Abstract
Introduction
Celiac disease (CD) is an autoimmune gastrointestinal disorder causes immune-mediated enteropathy against gluten. Gluten immunogenic peptides have the potential to trigger immune responses which leads to damage the small intestine. HLA-DQ2/DQ8 are major alleles that bind to epitope/antigenic region of gluten and induce celiac disease. There is a need to identify CD associated epitopes in protein-based foods and therapeutics.
Methods
In this study, computational tools have been developed to predict CD associated epitopes and motifs. Dataset used for training, testing and evaluation contain experimentally validated CD associated and non-CD associate peptides. We perform positional analysis to identify the most significant position of an amino acid residue in the peptide and checked the frequency of HLA alleles. We also compute amino acid composition to develop machine learning based models. We also developed ensemble method that combines motif-based approach and machine learning based models.
Results and Discussion
Our analysis support existing hypothesis that proline (P) and glutamine (Q) are highly abundant in CD associated peptides. A model based on density of P&Q in peptides has been developed for predicting CD associated peptides which achieve maximum AUROC 0.98 on independent data. We discovered motifs (e.g., QPF, QPQ, PYP) which occurs specifically in CD associated peptides. We also developed machine learning based models using peptide composition and achieved maximum AUROC 0.99. Finally, we developed ensemble method that combines motif-based approach and machine learning based models. The ensemble model-predict CD associated motifs with 100% accuracy on an independent dataset, not used for training. Finally, the best models and motifs has been integrated in a web server and standalone software package “CDpred”. We hope this server anticipate the scientific community for the prediction, designing and scanning of CD associated peptides as well as CD associated motifs in a protein/peptide sequence (https://webs.iiitd.edu.in/raghava/cdpred/).
Keywords: celiac disease, gluten immunogenic peptides, HLA-DQ2/DQ8, ensemble method, motif
1. Introduction
Celiac disease (CD) is an auto-immunological disorder which mainly affects the small intestine of the infected person (1). CD is a life-long disorder occurred due to the gluten associated foods which is found in various foods such as wheat, barley, spelt, kamut, and rye (2). The prevalence rate of CD is around 1.4% worldwide and it may vary with genetic and environmental factors. The occurrence of disease is significantly higher in children in comparison to adults (3). Various studies revealed that celiac disease patients develop inflammatory immune responses against gluten peptides. The innate immune responses cause toxic effects by gluten peptides on the intestinal epithelium due to increased production of cytokines such as interleukin-15 (4–7). However, the presence of certain class-II human leukocyte antigens (HLAs) molecules play a crucial role in the induction and regulation of immunological responses. The binding of gluten peptides with the HLA-DQ2/DQ8 receptors activates the adaptive immune responses (8). Whereas, HLA-DQ2 found in almost 94.5% of CD cases and HLA-DQ8 present in 2.7% of the cases (9). These binders are also linked with other autoimmunological disorders such as HLA-DQ8 associated with Type I diabetes (10).
As depicted in Figure 1 , the entry of gluten inside the lamina propria region of small intestine follows transcellular and paracellular pathways (11). In transcellular pathway, the entry of gluten is associated with the binding of secretory IgA (sIgA) in the apical region of intestine (12). However, in the paracellular pathway, the entry of gluten is associated with the binding of chemokine receptor 3 (CXCR3) present at enterocyte with the release of zonulin protein (13, 14). After entering inside the lamina propria region, a series of events trigger an inflammatory cascade which leads to the excessive release of antibodies (anti-tissue transglutaminase, anti-IgA antibodies and anti-endomysial antibodies) and cytokines (15) and ends with damage to the intestinal villi.
Figure 1.

Schematic representation of celiac disease pathogenesis and immune response.
Due to auto-inflammatory immune responses several gastrointestinal disorders like malabsorption, vomiting, bloating, diarrhoea, abdominal pain and distension occurred (16). Recently, a number of biological and genetic tests (such as detection of antibodies, intestinal tissue biopsy, HLA-typing and gluten challenge test) are available for the disease detection (1). It has been found in many studies that α-gliadin 33-mer peptide having the property of resistant to gastrointestinal cleavage and makes it highly immunogenic peptide (5, 17–19). Despite tremendous understanding of CD, effective treatment for the disease is life-long gluten free diet. In order to manage severity of CD effectively, it is important to identify CD associated epitopes or immunogenic peptides responsible for CD. Identification of CD associated epitopes/peptides is not only important for identifying CD free food/therapeutic proteins, it is also important for designing antigen-based immunotherapy against CD.
In the pilot study, we have developed a computational approach for the prediction of CD associated peptides. We have extracted the experimentally validated CD-associated peptides from the IEDB database. In order to create negative dataset, we have collected CD non-causing peptides and random peptides from IEDB and Swiss-Prot, respectively. We have identified highly conserved regions of disease-causing peptides using motif-based search. In addition, we have developed prediction models using composition-based features and machine learning algorithms. In order to facilitate the community, we have provided the webserver and standalone package for the prediction and scanning of CD causing protein/peptides using sequence information.
2. Material and methods
The complete architecture of our study is illustrated in Figure 2 . The detail of each step is described below.
Figure 2.
