Abstract
Innate immunity is one of the most ancient and conserved aspect of the immune system. It is responsible for an anti-infective response and has been intrinsically linked to the generation of inflammation. While the inflammatory response entails signaling to the adaptive immune system, it can be self-perpetuating and over-exaggerated, resulting in deleterious consequences, including cytokine storm, sepsis, and the development of inflammatory and autoimmune diseases. Cytokines are the defining features of the immune system. They are critical to mediation of inflammation and host immune defense, and are tightly regulated at several levels, including transcriptional and post-transcriptional levels. Recently, the role of post-transcriptional regulation in fine-tuning cytokine expression has become more appreciated. This interest has advanced our understanding of how various mechanisms are integrated and regulated to determine the amount of cytokine production in cells during inflammatory responses. Here, we would like to review how innate immunity recognizes and responds to pathogens by pattern-recognition receptors, and the molecular mechanisms regulating inflammatory responses, with a focus on the post-transcriptional regulations of inflammatory mediators by RNA-binding proteins, especially Regnase-1. Finally, we will discuss the regulatory mechanisms of Regnase-1 and highlight therapeutic strategies based on targeting Regnase-1 activity and its turnover as potential treatment options for chronic and autoimmune diseases.
Keywords: Innate immunity, Inflammation, Cytokines, Post-transcriptional regulations, RNA-binding proteins, Regnase-1
Introduction
Humans are continuously exposed to pathogens and debris from damaged host cells. Yet, only on some occasions can they cause infectious diseases, indicating that a delicate balance between induction and resolution of inflammation is critical for the host defense and survival. Failure to resolve inflammation results in the development of chronic inflammatory diseases, which are associated with irreversible tissue damage.
Since the discovery of pattern-recognition receptors (PRRs) in the mid-1990s (Janeway 1989), much progress has been made in unraveling the role of innate immunity in host defense. In particular, it has been revealed that the inflammatory process is initiated and/or propagated by a variety of endogenous molecules, including cytokines and chemokines, which are most often released during a short period of time upon recognition of conserved pathogen-associated molecular patterns (PAMPs) from microorganisms and damage-associated molecular patterns (DAMPs) from damaged host cells (Medzhitov and Janeway 1997). Upon PAMP recognition, PRRs initiate a series of signaling pathways that results in the activation of transcriptional factors, including nuclear factor kappa-B (NF-κB), activator protein 1 (AP-1), and interferon (IFN) regulatory factors (IRF). Activation of PRR-signaling pathways results in the transcription of a family of mRNAs encoding proinflammatory cytokines such as interleukin-6 (IL-6), IL-1β, and tumor necrosis factor (TNF), which are critical for initiating cell recruitment and local inflammation for clearance of many pathogens. However, the overproduction of cytokines by immune cells can be fatal and is the major driving cause of inflammatory and autoimmune diseases. Accordingly, mammalian systems have formulated ways to fine-tune cytokine expression at multiple levels, including gene transcription, mRNA translation, and mRNA degradation.
A growing body of evidence suggests that RNA-binding proteins (RBPs) are intimately involved in cytokine mRNA metabolism, specifically in the degradation of cytokine mRNAs by post-transcriptional mechanisms (Anderson 2008; Mino and Takeuchi 2013). RBPs bind to sequence and/or structural motifs in RNA via modular combinations of structurally well-defined RNA-binding domains such as RNA-recognition motif (RRM), Cys-Cys-Cys-His (CCCH) zinc finger (ZF) domain, or DEAD box helicase domain. Implicit in this characterization is the deficiency or dysregulation of RBP expression in the accumulation of cytokine transcripts and the development of autoinflammatory diseases. In this review, we summarize the PRR mechanisms underlying the activation and regulation of innate inflammatory responses. After that, we would like to review the regulatory mechanisms of the master transcriptional factors, NF-κB and IRF3/IRF7, as well as the post-transcriptional regulations that regulate cytokine mRNA turnover by RNA-binding proteins (RBPs) such as Regnase-1 and Roquin. Lastly, we will outline the post-transcriptional and post-translational regulations of Regnase-1 and therapeutic interventions which aim to specifically manipulate Regnase-1 stability and/or production.
Innate immunity-mediated pathogen elimination
Our immune system is composed of two types of immunity, innate and adaptive, to ensure a rapid and correct response to intruders. While the innate arm uses germline-encoded receptors to sense conserved molecular patterns of pathogens by PRRs, the adaptive arm comprises clonally selected lymphocytes. The hypothesis of PRRs and conserved molecular patterns was first proposed in the mid-1990s (Janeway 1989). Since then much progress has been made in unraveling the role of innate immunity in biological systems. The innate immune system in vertebrates is characterized by five major types of receptors based on protein domain homology; transmembrane Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), RIG-I-like receptors (RLRs), C-type lectin receptor (CLR), and absent in melanoma-2 (AIM2) like receptors (ALRs), each recognizing specific pattern derivatives of bacterial, fungal or virus, genotoxic stress or tissue damage (Akira et al. 2006; Takeuchi and Akira 2010; Broz and Dixit 2016; Kelley et al. 2019; Geijtenbeek and Gringhuis 2022). In general, PRRs are composed of ligand recognition domains, intermediate domains, and effector domains. Of note, the TLR family member is one of the best characterized PRR families, with each TLR harboring N-terminal leucine-rich repeats (LRRs, 20–30 amino acids), a transmembrane region followed by a cytoplasmic Toll/IL-1R (TIR) domain, which plays a major role in signal transduction. Because the recognition and binding of PRRs to their ligands is critical for the initiation of appropriate innate immune response, the location of receptors determines the molecules they are going to sense. For example, membrane-bound sensors survey molecules outside the cells or inside organelles, whereas cytosolic receptors are activated only when they encounter microbe/virus derivatives that have invaded the host cells. Based on their cellular localization and the pathogens they recognize, PRRs can be roughly categorized into three groups, (1) plasma membrane, (2) endosomal/lysosomal membrane, and (3) cytoplasm:
PRRs on the plasma membrane include TLR1, TLR2, TLR4, TLR5, TLR6 and the CLR
TLRs present on the plasma membrane mainly recognize distinct membrane component derived from pathogenic microorganisms, in the form of heterodimers or homodimers. TLR1/TLR2 and TLR2/TLR6 recognize lipoteichoic acids, TLR4 recognizes lipopolysaccharide (LPS) of Gram-negative bacteria, while TLR5 recognizes flagellin. Of note, stimulation of these TLR pathways results in the activation of NF-κB, mitogen-activated protein kinases (MAPK), and interferon (IFN)-regulatory factors, through associating with the cytoplasmic adaptor protein MyD88 (myeloid differentiation primary response gene 88) (Wesche et al. 1997; Medzhitov et al. 1998). In addition, activation of TLR4 not only results in the production of NF-κB-dependent transcription of proinflammatory cytokines, but also induces the generation of type I IFNs by recruiting the adaptor molecule TIR domain-containing adaptor inducing IFN-β (TRIF). On the other hand, CLRs recognize specific carbohydrates on the surface of pathogens through its carbohydrate recognition domain (CRD). Interestingly, CLRs are mainly expressed on antigen-presenting cells such as dendritic cells (DCs) and macrophages. Among the CLR family, dendritic cell-associated C-type lectin (dectin)-1 and dectin-2 detect β-1,3-glucans and mannan on fungal cell walls, respectively.
