Abstract
Background
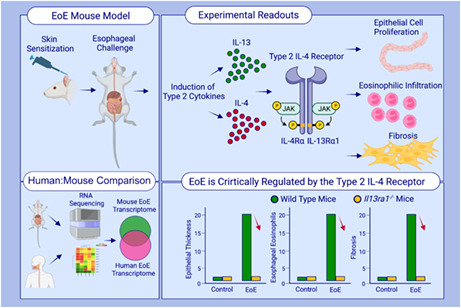
Eosinophilic esophagitis (EoE) is a chronic, food‐driven allergic disease, characterized by eosinophil‐rich inflammation in the esophagus. The histopathological and clinical features of EoE have been attributed to overproduction of the type 2 cytokines IL‐4 and IL‐13, which mediate profound alterations in the esophageal epithelium and neutralizing of their shared receptor component (IL‐4Rα) with a human antibody drug (dupilumab) demonstrates clinical efficacy. Yet, the relative contribution of IL‐4 and IL‐13 and whether the type II IL‐4 receptor (comprised of the IL‐4Rα chain in association with IL‐13Rα1) mediates this effect has not been determined.
Methods
Experimental EoE was induced in WT, Il13ra1 −/− , and Krt14 Cre /Il13ra1 fl/fl mice by skin‐sensitized using 4‐ethoxymethylene‐2‐phenyl‐2‐oxazolin (OXA) followed by intraesophageal challenges. Esophageal histopathology was determined histologically. RNA was extracted and sequenced for transcriptome analysis and compared with human EoE RNAseq data.
Results
Induction of experimental EoE in mice lacking Il13ra1 and in vivo IL‐13 antibody‐based neutralization experiments blocked antigen‐induced esophageal epithelial and lamina propria thickening, basal cell proliferation, eosinophilia, and tissue remodeling. In vivo targeted deletion of Il13ra1 in esophageal epithelial cells rendered mice protected from experimental EoE. Single‐cell RNA sequencing analysis of human EoE biopsies revealed predominant expression of IL‐13Rα1 in epithelial cells and that EoE signature genes correlated with IL‐13 expression compared with IL‐4.
Conclusions
We demonstrate a definitive role for IL‐13 signaling via IL‐13Rα1 in EoE. These data provide mechanistic insights into the mode of action of current therapies in EoE and highlight the type II IL‐4R as a future therapeutic target.
Keywords: allergy, eosinophilic esophagitis, eosinophils, IL‐13, IL‐13 receptor α1
This study described the development of an experimental model for EoE using skin sensitization followed by intraesophageal challenges of oxazolone. Experimental EoE recapitulates the major clinical features of human disease, including epithelial cell proliferation, intraepithelial eosinophilia, fibrosis, and marked transcriptional resemblance to human EoE. Il13ra1 −/− mice are completely protected from EoE demonstrating that the type 2 IL‐4 receptor (i.e., IL‐13Rα1) has a critical role in the pathogenesis of EoE.Abbreviations: EoE, eosinophilic esophagitis; IL‐4Rα, interleukin 4 receptor α; IL‐13α1, interleukin 13 receptor α1; JAK, Janus kinase
1. INTRODUCTION
Eosinophilic esophagitis (EoE) is a food antigen‐driven allergic disease, which is characterized by eosinophil‐predominant inflammation, esophageal dysfunction, 1 and expression of the hallmark Th2 cytokines, IL‐4, and IL‐13. 2 IL‐4 exerts its activities by interacting with the type I IL‐4R, a cell surface receptor comprising IL‐4 receptor (R) α and the common γ (γc) chain. 2 IL‐4 can also utilize the type II IL‐4R, which is comprised of IL‐4Rα and the IL‐13Rα1 chain. 2 , 3 , 4 The type II IL‐4R is also a functional receptor for IL‐13. 4 Thus, IL‐4 and IL‐13 can mediate their effects via a shared signaling component. Importantly, the differential expression of these receptor chains in distinct cells renders responsiveness to IL‐4, IL‐13 or both cytokines. 2 , 5 Additionally, IL‐13Rα2 can act either as a decoy receptor or as an independent signaling molecule that mediates IL‐13‐driven responses. 6 , 7 , 8 , 9 Thus, the relative contribution of IL‐4, IL‐13, and their respective receptor chains may significantly vary in a disease‐ and tissue‐specific fashion.
EoE displays prominent involvement of CD4+ T cells that co‐express IL‐4 and IL‐13 10 and epithelial cells, which respond to these cytokines. Esophageal biopsies and blood samples of patients with active EoE display increased levels of IL‐4 and IL‐13. 1 Furthermore, induction of epithelial‐associated IL‐13/IL‐4‐responsive genes (e.g., TSLP and calpain 4) 11 , 12 has been shown to promote EoE pathogenesis. 13 Lung overexpression of IL‐13 or intratracheal delivery of IL‐13 to mice induce an EoE‐like response, 14 , 15 producing an esophageal transcriptome that partially overlaps with the human EoE transcriptome. 15 Moreover, activation of human esophageal epithelial cells with IL‐13 induces a “transcriptome signature” that is remarkably similar to that observed in biopsies from EoE patients. 16 IL‐13 also contributes to EoE pathogenesis by inducing tissue remodeling, including collagen deposition and angiogenesis. 1 Furthermore, IL‐13 disrupts the epithelial barrier via a mechanism involving downregulation of desmoglein 1 (DSG1) and filaggrin. 17 IL‐4 inhibits the differentiation of esophageal epithelial cells from squamous toward columnar cells; 18 it alters mucosal integrity and increases epithelial proliferation. 18 Similar to IL‐13, IL‐4‐activated esophageal epithelial cells secrete eotaxins, 19 which promote eosinophil recruitment. 20 Finally, IL‐4 enhances the generation of IgE antibodies in response to food allergens. 21
Since IL‐4 and IL‐13 are increased in EoE and since these two cytokines can activate a type II IL‐4R‐dependent response, it is important to define the relative contribution of IL‐4/IL‐13 signaling via the type I or the type II IL‐4R. This is clinically relevant since drugs that target multiple components of this pathway are in use or under development. 22 For instance, anti‐IL‐13 treatment using QAX576 decreased esophageal eosinophil counts and normalized the EoE transcriptome. 23 Recently, RPC4046, an anti–IL‐13 monoclonal antibody, markedly decreased esophageal eosinophilia and improved histological and clinical features. 24 , 25 In addition, blockade of IL‐4Rα using dupilumab (which blocks the type I and type II IL‐4 receptors) improved endoscopic, histological, and clinical features of EoE in phase 2 and phase 3 studies. 26 , 27
Herein, we aimed to define the relative contribution of IL‐4 and IL‐13 in EoE via the type II IL‐4R. Our data demonstrate that esophageal epithelial and lamina propria thickening, basal cell proliferation, eosinophilia, and remodeling are regulated by IL‐13‐driven signaling via IL‐13Rα1. Collectively, we demonstrate a definitive role for IL‐13 signaling via the type II IL‐4R in EoE pathogenesis.
