Abstract
Gastric cancer (GC) screening is arguable in most Western countries. Liquid biopsies are a great promise to answer the unmet need for less invasive diagnostic biomarkers in GC. Thus, we aimed at systematically reviewing the current knowledge on liquid biopsy‐based biomarkers in GC screening. A systematic search on PubMed/MEDLINE and Scopus databases was performed on published articles reporting the use of non‐blood specimen (saliva, gastric juice [GJ], urine and stool) on GC diagnosis. 3208 records were retrieved by June 2022. After removal of duplicate records, 2379 abstracts were screened, and 84 full texts included in this systematic review. More than 90% of studies were reported on Asian populations. Overall, 9 studies explored stool‐, 12 saliva‐, and 29 urine‐derived biomarkers for GC detection. Additionally, 37 studies, representing the majority, analyzed GJ, focusing on nucleic acid molecules. Several miRNAs and lncRNA molecules have been associated with GC risk, particularly miR‐21 (area under the curve [AUC] = 0.97, 95% CI: 0.94–1.00). Considering salivary biomarkers, the best described model in validation sets included the soybean agglutinin and Vicia villosa agglutinin lectins (AUC = 0.89, 95% CI: 0.80–0.99). Most studies in urine carried out metabolomic approaches, with two discriminatory models presenting AUC values superior to 0.97. This systematic review emphasizes the potential role of non‐blood‐based biomarkers, although further validation, particularly in Western countries, is mandatory, namely for non‐invasive screening and/or monitoring, as well as the use of GJ as a tool to enhance upper gastrointestinal endoscopy accuracy.
Keywords: feces, gastric juice, saliva, stomach neoplasms, urine
INTRODUCTION
According to the International Agency for Research on Cancer (IARC), gastric cancer (GC) ranks fifth for incidence and fourth for mortality worldwide in 2020, being responsible for one in every 13 cancer‐related deaths globally. 1 The number of new GC cases is estimated to increase from 945,000 in 2020 to 1.48 million in 2040 in countries with high and very high human development index (HDI), emphasizing that this is not a resolved issue. 2
There is a particularly high incidence of GC in Asian countries, such as Japan and Korea, where population‐wide mass screening is implemented. 3 Upper gastrointestinal (GI) endoscopy remains the gold standard for GC diagnosis. However, in medium to low‐incidence countries, such as in Europe, this strategy as a standalone is unwarranted. 4
Liquid biopsies represent a great promise in precision medicine, as they are less invasive, improving patients' adherence to screening, and allow the monitoring of the real‐time tumor dynamics, critical for early diagnosis, prediction of disease prognosis and recurrence, and even assessment of therapy efficacy. 5 , 6 While most studies have been focusing on the sampling of blood as the standard concept of liquid biopsy, common bodily fluids, such as saliva, urine, and stool have demonstrated potential as a source of cancer biomarkers, having the potential to improve the cost‐effectiveness of GC screening in low to intermediate risk regions and empower citizens in their own personal risk, enabling a better management of (by default) limited resources. 6
Several blood‐based GC biomarkers used in clinical practice have been reported, with carcinoembryonic antigen (CEA) and cancer antigen (CA)19‐9 still being the most frequently used. 7 Other examples include CA72‐4, CA12‐5, alpha‐fetoprotein, BCA‐225, and pepsinogen I/II. 7 Nevertheless, in spite of being widely used for cancer screening in several countries, the former biomarkers have low sensitivity and have been shown to be inappropriate as a screening modality for GI cancer. 8 In 2020, GASTROClear, the world's first molecular blood test for early detection of GC, was made available in Singapore to assess the risk for this type of cancer in asymptomatic healthy people. With an accuracy of 87% higher than any other blood biomarkers previously assessed for detection of GC, namely Helicobacter pylori serology, serum pepsinogen, CEA and CA19‐9 tumor markers, this cancer test uses a clinically‐validated algorithm to detect a unique signature of 12 microRNA in the blood. 9 Recently, non‐invasive biomarkers for GC detection, including blood‐based, have been non‐systematically reviewed from a molecular perspective and using distinct methodology. 5 , 10
A non‐despicable rate of missed lesions (up to 10%) can still be expected during endoscopy. 11 This could be overcome by improved endoscopists training, appropriate surveillance programs, and maybe the development of functional endoscopy for the detection and diagnosis of GC. 12 Gastric juice (GJ), despite requiring access to the stomach through endoscopic examinations, is a renewable reservoir of potential biomarkers and could easily be obtained during those procedures without additional discomfort to the patient. 13 This biofluid is usually thrown away during upper GI endoscopy, while it could provide valuable information concerning patients' gastric conditions, potentially contributing to improving the accuracy of the endoscopic screening, through detection of missed lesions, monitoring, and surveillance. 13
Herein, using a systematic approach, we report the currently available evidence published in the last decade on the role of non‐blood‐based circulating biomarkers in GC detection, particularly targeting saliva, urine, and stool as non‐invasive liquid biopsies, aiming to summarize potential targets for early cancer detection. Furthermore, we also explore GJ as a potential source of valuable biomarkers that can enhance the accuracy of endoscopic procedures by reducing the rate of missing lesions.
METHODS
Search strategy and selection criteria
A systematic search was conducted following the PRISMA guidelines (Figure 1). 14 The MEDLINE (Pubmed) and Scopus databases were searched for studies reporting the role of saliva, GJ, urine, and stool‐derived biomarkers for GC screening or diagnosis, from January 2010 to June 2022. The search for records available online was performed using the following query: (( “liquid biops*” OR “body fluid” OR “bodily fluid” OR “Liquid biopsy” [Mesh]) OR (saliva OR salivary) OR (urine [Mesh] OR urinary) OR (fecal OR feces OR stool OR “fecal material” OR “faecal material”) OR (“gastric juice” OR “stomach juice” OR “gastric fluid” OR “stomach fluid” OR “gastric juice” [Mesh ])) AND ((gastroesophag* OR stomach OR gastric OR gastrointestin*) AND (cancer OR neoplasm OR neoplasia OR carcinoma OR tumor)) AND biomarker. This query was adjusted for Scopus and no filters besides publication date were applied. A complementary search was carried out in the reference lists of the included papers, as well as of two relevant reviews. 5 , 10
FIGURE 1.
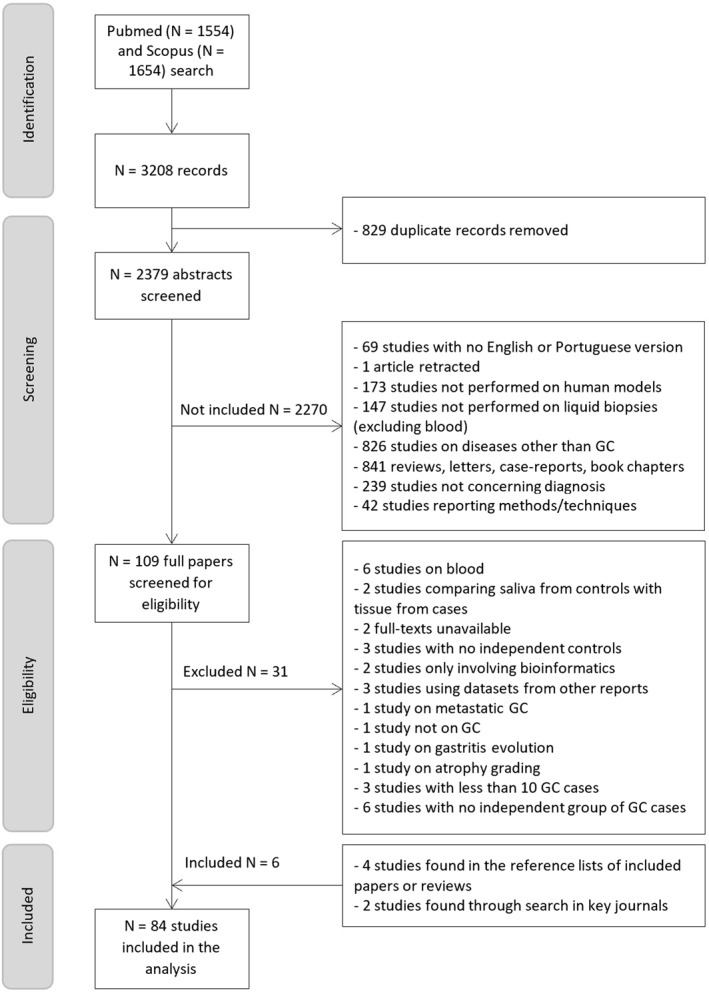
Flow chart of selection process. Of the 2379 records retrieved, 84 studies were included in the systematic review.
Eligibility criteria
After duplicate records removal, the abstracts were screened for eligibility by two independent researchers (Catarina Lopes and Jéssica Chaves). Case‐control or cohort studies identifying non‐invasive liquid biopsy‐derived biomarkers for GC detection were included. Studies with no English or Portuguese version, not enrolling human patients, not performed on saliva, GJ, urine or stool, with no independent controls, reporting on precancerous gastric lesions were excluded. Retracted records, reviews, letters, case‐reports, or book chapters were also excluded. Disagreements were analyzed and resolved by a third element (RO).
The full texts meeting the primary criteria were then reviewed by the same authors for final inclusion of all potential studies.
Data extraction
Data was extracted independently by two researchers (Catarina Lopes and Jéssica Chaves), including (i) study characteristics (i.e., first author, year, and reference); (ii) country; (iii) study design; (iv) sample size and percentage of females; (v) groups included in the analysis that are significant for this review; (vi) type of liquid biopsy analyzed; (vii) biomarker under study; (viii) method of detection; (ix) main results.
Quality assessment
Two review authors (Catarina Lopes and Jéssica Chaves) assessed the risk of bias of each included study independently, following a modified version of the Newcastle‐Ottawa quality assessment scale for case control studies. 15 Three domains were included: selection, comparability, and outcome. Selection was composed by five items, comparability by one item and outcomes by three items. 15 The article could receive one point in each item, getting a maximum to five points in selection, one or two in comparability, and a maximum of four in outcome. Thus, each article could receive a maximum of 11 points. Detailed information can be found in Online Resource.
If the information was missing or not described in the study, the corresponding authors were contacted by email. A third author was consulted to resolve disagreements when they could not be resolved by consensus.
RESULTS
Studies characteristics
Out of 2379 abstracts screened after duplicate records removal, 84 full papers were analyzed and considered eligible after applying the inclusion and exclusion criteria (Figure 1).
