Abstract
Enhancing the intracellular labile iron pool (LIP) represents a powerful, yet untapped strategy for driving ferroptotic death of cancer cells. Here, we show that NRF2 maintains iron homeostasis by controlling HERC2 (E3 ubiquitin ligase for NCOA4 and FBXL5) and VAMP8 (mediates autophagosome-lysosome fusion). NFE2L2/NRF2 knockout cells have low HERC2 expression, leading to a simultaneous increase in ferritin and NCOA4 and recruitment of apoferritin into the autophagosome. NFE2L2/NRF2 knockout cells also have low VAMP8 expression, which leads to ferritinophagy blockage. Therefore, deletion of NFE2L2/NRF2 results in apoferritin accumulation in the autophagosome, an elevated LIP, and enhanced sensitivity to ferroptosis. Concordantly, NRF2 levels correlate with HERC2 and VAMP8 in human ovarian cancer tissues, as well as ferroptosis resistance in a panel of ovarian cancer cell lines. Last, the feasibility of inhibiting NRF2 to increase the LIP and kill cancer cells via ferroptosis was demonstrated in preclinical models, signifying the impact of NRF2 inhibition in cancer treatment.
NRF2 regulates ferritin synthesis and degradation to maintain iron homeostasis and prevent ferroptotic cell death.
INTRODUCTION
The transcription factor nuclear factor erythroid 2–related factor 2 (NFE2L2/NRF2) was originally identified as a master regulator of cellular redox homeostasis and xenobiotic detoxification (1–3). Since then, NRF2 has been shown to regulate genes that are important for proteostasis, the pentose phosphate pathway, and the metabolism of amino acids and carbohydrates (4–9). In normal tissues, NRF2 protein levels are kept low through interaction with KEAP1 (an adaptor for an E3 ubiquitin ligase complex) that targets NRF2 for ubiquitylation and subsequent proteasomal degradation (10–13). Controlled activation of NRF2 in normal cells prevents cancer initiation induced by chemical carcinogens (14). However, recent evidence reveals that an oncogenic activity of NRF2, i.e., high NFE2L2/NRF2 expression, largely due to somatic mutation of Kelch-like ECH-associated protein 1 (KEAP1), promotes tumor progression, metastasis, and resistance to anticancer therapies (15–19). Clinically, high expression of NRF2 in patient tumors strongly correlates with a poor prognosis (20). Therefore, it is important to understand molecular mechanisms by which NRF2 helps cancer cells evade cell death to facilitate the development of innovative therapeutic strategies for effective treatment of cancer.
One hurdle in cancer treatment is acquired resistance, when the cancer cells are gradually desensitized to pro-apoptotic signals. In the hopes of discovering compounds to treat apoptosis-resistant Ras mutant cancer cells, a form of cell death termed ferroptosis was discovered (21, 22). Ferroptosis is an iron- and lipid peroxidation–driven form of cell death that is morphologically distinct from necrosis, apoptosis, and autophagy (22–25). The compounds discovered in this screen to induce ferroptosis, erastin, and RSL-3 were later found to inhibit the system xc-cystine/glutamate antiporter and key lipid peroxide reducing enzyme glutathione (GSH) peroxidase 4 (GPX4), respectively (26, 27). Over the past decade, many ferroptosis inducers [erastin, RSL-3, sorafenib, FIN-56, and sulfasalazine (SAS)] have been used to kill different cancer cell types in vitro (23). Imidazole ketone erastin (IKE), a modified form of erastin, was recently developed for in vivo use and shown to be pharmacologically stable, safe, and efficacious in the different murine cancer models tested (28, 29). Therefore, IKE has the potential to be used clinically to treat patients with aggressive, resistant, and metastatic cancer through ferroptotic cell death (30).
Ferroptosis is an oxidative form of cell death that is triggered when there is excessive lipid peroxidation. Many critical ferroptosis proteins identified, such as solute carrier family 7 member 11 (SLC7A11; a subunit of the cystine/glutamate antiporter xCT) and the catalytic and modifier subunits of glutamate cysteine ligase (GCLC/GCLM), both of which control the level of GSH (a cofactor for GPX4), are well-defined NRF2 target genes (23, 31). Hence, the anti-ferroptotic activity of NRF2 was naturally thought to be derived from its antioxidant target genes (23). However, another essential aspect of ferroptosis is the labile iron pool (LIP), as iron chelation abolishes ferroptotic cell death in cells treated with a ferroptosis inducer (22, 23). Although an increasing number of pro-ferroptotic factors that control lipid peroxidation have been revealed (32–34), information regarding dysregulation of iron homeostasis during ferroptotic cell death is limited. The current study was designed to dissect the detailed molecular mechanisms by which NRF2 protects cancer cells against ferroptotic cell death via mitigation of both lipid peroxidation and free iron. Our results demonstrate that NRF2 controls ferritin synthesis and degradation through HECT and RLD domain containing E3 ubiquitin protein ligase 2 (HERC2), vesicle-associated membrane protein 8 (VAMP8), and nuclear receptor coactivator 4 (NCOA4), thus altering the intracellular LIP and dictating cancer cell susceptibility to ferroptotic cell death. Furthermore, the importance of this role of NRF2 in controlling iron homeostasis to dictate ferroptosis was demonstrated by the fact that HERC2/VAMP8 double knockout (KO) cells behaved equivalently to NFE2L2/NRF2 KO cells. Also, NCOA4 KO in NFE2L2/NRF2 KO cells protected against ferroptotic cell death. These results clearly validate the antioxidant-independent function of NRF2 in mediating the ferroptotic response. We also demonstrated that cancer cells with high NRF2 levels were more resistant to ferroptosis and that NRF2 inhibition sensitized these cancer cells to ferroptosis inducers. Last, using preclinical models, we provide solid evidence that ovarian tumor spheroids, xenografts, and patient-derived tumors can be sensitized to ferroptotic cell death by enhancing toxic free iron accumulation through NRF2 inhibition. Collectively, this study not only offers basic knowledge regarding how NRF2 controls iron homeostasis but also indicates the translational value of inhibiting NRF2 to enhance the LIP and drive cancer cell death through ferroptosis.
RESULTS
NFE2L2/NRF2 deletion sensitizes ovarian cancer cells to ferroptosis
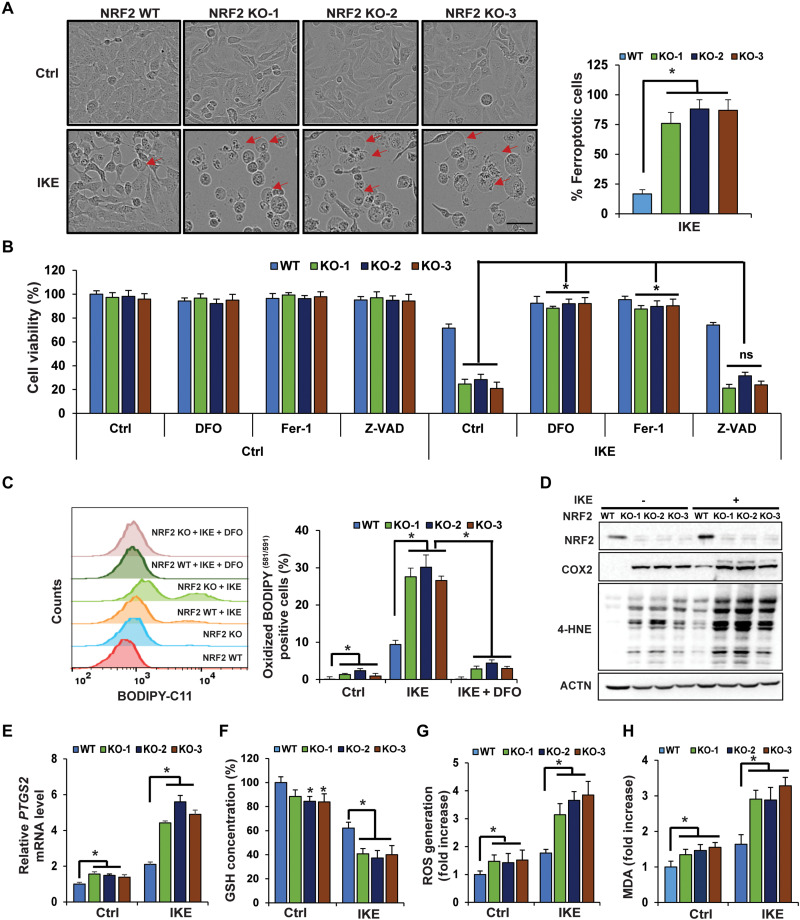
One of the many cancers that have been shown to have high levels of NRF2 is epithelial ovarian cancer, the most lethal gynecological cancer due to its aggressiveness, propensity to develop drug resistance, recurrence, and metastatic potential. Although more than 80% of ovarian cancer patients respond to first-line chemotherapy, the majority will develop recurrent tumors that are drug resistant and fatal. Treatment relapses of ovarian cancer patients continue to represent a major obstacle for its clinical management (35, 36). To determine the role of NRF2 in mediating ovarian cancer cell resistance to ferroptosis, CRISPR-Cas9 gene editing was used to establish NFE2L2/NRF2 KO SKOV-3 ovarian carcinoma cells, and three different NFE2L2/NRF2 KO clones, which were established using two different single-guide RNAs (sgRNAs), were chosen for further study. Successful deletion of NFE2L2/NRF2 was confirmed, with NRF2 protein levels being undetectable in all three KO cell lines, as well as lower mRNA and protein levels of several NRF2 target genes (fig. S1, A and B). Compared to wild type (WT), NFE2L2/NRF2 KO cells treated with IKE, a ferroptosis inducer that inhibits system xc-, exhibited a notable increase in the number of cells undergoing the “ballooning” morphology indicative of ferroptotic cell death (representative images of 24-hour treatment are presented in Fig. 1A: left panel shows ferroptotic cell morphology; right panel is the percentage of ferroptotic cells). IKE induced 18.66% ferroptotic cells in NFE2L2/NRF2 WT cells versus 76% in NFE2L2/NRF2 KO cells (Table 1). Consistent with the pro-ferroptotic change in morphology (Fig. 1A), IKE treatment resulted in an ~30% decrease in cell viability in NFE2L2/NRF2 WT, as compared to an ~75% decrease in NFE2L2/NRF2 KO cells at 24 hours, as measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 1B). Furthermore, IKE-induced ferroptotic cell death was completely abolished by deferoxamine (DFO; an iron chelator), ferrostatin 1 (Fer-1; an inhibitor of lipid peroxidation), but not Z-VAD-fmk (a pan-caspase inhibitor) (Fig. 1B), indicating the importance of free iron accumulation and lipid peroxidation in promoting ferroptosis. Consistent with a ferroptosis-dependent decrease in cell viability, higher levels of lipid peroxides were also observed in IKE-treated NFE2L2/NRF2 KO cells, which could be reversed when the cells were cotreated with DFO (Fig. 1C). Next, more ferroptotic cell death in IKE-treated NFE2L2/NRF2 KO cells compared to NFE2L2/NRF2 WT cells was further confirmed via assessment of key ferroptosis markers, including higher mRNA levels of prostaglandin endoperoxide synthase 2 (PTGS2) and its protein product cyclooxygenase 2 (COX2) (Fig. 1, D and E), as well as increased 4-hydroxynonenal (4-HNE) protein adduct formation (Fig. 1D). In addition, NFE2L2/NRF2 KO cells had decreased total GSH levels, higher levels of reactive oxygen species (ROS), and more malondialdehyde (MDA) formation under untreated conditions (Fig. 1, F to H); IKE treatment further exacerbated the differences in these measurements between NFE2L2/NRF2 WT and KO cells (Fig. 1, F to H). Similar results were obtained when a less specific ferroptosis inducer, SAS, was used (fig. S1, C to J).
Fig. 1. NFE2L2/NRF2 deletion sensitizes ovarian cancer cells to ferroptosis.
(A) WT or three individual CRISPR NFE2L2/NRF2 KO SKOV-3 ovarian cancer cell lines were left untreated or treated with IKE (10 μM), and cell growth was monitored using the IncuCyte imaging system. Results shown here were at 24 hours after treatment. Ferroptotic cells were identified on the basis of ferroptotic cell morphology (ballooning). More than 1000 cells were counted, and the percentage of ferroptotic cells was plotted. Scale bar, 50 μm. (B) NFE2L2/NRF2 WT and KO cell lines were cotreated with DFO (100 μM), Fer-1 (10 μM), or Z-VAD-fmk (20 μM), alone with DMSO or IKE (10 μM) for 24 hours. Cell viability was measured by MTT assay. ns, not significant. (C to H) NFE2L2/NRF2 WT and KO cell lines were treated with 10 μM IKE for 12 hours before the following end points were measured: (C) Lipid peroxide production was assessed by flow cytometry using C11-BODIPY581/591. DFO cotreatment: 100 μM, 12 hours. (D) 4-Hydroxynonenal (4-HNE) protein adducts and COX2 protein levels were measured by immunoblot analysis. (E) mRNA levels of PTGS2 were measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR). (F) Total intracellular GSH levels were measured by QuantiChrom GSH assay. (G) ROS were measured by electron paramagnetic resonance (EPR) spectroscopy. (H) Malondialdehyde (MDA) formation was detected colorimetrically using the thiobarbituric acid reactive substance (TBARS) assay. Data are represented as means ± SEM of three biological replicates. n = 3; *P < 0.05.
