Abstract
Vector-borne diseases (VBDs) are a worldwide critical concern accounting for 17% of the estimated global burden of all infectious diseases in 2020. Despite the various medicines available for the management, the deadliest VBD malaria, caused by Plasmodium sp., has resulted in hundreds of thousands of deaths in sub-Saharan Africa only. This finding may be explained by the progressive loss of antimalarial medication efficacy, inherent toxicity, the rise of drug resistance, or a lack of treatment adherence. As a result, new drug discoveries from uncommon sources are desperately needed, especially against multi-drug resistant strains. Marine organisms have been investigated, including sponges, soft corals, algae, and cyanobacteria. They have been shown to produce many bioactive compounds that potentially affect the causative organism at different stages of its life cycle, including the chloroquine (CQ)-resistant strains of P. falciparum. These compounds also showed diverse chemical structures belonging to various phytochemical classes, including alkaloids, terpenoids, polyketides, macrolides, and others. The current article presents a comprehensive review of marine-derived natural products with antimalarial activity as potential candidates for targeting different stages and species of Plasmodium in both in vitro and in vivo and in comparison with the commercially available and terrestrial plant-derived products, i.e., quinine and artemisinin.
Vector-borne diseases (VBDs) are a worldwide critical concern accounting for 17% of the estimated global burden of all infectious diseases in 2020.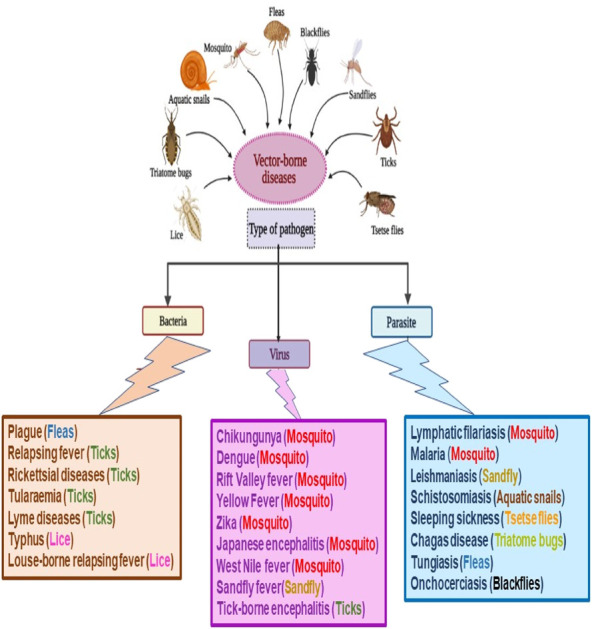
1. Introduction
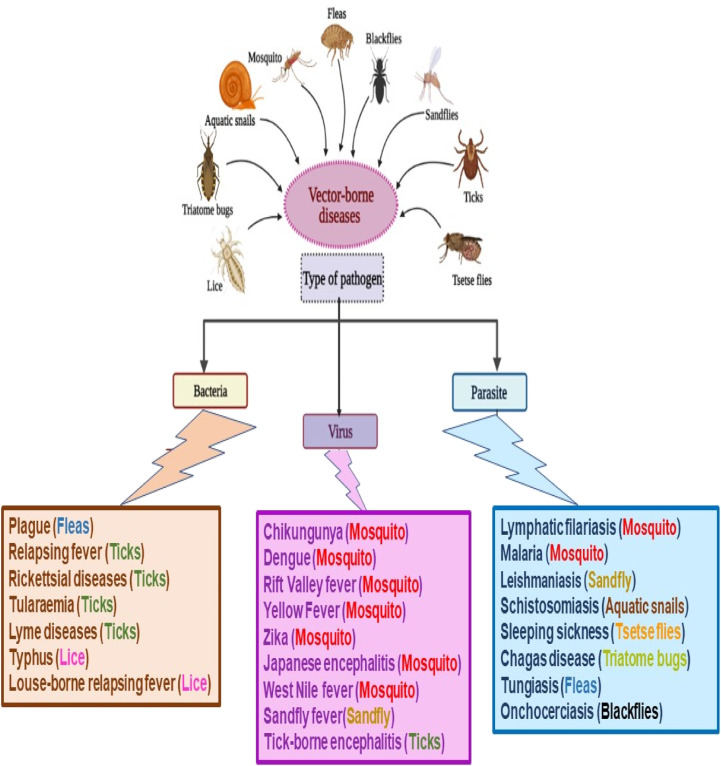
Vector-borne diseases (VBDs) are infectious diseases caused by parasites, bacteria, and viruses transmitted via vectors. About 700 000 deaths are reported officially by the World Health Organization (WHO) from these diseases per year worldwide, including malaria, dengue, schistosomiasis, human African trypanosomiasis, leishmaniasis, Chagas disease, chikungunya fever, Zika virus fever, yellow fever, West Nile fever, Japanese encephalitis, and onchocerciasis, Fig. 1. Based on the WHO reports released in 2020, VBDs are a worldwide concern that accounts for 17% of the estimated global burden of all infectious diseases.1
Fig. 1. Classification of vector-borne diseases (VBDs) according to pathogen types.
VBDs are commonly associated with weather and climate, where the incidence of these diseases is mainly in the tropics and subtropical regions. The low hygiene, sanitation, waste management, and housing in these urban areas help also spread such diseases between the world's poorest people, communities, and countries.2 Various native to these regions as arthropods, including mosquitoes, ticks, sand flies, triatomine bugs, cockroaches, lice, fleas, and aquatic snails, are involved as mediators transmitting VBDs.3,4
Particularly, malaria is the most challenging VBD that leads to health problems worldwide, especially in developing countries. It is a mosquito-borne infectious disease that affects humans and other animals. An estimated 405 000 malaria deaths worldwide were registered, along with 228 million cases in 2018, compared to 229 million cases and 409 000 deaths with more than 400 000 deaths in 2019, based on the WHO report.5,6 In other words, malaria accounts for more than 50% of VBDs deaths.
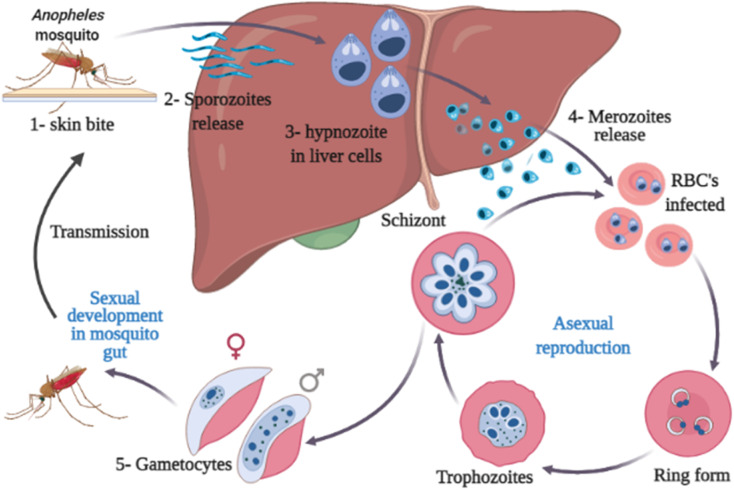
Malaria is transmitted through the bite of an infected Anopheles female mosquito. The infected mosquitoes carry one of several protozoans belonging to the genus Plasmodium (P. falciparum, P. ovale, P. vivax, P. knowlesi, and P. malariae).7,8 The parasite is then released into the bloodstream causing severe anemia and other signs and symptoms, including chills, fever, profuse sweating, headache, nausea, vomiting, abdominal pain, diarrhea, muscle pain, convulsions, coma, bloody stools.6,9,10 Serious complications or even death can occur in case of improper diagnosis or treatment.11–13 The life cycle of the malaria parasite of Plasmodium sp. is illustrated in Fig. 2. This figure is of great importance for helping drug discovery processes of novel drugs targeting critical stages in the parasite life cycle.
Fig. 2. Life cycle of the malaria parasite.
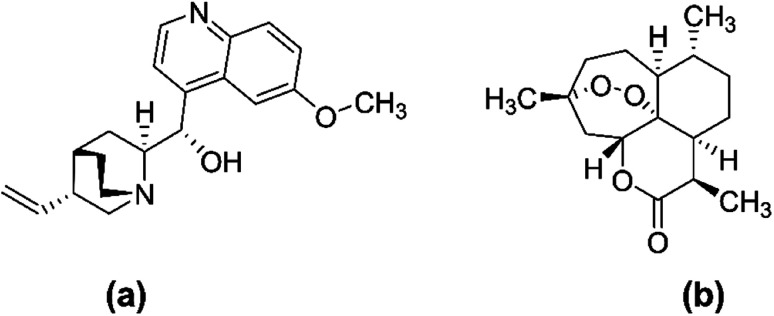
Chloroquine (CQ) and hydroxychloroquine are two existing chemical medicines that have limited usefulness and efficacy as antimalarials due to their high cost, unpleasant side effects, and evolution of multi-drug resistance associated with them.14 Hence, there has been an urgent need and continuous search for novel sources of more efficacious drugs to combat the disease. Nevertheless, natural products, including terrestrial medicinal plants, have a long history in the treatment of malaria owing to their relative efficacy, safety, reasonable cost, and availability.15,16 The two most successful antimalarial drugs; namely artemisinin and quinine (Fig. 3), were sourced from medicinal plants of cinchona qinghao (Artemisia annua, Family Asteraceae) and (Cinchona officinalis, Family Rubiaceae), respectively, and have been used for hundreds of years and before the mosquito cycle was explored. Even today, in the fight against malaria, both quinine and artemisinin are still of prime importance.17
Fig. 3. Chemical structure of quinine (a) and artemisinin (b).
In contrast to terrestrial plants, marine organisms do not have a remarkable history of use in traditional medicine. However, recent advances in marine biology and engineering have helped investigation and scientific exploration of the marine environment to identify and isolate novel compounds, which have proven their potential bioactivities against life-threatening diseases, including tumor and viral infections.18–20 More than 30 000 compounds have been identified from about 240 000 known species of marine organisms.21 Few of them have been approved by the Food and Drug Administration (FDA), including ziconotide (Prialt®) as a potent analgesic, trabectedin (Yondelis®), and cytarabine or ara-C (Cytosar-U®) as anti-tumor agents, vidarabine or ara-A (Vira-A®) and iota-carrageenan (Carragelose®) as an antiviral, and omega-3-acid ethyl ester (Lovaza®) for treating hypertriglyceridemia (Table 1).22–24
A list of some examples of approved marine-derived drugs currently on the market.
| Trade name | Scientific name | Source | Family | Indication | Ref. |
|---|---|---|---|---|---|
| Prialt® | Ziconotide | Cone snail species Conus magus | Conidae | potent analgesic | 22–24 |
| Yondelis® | Trabectedin | Candidatus Endoecteinascidia frumentensis | Unclassified family candidatus endolissoclinum | Anti-tumor | 22–24 |
| Cytosar-U® | Cytarabine or ara-C | Cryptotethia crypta sponge | Tethyidae | Anti-tumor | 22–24 |
| Carragelose® | Iota-carrageenan | Eucheuma denticulatum sponge | Solieriaceae | Antiviral | 22–24 |
| Vira-A® | Vidarabine or ara-A | Tectitethya crypta sponges | Tethyidae | Antiviral | 22–24 |
Recently, Nweze, et al. published a review article highlighting the potential of marine-derived natural products for the treatment of some examples of diseases for neglected communities, including malaria, leishmaniasis, and trypanosomiasis.25 Although some of the previous studies could not identify the chemical structure of bioactive components that acted significantly against malaria,26 the current article focuses on malaria. It reviews the different chemical classes, i.e., alkaloids, terpenoids, endoperoxides, phosphotriesters, peptides and depsipeptides, and macrolides, derived from marine organisms, including sponges, cyanobacteria, actinomycete bacteria, soft corals, and algae. These bioactive have been confirmed to be potential candidates for managing malaria compared to commercially available products by targeting various stages in Plasmodium sp. life cycle. Moreover, the half maximum cytotoxic (CC50) and inhibitory concentration (IC50) against the different stages of the malaria parasite shall be highlighted, in addition to the possible mechanism of action and structure-activity relationships (SAR) in previous reports investigated the antiplasmodium activity. Hence, the current review may open new frontiers for discovery and approval of novel potent drugs for this life-threatening disease.
