Abstract
Protein structures and mutagenesis studies have been instrumental in elucidating molecular mechanisms of ion channel function, but making informed choices about which residues to target for mutagenesis can be challenging. Therefore, we investigated the potential for using human population genomic data to further refine our selection of mutagenesis sites in TRPV1. Single nucleotide polymorphism data of TRPV1 from gnomAD 2.1.1 revealed a lower number of missense variants within buried residues of the ankyrin repeat domain and an increased number of variants between secondary structure elements of the transmembrane segments. We hypothesized that residues critical to interactions at interfaces between subunits or domains in the channel would exhibit a similar reduction in variants. We identified in the structure of ground squirrel TRPV1 (PDB: 7LQY) a possible electrostatic network between K155 and K160 in the N-terminal ankyrin repeat domain and E761 and D762 in the C-terminus (K-KED). Consistent with our hypothesis for residues at key interface sites, none of the four residues have any variants reported in gnomAD 2.1.1. Ca2+ imaging of TRPV1 K-KED mutants confirmed significant roles for these residues, but we found that the electrostatic interaction is not essential since channel function is still observed in total charge reversals on the C-terminal side of the interface (E761K/D762K). Interestingly, Ca2+ imaging responses for a charge swap experiment with K155D/D762K showed partially restored wild-type responses. Using electrophysiology, we found that charge reversals on either K155 or D762 increased the baseline currents of TRPV1, and the charge swapped double mutant, K155D/D762K, partially restored baseline currents to wild-type levels. We interpret these results to mean that contacts across residues in the K-KED interface shift the equilibria of conformations to closed pore states. Our study demonstrates the utility and applicability of a combined missense variant and structure targeted investigation of residues at TRPV1 subunit interfaces.
Significance
Ion channels have important roles across diverse aspects of physiology. A cornerstone of attaining mechanistic understanding of ion channels is derived through relating their physical structure to functional experiments. To the best of our knowledge, we apply a novel strategy to examine structure-function relationships of TRPV1 by augmenting structural models of the ion channel with human genome variant data. Our bioinformatic analysis highlighted a protein interaction surface between the N- and C-termini of the channel subunits. Results obtained in our Ca2+ imaging and electrophysiology experiments implicate the formation of a salt bridge across K155 and D762 as critically important for maintaining low basal TRPV1 activity.
Introduction
Transient receptor potential vanilloid 1 (TRPV1) is a nonselective cation channel expressed in nociceptors and known to mediate inflammatory pain (1,2). Numerous cell signaling pathways impinge on TRPV1 to modulate its sensitivity to various external stimuli, which range from noxious heat, acidity, oxidation, and exogenous ligands (3,4,5,6,7,8,9). The wide range of convergent inputs on TRPV1 and its role in pain have highlighted the channel as a promising target for health research, and recent advances in cryo-EM structural biology methods have made a more comprehensive understanding of the biophysical basis for TRPV1 activity possible (10,11,12). In fact, the number of high-resolution cryo-EM TRPV1 structures grows at an astonishing rate, and, at last count, it exceeds 50 entries in the rcsb.org database (RCSB database: http://www.rcsb.org).
Although the windfall of TRPV1 structures is broadly appreciated, one concern raised in a recent review is that resolution in the range of 3–4 Å for cryo-EM structures is sufficient to model the protein backbone, but confidence for exact atomic positions and side-chain orientations is not certain, and without cross-validation, there is no guarantee against overfitting the model to the cryo-EM map (13,14). Moreover, the resolution for most cryo-EM ion channel structures is “near atomic,” and local resolution for the density may also vary significantly, diminishing uniform confidence in a reconstructed model (15). Functional assays have been a reliable tool for testing hypotheses generated by structures and remain the litmus test for verifying interfacial contacts between nonsequential side chains. Nevertheless, identifying potential interaction partners via mutagenesis experiments may be time consuming in the absence of a well-resolved structure.
In lieu of high resolution at a structural interface, sequence alignments between species have proven practical for locating amino acids likely to be important for maintaining structural and functional integrity (16). The deep pool of sequences across various organisms has made alignments for conserved amino acid positions an invaluable tool, but conserved residues in proteins frequently persist across vast stretches of sequence even as the organisms become phylogenetically separated on an evolutionary tree (17). Adding more organisms to the genomic pool used for sequence alignments improves the odds of isolating candidate mutagenesis sites. However, by reaching across further removed phyla for sequence data, our confidence that all genes are functional orthologs is also reduced (18). Given the limitations of using multiple sequence alignments as a tool for locating residues for functional studies, we ask, “are there additional genomic resources to inform hypothesis generation?”
Here we leverage the Genome Aggregation Database (gnomAD database: https://gnomad.broadinstitute.org/) 2.1.1 collection of recorded single nucleotide polymorphisms (SNPs) in human TRPV1 and their respective genomic variant protein products to screen structurally interesting locations for site-directed mutagenesis experiments. Our approach innovates on methods that exploit information obtained from structure and maps genomic sequence data onto this information (19). The currently available structures of TRPV1 have allowed us to examine how the extension of the C-terminal domain of the channel (rTRPV1: 745–825) forms a potential interface with the N-terminal ankyrin repeat domain (ARD) of the neighboring subunit, and genomic SNP data highlight specific residue side chains that have reduced numbers of missense variants. Our Ca2+ imaging and electrophysiology studies of TRPV1 mutants generated from these structural/missense variant screens revealed two regions of critical importance for TRPV1 function located at this surface.
Materials and methods
For the cell culture, HEK293T/17 cells were incubated in Dulbecco’s Modified Eagle Medium with the addition of 10% fetal bovine serum, 50 μg streptomycin, and 50 units/mL penicillin at 37°C and 5% CO2. Cells were passaged onto 12 mm poly-L-lysine treated coverslips. Cells were left to settle onto the slips for at least 2 h before transfections with Lipofectamine 2000 (Life Technologies) as described in the manufacturer’s instructions. Optimal levels of channel expression were achieved by varying the ratio of channel DNA to GFP DNA and by varying the incubation times. Experiments were performed in the morning after transfections.
Regarding molecular biology, TRPV1-E761K/D762K and TRPV1-K155D/K160E/E761K/D762K were constructed by Genewiz (South Plainfield, NJ). All other TRPV1 constructs, were created using either the Quickchange (Agilent, Santa Clara, CA) or in vitro assembly (IVA) methods for site-directed mutagenesis (20). For all mutant constructs, our wild-type TRPV1.pcDNA3 vector was used as a template with either entirely (Quickchange) or partially (IVA) overlapping primers that contain the desired mutation. All multimutant constructs were created using sequential rounds of site-directed mutagenesis (Quickchange/IVA). The full sequence of each completed construct was confirmed using Sanger sequencing.
