Abstract
The most commonly used β-lactam antibiotics for the therapy of infectious diseases are penicillin and cephalosporin. Penicillin is produced as an end product by some fungi, most notably by Aspergillus (Emericella) nidulans and Penicillium chrysogenum. Cephalosporins are synthesized by both bacteria and fungi, e.g., by the fungus Acremonium chrysogenum (Cephalosporium acremonium). The biosynthetic pathways leading to both secondary metabolites start from the same three amino acid precursors and have the first two enzymatic reactions in common. Penicillin biosynthesis is catalyzed by three enzymes encoded by acvA (pcbAB), ipnA (pcbC), and aatA (penDE). The genes are organized into a cluster. In A. chrysogenum, in addition to acvA and ipnA, a second cluster contains the genes encoding enzymes that catalyze the reactions of the later steps of the cephalosporin pathway (cefEF and cefG). Within the last few years, several studies have indicated that the fungal β-lactam biosynthesis genes are controlled by a complex regulatory network, e.g., by the ambient pH, carbon source, and amino acids. A comparison with the regulatory mechanisms (regulatory proteins and DNA elements) involved in the regulation of genes of primary metabolism in lower eukaryotes is thus of great interest. This has already led to the elucidation of new regulatory mechanisms. Furthermore, such investigations have contributed to the elucidation of signals leading to the production of β-lactams and their physiological meaning for the producing fungi, and they can be expected to have a major impact on rational strain improvement programs. The knowledge of biosynthesis genes has already been used to produce new compounds.
The discovery of antibiotics for clinical use started with the discovery of the efficacy of a β-lactam compound and is perhaps the most important discovery in the history of therapeutic medicine. The application of antibiotics to the therapy of infectious diseases may conceivably have saved more lives than any other medical development (135). It began in 1929, when Alexander Fleming published his observation about the inhibition of growth of Staphylococcus aureus on an agar plate contaminated with Penicillium notatum (117). Three years later, it was shown that the growth inhibition was due to penicillin (71). The first clinical trials with penicillin were undertaken in 1941 (reviewed in reference 2). In parallel with efforts to provide penicillin in large amounts, its structure was elucidated in 1945, when Hodgkin and Low showed by X-ray crystallography analysis that it is composed of a β-lactam structure (reviewed in reference 1).
During the late 1940s, the fungus Cephalosporium acremonium (now renamed Acremonium chrysogenum) was isolated from the sea at Cagliari, Italy, by Guiseppi Brotzu (51). This fungus was first found to produce penicillin N; later, another antibiotic was discovered in the culture broth, which was found to consist of different derivatives of a β-lactam compound designated cephalosporin (reviewed in reference 2). The structure of cephalosporin C was described in 1961 by Abraham and Newton (3) and confirmed by X-ray crystallography analysis (141). The discovery of cephalosporin C generated a whole new group of clinically significant β-lactams.
The success of β-lactams in the treatment of infectious disease is due to their high specificity and their low toxicity. Despite a growing number of antibiotics and the incidence of penicillin-resistant isolates, β-lactams are still by far the most frequently used antibiotics (299).
However, it is only in the past 20 years that the biosynthetic pathways leading to penicillins (penams) and cephalosporins (ceph-3-ems) have been elucidated. This is partly because biosynthetic enzymes are often unstable and are present in the cell in only small amounts, making their purification difficult. In addition, industrial production of penicillin and cephalosporin was achieved with P. chrysogenum and A. chrysogenum, respectively. These fungi, however, belong to the deuteromycetes, which are in general difficult to analyze genetically. Currently, the greatest progress in elucidation of the molecular regulation of biosyntheses of β-lactams in fungi has been made in the penicillin producer Aspergillus (Emericella) nidulans, since this fungus is an ascomycete with a sexual cycle. Therefore, classical genetic techniques can be applied to A. nidulans (262) and hence a detailed genetic map is available (70). Together with molecular techniques, this facilitated a thorough analysis of the genetic regulation of metabolic pathways, including that of penicillin biosynthesis (reviewed in references 20, 49, and 204).
Since the biochemistry of penicillin and cephalosporin biosynthesis is rather well understood and recombinant techniques have been developed for some filamentous fungi, recent research has aimed at elucidating the molecular regulation of β-lactam biosynthesis. Within the last few years, several studies have indicated that the β-lactam biosynthesis genes are controlled by a complex regulatory network. A comparison with known regulatory proteins and DNA elements of eukaryotes involved in the regulation of genes of primary metabolism is thus of great interest. Such investigations might also provide hints about both the evolution of secondary metabolism and the signals leading to the production of β-lactams. Furthermore, the overexpression of regulatory genes will lead to higher yields of β-lactams in the respective production strains and knowledge of biosynthesis genes will allow the production of new compounds by combinatorial biology.
The biosynthesis of β-lactam compounds and their molecular genetics were the subject of several recent reviews (8, 45, 49, 152, 211, 299). In particular, the regulation of β-lactam biosynthesis in fungi has seen a tremendous increase in knowledge within the last years, and it is this aspect which is considered in most of the present review.
GENERAL ASPECTS OF β-LACTAM COMPOUNDS
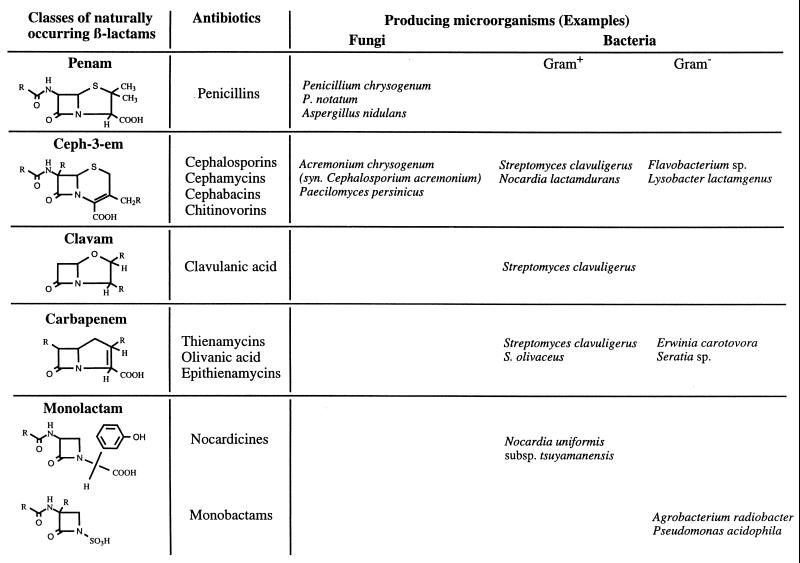
β-Lactams can be classified into five groups on the basis of their chemical structures (Fig. 1). All of these compounds have in common the four-membered β-lactam ring. Apart from the monolactams, which have a single ring only, β-lactams consist of a bicyclic ring system. The ability to synthesize β-lactams is widespread in nature. It has been found in some fungi but also in some gram-positive and gram-negative bacteria (Fig. 1). However, although organisms belonging to all three groups can synthesize the hydrophilic cephalosporin compounds (ceph-3-ems), the hydrophobic penicillins are produced as end products only by filamentous fungi (Fig. 1). For the remaining groups of β-lactams listed in Fig. 1, only bacterial producers have been reported so far. The number of prokaryotic and eukaryotic microorganisms recognized as being able to synthesize β-lactam antibiotics is continuously increasing (152).
FIG. 1.
Naturally occurring classes of β-lactam antibiotics essentially as compiled by O’Sullivan and Sykes (249). Modified from reference 8 with permission of the publisher.
BIOSYNTHESIS OF PENICILLINS AND CEPHALOSPORINS: AN OUTLINE
To give a brief overview of the complete biosynthetic pathways, references for fungal enzymes and genes have been omitted in the text and are given in the tables and the following sections.
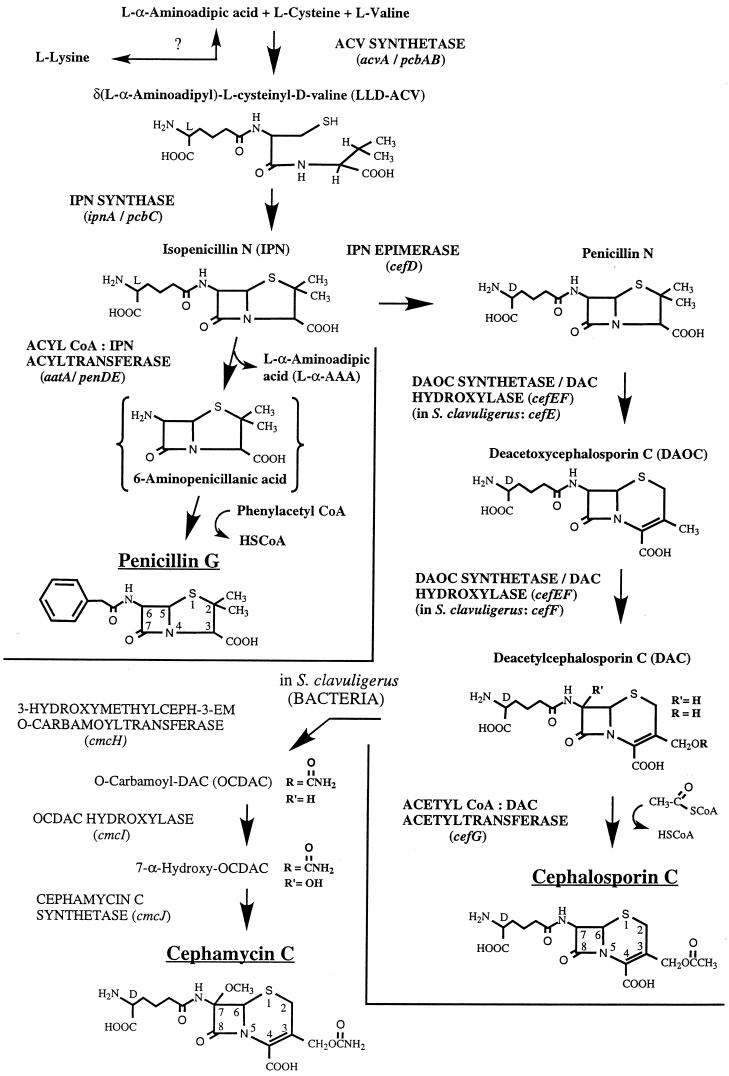
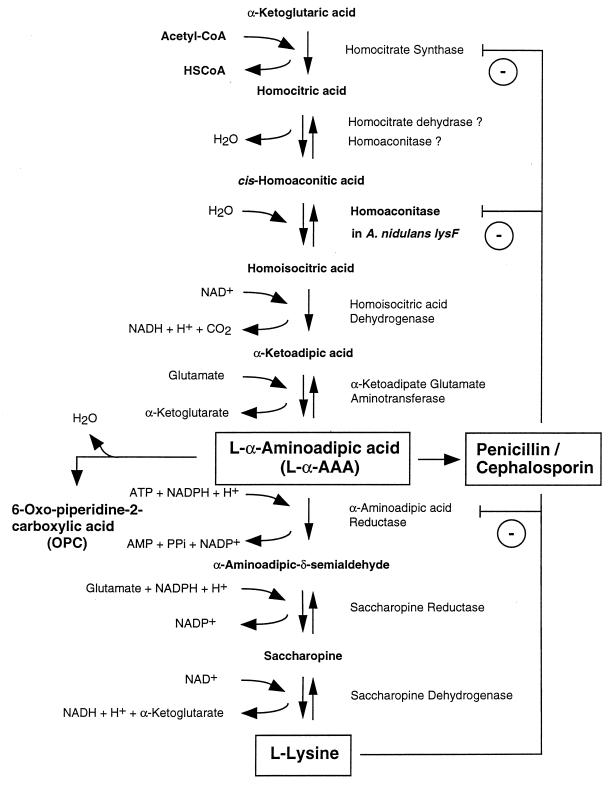
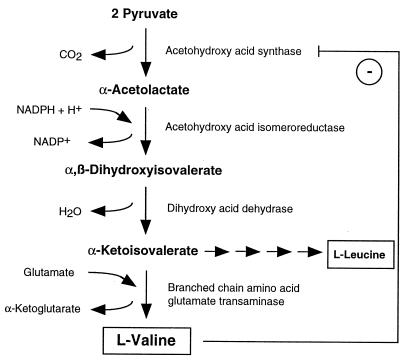
The biosyntheses of penicillins and cephalosporins have the first two steps in common (Fig. 2). All naturally occurring penicillins and cephalosporins are formed from the same three amino acids: l-α-aminoadipic acid (l-α-AAA), l-cysteine, and l-valine. l-α-AAA is a nonproteinogenic amino acid and is derived from the fungus-specific aminoadipate pathway which leads to the formation of l-lysine. It can also be provided, at least in A. nidulans and P. chrysogenum, by catabolic degradation of l-lysine, although the contribution of this pathway to penicillin biosynthesis in these fungi has not been clarified yet (see “Amino acids as precursors and mediators of regulation,” below).
FIG. 2.
Biosynthesis of penicillin, cephalosporin C, and cephamycin C. Gene and organism names are in italics, names of enzymes are in capital letters. l-α-AAA is an intermediate of the l-lysine biosynthetic pathway but can also be provided by catabolic degradation of l-lysine. The question mark indicates that the contribution of the latter to β-lactam production has not been clarified yet. The penicillin biosynthesis occurs in fungi only, whereas cephalosporins are synthesized in both fungi (e.g., cephalosporin C by A. chrysogenum) and bacteria (e.g., cephamycin C by S. clavuligerus) (Fig. 1). See the text for details.
In the first reaction cycle, the amino acid precursors are condensed to the tripeptide δ-(l-α-aminoadipyl)–l-cysteinyl–d-valine (ACV). All reactions which are required for the formation of this tripeptide, e.g., specific recognition of amino acids, their activation via the formation of aminoacyl adenylates, and formation of peptide bonds, are catalyzed by a single multifunctional enzyme designated according to the product formed, ACV synthetase (ACVS) (Fig. 2). ACVS is encoded by a single structural gene designated acvA (pcbAB) (Fig. 2; Table 1).
TABLE 1.
acvA (pcbAB) genes of the different fungi encoding ACVS
| Characteristic | A. nidulans |
P. chrysogenum strains
|
A. chrysogenum | |
|---|---|---|---|---|
| Oli13 | AS-P-78 | |||
| Genome DNA (bp) | 11,310 | 11,328b | 11,376 | 11,136 |
| No. of amino acids | 3,770 | 3,776b | 3,792 | 3,712 |
| Mr | 422,486 | 423,996b | 425,971 | 414,791 |
| Transcript size (kb) | >9.5 | NDa | 11.5 | 11.4 |
| Transcript start position | Major, −230; minor, −317, −195, −188 | ND | ND | ND |
| No. of introns | 0 | 0 | 0 | 0 |
| No. of domains | 3 | 3 | 3 | 3 |
| Reference(s) | 201, 203 | 305, 307 | 91 | 130 |
ND, not determined.
Correction of published sequence in reference 49. Translation start moved upstream by 90 bp. The discrepancy between published sequences of P. chrysogenum is now at the 3′ end.
In the second step, oxidative ring closure of the linear tripeptide leads to the formation of a bicyclic ring structure, i.e., the four-membered β-lactam ring fused to the five-membered thiazolidine ring, which is characteristic of all penicillins. The resulting compound, isopenicillin N (IPN), possesses weak antibiotic activity and is thus the first bioactive intermediate of both the penicillin and cephalosporin pathways. This reaction is catalyzed by isopenicillin N synthase (IPNS), encoded by the ipnA (pcbC) gene (Table 2). IPN is the branch point of the penicillin and cephalosporin biosyntheses.
TABLE 2.
Fungal ipnA (pcbC) genes encoding IPNS
| Characteristic | A. nidulans | P. chrysogenum | A. chrysogenum |
|---|---|---|---|
| Genomic DNA (bp) | 993 | 993 | 1,014 |
| No. of amino acids | 331 | 331 | 338 |
| Mr | 37,480 | 38,012 | 38,416 |
| Transcript size (kb) | ∼1.7 | 1.1 | ∼1.5, 1.15 |
| Transcript start positiona | Major, −106 | −11 | Major, −72 (−73), −56 (−51); minor, −78 (−80), −58 (−54), (−97) |
| No. of introns | 0 | 0 | 0 |
| References | 255, 257, 266, 339 | 34, 63, 304 | 129, 283, 303 |
Values for transcript starts were determined by primer extension or S1 mapping (in parentheses).
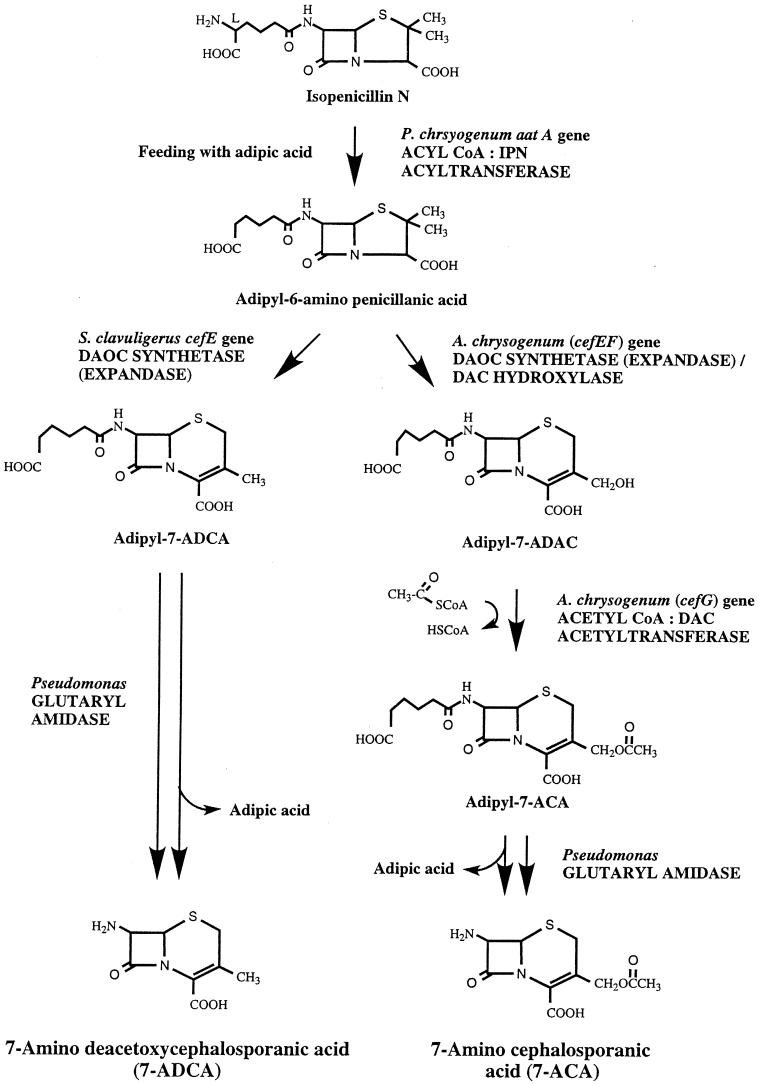
In the third and final step of penicillin biosynthesis, the hydrophilic l-α-AAA side chain of IPN is exchanged for a hydrophobic acyl group; the exchange is catalyzed by acyl coenzyme A (CoA):isopenicillin N acyltransferase (IAT). The corresponding gene has been designated aatA (penDE) (Table 3). In their natural habitats, penicillins DF, F, and K, which contain hexenoic acid, Δ3-hexenoic acid, and octenoic acid as side chains, respectively, are synthesized. By supplying the cultivation medium with phenoxyacetic or phenylacetic acid, the synthesis can be directed mainly toward penicillin V or penicillin G, respectively (Fig. 2, shown for penicillin G). The side chain precursors must be activated before they become substrates for the IAT. It is generally believed that the activated forms of the side chains consist of their CoA thioesters, but the mechanism behind this activation has still not been fully elucidated.
TABLE 3.
aatA (penDE) genes of different fungi encoding IAT
| Characteristic | A. nidulans | P. chrysogenum |
|---|---|---|
| Genomic DNA (bp) | 1,237 | 1,274 |
| No. of amino acids | 357 | 357 |
| Mr | 39,240 | 39,943 |
| Transcript size (kb) | NDa | 1.15 |
| Transcript start position | −61 (−60), −52, −82 | ND |
| No. of introns | 3 | 3 |
| Posttranslationally processed | + | + |
| References | 194, 234, 325, 341 | 35, 325, 334 |
ND, not determined.
The first step that commits the pathway to the production of cephalosporins is the isomerization of the l-α-AAA side chain of IPN to the d enantiomer to give penicillin N. This reaction is catalyzed by IPN epimerase (Fig. 2). Penicillin N is the precursor of antibiotics containing the ceph-3-em nucleus, i.e., cephalosporins and cephamycins (7-methoxycephalosporins), produced by fungi and bacteria, respectively (Fig. 1 and 2). Penicillin N is converted to deacetoxycephalosporin C (DAOC) by DAOC synthetase (expandase) activity (Fig. 2). This ring expansion step involves the oxidative opening of the penam thiazolidine ring to give, upon ring closure, the six-membered dihydrothiazine ring, which is characteristic of all ceph-3-ems. In the next step, the methyl group at C-3 of DAOC is hydroxylated and oxidized to form deacetylcephalosporin C (DAC) (Fig. 2). In A. chrysogenum, both reactions are catalyzed by a single enzyme, DAOC synthetase (expandase)/DAC hydroxylase, encoded by the cefEF gene, whereas in the bacterial cephalosporin producer Streptomyces clavuligerus, one enzyme has been found for each reaction, encoded by the two genes cefE and cefF (Table 4).
TABLE 4.
cefEF genes encoding DAOC/DAC
| Characteristic | A. chrysogenum cefEF |
S. clavuligerus
|
|
|---|---|---|---|
| cefE | cefF | ||
| Genomic DNA (bp) | 996 | 933 | 954 |
| No. of amino acids | 332 | 311 | 318 |
| Mr | 36,462 | 34,519 | 34,584 |
| Transcript size (kb) | 1.2 | ||
| Transcript start position | NDa | ||
| No. of introns | 0 | ||
| Reference(s) | 131, 284 | 177 | 175 |
ND, not determined.
In the last step of cephalosporin C biosynthesis, which is best studied in the fungus A. chrysogenum, an acetyl moiety from acetyl-CoA is transferred to the -OH group of DAC; this step is catalyzed by the product of cefG, acetyl CoA:DAC acetyltransferase (Fig. 2; Table 5). Several cephalosporins that differ from cephalosporin C in the substituent attached to the 3′ C oxygen have been isolated from a variety of microorganisms (152).
TABLE 5.
cefG gene encoding acetyl-CoA:DAC acetyltransferase
| Characteristic |
A. chrysogenum strains
|
||
|---|---|---|---|
| C10 | PLC | IS-5 | |
| Genomic DNA (bp) | 1,332b | 1,300b | 1,299b |
| No. of amino acids | 444b | 399b | 385b |
| Mr | 49,269b | 41,000b | |
| Transcript size (kb) | 1.4 | 1.5 | |
| Transcript start position | NDa | −71, −114 | ND |
| No. of introns | 2 | 2 | 2 |
| Reference(s) | 131 | 219 | 220, 221 |
ND, not determined.
Differences in the size of the published genes result in part from differences in fixation of the start ATG.
In addition, cephalosporins carrying a methoxy group at C-7 (7-methoxycephalosporin, or cephamycin) are produced by both S. clavuligerus (compound A-16884A [138]) and S. lipmanii (239, 265) (Fig. 2). The specific reactions leading to the formation of cephamycin C, which have been studied best in S. clavuligerus, start from the intermediate DAC (Fig. 2). A carbamoyl group is attached to DAC to give O-carbamoyl-DAC (OCDAC). This reaction is catalyzed by 3-hydroxymethyl ceph-3-em O-carbamoyltransferase, which is encoded by the cmcH gene (79) (Fig. 2). Then, C-7 is hydroxylated by the action of OCDAC hydroxylase, encoded by cmcI (343) (Fig. 2). In the final step of cephamycin biosynthesis, the hydroxy group at C-7 is methylated to form cephamycin C (7-methoxycephalosporin C); the reaction is catalyzed by cephamycin C synthetase, whose corresponding gene has been designated cmcJ (79).
GENETIC NOMENCLATURE
Before their identification, the putative genes encoding ACVS were designated pcbA (penicillin cephalosporin biosynthesis) and pcbB, because it was believed that two enzymes were involved in the formation of an l-(α-aminoadipyl)–l-cysteine (AC) dipeptide and the final ACV tripeptide, respectively (Fig. 1) (reviewed in references 147 and 245). Cloning and sequencing of the corresponding gene revealed, however, that a single polypeptide encoded by a single gene is responsible for the formation of the ACV tripeptide. Publications reporting the DNA sequence of the P. chrysogenum, A. nidulans, and A. chrysogenum genes named the gene acvA, which reflected the involvement of one genetic locus in the synthesis of ACVS (201, 203, 305, 307), or pcbAB, derived from the combination of pcbA and pcbB (91, 130). In the following discussion, the term acvA is used because it indicates that a single gene encodes ACVS. The same is relevant for the gene encoding IAT, which has been named penDE or aat. In this review, the gene is designated aatA, reflecting both the correct genetic nomenclature and the fact that one genetic locus encodes the enzyme. The IPNS gene has been named ipnA. The alternative names are shown in parentheses at the beginning of the relevant sections.
CLUSTERING OF BIOSYNTHESIS GENES
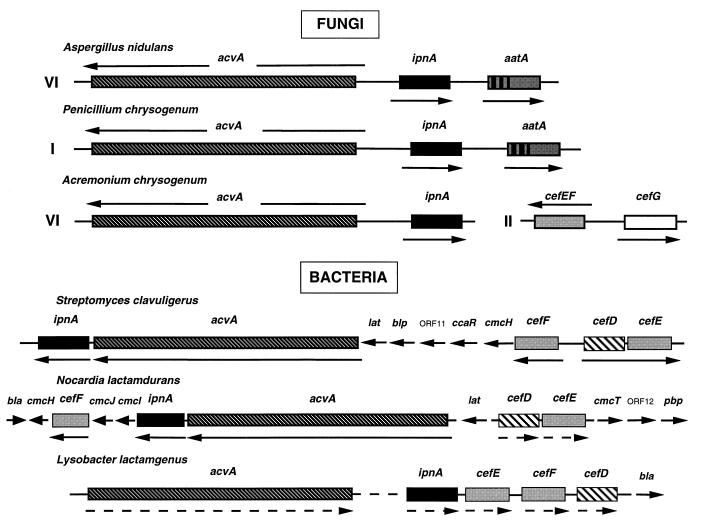
As far as we know, in bacteria and fungi all structural genes of β-lactam biosyntheses are clustered (Fig. 3). In fungi, first the penicillin biosynthesis genes of both A. nidulans and P. chrysogenum are tightly clustered (201, 305). In the following, in A. chrysogenum, two clusters containing the cephalosporin biosynthesis genes have been identified (130, 131, 219, 220), whereas in cephamycin C-producing bacteria, the genes are organized into a single cluster (Fig. 3).
FIG. 3.
β-Lactam biosynthesis gene cluster in fungi and bacteria. References to each fungal gene are listed in Tables 1 to 5 or in the text. The A. chrysogenum cefD gene has not been identified yet. The whole gene cluster of N. lactamdurans has been characterized (76–79). The organization of the L. lactamgenus gene cluster was taken from references 166 and 167. Roman numerals indicate the chromosomes (in fungi) on which the genes are localized (115, 201, 235, 300). The intergenic regions between acvA and ipnA of P. chrysogenum, A. nidulans, and A. chrysogenum are 1,107 bp (91, 307), 872 bp (203) and 1,233 bp (230), respectively. Bacterial genes with fungal homologs are boxed. The transcriptional orientation and the transcript units (Bacteria), as far as it has been determined, are indicated by arrows above or below the boxes. Arrows between boxes (Bacteria) and arrows with broken lines below boxes mark the orientation of genes. The function of ORF is unknown. Abbreviations (76–79, 258): cmcT, transmembrane protein; pbp, penicillin binding protein; bla, β-lactamase; blp, showing similarity to the extracellular β-lactamase inhibitory protein BLIP.
The linkage of antibiotic biosynthesis genes is a well-known phenomenon in many antibiotic-producing organisms. It has been speculated that linkage occurred during evolution owing to an ecological selective advantage (212). Seno and Baltz (294) suggested that coordinated regulation of antibiotic biosynthesis genes could be achieved by organizing the genes into large operons controlled by a single promoter. For example, genes of the actinorhodin biosynthetic pathway in Streptomyces coelicolor are clustered and expressed in several polycistronic mRNAs (207). In eukaryotic fungi, however, β-lactam biosynthesis genes are transcribed separately and are expressed from different promoters (reviewed in reference 49). Hence, in fungi, there is no obvious need for clustering, and it thus seems more likely that linkage reflects a common ancestral origin (see “Origin of β-lactam biosynthesis genes in fungi”). However, there is no evidence that the IAT gene (aatA) has a close relative in modern prokaryotes, even though it is part of the cluster. This fact supports the hypothesis that linkage might also confer an ecological advantage to the eukaryotic fungi in their natural habitat, although the reason for this is not yet understood.
STRUCTURAL GENES AND DEDUCED PROTEINS
Genes Common to Penicillin- and Cephalosporin-Producing Fungi
acvA (pcbAB), encoding ACVS.
The first reaction of the cephalosporin and penicillin biosynthesis is the formation of the ACV tripeptide, which was first found in 1960 as an intracellular component of P. chrysogenum (16). All the reactions required for synthesis of the tripeptide are catalyzed by a single enzyme, ACVS, which is encoded by the acvA (pcbAB) gene (reviewed in reference 169) (Fig. 2; Table 1). Thus, the ACV tripeptide is formed via a nonribosomal enzyme thiotemplate mechanism from its amino acid precursors. This is similar in many aspects to the synthesis of other microbial peptides (reviewed in reference 169).
Early studies with cell-free systems of P. chrysogenum (185) and A. chrysogenum (30) demonstrated the existence of ACVS activity. It was suggested that two separate enzymes may be involved in the formation of ACV. However, based on the observation that less ACV is formed from the dipeptide AC and l-valine than from the free amino acids, Banko et al. (31) proposed that a single multifunctional enzyme may be responsible for ACV synthesis. This proposal was supported by the first isolation of an ACVS protein by van Liempt et al. (333), who purified ACVS of A. nidulans 118-fold. Their results suggested that ACVS consists of a single polypeptide chain. Since then, ACVS enzymes have been purified from different organisms, including S. clavuligerus (25, 26, 157), A. chrysogenum (25, 26, 345), and Nocardia lactamdurans (74). Attempts to purify ACVS from P. chrysogenum have thus far been unsuccessful because the enzyme seems to be rapidly degraded during chromatographic purification (7).
Although not entirely clarified, it is believed that ACVS multienzymes are monomers with molecular masses of around 420 kDa and exhibit different catalytic activities, such as the specific recognition of the three amino acid precursors and their activation, peptide bond formation, isomerization of the l-valine moiety to the d form, and release of the peptide. As in ribosomal peptide biosynthesis, the carboxyl function of the amino acid is activated by the formation of a mixed anhydride with the α-phosphate of ATP, resulting in the release of pyrophosphate (PPi). This has been used to develop assays based on amino acid-dependent exchange of 32P between PPi and ATP (333).
After activation of an amino acid, the aminoacyl adenylate formed is cleaved by the action of an enzyme thiol, resulting in the formation of a thioester bond between the enzyme (at an appropriate location on the enzyme) and the amino acid and in the release of AMP. These thioesterified amino acids play the same role as the tRNA-bound amino acids in the ribosomal peptide biosynthesis. They are high-energy intermediates, which are the targets for nucleophilic attack by the amino group of a second amino acid, resulting in the formation of a peptide bond. As in the ribosome, the nascent peptide grows from the amino terminus to the carboxy terminus and the intermediate peptides remain bound (as thioesters) to the enzyme (169). The substrate specificity is less strict than in protein synthesis, since a variety of tripeptide analogs are known (28).
By assuming three independent activation sites, the dissociation constants for the S. clavuligerus ACVS have been estimated to be 1.25 and 1.5 mM for cysteine and ATP, respectively, and 2.4 and 0.25 mM for valine and ATP, respectively. No l-α-AAA-dependent ATP-PPi exchange was detected with the enzyme preparation used, although the amino acid binding to the enzyme was dependent on ATP (292). The reason for the lack of detection of l-α-AAA-dependent ATP-PPi exchange remains obscure, because fungal ACVS (from both A. nidulans and A. chrysogenum) drove radioactivity exchange that was dependent on all three amino acids (292, 333). Dissociation constants for aminoacyl-tRNA synthetases are much lower than those of S. clavuligerus ACVS, usually below 100 μM for their respective amino acids (64, 150, 161). This may be a way of guaranteeing the supply of amino acids to the primary metabolism and avoiding the depletion of vital cellular components by secondary metabolism (292). l-Valine is apparently epimerized to the d form at the tripeptide stage, since no d-valine intermediate has been detected (292).
