Abstract
Gene therapy is the manipulation of gene expression patterns in specific cells to treat genetic and pathological diseases. This manipulation is accomplished by the controlled introduction exogenous nucleic acids into target cells. Given the size and negative charge of these biomacromolecules, the delivery process is driven by the carrier vector, of which is dominated by the usage of viral vectors. Taking into account the limitations of viral vectors, nonviral alternatives have gained significant attention due to their flexible design, low cytotoxicity and immunogenicity, and their gene delivery efficacy. That stated, the field of nonviral vectors has been dominated by research dedicated to overcoming barriers in gene transfer. Unfortunately, these traditional nonviral vectors have failed to completely overcome the barriers required for clinical translation and thus, have failed to match the delivery outcomes of viral vector. This has consequently encouraged the development of new, more radical approaches that have the potential for higher clinical translation. In this review, we discuss recent advances in vector technology and nucleic acid chemistry that have challenged the current standing of nonviral systems. The diversity of these approaches highlights the numerous alternative avenues for overcoming innate and technical barriers associated with gene delivery.
Introduction
Gene therapy has emerged as a viable therapeutic option due to its capacity to address the causative factors of various disorders at the genetic level. Gene therapy is broadly defined as the delivery of genetic-based material (e.g., DNA or RNA) to specific cells to modify expression patterns (Figure 1). Manipulation of cellular gene expression patterns can be utilized in genetic vaccines, for example, to provide protection against difficult to treat ailments, such as cancer or AIDS, by enabling expression of associated disease markers to provoke an immune response.
Figure 1.
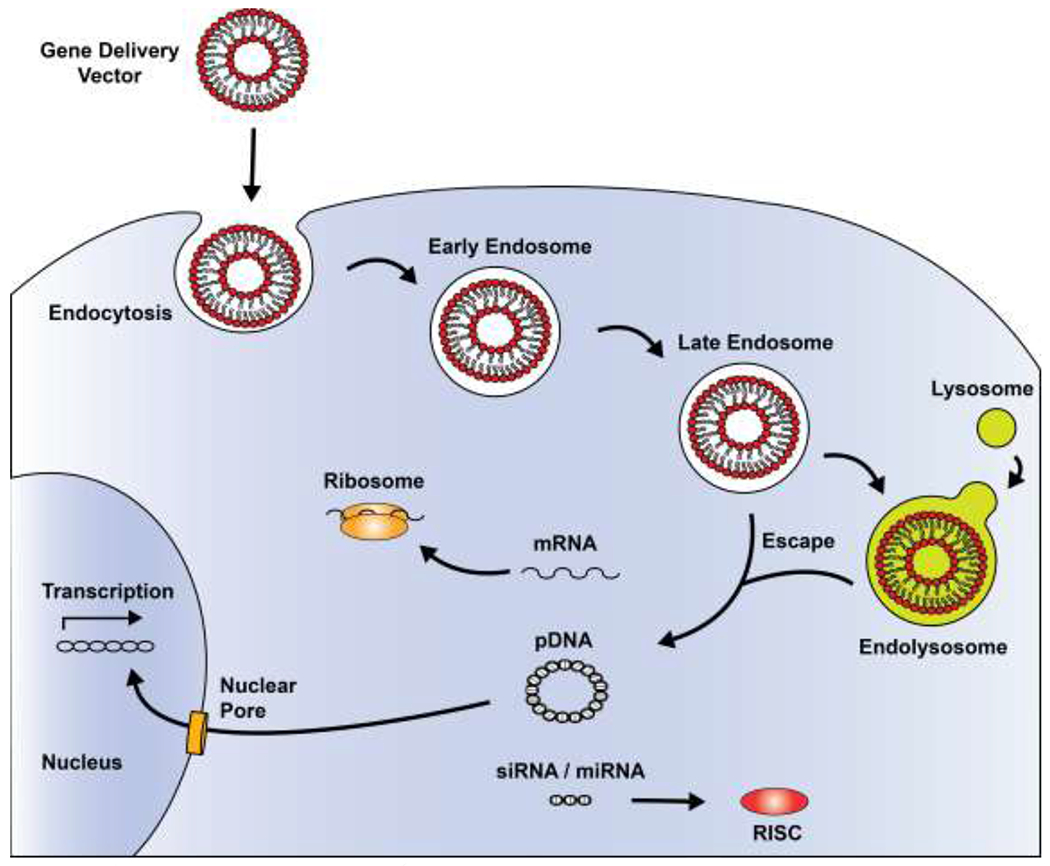
Generalized process for gene therapy. Nonviral vectors can be used to facilitate the delivery of DNA and RNA molecules by first mediating cellular uptake through endocytosis mechanisms. The vectors also provide protection to the genetic cargo and prompt compartmental escape as the endosome gradually acidifies. Once in the cytosol, siRNA-and miRNA-based cargo must be loaded into the RNA-induced silencing complex (RISC); whereas, mRNA must bind to cellular ribosomes to promote translation. Conversely, DNA requires translocation to the nucleus.
