Abstract
Insertion sequences (ISs) constitute an important component of most bacterial genomes. Over 500 individual ISs have been described in the literature to date, and many more are being discovered in the ongoing prokaryotic and eukaryotic genome-sequencing projects. The last 10 years have also seen some striking advances in our understanding of the transposition process itself. Not least of these has been the development of various in vitro transposition systems for both prokaryotic and eukaryotic elements and, for several of these, a detailed understanding of the transposition process at the chemical level. This review presents a general overview of the organization and function of insertion sequences of eubacterial, archaebacterial, and eukaryotic origins with particular emphasis on bacterial elements and on different aspects of the transposition mechanism. It also attempts to provide a framework for classification of these elements by assigning them to various families or groups. A total of 443 members of the collection have been grouped in 17 families based on combinations of the following criteria: (i) similarities in genetic organization (arrangement of open reading frames); (ii) marked identities or similarities in the enzymes which mediate the transposition reactions, the recombinases/transposases (Tpases); (iii) similar features of their ends (terminal IRs); and (iv) fate of the nucleotide sequence of their target sites (generation of a direct target duplication of determined length). A brief description of the mechanism(s) involved in the mobility of individual ISs in each family and of the structure-function relationships of the individual Tpases is included where available.
General Scope
Since the publication in 1989 of the volume Mobile DNA (29), the field of transposition has experienced some striking advances. Not the least of these has been the development of various in vitro transposition systems for both prokaryotic and eukaryotic elements and an understanding of the transposition process at the chemical level for several of these (for reviews, see references 66, 179, 239, and 266). Another development has been the veritable explosion in the number of different transposable elements isolated and characterized at the nucleotide sequence level. In the case of bacterial insertion sequences (IS), only approximately 50 had been analysed at this level in 1989 (107), compared to over 500 today. This is equally true of eukaryotic “insertion sequences” such as mariner, derivatives of which have been found in over 240 insect species in addition to fungi, mammals, fish, and plants (295), and related elements such as Tc1 (87, 274). This enormous diversity and distribution is astonishing. The number of different ISs has now become so great that there is a growing need for a framework to enable a systematic classification. We have attempted here to provide a classification of these elements by assigning them to various families or groups based on similarities and differences in structure, organization, and nucleotide and protein sequence relationships. We also include a brief description, where available, of the mechanism(s) involved in the mobility of individual ISs and of the structure-function relationships of the enzymes which mediate the transposition reactions, the recombinases/transposases (Tpases). Most of the elements of this framework are already available but scattered throughout the literature.
In assembling this review, we have limited our treatment to IS elements, which we loosely define as small (<2.5-kb), phenotypically cryptic segments of DNA with a simple genetic organization and capable of inserting at multiple sites in a target molecule. In this definition, we voluntarily eliminate several types of mobile genetic element. These include elements with RNA as intermediates such as the retroviruses, retrotransposons and retroposons (102, 315), DNA elements such as the conjugative transposons (306) which use a phage λ-type mechanism for their translocation, and elements such as bacteriophage Mu (239), Tn7 (66), and transposons of the Tn554 type (249), which are large and relatively complex. We have also eliminated an additional large and coherent group of elements, the type II transposons of the Tn3 family (373). Although several are small and might qualify as insertion sequences [e.g., Tn1000 (γδ) and IS101 (324) and IS1071 (86a, 251)], many are complicated in structure and include multiple antibiotic resistance genes carried by another type of transposable element, the integron (278).
Although they are not considered here in detail, many of these elements have significant functional similarities to ISs and have provided many of the important insights into transposition mechanism. Reference to these is included where they may prove useful to understanding the behavior of IS elements.
Nomenclature
Several systems of nomenclature are in operation. One, initiated in 1978 (201) and centralized by E. Lederberg (Stanford University), attributes a single number to an IS element (e.g., IS1). While adequate when only few ISs were known, this system does not include sufficient information and becomes less transparent with the large numbers of elements known today. A second system, which provides some information about the source of the element, includes the initials of the bacterial species from which it was isolated (e.g. ISRm1 for Rhizobium meliloti). At present, we are confronted by both types of nomenclature in the literature. In addition, closely related elements with only few differences at the nucleotide sequence level are in some cases designated isoforms of the parent element and sometimes attributed a specific number. Finally, some elements have been baptized with names that fit none of these rules (e.g., RSalpha-9). Where appropriate, we have simplified certain of the more complicated assignments by adopting the nomenclature which includes the initials of the host bacterial species.