Each ACVS is encoded by a single structural gene (designated acvA) of more than 11 kb (Table 1). The clustering of penicillin biosynthesis genes (Fig. 3) was first shown for A. nidulans (201, 305) and P. chrysogenum (305). The identification of the gene cluster was based on the assumption that biosynthesis genes for antibiotics are clustered, and information had accumulated about ipnA genes from several organisms. Hence, by cross-hybridization with ipnA genes as the probe, the acvA genes from both organisms were detected upstream from the ipnA genes, separated by about 1 kb (Fig. 3). In A. nidulans, the presence of an open reading frame (ORF) upstream of the ipnA gene was confirmed by disruption of the upstream region, which led to a non-penicillin-producing phenotype of the transformants (201, 305). Furthermore, MacCabe et al. (201) had purified A. nidulans ACVS and used it to generate partial amino acid sequence data, from which oligonucleotides were deduced and synthesized. They were used for Southern analysis, which showed that the ACVS-encoding gene is in fact upstream of the ipnA gene. In addition, the molecular mapping data obtained predicted a size of more than 11 kb for the acvA gene (201, 305). This was supported by MacCabe et al. (201), who first showed by Northern blot analysis that the acvA transcript is indeed larger than 9.5 kb. This finding was confirmed by the sequencing of the acvA genes of A. nidulans (203), P. chrysogenum (91, 307) and A. chrysogenum (130) and by a Northern blot analysis of the P. chrysogenum acvA gene (91) (Table 1). Subsequently, the corresponding acvA genes were also cloned and sequenced from bacterial cephamycin producers such as S. clavuligerus (Fig. 3).
Even in fungi, the acvA genes form a single ORF, which does not seem to be interrupted by introns, although this assumption has not been proved yet by sequencing of the respective cDNAs. The properties of genes and their deduced enzymes are summarized in Table 1. Fungal acvA genes are divergently oriented with respect to the ipnA genes (Fig. 3). The sizes of the intergenic regions between the genes vary slightly among the different fungi and are about 1 kb long (Fig. 3). In both A. nidulans and P. chrysogenum, the acvA mRNA starts within the intergenic region between acvA and ipnA (Table 1; Fig. 3). acvA expression is driven by a rather weak promoter that is probably located entirely in the intergenic region between acvA and ipnA (see “Expression of biosynthesis genes under standard fermentation conditions” below).
Amino acid sequences of ACVS proteins of all fungal and bacterial species so far identified contain three homologous regions of about 600 amino acids. These contain repeated domains that have extensive amino acid sequence similarities to each other, to the corresponding regions of the ACVS protein of other fungi and bacteria, and to the repeated domains identified for Bacillus brevis gramicidin S synthetases 1 and 2 and tyrocidine synthetase I (169, 329). Since all of these enzymes specifically recognize amino acids and form adenylates, it is most likely that the respective adenylate-forming domains (for the nomenclature of ACVS domains, see reference 169) recognize and adenylate one of the constituent amino acids. The order of the biosynthesis of the δ-(l-α-AAA)–l-Cys–d-Val tripeptide is believed to reflect the linear organization of the ACVS in AAA-, Cys- and Val-activating domains (reviewed in reference 169). A surprising result, however, was the observation of the formation of O-methyl-seryl-d,l-valine by ACVS upon replacement of cysteine by O-methylserine (297). This finding suggested that the second peptide bond is initially formed. Consequently, an order of peptide formation starting with Cys-Val and continuing with addition of l-α-AAA would thus be conceivable. To get more information about the order of peptide formation, Kallow et al. (162) investigated enzyme-bound intermediates by omitting the last amino acid, l-Val, in an in vitro reaction. Depending on the reaction mode, this would lead to the accumulation of either l-cysteinyl- or l-α-AAA–l-cysteinyl intermediates bound to the A. nidulans ACVS enzyme. In fact, the formation of the AC thioester in the absence of l-Val was observed. It was concluded that the first peptide bond is formed between the δ-carboxyl of l-α-AAA and l-Cys and that this is followed by addition of the dipeptidyl intermediate to l-Val. The formation of O-methylseryl-d,l-valine by ACVS previously observed (297) was suggested to be due either to a side reaction initiating peptide synthesis in position 2 of ACVS or to an N-terminal cleavage of the N-terminal aminoadipyl side chain of the tripeptide formed (162). This conclusion is also consistent with the result that l-Cys–d-Val is not a substrate for ACV biosynthesis (297) while δ-(l-AAA)–l-Cys is accepted as a substrate for adenylation and biosynthesis (28, 31; reviewed in reference 7).
Using a microbiological assay for detection of pantothenic acid, Baldwin et al. (25, 26) observed that 1 mol of pantothenic acid was liberated per mol of purified A. chrysogenum ACVS. This implied the presence of one phosphopantetheine molecule per ACVS molecule. It was therefore thought that ACVS follows the classical thiotemplate mechanism; i.e., after activation as aminoacyl adenylates, the intermediates bound as thioesters are assembled by one central swinging arm, the cofactor 4′-phosphopantetheine. Sequencing of the ACVS structural genes (Table 1) revealed, however, that in the three repeated regions of each ACVS, some similarity to 4′-phosphopantetheine attachment sites described for polyketide synthases (i.e., Asp-Ser-Leu) is evident (203). This may reflect the attachment of multiple cofactors to ACVS. Because a single phosphopantetheine arm is sufficient for the activity of fatty acid synthases, the finding of several phosphopantetheine attachment sites suggests a modified mechanism for the thiotemplate pathway to polypeptides (multiple-cofactor model) (203, 291, 313, 314). Although the relevance of all three pantetheine attachment sites of ACVS enzymes has not been proved experimentally, it is believed that peptide assembly is accomplished by the transfer of acyl intermediates between adjacent cofactors (313, 314). In the carboxyl-terminal region of the ACVS enzymes, sequence similarities to the thioesterase active-site region, GXSXG, have been found which would be required to release the generated tripeptide from the enzyme (203).
The current view of the thiotemplate mechanism of ACVS catalysis is summarized in detail by Zhang and Demain (347) and by Kleinkauf and von Döhren (169).
ACVS enzymes are especially interesting since they represent a route for peptide bond formation independent of the ribosome and allow the incorporation of many nonproteinogenic amino acids (28, 168). Furthermore, since different parts of peptide synthetases are specific for certain amino acids, this can be used to engineer genetically new peptide synthetases, and hence to produce new compounds, possibly with new pharmacological activities. This approach has been successfully used by Stachelhaus et al. (312) and is summarized in “Applications” below.
ipnA (pcbC), encoding IPNS.
The second step of the penicillin and cephalosporin biosynthesis, i.e., cyclization of the linear ACV tripeptide to the bicyclic IPN, is catalyzed by IPNS (cyclase), a nonheme-Fe(II)-dependent oxidase (106, 107, 174, 248, 340) (Fig. 2; Table 2). The enzyme has a molecular mass of about 38 kDa and formally catalyzes the removal of four hydrogen equivalents of the ACV tripeptide in a desaturative ring closure with concomitant reduction of dioxygen to water (reviewed in reference 279).
IPNS activity was first detected in cell extracts of A. chrysogenum (107, 174, 248). The IPNS reaction requires ferrous iron, molecular oxygen as the cosubstrate, and ascorbate as the electron donor to form the β-lactam and thiazolidine ring of IPN (151, 288, 340; reviewed in reference 245). It was shown that P. chrysogenum IPNS is strongly inhibited by glutathione and is also sensitive to cobalt inhibition (267). IPNS was purified to homogeneity from A. chrysogenum (27, 142, 251) and has subsequently been obtained from P. chrysogenum (267), A. nidulans (339), several actinomycetes such as S. clavuligerus (153), and the gram-negative bacterium Flavobacterium sp. (250). It was shown that two interconvertable forms of the enzyme exist, an oxidized state with a disulfide linkage and a reduced state (27).
Only the free thiol form of the tripeptide ACV serves as a substrate; the bis-disulfide dimer which is spontaneously formed is inactive (259). S. clavuligerus possesses a disulfide reductase that recognizes bis-ACV as a substrate (6). In P. chrysogenum, a broad-range disulfide reductase belonging to the thioredoxin family of oxidoreductases was found which efficiently reduced bis-ACV to the thiol monomer. When coupled to IPNS in vitro, it converted bis-ACV to IPN and was therefore suggested to play a role in penicillin biosynthesis (72). In enzyme assays in vitro, the thiol groups of both the ACV tripeptide and the IPNS enzyme are kept in a reduced state by the addition of ascorbate and dithiothreitol (see, e.g., reference 46). In these assays, the appearance of antibiotic activity due to the formation of IPNS from the antibiotically inactive ACV is measured with an indicator organism which is sufficiently sensitive (174). Alternatively, IPN can be monitored by high-pressure liquid chromatography (154).
Mössbauer, electron paramagnetic resonance, and optical spectroscopy suggested that ACV binds directly to the active-site iron of IPNS via the cysteinyl thiol of ACV (65). A six-coordinate metal center at the active site was proposed, with two or three endogenous histidine ligands, an aspartate, and sites for the thiolate of ACV, oxygen, and solvent (232). It was shown that the ACV sulfur atom binds to the active-site iron of the enzyme (247, 270). The crystal structure of the A. nidulans IPNS was recently solved at a resolution of 2.5 and 1.3 Å complexed with manganese (279) and with Fe2+ and substrate (280), respectively. The secondary structure of IPNS was found to consist of 10 helices and 16 β-strands. Eight of the β-strands fold to give a “jelly-roll” motif. The active-site structure shows the manganese ion attached to four protein ligands (His 214, Asp 216, His 270, and Gln 330) and bears two water molecules occupying coordination sites directed into a hydrophobic cavity within the protein (279). The Fe(II)-ACV-IPNS structure has one protein molecule with ferrous ion and ACV bound at the active site. The side chain of Gln 330, which coordinates the metal in the absence of substrate, is replaced by the ACV thiolate (280). In the substrate complex, three of the five coordination sites are occupied by protein ligands: His 214, His 270, and Asp 216 (43). The remaining two sites are occupied by a water molecule (at position 298) and the ACV thiolate (280). Such a structural characteristic (an iron-binding site within an unreactive hydrophobic substrate-binding cavity) is probably a requirement for this class of enzyme, since it results in the isolation of the reactive complex and subsequent intermediates from the external environment. Thus, the reaction can be channelled along a single path, avoiding the many side reactions potentially open to the highly reactive species resulting from the reduction of dioxygen at the metal (279). The role of enzymes in such processes has been designated negative catalysis (275). IPNS catalyzes a unique enzyme reaction with no precedent in chemistry (279).
All intact IPNS enzymes whose genes have been cloned to date have proline at position 285 in a highly conserved region (269; reviewed in reference 152). This Pro residue seems to be essential for activity because a mutant (N2) of A. chrysogenum (298) which did not produce cephalosporin encodes a defective ipnA gene, probably due to the mutation of a single base pair that results in a change from Pro (amino acid 285) to Leu (amino acid 269).
Baldwin and Wan (29) proposed a catalytic mechanism for IPNS which involves the formation of an intermediate carbon radical of the LLD form of ACV, but complete details of the reaction have yet to be determined. Additional data on the mechanism of the IPNS reaction suggests that initial formation of the β-lactam ring is followed by closure of the thiazolidine ring (24). A model was proposed by Roach et al. (279, 280).
The IPNS shows a broad substrate specificity, in particular with alterations in the l-α-AAA moiety and the valine residue of ACV. This finding has been ingeniously used to create novel penicillins from ACV analogs in vitro, although cyclization of unnatural tripeptides occurs at lower efficiency (342). Nevertheless, many new penicillins have been produced biosynthetically via this route (23), which is thus very promising for the generation of new β-lactam compounds in vivo.
The genes encoding IPNS enzymes are designated ipnA (pcbC). The ipnA gene from A. chrysogenum was the first gene encoding an enzyme of β-lactam biosynthesis to be cloned and sequenced (283). This was achieved by purification of the enzyme, determination of its N-terminal amino acid sequence, and subsequent cloning of the gene by reverse genetics. The gene was overexpressed in Escherichia coli. ipnA genes have since been cloned and sequenced from several different fungi and bacteria. Their features are summarized in Table 2.
The ipnA and acvA genes lie close together on the chromosome. In contrast to bacteria, in fungi ipnA and acvA are bidirectionally oriented (Fig. 3). For the fungal genes, it has been demonstrated that the ipnA transcripts start in the corresponding intergenic regions (Fig. 3). The fungal IPNS genes identified thus far do not possess introns (reviewed in reference 49).
Gene Specific for Penicillin Biosynthesis
aatA (penDE), encoding IAT.
The third and final reaction of penicillin biosynthesis, which does not occur in cephalosporin biosynthesis and has been found in fungi only, is catalyzed by IAT (10, 11, 54, 98, 123, 199, 263, 265, 311). The hydrophilic l-α-AAA side chain is exchanged for a hydrophobic acyl group, e.g., phenylacetyl in penicillin G (Fig. 2). IAT shows a broad substrate specificity (reviewed in references 198 and 211). By addition of appropriate precursor molecules, the fermentation can be directed toward a specific penicillin; e.g., for production of penicillin G, phenylacetic acid is added, whereas for production of penicillin V, phenoxyacetic acid is added. Once the precursor has been taken up, it must be activated to its CoA thioester. This reaction seems to be carried out by phenylacetyl-CoA ligase (53, 171). It is unclear, however, whether a specific enzyme is needed for this reaction, because a possible candidate is the acetyl-CoA synthetase (ACS), which has been purified from P. chrysogenum and whose structural gene, acuA, has been cloned (215, 216). It was shown that the ACS enzymes of both P. chrysogenum and A. nidulans have the capability to catalyze in vitro the activation (to their CoA thioesters) of some of the side chain precursors required for the production of several penicillins by these fungi (216).
A two-step enzymatic process for the conversion of IPN to penicillin G has been proposed (265). In the first step, IPN is deacylated to 6-aminopenicillanic acid (6-APA), and in the second step, 6-APA is acylated to penicillin G through the addition of a phenylacetyl group from its CoA derivative (Fig. 2). Thus, two enzymatic functions are required, an amidohydrolase and an acyl-CoA:6-APA acyltransferase function. For many years, it remained unclear whether two enzymes and 6-APA as an intermediate are involved in this step of penicillin biosynthesis. The cloning and sequencing of the gene (Table 3) helped to answer this question. It was found that IAT of P. chrysogenum has isopenicillin N amidohydrolase, 6-APA acyltransferase, and penicillin amidase activities, all of which are encoded by the single aatA gene (11). 6-APA remains bound to IAT when the enzyme is saturated with appropriate acyl-CoA substrates but is released in their absence. Significant amounts of 6-APA are produced when exogenous side chain precursors are not fed to P. chrysogenum (reviewed in reference 264).
The purification of P. chrysogenum IAT (9, 10, 199) led to the assumption that the active enzyme is a monomeric protein with a molecular mass of 29 kDa (9, 10). In several partially purified enzyme preparations, however, three proteins of about 40, 29, and 11 kDa were present in ratios that differed among experiments. The discrepancy in the size of IAT among the various purifications was solved by the cloning and sequencing of the corresponding P. chrysogenum and A. nidulans aatA genes independently by Barredo et al. (35) and Tobin et al. (325) (Fig. 3; Table 3). Both groups used purified P. chrysogenum IAT to determine the N-terminal sequence of the 29-kDa subunit. Based on this information, oligonucleotides were used to identify recombinant bacteriophage lambda EMBL-3 clones which carried a single gene, aatA, only. The deduced amino acid sequence showed that the ORF encoded a protein of about 40 kDa. Downstream of this sequence, however, the sequence matched the N-terminal sequence of the 29-kDa protein that was initially purified, implying that the protein was proteolytically processed. The A. nidulans gene was also cloned by both groups. By using oligonucleotides derived from the P. chrysogenum aatA gene, a bacteriophage lambda EMBL-3 clone which was known to contain the ipnA gene (325) and a whole bacteriophage lambda EMBL-3 genomic DNA bank of A. nidulans were screened (234). Computer analysis revealed that in contrast to the other penicillin biosynthesis genes (acvA and ipnA), the aatA genes contain three introns in both organisms at similar positions (Table 3) (35, 325). This was confirmed by analysis of their respective cDNAs (112, 325). No DNA sequence homologous to the aatA gene of P. chrysogenum was found in the genomes of three different strains of A. chrysogenum (129) and actinomycetes (211). This finding is consistent with the notion that 6-APA:acyltransferase activity which is also carried out by IAT, is lacking in A. chrysogenum and other cephalosporin producers (10). Therefore, these organisms do not produce penicillin G or any other penicillins with a hydrophobic side chain.
As mentioned above, the active form of the IAT enzyme was proposed to result from processing of the 40-kDa monomeric precursor to a heterodimer containing subunits of 11 and 29 kDa (10, 35, 325). Further evidence for processing of IAT was provided by Whiteman et al. (341), who purified both the A. nidulans and P. chrysogenum IAT. When the authors used solutions containing one of the P. chrysogenum 11- or 29-kDa proteins only, low levels of activity were measured. This activity could be attributed to an incomplete separation of the two fragments on a gel permeation column. A substantial increase (up to 15-fold) occurred, however, when the solutions containing the subunits were mixed before the assay, indicating that reassociation of the 29- and 11-kDa proteins is required for IAT activity. When both subunits from P. chrysogenum were expressed from two different plasmids in E. coli, production of either subunit in the absence of the other did not result in active recombinant IAT. However, cotransformation of E. coli with two plasmids, each encoding a different IAT subunit, produced recombinant IAT having acyl-CoA:6-APA acyltransferase activity, providing evidence that both subunits are required for activity (323).
To investigate the mechanism of IAT proteolysis in detail, P. chrysogenum IAT was overexpressed from the aatA gene in E. coli and recombinant active IAT was isolated. When purified to homogeneity, recombinant IAT was an α,β-heterodimer, composed of 11-kDa (α) and 29-kDa (β) subunits, derived from a 40-kDa precursor polypeptide by posttranslational cleavage. The processing event that generated the two subunits of recombinant IAT from the 40-kDa precursor polypeptide occurred between Gly 102 and Cys 103 (13). Mutation of aatA in the 11-kDa (α) subunit, resulting in replacement of Thr 105 with Asn, led to inactive and uncleaved recombinant IAT. However, coexpression of this mutant aatA with an aatA derivative encoding the β subunit in E. coli produced acyl-CoA:6-APA acyltransferase activity (323). These results suggest that the formation of recombinant IAT involves cooperative folding events between the subunits. In vitro transcription and translation were used to determine the origin of the IAT hydrolase activity that cleaved the 40-kDa precursor polypeptide. The appearance of a 29-kDa protein (and presumably the corresponding 11-kDa protein, although not observable) from the 40-kDa in vitro-translated protein provided evidence that IAT hydrolysis is an autocatalytic event (323).
Site-directed mutagenesis of the aatA gene and expression in E. coli revealed that Cys 103 is required for IAT proenzyme cleavage. Whether this requirement reflects a direct participation of Cys 103 in cleavage or as part of a cleavage recognition site has not been clarified yet. However, it cannot be entirely excluded yet that Cys 103 is involved in IAT enzyme activity, because all of these experiments were based on the detection of enzyme specific activity (324). The encoded amino acid sequence in the cleavage site is identical in P. chrysogenum and A. nidulans (Arg-Asp-Gly…Cys-Thr-Thr) (13, 14, 324).
IAT enzymes seem to be functionally similar to bacterial penicillin and cephalosporin acylases that catalyze the deacylation of acyl side chains of penicillins and cephalosporins to yield 6-APA and 7-aminocephalosporanic acid, respectively. The penicillin acylase from E. coli ATCC 11105 is a periplasmic enzyme which consists of two nonidentical subunits (40) that are produced by posttranslational processing from a precursor protein (41). This functional similarity between fungal IATs and bacterial acylases is striking. However, there is only very low sequence similarity (approximately 11% identical amino acids) between fungal IATs and the E. coli acylase.
Genes Specific for Cephalosporin Biosynthesis
cefD, encoding IPN epimerase.
The reaction catalyzed by IPN epimerase directs the pathway (Fig. 2) to the production of cephalosporins. IPN epimerase catalyzes the epimerization of the l-α-AAA side chain of IPN to the d enantiomer to give penicillin N, which is the precursor of antibiotics containing the ceph-3-em nucleus, i.e., cephalosporins and cephamycins (7-methoxycephalosporins) (151, 174) (Fig. 2). The purification of A. chrysogenum IPN epimerase proved to be difficult because the enzyme was extremely labile in cell-free preparations (174). So far, no further data on the fungal protein or gene are available. Therefore, it is still unknown whether the putative cefD gene is part of one of the A. chrysogenum clusters (Fig. 3).
The S. clavuligerus IPN epimerase was found to be more stable and was characterized biochemically (155). The corresponding gene (cefD) was cloned and sequenced. cefD is located immediately upstream of the gene encoding the DAOC synthetase (expandase) (cefE) (176) (Fig. 3). cefD encodes 398 amino acid residues with a deduced molecular mass of 43,497 Da. The cloning of the cefD genes of Streptomyces lipmanii (61, 299), Nocardia lactamdurans (77), and Lysobacter lactamgenus YK90 (166, 167) has been reported (Fig. 3).
P. chrysogenum expresses no IPN epimerase activity, and a probe of the S. lipmanii cefD gene did not hybridize to DNA of P. chrysogenum (61). However, some minor IPN epimerase activity exists in P. chrysogenum (12), because recombinant strains of P. chrysogenum expressing the S. clavuligerus cefE gene encoding DAOC synthetase were found to produce DAOC (12, 82) (Fig. 2). Since the DAOC synthetase shows no affinity toward IPN, isomerization of IPN into penicillin N must have occurred (94, 182). However, Alvi et al. (12) emphasized that IPN epimerase activity in P. chrysogenum could be due to some unspecific amino acid racemase. Hence, these experiments do not necessarily imply the presence of a cefD-like gene in P. chrysogenum.
cefEF, encoding DAOC synthetase (ring expandase)/DAC hydroxylase.
In cephalosporin biosynthesis, penicillin N is converted to DAOC by expansion of the five-membered thiazolidine ring to give the six-membered dihydrothiazine ring of DAOC (Fig. 2). This reaction is catalyzed by DAOC synthetase, which possesses the required expandase function (172). The enzyme was purified from A. chrysogenum (93, 182), S. clavuligerus (155), and S. lactamdurans (81). Fungal and bacterial expandases are stimulated by reducing agents, like dithiothreitol or glutathione, and show an absolute requirement for Fe2+, molecular oxygen, α-ketoglutarate, ascorbate, and possibly ATP. These unusual cofactor requirements are characteristic of the class of intermolecular dioxygenases which activate oxygen in the decomposition of equimolar amounts of α-ketoglutarate to form carbon dioxide and succinate (182; reviewed in reference 245).
In the following reaction step, the methyl group at C-3 of DAOC is hydroxylated and oxidized to give DAC (93, 290). This reaction is catalyzed by DAC hydroxylase, which is very similar to DAOC synthetase. The enzyme also belongs to the group of α-ketoglutarate-linked dioxygenases.
Ring expansion by DAOC synthetase and the hydroxylation reaction are both carried out by the same peptide in A. chrysogenum (93, 284, 290). Purification of this enzyme, determination of its N-terminal amino acid sequence, and reverse genetics allowed the cloning of the structural gene, designated cefEF (284). Expression of the cloned cefEF gene in E. coli (284) established unequivocally that in A. chrysogenum one protein is responsible for the ring expansion of penicillin N to DAOC and the 3′ hydroxylation of DAOC to DAC (Fig. 2; Table 4). In contrast, in S. clavuligerus, the two enzymatic activities were separated by anion-exchange chromatography (156) and were later found to be encoded by two genes, cefE and cefF (Table 4; Fig. 2).
The cefEF gene of A. chrysogenum is located on chromosome II (300). It is closely linked to the cefG gene but is transcribed in the opposite direction (Fig. 3). The intergenic region of about 1 kb is believed to contain the promoters for both genes (131). In S. clavuligerus, the cefF gene is closely linked to the cefD and cefE genes but is transcribed in the opposite orientation (Fig. 3) (175). In Lysobacter lactamgenus and Nocardia lactamdurans, different orders of genes were found (75, 77, 79, 166, 167) (Fig. 3).
The only data on regulation of the A. chrysogenum gene thus far reported is a Northern blot analysis which showed that a 1.2-kb transcript corresponding to the cefEF gene was detectable in cells after 48 h of growth in a defined production medium (131).
Gene Specific for Cephalosporin C Biosynthesis in Fungi
cefG, encoding acetyl-CoA:DAC acetyltransferase.
In the last step of the cephalosporin C biosynthesis, which is best studied in the fungus A. chrysogenum, an acetyl moiety from acetyl-CoA is transferred to the -OH group of DAC; this step is catalyzed by acetyl-CoA:DAC acetyltransferase (108, 122) (Fig. 2). The corresponding structural gene (cefG) of A. chrysogenum was cloned and sequenced independently by three groups (Table 5). The cefG gene was cloned based on its close linkage to cefEF. Sequencing of the region adjacent to cefEF led to the identification of an ORF, and a DNA fragment encoding this ORF allowed complementation of the A. chrysogenum mutant M40, which is deficient in acetyl-CoA:DAC acetyltransferase activity. Thus, the identified ORF most probably contained the cefG gene, which was confirmed by overexpression of the gene in A. niger, which naturally lacks such an activity (219). Gutiérrez et al. (131) screened an A. chrysogenum bacteriophage lambda library with a probe specific for the cefEF gene. Northern blotting and DNA sequence analysis revealed the existence of the cefG gene close to the cefEF gene. The authenticity of the cefG gene was proved by complementation of A. chrysogenum ATCC 20371, which lacks acetyl-CoA:DAC acetyltransferase activity. Matsuda et al. (220) cloned the gene by screening a cDNA library with oligonucleotides based on the N-terminal sequence of the enzyme. In addition, they proved the cloning of cefG by performing a gene disruption (replacement) experiment. The cefG-disrupted strains lacked the ability to produce cephalosporin C and accumulated its precursor, DAC, in the culture broth (221).
cefG contains two introns, as demonstrated by sequencing of its cDNA (219, 220). It is closely linked to cefEF but is transcribed in the opposite orientation. The size of the separating intergenic region is not clear, because Gutiérrez et al. (131) found 938 bp, Mathison et al. (219) found 1,077 bp, and Matsuda et al. (220) reported a 1,114-bp intergenic region (Fig. 3; Table 5).
Northern blot analysis of A. chrysogenum RNA showed a very weak transcript of about 1.4 kb, corresponding to the cefG gene, in cells grown in a defined production medium for 48 and 96 h (131, 219). These findings seem to agree with reports on the late conversion of DAC to cephalosporin C in cephalosporin fermentations (344). cefEF appears to be expressed at an earlier stage of the fermentation, suggesting that cefEF and cefG are expressed differently from their intergenic region (131) (Fig. 3).
COMPARTMENTALIZATION OF GENE PRODUCTS
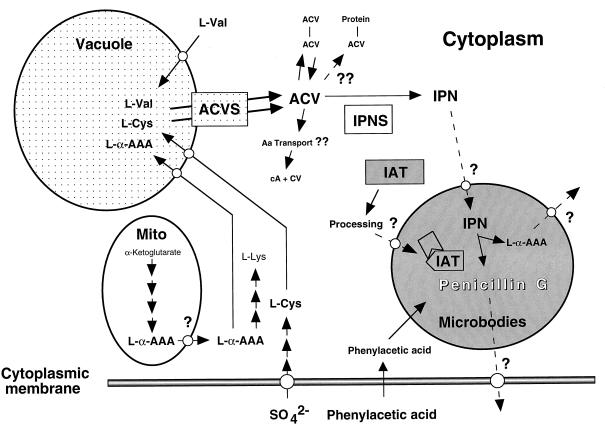
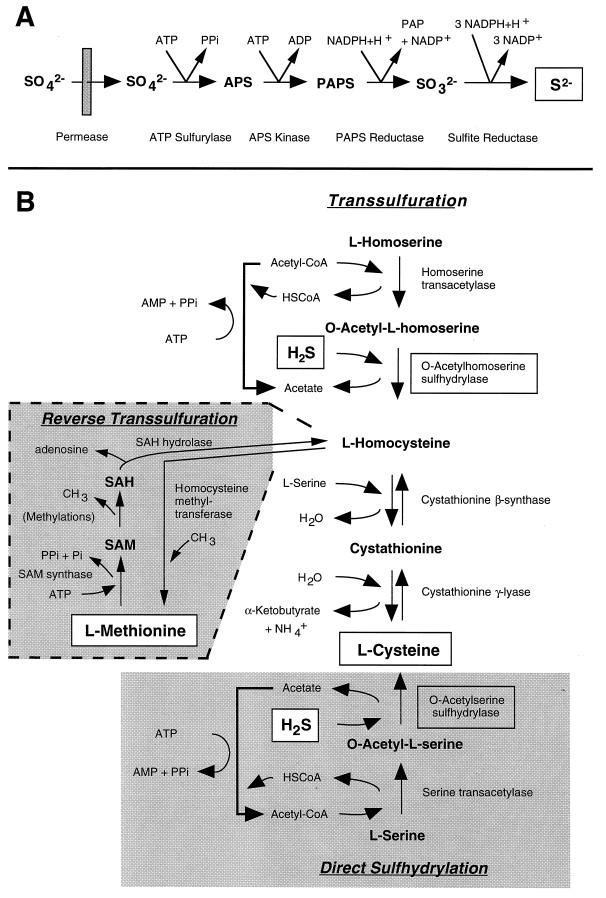
The penicillin biosynthesis pathway occurs in different compartments of the cell (Fig. 4). For localization of the ACVS protein, subcellular fractions obtained from protoplasts of a high-penicillin-producing P. chrysogenum strain (BC 1505) were analyzed. Because ACVS protein was detected in both the membrane and the soluble fraction of purified vacuoles, it was concluded that it is located either within or bound to the vacuolar membrane (187). In addition, a large portion of cellular l-α-AAA, which is most probably used for β-lactam synthesis, is also contained in the vacuoles and thus is sequestered from the cytosol (5, 143) (Fig. 4).
FIG. 4.
Compartmentalization of penicillin biosynthesis gene products. Enzyme names are boxed. Reactions which are hypothetical are labelled by two question marks. Most of the transport processes (indicated by a single question mark) which seem likely to exist because of the compartmentalization of the different enzymes have not been elucidated yet. The ACVS seems to be located within or bound to the vacuolar membrane. IPNS and IAT are located in the cytoplasm and in microbodies, respectively. See the text for details. The stage at which the processing of IAT occurs remains to be determined. Abbreviations: Aa, amino acids; cA, cyclized l-α-AAA (6-oxo-piperdine-2-carboxylic acid [Fig. 6]); CV, l-cysteinyl–d-valine; Mito, mitochondrion.
P. chrysogenum IPNS protein was found in the cytoplasm, whereas IAT was detected in organelles with a diameter of 200 to 800 nm, which were assumed to be microbodies (238) (Fig. 4). The latter result has been supported by the finding that the P. chrysogenum IAT contains a putative targeting signal sequence, a C-terminal alanine-arginine-leucine (34, 125). The importance of this sequence was proved by deleting it in vitro. After transformation of the P. chrysogenum npe6 strain lacking IAT specific activity, the mutated enzyme was located in vacuoles and the neighboring cytoplasm. Although IAT was produced in vivo, as shown by Western blot analysis and by measurement of IAT specific activity in vitro, the mutants did not produce penicillin (237), indicating that targeting of the enzyme to microbodies is essential for penicillin biosynthesis. Furthermore, a positive correlation between the capacity for penicillin production and the number of organelles per cell was observed when different P. chrysogenum strains were compared (238). Hence, the biogenesis of organelles and the genes responsible for this process might have an impact on the penicillin production. The localization of the penicillin biosynthesis in three different cellular compartments reflects the complexity of this biosynthetic pathway. Several transport steps are thus required to bring intermediates of the penicillin biosynthesis pathway together with the enzymes.
Although the presence in P. chrysogenum Wis54-1255 of a transport system for the side chain precursor phenylacetic acid was reported (113), subsequent investigations showed that phenylacetic acid passes the plasma membrane via passive diffusion of the protonated species (140).
