Abstract
Objective
To identify FN1 transcripts associated with OA pathophysiology and investigate the downstream effects of modulating FN1 expression and relative transcript ratio.
Methods
FN1 transcriptomic data was obtained from our previously assessed RNA-seq dataset of lesioned and preserved OA cartilage samples from the Research osteoArthritis Articular Cartilage (RAAK) study. Differential transcript expression analysis was performed on all 27 FN1 transcripts annotated in the Ensembl database. Human primary chondrocytes were transduced with lentiviral particles containing short hairpin RNA (shRNA) targeting full-length FN1 transcripts or non-targeting shRNA. Subsequently, matrix deposition was induced in our 3D in vitro neo-cartilage model. Effects of changes in the FN1 transcript ratio on sulphated glycosaminoglycan (sGAG) deposition were investigated by Alcian blue staining and dimethylmethylene blue assay. Moreover, gene expression levels of 17 cartilage-relevant markers were determined by reverse transcription quantitative polymerase chain reaction.
Results
We identified 16 FN1 transcripts differentially expressed between lesioned and preserved cartilage. FN1-208, encoding migration-stimulating factor, was the most significantly differentially expressed protein coding transcript. Downregulation of full-length FN1 and a concomitant increased FN1-208 ratio resulted in decreased sGAG deposition as well as decreased ACAN and COL2A1 and increased ADAMTS-5, ITGB1 and ITGB5 gene expression levels.
Conclusion
We show that full-length FN1 downregulation and concomitant relative FN1-208 upregulation was unbeneficial for deposition of cartilage matrix, likely due to decreased availability of the classical RGD (Arg-Gly-Asp) integrin-binding site of fibronectin.
Keywords: OA, cartilage, fibronectin, FN1, alternative splicing, chondrogenesis, migration-stimulating factor
Rheumatology key messages.
The truncated FN1 transcript encoding migration-stimulating factor was most significantly upregulated in lesioned vs preserved OA cartilage.
Downregulation of full-length FN1 is unbeneficial for deposition of cartilage matrix.
Introduction
Currently, OA is the most prevalent degenerative joint disease worldwide, associated with a high societal and economic burden [1]. A general hallmark of OA is the degeneration of articular cartilage [2, 3]. To date, no effective treatment to reverse or slow down the disease is available, except pain relief medication and joint replacement surgery. Therefore, more insight into the underlying pathophysiology of OA is necessary for the development of druggable targets.
In this regard, transcriptome-wide analyses of cartilage have been performed to identify underlying biological mechanisms driving OA [4–6]. Specifically, among the highest expressed and significantly upregulated genes in affected OA cartilage is fibronectin (FN1) [4, 6]. FN1 encodes a high molecular weight, dimeric glycoprotein that in articular cartilage is deposited by chondrocytes and mostly localized in the pericellular matrix directly surrounding the chondrocytes [7]. Fibronectin mediates a wide variety of cellular interactions with the extracellular matrix (ECM) by binding to matrix proteins via multiple binding domains, as well as interactions with chondrocytes via integrins that mediate intracellular signalling. The main integrin-binding domain of fibronectin is the RGD motif, which binds multiple integrin heterodimers, including the classic fibronectin receptor integrin α5β1 [8]. Recently it has been shown that fibronectin–α5β1 adhesion is critical for cartilage regeneration in mice [9]. More recently, our group identified a high-impact mutation in the gelatin-binding domain of FN1 in an early-onset OA family, resulting in decreased binding capacity of fibronectin to collagen type II [10]. Furthermore, fibronectin can be degraded by proteases and the resulting fibronectin fragments are known to amplify catabolic processes in the articular cartilage [11, 12]. Taken together, these studies show that proper binding of fibronectin to both ECM components and integrins is important for cartilage homeostasis.
The fibronectin gene is known to give rise to 20 different protein coding transcripts by virtue of alternative splicing as well as multiple non-protein coding transcripts [13]. Alternative splicing occurs at three major sites, called extra domain A (EDA), extra domain B (EDB) and variable region (V) [14, 15]. Splicing at the EDA and EDB domains results in inclusion or exclusion of one exon, whereas splicing at the V region can occur at multiple splice sites [14]. This splicing variation provides cells with the capacity to generate large numbers of protein isoforms with different binding properties to precisely alter the ECM composition in a developmental and tissue-specific manner. As a result, each isoform has a unique function in cell–ECM interactions [8]. Among FN1 splice variants is the intact 70 kDa N-terminus of the full length protein, also known as migration stimulating factor (MSF) [16]. MSF includes the heparin- and gelatin-binding domains, but does not have the classical RGD integrin-binding domain. Previously, EDB+, EDB−, EDA− and V+ transcript variants were shown to be present in multiple joint tissues, while EDA+ transcript variants were rarely detected [17].
It is still unknown which specific FN1 transcripts may be involved in the response to a healthy or disease state of cartilage, as well as what the effect of changed FN1 expression is in OA cartilage. Therefore we aimed to identify FN1 transcripts associated with the OA process by assessing our previously published RNA-seq dataset with paired lesioned and preserved OA cartilage samples [4]. Subsequently the downstream effects of modulating FN1 expression and the transcript ratio was investigated in our established human 3D in vitro OA cartilage model.
Materials and methods
Sample description
Macroscopically lesioned and preserved articular cartilage samples were obtained from patients who underwent joint replacement surgery due to OA in the Research Osteoarthritis and Articular Cartilage (RAAK) study, as described previously [18]. The RAAK study was approved by the Medical Ethics Committee of the Leiden University Medical Center (P08.239/P19.013) and written informed consent was obtained from all participants. In the current study, previously assessed RNA-seq data of 101 cartilage samples were used, of which 35 were paired samples between lesioned and preserved (25 knees, 7 hips) [4]. The replication cohort consisted of an additional 10 paired cartilage samples (5 knees, 5 hips). Primary articular chondrocytes obtained from knee replacement surgeries of six participants in the RAAK study were isolated and cultured to perform lentiviral transduction. For all sample characteristics, see Supplementary Table S1, available at Rheumatology online.