2. MATERIALS AND METHODS
2.1. Experimental EoE
Mice (male c57BL6 mice, 6–8 weeks) were skin sensitized (15 μl 1% OXA, Sigma #E0753, dissolved in acetone) on both ear flanks (60 μl per mouse). After six additional skin challenges (0.5% OXA in acetone), the mice were bled and serum IgE was quantitated (ELISA, BD Bioscience). On Day 18, the mice were intraesophageally challenged with 200 μl OXA (1% in a 1:2 ratio of olive oil and 95% alcohol, respectively) using a plastic feeding tube that reaches the mid esophagus (INSTECH #ITH‐FTP‐22‐25‐50, 22ga X 25 mm) and has been modified for intraesophageal administration by puncturing 8 holes with a 14G needle (Figure S1‐S12). On Day 36, the mice were euthanized. Additional methods and analyses can be found in Appendix S1 section.
3. RESULTS
3.1. Intraesophageal challenges of oxazolone to skin‐sensitized mice results in experimental EoE
To establish an experimental model of EoE, C57BL6 mice were skin‐sensitized [day 0, 1% oxazolone (OXA), Figure 1A]. Subsequently, the mice received six additional skin challenges (0.5% OXA, Figure 1A). Twenty‐four hours after the last challenge, serum was collected and IgE levels determined (Figure 1B). Thereafter, mice were challenged with seven intraesophageal challenges (OXA, 1%) and the esophagus (Day 36) was dissected into 4 segments representing proximal to distal areas of the esophagus (Figure 1C). Assessment of eosinophilic infiltration using anti‐MBP staining revealed marked eosinophilic influx into the esophageal lamina propria in all segments (Figure 1D,E). Intra‐epithelial eosinophils, a hallmark feature of human EoE, were readily observed (Figure 1F,G). Quantitation of esophageal epithelial and lamina propria thickness revealed markedly increased thickening of the lamina propria (Figure 1H,I) and epithelial layer (Figure 1J,K) in OXA‐challenged mice. Increased epithelial thickness was accompanied with increased basal cell hyperplasia as observed by Ki67 staining (Figure 1L,M). Masson's trichrome staining demonstrated increased levels of collagen deposition in OXA‐treated mice (Figure 1N,O).
FIGURE 1.
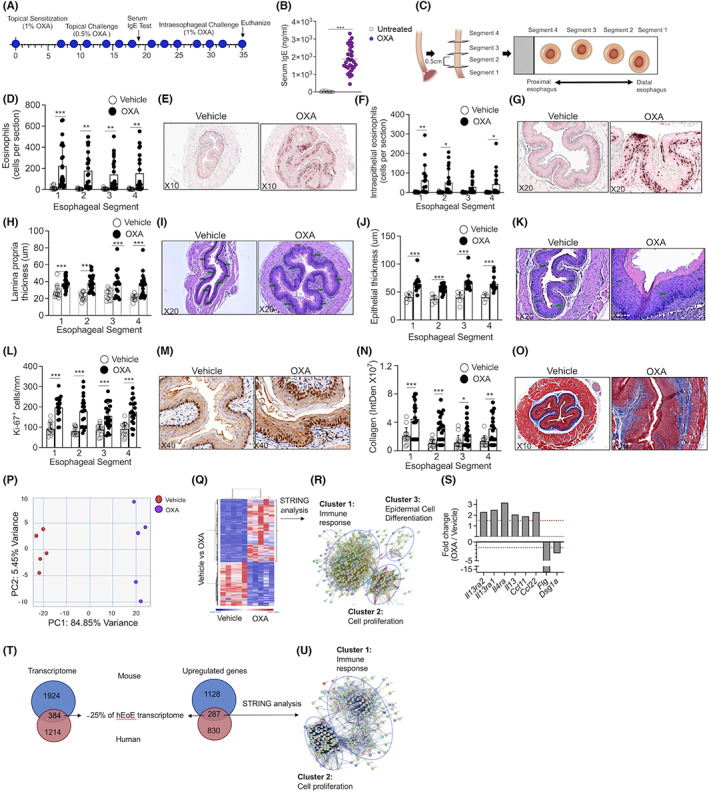
Experimental EoE is characterized by predominant type 2‐associated inflammation that overlaps with the human disease. Schematic representation of the experimental EoE protocol is shown (A). Allergen sensitization was confirmed by quantitation of serum IgE (B). Esophageal specimens (C) were stained with anti‐major basic protein (MBP, E,G) and total (D) as well as intraepithelial (F) eosinophils were quantified. The slides were stained with H&E (I,K), and lamina propria (H) and epithelial (J) thickness were determined. Epithelial cell proliferation (anti‐Ki67 staining, L,M) and fibrosis were determined (Masson's trichrome staining, N,O). Representative photomicrographs of anti‐MBP (E,G), H&E (I,K), anti Ki‐67 (M), and Masson's trichrome (O) are shown. Data are representative of n = 3 experiments; each circle represents one mouse. *‐p < 0.05, **‐p < 0.01, ***‐p < 0.001. RNA was extracted and subjected to RNA sequencing. PCA plot (P) and heat plot (Q) of vehicle‐ and OXA‐treated mice is shown. Top 500 upregulated transcripts (>two‐fold, p < 0.05) were analyzed using STRING analysis (R). Analysis of selected transcripts that are associated with type 2 immune responses (S). Dashed red line represents 1.5‐fold. Venn plot comparison between the expression of total differentially expressed transcripts (T), and differentially expressed transcripts that were upregulated in oxazolone‐induced experimental EoE and human EoE is shown. (U) The transcripts that were mutually increased in human and mouse EoE were subjected to STRING analysis
3.2. Experimental EoE is characterized by predominant type 2‐associated inflammation that overlaps with human disease
Esophageal RNA was subjected to bulk RNA sequencing. OXA‐challenged mice displayed a markedly different transcript signature from vehicle‐challenged mice (Figure 1P,Q). Unbiased STRING analysis 28 revealed that the transcriptome signature of experimental EoE was divided into three main clusters comprising of transcripts that are associated with (a) immune response (Table S1); (b) epidermal cell differentiation (Table S2); and (c) cell proliferation (Table S3, and Figure 1R). The expression of IL‐13, IL‐4Rα, IL‐13Rα1, and IL‐13Rα2 as well as the expression of IL‐13/IL‐4‐regulated chemokines such as Ccl11 and Ccl22 was increased (Figure 1S). Notably, expression of filaggrin and desmoglein that regulate barrier integrity in EoE 17 , 29 , 30 was decreased in experimental EoE (Figure 1S). Next, we examined the overlap between the transcriptome experimental EoE and of human EoE (obtained from 31 ). This analysis revealed an overlap of 384 transcripts which comprise 16% of the mouse EoE transcriptome and 25% of the human EoE transcriptome (Figure 1T, Table S4). Among the upregulated transcripts (Figure 1T), 287 transcripts were identified as commonly increased (Table S4). These 287 transcripts comprise 20% of the mouse transcriptome and 26% of the human upregulated transcripts (Figure 1T). STRING analysis of the commonly upregulated transcripts clustered into two distinct nodes corresponding with immune response and cell proliferation (Figure 1U, Tables [Link], [Link]).