2. Alkaloids
Various classes of marine-derived alkaloids have shown potent antimalarial activity. β-Carboline, indole, imidazole, and pyrrole alkaloids are mostly found. They showed bioactivities against different stages of the Plasmodium parasite with a unique mechanism of action. Fifteen classes represented by 67 compounds were reviewed. Among them are manzamine alkaloids which showed inhibitory activity against glycogen synthase 3 (GSK-3) topoisomerase. In addition, salinosporamide showed a potent protease inhibitory effect. Numerous marine-derived alkaloids shall be discussed in detail in the following sub-sections and Table 2, including their sources, IC50, chemical structures, SAR, and mechanism of action.
A list of marine-derived antimalarial alkaloids showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic sourcea.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Manzamine A (1) | W2 = 0.015 μM D6 = 0.0082 μM |
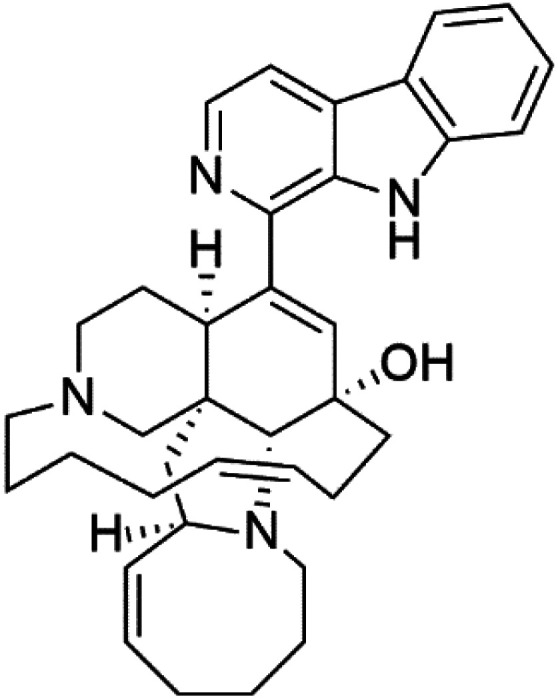
|
Okinawan Haliclona | Sponge | 27 |
| 8-Hydroxymanzamine (2) | W2 = 0.014 μM D6 = 0.010 μM |
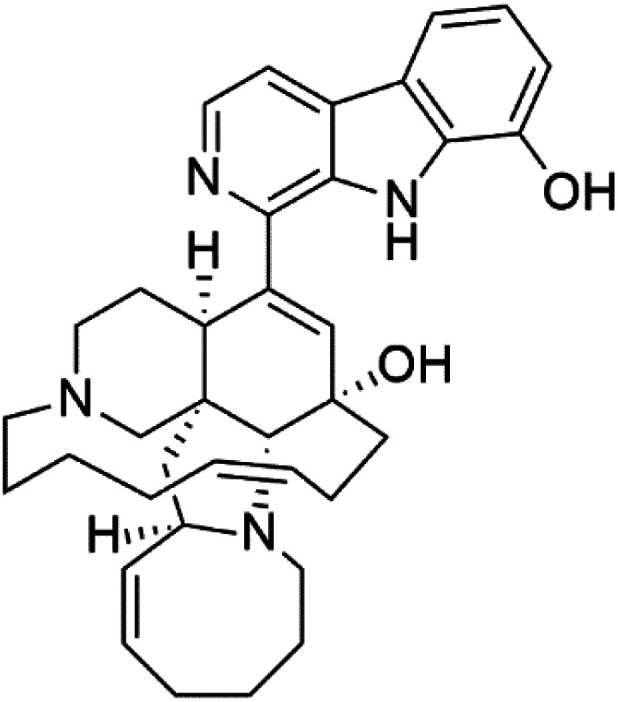
|
Sponge | 27 | |
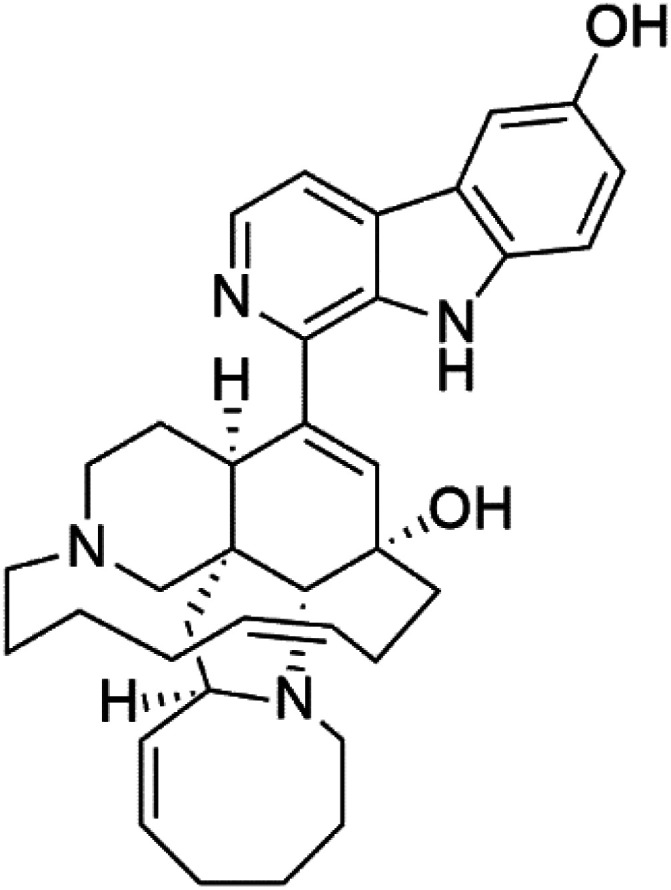
| Manzamine F (3) | W2 = 2.93 μM 6 = 1.34 μM |
|
Sponge | 27 | |
| 6-Hydroxymanzamine (4) | W2 = 1.5 μM D6 = 1.36 μM |
|
Sponge | 27 | |
| Neo-kauluamine (5) | D6 = 1.46 μM W2 = 2.41 μM |
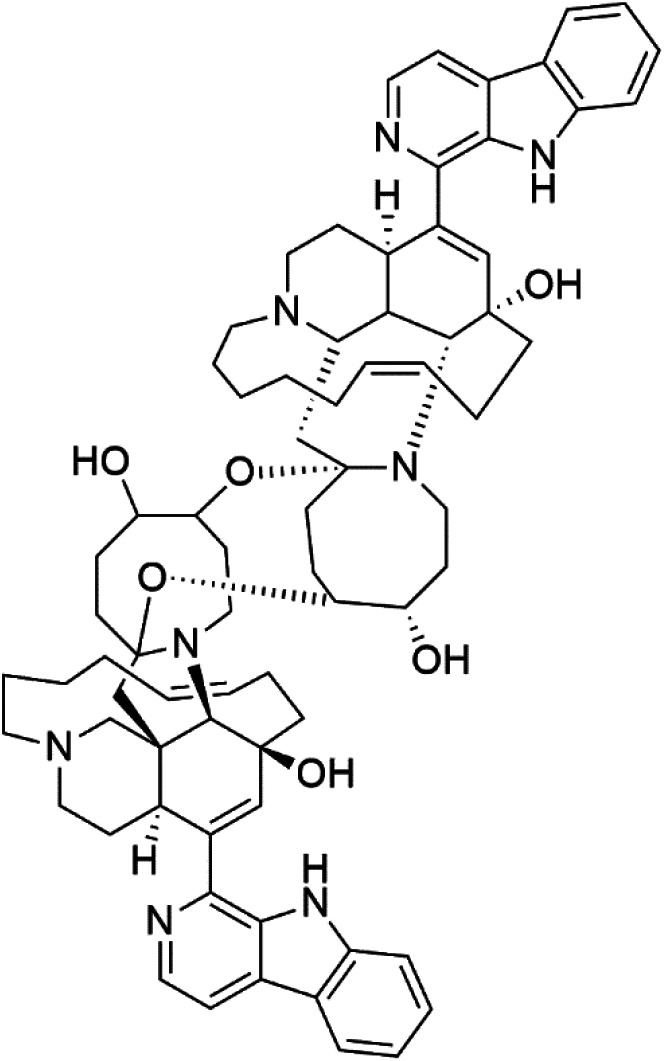
|
Indo-pacific sponge | Sponge | 28 |
| 12,34-Oxamanzamine A (6) | D6 = 8.97 μM |
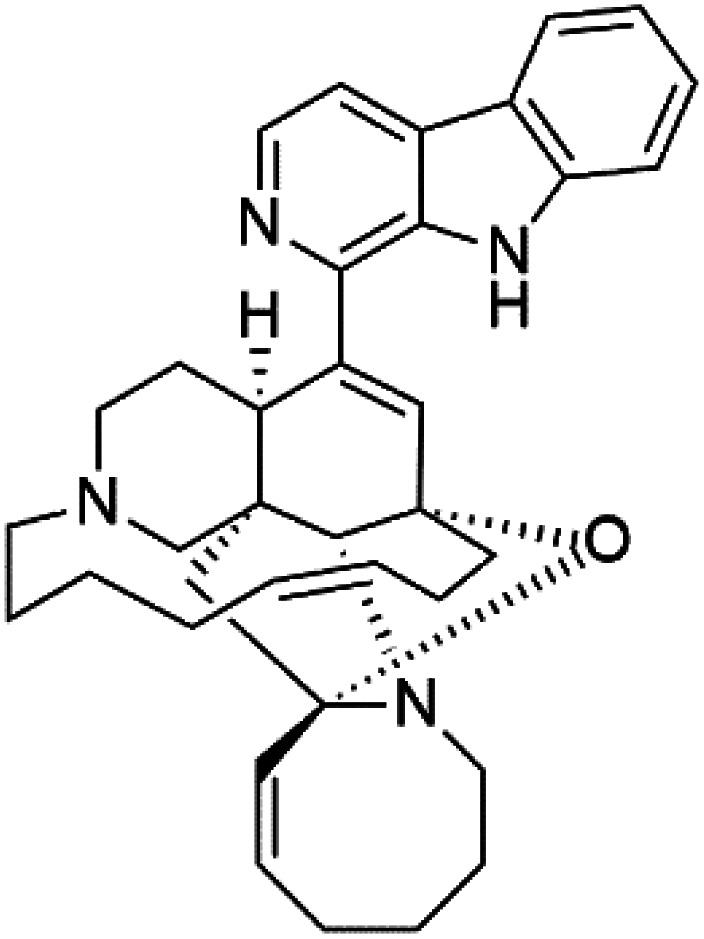
|
Sponge | 28 | |
| Zamamidine A (7) | 0.0008 to 0.016 μM |
|
Amphimedon sp. | Sponge | 35 |
| Zamamidine B (8) |
|
Sponge | |||
| Zamamidine C (9) |
|
Sponge | |||
| Zamamidine D (10) |
|
Sponge | |||
| Homofascaplysin (11) | K1 = 0.04 μM NF54 = 0.07 μM |
|
Hyrtios sponge | Sponge | 38 |
| Marinacarboline A (12) Marinacarboline B (13) Marinacarboline C (14) | 3D7 and Dd2 IC50 from 1.92 to 36.03 μM |
|
Marinactinospora thermotolerans | Actinomycete bacteria | 39 |
| Marinacarboline D (15) |
|
||||
| 13-N-Demethyl-methylpendolmycin (16) methylpendolmycin-14-O-α-glucoside (17) | 3D7 = 20.75 and 10.43 μM Dd2 = 18.67 and 5.03 μM |
|
Marinactinospora thermotolerans | Actinomycete bacteria | 38 |
| Crambescidin 800 (18) | 3D7 = 0.16 μM FCR3 = 0.24 μM |
|
Monanchora unguiculate | Sponge | 39 and 40 |
| Crambescidin 359 (19) crambescidin acid (20) | 3D7 = 20.75 and 10.43 μM Dd2 = 18.67 and 5.03 μM |
|
|||
| Fromiamycalin (21) | 3D7 = 0.24 μM |
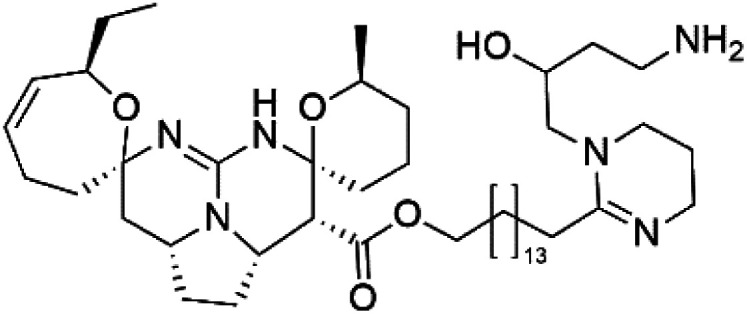
|
|||
| Unguiculin A (22) | 3D7 = 12.86 μM |
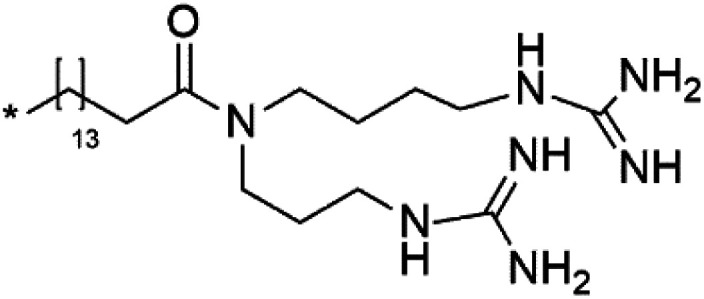
|
|||
| Ptilomycalins E (23) Ptilomycalins F (24) Ptilomycalins G (25) | 3D7 = 0.35, 0.23, and 0.46 μM |
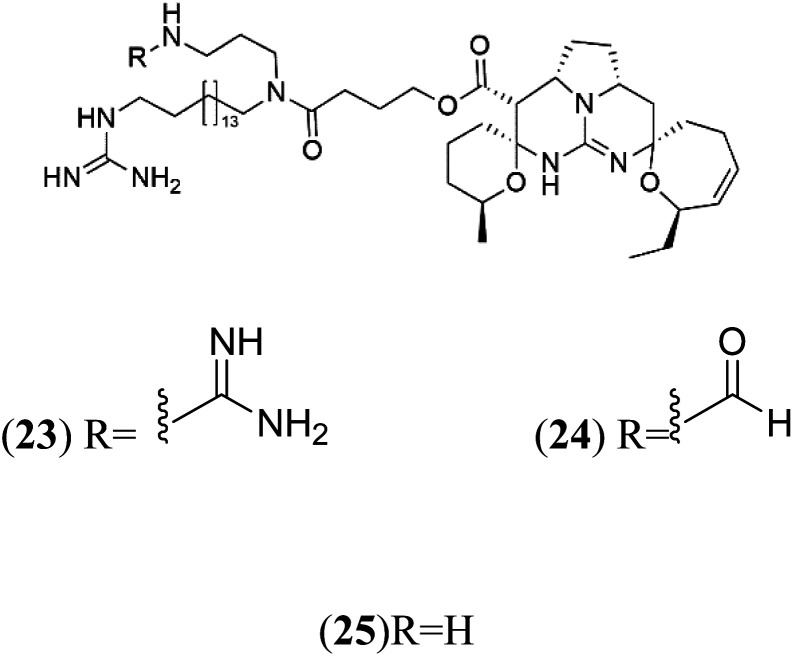
|
41 | ||
| Ptilomycalins H (26) | 3D7 = 0.46 μM |
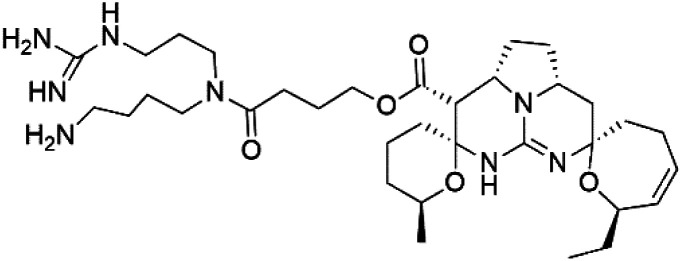
|
|||
| Opacaline B (27) Opacaline C (28) | K1 range of 2.5–14 μM |
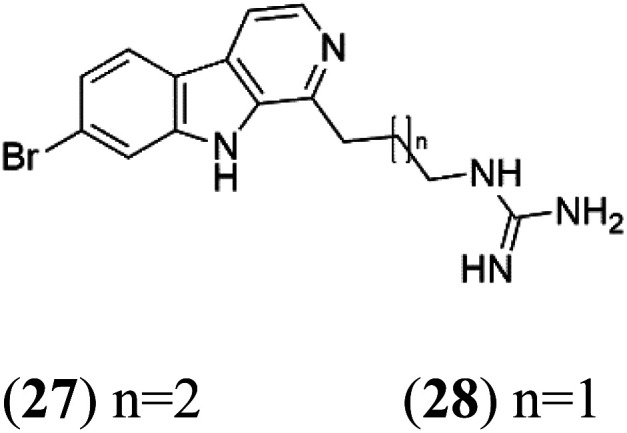
|
Pseudodistoma opacum | New Zealand ascidian | 42 |
| Didemnidine A (29) Didemnidine B (30) | K1 = 0.047 μM |
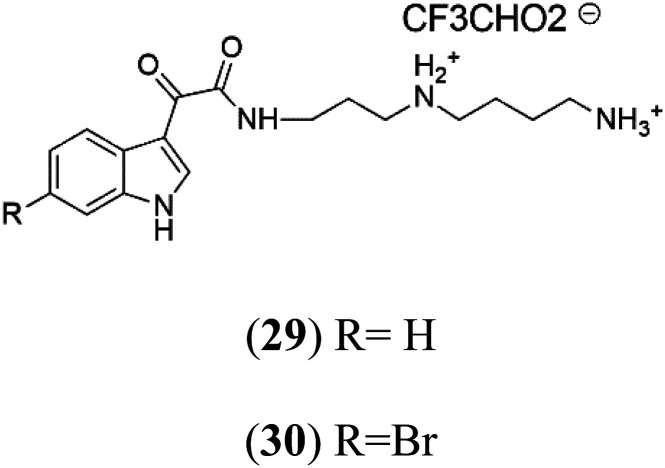
|
Ascidian Didemnum sp. | Marine tunicate | 43 |
| Salinosporamide A (31) | 3D7 = 11.4 nM FCB = 19.6 nM |
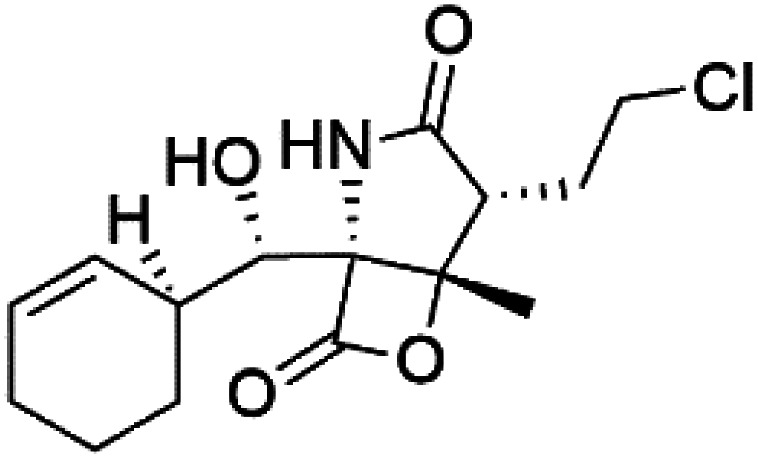
|
Salinispora tropica | Marine actinomycete bacteria | 44 |
| Psammaplysin H (32) Psammaplysin F (33) Psammaplysin G (34) | 3D7 = 0.41, 1.92, and 5.22 μM |
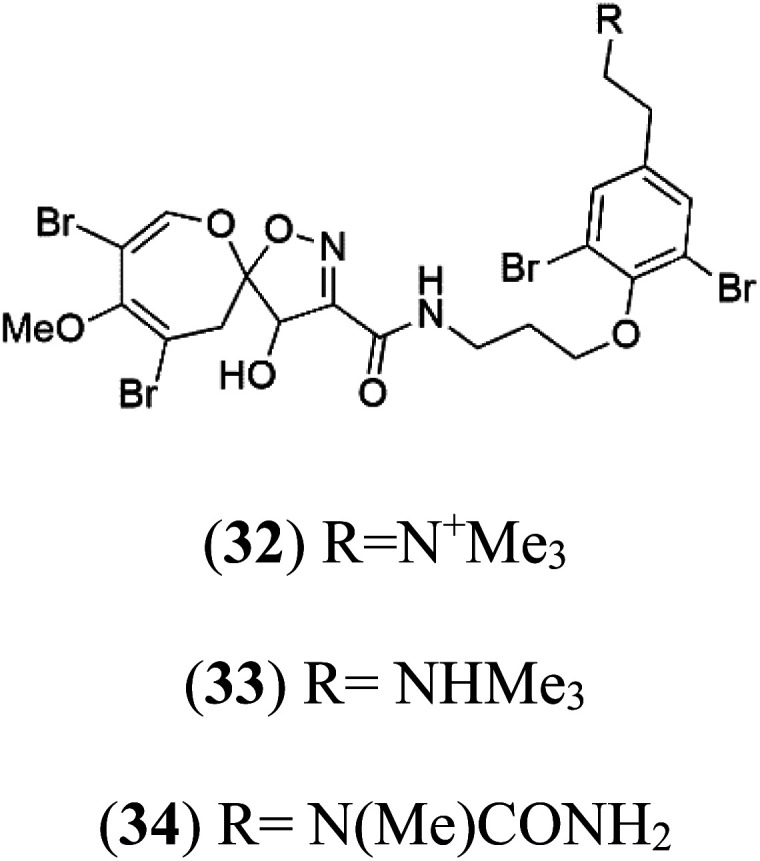
|
Aplysinella strongylata | Sponge | 52 |
| Ceratinadin E (35) | K1 = 0.9 μM |
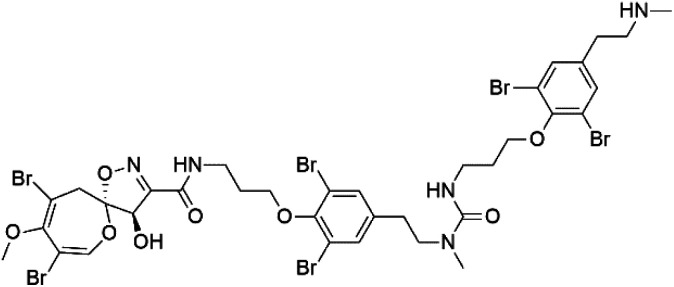
|
Okinawan Pseudoceratina | Sponge | 54 |
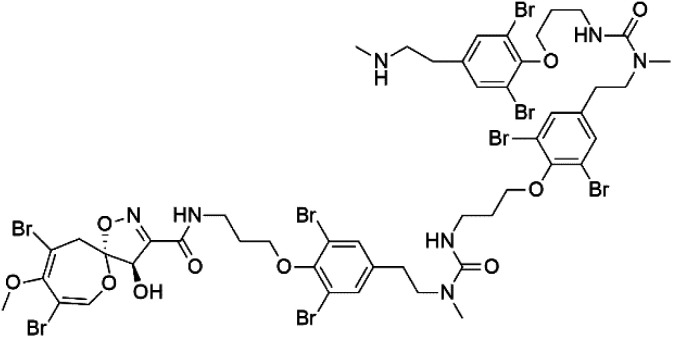
| Ceratinadin F (36) | K1 > 8.16 μM |
|
|||
| Tsitsikammamine C (37) | 3D7 = 13 nM Dd2 = 18 nM |
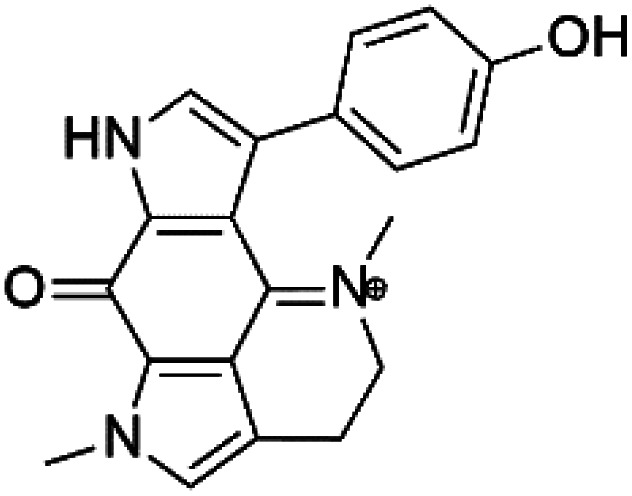
|
Zyzzya sp. | Sponge | 55 |
| Makaluvamine J (38) | 3D7 = 25 nM Dd2 = 22 nM |
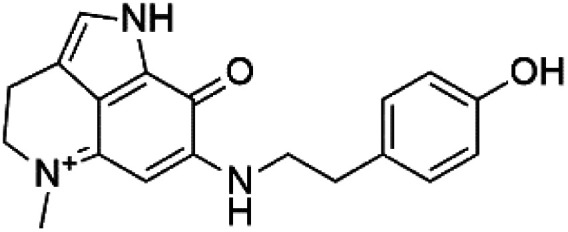
|
55 | ||
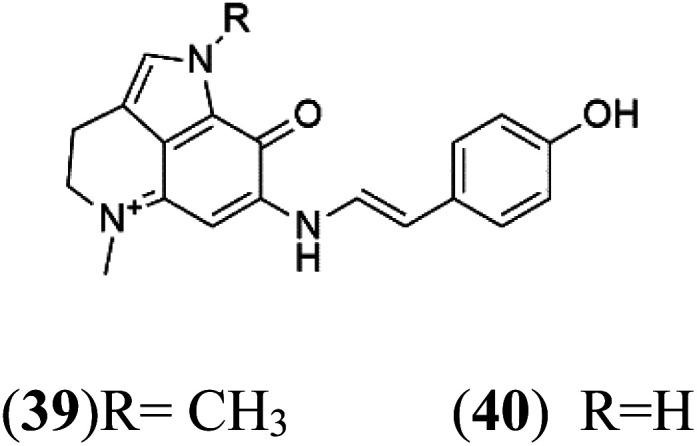
| Makaluvamine G (39) Makaluvamine L (40) | 3D7 = 36, 40 nM Dd2 = 39, 21 nM |
|
|||
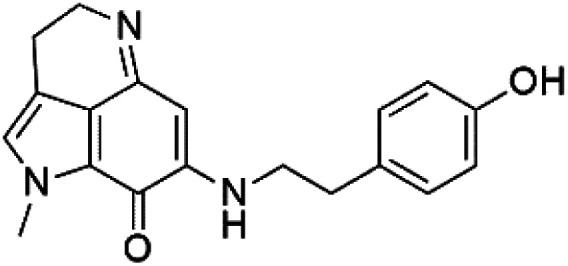
| Makaluvamine K (41) | 3D7 = 396 nM Dd2 = 300 nM |
|
|||
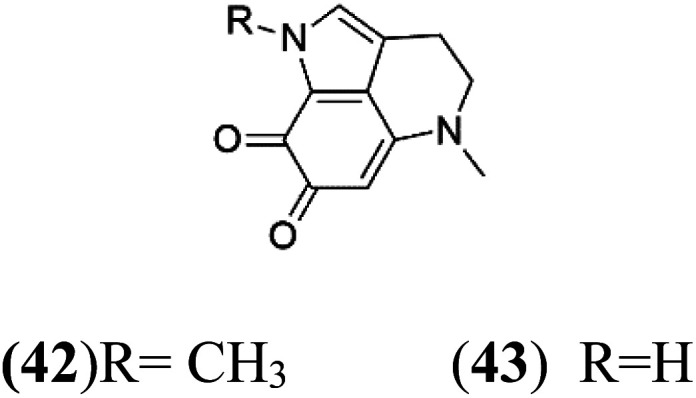
| Damirone A (42) Damirone B (43) | 3D7 = 1880 Dd2 = 360 nM |
|
|||
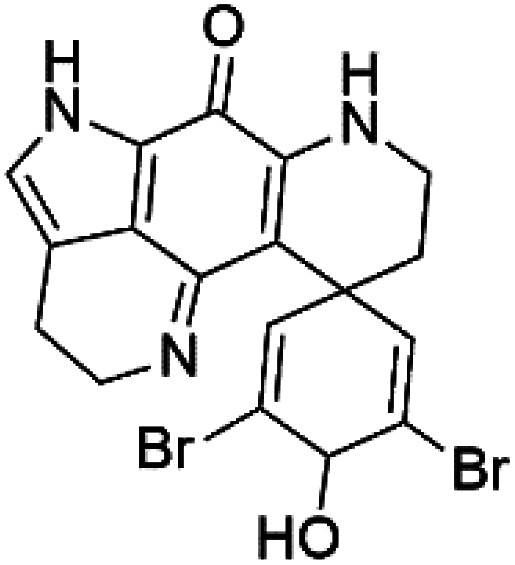
| Dihydrodiscorhabdin B (44) | D6 = 0.17 μM W2 = 0.13 μM |
|
Latrunculia sp. | Sponge | 56 |
| Discorhabdin Y (45) | EC50 = 0.5 μM |
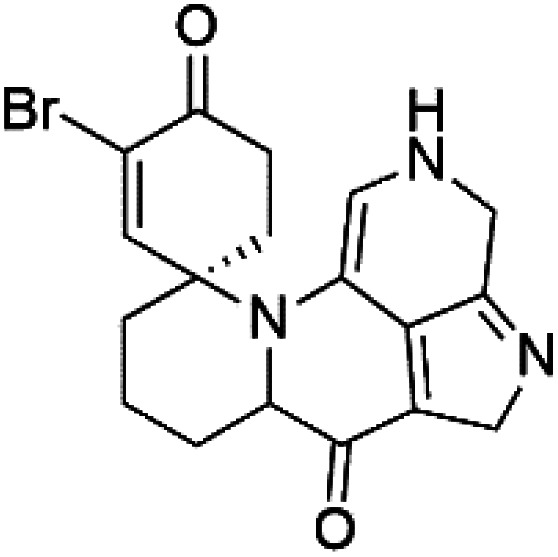
|
|||
| Discorhabdin A (46) | D6 = 0.05 μM W2 = 0.05 μM |
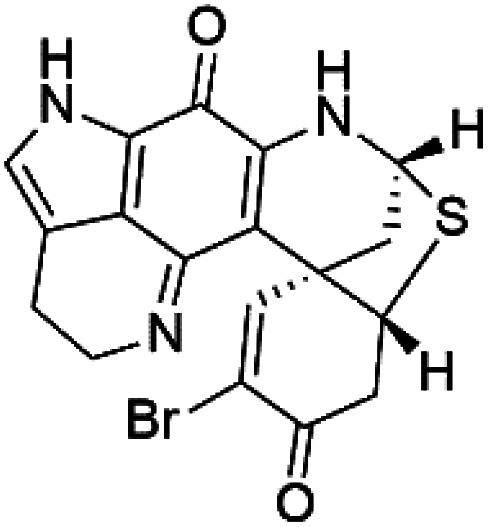
|
56 | ||
| Discorhabdin C (47) | D6 = 2.8 μM W2 = 2.0 μM |
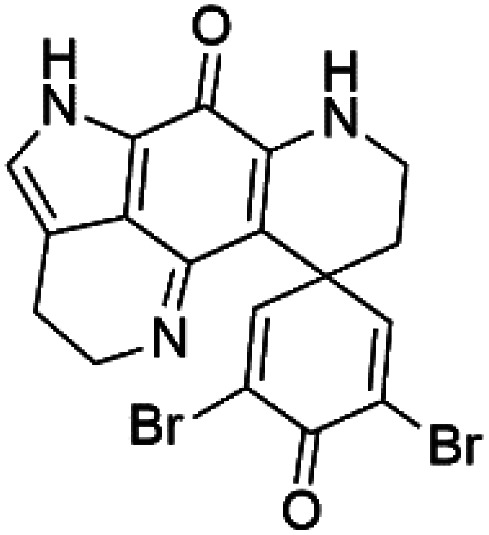
|
|||
| Discorhabdin E (48) | W2 = 0.2 μM |
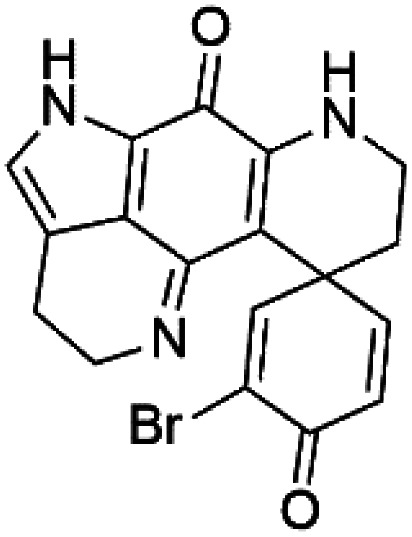
|
|||
| Discorhabdin L (49) | W2 = 0.13 μM |
|
|||
| Dihydrodiscorhabdin C (50) | D6 = 0.17 μM W2 = 0.13 μM |
|
|||
| Girolline (51) | FCM29 = 0.13 μM |
|
Cymbastela cantharella | Sponge | 58 |
| Thiaplakortone A (52) | 3D7 = 51 Dd2 = 6.6 nM |
|
Plakortis lita | Sponge | 59 |
| Thiaplakortone B (53) | 3D7 = 650 Dd2 = 92 nM |
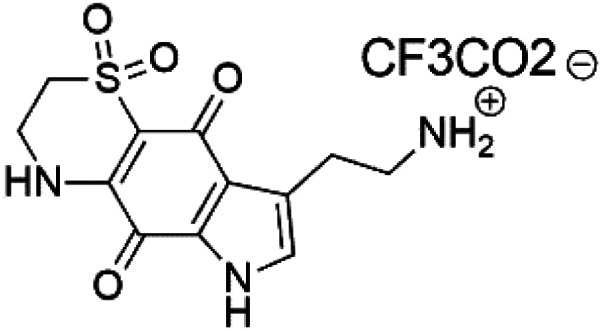
|
|||
| Thiaplakortone C (54) | 3D7 = 309 Dd2 = 171 nM |
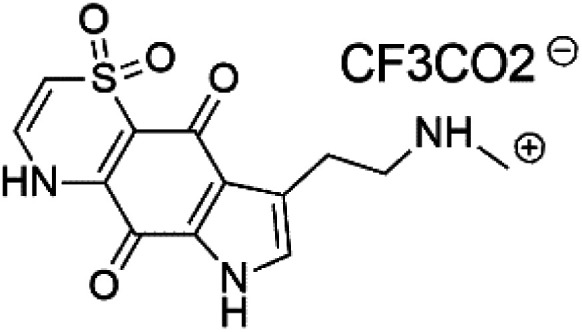
|
|||
| Thiaplakortone D (55) | 3D7 = 279 Dd2 = 159 nM |
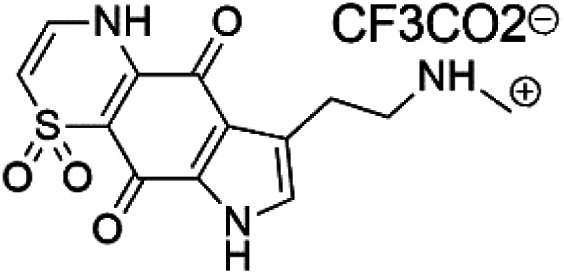
|
|||
| Monamphilectine A (56) | W2 = 600 nM |
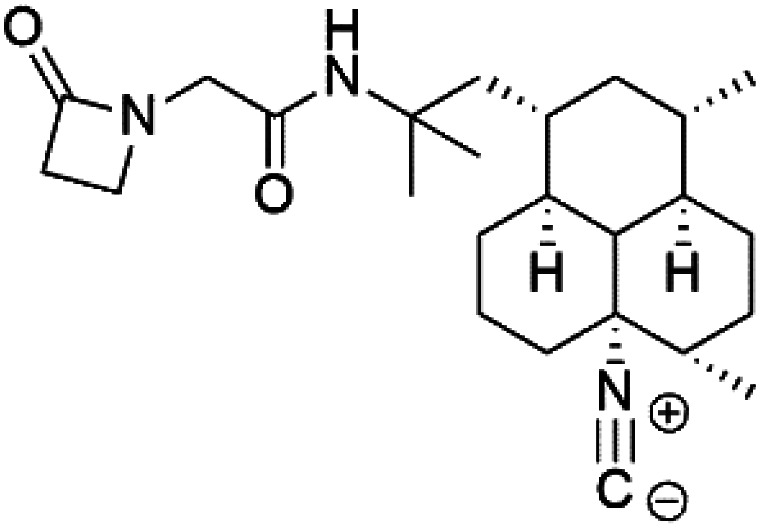
|
Hymeniacidon sp. | Sponge | 60 |
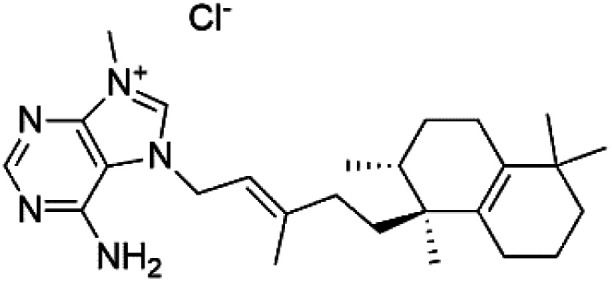
| Agelasine J (57) | FcB1 = 6.6 μM |
|
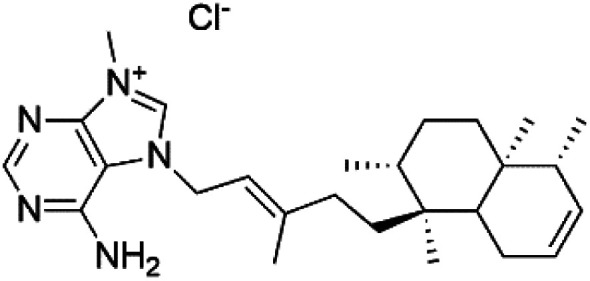
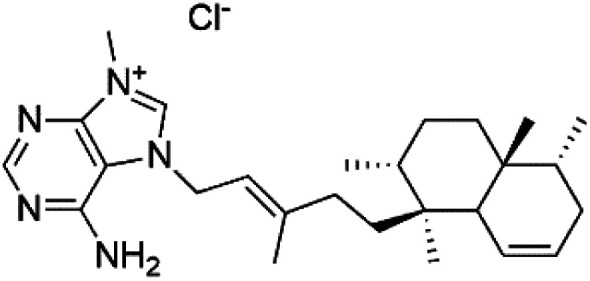
Agelas mauritiana | Sponge | 61 |
| Agelasine K (58) | FcB1 = 8.3 μM |
|
|||
| Agelasine L (59) | FcB1 = 18 μM |
|
|||
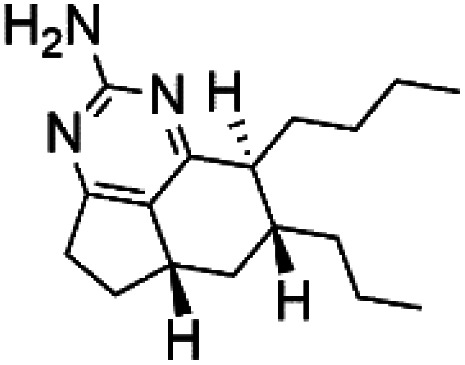
| Netamine G (60) | NA |
|
Biemna laboutei | Sponge | 62 |
| Netamine H (61) |
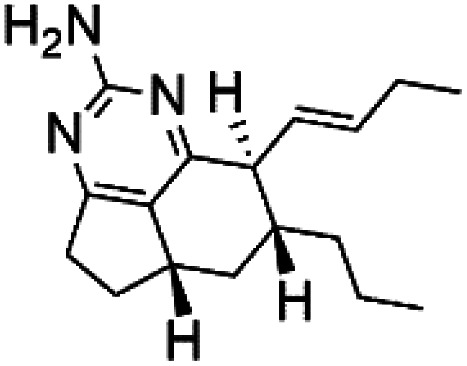
|
||||
| Netamine I (62) |
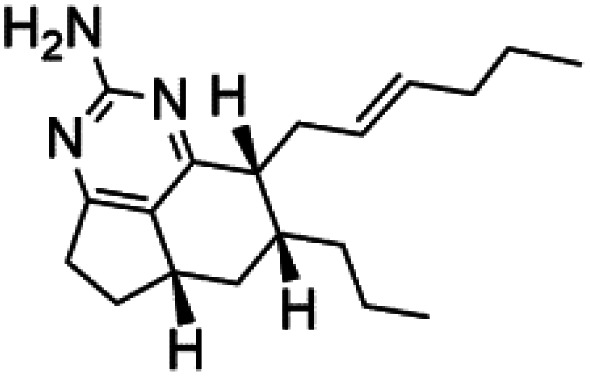
|
||||
| Netamine J (63) |
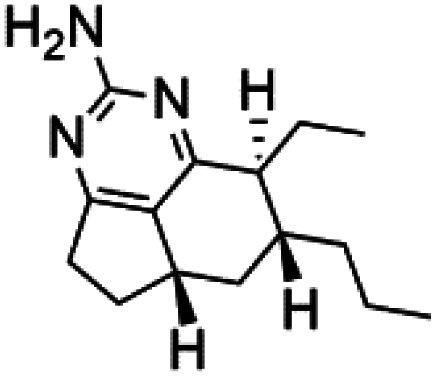
|
||||
| Netamine K (64) | (64) IC50 = 2.4 μM NA for the other compounds (65–67) |
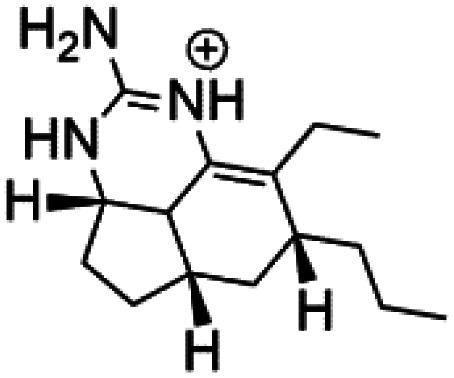
|
|||
| Netamine L (65) |
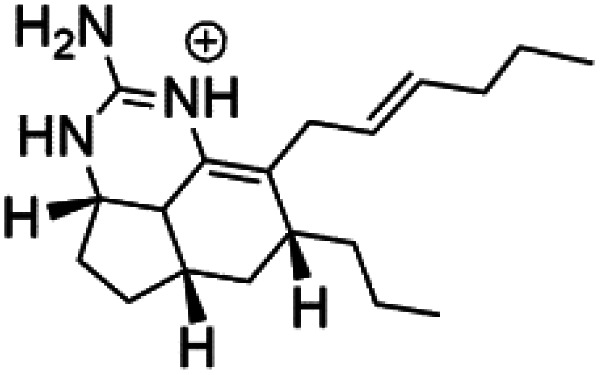
|
||||
| Netamine M (66) |
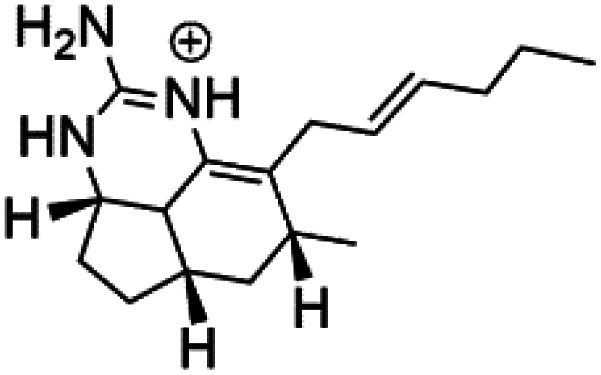
|
||||
| Netamine N (67) |
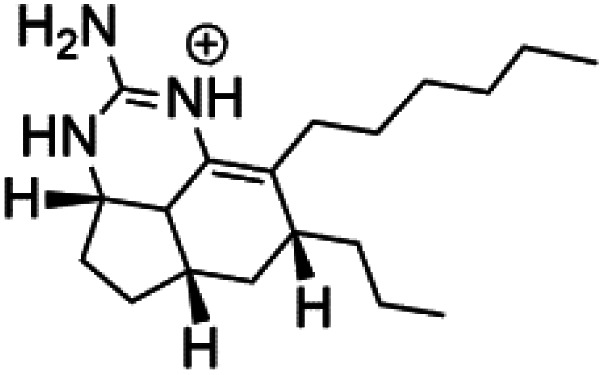
|
Biemna laboutei | Sponge |
NA:not available.
2.1. Manzamines
Manzamines are polycyclic (7–8 rings) alkaloids containing a β-carboline moiety. Manzamine A (1) was first reported from an Okinawan sponge belonging to the Haliclona species (family Chalinidae).27 They are one of the essential antimalarial alkaloids. In addition to the lack of in vivo toxicity, the manzamines demonstrated greater effectiveness as antimalarial agents than the commonly used drugs artemisinin and CQ.28 The mechanism of manzamine alkaloids is not completely understood. Still, authors described β-carboline alkaloids as micromolar inhibitors of glycogen GSK-3 by malaria parasites and inhibitors of topoisomerase DNA through intercalation in DNA-base pairs.
Hence, a complete SAR investigation of manzamine alkaloids is necessary to understand the importance of each moiety (β-carboline and pentacyclic ring) and the influence of different substituents on antimalarial activity. Manzamines SAR is summarized into two objectives. The first is the effect of various substitutions on the β-carboline nucleus, and the other is the effect of substitutions on the pentacyclic ring.29 The β-carboline moiety of manzamine alkaloids is responsible for antimalarial activity. 9-N alkylation of the β-carboline ring decreases antimalarial activity, indicating that 9-NH is necessary for their antimalarial activity. Hydroxyl-group substitution at position 8 of the β-carboline skeleton does not significantly affect its antimalarial activity.