Sources of Bacterial Insertion Sequences
Bacterial insertion sequences were initially identified during studies of model genetic systems by their capacity to generate mutations as a result of their translocation. Interest in antibiotic resistance and transmissible plasmids subsequently revealed an important role for these mobile elements in dissemination of resistance genes and in promotion of gene acquisition. In particular, it was observed that several different elements were often clustered in “islands” within plasmid genomes and served to promote plasmid integration and excision (46). In addition, two copies of certain ISs flanking a DNA segment were found to be able to act in concert, rendering the intervening region mobile. These structures were termed composite or compound transposons (29). The influence of ISs on bacterial pathogenicity and virulence could also have been anticipated at that time by the isolation of an IS1-based compound transposon specifying a heat-stable toxin (318). With the development of studies of the mechanisms of bacterial pathogenesis over recent years, the finding of association between ISs and many pathogenic and virulence functions has become increasingly frequent. Such associations have been observed in animal pathogens (e.g., Bacillus [225], Bordetella, Brucella, Campylobacter, Clostridium [45, 65], Escherichia [61, 109, 148, 318], Haemophilus [88], Neisseria [135], Vibrio [327], and Yersinia [99, 101, 122, 130]), plant pathogens (e.g., Agrobacterium [258], Erwinia, and Pseudomonas), and symbionts (e.g., Rhizobium [104]). Another area which has received increasing attention over recent years is that of bioremediation. Here, too, insertion sequences can be clearly associated with genes forming parts of degradative or catabolic pathways (49), an observation foreshadowed by the early identification of transposons carrying genes permitting citrate utilization (159). Association of ISs with these biological processes does not imply that the processes are in any way “special.” It presumably simply reflects the (evolving) interests of the investigator. What these examples do underscore, however, is the importance of transposition mechanisms in assembling sets of “accessory” functions in bacteria.
It should not be forgotten, of course, that ISs are involved in phenomena other than the acquisition of accessory functions. Many form an integral part of the chromosomes of most bacterial species, where they have been shown, for example, to participate in chromosome rearrangements (see, e.g., references 128, 215, and 301) and in plasmid integration (reference 215a and references therein). In certain cases, the localization of different specific IS elements at defined places in the chromosome is sufficiently stable to allow them to be used as markers in restriction fragment length polymorphism studies for species typing and for epidemiological purposes (e.g., IS6100 in Mycobacterium tuberculosis [316], IS1296 in Mycoplasma mycoides [58], IS200 in Salmonella [323], and IS1004 in Vibrio cholerae [35]). One exception, which has as yet exhibited no ISs, is the common laboratory strain of Bacillus subtilis (372), whose entire genomic sequence has recently been determined (189).
GENERAL FEATURES AND PROPERTIES OF INSERTION SEQUENCE ELEMENTS
Organization
In addition to being small, insertion sequences are genetically compact (Fig. 1). They generally encode no functions other than those involved in their mobility. These include factors required in cis, in particular recombinationally active DNA sequences which define the ends of the element, together with an enzyme, the Tpase, which recognizes and processes these ends. The Tpase is generally encoded by one or perhaps two open reading frames and consumes nearly the entire length of the element.
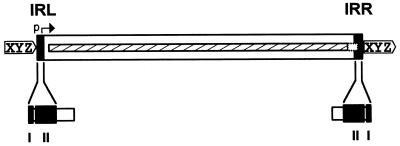
FIG. 1.
Organization of a typical IS. The IS is represented as an open box in which the terminal IRs are shown as grey boxes labelled IRL (left inverted repeat) and IRR (right inverted repeat). A single open reading frame encoding the transposase is indicated as a hatched box stretching along the entire length of the IS and extending within the IRR sequence. XYZ enclosed in a pointed box flanking the IS represents short DR sequences generated in the target DNA as a consequence of insertion. The Tpase promoter, p, which is partially localized in IRL, is shown by a horizontal arrow. A typical domain structure (grey boxes) of the IRs is indicated beneath. Domain I represents the terminal base pairs at the very tip of the element whose recognition is required for Tpase-mediated cleavage. Domain II represents the base pairs necessary for sequence-specific recognition and binding by the Tpase.