ORIGIN OF β-LACTAM BIOSYNTHESIS GENES IN FUNGI
β-Lactam biosynthesis genes have been found both in bacterial species and in fungi. The availability of sequence information about bacterial and fungal genes led to speculations about their evolutionary relationship. Based on several observations, a horizontal transfer of β-lactam biosynthesis genes from bacteria to fungi during evolution has been proposed by several authors (63, 184, 254, 339). This hypothesis was recently questioned by Smith et al. (308). The arguments in favor of a horizontal gene transfer are as follows. (i) ipnA genes of fungi and bacteria show high sequence similarities. More than 60% of the nucleotide bases and 50% of the deduced amino acids are identical. (ii) Bacterial as well as fungal β-lactam genes are organized in clusters. The β-lactam biosynthesis genes in bacteria are organized into a single cluster, as are the penicillin biosynthesis genes in fungi (Fig. 3). The cephalosporin biosynthesis genes in A. chrysogenum are organized into two clusters located on different chromosomes (Fig. 3). This finding led to the assumption that the β-lactam biosynthesis genes were transferred as a single cluster from an ancestral prokaryote to a common ancestor of the β-lactam-synthesizing fungi. In the eukaryotic ancestor, the biosynthesis genes were split between two chromosomes. One part encodes the early genes of β-lactam biosynthesis, and the other encodes the late genes. Later in the lineage, an ancestor of A. nidulans and P. chrysogenum diverged from A. chrysogenum and has presumably lost the second cluster with the genes for the late stage of cephalosporin biosynthesis (299) (Fig. 3). (iii) The G+C content in the third position of codons containing the ipnA gene of A. nidulans and P. chrysogenum is unusually high and could indicate an evolutionary origin from streptomycetes, which show G+C contents of greater than 70% (8). (iv) Fungal acvA and ipnA genes do not contain introns, indicating a bacterial origin of the genes.
In addition, Aharonowitz et al. (8) proposed that during the evolution of β-lactam biosynthesis genes, Streptomyces spp. must have evolved specific resistance mechanisms to avoid self-toxification. If the transfer had occurred from fungi to bacteria, it would have been lethal for bacteria. Hence, the transfer is more likely to have occurred from bacteria to fungi, which would not have been forced to evolve resistance mechanisms because of their lack of susceptibility. In contrast to the other penicillin biosynthesis genes, the aatA genes contain introns. On the basis of linkage of the aatA and ipnA genes (Fig. 3), Skatrud (299) suggested that a sequence functionally related to aatA was transferred together with the β-lactam genes and was later modified to its current functional form. Since IAT possesses amidohydrolase activity to deacylate IPN to 6-APA (11) (Fig. 2), which shows a weak antibiotic activity only, an ancestral amidohydrolase activity in the prokaryotic ancestor might have had a resistance function. Its corresponding gene might have been fused in fungi with a eukaryotic gene (299). Genetic linkage of antibiotic synthesis genes and resistance genes is common in prokaryotes (294). Based on the DNA sequences of ipnA genes from gram-positive streptomycetes and from fungi and a rate of nucleotide substitution of 10−9 nucleotide change per site per year (188), Weigel et al. (339) proposed that the transfer occurred 370 million years ago. The cloning and sequencing of an ipnA gene from a gram-negative bacterium, Flavobacterium sp., however, led to an extension and modification of the hypothesis of horizontal gene transfer. The ipnA gene of Flavobacterium sp. is 69% identical to the streptomycete gene and 64 to 65% identical to the fungal genes (A. chrysogenum and P. chrysogenum) (73). A recent reevaluation of the divergence times of organisms by using a protein clock suggested that gram-positive and gram-negative bacteria split about 2 billion years ago and that prokaryotes and a eukaryotic ancestor split about 3.2 to 3.8 billion years ago (111). If the gene transfer from streptomycetes to fungi had occurred only 370 million years ago, as proposed by Weigel et al. (339), it could be expected that the fungal and streptomycete genes would show a greater similarity than the gram-positive (streptomycete) and gram-negative (Flavobacterium sp.) bacterial genes. As outlined above, this is not the case (73). Hence, Aharonowitz et al. (8) suggested that multiple gene transfer events might have occurred from bacteria to fungi. It is difficult to imagine, however, why these multiple gene transfers then happened at about the same time as would be expected from the degree of similarity among the proteins of the various organisms. In addition, Smith et al. (308) argued against a horizontal transfer. The authors pointed out that the hypothesis of a horizontal gene transfer, e.g., of the ipnA gene, was made on the basis of a very limited data set and was based solely on assumptions about rates of change. They compared the similarity of both IPNS of A. nidulans, P. chrysogenum, A. chrysogenum, S. clavuligerus, S. anulatus, and Flavobacterium sp. and DAOC synthetase of S. clavuligerus and A. chrysogenum. Based on these similarities, they constructed a tree with conventional evolutionary descent. The authors argued that the simplest interpretation is that the genes for the two enzymes resulted from a duplication that occurred before the prokaryote-eukaryote divergence. The topology of the tree rooted with the duplicated enzymes, the depth of the bacterial branches, and the different orientations of the genes in fungi and eubacteria all appear to be consistent with an ordinary evolution for IPNS. However, if the genes appeared very early in the evolution, why have most of the eukaryotes and fungi lost the gene cluster? This question cannot be satisfactorily answered at the moment. Thus, the evolutionary origin of β-lactam biosynthesis genes remains speculative.
REGULATORY CIRCUITS AND REGULATORY GENES
Expression of Biosynthesis Genes under Standard Fermentation Conditions
Studies of the expression of penicillin biosynthesis genes were performed mainly with the E. coli reporter genes lacZ and uidA, encoding β-galactosidase (β-Gal) and β-glucuronidase (β-Glu), respectively (see, e.g., references 46, 124, and 194). Most of the results based on the analysis of gene fusions were supported by Northern or Western blot analysis or determination of enzyme specific activities and penicillin titers.
These studies led to the finding that the promoter strengths of penicillin biosynthesis genes are rather different. It was shown that in A. nidulans, aatA had lower expression than ipnA and threefold-higher expression than acvA (46, 194). A similar observation was made for the corresponding acvA and ipnA genes of both P. chrysogenum and A. chrysogenum. On the basis of reporter gene fusions, it became evident that in both fungi the expression of acvA was much weaker than that of ipnA (109, 230). The intergenic regions between acvA and ipnA thus seem to contain the information required for the remarkable difference in expression levels between acvA and ipnA. The low expression of acvA is, at least in wild-type strains of A. nidulans, rate limiting for penicillin production, because overexpression of acvA led to drastically increased production of penicillin (165) while similar overexpression of ipnA and aatA did not (112).
It seems reasonable to assume that the expression of penicillin biosynthesis genes is coordinated to ensure the synthesis of penicillin by the concomitant appearance of all gene products. But how is coordination achieved? Biosynthesis genes could be expressed simultaneously; i.e., the genes could be activated by the same regulatory factors. Alternatively, the expression of biosynthesis genes could be sequentially induced.
Ramos et al. (268) showed that a mutant of A. chrysogenum (N-2), incapable of producing the β-lactam cephalosporin, lacked IPNS, IPN epimerase, and DAOC synthetase (expandase) activities (Fig. 2). Subsequent investigations revealed that strain N-2 encodes an inactive IPNS caused by a single C-to-T mutation within the coding region of the ipnA gene. It was postulated that a functional IPNS or its biosynthetic product IPN might be necessary for the regulation of the later stages of the biosynthesis, i.e., induction of the cefD and cefEF expression, respectively (Fig. 2 and 3) (269). Furthermore, Hoskins et al. (145) disrupted the acvA gene of A. chrysogenum. Although the predicted alterations of the target gene were not detected, the authors demonstrated IPNS activity in non-cephalosporin-producing transformants. This suggested that the ipnA gene can be expressed without the presence of precursor tripeptide molecules. Hence, in A. chrysogenum, the ipnA gene seems to be coordinatedly regulated whereas the later genes of the cephalosporin pathway (cefD and cefEF) (Fig. 3) appear to be sequentially induced.
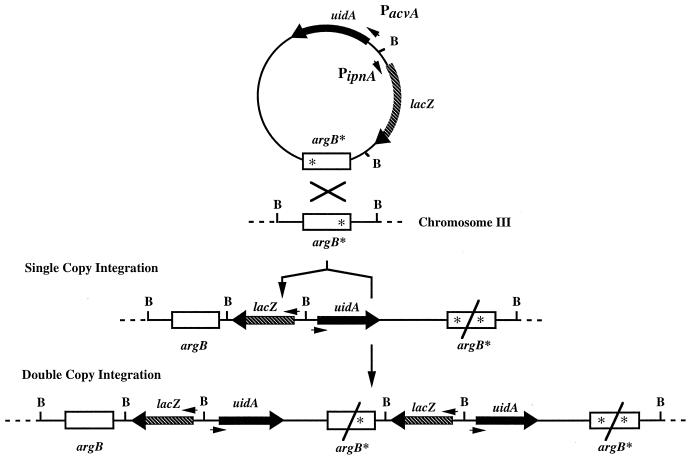
To further study these observations, acvA was disrupted in a strain of A. nidulans (47, 305). This strain had a disrupted acvA gene on chromosome VI and, in addition, reporter gene fusions of the penicillin biosynthesis genes integrated in single copy at the chromosomal argB gene locus on chromosome III. acvA, ipnA, and aatA gene fusions were expressed at the same level in this strain as in the nondisrupted strain (47, 194). This was confirmed by determining IPNS and IAT specific activities and by Western blot analysis of IPNS, which showed the presence of both enzymes in an acvA-disrupted strain (47, 194). Hence, the genes were expressed despite the lack of precursor ACV and IPN molecules, indicating that in contrast to the cephalosporin biosynthesis genes, none of the penicillin biosynthesis genes are sequentially induced and their expression is most probably coordinated. This is also supported by the similar time course in the expression of the A. nidulans acvA and ipnA genes (46) and by the appearance of transcripts of all three penicillin biosynthesis genes in P. chrysogenum at the same time during a fermentation run (273).
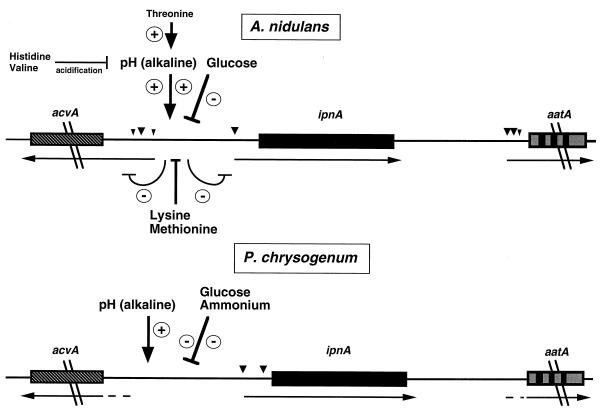
However, some observations make the view of a coordinated expression of the genes more complicated. Differential regulation of acvA, ipnA, and aatA in A. nidulans has been observed in response to exogenous signals. While the expression of A. nidulans acvA and ipnA genes was significantly repressed by l-lysine, aatA expression was not (48, 191). Furthermore, in fermentation medium, only ipnA was significantly repressed by glucose (46, 194).
It is interesting that in fermentation medium, acvA and ipnA gene fusions were expressed for up to 68 and 46 h, respectively (46). In contrast, aatA expression was detected for only about 24 h. In this respect, the temporal expression pattern of aatA is more similar to that of an oliC-lacZ gene fusion; oliC encodes subunit 9 of the A. nidulans mitochondrial ATP synthase complex and is regarded as a gene of the primary metabolism (46, 194).
In contrast to the other penicillin biosynthesis genes, the aatA genes of both A. nidulans and P. chrysogenum encode three introns (Table 3; Fig. 3). In theory, introns could play a role in regulating aatA expression. This was also suggested by the observation that when the inducible alcA promoter was used for the expression of aatA, the use of aatA cDNA gave higher IAT specific activity than did the use of the genomic gene carrying all three introns (112). However, a regulatory role of the first intron in the wild-type gene is unlikely, since there was no difference in the expression pattern of an A. nidulans aatA-lacZ fusion compared with an aatA-lacZ fusion, which had the first intron included (194). It is conceivable, however, that the second and/or third intron plays a regulatory role or, alternatively, that excision of introns becomes limiting in strains in which the steady-state level of aatA mRNA is strongly increased (112).
Previous studies of the formation of secondary metabolites in batch fermentations led to the definition of two phases: the growth phase (trophophase) and the period of secondary-metabolite production (idiophase) (reviewed in reference 210). A strict separation of the two phases, with production of secondary metabolites being virtually restricted to the idiophase, has been observed in antibiotic-producing bacterial cultures. It was first reported that the expression of the ipnA gene encoding IPNS in A. nidulans is temporally delayed in a similar manner to that of expression of bacterial secondary-metabolism genes (124). However, when lactose was used as the sole carbon source, there was no sharp separation of the trophophase and idiophase with respect to penicillin production or the expression of penicillin biosynthesis genes (46, 194). The previously observed delayed expression of ipnA is due to the use of glucose as the carbon source (101), which repressed transcription and expression of the ipnA gene (46, 101) (Fig. 5). For some secondary metabolites, it seems likely that their production in a clear-cut idiophase reflects the inhibiting effects of certain compounds in the medium (46, 101) rather than an intrinsic temporal delay in the product formation pattern. This view is consistent with the results of Renno et al. (273), who showed that in wild-type P. chrysogenum (NRRL 1951) and the high-penicillin producing strain P2, the highest steady-state level of mRNAs of all penicillin biosynthesis genes was observed during maximal growth in both shake flasks and 2-liter fermentors.
FIG. 5.
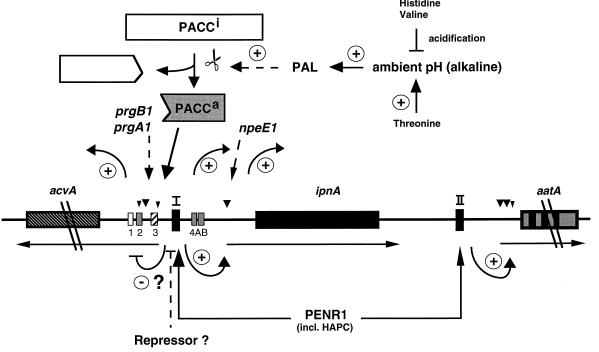
Regulatory circuits affecting the expression of the penicillin biosynthesis genes acvA and ipnA in A. nidulans and P. chrysogenum. Large and small arrows indicate major and minor transcription start sites, respectively. Transcript start sites of acvA and aatA in P. chrysogenum have not been reported yet. Until now, in P. chrysogenum the effect of alkaline pH has been shown for the ipnA gene (316). The addition of certain amino acids to the medium results in the indicated effects on gene expression. Some of these effects are mediated by the ambient pH. See the text for details. Regulatory circuits affecting the expression of the aatA gene in both fungi have not been identified yet.
However, some observations imply that in P. chrysogenum there might be a temporal expression of β-lactam biosynthesis genes. ipnA steady-state mRNA levels increased with the age of the culture, indicating preferential transcription of the gene at late growth times (316). This is consistent with the observation of Feng et al. (109) that expression of an ipnA-uidA gene fusion was detectable after only 24 h in fermentation medium with lactose as the carbon source whereas an acvA-uidA gene fusion seemed to be expressed from the beginning of a fermentation run. Hence, there might even be differences in the temporal expression of genes of the same cluster as well as in different fungi.
Promoter Structures of the A. nidulans Genes acvA and ipnA
So far, only two studies have analyzed the promoters of β-lactam biosynthesis genes in the homologous host in detail. Pérez-Esteban et al. (257) reported a comprehensive deletion analysis of the A. nidulans ipnA promoter in which the promoter region was fused in frame with the E. coli lacZ reporter gene (encoding β-Gal). Integration of such deletion constructs in single copy at the chromosomal argB gene locus and determination of the β-Gal specific activity of the different A. nidulans transformant strains revealed that several regions of the promoter were involved in the basal expression of ipnA and the repression by sucrose (see “Carbon source regulation” below).
In this deletion study, the expression of the divergently transcribed acvA gene (Fig. 3) was not determined. Divergently transcribed genes, however, provide an interesting case of regulation, because possible functional interactions between the promoters could exist and single cis-acting sites might have the potential to regulate two genes simultaneously. Hence, a moving-window analysis was carried out; i.e., nested deletions were introduced into the intergenic region between A. nidulans acvA and ipnA. The ATG start codons of both genes were fused in frame with the two E. coli reporter genes lacZ and uidA (encoding β-Gal and β-Glu), respectively, and these deletion constructs were integrated in single copy at the chromosomal argB gene locus. Hence, the effects of deletions on the expression of both genes were measured simultaneously within one transformant strain by determining β-Gal and β-Glu specific activities (319). These deletion analyses revealed that the intergenic region between acvA and ipnA contains several regions with cis-acting DNA elements. Thus far, no element was found in the intergenic region which affected the expression of one gene only, although differential expression of genes, e.g., as a result of the addition of amino acids to the medium, was observed. Taken together, these findings suggest that the promoters of both genes are, at least in part, physically overlapping and share common cis-acting elements (319).
Carbon Source Regulation
Industrial production of penicillin by P. chrysogenum was previously carried out with lactose as the C source, since it gave the highest penicillin titer. The use of excess glucose leads to a drastic reduction of the penicillin titer (309). Fructose, galactose, and sucrose also have a negative effect on penicillin production, but lactose does not (276). Nowadays, this problem has been partially overcome by feeding subrepressing doses of glucose and by using lactose as the C source (317). Since in general the fungus grows better with glucose than with lactose (46), the production of penicillin appears to be favored by suboptimal growth conditions.
C-source regulation seems to act at several points of the penicillin biosynthesis: (i) flux of l-α-AAA, (ii) activation of side chain precursors, (iii) transcription of penicillin biosynthesis genes (Fig. 5), and (iv) posttranscriptional regulation of penicillin biosynthesis genes.
The replacement of lactose by glucose reduced the l-α-AAA pool in mycelia of P. chrysogenum and thus probably the flux of l-α-AAA to ACV (144 (see “Amino acids as precursors and mediators of regulation” below). The formation of ACV and IPN was also repressed by glucose (277) (Fig. 2). Indirect measurements based on the incorporation of [14C]valine into ACV in vivo suggested that the glucose repression of penicillin biosynthesis involves repression but not inhibition of penicillin-synthesizing enzymes. Furthermore, the specific activity of IPNS but not of IAT was reduced in cells of a penicillin-producing strain (AS-P-78) grown on glucose. These results suggested that glucose represses the formation of ACVS and IPNS in P. chrysogenum (144, 276, 277). In agreement with these findings was the observation that the expression of both the acvA and ipnA genes of P. chrysogenum Q176, both measured by using the uidA reporter gene, was repressed by glucose (109) (Fig. 5). Renno et al. (273) claimed, however, that steady-state mRNA levels of all three P. chrysogenum penicillin biosynthesis genes were highest during rapid growth, when considerable levels of glucose were present. This shows that measurement of carbon regulation depends, at least in part, on the experimental approach used.
In A. nidulans, a similar phenomenon was observed. The use of repressing carbon sources such as glucose or sucrose in the fermentation medium reduced the amount of penicillin produced (46, 101). Results obtained with reporter gene fusions showed that the expression of the ipnA gene was repressed when glucose or sucrose was used instead of lactose as the C source during fermentation (46, 101) (Fig. 5). This was further supported by the finding that the IPNS specific activity was drastically reduced in glucose-grown mycelia (46). Repression of ipnA expression by repressing C sources occurs, at least in part, at the transcriptional level, because the steady-state level of ipnA mRNA decreased when mycelia were cultivated with repressing C sources such as sucrose (101).
Unexpectedly, the expression of both acvA and aatA reporter gene fusions was repressed only slightly, if at all, by glucose in fermentation medium (46, 194). However, the specific activity of the aatA gene product, IAT, was reduced in mycelia grown with glucose instead of lactose (46, 194). This suggests that the glucose regulation of IAT takes place, at least in part, posttranscriptionally. The effect of glucose on IAT specific activity could be reversed by washing cells and reincubating them in lactose-containing medium (194). It is unclear why glucose regulation occurs, in the case of ipnA, transcriptionally or, in the case of aatA, apparently posttranscriptionally.
The results obtained with A. chrysogenum seem to be similar to those obtained with A. nidulans. Cephalosporin C formation by A. chrysogenum depends on the C source used (85). C sources which support the most rapid growth, like glucose or glycerol, had a negative effect on β-lactam production (87, 223). Fermentations with different C sources showed that glucose exerted a much greater negative effect on cephalosporin C than on penicillin N formation, suggesting that it had a more pronounced effect on the later steps of the pathway (36). The ACVS level was not affected by higher concentrations of glucose, making a major repressive effect of glucose at the level of gene expression unlikely (349). However, ACVS specific activity measured in crude protein extracts was severely inhibited by glucose and glycerol. The inhibition could be reversed by the addition of ATP. Interestingly, the purified enzyme was not inhibited by glucose but was inhibited by glyceraldehyde-3-phosphate and glyceraldehyde (346). From these observations, the authors concluded that inhibition of ACVS specific activity in crude extracts, by glucose and some other sugars, is due to depletion of the cofactor ATP via sugar metabolism.
In contrast to the penicillin production strain P. chrysogenum AS-P-78, investigated by Revilla et al. (277), the IAT specific activity of both A. nidulans and the P. chrysogenum wild-type strain NRRL 1951 was clearly reduced in glucose-grown cultures (46). Glucose was also found to cause inactivation of P. chrysogenum acetyl-CoA synthetase, which can catalyze the activation (to their CoA thioesters) of some of the side chain precursors required for the production of several penicillins in vitro (216). The contributions to the overall reduction of the penicillin titer by each of the steps affected by glucose are not yet clear.
Previously, it was reported that the uptake of side chain precursors of phenylacetic acid was regulated by glucose (see, e.g., reference 113). However, it was recently shown that phenylacetic acid passes the plasma membrane via passive diffusion of the protonated species (140), thus excluding the possibility that the uptake could be regulated by the available C source.
An involvement of cyclic AMP (cAMP) in C-source regulation is controversial. In P. chrysogenum, cAMP levels were high during growth on lactose and decreased markedly (by about 75%) when glucose was added (178). In A. nidulans, however, a correlation between intracellular cAMP levels and the C source (lactose and glucose) was not detected (44).
To study the molecular basis of C-source regulation of ipnA expression, several mutants of A. nidulans carrying previously characterized loci affecting the glucose repression of several genes of the primary metabolism (creAd-1, creB304, and creC302) (21, 146) have been analyzed. In these mutants, penicillin production was still reduced by glucose (46, 49). However, Espeso and Peñalva (101) observed in mutants with extreme loss-of-function mutations in creA slightly derepressed ipnA steady-state transcript levels. This was consistent with a deletion analysis of the ipnA promoter, demonstrating that a cis-acting DNA region crucial to sucrose repression maps between −1334 and −966 relative to the transcriptional start site of the ipnA gene (257). A single CREA binding site was detected in this region, which was protected in DNase I footprint analysis by using a glutathione S-transferase (GST)–CREA protein which contained amino acids 35 to 240 of CREA (104). The full-length CREA polypeptide deduced from the gene consists of 415 amino acids (95). To test the functionality of this putative CREA binding site in vivo, the effect of its deletion was measured in an ipnA-lacZ gene fusion. The expression of the gene fusion with a deleted CREA binding site was still repressed when sucrose instead of lactose was used as the C source (104). Hence, it is unlikely that CREA plays a role in C-source repression of penicillin biosynthesis. This also agrees with the finding that acetate and glycerol, which are repressing and derepressing C sources, respectively, of some primary-metabolism genes in the creA-mediated circuit of carbon catabolite repression, behaved in the opposite manner to what would be expected from creA control. The use of acetate led to increased steady-state levels of ipnA transcript and penicillin titers, and the use of glycerol led to the opposite effects, i.e., decreased ipnA transcript levels and penicillin titers (104).
Previously, it was shown that in creB and creC mutants the production of penicillin was still reduced when glucose instead of lactose was used as the C source (46, 49). Additional experiments further excluded the possibility of a direct involvement of creB and creC mutations in C-source repression of ipnA transcription (100). Thus, the mechanism(s) of regulation of penicillin biosynthesis by repressing C sources remains to be elucidated.
An interesting observation was made by Kennedy and Turner (165). In A. nidulans strains such as WG355 or R21, glucose usually led to a reduction of the penicillin titer to about 30 to 50% (46, 101), whereas with strain G191, a reduction to about 10% was measured. This might represent an unexpected strain difference, with penicillin production in G191 being more sensitive to glucose repression (165). Strain G191, for example, carries a mutation in the uaY gene (uaY9), which encodes a transcriptional activator for a number of unlinked genes involved in purine utilization (315), or cryptic mutations could be present in G191 which could be responsible for this unexpected observation. It seems worth investigating the difference between strain G191 and other strains with respect to carbon regulation of penicillin biosynthesis.
pH Regulation Mediated by the Transcriptional Factor PACC
Penicillin production is subject to regulation by ambient pH (104, 295) (Fig. 5). Wild-type strains of A. nidulans can grow in media over the pH range of 2.5 to 10.5 (282). There was markedly more penicillin in the culture broth when the pH of the medium was kept constant at 8.1 than when it was kept constant at 6.5 or 5.1 (295). Control experiments excluded the possibility that the difference in penicillin yield measured was due to the higher stability of penicillin G at alkaline pH (104).
To study the molecular basis of this interesting phenomenon, mutations involved in the pH regulatory network of A. nidulans were isolated and analyzed (19, 92, 104, 295). Previously, Dorn (92) had identified mutations specifically affecting the pH regulation of the formation of alkaline and acidic phosphatases. These mutations were divided into two distinct groups (pal and pac) based on the phenotypes they encoded. Mutations in some of the pal genes (palA, palB, palC, palE, and palF), which are phenotypically indistinguishable, have been shown to mimic the effects of growth at acidic pH (57). Arst et al. (19) identified two new pal genes, palH and palI, and demonstrated that palB and palE actually represented the same gene. Thus far, there are a total of six discrete regulatory pal genes (palA, palB, palC, palF, palH, and palI) (19). The alkaline phosphatase and acidic phosphatase structural gene were designated palD and pacA, respectively (reviewed in reference 18). The regulatory pal mutations impair the pH regulation of the syntheses of extracellular enzymes, such as phosphatases and permeases. Extracellular enzymes and permeases are not protected by the intracellular pH homeostatic system; therefore, their synthesis is controlled by the external pH. This mode of regulation avoids the synthesis of, for example, secreted alkaline phosphatase in an acidic environment (57).
Mutations at the pacC locus result in the phenotypic mimicry of growth under alkaline conditions (57). It has also been noted that pacC mutations themselves can be divided into two classes defined by the phenotype when cultured at neutral pH: those that mimic growth under alkaline conditions (the type originally observed) and those (including a null allele) that mimic growth under acidic conditions (57, 322).
PacC mutants mimicking the effect of growth at alkaline pH produced about twice as much penicillin as did wild-type strains, whereas strains carrying pal mutations mimicking growth at acidic pH showed the lowest penicillin titers. These findings are consistent with the observation that penicillin titers were higher when wild-type strains were grown at alkaline pH in wild-type strains (295).
The pacC gene was cloned and sequenced by complementation of an argB2 pacCc14 (mimicking alkaline growth conditions) recipient by using a genomic A. nidulans library constructed in an argB+-containing plasmid. The DNA sequence of the pacC gene consists of 2,172 bp interrupted by two introns of 85 and 53 bp. The 678-residue-derived protein (Mr 72,939) revealed that PACC contains three putative Cys2His2 zinc fingers (322). At alkaline ambient pH, PACC activates the transcription of alkali-expressed genes (e.g., the alkaline phosphatase and protease genes palD and prtA, respectively) and also of the penicillin biosynthesis genes ipnA (322) and acvA (103, 318). The intergenic region between acvA and ipnA was found to contain four in vitro PACC binding sites designated ipnA1, ipnA2, ipnA3, and ipnA4AB, recognized by a GST-PACC (amino acids 31 to 195) fusion protein (see Fig. 10). The fusion protein was demonstrated to bind to the core consensus GCCARG (322). The mode of DNA recognition by PACC was studied in detail (105).
FIG. 10.
Regulatory genes involved in the regulation of the penicillin biosynthesis genes of A. nidulans. Addition of the indicated amino acids to the medium results in the indicated effects on gene expression (318). Some of these effects are mediated by the ambient pH, most probably via pal genes and the central regulatory protein PACC encoded by the pacC gene (318). PACCi symbolizes the inactive form, which is proteolytically cleaved upon the pH signal (alkalinity) into PACCa, i.e., the active shorter version of PACC (246, 322). The two identified PENR1 binding sites containing CCAAT sequences are marked with black boxes and roman numerals (I and II). One of the protein components of the PENR1 complex is HAPC (in parentheses below PENR1). The four PACC binding sites bound in vitro by PACC (322) are designated 1 (ipnA1), 2 (ipnA2), 3 (ipnA3), and 4AB (ipnA4AB) (102) and are boxed in the intergenic region between acvA and ipnA. PACC sites that were found to be functional in vivo for expression of ipnA-lacZ and/or acvA-uidA are indicated by hatched boxes. Site 3 seems to be of major importance for both ipnA-lacZ (102) and acvA-uidA (318) expression. Large and small arrows indicate major and minor transcription start sites, respectively. Names printed in italics indicate mutations which formally represent trans-acting genes with positive effects on the expression of the indicated genes. The existence of the repressor is hypothetical. Only two putative regulatory proteins have been described so far in P. chrysogenum. PACC most probably mediates the effect of pH on ipnA transcription (316), and NRE mediates the nitrogen regulation of acvA and ipnA (109, 132) (Fig. 5).
A mutation analysis of each of these sites by using ipnA-lacZ gene fusions revealed that in vivo the binding site ipnA3 was most important for PACC-dependent ipnA expression whereas ipnA2 and ipnA4AB were less important, although ipnA2 was bound with highest affinity by PACC in vitro. ipnA1 apparently was not required for PACC-dependent ipnA expression (102) (see Fig. 10). As observed for expression of an ipnA-lacZ gene fusion (104), expression of an acvA-uidA gene fusion was increased in a PacC5 mutant strain (318). Because pacC5 is an allele which is active irrespective of the ambient pH (322), this suggests that PACC also regulates the expression of acvA. This assumption was further supported by the observation that the steady-state levels of acvA mRNA were increased at alkaline ambient pH (103). Furthermore, the addition of histidine and valine to the culture medium led to its acidification and to reduced acvA-uidA expression. This effect was not observed in a deletion strain (Δ183–312) carrying a deletion spanning PACC binding site ipnA3 (at nucleotides 265 to 270) or in the PacC5 mutant strain with a constitutively active PACC protein. Taken together, these data suggest that PACC also regulates the acvA expression of A. nidulans predominantly from ipnA3 (318). Therefore, an external alkaline pH is sufficient to give rise to considerable transcriptional derepression of steady-state acvA and ipnA transcript levels, expression of both genes measured by using gene fusions, and penicillin titers.
In addition, at alkaline ambient pH, PACC prevents the transcription of acid-expressed genes (322; reviewed in reference 18). The full-length form of PACC, which predominates at acidic ambient pH or in PalB7 strains grown under acidic or neutral conditions, is not functional (90, 246). It must be specifically proteolyzed to yield the functional (for both positive and negative roles) version containing the N-terminal 40% of the protein (246) (see Fig. 10). The processed form is functional as both the activator and repressor (246; reviewed in reference 18). The pacC gene seems to be autogenously regulated, because the pacC transcript is strongest at alkaline pH, and the pacC promoter contains PACC binding sites (322).
PACC proteolysis occurs in response to a signal provided by the six regulatory pal gene products in alkaline environments (246, 322; reviewed in reference 18). Thus, the products of the palA, palB, palC, palF, palH, and palI genes constitute an alkaline ambient pH signal transduction pathway which is required for the conversion of PACC to its functional form (90).
Recently, both the palA and palB gene have been cloned and sequenced (90, 242). The amino acid sequence of the derived palA protein showed similarities to that of a hypothetical protein from the nematode Caenorhabditis elegans and also has possible yeast homologs. These data, however, did not allow speculation about the putative function of the protein (242). The predicted palB gene product has similarity to the catalytic domain of the calpain family of calcium-activated cysteine proteases. However, PALB does not catalyze the final step of proteolytic processing of PACC (90). The pal pathway is thought to introduce a modification of PACC at alkaline pH in wild-type strains, disrupting intramolecular interactions to allow activating proteolysis (246). The type of PACC modification mediated by the pal pathway in response to alkaline pH is not known, but an earlier (and more C-terminal) proteolytic cleavage of PACC resulting in susceptibility to further proteolysis might be mediated by PALB (90). Alternatively, it was suggested that PALB might proteolyze one of the other pal gene products in a signalling cascade (90).
