Abstract
Background
The poor outcome of patients with lung squamous cell carcinoma (LUSC) highlights the importance of the identification of novel effective prognostic markers and therapeutic targets. Long noncoding RNAs (lncRNAs) have generally been considered to serve important roles in tumorigenesis and the development of various types of cancer, including LUSC.
Methods
Here, we aimed to investigate the role of LINC02323 in LUSC and its potential mechanisms by performing comprehensive bioinformatic analyses.
Results
LINC02323 was elevated and positively associated with unfavorable prognosis of LUSC patients. LINC02323 exerted oncogenic function by competitively binding to miR‐1343‐3p and miR‐6783‐3p, thereby upregulating L1CAM expression. Indeed, we also determined that LINC02323 could interact with the RNA‐binding protein DDX3X, which regulates various stages of RNA expression and processing.
Conclusion
Taken together, we identified that LINC02323 and its indirect target L1CAM can act as novel biomarkers for determining the prognosis of patients with LUSC and thus deserves further study.
Keywords: DDX3X, L1CAM, LINC02323, miR‐1343‐3p, miR‐6783‐3p, lung squamous cell carcinoma
LINC02323 sponges miR‐1343‐3p and miR‐6783‐3p and upregulates L1CAM for promoting LUSC progression. An RNA‐binding protein DDX3X is proposed to modulate LINC02323 expression.
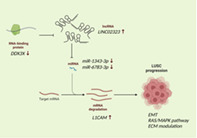
INTRODUCTION
Lung cancer is the leading cause of tumor‐related mortality worldwide. 1 Non‐small cell lung cancer (NSCLC), which accounts for more than 85% of all lung cancers, consists of two predominant subtypes: lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). The 5‐year overall survival (OS) rate of advanced NSCLC has greatly improved due to the targeted therapies and immune checkpoint inhibitors. 2 , 3 However, these agents preferentially benefit patients with LUAD, 5 and the current OS of LUSC remains 20%–30% less than that of LUAD. 6 , 7 , 8 These differences in available therapeutic strategies and clinical outcomes result from different molecular patterns according to tumor genomic aberration or transcript alterations. 4 Hence, elucidating the molecular mechanisms underlying tumorigenesis and progression might be conducive to developing more effective therapeutic targets for LUSC.
Noncoding RNAs (ncRNAs), especially microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), contribute to various biological processes in cancer tumorigenesis and progression. 5 , 6 LncRNA with a length >200 nucleotides may be involved in more diverse and complex mechanisms for regulating the biogenetic process because of the longer sequences and more complex spatial structures compared with small RNAs. 7 , 8 LncRNAs interact with DNAs, RNAs, and proteins to participate in numerous physiological and pathological processes at the transcriptional, translational, and epigenetic levels and other mechanisms such as chromatin remodeling, cell cycle regulation, splicing regulation, mRNA degradation, and translational regulation. 9 , 10 Several lncRNAs are expressed abnormally in lung cancer, particularly in LUAD, where they may act as oncogenes or tumor suppressor genes, suggesting lncRNA as novel tumorigenic drivers in lung cancer. LncRNA H19 interacts with mutant‐type p53 (R175H), leading to LUAD progression. 11 LncRNA‐MALAT1 promotes the spread and invasion of lung cancer by regulating the expressions of miR‐124/STAT3 and miR‐206/AKT. 12 LINC00261 decreases hypoxia‐induced tumor angiogenesis and its loss was associated with a poor prognosis for patients with LUAD. 13 However, the relationship between lncRNAs and the progression of LUSC remained unclear.
Systemic biology and bioinformatic approaches can facilitate the understanding of the pathogenesis of LUSC and the identification of potential novel biomarkers and druggable targets. 14 , 15 , 16 Many transcriptomic analyses and datasets of LUSC samples have been established. Nevertheless, compared with coding genes and microRNAs, the specific lncRNAs involved in the onset and development of LUSC remain unknown. The current study was designed to investigate LINC02323 expression (located between 545 253 and 102 592 425 on chromosome 14) and its prognostic value in LUSC. Our results showed that LINC02323 functioned as an oncogene in LUSC by sponging miR‐1343, miR‐6783, and DDX3X‐interacted axis, then boosting cell growth and maintaining CSC features. Our findings may be used to further investigate the characteristics of LUSC and are useful in determining the prognosis and therapy of patients with LUSC.
METHODS
Data collection
The gene expression quantification datasets of LUSC were extracted from samples of The Cancer Genome Atlas (TCGA) (available online: https://portal.gdc.cancer.gov, accessed on August 24, 2022). The criterion in the analysis was a p‐value <0.05 which was calculated using the University of Alabama at Birmingham CANcer (UALCAN) data analysis portal (available online: http://ualcan.path.uab.edu, accessed on August 24, 2022), the Encyclopedia of RNA Interactomes (ENCORI) (available online: https://starbase.sysu.edu.cn/index.php, accessed on October 9, 2022), or TNM plotter (available online: https://tnmplot.com/analysis/, accessed on August 24, 2022).
Survival analysis of LINC02323 , L1CAN, and DDX3X in LUSC
The survival analyses of the various candidate genes in LUSC were assessed via the Kaplan Meier (KM) plotter (available online: http://kmplot.com/analysis/, accessed on October 7, 2022). The hazard ratios (95% confidence intervals) were calculated using the Cox proportional model.
Survival analysis of miR‐1343‐3p and miR‐6783‐3p in LUSC
The Pan‐Cancer Analysis Platform of ENCORI was used to analyze the association of the miR‐1343‐3p and miR‐6783‐3p expression with overall survival (OS). Patients were divided into two groups by median value, which was computed with median survival.
Screening for targets of miR‐1343‐3p and miR‐6783‐3p
The target gene of miR‐1343‐3p and miR‐6783‐3p was predicted via miRDB (available online: http://mirdb.org/, accessed on October 7, 2022), which searched the target genes based on the conserved sites 8 mer and 7 mer sites matching the seed regions of miRNAs. Target score >90% was set as the cutoff criteria.