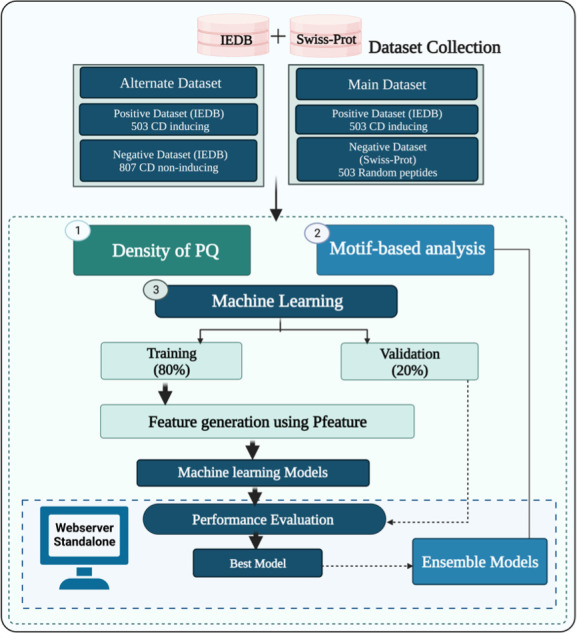
Overall architecture of the study.
2.1. Dataset collection and pre-processing
In this study, we have collected experimentally validated peptides from the immune epitope database (IEDB) (20). At first, we extracted a total of 521 unique celiac disease (CD) associated peptides from IEDB as a positive dataset. Further, we have selected unique peptides with a length of 9-20 amino-acid residues and got 503 CD associated peptides. Secondly, we extracted experimentally validated CD non-associated peptides from IEDB and random peptides from Swiss-Prot database (21). The main dataset incorporates 503 CD associated called positive peptides and 503 random peptides called negative peptides. The alternate dataset consists of 503 CD associated and 807 non-associated peptides (which can cause autoimmune disorders other than celiac disease). Finally, we obtained two datasets, i.e., the main dataset comprises an equal number of positive and negative peptides and alternate dataset 503 positive and 807 negative peptides.
2.2. Sequence logo
In order to understand the preference of amino-acid residues at a specific position, we have generated a one sample logo using WebLogo software (22). This tool needs a fixed length input sequence vector. Since, the minimum length of peptides in our datasets is 9 residues, so we have extracted 9-mers from N-terminal and 9-mers from C-terminal from each peptide. After that, we re-join both the regions in order to create a fixed length vector of 18 amino acids. The sequences of 18-residues were generated for all the peptides of both positive and negative datasets and used for the creation of one sample logo plots.
2.3. Amino-acid composition
We have used Pfeature (23) software for the computation of composition-based features. In this current study, we have computed amino acid composition based (AAC) features. In the case of AAC, the composition of each residue is computed in the peptide sequences and a vector od 20 length is generated using the equation 1.
| [1] |
Where, AACi is amino-acid composition of residue type i, Ri is the number of residues in i, and L is the length of peptide sequence.
2.4. Machine learning models
We have employed a number of machine learning algorithms for the classification of CD-causing peptides. Currently, we have used Scikit-learn (24) python library for the implementation of several classifiers including Decision Tree (DT), Random Forest (RF), XGBoost (XGB), Gaussian Naïve Bayes (GNB) Logistic Regression (LR), ExtraTree classifier (ET), and k-nearest neighbors (KNN).
2.5. Five-fold cross validation
In order to avoid overfitting, we have train, test and validate the machine learning models by employing five-fold cross validation technique as implemented in previous studies (25–28). At first, the complete dataset was divided into 80:20 ratio, where 80% dataset used for the training and 20% used for the external validation (29, 30). The five-fold cross-validation process is implemented on the 80% training dataset. In this process, the entire training dataset was divided into five equal sets, where each set is used for training and validation purpose. At first, four sets were used for training and fifth set was used for the testing, similarly the process is repeated five times so that each set can be used as testing dataset. Finally, we calculated the average performance of five sets which resulted after five iterations.
2.6. Model evaluation
In this study, we have used standard parameters for the evaluation of prediction models. Here, we have calculated both threshold dependent as well as independent parameters. In the case of threshold-dependent parameters we have computed, sensitivity (Sens), specificity (Spec), accuracy (Acc) and Matthews correlation coefficient (MCC) using the following equations (1-4). In addition, we have measured the performance of models with a well-established and threshold-independent parameter Area Under the Receiver Operating Characteristic (AUROC) curve.
| [2] |
| [3] |
| [4] |
| [5] |
| [6] |
Where, TP, TN, FP and FN stand for true positive, true negative, false positive and false negative, respectively.
2.7. Ensemble method
The ensemble method is a hybrid approach in which both motifs based, and machine learning methods combined to achieve better performance. In this method, first motif-based approach is used to identify the disease-causing peptides and then we use machine learning methods to predict those peptides which are not covered by the motif-based approach. Finally, we generate an ensemble method which is a combination of both motif-based approach and machine learning method.
2.8. Web implementation
We have developed a webserver named “CDpred” for the prediction of CD associated peptides. The webserver is implemented by HTML5, JAVA, CSS3 and PHP scripts and compatible on several devices such as iMac, desktop, tablet and mobile. The webserver provides five user-friendly modules such as predict, PQ density, motif scan, protein scan, and design.
3. Results
3.1. Positional conservation analysis
The specific position of a residue is important for specific role and structure arrangement of a particular peptide or protein. To identify the most significant position of an amino acid residue in the peptide, we perform the positional analysis of CD causing peptides and CD non-causing peptides by using WebLogo (See Figure 3 ). It is worth noting that the first nine locations correspond to peptide N-terminal residues, whereas the latter nine positions correspond to peptide C-terminus. Here, we found that the proline (P) and glutamine (Q) residues are highly prominent at every position while the Phenylalanine (F) and glutamic acid (E) are also found at some positions.