RNA sequencing
Total RNA isolation from articular cartilage, sequencing and quality control was performed as previously described [4]. Detailed information on the alignment, mapping and filtering is available in the Supplementary methods, available at Rheumatology online.
Differential expression analysis and replication
Differential expression analysis was performed between lesioned and preserved OA cartilage samples. Results were validated by means of visualizing exon count data and replicated by means of reverse transcription quantitative polymerase chain reaction (RT-qPCR). More detailed information is available in the Supplementary methods, available at Rheumatology online.
Lentiviral production and cell culture
For knockdown experiments, the pLKO.1-puro vector from the Sigma-Aldrich Mission short hairpin RNA (shRNA) library targeting all full-length protein coding transcripts of FN1 (TRCN0000286357) and non-targeting control virus particles (SHC002) were kindly provided by Martijn Rabelink (Department of Cell and Chemical Biology, Leiden University Medical Center, Leiden, The Netherlands). Detailed information on the lentiviral production and chondrocyte cell culture and transduction is available in the Supplementary methods, available at Rheumatology online. In vitro chondrogenesis was induced as previously described [18]. Neo-cartilage pellets and medium were collected following 3 days of chondrogenesis.
ELISA
Culture medium of neo-cartilage pellets of primary chondrocytes transduced with non-targeting shRNA (control) and FN1 targeting shRNA was collected following 3 days of chondrogenesis. Fibronectin concentration was determined using the Human Fibronectin ELISA kit (Thermo Fisher Scientific, Vienna, Austria) according to the manufacturer’s protocol. The absorbance was measured at 450 nm in a microplate reader (Spectramax iD3, Molecular Devices, San Jose, CA, USA).
Histology and immunohistochemistry
Neo-cartilage pellets were fixed in 4% formaldehyde overnight and stored in 70% ethyl alcohol at 4°C. Detailed information on histology and immunohistochemistry is available in the Supplementary methods, available at Rheumatology online.
RNA isolation and relative gene expression analysis
Two neo-cartilage pellets were pooled in 200 µl Trizol reagent (Thermo Fisher Scientific, Carlsbad, CA, USA) and homogenized using micropestles. Further details on RNA isolation and gene expression analysis is available in the Supplementary methods, available at Rheumatology online. Primer sequences are shown in Supplementary Table S2, available at Rheumatology online.
Sulphated glycosaminoglycan (sGAG) measurement
The sGAG content in the neo-cartilage pellets was measured with the 1,9-dimethylmethylene blue (DMMB) assay [19]. Pellets were digested with 200 µl Papain from papaya (Sigma Aldrich, Zwijndrecht, The Netherlands) at 60°C overnight. Shark chondroitin sulphate (Sigma Aldrich, Zwijndrecht, The Netherlands) was used as a reference standard. The absorbance was measured at 525 and 595 nm in a microplate reader (Spectramax iD3, Molecular Devices, San Jose, CA, USA).
FN1 downstream interactions
To identify potential new FN1 downstream interactions, Pearson correlations were calculated between FN1 and the previously reported significantly differentially expressed genes (n = 2387) [4] in the same lesioned (n = 44) and preserved (n = 57) OA cartilage samples (Supplementary Table S1D, available at Rheumatology online). Genes with r ≥ 0.8 were analysed for enrichment in protein–protein interactions with the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, version 11.0) [20].
Statistical analyses
Statistical analyses were performed using SPSS version 25 (IBM, Armonk, NY, USA). Data are either shown as mean (s.d.) or boxplots representing the 25th, 50th and 75th percentiles and whiskers representing the lowest and highest data point lying within 1.5 times the interquartile range. Individual samples are depicted by dots in each boxplot. The reported P-values of the lentiviral experiments were determined by applying generalized estimating equations (GEEs) to the experimental readout to adjust for dependencies of the different donors [21]. We followed a linear GEE model, with the readout data as a dependent variable and group and donor as covariate and exchangeable working matrix: Readout ∼ Group + (1|Donor). P-values <0.05 were considered statistically significant.
Data availability
The processed dataset generated and the code to reproduce the differential expression analysis is available from https://git.lumc.nl/mvanhoolwerff/fn1-transcripts.
Results
Characterization of FN1 transcripts in OA cartilage
To characterize the FN1 transcripts in cartilage we used our previously assessed RNA-seq data on 35 paired samples (28 knees, 7 hips) of lesioned and preserved OA cartilage (Supplementary Table S1A, available at Rheumatology online) [4]. Our in-house pipeline was applied to obtain transcriptomic FN1 data from the Ensembl database, which has 27 FN1 transcripts annotated (Supplementary Fig. S1, available at Rheumatology online). To robustly detect the FN1 transcripts expressed in OA cartilage, a cut-off was applied in the lesioned and preserved cartilage samples separately of an average of ≥10 counts per three samples. As a result, 22 transcripts were found to be expressed in cartilage, represented as the mean of normalized counts corrected for transcript length and sequencing depth, as shown in Table 1. Notably, the highest expressed protein coding transcripts were full-length transcripts FN1-211 (base mean counts = 430 585.6; quartile 4), which is EDB−, EDA− and V+, and FN1-209 (base mean counts = 120 949.2; quartile 4), which is EDB+, EDA− and V+. Expression levels of FN1-211 and FN1-209 represent 39.4% and 21.4%, respectively, of total base mean counts, indicating that these were the main protein coding FN1 transcripts transcribed in OA cartilage. Moreover, among the top 10 highest expressed transcripts, 2 are classified as retained introns and therefore not protein coding, namely FN1-227 (base mean counts = 50 673.0; quartile 4) and FN1-225 (base mean counts = 33 536.1; quartile 3), suggesting that these non-protein coding transcripts may be functional in OA cartilage.
Table 1.