Table 1. The percentage of ferroptotic cells following IKE treatment (10 μM, 24 hours).
| Cell line | % Ferroptotic cells |
|---|---|
| NRF2 WT | 18.66 ± 2.6 |
| NRF2 KO-1 | 71.15 ± 5.2 |
| NRF2 KO-2 | 79 ± 7.2 |
| NRF2 KO-3 | 78 ± 7.9 |
| NRF2 KO-1,2,3 | 76.05 ± 6.76 |
| VAMP8 KO | 40 ± 5.28 |
| HERC2 KO | 42 ± 6.35 |
| DKO | 71.25 ± 9.35 |
NFE2L2/NRF2 deletion impairs iron homeostasis and increases the LIP through depletion of its target gene HERC2
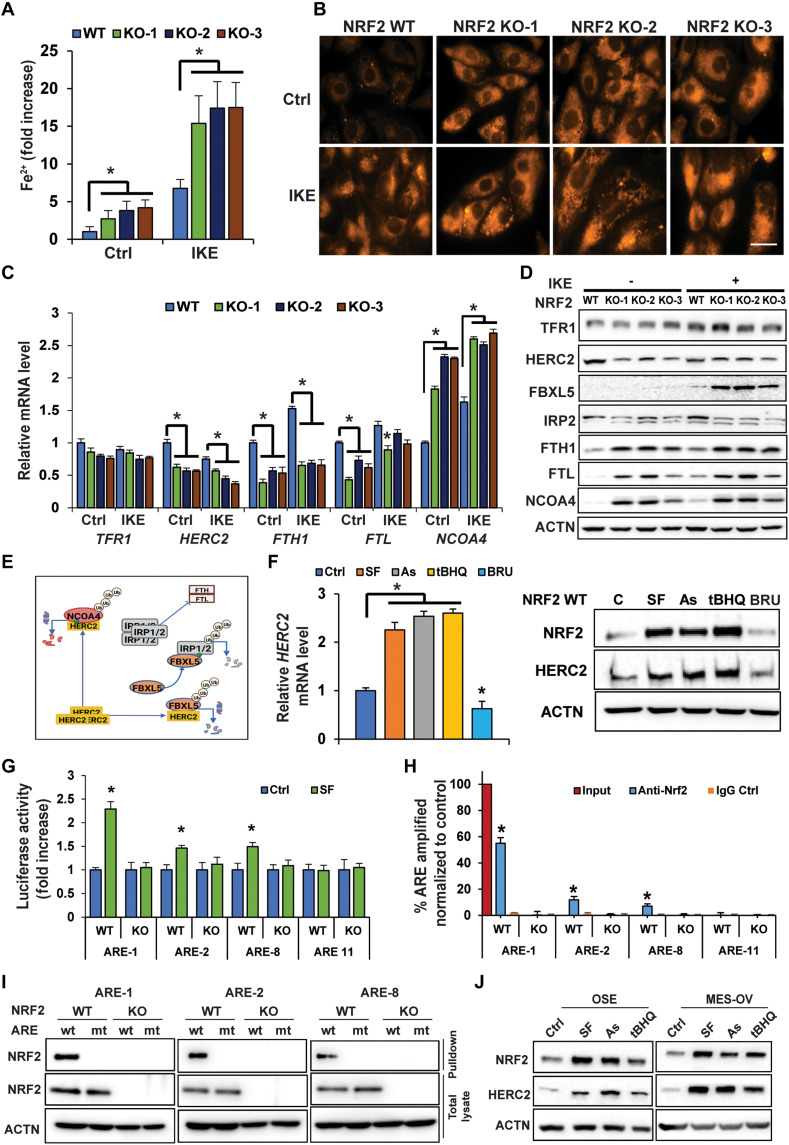
As free iron (ferrous iron, Fe2+) is required to drive ferroptotic cell death, the intracellular Fe2+ concentration was determined using a Ferene-S–based colorimetric assay. Basal intracellular Fe2+ levels increased ~3- to 5-fold in NFE2L2/NRF2 KO compared to NFE2L2/NRF2 WT cells; IKE treatment further enhanced free Fe2+ levels (Fig. 2A). Increased free iron levels in NFE2L2/NRF2 KO cells under both untreated and IKE-treated conditions were verified by immunofluorescence using the Fe2+-sensitive probe FerroOrange (Fig. 2B). Please note that IKE treatment led to an increase in free Fe2+ levels even in NFE2L2/NRF2 WT control cells, which is consistent with previous reports when cells were treated with erastin (22, 37). Next, expression of genes critical in maintaining iron homeostasis was compared in NFE2L2/NRF2 WT versus KO cells. Transferrin receptor 1 (TFRC/TFR1), the primary receptor controlling iron uptake, was moderately induced by IKE treatment at the protein but not mRNA level in all the cell lines, whereas there was a slight reduction in TFRC/TFR1 mRNA levels, but not protein, in all NFE2L2/NRF2 KO cells compared to NFE2L2/NRF2 WT cells (Fig. 2, C and D). In contrast, NFE2L2/NRF2 KO altered the expression of many proteins involved in ferritin synthesis and degradation [HERC2, F-box and leucine-rich repeat protein 5 (FBXL5), iron regulatory protein 2 (IRP2), and NCOA4] (Fig. 2, C and D), which in turn would affect the LIP. First, NFE2L2/NRF2 KO affected ferritin synthesis as evidenced by the following findings: NFE2L2/NRF2 KO cells exhibited altered levels of key iron sensor proteins constituting the HERC2-FBXL5-IRP1/2-FTH1/FTL pathway, including decreased HERC2, increased FBXL5, decreased IRP2, and increased ferritin heavy/light chains (FTH1 and FTL) (Fig. 2D). As shown in the model (Fig. 2E), HERC2 is an E3 ubiquitin ligase for both FBXL5 and NCOA4; however, HERC2 binds FBXL5 in the absence of free iron (Fe2+), whereas HERC2 binds NCOA4 only in the presence of free iron (38–41). In addition, IRP2 binds to the iron-responsive element in the 5′ untranslated region of FTH1 and FTL mRNA to repress the synthesis of ferritin during low free iron conditions. Thus, this NFE2L2/NRF2 deletion–mediated decrease in IRP2 should ultimately result in enhanced synthesis of ferritin in NFE2L2/NRF2-KO SKOV-3 cells. Consistent with this notion, increased protein levels of both FTH1 and FTL were observed (Fig. 2D) although their mRNA expression decreased (Fig. 2C), which was in accordance with FTH1/FTL being NRF2 target genes (42). Second, NFE2L2/NRF2 KO also affected ferritin degradation. NCOA4 is a critical cargo receptor that specifically recruits ferritin to the autophagosome for degradation, a process termed ferritinophagy (43). Both the mRNA and protein levels of NCOA4 were increased in NFE2L2/NRF2 KO cells (Fig. 2, C and D). The increased NCOA4 protein level seen in NFE2L2/NRF2 KO cells was partly due to decreased expression of HERC2, resulting in its stabilization (Fig. 2, D and E). Collectively, these results indicate that NFE2L2/NRF2 deletion impairs iron homeostasis by altering expression of proteins controlling ferritin synthesis and degradation, resulting in an increased LIP and enhanced sensitivity of cancer cells to ferroptosis.
Fig. 2. NFE2L2/NRF2 deletion impairs iron homeostasis and increases the LIP through depletion of its target gene HERC2.
(A and B) NFE2L2/NRF2 WT and KO SKOV-3 cell lines were treated with 10 μM IKE for 12 hours before the LIP was measured by (A) Ferene-S absorbance (*P < 0.05, n = 3) or (B) FerroOrange immunofluorescence. Scale bar, 25 μm. (C and D) mRNA or protein levels of genes involved in iron metabolism/storage were measured in NFE2L2/NRF2 WT and NFE2L2/NRF2 KO SKOV-3 cells treated with IKE (10 μM, 12 hours) by (C) qRT-PCR (*P < 0.05, n = 3) or (D) immunoblot analysis. (E) Model showing HERC2 regulation of ferritin synthesis and NCOA4-mediated ferritin degradation. Green dot = ferrous iron; Ub, ubiquitin. (F) HERC2 mRNA (right; *P < 0.05, n = 3) and protein levels (left) in NFE2L2/NRF2 WT SKOV-3 cells treated with DMSO (Ctrl), sulforaphane (SF; 5 μM), arsenic (As; 1 μM), tert-butylhydroquinone (tBHQ; 25 μM), or brusatol (BRU; 20 nM) for 12 hours. (G) NFE2L2/NRF2 WT SKOV-3 cells were transfected with the indicated antioxidant response element (ARE) firefly and thymidine kinase renilla luciferase vectors and then treated with DMSO or SF (5 μM) for 12 hours, and luciferase activity was measured. *P < 0.05, n = 3. (H) NRF2-ARE binding was determined via ChIP-PCR. *P < 0.05, n = 3. (I) Biotinylated ARE pulldown of three putative HERC2 WT or mutant (mt) AREs using cell lysate from NFE2L2/NRF2 WT or NFE2L2/NRF2 KO SKOV-3 cells. DNA binding proteins were pulled down using streptavidin beads, and NRF2 was detected by immunoblot analysis. (J) NRF2 and HERC2 protein levels in ovarian surface epithelial (OSE) and MES-OV ovarian cell lines following treatment with SF (5 μM), As (1 μM), or tBHQ (25 μM) for 12 hours.
Since both FBXL5-IRP2–mediated ferritin synthesis and NCOA4-dependent ferritin degradation through the autophagy-lysosome pathway can be controlled by the key E3 ubiquitin ligase HERC2 (Fig. 2E) (41, 43), coupled with the result described in Fig. 2D indicating a decrease in HERC2 protein levels in NFE2L2/NRF2 KO cells, the possibility of HERC2 being an NRF2 target gene was explored. Consistent with the observed positive correlation between the protein levels of NRF2 and HERC2 (Fig. 2D), a higher level of HERC2 mRNA in NFE2L2/NRF2 WT cells was observed in both untreated and IKE-treated conditions, compared to NFE2L2/NRF2 KO cells (Fig. 2C). The NRF2 activators sulforaphane (SF), arsenic (As), and tert-butylhydroquinone (tBHQ) or the NRF2 inhibitor brusatol (BRU) was able to increase or decrease both the mRNA and protein level of HERC2, respectively (Fig. 2F). In silico analysis identified 12 putative antioxidant response elements (AREs) in HERC2, which were individually cloned into the promoter region of a firefly luciferase vector, followed by measurement of firefly luciferase activity using a dual luciferase reporter assay (renilla was used as an internal control). Three functional AREs (ARE-1, ARE-2, and ARE-8) were identified as their luciferase activity was enhanced by SF treatment; one nonfunctional ARE (ARE-11) was also included for comparison (Fig. 2G). Endogenous binding of NRF2 to these three AREs was further verified to occur in WT, but not NFE2L2/NRF2 KO, cells by chromatin immunoprecipitation (ChIP)–polymerase chain reaction (PCR) (Fig. 2H). Last, using biotinylated 41–base pair (bp) DNA probes containing the WT ARE-1, ARE-2, ARE-8, or their mutated ARE sequences (mt-AREs), NRF2 was confirmed to specifically bind to ARE-1, ARE-2, and ARE-8, but not their mutant counterparts (Fig. 2I). These results demonstrate that the HERC2 promoter, intron1, and exon1 contain functional AREs that are regulated by NRF2. In addition, NRF2 activators were able to induce NRF2 and HERC2 protein levels in two additional ovarian cell lines with low basal levels of NRF2 (Fig. 2J). More broadly, a positive correlation between NRF2 and HERC2 was also observed in most ovarian cell lines tested where NFE2L2/NRF2 expression was reduced by transient knockdown using two different NFE2L2/NRF2 small interfering RNAs (siRNAs) (fig. S2).
VAMP8 down-regulation in NFE2L2/NRF2 KO cells results in a defect of ferritinophagy and autophagosomal accumulation of apoferritin/NCOA4
As shown in Fig. 2D, the protein levels of FTH1, FTL, and NCOA4 were increased in NFE2L2/NRF2 KO cells, indicative of a defect in ferritin degradation. Since ferritin is degraded by the autophagy-lysosome pathway, a process termed ferritinophagy (see a model illustration for ferritinophagy in Fig. 3A), this prompted us to examine the possibility that the observed increase in these proteins was a result of blockage of autophagic flux. NFE2L2/NRF2 KO cells exhibited a larger number of microtubule-associated protein 1A/1B-light chain 3 (LC3)–positive autophagosomes, but not autolysosomes, as assessed by monomeric–red fluorescent protein (mRFP)–green fluorescent protein (GFP)–LC3 tandem–based immunofluorescence (Fig. 3B). This result indicates that there was no defect in autophagosome formation, but instead a blockage at the later stage of the pathway in NFE2L2/NRF2 KO cells. Transmission electron microscopy (TEM) also indicated an accumulation of protein aggregates and double-membrane autophagosomes in NFE2L2/NRF2 KO cells (Fig. 3C). Next, the snap receptor (SNARE) complex [VAMP8, synaptosome-associated protein 29 (SNAP29), and syntaxin (STX17)], which plays a critical role in mediating autophagosome-lysosome fusion (44), was examined. Unexpectedly, the protein level of VAMP8, a lysosomal SNARE protein, was markedly reduced, while there were no obvious changes in STX17 or SNAP29 protein levels in NFE2L2/NRF2 KO cells (Fig. 3D). Furthermore, NFE2L2/NRF2 KO cells had ~30 to 40% lower VAMP8 mRNA levels compared to WT cells (Fig. 3E). Transcription of lysosomal genes has been shown to be regulated by transcription factor EB (TFEB), whose activity is controlled by serine phosphorylation. Previous studies indicate that phosphorylation at S142/S138 inactivates TFEB by enhancing its nuclear export, whereas S211 phosphorylation mediates cytosolic retention. Thus, the net effect of TFEB phosphorylation at these sites is less nuclear localization and reduced transcription of lysosomal genes (45–49). Therefore, the phosphorylation status of TFEB and mammalian target of rapamycin (mTOR), one of the upstream kinases for TFEB, were assessed. Basal phosphorylation of mTOR at S2448 was higher in NFE2L2/NRF2 KO cells, while the total mTOR level remained unchanged; TFEB phosphorylation was also enhanced at S211 and S142 with no changes in its total protein expression (Fig. 3F). Consistently, when cells with nuclear localization of TFEB were counted, significantly more cells with nuclear TFEB localization in NFE2L2/NRF2 WT cells than NFE2L2/NRF2 KO cells were identified (Fig. 3G). TFEB binds to DNA sequences containing an EBOX motif (CANNTG), and in silico analysis identified six putative EBOX motifs in the promoter region of VAMP8. Using a dual luciferase assay, one functional EBOX (−1932CACTTG−1927) was identified, as demonstrated by increased EBOX-driven firefly luciferase activity in the presence of mTOR inhibitors (torin and rapamycin) and adenosine monophosphate–activated protein kinase (AMPK) activator (AICAR), as well as a decreased luciferase activity following mTOR activation (amino acids) (Fig. 3H). These results indicate that NFE2L2/NRF2 KO leads to ferritinophagy blockage through TFEB phosphorylation and nuclear exclusion, which decreases the transcription of VAMP8, although the precise mechanism by which deletion of NFE2L2/NRF2 leads to activation of mTOR, or possibly other kinases that may regulate TFEB phosphorylation and nuclear localization, requires further investigation. The notion that mTOR was activated in NFE2L2/NRF2 KO cells, resulting in decreased VAMP8 transcription, was further supported by the results in fig. S3. Rapamycin treatment reduced phosphorylated mTOR levels in NFE2L2/NRF2 KO cells without affecting the total mTOR level, indicating inactivation of mTOR by rapamycin in NFE2L2/NRF2 KO cells. Rapamycin increased VAMP8 expression in NFE2L2/NRF2 KO cells, which partially rescued the blockage of ferritinophagy flux as indicated by the reduced level of NCOA4, p62, and LC3-II in rapamycin-treated NFE2L2/NRF2 KO cells (fig. S3C). Similar to DFO, rapamycin also reduced the number of ferroptotic cells (fig. S3A) and improved cell survival in response to SAS in NFE2L2/NRF2 KO cells (fig. S3B). Similar to HERC2, the positive correlation between NRF2 and VAMP8 in a variety of ovarian cell lines was also observed when NFE2L2/NRF2 expression was reduced by NFE2L2/NRF2 siRNAs (fig. S2). Immortalized normal ovarian surface epithelial (OSE) (50) and fallopian tube (FT-246) (51) cell lines had a low basal protein level of NRF2 and an undetectable level of VAMP8, while NFE2L2/NRF2 knockdown in the ovarian cancer cell lines tested all resulted in a decrease in VAMP8 (fig. S2).