Figure 3.
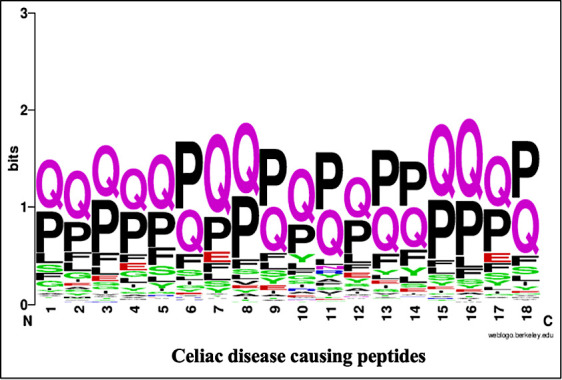
WebLogo of celiac disease-causing peptides.
3.2. Composition analysis
We compute the amino acid composition for main and alternate datasets. Figure 4 depicts the average composition of CD inducing and non-inducing peptides. In CD causing peptides, the average composition of Proline (P), Glutamine (Q) and Phenylalanine (F) is higher in comparison with disease non-causing peptides, negative random and general proteome.
Figure 4.
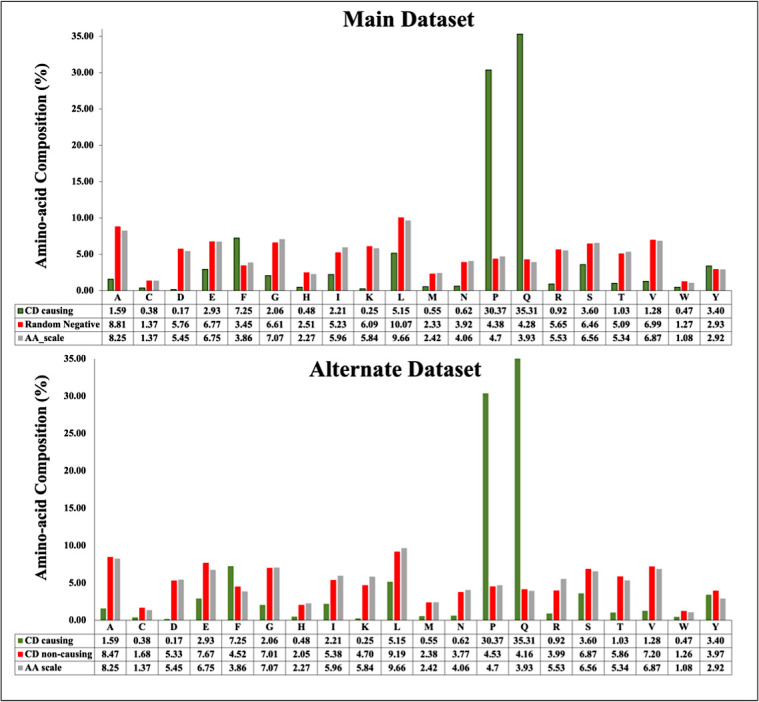
Average amino acid composition of peptides in main dataset and alternate dataset.
3.3. Frequency of HLA alleles
In the past, a number of studies report that celiac disease occurred due to the presence of certain HLA molecules such as HLA-DQ2 and HLA-DQ8 (29, 31, 32). Sallese et al. indicated that, in addition to HLA-DQ2 and HLA-DQ8, non-HLA variants are also associated to CD susceptibility (33). As shown in Table 1 , we observed that maximum CD-associated peptides are HLA-DQ2/DQ8 binders, while some of the CD-associated peptides are linked with other HLA-alleles. This shows that innate (non-HLA-DQ mediated) and adaptive (HLA-DQ mediated) immune responses can be caused by gluten peptides. The complete frequency distribution of HLA-alleles binders of CD causing and non-causing peptides are given in Supplementary Table S1 .
Table 1.
Distribution of HLA alleles in CD causing and non-causing peptides.
| HLA | CD causing (Positive) | CD non-causing (Negative) | |
|---|---|---|---|
| HLA-class I | HLA-A | 13 | 0 |
| HLA-class II | HLA-DQ2 | 263 | 148 |
| HLA-DQ8 | 18 | 110 | |
| HLA-DQ2/DQ8 | 24 | 9 | |
| HLA-DR | 3 | 402 | |
| Other | 182 | 138 | |
| Total | 503 | 807 | |
3.4. Motif-based analysis
Motifs are known as the specific regions of a protein sequence which helps to identify the amino acid arrangement shared by a family of protein. The motifs are identified in the CD causing peptide sequences by MERCI program. The MERCI program helps to identify the motif regions in a set of sequences. We utilized the MERCI tool to look for motifs seen only in CD-causing peptides and not in disease non-causing or random peptides. We also looked for motifs found only in disease non-causing and random peptides. Here, we found 50 motifs in CD causing peptides of different length in which P and Q residues are present in abundance in CD causing peptides. We also checked the common motifs found in disease causing, non-causing and random negative peptides. The list of motifs and their occurrence in all the three datasets are given in Table 2 .
Table 2.
