Abstract
An essential feature of bacterial plasmids is their ability to replicate as autonomous genetic elements in a controlled way within the host. Therefore, they can be used to explore the mechanisms involved in DNA replication and to analyze the different strategies that couple DNA replication to other critical events in the cell cycle. In this review, we focus on replication and its control in circular plasmids. Plasmid replication can be conveniently divided into three stages: initiation, elongation, and termination. The inability of DNA polymerases to initiate de novo replication makes necessary the independent generation of a primer. This is solved, in circular plasmids, by two main strategies: (i) opening of the strands followed by RNA priming (theta and strand displacement replication) or (ii) cleavage of one of the DNA strands to generate a 3′-OH end (rolling-circle replication). Initiation is catalyzed most frequently by one or a few plasmid-encoded initiation proteins that recognize plasmid-specific DNA sequences and determine the point from which replication starts (the origin of replication). In some cases, these proteins also participate directly in the generation of the primer. These initiators can also play the role of pilot proteins that guide the assembly of the host replisome at the plasmid origin. Elongation of plasmid replication is carried out basically by DNA polymerase III holoenzyme (and, in some cases, by DNA polymerase I at an early stage), with the participation of other host proteins that form the replisome. Termination of replication has specific requirements and implications for reinitiation, studies of which have started. The initiation stage plays an additional role: it is the stage at which mechanisms controlling replication operate. The objective of this control is to maintain a fixed concentration of plasmid molecules in a growing bacterial population (duplication of the plasmid pool paced with duplication of the bacterial population). The molecules involved directly in this control can be (i) RNA (antisense RNA), (ii) DNA sequences (iterons), or (iii) antisense RNA and proteins acting in concert. The control elements maintain an average frequency of one plasmid replication per plasmid copy per cell cycle and can “sense” and correct deviations from this average. Most of the current knowledge on plasmid replication and its control is based on the results of analyses performed with pure cultures under steady-state growth conditions. This knowledge sets important parameters needed to understand the maintenance of these genetic elements in mixed populations and under environmental conditions.
Plasmids are extrachromosomal DNA elements with characteristic copy numbers within the host. These replicons have been found in species from the three representatives of the living world, namely, the domains Archaea, Bacteria, and Eukarya (318). Plasmids may constitute a substantial amount of the total genetic content of an organism, representing more than 25% of the genetic material of the cell in some members of the Archaea (127, 331). They can incorporate and deliver genes by recombination or transposition, thus favoring genetic exchanges in bacterial populations. Since plasmids can be introduced into new hosts by a variety of mechanisms, they can be considered to be a pool of extrachromosomal DNA which is shared among populations. The wealth of genetic information carried by plasmids, their impact in the microbial communities, and the potential of these elements to act as natural cloning vectors have stimulated research into plasmids not only from the fundamental but also from the clinical, biotechnological, and environmental points of view. Three main factors have contributed to the development of plasmid research: (i) the genetic organization of these elements is apparently simple, (ii) they can be easily isolated and manipulated in vitro, and (iii) since plasmids are dispensable, their manipulation does not appear, in principle, to have adverse consequences to the hosts.
The feature that better defines plasmids is that they replicate in an autonomous and self-controlled way. The analysis of plasmid replication and its control has led to milestone discoveries, such as the existence of antisense RNAs, and has contributed to the unraveling of mechanisms of DNA replication, macromolecular interactions, and control of gene expression. The ability of some plasmids to pass across the so-called genetic barriers among different living organisms has posed questions about general mechanisms governing replication and about the communication between plasmid replication components and the host machinery involved in DNA replication. This plasmid-host communication has attracted the attention of researchers working in environmental and in evolutionary fields. Plasmid host range studies also have clear implications in clinical microbiology and in biotechnology. Despite their autonomous replication, plasmids extensively use the replication machinery of the host, and therefore plasmid replication studies facilitate the exploration of the mechanisms involved in chromosome replication.
PLASMID REPLICATION MECHANISMS
There are three general replication mechanisms for circular plasmids, namely, theta type, strand displacement, and rolling circle (RC). Historical development of research on plasmids has led to the idea that theta replication is more frequent in replicons from gram-negative than from gram-positive bacteria whereas the opposite is found for plasmids replicating by the RC mode. This belief is probably wrong. It is true, however, that present knowledge on theta-replicating plasmids stems from replicons from gram-negative bacteria and that on RC-replicating plasmids derives from replicons from gram-positive hosts. Strand displacement replication has been associated with broad-host-range plasmids from the IncQ family. The molecular interactions and the functional relationships that take place in these three types of replication mechanisms are the focus of this review. Linear plasmids have been found in both gram-positive and gram-negative bacteria, and their structure can be of two types: those having a hairpin at each end, and those having a protein covalently bound at their 5′ ends. Linear plasmids of the first group replicate via concatemeric intermediates, whereas those of the second group seem to replicate by a protein-priming mechanism, similar to that of bacteriophage φ29 (264). However, initiation of replication from an internal origin in a plasmid with a terminal protein has been reported (48). Linear plasmids have been reviewed previously (123), and they will not be discussed here. Replication of plasmids from gram-negative bacteria has been specifically addressed (168a).
Concerning their genetic structure, plasmids have an essential region which contains the genes or loci involved in replication and its control. The organization of this essential region corresponds, in general, to the one described in the replicon model. In addition, plasmids may bear genes that could be considered dispensable, although they could actually play an important role for the plasmid itself and/or for the host. Some of these so-called dispensable genes are involved in processes such as plasmid transfer and spread among bacteria, resistance to antibiotics and heavy metals, resistance to radiation, and transfer of DNA to higher eukaryotes. Within the plasmid essential region, several genes and sequences can be considered. (i) The first is the origin(s) of replication (generically termed ori), which is characteristic of each replicon. (ii) Although this is not a general feature, many plasmids encode a protein involved in the initiation of replication, usually termed Rep protein. (iii) The third is the plasmid-borne genes involved in the control of replication. The requirement of a plasmid-encoded initiator is reflected by the presence of DNA cognate sites in the origin of replication, where protein-DNA interactions take place. These specific sites are the hallmark of a class of replicons that are different from replicons that do not require specific initiators.
Replication by the Theta Mechanism
Replication by the theta-type mechanism has been most extensively studied among the prototype circular plasmids of gram-negative bacteria, although this replication mode has also been described for plasmids isolated from gram-positive bacteria, namely, the streptococcal/enterococcal Inc18 group (40), some lactococcal replicons (152), and at least one Bacillus subtilis plasmid (192). DNA replication through the theta mechanism involves melting of the parental strands, synthesis of a primer RNA (pRNA), and initiation of DNA synthesis by covalent extension of the pRNA (163). DNA synthesis is continuous on one of the strands (leading strand) and discontinuous on the other (lagging strand), although synthesis of the two strands seems to be coupled (reviewed in references 148 and 326). Theta-type DNA synthesis can start from one or from several origins, and replication can be either uni- or bidirectional. Under electron microscopy (EM), the replication intermediates are seen as typical Θ (“theta”)-shaped molecules that, when digested with enzymes that cleave within the replicated region, yield Y-shaped molecules (“forks”). The replication intermediates can also be monitored by one- or two-dimensional electrophoresis. These analyses provide information on the nature of the replication intermediates, direction of replication, location of the origin and terminus, and degree of coupling between leading- and lagging-strand synthesis.
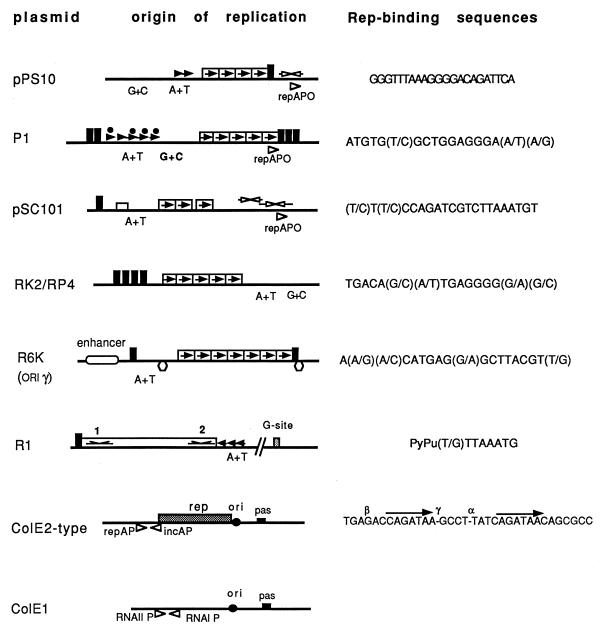
With some exceptions, plasmids using the theta mechanism of replication require a plasmid-encoded Rep initiator protein. Some replicons may require the host DNA polymerase I (DNA Pol I) during the early stages of leading-strand synthesis. Some features of various well-known replicons which are described here are depicted in Fig. 1.
FIG. 1.
Origins of replication and related regions of some representative theta-replicating plasmids from gram-negative bacteria. Plasmid names (left), origins of replication (center), and Rep initiator-binding sites (right) are indicated. The symbols used are as follows: solid boxed arrows correspond to repeated Rep-binding sequences (iterons); open arrows above maps indicate inverted repeats that have partial homology to the iterons; solid arrowheads indicate repeats found in AT-rich regions (A+T); for R1, arrows indicate the imperfect palindromes initially recognized by the RepA initiator protein. Promoters are indicated as open arrowheads below the maps. Other sites of interaction are as follows: IHF-binding sites (open rectangles), dnaA boxes (solid rectangles); FIS-binding sites (hexagons), dam methylation sites (solid circles), and primase assembly sites (pas). Other sites are indicated in the figure. Maps are based on the following references: pPS10 (104, 215); pSC101 (132, 284); P1 (6); RK2 (278); R6K (277); R1 (101); ColE1 (280, 301); ColE2-type plasmids (124).
Origins of replication.
Plasmid origins of replication can be defined as (i) the minimal cis-acting region that can support autonomous replication of the plasmid; (ii) the region where DNA strands are melted to initiate the replication process, or (iii) the base(s) at which leading-strand synthesis starts. Replication origins contain sites that are required for interactions of plasmid-encoded and/or host-encoded proteins.
(i) General features.
With some exceptions, initiation of plasmid DNA replication requires a specific plasmid-encoded Rep initiator protein. This is reflected by the presence, at the origin of replication, of specific sequences with which the Rep protein interacts. Additional features found in many origins of theta-replicating plasmids are (i) an adjacent AT-rich region containing sequence repeats, where opening of the strands and assembly of host initiation factors occur, and (ii) one or more sites (dnaA boxes) where the host DnaA initiator protein binds (30, 163). Multiple Dam methylation sequences, which are present in the origin of replication of the Escherichia coli chromosome, oriC, can also be found at the origin of replication of plasmids such as P1 (36, 38) and pSC101 (30). Methylation is not essential for replication, its role being primarily in postreplication (3). Dam methylation sequences are not present in other plasmid replicons.
Comparative analysis of the structural organization of the Pol I-independent origins of replication predicts that although the Rep-binding site is located within a potentially curved DNA region, the DNA within the repeats of the AT-rich region is essentially straight (81). Intrinsic DNA bends at the Rep-binding sites would favor additional curvatures of the origin induced by Rep proteins. The origins of replication can also contain sites for factors (e.g., the integration host factor, IHF, or the factor for inversion stimulation, FIS) that play an architectural role. These host-encoded proteins favor a topological proximity between different ori regions or even between different origins present in the same plasmid (as in plasmid R6K [see below]). The plasmid DNA sites are essential components of the origin of replication since they are required to organize a functional replisome (61, 62, 282). The presence of DNA sites for the binding of structural factors, found at the origin of replication of several plasmids (see below), resembles the situation found in oriC (317).
(ii) Iteron-containing origins.
In many cases, the origin of replication contains directly repeated sequences, termed iterons, which are the binding sites for the plasmid-encoded Rep proteins and which have control properties. As discussed below, iterons not only are essential for replication but also are key elements for the control of plasmid replication (reviewed in references 51, 87, 155, and 223). Among plasmids which restrict their establishment to a single or a few species of enterobacteria, iterons have been described for several replicons like P1 (5), F (209, 295), pSC101 (52), R6K (97, 98, 277, 278), Rts1 (144), and pColIV-K30 (247). Iterons are also found in theta-replicating broad-host-range plasmids such as RK2/RP4 (241, 279), pCU1 (164) and pSa (286), as well as in conditional broad-host-range plasmids such as pPS10 (85, 104, 215). It should be noted that the presence of directly repeated sequences to which Rep proteins bind is not restricted to plasmids replicating by the theta mechanism, since these sequences have been reported for plasmids using the strand-displacement mechanism or the RC mechanism (171, 176, 267). Iterons can also be found outside the origin region in some plasmids (P1, F, RK2, R6K, Rts1, and pColIV-K30). These iterons, unlike the origin iterons, are not required for initiation but play an important role in the control of replication, as the origin iterons do. In plasmids that do not have auxiliary iterons, the origin iterons are the only locus involved in control (see below).
Iterons can be adjacent or separated by intervening sequences. Iterons found in the origin region tend to be arranged as tandem repeats situated at a distance that is, in general, a multiple of 11 bp, i.e., close to the helical periodicity of the DNA double helix. This implies that the Rep-iteron nucleoprotein complexes roughly place the Rep molecules aligned on the same face of the DNA. In general, for a particular origin, the sequences of the different iterons are not identical, although they adjust to a consensus motif that defines the essential features of these sequences. However, the four 22-bp iterons of plasmid pPS10 are identical (215). Statistical analysis of the frequency of base changes within the iterons of plasmid P1, combined with the available footprinting data, have been performed (242). Three highly conserved sequence patches are found within the iterons of this replicon. The two outer patches are separated by one helix turn. Protection experiments indicated that the major groove sides of those patches are contacted by the RepA initiator protein of P1. The function of the middle patch is less clear, but it may contribute to a proper conformation of the RepA-binding site. It is remarkable that this pattern resembles the DNA-binding patterns of dimeric proteins, some of which are transcriptional repressors. Taking into account that some of these iterons are contacted by monomeric forms of the initiator proteins, this may reflect the presence of two DNA-binding domains in RepA (discussed in reference 51), a feature that may be extended to other plasmid-encoded Rep proteins (100). Alignment of iterons present in the origin of replication of different plasmids showed the conservation of the hexanucleotide TCAGPuG (86), which is directly involved in the binding of the π initiator protein to the ori-γ region of plasmid R6K (97, 98).
Multiple iterons are required for origin activity, although not all iterons present in a given origin have to be essential. For instance, removal of one of the seven iterons from ori-γ of plasmid R6K has no effect but deletion of two reduces the efficiency of replication and deletion of three or more abolishes plasmid replication (160). Interestingly, the deletions make ori-γ replication independent of DnaA (16a). In the case of P1, all five iterons seem to be required for replication in vivo, but deletion of one can be tolerated in an in vitro replication system (314).
Single iterons are present in the ori-α and ori-β origins of plasmid R6K and in the minimal origins of plasmids ColE2 and ColE3. In R6K, ori-α and ori-β contain just one iteron and half an iteron, respectively (87). This situation is compensated for by the presence of a cis-acting sequence (enhancer), which is located in a third origin (ori-γ) that contains seven iterons. The enhancer facilitates the transfer of the initiator π protein, assembled at the seven iterons of ori-γ, to ori-α and ori-β, and leads to initiation of DNA replication (see below). The smallest of all the prokaryotic origins described so far have been found in the ColE2 and ColE3 replicons (322). They consist of a stretch of 47 bp (ColE2) or 33 bp (ColE3) and contain two major directly repeated sequences.
(iii) Other origin configurations.
Origins of replication without iterons can be found in other well studied theta-replicating plasmids like R1 and ColE1, as well as in plasmid pLS20 from B. subtilis.
(a) Plasmid R1.
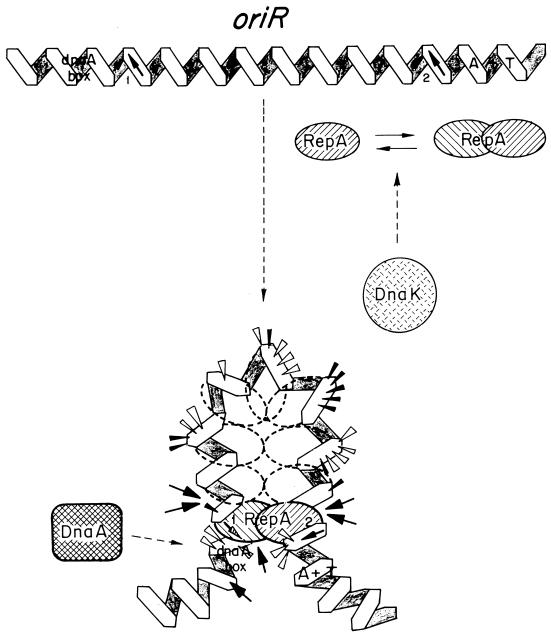
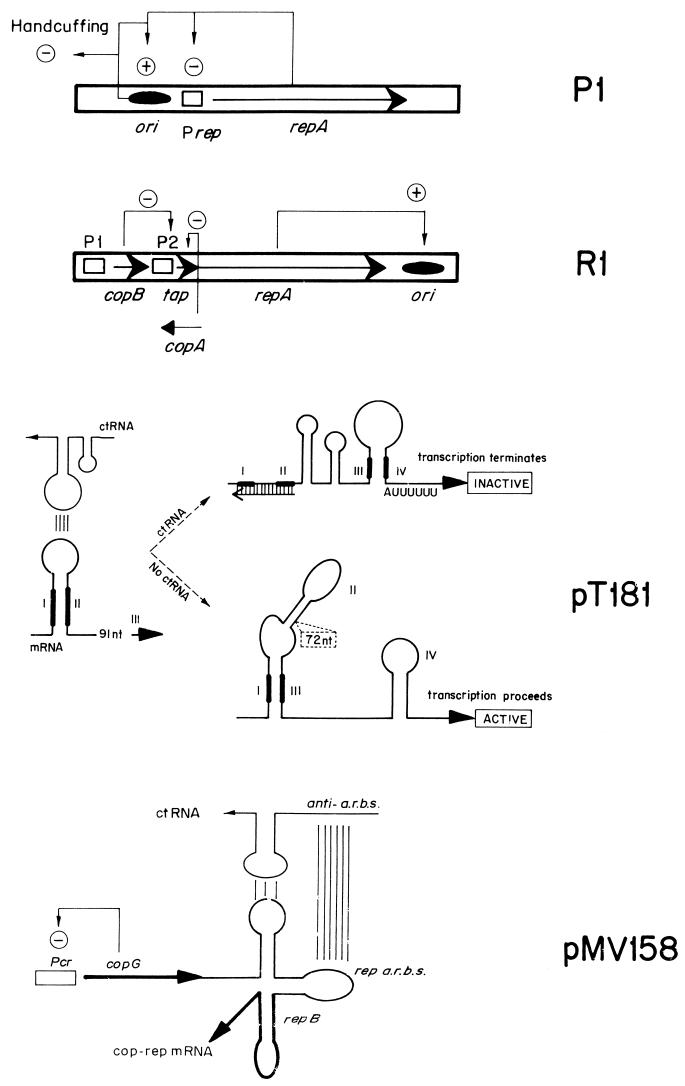
Initiation of replication of R1 is dependent on a plasmid-encoded initiator protein, RepA. The minimal region required for RepA-dependent replication (oriR) is included within a 188-bp DNA region (183) and comprises (i) a 9-bp dnaA box, (ii) a contiguous 100-bp region where RepA interacts, and (iii) an adjacent AT-rich region containing three 9-mers. A detailed study of the site of RepA interaction revealed two RepA-binding sites: a preferential RepA site, termed site 1 (5′-CAGTTAAATG-3′), which is adjacent to the dnaA box, and a related RepA binding sequence, site 2 (5′-TGTTTAAAAG-3′), for which the protein has a lower affinity. This second site is contiguous to the AT-rich region. Sites 1 and 2 share a core sequence (g/tTTAAA) that is an imperfect palindrome (101). The intervening sequence between the sites shows potential intrinsic curvature. The presence of the dnaA box optimizes the action of the DnaA protein at the origin, both in vivo and in vitro, but it is not absolutely required for the DnaA-dependent replication of R1 (233). EM of replicating intermediates obtained in vivo and in vitro shows that initiation of R1 replication occurs in a locus that is separated from the minimal origin region (78). A G-type priming signal, located 400 bp downstream of the RepA-binding sequences, has been identified as the site where initiation of the leading strand, primed by DnaG, occurs (186).
(b) Plasmid ColE1.
ColE1 is the prototype of a class of small multicopy plasmids that replicate by a theta-type mechanism. Unlike R1, ColE1 does not require a plasmid initiator protein but requires DNA Pol I to initiate replication. The origin of ColE1 replication spans a region of about 1 kb that includes (i) sequences promoting the synthesis of RNA II, the primer of the leading strand (298, 299); (ii) sequences that allow a stable hybridization of RNA II to DNA (139, 189); (iii) sequences that favor specific processing of this coupled complex by RNase H, which generates the 3′ end needed to prime leading-strand synthesis (122, 139); (iv) a primosome assembly site (pas or ssiA) that allows the loading of the DnaB helicase and DnaG primase to initiate the discontinuous priming of the lagging strand (28, 189, 220) (a dnaA box that is close to pas can be used as a DnaA-dependent DnaB-DnaC assembly site [269, 270]); and (v) a sequence for termination of lagging-strand synthesis, terH, which determines unidirectional replication (57, 198). The first two sequences are the most relevant, since they are required for ColE1 replication in the presence or absence of DNA Pol I and RNase H (57, 158, 211). The origin of ColE1 replication, defined as the transition point between RNA II primer and DNA synthesized by DNA Pol I, has been positioned 555 bp downstream of the start point of RNA II (24, 300). This transition point corresponds with data obtained in vivo for plasmid pMB1 (closely related to ColE1) (29). Analysis of replication intermediates of ColE1 by EM, located a single origin and showed that replication is unidirectional. At an early stage, leading-strand synthesis proceeds in the absence of lagging-strand synthesis (297, 298).
(c) Plasmid pLS20.
An interesting example of plasmid from gram-positive bacteria is the B. subtilis plasmid pLS20, for which a preliminary characterization has been reported (192). This plasmid replicates by the theta mechanism, and its replication is independent of DNA Pol I and of a Rep initiator protein. Several palindromes flanking a putative dnaA box are located within the origin of replication of pLS20.
Rep proteins.
Up to now, dozens of plasmids have been isolated from most bacteria, but not many of them have had their basic replicons dissected and characterized to the level of their nucleotide sequence, and even fewer replicons have been genetically and biochemically studied in detail. The classic way of classifying plasmids is to distribute them among incompatibility groups, whose members have very similar origin sequences and replication control mechanisms. However, due to the difficulty to cope with a complex experimentally based classification of each newly isolated replicon, a criterion based on sequence comparisons appears to be much more practical. Such a criterion could be the comparison of the amino acid sequences of Rep initiator proteins, since they are encoded by most of the plasmids and they share common functions. Rep proteins recognize specific sequences at the origin of replication, similar to the DnaA initiator protein in bacterial chromosomal replication, and they generate a nucleoprotein initiation complex in which essential macromolecular interactions take place (Rep-DNA, Rep-Rep, and Rep with other initiation proteins of the host) (30). In addition, many Rep proteins can generate complexes that negatively regulate their synthesis and the frequency of initiation.