Hence, 8-hydroxymanzamine (2) has the same effect as manzamine A, while manzamine F (3), a related derivative of manzamine A, is inactive. The eight-membered rings differ between the inactive manzamine F and the active manzamine A. The double bond reduction and/or the incorporation of a ketone group into the adjacent carbon is harmful to antimalarial activity. Likewise, hydroxyl group attachment at position 6 instead of position 8 has a negative effect on antimalarial activity, as shown by the lower potency of 6-hydroxy-manzamine A (4).27In vitro and in vivo studies, manzamines A and its 8-hydroxy derivative inhibited P. falciparum growth.30 Several total syntheses of manzamines have been accomplished.29,31
2.2. Neo-kauluamine
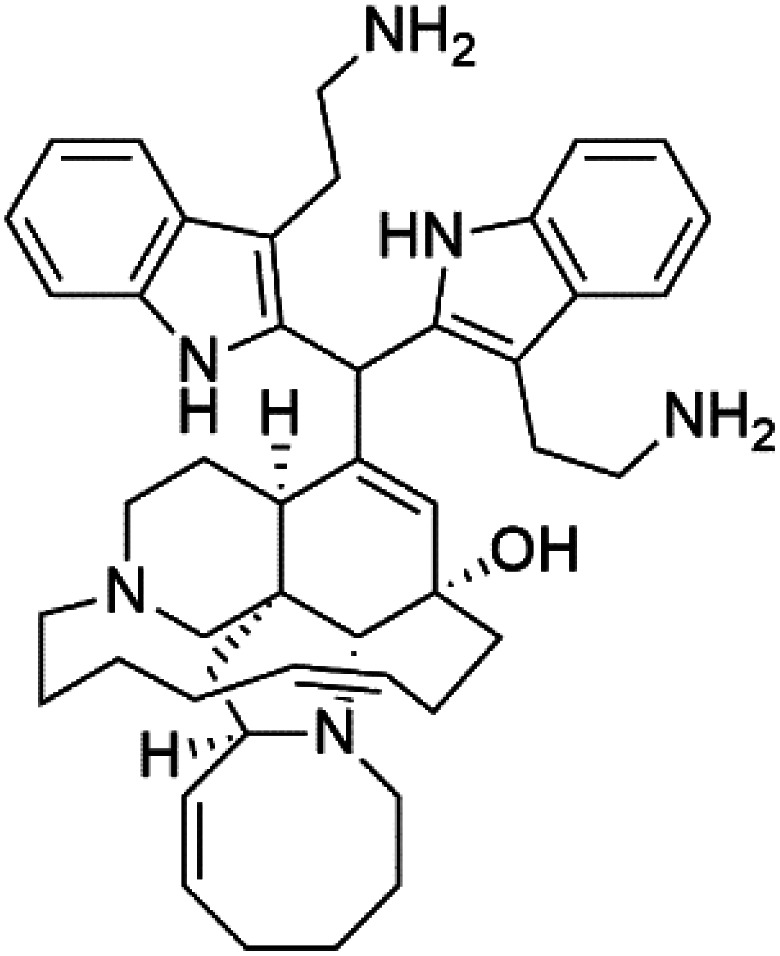
Neo-kauluamine (5) is a manzamine dimer, with a highly complex molecule composed of two units of manzamine fragmented by ether bonds between the eight-membered rings isolated from an unspecified genus of Indo-Pacific sponge (Petrosiidae, order Haplosclerida). Despite its structural complexity, neo-kauluamine displayed a strong efficacy in vivo and is considered an up-and-coming agent in malaria.27 Although this structure, like manzamine F (3), lacks the double bond in the eight-membered ring, it showed the same activity as manzamine A. The lack of antimalarial activity for 12,34-oxamanzamine A (6) suggested that the C-12 hydroxyl, the C-34 methine, or the 8-ring conformation are of great importance for the antimalarial activity.28
2.3. Zamamidines
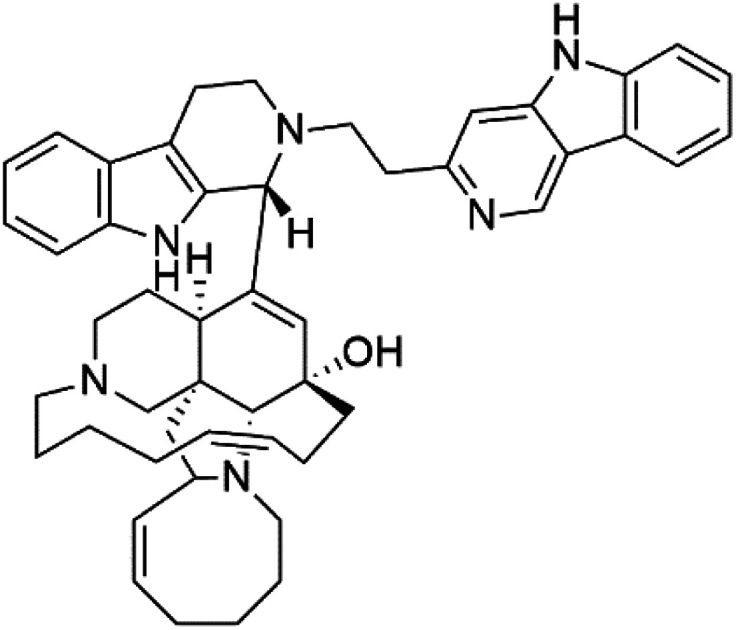
Zamamidines A–D (7–10) are also manzamine alkaloids obtained from Amphimedon sp. sponge (Niphatidae). They had inhibitory activities against P. falciparum (IC50 values from 0.0008 to 0.016 μM).32–34 Zamamidine C (9) is the most active one of the series. Zamamidine D (10) is the first manzamine alkaloid characterized by having a moiety of 2,2′-methylene bis-tryptamine instead of a unit of β-carboline.35
2.4. Homofascaplysin A
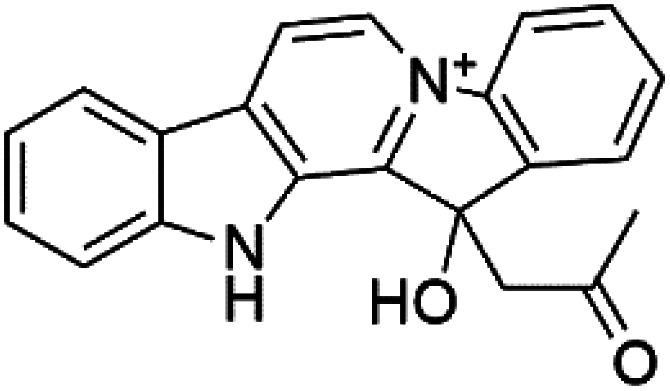
Homofascaplysin A (11) is also β-carboline alkaloid. It was extracted from the Hyrtios erecta sponge (Thorectidae).36 This alkaloid presented potent activity against CQ-resistant P. falciparum strains (IC50 = 0.07 μM) with approximately 10-fold less cytotoxicity.37 This potent antiplasmodial activity of this compound demonstrated its potential as a lead structure among antimalarial agents and became a synthesis target for the production of other similar analogues.38
2.5. Marinacarbolines
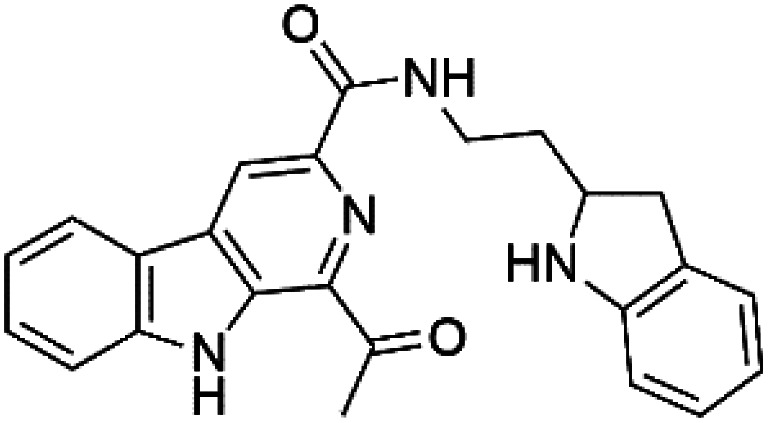
Marinacarbolines A–D (12–15), series of β-carboline alkaloids were obtained from the fermentation broth of the marine actinomycete bacteria Marinactinospora thermotolerans (Nocardiopsaceae). Marinacarbolines displayed antiplasmodial activities against 3D7 and Dd2 lines of P. falciparum, with IC50 from 1.92 to 36.03 μM.39
2.6. Indolactam alkaloids
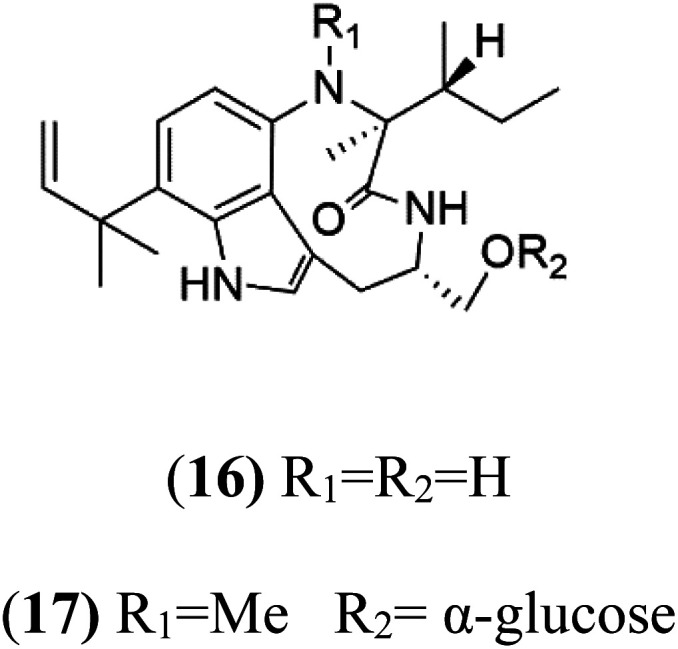
13-N-Demethyl-methylpendolmycin (16) and methylpendolmycin-14-O-α-glucoside (17) were derived from Marinactinospora thermotolerans (Nocardiopsaceae) fermentation broth. They were also found to exhibit moderate or weak activity against 3D7 (IC50 = 20.75 μM and 10.43 μM) and Dd2 (IC50 = 18.67 μM and 5.03 μM) strains of P. falciparum, respectively.39
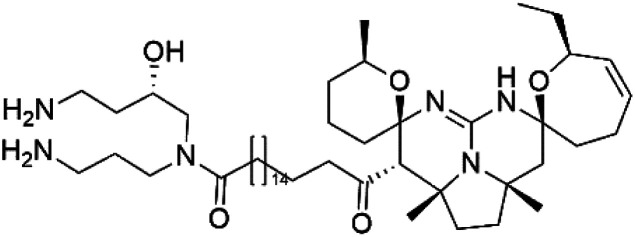
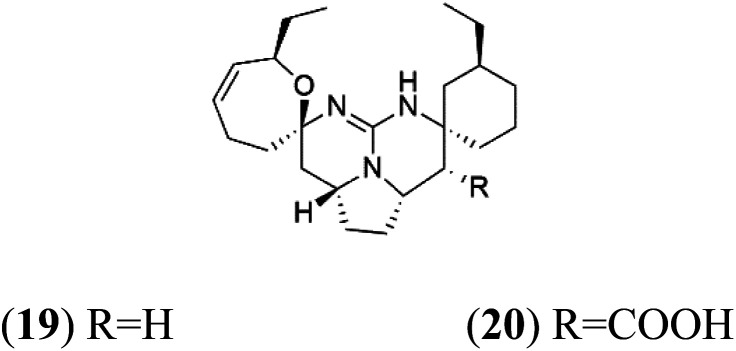
2.7. Crambescidins
Crambescidin 800 (18) was obtained from the Indonesian sponge (Mycophora sp. Crambeidae). Crambescidin 800 showed IC50 of 160 nM and 240 nM, respectively, against the 3D7 and FCR3 lines of P. falciparum.40 Also, other alkaloids, including crambescidin 359 (19), crambescidin acid (20), and fromiamycalin (21), were extracted from the sponge Monanchora unguiculate collected in Madagascar. Crambescidin 359 was active against the 3D7 line of P. falciparum (IC50 = 0.52 μM).41 Additionally, Unguiculin A (22), an acyclic guanidine alkaloid, was detected in this sponge. In addition to four pentacyclic alkaloids ptilomycalins E–H (23–26) were also isolated. Among them, fromiamycalin (IC50 = 0.24 μM), unguiculin A (IC50 = 12.86 μM) ptilomycalins E (IC50 = 0.35 μM), F (IC50 = 0.23 μM), ptilomycalins G and H mixture (IC50 = 0.46 μM), respectively exhibited promising activity against P. falciparum.41
2.8. Opacalines
Opacalines are alkyl guanidine-substituted β-carboline-containing metabolites obtained from the New Zealand ascidian Pseudodistoma opacum (Pseudodistomidae). Opacalines B (27) and C (28) displayed moderate activity against the CQ-resistant P. falciparum strain (IC50 range of 2.5–14 μM).42
2.9. Spermidine
Two indole alkaloids, Didemnidines A (29) and B (30), were obtained from the New Zealand ascidian Didemnum sp. (Didemnidae). Among them, Didemnidine B showed mild activity (IC50 = 0.047 μM) against P. falciparum.43
2.10. Salinosporamide
Salinosporamide A (31) is a simple γ-lactam spiro-alkaloid isolated from an actinomycete bacteria belonging to the genus Salinispora tropica (Micromonosporaceae).44 Salinosporamide A is also a cyclic depsipeptide (bicyclic β-lactone γ-lactam peptide).
Salinosporamide A showed potential as an antimalarial candidate. It exhibited a potent parasite proteasome inhibitor and antimalarial activity against P. falciparum in vitro (IC50 = 11.4 nM) and in vivo against P. yoelii.45,46 By controlling T cell proliferation and leading to cell cycle arrest, Salinosporamide A suppressed T cell activation and regulated the expression of cyclin-dependent kinases.47 Recently total synthesis for salinosporamide A molecule has been reported.46,48,49
2.11. Bromotyrosine alkaloid
Several psammaplysin derivatives were obtained from the Indonesian marine sponge Aplysinella strongylata (Aplysinellidae).50,51 Psammaplysin H (32) is a bromotyrosine alkaloid from a marine sponge Pseudoceratina sp. and displayed more than 97% antimalarial activity (0.41 μM concentration).52 Psammaplysins H, F (33), and G (34) exhibited antimalarial activity, while Psammaplysins F and G displayed antimalarial activities against the drug-resistant strains of Plasmodium falciparum.53 Ceratinadins were bromotyrosine alkaloids that had the 1,6-dioxa-2-azaspiro[4.6] undecane skeleton, Ceratinadins E (35) and F (36), obtained from an Okinawan marine sponge Pseudoceratina sp. (Pseudoceratinidae). Ceratinadins E showed antimalarial activities against drug-resistant and drug-sensitive K1 P. falciparum strains.54
2.12. Pyrrolo-iminoquinones
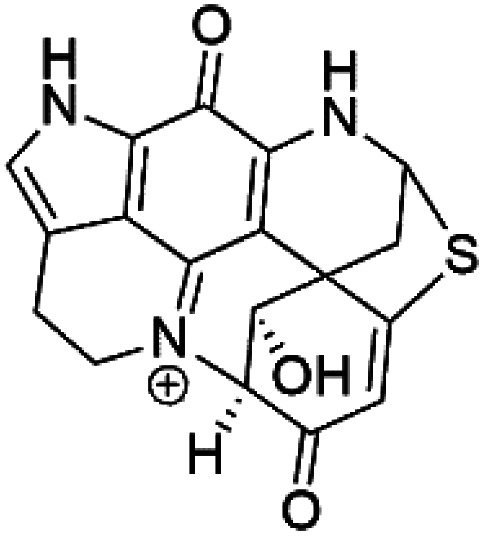
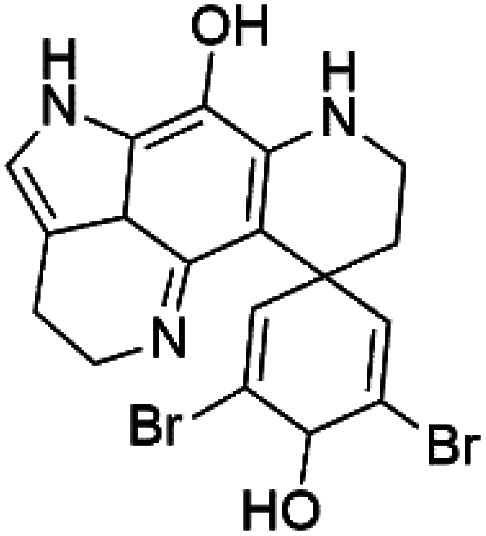
Pyrroloiminoquinone compounds, Tsitsikammamine C (37), makaluvamines J (38), G (39), L (40), K (41), damirones A (42), and B (43), were extracted from the Australian marine sponge Zyzzya sp. (Acarnidae, order Poecilosclerida). All compounds were investigated against 3D7 and Dd2 P. falciparum strains. Among them, tsitsikammamine C with IC50 = 13 and 18 nM, respectively, inhibited both ring and trophozoite stages of the malaria parasite life cycle. Makaluvamines J, G, and L showed a potent antimalarial activity (IC50 < 100 nM) in vitro against both strains.55 A class of two new brominated pyrroloiminoquinones; dihydrodiscorhabdin B (44) and discorhabdin Y (45), along with six pyrroloiminoquinone alkaloids, discorhabdins A (46), C (47), E (48), and L (49), dihydrodiscorhabdin C (50) were obtained from Alaskan sponge genus Latrunculia (Latrunculiidae) among them, discorhabdins A, C, and dihydrodiscorhabdin C displayed antimalarial in vitro activity against both D6 and W2 P. falciparum strains.56
2.13. Imidazole alkaloids
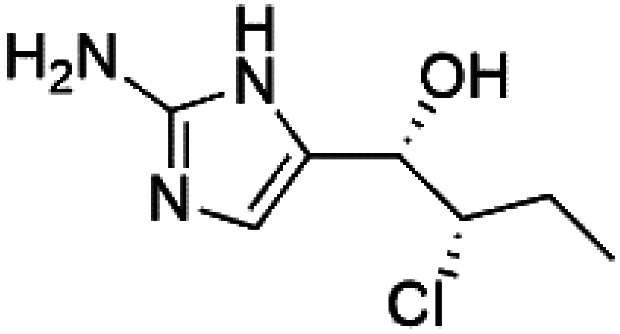
Girolline (51), a 2-aminoimidazol derivative initially isolated from a Caledonian sponge Cymbastela cantharella (Axinellidae)57 showed antimalarial activity against P. falciparum (FCM29) strain of (IC50 = 0.13 μM) a high in vivo activity in a P. vinckei petteri rodent model.58
2.14. Thiazine-derived alkaloids
Four thiazine-pyrroloquinone containing tricyclic quaternary alkaloids, Thiaplakortones A–D (52–55), were obtained from the Australian sponge Plakortis lita (Plakinidae). Thiaplakortones A–D exhibited potent inhibition against 3D7 CQ-sensitive (IC50 = 51 nM, 0.65, 0.309, and 0.279 μM) and Dd2 CQ-resistant P. falciparum (IC50 = 6.6 nM 0.092, 0.171, and 0.159 μM), respectively.59
2.15. Terpenoid alkaloids
Several terpenoid alkaloids had been showing reasonable antimalarial activity. For instance, Monamphilectine A (56), a diterpenoid-lactam alkaloid from sponge Hymeniacidon sp., displayed action against the P. falciparum W2 strain (IC50 = 0.60 μM).60 Agelasines J (57), K (58), and L (59), three adenine terpenoids, were obtained from the sponge Agelas mauritiana (Agelasidae). They have weak antimalarial activity against the P. falciparum Columbian FcB1 strain (IC50 = 6.6, 8.3, and 18 μM, respectively).61 Netamines G–N (60–67) and tricyclic alkaloids were obtained from the Madagascar sponge Biemna laboutei (Desmacellidae). Netamine K showed antimalarial activity (IC50 = 2.4 μM) against P. falciparum.62
3. Terpenoids
All terpenes or terpenoids have fundamental repeating five-carbon isoprene units. Terpenes are classified as hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), sesterterpenes (C25), triterpenes (C30), and tetraterpenes/carotenoids (C40).63 Marine-derived terpenoids have attracted potential interest similar to the terrestrial analogues represented by the sesquiterpene lactone artemisinin and isonitriles-containing terpenes.7,64,65 More than 30 compounds were isolated and showed antimalarial activity from marine organisms. Unique mechanisms were demonstrated, including the inhibitory activity against heme detoxification by isonitrile derivatives. They are discussed in the following subsections, and their chemical structures are shown in Table 3.
A list of marine-derived terpenes-containing antimalarial drugs showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic source.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Axisonitrile-1 (68) Axamide-1 (69) | W2 = 0.073 μM D6 = 0.61 μM |
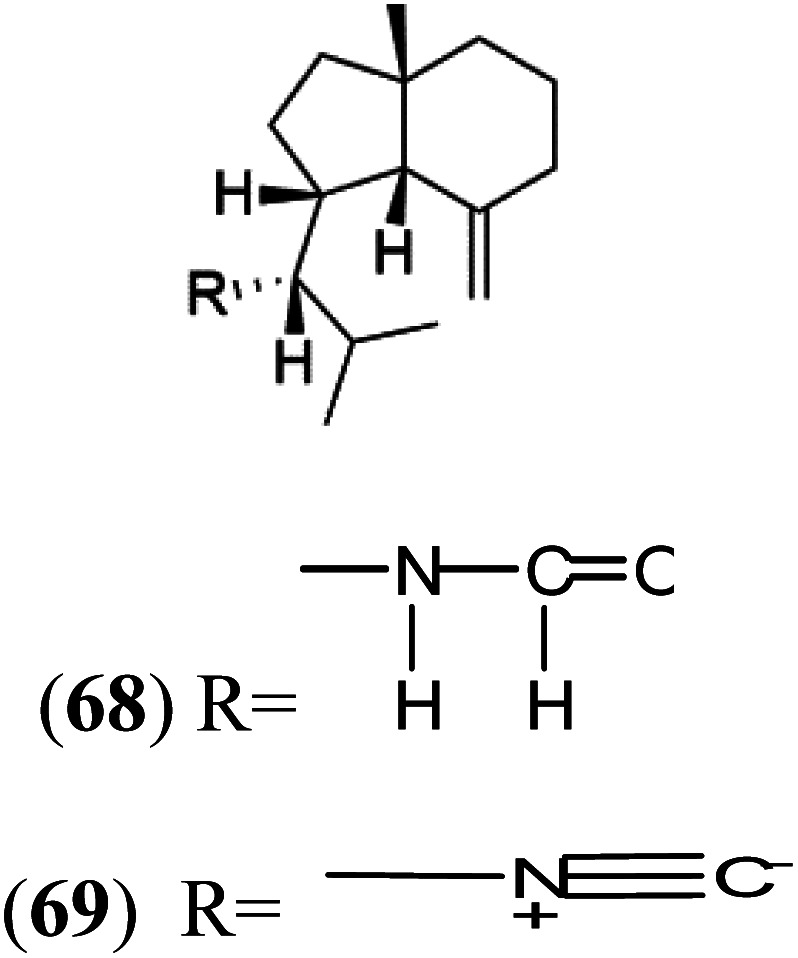
|
Axinella cannabina | Sponge | 67 and 69–72 |
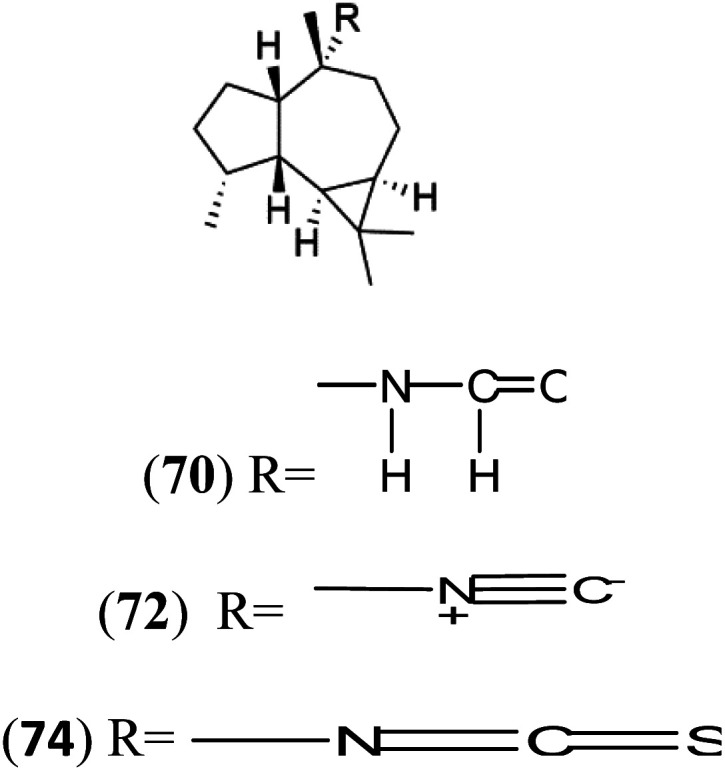
| Axamide-2 (70) Axisonitrile-2 (72) Axisothiocyanate-2 (74) |
|
||||
| Axamide-3 (71) Axisonitrile-3 (73) Axisothiocyanate-3 (75) |
|
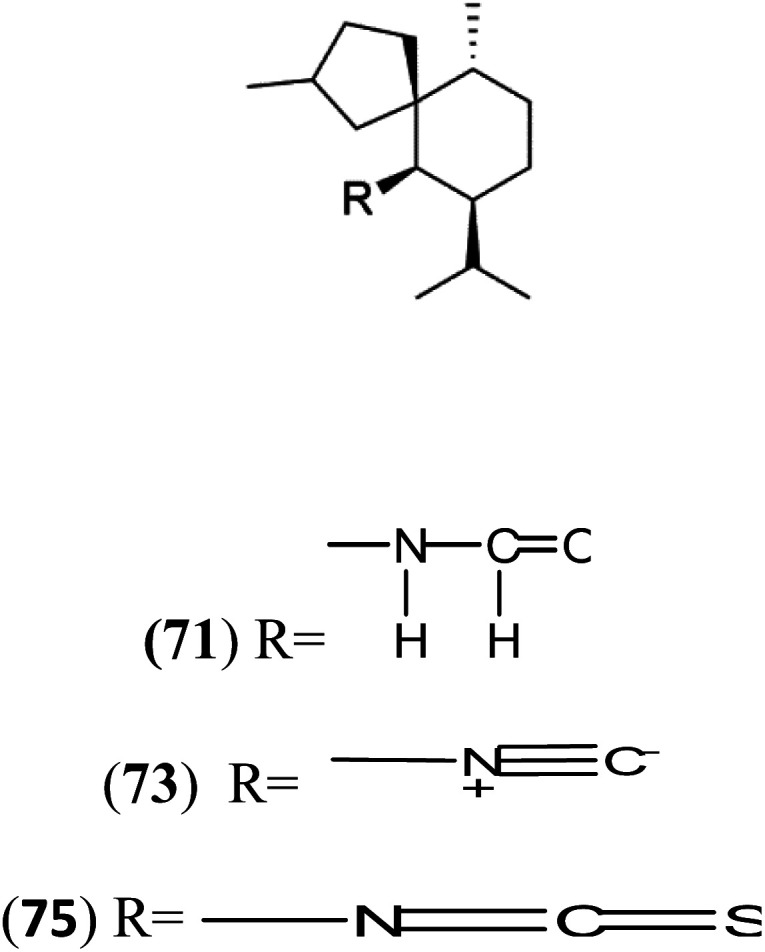
||||
