant alleles range from a loss of function and no enzyme activity (PM) to gene duplication/multiplication that can lead to increased activity (UM)[7]. An activity score is given to a specific allele of CYP2D6, which correlates with the enzyme function. These allelic activity scores are used to calculate the predicted activity score of the encoded protein[6,30]. Normal functional alleles(e.g. *1, *2, or *35) are considered to have an activity score of 1 whereas reduced functional alleles (e.g. *9, *17, *10 etc.) have an activity score of 0.25 or 0.5 and non-functional alleles (e.g. *3, *4 etc.) have activity score of 0 (Table 2)[5,6]. The diplotype score (sum of the activity scores of the two alleles) is considered the total activity score of CYP2D6 and is used to predict the phenotype of the enzyme. Diplotype scores of 1.25– 2.25 are considered a normal metabolizer (NM), a score of 0.25–1 as intermediate metabolizer (IM), a score of 0 as PM, and a score more than 2.25 as UM. The frequencies of PM, IM, and UM phenotypes based on genotypes range from 5–10%, 5–11%, and 3–29%, respectively, among various racial/ethnic groups[7,31]. Figure 1 shows the differences in rate and extent of metabolism among different CYP2D6 phenotypes.
Table 2.
CYP2D6 activity scores (AS) and phenotype adjustment for CYP2D6 inhibitors
| Alleles and assigned score | Alleles | Activity score | |
| Functional allele | *1, *2 or*35 | 1 | |
| Reduced functional allele | *9, *17, *29 or*41 | 0.5 | |
| *10 | 0.25 | ||
| Nonfunctional allele | *3 through*8, *11 or*15 | 0 | |
| AS considering only genotype: Genotype-based Activity Score=Sum of the scores of the alleles | |||
| AS considering both genotype and CYP2D6 inhibitor: CYP2D6 activity score= Inhibitor factor (IF) x genotype-based AS; IF= 0, 0.5 and 1 for strong, moderate and no CYP2D6 inhibitor | |||
| Phenotype based on genotype and inhibitors | Phenotype | CYP2D6 activity score | |
| Ultra-rapid metabolizers (UM) | >2.25 | ||
| Normal Metabolizers (NM) | 1.25–2.250 | ||
| Intermediate metabolizers (IM) | 0.25–1 | ||
| Poor Metabolizers (PM) | 0 | ||
Figure 1. Representative differences in rate and extent of metabolism among different CYP2D6 phenotypes.
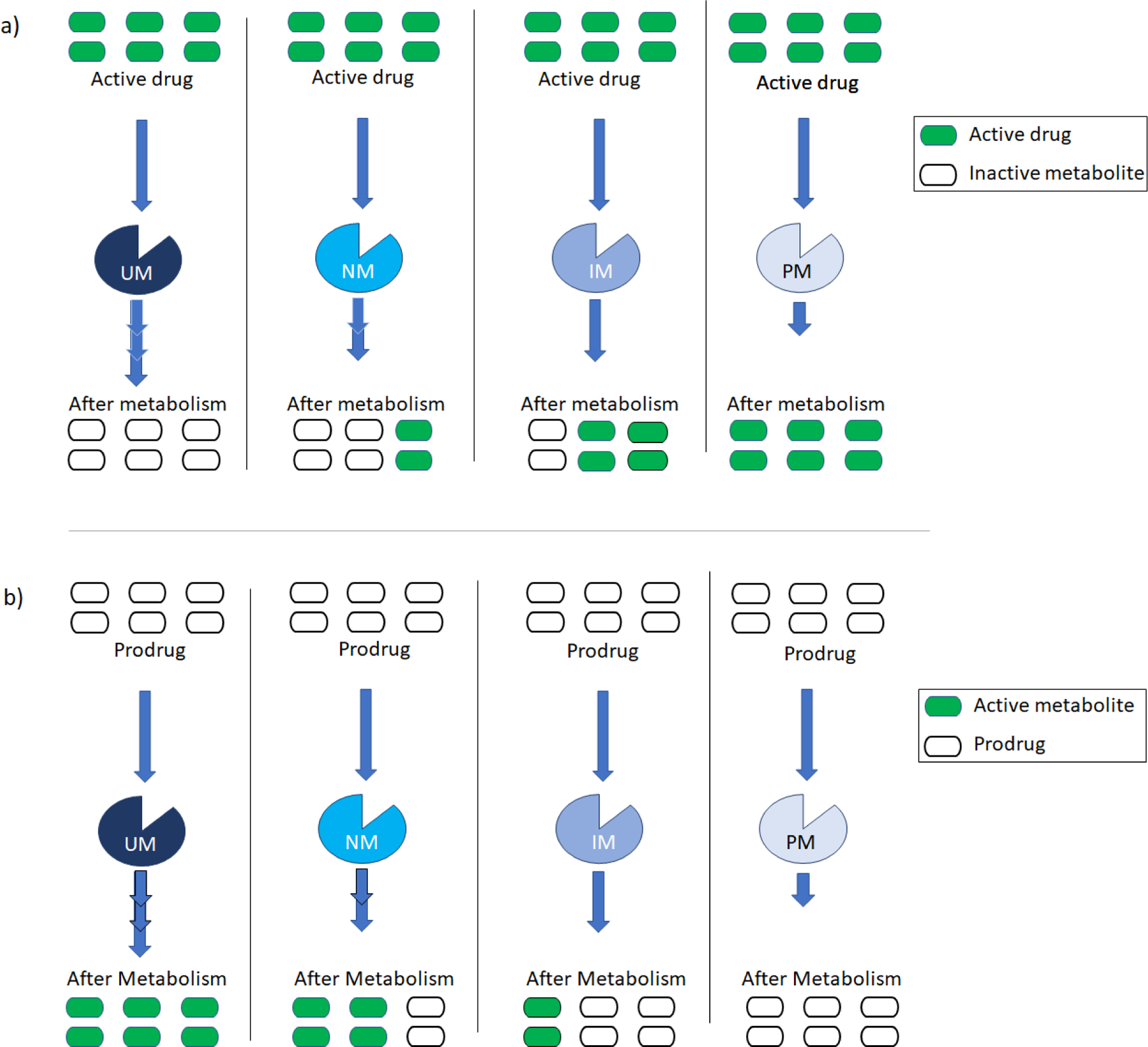
Figure 1a shows metabolism of active drug (green bars) to inactive metabolite (open bars) and Figure 1b shows the metabolism of a prodrug (inactive) (open bars) to active metabolite (green bars) in absence of CYP2D6 inhibitors. NM can metabolize the drugs in an expected standard rate (represented by two (>>) arrows) and extent (4 out of 6 units in a given time) to provide the pharmacological activity. UM, being more active and rapid, can metabolize the drugs at a higher rate (represented by three (>>>) arrows) and extent than NM (6 out of six units in a given time). IM is slower (represented by one (>) arrow) and can metabolize less(2 out of 6 units in each given time) than NM. PM has no metabolic activity and cannot metabolize a drug.
UM- ultrarapid metabolizer; NM- normal metabolizer; IM- intermediate metabolizer; PM- poor metabolizer.
Here we will discuss the impact of different CYP2D6 genotypes on metabolism and clinical outcomes of the most commonly studied CYP2D6 substrates including CYP2D6 metabolized opioids, antidepressants, antipsychotics, tamoxifen, atomoxetine, ondansetron, tropisetron, and metoprolol.
2.1. CYP2D6 pharmacogenetics and opioids
CYP2D6-metabolized opioids tramadol, codeine, and hydrocodone account for the majority of opioid prescriptions written in the U.S., accounting for more than 50 million opioid prescriptions dispensed in 2020 [32,33]. These opioids are widely used to treat moderate to severe pain[34]. CYP2D6 bioactivates codeine, tramadol, and hydrocodone into active metabolites, and the parent compounds have little to no analgesic activity compared to the active metabolites[7,35,36]. Thus, the active metabolites are entirely or largely responsible for the analgesic effects these specific opioids provide, and therefore the extent of analgesic activity of these opioids largely depends on the activity of CYP2D6[7,36]. Generally, it is expected that PMs and IMs would have reduced generation of the active metabolite and thus reduced pain control whereas UMs would be expected to generate excessive amounts of active metabolites and thus risk of toxicity.
In clinical trials, CYP2D6 PM and IM phenotypes were found to be associated with lower formation of morphine from codeine when compared to NM phenotype[37–42], and thus were associated with poor pain control after codeine administration[43]. In contrast CYP2D6 gene duplication/multiplication (UMs) causes higher plasma concentrations of morphine[44–46], which can lead to oversedation, respiratory depression, and even death in pediatric population. This is the reason why U.S. Food and Drug Administration (FDA) contraindicated the use of codeine to treat pain and cough in patients under the age of 12 in April, 2017[47].
In the case of tramadol, the effects of different CYP2D6 phenotypes are similar to that of codeine. Lower biotransformation of tramadol to active form O-desmethyl tramadol was observed among the CYP2D6 PMs and IMs when compared to NMs[48–54]. Tramadol’s effectiveness in pain control was worse among CYP2D6 PMs compared to NMs or IMs[55–59]. Increased frequency of adverse events, including respiratory depression, were observed in UMs compared to IMs or PMs [54,60,61] and the FDA has also contraindicated use of tramadol in children under 12, as of April, 2017 [47].
CYP2D6 PMs have also been shown to produce a decreased amount of hydromorphone from hydrocodone compared to NMs[62,63], whereas UMs are at increased risk of adverse effects[64]. The data from these three drugs clearly show the importance of considering CYP2D6 pharmacogenetics in the clinical setting while prescribing opioids, where such an approach has the potential to improve efficacy and decrease toxicity, in both cases typically through selection of a non-CYP2D6 metabolized opioid or non-opioid as alternative therapy[7].
Considering the evidence, CPIC provides recommendations for dosing of codeine, tramadol and hydrocodone based on CYP2D6 genotypes (Table 3).
Table 3.