Abundance of motifs in CD-causing, non-causing and random negative peptides.
| Motifs | Positive | Random Negative | Non-CD-causing |
|---|---|---|---|
| QPF | 276 | 0 | 4 |
| QQPF | 170 | 0 | 1 |
| PYP | 120 | 0 | 3 |
| PEQ | 56 | 0 | 4 |
| QPQ | 350 | 1 | 0 |
| PQPQ | 189 | 1 | 0 |
| QQPQ | 131 | 1 | 0 |
| PQL | 84 | 1 | 0 |
3.5. PQ density
On performing the compositional and motif analysis, it was found that (P) and (Q) are the most abundant residues in CD-causing peptides as compared to non-causing peptides. In order to classify the peptides based on the PQ density, we have first generated the overlapping patterns of window size ranging from 3 to 9 for each peptide, since 9 was the minimum length of the peptides, and calculated the composition of residues P and Q in each pattern. Each peptide in the dataset is represented by the maximum value of composition for the respective pattern size and found the optimal composition at which we can classify the peptides with balanced sensitivity and specificity. To find the optimal pattern size, we have varied the size from 3 to 9, and found out that window size 5 and 6 performed best among the other sizes for main and alternate datasets, respectively as shown in Table 3 .
Table 3.
Performance of the PQ abundance-based method on different window sizes.
| Main Data | |||||
|---|---|---|---|---|---|
| Window size | Threshold | Sensitivity | Specificity | Accuracy | AUROC |
| 3 | 0.670 | 85.686 | 98.807 | 92.247 | 0.971 |
| 4 | 0.510 | 91.650 | 96.620 | 94.135 | 0.977 |
| 5 | 0.410 | 93.837 | 94.235 | 94.036 | 0.978 |
| 6 | 0.340 | 95.427 | 92.445 | 93.936 | 0.978 |
| 7 | 0.290 | 96.421 | 91.650 | 94.036 | 0.979 |
| 8 | 0.380 | 93.241 | 97.018 | 95.129 | 0.981 |
| 9 | 0.340 | 94.235 | 96.620 | 95.427 | 0.981 |
| Alternate Data | |||||
| Window size | Threshold | Sensitivity | Specificity | Accuracy | AUROC |
| 3 | 0.670 | 85.686 | 98.761 | 93.740 | 0.970 |
| 4 | 0.510 | 91.650 | 97.770 | 95.420 | 0.977 |
| 5 | 0.410 | 93.837 | 96.159 | 95.267 | 0.979 |
| 6 | 0.340 | 95.427 | 94.796 | 95.038 | 0.980 |
| 7 | 0.290 | 96.421 | 94.300 | 95.115 | 0.981 |
| 8 | 0.260 | 97.018 | 92.937 | 94.504 | 0.983 |
| 9 | 0.340 | 94.235 | 98.017 | 96.565 | 0.982 |
3.6. Machine learning based prediction
Various machine learning classifiers such as RF, DT, GNB, XGB, KNN, ETN, SVCN and LR are used to develop a prediction model. For this, we have computed the features of disease causing and disease non-causing peptides using composition-based module of Pfeature.
3.6.1. Performance of AAC based features
Firstly, we have computed features of amino acid composition, using which we applied different machine learning techniques. As shown in Table 4 , ET achieves maximum performance in comparison to other models with AUROC 0.991 and 0.995 and accuracy 96.02 and 97.03 on both training and validation dataset with a good balance of sensitivity and specificity in main data. Similarly, ET achieves maximum performance in comparison to other models with AUROC 0.995 and 0.999 and accuracy 97.519 and 98.092 on both training and validation dataset with a good balance of sensitivity and specificity in alternate data.
Table 4.
The performance of machine learning classifiers on AAC based features on main and alternate dataset.
| Main dataset | ||||||||
|---|---|---|---|---|---|---|---|---|
| Training | Validation | |||||||
| Classifier | Sensitivity | Specificity | Accuracy | AUROC | Sensitivity | Specificity | Accuracy | AUROC |
| DT | 92.269 | 92.556 | 92.413 | 0.962 | 97.059 | 91.000 | 94.059 | 0.982 |
| RF | 95.262 | 95.533 | 95.398 | 0.989 | 98.039 | 97.000 | 97.525 | 0.994 |
| LR | 96.010 | 96.030 | 96.020 | 0.988 | 98.039 | 96.000 | 97.030 | 0.990 |
| XGB | 95.761 | 95.782 | 95.771 | 0.987 | 98.039 | 93.000 | 95.545 | 0.995 |
| KNN | 95.262 | 95.285 | 95.274 | 0.986 | 97.059 | 96.000 | 96.535 | 0.991 |
| GNB | 93.017 | 98.263 | 95.647 | 0.976 | 93.137 | 98.000 | 95.545 | 0.990 |
| ET | 96.010 | 96.030 | 96.020 | 0.991 | 98.039 | 96.000 | 97.030 | 0.995 |
| SVC | 95.761 | 95.782 | 95.771 | 0.987 | 97.059 | 96.000 | 96.535 | 0.991 |
| Alternate dataset | ||||||||
| Training | Validation | |||||||
| DT | 92.537 | 92.570 | 92.557 | 0.968 | 94.059 | 99.379 | 97.328 | 0.990 |
| RF | 97.015 | 97.368 | 97.233 | 0.995 | 98.020 | 97.516 | 97.710 | 0.998 |
| LR | 96.269 | 96.285 | 96.279 | 0.990 | 97.030 | 96.273 | 96.565 | 0.987 |
| XGB | 97.015 | 97.059 | 97.042 | 0.992 | 99.010 | 93.168 | 95.420 | 0.998 |
| KNN | 95.771 | 95.975 | 95.897 | 0.992 | 98.020 | 95.652 | 96.565 | 0.995 |
| GNB | 92.537 | 97.059 | 95.324 | 0.977 | 96.040 | 96.273 | 96.183 | 0.983 |
| ET | 97.512 | 97.523 | 97.519 | 0.995 | 98.020 | 98.137 | 98.092 | 0.999 |
| SVC | 97.015 | 96.904 | 96.947 | 0.993 | 98.020 | 96.894 | 97.328 | 0.996 |
# DT, Decision tress; RF, Random Forest; LR, Logistic regression; XGB, XGBoost; KNN, k-nearest neighbour; GNB, Gaussian naïve base; ET, Extra tree classifier; SVC, support vector classifier.
