Abstract
Several members of the aldehyde dehydrogenase (ALDH) family, especially ALDH1 isoenzymes, have been identified as biomarkers of cancer stem cells (CSCs), a small subpopulation of oncogenic cells with self-renewal and multipotency capability. Consistent with this contention, cell populations with high ALDH enzymatic activity exhibit greater carcinogenic potential. It has been reported that ALDH1, especially ALDH1A1, serves as a valuable biomarker for colon CSCs. However, the functional roles of ALDHs in CSCs and solid tumors of the colon tissue is not fully understood. The aim of the present study was to identify molecular signature associated with high ALDH activity in human colorectal adenocarcinoma (COLO320DM) cells by proteomics profiling. Aldefluor™ assay was performed to sort COLO320DM cells exhibiting high (ALDHhigh) and low (ALDHlow) ALDH activity. Label-free quantitative proteomics analyses were conducted on these two cell populations. Proteomics profiling revealed a total of 229 differentially expressed proteins (DEPs) in ALDHhigh relative to ALDHlow cells, of which 182 were down-regulated and 47 were up-regulated. In agreement with previous studies, ALDH1A1 appeared to be the principal ALDH isozyme contributing to the Aldefluor™ assay activity in COLO320DM cells. Ingenuity pathway analysis of the proteomic datasets indicated that DEPs were associated with mitochondrial dysfunction, sirtuin signaling, oxidative phosphorylation and nucleotide excision repair. Our proteomics study predicts that high ALDH1A1 activity may be involved in these cellular pathways to promote a metabolic switch and cellular survival of CSCs.
1. Introduction
The human ALDH superfamily consists of 19 isozymes that catalyze a wide spectrum of endogenous and exogenous aldehydes to their corresponding carboxylic acid in the cytoplasm, mitochondria or nucleus [1]. These enzymatic activities serve to protect cells against the toxic effects of aldehydes and to generate molecules with important physiological functions [2]. For instance, ALDH2 is the most efficient enzyme in detoxifying the acetaldehyde during alcohol metabolism. ALDH1 isozymes (including ALDH1A1, ALDH1A2, ALDH1A3 and ALDH1B1), are involved in the biosynthesis of retinoic acid, which are required for cell maintenance and differentiation in human [3]. In addition to the enzymatic properties, structural properties of some isozymes also endow these ALDHs with intrinsic non-enzymatic functions [4]. For example, ALDH16A1 which lacks the enzymatic Cys residue has been identified as a non-enzymatic member working as a binding protein [5, 6]. ALDH3A1 is a corneal crystallin which directly absorbs UV radiation to maintain corneal epithelial homeostasis [7]. The diverse functions of ALDH superfamily are also evidenced by mutations in ALDH genes leading to defective aldehyde metabolism, which are the molecular basis of several human diseases, including hyperprolinaemia, Parkinson’s disease, cancers and so on [1, 3, 8].
Accumulating evidence suggests that enhanced levels of ALDH expression and/or activity correlate with increased tumorigenesis, poor prognosis and chemoresistance of several different types of cancers [9–13]. The expression patterns of ALDH isozymes are tissue- and cancer-specific [3].There are several research reports documenting abnormally-high ALDH isozyme expressions in cancerous tissue and cells of the colon [14–20]. Increased expression of ALDH1A1 has been first proposed to serve as a biomarker for colonic cancer stem cells (CSCs), which exhibit a remarkable capacity for self-renewal, differentiation and self-protection [14, 21]. In SW403 colon cancer cells which constitutively have high ALDH1A1 expression, a subpopulation of cells with high ALDH activity have shown potent clonogenicity and proliferating capability [22]. Global gene expression profiling of SW403 cells indicates that ALDH activity plays a role in the preferential activation of mitogen-activated protein kinases, focal adhesion kinase and oxidative stress response signaling pathways [22]. In addition to ALDH1A1, previous studies from our group revealed that ALDH1B1 could be a more reliable biomarker of colon cancer, since not only ALDH2 and ALDH1B1 mRNA were highly expressed in human CRC tissues but only ALDH1B1 protein was consistently overexpressed throughout CRC tissues [23]. In line with this, in SW480 cells which do not express ALDH1A1 but have constitutively high ALDH1B1 expression, our group showed that shRNA-induced suppression of ALDH1B1 expression decreased the number and size of spheroids formation in vitro and xenograft tumor growth in vivo [24]. Wnt/β-catenin, Notch and PI3K/Akt-signaling pathways were down-regulated in SW480 cells in which ALDH1B1 expression had been suppressed [24]. Although these pathways are implicated in the modulation of distinct oncogenic mechanisms, they are critically involved in colon cancer progression [25].
The measurement of total cellular ALDH activity has been facilitated by the use of the Aldefluor™ assay [26]. The intracellular accumulation of a fluorescent metabolite of ALDH allows cells to be isolated by flow cytometry based on their ALDH enzymatic activity [26].
Subpopulations of high (ALDHhigh) and low (ALDHlow) ALDH activity cells have been tested for their tumorigenic potential in vitro and/or in vivo in numerous colon cancer studies [14, 17, 27]. These studies show that ALDHhigh subpopulations of HCT116, SW480 and SW640 colon cancer cell lines exhibit characteristics of CSCs (i.e., rapid growth rate and more colonies developed in vitro), while ALDHhigh subpopulation of COLO colon cells only showed the faster growth rate [17]. Importantly, these colon cancer cells present a considerably different pattern of ALDH mRNA expression in the two subpopulations sorted by the Aldefluor™ assay [17]. Recently, a cancer-type specific expression pattern of nine active ALDH isozymes was identified using the Aldefluor™ assay in breast cells and kidney cells [20]. However, a similar differential expression pattern of active ALDH isozymes in colon cancer cells has not been established. Elucidation of the identity of the ALDH enzymes that contribute to the Aldefluor™ identified enzymatic activity is important for understanding the precise role played by ALDH isozymes in colon carcinogenesis, as well as the elucidating the mechanisms by which ALDH activity affects colon cancer cell function.
COLO320DM cells, derived from a human colorectal cancer cell line, have been widely used for colon tumorigenesis studies. We have previously shown that mRNA and protein expression of ALDH1A1 was higher in these cells than ALDH2 and ALDH1B1 [23]. In the present study, we isolated ALDHhigh and ALDHlow subpopulations of COLO320DM cells and performed global proteomic profiling on them, with the intention of identifying the molecular signature associated with high cellular ALDH enzymatic activity and interpreting these data in the context of colon tumorigenesis.