PRRs on the endosomal/lysosomal membrane include TLR3, TLR7, TLR8, and TLR9
This family of TLRs is usually expressed in the form of homodimers, and mainly recognize nucleic acids from viruses and bacteria, as well as endogenous nucleic acids from damaged cells. Unmethylated DNA with CpG DNA motifs derived from bacteria and viruses, as well as single-stranded DNA (ssDNA), can be recognized by TLR9, while double-stranded RNA (dsRNA) and synthetic dsRNA analog polyinosinic polycytidylic acid (poly I:C) are detected by TLR3. In addition, TLR7 and TLR8 are responsible for the recognition of single-stranded RNA (ssRNA) from RNA viruses, as well as anti-viral small compounds imidazoquinolines (Diebold et al. 2004; Heil et al. 2004).
Intracellular PRRs include NLR, RLR, AIM2, and the cyclin GMP-AMP synthase (cGAS)
NLRs commonly possess protein-binding motifs such as pyrin domain (PYD), caspase activation and recruitment domains (CARDs) or baculovirus inhibitor of apoptosis protein repeat (BIR), both are members of the death domain superfamily—in their N-terminal regions (Inohara et al. 2005). Particularly, activation of the cytoplasmic NLRs by components of bacterial outer membranes (muramyl dipeptide) or cell wall component including diaminopimelic acid, triggers NLR oligomerization. Two of the best-characterized NLR family proteins are NOD1 and NOD2, which amplify the effect of anti-pathogen infection by recruiting adaptor molecules RIP2/RICK to drive NF-κB-mediated expression of proinflammatory cytokines (Hsu et al. 2008; Park et al. 2007). In addition, it was reported that NOD2 also stimulates IRF3-dependent expression of type I IFNs to combat with respiratory syncytial virus infection (Sabbah et al. 2009). Other NLRs, including NLRP1, NLRP3 and NLRC4, oligomerize to form multiprotein inflammasome complexes, which ultimately results in the recruitment of pro-caspase-1 and their activation through proteolytic cleavage (Faustin et al. 2007; Franchi et al. 2009; Kufer and Sansonetti 2011). The activated caspase-1 then cleaves the immature pro-inflammatory cytokines pro-IL-1β, pro-IL-18, and Gasdermin-D, which are responsible for subsequent inflammatory signaling event and pyroptotic cell death, respectively. The RLR family also recognizes viral nucleic acids and is composed of three major members, including RIG-I, melanoma differentiation factor 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (Yoneyama and Fujita 2008; Takeuchi and Akira 2009). While MDA5 recognizes relatively long dsDNA (> 1000 bp), RIG-I tends to detect short-chain dsRNA harboring 5’-triphosphate ends. Upon recognition of the corresponding viral RNA, activated RIG-I and MDA5 induce downstream signal transduction by associating with the adaptor protein mitochondrial antiviral signaling protein (MAVS) through the two N-terminal CARD domains, resulting in the IRF3/IRF7-mediated production of type I IFNs and type III IFNs, as well as the NF-κB-mediated production of target cytokines. In addition, sensing of intracellular dsDNA by AIM2 triggers the formation of inflammasomes and caspase-1, resulting in the maturation and release of IL-1β and IL-18. Alternatively, dsDNA can also be recognized by cGAS, the binding of which activates cGAS catalytic activity to produce 2’–3’-cyclic GMP-AMP (cGAMP) as a second messenger molecule to further trigger the STING (stimulator of interferon genes)-mediated response against viral infection through the production of type I IFNs and related gene products by activating IRF3 (Ishikawa et al. 2009; Ablasser et al. 2013; Decout et al. 2021).
Regulation of the activity of transcription factors
A major consequence of the PRR-induced innate immune response is the transcription of proinflammatory molecules through activation of distinct intracellular signaling pathways. In the case of TLR, for example, upon recognition of PAMPs and DAMPs, the TIR domains conduct signal transduction by binding to different adaptor molecules in the cytoplasmic region, thereby activating the downstream transcription factors including NF-κB and IRF, to promote the expression of proinflammatory cytokines, chemokines and type I IFNs. Notably, two of the well-characterized TIR-containing adaptor molecules, MyD88 and TRIF, mediate most, if not all, TLR signaling pathways, resulting in the activation of transcription factors NF-κB and IRF3/IRF7, respectively.
NF-κB
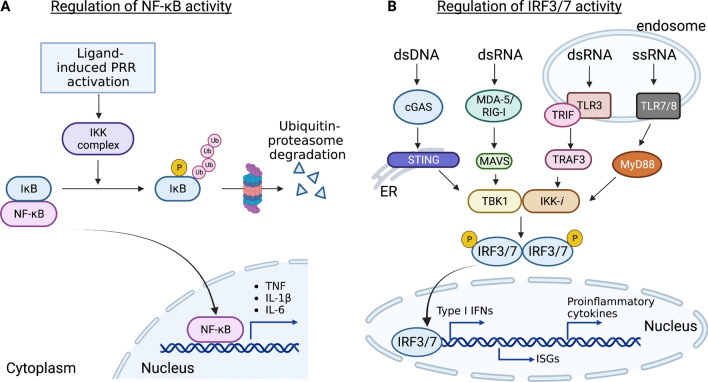
NF-κB is a well-known transcription factor in the regulation of innate and adaptive immune responses, lymphocyte function, and cell survival, and is widely expressed in different types of immune cells (Schulze-Luehrmann and Ghosh 2006; Liu et al. 2017). Because aberrant activation of NF-κB is associated with chronic inflammatory diseases such as rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, and asthma, activation of NF-κB is tightly controlled by its cellular localization by distinct inhibitor proteins. Under homeostatic conditions, NF-κB remains in an inactive form and is sequestered in the cytoplasm by IκB (inhibitor of κB) family members and related proteins through their ankyrin repeats. Canonical NF-κB pathway responds to stimuli from diverse immune receptors including PRRs (TLRs, NLRs, RLRs, and CLRs), cytokine receptors, as well as T cell-receptor (TCR) and B-cell receptor (BCR). The canonical pathway depends on site-specific phosphorylation of IκB at distinct N-terminal lysine residues (Lys 48) by the IκB kinase (IKK) complex, which ultimately results in the degradation of IκB molecules by the ubiquitination-proteasome pathway (Fig. 1A). In particular, IKK complex contains two catalytic subunits, IKKα and IKKβ, and a regulatory subunit known as NF-κB essential modulator (NEMO). Degradation of IκB allows NF-κB translocation to the nucleus for initiation of transcriptional activation of a wide range of genes that control immune responses, including proinflammatory cytokines (Il6, Il1β, Tnf and Il12p40), chemokines, as well as activation, differentiation and effector functions of inflammatory T cells. Importantly, it is reported that after its degradation by IKK-mediated phosphorylation, IκB is resynthesized via NF-κB-mediated induction of its gene expression, thus providing a negative feedback mechanism to terminate NF-κB responses in a timely manner (Sun et al. 1993). These findings together illustrate the intricacy and complexity of dynamic NF-κB shuttling between cytoplasm and nucleus, suggesting that better understanding of the mechanism is crucial for designing more specific and effective therapeutic agents for the treatment of inflammatory diseases.