Expression levels of the 22 FN1 transcripts that were robustly expressed in lesioned and preserved osteoarthritic cartilage samples
| Ensembl ID | Name | Biotype | Base mean | Quartile | EDA | EDB | V |
|---|---|---|---|---|---|---|---|
| ENST00000443816.5 | FN1-211 | Protein coding | 430 585.6 | 4 | No | No | Yes |
| ENST00000432072.6 | FN1-209 | Protein coding | 234 116.5 | 4 | No | Yes | No |
| ENST00000456923.5 | FN1-213 | Protein coding | 113 350.0 | 4 | No | Yes | Yes |
| ENST00000356005.8 | FN1-204 | Protein coding | 112 854.7 | 4 | No | No | Yes |
| ENST00000498719.1 | FN1-227 | Retained intron | 50 673.0 | 4 | |||
| ENST00000446046.5 | FN1-212 | Protein coding | 37 427.2 | 3 | Yes | No | Yes |
| ENST00000494446.1 | FN1-225 | Retained intron | 33 536.1 | 3 | |||
| ENST00000421182.5 | FN1-207 | Protein coding | 30 469.0 | 3 | No | No | Yes |
| ENST00000357867.8 | FN1-205 | Protein coding | 23 405.6 | 3 | No | No | No |
| ENST00000426059.1 | FN1-208 | Protein coding | 16 523.2 | 3 | No | No | No |
| ENST00000438981.1 | FN1-210 | Protein coding | 3472.7 | 2 | No | No | Yes |
| ENST00000461974.1 | FN1-215 | Retained intron | 2163.2 | 2 | |||
| ENST00000492816.6 | FN1-224 | Retained intron | 2028.0 | 2 | |||
| ENST00000471193.1 | FN1-217 | Retained intron | 903.3 | 2 | |||
| ENST00000460217.1 | FN1-214 | Retained intron | 447.5 | 2 | |||
| ENST00000480024.1 | FN1-220 | Retained intron | 277.2 | 2 | |||
| ENST00000473614.1 | FN1-218 | Retained intron | 155.2 | 1 | |||
| ENST00000496542.1 | FN1-226 | Retained intron | 87.6 | 1 | |||
| ENST00000474036.1 | FN1-219 | Retained intron | 68.0 | 1 | |||
| ENST00000469569.1 | FN1-216 | Retained intron | 41.5 | 1 | |||
| ENST00000480737.1 | FN1-221 | Retained intron | 38.8 | 1 | |||
| ENST00000490833.5 | FN1-223 | Processed transcript | 8.8 | 1 |
The splice variant of the protein coding transcripts is indicated.
Base mean: mean of normalized counts of all samples normalized for transcript length and sequencing depth; Quartile: expression in quartiles, with 1 being the lowest and 4 being the highest.
Differential expression of FN1 transcripts between lesioned and preserved OA cartilage
To identify specific FN1 transcripts associated with the OA process, differential expression analysis was performed on 22 previously identified expressed FN1 transcripts in paired lesioned and preserved OA cartilage samples, resulting in 16 significantly upregulated FN1 transcripts [false discovery rate (FDR) <0.05; Table 2]. The most significantly upregulated transcript was FN1-220 [fold change (FC) 2.8, FDR = 5.8 × 10−13), which is classified as a retained intron, but had relatively low expression levels (base mean counts = 277.2; quartile 2). Notably, the most significant upregulated protein coding transcript was FN1-208 (FC = 2.3, FDR = 4.9 × 10−6; Supplementary Fig. S2, available at Rheumatology online), which encodes migration stimulating factor. This truncated fibronectin protein contains the heparin- and gelatin-binding domain of fibronectin, but not the integrin-binding domain, and has been identified as a potent motogenic factor yet has not been previously identified in OA cartilage. To validate the differential expression, we analysed FN1 exon count data using DEXSeq, which separates the abundance of exons and parts of exons that are not the same for all transcripts in counting bins. We observed that the exon specific for FN1-208 was higher expressed in lesioned OA cartilage compared with preserved, consequently validating its differential expression (Supplementary Fig. S3, available at Rheumatology online). To replicate the identified upregulation of FN1-208 in lesioned cartilage, we performed RT-qPCR for FN1-208 in an independent cohort consisting of 10 paired cartilage samples (Supplementary Table S1B, available at Rheumatology online). The transcript was detected in all samples and showed significant upregulation (FC = 2.0, P = 9.2 × 10−3; Supplementary Fig. S4, available at Rheumatology online).
Table 2.
FDR significant differentially expressed FN1 transcripts between lesioned and preserved osteoarthritic cartilage samples
| Ensembl ID | Name | Biotype | Quartile | FC | FDR | EDA | EDB | V |
|---|---|---|---|---|---|---|---|---|
| ENST00000480024.1 | FN1-220 | Retained intron | 2 | 2.8 | 5.8 × 10−13 | |||
| ENST00000494446.1 | FN1-225 | Retained intron | 3 | 2.4 | 4.4 × 10−8 | |||
| ENST00000473614.1 | FN1-218 | Retained intron | 1 | 2.3 | 1.7 × 10−7 | |||
| ENST00000492816.6 | FN1-224 | Retained intron | 2 | 2.4 | 3.3 × 10−6 | |||
| ENST00000426059.1 | FN1-208 | Protein coding | 3 | 2.3 | 4.9 × 10−6 | No | No | No |
| ENST00000496542.1 | FN1-226 | Retained intron | 1 | 2.9 | 5.5 × 10−6 | |||
| ENST00000421182.5 | FN1-207 | Protein coding | 3 | 2.6 | 5.4 × 10−4 | No | No | Yes |
| ENST00000490833.5 | FN1-223 | Processed transcript | 1 | 2.3 | 1.1 × 10−3 | |||
| ENST00000460217.1 | FN1-214 | Retained intron | 2 | 2.2 | 2.1 × 10−3 | |||
| ENST00000471193.1 | FN1-217 | Retained intron | 2 | 1.8 | 2.1 × 10−3 | |||
| ENST00000498719.1 | FN1-227 | Retained intron | 4 | 2.3 | 5.2 × 10−3 | |||
| ENST00000469569.1 | FN1-216 | Retained intron | 1 | 1.7 | 1.5 × 10−2 | |||
| ENST00000461974.1 | FN1-215 | Retained intron | 2 | 1.8 | 1.7 × 10−2 | |||
| ENST00000446046.5 | FN1-212 | Protein coding | 3 | 2.1 | 3.0 × 10−2 | Yes | No | Yes |
| ENST00000356005.8 | FN1-204 | Protein coding | 4 | 2.0 | 5.0 × 10−2 | No | No | Yes |
| ENST00000432072.6 | FN1-209 | Protein coding | 4 | 2.1 | 5.0 × 10−2 | No | Yes | No |
The splice variant of the protein coding transcripts is indicated.