By cross-hybridization with a nearly full-length A. nidulans pacC gene as the probe and a genomic library of P. chrysogenum NRRL 1951, the P. chrysogenum pacC gene was cloned and sequenced (316). The gene complemented an A. nidulans strain carrying a null pacC mutation, thus showing that it was functional. The gene is 1,979 bp long and is interrupted by a single intron of 56 bp. The predicted 641-residue protein (Mr 68,681) exhibits most of the features described for the A. nidulans pacC protein, including three zinc fingers of the Cys2His2 class. Steady-state levels of the pacC transcript were elevated at alkaline pH, suggesting that the P. chrysogenum pacC transcription was also pH regulated (316).
A fusion protein of GST with amino acids 46 to 154 of P. chrysogenum PACC [GST-PACC(46–154)], overexpressed in E. coli and purified, bound in vitro to the intergenic region between P. chrysogenum acvA and ipnA. By computer analysis, seven PACC binding consensus sites (5′-GCCARG-3′) were found in the intergenic region. This is consistent with the finding that steady-state ipnA mRNA levels were increased at alkaline pH (316).
The upstream region of the P. chrysogenum aatA gene contains eight binding sites for PACC, whereas that of A. nidulans has just one such sequence, suggesting that these genes might be regulated by PACC as well (316).
In addition, several groups have characterized the intergenic region between the P. chrysogenum acvA and ipnA genes by band shift assays. Chu et al. (68) and Feng et al. (110) reported independently that partially purified crude extracts of P. chrysogenum bound in vitro to the sequences TGCCAAG and GCCAAGCC, respectively. Although the binding sites were located at different positions in the intergenic region, their degree of identity is striking. Furthermore, the proposed DNA binding proteins of P. chrysogenum appear to recognize the same sequence motif which is bound by the PACC proteins of both A. nidulans and P. chrysogenum (316, 322), which, for A. nidulans, has been reported to bind to GCCA(A/G)G (322). Hence, the binding sites identified almost certainly correspond to PACC binding sites (316).
In the intergenic region between acvA and ipnA of A. chrysogenum (230), there are three potential PACC binding sites, making it likely that these genes are also regulated by a PACC homolog. In addition, it will be interesting to analyze whether the genes of the second cephalosporin biosynthesis cluster are regulated by a PACC homolog in A. chrysogenum.
pacC genes are not confined to β-lactam-producing fungi, as shown by the recent cloning of the A. niger pacC gene (202). This gene encodes a three-zinc-finger protein of 667 amino acids (Mr 70,901; gene size of 2192 bp interrupted by two exons of 139 and 52 bp). The gene seems to be pH dependent and thus autogenously regulated. Hence, PACC seems to represent a wide-domain regulator which is involved in the regulation of expression of penicillin biosynthesis genes.
Because the use of glucose or sucrose as the C source leads to acidification of the medium (see, e.g., references 46, 101, and 104), it was conceivable that the glucose/sucrose effect was due to pH regulation (104). This was further supported by the observation that external alkaline pH could bypass the sucrose repression of steady-state ipnA transcript levels and penicillin titers. Inclusion of 100 mM Na2HPO4 in sucrose broth resulted in an initial pH of 8.0. Northern blot analysis revealed that the ipnA mRNA levels under these conditions were completely derepressed (104). Because buffering at alkaline pH with Tris-Cl increased penicillin titers in glucose media as well, this suggested that the increases were not caused by phosphate ions (104, 295). Additional experiments confirmed that alkaline pH is the factor that derepresses penicillin production in 3% sucrose broth (104). Furthermore, analysis of pacC mutants revealed that they bypassed carbon regulation of ipnA transcript levels, i.e., that pacC mutations caused derepression of steady-state levels of the ipnA mRNA in sucrose broth despite external acidic pH resulting from sucrose utilization (104). However, neither acidic external pH nor the palA1, palB7, or palF5 mutation mimicking the effects of growth at acidic pH prevented C-source derepression (104). Furthermore, the PACC binding sites determined in vitro by the use of a fusion polypeptide containing the PACC DNA binding domain are not located in the cis-acting region, which was shown to mediate C-source repression of ipnA-lacZ expression (104, 322; see “Carbon source regulation” above). Taken together, these data support the model of Espeso et al. (104) and Tilburn et al. (322) involving independent regulatory mechanisms, one mediating carbon source regulation and another mediating pH regulation through the pacC-encoded transcriptional regulator. Since alkaline pH values per se seem to derepress ipnA transcription, Espeso et al. (104) proposed that internal alkalinity represents a physiological signal which triggers penicillin biosynthesis.
However, the use of nonrepressing C sources with respect to ipnA steady-state transcript levels, such as 3% lactose, 1% l-arabinose, or 100 mM acetate, as the principal C source resulted in an increase of the external pH. Growth in repressing C sources, such as 3% sucrose, 1% d-glucose or 1% glycerol caused acidification of the media (104). The authors concluded that carbon limitation, either by using less favorable C sources or by reducing the concentration of favorable C sources, results in external alkalinization whereas the availability of sufficient amounts of a favorable C source causes external acidification. Thus, carbon and pH regulation normally act in concert although the mechanisms are different (104).
In contrast to the situation in A. nidulans, an alkaline ambient pH did not seem to override the negative effect of a repressing C source on ipnA transcription in P. chrysogenum, because full ipnA expression was dependent on carbon source derepression irrespective of the ambient pH (316).
The reason for the pH-mediated regulation of penicillin biosynthesis is unclear. Arst (18) suggested that it might be associated with the observation that β-lactams exhibit increased toxicity on at least some bacterial species at alkaline pH. Furthermore, bacterial competition with fungi may be more intense at alkaline pH (reviewed in reference 18). This agrees with the results of Espeso et al. (104), who concluded that for A. nidulans, penicillin biosynthesis in an alkaline environment might be advantageous even if the C source is not limiting, since alkaline pH appears to override C repression. On the other hand, limiting C sources seem to allow derepression of ipnA expression at any external pH; i.e., carbon derepression of ipnA transcription bypasses the requirement for PACC (104).
Nitrogen Regulation
The effect of the availability of nitrogen source on the penicillin biosynthesis has been discussed for a long time. Sanchez et al. (286) reported the inhibition of penicillin biosynthesis in P. chrysogenum by high levels of ammonium. This effect was correlated with low levels of glutamine synthetase. Mycelia grown at low NH4+ concentrations accumulated higher concentrations of glutamine in the free pool of amino acids and lower concentrations of glutamate. In contrast, increasing the concentration of NH4+ led to a decreased pool of glutamine, an increased glutamate concentration, and poor antibiotic production. It was found that ammonium concentrations [(NH4)2SO4] higher than 100 mM also strongly interfered with cephalosporin C production in A. chrysogenum. l-Asparagine and l-arginine were better nitrogen sources than ammonium with respect to antibiotic yield (296).
Recently, it was demonstrated that in P. chrysogenum, ammonium directly influenced the expression of penicillin biosynthesis genes. By using gene fusions of both penicillin biosynthesis genes ipnA and acvA with the E. coli reporter gene uidA and integration of these gene fusions in single copy at the P. chrysogenum niaD gene locus, it was shown that the expression of both genes was repressed by the addition of 40 mM (NH4)Cl to lactose-grown mycelia (109) (Fig. 5).
Nitrogen metabolite repression is generally regarded as a wide-domain regulatory system, which operates to ensure that a constant nitrogen supply is readily available for growth in response to widely variable or rapidly changing environments. In the absence of favored nitrogen sources, such as ammonium or glutamine, many secondary nitrogen sources, e.g., nitrate, purines, proteines, and amides, were used. These require the de novo synthesis of permeases and catabolic enzymes, whose expression is highly regulated by the nitrogen regulatory circuit (reviewed in references 58 and 218). In A. nidulans and Neurospora crassa, global nitrogen repression and derepression is mediated by the major positive control genes areA and nit-2, respectively (121, 180). Recently, by using a PCR-based approach, the homologous gene of P. chrysogenum, nre, was cloned and shown to complement Nit-2 mutants of N. crassa (132). Each of these three genes encodes regulatory factors with a single Cys-X2-Cys-X17-Cys-X2-Cys-type zinc finger, which, in combination with an immediate downstream basic region, constitutes a DNA binding domain. The overall amino acid sequences of these three regulatory proteins show only 30% identity, but they have 98% identity in their DNA binding domains. A similar zinc finger motif has been found in a wide range of eukaryotic organisms including yeast, plants, chickens, mice, and humans. All of these transcription factors recognize the consensus sequence GATA and can be grouped into a GATA protein family (reviewed in reference 218). The optimal binding sites for NIT-2 were found to consist of at least two GATA elements, which can face in the same or opposite directions, with a spacing which can vary from 3 to 30 bp (66).
A protein consisting of 181 amino acid residues of the 835-residue P. chrysogenum NRE (132), containing its zinc finger domain, fused to the N terminus of E. coli β-Gal was expressed in E. coli. Band shift assays with the purified β-Gal–NRE fusion protein showed binding with high affinity to a DNA fragment derived from the intergenic region between acvA and ipnA of P. chrysogenum. Although there are six GATA sequences in the intergenic region, missing-contact experiments with the β-Gal–NRE fusion protein revealed that NRE strongly interacts with a site that contains two of these GATA sequences (133). The same GATA sequences were also strongly bound by N. crassa NIT-2 (110). In this binding site, the two GATA core sequences are arranged in a head-to-head fashion and are separated by 27 bp. Therefore, it appears very likely that nitrogen metabolite regulation of penicillin biosynthesis genes is mediated through NRE and that these structural genes for secondary metabolism are regulated as members of a nitrogen control circuit. This suggests that the availability of favored nitrogen sources, and thus good growth conditions, leads to reduced penicillin synthesis by the fungus. However, in vivo studies are clearly needed to relate NRE binding to potential regulatory functions (133).
No evidence for nitrogen-dependent regulation of penicillin biosynthesis in A. nidulans has been reported so far. This is also consistent with the observation that the intergenic region between acvA and ipnA of A. nidulans contains only a single GATA motif whereas six GATA sequences are found in the corresponding P. chrysogenum region (133). Thus, it seems unlikely that a single GATA motif is bound by an A. nidulans GATA factor. However, it cannot be entirely ruled out, because members of the metazoan GATA transcription factor family, each containing two zinc fingers homologous to the single zinc finger motif of NRE, also bind to DNA fragments containing single GATN (where N is A, T, C, or G) motifs (170, 231). Interestingly, the P. chrysogenum promoter was shown to respond to nitrogen control when transformed in A. nidulans, indicating that the nitrogen-repressing system of A. nidulans acted on the heterologous promoter (173). Furthermore, it is worth noting that in the intergenic region of the corresponding A. chrysogenum genes (230), there are 15 GATA motifs. It is thus conceivable that these genes are also regulated by a GATA factor.
Amino Acids as Precursors and Mediators of Regulation
Radiochemical tracer studies during the 1940s and 1950s (reviewed in reference 80) revealed that penicillins and cephalosporins are naturally synthesized from the three amino acid precursors l-α-AAA, l-cysteine, and l-valine. Although only cysteine and valine are finally incorporated in the β-lactam nucleus, the requirement for l-α-AAA has long been established (reviewed in reference 86).
Cysteine and valine are ubiquitous, proteinogenic amino acids, whereas l-α-AAA is an intermediate of the l-lysine biosynthetic pathway, which occurs in this form only in higher fungi and euglenids (Euglena gracilis) (Fig. 6). The pathway starts from α-ketoglutaric acid and is named according to its key intermediate, α-aminoadipate (336; reviewed in references 38 and 331). Hence, l-α-AAA is a branch point between l-lysine and penicillin and cephalosporin biosynthesis (Fig. 6). Genetic data suggested, however, that in A. nidulans, l-α-AAA for penicillin biosynthesis is also provided by catabolic conversion of l-lysine by an as yet unidentified pathway. This was concluded from the observation that lysine-auxotrophic mutants defective in the lower part of the pathway leading to the formation of l-α-AAA produced penicillin when l-lysine was added to the medium (330) (Fig. 6).
FIG. 6.
Lysine biosynthetic pathway (aminoadipate pathway) with the intermediate l-α-AAA. It has not been clarified whether the reaction from homocitric acid to cis-homoaconitic acid is catalyzed by homoaconitase, as is the following reaction, or by a specific enzyme designated homocitrate dehydrase (38). Therefore, this reaction is labelled with a question mark. OPC is cyclized l-α-AAA and is found as by-product in the culture broth of penicillin fermentations. The minus signs indicate negative effects of l-lysine on the indicated reactions which were identified from either P. chrysogenum or A. nidulans. These might be mediated at the transcriptional level or posttranscriptionally. See the text for details.
For P. chrysogenum, a similar observation was made (99). A lysine-auxotrophic mutant blocked in a step before l-α-AAA was also able to synthesize penicillin when l-lysine was added to the medium. Additional biochemical studies with [U-14C]lysine revealed the formation of labeled saccharopine and l-α-AAA. Hence, l-α-AAA can also be obtained for penicillin synthesis by reversal of the last steps of the l-lysine biosynthetic pathway (99) (Fig. 6). In addition, the authors found a high lysine:α-ketoglutarate-ɛ-aminotransferase activity in P. chrysogenum, which converts l-lysine into 1-piperidine-6-carboxylic acid. This finding suggests the existence of at least two different pathways involved in the degradation of l-lysine (99). Taken together, these data demonstrate that l-α-AAA can be provided by both the l-lysine biosynthetic pathway and degradation of l-lysine. The contribution of the latter catabolic pathway to penicillin production, however, remains to be shown.
In bacteria, the situation is different, because bacteria and green plants synthesize l-lysine starting from aspartic acid via a different route, designated the diaminopimelic acid pathway. Since l-α-AAA is not an intermediate in this pathway, the question how cephalosporin-producing bacteria obtain the required l-α-AAA was investigated. In S. clavuligerus, a specific enzyme activity, lysine ɛ-aminotransferase, was discovered; this enzyme catalyzes the removal of an amino group of l-lysine to give cyclic 1-piperidine-6-carboxylic acid, which is then oxidized to l-α-AAA (205). By this means, β-lactam-producing streptomycetes can form the precursor amino acid l-α-AAA for β-lactam biosynthesis. The gene encoding lysine ɛ-aminotransferase was designated lat and cloned from both S. clavuligerus (206, 326) and N. lactamdurans (76). In both bacteria, it is located within the cephamycin biosynthesis gene cluster (Fig. 3). This supports the finding that it plays a role exclusively in β-lactam biosynthesis (76, 205, 206, 326).
A number of different by-products are commonly encountered during the production of penicillin by P. chrysogenum, one of which is the δ-lactam of l-α-AAA, 6-oxo-piperidine-2-carboxylic acid (OPC), formed by cyclization of l-α-AAA. This is the reason why net synthesis of l-α-AAA is required, although the l-α-AAA moiety of IPN can be reused (52, 120, 143, 183, 328) (Fig. 6).
The synthesis of cysteine for β-lactam biosynthesis is another crucial factor in the economics of β-lactam formation. Sulfate assimilation in fungi has been studied primarily in S. cerevisiae and N. crassa (reviewed in reference 217). Genetic and biochemical analyses have shown that the pathway in the β-lactam-producing A. nidulans requires a sulfate permease (sB) to transport sulfate into the cell, ATP sulfurylase (sC), adenosine-5-phosphosulfate (APS) kinase (sD), and 3-phosphoadenosine-5-phosphosulfate (PAPS) reductase (17, 127) (Fig. 7A). Sulfite is reduced to sulfide by the multienzyme complex sulfite reductase, which involves the transfer of six electrons from NADPH to the sulfur atom. The overall consumption of cofactors for the reduction of sulfate to sulfide is 4 mol of NADPH and 2 mol of ATP (Fig. 7A). Thus, the reduction of sulfate to sulfide accounts for a major part of the metabolic costs in the overall biosynthesis of l-cysteine (244). Some of the enzymes involved in β-lactam-producing fungi have been characterized, and the first genes have been cloned (42, 55, 69, 214, 241, 274, 293). The regulation of both cysteine and methionine biosynthesis appears to be complex and awaits further investigations. Recent work conducted on A. nidulans and P. chrysogenum has indicated an additional mode of regulation by allosteric inhibition, which is absent in yeast (42, 118). This regulation is thought to occur by the binding of PAPS to a pseudosubstrate site located in ATP sulfurylase, thus causing feedback inhibition when excess PAPS is present. At least four regulatory genes control sulfur metabolite repression in A. nidulans (241).
FIG. 7.
Pathways leading to the synthesis of l-cysteine. (A) Assimilation of sulfate. For reduction of sulfate to sulfide, several enzymes are required. Sulfate is transported into the cytoplasm by a specific permease, encoded by the sB gene in A. nidulans. See the text for details. (B) Starting from sulfide, three different routes leading to the formation of cysteine are used by the various β-lactam-producing fungi. They are designated transsulfuration, direct sulfhydration (grey background), and reverse transsulfuration (grey background bounded by broken lines). Although all the pathways seem to exist in A. chrysogenum, the fungus prefers to generate cysteine for optimal cephalosporin C synthesis by conversion of methionine to cysteine via reverse transsulfuration. In contrast, P. chrysogenum and A. nidulans synthesize cysteine mainly by direct sulfhydrylation starting from serine. See the text for details. Abbreviations: SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine.
Starting from sulfide, three different routes leading to cysteine biosynthesis have been described (reviewed in references 209, 327, and 331) (Fig. 7B). In the direct sulfhydrylation pathway, reduced sulfur is incorporated by O-acetylserine sulfhydrylase into O-acetyl-l-serine to give cysteine. In a different pathway, transsulfuration, sulfide incorporation is catalyzed by O-acetylhomoserine sulfhydrylase. The third possibility is reverse transsulfuration, in which the sulfur of methionine is transferred to cysteine (Fig. 7B). Interestingly, different routes leading to the formation of cysteine are used by the various β-lactam-producing fungi. This is apparently genetically determined. Although all the pathways seem to exist in A. chrysogenum (260, 327), the fungus prefers to generate cysteine for optimal cephalosporin C synthesis by conversion of methionine to cysteine via reverse transsulfuration. This finding well agrees with the observation that methionine at certain concentrations has a regulatory function in the cephalosporin biosynthesis of A. chrysogenum (see below). In contrast, P. chrysogenum and A. nidulans synthesize cysteine mainly from direct sulfhydrylation starting from serine (260, 327) (Fig. 7B). The direct sulfhydrylation pathway is energetically more favorable than the transsulfuration pathway (244) (Fig. 7B).
The biosynthesis of valine is closely connected to the biosynthesis of leucine. Valine biosynthesis proceeds by four enzymatic steps with two moles of pyruvate as precursor metabolites (Fig. 8) (reviewed in reference 245). The presence of pools of the three amino acids specific for β-lactam antibiotic biosynthesis (l-cysteine, l-valine, and l-α-AAA), separate from the main pool of each amino acid, has been proposed (144).
FIG. 8.
Biosynthetic pathway of l-valine. The minus sign indicates a negative effect of l-valine on the indicated reaction, as determined in P. chrysogenum and A. chrysogenum. See the text for details.
Because penicillin and cephalosporin are synthesized from amino acid precursors, it was conceivable that amino acids play a role in the regulation of their biosyntheses. This was supported by the observation that in both A. nidulans and P. chrysogenum the addition of l-lysine to fermentation medium led to reduced penicillin titers (48, 84). High levels of l-lysine also interfere with cephalosporin production in A. chrysogenum (227).
Since l-α-AAA is a branch point between l-lysine and the penicillin and cephalosporin biosynthetic pathways, l-lysine inhibition of penicillin biosynthesis was suggested to operate at one or more steps of the l-lysine pathway (reviewed in 209) (Fig. 6). This was based on the notion that l-lysine caused feedback inhibition of homocitrate synthase activity in P. chrysogenum (88, 119, 200) (Fig. 6). Homocitrate synthase catalyzes the initial reaction of l-lysine biosynthesis. In P. chrysogenum, 75% of this activity is in the cytoplasm and 25% is in mitochondria (148). Hence, compartmentalization might be important as well. Whether l-lysine also represses the synthesis of the enzyme is controversial (149, 200).
α-Aminoadipate reductase is also inhibited by l-lysine, and the inhibition occurs at physiological concentrations, i.e., typical cytosolic concentrations of l-lysine (4). It was demonstrated that in A. nidulans, another enzyme of the l-lysine biosynthetic pathway required for the synthesis of l-α-AAA, homoaconitase encoded by lysF, was reduced in l-lysine-grown mycelia (338). So far, lysF is the only gene of the l-lysine biosynthetic pathway cloned from a β-lactam-producing fungus (338). Taken together, it seems very likely that the l-α-AAA pool available for penicillin production is reduced by l-lysine through feedback inhibition and through repression of several l-lysine biosynthesis genes and enzymes (4, 143, 144, 310, 338). When the α-aminoadipate reductase activities (Fig. 6) of three penicillin-producing strains of P. chrysogenum were compared, α-aminoadipate (α-AAA) reductase from the lowest penicillin producer was found to be least sensitive to l-lysine inhibition. The activity in vivo of α-AAA reductase from superior penicillin producer strains of P. chrysogenum was more strongly inhibited, suggesting that this might explain the ability of high producers to accumulate increased amounts of l-α-AAA and consequently produce more penicillin (196). Furthermore, in A. nidulans, l-lysine led to repression of the expression of both acvA and ipnA gene fusions (48), suggesting, in addition, a more direct effect on the expression of penicillin biosynthesis genes (Fig. 5). Apparently, in the Panlab strain development program, there has been selection against inhibition and/or repression by l-lysine (186).
An important aspect with regard to l-lysine regulation is the compartmentation of the pathway. Biochemical data obtained with Saccharomyces cerevisiae suggest that the reactions of the first part of the pathway up to formation of l-α-AAA take place in the mitochondria whereas those of the second part are in the cytosol (37). Hence, l-α-AAA has to leave the mitochondria (Fig. 4). In addition, the major part of intracellular l-lysine is sequestered in the vacuole (38, 143).
It was reported that the addition of d,l-methionine to the A. chrysogenum medium (final concentration, 20 mM) led to a three- to fourfold increase in the production of cephalosporin C (96, 335). Early during the investigation of this methionine effect, the question arose whether the role of methionine was solely that of a sulfur donor, because the methionine analog norleucine, lacking a sulfur atom, mimicked the methionine inductive effect (289). Final and conclusive proof that the methionine effect acted at the gene expression level came from Velasco et al. (335). These authors reported that the increased production of cephalosporin C triggered by addition of methionine to the medium was paralleled by increased steady-state levels of the mRNAs of the cephalosporin biosynthesis genes acvA, ipnA, cefEF and, to a slight extent, cefG.
Lara et al. (185) reported an inductive effect of l-glutamate on penicillin biosynthesis by P. chrysogenum. However, for these experiments, ammonium chloride (8.5 mM) was used as the nitrogen source and was then replaced by l-glutamate (10 mM). With glutamate, cultures produced about five times the amount of penicillin as in the ammonium-grown controls. Recently, it was shown that the penicillin biosynthesis in P. chrysogenum was reduced when ammonium was used as the nitrogen source. This effect seems to be due, at least in part, to the repression of the penicillin biosynthesis genes acvA and ipnA by ammonium (109), which seems to be mediated by the nitrogen regulatory protein NRE (133). Therefore, it is conceivable that Lara et al. (185) measured nitrogen repression rather than an inducing effect of glutamate.
It was comprehensively tested whether the addition of different amino acids to the medium affected penicillin biosynthesis by A. nidulans (318). Differential effects due to various amino acids on the expression of the penicillin biosynthesis genes acvA and ipnA and penicillin production were measured. Interestingly, acvA expression seemed to be more subject to regulation by external amino acids than did ipnA expression. (i) Some amino acids (e.g., l-threonine, l-aspartate, l-glutamate, and l-cysteine) led to increased acvA-uidA expression but had no major effect on ipnA-lacZ expression. (ii) The amino acids l-methionine (at concentrations greater than 10 mM), l-leucine, l-isoleucine, l-phenylalanine, l-valine, l-histidine, and l-lysine led to repression of both acvA-uidA and ipnA-lacZ expression, which was dependent on the concentration used. (iii) The amino acids l-tyrosine, l-tryptophan, l-proline, and l-α-AAA had no major effect on acvA-uidA expression but led to repression of ipnA-lacZ expression. (iv) Two amino acids, l-serine and l-arginine, did not show any effect on the expression of either gene fusion at any of the concentrations tested.
l-Amino acids with a major negative effect on the expression of acvA-uidA and ipnA-lacZ gene fusions, i.e., histidine, valine, lysine, and methionine (only at concentrations greater than 10 mM), led to decreased penicillin titers and a decreased ambient pH during cultivation of the fungus. An analysis of deletion clones lacking binding sites of the pH-dependent transcriptional factor PACC (104, 246, 322) in the intergenic region between the acvA-uidA and ipnA-lacZ gene fusions and in a PacC5 mutant strain suggested that the negative effects of l-histidine and l-valine on acvA-uidA expression were due to reduced activation by PACC under the acidic conditions caused by these amino acids (see Fig. 10). These data also implied that PACC regulates the expression of acvA, predominantly through PACC binding site ipnA3 (see Fig. 10). The repressing effect caused by l-lysine and l-methionine on acvA, however, was even enhanced in one of the deletion clones and the pacC5 mutant strain, suggesting that these amino acids act independently of PACC by exerting effects on gene expression by unknown mechanisms (Fig. 5). For l-methionine, the analysis of deletion clones indicated that a major cis-acting DNA element responsible for its effect on acvA-uidA expression is located between nucleotides 433 and 569 of the intergenic region. For l-lysine, no DNA region which could be involved has yet been identified. The effects of l-methionine and l-lysine in the medium are mediated by regulatory mechanisms, which appear to differ even between these two amino acids (318). It is interesting that l-lysine and l-methionine are closely related to the precursor amino acids l-α-AAA and l-cysteine, respectively.
Velasco et al. (335) noticed several consensus CANNTG sequences in the intergenic region of the A. chrysogenum cephalosporin biosynthesis genes acvA and ipnA whose transcription was increased after addition of d,l-methionine. Members of the basic region-helix-loop-helix (bHLH) protein family recognize such a consensus motif. There are many bHLH proteins which are involved in the regulation of a number of genes. These proteins can bind as homo- or heterodimers to DNA (190). Some of these transcription factors are involved in the transcriptional control of the sulfur network in S. cerevisiae (22, 59, 229, 321). Thus, the authors suggested that a member of the bHLH proteins might mediate these effects. By analogy, it is conceivable that the effect of l-methionine on the corresponding A. nidulans genes is due to the action of a similar regulator. However, since there is no conserved CANNTG consensus motif in the region spanned by nucleotides 433 to 569 of the intergenic region between acvA and ipnA, it remains to be elucidated how the effect of methionine is mediated at the molecular level.
Influence of Phosphate and Oxygen
Excess phosphate was found to exert a negative effect on cephalosporin production in A. chrysogenum W53253 (181). The basis of this effect is still a matter of debate, and the molecular mechanism is not understood. It was suggested that phosphate increased the consumption rate of glucose, thus enhancing glucose repression (181), whereas resting-cell experiments indicated that in the absence of glucose, phosphate itself decreased the overall flux to formation of cephalosporin C (213). This was further supported by Zhang et al. (348), who found a direct negative effect of phosphate on formation of several enzymes (ACVS, IPNS, and DAOC synthetase/DAC hydroxylase) of the cephalosporin biosynthesis pathway. It was suggested that phosphate acted on IPNS and DAOC synthetase/DAC hydroxylase by complexing the iron needed for enzyme activity, because the inhibition of these two enzymes could be reversed by adding more ferrous iron (197). However, even inhibition of ACVS, which does not require ferrous iron for activity, was reversed by the addition of Fe2+ (349). Therefore, the cause of the inhibitory effect of phosphate is not yet fully understood.
The availability of oxygen is clearly important for penicillin production. Good aeration of mycelia with oxygen is a prerequisite for high β-lactam titers (139; reviewed in reference 317). Since several enzymes, such as IPNS and DAOC synthetase/DAC hydroxylase, require oxygen for their activity, it is conceivable that this is the reason for the oxygen requirement. The importance of oxygen is also supported by the possibility of increasing cephalosporin production genetically by introducing a bacterial oxygen binding protein into A. chrysogenum (89; see “Applications” below). However, there is a contradictory report which shows that reduction of oxygen led to increased acvA and ipnA expression of P. chrysogenum, possibly as part of a stress response (273).
The CCAAT Box Binding Protein Complex PENR1
Since several regulatory circuits affect the regulation of penicillin biosynthesis and presumably more remain to be discovered, a variety of cis-acting DNA elements and regulatory factors can be expected to be involved. Some of these have been found by analysis of mutants (pacC and pal) or by physiological and biochemical analyses (nre). For a comprehensive search of additional cis-acting DNA elements, a deletion analysis of the A. nidulans ipnA gene promoter (257) and a moving-window analysis of the intergenic region between the A. nidulans acvA and ipnA genes (319) were carried out. In the latter, the effects of deletions in the intergenic region were analyzed for the expression of both genes, acvA and ipnA, simultaneously.
Based on the results of this moving-window analysis and together with band shift and methyl interference assays, a CCAAT-containing DNA motif (box I) located 409 bp upstream of the ATG initiation codon of the A. nidulans acvA gene, which is bound by a protein complex designated PENR1 (penicillin regulator 1), was identified (319) (see Fig. 10). This CCAAT box I is of major importance for the regulation of both genes, since a 4-bp deletion within this site (ΔCCA-G) led to an eightfold increase of acvA expression and simultaneously to a reduction of ipnA expression to about 30% of the original level (319). Furthermore, scanning of the A. nidulans aatA promoter region by band shift assays, using protein fractions eluted with different KCl concentrations in the buffer from a heparin-agarose column, led to the identification of an additional CCAAT-containing DNA element (box II) located about 250 bp upstream of the transcriptional start sites of aatA. All biochemical data suggested that it was specifically bound by the same PENR1 regulatory protein complex. Substitution of the CCAAT core sequence by GATCC led to a fourfold reduction of expression of an aatA-lacZ gene fusion (195), indicating that the identified binding site was functional in vivo and positively influenced aatA expression (see Fig. 10).
Because PENR1 bound to a CCAAT-containing DNA sequence, this suggested that it belongs to the class of CCAAT binding proteins (195, 319). CCAAT boxes are present in the promoters of many eukaryotic genes. They are bound by distinct transcription factors, which may explain the variety of functions that have been attributed to the CCAAT sequences in the modulation of transcript levels in eukaryotic cells (see, e.g., references 32, 225, 236, 261, 272, and 287; reviewed in references 158 and 233). Several CCAAT binding factors have been found to bind to their target sequence as a complex of heterologous subunits, or heteromers (see, e.g., references 67, 134, and 272). For the time being, it is unknown how many different CCAAT binding proteins exist in filamentous fungi.
Some genes of A. nidulans have also been reported to contain functional CCAAT sequences in their promoter regions. Band shift experiments and deletion analysis showed that a CCAAT binding factor (AnCF) is involved in the regulation of the acetamidase gene amdS, which is required for the use of acetamide as the nitrogen and C source (189, 332). In addition, the intergenic region of the bidirectionally transcribed genes lamA and lamB (needed for utilization of lactams) and the promoter region of gatA (γ-amino butyric acid transaminase) contain CCAAT boxes which were bound by DNA binding factors (278). For the gatA CCAAT sequence, this also most probably seems to be AnCF (252). An additional CCAAT binding factor (AnCP) of A. nidulans was proposed (163, 240) which bound in vitro to the Taka-amylase gene promoter of A. oryzae. AnCP was later found to be most probably identical to AnCF (see below). Finally, deletion of a CCAAT box in the promoter of the developmentally regulated yA gene of A. nidulans led to elevated expression of a yA-lacZ gene fusion (15).
So far, the only CCAAT box binding factor that has been characterized in detail both genetically and biochemically in lower eukaryotes is the S. cerevisiae HAP complex, which consists of at least four subunits: HAP2, HAP3, and HAP5, which form a heterotrimeric complex that is essential for DNA binding, and HAP4, which is an acidic protein that acts as the transcriptional activation domain. Mutations in either of the hap genes led to the inability of yeast cells to grow on nonfermentable C sources such as lactate. It was shown that the HAP complex regulates, for example, the iso-1-cytochrome c (CYC1) gene required for respiration (see, e.g., reference 226; reviewed in reference 128).