A critical part of this process involves the choice of delivery mechanism, which greatly influences the type, duration, and outcome of the specific treatment. Furthermore, successful delivery of the genetic cargo requires both successful uptake and correct translocation to respective active sites (most often, DNA is transported to the nucleus and RNA to the cytoplasm). Traditionally, delivery has been facilitated using either physical administration techniques such as electroporation (i.e., the use of electrical current to render desired cells permeable to genetic material) or by vector-mediated delivery. Various gene delivery vectors have been developed that are generally divided into two classes: viral or nonviral systems. Historically, viral-based technologies have represented the standard for gene delivery due to their unsurpassed transfection efficiency (Jones et al., 2013a; Jones et al., 2014a). Despite delivery efficacy, viral vectors possess limitations including complex formulation, storage-related difficulties, and off-target effects (e.g., undesired toxicity, immunogenicity, and tumorigenicity) (Baum et al., 2006; Musacchio & Torchilin, 2013; Nayerossadat et al., 2012; Seow & Wood, 2009). Nonviral gene delivery vectors stand as alternatives and feature extensive design strategies; however, nonviral approaches have thus far failed to match the efficacy of their viral counterparts. This has consequently encouraged the development of new, more radical approaches (Pack et al., 2005). For example, vectors have been redesigned to deviate from various established delivery paradigms pertaining to formulation, storage, administration, circulation and cell targeting, and genetic processing. To describe the emerging shift from traditional nonviral vectors, the accompanying text will highlight alternative approaches being developed to overcome specific barriers associated with gene delivery.
Biologically-derived vectors
In a general sense, the extent of gene therapy using transfection is determined by the mechanism chosen to facilitate the delivery of the desired genetic cargo. Over the past few years, unique classes of delivery vectors have been developed that can influence gene delivery outcomes in ways that are unachievable using traditional approaches.
One such class includes hybrid delivery vectors that consist of biologically derived vectors (e.g., bacteria or viral alternatives) actively interfaced with biocompatible materials. A prominent example of this strategy is the synergistic combination of invasive (innate or engineered) bacteria with rationally designed biomaterial agents (Akin et al., 2007; Jones et al., 2015; Jones et al., 2014b). Both components of these hybrid vectors have been evaluated independently for their gene delivery efficacy, thus, the combination of these technologies enables the application of vector-specific engineering opportunities towards overcoming notoriously difficult delivery barriers.
Independently, bacterial vectors have demonstrated the ability to deliver diverse genetic cargo including plasmid DNA (pDNA) (Grillot-Courvalin et al., 2002; Jones et al., 2013b; Larsen et al., 2008; Parsa et al., 2008a; Parsa et al., 2008b; Schaffner, 1980), RNA-based constructs (Kruhn et al., 2009; Xiang et al., 2006; Xu et al., 2009), and larger genetic elements (Cheung et al., 2012; Laner et al., 2005). Additionally, bacterial vectors possess the potential to incorporate powerful genomic integration technologies, to be described below, that can impart permanent changes currently only achievable with viral delivery vectors. This potential arises from bacterial properties such as ideal sizing for uptake into antigen presenting cells (APCs; i.e., macrophages and dendritic cells), general cellular uptake through invasive properties, adjuvant-like compositional makeup, genetic cargo maintenance capability (removing a need to manufacture and purify genetic material), no theoretical limit to genetic cargo size, the ability to deliver both genetic and protein cargo, and evolutionarily-optimized and/or protein-based endosomal escape mechanisms. However, despite the demonstration of safe in vivo cytotoxicity panels (Chart el al., 2000; Jones el al., 2014b), concerns over biosafety impede widespread clinical translation relative to other non-biological vector formats.
Gene delivery vectors comprised of biomaterials include an expansive list of compositionally unique constructs that can be predominantly divided into two classes, polymers and lipids. Regardless of the material selected, this class of vectors can be readily synthesized and tailored to address application-specific problems and mediate delivery through physically-driven phenomena. Specifically, after packaging the genetic cargo (through electrostatically-driven complexation or physical encapsulation), these particulate systems are internalized through endocytosis by host cells (Jones et al., 2013a). Following cellular uptake, endosomal escape is required to ensure gene expression. An important design consideration is the manner in which endosomal escape is achieved, which differs significantly depending on the specific biomaterial vector. The most common escape mechanisms involve either the “proton sponge effect” or lipid mixing, which are mediated by cationic polymers or lipid-based systems, respectively (Khatri et al., 2012; Miller, 2013; Schlenk et al., 2013). However, biomaterial vectors possess potential limitations such as prohibitive scale-up costs, reduced genetic cargo capacity, limited colloidal stability, cytotoxicity, immunogenicity, and sub-biological endosomal escape and gene transfer efficacy (Jones et al., 2013a; Pack et al., 2005; Yin et al., 2014).