2. Materials and Methods
2.1. Cell lines and cell culture
The colorectal adenocarcinoma cancer cell line, COLO320DM (ATCC® CCL-220™), was kindly provided by Dr. David Ross (University of Colorado Skaggs School of Pharmacy & Pharmaceutical Sciences). The cells were grown in 100mm dishes containing RPMI-1640 medium (Life Technologies; Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1% antibiotic antimycotic (100X) (Gibco: Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humidified atmosphere of 5% CO2 in air maintained at 37°C. Upon attaining 70–80% confluence, cells were collected using 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
2.2. Aldefluor™ assay and fluorescence-activated cell sorting
The analysis and sorting of COLO320DM ALDHhigh and ALDHlow cells were performed using the Aldefluor™ kit (StemCell Technologies, Inc., Vancouver, BC, Canada), according to the manufacturer’s instructions. In brief, COLO320DM cells (1×106 cells/ sample) were suspended in 1 ml Aldefluor™ buffer maintained on ice. Five μl BODIPY-aminoacetaldehyde (a fluorescent ALDH reaction substrate) was then mixed with the cell suspension. Five hundred μl of the resultant cell suspension was immediately transferred to a control tube that contained 5 μl ALDH inhibitor (diethylaminobenzaldehyde (DEAB)) and mixed thoroughly; this served as the negative control. The cell suspension was incubated for 30 min at 37°C, and subsequently subjected to centrifugation (250xg) for 5 min. After discarding the supernatant, the pellet was resuspended in 0.5 mL Aldefluor™ assay buffer and the resulting cell suspension was subjected to fluorescence-activated cell sorting and analysis using BD SORP FACSAria II (BD Biosciences, New Jersey, USA) with standard doublet discrimination. The fluorescence intensity of the negative (DEAB) control was used to set the threshold of the selection window for the ALDHlow cell group. ALDHhigh cells were identifiable by having great fluorescence than cells in which the enzyme activity was inhibited by DEAB. This procedure allowed the detection and separation of cells with high (ALDHhigh) and low (ALDHlow) enzymatic activity based on the strength of the fluorescence signals. The isolated cells were collected, aliquoted, flash-frozen, and stored at −80 °C until further processing.
2.3. Protein sample preparation
Cell pellets were suspended in ice-cold Dulbecco’s phosphate-buffered saline (DPBS) supplemented with complete protease and phosphatase inhibitor cocktails (Roche Diagnostics GmbH, Mannheim, Germany) to a final concentration of 1×106 cells/ml. The resultant suspension was subjected to centrifugation at 10,000 rpm for 15 min at 4°C, and the supernatant was discarded. This process was repeated three times. The resulting pellets were frozen in −80°C and submitted to the Yale Mass Spectrometry (MS) & Proteomics Resource of the W.M. Keck Foundation Biotechnology Resource Laboratory for proteomics analysis.
2.4. Label-free quantitative proteomic analysis
Each sample (n=3/group) was subjected to three 15 sec bursts of sonication (Model 450, Branson Ultrasonics Corporation, Danbury, CT) on ice. The suspension was subjected to centrifugation at 14,000 rpm for 10 min at 4°C. One hundred μl of the supernatant was added to chloroform: methanol: water (at a ratio of 1:4:3%v/v, respectively) and subjected to centrifugation at 14,000 rpm for 1 min at 4°C. The supernatant was removed and 400 μl methanol was mixed with the remaining pellet by vertexing. The resultant solution was subjected to centrifugation at 14,000 rpm for 2 min at 4°C, the supernatant was discarded, and the pellet was dried using a SpeedVac (SPD111V Savant, Fisher Thermo Scientific). The dried protein pellet was resuspended in 25 μL RapiGest SF surfactant solution (Waters Corporation, Milford, MA) containing 50mM NH4HCO3, reduced by the addition of 5 μl dithiothreitol (45 mM) and further alkylated by the addition of 5μL iodoacetamide (100 mM). The resultant mixture was then digested by incubation with dual enzymatic LysC and trypsin for 20 hr at 37°C. The digestion incubation was subsequently quenched using 0.1% formic acid (FA) during the de-salting step with C18 UltraMicroSpin columns. The effluent from the de-salting step was dried and re-dissolved in a solution comprising 5 μL 70% FA and 35 μL 0.1% trifluoroacetic acid (TFA). A 2 μL aliquot of this digest solution was taken. To make a 1:10 dilution prior to sample injection, 3.2 μL Pierce Retention Time Calibration Mixture (Thermo Fisher Cat#88321) was added to each sample. To allow normalization of label-free quantitation data, 5 μL of the sample was injected into a UPLC system (Waters nanoACQUITY) (equipped with a C18 (180 μm × 20 mm) trap column (Waters Symmetry®) and a 1.7-μm, 75 μm × 250 mm UPLC column (Waters nanoACQUITY)) connected to a mass spectrometer (Thermo Scientific Orbitrap Fusion). Samples were evaluated as duplicates in a block randomized order.
To ensure a high level of identification and quantitation integrity, a resolution of 120,000 and 60,000 was utilized for MS and MS/MS scans, respectively. High-energy Collisional Dissociation (HCD) MS/MS spectra filtered by dynamic exclusion were acquired over a 3 sec duty cycle for charge states 2–8 with an m/z isolation window of 1.6. All MS (profile) and MS/MS (centroid) peaks were detected by the mass spectrometer. Trapping was carried out for 3 min at 5 μL/min in 97% Buffer A (0.1% FA in water) and 3% Buffer B (0.075% FA in acetonitrile (ACN)) prior to eluting with linear gradients that reached 6% B at 5 min, 35% B at 170 min, 50% B at 175 min, 97% B at 180 min (and maintained for 5 min), and then reduced to 3% B from 186 min to 201 min. Three blanks (1st 100% ACN, 2nd and 3rd Buffer A) followed each injection to avoid sample carry over.
2.5. Protein identification
The UPLC-MS/MS data was processed using Progenesis QI Proteomics software (Nonlinear Dynamics, version 2.0), with protein identification carried out using the Mascot search algorithm [28]. A normalization factor for each run was calculated to account for differences in sample load between injections, and for differences in ionization. The normalization factor was determined by comparing the total ion abundance among all samples, with the expectation that they were normalized based on an equal total amount of peptides being loaded onto the column. The algorithm calculated the tabulated raw and normalized abundances, maximum fold-change, and P-values for each feature in the data set. The MS/MS spectra were exported as Mascot generic files (.mgf) for database searching. The Mascot search results were exported as an ‘.xml’ file using a significance cutoff of false discovery rate (FDR) q<0.01 and then imported into the Progenesis QI software, wherein search hits were assigned to corresponding peak features that were extracted from the MS data. Proteins with two or more unique quantifiable peptides were filtered and used in downstream analyses. The expression level of a specific protein is reported as the average of normalized abundance. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011197.