CPIC dosing recommendations for different drugs based on CYP2D6 phenotype
| Drug class | Medications | CYP2D6 phenotypes | Implications | Recommendations | Classification of recommendation | Reference |
|---|---|---|---|---|---|---|
| Tricyclic antidepressants | Amitriptyline and nortriptyline | UM | Increased metabolism to less active compounds increasing the probability to lack of efficacy. | Avoid use of amitriptyline and nortriptyline. Consider alternative drug not metabolized by CYP2D6. | Strong | Hicks et. al., 2017 [16] |
| NM | Expected metabolism to less active compounds | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced metabolism to less active compounds increasing risk of adverse effects | Consider a 25% reduction of recommended starting dose | Moderate | |||
| PM | Greatly reduced metabolism to less active compounds increasing risk of adverse effects | Avoid use of amitriptyline and nortriptyline. Consider alternative drug not metabolized by CYP2D6. | Strong | |||
| Clomipramine, desipramine, doxepin, imipramine, and trimipramine | UM | Increased metabolism to less active compounds increasing the probability for lack of efficacy. | Avoid use of the drugs. Consider alternative drug not metabolized by CYP2D6. | Optional | ||
| NM | Expected metabolism to less active compounds | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced metabolism to less active compounds increasing risk of adverse effects | Consider a 25% reduction of recommended starting dose. | Optional | |||
| PM | Greatly reduced metabolism to less active compounds increasing risk of adverse effects | Avoid use of the drugs. Consider alternative drug not metabolized by CYP2D6. | Optional | |||
| Selective serotonin reuptake inhibitors | Paroxetine | UM | Increased metabolism to less active compounds increasing the probability for lack of efficacy. | Avoid paroxetine. Consider alternative drug not predominantly metabolized by CYP2D6 | Strong | Hicks et. al., 2015 [21] |
| NM | Expected metabolism to less active compounds | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced metabolism to less active compounds increasing risk of adverse effects | Initiate therapy with recommended starting dose. | Moderate | |||
| PM | Greatly reduced metabolism to less active compounds increasing risk of adverse effects | Avoid paroxetine. Consider alternative drug not predominantly metabolized by CYP2D6 | Optional | |||
| Fluvoxamine | UM | No data available for CYP2D6 ultrarapid metabolizers | No recommendation due to lack of evidence | Optional | ||
| NM | Expected metabolism to less active compounds | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced metabolism to less active compounds increasing risk of adverse effects | Initiate therapy with recommended starting dose. | Moderate | |||
| PM | Greatly reduced metabolism to less active compounds increasing risk of adverse effects | Consider a 25–50% reduction of recommended starting dose and titrate to response. Or, Consider alternative drug not predominantly metabolized by CYP2D6 | Optional | |||
| Antiemetic | Ondansetron and tropisetron | UM | Increased metabolism to less active compounds increasing the probability to lack of efficacy. | Avoid use of the drugs. Consider alternative drug not metabolized by CYP2D6. | Moderate | Bell et. al., 2017 [131] |
| NM | Expected metabolism to less active compounds | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Very limited data available | Initiate therapy with recommended starting dose. | No recommendation | |||
| PM | Very limited data available | Initiate therapy with recommended starting dose. | No recommendation | |||
| ADHD treatment | Atomoxetine | UM | Very limited data available | Initiate with a dose of 0.5mg/kg/day in children or 40 mg/day in adults and increase to 1.2mg/kg/day in children or 80 mg/ day in adults after 3 days. If no clinical response, consider titration to reach 400 ng/mL peak plasma concentration. Details here. | Moderate | Brown et. al., 2019 [14] |
| NM | Expected metabolism to less active compounds | Initiate with a dose of 0.5mg/kg/day in children or 40 mg/day in adults and increase to 1.2mg/kg/day in children or 80 mg/ day in adults after 3 days. If no clinical response consider titration to reach 400 ng/mL peak plasma concentration. Details here. If unacceptable side effects are present at any time, consider a reduction in dose. Details here. | Moderate | |||
| IM | Reduced metabolism to less active compounds increasing risk of discontinuation | Initiate with a dose of 0.5mg/kg/day in children or 40 mg/day in adults. If no clinical response, consider titration to reach 400 ng/mL peak plasma concentration. Details here. If unacceptable side effects are present at any time, consider a reduction in dose. Details here. | Moderate | |||
| PM | Greatly reduced metabolism to less active compounds increasing risk of adverse effects but may result in greater improvement of ADHD. | Initiate with a dose of 0.5mg/kg/day in children or 40 mg/day in adults. If no clinical response, consider titration to reach 400 ng/mL peak plasma concentration. Details here. If unacceptable side effects are present at any time, consider a reduction in dose. Details here. | Moderate | |||
| Opioids | Codeine | UM | Increase formation of morphine leading to higher risk of toxicity | Avoid use of codeine. Consider use of non-tramadol opioid or other analgesics | Strong | Crews et. al., 2021 [7] |
| NM | Expected metabolism to morphine | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced formation of morphine | Initiate therapy with recommended starting dose. If no response consider use of non-tramadol opioid or other analgesics | Moderate | |||
| PM | Greatly reduced formation of morphine leading to diminished analgesia | Avoid use of codeine. Consider use of non-tramadol opioid or other analgesics | Strong | |||
| Tramadol | UM | Increase formation of o-desmethyltramadol leading to higher risk of toxicity | Avoid use of tramadol. Consider use of non-codeine opioid or other analgesics | Strong | ||
| NM | Expected metabolism to o-desmethyltramadol | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced formation of o-desmethyltramadol | Initiate therapy with recommended starting dose. If no response consider use of non-codeine opioid or other analgesics | Optional | |||
| PM | Greatly reduced formation of o-desmethyltramadol leading to diminished analgesia | Avoid use of tramadol. Consider use of non-codeine opioid or other analgesics | Strong | |||
| Hydrocodone | UM | Very limited data available | No recommendation due to lack of evidence | No recommendation | ||
| NM | Expected metabolism to hydromorphone | Initiate therapy with recommended starting dose. | Strong | |||
| IM | Very limited data available | Initiate therapy with recommended starting dose. If no response consider use of non-codeine or non-tramadol opioid or other analgesics | Optional | |||
| PM | Reduced formation of hydromorphone, but there is insufficient evidence to determine if these effects on pharmacokinetics translate into decreased analgesia or side effects | Initiate therapy with recommended starting dose. If no response consider use of non-codeine or non-tramadol opioid or other analgesics | Optional | |||
| Antiestrogens | Tamoxifen | UM | Expected metabolism to endoxifen | Avoid moderate and strong inhibitors. Initiate therapy with recommended starting dose. | Strong | Goetz et. al., 2018 [15] |
| NM | Expected metabolism to endoxifen | Avoid moderate and strong inhibitors. Initiate therapy with recommended starting dose. | Strong | |||
| IM | Reduced formation of endoxifen. May increase the risk of breast cancer recurrence, event-free and recurrence-free survival | Avoid moderate and strong inhibitors. Consider hormonal therapy such as an aromatase inhibitor with/without ovarian function suppression. If aromatase inhibitor use is contraindicated, consideration should be given to use a higher but FDA approved tamoxifen dose (40 mg/day). Details here. | Moderate | |||
| PM | Reduced formation of endoxifen. May increase the risk of breast cancer recurrence, event-free and recurrence-free survival | Recommend alternative hormonal therapy such as an aromatase inhibitor with/without ovarian function suppression. If aromatase inhibitor use is contraindicated, consideration should be given to use a higher but FDA approved tamoxifen dose (40 mg/day). Details here. | Strong |
2.2. CYP2D6 pharmacogenetics and antidepressants
CYP2D6 is involved in metabolizing several antidepressants, including tricyclic antidepressants (e.g., amitriptyline, nortriptyline), selective serotonin reuptake inhibitors (SSRIs) (e.g., paroxetine, fluoxetine), and serotonin and norepinephrine reuptake inhibitors (SNRIs)(e.g., venlafaxine). CYP2D6 metabolizes the many antidepressants to their less or inactive metabolites ( and in some cases the parent and metabolite are active, e.g. with amitripline and its metabolism to nortriptyline). Active antidepressant drugs are expected to have higher concentrations in PMs and IMs and thus PMs and IMs are at risk of concentration related adverse effects. Conversely UMs are expected to have low concentrations of active drug, thus increasing the risk of inefficacy.
Multiple studies have shown decreased metabolism of amitriptyline[65–67], and nortriptyline[68] among CYP2D6 PMs and IMs when compared to NMs, leading to higher plasma concentrations of the active drugs. Increased plasma concentrations of were found to be associated with several dose-dependent severe adverse effects including cardiotoxicity among PMs, which required discontinuation of the treatment or dose reduction[66,68]. Conversely, amitriptyline discontinuation was recorded more among CYP2D6 UMs due to decreased response to the drug[69]. CYP2D6 also metabolizes paroxetine and fluoxetine to pharmacologically less active or inactive metabolites[70,71]. The most active alleles had the lowest paroxetine concentrations in plasma[72–74]. As CYP2D6 UMs have very high paroxetine clearance, studies found lower plasma concentration of paroxetine at a steady state in CYP2D6 UMs[75–77] that resulted in decreased response to paroxetine[75]. In contrast, CYP2D6 PMs had significantly higher plasma paroxetine concentrations at a steady state[75,78], contributing to adverse drug effects[79]. For fluvoxamine, lower plasma clearance and longer half-life were observed among CYP2D6 PMs versus NMs[80,81]. Increased exposure to nortriptyline[82–84] and venlafaxine[83,85,86] was also observed among CYP2D6 PMs and IMs. Considering the clinical evidence, CPIC recommends using CYP2D6 pharmacogenetics for prescribing with SSRIs (paroxetine, fluoxetine) and tricyclic antidepressants (e.g., amitriptyline, nortriptyline) (Table 3)[16,21].
2.3. CYP2D6 pharmacogenetics and antipsychotics
Several antipsychotics (e.g., aripiprazole, risperidone) are metabolized to their inactive metabolites by CYP2D6. In CYP2D6 PMs and IMs, due to lower catalytic activity, plasma concentrations of the active drugs are expected to be higher than in NMs, causing concentration-related adverse effects. On the other hand, in UMs, due to higher enzymatic activity, active drug concentration is anticipated to be lower and may result in reduced efficacy. As CYP2D6 is also present in the brain, different phenotypes of CYP2D6 determine drug response not only via drug biotransformation in the liver, but also via drug and endogenous substrate metabolism in the brain[29].
Several studies identified that CYP2D6 PMs and IMs had poor aripiprazole catabolizing capacity when compared to NMs, resulting in increased plasma aripiprazole concentrations among PMs and IMs[87–94]. PMs and IMs also exhibited higher Cmax and T1/2 [88,95],and the mean elimination half-life of aripiprazole was shown to increase from 75 hours to 146 hours among PMs [89,96]. A recent meta-analysis including 12 studies and 1038 patients found that CYP2D6 PMs and IMs had significantly increased exposure to aripiprazole (1.48 ratio of means in PM+IM vs. NM)[83]. Increased concentration of aripiprazole may lead to several adverse drug effects, such as somnolence, headache, insomnia, dizziness, restlessness, palpitations, among others [97].
Among PMs and IMs, risperidone plasma concentration was significantly higher than NMs[89,94,98–103]. In one study, CYP2D6 PMs and IMs also had 1.7-fold increased exposure to risperidone compared to NMs[83]. PMs for CYP2D6 showed a higher risperidone Cmax, AUC and T1/2 and a lower clearance[98,103]. Higher concentrations of risperidone among PMs and IMs may cause several adverse effects, such as somnolence, headache, and dizziness [98,104]. One study on 76 schizophrenic patients reported that CYP2D6 PM phenotype was associated with significant improvement[105] suggesting that the higher drug concentrations led to improved efficacy. Further, previous studies have reported greater side effects and treatment discontinuations in CYP2D6 PMs compared to NMs [106,107]. Plasma concentrations of haloperidol were also found to be significantly higher among PMs and IMs when compared to NMs[83,94,99].
2.4. CYP2D6 pharmacogenetics and tamoxifen
CYP2D6 catalyzes the anticancer antiestrogenic drug tamoxifen to its active form, endoxifen. CYP2D6 phenotypes are found to be associated with variations in plasma concentration of endoxifen [108–112]. CYP2D6 PMs and IMs have no to lower concentration of endoxifen when compared to NMs or UMs[15] and CYP2D6 phenotypes are essential predictors of response to tamoxifen therapy[108]. Several clinical studies reported that recurrence of breast cancer was observed more commonly among CYP2D6 IMs or PMs than among NMs[70,113–116]. Other clinical trial data suggest worse event-free survival among CYP2D6 PMs versus NMs treated with tamoxifen [116–118]. Considering the importance CYP2D6 genetics, CPIC recommends using CYP2D6 pharmacogenetics guidelines while prescribing tamoxifen(briefly described in Table 3)[15].
2.5. CYP2D6 pharmacogenetics and atomoxetine
A selective norepinephrine reuptake inhibitor, atomoxetine is widely used to treat ADHD among children and adults. CYP2D6 is involved in the metabolism of atomoxetine to an inactive metabolite. Decreased metabolism in CYP2D6 PMs causes increased plasma concentration of atomoxetine [119–121], which may cause several moderate to severe adverse drug effects, including an increase in pulse and blood pressure or decrease in body weight [119,120,122]. Higher plasma concentrations of atomoxetine in PMs is also associated with improvement in ADHD among them[119,120,123]. Based on the evidence, CPIC provides guidelines for dosing of atomoxetine based on the genotypes of CYP2D6(Table 3) [14].
2.6. CYP2D6 pharmacogenetics and ondansetron and tropisetron
Ondansetron and tropisetron are selective 5-HT3 receptor antagonists and are metabolized by CYP2D6 to inactive metabolites[124–126]. CYP2D6 PMs had higher plasma concentrations of tropisetron when compared to NMs[127]. CYP2D6 UMs is associated with decreased AUC of ondansetron[128] and tropisetron[129] and thus decreased response to those drugs[127,130] Considering the importance of CYP2D6 genotypes on those drugs, CPIC provides dosing recommendations based on CYP2D6 genotypes (Table 3) [131].
2.7. CYP2D6 pharmacogenetics and metoprolol
Metoprolol, a beta blocker, is widely used to treat hypertension, angina, and heart failure. Metoprolol is also metabolized by CYP2D6[132,133]. Individuals with reduced or CYP2D6 PM phenotype may have plasma metoprolol concentration up to almost five times higher than NMs, which may increase the risk of various side effects[134–137]. However, some studies clinical study reported that neither antihypertensive response rate, blood pressure changes nor adverse event rates significantly differed by activity scores or among different CYP2D6 phenotypes. This is likely the result of the dosing strategy of beta-blockers as doses are selected based on the titration to a heart rate response[138–140].