Interaction of RNA‐binding protein (RBP) and miRNAs with LINC02323
The miRNA or RBP interaction analyses with lncRNA (miRNA‐lncRNA, and RBP‐lncRNA) were performed using the ENCORI online database. The potential interactive RBPs with LINC02323 were searched using RBP‐lncRNA module, with a selection criterion of the stringency equal to, or more than 1, in the crosslinking immunoprecipitation (CLIP) database.
Functional analysis and the gene set variation analysis (GSVA) of gene sets
To investigate the role of L1 cell adhesion molecule (L1CAM) and DDX3X, the LUSC patients of TCGA were divided into L1CAM or DDX3X high‐ and low‐expression groups according to the highest and lowest quartiles, and gene set enrichment analysis (GSEA) was conducted to assess the enrichment of datasets between high‐ and low‐target gene groups. False discovery rate (FDR) < 0.05 and nominal p‐value <0.05 were set as the cutoff criteria. The gene set “c2.cp.kegg.v6.2.symbols.gmt” was chosen as the reference gene set. The correlation of the gene sets positively correlated with LINC02323, L1CAM, or DDX3X was also extracted from the UALCAN. The criteria in gene extraction were Pearsons correlation coefficient (CC) > 0.3 and p‐value <0.05, which was calculated using the UALCAN. The GSVA score of signaling pathway on survival rate was calculated using gene set cancer analysis (GSCA) (available online: http://bioinfo.life.hust.edu.cn/GSCA/#/, accessed on October 10, 2022). Metascape (available online: http://metascape.org/gp/index.html#/main/step1) and NetworkAnalyst (available online: https://www.networkanalyst.ca/, accessed on October 7, 2022) were used to visualize for KEGG of positively‐correlated genes of L1CAM.
Signaling pathway and network analysis
The significant enrichment analysis of the gene sets positively‐correlated with L1CAM was also assessed based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Available online: https://david.ncifcrf.gov/, accessed on October 10, 2022).
Statistical analysis
Spearman's correlation was applied for the analysis of the correlation. Most analyses were conducted using GraphPad Prism 9.0. Logistic regression, univariate, and multivariate analyses were used to assess the influence of clinical variables on patient survival. A p‐value less than 0.05 was considered statistically significant.
RESULTS
Analysis of LINC02323 expression in patients with lung cancer
We first analyzed LINC02323 expression in LUSC. As shown in Figure 1a, LINC02323 expressions were higher (5.5‐fold, p‐value = 2.6 × 10−39) in LUSC in the cancer genome atlas (TCGA) database collected from UALCAN. However, the expression did not depend on the progress of cancer stages and smoking/reformed smoking status in LUSC (Figure 1b, c). In addition, LINC02323 expression was significantly increased in an additional 13 cancer types other than lung cancer (acute myelogenous leukemia, bladder, colon, liver, ovary, pancreas, rectum, renal papillary cell carcinoma, stomach, testis, thyroid, uterine, and uterine endometrial carcinoma) on the TMN plotter website and seven types in TCGA cohort (bladder, urothelial carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, colon adenocarcinoma, esophageal carcinoma, head and neck squamous cell carcinoma, pancreatic adenocarcinoma, rectum adenocarcinoma, and stomach adenocarcinoma) (Figure 1e, f) compared with these in normal tissues. These data showed that LINC02323 may exert an oncogenic role in various cancers, including LUSC.
FIGURE 1.
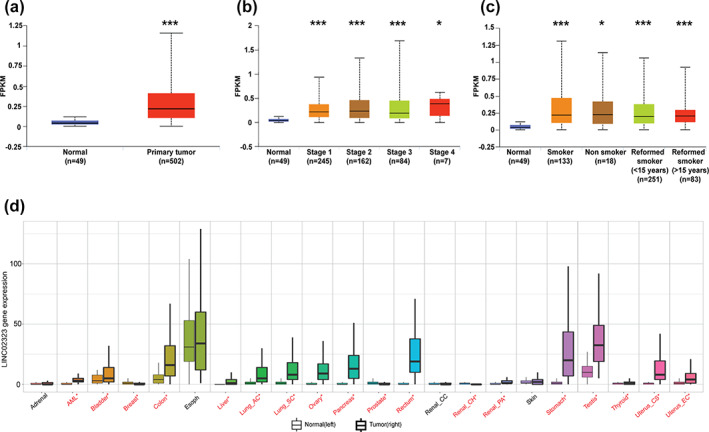
The expression of LINC02323 in cancers. (a) The expression of LINC02323 in TCGA lung squamous cell carcinoma (LUSC) dataset. The levels of LINC02323 in (b) the different stages and (c) smoking status. (d) The levels of LINC02323 in various cancers extracted from TNM plotter. **p < 0.01; ***p < 0.005; NS, not significant
Correlation between LINC02323 expression and the prognosis of patients with LUSC
Next, we investigated the correlation between LINC02323 expression and overall survival (OS) in the TCGA database. The results indicated that high LINC02323 expression was related to poor survival in LUSC in TCGA database (Figure 2a). These results were validated by data from the KM plotter website (Figure 2b). However, LINC02323 was not correlated with relapse‐free survival (RFS) in LUSC (Figure 2c). Disease ontology analysis by ENCORI website also showed that LINC02323 was associated with the occurrence of lung cancer, and also correlated with cell cycle‐related oncogenic genes, including E2F (Figure 2d). Pathway analysis also indicated that the targets of LINC02323 were involved in cell cycle regulation, DNA replication and repair, and chromosome maintenance (Figure 2e).
FIGURE 2.