Taken together, the combination of the aforementioned technologies enables the formulation of hybrid vectors that retain vector-specific advantages while compensating for their individual limitations. For example, our group developed a class of hybrid biosynthetic gene delivery vectors composed of an Escherichia coli inner-core and a cationic polymer outer-core (Jones et al., 2015; Jones et al., 2014b) which enabled APC cellular outcomes significantly improved relative to individual component vectors or commercially-available vectors in both in vitro and in vivo models. Although the current gene delivery platform was tailored for APC-specific activity, other studies have demonstrated that E. coli can be applied to various non-phagocytic cell lines through the expression of invasive proteins such as invasin (Critchley-Thorne et al., 2006; Critchley et al., 2004).
Another delivery strategy gathering momentum is the application of bacterial outer membrane vesicles (OMVs). These vectors are generated from the natural or induced budding of proteoliposomes from Gram-negative bacteria (Unal et al., 2011). Particles range in size from 50-250 μm and can be designed to display targeting ligands and additional application-specific proteins on the OMV surface using traditional molecular biology tools (Avila-Calderon et al., 2015; Gujrati et al., 2014). Despite wide usage in vaccination regimes, OMVs retain substantially smaller genetic cargo loads and require complex purification schemes prior to use. Similarly to OMVs, non-denatured, hollowed cell envelopes from Gram-negative bacteria, termed bacterial ghosts (BGs), can act as natural scaffolds for genetic cargo loading. BGs are produced by the heterologous expression of lytic proteins from bacteriophages (Paukner et al., 2005; Vilte et al., 2012). These vectors possess the intrinsic properties of OMVs while requiring simpler purification and possessing higher loading capacities (completed after purification) (Acevedo et al., 2014). Although OMVs and BGs have demonstrated success in both in vitro and in vivo models and possesses an accessible tool-set for vector engineering, biomaterial-functionalization has yet to be demonstrated.
Nontraditional biomaterial vectors
Biomaterial particle-based delivery vectors have been the standard of gene delivery research (Yin et al., 2014). Successful constructs have been generated using a myriad of biocompatible materials that can be readily tuned to fully engage desired cellular pathways. An underlying theme of this research is the search for compositional-driven solutions to current particulate-based limitations, that is, developing new synthetic schemes and reactions for tuning particular biophysical properties (i.e., degradation rates, solubility, and colloidal stability) for desirable outcomes. Studies surveying specifics of designing traditional biomaterial gene delivery vectors are reviewed in detail elsewhere (Jones et al., 2013a; Pack et al., 2005; Yin et al., 2014).
A limitation of using compositionally-driven approaches to overcome gene delivery barriers is the resulting biomaterial systems will ultimately be hindered by the innate properties of the chosen material and/or delivery strategy, meaning that certain drawbacks associated with biomaterial systems can only be mitigated, but not eliminated, when using current technology. Regardless, several studies have provided a foundation for new directions in biomaterial delivery systems. These studies range from re-tooling of biomaterial vectors away from traditional particulate-based systems to biomimicry technology. Specifically, by using a modified microneedle strategy (termed “polymer tattooing”), Irvine and coworkers developed skin biodegradable DNA delivery vectors capable of continuous and sustained in situ polyplex formation (DeMuth et al., 2013). This method utilized a layer-by-layer (LBL) formation approach for loading microneedles with releasable polymer films containing alternating layers of pDNA, polymer, and adjuvants. Since its advent, LBL patterning has been widely adapted and serially optimized (Bechler & Lynn, 2012; Flessner et al., 2011; Li et al., 2014; Santos et al., 2012; Saurer et al., 2010; Zou et al., 2014). A similar approach involves the application of particle replication in nonwetting templates (PRINT) technology for gene delivery, developed by DeSimone and coworkers (Xu et al., 2013). The technology results in highly conserved and reproducible batches of cylindrical particles by mixing the genetic cargo with bovine serum albumin (BSA), lactose, and glycerol in a PRINT mold. After heating, the particles can be complexed with either commercial transfection reagents or a mixture of lipids to provide an endosomal escape mechanism. The novelty associated with this class of delivery vectors is the particle uniformity and associated potential as a standardized research platform. Unlike most biomaterial-based agents, experimental variation (from researcher to researcher) is significantly reduced, facilitating more reproducible studies.
Another contemporary biomaterial approach is the development of delivery vectors that adopt and employ properties displayed by living organisms. This class of delivery vectors, termed biomimetic vectors, utilizes entirely biomaterial-based vectors that mimic biophysical properties (size, shape, immunogenic signals, and surface antigens) in the context of gene delivery (e.g., the mimicry of viral particles) (He et al., 2014; Kang et al., 2013; Xu et al., 2002). This strategy is marked by the assembly of biomaterial elements into viral-like structures that contain the appropriate decoration of viral proteins. For example, liposomes have been synthesized that self-assemble into multi-center lamellar nanostructures encapsulating pDNA and possessing surface decoration with transferrin (Xu et al., 2002). A different approach utilizes pH-sensitive nanogel systems to mimic a viral capsid-like structure through the use of hydrophobic cores protected by two accompanying layers of a hydrophilic shell with the surface grafting of serum albumin-linked poly(ethylene glycol) (Lee et al., 2008). Unlike the first viral mimicry strategy, these hydrogels have an additional endosomal escape mechanism via pH-sensitive swelling that occurs after the transition from physiological (pH 7.4) to endosomal (pH 6.4) conditions. To date, the majority of biomimicry vector studies have been conducted in vitro and their in vivo efficacy has not been well-established. However, by combining available biomaterial tool-sets and flexibility with the innate properties of viral structures, numerous opportunities can be envisioned in the development and modification of gene delivery strategies using this approach.
