Summary
Eukaryotic initiation factor-4A2 (EIF4A2) is an ATP-dependent RNA helicase and a member of the DEAD-box protein family that recognizes the 5ʹ cap structure of mRNAs, allows mRNA to bind to the ribosome, and plays an important role in microRNA-regulated gene repression. Here, we report on 15 individuals from 14 families presenting with global developmental delay, intellectual disability, hypotonia, epilepsy, and structural brain anomalies, all of whom have extremely rare de novo mono-allelic or inherited bi-allelic variants in EIF4A2. Neurodegeneration was predominantly reported in individuals with bi-allelic variants. Molecular modeling predicts these variants would perturb structural interactions in key protein domains. To determine the pathogenicity of the EIF4A2 variants in vivo, we examined the mono-allelic variants in Drosophila melanogaster (fruit fly) and identified variant-specific behavioral and developmental defects. The fruit fly homolog of EIF4A2 is eIF4A, a negative regulator of decapentaplegic (dpp) signaling that regulates embryo patterning, eye and wing morphogenesis, and stem cell identity determination. Our loss-of-function (LOF) rescue assay demonstrated a pupal lethality phenotype induced by loss of eIF4A, which was fully rescued with human EIF4A2 wild-type (WT) cDNA expression. In comparison, the EIF4A2 variant cDNAs failed or incompletely rescued the lethality. Overall, our findings reveal that EIF4A2 variants cause a genetic neurodevelopmental syndrome with both LOF and gain of function as underlying mechanisms.
Keywords: neurodevelopmental disorder, DEAD-box protein, transcription factor, epilepsy, Drosophila
15 individuals with mono-allelic or inherited bi-allelic variants in the DEAD-box gene EIF4A2 were identified with global developmental delay, intellectual disability, hypotonia, epilepsy, and structural brain anomalies. In Drosophila melanogaster, mono-allelic variants in EIF4A2 led to variant-specific behavioral and developmental defects, secondary to both loss- and gain-of-function mechanisms.
Introduction
Neurodevelopmental disorders (NDDs) encompass highly prevalent conditions such as autism, epilepsy, and intellectual disability (ID), and ID alone affects 1%–3% of the global population.1 NDDs are characterized by altered brain development and maturation that lead to impairments in cognition and adaptive behaviors.1,2 Growing evidence suggests that the majority of NDDs are secondary to a genetic etiology, and an increasing number of disease-associated genes are identified each year.3
Among known NDD-associated genes, there are several genes encoding members of the Asp-Glu-Ala-Asp (DEAD)-box family,4,5,6,7 which is composed of 50 well-conserved RNA helicases that use ATP hydrolysis to unwind RNA’s secondary structures.8 A DEAD-box protein is comprised of two helicase domains, a DEAD-box motif, and a conserved Q motif for ATP-binding and hydrolysis.9,10 DEAD-box proteins play varied roles in brain development from the regulation of neurogenesis, WNT (wingless-related integration site) signaling to global inhibition of protein translation and ribonucleoprotein processing body defects in mRNA turnover.4,5,11 De novo variants in DDX3X (MIM: 300160), DDX6 (MIM: 600326), DHX30 (MIM: 616423), DDX54 (MIM: 611665), DHX34 (MIM: 615475), and DHX16 (MIM: 603405) are associated with ID syndromes,4,5,9 and pathogenic variants in DDX3X alone account for 1%–3% of ID in females.6 Reduced expression of DEAD-box-protein-72 and DEAD-box-protein-1 was identified in the fetal brains of individuals with Down syndrome (MIM: 190685), an NDD characterized by abnormal brain morphology, which suggests that DEAD-box proteins are essential for the growth and differentiation during early brain development.12 Together, affected individuals with pathogenic variants in genes encoding DEAD-box proteins have a range of clinical phenotypes and disease severity that include ID, hypotonia, seizures, and variable structural brain abnormalities.4,5,6,7
One of the ubiquitously expressed DEAD-box proteins is eukaryotic initiation factor-4A2 encoded by EIF4A2 (MIM: 601102), which is part of the eIF4F-complex that binds to the 5ʹ cap of mRNAs to promote protein translation initiation.13,14 EIF4A2 is an essential regulator of protein translation, the recognition of the 5ʹ cap structure of mRNAs, and binding of mRNA to the ribosome.15 EIF4A2 also plays an important role in microRNA-regulated gene repression.16 Similar to its fellow DEAD-box protein, DDX6, EIF4A2 alters polyadenylation through interactions with the CCR4-NOT complex. EIF4A2 binding to the CCR4-NOT complex inhibits CNOT7 deadenylation and increases polyadenylation of the associated mRNA.17 This pathway is important for neurodevelopmental processes, and pathogenic DDX6 (MIM: 600326) and CNOT1 (MIM: 604917) variants are associated with ID, hypotonia, and epilepsy.5,18
EIF4A2 is well conserved from invertebrates to vertebrates and highly expressed in all tissues, especially in the brain and skeletal muscle. In porcine skeletal muscle, EIF4A2 expression increases throughout embryonic and neonatal development and is regulated by MyoD.19,20 Cellular assays show that MyoD binds to the EIF4A2 promoter and increases EIF4A2 transcription during myogenesis.20 In the Xenopus embryo, EIF4A2 is essential for neural induction and leads to upregulation of neural fold regulatory genes.21 In addition to vertebrate development, the invertebrate homologs are also key mediators of developmental processes. In Drosophila melanogaster (fruit fly), the homolog is eIF4A, which is 89% similar to the EIF4A2 gene.22 Prior fruit fly studies demonstrate that eIF4A negatively regulates the TGF-β/BMP pathway orthologue, decapentaplegic (dpp), and leads to Smad degradation.23 In the fly, dpp signaling regulates embryo patterning, eye and wing morphogenesis, and stem cell identity.24,25,26 In vertebrates, TGF-β/BMP signaling is critical for neuronal differentiation, development, and function.27,28 Consistent with these essential roles, the dysregulation of TGF-β/Smad signaling is associated with a broad range of human neurological and neurodevelopmental phenotypes.27,28,29
Here, we report on 15 individuals from 14 families with extremely rare EIF4A2 variants presenting with a spectrum of NDDs including global developmental delay (GDD) (9/15), ID (7/15), hypotonia (14/15), epilepsy (11/15), and structural brain alterations (10/15). Of these 15 individuals, ten have de novo EIF4A2 missense variants, two have de novo frameshift variants (germline and mosaic, one each), one has a bi-allelic single amino acid deletion, and two siblings are compound heterozygous for frameshift variants. The homozygous variant identified for individual, 2, p.Asp37del, was previously reported.30 To determine the pathogenicity and rescue potential of the EIF4A2 variants in vivo, we examined four of the de novo EIF4A2 missense variants, c.647C>T (p.Thr216Ile), c.728C>T (p.Thr243Ile), c.1032G>C (p.Leu344Phe), and c.1091G>A (p.Gly364Glu), in the fruit fly and identified variant-specific behavioral and developmental defects. Our loss-of-function (LOF) rescue assay demonstrated a pupal lethality phenotype induced by loss of eIF4A, which was fully rescued with human EIF4A2 wild-type (WT) cDNA expression. In comparison, the EIF4A2 variant cDNAs failed or incompletely rescued the lethality. Together, our in vivo functional findings reveal that EIF4A2 variants cause a syndromic NDD through both LOF and gain-of-function (GOF) mechanisms.
Subjects and methods
Human subjects
Clinical data were obtained after written informed consent was provided by a parent or legal guardian for all subjects in accordance with local institutional review boards (IRBs) of the participating centers. We used Matchmaker Exchange to form an international collaboration, allowing for comparison of individuals and their variants. Collection and analysis of the de-identified clinical cohort was approved by the Boston Children’s Hospital IRB. EIF4A2 mono-allelic and bi-allelic variants were identified by exome sequencing through each proband’s respective institution. We performed Sanger sequencing of the parental samples as necessary to confirm de novo dominant and recessive segregation for all probands. Paternity was confirmed by the inheritance of rare single-nucleotide polymorphisms (SNPs) from the parents. Sample swap was excluded. MRIs from individuals (I:4, I:5, I:8, I:10, and I:12) were obtained and independently reviewed by a pediatric neuroradiologist at Boston Children’s Hospital. The clinical phenotypes of these individuals and their EIF4A2 variants are depicted in Figure 1, Tables 1 and 2, and Table S1 and summarized below.
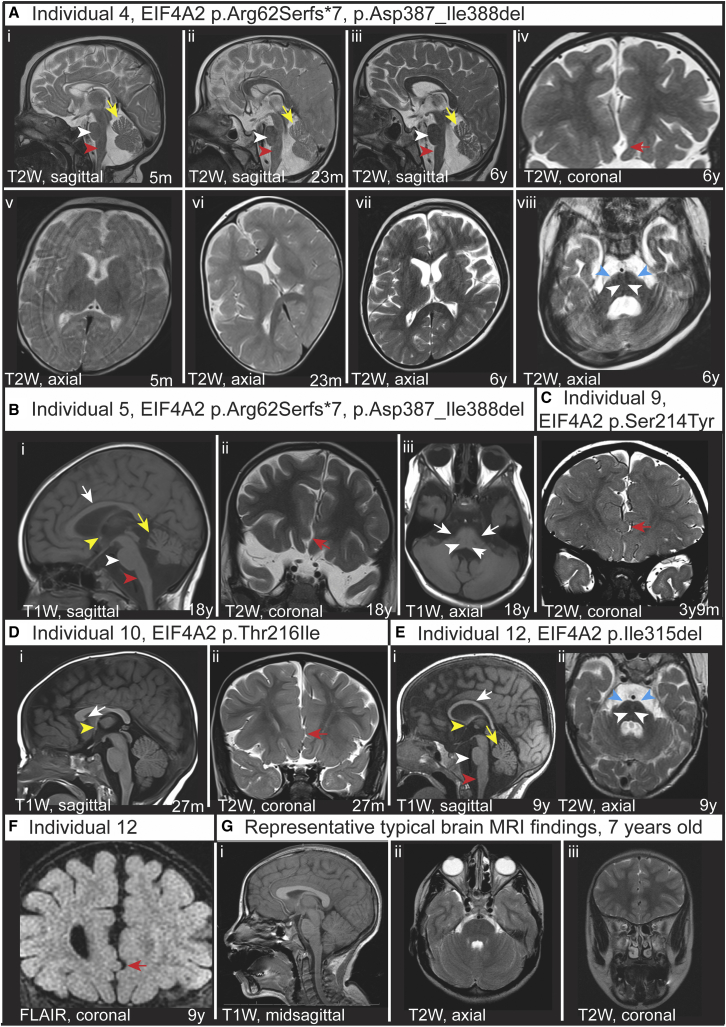
Figure 1.
Neuroanatomical MRI findings in individuals with EIF4A2 variants
(A) MRI images of individual 4 with compound heterozygous variants p.Arg62Serfs∗7 and p.Asp387_Ile388del at 5 months (i, v), 23 months (ii, vi), and 6 years (iii, iv, vii, viii) of age. (i–iii) are sagittal T2-weighted (T2W) images showing a rotated vermis with prominence of the fissures (yellow arrow) consistent with increasing volume loss of the vermis. The pons (white arrowhead) and medulla (red arrowhead) are small in each image. (iv) Coronal T2W image shows subtle interdigitation of the inferior midline frontal lobes (red arrow). (v–vii) are axial T2W images showing increasing sulcal and ventricular size consistent with diffuse parenchymal volume loss. (viii) Axial T2W image shows small pons (white arrowhead) and thin middle cerebellar peduncles (cyan arrowhead).