Fig. 3. VAMP8 down-regulation in NFE2L2/NRF2 KO cells results in a defect of ferritinophagy and autophagosomal accumulation of apoferritin/NCOA4.
(A) Illustration of the ferritinophagy pathway. (B) Immunofluorescence analysis of autophagy flux in NFE2L2/NRF2 WT and NFE2L2/NRF2 KO SKOV-3 cells transfected with an mRFP-GFP-LC3 tandem reporter for 24 hours. Yellow puncta, autophagosomes; red puncta, autolysosomes. Scale bar, 10 μm. (C) Representative TEM micrographs of WT and NFE2L2/NRF2 KO cells. Arrows indicate autophagosomes or protein aggregates. (D) Protein levels of three SNARE proteins that direct autophagosome-lysosome fusion. (E) VAMP8 mRNA levels in NFE2L2/NRF2 WT and KO cells. *P < 0.05, n = 3. (F) Total or phosphorylated protein levels of mTOR and TFEB. (G) Nuclear localization of TFEB was detected by indirect immunofluorescence (top); 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclear staining, and the number of TFEB nuclear-positive cells was counted and plotted (bottom). *P < 0.05, n = 3. Scale bar, 50 μm. (H) One functional EBOX in the promoter of VAMP8 was identified. The 41-bp EBOX-containing sequence was shown above. Relative EBOX luciferase activity was measured by dual luciferase assay, following treatment with mTOR inhibitors [torin, 100 nM, 24 hours; rapamycin (Rap), 1 μM, 24 hours; and AICAR, 1 mM, 4 hours] or mTOR activation by amino acids (AA). (I and J) Indirect immunofluorescence of (I) FTH1 and LC3, and (J) FTH1 and LAMP1 in NFE2L2/NRF2 WT or KO SKOV-3 cells. Inset shows magnified puncta. (K) Cells were treated with or without ferric ammonium citrate (FAC; 100 μM, 12 hours). Ferric iron deposits were assessed by Perls Prussian blue staining, followed by indirect immunofluorescence of FTH1/LC3. Scale bars, 10 μm.
We have shown that decreased HERC2 expression led to enhanced protein synthesis of FTH1/FTL and enhanced NCOA4 protein stability, and that decreased VAMP8 expression resulted in ferritinophagy blockage in NFE2L2/NRF2 KO cells. Next, the effect of NFE2L2/NRF2 deletion on autophagic ferritin turnover and intracellular free iron was assessed. Immunofluorescence analysis revealed that while WT cells had very little FTH1/FTL signal (less ferritin aggregates shown as puncta), NFE2L2/NRF2 KO cells had significantly more FTH1/FTL aggregates, most of which colocalized with LC3-positive puncta (Fig. 3I, Table 2, and fig. S4A). However, while FTH1/FTL colocalized with LC3, indicating recruitment of ferritin to the autophagosome, FTH1/FTL failed to be delivered to the lysosome, as very few FTH1/FTL aggregates colocalized with LAMP1-positive lysosomes in NFE2L2/NRF2 KO cells (Fig. 3J, Table 2, and fig. S4B). These results support a model whereby FTH1/FTL was recruited by NCOA4 into autophagosomes, which then accumulated due to the blockage of VAMP8-mediated autophagosome-lysosome fusion in NFE2L2/NRF2 KO cells. In addition, accumulation of FTH1 aggregates was observed in other ovarian cell lines where NFE2L2/NRF2 was knocked down via siRNA (fig. S4C). Paradoxically, enhanced ferritin synthesis and blockage of ferritin degradation (inhibition of ferritinophagy) in NFE2L2/NRF2 KO cells should have resulted in a decrease, rather than a significant increase, of the LIP (Fig. 2, A and B). We hypothesized that the availability of higher than normal NCOA4, a critical cargo receptor that recruits ferritin to the autophagosome, would overwhelm the other proteins, such as poly(RC) binding protein 1 (PCBP1), needed to deliver iron to ferritin (52), resulting in NCOA4-mediated recruitment of apoferritin into the autophagosome. As shown in Fig. 3K, NFE2L2/NRF2 WT cells showed an increase in Perls staining (dark dots in the cytosol) after ferric ammonium citrate (FAC) treatment, indicative of ferric iron stored in ferritin cages, while the NFE2L2/NRF2 KO cells had very limited Perls staining after FAC treatment, although these cells contained many ferritin aggregates colocalized with LC3 (Fig. 3K), indicating recruitment of apoferritin into autophagosomes in NFE2L2/NRF2 KO cells. This result suggests that the excessive amount of NCOA4 resulted in the rapid recruitment of apoferritin into autophagosomes before iron incorporation into the ferritin complex. Next, the effect of siRNA-mediated knockdown of NCOA4 on alleviating the increased LIP and ferroptosis observed in NFE2L2/NRF2 KO cells was tested. NCOA4-siRNA had a slight effect on the protein levels of FTH1/FTL (increased) and LC3-I/LC3-II (decreased) in both the WT and KO cell lines (fig. S4D). Knockdown of NCOA4 alleviated 4-HNE–adducted protein formation in IKE-treated NFE2L2/NRF2 KO cells (fig. S4, D and E). The protection afforded by NCOA4 knockdown in NFE2L2/NRF2 KO cells was also visualized by a reduction in FTH1 puncta formation, decreased Fe2+-sensitive FerroOrange fluorescence intensity, and fewer cells undergoing ferroptotic death (fig. S4F). Consistent with these results, NCOA4 knockdown significantly enhanced the survival of NFE2L2/NRF2 KO cells under IKE treatment (fig. S4G). In addition, the mRNA levels of ferroportin (SLC40A1) were significantly higher in the NFE2L2/NRF2 KO cells compared to WT (fig. S4H), demonstrating that the accumulation of free labile iron observed in the NFE2L2/NRF2 KO cells is most likely not due to changes in ferroportin expression, despite previous reports (53).
Table 2. Mander’s and Pearson’s coefficients for ferritin (FTH1/FTL) colocalization with autophagosome (LC3) versus lysosome (LAMP1).
| Mander’s coefficient | Pearson’s coefficient | ||
|---|---|---|---|
| LC3/FTH1 | FTH1/LC3 | ||
| NRF2 WT | 0.466 | 0.356 | r = 0.696 |
| NRF2 KO-1 | 0.543 | 0.684 | r = 0.825 |
| NRF2 KO-2 | 0.516 | 0.678 | r = 0.819 |
| LAMP1/FTH1 | FTH1/LAMP1 | ||
| NRF2 WT | 0.336 | 0.426 | r = 0.701 |
| NRF2 KO-1 | 0.187 | 0.325 | r = 0.305 |
| NRF2 KO-2 | 0.195 | 0.321 | r = 0.225 |
| LC3/FTL | FTL/LC3 | ||
| NRF2 WT | 0.358 | 0.222 | r = 0.676 |
| NRF2 KO-1 | 0.695 | 0.725 | r = 0.850 |
| NRF2 KO-2 | 0.622 | 0.756 | r = 0.785 |
| LAMP1/FTL | FTL/ LAMP1 | ||
| NRF2 WT | 0.329 | 0.252 | r = 0.621 |
| NRF2 KO-1 | 0.185 | 0.318 | r = 0.252 |
| NRF2 KO-2 | 0.105 | 0.345 | r = 0.325 |
NRF2 regulates iron homeostasis through HERC2, VAMP8, and NCOA4
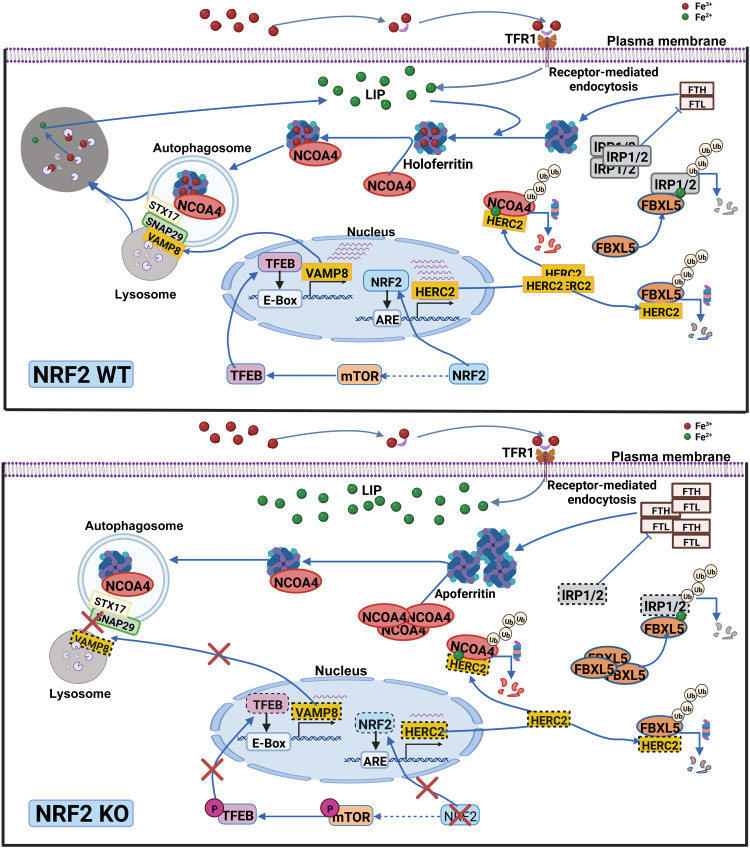
Collectively, our results have revealed a previously undiscovered function of NRF2, i.e., NRF2 regulates iron homeostasis and ferroptosis through HERC2, VAMP8, and NCOA4 (illustration in Fig. 4). By controlling their protein levels, NRF2 regulates both ferritin synthesis and degradation. First, HERC2, an E3 ubiquitin ligase for FBXL5 and NCOA4, is an NRF2 target gene; deletion of NRF2 results in reduced HERC2 expression, increased stability of FBXL5 and NCOA4, decreased IRP2 protein stability, and enhanced FTH synthesis. Second, NRF2 indirectly controls VAMP8 through the mTOR-TFEB axis. In NFE2L2/NRF2 KO cells, decreased TFEB-dependent transcription of VAMP8 results in blockage of autophagosome-lysosome fusion and inhibition of ferritinophagy. Third, excessive accumulation of NCOA4 in NFE2L2/NRF2 KO cells causes recruitment of apoferritin into autophagosomes, leading to autophagosomal accumulation of apoferritin/NCOA4, increased LIP, and enhanced sensitivity to ferroptotic cell death.
Fig. 4. NRF2 regulates iron homeostasis through HERC2, VAMP8, and NCOA4.
NRF2 regulates iron homeostasis by controlling both ferritin synthesis and degradation. First, HERC2, an E3 ubiquitin ligase for FBXL5 and NCOA4, is an NRF2 target gene; deletion of NFE2L2/NRF2 results in reduced HERC2 expression, increased stability of FBXL5 and NCOA4, decreased IRP2 protein stability, and enhanced FTH synthesis. Second, NRF2 indirectly controls VAMP8 through the mTOR-TFEB axis. In NFE2L2/NRF2 KO cells, decreased TFEB-dependent transcription of VAMP8 results in blockage of autophagosome-lysosome fusion and inhibition of ferritinophagy. Third, excessive accumulation of NCOA4 in NFE2L2/NRF2 KO cells causes recruitment of apoferritin into autophagosomes, leading to autophagosomal accumulation of apoferritin/NCOA4, increased LIP, and enhanced sensitivity to ferroptotic cell death. Green dot, ferrous iron; red dot, ferric iron.
