Abstract
Objective
The contribution of vitamin D (VD) deficiency to the pathogenesis of allergic diseases remains elusive. We aimed to define the impact of VD on oesophageal allergic inflammation.
Design
We assessed the genomic distribution and function of VD receptor (VDR) and STAT6 using histology, molecular imaging, motif discovery and metagenomic analysis. We examined the role of VD supplementation in oesophageal epithelial cells, in a preclinical model of IL-13-induced oesophageal allergic inflammation and in human subjects with eosinophilic oesophagitis (EoE).
Results
VDR response elements were enriched in oesophageal epithelium, suggesting enhanced VDR binding to functional gene enhancer and promoter regions. Metagenomic analysis showed that VD supplementation reversed dysregulation of up to 70% of the transcriptome and epigenetic modifications (H3K27Ac) induced by IL-13 in VD-deficient cells, including genes encoding the transcription factors HIF1A and SMAD3, endopeptidases (SERPINB3) and epithelial-mesenchymal transition mediators (TGFBR1, TIAM1, SRC, ROBO1, CDH1). Molecular imaging and chromatin immunoprecipitation showed VDR and STAT6 colocalisation within the regulatory regions of the affected genes, suggesting that VDR and STAT6 interactome governs epithelial tissue responses to IL-13 signalling. Indeed, VD supplementation reversed IL-13-induced epithelial hyperproliferation, reduced dilated intercellular spaces and barrier permeability, and improved differentiation marker expression (filaggrin, involucrin). In a preclinical model of IL-13-mediated oesophageal allergic inflammation and in human EoE, VD levels inversely associated with severity of oesophageal eosinophilia and epithelial histopathology.
Conclusions
Collectively, these findings identify VD as a natural IL-13 antagonist with capacity to regulate the oesophageal epithelial barrier functions, providing a novel therapeutic entry point for type 2 immunity-related diseases.
INTRODUCTION
The occurrence of allergy has risen globally over the past few decades and has reached epidemic levels.1 Epidemiological data show that vitamin D (VD) levels in the population have dramatically decreased, with 50% of the Western population being VD insufficient (serum levels<30 ng/mL).2 As VD is considered an essential factor required for healthy immune responses,3–5 given its pleiotropic effects on various tissues,6 including the epithelium,3 its deficiency may have profound effects on human health. Indeed, VD deficiency was linked with the risk and severity of numerous inflammatory diseases, including allergies7; yet, its role and mechanism are not yet agreed on.7–9
Eosinophilic oesophagitis (EoE) is a chronic allergic inflammatory disease of the oesophagus characterised by epithelial barrier dysfunction, basal zone hyperplasia (BZH), loss of epithelial differentiation, fibrosis and dilated intercellular spaces.10 The epithelium forms a protective defence barrier against environmental stimuli; on damage, antigens penetrate the epithelium and promote inflammation.11 Epithelial barrier dysfunction, such as loss of structural and cell junction proteins, has been proposed to promote antigen penetration and inflammation in EoE.12–14 The disease promoting pro-inflammatory type 2 cytokine IL-1310 is a potent inducer of epithelial barrier dysfunction, mediated by the STAT6 transcription factor.15 Notably, IL-13 induces loss of gene expression of epithelial structural differentiation markers, also implicated in EoE such as involucrin (IVL) and filaggrin (FLG).14 16 17 Interestingly, genetic ablation of the VD receptor (VDR) gene in mice leads to loss of skin IVL and FLG expression and severely impairs epithelial barrier function.18 Preliminary studies have reported that patients with EoE may have VD deficiency.19 Furthermore, low VD has been shown to increase the risk of allergic sensitisation.8 9 Taken together, functional and epidemiological data suggest that IL-13 and VD crosstalk may impact type 2 inflammation. Here, we aimed to mechanistically dissect VD involvement in allergic inflammation at the molecular level and assess its therapeutic potential for treating diseases related to IL-13-associated type 2 immunity.
RESULTS
VDR is a key transcription factor governing gene expression in oesophageal epithelium
To identify key transcription factors governing oesophageal epithelium gene expression, we compared it to the publicly available data using two complementary approaches. First, transcription factor binding site motif enrichment analysis using the HOMER software package20 and comparative analysis with reference motifs contained in the CIS-BP database21 indicated that the VDR binding site is highly enriched in histone 3 lysine (K) 27 acetylation (H3K27Ac) peaks in oesophageal epithelial cells (p≤10−20; #2 ranked motif class behind AP-1) (online supplemental table 1). Second, we compared a publicly available VDR chromatin immunoprecipitation sequencing (ChIPseq) dataset (GEO accession GSE31939) with a collection of 172 publicly available H3K27Ac ChIPseq datasets derived from a wide range of cell types and tissues using the regulatory element locus intersection (RELI) algorithm.22 The strongest overlap between VDR binding and H3K27Ac marks was obtained for H3K27Ac in oesophageal epithelial cells (GEO accession GSE57637) (online supplemental figure 1A; online supplemental table 1). Collectively, these results suggest enhanced VDR binding to functional gene enhancer and promoter regions (as indicated by H3K27Ac marks23) in oesophageal epithelial cells and implicate VDR as one of the primary transcription factors mediating oesophageal epithelium gene expression.
VD and IL-13 induce gene expression and epigenetic changes in oesophageal epithelium
On basis of the role that VDR likely has in transcriptome formation in the oesophageal epithelium and since oesophageal inflammation in EoE is mediated by an IL-13-driven epithelial cell transcriptional programme,10 we hypothesised that VD deficiency may amplify oesophageal epithelium responses to IL-13. We assessed the dynamics of transcription and epigenetic modifications (H3K27Ac marks) in IL-13-supplemented or VD-supplemented oesophageal epithelial cells compared with control cells (non-IL-13 treated or VD deficient) accordingly (online supplemental figure 1B–E). IL-13 induced expression of 749 genes and attenuated expression of 1400 genes (online supplemental figure 1B; online supplemental table 1). Similarly, VD induced expression of 1090 genes and attenuated expression of 1351 genes (online supplemental figure 1C; online supplemental table 2). Heatmaps of differential gene expression and corresponding epigenetic H3K27Ac marks within 100 kb of the differentially expressed genes (online supplemental figure 1B,C; online supplemental table 2) showed parallel changes (upregulated or down-regulated) of both transcription and epigenetic marks to either IL-13 (Spearman’s r 0.68, p≤0.0001) or VD (Spearman’s r 0.75, p≤0.0001). Furthermore, intersection of individually and bidirectionally affected genes (by either IL-13 or VD) showed a remarkable overlap of 1098 genes (online supplemental figure 1D; online supplemental table 2). Taken together, these data substantiate that both VD and IL-13 individually regulate the same approximately 30% of differentially expressed genes in the oesophageal epithelium due to VD or IL-13 treatment, respectively.
Clinical significance of IL-13-modified and VD-modified oesophageal transcriptomes
We examined the potential clinical relevance of the oesophageal epithelial cell transcriptional responses to IL-13 or VD (online supplemental figure 1B,C; online supplemental table 2) by comparing the transcriptional effects to the EoE transcriptome (online supplemental table 2). We intersected genes individually affected by either IL-13 or VD with the EoE transcriptome (GEO accession GSE58640) (online supplemental figure 1D). About 20% of the genes regulated by either VD or IL-13, individually, also overlapped with the EoE transcriptome (genes differentially expressed in EoE compared with controls—see the Methods section; 222 common genes; online supplemental figure 1D; online supplemental table 2). Among the common effector genes were some of the main drivers of epithelial barrier homoeostasis, such as desmoglein 1 (DSG1)13; synaptopodin (SYNPO)24; serine peptidase inhibitor, kazal type 7 (SPINK7)25; IVL and FLG,14 18 26 serine peptidase inhibitor B1, B3, B4, and B13 (SERPINB)10 27 and peptidylarginine deiminases (PADI)28 29 (online supplemental table 2). Biological pathway analysis demonstrated that the common genes regulate epithelial cell migration, adhesion, proteolytic functions and metabolic and apoptotic processes among other biological processes implicated in regulation of epithelium development and barrier functions (online supplemental figure 1E; online supplemental table 2). Taken together, these findings demonstrate dynamic transcriptional and epigenetic responses of epithelial cells to both VD and IL-13, individually, which likely combinatorically mediate diverse epithelial barrier functions in homeostatic and EoE states.