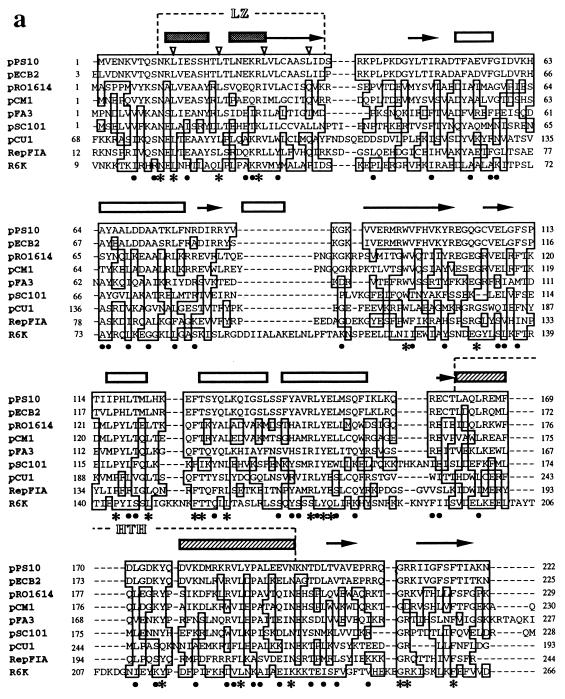
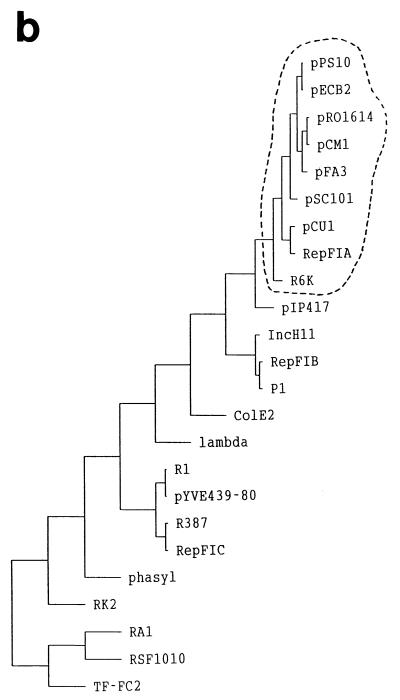
Based on amino acid sequence alignments of multiple Rep proteins from theta-replicating plasmids, it is possible to construct phylogenetic trees like the one depicted in Fig. 2b. It must be considered that for plasmid sequences, evolution can occur not only by mutation and selection but also by horizontal gene transfer. This constitutes an additional difficulty in establishing evolutionary relationships among plasmids. The phylogenetic tree groups replicons with similar replicative features: plasmids with Rep proteins binding to iterons (like pPS10, pSC101, R6K, and F) cluster apart from others whose initiators bind to nonrepeated sequences (R1 and its relatives), whereas broad-host-range plasmids (RK2, RA1, RSF10110, and TF-FC2) and replicons with dissimilar initiation mechanisms (phage lambda and phasyl) cluster in separated branches. Figure 2a shows an amino acid alignment of a large family of iteron-binding Rep initiators (encircled in Fig. 2b), comprising most of the best-characterized plasmids. The use of such alignments has allowed us the identification of Rep protein motifs, involved in protein-protein interaction (leucine zipper [LZ]) and in DNA binding (αhelix-turn-αhelix [HTH]) (93, 94, 103). A recent in vitro study performed with pPS10-RepA has revealed the existence of two globular domains, joined by a flexible linker, in a region of the protein located C-terminal to the LZ motif (102). Protein conformational changes are coupled to the dissociation of RepA dimers (which have a compact package of both domains) into monomers (with an elongated arrangement of the domains). The LZ motif and, to a lesser extent, the first globular domain mediate RepA dimerization. In the compact dimer, the second domain (including the HTH motif) binds to each arm of the operator sequence. In the elongated monomers, the second domain binds to the 3′ end of each iteron sequence whereas a DNA-binding activity in the first domain (previously cryptic) is responsible for additional recognition of the 5′ half. The sequence alignments in Fig. 2a support a similar structural organization for other Rep proteins of theta-replicating plasmids.
FIG. 2.
Sequence alignment and phylogenetic tree of Rep initiator proteins from theta-replicating plasmids. (a) Sequence alignment of the Rep initiator proteins of nine related plasmids of the iteron-containing class. Sequences were aligned with the program CLUSTAL W (version 1.5) by using, for pairwise alignment, gap opening and extension penalties of 10.0 and 0.1, respectively, and the protein weight matrix BLOSUM30. For multiple alignment, the delay for divergent sequences was set to 40% (294). The degree of sequence identity to the pPS10 initiator sequence for conserved residues is shown: ∗, identical residues in eight or nine of the total sequences; •, identical residues in five to seven sequences. Over the sequences is shown a secondary-structure prediction performed by the neural network algorithm PHD (260) on the output from CLUSTAL W: predicted α-helical regions (boxes) and β-strands (arrows). The two characteristic motifs found in the Rep initiators, LZ (hydrophobic heptad residues pointed to by open arrowheads) and HTH, are indicated. The EMBL database accession numbers are as follows: pPS10, X58896; pECB2, Y10829; pRO1614, L30112; pCM1, X86092; pFA3, M31727; pSC101, K00828; pCU1, Z11775; RepFIA, Y00547; R6K, M65025. (b) Phylogenetic tree for theta-type replicons from gram-negative bacteria, based on sequence alignments of their Rep proteins (such as the one shown in panel a for the pPS10 family, encircled in the tree with a dashed line). The sequences for 35 initiators were retrieved from databases, and a preliminary alignment was performed with CLUSTAL W (data not shown). Sequences that were virtually identical (pairwise scores, ≥90) were discarded, and a refined alignment was used as input data for the DISTANCES program of the Genetics Computer Group software package (95). Distance matrixes were corrected for multiple substitutions by the method of Kimura. The phylogenetic tree was built up according to the UPGMA method with the program GROWTREE (95). For further discussion, see the text.
(i) Protein-protein interactions: the leucine zipper-like motif.
A protein-protein interaction motif resembling the LZ is present in several plasmid-encoded Rep proteins. The LZ motif is responsible for dimerization in several eukaryotic regulatory proteins, through formation of two-stranded coiled coils (172). LZ-like motifs have been detected in the N-terminal region of the Rep proteins of several plasmids (103, 215) (Fig. 3). A mutational analysis has been carried out in the LZ-like motif of the RepA protein of plasmid pPS10 (94). Substitutions of the first two Leu residues of the LZ-like motif (d position according to a coiled-coil nomenclature) with Val resulted in a 13-fold decrease in the RepA association constant (as determined by sedimentation equilibrium analysis of maltose-binding protein–RepA fusions). This finding indicates that the LZ-like motif is a protein-protein interaction interface that regulates the equilibrium between monomers and dimers of the RepA protein. A conservative Ala→Val change in a different residue of the motif (b position) has no effect on monomer-dimer equilibrium, which points to a relevant and specific role in dimerization for the Leu residues of the motif. RepA mutants having Leu→Val substitutions were still able to interact in vitro with the iterons of the origin, indicating that the LZ-like motif is not directly involved in the binding of RepA to DNA. Further analyses indicated that RepA monomers bind to the iterons of the origin of replication whereas dimers of the protein bind to the repA promoter region, pointing to the functional relevance of the two forms of the RepA protein. Similar results have been obtained with the RepA protein of plasmid pSC101. This protein exists in a monomer-dimer equilibrium, although it is mainly in the monomeric form at the protein concentration present in cells harboring wild-type pSC101 (134). However, when the repA gene is overexpressed, replication is inhibited (133, 308). Inhibition under overexpression conditions was explained by assuming that elevated concentrations of RepA would promote its dimerization and that the RepA dimers would hinder the interaction of the active RepA monomeric forms with the iterons of the origin (134). Since overproduction of host DnaA protein can reverse inhibition by excess of RepA (133), an alternative explanation to understand inhibition by excess of initiation proteins involves titration of host replication factors.
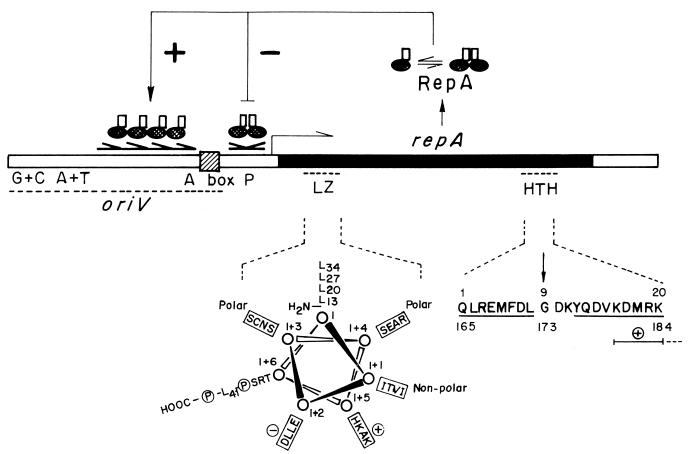
FIG. 3.
The theta-type replicon of the Pseudomonas plasmid pPS10. The iteron-containing origin (oriV) and motifs found in the replication initiator protein (RepA) are depicted. The minimal origin (oriV) of pPS10 plasmid is a good example for a “canonical” iteron-containing origin. It is composed of four contiguous and identical 22-bp iterons arranged as direct repeats (half arrows), flanked by a 9-bp dnaA box (hatched) and AT- and GC-rich sequences (215). The pPS10 replicon also contains the repA gene, encoding the RepA initiator protein (shadowed ovals). RepA is under a monomer-to-dimer equilibrium, which has consequences for protein activity: RepA dimers bind to an inverted repeat (with a sequence partially homologous to that of the iterons) that overlaps with the repA promoter (P), acting as self-repressor of repA expression, whereas RepA monomers bind to the iterons to form the initiation complex (94). Protein motifs found in RepA are depicted under the protein gene. The LZ motif, responsible for protein dimerization, is represented as a helical wheel projection, in which the hydrophobic spine of Leu residues and the chemical nature of the other displayed residues is indicated (103). For the HTH motif, involved in binding to DNA, the two proposed α-helices are underlined and a stretch of basic residues at the C end of the DNA recognition helix is indicated (+), whereas an arrow points to the Gly residue thought to start the turn (93).
A region of the pPS10-RepA protein probably involved in protein-protein interactions with host replication factors has been identified. This information was obtained from the analysis of mutations that allowed the establishment of pPS10 (originally a narrow-host-range plasmid from Pseudomonas savastanoi) in E. coli (85, 104). Three independently isolated mutations were located within the region encoding the LZ motif of RepA. They all resulted in the same Ala→Val change (I+5 position; Fig. 3). Other mutations that broaden the host range of pPS10 map in residues adjacent to the LZ-like motif (180), indicating that the RepA region responsible for this phenotype, although partially overlapping, is different from the LZ motif. Since some of the mutations broadened the pPS10 host range without altering RepA-RepA, RepA-oriV, or RepA-repA promoter interactions (94), it would appear that these changes in the pPS10 initiator should favor proper RepA interactions with host initiation proteins.
Genetic analyses revealed later that the LZ-like motifs found in other Rep proteins play a relevant role. For instance, a mutation that affects the LZ-like region of the R6K initiator protein π resulted in a protein that failed to activate the α or β origins of replication (199). Translation of the gene for the π protein of R6K, starting from an internal initiation codon, can give rise to shorter protein variants in which most of LZ is deleted. This could represent a mechanism for regulation of the level of active replication protein (87). Another example is found in the RepA protein of plasmid pSC101, in which a mutation located in the proximity of the region encoding the LZ-like motif increases the copy number of this plasmid (133).
The existence of protein-protein interfaces apart from the LZ motif in initiator proteins is supported by several lines of evidence. First, the initiator protein of plasmid R1, RepA, interacts cooperatively with DNA sequences at the origin of replication, oriR (see above). A mutation located at the 3′ end of repA results in a thermosensitive replication phenotype (232). The protein variant conserves its ability to interact specifically with oriR, but the mutation affects the cooperativity of these interactions (101). This indicates that the mutation has affected a protein-protein interface and suggests that this interface could be located in the C-terminal region of RepA. Mutations affecting RepA residues involved in binding to oriR have not been described. Second, a single-amino-acid change at the N-terminal end of the initiator protein π of plasmid R6K allows this protein to discriminate between palindromic and nonpalindromic binding sites (325). It has been proposed that the change alters a protein-protein interface which modulates interactions of π protein with differently arranged DNA target sequences. Third, RK2 is a broad-host-range plasmid, a characteristic that is related to the existence of two forms of the replication protein, TrfA-44 and TrfA-33. The larger form, TrfA-44, is required for replication in P. aeruginosa (80, 274). The shorter version, TrfA-33, starts in an internal initiation codon of trfA and promotes the establishment of RK2 in most of its hosts, including E. coli and P. putida. In addition, the origin requirements are different in the two cases (56, 142, 213, 241). A mutation at the 3′ end of the trfA gene (affecting the two versions of the protein) modifies the host range of RK2 without altering the binding of the protein to DNA (45, 175; also see reference 241). These results suggest that the C terminus of TrfA plays an important role in the interactions of the initiator protein(s) with host replication factors. Interactions of plasmid initiator proteins with host replication factors have been reported in different systems: (i) the DnaJ protein interacts with the initiation protein of plasmid P1 (312a) and with other chaperones, promoting the efficient binding of this initiator to the origin of replication; (ii) the DnaA protein requires a functional interaction with the RepA protein of plasmid R1 to enter the DnaA box present in the origin of replication (184) (this protein interaction seems to be sufficient to promote DNA replication in the absence of the DnaA box [233]); and (iii) most interestingly, the DnaA, DnaB, and DnaG proteins of the host interact with the π protein of plasmid R6K (16a, 258a) (mutations in the π protein that disrupt the interaction with the DnaA protein are defective in R6K replication [16a]; the specific regions of DnaB and π proteins involved in their interaction have been defined [258a]).
(ii) Specific binding of Rep proteins to DNA: the helix-turn-helix motif.
As mentioned above, Rep proteins are able to specifically recognize DNA sequences in the origin region. In addition to this, some of the Rep proteins autoregulate their own synthesis at the transcriptional level by binding to sequences in the rep promoter (operator) which show some degree of homology to those present in the origin region. When autoregulation exists, either a single species of the protein is involved in both regulation and replication, or different species of the protein, monomeric and dimeric, recognize the origin and the regulatory regions, respectively (discussed in reference 51). rep mutants leading to impaired Rep protein-DNA interactions have been found in various plasmids.
The Pseudomonas plasmid pPS10 contains four identical iterons in its origin of replication and an inverted repeat (IR) in the repA promoter region. The iterons and IR have partial sequence similarity (92). RepA variants that fail to repress the repA promoter had amino acid changes within or in the vicinity of an HTH motif located at the C-terminal end of the protein (93). This motif has been described in many prokaryotic DNA-binding proteins, where it is involved in binding to specific regulatory DNA regions (39, 235). The RepA proteins affected in the HTH motif failed to interact with both the repA promoter and the oriV, indicating that the motif is involved in interactions with both the DNA regions. A working model proposes that the RepA protein contacts the inverted repeats of the repA promoter region as a dimer, using the HTH motif (92, 93). This HTH motif also binds to a conserved 3′ region in the iterons, which in their 5′ ends would be bound by another region of the RepA protein (102). A similar model has been postulated for other plasmid Rep proteins, in which monomeric and dimeric forms of the protein are involved in replication and autoregulation, respectively (discussed in reference 51).
In plasmid RK2, mutations that lead to TrfA protein variants affected in binding to the origin were scattered over a trfA gene region encoding the 162-amino-acid C-terminal moiety of the protein (46). In plasmid pSC101, the last third of the RepA protein is not needed for binding to specific DNA sites (181), which contrasts with the role of the C-terminal region in other initiators.
Initiation and elongation of replication.
(i) DNA replication dependent on plasmid initiators.
Initiation of replication requires the assembly of the complete replication machinery including DNA polymerase III holoenzyme (DNA Pol III-HE), DnaB helicase, and primase at the plasmid origin. Once the checkpoint corresponding to the initiation of leading-strand synthesis is past, replication continues until completion, following a process catalyzed by DNA Pol III and other host proteins. Most of the theta-type replicons require, at least, a plasmid-encoded Rep protein and the host DnaA protein for the initiation step. The general organization of the origin region in these plasmids resembles the arrangement found in oriC (30). The plasmid ori includes not only the specific sequences where the Rep and DnaA proteins interact but also an AT-rich region containing direct repeats, analogous to the 13-mers in oriC, where the DNA strands are melted. The AT-rich repeats have also been involved in the transfer of the DnaB-DnaC complex to oriC. In the theta-replicating plasmids, the Rep protein binds to specific sequences in the origin, forming a nucleoprotein preinitiation complex analogous to the one formed by DnaA at oriC. The Rep-DNA complex, in combination with DnaA, facilitates the transfer of the DnaB-DnaC complex to the origin and the opening of the strands in the AT-rich region. The structural organization of the initiation complex could be facilitated by host factors such as HU, IHF, or FIS. The assembly of the preinitiation complex and details of the molecular interactions leading to the initiation of replication are well documented for a few theta-replicating plasmids (described below).
(a) Plasmid pSC101.
RepA, the initiator protein of plasmid pSC101, exists in a monomer-dimer equilibrium, which determines the efficiency of RepA in replication (134). Monomers and dimers of RepA are both functional, but they play different roles: monomers bind to the iterons at the origin, promoting initiation, whereas dimers bind to the adjacent inverted repeat, repressing transcription of the repA gene (181). However, interactions of the RepA dimers with the inverted repeat also play a role in replication in the absence of the par locus (197). Initiation of pSC101 replication requires, in addition to RepA (308), the DnaA host replication initiator (113), and IHF proteins (91, 283). Binding of IHF to its target, within the AT-rich region, leads to DNA bending (283), which promotes interactions between DnaA molecules bound to dnaA boxes separated by some 200 bp (282). Binding of RepA to the origin region further stabilizes DnaA contacts with the distant dnaA boxes (282). The RepA-DNA-DnaA complex plays a role in replication but also in partitioning of the plasmid (54). Stable plasmid inheritance requires the par locus, which is close to the origin region: this locus contains a site for DNA topoisomerase II and also determines the proper supercoiling at the origin region needed for initiation (53, 132).
(b) Plasmid P1.
Plasmid P1 replication is dependent, both in vivo and in vitro, on the specific initiator protein RepA (6, 313) and on the host DnaA protein (110, 313). Formation of the initiation complex requires the monomeric form of RepA (315), and RepA-DNA binding is stimulated by heat shock chaperones. The latter proteins could contribute to the dissociation of the RepA dimers into monomers, which is the form of the RepA protein that recognizes the five iterons of the origin (58, 315). However, growing evidence indicates that the chaperones are required to activate the monomers of RepA (50, 78a, 236). Binding of the activated RepA monomers to the five iterons of the origin results in wrapping of the DNA around RepA, presumably due to in-phase bending of DNA (206). RepA monomers contact each iteron through two consecutive major grooves on the same face of the DNA helix (242). RepA alone is unable to melt the origin; this role is performed or favored by DnaA, which also stimulates the DNA-binding activity of RepA (206). There is a set of two tandem dnaA boxes at one end and a set of three tandem dnaA boxes at the other end of the P1 origin. Although either of the sets, or even just one dnaA box that conforms exactly to the consensus, is sufficient to support DnaA-dependent replication (4, 36, 38), melting of the origin region by DnaA is maximally efficient when both sets are present, probably due to DNA looping mediated by DnaA bound to the two sets (206). The orientation of the dnaA boxes and the different sensitivity of the two strands to reagents specific for single-stranded DNA suggest that DnaA-dependent loading of DnaB preferentially occurs in one of the strands, which can account for the unidirectional mode of P1 replication (206). Efficient replication of P1 requires adenine methylation of the five GATC sites of the origin. These GATC sites are clustered in direct heptamer repeats which are separated from the RepA-binding site by a GC-rich spacer (1, 2, 37).
(c) Plasmid RK2.
Important information on the initial events of replication of plasmid RK2 has been obtained (161a). The ClpX chaperone yields monomers of the plasmid initiation protein, TrfA, which is the form that is active in binding to the five 17-bp iterons of the origin (161b). This binding promotes, in the presence of HU protein, local strand opening within the AT-rich region of the origin. Interactions of the DnaA protein of the host with four DnaA boxes present in this region are also required for initiation of plasmid replication. These interactions increase, but are not strictly required for, the opening of the strands. DnaA is required for the delivery of the DnaB helicase to the origin region and both DnaA and TrfA are required for DnaB-induced template unwinding. This suggests a role of TrfA in the repositioning and activation of the DnaB activity (79b). The requirement of particular DnaA boxes is host-dependent (79a). This is consistent with the plasticity of the RK2 origin with respect to structural requirements for replication in different bacterial hosts.
(d) Plasmid R6K.
As stated above, replication from the γ ori of R6K requires π protein (96, 160, 272, 278). This protein can recognize different types of DNA sequences: iterons, enhancer, inverted repeats in the promoter of the pir gene (encoding the π protein), and the AT-rich segment of the origin (174). The π initiator promotes the initiation of replication from three origins of replication: α, β, and γ (reviewed in reference 87). ori-γ is in a central position, separated by 3 and 1.2 kb, respectively, from ori-α and ori-β. ori-γ contains seven 22-bp iterons, flanked by two IHF-binding sites and two dnaA boxes. Contiguous to the iterons is an AT-rich region which contains one of the dnaA boxes and one of the IHF-binding sites (62). ori-β contains half an iteron, and ori-α one complete iteron that are essential for function. Under standard conditions, ori-α and ori-β are more active than ori-γ, but they depend on a distant enhancer for activity (146, 147). This enhancer partially overlaps ori-γ, but its activity and the origin function have been distinguished by mutational analysis.
A dimer of π seems to bind to each of the seven 22-bp iterons present in ori-γ (86). Protein π binds preferentially to one of the strands of the ori-γ (98) and bends the DNA, generating a wrapped nucleoprotein structure (205). DnaA protein is required for replication from this origin, and it can bind the two dnaA boxes that flank the iterons (146, 147). Although two IHF-binding sites are flanking the iterons of ori-γ, the preferential or unique binding site(s) is the one located within the AT-rich region (62, 145a). IHF protein binding to these sequences induces conformational changes that are important in the regulation of replication initiation (145a). This binding could also affect the interaction of the π protein with the DnaA initiation protein of the host (16a). In the presence of normal levels of π, IHF is required for replication from ori-γ (61). An active ori-γ requires the binding sites for π, DnaA, and IHF proteins in the correct geometrical alignment (147). Protein π binds efficiently to the iterons of the ori-γ but not to ori-β or ori-α. However, the enhancer favors the long-range activation of ori-β and ori-α by transfer of the initiator protein, and possibly other initiation factors, from ori-γ (199, 203, 204). The activation of ori-β, unlike ori-γ and ori-α, does not require DnaA protein (147). Three new R6K gene products that distort essential sequences of ori-α and ori-β have been described (89). However, these proteins have been identified as proteins needed for conjugative transfer rather than for plasmid replication (230a).
(e) Plasmid R1.
Plasmid R1 is the most extensively studied member of the IncFII family of plasmids. In vivo and in vitro replication of R1 requires the initiator protein, RepA (77, 159, 183, 305). oriR, defined as the minimal region required for RepA-dependent replication of R1 in vitro, is bound specifically by RepA (101, 183, 184). Unlike the above cases, oriR does not contain iterons. RepA protein, probably as a dimer, recognizes sequentially (albeit with different affinities) the cores of two partially palindromic sequences (Fig. 4) (101). These sequences are located on the same face of the DNA helix, within a region of potential curvature, and are 8 helical turns apart (79). Interactions between RepA molecules bound to each of the sites could be responsible for DNA looping at the ends of a 100-bp region within oriR that is protected by RepA against DNase I cleavage (101). Following formation of the initial complex, additional RepA molecules could bind to the intermediate region by cooperative protein-protein interactions, generating a high-order complex. These interactions are needed for replication, as indicated by the defective replication phenotype associated with a repA mutant that failed to generate high-order RepA-oriR complexes (101, 232). The RepA-DNA complex seems to melt the DNA strands in the AT-rich region (185). In vitro replication of R1 requires DnaA protein (184, 231). DnaA binds to a dnaA box that is adjacent to the RepA-binding region, but this binding does not occur, or is very inefficient, in the absence of RepA (184, 233). It is likely that RepA-DnaA contacts guide the entrance of the DnaA protein in oriR, because the dnaA box is not absolutely necessary for the DnaA-dependent replication of R1 (233). Surprisingly, although in vitro replication of R1 is dependent on DnaA, this protein is dispensable for the replication of IncFII plasmids in vivo (20, 290). In vivo replication in the absence of DnaA is inefficient, but plasmid copy mutants that increase the levels of RepA protein improve the efficiency of replication (20). These results show the essential role of RepA in origin activation and imply that RepA could promote melting of the DNA strands at the origin and loading of the DnaB-DnaC complexes.
FIG. 4.
RepA-oriR complexes in initiation of R1 plasmid replication. A 100-bp region in the oriR replication origin is continuously bound by the plasmid RepA protein to form the initiation complex. There are no iterons in oriR, but two partially palindromic 10-bp sequences (sites 1 and 2) are found at the ends of that 100-bp region. They are flanked, respectively, by a consensus dnaA box and by three AT-rich sequences. These three sequences are believed to be melted to allow the DnaBC complex access to the open origin. A RepA initiator (hatched oval) dimer binds with high affinity to site 1, and then, in a second binding event, a different RepA dimer would bind with lower affinity to the distal site 2 sequence. The DNA of the oriR region could be bent to facilitate the topological proximity of sites 1 and 2, which are disposed on the same face of DNA double helix. Binding of RepA dimers to sites 1 and 2 would generate a small DNA loop, held together by protein-protein interactions. The DNA loop would be filled afterwards with more RepA molecules that are brought to the complex mainly by protein-protein interactions. This model is based on experimental gel mobility shift assays and footprinting data with both wild-type and mutant RepA proteins and oriR sequences (101). Arrowheads indicate DNase I-hypersensitive sites (the size is proportional to the intensity of cleavage), whereas arrows point to strong cleavage sites for hydroxyl radicals. The interaction of DnaA host initiator with its DNA-binding site is dependent on the previous formation of the RepA-oriR nucleoprotein complex (233). A requirement for a DnaK chaperone has been described for R1 DNA replication (105). A hypothetical role for DnaK in modulating the aggregation and activation state of RepA dimers, inspired by the findings for P1 plasmid RepA initiator (316), is also shown.