3.6.2. Performance of ensemble model
In ensemble method, first we used the motif-based approach by identifying the coverage of motifs in the given protein/peptide sequences. Our motif-based approach achieves 81.71% accuracy in the independent dataset as shown in Table 5 . The rest sequences, which were not predicted using motif-based approach, were covered by using the machine learning method. By combining both approaches we achieve the highest performance of 100% accuracy on independent dataset. Our ensemble method is the best approach for predicting the CD associated peptides.
Table 5.
The table shows the occurrence of motif in positive sequences with their cumulative coverage.
| Motif | Occurrence | Percentage | Cumulative |
|---|---|---|---|
| QPF | 276 | 54.87 | 54.87 |
| PQQP | 41 | 8.15 | 63.02 |
| PYP | 33 | 6.56 | 69.58 |
| QPQQ | 28 | 5.57 | 75.15 |
| PFP | 14 | 2.78 | 77.93 |
| PEQ | 12 | 2.39 | 80.32 |
| FPQP | 4 | 0.8 | 81.11 |
| FPQQ | 2 | 0.4 | 81.51 |
| PQLP | 1 | 0.2 | 81.71 |
| ML Prediction | 92 | 18.29 | 100 |
3.7. Services to scientific community
We design a user-friendly prediction web server that incorporates several modules to determine CD-causing peptides in order to serve the scientific community. The prediction models used in the study are implemented in the web server. Based on the prediction models’ score at a different threshold, users can predict whether a query peptide causes CD or not. The web server comprises five major modules 1) Prediction, 2) PQ Density, 3), Motif 4) Scan and 5) Design. The user can classify CD-causing peptides from disease non-causing peptides using the ‘Predict’ module. The “PQ Density” module used to calculate PQ content in a given query sequence based upon the window size. Users can map or scan CD-causing motifs in the query sequence using the “Motif” module. We used the MERCI software to extract themes from CD-causing peptides that had been empirically confirmed. The “Scan” module was used to scan the amino-acid sequence for CD-causing areas. The user can generate all potential analogs of the input sequence using the “Design” module. The positive and negative datasets utilized in this work are also available for download, and the peptide sequence are available in FASTA format. HTML, JAVA, and PHP scripts were used to create the web server CDpred https://webs.iiitd.edu.in/raghava/cdpred/. The server is user-friendly and compatible with a variety of devices, including computers, Android phones, iPhones, and iPads. In addition, we provided a standalone package in the form of a Docker container.
4. Case study: Evaluation of CDpred on external dataset
In this study, we evaluate our model by utilizing a new dataset (i.e., CD-associated peptide sequences) obtained from AllergenOnline database (http://www.allergenonline.org) (34) under celiac disease section (35). A total of 1040 unique experimentally validated CD-associated peptide were collected from AllergenOnline. We found 265 common sequences with our dataset (used for developing CDpred), hence we removed those sequences and left with 775 unique CD-associated new sequences. After that, we evaluate the performance of CDpred on independent dataset of 775 peptides using default parameters of “Ensemble module” and achieved 100% accuracy. Our method predicts all 775 CD-associated peptides correctly, where 661 peptides were predicted using Motif based approach and 114 using machine learning based approach.
5. Discussion and conclusion
Celiac disease is a chronic, genetically predisposed enteropathy triggered by gluten showing a wide spectrum of clinical manifestations (5, 29). It can be associated to a number of diseases such as cirrhosis, autoimmune hepatitis, diabetes mellitus, gluten ataxia, peripheral neuropathies, etc (36, 37). Moreover, CD is not limited to gastrointestinal tract, in fact it is associated with a number of extra-intestinal manifestations and other autoimmune disorders (38–41). The origins of the onset and appearance of related diseases may vary; for example, type 1 diabetes mellitus (T1D), share a common genetic basis; while others may have similar pathogenic pathways. Granito et al., also showed a significant correlation between anti-microfilament IgA and severity of intestinal damage in CD patients (42). Recent studies reported that, celiac patients have an elevated risk of getting small bowel cancer and intestinal lymphomas (43, 44).
Some autoimmune neurological manifestations including cerebral ataxia, peripheral neuropathy, epilepsy, dementia, and depression are associated with CD (45–49). Volta et al., found that there is a significant correlation between anti-ganglioside antibodies and neurological disorders in CD patients (50). Cervio et al., observed that antigliadin and anti-tTG antibodies in CD patients are linked to the development of neurological disorders (51). Moreover, immune-related diseases are also occurred due to gluten intake for example, atopy (52). The only effective lifelong treatment of this disease is a gluten-free diet. Due to increased number of cases in worldwide a number of gluten-free products are available for celiac susceptible people (18, 53). Thus, it is essential to identify or eliminate gluten immunogenic peptides from the food products which can induce the celiac disease and sensitive to celiac patients.