VD counteracts the IL-13-induced transcriptome and epigenetic changes
We aimed to dissect the effect of VD supplementation on IL-13-induced epithelial barrier impairment. Prior reports of the IL-13 effect on oesophageal epithelium have been based on VD-deficient tissue culture conditions (0.5 nM VD in keratinocytes serum-free medium—see the Methods section).10 Accordingly, we compared the responses of VD-deficient (VD low; 0.5 nM VD—standard conditions—see the Methods section) and VD-sufficient cells (VD normal; supplemented with 100 nM VD) after IL-13 stimulation. We assessed the localisation of H3K27Ac marks and the distribution of binding sites for STAT6 and VDR, major effectors of IL-13 and VD pathways, respectively, by ChIPseq (H3K27Ac, VDR and STAT6; online supplemental table 3). Genes exhibiting transcriptional responses (as assessed by RNAseq) that were accompanied by change in H3K27Ac marks, VDR and/or STAT6 binding sites, were selected for further analysis (online supplemental table 3). VD supplementation (VD normal) counteracted expression of approximately 70% of genes otherwise affected by IL-13 in VD deficient (VD low) conditions (436 genes, antagonistic response; figure 1A; online supplemental table 3). Accordingly, the dynamic range of H3K27Ac epigenetic marks showed changes, which strongly correlated with the transcriptional response (Spearman’s r 0.72, p≤0.0001). Transcription and epigenetic marks in the 436 ‘antagonistic’ genes were restored to within the control range in the presence of VD (VD normal; figure 1A; online supplemental table 3). The ‘antagonistic’ response constituted a network enriched in oesophageal-specific genes10 (n=24; χ2=82.03; p≤0.0001; online supplemental table 2). Gene ontology analysis and a circa-plot breakdown of the major biological processes (clockwise, from high to low; figure 1B; online supplemental table 3) identified epithelial cell migration, proliferation, differentiation, and apoptotic and metabolic processes among other biological processes implicated in regulation and development of epithelial barrier function (figure 1B). The top 10 genes governing these processes (clockwise, from high to low; figure 1B) are transcription factors associated with EoE (eg, hypoxia-inducible factor 1A (HIF1A)30 and SMA-related and MAD-related protein 3 (SMAD3)31 32), genes implicated in the epithelial-mesenchymal transition (EMT)33 34 (eg, transforming growth factor beta receptor (TGFBR1) and neuropilin-1 (NRP1)35 36), T-lymphoma invasion and metastasis-1 (TIAM1), sarcoma kinase (SRC), roundabout guidance receptor 1 (ROBO1), E-cadherin (CDH1),37–40 and genes implicated in anti-peptidase activity (eg, SERPINB3),10 27 as well as the IL-1 family members (IL-1α, IL-1β).41–43 Taken together, these data demonstrate that VD can counteract IL-13-induced transcriptional and epigenetic changes, as well as EMT functional processes, by maintaining normal expression of oesophageal epithelial barrier genes.
Figure 1.
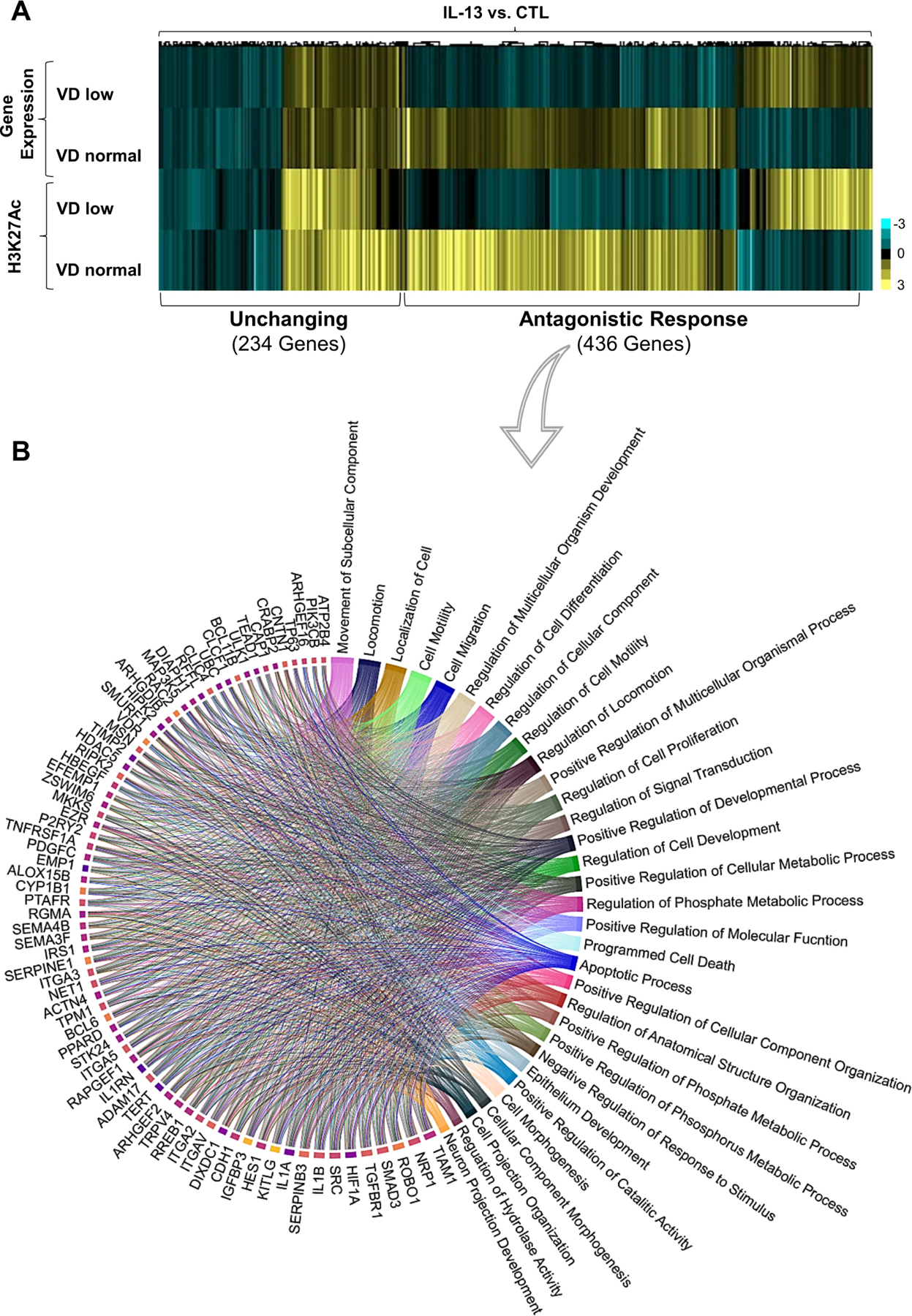
Effect of vitamin D (VD) on IL-13-induced epithelial gene and epigenetic marks expression. (A, B) Superconfluent organotypic cultures of oesophageal epithelial cells were maintained for 2 days in serum-free medium alone (0.5 nM VD; VD low) and supplemented with low VD medium or medium containing 100 nM VD (VD normal) for 2 hours and then stimulated with IL-13 (100 ng/mL) or vehicle (medium; control (CTL)) for an additional 6 hours. (A) The heatmap of H3K27Ac and differentially expressed genes fold changes; genes containing VDR and/or STAT6 binding sites as determined in ChIPseq (online supplemental table 3) in cells treated with IL-13 and VD compared with cells treated with IL-13 alone. Clustering revealed two main groups of genes where transcription and epigenetic marks are affected (antagonistic response) or not affected (unchanging) by VD supplementation. (B) Circa-plot of antagonistic response genes. Genes were functionally assessed using gene ontology (GO) analysis, and the top 76 genes driving their corresponding biological function were plotted clockwise–from higher to lower number of pathways per gene. Data are a summary of n=3 independent experiments. Each data point is a mean measurement for expression of an individual gene or gene-associated epigenetic mark. Statistics of differentially expressed genes by DESEQ2 (false discovery rate (FDR)-adjusted p≤0.05; see the Methods section).
VDR and STAT6 colocalisation in transcriptionally active sites
We tested the distribution of VDR and STAT6 response elements in the oesophageal epithelium following IL-13 stimulation in VD-deficient (VD low) and normal (VD normal) conditions. We first examined VDR and STAT6 colocalisation in the epithelial cell nuclei using confocal microscopy (figure 2) on the microscale within a 0.21 µM/pixel (~600 bp) range (figure 2B,C). We found that VDR and STAT6 exhibit a lower degree of colocalisation in VD-deficient cells with or without IL-13 stimulation (figure 2A,D). However, under normal VD conditions, colocalisation of STAT6 and VDR increased (figure 2C,D) in an IL-13-independent manner. The specificity of the STAT6/VDR interaction was substantiated by the lack of VDR localisation with the Lamin B (negative control) under any tested condition (figure 2E). Taken together, these data support that VDR and STAT6 interact with the same genomic fragments within a 600-bp range, corresponding to a range that could be localised within individual gene regulatory regions.
Figure 2.