Fig. 1.
Regulation of the activation of transcription factor NF-κB and IRF3/7. A IκB is a pivotal regulator of NF-κB activity. Under homeostatic condition, NF-κB remains inactive because it is sequestered in cytoplasm by IκB. Following stimulation of PRRs, IκB proteins are rapidly phosphorylated by the IκB kinases (IKKs), resulting in the ubiquitin-mediated proteasomal degradation (Lys48) of IκB. This enables NF-κB translocation to the nucleus for transcription activation of inflammation-mediated genes such as Il6, Il1b and Tnf. B Binding of double stranded DNA (dsDNA) by cGAS results in the synthesis of cGAMP, which then binds and activates the endoplasmic reticulum resident protein STING, while sensing of viral RNA by RLRs such as RIG-1 and MDA5, activates a signaling cascade through the adapter MAVS. This event stimulates the IKK-related kinase IKK-i/TBK1, which phosphorylate IRF3/7 to induce production of type I interferons (IFNs), interferon-stimulated genes (ISGs) and proinflammatory cytokines. Similarly, TLR7/8 senses single-stranded RNA (ssRNA) in the endosome and signals through the adaptor molecule MyD88, while TLR3 recognizes double-stranded RNA (dsRNA) and uses the adaptor protein TRIF to further activate TRAF3 and IKK-i/TBK1 and the phosphorylation of IRF3/7 and the nucleus translocation to activate IFN expressions. This figure is made with BioRender: https://biorender.com/
IRF3/IRF7
IRF3 and IRF7 are well-studied transcriptional factors which counteract viral infection by promoting the transcription of type I IFNs and IFN-inducible genes. IRF3 is constitutively expressed in various types of cells, whereas IRF7 is abundantly expressed in plasmacytoid DCs (pDCs) (Au et al. 1998). Of note, IRF expression is induced by type I IFNs, resulting in a positive feedback loop to maximally drive type I IFN expression (Marié et al. 1998). The activity of IRF3 is controlled by the phosphorylation and dimerization state, as well as its translocation ability from the cytoplasm to the nucleus. IRF3 is activated by phosphorylation by the Traf-family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1) and inducible IκB kinase (IKK-i), also known as IKKε, which changes its confirmation and triggers dimerization and translocation into the nucleus, where it associates with co-activator CREB-binding protein (CBP) and binds to the IFN-stimulated regulatory element (ISREs) of the promoters of IFN-α and IFN-β (Fitzgerald et al. 2003; Sharma et al. 2003) (Fig. 1B). While TLR3 and TLR4 use the adaptor protein TRIF to recruit IKK-i/TBK1 to endosomes and phagosomes, respectively, RIG-I/MDA5 recruit the adaptor protein MAVS to recruit and activate IKK-i/TBK1 at the mitochondrial membrane. Recently a growing body of evidence suggests that cytosolic DNA-detecting PRRs such as cGAS, utilize the adaptor protein STING to recruit and activate TBK1, and ultimately results in IRF3 phosphorylation and production of IFNs (Chen et al. 2016). Similarly, IRF7 is activated by TBK1/IKK-i downstream of cytosolic RNA/DNA sensors and TRIF dependent pathways. However, IRF7 can form a homodimer or heterodimer with IRF3 to induce IFN-α/β expression. Alternatively, stimulation of the TLR7/TLR8 pathway in pDCs results in phosphorylation and activation of IRF7 by the recruitment of MyD88 and the activation of IRAK1/2/4 signaling complex, and is independent on IKK-i/TBK1 pathway (Ikushima et al. 2013). Indeed, cells that are deficient of IRF3 and IRF7 are defective in IFN-α/β production, in response to viral infection (Honda et al. 2005). Thus, the coordinated activity of IRF3/7 downstream of the various PRRs determines the extent of type I IFN production and the pattern of cytokines induced. Given the importance of IRF3 and IRF7 in regulating the production of a wide range of antiviral and inflammatory mediators, it is hardly surprising that dysregulation of IRF3/IRF7 has been implicated in COVID-19 and the pathogenesis of autoimmune diseases including systemic lupus erythematosus (SLE). In particular, it is reported that SARS-CoV-2 uses the nucleocapsid protein to inhibit IRF3 nuclear translocation (Wang et al. 2021b) and RIG-I/MAVS signaling (Wang et al. 2021a), suggesting that targeting this process might be a useful therapeutic strategy to boost antiviral immunity against COVID-19.