2.6. Statistical and bioinformatics analyses
Differentially expressed proteins (DEPs) in ALDHhigh cells (relative to ALDHlow cells) were identified using a two-tailed Student’s unpaired t-test, with a cutoff p value < 0.05. The differential abundance of proteins was expressed by the fold change, which was calculated based on the ratio of the protein abundance in ALDHhigh cells versus that in ALDHlow cells. Specifically, if the ratio value is > 1, fold change equals to the ratio; if the ratio is between 0 and 1, fold change equals to −1*(1/ratio) [29]. Therefore, the fold changes of up-regulated proteins are positive numbers, whereas the fold changes of down-regulated proteins are negative numbers. Top upregulated and downregulated proteins were ranked based on their fold change values. Qlucore Omics Explorer 3.4 (Qlucore AB, Lund, Sweden) was used for data visualization and analysis, including heatmap generation based on the calculated prediction activation scores (z-score) in the range of ~ −2 < z >2. The expression of detected ALDH isozymes was compared between ALDHhigh and ALDHlow cells using Welch’s t-test on the triplicate measurements per group. The percentage of each ALDH isozyme was calculated as a proportion of the total ALDH isozyme expression in each group. For an overview of DEPs, UniProt Knowledgebase (UniProtKB) and Gene Ontology (GO) databases were used and the corresponding accession numbers were imported into Ingenuity Pathway Analysis (IPA) (IPA, version 01-20-04, Qiagen, CA). Fisher’s exact test was used to test for pathway significance (with P < 0.05 being considered significant) and Benjamin-Hochberg (B-H) FDR was used to correct for multiple comparisons to yield a network’s score and to rank networks according to their degree of association with our data set [30]. IPA automatically calculates the z-score based on differentially expressed proteins from our dataset with the information stored in IPA knowledge database [31]. Top pathways were selected based on the FDR value of enriched pathways with FDR q < 0.05. The network of top pathways was constructed in Cytoscape 3.9.1.
3. Results
1.1. Proteomic profiling revealed 229 proteins differentially expressed in ALDHhigh versus ALDHlow COLO320DM cells.
Isolation of COLO320DM cells was performed by fluorescence-activated cell sorting (FACS) based on Aldefluor™ assay activity. The cell viability was greater than 95% after resuspended the cells in the Aldefluor™ assay buffer. According to the threshold set by the negative control, isolated cells without fluorescence signal were assigned as ALDHlow cells while cells with bright fluorescence signals were assigned as ALDHhigh cells. This sorting showed an average of ≈10% of total cell population were ALDHhigh cells (Fig. 1A). Label-free quantitative proteomic analyses were performed in the two cell populations, which identified a total of 3,222 proteins, of which 229 were identified as being differentially-expressed in ALDHhigh cells. A heatmap analysis showed the relative differences in DEP abundance in the two cell populations (Fig. 1B). Of the identified DEPs, 47 were up-regulated and 182 were down-regulated in the ALDHhigh group. The DEPs included (in the descending order of percent of total DEPs) unclassified others, enzymes, transporters, transcription factors, kinase, peptidase, phosphatase, ion channel, transmembrane receptor, translation regulator and growth factor. (Fig. 1C). The top ten significantly up-regulated or down-regulated proteins ranked by their fold change values in ALDHhigh cells are presented in Supplemental Table 1.
Figure 1. Quantitative proteomic profiling in ALDHhigh and ALDHlow COLO320DM cells.
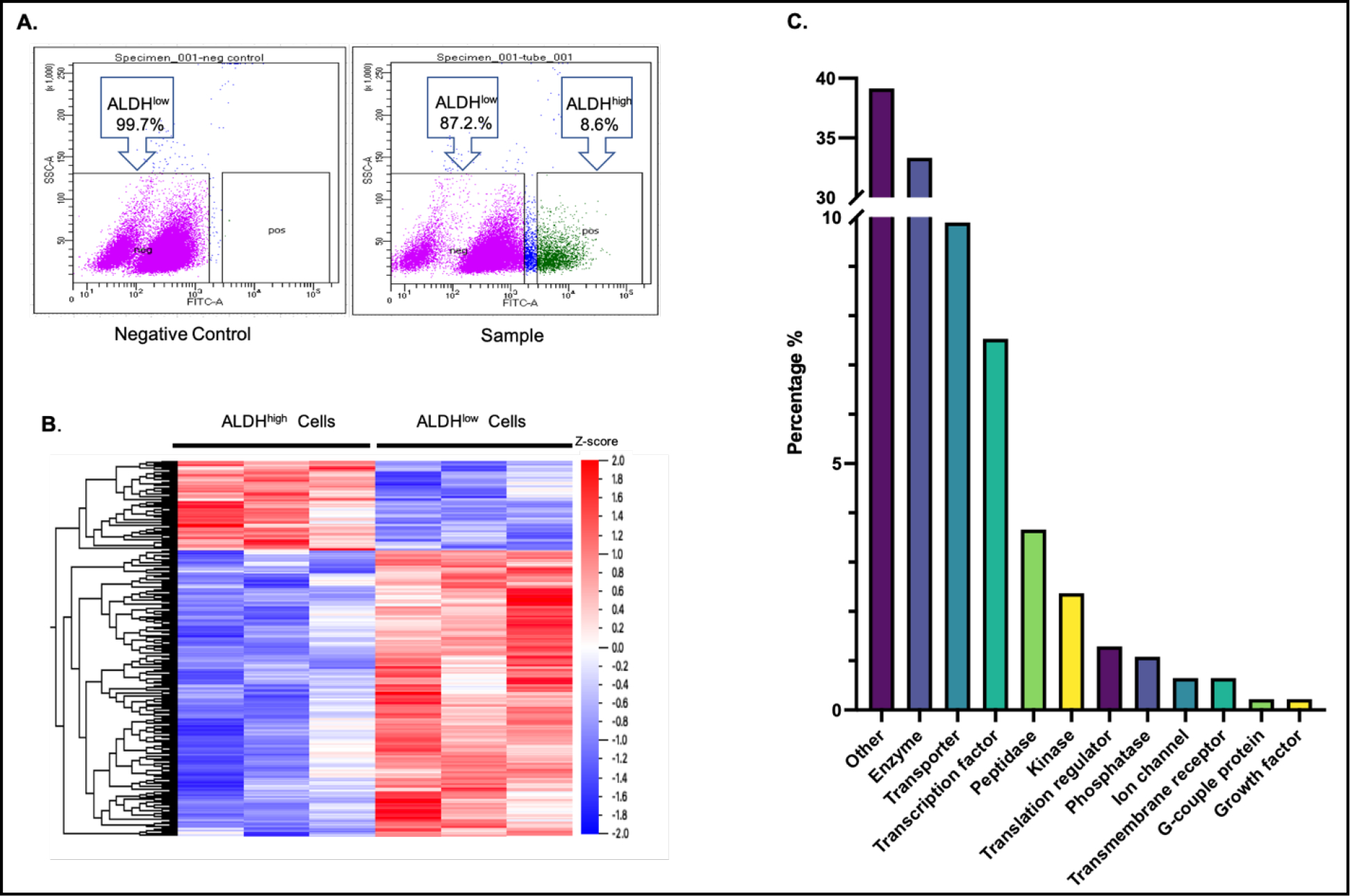
(A) Flow cytometry profiles of COLO320DM cells isolated by Aldefluor™ assay. ALDHhigh cells were distinguished from ALDHlow cells using fluorescence-activated cell sorting. Cells treated with the ALDH inhibitor, diethylaminobenzaldehyde (DEAB), served as a negative control, and cells without DEAB treatment were the sample group. The distribution of ALDHlow (pink dots) and ALDHhigh (green dots) cells are shown. Their corresponding proportion of thetotal cells population are indicated as percentages in the box. (B) Heat map of the relative abundance of 229 differentially expressed proteins (DEPs) in ALDHhigh and ALDHlow cells based on proteomic analysis. Hierarchical clustering and distance trees were constructed using Qlucore software with unsupervised hierarchical classification. Cell samples are shown in columns and DEPs are shown in rows. The hierarchical heat map scale of Z-scores ranges from −2 (blue) to 2 (red) with a midpoint of 0 (white). The intensity of color reflects the Z-score of protein expression. (C) Functional classifications of DEPs. The distribution of the 229 DEPs among different general functional categories.