Interactions of RepA protein with the oriR region promote, both in vitro and in vivo, initiation of leading-strand synthesis at a DnaG-priming site (the G site, resembling the bacteriophage G4 origin for complementary strand synthesis) (21, 186) that is located 400 bp downstream of oriR. It has been proposed that the G site is activated when synthesis of the lagging strand, which initiates at the AT-rich region, reaches the G4-like site. Initiation at the G site, promoted by DnaG, cannot progress toward the origin. This leaves a gap that is filled later in the replication cycle (186). The relevant role played by DnaG in initiation of R1 replication is consistent with the complete inhibition of the in vitro replication of the plasmid by anti-DnaG antibodies (231). Antibodies against the E. coli single-stranded DNA-binding protein (SSB) also block in vitro R1 replication (231), indicating the essential role of this protein at an early stage in R1 replication. Finally, it has been established, both in vivo and in vitro, that replication of R1 requires DnaK protein (105). RepA protein of R1 seems to interact initially with the two partially palindromic sequences at the oriR region as a dimer, but further binding to the intermediate region could be by monomers, since there are no symmetrical sequences (102). This could support the notion that DnaK plays a role in the activation of R1 replication similar to that proposed for P1 replication (58, 315).
(f) Plasmids ColE2 and ColE3.
As mentioned above, the smallest of all the prokaryotic origins described so far have been defined in the ColE2 and ColE3 replicons (322). These plasmids require for replication a plasmid-specific initiator, but, like ColE1 (see below), they also require DNA Pol I to initiate leading-strand synthesis. Initiation of ColE2 and ColE3 replication is dependent on the synthesis of specific primer by their Rep proteins (288). A single-strand initiation site (ssi) for the priming of DNA replication has been located near the ori of ColE2 (219).
(g) Plasmids of the pAMβ1 family.
Replication of the pAMβ1/pIP501 broad-host-range plasmid family from gram-positive bacteria (32, 41, 42) requires, like ColE2 and ColE3, DNA Pol I and Rep protein. A model based on the synthesis of a primer RNA catalyzed by the host RNA polymerase (RNAP), the specific cleavage of the primer at the origin region (perhaps mediated by the Rep protein), and the extension of the 3′ end catalyzed by DNA Pol I has been proposed (41).
(ii) Replication independent of plasmid-encoded initiator proteins.
The best-characterized replicon that is independent of plasmid-encoded initiator proteins is ColE1. Initiation of ColE1 replication involves the consecutive activities of RNAP, RNase H, DNA Pol I, and DNA Pol III-HE (reviewed in references 163, 182, 248, 280, and 301). Transcription mediated by the host RNAP is required to synthesize the preprimer for leading-strand synthesis. Specific cleavage by RNase H of the preprimer (termed RNA II) annealed to DNA provides a 3′ end that defines the starting point for leading-strand synthesis. This synthesis is initially carried out by DNA Pol I. Steric hindrance of the bulky DNA Pol III-HE by the folded RNA II (upstream of the RNA-to-DNA transition point) probably prevents extension of the primer by this polymerase (188). DNA Pol I synthesizes about 400 nucleotides of the leading strand, exposing, on the displaced strand, a primosome assembly site (pas). Once the primosome is assembled at the pas site, it translocates in the 5′→3′ direction, unwinding the helix and priming the discontinuous DNA synthesis. At this point, DNA Pol I is replaced by the highly processive DNA Pol III-HE. The switch between DNA Pol I and DNA Pol III-HE could be favored by helix-destabilizing proteins bound to the template of the leading strand, which has to be exposed by the DnaB helicase (discussed in reference 280). Leading-strand synthesis can occur uncoupled from lagging-strand synthesis, and DnaG, but not DnaB, is dispensable for this uncoupled synthesis (281). Lagging-strand synthesis, initiated at the pas site, extends toward the promoter of RNA II but is arrested at a site (terH) 17 bp upstream of the leading-strand initiation site (57). The mechanism of this arrest is unknown. These events determine the unidirectional pattern of ColE1 replication.
Termination of replication.
The points at which theta-type replication terminates can be actively determined by molecular interactions at particular sequences. The first replication arrest sequence, ter, was identified in plasmid R6K as a barrier to the unidirectional replication initiated in either ori-α or ori-β of this plasmid. Replication starts then from the initial origin in the opposite direction and progresses to completion (177). The R6K terminus acts as a temporal barrier to replication initiated in other replicons (160). The nucleotide sequence of the ter region was determined, and the replication terminus of R6K was cloned (17, 18). The organization of this sequence as two separable and polar terminus sites was recognized and verified (130). The recognition of the essential features of the ter site allowed the identification of similar sites in plasmids of the IncFII (R1 and R100) and IncFI (repFIC) groups, as well as in the chromosome of E. coli. The ter sequence is the binding site of Tus, a monomeric protein of E. coli that promotes the termination of plasmid replication (121, 275). The identification of ter homologous sites in the chromosome of E. coli triggered replication termination studies in this bacteria and also in B. subtilis, where DNA-arresting sequences (IR-I and IR-II) and a dimeric protein that promotes the termination of DNA replication, RTP, have been identified (reviewed in references 14, 19, and 120). Unlike Tus, which acts as a monomer, RTP acts as a tetramer of two dimers (261, 262).
A major step in understanding active termination of DNA replication was the determination of the three-dimensional structure of the RTP dimer at the atomic level by crystallographic methods (43). This determination allowed the identification of protein-protein interfaces in RTP and opened the way to comparative structural analyses which suggested that RTP folding is similar to the “winged-helix” domain found in a family of DNA-binding proteins (43, 285). The polar arrest of the replication fork caused by RTP protein has been proposed to be due to specific interactions between the terminator protein and the DnaB helicase (116, 151, 172a, 261). Like the B. subtilis RTP-IR complex, the E. coli Tus-ter complex interferes with the helicase activity of the replisome complex in an orientation-dependent manner (151). Following the determination of the RTP structure, the structure of the Tus-ter complex was also solved by X-ray crystallography (143). This structure provided information on the singular architecture of the Tus protein and on the Tus-ter protein-DNA interactions involved. In addition, these studies support the speculation that the polar arrest of the replication fork, occurring at the Tus-ter complex, could be due to the polar inaccessibility of the helicase to the protein DNA-binding site. The termination of DNA replication is a regulated process, as indicated by the identification of a small protein of E. coli that binds to a terminator site and prevents replication fork arrest mediated by the Tus protein (214).
Sequences arresting lagging-strand synthesis, called terH, have been found upstream of and close to the pas site of ColE1; the arrest seems to be caused by the nonhybridized portion of RNA II (57, 212). It has been found that in multimers containing ColE1-type replicons oriented head-to-head, one origin of replication acts as a polar pausing site for replication initiated in the other origin (307). It is possible that this pausing is due to the stalling of the replication fork by the unhybridized portion of RNA II. Alternatively, the replisome could be transiently stalled at a protein-DNA complex, such as the pas site (discussed in reference 307). The potential role of an initiator protein to arrest replication progressing toward the origin has been reported in plasmid R1 (168). Active stalling of replication forks could be important for determining the direction of replication and for accurate termination and may modulate the efficiency of replication or the coupling between replication and cell division.
During the final stages of plasmid replication, catenates containing gaps in both daughter strands can be originated (212). These catenates can be resolved by either type I or type II topoisomerases. Genetic analysis revealed the involvement of a specific type II topoisomerase, Topo IV, in the segregation of plasmids and bacterial nucleoids (145). A two-stage model for the segregation of the replication products has been proposed (8, 9), with DNA gyrase reducing the linking number during elongation of DNA synthesis (stage I) and Topo IV resolving the supercoiled catenates which are the products of replication (stage II). Although cross-activities of DNA gyrase in decatenation and of Topo IV in supporting fork progression can be detected, in vivo and in vitro data confirm the specialized role of Topo IV in unlinking daughter replicons (117, 245, 246, 327). Maturation of the open-circular forms arising from the decatenation by Topo IV into supercoiled molecules can be efficiently carried out by DNA gyrase. Data obtained with plasmid R1 indicate that maturation of newly replicated DNA molecules is a slow process, which prevents rapid reutilization of the last replicated molecules (221). As the result of an odd number of homologous recombination events, dimers can arise. Replication intermediates can provide ideal substrates for these homologous recombination events. The resolution of DNA dimers by specialized systems can also be considered part of the replication termination process (discussed in reference 19).
Synopsis.
Replication by the theta-type mechanism is widespread among plasmids from gram-negative bacteria and has also been reported in plasmids from gram-positive bacteria. EM shows that replicating intermediates appear as bubbles (early stages) that, when they increase in size, result in theta-shaped molecules. Two early events in this mode of replication are the opening of the strands at specific sequences (the origin of replication) and the synthesis of RNA primers. Opening of the strands is catalyzed by specific initiators (Rep and DnaA proteins) and/or by transcription by RNAP. Initiation proteins promote, at the origin of replication, the sequential assembly of components of the replisome complex. The main replicative helicase of the cell catalyzes further unwinding of the strands. RNA primers are synthesized either by RNAP or by bacterial or plasmid primases. DNA synthesis of both strands is coupled and occurs continuously on one of them (leading strand) and discontinuously on the other (lagging strand). DNA Pol III is required for elongation of plasmid DNA replication. In addition, DNA Pol I can participate in the early synthesis of the leading strand (ColE1 and pAMβ1). Theta-type replication is, in most cases, unidirectional. Topoisomers are originated at termination (right-handed catenates), and their resolution requires the participation of Topo IV. Termination of DNA replication is determined in some plasmids by specific protein-DNA complexes.
Strand Displacement Replication
The best-known examples of plasmids replicating by the strand displacement mechanism are the promiscuous plasmids of the IncQ family, whose prototype is RSF1010. Members of this family require three plasmid-encoded proteins for initiation of DNA replication. These proteins promote initiation at a complex origin region, and replication then proceeds in either direction by a strand displacement mechanism (266; reviewed in reference 263).
Origins of replication.
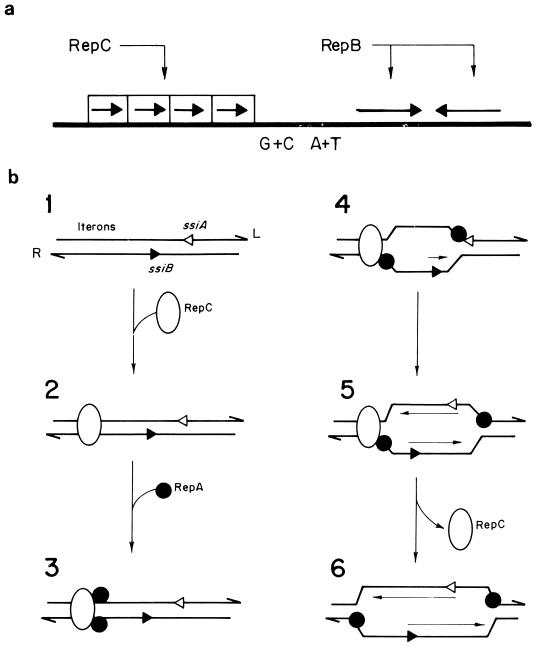
The origin of replication of plasmid RSF1010 has been defined as the minimal region able to support bidirectional replication when the RSF1010 replication proteins (RepA, RepB, and RepC) are supplied in trans by a second plasmid (266). This region also contains the origin of replication, as defined by EM analysis of replication intermediates obtained in vivo (59) and in vitro (266). The minimal ori region includes three identical 20-bp iterons plus a 174-bp region that contains a GC-rich stretch (28 bp) and an AT-rich segment (31 bp). The origin extends further with a nonessential region and two small palindromic sequences containing the ssiA and ssiB sites located in opposite strands (Fig. 5a). Iterons are the RepC-binding sites (111, 112). The inverted repeats at the ssi sites could favor the formation of hairpins. In these hairpins, base complementarity in the upper part of the putative stem is essential for replication, while base complementarity and sequence specificity in the lower part of the stem are important for primer synthesis (195). The ssiA and ssiB sequences are specifically recognized by the plasmid-encoded RepB primase, which primes continuous replication from these sequences (111, 128, 129). Genetic analysis indicated that a single ssi, in an orientation that favors priming and chain elongation away from the iterons, is sufficient for RSF1010 replication (118). This organization suggests that the origin of replication of RSF1010 can be separated into three functional loci: the iterons, the ssiA region and the ssiB region. The iterons and the adjacent AT-rich region function as a duplex-opening region, and the ssiA and ssiB sites form a priming region (263).
FIG. 5.
Replication of plasmid RSF1010 by the strand displacement mechanism. (a) Origin of replication and related regions. The interaction sites of RepB (inverted repeat [convergent arrows]) and of RepC (iterons [boxed arrows]) are indicated. GC- and AT-rich regions are also depicted. (b) Model for initiation of replication by the strand displacement mechanism in plasmid RSF1010 (266). Replication occurs with opposite polarities from two origins (ssiA and ssiB), which are independently used. Interactions between the plasmid-encoded proteins RepC and RepA are indicated. Priming is catalyzed by RepB′ (not shown). Thin lines indicate newly synthesized DNA, with the direction of synthesis indicated by arrowheads. See the text for details.
Rep proteins.
As indicated above, replication of RSF1010 is promoted by the joint activity of three plasmid-encoded proteins, RepA, RepB, and RepC, that have, respectively, 5′→3′ helicase, primase, and initiator activity (111, 263). The RepC protein, a dimer of 31-kDa subunits, interacts specifically with the iterons of the origin (111, 112) and probably with the RepA helicase, promoting the exposure of the ssi sites in a single-stranded DNA (ssDNA) configuration (129, 266, 289). The RepA protein is a hexamer of 30-kDa subunits, and it contains two activities: an ssDNA-dependent ATPase and a 5′→3′ DNA helicase. Expression of repB from two in-frame alternative start codons results in two polypeptides of 36 and 38 kDa, which correspond to two functional forms of the RepB primase: RepB and RepB′ (111, 267). The 38-kDa RepB′ was shown to be identical to the RSF1010-encoded MobA protein (involved in conjugative mobilization) (266).
Replication mechanism.
Replication of RSF1010 DNA is independent of the host-encoded DnaA, DnaB, DnaC, and DnaG proteins, whose roles are played by the combined action of the plasmid-encoded RepA, RepB, and RepC proteins (90, 111, 265). The template for initiation of RSF1010 replication is supercoiled plasmid DNA (78, 266). DNA Pol III-HE and SSB are required for replication. Figure 5b outlines a model for initiation of RSF1010 replication, proposed by Scherzinger et al. (266). The first stage of this process involves the binding of the RepC protein to the iterons of the origin. It is assumed that the RepA helicase binds to both DNA strands in the AT-rich region, close to the site of interaction of RepC. Subsequent translocation in the 5′→3′ direction of the RepA helicase bound to the L strand (the DNA strand which has the same sequence as the mRNAs coding for 10 of the 11 known RSF1010 proteins) (267) melts the duplex, exposing and activating the ssi sites. Alternatively, the interaction of RepC with the iterons could induce the opening of the duplex near the ssi sites. The exposure of the stem-loop structure in the ssi sites is probably required for the assembly of the RepB-primase to initiate replication (195). Initiation at either ssi site can occur independently, and replication proceeds continuously, with the RepA helicase facilitating displacement of the nonreplicated parental strand as a D loop. Continuous replication from each ssi signal in opposite directions would originate a double-stranded DNA theta-shaped structure in the overlapping region and two D loops beyond this region. The helicase activity of the RepA protein is required during the elongation of RSF1010 replication, and this protein cannot be replaced by the host DnaB helicase. The RepA helicase of RSF1010 works in the 5′→3′ direction, which implies that it is working while bound to the displaced strand. The end products of the strand-displacement replication mechanism are ss-displaced circles and double-stranded supercoiled circles. The ssDNA molecules could correspond to either DNA strand and therefore could contain either the ssiA or ssiB sequences. These sequences are used to initiate synthesis of the complementary strand, which converts the ssDNA templates into double-stranded supercoiled circles. Therefore, double-stranded DNA (dsDNA) molecules, displaced single-stranded circular molecules, and partial double-stranded circles can be formed in this mode of replication.
Synopsis.
IncQ plasmids (typically RSF1010) are replicons that can be propagated in many different hosts. Replication of RSF1010 occurs from two symmetrical and adjacent single-stranded origins (ssiA and ssiB) positioned one on each DNA strand. Replication starts when these origins are exposed as single-stranded regions. The melting of the DNA strand is dependent on two plasmid replication proteins, RepC and RepA, and is facilitated by an AT-rich region that precedes the ssiA and ssiB regions. RepC recognizes directly repeated sequences of the origin adjacent to the AT-rich region, and RepA is a DNA helicase. Priming of DNA synthesis at these origins is catalyzed by the plasmid-specific primase (RepB). Synthesis of each one of the strands occurs continuously and results in the displacement of the complementary strand. Replication of this displaced strand is initiated at the exposed ssi origin. Due to the activities of the three plasmid replication proteins (RepA, RepB, and RepC), initiation of RSF1010 replication is independent of transcription by host RNAP and of host replication factors acting at the early replication stages (DnaA, DnaB, DnaC, and DnaG). This independence may account for the broad-host-range character of the IncQ replicons.
Rolling-Circle Replication
Replication by the RC mechanism has to be unidirectional, and it is considered to be an asymmetric process because synthesis of the leading strand and synthesis of the lagging strand are uncoupled (reviewed in references 69, 84, 108, 150, 150a, and 228). One of the most relevant features of RC replication is that the newly synthesized leading plus strand remains covalently bound to the same parental plus strand. RC replication was originally thought to be limited to ssDNA coliphages and to small multicopy plasmids isolated from gram-positive bacteria. However, there are known instances of plasmids isolated from gram-negative bacteria, from cyanobacteria, and from species of Archaea that use the RC mode for replication (83, 99, 127, 156, 324, 331). Although most of the RC-replicating plasmids so far described are smaller than 10 kb, all small plasmids do not necessarily replicate by the RC mode. For example, small plasmids like pRJF1 (2.6 kb) and pWV02 (3.8 kb), isolated from gram-positive bacteria, replicate by the theta mode (115, 152). Studies on the molecular mechanisms underlying RC replication have been done mainly with the staphylococcal plasmids pT181 (228), pC221 (292), pUB110, and pC194 (108) and with the streptococcal plasmid pMV158 and its Δmob derivative pLS1 (69).
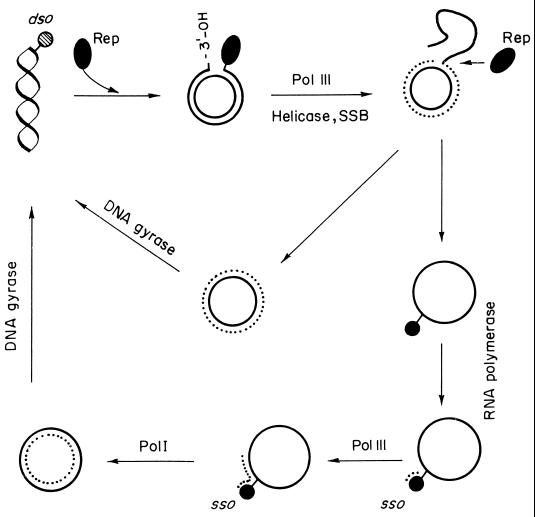
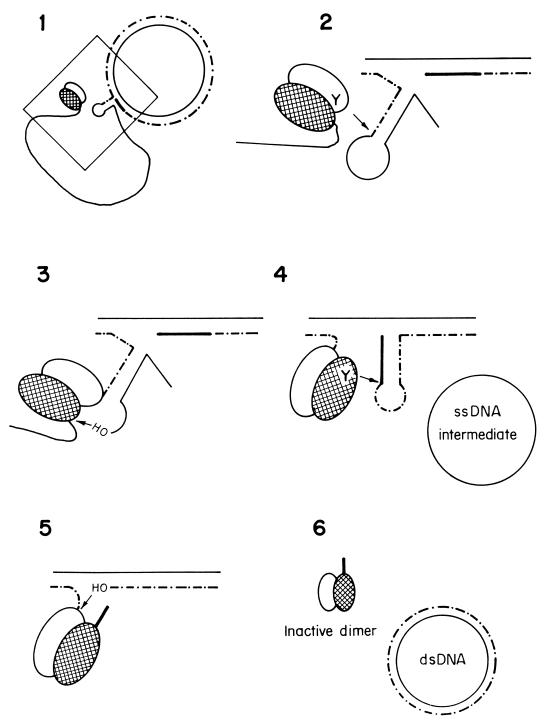
The current model for RC replication involves several experimentally distinguishable stages (Fig. 6). Replication is initiated by the plasmid-encoded Rep protein, which introduces a site-specific nick on the plus strand, at a region termed double-stranded origin (dso). The nick leaves a 3′-OH end that is used as a primer for leading-strand synthesis, which most probably involves host replication proteins (at least DNA Pol III, SSB, and a helicase). Elongation from the 3′-OH end, accompanied by the displacement of the parental plus strand, continues until the replisome reaches the reconstituted dso, where a DNA strand transfer reaction(s) takes place to terminate leading-strand replication (see below). Thus, the end products of leading-strand replication are a dsDNA molecule constituted by the parental minus strand and the newly synthesized plus strand, and a ssDNA intermediate which corresponds to the parental plus strand. Unlike replication by the strand displacement mechanism, the ssDNA intermediates generated by the RC replication mode correspond to only one of the plasmid DNA strands. The pioneering work in Ehrlich’s laboratory showed that generation of ssDNA is the hallmark of plasmids replicating by the RC mechanism (108, 291). Finally, the parental plus strand is converted into dsDNA forms by host proteins initiating at the single-strand origin (sso), which is physically distant from the dso. The last step would be the supercoiling of the replication products by the host DNA gyrase.
FIG. 6.
Model for RC replication. The plasmid-encoded Rep protein recognizes the dso on supercoiled DNA and introduces a site-specific nick generating a free 3′-OH end. This end is elongated by host proteins as the parental strand is being displaced. When the replication fork reaches the reconstituted dso, Rep protein catalyzes a strand transfer reaction, releasing an ssDNA intermediate and a dsDNA molecule with a parental and a newly synthesized (dotted) strand. Lagging-strand synthesis on the ssDNA molecule is initiated at the sso signal by the host RNA polymerase. This enzyme would synthesize a short primer RNA, and lagging-strand synthesis is performed by host DNA polymerases. The end products are supercoiled plasmid DNA molecules.
Origins of leading-strand synthesis.
RC-replicating plasmids are made up of interchangeable gene modules (253). There is only one essential module. This harbors the functions for plasmid replication and includes the dso, the rep gene, and the plasmid elements involved in replication control. This module has been named the leading-strand initiation and control (LIC) region (69). In addition, plasmids may include an antibiotic resistance determinant, a gene involved in conjugative mobilization (mob), and one or two sso regions. Based on the homologies observed in the essential LIC module, four main plasmid families have been defined, their prototypes being pT181, pC194, pMV158, and the staphylococcal plasmid pSN2 (69, 84, 108, 228). Little information is available on the fourth plasmid family, and newly described plasmids from Pyrococcus abyssi and from extreme halophiles seem to belong to one of these families or to fall within a different, less well characterized, fifth family (83, 127). An interesting replicon is the natural phage-plasmid hybrid replicon, termed phasyl, which has been isolated from E. coli. It contains two functional origins: one has homology to the viral and complementary origins of the ssDNA coliphage M13, whereas the other origin requires activation by the phasyl-encoded Arp protein. Iterons or dnaA boxes typical of the theta-type replicons have not been found in phasyl (271).