3. CYP2D6 phenoconversion
Phenoconversion occurs when the individual’s genotype-based prediction of phenotype for drug metabolism does not match with the true capacity of metabolizing the drugs caused by some extrinsic nongenetic factors[8]. In other words, CYP2D6 inhibitors can cause the conversion of CYP2D6 genotypic ultra or normal metabolizers (UMs or NMs) into phenotypic intermediate or poor metabolizers (IMs or PMs) and this phenomenon is called phenoconversion. In the context of this review, phenoconversion is the result of drug-drug interactions, whose implications vary by genotype, also known as drug-drug-gene interactions.
The FDA publishes a list of strong, moderate or weak CYP2D6 inhibitors [9], and of them, strong and moderate inhibitors may cause a significant reduction in CYP2D6-mediated metabolism ranging from half (by moderate inhibitors) to null (by strong inhibitors), converting the phenotype from UM or NM to IM or PM[7,141]. Figure 2 shows the phenoconversion in the presence of strong (SI) or moderate (MI) inhibitors and the extent of metabolism of drugs by CYP2D6 phenotypes. Although multiple sources for CYP2D6 inhibitors are available, we contend that the FDA list is the most appropriate for considering clinical implications [17]. This is because FDA categorizes the inhibitors based on strong, moderate, and weak status, defined by the increase in AUC of sensitive index substrates. Specifically, a strong inhibitor increases plasma concentration (i.e. AUC) ≥5-fold, whereas moderate and weak inhibitors increase AUC by ≥2 to <5-fold, and ≥1.25 to <2-fold, respectively [9]. Knowledge of the strength of the inhibitor (moderate or strong) is vital to understanding the degree of phenoconversion, which along with genotype is used in defining the phenotype [142,143]. There is consensus among experts that weak inhibitors are not clinically important to consider because they minimally impact the area under the curve [17]. Table 4 lists the FDA strong and moderate inhibitors that can significantly inhibit CYP2D6. Consideration of phenoconversion cannot be ignored as concomitant use of the CYP2D6 inhibitors is very common among the patients prescribed CYP2D6 metabolized medications. Specifically, data indicate that up to 20– 70% of the patients who are on CYP2D6 metabolized drugs are at risk of CYP2D6 phenoconversion by concomitant medications[6,11,12].
Figure 2. Phenoconversion of CYP2D6 in presence of strong (SI) or moderate (MI) inhibitors and metabolism of drugs by CYP2D6 phenotypes.
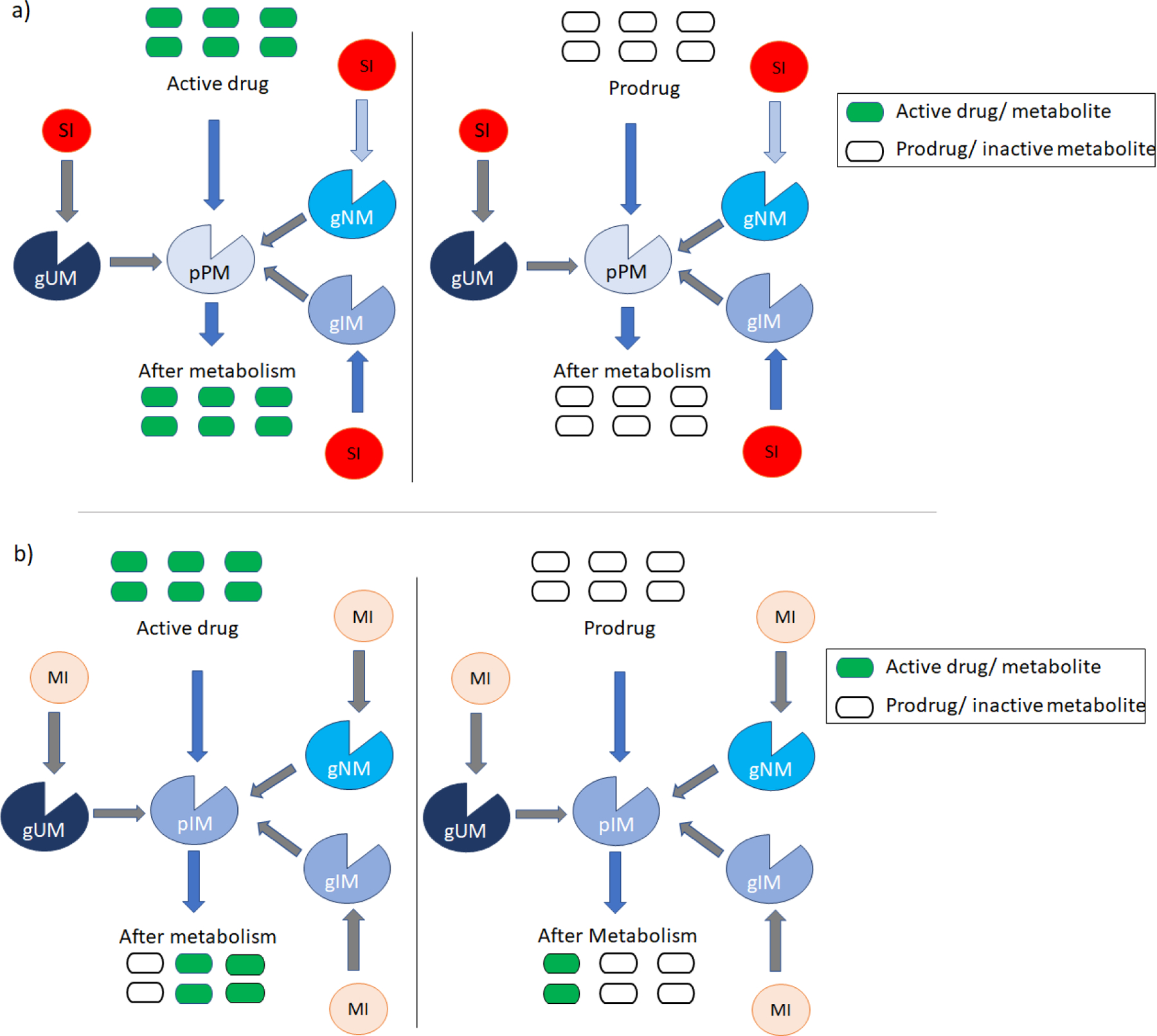
Figure 2a shows the phenoconversion in presence of SI and metabolism of active drug (green bars) to inactive metabolite (open bars) and prodrug (open bars) to active metabolite (green bars) in phenoconverted CYP2D6 phenotypic PMs(pPM). SI can decrease the activity of the CYP2D6 enzymes to null and phenoconvert the gUM, gNM or gIM to pPM. Thus, the pPM cannot metabolize the drug.
Figure 2b shows the phenoconversion in presence of MI and metabolism of active drug (green bars) to inactive metabolite (open bars) and prodrug (open bars) to active metabolite (green bars) in phenoconverted CYP2D6 phenotypic IMs(pIM). MI can decrease the activity of the CYP2D6 enzymes to nearly half and can phenoconvert the gUM, gNM or gIM to pIM. Thus, the pIM metabolizes the drug very slowly and to a lower extent (2 out of 6 units).
Note: For gUMs, in presence of three or more normal functioning alleles MI can phenoconvert them to NM or UM (not shown in the figure).
SI- strong inhibitors; MI- moderate inhibitor; gUM- genotypic ultrarapid metabolizer; gNM- genotypic normal metabolizer; gIM- genotypic intermediate metabolize; pPM- phenotypic PM; pIM- phenotypic IM; PM- poor metabolizer; IM- intermediate metabolizer.
Table 4.
Strong and moderate CYP2D6 inhibitors according to US FDA[9]
| Type of inhibitors | CYP2D6 inhibitors |
|---|---|
| Strong inhibitors | bupropion, fluoxetine, paroxetine, terbinafine, quinidine |
| Moderate inhibitors | abiraterone, cinacalcet, mirabegron, duloxetine, lorcaserin, rolapitant |
The impact of CYP2D6 inhibitors in combination with CYP2D6 genotype has gotten increasing attention in recent years. One study suggested that patients co-administered another CYP2D6-metabolized drug were 9.5 times more likely to have genotype-phenotype discordance[18]. Phenoconversion to IMs or PMs can affect either the metabolism of the active drug to its inactive metabolite or the metabolism of the inactive drug to its active metabolite[144]. In general, whether the patient is a PM or IM due to genetics, CYP2D6 inhibitor or both, the impact on drug metabolism and thus active drug concentration is the same. For example, coadministration of paroxetine (a CYP2D6 inhibitor) with tramadol (an inactive drug metabolized to an active metabolite by CYP2D6) may result in a lack of proper pain control in a CYP2D6 *1/*1 (NM) patient. This is because paroxetine phenoconverts the NM to a PM resulting in decreased formation of morphine (the active metabolite of codeine) that provides the analgesic effect. Similarly, coadministration of bupropion (a CYP2D6 inhibitor) with aripiprazole (an active drug metabolized to an inactive metabolite by CYP2D6) may lead to excessively high aripiprazole concentrations, and potential for discontinuation of the treatment due to adverse drug effects (ADEs).
Considering the importance of phenoconversion, CPIC guidelines for CYP2D6-metabolized drug therapy suggest considering the concomitant use of CYP2D6 inhibitors while calculating the CYP2D6 activity score[7,14–16]. It is recommended that the genotype-based activity score of CYP2D6 is adjusted based on the use of inhibitors, with the specific adjustment for phenoconversion depending on the strength of the inhibitor (moderate or strong)[142,143]. If the individual is taking one of the strong inhibitors concomitantly, then the genotype-based activity score is multiplied by 0, and if the individual is taking moderate inhibitors concomitantly, then the genotype-based activity score is multiplied by 0.5[7]. Details of the activity score of the alleles, calculation of genotype-based score, and adjustment of score based on the use of CYP2D6 inhibitors are summarized in Table 2.
The impact of inhibition by a CYP2D6 inhibitor may differ according to genotype. A CYP2D6 PM by genotype will be unaffected by the concomitant use of an inhibitor since, in PMs, there is no active enzyme to be inhibited [143,145,146]. On the other hand, CYP2D6 UMs, NMs and IMs may have a significant reduction in their metabolic activity by the same inhibitor[8,143,145,146]. Some studies suggest CYP2D6 IMs are likely to be more susceptible to clinically important phenoconversion due to their already compromised activity by the genotype [8,146].
Consideration of CYP2D6 phenoconversion is an integral part of CYP2D6 pharmacogenetics-based personalized therapy. CYP2D6 genetics alone may not result in a better prediction of drug effectiveness or drug safety [8,147–152]. Furthermore, phenoconversion could clarify many of the inconsistent pharmacogenetic findings and might explain the failures of replication of the published results. [143,150,153,154]. Some studies reported inconsistent findings in pharmacogenetic studies on tamoxifen. [155,156]. Kiyotani et al. [150] reported that this discrepancies in the results may happen due to the concomitant medications the patients received. They observed that recurrence free survival was not associated with CYP2D6 genotypes if breast cancer patients who received other medications in addition to tamoxifen were included in the cohort. Moreover, positive association was found in the subgroup analysis where patients who were only on tamoxifen were included.
3.1. CYP2D6 phenoconversion in various clinical studies
Several clinical studies have been conducted to understand the importance of phenoconversion in the clinical setting. These studies mainly focus on the extent of phenoconversion (drug-drug or drug-drug-gene interaction) and impact on pharmacokinetics and clinical outcome of different medications. Here we will discuss several different clinical studies that addressed the clinical relevance of phenoconversion.