Terminal inverted repeats.
With several notable exceptions (the IS91, IS110, and IS200/605 families [see Table 2]), the majority of ISs exhibit short terminal inverted-repeat sequences (IR) of between 10 and 40 bp. In cases examined experimentally, the IRs can be divided into two functional domains (Fig. 1). One (II) is positioned within the IR and is involved in Tpase binding. The other (I), which includes the terminal 2 or 3 bp, is involved in the cleavages and strand transfer reactions leading to transposition of the element (82, 83, 152, 166, 227, 378). A similar organization has also been proposed for the transposon Tn3 (153). The simple single terminal Tpase binding sites of ISs are to be contrasted with the multiple and asymmetric protein binding sites of bacteriophage Mu (69) and transposons Tn7 (67) and probably Tn552 (242). Multiple protein binding sites are also a characteristic of the complex En/Spm and Ac elements of maize (114, 191). By accommodating different binding patterns at each end, such an arrangement can provide a functional distinction between the ends either in the assembly or in the activity of the synaptic complex. In addition, indigenous IS promoters are often located partially within the IR sequence upstream of the Tpase gene, by convention known as IRL. This arrangement may provide a mechanism for autoregulation of Tpase synthesis by Tpase binding. Binding sites for host-specified proteins are also often found within or close to the terminal IRs, and these proteins may play a role in modulating transposition activity or Tpase expression.
TABLE 2.
Major features of prokaryote IS families
| Family | Group(s) | Size range (bp)a | DR (bp)b | ENDSc | IRd | No. of ORFse | Commentsf |
|---|---|---|---|---|---|---|---|
| IS1 | 770 | 9 (8–11) | GGT | Y | 2 | Phage λ integrase? | |
| IS3 | IS2 | 1,300–1,350 | 5 | TGA | Y | 2 | DD(35)E |
| IS3 | 1,200–1,300 | 3 (4) | 2 | ||||
| IS51 | 1,300–1,400 | 3 (4) | 2 | ||||
| IS150 | 1,400–1,550 | 3–5 | 2 | ||||
| IS407 | 1,200–1,250 | 4 | 2 | ||||
| IS4 | 1,300–1,950 | 9–12 | C(A) | Y | 1 | DDE | |
| IS5 | IS5 | 1,100–1,350 | 4 | GG | Y | 1 | DDE |
| IS427 | 800–1,000 | 2–3 | Ga/g | 2 | |||
| IS903 | 1,000–1,100 | 9 | GGC | 1 | |||
| IS1031 | 850–950 | 3 | GAG | 1 | |||
| ISH1 | 900–1,150 | 8 | 1 | ||||
| ISL2 | 800–1,100 | 2–3 | 1 | ||||
| IS6 | 750–900 | 8 | GG | Y | 1 | DD(34)E | |
| IS21 | 1,950–2,500 | 4 (5, 8) | TG | Y | 2 | DDE | |
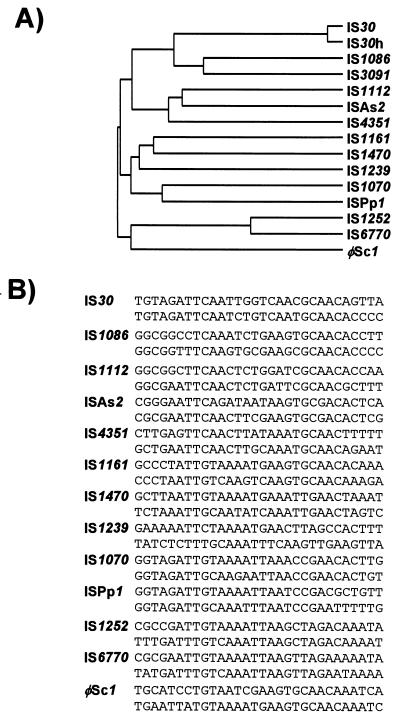
| IS30 | 1,000–1,250 | 2–3 | Y | 1 | DD(33)E | ||
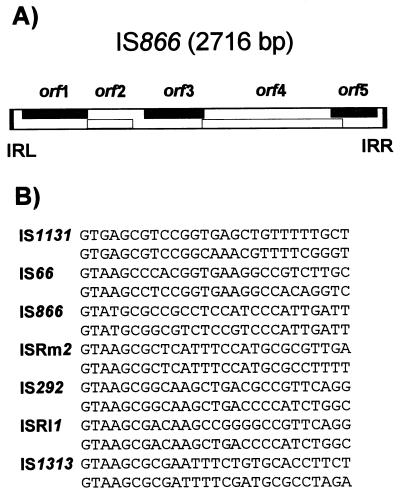
| IS66 | 2,500–2,700 | 8 | GTA | Y | >3 | ||
| IS91 | 1,500–1,850 | 0 | N | 1 | ssDNA Rep | ||
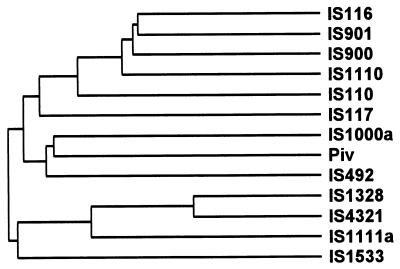