Quartile: expression in quartiles, with 1 being the lowest and 4 being the highest.
To explore whether joint-specific FN1 transcripts could be identified, we performed stratified analyses for knee (28 pairs) and hip samples (7 pairs). After filtering, we identified 22 transcripts to be robustly expressed in knee OA cartilage samples, while 19 transcripts were robustly expressed in hip samples. This difference could be partly explained by the lower number of hip samples in the analysis. Upon performing differential expression analysis on the knee samples, we identified 16 significantly differentially expressed FN1 transcripts (Supplementary Table S3, available at Rheumatology online). In the hip samples, we also identified 16 significantly differentially expressed FN1 transcripts (Supplementary Table S4, available at Rheumatology online). Notably, we identified one differentially expressed FN1 transcript (ENST00000490833.5, FN1-223) only present in the knee stratified analysis. However, its expression was very low (base mean 8.8, quartile 1); as a result, it was likely removed in the hip analysis due to filtering. Furthermore, we identified one transcript (ENST00000357867.8, FN1-205) that was only significantly upregulated in the hip stratified analysis. However, upon closer inspection this transcript was upregulated in the knee samples, but with a P-value slightly more than the cut-off value of 0.05, namely 0.051. Consequently we could not identify joint-specific FN1 transcripts in OA cartilage.
Effect of modulation of FN1 and FN1-208 ratio levels on matrix deposition
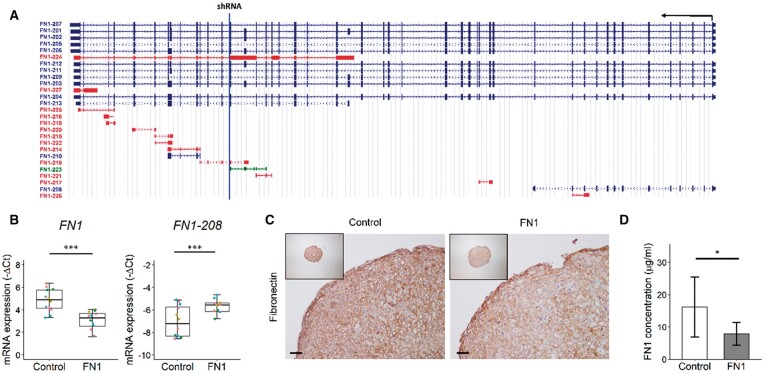
Next, we aimed to study downstream effects of observed FN1 transcript changes, including FN1-208, in our established human 3D in vitro neo-cartilage model. Since overall FN1 expression is inherently high in cartilage, we could not obtain big changes by overexpressing FN1-208 (Supplementary Fig. S5, available at Rheumatology online), therefore we aimed to downregulate full-length FN1. To this end, primary chondrocytes were lentivirally transduced with FN1 targeting shRNA, as depicted in Fig. 1A. The shRNA targets all full-length protein coding FN1 transcripts but not FN1-208. Human primary chondrocytes of six donors were transduced (Supplementary Table S1C, available at Rheumatology online), after which in vitro chondrogenesis was induced in 3D pellet culture for 3 days. Consequently we observed a downregulation of the full-length transcripts (FN1; FC = 0.3, P = 7.6 × 10−7), but since FN1-208 is not targeted, the FN1-208 expression relative to full-length transcripts was increased (FC = 3.5, P = 6.0 × 10−6; Fig. 1B), thereby mimicking lesioned OA cartilage status. The overall downregulation of fibronectin was also observed at the protein level, both in the neo-cartilage pellets (Fig. 1C) and culture medium (Fig. 1D). The fibronectin concentration was decreased 50% in the FN1 group (P = 1.0 × 10−2) compared with the control group, as determined by ELISA.
Fig. 1.
Downregulation of FN1 gene and protein expression in neo-cartilage pellets after 3 days of chondrogenesis
(A) Schematic representation of FN1 transcripts, which are transcribed from the antisense strand, represented by the black arrow. The blue line represents the location of the target sequence of the shRNA targeting FN1 transcripts. Blue transcripts are protein coding, red and green transcripts are non-protein coding. Source: https://genome.ucsc.edu/. (B) Gene expression levels depicted by boxplots of −ΔCt values of FN1 and FN1-208 ratio relative to all full-length FN1 transcripts in neo-cartilage pellets of primary chondrocytes transduced with non-targeting shRNA (control) and FN1 targeting shRNA (FN1). Individual samples are represented by coloured dots; colours of dots represent the different donors (n = 12). (C) Representative images of fibronectin staining of control and FN1 downregulated pellets, confirming FN1 downregulation on the protein level. Scale bar = 50 µm. (D) Fibronectin concentration in conditioned medium in the control (n = 6) and FN1 group (n = 5), as determined by ELISA. Data are mean (s.d.). P-values were determined by GEEs, with experimental readout as the dependent variable and donor and group as covariates. *P < 0.05, ***P < 0.005. Colour version is available at Rheumatology online.
