Abstract
The aerobic anoxygenic phototrophic bacteria are a relatively recently discovered bacterial group. Although taxonomically and phylogenetically heterogeneous, these bacteria share the following distinguishing features: the presence of bacteriochlorophyll a incorporated into reaction center and light-harvesting complexes, low levels of the photosynthetic unit in cells, an abundance of carotenoids, a strong inhibition by light of bacteriochlorophyll synthesis, and the inability to grow photosynthetically under anaerobic conditions. Aerobic anoxygenic phototrophic bacteria are classified in two marine (Erythrobacter and Roseobacter) and six freshwater (Acidiphilium, Erythromicrobium, Erythromonas, Porphyrobacter, Roseococcus, and Sandaracinobacter) genera, which phylogenetically belong to the α-1, α-3, and α-4 subclasses of the class Proteobacteria. Despite this phylogenetic information, the evolution and ancestry of their photosynthetic properties are unclear. We discuss several current proposals for the evolutionary origin of aerobic phototrophic bacteria. The closest phylogenetic relatives of aerobic phototrophic bacteria include facultatively anaerobic purple nonsulfur phototrophic bacteria. Since these two bacterial groups share many properties, yet have significant differences, we compare and contrast their physiology, with an emphasis on morphology and photosynthetic and other metabolic processes.
For decades, it was widely assumed that purple bacterial anoxygenic photosynthesis is fundamentally an anaerobic metabolic process, resulting in growth under illuminated anaerobic conditions (72, 76, 139). The function of the anoxygenic photosynthetic apparatus is the transformation of light energy into an electrochemical gradient of protons across the photosynthetic membrane (PM), which can be used for ATP production, active transport, motility, and other energy-consuming processes. Oxygen partial pressure is the major factor that regulates the formation of the photosynthetic apparatus and the cell differentiation of most facultative purple phototrophic bacteria capable of respiratory and photosynthetic modes of energy transduction (38), although two species which form the photosynthetic apparatus under both aerobic and anaerobic conditions are exceptions to this generalization (65, 124).
However, many representatives of a new physiological group of bacteria that produce bacteriochlorophyll (Bchl) a and carotenoid pigments have been isolated relatively recently and designated aerobic anoxygenic phototrophic bacteria (48, 146, 148, 208, 212, 214–216, 223, 231). The novel aspect of this increasingly large group of bacteria is the inability to use Bchl for anaerobic growth. It is astounding that, of all the species which synthesize Bchl that have been isolated, there is a marked discontinuity in respect to photosynthetic energy transduction. That is, either an isolate grows robustly in a light-dependent fashion under anaerobic conditions (in which case it is thought to be a “typical” anoxygenic phototrophic bacterium), or it is incapable of anaerobic photosynthesis and light stimulates at best a transient enhancement of aerobic growth after a shift from the dark to illumination (in which case it is grouped with the aerobic anoxygenic phototrophic bacteria).
Although the composition of the photosynthetic apparatus and electron transfer carriers (51, 130, 131, 210, 211, 226), as well as the amino acid sequences of the reaction center (RC) and light-harvesting (LH) I polypeptides (104) of aerobic phototrophic bacteria, is similar to those of anaerobic purple phototrophic bacteria, efficient photoinduced electron transfer is operative only under aerobic conditions in the aerobic phototrophic bacteria (51, 131, 226).
For simplicity and clarity, we frequently refer to anoxygenic purple phototrophic bacteria that are capable of anaerobic photosynthesis as anaerobic phototrophic bacteria and to aerobic bacteria that contain Bchl and photosynthetic complexes as aerobic phototrophic bacteria (see “Conclusions and perspectives” for a discussion of trivial nomenclature).
The presence of Bchl a has also been detected in some physiologically distinct bacterial groups such as aerobic methylotrophic bacteria and rhizobia (43, 149, 184, 205). A review on rhizobial photosynthetic bacteria was recently published (see Addendum in Proof). In this article, only nonmethylotrophic and nonsymbiotic species, the so-called “Erythrobacteria” isolated from aquatic environments, are reviewed. Because of the rich history of studies on facultative anaerobic purple nonsulfur phototrophic bacteria, where appropriate this review compares and contrasts metabolic processes and the physiology and phylogeny of this group with those of the aerobic phototrophic bacteria.
GENERAL CHARACTERISTICS OF THE AEROBIC PHOTOTROPHIC BACTERIA
Habitats
The discovery of obligately aerobic bacteria containing Bchl a was first reported by T. Shiba et al. (148), in whose work the isolation and enumeration of these microorganisms on seaweed and in seawater, sand, and bottom sediments of Tokyo Bay and adjacent areas were described. Sixteen strains of aerobic pink or orange bacteria that contained Bchl a were isolated from these aerobic marine environments with a rich medium (148) and were found to be abundant on thalli of Enteromorpha linza and Sargassum horneri and in beach sand. The proportions of these bacteria among the species that formed colonies on the medium employed ranged from 0.9 to 1.1% in the seaweed samples and from 1.2 to 6.3% in the beach sand samples (148).
Subsequent reports broadened the geographical area and ecological niches in which obligately aerobic Bchl a-containing bacteria are found. The presence of aerobic heterotrophic Bchl a-synthesizing strains in high proportions (10 to 30%) of the total heterotrophic bacterial strains cultivated was described for marine environments on the west and east coasts of Australia (146) and at the Pacific Ocean inlet English Bay, in Vancouver, Canada (209).
Investigation of samples taken from freshwater cyanobacterial mats in hot springs of the Bol’shoi River valley (Lake Baykal region) and from Neskuchninskii Spring situated on Southern Kurily in Russia led to the discovery of freshwater strains of obligately aerobic bacteria that synthesize Bchl a (212, 214–216). The isolation and growth of freshwater species were obtained in a rich organic medium (212). The mats from which these isolates came were largely composed of cyanobacteria (e.g., Oscillatoria subcapitata), diatoms, and the purple phototrophic bacteria Thiocapsa roseopersicina and Rhodopseudomonas palustris. The mats were located at the boundary of anaerobic and aerobic zones at alkaline pH values ranging from 8.0 to 9.4 and hydrogen sulfide concentrations from 0.6 to 7.4 mg/liter, depending on the spring (213, 217). The samples of these mats contained up to 106 cells of aerobic bacteria containing Bchl a per ml. Some strains were isolated from environments with considerably hot temperatures: strain KR-99 was isolated from an environment with a temperature of about 40°C and strains RB3 and RB7 came from a site with a temperature of 54°C (214, 215). However, in pure laboratory cultures all of these strains demonstrated typical mesophilic properties and grew optimally at 28 to 30°C (214, 215). Such thermotolerancy has been found for purple nonsulfur bacteria such as R. palustris, Rhodomicrobium vannielii, and Rhodopseudomonas viridis, which have also been detected in high-temperature environments and which have temperature optima of 30 to 35°C in pure culture (21, 57, 58). Why and how these latter species and obligately aerobic species survive in thermal environments are unclear.
Mildly thermophilic (growth temperature optima at 42 to 45°C) species of purple phototrophic bacteria, Rhodospirillum centenum, Rhodopseudomonas cryptolactis, and Rhodopseudomonas strain G1, have been isolated (21). Two moderately thermophilic or thermotolerant aerobic anoxygenic representatives, strain OT3 and strain JF-1, were discovered recently (63, 228). Strain OT3 was isolated from bacterial mats in the brackish Usami hot spring (Japan). The temperature at the sampling site was 42.7°C, the pH was 5.8, and the bacterial mat consisted mainly of a dark green layer of thermophilic filamentous cyanobacteria. The new isolate OT3 grew at temperatures up to 50°C, and optimal growth occurred at 40 to 48°C (63). Aerobic anoxygenic phototrophic strains containing Bchl a were discovered in hydrothermal black smoker plume waters of the Juan de Fuca Ridge in the Pacific Ocean (208). Water samples taken from about 2,000 m beneath the ocean surface were found to contain aerobic bacteria producing Bchl a in numbers of 20 to 40 cells/ml of the samples, about 30% of the pigmented strains that formed colonies on the rich medium used. The representative strain JF-1 revealed a broad tolerance for culture conditions such as salinity, temperature, and pH. Thus, growth was obtained in a freshwater medium and a medium supplemented with 10% NaCl, at temperatures ranging from 5 to 42°C and at pH values of 5.5 to 10.0. Therefore, JF-1 is a salt- and pH-tolerant and thermotolerant strain (222).
Several strains of pelagic bacteria were purified from the surface of a freshwater subtropical pond in Australia (48), and acidophilic heterotrophic bacteria that synthesize Bchl a were isolated from an acidic mine drainage system (190). Aerobic phototrophic bacteria were detected in high numbers relative to the numbers of other heterotrophic strains in the North Adriatic Sea, where they comprised 5 to 55% of the total cells cultivated (103).
The strains isolated from freshwater cyanobacterial mats in Russia are obligately freshwater species. For example, the growth of strain RB16-17 was strongly inhibited by salt concentrations higher than 1% (212). Salt-tolerant strains (T1 through T7) were isolated from a cyanobacterial mat located in the supralittoral zone on the West Frisian island of Texel in The Netherlands and comprised 2 to 23% of the aerobic pigmented strains that formed colonies on the medium used (231). This microbial mat was known to be flooded twice a month by the North Sea. Because of alternating heavy rainfall and evaporation, the salinity of this environment varies from 8 to 10‰ to more than 100‰. The organisms isolated from this mat are able to grow over a broad salinity range, from 5‰ (freshwater) to 96‰ (Table 1). This ability may reflect an adaptation to an environment with fluctuating salinity. Similarly salt-tolerant strains of obligately aerobic Bchl a-containing bacteria (strains 15s.b. and 23s.b.) were isolated from English Bay in Vancouver. The strains were found on the surfaces of seaweeds and sand alternately exposed to air or covered by water during low or high tides, respectively. During summer low tides, the bacterial environment is dried for several hours, consequently presenting an econiche with fluctuating salinity (207). Such salt-tolerant strains can be described as facultatively marine or freshwater organisms.
TABLE 1.
Effect of salinity on doubling time and final yield of the aerobic photosynthetic strain T2 isolated from the microbial mat on Texel
| Salinity (‰) | Doubling time (h) | Final yield (OD660)a |
|---|---|---|
| 5 | 3.78 | 0.370 |
| 15 | 3.37 | 0.410 |
| 51 | 3.97 | 0.374 |
| 78 | 6.09 | 0.346 |
| 87 | 7.07 | 0.300 |
| 96 | 8.80 | 0.200 |
| 141 | No growth | |
| 186 | No growth |
OD660, optical density at 660 nm.
In summary, most strains of aerobic anoxygenic phototrophic bacteria isolated so far inhabit a wide variety of eutrophic aquatic environments and seem to comprise a significant part of the aerobic heterotrophic bacterial population. An exception is strain JF-1, isolated from apparently oligotrophic deep-sea hydrothermal vent plume waters. In spite of the broad geographical distribution of aerobic phototrophic bacteria in different ecological niches and their presence in high numbers, the ecological importance of this group of organisms (their role in microbial populations) has not been studied.
Isolation, Enrichment, and Maintenance
No selective medium has been developed to isolate aerobic phototrophic bacteria, and many nonphototrophic microorganisms grow well on rich organic media solidified with agar. Instead, the color of bacterial colonies caused by the presence of carotenoids has been used as an initial indication of aerobic phototrophic bacteria, which are subsequently screened for the presence of Bchl in absorption spectra.
A wide variety of media rich in organic components such as yeast extract, peptone, Casamino Acids, salts of tricarboxylic acids (TCAs), or sugars have been used to isolate pure cultures of different aerobic phototrophic species (48, 63, 146–148, 189, 208, 212, 230). They can be isolated by direct inoculation of water samples (for free-floating strains) or homogenized mat or sand samples (for the strains found in cyanobacterial communities or on solid surfaces) with dilutions on agar plates of rich organic media (48, 63, 146–148, 189, 208, 212, 230). As a rule, inoculated plates have been incubated in the dark at temperature and pH values similar to those of the environment from which samples were collected. Pigmented colonies are streaked on agar plates to obtain pure isolates. Therefore, pure cultures are easily obtainable. When a pure culture is obtained, a single colony is transferred into liquid medium and cultivated aerobically in the dark. Aerobic phototrophic strains are distinguished from other heterotrophic bacteria by the presence of Bchl a, as indicated by absorption peaks in the region from 800 to 880 nm in cell suspensions or by an absorption peak around 770 nm in acetone-methanol extracts of cells.
It has been found that liquid (taken from late-logarithmic growth phase) and agar surface cultures of most aerobic phototrophic species remain viable after storage at 4°C for at least 2 months (207). Long-term preservation is possible by storage in liquid nitrogen or freezing at −70°C. For this purpose, dense cell suspensions of liquid cultures (mid-logarithmic growth phase) are supplemented with glycerol (30%) as a cryoprotective agent. Lyophilization can also be used as a method of preservation.
Taxonomy and Phylogeny
At present, aerobic phototrophic bacteria are taxonomically classified in the two marine genera Erythrobacter and Roseobacter (144, 147) and the six freshwater genera Erythromicrobium, Roseococcus (214–216, 223, 230), Porphyrobacter (48), Acidiphilium (190), Erythromonas, and Sandaracinobacter (229) (Table 2).
TABLE 2.
Determinative characteristics of aerobic anoxygenic phototrophic genera
| Characteristic | Sandaracinobacter | Erythromonas | Erythromicrobium | Roseococcus | Porphyrobacter | Acidiphilium | Erythrobacter | Roseobacter |
|---|---|---|---|---|---|---|---|---|
| Environment | Freshwater | Freshwater | Freshwater | Freshwater | Freshwater | Freshwater | Marine | Marine |
| Cell shape and size (μm) | Thin, long rods (0.3–0.5 by 1.5–2.5) | Ovoid (0.8–1.0 by 1.3–2.6) | Rods, branched (0.7–1.0 by 1.6–2.5) | Coccoid (0.9–1.3 by 1.3–1.6) | Pleomorphic (0.4–0.8 by 1.1–2.0) | Rods (0.6 by 2.0) | Rods (0.4–0.5 by 1.0–5.0) | Ovoid (0.6–0.9 by 1.0–2.0) |
| Color | Yellow-orange | Orange-brown | Red-orange | Pink-red | Orange-red | Red-pink | Orange | Pink |
| Carotenoid in vivo peaks (nm) | 424, 450, 474 | 430, 458, 485 | 466, 478 | 482, 510, 538 | 464, 491 | 465, 492, 525 | 470 | 510 |
| Bchl a in vivo peaks (nm) | 800, 867 | 800, 867 | 798, 832, 868 | 800, 858 | 799, 869 | 792, 864 | 800, 869 | 806, 868 |
| DNA G+C content (mol%) | 68.5 | 65.4 | 64.2 | 70.4 | 65–66 | 63.2 | 60–64 | 59.6 |
The discovery of obligately aerobic heterotrophs that synthesize Bchl a (148) stimulated research on their taxonomic and phylogenetic positions, to address the questions of their closest relatives and their evolutionary origin. Preliminary analyses of marine strains (144, 145, 147) and strains isolated from freshwater environments (223) used DNA GC content and DNA-DNA hybridization. These analyses showed that purple nonsulfur phototrophic bacteria seem to be the closest relatives. The GC content of DNA of aerobic phototrophic bacteria calculated from thermal melting points ranges from 57 to 60 mol% in Erythrobacter species to 70.4 mol% in Roseococcus thiosulfatophilus (Table 2). Although similar GC composition does not necessarily indicate relatedness, a high GC content is characteristic of DNA purified from purple nonsulfur bacteria, ranging from 59 to 60 mol% for Rhodoferax fermentans to 70 to 72 mol% for Rubrivivax gelatinosus (73). DNA-DNA hybridization in combination with morphological and physiological observations led to proposals for the establishment of four new genera: the marine Erythrobacter and Roseobacter (144, 147) and the freshwater Erythromicrobium and Roseococcus (214–216, 223).
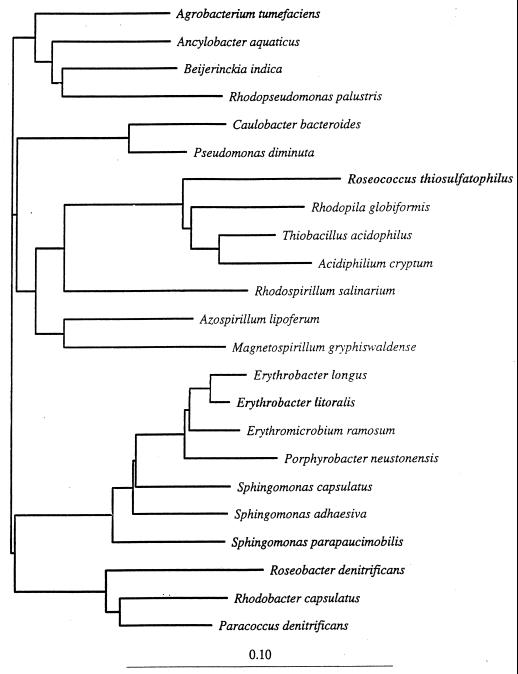
The isolation of new strains, and detailed biochemical, physiological, and molecular biological analyses (e.g., 5S rRNA and 16S ribosomal DNA [rDNA] sequence comparisons) in comparison to existing strains, resulted in the current classification of aerobic phototrophic bacteria and phylogenetic relations with other bacterial taxa (48, 80, 159, 183, 229, 230). Phylogenetically, all aerobic phototrophic species are associated with members of the α subclass of the class Proteobacteria. R. thiosulfatophilus is a member of subclass α-1 and is moderately related to Rhodopila globiformis, Thiobacillus acidophilus, and members of the genus Acidiphilium (Fig. 1). Erythromicrobium, Erythrobacter, and Porphyrobacter are very closely related genera and are clustered in the α-4 subclass, more distant from other aerobic phototrophs. The relatively isolated position of this subgroup in regard to other aerobic phototrophic species and its placement close to the branching point of the α subclass from the other subclasses of the Proteobacteria were shown in a study of Porphyrobacter strains (48) and subsequently (230) in a study of Erythromicrobium species (Fig. 1). The 16S rRNA sequence data placed Roseobacter denitrificans in a branch separate from α-4 and α-1 representatives and in a relatively close phylogenetic relationship with Rhodobacter sphaeroides and Rhodobacter capsulatus (Fig. 1) (48, 80, 159, 229, 230). A phylogenetic study performed on a psychrophilic gas vacuolate bacterium isolated from polar sea ice, Octadecabacter sp., placed this organism as a close nonphototrophic relative of Roseobacter species (59) (Fig. 2). Recently, the nonphotosynthetic Sphingomonas group was included in the α-4 subclass (201), such that the Erythromicrobium-Erythrobacter-Porphyrobacter cluster is most closely related to members of the genus Sphingomonas (230).
FIG. 1.
16S rDNA phylogenetic positions of representatives of the α subclass of the Proteobacteria, as determined by the neighbor-joining method. Scale bar = 10% difference in nucleotide sequences. The total distance between two organisms is the sum of the horizontal branch lengths (230).
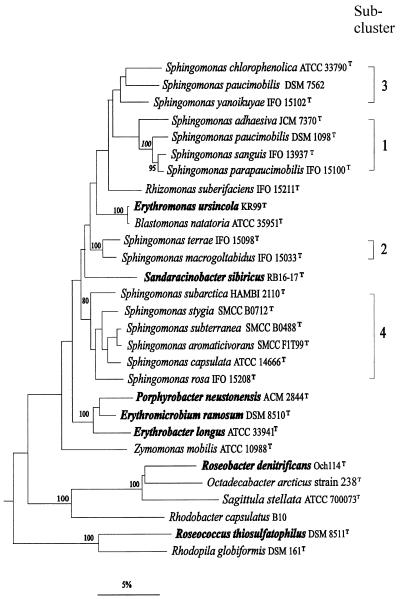
FIG. 2.
16S rDNA dendrogram of relatedness showing the phylogenetic positions of E. ursincola and S. sibiricus released from the genus Erythromicrobium, within the radiation of members of the genus Sphingomonas and related taxa. Numbers refer to bootstrap values, of which only those above 80% are shown. Bar = 5% inferred sequence divergence. (This tree was created by E. Stackebrandt.)
Based on phenotypic similarities, the five isolated strains RB16-17, KR-99, E1, E4(1), and E5 were placed in the same genus, Erythromicrobium, and described as Erythromicrobium sibiricum, Erythromicrobium ursincola, Erythromicrobium ezovicum, Erythromicrobium hydrolyticum, and Erythromicrobium ramosum, respectively. However, DNA-DNA hybridization showed that DNA from the species E. sibiricum and E. ursincola had low homology (11 to 17%) with the other three species of this genus (223). Additionally, an analysis of 5S rRNA sequences indicated a phylogenetic heterogeneity of the Erythromicrobium genus (183). Combination and comparison of new results on the morphology, physiology, biochemistry, molecular biology, and phylogenetic relationships of five Erythromicrobium representatives resulted in the elevation of the tentative species “E. sibiricum” and “E. ursincola” to type species of two new genera: Sandaracinobacter sibiricus and Erythromonas ursincola, respectively (229). These two genera form two separate sublines within the radiation of Sphingomonas species (Fig. 2). The branching point of Erythromonas is between Sphingomonas subclusters 1 and 3 and subcluster 2, while that of Sandaracinobacter is between subclusters 1, 2, 3, and 4. The closest relative of E. ursincola is the nonphotosynthetic Blastomonas natatoria, whereas S. sibiricus stands phylogenetically isolated, with less than 93.5% 16S rDNA sequence identity with any of the reference organisms (Fig. 2) (229).
Phylogenetic 16S rDNA sequence comparisons determined that E. ursincola clusters with B. natatoria (99.8% sequence identity) (229). However, significant physiological differences that exist between E. ursincola and B. natatoria preclude their assignment to the same genus. B. natatoria contains carotenoid pigments but lacks Bchl, whereas E. ursincola is physiologically similar to aerobic anoxygenic phototrophic bacteria because it contains carotenoids and Bchl a. E. ursincola contains Bchl a incorporated into a photochemically active RC and LH complexes and contains electron transfer components of a cyclic photosynthetic pathway (such as a cytochrome [cyt] c bound to the RC, a soluble cyt c2, the RC quinone primary electron acceptor [QA], and the special pair P of the RC). The high 16S rDNA sequence similarity between E. ursincola and B. natatoria indicates a close phylogenetic relationship and a common ancestor. However, due to the existence of significant physiological differences (photosynthesis is a restricted mode of energy generation), they were not designated as members of the same genus (229).
Although 16S rDNA sequences of E. ursincola and B. natatoria had a high level of identity (99.8%), DNA-DNA hybridization of their entire DNA indicated a low level of homology, 40 to 43% (158). This result shows that the taxonomic designation of Erythromonas and Blastomonas as separate genera is correct and that DNA-DNA hybridization analysis should be used in similarly questionable situations.
At present, the taxonomic importance of photosynthetic pigments is controversial. On the one hand, because 16S rRNA sequence analysis indicates close relationships between phototrophic and nonphototrophic bacteria, it has been proposed that a taxonomic rearrangement of phototrophic bacteria which would include phototrophs and nonphototrophs in the same genus should be performed (80). Some authors speculate that, because 16S rRNA differences between phototrophic and nonphototrophic species are very small, this may indicate that independent loss or gain of an essential photosynthesis gene, or cluster coding for the photosynthetic apparatus, occurred in one of two otherwise “identical” species (159). On the other hand, the phylogenetic congruence of most of the genera defined by photosynthetic organisms favors the traditional taxonomic emphasis on this property (74, 159).