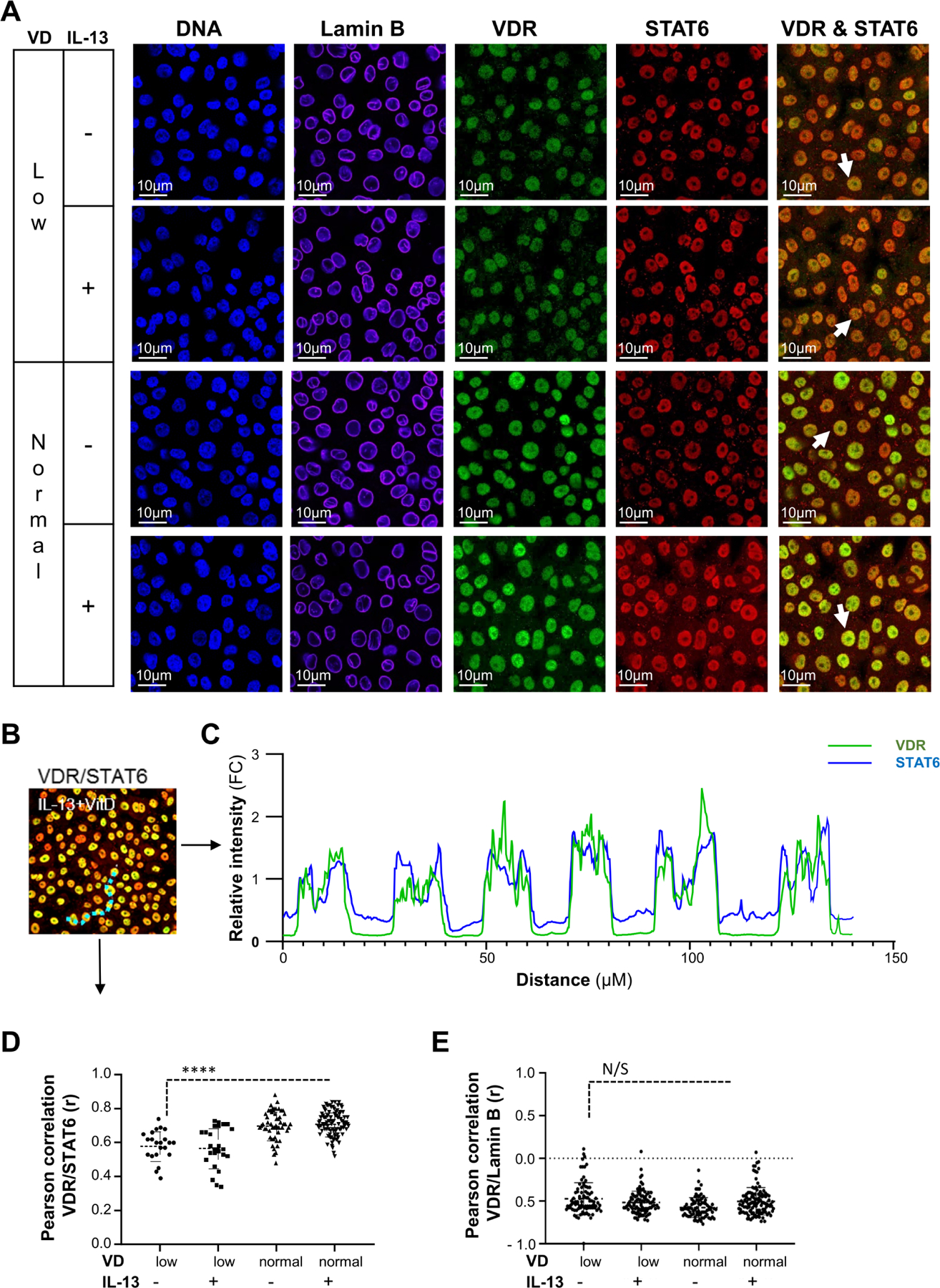
Analysis of VDR and STAT6 colocalisation in cell nuclei. (A–E) Superconfluent organotypic cultures of oesophageal epithelial cells were maintained for 2 days in serum-free medium alone (0.5 nM VD; VD low) and supplemented with low VD medium or medium containing 100 nM VD (VD normal) for 2 hours and then stimulated with IL-13 (100 ng/mL) or vehicle (medium; control (CTL)) for an additional 6 hours. (A) Immunofluorescent detection of cell nuclei (DNA; DAPI), lamin B, VDR, and STAT6 or staining overlay (VDR & STAT6; indicated with arrowheads). (B) Representative image of VDR and STAT6 overlay in VD- and IL-13-treated cells. A dotted line (light blue) was drawn across six individual cell nuclei as an example of the nuclear colocalisation of VDR and STAT6. (C) Relative fluorescence intensity of any individual signal per pixel as a function of distance between six individual nuclei (B; dotted line). (D–E) Pearson correlation of VDR and STAT6 (D) or VDR and lamin B (E) colocalisation. Data are representative (A) or a summary (B–E) of n=3 independent experiments. Each data point is a mean of a technical duplicate ±SD of individual in vitro assays measurements. Statistics by one-way ANOVA with Tukey’s multiple comparisons test: ****p≤0.0001; ANOVA, analysis of variance; N/S, not significant; VDR, vitamin D receptor.
To explain how VD and IL-13 might regulate the same genes, we examined VDR and STAT6 peak binding distributions around the transcription start site (TSS) of the affected ‘antagonistic’ versus ‘unchanged’ genes (figure 1A; online supplemental table 3). To this end, we measured the enrichment of overlapping VDR and STAT6 ChIPseq peaks proximal to ‘antagonistic’ vs ‘unchanged’ genes using the RELI algorithm.22 This analysis revealed that antagonistic genes are more highly enriched for overlapping STAT6 and VDR peaks within 100 kb of their promoters than were unchanged genes (3.1 fold; p=10−12 vs 2.2 fold; p=1, respectively). These results indicate that differential binding of STAT6 and VDR proximal to affected genes might have a key role in controlling the observed ‘antagonistic’ gene expression and epigenetic pattern, counteracting IL-13’s effect on the oesophageal epithelium.
VD deficiency exaggerates IL-13-mediated oesophageal epithelial barrier dysfunction
Based on the data showing that VD can counteract the effect of IL-13 on the oesophageal epithelium, we hypothesised that VD deficiency may promote oesophageal epithelial barrier dysfunction. To test this hypothesis, we maintained oesophageal epithelial cells in air-liquid interface (ALI) organotypic cultures in standard VD-deficient conditions (0.5 nM, VD low—see the Methods section) or supplemented with physiological concentration of 100 nM VD (VD normal). Next, organotypic cultures were stimulated with IL-13 to initiate an EoE-like epithelial barrier impairment. Supplementation of VD alone (CTL VD low vs CTL VD normal; figure 3) did not influence epithelium thickness (hyperproliferation), dilated intercellular spaces, transepithelial electrical resistance (TEER) nor low- or high-molecular-weight dextran flux (barrier permeability; figure 3A–F). IL-13-treated, VD normal organotypic cultures did not develop significant histopathological changes (figure 3A–C) compared with IL-13 untreated organotypic cultures. Similarly, IL-13 treatment did not substantially affect TEER nor dextran flux in VD normal conditions (figure 3D–F). However, there was increased epithelial thickness (figure 3A,B), dilated intercellular spaces (figure 3A,C), lower TEER (figure 3D) and increased low-molecular-weight and high-molecular-weight dextran flux (figure 3E,F) in VD low conditions as compared with VD normal organotypic cultures treated with IL-13. Indeed, VD low, but not VD normal organotypic cultures exhibited IL-13-mediated loss of the major epithelial structural differentiation markers IVL and FLG14 16 17 in the upper differentiated layers as shown by immunofluorescence analysis (figure 3G; individual and merged images). IL-13 treatment of VD normal organotypic cultures, or VD supplementation alone (VD normal; control), did not have any substantial effects on IVL and FLG expression (figure 3G). Taken together, these data demonstrate that VD deficiency exaggerates IL-13-mediated epithelial barrier dysfunction.
Figure 3.