Recently, Papagiannopoulos et al. (252) reported the cloning of a gene from A. nidulans designated hapC that exhibits significant similarity to the S. cerevisiae HAP3 gene. In a hapC deletion strain (ΔhapC), the activity of the CCAAT binding factor AnCF, which is present in wild-type strains and was detected by the binding of AnCF to CCAAT boxes present in the promoter regions of the genes gatA and amdS, was missing. In addition, the ΔhapC strain hardly grew on acetamide as the sole nitrogen and C source, indicating that HAPC in fact plays a role in regulating amdS expression. This was confirmed by analyzing an amdS-lacZ gene fusion, which showed reduced expression in a ΔhapC background. Hence, it seems very likely that HAPC is a member of the proposed AnCF complex that binds at least to the CCAAT motif in the gatA and amdS promoters. Furthermore, band shift and supershift experiments with polyclonal antibodies against HAPC revealed that AnCP also contains the HAPC protein (164).
For the CCAAT-binding complex PENR1, its binding consensus motif was determined by band shift assays as RRCCAAT(C/A)RCR (320). The deviation of a single base from the consensus motif reduced the percentage of bound DNA to at least 40% (for some bases even to 2 to 10%). Since the surrounding bases of the CCAAT site in the amdS promoter (AGCCAATCACC) match the PENR1 binding consensus motif except for the last cytosine, it was conceivable that HAPC is also part of PENR1 and, consequently, that AnCF and PENR1 share at least some protein components. Partial purification of PENR1 and Western blotting with anti-HAPC antibodies indicated that HAPC was highly enriched during purification of PENR1. Furthermore, band shift assays with partially purified crude extracts of a ΔhapC mutant strain revealed that in such crude extracts no PENR1 activity was detectable. Immunoprecipitation and supershift experiments with anti-HAPC antibodies provided evidence that HAPC is a component of the PENR1 complex (193). PENR1 thus represents a HAP-like transcriptional complex, as does AnCF.
Deletion or mutagenesis of the PENR1 binding sites had opposite effects; the expression of acvA was increased eightfold (319), while the expression of ipnA (319) and aatA (195) was reduced (see Fig. 10). Formally, the CCAAT box I mediated a negative effect on acvA, but a positive effect on ipnA, and box II a positive effect on aatA expression (see Fig. 10). Consistent with data obtained by deletion of the PENR1 binding site, expression of an ipnA-lacZ gene fusion in a ΔhapC background was reduced to about 15% compared with its expression in the wild type. A similar effect was observed for the expression of an aatA-lacZ gene fusion, which was reduced to 10% in the ΔhapC strain. Hence, PENR1 is of major importance as a positively acting factor of ipnA and aatA expression. Interestingly, the expression of ipnA and aatA gene fusions in the ΔhapC strains was even more reduced than in strains which had a deletion or substitution of the relevant CCAAT boxes. This might suggest that in addition to the identified CCAAT boxes I and II, other CCAAT boxes are important for the activity of PENR1 in vivo. Both in the intergenic region between acvA and ipnA and in the aatA promoter, there are additional CCAAT boxes. However, the analysis of the whole aatA promoter by band shift assays did not reveal another site apart from box II which was bound by PENR1 in vitro (195). This finding does not entirely exclude, however, that another CCAAT site which is upstream of box II might be relevant to a certain extent in vivo, although it can only be bound with low affinity by PENR1 in vitro.
Penicillin titers were reduced in a ΔhapC background as well, but only by about 30%. In addition, expression of an acvA-uidA gene fusion was hardly affected by the ΔhapC mutation (193). The minor effect of lack of PENR1 on penicillin production is consistent with the view that acvA expression is rate limiting in A. nidulans wild-type strains (165). Whereas overexpression of the acvA gene led to a massively increased penicillin production (165), simultaneous overexpression of both ipnA and aatA had little effect on penicillin production (112). In contrast, decreased expression of ipnA and aatA even by a factor of 5, as observed in the ΔhapC strain, results in a reduction of penicillin production of only about 30% when the expression of the acvA gene is only marginally affected.
The observation that acvA expression was less strongly affected in a ΔhapC strain was unexpected because it was shown that specific deletion of the four nucleotides CCA plus the 3′ G of the PENR1 binding site between acvA and ipnA (box I) resulted in a strong increase of acvA expression (319). It thus appears likely that in addition to PENR1, a repressor protein binds closely to or overlaps the PENR1 binding site, which would explain why the PENR1 binding site exhibits a repressing effect on acvA expression in the wild type. Consistent with this view, lack of PENR1 binding in the ΔhapC mutant did not prevent binding of this putative repressor protein and hence acvA-uidA expression was not increased. Deletion of the PENR1 binding site, however, prevented binding of both PENR1 and the repressor, causing the phenotype of increased acvA expression. However, the existence of such a putative repressor is hypothetical and remains to be shown experimentally. Proteins binding overlapping or adjacent to CCAAT boxes have been previously noted for several CCAAT-containing sequences in vertebrates and fungi (32, 97, 332). It would thus be very interesting to elucidate whether this also applies to the PENR1 binding sites (193). In summary, PENR1 represents a HAP-like complex which shares core proteins, at least HAPC, with AnCF. Because PENR1 does not bind to all CCAAT-containing sequences in vitro (195, 319) and, in addition, it is difficult to imagine that the amdS gene is regulated by the identical complex to that regulating the penicillin biosynthesis genes (and thus belongs to the same regulatory circuit), it seems likely that PENR1 differs from AnCF in an accessory protein(s). A. nidulans genes designated hapE (253) and hapB (192), exhibiting similarity to the S. cerevisiae hap5 and hap2 genes, respectively, have been cloned and sequenced. The availability of these genes will help to elucidate both PENR1 and its relation to the AnCF complex of A. nidulans and, in addition, to search for further proteins that may be part of the PENR1 complex.
As a result of PENR1 binding, a DNA fragment spanning the CCAAT box I between acvA and ipnA was distorted in vitro by more than 94°. The center of the flexure was predicted to be near or at the CCAAT sequence (193). A similar observation was made for the murine NF-Y complex, which is analogous to the S. cerevisiae HAP complex (281). Binding of NF-Y resulted in distortion of DNA to a degree dependent on the composition of neighboring sequences, varying between 62 and 82°. The center of the bend was also the CCAAT box (281).
The physiological function of HAP-like regulatory factors in lower eukaryotes remains obscure. In yeast, the HAP complex activates the expression of genes whose products are required for respiration. Hence, Hap mutants are not able to grow on nonfermentable carbon sources (see, e.g., reference 226; reviewed in reference 128). Because A. nidulans, however, is an aerobic fungus, S. cerevisiae may not be a good model for the role of HAP-like complexes in aerobic eukaryotes. Furthermore, the lack of a functional HAP complex (ΔhapC strain) is not lethal for A. nidulans (252). In addition, the HAP-like PENR1 complex in A. nidulans regulates secondary-metabolism genes (penicillin biosynthesis genes). Hence, it will be interesting to elucidate whether this particular function of a HAP-like complex requires as yet unknown accessory proteins, or, alternatively, which mechanism is involved in allowing a HAP-like complex to regulate certain sets of genes.
It is interesting that deletion of 4 bp of the PENR1 binding site led to increased acvA expression and reduced ipnA expression (319). It has not been clarified whether this increase of acvA and decrease of ipnA expression is of physiological relevance. It is tempting to speculate, however, that under certain physiological conditions an increase of acvA expression and a simultaneous reduction in the expression of ipnA and aatA could lead to larger cellular amounts of ACV. The tripeptide could have, apart from being the precursor of β-lactams, additional functions, e.g., in amino acid transport, as suggested by Del Carmen Mateos and Sanchez (83). ACV is structurally very similar to glutathione (GSH), consisting of the tripeptide γ-(l-glutamyl)-l-cysteinyl-glycine. Both ACV and GSH contain the free thiol group of the cysteine residue. They also have one unusual peptide bond between the δ-carbon atom of l-α-AAA and the γ-carbon atom of glutamate and the cysteine residue in ACV and GSH, respectively. In higher eukaryotes, GSH is a part of the γ-glutamyl cycle, which, among other functions, serves to transport extracellular amino acids across the plasma membrane (228). Experiments with resting cells of P. chrysogenum implied that the ACV tripeptide might be important for the uptake of glutamine from the medium (83). Consistent with this hypothesis is the observation that early during a fermentation run, both ACV tripeptide and the δ-lactam OPC (cyclized l-α-AAA) are found in the fermentation broth (52, 159). OPC appears very similar to 5-oxoproline, the cyclized product of the γ-glutamyl moiety of GSH, produced during GSH-dependent amino acid transport. Under certain physiological conditions, it might thus be advantageous for the fungus to produce the tripeptide first (e.g., for amino acid transport). Then, when binding of PENR1 and/or a putative repressor causes the expression of acvA to be reduced and simultaneously the expression of ipnA and aatA to be increased, the tripeptide is channelled towards penicillin. In addition, ACV might serve the function of fixing cysteine, which, at high levels, is toxic. Alternatively, the presence of a repressor could provide an additional possibility to fine-tune acvA expression. These assumptions, however, are speculative and need to be tested in the future.
Since HAPC is part of PENR1, it seems very likely that the PENR1 complex is conserved among the industrially important β-lactam-producing fungi. This assumption is supported by the observation that DNA fragments spanning the corresponding intergenic regions between acvA and ipnA of P. chrysogenum and A. chrysogenum and the promoter region of the P. chrysogenum aatA gene were able to dilute the complexes of the corresponding A. nidulans probes and A. nidulans PENR1 protein (195, 319). Computer analysis showed that DNA elements with a high degree of sequence identity to the A. nidulans PENR1 site reside within the intergenic regions of both P. chrysogenum and A. chrysogenum and the aatA promoter region of P. chrysogenum. These sites could be potential targets of homologous PENR1 complexes in P. chrysogenum and A. chrysogenum.
trans-Acting Mutations Affecting the Expression of Penicillin Biosynthesis Genes
The molecular approaches so far described led to the identification of DNA binding proteins involved in the regulation of penicillin biosynthesis genes. These regulatory proteins seem to represent wide-domain regulators (PACC and NRE). For the HAP-like complex PENR1, this cannot be predicted with certainty at the moment, although it seems likely. However, by analogy to the regulation of biosynthesis of secondary metabolites in streptomycetes (reviewed in reference 39), it can be expected that pathway specific regulatory genes exist.
To identify both such putative genes and regulatory genes not belonging to the family of DNA binding proteins (such as, e.g., coactivator proteins or members of a signal transduction cascade), a combination of classical and molecular techniques was independently used by Pérez-Esteban et al. (256) and Brakhage and Van den Brulle (50). The aim was the isolation of mutants of A. nidulans carrying mutations which are specifically involved in trans in the regulation of expression of the penicillin biosynthesis genes. The rationale of the approach can be summarized as follows. A. nidulans strains carrying ipnA-lacZ gene fusions integrated in double-copy at the chromosomal argB gene locus were used (Fig. 9). On minimal agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d- galactopyranoside (X-Gal) as an indicator for β-galactosidase activity, colonies of such a strain stained blue, indicative of ipnA-lacZ expression. After mutagenesis, colonies which did not produce a blue color or produced only a faint blue color were isolated. Cells of such colonies most probably carried mutations affecting the expression of ipnA-lacZ gene fusions in trans. The identification of cis-acting mutations, i.e., in the ipnA promoter or the lacZ gene, seemed to be unlikely, because such mutations would probably not be detected due to the second gene fusion located on the chromosome (Fig. 9).
FIG. 9.
Diagrammatic representation of possible homologous recombination events of a plasmid (pAXB4A) carrying acvA-uidA and ipnA-lacZ gene fusions integrated in single or double copy at the chromosomal argB gene locus of A. nidulans. The restriction map around the argB locus with and without integrated plasmid is shown. By using an appropriate probe, transformants carrying the plasmid integrated in double copy were identified by Southern blot analysis (46, 50). Essentially the same system with an ipnA-lacZ gene fusion only was independently used by Pérez-Esteban et al. (256) to identify the npeE1 mutation. Abbreviations: argB*, mutated argB allele which can complement the chromosomal argB2 mutation by homologous recombination; B, BamHI restriction site; lacZ, E. coli β-galactosidase gene; PacvA, promoter region of the acvA gene; PipnA, promoter region of the ipnA gene; uidA, E. coli β-glucuronidase gene. Modified from reference 50 with permission of the publisher.
By using this approach, mutants carrying recessive mutations were identified, and the genes were designated prg (penicillin regulation) (50) and npeE1 (impaired in penicillin biosynthesis) (256). In penicillin production medium, the mutants exhibited only 5 to 50% of the level of ipnA-lacZ expression and produced only 10 to 30% of the amount of penicillin compared with the wild-type strain. It was demonstrated that mutants Prg-1 (prgA1) and Prg-6 (prgB1) also differed in acvA-uidA expression levels from the wild type and, by Western blot analysis, that these mutants contained reduced amounts of the ipnA gene product, i.e., IPNS (50). Segregation analysis indicated that in both mutants the Prg phenotype resulted from mutation of a single gene. Two different complementation groups were identified, which were designated prgA1 (mutant strain Prg1) and prgB1 (mutant strain Prg6). However, the specific activity of the aatA gene product was essentially the same in the prgA1 and prgB1 mutants as in the wild type, implying that the last step of the penicillin biosynthetic pathway might not be affected, at least to the same extent, by these trans-acting mutations.
Genetic analysis showed that the npeE1 gene is located on linkage group IV (256). To date, it has not been clarified whether npeE1 differs from prgA1 and prgB1. The results obtained by genetic and biochemical analyses indicated that the mutants isolated most probably carry mutations in positively acting regulatory genes, which in the case of prgA1 and prgB1 specifically affect both acvA and ipnA and in the case of npeE1 affect at least ipnA expression (Fig. 10). Cloning of the corresponding genes will show to which kind of putative regulatory genes these mutations correspond and to which regulatory circuits they belong.
Using nitrosoguanidine, Cantoral et al. (60) isolated nine mutants of P. chrysogenum Wis54-1255 impaired in penicillin production. Biochemical and genetic analysis suggested that two of these (mutants Npe2 and Npe3) carry mutations in regulatory genes affecting the expression of the entire penicillin biosynthesis gene cluster.
Posttranscriptional Regulation
Discrepancies observed between the expression of structural genes and enzyme specific activities of the corresponding proteins suggested that besides transcriptional regulation, posttranscriptional regulation of penicillin biosynthesis genes occurs (47, 194, 346).
Mycelia of A. nidulans grown with glucose showed reduced IAT specific activities compared with those grown with lactose. When mycelia were first grown with glucose for 12 h, washed to remove remaining glucose, and subsequently transferred to medium with lactose, the IAT specific activities of mycelia rapidly increased, and reached even higher values after 24 h than did control cultures incubated with lactose only (194). This suggested that the effect of glucose on IAT specific activity was reversible. When cultures were first grown with lactose and then transferred to medium containing glucose, the IAT specific activity decreased compared with values measured in mycelia which were further incubated with lactose. In contrast, the expression of an aatA-lacZ gene fusion was only marginally reduced in glucose-grown mycelia compared with lactose-grown mycelia. Taken together, these results suggested that the effect of glucose on IAT specific activity was posttranscriptionally mediated (194).
For the A. chrysogenum ACVS, some posttranslational regulation was also observed with respect to its reduced activity measured in crude extracts obtained from glucose-grown mycelia (346, 349) (see “Carbon source regulation” above).
REGULATION OF β-LACTAM BIOSYNTHESIS IN FUNGAL PRODUCTION STRAINS
Apart from the academic interest in elucidating the molecular regulation of biosynthesis of secondary metabolites in lower eukaryotes, there is a strong interest from an industrial point of view because β-lactam compounds are still the most widely sold antibiotics in the world’s antibiotic market (reviewed in reference 211). Hence, it is desirable to analyze highly producing production strains which are highly mutated and have been derived from several different strain development programs. This will help to elucidate the molecular basis of deregulation and of the high production and also any remaining bottlenecks.
Nowadays, industrial penicillin and cephalosporin production is carried out mainly with P. chrysogenum and A. chrysogenum, respectively. Most of these strains have been produced by mutagenesis followed by screening or selection. In 1972, the initial Panlabs Inc. P. chrysogenum strain made 20,000 U of penicillin per ml in 7 days (an activity equivalent to 12 mg of pure penicillin G, Na salt, per ml [317]). In 1990, the improved strain made 70,000 U per ml in 7 days. Penicillin titers in industry in 1993 were as high as 100,000 U per ml (reviewed in references 211 and 219).
Commercial production of penicillin began in 1941 with P. notatum NRRL 1249-B21 (Northern Regional Research Laboratory, Peoria, Ill.) in surface culture fermentation. The search for superior production strains led to the isolation of P. chrysogenum NRRL 1951 (271). This strain became the ancestor of strains developed at the University of Wisconsin, which resulted in a family of strains (Wis strains) producing ever higher titers of penicillin. Strains descended from members of this series continue to be isolated by pharmaceutical companies (243).
Two important genetic features of P. chrysogenum production strains have been identified: (i) amplification of structural genes and (ii) their massively increased steady-state mRNA levels.
Slot blot and densitometry analysis of genomic DNA from the highly producing strain P. chrysogenum BW1890 (Beecham Pharmaceuticals; derived from the Wisconsin series of strains [reviewed in reference 244]) probed with the ipnA gene indicated that between 8 and 16 copies are present in that strain (304). Furthermore, Southern analysis demonstrated that the entire 39-kb insert of cosmid pCX3.2 carrying all penicillin biosynthesis structural genes (305) is amplified in this high-titer production strain. Northern blot analysis established that the steady-state level of ipnA mRNA in strain BW1890 was 32- to 64-fold that of NRRL 1951, an increase too great to be due to the amplification alone (304). Independently, Barredo et al. (33) showed that in the P. chrysogenum high-titer-producing strains P-2 (a highly producing strain from Nippon Kayaku Co. which was used as the ancestor of many strains in the Panlab series [reviewed in reference 244]) and AS-P-78 (old production strain used by Antibióticos [reviewed in reference 244]), the copy numbers of penicillin biosynthesis genes are approximately nine- and sixfold, respectively.
Fierro et al. (114) showed that in the high-titer P. chrysogenum strains E1 and AS-P-78, the amplifications are organized in tandem repeats. A conserved TTTACA hexanucleotide sequence was suggested to be involved in their generation. This TTTACA sequence borders the 106.5-kb penicillin biosynthesis gene cluster in the wild-type strain NRRL 1951 and also in P. notatum ATCC 9478 (Fleming’s isolate). Furthermore, three non-penicillin-producing P. chrysogenum mutants were independently isolated. It was shown that in all of these mutants deletion of the penicillin biosynthesis gene cluster had occurred at a specific site within the conserved hexanucleotide sequence (116). The authors suggested that this site may represent a hot spot for site-specific recombination after mutation with nitrosoguanidine, with the process possibly being part of a fungal SOS system similar to that found in E. coli (114, 116).
This analysis was extended to the characterization of other members of the SmithKline Beecham Pharmaceuticals strain improvement series (243). It was found that the amplicon was 57.5 kb long in these strains. Sequence analysis of the promoter regions of the acvA, ipnA, and aatA genes in the high titer strain BW1901 and comparison with wild-type sequences have not identified any potentially titer-enhancing mutations. Furthermore, cDNA screening has not identified any further transcribed elements within the coamplified region apart from those derived from the structural penicillin biosynthesis genes. The homogeneity of hybridization patterns and the identification and analysis of a single-copy revertant have shown that the amplification is of a direct tandem nature. It was proposed that whether or not the initial recombination event was homologous, once duplication had been achieved, a large stretch of homologous DNA was available for misalignment and hence further amplification or indeed deletion (243).
It appears that in both the SmithKline Beecham strain series and strain E1 (114), the amplified regions are bounded by the same nonamplified BamHI DNA fragments, although the amplicons are apparently not identical (243). Taken together, these data indicated the presence of recombinogenic regions flanking the penicillin biosynthesis gene cluster (114, 243).
Since sequence analysis of promoter regions has shown that no mutations have been generated within the promoter regions of the penicillin biosynthesis structural genes (243), these data suggest that the amplification of gene clusters may be accompanied by changes in trans-acting factors. This is also consistent with the observation that in the high-titer-producing P. chrysogenum strain BW1890 the increase in the steady-state level of ipnA mRNA was too great to be due to the amplification alone (304). However, data obtained with several production strains indicate that penicillin titers were not proportionally increased with copy number. Hence, in these strains the effect of other limiting factors, such as primary metabolic pathways or, in these cases, trans-acting regulatory factors, must be alleviated before the full potential of the extra copies is realized (243). Therefore, it will be of considerable interest to compare regulatory genes already found in both A. nidulans and P. chrysogenum between P. chrysogenum wild-type and production strains.
Furthermore, during the later stages of the fermentation in high-penicillin-producing strain BW1890, a high level of IPNS specific activity was maintained which was not due to increased transcription (304). The stability of IPNS might have increased, because results with A. nidulans imply that IPNS might be inactivated during its catalytic reaction (47).
The IAT specific activity was reduced in cells of A. nidulans and wild-type P. chrysogenum NRRL 1951 grown in medium containing glucose (46) but not in glucose-grown cells of the P. chrysogenum production strain AS-P-78 (277). The loss of the effect of glucose on IAT might be important for P. chrysogenum production strains.
In contrast to the gene amplification of structural genes reported in P. chrysogenum production strains, the β-lactam biosynthesis genes seem to be present in single copy in the cephalosporin C production strain A. chrysogenum LU4-79-6 (299, 301). Kück et al. (179) showed that A. chrysogenum DSM produces a considerably larger amount of ipnA transcript and of cephalosporin C (10-fold larger amount) than does strain ATCC 140553. Hence, as already demonstrated for penicillin production, increased transcription and consequently expression of biosynthesis genes seems to be an important feature of A. chrysogenum production strains.
Several high-cephalosporin C-producing A. chrysogenum strains contained chromosome rearrangements which partly involve the chromosome carrying the acvA and ipnA gene (300, 302, 337). However, for the time being, it is unclear whether rearrangements have an impact on the cephalosporin biosynthesis in production strains.
There are certainly numerous other mutations involved which lead to a high-producer phenotype. These also concern factors such as deregulation of enzymes involved in amino acid biosynthesis pathways and hence the amount of precursor amino acids produced. It was shown, for instance, that improved P. chrysogenum penicillin production strains have reduced catabolism of l-α-AAA and flux of l-α-AAA to l-lysine (143). In addition, α-aminoadipate reductase activity from the lowest penicillin producer of P. chrysogenum was least sensitive to l-lysine inhibition whereas the enzyme activity of superior penicillin producer strains was more strongly inhibited. This might explain the ability of highly producing strains to accumulate increased amounts of l-α-AAA, and consequently more penicillin (196) (see “Amino acids as precursors and mediators of regulation” above). Goulden and Chattaway (126) found that a high-producing P. chrysogenum strain had twice the acetohydroxy acid synthase activity of an ancestral strain and that the enzyme in the superior strain was deregulated to valine feedback inhibition (Fig. 8). A similar observation was reported for an A. chrysogenum highly producing mutant, in which acetohydroxy acid synthase was found to be partially desensitized to valine feedback inhibition (224). These alterations seem to be required to provide the large amounts of valine needed for penicillin production. Furthermore, transport of the intermediates of penicillin biosynthesis between organelles and the number of organelles can be predicted to also be important for penicillin production strains.
APPLICATIONS
The increasing knowledge of the molecular genetics of β-lactam biosynthesis has opened up new possibilities to rationally improve β-lactam production strains and to engineer new biosynthetic pathways. This leads to the question, however, whether an improvement of productivity is still possible. Cephalosporin C production by A. chrysogenum is well below the productivity reached with P. chrysogenum for penicillin. Therefore, much effort is needed to increase cephalosporin C production. For penicillin, several theoretical models have been established based on the available experimental data (summarized in reference 244). By using detailed stoichiometric models, the theoretical yield was calculated to be 0.47 to 0.50 mol of penicillin per mol of glucose (136, 160). Up to now, the maximum theoretical yields calculated are 8 to 10 times higher than the overall yields observed in fed-batch cultures, and there is therefore a considerable potential for further improvement of the process. However, it seems unlikely that the maximum theoretical yields can be reached in a real process, since the penicillin biosynthesis is indirectly coupled to other cellular reactions. Therefore, taking this into account, it was estimated that it should still be possible to improve the current yields by a factor of 4 to 5, which would have a dramatic effect on the economy of the process (244).
Several molecular strategies have been followed to improve or alter β-lactam production. (i) β-Lactam biosynthesis genes have been overexpressed by introducing additional copies or by using strong promoters. Manipulation of regulatory genes has not been reported yet because the regulatory genes have only recently been identified. (ii) β-Lactam biosynthetic pathways have been metabolically engineered by expression of heterologous genes, e.g., production of cephalosporin precursors in P. chrysogenum. (iii) The increasing knowledge about peptide synthetase genes, such as those for ACVS enzymes, has been used to produce novel compounds by genetic engineering (combinatorial biology [208, 312], as also discussed for polyketide synthetases [137]).
Increase of Expression of β-Lactam Biosynthesis Genes
Kennedy and Turner (165) demonstrated that acvA expression in A. nidulans is rate limiting for penicillin production. The weak acvA gene promoter was replaced by the strong inducible ethanol dehydrogenase promoter (alcAp). The expression level of alcAp was determined by using a strain in which the reporter gene, lacZ, was under the control of alcAp. It was found to be up to 100 times higher than that from the acvA promoter when induced under fermentation conditions with the artificial inducer cyclopentanone (10 mM). Penicillin yields were increased by as much as 30-fold when the acvA gene was overexpressed by induction of the alcA promoter. Glucose, which strongly represses transcription from alcAp, also repressed penicillin biosynthesis in the overproducing strain.
Fernández-Cañón and Peñalva (112) overexpressed both the ipnA and aatA genes of A. nidulans. For this purpose, they placed the promoters of these genes under the control of the alcA promoter. Transcriptional induction of the chimeric genes resulted in 10-fold-higher levels of ipnA or aatA transcripts than of those resulting from transcription of the corresponding endogenous genes. This increase caused a 40-fold rise in IPNS activity or an 8-fold rise in IAT activity. Despite this rise in enzyme levels, forced expression of the ipnA gene resulted in only a modest increase in levels of exported penicillin (increase by about 25%), whereas forced expression of the aatA gene even reduced penicillin production (a decrease of about 10 to 30%), showing that neither of these enzymes is rate limiting for penicillin biosynthesis. These findings agree well with those of Kennedy and Turner (165) that the expression of acvA is rate limiting for penicillin production in A. nidulans. They are also consistent with the observation that the massive reduction of ipnA and aatA expression caused by deletion of the hapC gene, whose product is part of PENR1, resulted in only a moderate reduction of the penicillin titer because acvA expression was only marginally, if at all, affected by deletion of the hapC gene (193).
Furthermore, a genomic version of the alcAp-aatA fusion in which the aatA gene is interrupted by three introns was inducible by threonine (an inducer of alcAp) to a lesser extent (as determined by both aatA mRNA levels and IAT enzyme levels) than was the corresponding cDNA version. This finding suggests that processing of the introns present in the primary transcript may limit aatA overexpression (112).
Consistent with data obtained with A. nidulans, attempts to increase cephalosporin C yields in A. chrysogenum 394-4 (improved cephalosporin C production strain) by inserting multiple copies of the ipnA gene were unsuccessful (301). However, Veenstra et al. (334) transformed P. chrysogenum Wis 54-1255 (an early strain of the strain improvement series) with a DNA fragment containing the P. chrysogenum Wis 54-1255 ipnA and aatA genes. Some of the resulting transformants produced up to 40% more penicillin V, indicating that in this particular P. chrysogenum strain the products of aatA, ipnA, or both can be used to improve penicillin production.
Skatrud et al. (301) reported a 15% increase in cephalosporin C production in an industrial strain of A. chrysogenum (394-4) by transformation of the strain with an extra copy of the DAOC synthetase/DAC hydroxylase gene (cefEF) (to give strain LU4-97-6) (Fig. 2). However, when the experiments were carried out, it was not known that cefG encoding acetyl-CoA:DAC acetyltransferase is closely linked to cefEF (Fig. 3) and was also present on the plasmid used for transformation. Then it was demonstrated that transformants of wild-type A. chrysogenum, carrying additional copies of the cefG gene only, showed a direct relationship between cefG copy number, cefG mRNA levels, and cephalosporin C titers, suggesting that this enzyme might be a rate-limiting step in cephalosporin C production (219, 221). Hence, the effect of the cefEF gene alone in increasing cephalosporin C production remains unclear.
Taken together, these data show that in a wild-type strain of A. nidulans, expression of biosynthesis genes, at least of the acvA gene, is rate limiting. It remains to be elucidated, however, whether this holds for penicillin and cephalosporin production strains. Nevertheless, the expression level of biosynthesis genes, as shown for the cephalosporin C production, is of great importance.
Genetic Engineering of β-Lactam Biosynthetic Pathways
Cloned β-lactam biosynthesis genes can already be used for rational improvement of β-lactam production. DAOC and cephalosporin C can be enzymatically deacylated to form 7-aminodeacetoxycephalosporanic acid (7-ADCA) and 7-aminocephalosporanic acid (7-ACA), respectively; these are important intermediates in the manufacturing of oral cephalosporin antibiotics (Fig. 11). Medically important oral cephalosporins, e.g., cephalexin and cephradine, are synthesized by derivatizing the 7-amino group of 7-ADCA or 7-ACA with appropriate side chain moieties (56). Although cephalosporins are superior antibiotics compared to penicillins, their production is limited because the process for producing 7-ADCA and 7-ACA is complex. Removal of the natural d-α-aminoadipyl side chain from cephalosporins is inefficient (82, 222). Hence, there have been efforts in both directions: isolation of superior enzymes to remove the side chain directly and development of alternative biosynthetic routes to 7-ACA and/or 7-ADCA (82, 222).
FIG. 11.
Production of cephalosporin intermediates via production of adipyl-cephalosporins by using recombinant strains of P. chrysogenum (82). The single arrows indicate reactions of the recombinant P. chrysogenum strains, and the double arrows indicate contact of the culture filtrate with immobilized Pseudomonas amidase. Modified from reference 82 with permission of the publisher.
At Eli Lilly & Co., clinically important cephalosporins were apparently produced by isolating penicillin G from P. chrysogenum and by chemical ring expansion of this natural product, which requires protection and sulfoxidation reactions followed by chemical deprotection (62, 264). The resulting deacetoxycephalosporin G is enzymatically deacylated to yield 7-ADCA (61, 62). Cantwell et al. (62) proposed a biological process in which penicillin V was used instead of penicillin G because P. chrysogenum strains that produce large amounts of penicillin V are available. For expansion of the thiazolidine ring of penicillin V to give a dihydrothiazine ring characteristic of ceph-3-ems, a modified penicillin N expandase from S. clavuligerus with a changed substrate specificity would be required. This enzyme should recognize and convert penicillin V to deacetoxycephalosporin V by ring expansion better than the wild-type enzyme does. In the last step, deacetoxycephalosporin V is enzymatically hydrolyzed to 7-ADCA with a penicillin amidase (62). Although this has apparently not been achieved experimentally yet, initial experiments showed that it is possible to express an S. clavuligerus penicillin N expandase gene (cefE) in P. chrysogenum. The cefEF gene of A. chrysogenum would not be useful for this purpose because it also encodes the hydroxylase function (61) (Table 4). 3′-hydroxylated cephalosporins, however, are not useful for the manufacturing of 7-ADCA (62). Based on this, Crawford et al. (82) expressed the S. clavuligerus cefE (expandase) or A. chrysogenum cefEF (expandase-hydroxylase) gene, with and without the acetyltransferase gene (cefG), in P. chrysogenum by fusing the genes to the P. chrysogenum ipnA or the β-tubulin gene promoter (Fig. 11). Feeding of such transformants with adipic acid led to the production of cephalosporins having an adipyl side chain (Fig. 11). This proved that adipyl-6-APA is a substrate for either enzyme in vivo. Transformants expressing cefE (expandase) produced adipyl-7-ADCA, whereas transformants expressing cefEF (expandase-hydroxylase) produced both adipyl-7-ADCA and adipyl-7-aminodeacetylcephalosporanic acid (adipyl-7-ADAC). Transformants expressing cefEF and cefG (acetyltransferase) produced adipyl-7-ADCA, adipyl-7-ADAC, and adipyl-7-ACA. The adipyl side chain of these cephalosporins was easily removed with a Pseudomonas-derived glutaryl amidase to yield the cephalosporin intermediates (Fig. 11). Hence, by these measures, an important step in cephalosporin manufacturing could be improved (82).