Contribution of the NRF2-HERC2 and NRF2-VAMP8/NCOA4 axes in controlling iron homeostasis and dictating ferroptosis sensitivity
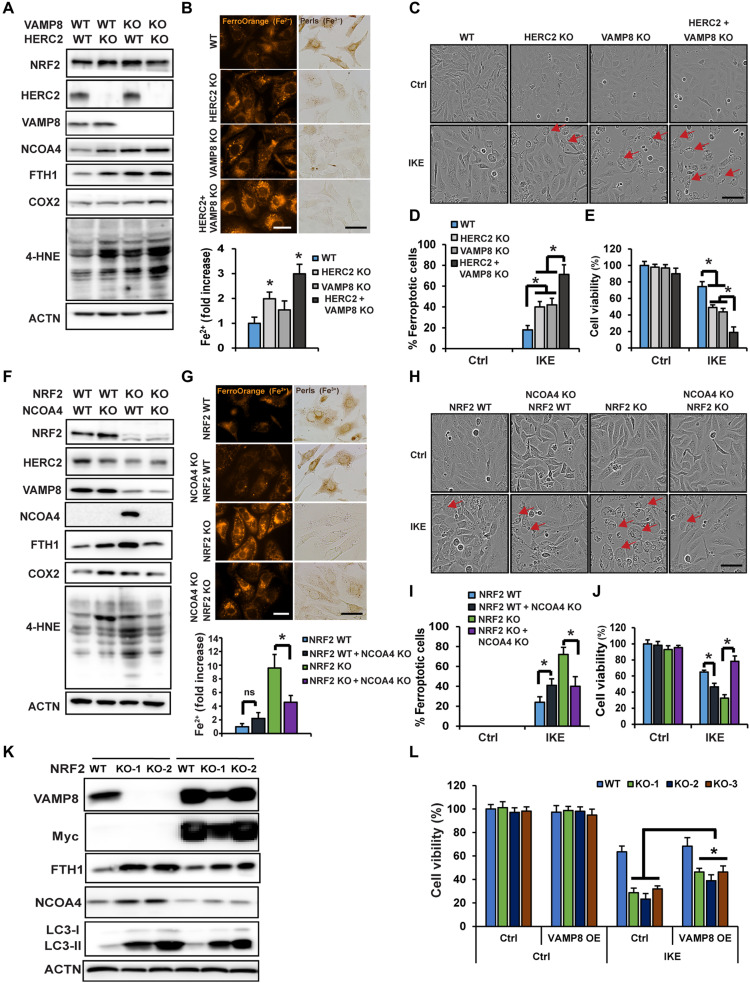
To further validate the contribution of the newly identified function of NRF2 in iron homeostasis, and to ensure the anti-ferroptotic function of NRF2 extends beyond just its mediation of redox balance, HERC2 and VAMP8 single or double KO cells were generated. Like what was observed in NFE2L2/NRF2 KO cells, KO of HERC2 or VAMP8 resulted in increased protein levels of FTH1 and NCOA4, as well as the ferroptosis markers COX2 and 4-HNE, with double- KO of HERC2/VAMP8 exhibiting a more pronounced phenotype than single KO (Fig. 5A). Also, KO of HERC2 or double KO significantly increased free iron levels as measured by both FerroOrange (Fig. 5B, FerroOrange panel) and Ferene-S colorimetric assay (Fig. 5B, bar graph, bottom), but decreased ferric iron–loaded ferritin cages (Fig. 5B, Perls staining). Consistently, KO enhanced ferroptotic cell death (Fig. 5, C and D) and reduced cell survival (Fig. 5E). The percentage of ferroptotic cells was ~20% in IKE-treated WT cells versus ~75% in the HERC2/VAMP8 double KO cells (Fig. 5D), with an ~30% decrease in cell viability in NFE2L2/NRF2 WT compared to an ~80% decrease in HERC2/VAMP8 double KO cells at 24 hours (Fig. 5E). A comparison of the percentage of ferroptotic cells in NFE2L2/NRF2 WT and the different KO cell lines following IKE treatment shows that cells with double KO of HERC2/VAMP8 exhibit an equivalent ferroptosis sensitivity as NFE2L2/NRF2 KO cells (Table 1). These results demonstrated that HERC2/VAMP8 double KO cells behaved like NFE2L2/NRF2 KO cells, as indicated by a similar accumulation of apoferritin/NCOA4, increased LIP, enhanced ferroptosis markers (COX2 and 4-HNE), and increased sensitivity to ferroptotic cell death.
Fig. 5. Contribution of the NRF2-HERC2 and NRF2-VAMP8/NCOA4 axes in controlling iron homeostasis and dictating ferroptosis sensitivity.
HERC2 and VAMP8 single or double KO cells were established. (A) Status of HERC2/VAMP8 KO was determined by immunoblot analysis, along with the levels of the other indicated proteins. (B) These four cell lines were used to measure free iron using FerroOrange (upper left) and Ferene-S colorimetic assay (bottom), or ferric iron–ferritin cages by Perls staining after FAC treatment (100 μM, 12 hours) (upper right). Scale bars, 25 μm. (C) These four cell lines were left untreated or treated with IKE (10 μM), and cell growth was monitored using the IncuCyte imaging system. Results shown here were at 24 hours after treatment of IKE. Scale bar, 50 μm. (D) Percentage of ferroptotic cells from (C). (E) Cell viability measured by MTT assay. (F and G) Pooled NCOA4 KO cells in the NFE2L2/NRF2 WT or KO background were established and (F) the levels of the indicated proteins were determined by immunoblot analysis, and (G) free iron was measured using FerroOrange (upper left) or Ferene-S (bottom), or ferric iron–ferritin cages by Perls staining after FAC treatment (100 μM, 12 hours) (upper right). Scale bars, 25 μm. (H) These four cell lines were left untreated or treated with IKE (10 μM), and cell growth was monitored using the IncuCyte imaging system. Images shown were at 24 hours after treatment with IKE. Scale bar, 50 μm. (I) Percentage of ferroptotic cells from (H). *P < 0.05, n = 3. (J) Cell viability measured by MTT assay. *P < 0.05, n = 3. (K) FTH1, NCOA4, and LC3-I/II protein levels in NFE2L2/NRF2 WT or KO cells overexpressing VAMP8 were determined by immunoblot analysis. (L) Cell viability measured by MTT assay.
As shown in our model (Fig. 4), NFE2L2/NRF2 KO cells exhibit excessive accumulation of NCOA4, which is a critical factor in mediating the recruitment of apoferritin into autophagosomes, leading to the observed increase in the LIP and enhanced sensitivity to ferroptotic cell death. To fully confirm this mechanism, and also validate the NCOA4 siRNA knockdown results presented in fig. S4 (D to G), CRISPR NCOA4 KO NFE2L2/NRF2 WT and NFE2L2/NRF2 KO cell lines were generated. NCOA4 KO in NFE2L2/NRF2 WT cells enhanced ferroptosis as indicated by increased COX2 and 4-HNE (Fig. 5F), which was accompanied by increased ferroptosis and decreased survival (Fig. 5, H, I, and J). A marked contrast was observed when NCOA4 was knocked out in NFE2L2/NRF2 KO cells, as NCOA4 deletion reduced FTH1 protein levels, reduced ferroptosis markers COX2 and 4-HNE (Fig. 5F), decreased LIP as measured by both FerroOrange (Fig. 5G, FerroOrange panel) and Ferene-S colorimetic assay (Fig. 5G, bar graph, bottom), increased ferric iron–loaded ferritin cages (Fig. 5G, Perls staining), decreased percentage of ferroptotic cells (Fig. 5, H and I), and restored NFE2L2/NRF2 KO cell survival following IKE treatment to that of NFE2L2/NRF2 WT cells (Fig. 5J). In addition, overexpression of VAMP8 in NFE2L2/NRF2 KO SKOV3 cells alleviated autophagy blockage, as indicated by reduced protein levels of FTH1, NCOA4, and LC3-I/II (Fig. 5K), as well as reduced ferroptosis sensitivity in IKE-treated NFE2L2/NRF2 KO but not NFE2L2/NRF2 WT cells (Fig. 5L). Together, these results demonstrate the significance of NRF2-mediated iron sensing, by controlling ferritin synthesis/degradation, in determining a cancer cell’s response to ferroptosis inducers.
NRF2 expression correlates with HERC2 and VAMP8 levels in human cancer tissues, as well as ferroptosis resistance of cancer cell lines
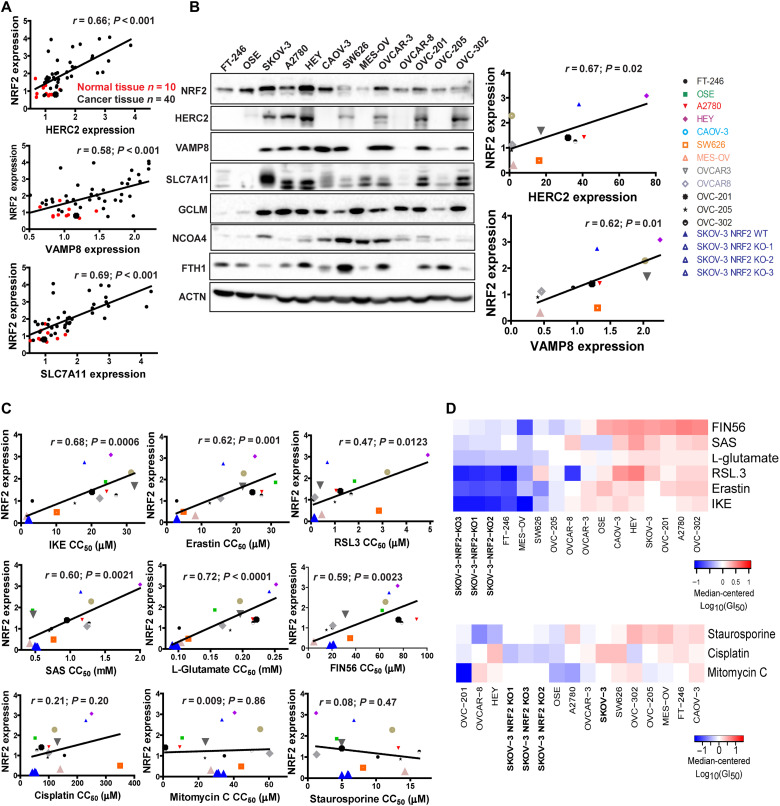
The translational relevance of this study was evaluated using human ovarian patient tissues to test whether there is a correlation between the expression of HERC2, VAMP8, and NRF2. NRF2 protein levels correlated strongly with HERC2 and VAMP8 (SLC7A11, a known NRF2 target gene, was used as a positive control) (Fig. 6A and fig. S5A). Notably, NRF2 expression was low in normal tissues but up-regulated in tumors (Fig. 6A, red versus black dots). Similarly, there was a positive correlation between NRF2 and HERC2, as well as NRF2 and VAMP8 across a wide variety of different ovarian cancer cell lines (Fig. 6B). Normal ovarian cell lines FT-246 and OSE were excluded in the correlation curve because of the low levels of HERC2 and VAMP8 (Fig. 6B). In support of our model, most cancer cell lines showed a negative correlation between the levels of NRF2 and either FTH1 or NCOA4 (Fig. 6B). Next, the response of these cell lines to ferroptosis and other mode of cell death inducers was measured (fig. S5B) and the 50% growth inhibition concentration (GI50) for each individual compound was calculated (Table 3). As expected, a positive correlation between NRF2 expression and GI50 values of the different ferroptosis inducers among these cell lines was observed (Fig. 6C, top two rows). In contrast, there was no correlation between NRF2 and GI50 values of the other cytotoxic compounds that do not induce ferroptosis, including chemotherapeutic drugs (cisplatin and mitomycin C) and the apoptosis inducer staurosporine (Fig. 6C, bottom row). The finding that NRF2 determines the resistance of ovarian cancer cell lines to ferroptosis inducers was further illustrated in a heatmap indicating the GI50 profiles of these cell lines in response to ferroptotic inducers, as the three NFE2L2/NRF2 KO SKOV-3 cell lines were clustered as the most sensitive to all ferroptotic inducers, far apart from WT SKOV-3 (Fig. 6D, top). In contrast, the sensitivity of the three NFE2L2/NRF2 KO SKOV-3 cell lines to other compounds in relation to the other cell lines tested was only increased slightly (Fig. 6D, bottom).
Fig. 6. NRF2 expression correlates with HERC2 and VAMP8 levels in human cancer tissues, as well as ferroptosis resistance.
(A) Correlation between NRF2 expression and HERC2 or VAMP8 levels in human ovarian tissues was determined by immunohistochemistry analysis. HERC2, VAMP8, and SLC7A11 expression were imaged (see fig. S4A), and average intensity was measured and plotted against NRF2. Normal tissue (n = 10) plotted as red dots; cancer tissue (n = 40) plotted as black dots. (B) NRF2 and ferroptosis-related protein levels in various ovarian cancer cell lines were measured by immunoblot analysis (left). The expression of HERC2, VAMP8, and SLC7A11 was quantified and plotted against NRF2 in each cell line (right). (C) Correlation between NRF2 expression and the GI50 of ferroptosis-inducing compounds (IKE, erastin, RSL3, SAS, l-glutamate, and FIN56) and apoptosis-inducing compounds (cisplatin, mitomycin C, or staurosporine). Each cell line was treated with eight doses of the indicated compound for 24 hours, and cell viability was measured by MTT assay. GI50 values were calculated by log-logistic fitting (Table 3). (D) Heatmap GI50 profiles were created using median-centered z-score analysis of the GI50 values.
Table 3. The 50% growth inhibition concentration (GI50) for each individual compound.