3.3. Development of EoE is dependent on the type II IL‐4R
A valuable way to distinguish the role of the type I vs. the type II IL‐4R is by genetic deletion of the IL‐13Rα1 chain. 5 , 32 , 33 Such genetically engineered mice would harbor a functional deletion of the type II IL‐4R but have an intact type I IL‐4R. Consequently, experimental EoE was induced in Il13ra1 −/− mice. As previously shown, 32 IgE levels of skin‐sensitized Il13ra1 −/− mice were increased (Figure S2). OXA‐challenged Il13ra1 −/− mice were markedly protected from experimental EoE as assessed by lamina propria and epithelial layer thickening (Figure 2A,C,D). Assessment of epithelial cell proliferation by Ki67 staining showed that OXA‐induced proliferation of the esophageal epithelial basal cell layer was dependent on IL‐13Rα1 (Figure 2B, Figure 2E). Anti‐eosinophil major basic protein staining demonstrated that OXA‐induced esophageal eosinophilia and intraepithelial eosinophils were markedly decreased in OXA‐challenged Il13ra1 −/− mice (Figure 2F, Figure 2H,I). Furthermore, Il13ra1 −/− mice were protected from OXA‐induced subepithelial fibrosis (Figure 2G, Figure 2J).
FIGURE 2.
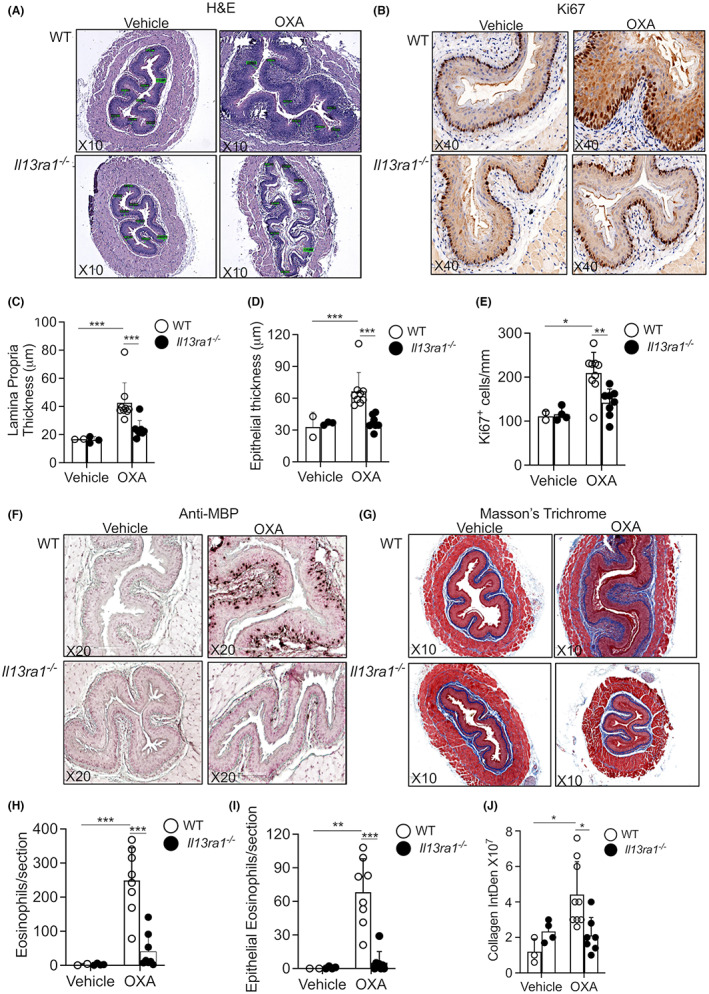
IL‐13Rα1 critically governs EoE pathology. Experimental EoE was induced in wild‐type (WT) and Il13ra1 −/− mice. H&E‐stained slides (A) were analyzed for lamina propria (C) and epithelial (D) thickness. Epithelial cell proliferation was determined using anti‐Ki67 staining (B,E). The slides were stained with anti‐major basic protein (MBP, F), and total (H) and intraepithelial eosinophils were quantified (I). Esophageal fibrosis was determined using Masson's trichrome staining (G,J). Representative photomicrographs of H&E (A), anti‐Ki67 (B), anti‐MBP (F), and Masson's trichrome (G) are shown. Data are representative of n = 3 experiments where each circle represents one mouse. *‐p < 0.05, **‐p < 0.01, ***‐p < 0.001
Given the recent FDA approval of dupilumab for the treatment of EoE patients, we were interested to determine the effects of blocking IL‐4Rα on the epithelium in experimental EoE. Blockade of IL‐4Rα decreased epithelial cell proliferation and subsequent epithelial thickness (Figure S3).
3.4. Identification of type II IL‐4R‐dependent transcripts in the esophagus
Next, global RNA sequencing was conducted on vehicle‐ and OXA‐challenged WT and Il13ra1 −/− mice. Under baseline conditions, Il13ra1 −/− mice displayed alteration of only 30 transcripts in the esophagus (Figure S4). Principal component analysis of vehicle‐ and OXA‐challenged WT and Il13ra1 −/− mice identified that the majority of transcript variation (67% variance, PC1, Figure 3A) was due to the difference between vehicle‐challenged WT mice, vehicle‐challenged Il13ra1 −/− mice, and OXA‐challenged Il13ra1 −/− to OXA‐challenged WT mice (Figure 3A). Gene set enrichment analysis (GSEA) of the IL‐13Rα1‐dependent EoE transcriptome demonstrated that IL‐13Rα1 regulated key pathways involved in EoE pathogenesis including the G2M cell cycle checkpoint, inflammatory responses, angiogenesis, and epithelial‐to‐mesenchymal transition (Figure 3B, Tables [Link], [Link]). Specifically, OXA‐challenged WT mice displayed increased expression of 904 transcripts (Tables S9) and downregulation of 1313 transcripts (Tables S10) in comparison with OXA‐challenged Il13ra1 −/− mice (Figure 3C,D). Over‐representation gene ontology analysis of the biological processes that were regulated by IL‐13Rα1 revealed that OXA‐challenged Il13ra1 −/− mice displayed decreased enrichment of cell division, regulation of immune system activation, keratinization, and locomotion (Figure 3E, Figure S5A, Tables [Link], [Link]). In contrast, OXA‐challenged Il13ra1 −/− mice showed elevation of various metabolic pathways and epidermal development (Figure 3F, Figure S5B, Tables [Link], [Link]). Hierarchical clustering and heat plot analysis of all differentially expressed transcripts showed that OXA‐challenged WT mice clustered together (Figure 3G). Vehicle‐challenged WT and Il13ra1 −/− mice, and OXA‐challenged Il13ra1 −/− clustered together, indicating that they are indistinguishable. For example, OXA‐treatment induced the expression of various cytokine receptors (e.g., Il4r, Il2rb, Il7r, and Il10ra) and inhibitory receptors (e.g., Cd300a, Lilrb3, and Lair1), which were either unaltered or increased to a lesser extent in OXA‐challenged Il13ra1 −/− and vehicle control‐treated mice (Figure 3H). In addition, induction of multiple cytokines and chemokines including Ccl5, Ccl22, Cxcl6, Il1b, Ccl7, and Il19 was abrogated in OXA‐challenged Il13ra1 −/− mice (Figure 3I). Similarly, induction of multiple transcription factors such as Nfkb1, Nfkbiz, Runx1, Klf7, and Rela was absent in Il13ra1 −/− mice (Figure 3J). Finally, expression of extracellular matrix components including Adam8, Fn1, and Timp1 was reduced in OXA‐challenged Il13ra1 −/− mice (Figure 3K). These data demonstrate that the development and transcriptome signature of EoE are critically regulated by the type II IL‐4R.