Cephalosporin C biosynthesis is regulated by the oxygen content of the medium (139). The overall rate of cephalosporin C synthesis is severely reduced under conditions of low oxygen. Reduction of the oxygen supply leads to the accumulation of penicillin N (Fig. 2). The mechanism of oxygen control is not understood. It may be connected to the oxygen-requiring reactions involved in cephalosporin biosynthesis (see “Influence of phosphate and oxygen” above). DeModena et al. (89) improved aerobic metabolism in A. chrysogenum by using the oxygen binding heme protein hemoglobin from the bacterium Vitreoscilla. Its structural gene (VHb) was fused with the strong constitutive TR1 promoter from Trichoderma reseii. After transformation of A. chrysogenum C10, the protein was produced as demonstrated by Western blotting. Several transformants produced significantly higher yields of cephalosporin C than did control strains in batch culture experiments.
Generation of Novel Compounds by Genetic Engineering of Peptide Synthetases
In general, peptide synthetases such as ACVS enzymes possess a highly conserved domain structure. The arrangement of these domains determines the number and order of the amino acid constituents of the peptide product (reviewed in reference 169). Hence, the shuffling of domains, i.e., reprogramming of the protein template, should result in the synthesis of new peptides exhibiting novel amino acid orders. This was achieved by Stachelhaus et al. (312), who exchanged domain-coding regions of bacterial and fungal origin. The authors developed a system allowing targeted substitution of amino acid-activating domains within the srfA operon, which encodes the protein templates for the synthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. By this method, new hybrid genes were produced whose products showed altered amino acid specificities. The genes were expressed in B. subtilis and led to the production of novel peptides, indicating that the rational design of bioactive peptides by recombinatorial biology is feasible (reviewed in reference 208).
CONCLUSIONS AND FUTURE DIRECTIONS
Research on the regulation of biosyntheses of β-lactam antibiotics is heading toward the elucidation of (i) further regulatory circuits involved, (ii) inducing and repressing signals, (iii) signal transduction pathways (missing links between regulatory circuits and regulatory genes), (iv) additional regulators (transcriptional factors, coactivators, and corepressors), and (v) the mode of action of these regulatory proteins. Understanding these aspects will also help to elucidate the physiological and ecological functions of β-lactams for the producing fungi. The application of this knowledge will contribute not only to a further increase of β-lactam production and the production of novel related compounds but also to the identification of new β-lactam-producing organisms by genetic means. For some of these aspects, a considerable increase in our knowledge has already been accumulated; the following discussion summarizes the underlying results and their future directions.
β-Lactam biosynthesis genes are transcribed from specific promoters and regulated transcriptionally and very probably posttranscriptionally. Transcription of biosynthesis genes is subject to sophisticated control by nutritional factors (e.g., glucose, nitrogen which is putatively mediated by NRE in P. chrysogenum), the amino acids lysine and methionine, and ambient pH. It can be assumed that there are other regulatory circuits which remain to be discovered.
With the exception of PACC, which mediates pH-dependent transcription, the regulatory factors responsible for the other regulatory circuits remain unknown. For the repressing effects on A. nidulans gene expression mediated by both l-lysine and l-methionine, their mechanistic basis (signals, regulatory factors) is still obscure. The same applies to the effect of methionine in A. chrysogenum. The effects are regulated at the transcriptional level, and for the methionine effect in A. nidulans a region probably containing the cis-acting DNA element responsible could be located. Nevertheless, the underlying mechanisms are unknown and even seem to differ for l-lysine and l-methionine.
For both penicillin and cephalosporin biosynthesis, the carbon source is important. Glucose and sucrose lead to reduced β-lactam production. It was shown that some β-lactam biosynthesis genes are repressed by these C sources. Interestingly, the repressor CREA, which mediates C-source-dependent repression of some A. nidulans genes, is not involved. This implies that C-source regulation of β-lactam biosynthesis genes does not follow the general mechanism of C-source regulation.
Nitrogen regulation of the penicillin biosynthesis genes in P. chrysogenum is most probably due to the action of NRE, which appears to represent a wide-domain nitrogen regulator homologous to the A. nidulans AREA factor. However, the involvement of NRE in nitrogen regulation of P. chrysogenum penicillin biosynthesis genes remains to be confirmed by in vivo studies.
Major questions concern the mechanistic basis of the regulatory circuits identified, and in the case of the HAP-like regulatory complex PENR1, the physiological signals activating this factor.
Much of our knowledge about the molecular regulation of penicillin biosynthesis genes has resulted from work on the fungal pH regulatory system (104, 246, 295, 322). This regulation is particularly interesting because it involves the proteolytic activation of a transcription factor by unknown molecular signals which are triggered by an alkaline ambient pH. Furthermore, several components of the regulation have already been elucidated, i.e., the regulatory circuit including the signal (alkaline ambient pH) and the mechanistic basis (transcriptional factor PACC). Based on the identified and available regulatory pal mutants, the tools are available to investigate the underlying signal transduction pathway.
It is interesting that although the overall patterns of penicillin production are similar in A. nidulans and P. chrysogenum, it appears that differences exist. Suarez and Peñalva (316) proposed that in P. chrysogenum the genes seem to be regulated more strictly than in A. nidulans, e.g., with respect to pH regulation but also with respect to C source and nitrogen regulation. The authors proposed that penicillin production might be more important for the competitiveness and survival of P. chrysogenum. This seems to be supported by the fact that wild-type P. chrysogenum isolates generally produce more penicillin than A. nidulans. However, this cannot be concluded with certainty because for screening of penicillin-producing isolates, only strains which showed increased penicillin production were selected. Thus, this might represent the (pathological) exception rather than the usual production level of P. chrysogenum.
Another question of particular interest is whether the genes are regulated by pathway-specific or wide-domain regulators. This question cannot be answered yet. However, the participation of PACC and possibly NRE, which are also present in non-penicillin-producing fungi, and the fact that the genes are expressed in the non-β-lactam producer A. niger (194, 306) indicate that they are regulated, at least in part, by wide-domain regulatory genes. For the HAP-like complex PENR1, this has not been confirmed yet, although it seems likely. PENR1 consists of subunits which are also involved in the regulation of other than β-lactam biosynthesis genes, such as amdS. However, since not all components of PENR1 have been determined yet, it is unclear whether this particular complex only regulates penicillin biosynthesis. Since PENR1 binds to only certain CCAAT sequences with high affinity in vitro, it appears possible that PENR1 regulates only some genes. The physiological function of HAP-like regulatory complexes in lower eukaryotes remains obscure. In S. cerevisiae, the HAP complex activates the expression of genes whose products are required for respiration. S. cerevisiae predominantly ferments carbon sources. Therefore, hap mutants are not able to grow on nonfermentable carbon sources (reviewed in reference 128). However, because A. nidulans is an aerobic fungus, S. cerevisiae may not be a good model for the role of HAP-like complexes in aerobic eukaryotes. Hence, further characterization of the regulatory complexes PENR1 and AnCF, which at least have common protein components, even if they are not identical, is needed to elucidate the structure and function of these complexes. If PENR1 regulates other genes than the penicillin biosynthesis genes, such as amdS and gatA, it will be very interesting to elucidate why these genes belong to a common regulatory circuit. Candidates for pathway-specific genes, however, might be found among the recessive mutations identified (prgA1, prgB1, and npeE1) in A. nidulans and by the mutations carried by the P. chrysogenum mutants Npe2 and Npe3, which seem to specifically affect the expression of penicillin biosynthesis genes.
In addition to positively acting regulators, all data are consistent with the involvement of negatively acting regulatory factors. A repressor of acvA expression has been proposed in A. nidulans which binds close to or overlaps the PENR1-binding site (box I) (Fig. 10).
Also, because fungi are eukaryotes, it can be expected that the regulation of β-lactam genes differs from that in prokaryotes. This view is consistent with the finding that all regulators identified in fungal β-lactam biosynthesis which are summarized here have no counterpart in prokaryotes and vice versa. A regulatory gene (ccaR) of both cephamycin and clavulanic acid biosynthesis was found to be located in the cephamycin biosynthesis gene cluster of S. clavuligerus (Fig. 3). This gene shows high similarity to regulatory genes involved in the biosynthesis of other secondary metabolites in streptomycetes, such as actinorhodin biosynthesis (258). Furthermore, some fungal regulators might be part of larger complexes, as proposed for the HAP-like transcriptional complex PENR1. This would also have some effect on strain improvement programs, because it might be required to overproduce all components of such complexes in order to obtain overexpression of β-lactam biosynthesis genes and thus overproduction of β-lactams.
So far, only alkaline ambient pH has been identified as a physiological signal which triggers penicillin biosynthesis (104). It appears that the expression of genes is to a certain extent intrinsically constitutive, although this cannot be concluded with absolute certainty. It might well be that signals that have not yet been identified, such as cell density, which was proposed to stimulate β-lactam production in streptomycetes (285), are important. In addition, despite significant levels of gene expression in minimal medium, there is only little penicillin production, even when sufficient amounts of side chain precursor molecules are fed. This might imply that not only is the production of penicillin of relevance, but also the production of the ACV and/or IPN precursor molecule, which could serve additional physiological functions, is relevant. Nevertheless, the presence of a basal gene expression level implies that the presence of penicillin and/or precursor molecules is advantageous to the fungus under different physiological conditions, although their production consumes energy. This basal gene expression level seems to be modulated in response to the different environmental and/or endogenous stimuli; e.g., growth is maximal in the presence of rapidly metabolizable carbon sources whereas gene expression and production of penicillin is reduced. Possibly, fungi do not need to produce large amounts of antibiotics to compete with bacteria when their growth rate is maximal as a result of the availability of good C and N sources. In general, β-lactam production seems to be optimal when the growth of fungi is suboptimal. Therefore, the elucidation of regulatory genes might also contribute to our understanding of such questions related to the physiological function of these compounds for the producing fungus.
It is unclear whether β-lactams serve another function such as defending the habitat against bacteria. It was suggested that the ACV tripeptide could have a function in amino acid transport or could help to fix cysteine, which is toxic at high concentrations. Alternatively, because the last step of penicillin biosynthesis occurs in microbodies and can lead to the attachment of aromatic rings to IPN, this might represent a mechanism of detoxifying these compounds and excreting them to the environment. There are numerous such compounds in the environment of penicillin-producing fungi, e.g., derived from the degradation of lignin. All of these theories, however, have yet to be proved experimentally.
Since gene expression has a major effect on the production of β-lactam antibiotics in production processes, the knowledge of the molecular regulation of their biosynthesis genes can be expected to be of great importance for rational strain improvement programs. This is particularly relevant because theoretical calculations based on flux analyses have indicated that there is still an enormous potential to increase β-lactam productivity. Therefore, overexpression of regulatory genes that positively affect the expression of both the β-lactam biosynthesis genes and precursor amino acid biosynthesis genes will be attempted and negatively acting regulatory genes will be inactivated.
Although considerable progress has been made in the understanding of the molecular regulation of penicillin and cephalosporin biosynthesis in fungi, our picture is far from complete.
ACKNOWLEDGMENTS
The members of my laboratory, in particular Katharina Then Bergh, Olivier Litzka, Stefan Steidl, Jan Van den Brulle, and Gerhard Weidner, who have generated some of the results summarized here, are gratefully acknowledged for their dedicated work and helpful discussions. I thank Geoffrey Turner, Hans von Döhren, and Arnold L. Demain for comments on the manuscript and Roger Newbert, Miguel A. Peñalva, Norihiro Tsukagoshi, and Geoffrey Turner for kindly communicating results prior to publication. Thanks are due to Natalie Domke for excellent technical assistance and Kim Langfelder for help in improving the manuscript. I am indebted to August Böck for generous support and valuable discussions.
Research in my laboratory was supported by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 369) and the European Union (BIO CT 94-2100 and EUROFUNG).
REFERENCES
- 1.Abraham E P. History of β-lactam antibiotics. In: Demain A L, Solomon N A, editors. Antibiotics containing the β-lactam structure. I. Berlin, Germany: Springer-Verlag KG; 1983. pp. 1–14. [Google Scholar]
- 2.Abraham E P. Selective reminiscences of β-lactam antibiotics: early research on penicillin and cephalosporins. Bioessays. 1990;12:601–606. doi: 10.1002/bies.950121208. [DOI] [PubMed] [Google Scholar]
- 3.Abraham E P, Newton G G F. The structure of cephalosporin C. Biochem J. 1961;79:377–393. doi: 10.1042/bj0790377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Affenzeller K, Jaklitsch W M, Hönlinger C, Kubicek C P. Lysine biosynthesis in Penicillium chrysogenum is regulated by feedback inhibition of α-aminoadipate reductase. FEMS Microbiol Lett. 1989;58:293–298. doi: 10.1016/0378-1097(89)90056-6. [DOI] [PubMed] [Google Scholar]
- 5.Affenzeller K, Kubicek C P. Evidence for a compartmentation of penicillin biosynthesis in a high- and a low-producing strain of Penicillium chrysogenum. J Gen Microbiol. 1991;137:1653–1660. doi: 10.1099/00221287-137-7-1653. [DOI] [PubMed] [Google Scholar]
- 6.Aharonowitz Y, Av-Gay Y, Schreiber R, Cohen G. Characterization of a broad-range disulfide reductase from Streptomyces clavuligerus and its possible role in β-lactam antibiotic biosynthesis. J Bacteriol. 1993;175:623–629. doi: 10.1128/jb.175.3.623-629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aharonowitz Y, Bergmeyer J, Cantoral J M, Cohen G, Demain A L, Fink U, Kinghorn J R, Kleinkauf H, MacCabe A, Palissa H, Pfeifer E, Schwecke T, van Liempt H, von Döhren H, Wolfe S, Zhang J. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase, the multienzyme integrating the four primary reactions in β-lactam biosynthesis, as a model peptide synthetase. Bio/Technology. 1993;11:807–810. doi: 10.1038/nbt0793-807. [DOI] [PubMed] [Google Scholar]
- 8.Aharonowitz Y, Cohen G, Martin J F. Penicillin and cephalosporin biosynthetic genes: structure, organisation, regulation, and evolution. Annu Rev Microbiol. 1992;46:461–495. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- 9.Alonso M J, Bermejo F, Reglero A, Fernández-Cañón J M, González de Buitrago G, Luengo J M. Enzymatic synthesis of penicillins. J Antibiot. 1988;XLI:1074–1084. doi: 10.7164/antibiotics.41.1074. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez E, Cantoral J M, Barredo J L, Diez B, Martin J F. Purification to homogeneity and characterization of acyl coenzyme A:6-amino penicillanic acid acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother. 1987;31:1675–1682. doi: 10.1128/aac.31.11.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez E, Meesschaert B, Montenegro E, Gutiérrez S, Diez B, Barredo J L, Martin J F. The isopenicillin N acyltransferase of Penicillium chrysogenum has isopenicillin N amidohydrolase, 6-aminopenicillanic acid acyltransferase and penicillin amidase activities, all of which are encoded by the single penDE gene. Eur J Biochem. 1993;215:323–332. doi: 10.1111/j.1432-1033.1993.tb18038.x. [DOI] [PubMed] [Google Scholar]
- 12.Alvi K A, Reeves C D, Peterson J, Lein J. Isolation and identification of a new cephem compound from Penicillium chrysogenum strains expressing deacetoxycephalosporin synthase activity. J Antibiot. 1995;48:338–340. doi: 10.7164/antibiotics.48.338. [DOI] [PubMed] [Google Scholar]
- 13.Aplin R T, Baldwin J E, Cole S C, Sutherland J D, Tobin M B. On the production of alpha, beta-heterodimeric acyl-coenzyme A:isopenicillin N-acyltransferase of Penicillium chrysogenum. Studies using a recombinant source. FEBS Lett. 1993;319:166–170. doi: 10.1016/0014-5793(93)80060-8. [DOI] [PubMed] [Google Scholar]
- 14.Aplin R T, Baldwin J E, Roach P L, Robinson C V, Schofield C J. Investigations into the post-translational modification and mechanism of isopenicillin N:acyl-CoA acyltransferase using electrospray mass spectrometry. Biochem J. 1993;294:357–363. doi: 10.1042/bj2940357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aramayo R, Timberlake W E. The Aspergillus nidulans yA gene is regulated by abaA. EMBO J. 1993;12:2039–2048. doi: 10.1002/j.1460-2075.1993.tb05853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnstein H R V, Morris D. The structure of a peptide containing α-aminoadipic acid, cysteine and valine, present in the mycelium of Penicillium chrysogenum. Biochem J. 1960;76:357–361. doi: 10.1042/bj0760357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arst H N., Jr Genetic analysis of the first steps of sulphate metabolism in Aspergillus nidulans. Nature. 1968;219:268–270. doi: 10.1038/219268a0. [DOI] [PubMed] [Google Scholar]
- 18.Arst H N., Jr . Regulation of gene expression by pH. In: Brambl R, Marzluf G A, editors. The Mycota. III. Biochemistry and molecular biology. Berlin, Germany: Springer-Verlag KG; 1996. pp. 235–240. [Google Scholar]
- 19.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 20.Arst H N, Jr, Scazzocchio C. Formal genetic methodology of Aspergillus nidulans as applied to the study of control systems. In: Bennett J W, Lasure L L, editors. Gene manipulations in fungi. New York, N.Y: Academic Press, Inc.; 1985. pp. 309–343. [Google Scholar]
- 21.Bailey C, Arst H N., Jr Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem. 1975;51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- 22.Baker R E, Masison D C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere binding protein CP1. Mol Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin J E, Abraham E P. The biosynthesis of penicillins and cephalosporins. Nat Prod Rep. 1988;5:129–145. doi: 10.1039/np9880500129. [DOI] [PubMed] [Google Scholar]
- 24.Baldwin J E, Adlington R M, Moroney S E, Field L D, Ting H-H. Stepwise ring closure in penicillin biosynthesis. Initial β-lactam formation. J Chem Soc Chem Commun. 1984;1984:984–986. [Google Scholar]
- 25.Baldwin J E, Bird J W, Field R A, O’Callaghan N M, Schofield C J. Isolation and partial characterization of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. J Antibiot. 1990;XLIII:1055–1057. doi: 10.7164/antibiotics.43.1055. [DOI] [PubMed] [Google Scholar]
- 26.Baldwin J E, Bird J W, Field R A, O’Callaghan N M, Schofield C J, Willis A C. Isolation and partial characterization of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. Evidence for the presence of phosphopantothenate in ACV synthetase. J Antibiot. 1991;44:241–248. doi: 10.7164/antibiotics.44.241. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin J E, Gagnon J, Ting H. N-terminal amino acid sequence and some properties of isopenicillin-N synthetase from Cephalosporium acremonium. FEBS Lett. 1985;188:253–256. doi: 10.1016/0014-5793(85)80382-3. [DOI] [PubMed] [Google Scholar]
- 28.Baldwin J E, Shiau C-Y, Byford M F, Schofield C J. Substrate specificity of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase from Cephalosporium acremonium: demonstration of the structure of several unnatural tripeptide products. Biochem J. 1994;301:367–372. doi: 10.1042/bj3010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldwin J E, Wan T E. Penicillin biosynthesis. Retention of configuration at C-3 of valine during its incorporation into the Arnstein tripeptide. Tetrahedron. 1981;37:1589–1595. [Google Scholar]
- 30.Banko G, Demain A L, Wolfe S. Cell-free synthesis of δ-(l-α-aminoadipyl)-l-cysteine, the first intermediate of penicillin and cephalosporin biosynthesis. Biochem Biophys Res Commun. 1986;137:528–535. doi: 10.1016/0006-291x(86)91242-8. [DOI] [PubMed] [Google Scholar]
- 31.Banko G, Demain A L, Wolfe S. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase (ACV synthetase): a multifunctional enzyme with broad substrate specificity for the synthesis of penicillin and cephalosporin precursors. J Am Chem Soc. 1987;109:2858–2860. [Google Scholar]
- 32.Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 33.Barredo J L, Diez B, Alvarez E, Martin J F. Large amplification of a 35-kb DNA fragment carrying two penicillin biosynthetic genes in high penicillin producing strains of P. chrysogenum. Curr Genet. 1989;16:453–459. doi: 10.1007/BF00340725. [DOI] [PubMed] [Google Scholar]
- 34.Barredo J L, Cantoral J M, Alvarez E, Diez B, Martin J F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989;216:91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- 35.Barredo J L, van Solingen P, Diez B, Alvarez E, Cantoral J M, Kattevilder A, Smaal E B, Groenen M A M, Veenstra A E, Martin J F. Cloning and characterization of the acyl-coenzyme A:6- amino-penicillanic-acid-acyltransferase gene of Penicillium chrysogenum. Gene. 1989;83:291–300. doi: 10.1016/0378-1119(89)90115-7. [DOI] [PubMed] [Google Scholar]
- 36.Behmer C J, Demain A L. Further studies on carbon catabolite regulation of β-lactam antibiotic synthesis in Cephalosporium acremonium. Curr Microbiol. 1983;8:107–114. [Google Scholar]
- 37.Betterton H, Fjellstedt T, Matsuda M, Ogur M, Tate R. Localisation of the homocitrate pathway. Biochim Biophys Acta. 1968;170:459–461. doi: 10.1016/0304-4165(68)90036-6. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharjee J K. Evolution of α-aminoadipate pathway for the synthesis of lysine in fungi. In: Mortlock R P, editor. The evolution of metabolic function. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 47–80. [Google Scholar]
- 39.Bibb M. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 40.Böck A, Wirth R, Schmid G, Schumacher G, Lang G, Buckel P. The penicillin acylase from Escherichia coli ATCC11105 consists of two dissimilar subunits. FEMS Microbiol Lett. 1983;135:135–139. [Google Scholar]
- 41.Böck A, Wirth R, Schmid G, Schumacher G, Lang G, Buckel P. The two subunits of penicillin acylase are processed from a common precursor. FEMS Microbiol Lett. 1983;20:141–144. [Google Scholar]
- 42.Borges-Walmsley M I, Turner G, Bailey A M, Brown J, Lehmbeck J, Clausen I G. Isolation and characterisation of genes for sulphate activation and reduction in Aspergillus nidulans: implications for evolution of an allosteric control region by gene duplication. Mol Gen Genet. 1995;247:423–429. doi: 10.1007/BF00293143. [DOI] [PubMed] [Google Scholar]
- 43.Borovok I, Landman O, Kreisberg-Zakarin R, Aharonowitz Y, Cohen G. Ferrous active site of isopenicillin N synthase: genetic and sequence analysis of the endogenous ligands. Biochemistry. 1996;35:1981–1987. doi: 10.1021/bi951534t. [DOI] [PubMed] [Google Scholar]
- 44.Brakhage, A. A. Unpublished data.
- 45.Brakhage A A. Molecular regulation of the penicillin biosynthesis in Aspergillus nidulans. FEMS Microbiol Lett. 1997;148:1–10. doi: 10.1111/j.1574-6968.1997.tb10258.x. [DOI] [PubMed] [Google Scholar]
- 46.Brakhage A A, Browne P, Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992;174:3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brakhage A A, Browne P, Turner G. Analysis of penicillin biosynthesis and the expression of penicillin biosynthesis genes of Aspergillus nidulans by targeted disruption of the acvA gene. Mol Gen Genet. 1994;242:57–64. doi: 10.1007/BF00277348. [DOI] [PubMed] [Google Scholar]
- 48.Brakhage A A, Turner G. l-Lysine repression of penicillin biosynthesis and expression of penicillin biosynthesis genes acvA and ipnA in Aspergillus nidulans. FEMS Microbiol Lett. 1992;98:123–128. doi: 10.1016/0378-1097(92)90142-b. [DOI] [PubMed] [Google Scholar]
- 49.Brakhage A A, Turner G. Biotechnical genetics of antibiotic biosynthesis. In: Kück U, editor. The Mycota. II. Genetics and biotechnology. Berlin, Germany: Springer-Verlag KG; 1995. pp. 263–285. [Google Scholar]
- 50.Brakhage A A, Van den Brulle J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J Bacteriol. 1995;177:2781–2788. doi: 10.1128/jb.177.10.2781-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brotzu G. Richerche su di un nuovo antibiotico. Lav. Ist. d’Igiene Cagliari; 1948. 1948:1–11. [Google Scholar]
- 52.Brundidge S P, Gaeta F C A, Hook D J, Sapino C, Jr, Elander R P, Morin R B. Association of 6-oxo-piperidine-2-carboxylic acid with penicillin V production in Penicillium chrysogenum fermentations. J Antibiot. 1980;33:1348–1351. doi: 10.7164/antibiotics.33.1348. [DOI] [PubMed] [Google Scholar]
- 53.Brunner R, Röhr M. Phenylacyl:CoA ligase. Methods Enzymol. 1975;43:476–481. doi: 10.1016/0076-6879(75)43107-x. [DOI] [PubMed] [Google Scholar]
- 54.Brunner R, Zinner M. Zur Biosynthese des Penicillins. Hoppe-Seyler’s Z Physiol Chem. 1968;349:95–103. doi: 10.1515/bchm2.1968.349.1.95. [DOI] [PubMed] [Google Scholar]
- 55.Brzywczy J, Yamagata S, Paszewski A. Comparative studies on O-acetylhomoserine sulfhydrylase: physiological role and characterization of the Aspergillus nidulans enzyme. Acta Biochim Pol. 1993;40:421–428. [PubMed] [Google Scholar]
- 56.Bunnel C A, Luke W D, Perry F M., Jr . Industrial manufacture of cephalosporins. In: Queener S F, Webber J A, Queener S W, editors. β-Lactam antibiotics for clinical use. New York, N.Y: Marcel Dekker Inc.; 1986. pp. 255–284. [Google Scholar]
- 57.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 58.Caddick M X, Peters D, Platt A. Nitrogen regulation in fungi. Antonie Leeuwenhoek. 1994;65:169–177. doi: 10.1007/BF00871943. [DOI] [PubMed] [Google Scholar]
- 59.Cai M, Davis R W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 60.Cantoral J M, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H, Martin J F. Biochemical characterisation and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem. 1993;268:737–744. [PubMed] [Google Scholar]
- 61.Cantwell C, Beckmann R, Whiteman P, Queener S W, Abraham E P. Isolation of deacetoxycephalosporin C from fermentation broths of Penicillium chrysogenum transformants: construction of a new fungal biosynthetic pathway. Proc R Soc London Ser B. 1992;248:283–289. doi: 10.1098/rspb.1992.0073. [DOI] [PubMed] [Google Scholar]
- 62.Cantwell C A, Beckmann R J, Dotzlaf R J, Fisher D L, Skatrud P L, Yeh W-K, Queener S W. Cloning and expression of a hybrid S. clavuligerus cefE gene in P. chrysogenum. Curr Genet. 1990;17:213–221. doi: 10.1007/BF00312612. [DOI] [PubMed] [Google Scholar]
- 63.Carr L G, Skatrud P L, Scheetz II M E, Queener S W, Ingolia T D. Cloning and expression of the isopenicillin N synthetase gene from Penicillium chrysogenum. Gene. 1986;48:257–266. doi: 10.1016/0378-1119(86)90084-3. [DOI] [PubMed] [Google Scholar]
- 64.Charlier J. Isoleucyl-transfer ribonucleic acid synthetase from Bacillus stearothermophilus. 2. Dynamics of the enzyme-substrate interactions. Eur J Biochem. 1972;25:175–180. doi: 10.1111/j.1432-1033.1972.tb01682.x. [DOI] [PubMed] [Google Scholar]
- 65.Chen V J, Orville A M, Harpel M R, Frolik C A, Surerus K K, Münck E, Lipscomb J D. Spectroscopic studies of isopenicillin N synthase. A mononuclear nonheme Fe2+ oxidase with metal coordination sites for small molecules and substrate. J Biol Chem. 1989;264:21677–21681. [PubMed] [Google Scholar]
- 66.Chiang T Y, Marzluf G A. DNA recognition by the NIT2 nitrogen regulatory protein: importance of the number, spacing, and orientation of GATA core elements and their flanking sequences upon NIT2 binding. Biochemistry. 1994;33:576–582. doi: 10.1021/bi00168a024. [DOI] [PubMed] [Google Scholar]
- 67.Chodosh L A, Olesen J, Hahn S, Baldwin A S, Guarente L, Sharp P A. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988;53:25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- 68.Chu Y-W, Renno D, Saunders G. Detection of a protein which binds specifically to the upstream region of the pcbAB gene in Penicillium chrysogenum. Curr Genet. 1995;27:184–189. doi: 10.1007/BF00315786. [DOI] [PubMed] [Google Scholar]
- 69.Clarke D L, Newbert R W, Turner G. Cloning and characterisation of the adenosyl phosphosulphate kinase gene from Aspergillus nidulans. Curr Genet. 1997;32:408–412. doi: 10.1007/s002940050295. [DOI] [PubMed] [Google Scholar]
- 70.Clutterbuck A J. Aspergillus nidulans, nuclear genes. In: O’Brien S J, editor. Genetic maps. 6th ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. pp. 371–384. [Google Scholar]
- 71.Clutterbuck P W, Lovell R, Raistrick R. The formation from glucose by members of the Penicillium chrysogenum series of a pigment, an alkali-soluble protein and penicillin—the antibacterial substance of Fleming. Biochem J. 1932;26:1907–1918. doi: 10.1042/bj0261907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cohen G, Argaman A, Schreiber R, Mislovati M, Aharonowitz Y. The thioredoxin system of Penicillium chrysogenum and its possible role in penicillin biosynthesis. J Bacteriol. 1994;176:973–984. doi: 10.1128/jb.176.4.973-984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen G, Shiffman D, Mevarech M, Aharonowitz Y. Microbial isopenicillin N synthase genes: structure, function, diversity and evolution. Trends Biotechnol. 1990;8:105–111. doi: 10.1016/0167-7799(90)90148-q. [DOI] [PubMed] [Google Scholar]
- 74.Coque J-J R, De La Fuente J L, Liras P, Martin J F. Overexpression of the Nocardia lactamdurans α-aminoadipyl-cysteinyl-valine synthetase in Streptomyces lividans. The purified multienzyme uses cystathionine and 6-oxopiperidine 2-carboxylate as substrates for synthesis of the tripeptide. Eur J Biochem. 1996;242:264–270. doi: 10.1111/j.1432-1033.1996.0264r.x. [DOI] [PubMed] [Google Scholar]
- 75.Coque J-J R, Enguita F J, Martin J F, Liras P. A two-protein component 7 alpha-cephem-methoxylase encoded by two genes of the cephamycin C cluster converts cephalosporin C to 7-methoxycephalosporin C. J Bacteriol. 1995;177:2230–2235. doi: 10.1128/jb.177.8.2230-2235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coque J-J R, Liras P, Laiz L, Martin J F. A gene encoding lysine 6-aminotransferase, which forms the β-lactam precursor α-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J Bacteriol. 1991;173:6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coque J-J R, Liras P, Martin J F. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993;12:631–639. doi: 10.1002/j.1460-2075.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coque J-J R, Martin J F, Calzada J G, Liras P. The cephamycin biosynthetic genes pcbAB, encoding a large multidomain peptide synthetase, and pcbC of Nocardia lactamdurans are clustered together in an organization different from the same genes in Acremonium chrysogenum and Penicillium chrysogenum. Mol Microbiol. 1991;5:1125–1133. doi: 10.1111/j.1365-2958.1991.tb01885.x. [DOI] [PubMed] [Google Scholar]
- 79.Coque J-J R, Perez-Llarena F J, Enguita F J, Fuente J L, Martin J F, Liras P. Characterization of the cmcH genes of Nocardia lactamdurans and Streptomyces clavuligerus encoding a functional 3′-hydroxymethylcephem O-carbamoyltransferase for cephamycin biosynthesis. Gene. 1995;162:21–27. doi: 10.1016/0378-1119(95)00308-s. [DOI] [PubMed] [Google Scholar]
- 80.Corbett K. The history of antibiotic production. Biochemist. 1990;12:8–13. [Google Scholar]
- 81.Cortes J, Martin J F, Castro J M, Laiz L, Liras P. Purification and characterization of a 2-oxoglutarate-linked ATP-independent deacetoxycephalosporin C synthase of Streptomyces lactamdurans. J Gen Microbiol. 1987;133:3165–3174. doi: 10.1099/00221287-133-11-3165. [DOI] [PubMed] [Google Scholar]
- 82.Crawford L, Stepan A M, McAda P C, Rambosek J A, Conder M J, Vinci V A, Reeves C D. Production of cephalosporin intermediates by feeding adipic acid to recombinant Penicillium chrysogenum strains expressing ring expansion activity. Bio/Technology. 1995;13:58–62. doi: 10.1038/nbt0195-58. [DOI] [PubMed] [Google Scholar]
- 83.Del Carmen Mateos R, Sanchez S. Transport of neutral amino acids and penicillin formation in Penicillium chrysogenum. J Gen Microbiol. 1990;136:1713–1716. [Google Scholar]
- 84.Demain A L. Inhibition of penicillin formation by lysine. Arch Biochem Biophys. 1957;67:244–245. doi: 10.1016/0003-9861(57)90265-5. [DOI] [PubMed] [Google Scholar]
- 85.Demain A L. Synthesis of cephalosporin C by resting cells of Cephalosporium sp. Clin Med. 1963;70:2045–2051. [PubMed] [Google Scholar]
- 86.Demain A L. Biosynthesis of β-lactam antibiotics. In: Demain A L, Solomon N A, editors. Antibiotics containing the β-lactam structure. I. New York, N.Y: Springer-Verlag; 1983. pp. 189–228. [Google Scholar]
- 87.Demain A L, Kennel Y M, Aharonowitz Y. Carbon catabolite regulation of secondary metabolism. In: Bull A T, Ellwood D C, Ratledge C, editors. Microbial technology: current state, future prospects. Vol. 29. Cambridge, United Kingdom: Cambridge University Press; 1979. pp. 163–185. [Google Scholar]
- 88.Demain A L, Masurekar P S. Lysine inhibition of in vivo homocitrate synthesis in Penicillium chrysogenum. J Gen Microbiol. 1974;82:143–151. doi: 10.1099/00221287-82-1-143. [DOI] [PubMed] [Google Scholar]
- 89.DeModena J A, Gutiérrez S, Velasco J, Fernández F J, Fachini R A, Galazzo J L, Hughes D E, Martin J F. The production of cephalosporin C by Acremonium chrysogenum is improved by the intracellular expression of a bacterial hemoglobin. Bio/Technology. 1993;11:926–929. doi: 10.1038/nbt0893-926. [DOI] [PubMed] [Google Scholar]
- 90.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 91.Diez B S, Gutiérrez S, Barredo J L, van Solingen P, van der Voort L H M, Martin J F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the α-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 92.Dorn G. Genetic analysis of the phosphatases in Aspergillus nidulans. Genet Res. 1965;6:13–26. doi: 10.1017/s0016672300003943. [DOI] [PubMed] [Google Scholar]
- 93.Dotzlaf J E, Yeh W-K. Copurification and characterization of deacetoxycephalosporin C synthase/hydroxylase from Cephalosporium acremonium. J Bacteriol. 1987;169:1611–1618. doi: 10.1128/jb.169.4.1611-1618.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dotzlaf J E, Yeh W-K. Purification and properties of deacetoxycephalosporin C synthase from recombinant Escherichia coli and its comparison with the native enzyme purified from Streptomyces clavuligerus. J Biol Chem. 1989;264:10219–10227. [PubMed] [Google Scholar]
- 95.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drew S W, Demain A L. Methionine control of cephalosporin C formation. Biotechnol Bioeng. 1973;15:743–754. doi: 10.1002/bit.260150408. [DOI] [PubMed] [Google Scholar]
- 97.El-Hodiri H M, Perry M. Interaction of the CCAAT displacement protein with shared regulatory elements required for transcription of paired histone genes. Mol Cell Biol. 1995;15:3587–3596. doi: 10.1128/mcb.15.7.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Erickson R C, Dean L D. Acylation of 6-aminopenicillanic acid by Penicillium chrysogenum. Appl Microbiol. 1966;14:1047–1048. doi: 10.1128/am.14.6.1047-1048.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Esmahan C, Alvarez E, Montenegro E, Martin J F. Catabolism of lysine in Penicillium chrysogenum leads to formation of 2-aminoadipic acid, a precursor of penicillin biosynthesis. Appl Environ Microbiol. 1994;60:1705–1710. doi: 10.1128/aem.60.6.1705-1710.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Espeso E A, Fernández-Cañón J M, Peñalva M A. Carbon regulation of penicillin biosynthesis in Aspergillus nidulans: a minor effect of mutations in creB and creC. FEMS Microbiol Lett. 1995;126:63–68. doi: 10.1016/0378-1097(94)00527-x. [DOI] [PubMed] [Google Scholar]
- 101.Espeso E A, Peñalva M A. Carbon catabolite repression can account for temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol Microbiol. 1992;6:1457–1465. doi: 10.1111/j.1365-2958.1992.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 102.Espeso E A, Peñalva M A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J Biol Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 103.Espeso, E. A., and M. A. Peñalva. Unpublished data.