| Cell lines | IKE (μM) | Erastin (μM) | RSL-3 (μM) | SAS (μM) | l-Glutamate (μM) | FIN46 (μM) | Cisplatin (μM) | Staurosporine (μM) | Mitomycin C (μM) |
|---|---|---|---|---|---|---|---|---|---|
| HEY | 25.5 ± 0.48 | 225.32 ± 0.22 | 3.68 ± 0.09 | 2,000 ± 30.6 | 252,020 ± 117 | 75.25 ± 0.48 | 250 ± 1.93 | 2.22 ± 0.29 | 40.51 ± 0.40 |
| SKOV-3 | 18 ± 0.16 | 16.23 ± 0.16 | 1.85 ± 0.01 | 1,250 ± 9.73 | 241,300 ± 62.9 | 68 ± 0.16 | 230 ± 1.92 | 6.75 ± 0.59 | 33.29 ± 0.33 |
| CAOV-3 | 31.25 ± 0.20 | 22.1 ± 0.02 | 3.2 ± 0.25 | 1,300 ± 10.64 | 195,690 ± 97.7 | 65.02 ± 0.21 | 120 ± 3.21 | 12.35 ± 0.99 | 55.01 ± 1.60 |
| OSE | 24 ± 0.31 | 31.25 ± 0.31 | 1.48 ± 0.03 | 460 ± 8.70 | 156,700 ± 88.8 | 62.26 ± 1.71 | 52 ± 1.73 | 4.24 ± 0.13 | 10.8 ± 0.18 |
| OVCAR-3 | 28 ± 0.01 | 21.8 ± 0.01 | 0.98 ± 0.02 | 470 ± 20.1 | 196,150 ± 100 | 31.25 ± 0.19 | 130 ± 3.25 | 4.97 ± 0.54 | 23.96 ± 0.23 |
| A2780 | 24.3 ± 0.02 | 27.3 ± 0.98 | 1.65 ± 0.07 | 1,170 ± 11.5 | 215,510 ± 136 | 91.25 ± 0.27 | 100 ± 1.52 | 13.25 ± 0.87 | 9.44 ± 0.45 |
| OVC-201 | 20.15 ± 0.02 | 24.6 ± 1.16 | 1.23 ± 0.09 | 952 ± 30.42 | 221,343 ± 157 | 75.96 ± 0.23 | 74 ± 1.55 | 4.99 ± 0.56 | 1.136 ± 0.09 |
| OVC-302 | 27 ± 0.97 | 27.2 ± 0.02 | 1.68 ± 0.09 | 1,270 ± 9.13 | 216,090 ± 142 | 78.7 ± 0.97 | 58 ± 0.98 | 16.32 ± 1.82 | 50.19 ± 0.98 |
| OVCAR-8 | 22.35 ± 0.02 | 15.5 ± 0.03 | 0.16 ± 0.03 | 1,240 ± 25.7 | 168,570 ± 71.2 | 20.71 ± 0.22 | 99 ± 2.01 | 1.3 ± 0.39 | 60.32 ± 1.05 |
| FT-246 | 3.2 ± 0.03 | 2.86 ± 0.06 | 0.112 ± 0.02 | 730 ± 24.3 | 101,300 ± 106 | 20.35 ± 0.35 | 155 ± 4.41 | 10.25 ± 0.82 | 34.7 ± 0.88 |
| OVC-205 | 15.26 ± 0.11 | 12.36 ± 0.30 | 1 ± 0.04 | 700 ± 13.6 | 180,700 ± 103 | 15.25 ± 0.11 | 96 ± 1.73 | 13.57 ± 0.67 | 23.69 ± 0.48 |
| SW626 | 10.15 ± 1.09 | 4.85 ± 0.02 | 1.65 ± 0.25 | 760 ± 17.5 | 115,610 ± 157 | 20 ± 1.09 | 240 ± 4.68 | 8.09 ± 0.37 | 44.25 ± 0.78 |
| MES-OV | 1.6 ± 0.04 | 8.21 ± 0.03 | 0.45 ± 0.05 | 420 ± 25.2 | 103,960 ± 44.4 | 4.65 ± 0.42 | 140 ± 2.51 | 14.21 ± 0.51 | 27.04 ± 0.27 |
| SKOV-3 NRF2 KO-1 | 2.01 ± 0.03 | 2.8 ± 0.05 | 0.15 ± 0.03 | 521 ± 4.87 | 95,052 ± 31.9 | 20.56 ± 0.31 | 42 ± 0.21 | 5.99 ± 0.16 | 30.25 ± 0.17 |
| SKOV-3 NRF2 KO-2 | 2.32 ± 0.01 | 3.01 ± 0.04 | 0.16 ± 0.02 | 533 ± 5.63 | 98,665 ± 70.2 | 18.22 ± 0.18 | 45 ± 0.63 | 4.99 ± 0.26 | 34.25 ± 0.10 |
| SKOV-3 NRF2 KO-3 | 2.11 ± 0.03 | 2.95 ± 0.03 | 0.13 ± 0.04 | 495 ± 9.33 | 90,355 ± 42.5 | 21.25 ± 0.32 | 51 ± 0.5 | 5.86 ± 0.44 | 31.25 ± 0.16 |
Genetic or pharmacological NRF2 inhibition enhanced sensitivity to ferroptotic cell death in preclinical models
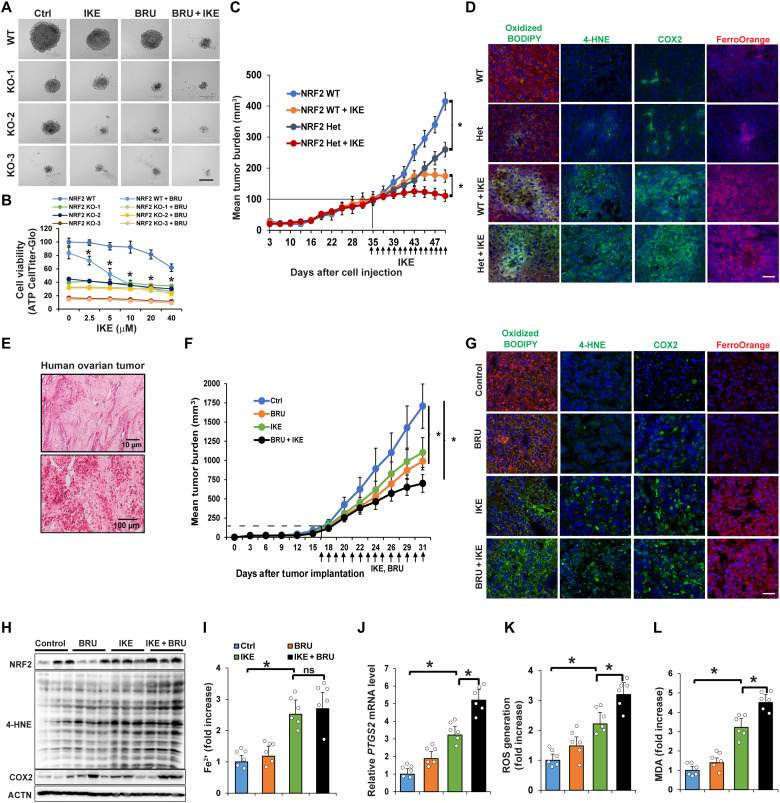
The results shown in Fig. 6 (C and D) indicated that NRF2 effectively protected ovarian cancer cells from ferroptosis-inducing compounds; thus, the idea of killing cancer cells using a combined treatment of an NRF2 inhibitor and ferroptosis inducer was explored using three preclinical models. (i) Three-dimensional (3D) tumor spheroids: In NFE2L2/NRF2-WT SKOV-3 cells, IKE resulted in a dose-dependent decrease in tumor spheroid size and cell viability, both of which were further decreased by adding BRU, an NRF2 inhibitor (24-hour IKE treatment data; Fig. 7, A and B). However, the same number of NFE2L2/NRF2-KO SKOV-3 cells formed much smaller spheroids with a lower baseline viability even in the absence of IKE treatment. Similar results were obtained using two other ovarian cancer cell lines (OVCAR-8 and OVC-201) (fig. S6A). (ii) NFE2L2/NRF2 WT and heterozygous SKOV-3 xenografts: NFE2L2/NRF2 WT xenografts implanted in NSG (nonobese diabetic severe combined immunodeficient gamma) mice grew quickly, and tumor growth was significantly inhibited by treatment with IKE (Fig. 7C). The NFE2L2/NRF2 heterozygous tumor growth curve before the tumor size reached 100 to 125 mm3 was the same as NFE2L2/NRF2 WT; however, the growth of NFE2L2/NRF2 heterozygous tumors after they reached 125 mm3 slowed as the tumor size increased. Moreover, NRF2 heterozygous tumor size was significantly smaller than NFE2L2/NRF2 WT tumor size after daily injection of IKE (40 mg/kg) for 14 days (Fig. 7C). No obvious weight changes in the treated mice were observed (fig. S6B). Furthermore, IKE-treated NFE2L2/NRF2 WT and heterozygous tumor tissues showed increased lipid droplet formation [arrowheads in hematoxylin and eosin (H&E) and Nile Red] and necrotic areas (arrows in H&E) (fig. S6C). WT and NFE2L2/NRF2 heterozygous tumors treated with IKE exhibited increased markers of ferroptosis, including C11-BODIPY581/591 staining, 4-HNE, COX2 (immunifluorescence and immunoblots), PTGS2 mRNA level), LIP (FerroOrange staining and Ferene-S assay), MDA, and ROS (Fig. 7D and fig. S6, D to H). Several ferroptosis markers, such as 4-HNE protein adduct formation, MDA production, and PTGS2 mRNA levels, were significantly higher in IKE-treated NFE2L2/NRF2 heterozygous than in IKE-treated WT tumors. (iii) Poorly differentiated ovarian carcinoma patient-derived xenografts (PDX): We established an ovarian PDX tumor model using second passage patient tissues implanted into an NSG mouse. Immunohistochemistry analysis of the patient tissue before implantation confirmed the clinical diagnosis that the case appeared to have originated from the ovary and was poorly differentiated (Fig. 7E). PDX tumor growth decreased significantly in the presence of BRU, IKE, or both in combination, with the dual treatment having the greatest effect (Fig. 7F). None of the treatment groups exhibited a significant decrease in mouse body weight (fig. S6I). Lipid droplet formation (H&E and Nile Red staining) was increased in both the IKE and IKE + BRU groups compared to BRU alone or control (fig. S6J). All specific ferroptosis markers measured, such as 4-HNE adduct formation, COX2/PTGS2, and MDA/ROS generation, were significantly increased in the presence of IKE + BRU compared to IKE alone (Fig. 7, G to L). Collectively, these results from three different preclinical models demonstrate the effectiveness of combined NRF2 inhibition and ferroptosis induction in treating cancer.
Fig. 7. Genetic or pharmacological NRF2 inhibition enhanced sensitivity to ferroptotic cell death in preclinical models.
(A) NFE2L2/NRF2 WT or KO SKOV-3 cell lines were grown for 3 to 5 days until 3D sphere formation. Spheroids were treated with IKE (10 μM), BRU (20 nM), or IKE + BRU for 24 hours. Scale bar, 50 μm. (B) Cell viability of NFE2L2/NRF2 WT and KO cell lines at 24-hour treatment assessed by CellTiter-Glo assay. *P < 0.05, n = 8. (C) NFE2L2/NRF2 WT or heterozygous (Het) SKOV-3 cells were subcutaneously injected; once tumors reached 100 mm3, mice were intraperitoneally injected with IKE (40 mg/kg) or vehicle control for 14 days. Tumor volume was measured and plotted as mean tumor burden (mm3). *P < 0.05, n = 15 per group. (D) Tumor tissues were probed with C11-BODIPY581/591, 4-HNE and COX2 antibodies, or FerroOrange. Scale bar, 50 μm. (E) H&E of human ovarian tissue (P0) used for the PDX model. (F) PDX tissue (P1) was implanted; once tumor size reached ~100 mm3, mice were intraperitoneally injected with vehicle, BRU (0.25 mg/kg), IKE (20 mg/kg), or IKE + BRU for 14 days. Tumor size was measured and plotted same as (C). *P < 0.05, n = 10 per group. (G) Tumor tissues were probed with C11-BODIPY581/591, 4-HNE and COX2 antibodies, or FerroOrange. Scale bar, 50 μm. (H) 4-HNE adduct and COX2 protein levels measured by immunoblot analysis. (I) LIP measured using Ferene-S. (J) PTGS2 mRNA levels measured by RT-PCR. (K) ROS levels measured by EPR spectroscopy. (L) MDA detected by TBARS assay. Data in (I) to (L) are represented as means ± SEM of tumor tissues from six individual mice (n = 6). *P < 0.05.
DISCUSSION
Cancer cells can thrive under high ROS conditions and excessive iron accumulation through metabolic reprogramming. Along these lines, many redox and iron regulatory signaling pathways have been shown to be altered, with cancer cells having developed an enhanced dependence on these changes (54). NRF2, one of several key players responsible for metabolic reprogramming, is up-regulated in many types of cancers to cope with the increased metabolic demands of the cancer cell (3, 20, 55). Since NRF2-addicted cancer cells have been shown to be highly resistant to classical chemotherapeutic drugs, we believe that enhancing toxic free iron accumulation to drive cancer cell death through ferroptosis would be the most effective way to kill these resistant cancer cells. However, there has been no effective and safe way to enhance the intracellular LIP, as iron administration would cause damage to normal organs or cells, as reported previously (56, 57). In this study, we have demonstrated that NRF2 inhibition can boost free iron accumulation and sensitize cancer cells and xenograft tumors to the ferroptosis inducer IKE. In contrast to feeding mice a high iron-containing diet that causes organ damage, NRF2 inhibition would selectively enhance free iron to toxic levels only in tumors with high NRF2 expression, and not in normal tissues. This creates an exploitable sensitivity of tumors to ferroptosis, thus providing a therapeutic window. The notion that specific NRF2 inhibition preferentially kills cancer cells without harming normal cells is also supported by these previous findings: First, the basal level of NRF2 in normal cells/tissues is low, whereas most cancer cells/tumors have higher levels of NRF2 to survive the stressful tumor microenvironment, which was also observed in the current study (Fig. 6, A and B). Second, NRF2 KO cells and mice grow and develop normally, indicating that NRF2 is not essential for normal cell growth or mouse development.
Over the years, an increasing number of NRF2 target genes have been identified. NRF2 has been recognized to be crucial for promoting cancer cell survival by controlling cellular redox homeostasis (classical function), proteostasis, and metabolic stress (2–9). In this study, we discovered a signaling network through which NRF2 controls iron homeostasis through ferritin synthesis/degradation, thus dictating the intracellular LIP, and the sensitivity of cancer cells to ferroptotic cell death. Mechanistically, NRF2 regulates iron homeostasis via the following signaling pathways (Fig. 4): (i) NRF2-HERC2-FBXL5-IRP1/2–dependent ferritin synthesis, (ii) NRF2-TFEB-VAMP8–driven ferritinophagy for ferritin degradation, and (iii) NRF2-HERC2-NCOA4–mediated recruitment of ferritin to the autophagosome. Through these signaling axes, NFE2L2/NRF2 deletion disrupted ferritin metabolism through excessive ferritin synthesis and the accumulation of apoferritin-NCOA4 complexes in autophagosomes, leading to the massive toxic free iron accumulation that rendered NFE2L2/NRF2 KO cells more susceptible to ferroptotic cell death. It is worth emphasizing that our finding that NRF2 controls ferritin synthesis and degradation at the protein level is distinct from previous reports that NRF2 controls the mRNA levels of FTH1 and FTL. As shown in Fig. 2, the protein levels of FTH1 and FTL are higher in NRF2- KO cells (Fig. 2D), despite lower FTH1 and FTL mRNA levels (Fig. 2C), which may be due to the fact that the formation of the ferritin complex is primarily controlled by the UPS (HERC2, FBXL5, NCOA4, and IRP1/2), lysosomal degradation (FTH1, FTL, and NCOA4), and protein synthesis (FTH1 and FTL). Furthermore, we demonstrated the important contribution of this newly discovered NRF2-dependent regulation of iron metabolism in controlling cancer cell susceptibility to ferroptotic cell death, as double KO of HERC2 and VAMP8 in WT cells recapitulated the ferroptotic response observed in NFE2L2/NRF2 KO cells. In addition, while NCOA4 KO in NFE2L2/NRF2 WT enhanced ferroptosis, its KO in NFE2L2/NRF2 KO cells was protective. Furthermore, overexpression of VAMP8 in NFE2L2/NRF2 KO SKOV3 cells reduced their ferroptosis sensitivity, while it had no effect in WT cells. These results argue that NRF2-mediated ferritin metabolism and maintenance of iron homeostasis is critical in determining a cancer cell’s response to ferroptosis inducers.