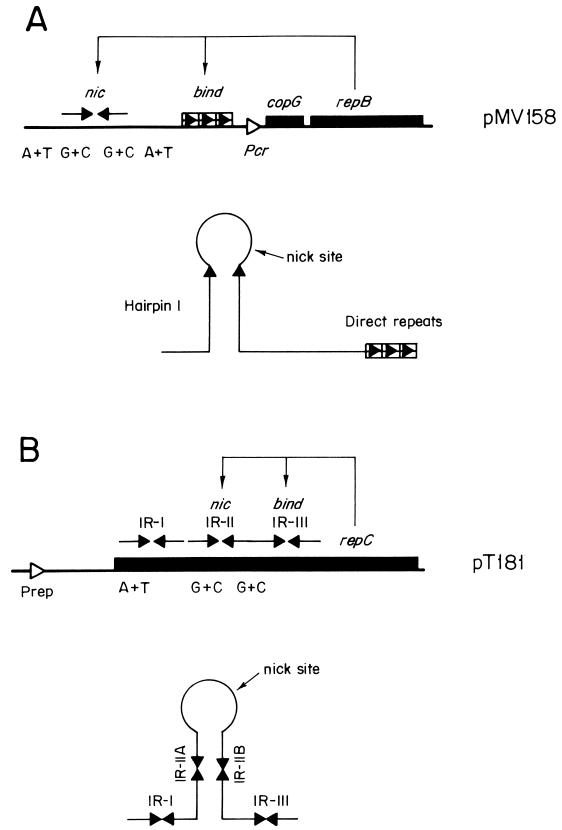
Two loci have been defined within the dso, namely, the bind and nic regions (70). The former locus includes the sequences needed for the Rep protein to bind to the plasmid DNA, whereas the latter contains the site where the Rep protein introduces the initial nick (Fig. 7). The two loci can be contiguous (pT181 family) or can be separated by up to about 100 bp (pMV158 family). A typical feature of the RC-replicating plasmids is that the nic regions are highly conserved among replicons of the same family whereas differences are found at the bind loci. This fact shows that Rep proteins of plasmids from the same family must have a common catalytic domain for phosphodiester bond cleavage and sealing whereas replicon specificity (i.e., Rep binding to the bind region) would be located in a different, nonconserved domain (see below). The DNA sequences of the bind regions are either an inverted repeat contiguous to the nick site (IR-III in the pT181 and pC194 families [Fig. 7]) or a set of two or three direct repeats separated from the nick site by intervening sequences (pMV158 family [Fig. 7]). The essential regions of the pT181-dso (IR-II and IR-III) involved in the interaction with the plasmid RepC initiator protein have been defined in a systematic study (312). IR-II (nic region) contains the RepC nick site, and the IR-III (bind region) is contiguous to it (Fig. 7 and 8). Whereas IR-II is conserved among plasmids of the pT181 family, IR-III is not, suggesting that the origin specificity is provided by IR-III. The proximal half of IR-III is more important for RepC recognition of the dso than is the distal half. In addition, the spacing and phasing between IR-II and IR-III are important for dso function (312). A similar picture is found in plasmids of the pC194 family. Conservation of the nic locus and divergence in the bind region can also be observed in the plasmids of the pMV158 family. In these plasmids, the distance between the conserved nick site and the direct repeats (the nonconserved bind locus) ranges between 14 and 95 nucleotides. The RepB protein of pMV158 binds in vitro to a dsDNA fragment containing the direct repeats (60). However, unlike the plasmids with iteron-containing origins described above, the pMV158 direct repeats do not constitute an incompatibility determinant toward pMV158 (70). These direct repeats seem to be essential for plasmid replication in vivo but not for in vitro relaxation of supercoiled DNA mediated by the plasmid Rep protein (201).
FIG. 7.
Origins of replication and related regions of plasmids replicating by the RC mode, as exemplified by plasmids pMV158 (A) and pT181 (B). The bind and nic regions of the dso are indicated. Other symbols are as in Fig. 1.
FIG. 8.
Model for termination of RC replication based on results from Novick’s laboratory (140, 141, 255–257). Nucleophilic attacks exerted by the OH groups of the Tyr residue of Rep (Y) or by 3′-OH groups of the DNA (OH) are indicated by arrows. Solid lines, parental DNA; broken lines, newly synthesized DNA. The thick solid line indicates the nucleotides that are newly synthesized past the reconstituted dso and that will remain covalently bound to the Rep protein to generate a Rep/Rep* inactive dimer. The two subunits of the RepC dimer are differently depicted.
The nic region contains inverted repeats able to generate one or two hairpin structures (65, 216, 254). Three different types of nic regions, belonging to (i) the pT181 family, similar to that of M13, (ii) the pC194 family, exhibiting similarities to φX174, and (iii) the pMV158 family, with no relevant homologies to nic regions from known ssDNA coliphages, have been reported (Table 1) (69, 108, 228). The development of a system in which DNA strand discontinuities can be mapped with nucleotide resolution in vivo has allowed the identification of the nick sites of other plasmids of the pMV158 family, namely, pE194, and pFX2 (106, 328). The DNA sequences recognized by the Rep proteins to introduce the nick would be located on unpaired regions within these hairpins (IR-II in pT181; hairpin I in pMV158 [Fig. 7]), which accounts for the absolute requirement of supercoiled DNA as the substrate for replication (64, 201, 216). Genetic analysis pointed to the existence of a stem-loop structure within the nic region of pC194 (196). However, for the related plasmid pUB110, no transient hairpins seem to be required to initiate replication (10). In vitro analyses have shown that IR-II of plasmid pC221 (closely related to pT181) or hairpin I of pMV158 are sufficient for the nicking-closing activity of their respective Rep proteins. This reaction is not plasmid specific, since cognate nic regions are also cross-recognized by these Rep proteins (201, 292, 332). In addition, in vitro replication of plasmid pC221 is reduced if a competing pT181-dso is present (332). In spite of such an in vitro recognition and extensive homologies of the Rep proteins and the dso of pT181 and pC221, there is no cross-reactivity between the Rep proteins and the dso of these plasmids in vivo, unless the Rep proteins are overexpressed (135).
TABLE 1.
nic regions in RC replicons
| Plasmid or phage | Sequence of nic regiona |
|---|---|
| pT181 | AAAACCGGCTACTCT/AATAGCCGGTT |
| pC221 | AAAACCGGCTACTCT/AATAGCCGGTT |
| pS194 | AAAACCGGaTACTCT/AATAGCCGGTT |
| fd | gAgtCCacgTtCTtT/AATAGtgGacT |
| pC194 | TCTTTCTTATCTTG/ATAATA |
| pUB110 | TCTTTCTTATCTTG/ATAcat |
| φX174 | gCTccCccAaCTTG/ATAtTA |
| pMV158 | GGGGGGCTACTACG/ACCCCCCC |
| pE194 | aGGGGGgTACTACG/ACCtCCCC |
| pFX2 | gcttccgTACTACG/ACCCCCCa |
Capital letters denote conserved bases. Slashes denote the sites of cleavage by the initiator protein. The positions of cleavage in pS194 and in pUB110 have not been unambiguously determined, and they are derived from homologies to plasmids of the same family.
Rep proteins.
Rep proteins of the RC-replicating plasmids have DNA strand transferase enzymatic activity, so that they are able to cleave and to join plasmid DNA in a type I topoisomerase-like fashion. RepC protein of plasmid pT181 was purified and shown to have nicking activity on the plasmid dso (156b). This finding, together with the presence of intracellular plasmid ssDNA and with the lack of requirement of a primer RNA for leading-strand synthesis, provided the first indications of the RC mechanism for plasmid replication (156a, 291). Rep-mediated nicking participates in the initiation of replication because the Rep proteins cleave supercoiled DNA within an unpaired sequence of the nic region. Cleavage and joining are essential for termination of replication (see below). Rep proteins encoded by RC-replicating plasmids have several conserved motifs which are shared with the Tra and Mob proteins (involved in plasmid transfer or conjugative mobilization) and with the Rep proteins of ssDNA coliphages and geminiviruses (131, 162, 239, 310). The two more relevant motifs correspond to the enzymatic activity domain and to a putative metal-binding domain (Table 2). Curiously, this last motif is not present in plasmids of the pT181 family or in ssDNA filamentous phages. In plasmids pT181 and pC221, the phosphodiester bond 5′-ApT-3′ is cleaved by Tyr-191 or Tyr-188, respectively, of the corresponding Rep proteins, which remain covalently bound to the 5′-phosphate end generated by the cleavage reaction (292). In the RepA protein of pC194, mutations in Tyr-214 abolished catalytic activity without affecting RepA-binding affinity. Furthermore, two other residues (Glu-142 and Glu-210) are also required for the catalytic activity (217). This interesting finding led to the proposal that the pC194-RepA protein contains two different catalytic residues, one (Tyr-214) involved in the covalent RepA-DNA binding for initiation of replication through transesterification and the second (Glu-210) participating in the termination step by directing the hydrolysis of a phosphodiester bond (217). The RepA Glu-210 residue was replaced by Tyr, so that termination of replication was shifted from hydrolysis to transesterification. This elegant approach led pC194-based plasmids to reinitiate after one round of replication, in a similar fashion to the replication of φX174 DNA (218). Stable covalent Rep-DNA linkage does not seem to be a general feature, since the RepB protein of plasmid pMV158 does not generate a stable covalent tyrosyl-phosphodiester bond with its DNA target (200). However, a transient bond between RepB and a DNA sequence containing the cleavage site of RepB (5′-GpA-3′) could mediate the initiation of replication, resembling the situation found in the ssDNA coliphage f1 (163, 200). Analysis of the chirality of the phosphate involved in the cleavage reaction has shown that the nicking and closing reaction mediated by RepB is exerted through an even number of steps, suggesting that a covalent bond, albeit a transient one, between RepB and its target exists (202).
TABLE 2.
Conserved motifs in initiator proteins of RC repliconsa
| Familyb | Sequence ofc:
|
|
|---|---|---|
| Motif 2 | Motif 3 | |
| φX174 | GRLHFHAVHFM | VGFYVAKYVNKKSDM |
| cons | gxuHUHuxuux | ugxYuakYuxkxxxx |
| pC194 | YNPHFHVLIAV | ELYEMAKYSGKDSDY |
| cons | yxxHUHvLUxV | xxxExxKYxxKxxDU |
| pMV158 | KKAHYHVLYIA | NVENMYLYLTHESKD |
| cons | KkxHYHUUUxx | xxxgxUxYUtHxxxD |
| pT181 | SNRFIRIYNKKQERK | |
| cons | SdRFIRIYNKKqERK | |
The two motifs found in Rep proteins of plasmids representing the main plasmid families, and their comparison with those found in the initiator proteins of ssDNA coliphages (162).
Below each Rep protein, a derived consensus is shown.
Motif 2, termed the HUH motif, is thought to be the metal-binding domain of the Rep proteins. Note that this motif is not found in the Rep proteins of plasmids of the pT181 family or in the initiator gpII protein of filamentous coliphages. Motif 3 (the catalytic motif) contains the conserved Tyr residue involved in the nucleophilic attack to the plasmid DNA at initiation of replication. Capital letters denote totally conserved residues; lowercase letters denote residues conserved in many but not all proteins within the family; U and u denote hydrophobic residues; x denotes any residue.
For plasmids of the pT181 family, studies with hybrid initiator proteins have demonstrated that a stretch of 6 amino acids, located at the C terminus of the Rep proteins, is sufficient to confer dso specificity, i.e., to interact with the nonconserved bind region of the cognate dso (73, 311). Although both the DNA-binding and -nicking activities of the pT181-RepC protein are required for replication, these activities are mutationally separable (73a). Protein-DNA cross-linking methods have shown that a carboxyl-terminal proteolytic fragment of the RepD from pC221 makes a specific noncovalent interaction with the bind region of its cognate replication origin (293). Such a recognition domain of this Rep protein is separated by some 80 residues from the active Tyr residue, whereas the nic and the bind sites within the dso are contiguous. These findings indicate that at least two separate domains of the Rep proteins could fold together in the native structure of the protein (311, 312). Resolution of the crystal structure of the Rep proteins of pC221 and/or pT181 will define the nature of the active center of these proteins. In the plasmids of the pMV158 family, the higher degree of homology found at the N-terminal region than at the C terminus of their Rep proteins suggested that their conserved N-terminal moiety would be involved in nicking activities whereas the C termini would be involved in specific dso recognition (69).
RepC of pT181 has been shown to function as a multimer, by demonstrating intragenic complementation between two repC mutant alleles. Gel filtration studies have shown that the apparent molecular weight of RepC is consistent with dimer formation (257). In RepB from pMV158, the use of glycerol gradients and analytical ultracentrifugation has shown that the protein is purified as an hexamer (60, 200), although the configuration of the protein bound to DNA has not yet been determined. RepK protein of plasmid pKYM (belonging to the pC194 plasmid family) binds the dso as a monomer (234).
Many of the rep genes have a weak Shine-Dalgarno (SD) sequence or do not have it at all. In the repB gene of pMV158, the existence of an atypical ribosome-binding site sequence, possibly used in Streptococcus pneumoniae and in E. coli, has been postulated (60, 72, 171). In vitro and in vivo indications that the atypical ribosome-binding site is involved in the translation of the pMV158 repB gene has been obtained (70). Whether those features of the translation initiation signals of the rep genes reflect an additional control mechanism to keep a low level of the Rep protein is not known. It is interesting that the amount of RepC protein of pT181 has been estimated to be one dimer molecule per plasmid replication event (16). Furthermore, once a RepC dimer has been used, it becomes inactive for a new round of replication because of the attachment of an oligonucleotide to one of its subunits, generating a heterodimer, RepC/C* (255–257). Inactivation through generation of heterodimers has also been shown for pT181-related plasmids (256) and has been suggested to exist for the RepU protein of pUB110 (207). The footprints generated on supercoiled DNA by the RepC/C homodimer are different from those formed by the RepC/C* heterodimer, and whereas RepC/C was able to enhance cruciform extrusion at the dso, RepC/C* was not (140). Nevertheless, biochemical analysis has shown that RepC and RepC* appeared to be identical, except for the absence of the active Tyr residue in the modified protein (258).
None of the Rep proteins seems to have the HTH motif typical of many DNA-binding proteins, although a putative LZ motif exists in the RepB protein of plasmid pMV158 (60). Inspection of the DNA sequence of various rep genes shows two possible translation initiation signals, which would give rise to Rep and Rep′ proteins (similar to the A and A* initiator proteins, respectively, of the ssDNA coliphage φX174) (13). However, genetic evidence has shown that, at least for RepB of pMV158, the putative shorter RepB′ protein is not sufficient to conduct replication in vivo (70). Plasmid pUB110 and its relative pRBH1 were reported to contain two putative SD sequences followed by a Met residue (178, 208). Overproduction of the pUB110-Rep protein showed only one major product, the one translated from the first Met residue (178). Thus, the significance of these putative Rep′ proteins (if any) remains to be clarified.
Initiation and elongation of leading-strand synthesis.
Little information is available on the initiation complex in RC-replicating plasmids. As stated above, the Rep proteins of pT181 and pC221 nick the dso and become covalently attached, by a phosphotyrosine bond, to the thymidilate residue at 3′ of the nick. A strict requirement for supercoiled DNA as the substrate to initiate replication, most probably for cruciform extrusion, has been proposed (10, 64, 65, 216). In pMV158, in vitro RepB-dependent cleavage of the nick sequence absolutely requires ssDNA as the substrate (200, 201) and the hairpin corresponding to the nic region has been shown to extrude on supercoiled plasmid DNA (65, 254). For pT181, at low to moderate RepC/pT181 origin ratios, only supercoiled molecules can be used as the substrate for replication initiation (149), although specific RepC nicking activity has been demonstrated in vitro on linearized plasmid DNA (157). Once the Rep-DNA complex has formed, several host proteins would be required for leading-strand synthesis. To generate the replication initiation complex of pT181, the product of the Staphylococcus aureus chromosomal pcrA gene is required. PcrA has homology to helicase II and Rep helicase of E. coli (136). The PcrA protein is essential for cell viability and for replication of plasmid pT181. A mutation (pcrA3) found in the pcrA gene led to an increase in the proportion of nicked DNA plasmid forms, which were identified as replication initiation complexes (138). The copy number of pT181 was reduced in an S. aureus pcrA3 background, and suppressor mutations were shown to map in the plasmid repC gene, indicating a direct interaction between RepC and PcrA (137). In addition, it is assumed that DNA Pol III-HE extends the leading strand and that due to the generation of ssDNA intermediates, SSB also participates in the replication complex.
Termination of leading-strand synthesis.
Specific recognition between the Rep protein and the dso is required not only for initiation of RC replication but also for the termination step. Rep-dso recognition seems to be more stringent for initiation than for termination, because (i) abortive termination can occur at sequences similar but not identical to the origin (15, 196); (ii) part of the dso seems to be sufficient for termination of replication of pC194 (107); and (iii) an 18-bp sequence is long enough for termination of pT181 replication, but initiation requires a larger region which includes the RepC binding site (329). Mutational analyses have shown that the nucleotides of the right arm of the inverted repeat IR-II must be conserved for termination to occur whereas changes are allowed in some of the nucleotides of the left arm of the IR-II (330). Genetic approaches suggest that a DNA sequence, probably located 3′ of the nic region of pUB110, is required for efficient termination of replication (10). Thus, two partially overlapping sequences, one required for initiation and the other required for termination, should be present in pUB110. For this plasmid, a single amino acid change in its Rep protein appears to reduce the ability of the Rep protein to terminate replication specifically (22).
The mechanistic information on termination of RC replication has arisen from the elegant set of experiments performed in Novick’s laboratory with pT181 (141, 255, 257, 311, 312). This information has been extended for other plasmids of the pT181 family (256), as well as for pUB110 (207). The current model for RepC-catalyzed initiation and termination of pT181 replication (schematized in Fig. 8; see also Fig. 6 and Table 2) postulates that Tyr-191 of one of the subunits of a RepC homodimer exerts a nucleophilic attack at the dso, generating a RepC::RepC-DNA covalent complex (141, 255). Replication of the leading strand proceeds more than one full round and beyond the nick site (Fig. 8, stage 1). This would permit the IR-II hairpin at the dso to be exposed in both the displaced strand and the newly synthesized strand. Then the RepC subunit which was not used at the initiation step would cleave the regenerated dso, being covalently bound to the 5′ end of the newly synthesized DNA strand (stage 2). The 3′-OH end released in this reaction (belonging to the parental strand) would exert a nucleophilic attack on the tyrosyl-phosphodiester bond generated at the initiation step (stage 3). This last reaction will release the ssDNA intermediate. The free OH group of the Tyr-191 residue of the RepC subunit involved in the initiation step attacks the nick site generated on the newly synthesized strand by the further extension of DNA synthesis (stage 4). Finally, the 3′-OH end generated would attack the tyrosyl-phosphodiester bond generated between RepC and DNA in stage 2. The end products of this reaction would be a dsDNA plasmid molecule (containing the newly synthesized plus strand) and a RepC/C* heterodimer composed of one intact RepC molecule and one modified subunit which has a short oligonucleotide covalently bound. This oligonucleotide is 12-mer on average, and it contains the 3′ half of the IR-II (255). Conversion of RepC/C homodimer to RepC/C* heterodimer has been shown to be replication dependent (257). The RepC/C* heterodimer is not able to relax supercoiled pT181 DNA or to reinitiate replication, indicating that inactivation of one subunit (RepC*) may inactivate the whole molecule and that a RepC dimer is used only once in a replication cycle (255, 257). A RepU/U* heterodimer has also been found in pUB110, which would point to a general inactivation mechanism for plasmids replicating by the RC mode (208). Lack of recycling of the Rep initiator is a critical feature for a plasmid, since it would prevent overreplication (255). In pMV158, the pT181 model for termination of replication does not seem to be applicable, since RepB does not remain covalently bound to DNA (200). However, no alternative model for leading-strand termination has been proposed, and inactivation of the RepB protein has not been analyzed. A model, similar to the one proposed by Novick for pT181 RC replication, has been recently proposed for termination of ssDNA conjugal transfer mediated by a dimer of the TraI protein of the IncPα plasmid RP4 (240).
Replication of the lagging strand.
The last stage of RC replication is the conversion of the ssDNA intermediate into a duplex plasmid DNA molecule. Lagging-strand synthesis initiates and terminates at the sso, which is a specific noncoding region with the potential to generate imperfect stem-loop structures. These secondary structures function in an orientation-dependent manner (109). This suggests that the ssDNA→dsDNA conversion requires unpaired sequences within the secondary structures that constitute the sso (65, 68, 109). Based on sequence homologies, various types of sso have been reported (228). The best characterized are ssoA (pT181 and pC194), ssoU (pUB110), ssoT (pTA1060 and pBAA1) (75, 193), and ssoW (pWV01 and closely related replicons) (268). The streptococcal plasmid pMV158 has the interesting feature of bearing both ssoA and ssoU (68, 171, 250, 306). Deletion of plasmid sso regions leads to reduction in the plasmid copy number (measured as dsDNA), accumulation of ssDNA molecules, and rapid loss of the plasmid in the absence of selective pressure (65, 109). Such an instability does not depend on the average number of copies of double-stranded plasmid DNA, since it cannot be overcome by mutations increasing the plasmid copy number of these deleted derivatives (68). Analysis of the DNA sequence and structure of the ssoA of various plasmids (228) showed the presence of two conserved unpaired regions. One of them includes part of recombination site B (RSB), which is highly conserved in almost all of the reported ssoA sequences. In the center of the palindromic sequence of the ssoA regions, there is another unpaired region, which is conserved only among the members of each one of two different groups of ssoA sequences, represented by pT181-ssoA and pMV158-ssoA (65, 228). In the ssoA of the plasmid group represented by pMV158, the central conserved sequence, termed CS-6, is 5′-TAGCGt/a-3′ (65). The CS-6 has been shown to function as a transcriptional terminator for the synthesis of an RNA primer (see below). Mutations affecting the CS-6 sequence of the pMV158-ssoA moderately increase the intracellular amount of plasmid ssDNA, although the effect is not as dramatic as that of the deletion of the entire ssoA (165, 167).
The mechanisms of ssDNA→dsDNA plasmid conversion are poorly understood, and no known plasmid-encoded functions seem to be involved. In analogy to ssDNA coliphages, a pRNA is assumed to be needed to initiate lagging-strand synthesis. Experimental evidence suggests that in most cases, ssDNA→dsDNA conversion is mediated by RNAP, since ssDNA accumulates in vivo in the presence of rifampin (27, 74, 165). In vitro studies have shown that an ssDNA fragment containing the pMV158-ssoA was able to promote RNAP-directed synthesis of a 20-nucleotide pRNA. This ssDNA fragment specifically bound RNAP, with this binding requiring an intact RSB sequence (166).
The ssoW of plasmid pWV01 is made up of two inverted repeats (IR1 and IR2). Conversion of ssDNA→dsDNA from an intact ssoW seems to require the host RNAP (173). However, deletion of the IR2 resulted in a reduced and RNAP-independent conversion. These findings, together with the homologies between the ssoW-IR1 and the lagging-strand origin of φX174, led to the proposal of an alternative mechanism of pWV01 lagging-strand priming mediated by the host primosome (173, 268).
In addition to the host RNAP, DNA Pol I is involved in lagging-strand replication both in initiation and in termination (76, 166). Detection of ssDNA intermediates may be masked because the amount of ssDNA accumulated depends upon the specific host-plasmid pair and, in their natural hosts, single-stranded→double-stranded plasmid DNA conversion is so efficient that the half-lives of ssDNA intermediates are too short to be detectable (68, 108, 306). Nevertheless, implementation of an in vitro system for replication of ssDNA (23, 210) facilitated the determination of the RNA-DNA transition points, which corresponded to regions including the above-mentioned conserved central sequences (CS-6) of the secondary structure of the sso regions (74). The use of pMV158-based vectors with no sso indicated the existence of an alternative pathway of conversion in which plasmid-encoded antisense RNAs, annealed to the complementary region on the ssDNA intermediates, may act as primers in a process mediated by the host RecA protein (194).
Synopsis.
Replication by the RC mechanism is widespread among multicopy plasmids from the Archaea and Bacteria. The 3′-OH end required for initiation of replication is provided by the site-specific nicking activity of the plasmid-encoded Rep protein on one of the parental plasmid strands. The DNA substrate for the Rep-mediated nicking has to be in a single-stranded configuration. This can be achieved by Rep-facilitated extrusion of a cruciform structure encompassing the nic region of the origin, where the nick site is unpaired. Leading-strand synthesis is terminated by various specific strand transfer reactions, also mediated by the Rep protein. After completion of leading-strand synthesis, the Rep protein is inactivated and plasmid ssDNA intermediates are generated. RNAP-directed synthesis of a short RNA primer initiates lagging-strand replication. In this step, only host-encoded proteins are involved.
CONTROL OF PLASMID REPLICATION
Although plasmid copy number may vary in different bacteria, within a given host, and under fixed growth conditions, any particular plasmid has a characteristic copy number. This is achieved by plasmid-encoded control elements that regulate the initiation of the replication step. Control systems maintain the rate of replication in the steady state at an average of one replicative event per plasmid copy and cell cycle by correcting deviations from the average copy number in individual cells. To define and to maintain the copy number, plasmids use negative regulatory circuits (251). The genetic analysis of replication control mechanisms was first performed for plasmid R1 through the isolation of mutants which exhibited an increased copy number (225). Characterization of these mutants indicated that the determinants of copy number control resided in the plasmid itself and that negative regulators (inhibitors), acting at the initiation step, were involved in this control. A model of control of replication modulated by negative effectors was first proposed and described in quantitative terms by Pritchard et al. (252). When a plasmid colonizes a new host, the concentration of these negative regulators should be negligible. This seems desirable for successful establishment, since unhindered plasmid replication would permit the normal copy number to be reached in a short time. However, once the characteristic copy number is reached, keeping the average copy number in the population requires adjustments to fluctuations in this value in individual cells. The control systems do just that by either increasing or decreasing the rate of replication per plasmid copy and cell cycle. Individual plasmid copies are selected for replication at random from a pool that includes replicated and nonreplicated copies. However, mechanisms that counterselect newly replicated molecules exist, e.g., hemimethylation and supercoiling (3, 224).