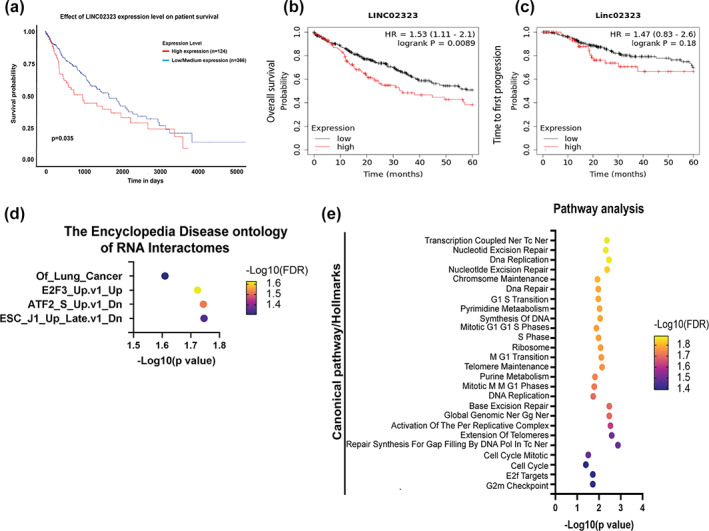
The correlation of LINC02323 with survival of lung squamous cell carcinoma (LUSC) patients. LINC02323 is associated with a poor prognosis in LUSC. The association of LINC02323 expression and survival time through data extracted from the Kaplan‐Meier plotter website on overall survival disadvantage when (a) expression is high with divergent trends (p < 0.05), (b) time to first progression, and (c) post‐progression survival. Red and black lines: High‐ and low‐expressed LINC02323, respectively. HR, hazard ratio. (d) Disease ontology (DO) annotated LINC02323 was associated with lung cancer occurrence. (e) Pathway analysis of LINC02323, as assessed by ENCORI website
Interactions between LINC02323 with miR‐1343‐3p/miR‐6783‐3p‐L1CAM axis in LUSC
LncRNAs are known to act as a sponge of miRNAs, thereby inhibiting the degradation of mRNAs targeted by miRNAs. We thus predicted the interactions of LINC02323 using the ENCORI website. The results showed that miR‐1343‐3p and miR‐6783‐3p were complementary to LINC02323 (Figure 3a). The levels of miR‐1343‐3p and miR‐6783‐3p were higher in LUSC by 3.5‐ (p‐value = 0.031) and 3.84‐fold (p‐value = 0.0011) compared with these in normal tissues, respectively (Figure 3b, c). However, these two elevated miRNAs had no significant effect on the OS of LUSC patients (Figure 3d, e). We further assessed the potential targets of miR‐1343‐3p and miR‐6783‐3p predicted by miRDB (Supporting Information, Table S1, Figure 3f). According to the screening by bioinformatics, L1CAM was the target for both miR‐1343‐3p and miR‐6783‐3p, which have high affinity on the binding of 3'UTR of L1CAM (Figure 3g, h). In addition, cross‐analysis revealed that the HR of LINC02323 for survival was decreased from 1.6 (with higher level of L1CAM) to 0.64 (with lower level of L1CAM) in LUSC (Figure 3i).
FIGURE 3.
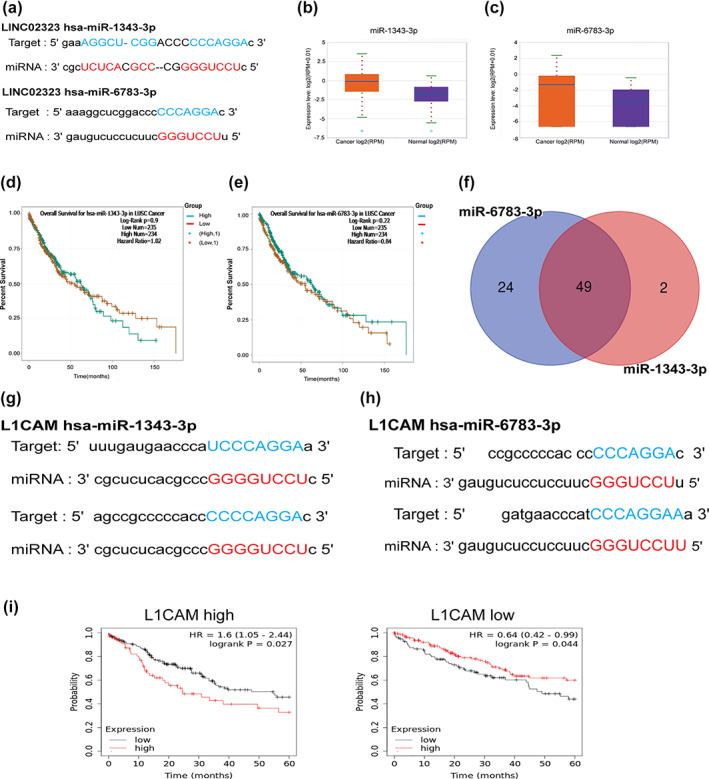
The interaction of LINC02323 with miR‐1343‐3p or miR‐6783‐3p. (a) The binding of miR‐1343 and miR‐6783 on the sequence of LINC02323. (b) The expression of miR‐1343‐3p and (c) miR‐6783‐3p in lung squamous cell carcinoma (LUSC). (d) The association of miR‐1343‐3p and (e) miR‐6783‐3p on OS of LUSC patients. (f) The common targets of miR‐1343 and miR‐6783. (g) The binding sites of mi‐1343‐3p and (h) miR‐6783‐3p in the 3'UTR of L1CAM. (i) The cross‐analysis of LINC02323 with L1CAM‐high and L1CAM‐low LUSC patients. *p < 0.05; **p < 0.01; ***p < 0.005; NS, not significant
Upregulation of L1CAM conferred poor survival in LUSC
To identify whether L1CAM expression was correlated with LUSC development, we assessed the L1CAM expression in normal and LUSC tumor tissue using a TCGA cohort. The results showed that L1CAM expression was increased (5.01‐fold; p‐value = 0.0037; FDR = 0.0087) in TCGA cohort (Figure 4a), although expression did not depend on the progress of cancer stages and smoking status in LUSC (Figure 4b, c). The elevated L1CAM was also correlated with poor OS and RFS in patients with LUSC (Figure 4d, e). To elucidate the prognostic value of coexpression gene sets of L1CAM in LUSC development, we extracted the gene sets with a positive correlation of L1CAM in LUSC and assessed by GCSC website. We found that 54 genes (r > 0.3, p < 0.05) were strongly associated with L1CAM expression (Supporting Information, Table S2). The higher GSVA score of this gene set was linked to poor prognosis, including shorter OS (HR = 1.52, p‐value = 0.002), PFS (HR = 1.42, p‐value = 0.005), and disease‐specific survival (DSS) (HR = 1.62, p‐value = 0.02), but not disease‐free interval (DFI) (HR = 0.71, p‐value = 0.19) (Figure 4f–i).