While prescribing opioids, consideration of CYP2D6 strong and moderate inhibitors is very important, as 20 – 30% of patients treated for pain are also prescribed a CYP2D6 inhibitor [6,12] which can lead to increased incidences of phenoconversion among opioid users. In studies of acute[157] and chronic pain[158], the percentage classified as PMs went up from 6% to 17% and 5.3% to 19.2%, respectively, when CYP2D6 inhibitors were considered. In one of the first studies on phenoconversion was published in 1991, in a randomized cross-over study, Dayer et al. demonstrated that quinidine, a strong CYP2D6 inhibitor can convert the genotypic NM to a phenotypic PM, and if quinidine was administered with codeine, there was very little to no morphine (active metabolite of codeine) in the genotypic NM[159,160]. Consequently poor response to opioid was observed in genotypic NMs because of their phenoconversion to phenotypic PMs by a CYP2D6 inhibitor [160]. Several other studies compared pain control, and morphine milligram equivalents to manage pain among CYP2D6 inhibitor users vs non-users. Individuals in the CYP2D6 inhibitor user group required significantly more breakthrough morphine milligram equivalents per day compared with patients in CYP2D6 inhibitor non-user group (geometric mean ± SD 18.2 ± 6.3 vs 5.7 ± 6.7 mg morphine milligram equivalents, p<0.001)[141]. Another study compared association between pain control vs activity score based on genotype alone (DGI-drug-gene interaction model), and inhibitor and genotype both (DDI + DGI- drug-drug interaction and drug-gene interaction model). In the DDI + DGI model a significant association between activity score and uncontrolled pain was observed whereas in the DGI model no significant association with uncontrolled pain was observed. Among individuals in DDI+DGI model with an activity score of 0.5 or less, approximately 41% complained of uncontrolled pain while only 15.7% of the individuals with an activity score of 1 or greater relayed similar complaints[161]. Furthermore, in a study performed with data from more than 50,000 adults, it was reported that the average expenses for healthcare related services were higher for opioid users with drug-drug interactions (DDI) compared to those without DDIs ($7841 vs. $5625)[162]. Thus, a CYP2D6-guided approach considering both genotype and inhibitor use is recommended by CPIC when prescribing these opioids[7].
When CYP2D6 genotype and phenoconversion are both considered, improved pain control by CYP2D6-metabolized opioids was observed. One study reported that 24% of CYP2D6-phenotype guided (considering both genotype and phenoconversion) vs. 0% of usual care participants reported greater than 30% reduction in pain score of, which is often considered the clinically significant level of pain improvement. As there were no differences in prescribing between IMs/PMs and NMs, the greater pain control in the CYP2D6-phenotype guided group was due to the intervention. [158]. Another study on postoperative pain management reported that if phenoconversion is considered in addition to CYP2D6 genotypes, fewer morphine milligram equivalents(MME) of opioid provides a similar level of pain control when compared with the usual care arm (200 vs. 230 MME; p = 0.047)[157]. The IGNITE network, funded by the NIH, is conducting a large multicenter randomized pragmatic trial to determine the effect of CYP2D6-guided opioid prescribing on chronic pain and post-operative pain control and opioid usage[163].
Phenoconversion was also found to be the limiting factor for predicting clinical outcome of aripiprazole treatment based on the CYP2D6 genotype[106]. A significant percentage of the patients prescribed aripiprazole are also prescribed a CYP2D6 inhibitor. In a study of aripiprazole tolerability in 277 pediatric patients with mood disorders, 72% of the cohort were concomitantly taking a CYP2D6 inhibitor. Consideration of the inhibitors while phenotyping CYP2D6 decreased the NMs from 57% to 27% while increasing the PMs from 6% to 49%[164]. Those phenoconverted IMs and PMs have lower CYP2D6 metabolic activity that caused the accumulation of active aripiprazole in the blood. [106,142]. In one study, among the NM patients who were taking inhibitors concomitantly, aripiprazole concentrations were about 50% higher than the NMs who were not taking the inhibitors [142]. 37% of the patients of a study discontinued treatment for adverse effects where most of them (67%) are phenotypic PMs[164]. The data highlight the importance of considering CYP2D6 inhibitors in patients prescribed CYP2D6 substrates like aripiprazole.
The antiestrogenic clinical activity of tamoxifen is provided by endoxifen that is produced from hydroxylation of N-Desmethyl tamoxifen (primary metabolite of tamoxifen) by CYP2D6[165]. CYP2D6 NMs in the presence of strong or moderate inhibitors can phenoconvert to IMs or PMs and cannot produce adequate active metabolite endoxifen. Endoxifen concentrations can be decreased by 64% (95% CI = 39% to 89%) in women with a NM CYP2D6 genotype after paroxetine coadministration[166]. One study reported that plasma concentrations of endoxifen were significantly lower in CYP2D6 NMs who were taking strong CYP2D6 inhibitors than NMs who were not taking CYP2D6 inhibitors (23.5 nmol/L vs. 84.1 nmol/L, P < .001) [165]. For 25%, 50%, and 75% increases in percent overlap days between paroxetine and tamoxifen, hazard ratios for subsequent breast cancer risk were 1.06, 1.13, and 1.20 respectively[167]. Concomitant use of paroxetine resulted in increased risk of death from breast cancer. One study demonstrated that 25%, 50%, and 75% increase in the overlapping time of paroxetine and tamoxifen increased the risk of death from breast cancer to 24%, 54%, and 91% respectively (P<0.05 for each comparison) [168]. Therefore, according to the CPIC guidelines, it is strongly recommended not to use any CYP2D6 strong or moderate inhibitors when prescribing tamoxifen among CYP2D6 NMs and UMs[15].
Metabolism of atomoxetine by CYP2D6 is also decreased when coadministered with CYP2D6 inhibitors, which leads to change in pharmacokinetic parameters of atomoxetine[169–173]. With concomitant paroxetine treatment, AUC0−24 of atomoxetine was increased by 2.3-, 1.7-, and 1.3-fold, in CYP2D6*wt/*wt, CYP2D6*wt/*10, and CYP2D6*10/*10 respectively when compared to atomoxetine concentration without paroxetine coadministration[169]. Atomoxetine Cmax was also increased from 221to 373 ng/mL[172] with concomitant paroxetine. Similar results were also reported for bupropion coadministration where systemic exposure to atomoxetine was increased (5.1-fold) and exposure to its main metabolite was decreased (1.5-fold) when compared to exposure to atomoxetine without bupropion administration[173]. Cardiovascular side effects were observed after atomoxetine and fluoxetine comedication[171,174]. Considering the importance of phenoconversion, CPIC suggests considering CYP2D6 inhibitors when prescribing atomoxetine[14].
4. Implementation in clinical practice
Utilization of CYP2D6 pharmacogenetics for personalized therapy is uncommon in clinical practice and consideration of the effect of CYP2D6 inhibitors is even more rarely considered. The data summarized herein highlight that CYP2D6 phenotype (as determined by genotype and/or drug interactions) have clinically important implications on the efficacy and/or toxicity of many CYP2D6-metabolized drugs. Thus clinical implementation of CYP2D6 pharmacogenetics and phenoconversion in clinical practice is a critical component to providing personalized drug therapy.
CPIC currently has pharmacogenetics guidelines for tramadol, codeine, hydrocodone, atomoxetine, tamoxifen, paroxetine, fluoxetine, fluvoxamine, amitriptyline, nortriptyline, ondansetron and tropisetron[7,14–16,21,131]. These guidelines address the effects of CYP2D6 inhibitor on metabolism of those drugs except for ondansetron and tropisetron. Lack of data and/or lack of a recent update might be the reason for not including the phenoconversion in the CPIC guidelines of ondansetron and tropisetron[131]. New CPIC pharmacogenetic guidelines for additional CYP2D6 substrate drugs including antipsychotics(e.g., aripiprazole, brexpiprazole, pimozide etc.), antidepressants (e.g., SNRIs), beta-blockers (e.g., carvedilol, metoprolol etc.) and one update on guidelines for SSRIs are now in progress[175].
To benefit patients based on the recommendations in CYP2D6 pharmacogenetics guidelines, CYP2D6 pharmacogenetic testing can be ordered if not already available in the EHR [7,14–16,21,131]. As pharmacogenetics results are lifelong results it is not required to be retested again, unless additional clinically important alleles are discovered[176]. The test results often include the CYP2D6 genotypes (e.g., CYP2D6 *1/*4) and activity score (AS) of CYP2D6 (e.g., AS 1, intermediate metabolize) or both. To get the actual clinical phenotype of the patient, CYP2D6 genotypic phenotype and concomitant inhibitor medication should both be taken into consideration. Based on the type of inhibitor, activity score should be modified and the clinical phenotype can be determined. Details of the activity score of specific alleles, calculation of the activity score for genotype alone and adjustment for CYP2D6 inhibitors and interpretation of clinical phenotype have been described in Table 2. CYP2D6 can regain its normal activity after 5–7 days of discontinuation of CYP2D6 inhibitors[177]. So, concomitant medications need to be considered routinely since the phenotype based on inhibitor presence or absence can change over time.
As no tool is currently available in the EHR to assist in consideration of CYP2D6 phenotype based on both genotype and drug interactions, adjustment of the activity score needs to be done manually for patients. To make this process easier and to implement phenoconversion in practice, a CYP2D6 phenoconversion calculator tool was developed (https://precisionmedicine.ufhealth.org/phenoconversion-calculator/) [17]. This calculator needs genotype information (e.g., CYP2D6 *1/ *4), number of alleles present, and comedication information to provide the predicted clinical CYP2D6 phenotype of the individual. Using the clinical phenotype and pharmacogenetics guidelines provided by CPIC, clinicians can provide the best available personalized therapy to the patients. While this is a valuable tool to assist clinicians, widespread adoption of personalized CYP2D6 treatments will require and approach that is automated within the EHR.
There are several challenges to incorporate CYP2D6 pharmacogenetics and phenoconversion into clinical practice, which have been highlighted by several groups who have undertaken such a clinical implementation [178–181]. Of those, one of the most important challenges is out-of-pocket cost of pharmacogenetic testing. Presenting the reimbursement data to patients and prior authorizations to cover the pharmacogenetic tests costs can help to overcome the barrier[179]. As data emerge on the clinical impact of using CYP2D6 phenotype to guide drug therapy, it is anticipated that more payers will provide coverage for the pharmacogenetic testing. Another barrier to proper implementation is lack of use of clinical decision support tools to provide proper guidance and knowledge on genotype-based actions[180]. The use of clinical decision support tools can help integrate pharmacogenetics into clinical practice, but incorporation of phenoconversion along with genotype is more difficult and there are not EHR-based tools available yet to support this. In the clinical setting, an updated list of current medications is essential to determine whether phenoconversion is present for individuals who are also taking a CYP2D6-relevant drug. So routine update of the medication list in the medical record is a prerequisite. Moreover, phenoconversion should be continually evaluated at least before every prescription as concurrent medication may change. Manual calculation of the activity score adjusted for phenoconversion can also be challenging though utilization of the phenoconversion calculator tool can ease this process for healthcare providers [17]. Most of the resources commonly used by prescribers (e.g., Drug.com, RxList, or other drug interaction checkers) include limited information about CYP2D6 genetics, drug interactions and guidelines for incorporating CYP2D6 pharmacogenetics and phenoconversion in clinical practice [144]. Proper clinical integration and training can help to disseminate the importance of CYP2D6 pharmacogenetics and phenoconversion among clinicians.
5. Conclusion
Considering the extent and impact of genetic variability of CYP2D6, vulnerability of CYP2D6 after co-administration of inhibitor drugs, and significance of CYP2D6 in drug metabolism, CYP2D6 pharmacogenetics is highly important for designing and implementing personalized drug therapy to ensure drug safety and efficacy. Proper genetic testing and interpretation of the results, and adjustment of the phenotype considering the concomitant CYP2D6 inhibitors is essential for CYP2D6 pharmacogenetics-based personalized therapy. Approaches to overcome the challenges including appropriate use of clinical decision support tools, integration of CYP2D6 phenoconversion into clinical care, reimbursement of pharmacogenetic testing costs, and a local clinical expert to assist other clinicians can help achieve proper implementation of CYP2D6 pharmacogenetics and phenoconversion in the clinical setting to improve drug therapy outcomes in patients.