| IS110 | 1,200–1,550 | 0 | N | 1 | Site-specific recombinase | ||
| IS200/IS605 | 700–2,000 | 0 | N | 1 (2) | Complex organization | ||
| IS256 | 1,300–1,500 | 8–9 | Gg/a | Y | 1 | DDE, eukaryotic relatives | |
| IS630 | 1,100–1,200 | 2 | Y | 1 | DDE, eukaryotic relatives | ||
| IS982 | 1,000 | NDg | AC | Y | 1 | DDE | |
| IS1380 | 1,650 | 4 | Cc/g | Y | 1 | ||
| ISAs1 | 1,200–1,350 | 8 | C | Y | 1 | ||
| ISL3 | 1,300–1,550 | 8 | GG | Y | 1 |
Size range represents the typical range of each group.
Length of direct target repeats. Less frequently observed lengths are included in parentheses.
Conserved terminal base pairs. Capital leters (and capital letters within parentheses) refer to mostly (and often) conserved bases. Lowercase letters separated by slashes indicate alternative conservation at that position.
Presence (Y) or absence (N) of terminal inverted repeats.
ORF, open reading frame. Number in parentheses indicates the possible involvement of a second ORF in the transposition process.
DDE represents the common acidic triad presumed to be part of the active site of the transposase. ssDNA, single-stranded DNA.
ND, not determined.
Domain structure of Tpases.
A general pattern for the functional organization of Tpases appears to be emerging from the limited number which have been analyzed. The sequence-specific DNA binding activities of the proteins are generally located in the N-terminal region, while the catalytic domain is often localized toward the C-terminal end (IS1 [221, 377], IS30 [322], Mu [199], Tn3 [224], IS50 [365], and IS911 [270]). One functional interpretation of this arrangement for prokaryotic elements is that it may permit the interaction of a nascent protein molecule with its target sequences on the IS, thus coupling expression and activity. This notion is reinforced by the observation that the presence of the C-terminal region of both the IS50 and IS10 Tpases appears to mask the DNA binding domain and reduce binding activity (162, 357). This arrangement would favor the activity of the protein in cis, a property shared by several Tpases (see “Activity in cis” below). Similar masking appears to occur with the IS1 (374b) and the IS911 (139) Tpases. In several cases, these domains are assembled into a single protein from consecutive orfs by translational frameshifting (see “Programmed translational frameshifting” below).
An additional characteristic of some, if not all, Tpases is the capacity to generate multimeric forms essential for their activity (266). This is true of both prokaryotic elements such as bacteriophage Mu (52), IS50 (357), and IS911 (139) (but apparently not IS10 [39]) and eukaryotic elements such as the retroviruses (171) and the mariner-like element, Mos1 (213).