(B) MRI images of individual 5 with compound heterozygous variants p.Arg62Serfs∗7 and p.Asp387_Ile388del at 18 years of age. (i) Sagittal T1-weighted (T1W) image shows small volume corpus callosum (white arrow) and thin anterior commissure (yellow arrowhead) as well as small pons (white arrowhead), small medulla (red arrowhead), and rotated vermis with prominent fissures (yellow arrow). (ii) Coronal T2W image shows subtle interdigitation of the inferior midline frontal lobes (red arrow). (iii) Axial T1W image shows small pons (white arrowhead) and thin middle cerebellar peduncles (white arrows).
(C) MRI images of individual 7 with variant p.Se214Tyr at 3 years 9 months of age. Interdigitation of the inferior midline frontal lobes is noted (red arrow).
(D) MRI images of individual 10 with variant p.Thr216Ile at 27 months of age. (i) Sagittal T1W image with an incompletely formed rostrum of the corpus callosum (white arrow points to termination of the rostrum, which typically connects to the lamina terminalis) and thin anterior commissure (yellow arrowhead). (ii) Coronal T2W image shows subtle interdigitation of the inferior midline frontal lobes (red arrow).
(E) MRI images of individual 12 with variant p.Ile315del at 9 years of age. (i) Sagittal T1W image shows small volume corpus callosum (white arrow) and thin anterior commissure (yellow arrowhead) as well as small pons (white arrowhead), small medulla (red arrowhead), and mildly rotated vermis with mildly prominent fissures (yellow arrow). (ii) Axial T2W image shows small pons (white arrowhead) and thin middle cerebellar peduncles (cyan arrowhead).
(F) MRI images of individual 12 with variant p.Ile315del at 9 years of age. Coronal FLAIR image shows interdigitation of the inferior midline frontal lobes (red arrow).
(G) Representative typical MRI brain findings in a 7 year-old female. (i) T1W mid-sagittal, (ii) T2W axial, and (iii) T2W coronal.
Table 1.
Clinical and genetic findings of individuals with variants in EIF4A2 (individuals 1–7)
| Individual # | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| EIF4A2 variant information | |||||||
| Inheritance | de novo | autosomal recessive | de novo (mosaic) | autosomal recessive | autosomal recessive | de novo | de novo |
| Genomic (GRCh38) | chr3: 186,783,615C:G | chr3: 186,784,592_186,502,381TTGA:T | chr3: 186,784,619_186,784,620del | chr3: 186,784,674_186,784,675del, 186,789,206_186789211del | chr3: 186,784,674_186,784,675del, 186,789,206_186,789,211del | chr3: 186,786,015:G:T | GRCh38:3: 186,786,220:G:A |
| cDNA (GenBank: NM_001967.3) | c.5C>G | c.109_111del | c.131_132delTC | c.186_187del, c.1161_1166del | c.186_187del, c.1161_1166del | c.481G>T | c.574G>A |
| Protein | p.Ser2Cys | p.Asp37del | p.Leu44Profs∗10 | p.Arg62Serfs∗7, p.Asp387_Ile388del | p.Arg62Serfs∗7, p.Asp387_Ile388del | p.Gly161Trp | p.Gly192Ser |
| Sequencing method | research-based exome sequencing (trio), Sanger confirmed | research-based exome sequencing (single) | clinical-based exome sequencing (trio), Sanger confirmed | research-based exome sequencing (single), Sanger confirmed, Sanger carrier testing of parents and sister | research-based Sanger sequencing after individual 4’s diagnostic exome sequencing | clinical exome sequencing (trio), research Sanger pending | research-based genome sequencing, Sanger confirmed |
| gnomAD frequency | 3.976 × 10−6 | 3.976 × 10−6 | 0 | 0 | 0 | 0 | 0 |
| CADD score | 29.7 | 23.1 | 32 | 33, 20.7 | 33, 20.7 | 32 | 29.9 |
| Polyphen-2 | possibly damaging, 0.880 | – | – | – | – | probably damaging, 1.00 | probably damaging, 0.995 |
| Other genetic variants | – | – | – | – | – | (1) IFIH1 c.1358T>G (maternally inherited), (2) MOCS2 c.26C>T (paternally inherited), (3) PHGDH c.1406G>A (paternally inherited | TSC2 exon 34 c.4225C>T heterozygous, shared with unaffected mother |
| Patient information | |||||||
| Sex | female | female | female | male | female | male | female |
| Ethnicity | European descent | Syrian | European descent | European descent | European descent | European descent | European descent |
| Institution | CHU de Nantes, Straboug, France | University of Leipzig, Institut für Humangenetik | Children’s Hospital of Philadelphia | Carl-Thiem-Klinikum Cottbus, Germany | Carl-Thiem-Klinikum Cottbus, Germany | Texas Children’s Hospital | Manchester Center for Genomic Medicine |
| Family history | – | parents are cousins once removed | brother with laryngomalacia and tracheomalacia | sibling affected with same compound heterozygous variants; parents asymptomatic | sibling affected with same compound heterozygous variants; parents asymptomatic | older sister with simple febrile seizures, otherwise healthy | – |
| Current age | 12 years | unknown | 3 years 7 months | 11 years | 18 years | 2 years | 2 years 10 months |
| Gestational age at birth | full term | unknown | 37 weeks | 38 weeks | 40 weeks | full term | 41 4/7 weeks |
| Birth weight (with percentiles or Z score) | 3.3 kg | unknown | 5 lbs 11 oz (5th%ile) | 3,160 g | 3,640 g | unknown | 2.97 kg (−0.59 SD) |
| Birth length (with percentiles or Z score) | 50 cm | unknown | 19 inches (25th%ile) | 53 cm | 53 cm | unknown | unknown |
| OFC birth (with percentiles or Z score) | 33.5 cm | unknown | 34 cm (30th%ile) | 33.5 cm | 34.5 cm | unknown | unknown |
| Neurologic features | |||||||
| Delayed speech development | + | + | – | +, absent speech | +, absent speech | + | +, absent speech |
| Delayed gross motor development | – | – | + | +, severe, does not sit | + | + | + |
| Delayed fine motor skills | + | – | + | + | + | + | unknown |
| Developmental delay/intellectual disability (IQ if available) | GDD, borderline/mild ID (IQ 78) | mild ID | GDD, mild FSIQ 87 | severe neurodegenerative disease since infancy; severe ID, no IQ measurable | severe neurodegenerative disease since infancy; severe ID, no IQ measurable | mild GDD | severe GDD |
| Seizures | – | – | – | +, myoclonic seizures starting at 1 year | +, myoclonic seizures | +, generalized-tonic clonic, onset 19 months | starting at 3 months of age; epileptic spasms, tonic seizures with clonic component |
| Autism | – | – | – | – | – | – | – |
| Hypotonia | – | + | + | + | + | + | + |
| Structural brain abnormalities on MRI | normal | ND | ND | small anterior commissure, subtle interdigitation of interomedial frontal gyri, rotated small cerebellar hemispheres rotated small vermis, small pons, medulla and middle cerebellar peduncles, small rounded hippocampi, plagiocephaly, progressive mild cerebral volume loss, slightly small optic chiasm and tracts, increasing size of cisterna magna | decreased volume of corpus callosum, small anterior commissure, prominent ventricles, subtle interdigitation of interomedial frontal gyri, rotated small cerebellar hemispheres rotated small vermis, small pons, medulla and middle cerebellar peduncles, small rounded hippocampi, plagiocephaly, small temporal tips, uncovered insula | retrocerebellar arachnoid cysts | short corpus callosum with disproportionately small splenium |
| Behavior abnormalities | – | – | +, impulsivity, inattention, ADHD | – | – | overly friendly in comparison with two older sisters | – |
| Other clinical findings | |||||||
| Failure to thrive | – | – | – | +, early | +, in infancy | – | – |
| Feeding issues | – | – | + | +, G-tube | +, G-tube | – | G-tube, GER |
| Dysmotility | – | – | – | +, difficulty swallowing, constipation | +, difficulty swallowing, constipation | – | – |
| Hypoventilation | – | – | – | + | +, nasal cannula while asleep | – | – |
| Vision/eye abnormalities | – | – | – | visual impairment since 1 year of age (suspected first at 4 months) | blindness impairment since 6 years of age | – | cortical blindness |
| Hearing impairment | – | – | – | +, bilateral deafness, deaf aid | +, since 1 year of age (deaf aid) | – | – |
| Dysmorphic features | epicanthal folds, small ears, anteverted nares, small mouth | microcephaly, short stature | mild synophrys, mild hirsutism on back, bilateral 5th finger clinodactyly | – | no, only secondary due to spasticity | – | – |
| Congenital anomalies | – | – | – | – | – | – | – |
| Muscle findings (biopsy results if applicable) | ND | ND | ND | ND | high variation of the fiber size and round structures in nerve fascicles, expression of MHC-neonatal | – | ND |
| Other | – | ataxia | – | – | estrogen deficiency, no menarche; 2012 skin biopsy (neuropathology Charité Berlin): suspicion of Morbus Cori (glycogenosis 3), Lafora disease (unconfirmed) | had ataxic episode lasting few days in the context of a URI, microcytic anemia, mild lymphopenia, EEGs showing midline epileptiform discharges, intermittent focal slow R > L hemisphere and midline central regions | poor sleep—required melatonin and phenergen as well as trial of Chloral hydrate |
Table shows personal characteristics, genetic variant information, congenital anomalies and neurologic, musculature, feeding, respiratory, vision, and hearing findings for each of the individuals. Abbreviations: CP, cerebral palsy; IQ, intellectual quotient; ID, intellectual disability; GDD, global developmental delay; FSIQ, full scale intelligence quotient; ADHD, attention deficit and hyperactivity disorder; ND, not done; s/p, status/post; hx, history of; G-tube, gastrostomy tube; J-tube, jejunostomy tube; GER, gastroesophageal reflux; PDA, patent ductus arteriosus; EM, electron microscopy; ASD, atrial septal defect.
Table 2.