For Ca2+ imaging, cells were grown on 25-mm coverslips and then incubated for 30 min at room temperature with Fluo-4 (AM; Thermofisher) at a concentration of 3.2 μM. Then the cells were rinsed with Hepes buffered Ringer's (HBR) solution (in mM, 140 NaCl, 4 KCl, 1.5 MgCl2, 5 D-glucose, 10 HEPES, and 1.8 CaCl2 and pH adjusted to 7.4 with NaOH) and allowed to rest for another 30 min in HBR at room temperature. The cells were imaged on a Nikon Eclipse Ti microscope using a 10× objective. For each slip, a brightfield image, a fluo-4 image, and a 3-min fluo-4 fluorescence movie with exposures of 100-ms and 0.5-s intervals were captured. During the time sequence, HBR is initially being perfused throughout the chamber. Perfusion is switched to 500 nM capsaicin in HBR at 30 s, and at the 2 min mark, 3 μM ionomycin is added to the chamber. The HBR and 500 nM capsaicin in HBR were perfused into the chamber using open, gravity-driven reservoirs, and 500 uL of 3 μM ionomycin was Pipetted into the chamber via micro-Pipette. The data obtained during these experiments were analyzed using Image J and Matlab. For each time sequence, ROIs sized 10 × 10 pixels were placed over approximately seven responding cells where their level of fluorescence was tracked throughout the experiment. For each ROI, using the basal fluorescence during HBR perfusion as a baseline, the proportion of the maximal fluorescence (which was achieved during ionomycin application) that was reached during capsaicin application was calculated. The Mann-Whitney U test was used to determine significance at p = 0.05.
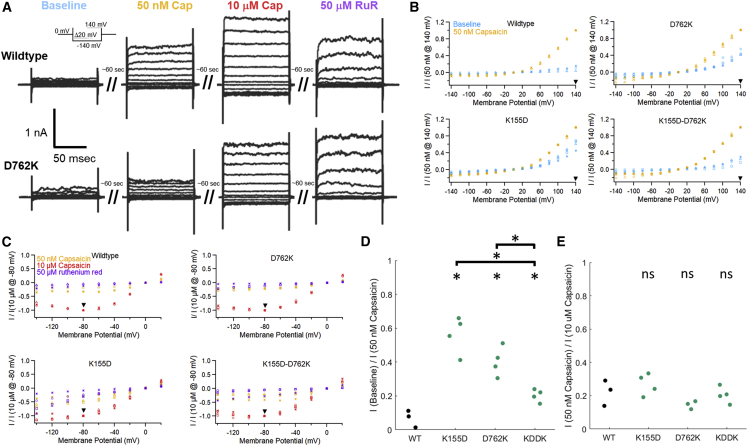
For whole-cell electrophysiology, whole-cell patches were obtained using filamented borosilicate glass Pipettes (Sutter BF150-86-10) that were pulled and polished to openings with 3–6 MOhm resistance. Once whole-cell configuration was achieved and series resistance was compensated to 40% or better, recordings were low-pass filtered to 5 kHz and acquired at 50 kHz using an Axopatch 200B amplifier (Axon Instruments) with PatchMaster v2x73.2 (HEKA Elektronik). Experiments were conducted at room temperature in a Ca2+-free HBR solution (in mM, 140 NaCl, 4 KCl, 1.5 MgCl2, 5 D-glucose, 10 HEPES, and 1 EGTA and pH adjusted to 7.4 with NaOH) with a cesium-chloride intracellular Pipette solution (in mM, 120 CsCl, 10 NaCl, 1 MgCl2, 10 EGTA, 5 HEPES, 3 Mg-ATP, and 1 Na-GTP pH adjusted to 7.2 with CsOH). Capsaicin solutions were made from EtOH stocks and diluted into Ca2+-free HBR. Solutions containing 50 nM capsaicin, 10 μM capsaicin, and 50 μM ruthenium red were applied through bath solution exchanges with the use of an eight-port perfusion manifold in order of increasing concentration during experiments followed lastly by the 50 μM ruthenium red block treatment. To monitor equilibration of capsaicin and ruthenium red between solution and cell membrane, we used a (−80/20 mV) voltage pulse protocol with holding potential at 0 mV during solution exchange, which completed on the order of ∼60 s. The voltage protocol used for testing responses to different treatments was a 100-ms voltage step range from −140 mV to 200 mV in 20-mV increments with a 260-ms sweep interval at holding potential of 0 mV. However, Fig. 5 only shows the range of −140 to 140 mV. The mutant K155E used a similar protocol with a voltage range of −120 mV to 200 mV. The possibility that a reduced sensitivity to capsaicin would introduce an artifactual baseline activity in our K155D and D762K mutants was ruled out by evaluating the limited responses of the mutants at −80 mV to 50 nM capsaicin normalized to saturating doses at 10 μM. The negative potential was chosen because TRPV1 currents are reduced by the weak voltage dependence of activation, and we avoid the complication of outward currents becoming saturated by the series resistance of the whole-cell patch-clamp configuration, even if we partially compensate for this with the amplifier. As was previously shown by Jendryke et al., the TRPV1 capsaicin dose-response curve acquired with inward currents closely approximates the dose-response curve derived from outward currents with a positive potential (21). The rTRPV1 mutants K155D and D762K were transfected with the addition of 5 μM ruthenium red in the cell culture media to prevent Ca2+ cytotoxicity and allow for sufficient channel expression in the plasma membrane. Coverslips were transferred in Ca2+-free HBR and rinsed with Ca2+-free HBR in the experimental chamber to remove any trace amounts of ruthenium red.
Figure 5.