With the results of an increasing number of structural studies of these types of enzymes, it will be of great interest to compare the overall similarities of equivalent functional domains, as has recently become possible for the catalytic domains of retroviral integrases, Mu transposase, and other polynucleotidyl transferases such as the Holiday resolvase, RuvC, and RNase H (121, 289).
Direct target repeats.
Another general feature of IS elements is that on insertion, most generate short directly repeated sequences (DRs) of the target DNA flanking the IS. Attack of each DNA strand at the target site by one of the two transposon ends in a staggered way during insertion provides an explanation for this observation. The length of the DR, between 2 and 14 bp, is characteristic for a given element, and a given element will generally generate a duplication of fixed length. Certain ISs have been shown to generate DRs of atypical length at a low frequency, presumably reflecting small variations in the geometry of the transposition complex (reference 107 and references therein). Although some notable exceptions exist in which there is a systematic absence of DRs (either within a given family or in several independent transposition events of a given element), care should be taken in interpreting the absence of DRs in isolated cases. A lack of DRs can simply result from homologous inter- or intramolecular recombination between two IS elements, each with a different DR. This would result in a hybrid element carrying one DR of each parent. It can also arise from the formation of adjacent deletions resulting from duplicative intramolecular transposition. In this case, a single copy of the DR is located on each of the reciprocal deletion products (see, for example, references 342 and 356).
Control of Neighboring-Gene Expression
Many IS elements have been shown to activate the expression of neighboring genes. A nonexhaustive list includes IS1, IS2, and IS5 (107) and, more recently, elements such as IS406 (305), IS1186 (262), IS481 (85), IS928B (214), ISSg1 (78), IS1490 (150), and ISVa1 (336). Many other examples can be found in the literature.
It has been known for some time from experimental observation that elements such as IS1, IS2, and IS5 possess outwardly directed −35 promoter hexamers located in the terminal IRs. When placed (by transposition) at the correct distance from a resident −10 hexamer, new promoters capable of driving the expression of neighboring genes can be created. Potential −35 hexamers were detected within the terminal IRs of many ISs (107). The list of elements which have been demonstrated experimentally to carry functional −35 hexamers is now extensive and includes IS21 (283), IS30 (70), IS257 (203), IS2 (330), IS911 (338), and IS982 in Lactococcus lactis (214).
It is interesting that in several cases, an inwardly directed −10 hexamer has also been detected in the IRL of several elements. When two ends of such an element are juxtaposed, by formation of head-to-tail dimers or of circular copies of the IS, the combination of the −10 hexamer with a −35 hexamer resident in the neighboring right end can generate relatively strong promoters (IS21 [283], IS30 [70], and IS911 [338]). This arrangement can lead to high Tpase expression and consequent increases in transposition activity (see “IS3 family,” “IS21 family,” and “IS30 family” below).
Other elements have been reported to influence the expression of neighboring genes by endogenous transcription “escaping” the IS and traversing the terminal IR (e.g., IS3 [56], IS10 [60], IS481 [85], and IS982 in Escherichia coli [214]).
An additional type of control of neighboring genes is illustrated by the (normally cryptic) bgl operon of E. coli. Activation of the operon can be accomplished in several ways, including insertion of either IS1 or IS5 upstream or downstream of the promoter (284, 285). Although a detailed explanation of the effect is not available, it has been suggested that activation involves changes in DNA structure (e.g., changes in curvature or topology), since mutations in the cap, topI, and hns genes have a similar activation effect. For IS5, activation is abolished by internal deletions, leaving only 25 bp of IRL and 32 bp of IRR, but is restored by providing an IS5-encoded gene product, Ins5A, necessary for transposition in trans (303). The implication of these results is that interaction of Ins5A with the IS5 ends in some way changes the topology of the bgl promoter region. At present, no other examples of such control mechanisms are available.
Control of Transposition Activity
Transposition activity is generally maintained at a low level. An often cited reason for this is that high activities and the accompanying mutagenic effect of genome rearrangements would be detrimental to the host cell (89). Tpase promoters are generally weak, and many are partially located in the terminal IRs, enabling their autoregulation by Tpase binding.
Tpase expression and activity.