FIGURE 3.
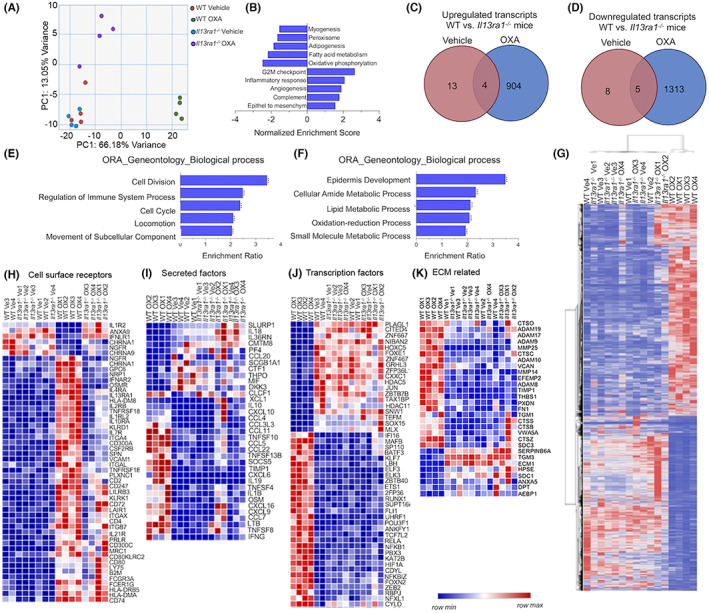
IL‐13Rα1 regulates the transcriptome signature of EoE. Experimental EoE was induced in wild‐type (WT) and Il13ra1 −/− mice. Thereafter, RNA was extracted and subjected to RNA sequencing. PCA plot of vehicle‐ and OXA‐treated WT and Il13ra1 −/− mice is presented (A). Transcripts were submitted to Gene Set Enrichment Analysis (GSEA), and comparison of OXA‐challenged WT mice vehicle‐ and OXA‐challenged WT and Il13ra1 −/− mice was performed. The top 5 up‐ and down‐regulated pathways in OXA‐challenged WT mice, which were regulated by IL‐13Rα1, are presented (B). Differentially expressed genes (fold>2, p < 0.05) were subjected to Venn plot analysis. Upregulated (C) and downregulated (D) genes in OXA‐challenged WT mice in comparison with o Il13ra1 −/− mice are shown. Differentially expressed genes were subjected to pathway analysis (E,F). Heat plot analysis of differentially expressed genes (fold>2, p < 0.05) (G), as well as heat plots of the top 50 differentially expressed cell surface receptors (fold>2, p < 0.000183, H), secreted factors (fold>2, p < 0.05, I), transcription regulators (fold>2, p < 9.74E‐09, J), and transcription factors (fold>2, p < 0.05, K)
3.5. Pathogenesis of experimental EoE is dependent on IL‐13
IL‐4 and IL‐13 can deliver signals via the type II IL‐4R. 34 Thus, we aimed to determine the relative contribution of each cytokine to EoE pathogenesis. Skin‐sensitized mice were treated with neutralizing anti‐IL‐13, anti‐IL‐4, or control antibodies. Neutralization of IL‐13 reduced lamina propria thickening (Figure 4A,E), epithelial cell thickening (Figure 4A,F), and proliferation (Figure 4B,G). In addition, anti‐IL‐13 treatment resulted in decreased total eosinophil infiltration (Figure 4C, H) and decreased intraepithelial eosinophilia (Figure 4I). Moreover, anti‐IL‐13 treatment resulted in markedly decreased esophageal fibrosis (Figure 4D,J). Anti‐IL‐4 treated mice displayed overall similar pathology as isotype control‐treated mice (Figure S6). Interestingly, while OXA‐induced lamina propria thickening was similar between anti‐IL‐4 and isotype control‐treated mice (Figure 4K), epithelial thickness was increased and a trend toward increased epithelial cell proliferation was observed (Figure 4L,M, Figure S6A‐E, respectively). Total and intraepithelial eosinophilia was similar in OXA‐challenged isotype control‐treated and anti‐IL‐4‐treated WT mice. Furthermore, no difference was observed in subepithelial fibrosis of anti‐IL‐4‐treated OXA‐challenged mice (Figure S6F,G). Anti‐IL‐4 treatment abrogated the induction of IgE (Figure S7) and serum IL‐4 expression (Figure S8), indicating that anti‐IL‐4 antibody treatment was functional. Furthermore, inability of anti‐IL‐4 treatment to decrease esophageal pathology was not due to lack of IL‐4 in the esophagus since esophageal protein expression of IL‐4 and the IL‐4‐associated chemokine CCL24 were increased in the esophagus of oxazolone‐treated mice (Figure S9). Furthermore, increased IL‐4 was observed in eosinophils, CD4+ T cells, and NKT cells that were isolated from the esophagus upon treatment with OXA (Figure S10). These data suggest a major contribution for IL‐13 via the type II IL‐4R in EoE pathogenesis.
FIGURE 4.
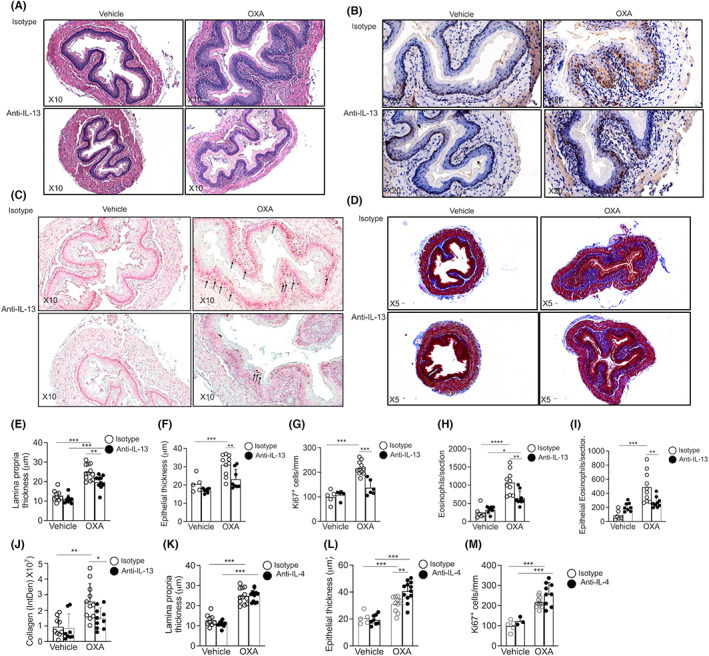
IL‐13 signaling via IL‐13Rα1 regulates the development of EoE. Wild‐type (WT) mice were subjected to experimental EoE. Starting on Day 17, the mice were treated with isotype control, anti‐IL‐13 (A–J), or anti‐IL‐4 (K–M). Thereafter, histopathological analysis of the esophagus was performed. The slides were stained with H&E (A), and lamina propria (E,K) and epithelial (F,L) thickness were determined. Epithelial cell proliferation was determined using anti‐Ki67 staining (B,G,M). The slides were stained with anti‐major basic protein (MBP, C), and total (H) and intraepithelial eosinophils (I) were quantified. Esophageal fibrosis was determined using Masson's trichrome staining (D,J). Representative photomicrographs of H&E (A), anti‐Ki67 (B), anti‐MBP (F), and Masson's trichrome (D) are shown. Data are representative of n = 3 experiments where each circle represents one mouse. *‐p < 0.05, **‐p < 0.01, ***‐p < 0.001
3.6. Expression of the type II IL‐4R in human esophageal epithelial cells
To gain insight regarding the cellular expression of IL‐13Rα1 in human EoE, we analyzed single‐cell RNA sequencing data that were obtained from esophageal biopsies. 35 This analysis identified 14 unique cell populations across the input of 10 samples. Top marker genes with high specificity were used to classify cell markers into specific cell types (“bimod” test with a threshold set to p < 0.05) (Figure 5A). Assessment of the expression pattern of the different receptor chains comprising the type I and type II IL‐4 receptors (i.e., Il2rg, Il4ra, and Il13ra1) revealed predominant expression of Il13ra1 in epithelial cells. In control patients and patients with inactive EoE, Il13ra1 was primarily expressed by differentiated epithelial cells, with lower expression in endothelial cells, fibroblasts, and myeloid cells (Figure 5B,C). Il4ra was predominantly expressed by T cells, mast cells, and epithelial cells and to lesser extent by endothelial cells (Figure 5D–F). Il2rg was highly expressed by T cells and mast cells and to lesser extent by epithelial cells and myeloid cells (Figure 5G–I). Collectively, these data suggest predominant expression of the type II IL‐4 receptor in human esophageal epithelial cells.