Post-transcriptional regulation of inflammatory messenger RNAs
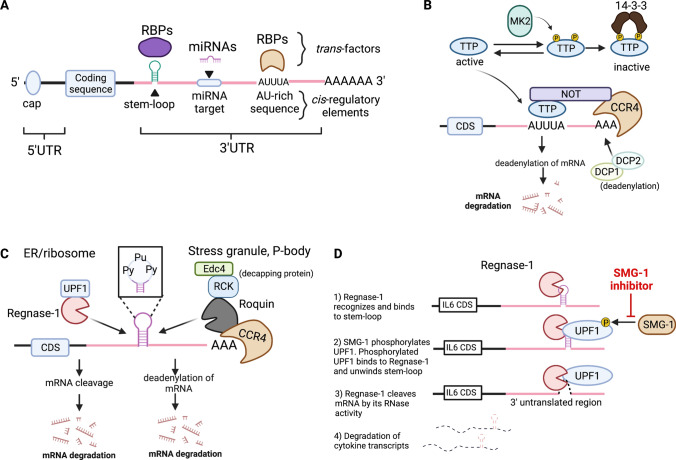
Thus far, we have described the host receptors and signaling pathways of key transcription factors such as NF-κB and IRF3/7, and how they act as an accelerator to produce inflammatory molecules to protect the host from infection. Equally important to this is the rapid elimination of the inflammatory transcripts to prevent further tissue damage and maintain immune homeostasis. Post-transcriptional regulation constitutes an essential layer of regulation of innate inflammatory signaling via affecting the stability of inflammatory mediators at the mRNA level including splicing, capping, polyadenylation, export, localization, translation and decay. Emerging evidence suggests that CCCH-zinc finger proteins are intimately involved in mRNA metabolism, specifically in the degradation of cytokine mRNAs (Hall 2005; Maeda and Akira 2017), and have crucial roles in the regulation of cytokine production, immune cell activation, immune homeostasis and antiviral innate immune responses (Fu and Blackshear 2016; Mino and Takeuchi 2018; Uehata and Takeuchi 2021). Here, we describe how RBPs, particularly tristetraprolin (TTP) and two well-characterized ribonucleases namely Regnase-1 and Roquin, contribute to promoting the resolution of inflammation through recognizing specific cis-elements such as adenine/uridine-rich elements (AREs) and stem-loop structures in the 3’ untranslated regions (UTRs) to promote mRNA decay of inflammatory molecules (Fig. 2A).
Fig. 2.
Post-transcriptional regulation of inflammatory mRNAs by different RBPs. A Schematic drawing of different cis-regulatory elements in a mRNA. The cis-regulatory elements in the 3’ UTR include stem-loop secondary structure, AU-rich elements and miRNA target sequence. These elements are recognized by RNA-binding proteins or miRNA through cognate RNA-binding domains and distinct sequence, respectively. B TTP recognizes the AU-rich elements in the 3’ UTR of target genes and induces exonucleolytic mRNA decay by recruiting the CCR4-NOT deadenylase complex and DCP1/2 decapping enzymes. Under homeostatic condition, MK2 phosphorylates TTP at two distinct serine residues, and the phosphorylated TTP is sequestered by the 14-3-3 protein, making it inaccessible to the target mRNAs. Upon PRR stimulation, however, its phosphorylation is readily reversible by a protein phosphatase, allowing it to access to the inflammatory cytokine transcripts. C Regnase-1 and Roquin are well-characterized RBPs that recognize stem-loop structures located at the 3’ UTR of common target transcripts. A pyrimidine-purine-pyrimidine (Py-Pu-Py) tri-nucleotide sequence at the stem-loop region preferentially associates with Regnase-1 and Roquin. While Regnase-1 degrades mRNAs with the association of the helicase UPF1 in the ribosome and endoplasmic reticulum (ER), Roquin targets transcripts by recruiting a CCR4-NOT deadenylase complex and Edc4/RCK decapping proteins in the possessing body (PB) and stress granules (SG). CDS, coding sequence. D Sequential dynamics of Regnase-1 degradation of mRNA is mediated by unwinding of stem-loop structures by the helicase UPF1. Small molecule SMG-1 inhibitor blocks SMG1-mediated phosphorylation of UPF1. This figure is made with BioRender: https://biorender.com/
TTP, also known as ZFP36, plays important role in the regulation of immune functions (Taylor et al. 1996; Sanduja et al. 2011). TTP-deficient mice spontaneously develop severe inflammatory and autoimmune phenotypes characterized by excessive accumulation of TNF and high titers of anti-DNA and antinuclear antibodies in periphery (Taylor et al. 1996; Carballo et al. 1998). Structural studies indicated that TTP harbors two CCCH ZF domains, which are responsible for the destabilization of TNF mRNA (Fabian et al. 2013). Subsequent biochemical studies illustrated that through the association to AU-rich elements (AREs) (AUUUA sequence) in the TNF 3’ UTR, TTP promotes the removal of poly(A) tail by recruiting CCR4-NOT deadenylase and decapping enzymes DCP1 and DCP2 to further trigger exonucleolytic mRNA decay (Carballo et al. 1998; Lai et al. 1999; Lykke-Andersen and Wagner 2005). Consistently, deletion of the ARE sequence in TNF 3’ UTR results in a similar phenotype in mice, indicating the importance of TTP-ARE recognition in controlling TNF mRNA decay (Kontoyiannis et al. 1999). Besides TNF, TTP can destabilize a variety of inflammatory cytokine mRNAs including IL2, IL6 and GMCSF (granulocyte–macrophage colony-stimulating factor) by associating to the AREs in their 3’ UTRs (Carballo et al. 2000; Zhao et al. 2011). TTP expression is inducible by several inflammatory modulators including TNF, LPS, glucocorticoids, insulin and IFN-γ. Upon TLR stimulation, however, TTP becomes phosphorylated at site Ser-52 and Ser-178 by p38/MAPK-activated protein kinase 2 (MK2) (Mahtani et al. 2001). Phosphorylated TTP is then recognized by 14-3-3 proteins, which sequester TPP from recruiting CCR4-NOT complex for destabilization of inflammatory mRNAs (Chrestensen et al. 2004) (Fig. 2B). Interestingly, mutation of the two TPP phosphorylation sites to alanine residues results in potent suppression of inflammatory responses in mice (Ross et al. 2015), suggesting that small molecules specifically blocking TPP phosphorylation might provide therapeutic benefits for the treatment of inflammatory diseases and thus warrants further investigation.
Regnase-1, also known as Zc3h12a and Mcpip1, is an endoribonuclease that plays important roles in the regulation of immune response, iron metabolism, and virus infection (Matsushita et al. 2009; Lin et al. 2013, 2014; Uehata et al. 2013; Cui et al. 2017; Yoshinaga et al. 2017; Li et al. 2018; Nakatsuka et al. 2018). Structural study reveals that Regnase-1 contains four domains: an N-terminal domain (NTD), a CCCH ZF domain, and a PIN domain-like RNase domain and a C-terminal domain (CTD) (Yokogawa et al. 2016). Accumulating evidence suggests that Regnase-1 expression is inducible by a variety of stimulus, including LPS, IL-1β and IL-17A (Iwasaki et al. 2011; Ruiz-Romeu et al. 2016; Takeuchi 2018; Tanaka et al. 2019). Previous reports showed that mice with Regnase-1 deficiency (Regnase-1–/–) displayed increased cytokine, serum immunoglobin and autoantibody levels, and spontaneously die within 8–12 weeks after birth (Matsushita et al. 2009). Further analysis demonstrate that Regnase-1 deficient mice display severe autoimmune phenotypes characterized by lymphadenopathy and splenomegaly, massive infiltration of lymphocytes in the lungs and liver, and increased levels of effector/memory T cells and plasma cell counts. In addition, increased deposition of IL-6 and IL-12p40 in Regnase-1–/– macrophages was observed in response to LPS stimulation.