Clinical and genetic findings of individuals with variants in EIF4A2 (individuals 8–15)
| Individual # | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|
| EIF4A2 variant information | ||||||||
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo |
| Genomic (GRCh38) | ch3: 186,786,515C:A | chr3: 186,786,520A:G | chr3: 186,786,521C:T | chr3: 186,786,602C:T | chr3: 186,787,530_186,787,532del | chr3:186,787,835G:C | GRCh38:3: 186,789,129:G:A | chr3: 186,789,136G:A |
| cDNA (GenBank: NM_001967.3) | c.641C>A | c.646A>G | c.647C>T | c.728C>T | c.945_947delCAT | c.1032G>C | c.1084G>A | c.1091G>A |
| Protein | p.Ser214Tyr | p.Thr216Ala | p.Thr216Ile | p.Thr243Ile | p.Ile315del | p.Leu344Phe | p.Gly362Ser | p.Gly364Glu |
| Sequencing method | exome sequencing (trio), Sanger confirmed | clinical-based exome sequencing (trio) | clinical-based exome sequencing (trio), Sanger confirmed | research-based exome sequencing (trio), Sanger confirmed | exome sequencing (trio), Sanger confirmed | research-based exome sequencing (trio), Sanger confirmed | research-based genome sequencing, Sanger confirmed | clinical-based exome sequencing (trio) |
| gnomAD frequency | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CADD score | 28.7 | 26.4 | 26.5 | 26.1 | 22.2 | 25.4 | 33 | 28.5 |
| Polyphen-2 | probably damaging, 1.00 | probably damaging, 0.987 | probably damaging, 1.00 | probably damaging, 0.993 | – | probably damaging, 0.996 | probably damaging, 0.994 | possibly damaging, 0.770 |
| Other genetic variants | (1) ARFGEF3 homozygous c.1391G>T (p.Gly464Val); (2) GLDC homozygous c.2607C>A (p.Pro869Pro) | – | PIEZO2, c.4807dupA (p.Arg1603LysfsX27); shared with unaffected mother | – | – | (1) IPO4 C2294G>A (p.Arg765His); (2) IPO4 c.1337C>T (p.Ser446Leu) | – | – |
| Patient information | ||||||||
| Sex | male | male | male | female | male | male | female | male |
| Ethnicity | Arab | European descent | European descent | European descent | European descent | Lithuanian | European descent | European descent |
| Institution | Sheba Medical Center, Israel | Kaiser Permanente San Francisco | Cook Children’s Hospital, Texas | Federico II University Hospital | Seattle Children’s Hospital, Washington | Boston Children’s Hospital | Chelsea and Westminster, London, UK | Bambino Gesù Children’s Hospital, Rome, Italy |
| Family history | – | – | siblings with ADHD, dyslexia, tics; maternal aunt with CP, ID, hydrocephalus, and seizures | – | – | father with PDA, diagnosed at 5 years old | non-consanguineous parents, both have mild learning difficuties, maternal dyslexia, strong maternal Fhx of epilepsy | – |
| Current age | 3 years 9 months | 3 years | 48 months | 9.5 years | 9 years | 9 years | 3 years 7 months | 5 years |
| Gestational age at birth | full term | 38 6/7 weeks | 39 weeks | 41 weeks | 39+1 week | 37 weeks | full term | unknown |
| Birth weight (with percentiles or Z score) | 3kg (50th%ile) | 2,435 g (3.81%ile) | 3.07 kg (28th%ile, Z score −0.58) | 2,810 g | unknown | 2,660 g | unknown | 3,160 g |
| Birth length (with percentiles or Z score) | unknown | 45.7 cm (5.6%ile) | 53.3 cm (96th%ile, Z score +1.8) | 49 cm | unknown | 48 cm | unknown | 50 cm |
| OFC birth (with percentiles or Z score) | 34 cm (50th%ile) | 34.3 cm (22.42%ile) | 35 cm (66th%ile, Z score +0.42) | 33 cm | unknown | unknown | unknown | 34 cm |
| Neurologic features | ||||||||
| Delayed speech development | +, absent speech | + | + | +, absent speech | +, absent speech | + | +, absent speech | +, absent speech |
| Delayed gross motor development | + | + | + | +, severe | + | + | + | +, spastic tetraparesis |
| Delayed fine motor skills | +, severe | + | + | + | – | + | – | + |
| Developmental delay/intellectual disability (IQ if available) | severe ID | GDD | GDD | severe GDD, ID | GDD | severe, GDD | severe GDD | ID |
| Seizures | + | – | Lennox Gastaut syndrome with tonic seizures and atypical absence seizures | +, severe and present in infancy, sitting position never acquired | +, focal, well controlled; starting at 3 years | +, West syndrome (infantile spasms, onset 6months) | starting at 5 months: infantile spasms and myoclonic seizures, diagnosed with West syndrome; now with tonic, focal, gelastic, and absence seizures and diagnosed with Lennox-Gastaut syndrome | +, EEG at birth showed pathological electrical activity |
| Autism | + | + | not tested | – | + | – | – | – |
| Hypotonia | +, mild | +, history of hypotonia, now possible hypertonia | +, diffuse | +, severe and present in infancy, sitting position never acquired | +, central | + | present in infancy, now with hypertonia of extremities | +, severe |
| Structural brain abnormalities on MRI | subtle interdigitation of inferomedial frontal gyri | scattered foci of bifrontal subcortical and hazy periatrial signal abnormality, nonspecific, possible sequela of prior injury | decreased volume of corpus callosum, small anterior commissure, subtle interdigitation of the inferomedial frontal gyri | thinned corpus callosum, enlarged lateral and 3rd ventricles | normal | small temporal tips and uncovered insula, decreased volume but fully formed corpus callosum; small anterior commissure, decreased cerebral volume predominantly white matter and most prominent right greater than left frontal lobe and peritrigonal regions with probably secondary decreased thalamic and cerebral peduncle volume, gliosis in the occipital periventricular white matter, interdigitation of the inferomedial frontal gyri, small fornices, small optic nerves, chiasm and tracts, mildly rotated, slightly small vermis, mild prominence of the cerebellar and vermian fissures suggesting gray matter volume loss, small pons, medulla and middle cerebellar peduncles | normal | hypoplastic cerebellar vermis, thin corpus callosum, hypoplastic pons |
| Behavior abnormalities | – | head banging, repetitive motions | – | very limited interactions | episodes of rage, ABA therapy | – | unsettled behavioral episodes | |
| Other clinical findings | ||||||||
| Failure to thrive | – | – | – | + | +, in infancy | – | – | – |
| Feeding issues | – | +, hx of NG after birth | + | +, G-tube | +, G-tube | +, G-tube | + | +, no suckling reflex, G- and J-tubes |
| Dysmotility | – | – | +, dysphagia, constipation | feeding difficulties, dysphagia | + | – | +, dysphagia, constipation | – |
| Hypoventilation | – | – | – | – | – | +, tracheostomy in place | – | +, tracheostomy |
| Vision/eye abnormalities | – | – | bilateral exotropia | central blindness | – | – | – | cortical blindness |
| Hearing impairment | – | – | – | – | – | – | – | – |
| Dysmorphic features | long palpebral fissures, small hands and feet | macrocephaly, hx sagittal craniosynostosis s/p repair | relative macrocephaly | microcephaly | – | – | micrognathia, bilaterally inverted nipples | epicanthus, upslanting palpebral fissures, broad nasal root, short philtrum, curved upper lip |
| Congenital anomalies | – | sagittal craniosynostosis s/p repair | penile chordee with phimosis | – | – | PDA s/p closure | – | ostium secundum ASD |
| Muscle findings (biopsy results if applicable) | ND | ND | ND | normal | complex I defect, muscle structure normal on histochemical and EM analysis | type I and II atrophic myocytes, ∼80% of myocytes express lipid globules compilations | ND | ND |
| Other | ataxia | sialorrhea | episodes of dysautonomic storm | – | severe dysautonomia (severe GI dysmotility, heart rate, temperature dysregulation), migraine headaches; episodic elevation of NH4 and ALT levels | – | – | osteopenia |
Table shows personal characteristics, genetic variant information, congenital anomalies and neurologic, musculature, feeding, respiratory, vision, and hearing findings for each of the individuals. Abbreviations: CP, cerebral palsy; IQ, intellectual quotient; ID, intellectual disability; GDD, global developmental delay; FSIQ, full scale intelligence quotient; ADHD, attention deficit and hyperactivity disorder; ND, not done; s/p, status/post; hx, history of; G-tube, gastrostomy tube; J-tube, jejunostomy tube; GER, gastroesophageal reflux; PDA, patent ductus arteriosus; EM, electron microscopy; ASD, atrial septal defect.
Molecular modeling
The EIF4A2 protein structure model is built on the crystal structure of EIF4A1 (PDB: 5ZC9), as EIF4A2 and EIF4A1 share high sequence conservation. The protein structure illustration and amino acid mutations are generated by PyMOL (The PyMOL Molecular Graphics System, version 2.0 Schrödinger).
Drosophila melanogaster stocks and maintenance
All the fruit fly stocks used in this study were reared in standard cornmeal- and molasses-based fly food at room temperature (20°C–21°C) unless otherwise noted. The fruit fly stocks used in the study were either obtained from stock centers (Bloomington Drosophila Stock Center [BDSC], Vienna Drosophila Research Center [VDRC], and FlyORF) or generated at the Jan and Dan Duncan Neurological Research Institute. We generated transgenic fly alleles as previously described31 by utilizing the pUASg-HA-attB vector32 to express the human wild-type (WT) EIF4A2 and variant cDNAs with a C-terminal hemagglutinin (HA) tag under the control of upstream activating system (UAS) elements by Gateway LR Cloning (LR Clonase II, Thermo Fisher Scientific, Cat #11791020). To generate the EIF4A2 variants, we utilized the human full-length cDNA of the most abundant isoform of EIF4A2 (GenBank: NM_001967.3). EIF4A2 p.Gly364Glu (c.1091G>A [GenBank: NM_001967.3]), EIF4A2 p.Leu344Phe (c.1032G>C [GenBank: NM_001967.3]), EIF4A2 p.Thr243Ile (c.728C>T [GenBank: NM_001967.3]), and EIF4A2 p.Thr216Ile (c.647C>T [GenBank: NM_001967.3]) were generated by Q5 site-directed mutagenesis (New England Biolabs, Cat #M0491S) in the pDONR221 Gateway compatible donor vector. The constructs were confirmed by Sanger sequencing. Primers for site-directed mutagenesis and Sanger sequencing are listed in Table S2. Human EIF4A2 WT and variant cDNAs were inserted into the chromosome-3 VK33 (PBac{y[+]-attP}VK00033) docking site by φC31-mediated recombination for fruit fly transgenesis.32
Transgenic UAS fly alleles generated in this study include UAS-EIF4A2-WT-HA, UAS-EIF4A2-Gly364Glu-HA, UAS-EIF4A2-Leu344Phe-HA, UAS-EIF4A2-Thr243Ile-HA, and UAS-EIF4A2-Thr216Ile-HA. Fly alleles available from the stock centers include the following: UAS-eIF4A (FlyORF-F001132), UAS-eIF4A RNAi (VDRC#42201), UAS-eIF4A RNAi (BDSC#33970), UAS-empty-VK33 (BDSC#9750), UAS-dpp GFP (BDSC#53716), Nubbin-GAL4 (BDSC#51635), and Elav-GAL4 (BDSC#8765). The GMR-GAL4 was obtained from Dr. Hugo J. Bellen.33
Larval imaginal discs immunostaining and confocal microscopy
Fruit fly larval brains or wing discs were dissected from third instar wandering larvae in ice-cold 1X-PBS and fixed in 4% paraformaldehyde for 20–30 min at room temperature. The tissues were washed four times in Tri-PBS (1X-PBS + 0.2% Triton X-100) with 1% BSA for 15 min each followed by incubation in blocking solution (Tri-PBS with 0.1% BSA and 8% normal donkey serum) for 30 min. Primary antibodies, rat anti-HA (1:50, clone 3F10, Millipore Sigma, Cat# 11867423001), mouse anti-elav (1:100, Developmental Studies Hybridoma Bank, Cat# 9F8A9), and rabbit anti-Phospho-Smad1/5 (pMad) (1:75, Cell signaling technology, Cat# 9516) were diluted in blocking solution and added to the tissues and incubated overnight at 4°C. The tissues were rinsed three times in Tri-PBS with 1% BSA for 15 min each followed by incubation in blocking solution for 30 min. The secondary antibodies, donkey anti-rat IgG antibody (Cy3) (1:300, Jackson Immunoresearch, Cat# 712-165-153), Alexa Flour 488 Affinipure donkey anti-rabbit IgG (H + L) (1:300, Jackson Immunoresearch, Cat# 711-545-152), and Alexa Flour 488 Affinipure donkey anti-mouse IgG (H + L) (1:300, Jackson Immunoresearch, Cat# 712-545-151), were diluted in blocking solution and added to the tissues for 90 min incubation at room temperature on a rocker. After removing the secondary antibody, tissues were washed three times in Tri-PBS with 1% BSA for 15 min each. The tissues were then rinsed in 1X-PBS followed by incubation in 406-diamidino-2-phenylindole dihydrochloride (DAPI, 1 mg/mL, Cayman Chemical, Cat# 14285) for 30 min at room temperature. After removing DAPI, a final wash was completed with 1X-PBS for 15 min at room temperature. The tissues were mounted in Prolong Glass anti-fade mountant (Thermo Fisher Scientific, Cat#36984). Images were acquired on a Leica S-P8 laser-scanning confocal microscope. The same settings for laser power and detector gain were used for all genotypes. Third instar larval brain images were acquired as a z stack with a z-step of 1 μm and line average of four at 400 Hz with a 20× objective at 1,024 × 1,024 pixel resolution. Maximum intensity projections were created from the z stack in ImageJ. All images were processed and assembled with ImageJ and Adobe Illustrator.