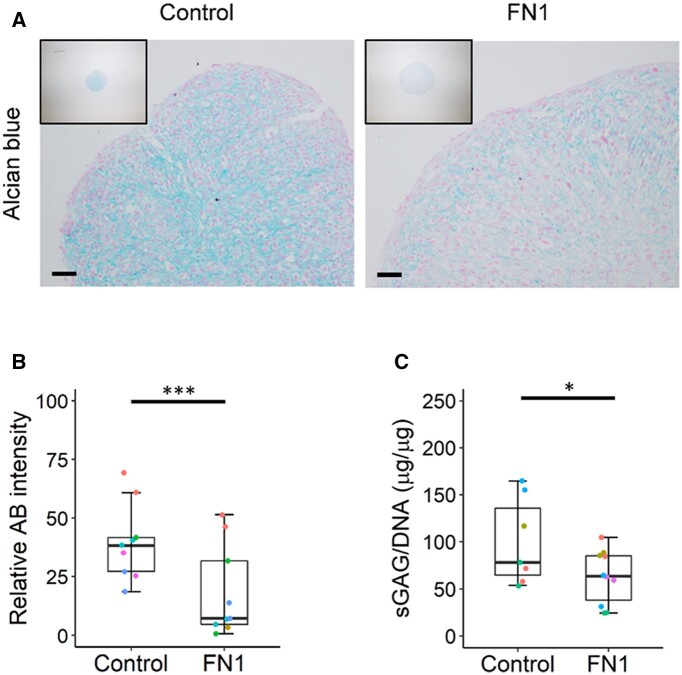
Subsequently the effect of FN1 downregulation on neo-cartilage deposition was investigated by Alcian blue staining, where we observed a decreased deposition of sGAG in the FN1 downregulated pellets (Fig. 2A). Quantification of the Alcian blue staining showed this decreased matrix deposition was 54% (P = 1.3 × 10−9; Fig. 2B). Furthermore, quantification of sGAG content normalized to DNA with the DMMB assay confirmed there was a significant decrease (P = 2.6 × 10−2) in the FN1 downregulated group compared with controls (Fig. 2C). These data imply that FN1 downregulation and concomitant relative upregulation of FN1-208 have a negative effect on neo-cartilage deposition.
Fig. 2.
Decreased overall FN1 expression and change of FN1 transcript ratios results in decreased matrix deposition
(A) Representative images of Alcian blue staining of neo-cartilage pellets of primary chondrocytes transduced with non-targeting shRNA (control) and FN1 targeting shRNA (FN1). (B) Quantification of Alcian blue (AB) pixel intensity staining of control and FN1 targeting shRNA transduced pellets (n = 9). Colours of dots represent the different donors. (C) sGAG content normalized to DNA content in pellets of the control (n = 7) and FN1 group (n = 11) analysed by dimethylmethylene blue assay. P-values were determined by GEEs, with experimental readout as the dependent variable and donor and group as covariates. *P < 0.05, ***P < 0.005. Colour version is available at Rheumatology online.
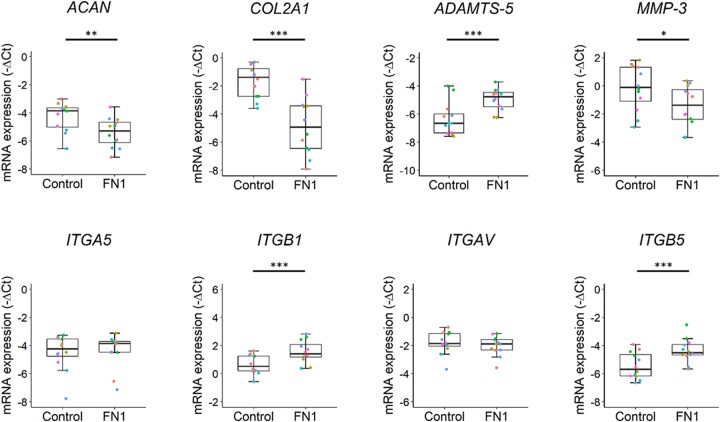
To investigate the effect of changes in FN1 transcript ratios on gene expression, RT-qPCR was performed on 17 cartilage-relevant genes (Supplementary Table S5, available at Rheumatology online). As shown in Fig. 3, both ACAN (FC = 0.5, P = 5.7 × 10−3) and COL2A1 (FC = 0.1, P = 7.0 × 10−7) were strongly downregulated in the FN1 group compared with controls. Moreover, ADAMTS-5 was significantly upregulated (FC = 2.6, P = 9.0 × 10−6), while MMP-3 was significantly downregulated (FC = 0.4, P = 2.9 × 10−2). These data imply that decreased FN1 expression results in a more catabolic response of the chondrocytes via ADAMTS-5. Subsequently we aimed to investigate the downstream effects of FN1 downregulation on the fibronectin-binding chondrocyte transmembrane integrin receptors. Gene expression levels of ITGB1 (FC = 2.2, P = 1.7 × 10−4) and ITGB5 (FC = 2.9, P = 1.9 × 10−4) were significantly upregulated in the FN1 group compared with controls (Fig. 3).
Fig. 3.
Decreased FN1 expression and change of FN1 transcript ratios results in catabolic chondrocyte metabolism.
Boxplots of −ΔCt values of cartilage matrix–relevant genes ACAN, COL2A1, ADAMTS-5, MMP-3, ITGA5, ITGB1, ITGAV and ITGB5 in neo-cartilage pellets of primary chondrocytes transduced with non-targeting shRNA (control) (n = 12) and FN1 targeting shRNA (FN1) (n = 12). Individual samples are represented by coloured dots; colours of dots represent the different donors. P-values were determined by GEEs, with experimental readout as the dependent variable and donor and group as covariates. *P < 0.05, **P < 0.01, ***P < 0.005. Colour version is available at Rheumatology online.