Genetic cargo engineering strategies
In its simplest form, development of gene delivery strategies can be divided into two areas: the design and synthesis of a delivery vector and the type of genetic cargo to be delivered. Conventional gene therapy applications are predicated on the delivery of temporarily-active molecules such as pDNA, siRNA, shRNA, miRNA, and larger genetic constructs. While apt in vaccine-based applications, a shared limitation of all the aforementioned genetic cargo is their native expression brevity. One solution gathering attention is the delivery of genome editing systems using nonvrial vectors to enable the permanent correction of genetic disease markers such as point mutations and deletions (Figure 2). This has been achieved through the use of sequence-specific zinc-finger-proteins (ZFPs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPRs)-Cas systems. ZFPs and TALENs can be designed to target specific genomic sequences through modification of their DNA-binding domains (Boch et al., 2009; Moscou & Bogdanove, 2009; Urnov et al., 2010). Distinct from these strategies, the bacterial-derived CRISPR-Cas system can recognize and induce genomic modification through a synthetic guide RNA that hybridizes with target sequences (Cho et al., 2013; Mali et al., 2013). The RNA-guided nature of this strategy enables faster and simpler implementation than either ZFNs or TALENs and has the potential to simultaneously modify multiple genomic sites with a single administration. Due to the potential of the CRISPR-Cas system, significant efforts have been invested to improve specificity and efficiency and to reduce off-target effects (Farboud & Meyer, 2015; Ran et al., 2013; Sanjana et al., 2014).
Figure 2.
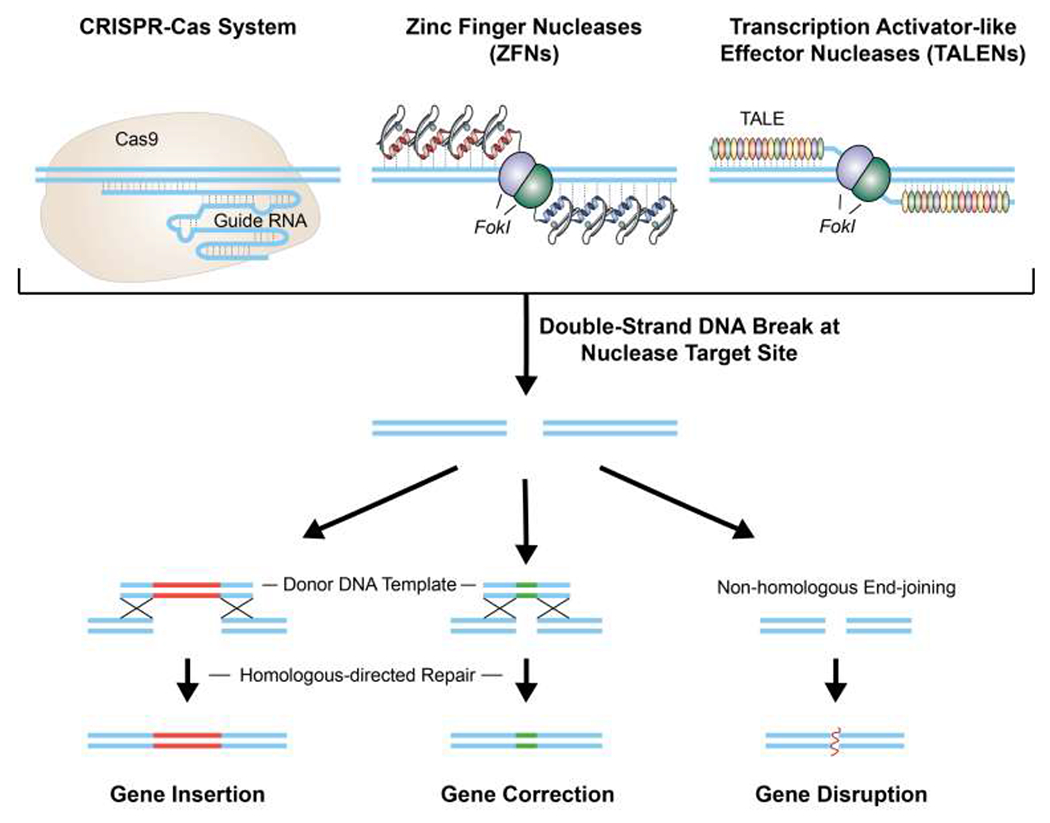
Genome editing systems. Clustered regularly interspaced short palindromic repeat-Cas9 (CRISPR-Cas), zinc-finger nuclease (ZFNs), and transcription activator-like effector nucleases (TALENs) are systems that can manipulate mammalian genomes with high precision and high efficiency by mediating double-strand breaks or single nicks (one strand) in a targeted sequence. The double-stand breaks are repaired by either homology-directed recombination, if a genetic donor template is available, or non-homologous end-joining.