It is important to note that the key is cellular localization/compartmentalization. The free LIP, as well as iron-loaded ferritin cages, is typically located in the cytosol. In normal cells, low iron leads to increased protein levels of NCOA4, which recruits ferritin to the autophagosome for autophagic degradation and release of free iron into the cytosol. In NRF2 KO cells, NRF2-dependent expression of HERC2, which degrades NCOA4, is low, resulting in constitutively higher levels of NCOA4 and recruitment of empty ferritin (apoferritin) to the autophagosome. This decoupling of NCOA4 degradation from the LIP, combined with blockage of autophagy flux (low VAMP8), results in ferritin sequestration away from labile iron in the cytosol, hence the observed increase in the LIP. It is also worth noting that this study is not merely claiming that NRF2 has anti-ferroptotic functions, which as mentioned above is already generally accepted because of the fact that several NRF2 target genes, including SLC7A11 (xCT) and GCLM, play a critical role in modulating intracellular GSH levels and thus GPX4 activity and sensitivity to inducers of ferroptosis. The important finding from this study is that the anti-ferroptosis function of NRF2 extends beyond its regulation of the antioxidant response, which is supported by our results (Fig. 5) showing the contribution of the NRF2-HERC2 and NRF2-VAMP8/NCOA4 axes to controlling iron homeostasis and dictating ferroptosis sensitivity. Another interesting observation is that the double-KO HERC2/VAMP8 cells behaved similarly to NFE2L2/NRF2 single KO cells, as evidenced by a similar accumulation of apoferritin/NCOA4, increased LIP, higher levels of ferroptosis markers (COX2 and 4-HNE adducts), and enhanced sensitivity to ferroptotic cell death. Although ferroptosis has been defined as both lipid peroxidation and iron dependent, whether lipid peroxidation, iron, or both are essential for ferroptosis induction is unclear. The fact that iron chelation can block ferroptosis induction argues that the accumulation of free iron alone is essential and sufficient to induce ferroptosis; however, it is hard to totally isolate the ferroptotic effect of labile iron from lipid peroxidation, since labile iron itself is redox active, the accumulation of which would be expected to lead to increased lipid peroxidation.
In summary, we not only identified a previously undiscovered function of NRF2 in iron signaling by controlling ferritin synthesis/degradation and maintenance of iron homeostasis through HERC2, VAMP8, and NCOA4 but also provided evidence that NRF2 inhibition can enhance free LIP and make cancer cells with high NFE2L2/NRF2 expression specifically vulnerable to ferroptotic inducers. Furthermore, we have also provided proof of concept for the translational value of our findings using three preclinical models, suggesting that a combination therapy tailored around NRF2 inhibition and ferroptosis induction represents an effective therapeutic strategy to treat cancer patients. This study signifies a critical step before ferroptotic inducers can be successfully used in the clinic as the next generation of cancer therapy to treat resistant, refractory, and recurrent cancers.
MATERIALS AND METHODS
Chemicals and reagents
IKE and BRU were synthesized and used as described previously (29, 58). SAS, SF, bixin, tBHQ, sodium (meta)arsenite (S7400), Fer-1, and MTT were purchased from Sigma-Aldrich. Z-VAD-fmk was purchased from ApexBio (A1902). 1-Hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidine (CMH; NOX-0.2), DFO, diethyldithiocarbamic acid (DETC), and Krebs Hepes buffer (NOX-07.6.1) were purchased from Noxygen. Protein A-agarose beads (15918014) were obtained from Invitrogen.
Cell culture and generation of NRF2−/−, HERC2−/−, VAMP8−/−, NCOA4−/−, and HERC2−/−/VAMP8−/− cell lines
SKOV-3, CAOV-3, MES-OV, SW-626, OVCAR-3, and OVCAR-8 ovarian cancer cells were purchased from the American Type Culture Collection. HEY and A2780 cells were obtained from MD Anderson. OSE, FT-246, OVC-201, OVC-205, OVC-302, and OVC-205-mCherry cells were patient-derived cell lines generated and reported previously (59). Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (SKOV-3, CAOV-3, MES-OV, and SW-626), DMEM:F12 (OSE and FT-246), or RPMI (A2780, HEY, OVCAR-3, OVCAR-8, OVC-201, OVC-205, and OVC-302) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
CRISPR KO cell lines were generated using CRISPR-Cas9–mediated gene editing. We used sgRNA species previously designed and published by Wang et al. (60) as an activity optimized pooled sgRNA library. In this pooled library, there are 10 different species of sgRNAs targeting each gene. For each gene, all 10 sgRNA species were cloned into the pSpCas9(BB)-GFP plasmid (61), and genome targeting efficiency of each sgRNA construct was determined by T7 endonuclease I assay. One or two of the highest efficiency sgRNAs were selected for making KO cell lines. SKOV-3 cells were transfected with the selected sgRNA plasmid for 24 hours, and GFP-positive cells were isolated using fluorescence-activated cell sorting and subsequently plated at low confluency. Once individual colonies were visible to the naked eye, they were isolated using a clonal ring and expanded individually. Successful homozygous/heterozygous KO/knockdown of a particular gene was confirmed by Sanger sequencing the sgRNA binding region (around 500 bp flanking the sgRNA binding region) and by immunoblot analysis.
For pooled KO cell lines, for each gene, the top two most efficient sgRNA species (as determined by T7 endonuclease I assay) were cloned into pLentiCRISPRV2 (60). The resulting lentivirus vectors were packaged and transduced into target cell lines at high multiplicity of infection. Positively transduced cell lines were selected for puromycin resistance. The resulting KO cell lines were used as a pooled population. Status of KO was determined by immunoblot analysis.
The best sgRNAs used for making the NRF2−/−, HERC2−/−, VAMP8−/−, NCOA4−/−, and HERC2−/−/VAMP8−/− cell lines are listed below: NRF2: sgRNA-1, 5′-TATTTGACTTCAGTCAGCGA-3′ and sgRNA-2, 5′-TAGTTGTAACTGAGCGAAAA-3′; HERC2: sgRNA-1, 5′-CACCGAACAGCCTGAAGCAGACGG-3′ and sgRNA-2, 5′-CACCGCAGCTGGCGGCACTTCGCGG-3′; VAMP8: sgRNA-1, 5′-CACCGACATCTCCGCAACAAGACAG-3′; NCOA4: sgRNA-1, 5′-CACCGGTATGGCTGTATGAACAGG-3′ and sgRNA-2, 5′-CACCGGAATGTCTTAGAAGCCGTG-3′.
Transfection and live cell fluorescent imaging
Cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific, L3000150) according to the manufacturer’s instructions. The pCMV-myc-VAMP8 plasmid were purchased from Addgene. pcDNA5 FRT/TO SF-HERC2 (ShB-R) was a gift from D. Chan (Addgene plasmid #55613). The plasmid containing mRFP-GFP-LC3 was obtained from E. White (Cancer Institute of New Jersey, Rutgers, NJ). For imaging, SKOV-3 cells were seeded in glass-bottom 35-mm dishes. Cells were transfected with mRFP-GFP-LC3 using Lipofectamine 3000. Twenty-four hours later, cells were gently washed with 1× phosphate-buffered saline (PBS), and DMEM without phenol red was added. Images were taken with a Zeiss Observer Z1 microscope using the Slidebook 4.2.0.11 software (Intelligent Imaging Innovations Inc.).
siRNA-mediated knockdown of NRF2, NCOA4, and VAMP8
To knock down protein levels of NRF2, NCOA4, and VAMP8, 1 × 105 cells were seeded in six-well plates and, 24 hours later, treated with 5 nM nontargeted control (control), NFE2L2/NRF2, NCOA4, or VAMP8 siRNA using Qiagen Hiperfect according to the manufacturer’s instructions. Four siRNA constructs against each target of interest were obtained from Qiagen (Flexitube), and the construct that obtained the maximum knockdown was used for further study. Following 72 hours of transfection, knockdown cells were subjected to immunoblot or immunofluorescence analysis.
mRNA extraction and real-time qRT-PCR
Total mRNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA was then synthesized using 2 μg of mRNA and a Transcriptor first-strand cDNA synthesis kit (Promega). Real-time quantitative PCR (qPCR) was then performed. GAPDH was used for qPCR normalization, and all experiments were measured in triplicate. Primer sequences are as follows: human-VAMP8: 5′-AATGATCGTGTGCGGAACCT-3′ (forward) and 5′-GTGCTCAGATGTGGCTTCCA-3′ (reverse); human-TFR1: 5′-TCGGAGAAACTGGACAGCAC-3′ (forward) and 5′-ATCACGCCAGACTTTGCTGA-3′ (reverse); human-ACSL4: 5′-GGAATGACAGGCCAGTGTGA-3′ (forward) and 5′-TCACCAGTGCAAAACCACCT-3′ (reverse); human-HERC2: 5′-GCCTCGACTCCAAATGGTTG-3′ (forward) and 5′-GACTCCTGCAACAGCTCACT-3′ (reverse); human-NCOA4: 5′-GGGCAACCTCAGCCAGTTAT-3′ (forward) and 5′-GGGATCTGAAAATTCCCAACGG-3′ (reverse); human-PTGS2: 5′-GTTCCACCCGCAGTACAGAA-3′ (forward) and 5′-AGGGCTTCAGCATAAAGCGT-3′ (reverse); human-FTH1: 5′-CTTTGACCGCGATGATGTGG-3′ (forward) and 5′-CCTGAAGGAAGATTCGGCCA-3′ (reverse); human-FTL: 5′-CAGGCCTCCTACACCTACCT-3′ (forward) and 5′-AGGGCCTGGTTCAGCTTTTT-3′ (reverse); human-SLC7A11: 5′-TGCCCAGATATGCATCGTCC-3′ (forward) and 5′-TCTTCTTCTGGTACAACTTCCAGT-3′ (reverse); human-AKR1C1: 5′-CATGCCTGTCCTGGGATTT-3′ (forward) and 5′-AGAATCAATATGGCGGAAGC-3′ (reverse); human-GCLM: 5′-GACAAAACACAGTTGGAACAGC-3′ (forward) and 5′-CAGTCAAATCTGGTGGCATC-3′ (reverse); human-NQO1: 5′-AACTTTCAGAAGGGCCAGGT-3′ (forward) and 5′-CTGGGCTCTCCTTGTTGC-3′ (reverse); human-GAPDH: 5′-CTGACTTCAACAGCGACACC-3′ (forward) and 5′-TGCTGTAGCCAAATTCGTTGT-3′ (reverse).
Chromatin immunoprecipitation–PCR
A ChIP assay was performed according to the manufacturer’s instructions (EZ-CHIPTM, Merck, Germany). Briefly, SKOV-3 WT or NFE2L2/NRF2 KO cells were treated with 1% formaldehyde in DMEM for 10 min to cross-link DNA-protein complexes. The cells were then lysed using SDS lysis buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1% protease inhibitor cocktail (Sigma-Aldrich). Solubilized chromatin was then incubated with anti-NRF2 antibody (Santa Cruz Biotechnology) or normal rabbit IgG (Santa Cruz Biotechnology) for 16 hours at 4°C with rotation, and DNA-protein complexes were pulled down using Protein G-agarose beads (Sigma-Aldrich). DNA from the immunoprecipitated complexes and total chromatin input were extracted via ethanol precipitation, and 1 μl of purified DNA was amplified and run on an agarose gel. Primers were as follows: HERC2-ARE-1: CTCCATCTCTTAAGTGGTGGTGGC (forward) and GGGAAGGGAAAGCCTCCTTCCAAG (reverse); HERC2-ARE-2: GAGCAGCCTCGTTGCGATTG (forward) and CCACCACCACTTAAGAGATGGAGAGAG (reverse); HERC2-ARE-8: CAGTCTGGTCTCGAACTCCTGACG (forward) and CCCAAAATGCTGGGACTACAGACA (reverse); HERC2-ARE-11: GACGAAGTCTTGCTGTGTCGCCCA (forward) and GATCAGGAAGTCAGGAGATTGAGACC (reverse).
Dual luciferase assay
For the dual luciferase assay, a 41-bp portion of the human HERC2 promoter containing the putative ARE sequence for NRF2 and the human VAMP8 promoter containing the putative Coordinated Lysosomal Expression and Regulation (CLEAR) sequence for TFEB were amplified by PCR and then cloned into the pGL4.22 luciferase vector (Promega). Next, SKOV-3 WT or NFE2L2/NRF2 KO cells were cotransfected with 0.5 μg of a plasmid encoding a Firefly luciferase under the control of either a HERC2-ARE– or VAMP8-CLEAR–driven promoter, as well as 0.05 μg of a Renilla luciferase plasmid under the control of a universal promoter as an internal control. Luciferase activity was measured using the dual luciferase reporter assay system (Promega). For relative luciferase activity analysis, the value of Firefly luciferase was normalized to the value of Renilla luciferase. The experiment was repeated three times. The data are expressed as means ± SEM.
VAMP8-CLEAR; 5′-GCTGAAGCAGGAGAATCACTTGAACCTGGGAGGCGGAGGTT-3′
Biotinylated ARE pulldown
Biotin-DNA pulldown was performed as reported previously (62). Briefly, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 1 mM dithiothreitol, 1 mM PMSF, and 1% protease inhibitor cocktail (Sigma-Aldrich). The cell lysates were precleared with streptavidin beads and incubated with 2 μg of biotinylated DNA probes that spanned the ARE-containing sequences in the promoter region of HERC2. The DNA-protein complexes were pulled down by streptavidin beads, and complexes were washed three times, resolved on an SDS–polyacrylamide gel electrophoresis (PAGE) gel, and subjected to immunoblot analysis. The sequences of the 41-bp biotinylated DNA probes used are as follows: HERC2-ARE-1, 5′-CTGTGGGGTAAAGCGGCAGATTCATGCTGCTGTCATTTGTC-3′; HERC2-mtARE-1, 5′-CTGTGGGGTAAAGCGCGAGATAGTTGCTGCTGTCATTTGTC-3′; HERC2-ARE-2, 5′-ATCGCGGGCGCCGGCTGAGCCAGCGGCTCTTGGGAGGCTGC-3′; HERC2-mtARE-2, 5′-ATCGCGGGCGCCGGCACTGCCACGGGCTCTTGGGAGGCTGC-3′; HERC2-ARE-8, 5′-ATGAGGCATGATGCCTGACCCAGCCTTTTTTAAAATGAAGG-3′; HERC2-mtARE-8, 5′-ATGAGGCATGATGCCACTCCCACGCTTTTTTAAAATGAAGG-3′.
Cell viability and ferroptotic cell count
Cell viability was detected using an MTT assay. Cells were seeded in a 96-well plate. Twenty-four hours after treatment, 20 μl of MTT (5 mg/ml) in PBS solution was added directly to the cell culture medium and allowed to incubate for 3 hours. The medium was then removed, and solvent (isopropanol/HCl) was added to the cells and absorbance was measured at 570 nm with the SpectraMax iD5 Multi-Mode Microplate Reader (Molecular Devices). Morphology and ferroptotic cells (bubbles) were detected via IncuCyte (Essen Biosciences), and then ferroptotic cells were counted manually.