On the other hand, Roquin, comprised of Roquin-1 and Roquin-2, is a E3 ubiquitin ligase originally discovered to be associated with the pathogenesis of SLE-like symptoms in mice (Vinuesa et al. 2005). In addition to a CCCH ZF domain, Roquin harbors a RING finger domain, a ROQ domain and a proline-rich domain (Schlundt et al. 2016). The ROQ domain of Roquin proteins is responsible for binding with target mRNAs harboring a stem-loop structure at the 3’ UTRs. While deletion of Roquin leads to postnatal mortality in mice, dominant negative mutation of this RBP results in spontaneous activation of myeloid cells and T cells, increased autoantibody production and excessive T follicular helper cell activation (Vinuesa et al. 2005; Yu et al. 2007; Linterman et al. 2009). Consistently, T cell-specific ablation of Requin-1 and Roquin-2 led to the development of lupus-like autoimmune disease in mice due to the activation of T cells (Vogel et al. 2013). Further studies showed that Roquin post-transcriptionally represses the expression of inducible costimulatory (ICOS) in T cells, as well as the NF-κB pathway regulatory molecules IκBα and A20, thereby modulating the activity and the intensity of the IKK/NF-κB signaling pathway (Murakawa et al. 2015).
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) studies revealed that Regnase-1 and Roquin recognize overlapping sets of mRNAs harboring stem-loop secondary structures located in the 3’ UTRs (Mino et al. 2015). Using HITS-CLIP analysis, Mino and colleagues further showed that the sequence motifs shared by these stem-loop structures contain a common pyrimidine-purine-pyrimidine (Py-Pu-Py) tri-loop sequence, a well-studied cis-element recognized by a set of RBPs for controlling the stability of target mRNAs (Fig. 2C). Importantly, these stem-loop structures are located in the 3’ UTR of immune-related mRNAs including Il6, Cox2 (also known as Ptgs2), Cxcl, Nfkbiz and so on, highlighting the importance of these structural elements in controlling gene expression at the post-transcriptional level (Leppek et al. 2013; Mino et al. 2015). Further investigation showed that Regnase-1-mediated mRNA decay of inflammatory transcripts requires the assistance of the helicase upstream frameshift 1 (UPF1) in a manner reminiscent of nonsense-mediated mRNA decay (Mino et al. 2015, 2019) (Fig. 2C). More specifically, upon phosphorylation at the Thr28 position by a PI3K-related protein kinase, SMG-1, two points of interaction occur between UPF1 and Regnase-1 to promote the unwinding activity of the UPF1 helicase, thus allowing Regnase-1 to endonucleolytically cleave target mRNAs (Fig. 2D). Indeed, depletion of UPF1 in bone marrow-derived macrophages resulted in the accumulation of Regnase-1 targeting proinflammatory transcripts following LPS stimulation (Mino et al. 2019). On the other hand, Roquin promotes mRNA decay by recruiting the CCR4-NOT deadenylase complex (Leppek et al. 2013) or Rck/EDC4 decapping protein (Glasmacher et al. 2010) (Fig. 2C). Importantly, although Regnase-1 and Roquin share overlapping binding motifs, Regnase-1 degrades target mRNAs following the pioneer rounds of translation in the endoplasmic reticulum (ER) or cytoplasm, while Roquin induces mRNA degradation located in the stress granules and possessing bodies (Mino et al. 2015, 2019). These findings together showed that these RBPs function spatiotemporally to fine-tune inflammatory responses and highlight the importance of RBPs as important “brakes” to halt inflammation through controlling the stability of inflammatory mRNAs.
Regulation of Regnase-1 by multiple mechanisms and its implication in therapeutic interventions
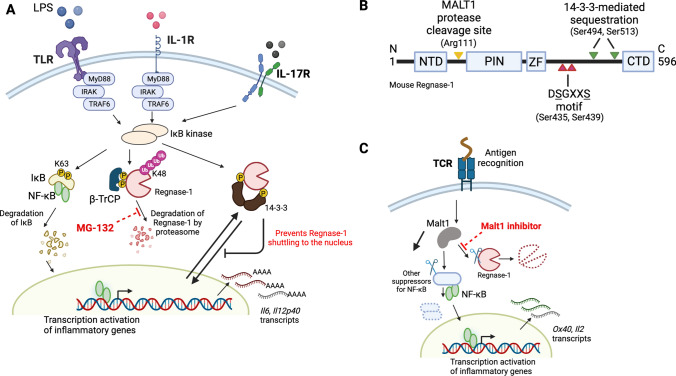
Regnase-1 activation and expression are dynamically regulated in the homeostatic and inflammatory conditions to achieve appropriate immune reactions. Upon TLR or IL-1 receptor (IL-1R) stimulation, Regnase-1 protein is rapidly degraded (Matsushita et al. 2009; Iwasaki et al. 2011) (Fig. 3A). Subsequent biological studies revealed that IKK complex cooperates with the kinase IRAK1 to phosphorylate Regnase-1 at the Ser435 and Ser439 on the canonical DSGXXS motif (Fig. 3B). Phosphorylation of Regnase-1 was also induced by IL-17 stimulation depending on the Act1 signaling pathway (Tanaka et al. 2019). Because phosphorylated Regnase-1 is inherently unstable, it is later subjected to ubiquitin-dependent proteasome degradation via the E3 ligase β-TrCP complex (Iwasaki et al. 2011). Interestingly, further investigation suggests that proteasome inhibitor MG-132 results in the suppression of Regnase-1 degradation. Additionally, a recent study by Akaki et al. revealed an extra layer of post-translational regulation of Regnase-1 (Akaki et al. 2021). Analysis of the Regnase-1 interactome shows dynamic interaction between Regnase-1 and 14-3-3 complex through phosphorylation of Regnase-1 at Ser494 and Ser513, upon TLR/IL-1R activation (Fig. 3A). Although the formation of the Regnase-1-14-3-3 complex protects its degradation by β-TrCP, it abolishes Regnase-1-medaited mRNA decay by sequestering Regnase-1 in the cytoplasm, thereby preventing Regnase-1 nuclear-cytoplasmic shutting and its recognition of inflammatory mRNAs in the nucleus. Thus phosphorylation-dependent 14-3-3-mediated cytoplasmic sequestration of Regnase-1 thereby inhibits its degradation of inflammatory mRNA in the nucleus (Akaki et al. 2021). In the case of T cells, however, stimulation of T cell receptor (TCR) results in the activation of mucosa-associated lymphoid tissue 1 (MALT1), which is the only paracaspase in mammalian cells, and the activation of which results in proteolytic cleavage of Regnase-1 at site Arg111 (Uehata et al. 2013) (Fig. 3C). Importantly treatment of cells with the MALT1 protease inhibitor suppressed Regnase-1 degradation, highlighting its potential role in the treatment of diseases where aberrant activation of T cells is a driving force.