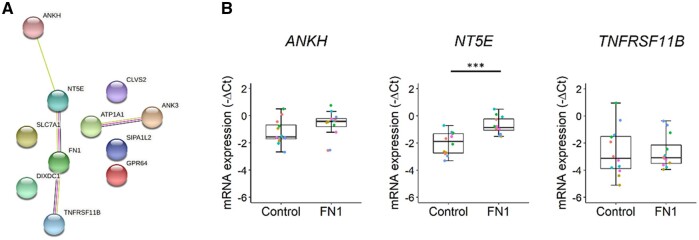
Finally, we aimed to identify novel FN1 downstream pathways in OA cartilage. To this end we calculated correlations between our previously reported differentially expressed genes (n = 2378) and FN1 in lesioned and preserved OA cartilage (n = 101) samples (Supplementary Table S1D, available at Rheumatology online) [4]. As a result, we found 60 genes to be highly correlated (r > 0.7) (Supplementary Table S6, available at Rheumatology online). Pathway enrichment analysis between these highly correlating genes (r > 0.7) and FN1 using STRING resulted in five FDR significantly enriched Gene Ontology (GO) terms: cell surface (GO 0009986), plasma membrane (GO 0005886), membrane (GO 0016020), vesicle (GO 0031982) and intrinsic component of membrane (GO 0031224) (Supplementary Table S7, available at Rheumatology online). These processes are mainly characterized by ITGA5, NT5E, BCAM, CD55 and CD109, indicating the relation of fibronectin with basic cellular processes such as cell adhesion and cell growth. Of the 60 highly correlated genes, 10 showed correlations >0.8. When analysing for interaction among these 10 genes and FN1, 3 genes were either directly or indirectly connected to FN1, namely ANKH, NT5E and TNFRSF11B (Fig. 4A). As shown in Fig. 4B, only NT5E (FC = 2.5, P = 3.0 × 10−6) was significantly differentially expressed in the FN1 group compared with controls.
Fig. 4.
Identification of new FN1 downstream genes.
(A) Protein–protein interactions between the genes with correlations r > 0.8 with FN1 in preserved and lesioned OA cartilage samples, as determined with STRING. (B) Boxplots of −ΔCt values of connected genes to FN1, ANKH, NT5E and TNFRSF11B in neo-cartilage pellets of primary chondrocytes transduced with non-targeting shRNA (control) (n = 12) and FN1 targeting shRNA (FN1) (n = 12). Individual samples are represented by coloured dots; colours of dots represent the different donors. P-values were determined by GEEs, with experimental readout as the dependent variable and donor and group as covariates. ***P < 0.005. Colour version is available at Rheumatology online.
Discussion
To the best of our knowledge, we are the first to use RNA sequencing to characterize the FN1 transcriptome in OA cartilage. As a result, we identified 16 FN1 transcripts FDR significantly upregulated in lesioned OA cartilage, of which 5 were protein coding and 11 non-protein coding. These results show that considerable changes occur in the FN1 transcriptome during OA, likely affecting proper function. Moreover, we identified the truncated protein coding transcript FN1-208 as significantly upregulated in lesioned OA cartilage. Upon downregulation of full-length FN1 in our human 3D in vitro OA cartilage model, we generated an increased ratio of FN1-208 relative to the full-length FN1 transcripts, as such mimicking cartilage in an OA-affected state. This resulted in decreased cartilage deposition compared with the control group, with upregulation of the β1 and β5 integrin subunits, suggesting a change of integrin heterodimers. Together, our results show that downregulation of full-length FN1 is unbeneficial for neo-cartilage deposition, while also highlighting the importance of balance of FN1 transcripts for healthy cartilage homeostasis.
We identified FN1-208, known as MSF, as the most significantly upregulated protein coding transcript, which has not been previously associated with OA. MSF contains the functional heparin- and gelatin-binding domain of full-length fibronectin and cannot bind to transmembrane integrins via the classical RGD binding site. As a result, MSF has distinctive bioactivity compared with full-length fibronectin. MSF has been shown to be a potent motogenic factor and has been associated with cancer pathogenesis as a potential driver of tumour progression by inducing angiogenesis [16, 22]. Furthermore, blocking of MSF suppressed tumour growth through inhibition of tumour-related angiogenesis in an in vitro oesophageal cancer model [23]. Angiogenesis contributes to OA pathology, as blood vessels from the subchondral bone invade the articular cartilage, thereby disrupting homeostasis of the chondrocytes. Therefore we aimed to study whether the identified upregulation of FN1-208 is beneficial or unbeneficial to the OA process in cartilage. Since FN1 is already highly expressed in our established in vitro neo-cartilage model, we could not obtain a significant upregulation of FN1-208. Therefore we downregulated full-length FN1 expression in our model and as a result we obtained an increased FN1-208 ratio, thus mimicking an OA-related upregulation. Consequently, in our study design we cannot distinguish between the effect of downregulating FN1 and increasing relative amounts of the OA-sensitive transcript FN1-208. Nonetheless, the shift in FN1-208 ratio relative to the total protein coding transcripts resulted in decreased neo-cartilage deposition, as well as a catabolic state of the chondrocytes. Moreover, we observed upregulation of ITGB1 and ITGB5 gene expression levels. It has been shown that the β1 integrin is upregulated in osteoarthritic compared with normal cartilage [24]. Therefore the upregulation of ITGB1 and ITGB5 may represent a higher disease state of the chondrocytes in the FN1 downregulated pellets. Together, these data show that decreased availability of the classical integrin-binding site of fibronectin to the cells is detrimental for chondrogenesis, which is likely mediated via β1 and β5 integrin subunits. However, this should be confirmed by quantifying protein expression of the integrin subunits, e.g. by western blot. Furthermore, investigating changes in integrin downstream signalling can shed light on the effects of ITGB1 and ITGB5 upregulation. The retained intron transcripts FN1-225 and FN1-227 were relatively highly expressed and significantly differentially expressed between lesioned and preserved OA cartilage, suggesting they may play a role in OA pathophysiology. Intron retention as an alternative splicing mechanism has recently been getting more attention regarding potential regulatory functions as opposed to being merely a consequence of mis-splicing [25]. Intron retention is mostly associated with downregulation of gene expression via nonsense-mediated decay of the intron-retaining transcript, which has been shown to be a physiological mechanism of gene expression control regulating granulocyte differentiation [26]. However, intron retention has been suggested to potentially regulate non-coding RNAs contained within such introns [27]. Future studies regarding the function of retained intron FN1 transcripts should address whether they regulate gene expression levels of the protein coding FN1 transcripts, e.g. via expression or regulation of non-coding RNAs such as micro-RNAs or long non-coding RNAs.