Whole-cell electrophysiology experiments with TRPV1 mutants D762K, K155D, and K155D/D762K (KDDK). (A) Representative current traces of voltage protocol used to measure baseline current (blue), 50 nM capsaicin elicited current (yellow), 10 μM capsaicin evoked current (red), and currents blocked by 50 μM ruthenium red (purple). Wild-type and D762K TRPV1 are shown with intervals of approximately ∼60 s between traces to allow for cell equilibration with bath solution. Note that although wash-in of capsaicin is rapid, 10 μM capsaicin does not wash out extensively before 50 μM ruthenium red is applied to check for leak currents at negative potentials. Large outward currents through TRPV1 persist at 60 mV and greater in the presence of ruthenium red because its pore blocking action is voltage dependent. (B) Comparison of currents recorded from baseline activity to the currents evoked by 50 nM capsaicin with all currents normalized by the 50 nM capsaicin response at +140 mV (arrowhead) in the same cell for wild-type (WT), K155D-D762K, K155D, and D762K respectively. (C) Comparison of wild-type (WT), K155D-D762K, K155D, and D762K currents evoked by 50 nM capsaicin normalized to the 10 μM capsaicin response at −80 mV (arrowhead) in the same cell (see Fig. S7). (D) Summary plot of leak-subtracted baseline activity compared with 50 nM capsaicin activity at 140 mV (see materials and methods). The mutants, K155D-D762K (p < 0.022), K155D (p < 0.0009), and D762K (p < 0.002) differed significantly from wild-type activity. The double-swap, or predicted rescue mutant, K155D-D762K, differed significantly from the other mutants, K155D (p < 0.0045), and D762K (p < 0.013). See Fig. S6 for additional K155E data not shown here. Statistical significance at p < 0.05 indicated with “∗.” Isolated “∗” indicates comparison to wild-type, and brackets show additional significance tests. (E) Summary plot of leak-subtracted 50 nM capsaicin evoked responses normalized to 10 μM capsaicin activity at −80 mV. None of the mutants showed a significant difference from wild-type TRPV1 at the p < 0.05 level. Significance tests determined by Student's t-test for two samples with unequal variance. To see this figure in color, go online.
For data analysis, the electrophysiology data collected in PatchMaster was exported to, analyzed with, and plotted with IGOR Pro 8, version 8.0.4.2 32-bit. Any data recordings that did not show characteristic TRPV1 rectification were discarded. The values displayed in the plots in Figs. 5 B, C and S6a were collected from the average of the last 20 ms of each voltage step to ensure only steady-state currents were used. Leak subtraction in summary data of Fig. 5 d was done by removing the absolute value of the baseline inward current at −140 mV from baseline and 50 nM capsaicin outward current at 140 mV before normalization. Leak subtraction in summary data of Fig. 5 E was done by removing the absolute value of the ruthenium red inward current at −80 mV from the absolute value of 50 nM and 10 μM capsaicin inward current at −80 mV before normalization. Leak subtraction in summary data of Fig. S6b was done by removing the absolute value of the baseline inward current at −120 mV from baseline and 50 nM capsaicin outward current at 120 mV before normalization. Student's t-tests across samples were done in Excel (Microsoft).
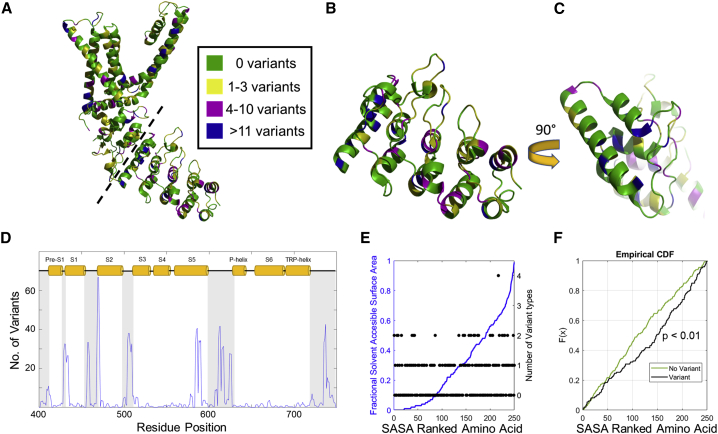
Regarding computational methods, human genomic variant data from gnomAD 2.1.1. was downloaded directly from gnomAD servers (http://gnomad.broadinstitute.org; “export variants to CSV” option). Spreadsheet data was formatted in Excel (Microsoft) to read in the variant data to a Matlab (Mathworks, MA) script that generated a frequency plot for the number of missense variants at each residue position (See Figs. 1 D and S4). Silent mutations are ignored, and each occurrence of a missense SNP is recorded as a missense variant in our counts. Residues with >100 variants were truncated to 100 counts, and positional frequency was also smoothed with a boxcar average of three residues. Alignment with secondary structure was done with respect to rat TRPV1 residue positions, and structural data was defined by Fig. S3 in Nadezhdin et al. (22).
Figure 1.
Human variant analysis of TRPV1 and structural correlation with variant residues. (A) A subunit of rat TRPV1 structure, PDB: 3J5P, with backbone positions of residues color coded by gnomAD 2.1.1 variant frequency. Green: 0 variants; yellow: 1–3 variants; magenta: 4–10 variants; blue: >11 variants. (B) Residues 111–320 of rat TRPV1 structure PDB: 3J5P separated from subunit in (A) along dashed line. (C) Structure in (B) rotated 90° toward viewer. (D) Variant frequency in human TRPV1 sequence 400–750 aligned with secondary structure of rat TRPV1 represented as orange cylinders (defined by Fig. S3 in Nadezhdin et al. [22]). (E) Fractional solvent accessible surface area (SASA: blue) plotted against SASA-ranked order of residues 111–361 from the ARD of rat TRPV1 structure PDB: 3J5P. Filled circle markers (black) and the y-axis on the right indicate the number of different types of amino acids arising from missense mutations at a given position. (F) Empirical cumulative distribution function (CDF) for the distribution of residues without variants and residues with variant amino acids plotted against SASA-ranked amino acid of the ARD. A comparison of the two distributions was made with the Kolmogorov-Smirnov test for two samples, and the p-value is 0.0087.
Fractional solvent accessible surface area (SASA) was calculated in PyMOL with the “get_SASA_relative” function on the region of interest in the rat TRPV1 model PDB: 3J5P. This function calculates the fully exposed solvated surface area by removing all the residues except for the neighbor residues (i + 1, i – 1) of the target. Then it uses this fully exposed value to normalize the solvent accessible area detected in the model.
We also examined the number of types of amino acids at variant residues and how this is affected from lower to higher SASA in Fig. 1 E. Two distributions were generated by extracting the number of types of amino acids for each variant and ranking the residue according to its SASA score: one set contained all the residue positions without variants, and a second set contained all the residue positions with a different amino acid than in the standard human TRPV1 sequence. A statistical examination of the difference between these SASA-ranked residue sets was done with the nonparametric Kolmogorov-Smirnov test for two samples and is shown in Fig. 1 F.