Control of replication by negative regulators and random selection of plasmids for replication have an additional consequence: two plasmids with identical replicons are unable to stably coexist within a given cell in the absence of selective pressure. This leads to segregation of plasmids within the host population, a phenomenon known as plasmid incompatibility. The inhibition of plasmid replication, associated with an increase in the gene dosage of copy number control genes, has been used to identify these genes (reviewed in references 12, 51, 155, 222, 227, and 251).
Control of replication by inhibitors would require that they could “measure” the concentration of plasmid copies within the cell. This could be achieved by an unstable inhibitor expressed constitutively or by a stable inhibitor synthesized shortly after each initiation event (252). Any of these alternatives would reduce the initiation frequency after each initiation event and would increase or decrease the rate of initiation of replication when the average copy number is, respectively, lower or higher than required. When the frequency of initiation is determined by the level of an initiator protein, one mechanism for controlled plasmid replication (unlike phage replication) would be to inactivate the initiator protein after each replication event (255, 319).
Mechanisms controlling replication have been studied in various systems (reviewed in reference 293a), and several types of inhibitors have been detected: (i) antisense RNA (ColE1, R1, and pT181); (ii) both an antisense RNA and a protein (pMV158 and pIP501), and (iii) DNA sites for binding initiator proteins (F, P1, RK2, and R6K). A schematic representation of these regulatory loops is depicted in Fig. 9. Control of replication by a protein (lambda-dv) is well documented and quantified (222), but it has not been described in natural plasmids and is not discussed here. Accessory elements that can also modulate replication but that are not directly involved in controlling the initiation frequency are not discussed either.
FIG. 9.
Control of plasmid DNA replication. Examples of control by Rep DNA-binding sites (plasmid P1), by antisense RNAs (plasmids R1 and pT181), and by a dual system (plasmid pMV158) are depicted. Transcripts are shown as continuous lines, with arrowheads indicating the direction of synthesis. mRNAs are shown with thicker lines than antisense RNAs; countertranscribed RNAs (thin lines with small arrowheads) are indicated. Other symbols as follows: solid ellipses, origins; rectangles, promoters; a.r.b.s., atypical ribosome-binding site; parallel vertical lines, mRNA-ctRNA interactions; plus, positive interactions; minus, and inhibitory RNA-RNA or protein-DNA interactions.
Control by Antisense RNA
Interactions between RNAs can modulate the availability of either a pRNA or the level of a rate-limiting Rep protein, which are essential for replication. Antisense RNAs controlling plasmid copy number are complementary to a region in the 5′ end of the target transcript (preprimer RNA for replication or rep mRNA) and are termed “countertranscribed RNAs” (ctRNAs) (227, 228, 309). In some cases, inhibition by ctRNAs is achieved in the region of pairing with the target RNA. In other cases, formation of the inhibitor-target duplex leads to a conformational change in the target transcript, which is rendered inactive. Examples of these mechanisms of control are described below.
Control of primer RNA processing: plasmid ColE1.
ColE1 was the first example where control of replication was found to be exerted by RNA (170, 296). In fact, the analyses of copy number control circuits of ColE1 led to the identification of antisense RNAs, a milestone discovery in molecular biology. trans-acting antisense RNAs permit the silencing of specific genes by pairing to mRNA molecules. The bases of the control of ColE1 replication are well known (reviewed in references 47 and 82). Initiation of replication is primed by a 550-nucleotide transcript, RNA II, which is made by RNAP and is specifically processed by RNase H. This processing is required for the transition of RNA to DNA synthesis, since it generates a 3′-OH end on which Pol I initiates the synthesis of the leading strand. The availability of this 3′-OH end is rate limiting for initiation and is modulated by an RNA of 108 nucleotides, the antisense RNA I, which is entirely complementary to a region in the 5′ end of the pRNA II. Hybrid formation between the antisense and the preprimer RNA alters the overall secondary structure of the preprimer. As a consequence of this conformational change, the 3′ end of the preprimer cannot hybridize to the DNA. This, in turn, inhibits the initiation of replication because, in the absence of DNA-RNA hybrid formation, the preprimer is not processed by RNase H. Genetic and biochemical analyses showed that interactions of the antisense RNA with the complementary sequences of the preprimer occur initially at regions exposed as single-stranded loops on both folded RNA molecules. Formation of this initial contact (or “kissing” complex) is the rate-limiting step of the interaction and leads to the full annealing of the antisense RNA I with the preprimer RNA II, in a process that starts at the 5′ end of the RNA I. The RNA I is synthesized from a constitutive promoter (therefore its level is proportional to the plasmid copy number) and is unstable. An auxiliary role is provided by a small basic protein, termed Rop or Rom (276, 304). Rop interacts with the antisense RNA and with the preprimer, increasing the efficiency of formation of the “kissing” complex and thus decreasing the efficiency of replication. The gene encoding this protein is not an incompatibility determinant, but it can reinforce the antisense RNA I-mediated incompatibility.
Copy number control of plasmid R1.
Copy number control in R1 is exerted at the level of RepA synthesis, which is modulated by the products of the copy number control genes, copB and copA (reviewed in references 224 and 309). The products of copB and of copA inhibit repA expression at the transcriptional and posttranscriptional levels, respectively. RepA is rate limiting for replication, acts preferentially in cis, and cannot be reused. The main copy number control gene is copA, while copB plays an accessory role. copA is transcribed from a constitutive promoter, and its product is an unstable (half-life, 2 min) RNA, CopA, which is complementary to a leader region (termed CopT) of the repA mRNA. Hybrid formation between CopA and its CopT target is also achieved through formation of a “kissing” complex. The CopA-CopT hybrid inhibits RepA synthesis by hindering the translation of a leader peptide-encoding gene, tap, which is translationally coupled to repA (26). The efficiency of inhibition depends on the affinity of kissing-complex formation (reviewed in reference 226). Mutations in copA that decrease the efficiency of the complex formation increase the copy number, but replication still is controlled. However, complete inactivation of CopA leads to uncontrolled (runaway) replication (reviewed in references 224 and 226). The duplex CopA-CopT is specifically cleaved by RNase III, and this cleavage seems to be an important step in the control of RepA synthesis (25). Mutational analysis performed at copA showed that the structure of the major stem-loop in CopA is important for its function. The presence of bulges in the stem region of CopA is required for rapid duplex formation with its CopT target in vitro, as well as for effective inhibition in vivo (126). In addition, the presence of bulged nucleotides in the CopA stem seems to result in a structural destabilization that plays a protective role against degradation by RNase III (125). Structural and functional studies on CopA have permitted us to draw a direct correlation between gene dosage (plasmid copy number) and CopA levels. This determines a constant rate of replication, independent of the copy number of the plasmid or the growth rate of the host. Two consequences derive from these results: (i) the frequency of plasmid replication per copy is >1 when the number of plasmid copies per cell is below the average and <1 when the number of copies is above the average; and (ii) the copy number of R1 increases with decreasing host growth rate. A constant rate of replication per cell, independent of copy number, will slowly correct deviations from the average frequency of plasmid replication (one per plasmid copy per cell cycle), and therefore it will actively maintain the average copy number constant (222). The second regulatory element in R1, the CopB protein, plays an auxiliary role under normal circumstances. This small dimeric and basic protein represses transcription of repA from a promoter, P2, which is located downstream of copB (Fig. 9). Under steady-state conditions, CopB is present at saturating concentrations, fully repressing transcription from P2 to a basal level. Under these conditions, the repA mRNA is synthesized mainly as a polycistronic copB-tap-repA RNA. However, under conditions in which the copy number drops or is low because of the early stages of plasmid establishment, the CopB-mediated repression is not efficient. In these circumstances, the P2 promoter is derepressed and repA is also transcribed as a tap-repA mRNA. This leads to a transient increase in the transcription rate of repA and, as a consequence, to a temporal increase in the frequency of plasmid replication. Thus, derepression of the P2 promoter would play an important role during the early phase of plasmid establishment or in situations in which the copy number falls below the wild-type level (224, 320). However, a computer-based simulation suggested that the auxiliary control loop mediated by CopB should have only a marginal effect not only under steady-state conditions but also during plasmid establishment (259).
Other instances of control by antisense RNAs.
There are other examples in which replication is controlled by antisense RNA. For instance, in plasmids of the IncIα and IncIβ groups, control of replication is modulated by a short ctRNA that inhibits, as in R1, synthesis of a rate-limiting Rep protein, which, in turn, is translationally coupled to the synthesis of a leader peptide. Inhibition by ctRNA is exerted at the posttranscriptional level by hindering the synthesis of the leader peptide. In addition, ctRNA prevents the formation of an activator RNA pseudoknot that is required for efficient synthesis of the Rep protein (reviewed in reference 309).
Antisense RNA has been involved in the silencing of the ori-γ of plasmid R6K (243, 244). Replication from this origin requires the synthesis of an activator RNA that starts within the seven iterons of the origin and is silenced by the antisense RNA. The replication initiator protein, π, seems to favor interactions between these transcripts. This regulatory system is independent of the one modulated by iterons. R6K is not the only theta-replicating iteron-containing plasmid in which antisense RNA plays a role in regulating replication. In plasmid ColE2, a ctRNA complementary to the leader rep mRNA has been involved in the control of replication at the posttranscriptional level through generation of a ctRNA-mRNA hybrid that sequesters a sequence(s) essential for Rep synthesis other than the SD region (287, 323).
Direct inhibition of Rep synthesis: blocking rep translation.
A direct way to control plasmid copy number by a ctRNA is the inhibition of Rep synthesis by blocking the accessibility of the ribosomes to the rep mRNA translation initiation sequences. This type of control has been proposed for some plasmids replicating by the mechanisms of strand displacement (R1162) (154) and RC replication (pMV158) (67). In both instances, the main inc determinant is located in a region encoding a ctRNA, and the putative signals for initiation of rep translation are placed on a predicted loop of the rep mRNA. In the case of R1162/RSF1010, the frequency of initiation is determined by the level of the RepC DNA-binding protein (112, 153). Expression of repC is regulated at the transcriptional level by the first gene of the repC operon (179) and at the translational level by a ctRNA that is complementary to the translation initiation signals of repC (154). Which of these two regulatory mechanisms makes the RepC synthesis rate limiting is not yet well understood. In the RC-replicating plasmid pMV158, a ctRNA has been proposed to inhibit the translation of the repB gene by direct interaction with the initiation of translation sequences present in the repB mRNA (67, 71, 72). Secondary structures in the ctRNA, other than that corresponding to the transcriptional terminator, have not been found. Plasmids lacking the DNA region encoding this terminator and the complementary structure on the cop-rep mRNA are still sensitive to the wild-type ctRNA supplied in trans, suggesting that formation of a kissing complex may be important but is not essential for the generation of the hybrid ctRNA-mRNA. A more complex regulatory loop in pMV158, which involves the joint participation of the ctRNA and of a repressor Cop protein, was described later (see below). Analogous genetic structures in the control-of-replication region have been found in plasmids of the pMV158 family (69). A two-component system (Cop protein and antisense RNA) controlling the replication of plasmid pIP501 has also been proposed (35; see below). For plasmid pUB110, synthesis of the initiator RepU protein seems to be controlled by two antisense ctRNAs that interfere with repU translation, since the putative ctRNAs are complementary to repU initiation of translation sequences (178), although a mechanism of control similar to that of pT181 has also been suggested (11, 249). However, since the synthesis of RepU is autoregulated (207), a more complex control level may have to be considered.
Transcriptional attenuation: the pT181 paradigm.
A singular copy number control mechanism, involving ctRNAs, has been well established for pT181 (119, 169, 227, 228). The current model for the control of replication of pT181 (references 227 and 228 and references therein) proposes that synthesis of the Rep initiator is limited by two small ctRNAs (having the same 5′ end but different 3′ ends), which are complementary to the untranslated 5′ end of the rep mRNA (44, 119, 227, 229). The initial kissing-complex formation between ctRNAs and rep mRNA would take place through nucleotides exposed in complementary loops of hairpin structures that can be formed on both RNA species. Formation of the ctRNA-mRNA hybrid would lead to a conformational change in a region of the mRNA far apart from the region of complementarity. The new mRNA configuration would be such that a new hairpin ending in an A(U)6 sequence could form. This secondary structure resembles a Rho-independent transcription terminator and therefore can truncate the rep mRNA (230). Thus, transcriptional attenuation (as found in the control of many biosynthetic operons) (161), rather than blocking translation initiation, is the mechanism for controlling the copy number of pT181. A similar control, through premature termination of the rep mRNA promoted by the indirect action of a ctRNA, has been proposed for plasmids of the pIP501/pAMβ1 family (33). For pIP501, it has been shown that steps occurring before stable ctRNA-mRNA duplex formation are sufficient to attenuate mRNA synthesis (34).
Control by both Transcriptional Repressor and Antisense RNA
Control by the joint action of a transcriptional repressor and an antisense RNA has been characterized for plasmids R1, pMV158, and, more recently, pIP501, although differences in the role of the components have been found (35, 69, 71, 72, 224). As described above, the main control in R1 is exerted by the ctRNA CopA whereas CopB plays an auxiliary role. In fact, deletion of copB leads only to a moderate increase in plasmid copy number, and the copB gene cloned into a compatible replicon does not exhibit incompatibility toward R1.
In the case of pMV158, a transcriptional repressor, CopG, binds to and represses transcription from a single promoter for both the copG and repB genes (66). Mutations or deletions in copG lead to an increase in the plasmid copy number (7), and copG exerts only a weak incompatibility toward plasmids with the pMV158 replicon when cloned, under its own transcription and translation signals, into a high-copy-number compatible vector (67). A second element involved in copy number control is a small countertranscribed RNA, RNA II, which is complementary to a region of the cop-rep mRNA between genes copG and repB. rnaII is a strong inc determinant when supplied in trans at high gene dosage. Mutations that abolish the synthesis of the ctRNA also lead to an increase in copy number, but replication is still controlled by CopG. However, in this case, great fluctuations in copy number within individual cells were observed (71). No significant incompatibility toward plasmids with the pMV158 replicon was found when the rnaII gene was cloned in a low-copy-number plasmid (71). The control circuit elements (copG and/or rnaII) have been cloned at a physiological gene dosage (i.e., at a copy number similar to that of the wild-type plasmid). In this case, a strong incompatibility toward pMV158 was found when the entire circuit was provided in trans whereas either component cloned separately exhibited weak (ctrnaII) or no (copG) incompatibility. The hypothesis derived from these experiments was that the control of replication of pMV158 is exerted by the entire regulatory circuit rather than by a hierarchy of any of the components (71). Due to the genetic organization of the plasmids of the pMV158 family, a similar regulatory circuit seems to exist in all of them (61).
The theta-replicating plasmid pIP501 seems to share features with R1 and pMV158 in their replication control circuit. Similarly to R1, the CopR protein of pIP501 is not a regulator of its own synthesis but represses transcription from the essential downstream rep promoter (31). However, and unlike R1, the CopR-inhibited rep promoter is not fully repressed, a situation resembling pMV158. The second element regulating pIP501 copy number is the unusually long-lived antisense RNA III (35). The stability of this antisense RNA III poses the problem of corrections of down-fluctuations in copy number, since a decrease in the number of plasmid copies will not be rapidly followed by a concomitant reduction in the levels of the inhibitor. This would lead to plasmid loss, which has not been observed (139a). To solve this apparent paradox, a model similar to the one proposed for pMV158 (71), in which a reduction in the pIP501 copy number would lead to derepression of the rep promoter due to reduction in the CopR levels, has been postulated (35).
The recent finding that the RepU protein of plasmid pUB110 regulates its own synthesis, thus introducing an additional control for plasmid copy number (207), would represent another example of dual control (transcriptional and translational regulation). Mechanistically, the situations for pUB110 and the pMV158 may be indistinguishable, which may point to a more general control of replication mechanism in these plasmids replicating by the RC mode.
Control by Iterons
The Rep proteins of the theta-replicating iteron-containing plasmids are either autoregulated or under transcriptional control, and the iterons play a role in the control of plasmid copy number (reviewed in references 51, 155, and 223). The role of the iterons was initially described for plasmid F, based on the observation that oriS-dependent replication was inhibited when the iterons of oriS were cloned on a compatible plasmid (295). Similar situations were also found for other iteron-containing plasmids, such as P1 or R1162/RSF1010. The extent of inhibition in P1 was found to be proportional to the number of cloned iterons (238) whereas in R1162 the inhibition was abolished by overproduction of the rate-limiting Rep protein (153). These observations led to the formulation of the titration model (303). The model assumed that the Rep protein is rate limiting for initiation and that iterons titrate the Rep protein, thus limiting the frequency of initiation. A paradox developed when the Rep proteins of most iteron-containing plasmids were found to be autoregulated, which implies that titration could be overcome by derepression (49). Furthermore, in other systems (R6K and RK2), the amount of Rep protein within the cell was apparently too large to be limiting for saturation of iteron binding (88, 190). This led to the modification of the initial model and to the proposal that different forms of the Rep protein were involved in autoregulation and in initiation, so that initiator titration does not cause derepression (302). A computer model that adjust to this hypothesis has been developed (321).
A second solution to the titration/autoregulation paradox was provided by the model that Rep molecules bound to the P1 iterons were still able to repress the repA promoter (49). The model was based on the ability of Rep proteins to bind to two iterons simultaneously (in both P1 and R6K) (204). This property of the Rep proteins also provided the basis for a new model of negative control of replication called the “steric hindrance” (238) or “handcuffing” (155, 191) model. According to this model, when Rep proteins bind to and saturate the iterons of the origin, initiation occurs if the plasmid copy number is low. As the number of copies increases, Rep molecules bound to the iterons of one origin begin to interact with similar complexes generated on other origins. As a consequence, plasmid molecules pair through Rep-Rep interactions, causing a steric hindrance to the function of both origins (“handcuffing”). The plasmid pairs are apparently separated during subsequent cellular growth, and the initiation potential of individual molecules is restored. According to this model, it is the iteron concentration, rather than the level of Rep expression, that determines the rate of replication. The question of the role of autoregulation remains open, and no simple answer has been provided. As mentioned above, in RK2 and R6K, a substantial decrease in the levels of Rep protein has no effect on the rate of plasmid replication. However, in P1, a twofold reduction in the level of the Rep protein abolishes replication and a fourfold increase can lead to inhibition of replication (238). Similarly, in R6K, a twofold increase in initiator concentration is inhibitory to initiation. These data indicate that autoregulation is important, at least in some cases, to keep an optimal concentration of the initiator protein. It has recently been suggested that in particular plasmids in which initiators are limiting, initiator titration and initiator pairing (handcuffing) could be working together (51).
Some of the replicons containing iterons in the origin also contain iterons outside this region, which express incompatibility and further reduce the rate of replication (237). In principle, the probability of Rep-mediated iteron pairing greatly increases in these replicons, since it can occur in cis by DNA looping and in various combinations in trans. Since control systems should measure the copy number, and since intramolecular pairing is independent of this parameter, this pairing cannot correct deviations from the average copy number. However, intramolecular pairing can regulate the level at which the copy control operates (discussed in reference 223). A different question is whether the iterons within and outside the origin region are equally effective as replication control elements. Recent data on RK2 and P1 plasmids indicate that iterons outside of ori are more effective as negative regulators of replication than are iterons present in the origin region, probably due to differences in the quantitative and structural features of the two sets (5, 273). It has been proposed that in RK2 these auxiliary iterons stimulate initial pairing between origins. The data for P1 indicate that the concentration of the auxiliary iterons rather than their relative arrangements is the deterministic factor for replication control (51).
The role played by Rep proteins of iteron-containing plasmids in the control of replication and in autoregulation implies that specific rep mutations affecting protein-protein or protein-DNA interactions could increase the plasmid copy number. In R6K, copy-up mutations affecting the plasmid initiator protein have been obtained (87). Most of the copy-up mutations affected a 32-amino-acid region placed between the LZ motif and the putative DNA-binding domain of the π protein, a region that seems to be involved in high-order oligomerization of the initiator protein (199). Mutations that affect the LZ motif of the Rep protein and lead to copy-up phenotypes in pSC101 have been described (133). Copy-up mutants of plasmid RK2 containing mutations encoding TrfA variants defective in binding to DNA have been isolated, indicating that copy number control is modulated by TrfA-DNA interactions (46). Copy-up mutations in the trfA gene have also been selected as intragenic suppressors of thermosensitive trfA mutations. Some of these copy-up mutations were proposed to reduce a strong protein-protein inhibitory interaction induced, at the restrictive temperature, by the thermosensitive mutation (114).
Hemimethylation and Regulation of Plasmid Replication
The heptamer repeats containing the methylation sites in the origin of replication of plasmid P1 constitute the target of the host protein SeqA, involved in sequestering hemimethylated oriC into the bacterial membrane (37). Methylation seems to regulate replication at two different levels. First, hemimethylation would exert a negative regulation through ori-membrane interactions. The newly replicated, hemimethylated DNA is sequestered in the cell membrane, thus preventing reinitiation. The sequestration is relieved as the hemimethylated DNA undergoes new methylation (51). At a second level, methylation increases the initiation efficiency both in vivo and in vitro. This stimulation could be due to enhanced bending or unwinding of the DNA or to recycling of initiators (3, 51).
Synopsis
Plasmid DNA replication occurs coupled to the cell cycle of the bacterial host in such a way that a fixed concentration of plasmids is maintained in the bacterial population. This is achieved by mechanisms that monitor the initiation frequency and adjust this frequency to an average of one replication per plasmid copy and cell cycle. The control of plasmid replication is plasmid encoded and is performed by molecules (antisense RNA, proteins, or DNA sequences) that inhibit the initiation of replication in a dose-dependent way. This inhibition can limit the level of active Rep protein, the availability of the primer for the leading strand, or the number of active origins. Control of plasmid replication may be exerted (i) by a single inhibitor, (ii) by a main controller and an auxiliary element, or (iii) by the concerted action of two inhibitors (a protein and an antisense RNA or two sets of iterons). In general, any model on control of plasmid replication should explain how the average plasmid copy number is maintained in a growing population and how fluctuations on this average are corrected.
SUMMING UP: DIFFERENCES AND SIMILARITIES IN PLASMID REPLICATION MECHANISMS
In spite of the progress that has been made in the study of plasmid replication and its control, there are major gaps in our knowledge of the various replication systems. Initiation of replication demands melting of the origin and interactions between plasmid-encoded and host-encoded factors, which are in general only poorly understood. Melting of the origin can be stabilized by either manifesting the cryptic ssDNA-binding activity of the initiation proteins (DnaA and R6K-π) or by the formation of intramolecular secondary structures (e.g., cruciform extrusion). The configuration of the plasmid origins indicates that DNA curvature and Rep-enhanced origin bending seem to be a common but yet not fully explored feature (81, 156c, 205, 206, 283). DNA deformations at the origin may provide an appropriate configuration for the initiation event: cruciform extrusion and nicking by the initiator for RC-replicating plasmids versus assembly of the initiation complex and synthesis of an RNA primer for the other replicons. The role of host-encoded proteins in the generation of the initiation complex is relatively well defined for several plasmids replicating by the theta mechanism, but little is known for plasmids using the RC mode (apart from the PcrA helicase from S. aureus in pT181 replication) (135–138). Definition of the specificity of the interactions between Rep proteins and host replication factors is relevant to our understanding of the ability of plasmids to colonize different hosts. The convergence of structural studies with the functional analysis of initiators and inhibitors of DNA replication is an important area of future development. Host replication factors and plasmid initiators form part of the nucleoprotein complex that initiates plasmid replication. The structural definition of these macromolecular assemblies will provide mechanistic information relevant to our understanding of the initiation of plasmid replication and the interrelations between plasmids and their hosts. Since the continuity of the parental DNA strands is kept in plasmids replicating by the theta and strand displacement mechanisms, superhelical tensions accumulate during the elongation stage. A role for topoisomerases in the elongation of DNA synthesis and in the separation of both catenated daughter molecules has been documented (8, 9, 117, 245, 246, 327). Such a role may not be required for plasmids replicating by the RC mechanism, although the relaxed DNA molecules (which are the products of RC replication) must be supercoiled by DNA gyrase before they become a substrate for a new round of replication. Finally, information is scarce on three important plasmid replication events: (i) the role of transcription through origins to create waves of supercoiling, (ii) the structural changes introduced by chaperones that result in activation of Rep proteins, and (iii) the mechanisms of possible Rep inactivation, except for several RC-replicating plasmids (255, 256).