Previously Scanzello et al. [17] investigated fibronectin splice variants in joint tissues, including cartilage. EDB+, EDB−, and EDA− variants were found to be present in cartilage, while EDA+ variants were barely detected, as determined by RT-PCR. In line with these observations, we identified FN1-211 and FN1-209 to be the highest expressed transcripts in OA cartilage, which are both EDA−. The only EDA+ transcript that we identified to be robustly expressed in OA cartilage was FN1-212 (base mean count = 37 427.2; quartile 3), which represented only 3.4% of the total transcripts. The EDA domain has been associated with many of the functions ascribed to fibronectin, including cell adhesion, matrix assembly and dimer formation [13, 28]. However, given its low expression, our results suggest that the EDA domain is not essential for proper functioning of fibronectin in cartilage. On a different note, we observed FN1-213 to be highly expressed (base mean count = 113 350.0; quartile 4), which is a 5′-truncated transcript, that has not been previously identified in cartilage.
All in all, we showed that RNA sequencing is a powerful technique for identifying involvement of the known FN1 transcripts in OA cartilage. However, the previously identified cartilage-specific (V+III−15+I−10)−, (V+I−10)− and (V+III−15)− variants were not present in the Ensembl database and were therefore not detected in our analysis [29, 30]. To circumvent this issue, de novo transcriptome assembly could be performed. Nonetheless, in the current study we prioritized reporting previously unknown FN1 transcripts present in the Ensembl database involved in OA pathophysiology as opposed to reporting on previously identified FN1 transcripts. A drawback of our study design is that we only investigated end-stage OA cartilage. Consequently, we cannot identify FN1 transcripts that are specific for OA cartilage compared with healthy cartilage and thereby potentially involved in the early phase of OA pathophysiology. Nonetheless, our paired analysis allows for identification of FN1 transcripts specific to the pathophysiologic process of OA, independent of confounding factors such as sex and age.
To explore potential FN1 downstream pathways, correlations were calculated between FN1 and differentially expressed genes in OA cartilage. Among the highest correlated genes, three genes were interconnected in a protein network. Among these three genes we identified NT5E as a novel FN1 downstream signalling gene in cartilage. NT5E encodes the protein 5′-nucleotidase, also known as CD73, which is a plasma protein that catalyses the conversion of extracellular nucleotides to membrane-permeable nucleosides and is a marker for mesenchymal stromal cells [31]. In addition to the enzymatic function, CD73 also functions as a receptor molecule that can interact with ECM components [32]. Defects in NT5E resulting in CD73 deficiency have been shown to facilitate calcification of joints [33, 34], whereas NT5E was significantly upregulated between lesioned and preserved OA cartilage [4]. These data imply that its upregulation as a result of FN1 downregulation is a compensatory mechanism as a response to the OA disease state.
In conclusion, we identified multiple novel FN1 transcripts associated with OA pathophysiology while showing the potential role of FN1-208. We show that downregulation of full-length FN1 was unbeneficial for neo-cartilage deposition and resulted in upregulation of integrin β1 and β5 expression levels, likely via decreased availability of the classical RGD integrin-binding site of fibronectin.
Supplementary Material
Acknowledgements
We thank all the participants of the RAAK study (supported by Leiden University Medical Center). We thank all the members of the MolEpi Osteoarthritis group for valuable discussion and feedback. We also thank Demiën Broekhuis, Robert van der Wal, Anika Rabelink-Hoogenstraaten, Peter van Schie, Shaho Hasan, Maartje Meijer, Daisy Latijnhouwers and Geert Spierenburg for collecting the RAAK material. We thank Martijn Rabelink for kindly providing us with the lentiviral shRNA plasmids and virus from the Mission shRNA library and performing the lentiviral p24 ELISA. Data were generated within the scope of the Medical Delta programs Regenerative Medicine 4D: Generating complex tissues with stem cells and printing technology and Improving Mobility with Technology.
Funding: The study was funded by the Dutch Research council/NWO/ZonMW VICI scheme (91816631/528) and Dutch Arthritis Society (grant DAF-16-1-405).
Disclosure statement: The authors have declared no conflicts of interest.
Contributor Information
Marcella van Hoolwerff, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Margo Tuerlings, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Imke J L Wijnen, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
H Eka D Suchiman, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Davy Cats, Sequencing Analysis Support Core.
Hailiang Mei, Sequencing Analysis Support Core.
Rob G H H Nelissen, Department of Orthopaedics, Leiden University Medical Center, Leiden, The Netherlands.
Henrike M J van der Linden–van der Zwaag, Department of Orthopaedics, Leiden University Medical Center, Leiden, The Netherlands.
Yolande F M Ramos, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Rodrigo Coutinho de Almeida, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Ingrid Meulenbelt, Department of Biomedical Data Sciences, Section Molecular Epidemiology.