Results
Genomic variation in H. Sapiens TRPV1 protein sequence
The gnomAD is a publicly accessible collection of genome variation data collected from unrelated individuals in the human population (23). By highlighting the individual amino acids according to the total number of missense variants occurring at that position and projecting this information onto the apo-structure of a rat TRPV1 subunit, we initially intended to reveal novel interaction surfaces through visual inspection. A brief survey of the different types of mutations and number of SNPs listed in the gnomAD prompted us to represent the frequency of TRPV1 missense variants at each amino acid in a color-coded format. On a first impression, the overlayed pattern of gnomAD variants on the structure of TRPV1 appears to be random in both distribution and degree (Fig. 1 A). Since the variant positions may be better tolerated at sites of poor conservation, we compared the sequence variant positions to the conservation of TRPV1 residues across several orthologs and found that sequence variants extended to within highly conserved regions across the entire TRPV1 protein sequence (Figs. S1, S2, and S3). Using a hybrid approach of sequence and secondary structure information, we generated a variant alignment overlaid with secondary structural content of TRPV1 transmembrane regions (Pre-S1 through TRP-helix). Thus, a correlation emerged between variant suppression and structured segments, which are indicated as white regions in the plot of Fig. 1 D (sequence analysis extended to N-/C-terminus in Fig. S4). We were encouraged by our discovery that an anticorrelated relationship exists between variant location and secondary structure and hypothesized that residues with buried side chains also exhibited fewer variants. We therefore sought to compare how often a variant occurs against SASA of each amino acid side chain. We limited our analysis to the cytosolic, N-terminal ARD resolved in the PDB: 3J5P structure of TRPV1 and observed an increasing trend of variant types for each residue as fractional SASA becomes greater (Fig. 1 E). To confirm our observation that side-chain accessibility to solvent correlated with a tolerance to different types of amino acid substitutions in variants, we compared the set of residues in the ARD without variants to the set of ARD residues with an alternative amino acid as defined by gnomAD data. The cumulative distribution function of these sets, each with residues ranked according to fractional SASA, reveals an unambiguous difference, which the Kolmogorov-Smirnov test also showed as significant (Fig. 1 F). Similarly, a cross-sectional cut through the TRPV1 subunit, indicated as a dashed line in Fig. 1 A, exposes a buried surface of the channel structure (Fig. 1 B). This surface, shown in Fig. 1 C, which is characterized as having limited solvent accessibility, also exhibited no variant sites. Both a reduced number of types of variants in lower SASA scoring residues as well as reduced variant frequency in residues of buried interfaces led us to hypothesize that any continuous surfaces of the TRPV1 structure with a reduced collection of variant residues may be the hallmark of a critical interaction surface.
Variant analysis of TRPV1 ARD locates interaction surfaces
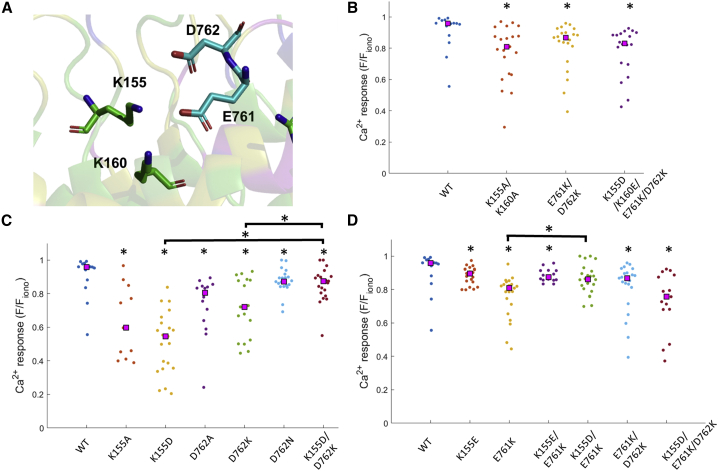
The ARD of the PDB: 3J5P TRPV1 structure extends radially away from the central pore axis of the channel. Apart from a beta-sheet that forms a point of contact between adjacent ARDs, there are limited interactions across ARDs and neighboring subunits observed in this structure of TRPV1. We generated a structural map of putative reduced variant surfaces by rendering the surface of the ARD fragment (residues 111–320) with color coding that conveys variant frequency (Fig. 2 A–C). We complemented the static rendering of surface variants in the ARD with a dynamic variant rendering scheme, which moves the cutoff value for coloring a variant from higher to lower thresholds (Fig. 2 B and C respectively, Videos S1 and S2). One continuous, reduced variant surface region identified by our variant analysis overlaps with a putative ATP interaction site in structure studies of the isolated ARD (24). Alanine mutagenesis of K155, K160, and Y199, all of which are circumscribed by the patch in the ARD with reduced variants, altered the gating properties of TRPV1, and a recent ground squirrel TRPV1 structure with a resolved C-terminal region of the channel showed that the same ARD surface is in extensive contact with the C-terminus (Fig. 2 D) (22). Based on our variant colorized rendering of the interaction surface, we hypothesize that residues K155 and K160, functionally characterized by Lishko et al., may form critical salt bridge interactions with D762 and E761, respectively, across the N- and C-termini (Fig. 2 D and E). All four residues are without variants in the gnomAD 2.1.1 data set, and we selected this site for further functional studies. At a separate site located along the same N-/C-terminus interaction surface but closer to the channel pore, we observed a possible cation-π interaction between the R212 residue of the ARD and W752 of the C-terminus (Fig. 3 A). We also selected this site for further functional studies since the arginine was identified as having 19 variants in the gnomAD 2.1.1 data set. The prevailing logic for this decision was an expectation that the increased variability for the R212 site would translate to less impact on channel function if the site were mutated. If mutation of R212 has negligible impact on channel function, then high variant sites could be marked as having decreased functional consequences if mutated.
Figure 2.
Variant analysis of TRPV1 surface residues exposes N-terminal/C-terminal interface. (A) Ribbon representation of rat TRPV1 residues 111–320 from structure PDB: 3J5P. Variant frequency color scheme adopted from Fig. 1. (B) Surface representation of TRPV1 fragment shown in (A). (C) Structure from (B) rotated 90° as indicated. (D) TRPV1 C-terminal region (PDB: 7LQY) residues 746–775 shown with variant coloring scheme (left) and aligned with structure PDB: 3J5P overlaid onto N-terminal ARD (right, cyan). (E) Inset of box indicated by dashed outline in (D) with highlighted residues from N-terminal ARD shown in green (K155, K160, R211) and blue (R212) in accordance with variant coloring scheme and highlighted residues in C-terminal region shown in cyan (W752, E761, D762).
Figure 3.