Fruit fly behavioral assays
For the climbing assay, 10–11-day-old flies of both genders were anesthetized 24 h prior to being tested and two to three flies were housed in food-containing vials at room temperature. At the time of assessment, these flies were transferred to a clear graduated cylinder with a 15 cm mark. The flies were tapped three times to the bottom of the cylinder to examine negative geotaxis (climbing upward). The cutoff time to reach the 15 cm mark was 30 s. A total of 50–75 flies were tested for each genotype. Crosses for the climbing assay were set up at 25°C and the assay was performed at 20°C–21°C.
Adult fruit fly wing mounting
Adult wings were dissected from 4- to 5-day-old flies and washed in isopropanol for 20–30 s at room temperature. The wings were mounted with CMCP-10 High Viscosity Mountant (D/S259) (Electron Microscopy Sciences, Cat#18004-02). Wing images were taken with the Leica MZ16 stereomicroscope. Images were processed and assembled with Adobe Photoshop CS5.1 and Adobe illustrator. Crosses were set up at 20°C–21°C.
Adult eye imaging and nail polish imprinting
Anesthetized flies were placed on a glass slide and the eye images were taken with the Leica MZ10F stereomicroscope. Ommatidia defects in the adult eyes were analyzed with nail polish imprinting.34,35 Adult heads were dissected, and a drop of transparent nail polish was poured on the heads. After 2 min, the layer of nail polish was removed from the head and allowed to dry at room temperature. Before drying out completely, the nail polish on the eye field was carefully sliced off and mounted on a glass slide with a coverslip. Images of the nail polish imprints were taken on the Zeiss 880 Airyscan confocal microscope. The images were processed with ZEN blue and Adobe Photoshop CS5.1 and assembled with Adobe Illustrator. Crosses were set up at 20°C–21°C.
Fruit fly gene expression analysis by quantitative real-time PCR
Total RNA from ten pharate heads was extracted with Zymo Direct-zol RNA Miniprep (Zymo Research, Cat# R2052) as per manufacturer’s instructions. RNA was resuspended in nuclease-free water and quantified with the Nanodrop (Thermo Fisher Scientific-Nanodrop One C). We used an equal amount (1,000 ng) of DNase-treated RNA for each genotype to synthesize cDNA by using BioRad iScript Reverse Transcription supermix as per manufacturer’s instructions (Cat#1708840). Quantitative real-time PCR was performed with the BioRad SsoAdvanced Universal SYBR-Green supermix (Cat#1725274) and a BioRad CFX96 Touch Real-Time PCR detection system. The experiments were carried out in triplicate for each dataset and repeated for three biological replicates. The relative change in gene expression was determined by the Livak method and fold changes were calculated with the 2−ΔΔCT formula.36 Fly eif4A expression levels were normalized to the expression of the endogenous reference gene rps17 and plotted as fold-change relative to the control. Crosses were set up at 20°C–21°C. Primers for quantitative real-time PCR analysis are listed in Table S2.
Results
Identification of EIF4A2 variants in individuals with global developmental delay, intellectual disability, hypotonia, and epilepsy
An international collaboration through Matchmaker Exchange37,38,39 facilitated the identification of 15 individuals with extremely rare de novo mono-allelic or inherited bi-allelic variants in EIF4A2. The variants from all probands were identified through exome sequencing and a majority (11/15) were confirmed with Sanger sequencing (Tables 1 and 2). Twelve of the individuals harbored de novo mono-allelic (I:1, I:3, and I:6–I:15) variants, while three had bi-allelic variants (I:2, I:4, and I:5). Ten of the 12 de novo mutations were missense, while one was frameshift and one deletion of a single amino acid. A pair of siblings (I:4 and I:5) had the same bi-allelic frameshift deletions, each variant inherited from asymptomatic parents, and another individual (I:2) was homozygous for a single amino acid deletion inherited from asymptomatic consanguineous first cousin parents. The combined annotation-dependent depletion (CADD) score for the variants ranged from 22.2 to 33.40 Thirteen out of 15 of the variants were absent from the Genome Aggregation Database (gnomAD). Two of the variants, p.Ser2Cys (I:1) and p.Asp37del (I:2), had a frequency of 3.976 × 10−6 (1/251,478) in gnomAD; both variants were identified in individuals with mild clinical phenotypes in the cohort: p.Asp37del was identified as a homozygous variant.
In a statistical model of de novo variants for autism spectrum and ID disorders (ASD/ID), the model identified EIF4A2 as one of ∼1,000 genes significantly lacking functional variation in non-ASD/ID-affected individuals but are enriched with de novo variants in ASD/ID-affected individuals.41 Furthermore, analysis of gnomAD revealed that EIF4A2 has a high probability of LOF intolerance (pLI = 1.0), as 23.1 LOF variants were expected given the gene size and GC content but only one LOF variant was observed.42 EIF4A2 is also a highly constrained gene with a Z score of 3.89, suggesting intolerance to missense variation. Together, these findings provide strong statistical evidence that the de novo mono-allelic and inherited bi-allelic EIF4A2 variants cause the neurodevelopmental phenotypes observed in these 15 individuals.
All individuals in the cohort had GDD and ID, while 14 out of 15 had hypotonia (Tables 1 and 2). Epilepsy was a prevalent finding (12/15, I:4–I:8 and I:10–I:15), including infantile spasms reported in three and myoclonic epilepsy reported in four of the individuals. No consistent antiepileptic medication regimen was found to be effective for seizure control (Table S1). Ten out of 15probands had brain structural changes detected by MRI, five of which were closely reviewed and compared by a pediatric radiologist (I:4, I:5, I:8, I:10, and I:12). Brain MRIs were notable for decreased volume of the corpus collosum (6/10), small anterior commissures (4/10), small vermis (4/10), small pons, medulla and middle cerebellar peduncles (3/10), and slightly small optic chiasms and tracts (3/10) (Figure 1 and Tables 1 and 2). Delayed speech development was present in 14 out of 15 of the individuals with absent expressive language in eight individuals. Seven of the 15 individuals were gastrostomy tube dependent and four had respiratory complications with two requiring tracheostomies. Vision was impaired in six of the individuals, including three individuals diagnosed with cortical blindness. Gastrointestinal (GI) dysmotility characterized by constipation and dysphagia was present in six out of 15 of the individuals. Three individuals had a history of ataxia and one had severe dysautonomia. Dysmorphic facial features were present in nine of the individuals and included epicanthal folds, long palpebral fissures, short philtrum, and micro- or macrocephaly (Figure 2 and Tables 1 and 2).
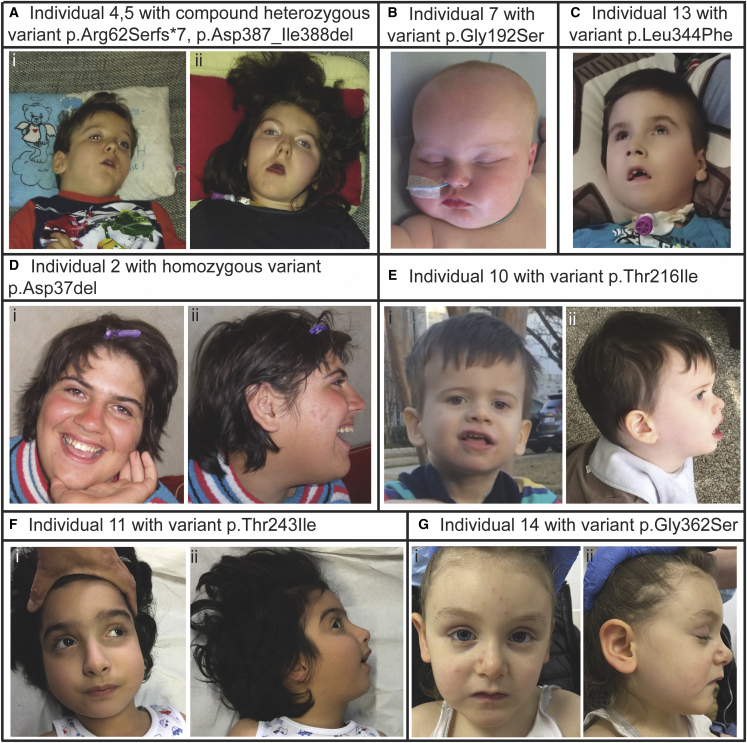
Figure 2.
Photographs of individuals with variants in EIF4A2
Representative photographs of individuals with compound heterozygous or de novo EIF4A2 variants.
(A) Siblings with compound variants p.Arg62Serfs∗7 and p.Asp387_Ile388del with individual 4 in (i) and individual 5 in (ii).
(B) Individual 7 with de novo p.Gly192Ser variant.
(C) Individual 13 with de novo p.Leu344Phe variant, (i) frontal and (ii) profile images.
(D) Individual 2 with autosomal recessive p.Asp37del variant, (i) frontal and (ii) profile images.
(E) Individual 10 with de novo p.Thr216Ile variant, (i) frontal and (ii) profile images.
(F) Individual 11 with de novo p.Thr243Ile variant, (i) frontal and (ii) profile images.
(G) Individual 14 with de novo p.Gly362Ser variant, (i) frontal and (ii) profile images.
In vivo functional analysis of EIF4A2 missense variants in fruit flies
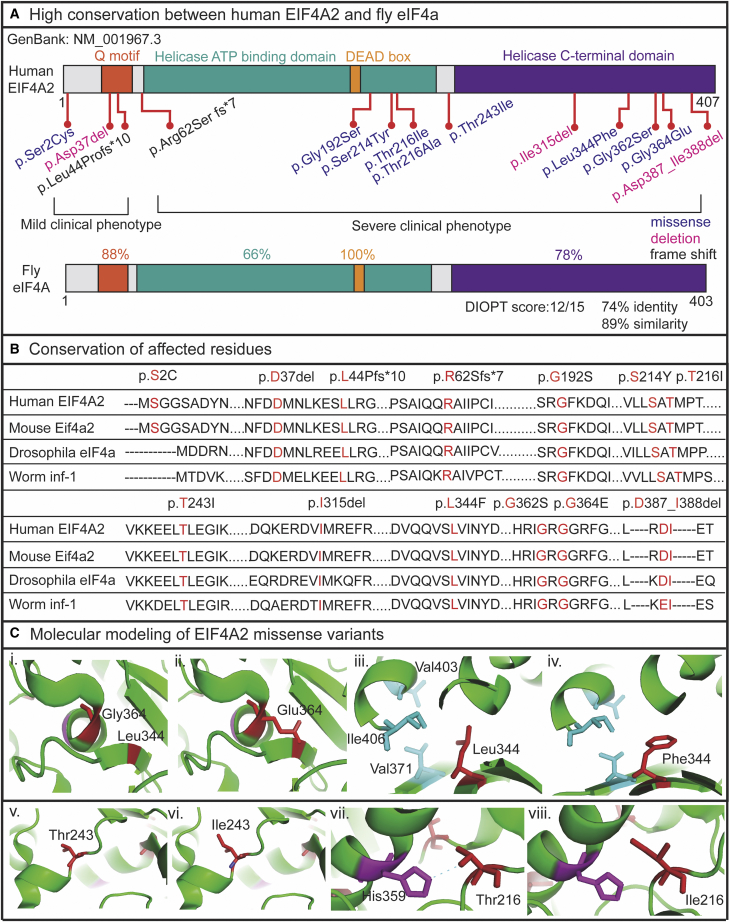
To validate the pathogenicity of the EIF4A2 variants in vivo, we used fruit flies to model the human variants. The fly homolog of EIF4A2 is eIF4A and the fly protein shows an overall 74% identity and 89% similarity with the human protein (Figure 3A). The four domains of eIF4A share significant homology with EIF4A2, including 88% identity for the Q motif, 100% identity for the DEAD-box, 66% identity for the helicase ATP-binding domain, and 78% identity for the helicase C-terminal domain (Figure 3A). Conservation analysis of affected residues reveals that the affected residues in the Q motif, p.Asp37 and p.Leu44, are well conserved in mouse, fruit fly, and worm (Figure 3B). The affected residues in the helicase ATP-binding domain, p.Gly192, p.Ser214, p.Thr216, and p.Arg62, which is near this domain, are conserved across species. Similarly, the affected residues in the helicase C-terminal domain, p.Ile315, p.Leu344, p.Gly362, p.Gly364, and p.Thr243, which is near this domain, are conserved across species. In contrast, the residues p.Asp387 and p.Ile388 are conserved in mouse and fruit flies but not in worms (Figure 3B).