Most studies explored GJ (n = 36) and urine (n = 28), with only 10 and seven studies reporting on saliva or tongue coating and stool, respectively. Additionally, one study reported on tongue coating and gastric fluid, one study used oral swab and stool samples and one study used stool and urine. Despite the interest in early GC detection, few studies committed to earlier stages of the disease. In fact, only two studies assessed salivary biomarkers in patients with atrophic gastritis, and one and three studies focused on early‐stage patients to address urinary and GJ biomarkers, respectively.
All the included studies were written in English and their baseline characteristics and study design variables are summarized in Table S1 of Online Resource. Studies were conducted in 12 countries: seven in Asia (China, Iran, Japan, Singapore, South Korea, Taiwan, Turkey), three in Europe (Germany, Italy, Russia), two in America (Canada and United States of America), and one in Africa (Zambia). Sample sizes ranged from 22 to 1506 participants (median: 135 participants). Regarding quality control assessment, over 85% of the included studies scored under 5.5/11 points, which means most studies have some degree of risk of bias. Worth highlighting, the risk of selection bias associated with the representativeness of controls, which failed to be reported in over 95% of the studies. Another potential weakness of most studies is the lack of statistical power or its adequate report.
Non‐blood non‐invasive liquid biopsies in GC screening
Saliva
We included 12 eligible studies performed in saliva, tongue coating or oral swab samples in this review, exclusively performed in Asian populations: nine from China, 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 two from South Korea, 25 , 26 and one from Iran. 27 Specifically, studies by Xu et al. 28 and Cui et al. 24 used tongue coating samples and focused on oral microbiota analysis by polymerase chain reaction (PCR) and sequencing. In saliva, microbiota was assessed in three studies. 18 , 23 , 29 Serine peptidase inhibitor kazal type 7 (SPINK7), periplakin (PPL), semaphoring 4B (SEMA4B), and SMAD family member 4 (SMAD4) messenger RNAs (mRNAs) were assessed in two studies, 21 , 25 as well as salivary glycans, 19 , 20 and proteins, namely CSTB and DMTB1. 26 , 27 Amino acids were characterized only once. 16 Table 1 summarizes the identified salivary biomarkers with reported C‐statistic values for GC and atrophic gastritis detection. The biomarkers with best discriminatory power are Aleuria aurantia lectin (AAL, area under the curve [AUC] = 0.98) and Vicia villosa agglutinin (VVA, AUC = 0.96), although not validated in independent cohorts. 20 Considering only studies with a validation set, two models reached AUC values equal or superior to 0.87: one including two lectins, soybean agglutinin (SBA) and VVA (AUC = 0.89, 95% CI: 0.80–0.99), 20 and another including three mRNA molecules (SPINK7, PPL, and SEMA4B), two microRNAs (miR‐140‐5p and miR‐301a), and demographic factors (AUC = 0.87, 95% CI: 0.80–0.93). 25 Focusing on precancerous conditions, the biomarkers with the best performance for the detection of atrophic gastritis were lectins Datura stramonium agglutinin (DSA, AUC = 0.97, 95% CI: 0.95–1.00) and Lycopersicon esculentum lectin (LEL, AUC = 0.96, 95% CI: 0.93–1.00), but similarly to GC they were not validated. 20 On the other hand, a model including the lectins DSA and VVA was independently validated and reached an AUC of 0.83. 20
TABLE 1.
Salivary and tongue coating biomarkers for gastric cancer and atrophic gastritis detection with available expression and AUC data
| Gastric cancer detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Saliva | |||||||
| Lectins | AAL 20 | ↓ | 0.98 (0.97–1.00) | <0.001 | 92.0 | 95.0 | No |
| ECA 20 | ↑ | 0.86 (0.80–0.93) | <0.001 | 72.0 | 88.0 | No | |
| VVA 20 | ↑ | 0.96 (0.93–0.99) | <0.001 | 85.0 | 94.0 | No | |
| RNA | mRNA PPL 21 | ↓ | 0.72 | NA | 63.0 | 64.0 | Validation set |
| miR‐140‐5p 25 | ↓ | 0.70 (0.64–0.78) | NA | NA | NA | Validation set | |
| Proteins | CSTB 27 | ↓ | 0.73 (0.60–0.83) | 0.002 | 83.9 | 71.0 | No |
| DMBT1 27 | ↑ | 0.74 (0.61–0.84) | <0.001 | 80.7 | 64.5 | No | |
| ModelsRef. | |||||||
| 10 amino acids SERS spectra bands 16 | NA | NA | 94.8 | 90.2 | No | ||
| Bacterial genera 18 | 0.91 (0.78–0.99) | NA | NA | NA | CV | ||
| 2 lectins (SBA and VVA) 20 | 0.89 (0.80–0.99) | <0.001 | 96.0 | 80.0 | Validation set | ||
| 3 mRNAs and 2 miRNAs a 25 | 0.81 (0.72–0.89) | NA | 75.0 | 83.0 | Validation set | ||
| 3 mRNAs, 2 miRNAs a and demographic factors 25 | 0.87 (0.80–0.93) | NA | 82.0 | 77.0 | Validation set | ||
| 3 proteins (CSTB, TPI1, DMBT1) 26 | 0.93 | NA | 85.0 | 80.0 | Pre‐validation | ||
| Oral swab | |||||||
| Oral microbiota (13 OTUs) 23 | 0.82 (0.73–0.92) | NA | NA | NA | Pre‐validation | ||
| Tongue coating | |||||||
| 6 bacterial genera b 28 | 0.88 (0.80–0.95) | NA | NA | NA | Pre‐validation | ||
| Atrophic gastritis detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Saliva | |||||||
| Lectins | DSA 20 | ↓ | 0.97 (0.95–1.00) | < 0.001 | 87.0 | 96.0 | No |
| LEL 20 | ↑ | 0.96 (0.93–1.00) | < 0.001 | 93.0 | 92.0 | No | |
| VVA 20 | ↑ | 0.81 (0.71–0.91) | <0.001 | 70.0 | 91.0 | Validation set | |
| ModelsRef. | |||||||
| Bacterial genera 18 | 0.76 | NA | NA | NA | CV | ||
| 2 lectins (DSA and VVA) 20 | 0.83 (0.71–0.94) | <0.001 | 70.0 | 91.0 | Validation set | ||
Note: Only molecules with reported relative abundance and reaching AUC >0.70 were included (suggested by 38 as moderate accuracy). In bold are AUC values > 0.90. Pre‐validation means the biomarkers have been validated using the same population, whereas CV is a resampling validation technique that tests and trains a model using different portions of the data.
Abbreviations: AUC, area under the receiver operating characteristic (ROC) curve; CI, confidence interval; CV, cross‐validation; GC, gastric cancer; OTU, operational taxonomic units.
SPINK7, PPL, SEMA4B, miR‐140‐5p, and miR‐301a.
Fusobacterium, Peptococcus, Peptostreptococcus, Porphyromonas, Megamonas, and Rothia.
Stool
All the included reports in stool samples were performed in Asian populations, seven in China 23 , 30 , 31 , 32 , 33 , 34 , 35 and one in Iran, 36 and were case‐control studies. Overall, four biomarkers were explored: (a) fecal calprotectin in two studies 33 , 36 ; (b) B‐cell activating factor (BAFF) protein in one study 33 ; (c) telomerase reverse transcriptase (TERT) gene promoter methylation in one population 30 and (d) the fecal microbiota was explored in five Chinese populations, 23 , 31 , 32 , 34 , 35 either targeting bacterial 16S ribosomal RNA (rRNA) and fungal 18S rRNA or bacterial DNA. Stool‐based biomarkers with reported expression in GC, as well as AUC values, are summarized in Table 2. The bacterial genera Desulfovibrio, Escherichia, Faecalbacterium, and Oscillospira exhibit the best diagnostic performance, achieving AUC values ≥ 0.90. 31 Those results involved a five‐fold cross‐validation for data preparation of a random forest mode in a single study gathering 73 participants with 76% of patients diagnosed at more advanced stages of the disease. Additionally, two models including fecal microbiota achieved AUC values of 0.97 and 0.94, the latter involving cross‐validation for data preparation. 23 , 34 None of the remaining biomarkers included in Table 2 were validated.
TABLE 2.
Stool‐based biomarkers for gastric cancer detection with available expression and AUC data
| Stool | |||||
|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p Value | With validation |
| Protein | Fecal calprotectin 36 | ↑ | 0.71 b | NA | No |
| DNA (TERT promoter methylation) | CpG site 1 30 | ↑ | 0.77 (0.66–0.88) | NA | No |
| CpG site 2 30 | ↑ | 0.79 (0.69–0.88) | NA | No | |
| Mean of sites 1 and 2 30 | ↑ | 0.80 (0.70–0.90) | NA | No | |
| Microbiota genera a | Desulfovibrio 31 , 32 | ↑ | 0.90 | NA | CV |
| ↑ | 0.71 (0.60–0.82) | 0.001 | No | ||
| Escherichia 31 | ↑ | 0.90 | NA | CV | |
| Faecalbacterium 31 | ↓ | 0.92 | NA | CV | |
| Megasphaera 32 | ↑ | 0.75 (0.64–0.85) | <0.001 | No | |
| Oscillospira 31 | ↑ | 0.90 | NA | CV | |
| Prevotella 7 32 | ↑ | 0.74 (0.64–0.84) | <0.001 | No | |
| Veillonela 32 | ↑ | 0.86 (0.77–0.94) | <0.001 | No | |
| Streptococcus 1 35 | ↑ | 0.77 | <0.001 | No | |
| Streptococcus 2 35 | ↑ | 0.84 | <0.001 | No | |
| Microbiota species a | Bifidobacterium dentium 32 | ↑ | 0.74 (0.64–0.85) | <0.001 | No |
| Lactobacillus salivarius 32 | ↑ | 0.71 (0.59–0.82) | 0.001 | No | |
| Streptococcus mitis 32 | ↓ | 0.72 (0.61–0.83) | <0.001 | No | |
| Streptococcus salivarius subsp. Salivarius 32 | ↑ | 0.74 (0.63–0.84) | <0.001 | No | |
| ModelsRef. | |||||
| Fecal microbiota (9 OTUs) 34 | 0.97 (0.94–1.00) | NA | No | ||
| Fecal microbiota (13 OTUs) 23 | 0.94 (0.88–1.00) | NA | CV | ||
| Stool pellet‐based model c 37 | 0.76 | NA | No | ||
Note: In bold are AUC values > 0.90. CV is a resampling validation technique that tests and trains a model using different portions of the data.