While many of the classical mechanisms of controlling gene expression, such as the production of transcriptional repressors (IS1 [95, 221, 377] and IS2 [147]) or translational inhibitors (antisense RNA in IS10 [179]), are known to operate in Tpase expression, several other mechanisms have also been uncovered.
(i) Sequestration of translation initiation signals.
Protection of certain elements from activation by impinging transcription following insertion into highly expressed genes has been shown to operate at the level of translation initiation. In these elements, internal IR sequences are located close to the left ends and contain the ribosome binding site or translation initiation codon for the Tpase gene. Transcripts from the resident promoter include only the distal repeat unit, while transcripts from neighboring DNA include both repeats and would generate secondary structures in the mRNA which would sequester translation initiation signals (74, 182). This has been demonstrated experimentally for IS10 and IS50, but several additional ISs carry such potential structures and might be expected to exhibit a similar mechanism (287).
(ii) Programmed translational frameshifting.
A second mechanism acts at the level of translation elongation and involves programmed translational frameshifting between two consecutive open reading frames. Typically a −1 frameshift is observed in which the translating ribosome slides 1 base upstream and resumes in the alternative phase. This generally occurs at the position of so-called slippery codons in a heptanucleotide sequence of the type Y YYX XXZ in phase 0 (where the bases paired with the anticodon are underlined), which is read as YYY XXX Z in the shifted −1 phase (see, e.g., reference 55, 96, and 110). The sequence A AAA AAG is a common example of this type of heptanucleotide. Ribosomal shifting of this type is stimulated by structures in the mRNA which tend to impede the progression of the ribosome, such as potential ribosome binding sites upstream or secondary structures (stem-loop structures and potential pseudoknots) downstream of the slippery codons.
Translational control of transposition by frameshifting has been demonstrated both for IS1 (95, 217, 310) and for members of the IS3 family (268; see also reference 55), but it may also occur in several other IS elements (see, for example, “IS5 family” below). In these cases, the upstream frame appears to carry a DNA recognition domain whereas the downstream frame encodes the catalytic site. While the product of the upstream frame alone acts as a modulator of activity, presumably by binding to the IR sequences, frameshifting assembles both domains into a single protein, the Tpase, which directs the cleavages and strand transfer necessary for mobility of the element. The frameshifting frequency is thus critical in determining the overall transposition activity. This is treated in more detail in the sections describing the IS1 and IS3 families. Although it has yet to be explored in detail, frameshifting could be influenced by host physiology thus coupling transposition activity to the state of the host cell.
(iii) Translation termination.
A third potential mechanism derives from the observation that the translation termination codon of Tpase genes of certain elements is located within their IRs. Although, to our knowledge, no extensive analysis of the significance of this arrangement has yet been undertaken, it seems possible that in some manner it couples translation termination, Tpase binding, and transposition activity.
The Tpase gene of several elements does not possess a termination codon. These include IS240C, a member of the IS6 family (57a); two members of the IS5 family, IS427 (77) and ISMk1 (228); and various members of the IS630 family, including IS870 and ISRf1 (103). Instead, some of these elements insert into a relatively specific target sequence in which the target DR produced on insertion itself generates the Tpase termination codon (see “IS630 family” below). The relevance of this as a control mechanism has yet to be explored.
(iv) Impinging transcription.
Early studies of several elements demonstrated that impinging transcription from outside reduces transposition activity. Transposition of both IS1 and IS50 was shown to be sensitive, although other elements have, to our knowledge, not been examined (107). In bacteriophage Mu, transcription originating from within the element and impinging on the left end also reduces activity (116). It is possible that transcription disrupts the formation of intermediates including Tpase and one or both Mu ends, which normally lead to stable transposition complexes.
(v) Tpase stability.
Tpase stability can also contribute to control of transposition activity. This has been demonstrated for IS903, where the Tpase is sensitive to the E. coli Lon protease (83). This sensitivity limits the activity of the Tpase both temporally and spatially and this may provide an explanation for the observation that several Tpases function preferentially in cis (see below). Indeed, mutant IS903 Tpase derivatives have been isolated which exhibit an increased capacity to function in trans. These are more refractive to Lon degradation than is the wild-type protein (80). Some evidence that Lon may also be involved in regulating Tn5 (IS50) transposition has also been presented (181). An observation which might also reflect Tpase instability is the temperature-sensitive nature of IS1-mediated adjacent deletions in vivo (279) and of IS911 intramolecular recombination both in vivo and in vitro (138). For IS911, incubation of the Tpase at 42°C results in an irreversible loss in activity.