| (−)-8,15-Diisocyano-11(20)-amphilectene (76) | W2 = 15 nM D6 = 16 nM K1 = 90 nM |
|
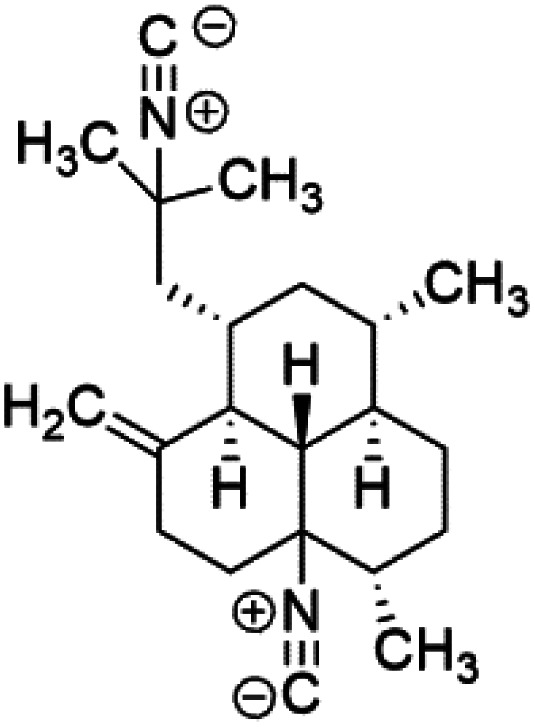
Hymeniacidon amphilecta, Venzea flava | Sponge | 73 and 74 |
| Kalihinol A (77) | FCR-3 = 12 μM |
|
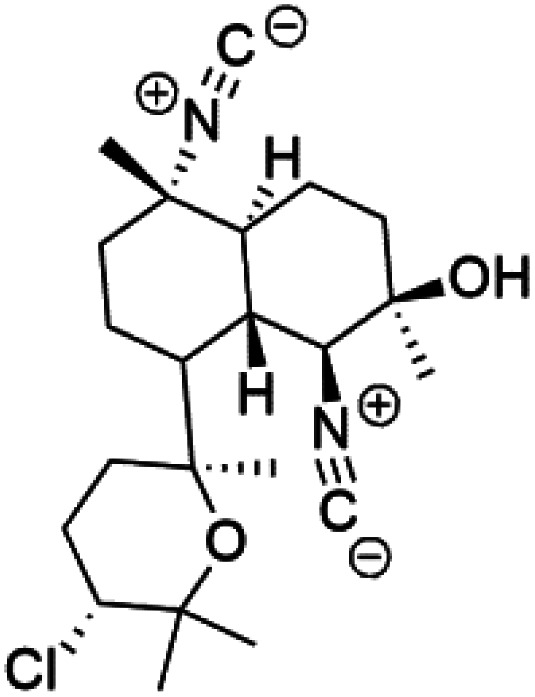
Acanthella sp. | Sponge | 68 |
| (8R)-8-Bromo-10-epi-β-snyderol (78) | D6 = 0.012 and W2 = 0.017 μM |
|
Laurencia obtusa | Red alga | 77 |
| Chloroaureol (79) Aureol (80) Aureol acetate (81) | (79) D6 = 9.74 μM (80) and (81) NA |
|
Smenospongia aurea | Sponge | 78 |
| Pelorol (82) | NF54 = 0.005 and K1 = 0.0019 μM |
|
Dactylospongia elegans | Sponge | 79 |
| Ilimaquinone (83) | NF54 = 0.0026 K1 = 0.0048 μM |
|
79 | ||
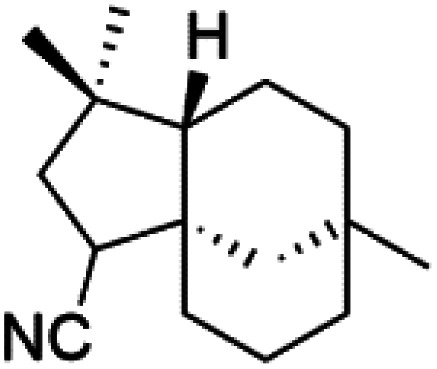
| Isocyanoclovane (84) | 3D7 = 300, 290, and 260 nM Dd2 = 360, 830, and 870 nM |
|
Phyllidia ocellate | Nudibranch | 80 |
| 2-Isocyanoclovene (85) |
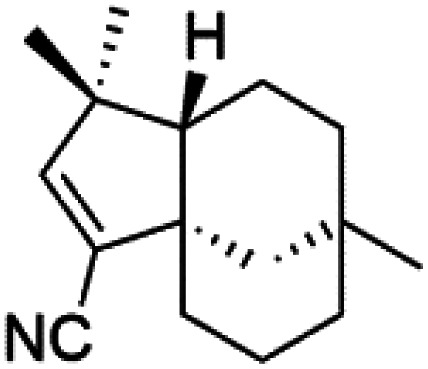
|
||||
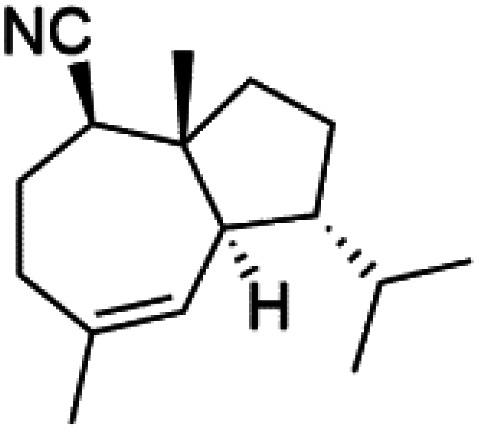
| 4,5-Epi-10-isocyanoisodauc-6-ene (86) |
|
||||
| 8a,11-Dihydroxypachydictoyl A (87) | K1 = 10.0 μM |
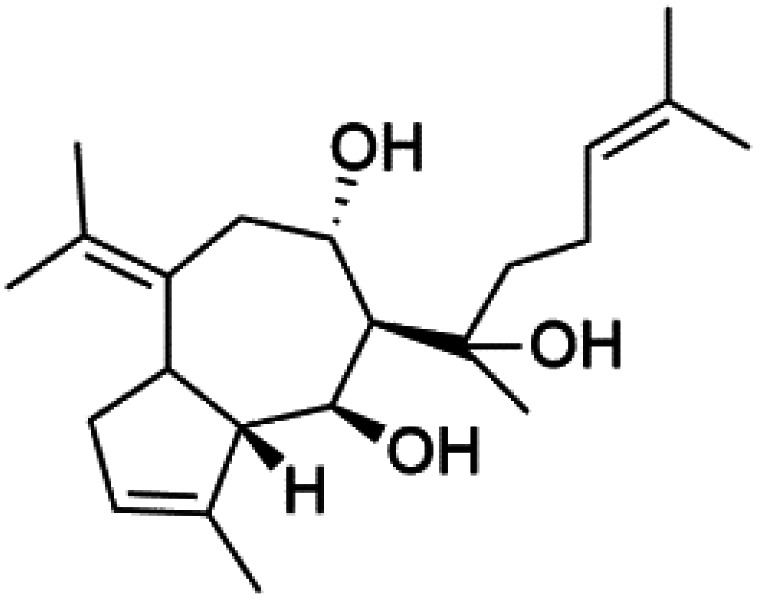
|
Dictoyta sp. | Brown alga | 83 |
| 4,18-Dihydroxydictyolactone (88) |
|
||||
| Laevigatol A (89) | Dd2 IC50 < 5.0 μM |
|
Pseudopterogorgia elisabethea | Soft coral | 84 and 85 |
| Pseudopterosin V (90) | Dd2 = 2.2 μM |
|
Pseudopterogorgia elisabethea | Soft coral | 86 |
| Dorisenone D (91) | K1 = 1.3 μM |
|
Dysidea arenaria | Sponge | 87 |
3.1. Sesquiterpene isonitriles
Sesquiterpenoid metabolites containing isonitrile, isothiocyanate, and formamide were isolated for the first time from Axinella cannabina sponge (Axinellidae).66,67 Isonitrile-containing compounds do their activity by blocking heme (FP) detoxification. The interaction of isonitrile with free heme was demonstrated by forming a coordination complex with the iron center. The drug carrier must possess a solid lipophilic molecular nucleus with at least a tricyclic structure that carries an isonitrile group and establishes further hydrophobic reactions above the ring plane. The interaction of marine isonitrile derivatives with heme can suppress the sequestration of FP into beta-hematin and prevent the peroxidative and glutathione-mediated destruction of FP under conditions designed to imitate the environment inside the malaria parasite.68 Axisonitrile-1 (68) was the parent compound of the class of isonitrile containing sesquiterpenoids marine secondary metabolites obtained from A. cannabina sponge.67,69
Isolation of axisonitrile-1 was followed by other related sesquiterpenoids from the same sponge, as axamide-1 (69), axamide-2 (70), axamide-3 (71), axisonitrile-2 (72), axisonitrile-3 (73), axisothiocyanate-2 (74), axisothiocyanate-3 (75).70–72 In 1978, (−)-8,15-diisocyano-11(20)-amphilectene (76) was first reported from Hymeniacidon amphilecta (Halichondriidae) and was subsequently shown to demonstrate potent anti-infective activity in vitro.73,74 The closely related axisothiocyanate-3 was inactive, indicating that the activity depends not only on the carbon skeleton's structural characteristics but also directly on the existence of the isonitrile group. The action was confirmed by comparison among the activities of related compounds. It showed that the biological activity generally depends on the isocyanide functionality and the carbon skeleton structural features. The isocyanide group's location also has a pivotal role.75
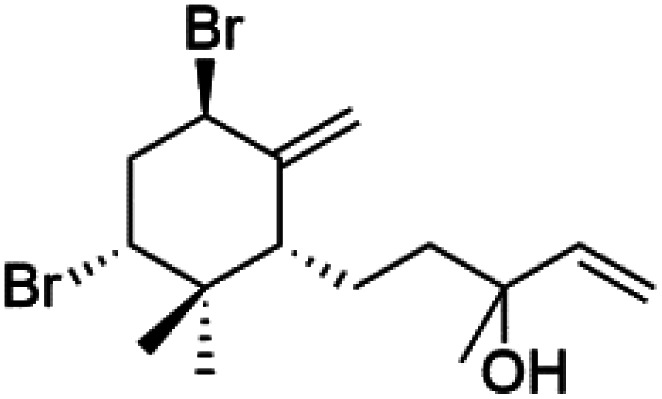
Bis-isonitrile-containing product and isocyanoterpene members, Kalihinols, were isolated from a Guam sponge, Acanthella sp.68,76 Many natural Kalihinol products are potent inhibitors of P. falciparum. Hence its importance, several Kalihinol analogs were synthesized and investigated using drug-sensitive and resistant P. falciparum for blood-stage antimalarial activity.68 Kalihinol A (77) showed increased potency and activity against the FCR-3 strain (Kalihinol A EC50 = 1.2 nM).67. (8R)-8-Bromo-10-epi-β-snyderol (78), a sesquiterpene isolated from Laurencia obtuse (Rhodomelaceae) red alga displayed activity against P. falciparum D6 and W2 strains (D6 = 0.012 and W2 = 0.017 μM), respectively.77
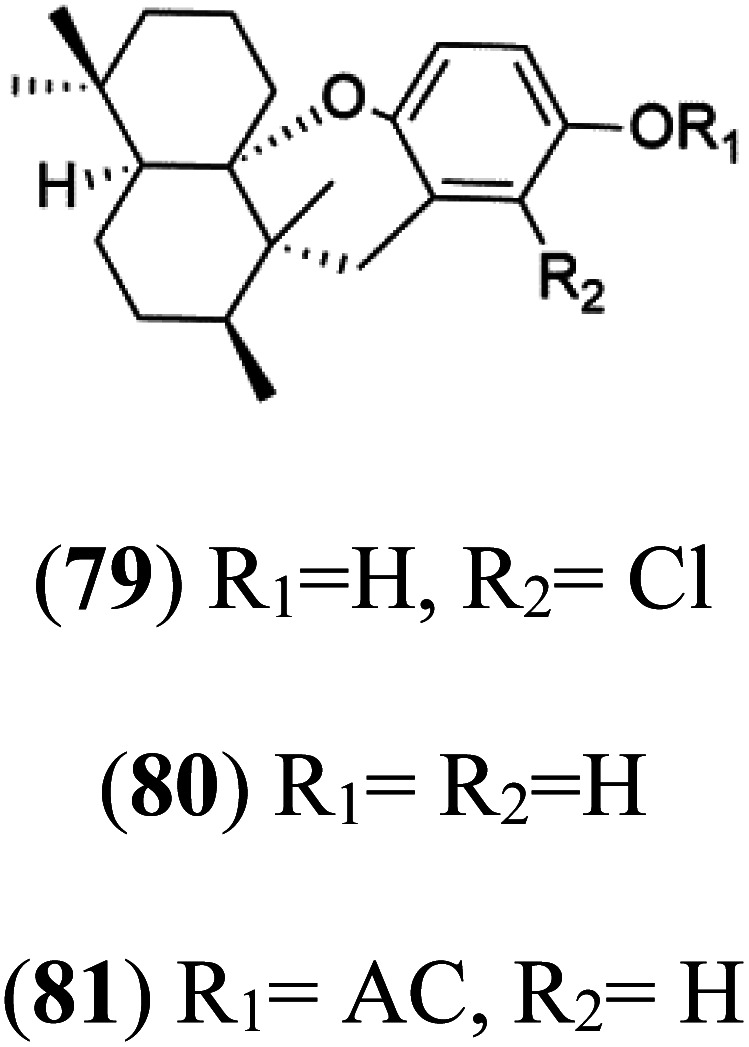
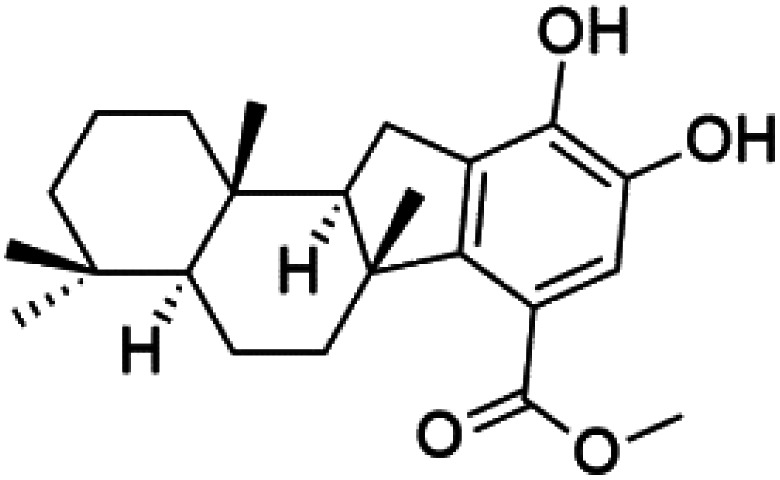
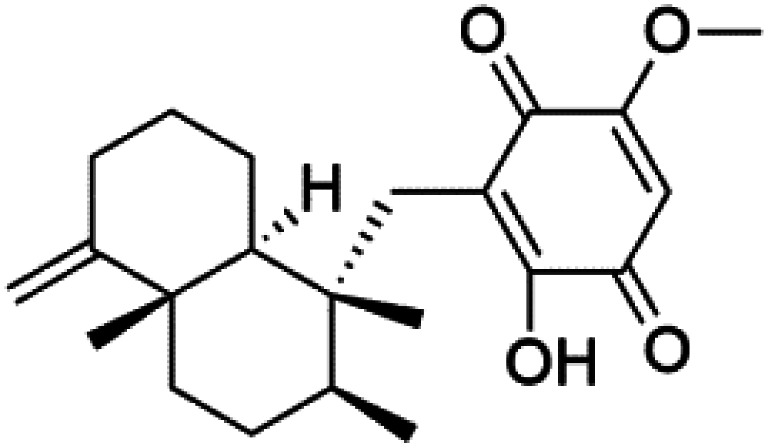
6′-Chloroaureol (79), aureol (80) and aureol acetate (81), sesquiterpene-phenol were obtained from Smenospongia aurea sponge.78 These derivatives displayed reasonable activity against P. falciparum D6 strain. Another marine sesquiterpene, pelorol (82), and sesquiterpene quinone, ilimaquinone (83), were extracted from the sponge Dactylospongia elegans (Thorectidae). Both showed moderate antiplasmodial activity against K1 and NF54 strains of P. falciparum.79
Sesquiterpenes, 2-isocyanoclovene (84) and 2-isocyanoclovane (85) were obtained from the Australian nudibranch Phyllidia ocellate (Phyllidiidae). They showed activity against 3D7 (IC50 = 300, 290, and 260 nM, respectively) and Dd2 P. falciparum clones (IC50 = 360, 830, and 870 nM, respectively).80
3.2. Diterpene isonitriles
A phytochemical investigation of the Cymbastela hooperi sponge provided 15 diterpenes containing isonitriles isocyanate, isothiocyanate, and isonitrile functionalities, which showed higher antimalarial effect and moderate toxicity.81 The activity of analogues with isocyanate and isothiocyanate functionality was up to ten times lower, demonstrating that the isonitrile group improved activity. An analogue containing only the formamide functional group but no isonitrile group was also ineffective against P. falciparum, implying that the formamide group isn't necessary for antiplasmodial efficacy.82 Monamphilectine A (56) was a diterpenoid β-lactam alkaloid separated from a Hymeniacidon sp. sponge (Halichondriidae). Monamphilectine A displayed a potent antimalarial activity,.60 While (−)-8,15-diisocyano-11(20)-amphilectene (86), re-isolated from the Svenzea flava sponge (Scopalinidae), was used as a precursor to synthesize five new products, all of which were tested against laboratory colonies of P. falciparum and Mycobacterium tuberculosis H37Rv.75
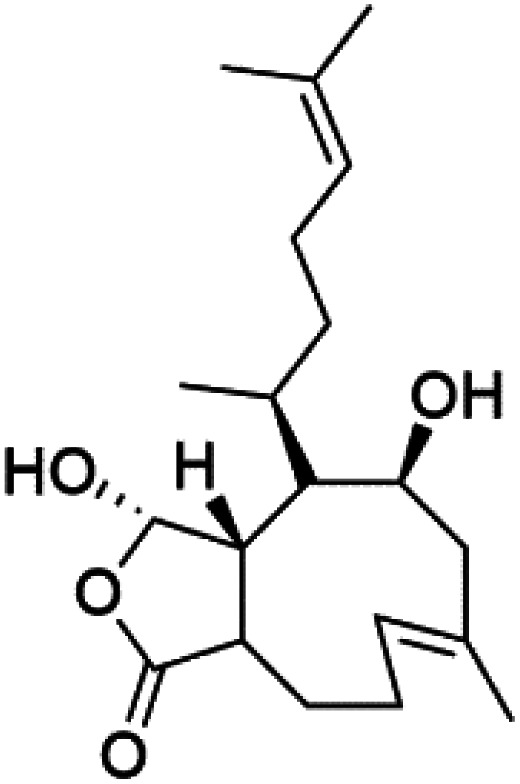
In addition, 8a,11-dihydroxypachydictoyl A (87), and 4,18-dihydroxydictyolactone (88) were diterpenoids isolated from Dictoyta sp. of the brown alga, which displayed antimalarial activity (IC50 = 10.0 μM) against K1 strain of P. falciparum.83
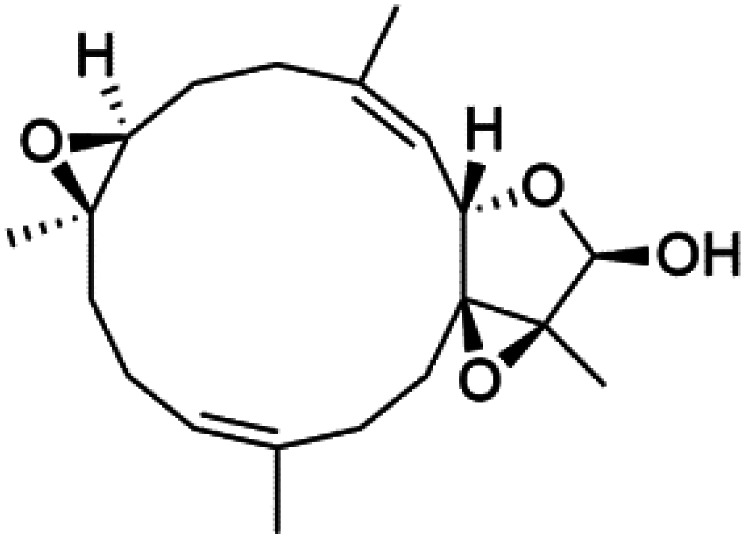
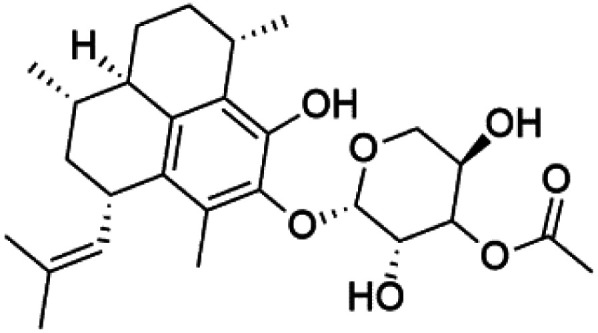
Moreover, soft corals and echinoderms living in Vietnamese seas provided several diterpenes. Among them is laevigatol A (89), which had a moderately antiplasmodial activity with an IC50 < 5.0 μM.84,85 Also, a series of diterpene glycosides was obtained from the Caribbean soft coral Pseudopterogorgia elisabethea (Gorgoniidae). Among them pseudopterosin V (90) exhibited an antimalarial activity (IC50 = 2.2 μM) against CQ-resistant P. falciparum colonies.86
3.3. Other terpenoids
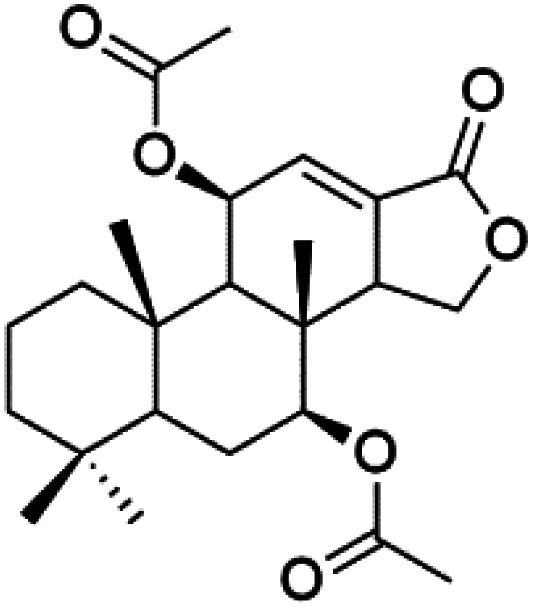
Screening of marine sponges extracts from Spongia, and Ircinia genera revealed thr presence of broad-spectrum antiprotozoal meroterpenes, linear triterpenoid, and squalene, with inhibitory effects on P. falciparum and Trypanosoma. The dorisenone D (91), a dimeric C21 meroterpenoid obtained from Dysidea arenaria sponge (Dysideidae), may become a promising antiplasmodial compound.87
4. Endoperoxide-containing compounds
One of the most fundamental advances in malaria chemotherapy was the discovery and manufacturing of endoperoxide-containing drugs. Undoubtedly, the artemisinin discovery was the beginning of research in this area. Artemisinin is a cadinane sesquiterpene lactone establishing a 1,2,4-trioxane moiety isolated from sweet wormwood Artemisia annua L. leaves (Asteraceae). Artemisinin showed nanomolar potency against CQ-resistant Plasmodium strains. The endoperoxide linkage is essential for antimalarial activity. One of the artemisinin derivatives lacking the endoperoxide bridge showed no antimalarial activity.88
These drugs containing endoperoxide were purported to interact via endoperoxide bond with the iron(ii) center of the heme unit released in the food vacuole during the digestion of hemoglobin and lead to peroxide bridge cleavage the consequent formation of oxygen-centered radicals. Because of an intramolecular rearrangement, these reactive species were converted into free C-centered radicals, toxic to the parasite through the alkylation of sensitive macromolecular targets. A sarco-endoplasmic reticulum Ca2+ dependent ATPase of P. falciparum has been proposed as a possible target for these active species.89,90 Although, most likely, artemisinin activity is not mediated by interaction with a single enzyme. It had been proposed that the Fe2+-containing species interacting with the endoperoxide bond is not heme.91 The marine antimalarial drug-containing endoperoxide was divided according to their structural feature into peroxyketal and non-peroxyketal, as demonstrated below, and the chemical structures are listed in Table 4.
A list of marine-derived endoperoxide-containing antimalarial drugsa.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Peroxyplakoric acids A3 (92) | FCR3 = 150 and 120 nM, respectively |
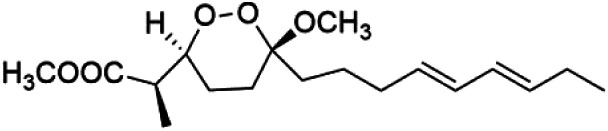
|
Plakortis sp. | Sponge | 92 and 93 |
| Peroxyplakoric acid B3 (93) |
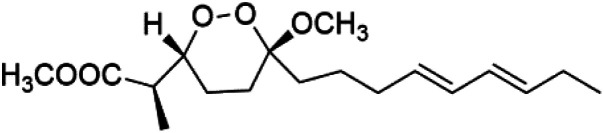
|
||||
| Plakortin (94) | W2 = 0.16 nM |
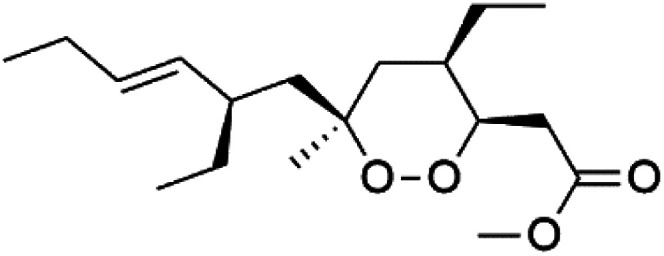
|
Plakortis halichondroides, Plakortis simplex | Sponge | 96 |
| Dihydroplakortin (95) | D10 = 1.12 μM W2 = 0.76 μM |
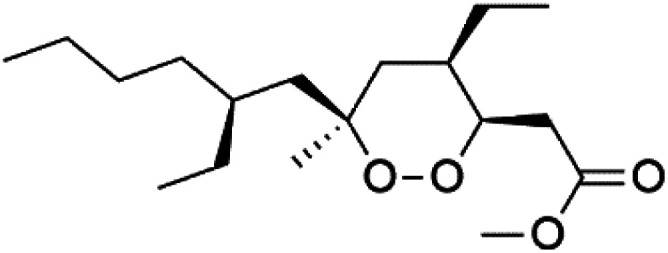
|
|||
| 3-Epiplakortin (96) | NA |
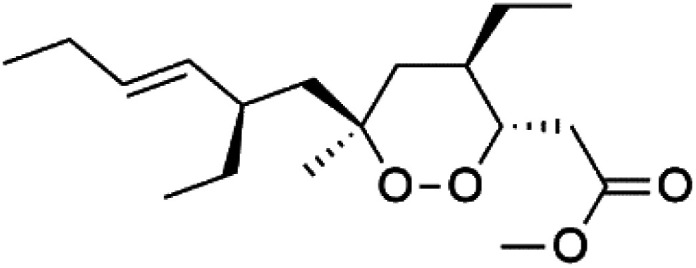
|
|||
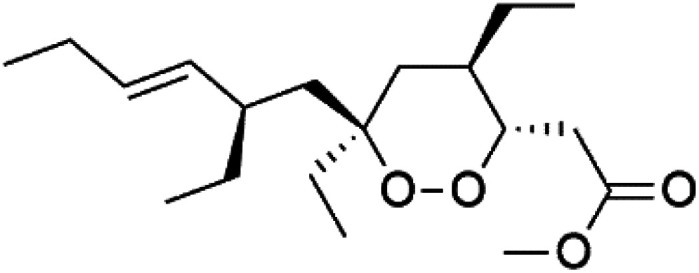
| Plakortide Q (97) | D10 = 1000 nM W2 = 520 nM |
|
|||
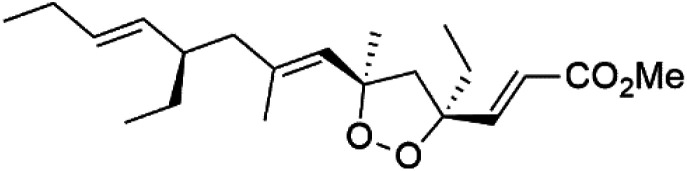
| Plakortide E (98) | D6 = 1.37 nM W2 = 1.11 nM |
|
|||
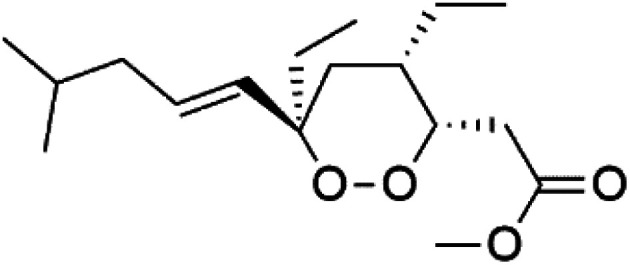
| Plakortide L (99) | NA |
|
Plakortis sp. | Sponge | 97 and 98 |
| Plakortide O (100) | NA |
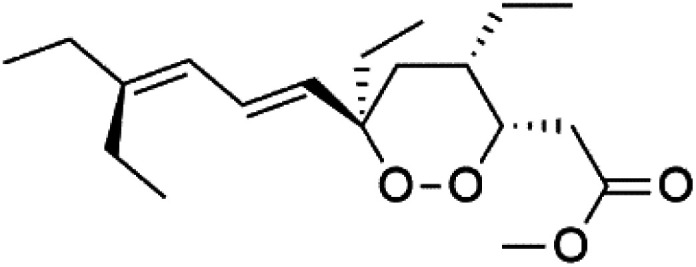
|
|||
| Plakortide P (101) | NA |
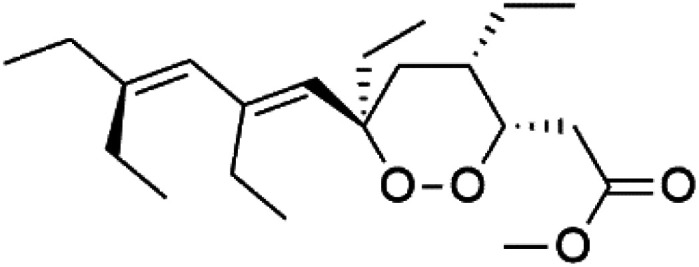
|
|||
| Plakortin diol (102) | FcM29 = 800 nM |
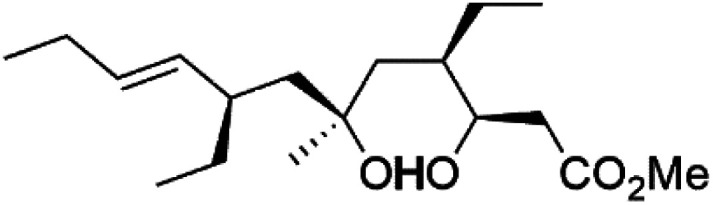
|
101 | ||
| Plakortin hydroxyl (103) Plakortin methoxy (104) Plakortin acetoxy (105) | D10 = 1.26 nM |
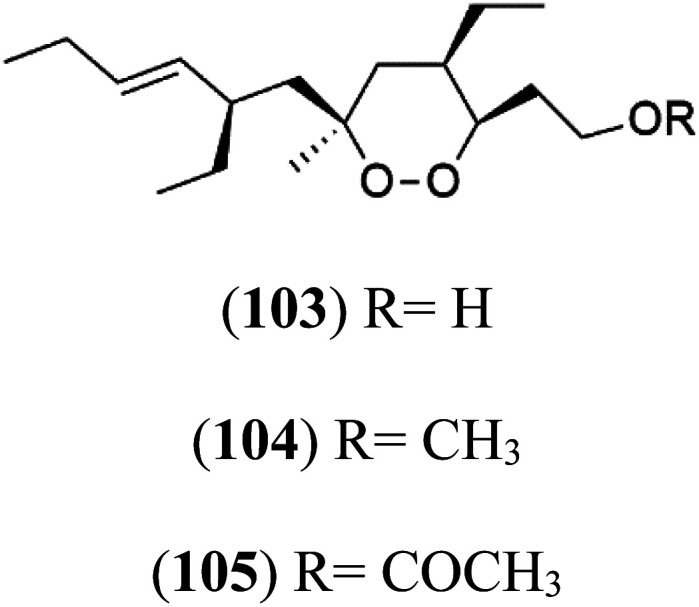
|
NA: not available.