FIGURE 4.
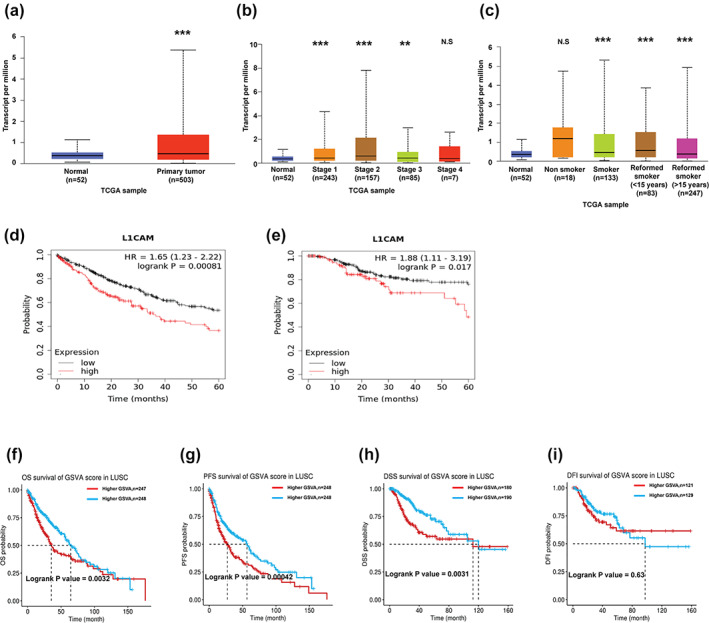
The correlation of L1CAM in the survival rate of lung squamous cell carcinoma (LUSC) patients. (a) L1CAM expression in LUSC. The levels of L1CAM in (b) different stages and (c) smoking status. (d) Elevated L1CAM was also correlated with poor OS and (e) PRF in the patients with LUSC. (f) The influence of gene set positively correlated with L1CAM levels on the OS, (g) PFS and (h) disease‐specific survival (DSS) and (i) disease‐free interval (DFI). *p < 0.05; **p < 0.01; ***p < 0.005; NS, not significant
Biological functions of LINC02323
For biological analysis, the gene set which was positively correlated with L1CAM expression had a positive correlation with the epithelial to mesenchymal transition (EMT) and RAS‐dependent mitogen‐activated protein kinase (RAS/MAPK) pathways (Figure 5a, b). To validate the functional analysis of L1CAM, we also divided LUSC patients in the TCGA cohort into L1CAM higher and lower expression groups, and then applied the gene expression profiles onto GSEA analysis. The results showed that L1CAM and its regulated gene sets in LUSC were mainly involved in collagen formation and FGFR3 activation (Figure 5c, d). KEGG pathway analysis also supported that the gene set was associated with the process of ECM, including focal adhesion, proteoglycans in cancer and ECM‐receptor interaction (Figure 5e). The hub genes of the L1CAM‐positive gene set were ITGA1, ITGB3, TGFBI, AXL, XDH, PLNCDH3, WNT7A, CHRNB1, and COL17A1 (Figure 5f).
FIGURE 5.
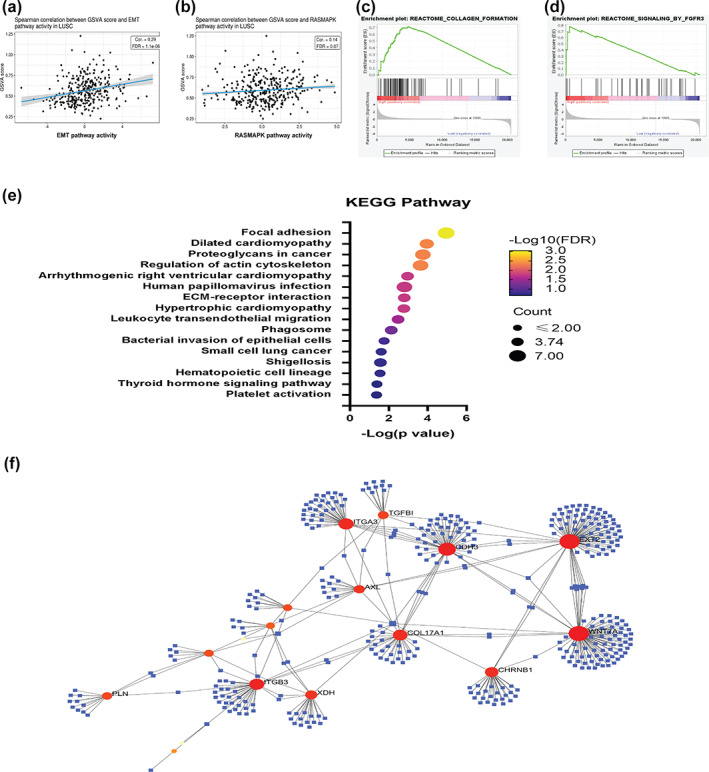
Elevated L1CAM expression is associated with extracellular matrix (ECM) remodeling and RAS activation. The gene set positively correlated with L1CAM levels associated with (a) EMT and (b) RAS/MAPK pathway. The transcriptomes of LUSC patients with high L1CAM expression are associated with (c) collagen formation and (d) FGFR3 activation. (e) Pathway and (f) hub gene analysis of the transcriptomes of LUSC patients with high L1CAM expression. ***p < 0.005
Interaction of LINC02323 with RNA‐binding proteins (RBPs)
RBPs are known to be abnormally expressed in cancer and regulate the function of large noncoding RNAs. To better understand the potential regulatory mechanisms of LINC02323 in LUSC, we examined the expression of RBPs, which interacts with LINC02323 in LUSC. We used the ENCORI database to explore possible RBPs for LINC02323. The results showed that 52 RBPs might modulate LINC02323 function. A total of 43 RBPs were upregulated and four RBPs were downregulated in LUSC. Among them, DDX3X expression was reduced and associated with better OS in LUSC. In contrast, the levels of FAM120A, LSM11, and XRN2 were increased and correlated with shorter OS in LUSC (Table 1). To identify the RBP which most probably regulates the LINC02323, we conducted a cross‐analysis of LINC02323 with these four RBPs. Cross‐analysis of DDX3X with LINC02323 on OS showed that the hazard ratio (HR) of LINC02323 declined in the high DDX3X expression group (HR = 0.74, p = 0.16), compared with that in low DDX3X expression LUSC patients (HR = 1.9, p = 0.0034) (Figure 6a). In contrast, the effect of LINC02323 only slightly, or insignificantly, decreased the OS of LUSC patients with lower expressions of FAM120A, LSM11, and XRN2 in LUSC (Figure 6b–d). These results showed that DDX3X could be a RBP of LINC02323, contributing to the development of LUSC.