Abbreviations: AUC, area under the curve; CI, confidence interval; CV, cross validation; GC, gastric cancer; OTU, operational taxonomic units; TERT, telomerase reverse transcriptase.
Only microbiota genera and species with reported relative abundance and reaching AUC >0.70 were included (suggested by 38 as moderate accuracy).
P < 0.001.
Klebsiella, Subdoligranulum, Prevotella 9, Streptococcus, Ruminiclostridium 9.
Urine
Urine was the second most explored liquid biopsy in GC detection over the last decade, with 29 eligible studies included in this systematic review. Similarly to that observed with saliva and stool, most studies were performed in Asian populations: 12 from China, 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 five from Japan, 51 , 52 , 53 , 54 , 55 four from Taiwan, 56 , 57 , 58 , 59 three from South Korea, 37 , 60 , 61 and one from Iran. 62 The remaining four studies were carried‐out in Canada, 63 European portion of Russia 64 , 65 and Zambia. 66 Most urine‐based reports focused on metabolites, mainly using metabolomic approaches. 39 , 41 , 44 , 45 , 46 , 47 , 50 , 56 , 59 , 60 , 61 , 62 , 63 , 66 Six studies focused on protein analysis 42 , 43 , 48 , 54 , 55 , 65 and three on RNA molecules, particularly miRNAs, quantified in urine samples by reverse transcription‐real time PCR (RT‐qPCR). 52 , 57 , 58 Other biomarkers include nitrate, nitrite, and N‐nitroso compounds, 49 nematode‐NOSE (N‐NOSE), 53 analyzed by the olfactory behavior of C. elegans according to chemotaxis value, oxidative modifications, such as 8‐hydroxydeoxyguanosine (8‐OHdG) and 8‐hydroxyguanosine (8‐OHG), 40 glycans, 51 and volatile organic compounds (VOCs), 64 all reported once. Table 3 includes the urinary biomarkers able to distinguish between GC patients, including early GC, and controls. Most studies included training and validation sets although from the same sample of recruited participants. Considering all biomarkers independently, endothelial lipase presented the highest AUC value of 0.97. 42 On the other hand, several models, mostly including metabolites, reached C‐statistics superior to 0.95, particularly a model including age, β‐(pyrazol‐1‐yl)‐L‐alanine (L‐PA), D‐isoleucine, and D‐serine (AUC = 0.98, 95% CI: 0.95–1.00). 44 Furthermore, only one study reporting a model of 17 metabolites included a validation set (AUC = 0.97). 60 Focusing on early GC detection, two miRNAs, miR‐6807‐5p and miR‐6856‐5p, were assessed and a model including both molecules and H. pylori infection reached an AUC value of 0.75. 52
TABLE 3.
Urinary biomarkers for gastric cancer and early gastric cancer detection with available expression and AUC data
| Gastric cancer detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Urine | |||||||
| Metabolites | 3‐Hydroxybutyrate 61 | ↓ | 0.71 (0.63–0.79) | NA | NA | NA | Pre‐validation |
| Ala 41 , 61 | ↑ | 0.80 | NA | 78.3 | 67.8 | Pre‐validation | |
| ↑ | 0.75 (0.65–0.83) | NA | 70.0 | 80.0 | Pre‐validation | ||
| Benzylmalonic acid 41 | ↑ | 0.67 | NA | 47.2 | 83.9 | Pre‐validation | |
| Citrate 61 | ↑ | 0.63 (0.53–0.72) | NA | 50.0 | 70.0 | Pre‐validation | |
| Creatine 61 | ↑ | 0.86 (0.79–0.92) | NA | 80.0 | 90.0 | Pre‐validation | |
| Creatinine 61 | ↓ | 0.72 (0.63–0.80) | NA | 60.0 | 80.0 | Pre‐validation | |
| Glycerol 61 | ↓ | 0.94 (0.89–0.98) | NA | 90.0 | 90.0 | Pre‐validation | |
| Gly 41 | ↑ | 0.74 | NA | 91.5 | 41.4 | Pre‐validation | |
| Hippurate 61 | ↑ | 0.74 (0.65–0.82) | NA | 70.0 | 70.0 | Pre‐validation | |
| D‐Ile 44 | ↑ | 0.76 (0.65–0.87) | NA | 58.1 | 95.0 | Pre‐validation a | |
| Ethyl 2‐methylacetoacetate 41 | ↑ | 0.72 | NA | 53.8 | 80.4 | Pre‐validation | |
| Ile 41 | ↑ | 0.77 | NA | 67.0 | 72.4 | Pre‐validation | |
| Met 41 | ↑ | 0.78 | NA | 67.9 | 78.2 | Pre‐validation | |
| Levulinic acid 41 | ↑ | 0.67 | NA | 65.1 | 64.4 | Pre‐validation | |
| L‐PA 44 | ↓ | 0.89 (0.82–0.96) | NA | 83.7 | 77.5 | Pre‐validation a | |
| p‐cresol 41 | ↑ | 0.70 | NA | 72.6 | 62.1 | Pre‐validation | |
| Phe 61 | ↑ | 0.80 (0.74–0.88) | NA | 80.0 | 70.0 | Pre‐validation | |
| Pro 41 | ↑ | 0.79 | NA | 84.0 | 63.2 | Pre‐validation | |
| D‐Ser 44 | ↑ | 0.78 (0.68–0.88) | NA | 58.1 | 87.5 | Pre‐validation a | |
| Ser 41 | ↑ | 0.81 | NA | 72.6 | 75.9 | Pre‐validation | |
| Taurine 61 | ↑ | 0.76 (0.66–0.84) | NA | 80.0 | 70.0 | Pre‐validation | |
| Thr 41 | ↑ | 0.82 | NA | 81.1 | 67.8 | Pre‐validation | |
| Trp 41 | ↑ | 0.70 | NA | 82.1 | 51.7 | Pre‐validation | |
| Tyr 41 | ↑ | 0.69 | NA | 85.8 | 47.2 | Pre‐validation | |
| Val 41 | ↑ | 0.73 | NA | 62.3 | 73.5 | Pre‐validation | |
| Modified nucleosides spectra 46 | ↑ | 0.95 | NA | 84.0 | 95.8 | No | |
| N‐NOSE 53 | ↑ | 0.87 (0.82–0.93) | <0.001 | NA | NA | No | |
| Oxidative modification | DNA (8‐OHdG) 40 | ↑ | 0.78 (0.70–0.86) | NA | NA | NA | No |
| RNA (8‐OHG) 40 | ↑ | 0.84 (0.77–0.91) | NA | NA | NA | No | |
| Proteins | ADAM12 54 , 55 | ↑ | 0.76 (0.64–0.88) | <0.001 | NA | NA | No |
| ↑ | 0.70 (0.59–0.81) | 0.001 | NA | NA | Pre‐validation | ||
| Endothelial lipase 42 , 43 | ↓ | 0.97 (0.94–0.99) | NA | 79.0 | 100.0 | No | |
| ↓ | > 0.90 | NA | NA | NA | No | ||
| MMP‐9/NGAL 55 | ↑ | 0.66 (0.53–0.79) | 0.02 | NA | NA | No | |
| TFF1 54 | ↑ | 0.85 (0.77–0.93) | <0.001 | NA | NA | Pre‐validation | |
| TFF3 48 | ↑ | 0.87 | <0.001 | 80.4 | 80.1 | No | |
| RNA | miR‐376c 57 | ↑ | 0.70 | NA | 60.0 | 64.0 | No |
| miR‐6807‐5p 52 | ↑ | 0.87 (0.80–0.99) | NA | NA | NA | Pre‐validation | |
| miR‐6856‐5p 52 | ↑ | 0.71 (0.60–0.81) | NA | NA | NA | Pre‐validation | |
| ModelsRef. | |||||||
| Age and amino acids (L‐PA, D‐Ile, D‐Ser) 44 | 0.98 (0.95–1.00) | NA | 90.7 | 95.0 | Pre‐validation | ||
| Metabolites (2‐HIB, 3‐IS, Ala) 63 | 0.95 (0.86–0.99) | NA | 95.0 | 80.0 | No | ||
| 2 metabolites 39 | 1.00 | NA | NA | NA | No | ||
| 5 metabolites b 47 | 0.96 (0.92–1.00) | NA | 85.7 | 90.3 | Pre‐validation | ||
| 14 metabolites 41 | 0.89 | NA | 77.4 | 85.1 | Pre‐validation | ||
| 17 metabolites 60 | 0.97 | <0.001 | NA | NA | Validation set | ||
| Microbiota c 37 | 0.82 | NA | 67.7 | 84.9 | No | ||
| Urine | |||||||
| miRNAs (miR‐6807‐5p and miR‐6856‐5p) 52 | 0.87 (0.81–0.94) | NA | NA | NA | Pre‐validation | ||
| miRNAs (miR‐6807‐5p and miR‐6856‐5p) and H. pylori 52 | 0.89 (0.82–0.95) | NA | NA | NA | Pre‐validation | ||
| Proteins (ADAM12 and TFF1) 54 | 0.81 (0.72–0.90) | <0.001 | NA | NA | Pre‐validation | ||
| Proteins (ADAM12 and TFF1) and H. pylori 54 | 0.87 (0.79–0.95) | <0.001 | NA | NA | Pre‐validation | ||
| Proteins (MMP‐9/NGAL and ADAM12) 55 | 0.83 (0.72–0.93) | <0.001 | NA | NA | No | ||
| Early gastric cancer detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Urine | |||||||
| RNA | miR‐6807‐5p 52 | ↑ | 0.68 (0.62–0.75) | NA | NA | NA | Pre‐validation |
| miR‐6856‐5p 52 | ↑ | 0.64 (0.57–0.71) | NA | NA | NA | Pre‐validation | |
| ModelsRef. | |||||||
| 2 miRNAs (miR‐6807‐5p and miR‐6856‐5p) 52 | 0.68 (0.62–0.75) | NA | NA | NA | Pre‐validation | ||
| 2 miRNAs (miR‐6807‐5p and miR‐6856‐5p) and H. pylori 52 | 0.75 (0.68–0.81) | NA | NA | NA | Pre‐validation | ||
Note: In bold are AUC values > 0.90. Pre‐validation means the biomarkers have been validated using the same population.