Calcium imaging of TRPV1 R211 mutants and testing of putative cation-π interaction between R212-W752. (A) Stick representation of R211, R212 from structure PDB: 3J5P, and W752 from structure PDB: 7LQY. Color scheme for R211 and R212 follows for variant descriptions: green: 0 variants; yellow: 1–3 variants; magenta: 4–10 variants; blue: >11 variants. W752 has no variants and is colored cyan to distinguish this residue from those on the N-terminus. (B) Putative cation-π interaction tested with Ca2+ responses of wild-type (WT) and various R211/212 mutants to 500 nM capsaicin. (C) Stick representation of R211, K155, and K160 from structure PDB: 3J5P, and E761, D762 as well as beta-sheet extension (W372–H378) from neighboring subunit in structure PDB: 7LQY. Interatomic distances between R211 nitrogens and oxygens in backbone carbonyl and E761 side chain are shown. Color scheme for R211, K155, and K160 follows for variant descriptions as in (A). C-terminal residues E761, D762 and beta-sheet are colored cyan to distinguish these residues from N-terminus. (D) Electrostatic interaction between R211 and E761 tested with Ca2+ responses of various R211 and E761 mutants to 500 nM capsaicin. Statistical significance (∗) at p < 0.05 conducted with Mann-Whitney U test. Not significant indicated with “ns.” NR: no response. To see this figure in color, go online.
Surface rendering from Fig. 2B in green with variant number threshold decreasing and shown as magenta
Surface rendering from Fig. 2C in green with variant number threshold decreasing and shown as magenta
Ca2+ imaging of TRPV1 N-/C-terminal interface mutants
We used Ca2+ imaging to test the function of TRPV1 N-/C-terminus mutants in HEK 293T cells. Changes to the Ca2+ response in our mutants compared with wild-type TRPV1 were interpreted as site-directed mutants that altered channel function or expression. We first tested our hypothesis that the number of variants at position R212 diminished the importance of this site for channel function or expression and confirmed that mutating R212 to alanine did not significantly affect Ca2+ responses to capsaicin (Fig. 3 A and B). Only one amino acid adjacent to R212 is R211, which happens to have no variants. The local resolution of the gsTRPV1 structure near R211 is less than the structural average (3.19 Å), and we wanted to be certain that a register shift had not led us to assign the incorrect arginine to the putative cation-π interaction. When tested by mutation to alanine, Ca2+ responses from R211A deviated significantly from wild-type responses. Because of the significant response at R211, we made site-directed mutants that switched residues identities between R211 and W752 but which preserved the cation-π interaction. To alleviate any concern that the side chain of R212 may compensate for changes made in R211, we made our site-directed mutations in the R212A background. Substituting tryptophan at R211 in the R212A background gave us no response in our experiments. Although the R212A/W752R mutation gave us Ca2+ responses similar to wild-type, the fully exchanged mutant R212A/W752R/R211W was nonresponsive (Fig. 3 B). Based on these functional data, we surmised that a critical cation-π interaction does not extend from R211 or R212 to W752. Nevertheless, a critical role for R211 was uncovered in our Ca2+ imaging experiments, and we therefore examined the structures for evidence of any interactions involving R211 that span the N-/C-terminus interface. There are two carbonyl oxygens across the interface that lie within ∼4 Å of the side-chain amines on R211 (Fig. 3 C). These carbonyls, which are located on the beta finger of the neighboring subunit as well as the interfacial C-terminus, may form critical interactions with the R211 amines. Limited by our use of mutagenesis to probe potential interaction partners, the nearest residue that could be tested for a potential side-chain electrostatic interaction is E761. Although the PDB: 7LQY structure places the R211 amine groups at ∼7 Å from the terminal oxygen of E761, we expect that surface position fluctuations of E761 would bring the carboxylic group in closer proximity to R211 (Fig. 3 C). We hypothesized the existence of a channel state with a salt bridge between R211 and E761 and designed TRPV1 constructs to test this with a charge swap mutant. Although either R211E and E761R significantly reduced the Ca2+ response in a manner consistent with these mutants affecting channel function or expression, the charge swap mutant R211E/E761R, which would preserve the electrostatic interaction of a salt bridge, showed no Ca2+ responses in transfected cells (Fig. 3 D). We deduced that the additive effect of the single mutations gave a nonfunctional channel and that a stable, electrostatic interaction between R211 and E761 is unlikely. Moreover, the absence of a response in our charge swap mutants prevented us from further investigating a transient, state-dependent interaction between these two residues.
We next turned our attention to the putative electrostatic interaction network between K155 and K160 in the N-terminal ARD and E761 and D762 of the C-terminus shown in Fig. 4 A. In a simple brute-force scheme to exchange all the basic residues on the ARD with the acidic residues of the C-terminus, we conducted a broad charge swap across both sides of the interface to observe whether Ca2+ responses would initially decrease with charges altered on only one side (E761K/D762K) before recovering wild-type responses in a quadruple mutant rescue (K155D/K160E/E761K/D762K). Although the initial double mutant (E761K/D762K) showed a significant reduction in the Ca2+ response compared with wild-type, the quadruple mutant that would preserve the electrostatic network was not able to restore the wild-type response (Fig. 4 B). We deduced that the electrostatic network could be susceptible to small changes in side-chain size and position and sought to study the interaction surface with less impactful perturbations. Beginning with K155 and D762, we observed a significant loss of Ca2+ responses when both positions are individually mutated to alanine compared with wild-type. Ca2+ responses were further diminished by switching the amino acid at K155 and D762 to the cognate partner amino acid across the interface. Simultaneously swapping the charges in a double mutant, K155D/D762K, partially recovered Ca2+ responses observed in wild-type, and the Ca2+ response in the double mutant was indistinguishable from a D762N mutant that would partially disrupt an electrostatic interaction (Fig. 4 C).
Figure 4.
Calcium imaging of TRPV1 and putative electrostatic interaction between K155, K160, E761, and D762. (A) Stick representation of K155, K160, E761, and D762 from structures PDB: 3J5P and PDB: 7LQY. Color scheme for K155 and K160 follows for variant descriptions: green: 0 variants; yellow: 1–3 variants; magenta: 4–10 variants; blue: >11 variants. E761 and D762 are colored cyan to distinguish these residues from the C-terminus. (B) Ca2+ responses to 500 nM capsaicin of wild-type (WT) and double or quadruple mutants. (C) Partial charge reversal at positions K155 and D762 and charge swap double mutant K155D/D762K tested with Ca2+ response to 500 nM capsaicin. (D) Partial charge reversal at positions K155 and E761 and charge swap double mutants K155(D,E)/E761K or triple mutant K155D/E761K/D762K tested with Ca2+ response to 500 nM capsaicin. Statistical significance (∗) at p < 0.05 conducted with Mann-Whitney U test. Isolated “∗” indicates comparison to wild-type, and brackets show additional significance tests. To see this figure in color, go online.