In this study, we have made a systematic attempt for the prediction of peptides responsible for causing the disease. We have collected the dataset from IEDB and Swiss-Prot databases. We have created two datasets for the analysis and prediction of CD causing peptides. The positive dataset contain experimentally validated peptides obtained from IEDB that are responsible for celiac disease. These peptides are not only gluten peptides (high frequency of P & Q) but also associated with celiac disease.
In addition, we have created alternate dataset where we have taken negative set contain peptides which cause diseases other than celiac disease. This is not necessary that all the gluten peptides are responsible for the celiac disease. There are few gluten peptides which showed toxic effect on intestinal epithelium cells and induce innate immune response (4). L. Maiuri et al. also showed the effect of gluten peptides on mucosal surface of the celiac patients and healthy individuals (54, 55). In this study, we observed that amino acid residues (P and Q) are highly abundant in CD causing peptides in comparison with negative and random peptides. The similar findings are supported by the previous studies where they have shown the abundance of P and Q amino acids in gluten proteins (56, 57). From the motif-based approach we identified certain motifs (QPQ, QPF, PQPQ, QQPF, QQPQ, PYP), which are highly conserved in CD causing peptides in comparison with CD non-causing peptides. So, we performed PQ based analysis where we calculate the abundance of PQ residues in the CD causing and non-causing peptides. In addition, we have developed prediction models using amino-acid composition-based features. We achieved maximum performance with AUROC of 0.99 on the training and validation datasets, respectively. We have also developed an ensemble method by combining both motif-based approach and machine learning based models. This ensemble approach provides the 100% accuracy on independent dataset. In addition, we have developed a webserver named CDpred (https://webs.iiitd.edu.in/raghava/cdpred/), standalone package (https://webs.iiitd.edu.in/raghava/cdpred/standalone.php) and GitLab (https://gitlab.com/raghavalab/cdpred) for the prediction of CD causing peptides.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author contributions
RT, AD and GR collected and processed the datasets. RT, SP and GPSR implemented the algorithms and developed the prediction models. RT, AD, SP and GR analysed the results. RT and SP created the back-end of the web server the front-end user interface. RT, AD, and GR penned the manuscript. GPSR conceived and coordinated the project. All authors contributed to the article and approved the submitted version.
Acknowledgments
Authors are thankful to the Department of Bio-Technology (DBT) and Department of Science and Technology (DST-INSPIRE) for fellowships and the financial support and Department of Computational Biology, IIITD New Delhi for infrastructure and facilities.
Funding Statement
The current work has received grant from the Department of Bio-Technology (DBT), Govt. of India, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1056101/full#supplementary-material
References
- 1. Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, et al. Coeliac disease. Nat Rev Dis Primers (2019) 5:3. doi: 10.1038/s41572-018-0054-z [DOI] [PubMed] [Google Scholar]
- 2. Caio G, Volta U, Sapone A, Leffler DA, De Giorgio R, Catassi C, et al. Celiac disease: a comprehensive current review. BMC Med (2019) 17:142. doi: 10.1186/s12916-019-1380-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, et al. Global prevalence of celiac disease: Systematic review and meta-analysis. Clin Gastroenterol Hepatol (2018) 16:823–836 e822. doi: 10.1016/j.cgh.2017.06.037 [DOI] [PubMed] [Google Scholar]
- 4. Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J, et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity (2004) 21:367–77. doi: 10.1016/j.immuni.2004.06.018 [DOI] [PubMed] [Google Scholar]
- 5. Ciccocioppo R, Di Sabatino A, Corazza GR. The immune recognition of gluten in coeliac disease. Clin Exp Immunol (2005) 140:408–16. doi: 10.1111/j.1365-2249.2005.02783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abadie V, Jabri B. IL-15: a central regulator of celiac disease immunopathology. Immunol Rev (2014) 260:221–34. doi: 10.1111/imr.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunne MR, Byrne G, Chirdo FG, Feighery C. Coeliac disease pathogenesis: The uncertainties of a well-known immune mediated disorder. Front Immunol (2020) 11:1374. doi: 10.3389/fimmu.2020.01374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monsuur AJ, Wijmenga C. Understanding the molecular basis of celiac disease: What genetic studies reveal. Ann Med (2006) 38:578–91. doi: 10.1080/07853890600989054 [DOI] [PubMed] [Google Scholar]