6. Expert opinion
CYP2D6 is involved in metabolizing 20–25% of all the drugs available on the market. This enzyme can either metabolize the active drug to inactive metabolite(s), facilitating the excretion of the drug, or metabolize the inactive prodrug to the active drug, assisting in exerting the proper pharmacological activity of the drug. Reduced or absent (IM or PM) CYP2D6 can lead to increased plasma concentrations for the active drug for a longer time, increasing risk of adverse drug effects and non-adherence. Conversely, decreased conversion of prodrugs to active drugs in those with impaired CYP2D6 activity is likely to impair efficacy. Conversely, gene duplication/multiplication or an increase in the activity of CYP2D6 (UM) may inactivate an active drug very quickly, leading to reduced efficacy whereas in the case of a prodrug, the excessive conversion to an active metabolite may cause serious adverse drug effects. While the UM phenotype can only be accomplished through genetic variation, the PM and IM phenotypes may occur based on genotype, drug interactions or the combination. The large number of drugs metabolized by CYP2D6, the frequency of genetic variation in CYP2D6 that leads to a non-normal phenotype, and the common clinical use of CYP2D6 inhibitors, which can alter the CYP2D6 phenotype makes attention to CYP2D6 phenotype essential in the quest to optimize therapy through personalized approaches.
Recent developments within pharmacogenetics and various guidelines from CPIC have promoted personalized therapy, ensuring drug safety and efficacy, especially for the medications metabolized by CYP2D6 (including tramadol, codeine, hydrocodone, atomoxetine, tamoxifen, paroxetine, fluoxetine, fluvoxamine, amitriptyline, nortriptyline, ondansetron and tropisetron). Although genotype-guided therapy for CYP2D6 metabolized drugs has gained popularity in the last decade, use of CYP2D6 pharmacogenetics while prescribing in clinical practice is not yet standard of care. And in the absence of genetic data, even the consideration of phenoconversion while prescribing is extremely limited. The impact of CYP2D6 inhibitors on the efficacy and safety of CYP2D6-metabolized drugs and percentages of concomitant use of the CYP2D6 inhibitors can easily explain the need to implement phenoconversion along with CYP2D6 pharmacogenetics to lead to optimal therapeutic outcomes. And in the absence of genetic data, clinicians should still consider the impact of a concomitant CYP2D6 inhibitor on the therapeutic outcomes. Concomitant use of CYP2D6 inhibitors can make the genotypic NM act like a PM, although the individual doesn’t carry any non-functional allele. Moreover, data indicate that approximately 20– 70% of patients on CYP2D6 metabolized medications are at risk of CYP2D6 phenoconversion by concomitant medications. Thus, consideration of CYP2D6 genotype alone is insufficient in the quest for personalizing therapy with CYP2D6 substrates. This approach can only be optimal if CYP2D6 genotype and drug interactions are considered together in assigning a CYP2D6 phenotype.
There are many challenges to implementing use of CYP2D6 phenotype to guide prescribing. These include proper interpretation of genotype-based phenotype from CYP2D6 alleles or activity score, adjustment of phenotype considering the concomitant administration of CYP2D6 inhibitor medications, routinely checking concomitant medications for phenotype adjustment, reimbursement of genetic testing costs, proper training of the clinicians among other challenges. For widespread utilization of personalized approaches for drugs metabolized by CYP2D6, proper integration of CYP2D6 pharmacogenetics and phenoconversion in the EHR is necessary. While much progress has been made in recent years in optimizing the reporting of CYP2D6 genotype in a meaningful way in the EHR, we are aware of no EHR system that does this along with incorporating the drug interaction effectively to provide the clinician with a predicted phenotype. Laboratory CYP2D6 genotyping reports with appropriate interpretations of the CYP2D6 phenotype and quick access to evaluate the risks of phenoconversion could ease the path to personalized medicine in clinical care. In addition to integrating those support tools, increasing awareness among prescribers of the clinical implications of CYP2D6 inhibitors and their ability to create phenoconversion is also critical to optimal use of drug therapy.
In summary, the utilization of proper guidelines incorporating both CYP2D6 pharmacogenetics and phenoconversion in clinical care is essential to ensure the most benefits of personalized medicine.
Article highlights:
CYP2D6 is estimated to contribute to the metabolism of approximately 20–25% of drugs. CYP2D6 is involved not only in metabolizing active drugs into its inactive metabolites (e.g., paroxetine, aripiprazole) but also in metabolizing inactive drugs into its active metabolite including (e.g., codeine, tamoxifen). Therefore, the metabolic activity of CYP2D6 is associated with adverse drug reactions or drug ineffectiveness.
The gene encoding CYP2D6 is highly polymorphic and around 100 alleles of CYP2D6 with different impact on function of the encoded protein have been identified. An activity score is given to a specific allele of CYP2D6 and summing the activity scores of the two (or more) alleles leads to the diplotype activity score. Using the diplotype score, genotypic phenotype of CYP2D6 is estimated, with resulting phenotypes including poor metabolizer (PM), intermediate metabolizer (IM), normal metabolizer (NM), ultra-rapid metabolizer (UM).
Strong (e.g., paroxetine, bupropion) and moderate (e.g., duloxetine, sertraline) inhibitors can also decrease the CYP2D6 activity, phenoconverting the genotypic NMs or UMs into phenotypic PMs or IMs. So, concomitant use of those drugs cannot be overlooked while predicting the CYP2D6 phenotype. Moreover, data indicate that up to 20– 70% of the patients who are on CYP2D6 metabolized drugs are at risk of CYP2D6 phenoconversion by concomitant medications.
It is important to adjust the activity score of CYP2D6 based on use of CYP2D6 inhibitors. If the individual is taking one of the strong or moderate inhibitors concomitantly, then the genotype-based activity score should be multiplied by 0 or 0.5 respectively. Then the actual clinical phenotype can be estimated based on the adjusted activity score.
Pharmacogenetic-based drug therapy guidelines have been developed for at least 50 CYP2D6-metabolized drugs based on the activity of CYP2D6. Considering the importance of phenoconversion, Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines suggest incorporating the effects of CYP2D6 inhibitors while calculating the activity score of CYP2D6 for CYP2D6 metabolized opioids, tricyclic antidepressants, tamoxifen and atomoxetine.
There are many challenges to implementing use of CYP2D6 pharmacogenetics and phenoconversion to guide prescribing. These include proper interpretation of genotype-based phenotype from CYP2D6 alleles or activity score, adjustment of phenotype considering the concomitant administration of CYP2D6 inhibitor medications, routinely checking of concomitant medications for phenotype adjustment, reimbursement of genetic testing costs, proper training of the clinicians etc.
Funding
This paper was funded by the National Institutes of Health (U01-HG007269) to JA Johnson and an American College of Clinical Pharmacy Foundation Futures Grant to NA Nahid.
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as: * of interest or ** of considerable interest
- 1.Abbasi J Getting Pharmacogenomics Into the Clinic. JAMA 2016. Oct 18;316(15):1533–1535. [DOI] [PubMed] [Google Scholar]
- 2.Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. Lancet 2019. 08 10;394(10197):521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. pharmgkb.org. https://www.pharmgkb.org/guidelineAnnotations.
- 4.PharmVar.org. Pharmacogene Variation Consortium (PharmVar) at www.PharmVar.org (Gaedigk et al. 2018, CPT 103:399; Gaedigk et al. 2019, CPT 105:29) Last accessed on 08/06/2022.
- 5.Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 2020. 01;13(1):116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaedigk A, Simon SD, Pearce RE, et al. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 2008. Feb;83(2):234–42. [DOI] [PubMed] [Google Scholar]
- 7. Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther 2021. 10;110(4):888–896. *CPIC guidelines for tramadol, codeine and hydrocodone.
- 8. Shah RR, Smith RL. Addressing phenoconversion: the Achilles’ heel of personalized medicine. Br J Clin Pharmacol 2015. Feb;79(2):222–40. **One of the first review papers that focuses primarily on co-medication-induced phenoconversion and discusses the impact of phenoconversion in personalized medicine.
- 9.fda.gov. U.S. Food and Drug Administration. Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers https://www.fda.gov/drugs/developmentapprovalprocess/developmentresources/druginteractionslabeling/ucm093664.htm Accessed on 08/06/2022.
- 10.rxsaver.com. https://www.rxsaver.com/blog/top-50-prescription-drugs-filled Last accessed on October 11, 2022.
- 11.Chanfreau-Coffinier C, Tuteja S, Hull LE, et al. Drug-drug-gene interaction risk among opioid users in the U.S. Department of Veterans Affairs. Pain 2022. Mar 23. [DOI] [PubMed]
- 12.Tirkkonen T, Laine K. Drug interactions with the potential to prevent prodrug activation as a common source of irrational prescribing in hospital inpatients. Clin Pharmacol Ther 2004. Dec;76(6):639–47. [DOI] [PubMed] [Google Scholar]
- 13.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011. Mar;89(3):464–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown JT, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin Pharmacol Ther. 2019. 07;106(1):94–102. *CPIC Guideline for CYP2D6 genotype and atomoxetine therapy.
- 15. Goetz MP, Sangkuhl K, Guchelaar HJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther 2018. 05;103(5):770–777. *CPIC Guideline for CYP2D6 genotype and tamoxifen therapy.
- 16. Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 2017. 07;102(1):37–44. *CPIC Guideline for CYP2D6 genotype and dosing of tricyclic antidepressants(e.g. amitriptyline, nortriptyline).
- 17. Cicali EJ, Elchynski AL, Cook KJ, et al. How to Integrate CYP2D6 Phenoconversion Into Clinical Pharmacogenetics: A Tutorial. Clin Pharmacol Ther 2021. 09;110(3):677–687. **Provides tool to integrate phenoconversion in HER.
- 18.Monte AA, West K, McDaniel KT, et al. CYP2D6 Genotype Phenotype Discordance Due to Drug-Drug Interaction. Clin Pharmacol Ther 2018. 11;104(5):933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Botton MR, Scott ER, et al. Sequencing the CYP2D6 gene: from variant allele discovery to clinical pharmacogenetic testing. Pharmacogenomics 2017. May;18(7):673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis JP, Peter AP, Shaman JA. Consequences of. Front Psychiatry 2019;10:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 2015. Aug;98(2):127–34. *CPIC Guideline for CYP2D6 genotype and dosing of SSRIs(e.g. paroxetine, fluvoxamine).
- 22.Gaedigk A Complexities of CYP2D6 gene analysis and interpretation. Int Rev Psychiatry 2013. Oct;25(5):534–53. [DOI] [PubMed] [Google Scholar]
- 23.Beoris M, Amos Wilson J, Garces JA, et al. CYP2D6 copy number distribution in the US population. Pharmacogenet Genomics 2016. Feb;26(2):96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleeman N, Dundar Y, Dickson R, et al. Cytochrome P450 testing for prescribing antipsychotics in adults with schizophrenia: systematic review and meta-analyses. Pharmacogenomics J 2011. Feb;11(1):1–14. [DOI] [PubMed] [Google Scholar]
- 25.Taylor C, Crosby I, Yip V, et al. A Review of the Important Role of. Genes (Basel) 2020. 10 30;11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gressier F, Ellul P, Dutech C, et al. Serotonin toxicity in a CYP2D6 poor metabolizer, initially diagnosed as a drug-resistant major depression. Am J Psychiatry 2014. 08;171(8):890. [DOI] [PubMed] [Google Scholar]
- 27.Koren G, Cairns J, Chitayat D, et al. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 2006. Aug 19;368(9536):704. [DOI] [PubMed] [Google Scholar]
- 28.Suzumura T, Kimura T, Kudoh S, et al. Reduced CYP2D6 function is associated with gefitinib-induced rash in patients with non-small cell lung cancer. BMC Cancer 2012. Dec 04;12:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel WA, Bromek E, Danek PJ, et al. The mechanisms of interactions of psychotropic drugs with liver and brain cytochrome P450 and their significance for drug effect and drug-drug interactions. Biochem Pharmacol. 2022. 05;199:115006. [DOI] [PubMed] [Google Scholar]
- 30.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 2017. 02;19(2):215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, et al. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 2017. 01;19(1):69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.imshealth.com. IMS Institute for Healthcare Informatics. Medication use and the shifting cost of healthcare - a review of use of medicines in the United States in 2013 http://www.imshealth.com/deployedfiles/imshealth/Global/Content/Corporate/IMS%20Health%20Institute/Reports/Secure/IIHI_US_Use_of_Meds_for_2013.pdf. Accessed 08/06/2020..