Fig. 3.
Regulation of Regnase-1 turnover by various mechanisms. A Activation of TLR, IL-1R and IL-17R by respective ligands not only results in IκB degradation and NF-κB migration to the nucleus, but also phosphorylates Regnase-1 at two distinct serine residues and results in its recognition by βTRCP, which further induces K48-linked polyubiquitination of Regnase-1, followed by proteasome-mediated degradation. Concurrently, blockage of proteasome degradation by MG-132 inhibits Regnase-1 degradation. Thirdly, activated IKKs can phosphorylate Regnase-1 at alternative serine resides, which results in 14-3-3-medaited sequestration from the nucleus, rendering Regnase-1 unable to binding inflammatory mRNAs. B Schematic diagram of mouse Regnase-1. NTD: N-terminal domain, PIN: PilT N-terminus like domain, ZF: zinc finger domain, and CTD: C-terminal domain. The cleavage site by MALT-1 protease, Arg111, is indicated in yellow arrowhead. The two IKK-mediated phosphorylated sites, Ser435 and Ser439, in the DSGXXS motif are indicated in red arrowheads. The phosphorylation sites responsible for 14-3-3 binding and sequestration are highlighted with green arrowheads (Ser494 and Ser513). C Alternatively, stimulation of T-cell receptor activates the MALT1 protease to cleave the serine-arginine sequence (Arg111) at the N-terminus of Regnase-1. Interestingly, treatment of cells with Malt1 inhibitor prevents Regnase-1 cleavages upon TCR activation. This figure is made with BioRender: https://biorender.com/
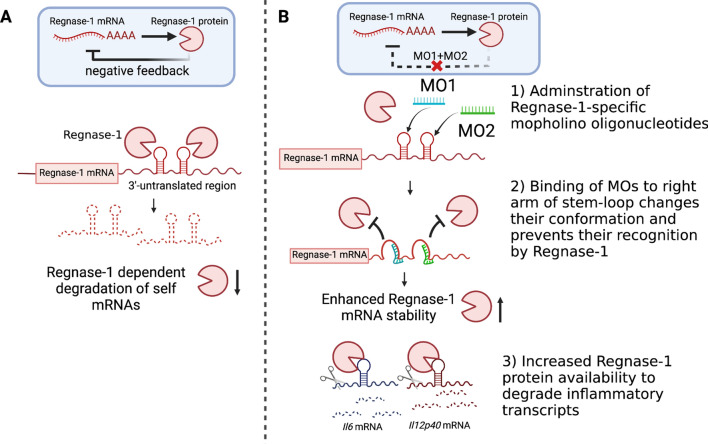
In addition to Regnase-1 protein, Regnase-1 mRNA level is also tightly regulated through different mechanisms. For example, it was reported that microRNA-9 downregulates Regnase-1 expression by associating to the 3’ UTR of Regnase-1 mRNA, thereby promoting activation of microglial cells (Yao et al. 2014). Alternatively, accumulating reports show that similar stem-loop secondary structures were found in the 3’ UTR of Regnase-1 mRNA, suggesting Regnase-1 can degrade its own mRNA in an autoregulatory fashion (Iwasaki et al. 2011; Tse et al. 2022) (Fig. 4A).
Fig. 4.
Regnase-1 self-regulatory mechanisms and therapeutic strategy manipulating such pathway. A Regnase-1 harbors two distinct stem-loop structures at its 3’ UTR, which are responsible for Regnase-1-dependent degradation of self mRNAs. Such negative-feedback pathway may be a backup mechanism to regulate Regnase-1 abundance. B Targeting of two morpholino oligonucleotides (MO) to the right-arm of each stem-loop structures in Regnase-1 3’ UTR interferes the stem-loop conformation, which blocks Regnase-1 association to its own mRNA and thereby increases its availability. The increased Regnase-1 protein abundance enables heightened degradation of factors related to inflammation. This figure is made with BioRender: https://biorender.com/
We have recently demonstrated that enhancing Regnase-1 expression by blocking its autoregulation was therapeutically effective in alleviating inflammatory and autoimmune diseases (Tse et al. 2022). Our strategy focuses on blocking the binding interaction between Regnase-1 and the two stem-loop structures in its 3’ UTR by using two antisense phosphorodiamidate morpholino oligonucleotides (MOs). MOs are short synthetic nucleic acids that bind to mRNA via complementary base-pairing interactions to prevent mRNA translation or alter splicing events (Crooke et al. 2021). Contrary to the common usages of MO which target the coding region to downregulate mRNA expression, we aimed to direct these antisense MOs to the self-regulatory elements in the untranslated region of Regnae-1 mRNAs (Fig. 4B). Further proof-of-concept studies revealed that Regnase-1-targeting MOs increase Regnase-1 mRNA transcript stability and therefore its protein abundance. In addition, the increased Regnase-1 availability is able to potentiate its degradation of target inflammatory cytokines, leading to a lower level of inflammatory cytokine deposition in the BMDMs stimulated with LPS, and in primary monocytes which have undergone IL-1β stimulation. The therapeutic efficacy of tissue-targeted delivery of Regnase-1 specific MOs was demonstrated in several animal models, and we found that the acute respiratory inflammation and chronic fibrosis in the lung, as well as the experimental autoimmune encephalitis symptoms in the nervous system were attenuated by the local treatment of mice with the Regnase-1 specific MOs. Of note, increasing Regnase-1 abundance by the MO approach has benefits over current biological therapy because it can simultaneously reduce the production of multiple inflammatory cytokines and chemokines that are directly targeted by Regnase-1, including IL-6, IL-1β and CXCL1.
The above methods aim to enhance Regnase-1 availability by blocking the signal-induced degradation or cleavage as well as its autoregulation. They are anticipated to have therapeutic potential in diseases where dysregulation of Regnase-1 was reported as a major driving force (Cui et al. 2017; Yoshinaga et al. 2017; Nakatsuka et al. 2018, 2020; Yaku et al. 2022). However, in some cases, downregulation of Regnase-1, rather than its upregulation, is more therapeutically favorable. One example is given by Wei and colleagues, where they show that targeting Regnase-1 was a potential strategy to improve the persistence and effector function of CD8+ T cells (Wei et al. 2019). In particular, they found that ablation of murine REGNASE-1 in CD8+ T cells prolonged the survival and enhanced CD8+ T cytotoxic function, by stabilizing the BATF (basic leucine zipper ATF-like transcription factor) mRNA, which is a key regulator of CD8+ T cell differentiation. Interestingly, subsequent animal experiments demonstrated that Regnase-1 deficient CD8+ T cells had enhanced anticancer efficacy in melanoma and acute lymphocytic leukemia models. Approaches to specifically attenuate Regnase-1 expression, including usage of small interfering RNA-based gene knockdown or CRISPR-Cas9 gene-editing technology are preferable to respectively reduce or delete Regnase-1 expression in a cell-specific manner. Alternatively, given the importance of Regnase-1-UPF1 axis in mediating inflammatory mRNA decay, and that phosphorylation at the Thr28 position of UPF1 helicase by a PI3K-related protein kinase, SMG1, is necessary for Regnase-1 binding, it has been shown that inhibition of SMG1 kinase activity by small molecule could abrogate Regnase-1-medaited mRNA decay and therefore, potentiate the activation of a variety of immune cells (Fig. 2D) (Mino et al. 2019).