Electron paramagnetic resonance spectroscopy
Electron paramagnetic resonance was performed as described previously (63). Briefly, relative production of ROS is represented as the nanomolar concentration of oxidized spin trap divided by the time of trap incubation time, then normalized to the total milligrams of protein per well. Cells were plated in 24-well plates and, after appropriate treatment, incubated with spin trap in the presence of metal chelators (200 μM CMH, 25 μM DFO, and 5 μM DETC in filtered Krebs-Hepes buffer) (Noxygen) for 30 min before collection and measurement. Cells were harvested in RIPA buffer [10 mM sodium phosphate (pH 7.2), 150 mM NaCl, 1% sodium deoxycholate, 2 mM EDTA, 0.1% SDS, and 1% NP-40], and protein concentration was determined with a BCA kit (Pierce; 23227). For xenograft tumor tissues, samples were incubated with spin trap in the presence of metal chelators for 30 min before collection and measurement. The final data were normalized by tissue weight.
GSH assay
Intracellular GSH concentrations were measured using the QuantiChrom GSH assay kit from BioAssay Systems (DIGT-250). The procedure was performed according to the manufacturer’s instructions. Briefly, 5,5′-dithiobis(2-nitrobenzoic acid) reacts with intracellular GSH to form a yellow product. Absorbance was measured at 412 nm, and GSH in cellular extracts was quantified via comparison to a calibration curve generated using GSH as a standard. Data were normalized to cellular protein content. Three independent experiments were performed for the calculation of means and SDs.
Lipid peroxidation assay
The relative intracellular MDA concentration was assessed using the Lipid Peroxidation (MDA) Assay Kit from Sigma-Aldrich (MAKO85). Briefly, MDA in the sample reacts with thiobarbituric acid (TBA) to generate an MDA-TBA adduct. The MDA-TBA adduct can be easily quantified colorimetrically (optical density = 532 nm). C11-BODIPY581/591 dye (Thermo Fisher Scientific, D3861) was used to detect lipid peroxidation in live cells. For imaging, cells were incubated with 2 μM C11-BODIPY581/591 and fluorescence was measured on a fluorescence microscope at ~510 nm. For flow cytometry, 5 × 105 cells were seeded in six-well plates and allowed to adhere overnight at 37°C. Then, cells were treated with dimethyl sulfoxide (DMSO) or SAS (2.5 mM)for 12 hours. Cells were then trypsinized, washed, suspended in Hanks’ balanced salt solution (HBSS) containing 2 μMC11-BODIPY581/591, and incubated at 37°C for 15 min. Cells were pelleted and resuspended in HBSS. Oxidation of the polyunsaturated butadienyl portion of the dye results in a shift of the fluorescence emission peak from ∼590 nm to ∼510 nm.
Iron measurement
Intracellular ferrous (Fe2+) iron levels were determined using the iron assay kit from Sigma-Aldrich (MAKO25). In this assay, iron is released by the addition of an acidic buffer. Ferrous iron is reacted with a chromagen resulting in a colorimetric (593 nm) product, proportional to the iron present. Intracellular chelatable iron was determined using the fluorescent probe FerroOrange from Dojindo (F374). FerroOrange is a fluorescent probe that enables live-cell fluorescence imaging of intracellular Fe2+. Cells were incubated with 1 μM for 20 min, and images were captured using a fluorescence microscope at excitation of 543 nm and emission of 580 nm. For the Perls staining, cells were fixed with 2% paraformaldehyde (PFA), incubated in Perls solution [1% K4Fe(CN)6 and 1% HCl] for 15 min, and then incubated with 0.05% diaminobenzidine (DAB) and hydrogen peroxide (0.5%) for an additional 15 min. For coimmunostaining, cells were first stained with Perl’s solution and DAB and then incubated with anti-LC3, followed by incubation with an Alexa Fluor 594–conjugated anti-rabbit secondary antibody.
Indirect immunofluorescence
Cells were fixed in 2% PFA for 20 min and then washed in PBS. Cells were blocked with 10% FBS for 1 hour and then incubated with primary antibodies: anti-FTH1 (1:250; sc-376594), anti-FTL (1:250; sc-74513), anti-LC3 (1:1000; Sigma-Aldrich, L7543), anti-LAMP1 (1:1000; Cell Signaling Technology, #9091S), and anti-TFEB (1:1000; ab-220695) at 4°C for 16 hours. Cells were incubated with either an anti-mouse Alexa Fluor 488 or anti-rabbit Alexa Fluor 594 (1:1000) secondary antibody for 1 hour. Images were acquired with a Zeiss Observer Z1 fluorescence microscope using the Slidebook 4.2.0.11 software (Intelligent Imaging Innovations Inc.). Mander’s (M1 and M2) and Pearson’s coefficient values for signal overlap were determined using ImageJ [National Institutes of Health (NIH)]. The threshold values were calculated using a Costes approach. For colocalization of FTH1/LC3, FTL/LC3, FTH1/LAMP1, and FTL/LAMP1, >25 cells from three independent experiments were analyzed and presented in Table 2.
Immunoblot analysis
Cells were harvested in 1× sample buffer [62.5 mM tris-HCl (pH 6.9), 3% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.1% brilliant blue]. Lysates were boiled, sonicated, and resolved by SDS-PAGE and then subjected to the following primary antibodies: anti-NCOA4 (1:2000; Bethyl Laboratories, ARA70), anti-FTH1 (1:1000; sc-376594), anti-NRF2 (1:1000; sc-13032), anti-FTL (1:1000; sc-74513), anti-GPX4 (1:1000; sc-166570), anti AKR1C1 (1:1000; sc-398596), anti-GCLM (1:1000; sc-166603), anti-NQO1 (1:1000; sc-32793), anti-4-HNE (1:2000; ab-46545), anti-COX2 (1:1000: sc-166475), anti-LC3 (1:2000; Sigma-Aldrich, L7543), anti-HERC2 (1:1000; Bethyl Laboratories, A301-905), anti-TFR1 (1:1000; ab-84036), anti-IRP2 (1:1000; sc-33682), anti-FBXL5 (1:500; Sigma-Aldrich, AV43104), anti-TFEB (1:1000; ab-220695), anti–phospho-TFEB (S142, 1:1000; Millipore, ABE197), anti–phospho-mTOR (S2448, 1:1000; Cell Signaling Technology, #5536), anti-mTOR (1:1000; Cell Signaling Technology, #2983), anti-SLC7A11 (1:1000; Cell Signaling Technology, #12691), anti-VAMP8 (1:1000. Ab-76021), anti-STX17 (1:1000; Invitrogen, PA5-40127), anti-SNAP29 (1:2000; ProteinTech, 12704-1-AP), and anti–β-actin (1:5000; Sigma-Aldrich, A2066) at 4°C for overnight. Then, membranes were incubated with anti-mouse or anti-rabbit horseradish peroxidase (HRP)–conjugated secondary antibodies (1:3000; Sigma-Aldrich) for 1 hour. All immunoblot images were taken using the Azure 600 system (Azure Biosystems) and analyzed using ImageJ (NIH).
Immunohistochemistry
Slides containing fixed normal (N = 10) and ovarian cancer (N = 40) tissue were processed for NRF2, HERC2, SLC7A11, and VAMP8 expression as described previously (64). Briefly, sodium citrate buffer (0.01 M, pH 6.0) was used for antigen retrieval, and endogenous peroxidase activity was blocked using 0.3% H2O2. Slides were blocked with 5% bovine serum albumin for 30 min and then incubated with primary antibodies against NRF2 (1:100), HERC2 (1:500), SLC7A11 (1:500), and VAMP8 (1:200) overnight at 4°C. The next day, slides were washed with PBS and stained using the EnVision + System-HRP Kit (Dako) as per the manufacturer’s instructions. Images were taken on a Nikon Eclipse 50i microscope using NIS Elements software v4.0. The expression of NRF2, HERC2, VAMP8, and SLC7A11 was measured as the mean intensity of immunoreactivity using ImageJ.
Tumor spheroid assay
Ovarian cancer cell lines were trypsinized and resuspended in complete growth medium, and 20 μl of cell suspension containing 250 to 500 cells was added to each well of the hanging drop array plates from XCENTRIC. The water reservoir on the hanging drop array plates was filled with 2.5 ml of sterile deionized water (DIW). Hanging drop array plates were then placed on top of a six-well plate containing sterile DIW, and the two plates were wrapped in parafilm and placed in a humidified 37°C carbon dioxide incubator. Two to five microliters of fresh growth medium were added to the hanging drops every other day to maintain the 20-μl drop volume. Once spheroids were formed, cells were treated with IKE and BRU at the indicated concentrations for 24 hours and cell viability was measured by ATP CellTiter-Glo assay (Promega, G9682).
Xenograft/PDX models
NSG (NOD.Cg-Prkdcscid⋅Il2rgtm1Wjl/SzJ) female mice, 7 to 8 weeks old, were injected subcutaneously with 2 × 107 SKOV-3 NFE2L2/NRF2 WT or KO cells. The mice were treated after the tumor size reached 100 mm3. Mice were randomly allocated into treatment groups and dosed with IKE (40 mg/kg) or vehicle control (5% DMSO/95% HBSS at pH 4) once daily by intraperitoneal injection for 14 days. IKE concentration was determined on the basis of previous in vivo pharmacokinetic studies (29). Tumor volume was measured daily with an electronic caliper and calculated using the following formula: 0.5 × length × width2. Three hours after the final dosage, mice were euthanized and tumor tissue was dissected, weighed, divided into four to six segments, frozen, and stored at −80°C. Tumor volume change was analyzed in GraphPad Prism, with significance determined by one-way analysis of variance (ANOVA).
PDX model: Patient tumor samples were obtained from the Biospecimen Repository at the University of Arizona Cancer Center following Institutional Review Board approval. Patient tumor tissues were cleaned, cut into 3-mm3 pieces, and implanted aseptically into the mammary pads of female NSG mice for P1 tumors. Once P1 tumors grew to 1.5 to 2 cm3, they were isolated and cut into 3-mm3 pieces for implantation into the experimental mice (P2 tumor). Treatment started once the tumor size reached 100 mm3 using the following four groups: vehicle control, BRU (0.25 mg/kg), IKE (20 mg/kg), or IKE + BRU once daily for 14 days.
Transmission electron microscopy
For TEM, 6 × 105 WT or NFE2L2/NRF2 KO SKOV-3 cells were seeded in six-well plates and, 24 hours later, processed as described previously (65). Briefly, cells were fixed in 2.5% glutaraldehyde + 2% PFA in 0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) or Pipes buffer (pH 7.4) for 1 hour at room temperature. Samples were postfixed in 1% osmium tetroxide in Pipes buffer for 1 hour following a wash in 0.05 M Pipes buffer + 0.05 M glycine and 2 × 10 min washes in 0.1 M Pipes. Following two more 10-min washes in DIW, samples for TEM were block-stained in 2% aqueous uranyl acetate, washed in DIW, dehydrated through a graded series of alcohols, infiltrated with 1:1 alcohol and Spurr’s resin overnight, and then embedded in 100% Spurr’s resin overnight at 60°C. Sections were viewed using a FEI Tecnai Spirit electron microscope (FEI Company, Hillsboro, OR) operated at 100 kV. Eight-bit TIFF images were captured through an AMT 4-megapixel camera.
Statistical analysis
Experimental replicates were independent and performed on separate days. Collected data were analyzed using an unpaired Student’s t test or one-way/two-way ANOVA, and the appropriate post hoc test using GraphPad Prism or the SigmaPlot/Stat package. Data were plotted as means ± SD or SEM using the same package for statistical analysis. Relative densitometry analysis of Western blots was performed using ImageJ (NIH).
Acknowledgments
Funding: Research reported was supported by the following grants from the NIH: R35ES031575 and P42ES004940 to D.D.Z.; R01CA226920 to A.O.; R01ES031463 to E.C.; P01CA87497 and R35CA209896 to B.R.S.; center grants P30 ES006694 and P30CA023074.