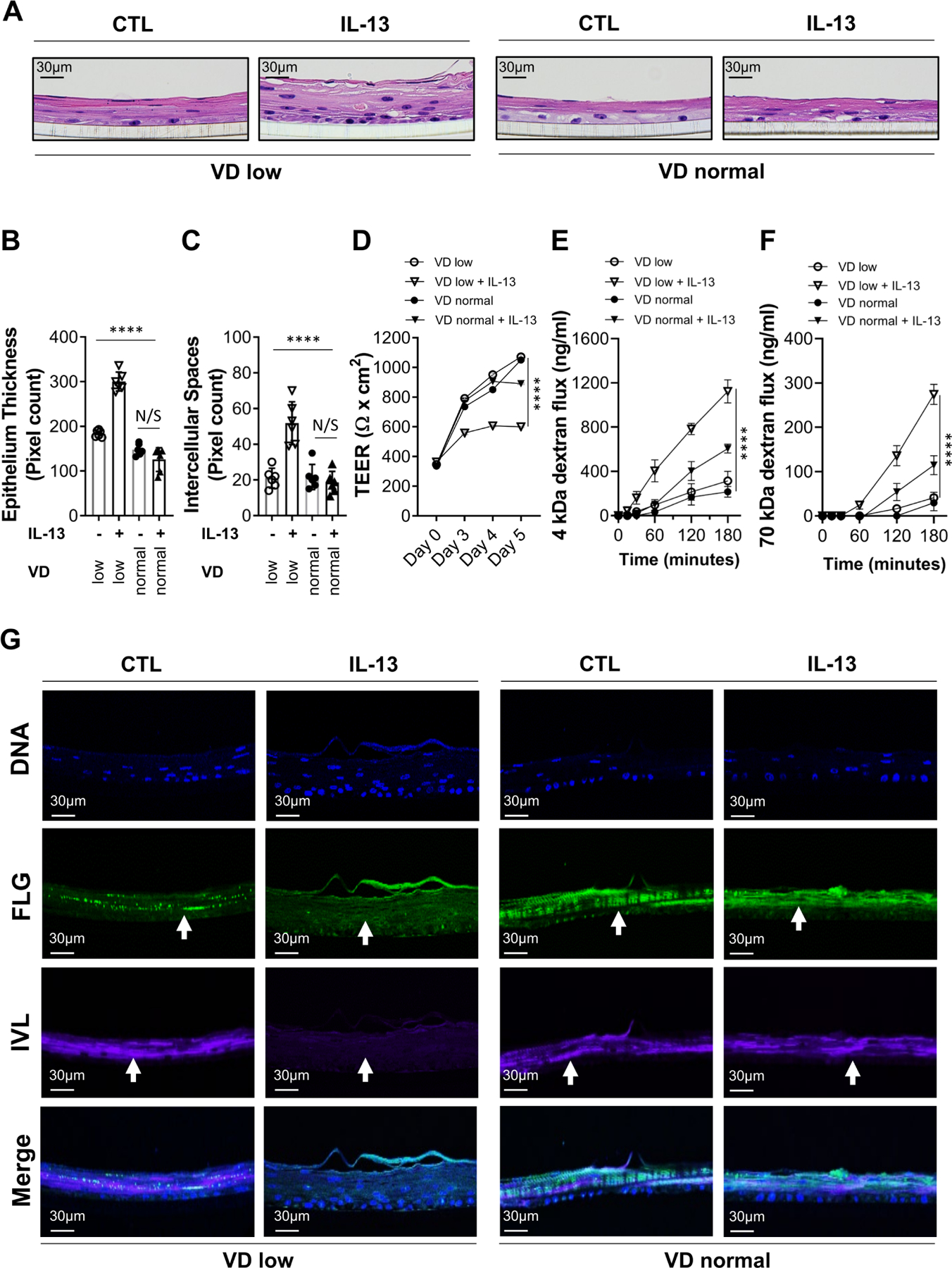
Effect of vitamin D (VD) on IL-13-dependent epithelial barrier dysfunction. (A–E) The oesophageal epithelial cell air-liquid interface (ALI) organotypic culture was maintained in VD low (0.5 nM) or normal (100 nM) conditions and supplemented with IL-13 (100 ng/mL) or medium alone (control; CTL) for 5 days. (A) ALI cultures were harvested on day 5 and H&E stained. (B–C) Epithelial barrier histopathological quantification of epithelium thickness (B) and intercellular spaces (C) based on H&E staining. (D–F) Epithelial barrier permeability quantification of transepithelial electrical resistance (TEER) (D), 4 kDa FITC-dextran (E) and 70 kDa Texas Red-dextran flux (G) were performed as indicated. (G) Immunofluorescent detection of individual and merged staining of FLG and IVL epithelial structural differentiation markers and cell nuclei (DNA; DAPI). Arrows show marker staining in the ALI culture as indicated. Data are representative (A, G) of n=3 independent experiments or a summary of n=3 (B–D) and n=2 (E, F) independent experiments. Each data point is a mean of a technical duplicate ±SD of individual in vitro assay measurements. Statistics by two-way ANOVA with Tukey’s multiple comparisons test: ****p≤0.0001; ANOVA, analysis of variance; FLG, filaggrin; IVL, involucrin; N/S, not significant.
VD deficiency amplifies pathology in a preclinical model of EoE
We employed a model of EoE mediated by overexpression of IL-13 (IL-13 transgenic (TG)) to test the effect of VD deficiency on allergic inflammation in the oesophagus. IL-13 TG and control mice were fed a low (0.1 IU/g) or normal (1 IU/g) VD diet for 2 weeks, and then IL-13 expression was induced for an additional 2 weeks while maintaining the same VD diet (see Methods). We assessed serum VD and IL-13 levels in all experimental groups (online supplemental figure 2); ~15 ng/mL VD serum levels indicated that mice fed with a low VD diet developed VD deficiency; in contrast, mice fed a normal VD diet had serum VD levels considered normal (~30 ng/mL) (online supplemental figure 2). IL-13 TG mice showed similar ~1 ng/mL IL-13 serum levels independent of the VD diet type (online supplemental figure 2). We subsequently examined the oesophageal epithelial tissue in both VD-deficient (VD low) and VD-sufficient (VD normal) mice using H&E (morphology), trichrome (morphology and fibrosis) and antimajor basic protein (MBP; eosinophil marker) staining (figure 4A). Histopathological assessment revealed that VD-deficient mice (VD low) developed increased eosinophilia (figure 4A,B) and fibrosis, as well as dilated intercellular spaces and epithelial thickness (hyper-proliferation), whereas VD-sufficient (VD normal) counterparts had significantly less severe corresponding histopathology score (figure 4A, C–E). Furthermore, the histopathology strongly correlated with serum VD levels, with lower VD levels corresponding to worse histopathology score (figure 4F–I). Taken together, these data demonstrate that VD deficiency amplifies IL-13-mediated oesophageal pathology, whereas VD sufficiency protects in an IL-13-driven EoE preclinical model.
Figure 4.
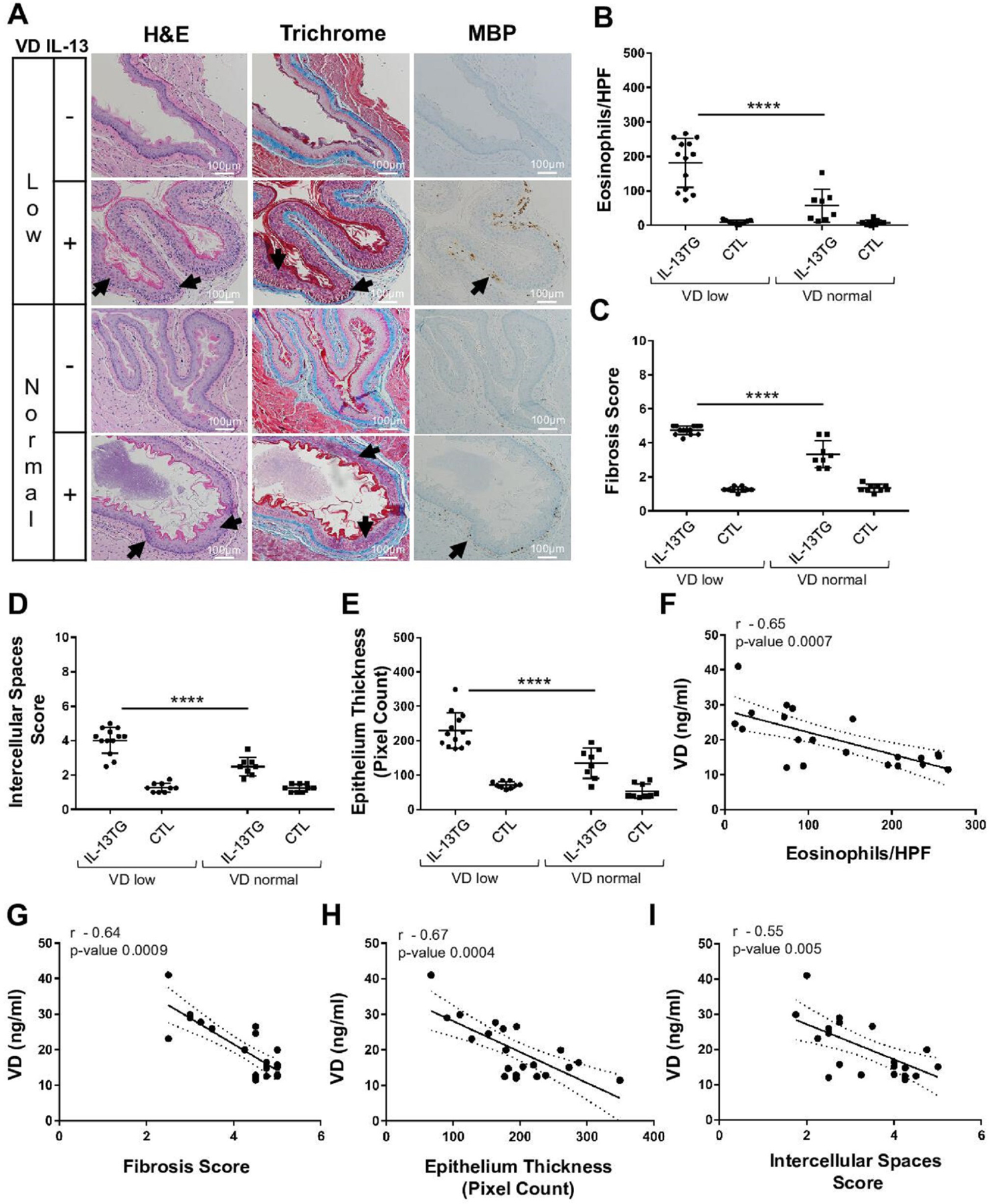
Effect of vitamin D (VD) on allergic oesophageal inflammation in vivo. (A–I) Wildtype controls (CTL) and IL-13 transgenic (IL-13TG) mice were maintained on low or normal VD diets (see the Methods section). (A) Mouse oesophageal tissue samples were stained for H&E, trichrome and anti-MBP (eosinophil marker). Arrows indicate hyperproliferation (H&E), fibrosis (trichrome), intercellular spaces (H&E; trichrome) and eosinophils (anti-MBP). (B–E) Quantification of eosinophils (B), fibrosis (C), intercellular spaces (D), and hyperproliferation (epithelial thickness; E). (F–I) Spearman correlation of serum VD levels (Supplemental Figure 2) with oesophageal histopathological markers of EoE (IL-13 TG group): eosinophil counts (F), fibrosis (G), hyperproliferation (epithelium thickness; H) and intercellular spaces (I). Data are representative (A) or a summary (B–I) of n=3 independent experiments. Each data point is a mean of a technical duplicate ±SD of in vivo (individual mouse tissue sample) assays. Statistics by one-way ANOVA with Tukey’s multiple comparisons test (B–E): ****p≤0.0001; Spearman correlation coefficient (r) and p values are as indicated (F–I). ANOVA, analysis of variance; EoE, eosinophilic oesophagitis.