In our opinion, the photosynthetic nature of bacteria should be considered a valid and irrefutable taxonomic marker in bacterial classification. Of course, if it is clear that only a small part of the genome, as much as a photosynthesis gene cluster, was lost by a progenitor of a species and that the majority of the genome is still identical, the relationship between two otherwise largely isogenic isolates should be designated taxonomically. The distinction between loss (or gain) of a relatively small part (∼1.3%) of a genome and genuinely large differences in genotype (and phenotype) comes down to an exercise in hair splitting. Because of the almost seamless division between some species, it seems that there will always be a certain element of controversy in the assignment of new isolates which are closely related to previous isolates either by taxonomic or by phylogenetic criteria to specific genera. Because the presence of photosynthetic pigments not only results in colors visible to the eye (the presence of absorption peaks is also readily obtained in absorption spectra) but also usually determines the ability of a species to utilize a restricted mode of energy generation, the presences of Bchl, RC and LH pigment-protein complexes, and related cyclic electron transfer components should remain as valid taxonomic criteria.
In the context of this discussion, we think that the assignment of a recently isolated strain to the genus Roseobacter, designated as Roseobacter algicola (99), is not appropriate. If a major phenotypic difference exists between two strains, they should be in different genera—this is the rule generally followed. The rules of nomenclature require that the description of a new species of an existing genus should correspond to the main genus characteristics previously published in an original article or in Bergey’s Manual of Determinative Bacteriology. Although the genus Roseobacter is described as a genus of the aerobic anoxygenic phototrophic bacteria containing Bchl a and carotenoids, with a photosynthetic apparatus that functions under aerobic conditions (51, 130, 131, 144, 149), the physiologically and morphologically different (as shown in electron micrographs [EMs]) nonphotosynthetic strain, which does not synthesize Bchl or carotenoids, was described as a new member of the genus Roseobacter (99). The main reason given for this unification was the high similarity of 16S rRNA subunit sequences of R. denitrificans to the sequence of the newly isolated strain, although DNA G+C content was not determined. Therefore, we think the designation of “R. algicola” as a Roseobacter species was an inappropriate taxonomic assignment. Although in many cases there is reason to assign two phylogenetically closely related bacteria to the same genus, the two species should not be greatly different with respect to their physiology for determinative and taxonomic purposes. Since there is no standard phylogenetic distance that defines taxonomic ranks, classical taxonomic criteria must be used. Therefore, the new isolate “R. algicola” may be a close phylogenetic neighbor to Roseobacter, but due to the existence of significant physiological differences in comparison to genuine Roseobacter species, this isolate should not be designated as a member of this genus.
In summary, phototrophy is an important and easily recognizable taxonomic marker which should continue to play a significant determinative role in bacterial classification.
Morphological Diversity
Although all aerobic phototrophic bacteria so far examined possess a gram-negative cell wall, their morphologies are very diverse (Table 2; Fig. 3 and 4). Erythrobacter and Sandaracinobacter species are typical rods (very thin in the case of S. sibiricus: 0.3 to 0.5 by 1.5 to 2.5 μm) and produce long chains of up to 10 cells (147, 212, 229, 230) (Fig. 3). The genus Roseococcus contains bacteria with coccoid cells, 0.9 to 1.3 by 1.3 to 1.6 μm in size. The mode of cell division in these three genera is binary fission (147, 212, 214). The cell shape of species in the genera Roseobacter (0.6 to 0.9 by 1.2 to 2.0 μm), Porphyrobacter (0.4 to 0.8 by 1.1 to 2.0 μm), and Erythromonas (0.8 to 1.0 by 1.3 to 2.6 μm) is ovoid rod. Budding in addition to binary division occurs in Porphyrobacter neustonensis and E. ursincola (48, 144, 229). The representatives of Erythromicrobium are very long rods producing characteristic thread-like cells, dividing by symmetric or asymmetric constrictions (Fig. 4). For E. ramosum and E. hydrolyticum, ternary fission and branching were demonstrated (215, 229, 230).
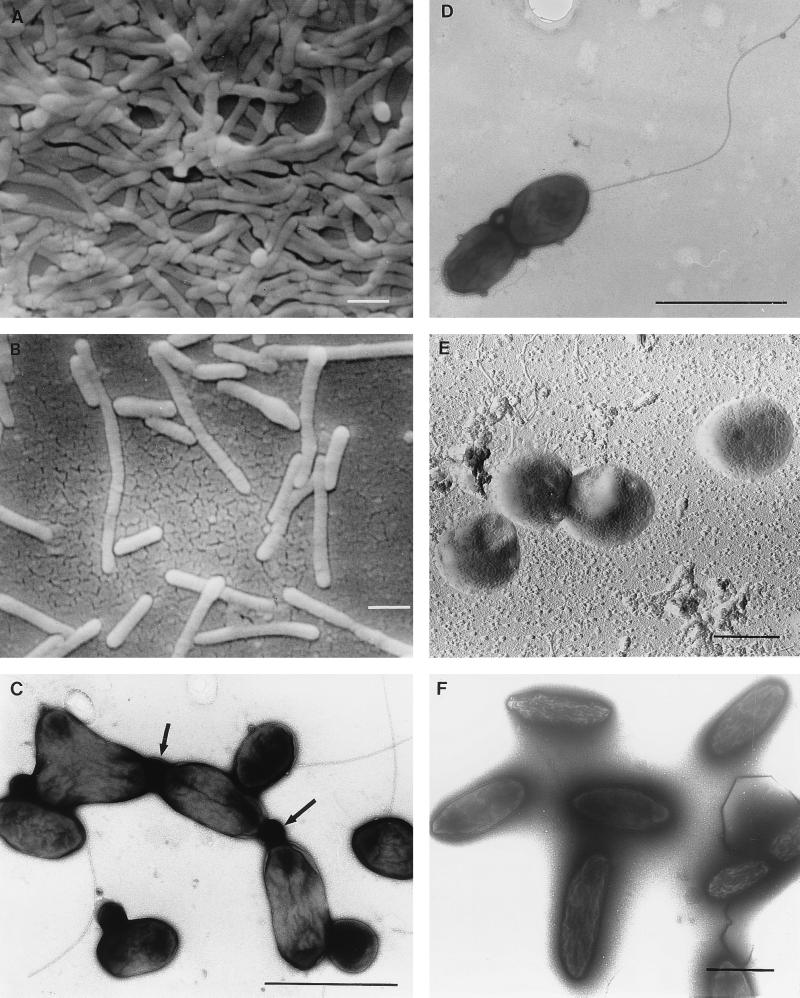
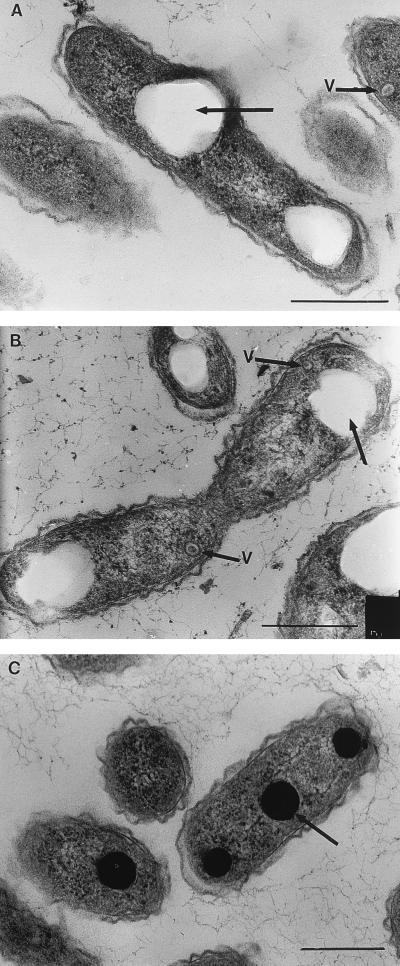
FIG. 3.
EMs showing the morphological diversity of aerobic anoxygenic phototrophic bacteria. (A) Distribution of S. sibiricus cells in a microcolony. (B) Single and thread-like cells of S. sibiricus. (C) Pleomorphic cells of strain JF-1 are connected by membranous material (indicated by arrows). (D) A cell of strain JF-1 containing a single flagellum. (E) Coccus cells of R. thiosulfatophilus. (F) Nonmotile cells of the strain 15s.b. embedded in a capsule-like matrix. (A, B, and E) Scanning EMs of carbon-shadowed cells. (C, D, and F) Transmitting EMs of negatively stained cells. Bars, 1 μm.
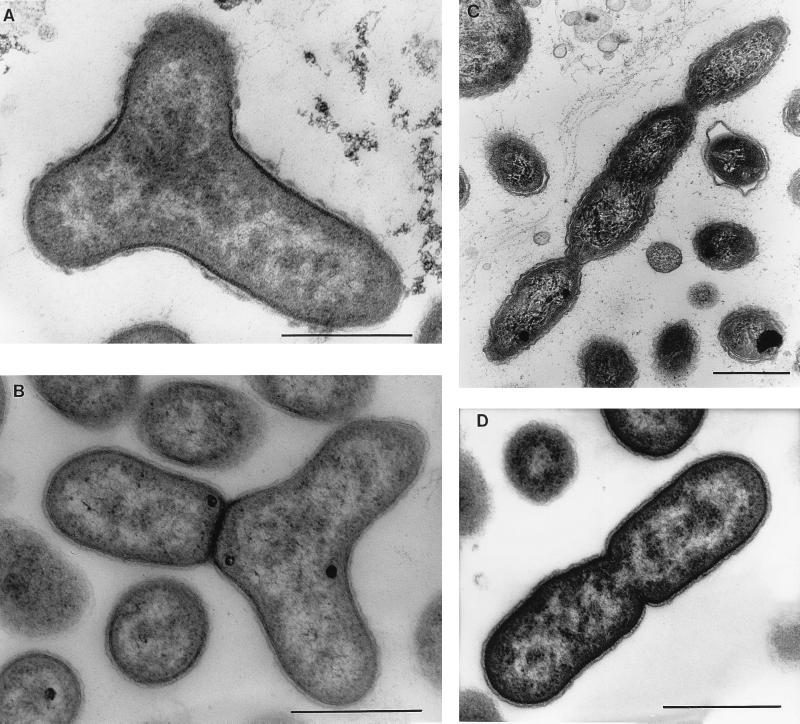
FIG. 4.
Different types of cell division revealed by electron microscopy of thin sections of aerobic anoxygenic phototrophic species. (A) A strain JF-1 Y cell presumably preceding division to form three daughter cells. The nucleoid is seen as light zones of the section, distributed in three directions. (B) A later stage of Y-cell division. One daughter cell is separated by the cell wall from two as-yet-unseparated nascent cells. (C) E. ezovicum dividing by constrictions. (D) Binary division of the strain JF-1. Bars, 0.5 μm.
The new strain (JF-1) of aerobic anoxygenic phototrophic bacteria isolated recently from deep-ocean hydrothermal vent plume waters is unusually pleomorphic. Depending on the age of cultures and composition of liquid medium, the cells can be found as almost coccoid (0.4 to 0.5 by 0.5 to 0.8 μm), as ovoid rods (0.4 to 0.5 by 1.0 to 1.2 μm), as bean shaped, or as thread-like formations of up to five cells. This microorganism is very flexible in its character of cell division, since budding, ternary fission, binary division, and symmetric and asymmetric constrictions were observed. Strain JF-1 forms Y cells, a rare type of bacterial multiplication, which results in the possibility of three daughter cells being produced from one mother cell (Fig. 3 and 4). Cells often remain attached after division, perhaps by a membranous connective material. Therefore, individual cells remain in close contact after division within a free-floating population (Fig. 3) (208).
Most species of aerobic phototrophic bacteria are motile, usually by means of one polar or subpolar flagellum (147, 208, 214–216, 230). R. denitrificans and S. sibiricus (formerly E. sibiricum) have up to three subpolar flagella (144, 212, 229).
The strains 15s.b. and 23s.b., recently discovered at English Bay, Vancouver, Canada (209), and Porphyrobacter tepidarius (63) are the only nonmotile aerobic phototrophic bacteria. The strains 15s.b. and 23s.b. form ovoid cells (0.4 to 0.6 by 1.2 to 1.5 μm) surrounded by capsules and frequently produce a matrix in which cells are embedded (Fig. 3). Therefore, these strains are nonmotile and highly agreggative in liquid culture (209).
In summary, aerobic phototrophic bacteria differ greatly in morphology and in their mode of cell division. However, the diversity of bacterial morphology is not yet exhausted by the group, and the discovery of new strains with vibrio or spirillum morphologies, and conceivably of gram-positive species, is possible.
PIGMENTS AND PHOTOSYNTHETIC PIGMENT-PROTEIN COMPLEXES
Carotenoids and Bacteriochlorophyll
Carotenoids comprise a diverse class of pigments found in photosynthetic and nonphotosynthetic prokaryotic and eukaryotic organisms. The functions of carotenoids in protection from photooxidative damage and in light absorption and as a structural component of the PM in anoxygenic phototrophic bacteria have been reviewed previously (4, 23).
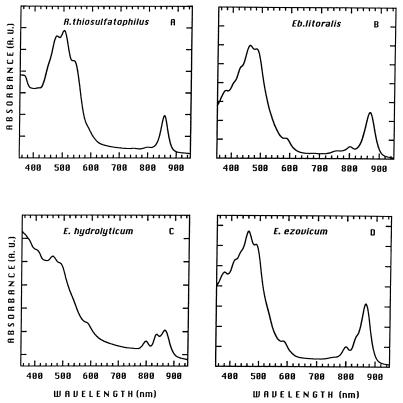
All species of aerobic phototrophic bacteria synthesize large amounts of carotenoid pigments, which determine the color of the organism and give peaks in the blue and green regions (420 to 550 nm) of absorption spectra (Fig. 5) (48, 144, 147, 212, 214–216, 229–231). The carotenoid composition is species specific, often indicating a large number of different carotenoids of unusual chemical structure (167–170, 211).
FIG. 5.
Absorption spectra of membranes isolated from R. thiosulfatophilus (A), E. litoralis (B), E. hydrolyticum (C), and E. ezovicum (D). Cells were cultivated under the same dark-aerobic condition. A. U., absorbance units.
About 20 different carotenoids have been found in the red-orange E. ramosum (207), of which the 10 predominant ones were purified and structurally characterized (Table 3) (211). All of these purified carotenoids were identified as C40 carotenoids, which were classified into four groups: (i) bicyclic carotenoids (β-carotene and hydroxyl derivatives such as zeaxanthin, adonixanthin, caloxanthin, and nostoxanthin), (ii) the monocyclic carotenoid bacteriorubixanthinal, (iii) the acyclic spirilloxanthin, and (iv) the polar carotenoid erythroxanthin sulfate (Fig. 6B). A carotenoid composition similar to that of E. ramosum was described for Erythrobacter longus (168–170) and Erythrobacter litoralis (230), with the exception of adonixanthin and 2,3,2′,3′-tetrahydroxy-β,β-carotene-4-one. Zeaxanthin is a major carotenoid in Erythrobacter, whereas in Erythromicrobium, zeaxanthin is a minor component and bacteriorubixanthinal, erythroxanthin sulfate, and 2,3,2′,3′-tetrahydroxy-β,β-carotene-4-one are the major compounds (211).
TABLE 3.
Carotenoids in E. ramosum (in order of polarity)a
| No. | Carotenoid | Common name | Chemical formula |
|---|---|---|---|
| 1 | β,β-Carotene | β-Carotene | C40H56 |
| 2 | 1,1′-Dimethoxy-3,4,3′4′-tetradehydro-1,2,1′,2′-tetrahydro-ψ,ψ-carotene | Spirilloxanthin | C42H60O2 |
| 3 | 3-Hydroxy-1′-methoxy-3′,4′-didehydro-1′,2′-dehydro-β,ψ-caroten-19′-al | Bacteriorubixanthinal | C41H56O3 |
| 4 | 3,3′-Dihydroxy-β,β-carotene | Zeaxanthin | C40H56O2 |
| 5 | 3,3′-Dihydroxy-β,β-carotene-4-one | Adonixanthin | C40H54O3 |
| 6 | 2,3,3′-Trihydroxy-β,β-carotene | Caloxanthin | C40H56O3 |
| 7 | 3,2′,3′-Trihydroxy-β,β-carotene-4-one (probable structure) | None | C40H54O4 |
| 8 | 2,3,2′,3′-Tetrahydroxy-β,β-carotene | Nostoxanthin | C40H56O4 |
| 9 | 2,3,2′,3′-Tetrahydroxy-β,β-carotene-4-one | None | C40H54O5 |
| 10 | 3,2′,3′-Trihydroxy-β,β-carotene-4-one-3-sulfate | Erythroxanthin sulfate | C40H54O7S |
Modified from reference 211.
FIG. 6.
Chemical structures of highly polar carotenoids (211). (A) C30 carotenoid (4,4′-diapocarotene-4,4′-dioate) (compound I) and the corresponding diglucosyl ester (compound II) of R. thiosulfatophilus. (B) Erythroxanthin sulfate found in cells of E. ramosum, E. longus, and E. litoralis.
Bicyclic carotenoids such as β-carotene and its hydroxyl derivatives were found in Erythrobacter and Erythromicrobium species, and the color of Erythromonas and Sandaracinobacter indicates that these bacteria also contain carotene carotenoids, which are rarely present in purple phototrophic bacteria (small amounts of β-carotene were detected in R. vannielii [17, 136]). Zeaxanthin and β-carotene are widely distributed among oxygenic phototrophs, green plants, cyanobacteria and algae, and some strains of Flavobacterium (90). The highly polar carotenoid sulfates have hitherto been found exclusively in the carotenoids of the aerobic phototrophic bacteria (168, 211, 230) and were recently described as carotenoids of novel structures (168).
The carotenoid composition of two other Erythromicrobium representatives, E. ezovicum and E. hydrolyticum, has not yet been analyzed in detail. Nevertheless, in vivo absorption spectra revealed carotenoid absorption peaks at 466 and 478 nm, as in E. ramosum, indicating similar carotenoid compositions of these species, also apparent in the color of liquid cultures (intensely red-orange) (Table 2) (229).
The carotenoid composition of the pink-red Roseococcus species is not as rich as those of Erythrobacter and Erythromicrobium species but nevertheless is unusual. R. thiosulfatophilus contains mainly two very polar red pigments, C30 carotene-dioate (4,4′-diapocarotene-4,4′-dioate) and the respective diglucosyl ester (di[β-d-glucopyranosyl]-4,4′-diapocarotene-4,4′-dioate) (Fig. 6A). Together, they contribute 95% of the total carotenoid content (211). Such highly polar C30 carotenoid glycosides have never before been observed in purple phototrophic bacteria, although the same carotenoid and its diglucosylated form have previously been postulated to exist in Methylobacterium rhodinum (formerly Pseudomonas rhodos) (84).
The most abundant carotenoid species detected in Roseobacter sp. is spheroidenone, which is the major carotenoid of anaerobic purple bacteria such as Rhodobacter species (68, 144, 167).
The only carotenoid of Acidiphilium rubrum is spirilloxanthin, which is found in Rhodospirillum rubrum and several other purple phototrophic bacteria (191).
Bchls have long-lived excited states and appropriate oxidation-reduction potentials that are suited to their function in LH energy transfer and electron transfer reactions. The only Bchl found in aerobic phototrophic bacteria thus far is Bchl a, on the basis of in vivo absorption spectra of intact cells and from organic solvent extracts. Since Bchl a is incorporated into species-specific types of pigment-protein complexes (see “Light-harvesting systems” and “Reaction center”), the corresponding in vivo absorption peaks are in the near-infrared region from about 800 to 870 nm (Fig. 5). Upon extraction with organic solvent, a far-red absorption peak at about 770 nm is obtained, as well as a peak at 370 to 390 nm (48, 63, 144, 145, 147, 148, 190, 212, 214–216, 228–231), consistent with the identity of this pigment as Bchl a.
Typically, cells of aerobic phototrophic bacteria contain small amounts of Bchl relative to the abundance of carotenoids (compared to anaerobic purple phototrophic bacteria). For example, the anaerobic phototrophic bacterium R. sphaeroides may yield about 20 nmol of Bchl/mg (dry weight) of cells, whereas the Bchl content of obligately aerobic species was found to be as follows: E. longus, 2.0 nmol/mg (dry weight) of cells; S. sibiricus and E. hydrolyticum, 1.0 to 4.0 nmol/mg of protein; A. rubrum, 0.7 nmol/mg (dry weight) of cells; R. thiosulfatophilus, 0.1 to 1.0 nmol/mg of protein. Therefore, the ratio of Bchl to carotenoid peaks in whole cells of aerobic phototrophic species is typically about 1:8 to 1:10 (66, 149, 177, 190, 206, 214, 216, 230, 232). (However, see “The influence of light on growth and pigment formation” and “Effect of oxygen on growth and pigment synthesis” for a discussion of oxygen and light effects on Bchl content of cells.)
Bchl a purified from E. longus, R. denitrificans, and Acidiphilium species was found to be Bchl ap, which contains phytol as the esterifying alcohol, the most common Bchl a form found in purple phototrophic bacteria (66, 70, 90, 97, 190). No Bchl a esterified to geranylgeraniol, as it is in the anaerobic phototrophic bacterium R. rubrum (19), has been detected in aerobic phototrophic species. However, the ester moiety of Bchl from most species of Erythromicrobium, Erythrobacter, Roseococcus, Sandaracinobacter, and Erythromonas has not yet been determined.
Until recently, all natural chlorophylls were thought to be porphyrin derivatives containing a magnesium atom at the center of a chlorin macrocyclic ring. Among the semisynthetic chlorophyll derivatives containing metals other than Mg, only Zn-containing chlorophylls have photochemical properties comparable to those of Mg-chlorophylls (195). Zn-containing Bchl a was introduced artificially into isolated antenna or RC proteins to replace Mg-Bchl a (122, 142), but until recently, natural photosynthesis without Mg-chlorophylls was unknown. However, a natural Zn-containing Bchl a was discovered in the aerobic acidophilic bacterium A. rubrum (191). This Zn-containing Bchl a is esterified with phytol (Zn-Bchl ap). Chemical analysis of A. rubrum cell extracts yielded a 13:2:1 molar ratio of Zn-Bchl to Mg-Bchl to bacteriopheophytin, and most of these pigments were determined to be photochemically active (191).
In summary, the aerobic phototrophic bacteria contain an unusually diverse variety of carotenoids, but at present Bchl a (containing either Mg or Zn) is the only chlorophyll that has been found. The discovery of Zn-Bchl in A. rubrum raises the possibility that other Bchls or novel chlorins might exist in species that have not yet been carefully analyzed or discovered.
Development of Photosynthetic Membranes
Most of the anaerobic purple phototrophic bacteria have, in addition to the cytoplasmic membrane (CM), an intracytoplasmic membrane (ICM) system, of species-specific morphology (38, 129). It is thought that the ICM is derived from and is contiguous with the CM. The ICM may form vesicles, tubules, or thylakoid-like sheets (38). The pigment-protein complexes of the photosynthetic apparatus of anaerobic phototrophic bacteria are incorporated into the ICM or, in a few cases, seem to be located in the CM (37, 192, 193). In most anaerobic phototrophic bacteria, light and oxygen tension regulate the formation of the ICM, such that ICM formation is induced when the oxygen tension is lowered and the most extensive ICM development occurs during anaerobic growth.
Aerobic phototrophic bacteria contain significantly lower amounts of Bchl than do typical anaerobic phototrophic bacteria (see “Carotenoids and bacteriochlorophyll”). As noted above, Bchl content commonly differs between anaerobic phototrophic and aerobic phototrophic bacteria by factors of 10 to 20. With such a low number of photosynthetic units (RC plus LH system), the absence of an extensive ICM system in obligately aerobic species is not surprising. An ICM system was not detected in thin-section EMs of Erythrobacter, Erythromicrobium, Roseococcus, Porphyrobacter, Erythromonas, or strain JF-1 (48, 63, 208, 214–216, 218, 229, 230). Occasionally, chromatophore-like vesicle structures have been observed in R. denitrificans (69, 149). Thin-section EMs of S. sibiricus (renamed E. sibiricum) revealed rare vesicular or loop-like CM invaginations (225).