TABLE 1.
The potential RBPs of LINC02323
| Fold change (tumor/normal) | HR of OS | Fold change (tumor/normal) | HR of OS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | FC | p‐value | HR | p‐value | Gene | FC | p‐value | HR | p‐value |
| AUH | 0.80 | <0.001 | 1.62 | 0.01 | ILF3 | 1.71 | <0.001 | 0.78 | 0.10 |
| BCCIP | 1.63 | <0.001 | 0.62 | 0.04 | KHDRBS1 | 1.36 | <0.001 | 0.78 | 0.11 |
| BUD13 | 1.11 | <0.001 | 0.73 | 0.04 | LIN28B | n.a | 1.20 | 0.23 | |
| CSTF2T | 1.03 | 0.004 | 0.69 | 0.04 | LSM11 | 1.12 | <0.001 | 1.28 | 0.11 |
| DDX3X | 0.87 | 0.001 | 0.85 | 0.27 | NONO | 1.74 | <0.001 | 0.74 | 0.04 |
| DDX42 | 1.18 | <0.001 | 0.86 | 0.34 | NUMA1 | 0.77 | 0.06 | 0.74 | 0.04 |
| DGCR8 | 1.49 | <0.001 | 0.76 | 0.13 | PCBP2 | 1.27 | <0.001 | 0.67 | 0.01 |
| DKC1 | 3.59 | <0.001 | 0.67 | 0.02 | PRPF8 | 0.77 | <0.001 | 1.23 | 0.18 |
| EIF4A3 | 2.21 | <0.001 | 0.81 | 0.17 | PTBP1 | 1.72 | <0.001 | 0.84 | 0.26 |
| EIF4G2 | 1.05 | <0.001 | 1.49 | 0.01 | RBFOX2 | n.a. | 0.75 | 0.05 | |
| FAM120A | 1.18 | <0.001 | 1.27 | 0.15 | RBM27 | 1.09 | 0.006 | 0.74 | 0.05 |
| FBL | 2.85 | <0.001 | 0.72 | 0.03 | SAFB2 | 1.01 | 0.02 | 0.74 | 0.08 |
| FKBP4 | 4.09 | <0.001 | 0.70 | 0.02 | SF3A3 | 1.25 | <0.001 | 0.77 | 0.11 |
| FMR1 | 1.06 | <0.001 | 0.69 | 0.02 | SF3B4 | 1.84 | <0.001 | 0.70 | 0.02 |
| FUS | 1.36 | <0.001 | 0.68 | 0.03 | SLTM | 1.06 | 0.01 | 1.23 | 0.17 |
| FXR1 | 2.61 | <0.001 | 0.71 | 0.02 | SMNDC1 | 1.50 | <0.001 | 0.78 | 0.09 |
| FXR2 | 0.76 | <0.001 | 1.31 | 0.08 | SND1 | 1.33 | <0.001 | 1.26 | 0.19 |
| GTF2F1 | 1.12 | <0.001 | 0.72 | 0.03 | SRSF1 | n.a. | 0.72 | 0.04 | |
| HNRNPA1 | 1.34 | <0.001 | 0.75 | 0.05 | SRSF7 | n.a. | 0.79 | 0.12 | |
| HNRNPC | 1.69 | <0.001 | 0.72 | 0.06 | SRSF9 | n.a. | 0.83 | 0.26 | |
| HNRNPK | 1.50 | <0.001 | 0.78 | 0.10 | TAF15 | 1.27 | <0.001 | 0.88 | 0.42 |
| HNRNPM | 1.34 | <0.001 | 0.73 | 0.03 | TARBP2 | 1.91 | <0.001 | 0.81 | 0.19 |
| HNRNPU | 1.31 | <0.001 | 0.64 | 0.01 | TNRC6A | 1.08 | 0.001 | 0.70 | 0.02 |
| HNRNPUL1 | 1.25 | <0.001 | 0.70 | 0.02 | TRA2A | 1.02 | 0.02 | 0.70 | 0.03 |
| IGF2BP2 | 4.91 | <0.001 | 0.75 | 0.05 | U2AF2 | 1.68 | <0.005 | 0.69 | 0.05 |
| IGF2BP3 | 29.88 | <0.001 | 0.69 | 0.02 | XRN2 | 1.31 | <0.005 | 1.17 | 0.36 |
Abbreviations: FC, fold change; HR, hazard ratio; n.a., not available; OS, overall survival; RBPs, RNA‐binding proteins.
FIGURE 6.