Figure 3.
The EIF4A2 variants are well conserved across different species
(A) EIF4A2 variants are positioned in the corresponding protein domains (missense variants are shown in purple, deletion variants are shown in pink, and the frameshift variants are shown in black). The fruit fly homolog, eIf4A shows 74% identity and 89% similarity with 88% matching in Q motif, 66% matching in helicase ATP-binding domain, 100% matching in DEAD box, and 78% matching in helicase C-terminal domain.
(B) Conservation analysis of affected residues in different species are shown.
(C) Molecular modeling of four of the missense variants are shown and the critical residues are colored in red. (i, iii, v, vii) shows the WT residues and (ii, iv, vi, viii) shows the variant residues.
Molecular modeling was completed for each of the missense variants. Three of the variants, p.Leu344Phe (I:12), p.Gly362Ser (I:13), and p.Gly364Glu (I:14), are located in the C-terminal domain (CTD) and a fourth variant, p.Thr243Ile (I:11), is located adjacent to the CTD (Figure 3A). Three variants, p.Gly192Ser (I:7), p.Ser214Tyr (I:8), and p.Thr216Ile (I:10), are located in the N-terminal ATP-helicase domain (NTD) (Figure 3A). The residue p.Thr216 forms a hydrogen bond with the CTD residue p.His359 (Figure 3C). The variant p.Thr216Ile disrupts the polar interaction between p.His359 and p.Thr216, which is predicted to perturb the interaction dynamics between the NTD and CTD (Figure 3C). The variant p.Ser214Tyr introduces an aromatic ring, which may disrupt the interaction between p.His359 and p.Thr216 (Figure S1). Residue p.Gly192 is located at the binding interface between EIF4A2 and RNA (Figure S1). The variant p.Gly192Ser increases the interaction between EIF4A2 and RNA, which is predicted to impact the enzymatic activity of EIF4A2. Residue p.Leu344 is located in a hydrophobic core composed of p.Val403, p.Ile406, and p.Val371 (Figure 3C). The variant p.Leu344Phe introduces a bulk side chain into this core (Figure 3C) and may destabilize it. p.Gly362 and p.Gly364 are both located in a helix within the CTD (Figures S1 and 3C); the variants p.Gly362Ser and p.Gly364Glu both introduce bulky side chains into the compact helix and may disrupt its conformational stability (Figures S1 and 3C). Finally, residue p.Thr216 is located on the protein surface (Figure 3C). The hydrophobic side chain of the variant p.Thr243Ile is predicted to potentially affect the hydroshell of the protein (Figure 3C). Together, the molecular modeling suggests the missense variants may perturb EIF4A2 protein function by disrupting protein conformation and interactions with RNA.
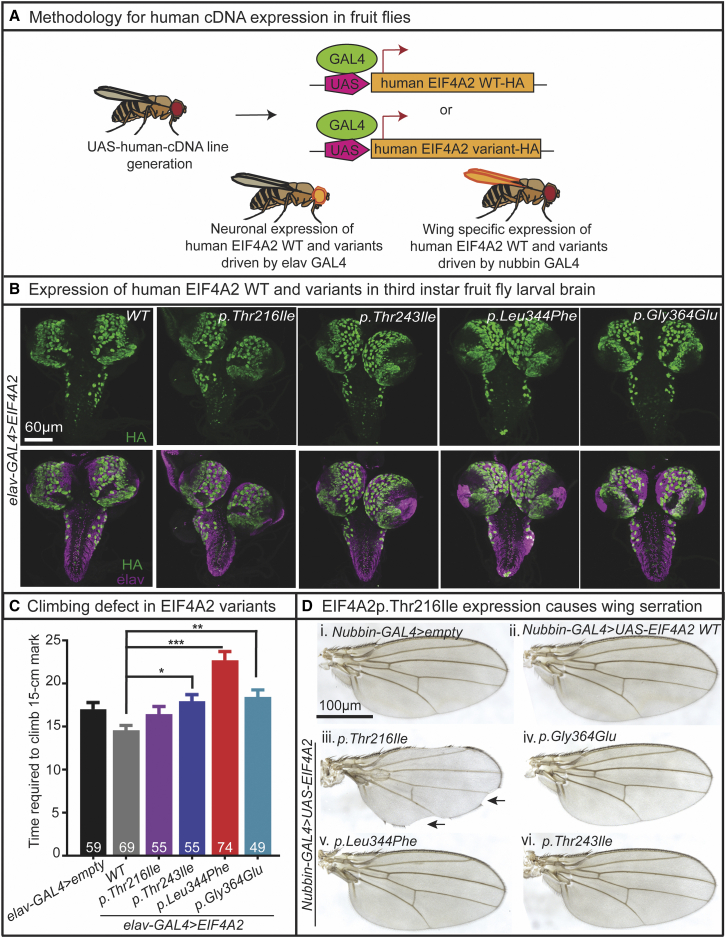
To study the functional consequences of EIF4A2 variants in vivo, we selected four of the missense mono-allelic variants to assess a dominant negative effect in a WT fly genetic background. We generated UAS-EIF4A2-WT-HA, UAS-EIF4A2-p.Gly364Glu-HA, UAS-EIF4A2-p.Leu344Phe-HA, UAS-EIF4A2-p.Thr243Ile-HA, and UAS-EIF4A2-p.Thr216Ile-HA transgenic fly alleles. We used the UAS-GAL4 expression system to express these variants and WT EIF4A2 cDNAs under the spatiotemporal regulation of the transactivator protein GAL4 (Figure 4A). We used a pan-neuronal driver on the second chromosome, elav-GAL4, to express C-terminal HA-tagged WT and variant EIF4A2 cDNAs in neurons. Neuronal expression of the HA-tagged cDNAs were confirmed in the third instar larval brain by immunostaining (Figure 4B). To determine whether the neuronal expression of EIF4A2 WT and missense variants cause motor defects, we performed a climbing assay to assess negative geotaxis. The normal behavior of the flies is to climb upward, and increased time to climb represents a motor defect. We found that EIF4A2 WT-expressing flies had motor function similar to controls, whereas EIF4A2 p.Gly364Glu-, p.Leu344Phe-, and p.Thr243Ile-expressing flies exhibited climbing defects (Figure 4C). In contrast, EIF4A2 p.Thr216Ile-expressing flies did not show any defects in climbing ability (Figure 4C).
Figure 4.
UAS-GAL4-mediated expression of EIF4A2 variants results in behavioral and anatomical defects in fruit flies
(A) Methodology for expressing UAS-EIF4A2-WT and variant cDNAs in the fruit fly CNS and wings using elav-GAL4 and Nubbin-GAL4 is shown.
(B) elav-GAL4-mediated expression of EIF4A2-WT and four missense variants are shown in the third instar larval brain using immunostaining against HA (green). Elav (purple) marks the neurons in the third instar larval brain.
(C) Neuronal expression of EIF4A2 p.Thr243Ile, EIF4A2 p.Leu344Phe, and EIF4A2 p.Gly364Glu resulted in climbing defect compared to the expression of EIF4A2 WT cDNA. Two to three flies were housed in clear cylinders and then tapped three times to the bottom of the cylinder to examine negative geotaxis (climbing upward). The cutoff time to reach the 15 cm mark was 30 s. Flies that failed cross the 15 cm mark were given the score 30 s. One-way ANOVA followed by Tukey’s post hoc test was performed for the statistical analysis. Data shown as mean ± SE of mean (SEM) with the sample size of total number of male and female flies shown in figure. Significance shown as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Nubbin-GAL4-mediated wing-specific (iii) expression of EIF4A2 p.Thr216Ile resulted in wing margin serrations (black arrows) compared to the (i) empty and (ii) EIF4A2 WT controls. Expression of (iv–vi) EIF4A2 p.Thr243Ile, EIF4A2 p.Leu344Phe, and EIF4A2 p.Gly364Glu did not induce any wing specific phenotypes.
To assess whether overexpression of EIF4A2 WT and variants affected developmental processes, we used Nubbin-GAL4 to drive expression of the UAS-EIF4A2 cDNA alleles in the third instar larval wing pouch and pupal wing blade.43,44 We found that EIF4A2 WT expression did not perturb the development of adult wings. In contrast, EIF4A2 p.Thr216Ile caused wing margin serrations (Figure 4Diii), which is similar to eIF4A GOF mechanisms.23 However, expression of EIF4A2 p.Gly364Glu, p.Leu344Phe, and p.Thr243Ile did not induce any wing-specific phenotypes (Figures 4Div and 4Dvi). As eIF4a GOF interrupts the dpp signaling in wing discs,23 we tested the expression of pMad as a readout for dpp signaling in Nubbin-GAL4-driven larval wing discs to see the effect of EIF4A2 p.Thr216Ile expression. We found that similar to the eIF4a overexpression, EIF4A2 p.Thr216Ile expression also reduced the expression of pMad in the wing pouch (Figures S2A and S2B). Both eIF4A and EIF4A2 p.Thr216Ile overexpression perturb the adult wing development leading to blister formation and reduced wing size with variable wing margin serration (Figure S2C). Together, the elav-GAL4 and Nubbin-Gal4 studies reveal that overexpression of EIF4A2 p.Gly364Glu, p.Leu344Phe, and p.Thr243Ile variants in neurons and expression of EIF4A2 p.Thr216Ile in the developing wing have dominant effects. This is consistent with the autosomal dominant pattern identified for these mono-allelic missense variants in our cohort.