Two kinds of intracellular membrane fragments produced by French press disruption of cells were designated in R. thiosulfatophilus and E. ramosum (Table 4) (210, 211), on the basis of two distinct membrane fractions separated in sucrose gradients. Both fractions isolated from R. thiosulfatophilus and E. ramosum were free of peptidoglycan and active in NADH dehydrogenase, confirming the CM nature of these fractions. One fraction banded at a sucrose concentration of 1.0 to 1.2 M (fraction I), and a second fraction banded at 1.2 to 1.5 M (fraction II). The RC and LH complexes were located mainly in the 1.0 to 1.2 M fraction of R. thiosulfatophilus and exclusively in the 1.2 to 1.5 M fraction of E. ramosum, whereas carotenoids were found in both fractions (211). The ratio of Bchl to carotenoids detected in whole cells (1:9 in both species) decreased to 1:4 in purified PMs of R. thiosulfatophilus and to 1:7 in the E. ramosum PMs. Most of the carotenoids were not bound to the PM but were located in cell wall and other peripheral membrane fractions (Table 4) (211). Membrane preparations from cell suspensions of Erythrobacter, Erythromicrobium, Erythromonas, and Sandaracinobacter species also gave rise to two membrane fractions in sucrose gradients (233). It is not clear how the two fractions relate to CM or ICM differentiation, but the data indicate a discontinuous organization of membranes in these species.
TABLE 4.
Enzyme activities, muramic acid, diaminopimelic acid, Bchl, and carotenoid content of membrane fractions purified by sucrose density gradient centrifugationa
| Sp. and membrane fractionb | Activity (μmol/min/mg of protein)
|
Content (nmol/mg of protein)
|
||||
|---|---|---|---|---|---|---|
| NADH-dehydrogenase | NADH oxidase | DAP | Mu | Bchl | Car | |
| R. thiosulfatophilus | ||||||
| I | 0.7 | 0.05 | 0 | 0 | 1.3 | 5.4 |
| II | 0.26 | 0.03 | 0 | 0 | 1.1 | 11.4 |
| III (pellet) | 0.01 | 0.07 | 0.2 | 0.1 | 0.2 | 5.9 |
| E. ramosum | ||||||
| I | 0.4 | 0.005 | 0 | 0 | 0.04 | 0.64 |
| II | 0.17 | 0.02 | 0 | 0 | 1.5 | 10.8 |
| III (pellet) | 0.006 | 0.005 | 0.2 | 0.1 | 0.25 | 4.6 |
Abbreviations: Mu, muramic acid; DAP, diaminopimelic acid; Car, carotenoid.
For R. thiosulfatophilus, fraction I was isolated from 1.0 M sucrose layer and fraction II was isolated from 1.5 M sucrose layer. For E. ramosum, fraction I was isolated from 1.0 M sucrose layer and fraction II was isolated from 1.2 M sucrose layer.
In conclusion, it is evident that the photosynthetic apparatus of aerobic phototrophic bacteria is present in lower amounts than that of typical anaerobic phototrophic bacteria and probably is located mainly in the CM or ICM structures that are not visible in thin-section EMs of cells.
Light-Harvesting Systems
The major light-absorbing pigments in anaerobic phototrophic bacteria are Bchl and carotenoids. These pigments are noncovalently attached to two types of integral membrane proteins, forming on the one hand the photochemical RC and on the other hand the LH (antenna) complexes (24). Purple bacteria have a relatively simple LH system consisting of a core antenna protein complex (LHI) closely associated with the RC and, in many species, one or more peripheral antenna complexes (LHII), all of which are located in the ICM (236). The LH complexes absorb light quanta, and the energy migrates through the pigments of the antenna system to the RC (24). The LH complexes are sometimes designated by their long-wavelength light absorption maxima, for example, B870 (LHI) and B800-850 (LHII) in R. capsulatus.
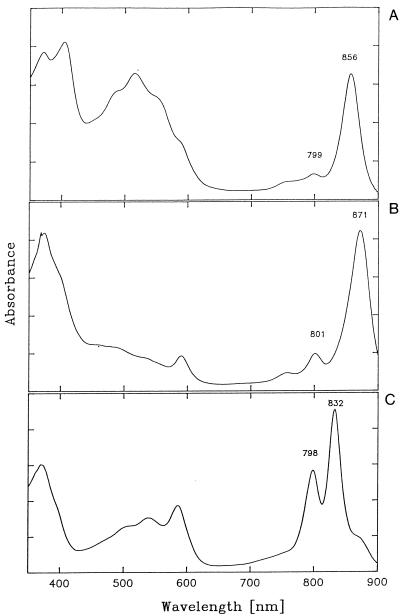
The isolation and characterization of new aerobic phototrophic species have led to the discovery of LH complexes with unusual absorption maxima. For example, in vivo absorption spectra of R. thiosulfatophilus cells yielded a major peak of Bchl a at 859 nm (214), and spectra of E. ramosum yielded two major peaks at 836 and 871 nm (215). Subsequent purification of LH complexes from these bacteria by detergent treatment of membranes and sucrose density gradient centrifugation revealed the existence of an R. thiosulfatophilus LHI complex with an absorption peak at 856 nm and an E. ramosum LHII complex with peaks at 798 and 832 nm (Fig. 7) (210). E. ramosum additionally contains an LHI complex with absorption characteristics (maximum at 871 nm) similar to those measured in many anaerobic phototrophic bacteria and other aerobic phototrophic bacteria.
FIG. 7.
Absorption spectra of isolated pigment-protein complexes recorded at room temperature (210). (A) LHI-RC complex of R. thiosulfatophilus. (B) E. ramosum LHI-RC. (C) LHII complex of E. ramosum.
The unusual absorption properties of the LHI (B856) complex isolated from R. thiosulfatophilus together with the polypeptide pattern (four polypeptides; two of about 8.0 kDa and two of about 7.0 kDa) indicated an unusual protein environment of Bchl. Preliminary Raman spectroscopy analysis of the R. thiosulfatophilus blue-shifted RC-B856 core complex suggested that the presence of free 2-acetyl carbonyl groups of Bchl may be responsible for the blueshifting in absorbancy (50). However, a blueshift of LHI Bchl a absorbancy in whole cells could be due to the presence of Zn-Bchl instead of Mg-Bchl, giving a blueshift of 5 to 15 nm, as reported for A. rubrum (191) (see “Carotenoids and bacteriochlorophyll”). Although this blueshift could result from a complex, simultaneous effect of several factors, the possibility of the presence of Zn-Bchl in R. thiosulfatophilus should be investigated.
The LHII complex (B798-832) of E. ramosum is composed of three polypeptides of about 16.0, 9.0, and 8.0 kDa (210). Three proteins, designated α, β, and γ, with molecular weights of 14,000, 7,000, and 5,000, were previously found to copurify with the LHII (B800-850) complex from the purple nonsulfur bacterium R. capsulatus (36, 155, 165). According to preliminary Raman spectroscopy results, the absorption features of the E. ramosum B798-832 LHII complex indicate the presence of H bonds to the 2-acetyl substituents of both Bchl molecules, which would explain the most redshifted electronic transition (36, 155, 165).
Because of the unusual properties of LH complexes of aerobic phototrophic bacteria, it would be of interest to determine the amino acid sequences of these antenna polypeptides. It also would be interesting to determine if E. ramosum forms a variety of LHII complexes dependent on temperature, oxygen, and light conditions, as do the anaerobic phototrophic bacteria Rhodopseudomonas acidophila and R. palustris (12, 42, 54, 166).
In a recent investigation, E. hydrolyticum and E. ezovicum were found to possess LHI and LHII complexes with absorption characteristics similar to those of E. ramosum (229) (Table 5).
TABLE 5.
Comparative data on the photosynthetic apparatus and electron carriersa
| Sp. | RC | Absorption peak(s) (nm)
|
RC-bound cyt c (molecular mass in kDa) | No. and molecular mass (kDa) of soluble cyt c | No. and molecular mass (kDa) of membrane-bound cyt c | Ubiquinone (mmol/g of dry cells)
|
||
|---|---|---|---|---|---|---|---|---|
| LHI | LHII | Q9 | Q10 | |||||
| Sandaracinobacter sibiricus | + | 867 | Absent | 37.0 | 1 (14.0) | 2 (30.0, 37.0) | 0.06 | 0.71 |
| Erythromonas ursincola | + | 867 | Absent | 40.0 | 3 (6.5, 9.0, 14.0) | 4 (14.3, 21.0, 24.0, 40.0) | ND | 0.11 |
| Roseococcus thiosulfatophilus | + | 856 | Absent | 44.0 | 2 (4.0, 6.5) | 4 (21.5, 23.0, 26.0, 44.0) | NA | NA |
| Roseobacter denitrificans | + | 870 | 806 | 42.0 | 2 (13.5, 14.5) | NA | ND | + |
| Erythromicrobium ramosum | + | 868 | 798, 832 | Absent | 2 (8.0, 14.3) | 3 (8.0, 26.0, 30.0) | 0.09 | 0.19M |
| Erythromicrobium ezovicum | + | 868 | 800, 832 | Absent | 2 (8.0 14.3) | 2 (30.0, 34.0) | 0.02 | 0.3 |
| Erythromicrobium hydrolyticum | + | 866 | 799, 833 | Absent | 1 (14.3) | 2 (21.0, 30.0) | 0.02 | 0.01M |
| Erythrobacter litoralis | + | 868 | Absent | Absent | 4 (14.0, 21.5, 24.0, 26.0) | 2 (30.0, 35.0) | NA | NA |
| Erythrobacter longus | + | 870 | Absent | Absent | 2 (12.5, 17.0) | NA | ND | + |
The data for P. neustonensis and A. rubrum are not available. Species were cultivated under dark-aerobic conditions. Symbols and abbreviations: +, present; ND, not detected; NA, data not available; M, in addition to ubiquinone Q10 the methylated form was revealed.
R. denitrificans has a unique type of antenna in addition to the RC-LHI core (RC-B870). This complex exhibits one peak at 806 nm, is composed of two polypeptides (5.0 and 7.0 kDa), and is considered to be a peripheral antenna (LHII), since detergent-sucrose gradient-purified preparations that lacked RC showed a Raman spectrum similar to those of LHII complexes of purple bacteria and transferred energy to the RC-LHI complex with high efficiency (145, 150, 151, 153).
The LHI-RC core complex has been purified from E. longus (151), E. litoralis (233), E. ursincola, and S. sibiricus (229) (Table 5). These LHI-RC core complexes are similar to analogous core complexes of anaerobic phototrophic bacteria on the basis of absorption spectroscopy. A. rubrum produces only an LHI complex with absorption characteristics similar to those of the corresponding complexes of anaerobic phototrophic bacteria. The main difference is a blue-shifted absorption maximum of the main peak in the near-infrared region due to the presence of Zn-Bchl a in this complex (152, 191). The absorption spectra of intact cells or membranes of Porphyrobacter strains indicated the absence of LHII complexes, although pigment-protein complexes have not yet been purified from these bacteria (48, 190).
In summary, the LH systems of aerobic phototrophic bacteria are very diverse in regard to their light absorption properties and frequently differ from analogous complexes found in anaerobic phototrophic bacteria. However, the major principles of LH system organization seem to be similar between these two groups.
Reaction Center
The photosynthetic RC is defined as the minimal functional unit that catalyzes light-induced electron transfer processes, leading to a stable charge separation, and, as C. R. D. Lancaster and H. Michel (100) nicely wrote, “their function lies at the heart of the photosynthetic process of converting solar energy into biochemically amenable energy” (for reviews, see references 41, 44, 100, and 133). The purple bacterial anoxygenic RC is an integral membrane pigment-protein complex that contains three protein subunits (L, M, and H) and the following cofactors: four Bchls, two bacteriopheophytins, two quinones, and one nonheme high-spin Fe2+ (100). The RC of anaerobic phototrophic bacteria is at present the structurally and functionally best-characterized membrane protein complex. High-resolution three-dimensional structures have been determined by X-ray crystallography for the RC of R. viridis and R. sphaeroides (160), and structure-function questions have been addressed by site-directed mutagenesis (200).
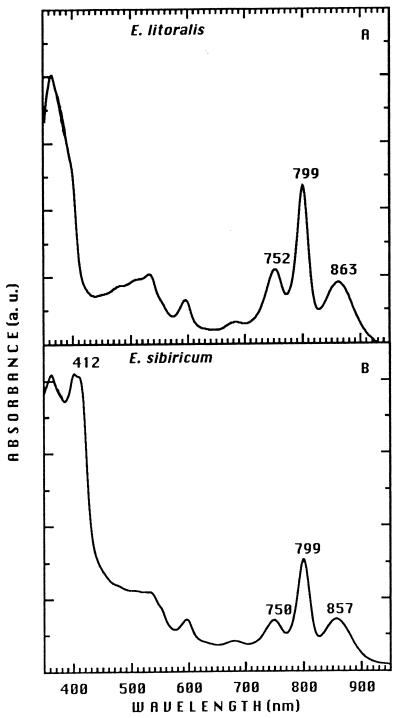
The presence of a functional RC in aerobic phototrophic bacteria was first shown for R. denitrificans and E. longus on the basis of light-induced absorption changes (69), and a purified RC preparation from R. denitrificans was described previously (175). The purification of an RC from Erythrobacter, Erythromicrobium, Sandaracinobacter, and Roseococcus species has been difficult. Explorations of modified techniques allowed successful RC purification from E. litoralis T4, E. ursincola KR99, and S. sibiricus RB16-17 (renamed E. sibiricum) (Fig. 8). The content and overall organization of chromophores in these preparations, as determined by linear dichroism analysis, appear to be very similar to those of anaerobic phototrophic bacteria (224).
FIG. 8.
Absorption spectra of the RC purified from E. litoralis (A) and S. sibiricus (renamed E. sibiricum) (B). a. u., absorbance units.
An RC preparation purified from E. litoralis did not contain a tightly bound cyt, whereas the RCs of S. sibiricus and E. ursincola possess tetraheme cyts c. Each of the tetraheme cyts contains two high-potential hemes (+330 and +305 mV for E. ursincola and +380 and +300 mV for S. sibiricus) and two low-potential hemes (+40 and −40 mV for E. ursincola and +30 and −40 mV for S. sibiricus) (224).
An RC preparation containing a bound cyt c was isolated from R. denitrificans, and absorption spectra of reduced and oxidized forms of the RC were similar to those of the RC of R. sphaeroides except for the contributions of cyt c and carotenoids (175). The cofactors of R. denitrificans RC were the same as those of the RC of anaerobic purple bacteria and contained the following numbers of molecules (per RC): four of Bchl, two of bacteriopheophytin, four of cyt c554, and two of ubiquinone-10 and carotenoid(s); the cofactors also contained four different polypeptides of 26, 30, 32, and 42 kDa. The 42-kDa protein corresponds to tetraheme cyt c (175). The heme which possesses the highest redox midpoint potential (+290 mV, designated H1) has an α band absorption of 555 nm. The second high-potential heme (+240 mV [H2]) exhibits an α peak at 554 nm. The two low-potential hemes, L1 and L2, had similar high redox midpoint potentials (about +90 mV), with α bands at 553 and 550 nm, respectively (51). The values of the midpoint potentials of hemes H1 and H2 are lower than those determined for anaerobic purple phototrophic bacteria (+370 and +320 mV for R. viridis [188]); +350 and +320 mV for Chromatium vinosum [39]). These values are also lower than the values calculated for the H1 and H2 hemes of the RC-bound cyt in the obligately aerobic species E. ursincola and S. sibiricus (see above). Furthermore, the values of the low-potential hemes are relatively high (+90 mV). Similarly high midpoint potentials were measured in R. gelatinosus (39) and Chloroflexus aurantiacus (47, 186).
The RC of anaerobic phototrophic bacteria is thought to be surrounded by a ring-shaped LHI complex at a fixed stoichiometry relative to the RC (24). A constant number of about 30 LHI Bchl molecules per RC has been measured for seven aerobic phototrophic species (233). These results are in good accordance with data reported for anaerobic phototrophic bacteria (1, 32, 46) and suggest a similar overall organization of the photosynthetic apparatus in aerobic and anaerobic phototrophic bacteria. Nevertheless, the LH complexes of some aerobic phototrophic bacteria have unusual spectral properties presumably due to different protein environments of the Bchl (see “Light-harvesting systems”).
ELECTRON TRANSFER SYSTEM AND PHOTOSYNTHESIS
Quinones
Quinones are found in many bacteria, plants, and animals (9). The characteristic feature of quinones is their function as redox carriers in electron transport within membranes in metabolic processes such as aerobic and anaerobic respiration and photosynthesis (79, 187, 235). The composition of quinones was shown to vary among different representatives of phototrophs. Some species of phototrophic bacteria contain only ubiquinone Q10, whereas other species contain Q8 or Q9 as well as the menaquinone MK8 or MK9 (25, 75).
Most studies of quinone function in phototrophic bacteria have been conducted on anaerobic phototrophic bacteria, and very little is known about the structure, function, and synthesis of quinones in aerobic phototrophic bacteria. Freshwater species of aerobic phototrophs from the genera Erythromicrobium, Sandaracinobacter, Erythromonas, and Acidiphilium (56, 189) and marine species of Erythrobacter and Roseobacter (144, 147, 149) possess Q10 as the major quinone species. No menaquinones or rhodoquinones have been found in these species. The ubiquinone Q9 was detected as a minor quinone in addition to Q10 in S. sibiricum, E. ezovicum, and E. ramosum. However, in E. hydrolyticum the content of Q9 was twice as high as that of Q10 (Table 5). The quinone Q10 of E. hydrolyticum and E. ramosum seems to exist as a methylated form (56). The total average amount of ubiquinones in obligately aerobic species (0.02 and 0.7 μmol/g of dry cells in E. hydrolyticum and S. sibiricus, respectively) is much lower than that determined for anaerobic purple bacteria (2 to 4 μmol/g of dry cells in Rhodobacter species) (56). No effect of light on the quinone composition and content of obligately aerobic freshwater species was detected (56).
Cytochrome Composition
cyts are found in most organisms that carry out electron transport through membrane-bound chains of carriers, regardless of the ultimate oxidant. Thus, cyts are not only present in chloroplasts and mitochondrial and aerobic bacterial respiratory chains but are also found in facultative anaerobes, obligate anaerobes, facultative photoheterotrophs, and the cyanobacteria (60, 93, 117, 127, 187, 235).
A fundamental process in the transformation of light into chemical energy in anaerobic phototrophic bacteria is a cyclic series of electron transfer reactions linked to the transport of protons across a membrane. This process involves two highly conserved integral membrane multisubunit complexes, the photosynthetic RC (see “Reaction center”) and the cyt bc1 complex. Two diffusible components, ubiquinone in the hydrophobic domain of the membrane and cyt c2 in the periplasmic space, usually connect these two transmembrane complexes on the acceptor and donor sides of the RC, respectively (30, 139). Electron transfer from the bc1 complex to the RC may be mediated by more than one type of mobile cyt c in R. sphaeroides, R. capsulatus, R. rubrum, and R. palustris (117). In most species of anaerobic phototrophic bacteria studied, however, a cyt c does not directly reduce the photooxideized special pair (P+) of RC but instead donates an electron to a multiheme cyt bound to the RC, which is the immediate electron donor to P+ (127). The photosynthetic electron transfer system shares some carriers, such as mobile cyts c, quinone molecules, and the cyt bc1 complex, with the respiratory electron transfer system (60).
Analyses of light-induced difference spectra in the presence of oxygen in whole cells of Erythromicrobium, Sandaracinobacter, Roseobacter, Roseococcus, Erythromonas, and Erythrobacter species, as well as redox titrations and gel electrophoresis of soluble, membrane, and LHI-RC purified fractions, resulted in the distinction of two different groups on the basis of electron transfer properties and cyt compositions (Table 5). The first group contains R. denitrificans, R. thiosulfatophilus, E. ursincola, and S. sibiricus (175, 226). These species possess a cyt c tightly bound to the RC that serves as the immediate electron donor to the photooxidized RC. On the basis of sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the molecular masses of these cyt species are 42, 44, 37, and 40 kDa in R. denitrificans, R. thiosulfatophilus, S. sibiricus, and E. ursincola, respectively, which is similar to that of the analogous tetraheme cyt c of anaerobic phototrophic bacteria (175, 224, 226, 227). The second group includes E. longus, E. litoralis, E. ramosum, E. hydrolyticum, and E. ezovicum, which contain an RC that is directly reduced by a soluble cyt c, apparently a cyt c2 (172, 226, 229). The RC of A. rubrum seems to contain tetraheme bound cyt c, which is readily detached in vitro (77).
A comparison of the soluble and membrane-bound cyt c populations of aerobic phototrophic bacteria reveals that all species contain a complex mixture of these two general classes of cyt c (Table 5). In two species (S. sibiricus and E. hydrolyticum), a single soluble cyt c with a midpoint potential of 295 mV was present in the soluble protein fraction and, therefore, probably involved in both respiration and photosynthesis (226). Only one soluble cyt c possesses a redox potential high enough (Em = 210 mV) to sustain respiration and photosynthesis in R. thiosulfatophilus. Several soluble cyts c are found in E. litoralis, E. ursincola, E. ramosum, and E. ezovicum with midpoint potentials ranging from 340 to 275 mV, and so they could participate in both the respiratory and photosynthetic pathways. Unusually small soluble cyts c were isolated from R. thiosulfatophilus (cyt c549 [6.5 kDa] and c552 [4.0 kDa]) and E. ursincola (cyt c550 of 6.5 kDa) (Table 5) (226). Similarly small cyts c have so far been purified from only Hydrogenobacter thermophilus (cyt c550 of 6.0 kDa) and Methylomonas strain A4 (cyt c554 of 4.0 kDa) (202).
No evidence of cyt cd1 in soluble fractions of freshwater species was found, consistent with the inability of these bacteria to reduce nitrite (212, 214–216, 229, 230). In contrast, the soluble fraction of R. denitrificans (which is capable of denitrification) contains a cyt cd1 reductase (34). Detailed studies showed that there are two types of cyt cd1, which have slightly different absorption spectra. Purified cyts cd1 had cyt c oxidase and nitrite reductase activity, although the nitrite reductase activity was much lower than the oxidase activity (172).
In some species of aerobic phototrophic bacteria, several membrane-bound cyts were revealed by redox titration and sodium dodecyl sulfate-polyacrylamide gel electrophoresis in addition to the RC-bound tetraheme cyt c (49, 171, 226). The cyt bc1 complex was shown to be present in R. denitrificans, E. litoralis, E. hydrolyticum, E. ramosum, and E. ezovicum (172, 226). In several species, the Em values for the high- and low-potential cyt b ranged from 70 to 30 mV and from 20 to −150 mV, respectively, in purified membranes (226). Such low values for the Em of a bc1 complex cyt b heme were reported for the thermophilic bacterium PS3 and the photosynthetic Heliobacillus species (92). However, in these two organisms the quinone pool is composed of menaquinone, a low-potential quinone, which is not the case in aerobic bacteria, in which this pool is mainly composed of ubiquinone Q10 (see “Quinones”). Therefore, it would be useful to confirm these low Em values for the cyt b of these aerobic phototrophic bacteria by using purified cyt bc1 complexes.