Abbreviations: 2‐HIB, 2‐hydroxyisobutyrate; 3‐IS, 3‐indoxylsulfate; AUC, area under the receiver operating characteristic (ROC) curve; CI, confidence interval; GC, gastric cancer.
Values relative to training set.
Myo‐inositol, lactic acid, 3‐indoxylsulfate, glutamine, and 1‐methylnicotinamide.
Peptoniphilus, Diaphorobacter, Neisseria, Staphylococcus, Bifidobacterium, Corynebacterium 1, Actinomyces, Acinetobacter, Sphingomonas.
Gastric juice as a tool to enhance upper gastrointestinal endoscopy accuracy
In the last decade, 45% of published studies that explored the role of non‐blood‐based circulating biomarkers in GC detection characterized GJ samples following a case‐control study design and mostly including Han Chinese participants (23 of 36 reports). 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 89 A high level of heterogeneity regarding the class of analyzed biomarkers was observed: (a) 17 studies focused on distinct nucleic acids, including mRNA, miRNA, long non‐coding RNA (lncRNA), long intergenic non‐coding RNA (lincRNA), piwi‐interacting RNA (piRNA), circular RNA, and DNA 67 , 68 , 71 , 74 , 75 , 76 , 77 , 78 , 79 , 81 , 82 , 83 , 84 , 85 , 86 , 88 , 90 ; (b) nine studies on proteins or amino acids 69 , 70 , 72 , 80 , 91 , 92 , 93 , 94 ; (c) four studies primarily on metabolites 95 , 96 , 97 , 98 ; (d) two studies focused on glycoproteins, including CEA, CA19‐9, CA72‐4, and CA50 99 , 100 ; (e) two studies on microbiota 89 , 101 ; (f) one study on intrinsic fluorescence spectra 87 ; (g) one study on blood. 102 GC‐associated biomarkers found in GJ with reported expression, AUC, or sensitivity and specificity values, are summarized in Table 4. Overall, proteins pepsin A, α1‐antitrypsin, and gastricsin had the best discriminatory power, with AUC values of 0.96, 0.96, and 0.94, along with miRNA molecules miR‐21 and miR‐133a, reaching 0.97 and 0.91, respectively. 68 , 75 , 91 , 93 Moreover, three models of amino acids reached AUC values equal or superior to 0.90, one including 14 GJ free amino acids (AUC = 0.90, 95% CI: 0.85–0.96), another including six (AUC = 0.91, 95% CI: 0.85–0.98), and a third including three non‐aromatic amino acids (AUC = 0.90, 95% CI: 0.83–0.97). 73 A few studies reported the capability of GJ biomarkers distinguishing between early GC cases and controls and that data is also displayed in Table 4. The better model includes the same six amino acids as previously mentioned, leucine, threonine, serine, tyrosine, phenylalanine, and tryptophan, with an AUC superior to 0.90. 73 Most of reported biomarkers were either not validated in an independent population or were pre‐validated, that is, validated using a subset from the same pool of participants.
TABLE 4.
Gastric juice‐based biomarkers for gastric cancer and early gastric cancer detection with available expression and AUC data
| Gastric cancer detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Gastric juice | |||||||
| Amino acids | Leu 73 | ↑ | 0.87 (0.79–0.95) | NA | NA | NA | Pre‐validation |
| Phe 70 , 72 , 73 | ↑ | 0.86 (0.76–0.96) | <0.001 | 87.9 | 79.4 | No | |
| ↑ | 0.83 (0.73–0.93) | NA | NA | NA | Pre‐validation | ||
| ↑ | 0.76 (0.65–0.88) | NA | NA | NA | Pre‐validation | ||
| Ser 73 | ↑ | 0.87 (0.79–0.96) | NA | NA | NA | Pre‐validation | |
| Thr 73 | ↑ | 0.90 (0.83–0.97) | NA | NA | NA | Pre‐validation | |
| Trp 70 , 72 , 73 | ↑ | 0.85 (0.76–0.94) | NA | NA | NA | Pre‐validation | |
| ↑ | 0.82 (0.72–0.91) | <0.001 | 60.6 | 94.1 | No | ||
| ↑ | 0.77 (0.65–0.89) | NA | NA | NA | Pre‐validation | ||
| Tyr 70 , 72 , 73 | ↑ | 0.84 (0.74–0.94) | <0.001 | 63.6 | 94.1 | No | |
| ↑ | 0.84 (0.75–0.93) | NA | NA | NA | Pre‐validation | ||
| ↑ | 0.78 (0.67–0–89) | NA | NA | NA | Pre‐validation | ||
| Fluorescence intensity 70 | ↑ | 0.73 (0.62–0.85) | <0.001 | 69.7 | 72.1 | No | |
| Glycoproteins | CA19‐9 99 | NA | <0.002 | 51.4 | 93.3 | No | |
| CA50 99 | NA | <0.01 | 48.4 | 96.7 | No | ||
| CA72‐4 86 , 99 | 0.67 (0.58–0.77) | 0.05 | 65.4 | 62.1 | No | ||
| NA | 0.07 | 30.6 | 96.7 | No | |||
| CEA 86 , 99 | 0.64 (0.54–0.74) | 0.005 | 81.8 | 57−6 | No | ||
| NA | 0.36 | 88.5 | 34.3 | No | |||
| Intrinsic fluorescence spectra 87 | ↑ | 0.87 | NA | 83.2 | 80.7 | Pre‐validation | |
| pH 72 | ↑ | 0.79 (0.69–0.90) | NA | NA | NA | Pre‐validation | |
| Proteins | α1‐antitrypsin 93 | ↑ | 0.84 (0.72–0.95) | <0.001 | 74.0 | 88.0 | Validation set |
| Cystatin D 91 | ↓ | 0.79 | NA | 91.4 | 64.7 | Pre‐validation | |
| Elastase 3A 91 | ↑ | 0.85 | NA | 72.9 | 88.2 | Pre‐validation | |
| Gastric lipase 91 | ↓ | 0.89 | NA | 78.6 | 94.1 | Pre‐validation | |
| Gastricsin 91 | ↓ | 0.93 | NA | 88.6 | 94.1 | Pre‐validation | |
| GIF 94 | ↓ | 0.75 | 0.003 | NA | NA | Validation set | |
| Pepsin A 91 | ↓ | 0.96 | NA | 90.0 | 100.0 | Pre‐validation | |
| S100A9 94 | ↑ | 0.77 | 0.001 | NA | NA | Validation set | |
| Total protein 70 | ↑ | 0.71 (0.60–0.82) | 0.001 | 81.8 | 55.9 | No | |
| RNA | lncRNA‐AA174084 76 | ↑ | 0.85 (0.78–0.92) | <0.001 | 46.0 | 93.0 | No |
| lncRNA‐ABHD11‐AS1 81 | ↑ | 0.65 (0.54–0.77) | <0.01 | 41.0 | 93.4 | No | |
| lncRNA‐RMRP 79 | ↑ | 0.70 (0.59–0.81) | <0.001 | 56.4 | 75.4 | No | |
| miR‐106a 68 | ↓ | 0.87 (0.80–0.95) | <0.001 | 73.8 | 89.3 | No | |
| miR‐106a + miR21 68 | ↓ | 0.98 | NA | NA | NA | No | |
| miR‐129‐1‐3p 82 | ↓ | 0.64 (0.54–0.74) | 0.009 | 45.2 | 83.8 | No | |
| miR‐129‐2‐3p 82 | ↓ | 0.65 (0.55–0.75) | 0.005 | 42.9 | 85.9 | No | |
| miR‐129‐1‐3p + miR‐129‐2‐3p 82 | ↓ | 0.66 (0.56–0.76) | 0.003 | 68.7 | 71.9 | No | |
| miR‐133a 75 | ↓ | 0.91 (0.86–0.96) | < 0.001 | 85.9 | 84.8 | No | |
| miR‐21 68 | ↓ | 0.97 (0.94–1.00) | < 0.001 | 85.7 | 97.8 | No | |
| miR‐421 84 | ↓ | 0.77 (0.68–0.85) | <0.001 | 71.4 | 72.7 | No | |
| piR‐1245 86 | ↑ | 0.89 (0.83–0.94) | <0.0001 | 90.9 | 74.2 | No | |
| ModelsRef. | |||||||
| Gender, pH, Tyr, Phe, Trp 72 | 0.81 (0.70–0.91) | NA | NA | NA | Pre‐validation | ||
| 3 aromatic amino acids (Phe, Trp, Tyr) 73 | 0.85 (0.76–0.94) | NA | NA | NA | Pre‐validation | ||
| 3 non‐aromatic amino acids (Leu, Ser, Thr) 73 | 0.90 (0.83–0.97) | NA | NA | NA | Pre‐validation | ||
| 6 amino acids a 73 | 0.91 (0.85–0.98) | NA | 71.9 | 97.4 | Pre‐validation | ||
| 14 amino acids b 73 | 0.90 (0.85–0.96) | NA | 85.1 | 89.2 | No | ||
| Early gastric cancer detection | |||||||
|---|---|---|---|---|---|---|---|
| Type of biomarker | BiomarkerRef. | Expression in GC | AUC (95% CI) | p‐value | Sensitivity (%) | Specificity (%) | With validation |
| Amino acids | Leu 73 | ↑ | 0.90 (0.78–1.00) | NA | NA | NA | Pre‐validation |
| Phe 69 , 73 | ↑ | 0.84 (0.69–0.99) | NA | NA | NA | Pre‐validation | |
| ↑ | 0.83 (0.75–0.91) | <0.001 | NA | NA | No | ||
| Ser 73 | ↑ | 0.87 (0.71–1.00) | NA | NA | NA | Pre‐validation | |
| Thr 73 | ↑ | 0.90 (0.77–1.00) | NA | NA | NA | Pre‐validation | |
| Trp 69 , 73 | ↑ | 0.87 (0.72–1.00) | NA | NA | NA | Pre‐validation | |
| ↑ | 0.82 (0.74–0.90) | <0.001 | NA | NA | No | ||
| Tyr 69 , 73 | ↑ | 0.87 (0.73–1.00) | NA | NA | NA | Pre‐validation | |
| ↑ | 0.79 (0.70–0.88) | <0.001 | NA | NA | No | ||
| Proteins | AAT 94 | ↑ | 0.71 | 0.03 | NA | NA | Validation set |
| S100A9 94 | ↑ | 0.75 | 0.01 | NA | NA | Validation set | |
| AAT + S100A9 94 | ↑ | 0.81 | 0.001 | NA | NA | Validation set | |
| Total protein 69 | ↑ | 0.72 (0.62–0.81) | <0.001 | 59.2 | 81.4 | No | |
| ModelsRef. | |||||||
| 3 aromatic amino acids (Phe, Trp, Tyr) 73 | 0.87 (0.72–1.00) | NA | NA | NA | Pre‐validation | ||
| 3 non‐aromatic amino acids (Leu, Ser, Thr) 73 | 0.90 (0.79–1.00) | NA | NA | NA | Pre‐validation | ||
| 6 amino acids a 73 | 0.91 (0.81–1.00) | NA | 72.7 | 97.4 | Pre‐validation | ||
| 14 amino acids b 73 | 0.88 (0.79–0.97) | NA | NA | NA | No | ||
Note: In bold are AUC values ≥ 0.90. Pre‐validation means the biomarkers have been validated using the same population, whereas CV is a resampling validation technique that tests and trains a model using different portions of the data.