Alternatively, in the context of expression-based gene therapy (i.e., pDNA), techniques have been developed to substantially extend the duration of expression compared to that of a typical expression vector (Figure 3) (Dietz et al., 2013; Kay et al., 2010; Keeney et al., 2013; Lu et al., 2013). One such approach involves removing extraneous genetic elements outside the transcriptional unit (Lu et al., 2012). For example, minicircle DNA (MC) vectors are one class of plasmids that are completely devoid of the normally-associated bacterial plasmid backbone and have demonstrated in vivo sustained efficacy in quiescent tissue as compared to regular pDNA (Chen et al., 2003; Lu et al., 2013). Even though protocols and manufacturing have been established for MC vectors (Kay et al., 2010), their production is still substantially more tedious and complicated as compared to traditional pDNA. To overcome manufacturing-related problems, Kay and co-workers developed an expression vector, termed the mini-intronic plasmid (MIP), which locates the bacterial origin of replication and antibiotic-free selection markers to an intron inside the transgene expression cassette (Lu et al., 2013). By preserving the extraneous elements required for bacterial propagation, but in a concealed manner, the resulting vector demonstrated improved efficacy over both pDNA and MC vectors in in vivo studies.
Figure 3.
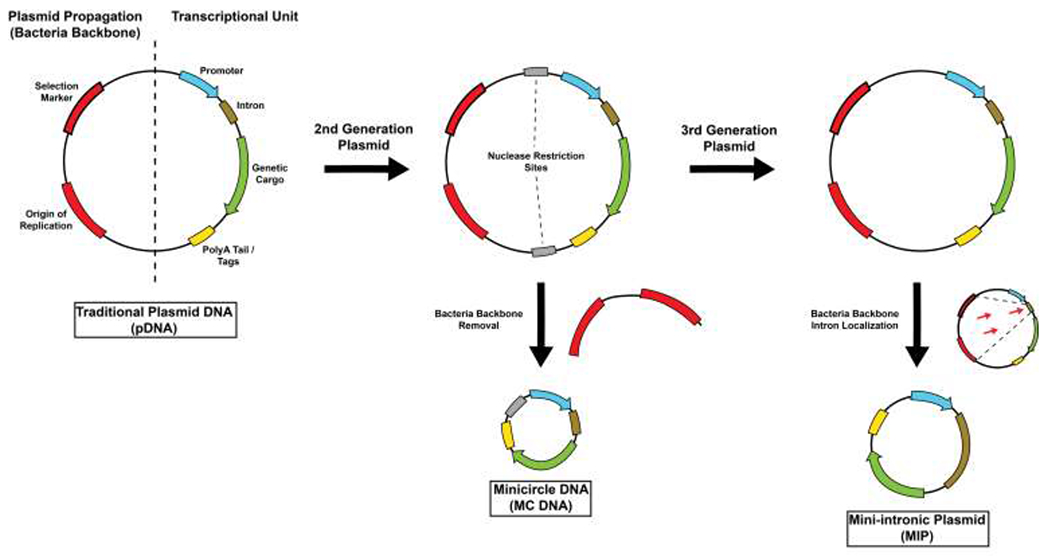
Generational changes of plasmid expression systems. Traditional plasmid DNA (pDNA) design usually contains two regions, one dedicated to plasmid propagation and the other to genetic cargo activity. However, in second generation plasmids, the vector is propagated and then processed with nucleases and ligated to remove the bacterial backbone. Alternatively, in third generation plasmids, no additional processing is required, as the plasmid propagation region is located to the intron in the transcriptional unit.
However, the continuous presence of heterologous genetic content is associated with potential side-effects. As such, in some applications, the desired cellular response falls in a range between typical transient expression vectors and the permanent genome editing strategies detailed above. Recently, proof-of-concept studies demonstrated the development of an expression construct that contains a CRISPR-based self-cleaving mechanism (Figure 4) (Moore et al., 2015). Specifically, upon delivery, Cas9 activity is directed against strategically placed targets thereby inactivating a co-expressed gene of interest. Thus, by combining various tools, one can envision extended expression with a controllable self-destruction mechanism in order to provide gene therapy outcomes across any time scale.
Figure 4.