- 104.Espeso E A, Tilburn J, Arst H N, Jr, Peñalva M A. pH regulation is a major determinant in expression of a fungal biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Espeso E A, Tilburn J, Sánchez-Pulido L, Brown C V, Valencia A, Arst H N, Jr, Peñalva M A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J Mol Biol. 1997;274:466–480. doi: 10.1006/jmbi.1997.1428. [DOI] [PubMed] [Google Scholar]
- 106.Fawcett P A, Usher J J, Abraham E P. Behaviour of tritium-labelled isopenicillin N and 6-aminopenicillanic acid as potential penicillin precursors in an extract of Penicillium chrysogenum. Biochem J. 1975;151:741–746. doi: 10.1042/bj1510741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fawcett P A, Usher J J, Huddleston J A, Bleaney R C, Nisbet J J, Abraham E P. Synthesis of δ(α-aminoadipyl)cysteinylvaline and its role in penicillin biosynthesis. Biochem J. 1976;157:651–660. doi: 10.1042/bj1570651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Felix H R, Nüesch J, Wehrli W. Investigation of the two final steps on the biosynthesis of cephalosporin C using permeabilised cells of Cephalosporium acremonium. FEMS Microbiol Lett. 1980;8:55–58. [Google Scholar]
- 109.Feng B, Friedlin E, Marzluf G A. A reporter gene analysis of penicillin biosynthesis gene expression in Penicillium chrysogenum and its regulation by nitrogen and glucose catabolite repression. Appl Environ Microbiol. 1994;60:4432–4439. doi: 10.1128/aem.60.12.4432-4439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng B, Friedlin E, Marzluf G A. Nuclear DNA-binding proteins which recognize the intergenic control region of penicillin biosynthetic genes. Curr Genet. 1995;27:351–358. doi: 10.1007/BF00352104. [DOI] [PubMed] [Google Scholar]
- 111.Feng D-F, Cho G, Doolittle R F. Determining divergence times with a protein clock: update and reevaluation. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fernández-Cañón J M, Peñalva M A. Overexpression of two penicillin structural genes in Aspergillus nidulans. Mol Gen Genet. 1995;246:110–118. doi: 10.1007/BF00290139. [DOI] [PubMed] [Google Scholar]
- 113.Fernández-Cañón J M, Reglero A, Martinez-Blanco H, Luengo J M. I. Uptake of phenylacetic acid by Penicillium chrysogenum Wis54-1255: a critical regulatory point in benzylpenicillin biosynthesis. J Antibiot. 1989;XLII:1389–1409. doi: 10.7164/antibiotics.42.1398. [DOI] [PubMed] [Google Scholar]
- 114.Fierro F, Barredo J L, Diez B, Gutiérrez S, Fernández F J, Martin J F. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc Natl Acad Sci USA. 1995;92:6200–6204. doi: 10.1073/pnas.92.13.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fierro F, Gutiérrez S, Diez B, Martin J F. Resolution of four large chromosomes in penicillin-producing filamentous fungi: the penicillin gene cluster is located on chromosome II (9.6 Mb) in Penicillium notatum and chromosome I (10.4 Mb) in Penicillium chrysogenum. Mol Gen Genet. 1993;241:573–579. doi: 10.1007/BF00279899. [DOI] [PubMed] [Google Scholar]
- 116.Fierro F, Montenegro E, Gutiérrez S, Martin J F. Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl Microbiol Biotechnol. 1996;44:597–604. doi: 10.1007/BF00172491. [DOI] [PubMed] [Google Scholar]
- 117.Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenza. Br J Exp Pathol. 1929;10:226–236. [Google Scholar]
- 118.Foster B A, Thomas S M, Mahr J A, Renosto F, Patel H C, Segel I H. Cloning and sequencing of ATP sulfurylase from Penicillium chrysogenum: identification of a likely allosteric domain. J Biol Chem. 1994;269:19777–19786. [PubMed] [Google Scholar]
- 119.Friedrich C G, Demain A L. Homocitrate synthase as the crucial site of the lysine effect on penicillin biosynthesis. J Antibiot. 1977;XXX:760–761. doi: 10.7164/antibiotics.30.760. [DOI] [PubMed] [Google Scholar]
- 120.Friedrich C G, Demain A L. Uptake and metabolism of α-aminoadipic acid by Penicillium chrysogenum Wis 54-1255. Arch Microbiol. 1978;119:43–47. doi: 10.1007/BF00407926. [DOI] [PubMed] [Google Scholar]
- 121.Fu Y H, Marzluf G A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA. 1990;87:5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fujisawa Y, Kanzaki T. Role of acetyl-CoA:deacetylcephalosporin C acetyltransferase in cephalosporin C biosynthesis in Cephalosporium acremonium. Agric Biol Chem. 1975;39:2043–2048. [Google Scholar]
- 123.Gatenbeck S, Brunsberg H. Biosynthesis of penicillins. I. Isolation of 6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum. Acta Chem Scand. 1968;22:1059–1061. doi: 10.3891/acta.chem.scand.22-1059. [DOI] [PubMed] [Google Scholar]
- 124.Gómez-Pardo E, Peñalva M A. The upstream region of the IPNS gene determines expression during secondary metabolism in Aspergillus nidulans. Gene. 1990;89:109–115. doi: 10.1016/0378-1119(90)90212-a. [DOI] [PubMed] [Google Scholar]
- 125.Gould S J, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goulden S A, Chattaway F W. End product control of acetohydroxy acid synthetase by valine in Penicillium chrysogenum Q-176 and a high penicillin-yielding mutant. J Gen Microbiol. 1969;59:111–118. doi: 10.1099/00221287-59-1-111. [DOI] [PubMed] [Google Scholar]
- 127.Gravel R A, Kafer E, Niklewicz-Borkenhagen A, Zambryski P. Genetic accumulation studies in sulfite-requiring mutants of Aspergillus nidulans. Can J Genet Cytol. 1970;12:831–840. doi: 10.1139/g70-107. [DOI] [PubMed] [Google Scholar]
- 128.Guarente L. Messenger RNA transcription and its control in Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. 2. Gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 49–98. [Google Scholar]
- 129.Gutiérrez S, Diez B, Alvarez E, Barredo J L, Martin J F. Expression of the penDE gene of Penicillium chrysogenum encoding isopenicillin N acyltransferase in Cephalosporium acremonium: production of benzylpenicillin by the transformants. Mol Gen Genet. 1991;225:56–64. doi: 10.1007/BF00282642. [DOI] [PubMed] [Google Scholar]
- 130.Gutiérrez S, Diez B, Montenegro E, Martin J F. Characterization of the Cephalosporium acremonium pcbAB gene encoding alpha-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol. 1991;173:2354–2365. doi: 10.1128/jb.173.7.2354-2365.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gutiérrez S, Velasco J, Fernandez F J, Martin J F. The cefG gene of Cephalosporium acremonium is linked to the cefEF gene and encodes a deacetylcephalosporin C acetyltransferase closely related to homoserine O-acetyltransferase. J Bacteriol. 1992;174:3056–3064. doi: 10.1128/jb.174.9.3056-3064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Haas H, Bauer B, Redl B, Stöffler G, Marzluf G A. Molecular cloning and analysis of nre, the major nitrogen regulatory gene of Penicillium chrysogenum. Curr Genet. 1995;27:150–158. doi: 10.1007/BF00313429. [DOI] [PubMed] [Google Scholar]
- 133.Haas H, Marzluf G A. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr Genet. 1995;28:177–183. doi: 10.1007/BF00315785. [DOI] [PubMed] [Google Scholar]
- 134.Hahn S, Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988;240:317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- 135.Heatley N G. Early work at Oxford on penicillin. Biochemist. 1990;12:4–7. [Google Scholar]
- 136.Hersbach G J M, van der Beek C P, van Dijck P W M. The penicillins: properties, biosynthesis and fermentation. In: Vandamme E J, editor. Biotechnology of industrial antibiotics. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 45–140. [Google Scholar]
- 137.Hershberger C L. Metabolic engineering of polyketide biosynthesis. Curr Opin Biotechnol. 1996;7:560–562. doi: 10.1016/s0958-1669(96)80062-0. [DOI] [PubMed] [Google Scholar]
- 138.Higgens C E, Hamill R C, Sands T H, Hoehn M M, Davis N E. The occurrence of deacetoxycephalosporin C in fungi and Streptomycetes. J Antibiot. 1974;27:298–300. doi: 10.7164/antibiotics.27.298. [DOI] [PubMed] [Google Scholar]
- 139.Hilgendorf P, Heiser V, Diekmann H, Thoma M. Constant dissolved oxygen concentrations in cephalosporin C fermentation: applicability of different controllers and effect on fermentation parameters. Appl Microbiol Biotechnol. 1987;XXVII:247–251. [Google Scholar]
- 140.Hillenga D J, Versantvoort H J M, van der Molen S, Driessen A J M, Konings W N. Penicillium chrysogenum takes up the penicillin G precursor phenylacetic acid by passive diffusion. Appl Environ Microbiol. 1995;61:2589–2595. doi: 10.1128/aem.61.7.2589-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hodgkin D C, Maslen E N. The X-ray analysis of the structure of cephalosporin C. Biochem J. 1961;79:393–402. doi: 10.1042/bj0790393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hollander I J, Shen V Q, Heim J, Demain A L, Wolfe S. A pure enzyme catalyzing penicillin biosynthesis. Science. 1984;224:610–612. doi: 10.1126/science.6546810. [DOI] [PubMed] [Google Scholar]
- 143.Hönlinger C, Kubicek C P. Metabolism and compartmentation of α-aminoadipic acid in penicillin-producing strains of Penicillium chrysogenum. Biochim Biophys Acta. 1989;993:204–211. [Google Scholar]
- 144.Hönlinger C, Kubicek C P. Regulation of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine and isopenicillin N biosynthesis in Penicillium chrysogenum by the α-aminoadipate pool size. FEMS Microbiol Lett. 1989;65:71–76. doi: 10.1016/0378-1097(89)90368-6. [DOI] [PubMed] [Google Scholar]
- 145.Hoskins J A, O’Callaghan N, Queener S W, Cantwell C A, Wood J S, Chen V J, Skatrud P L. Gene disruption of the pcbAB gene encoding ACV synthetase in Cephalosporium acremonium. Curr Genet. 1990;18:523–530. doi: 10.1007/BF00327023. [DOI] [PubMed] [Google Scholar]
- 146.Hynes M J, Kelly J. Pleiotropic mutants of Aspergillus nidulans altered in carbon source metabolism. Mol Gen Genet. 1977;150:193–204. doi: 10.1007/BF00695399. [DOI] [PubMed] [Google Scholar]
- 147.Ingolia T D, Queener S W. β-Lactam biosynthetic genes. Med Res Rev. 1989;9:245–264. doi: 10.1002/med.2610090206. [DOI] [PubMed] [Google Scholar]
- 148.Jaklitsch W M, Kubicek C P. Homocitrate synthase from Penicillium chrysogenum. Biochem J. 1990;269:247–253. doi: 10.1042/bj2690247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jaklitsch W M, Röhr M, Kubicek C P. Lysine biosynthesis in Penicillium chrysogenum: regulation by general amino acid control and absence of lysine repression. Exp Mycol. 1987;11:141–149. [Google Scholar]
- 150.Jakubowski H, Pawelkiewicz J. Fractionation of plant aminoacyl-tRNA synthetases on t-RNA-sepharose columns. Eur J Biochem. 1975;52:301–310. doi: 10.1016/0005-2787(75)90286-5. [DOI] [PubMed] [Google Scholar]
- 151.Jayatilake S, Huddleston J A, Abraham E P. Conversion of isopenicillin N into penicillin N in cell-free extracts of Cephalosporium acremonium. Biochem J. 1981;194:645–647. doi: 10.1042/bj1940645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jensen S E, Demain A L. Beta-lactams. In: Vining L C, Stuttard C, editors. Genetics and biochemistry of antibiotic production. Newton, Mass: Butterworth-Heinemann; 1995. pp. 239–268. [Google Scholar]
- 153.Jensen S E, Leskiw B K, Vining L C, Aharonowitz Y, Westlake D W S. Purification of isopenicillin N synthetase from Streptomyces clavuligerus. Can J Microbiol. 1986;32:953–958. doi: 10.1139/m86-176. [DOI] [PubMed] [Google Scholar]
- 154.Jensen S E, Westlake D W S, Wolfe S. High performance liquid chromatographic assay of cyclisation activity in cell-free systems from Streptomyces clavuligerus. J Antibiot. 1982;XXXV:1026–1032. doi: 10.7164/antibiotics.35.1026. [DOI] [PubMed] [Google Scholar]
- 155.Jensen S E, Westlake D W S, Wolfe S. Partial purification and characterization of isopenicillin N epimerase activity from Streptomyces clavuligerus. Can J Microbiol. 1983;29:1526–1531. doi: 10.1139/m83-234. [DOI] [PubMed] [Google Scholar]
- 156.Jensen S E, Westlake D W S, Wolfe S. Deacetoxycephalosporin C synthetase and deacetoxycephalosporin C hydroxylase are two separate enzymes in Streptomyces clavuligerus. J Antibiot. 1985;XXXVIII:263–265. doi: 10.7164/antibiotics.38.263. [DOI] [PubMed] [Google Scholar]
- 157.Jensen S E, Wong A, Rollins M J, Westlake D W S. Purification and partial characterization of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase from Streptomyces clavuligerus. J Bacteriol. 1990;172:7269–7271. doi: 10.1128/jb.172.12.7269-7271.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Johnson P F, McKnight S L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 159.Jorgensen H S, Nielsen J, Villadsen J, Molgaard H. Analysis of the penicillin V biosynthesis during fed-batch cultivations with a high yielding strain of Penicillium chrysogenum. Appl Microbiol Biotechnol. 1995;43:123–130. doi: 10.1007/BF00170633. [DOI] [PubMed] [Google Scholar]
- 160.Jorgensen H S, Nielsen J, Villadsen J, Molgaard H. Metabolic flux distributions in Penicillium chrysogenum during fed-batch cultivations. Biotechnol Bioeng. 1995;46:117–131. doi: 10.1002/bit.260460205. [DOI] [PubMed] [Google Scholar]
- 161.Kakitani M, Tonomura B, Hiromi K. Order of binding of substrate to valyl-tRNA synthetase from Bacillus stearothermophilus in amino acid activation reaction. Biochemistry. 1987;14:597–603. [PubMed] [Google Scholar]
- 162.Kallow W, Neuhof T, Arezi B, Jungblut P, von Döhren H. Penicillin biosynthesis: intermediates of biosynthesis of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine formed by ACV synthetase from Acremonium chrysogenum. FEBS Lett. 1997;414:74–78. doi: 10.1016/s0014-5793(97)00977-0. [DOI] [PubMed] [Google Scholar]
- 163.Kato M, Aoyama A, Naruse F, Kobayashi T, Tsukagoshi N. An Aspergillus nidulans nuclear protein, AnCP, involved in enhancement of Taka-amylase A gene expression, binds to the CCAAT-containing taaG2, amdS, and gatA promoters. Mol Gen Genet. 1997;254:119–126. doi: 10.1007/s004380050399. [DOI] [PubMed] [Google Scholar]
- 164.Kato M, Aoyama A, Naruse F, Tateyama Y, Hayashi K, Miyazaki M, Papagiannopoulos P, Davis M A, Hynes M J, Kobayashi T, Tsukagoshi N. The Aspergillus nidulans CCAAT-binding factor, AnCP/AnCF, is a heteromeric protein analogous to the HAP complex of Saccharomyces cerevisiae. Mol Gen Genet. 1998;257:404–411. doi: 10.1007/s004380050664. [DOI] [PubMed] [Google Scholar]
- 165.Kennedy J, Turner G. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol Gen Genet. 1996;253:189–197. doi: 10.1007/s004380050312. [DOI] [PubMed] [Google Scholar]
- 166.Kimura H, Izawa M, Sumino Y. Molecular analysis of the gene cluster involved in cephalosporin biosynthesis from Lysobacter lactamgenus YK90. Appl Microbiol Biotechnol. 1996;44:589–596. doi: 10.1007/BF00172490. [DOI] [PubMed] [Google Scholar]
- 167.Kimura H, Miyashita H, Sumino Y. Organization and expression in Pseudomonas putida of the gene cluster involved in cephalosporin biosynthesis from Lysobacter lactamgenus YK90. Appl Microbiol Biotechnol. 1996;45:490–501. doi: 10.1007/BF00578461. [DOI] [PubMed] [Google Scholar]
- 168.Kleinkauf H, von Döhren H. Biosynthesis of peptide antibiotics. Annu Rev Microbiol. 1987;41:259–289. doi: 10.1146/annurev.mi.41.100187.001355. [DOI] [PubMed] [Google Scholar]
- 169.Kleinkauf H, von Döhren H. A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 170.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kodekar R, Deshpande V D. Biosynthesis of penicillin in vitro. Purification and properties of phenyl/phenoxyacetic acid activating enzyme. Indian J Biochem Biophys. 1982;19:257–261. [PubMed] [Google Scholar]
- 172.Kohsaka M, Demain A L. Conversion of penicillin N to cephalosporin(s) by cell-free extracts of Cephalosporium acremonium. Biochem Biophys Res Commun. 1976;70:465–473. doi: 10.1016/0006-291x(76)91069-x. [DOI] [PubMed] [Google Scholar]
- 173.Kolar M, Holzmann K, Weber G, Leitner E, Schwab H. Molecular characterization and functional analysis in Aspergillus nidulans of the 5′-region of the Penicillium chrysogenum isopenicillin N synthetase gene. J Biotechnol. 1991;17:67–80. doi: 10.1016/0168-1656(91)90027-s. [DOI] [PubMed] [Google Scholar]
- 174.Konomi T, Herchen S, Baldwin J E, Yoshida M, Hunt N A, Demain A L. Cell-free conversion of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine into antibiotic with the properties of isopenicillin N in Cephalosporium acremonium. Biochem J. 1979;184:427–430. doi: 10.1042/bj1840427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kovacevic S, Miller J R. Cloning and sequencing of the β-lactam hydroxylase gene (cefF) from Streptomyces clavuligerus: gene duplication may have led to separate hydroxylase and expandase activities in the actinomycetes. J Bacteriol. 1991;173:398–400. doi: 10.1128/jb.173.1.398-400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kovacevic S, Tobin M B, Miller J R. The β-lactam biosynthesis genes for isopenicillin N epimerase and deacetoxycephalosporin C synthetase are expressed from a single transcript in Streptomyces clavuligerus. J Bacteriol. 1990;172:3952–3958. doi: 10.1128/jb.172.7.3952-3958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kovacevic S, Weigel B J, Tobin M B, Ingolia T D, Miller J R. Cloning, characterization, and expression in Escherichia coli of the Streptomyces clavuligerus gene encoding deacetoxycephalosporin C synthetase. J Bacteriol. 1989;171:754–760. doi: 10.1128/jb.171.2.754-760.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kozma J, Bartok G, Szentirmai A. Fructose-2,6-bisphosphate level and β-lactam formation in Penicillium chrysogenum. Int J Basic Microbiol. 1993;33:27–34. doi: 10.1002/jobm.3620330106. [DOI] [PubMed] [Google Scholar]
- 179.Kück U, Walz M, Mohr G, Mracek M. The 5′-sequence of the isopenicillin N-synthetase gene (pcbC) from Cephalosporium acremonium directs the expression of the prokaryotic hygromycin B phosphotransferase gene (hph) in Aspergillus niger. Appl Microbiol Biotechnol. 1989;31:358–365. [Google Scholar]
- 180.Kudla B, Caddick M X, Langdon T, Martinez-Rossi N M, Benett C F, Silbey S, Davis R W, Arst H N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Küenzi M T. Regulation of cephalosporin synthesis in Cephalosporium acremonium by phosphate and glucose. Arch Microbiol. 1980;128:78–81. doi: 10.1007/BF00422309. [DOI] [PubMed] [Google Scholar]
- 182.Kupka J, Shen Y-Q, Wolfe S, Demain A L. Partial purification and properties of the α-ketoglutarate-linked ring expansion enzyme of β-lactam biosynthesis. FEMS Microbiol Lett. 1983;16:1–6. [Google Scholar]
- 183.Kurzatkowski W, Kurzatkowski J D, Filipek J, Solecka J, Holska W, Kurylowicz W. Reversion of l-lysine inhibition of penicillin G biosynthesis by 6-oxopiperidine-2-carboxylic acid in Penicillium chrysogenum PQ96. Appl Microbiol Biotechnol. 1990;34:397–398. doi: 10.1007/BF00170067. [DOI] [PubMed] [Google Scholar]
- 184.Landan G, Cohen G, Aharonowitz Y, Shuali Y, Graur D, Shiffman D. Evolution of isopenicillin N synthase genes may have involved horizontal gene transfer. Mol Biol Evol. 1990;7:399–406. doi: 10.1093/oxfordjournals.molbev.a040615. [DOI] [PubMed] [Google Scholar]
- 185.Lara F, del Carmen Mateos R, Vazquez D, Sanchez S. Induction of penicillin biosynthesis by glutamate in Penicillium chrysogenum. Biochem Biophys Res Commun. 1982;105:172–178. doi: 10.1016/s0006-291x(82)80027-2. [DOI] [PubMed] [Google Scholar]
- 186.Lein J. The Panlabs penicillin strain improvement program. In: Vanek Z, Hostálek Z, editors. Overproduction of microbial metabolites. Boston, Mass: Butterworths; 1986. pp. 105–139. [Google Scholar]
- 187.Lendenfeld T, Ghali D, Wolschek M, Kubicek-Pranz E M, Kubicek C P. Subcellular compartmentation of penicillin biosynthesis in Penicillium chrysogenum. J Biol Chem. 1993;268:665–671. [PubMed] [Google Scholar]
- 188.Li W-H, Luo C-C, Wu C-I. Evolution of DNA sequences. In: MacIntyre R J, editor. Molecular evolutionary genetics. New York, N.Y: Plenum Press; 1985. pp. 1–94. [Google Scholar]
- 189.Littlejohn T G, Hynes M J. Analysis of the site of action of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet. 1992;235:81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- 190.Littlewood T, Evan G I. Protein profile. Transcription factors 2: helix-loop-helix. 1, issue 6. London, United Kingdom: Academic Press, Ltd.; 1994. [PubMed] [Google Scholar]
- 191.Litzka, O., and A. A. Brakhage. Unpublished data.
- 192.Litzka, O., and A. A. Brakhage. Unpublished data. EMBL accession no. Y13768.