The study of specific signals involved in the termination of replication of theta-type plasmid replicons is a matter of growing interest. Elucidation of the structure of termination proteins (RTP and Tus) acting at specific termination sites has opened new avenues to perform detailed mechanistic analysis of this stage of replication (19).
Concerning lagging-strand synthesis, only host factors are thought to be required in plasmids replicating by the RC and theta modes. This contrasts with the situation for plasmids replicating by the strand displacement mechanism, in which the same plasmid-encoded primase and helicase proteins are involved in replication of both strands, in a continuous process that starts in two symmetrical origins and proceeds in opposite directions (263, 266). In this latter plasmid category, uncoupling synthesis of both DNA strands may occur, leading to the generation of ssDNA plasmid intermediates. However, unlike plasmids replicating by the RC mechanism, there is no strand specificity in the ssDNA intermediates generated during the strand displacement replication.
Control of plasmid replication is one of the key features of these extrachromosomal elements. Such control is always exerted at the initiation stage, perhaps because the initiation events, as opposed to the elongation and termination steps, are invariably replicon specific. The role of RNA inhibitors in the control of replication is relatively well understood (226, 309). However, replication control modulated by iterons is less well known and is an object of intense research (51, 155). Many of the initiators that bind to iterons autoregulate their own synthesis. How this autoregulation influences the frequency of initiation is important to a full understanding of control of replication in these plasmids. Interestingly, the strategies used to control plasmid replication do not correlate with the mechanism by which the plasmid replication initiates: replication initiation by different mechanisms may have similarities in control circuits (pIP501 and pMV158; pIP501 and pT181; and R1162 and pMV158). Finally, although the elements controlling replication have been identified in many instances and their modes of action have been described, there has not been much efforts to understand the control in terms of kinetics, except for a few cases (12, 223).
CONCLUDING REMARKS AND PROSPECTS
The main emphasis of this review has been to show that plasmids are important models with which the combination of genetic, biochemical, and molecular approaches have revealed many biological phenomena. An important focus for molecular biologists studying plasmids has been on the mechanisms involved in their replication and the control of this process. Replication studies have revealed a variety of mechanisms that are used to replicate circular or linear plasmids. Plasmids contribute with specialized sequences and functions to their own replication but, in general, use extensively the replication machinery of the host. Replication of these genetic elements therefore provides a window to gain information on the communication between plasmid- and host-specific replication machinery. Buried in these interrelationships is the ability of plasmids to colonize different hosts (i.e., their host range), which has important ecological and biotechnological implications.
Plasmid initiator proteins have drawn much interest, and the studies on these activators have been focused on the specific protein-protein and protein-DNA interactions involved in the formation of an active initiation complex. Although genetic and biochemical analyses have allowed the isolation and characterization of Rep variants affected in their interaction with DNA, the relevant protein domains involved in contacting DNA are not clearly known in most cases. Concerning protein-protein interaction, the available information indicates that the initiation complex is formed by multiple copies of the initiator protein interacting not only with the DNA in the origin region but also with each other and with host initiation factors, not to mention the role of membranes in DNA replication. The analysis of the Rep protein of plasmid pPS10 has given important clues on protein motifs involved in these interactions, although the picture is far from complete. Atomic resolution of the structure of an initiator and of a multiprotein-DNA initiation complex will be an important step toward a clear understanding of these interactions and the changes introduced in the target DNA. This could be done not only through the resolution of crystal structures of protein-DNA complexes but also through the use of many other available physicochemical approaches (such as nuclear magnetic resonance, light and neutron scattering, circular dichroism, and analytical ultracentrifugation, to name a few). Some steps in these directions have recently been taken for the RepA protein of plasmid P1 (50).
Studies on the control of plasmid replication have revealed the role played by small antisense RNAs in the processing of RNAs or in the specific inhibition of Rep-protein synthesis at the transcriptional or translational level. The biotechnological and clinical implication of this important discovery is well known and reveals, once more, the important practical implication of basic studies. The different ways in which antisense RNAs modulate the control of replication are well established. A less well known mechanism is the control of replication modulated by iterons, and this is an active field of research.
In contrast to molecular biologists, microbial ecologists have focused their attention mostly on lateral spread and persistence of plasmids in natural environments. However, in this area there is a lack of application of molecular biology techniques, which may result in a biased identification and characterization of plasmids isolated from new environments or from environments which have not been subjected to extreme selective pressures. Molecular analysis of plasmid replication and its control occurring in bacterial populations growing under natural conditions should be considered. Growth phases other than the exponential phase should also be considered and analyzed, and new methods to study plasmid replication in mixed bacterial populations and in bacteria growing on microfilms should be developed. Analysis of the so-called cryptic functions harbored by plasmids may reveal new areas of potential interest. A study on the presence of plasmids in the nonculturable bacteria, the vast majority of the microorganisms contributing to the biodiversity, remains to be done. The interests of biotechnologists in plasmids harboring “food-grade” selectable markers, in the mechanisms of stable inheritance, and in microbially aided improved crops are very different from those of people working in hospitals, who are more concerned with clinical outbreaks of pathogenic strains harboring antibiotic resistance plasmids and in the mechanisms of their elimination. The recent report on gene transfer from plasmid-harboring bacteria to mammalian cells opens the possibility of targeted gene therapy of human diseases (55).
Be that as it may, a wealth of information has been accumulated in recent years on the genetic and functional organization of plasmids, which can now be applied to study the mechanisms involved in the persistence and spread of plasmids in the environment. The incorporation of plasmid-encoded host-killing systems is an interesting feature in the design of microbes to be released into the environment. Knowledge of mechanisms which can specifically hinder plasmid replication, as well as of phenomena underlying plasmid population kinetics, is an important tool for microbial pathologists. In general, we may conclude that although a great deal has been achieved, the new stages in the research of plasmid biology should be based on the confluence of the areas of research outlined here and, ultimately, on the study of plasmids within their natural hosts and in more natural environments.
ACKNOWLEDGMENTS
We are indebted to Deepak Bastia, Dhruba Chattoraj, Don Helinski, and Martine Couturier for critical reading of the manuscript and many useful comments and to Saleem Khan for many interesting discussions. We thank all the colleagues who provided us with unpublished information, and we apologize to those colleagues who feel that their work is insufficiently cited. Thanks are due also to our past and present collaborators.
During the writing of this review, our labs were financed by Comisión Interministerial de Ciencia y Tecnología, grants PB94-0127 and PB96-0917 (to R.D.) and grant BIO97-0347 (to M.E.).
REFERENCES
- 1.Abeles A L, Austin S J. P1 plasmid replication requires methylated DNA. EMBO J. 1987;10:3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeles A L, Austin S J. P1 plasmid replication requires Escherichia coli Dam-methylated DNA. Gene. 1988;74:185–186. doi: 10.1016/0378-1119(88)90281-8. [DOI] [PubMed] [Google Scholar]
- 3.Abeles A L, Brendler T, Austin S J. Evidence of two levels of control of P1 oriR and host oriC replication origins by DNA adenine methylation. J Bacteriol. 1993;175:7801–7807. doi: 10.1128/jb.175.24.7801-7807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeles A L, Reaves L D, Austin S J. A single DnaA box is sufficient for initiation from the P1 plasmid origin. J Bacteriol. 1990;172:4386–4391. doi: 10.1128/jb.172.8.4386-4391.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abeles A L, Reaves L D, Youngre-Grimes B, Austin S J. Control of P1 plasmid replication by iterons. Mol Microbiol. 1995;18:903–912. doi: 10.1111/j.1365-2958.1995.18050903.x. [DOI] [PubMed] [Google Scholar]
- 6.Abeles A L, Snyder K M, Chattoraj D K. P1 plasmid replication: replicon structure. J Mol Biol. 1984;173:307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- 7.Acebo P, Alda M T, Espinosa M, del Solar G. Isolation and characterization of pLS1 plasmid mutants with increased copy numbers. FEMS Microbiol Lett. 1996;140:85–91. doi: 10.1111/j.1574-6968.1996.tb08319.x. [DOI] [PubMed] [Google Scholar]
- 8.Adams D E, Shektman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 9.Adams D E, Bliska J B, Cozzarelli N R. Cre-lox recombination in Escherichia coli cells. Mechanistic differences from the in vitro reaction. J Mol Biol. 1992;226:661–673. doi: 10.1016/0022-2836(92)90623-r. [DOI] [PubMed] [Google Scholar]
- 10.Alonso J C, Leonhardt H, Stiege C. Functional analysis of the leading strand replication origin of plasmid pUB110 in Bacillus subtilis. Nucleic Acids Res. 1988;16:9127–9145. doi: 10.1093/nar/16.19.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso J C, Taylor R H. Initiation of plasmid pC194 replication and its control in Bacillus subtilis. Mol Gen Genet. 1987;210:476–484. doi: 10.1007/BF00327200. [DOI] [PubMed] [Google Scholar]
- 12.Austin S J, Nordström K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 13.Baas P D, Jansz H S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- 14.Baker T A. Replication arrest. Cell. 1995;80:521–524. doi: 10.1016/0092-8674(95)90504-9. [DOI] [PubMed] [Google Scholar]
- 15.Ballester S, López P, Espinosa M, Alonso J C, Lacks S A. Plasmid structural instability associated with pC194 replication functions. J Bacteriol. 1989;171:2271–2277. doi: 10.1128/jb.171.5.2271-2277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bargonetti J, Wang P Z, Novick R P. Measurement of gene expression by translational coupling: effect of copy mutations on pT181 initiator synthesis. EMBO J. 1993;12:3659–3667. doi: 10.1002/j.1460-2075.1993.tb06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Bastia, D. Personal communication.
- 17.Bastia D, Germino J, Crossa J, Ram J. The nucleotide sequence surrounding the replication terminus of R6K. Proc Natl Acad Sci USA. 1981;78:2095–2099. doi: 10.1073/pnas.78.4.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastia D, Germino J, Crossa J, Hale P. Molecular cloning of the replication terminus of the plasmid R6K. Gene. 1981;14:81–89. doi: 10.1016/0378-1119(81)90150-5. [DOI] [PubMed] [Google Scholar]
- 19.Bastia D, Mohanthy B K. Mechanisms for completing DNA replication. In: DePanphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 177–215. [Google Scholar]
- 20.Bernander R, DasGupta S, Nordström K. The Escherichia coli cell cycle and the plasmid R1 replication cycle in the absence of DnaA protein. Cell. 1991;64:1145–1153. doi: 10.1016/0092-8674(91)90269-5. [DOI] [PubMed] [Google Scholar]
- 21.Bernander R, Krabbe M, Nordström K. Mapping of the in vivo start site for leading strand DNA synthesis in plasmid R1. EMBO J. 1992;11:4481–4487. doi: 10.1002/j.1460-2075.1992.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bidnenko V E, Gruss A, Ehrlich S D. Mutation in the plasmid pUB110 Rep protein affects termination of rolling circle replication. J Bacteriol. 1993;175:5611–5616. doi: 10.1128/jb.175.17.5611-5616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birch P, Khan S. Replication of single-stranded plasmid pT181 DNA in vitro. Proc Natl Acad Sci USA. 1992;89:290–294. doi: 10.1073/pnas.89.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird R E, Tomizawa J. Ribonucleotide-deoxyribonucleotide linkages at the origin of DNA replication of colicin E1 plasmid. J Mol Biol. 1978;120:137–143. doi: 10.1016/0022-2836(78)90300-5. [DOI] [PubMed] [Google Scholar]
- 25.Blomberg P, Wagner E G H, Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9:2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blomberg P, Nordström K, Wagner E G H. Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 1992;11:2675–2683. doi: 10.1002/j.1460-2075.1992.tb05333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boe L, Gros M-F, te Riele H, Ehrlich S D, Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989;171:3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böldicke T W, Lanka E, Staudenbauer W L. Rifampicin-resistant initiation of DNA synthesis on the isolated strands of ColE1 plasmid DNA. Nucleic Acids Res. 1981;20:5215–5231. doi: 10.1093/nar/9.20.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolivar F, Rodriguez R I, Betlach M C, Boyer H W. Ampicillin-resistant derivatives of the plasmid pMB9: construction and characterization of new cloning vehicles. I. Proc Natl Acad Sci USA. 1977;74:5265–5269. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- 30.Bramhill D, Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988;54:915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 31.Brantl S. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol Microbiol. 1994;14:473–483. doi: 10.1111/j.1365-2958.1994.tb02182.x. [DOI] [PubMed] [Google Scholar]
- 32.Brantl S, Behnke D, Alonso J C. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAMβ1 and pSM19035. Nucleic Acids Res. 1990;18:4783–4789. doi: 10.1093/nar/18.16.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brantl S, Birch-Hirschfeld E, Behnke D. Rep protein expression on plasmid pIP501 is controlled by an antisense RNA-mediated transcription attenuation mechanism. J Bacteriol. 1993;175:4052–4061. doi: 10.1128/jb.175.13.4052-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brantl S, Wagner E G H. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 1994;13:3599–3607. doi: 10.1002/j.1460-2075.1994.tb06667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brantl S, Wagner E G H. An unusually long-lived antisense RNA in plasmid copy number control: in vivo RNAs encoded by the streptococcal plasmid pIP501. J Mol Biol. 1996;255:275–288. doi: 10.1006/jmbi.1996.0023. [DOI] [PubMed] [Google Scholar]
- 36.Brendler T, Abeles A, Austin S J. Critical sequences in the core of the P1 plasmid replication origin. J Bacteriol. 1991;173:3935–3942. doi: 10.1128/jb.173.13.3935-3942.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brendler T, Abeles A, Austin S J. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brendler T G, Abeles A L, Reaves L D, Austin S J. Unique sequence requirements for the P1 plasmid replication origin. Res Microbiol. 1991;142:209–216. doi: 10.1016/0923-2508(91)90032-6. [DOI] [PubMed] [Google Scholar]
- 39.Brennan R G, Matthews B W. The helix-turn-helix DNA-binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 40.Bruand C, Ehrlich S D, Jannière L. Unidirectional replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 1991;10:2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruand C, Le Chatelier E, Ehrlich S D, Jannière L. A fourth class of theta-replicating plasmids: the pAMβ1 family from Gram-positive bacteria. Proc Natl Acad Sci USA. 1993;90:11668–11672. doi: 10.1073/pnas.90.24.11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruand C, Ehrlich S D, Jannière L. Primosome assembly site in Bacillus subtilis. EMBO J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussiere D E, Bastia D, White S. Crystal structure of the replication terminator protein from Bacillus subtilis at 2.6 A. Cell. 1995;80:651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- 44.Carleton S M, Projan S J, Highlander S K, Moghazeh S M, Novick R P. Control of pT181 replication. II. Mutational analysis. EMBO J. 1984;3:2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cereghino J L, Helinski D R. Essentiality of the three carboxyl-terminal amino acids of the plasmid RK2 replication initiation protein TrfA for DNA binding and replication activity in gram-negative bacteria. J Biol Chem. 1993;33:24926–24933. [PubMed] [Google Scholar]
- 46.Cereghino J L, Helinski D R, Toukdarian A E. Isolation and characterization of DNA-binding mutants of a plasmid replication initiation protein utilizing an in vivo binding assay. Plasmid. 1994;31:89–99. doi: 10.1006/plas.1994.1009. [DOI] [PubMed] [Google Scholar]
- 47.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 48.Chang P C, Cohen S N. Bidirectional replication from an internal origin in a linear streptomyces plasmid. Science. 1994;265:952–954. doi: 10.1126/science.8052852. [DOI] [PubMed] [Google Scholar]
- 49.Chattoraj D K, Mason R J, Wickner S H. Mini-P1 plasmid replication: the autoregulation-sequestration paradox. Cell. 1988;52:551–557. doi: 10.1016/0092-8674(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 50.Chattoraj D K, Ghirlando R, Park K, Dibbens J A, Lewis M S. Dissociation kinetics of RepA dimers: implications for mechanisms of activation of DNA binding by chaperones. Genes Cells. 1996;1:189–199. doi: 10.1046/j.1365-2443.1996.d01-235.x. [DOI] [PubMed] [Google Scholar]
- 51.Chattoraj D K, Schneider T D. Replication control of plasmid P1 and its host chromosome: the common ground. Prog Nucleic Acid Res Mol Biol. 1997;57:145–186. doi: 10.1016/s0079-6603(08)60280-9. [DOI] [PubMed] [Google Scholar]
- 52.Churchward G, Linder P, Caro L. The nucleotide sequence of replication and maintenance functions encoded by plasmid pSC101. Nucleic Acids Res. 1983;11:5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conley D L, Cohen S N. Effects of the pSC101 partition (par) locus on in vivo DNA supercoiling near the plasmid replication origin. Nucleic Acids Res. 1995;23:701–707. doi: 10.1093/nar/23.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conley D L, Cohen S N. Isolation and characterization of plasmid mutations that enable partitioning of pSC101 replicons lacking the partition (par) locus. J Bacteriol. 1995;177:1086–1089. doi: 10.1128/jb.177.4.1086-1089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Courvalin P, Goussard S, Grillot-Courvalin C. Gene transfer from bacteria to mammalian cells. C R Acad Sci Paris. 1995;318:1207–1212. [PubMed] [Google Scholar]
- 56.Cross M A, Warne S R, Thomas C M. Analysis of the vegetative replication origin of broad-host-range plasmid RK2 by transposon mutagenesis. Plasmid. 1986;15:136–146. doi: 10.1016/0147-619x(86)90049-1. [DOI] [PubMed] [Google Scholar]
- 57.DasGupta S, Masukata H, Tomizawa J I. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987;51:1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- 58.DasGupta S, Mukhopadhyay G, Papp P P, Lewis M S, Chattoraj D K. Activation of DNA binding by the monomeric form of the P1 replication initiator RepA by heat shock proteins DnaJ and DnaK. J Mol Biol. 1993;232:23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- 59.de Graaff J, Crosa J H, Heffron F, Falkow S. Replication of the nonconjugative plasmid RSF1010 in Escherichia coli K-12. J Bacteriol. 1978;134:1117–1122. doi: 10.1128/jb.134.3.1117-1122.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de la Campa A G, del Solar G, Espinosa M. Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J Mol Biol. 1990;213:247–262. doi: 10.1016/S0022-2836(05)80188-3. [DOI] [PubMed] [Google Scholar]
- 61.Dellis S, Filutowicz M. Integration host factor of Escherichia coli reverses the inhibition of R6K plasmid replication by pi initiator protein. J Bacteriol. 1991;173:1279–1286. doi: 10.1128/jb.173.3.1279-1286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dellis S, Schatz T, Rutlin K, Inman R B, Filutowicz M. Two alternative structures can be formed by IHF protein binding to the plasmid R6K gamma origin. J Biol Chem. 1992;267:24426–24432. [PubMed] [Google Scholar]
- 63.del Solar, G. Unpublished data.
- 64.del Solar G, Díaz R, Espinosa M. Replication of the streptococcal plasmid pMV158 and derivatives in cell-free extracts of Escherichia coli. Mol Gen Genet. 1987;206:428–435. doi: 10.1007/BF00428882. [DOI] [PubMed] [Google Scholar]
- 65.del Solar G, Puyet A, Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987;15:5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Solar G, de la Campa A G, Pérez-Martín J, Choli T, Espinosa M. Purification and characterization of RepA, a protein involved in the copy number control of plasmid pLS1. Nucleic Acids Res. 1989;17:2405–2420. doi: 10.1093/nar/17.7.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.del Solar G, Espinosa M. The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein RepA. Mol Microbiol. 1992;6:83–94. doi: 10.1111/j.1365-2958.1992.tb00840.x. [DOI] [PubMed] [Google Scholar]
- 68.del Solar G, Kramer G, Ballester S, Espinosa M. Replication of the promiscuous plasmid pLS1: a region encompassing the minus origin of replication is associated with stable plasmid inheritance. Mol Gen Genet. 1993;241:97–105. doi: 10.1007/BF00280206. [DOI] [PubMed] [Google Scholar]
- 69.del Solar G, Moscoso M, Espinosa M. Rolling circle-replicating plasmids from gram-positive and -negative bacteria: a wall falls. Mol Microbiol. 1993;8:789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]
- 70.del Solar G, Moscoso M, Espinosa M. In vivo definition of the functional origin of replication (ori(+)) of the promiscuous plasmid pLS1. Mol Gen Genet. 1993;237:65–72. doi: 10.1007/BF00282785. [DOI] [PubMed] [Google Scholar]
- 71.del Solar G, Acebo P, Espinosa M. Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol Microbiol. 1995;18:913–924. doi: 10.1111/j.1365-2958.1995.18050913.x. [DOI] [PubMed] [Google Scholar]
- 72.del Solar G, Acebo P, Espinosa M. Replication control of plasmid pLS1: the antisense RNA II and the compact rnaII region are involved in translational regulation of the initiator RepB synthesis. Mol Microbiol. 1997;23:95–108. doi: 10.1046/j.1365-2958.1997.1981561.x. [DOI] [PubMed] [Google Scholar]
- 73.Dempsey L A, Birch P, Khan S A. Six amino acids determine the sequence-specific DNA binding and replication specificity of the initiator proteins of the pT181 family. J Biol Chem. 1992;267:24538–24543. [PubMed] [Google Scholar]
- 73a.Dempsey L A, Birch P, Khan S A. Uncoupling of the DNA topoisomerase and replication activities of an initiator protein. Proc Natl Acad Sci USA. 1992;89:3083–3087. doi: 10.1073/pnas.89.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dempsey L A, Zhao A C, Khan S A. Localization of the start sites of lagging-strand replication of rolling-circle plasmids from gram-positive bacteria. Mol Microbiol. 1995;15:679–687. doi: 10.1111/j.1365-2958.1995.tb02377.x. [DOI] [PubMed] [Google Scholar]
- 75.Devine K M, Hogan S T, Higgins D G, McConnell D J. Replication and segregational stability of the Bacillus plasmid pBAA1. J Bacteriol. 1989;171:1166–1172. doi: 10.1128/jb.171.2.1166-1172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Díaz A, Lacks S A, López P. Multiple roles for DNA polymerase I in establishment and replication of the promiscuous plasmid pLS1. Mol Microbiol. 1994;14:773–783. doi: 10.1111/j.1365-2958.1994.tb01314.x. [DOI] [PubMed] [Google Scholar]
- 77.Díaz R, Nordström K, Staudenbauer W L. Plasmid R1 DNA replication dependent on protein synthesis in cell-free extracts of E. coli. Nature. 1981;289:326–328. doi: 10.1038/289326a0. [DOI] [PubMed] [Google Scholar]
- 78.Díaz R, Staudenbauer W L. Origin and direction of mini-R1 plasmid DNA replication in cell extracts of Escherichia coli. J Bacteriol. 1982;150:1077–1084. doi: 10.1128/jb.150.3.1077-1084.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78a.Dibbens J A, Muraiso K, Chattoraj D K. Chaperone-mediated reduction of RepA dimerization is associated with RepA conformational change. Mol Microbiol. 1997;26:185–195. doi: 10.1046/j.1365-2958.1997.5691920.x. [DOI] [PubMed] [Google Scholar]
- 79.Dong X, Rovillard K P, Womble D D, Rownd R H. DNA bending near the replication origin of Inc FII plasmid NR1. J Bacteriol. 1989;171:703–707. doi: 10.1128/jb.171.2.703-707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79a.Doran, K. S., D. R. Helinski, and I. Konieczny. Unpublished data.