Conducting a charge swap with the neighboring sites, K160E/E761K does not recover any part of the Ca2+ response signal recorded in the individual K160E or E761K mutants (Fig. S5). Our initial interpretation for the effects observed in Ca2+ responses with K160, E761 mutations is that a simple interaction across the amino acid pair is unlikely. Because we observed a recovery in K155D/D762K Ca2+ responses relative to K155D, we decided to interrogate whether the interaction of K155 with the C-terminus extended to E761. We first tested the responses of K155E, and to our surprise, we noticed that the signal was far different from the K155D mutant, although the same acidic, functional group was introduced albeit with a longer side chain. Completing the charge swap with K155E/E761K showed a modest improvement over the Ca2+ response in E761K alone (Fig. 4 D). The diminished Ca2+ response of K155D was also restored by adding the E761K mutation (K155D versus K155D/E761K: p = 1.4E-7), further supporting the case for an electrostatic interaction across residues K155, E761, and D762. We hypothesized that an electrostatic network across three side chains, with K155 on the N-terminal domain and E761/D762 on the C-terminal domain, could be partially maintained in a mutant that exhibited the same network architecture of two salt bridges between three side chains. The triple mutant K155D/E761K/D762K failed to fully recover the Ca2+ response observed in wild-type (Fig. 4 D). Based on our observations of K155E and the quadruple mutant, the structural perturbations caused by side-chain substitutions may complicate a complete interpretation of our Ca2+ imaging results and how the salt bridge across the N-terminal/C-terminal interface influences channel function. To develop our understanding of how the nuances of this interaction influence TRPV1 function, we used whole-cell electrophysiology to explore the functional responses in mutants of the interface.
Electrophysiological recordings of K155D, D762K, and K155D/D762K
Our initial findings that K155D/D762K showed modest recovery of the Ca2+ imaging response that was lost in either K155D or D762K alone implicated the double mutant as a functional rescue of the individual mutants. However, Ca2+ imaging data does not distinguish between reduced expression or channel sensitivity to agonist, which gives us functional information. We therefore used whole-cell electrophysiology to determine the functional consequences of introducing mutations at K155 and D762K. Whole-cell electrophysiological recordings reveal the characteristic outward rectification of TRPV1 currents even in the absence of stimuli (Fig. 5 A). TRPV1 voltage-dependent activation is weak but noticeable at voltages above 100 mV. Based on prior evidence that the mutation K155E increases the basal activity of TRPV1, we compared the activity at 140 mV of unstimulated mutants K155D, D762K, and K155D/D762K compared with wild-type TRPV1 at 140 mV (25). Normalization was accomplished by recording the responses to 50 nM capsaicin at 140 mV. Similar to K155E at 120 mV, both K155D and D762K show significantly increased basal channel currents at 140 mV when compared with wild-type (Figs. 5 B, D and S6). Our rescue double mutant K155D/D762K showed a partial recovery of the baseline activity that was observed in wild-type TRPV1. Although the baseline currents of K155D/D762K still differed significantly from wild-type (p < 0.022), there was a significant difference observed between the rescue mutant and K155D (p < 0.0045) or D762K (p < 0.013).
We considered two possible reasons for increased baseline activity in K155D and D762K: 1) the mutants exhibit increased activity in comparison to wild-type TRPV1; 2) mutants are less sensitive to the agonist capsaicin compared with wild-type TRPV1. In the latter case, similar levels of spontaneous activity in the absence of agonist would be present in mutants and wild-type; however, normalizing baseline currents by relatively smaller capsaicin responses in the mutant would show raised baseline activity compared with wild-type. To address this, we compared mutant and wild-type responses to 50 nM capsaicin at −80 mV normalized to saturating (10 μM) capsaicin responses (Figs. 5 C and S7; see materials and methods). Our 50 nM and 10 μM current responses obtained from wild-type, K155D, D762K, and K155D/D762K show that the fraction of currents activated by 50 nM capsaicin relative to a saturating concentration is similar in wild-type and mutant channels. There was no indication that the mutants exhibited strongly reduced (>two-fold) responses to 50 nM capsaicin compared with wild-type (Fig. 5 E).
Discussion
Our investigations of the association between TRPV1 genomic variants and structure initially compared the known secondary structure of the ion channel with variant frequency in the raw amino acid sequence. An alignment between alpha helices in the transmembrane domain and amino acid sequence revealed a correlation between variant frequency and intracellular or extracellular loops connecting transmembrane segments (Fig. 1 D). Higher conservation in the transmembrane segments compared with the connecting intracellular and extracellular loops is a defining trait of ion channels that appears to be maintained in the pattern of variant residues. We expect that future studies of this pattern along transmembrane regions will be a promising area for hypothesis generation in function structure studies of ion channels and integral membrane proteins. Furthermore, when the fractional SASA is calculated for the amino acids in the rTRPV1 ARD (111–360: PDB: 3J5P), we observed an increasing trend for the number of types of amino acids in variant residues with greater fractional SASA of the side-chain (Fig. 1 E and F).
The ARD appears to be in contact with a neighboring subunit via a beta-sheet extension, and our variant analysis of the ARD revealed a reduction in variant frequency across surface residues near the beta-sheet contact point and extending out to the N-terminus of the ARD. This surface of the ARD included residues such as K155 and K160 with previously known functional importance when mutated (24). Publication of the PDB: 7LQY gsTRPV1 structure and detailed resolution of the C-terminal region that interacts with the N-terminal ARD places the contact interface precisely over the bounds that we proposed with our rendering of variant surface residues (Fig. 2 D), and more recent TRPV1 and related TRPV3 structures further confirmed the significance of this surface in forming a contact interface (26,27,28).
We first interrogated a possible cation-π interaction between R212 in the N-terminus and W752 in the C-terminus inferred from the ground squirrel TRPV1 structure (Fig. 3 A). The mutation R212A did not significantly alter the Ca2+ responses of cells expressing the mutant construct, and we interpreted this result as a reflection of the large number of variants at position 212 in the human TRPV1 sequence. In essence, more missense mutations for a specific amino acid within the general population of humans indicates a greater tolerance for mutations introduced at the same position because normal channel function would remain intact. However, in contrast to R212, the absence of variants at R211 foreshadowed a significant defect in Ca2+ imaging experiments with mutations to this residue, and we speculate that R211 is critically important for maintaining the interface contacts (Fig. 3 B and D). A second N-/C-terminus interaction site that we deemed worth studying was a putative electrostatic network at K155/K160/E761/D762K (K-KED). Although complete exchange of the electrostatic charges across the interface does not adequately recover Ca2+ imaging responses from losses incurred by charge reversals in the C-terminus with E761K/D762K, paired site-directed mutagenesis experiments more concisely interrogated an electrostatic interaction mechanism (Fig. 4 B, C, and D).