4.1. Peroxyketal
Peroxyplakoric acids are the parent compound in the class of peroxyketals/3-alkoxy-1,2-dioxane derivatives, and its methyl esters; peroxyplakoric acids A3 (92) and B3 (93). It was extracted from the Okinawan sponge of Plakortis sp. (Plakinidae).92,93 Peroxyketals derivatives showed potent activity in vitro (IC50 = 150 and 120 nM against P. falciparum FCR3) and a good selective toxicity index. The long alkyl side chain in these derivatives is important for antimalarial activity as the synthetic analog containing methyl group instead of the nonadienyl group was completely nonactive. It has been observed that transforming the ester group into an amide group increases in vivo antimalarial potency.94
4.2. Non-peroxyketal
Plakortin (94) is a simple 1,2-dioxane metabolite, and was isolated from Plakortis halichondroides.95 Plakortin and its analogues, named dihydroplakortin (95), 3-epiplakortin (96), plakortide Q (97), plakortide E (98), were re-isolated from the Caribbean sponge Plakortis simplex.96 All these compounds, except plakortide E, displayed good antimalarial activity against (D10) CQ-sensitive and (W2) CQ-resistant strains of P. falciparum, with no cytotoxicity and a more potent activity on the (W2) strain (IC50 in D6 = 1.37 nM, and W2 = 1.11 nM). Plakortide E was found to be inactive. It could be ascribed to a five-membered ring presence instead of a six-membered ring and/or the crowded substituents at carbons flanking the endoperoxide linkage.97,98 Currently, plakortin is among pre-clinically investigated antiplasmodial candidates.99
On the other side, plakortide L (99), was extracted from a Jamaican sponge Plakortis sp.97,98 Plakortide O (100) and plakortide P (101), were isolated from plakortis halichondrioides, presented mild antimalarial activity in vitro (IC50 > 0.023 μM). Despite of their similarities with the plakortin, the configurational changes around the dioxane ring and/or the differences in the alkyl side chains are responsible for the observed decrease of activity.100 It was established that the role of the endoperoxide system in the antimalarial activity of plakortin derivatives is pivotal, as the diol derivative (102) with an open peroxide ring exhibited no antimalarial activity, when the ester group substitution to the corresponding hydroxyl (103), methoxy (104), or acetoxy (105) derivatives affected the antimalarial activity or the selectivity of these derivatives.101
5. Quinones, polyketides, phenols, and acids
Several phenolic compounds with antimalarial activity have been isolated from different marine sources. For instance, from Hyrtios sponge (Thorectidae), 5-oxopuupehenol (106) was isolated. It exhibited antimalarial activity against D6 and W2 clones of P. falciparum.102 In addition, three xanthones were isolated from the marine fungi Chaetomium sp. (Chaetomiaceae), i.e., chaetoxanthones A (107), B (108), and C (109). Chaetoxanthones A and B contain a dioxane-tetrahydropyran moiety, while chaetoxanthone C is a chlorinated xanthone containing a tetrahydropyran ring. Among them, chaetoxanthone B exhibited a significant antimalarial activity (IC50 = 1.4 nM).103
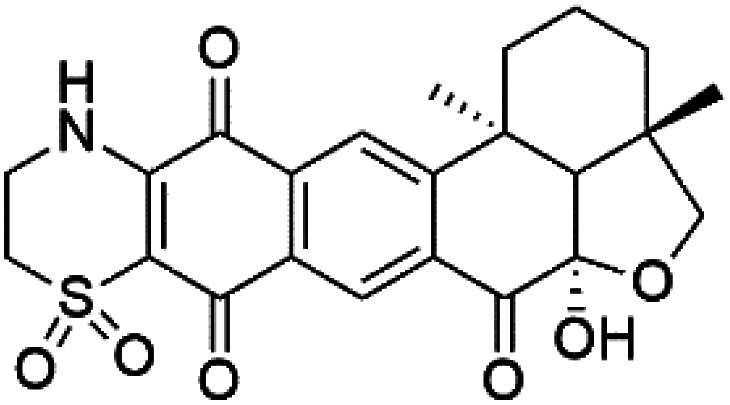
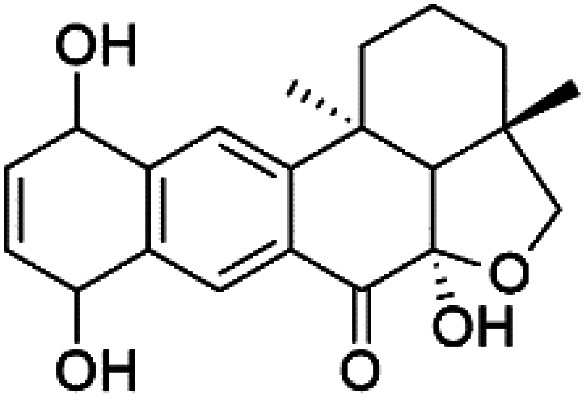
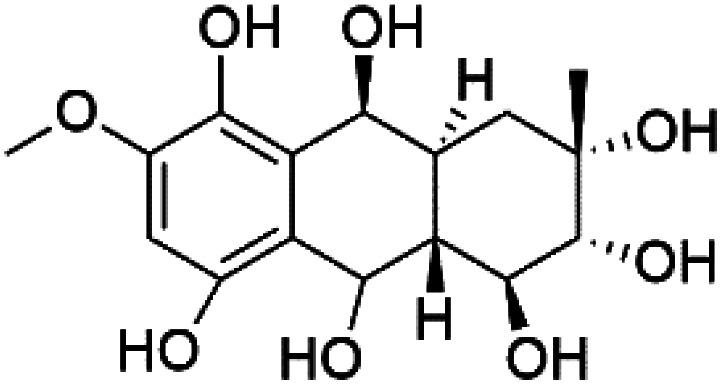
A family of monoterpene-quinones, xestoquinone, and alisiaquinones, showed promising antimalarial activities. Xestoquinone (110) was obtained from a Vanuatu Pacific marine sponge Xestospongia sp., while alisiaquinone A (111), B (112), C (113), and alisiaquinol (114) were extracted from an unidentified deep-water Caledonian sponge. Xestoquinone was a selective active inhibitor of a protein kinase (Pfnek-1) of P. falciparum with IC50 = 1 μM. Alisiaquinone C presented activity against F32, FcB1, and FcM29 of P. falciparum (IC50 = 0.15, 0.21, and 0.08 μM, respectively).104,105 From the marine-derived fungus Fusarium sp., a series of secondary metabolites such as 9α-hydro-xyhalorosellinia A (115), bostrycin (116), nigrosporin B (117), javanicin (118), and anhydrofusarubin (119) were isolated.106
Among marine fungi, phytochemical investigation of Halorosellinia oceanica BCC 5149 resulted in isolation of different compounds such as 2-hexylidene-3-methylsuccinic acid, cytochalasin Q, 5-carboxymellein, 2-hexylidene-3-methylsuccinic acid 4-methyl ester, and halorosellinic acid. Cytochalasin Q, 5-carboxymellein, halorosellinic acid, and its acetonide derivative showed antimalarial activity with IC50 values of 17, 4, 13, and 19 μg mL−1, respectively.107 Also, the tetramic acid called vermelhotin was isolated from an unidentified fungus CRI247-01 (a member of the order Pleosporales). Vermelhotin exhibited moderate antiplasmodial activity 1–10 μg mL−1.108 Besides, marine sponges (e.g., a Vanuatu marine sponge Pseudoceratina sp.) produced homogentisic acid derivative acting as protein kinase inhibitors Pfnek-1 with an IC50 about 1.8 μM and moderately active in vitro against a FcB1 P. falciparum strain (IC50 = 12 μM).109 Homogentisic acid is among pre-clinically investigated antiplasmodial candidates.99
Furthermore, (S)-curcuphenol (120) is a sesquiterpene phenol isolated from the Jamaican sponge Didiscus oxeata, displayed in vitro activity against D6 and W2 strains of P. falciparum (MIC = 0.017 and 0.008 μM, respectively).110 (2Z,6R,8R,9E)[3-ethyl-5-(2-ethyl-hex-3-enyl)-6-methyl-5H-furan-2-ylidene]-acetic acid methyl ester (121), a polyketide was isolated from Plakortis angulospiculatus sponge. It showed mild antimalarial activity against D6 and W2 P. falciparum colonies (IC50 = 6.6 nM on both strains).111,112
Also, gracilioether A–C (122–124), a polyketal peroxide obtained from the crude extract of the deep sea Agelas gracilis sponge (Agelasida), presented an antimalarial activity (IC50 = 28.22 μM).113
Recently, the angucyclines Actinosporins E, H, G (125–127), Tetrangulol (128), and Capillasterquinone B (129) have been isolated based on antimalarial guided fractionation of the co-cultured fermentation of the marine bacterium Actinokineospora spheciospongiae (Pseudonocardiaceae). Nevertheless, these compounds have not been reported under axenic conditions. Upon antimalarial screening, they displayed activity ranging from IC50 = 0.019 to 0.028 μM.114 The chemical structure and source are summarized in Table 5.
A list of marine-derived quinones, polyketides, and phenols antimalarial drugs showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic source.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| 5-Oxopuupehenol (106) | D6 = 5.8 μM W2 = 3.8 μM |
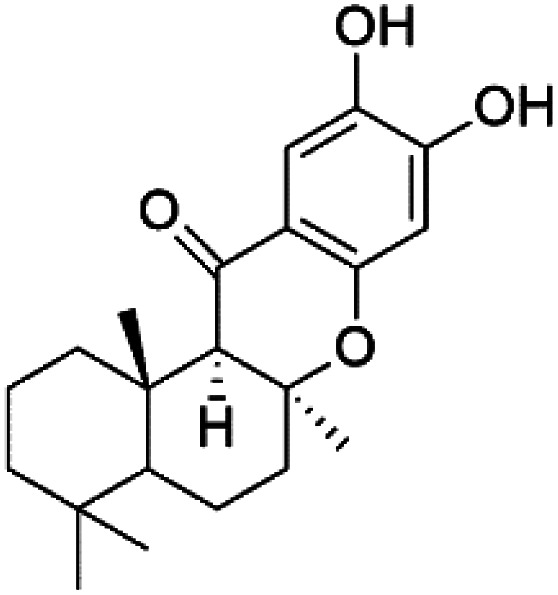
|
Hyrtios sp. | Sponge | 102 |
| Chaetoxanthone A (107) Chaetoxanthone B (108) | K1 = 1.4 nM |
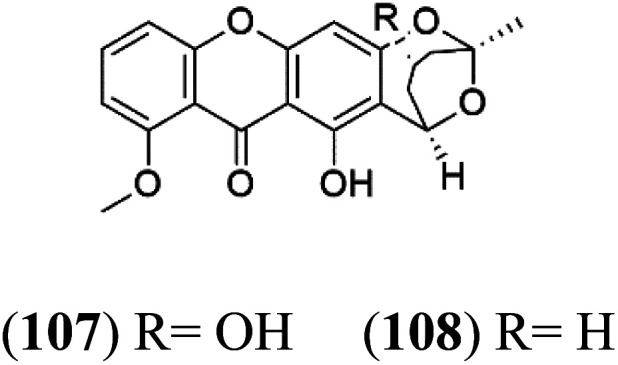
|
Chaetomium sp. | Marine fungi | 103 |
| Chaetoxanthone C (109) | NA |
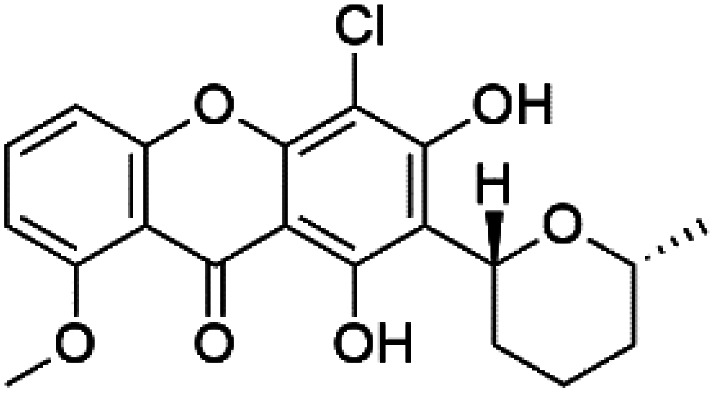
|
|||
| Xestoquinone (110) | FcB1 = 3.0 μM |
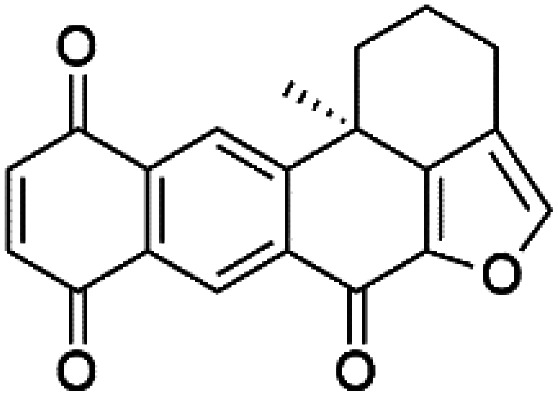
|
Xestospongia sp. | Sponge | 104 and 105 |
| Alisiaquinone A (111) alisiaquinone B (112) | FcMC29 = 8.50, 2.60 μM FcB1 = 7.40, 8.40 μM F32 = 9.10, 7.10 Mm, respectively |
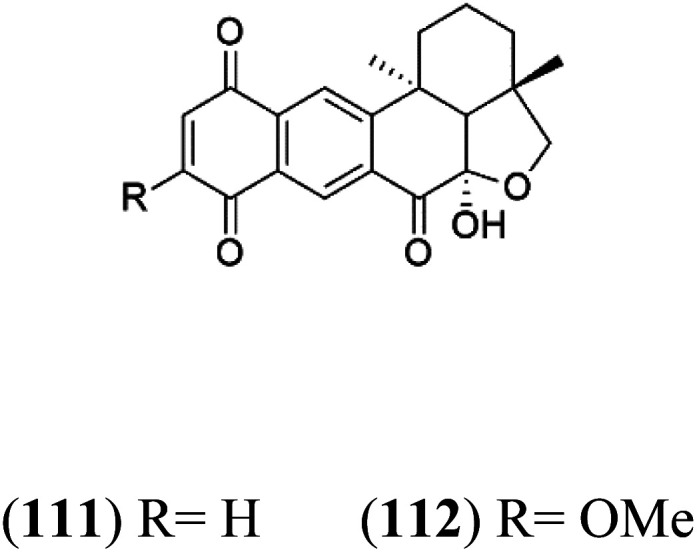
|
Unidentified deep-water caledonian sponge | Sponge | 104 and 105 |
| Alisiaquinone C (113) | FcMC29 = 0.08 μM FcB1 = 0.21 μM F32 = 0.15 μM |
|
|||
| Alisiaquinol (114) | FcMC29 = 7.90 μM FcB1 = 6.40 μM F32 = 9.90 μM |
|
|||
| 9α-Hydro-xyhalorosellinia A (115) | K1 = 25 μM |
|
Fusarium sp. | Marine fungi | 106 |
| Bostrycin (116) Nigrosporin B (117) | K1 = 9.8 μM (116) NA for (117) |
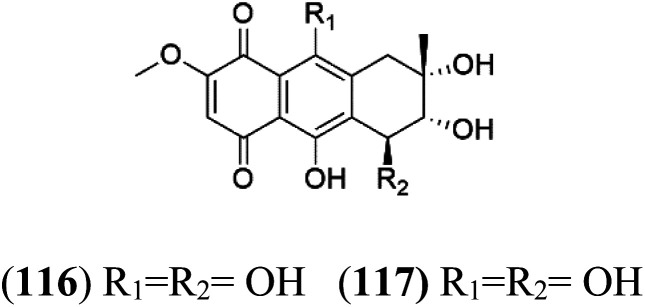
|
|||
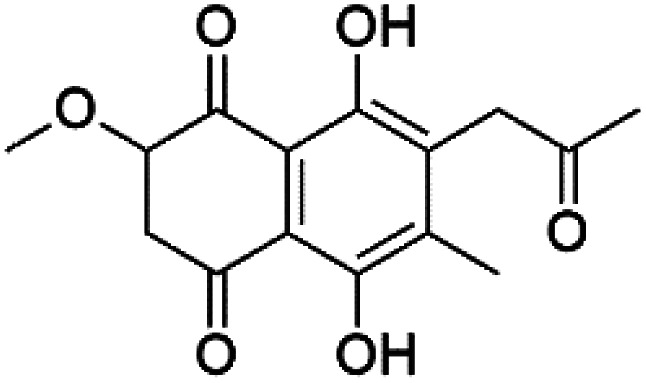