EIF4A2 variants modify the dpp-induced eye phenotypes and fail to rescue lethality as a result of complete loss of fly eIF4A function
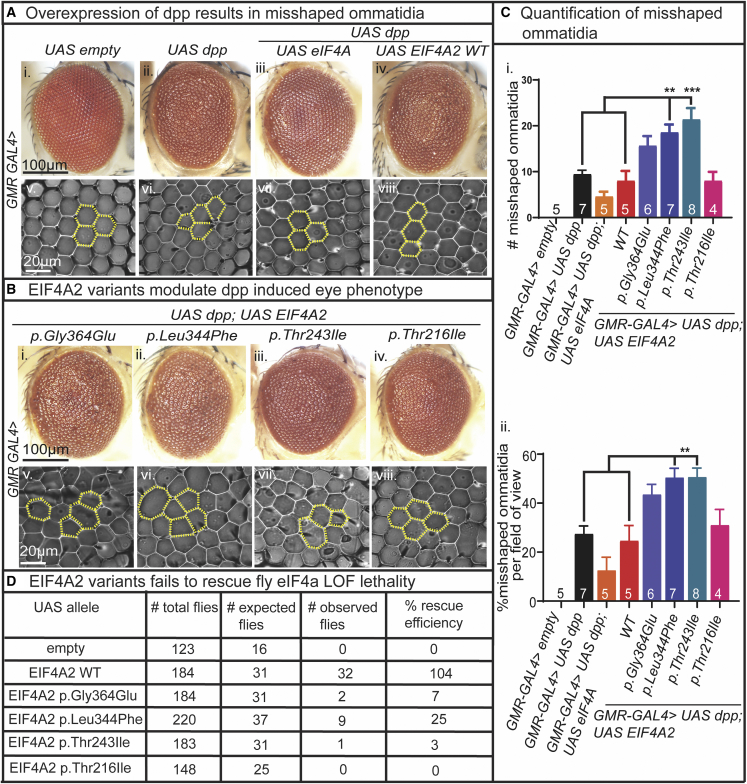
The fruit fly homolog eIF4A negatively regulates dpp signaling. Prior studies showed increased dpp signaling in eIF4A LOF fly embryos, whereas eIF4A GOF recapitulated dpp LOF phenotypes.23 In the adult fruit fly eye, dpp overexpression disrupts ommatidia formation, leading to a roughened eye surface that is rescued with increased eIF4A expression.23 In contrast, expression of eIF4A LOF fly alleles in the background of dpp overexpression caused lethality and severe eye roughening in fly escapers.23 Together, these findings revealed that eIF4A is a critical negative-regulator of dpp signaling and eIF4A LOF causes a toxic upregulation of dpp signaling.
To determine whether the EIF4A2 missense variants cause LOF, we expressed EIF4A2 WT and variant cDNAs in the background of GMR-GAL4-mediated overexpression of dpp in adult fly eyes. GMR-GAL4-mediated overexpression of dpp resulted in a roughened eye surface with misshaped and disorganized ommatidia (Figures 5Aii and 5Avi) compared to the typical hexagonal arrangement of ommatidia in the control (Figures 5Ai and 5Av). Expression of either fly eIF4A WT cDNA (Figures 5Aiii and 5Avii) or human EIF4A2 WT cDNA (Figures 5Aiv and 5Aviii) in the background of dpp overexpression did not alter the dpp-associated roughened eye phenotype (Figures 5Ci and 5Cii). However, we found that GMR-GAL4 mediated overexpression of p.Leu344Phe (Figures 5Bii and 5Bvi) and p.Thr243Ile (Figures 5Biii and 5Bvii) exacerbated the dpp-induced ommatidial disorganization with a significant increase in the number and percentage of misshaped ommatidia (Figures 5Ci and 5Cii). This finding suggests these missense variants cause strong EIF4A2 LOF and perturb eye development. In contrast, the GMR-GAL4-induced expression of EIF4A2 p.Gly364Glu and p.Thr216Ile did not cause any significant changes to the dpp-induced eye phenotypes.
Figure 5.
EIF4A2 variants differently modulate dpp-induced eye phenotypes and failed to rescue the fly eIF4A LOF lethality
(A) Overexpression of dpp in the fruit fly eye using GMR-GAL4. (i) Empty control eye shows smooth eye surface and (v) hexagonal arrangement of ommatidia in the imprint. (ii) Expression of dpp results in roughened eye surface and (vi) disorganized ommatidial arrangement. (iii) Expression of fly eIF4A in the background of GMR-GAL4>dpp shows slight reduction in roughened eye surface (iv) and better ommatidial arrangement. (v) Expression of EIF4A2 WT in the background of GMR-GAL4>dpp did not make any changes to the roughened eye surface (iv) and ommatidial arrangement.
(B) Expression of EIF4A2 variants in the background of GMR-GAL4>dpp. (i) Expression of EIF4A2 p.Gly364Glu did not make significant changes to the roughened eye surface and (v) disorganized ommatidial arrangement in the imprint. (ii and iii) Expression of EIF4A2 p.Leu344Phe and EIF4A2 p.Thr243Ile show roughened eye surface (vi and vii) and exacerbates the disorganized ommatidial arrangement. (iv) Expression of EIF4A2 p.Thr216Ile did not made any changes to the roughened eye surface (viii) and ommatidial arrangement.
(C) Quantification of misshaped ommatidia. (i and ii) Expression of EIF4A2 p.Leu344Phe and EIF4A2 p.Thr243Ile shows significant increase in the total number of misshaped ommatidia and percentage of misshaped ommatidia per field of view compared to the GMR-GAL4>dpp and GMR-GAL4>dpp; EIF4A2 WT controls. One-way ANOVA followed by Tukey’s post hoc test was performed for the statistical analysis. Data shown mean ± SEM with sample size of total number of male and female flies shown in figure. Significance shown as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Knockdown of eIF4A in the GMR domain results in pupal lethality. Expression of human EIF4A2 WT in this background results in complete rescue of pupal lethality, whereas the variants fail to rescue the lethality.
Finally, we performed in vivo rescue experiments with eIF4A RNAi fly alleles to determine the functional nature of the human EIF4A2 variants in the background of absent fly eIF4A. Previous studies showed that complete loss of eIF4A function is embryonic lethal45 and tissue-specific RNAi-mediated knockdown of eIF4A resulted in either embryonic or pupal lethality.45 We used two different UAS-eIF4A RNAi lines to verify the eIF4A LOF phenotypes. Expression of eIF4A RNAi (VDRC#42201) with both GMR-GAL4 and Nubbin-GAL4 at 25°C resulted in embryonic lethality (Figure S3A). In contrast, expression of eIF4A RNAi (BDSC#33970) with GMR-GAL4 and Nubbin-GAL4 at 25°C resulted in late pupal death (Figure S3A). When we performed these experiments at 20°C, GMR-GAL4-driven expression of both eIF4A RNAi alleles (GMR-GAL4>eIF4A RNAi [VDRC] and GMR-GAL4>eIF4A RNAi [BDSC)]) resulted in pupal lethality at 20°C (Figure S3A). The qPCR analysis of eIF4A transcript levels in pharate heads reveals a 50% reduction in eIF4A transcript levels from the eIF4A RNAi (BDSC#33970) and 60% reduction from the eIF4a RNAi (VDRC#42201) (Figure S3B); therefore, we selected the eIF4A RNAi (VDRC) for the rescue assessment as a result of the higher RNAi-knockdown efficiency. The fly crosses set up for rescue study are shown in Table S5.
We expressed the human EIF4A2 WT or variant cDNAs in the background of GMR-GAL4>eIF4A RNAi (VDRC) at 20°C and assessed for rescue of pupal lethality. Expression of EIF4A2 WT completely rescued the pupal lethality, which indicates a strong functional conservation in fruit flies (Figure 5D). In contrast, expression of the EIF4A2 variant cDNAs had differential rescue efficiencies: EIF4A2 p.Leu344Phe showed 25% rescue, p.Thr243Ile showed 7% rescue, and p.Gly364Glu showed 3% rescue in over 100 scored flies. In contrast, the expression of EIF4A2 p.Thr216Ile completely failed to rescue the pupal lethality.
Together, the in silico protein molecular modeling and in vivo fly functional assessments demonstrate that our probands’ missense EIF4A2 variants result primarily in loss of EIF4A2 function (p.Leu344Phe, p.Thr243Ile, and p.Gly364Glu) but can also cause toxic gain of EIF4A2 function (p.Thr216Ile). These findings and our clinical characterizations show that EIF4A2 de novo mono-allelic and inherited bi-allelic variants in key functional domains lead to a syndromic neurodevelopmental disorder.
Discussion
We present 15 individuals with NDDs and extremely rare variants in EIF4A2, encoding a DEAD-box-containing protein, which has not been previously associated with human disease. Each variant was identified by clinical or research analysis of whole-exome or genome sequencing. The results of our clinical and molecular characterizations with in silico protein predictions and in vivo fruit fly modeling show that mono-allelic and bi-allelic EIF4A2 variants in key functional domains lead to a syndromic neurodevelopmental disorder comprised of GDD, ID, hypotonia, epilepsy, and structural brain alterations. Molecular modeling of the EIF4A2 missense variants revealed that three of the missense variants, p.Leu344Phe, p.Gly362Ser, and p.Gly364Glu, are located in the C-terminal helicase domain of EIF4A2 and the variant p.Thr243Ile is very close to this domain. The other four missense variants in individuals with severe clinical phenotypes, as defined by co-morbid epilepsy and significantly delayed development, p.Gly192Ser, p.Ser214Tyr, p.Thr216Ile, and p.Thr216Ala, are located in the N-terminal helicase ATP-binding domain. Both these domains in EIF4A2 are critical for the RNA helicase activity. A prior study showed that variants located in either of the two helicase domains of another DEAD-box-containing protein, DDX3X, are associated with a more severe developmental phenotype due to decreased helicase activity, which supports the critical function of these domains in the DEAD-box family.11 Together, our detailed in silico analysis suggests that these missense EIF4A2 variants disrupt the interaction between key residues and introduces new bulky sidechains that may perturb protein interactions. These changes are predicted to impact the enzymatic activity of EIF4A2, disrupt the stability of the protein, and hinder EIF4A2 protein function.
Comparing genotypes and phenotypes within the cohort revealed several findings of interest, suggesting a genotype-phenotype correlation with the affected protein domain. Clinical severity ranged from mild ID to profound cognitive impairment with almost absent developmental progress and the severity correlated with the variant’s location in key protein domains (Figure 3A). Mild clinical phenotypes were seen in three individuals (I:1–I:3) with EIF4A2 variants upstream of the helicase domains, p.Ser2Cys, p.Asp37del, and p.Leu44Profs∗10, respectively (Tables 1 and 2). The variant p.Leu44Profs∗10 was found to be mosaic for individual I:3, which may also contribute to the milder clinical presentation. All three individuals presented with mild ID, normal vision, hearing, respiratory status, GI motility, and had no history of seizures. Hypotonia was identified in two of these individuals. In contrast, severe NDDs were seen in 12 individuals (I:4–I:15) (Tables 1 and 2). Individuals I:4–I:10 had variants present in the helicase ATP-binding domain with p.Arg62Serfs∗7 (variant 1 for I:4 and I:5), p.Gly161Trp, p.Gly192Ser, p.Ser214Tyr, p.Thr216Ala, and p.Thr216Ile (I:6–I:10). Individuals I:12–I:15 had variants present in the helicase C-terminal domain with p.Ile315del, p.Leu344Phe, p.Gly362Ser, p.Gly364Glu, and p.Asp387_Ile388del (variant 2 for I:4 and I:5). Finally, individual I:11 had a variant immediately adjacent to the helicase C-terminal domain, p.Thr243Ile. All these 12 individuals had hypotonia and seizures, and 11 out of 12 of them had severe to profound GDD or ID. Ten of the 12 individuals had feeding difficulties with dysmotility present in five and g-tube dependence in seven individuals.