VD in human allergic inflammation
We next tested the hypothesis that VD levels may influence the phenotype of human EoE. To assess the potential effect of VD serum levels in EoE, we examined serum VD levels in relationship to the degree of EoE histopathological changes as assessed by the EoE Histology Scoring System (HSS). VD serum levels ranged from severe deficiency (15 ng/mL) to normal levels (>30 ng/mL) and inversely correlated with oesophageal eosinophilia (figure 5A) and the degree of HSS total score (figure 5B). Eosinophilic inflammation (EI) and IL-13-associated oesophageal epithelial remodelling and inflammation markers,10 44 such as BZH, dyskeratotic epithelial cells (DECs) and laminal propria fibrosis (LPF), all inversely correlated with VD serum levels (figure 5C). Furthermore, mean serum VD levels in patients with active EoE (~25 ng/mL) were significantly lower than that of controls (~35 ng/mL; p≤0.01; figure 5D). In summary, these data demonstrate that VD deficiency may exacerbate disease severity in EoE, amplifying eosinophilia and epithelial tissue remodelling (figure 5E).
Figure 5.
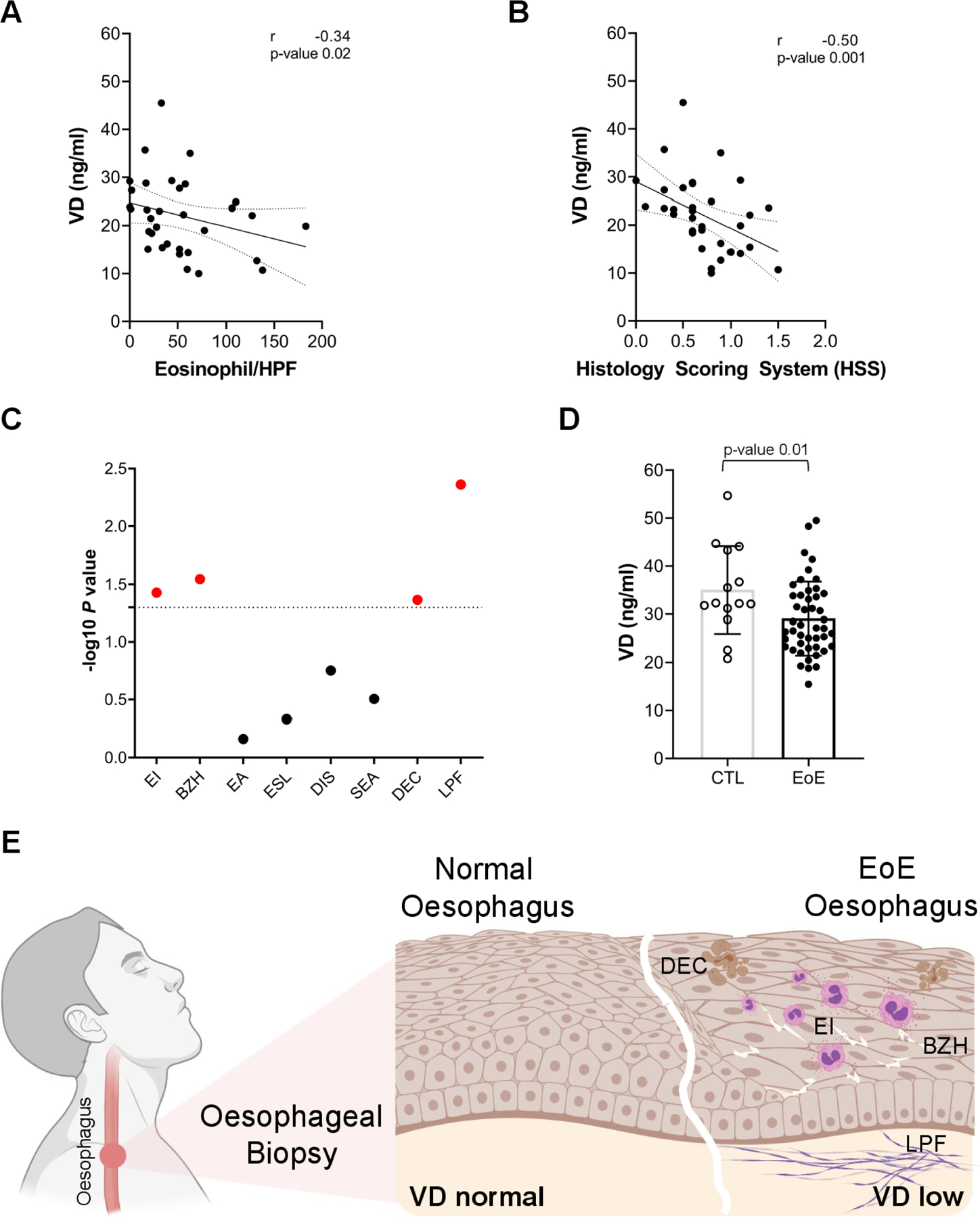
Role of vitamin D (VD) in eosinophilic oesophagitis. (A) Correlation of oesophageal eosinophilia with serum VD levels. (B) Correlation of EoE total Histology Scoring System (HSS) values with serum VD levels. (C) Significance (-log p value) of correlations of serum VD levels with individual HSS scores. Significant correlation in red. (D) Serum VD levels in patients with active EoE and controls (CTL). (E) Model of the oesophageal epithelium in EoE and control individuals. VD levels confer the disease state in which low VD levels induce EI, DEC, BZH and LPF (Created by BioRender). (A–D) Each data point is a mean of a technical duplicate ±SD of individual measurements. Statistics by unpaired one-tailed t-test (A) and Spearman correlation (B–D) for given p values (A–D) and Spearman correlation coefficients (r; A, B). BZH, basal zone hyperplasia; DEC, dyskeratotic epithelial cells; DIS, dilated intercellular spaces; EA, eosinophil abscess; EI, eosinophilic inflammation; EoE, eosinophilic oesophagitis; ESL, eosinophil surface layering; LPF, lamina propria fibrosis; SEA, surface epithelial alteration.
DISCUSSION
IL-13-induced pathways and genes are fundamental components driving the manifestations of type 2 immune responses, such as epithelial tissue inflammation in EoE. Accordingly, therapeutic agents that antagonise IL-13 are essential to alleviate allergic disease. Despite recent advances in monoclonal antibody therapies blocking IL-13 (eg, recently FDA approved Dupilumab45), the efficacy of these therapies is rarely achieving more than 50% improvement.45 One of the potential limitations is the inability of monoclonal antibodies to penetrate epithelial tissue where IL-13 has the most effect.10 14 44 New strategies involving small molecule drugs that can penetrate the tissues and modify IL-13 responses with long-lasting effects are needed. Here, we describe an unexpected protective function of VD in oesophageal allergic inflammation. We identify VD as a natural IL-13 antagonist with capacity to regulate the oesophageal epithelial barrier and immune functions, providing a novel therapeutic entry point for type 2 immunity-related diseases. We assessed the genomic distribution of VDR and found high enrichment in oesophageal epithelium. Mechanistically, we demonstrated that the VDR and STAT6 transcription factors are colocalised within the regulatory regions of the affected genes, suggesting that VDR and STAT6 interactome governs epithelial tissue responses to IL-13 signalling. Indeed, VD supplementation reversed IL-13-induced dysregulation of the transcriptomic and epigenetic modifications, greatly improving epithelial barrier inflammation scores and alleviating eosinophilic inflammation in preclinical models. Furthermore, VD supplementation obliterated IL-13-induced epithelial eotaxin 3 expression (online supplemental figure 3), which likely explains the reduction in tissue eosinophilia. On the gene level, VD antagonised responses that were associated with interleukin 1 beta (IL-1β) and TGF-β canonical pathways.43 46 Notably, VD supplementation reversed IL-13-induced dysregulation of EMT markers (TGFBR1, NRP1, TIAM1, SRC, ROBO1, CDH1)33 35 36 38 39 and epithelial structural differentiation markers (IVL, FLG, SERPINs, PADI).10 14 16 17 29 These data suggest a crucial role for VD in reducing EMT and attenuating oesophageal epithelial barrier dysfunction through balancing tissue proteolytic activity, epithelial barrier protein expression and post-translational modifications.