Author contributions: D.D.Z. and A.A. conceived the project. A.A., M.D., A.S., J.C., P.L., and Y.W. conducted the experiments and analyzed the data. H.T. and Q.W. synthesized IKE. Z.J. and K.Y. performed the 3D tumor spheroid experiments. J.G.N.G., S.K.C., E.C., A.O., Y.Y.-H., and B.R.S. provided reagents, discussed the project, and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Imidazole ketone erastin (IKE) can be provided by B.R.S. pending scientific review and a completed material transfer agreement. Requests for IKE should be submitted to B.R.S.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.D. D. Zhang, Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 38, 769–789 (2006). [DOI] [PubMed] [Google Scholar]
- 2.J. D. Hayes, A. T. Dinkova-Kostova, K. D. Tew, Oxidative stress in cancer. Cancer Cell 38, 167–197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Rojo de la Vega, E. Chapman, D. D. Zhang, NRF2 and the hallmarks of cancer. Cancer Cell 34, 21–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Tao, P. Liu, G. Luo, M. Rojo de la Vega, H. Chen, T. Wu, J. Tillotson, E. Chapman, D. D. Zhang, p97 negatively regulates NRF2 by extracting ubiquitylated NRF2 from the KEAP1-CUL3 E3 complex. Mol. Cell. Biol. 37, e00660-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Mitsuishi, K. Taguchi, Y. Kawatani, T. Shibata, T. Nukiwa, H. Aburatani, M. Yamamoto, H. Motohashi, Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012). [DOI] [PubMed] [Google Scholar]
- 6.G. M. DeNicola, P.-H. Chen, E. Mullarky, J. A. Sudderth, Z. Hu, D. Wu, H. Tang, Y. Xie, J. M. Asara, K. E. Huffman, I. I. Wistuba, J. D. Minna, R. J. DeBerardinis, L. C. Cantley, NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. Liu, M. Dodson, H. Li, C. J. Schmidlin, A. Shakya, Y. Wei, J. G. N. Garcia, E. Chapman, P. R. Kiela, Q.-Y. Zhang, E. White, X. Ding, A. Ooi, D. D. Zhang, Non-canonical NRF2 activation promotes a pro-diabetic shift in hepatic glucose metabolism. Mol. Metab. 51, 101243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. K. Kwak, N. Wakabayashi, J. L. Greenlaw, M. Yamamoto, T. W. Kensler, Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol. Cell. Biol. 23, 8786–8794 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Pajares, N. Jiménez-Moreno, A. J. Garcia-Yagüe, M. Escoll, M. L. de Ceballos, F. Van Leuven, A. Rábano, M. Yamamoto, A. I. Rojo, A. Cuadrado, Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy 12, 1902–1916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. Itoh, N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, M. Yamamoto, Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D. D. Zhang, S.-C. Lo, J. V. Cross, D. J. Templeton, M. Hannink, Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941–10953 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. McMahon, N. Thomas, K. Itoh, M. Yamamoto, J. D. Hayes, Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a "tethering" mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 281, 24756–24768 (2006). [DOI] [PubMed] [Google Scholar]
- 13.K. I. Tong, A. Kobayashi, F. Katsuoka, M. Yamamoto, Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 387, 1311–1320 (2006). [DOI] [PubMed] [Google Scholar]
- 14.M. Ramos-Gomez, M. K. Kwak, P. M. Dolan, K. Itoh, M. Yamamoto, P. Talalay, T. W. Kensler, Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 98, 3410–3415 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.G. M. DeNicola, F. A. Karreth, T. J. Humpton, A. Gopinathan, C. Wei, K. Frese, D. Mangal, K. H. Yu, C. J. Yeo, E. S. Calhoun, F. Scrimieri, J. M. Winter, R. H. Hruban, C. Iacobuzio-Donahue, S. E. Kern, I. A. Blair, D. A. Tuveson, Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475, 106–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. M. Chio, M. Jafarnejad, Y. Ponz-Sarvise, K. Park, W. Rivera, J. Palm, V. Wilson, Y. Sangar, D. Hao, K. Ohlund, D. Wright, E. J. Filippini, B Da Lee, C. Silva, J. E. Schoepfer, J. M. Wilkinson, G. M. Buscaglia, H. DeNicola, M. Tiriac, H. C. Hammell, E. E. Crawford, C. B. Schmidt, D. J. Thompson, N. Pappin, S. A. Sonenberg, NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell 166, 963–976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.H. Wang, X. Liu, M. Long, Y. Huang, L. Zhang, R. Zhang, Y. Zheng, X. Liao, Y. Wang, Q. Liao, W. Li, Z. Tang, Q. Tong, X. Wang, F. Fang, M. Rojo de la Vega, Q. Ouyang, D. D. Zhang, S. Yu, H. Zheng, NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 8, 334ra351 (2016). [DOI] [PubMed] [Google Scholar]
- 18.X. J. Wang, Z. Sun, N. F. Villeneuve, S. Zhang, F. Zhao, Y. Li, W. Chen, X. Yi, W. Zheng, G. T. Wondrak, P. K. Wong, D. D. Zhang, Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. Lignitto, S. E. LeBoeuf, H. Homer, S. Jiang, M. Askenazi, T. R. Karakousi, H. I. Pass, A. J. Bhutkar, A. Tsirigos, B. Ueberheide, V. I. Sayin, T. Papagiannakopoulos, M. Pagano, Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.H. Kitamura, H. Motohashi, NRF2 addiction in cancer cells. Cancer Sci. 109, 900–911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. J. Wolpaw, K. Shimada, R. Skouta, M. E. Welsch, U. D. Akavia, D. Pe’er, F. Shaik, J. C. Bulinski, B. R. Stockwell, Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 108, E771–E780 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. J. Dixon, K. M. Lemberg, M. R. Lamprecht, R. Skouta, E. M. Zaitsev, C. E. Gleason, D. N. Patel, A. J. Bauer, A. M. Cantley, W. S. Yang, B. Morrison III, B. R. Stockwell, Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. R. Stockwell, J. P. Friedmann Angeli, H. Bayir, A. I. Bush, M. Conrad, S. J. Dixon, S. Fulda, S. Gascon, S. K. Hatzios, V. E. Kagan, K. Noel, X. Jiang, A. Linkermann, M. E. Murphy, M. Overholtzer, A. Oyagi, G. C. Pagnussat, J. Park, Q. Ran, C. S. Rosenfeld, K. Salnikow, D. Tang, F. M. Torti, S. V. Torti, S. Toyokuni, K. A. Woerpel, D. D. Zhang, Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. J. Dixon, B. R. Stockwell, The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 10, 9–17 (2014). [DOI] [PubMed] [Google Scholar]
- 25.W. S. Yang, K. J. Kim, M. M. Gaschler, M. Patel, M. S. Shchepinov, B. R. Stockwell, Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 113, E4966–E4975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. J. Dixon, D. N. Patel, M. Welsch, R. Skouta, E. D. Lee, M. Hayano, A. G. Thomas, C. E. Gleason, N. P. Tatonetti, B. S. Slusher, B. R. Stockwell, Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 3, e02523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.W. S. Yang, R. SriRamaratnam, M. E. Welsch, K. Shimada, R. Skouta, V. S. Viswanathan, J. H. Cheah, P. A. Clemons, A. F. Shamji, C. B. Clish, L. M. Brown, A. W. Girotti, V. W. Cornish, S. L. Schreiber, B. R. Stockwell, Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. F. Ye, K. R. Chaudhary, F. Zandkarimi, A. D. Harken, C. J. Kinslow, P. S. Upadhyayula, A. Dovas, D. M. Higgins, H. Tan, Y. Zhang, M. Buonanno, T. J. C. Wang, T. K. Hei, J. N. Bruce, P. D. Canoll, S. K. Cheng, B. R. Stockwell, Radiation-induced lipid peroxidation triggers ferroptosis and synergizes with ferroptosis inducers. ACS Chem. Biol. 15, 469–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Y. Zhang, H. Tan, J. D. Daniels, F. Zandkarimi, H. Liu, L. M. Brown, K. Uchida, O. A. O’Connor, B. R. Stockwell, Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 26, 623–633.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Yi, A. M. Minikes, X. Jiang, Aiming at cancer in vivo: Ferroptosis-inducer delivered by nanoparticles. Cell Chem. Biol. 26, 621–622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. Anandhan, M. Dodson, C. J. Schmidlin, P. Liu, D. D. Zhang, Breakdown of an ironclad defense system: The critical role of NRF2 in mediating ferroptosis. Cell Chem. Biol. 27, 436–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. Bersuker, J. Hendricks, Z. Li, L. Magtanong, B. Ford, P. H. Tang, M. A. Roberts, B. Tong, T. J. Maimone, R. Zoncu, M. C. Bassik, D. K. Nomura, S. J. Dixon, J. A. Olzmann, The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Doll, F. P. Freitas, R. Shah, M. Aldrovandi, M. C. da Silva, I. Ingold, A. G. Grocin, T. N. Xavier da Silva, E. Panzilius, C. Scheel, A. Mourao, K. Buday, M. Sato, J. Wanninger, T. Vignane, V. Mohana, M. Rehberg, A. Flatley, A. Schepers, A. Kurz, D. White, M. Sauer, M. Sattler, E. W. Tate, W. Schmitz, A. Schulze, V. O’Donnel, B. Proneth, G. M. Popowicz, D. Pratt, J. P. F. Angeli, M. Conrad, FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 34.C. Mao, X. Liu, Y. Zhang, G. Lei, Y. Yan, H. Lee, P. Koppula, S. Wu, L. Zhuang, B. Fang, M. V. Poyurovsky, K. Olszewski, B. Gan, DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 593, 586–590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S. K. Chambers, H. H. Chow, M. F. Janicek, J. M. Cragun, K. D. Hatch, H. Cui, C. Laughren, M. C. Clouser, J. L. Cohen, H. M. Wright, N. Abu Shahin, D. S. Alberts, Phase I trial of intraperitoneal pemetrexed, cisplatin, and paclitaxel in optimally debulked ovarian cancer. Clin. Cancer Res. 18, 2668–2678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.G. H. Giornelli, Management of relapsed ovarian cancer: A review. Springerplus 5, 1197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. Venkatesh, N. A. O’Brien, F. Zandkarimi, D. R. Tong, M. E. Stokes, D. E. Dunn, E. S. Kengmana, A. T. Aron, A. M. Klein, J. M. Csuka, S. H. Moon, M. Conrad, C. J. Chang, D. C. Lo, A. D’Alessandro, C. Prives, B. R. Stockwell, MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 34, 526–543 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. A. Vashisht, K. B. Zumbrennen, X. Huang, D. N. Powers, A. Durazo, D. Sun, N. Bhaskaran, A. Persson, M. Uhlen, O. Sangfelt, C. Spruck, E. A. Leibold, J. A. Wohlschlegel, Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326, 718–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. A. Salahudeen, J. W. Thompson, J. C. Ruiz, H.-W. Ma, L. N. Kinch, Q. Li, N. V. Grishin, R. K. Bruick, An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326, 722–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.N. C. Andrews, An iron-clad role for proteasomal degradation. Cell Metab. 14, 281–282 (2011). [DOI] [PubMed] [Google Scholar]
- 41.T. Moroishi, T. Yamauchi, M. Nishiyama, K. I. Nakayama, HERC2 targets the iron regulator FBXL5 for degradation and modulates iron metabolism. J. Biol. Chem. 289, 16430–16441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.E. C. Pietsch, J. Y. Chan, F. M. Torti, S. V. Torti, Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. J. Biol. Chem. 278, 2361–2369 (2003). [DOI] [PubMed] [Google Scholar]
- 43.J. D. Mancias, L. Pontano Vaites, S. Nissim, D. E. Biancur, A. J. Kim, X. Wang, Y. Liu, W. Goessling, A. C. Kimmelman, J. W. Harper, Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.E. Itakura, C. Kishi-Itakura, N. Mizushima, The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012). [DOI] [PubMed] [Google Scholar]
- 45.M. Sardiello, M. Palmieri, A. di Ronza, D. L. Medina, M. Valenza, V. A. Gennarino, C. Di Malta, F. Donaudy, V. Embrione, R. S. Polishchuk, S. Banfi, G. Parenti, E. Cattaneo, A. Ballabio, A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 46.G. Napolitano, A. Esposito, H. Choi, M. Matarese, V. Benedetti, C. Di Malta, J. Monfregola, D. L. Medina, J. Lippincott-Schwartz, A. Ballabio, mTOR-dependent phosphorylation controls TFEB nuclear export. Nat. Commun. 9, 3312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Settembre, R. Zoncu, D. L. Medina, F. Vetrini, S. Erdin, S. Erdin, T. Huynh, M. Ferron, G. Karsenty, M. C. Vellard, V. Facchinetti, D. M. Sabatini, A. Ballabio, A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Roczniak-Ferguson, C. S. Petit, F. Froehlich, S. Qian, J. Ky, B. Angarola, T. C. Walther, S. M. Ferguson, The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. A. Martina, Y. Chen, M. Gucek, R. Puertollano, MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. B. Alvero, D. A. Fishman, M. B. Qumsiyeh, M. Garg, B. M. Kacinski, E. Sapi, Telomerase prolongs the lifespan of normal human ovarian surface epithelial cells without inducing neoplastic phenotype. J. Soc. Gynecol. Investig. 11, 553–561 (2004). [DOI] [PubMed] [Google Scholar]
- 51.A. M. Karst, R. Drapkin, Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nat. Protoc. 7, 1755–1764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. V. Torti, F. M. Torti, Iron and cancer: 2020 vision. Cancer Res. 80, 5435–5448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. Namgaladze, D. C. Fuhrmann, B. Brüne, Interplay of Nrf2 and BACH1 in inducing ferroportin expression and enhancing resistance of human macrophages towards ferroptosis. Cell Death Discov. 8, 327 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. H. Manz, N. L. Blanchette, B. T. Paul, F. M. Torti, S. V. Torti, Iron and cancer: Recent insights. Ann. N. Y. Acad. Sci. 1368, 149–161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. S. Lawrence, P. Stojanov, C. H. Mermel, J. T. Robinson, L. A. Garraway, T. R. Golub, M. Meyerson, S. B. Gabriel, E. S. Lander, G. Getz, Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Yu, L. Jiang, H. Wang, Z. Shen, Q. Cheng, P. Zhang, J. Wang, Q. Wu, X. Fang, L. Duan, S. Wang, K. Wang, P. An, T. Shao, R. T. Chung, S. Zheng, J. Min, F. Wang, Hepatic transferrin plays a role in systemic iron homeostasis and liver ferroptosis. Blood 136, 726–739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.H. Wang, P. An, E. Xie, Q. Wu, X. Fang, H. Gao, Z. Zhang, Y. Li, X. Wang, J. Zhang, G. Li, L. Yang, W. Liu, J. Min, F. Wang, Characterization of ferroptosis in murine models of hemochromatosis. Hepatology 66, 449–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D. Ren, N. F. Villeneuve, T. Jiang, T. Wu, A. Lau, H. A. Toppin, D. D. Zhang, Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. U.S.A. 108, 1433–1438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.F. Bi, Z. Jiang, W. Park, T. M. P. Hartwich, Z. Ge, K. Y. Chong, K. Yang, M. J. Morrison, D. Kim, J. Kim, W. Zhang, L. M. Kril, D. S. Watt, C. Liu, Y. Yang-Hartwich, A benzenesulfonamide-based mitochondrial uncoupler induces endoplasmic reticulum stress and immunogenic cell death in epithelial ovarian cancer. Mol. Cancer Ther. 20, 2398–2409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.T. Wang, K. Birsoy, N. W. Hughes, K. M. Krupczak, Y. Post, J. J. Wei, E. S. Lander, D. M. Sabatini, Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.F. A. Ran, P. D. Hsu, J. Wright, V. Agarwala, D. A. Scott, F. Zhang, Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.T. Wu, X. J. Wang, W. Tian, M. C. Jaramillo, A. Lau, D. D. Zhang, Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic. Biol. Med. 67, 69–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.M. Dodson, M. R. de la Vega, B. Harder, R. Castro-Portuguez, S. D. Rodrigues, P. K. Wong, E. Chapman, D. D. Zhang, Low-level arsenic causes proteotoxic stress and not oxidative stress. Toxicol. Appl. Pharmacol. 341, 106–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P. Liu, M. Rojo de la Vega, S. Sammani, J. B. Mascarenhas, M. Kerins, M. Dodson, X. Sun, T. Wang, A. Ooi, J. G. N. Garcia, D. D. Zhang, RPA1 binding to NRF2 switches ARE-dependent transcriptional activation to ARE-NRE-dependent repression. Proc. Natl. Acad. Sci. U.S.A. 115, E10352–E10361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.A. Anandhan, N. Nguyen, A. Syal, L. A. Dreher, M. Dodson, D. D. Zhang, L. Madhavan, NRF2 loss accentuates parkinsonian pathology and behavioral dysfunction in human α-synuclein overexpressing mice. Aging Dis. 12, 964–982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6