These affected residues in our cohort are highly conserved in both vertebrate and invertebrate species, suggesting they are critical for the protein’s conserved functions. Our functional analysis in flies reveal an important role for EIF4A2 in mediating critical developmental processes. EIF4A2 and its protein domains are well conserved in the fly homolog, eIF4A, which is ubiquitously expressed during fly embryogenesis, larval wing imaginal disc, haltere, leg, and eye discs.46 A recent study showed that eIF4A has an indispensable role in sensory dendrite pruning in flies.45 The dendrite pruning of fly sensory neurons occurs during ecdysone-mediated metamorphosis through a defined transcriptional program. eIF4A is one of the proteins required for the translation of ecdysone receptor target genes to regulate dendrite pruning.45 Another critical development role for eIF4A is the negative regulation of dpp signaling. In flies, eIF4A physically interacts with the Smad proteins, Mad and Medea, to promote their degradation, which leads to decreased dpp signaling.23 Together, these prior findings show that eIF4A regulates key pathways required for nervous system development and function.
We found that neuron-specific expression of the EIF4A2 missense variants, p.Gly364Glu, p.Leu344Phe, and p.Thr243Ile, resulted in fly climbing defects, revealing these are dominant variants with potentially LOF effects. Interestingly, expression of EIF4A2 p.Thr216Ile in the wing pouch and blade resulted in wing margin serrations and reduced pMad expression that is similar to the effects of fly eIF4A overexpression in the wing margin.23 This result reveals that EIF4A2 p.Thr216Ile is a potential dominant GOF allele. As eIF4A is shown to negatively regulate dpp signaling in fruit flies, we examined the effects of EIF4A2 variants in the background of increased dpp signaling. In flies, overexpression of dpp in the eye results in a roughened eye surface. We found that EIF4A2 p.Leu344Phe and p.Thr243Ile expression in the dpp-overexpression background worsened the dpp-induced eye phenotype and led to disorganized ommatidial arrangement. Together, these results reveal that EIF4A2 p.Leu344Phe, p.Thr243Ile, and p.Gly364Glu variants in or adjacent to the CTD act as dominant LOF alleles, while the EIF4A2 p.Thr216Ile variant in the NTD acts as a GOF allele.
To determine whether there is a functional difference between the EIF4A2 missense variants, we examined the ability of these variants to rescue the pupal lethality resulting from GMR-Gal4-mediated loss of over 50% of eIF4A function. Our rescue assessment showed that the human EIF4A2 WT fully rescued the pupal lethality caused by reduced eIF4A expression in the GMR domain, but there was reduced (p.Leu344Ph, p.Gly364Glu, p.Thr243Ile) to absent (p.Thr216Ile) rescue efficiency for the missense variants. These findings further support that EIF4A2 p.Leu344Phe, p.Gly364Glu, and p.Thr243Ile are potentially LOF variants, whereas p.Thr216Ile is a GOF variant with most likely toxic effect upon expression in fruit flies. Together, our overexpression and rescue fly assays reveal that EIF4A2 variants contribute to human neurological phenotypes through both LOF and GOF mechanisms.
In addition to the de novo mono-allelic EIF4A2 variants identified in 12 individuals in our cohort, three other individuals had inherited bi-allelic EIF4A2 variants. These mono-allelic and bi-allelic EIF4A2 variants reported here lead to a distinct overlapping phenotypic spectrum. To date, over 30 disease genes have been identified with both dominant (mono-allelic) and recessive (bi-allelic) inheritance patterns.47,48 Such allelic heterogeneity may result from variability in the functional consequences of the pathogenic variants resulting in dominant negative, LOF, or GOF mechanisms,49 which is consistent with our findings that EIF4A2 p.Leu344Phe, p.Gly364Glu, and p.Thr243Ile are likely LOF variants, whereas p.Thr216Ile is a GOF variant. Several of the known disease genes that are associated with both recessive and dominant inheritance patterns are found to have a pLI score of 1.50 Despite this apparent lack of tolerance for LOF variants, the human disease is associated with asymptomatic parents who are heterozygous carriers for the LOF variant and simultaneously there are symptomatic individuals with de novo mono-allelic missense variants with likely LOF effects.47,50 These results suggest that the predicted functional consequences of potentially pathogenic variant alleles cannot fully explain the allelic heterogeneity alone and that other factors, such as epigenetic regulation and protein-protein interactions, may also be contributory.
Of our three individuals with inherited bi-allelic EIF4A2 variants, one individual (I:2) is homozygous for p.Asp37del and two individuals are siblings (I:4 and I:5) with p.Arg62Serfs∗7 and p.Asp387_Ile388del bi-allelic variants. I:2’s homozygous variant, p.Asp37del, causes a single amino acid deletion in the N-terminus of EIF4A2 that is within the Q motif upstream of the ATPase domains. The Q motif is an important regulatory element within DEAD-box proteins that plays a role in ATP binding, hydrolysis, and RNA recruitment.51 This individual has a milder neurodevelopmental phenotype that is characterized by mild ID and hypotonia with an absence of known structural brain abnormalities and seizures. This variant was inherited from asymptomatic consanguineous parents. A single amino acid deletion may not lead to a disease state in a mono-allelic state, as is seen with the common CFTR p.Phe508del variant in cystic fibrosis (MIM: 602421) where one out of every 25 individuals of European ancestry are mono-allelic asymptomatic carriers. However, cystic fibrosis occurs in the presence of bi-allelic CFTR p.Phe508del variants as a result of a protein processing defect that significantly decreases protein levels and impairs the function of the residual abnormally folded protein.52 Therefore, it is possible that EIF4A2 p.Asp37del may have a similar effect with an adequate amount of functional protein present in the mono-allelic state, but in the bi-allelic form there may be pronounced impairments in ATP binding, hydrolysis, or RNA recruitment at the Q motif.
In contrast, I:4 and I:5 have inherited bi-allelic EIF4A2 variants leading to both a frameshift (p.Arg62Serfs∗7) and a deletion of two amino acids (p.Asp387_Ile388del). Both variants are inherited from asymptomatic parents with a mono-allelic variant. It is possible that an adequate amount of functional protein is present in a mono-allelic state, but the disease pathogenesis occurs in the bi-allelic state because of pronounced loss of functional protein. This graded sensitivity to the functional dosage of EIF4A2 protein function may be consistent with the more severe clinical findings in I:4 and I:5. While their clinical findings do overlap with the 11 affected individuals in our cohort with de novo mono-allelic variants, I:4 and I:5 have progressive neurodegeneration that is not seen in the individuals with mono-allelic variants. They also have severe ID, absent speech, hypotonia, intractable seizures, and structural brain abnormalities. Our fruit fly findings support a linear correlation between disease severity and the functional dosage of EIFA2. In our eIF4A RNAi-mediated knockdown study, we found that eIF4A knockdown at a higher temperature (25°C) has a more severe phenotype (lethality at embryonic stage) compared to the lower temperature (20°C–21°C) phenotype (lethality at pupal stage) (Figure S2). The difference in phenotypic severity suggests a dose-dependent activity for eIF4A in the fruit flies resulting from the temperature sensitivity of the UAS-GAL4 system. This temperature sensitivity is presumably due to increased GAL4-dependent transcriptional activation of the UAS allele at higher temperatures and lower transcriptional activation at lower temperatures.53 Therefore, at 25°C the eIF4A knockdown is stronger as a result increased RNAi dosage than at 20°C–21°C, which demonstrates that relatively milder loss of eIF4A function leads to a relatively milder phenotype compared to stronger loss of eIF4A function. Longitudinal follow-up and new prospective recruitment efforts will further refine the EIF4A2 genotype-phenotype correlations, which may advance our understanding of mono-allelic and bi-allelic variants in disease pathogenesis.
In conclusion, our study provides evidence that both mono-allelic and bi-allelic EIF4A2 variants result in a syndromic neurodevelopmental disorder characterized by variable developmental delays, epilepsy, hypotonia, cognitive impairments, GI dysmotility, respiratory complications, and visual impairments. Although a comprehensive interpretation of genotype to phenotype severity is limited by the small sample size, our clinical and molecular findings suggest that variants upstream of the helicase domains are less damaging than variants in the helicase domains. Furthermore, the in vivo functional modeling in fruit flies reveal that EIF4A2 variants may contribute to disease pathogenesis through both LOF and GOF mechanisms. A larger sample size and longitudinal monitoring of neurodevelopment would be beneficial for determining the genotype-phenotype correlations between affected protein domains and the inheritance pattern (recessive or dominant).
Acknowledgments
We thank the families and clinical staff at each location for participation in this study. P.B.A.’s research work is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR068429-01), National Human Genome Research Institute (1R01HG011798-01A1), and "Because of Bella" foundation. H.T.C. is funded from the McNair Medical Institute at Robert and Janice McNair Foundation, Child Neurology Foundation and Society, The Gordon and Mary Cain Foundation, Annie and Bob Graham, The Elkins Foundation, and the Mark A. Wallace Endowment Award. M.S.P.’s research effort is supported in part by the National Ataxia Foundation and the Burroughs Wellcome Fund. A.R.D.’s research effort was supported by the National Institutes of Health, (T32HD098061). D.G.C.’s research effort was supported by Muscular Dystrophy Association Development grant 873841 (https://doi.org/10.55762/pc.gr.147552), Chao Physician-Scientist Award, and 5T32GM007526 Medical Genetics Research Program. This work was also supported by the Manton Center for Orphan Disease Research, Boston Children’s Hospital IDDRC Molecular Genetics Core Facility (U54HD090255), Broad Institute of MIT and Harvard Center for Mendelian Genomics (R01HG009141 and UM1HG008900), Strasbourg’s Interdisciplinary Thematic Institute (ITI) for Precision Medicine, IdEx Unistra (ANR-10-IDEX-0002) and SFRI-STRAT’US (ANR-20-SFRI-0012), Fondazione Telethon, Telethon Undiagnosed Diseases Program (TUDP, GSP15001), NIHR Oxford Biomedical Research Centre Program, the Wellcome Trust (203141/Z/16/Z), Solve-RD (to A.J.), Ricerca Corrente 2021, Ministero della Salute (to E.A. and A.N.), Deutsche Forschungsgemeinschaft (DFG, German Research Foundation to T.B.H.), German Federal Ministry of Education and Research (grant no. FKZ 01ZX1405C to B.A.), and the German Network for mitochondrial disorders (mitoNET, 01GM1906D to B.A.). We thank Drs. Hugo Bellen and Hamed Jafar-Nejad for providing Drosophila melanogaster stocks. Please see supplemental information for complete acknowledgments.
Author contributions
M.S.P., A.R.D., P.B.A., and H.T.C. conceived and designed the study, acquired and analyzed the data, and drafted the manuscript and figures. C.A.G., P.E.G., J.S., M.P., N.B.-P., A.G.-F., R.P.S., G.Z., C.L., E.A., A.N., U.B., T.B.H., W.H., E.M., B.A., R.A.J., T.B., S.H., R.C., B.I., S.B., A.R., K.I., B.P.S., O.B., B.B.Z., A.B., D.A.C., S.V.M., T.B.P., D.G.C., K.S., A.R.P.A., R.T., G.E.R.C., S.B., S.D., A.J., H. Pan, N.I., and H.Pirt contributed to the acquisition and analysis of the data and reviewed and edited the manuscript.
Declaration of interests
The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the clinical exome and genome sequencing offered at Baylor Genetics. P.B.A. is on the Scientific Advisory Board of Illumina, Inc., and GeneDx. S.V.M., D.A.C., T.B.P. and A.B. are employees of GeneDx. H. Pirt is currently employed by Illumina. N.B.P. receives consult fees from Genespire and Pfizer for work unrelated to this project.
Published: December 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.11.011.
Contributor Information
Hsiao-Tuan Chao, Email: hc140077@bcm.edu.
Pankaj B. Agrawal, Email: pankaj.agrawal@enders.tch.harvard.edu.
Supplemental information
Data and code availability
All variants have been submitted to the public databases. Variants for individuals I:1–I:5, I:8–I:13, and I:15 were submitted to ClinVar. Variants for individuals I:7 and I:14 were submitted to DECIPHER.