- 79b.Doran, K. S., I. Konieczny, and D. R. Helinski. Replication origin of the broad host range plasmid RK2: positioning of various motifs is critical for initiation of replication. J. Biol. Chem., in press. [DOI] [PubMed]
- 80.Durland R H, Helinski D R. The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid. 1987;18:164–169. doi: 10.1016/0147-619x(87)90044-8. [DOI] [PubMed] [Google Scholar]
- 81.Eckdahl T T, Anderson J N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990;18:1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eguchi Y, Itoh T, Tomizawa J. Antisense RNA. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- 83.Erauso G, Marsin S, Benbouzid-Rollet N, Baucher M F, Barbeyron T, Zivanovic Y, Prieur D, Fonterre P. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J Bacteriol. 1996;178:3232–3237. doi: 10.1128/jb.178.11.3232-3237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Espinosa M, del Solar G, Rojo F, Alonso J C. Plasmid rolling circle replication and its control. FEMS Microbiol Lett. 1995;130:111–120. doi: 10.1111/j.1574-6968.1995.tb07707.x. [DOI] [PubMed] [Google Scholar]
- 85.Fernández-Tresguerres M E, Martín M, García de Viedma D, Giraldo R, Díaz-Orejas R. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol. 1995;177:4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Filutowicz M, Uhlenhopp E, Helinski D R. Binding of purified wild-type and mutant Pi initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1985;187:225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- 87.Filutowicz M, Dellis S, Levchenko I, Urh M, Wu F, York D. Regulation of replication in an iteron-containing DNA molecule. Prog Nucleic Acid Res Mol Biol. 1994;68:239–273. doi: 10.1016/s0079-6603(08)60857-0. [DOI] [PubMed] [Google Scholar]
- 88.Filutowicz M, MacEachern M J, Helinski D R. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci USA. 1986;83:9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flashner Y, Shlomai J, Shafferman A. Three novel plasmid R6K proteins act in concert to distort DNA within the alpha and beta origins of DNA replication. Mol Microbiol. 1996;19:985–996. doi: 10.1046/j.1365-2958.1996.428960.x. [DOI] [PubMed] [Google Scholar]
- 90.Frey J, Bagdasarian M. The molecular biology of IncQ plasmids. In: Thomas C M, editor. Promiscuous plasmids of Gram-negative bacteria. London, United Kingdom: Academic Press, Ltd.; 1989. pp. 79–94. [Google Scholar]
- 91.Gamas P, Burger A C, Churchward G, Caro L, Galas D, Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol Gen Genet. 1986;204:85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- 92.García de Viedma D, Giraldo R, Ruiz-Echevarria M J, Lurz R, Diaz-Orejas R. Transcription of repA, the gene of the initiation protein of the Pseudomonas plasmid pPS10, is autoregulated by interactions of the RepA protein at a symmetrical operator. J Mol Biol. 1995;247:211–223. doi: 10.1006/jmbi.1994.0134. [DOI] [PubMed] [Google Scholar]
- 93.García de Viedma D, Serrano-Lopez A, Diaz-Orejas R. Specific binding of the replication protein of plasmid pPS10 to direct and inverted repeats is mediated by an HTH motif. Nucleic Acids Res. 1995;23:5048–5054. doi: 10.1093/nar/23.24.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.García de Viedma D, Giraldo R, Rivas G, Fernandez-Tresguerres E, Diaz-Orejas R. A leucine zipper motif determines different functions in a DNA replication protein. EMBO J. 1996;15:925–934. [PMC free article] [PubMed] [Google Scholar]
- 95.Genetics Computer Group. Wisconsin sequence analysis package, version 8.1. Madison, Wis: Genetics Computer Group; 1995. [Google Scholar]
- 96.Germino J, Bastia D. Primary structure of the replication initiation protein of plasmid R6K. Proc Natl Acad Sci USA. 1982;79:5475–5479. doi: 10.1073/pnas.79.18.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Germino J, Bastia D. The replication initiator protein of plasmid R6K tagged with beta-galactosidase shows sequence-specific DNA-binding. Cell. 1983;32:131–140. doi: 10.1016/0092-8674(83)90503-2. [DOI] [PubMed] [Google Scholar]
- 98.Germino J, Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983;34:125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- 99.Gielow A, Diederich L, Messer W. Characterization of a phage-plasmid hybrid (Phasyl) with two independent origins of replication isolated from Escherichia coli. J Bacteriol. 1991;173:73–79. doi: 10.1128/jb.173.1.73-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giraldo, R. Unpublished data.
- 101.Giraldo R, Díaz R. Differential binding of wild-type and mutant RepA protein to oriR sequence suggest a model for the initiation of plasmid R1 replication. J Mol Biol. 1992;228:787–802. doi: 10.1016/0022-2836(92)90864-g. [DOI] [PubMed] [Google Scholar]
- 102.Giraldo, R., J. M. Andreu, and R. Díaz. Unpublished data.
- 103.Giraldo R, Nieto C, Fernández-Tresguerres M E, Díaz R. Bacterial zipper. Nature. 1989;342:866. doi: 10.1038/342866a0. [DOI] [PubMed] [Google Scholar]
- 104.Giraldo R, Martín M, Fernández-Tresguerres M E, Nieto C, Díaz R. Mutations within the minimal replicon of plasmid pPS10 increase its host range. In: Hughes P, Fanning E, Kohiyama M, editors. DNA replication: the regulatory mechanisms. Berlin, Germany: Springer-Verlag KG; 1992. pp. 225–237. [Google Scholar]
- 105.Giraldo-Suárez R, Fernández-Tresguerres M E, Díaz-Orejas R, Malki A, Kohiyama M. The heat-shock DnaK protein is required for plasmid R1 replication and it is dispensable for plasmid ColE1 replication. Nucleic Acids Res. 1993;21:5495–5499. doi: 10.1093/nar/21.23.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grohmann E, Zechner E L, Espinosa M. Determination of specific DNA strand discontinuities with nucleotide resolution in exponentionally growing bacteria harbouring rolling circle-replicating plasmids. FEMS Microbiol Lett. 1997;152:363–369. doi: 10.1111/j.1574-6968.1997.tb10453.x. [DOI] [PubMed] [Google Scholar]
- 107.Gros M-F, te Riele H, Ehrlich S D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987;6:3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gruss A D, Ehrlich S D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gruss A D, Ross H, Novick R P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci USA. 1987;84:2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hansen E B, Yarmolinsky M B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci USA. 1986;83:4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haring V, Scherzinger E. Replication proteins of the IncQ plasmids. In: Thomas C M, editor. Promiscuous plasmids of Gram-negative bacteria. London, United Kingdom: Academic Press, Ltd.; 1989. pp. 95–124. [Google Scholar]
- 112.Haring V, Scholz P, Scherzinger E, Frey J, Derbyshire K, Hatfull G, Willetts N S, Bagdasarian M. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc Natl Acad Sci USA. 1985;82:6090–6094. doi: 10.1073/pnas.82.18.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hasunuma K, Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977;154:225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- 114.Haugan K, Karunakan P, Blatny J M, Valla S. The phenotypes of temperature-sensitive mini-RK2 replicons carrying mutations in the replication control gene trfA are suppressed nonspecifically by intragenic cop mutations. J Bacteriol. 1992;174:7026–7032. doi: 10.1128/jb.174.21.7026-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hefford M A, Teather R M, Forster R J. The complete nucleotide sequence of a small cryptic plasmid from a rumen bacterium of the genus Butyrivibrio. Plasmid. 1993;29:63–69. doi: 10.1006/plas.1993.1008. [DOI] [PubMed] [Google Scholar]
- 116.Hiasa H, Marians K J. Differential inhibition of the DNA translocation and DNA unwinding activities of DNA helicases by the Escherichia coli Tus protein. J Biol Chem. 1992;267:11379–11385. [PubMed] [Google Scholar]
- 117.Hiasa H, Marians K J. Topoisomerase IV can support oriC DNA replication in vitro. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 118.Higashi A, Sakai H, Honda Y, Tanaka T, Miao D M, Nakamura T, Taguchi Y, Komano T, Bagdasarian M. Functional features of oriV of the broad host range plasmid RSF1010 in Pseudomonas aeruginosa. Plasmid. 1994;31:196–200. doi: 10.1006/plas.1994.1020. [DOI] [PubMed] [Google Scholar]
- 119.Highlander S K, Novick R P. Mutational and physiological analyses of plasmid pT181 functions expressing incompatibility. Plasmid. 1990;23:1–15. doi: 10.1016/0147-619x(90)90039-f. [DOI] [PubMed] [Google Scholar]
- 120.Hill T M. Arrest of bacterial DNA replication. Annu Rev Microbiol. 1992;46:603–633. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 121.Hill T M, Tecklenburg M, Pelletier A, Kuempel P L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc Natl Acad Sci USA. 1989;86:1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hillenbrand G, Staudenbauer W L. Discriminatory function of ribonuclease H in the selective initiation of plasmid DNA replication. Nucleic Acids Res. 1982;10:834–853. doi: 10.1093/nar/10.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hinnebusch J, Tilly K. Linear plasmids and chromosomes in bacteria. Mol Microbiol. 1993;10:917–922. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 124.Hiraga S I, Sugiyama T, Itoh T. Comparative analysis of the replicon regions of eleven ColE2-related plasmids. J Bacteriol. 1994;176:7233–7234. doi: 10.1128/jb.176.23.7233-7243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hjalt T A H, Wagner E G H. Bulged-out nucleotides protect an antisense RNA from RNase III cleavage. Nucleic Acids Res. 1995;23:571–579. doi: 10.1093/nar/23.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hjalt T A H, Wagner E G H. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 1995;23:580–587. doi: 10.1093/nar/23.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Holmes M L, Pfeifer F, Dyall-Smith M L. Analysis of the halobacterial plasmid pHK2 minimal replicon. Gene. 1995;153:117–121. doi: 10.1016/0378-1119(94)00761-g. [DOI] [PubMed] [Google Scholar]
- 128.Honda Y, Sakai H, Komano T, Bagdasarian M. RepB′ is required in trans for the two single-strand DNA initiation signals in oriV of plasmid RSF1010. Gene. 1989;80:155–159. doi: 10.1016/0378-1119(89)90261-8. [DOI] [PubMed] [Google Scholar]
- 129.Honda Y, Sakai H, Hiasa H, Tanaka K, Komano T, Bagdasarian M. Functional division and reconstruction of a plasmid replication origin: molecular dissection of the oriV of the broad host-range plasmid RSF1010. Proc Natl Acad Sci USA. 1991;88:179–183. doi: 10.1073/pnas.88.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Horiuchi T, Hidaka M. Core sequence of two separable terminus sites is a 20 bp inverted repeat. Cell. 1988;54:515–523. doi: 10.1016/0092-8674(88)90073-6. [DOI] [PubMed] [Google Scholar]
- 131.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes, and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ingmer H, Cohen S N. The pSC101 par locus alters protein-DNA interactions in vivo at the plasmid replication origin. J Bacteriol. 1993;175:6046–6048. doi: 10.1128/jb.175.18.6046-6048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ingmer H, Cohen S N. Excess intracellular concentration of pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J Bacteriol. 1993;175:7834–7841. doi: 10.1128/jb.175.24.7834-7841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ingmer H, Fong E L, Cohen S N. Monomer-dimer equilibrium of the pSC101 RepA protein. J Mol Biol. 1995;250:309–314. doi: 10.1006/jmbi.1995.0378. [DOI] [PubMed] [Google Scholar]
- 135.Iordanescu S. Specificity of the interactions between the Rep proteins and the origins of replication of Staphylococcus aureus plasmids pT181 and pC221. Mol Gen Genet. 1989;217:481–487. doi: 10.1007/BF02464921. [DOI] [PubMed] [Google Scholar]
- 136.Iordanescu S. Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol Gen Genet. 1993;241:185–192. doi: 10.1007/BF00280216. [DOI] [PubMed] [Google Scholar]
- 137.Iordanescu S. Plasmid pT181-linked suppressors of the Staphylococcus aureus pcrA3 chromosomal mutation. J Bacteriol. 1993;175:3916–3917. doi: 10.1128/jb.175.12.3916-3917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iordanescu S, Basheer R. The Staphylococcus aureus mutation pcrA3 leads to the accumulation of pT181 replication initiation complexes. J Mol Biol. 1991;221:1183–1189. doi: 10.1016/0022-2836(91)90927-x. [DOI] [PubMed] [Google Scholar]
- 139.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci USA. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139a.Jannière L, Bruand C, Ehrlich S D. Structurally stable DNA cloning vectors. Gene. 1990;87:53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- 140.Jin R, Zhou X, Novick R P. The inactive pT181 initiator heterodimer, RepC/C*, binds but fails to induce melting of the plasmid replication origin. J Biol Chem. 1996;271:31086–31091. doi: 10.1074/jbc.271.49.31086. [DOI] [PubMed] [Google Scholar]
- 141.Jin R, Rasooly A, Novick R P. In vitro inhibitory activity of RepC/C*, the inactivated form of the pT181 plasmid initiation protein, RepC. J Bacteriol. 1997;179:141–147. doi: 10.1128/jb.179.1.141-147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jovanovic O S, Ayres E K, Figurski D H. Host-inhibitory functions encoded by promiscuous plasmids. Transient arrest of Escherichia coli segregants that fail to inherit plasmid RK2. J Mol Biol. 1994;237:52–64. doi: 10.1006/jmbi.1994.1208. [DOI] [PubMed] [Google Scholar]
- 143.Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Structure of a replication-terminator protein complexed with DNA. Nature. 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- 144.Kamio Y, Terawaki Y. Nucleotide sequence of an incompatibility region of mini-Rts1 that contains five direct repeats. J Bacteriol. 1983;155:1185–1191. doi: 10.1128/jb.155.3.1185-1191.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. Gene organization in a region containing a new gene involved in chromosome partitioning in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145a.Kelley W L, Bastia D. Conformational changes induced by integration host factor at origin gamma of R6K and copy control. J Biol Chem. 1991;266:15924–15937. [PubMed] [Google Scholar]
- 146.Kelley W L, Bastia D. Activation in vivo of the minimal replication origin beta of plasmid R6K requires a small target sequence essential for DNA looping. New Biol. 1992;4:569–580. [PubMed] [Google Scholar]
- 147.Kelley W L, Patel I, Bastia D. Structural and functional analysis of a replication enhancer: separation of the enhancer activity from origin function by mutational dissection of the replication origin gamma of plasmid R6K. Proc Natl Acad Sci USA. 1992;89:5078–5082. doi: 10.1073/pnas.89.11.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 149.Khan S A, Carleton S M, Novick R P. Replication of plasmid pT181 in vitro: requirement for a plasmid-encoded product. Proc Natl Acad Sci USA. 1981;78:4902–4906. doi: 10.1073/pnas.78.8.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Khan S A. Mechanism of replication and copy number control of plasmids in Gram-positive bacteria. Genet Eng. 1996;18:183–201. doi: 10.1007/978-1-4899-1766-9_11. [DOI] [PubMed] [Google Scholar]
- 150a.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khatri G S, MacAllister T, Sista P R, Bastia D. The replication terminator protein of Escherichia coli is a DNA sequence-specific contra-helicase. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- 152.Kiewiet R, Bron S, de Jonge K, Venema G, Seegers J F M L. Theta replication of the lactococcal plasmid pWVO2. Mol Microbiol. 1993;10:319–327. [PubMed] [Google Scholar]
- 153.Kim K, Meyer R J. Copy number of the broad host-range plasmid R1162 is determined by the amount of essential plasmid-encoded proteins. J Mol Biol. 1985;185:755–767. doi: 10.1016/0022-2836(85)90060-9. [DOI] [PubMed] [Google Scholar]
- 154.Kim K, Meyer R J. Copy number of broad host-range plasmid R1162 is regulated by a small RNA. Nucleic Acids Res. 1986;14:8027–8046. doi: 10.1093/nar/14.20.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kittell B L, Helinski D R. Plasmid incompatibility and replication control. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1992. pp. 223–242. [Google Scholar]
- 156.Kleanthous H, Clayton C L, Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in Gram-positive bacteria. Mol Microbiol. 1991;5:2377–2389. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- 156a.Koepsel R R, Murray R W, Khan S A. Sequence-specific interaction between the replication initiator protein of plasmid pT181 and its origin of replication. Proc Natl Acad Sci USA. 1986;83:5484–5488. doi: 10.1073/pnas.83.15.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156b.Koepsel R R, Murray R W, Rosenblum W D, Khan S A. Purification of pT181-encoded RepC protein required for the initiation of plasmid replication. J Biol Chem. 1985;260:8571–8577. [PubMed] [Google Scholar]
- 156c.Koepsel R R, Khan S A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986;233:1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- 157.Koepsel R R, Khan S A. Cleavage of single-stranded DNA by plasmid pT181-encoded RepC protein. Nucleic Acids Res. 1987;15:4085–4097. doi: 10.1093/nar/15.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kogoma T. Absence of RNase H allows replication of pBR322 in Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7845–7849. doi: 10.1073/pnas.81.24.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kollek R, Oertel W, Goebel W. Isolation and characterization of the minimal fragment required for autonomous replication (“basic replicon”) of a copy mutant (pKN102) of the antibiotic resistance factor R1. Mol Gen Genet. 1978;162:51–57. doi: 10.1007/BF00333850. [DOI] [PubMed] [Google Scholar]
- 160.Kolter R, Helinski D. The activity of the replication terminus of plasmid R6K in hybrid replicons in Escherichia coli. J Mol Biol. 1978;124:425–441. doi: 10.1016/0022-2836(78)90180-8. [DOI] [PubMed] [Google Scholar]
- 161.Kolter R, Yanowsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- 161a.Konieczny I, Doran K S, Helinski D R, Blasina A. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J Biol Chem. 1997;272:20173–20178. doi: 10.1074/jbc.272.32.20173. [DOI] [PubMed] [Google Scholar]
- 161b.Konieczny, I., and D. R. Helinski. Personal communication.
- 162.Koonin E V, Ilyina T V. Computer-assisted dissection of rolling circle DNA replication. BioSystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 163.Kornberg A, Baker T. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 164.Kozlowski M, Thatte V, Lau P C K, Visentin L P, Iyer V N. Isolation and structure of the replicon of the promiscuous plasmid pCU1. Gene. 1987;58:217–228. doi: 10.1016/0378-1119(87)90377-5. [DOI] [PubMed] [Google Scholar]
- 165.Kramer M G, del Solar G, Espinosa M. Lagging-strand origins of the promiscuous plasmid pMV158: physical and functional characterization. Microbiology. 1995;141:655–662. doi: 10.1099/13500872-141-3-655. [DOI] [PubMed] [Google Scholar]
- 166.Kramer M G, Khan S A, Espinosa M. Plasmid rolling circle replication: identification of the RNA polymerase-directed primer RNA and requirement of DNA polymerase I for lagging strand initiation. EMBO J. 1997;16:5784–5795. doi: 10.1093/emboj/16.18.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kramer M G, Khan S A, Espinosa M. Lagging-strand replication from the ssoA origin of plasmid pMV158 in Streptococcus pneumoniae: in vivo and in vitro influences of mutations in two conserved ssoA regions. J Bacteriol. 1998;180:83–89. doi: 10.1128/jb.180.1.83-89.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kubota Y-H, Arai K-I, Masai H. Roles of the G-site and φX174-type primosome assembly site in priming of leading-strand synthesis: initiation by a mobile primosome and replication-fork arrest by RepA protein bound to oriR. Gene. 1993;126:9–16. doi: 10.1016/0378-1119(93)90584-p. [DOI] [PubMed] [Google Scholar]
- 168a.Kües U, Stahl U. Replication of plasmids in gram-negative bacteria. Microbiol Rev. 1989;53:491–516. doi: 10.1128/mr.53.4.491-516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kumar C, Novick R P. Plasmid pT181 replication is regulated by two countertranscripts. Proc Natl Acad Sci USA. 1985;82:638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lacatena R M, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 171.Lacks S A, López P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 172.Landschultz W H, Johnson P F, Mcknight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 172a.Lee E H, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc Natl Acad Sci USA. 1989;86:9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leenhouts K J, Tolner B, Bron S, Kok J, Venema G, Seegers J F M L. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid. 1991;26:55–66. doi: 10.1016/0147-619x(91)90036-v. [DOI] [PubMed] [Google Scholar]
- 174.Levchenco I, Filutowicz M. Initiator protein Pi can bind independently to two domains of the gamma origin core of plasmid R6K: the direct repeats and the A+T-rich segment. Nucleic Acids Res. 1996;24:1936–1942. doi: 10.1093/nar/24.10.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin J, Helinski D R. Analysis of mutations in trfA, the replication initiation gene of the broad-host-range plasmid RK2. J Bacteriol. 1992;174:4110–4119. doi: 10.1128/jb.174.12.4110-4119.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lin L S, Meyer R J. Directly repeated, 20 bp sequence of plasmid R1162 is required for replication, expression of incompatibility, and copy-number control. Plasmid. 1986;15:35–47. doi: 10.1016/0147-619x(86)90012-0. [DOI] [PubMed] [Google Scholar]
- 177.Lovett M A, Sparks R B, Helinski D R. Bidirectional replication of plasmid R6K DNA in Escherichia coli: correspondence between origin of replication and position of single-stranded break in relaxed complex. Proc Natl Acad Sci USA. 1975;72:2905–2909. doi: 10.1073/pnas.72.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Maciag I E, Viret J-F, Alonso J C. Replication and incompatibility properties of plasmid pUB110 in Bacillus subtilis. Mol Gen Genet. 1988;212:232–240. doi: 10.1007/BF00334690. [DOI] [PubMed] [Google Scholar]
- 179.Maeser S, Scholz P, Otto S, Scherzinger E. Gene F of plasmid RSF1010 codes for a low-molecular-weight repressor protein that autoregulates expression of the repAC operon. Nucleic Acids Res. 1990;18:6215–6222. doi: 10.1093/nar/18.21.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Maestro, B. Personal communication.