FIGURE 5.
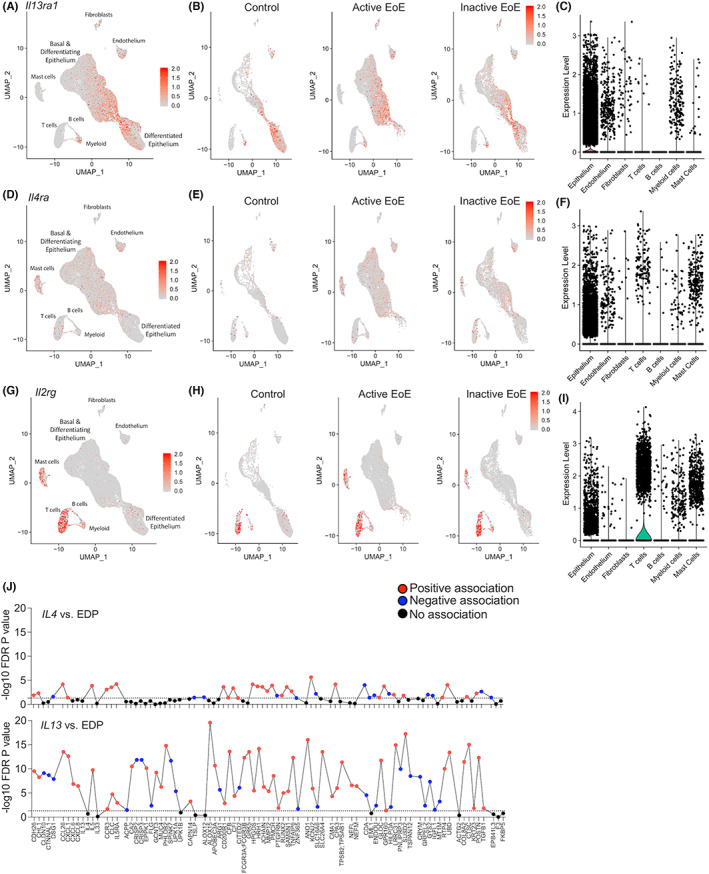
Expression of the receptor chains comprising the type I and type II IL‐4R in human EoE. Esophageal biopsies from control, active EoE patients (active EoE), and EoE patients in remission (inactive EoE) were subjected to single‐cell RNA sequencing. Data were subjected to Uniform Manifold Approximation and Projection (UMAP) analysis. Top marker genes were used to classify cell markers into known cell types: epithelium, fibroblast, endothelium, lymphocytes, mast cells, and antigen‐presenting cells (APCs) (A,D,G). The cells that express Il13ra1 (B), Il4ra (D) and Il2rg (G) were identified and marked (red dots). Violin plots of relative expression values per cell type are shown (C,F,I). Spearman correlations between IL4 or IL13 transcript levels and a diagnostic subset of genes from the EoE transcriptome (EDP) are shown (J, n = 146 patients). The dashed line indicates an FDR p value of 0.05
Next, we were interested to dissect the relative contribution of IL‐4 vs. IL‐13 in human EoE. Using a cohort of active EoE patients (n = 146), we examined the association between IL‐4 and IL‐13 transcript levels, and a set of 84 representative EoE‐associated transcripts that collectively comprise an EoE molecular diagnostic panel (EDP). 36 IL13 was markedly associated with the EoE transcriptome based on the magnitude of the genes and the statistical significance compared with IL‐4 (Figure 5J). Specifically, epithelial‐ and remodeling‐related genes were strongly associated with IL13 as IL13 positively associated with 91% of epithelial‐related and 83% of remodeling‐related genes (11/12 and 5/6, respectively).
3.7. The type II IL‐4R on esophageal epithelial cells mediates experimental EoE
The epithelial cell‐associated expression pattern for IL‐13Rα1 in human EoE and its association with IL‐13 led us to hypothesize that IL‐13 signaling via epithelial cell‐expressed type II IL‐4R has an important role in EoE pathogenesis. First, we validated that mouse epithelial cells express IL‐13Rα1. Under baseline conditions, IL‐13Rα1 was predominantly expressed by esophageal epithelial cells (Figure S11). Following OXA challenge, expression of IL‐13Rα1 in esophageal epithelial cells increased and additional IL‐13Rα1+ cells in the lamina propria were observed (Figure S11). Next, Il13ra1 fl/fl mice were generated (Figure S12) and mated with Krt14‐Cre mice in order to derive mice with a specific deletion of Il13ra1 in squamous epithelial cells. Thereafter, the mice were skin sensitized and challenged with OXA. OXA‐challenged Il13ra1 fl/fl ‐K14‐Cre mice displayed decreased lamina propria thickening (Figure 6A,E), decreased epithelial thickening (Figure 6A,F), and decreased epithelial cell proliferation (Figure 6B,G). In addition, they displayed decreased eosinophilic infiltration (Figure 6C–H). Interestingly, Il13ra1 fl/fl ‐K14‐Cre displayed increased levels subepithelial fibrosis at baseline which was enhanced following challenge with oxazolone (Figure 6D–I).
FIGURE 6.