- 33.clincalc.com. https://clincalc.com/DrugStats/Top200Drugs.aspx last accessed on October 11, 2022.
- 34.Persons APoPPiO. The management of persistent pain in older persons. J Am Geriatr Soc 2002. Jun;50(6 Suppl):S205–24. [DOI] [PubMed] [Google Scholar]
- 35.Foster A, Mobley E, Wang Z. Complicated pain management in a CYP450 2D6 poor metabolizer. Pain Pract 2007. Dec;7(4):352–6. [DOI] [PubMed] [Google Scholar]
- 36.Volpe DA, McMahon Tobin GA, Mellon RD, et al. Uniform assessment and ranking of opioid μ receptor binding constants for selected opioid drugs. Regul Toxicol Pharmacol 2011. Apr;59(3):385–90. [DOI] [PubMed] [Google Scholar]
- 37.Chen ZR, Somogyi AA, Bochner F. Polymorphic O-demethylation of codeine. Lancet Vol. 2. England1988. p. 914–5. [DOI] [PubMed] [Google Scholar]
- 38.Lötsch J, Skarke C, Schmidt H, et al. Evidence for morphine-independent central nervous opioid effects after administration of codeine: contribution of other codeine metabolites. Clin Pharmacol Ther 2006. Jan;79(1):35–48. [DOI] [PubMed] [Google Scholar]
- 39.Vevelstad M, Pettersen S, Tallaksen C, et al. O-demethylation of codeine to morphine inhibited by low-dose levomepromazine. Eur J Clin Pharmacol 2009. Aug;65(8):795–801. [DOI] [PubMed] [Google Scholar]
- 40.Yue QY, Svensson JO, Alm C, et al. Codeine O-demethylation co-segregates with polymorphic debrisoquine hydroxylation. Br J Clin Pharmacol 1989. Dec;28(6):639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frost J, Løkken TN, Helland A, et al. Post-mortem levels and tissue distribution of codeine, codeine-6-glucuronide, norcodeine, morphine and morphine glucuronides in a series of codeine-related deaths. Forensic Sci Int 2016. May;262:128–37. [DOI] [PubMed] [Google Scholar]
- 42.Haffen E, Paintaud G, Berard M, et al. On the assessment of drug metabolism by assays of codeine and its main metabolites. Ther Drug Monit 2000. Jun;22(3):258–65. [DOI] [PubMed] [Google Scholar]
- 43.Radford H, Simpson KH, Rogerson S, et al. A Single Site Population Study to Investigate CYP2D6 Phenotype of Patients with Persistent Non-Malignant Pain. Medicina (Kaunas) 2019. May 28;55(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He YJ, Brockmöller J, Schmidt H, et al. CYP2D6 ultrarapid metabolism and morphine/codeine ratios in blood: was it codeine or heroin? J Anal Toxicol 2008. Mar;32(2):178–82. [DOI] [PubMed] [Google Scholar]
- 45.Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J 2007. Aug;7(4):257–65. [DOI] [PubMed] [Google Scholar]
- 46.Lötsch J, von Hentig N, Freynhagen R, et al. Cross-sectional analysis of the influence of currently known pharmacogenetic modulators on opioid therapy in outpatient pain centers. Pharmacogenet Genomics 2009. Jun;19(6):429–36. [DOI] [PubMed] [Google Scholar]
- 47.fda.gov. https://www.fda.gov/consumers/consumer-updates/codeine-and-tramadol-can-cause-breathing-problems-children#:~:text=The%20FDA%20is%20warning%20that,tramadol%20for%20use%20in%20children. Last accessed on October 15, 2022.
- 48.Paar WD, Poche S, Gerloff J, et al. Polymorphic CYP2D6 mediates O-demethylation of the opioid analgesic tramadol. Eur J Clin Pharmacol 1997;53(3–4):235–9. [DOI] [PubMed] [Google Scholar]
- 49.Haage P, Kronstrand R, Josefsson M, et al. Enantioselective pharmacokinetics of tramadol and its three main metabolites; impact of. Pharmacol Res Perspect 2018. 07;6(4):e00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slanar O, Nobilis M, Kvetina J, et al. Miotic action of tramadol is determined by CYP2D6 genotype. Physiol Res 2007;56(1):129–136. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka H, Naito T, Sato H, et al. Impact of CYP genotype and inflammatory markers on the plasma concentrations of tramadol and its demethylated metabolites and drug tolerability in cancer patients. Eur J Clin Pharmacol 2018. Nov;74(11):1461–1469. [DOI] [PubMed] [Google Scholar]
- 52.Bastami S, Haage P, Kronstrand R, et al. Pharmacogenetic aspects of tramadol pharmacokinetics and pharmacodynamics after a single oral dose. Forensic Sci Int 2014. May;238:125–32. [DOI] [PubMed] [Google Scholar]
- 53.Lane K, Dixon JJ, McKeown D, et al. Using tramadol to measure CYP2D6 metabolism in critically ill adults. Intensive Care Med 2014. Aug;40(8):1177–8. [DOI] [PubMed] [Google Scholar]
- 54.Kirchheiner J, Keulen JT, Bauer S, et al. Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol 2008. Feb;28(1):78–83. [DOI] [PubMed] [Google Scholar]
- 55.Susce MT, Murray-Carmichael E, de Leon J. Response to hydrocodone, codeine and oxycodone in a CYP2D6 poor metabolizer. Prog Neuropsychopharmacol Biol Psychiatry 2006. Sep 30;30(7):1356–8. [DOI] [PubMed] [Google Scholar]
- 56.Tillman EM, Skaar TC, Eadon MT. Nephrotoxicity in a Patient With Inadequate Pain Control: Potential Role of Pharmacogenetic Testing for Cytochrome P450 2D6 and Apolipoprotein L1. Front Pharmacol 2019;10:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamer UM, Lehnen K, Höthker F, et al. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain 2003. Sep;105(1–2):231–8. [DOI] [PubMed] [Google Scholar]
- 58.Stamer UM, Musshoff F, Kobilay M, et al. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther 2007. Jul;82(1):41–7. [DOI] [PubMed] [Google Scholar]
- 59.Zhao Q, Sun J, Tao Y, et al. A logistic equation to determine the validity of tramadol from related gene polymorphisms and psychological factors. Pharmacogenomics 2014. Mar;15(4):487–95. [DOI] [PubMed] [Google Scholar]
- 60.Befort K, Filliol D, Decaillot FM, et al. A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem 2001. Feb 02;276(5):3130–7. [DOI] [PubMed] [Google Scholar]
- 61.Stamer UM, Stüber F, Muders T, et al. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth Analg 2008. Sep;107(3):926–9. [DOI] [PubMed] [Google Scholar]
- 62.Stauble ME, Moore AW, Langman LJ, et al. Hydrocodone in postoperative personalized pain management: pro-drug or drug? Clin Chim Acta 2014. Feb 15;429:26–9. [DOI] [PubMed] [Google Scholar]
- 63.Kaplan HL, Busto UE, Baylon GJ, et al. Inhibition of cytochrome P450 2D6 metabolism of hydrocodone to hydromorphone does not importantly affect abuse liability. J Pharmacol Exp Ther 1997. Apr;281(1):103–8. [PubMed] [Google Scholar]
- 64.de Leon J, Dinsmore L, Wedlund P. Adverse drug reactions to oxycodone and hydrocodone in CYP2D6 ultrarapid metabolizers. J Clin Psychopharmacol 2003. Aug;23(4):420–1. [DOI] [PubMed] [Google Scholar]
- 65.de Vos A, van der Weide J, Loovers HM. Association between CYP2C19*17 and metabolism of amitriptyline, citalopram and clomipramine in Dutch hospitalized patients. Pharmacogenomics J 2011. Oct;11(5):359–67. [DOI] [PubMed] [Google Scholar]
- 66.Halling J, Weihe P, Brosen K. The CYP2D6 polymorphism in relation to the metabolism of amitriptyline and nortriptyline in the Faroese population. Br J Clin Pharmacol 2008. Jan;65(1):134–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith JC, Curry SC. Prolonged toxicity after amitriptyline overdose in a patient deficient in CYP2D6 activity. J Med Toxicol 2011. Sep;7(3):220–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bijl MJ, Visser LE, Hofman A, et al. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br J Clin Pharmacol 2008. Apr;65(4):558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peñas-Lledó EM, Trejo HD, Dorado P, et al. CYP2D6 ultrarapid metabolism and early dropout from fluoxetine or amitriptyline monotherapy treatment in major depressive patients. Mol Psychiatry 2013. Jan;18(1):8–9. [DOI] [PubMed] [Google Scholar]
- 70.Mandrioli R, Mercolini L, Saracino MA, et al. Selective serotonin reuptake inhibitors (SSRIs): therapeutic drug monitoring and pharmacological interactions. Curr Med Chem 2012;19(12):1846–63. [DOI] [PubMed] [Google Scholar]
- 71.Tang SW, Helmeste D. Paroxetine. Expert Opin Pharmacother 2008. Apr;9(5):787–94. [DOI] [PubMed] [Google Scholar]
- 72.Feng Y, Pollock BG, Ferrell RE, et al. Paroxetine: population pharmacokinetic analysis in late-life depression using sparse concentration sampling. Br J Clin Pharmacol 2006. May;61(5):558–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Findling RL, Nucci G, Piergies AA, et al. Multiple dose pharmacokinetics of paroxetine in children and adolescents with major depressive disorder or obsessive-compulsive disorder. Neuropsychopharmacology 2006. Jun;31(6):1274–85. [DOI] [PubMed] [Google Scholar]
- 74.Sawamura K, Suzuki Y, Someya T. Effects of dosage and CYP2D6-mutated allele on plasma concentration of paroxetine. Eur J Clin Pharmacol 2004. Oct;60(8):553–7. [DOI] [PubMed] [Google Scholar]
- 75.Charlier C, Broly F, Lhermitte M, et al. Polymorphisms in the CYP 2D6 gene: association with plasma concentrations of fluoxetine and paroxetine. Ther Drug Monit 2003. Dec;25(6):738–42. [DOI] [PubMed] [Google Scholar]
- 76.Gex-Fabry M, Eap CB, Oneda B, et al. CYP2D6 and ABCB1 genetic variability: influence on paroxetine plasma level and therapeutic response. Ther Drug Monit 2008. Aug;30(4):474–82. [DOI] [PubMed] [Google Scholar]
- 77.Lam YW, Gaedigk A, Ereshefsky L, et al. CYP2D6 inhibition by selective serotonin reuptake inhibitors: analysis of achievable steady-state plasma concentrations and the effect of ultrarapid metabolism at CYP2D6. Pharmacotherapy 2002. Aug;22(8):1001–6. [DOI] [PubMed] [Google Scholar]
- 78.Sindrup SH, Brøsen K, Gram LF, et al. The relationship between paroxetine and the sparteine oxidation polymorphism. Clin Pharmacol Ther 1992. Mar;51(3):278–87. [DOI] [PubMed] [Google Scholar]
- 79.Sato A, Okura Y, Minagawa S, et al. Life-threatening serotonin syndrome in a patient with chronic heart failure and CYP2D6*1/*5. Mayo Clin Proc 2004. Nov;79(11):1444–8. [DOI] [PubMed] [Google Scholar]
- 80.Carrillo JA, Dahl ML, Svensson JO, et al. Disposition of fluvoxamine in humans is determined by the polymorphic CYP2D6 and also by the CYP1A2 activity. Clin Pharmacol Ther 1996. Aug;60(2):183–90. [DOI] [PubMed] [Google Scholar]
- 81.Spigset O, Granberg K, Hägg S, et al. Relationship between fluvoxamine pharmacokinetics and CYP2D6/CYP2C19 phenotype polymorphisms. Eur J Clin Pharmacol 1997;52(2):129–33. [DOI] [PubMed] [Google Scholar]
- 82.Lee SY, Sohn KM, Ryu JY, et al. Sequence-based CYP2D6 genotyping in the Korean population. Ther Drug Monit 2006. Jun;28(3):382–7. [DOI] [PubMed] [Google Scholar]
- 83. Milosavljevic F, Bukvic N, Pavlovic Z, et al. Association of CYP2C19 and CYP2D6 Poor and Intermediate Metabolizer Status With Antidepressant and Antipsychotic Exposure: A Systematic Review and Meta-analysis. JAMA Psychiatry 2021. 03 01;78(3):270–280. *Meta-analysis of 12 studies on CYPD6 pharamcogenetics and aripiprazole.