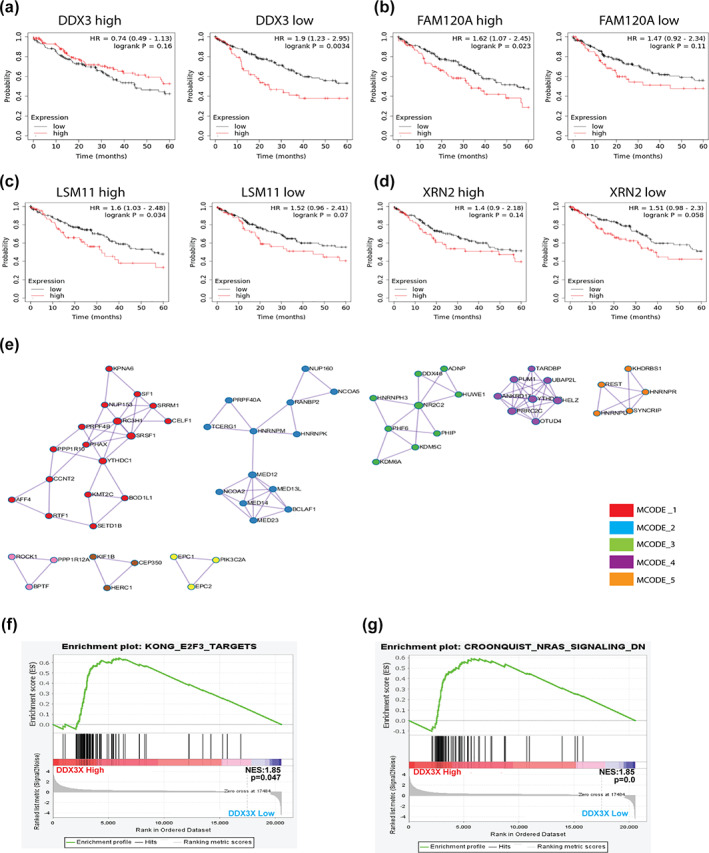
DDX3X is the RNA‐binding protein (RBP) of LINC02323 in LUSC. The cross‐analysis of LINC02323 in (a) DDX3X, (b) FAM120A, (c) LSM11, and (d) XRN on the OS of LUSC patients. (e) Pathway analysis of the transcriptomes of LUSC patients with high DDX3X expression. (f) The gene set positively correlated with DDX3X levels associated with (g) E2F3 and (h) N‐RAS pathway in LUSC patients. *p < 0.05; **p < 0.01; ***p < 0.005; NS, not significant
For biological analysis, we extracted the gene sets which were positively correlated with DDX3X expression and assessed it using Metascape. The results showed the gene sets were grouped into functional mcodes, which were involved in RNA splicing, mRNA destabilization and metabolism, histone modification, and stress granule assembly (Figure 6e). In addition, GSEA analysis also supported that DDX3X was associated with E2F targets, as the pathway analysis of LINC02323 (Figure 6f). In addition, the NRAS pathway was also involved in the gene enrichment analysis of DDX3X, as it was in the analysis of L1CAM, a downstream factor of LINC02323 (Figure 6g).
DISCUSSION
Based on the poor outcome, improving the prognosis of LUSC patients by finding novel biomarkers and targets for drug development is urgently required. The association between the dysregulated lncRNAs and the prognosis of patients has been reported in various malignancies. 17 Zhang et al. has reported that LINC02323 increased metastasis of LUAD by promoting EMT. 18 The correlations of LINC02323 expression have not yet been described in LUSC. In our results, LINC02323 expression was elevated in LUSC tissues. Survival analysis showed that high expression of LINC02323 was positively correlated with the poor prognosis of patients with LUSC. This is the first comprehensive evaluation of LINC02323 that implicates its potential oncogenic role in LUSC.
LncRNAs can act as sponges to regulate gene expression by competitively binding miRNAs. 19 LINC02323 as an oncogenic gene has been found in some studies, and it is linked to numerous malignancies, such as ovarian cancer, liver cancer, and LUAD. 20 Recent evidence showed that LINC02323 upregulates E2F1 expression by competitively binding to miR‐29c‐3p, thereby changing the immune microenvironment and chemoresistance in hepatocellular carcinoma (HCC). 21 miR‐129‐5p has also been found to inhibit the proliferation, invasion, and migration of gastric cancer cells by selectively decreasing COL1A1 expression; miR‐1343‐3p have been indicated to involve LINC02323‐mediated cancer growth and progression by suppression of TGF‐β receptor in LUAD and ovarian cancer. 18 , 20 Chen et al. reported that LINC00173.v1 binds to miR‐511‐5p to regulate VEGFA expression, thereby promoting angiogenesis and development in LUSC. Animal experiments have demonstrated that LINC00173.v1 increases the therapeutic sensitivity of LUSC cells to cisplatin and that targeting LINC00173.v1 could be a potential treatment for combating LUSC. The current study identified two miRNAs, miR‐1343‐3p and miR‐6783‐3p, associated with various cancers, including colorectal cancer, glioma, HCC, and LUAD. 22 , 23 , 24 Regardless of high expression of miR‐1343‐3p and miR‐6783‐3p in LUSC, they were not significantly associated with an OS in LUSC, suggesting the functions of miRNAs were probably via a regulatory network. Indeed, the target gene of miR‐1343‐3p and miR‐6783‐3p, of L1 cell adhesion molecule (L1CAM), was upregulated in LUSC and also positively correlated with LINC02323 expression in LUSC (R = 0.18, p < 0.05), supporting the findings that LINC02323 inhibited the function of miR‐1343‐3p and miR‐6783‐3p. However, the network of LINC02323‐miR‐1343‐3p and LINC02323‐miR‐6783‐3p interactions warrants further experimental studies in LUSC.
L1CAM is a transmembrane glycoprotein that plays a critical role in cell adhesion, cell migration and cancer development, and is associated with poor prognosis in patients with cancer. 25 L1CAM is also regarded as a potential important gene as a predictor of platinum response in high‐risk endometrial carcinoma. 26 LUAD patients with high L1CAM expression in surgically‐resected brain metastases have been reported to have an unfavorable overall survival time. 27 Antagonistic mimetic compounds targeting L1CAM decreases glioblastoma cell migration. 28 Specifically, increased L1CAM expression has been observed in LUSC patients and has a close correlation with an adverse prognosis in LUSC. Functional enrichment analysis showed that the upregulated genes correlated with L1CAM were primarily implicated in various tumor microenvironment remodeling and growth factor‐related pathway, including the EMT process, collagen formation and NRAS pathway. Particularly, the upregulated gene set also predicted an adverse prognosis in LUSC patients, including OS, PRF, and DSS. Additional studies and clinical trials are needed to evaluate whether L1CAM blockade may have significant therapeutic efficacy to improve the treatment of LUSC.