(vi) Activity in cis.
Early studies on several transposable elements indicated that transposition activity was more efficient if the transposase was provided by the element itself or by a Tpase gene located close by on the same DNA molecule. This preferential activity in cis reduced the probability that Tpase expression from a given element would activate transposition of related copies elsewhere in the genome. The effect can be of several orders of magnitude and has been observed for a variety of elements including IS1 (223, 271), IS10 (245), IS50 (156), and IS903 (118, 120). This property presumably reflects a facility of the cognate Tpases to bind to transposon ends close to their point of synthesis and is likely to be the product of several phenomena.
For IS903, increased stability (83) and expression (80) have been shown to increase the capacity for Tpase activity in trans. Likewise, for IS10, mutations which increase translation of the Tpase also decrease the cis preference of the enzyme, and it has been suggested that the cis preference is strongly dependent on the half-life of the Tpase mRNA and the rate at which transcripts are released from their templates (162).
An additional consideration which may promote preferential activity in cis is reflected in the domain structure of known Tpases. In most of these cases, the DNA binding domain is located at the N-terminal end of the protein. This arrangement would permit preferential binding of nascent Tpase polypeptides to neighboring binding sites (see “Domain structure of Tpases” above). Moreover, in several cases it has been shown that the N-terminal portion of the protein exhibits a higher affinity for the ends than does the entire Tpase molecule, suggesting that the C-terminal end may in some way mask the DNA binding activity of the N-terminal portion. This is discussed in the sections below, which deal with the individual insertion sequences.
Host Factors
Transposition activity is frequently modulated by various host factors. These effects are generally specific for each element. A nonexhaustive list of such factors includes the DNA chaperones (or histone-like proteins) integration host factor (IHF), HU, HNS, and FIS; the replication initiator DnaA; the protein chaperone/proteases ClpX, ClpP, and ClpA; the SOS control protein LexA; and the Dam DNA methylase. In addition, proteins which govern DNA supercoiling in the cell might influence transposition.
The DNA chaperones may play roles in ensuring the correct three-dimensional architecture in the evolution of various nucleoprotein complexes necessary for productive transposition. They may also be involved in regulating Tpase expression. Several elements carry specific binding sites for IHF within or close to their terminal IRs. These can lie within or close to the Tpase promoter (e.g., IS1 [108], IS903 [118], and IS10 [152]). IHF, HU, HNS, and FIS have all been variously implicated in the case of bacteriophage Mu, either in the control of Mu gene expression or directly in the transposition process itself (see reference 52 for a review). IHF appears to influence the nature of IS10 transposition products by binding to a site 43 bp from one end (314). It also stimulates Tpase binding to the ends of the Tn3 family member Tn1000 (γδ) (362). Ironically, although IS1 was the first element in which IHF sites were identified (one within each IR), conditions have not yet been found in which IHF shows a clear effect on transposition or gene expression (374b). For IS50, an element of the same family as IS10, both the protein Fis and the replication initiator protein DnaA have been reported to intervene in transposition (286).
Although their mode of action is at present unknown, several other host proteins with otherwise entirely different functions have been implicated in transposition. Acyl carrier protein was independently shown to stimulate 3′-end cleavage of Tn3 by its cognate Tpase (224a) and, together with ribosomal protein L29, to greatly increase the binding of TnsD (a protein involved in Tn7 target selection) to the chromosomal insertion site, attTn7 (313a). Moreover ACP and L29 moderately stimulate Tn7 transposition in vitro, while L29 alone has a significant stimulatory effect in vivo (313a).