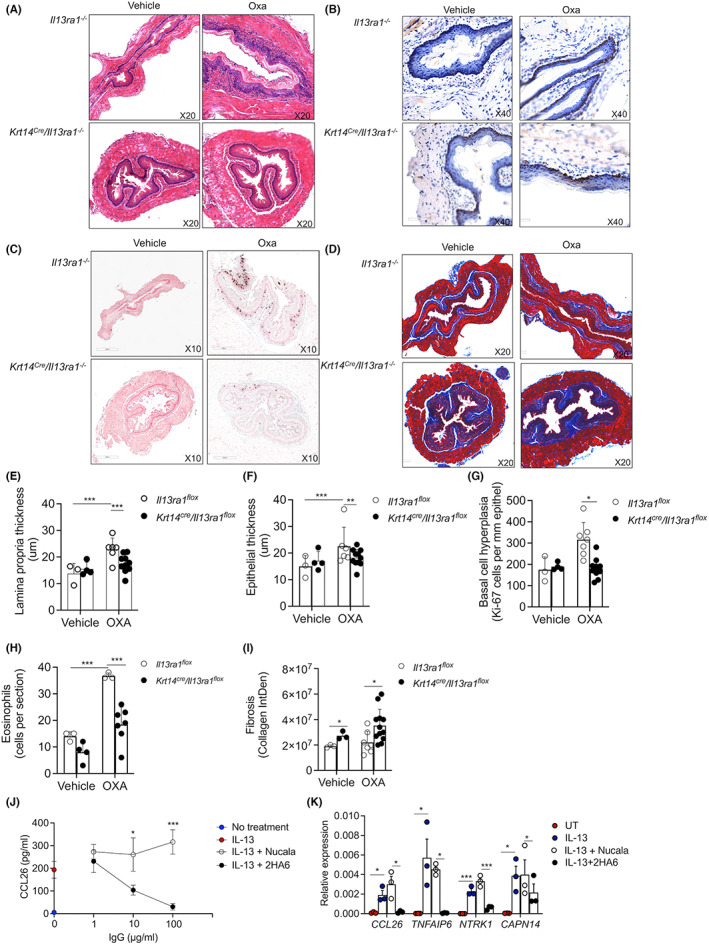
Key role for epithelial cell‐expressed IL‐13Rα1 in EoE. Il13ra1 flox and Krt14 cre /Il13ra1 flox were subjected to experimental EoE. Thereafter, histopathological analysis of the esophagus was performed. The slides were stained with H&E (A), and lamina propria (E) and epithelial (F) thickness were determined. Epithelial cell proliferation was determined using anti‐Ki67 staining (B,G). The slides were stained with anti‐major basic protein (MBP, C), and total esophageal eosinophils were quantified (H). Esophageal fibrosis was determined using Masson's trichrome staining (D, I). Representative photomicrographs of H&E (A), anti‐Ki67 (B), anti‐MBP (C), and Masson's trichrome (D) are shown. Data are representative of n = 2 experiments where each circle represents one mouse. *p < 0.05, **p < 0.01, ***p < 0.001. The ability of anti‐IL‐13Rα1 (clone 2hA6) to block IL‐13‐induced CCL26 secretion (J) and IL‐13‐induced mRNA expression of IL‐13‐target genes was determined in esophageal epithelial EPC2 cells (K). Nucala was used as a control antibody. Data are representative of n = 3 where each dot represents a different experiment, *‐p < 0.05, **‐p < 0.01, ***‐p < 0.001
3.8. Antibody‐mediated blockade of IL‐13Rα1 abrogates IL‐13‐induced effects in esophageal epithelial cells
To determine whether pharmacological targeting of IL‐13Rα1 could block IL‐13‐induced activation of human esophageal epithelial cells, primary human epithelial cells were stimulated with IL‐13 and the secretion of CCL26 was assessed (Figure 6J). Treatment with an anti‐IL‐13Rα1 antibody (2HA6) 32 abrogated IL‐13‐induced CCL26 secretion in a dose‐dependent fashion (Figure 6J). Anti‐IL‐13Rα1 treatment abrogated additional IL‐13‐induced genes including Tnfaip, Ntrrk1, and Capn14 whereas human IgG antibody control [anti‐IL‐5 (Nucala)] had no impact (Figure 6K).
4. DISCUSSION
Emerging clinical and experimental evidence underscore the type 2 cytokines IL‐4 and IL‐13 as well as the esophageal epithelium at the center of EoE pathogenesis. 13 Nonetheless, the contribution of each cytokine and the cellular compartment, which governs the development of EoE by responding to IL‐4 and/or IL‐13, is still unclear. Herein, we describe an experimental model for EoE that resembles multiple features of human disease including intraepithelial eosinophilic infiltration, esophageal remodeling, and disease transcriptome. Unbiased global RNA sequencing showed a 25% overlap between the genetic signature of human and experimental EoE. This included increased expression of key Th2‐associated cytokines and their receptors (e.g., IL‐13, IL‐13Rα1, and IL‐4Rα), eosinophil‐associated chemokines (e.g., CCL11), and downregulation of EoE epithelial‐associated susceptibility genes (e.g., DSMG1 and Fillagrin). The mouse‐human transcriptome overlap is striking especially since the composition of the human biopsies is markedly different than the mouse biopsies. While in humans, the biopsy primarily contains the esophageal epithelial layer, in mice the entire esophagus including the lamina propria and muscularis mucosa was sequenced. This relatively simple mouse model could now serve as a pre‐clinical model for drug screening and for further mechanistic studies that are limited in human samples.
IL‐13 is a key effector cytokine that is involved in the pathogenesis of EoE and additional allergic diseases such as atopic dermatitis (AD). Activation of human esophageal epithelial cells with IL‐13 induces a “transcriptome signature” that is remarkably similar to that observed in biopsies from EoE patients. 16 Furthermore, loss‐of‐function mutations in the IL‐13‐regulated genes, FLG (fillagrin), 29 and DSG1 (desmoglein 1), 17 , 37 are associated with increased risk for AD and EoE. 38 Candidate‐gene identification and genome‐wide association studies (GWAS) identified multiple genes that likely contribute to the development of EoE, including TSLP (encoding thymic stromal lymphopoietin), 11 CAPN14 (encoding calpain 14), 39 and STAT6 (encoding signal transducer and activator of transcription 6). 39 STAT6 is a key transcription factor involved in Th2 cell development, and is a critical signaling molecule that mediates IL‐13‐ and IL‐4‐induced responses. 2 CAPN14 is a intracellular proteolytic enzyme that is specific to the esophagus and induced by IL‐13. 37 Polymorphisms in the gene encoding IL‐13 have also been shown to be genetically associated with EoE in a phenome‐wide association study. 12 Similar to IL‐13, IL‐4 can also induce de‐differentiation of epithelial cells and induce the synthesis and secretion of eosinophil‐promoting chemokines. 18 , 19 These data highlight a central role for IL‐4 and IL‐13 in the pathogenesis of EoE. While our experimental data imply a predominant role for IL‐13 signaling via IL‐13Rα1 in mediating the pathology of EoE, we cannot rule out a role for IL‐4 or IL‐13Rα2. Our studies are limited by the fact that we did not demonstrate that anti‐IL‐4 treatment neutralized IL‐4 production or expression in the esophagus. Nonetheless, neutralization of IL‐4 in experimental EoE induced increased epithelial layer thickness. This likely reflects the fact that IL‐4 and IL‐13 compete with each other on binding the type 2 IL‐4R. In the absence of IL‐4 (due to neutralization), IL‐13 is available to induce more proliferation. Furthermore, IL‐13Rα2, which binds IL‐13 with high affinity, was previously shown to promote cellular signaling and epithelial cell proliferation. 9 , 40 Whether an interplay between IL‐13Rα1 and IL‐13Rα2 exists in EoE, remains to be defined.