Notably, VD can accumulate in the tissue, and it levels may vary as compared with serum levels as a function of adipose tissue accumulation and malnutrition in patients with eosinophilic gastrointestinal disorders.47 Indeed, there is a positive association between body mass index and atopy rates.48 Furthermore, VD may exert pleiotropic effects on various immune cells, including reducing eosinophil activation,49 which may have additive effects on the VD-mediated attenuation of inflammation in eosinophilic gastrointestinal disorders, prompting further investigation of tissue-specific and cell-specific responses to VD in patients with eosinophilic gastrointestinal disorders. We propose that the oesophagus is normally equipped with VD-mediated molecular machinery that allows it to successfully respond to the variety of antigenic and biophysical stimuli encountered as environmental triggers. Accordingly, VD deficiency may be concurrent with IL-13-driven imbalances of oesophageal homeostatic processes, thereby contributing to the development of type 2 immunity. Human clinical data showed that VD serum levels were significantly lower in individuals with EoE than non-EoE control individuals, consistent with prior reports.50 51 Remarkably, VD serum levels inversely associated with severity of oesophageal histopathology, substantiating a previously unappreciated role for VD in IL-13-induced type 2 responses in the oesophagus. Indeed, lower VD levels were associated with higher grade eosinophilia, oesophageal epithelial tissue remodelling, and inflammatory processes including BZH, DECs and especially LPF. Notably, fibrostenosis, the most serious EoE complication,52 is mediated by IL-13-induced EMT.33 34 52 Our data mechanistically link VD deficiency with EMT and provides a rationale for considering VD supplementation in the context of anti-IL-13 and anti-IL-4 monoclonal antibody therapies.33 53 54
In summary, we have uncovered a molecular link between VD and IL-13-mediated type 2 allergic inflammation converging on functional colocalisation of VDR and STAT6 transcription factors within the regulatory regions of key response genes. Indeed, the preclinical data identify VD as a key regulator of oesophageal epithelial barrier function, where VD supplementation attenuated eosinophilia, epithelial tissue remodelling and fibrosis. Clinical findings substantiated the significance of these data and demonstrated an inverse correlation of VD levels with oesophageal eosinophilia and epithelial histopathology. Collectively, the findings presented here suggest that VD supplementation, topical or systemic, alone or in combination with anti-IL-13 and anti-IL-4 monoclonal antibodies therapies, may therapeutically alleviate IL-13-induced epithelial barrier dysfunction and potentially provide a novel therapeutic entry point for type 2 immunity-related diseases using small molecules, namely VDR agonists.
MATERIALS AND METHODS
Cell culture
EPC2 cells from an hTERT-immortalised, normal, human, oesophageal epithelial cell line (gift from Anil Rustgi, PhD, Columbia University, New York, New York, USA; used in figures 1–3, online supplemental figures 1; 3) and primary oesophageal epithelial cells were obtained as previously described55 (see procurement and processing of oesophageal biopsies below; used in figure 3A–D,G) and were cultured in standard keratinocyte serum-free media supplemented with bovine pituitary extract (BPE; 4 µL/mL) and epidermal growth factor (1 ng/mL) (Life Technologies). These standard cell culture conditions are VD deficient as BPE supplementation is the solemn source of VD (0.5 nM). In monolayer culture experiments, cells were maintained at a super-confluent state (1×106 cells/cm2) to induce organotypic epithelial barrier formation. Cells were treated with recombinant human IL-13 (PeproTech, Cat. No. 200–13; 100 ng/mL) and/or VD (1,25-dihydroxycholecalciferol; StemCell, Cat. No. 72412; low (0.5 nM) and normal (100 nM)) as indicated. Air-liquid interface (ALI) culture system was performed as previously described.13 ALI cultures’ barrier function was assessed by the means of TEER acquisition and dextran flux assays using FITC (Sigma FD4) and Texas Red-conjugated dextran (ThermoFisher D1864), of average molecular weight 4 kDa and 70 kDa, respectively. TEER and dextran flux were measured after ALI induction as indicated using an EVOM (World Precision Instruments, Sarasota, Florida, USA) and on day 5 using a fluorescence plate reader (BioTek, Winooski, VT; using red (520/40 excitation 620/640 emission) and green (420/485 excitation, 520/528 emission) excitation/emission filters), respectively. Low-molecular-weight and high-molecular-weight dextran flux and TEER measurement are applied for assessing the epithelial barrier permeability of both paracellular and transcellular paths, respectively.56 Cells passed authentication and tested negative for mycoplasma. Cell cultures were maintained at 37°C in a humidified 5% CO2 incubator.
Animals and induction of IL-13-dependent model of EoE
All experiments were performed in accordance with the Institutional Biosafety and Animal Care and Use Committees. BALB/c mice were obtained from Charles River Labs. IL-13 transgenic mice (IL-13 TG; CC10-rtta, TetO-IL-13) were generated in a manner allowing doxycycline-inducible IL-13 expression as previously described.44 IL-13 TG and control WT BALB/c mice were fed an AIN-93G-based diet containing 0.1 or 1 IU/g VD (TestDiet) for 2 weeks and received pure drinking water. Next, drinking water was supplemented with 2 mg/mL doxycycline and 1% sucrose for another 2 weeks to induce IL-13 expression, while maintaining the VD diet as described above. Mice were tail-bled, and serum IL-13 and VD levels were determined following the manufacturer’s protocol (R&D DY413 and RDKAP1971, respectively).
Histology and microscopy
Murine tissues and ALI membranes were fixed with formalin and embedded in paraffin. Antigen retrieval, H&E, major basic protein (MBP; eosinophil marker), trichrome, IVL (Abcam antibody ab53112), FLG (SantaCruz antibody sc-66192), STAT6 (Cell Signalling antibody 5397) and VDR (Cell Signalling antibody 12550) staining were performed according to previously published protocols.25 Histological scoring of fibrosis and inter-cellular spaces in murine oesophageal tissue was derived from trichrome figure 3-colour deconvolution and scoring of staining for collagen or acellular spacing penetration from the submucosa (1) to the basal layer (2), spinous layer (3), granular layer (4) and stratum corneum (5). Hyperproliferation was assessed as the number of epithelial layers from the basal membrane to the stratum corneum. In ALI cultures, epithelium thickness was measured as the mean of the multilayer epithelial cell culture thickness (from the membrane to the top; in pixel); intercellular spaces were measured as the mean width of the cell-to-cell spaces (in pixel). Immunofluorescence imaging and colocalisation assessment were performed using a Nikon A1R LUN-V inverted confocal microscope and Nikon Imaging Solutions Elements software. All images were analysed with ImageJ (Fiji open-source Java image processing programme).
Procurement and processing of oesophageal biopsies
Informed patient assent (17 years of age), patient consent (18 years of age) and legal guardian consent (patients 2–17 years of age) were obtained for patients to donate tissue samples for research and to have their clinical information entered into the Cincinnati Center for Eosinophilic Disorders (CCED) database. To correlate serum VD levels to the disease status (figure 5A–C), we examined a cohort of 17 patients with active EoE who were 4–17 years of age, comprising 13 males and 4 females. The data included a total of 34 longitudinal samples (before and after) that were used to examine serum VD levels and corresponding EoE histological scoring data as a function of disease severity. The CCED database samples were composed of 2–18-year-old patients for the comparative study designed to measure serum VD levels (figure 5D). Patients were 14 control individuals (9 males; 5 females) and 52 individuals with active EoE (40 males; 12 females). Serum VD levels were determined following the manufacturer’s protocol (R&D RDKAP1971). Patients with active EoE were defined as those having ≥15 oesophageal eosinophils per 400X high-power microscopic field at the time of biopsy and not receiving swallowed glucocorticoid nor dietary treatment at the time of endoscopy.