A high-potential membrane-bound cyt c (350 mV) was observed in E. hydrolyticum, E. ramosum, and E. ezovicum (226). Since no cyt photooxidation was detected in isolated membranes of these three species, reduction of the photooxidized primary donor of RC seems to involve a soluble cyt c and not this high-potential membrane-bound electron carrier, as postulated for Protaminobacter ruber (173).
cyt oxidases were purified from E. longus (49) and R. denitrificans (34). The E. longus cyt c oxidase is a cyt aa3 type, which is unusual since previously R. sphaeroides was thought to be the only purple photosynthetic bacterial species that contains a cyt aa3 oxidase, composed of three polypeptides (53). The E. longus cyt aa3 is composed of two proteins of the same molecular weight.
In summary, the cyt content and composition of soluble and membrane fractions of aerobic phototrophic bacteria are highly diverse and species specific.
Photosynthetic Electron Transfer
In anoxygenic photosynthetic energy transduction, absorption of photons in the antenna system results in energy transfer to the RC, where the primary donor P (the special pair of Bchl molecules) is excited. Excited P* is a strong reductant. An electron is rapidly transferred (perhaps through an accessory Bchl) from the excited primary donor to a bacteriopheophytin (H). The electron is transferred from H− to the quinone QA within 220 ps, which results in transformation of the relatively unstable excited state into a relatively stable electrical potential across the membrane (Δψ, outside positive, inside negative). The negatively charged primary donor may be directly reduced by a reduced cyt c2 or by an RC-bound tetraheme cyt (see “Cytochrome composition”) (41, 100, 133).
The photochemical activity of the aerobic bacterial photosynthetic apparatus has been analyzed independently in several laboratories by using different species and techniques (51, 63, 131, 132, 174, 191, 226). The results indicate that the photosynthetic apparatus of aerobic phototrophic bacteria, although it has some peculiarities, is functional in terms of a cyclic electron transfer system.
In species of the genera Erythrobacter, Roseobacter, Roseococcus, Erythromicrobium, Erythromonas, and Sandaracinobacter, photoinduced cyclic electron transfer occurs only under relatively oxidized (aerobic) conditions, as elucidated by light-induced absorbance changes in whole cells (51, 131, 226). Under relatively reduced (anaerobic) conditions, no light-induced RC absorbance changes were observed. The lack of photochemistry under anaerobic conditions is consistent with the inability of these bacteria to grow by light-dependent photophosphorylation in the absence of oxygen (130, 144, 145, 147, 212, 214–216, 229, 230).
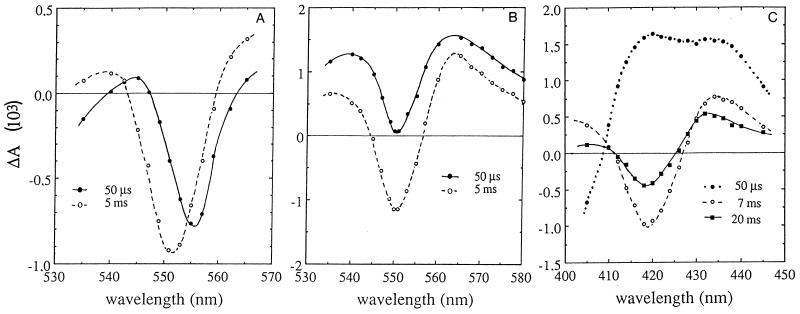
The very fast cyt c photooxidation observed after flash excitation in E. ursincola, S. sibiricus, R. thiosulfatophilus, and R. denitrificans (51, 131, 226) confirmed that the immediate electron donor to the RC in these species is a cyt c tightly bound to the RC. The difference spectrum measured at 50 μs was centered at 556 nm and correlates with the wavelength position of high-potential hemes of RC-bound cyts c of various photosynthetic bacteria (Fig. 9A) (35, 39, 51, 52). The wavelength of maximum bleaching at 5 ms was centered at 551 nm and corresponds to the photooxidation of soluble cyt c species, as reported for whole cells of the anaerobic phototrophic bacterium R. viridis (52). A large bleaching of cyt c attributed to photooxidation of soluble cyt c was induced in aerobic phototrophic bacterial species under continuous illumination (227).
FIG. 9.
Difference absorption spectra obtained for intact cells of aerobic anoxygenic phototrophic bacteria suspended in growth medium under aerobic conditions. (A) E. ursincola, determined at 50 μs and 5 ms after a saturating flash. (B) E. hydrolyticum, determined at 50 μs and 5 ms. (C) E. litoralis, determined at 50 μs and 7 and 20 ms.
As noted above, E. ramosum, E. hydrolyticum, E. ezovicum (226), and E. longus (132) do not possess a cyt c bound to the RC, and their photochemistry is similar to that observed in R. sphaeroides (16). The light-induced difference spectrum detected at 50 μs corresponds to a cyt c2 photooxidation, with an absorption band at 550 nm superimposed on absorption changes linked to the photooxidation of the RC special pair (Fig. 9B). There was no detectable spectral shift between 50 μs and 5 ms. As observed in cells of R. sphaeroides (78, 135), the cyt c photooxidation detected at 551 to 542 nm consists of two phases: a fast phase (with a half-time of less than 50 μs, not resolved in experiments on obligately aerobic species) and a slower phase with a half-time of 250 μs. The half-time of the subsequent cyt c reduction was about 40 ms (226). Continuous illumination caused the oxidation of a large amount of a soluble cyt c, about five times more than the amount detected after one saturating flash (227).
E. litoralis also does not possess a cyt c tightly bound to RC. However, the time-resolved photochemistry of this species was different from that of E. longus, E. ramosum, E. hydrolyticum, and E. ezovicum, reminiscent of that described for the anaerobic purple nonsulfur species R. rubrum (185). A light-induced difference spectrum detected at 50 μs indicates photooxidation of the RC primary donor, and complete cyt c oxidation was observed only after 10 ms (Fig. 9C). No fast phase of cyt c photooxidation has been observed in E. litoralis cells. Under continuous illumination, a large extent of cyt b reduction was observed (227).
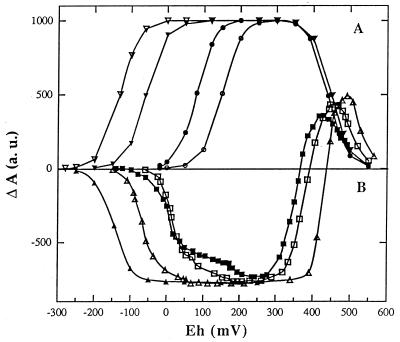
The above results indicate that a photosynthetic electron transfer system is operative in aerobic phototrophic bacteria under aerobic conditions. However, it was demonstrated that the photosynthetic electron transfer system of R. denitrificans, E. litoralis, E. ramosum, E. ursincola, S. sibiricus, and R. thiosulfatophilus (131, 226, 227) is inoperative in anaerobic cells, presumably due to the high midpoint potential (Em) of the RC primary acceptor QA (Fig. 10). The values of QA midpoint potential determined at pH 7.8 were +150, +80, +25, and +5 mV for E. litoralis, E. ramosum, E. ursincola, and S. sibiricus, respectively. These are much higher values than those detected in anaerobic phototrophic bacteria, such as R. sphaeroides, R. rubrum, Rhodocyclus tenuis, and R. viridis, which yield values in the negative region of the Eh (ambient redox potential) scale (Fig. 10). The Em values of the primary electron acceptor measured above the pK (pH value above which the midpoint potential of an electron carrier is not affected by pH) for E. litoralis, R. denitrificans, S. sibiricus, and E. ursincola are equal to −30, −44, −55, and −85 mV, respectively. These values are 65 to 120 mV more positive than those observed in anaerobic phototrophic bacteria (175, 227). Therefore, it is likely that the QA of aerobic phototrophic bacteria is in the reduced state (i.e., dihydroquinol) under anaerobic conditions and that acceptance of an electron from the special pair cannot occur unless an oxidant such as O2 is provided to maintain QA in the quinone form, which is capable of acting as an electron acceptor.
FIG. 10.
Redox titration curves of the RC primary acceptor QA midpoint potential determination performed at pH 7.8. The light-induced absorption changes were detected in membranes 1 ms after the excitation flash. (A) Species lacking an RC-bound cyt c: E. litoralis (○), E. ramosum (•), R. sphaeroides (▾), and R. rubrum (▿), measured at 603 nm. (B) Species containing an RC-bound cyt c: E. ursincola (□), S. sibiricus (■), R. tenuis (▵), and R. viridis (▴), measured at 555 nm. At this wavelength, the light-induced absorption changes are positive at high Eh due to the spectral contribution of the photooxidized primary donor and negative when the ambient potential is lowered due to the absorption changes linked to the RC-bound cyt c (224). (This figure was created with the help of L. Menin.) a. u., absorbance units.
It was shown that cyclic electron transfer between the bc1 and RC complexes in R. denitrificans is mediated by a soluble cyt c551 that is not tightly bound to the RC, and so the reduction of this cyt c551 by the bc1 complex is dependent on cyt diffusion. Under dark, semiaerobic conditions, the low-potential hemes of the RC-bound cyt c were reduced. These hemes were photooxidized under illumination but only slowly rereduced. Thus, it was proposed that this cyt redox state is a second possible explanation for the aerobic dependence of photochemical reactions in the RC of R. denitrificans (51).
Transfer of Excitation Energy from Carotenoids to Bacteriochlorophyll
Membranes of aerobic phototrophic bacteria are highly abundant in carotenoids. For example, the molar ratio of Bchl to carotenoid content in membranes of R. thiosulfatophilus and E. ramosum is 1:4 and 1:7, respectively (see “Carotenoids and bacteriochlorophyll”). However, in purified photosynthetic pigment-protein complexes this ratio was 1:1.4 in an enriched RC-LHI core complex of R. thiosulfatophilus, from 1:0.1 to 1 in the purified RC-LHI, and from 1:0.3 to 1 in the purified LHII complex of E. ramosum (211). The RC-LHI core complex of R. thiosulfatophilus contained only the C30 carotenoid diglucosyl ester (di[β-d-glucopyranosyl]-4,4′-diapocarotene-4,4′-dioate). Bacteriorubixanthinal is the major carotenoid in the pigment-protein complexes (LH and RC) of E. ramosum, along with small amounts of spirilloxanthin (RC-LHI) and zeaxanthin (LHII) (211).
The quantum yields of singlet energy transfer between carotenoids and Bchl (an LH function) calculated from comparison of absorption and fluorescence excitation spectra indicated that the majority of the carotenoids in the membrane of E. ramosum and R. thiosulfatophilus do not contribute to the LH function (210). The function of such large amounts of carotenoids in these cells is unclear. Carotenoids could play a role in scavenging singlet oxygen and/or free radicals, processes observed for several carotenoids in organic solvents (94, 95, 134), or perhaps in screening cells from high intensities of blue light.
Cells of E. longus are also abundant in such photosynthetically uncoupled carotenoids, as more than 70% of the total amount of carotenoids do not function as LH pigments (128). R. denitrificans has a qualitatively limited carotenoid composition compared to that of other aerobic phototrophic bacteria. The cells of this species produce the carotenoid spheroidenone, which is also the major carotenoid in semiaerobically grown Rhodobacter species (144, 151). A significant fraction of spheroidenone in R. denitrificans is present as the reduced 3,4- dihydrospheroidenone under illuminated anaerobic conditions, changing the culture color from pink to yellow. It was proposed that this effect was due to chemical reduction of redox components of the photosynthetic apparatus, which would interfere with normal photosynthetic electron transfer as well as the photoreduction of the C⩵C double bond at the 3,4- position of spheroidenone (167). The chemical structures of other carotenoids detected only in anaerobically illuminated cells of R. denitrificans are unknown. Preliminary studies suggested that they were not oxidized products of spheroidenone (167).
The Influence of Light on Growth and Pigment Formation
It is well established that in various anaerobic phototrophic bacteria light intensity affects the numbers and size of the photosynthetic unit (1, 38). The size of the core complex (RC-LHI) seems to remain constant at about 30 LHI Bchl molecules per RC under all light intensities (1), whereas the amount of the core complex and the relative amount of the LHII complex increase with decreased light intensity. For example, in R. capsulatus after a shift from a high to a low intensity of light, the size of the photosynthetic unit may increase two- to fivefold because of increases in the relative concentration of LHII (8).
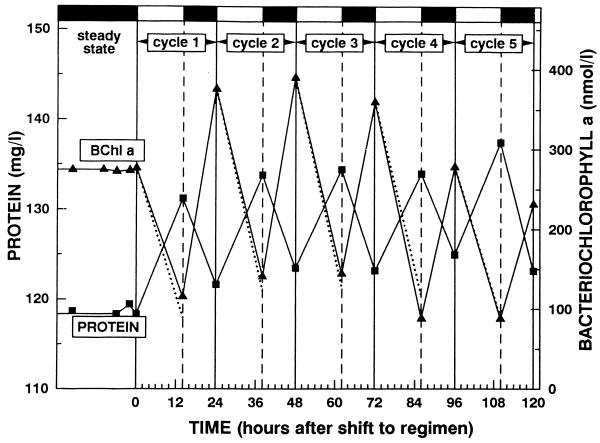
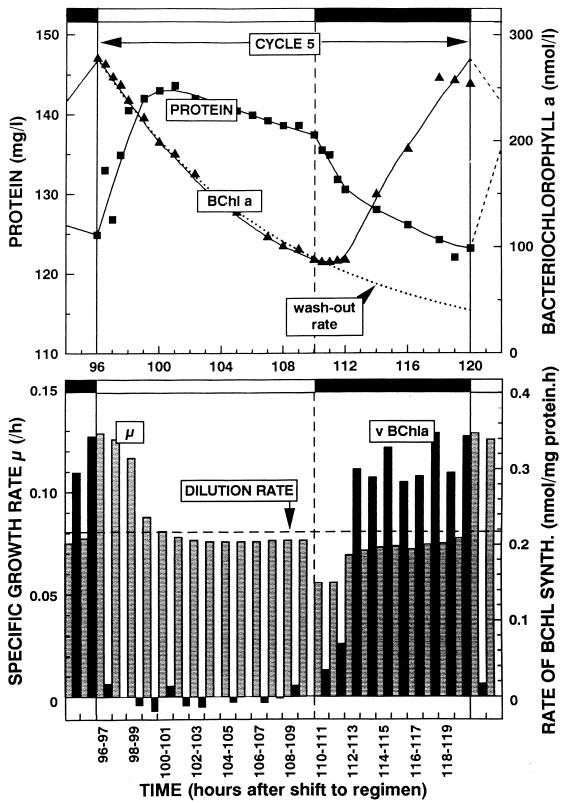
The effect of light on pigment synthesis in the aerobic phototrophic bacteria is roughly qualitatively similar to the light effect on the anaerobic phototrophic bacteria, but there are quantitative differences. In common are the light stimulation of growth and inhibition of aerobic respiration, suggesting the operation of a photosynthetic electron transport system that shares components with a respiratory system in both groups (67, 69, 220). Differences are described below. In anaerobic phototrophic bacteria, high light intensities repress the synthesis of Bchl, whereas at low light intensities Bchl synthesis is maximal (8). In aerobic phototrophic bacteria, the influence of light intensities as low as 20 μE/m2/s was found to be strongly inhibitory and abolished Bchl synthesis (232). Transient light stimulation of growth and complete inhibition of Bchl synthesis were demonstrated in batch culture experiments on E. longus, R. denitrificans, E. hydrolyticum, and S. sibiricus (67, 69, 104, 220, 232). However, the clearest results were obtained with continuous (chemostat) cultures of E. hydrolyticum (232). This is because experiments in which the light intensity is not influenced by the continuously changing cell density (due to self-shading), and physiological adaptation due to changes in medium composition, are difficult to carry out in batch cultures. Chemostat cultivation allows the maintenance of all culture conditions constant (except for one, limiting factor) and the analysis of the effects of changes in one variable parameter.
When a steady-state, dark-grown (acetate-limited) chemostat culture of E. hydrolyticum was illuminated with tungsten filament incandescent lamps (20 or 40 μE · m−2 · s−1), while the dilution rate was maintained constant, the Bchl a concentration of 219 nM progressively decreased to become 11 nM after 52 h of illumination. The decrease of Bchl followed the theoretical washout curve, suggesting neither synthesis nor degradation of the pigment in the light. During the first 5 h of illumination, the growth rate increased and the concentration of protein increased to a maximum of 127 mg/liter, compared to 114 mg/liter in the dark steady state. This increase in protein concentration was attributed to photosynthetic energy transduction dependent on the presence of dark-synthesized Bchl, which contributed to the formation of biomass by photosynthesis (232).
In order to simulate natural conditions, experiments were done in which E. hydrolyticum was exposed to a light-dark regimen (14 h of light and then 10 h of dark), employing the low-intensity irradiance of 20 μE · m−2 · s−1 (Fig. 11). Again, the Bchl values observed at the end of the light periods were consistent with the idea that Bchl was neither synthesized nor degraded in the light. In the dark periods, two- to threefold increases in the concentration of Bchl were observed. After the initiation of illumination, the concentration of protein initially increased (growth > dilution rate), but it started to decrease after 5 h when the specific content of Bchl decreased to 1.3 nmol/mg of protein (Fig. 12). The maximum concentration of protein reached during the period of illumination was 15% higher than that at the start of the light period. After the shift to the dark, the concentration of Bchl remained approximately constant for about 2 h (and thus diverged from the theoretical washout curve), indicating that synthesis of Bchl started immediately upon cessation of illumination. A high rate of Bchl synthesis was maintained during the dark periods, resulting in the accumulation of 2.1 nmol of Bchl/mg of protein at the end of a dark period (232).
FIG. 11.
Fluctuation during cycles 1 to 5 of the concentration of protein (closed squares) and Bchl a (closed triangles) in a continuous culture (D = 0.081/h) of E. hydrolyticum subjected to a 14-h light–10-h dark regimen. The irradiance employed was 20 μE/m2/s. Dotted lines indicate the theoretical washout rate of Bchl at zero rate of synthesis (232).
FIG. 12.
Time course during cycle 5 of protein (upper panel, closed squares) and Bchl a (upper panel, closed triangles) and the specific rate of growth (lower panel, gray bars; calculated from the curve shown in upper panel) and specific rate of synthesis of Bchl (lower panel, black bars; calculated from the curve shown in upper panel) in a continuous culture (D = 0.081/h) of E. hydrolyticum subjected to a 14-h light–10-h dark regimen. The irradiance employed was 20 μE/m2/s. Dotted lines in the upper panel indicate the theoretical washout rate of Bchl at zero rate of synthesis. The horizontal dashed line in the lower panel indicates the dilution rate (232).
Therefore, Bchl synthesis in E. hydrolyticum is regulated differently from that in anaerobic phototrophic bacteria, because even low intensities of light completely inhibit synthesis. Because of the increase in protein content of the culture and the growth rate of cells during the first 5 h after the start of illumination (Fig. 12), these experiments show that light provided an additional source of energy that stimulated growth in light-dark cycles.
The action spectrum of the light inhibitory effect on photosynthetic pigment accumulation in R. denitrificans (71, 176) indicated that even in the absence of molecular oxygen (during anaerobic trimethylamine N-oxide respiration) the greatest inhibition of photosynthetic pigment (Bchl a and carotenoids) accumulation was found with blue light of about 400 nm. The authors suggested that a blue light-absorbing pigment exists in this bacterium, a pigment which regulates the biosynthesis of Bchl and carotenoids and/or the production of Bchl-protein complex apoproteins. The transcription of genes that encode LH and RC apoproteins was proposed to indirectly regulate the accumulation of Bchl and carotenoids, controlled by a signal transduction system associated with an unidentified blue light-absorbing pigment (176). However, our preliminary results (222) on the light action spectrum of E. ramosum E5 are different. As described in “Carotenoids and bacteriochlorophyll,” E. ramosum contains at least 20 different carotenoids, whereas R. denitrificans mainly contains spheroidenone. In contrast to the results obtained with R. denitrificans, we did not detect an inhibition of carotenoid synthesis by light of any wavelength (350 to 1,100 nm) in E. ramosum (222). The inhibition of Bchl synthesis in E. ramosum also seems to respond to different wavelengths from those shown for R. denitrificans. It seems that the biosynthetic pathways of Bchl and carotenoids are independently regulated in E. ramosum, in contrast to R. denitrificans, and so it will be interesting to do additional experiments on different species to see if a common pattern of light regulation emerges.
An investigation of the light effect on the amounts of puf operon mRNA in R. denitrificans grown under aerobic conditions (oxygen concentration of about 78% of saturation) in comparison with that in R. sphaeroides grown under semiaerobic conditions (oxygen concentration of about 2% of saturation) revealed that light inhibited the steady-state concentration of puf operon messages in R. denitrificans more than in R. sphaeroides (126). Therefore, although transcription of the R. denitrificans puf operon seems to be insensitive to oxygen, it is more sensitive to light. In darkness, the levels of total puf mRNA in R. denitrificans were about 1.3 times those of R. sphaeroides at low concentrations of oxygen. The oxygen tension, up to 94% saturation of dissolved oxygen, did not affect the levels of puf transcripts in R. denitrificans, whereas those in R. sphaeroides were reduced to 55% of the maximum level at 50% oxygen saturation (126). These different sensitivities to oxygen and light of puf operon mRNA levels in R. denitrificans and R. sphaeroides were interpreted as a mode of adaptation that allowed R. sphaeroides to avoid photodynamic damage by light under highly aerobic conditions (126). However, in our opinion the available data on light and oxygen regulatory systems in aerobic phototrophic bacteria are insufficient to make general conclusions, and so more species have to be analyzed in more detail.
Effect of Oxygen on Growth and Pigment Synthesis
Oxygen partial pressure is the major environmental factor that regulates the formation of the photosynthetic apparatus and membrane differentiation in most of the facultatively anaerobic phototrophic bacteria (8, 86, 87, 114). For example, the presence of an atmospheric level (ca. 20%) of oxygen results in an almost complete suppression of photopigment production in R. capsulatus. In contrast, growth under conditions of reduced oxygen tension in either the presence or the absence of light results in formation of the ICM enriched in photosynthetic pigment-protein complexes (8).
Since, of the aerobic phototrophic bacteria, only R. denitrificans is capable of anaerobic growth with nitrate or trimethylamine N-oxide as an electron acceptor (3, 154), the survival of most aerobic phototrophic bacteria seems to depend on the presence of oxygen. For example, molecular oxygen stimulates heterotrophic consumption of organic substrates, enhances the growth rate, and increases other metabolic activities (e.g., thiosulfate oxidation as an additional source of energy [see “Metabolism of sulfur compounds”] that results in ATP generation). This dependence on molecular oxygen for growth, and thus the synthesis of Bchl and carotenoids, is in contrast to the behavior of most species of anaerobic phototrophic bacteria. At present, only the purple nonsulfur species R. centenum and Rhodovulum sulfidophilum are known to synthesize a functionally significant photosynthetic apparatus under high concentrations of molecular oxygen (65, 203). In contrast, photophosphorylation and photosynthetic ATP synthesis in cells of R. denitrificans were reported to occur only under aerobic conditions. A decrease in oxygen concentration resulted in a decrease in ATP concentration, regardless of the presence of light (130).
As noted in “Photosynthetic electron transfer,” light-induced electron transport through cyts and the RC requires aerobiosis in membrane preparations and intact cells of aerobic phototrophic bacteria (51, 70, 226). Moreover, it seems likely that, due to the high midpoint potential of the RC primary acceptor QA, oxygen is necessary for aerobic phototrophic bacterial photosynthesis (see “Photosynthetic electron transfer”). Thus, these bacteria may have evolved to become aerobes that have retained photosynthesis as an accessory metabolic process that enhances growth under conditions of alternating exposure to light.