Abbreviations: AUC, area under the receiver operating characteristic (ROC) curve; CI, confidence interval; GC, gastric cancer.
Including leucine, threonine, serine, tyrosine, phenylalanine, and tryptophan.
Including threonine, serine, alanine, valine, methionine, isoleucine, leucine, tyrosine, phenylalanine, lysine, arginine, phosphoserine, ethanolamine phosphate, and urea. To note that the AUC of these differential GJ free amino acids ranged from 0.65 to 0.86.
DISCUSSION
By 2040, an increase of at least 30% is expected in GC incidence and mortality in Western countries, if no new strategies are implemented. 2 The early screening and diagnosis of GC is of paramount importance to improve the poor overall survival currently reported, of no more than 31% at 5 years, associated with diagnosis at advanced stages of the disease. 103
The concept of liquid biopsy has been widely associated with the study of blood samples. In fact, the National Cancer Institute definition of “liquid biopsy” states: “A test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood”. 104 Although some authors considered it a noninvasive source of biomarkers, 105 that definition is not consensual, as blood is primarily drawn by venipuncture, leading to a poorly empowered citizen since it must be performed by a health care provider and normally in a laboratory or clinical setting. 106 , 107 Therefore, the focus of this systematic review was to explore the role of circulating biomarkers derived from non‐invasive liquid biopsies, namely saliva, urine, and stool, in GC detection over the last decade.
While these samples can be non‐invasively collected by the patients themselves, without the need of medical assistance, potentially reducing inequalities in access to health care, GJ requires access to the stomach in a hospital setting. As a liquid biopsy, GJ represents a renewable reservoir of potential biomarkers, reflecting the functional state of the stomach through its direct contact with the gastric epithelium. 13 It avoids the lack of specificity and dilution of other circulating biomarkers, as it represents a fluid exclusively found in the stomach. 108 This liquid biopsy is not meant to substitute endoscopy as the gold standard for GC detection, but rather complement and enhance its accuracy, as it can be easily obtained during endoscopy without enduring additional discomfort to the patient, improving this one‐stop‐shop approach. 13
Whereas tissue biopsies fail to capture tumor clonal heterogeneity, liquid biopsies allow the analysis of a variety of circulating biological factors shed by the tumor and across distinct tumor‐cell subpopulations. 6 Moreover, they allow the serial sampling of proteomic, transcriptomic, genomic, and epigenetic cancer‐associated alterations. 6
The identification of minimally invasive or noninvasive biomarkers for the early detection of GC has been an emerging field in the last few years. Eighty‐four studies were included in this systematic review, published over the last decade on non‐invasive liquid biopsies‐derived biomarkers for GC detection, approximately a 10th of the blood‐based studies found in the literature, and most were reported on GJ and urine samples. Saliva has only been characterized in 12 studies; however, it is important to note that this liquid biopsy started being explored more recently, with the first report included in this systematic review published in 2016.
Over 10 classes of molecules were analyzed, including more than 60 circulating biomarkers. However, it is important to note the lack of representativeness of early lesions in the included studies. Overall, promising results have been published identifying biomarkers with high sensitivity and specificity for GC detection, irrespectively of sample type and biomarker class (Figure 2), although mostly deriving from single studies with no independent validation. Several molecules reached AUC values equal or superior to 0.90, the best ones being AAL lectin in saliva (AUC = 0.98, 95% CI: 0.97–1.00) 20 and a model including age and three amino acids in urine (L‐PA, D‐isoleucine, and D‐serine, AUC = 0.98, 95% CI: 0.95–1.00). 44 The most promising model with independent a validation set in the same study includes 17 metabolites and exhibited a discrimination capacity of 0.97 for GC. 60
FIGURE 2.
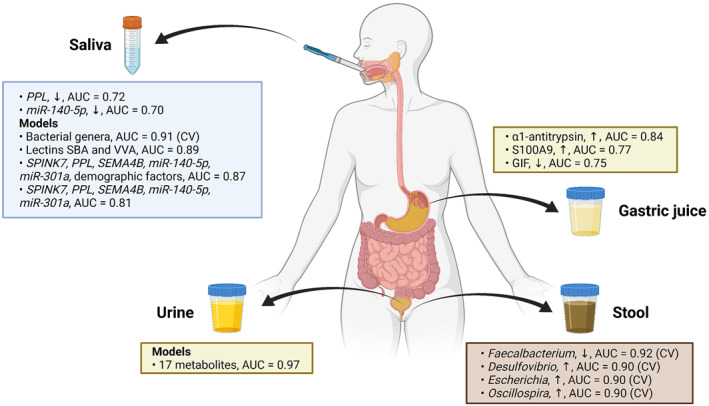
Validated biomarkers with high sensitivity and specificity for gastric cancer detection across all the analyzed liquid biopsy samples, including saliva, gastric juice, urine, and stool, including their expression in cancer (upregulated, ↑, or downregulated, ↓), and area under the curve (AUC) values (created with Biorender.com).
When applying a modified version of the Newcastle‐Ottawa quality assessment scale to each study, several quality concerns were highlighted, mostly associated with undetailed information on (1) sample size or statistical power estimations; (2) comparativeness of groups; (3) representativeness of cases; (4) definition and selection of controls, with most articles not providing information on the screening method used to define the outcome. If in fact biases in study design exist, particularly associated with representativeness of the population, an overestimation of the true association can be assumed, overrating the diagnostic value of the assessed biomarkers, and impairing the results of this systematic review. As highlighted in a review by Herrera‐Pariente et al., 10 there is a need to standardize the methodology for liquid biopsy sample collection and processing, as there is a high heterogeneity, as well as the statistical methods performed in similar studies, in order to increase comparability and implementation in a clinical setting. For example, for detection of salivary biomarkers, in most reports, participants were asked to refrain from eating or drinking for at least 30 min before sampling and whole saliva was collected in the morning. Centrifugation varied between 2600 xg and 13,000 xg during 10–30 min at 4°C. Concerning stool, samples were either preserved at 4°C, −20°C or −80°C until use. Most studies using urine involved collection in the morning before any treatment and storage at −80°C. GJ samples were collected after fasting and centrifuged in most studies, varying between 1000 xg and 10,000 xg from 10 to 30 min before storage. Regarding techniques, most heterogeneity was noticed across different classes of biomarkers: microbiota has been analyzed by sequencing, a high‐throughput technique; RNA molecules by RT‐qPCR, a more targeted approach; glycans and proteins mostly by mass spectrometry, microarrays, ELISA, and western blot; and metabolites by mass spectrometry, from gas chromatography‐mass spectrometry to high performance liquid chromatography‐tandem mass spectrometry, as well as proton nuclear magnetic resonance, which allow the possibility of large‐scale analysis. 109 Overall, a difference in the techniques used was not clearly noticed over time. 27 , 93 However, radioimmunoassay for protein or metabolite detection has only been performed in studies from 2011. 80 , 97
In this systematic review, most included studies were performed in high‐risk Eastern countries, namely China, Japan and South Korea. Translation of those findings in Western populations, without previous validation studies, should be taken with caution due to reported differences in biology, such as the lower proportions of signet ring histology and proximal stomach involvement in Eastern GC patients. 110 A few studies have reported the validation of Eastern survival nonograms or overall survival prediction models in Western populations, particularly American, 111 , 112 and Turkish, who constitute a bridge between East and West. 113 In fact, the Western nonogram was found to be more effective in the Turkish population to estimate the 5‐year overall probability of survival compared to the Eastern nonogram. 113 More recently, Pereira et al. 114 compared Eastern and Western cohorts and disparities reported in survival outcomes appeared not to be molecularly driven, although this study only targeted the E‐cadherin and CD44v6 protein expression. An anticipation in the diagnosis (8 years on average) and more extensive surgical procedures were reported in Eastern populations. 114 Interestingly, a study by Lin et al. 115 compared gene expression profiles from Asian and non‐Asian GC cohorts and found differentially expressed gene signatures related to inflammation and immune function. Genomic and clinical similarities between esophageal and gastric adenocarcinomas located in the cardia have been reported in the literature. 116 Non‐cardia GC is more prevalent globally and shows higher incidence in Asian countries, 117 which represent over 90% of the study population included in this systematic review. In fact, and although only 21 out of 84 studies offered data on tumor location, between 52% and 95% of tumors were ascribed to the non‐cardia site. Future studies should not only report on tumor location but also provide stratified data on unique molecular signatures.
The promise of precision oncology to improve diagnosis and treatment of cancer relies on the molecular profiling of tumors, that mostly depend on invasive sampling procedures that are not always feasible or prone to serial monitoring. An increasingly shift towards liquid biopsies has been observed in the last decade, with several promising non‐blood‐based circulating biomarkers here highlighted. Future research should consider the standardization of preanalytical variables and statistical methods together with adequate reporting, the design of multicenter studies with large enough and independent study populations, to facilitate the comparability of results and demonstration of both the clinical validity and clinical utility, the first step in the clinical adoption of a liquid biopsy test.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supporting Information S1
ACKNOWLEDGMENTS
This article is a result of the project NORTE‐01‐0145‐FEDER‐000050, supported by Norte Portugal Regional Operational Program (NORTE 2020), under the PORTUGAL 2020 partnership agreement, through the European Regional Development Fund (ERDF). Catarina Lopes (UI/BD/151488/2021) and Carina Pereira (SFRH/BPD/114803/2016) are research fellowship holders supported by Fundação para a Ciência e Tecnologia (FCT), co‐financed by European Social Funds (ESF) and national funds of MCTES under the Human Strategic Reference Framework (POCH).