- 9. Stankovic B, Radlovic N, Lekovic Z, Ristic D, Radlovic V, Nikcevic G, et al. HLA genotyping in pediatric celiac disease patients. Bosn J Basic Med Sci (2014) 14:171–6. doi: 10.17305/bjbms.2014.3.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Z, Reyes-Vargas E, Escobar H, Rudd B, Rockwood AL, Delgado JC, et al. Type 1 diabetes associated HLA-DQ2 and DQ8 molecules are relatively resistant to HLA-DM mediated release of invariant chain-derived CLIP peptides. Eur J Immunol (2016) 46:834–45. doi: 10.1002/eji.201545942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khaleghi S, Ju JM, Lamba A, Murray JA. The potential utility of tight junction regulation in celiac disease: focus on larazotide acetate. Therap Adv Gastroenterol (2016) 9:37–49. doi: 10.1177/1756283X15616576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heyman M, Menard S. Pathways of gliadin transport in celiac disease. Ann N Y Acad Sci (2009) 1165:274–8. doi: 10.1111/j.1749-6632.2009.04032.x [DOI] [PubMed] [Google Scholar]
- 13. Sander GR, Cummins AG, Henshall T, Powell BC. Rapid disruption of intestinal barrier function by gliadin involves altered expression of apical junctional proteins. FEBS Lett (2005) 579:4851–5. doi: 10.1016/j.febslet.2005.07.066 [DOI] [PubMed] [Google Scholar]
- 14. Drago S, El Asmar R, Di Pierro M, Grazia Clemente M, Tripathi A, Sapone A, et al. Gliadin, zonulin and gut permeability: Effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand J Gastroenterol (2006) 41:408–19. doi: 10.1080/00365520500235334 [DOI] [PubMed] [Google Scholar]
- 15. Gujral N, Freeman HJ, Thomson AB. Celiac disease: Prevalence, diagnosis, pathogenesis and treatment. World J Gastroenterol (2012) 18:6036–59. doi: 10.3748/wjg.v18.i42.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taylor AK, Lebwohl B, Snyder CL, Green PHR. “Celiac disease,”. In: Adam MP, Everman DB, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews((R)). (Seattle, WA:IntechOpen; ) (1993). [Google Scholar]
- 17. Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot (2002) 53:947–58. doi: 10.1093/jexbot/53.370.947 [DOI] [PubMed] [Google Scholar]
- 18. Bascunan KA, Vespa MC, Araya M. Celiac disease: understanding the gluten-free diet. Eur J Nutr (2017) 56:449–59. doi: 10.1007/s00394-016-1238-5 [DOI] [PubMed] [Google Scholar]
- 19. Schalk K, Lang C, Wieser H, Koehler P, Scherf KA. Quantitation of the immunodominant 33-mer peptide from alpha-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci Rep (2017) 7:45092. doi: 10.1038/srep45092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. R. V, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res (2019) 47:D339–43. doi: 10.1093/nar/gky1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res (2003) 31:365–70. doi: 10.1093/nar/gkg095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res (2004) 14:1188–90. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pande A, Patiyal S, Lathwal A, Arora C, Kaur D, Dhall A, et al. Computing wide range of protein/peptide features from their sequence and structure. bioRxiv (2019) 599126-599126. doi: 10.1101/599126 [DOI] [Google Scholar]
- 24. Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res (2011) 12:2825–30. doi: 10.48550/arXiv.1201.0490 [DOI] [Google Scholar]
- 25. Dhall A, Patiyal S, Kaur H, Bhalla S, Arora C, Raghava GPS. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front Genet (2020) 11:221. doi: 10.3389/fgene.2020.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patiyal S, Agrawal P, Kumar V, Dhall A, Kumar R, Mishra G, et al. NAGbinder: An approach for identifying n-acetylglucosamine interacting residues of a protein from its primary sequence. Protein Sci (2020) 29:201–10. doi: 10.1002/pro.3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dhall A, Patiyal S, Sharma N, Usmani SS, Raghava GPS. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Brief Bioinform (2021) 22:936–45. doi: 10.1093/bib/bbaa259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patiyal S, Dhall A, Raghava GPS. A deep learning-based method for the prediction of DNA interacting residues in a protein. Brief Bioinform (2022) 23(5):bbac322. doi: 10.1093/bib/bbac322 [DOI] [PubMed] [Google Scholar]
- 29. Sharma N, Patiyal S, Dhall A, Pande A, Arora C, Raghava GPS. AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes. Brief Bioinform (2020) 22(4):bbaa294. doi: 10.1093/bib/bbaa294 [DOI] [PubMed] [Google Scholar]
- 30. Dhall A, Patiyal S, Sharma N, Devi NL, Raghava GPS. Computer-aided prediction of inhibitors against STAT3 for managing COVID-19 associated cytokine storm. Comput Biol Med (2021) 137:104780. doi: 10.1016/j.compbiomed.2021.104780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in celiac disease predisposition: practical implications of the HLA molecular typing. J BioMed Sci (2012) 19:88. doi: 10.1186/1423-0127-19-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siddiqui K, Uqaili AA, Rafiq M, Bhutto MA. Human leukocyte antigen (HLA)-DQ2 and -DQ8 haplotypes in celiac, celiac with type 1 diabetic, and celiac suspected pediatric cases. Med (Baltimore) (2021) 100:e24954. doi: 10.1097/MD.0000000000024954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sallese M, Lopetuso LR, Efthymakis K, Neri M. Beyond the HLA genes in gluten-related disorders. Front Nutr (2020) 7:575844. doi: 10.3389/fnut.2020.575844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goodman RE, Ebisawa M, Ferreira F, Sampson HA, Van Ree R, Vieths S, et al. AllergenOnline: A peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol Nutr Food Res (2016) 60:1183–98. doi: 10.1002/mnfr.201500769 [DOI] [PubMed] [Google Scholar]