We report that IL‐13Rα1 was responsible for all of the pathological features of EoE. Consistent with our previous findings, skin sensitization with oxazolone resulted in increased serum IgE production in Il13ra1 −/− mice compared to WT mice. This likely reflects that induction of IgE is dependent on the route of sensitization, the nature of the antigen, and the relative induction of IL‐4 in comparison with IL‐13. 32 , 41 This also suggests that decreased esophageal pathology in the absence of Il13ra1 −/− is not due to altered skin sensitization. Tissue eosinophilia was also regulated by IL‐13 signaling and the type II IL‐4R. This is especially interesting since previous studies using Il13ra1 −/− mice have shown that lung and skin eosinophilia is predominantly driven by IL‐4 signaling through the type I IL‐4R. 5 , 32 , 41 In our experimental model, RNA sequencing analyses revealed that induction of CCL11 was entirely dependent on the type II IL‐4R. These data are well‐suited with our previous reports that demonstrate a key role for IL‐13 signaling via the type II IL‐4R in induction of CCL11. 5 , 32 Importantly, human EoE is characterized by a marked induction of CCL26, 42 a chemokine that is not expressed in mice. Since the cellular source for CCL26 is IL‐13‐stimulated esophageal epithelial cells and since IL‐13Rα1 is primarily expressed by epithelial cells in EoE, it is likely that eosinophilia in human EoE is also driven by IL‐13 signaling and the type II IL‐4R. Certainly, we show that neutralization of IL‐13Rα1 in cultured esophageal epithelial cells blocked IL‐13‐induced secretion of CCL26.
Single‐cell RNAseq analyses in human EoE supported predominant expression of the type II IL‐4R in non‐hematopoietic esophageal cells such as epithelial cells. Furthermore, we demonstrate that in human EoE IL‐13 (but not IL‐4) was found to be strongly associated with epithelial cell pathology and remodeling. This raised the hypothesis that IL‐13 signaling via the type II IL‐4R in esophageal cells mediates EoE pathogenesis. Using mice that specifically lack expression of the type II IL‐4R in esophageal cells, we show that esophageal epithelial cells are primary responders to IL‐13 and key drivers of EoE pathology. This is evident by decreased lamina propria thickening, decreased epithelial cell‐associated pathology and in decreased eosinophilia. Indeed, emerging evidence places the esophageal epithelium at the center of EoE pathogenesis. 13 The main EoE disease susceptibility loci at chromosome 2p23 and 5q22 encode for esophageal epithelium‐associated genes (i.e., CAPN14 and TSLP, respectively). 11 , 37 , 39 , 43 Furthermore, EoE is associated with loss of differentiation and increased proliferation of esophageal epithelial cells. 29 For example, numerous transcripts, including filaggrin and several small proline‐rich protein (SPRR) family members, that comprise an epidermal differentiation complex are decreased in human EoE biopsies and downregulated by IL‐13. 29 Our data strengthen the centrality of epithelial cells in EoE pathogenesis and highlight the type II IL‐4R receptor as a key regulator of their activities. Our results further establish a fundamental role for IL‐13 in EoE pathogenesis that is mediated by its interactions with the type II IL‐4R in epithelial cells.
These data suggest that pharmacological targeting of the type 2 IL‐4R via blockade of IL‐13Rα1 may serve as a target for the treatment of EoE and perhaps additional allergic diseases.
AUTHOR CONTRIBUTIONS
These authors contributed as follows: SA and AM: Conception and/or design of the work. SA, GS, NR, MI, AD, IH, SGT, YG, TS, AB, NMB, MR, YD, LN, and AB: Data collection. SA, GS, NR, AB, IB, CV, MER, and AM: Data analysis and interpretation. SA, MER, and AM: Drafting the article. SA, GS, NR, MI, AD, IH, SGT, YG, TS, AB, NMB, MR, YD, LN, AB, IB, CV, ZH, MER, and AM: Critical revision of the article. AM: Final approval of the version to be published.
FUNDING INFORMATION
Ariel Munitz is supported by the US‐Israel Bi‐national Science Foundation (grant no. 2015163), by the Israel Science Foundation (grants no. 886/15 and 542/20), the Israel Cancer Research Fund, the Richard Eimert Research Fund on Solid Tumors (TAU), the Israel Cancer Association Avraham Rotstein Donation, and the Cancer Biology Research Center (TAU). C.V. and A.M. are supported by the Azrieli Foundation Canada‐Israel. Marc E. Rothenberg was supported by grants [R37AI045898, R01AI124355, U19AI070235, and P30DK078392 (Gene and Protein Expression Core); the Campaign Urging Research for Eosinophilic Disease (CURED); the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning].
CONFLICT OF INTEREST
Ariel Munitz is a consultant for Glaxo Smith Kline, Astra Zeneca, Sanofi, Oravax, and Sartorious and is an inventor of patents owned by the Tel Aviv University. Itai Benhar and Almog Bitton are inventors of patents owned by the Tel Aviv University. Marc E. Rothenberg is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, Allakos, Celgene, Astra Zeneca, Arena Pharmaceuticals, Glaxo Smith Kline, Revolo Biotherapeutics, and Guidepoint and has an equity interest in the Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, and Allakos and royalties from Reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust), and UpToDate and is an inventor of patents owned by Cincinnati Children's Hospital. The remaining authors disclose no conflicts.
Supporting information
Figures S1‐S12
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Table S10
Table S11
Table S12
Table S13
Table S14
Table S15
Table S16
Table S17
Table S18
Table S19
ACKNOWLEDGMENTS
We wish to thank all patients who participated in the study and Guy Shapira for assisting with handling the RNAseq data.
Avlas S, Shani G, Rhone N, et al. Epithelial cell‐expressed type II IL‐4 receptor mediates eosinophilic esophagitis. Allergy. 2023;78:464‐476. doi: 10.1111/all.15510
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in GSE at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE194316, reference number GSE194316.