LncRNAs can specifically bind to RBPs and influence the biological functions of RBPs. Conversely, certain specific RBPs can bind to lncRNAs to influence the function of lncRNAs on the regulation of downstream gene expression. On the other hand, RBPs can also regulate the expression of lncRNAs at the transcriptional level. DDX3X, one of the members of the DEAD‐box helicase family, is able to control most stages of RNA metabolism from transcription, splicing, RNA export to translation. 29 , 30 DDX3X can bind to lncRNA TUG1 and increases the stabilization of Bcl‐2‐associated athanogene 5. 31 LncRNA Rncr4 (retinal noncoding RNA 4) regulates the inhibitory function of DDX3X to mouse retina architecture. 32 The role of DDX3X in lung cancer is controversial. DDX3X is the RBP of LINC00673‐v4, which promotes LUAD progression by activating the Wnt pathway. 33 Inhibition of DDX3X by chemical inhibitor RK‐33 has been reported to induce apoptosis and increased radiation sensitivity in LUAD A549 cell and NSCLC H1299 cells. 34 In contrast, DDX3X has been reported to be a target of tumor suppressor p53, which enhances the cyclin‐dependent kinase inhibitor p21, resulting in cell cycle arrest in lung cancer. Indeed, loss of p53 decreased the expression of DDX3X which in turn contributes to cancer progression and poor patient outcome in NSCLC. Loss of DDX3X decreases E‐cadherin expression by MDM2‐Slug pathway expression and promotes tumor metastasis. 35 The present study revealed that DDX3X could be a RBP of LINC02323 due to the loss of LINC02323 impact on OS in the patient with lower level of DDX3X. Reduced expression of DDX3X predicts adverse outcomes and is related to a poor prognosis in patients with LUSC. The consistency between upregulated LINC02323 and downregulated DDX3X was associated with E2F targets and cell cycle regulation, indicating a negative feedback existed between LINC02323 and DDX3X. However, research on the impact of LINC02323 on the biological function of DDX3X is still limited, and further exploration of the associated regulatory mechanism is required.
The limitations of our study are as follows: Biological experiments are required to confirm our results because the study was based on a bioinformatic analysis, and the characteristic details (age, race, tumor grade, size, and therapy) were not taken into account due to our study mainly concentrating on the genes and those commonly identified as significantly changed in TCGA datasets. Therefore, biological information may be needed for further validation.
This study found that the LINC002323‐miR‐1343/6783‐3p‐L1CAM and LINC02323‐DDX3X axis might be the critical network for determining important molecular mechanisms in the progression of LUSC and they might be valuable prognostic markers and potential therapeutic targets in the progression of LUSC.
AUTHORS' CONTRIBUTIONS
KLW and JYH conceptualized the entire study. YCH, YYW, and CYC provided the technical support, performed the experiments and acquired the data. CYC provided the software management and analyzed the data. YWL and YCH validated the results. YMT and YCH executed the formal analysis. JYH pursued the investigation and provided the resources. YMT, KLW, and JYH performed data curation and interpreted the data. YMT and KLW wrote original draft. JYH and YLH wrote, reviewed and edited the final manuscript. JYH supervised the study, was the project administrator and acquired the funding. All authors read and confirmed the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Table S1. The common targets of miR‐1343‐3p and miR‐6783‐3p
Table S2. The positively correlated genes with L1CAM
ACKNOWLEDGMENTS
The authors would like to thank the Center for Research Resources and Development at Kaohsiung Medical University for their technical support. The genetic expression analyses are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. The authors would like to acknowledge the investigators of TCGA Research Network.
Wu K‐L, Tsai Y‐M, Huang Y‐C, Wu Y‐Y, Chang C‐Y, Liu Y‐W, et al. LINC02323 facilitates development of lung squamous cell carcinoma by miRNA sponge and RBP dysregulation and links to poor prognosis. Thorac Cancer. 2023;14(4):407–418. 10.1111/1759-7714.14760
Funding information Kaohsiung Medical University Chung‐Ho Memorial Hospital, Grant/Award Numbers: KMUH‐110‐0R14, KMUH‐110‐0R17; Ministry of Science and Technology, Taiwan, Grant/Award Numbers: MOST 110‐2314‐B‐037‐124‐MY3, MOST 110‐2314‐B‐037‐126‐MY2
DATA AVAILABILITY STATEMENT
Datasets of the present study are not available in public based on some ongoing studies in our laboratory. However, it is available from the corresponding author on reasonable request.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung‐cancer treatment on population mortality. N Engl J Med. 2020;383:640–9. 10.1056/NEJMoa1916623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323–58. 10.1016/j.jtho.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 4. Socinski MA, Obasaju C, Gandara D, Hirsch FR, Bonomi P, Bunn P, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11:1411–22. 10.1016/j.jtho.2016.05.024 [DOI] [PubMed] [Google Scholar]
- 5. Aprile M, Costa V, Cimmino A, Calin GA. Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int J Cancer. 2022. 10.1002/ijc.34282 [DOI] [PubMed] [Google Scholar]
- 6. Yu P, He X, Lu F, Li L, Song H, Bian X. Research progress regarding long‐chain non‐coding RNA in lung cancer: a narrative review. J Thorac Dis. 2022;14:3016–29. 10.21037/jtd-22-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Welsh SA, Gardini A. Genomic regulation of transcription and RNA processing by the multitasking integrator complex. Nat Rev Mol Cell Biol. 2022. 10.1038/s41580-022-00534-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Somasundaram K, Gupta B, Jain N, Jana S. LncRNAs divide and rule: the master regulators of phase separation. Front Genet. 2022;13:930792. 10.3389/fgene.2022.930792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singh AK, Aryal B, Zhang X, Fan Y, Price NL, Suárez Y, et al. Posttranscriptional regulation of lipid metabolism by non‐coding RNAs and RNA binding proteins. Semin Cell Dev Biol. 2018;81:129–40. 10.1016/j.semcdb.2017.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nasiri‐Aghdam M, Garcia‐Garduño TC, Jave‐Suárez LF. CELF family proteins in cancer: highlights on the RNA‐binding protein/noncoding RNA regulatory Axis. Int J Mol Sci. 2021;22:11056. 10.3390/ijms222011056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Y, Xia Q. LncRNA H19 promotes lung adenocarcinoma progression via binding to mutant p53 R175H. Cancer. 2022;14:4486. 10.3390/cancers14184486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang Y, Xiao G, Chen Y, Deng Y. LncRNA MALAT1 promotes migration and invasion of non‐small‐cell lung cancer by targeting miR‐206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29:725–35. 10.1097/cad.0000000000000650 [DOI] [PubMed] [Google Scholar]