Concluding remarks
In the past decades, the interest in innate immunity has grown enormously and tremendous progress has been made in understanding how pathogens detection is achieved by PRRs. This review started with a revision of different PRRs and how the sensing of different pathogens and cellular components triggers activation of distinct signaling pathways and results in inflammation. Following this we have described the activation and regulation of transcription factors, NF-κB and IRF3/7, and their subsequent production of cytokines and type I IFNs through the association of different adaptor proteins, including MyD88 and TRIF.
In the second part of this review, we discussed about the importance of post-transcriptional regulation in controlling the production of inflammatory mediators. More specifically, we summarized how RBPs, especially the members in the CCCH zinc finger proteins (TTP, Roquin and Regnase-1), participate in the resolution of inflammation through destabilizing inflammatory cytokine mRNAs by recognizing distinct cis-elements in the 3’ UTR of inflammatory mRNAs, including stem-loop structures and AU-rich elements. While not discussed in this review, there exists a diversity of RBPs that are not well-characterized, thus further studies on the function and mechanisms of these less-characterized proteins will provide valuable information in understanding the regulatory mechanisms underlying the post-transcriptional regulation of immune reactions. Lastly, we outlined the functional termination of Regnase-1 through multiple mechanisms and highlighted the therapeutic interventions by enhancing or repressing its expression and/or function in treating inflammatory diseases as well as cancer.
In conclusion, the role of RBP-mediated post-transcriptional regulation of cytokine mRNAs has an inevitable impact on the physiological well-being of biological systems. Future work will extend this field of research and provide novel insight into new therapeutics for autoimmunity and other inflammatory diseases.
Acknowledgements
The authors thank all members of our laboratory for discussions. This work is supported by the Japan Society for the Promotion of Science KAKENHI (18H05278), and the Japan Agency for Medical Research and Development (AMED) (JP20gm4010002 and JP21ae0121030).
Data availability
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest to report.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner K-P, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaki K, Ogata K, Yamauchi Y, Iwai N, Tse K, Hia F, Mochizuki A, Ishihama Y, Mino T, Takeuchi O. IRAK1-dependent Regnase-1-14-3-3 complex formation controls Regnase-1-mediated mRNA decay. Elife. 2021 doi: 10.7554/eLife.71966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- Au WC, Moore PA, LaFleur DW, Tombal B, Pitha PM. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ, et al. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. doi: 10.1182/blood.V95.6.1891. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14–3-3 binding. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- Crooke ST, Baker BF, Crooke RM, Liang XH. Antisense technology: an overview and prospectus. Nat Rev Drug Discov. 2021;20:427–453. doi: 10.1038/s41573-021-00162-z. [DOI] [PubMed] [Google Scholar]
- Cui X, Mino T, Yoshinaga M, Nakatsuka Y, Hia F, Yamasoba D, Tsujimura T, Tomonaga K, Suzuki Y, Uehata T, Takeuchi O. Regnase-1 and roquin nonredundantly regulate Th1 differentiation causing cardiac inflammation and fibrosis. J Immunol. 2017;199:4066–4077. doi: 10.4049/jimmunol.1701211. [DOI] [PubMed] [Google Scholar]
- Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol. 2016;17:130–143. doi: 10.1038/nri.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijtenbeek TBH, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2022;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, Karin M. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci USA. 2008;105:7803–7806. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- Inohara C, McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, Akira S. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12:1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019 doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/MCB.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Li M, Yan K, Wei L, Yang Y, Qian Q, Xu W. MCPIP1 inhibits coxsackievirus B3 replication by targeting viral RNA and negatively regulates virus-induced inflammation. Med Microbiol Immunol. 2018;207:27–38. doi: 10.1007/s00430-017-0523-0. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Chien HL, Lin SY, Chang BL, Yu HP, Tang WC, Lin YL. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 2013;41:3314–4426. doi: 10.1093/nar/gkt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Chu JS, Chien HL, Tseng CH, Ko PC, Mei Y, Tang WC, Kao YT, Cheng HY, Liang YC, Lin SY. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol. 2014 doi: 10.4049/jimmunol.1400337. [DOI] [PubMed] [Google Scholar]
- Linterman MA, Rigby RJ, Wong R, Silva D, Withers D, Anderson G, Verma NK, Brink R, Hutloff A, Goodnow CC, Vinuesa CG. Roquin differentiates the specialized functions of duplicated T cell costimulatory receptor genes CD28 and ICOS. Immunity. 2009;30:228–241. doi: 10.1016/j.immuni.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:1–9. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Akira S. Regulation of mRNA stability by CCCH-type zinc-finger proteins in immune cells. Int Immunol. 2017;29:149–155. doi: 10.1093/intimm/dxx015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA. Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998 doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- Mino T, Takeuchi O. Post-transcriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation. Biotechnol Genet Eng Rev. 2013 doi: 10.1080/02648725.2013.801236. [DOI] [PubMed] [Google Scholar]