| Javanicin (118) | K1 = 12 μM |
|
|||
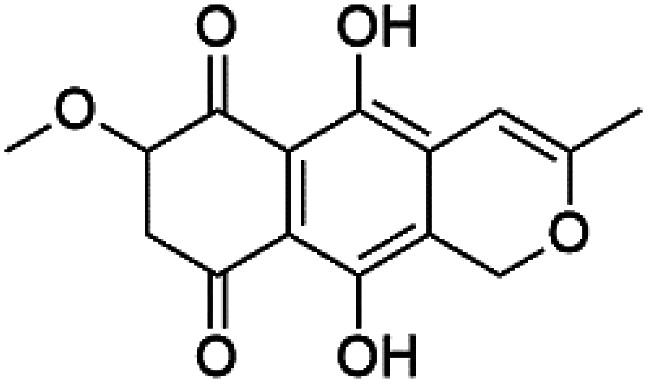
| Anhydrofusarubin (119) | K1 = 14 μM |
|
|||
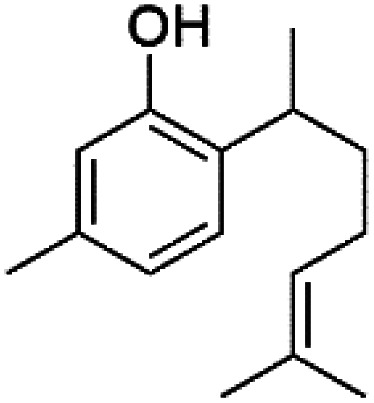
| (S)-Curcuphenol (120) | D6 = 0.017 μM W2 = 0.008 μM |
|
Didiscus oxeata | Sponge | 110 |
| (2Z,6R,8R,9E)[3-ethyl-5-(2-ethyl-hex-3-enyl)-6-methyl-5H-furan-2-ylidene]-acetic acid methyl ester (121) | D6 = 6.6 nM W2 = 6.6 nM |
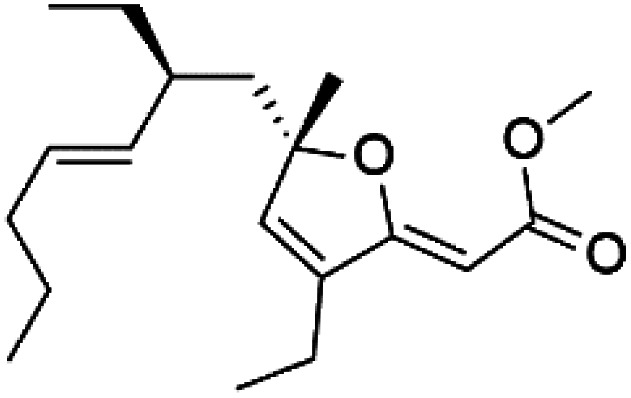
|
Plakortis angulospiculatus | Sponge | 111 and 112 |
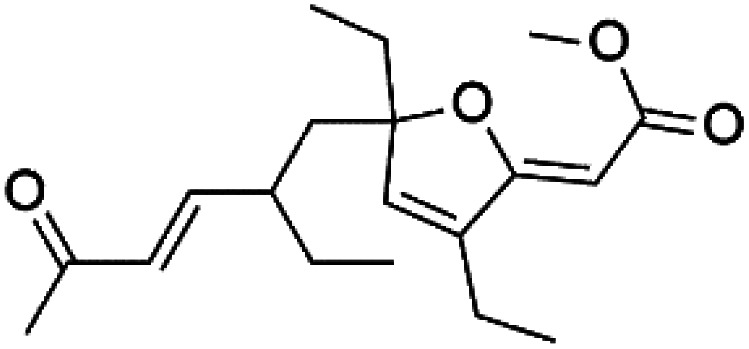
| Gracilioether A (122) | ItG = 28.22 μM |
|
Agelas gracilis | Sponge | 113 |
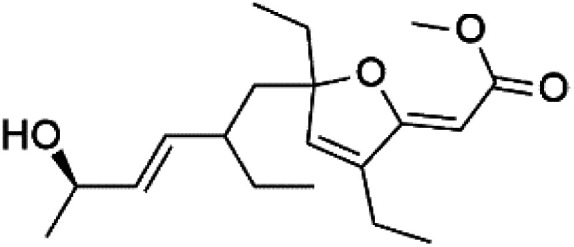
| Gracilioether B (123) | ItG = 1.56 μM |
|
|||
| Gracilioether C (124) | ItG = 31.02 μM |
|
|||
| Actinosporin E (125) | IC50 = 0.019 to 0.028 μM |
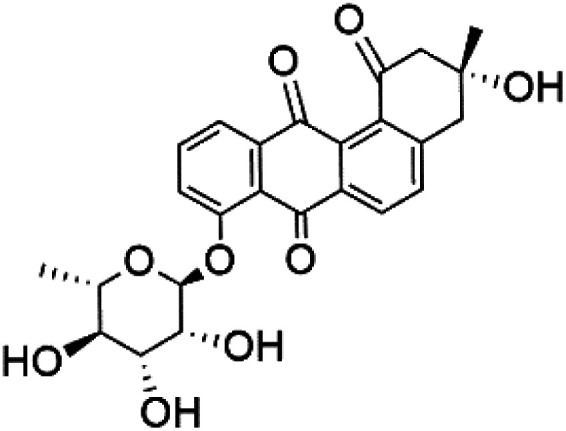
|
Actinokineospora spheciospongiae | Actinomycete bacteria | 114 |
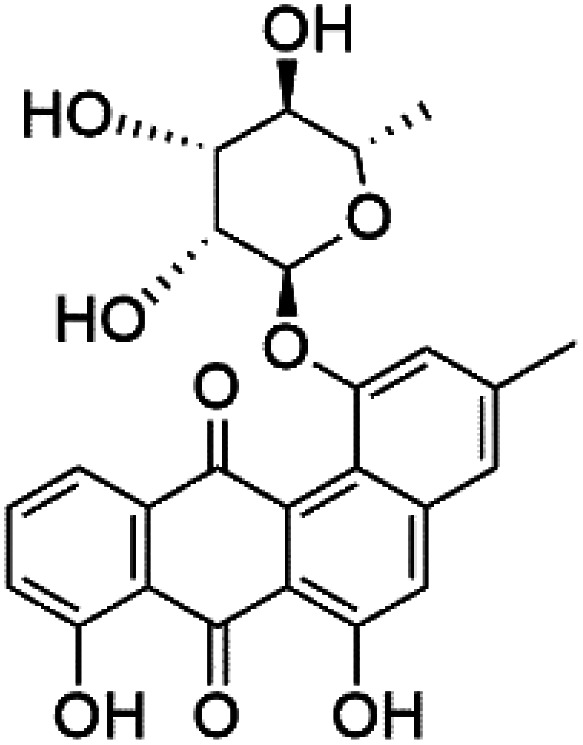
| Actinosporin H (126) |
|
||||
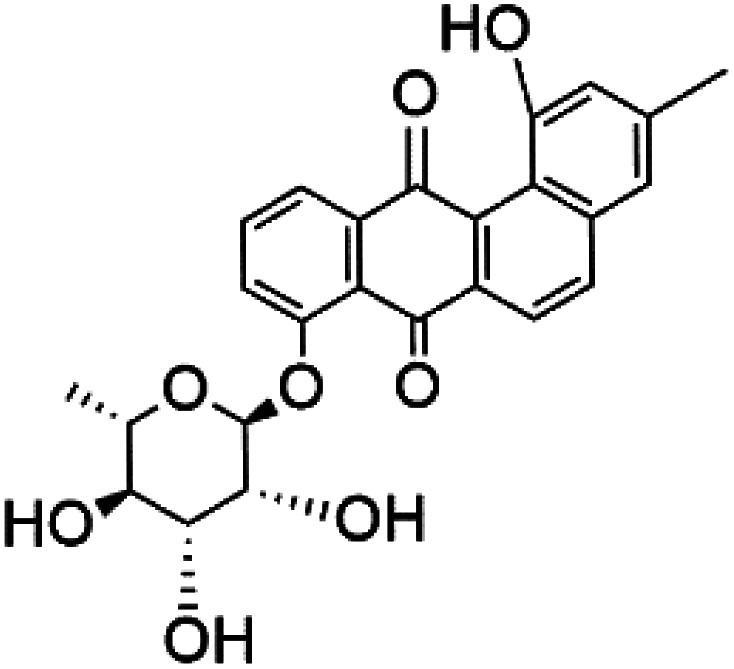
| Actinosporin G (127) |
|
||||
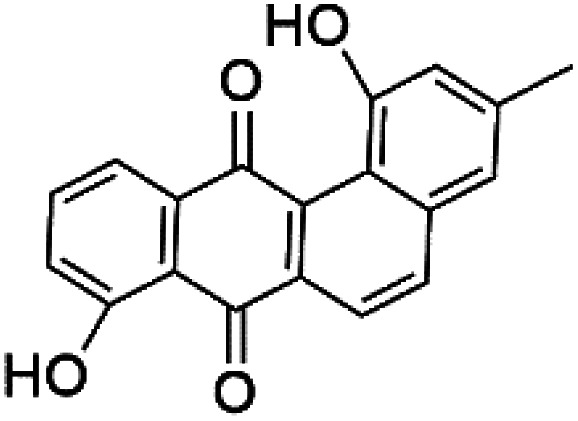
| Tetrangulol (128) |
|
||||
| Capillasterquinone B (129) |
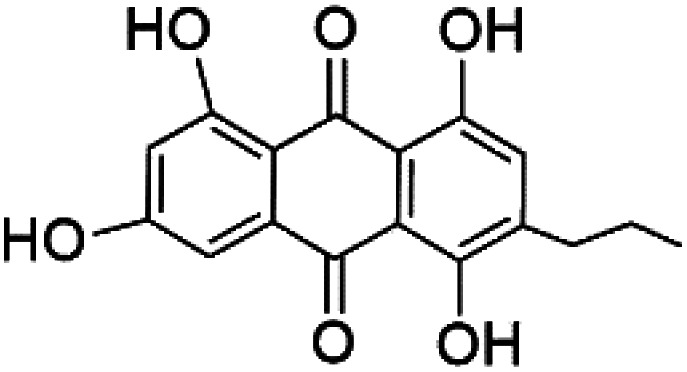
|
6. Macrolides
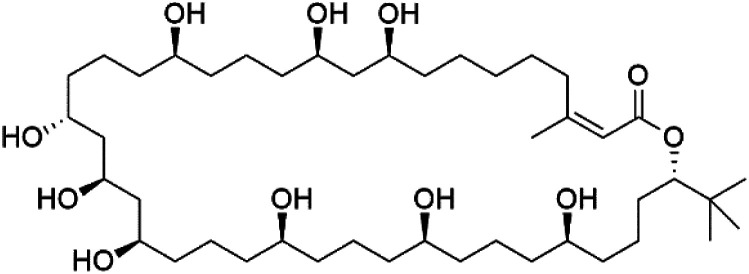
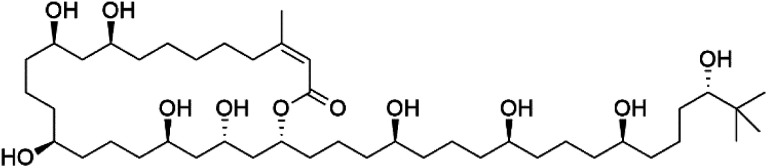
Bastimolide A, a 40-membered ring polyhydroxy macrolide with 10 stereocentres and a rare tert-butyl terminus (130), was obtained from Okeania hirsuta marine Cyanobacterium. It displayed highly potent activity against four multidrug-resistant TM90-C2A, TM90-C2B, W2, and TM91-C235 (IC50 = 0.089, 0.11, 0.18, 0.34 nM, respectively) P. falciparum strains.115 Bastimolide B (131), polyhydroxy macrolide with a long aliphatic chain containing terminal tert-butyl group. It exhibited strong antimalarial activity against CQ-sensitive P. falciparum strain HB3 (IC50 = 5.7 μM).116 X-ray crystallography was used to determine the macrocyclic lactone's planar structure and absolute configuration, which consists of a 1,3-diol, one 1,3,5-triol, and six 1,5-diols. The rare tert-butyl group near the lactone ester in the bastimolide structure is thought to protect the lactone ring against hydrolysis.117
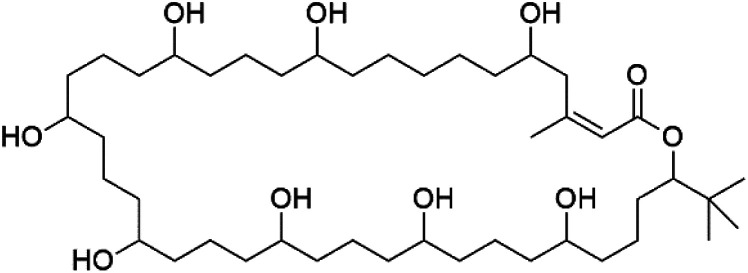
In addition, Palstimolide A, a polyhydroxy macrolide, was obtained from a tropical cyanobacterium Leptolyngbya sp. (Leptolyngbyaceae). Palstimolide A (132) had structural similarities to bastimolides. It displayed potent antimalarial activity against the blood stage of P. falciparum Dd2 strain (IC50 = 172.5 nM).118
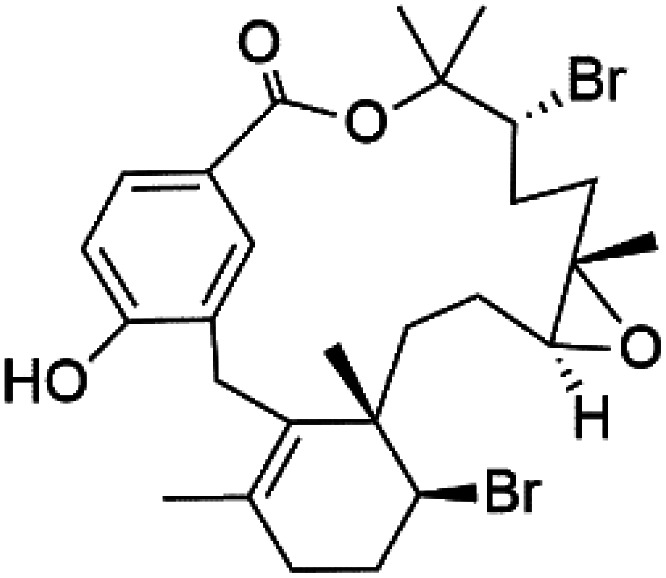
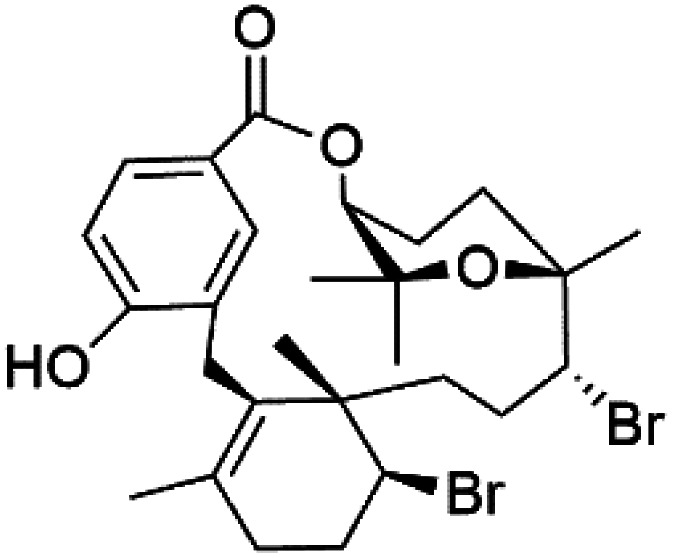
Moreover, bromophycolides A (133), D (134), R (135), S (136), T (137), and U (138), diterpene benzoates macrolides were obtained from Callophycus serratus red algae (Onagraceae). Bromophycolides having 15- and 16-membered rings have been demonstrated to be effective antimalarials. No significant effects of the lactone ring size on activity were observed between 15- and 16-membered lactone rings. Some bromophycolides target haem crystallization, implying that haemozoin production is inhibited as a mechanism of action.119 Bromophycolide A presented good activity against drug-resistant Dd2 strain (IC50 = 377 nM), drug-sensitive 3D7, and HB3 strains (IC50 = 499 and 493 nM), respectively.120 The chemical structures of these compounds belonging to macrolides are traced in Table 6. Bromophycolides have been mentioned among drugs that are investigated in pre-clinical trials.99
A list of marine-derived macrolides antimalarial drugs showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic source.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Bastimolide A (130) | TM90-C2A = 0.089TM90-C2B = 0.11 W2 = 0.18 TM91-C235 = 0.34 nM |
|
Okeania hirsuta | Cyanobacteria | 115 |
| Bastimolide B (131) | HB3 = 5.7 μM |
|
116 | ||
| Palstimolide A (132) | Dd2 = 172.5 nM |
|
Leptolyngbya sp. | Cyanobacteria | 118 |
| Bromophycolide A (133) | Dd2 = 377 nM, 3D7 = 499 nM HB3 = 493 nM |
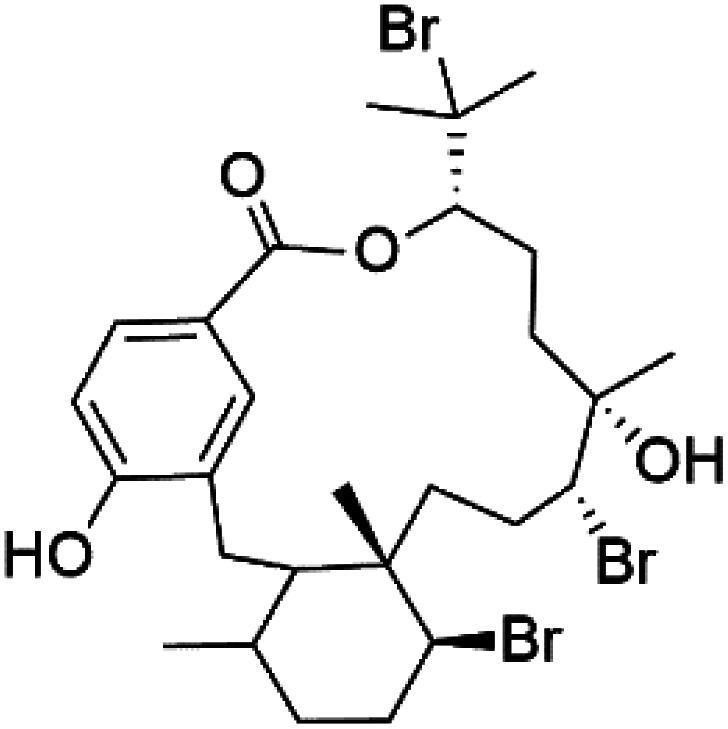
|
Callophycus serratus | Red alga | 119 |
| Bromophycolide D (134) |
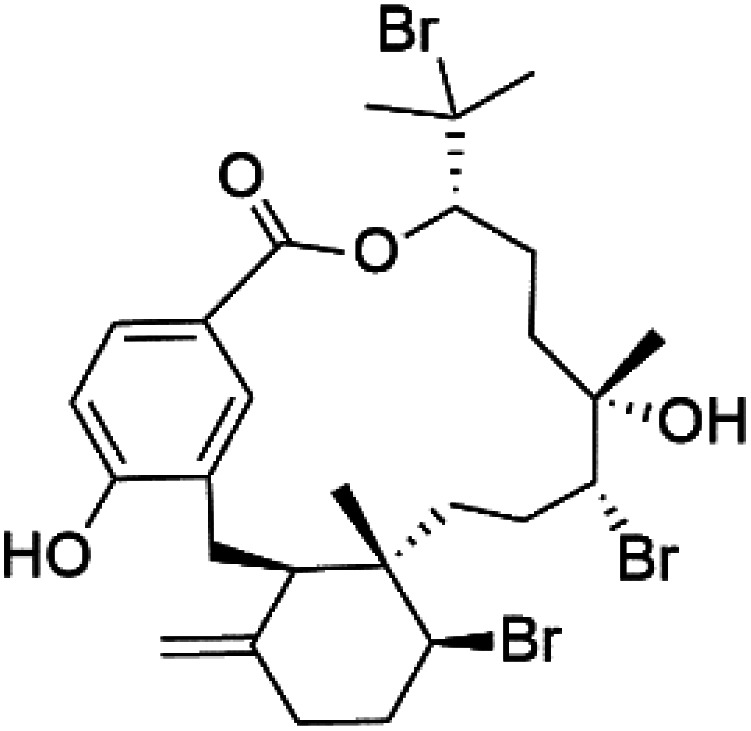
|
||||
| Bromophycolide R (135) |
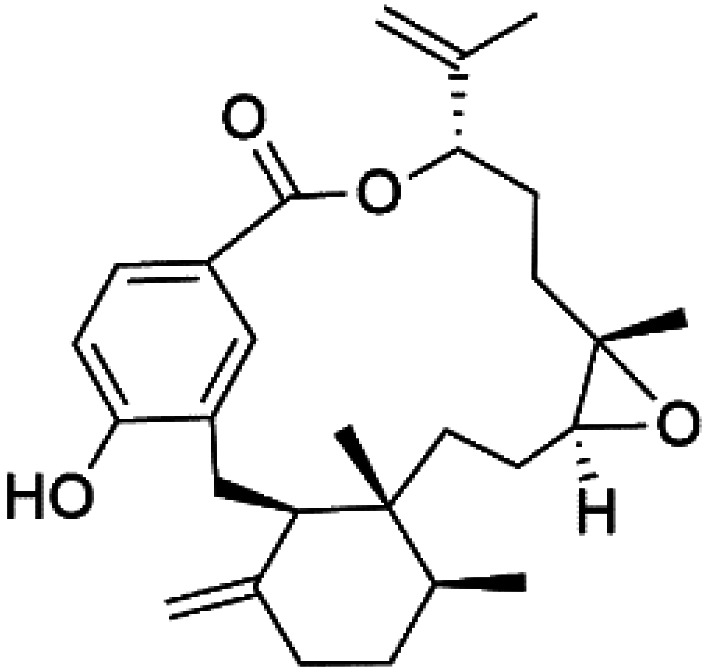
|
||||
| Bromophycolide S (136) |
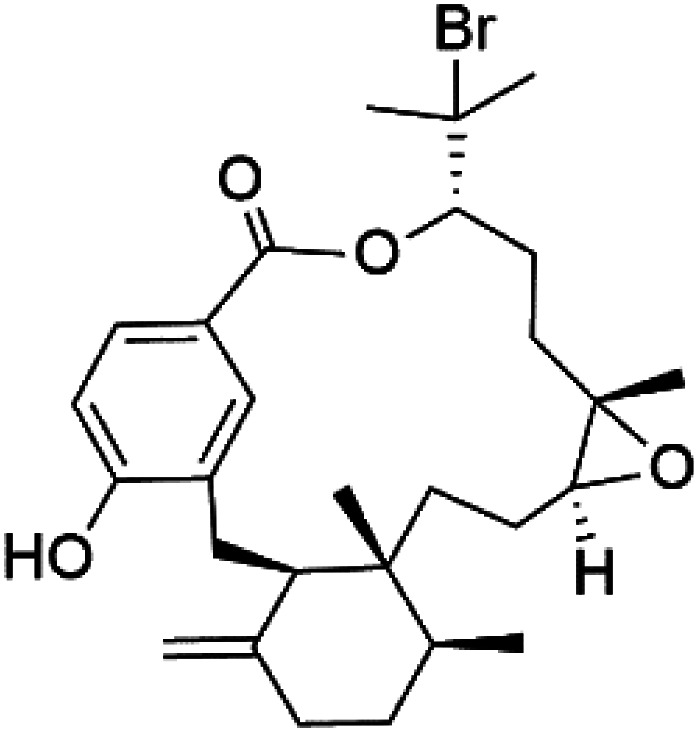
|
||||
| Bromophycolide T (137) |
|
||||
| Bromophycolide U (138) |
|
7. Peptides and depsipeptides
Peptides are short cyclic or acyclic chains between two and fifty amino acids, linked by amide covalent bonds (peptidic bonds). At the same time, depsipeptides are cyclic or acyclic compounds of α-amino and α-hydroxycarboxylic acids linked to each other by esters and amides units.121 Several peptides and depsipeptides from marine sources were reported for antiplasmodial activity are, summarized in Table 7.
A list of marine-derived peptides and depsipeptides antimalarial drugs showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic sourcea.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Balgacyclamide A (139) | K1 IC50 = of 9.0 and 8.2 μM, respectively |
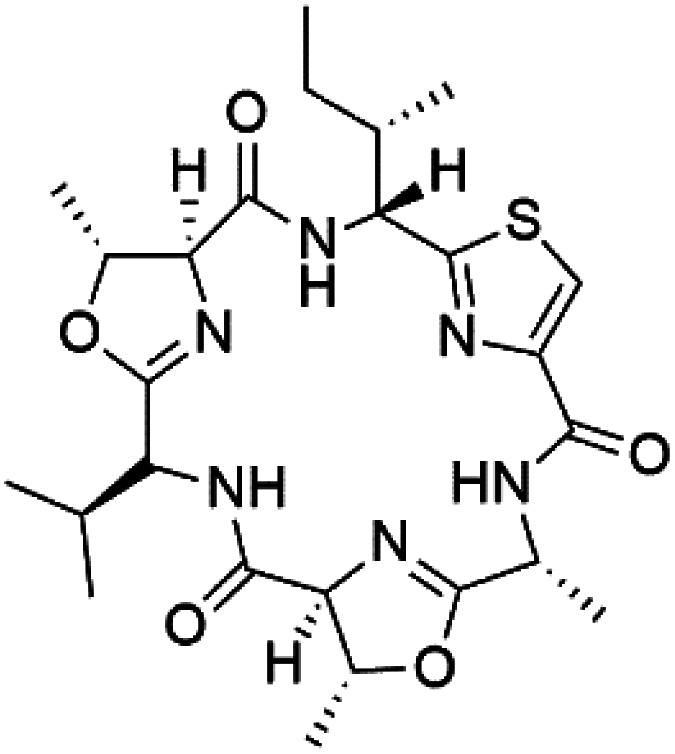
|
Microcystis aeruginosa | Cyanobacteria | 122 |
| Balgacyclamide B (140) |
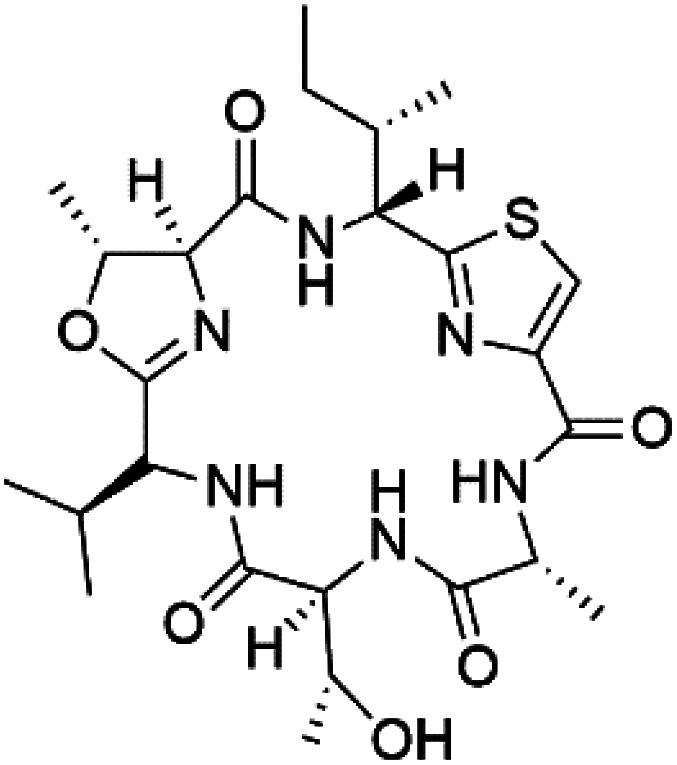
|
||||
| Balgacyclamide C (141) | NA |
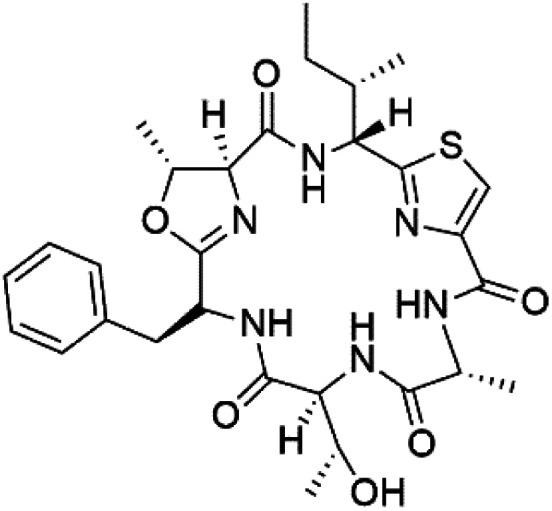
|
|||
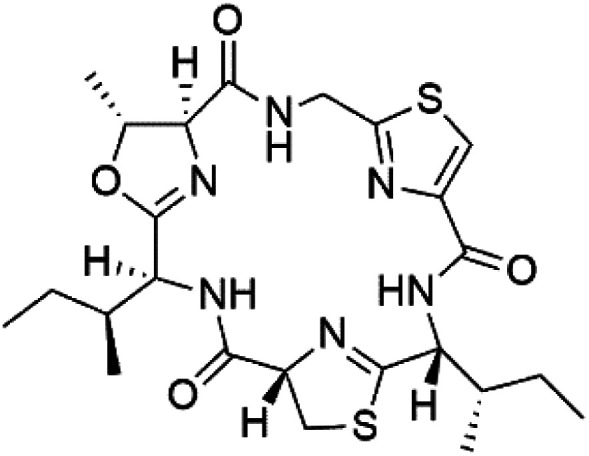
| Aerucyclamide A (142) | K1 = 5.0, 0.7, 2.3, and 6.3 μM, respectively |
|
Microcystis aeruginosa | Cyanobacteria | 123 and 124 |
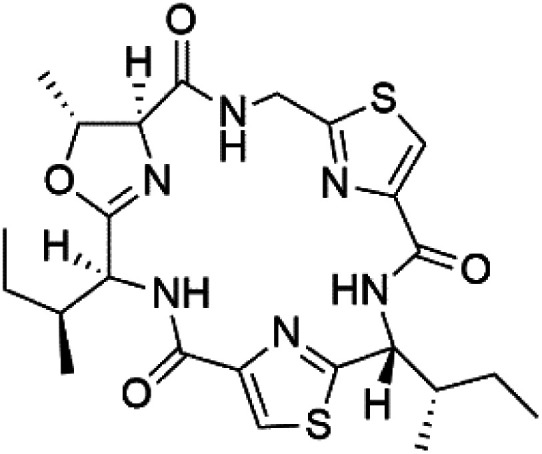
| Aerucyclamide B (143) |
|
||||
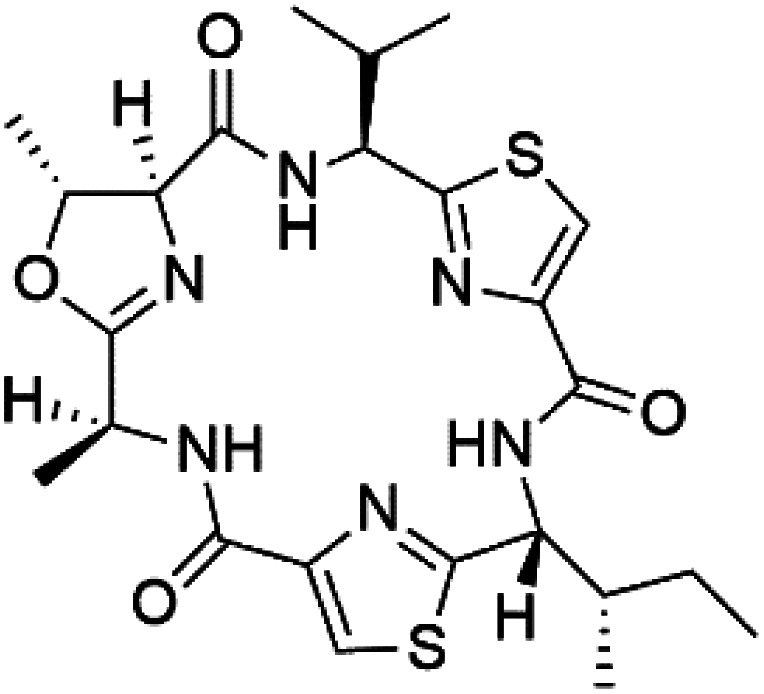
| Aerucyclamide C (144) |
|
||||
| Aerucyclamide D (145) |
|
||||
| Mollamide B (146) | D6 = 0.0029 μM W2 = 0.003 μM |
|
Didemnum mole | Tunicate | 125 |
| Dolastatin 10 (147) | FCH5·C2 = 0.1 nM |
|
Symploca sp. | Cyanobacteria | 126 and 127 |
| Dragonamide A (148) | 3D7 = 7.7, 7.0, and 6.0 μM |
|
Moorea producens | Cyanobacteria | 128 and 129 |
| Dragonamide B (149) |
|
||||
| Dragomabin (150) |
|
||||
| Carmabin A (151) | 3D7 = 4.3 μM |
|
|||
| Malyngamide X (152) | K1 ED50 = 5.44 μM |
|
Bursatella leachii | Thai sea hare | 130 |
| Romidepsin (153) | ∼150 nM |
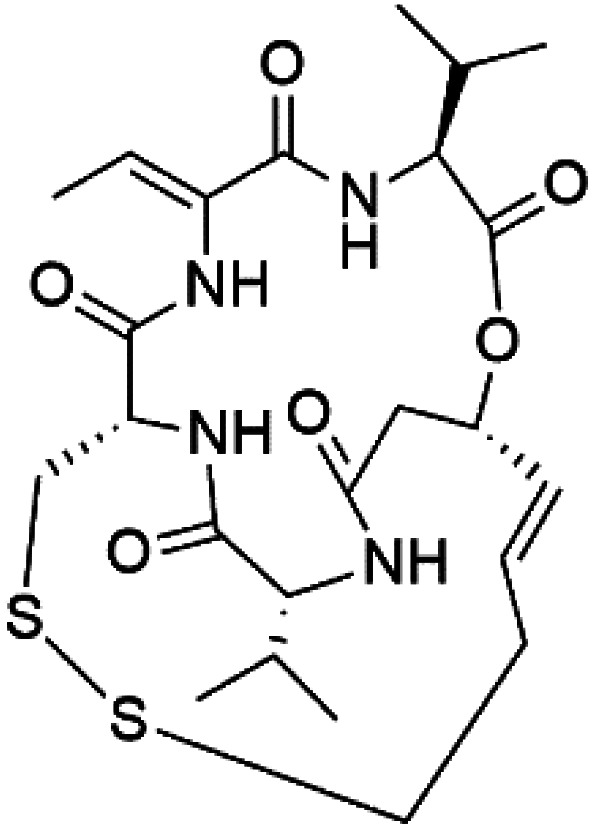
|
Violaceous sp | Cyanobacteria | 131 and 132 |
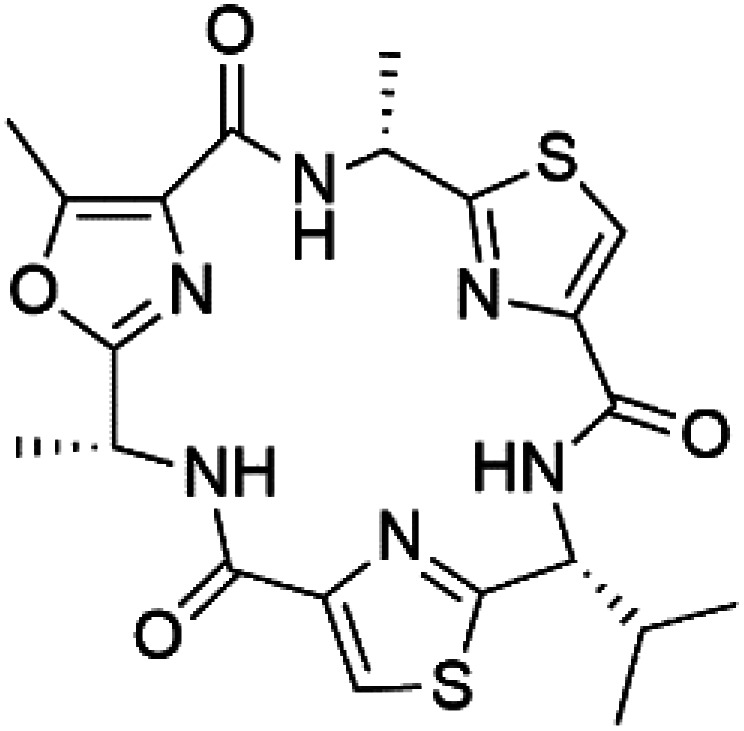
| Venturamide A (154) | W2 = 8.2 and 5.2 μM |
|
Oscillatoria sp. | Cyanobacteria | 133 |
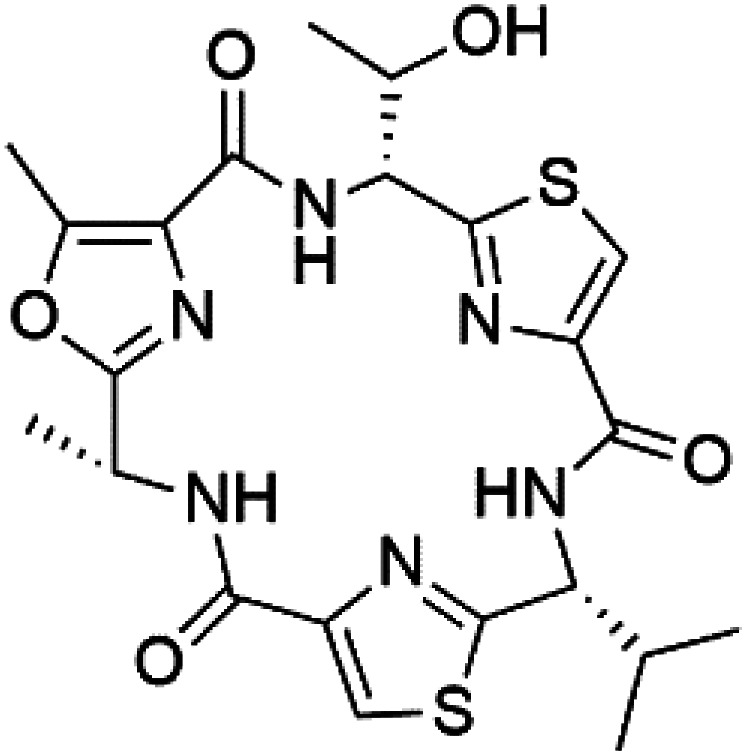
| Venturamide B (155) |
|
||||
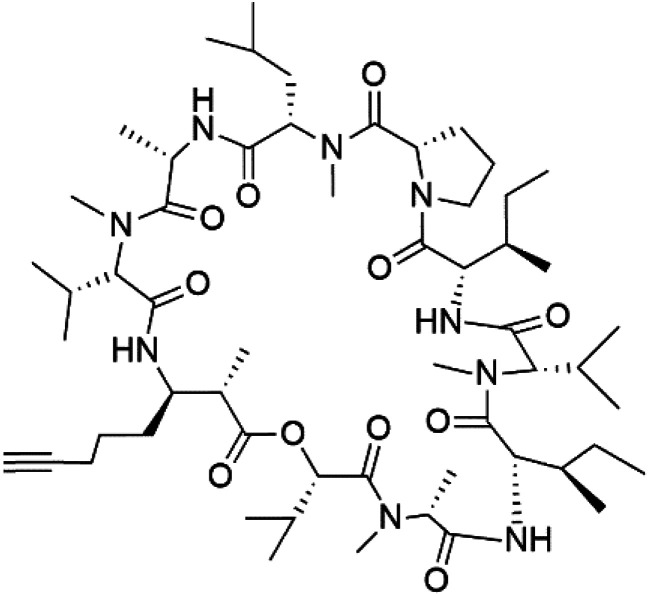
| Companeramide A (156) | D6 = 570 and 220 nM Dd2 = 1000 and 230 nM |
|
Panamanian marine Cyanobacterium sp. | Cyanobacteria | 134 |
| Companeramide B (157) |
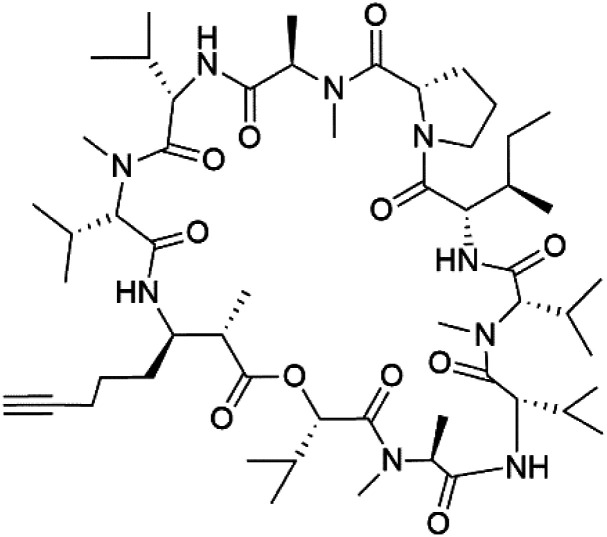
|
||||
| Lagunamide A (158) | NF54 = 190, 910 and 290 nM |
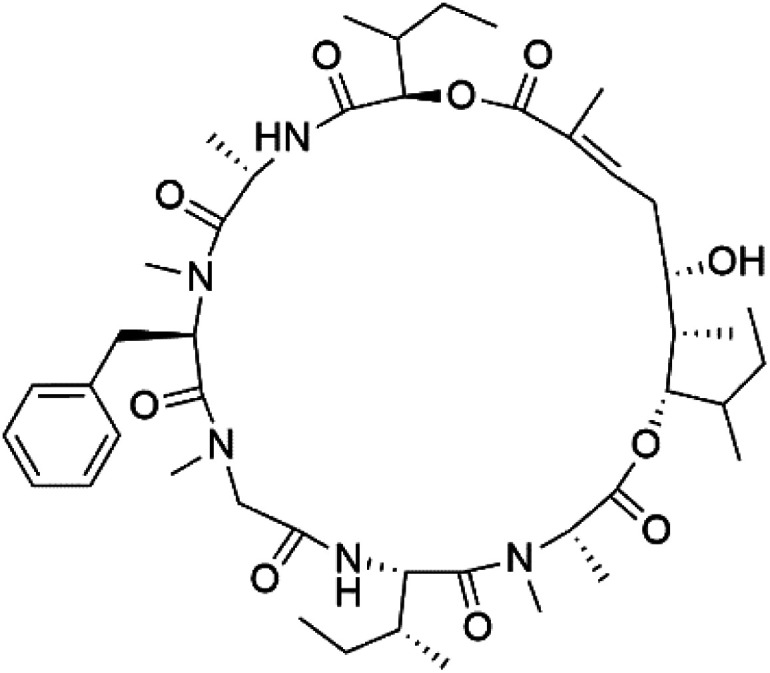
|
Lyngbya majuscule | Cyanobacteria | 135 and 136 |
| Lagunamide B (159) |
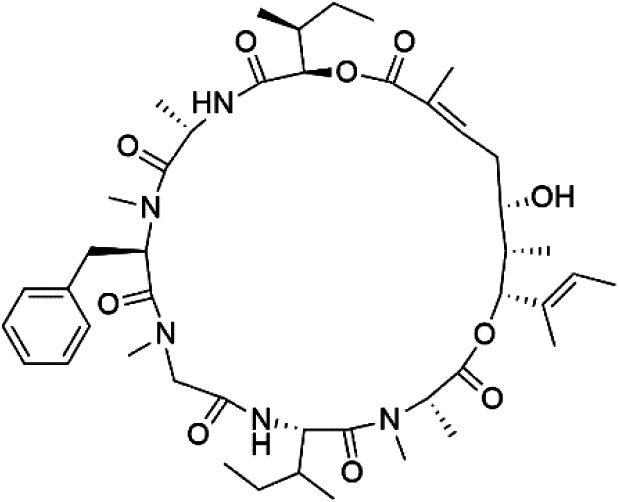
|
||||
| Lagunamide C (160) |
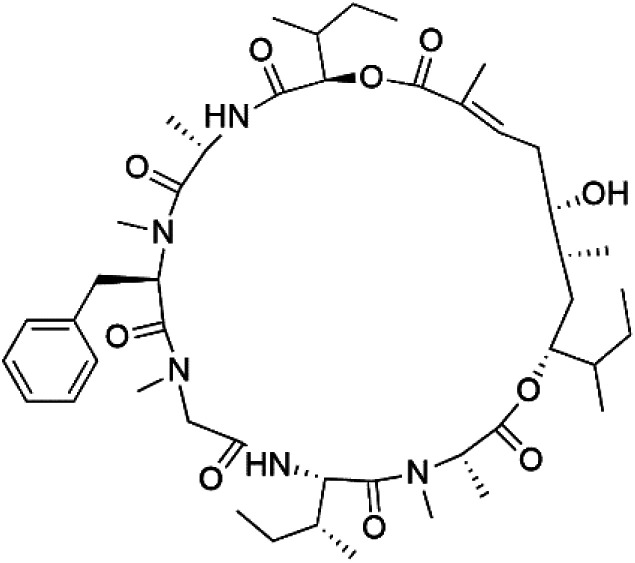
|
||||
| Mollemycin A (161) | 3D7 = 7 nM Dd2: 9 nM |
|
Streptomyces sp. | Marine actinomycetes | 137 |
| Symplocamide A (162) | W2 = 0.95 μM |
|
Streptomyces ballenaensis and S. bangulaensis | Marine actinomycetes | 138 and 139 |
| Actinoramide A (163) | HB3 = 190 nM Cp250 = 210 nM Dd2 = 220 nM 7G8 = 160 nM GB4 = 340 nM |
|
140 | ||
| Symplostatin 4 (164) | 3D7 ED50 = 74 nM |
|
Symploca sp | Cyanobacteria | 141 and 142 |
| Gallinamide A (165) | 3D7 = 50.1 ± 7.6 nM W2 = 8.4 μM |
|
Schizothrix sp. | Cyanobacteria | 144 |
| Viridamide A (166) | W2 = 5.8 μM |
|
Oscillatoria nigro-viridis | Cyanobacteria | 145 |
NA: not available.