REFERENCES
- 1. O'Shea KM, Aceves SS, Dellon ES, et al. Pathophysiology of eosinophilic esophagitis. Gastroenterology. 2018;154(2):333‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wynn TA. IL‐13 effector functions. Annu Rev Immunol. 2003;21:425‐456. [DOI] [PubMed] [Google Scholar]
- 3. Matthews DJ, Hibbert L, Friedrich K, Minty A, Callard RE. X‐SCID B cell responses to interleukin‐4 and interleukin‐13 are mediated by a receptor complex that includes the interleukin‐4 receptor alpha chain (p140) but not the gamma c chain. Eur J Immunol. 1997;27(1):116‐121. [DOI] [PubMed] [Google Scholar]
- 4. Miloux B, Laurent P, Bonnin O, et al. Cloning of the human IL‐13R alpha1 chain and reconstitution with the IL4R alpha of a functional IL‐4/IL‐13 receptor complex. FEBS Lett. 1997;401(2–3):163‐166. [DOI] [PubMed] [Google Scholar]
- 5. Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL‐13 and IL‐4 via IL‐13 receptor alpha1 and the type II IL‐4 receptor in asthma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(20):7240‐7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fichtner‐Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL‐13 signaling through the IL‐13alpha2 receptor is involved in induction of TGF‐beta1 production and fibrosis. Nat Med. 2006;12(1):99‐106. [DOI] [PubMed] [Google Scholar]
- 7. Yang SJ, Allahverdian S, Saunders ADR, Liu E, Dorscheid DR. IL‐13 signaling through IL‐13 receptor alpha2 mediates airway epithelial wound repair. FASEB J. 2019;33(3):3746‐3757. [DOI] [PubMed] [Google Scholar]
- 8. Newman JP, Wang GY, Arima K, et al. Interleukin‐13 receptor alpha 2 cooperates with EGFRvIII signaling to promote glioblastoma multiforme. Nat Commun. 2017;8(1):1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee CM, He CH, Nour AM, et al. IL‐13Ralpha2 uses TMEM219 in chitinase 3‐like‐1‐induced signalling and effector responses. Nat Commun. 2016;7:12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wen T, Aronow BJ, Rochman Y, et al. Single‐cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. 2019;129(5):2014‐2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rothenberg ME, Spergel JM, Sherrill JD, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kottyan LC, Davis BP, Sherrill JD, et al. Genome‐wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895‐900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rochman M, Azouz NP, Rothenberg ME. Epithelial origin of eosinophilic esophagitis. J Allergy Clin Immunol. 2018;142(1):10‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pope SM, Brandt EB, Mishra A, et al. IL‐13 induces eosinophil recruitment into the lung by an IL‐5‐ and eotaxin‐dependent mechanism. J Allergy Clin Immunol. 2001;108(4):594‐601. [DOI] [PubMed] [Google Scholar]
- 15. Zuo L, Fulkerson PC, Finkelman FD, et al. IL‐13 induces esophageal remodeling and gene expression by an eosinophil‐independent, IL‐13R alpha 2‐inhibited pathway. J Immunol. 2010;185(1):660‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanchard C, Mingler MK, Vicario M, et al. IL‐13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292‐1300. [DOI] [PubMed] [Google Scholar]
- 17. Sherrill JD, Kc K, Wu D, et al. Desmoglein‐1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7(3):718‐729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shan J, Oshima T, Farre R, Fukui H, Watari J, Miwa H. IL‐4 induces columnar‐like differentiation of esophageal squamous epithelium through JAK/PI3K pathway: possible role in pathogenesis of Barrett's esophagus. Am J Physiol Gastrointest Liver Physiol. 2014;306(8):G641‐G649. [DOI] [PubMed] [Google Scholar]
- 19. Neilsen CV, Bryce PJ. Interleukin‐13 directly promotes oesophagus production of CCL11 and CCL24 and the migration of eosinophils. Clin Exp Allergy. 2010;40(3):427‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conroy DM, Williams TJ. Eotaxin and the attraction of eosinophils to the asthmatic lung. Respir Res. 2001;2(3):150‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. 2011;50(1):87‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karo‐Atar D, Bitton A, Benhar I, Munitz A. Therapeutic targeting of the interleukin‐4/interleukin‐13 signaling pathway: in allergy and beyond. BioDrugs. 2018;32(3):201‐220. [DOI] [PubMed] [Google Scholar]
- 23. Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti‐IL‐13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500‐507. [DOI] [PubMed] [Google Scholar]
- 24. Hirano I, Collins MH, Assouline‐Dayan Y, et al. RPC4046, a monoclonal antibody against il13, reduces histologic and endoscopic activity in patients with eosinophilic esophagitis. Gastroenterology. 2019;156(3):592‐603 e510. [DOI] [PubMed] [Google Scholar]
- 25. Dellon ES, Collins MH, Rothenberg ME, et al. Long‐term efficacy and tolerability of RPC4046 in an open‐label extension trial of patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2021;19(3):473‐483. [DOI] [PubMed] [Google Scholar]
- 26. Hirano I, Dellon ES, Hamilton JD, et al. Efficacy of dupilumab in a phase 2 randomized trial of adults with active eosinophilic esophagitis. Gastroenterology. 2019;158(1):111‐122. [DOI] [PubMed] [Google Scholar]
- 27. Rothenberg ME, Hirano I, Dellon ES, et al. Dupilumab reduces biomarkers of type 2 inflammation in adult and adolescent patients with eosinophilic esophagitis: results from parts A and C of a three‐part, phase 3 LIBERTY EoE TREET study. J Allergy Clin Immunol. 2022;149(2):AB210. [Google Scholar]
- 28. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362‐D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blanchard C, Stucke EM, Burwinkel K, et al. Coordinate interaction between IL‐13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033‐4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simon D, Page B, Vogel M, et al. Evidence of an abnormal epithelial barrier in active, untreated and corticosteroid‐treated eosinophilic esophagitis. Allergy. 2018;73(1):239‐247. [DOI] [PubMed] [Google Scholar]
- 31. Sherrill JD, Kiran KC, Blanchard C, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15(6):361‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bitton A, Avlas S, Reichman H, et al. A key role for IL‐13 signaling via the type 2 IL‐4 receptor in experimental atopic dermatitis. Sci Immunol. 2020;5(44):eaaw2938. [DOI] [PubMed] [Google Scholar]
- 33. Ramalingam TR, Pesce JT, Sheikh F, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9(1):25‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gour N, Wills‐Karp M. IL‐4 and IL‐13 signaling in allergic airway disease. Cytokine. 2015;75(1):68‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Azouz NP, Klingler AM, Pathre P, et al. Functional role of kallikrein 5 and proteinase‐activated receptor 2 in eosinophilic esophagitis. Sci Transl Med. 2020;12(545):eaaz7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289‐1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis BP, Stucke EM, Khorki ME, et al. Eosinophilic esophagitis‐linked calpain 14 is an IL‐13‐induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simon D, Simon HU. Relationship of skin barrier breakdown and eosinophilic esophagitis. J Allergy Clin Immunol. 2020;145(1):90‐92. [DOI] [PubMed] [Google Scholar]
- 39. Sleiman PM, Wang ML, Cianferoni A, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kawashima R, Kawamura YI, Kato R, Mizutani N, Toyama‐Sorimachi N, Dohi T. IL‐13 receptor alpha2 promotes epithelial cell regeneration from radiation‐induced small intestinal injury in mice. Gastroenterology. 2006;131(1):130‐141. [DOI] [PubMed] [Google Scholar]
- 41. Rothenberg ME, Wen T, Shik D, Cole ET, Mingler MM, Munitz A. IL‐13 Receptor {alpha}1 differentially regulates aeroallergen‐induced lung responses. J Immunol. 2011;187(9):4873‐4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blanchard C, Wang N, Stringer KF, et al. Eotaxin‐3 and a uniquely conserved gene‐expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kottyan LC, Rothenberg ME. Genetics of eosinophilic esophagitis. Mucosal Immunol. 2017;10(3):580‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1‐S12
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Table S7
Table S8
Table S9
Table S10
Table S11
Table S12
Table S13
Table S14
Table S15
Table S16
Table S17
Table S18
Table S19
Data Availability Statement
The data that support the findings of this study are openly available in GSE at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE194316, reference number GSE194316.