A yeast forwards genetics study to identify potentiating mutations in TRPV1 led to the discovery of the K155E mutation, and electrophysiological experiments in oocytes determined that the mutation increases the baseline activity of TRPV1 (25). We evaluated the K155E mutation alongside K155D and D762K by electrophysiology for increased baseline activity at depolarized potentials and determined that baseline activity is greater in these mutants compared with the wild-type channel (Fig. 5 B). Moreover, the double mutant K155D/D762K, which partially restores the electrostatic interactions across K-KED, significantly reduces the baseline activity observed in either K155D or D762K. This result appears at odds with our Ca2+ imaging data, which showed reduced responses in all K-KED single mutants relative to wild-type. To explain this apparent discrepancy, we propose that potentiation in K-KED mutants may be harmful to cell function, or there is an expression defect. Our Ca2+ imaging data implicate diminished channel accumulation in the plasma membranes of cells. An expression defect is supported by our transfection methods for K155D and D762K TRPV1, which necessitated a 5:1 ratio of channel plasmid to GFP plasmid in comparison to a 1:5 ratio of wild-type TRPV1 to GFP plasmid. However, we also supplemented our cell growth media with 5 μM ruthenium red to increase TRPV1-K155D channel currents, which was indicative of cell toxicity from excessive intracellular Ca2+ in our mutants (See materials and methods). Our tactics to increase surface expression of the K155D mutant by introducing the pore-blocker ruthenium red to the cell culture media supports our conclusion that K-KED mutants have greater spontaneous baseline activity compared with wild-type, and surface expression is reduced because of Ca2+-based cytotoxicity. This finding offers some insight into the use of variant analysis in our gene of interest. When taking into consideration that TRPV1 spontaneous channel activity is cytotoxic and that TRPV1 knock-out in the mouse model is nonlethal, TRPV1 residues without variants in the gnomAD database could be biased toward gain of function when mutated rather than loss of function. This would be less the case for a lethal knockout gene, in which a mutation to a residue without variants is more likely to be damaging to the organism through loss of function.
Our Ca2+ imaging survey of myriad K-KED mutations brought to light two important considerations: 1) the channel is still functional if the network is completely disrupted by the double mutant E761K/D762K, implying that the salt bridge is not essential; 2) unique responses between K155D and K155E, which differ by only one additional carbon bond, implicate sensitivity to side-chain packing at the interface. The latter concern may account for why out of the numerous salt bridge interactions tested in the K-KED site, only K155 and D762 could show partial restoration of wild-type TRPV1 Ca2+ responses. Moreover, the D762N mutation, which maintains Ca2+ responses similar to the K155D/D762K mutant, reduces the electrostatic interaction across the interface but maintains the excluded volume compared with a lysine substitution at this site. In summary, we have used structure/variant analysis of residue-specific interactions that is a valuable tool for mutagenesis studies. We limited our studies to sites of interaction with a high correlation between sequence conservation across functional orthologs and reduced variants in the human genomic data set. Nevertheless, we put forth that the power of variant analysis approaches can be expanded to evaluate specific functional consequences across species. In Laursen et al., the authors identified partially conserved residues at positions S124 and Q188 of TRPV1, which have a well-defined effect on temperature-dependent activation (29). These two residues have no variants in the gnomAD 2.1.1. data set. We propose that correlation between decreased sequence conservation and genomic nonvariant sites may offer mechanistic insight giving rise to functional properties unique to the species.
We interpret these results to mean that the stability of the N-/C-terminal interface across K-KED influences the equilibrium between channel open and closed states. If the salt bridge interaction between K155 and D762 is intact, the channel is more likely to assume a closed pore conformation, and a disruption to this interaction shifts the equilibrium to open states. Evidence from several 3D class structures of TRPV1 with different arrangements of the K-KED side chains support a switch-like role for this interface, which has been proposed for TRPV3 (26,27,30). In the case of TRPV3, functional experiments have shown that mutations of residues located at the N-/C-terminus interface shift the equilibrium of the pore toward an open state (31,32). Although it remains to be seen whether K-KED interface disruption is an essential step toward channel gating, prior experiments with TRPV1 constructs that truncate the channel beyond residue 777 or delete C-terminal segment 767–801 demonstrate that gating modulation is arbitrated by sites even further distal to E761 and D762 (33,34). We therefore speculate that the interface switch at K-KED may be coupled to modulatory input from events occurring at downstream segments of the C-terminus.
Conclusions
This study demonstrates the utility of combining structural and genomic variant data to screen for critical residues in protein interfaces. We have chosen the ion channel TRPV1 as a model to test the feasibility of using gnomAD 2.1.1 SNP data to 1) locate candidate protein-protein interfaces and 2) for screening residues in a site-directed mutagenesis functional study. Our bioinformatics findings suggested that points of contact along protein surfaces were less likely to tolerate missense mutations, identifiable in the gnomAD 2.1.1 data set. Therefore, we examined the surface of the rTRPV1 structure ARD for contiguous regions with lower variant frequency. Based on our structure and genomic variant screening strategy, we used Ca2+ imaging and site-directed mutagenesis experiments to test interfacial contacts between the N- and C-terminus of TRPV1. Ca2+ imaging experiments of N-terminal R212 and C-terminal W752 did not support the existence of an essential cation-π interaction between these residues. The increased number of variants for R212 matched our expectations that the site was not critical to channel function. Our Ca2+ imaging experiments at a second site in the interface termed K-KED isolated a potential salt bridge between K155 and D762, which we tested with further electrophysiological experiments. Current recordings obtained from the charge reversal mutants K155D and D762K showed an increase in baseline activity over wild-type TRPV1, and if the charges are swapped in a K155D/D762K mutant, we observed partial restoration of wild-type currents. Our electrophysiological experiments with mutations to K155 and D762, which lie juxtaposed at an N-/C- terminal interface, implicate the formation of a salt bridge that is critical to maintaining the pore of TRPV1 in a closed state in the absence of a stimulus.