Lopes C, Chaves J, Ortigão R, Dinis‐Ribeiro M, Pereira C. Gastric cancer detection by non‐blood‐based liquid biopsies: a systematic review looking into the last decade of research. United European Gastroenterol J. 2023:11(1):114–30. 10.1002/ueg2.12328
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Ferlay JLM, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global cancer observatory: cancer tomorrow. Lyon: International Agency for Research on Cancer. https://gco.iarc.fr/tomorrow. Accessed 21 Dec 2021. [Google Scholar]
- 3. Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int J Mol Sci. 2020;21(11):4012. 10.3390/ijms21114012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petryszyn P, Chapelle N, Matysiak‐Budnik T. Gastric cancer: where are we heading? Dig Dis. 2020;38(4):280–5. 10.1159/000506509 [DOI] [PubMed] [Google Scholar]
- 5. Yamamoto H, Watanabe Y, Sato Y, Maehata T, Itoh F. Non‐invasive early molecular detection of gastric cancers. Cancers (Basel). 2020;12(10):2880. 10.3390/cancers12102880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–86. 10.1016/j.tips.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 7. Matsuoka T, Yashiro M. Biomarkers of gastric cancer: current topics and future perspective. World J Gastroenterol. 2018;24(26):2818–32. 10.3748/wjg.v24.i26.2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sekiguchi M, Matsuda T. Limited usefulness of serum carcinoembryonic antigen and carbohydrate antigen 19‐9 levels for gastrointestinal and whole‐body cancer screening. Sci Rep. 2020;10(1):18202. 10.1038/s41598-020-75319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. So JBY, Kapoor R, Zhu F, Koh C, Zhou L, Zou R, et al. Development and validation of a serum microRNA biomarker panel for detecting gastric cancer in a high‐risk population. Gut. 2021;70(5):829–37. 10.1136/gutjnl-2020-322065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herrera‐Pariente C, Montori S, Llach J, Bofill A, Albeniz E, Moreira L. Biomarkers for gastric cancer screening and early diagnosis. Biomedicines. 2021;9(10):1448. 10.3390/biomedicines9101448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pimenta‐Melo AR, Monteiro‐Soares M, Libânio D, Dinis‐Ribeiro M. Missing rate for gastric cancer during upper gastrointestinal endoscopy: a systematic review and meta‐analysis. Eur J Gastroenterol Hepatol. 2016;28(9):1041–9. 10.1097/meg.0000000000000657 [DOI] [PubMed] [Google Scholar]
- 12. Sekiguchi M, Oda I. High miss rate for gastric superficial cancers at endoscopy: what is necessary for gastric cancer screening and surveillance using endoscopy? Endosc Int Open. 2017;05(08):E727–8. 10.1055/s-0043-112245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu W, Chung MC. The gastric fluid proteome as a potential source of gastric cancer biomarkers. J Proteomics. 2013;90:3–13. 10.1016/j.jprot.2013.04.035 [DOI] [PubMed] [Google Scholar]
- 14. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst Rev. 2021;10(1):89. 10.1186/s13643-021-01626-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wells GSB, O'Connell D, Robertson J, Peterson J, Welch V, Losos M, et al. The Newcastle‐Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta‐analysis. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp2000. Accessed 28 Sept 2021.
- 16. Aslam MA, Xue C, Liu M, Wang K, Cui D. Classification and prediction of gastric cancer from saliva diagnosis using artificial neural network. Eng Lett. 2021;29(1):10–24. [Google Scholar]
- 17. Chen Y, Cheng S, Zhang A, Song J, Chang J, Wang K, et al. Salivary analysis based on surface enhanced Raman scattering sensors distinguishes early and advanced gastric cancer patients from healthy persons. J Biomed Nanotechnol. 2018;14(10):1773–84. 10.1166/jbn.2018.2621 [DOI] [PubMed] [Google Scholar]
- 18. Huang K, Gao X, Wu L, Yan B, Wang Z, Zhang X, et al. Salivary microbiota for gastric cancer prediction: an exploratory study. Front Cell Infect Microbiol. 2021;11. 10.3389/fcimb.2021.640309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shu J, Yu H, Du H, Zhang J, Zhang K, Li X, et al. Identification of N‐ and O‐linked glycans recognized by AAL in saliva of patients with atrophic gastritis and gastric cancer. Cancer Biomark. 2018;22(4):669–81. 10.3233/cbm-171087 [DOI] [PubMed] [Google Scholar]
- 20. Shu J, Yu H, Li X, Zhang D, Liu X, Du H, et al. Salivary glycopatterns as potential biomarkers for diagnosis of gastric cancer. Oncotarget. 2017;8(22):35718–27. 10.18632/oncotarget.16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu F, Jiang M. Evaluation of predictive role of carcinoembryonic antigen and salivary mRNA biomarkers in gastric cancer detection. Medicine (Baltim). 2020;99(22):e20419. 10.1097/md.0000000000020419 [DOI] [PubMed] [Google Scholar]
- 22. Yao Y, Xu M, Liang L, Zhang H, Xu R, Feng Q, et al. Genome‐wide analysis of Epstein‐Barr virus identifies variants and genes associated with gastric carcinoma and population structure. Tumour Biol. 2017;39(10). 10.1177/1010428317714195 [DOI] [PubMed] [Google Scholar]
- 23. Zhang C, Hu A, Li J, Zhang F, Zhong P, Li Y, et al. Combined non‐invasive prediction and new biomarkers of oral and fecal microbiota in patients with gastric and colorectal cancer. Front Cell Infect Microbiol. 2022;12. 10.3389/fcimb.2022.830684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui J, Cui H, Yang M, Du S, Li J, Li Y, et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell. 2019;10(7):496–509. 10.1007/s13238-018-0596-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li F, Yoshizawa JM, Kim KM, Kanjanapangka J, Grogan TR, Wang X, et al. Discovery and validation of salivary extracellular RNA biomarkers for noninvasive detection of gastric cancer. Clin Chem. 2018;64(10):1513–21. 10.1373/clinchem.2018.290569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao H, Zhang Y, Kim Y, Kim S, Kim JJ, Kim KM, et al. Differential proteomic analysis of human saliva using tandem mass tags quantification for gastric cancer detection. Sci Rep. 2016;6(1):22165. 10.1038/srep22165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koopaie M, Ghafourian M, Manifar S, Younespour S, Davoudi M, Kolahdooz S, et al. Evaluation of CSTB and DMBT1 expression in saliva of gastric cancer patients and controls. BMC Cancer. 2022;22(1):473. 10.1186/s12885-022-09570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu S, Xiang C, Wu J, Teng Y, Wu Z, Wang R, et al. Tongue coating bacteria as a potential stable biomarker for gastric cancer independent of lifestyle. Dig Dis Sci. 2020;66(9):2964–80. 10.1007/s10620-020-06637-0 [DOI] [PubMed] [Google Scholar]
- 29. Sun JH, Li XL, Yin J, Li YH, Hou BX, Zhang Z. A screening method for gastric cancer by oral microbiome detection. Oncol Rep. 2018;39(5):2217–24. 10.3892/or.2018.6286 [DOI] [PubMed] [Google Scholar]
- 30. Liu L, Liu C, Fotouhi O, Fan Y, Wang K, Xia C, et al. TERT promoter hypermethylation in gastrointestinal cancer: a potential stool biomarker. Oncol. 2017;22(10):1178–88. 10.1634/theoncologist.2017-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu S, Dai J, Lan X, Fan B, Dong T, Zhang Y, et al. Intestinal bacteria are potential biomarkers and therapeutic targets for gastric cancer. Microb Pathog. 2021;151:104747. 10.1016/j.micpath.2021.104747 [DOI] [PubMed] [Google Scholar]
- 32. Wu J, Zhang C, Xu S, Xiang C, Wang R, Yang D, et al. Fecal microbiome alteration may Be a potential marker for gastric cancer. Dis Markers. 2020;2020:1–17. 10.1155/2020/3461315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie C, Quan R, Wang L, Chen C, Yan W, Fu Y. Diagnostic value of fecal B cell activating factor in patients with abdominal discomfort. Clin Exp Immunol. 2019;198(2):131–40. 10.1111/cei.13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li N, Bai C, Zhao L, Ge Y, Li X. Characterization of the fecal microbiota in gastrointestinal cancer patients and healthy people. Clin Transl Oncol. 2022;24(6):1134–47. 10.1007/s12094-021-02754-y [DOI] [PubMed] [Google Scholar]
- 35. Yu D, Yang J, Jin M, Zhou B, Shi L, Zhao L, et al. Fecal Streptococcus alteration is associated with gastric cancer occurrence and liver metastasis. mBio. 2021;12(6). 10.1128/mBio.02994-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khoshbaten M, Pishahang P, Nouri M, Lashkari A, Alizadeh M, Rostami‐Nejad M. Diagnostic value of fecal calprotectin as a screening biomarker for gastrointestinal malignancies. Asian Pac J Cancer Prev. 2014;15(4):1667–70. 10.7314/apjcp.2014.15.4.1667 [DOI] [PubMed] [Google Scholar]
- 37. Park JY, Kang CS, Seo HC, Shin JC, Kym SM, Park YS, et al. Bacteria‐derived extracellular vesicles in urine as a novel biomarker for gastric cancer: integration of liquid biopsy and metagenome analysis. Cancers (Basel). 2021;13(18):4687. 10.3390/cancers13184687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fischer JE, Bachmann LM, Jaeschke R. A readers' guide to the interpretation of diagnostic test properties: clinical example of sepsis. Intensive Care Med. 2003;29(7):1043–51. 10.1007/s00134-003-1761-8 [DOI] [PubMed] [Google Scholar]
- 39. Chen JL, Fan J, Lu XJ. CE‐MS based on moving reaction boundary method for urinary metabolomic analysis of gastric cancer patients. Electrophoresis. 2014;35(7):1032–9. 10.1002/elps.201300243 [DOI] [PubMed] [Google Scholar]