1.2. ALDH1A1 was identified as the exclusive ALDH isozyme differentially expressed in ALDHhigh versus ALDHlow COLO320DM cells.
Nine ALDH proteins were detected in ALDHhigh and ALDHlow cells, with the levels of the individual isozymes, in general, being comparable between these two cell populations (Fig. 2A). Of all of the ALDH isozymes, ALDH1A1 was the most highly expressed, accounting for 49% and 43% of total ALDH protein content in ALDHhigh and ALDHlow cells, respectively (Fig. 2B). Notably, ALDH1A1 was the only ALDH protein whose expression was elevated in ALDHhigh relative to ALDHlow cells (P < 0.0001) (Fig. 2A). ALDH2 and ALDH1B1 were the next most abundant isozymes, together amounting to ≈ 30% of the total ALDH proteins (Fig. 2A); there was a trend of increased abundance of ALDH2 in ALDHlow cells (P = 0.08) (Fig. 2A). There were no other differences between the two cell populations regarding the expression of the other lower abundance isozymes, i.e., ALDH18A1, ALDH6A1, ALDH7A1, ALDH9A1, ALDH3A2 and ALDH4A1 (Fig. 2B).
Figure 2. Expression of ALDH isozymes in ALDHhigh and ALDHlow COLO320DM cells.
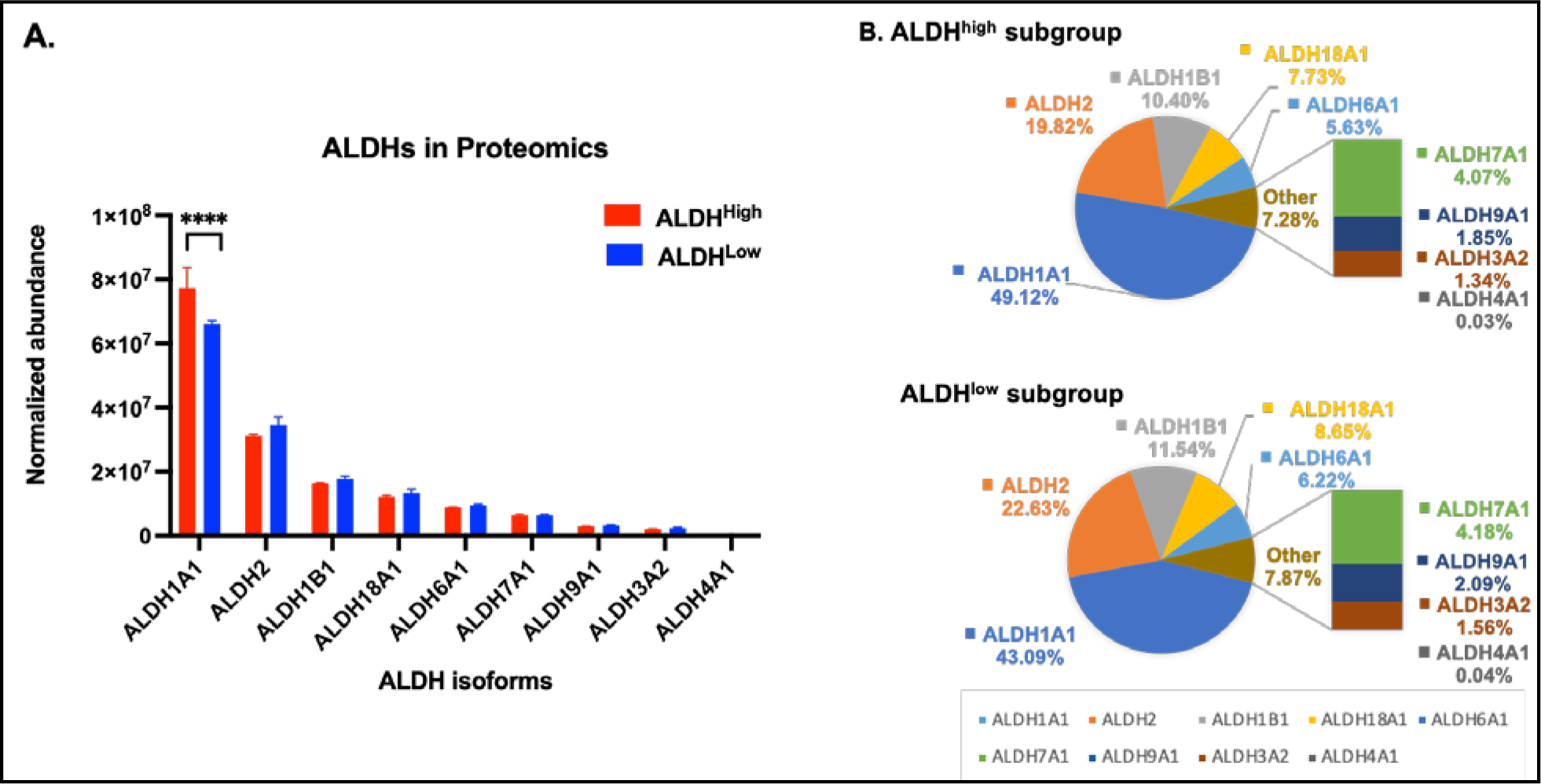
(A) Protein abundance of the nine ALDH isozymes detected by proteomics profiling in ALDHhigh (red bars) and ALDHlow (blue bars) COLO320DM cells. Data are presented as the mean and associated standard deviation from triplicate measurements. **** P< 0.0001, Welch’s t-test. (B) The percentage distribution of the ALDH isozymes identified in ALDHhigh and ALDHlow cells.