- 84.Yue QY, Zhong ZH, Tybring G, et al. Pharmacokinetics of nortriptyline and its 10-hydroxy metabolite in Chinese subjects of different CYP2D6 genotypes. Clin Pharmacol Ther 1998. Oct;64(4):384–90. [DOI] [PubMed] [Google Scholar]
- 85.Hermann M, Hendset M, Fosaas K, et al. Serum concentrations of venlafaxine and its metabolites O-desmethylvenlafaxine and N-desmethylvenlafaxine in heterozygous carriers of the CYP2D6*3, *4 or *5 allele. Eur J Clin Pharmacol 2008. May;64(5):483–7. [DOI] [PubMed] [Google Scholar]
- 86.Whyte EM, Romkes M, Mulsant BH, et al. CYP2D6 genotype and venlafaxine-XR concentrations in depressed elderly. Int J Geriatr Psychiatry 2006. Jun;21(6):542–9. [DOI] [PubMed] [Google Scholar]
- 87.Azuma J, Hasunuma T, Kubo M, et al. The relationship between clinical pharmacokinetics of aripiprazole and CYP2D6 genetic polymorphism: effects of CYP enzyme inhibition by coadministration of paroxetine or fluvoxamine. Eur J Clin Pharmacol 2012. Jan;68(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belmonte C, Ochoa D, Román M, et al. Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin Pharmacol Toxicol 2018. Jun;122(6):596–605. [DOI] [PubMed] [Google Scholar]
- 89.Jukic MM, Smith RL, Haslemo T, et al. Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: a retrospective, cohort study. Lancet Psychiatry 2019. May;6(5):418–426. [DOI] [PubMed] [Google Scholar]
- 90.Koller D, Belmonte C, Lubomirov R, et al. Effects of aripiprazole on pupillometric parameters related to pharmacokinetics and pharmacogenetics after single oral administration to healthy subjects. J Psychopharmacol 2018. 11;32(11):1212–1222. [DOI] [PubMed] [Google Scholar]
- 91.Nagai G, Mihara K, Nakamura A, et al. Prolactin concentrations during aripiprazole treatment in relation to sex, plasma drugs concentrations and genetic polymorphisms of dopamine D2 receptor and cytochrome P450 2D6 in Japanese patients with schizophrenia. Psychiatry Clin Neurosci 2012. Oct;66(6):518–24. [DOI] [PubMed] [Google Scholar]
- 92.Suzuki T, Mihara K, Nakamura A, et al. Effects of the CYP2D6*10 allele on the steady-state plasma concentrations of aripiprazole and its active metabolite, dehydroaripiprazole, in Japanese patients with schizophrenia. Ther Drug Monit 2011. Feb;33(1):21–4. [DOI] [PubMed] [Google Scholar]
- 93.Suzuki T, Mihara K, Nakamura A, et al. Effects of genetic polymorphisms of CYP2D6, CYP3A5, and ABCB1 on the steady-state plasma concentrations of aripiprazole and its active metabolite, dehydroaripiprazole, in Japanese patients with schizophrenia. Ther Drug Monit 2014. Oct;36(5):651–5. [DOI] [PubMed] [Google Scholar]
- 94.van der Weide K, van der Weide J. The Influence of the CYP3A4*22 Polymorphism and CYP2D6 Polymorphisms on Serum Concentrations of Aripiprazole, Haloperidol, Pimozide, and Risperidone in Psychiatric Patients. J Clin Psychopharmacol 2015. Jun;35(3):228–36. [DOI] [PubMed] [Google Scholar]
- 95.Kinghorn WA, McEvoy JP. Aripiprazole: pharmacology, efficacy, safety and tolerability. Expert Rev Neurother 2005. May;5(3):297–307. [DOI] [PubMed] [Google Scholar]
- 96.dailymed.nlm.nih.gov. ARIPIPRAZOLE- aripiprazole tablet [package insert] Princeton, NJ, USA: Dr.Reddy’s Pharmaceutical Inc; 2020. Available from: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=0aa7e178-456a-4942-93aa-9ec18a58939f. [Google Scholar]
- 97.Koller D, Almenara S, Mejía G, et al. Safety and cardiovascular effects of multiple-dose administration of aripiprazole and olanzapine in a randomised clinical trial. Hum Psychopharmacol 2021. 01;36(1):1–12. [DOI] [PubMed] [Google Scholar]
- 98.Cabaleiro T, Ochoa D, López-Rodríguez R, et al. Effect of polymorphisms on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. Hum Psychopharmacol 2014. Sep;29(5):459–69. [DOI] [PubMed] [Google Scholar]
- 99.Gassó P, Papagianni K, Mas S, et al. Relationship between CYP2D6 genotype and haloperidol pharmacokinetics and extrapyramidal symptoms in healthy volunteers. Pharmacogenomics 2013. Oct;14(13):1551–63. [DOI] [PubMed] [Google Scholar]
- 100.Troost PW, Lahuis BE, Hermans MH, et al. Prolactin release in children treated with risperidone: impact and role of CYP2D6 metabolism. J Clin Psychopharmacol 2007. Feb;27(1):52–7. [DOI] [PubMed] [Google Scholar]
- 101.Leon J, Susce MT, Pan RM, et al. A study of genetic (CYP2D6 and ABCB1) and environmental (drug inhibitors and inducers) variables that may influence plasma risperidone levels. Pharmacopsychiatry 2007. May;40(3):93–102. [DOI] [PubMed] [Google Scholar]
- 102.Novalbos J, López-Rodríguez R, Román M, et al. Effects of CYP2D6 genotype on the pharmacokinetics, pharmacodynamics, and safety of risperidone in healthy volunteers. J Clin Psychopharmacol 2010. Oct;30(5):504–11. [DOI] [PubMed] [Google Scholar]
- 103.Jovanović N, Božina N, Lovrić M, et al. The role of CYP2D6 and ABCB1 pharmacogenetics in drug-naïve patients with first-episode schizophrenia treated with risperidone. Eur J Clin Pharmacol 2010. Nov;66(11):1109–17. [DOI] [PubMed] [Google Scholar]
- 104.Oshikoya KA, Neely KM, Carroll RJ, et al. CYP2D6 genotype and adverse events to risperidone in children and adolescents. Pediatr Res 2019. 04;85(5):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Almoguera B, Riveiro-Alvarez R, Lopez-Castroman J, et al. CYP2D6 poor metabolizer status might be associated with better response to risperidone treatment. Pharmacogenet Genomics 2013. Nov;23(11):627–30. [DOI] [PubMed] [Google Scholar]
- 106.Lisbeth P, Vincent H, Kristof M, et al. Genotype and co-medication dependent CYP2D6 metabolic activity: effects on serum concentrations of aripiprazole, haloperidol, risperidone, paliperidone and zuclopenthixol. Eur J Clin Pharmacol 2016. Feb;72(2):175–84. [DOI] [PubMed] [Google Scholar]
- 107.Molden E, Waade RB, Hoff M, et al. Impact of Ageing on Serum Concentrations of Risperidone and Its Active Metabolite in Patients with Known CYP2D6 Genotype. Basic Clin Pharmacol Toxicol 2016. Nov;119(5):470–475. [DOI] [PubMed] [Google Scholar]
- 108.Schroth W, Winter S, Mürdter T, et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front Pharmacol 2017;8:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brauch H, Schwab M. Prediction of tamoxifen outcome by genetic variation of CYP2D6 in post-menopausal women with early breast cancer. Br J Clin Pharmacol 2014. Apr;77(4):695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jung JA, Lim HS. Association between CYP2D6 genotypes and the clinical outcomes of adjuvant tamoxifen for breast cancer: a meta-analysis. Pharmacogenomics 2014. Jan;15(1):49–60. [DOI] [PubMed] [Google Scholar]
- 111.Lim HS, Ju Lee H, Seok Lee K, et al. Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol 2007. Sep 01;25(25):3837–45. [DOI] [PubMed] [Google Scholar]
- 112.Saladores P, Mürdter T, Eccles D, et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenomics J 2015. Feb;15(1):84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mwinyi J, Vokinger K, Jetter A, et al. Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy. Cancer Chemother Pharmacol 2014. Jun;73(6):1181–8. [DOI] [PubMed] [Google Scholar]
- 114.Rae JM, Drury S, Hayes DF, et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 2012. Mar 21;104(6):452–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Teh LK, Mohamed NI, Salleh MZ, et al. The risk of recurrence in breast cancer patients treated with tamoxifen: polymorphisms of CYP2D6 and ABCB1. AAPS J 2012. Mar;14(1):52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Damodaran SE, Pradhan SC, Umamaheswaran G, et al. Genetic polymorphisms of CYP2D6 increase the risk for recurrence of breast cancer in patients receiving tamoxifen as an adjuvant therapy. Cancer Chemother Pharmacol 2012. Jul;70(1):75–81. [DOI] [PubMed] [Google Scholar]
- 117.Dezentjé VO, van Schaik RH, Vletter-Bogaartz JM, et al. CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. Breast Cancer Res Treat 2013. Jul;140(2):363–73. [DOI] [PubMed] [Google Scholar]
- 118.Thompson AM, Johnson A, Quinlan P, et al. Comprehensive CYP2D6 genotype and adherence affect outcome in breast cancer patients treated with tamoxifen monotherapy. Breast Cancer Res Treat 2011. Jan;125(1):279–87. [DOI] [PubMed] [Google Scholar]
- 119.Michelson D, Read HA, Ruff DD, et al. CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J Am Acad Child Adolesc Psychiatry 2007. Feb;46(2):242–51. [DOI] [PubMed] [Google Scholar]
- 120.Trzepacz PT, Williams DW, Feldman PD, et al. CYP2D6 metabolizer status and atomoxetine dosing in children and adolescents with ADHD. Eur Neuropsychopharmacol 2008. Feb;18(2):79–86. [DOI] [PubMed] [Google Scholar]
- 121.Byeon JY, Kim YH, Na HS, et al. Effects of the CYP2D6*10 allele on the pharmacokinetics of atomoxetine and its metabolites. Arch Pharm Res 2015. Nov;38(11):2083–91. [DOI] [PubMed] [Google Scholar]
- 122.Fijal BA, Guo Y, Li SG, et al. CYP2D6 predicted metabolizer status and safety in adult patients with attention-deficit hyperactivity disorder participating in a large placebo-controlled atomoxetine maintenance of response clinical trial. J Clin Pharmacol 2015. Oct;55(10):1167–74. [DOI] [PubMed] [Google Scholar]
- 123.Ramoz N, Boni C, Downing AM, et al. A haplotype of the norepinephrine transporter (Net) gene Slc6a2 is associated with clinical response to atomoxetine in attention-deficit hyperactivity disorder (ADHD). Neuropsychopharmacology 2009. Aug;34(9):2135–42. [DOI] [PubMed] [Google Scholar]
- 124.Dixon CM, Colthup PV, Serabjit-Singh CJ, et al. Multiple forms of cytochrome P450 are involved in the metabolism of ondansetron in humans. Drug Metab Dispos. 1995. Nov;23(11):1225–30. [PubMed] [Google Scholar]
- 125.Firkusny L, Kroemer HK, Eichelbaum M. In vitro characterization of cytochrome P450 catalysed metabolism of the antiemetic tropisetron. Biochem Pharmacol 1995. Jun 16;49(12):1777–84. [DOI] [PubMed] [Google Scholar]
- 126.Fischer V, Vickers AE, Heitz F, et al. The polymorphic cytochrome P-4502D6 is involved in the metabolism of both 5-hydroxytryptamine antagonists, tropisetron and ondansetron. Drug Metab Dispos 1994 1994. Mar-Apr;22(2):269–74. [PubMed] [Google Scholar]
- 127.Kaiser R, Sezer O, Papies A, et al. Patient-tailored antiemetic treatment with 5-hydroxytryptamine type 3 receptor antagonists according to cytochrome P-450 2D6 genotypes. J Clin Oncol 2002. Jun 15;20(12):2805–11. [DOI] [PubMed] [Google Scholar]
- 128.Stamer UM, Lee EH, Rauers NI, et al. CYP2D6- and CYP3A-dependent enantioselective plasma concentrations of ondansetron in postanesthesia care. Anesth Analg 2011. Jul;113(1):48–54. [DOI] [PubMed] [Google Scholar]
- 129.Kim MK, Cho JY, Lim HS, et al. Effect of the CYP2D6 genotype on the pharmacokinetics of tropisetron in healthy Korean subjects. Eur J Clin Pharmacol 2003. Jun;59(2):111–6. [DOI] [PubMed] [Google Scholar]
- 130.Candiotti KA, Birnbach DJ, Lubarsky DA, et al. The impact of pharmacogenomics on postoperative nausea and vomiting: do CYP2D6 allele copy number and polymorphisms affect the success or failure of ondansetron prophylaxis? Anesthesiology 2005. Mar;102(3):543–9. [DOI] [PubMed] [Google Scholar]
- 131. Bell GC, Caudle KE, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 2017. 08;102(2):213–218. *CPIC guideline for CYP2D6 and use of ondansetron and tropisetron.