- 40. Chen Q, Hu Y, Fang Z, Ye M, Li J, Zhang S, et al. Elevated levels of oxidative nucleic acid modification markers in urine from gastric cancer patients: quantitative analysis by ultra performance liquid chromatography‐tandem mass spectrometry. Front Chem. 2020;8. 10.3389/fchem.2020.606495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen Y, Zhang J, Guo L, Liu L, Wen J, Xu L, et al. A characteristic biosignature for discrimination of gastric cancer from healthy population by high throughput GC‐MS analysis. Oncotarget. 2016;7(52):87496–510. 10.18632/oncotarget.11754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dong X, Wang G, Zhang G, Ni Z, Suo J, Cui J, et al. The endothelial lipase protein is promising urinary biomarker for diagnosis of gastric cancer. Diagn Pathol. 2013;8(1):45. 10.1186/1746-1596-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong CS, Cui J, Ni Z, Su Y, Puett D, Li F, et al. A computational method for prediction of excretory proteins and application to identification of gastric cancer markers in urine. PLoS One. 2011;6(2):e16875. 10.1371/journal.pone.0016875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang R, Shen K, He Q, Hu Y, Sun C, Guo C, et al. Metabolic profiling of urinary chiral amino‐containing biomarkers for gastric cancer using a sensitive chiral chlorine‐labeled probe by HPLC‐MS/MS. J Proteome Res. 2021;20(8):3952–62. 10.1021/acs.jproteome.1c00267 [DOI] [PubMed] [Google Scholar]
- 45. Liang Q, Wang C, Li B. Metabolomic analysis using liquid chromatography/mass spectrometry for gastric cancer. Appl Biochem Biotechnol. 2015;176(8):2170–84. 10.1007/s12010-015-1706-z [DOI] [PubMed] [Google Scholar]
- 46. Lin X, Wang L, Lin H, Lin D, Lin J, Liu X, et al. A novel urine analysis technique combining affinity chromatography with Au nanoparticle based surface enhanced Raman spectroscopy for potential applications in non‐invasive cancer screening. J Biophotonics. 2019;12(4):e201800327. 10.1002/jbio.201800327 [DOI] [PubMed] [Google Scholar]
- 47. Shu Z, Ding D, Li Y. Potential urinary metabolite markers for diagnosing gastric cancer. Int J Clin Exp Pathol. 2017;10(6):6743–50. [Google Scholar]
- 48. Xiao L, Liu YP, Xiao CX, Ren JL, Guleng B. Serum TFF3 may be a pharamcodynamic marker of responses to chemotherapy in gastrointestinal cancers. BMC Clin Pathol. 2014;14(1):26. 10.1186/1472-6890-14-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu L, Qu YH, Chu XD, Wang R, Nelson HH, Gao YT, et al. Urinary levels of N‐nitroso compounds in relation to risk of gastric cancer: findings from the shanghai cohort study. PLoS One. 2015;10(2):e0117326. 10.1371/journal.pone.0117326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo C, Hu Y, Cao X, Wang Y. HILIC‐MS/MS for the determination of methylated adenine nucleosides in human urine. Anal Chem. 2021;93(51):17060–8. 10.1021/acs.analchem.1c03829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanzawa K, Tanaka‐Okamoto M, Murakami H, Mukai M, Takahashi H, Omori T, et al. Investigation of acidic free‐glycans in urine and their alteration in cancer. Glycobiology. 2021;31(4):391–409. 10.1093/glycob/cwaa100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iwasaki H, Shimura T, Yamada T, Okuda Y, Natsume M, Kitagawa M, et al. A novel urinary microRNA biomarker panel for detecting gastric cancer. J Gastroenterol. 2019;54(12):1061–9. 10.1007/s00535-019-01601-w [DOI] [PubMed] [Google Scholar]
- 53. Kusumoto H, Tashiro K, Shimaoka S, Tsukasa K, Baba Y, Furukawa S, et al. Efficiency of gastrointestinal cancer detection by nematode‐NOSE (N‐NOSE). In Vivo. 2020;34(1):73–80. 10.21873/invivo.11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shimura T, Dayde D, Wang H, Okuda Y, Iwasaki H, Ebi M, et al. Novel urinary protein biomarker panel for early diagnosis of gastric cancer. Br J Cancer. 2020;123(11):1656–64. 10.1038/s41416-020-01063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shimura T, Dagher A, Sachdev M, Ebi M, Yamada T, Yamada T, et al. Urinary ADAM12 and MMP‐9/NGAL complex detect the presence of gastric cancer. Cancer Prev Res. 2015;8(3):240–8. 10.1158/1940-6207.Capr-14-0229 [DOI] [PubMed] [Google Scholar]
- 56. Hsu WY, Chen CJ, Huang YC, Tsai FJ, Jeng LB, Lai CC. Urinary nucleosides as biomarkers of breast, colon, lung, and gastric cancer in Taiwanese e81701. PLoS One. 2013;8(12):e81701. 10.1371/journal.pone.0081701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hung PS, Chen CY, Chen WT, Kuo CY, Fang WL, Huang KH, et al. miR‐376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12(5):e0177346. 10.1371/journal.pone.0177346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kao HW, Pan CY, Lai CH, Wu CW, Fang WL, Huang KH, et al. Urine miR‐21‐5p as a potential non‐invasive biomarker for gastric cancer. Oncotarget. 2017;8(34):56389–97. 10.18632/oncotarget.16916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lo WY, Jeng LB, Lai CC, Tsai FJ, Lin CT, Chen WT. Urinary cytidine as an adjunct biomarker to improve the diagnostic ratio for gastric cancer in Taiwanese patients. Clin Chim Acta. 2014;428:57–62. 10.1016/j.cca.2013.10.008 [DOI] [PubMed] [Google Scholar]
- 60. Jung J, Jung Y, Bang EJ, Cho SI, Jang YJ, Kwak JM, et al. Noninvasive diagnosis and evaluation of curative surgery for gastric cancer by using NMR‐based metabolomic profiling. Ann Surg Oncol. 2014;21(S4):736–42. 10.1245/s10434-014-3886-0 [DOI] [PubMed] [Google Scholar]
- 61. Kwon HN, Lee H, Park JW, Kim Y.‐H, Park S, Kim JJ. Screening for early gastric cancer using a noninvasive urine metabolomics approach. Cancers. 2020;12(10):2904. 10.3390/cancers12102904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mokhtari M, Rezaei A, Ghasemi A. Determination of urinary 5‐hydroxyindoleacetic acid as a metabolomics in gastric cancer. J Gastrointest Cancer. 2015;46(2):138–42. 10.1007/s12029-015-9700-9 [DOI] [PubMed] [Google Scholar]
- 63. Chan AW, Mercier P, Schiller D, Bailey R, Robbins S, Eurich DT, et al. (1)H‐NMR urinary metabolomic profiling for diagnosis of gastric cancer. Br J Cancer. 2016;114(1):59–62. 10.1038/bjc.2015.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rozhentsov AA, Koptina AV, Mitrakov AA, Sharipova T, Tsapaev I, Ryzhkov VL, et al. A new method to diagnose cancer based on image analysis of mass chromatograms of volatile organic compounds in urine. Sovremennye Tehnologii v Medicine. 2014; 6 (4):151–7. [Google Scholar]
- 65. Albakova Z, Norinho DD, Mangasarova Y, Sapozhnikov A. Heat shock proteins in urine as cancer biomarkers. Front Med. 2021;8. 10.3389/fmed.2021.743476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Asombang AW, Kayamba V, Mwanza‐Lisulo M, Colditz G, Mudenda V, Yarasheski K, et al. Gastric cancer in Zambian adults: a prospective case‐control study that assessed dietary intake and antioxidant status by using urinary isoprostane excretion. Am J Clin Nutr. 2013;97(5):1029–35. 10.3945/ajcn.112.051284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen JS, Wang YF, Zhang XQ, Lv JM, Li Y, Liu XX, et al. H19 serves as a diagnostic biomarker and up‐regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016; 63(2):223–30. 10.4149/207_150821n454 [DOI] [PubMed] [Google Scholar]
- 68. Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, et al. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119(9):1618–26. 10.1002/cncr.27903 [DOI] [PubMed] [Google Scholar]
- 69. Deng K, Lin S, Zhou L, Li Y, Chen M, Wang Y, et al. High levels of aromatic amino acids in gastric juice during the early stages of gastric cancer progression. PLoS One. 2012;7(11):e49434. 10.1371/journal.pone.0049434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Deng K, Lin S, Zhou L, Geng Q, Li Y, Xu M, et al. Three aromatic amino acids in gastric juice as potential biomarkers for gastric malignancies. Anal Chim Acta. 2011;694(1‐2):100–7. 10.1016/j.aca.2011.03.053 [DOI] [PubMed] [Google Scholar]
- 71. Fei ZH, Yu XJ, Zhou M, Su HF, Zheng Z, Xie CY. Upregulated expression of long non‐coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancer. Tumour Biol. 2016;37(2):1983–93. 10.1007/s13277-015-3979-9 [DOI] [PubMed] [Google Scholar]
- 72. Liu J, Lin SR, Li ZP, Zhou LY, Xue Y, Yan XE, et al. A novel gastric juice index model for detecting early gastric cancer. Int J Clin Exp Med. 2017; 10(10):14425–35. [Google Scholar]
- 73. Liu J, Lin S, Li Z, Zhou L, Yan X, Xue Y, et al. Free amino acid profiling of gastric juice as a method for discovering potential biomarkers of early gastric cancer. Int J Clin Exp Pathol. 2018; 11(5):2323–36. [PMC free article] [PubMed] [Google Scholar]
- 74. Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, et al. Increased expression of long intergenic non‐coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35(6):5441–7. 10.1007/s13277-014-1709-3 [DOI] [PubMed] [Google Scholar]
- 75. Shao J, Fang PH, He B, Guo LL, Shi MY, Zhu Y, et al. Downregulated MicroRNA‐133a in gastric juice as a clinicopathological biomarker for gastric cancer screening. Asian Pac J Cancer Prev. 2016; 17 (5):2719–22. [PubMed] [Google Scholar]
- 76. Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–8. 10.1002/cncr.28882 [DOI] [PubMed] [Google Scholar]
- 77. Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–80. 10.1002/cam4.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26(3):1475–82. 10.1007/s12253-019-00716-y [DOI] [PubMed] [Google Scholar]