- 35. Amnuaycheewa P, Abdelmoteleb M, Wise J, Bohle B, Ferreira F, Tetteh AO, et al. Development of a sequence searchable database of celiac disease-associated peptides and proteins for risk assessment of novel food proteins. Front Allergy (2022) 3:900573. doi: 10.3389/falgy.2022.900573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jabri B. Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nat Rev Immunol (2009) 9:858–70. doi: 10.1038/nri2670 [DOI] [PubMed] [Google Scholar]
- 37. Lauret E, Rodrigo L. Celiac disease and autoimmune-associated conditions. Biomed Res Int (2013) 2013:127589. doi: 10.1155/2013/127589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rothfuss KS, Stange EF, Herrlinger KR. Extraintestinal manifestations and complications in inflammatory bowel diseases. World J Gastroenterol (2006) 12:4819–31. doi: 10.3748/wjg.v12.i30.4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laurikka P, Nurminen S, Kivela L, Kurppa K. Extraintestinal manifestations of celiac disease: Early detection for better long-term outcomes. Nutrients (2018) 10:1015. doi: 10.3390/nu10081015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nardecchia S, Auricchio R, Discepolo V, Troncone R. Extra-intestinal manifestations of coeliac disease in children: Clinical features and mechanisms. Front Pediatr (2019) 7:56. doi: 10.3389/fped.2019.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Popp A, Maki M. Gluten-induced extra-intestinal manifestations in potential celiac disease-celiac trait. Nutrients (2019) 11(2):320. doi: 10.3390/nu11020320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Granito A, Muratori P, Cassani F, Pappas G, Muratori L, Agostinelli D, et al. Anti-actin IgA antibodies in severe coeliac disease. Clin Exp Immunol (2004) 137:386–92. doi: 10.1111/j.1365-2249.2004.02541.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Gils T, Nijeboer P, Overbeek LI, Hauptmann M, Castelijn DA, Bouma G, et al. Risks for lymphoma and gastrointestinal carcinoma in patients with newly diagnosed adult-onset celiac disease: Consequences for follow-up: Celiac disease, lymphoma and GI carcinoma. United Eur Gastroenterol J (2018) 6:1485–95. doi: 10.1177/2050640618800540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marafini I, Monteleone G, Stolfi C. Association between celiac disease and cancer. Int J Mol Sci (2020) 21(11):4155. doi: 10.3390/ijms21114155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gobbi G, Bouquet F, Greco L, Lambertini A, Tassinari CA, Ventura A, et al. Coeliac disease, epilepsy, and cerebral calcifications. the Italian working group on coeliac disease and epilepsy. Lancet (1992) 340:439–43. doi: 10.1016/0140-6736(92)91766-2 [DOI] [PubMed] [Google Scholar]
- 46. Alaedini A, Green PH, Sander HW, Hays AP, Gamboa ET, Fasano A, et al. Ganglioside reactive antibodies in the neuropathy associated with celiac disease. J Neuroimmunol (2002) 127:145–8. doi: 10.1016/S0165-5728(02)00102-9 [DOI] [PubMed] [Google Scholar]
- 47. Chin RL, Sander HW, Brannagan TH, Green PH, Hays AP, Alaedini A, et al. Celiac neuropathy. Neurology (2003) 60:1581–5. doi: 10.1212/01.WNL.0000063307.84039.C7 [DOI] [PubMed] [Google Scholar]
- 48. Sander HW, Magda P, Chin RL, Wu A, Brannagan TH, 3rd, Green PH, et al. Cerebellar ataxia and coeliac disease. Lancet (2003) 362:1548. doi: 10.1016/S0140-6736(03)14743-5 [DOI] [PubMed] [Google Scholar]
- 49. Pengiran Tengah DS, Holmes GK, Wills AJ. The prevalence of epilepsy in patients with celiac disease. Epilepsia (2004) 45:1291–3. doi: 10.1111/j.0013-9580.2004.54104.x [DOI] [PubMed] [Google Scholar]
- 50. Volta U, De Giorgio R, Granito A, Stanghellini V, Barbara G, Avoni P, et al. Anti-ganglioside antibodies in coeliac disease with neurological disorders. Dig Liver Dis (2006) 38:183–7. doi: 10.1016/j.dld.2005.11.013 [DOI] [PubMed] [Google Scholar]
- 51. Cervio E, Volta U, Verri M, Boschi F, Pastoris O, Granito A, et al. Sera of patients with celiac disease and neurologic disorders evoke a mitochondrial-dependent apoptosis in vitro . Gastroenterology (2007) 133:195–206. doi: 10.1053/j.gastro.2007.04.070 [DOI] [PubMed] [Google Scholar]
- 52. Zauli D, Grassi A, Granito A, Foderaro S, De Franceschi L, Ballardini G, et al. Prevalence of silent coeliac disease in atopics. Dig Liver Dis (2000) 32:775–9. doi: 10.1016/S1590-8658(00)80354-0 [DOI] [PubMed] [Google Scholar]
- 53. Rai S, Kaur A, Chopra CS. Gluten-free products for celiac susceptible people. Front Nutr (2018) 5:116. doi: 10.3389/fnut.2018.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maiuri L, Troncone R, Mayer M, Coletta S, Picarelli A, De Vincenzi M, et al. In vitro activities of a-gliadin-related synthetic peptides: damaging effect on the atrophic coeliac mucosa and activation of mucosal immune response in the treated coeliac mucosa. Scand J Gastroenterol (1996) 31:247–53. doi: 10.3109/00365529609004874 [DOI] [PubMed] [Google Scholar]
- 55. Molberg O, Mcadam SN, Korner R, Quarsten H, Kristiansen C, Madsen L, et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med (1998) 4:713–7. doi: 10.1038/nm0698-713 [DOI] [PubMed] [Google Scholar]
- 56. Alves TO, D’almeida CTS, Ferreira MSL. Determination of gluten peptides associated with celiac disease by mass spectrometry. IntechOpen; (2017) 2017. [Google Scholar]
- 57. Jitendra Kumar, Kumar M, Pandey R, Chauhan NS. Physiopathology and management of gluten-induced celiac disease. Journal of food science (2017) 82(2):270–77. doi: 10.1111/1750-3841.13612 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.