- 193.Litzka O, Papagiannopoulos P, Davis M A, Hynes M J, Brakhage A A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur J Biochem. 1998;251:758–767. doi: 10.1046/j.1432-1327.1998.2510758.x. [DOI] [PubMed] [Google Scholar]
- 194.Litzka O, Then Bergh K, Brakhage A A. Analysis of the regulation of Aspergillus nidulans penicillin biosynthesis gene aat (penDE) encoding acyl coenzyme A:6-aminopenicillanic acid acyltransferase. Mol Gen Genet. 1995;249:557–569. doi: 10.1007/BF00290581. [DOI] [PubMed] [Google Scholar]
- 195.Litzka O, Then Bergh K, Brakhage A A. The Aspergillus nidulans penicillin biosynthesis gene aat (penDE) is controlled by a CCAAT containing DNA element. Eur J Biochem. 1996;238:675–682. doi: 10.1111/j.1432-1033.1996.0675w.x. [DOI] [PubMed] [Google Scholar]
- 196.Lu Y, Mach R L, Affenzeller K, Kubicek C P. Regulation of α-aminoadipate reductase from Penicillium chrysogenum in relation to the flux from α-aminoadipate into penicillin biosynthesis. Can J Microbiol. 1992;38:758–763. doi: 10.1139/m92-123. [DOI] [PubMed] [Google Scholar]
- 197.Lübbe C, Jensen S E, Demain A L. Prevention of phosphate inhibition of cephalosporin synthetases by ferrous ion. FEMS Microbiol Lett. 1984;25:75–79. [Google Scholar]
- 198.Luengo J M. Enzymatic synthesis of hydrophobic penicillins. J Antibiot. 1995;48:1195–1212. doi: 10.7164/antibiotics.48.1195. [DOI] [PubMed] [Google Scholar]
- 199.Luengo J M, Iriso J L, López-Nieto M J. Direct evaluation of phenylacetyl-CoA:6-aminopenicillanic acid acyltransferase of Penicillium chrysogenum by bioassay. J Antibiot. 1986;XXXIX:1565–1573. doi: 10.7164/antibiotics.39.1565. [DOI] [PubMed] [Google Scholar]
- 200.Luengo J M, Revilla G, López-Nieto M J, Villanueva J R, Martin J F. Inhibition and repression of homocitrate synthetase by lysine in Penicillium chrysogenum. J Bacteriol. 1980;144:869–876. doi: 10.1128/jb.144.3.869-876.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.MacCabe A P, Riach M B R, Unkles S E, Kinghorn J R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.MacCabe A P, van den Hombergh J P T W, Tilburn J, Arst H N, Jr, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 203.MacCabe A P, van Liempt H, Palissa H, Unkles S E, Riach M B R, Pfeifer E, von Döhren H, Kinghorn J R. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase from Aspergillus nidulans—molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266:12646–12654. [PubMed] [Google Scholar]
- 204.Macdonald K D, Holt G. Genetics of biosynthesis and overproduction of penicillin. Sci Prog Oxford. 1976;63:547–573. [PubMed] [Google Scholar]
- 205.Madurri K, Stuttard C, Vining L C. Lysine catabolism in Streptomyces spp. is primarily through cadaverine: β-lactam producers also make α-aminoadipate. J Bacteriol. 1989;171:299–302. doi: 10.1128/jb.171.1.299-302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Madurri K, Stuttard C, Vining L C. Cloning and location of a gene governing lysine epsilon-aminotransferase, an enzyme initiating beta-lactam biosynthesis in Streptomyces spp. J Bacteriol. 1991;173:985–988. doi: 10.1128/jb.173.3.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Malpartida F, Hopwood D A. Physical and genetic characterization of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2) Mol Gen Genet. 1986;205:66–73. doi: 10.1007/BF02428033. [DOI] [PubMed] [Google Scholar]
- 208.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 209.Martin J F, Aharonowitz Y. Regulation of biosynthesis of β-lactam antibiotics. In: Demain A L, Solomon N A, editors. Antibiotics containing the β-lactam structure. I. Berlin, Germany: Springer-Verlag, KG; 1983. pp. 229–254. [Google Scholar]
- 210.Martin J F, Demain A L. Control of antibiotic biosynthesis. Microbiol Rev. 1980;44:230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Martin J F, Gutiérrez S, Demain A L. β-Lactams. In: Anke T, editor. Fungal biotechnology. Antibiotics. Weinheim, Germany: Chapman & Hall; 1997. pp. 91–127. [Google Scholar]
- 212.Martin J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 213.Martin J F, Revilla G, Zanca D M, López-Nieto M J. Carbon catabolite regulation of penicillin and cephalosporin biosynthesis. In: Umezawa H, Demain A L, Hata T, Hutchinson C R, editors. Trends in antibiotic research. Tokyo, Japan: Japanese Antibiotics Research Association; 1982. pp. 258–268. [Google Scholar]
- 214.Martin R L, Daley L A, Lovric Z, Wailes L M, Renosto F, Segel I H. The “regulatory” sulfhydryl group of Penicillium chrysogenum ATP sulfurylase. J Biol Chem. 1989;264:11768–11775. [PubMed] [Google Scholar]
- 215.Martinez-Blanco H, Orejas M, Reglero A, Luengo J M, Peñalva M A. Characterisation of the gene encoding acetyl-CoA synthetase in Penicillium chrysogenum: conservation of intron position in plectomycetes. Gene. 1993;130:265–270. doi: 10.1016/0378-1119(93)90429-7. [DOI] [PubMed] [Google Scholar]
- 216.Martinez-Blanco H, Reglero A, Fernández-Valverde M, Ferrero M A, Moreno M A, Peñalva M A, Luengo J M. Isolation and characterisation of the acetyl-CoA synthetase from Penicillium chrysogenum. Involvement of this enzyme in the biosynthesis of penicillins. J Biol Chem. 1992;267:5474–5481. [PubMed] [Google Scholar]
- 217.Marzluf G A. Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu Rev Microbiol. 1993;47:31–55. doi: 10.1146/annurev.mi.47.100193.000335. [DOI] [PubMed] [Google Scholar]
- 218.Marzluf G A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mathison L, Soliday C, Stepan T, Aldrich T, Rambosek J. Cloning, characterization, and use in strain improvement of the Cephalosporium acremonium gene cefG encoding acetyl transferase. Curr Genet. 1993;23:33–41. doi: 10.1007/BF00336747. [DOI] [PubMed] [Google Scholar]
- 220.Matsuda A, Sugiura H, Matsuyama K, Matsumoto H, Ichikawa S, Komatsu K-I. Molecular cloning of acetyl coenzyme A:deacetylcephalosporin C O-acetyltransferase cDNA from Acremonium chrysogenum: sequence and expression of catalytic activity in yeast. Biochem Biophys Res Commun. 1992;182:995–1001. doi: 10.1016/0006-291x(92)91830-j. [DOI] [PubMed] [Google Scholar]
- 221.Matsuda A, Sugiura H, Matsuyama K, Matsumoto H, Ichikawa S, Komatsu K-I. Cloning and disruption of the cefG gene encoding acetyl coenzyme A:deacetylcephalosporin C O-acetyltransferase from Acremonium chrysogenum. Biochem Biophys Res Commun. 1992;186:40–46. doi: 10.1016/s0006-291x(05)80772-7. [DOI] [PubMed] [Google Scholar]
- 222.Matsumoto K. Production of 6-APA, 7-ACA and 7-ADCA by immobilised penicillin and cephalosporin amidases. In: Tanaka A, Tosa T, Kobayashi T, editors. Industrial application of immobilized biocatalysts. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 67–88. [PubMed] [Google Scholar]
- 223.Matsumura M, Imanaka T, Yoshida T, Taguchi H. Effect of glucose and methionine consumption rates on cephalosporin C production by Cephalosporium acremonium. J Ferment Technol. 1978;56:345–353. [Google Scholar]
- 224.Matsumura S, Suzuki M. Feedback regulation of acetohydroxy acid synthase in Cephalosporium acremonium M8650 and a high cephalosporin-producing mutant. Agric Biol Chem. 1986;50:505–507. [Google Scholar]
- 225.McKnight S, Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986;46:795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- 226.McNabb D S, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 227.Mehta R J, Speth J L, Nash C H. Lysine stimulation of cephalosporin C synthesis in Cephalosporium acremonium. Eur J Appl Microbiol Biotechnol. 1979;8:177–182. [Google Scholar]
- 228.Meister A, Anderson M E. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 229.Mellor J, Jiang W, Funk M, Rathjen J, Barnes J C, Hinz T, Hegeman J H, Philippsen P. CBF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Menne S, Walz M, Kück U. Expression studies with the bidirectional pcbAB-pcbC promoter region from Acremonium chrysogenum using reporter gene fusions. Appl Microbiol Biotechnol. 1994;42:57–66. doi: 10.1007/BF00170225. [DOI] [PubMed] [Google Scholar]
- 231.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins SP1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ming L-J, Que L, Jr, Kriauciunas A, Frolik C A, Chen V A. Coordination chemistry of the metal binding site of isopenicillin N synthase. Inorg Chem. 1990;29:1111–1112. [Google Scholar]
- 233.Mitchel P J, Tjian R. Transcriptional regulation in mammalian cells by sequence specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 234.Montenegro E, Barredo J L, Gutiérrez S, Diez B, Alvarez E, Martin J F. Cloning, characterization of the acyl-CoA:6-aminopenicillanic acid acyltransferase gene of Aspergillus nidulans and linkage to the isopenicillin N synthase gene. Mol Gen Genet. 1990;221:322–330. doi: 10.1007/BF00259395. [DOI] [PubMed] [Google Scholar]
- 235.Montenegro E, Fierro F, Fernandez F J, Gutiérrez S, Martin J F. Resolution of chromosomes III and VI of Aspergillus nidulans by pulsed-field gel electrophoresis shows that the penicillin biosynthetic pathway genes pcbAB, pcbC, and penDE are clustered on chromosome VI (3.0 megabases) J Bacteriol. 1992;174:7063–7067. doi: 10.1128/jb.174.21.7063-7067.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Morgan W D, Williams G T, Morimoto R I, Greene J, Kingston R E, Tjian R. Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol Cell Biol. 1987;7:1129–1138. doi: 10.1128/mcb.7.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Müller W H, Bovenberg R A L, Groothuis M H, Kattevilder F, Smaal E B, van der Voort L H M, Verkleij A. Involvement of microbodies in penicillin biosynthesis. Biochim Biophys Acta. 1992;1116:210–213. doi: 10.1016/0304-4165(92)90118-e. [DOI] [PubMed] [Google Scholar]
- 238.Müller W H, van der Krift T P, Krouwer A J J, Wösten H A B, van der Voort L H M, Smaal E B, Verkleij A J. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 1991;10:489–495. doi: 10.1002/j.1460-2075.1991.tb07971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nagarajan R, Boeck L D, Gorman M, Hamill R L, Higgens C E, Hoehn M M, Stark W M, Whitney J G. Beta-lactam antibiotics from Streptomyces. J Am Chem Soc. 1971;93:2308–2310. doi: 10.1021/ja00738a035. [DOI] [PubMed] [Google Scholar]
- 240.Nagata O, Takashima T, Tanaka M, Tsukagoshi N. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible Taka-amylase A gene. Mol Gen Genet. 1993;237:251–260. doi: 10.1007/BF00282807. [DOI] [PubMed] [Google Scholar]
- 241.Natorff R, Balinska M, Paszewski A. At least four regulatory genes control sulfur metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1993;238:185–192. doi: 10.1007/BF00279546. [DOI] [PubMed] [Google Scholar]
- 242.Negrete-Urtasun S, Denison S H, Arst H N., Jr Characterization of the pH signal transduction pathway palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Newbert R W, Barton B, Greaves P, Harper J, Turner G. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J Ind Microbiol Biotechnol. 1997;19:18–27. doi: 10.1038/sj.jim.2900411. [DOI] [PubMed] [Google Scholar]
- 244.Nielsen J. Physiological engineering aspects of Penicillium chrysogenum. Lyngby, Denmark: Polyteknisk Forlag; 1995. [Google Scholar]
- 245.Nüesch J, Heim J, Treichler H-J. The biosynthesis of sulfur-containing β-lactam antibiotics. Annu Rev Microbiol. 1987;41:51–75. doi: 10.1146/annurev.mi.41.100187.000411. [DOI] [PubMed] [Google Scholar]
- 246.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N, Jr, Peñalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 247.Orville A M, Chen V J, Kriauciunas A, Harpel M R, Fox B G, Munck E, Lipscomb J D. Thiolate ligation of the active site Fe2+ of isopenicillin N synthase derives from substrate rather than endogenous cysteine: spectroscopic studies of site-specific Cys→Ser mutated enzymes. Biochemistry. 1992;31:4602–4612. doi: 10.1021/bi00134a010. [DOI] [PubMed] [Google Scholar]
- 248.O’Sullivan J, Bleaney R C, Huddleston J A, Abraham E P. Incorporation of 3H from δ-(l-α-amino[4,5-3H]adipyl)-l-cysteinyl-d-[4,4-3H]valine into isopenicillin N. Biochem J. 1979;184:421–426. doi: 10.1042/bj1840421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.O’Sullivan J, Sykes R B. β-Lactam antibiotics. In: Pape H, Rehm H-J, editors. Biotechnology, a comprehensive treatise in 8 volumes. Vol. 4. Weinheim, Germany: VCH Verlagsgesellschaft; 1986. pp. 247–281. [Google Scholar]
- 250.Palissa H, von Döhren H, Kleinkauf H, Ting H H, Baldwin J E. β-Lactam biosynthesis in a gram-negative eubacterium: purification and characterization of isopenicillin N synthase from Flavobacterium sp. strain SC 12.154. J Bacteriol. 1989;171:5720–5728. doi: 10.1128/jb.171.10.5720-5728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pang C P, Chakravarti B, Adlington R M, Ting H H, White R L, Jayatilake G S, Baldwin J E, Abraham E P. Purification of isopenicillin N synthetase. Biochem J. 1984;222:789–795. doi: 10.1042/bj2220789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Papagiannopoulos P, Andrianopoulos A, Sharp J A, Davis M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 253.Papagiannopoulos, P., M. A. Davis, and M. J. Hynes. Unpublished data. GenBank accession no. U96847.
- 254.Peñalva M A, Moya A, Dopazo J, Ramon D. Sequences of isopenicillin N synthetase genes suggest horizontal gene transfer from prokaryotes to eukaryotes. Proc R Soc London Ser B. 1990;241:164–169. doi: 10.1098/rspb.1990.0081. [DOI] [PubMed] [Google Scholar]
- 255.Peñalva M A, Vian A, Patino C, Perez-Aranda A, Ramon D. Molecular biology of penicillin production in Aspergillus nidulans. In: Hershberger C L, Queener S W, Hegeman G, editors. Genetics and molecular biology of industrial microorganisms. Washington, D.C: American Society for Microbiology; 1989. pp. 256–261. [Google Scholar]
- 256.Pérez-Esteban B, Gómez-Pardo E, Peñalva M A. A lacZ reporter fusion method for the genetic analysis of regulatory mutations in pathways of fungal secondary metabolism and its application to the Aspergillus nidulans penicillin pathway. J Bacteriol. 1995;177:6069–6076. doi: 10.1128/jb.177.21.6069-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pérez-Esteban B, Orejas M, Gómez-Pardo E, Peñalva M A. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthetase gene is modulated by upstream negative elements. Mol Microbiol. 1993;9:881–895. doi: 10.1111/j.1365-2958.1993.tb01746.x. [DOI] [PubMed] [Google Scholar]
- 258.Pérez-Llarena F J, Liras P, Rodriguez-Garcia A, Martin J F. A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both β-lactam compounds. J Bacteriol. 1997;179:2053–2059. doi: 10.1128/jb.179.6.2053-2059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Perry D, Abraham E P, Baldwin J E. Factors affecting the isopenicillin N synthetase reaction. Biochem J. 1988;255:345–351. [PMC free article] [PubMed] [Google Scholar]
- 260.Pieniazek N P, Stepien P, Paszewski A. An Aspergillus nidulans mutant lacking cystathionine-synthase: identity of l-serine sulfhydrylase with cystathionine-synthase and its distinctness from O-acetyl-l-serine sulfhydrylase. Biochim Biophys Acta. 1973;297:37–47. doi: 10.1016/0304-4165(73)90047-0. [DOI] [PubMed] [Google Scholar]
- 261.Pinkham J L, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pontecorvo G, Roper J A, Hemmons L M, MacDonald K D, Bufton A W J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 263.Pruess D L, Johnson M J. Penicillin acyltransferase in Penicillium chrysogenum. J Bacteriol. 1967;94:1502–1508. doi: 10.1128/jb.94.5.1502-1508.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Queener S W. Molecular biology of penicillin and cephalosporin biosynthesis. Antimicrob Agents Chemother. 1990;34:943–948. doi: 10.1128/aac.34.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Queener S W, Neuss N. Biosynthesis of β-lactam antibiotics. In: Morin R B, Gorman M, editors. Chemistry and biology of β-lactam antibiotics. Vol. 3. London, United Kingdom: Academic Press, Ltd.; 1982. pp. 1–81. [Google Scholar]
- 266.Ramon D, Carramolino L, Patino C, Sanchez F, Peñalva M A. Cloning and characterization of the isopenicillin N synthetase gene mediating the formation of the β-lactam ring in Aspergillus nidulans. Gene. 1987;57:171–181. doi: 10.1016/0378-1119(87)90120-x. [DOI] [PubMed] [Google Scholar]
- 267.Ramos F R, López-Nieto M J, Martin J F. Isopenicillin N synthetase of Penicillium chrysogenum, an enzyme that converts δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine to isopenicillin N. Antimicrob Agents Chemother. 1985;27:380–387. doi: 10.1128/aac.27.3.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ramos F R, López-Nieto M J, Martin J F. Coordinate increase of isopenicillin N synthetase, isopenicillin N epimerase and deacetoxycephalosporin C synthetase in a high cephalosporin-producing mutant of Acremonium chrysogenum and simultaneous loss of the three enzymes in a non-producing mutant. FEMS Microbiol Lett. 1986;35:123–127. [Google Scholar]
- 269.Ramsden M, McQuade B A, Saunders K, Turner M K, Harford S. Characterization of a loss-of-function mutation in the isopenicillin N synthetase gene of Acremonium chrysogenum. Gene. 1989;85:267–273. doi: 10.1016/0378-1119(89)90493-9. [DOI] [PubMed] [Google Scholar]
- 270.Randall R C, Zang Y, True A E, Que L, Jr, Charnock J M, Garner C D, Fujishima Y, Schofield C J, Baldwin J E. X-ray absorption studies of the ferrous active site of isopenicillin N synthase and related model complexes. Biochemistry. 1993;32:6664–6673. doi: 10.1021/bi00077a020. [DOI] [PubMed] [Google Scholar]
- 271.Raper K B, Alexander D F, Coghill R D. Penicillin: natural variation and penicillin production in Penicillium notatum and allied species. J Bacteriol. 1944;48:639–659. doi: 10.1128/jb.48.6.639-659.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Raymondjean M, Cereghini S, Yaniv M. Several distinct “CCAAT” box binding proteins coexist in eukaryotic cells. Proc Natl Acad Sci USA. 1988;85:757–761. doi: 10.1073/pnas.85.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Renno D V, Saunders G, Bull A T, Holt G. Transcript analysis of penicillin genes from Penicillium chrysogenum. Curr Genet. 1992;21:49–54. doi: 10.1007/BF00318654. [DOI] [PubMed] [Google Scholar]
- 274.Renosto F, Martin R L, Wailes L M, Dailey L A, Segel I H. Regulation of inorganic sulfate activation in filamentous fungi. J Biol Chem. 1990;265:10300–10308. [PubMed] [Google Scholar]
- 275.Retey J. Reaktionsselektivität von Enzymen durch negative Katalyse oder wie gehen Enzyme mit hochreaktiven Intermediaten um? Angew Chem. 1990;102:373–379. . (Angew. Chem. Int. Ed. 29:355.) [Google Scholar]
- 276.Revilla G, López-Nieto M J, Luengo J M, Martin J F. Carbon catabolite repression of penicillin biosynthesis by Penicillium chrysogenum. J Antibiot. 1984;XXXVII:781–789. doi: 10.7164/antibiotics.37.781. [DOI] [PubMed] [Google Scholar]
- 277.Revilla G, Ramos F R, López-Nieto M J, Alvarez E, Martin J F. Glucose represses formation of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine and isopenicillin N synthase but not penicillin acyltransferase in Penicillium chrysogenum. J Bacteriol. 1986;168:947–952. doi: 10.1128/jb.168.2.947-952.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Richardson I B, Katz M E, Hynes M J. Molecular characterization of the lam locus and sequences involved in the regulation of the AmdR protein of Aspergillus nidulans. Mol Cell Biol. 1992;12:337–346. doi: 10.1128/mcb.12.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Roach P L, Clifton I J, Fülöp V, Harlos K, Barton G J, Hajdu J, Andersson I, Schofield C J, Baldwin J E. Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature. 1995;375:700–704. doi: 10.1038/375700a0. [DOI] [PubMed] [Google Scholar]
- 280.Roach P L, Clifton I J, Hensgens C M H, Shibata N, Schofield C J, Hajdu J, Baldwin J E. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 281.Ronchi A, Bellorini M, Mongelli N, Mantovani R. CCAAT-box binding protein NF-Y (CBF, CP1) recognizes the minor groove and distorts DNA. Nucleic Acids Res. 1995;23:4565–4572. doi: 10.1093/nar/23.22.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Rossi A, Arst H N., Jr Mutants of Aspergillus nidulans able to grow at extremely acidic pH acidify the medium less than wild type when grown at more moderate pH. FEMS Microbiol Lett. 1990;66:51–53. doi: 10.1016/0378-1097(90)90257-q. [DOI] [PubMed] [Google Scholar]
- 283.Samson S M, Belagaje R, Blankenship D T, Chapman J L, Perry D, Skatrud P L, van Frank R M, Abraham E P, Baldwin J E, Queener S W, Ingolia T D. Isolation, sequence determination and expression in Escherichia coli of the isopenicillin N synthetase gene from Cephalosporium acremonium. Nature. 1985;318:191–194. doi: 10.1038/318191a0. [DOI] [PubMed] [Google Scholar]
- 284.Samson S M, Dotzlaf J E, Slisz M L, Becker G W, van Frank R M, Veal L E, Yeh W-K, Miller J R, Queener S W, Ingolia T D. Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Bio/Technology. 1987;5:1207–1214. [Google Scholar]
- 285.Sánchez L, Braña A F. Cell density influences antibiotic biosynthesis in Streptomyces clavuligerus. Microbiology. 1996;142:1209–1220. doi: 10.1099/13500872-142-5-1209. [DOI] [PubMed] [Google Scholar]
- 286.Sanchez S, Flores M E, Demain A L. Nitrogen regulation of penicillin and cephalosporin fermentations. In: Sanchez-Esquival S, editor. Nitrogen source control of microbial processes. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 121–136. [Google Scholar]
- 287.Santoro C, Mermod N, Andrews P C, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 288.Sawada Y, Baldwin J E, Singh P D, Solomon N A, Demain A L. Cell-free cyclization of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine to isopenicillin N. Antimicrob Agents Chemother. 1980;18:465–470. doi: 10.1128/aac.18.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sawada Y, Konomi T, Solomon N A, Demain A L. Increase in activity of β-lactam synthetases after growth of Cephalosporium acremonium with methionine or norleucine. FEMS Microbiol Lett. 1980;9:281–284. [Google Scholar]
- 290.Scheidegger A, Küenzi M T, Nüesch J. Partial purification and catalytic properties of a bifunctional enzyme in the biosynthetic pathway of β-lactams in Cephalosporium acremonium. J Antibiot. 1984;XXXVII:522–531. doi: 10.7164/antibiotics.37.522. [DOI] [PubMed] [Google Scholar]
- 291.Schlumbohm W, Stein T, Ullrich C, Vater J, Krause M, Marahiel M A, Kruft V, Wittmann-Liebold B. An active serine is involved in covalent substrate amino acid binding at each reaction center of gramicidin S synthetase. J Biol Chem. 1991;266:23135–23141. [PubMed] [Google Scholar]
- 292.Schwecke T, Aharonowitz Y, Palissa H, von Döhren H, Kleinkauf H, van Liempt H. Enzymatic characterisation of the multifunctional enzyme δ-[l-α-aminoadipyl]-l-cysteinyl-d-valine synthetase from Streptomyces clavuligerus. Eur J Biochem. 1992;205:687–694. doi: 10.1111/j.1432-1033.1992.tb16830.x. [DOI] [PubMed] [Google Scholar]
- 293.Segel I H, Renosto F, Seubert P A. Sulfate-activating enzymes. Methods Enzymol. 1987;143:334–349. doi: 10.1016/0076-6879(87)43061-9. [DOI] [PubMed] [Google Scholar]
- 294.Seno E T, Baltz R H. Structural organization and regulation of antibiotic biosynthesis and resistance genes in actinomycetes. In: Shapiro S, editor. Regulation of secondary metabolism in actinomycetes. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 1–48. [Google Scholar]
- 295.Shah A J, Tilburn J, Adlard M W, Arst H N., Jr pH regulation of penicillin production in Aspergillus nidulans. FEMS Microbiol Lett. 1991;77:209–212. doi: 10.1016/0378-1097(91)90553-m. [DOI] [PubMed] [Google Scholar]
- 296.Shen Y-Q, Heim J, Solomon N A, Wolfe S, Demain A L. Repression of β-lactam production in Cephalosporium acremonium by nitrogen sources. J Antibiot. 1984;37:503–511. doi: 10.7164/antibiotics.37.503. [DOI] [PubMed] [Google Scholar]
- 297.Shiau C-Y, Baldwin J E, Byford M F, Sobey W J, Schofield C J. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase: the order of peptide bond formation and timing of the epimerisation reaction. FEBS Lett. 1995;358:97–100. doi: 10.1016/0014-5793(94)01320-z. [DOI] [PubMed] [Google Scholar]
- 298.Shirafuji H, Fujisawa Y, Kida M, Kanzaki T, Yoneda M. Accumulation of tripeptide derivatives by mutants of Cephalosporium acremonium. Agric Biol Chem. 1979;43:155–160. [Google Scholar]
- 299.Skatrud P L. Molecular biology of the β-lactam-producing fungi. In: Bennett J W, Lasure L L, editors. More gene manipulations in fungi. New York, N.Y: Academic Press, Inc.; 1991. pp. 364–395. [Google Scholar]
- 300.Skatrud P L, Queener S W. An electrophoretic molecular karyotype for an industrial strain of Cephalosporium acremonium. Gene. 1989;79:331–338. doi: 10.1016/0378-1119(89)90235-7. [DOI] [PubMed] [Google Scholar]
- 301.Skatrud P L, Tietz A J, Ingolia T D, Cantwell C A, Fisher D L, Chapman J L, Queener S W. Use of recombinant DNA to improve production of cephalosporin C by Cephalosporium acremonium. Bio/Technology. 1989;7:477–485. [Google Scholar]
- 302.Smith A W, Collis K, Ramsden M, Fox H M, Peberdy J F. Chromosome rearrangements in improved cephalosporin C-producing strains of Acremonium chrysogenum. Curr Genet. 1991;18:235–237. doi: 10.1007/BF00336492. [DOI] [PubMed] [Google Scholar]
- 303.Smith A W, Ramsden M, Dobson M J, Harford S, Peberdy J F. Regulation of isopenicillin N synthetase (IPNS) gene expression in Acremonium chrysogenum. Bio/Technology. 1990;8:237–240. doi: 10.1038/nbt0390-237. [DOI] [PubMed] [Google Scholar]
- 304.Smith D J, Bull J H, Edwards J, Turner G. Amplification of the isopenicillin N synthetase gene in a strain of Penicillium chrysogenum producing high levels of penicillin. Mol Gen Genet. 1989;216:492–497. doi: 10.1007/BF00334395. [DOI] [PubMed] [Google Scholar]
- 305.Smith D J, Burnham M R K, Bull J H, Hodgson J E, Ward J M, Browne P, Brown J, Barton B, Earl A J, Turner G. β-Lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990;9:741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Smith D J, Burnham M R K, Edwards J, Earl A J, Turner G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillium chrysogenum. Bio/Technology. 1990;8:39–41. doi: 10.1038/nbt0190-39. [DOI] [PubMed] [Google Scholar]
- 307.Smith D J, Earl A J, Turner G. The multifunctional peptide synthetase performing the first step of penicillin biosynthesis is a 421 073 dalton protein similar to Bacillus brevis peptide antibiotic synthetases. EMBO J. 1990;9:2743–2750. doi: 10.1002/j.1460-2075.1990.tb07461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Smith M W, Feng D-F, Doolittle R F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 309.Soltero F V, Johnson M J. The effect of the carbohydrate nutrition on penicillin production by Penicillium chrysogenum Q-176. Appl Microbiol. 1952;1:52–57. doi: 10.1128/am.1.1.52-57.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Somerson N L, Demain A L, Nunheimer T D. Reversal of lysine inhibition of penicillin production by α-aminoadipic or adipic acid. Arch Biochem Biophys. 1961;93:238–241. [Google Scholar]
- 311.Spencer B, Maung C. Multiple activities of penicillin acyltransferase of Penicillium chrysogenum. Biochem J. 1970;118:29–30. doi: 10.1042/bj1180029p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Stachelhaus T, Schneider A, Marahiel M A. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 313.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 314.Stein T, Vater J, Kruft V, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. Detection of 4′-phosphopantetheine at the thioester binding site for l-valine of gramicidin S synthetase 2. FEBS Lett. 1994;340:39–44. doi: 10.1016/0014-5793(94)80169-x. [DOI] [PubMed] [Google Scholar]
- 315.Suárez T, de Queiroz M V, Oestreicher N, Scazzocchio C. The sequence and binding specificity of UaY, the specific regulator of the purine utilisation pathway in Aspergillus nidulans, suggest an evolutionary relationship with the PPR1 protein of Saccharomyces cerevisiae. EMBO J. 1995;14:1453–1467. doi: 10.1002/j.1460-2075.1995.tb07132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Suárez T, Peñalva M A. Characterisation of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol Microbiol. 1996;20:529–540. doi: 10.1046/j.1365-2958.1996.5421065.x. [DOI] [PubMed] [Google Scholar]
- 317.Swartz R W. Penicillins. In: Blanch H W, Drew S, Wang D I C, editors. Comprehensive biotechnology. The principles, applications and regulations of biotechnology in industry, agriculture and medicine. 3. The practice of biotechnology: current commodity products. Oxford, United Kingdom: Pergamon Press; 1985. pp. 7–47. [Google Scholar]
- 318.Then Bergh K, Brakhage A A. Regulation of the Aspergillus nidulans penicillin biosynthesis gene acvA (pcbAB) by amino acids: implication for involvement of transcription factor PACC. Appl Environ Microbiol. 1998;64:843–849. doi: 10.1128/aem.64.3.843-849.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Then Bergh K, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Then Bergh, K., O. Litzka, and A. A. Brakhage. Unpublished data.
- 321.Thomas D, Jacquemin I, Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere binding factor I are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidic- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Tobin M B, Baldwin J E, Cole S C J, Miller J R, Skatrud P L, Sutherland J D. The requirement for subunit interaction in the production of Penicillium chrysogenum acyl-coenzyme A: isopenicillin N acyltransferase in Escherichia coli. Gene. 1993;132:199–206. doi: 10.1016/0378-1119(93)90196-a. [DOI] [PubMed] [Google Scholar]
- 324.Tobin M B, Cole S C J, Miller J R, Baldwin J E, Sutherland J D. Amino-acid substitutions in the cleavage site of acyl-coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum: effect on proenzyme cleavage and activity. Gene. 1995;162:29–35. doi: 10.1016/0378-1119(95)00369-h. [DOI] [PubMed] [Google Scholar]
- 325.Tobin M B, Fleming M D, Skatrud P L, Miller J R. Molecular characterization of the acyl-coenzyme A:isopenicillin N acyltransferase gene (penDE) from Penicillium chrysogenum and Aspergillus nidulans and activity of recombinant enzyme in Escherichia coli. J Bacteriol. 1990;172:5908–5914. doi: 10.1128/jb.172.10.5908-5914.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Tobin M B, Kovacevic S, Madduri K, Hoskins J A, Skatrud P L, Vining L C, Stuttard C, Miller J R. Localization of the lysine ɛ-aminotransferase (lat) and δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine synthetase (pcbAB) genes from Streptomyces clavuligerus and production of lysine ɛ-aminotransferase activity in Escherichia coli. J Bacteriol. 1991;173:6223–6229. doi: 10.1128/jb.173.19.6223-6229.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Treichler H J, Liersch M, Nüesch J, Döbeli H. Role of sulfur metabolism in cephalosporin C and penicillin biosynthesis. In: Sebeck O K, Laskin A I, editors. Genetics of industrial microorganisms. Washington, D.C: American Society for Microbiology; 1979. pp. 97–104. [Google Scholar]
- 328.Trown P W, Smith B, Abraham E P. Biosynthesis of cephalosporin C from amino acids. Biochem J. 1963;86:284–291. doi: 10.1042/bj0860284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Turgay K, Krause M, Marahiel M A. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 330.Turner G, Browne P E, Brakhage A A. Expression of genes for the biosynthesis of penicillin. NATO ASI Ser Ser H. 1993;69:125–138. [Google Scholar]
- 331.Umbarger H E. Amino acid biosynthesis and its regulation. Annu Rev Microbiol. 1978;47:533–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- 332.Van Heeswijck R, Hynes M J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.van Liempt H, von Döhren H, Kleinkauf H. δ-[l-α-Aminoadipyl]-l-cysteinyl-d-valine synthetase from Aspergillus nidulans. J Biol Chem. 1989;264:3680–3684. [PubMed] [Google Scholar]
- 334.Veenstra A E, van Solingen P, Bovenberg R A L, van der Voort L H M. Strain improvement of Penicillium chrysogenum by recombinant DNA techniques. J Biotechnol. 1991;17:81–90. doi: 10.1016/0168-1656(91)90028-t. [DOI] [PubMed] [Google Scholar]
- 335.Velasco J, Gutiérrez S, Fernandez F J, Marcos A T, Arenos C, Martin J F. Exogenous methionine increases levels of mRNAs transcribed from pcbAB, pcbC, and cefEF genes, encoding enzymes of the cephalosporin biosynthetic pathway, in Acremonium chrysogenum. J Bacteriol. 1994;176:985–991. doi: 10.1128/jb.176.4.985-991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Vogel H J. Two modes of lysine synthesis among lower fungi: evolutionary significance. Biochim Biophys Acta. 1960;41:172–173. [Google Scholar]
- 337.Walz M, Kück U. Polymorphic karyotypes in related Acremonium strains. Curr Genet. 1991;19:73–76. doi: 10.1007/BF00326285. [DOI] [PubMed] [Google Scholar]
- 338.Weidner G, Steffan B, Brakhage A A. The Aspergillus nidulans lysF gene encodes homoaconitase, an enzyme involved in the fungus specific lysine biosynthesis. Mol Gen Genet. 1997;255:237–247. doi: 10.1007/s004380050494. [DOI] [PubMed] [Google Scholar]
- 339.Weigel B J, Burgett S G, Chen V J, Skatrud P L, Frolik C A, Queener S W, Ingolia T D. Cloning and expression in Escherichia coli of isopenicillin N synthetase genes from Streptomyces lipmanii and Aspergillus nidulans. J Bacteriol. 1988;170:3817–3826. doi: 10.1128/jb.170.9.3817-3826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.White R L, John E M, Baldwin J E, Abraham E P. Stoichiometry of oxygen consumption in the biosynthesis of isopenicillin from a tripeptide. Biochem J. 1982;203:791–793. doi: 10.1042/bj2030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Whiteman P A, Abraham E P, Baldwin J E, Fleming M D, Schofield C J, Sutherland J D, Willis A C. Acyl coenzyme A:6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum and Aspergillus nidulans. FEBS Lett. 1990;262:342–344. doi: 10.1016/0014-5793(90)80224-7. [DOI] [PubMed] [Google Scholar]
- 342.Wolfe S, Demain A L, Jensen S E, Westlake D W S. Enzymatic approach to synthesis of unnatural β-lactams. Science. 1984;226:1386–1392. doi: 10.1126/science.6390683. [DOI] [PubMed] [Google Scholar]
- 343.Xiao X, Hintermann G, Hausler A, Barker P J, Foor F, Demain A L, Piret J. Cloning of a Streptomyces clavuligerus DNA fragment encoding the cephalosporin 7 alpha-hydroxylase and its expression in Streptomyces lividans. Antimicrob Agents Chemother. 1993;37:84–88. doi: 10.1128/aac.37.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zanca D M, Martin J F. Carbon catabolite regulation of the conversion of penicillin N into cephalosporin C. J Antibiot. 1983;XXXVI:700–708. doi: 10.7164/antibiotics.36.700. [DOI] [PubMed] [Google Scholar]
- 345.Zhang J, Demain A L. Purification from Cephalosporium acremonium of the initial enzyme unique to the biosynthesis of penicillins and cephalosporins. Biochem Biophys Res Commun. 1990;169:1145–1152. doi: 10.1016/0006-291x(90)92015-r. [DOI] [PubMed] [Google Scholar]
- 346.Zhang J, Demain A L. Regulation of ACV synthetase activity by carbon sources and their metabolites. Arch Microbiol. 1992;158:364–369. [Google Scholar]
- 347.Zhang J, Demain A L. ACV synthetase. Crit Rev Biotechnol. 1992;12:245–260. doi: 10.3109/07388559209069194. [DOI] [PubMed] [Google Scholar]
- 348.Zhang J, Wolfe S, Demain A L. Phosphate repressible and inhibitible β-lactam synthetases in Cephalosporium acremonium strain C-10. Appl Microbiol Biotechnol. 1988;29:242–247. [Google Scholar]
- 349.Zhang J, Wolfe S, Demain A L. Carbon source regulation of the ACV synthetase in Cephalosporium acremonium C-10. Curr Microbiol. 1989;18:361–367. [Google Scholar]