Data availability statement
The processed dataset generated and the code to reproduce the differential expression analysis are available from https://git.lumc.nl/mvanhoolwerff/fn1-transcripts.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Litwic A, Edwards MH, Dennison EM, Cooper C.. Epidemiology and burden of osteoarthritis. Br Med Bull 2013;105:185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Loeser RF, Goldring SR, Scanzello CR, Goldring MB.. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum 2012;64:1697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldring SR, Goldring MB.. Changes in the osteochondral unit during osteoarthritis: structure, function and cartilage-bone crosstalk. Nat Rev Rheumatol 2016;12:632–44. [DOI] [PubMed] [Google Scholar]
- 4. Coutinho de Almeida R, Ramos YFM, Mahfouz A. et al. RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann Rheum Dis 2019;78:270–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aki T, Hashimoto K, Ogasawara M, Itoi E.. A whole-genome transcriptome analysis of articular chondrocytes in secondary osteoarthritis of the hip. PLoS One 2018;13:e0199734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramos YF, den Hollander W, Bovee JV. et al. Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLoS One 2014;9:e103056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh P, Carraher C, Schwarzbauer JE.. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol 2010;26:397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pankov R, Yamada KM.. Fibronectin at a glance. J Cell Sci 2002;115:3861–3. [DOI] [PubMed] [Google Scholar]
- 9. Almonte-Becerril M, Gimeno LI, Villarroya O. et al. Genetic abrogation of the fibronectin-alpha5beta1 integrin interaction in articular cartilage aggravates osteoarthritis in mice. PLoS One 2018;13:e0198559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Hoolwerff M, Rodriguez Ruiz A, Bouma M. et al. High-impact FN1 mutation decreases chondrogenic potential and affects cartilage deposition via decreased binding to collagen type II. Sci Adv 2021;7:eabg8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Homandberg GA. Potential regulation of cartilage metabolism in osteoarthritis by fibronectin fragments. Front Biosci 1999;4:D713–30. [DOI] [PubMed] [Google Scholar]
- 12. Reed KSM, Ulici V, Kim C. et al. Transcriptional response of human articular chondrocytes treated with fibronectin fragments: an in vitro model of the osteoarthritis phenotype. Osteoarthritis Cartilage 2021;29:235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. White ES, Muro AF.. Fibronectin splice variants: understanding their multiple roles in health and disease using engineered mouse models. IUBMB Life 2011;63:538–46. [DOI] [PubMed] [Google Scholar]
- 14. Schwarzbauer JE, DeSimone DW.. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol 2011;3:a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwarzbauer JE. Alternative splicing of fibronectin: three variants, three functions. Bioessays 1991;13:527–33. [DOI] [PubMed] [Google Scholar]
- 16. Schor SL, Ellis IR, Jones SJ. et al. Migration-stimulating factor: a genetically truncated onco-fetal fibronectin isoform expressed by carcinoma and tumor-associated stromal cells. Cancer Res 2003;63:8827–36. [PubMed] [Google Scholar]
- 17. Scanzello CR, Markova DZ, Chee A. et al. Fibronectin splice variation in human knee cartilage, meniscus and synovial membrane: observations in osteoarthritic knee. J Orthop Res 2015;33:556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bomer N, den Hollander W, Ramos YF. et al. Underlying molecular mechanisms of DIO2 susceptibility in symptomatic osteoarthritis. Ann Rheum Dis 2015;74:1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farndale RW, Buttle DJ, Barrett AJ.. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta 1986;883:173–7. [DOI] [PubMed] [Google Scholar]
- 20. Szklarczyk D, Gable AL, Lyon D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeger SL, Liang KY.. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986;42:121–30. [PubMed] [Google Scholar]
- 22. Schor AM, Schor SL.. Angiogenesis and tumour progression: migration-stimulating factor as a novel target for clinical intervention. Eye (Lond) 2010;24:450–8. [DOI] [PubMed] [Google Scholar]
- 23. Hu H, Ran Y, Zhang Y. et al. Antibody library-based tumor endothelial cells surface proteomic functional screen reveals migration-stimulating factor as an anti-angiogenic target. Mol Cell Proteomics 2009;8:816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loeser RF, Carlson CS, McGee MP.. Expression of β1 integrins by cultured articular chondrocytes and in osteoarthritic cartilage. Exp Cell Res 1995;217:248–57. [DOI] [PubMed] [Google Scholar]
- 25. Jacob AG, Smith CWJ.. Intron retention as a component of regulated gene expression programs. Hum Genet 2017;136:1043–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong JJ, Ritchie W, Ebner OA. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 2013;154:583–95. [DOI] [PubMed] [Google Scholar]
- 27. Wong JJ, Au AY, Ritchie W, Rasko JE.. Intron retention in mRNA: no longer nonsense: known and putative roles of intron retention in normal and disease biology. Bioessays 2016;38:41–9. [DOI] [PubMed] [Google Scholar]
- 28. Manabe R, Oh-e N, Sekiguchi K.. Alternatively spliced EDA segment regulates fibronectin-dependent cell cycle progression and mitogenic signal transduction. J Biol Chem 1999;274:5919–24. [DOI] [PubMed] [Google Scholar]
- 29. Parker AE, Boutell J, Carr A, Maciewicz RA.. Novel cartilage-specific splice variants of fibronectin. Osteoarthritis Cartilage 2002;10:528–34. [DOI] [PubMed] [Google Scholar]
- 30. MacLeod JN, Burton-Wurster N, Gu DN, Lust G.. Fibronectin mRNA splice variant in articular cartilage lacks bases encoding the V, III-15, and I-10 protein segments. J Biol Chem 1996;271:18954–60. [DOI] [PubMed] [Google Scholar]
- 31. Dominici M, Le Blanc K, Mueller I. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006;8:315–7. [DOI] [PubMed] [Google Scholar]
- 32. Andrade CM, Lopez PL, Noronha BT. et al. Ecto-5′-nucleotidase/CD73 knockdown increases cell migration and mRNA level of collagen I in a hepatic stellate cell line. Cell Tissue Res 2011;344:279–86. [DOI] [PubMed] [Google Scholar]
- 33. Ichikawa N, Taniguchi A, Kaneko H. et al. Arterial calcification due to deficiency of CD73 (ACDC) as one of rheumatic diseases associated with periarticular calcification. J Clin Rheumatol 2015;21:216–20. [DOI] [PubMed] [Google Scholar]
- 34. St Hilaire C, Ziegler SG, Markello TC. et al. NT5E mutations and arterial calcifications. N Engl J Med 2011;364:432–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed dataset generated and the code to reproduce the differential expression analysis is available from https://git.lumc.nl/mvanhoolwerff/fn1-transcripts.
The processed dataset generated and the code to reproduce the differential expression analysis are available from https://git.lumc.nl/mvanhoolwerff/fn1-transcripts.