- Mino T, Takeuchi O. Post-transcriptional regulation of immune responses by RNA binding proteins. Proc Jpn Acad B. 2018;94:248–258. doi: 10.2183/pjab.94.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, Vinuesa CG, Ohler U, Standley DM, Landthaler M, Fujiwara T, Takeuchi O. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161:1058–1073. doi: 10.1016/j.cell.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Mino T, Iwai N, Endo M, Inoue K, Akaki K, Hia F, Uehata T, Emura T, Hidaka K, Suzuki Y, Standley DM, Okada-Hatakeyama M, Ohno S, Sugiyama H, Yamashita A, Takeuchi O. Translation-dependent unwinding of stem-loops by UPF1 licenses Regnase-1 to degrade inflammatory mRNAs. Nucleic Acids Res. 2019;47:8838–8859. doi: 10.1093/nar/gkz628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa Y, Hinz M, Mothes J, Schuetz A, Uhl M, Wyler E, Yasuda T, Mastrobuoni G, Friedel CC, Dölken L, Kempa S, Schmidt-Supprian M, Blüthgen N, Backofen R, Heinemann U, Wolf J, Scheidereit C, Landthaler M. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat Commun. 2015;6:1–14. doi: 10.1038/ncomms8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka Y, Vandenbon A, Mino T, Yoshinaga M, Uehata T, Cui X, Sato A, Tsujimura T, Suzuki Y, Sato A, Handa T, Chin K, Sawa T, Hirai T, Takeuchi O. Pulmonary Regnase-1 orchestrates the interplay of epithelium and adaptive immune systems to protect against pneumonia. Mucosal Immunol. 2018;11:1203–1218. doi: 10.1038/s41385-018-0024-5. [DOI] [PubMed] [Google Scholar]
- Nakatsuka Y, Yaku A, Handa T, Vandenbon A, Hikichi Y, Motomura Y, Sato A, Yoshinaga M, Tanizawa K, Watanabe K, Hirai T, Chin K, Suzuki Y, Uehata T, Mino T, Tsujimura T, Moro K, Takeuchi O. Profibrotic function of pulmonary group 2 innate lymphoid cells is controlled by Regnase-1. Eur Respir J. 2020 doi: 10.1183/13993003.00018-2020. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Núñez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- Ross EA, Smallie T, Ding Q, O'Neil JD, Cunliffe HE, Tang T, Rosner DR, Klevernic I, Morrice NA, Monaco C, Cunningham AF, Buckley CD, Saklatvala J, Dean JL, Clark AR. Dominant suppression of inflammation via targeted mutation of the mRNA destabilizing protein tristetraprolin. J Immunol. 2015;195:265–276. doi: 10.4049/jimmunol.1402826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Romeu E, Ferran M, Giménez-Arnau A, Bugara B, Lipert B, Jura J, Florencia EF, Prens EP, Celada A, Pujol RM, Santamaria-Babí LF. MCPIP1 RNase is aberrantly distributed in psoriatic epidermis and rapidly induced by IL-17A. J Invest Dermatol. 2016;136:1599–1607. doi: 10.1016/j.jid.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlundt A, Niessing D, Heissmeye V, Sattle M. RNA recognition by Roquin in posttranscriptional gene regulation. Wiley Interdiscip Rev RNA. 2016;7:455–469. doi: 10.1002/wrna.1333. [DOI] [PubMed] [Google Scholar]
- Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993 doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- Takeuchi O. Endonuclease Regnase-1/monocyte chemotactic protein-1-induced protein-1 (MCPIP1) in controlling immune responses and beyond. Wiley Interdiscip Rev RNA. 2018 doi: 10.1002/wrna.1449. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Arima Y, Kamimura D, Tanaka Y, Takahashi N, Uehata T, Maeda K, Satoh T, Murakami M, Akira S. Phosphorylation-dependent Regnase-1 release from endoplasmic reticulum is critical in IL-17 response. J Exp Med. 2019;216:1431–1449. doi: 10.1084/jem.20181078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Tse KM, Vandenbon A, Cui X, Mino T, Uehata T, Yasuda K, Sato A, Tsujimura T, Hia F, Yoshinaga M, Kinoshita M, Okuno T, Takeuchi O. Enhancement of Regnase-1 expression with stem loop-targeting antisense oligonucleotides alleviates inflammatory diseases. Sci Transl Med. 2022;14:eabo2137. doi: 10.1126/scitranslmed.abo2137. [DOI] [PubMed] [Google Scholar]
- Uehata T, Takeuchi O. Post-transcriptional regulation of immunological responses by Regnase-1-related RNases. Int Immunol. 2021;33:859–865. doi: 10.1093/intimm/dxab048. [DOI] [PubMed] [Google Scholar]
- Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, Roberts IS, Copley RR, Bell JI, Cornall RJ, Goodnow CC. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, Zöller J, Warth SC, Hoefig KP, Lohs C, Neff F, Kremmer E, Schick J, Repsilber D, Geerlof A, Blum H, Wurst W, Heikenwälder M, Schmidt-Supprian M, Heissmeyer V. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Wang S, Dai T, Qin Z, Pan T, Chu F, Lou L, Zhang L, Yang B, Huang H, Lu H, Zhou F. Targeting liquid–liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat Cell Biol. 2021;23:718–732. doi: 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhou Z, Xiao X, Tian Z, Dong X, Wang C, Li L, Ren L, Lei X, Xiang Z, Wang J. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol. 2021;18:945–953. doi: 10.1038/s41423-020-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Long L, Zheng W, Dhungana Y, Lim SA, Guy C, Wang Y, Wang YD, Qian C, Xu B, Kc A, Saravia J, Huang H, Yu J, Doench JG, Geiger TL, Chi H. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 2019;576:471–476. doi: 10.1038/s41586-019-1821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- Yaku A, Inagaki T, Asano R, Okazawa M, Mori H, Sato A, Hia F, Masaki T, Manabe Y, Ishibashi T, Vandenbon A, Nakatsuka Y, Akaki K, Yoshinaga M, Uehata T, Mino T, Morita S, Ishibashi-Ueda H, Morinobu A, Tsujimura T, Ogo T, Nakaoka Y, Takeuchi O. Regnase-1 prevents pulmonary arterial hypertension through mRNA degradation of interleukin-6 and platelet-derived growth factor in alveolar macrophages. Circulation. 2022;146:1006–1022. doi: 10.1161/CIRCULATIONAHA.122.059435. [DOI] [PubMed] [Google Scholar]
- Yao H, Ma R, Yang L, Hu G, Chen X, Duan M, Kook Y, Niu F, Liao K, Fu M, Hu G, Kolattukudy P, Buch S. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M, Tsushima T, Noda NN, Kumeta H, Enokizono Y, Yamashita K, Standley DM, Takeuchi O, Akira S, Inagaki F. Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions. Sci Rep. 2016 doi: 10.1038/srep22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Yoshinaga M, Nakatsuka Y, Vandenbon A, Ori D, Uehata T, Tsujimura T, Suzuki Y, Mino T, Takeuchi O. Regnase-1 maintains iron homeostasis via the degradation of transferrin receptor 1 and prolyl-hydroxylase-domain-containing protein 3 mRNAs. Cell Rep. 2017;19:1614–1630. doi: 10.1016/j.celrep.2017.05.009. [DOI] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, Goodnow CC, Vinuesa CG. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- Zhao W, Liu M, D'Silva NJ, Kirkwood KL. Tristetraprolin regulates interleukin-6 expression through p38 MAPK-dependent affinity changes with mRNA 3' untranslated region. J Interferon Cytokine Res. 2011;31:629–637. doi: 10.1089/jir.2010.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.