HETEROTROPHIC CARBON METABOLISM AND NUTRITION
Substrate Utilization in Heterotrophic Growth
Most of the aerobic phototrophic bacteria are capable of oxidation of a great diversity of organic carbon sources to support chemotrophic growth. Accordingly, media that contain a complex composition of organic sources of carbon such as yeast extract, peptone, casein hydrolysate, potato broth, or soytone result in the highest growth yields (48, 63, 144, 145, 147, 212, 214–216). Most species can metabolize sugars, TCAs, fatty acids, and amino acids. Some species of Erythromicrobium use ethanol in low concentrations as the sole carbon source (215, 230). A lipolytic activity was established for many species on the basis of the ability to hydrolyze Tweens (144, 147, 212, 215, 230), and some species hydrolyze gelatin or starch (147, 215).
The active growth of most aerobic phototrophic species in complex media containing high concentrations of organic compounds (48, 144, 147, 212, 214–216, 231) is in keeping with the high organic matter content of the eutrophic environments from which they were isolated (48, 146, 148, 206, 217, 231). The main exception is Acidiphilium, which grows best with relatively low concentrations of nutrients (190).
The aerobic phototrophic strain JF-1, isolated recently from the presumably oligotrophic environment of black smoker plume waters, grows on an unusually low (for this physiological group) number of substrates. Glutamate, butyrate, and yeast extract are the best carbon sources for JF-1, and acetate and glucose support weak growth (222).
Operation of the Tricarboxylic Acid Cycle
The utilization of organic carbon sources by purple nonsulfur bacteria in aerobic dark growth is usually connected with oxidation through the TCA cycle (2, 22, 90, 164). Most purple phototrophic bacteria grow on acetate, although the pathways and reactions used to metabolize this substrate vary in different species (111, 164).
The activity of key enzymes of the TCA and glyoxylate cycles was measured in extracts of S. sibiricus (renamed E. sibiricum) cells grown aerobically in the dark on acetate (Table 6) (219). The activity of enzymes of the TCA cycle with the exception of 2-oxoglutarate dehydrogenase was determined in E. longus (Table 6) (219). The presence of isocitrate lyase and malate synthase in both of these species indicates the presence of the glyoxylate shunt (14, 15, 90, 164, 219). When glucose was the sole source of carbon, the activity of two key enzymes of the TCA cycle, citrate synthase and 2-oxoglutarate dehydrogenase, significantly decreased, and fumarate hydratase and the glyoxylate cycle enzyme isocitrate lyase activities decreased to undetectable levels (Table 6). The investigation of TCA cycle enzyme activities in acetate-grown cultures of the freshwater aerobic phototrophic bacteria R. thiosulfatophilus, E. ursincola, E. ezovicum, E. hydrolyticum, and E. ramosum revealed that the TCA cycle was complete under these conditions, as it is in S. sibiricus (206, 214–216, 230). The lack of 2-oxoglutarate dehydrogenase activity in E. longus, converting the TCA cycle into a branched pathway, is probably specific to the genus Erythrobacter. However, more species of this genus should be analyzed, and additional metabolic studies need to be done. The glyoxylate cycle enzyme isocitrate lyase was present in most of the species studied (206).
TABLE 6.
Activity of TCA and glyoxylate cycle enzymes in S. sibiricus (renamed E. sibiricum) and E. longusa
| Enzyme | Activity (nmol/min/mg of protein)
|
||
|---|---|---|---|
|
S. sibiricus
|
E. longus on acetate | ||
| On acetate | On glucose | ||
| Citrate synthase | 100 | 16 | 33 |
| Aconitate hydratase | 41 | 36 | 63 |
| Isocitrate dehydrogenase | 164 | 141 | 30 |
| 2-Oxoglutarate dehydrogenase | 20 | 5 | 0 |
| Succinate dehydrogenase | 7 | 4 | —b |
| Fumarate hydratase | 55 | 0 | 2 |
| Malate dehydrogenase | 707 | 508 | 554 |
| Isocitrate lyase | 35 | 0 | 5 |
| Malate synthase | 21 | 20 | 20 |
| Aceto-coenzyme A-synthase | 39 | — | — |
| Acetokinase | 5 | — | — |
Modified from reference 219.
—, not determined.
Most species of the aerobic phototrophic bacteria share with the purple nonsulfur bacteria the trait of being highly metabolically versatile in terms of heterotrophic carbon metabolism. However, these two bacterial groups are different regarding their ability for autotrophic growth and CO2 fixation. None of the obligately aerobic species has yet been grown autotrophically, and the key enzyme of Calvin cycle ribulose-bisphosphate carboxylase has not been found in any species (149, 206, 214, 215, 230). Experiments on S. sibiricus with radiolabeled 14CO2 indicated a low level of CO2 fixation (0.4%) in both illuminated and dark conditions, consistent with heterotrophic CO2 fixation (18, 61) attributed to the enzyme phosphoenolpyruvate carboxylase (206). Nonetheless, light stimulation of CO2 uptake was detected for several species: E. longus (143, 206), R. denitrificans (145), S. sibiricus (206), and A. rubrum (83). This amount of CO2 fixation, too low to maintain autotrophic growth, could provide additional organic carbon intermediates for an otherwise essentially heterotrophic metabolism. This view is congruent with the observation that light-stimulated CO2 uptake occurs in concert with light-activated consumption of acetate and stimulation of growth in chemostat cultures of E. hydrolyticum (232).
Pathways of Sugar Utilization
All aerobic phototrophic bacteria are able to utilize one or more of the sugars fructose, glucose, and sucrose (48, 63, 147, 212, 214–216, 229, 230). In some cases, the ability or inability to use fructose is a specific characteristic of the species. For example, R. thiosulfatophilus grows on glucose as the sole carbon source, whereas fructose does not support growth (214, 230).
The catabolism of glucose by aerobic phototrophic bacteria was studied by analysis of enzyme activities for two species, S. sibiricus RB16-17 (renamed E. sibiricum) and E. longus OCh101 (Table 7) (219). Both species possess glucose-6-phosphate dehydrogenase and 2-keto-3-deoxygluconate-aldolase. The high activity of these two enzymes indicates that glucose utilization is mainly via the Entner-Doudoroff pathway (EDP) (113). A low activity of the key enzyme of the Embden-Meyerhof pathway (EMP), fructose-diphosphate-aldolase, was detected, and it was concluded that this enzyme functions in biosynthesis (219).
TABLE 7.
Activity of sugar metabolism enzymes in cells of S. sibiricus (renamed E. sibiricum) and E. longusa
| Enzyme | Activity (nmol/min/mg of protein)
|
|
|---|---|---|
| S. sibiricus | E. longus | |
| Hexokinase | 7 | 63 |
| Phosphoglucomutase | 0 | 0 |
| Phosphofructose isomerase | 434 | 1,022 |
| Glucose-6-phosphate dehydrogenase | 103 | 240 |
| 6-Phosphogluconate dehydrogenase | 0 | 0.4 |
| 2-Keto-3-deoxy-6-phosphogluconate-aldolase | 6 | 145 |
| Fructosediphosphate aldolase | 6 | 39 |
| Phosphofructokinase | 1 | 0.3 |
| Pyruvate kinase | 68 | 25 |
Modified from reference 219.
No 6-phosphogluconate dehydrogenase was detected in S. sibiricus, whereas E. longus cells contained trace amounts of 6-phosphogluconate dehydrogenase, suggesting that the pentose monophosphate pathway exists in E. longus (219).
Glucose catabolism via the EDP is common in gram-negative bacteria, and the EDP enzymes are present in anaerobic phototrophic bacteria (26–29, 31, 113, 164). Pathway switching was shown for R. capsulatus, which catabolizes glucose via the EDP and fructose via the EMP (27). In R. sphaeroides, fructose is catabolized by the concomitant operation of both the EMP and the EDP, whereas glucose is catabolized via the EDP (27). Since E. longus and S. sibiricus catabolize several sugars, the presence of key enzymes of other sugar-degrading pathways (EMP or pentose monophosphate pathway) in cells during growth on glucose suggests that these enzymes function concomitantly or in pathway switching in the presence of other sugars such as fructose.
Storage Materials
A variety of cytoplasmic inclusions are found in bacteria. Some inclusion bodies are metabolically active structures, and some simply contain storage products, whereas gas vesicles have an environmentally adaptive function. Many inclusion bodies function in the storage of energy-rich substrates or serve as a reservoir of structural building blocks, so as to provide a selective advantage if the stored substance becomes depleted in the extracellular environment (113, 115).
Aerobic phototrophic bacteria accumulate polysaccharides (e.g., glycogen), polyhydroxyalkoanates, and/or polyphosphates, depending on the growth conditions (Fig. 13) (214–216, 225, 230).
FIG. 13.
Intracytoplasmic components of S. sibiricus revealed by electron microscopy of ultrathin sections. (A and B) Big electron-clear granules of presumed PHB (indicated by arrows) often occupied up to 40 to 50% of the total cell volume, deforming the cell shape. Rare ICM vesicles are indicated by V. (C) Electron-dense granules of presumed polyphosphate (indicated by arrow) occupied up to 30 to 40% of the total cell volume. Bars, 0.5 μm.
Polyphosphates consist of linear polymers of orthophosphate that provide a reserve of inorganic phosphate (96, 198). Cells of S. sibiricus accumulated heavily osmium-stained granules of presumably polyphosphates under nearly all experimental conditions studied: in the light and the dark; with high or low aeration; and during growth with acetate, butyrate, or sucrose as the sole organic carbon source (225). The highest amount of polyphosphate was accumulated in a growth medium supplemented with sucrose. Under such conditions, polyphosphate granules occupied about 30 to 40% of the total cell volume (Fig. 13).
One of the most common inclusion bodies in prokaryotic organisms consists of polyhydroxyalkanoate compounds (hereafter referred to as PHB, a lipid-like compound that is formed from β-hydroxybutyric acid units) (113). Electron-transparent granules presumed to be PHB were found in several species of the genera Roseococcus, Erythromicrobium, and Sandaracinobacter (206, 214–216, 225), with S. sibiricus RB16-17 as the greatest PHB accumulator. PHB formation occurred when cells were grown in media not balanced for nitrogen (urea as a nitrogen source), as well as during incubation in a medium lacking fixed nitrogen. Replacement of ammonia with nitrate as the source of nitrogen also resulted in pronounced formation of PHB granules. Under these conditions, large PHB granules occupied about 40 to 50% of the total cell volume, to the extreme of cell deformation (Fig. 13).
Although storage compounds have been revealed in many aerobic phototrophic bacteria, little is known about conditions that promote accumulation or consumption of these cytoplasmic inclusions. It would be interesting to investigate these bacteria more carefully to elucidate this aspect of their physiology. The possibility of nitrogen fixation in aerobic phototrophic bacteria has been neither clearly confirmed nor definitely disproved. However, the growth of S. sibiricus in medium lacking fixed nitrogen with enhanced PHB accumulaton indicates the ability of this species to fix nitrogen under aerobic conditions. The confirmation of nitrogen fixation in S. sibiricus would undoubtedly strengthen its ecological and environmental roles, and this property should be investigated with other species.
METABOLISM OF SULFUR COMPOUNDS
Several species of purple nonsulfur phototrophic bacteria of the α-subclass of the Proteobacteria, the closest phylogenetic relatives of the aerobic phototrophic bacteria (48, 149, 159, 196, 197, 229, 230), use reduced sulfur compounds in a dissimilatory metabolism (20, 64, 82, 88, 112, 123, 140, 179, 181, 182). All purple sulfur phototrophic bacteria grow photosynthetically with an inorganic sulfur compounds (i.e., sulfide, sulfur, polysulfides, thiosulfate, or sulfite) as electron donor(s) for CO2 fixation (20, 89, 181, 182). However, some purple sulfur as well as nonsulfur phototrophic species exhibit the capacity for oxidation of inorganic sulfur compounds in the dark in the presence of organic compounds and oxygen (88, 180). Although aerobic phototrophic bacteria are incapable of autotrophic growth and CO2 fixation (see “Operation of the tricarboxylic acid cycle”), the aerobic oxidation of inorganic sulfur compounds in the presence of an organic carbon source was investigated (221).
Six species, R. thiosulfatophilus, E. ramosum, E. hydrolyticum, E. ezovicum, E. ursincola, and S. sibiricus (renamed E. sibiricum), were analyzed for the use of sulfide or thiosulfate for growth with these compounds as the sole sulfur source in a defined medium containing acetate as the sole source of carbon. None of these species oxidized sulfide, whereas a thiosulfate-oxidizing activity was detected with E. hydrolyticum E4(1) and R. thiosulfatophilus RB-7. Utilization of thiosulfate by both species was dependent on the presence of an organic carbon substrate and aeration. An increase in aeration increased the growth rate, which in turn provoked faster thiosulfate oxidation and the accumulation of the oxidized sulfur product tetrathionate or sulfate in the growth medium of E. hydrolyticum or R. thiosulfatophilus, respectively (221).
In E. hydrolyticum, no further oxidation of tetrathionate to sulfate occurred, as reported for C. vinosum (157) or R. palustris (140, 180). Thus, this oxidation pathway seems to be a dead end in E. hydrolyticum, similar to Rhodopseudomonas globiformis (179). However, it is possible that accumulated tetrathionate inhibits the growth of E. hydrolyticum and, consequently, oxidation of thiosulfate (206). It has been reported that tetrathionate inhibits the complete oxidation of thiosulfate to sulfate as well as growth of C. vinosum (157).
Although in accord with positive light effect on the growth rate and constructive metabolism of aerobic phototrophic bacteria (see “The influence of light on growth and pigment formation”), the light stimulation of thiosulfate oxidation in these bacteria does not seem to be surprising; the higher dark-grown culture accumulation of sulfate or tetrathionate resulting from thiosulfate oxidation, compared to that of illuminated cultures, indicates that thiosulfate, the sole sulfur source, is used not only in energetic metabolism but also for biosynthesis and that this activity is pronounced in the light (221).
E. hydrolyticum and R. thiosulfatophilus contained enzyme activities characteristic of thiosulfate metabolism (Table 8) (221). Since only tetrathionate was found as a product of thiosulfate oxidation by E. hydrolyticum and thiosulfate-dependent cyt c oxidoreductase activity was detected in cell extracts, the following equation for dissimilatory metabolism of thiosulfate was proposed (137, 221): 2S2O32−→S4O62− + 2e−, with the electrons used for oxidative phosphorylation. A thiosulfate-dependent cyt c oxidoreductase was isolated from the anaerobic phototrophic bacteria R. palustris, Chlorobium limicola f. sp. thiosulfatophilum, R. globiformis, and C. vinosum and proposed to be responsible for thiosulfate oxidation to tetrathionate and for donation of electrons to cyt c (98, 137, 157, 179, 181). During the formation of tetrathionate, only one electron per molecule of thiosulfate is released, which is probably insufficient to support photolithotrophic growth (as in the case of R. globiformis [179]). Nevertheless, organisms such as E. hydrolyticum and R. palustris effectively use this electron source as a supplement to heterotrophic pathways of oxidation (140, 180, 221). The high activity of thiosulfate reductase found in E. hydrolyticum cells is unclear (221).
TABLE 8.
Activity of thiosulfate metabolism enzymes in R. thiosulfatophilus and E. hydrolyticum cellsa
| Enzyme | Activity (nmol/min/mg of protein)
|
|
|---|---|---|
| R. thiosulfatophilus | E. hydrolyticum | |
| Thiosulfate-cyt c oxidoreductase | 75 | 86 |
| Rhodanese | 16 | 23 |
| Thiosulfate reductase | 640 | 2,800 |
| Sulfite-cyt c oxidoreductase | 346 | 47.3 |
| Adenosine-5-phosphosulfate reductase | NDb | NDb |
Modified from reference 221.
ND, not detected.
E. hydrolyticum and R. thiosulfatophilus were found to contain the enzyme rhodanese (Table 8). Rhodanese transfers the sulfane sulfur atom of thiosulfate to CN−, generating SO32− and SCN− (204). Thus, it was proposed that the rhodanese in these two bacteria functions as follows: S2O32− + CN−→CNS− + SO32− (221). However, rhodaneses are widely distributed enzymes with a number of functions and are found in plant cells and bacterial species incapable of thiosulfate oxidation (20, 141, 204). The purple nonsulfur phototrophic bacteria R. rubrum and R. capsulatus do not oxidize thiosulfate, although they produce a rhodanese (181). Therefore, it is possible that in R. thiosulfatophilus and E. hydrolyticum the rhodanese function is not (or not only) associated with thiosulfate metabolism (221).
Thiosulfate reductase and sulfite-dependent cyt c oxidoreductase activities were found in R. thiosulfatophilus (Table 8) (221). Since sulfate is the product of thiosulfate oxidation, it was proposed that thiosulfate reductase catalyzes the reductive splitting of thiosulfate into sulfide and sulfite: S2O32− + 2e−→S2− + SO32− (221). This enzymatic activity was demonstrated in cells of the anaerobic phototrophic bacteria T. roseopersicina, C. vinosum, Chromatium minus, R. rubrum, and R. palustris (10, 82, 137).
Sulfite can be further oxidized to sulfate in a reaction catalyzed by a sulfate-dependent cyt c oxidoreductase (81, 182, 221): SO32− + H2O→SO42− + 2H+ + 2e−. Sulfite oxidoreductase and adenosine-5-phosphosulfate reductase have been found in almost all phototrophic bacteria examined (45, 81), but adenosine-5-phosphosulfate reductase was not detected in E. hydrolyticum and R. thiosulfatophilus, and the transformation of the sulfide ion was not analyzed in detail (206).
Thus, E. hydrolyticum and R. thiosulfatophilus use thiosulfate as an electron donor, oxidizing the compound by different modes. These species do not grow autotrophically or produce ribulose-bisphosphate carboxylase, although a phosphoenolpyruvate carboxylase activity was found (see “Operation of the tricarboxylic acid cycle”). The available data indicate that E. hydrolyticum and R. thiosulfatophilus are capable of chemolithoheterotrophy, that is, enhancement of heterotrophic growth by oxidation of thiosulfate.
REDUCTION OF HEAVY METAL OXIDES AND POTENTIAL FOR BIOREMEDIATION
The study of microbial oxidation or reduction of elements is of great importance for our understanding of biogeochemical cycling of these elements in nature. Many transformations of metals in anaerobic and aerobic environments are the result of the direct enzymatic activity of bacteria (6, 33, 101, 102, 108, 109, 116, 234).
Since the industrial revolution, human activities have resulted in the release of an enormous amount of toxic chemicals into the environment, which contribute to serious pollution problems (199). Bacterial bioremediation is a potentially attractive and ecologically sound method of removing environmental contaminants. Many microorganisms have developed mechanisms of resistance to high concentrations of toxic metals such as Hg, As, V, U, Cd, Se, and Te. These mechanisms involve the detoxification of heavy metals by their precipitation, adsorption, or transformation to less toxic forms (40).
Several microorganisms can use certain metals and metalloids as terminal electron acceptors for respiratory growth in the absence of oxygen. Examples of these metals include Fe(III), Mn(IV), U(VI), Se(VI), V(V), and As(V) (102, 108–110, 234). As an alternative to genuine dissimilatory metal reduction, some bacteria are thought to use metal oxide reduction for disposal of electrons (e.g., reoxidation of NADH, NADPH, FADH2, reduced cytochromes, or quinones), thereby maintaining an appropriate redox poise in vivo, as well as detoxification of the environment (118, 119).
The heavy metal oxide tellurite is toxic to many microorganisms at concentrations as low as 1 μg/ml (161, 162, 194). The toxicity of tellurite to microorganisms is believed to be a consequence of its properties as a strong oxidant (161). Seven species of aerobic phototrophic bacteria (E. litoralis, R. thiosulfatophilus, E. ramosum, S. sibiricus, E. ursincola, E. ezovicum, and E. hydrolyticum) were recently found to possess a high level of resistance (HLR) to tellurite and the ability to reduce TeO32− to metallic tellurium (Table 9) (218). The tellurite MICs for aerobic phototrophic bacteria were found to be significantly higher than those determined for other representatives of the α subclass of the Proteobacteria (Table 10). The highest MICs of tellurite described for R. capsulatus and R. sphaeroides were 800 and 900 μg/ml, respectively (118, 119), whereas MICs of 2,300, 2,500, and 2,700 μg/ml were found for the aerobic E. ramosum, E. hydrolyticum, and E. ursincola, respectively (218). This reduction of tellurite to the relatively inert metallic tellurium, with accumulation as intracellular deposits, seems to be one way that bacteria detoxify tellurite (107, 118, 178). However, HLR without metal accumulation was observed for R. thiosulfatophilus grown with l-glutamine, succinate, malate, tartrate, or acetate and for E. ezovicum grown with acetate as the organic carbon source (Table 9). These results imply that tellurium accumulation is not essential to confer TeO32− resistance and that another mechanism such as continuous tellurite efflux, complexing, or methylation could specify this resistance.
TABLE 9.
Reduction of K2TeO3 by different species of aerobic photosynthetic bacteria depending on organic carbon sourcea
| C source |
E. litoralis
|
E. hydrolyticum
|
E. ursincola
|
E. ramosum
|
S. sibiricus
|
R. thiosulfatophilus
|
E. ezovicum
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| ROb | + | 500 | 1,200 | + | 500 | 1,200 | + | 500 | 1,000 | + | 50 | 750 | + | 50 | 750 | + | 100 | 500 | + | NR | 5 |
| Yeast extract | + | 250 | 1,200 | + | 500 | 2,000 | + | 500 | 2,000 | + | 250 | 1,500 | + | 500 | 1,200 | + | 250 | 1,000 | + | NR | 5 |
| l-Glutamine | + | 500 | 1,500 | + | 750 | 1,200 | + | 1,000 | 2,000 | + | 250 | 1,200 | + | 100 | 1,000 | + | NR | 1,200 | + | 1,000 | 2,000 |
| Succinate | + | 750 | 1,200 | + | 750 | 1,200 | + | 750 | 1,200 | + | 500 | 1,200 | − | + | NR | 1,200 | + | 750 | 2,000 | ||
| Malate | − | + | 750 | 1,200 | + | 500 | 1,200 | + | 250 | 1,200 | − | + | NR | 1,200 | + | 500 | 1,200 | ||||
| Tartrate | − | ± | NR | 5 | − | ± | 100 | 500 | − | + | NR | 1,200 | ± | NR | 5 | ||||||
| Acetate | + | 750 | 1,200 | + | 1,000 | 2,500 | + | 250 | 2,700 | + | 1,000 | 2,300 | + | 500 | 1,200 | + | NR | 1,200 | + | NR | 500 |
| Ethanol | ± | 250 | 750 | + | 100 | 250 | − | + | 250 | 1,000 | − | − | + | NR | 5 | ||||||
Modified from reference 218. Columns: 1, growth without tellurite addition; +, good growth; ±, weak growth; −, inability to use substrate; 2, the highest rate and completeness of K2TeO3 reduction (micrograms per milliliter). 3, MIC (micrograms per milliliter). NR, not reduced.
RO, rich organic medium.
TABLE 10.
Resistance to tellurite by some representative members of the Proteobacteria
| Sp. | Phylogenetic subgroup | MIC of K2TeO3a (μg/ml) |
|---|---|---|
| Aerobic photosynthetic | ||
| Erythromicrobium hydrolyticum | α-4 | 2,500 |
| Erythromicrobium ramosum | α-4 | 2,300 |
| Erythromicrobium ezovicum | α-4 | 2,000 |
| Erythromonas ursincola | α-4 | 2,700 |
| Erythrobacter litoralis | α-4 | 1,500 |
| Sandaracinobacter sibiricus | α-4 | 1,200 |
| Roseococcus thiosulfatophilus | α-1 | 1,200 |
| Anaerobic photosynthetic purple | ||
| Rhodospirillum rubrum | α-1 | 20 |
| Rhodopseudomonas palustris | α-2 | 200 |
| Rhodopseudomonas viridis | α-2 | 80 |
| Rhodobacter sphaeroides | α-3 | 900 |
| Rhodobacter capsulatus | α-3 | 800 |
| Rhodocyclus gelatinosus | β-1 | 5 |
| Nonphotosynthetic | ||
| Escherichia coli | γ-3 | <5 |
| Agrobacterium tumefaciens | α-2 | 50 |
| Pseudomonas putida | γ-3 | 40 |
Aerobic growth.