Even though a good number of peptides and depsipeptides have presented good antimalarial activity, their mechanism of action is not well understood. Some of them displayed a strong inhibitory effect on some key enzymes present in the malaria parasite; moreover, the relation between the inhibition and their antimalarial activity remains unestablished.58
7.1. Cyclic peptides
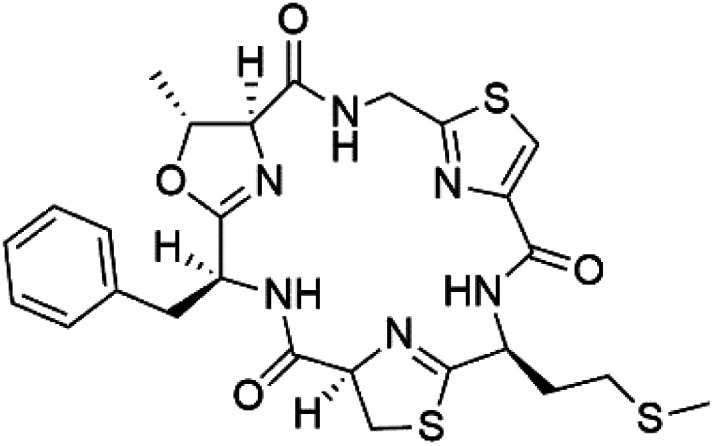
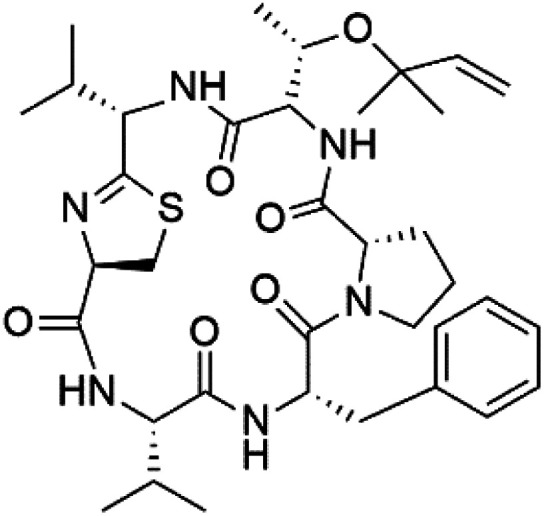
Three new heterocyclic macrocyclic peptides, Balgacyclamides A–C (139–141), were isolated from Microcystis aeruginosa (Microcystaceae). Balgacyclamides A and B displayed potent activity against the CQ-resistant strain K1 of P. falciparum (IC50 = of 9.0 and 8.2 μM), respectively.122 A class of modified hexacyclopeptides, aerucyclamides A–D (142–145) were obtained from the cyanobacterium Microcystis aeruginosa. They presented a potent activity against the K1 strain of P. falciparum (IC50 = 5.0, 0.7, 2.3, and 6.3 μM, respectively).123,124 Mollamide B (146), thiazoline hexapeptides found in the Indonesian tunicate Didemnum mole (Didemnidae). It showed a moderate antimalarial activity against D6 and W2 strains (IC50 = 0.0029 and 0.003 μM, respectively) of P. falciparum.125
7.2. Acyclic peptides
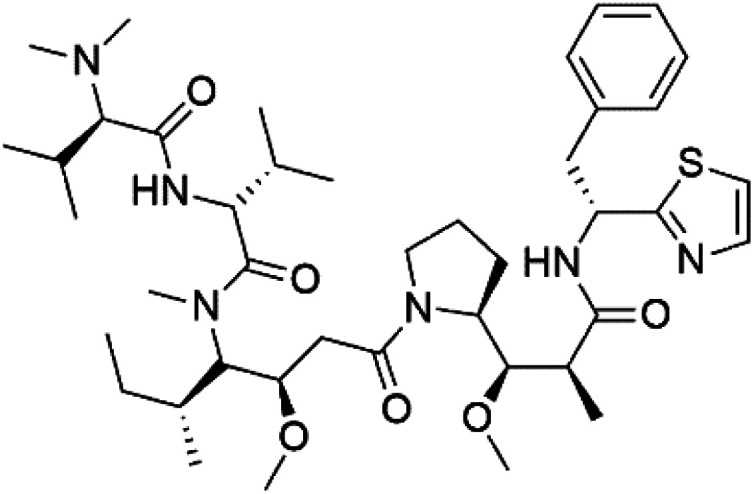
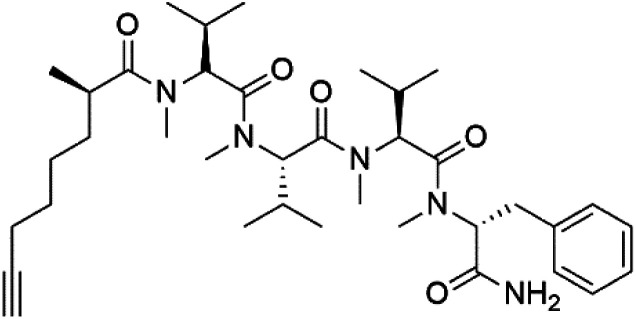
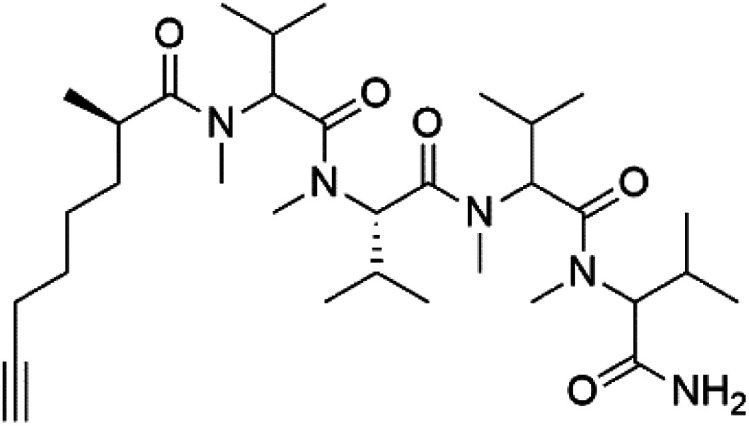
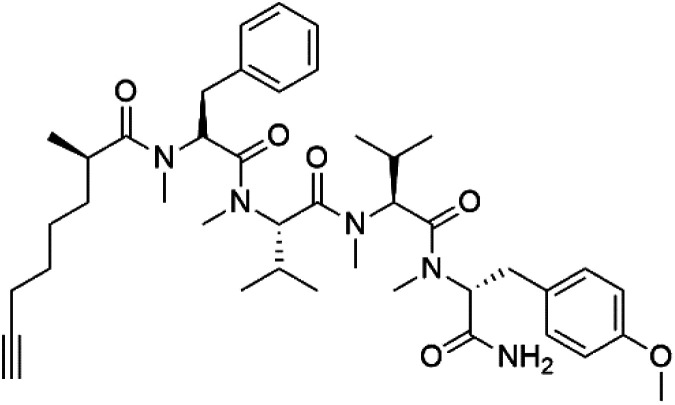
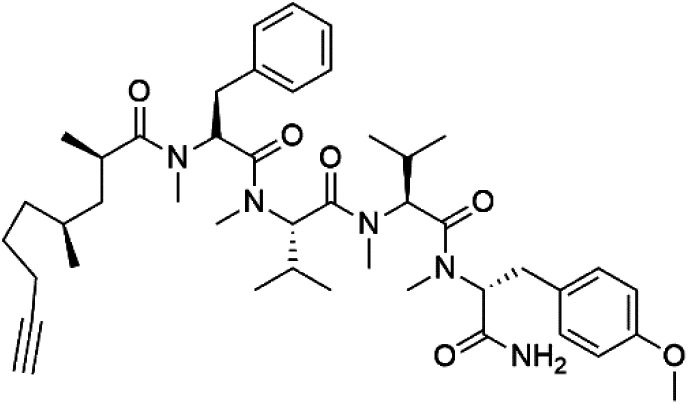
Dolastatin 10 (147), an acyclic peptide extracted from cyanobacterium Symploca species126,127 exhibited strong activity against P. falciparum 3D7 colonies, with IC50 = 74 nM. In addition, four acyclic lipopeptides, dragonamides A (148), B (149), dragomabin (150), and carmabin A (151) have been isolated from the cyanobacterium Moorea producens (Cyanobacteriaceae) (formerly Lyngbya majuscula). dragomabin, carmabin A and dragonamide A displayed good antimalarial activity (IC50 = 6.0, 4.3 and 7.7 μM, respectively).128,129
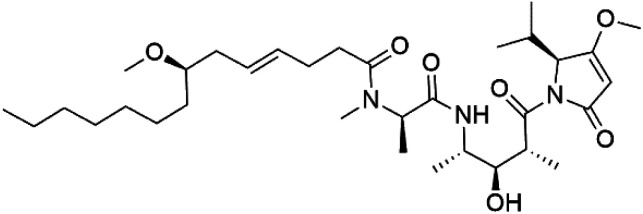
Morover, malyngamide X (152) is the first (7R)-lyngbic acid connected to a new tripeptide backbone. It was obtained from Bursatella leachii (Aplysiidae), a Thai sea hare, presented a moderate antimalarial activity with a half effective dose (ED50) = 5.44 μM against P. falciparum (K1) multidrug-resistant strain.130
7.3. Cyclic depsipeptides
Romidepsin (153), a cyclic depsipeptide histone deacetylase (HDAC) inhibitor, is responsible for the observed anti-Plasmodium activity of Chromobacterium species.131,132 Venturamides A (154) and B (155), two compounds from thiazole and oxazole cyclodepsipeptides class obtained from the cyanobacterium Oscillatoria sp., presented activity against (IC50 = 8.2 and 5.2 μM, respectively) P. falciparum W2 strain.133
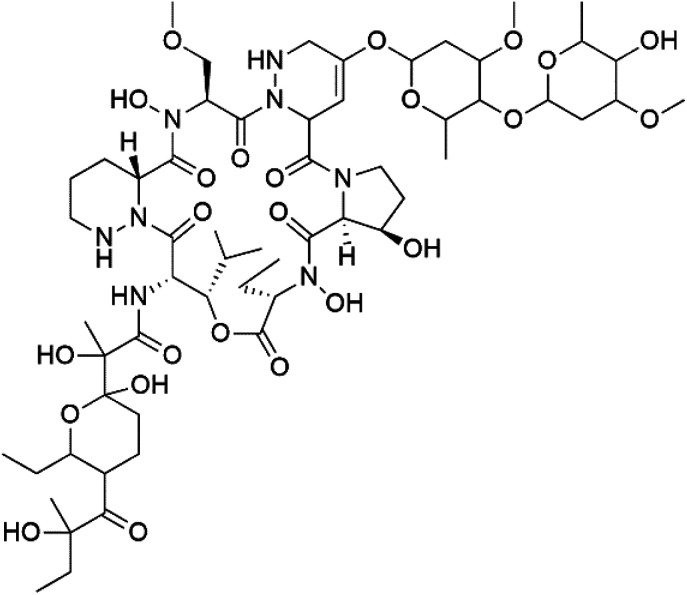
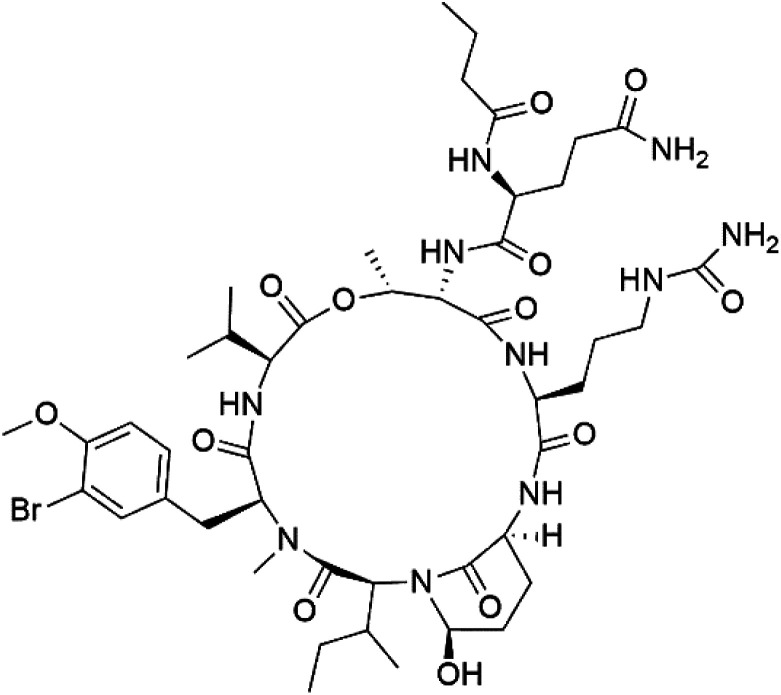
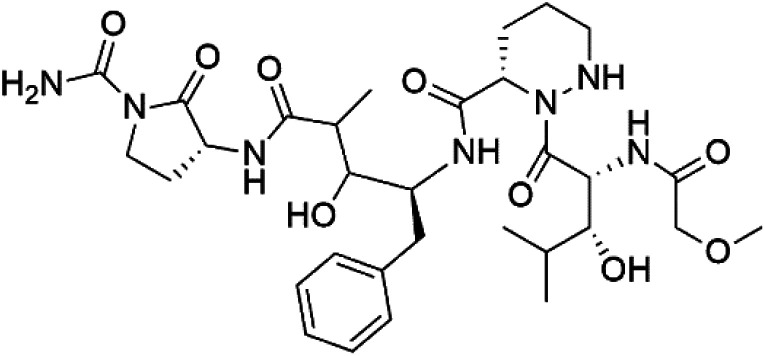
Two cyclodepsipeptides, companeramides A (156) and B (157), were obtained from a Panamanian marine Cyanobacterium sp. (Cyanobacteriaceae). Exhibited strong antiplasmodial activity against D6 strain in vitro (IC50 = 0.57 and 0.22 μM, respectively).134 Three cytotoxic cyclic depsipeptides, lagunamides A–C (158–160), were isolated from Lyngbya majuscule (Oscillatoriaceae). The planar lagunamide macrocyclic scafold consists of peptide and polyketide substructures, and the main diferences are in the polyketide part. Lagunamides A and B are 26-membered macrocycles, while lagumanide C has an additional methylene carbon in the polyketide structure. Lagunamides A–C, showed potent activity against P. falciparum NF54 strain (IC50 = 0.19, 0.91, and 0.29 μM, respectively).135,136 The double bond in the side chain of lagunamide B might be responsible for the lower activity.117 Mollemycin A (161), a glycol-hexadepsipeptide-polyketide isolated from a marine-derived Streptomyces sp. CMBM0244 (Streptomycetaceae), exhibited a potent and selective growth inhibitory activity against drug-sensitive 3D7 and multidrug-resistant Dd2 clones of P. falciparum (IC50 = 7.0 and 9.0 nM, respectively).137 Symplocamide A (162), cyclodepsipeptide was extracted from the marine Cyanobacterium symploca sp. (Cyanobacteriaceae), showed potent antimalarial activity against W2 strain (IC50 = 0.95 μM) of P. falciparum.138,139 Cyclic tetrapeptide, actinoramide A (163) was isolated from marine actinomycetes Streptomyces ballenaensis, and S. bangulaensis (Streptomycetaceae). This tetrapeptide had potent activity against clones of drug-resistant P. falciparum including Cp250, Dd2, 7G8, and GB4 (IC50 = 210, 220, 160, and 340 nM, respectively) and drug-sensitive HB3 clone (IC50 = 190 nM).140
7.4. Acyclic depsipeptides
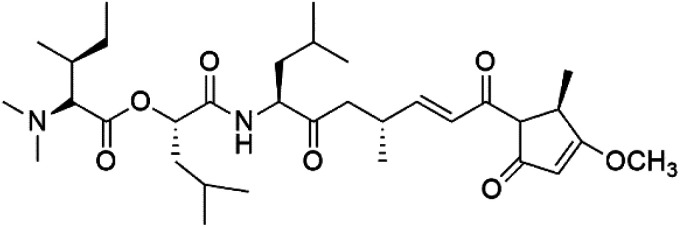
Symplostatin 4 (164), an acyclic depsipeptide extracted from Symploca sp. (Microcoleaceae). Symplostatin 4 showed a significant activity against 3D7 strain (ED50 = 74 nM) of P. falciparum.141,142 It displayed its activity on P. falciparum falcipains in infected red blood cells, indicating inhibition of the hemoglobin degradation pathway as a possible mode of action.143
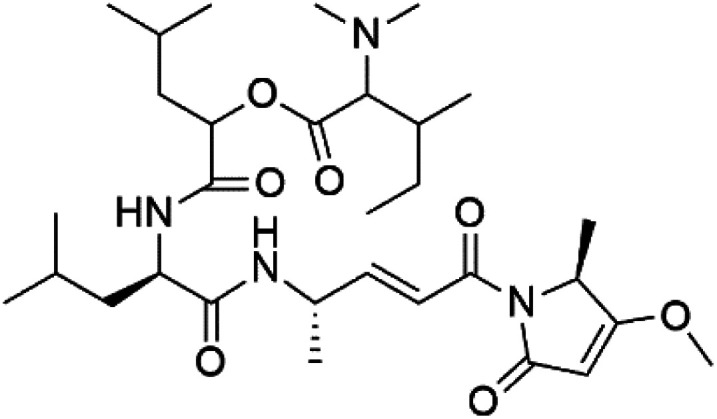
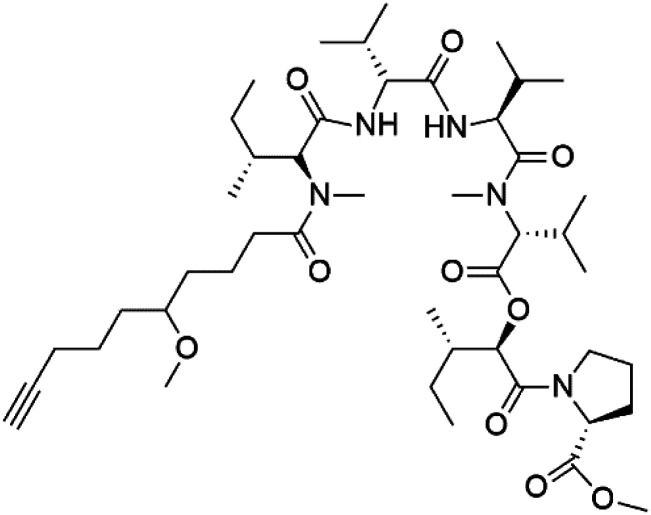
A further cyanobacterial acyclic depsipeptide derivative, named gallinamide A (165) obtained from tropical reef Schizothrix sp. (Cyprinidae). Gallinamide A showed a moderate in vitro antimalarial activity against the W2 strain (IC50 = 8.4 μM) of P. falciparum.144 Viridamide (166), a lipodepsipeptide obtained from the cyanobacterium Oscillatoria nigro-viridis (Oscillatoriaceae), showed the activity against P. falciparum (IC50 = 5.8 μM).145
8. Phosphotriesters
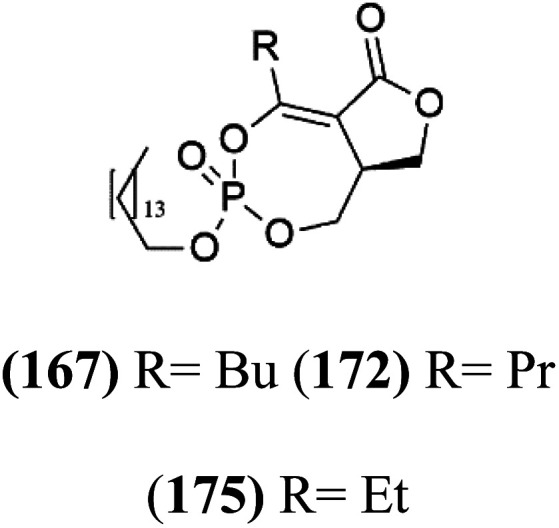
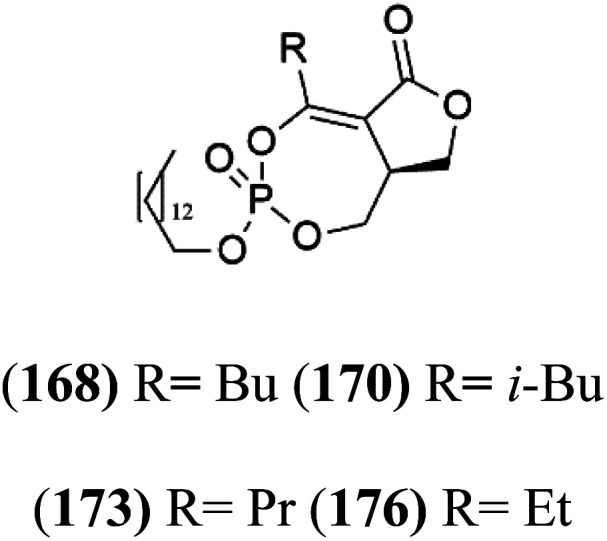
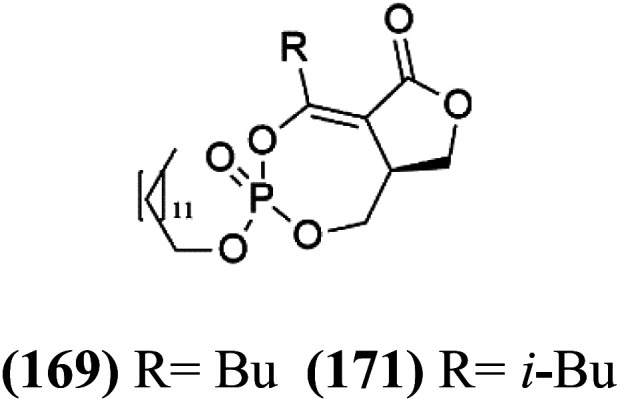
A new class of antimalarials with long-chain bicyclic phosphotriesters, salinipostins A–K (167–177), Table 8, were obtained from Salinospora sp. bacteria (Micromonosporaceae). SAR fndings indicated that an increase in alkyl chain length attached to the phosphoester oxygen and vinyl carbon led to increased activity while branching of the alkyl causes a slight reduction in activity. The most active compound salinipostin A, did not afect parasite schizonts, indicating that it acts by disrupting the processes required for the establishment or growth of intracellular parasites. Salinispostin A did not inhibit haemozoin formation but cause cellular disorganization and disintegration of internal structure.146 These compounds showed different activity against P. falciparum W2 strain. Salinipostins A and D displayed the most potent activity (IC50 = 50 and 82 nM) followed by salinipostins I, B, F, C, G, E and H (IC50 = 0.126, 0.139, 0.266, 0.1415, 1.52, 3.22, 8.70 μM), respectively. Only, salinipostins K and J displayed weak activity.146
A list of marine-derived phosphotriesters, polyether, and steroidal glycosides antimalarial drugs showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic sourcea.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Salinipostin A (167) Salinipostin F (172) Salinipostin I (175) | W2 = 50, 266, and 126 nM |
|
Salinospora sp. | Marine actinomycetes | 146 |
| Salinipostin B (168) Salinipostin D (170) Salinipostin G (173) Salinipostin J (176) | W2 = 139, 82, 1.52 μM, respectively (176) NA |
|
|||
| Salinipostin C (169) Salinipostin E (171) Salinipostin H (174) Salinipostin K (177) | W2 = 415, 3.22, 8.70 μM, respectively (177) NA |
|
|||
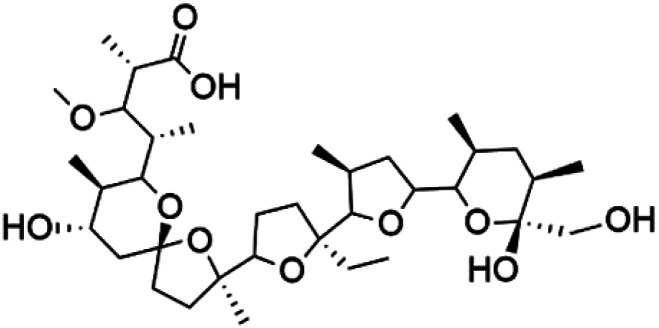
| Monensin (178) | D6 and W2 0.15 to 0.3 nM |
|
Streptomyces cinnamonensis | Marine actinomycetes | 147 and 148 |
| Pandaroside E (179) | W2 = 0.78.0.05 and 0.038 μM |
|
Pandaros acanthifolium | Sponge | 150 |
| Pandaroside G (180) |
|
||||
| Pandaroside H (181) |
|
NA: not available.
9. Miscellaneous compounds
9.1. Polyethers
A polyether ionophore isolated from Streptomyces cinnamonensis (Streptomycetaceae) named Monensin (178), Table 8. Monensin has been displayed a strong antimalarial activity against P. falciparum.147,148 In a recent study, a polyether metabolite was extracted from Streptomyces sp. strain H668. This polyether displayed in vitro antimalarial activity against both D6 and W2 strains of P. falciparum with IC50 values from 0.15 to 0.3 nM.149
9.2. Steroid glycosides
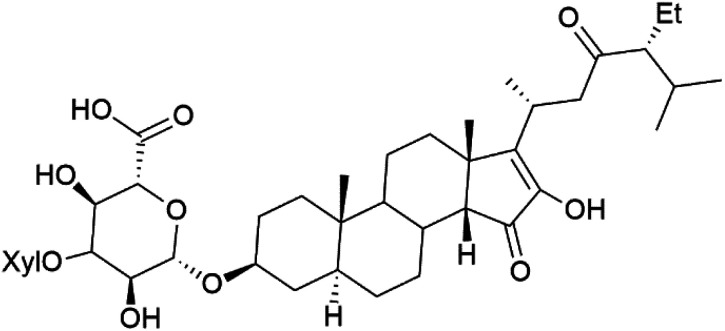
The steroidal glycosides pandaroside E (179), G (180), and H (181) (Table 8), had been isolated from the Caribbean sponge Pandaros acanthifolium (Microcionidae). They strongly inhibited the growth of P. falciparum at low sub-micromolar concentrations.150
10. Conclusion
Malaria is among the crucial VBDs affecting the global health, based on the WHO official reports. It is easily progressed to impairment of important human body organs, including the liver, and death in case improper diagnosis and treatment. This emergency has acquired a special interest among health care providers and researchers to find more effective and safer medicaments, especially against the multi-resistant strain of Plasmodium sp. for the people of the developing countries. Particularly, natural-derived treatment of infectious diseases, including malaria is still the most convenient, safe, effective, and diverse. Marine organisms have attracted great potential in the last few decades as a promising and non-traditional source of bioactive compounds. Moreover, recent technological advances have led to isolate and identify thousands of marine-derived compounds belonging to various chemical classes. A total of 181 compounds derived from different marine sources, including sponges, cyanobacteria, marine algae, and actinomycetes, were reviewed in the current research possessing potential antimalarial activities with unique SAR and targeting different growth stages, including ring and trophozoite stage. More than half of the compounds belong to three major chemical classes comprising alkaloids, terpenoids, and polyketides. Such chemical diversity, potency, and less cytotoxicity are recognized as great start point for further SAR and clinical investigations of antimalarial candidates. The current article assumed that marine-derived natural products can also open up novel resources of bioactive compounds for novel candidates for management of other infectious diseases, exploring the oceans and seas treasures. Three compounds, including bromophycolides, plakortin, and homogentisic acid, are investigated as antimalarial drugs in pre-clinical trials and may be approved and marketed soon.