Chromatin immunoprecipitation and sequencing
Cell fixation and chromatin preparation were performed as described elsewhere.57 Briefly, ~5 µg of chromatin, corresponding to 1–2 million cells and 5 µl of Protein A Dynabeads (Thermo Fisher Scientific) were used per reaction. ChIPseq experiments were carried out in an IP-Star Compact automation system (Diagenode) using anti-H3K27ac antibody (pAb-196–050; Diagenode), anti-STAT6 antibody (5397, Cell Signalling), and anti-VDR antibody (12550, Cell Signalling). Library preparation and sequencing were conducted per standard Illumina protocol. Data analysis was performed in Scientific Data Analysis Platform (SciDAP; https://scidap.com).58 Briefly, reads were mapped with BowTie and peaks called with MACS2. Peaks were assigned to the nearest gene for further analysis. Differential binding was analysed with MAnorm. Dockerised CWL pipelines are available at https://github.com/datirium/workflows.
RNA isolation and sequencing
RNA isolation and sequencing were performed as described elsewhere.10 Library preparation and sequencing were conducted per standard Illumina protocol. Data analysis was performed in SciDAP (https://scidap.com).58 Briefly, RNA-Seq reads were aligned using RNA-STAR to GRCh38 genome assembly with RefSeq transcriptome and assigned to genes for expression quantification. Differential expression analysis was performed with DESeq2. Dockerised CWL pipelines are available at https://github.com/datirium/workflows.
Data analysis and statistics
RNAseq and ChIPseq data analysis was performed in SciDAP (https://scidap.com).58 Dockerised CWL pipelines are available at https://github.com/datirium/workflows. To assess effect of VD or IL-13 stimuli, we first identified dysregulated genes in the cells treated with the individual stimuli (IL-13 or VD) vs untreated cells. We further refined this list by identifying directly regulated genes that were epigenetically modified by H3K27Ac (MAnorm-adjusted p≤0.05, M≥1 or M≤−1) and in which transcriptional changes coincided with the epigenetic response, namely transcriptional upregulation and downregulation were accompanied by increased and decreased levels of H3K27Ac, respectively. For these analyses, we examined ChIPseq peaks located within 100 kb of each gene TSS. To assess the effect of VD in the context of IL-13 stimulation, we intersected the genes that were regulated by VD or IL-13 individually with the genes significantly dysregulated (DESEQ2, adjusted p≤0.05) in the cells treated with IL-13 +VD (IL-13-treated cells in the presence of VD) compared with IL-13 alone. We further refined this list by identifying genes that 1) were epigenetically modified by H3K27Ac (MAnorm-adjusted p≤0.05, M≥1 or M≤−1) following IL-13 +VD treatment compared with IL-13 alone 2) had transcriptional changes coinciding with the epigenetic response, namely transcriptional upregulation and downregulation were accompanied by increased and decreased levels of H3K27Ac, respectively, and 3) had STAT6 and/or VDR bound in the vicinity of the gene. RELI and HOMER tools were used to assess enrichment of epigenetic marks and transcription factors as previously described.22 Quantitative real-time PCR analysis (online supplemental figure 3) was performed using SYBR Green (Applied Biosystems A25741) and specific primers (GAPDH forward GGAAGGACTCATGACCAC and reverse CAGCGCCAGTAGAGGCAG primers; CCL26 forward CTCC CAGCGGGCTGTGATATTC and reverse GGGTCCAAGC-GTCCTCGGA primers). Data were obtained with QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) and quantified using ddCT (2–∆∆Ct) method. GraphPad Prism software was used for the indicated statistical analyses. P values and false discovery rates (where applicable) ≤0.05 were considered statistically significant.
Supplementary Material
WHAT IS ALREADY KNOWN ON THIS TOPIC
Interleukin (IL)-13 is a Th2 cytokine that governs epithelial tissue inflammation in allergic disease and represents a critical factor in the development and persistence of asthma, atopic dermatitis, rhinosinusitis, eosinophilic oesophagitis and other allergic disease.
Despite recent advances in monoclonal antibody therapies blocking IL-13 signalling, the efficacy of these therapies is limited. New strategies, involving small molecule drugs that can modify IL-13 responses with long-lasting effects, are needed.
WHAT THIS STUDY ADDS
The study identifies vitamin D (VD) as a natural IL-13 antagonist with capacity to regulate the epithelial barrier and immune functions. Indeed, VD receptor (VDR) and STAT6 transcription factors driving responses to VD and IL-13, respectively, are colocalised within the regulatory regions of the affected genes.
VD supplementation reverses IL-13-induced dysregulation of the transcriptome and epigenetic modifications, greatly improving epithelial barrier inflammation scores and alleviating eosinophilic inflammation in preclinical models.
Human clinical data demonstrate that VD levels inversely associated with severity of oesophageal eosinophilia and epithelial histopathology.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This discovery provides a novel therapeutic entry point for type 2 immunity-related diseases and makes a significant advance in our understanding of the role and mechanism of VD in allergic inflammation and how IL-13 responses can be targeted using small molecules, namely VDR agonists.
Acknowledgements
The authors thank Sajjeev Jagannathan for assistance with ChIPseq experiments, Margaret Collins for Histology Scoring System (HSS) scoring and assistance and Shawna Hottinger for editorial assistance.
Funding
This work was supported in part by NIH R01AI045898, R01AI130033, R01AI123176, and R01AI113125; the Campaign Urging Research for Eosinophilic Disease (CURED); and the Sunshine Charitable Foundation and its supporters, Denise and David Bunning (M.E.R.).
Footnotes
Competing interests MER is a consultant for Pulm One, Spoon Guru, ClostraBio, Serpin Pharm, Allakos, Celldex, Nextstone One, Bristol Myers Squibb, Astra Zeneca, Ellodi Pharma, Glaxo Smith Kline, Regeneron/Sanofi, Revolo Biotherapeutics and Guidepoint and has an equity interest in the first seven listed and royalties from reslizumab (Teva Pharmaceuticals), PEESSv2 (Mapi Research Trust), and UpToDate. MER is an inventor of patents owned by Cincinnati Children’s Hospital Medical Center. AB is a cofounder of Datirium. MK and AB are inventors on intellectual property owned by Cincinnati Children’s Hospital Medical Center. The remaining authors declare no competing interests.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication Consent obtained from parent(s)/guardian(s)
Ethics approval This study involves human participants and was performed with the approval of the Institutional Review Board of Cincinnati Children’s Hospital Medical Center. Participants gave informed consent to participate in the study before taking part.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Raw sequencing data sets are publicly available in the GEO database under GEO accession GSE184868. Sequencing data for oesophageal biopsies of patients with EoE or controls is available under GEO accession GSE58640. All figures have associated raw data. The additional data that support the findings of this study are available from the corresponding author by request.