However, in most cases HLR to tellurite is correlated with its reduction to metallic tellurium. As revealed by electron microscopy, elemental tellurium accumulates inside the cells of seven species of the aerobic phototrophic bacteria tested (Fig. 14) (218). The tellurium crystals were located in the cytoplasm, except for E. litoralis, in which tellurium was observed in both the cytoplasm and the periplasm, suggesting the existence of two distinct pathways for tellurite reduction and possibly more than one tellurite reductase (207).
FIG. 14.
Electron microscopy of ultrathin sections. Shown are the intracellular localizations of presumed tellurium (indicated by arrows) as a product of tellurite reduction. (A) Two daughter cells of E. ramosum with tellurium crystals apparently interfering with cell division. (B) In E. litoralis, tellurium crystals sometimes occupy as much as 20 to 30% of the cell volume. (C) R. thiosulfatophilus accumulates relatively small Te crystals similar to Te deposits observed in E. coli or Rhodobacter species. Bars, 0.5 μm.
The maximum size and total amount of tellurium crystals in most of the aerobic phototrophic bacteria were much greater than those observed in Escherichia coli or Pseudomonas or Rhodobacter species (106, 118, 119, 163, 178, 218). In some cells of E. ramosum and E. litoralis, the metal crystals occupied 20 to 30% of the cell volume. The only exception was R. thiosulfatophilus, which accumulated small metallic deposits, similar to E. coli or R. sphaeroides (Fig. 14) (218).
Tellurium (especially of valence IV) is very toxic not only for microbes but also for other organisms, including humans. Therefore, the microbial reduction of soluble Te(IV) to solid Te(0) could be an important mechanism for the removal of this poison from polluted sites. In this context, the development of microbiological methods for environmental remediation of locales contaminated with tellurium oxides could utilize aerobic phototrophic bacteria, which are able to transform very high concentrations of Te(IV) to Te(0). Multiresistance to several different toxic metal oxides and the ability to reduce them have been detected for some bacterial species (102, 108, 125). Resistance to tellurite, selenite, and at least 15 other rare-earth oxides and oxyanions was shown for the facultative phototroph R. sphaeroides (118). Species of the genera Erythromicrobium, Erythrobacter, and Erythromonas were found to be resistant not only to tellurite but also to selenite (207). Therefore, it would be of interest to analyze the specificity of heavy metal reductases of aerobic phototrophic bacteria for other toxic metal oxides as well as their capacity for biotransformation of other compounds such as selenate and tellurate and different valencies of vanadium, mercurium, molybdenum, and arsenic. Such studies may increase the importance of these bacteria as prospective candidates for the bioremediation of industrial wastewaters polluted by a combination of several different heavy metal ions.
Microbial activities are used in the concentration of metals from natural ores and from mining tailings with metal levels too low for smelting. Bioleaching by Thiobacillus ferrooxidants and related thiobacilli, for example, results in the recovery of up to 70% of the copper in low-grade ores. About 10% of U.S. copper comes from leaching ore through the activity of bacteria such as Thiobacillus and Leptospirillum species (138). Therefore, tellurium accumulation by aerobic phototrophic bacteria could be helpful in tellurium extraction from ores or for recycling of tellurium oxides. Native tellurium is uncommon (an abundance in the lithosphere of 2 × 10−7%), usually occurring in conjunction with elemental sulfur. Tellurium is present in minerals as tellurides of lead, copper, silver, gold, and antimony (5). The extraction of tellurium is difficult because of this low content in natural ores, and tellurium compounds are usually obtained as by-products of metal refining processes (85). Aerobic phototrophic bacteria, which are easy to cultivate and which accumulate high amounts of Te in cells, could be used in microbiological metallurgy for tellurium concentration.
CONCLUSIONS AND PERSPECTIVES
The aerobic phototrophic bacteria are a relatively recently discovered physiological group, which carry out anoxygenic photosynthesis under aerobic conditions. Although the presence of Bchl in cells is a property shared with the anaerobic phototrophic bacteria, the aerobic phototrophic bacteria cannot use light as the sole source of energy under any laboratory growth conditions that have been tested with pure cultures. Most species are strict aerobes, and the best growth is obtained with organic substrates as the main source of carbon and energy. During the last 10 to 15 years, the number of aerobic phototrophic species characterized has increased greatly, resulting in two marine and six freshwater genera, all of which are phylogenetically located in the α subclass of the Proteobacteria. A better understanding of the evolutionary origin and diversity of the physiological properties of this group depends on continued study of existing species and isolation of new strains.
Recently created phylogenetic trees show that aerobic phototrophic genera do not present a homogeneous cluster but are distributed among photosynthetic and nonphotosynthetic species, and so some species are not closely related (Fig. 1 and 2). This situation is similar to the phylogeny of anaerobic phototrophic bacteria and is the basis of the proposal that nonphotosynthetic proteobacteria had a photosynthetic ancestor with photosynthesis genes lost during evolution (196). The existing data are far from sufficient to allow conclusions about the evolutionary origin of aerobic phototrophic bacteria, but two theories have been proposed.
One scenario focuses on the idea that the 16S rRNA phylogenetic heterogeneity as well as differences in the photosynthetic apparatus can be explained by the origin of aerobic phototrophic bacteria as branches from different species of anaerobic phototrophic bacterial ancestors. Thus, aerobic phototrophic bacteria could represent an intermediate phase of evolution from anaerobic purple bacteria to nonphotosynthetic aerobes (197).
A second possibility is that lateral transfer of photosynthesis genes (13, 120, 121) might have resulted in the acquisition of a photosynthesis gene cluster by nonphotosynthetic aerobes. However, the relatively recent acquisition of multigene-dependent photosynthetic complexes by lateral gene transfer to yield all aerobic phototrophic bacteria seems to be less likely.
The recent discovery of the aerobic phototrophic strain JF-1 in a deep-sea environment associated with hydrothermal vents could provide insight into the evolution and diversity of photosynthesis. The idea that photosynthesis may have originated at deep-sea hydrothermal vents and then dispersed to favorable refuges in shallow-water habitats raises the possibility that extant vent phototrophs may exhibit physiological properties representative of primitive phototrophs (7, 105, 156). Although it is not clear whether strain JF-1 evolved in proximity to hydrothermal vent plume waters or more later came to colonize this environment, the abundance of this bacterium (10% of cells that grew on the medium used) (208) indicates that the ability to produce Bchl is of selective advantage. It will be interesting to see if it is possible to differentiate between the evolution of photosynthesis at deep hydrothermal vents and that in solarly irradiated environments. Perhaps the isolate JF-1 exhibits some photosynthetic properties of evolutionarily early photosynthetic bacteria and the respiratory dependence of this species was developed during subsequent evolution. Although the evolutionary significance of the inability to grow anaerobically photosynthetically despite the presence of a photosynthetic apparatus is unclear, further investigations of aerobic phototrophs will ultimately solve this puzzle and contribute to our understanding of fundamental aspects of the evolution of photosynthesis.
A remarkable feature of aerobic phototrophic bacteria is their carotenoid composition. The high concentrations of carotenoids that seem to be uncoupled from photosynthetic energy transfer in members of Erythrobacter, Erythromicrobium, and Roseococcus are mainly composed of highly polar molecules. It was suggested that, due to the highly polar nature of these carotenoids, they are nonphotosynthetic and are separate from the photosynthetic apparatus (149). However, analyses of carotenoid composition and localization in the cell fractions of R. thiosulfatophilus have shown that the highly polar C30 diglucosyl ester is a major carotenoid component of the RC-LHI core complex (211) (see “Carotenoids and bacteriochlorophyll,” “Light-harvesting systems,” and “Transfer of excitation energy from carotenoids to bacteriochlorophyll”). Furthermore, absorption and fluorescence excitation spectra indicated that these polar carotenoids are functionally associated with the RC-LHI core complex and participate in LH transmission of light energy to Bchl molecules, although the major fraction of carotenoids is not photosynthetically active (210). Therefore, a high polarity of a carotenoid species does not preclude an LH function.
High concentrations of extrinsic carotenoids have also been observed in algae. It has been suggested that extrachloroplastic high concentrations of astaxanthin ester-containing lipid globules function as a sunscreen to minimize photodamage during periods of exposure to intense solar radiation of snow algae (e.g., Chlamydomonas nivalis [11]). In addition to the proposed sunscreen function, these secondary carotenoids may act as inhibitors of photodynamically induced damage (62). However, it remains to be seen if nonphotosynthetic carotenoids in cells of aerobic phototrophic bacteria provide similar protection.
The photosynthetic activity of aerobic phototrophic bacteria might seem to be negligible. However, under aerobic alternating light and dark conditions the photosynthetic electron transfer chain might significantly contribute to an increase of the proton gradient and help to maintain an electrochemical potential across the CM. This increase could be used for ATP production or for transport of substrates. For example, it is thought that cells spend a large portion of their total energy budget for active transport (113). Therefore, the use of light energy to enhance substrate transport could increase the efficiency of organic substrate consumption for biosynthesis. Such speculations are supported by results obtained with continuous cultures of E. hydrolyticum (232). Thus, the simultaneous operation of photosynthesis with heterotrophic respiration may be of advantage in competition with heterotrophs that lack photopigments.
Although the general physiological significance of the presence of the tetraheme cyt c subunit of the RC of purple bacteria is not clear, the presence of this subunit in aerobic phototrophic bacteria might provide a clue to its significance. It was shown that cyt photooxidation occurred much faster in intact cells of species that possess an RC-bound cyt c than in species lacking such an RC subunit (see “Photosynthetic electron transfer”). The presence of a tetraheme cyt bound to the RC could allow rapid reduction of the positively charged special pair, preventing a back-reaction from a negatively charged quinone, and thus could enhance the efficiency of the photosynthetic apparatus of aerobic phototrophic bacteria under conditions in which complete reduction (i.e., acquisition of 2e− and 2H+) of QB was slow.
The cyt composition and location have been investigated for several species of the aerobic phototrophic bacteria. However, additional purification and reconstitution experiments with isolated membrane fractions are necessary to elucidate the precise roles of the large number of cyt species in photosynthetic and respiratory electron transfer chains. Although the participation of a cyt c in light-driven electron transport was indicated by light-induced absorption changes in Roseobacter, Roseococcus, Erythrobacter, Erythromicrobium, Erythromonas, and Sandaracinobacter, the involvement of the bc1 complex in these bacteria has not been shown directly.
Since light seems to be used by aerobic phototrophic bacteria as a supplementary source of energy, these bacteria could be called photoheterotrophs. However, the sense of the term “photoheterotrophy” for anaerobic phototrophic bacteria and for the aerobic phototrophic bacteria differs significantly. Light is thought to be the major source of energy (ATP) for anaerobic purple phototrophic bacteria during anaerobic photoheterotrophic growth, whereas organic compounds are the source of substrates oxidized to provide reducing power and intermediates for biosynthetic anabolism. Furthermore, most anaerobic purple phototrophic bacteria are capable of photoautotrophic growth, whereas aerobic phototrophic bacteria are not. All the available data indicate that in aerobic phototrophic bacteria an organic substrate is used as the main source of energy, reducing power and carbon for biosynthesis independently of the presence of light.
Several suggestions concerning the trivial nomenclature of the aerobic phototrophic bacteria as a group have been proposed: “Erythrobacteria” or “aerobic Bchl-containing bacteria” (148), or “quasiphotosynthetic bacteria” or “paraphotosynthetic bacteria” (55). We agree with Gest (55) that Halobacterium and related species are fundamentally different from the classical photosynthetic bacteria in the biophysical and biochemical details of light energy transduction. However, we think it is premature to conclude that the aerobic phototrophic bacteria should be considered nonphotosynthetic, in contrast to other photosynthetic bacteria. Firstly, both groups of bacteria have many common features of the structure, function, and macromolecular organization of LH and RC complexes, indicative of a high degree of similarity of these photosynthetic complexes and the genes that encode them. The light-induced oxidation and reduction of the RC and other components of the photosynthetic apparatus in several representatives of this group have been demonstrated (see “Photosynthetic electron transfer,” “Transfer of excitation energy from carotenoids to bacteriochlorophyll,” and “The influence of light on growth and pigment formation”). Secondly, it is possible that a single combination of light wavelength, intensity, and other conditions will be found to enhance the photosynthetic activities of aerobic phototrophic bacteria. Therefore, we agree that the designation “aerobic anoxygenic phototrophic bacteria” made by Shimada (149) is the most suitable at this time.
Against the background of uncertainties regarding the aerobic phototrophic bacteria (see above), the question of why these bacteria are unable to grow by anaerobic photosynthesis despite the presence of a photosynthetic apparatus is fundamental. We consider below several explanations.
(i) Because the protein environment of the RC primary acceptor QA seems to produce a high midpoint potential, this results in nearly complete reduction of QA under anaerobic conditions, preventing light-driven charge separation (see “Photosynthetic electron transfer”).
(ii) Under low aeration, the low-potential hemes of the RC-bound cyt c are reduced and photooxidized under illumination but are rereduced extremely slowly. This result was invoked to explain the R. denitrificans requirement for aerobic conditions (51), but this does not explain the oxygen requirement of species lacking an RC-bound tetraheme cyt c (see “Photosynthetic electron transfer”).
(iii) Light-induced anaerobic changes of carotenoid pigments found in R. denitrificans cells impelled another hypothesis (167). This proposal features a reduction of photosynthetic apparatus electron transfer components that interferes with photosynthetic electron transfer and results in photoreduction of the C⩵C double bond at the 3,4- position of spheroidenone (see “Transfer of excitation energy from carotenoids to bacteriochlorophyll”).
(iv) Probably the most significant obstacle to prolonged, continuous photosynthetic growth is the dramatic inhibition of Bchl synthesis by light (see “The influence of light on growth and pigment formation”).
In our opinion, the inability of aerobic phototrophic bacteria to grow photosynthetically anaerobically is most likely a combination of the protein environment around the RC QA and an extraordinarily sensitive light-dependent regulatory system that shuts off Bchl synthesis. It would be interesting to isolate mutants that continue to synthesize Bchl under illumination and to study the nature of such mutations and the physiology of such mutants.
The study of the genetics of aerobic phototrophic bacteria is in its infancy, although the puf operon of R. denitrificans was cloned, sequenced, and showed high amino acid sequence similarity (up to 70%) with Rhodobacter species (104). Recently, the puf operon of R. denitrificans was expressed in an R. capsulatus puf-puc double-deletion mutant (91). The strongest expression of the R. denitrificans puf operon was observed under the control of the R. capsulatus puf promoter, in the presence of R. capsulatus pufQ and pufX genes, and in the absence of the R. denitrificans pufC gene. RC charge separation and recombination between the primary donor P+ and the primary acceptor QA− was observed for this strain, showing that the RC was correctly assembled. However, this strain did not grow photosynthetically under anaerobic conditions. The authors explained the absence of photosynthetic electron transport under anaerobic conditions by the presence of M and L subunits from R. denitrificans and the H subunit from R. capsulatus, which did not interact so as to stabilize the QB site or allow electron transport from QA to QB (91). The uncertainties surrounding the photosynthetic metabolism of aerobic phototrophic bacteria will undoubtedly be clarified by additional molecular biological investigations.
The recent discoveries of new species of aerobic phototrophic bacteria, the diversity of their LH systems and carotenoids, and the existence of Bchl a-containing Zn in A. rubrum have increased our appreciation of the great variability of biological photosynthesis and emphasize our poor understanding of how photosynthesis arose and diverged to result in the (undoubtedly incomplete) plethora of species that have so far been isolated. The answers to the questions raised in this section will enhance our understanding of the photometabolism of diverse phototrophic bacteria and the evolution of photosynthesis.
ACKNOWLEDGMENTS
We thank the following scientists for generously providing reprints and preprints and for helpful discussions: J. A. Fuerst, D. Garcia, H. Gest, V. Gorlenko, S. Hanada, S. Itoh, J. F. Imhoff, P. Lebaron, K. Shimada, E. Stackebrandt, K.-I. Takamiya, A. Vermeglio, and N. Wakao.
ADDENDUM IN PROOF
After this review was accepted for publication, we discovered a review on photosynthetic rhizobia by D. Fleischman and D. Kramer (Biochim. Biophys. Acta 1364:17–36, 1998).
REFERENCES
- 1.Aagaard J, Sistrom W R. Control of synthesis of reaction center bacteriochlorophyll in photosynthetic bacteria. Photochem Photobiol. 1972;15:209–225. doi: 10.1111/j.1751-1097.1972.tb06240.x. [DOI] [PubMed] [Google Scholar]
- 2.Albers H, Gottschalk G. Acetate metabolism in Rhodopseudomonas gelatinosa and several other Rhodospirillaceae. Arch Microbiol. 1976;111:45–49. doi: 10.1007/BF00446548. [DOI] [PubMed] [Google Scholar]
- 3.Arata H, Serikawa Y, Takamiya K I. Trimethylamine-N-oxide respiration by aerobic photosynthetic bacterium, Erythrobacter sp. OCh114. J Biochem. 1988;103:1011–1015. doi: 10.1093/oxfordjournals.jbchem.a122371. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong G A. Genetic analysis and regulation of carotenoid biosynthesis: structure and function of the crt genes and gene products. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1135–1157. [Google Scholar]
- 5.Bagnall K W. Selenium, tellurium, and polonium. In: Schmidt M, Siebert W, Bagnall K W, editors. The chemistry of sulfur, selenium, tellurium and polonium. New York, N.Y: Pergamon Press; 1975. pp. 935–1008. [Google Scholar]
- 6.Barbieri P, Bestetti G, Raniero D, Galli E. Mercury resistance in aromatic compound degrading Pseudomonas strains. FEMS Microbiol Ecol. 1996;20:185–194. [Google Scholar]
- 7.Baross J A. Hyperthermophilic Archaea: implications for the origin and early evolution of life at submarine hydrothermal vents. EOS Suppl. 1991;72:59–60. [Google Scholar]
- 8.Bauer C E, Buggy J, Mosley C. Control of photosystem genes in Rhodobacter capsulatus. Trends Genet Rev. 1993;9:56–60. doi: 10.1016/0168-9525(93)90188-N. [DOI] [PubMed] [Google Scholar]
- 9.Bentley R, Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol Rev. 1982;45:241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bias U, Truper H G. Species specific release of sulfur from adenylyl sulfate by ATP sulfurylase or ADP sulfurylase in the green sulfur bacteria Chlorobium limicola and Chlorobium vibrioforme. Arch Microbiol. 1987;147:406–410. [Google Scholar]
- 11.Bidigare R R, Ondrusek M E, Kennicutt M C, Iturriaga R, Harvey H R, Honam R W, Macko S A. Evidence for a photoprotective function for secondary carotenoids of snow algae. J Phycol. 1993;29:427–434. [Google Scholar]
- 12.Bissig I, Wagner-Huber R V, Brunisholz B A, Zuber H. Multiple antenna complex in various purple photosynthetic bacteria. In: Drews G, Dawes E A, editors. Molecular biology of membrane bound complexes in phototrophic bacteria. New York, N.Y: Plenum Press; 1990. pp. 199–210. [Google Scholar]
- 13.Blankenship R E. Origin and early evolution of photosynthesis. Photosynth Res. 1992;33:91–111. [PubMed] [Google Scholar]
- 14.Blasco R, Cardenas J, Castillo F. Acetate metabolism in purple non-sulfur bacteria. FEMS Microbiol Lett. 1989;58:129–132. [Google Scholar]
- 15.Blasco R, Cardenas J, Castillo F. Regulation of isocitrate lyase in Rhodobacter capsulatus E1F1. Curr Microbiol. 1991;22:73–76. [Google Scholar]
- 16.Bowyer J R, Crofts A R. On the mechanism of photosynthetic electron transfer in Rhodopseudomonas capsulata and Rhodopseudomonas sphaeroides. Biochim Biophys Acta. 1981;636:218–233. doi: 10.1016/0005-2728(81)90096-7. [DOI] [PubMed] [Google Scholar]
- 17.Britton G, Singh R K, Goodwin T W, Ben-Aziz A. The carotenoids of Rhodomicrobium vannielii (Rhodospirillaceae) and the effect of diphenylamine on the carotenoid composition. Phytochemistry. 1975;14:2427–2433. [Google Scholar]
- 18.Brock T D, Madigan M T, Martinko J M, Parker J. Biology of microorganisms. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1994. [Google Scholar]
- 19.Brockmann H, Knobloch G, Schweer J, Trowitzsch W. Die Alkoholkomponente des Bakteriochlorophill a aus Rhodospirillum rubrum. Arch Microbiol. 1973;90:161–164. [PubMed] [Google Scholar]
- 20.Brune D C. Sulfur compounds as photosynthetic electron donors. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 847–870. [Google Scholar]
- 21.Castenholz R W, Pierson B K. Ecology of thermophilic anoxygenic phototrophs. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 87–103. [Google Scholar]
- 22.Chernyad’ev I, Kondratieva E N, Doman N G. Assimilation of acetate by Rhodopseudomonas palustris. Mikrobiologiya. 1970;39:24–29. . (In Russian.) [PubMed] [Google Scholar]
- 23.Cogdell R J, Frank H A. How carotenoids function in photosynthetic bacteria. Biochim Biophys Acta. 1987;895:63–79. doi: 10.1016/s0304-4173(87)80008-3. [DOI] [PubMed] [Google Scholar]
- 24.Cogdell R J, Tyfe P K, Barret S J, Prince S M, Freer A A, Isaacs N W, McGlynn P, Hunter C N. The purple bacterial photosynthetic unit. Photosynth Res. 1996;48:55–63. doi: 10.1007/BF00040996. [DOI] [PubMed] [Google Scholar]
- 25.Collins M D, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol Rev. 1981;45:316–354. doi: 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad R, Schlegel H G. Different pathways for fructose and glucose utilization in Rhodopseudomonas capsulata and demonstration of I-phosphofructokinase in phototrophic bacteria. Biochim Biophys Acta. 1974;358:221–225. doi: 10.1016/0005-2744(74)90273-3. [DOI] [PubMed] [Google Scholar]
- 27.Conrad R, Schlegel H G. Different degradation pathways for glucose and fructose in Rhodopseudomonas capsulata. Arch Microbiol. 1977;112:39–48. doi: 10.1007/BF00446652. [DOI] [PubMed] [Google Scholar]
- 28.Conrad R, Schlegel H G. Influence of aerobic and phototrophic growth conditions on the distribution of glucose and fructose carbon into the Entner-Doudoroff and Embden-Meyerhof pathways in Rhodopseudomonas sphaeroides. J Gen Microbiol. 1977;101:277–290. [Google Scholar]
- 29.Conrad R, Schlegel H G. Metabolism of fructose in Thiocapsa roseopersicina. Z Allg Mikrobiol. 1978;18:309–320. doi: 10.1002/jobm.3630180502. [DOI] [PubMed] [Google Scholar]
- 30.Crofts A R, Wraight C A. The electrochemical domain of photosynthesis. Biochim Biophys Acta. 1983;726:149–185. [Google Scholar]
- 31.Dawes E A. Microbial energetics. Glasgow, United Kingdom: Blackie and Sons, Ltd.; 1986. [Google Scholar]
- 32.Dawkins D J, Ferguson L A, Cogdell R J. The structure of the core of the purple bacterial photosynthetic unit. In: Scheer H, Schneider S, editors. Photosynthetic light-harvesting systems. Berlin, Germany: Walter de Gruyter & Co.; 1988. pp. 115–127. [Google Scholar]