1.3. Functional enrichment analyses revealed four canonical pathways enriched by DEPs.
The 229 DEPs were subjected to Ingenuity Canonical Pathway analysis to elucidate the pathways and biological functions related to the proteome data. Four canonical pathways were identified to be significantly enriched by 30 of the DEPs in ALDHhigh cells, i.e., the pathways of mitochondrial dysfunction (FDR q =3.16E-7), sirtuin signaling (FDR q =3.16E-7), oxidative phosphorylation (OXPHOS) (FDR q = 2.81E-04), and nucleotide excision repair (NER) (FDR q = 0.01). Cytoscape was used to visualize the network connecting the DEPs and the pathways (Fig. 3A). This revealed clusters of proteins involved in the top three pathways, including NADH-ubiquinone oxidoreductase subunits (NDUFB1, NDUFB5, NDUFB6, NDUFS3, NDUFS8 and ATP5F1C), voltage dependent anion channel proteins (VDAC1 and VDAC3) and cytochrome c oxidase subunits (COX5A and COX15) (Fig. 3A). Meanwhile, xeroderma pigmentosum complementation group C (XPC) connected sirtuin and NER pathway (Fig. 3A). The majority of the proteins involved in these pathways were down-regulated, except for COX5A, translocase of inner mitochondrial membrane 9 (TIMM9) and 13 (TIMM13), and DNA polymerase epsilon 3 (POLE3) which were up-regulated in the ALDHhigh cells. Using Z-score calculations, the sirtuin signaling pathway was predicted to be activated (Z-score = 1.414), whereas the OXPHOS (Z-score = −2.828) and the NER (Z-score = −2.449) pathways were predicted to be inhibited in the ALDHhigh cells; no prediction was available from IPA for mitochondrial dysfunction pathway. Based on the protein annotation from Gene Ontology, all identified pathway proteins were located in mitochondrial or nucleus (Fig. 3B).
Figure 3. Functional enrichment analysis of DEPs.
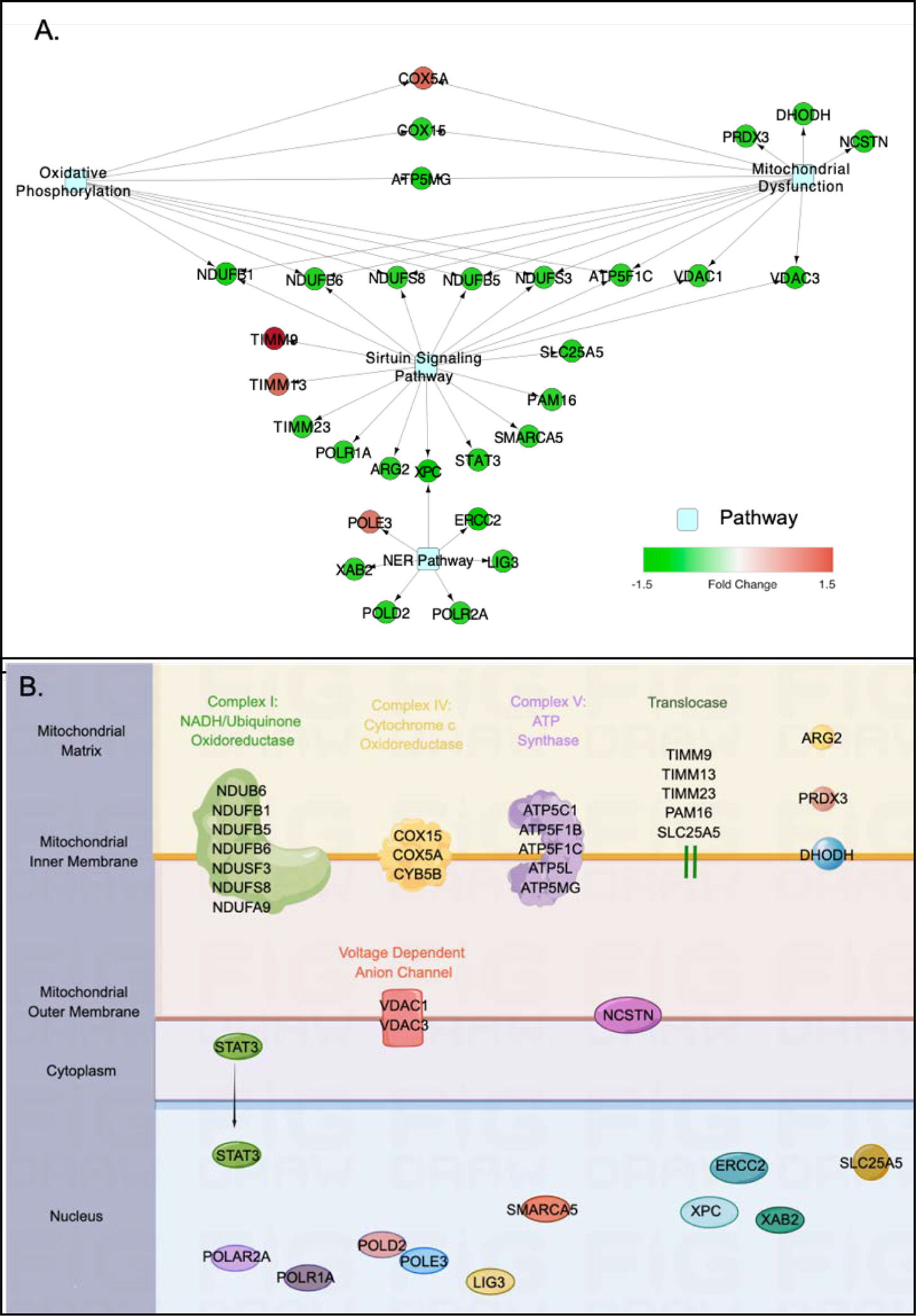
(A) Network of four canonical pathways significantly enriched by DEPs in ALDHhigh cells. Significant canonical pathways were identified by IPA functional enrichment analysis and a network was generated by Cytoscape using DEPs shared by the pathways. The fold changes of proteins in ALDHhigh cells (relative to in ALDHlow cells) are depicted by the intensity of the color code, with red representing up-regulation and green representing down-regulation. Light blue shaded nodes represent canonical pathways. (B) Subcellular localization of DEPs involved in the four identified pathways. Annotations of protein subcellular localization were based on Gene Ontology. A full list of abbreviations is provided in supplemental Table 2.