- 181.Manen D, Upegui-Gonzalez L C, Caro L. Monomers and dimers of the RepA protein in plasmid pSC101 replication: domains in RepA. Proc Natl Acad Sci USA. 1992;89:8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Marians K J. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 183.Masai H, Kaziro Y, Arai K-I. Definition of oriR, the minimum DNA segment essential for initiation of R1 plasmid replication in vivo. Proc Natl Acad Sci USA. 1983;80:6814–6818. doi: 10.1073/pnas.80.22.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Masai M, Arai K-I. RepA and DnaA proteins are required for initiation of R1 plasmid replication in vitro and interact with the oriR sequence. Proc Natl Acad Sci USA. 1987;84:4781–4785. doi: 10.1073/pnas.84.14.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Masai M, Arai K I. R1 plasmid replication in vitro: RepA- and DnaA-dependent initiation at oriR. In: Moses R E, Sammers W C, editors. DNA replication and mutagenesis. Washington, D.C: American Society for Microbiology; 1988. pp. 113–121. [Google Scholar]
- 186.Masai H, Arai K I. Leading strand synthesis of R1 plasmid replication in vitro is primed by primase alone at a specific site downstream of oriR. J Biol Chem. 1989;264:8082–8090. [PubMed] [Google Scholar]
- 187.Masai H, Nomura N, Kubota Y, Arai K I. Roles of φX174 type primase-dependent priming in initiation of lagging and leading strand synthesis of DNA replication. J Biol Chem. 1990;265:15124–15133. [PubMed] [Google Scholar]
- 188.Masukata H, Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984;36:513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- 189.Masukata H, Tomizawa J. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell. 1990;62:331–338. doi: 10.1016/0092-8674(90)90370-t. [DOI] [PubMed] [Google Scholar]
- 190.McEachern M J, Filutowicz M, Yang S, Greener A, Mukhopadyay P, Helinski D R. Elements involved in the copy number regulation of the antibiotic resistance plasmid R6K. Banbury Rep. 1986;24:195–204. [Google Scholar]
- 191.McEachern M J, Bott M A, Tooker P A, Helinski D R. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci USA. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Meijer W J J, de Boer A J, van Tongeren S, Venema G, Bron S. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 1995;23:3214–3223. doi: 10.1093/nar/23.16.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Meijer W J J, Venema G, Bron S. Characterization of single strand origins of cryptic rolling-circle plasmids from Bacillus subtilis. Nucleic Acids Res. 1995;23:612–619. doi: 10.1093/nar/23.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Meijer W J J. Alternative, RecA-mediated pathway for the initiation of lagging strand synthesis of the broad-host-range rolling circle plasmid pMV158 in Bacillus subtilis. Ph.D. thesis. Groningen, The Netherlands: R.U. University Groningen; 1995. [Google Scholar]
- 195.Miao D-M, Honda Y, Tanaka K, Higashi A, Nakamura T, Taguchi Y, Sakai H, Komano T, Bagdasarian M. A base-paired hairpin structure essential for the functional priming signal for DNA replication of the broad host-range plasmid RSF1010. Nucleic Acids Res. 1993;21:4900–4903. doi: 10.1093/nar/21.21.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Michel B, Ehrlich S D. Illegitimate recombination at the replication origin of the plasmid pC194. EMBO J. 1986;5:3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Miller C A, Ingmer H, Cohen S N. Boundaries of the pSC101 minimal replicon are conditional. J Bacteriol. 1995;177:4865–4871. doi: 10.1128/jb.177.17.4865-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Minden J S, Marians K J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985;261:11906–11917. [PubMed] [Google Scholar]
- 199.Miron A, Mukherjee S, Bastia D. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J. 1992;11:1205–1206. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Moscoso M, del Solar G, Espinosa M. Specific nicking-closing activity of the initiator of replication protein RepB of plasmid pMV158 on supercoiled or single-stranded DNA. J Biol Chem. 1995;270:3772–3779. doi: 10.1074/jbc.270.8.3772. [DOI] [PubMed] [Google Scholar]
- 201.Moscoso M, del Solar G, Espinosa M. In vitro recognition of the replication origin of pLS1 and of plasmids of the pLS1 family by the RepB initiator protein. J Bacteriol. 1995;177:7041–7049. doi: 10.1128/jb.177.24.7041-7049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moscoso M, Eritja R, Espinosa M. Initiation of replication of plasmid pMV158: mechanisms of DNA strand transfer reactions mediated by the initiator RepB protein. J Mol Biol. 1997;268:840–856. doi: 10.1006/jmbi.1997.1012. [DOI] [PubMed] [Google Scholar]
- 203.Mukherjee S, Erickson H, Bastia D. Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci USA. 1988;85:6287–6291. doi: 10.1073/pnas.85.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mukherjee S, Erickson H, Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- 205.Mukherjee S, Patel I, Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985;43:189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- 206.Mukhopadhyay G, Carr K M, Kaguni J M, Chattoraj D K. Open-complex formation by the host initiator, DnaA, at the origin of P1 plasmid replication. EMBO J. 1993;12:4547–4554. doi: 10.1002/j.1460-2075.1993.tb06143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Müller A K, Rojo F, Alonso J C. The level of the pUB110 replication initiator protein is autoregulated, which provides an additional control for plasmid copy number. Nucleic Acids Res. 1995;23:1894–1900. doi: 10.1093/nar/23.11.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Müller R E, Ano T, Imanaka T, Aiba S. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1, and its copy mutants. Mol Gen Genet. 1986;202:169–171. doi: 10.1007/BF00330534. [DOI] [PubMed] [Google Scholar]
- 209.Murotsu T, Matsubara K, Sugisaki H, Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981;15:257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- 210.Murray R W, Koepsel R R, Khan S A. Synthesis of single-stranded plasmid pT181 DNA in vitro. J Biol Chem. 1989;264:1051–1057. [PubMed] [Google Scholar]
- 211.Naito S, Uchida H. RNase H and replication of ColE1 DNA in Escherichia coli. J Bacteriol. 1986;166:143–147. doi: 10.1128/jb.166.1.143-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nakasu S, Tomizawa J. Structure of the ColE1 DNA molecule before segregation to daughter molecules. Proc Natl Acad Sci USA. 1992;89:10139–10143. doi: 10.1073/pnas.89.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nash J, Krishnapillai V. DNA sequence analysis of host range mutants of the promiscuous incP-1 plasmid R18 and R86 with Tn7. Plasmid. 1987;18:35–45. doi: 10.1016/0147-619x(87)90076-x. [DOI] [PubMed] [Google Scholar]
- 214.Natarajan S, Kaul S, Miron A, Bastia D. A 27 kd protein of Escherichia coli promotes antitermination of replication in vitro at a sequence-specific replication terminus. Cell. 1993;72:113–120. doi: 10.1016/0092-8674(93)90055-u. [DOI] [PubMed] [Google Scholar]
- 215.Nieto C, Giraldo R, Fernández-Tresguerres E, Díaz R. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae pathovar savastanoi. J Mol Biol. 1992;223:415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- 216.Noirot P, Bargonetti J, Novick R P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci USA. 1990;87:8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Noirot-Gros M F, Bidnenko V, Ehrlich S D. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 1994;13:4412–4420. doi: 10.1002/j.1460-2075.1994.tb06761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Noirot-Gros M F, Ehrlich S D. Change of a catalytic reaction carried out by a DNA replication protein. Science. 1996;274:777–780. doi: 10.1126/science.274.5288.777. [DOI] [PubMed] [Google Scholar]
- 219.Nomura N, Masai H, Inuzuka M, Miyazaki C, Ohtsubo E, Itoh T, Sasamoto S, Matsui M, Ishizaki R, Arai K I. Identification of eleven single-strand initiation sequences (ssi) for priming of DNA replication in the F, R6K, R100, and ColE2 plasmids. Gene. 1991;108:15–22. doi: 10.1016/0378-1119(91)90482-q. [DOI] [PubMed] [Google Scholar]
- 220.Nomura N, Ray D S. Expression of DNA strand initiation sequence of ColE1 plasmid in a single-stranded DNA phage. Proc Natl Acad Sci USA. 1980;77:6566–6570. doi: 10.1073/pnas.77.11.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nordström K. Control of plasmid replication. Plasmid. 1983;9:1–7. doi: 10.1016/0147-619x(83)90026-4. [DOI] [PubMed] [Google Scholar]
- 222.Nordström K. Control of plasmid replication: theoretical considerations and practical solutions. In: Helinski D R, Cohen S N, Clewell D B, Jackson D A, Hollaender A, editors. Plasmids in bacteria. New York, N.Y: Plenum Press; 1985. pp. 189–214. [DOI] [PubMed] [Google Scholar]
- 223.Nordström K. Control of plasmid replication. How do DNA iterons set the replication frequency? Cell. 1990;63:1121–1124. doi: 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- 224.Nordström K, Molin S, Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984;12:71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- 225.Nordström K, Ingram L C, Lundbäck A. Mutations in R factors of Escherichia coli causing an increased number of R-factor copies per chromosome. J Bacteriol. 1972;110:562–569. doi: 10.1128/jb.110.2.562-569.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nordström K, Wagner E G H. Kinetic aspect of control of plasmid replication by antisense RNA. Trends Biochem Sci. 1994;19:294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 227.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Novick R P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 229.Novick R P, Adler G K, Projan S J, Carleton S M, Highlander S K, Gruss S A, Khan S A, Iordanescu S. Control of pT181 replication. I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984;3:2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Novick R P, Iordanescu S, Projan S J, Kornblum J, Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989;59:395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 230a.Núñez B, Avila P, de la Cruz F. Genes involved in conjugative DNA processing of plasmid R6K. Mol Microbiol. 1997;24:1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 231.Ortega S, Lanka E, Díaz R. The involvement of host replication proteins and of specific origin sequences in the in vitro replication of miniplasmid R1 DNA. Nucleic Acids Res. 1986;14:4865–4879. doi: 10.1093/nar/14.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ortega S, de Torrontegui G, Díaz R. Isolation and characterization of a conditional replication mutant of the antibiotic resistance factor R1 affected in the gene of the replication protein repA. Mol Gen Genet. 1989;217:111–117. doi: 10.1007/BF00330949. [DOI] [PubMed] [Google Scholar]
- 233.Ortega-Jiménez S, Giraldo-Suárez R, Fernández-Tresguerres M E, Berzal-Herranz A, Díaz-Orejas R. DnaA dependent replication of plasmid R1 occurs in the presence of point mutations that disrupt the dnaA box of oriR. Nucleic Acids Res. 1992;20:2547–2551. doi: 10.1093/nar/20.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ozaki E, Yashubara H, Masamune Y. Purification of pKYM-encoded RepK, a protein required for the initiation of plasmid replication. J Gen Appl Microbiol. 1994;40:365–375. [Google Scholar]
- 235.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 236.Pak M, Wickner S. Mechanism of protein remodeling by ClpA chaperone. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Pal S, Mason R J, Chattoraj D K. Role of initiator titration in copy number control. J Mol Biol. 1986;192:275–285. doi: 10.1016/0022-2836(86)90364-5. [DOI] [PubMed] [Google Scholar]
- 238.Pal S K, Chattoraj D K. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988;170:3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Pansegrau W, Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991;19:3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pansegrau W, Lanka E. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPa plasmid RP4. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 241.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J Mol Biol. 1995;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 242.Papp P P, Chattoraj D K, Schneider T D. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993;233:219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- 243.Patel I, Bastia D. A replication origin is turned off by an origin “silencer” sequence. Cell. 1986;47:785–792. doi: 10.1016/0092-8674(86)90521-0. [DOI] [PubMed] [Google Scholar]
- 244.Patel I, Bastia D. A replication initiator protein enhances the rate of hybrid formation between a silencer RNA and an activator RNA. Cell. 1987;51:455–462. doi: 10.1016/0092-8674(87)90641-6. [DOI] [PubMed] [Google Scholar]
- 245.Peng H, Marians K J. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc Natl Acad Sci USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Peng H, Marians K J. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 247.Pérez-Casal J, Gammie A E, Crosa J H. Nucleotide sequence analysis and expression of the minimum RepI replication region and incompatibility determinants of pCo9lV-K30. J Bacteriol. 1989;171:2195–2201. doi: 10.1128/jb.171.4.2195-2201.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 249.Pouwels P H, van Luijk N, Leer R J, Posno M. Control of replication of the Lactobacillus pentosus plasmid p353-2: coding for the replication protein. Mol Gen Genet. 1994;242:614–622. doi: 10.1007/BF00285285. [DOI] [PubMed] [Google Scholar]
- 250.Priebe S D, Lacks S A. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J Bacteriol. 1989;171:4778–4784. doi: 10.1128/jb.171.9.4778-4784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pritchard R H. Control of DNA replication in bacteria. In: Molineux I, Kohiyama M, editors. DNA synthesis: present and future. New York, N.Y: Plenum Press; 1978. pp. 1–22. [Google Scholar]
- 252.Pritchard R H, Barth P T, Collins J. Control of DNA synthesis in bacteria. Symp Soc Gen Microbiol. 1969;19:263–297. [Google Scholar]
- 253.Projan S J, Novick R P. Comparative analysis of five related staphylococcal plasmids. Plasmid. 1988;19:203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 254.Puyet A, del Solar G, Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucleic Acids Res. 1988;16:115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rasooly A, Novick R P. Replication-specific inactivation of the pT181 plasmid initiator protein. Science. 1993;262:1048–1050. doi: 10.1126/science.8235621. [DOI] [PubMed] [Google Scholar]
- 256.Rasooly A, Projan S J, Novick R P. Plasmids of the pT181 family show replication-specific initiator protein modification. J Bacteriol. 1994;176:2450–2453. doi: 10.1128/jb.176.8.2450-2453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rasooly A, Wang P-Z, Novick R P. Replication-specific conversion of the Staphylococcus aureus pT181 initiator protein from an active homodimer to an inactive heterodimer. EMBO J. 1994;13:5245–5251. doi: 10.1002/j.1460-2075.1994.tb06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Rasooly A, Rasooly R S. Modification of the plasmid initiator protein RepC active site during replication. FEMS Microbiol Lett. 1996;145:245–253. doi: 10.1111/j.1574-6968.1996.tb08585.x. [DOI] [PubMed] [Google Scholar]
- 258a.Ratnakar P V, Mohanty B K, Lobert M, Bastia D. The replication initiator protein Pi of plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc Natl Acad Sci USA. 1996;86:3026–3030. doi: 10.1073/pnas.93.11.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rosenfeld R, Grover N B. Control of mini-R1 plasmid replication: a computer simulation. Plasmid. 1993;29:94–116. doi: 10.1006/plas.1993.1012. [DOI] [PubMed] [Google Scholar]
- 260.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 261.Sahoo T, Mohanty B K, Manna A, Lobert M, Bastia D. The contrahelicase activities of the replication terminator proteins of Escherichia coli and Bacillus subtilis are helicase-specific and impede both helicase translocation and authentic DNA unwinding. J Biol Chem. 1995;270:29138–29144. doi: 10.1074/jbc.270.49.29138. [DOI] [PubMed] [Google Scholar]
- 262.Sahoo T, Mohanty B K, Patel I, Bastia D. Termination of DNA replication in vitro. Requirement for stereospecific interactions between two dimers of the replication termination protein of Bacillus subtilis and with the terminator site to elicit polar contrahelicase and fork impedence. EMBO J. 1995;14:619–628. doi: 10.1002/j.1460-2075.1995.tb07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sakai H, Komano T. DNA replication of the IncQ broad-host-range plasmids in gram-negative bacteria. Biosci Biotechnol Biochem. 1996;60:377–382. doi: 10.1271/bbb.60.377. [DOI] [PubMed] [Google Scholar]
- 264.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 265.Scherzinger E, Bagdasarian M M, Scholz P, Lurz R, Rückert B, Bagdasarian M. Replication of the broad host-range plasmid RSF1010: requirement for three plasmid encoded proteins. Proc Natl Acad Sci USA. 1984;81:654–658. doi: 10.1073/pnas.81.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Scherzinger E, Haring V, Lurz R, Otto S. Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA, RepB and RepC proteins. Nucleic Acids Res. 1991;19:1203–1211. doi: 10.1093/nar/19.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–288. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 268.Seegers J F M L, Zhao A C, Meijer W J J, Khan S A, Venema G, Bron S. Structural and functional analysis of the single-strand origin of replication from the lactococcal plasmid pWV01. Mol Gen Genet. 1995;249:43–50. doi: 10.1007/BF00290234. [DOI] [PubMed] [Google Scholar]
- 269.Seufert W, Dobrinski B, Lurz R, Messer W. Functionality of DnaA protein binding site in DNA replication is orientation-dependent. J Biol Chem. 1988;263:2719–2723. [PubMed] [Google Scholar]
- 270.Seufert W, Messer W. DnaA protein binding to the plasmid origin region can substitute for primosome assembly during replication of pBR322 in vitro. Cell. 1987;48:73–78. doi: 10.1016/0092-8674(87)90357-6. [DOI] [PubMed] [Google Scholar]
- 271.Seufert W, Lurz R, Messer W. A novel replicon occurring naturally in Escherichia coli is a phage-plasmid hybrid. EMBO J. 1988;7:4005–4010. doi: 10.1002/j.1460-2075.1988.tb03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Shafferman A, Helinski D R. Structural properties of the beta origin of replication of plasmid R6K. J Biol Chem. 1983;258:4083–4090. [PubMed] [Google Scholar]
- 273.Shah D S, Cross M A, Porter D, Thomas C M. Dissection of the core and auxiliary sequences in the vegetative replication origin of promiscuous plasmid RK2. J Mol Biol. 1995;254:608–622. doi: 10.1006/jmbi.1995.0642. [DOI] [PubMed] [Google Scholar]
- 274.Shingler V, Thomas C M. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984;175:229–249. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- 275.Sista P R, Mukherjee S, Patel P, Khatri G S, Bastia D. A host-encoded DNA-binding protein promotes termination of plasmid replication at a sequence-specific replication terminus. Proc Natl Acad Sci USA. 1989;86:3026–3030. doi: 10.1073/pnas.86.9.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Som T, Tomizawa J. Regulatory regions of ColE1 that are involved in determination of plasmid copy number. Proc Natl Acad Sci USA. 1983;80:2257–2261. doi: 10.1073/pnas.80.11.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Stalker D M, Kolter R, Helinski D R. Nucleotide sequence of the region of an origin of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci USA. 1979;76:1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Stalker D M, Kolter R, Helinski D R. Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J Mol Biol. 1982;161:33–43. doi: 10.1016/0022-2836(82)90276-5. [DOI] [PubMed] [Google Scholar]
- 279.Stalker D M, Thomas C M, Helinski D R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181:8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- 280.Staudenbauer W L. Structure and replication of the colicin E1 plasmid. Curr Top Microbiol Immunol. 1978;83:93–156. doi: 10.1007/978-3-642-67087-9_3. [DOI] [PubMed] [Google Scholar]
- 281.Staudenbauer W L, Scherzinger H, Lanka E. Replication of the colicin E1 plasmid in extracts of Escherichia coli: uncoupling of leading strand from lagging strand synthesis. Mol Gen Genet. 1979;177:113–120. doi: 10.1007/BF00267260. [DOI] [PubMed] [Google Scholar]
- 282.Stenzel T T, MacAllister T, Bastia D. Cooperativity at a distance promoted by the combined action of two replication initiator proteins and a DNA bending protein at the replication origin of pSC101. Genes Dev. 1991;5:1453–1463. doi: 10.1101/gad.5.8.1453. [DOI] [PubMed] [Google Scholar]
- 283.Stenzel T T, Patel P, Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987;49:709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- 284.Sugiura S, Ohkubo S, Yamaguchi K. Minimal essential origin of plasmid pSC101 replication: requirement of a region downstream of iterons. J Bacteriol. 1993;175:5993–6001. doi: 10.1128/jb.175.18.5993-6001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Swindells M B. Identification of a common fold in the replication terminator protein suggests a possible model for DNA binding. Trends Biochem Sci. 1995;20:300–302. doi: 10.1016/s0968-0004(00)89055-6. [DOI] [PubMed] [Google Scholar]
- 286.Tait R C, Kado C I, Rodriguez R L. A comparison of the origin of replication of pSa with R6K. Mol Gen Genet. 1983;192:32–38. doi: 10.1007/BF00327643. [DOI] [PubMed] [Google Scholar]
- 287.Takechi S, Yasueda H, Itoh T. Control of ColE2 plasmid replication: regulation of Rep expression by a plasmid-coded antisense RNA. Mol Gen Genet. 1994;244:49–56. doi: 10.1007/BF00280186. [DOI] [PubMed] [Google Scholar]
- 288.Takechi S, Matsui H, Itoh T. Primer RNA synthesis by plasmid-specific Rep protein for initiation of ColE2 DNA replication. EMBO J. 1995;14:5141–5147. doi: 10.1002/j.1460-2075.1995.tb00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tanaka K, Kino K, Taguchi Y, Miao D M, Honda Y, Sakai H, Komano T, Bagdasarian M. Functional difference between the two oppositely oriented priming signals essential for the initiation of the broad host-range plasmid RSF1010 DNA replication. Nucleic Acids Res. 1994;22:767–772. doi: 10.1093/nar/22.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tang X B, Womble D D, Rownd R H. DnaA protein is not essential for replication of incF II plasmid NR1. J Bacteriol. 1989;171:5290–5295. doi: 10.1128/jb.171.10.5290-5295.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.te Riele H, Michel B, Ehrlich S D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci USA. 1986;83:2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thomas C D, Balson D F, Shaw W V. In vitro studies of the initiation of staphylococcal plasmid replication. J Biol Chem. 1990;265:5519–5530. [PubMed] [Google Scholar]
- 293.Thomas C D, Nikiforov T T, Connolly B A, Shaw W V. Determination of sequence specificity between a plasmid replication initiator protein and the origin of replication. J Mol Biol. 1995;254:381–391. doi: 10.1006/jmbi.1995.0625. [DOI] [PubMed] [Google Scholar]
- 293a.Thomas C M. Recent studies on the control of plasmid replication. Biochim Biophys Acta. 1988;949:253–263. doi: 10.1016/0167-4781(88)90150-9. [DOI] [PubMed] [Google Scholar]
- 294.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Tolun A, Helinski D R. Direct repeats of the plasmid incC region express F incompatibility. Cell. 1981;24:687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- 296.Tomizawa J, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA1 with primer transcript. Proc Natl Acad Sci USA. 1981;78:6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tomizawa J, Sakakibara Y, Kakefuda T. Replication of colicin E1 plasmid DNA in cell extracts. Origin and direction of replication. Proc Natl Acad Sci USA. 1974;71:2260–2264. doi: 10.1073/pnas.71.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tomizawa J, Sakakibara Y, Kakefuda T. Replication of colicin E1 plasmid DNA added to cell extracts. Proc Natl Acad Sci USA. 1975;72:1050–1054. doi: 10.1073/pnas.72.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Tomizawa J. Two distinct mechanisms of synthesis of DNA fragments on colicin E1 plasmid DNA. Nature. 1975;257:253–254. doi: 10.1038/257253a0. [DOI] [PubMed] [Google Scholar]
- 300.Tomizawa J, Ohmori H, Bird R E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci USA. 1977;74:1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Tomizawa J. Replication of colicin E1 plasmid DNA in vitro. In: Molineux I, Kohiyama M, editors. DNA synthesis: present and future. New York, N.Y: Plenum Publishing Co.; 1978. pp. 797–826. [Google Scholar]
- 302.Trawick J D, Kline C. A two-stage molecular model for control of mini-F replication. Plasmid. 1985;13:59–69. doi: 10.1016/0147-619x(85)90056-3. [DOI] [PubMed] [Google Scholar]
- 303.Tsutsui H, Fujiyama A, Murotsu T, Matsubara K. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J Bacteriol. 1983;155:337–344. doi: 10.1128/jb.155.1.337-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Twigg A J, Sherrat D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980;283:216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- 305.Uhlin B E, Nordström K. A runaway-replication mutant of plasmid R1drd-19: temperature-dependent loss of copy number control. Mol Gen Genet. 1978;165:167–179. doi: 10.1007/BF00269904. [DOI] [PubMed] [Google Scholar]
- 306.van der Lelie D, Bron S, Venema G, Oskam L. Similarity of minus origins of replication and flanking open reading frames of plasmid pUB110, pTB913 and pMV158. Nucleic Acids Res. 1989;17:7283–7294. doi: 10.1093/nar/17.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Viguera E, Hernandez P, Krimer D B, Boistov A S, Lurz R, Alonso J C, Schvartzman J B. The ColE1 unidirectional origin acts as polar replication fork pausing site. J Biol Chem. 1996;271:22414–22421. doi: 10.1074/jbc.271.37.22414. [DOI] [PubMed] [Google Scholar]
- 308.Vocke C, Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983;35:495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- 309.Wagner E G H, Simons R W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 310.Waters V L, Guiney D G. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol. 1993;9:1123–1130. doi: 10.1111/j.1365-2958.1993.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 311.Wang P-Z, Projan S J, Henriquez V, Novick R P. Specificity of origin recognition by replication initiator protein in plasmids of the pT181 family is determined by a six amino acid residue element. J Mol Biol. 1992;223:145–158. doi: 10.1016/0022-2836(92)90722-v. [DOI] [PubMed] [Google Scholar]
- 312.Wang P-Z, Projan S J, Henriquez V, Novick R P. Origin recognition specificity in pT181 plasmid is determined by a functionally asymmetric palindromic DNA element. EMBO J. 1993;12:45–52. doi: 10.1002/j.1460-2075.1993.tb05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312a.Wickner S. Three Escherichia coli heat shock proteins are required for P1 plasmid DNA replication: formation of an active complex between E. coli DnaJ protein and the P1 initiator protein. Proc Natl Acad Sci USA. 1990;87:3690–3694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Wickner S, Chattoraj D. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Wickner S, Hoskins J, Chattoraj D, McKenney K. Deletion analysis of the mini-P1 plasmid origin of replication and the role of Escherichia coli DnaA protein. J Biol Chem. 1990;165:11622–11627. [PubMed] [Google Scholar]
- 315.Wickner S, Hoskins J, McKenney K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature. 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 316.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Woelker B, Messer W. The structure of the initiation complex at the replication origin, oriC, of Escherichia coli. Nucleic Acids Res. 1993;21:5025–5033. doi: 10.1093/nar/21.22.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms. Proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 320.Womble D D, Rownd R H. Regulation of IncFII plasmid DNA replication. A quantitative model for control of plasmid NR1 replication in the bacterial cell division cycle. J Mol Biol. 1986;192:529–548. doi: 10.1016/0022-2836(86)90274-3. [DOI] [PubMed] [Google Scholar]
- 321.Womble D D, Rownd R H. Regulation of mini-F plasmid DNA replication. A quantitative model for control of plasmid mini-F replication in the bacterial cell division cycle. J Mol Biol. 1987;195:99–113. doi: 10.1016/0022-2836(87)90330-5. [DOI] [PubMed] [Google Scholar]
- 322.Yasueda H, Horii T, Itoh T. Structural and functional organization of ColE2 and ColE3 replicons. Mol Gen Genet. 1989;215:209–216. doi: 10.1007/BF00339719. [DOI] [PubMed] [Google Scholar]
- 323.Yasueda H, Takechi S, Sugiyama T, Itoh T. Control of ColE2 plasmid replication: negative regulation of the expression of the plasmid-specified initiator protein, Rep, at a posttranscriptional step. Mol Gen Genet. 1994;244:41–48. doi: 10.1007/BF00280185. [DOI] [PubMed] [Google Scholar]
- 324.Yasukawa H, Hase T, Sakai A, Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a Gram-negative bacterium. Proc Natl Acad Sci USA. 1991;88:10282–10286. doi: 10.1073/pnas.88.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.York D, Filutowicz M. Autoregulation-deficient mutant of the plasmid R6K-encoded pi protein distinguishes between palindromic and nonpalindromic binding sites. J Biol Chem. 1993;29:21854–21861. [PubMed] [Google Scholar]
- 326.Zavitz K H, Marians K J. Dissecting the functional role of PriA protein-catalyzed primosome assembly in Escherichia coli DNA replication. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 327.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 328.Zechner E L, Prüger H, Grohmann E, Espinosa M, Högenauer G. Specific cleavage of chromosomal and plasmid DNA strands in Gram-positive and Gram-negative bacteria can be detected with nucleotide resolution. Proc Natl Acad Sci USA. 1997;94:7435–7440. doi: 10.1073/pnas.94.14.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhao A C, Khan S A. An 18-base pair sequence is sufficient for termination of rolling-circle replication of plasmid pT181. J Bacteriol. 1996;178:5222–5228. doi: 10.1128/jb.178.17.5222-5228.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Zhao A C, Khan S A. Sequence requirements for the termination of rolling-circle replication of plasmid pT181. Mol Microbiol. 1997;24:535–544. doi: 10.1046/j.1365-2958.1997.3641730.x. [DOI] [PubMed] [Google Scholar]
- 331.Zillig W, Kletzin A, Schleper C, Holz I, Janekovick D, Hain J, Lanzendörfer M, Kristjansson J K. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol. 1994;16:609–628. [Google Scholar]
- 332.Zock J M, Birch P, Khan S A. Specificity of RepC protein in plasmid pT181 DNA replication. J Biol Chem. 1990;265:3484–3488. [PubMed] [Google Scholar]