Author contributions
T.M.M, J.S.I., and E.N.S. designed research; T.M.M, J.S.I., and E.N.S. performed experiments; T.M.M, J.S.I., N.K. and E.N.S. analyzed data; T.M.M, J.S.I, and E.N.S. wrote the paper.
Acknowledgments
We thank Dr. Sharona Gordon for the pcDNA3.1 TRPV1 wild-type and Sec61b-GFP constructs. We thank Dr. Marcel Goldschen-Ohm and Dr. Andres Jara-Oseguera for critical reading of the manuscript. We thank members of the Aldrich Lab for critical feedback throughout the course of this study. This study was funded in part by start up funds from the University of Texas (to E.N.S.) and National Science Foundation grant no. 2129209 (to E.N.S.)
Declaration of interests
The authors have no competing interests to disclose.
Editor: Valeria Vasquez.
Footnotes
Taylor M. Mott and Jordan S. Ibarra contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.12.012.
Supporting material
References
- 1.Caterina M.J., Schumacher M.A., et al. Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 2.Davis J.B., Gray J., et al. Sheardown S.A. Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature. 2000;405:183–187. doi: 10.1038/35012076. [DOI] [PubMed] [Google Scholar]
- 3.Yao J., Liu B., Qin F. Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys. J. 2010;99:1743–1753. doi: 10.1016/j.bpj.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F., Jara-Oseguera A., et al. Swartz K.J. Heat activation is intrinsic to the pore domain of TRPV1. Proc. Natl. Acad. Sci. USA. 2018;115:E317–E324. doi: 10.1073/pnas.1717192115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jara-Oseguera A., Bae C., Swartz K.J. An external sodium ion binding site controls allosteric gating in TRPV1 channels. Elife. 2016;5 doi: 10.7554/eLife.13356. e13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto-Posadas A., Picazo-Juárez G., et al. Rosenbaum T. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nat. Chem. Biol. 2012;8:78–85. doi: 10.1038/nchembio.712. [DOI] [PubMed] [Google Scholar]
- 7.Chuang H.H., Prescott E.D., et al. Julius D. Bradykinin and nerve growth factor release the capsaicin receptor from PtdIns(4, 5)P2-mediated inhibition. Nature. 2001;411:957–962. doi: 10.1038/35082088. [DOI] [PubMed] [Google Scholar]
- 8.Chuang H.H., Lin S. Oxidative challenges sensitize the capsaicin receptor by covalent cysteine modification. Proc. Natl. Acad. Sci. USA. 2009;106:20097–20102. doi: 10.1073/pnas.0902675106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbaum T., Gordon-Shaag A., et al. Gordon S.E. Ca2+/calmodulin modulates TRPV1 activation by capsaicin. J. Gen. Physiol. 2004;123:53–62. doi: 10.1085/jgp.200308906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao E., Liao M., et al. Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao M., Cao E., et al. Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao E. Structural mechanisms of transient receptor potential ion channels. J. Gen. Physiol. 2020;152 doi: 10.1085/jgp.201811998. e201811998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singer A., Sigworth F.J. Computational methods for single-particle electron cryomicroscopy. Annu. Rev. Biomed. Data Sci. 2020;3:163–190. doi: 10.1146/annurev-biodatasci-021020-093826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMaio F., Zhang J., et al. Baker D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 2013;22:865–868. doi: 10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beckers M., Mann D., Sachse C. Structural interpretation of cryo-EM image reconstructions. Prog. Biophys. Mol. Biol. 2021;160:26–36. doi: 10.1016/j.pbiomolbio.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Valdar W.S.J. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 17.Cooper G.M., Brown C.D. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008;18:201–205. doi: 10.1101/gr.7205808. [DOI] [PubMed] [Google Scholar]
- 18.Gabaldón T., Koonin E.V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma B., Elkayam T., et al. Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Nafría J., Watson J.F., Greger I.H. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci. Rep. 2016;6:27459. doi: 10.1038/srep27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jendryke T., Prochazkova M., et al. Wetzel C.H. TRPV1 function is modulated by Cdk5-mediated phosphorylation: insights into the molecular mechanism of nociception. Sci. Rep. 2016;6:22007. doi: 10.1038/srep22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadezhdin K.D., Neuberger A., et al. Sobolevsky A.I. Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Nat. Commun. 2021;12:2154. doi: 10.1038/s41467-021-22507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karczewski K.J., Francioli L.C., et al. Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lishko P.V., Procko E., et al. Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Myers B.R., Bohlen C.J., Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/j.neuron.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K., Julius D., Cheng Y. Structural snapshots of TRPV1 reveal mechanism of polymodal functionality. Cell. 2021;184:5138–5150.e12. doi: 10.1016/j.cell.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zubcevic L., Hsu A.L., et al. Lee S.Y. Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife. 2019;8 doi: 10.7554/eLife.47746. e45779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon D.H., Zhang F., et al. Lee S.Y. Heat-dependent opening of TRPV1 in the presence of capsaicin. Nat. Struct. Mol. Biol. 2021;28:554–563. doi: 10.1038/s41594-021-00616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laursen W.J., Schneider E.R., et al. Garcheva E.O. Low-cost functional plasticity of TRPV1 supports heat tolerance in squirrels and camels. Proc. Natl. Acad. Sci. USA. 2016;113:11342–11347. doi: 10.1073/pnas.1604269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Z., Maksaev G., et al. Yuan P. Gating of human TRPV3 in a lipid bilayer. Nat. Struct. Mol. Biol. 2020;27:635–644. doi: 10.1038/s41594-020-0428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H., Yang P., et al. Zhu M.X. Mechanisms of proton inhibition and sensitization of the cation channel TRPV3. J. Gen. Physiol. 2021;153 doi: 10.1085/jgp.202012663. e202012663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macikova L., Vyklicka L., et al. Vlachova V. Cytoplasmic inter-subunit interface controls use-dependence of thermal activation of TRPV3 channel. Int. J. Mol. Sci. 2019;20:3990. doi: 10.3390/ijms20163990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prescott E.D., Julius D. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 2003;300:1284–1288. doi: 10.1126/science.1083646. [DOI] [PubMed] [Google Scholar]
- 34.Numazaki M., Tominaga T., et al. Tominaga M. Structural determinant of TRPV1 desensitization interacts with calmodulin. Proc. Natl. Acad. Sci. USA. 2003;100:8002–8006. doi: 10.1073/pnas.1337252100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Surface rendering from Fig. 2B in green with variant number threshold decreasing and shown as magenta
Surface rendering from Fig. 2C in green with variant number threshold decreasing and shown as magenta