References
- 1.Moeschler J.B., Shevell M. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134 doi: 10.1542/peds.2014-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertelli M.O., Munir K., Harris J., Salvador-Carulla L. “Intellectual developmental disorders”: reflections on the international consensus document for redefining “mental retardation-intellectual disability” in ICD-11. Adv. Ment. Heal. Intellect. Disabil. 2016;10:36–58. doi: 10.1108/AMHID-10-2015-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 4.Reinders M.R.F., Schob C., Küry S., Harel T., Eldomery M.K., Coban-Akdemir Z., Denecke J., Edvardson S., Colin E., Stegmann A.P. De novo missense mutations in DHX30 impair global translation and cause a neurodevelopmental disorder. Am. J. Hum. Genet. 2017;101:716–724. doi: 10.1016/j.ajhg.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balak C., Bénard M., Schaefer E., Iqbal S., Ramsey K., Ernoult-Lange M., Mattioli F., Llaci L., Geoffroy V., Courel M., Naymik M. Rare De novo missense variants in RNA helicase DDX6 cause intellectual disability and dysmorphic features and lead to P-Body defects and RNA dysregulation. Am. J. Hum. Genet. 2019:509–525. doi: 10.1016/j.ajhg.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blok L.S., Madsen E., Juusola J., Gilissen C., Baralle D., Reijnders M.R., Venselaar H., Helsmoortel C., Cho M.T., Hoischen A., Vissers L.E. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on wnt signaling. Am. J. Hum. Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paine I., Posey J.E., Grochowski C.M., Jhangiani S.N., Rosenheck S., Kleyner R., Marmorale T., Yoon M., Wang K., Robison R., Cappuccio G. Paralog studies augment gene discovery : DDX and DHX genes. Am. J. Hum. Genet. 2019;105:302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelhaleem M., Maltais L., Wain H. The human DDX and DHX gene families of putative. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 9.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 10.Tanner N.K., Cordin O., Banroques J., Doère M., Linder P. The Q motif : a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 11.Lennox A.L., Hoye M.L., Jiang R., Johnson-Kerner B.L., Suit L.A., Venkataramanan S., Sheehan C.J., Alsina F.C., Fregeau B., Aldinger K.A., et al. Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron. 2020;106:404–420.e8. doi: 10.1016/j.neuron.2020.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher S.G., Kim S.H., Fountoulakis M., Lubec G. Reduced levels of DEAD-Box proteins DBP-RB and p72 in fetal down syndrome brains. Neurochem. Res. 2002;27:1141–1146. doi: 10.1023/a:1020921324871. [DOI] [PubMed] [Google Scholar]
- 13.Merrick W.C. eIF4F: A retrospective. J. Biol. Chem. 2015;290:24091–24099. doi: 10.1074/jbc.R115.675280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitken C.E., Lorsch J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 15.Rogers G.W., Jr, Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 16.Meijer H.A., Kong Y.W., Lu W.T., Wilczynska A., Spriggs R.V., Robinson S.W., Godfrey J.D., Willis A.E., Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 17.Meijer H.A., Schmidt T., Gillen S.L., Langlais C., Jukes-Jones R., de Moor C.H., Cain K., Wilczynska A., Bushell M. DEAD-box helicase eIF4A2 inhibits CNOT7 deadenylation activity. Nucleic Acids Res. 2019;47:8224–8238. doi: 10.1093/nar/gkz509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vissers L.E., Kalvakuri S., de Boer E., Geuer S., Oud M., van Outersterp I., Kwint M., Witmond M., Kersten S., Polla D.L., et al. De novo variants in CNOT1 , a central component of the CCR4-NOT complex involved in gene expression and RNA and protein stability , cause neurodevelopmental delay. Am. J. Hum. Genet. 2020;107:164–172. doi: 10.1016/j.ajhg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Wang H., Zhu Z., Yang S., Li K. Molecular cloning, mapping, and expression analysis of the EIF4A2 gene in pig. Biochem. Genet. 2007;45:51–62. doi: 10.1007/s10528-006-9065-7. [DOI] [PubMed] [Google Scholar]
- 20.Galicia-Vázquez G., Di Marco S., Lian X.J., Ma J.F., Gallouzi I.E., Pelletier J. Regulation of eukaryotic initiation factor 4AII by MyoD during murine myogenic cell differentiation. PLoS One. 2014;9:e87237. doi: 10.1371/journal.pone.0087237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan R., Sargent M.G. The role in neural patterning of translation initiation factor eIF4AII; induction of neural fold genes. Development. 1997;124:2751–2760. doi: 10.1242/dev.124.14.2751. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Li W.X. A novel function of Drosophila eIF4a as a negative regulator of Dpp/BMP signalling that mediates SMAD degradation. Nat. Cell Biol. 2006;8:1407–1414. doi: 10.1038/ncb1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamaratoglu F., Affolter M., Pyrowolakis G. Seminars in Cell & Developmental Biology Dpp/BMP signaling in flies : From molecules to biology. Semin. Cell Dev. Biol. 2014;32:128–136. doi: 10.1016/j.semcdb.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 25.Fried P., Sánchez-aragón M., Aguilar-hidalgo D., Lehtinen B. A model of the spatio-temporal dynamics of drosophila eye disc development. PLoS Comput. Biol. 2016;12:1–23. doi: 10.1371/journal.pcbi.1005052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firth L.C., Bhattacharya A., Baker N.E. Cell cycle arrest by a gradient of Dpp signaling during Drosophila eye development. BMC Dev. Biol. 2010;10:28. doi: 10.1186/1471-213X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashima R., Hata A. The role of TGF- β superfamily signaling in neurological disorders. Acta Biochim. Biophys. Sin. 2018;50:106–120. doi: 10.1093/abbs/gmx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers E.A., Kessler J.A. TGF-β family signaling in neural and neuronal differentiation, development, and function. Cold Spring Harb. Perspect. Biol. 2017;9:a022244. doi: 10.1101/cshperspect.a022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiew L.F., Poon C.H., You H.Z., Lim L.W. Tgf-β/smad signalling in neurogenesis: implications for neuropsychiatric diseases. Cells. 2021;10:1382. doi: 10.3390/cells10061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuter M.S., Tawamie H., Buchert R., Hosny Gebril O., Froukh T., Thiel C., Uebe S., Ekici A.B., Krumbiegel M., Zweier C., et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psychiatr. 2017;74:293–299. doi: 10.1001/jamapsychiatry.2016.3798. [DOI] [PubMed] [Google Scholar]
- 31.Chao H.T., Davids M., Burke E., Pappas J.G., Rosenfeld J.A., McCarty A.J., Davis T., Wolfe L., Toro C., Tifft C., et al. A syndromic neurodevelopmental disorder caused by de novo variants in EBF3. Am. J. Hum. Genet. 2017;100:128–137. doi: 10.1016/j.ajhg.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bischof J., Sheils E.M., Björklund M., Basler K. Generation of a transgenic ORFeome library in Drosophila. Nat. Protoc. 2014;9:1607–1620. doi: 10.1038/nprot.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutta D., Briere L.C., Kanca O., Marcogliese P.C., Walker M.A., High F.A., Vanderver A., Krier J., Carmichael N., Callahan C., et al. De novo mutations in TOMM70, a receptor of the mitochondrial import translocase, cause neurological impairment. Hum. Mol. Genet. 2020;29:1568–1579. doi: 10.1093/hmg/ddaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arya R., Lakhotia S.C. A simple nail polish imprint technique for examination of external morphology of Drosophila eyes. Curr. Sci. 2006;90:1179–1180. [Google Scholar]
- 35.Paul M.S., Dutta D., Singh A., Mutsuddi M., Mukherjee A. Regulation of Notch signaling in the developing Drosophila eye by a T-box containing transcription factor, Dorsocross. Genesis. 2018;56:232511–e23315. doi: 10.1002/dvg.23251. [DOI] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Arachchi H., Wojcik M.H., Weisburd B., Jacobsen J.O.B., Valkanas E., Baxter S., Byrne A.B., O'Donnell-Luria A.H., Haendel M., Smedley D., et al. An open-source tool for patient matching via the Matchmaker Exchange. Hum. Mutat. 2018;39:1827–1834. doi: 10.1002/humu.23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., et al. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2016;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:886–894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A., et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Gauthier L.D. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of- function intolerance across human protein-coding genes. bioRxiv. 2019 doi: 10.1101/531210. Preprint at. [DOI] [Google Scholar]
- 43.Csordas G., Grawe F., Uhlirova M. Eater Cooperates with Multiplexin to Drive the Formation of Hematopoietic Compartments. eLife. 2020;9:e57297. doi: 10.7554/eLife.57297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iyer K.V., Piscitello-Gómez R., Paijmans J., Jülicher F., Eaton S. Epithelial viscoelasticity is regulated by mechanosensitive E-cadherin turnover. Curr. Biol. 2019;29:578–591.e5. doi: 10.1016/j.cub.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 45.Rode S., Ohm H., Anhäuser L., Wagner M., Rosing M., Deng X., Sin O., Leidel S.A., Storkebaum E., Rentmeister A., et al. Differential requirement for translation initiation factor pathways during ecdysone-dependent neuronal remodeling in drosophila. Cell Rep. 2018;24:2287–2299.e4. doi: 10.1016/j.celrep.2018.07.074. [DOI] [PubMed] [Google Scholar]
- 46.Hernández G., Lalioti V., Vandekerckhove J., Sierra J.M., Santarén J.F. Identification and characterization of the expression of the translation initiation factor 4A (eIF4A) from Drosophila melanogaster. Proteomics. 2004;4:316–326. doi: 10.1002/pmic.200300555. [DOI] [PubMed] [Google Scholar]
- 47.Dworschak G.C., Punetha J., Kalanithy J.C., Mingardo E., Erdem H.B., Akdemir Z.C., Karaca E., Mitani T., Marafi D., Fatih J.M., et al. Biallelic and monoallelic variants in PLXNA1 are implicated in a novel neurodevelopmental disorder with variable cerebral and eye anomalies. Genet. Med. 2021;23:1715–1725. doi: 10.1038/s41436-021-01196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posey J.E., O’Donnell-Luria A.H., Chong J.X., Harel T., Jhan-giani S.N., Coban Akdemir Z.H., Buyske S., Pehlivan D., Car-valho C.M.B., Baxter S., et al. Insights into genetics, human biology and disease gleaned from family based genomic studies. Genet. Med. 2019;21:798–812. doi: 10.1038/s41436-018-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White J., Beck C.R., Harel T., Posey J.E., Jhangiani S.N., Tang S., Farwell K.D., Powis Z., Mendelsohn N.J., Baker J.A., et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3–11. doi: 10.1186/s13073-015-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harel T., Yesil G., Bayram Y., Coban-Akdemir Z., Charng W.L., Karaca E., Al Asmari A., Eldomery M.K., Hunter J.V., Jhangiani S.N., et al. Monoallelic and Biallelic variants in EMC1 identified in individuals with global developmental delay, hypotonia, scoliosis, and cerebellar atrophy. Am. J. Hum. Genet. 2016;98:562–570. doi: 10.1016/j.ajhg.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordin O., Tanner N.K., Doère M., Linder P., Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cutting G.R. Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kramer J.M., Staveley B.E. GAL4 causes developmental defects and apoptosis when expressed in the developing eye of Drosophila melanogaster. Genet. Mol. Res. 2003;2:43–47. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants have been submitted to the public databases. Variants for individuals I:1–I:5, I:8–I:13, and I:15 were submitted to ClinVar. Variants for individuals I:7 and I:14 were submitted to DECIPHER.