2. Discussion
In the present study, we compared the proteomic profiles of COLO320DM cells exhibiting high (ALDHhigh) and low (ALDHlow) ALDH enzymatic activity. Globally, 229 proteins were found to be differentially expressed in the two populations of COLO320DM cells. Of these, ≈80% were expressed to a lesser extent in ALDHhigh cells. Among the nine detectable ALDH isozymes, ALDH1A1 was the most highly expressed, with the expression level in ALDHhigh cells being ≈20% higher than in ALDHlow cells. The canonical pathways enriched by DEPs include mitochondrial dysfunction, sirtuin signaling, OXPHOS and NER pathways. This study is the first report of molecular features at the proteome level in COLO320DM cells possessing high ALDH activity.
The ALDH activity measured using the Aldefluor™ assay reflects the combined activity of multiple ALDH isozymes and the contribution of each isozyme can vary depending on the tissue of origin and cancer type [20, 32–34]. The present study shows that ALDH1A1 is the key ALDH isozyme contributing to ALDH enzymatic activity in ALDHhigh COLO320DM cells because it was the only isozyme that was expressed to a greater extent in ALDHhigh cells. This finding is consistent with a previous report implicating ALDH1A1 as the main isozyme responsible for Aldefluor™ activity in CRC cell lines [17]. Other isozymes that have been reported as contributors to Aldefluor™ activity in other cancer cell types were either not detected (i.e., ALDH1A2 and ALDH1A3) [24, 25] or showed no change at the protein level (i.e., ALDH2 and ALDH1B1) [26–28] after Aldefluor™-based cell sorting. A failure to show any differences in the expression of ALDH18A1, ALDH6A1, ALDH7A1, ALDH4A1 and ALDH9A1 in ALDHhigh cells may have been predicted, given that these isozymes show no enzymatic activity in the Aldefluor™ assay [20]. ALDH3A2 had been detected in both subgroups and sustained a comparable low expression in two subpopulations of COLO320DM cells, which is consistent with previous finding that ALDH3A2 is one of the minority ALDH isozymes which is poorly affected by DEAB inhibition [20].
Using three aldehyde substrates to determine specific enzymatic activities of ALDH1A1 (propionaldehyde), ALDH1B1 and ALDH2 (acetaldehyde), and ALDH3A1 (benzaldehyde), a previous study showed that the expression levels of ALDH1A1 protein (measured by Western blot) correlated well with ALDH1A1 enzymatic activities in COLO320DM cells [23]. An interesting observation in the present study was that a modest (≈20%) increase in ALDH1A1 protein abundance was associated with a disproportionately large (≈80%) change in enzymatic activity (i.e., the increased Aldefluor™ signal in ALDHhigh cells). This apparent disparity between protein levels and activity may be due to the ALDH isozymes in ALDHhigh cells being more enzymatically active than that in ALDHlow cells, possibly due to post-translational modifications of ALDH protein(s) [35, 36] or differences in the availability of cofactors in ALDHhigh cells. Alternatively, there may be subpopulation of ALDHhigh cells (e.g., CD44+ cells) that disproportionately contribute to the ALDH enzymatic activity [23]. It is also possible that UPLC-MS/MS analysis may have underestimated the magnitude of the change in ALDH1A1 protein. While it would be valuable to confirm the relative expression of ALDH1A1 protein and the ALDH1A1-specific enzymatic activity in ALDHhigh and ALDHlow cells using Western blot analysis and proprionaldehyde oxidation, respectively, such approaches are challenging to conduct due to the limited yield of ALDHhigh cells from the COLO320DM population. These considerations notwithstanding, additional studies will be needed to discern the processes underlying the observed results.
The Aldefluor™ assay has been widely used to identify stem or progenitor cells (e.g., CSCs) in numerous cancerous tissues [32, 37, 38]. The function of high ALDH enzymatic activity in these cells remains to be established. In the present study, ALDHhigh COLO320DM cells differentially expressed proteins involved in three cellular pathways, the changes in which would be predicted positively influence CSC function. First, the OXPHOS pathway was predicted to be inhibited in ALDHhigh cells based on down-regulation of mitochondrial electron transport chain proteins and VDAC3 (a mitochondrial outer membrane protein that acts as a global regulator of mitochondrial metabolism) [39–41]. Decreases in the expression of these proteins have been shown to trigger a metabolic shift from OXPHOS to glycolysis [41–44]. Such a metabolic transition has been documented in cancer cells and stem cells [45]. It promotes self-renewal of cancer stem cells [42, 43]. All-trans retinoic acid, a product derived all-trans retinaldehyde, has been shown to redirect cellular production of ATP from the OXPHOS pathway to the aerobic glycolysis pathway in leukemia cells [46]. Given that ALDH1A1 metabolizes all-trans retinaldehyde to all-trans retinoic acid [47], it is possible that the higher ALDH1A1 levels in CSCs to further drive metabolism to the glycolytic pathway. In addition, it has been shown that ALDH1A1 promotes glucose uptake by uterine endometrial cancer stem cells by increasing expression of the glucose transporter GLUT1 [48]. As such, ALDH1A1 may enhance the effectiveness of the metabolic shift by increasing available substrates. Second, the NER pathway, the most versatile repair mechanism for carcinogen-induced DNA damage [49], was predicted to be inhibited in ALDHhigh cells based on the down-regulation of a panel of nuclear proteins that function in DNA damage repair, including ERCC2, LIG3, POLR2A, POLD2, XAB2 and XPC. ALDH enzymes play an important role in the metabolism of endogenous and exogenous aldehydes and thereby mitigate cellular oxidative or electrophilic stress [50]. It is possible that inhibition of the NER pathway in ALDHhigh cells may result from a declining cellular need for DNA repair due to accelerated clearance of genotoxic metabolites by high ALDH1A1 expression. In addition, reduced expression of XPC, a damage recognition protein that initiates the NER pathway [51], has been found to be associated with poor differentiation of cancerous tissues while XPC overexpression significantly increases the susceptibility of colon cancer cells to chemotherapy and radiation [52]. Thus, reduced capacity of the NER pathway may alter cellular behavior and promote survival of CSCs, i.e., ALDHhigh cells. Through such mechanisms, ALDH1A1 may promote CSC survival. Third, the sirtuin signaling pathway was predicted to be activated in ALDHhigh cells. As members of class III histone deacetylase family, sirtuins play diverse roles in carcinogenesis by affecting the response to genomic instability, regulating cancer-associated metabolism, and modifying the tumor microenvironment [53–56]. Mounting evidence suggests that sirtuins can function either as oncoproteins or tumor suppressors, depending on the genetic background or presence/absence of specific coexisting biochemical defects [57]. It is noteworthy that ALDH1A1 is a target of lysine acetylation [58]. Furthermore, Sirt2 induction has been shown to activate ALDH1A1 activity through deacetylation, which promoted the self-renewal of breast cancer stem cells [35]. It is conceivable that activation of the sirtuin pathway may be involved in the induction of ALDH1A1 enzymatic activity in ALDHhigh cells. Whether ALDH1A1 impacts the sirtuin pathway remains to be elucidated. Collectively, the pathway changes associated with high ALDH activity may impact cellular metabolism and behavior that would favor CSC survival and self-renewal. Given that cancers rely on the existence and persistence of CSCs [59], ALDH1A1 may play a critical role in the development of colon cancers.