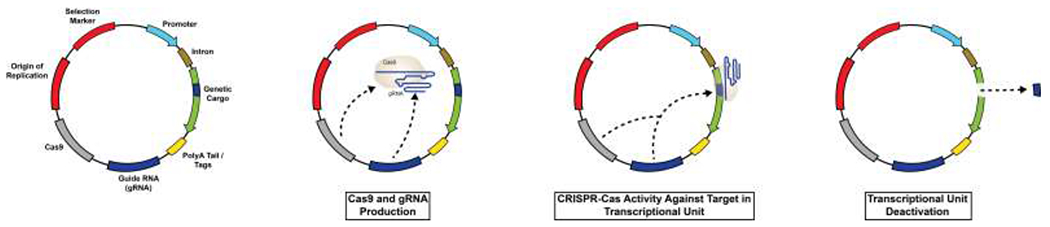
CRISPR-Cas mediated plasmid self-destruct mechanism. Internal production of Cas9 and guide RNA (gRNA) target DNA in the transcriptional unit. By self-cleaving, the plasmid will moderate outcomes in the host cell.
In summary, these genetic-based delivery strategies improve and expand upon current gene therapy technologies. Considering the current limitations associated with nonviral delivery vectors, universal usage is currently not feasible. However, by combining delivery and expression vector advancements, gene therapy, as a field, is becoming better equipped to systemically overcome notoriously difficult disease-specific barriers.
Conclusion
The field of gene therapy has historically been predicated on the use of both viral and nonviral delivery vectors to achieve systemic outcomes. However, due to various innate and technical limitations, few traditional delivery strategies have been utilized clinically. Moreover, recent advancements in various areas related to gene therapy have enabled the development of novel nonviral delivery vectors that allow new genetic outcome opportunities. From the re-envisioning of current biomaterial principles to the recapitulation of biological phenomena, these approaches collectively highlight the transition from standard particulate vectors to delivery platforms that are better equipped to traverse the barriers associated with achieving clinical relevancy. In addition, the development of advanced genetic cargo should increase the safety and scope of gene therapy to treat a wider range of adverse maladies. That stated, fulfillment of the immense therapeutic potential of these contemporary approaches in gene delivery remains on the horizon.
Funding Sources
The authors recognize support from NIH award AI088485 (BAP) and a SUNY-Buffalo Schomburg fellowship (CHJ).
REFERENCES
- Acevedo R, Fernandez S, Zayas C, Acosta A, Sarmiento ME, Ferro VA, Rosenqvist E, Campa C, Cardoso D, Garcia L, Perez JL. Bacterial outer membrane vesicles and vaccine applications. Front Immunol 5:121, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin D, Sturgis J, Ragheb K, Sherman D, Burkholder K, Robinson JP, Bhunia AK, Mohammed S, Bashir R. Bacteria-mediated delivery of nanoparticles and cargo into cells. Nat Nanotechnol 2(7):441–449, 2007. [DOI] [PubMed] [Google Scholar]
- Avila-Calderon ED, Araiza-Villanueva MG, Cancino-Diaz JC, Lopez-Villegas EO, Sriranganathan N, Boyle SM, Contreras-Rodriguez A. Roles of bacterial membrane vesicles. Arch Microbiol 197(1):1–10, 2015. [DOI] [PubMed] [Google Scholar]
- Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther 17(3):253–263, 2006. [DOI] [PubMed] [Google Scholar]
- Bechler SL, Lynn DM. Characterization of degradable polyelectrolyte multilayers fabricated using DNA and a fluorescently-labeled poly(beta-amino ester): shedding light on the role of the cationic polymer in promoting surface-mediated gene delivery. Biomacromolecules 13(2):542–552, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959):1509–1512, 2009. [DOI] [PubMed] [Google Scholar]
- Chart H, Smith HR, La Ragione RM, Woodward MJ. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5alpha and EQ1. J of Appl Microbiol 89(6):1048–1058, 2000. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther 8(3):495–500, 2003. [DOI] [PubMed] [Google Scholar]
- Cheung W, Kotzamanis G, Abdulrazzak H, Goussard S, Kaname T, Kotsinas A, Gorgoulis VG, Grillot-Courvalin C, Huxley C. Bacterial delivery of large intact genomic-DNA-containing BACs into mammalian cells. Bioeng Bugs 3(2):86–92, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31(3):230–232, 2013. [DOI] [PubMed] [Google Scholar]
- Critchley-Thorne RJ, Stagg AJ, Vassaux G. Recombinant Escherichia coli expressing invasin targets the Peyer’s patches: the basis for a bacterial formulation for oral vaccination. Mol Ther 14(2):183–191, 2006. [DOI] [PubMed] [Google Scholar]
- Critchley RJ, Jezzard S, Vassaux G. Genetically engineered E. coli as a protein delivery vehicle for killing cancer cells. Discov Med 4(22):194–197, 2004. [PubMed] [Google Scholar]