- 33.DeMoll-Decker H, Macy J M. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch Microbiol. 1993;160:241–247. [Google Scholar]
- 34.Doi M, Shioi Y. Two types of cytochrome cd1 in the aerobic photosynthetic bacterium, Erythrobacter sp. OCh114. Eur J Biochem. 1989;184:521–527. doi: 10.1111/j.1432-1033.1989.tb15045.x. [DOI] [PubMed] [Google Scholar]
- 35.Dracheva S M, Drachev L A, Konstantinov A A, Semenov A Y, Skulachev V D, Arvyjunjan A M, Shuvalov V A, Zabereshnaya S M. Electrogenic steps in the redox reactions catalysed by photosynthetic reaction-center complex from R. viridis. Eur J Biochem. 1988;171:253–264. doi: 10.1111/j.1432-1033.1988.tb13784.x. [DOI] [PubMed] [Google Scholar]
- 36.Drews G. Structure and functional organization of light-harvesting complexes and photochemical reaction centers in membranes of phototrophic bacteria. Microbiol Rev. 1985;49:59–70. doi: 10.1128/mr.49.1.59-70.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drews G. Energy transduction in phototrophic bacteria. In: Schlegel H G, Bowien B, editors. Biology of autotrophic bacteria. Madison, Wis: Science and Technology Publications; 1989. pp. 461–480. [Google Scholar]
- 38.Drews G, Golecki J R. Structure, molecular organization and biosynthesis of membranes of purple bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 231–257. [Google Scholar]
- 39.Dutton P L. Oxidation-reduction potential dependence of the interaction of cytochromes, bacteriochlorophyll and carotenoids at 77K in chromatophores of Chromatium D and Rhodopseudomonas gelatinosa. Biochim Biophys Acta. 1971;226:63–80. doi: 10.1016/0005-2728(71)90178-2. [DOI] [PubMed] [Google Scholar]
- 40.Ehrlich H L. Microbial life in presence of heavy metals. In: Ehrlich H L, editor. Microbial life in extremal conditions. Mir Publishing, Moscow, Russia. (Russian translation from English.) 1981. pp. 440–469. [Google Scholar]
- 41.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure of the photosynthetic reaction center from Rhodobacter sphaeroides at 2.65 Å resolution: cofactors and protein-cofactor interactions. Structure. 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 42.Evans M B, Hawthornthwaite A M, Cogdell R J. Isolation and characterization of the different B800-850 light-harvesting complexes from low- and high-light grown cells of Rhodopseudomonas palustris, strain 2.1.6. Biochim Biophys Acta. 1990;1016:71–76. [Google Scholar]
- 43.Evans W R, Fleischman D E, Calvert H E, Pyati P V, Alter G M, Rao N S S. Bacteriochlorophyll and photosynthetic reaction centers in Rhizobium strain BTA:1. Appl Environ Microbiol. 1990;56:3445–3449. doi: 10.1128/aem.56.11.3445-3449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feher G, Allen J P, Okamura M Y, Rees D C. Structure and function of bacterial photosynthetic reaction centers. Nature (London) 1989;339:111–116. [Google Scholar]
- 45.Fischer U. Enzymatic steps and dissimilatory sulfur metabolism by whole cells of anoxyphotobacteria. In: Satzman E S, Cooper W J, editors. Biogenic sulfur in the environment. Washington, D.C: American Chemical Society; 1989. pp. 262–279. [Google Scholar]
- 46.Francke C, Amesz J. The size of the photosynthetic unit in purple bacteria. Photosynth Res. 1995;46:347–352. doi: 10.1007/BF00020450. [DOI] [PubMed] [Google Scholar]
- 47.Freeman J C, Blankenship R E. Isolation and characterization of the membrane-bound cytochrome c-554 from the thermophilic green photosynthetic bacterium Chloroflexus aurantiacus. Photosynth Res. 1990;23:29–38. doi: 10.1007/BF00030060. [DOI] [PubMed] [Google Scholar]
- 48.Fuerst J A, Hawkins J A, Holmes A, Sly L I, Moore C J, Stackebrandt E. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from freshwater. Int J Syst Bacteriol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 49.Fukumori Y, Yamanaka T. Cytochrome aa3 from the aerobic photoheterotroph Erythrobacter longus: purification and enzymatic and molecular features. J Biochem. 1987;102:777–784. doi: 10.1093/oxfordjournals.jbchem.a122115. [DOI] [PubMed] [Google Scholar]
- 50.Gall A, Yurkov V, Cogdell R J, Vermeglio A, Robert B. The pigment-protein interactions of some unusual light-harvesting antennae: a Raman study. In: Mathis P, editor. Photosynthesis: from light to biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 251–254. [Google Scholar]
- 51.Garcia D, Richaud P, Breton J, Vermeglio A. Structure and function of the tetraheme cytochrome associated to the reaction centers of Roseobacter denitrificans. Biochimie. 1994;76:666–673. doi: 10.1016/0300-9084(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 52.Garcia D, Richaud P, Vermeglio A. The photoinduced cyclic electron transfer in whole cells of Rhodopseudomonas viridis. Biochim Biophys Acta. 1993;1144:295–301. [Google Scholar]
- 53.Garcia-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardiner A T, MacKenzie R C, Barrett S J, Kaiser K, Cogdell R J. The genes for the peripheral antenna complex apoproteins from Rhodopseudomonas acidophila 7050 form a multigene family. In: Murata N, editor. Research in photosynthesis. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 77–80. [Google Scholar]
- 55.Gest H. Photosynthetic and quasi-photosynthetic bacteria. FEMS Microbiol Lett. 1993;112:1–6. [Google Scholar]
- 56.Gogotov I N, Gorlenko V M. Influence of cultivation conditions on the composition of quinones in purple bacteria and freshwater Erythrobacteria. Microbiology (New York) 1995;64:654–656. [Google Scholar]
- 57.Gorlenko V M, Bonch-Osmolovskaya E A, Kompantseva E I, Starynin D A. Differentiation of microbial communities in connection with a change in the physicochemical conditions in thermophile springs. Microbiology (New York) 1987;56:314–322. [Google Scholar]
- 58.Gorlenko V M, Kompantseva E I, Puchkova N N. Influence of temperature on the prevalence of phototrophic bacteria in hot springs. Microbiology (New York) 1985;54:848–853. [Google Scholar]
- 59.Gosink J J, Herwig R P, Staley J T. Octadecabacter arcticus gen. nov., sp. nov., and O. antarcticus, sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from Polar Sea ice and water. Syst Appl Microbiol. 1997;20:356–365. [Google Scholar]
- 60.Gray K A, Daldal F. Mutational studies of the cytochrome bc1 complexes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 747–774. [Google Scholar]
- 61.Gusev M V, Mineeva L A. Mikrobiologiya. Moscow, Russia: Moscow University Publications; 1985. [Google Scholar]
- 62.Hagen C, Braune W, Greulich F. Functional aspects of secondary carotenoids in Haematococcus lacustis. IV. Protection from photodynamic damage. J Photochem Photobiol B Biol. 1993;20:153–160. [Google Scholar]
- 63.Hanada S, Kawase Y, Hiraishi A, Takaichi S, Matsuura K, Shimada K, Nagashima K V P. Porphyrobacter tepidarius sp. nov., a moderately thermophilic aerobic photosynthetic bacterium isolated from a hot spring. Int J Syst Bacteriol. 1997;47:408–413. doi: 10.1099/00207713-47-2-408. [DOI] [PubMed] [Google Scholar]
- 64.Hansen T A, van Gemerden H. Sulfide utilization by purple nonsulfur bacteria. Arch Microbiol. 1972;86:49–56. doi: 10.1007/BF00412399. [DOI] [PubMed] [Google Scholar]
- 65.Hansen T A, Veldkamp H. Rhodopseudomonas sulfidophila nov. sp., a new species of the purple nonsulfur bacteria. Arch Microbiol. 1973;92:45–58. doi: 10.1007/BF00409510. [DOI] [PubMed] [Google Scholar]
- 66.Harashima K, Hayasaki J, Ikari T, Shiba T. O2-stimulated synthesis of bacteriochlorophyll and carotenoids in marine bacteria. Plant Cell Physiol. 1980;21:1283–1294. [Google Scholar]
- 67.Harashima K, Kawazoe K, Yoshida I, Kamata H. Light-stimulated aerobic growth of Erythrobacter species OCh114. Plant Cell Physiol. 1987;28:365–374. [Google Scholar]
- 68.Harashima K, Nakada H. Carotenoids and ubiquinone in aerobically grown cells of an aerobic photosynthetic bacterium, Erythrobacter species OCh114. Agric Biol Chem. 1983;47:1057–1063. [Google Scholar]
- 69.Harashima K, Nakagava M, Murata N. Photochemical activity of bacteriochlorophyll in aerobically grown cells of heterotrophs, Erythrobacter species (OCh114) and Erythrobacter longus (OCh101) Plant Cell Physiol. 1982;23:185–193. [Google Scholar]
- 70.Harashima K, Takamiya K. Photosynthesis and photosynthetic apparatus. In: Harashima K, Shiba T, Murata N, editors. Aerobic photosynthetic bacteria. Berlin, Germany: Springer-Verlag; 1989. pp. 39–72. [Google Scholar]
- 71.Iba K, Takamiya K I. Action spectra for inhibition by light of accumulation of bacteriochlorophyll and carotenoid during aerobic growth of photosynthetic bacteria. Plant Cell Physiol. 1989;30:471–477. [Google Scholar]
- 72.Imhoff J F. Anoxygenic phototrophic bacteria. In: Austin B, editor. Methods in aquatic bacteriology. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 207–240. [Google Scholar]
- 73.Imhoff J F. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 74.Imhoff, J. F. 1997. Personal communication.
- 75.Imhoff J F, Bias-Imhoff U. Lipids, quinones and fatty acids of anoxygenic phototrophic bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 179–205. [Google Scholar]
- 76.Imhoff J F, Trüper H G. Purple nonsulfur bacteria. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams & Wilkins; 1989. pp. 1658–1661. [Google Scholar]
- 77.Itoh, S. 1997. Personal communication.
- 78.Joliot P, Vermeglio A, Joliot A. Evidence for supercomplexes between reaction centers, cytochrome c2 and cytochrome bc1 complex in Rhodobacter sphaeroides whole cells. Biochim Biophys Acta. 1989;975:336–345. [Google Scholar]
- 79.Kato S I, Urakami T, Komagata K. Quinone systems and cellular fatty acid composition in species of Rhodospirillaceae genera. J Gen Appl Microbiol. 1985;31:381–398. [Google Scholar]
- 80.Kawasaki H, Hoshino Y, Yamasato K. Phylogenetic diversity of phototrophic purple non-sulfur bacteria in the proteobacteria α group. FEMS Microbiol Lett. 1993;112:61–66. [Google Scholar]
- 81.Kelly D P. Physiology and biochemistry of unicellular sulfur bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Berlin, Germany: Springer-Verlag; 1987. pp. 193–217. [Google Scholar]
- 82.Keppen O I, Pedan L V, Rodova N A. Oxidation of thiosulfate by purple nonsulfur bacteria. Mikrobiologiya. 1980;49:682–685. . (In Russian.) [PubMed] [Google Scholar]
- 83.Kishimoto N, Fukaya F, Inagaki K, Sugio T, Tanaka H, Tano T. Distribution of bacteriochlorophyll a among aerobic and acidophilic bacteria and light-enhanced CO2-incorporation in Acidiphilium rubrum. FEMS Microbiol Ecol. 1995;16:291–296. [Google Scholar]
- 84.Kleinig H, Schmitt R, Meister W, Englert G, Thommen H. New C30-carotenoic acid glucosyl esters from Pseudomonas rhodos. Z Naturforsch. 1979;34:181–185. [Google Scholar]
- 85.Klevay L M. Pharmacology and toxicology of heavy metals: tellurium. Pharmacol Ther. 1976;1:223–229. [Google Scholar]
- 86.Klug G. Regulation of expression of photosynthesis genes in anoxygenic photosynthetic bacteria. Arch Microbiol. 1993;159:397–404. doi: 10.1007/BF00288584. [DOI] [PubMed] [Google Scholar]
- 87.Klug G, Gad’on N, Joch S, Narro M L. Light and oxygen effects share a common regulatory DNA sequence in Rhodobacter capsulatus. Mol Microbiol. 1991;5:1235–1239. doi: 10.1111/j.1365-2958.1991.tb01897.x. [DOI] [PubMed] [Google Scholar]
- 88.Kondratieva E N. Interrelation between modes of carbon assimilation and energy production in phototrophic purple and green bacteria. In: Quayle J R, editor. Microbial biochemistry. Baltimore, Md: University Park Press; 1979. pp. 117–249. [Google Scholar]
- 89.Kondratieva E N, Ivanovsky R N, Krasilnikova E N. Light and dark metabolism in purple sulfur bacteria. Sov. Sci. Rev. Biol. Rev. Sect. D. 1981. pp. 326–365. [Google Scholar]
- 90.Kondratieva E N, Maksimova I V, Samuilov V D. Photosynthetic microorganisms. Moscow, Russia: Moscow University Publications; 1989. . (In Russian.) [Google Scholar]
- 91.Kortluke C, Breese K, Gad’on N, Labahn A, Drews G. Structure of the puf operon of the obligately aerobic, bacteriochlorophyll a-containing bacterium Roseobacter denitrificans OCh114 and its expression in a Rhodobacter capsulatus puf puc deletion mutant. J Bacteriol. 1997;179:5247–5258. doi: 10.1128/jb.179.17.5247-5258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kramer D M, Schoepp B, Liebl U, Nitschke W. Cyclic electron transfer in Heliobacillus mobilis involving a menaquinol-oxidizing cytochrome bc complex and an RC1-type reaction center. Biochemistry. 1997;36:4203–4211. doi: 10.1021/bi962241i. [DOI] [PubMed] [Google Scholar]
- 93.Kranz R G, Beckman D L. Cytochrome biogenesis. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 709–723. [Google Scholar]
- 94.Krinsky N I. Carotenoid protection against oxidation. Pure Appl Chem. 1979;51:649–660. [Google Scholar]
- 95.Krinsky N I. Antioxidant functions of carotenoids. Free Radic Biol Med. 1989;7:617–635. doi: 10.1016/0891-5849(89)90143-3. [DOI] [PubMed] [Google Scholar]
- 96.Kulaev I S, Vagabov V M. Polyphosphate metabolism in microorganisms. Adv Microbiol. 1983;15:731–738. [Google Scholar]
- 97.Kuntzler A, Pfennig N. Das Vorkommen von Bacteriochlorophyll ap und aGg in Stammen aller Arten der Rhodospirillaceae. Arch Mikrobiol. 1973;91:83–86. [PubMed] [Google Scholar]
- 98.Kusai A, Yamanaka T. The oxidation mechanism of thiosulfate and sulfide in Chlorobium thiosulfatophilum: roles of cytochrome c-551 and cytochrome c-553. Biochim Biophys Acta. 1973;325:304–314. doi: 10.1016/0005-2728(73)90106-0. [DOI] [PubMed] [Google Scholar]
- 99.Lafay B, Ruimy R, Rausch de Traubenberg C, Breitmayer V, Gauthier M J, Christen R. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellata Prorocentum lima. Int J Syst Bacteriol. 1995;45:290–296. doi: 10.1099/00207713-45-2-290. [DOI] [PubMed] [Google Scholar]
- 100.Lancaster C R D, Michel H. Three-dimensional structures of photosynthetic reaction centers. Photosynth Res. 1996;48:65–74. doi: 10.1007/BF00040997. [DOI] [PubMed] [Google Scholar]
- 101.Langenhoff A A M, Bronwers-Ceiler D L, Engelberting J H L, Quist J J, Wolkenfelt J J P N, Zehnder A J B, Schraa G. Microbial reduction of manganese coupled to toluene oxidation. FEMS Microbiol Ecol. 1997;22:119–127. [Google Scholar]
- 102.Laverman A M, Blum J S, Schaefer J K, Phillips E J P, Lovley D R, Oremland R S. Growth of strain SES-3 with arsenate and other diverse electron acceptors. Appl Environ Microbiol. 1995;61:3556–3561. doi: 10.1128/aem.61.10.3556-3561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lebaron, P. 1997. Personal communication.
- 104.Liebetanz R, Hornberger U, Drews G. Organization of the genes coding for the reaction-centre L and M subunits and B870 antenna polypeptides α and β from the aerobic photosynthetic bacterium Erythrobacter species OCh114. Mol Microbiol. 1991;5:1459–1468. doi: 10.1111/j.1365-2958.1991.tb00792.x. [DOI] [PubMed] [Google Scholar]
- 105.LITE and Workshop Participants. RIDGE program report. Woods Hole, Mass: Oceanographic Institution; 1994. Light in thermal environments; p. 44. [Google Scholar]
- 106.Lloyd-Jones G, Osborn A M, Ritchie D A, Strike P, Hobman J L, Brown N L, Rouch D A. Accumulation and intracellular fate of tellurite in tellurite-resistant Escherichia coli: a model for the mechanism of resistance. FEMS Microbiol Lett. 1994;118:113–120. doi: 10.1111/j.1574-6968.1994.tb06812.x. [DOI] [PubMed] [Google Scholar]
- 107.Lloyd-Jones G, Ritchie D A, Strike P. Biochemical and biophysical analysis of plasmid pMJ600-encoded tellurite (TeO32−) resistance. FEMS Microbiol Lett. 1991;81:19–24. doi: 10.1016/0378-1097(91)90464-l. [DOI] [PubMed] [Google Scholar]
- 108.Lovley D R, Phillips E J P, Gorby Y A, Landa E R. Microbial reduction of uranium. Nature (London) 1991;350:413–416. [Google Scholar]
- 109.Lyalikova N N, Yurkova N A. Role of microorganisms in vanadium concentration and dispersion. Geomicrobiol J. 1992;10:15–26. [Google Scholar]
- 110.Macy J M, Lawson S, DeMoll-Decker H. Bioremediation of selenium oxyanions in San Joaquin drainage water using Thauera selenatis in a biological reactor system. Appl Microbiol Biotechnol. 1993;40:588–594. [Google Scholar]
- 111.Madigan M T. Microbiology, physiology and ecology of phototrophic bacteria. In: Zehnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley and Sons, Inc.; 1988. pp. 39–111. [Google Scholar]
- 112.Madigan M T, Gest H. Growth of the photosynthetic bacterium Rhodopseudomonas capsulata chemolithotrophically in darkness with H2 as the energy source. J Bacteriol. 1979;137:524–530. doi: 10.1128/jb.137.1.524-530.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Madigan M T, Martinko J M, Parker J. Brock biology of microorganisms. 8th ed. Upper Saddle River, N.J: Prentice-Hall; 1997. [Google Scholar]
- 114.Marrs B, Gest H. Regulation of bacteriochlorophyll synthesis by oxygen in respiratory mutants of Rhodopseudomonas capsulata. J Bacteriol. 1973;114:1052–1057. doi: 10.1128/jb.114.3.1052-1057.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mas J, van Gemerden H. Storage products in purple and green sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 973–990. [Google Scholar]
- 116.Mergeay M, Houba C, Gerits J. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, and zinc ions: evidence from curing in Pseudomonas. Arch Int Physiol Biochim. 1978;86:440–441. [PubMed] [Google Scholar]
- 117.Meyer T E, Donohue T J. Cytochromes, iron-sulfur and copper proteins mediating electron transfer from the cyt bc1 complex to photosynthetic reaction center complex. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 725–745. [Google Scholar]
- 118.Moore M D, Kaplan S. Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1505–1514. doi: 10.1128/jb.174.5.1505-1514.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moore M D, Kaplan S. Members of the family Rhodospirillaceae reduce heavy-metal oxyanions to maintain redox poise during photosynthetic growth. ASM News. 1994;60:17–23. [Google Scholar]
- 120.Nagashima K V P, Shimada K, Matsuura K. Phylogenetic analysis of photosynthetic genes of Rhodocyclus gelatinosus: possibility of horizontal gene transfer in purple bacteria. Photosynth Res. 1993;36:185–191. doi: 10.1007/BF00033037. [DOI] [PubMed] [Google Scholar]
- 121.Nagashima K V P, Hiraishi A, Shimada K, Matsuura K. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J Mol Evol. 1997;45:131–136. doi: 10.1007/pl00006212. [DOI] [PubMed] [Google Scholar]
- 122.Naveke A, Scheer H, Robert B, Sturgis J. Influence of metal exchange on absorption spectra of the light harvesting complex of Rhodospirillum rubrum. In: Mathis P, editor. Photosynthesis: from light to biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 259–262. [Google Scholar]
- 123.Neutzling O, Pfleiderer C, Truper H. Dissimilatory sulfur metabolism in phototrophic non-sulfur bacteria. J Gen Microbiol. 1985;131:791–798. [Google Scholar]
- 124.Nickens D, Fry C J, Ragatz L, Bauer C E, Gest H. Biotype of the purple nonsulfur photosynthetic bacterium, Rhodospirillum centenum. Arch Microbiol. 1996;165:91–96. doi: 10.1007/BF00262196. [DOI] [PubMed] [Google Scholar]
- 125.Nies D, Mergeay M, Friedrich B, Schlegel H G. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol. 1987;169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nishimura K, Shimada H, Ohta H, Masuda T, Shioi Y, Takamiya K I. Expression of the puf operon in an aerobic photosynthetic bacterium, Roseobacter denitrificans. Plant Cell Physiol. 1996;37:153–159. doi: 10.1093/oxfordjournals.pcp.a028926. [DOI] [PubMed] [Google Scholar]
- 127.Nitschke W, Dracheva S M. Reaction center associated cytochromes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 775–805. [Google Scholar]
- 128.Noguchi T, Hayashi H, Shimada K, Takaichi S, Tasumi M. In vivo states and function of carotenoids in an aerobic photosynthetic bacterium, Erythrobacter longus. Photosynth Res. 1992;31:21–30. doi: 10.1007/BF00049533. [DOI] [PubMed] [Google Scholar]
- 129.Oelze J, Drews G. Membranes of photosynthetic bacteria. Biochim Biophys Acta. 1972;265:209–239. doi: 10.1016/0304-4157(72)90003-2. [DOI] [PubMed] [Google Scholar]
- 130.Okamura K, Mitsuori F, Ito O, Takamiya K, Nishimura M. Photophosphorylation and oxidative phosphorylation in intact cells and chromatophores of an aerobic photosynthetic bacterium, Erythrobacter sp. strain OCh114. J Bacteriol. 1986;168:1142–1146. doi: 10.1128/jb.168.3.1142-1146.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Okamura K, Takamiya K, Nishimura M. Photosynthetic electron transfer system is inoperative in anaerobic cells of Erythrobacter species strain OCh114. Arch Microbiol. 1985;142:12–17. [Google Scholar]
- 132.Okamura K, Takamiya K I, Nishimura M. Photosynthetic and respiratory electron transfer systems in an aerobic photosynthetic bacterium Erythrobacter sp. strain OCh114. Adv Photosynth Res. 1984;1:641–644. [Google Scholar]
- 133.Okamura M Y, Feher G. Proton transfer in reaction centers from photosynthetic bacteria. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- 134.Oliveros E, Murasecco-Suardi P, Braun A M, Hansen H J. Efficiency of singlet oxygen quenching by carotenoids measured by near-infrared steady-state luminescence. Methods Enzymol. 1992;213:420–429. [Google Scholar]
- 135.Overfield R E, Wraight C A, De Vault D. Microsecond photooxidation kinetics of cytochrome c2 from Rhodopseudomonas sphaeroides: in vivo and solution studies. FEBS Lett. 1979;105:137–142. doi: 10.1016/0014-5793(79)80903-5. [DOI] [PubMed] [Google Scholar]
- 136.Patel N S, Britton G, Goodwin T W. Use of deuterium labelling from deuterium oxide to demonstrate carotenoid transformations in photosynthetic bacteria. Biochim Biophys Acta. 1983;760:92–96. [Google Scholar]
- 137.Petushkova Y P, Ivanovsky R N. Enzymes involved in thiosulfate metabolism in Thiocapsa roseopersicina during various conditions of growth. Mikrobiologiya. 1976;45:960–965. . (In Russian.) [PubMed] [Google Scholar]
- 138.Prescott L M, Harley J P, Klein D A. Industrial microbiology and biotechnology. In: Kane K, editor. Microbiology. Dubuque, Iowa: WCB Publishing; 1993. pp. 887–911. [Google Scholar]
- 139.Prince R C. Bacterial photosynthesis: from photons to Δp. The Bacteria. 1990;12:111–149. [Google Scholar]
- 140.Rodova N A, Pedan L V. Metabolism of thiosulfate in Rhodopseudomonas palustris. Mikrobiologiya. 1980;49:221–226. . (In Russian.) [PubMed] [Google Scholar]
- 141.Roy A B, Trudinger R A. The biochemistry of inorganic compounds of sulfur. Cambridge, United Kingdom: Cambridge University Press; 1970. [Google Scholar]
- 142.Scheer H, Hartwich G. Bacterial reaction centers with modified tetrapyrrole chromophores. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 649–663. [Google Scholar]
- 143.Shiba T. Utilization of light energy by the strictly aerobic bacterium Erythrobacter sp. OCh114. J Gen Appl Microbiol. 1984;30:239–244. [Google Scholar]
- 144.Shiba T. Roseobacter litoralis gen. nov., sp. nov. and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst Appl Microbiol. 1991;14:140–145. [Google Scholar]
- 145.Shiba T, Harashima K. Aerobic photosynthetic bacteria. Microbiol Sci. 1986;3:376–378. [PubMed] [Google Scholar]
- 146.Shiba T, Shioi Y, Takamiya K I, Sutton D C, Wilkinson C R. Distribution and physiology of aerobic bacteria containing bacteriochlorophyll a on the east and west coasts of Australia. Appl Environ Microbiol. 1991;57:295–300. doi: 10.1128/aem.57.1.295-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shiba T, Simidu U. Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a. Int J Syst Bacteriol. 1982;32:211–217. [Google Scholar]
- 148.Shiba T, Simidu U, Taga N. Distribution of aerobic bacteria which contain bacteriochlorophyll a. Appl Environ Microbiol. 1979;38:43–45. doi: 10.1128/aem.38.1.43-45.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shimada K. Aerobic anoxygenic phototrophs. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 105–122. [Google Scholar]
- 150.Shimada K, Hayashi H, Noguchi T, Tasumi M. Excitation and emission spectroscopy of membranes and pigment-protein complexes of an aerobic photosynthetic bacterium, Erythrobacter sp. Och114. Plant Cell Physiol. 1990;31:395–398. [Google Scholar]
- 151.Shimada K, Hayashi H, Tasumi M. Bacteriochlorophyll-protein complexes of aerobic bacteria, Erythrobacter longus and Erythrobacter species OCh114. Arch Microbiol. 1985;143:244–247. [Google Scholar]
- 152.Shimada K, Matsuura K, Itoh S, Iwaki M, Hiraishi A, Takaichi S, Kobayashi M, Wakao N. IX International Symposium on Phototrophic Prokaryotes, 1997, Vienna, Austria. 1997. Pigment-proteins of “Zn-bacteriochlorphyll” containing bacterium Acidiphilium rubrum, abstr. 102A; p. 102. [Google Scholar]
- 153.Shimada K, Yamazaki I, Tamai N, Mimuro M. Excitation energy flow in a photosynthetic bacterium lacking B850. Fast energy transfer from B806 to B870 in Erythrobacter sp. strain OCh114. Biochim Biophys Acta. 1990;1016:266–271. [Google Scholar]
- 154.Shioi Y, Doi M, Arata H, Takamiya K. A denitrifying activity in an aerobic photosynthetic bacterium, Erythrobacter sp. strain OCh114. Plant Cell Physiol. 1988;29:861–865. [Google Scholar]
- 155.Shiozawa J A, Welte W, Hodapp N, Drews G. Studies on the size and composition of the isolated light-harvesting B800-850 pigment protein complex of Rhodopseudomonas capsulata. Arch Biochem Biophys. 1982;213:473–485. doi: 10.1016/0003-9861(82)90573-2. [DOI] [PubMed] [Google Scholar]
- 156.Sleep N H. Projective impacts and life on early earth. EOS. 1991;72:59. [Google Scholar]
- 157.Smith A J. The role of tetrathionate in the oxidation of thiosulfate by Chromatium sp. strain D. J Gen Microbiol. 1966;42:371–380. doi: 10.1099/00221287-42-3-371. [DOI] [PubMed] [Google Scholar]
- 158.Stackebrandt, E. 1998. Personal communication.