3. Conclusions
Taken together, the current proteomics study utilizing ALDHhigh and ALDHlow COLO320DM cells has provided valuable insights into the molecular features associated with high ALDH activity in colon cancer cells, a widely used marker for cancer stem cells. Our results provide evidence that high ALDH enzymatic activity is associated with altered expression of proteins that participate in oxidative phosphorylation, DNA damage repair, and sirtuin signaling. We speculate that these molecular features are mechanistically linked to the stemness properties of ALDHhigh COLO320DM cells by promoting a metabolic switch and survival of these cancer cells. The present results lend further support to the targeting of ALDH1A1 in colon cancer stem cells as a novel colon cancer therapy. Additional studies are warranted to further define the function(s) of ALDH1A1 in the colon cancer stem cells.
Supplementary Material
Acknowledgements
This work was supported in parts by National Institutes of Health (NIH) Grants AA021724, AA022057 and AA028859. The Orbitrap Fusion mass spectrometer at the Yale MS & Proteomics Resource was funded in part by NIH SIG from the Office of The Director, National Institutes of Health under Award Number (S10OD018034). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Wendy Wang for and Jean Kanyo for preparing the samples and for LC-MS/MS data collection, respectively.
References
- [1].Jackson B, Thompson DC,Black W, Vasiliou K, Nebert DW, Vasiliou V, Update on the aldehyde dehydrogenase gene (ALDH) superfamily, Human Genomics 5(2011) 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Marchitti SA, Brocker C, Stagos D, Vasiliou V, Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily, Expert Opin Drug Metab Toxicol, 4 (2008) 697–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vassalli G, Aldehyde dehydrogenases: not just markers, but functional regulators of stem cells, Stem Cells Int, 2019 (2019) 3904645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shortall K, Djeghader A, Magner E, Soulimane T, Insights into aldehyde dehydrogenase enzymes: a structural perspective, Front Mol Biosci, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vasiliou V, Sandoval M, Backos DS, Jackson BC, Chen Y, Reigan P, Lanaspa MA, Johnson RJ, Koppaka V, Thompson DC, ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein-protein interactions with HPRT1, Chem Biol Interact, 202 (2013) 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu L-K, Tanner JJ, Crystal structure of aldehyde dehydrogenase 16 reveals trans-hierarchical structural similarity and a new dimer, J Mol Biol, 431 (2019) 524–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Estey T, Piatigorsky J, Lassen N, Vasiliou V, ALDH3A1: a corneal crystallin with diverse functions, Exp Eye Res, 84 (2007) 3–12. [DOI] [PubMed] [Google Scholar]
- [8].Grünblatt E, Riederer P, Aldehyde dehydrogenase (ALDH) in Alzheimer’s and Parkinson’s disease, J Neural Transm, 123 (2016) 83–90. [DOI] [PubMed] [Google Scholar]
- [9].Ruscito I, Darb-Esfahani S, Kulbe H, Bellati F, Zizzari IG, Rahimi Koshkaki H, Napoletano C, Caserta D, Rughetti A, Kessler M, Sehouli J, Nuti M, Braicu EI, The prognostic impact of cancer stem-like cell biomarker aldehyde dehydrogenase-1 (ALDH1) in ovarian cancer: A meta-analysis, Gynecol Oncol, 150 (2018) 151–157. [DOI] [PubMed] [Google Scholar]
- [10].Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL, Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer, Mol Cancer Res, 7 (2009) 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ma F, Li H, Li Y, Ding X, Wang H, Fan Y, Lin C, Qian H, Xu B, Aldehyde dehydrogenase 1 (ALDH1) expression is an independent prognostic factor in triple negative breast cancer (TNBC), Medicine 96 (2017) e6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ginestier MHHC, Jauffret EC, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer C, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G, ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome, Cell Stem Cell, 1 (2007) 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nakahata K, Uehara S, Nishikawa S, Kawatsu M, Zenitani M, Oue T, Okuyama H, Aldehyde dehydrogenase 1 (ALDH1) is a potential marker for cancer stem cells in embryonal rhabdomyosarcoma, PLoS One, 10 (2015) e0125454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM, Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis, Cancer Res, 69 (2009) 3382–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shenoy A, Butterworth E, Huang EH, ALDH as a marker for enriching tumorigenic human colonic stem cells, Methods Mol Biol, 916 (2012) 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V, Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer, Biochem Biophys Res Commun, 405 (2011) 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Feng H, Liu Y, Bian X, Zhou F, Liu Y, ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status, Br J Cancer, 118 (2018) 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chu P, Clanton DJ, Snipas TS, Lee J, Mitchell E, Nguyen M-L, Hare E, Peach RJ, Characterization of a subpopulation of colon cancer cells with stem cell-like properties, Int J Cancer, 124 (2009) 1312–1321. [DOI] [PubMed] [Google Scholar]
- [19].Kahlert C, Gaitzsch E, Steinert G, Mogler C, Herpel E, Hoffmeister M, Jansen L, Benner A, Brenner H, Chang-Claude J, Rahbari N, Schmidt T, Klupp F, Grabe N, Lahrmann B, Koch M, Halama N, Buchler M, Weitz J, Expression analysis of aldehyde dehydrogenase 1A1 (ALDH1A1) in colon and rectal cancer in association with prognosis and response to chemotherapy, Ann Surg Oncol, 19 (2012) 4193–4201. [DOI] [PubMed] [Google Scholar]
- [20].Zhou L, Sheng D, Wang D, Ma W, Deng Q, Deng L, Liu S, Identification of cancer-type specific expression patterns for active aldehyde dehydrogenase (ALDH) isoforms in ALDEFLUOR assay, Cell Biol Toxicol, 35 (2019) 161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tomita KTH, Tanaka T, Hara A, Aldehyde dehydrogenase 1A1 in stem cells and cancer, Oncotarget, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vishnubalaji MMR, Fahad M, Hamam R, Alfayez M, Kassem M, Aldahmash A, Alaiez NM, Molecular profiling of ALDH1+ colorectal cancer stem cells reveals preferential activation of MAPK, FAK, and oxidative stress pro-survival signalling pathways, Oncotarget, 9 (2018) 13551–13564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Matsumoto A, Arcaroli J, Chen Y, Gasparetto M, Neumeister V, Thompson DC, Singh S, Smith C, Messersmith W, Vasiliou V, Aldehyde dehydrogenase 1B1: a novel immunohistological marker for colorectal cancer, Br J Cancer, 117 (2017) 1537–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, Vasiliou V, ALDH1B1 is crucial for colon tumorigenesis by modulating wnt/β-catenin, notch and PI3K/Akt signaling pathways, PloS One, 10 (2015) e0121648–e0121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, Ghaedi K, Signaling pathways involved in colorectal cancer progression, Cell Biosci, 9 (2019) 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C, Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity, Proc Natl Acad Sci USA, 96 (1999) 9118–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH, Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer, Cancer Res, 69 (2009) 8208–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Matthiesen R, Jensen ON, Analysis of mass spectrometry data in proteomics, in: Keith JM (Ed.) Bioinformatics, Humana Press, Totowa, NJ, 2008, pp. 105–122. [DOI] [PubMed] [Google Scholar]