- Demuth PC, Min Y, Huang B, Kramer JA, Miller AD, Barouch DH, Hammond PT, Irvine DJ. Polymer multilayer tattooing for enhanced DNA vaccination. Nat Mater 12(4):367–376, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz WM, Skinner NE, Hamilton SE, Jund MD, Heitfeld SM, Litterman AJ, Hwu P, Chen ZY, Salazar AM, Ohlfest JR, Blazar BR, Pennell CA, Osborn MJ. Minicircle DNA is superior to plasmid DNA in eliciting antigen-specific CD8+ T-cell responses. Mol Ther 21(8):1526–1535, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Meyer BJ. Dramatic Enhancement of Genome Editing by CRISPR/Cas9 Through Improved Guide RNA Design. Genetics 199(4):959–971, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flessner RM, Jewell CM, Anderson DG, Lynn DM. Degradable polyelectrolyte multilayers that promote the release of siRNA. Langmuir 27(12):7868–7876, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillot-Courvalin C, Goussard S, Courvalin P. Wild-type intracellular bacteria deliver DNA into mammalian cells. Cell Microbiol 4(3):177–186, 2002. [DOI] [PubMed] [Google Scholar]
- Gujrati V, Kim S, Kim S-H, Min JJ, Choy HE, Kim SC, Jon S. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 8(2):1525–1537, 2014. [DOI] [PubMed] [Google Scholar]
- He Y, Nie Y, Cheng G, Xie L, Shen Y, Gu Z. Viral mimicking ternary polyplexes: a reduction-controlled hierarchical unpacking vector for gene delivery. Adv Mater 26(10):1534–1540, 2014. [DOI] [PubMed] [Google Scholar]
- Jones CH, Chen CK, Chen M, Ravikrishnan A, Zhang H, Gollakota A, Chung T, Cheng C, Pfeifer BA. PEGylated cationic polylactides for hybrid biosynthetic gene delivery. Mol Pharm 12(3):846–856, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming Nonviral Gene Delivery Barriers: Perspective and Future. Mol Pharm 10(11):4082–4098, 2013a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Hakansson AP, Pfeifer BA. Biomaterials at the interface of nano-and micro-scale vector-cellular interactions in genetic vaccine design. J of Mater Chem B 2(46):8053–8068, 2014a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Rane S, Patt E, Ravikrishnan A, Chen CK, Cheng C, Pfeifer BA. Polymyxin B treatment improves bactofection efficacy and reduces cytotoxicity. Mol Pharm 10(11):4301–4308, 2013b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Ravikrishnan A, Chen M, Reddinger R, Kamal Ahmadi M, Rane S, Hakansson AP, Pfeifer BA. Hybrid biosynthetic gene therapy vector development and dual engineering capacity. Proc Natl Acad Sci US A 111(34):12360–12365, 2014b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Lu K, Leelawattanachai J, Hu X, Park S, Park T, Min IM, Jin MM. Virus-mimetic polyplex particles for systemic and inflammation-specific targeted delivery of large genetic contents. Gene Ther 20(11):1042–1052, 2013. [DOI] [PubMed] [Google Scholar]
- Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nat Biotechnol 28(12):1287–1289, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney M, Ong SG, Padilla A, Yao Z, Goodman S, Wu JC, Yang F. Development of poly(beta-amino ester)-based biodegradable nanoparticles for nonviral delivery of minicircle DNA. ACS Nano 7(8):7241–7250, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri N, Rathi M, Baradia D, Trehan S, Misra A. In vivo delivery aspects of miRNA, shRNA and siRNA. Crit Rev Ther Drug Carrier Syst 29(6):487–527, 2012. [DOI] [PubMed] [Google Scholar]
- Kruhn A, Wang A, Fruehauf JH, Lage H. Delivery of short hairpin RNAs by transkingdom RNA interference modulates the classical ABCB1-mediated multidrug-resistant phenotype of cancer cells. Cell Cycle 8(20):3349–3354, 2009. [DOI] [PubMed] [Google Scholar]
- Laner A, Goussard S, Ramalho AS, Schwarz T, Amaral MD, Courvalin P, Schindelhauer D, Grillot-Courvalin C. Bacterial transfer of large functional genomic DNA into human cells. Gene Ther 12(21):1559–1572, 2005. [DOI] [PubMed] [Google Scholar]
- Larsen MD, Griesenbach U, Goussard S, Gruenert DC, Geddes DM, Scheule RK, Cheng SH, Courvalin P, Grillot-Courvalin C, Alton EW. Bactofection of lung epithelial cells in vitro and in vivo using a genetically modified Escherichia coli. Gene Ther 15(6):434–442, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Kim D, Youn YS, Oh KT, Bae YH. A virus-mimetic nanogel vehicle. Angew Chem Int Ed Engl 47(13):2418–2421, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Puhl S, Meinel L, Germershaus O. Silk fibroin layer-by-layer microcapsules for localized gene delivery. Biomaterials 35(27):7929–7939, 2014. [DOI] [PubMed] [Google Scholar]