- 159.Stackebrandt E, Rainey F A, Ward-Rainey N. Anoxygenic phototrophy across the phylogenetic spectrum: current understanding and future perspectives. Arch Microbiol. 1996;166:211–223. doi: 10.1007/s002030050377. [DOI] [PubMed] [Google Scholar]
- 160.Stowell M H B, McPhillips T M, Rees D C, Soltis S M, Abresch E, Feher G. Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science. 1997;276:812–816. doi: 10.1126/science.276.5313.812. [DOI] [PubMed] [Google Scholar]
- 161.Summers A O, Jacoby G A. Plasmid-determined resistance to tellurium compounds. J Bacteriol. 1977;129:276–281. doi: 10.1128/jb.129.1.276-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Summers A O, Silver S. Microbial transformation of metals. Annu Rev Microbiol. 1978;32:637–672. doi: 10.1146/annurev.mi.32.100178.003225. [DOI] [PubMed] [Google Scholar]
- 163.Suzina N E, Duda V I, Anisimova L A, Dmitriev V V, Boronin A M. Cytological aspects of resistance to potassium tellurite conferred on Pseudomonas cells by plasmids. Arch Microbiol. 1995;163:282–285. doi: 10.1007/BF00393381. [DOI] [PubMed] [Google Scholar]
- 164.Tabita F R. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 885–914. [Google Scholar]
- 165.Tadros M H, Garcia A F, Drews G, Gad’on N, Skatchkov M P. Isolation and characterization of a light-harvesting complex II lacking the γ-polypeptide from Rhodobacter capsulatus. Biochim Biophys Acta. 1990;1019:245–249. [Google Scholar]
- 166.Tadros M H, Katsiou E, Hoon M, Yurkova N, Ramji D P. Cloning of a new antenna gene cluster and expression analysis of the antenna gene family of Rhodopseudomonas palustris. Eur J Biochem. 1993;217:867–875. doi: 10.1111/j.1432-1033.1993.tb18315.x. [DOI] [PubMed] [Google Scholar]
- 167.Takaichi S, Furihata K, Harashima K. Light-induced changes of carotenoid pigments in anaerobic cells of the aerobic photosynthetic bacterium, Roseobacter denitrificans (Erythrobacter species OCh114): reduction of spheroidenone to 3,4-dihydrospheroidenone. Arch Microbiol. 1991;155:473–476. [Google Scholar]
- 168.Takaichi S, Furihata K, Ishidsu J I, Shimada K. Carotenoid sulphates from the aerobic photosynthetic bacterium, Erythrobacter longus. Phytochemistry. 1991;30:3411–3415. [Google Scholar]
- 169.Takaichi S, Shimada K, Ishidsu J I. Monocyclic cross-conjugated carotenal from an aerobic photosynthetic bacterium, Erythrobacter longus. Phytochemistry. 1988;27:3605–3609. [Google Scholar]
- 170.Takaichi S, Shimada K, Ishidsu J I. Carotenoids from the aerobic photosynthetic bacterium, Erythrobacter longus: β-carotene and its hydroxyl derivatives. Arch Microbiol. 1990;153:118–122. [Google Scholar]
- 171.Takamiya K, Shioi Y, Morita M, Arata H, Shimizu M, Doi M. Some properties and occurrence of cytochrome c-552 in the aerobic photosynthetic bacterium Roseobacter denitrificans. Arch Microbiol. 1993;159:51–56. doi: 10.1007/BF00244264. [DOI] [PubMed] [Google Scholar]
- 172.Takamiya K I. Cytochromes and respiratory systems. In: Harashima K, Shiba T, Murata N, editors. Aerobic photosynthetic bacteria. Berlin, Germany: Springer-Verlag; 1989. pp. 73–90. [Google Scholar]
- 173.Takamiya K I. Occurrence of cytochrome c-554 in a facultative methylotroph, Protaminobacter ruber strain NR-1, during bacteriochlorophyll formation. Arch Microbiol. 1995;143:15–19. [Google Scholar]
- 174.Takamiya K I, Arata H, Shioi Y, Doi M. Restoration of the optimal redox state for the photosynthetic electron transfer system by auxiliary oxidants in an aerobic photosynthetic bacterium, Erythrobacter sp. OCh114. Biochim Biophys Acta. 1988;935:26–33. [Google Scholar]
- 175.Takamiya K I, Iba K, Okamura K. Reaction center complex from an aerobic photosynthetic bacterium, Erythrobacter species OCh114. Biochim Biophys Acta. 1987;890:127–133. [Google Scholar]
- 176.Takamiya K I, Shioi Y, Shimada H, Arata H. Inhibition of accumulation of bacteriochlorophyll and carotenoids by blue light in an aerobic photosynthetic bacterium Roseobacter denitrificans, during anaerobic respiration. Plant Cell Physiol. 1992;33:1171–1174. [Google Scholar]
- 177.Takemoto J, Kao M Y. Effects of incident light levels on photosynthetic membrane polypeptide composition and assembly in Rhodopseudomonas sphaeroides. J Bacteriol. 1977;129:1102–1109. doi: 10.1128/jb.129.2.1102-1109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Taylor D E, Walter E G, Sherburne R, Bazett-Jones D P. Structure and location of tellurium deposited in Escherichia coli cells harboring tellurite resistance plasmids. J Ultrastruct Mol Struct Res. 1988;99:18–26. doi: 10.1016/0889-1605(88)90029-8. [DOI] [PubMed] [Google Scholar]
- 179.Then J, Truper H G. The role of thiosulfate in sulfur metabolism of Rhodopseudomonas globiformis. Arch Microbiol. 1981;130:143–146. [Google Scholar]
- 180.Truper H. Phototrophic bacteria and their sulfur metabolism. In: Muller A, Krebs B, editors. Sulfur, its significance for chemistry, for the geo-, bio-, and cosmosphere and technology. Amsterdam, The Netherlands: Elsevier; 1984. pp. 367–382. [Google Scholar]
- 181.Truper H G. Photolithotrophic sulfur oxidation. In: Bothe H, Trebst A, editors. Biology of inorganic nitrogen and sulfur. Berlin, Germany: Springer-Verlag; 1981. pp. 199–211. [Google Scholar]
- 182.Truper H G. Physiology and biochemistry of phototrophic bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Berlin, Germany: Springer-Verlag; 1987. pp. 267–281. [Google Scholar]
- 183.Turova T P, Burkal’tseva M V, Bulygina E S, Gorlenko V M. Phylogenetic position of freshwater Erythrobacteria studied by 5S rRNA analysis. Microbiology (New York) 1995;64:662–666. [Google Scholar]
- 184.Urakami T, Komagata K. Protomonas, a new genus of facultatively methyltrophic bacteria. Int J Syst Bacteriol. 1984;34:188–201. [Google Scholar]
- 185.Van Grondelle R, Duysens L N M, Van der Wal H N. Function of three cytochromes in photosynthesis of whole cells of Rhodospirillum rubrum as studied by flash spectroscopy. Evidence for two types of reaction center. Biochim Biophys Acta. 1976;449:169–187. doi: 10.1016/0005-2728(76)90131-6. [DOI] [PubMed] [Google Scholar]
- 186.Van Vliet P, Zannoni D, Nitschke W, Rutherford A W. Membrane-bound cytochromes in Chloroflexus aurantiacus studied by EPR. Eur J Biochem. 1991;199:317–323. doi: 10.1111/j.1432-1033.1991.tb16127.x. [DOI] [PubMed] [Google Scholar]
- 187.Vermeglio A, Joliot P, Joliot A. Organization of electron transfer components and supercomplexes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 279–295. [Google Scholar]
- 188.Vermeglio A, Richaud P, Breton J. Orientation and assignment of the four cytochrome hemes in Rhodopseudomonas viridis reaction centers. FEBS Lett. 1989;243:259–263. [Google Scholar]
- 189.Wakao N, Nagasawa N, Matsuura T, Matsukura H, Matsumoto T, Hiraishi A, Sakurai Y, Shiota H. Acidiphilium multivorum sp. nov., an acidophilic chemoorganotrophic bacterium from pyritic acid mine drainage. J Gen Appl Microbiol. 1994;40:143–159. [Google Scholar]
- 190.Wakao N, Shiba T, Hiraishi A, Ito M, Sakurai Y. Distribution of bacteriochlorophyll a in species of the genus Acidiphilium. Curr Microbiol. 1993;27:277–279. [Google Scholar]
- 191.Wakao N, Yokoi N, Isoyama N, Hiraishi A, Shimada K, Kobayashi M, Kise H, Iwaki M, Itoh S, Takaichi S, Sakurai Y. Discovery of natural photosynthesis using Zn-containing bacteriochlorophyll in an aerobic bacterium Acidiphilium rubrum. Plant Cell Physiol. 1996;37:889–896. [Google Scholar]
- 192.Wakim B, Golecki J R, Oelze J. The unusual mode of altering the cellular membrane content by Rhodospirillum tenue. FEMS Microbiol Lett. 1978;4:199–201. [Google Scholar]
- 193.Wakim B, Oelze J. The unique mode of adjusting the composition of the photosynthetic apparatus to different environmental conditions by Rhodospirillum tenue. FEMS Microbiol Lett. 1980;7:221–223. [Google Scholar]
- 194.Walter E G, Taylor D E. Plasmid-mediated resistance to tellurite: expressed and cryptic. Plasmid. 1992;27:52–64. doi: 10.1016/0147-619x(92)90006-v. [DOI] [PubMed] [Google Scholar]
- 195.Watanabe T, Kobayashi M. Electrochemistry of chlorophylls. In: Scheer H, editor. Chlorophylls. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 287–315. [Google Scholar]
- 196.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Woese C R, Stackebrandt E, Weisburg W G, Paster B J, Madigan M T, Fowler V J, Hahn C M, Blanz P, Gupta R, Nealson K H, Fox G E. The phylogeny of purple bacteria: the alpha subdivision. Syst Appl Microbiol. 1984;5:315–326. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- 198.Wood H G, Clark J E. Biological aspects of inorganic polyphosphates. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 199.Wood J M. Biological cycles for toxic elements in the environment. Science. 1974;4129:1049–1052. doi: 10.1126/science.183.4129.1049. [DOI] [PubMed] [Google Scholar]
- 200.Woodbury N W, Allen J P. The pathway, kinetics and thermodynamics of electron transfer in wild type and mutant reaction centers of purple nonsulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 527–557. [Google Scholar]
- 201.Yabuuchi E, Yano E, Oyizu H, Hashimoto Y, Ezaki T, Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov., and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34:99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 202.Yamanaka T. Group c cytochromes. In: Yamanaka T, editor. The biochemistry of bacterial cytochromes. Tokyo, Japan: Japanese Scientific Society Press; 1992. pp. 91–168. [Google Scholar]
- 203.Yildiz F H, Gest H, Bauer C E. Attenuated effect of oxygen on photopigment synthesis in Rhodospirillum centenum. J Bacteriol. 1991;173:5502–5506. doi: 10.1128/jb.173.17.5502-5506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yoch D C, Lindstrom E S. Survey of the photosynthetic bacteria for rhodanese (thiosulfate: cyanide sulfur transferase) activity. J Bacteriol. 1971;106:700–701. doi: 10.1128/jb.106.2.700-701.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Young J P W, Downer H L, Eardly B D. Phylogeny of the phototrophic Rhizobium strain BTA:1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yurkov V. Ph.D. thesis. Moscow, Russia: Academy of Sciences; 1990. [Google Scholar]
- 207.Yurkov, V. 1998. Unpublished data.
- 208.Yurkov V, Beatty J T. Isolation of aerobic anoxygenic photosynthetic bacteria from black smoker plume waters of the Juan de Fuca Ridge in the Pacific Ocean. Appl Environ Microbiol. 1998;64:337–341. doi: 10.1128/aem.64.1.337-341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yurkov, V., and T. Beatty. 1998. Unpublished data.
- 210.Yurkov V, Gad’on N, Angerhofer A, Drews G. Light-harvesting complexes of aerobic bacteriochlorophyll-containing bacteria Roseococcus thiosulfatophilus, RB3 and Erythromicrobium ramosum, E5 and the transfer of excitation energy from carotenoids to bacteriochlorophyll. Z Naturforsch. 1994;49(c):579–586. [Google Scholar]
- 211.Yurkov V, Gad’on N, Drews G. The major part of polar carotenoids of the aerobic bacteria Roseococcus thiosulfatophilus, RB3 and Erythromicrobium ramosum, E5 is not bound to the bacteriochlorophyll a complexes of the photosynthetic apparatus. Arch Microbiol. 1993;160:372–376. [Google Scholar]
- 212.Yurkov V, Gorlenko V M. Erythrobacter sibiricus sp. nov., a new freshwater aerobic bacterial species containing bacteriochlorophyll a. Microbiology (New York) 1990;59:85–89. [Google Scholar]
- 213.Yurkov V, Gorlenko V M. Ecophysiological peculiarities of phototrophic microbial communities of Bolsherechensky thermal springs. Microbiology (New York) 1992;61:115–122. [Google Scholar]
- 214.Yurkov V, Gorlenko V M. A new genus of freshwater aerobic, bacteriochlorophyll a-containing bacteria, Roseococcus gen. nov. Microbiology (New York) 1992;60:628–632. [Google Scholar]
- 215.Yurkov V, Gorlenko V M. New species of aerobic bacteria from the genus Erythromicrobium containing bacteriochlorophyll a. Microbiology (New York) 1993;61:163–168. [Google Scholar]
- 216.Yurkov V, Gorlenko V M, Kompantseva E I. A new type of freshwater aerobic orange-coloured bacterium containing bacteriochlorophyll a, Erythromicrobium gen. nov. Microbiology (New York) 1992;61:169–172. [Google Scholar]
- 217.Yurkov V, Gorlenko V M, Mityushina L L, Starynin D A. Effect of limiting factors on the structure of phototrophic associations in thermal springs. Microbiology (New York) 1992;60:129–138. [Google Scholar]
- 218.Yurkov V, Jappe J, Vermeglio A. Tellurite resistance and reduction by obligately aerobic photosynthetic bacteria. Appl Environ Microbiol. 1996;62:4195–4198. doi: 10.1128/aem.62.11.4195-4198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yurkov V, Krasilnikova E N, Gorlenko V M. Enzymes involved in heterotrophic carbon metabolism of aerobic Erythrobacter sibiricus and Erythrobacter longus, bacteria containing bacteriochlorophyll a. Microbiology (New York) 1992;60:401–403. [Google Scholar]
- 220.Yurkov V, Krasilnikova E N, Gorlenko V M. Effect of light and oxygen on metabolism of the aerobic bacterium Erythromicrobium sibiricum. Microbiology (New York) 1993;62:35–38. [Google Scholar]
- 221.Yurkov V, Krasilnikova E N, Gorlenko V M. Thiosulfate metabolism in aerobic bacteriochlorophyll-a containing bacteria. Microbiology (New York) 1994;63:181–188. [Google Scholar]
- 222.Yurkov, V., S. Krieger, and T. Beatty. 1998. Unpublished data.
- 223.Yurkov V, Lysenko A M, Gorlenko V M. Hybridization analysis of the classification of bacteriochlorophyll a-containing freshwater aerobic bacteria. Microbiology (New York) 1991;60:362–366. [Google Scholar]
- 224.Yurkov, V., L. Menin, B. Schoepp, and A. Vermeglio. Purification and characterization of reaction centers of the obligately aerobic anoxygenic phototrophic bacteria Erythrobacter litoralis, Erythromicrobium ursincola and Erythromicrobium sibiricum. Photosynth. Res., in press.
- 225.Yurkov V, Mityushina L L, Gorlenko V M. Ultrastructure of the aerobic bacterium Erythrobacter sibiricus, which contains bacteriochlorophyll a. Microbiology (New York) 1991;60:234–238. [Google Scholar]
- 226.Yurkov V, Schoepp B, Vermeglio A. Electron transfer carriers in obligately aerobic photosynthetic bacteria from genera Roseococcus and Erythromicrobium. In: Matthis P, editor. Photosynthesis: from light to biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 543–546. [Google Scholar]
- 227.Yurkov, V., B. Schoepp, and A. Vermeglio. Comparative study of the photosynthetic apparatus and electron carriers among obligate aerobic photosynthetic bacteria from genera Erythromicrobium, Erythromonas, Erythrobacter, Roseococcus, and Sandaracinobacter. Photosynth. Res., in press.
- 228.Yurkov V, Stackebrandt E, Beatty T. IX International Symposium on Phototrophic Prokaryotes, 1997, Vienna, Austria. 1997. New obligately aerobic anoxygenic photosynthetic bacteria isolated from “black smoker” plume waters of the Juan de Fuca Ridge in the Pacific Ocean, abstr. 190B; p. 190. [Google Scholar]
- 229.Yurkov V, Stackebrandt E, Buss O, Vermeglio A, Gorlenko V M, Beatty J T. Reorganization of the genus Erythromicrobium: description of “Erythromicrobium sibiricum” as Sandaracinobacter sibiricus, gen. nov., sp. nov., and “Erythromicrobium ursincola” as Erythromonas ursincola, gen. nov., sp. nov. Int J Syst Bacteriol. 1997;47:1172–1178. doi: 10.1099/00207713-47-4-1172. [DOI] [PubMed] [Google Scholar]
- 230.Yurkov V, Stackebrandt E, Holmes A, Fuerst J A, Hugenholtz P, Golecki J, Gad’on N, Gorlenko V M, Kompantseva E I, Drews G. Phylogenetic positions of novel aerobic, bacteriochlorophyll a-containing bacteria and description of Roseococcus thiosulfatophilus gen. nov., sp. nov., Erythromicrobium ramosum gen. nov., sp. nov., and Erythrobacter litoralis sp. nov. Int J Syst Bacteriol. 1994;44:427–434. doi: 10.1099/00207713-44-3-427. [DOI] [PubMed] [Google Scholar]
- 231.Yurkov V, van Gemerden H. Abundance and salt tolerance of obligately aerobic, phototrophic bacteria in a microbial mat. Neth J Sea Res. 1993;31:57–62. [Google Scholar]
- 232.Yurkov V, van Gemerden H. Impact of light/dark regime on growth rate, biomass formation and bacteriochlorophyll synthesis in Erythromicrobium hydrolyticum. Arch Microbiol. 1993;159:84–89. [Google Scholar]
- 233.Yurkov, V. V., and A. Vermeglio. 1998. Unpublished data.
- 234.Yurkova N A, Lyalikova N N. New vanadate-reducing facultative chemolithotrophic bacteria. Mikrobiologiya. 1990;59:968–975. . (In Russian.) [Google Scholar]
- 235.Zannoni D. Aerobic and anaerobic electron transport chains in anoxygenic phototrophic bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 949–971. [Google Scholar]
- 236.Zuber H, Cogdell R G. Structure and organization of purple bacterial antenna complexes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 315–348. [Google Scholar]