- 132.Brodde OE, Kroemer HK. Drug-drug interactions of beta-adrenoceptor blockers. Arzneimittelforschung 2003;53(12):814–22. [DOI] [PubMed] [Google Scholar]
- 133.Lennard MS, Silas JH, Freestone S, et al. Oxidation phenotype--a major determinant of metoprolol metabolism and response. N Engl J Med 1982. Dec 16;307(25):1558–60. [DOI] [PubMed] [Google Scholar]
- 134.Blake CM, Kharasch ED, Schwab M, et al. A meta-analysis of CYP2D6 metabolizer phenotype and metoprolol pharmacokinetics. Clin Pharmacol Ther 2013. Sep;94(3):394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gilbert EM, Abraham WT, Olsen S, et al. Comparative hemodynamic, left ventricular functional, and antiadrenergic effects of chronic treatment with metoprolol versus carvedilol in the failing heart. Circulation 1996. Dec 01;94(11):2817–25. [DOI] [PubMed] [Google Scholar]
- 136.Jin SK, Chung HJ, Chung MW, et al. Influence of CYP2D6*10 on the pharmacokinetics of metoprolol in healthy Korean volunteers. J Clin Pharm Ther 2008. Oct;33(5):567–73. [DOI] [PubMed] [Google Scholar]
- 137.Rau T, Wuttke H, Michels LM, et al. Impact of the CYP2D6 genotype on the clinical effects of metoprolol: a prospective longitudinal study. Clin Pharmacol Ther 2009. Mar;85(3):269–72. [DOI] [PubMed] [Google Scholar]
- 138.Zineh I, Beitelshees AL, Gaedigk A, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther 2004. Dec;76(6):536–44. [DOI] [PubMed] [Google Scholar]
- 139.Hamadeh IS, Langaee TY, Dwivedi R, et al. Impact of CYP2D6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin Pharmacol Ther 2014. Aug;96(2):175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Terra SG, Pauly DF, Lee CR, et al. beta-Adrenergic receptor polymorphisms and responses during titration of metoprolol controlled release/extended release in heart failure. Clin Pharmacol Ther 2005. Mar;77(3):127–37. [DOI] [PubMed] [Google Scholar]
- 141.Frost DA, Soric MM, Kaiser R, et al. Efficacy of Tramadol for Pain Management in Patients Receiving Strong Cytochrome P450 2D6 Inhibitors. Pharmacotherapy 2019. 06;39(6):724–729. [DOI] [PubMed] [Google Scholar]
- 142.Kiss Á, Menus Á, Tóth K, et al. Phenoconversion of CYP2D6 by inhibitors modifies aripiprazole exposure. Eur Arch Psychiatry Clin Neurosci 2020. Feb;270(1):71–82. [DOI] [PubMed] [Google Scholar]
- 143.Klomp SD, Manson ML, Guchelaar HJ, et al. Phenoconversion of Cytochrome P450 Metabolism: A Systematic Review. J Clin Med 2020. Sep 07;9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Malki MA, Pearson ER. Drug-drug-gene interactions and adverse drug reactions. Pharmacogenomics J 2020. 06;20(3):355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lesche D, Mostafa S, Everall I, et al. Impact of CYP1A2, CYP2C19, and CYP2D6 genotype- and phenoconversion-predicted enzyme activity on clozapine exposure and symptom severity. Pharmacogenomics J 2020. 04;20(2):192–201. [DOI] [PubMed] [Google Scholar]
- 146.Storelli F, Matthey A, Lenglet S, et al. Impact of CYP2D6 Functional Allelic Variations on Phenoconversion and Drug-Drug Interactions. Clin Pharmacol Ther 2018. 07;104(1):148–157. [DOI] [PubMed] [Google Scholar]
- 147.Monte AA, Heard KJ, Vasiliou V. Prediction of drug response and safety in clinical practice. J Med Toxicol 2012. Mar;8(1):43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ahern TP, Hertz DL, Damkier P, et al. Cytochrome P-450 2D6 (CYP2D6) Genotype and Breast Cancer Recurrence in Tamoxifen-Treated Patients: Evaluating the Importance of Loss of Heterozygosity. Am J Epidemiol 2017. 01 15;185(2):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med 2014. Aug;21(8):879–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kiyotani K, Mushiroda T, Hosono N, et al. Lessons for pharmacogenomics studies: association study between CYP2D6 genotype and tamoxifen response. Pharmacogenet Genomics 2010. Sep;20(9):565–8. [DOI] [PubMed] [Google Scholar]
- 151.Gloor Y, Lloret-Linares C, Bosilkovska M, et al. Drug metabolic enzyme genotype-phenotype discrepancy: High phenoconversion rate in patients treated with antidepressants. Biomed Pharmacother 2022. Aug;152:113202. [DOI] [PubMed] [Google Scholar]
- 152.Preskorn SH, Kane CP, Lobello K, et al. Cytochrome P450 2D6 phenoconversion is common in patients being treated for depression: implications for personalized medicine. J Clin Psychiatry 2013. Jun;74(6):614–21. [DOI] [PubMed] [Google Scholar]
- 153.Ferraldeschi R, Newman WG. The Impact of CYP2D6 Genotyping on Tamoxifen Treatment. Pharmaceuticals (Basel) 2010. Apr 15;3(4):1122–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kiss Á, Vaskó D, Déri MT, et al. Combination of CYP2C19 genotype with non-genetic factors evoking phenoconversion improves phenotype prediction. Pharmacol Rep 2018. Jun;70(3):525–532. [DOI] [PubMed] [Google Scholar]
- 155.de Souza JA, Olopade OI. CYP2D6 genotyping and tamoxifen: an unfinished story in the quest for personalized medicine. Semin Oncol 2011. Apr;38(2):263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kelly CM, Pritchard KI. CYP2D6 genotype as a marker for benefit of adjuvant tamoxifen in postmenopausal women: lessons learned. J Natl Cancer Inst 2012. Mar 21;104(6):427–8. [DOI] [PubMed] [Google Scholar]
- 157. Thomas CD, Parvataneni HK, Gray CF, et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet Med 2021. 04;23(4):621–628. *Includes CYP2D6 phenoconversion and pharmacogentics while predicting phenotype and compared treatment outcome with usual care group.
- 158. Smith DM, Weitzel KW, Elsey AR, et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med 2019. 08;21(8):1842–1850. *Includes CYP2D6 phenoconversion and pharmacogenetics while predicting phenotype and compares treatment outcome with usual care group.
- 159.Desmeules J, Gascon MP, Dayer P, et al. Impact of environmental and genetic factors on codeine analgesia. Eur J Clin Pharmacol 1991;41(1):23–6. [DOI] [PubMed] [Google Scholar]
- 160.Matos A, Bankes DL, Bain KT, et al. Opioids, Polypharmacy, and Drug Interactions: A Technological Paradigm Shift Is Needed to Ameliorate the Ongoing Opioid Epidemic. Pharmacy (Basel) 2020. Aug 25;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fulton CR, Zang Y, Desta Z, et al. Drug-gene and drug-drug interactions associated with tramadol and codeine therapy in the INGENIOUS trial. Pharmacogenomics 2019. 04;20(6):397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Michaud V, Bikmetov R, Smith MK, et al. Use of Drug Claims Data and a Medication Risk Score to Assess the Impact of CYP2D6 Drug Interactions among Opioid Users on Healthcare Costs. J Pers Med 2021. Nov 10;11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cavallari LH, Cicali E, Wiisanen K, et al. Implementing a pragmatic clinical trial to tailor opioids for acute pain on behalf of the IGNITE ADOPT PGx investigators. Clin Transl Sci 2022. Jul 28. [DOI] [PMC free article] [PubMed]
- 164.Jallaq SA, Verba M, Strawn JR, et al. CYP2D6 Phenotype Influences Aripiprazole Tolerability in Pediatric Patients with Mood Disorders. J Child Adolesc Psychopharmacol 2021. 02;31(1):56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther 2006. Jul;80(1):61–74. [DOI] [PubMed] [Google Scholar]
- 166.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 2003. Dec 03;95(23):1758–64. [DOI] [PubMed] [Google Scholar]
- 167.Haque R, Shi J, Schottinger JE, et al. Tamoxifen and Antidepressant Drug Interaction in a Cohort of 16,887 Breast Cancer Survivors. J Natl Cancer Inst 2016. Mar;108(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kelly CM, Juurlink DN, Gomes T, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ 2010. Feb 08;340:c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jung EH, Lee YJ, Kim DH, et al. Effects of paroxetine on the pharmacokinetics of atomoxetine and its metabolites in different CYP2D6 genotypes. Arch Pharm Res 2020. Dec;43(12):1356–1363. [DOI] [PubMed] [Google Scholar]
- 170.Kim YH, Bae JW, Jeong CH, et al. Drug-Drug Interaction of Paroxetine and Atomoxetine in Different CYP2D6 Genotypes. Clin Ther 2016. Oct 06;38(10S):e23. [DOI] [PubMed] [Google Scholar]
- 171.Paulzen M, Clement HW, Gründer G. Enhancement of atomoxetine serum levels by co-administration of paroxetine. Int J Neuropsychopharmacol 2008. Mar;11(2):289–91. [DOI] [PubMed] [Google Scholar]
- 172.Todor I, Popa A, Neag M, et al. The influence of paroxetine on the pharmacokinetics of atomoxetine and its main metabolite. Clujul Med 2015;88(4):513–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Todor I, Popa A, Neag M, et al. Evaluation of a Potential Metabolism-Mediated Drug-Drug Interaction Between Atomoxetine and Bupropion in Healthy Volunteers. J Pharm Pharm Sci 2016 2016. Apr-Jun;19(2):198–207. [DOI] [PubMed] [Google Scholar]
- 174.Naguy A, Al-Mutairi H, Al-Tajali A. Atomoxetine-related Takotsubo Cardiomyopathy. J Psychiatr Pract 2016. 05;22(3):232–3. [DOI] [PubMed] [Google Scholar]
- 175.cpicpgx.org. https://cpicpgx.org/prioritization-of-cpic-guidelines/ Last accessed on October 11, 2022.
- 176.Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn 2021. 09;23(9):1047–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Cicali EJ, Wiisanen K. The importance of phenoconversion when using the CYP2D6 genotype in clinical practice. Pharmacogenomics 2022. 09;23(14):749–752. **Provides importance and steps of considering phenoconversion to guide prescribing.
- 178.Arwood MJ, Chumnumwat S, Cavallari LH, et al. Implementing Pharmacogenomics at Your Institution: Establishment and Overcoming Implementation Challenges. Clin Transl Sci 2016. Oct;9(5):233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arwood MJ, Dietrich EA, Duong BQ, et al. Design and Early Implementation Successes and Challenges of a Pharmacogenetics Consult Clinic. J Clin Med 2020. Jul 17;9(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cicali EJ, Weitzel KW, Elsey AR, et al. Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene-drug pairs across ambulatory care settings. Genet Med 2019. 10;21(10):2264–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Weitzel KW, Duong BQ, Arwood MJ, et al. A stepwise approach to implementing pharmacogenetic testing in the primary care setting. Pharmacogenomics 2019. 10;20(15):1103–1112. **Provides step by step approach for implementing pharmacogeneitcs in clinical setting.