- [29].Aguilan JT, Kulej K, Sidoli S, Guide for protein fold change and p-value calculation for non-experts in proteomics, Mol Omics, 16 (2020) 573–582. [DOI] [PubMed] [Google Scholar]
- [30].Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing, J R Stat Soc Series B Stat Methodol, 57 (1995) 289–300. [Google Scholar]
- [31].Krämer A, Green J, Pollard J Jr., Tugendreich S, Causal analysis approaches in Ingenuity Pathway Analysis, Bioinformatics, 30 (2014) 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marcato P, Dean CA, Giacomantonio CA, Lee PW, Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform, Cell Cycle, 10 (2011) 1378–1384. [DOI] [PubMed] [Google Scholar]
- [33].Opdena LM, Modarai SR, Boman BM, The proportion of ALDEFLUOR-positive cancer stem cells changes with cell culture density due to the expression of different ALDH isoforms, Cancer Stud Mol Med, 2 (2015) 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Saw Y-T, Yang J, Ng S-K, Liu S, Singh S, Singh M, Welch WR, Tsuda H, Fong W-P, Thompson D, Vasiliou V, Berkowitz RS, Ng S-W, Characterization of aldehyde dehydrogenase isozymes in ovarian cancer tissues and sphere cultures, BMC Cancer, 12 (2012) 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, Xiong Y, Guan KL, Lei QY, NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells, J Clin Invest, 124 (2014) 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xiong Y, Guan KL, Mechanistic insights into the regulation of metabolic enzymes by acetylation, J Cell Biol, 198 (2012) 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Masciale V, Grisendi G, Banchelli F, D’Amico R, Maiorana A, Sighinolfi P, Stefani A, Morandi U, Dominici M, Aramini B, Isolation and identification of cancer stem-like cells in adenocarcinoma and squamous cell carcinoma of the lung: a pilot study, Front Oncol, 9 (2019) 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mele L, Liccardo D, Tirino V, Evaluation and isolation of cancer stem cells using ALDH activity assay, in: Papaccio G, Desiderio V (Eds.) Cancer Stem Cells, Springer New York, New York, NY, 2018, pp. 43–48. [DOI] [PubMed] [Google Scholar]
- [39].Mazure NM, VDAC in cancer, Biochim Biophys Acta Bioenerg, 1858 (2017) 665–673. [DOI] [PubMed] [Google Scholar]
- [40].Lemasters JJ, Holmuhamedov E, Voltage-dependent anion channel (VDAC) as mitochondrial governator--thinking outside the box, Biochim Biophys Acta, 1762 (2006) 181–190. [DOI] [PubMed] [Google Scholar]
- [41].Heslop KA, Milesi V, Maldonado EN, VDAC modulation of cancer metabolism: advances and therapeutic challenges, Front Physiol, 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vyas S, Zaganjor E, Haigis MC, Mitochondria and cancer, Cell, 166 (2016) 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liberti MV, Locasale JW, The warburg effect: how does it benefit cancer cells?, Trends Biochem Sci, 41 (2016) 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, Rickardson L, D’Arcy P, Gullbo J, Nygren P, Larsson R, Linder S, Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments, Nat Commun, 5 (2014) 3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Intlekofer AM, Finley LWS, Metabolic signatures of cancer cells and stem cells, Nat Metab, 1 (2019) 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kusakabe M, Onishi Y, Tada H, Kurihara F, Kusao K, Furukawa M, Iwai S, Yokoi M, Sakai W, Sugasawa K, Mechanism and regulation of DNA damage recognition in nucleotide excision repair, Genes Environ, 41 (2019) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Russo J, Barnes A, Berger K, Desgrosellier J, Henderson J, Kanters A, Merkov L, 4-(N,N-dipropylamino)benzaldehyde inhibits the oxidation of all-trans retinal to all-trans retinoic acid by ALDH1A1, but not the differentiation of HL-60 promyelocytic leukemia cells exposed to all-trans retinal, BMC Pharmacol, 2 (2002) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mori Y, Yamawaki K, Ishiguro T, Yoshihara K, Ueda H, Sato A, Ohata H, Yoshida Y, Minamino T, Okamoto K, Enomoto T, ALDH-dependent glycolytic activation mediates stemness and paclitaxel resistance in patient-derived spheroid models of uterine endometrial cancer, Stem Cell Rep, 13 (2019) 730–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Torgovnick A, Schumacher B, DNA repair mechanisms in cancer development and therapy, Front Genet, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V, Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress, Free Radic Biol Med, 56 (2013) 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lubin A, Zhang L, Chen H, White VM, Gong F, A human XPC protein interactome--a resource, Int J Mol Sci, 15 (2013) 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hu LB, Chen Y, Meng XD, Yu P, He X, Li J, Nucleotide excision repair factor XPC ameliorates prognosis by increasing the susceptibility of human colorectal cancer to chemotherapy and ionizing radiation, Front Oncol, 8 (2018) 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bhatti JS, Bhatti GK, Reddy PH, Mitochondrial dysfunction and oxidative stress in metabolic disorders - A step towards mitochondria based therapeutic strategies, Biochim Biophys Acta Mol Basis Dis, 1863 (2017) 1066–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chalkiadaki A, Guarente L, The multifaceted functions of sirtuins in cancer, Nat Rev Cancer, 15 (2015) 608–624. [DOI] [PubMed] [Google Scholar]
- [55].Yu J, Auwerx J, The role of sirtuins in the control of metabolic homeostasis, Ann N Y Acad Sci, 1173 Suppl 1 (2009) E10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, Wang F, Yang L, Xiang Z, Cui H, The roles of sirtuins family in cell metabolism during tumor development, Semin Cancer Biol, 57 (2019) 59–71. [DOI] [PubMed] [Google Scholar]
- [57].Zhang H-F, Lai R, STAT3 in cancer—friend or foe?, Cancers, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Poulose N, Raju R, Sirtuin regulation in aging and injury, Biochim Biophys Acta, 1852 (2015) 2442–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, Sastry N, Hu B, Cheng SY, Stem cell programs in cancer initiation, progression, and therapy resistance, Theranostics, 10 (2020) 8721–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