- 13. Xue J, Song Y, Xu W, Zhu Y. The CDK1‐related lncRNA and CXCL8 mediated immune resistance in lung adenocarcinoma. Cell. 2022;11:2688. 10.3390/cells11172688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang R, Wang W, Zhang N, Chen X, Liu W, Zhang L, et al. Systematic pan‐cancer analysis identifies RBM39 as an immunological and prognostic biomarker. J Cell Mol Med. 2022;26:4859–71. 10.1111/jcmm.17517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miao TW, Yang DQ, Chen FY, Zhu Q, Chen X. A ferroptosis‐related gene signature for overall survival prediction and immune infiltration in lung squamous cell carcinoma. Biosci Rep. 2022;42:BSR20212835. 10.1042/bsr20212835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W, Zhang Q, Che L, Xie Z, Cai X, Gong L, et al. Using biological information to analyze potential miRNA‐mRNA regulatory networks in the plasma of patients with non‐small cell lung cancer. BMC Cancer. 2022;22:299. 10.1186/s12885-022-09281-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X, Han J, Du L, Li X, Hao J, Wang L, et al. Unique metastasis‐associated lncRNA signature optimizes prediction of tumor relapse in lung adenocarcinoma. Thorac Cancer. 2020;11:728–37. 10.1111/1759-7714.13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, Du L, Han J, Li X, Wang H, Zheng G, et al. Novel long non‐coding RNA LINC02323 promotes epithelial‐mesenchymal transition and metastasis via sponging miR‐1343‐3p in lung adenocarcinoma. Thorac Cancer. 2020;11:2506–16. 10.1111/1759-7714.13562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu W, Zhang S, He J. The mechanism of long non‐coding RNA in cancer Radioresistance/Radiosensitivity: a systematic review. Front Pharmacol. 2022;13:879704. 10.3389/fphar.2022.879704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Zhao Z, Sun D, Li Y. Novel long noncoding RNA LINC02323 promotes cell growth and migration of ovarian cancer via TGF‐β receptor 1 by miR‐1343‐3p. J Clin Lab Anal. 2021;35:e23651. 10.1002/jcla.23651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dong W, Zhan C. Bioinformatic‐based mechanism identification of E2F1 ‐related ceRNA and E2F1 immunoassays in hepatocellular carcinoma. J Gastrointest Oncol. 2022;13:1915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Liu J, Lai Y, Huang S, Zheng L, Fan N. LINC01559 promotes colorectal cancer via sponging miR‐1343‐3p to modulate PARP1/PTEN/AKT pathway. Pathol Res Pract. 2021;224:153521. 10.1016/j.prp.2021.153521 [DOI] [PubMed] [Google Scholar]
- 23. Qi J, Wang Z, Zhao Z, Liu L. EIF3J‐AS1 promotes glioma cell growth via up‐regulating ANXA11 through sponging miR‐1343‐3p. Cancer Cell Int. 2020;20:428. 10.1186/s12935-020-01487-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR‐6783‐3p/DKK1 axis and inactivating Wnt/β‐catenin signaling pathway. Biochem Biophys Res Commun. 2019;508:37–45. 10.1016/j.bbrc.2018.11.079 [DOI] [PubMed] [Google Scholar]
- 25. Barnhill R, van Laere S, Vermeulen P, Roman‐Roman S, Gardrat S, Alsafadi S, et al. L1CAM and laminin vascular network: association with the high‐risk replacement histopathologic growth pattern in uveal melanoma liver metastases. Lab Invest. 2022;102:1214–24. 10.1038/s41374-022-00803-w [DOI] [PubMed] [Google Scholar]
- 26. Romani C, Capoferri D, Reijnen C, Lonardi S, Ravaggi A, Ratti M, et al. L1CAM expression as a predictor of platinum response in high‐risk endometrial carcinoma. Int J Cancer. 2022;151:637–48. 10.1002/ijc.34035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang JW, Wang SQ, Wu ZY, Liu Q, Yuan Q, Cai HQ, et al. L1 cell adhesion molecule high expression is associated with poor prognosis in surgically resected brain metastases from lung adenocarcinoma. Clinics (Sao Paulo, Brazil). 2022;77:100040. 10.1016/j.clinsp.2022.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagaraj V, Mikhail M, Baronio M, Gatto A, Nayak A, Theis T, et al. Antagonistic L1 adhesion molecule mimetic compounds inhibit glioblastoma cell migration In vitro. Biomolecules. 2022;12:439. 10.3390/biom12030439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mo J, Liang H, Su C, Li P, Chen J, Zhang B. DDX3X: structure, physiologic functions and cancer. Mol Cancer. 2021;20:38. 10.1186/s12943-021-01325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Secchi M, Lodola C, Garbelli A, Bione S, Maga G. DEAD‐box RNA helicases DDX3X and DDX5 as oncogenes or oncosuppressors: a network perspective. Cancer. 2022;14:3820. 10.3390/cancers14153820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lei X, Fang X, Chen T, Pu C, Yang J, Liu H. LncRNA TUG1 promoted stabilization of BAG5 by binding DDX3X to exacerbate ketamine‐induced neurotoxicity. Neurotox Res. 2022. 10.1007/s12640-022-00580-w [DOI] [PubMed] [Google Scholar]
- 32. Krol J, Krol I, Alvarez CPP, Fiscella M, Hierlemann A, Roska B, et al. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat Commun. 2015;6:7305. 10.1038/ncomms8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guan H, Zhu T, Wu S, Liu S, Liu B, Wu J, et al. Long noncoding RNA LINC00673‐v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β‐catenin signaling. Proc Natl Acad Sci U S A. 2019;116:14019–28. 10.1073/pnas.1900997116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7:648–69. 10.15252/emmm.201404368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu DW, Lee MC, Wang J, Chen CY, Cheng YW, Lee H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/slug/E‐cadherin pathway and poor patient outcome in non‐small‐cell lung cancer. Oncogene. 2014;33:1515–26. 10.1038/onc.2013.107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The common targets of miR‐1343‐3p and miR‐6783‐3p
Table S2. The positively correlated genes with L1CAM
Data Availability Statement
Datasets of the present study are not available in public based on some ongoing studies in our laboratory. However, it is available from the corresponding author on reasonable request.