- 79. Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, et al. LncRNA‐RMRP promotes carcinogenesis by acting as a miR‐206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812–24. 10.18632/oncotarget.9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ruan HL, Hong RT, Xie HJ, Hu NZ, Xu JM, Zhang W. Significance of elevated levels of collagen type IV and hyaluronic acid in gastric juice and serum in gastric cancer and precancerous lesion. Dig Dis Sci. 2011;56(7):2001–8. 10.1007/s10620-011-1571-8 [DOI] [PubMed] [Google Scholar]
- 81. Yang Y, Shao Y, Zhu M, Li Q, Yang F, Lu X, et al. Using gastric juice lncRNA‐ABHD11‐AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 2016;37(1):1183–8. 10.1007/s13277-015-3903-3 [DOI] [PubMed] [Google Scholar]
- 82. Yu X, Luo L, Wu Y, Yu X, Liu Y, Yu X, et al. Gastric juice miR‐129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30(1):365. 10.1007/s12032-012-0365-y [DOI] [PubMed] [Google Scholar]
- 83. Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63(03):442–9. 10.4149/314_150825n45 [DOI] [PubMed] [Google Scholar]
- 84. Zhang X, Cui L, Ye G, Zheng T, Song H, Xia T, et al. Gastric juice microRNA‐421 is a new biomarker for screening gastric cancer. Tumour Biol. 2012;33(6):2349–55. 10.1007/s13277-012-0497-x [DOI] [PubMed] [Google Scholar]
- 85. Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17(8):640–6. 10.1007/s12094-015-1290-2 [DOI] [PubMed] [Google Scholar]
- 86. Zhou X, Liu J, Meng A, Zhang L, Wang M, Fan H, et al. Gastric juice piR‐1245: a promising prognostic biomarker for gastric cancer. J Clin Lab Anal. 2020;34(4). 10.1002/jcla.23131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhou LY, Lin SR, Li Y, Geng QM, Ding SG, Meng LM, et al. The intrinsic fluorescence spectrum of dilute gastric juice as a novel diagnostic tool for gastric cancer. J Dig Dis. 2011;12(4):279–85. 10.1111/j.1751-2980.2011.00507.x [DOI] [PubMed] [Google Scholar]
- 88. Song J, Yu S, Zhong D, Yang W, Jia Z, Yuan G, et al. The circular RNA hsa_circ_000780 as a potential molecular diagnostic target for gastric cancer. BMC Med Genomics. 2021;14(1):282. 10.1186/s12920-021-01096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sun QH, Zhang J, Shi YY, Zhang J, Fu WW, Ding SG. Microbiome changes in the gastric mucosa and gastric juice in different histological stages of Helicobacter pylori‐negative gastric cancers. World J Gastroenterol. 2022;28(3):365–80. 10.3748/wjg.v28.i3.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yamamoto H, Watanabe Y, Oikawa R, Morita R, Yoshida Y, Maehata T, et al. BARHL2 methylation using gastric wash DNA or gastric juice exosomal DNA is a useful marker for early detection of gastric cancer in an H. pylori‐independent manner. Clin Transl Gastroenterol. 2016;7(7):e184. 10.1038/ctg.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wu W, Yong WW, Chung MC. A simple biomarker scoring matrix for early gastric cancer detection. Proteomics. 2016;16(22):2921–30. 10.1002/pmic.201600194 [DOI] [PubMed] [Google Scholar]
- 92. Choi JM, Park WS, Song KY, Lee HJ, Jung BH. Development of simultaneous analysis of tryptophan metabolites in serum and gastric juice ‐ an investigation towards establishing a biomarker test for gastric cancer diagnosis. Biomed Chromatogr. 2016;30(12):1963–74. 10.1002/bmc.3773 [DOI] [PubMed] [Google Scholar]
- 93. Hsu PI, Chen CH, Hsiao M, Wu DC, Lin CY, Lai KH, et al. Diagnosis of gastric malignancy using gastric juice alpha1‐antitrypsin. Cancer Epidemiol Biomarkers Prev. 2010;19(2):405–11. 10.1158/1055-9965.Epi-09-0609 [DOI] [PubMed] [Google Scholar]
- 94. Wu W, Juan WC, Liang CR, Yeoh KG, So J, Chung MC. S100A9, GIF and AAT as potential combinatorial biomarkers in gastric cancer diagnosis and prognosis. Proteomics Clin Appl. 2012;6(3‐4):152–62. 10.1002/prca.201100050 [DOI] [PubMed] [Google Scholar]
- 95. Akyüz M, Ata Ş, Dinç E. A chemometric optimization of method for determination of nitrosamines in gastric juices by GC‐MS. J Pharm Biomed Anal. 2016;117:26–36. 10.1016/j.jpba.2015.08.021 [DOI] [PubMed] [Google Scholar]
- 96. Lee W, Um J, Hwang B, Lee YC, Chung BC, Hong J. Assessing the progression of gastric cancer via profiling of histamine, histidine, and bile acids in gastric juice using LC‐MS/MS. J Steroid Biochem Mol Biol. 2020;197:105539. 10.1016/j.jsbmb.2019.105539 [DOI] [PubMed] [Google Scholar]
- 97. Dias A, Garcia C, Majewski M, Wallner G, McCallum RW, Poplawski C, et al. Gastric juice prostaglandins and peptide growth factors as potential markers of chronic atrophic gastritis, intestinal metaplasia and gastric cancer: their potential clinical implications based on this pilot study. Dig Dis Sci. 2011;56(11):3220–5. 10.1007/s10620-011-1758-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang YK, Chiang WC, Kuo FC, Wu MC, Shih HY, Wang SSW, et al. Levels of malondialdehyde in the gastric juice: its association with Helicobacter pylori infection and stomach diseases. Helicobacter. 2018;23(2):e12460. 10.1111/hel.12460 [DOI] [PubMed] [Google Scholar]
- 99. Virgilio E, Giarnieri E, Montagnini M, D''Urso R, Proietti A, Mesiti A, et al. Analyzing gastric lavage of gastric cancer patients: a prospective observational study on cytopathology and determination of intragastric CEA, CA 19.9, CA 72.4, and CA 50. Acta Cytol. 2016;60(2):161–6. 10.1159/000445765 [DOI] [PubMed] [Google Scholar]
- 100. Keller J, Reiss‐Sklan E, Refael M, Andresen V, Levy‐Herman Y, Ruvinsky I. Ca72‐4 may contribute to real‐time reconnaissance of gastric cancer. F1000Research. 2012;1:33. 10.12688/f1000research.1-33.v1 [DOI] [Google Scholar]
- 101. Park JY, Seo H, Kang CS, Shin TS, Kim JW, Park JM, et al. Dysbiotic change in gastric microbiome and its functional implication in gastric carcinogenesis. Sci Rep. 2022;12(1):4285. 10.1038/s41598-022-08288-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kayamba V, Chomba M, Kelly P. Short term micronutrient‐antioxidant supplementation has no impact on a serological marker of gastric atrophy in Zambian adults: retrospective analysis of a randomised controlled trial. BMC Gastroenterol. 2014;14(1):52. 10.1186/1471-230x-14-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Libânio D, Ortigão R, Pimentel‐Nunes P, Dinis‐Ribeiro M. Improving the diagnosis and treatment of early gastric cancer in the West. GE ‐ Portuguese J Gastroenterol. 2021;29(5):299–310. 10.1159/000520529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. NCI dictionary of cancer terms. https://www.cancer.gov/publications/dictionaries/cancer‐terms. Accessed 21 Dec 2021.
- 105. Jing R, Liu S, Jiang Y, Zong W, Ju S, Cui M. Determination of serum RP11‐731F5.2 as a noninvasive biomarker for gastric cancer diagnosis and prognosis. Pathol Res Pract. 2020;216(12):153261. 10.1016/j.prp.2020.153261 [DOI] [PubMed] [Google Scholar]
- 106. Buowari OY. Complications of venepuncture. Adv Biosci Biotechnol. 2013;04(01):126–8. 10.4236/abb.2013.41A018 [DOI] [Google Scholar]
- 107. Adler A, Geiger S, Keil A, Bias H, Schatz P, de Vos T, et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14(1):183. 10.1186/1471-230x-14-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kon OL, Yip T.‐T, Ho MF, Chan WH, Wong WK, Tan SY, et al. The distinctive gastric fluid proteome in gastric cancer reveals a multi‐biomarker diagnostic profile. BMC Med Genomics. 2008;1(1):54. 10.1186/1755-8794-1-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou J, Yin Y. Strategies for large‐scale targeted metabolomics quantification by liquid chromatography‐mass spectrometry. Analyst. 2016;141(23):6362–73. 10.1039/C6AN01753C [DOI] [PubMed] [Google Scholar]
- 110. Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA: A Cancer J Clinicians. 2021;71(3):264–79. 10.3322/caac.21657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Woo Y, Goldner B, Son T, Song K, Noh SH, Fong Y, et al. Western validation of a novel gastric cancer prognosis prediction model in US gastric cancer patients. J Am Coll Surg. 2018;226(3):252–8. 10.1016/j.jamcollsurg.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 112. Liu J, Geng Q, Liu Z, Chen S, Guo J, Kong P, et al. Development and external validation of a prognostic nomogram for gastric cancer using the national cancer registry. Oncotarget. 2016;7(24):35853–64. 10.18632/oncotarget.8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Akgül Ö, Ocak S, Gündoğdu SB, Yalaza M, Güldoğan CE, Tez M. Comparison of East and West survival nomograms in Turkish gastric cancer patients who underwent radical surgery. Scand J Surg. 2018;107(4):308–14. 10.1177/1457496918766724 [DOI] [PubMed] [Google Scholar]
- 114. Pereira C, Park JH, Campelos S, Gullo I, Lemos C, Solorzano L, et al. Comparison of East‐Asia and West‐Europe cohorts explains disparities in survival outcomes and highlights predictive biomarkers of early gastric cancer aggressiveness. Int J Cancer. 2021;150(5):868–80. 10.1002/ijc.33872 [DOI] [PubMed] [Google Scholar]
- 115. Lin SJ, Gagnon‐Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, et al. Signatures of tumour immunity distinguish Asian and non‐Asian gastric adenocarcinomas. Gut. 2015;64(11):1721–31. 10.1136/gutjnl-2014-308252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–75. 10.1038/nature20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kumar S, Mantero A, Delgado C, Dominguez B, Nuchovich N, Goldberg DS. Eastern European and Asian‐born populations are prone to gastric cancer: an epidemiologic analysis of foreign‐born populations and gastric cancer. Ann Gastroenterol. 2021;34:1–9. 10.20524/aog.2021.0640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