REFERENCES
- 1.Lambrecht BN, Hammad H. The immunology of the allergy epidemic and the hygiene hypothesis. Nat Immunol 2017;18:1076–83. [DOI] [PubMed] [Google Scholar]
- 2.Matsui T, Tanaka K, Yamashita H, et al. Food allergy is linked to season of birth, sun exposure, and vitamin D deficiency. Allergol Int 2019;68:172–7. [DOI] [PubMed] [Google Scholar]
- 3.Muehleisen B, Bikle DD, Aguilera C, et al. PTH/PTHrP and vitamin D control antimicrobial peptide expression and susceptibility to bacterial skin infection. Sci Transl Med 2012;4:135ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryoo E, Kumar R, Kita H, et al. Serum 25-hydroxyvitamin D concentrations and waning pneumococcal antibody titers among individuals with atopy. Allergy Asthma Proc 2013;34:370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrams SA, Coss-Bu JA, Tiosano D. Vitamin D: effects on childhood health and disease. Nat Rev Endocrinol 2013;9:162–70. [DOI] [PubMed] [Google Scholar]
- 6.Carlberg C, Bendik I, Wyss A, et al. Two nuclear signalling pathways for vitamin D. Nature 1993;361:657–60. [DOI] [PubMed] [Google Scholar]
- 7.Muehleisen B, Gallo RL. Vitamin D in allergic disease: shedding light on a complex problem. J Allergy Clin Immunol 2013;131:324–9. [DOI] [PubMed] [Google Scholar]
- 8.Hollams EM, Teo SM, Kusel M, et al. Vitamin D over the first decade and susceptibility to childhood allergy and asthma. J Allergy Clin Immunol 2017;139:472–81. [DOI] [PubMed] [Google Scholar]
- 9.Bordon Y. Asthma and allergy: vitamin D primes neonatal immune system. Nat Rev Immunol 2017;17:467. [DOI] [PubMed] [Google Scholar]
- 10.Rochman M, Travers J, Miracle CE, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol 2017;140:738–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 2014;14:141–53. [DOI] [PubMed] [Google Scholar]
- 12.Ravi A, Marietta EV, Geno DM, et al. Penetration of the esophageal epithelium by dust mite antigen in patients with eosinophilic esophagitis. Gastroenterology 2019;157:255–6. [DOI] [PubMed] [Google Scholar]
- 13.Sherrill JD, Kc K, Wu D, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol 2014;7:718–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard C, Stucke EM, Burwinkel K, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J.i 2010;184:4033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol 2000;105:1063–70. [DOI] [PubMed] [Google Scholar]
- 16.Nagano N, Araki A, Ishikawa N, et al. Immunohistochemical expression of filaggrin is decreased in proton pump inhibitor non-responders compared with proton pump inhibitor responders of eosinophilic esophagitis. Esophagus 2021;18:362–71. [DOI] [PubMed] [Google Scholar]
- 17.Oshima N, Ishihara S, Fukuba N, et al. Epidermal differentiation complex protein involucrin is down-regulated in eosinophilic esophagitis. Esophagus 2017;14:171–7. [Google Scholar]
- 18.Xie Z, Komuves L, Yu Q-C, et al. Lack of the vitamin D receptor is associated with reduced epidermal differentiation and hair follicle growth. J Invest Dermatol 2002;118:11–16. [DOI] [PubMed] [Google Scholar]
- 19.Teixeira TL, Linden MA, Lomazi EA, et al. Case-Control study on vitamin D status in children and adolescents with eosinophilic esophagitis. Arq Gastroenterol 2020;57:409–15. [DOI] [PubMed] [Google Scholar]
- 20.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert SA, Yang AWH, Sasse A, et al. Similarity regression predicts evolution of transcription factor sequence specificity. Nat Genet 2019;51:981–9. [DOI] [PubMed] [Google Scholar]
- 22.Harley JB, Chen X, Pujato M, et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet 2018;50:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008;40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochman M, Travers J, Abonia JP, et al. Synaptopodin is upregulated by IL-13 in eosinophilic esophagitis and regulates esophageal epithelial cell motility and barrier integrity. JCI Insight 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azouz NP, Ynga-Durand MA, Caldwell JM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furue M Regulation of filaggrin, loricrin, and involucrin by IL-4, IL-13, IL-17A, IL-22, AhR, and Nrf2: pathogenic implications in atopic dermatitis. Int J Mol Sci 2020;21:5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akei HS, Mishra A, Blanchard C, et al. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology 2005;129:985–94. [DOI] [PubMed] [Google Scholar]
- 28.Méchin M-C, Takahara H, Simon M. Deimination and peptidylarginine deiminases in skin physiology and diseases. Int J Mol Sci 2020;21:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nachat R, Méchin M-C, Takahara H, et al. Peptidylarginine deiminase isoforms 1–3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J Invest Dermatol 2005;124:384–93. [DOI] [PubMed] [Google Scholar]
- 30.Masterson JC, Biette KA, Hammer JA, et al. Epithelial HIF-1α/claudin-1 axis regulates barrier dysfunction in eosinophilic esophagitis. J Clin Invest 2019;129:3224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho JY, Doshi A, Rosenthal P, et al. Smad3-deficient mice have reduced esophageal fibrosis and angiogenesis in a model of egg-induced eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2014;59:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang X, March M, Mentch F, et al. A genome-wide association meta-analysis identifies new eosinophilic esophagitis loci. J Allergy Clin Immunol 2021. [DOI] [PMC free article] [PubMed]
- 33.Gann PH, Deaton RJ, McMahon N, et al. An anti–IL-13 antibody reverses epithelial-mesenchymal transition biomarkers in eosinophilic esophagitis: phase 2 trial results. J Allergy Clin Immunol 2020;146:367–76. [DOI] [PubMed] [Google Scholar]
- 34.Kagalwalla AF, Akhtar N, Woodruff SA, et al. Eosinophilic esophagitis: epithelial mesenchymal transition contributes to esophageal remodeling and reverses with treatment. J Allergy Clin Clinical Immunology 2012;129:1387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aspalter IM, Gordon E, Dubrac A, et al. Alk1 and ALK5 inhibition by NRP1 controls vascular sprouting downstream of Notch. Nat Commun 2015;6:7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawakita T, Espana EM, Higa K, et al. Activation of Smad-mediated TGF-β signaling triggers epithelial-mesenchymal transitions in murine cloned corneal progenitor cells. J Cell Physiol 2013;228:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Liu L, Deng X, et al. MicroRNA 483–3p targets Pard3 to potentiate TGF-β1-induced cell migration, invasion, and epithelial–mesenchymal transition in anaplastic thyroid cancer cells. Oncogene 2019;38:699–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan Q, Liang X-J, Lin S-M, et al. Engagement of Robo1 by SLIT2 induces formation of a trimeric complex consisting of Src-Robo1-E-cadherin for E-cadherin phosphorylation and epithelial-mesenchymal transition. Biochem Biophys Res Commun 2020;522:757–62. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Qin Y, Wu G, et al. PRRG4 promotes breast cancer metastasis through the recruitment of NEDD4 and downregulation of Robo1. Oncogene 2020;39:7196–208. [DOI] [PubMed] [Google Scholar]
- 40.Marlow R, Binnewies M, Sorensen LK, et al. Vascular Robo4 restricts proangiogenic VEGF signaling in breast. Proc Natl Acad Sci U S A 2010;107:10520–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen I, Rider P, Vornov E, et al. IL-1α is a DNA damage sensor linking genotoxic stress signaling to sterile inflammation and innate immunity. Sci Rep 2015;5:14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naik S, Larsen SB, Gomez NC, et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 2017;550:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki H, Ohnishi H, Hama K, et al. Autocrine loop between TGF-β 1 and IL-1β through Smad3- and ERK-dependent pathways in rat pancreatic stellate cells. Am J Physiol Cell Physiol 2006;290:C1100–8. [DOI] [PubMed] [Google Scholar]
- 44.Zuo L, Fulkerson PC, Finkelman FD, et al. Il-13 induces esophageal remodeling and gene expression by an Eosinophil-Independent, IL-13Rα2–Inhibited pathway. J.i 2010;185:660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirano I, Dellon ES, Hamilton JD, et al. Efficacy of Dupilumab in a phase 2 randomized trial of adults with active eosinophilic esophagitis. Gastroenterology 2020;158:111–22. [DOI] [PubMed] [Google Scholar]
- 46.Hou C, Yang Z, Kang Y, et al. MiR-193b regulates early chondrogenesis by inhibiting the TGF-beta2 signaling pathway. FEBS Lett 2015;589:1040–7. [DOI] [PubMed] [Google Scholar]
- 47.Votto M, De Filippo M, Olivero F, et al. Malnutrition in eosinophilic gastrointestinal disorders. Nutrients 2020;13:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Mutius E, Schwartz J, Neas LM. Relation of body mass index to asthma and atopy in children: the National health and nutrition examination study III. Thorax 2001;56:835–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ethier C, Yu Y, Cameron L, et al. Calcitriol reduces eosinophil necrosis which leads to the diminished release of cytotoxic granules. Int Arch Allergy Immunol 2016;171:119–29. [DOI] [PubMed] [Google Scholar]
- 50.Fissinger A, Mages KC, Solomon AB. Vitamin deficiencies in pediatric eosinophilic esophagitis: a systematic review. Pediatr Allergy Immunol 2020;31:835–40. [DOI] [PubMed] [Google Scholar]
- 51.Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol 2014;133:291–307. [DOI] [PubMed] [Google Scholar]
- 52.Chehade M, Jones SM, Pesek RD, et al. Phenotypic Characterization of Eosinophilic Esophagitis in a Large Multicenter Patient Population from the Consortium for Food Allergy Research. J Allergy Clin Immunol 2018;6:1534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti–IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. Journal of Allergy and Clinical Immunology 2015;135:500–7. [DOI] [PubMed] [Google Scholar]
- 54.Hirano I, Collins MH, Assouline-Dayan Y, et al. RPC4046, a Monoclonal Antibody Against IL13, Reduces Histologic and Endoscopic Activity in Patients With Eosinophilic Esophagitis. Gastroenterology 2019;156:592–603. [DOI] [PubMed] [Google Scholar]
- 55.Blanchard C, Mingler MK, Vicario M, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. Journal of Allergy and Clinical Immunology 2007;120:1292–300. [DOI] [PubMed] [Google Scholar]
- 56.Chen S, Einspanier R, Schoen J. TEER: a functional parameter to monitor the quality of oviduct epithelial cells cultured on filter supports. Histochem Cell Biol 2015;144:509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Schones DE, Malicet C, et al. High mobility group protein N5 (HMGN5) and lamina-associated polypeptide 2α (LAP2α) interact and reciprocally affect their genome-wide chromatin organization. J Biol Chem 2013;288:18104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotliar M, Kartashov AV, Barski A. CWL-Airflow: a lightweight pipeline manager supporting common workflow language. Gigascience 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on reasonable request. Raw sequencing data sets are publicly available in the GEO database under GEO accession GSE184868. Sequencing data for oesophageal biopsies of patients with EoE or controls is available under GEO accession GSE58640. All figures have associated raw data. The additional data that support the findings of this study are available from the corresponding author by request.