Conflicts of interest
Authors declare that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
List of abbreviations
- CQ
Chloroquine
- EC50
Half maximal effective concentration
- ED50
Half maximal effective dose
- FB
Iron-protoporphyrin IX
- FDA
Food and Drug Administration
- GSK-3P
Glycogen synthase 3
- IC50
Half-maximal inhibitory concentration
- MIC
minimum inhibitory concentration
- SAR
Structure-activity relationships
- VBDs
Vector-borne diseases
- P. falciparum D6
A West African clone
- P. falciparum W2
The Indochina clone W2
- WHO
World Health Organization
Supplementary Material
References
- WHO, Global Vector borne diseases, World Health Organization, 2020 [Google Scholar]
- Degroote S. Zinszer K. Ridde V. Infect. Dis. Poverty. 2018;7:96. doi: 10.1186/s40249-018-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnikov T. R., in Autoethnographies on the Environment and Human Health, Springer International Publishing, Cham, 2018, pp. 113–126, 10.1007/978-3-319-69026-1_9 [DOI] [Google Scholar]
- Lafferty K. D. Ecology. 2009;90:888–900. doi: 10.1890/08-0079.1. [DOI] [PubMed] [Google Scholar]
- World Health Organization WHO, World malaria report, World Health Organization, Geneva, Switzerland, Licence: CC BY-NC-SA 3.0 IGO, 2019 [Google Scholar]
- WHO, Key facts; news room, Malaria, 2020 [Google Scholar]
- Fattorusso E. Taglialatela-Scafati O. Mar. Drugs. 2009;7:130–152. doi: 10.3390/md7020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C. J. Tanomsing N. Nolder D. Oguike M. Jennison C. Pukrittayakamee S. Dolecek C. Hien T. T. Do Rosário V. E. Arez A. P. J. Infect. Dis. 2010;201:1544–1550. doi: 10.1086/652240. [DOI] [PubMed] [Google Scholar]
- Fançony C. Soares Â. Lavinha J. Barros H. Brito M. Int. J. Environ. Res. Public Health. 2019;16:466. doi: 10.3390/ijerph16030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gennaro F. Marotta C. Locantore P. Pizzol D. Putoto G. Trop. Med. Int. Health. 2020;5:141. doi: 10.3390/tropicalmed5030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleman S. Beyene Tufa T. Kebebe D. Belew S. Mekonnen Y. Gashe F. Mussa S. Wynendaele E. Duchateau L. De Spiegeleer B. J. Ethnopharmacol. 2018;213:262–279. doi: 10.1016/j.jep.2017.10.034. [DOI] [PubMed] [Google Scholar]
- Mpimbaza A. Nayiga S. Ndeezi G. Rosenthal P. J. Karamagi C. Katahoire A. PLoS One. 2019;14:e0217262. doi: 10.1371/journal.pone.0217262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G. M. McCall M. B. B. Dik W. A. Urbanus R. T. Wammes L. J. Koelewijn R. Sauerwein R. W. Verbon A. van Hellemond J. J. van Genderen P. J. J. Cytokine. 2020;125:154838. doi: 10.1016/j.cyto.2019.154838. [DOI] [PubMed] [Google Scholar]
- Ochsendorf F. R. J. Dtsch Dermatol. Ges. 2010;8:829–845. doi: 10.1111/j.1610-0387.2010.07490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzor P. F. Prasasty V. D. Agubata C. O. J. Evidence-Based Integr. Med. 2020;2020:9385125. doi: 10.1155/2020/9385125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston D. G. I. Cassera M. B. Prog. Chem. Org. Nat. Prod. 2022;117:1–106. doi: 10.1007/978-3-030-89873-1_1. [DOI] [PubMed] [Google Scholar]
- Mojab F. Avicenna J. Phytomed. 2012;2:52. [PMC free article] [PubMed] [Google Scholar]
- Alves C. Silva J. Pinteus S. Gaspar H. Alpoim M. C. Botana L. M. Pedrosa R. Front. Pharmacol. 2018;9:777. doi: 10.3389/fphar.2018.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A. Hahn T. Finkelmeier D. Burger-Kentischer A. Rupp S. Krämer R. Ulber R. Process Biochem. 2019;81:182–187. [Google Scholar]
- Zayed A. Muffler K. Hahn T. Rupp S. Finkelmeier D. Burger-Kentischer A. Ulber R. Mar. Drugs. 2016;14:79. doi: 10.3390/md14040079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindequist U. Biomol. Ther. 2016;24:561–571. doi: 10.4062/biomolther.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D. G. Weiskirchen R. Al-Musharafi S. K. Acta Pharmacol. Sin. 2015;36:158–170. doi: 10.1038/aps.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyshlovoy S. A. Honecker F. Mar. Drugs. 2020;18(1):20. doi: 10.3390/md18010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A., PhD thesis, TU Kaiserslautern, Kaiserslautern, Germany, 2018 [Google Scholar]
- Nweze J. A. Mbaoji F. N. Li Y.-M. Yang L.-Y. Huang S.-S. Chigor V. N. Eze E. A. Pan L.-X. Zhang T. Yang D.-F. Infect. Dis. Poverty. 2021;10:9. doi: 10.1186/s40249-021-00796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy P. J. Roberts B. F. Carbonell A. Roberts J. Wright A. E. Chakrabarti D. Trop. Med. Int. Health. 2019;4:103. doi: 10.3390/tropicalmed4030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T. Kurimoto S.-i. Kobayashi J. i. Alkaloids Chem. Biol. 2020;84:1–124. doi: 10.1016/bs.alkal.2020.03.001. [DOI] [PubMed] [Google Scholar]
- Dai J. Dan W. Schneider U. Wang J. Eur. J. Med. Chem. 2018;157:622–656. doi: 10.1016/j.ejmech.2018.08.027. [DOI] [PubMed] [Google Scholar]
- Ashok P. Ganguly S. Murugesan S. Drug Discovery Today. 2014;19:1781–1791. doi: 10.1016/j.drudis.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Skropeta D. Pastro N. Zivanovic A. Mar. Drugs. 2011;9:2131–2154. doi: 10.3390/md9102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok P. Lathiya H. Murugesan S. Eur. J. Med. Chem. 2015;97:928–936. doi: 10.1016/j.ejmech.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Shady N. H. Fouad M. A. Salah Kamel M. Schirmeister T. Abdelmohsen U. R. Mar. Drugs. 2019;17:19. doi: 10.3390/md17010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y. Kubota T. Fromont J. Kobayashi J. i. Org. Lett. 2009;11:21–24. doi: 10.1021/ol802251q. [DOI] [PubMed] [Google Scholar]
- Yamada M. Takahashi Y. Kubota T. Fromont J. Ishiyama A. Otoguro K. Yamada H. Ōmura S. Kobayashi J. i. Tetrahedron. 2009;65:2313–2317. [Google Scholar]
- Kubota T. Nakamura K. Kurimoto S.-i. Sakai K. Fromont J. Gonoi T. Kobayashi J. i. J. Nat. Prod. 2017;80:1196–1199. doi: 10.1021/acs.jnatprod.6b01110. [DOI] [PubMed] [Google Scholar]
- Shady N. H. Fouad M. A. Ahmed S. Pimentel-Elardo S. M. Nodwell J. R. Kamel M. S. Abdelmohsen U. R. J. Antibiot. 2018;71:1036–1039. doi: 10.1038/s41429-018-0092-5. [DOI] [PubMed] [Google Scholar]
- Kirsch G. König G. M. Wright A. D. Kaminsky R. J. Nat. Prod. 2000;63:825–829. doi: 10.1021/np990555b. [DOI] [PubMed] [Google Scholar]
- Zhidkov M. E. Sidorova M. A. Lyakhova I. A. Tetrahedron Lett. 2018;59:1417–1420. [Google Scholar]
- Huang H. Yao Y. He Z. Yang T. Ma J. Tian X. Li Y. Huang C. Chen X. Li W. J. Nat. Prod. 2011;74:2122–2127. doi: 10.1021/np200399t. [DOI] [PubMed] [Google Scholar]
- Shi Y. Moazami Y. Pierce J. G. Bioorg. Med. Chem. 2017;25:2817–2824. doi: 10.1016/j.bmc.2017.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos P.-E. Wolfender J.-L. Queiroz E. F. Marcourt L. Al-Mourabit A. Frédérich M. Bordignon A. De Voogd N. Illien B. Gauvin-Bialecki A. J. Nat. Prod. 2017;80:1404–1410. doi: 10.1021/acs.jnatprod.6b01079. [DOI] [PubMed] [Google Scholar]
- Chan S. T. Pearce A. N. Page M. J. Kaiser M. Copp B. R. J. Nat. Prod. 2011;74:1972–1979. doi: 10.1021/np200509g. [DOI] [PubMed] [Google Scholar]
- Finlayson R. Pearce A. N. Page M. J. Kaiser M. Bourguet-Kondracki M.-L. Harper J. L. Webb V. L. Copp B. R. J. Nat. Prod. 2011;74:888–892. doi: 10.1021/np1008619. [DOI] [PubMed] [Google Scholar]
- Dineshkumar K. Vasudevan A. Hopper W. Comb. Chem. High Throughput Screening. 2017;20:3–19. doi: 10.2174/1386207319666161215154128. [DOI] [PubMed] [Google Scholar]
- Prudhomme J. McDaniel E. Ponts N. Bertani S. Fenical W. Jensen P. Le Roch K. PLoS One. 2008;3:e2335. doi: 10.1371/journal.pone.0002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. Phat C. Hong S.-C. Peptides. 2017;95:94–105. doi: 10.1016/j.peptides.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Lee H.-S. Jeong G.-S. Molecules. 2020;25:5031. [Google Scholar]
- Marx L. B. Burton J. W. Chem.–Eur. J. 2018;24:6747–6754. doi: 10.1002/chem.201800046. [DOI] [PubMed] [Google Scholar]
- Gholami H. Kulshrestha A. Favor O. K. Staples R. J. Borhan B. Angew. Chem. 2019;131:10216–10219. doi: 10.1002/anie.201900340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwabusola E. T. Tabudravu J. N. Al Maqbali K. S. Annang F. Pérez-Moreno G. Reyes F. Jaspars M. Chem. Biodiversity. 2020;17:e2000335. doi: 10.1002/cbdv.202000335. [DOI] [PubMed] [Google Scholar]
- Hikmawan B. D. Wahyuono S. Setyowati E. P. J. Appl. Pharm. Sci. 2020;10:142–157. [Google Scholar]
- Xu M. Andrews K. T. Birrell G. W. Tran T. L. Camp D. Davis R. A. Quinn R. J. Bioorg. Med. Chem. Lett. 2011;21:846–848. doi: 10.1016/j.bmcl.2010.11.081. [DOI] [PubMed] [Google Scholar]
- Yang X. Davis R. A. Buchanan M. S. Duffy S. Avery V. M. Camp D. Quinn R. J. J. Nat. Prod. 2010;73:985–987. doi: 10.1021/np900834g. [DOI] [PubMed] [Google Scholar]
- Kurimoto S.-i. Ohno T. Hokari R. Ishiyama A. Iwatsuki M. Ōmura S. Kobayashi J. i. Kubota T. Mar. Drugs. 2018;16:463. doi: 10.3390/md16120463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. A. Buchanan M. S. Duffy S. Avery V. M. Charman S. A. Charman W. N. White K. L. Shackleford D. M. Edstein M. D. Andrews K. T. J. Med. Chem. 2012;55:5851–5858. doi: 10.1021/jm3002795. [DOI] [PubMed] [Google Scholar]
- Bilan M. I. Grachev A. A. Shashkov A. S. Kelly M. Sanderson C. J. Nifantiev N. E. Usov A. I. Carbohydr. Res. 2010;345:2038–2047. doi: 10.1016/j.carres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Singh B., Mal G., Gautam S. K. and Mukesh M., in Advances in Animal Biotechnology, Springer, 2019, pp. 429–439 [Google Scholar]
- Fotie J., in Discovery and Development of Therapeutics from Natural Products Against Neglected Tropical Diseases, Elsevier, 2019, pp. 7–47 [Google Scholar]
- Davis R. A. Duffy S. Fletcher S. Avery V. M. Quinn R. J. J. Org. Chem. 2013;78:9608–9613. doi: 10.1021/jo400988y. [DOI] [PubMed] [Google Scholar]
- Avilés E. Rodríguez A. D. Org. Lett. 2010;12:5290–5293. doi: 10.1021/ol102351z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balansa W. Wodi S. I. M. Rieuwpassa F. J. Ijong F. G. Biodiversitas. 2020;21(2):699–706. [Google Scholar]
- Gros E. Al-Mourabit A. Martin M.-T. r. s. Sorres J. Vacelet J. Frederich M. Aknin M. Kashman Y. Gauvin-Bialecki A. J. Nat. Prod. 2014;77:818–823. doi: 10.1021/np4009283. [DOI] [PubMed] [Google Scholar]
- Ludwiczuk A., Skalicka-Woźniak K. and Georgiev M., in Pharmacognosy, Elsevier, 2017, pp. 233–266 [Google Scholar]
- Emsermann J. Kauhl U. Opatz T. Mar. Drugs. 2016;14:16. doi: 10.3390/md14010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achan J. Talisuna A. O. Erhart A. Yeka A. Tibenderana J. K. Baliraine F. N. Rosenthal P. J. D’Alessandro U. Malar. J. 2011;10:144. doi: 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumarna S. Azis M. Sangadji S. S. Idrus I. N. Techno: Jurnal Penelitian. 2019;8:328–333. [Google Scholar]
- Kyei-Baffour K. Davis D. C. Boskovic Z. Kato N. Dai M. Bioorg. Med. Chem. 2020;28:115678. doi: 10.1016/j.bmc.2020.115678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub M. E. Prudhomme J. Ben Mamoun C. Le Roch K. G. Vanderwal C. D. ACS Med. Chem. Lett. 2017;8:355–360. doi: 10.1021/acsmedchemlett.7b00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bideau F. Kousara M. Chen L. Wei L. Dumas F. o. Chem. Rev. 2017;117:6110–6159. doi: 10.1021/acs.chemrev.6b00502. [DOI] [PubMed] [Google Scholar]
- Di Blasio B. Fattorusso E. Magno S. Mayol L. Pedone C. Santacroce C. Sica D. Tetrahedron. 1976;32:473–478. [Google Scholar]
- Fattorusso E. Magno S. Mayol L. Santacroce C. Sica D. Tetrahedron. 1974;30:3911–3913. [Google Scholar]
- Fattorusso E. Magno S. Mayol L. Santacroce C. Sica D. Tetrahedron. 1975;31:269–270. [Google Scholar]
- Manivel N. Shukla S. K. Muthuraman S. J. Mar. Biotechnol. 2020;4:2365–2409. [Google Scholar]
- Avilés E. Prudhomme J. Le Roch K. G. Franzblau S. G. Chandrasena K. Mayer A. M. S. Rodríguez A. D. Bioorg. Med. Chem. Lett. 2015;25:5339–5343. doi: 10.1016/j.bmcl.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves K. Prudhomme J. Le Roch K. G. Franzblau S. G. Rodríguez A. D. Bioorg. Med. Chem. Lett. 2016;26:854–857. doi: 10.1016/j.bmcl.2015.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilrukshi M., Hettiarachi S. and Edirisinghe E., 10th International Research Conference 2017, General Sir John Kotelawala Defence University, 2017, pp. 452–459 [Google Scholar]
- Cabrita M. T. Vale C. Rauter A. P. Mar. Drugs. 2010;8:2301–2317. doi: 10.3390/md8082301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha N. Silakari O. Eur. J. Med. Chem. 2017;134:159–184. doi: 10.1016/j.ejmech.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Ebada S. S. de Voogd N. Kalscheuer R. Müller W. E. Proksch P. Phytochem. Lett. 2017;22:154–158. [Google Scholar]
- White A. M. Pierens G. K. Skinner-Adams T. Andrews K. T. Bernhardt P. V. Krenske E. H. Mollo E. Garson M. J. J. Nat. Prod. 2015;78:1422–1427. doi: 10.1021/acs.jnatprod.5b00354. [DOI] [PubMed] [Google Scholar]
- Wright A. D. McCluskey A. Robertson M. J. MacGregor K. A. Gordon C. P. Guenther J. Org. Biomol. Chem. 2011;9:400–407. doi: 10.1039/c0ob00326c. [DOI] [PubMed] [Google Scholar]
- Avilés E. Prudhomme J. Le Roch K. G. Rodríguez A. D. Tetrahedron. 2015;71:487–494. doi: 10.1016/j.tet.2014.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongaramruong J. Kongkam N. J. Asian Nat. Prod. Res. 2007;9:743–751. doi: 10.1080/10286020701189203. [DOI] [PubMed] [Google Scholar]
- Thao N. P. No J. H. Luyen B. T. T. Yang G. Byun S. Y. Goo J. Kim K. T. Cuong N. X. Nam N. H. Van Minh C. Molecules. 2014;19:7869–7880. doi: 10.3390/molecules19067869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao N. P. Luyen B. T. T. Brun R. Kaiser M. Van Kiem P. Van Minh C. Schmidt T. J. Kang J. S. Kim Y. H. Molecules. 2015;20:12459–12468. doi: 10.3390/molecules200712459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez I. I. Shi Y.-P. García O. J. Rodríguez A. D. Mayer A. M. Sánchez J. A. Ortega-Barria E. González J. J. Nat. Prod. 2004;67:1672–1680. doi: 10.1021/np049802o. [DOI] [PubMed] [Google Scholar]
- Jiao W.-H. Cheng B.-H. Chen G.-D. Shi G.-H. Li J. Hu T.-Y. Lin H.-W. Org. Lett. 2018;20:3092–3095. doi: 10.1021/acs.orglett.8b01148. [DOI] [PubMed] [Google Scholar]
- Liu C.-x. Chin. Herb. Med. 2017;9:101–114. [Google Scholar]
- O’neill P. M. Barton V. E. Ward S. A. Molecules. 2010;15:1705–1721. doi: 10.3390/molecules15031705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. L. Ji Y. B. Yang B. Exp. Ther. Med. 2020;20:630–636. doi: 10.3892/etm.2020.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattorusso E. Taglialatela-Scafati O. Phytochem. Rev. 2010;9:515–524. [Google Scholar]
- Han B.-N., Hong L.-L., Gu B.-B., Sun Y.-T., Wang J., Liu J.-T. and Lin H.-W., in Symbiotic Microbiomes of Coral Reefs Sponges and Corals, Springer, 2019, pp. 329–463 [Google Scholar]
- Triningsih D. W. Tanaka J. Trianto A. Res. J. Biotechnol. 2019;14:126–130. [Google Scholar]
- Murakami N. Kawanishi M. Mostaqul H. M. Li J. Itagaki S. Horii T. Kobayashi M. Bioorg. Med. Chem. Lett. 2003;13:4081–4084. doi: 10.1016/j.bmcl.2003.08.073. [DOI] [PubMed] [Google Scholar]
- Schirmeister T. Oli S. Wu H. Della Sala G. Costantino V. Seo E.-J. Efferth T. Mar. Drugs. 2017;15:63. doi: 10.3390/md15030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnuolo C. Fattorusso E. Romano A. Taglialatela-Scafati O. Basilico N. Parapini S. Taramelli D. Eur. J. Org. Chem. 2005:5077–5083. [Google Scholar]
- Fattorusso E. Parapini S. Campagnuolo C. Basilico N. Taglialatela-Scafati O. Taramelli D. J. Antimicrob. Chemother. 2002;50:883–888. doi: 10.1093/jac/dkg008. [DOI] [PubMed] [Google Scholar]
- Oli S. Abdelmohsen U. R. Hentschel U. Schirmeister T. Mar. Drugs. 2014;12:2614–2622. doi: 10.3390/md12052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar C. Thakur N. Chakraborti S. Hussain S. S. A. Vashisht K. Pandey K. C. Front. Drug Discovery. 2022;2:1065231. [Google Scholar]
- Jiménez M. Garzon S. J. Nat. Prod. 2003;66(5):655–661. doi: 10.1021/np030021h. [DOI] [PubMed] [Google Scholar]
- Fattorusso C. Campiani G. Catalanotti B. Persico M. Basilico N. Parapini S. Taramelli D. Campagnuolo C. Fattorusso E. Romano A. J. Med. Chem. 2006;49:7088–7094. doi: 10.1021/jm060899g. [DOI] [PubMed] [Google Scholar]
- Wang H.-S. Li H.-J. Nan X. Luo Y.-Y. Wu Y.-C. J. Org. Chem. 2017;82:12914–12919. doi: 10.1021/acs.joc.7b02413. [DOI] [PubMed] [Google Scholar]
- Pontius A. Krick A. Kehraus S. Brun R. König G. M. J. Nat. Prod. 2008;71:1579–1584. doi: 10.1021/np800294q. [DOI] [PubMed] [Google Scholar]
- Desoubzdanne D. Marcourt L. Raux R. Chevalley S. Dorin D. Doerig C. Valentin A. Ausseil F. Debitus C. J. Nat. Prod. 2008;71:1189–1192. doi: 10.1021/np8000909. [DOI] [PubMed] [Google Scholar]
- Laurent D. Jullian V. Parenty A. Knibiehler M. Dorin D. Schmitt S. Lozach O. Lebouvier N. Frostin M. Alby F. Bioorg. Med. Chem. 2006;14:4477–4482. doi: 10.1016/j.bmc.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Trisuwan K. Khamthong N. Rukachaisirikul V. Phongpaichit S. Preedanon S. Sakayaroj J. J. Nat. Prod. 2010;73:1507–1511. doi: 10.1021/np100282k. [DOI] [PubMed] [Google Scholar]
- Chinworrungsee M. Kittakoop P. Isaka M. Rungrod A. Tanticharoen M. Thebtaranonth Y. Bioorg. Med. Chem. Lett. 2001;11:1965–1969. doi: 10.1016/s0960-894x(01)00327-4. [DOI] [PubMed] [Google Scholar]
- Kasettrathat C. Ngamrojanavanich N. Wiyakrutta S. Mahidol C. Ruchirawat S. Kittakoop P. Phytochemistry. 2008;69:2621–2626. doi: 10.1016/j.phytochem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Lebouvier N. Jullian V. Desvignes I. Maurel S. Parenty A. Dorin-Semblat D. Doerig C. Sauvain M. Laurent D. Mar. Drugs. 2009;7:640–653. doi: 10.3390/md7040640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M. Acharya D. Ali Bhat Z. Kumar D. Nat. Prod. J. 2017;7:84–96. [Google Scholar]
- Liu X.-F. Shen Y. Yang F. Hamann M. T. Jiao W.-H. Zhang H.-J. Chen W.-S. Lin H.-W. Tetrahedron. 2012;68:4635–4640. doi: 10.1016/j.tet.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epifanio R. d. A. Pinheiro L. S. Alves N. C. J. Braz. Chem. Soc. 2005;16:1367–1371. [Google Scholar]
- Ueoka R. Nakao Y. Kawatsu S. Yaegashi J. Matsumoto Y. Matsunaga S. Furihata K. van Soest R. W. Fusetani N. J. Org. Chem. 2009;74:4203–4207. doi: 10.1021/jo900380f. [DOI] [PubMed] [Google Scholar]
- Alhadrami H. A. Thissera B. Hassan M. H. Behery F. A. Ngwa C. J. Hassan H. M. Pradel G. Abdelmohsen U. R. Rateb M. E. Mar. Drugs. 2021;19:109. doi: 10.3390/md19020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C.-L. Linington R. G. Balunas M. J. Centeno A. Boudreau P. Zhang C. Engene N. Spadafora C. Mutka T. S. Kyle D. E. J. Org. Chem. 2015;80:7849–7855. doi: 10.1021/acs.joc.5b01264. [DOI] [PubMed] [Google Scholar]
- Shao C.-L. Mou X.-F. Cao F. Spadafora C. Glukhov E. Gerwick L. Wang C.-Y. Gerwick W. H. J. Nat. Prod. 2018;81:211–215. doi: 10.1021/acs.jnatprod.7b00917. [DOI] [PubMed] [Google Scholar]
- Tajuddeen N. Van Heerden F. R. Malar. J. 2019;18:1–62. doi: 10.1186/s12936-019-3026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. Siqueira-Neto J. L. Souza J. M. Eribez K. LaMonte G. M. Smith J. E. Gerwick W. H. Molecules. 2020;25:1604. doi: 10.3390/molecules25071604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout E. P. Cervantes S. Prudhomme J. France S. La Clair J. J. Le Roch K. Kubanek J. ChemMedChem. 2011;6:1572. doi: 10.1002/cmdc.201100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.-S. Stout E. P. Prudhomme J. Roch K. L. Fairchild C. R. Franzblau S. G. Aalbersberg W. Hay M. E. Kubanek J. J. Nat. Prod. 2010;73:275–278. doi: 10.1021/np900686w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Mishra A. Puri N. J. Biotechnol. 2017;259:148–159. doi: 10.1016/j.jbiotec.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann C. Sieber S. Wirthensohn S. Blom J. F. Da Silva L. Baudat E. Kaiser M. Brun R. Gademann K. J. Nat. Prod. 2014;77:557–562. doi: 10.1021/np400814w. [DOI] [PubMed] [Google Scholar]
- Portmann C. Blom J. F. Gademann K. Jüttner F. J. Nat. Prod. 2008;71:1193–1196. doi: 10.1021/np800118g. [DOI] [PubMed] [Google Scholar]
- Portmann C. Blom J. F. Kaiser M. Brun R. Jüttner F. Gademann K. J. Nat. Prod. 2008;71:1891–1896. doi: 10.1021/np800409z. [DOI] [PubMed] [Google Scholar]
- Donia M. S. Wang B. Dunbar D. C. Desai P. V. Patny A. Avery M. Hamann M. T. J. Nat. Prod. 2008;71:941–945. doi: 10.1021/np700718p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. T. Phyo M. Y. Molecules. 2020;25:2197. doi: 10.3390/molecules25092197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowruzi B. Sarvari G. Blanco S. InHandb. Algal Sci. Microbiol. Technol. Med. 2020:441–454. [Google Scholar]
- Mi Y. Zhang J. He S. Yan X. Mar. Drugs. 2017;15:132. doi: 10.3390/md15050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engene N. Rottacker E. C. Kaštovský J. Byrum T. Choi H. Ellisman M. H. Komárek J. Gerwick W. H. Int. J. Syst. Evol. Microbiol. 2012;62:1171. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntornchashwej S. Suwanborirux K. Koga K. Isobe M. Chem. – Asian J. 2007;2:114–122. doi: 10.1002/asia.200600219. [DOI] [PubMed] [Google Scholar]
- Saraiva R. G. Huitt-Roehl C. R. Tripathi A. Cheng Y.-Q. Bosch J. Townsend C. A. Dimopoulos G. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-24296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderMolen K. M. McCulloch W. Pearce C. J. Oberlies N. H. J. Antibiot. 2011;64:525–531. doi: 10.1038/ja.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linington R. G. González J. Urena L.-D. Romero L. I. Ortega-Barría E. Gerwick W. H. J. Nat. Prod. 2007;70:397–401. doi: 10.1021/np0605790. [DOI] [PubMed] [Google Scholar]
- Vining O. B. Medina R. A. Mitchell E. A. Videau P. Li D. Serrill J. D. Kelly J. X. Gerwick W. H. Proteau P. J. Ishmael J. E. J. Nat. Prod. 2015;78:413–420. doi: 10.1021/np5007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A. Puddick J. Prinsep M. R. Rottmann M. Chan K. P. Chen D. Y.-K. Tan L. T. Phytochemistry. 2011;72:2369–2375. doi: 10.1016/j.phytochem.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Tripathi A. Puddick J. Prinsep M. R. Rottmann M. Tan L. T. J. Nat. Prod. 2010;73:1810–1814. doi: 10.1021/np100442x. [DOI] [PubMed] [Google Scholar]
- Raju R. Khalil Z. G. Piggott A. M. Blumenthal A. Gardiner D. L. Skinner-Adams T. S. Capon R. J. Org. Lett. 2014;16:1716–1719. doi: 10.1021/ol5003913. [DOI] [PubMed] [Google Scholar]
- Linington R. G. Edwards D. J. Shuman C. F. McPhail K. L. Matainaho T. Gerwick W. H. J. Nat. Prod. 2008;71:22–27. doi: 10.1021/np070280x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Zhang Y. Dong L. Gao Q. Yin L. Quan H. Chen R. Fu X. Lin D. J. Ethnopharmacol. 2018;213:280–301. doi: 10.1016/j.jep.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Cheng K. C.-C. Cao S. Raveh A. MacArthur R. Dranchak P. Chlipala G. Okoneski M. T. Guha R. Eastman R. T. Yuan J. J. Nat. Prod. 2015;78:2411–2422. doi: 10.1021/acs.jnatprod.5b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy T. Guo J. T. Hunt N. H. Payne R. J. Org. Lett. 2010;12:5576–5579. doi: 10.1021/ol1024663. [DOI] [PubMed] [Google Scholar]
- Luesch H. Yoshida W. Y. Moore R. E. Paul V. J. Mooberry S. L. Corbett T. H. J. Nat. Prod. 2002;65:16–20. doi: 10.1021/np010317s. [DOI] [PubMed] [Google Scholar]
- Stolze S. C. Deu E. Kaschani F. Li N. Florea B. I. Richau K. H. Colby T. van der Hoorn R. A. Overkleeft H. S. Bogyo M. Chem. Biol. 2012;19:1546–1555. doi: 10.1016/j.chembiol.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linington R. G. Clark B. R. Trimble E. E. Almanza A. Ureña L.-D. Kyle D. E. Gerwick W. H. J. Nat. Prod. 2009;72:14–17. doi: 10.1021/np8003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons T. L. Engene N. Ureña L. D. Romero L. I. Ortega-Barría E. Gerwick L. Gerwick W. H. J. Nat. Prod. 2008;71:1544–1550. doi: 10.1021/np800110e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze C. J. Navarro G. Ebert D. DeRisi J. Linington R. G. J. Org. Chem. 2015;80:1312–1320. doi: 10.1021/jo5024409. [DOI] [PubMed] [Google Scholar]
- Lin C.-Y. Zhang Y. Wu J.-H. Xie R.-H. Qiao J. Zhao G.-R. Microorganisms. 2020;8:271. doi: 10.3390/microorganisms8020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran V. Ilamathi H. S. Dutt S. Lakshminarayana T. Ghosh P. C. Curr. Top. Med. Chem. 2018;18:1976–1986. doi: 10.2174/1568026619666181129141151. [DOI] [PubMed] [Google Scholar]
- Na M. Meujo D. A. Kevin D. Hamann M. T. Anderson M. Hill R. T. Tetrahedron Lett. 2008;49:6282–6285. doi: 10.1016/j.tetlet.2008.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalado E. L. Tasdemir D. Kaiser M. Cachet N. Amade P. Thomas O. P. J. Nat. Prod. 2010;73:1404–1410. doi: 10.1021/np100348x. [DOI] [PubMed] [Google Scholar]