- Lu J, Zhang F, Kay MA. A Mini-intronic Plasmid (MIP): a novel robust transgene expression vector in vivo and in vitro. Mol Ther 21(5):954–963, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhang F, Xu S, Fire AZ, Kay MA. The extragenic spacer length between the 5’ and 3’ ends of the transgene expression cassette affects transgene silencing from plasmid-based vectors. Mol Ther 20(11):2111–2119, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 339(6121):823–826, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD. Delivery of RNAi therapeutics: work in progress. Expert Rev Med Devices 10(6):781–811, 2013. [DOI] [PubMed] [Google Scholar]
- Moore R, Spinhirne A, Lai MJ, Preisser S, Li Y, Kang T, Bleris L. CRISPR-based self-cleaving mechanism for controllable gene delivery in human cells. Nucleic Acids Res 43(2):1297–1303, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 326(5959):1501, 2009. [DOI] [PubMed] [Google Scholar]
- Musacchio T, Torchilin VP. siRNA delivery: from basics to therapeutic applications. Front Biosci (Landmark Ed) 18:58–79, 2013. [DOI] [PubMed] [Google Scholar]
- Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res 1:27, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discovery 4(7):581–593, 2005. [DOI] [PubMed] [Google Scholar]
- Parsa S, Wang Y, Fuller J, Langer R, Pfeifer BA. A comparison between polymeric microsphere and bacterial vectors for macrophage p388d1 gene delivery. Pharm Res 25:1202–1208, 2008a. [DOI] [PubMed] [Google Scholar]
- Parsa S, Wang Y, Rines K, Pfeifer BA. A high-throughput comparison of recombinant gene expression parameters for E. coli-mediated gene transfer to P388D1 macrophage cells. J Biotechnol 137(1–4):59–64, 2008b. [DOI] [PubMed] [Google Scholar]
- Paukner S, Kudela P, Kohl G, Schlapp T, Friedrichs S, Lubitz W. DNA-loaded bacterial ghosts efficiently mediate reporter gene transfer and expression in macrophages. Mol Ther 11(2):215–223, 2005. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6):1380–1389, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11(8):783–784, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JL, Nouri A, Fernandes T, Rodrigues J, Tomas H. Gene delivery using biodegradable polyelectrolyte microcapsules prepared through the layer-by-layer technique. Biotechnol Prog 28(4):1088–1094, 2012. [DOI] [PubMed] [Google Scholar]
- Saurer EM, Flessner RM, Sullivan SP, Prausnitz MR, Lynn DM. Layer-by-layer assembly of DNA-and protein-containing films on microneedles for drug delivery to the skin. Biomacromolecules 11(11):3136–3143, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W Direct transfer of cloned genes from bacteria to mammalian cells. Proc Natl Acad Sci U S A 77(4):2163–2167, 1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk F, Grund S, Fischer D. Recent developments and perspectives on gene therapy using synthetic vectors. Ther Deliv 4(1):95–113, 2013. [DOI] [PubMed] [Google Scholar]
- Seow Y, Wood MJ. Biological gene delivery vehicles: beyond viral vectors. Mol Ther 17(5):767–777, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal CM, Schaar V, Riesbeck K. Bacterial outer membrane vesicles in disease and preventive medicine. Semin Immunopathol 33(5):395–408, 2011. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11(9):636–646, 2010. [DOI] [PubMed] [Google Scholar]
- Vilte DA, Larzabal M, Mayr UB, Garbaccio S, Gammella M, Rabinovitz BC, Delgado F, Meikle V, Cantet RJ, Lubitz P, Lubitz W, Cataldi A, Mercado EC. A systemic vaccine based on Escherichia coli O157:H7 bacterial ghosts (BGs) reduces the excretion of E. coli O157:H7 in calves. Vet Immunol Immunopathol 146(2):169–176, 2012. [DOI] [PubMed] [Google Scholar]
- Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol 24(6):697–702, 2006. [DOI] [PubMed] [Google Scholar]
- Xu DQ, Zhang L, Kopecko DJ, Gao L, Shao Y, Guo B, Zhao L. Bacterial delivery of siRNAs: a new approach to solid tumor therapy. Methods Mol Biol 487:161–187, 2009. [DOI] [PubMed] [Google Scholar]
- Xu J, Luft JC, Yi X, Tian S, Owens G, Wang J, Johnson A, Berglund P, Smith J, Napier ME, Desimone JM. RNA replicon delivery via lipid-complexed PRINT protein particles. Mol Pharm 10(9):3366–3374, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Frederik P, Pirollo KF, Tang WH, Rait A, Xiang LM, Huang W, Cruz I, Yin Y, Chang EH. Self-assembly of a virus-mimicking nanostructure system for efficient tumor-targeted gene delivery. Hum Gene Ther 13(3):469–481, 2002. [DOI] [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet 15(8):541–555, 2014. [DOI] [PubMed] [Google Scholar]
- Zou Y, Xie L, Carroll S, Muniz M, Gibson H, Wei WZ, Liu H, Mao G. Layer-by-layer films with bioreducible and nonbioreducible polycations for sequential DNA release. Biomacromolecules 15(11):3965–3975, 2014. [DOI] [PubMed] [Google Scholar]