Certain factors involved in protein “management,” such as ClpX, ClpP, and Lon, have been implicated in transposition. ClpX is essential for Mu growth (237), where it is required for disassembling the transpososome strand transfer complex and promoting installation of the phage replication machinery (184, 206). Recognition of Mu Tpase, pA, by ClpX requires the terminal 10 amino acids of pA (207). Together with ClpP, ClpX also plays a role in proteolysis of the Mu repressor (193, 360). As indicated above, the Lon protease is implicated in proteolysis of the IS903 Tpase (80, 83). At present, the involvement of these proteins in the transposition of other elements has not been well documented.
The third class of host factor includes host cell systems which act to limit DNA damage and maintain chromosome integrity. Studies with IS10 (179) and IS1 (198) have demonstrated that high levels of Tpase in the presence of suitable terminal IRs lead to the induction of the host SOS system. Some controversy exists for Tn5 (IS50). Reznikoff and colleagues have provided genetic evidence that transposition is inhibited by induction of the SOS system in a manner which does not require the proteolytic activity of RecA (358). On the other hand, Tessman and collaborators (185–187), using a different transposition assay, have found that constitutive SOS conditions actually enhance Tn5 transposition. Moreover, using yet another assay system, Ahmed (6) has concluded that intermolecular transposition of Tn5 is stimulated in the presence of RecA. Further investigation is clearly required to understand these apparently incompatible results.
Ahmed has also concluded that intermolecular transposition of the IS1-based transposon Tn9 behaves in a similar way to that of Tn5 with respect to the recA allele (6). In contrast, however, the frequency of adjacent deletions mediated by IS1 was significantly increased in the absence of RecA. This has received some independent support in a physical assay, where it was shown that deletion products accumulate in a recA host but not in a wild-type host and, moreover, that like IS1 induction of the SOS system, accumulation of such adjacent deletions was dependent on recBC (374a). It should be noted that the recBC genes have also been implicated in the behavior of other transposons such as Tn10 and Tn5 (216), where they affect precise and imprecise excision. However, this process is independent of transposition. It is more pronounced with composite transposons in which the component insertion sequences IS10 and IS50 are present in direct repeat and is stimulated when the transposon is carried by a transfer-proficient conjugative plasmid. It seems probable that such excisions occur by a process involving replication fork slippage (see reference 107 for further discussion).
Early studies implicated both DNA polymerase I (300, 329) and DNA gyrase (157, 325) in the transposition of Tn5. While the effect of gyrase may reflect a requirement for optimal levels of supercoiling, the role of DNA polymerase I remains a matter of speculation. It may be involved in DNA synthesis which is necessary to repair the single-strand gaps resulting from staggered cleavage of the target and which presumably gives rise to the direct target repeats. DNA gyrase has also been shown to be important in the transposition of bacteriophage Mu (259).
Another host function, the Dam DNA methylase, can be important in modulating both Tpase expression and activity. IS10, IS50, and IS903 all carry methylation sites (GATC) in the Tpase promoter regions, and in each case, promoter activity is increased in a dam mutant host (292, 369). Additional evidence has been presented that the methylation status of GATC sites within the terminal IRs also modulates the activity of these ends (292). Similar sites have been previously observed in IS3, IS4, and IS5. A survey of the elements included in the database has shown that most groups or families (data not shown) contain members which have GATC sites within the first 50 bp of one or both extremities. The most numerous are members of the IS3 (20 of 82 elements), IS5 (32 of 68), and IS256 (12 of 33) families. Except for IS3 itself, where strong stimulation of transposition has been observed in a dam host (320), in most of these cases the biological relevance of these sites is unknown. Moreover, it should be pointed out that the probability that any 100-bp DNA sequence carries the GATC tetranucleotide is about 40%. The role of Dam methylation in IS10 and IS50 transposition is described in detail in the appropriate sections dealing with these elements.
Reaction Mechanisms
A detailed examination of the reaction mechanisms involved in transposition is outside the scope of this review and has been treated in depth elsewhere (238, 239). However, since such mechanisms are pertinent to an understanding of the various behaviors of the IS elements described below, we include here a brief and simplified description. The process can be divided into several defined steps, generally comprising binding of the recombinase to the ends; elaboration of a synaptic complex involving the recombinase, perhaps accessory proteins, and both transposon ends—this step involves either concomitant or subsequent (depending on the element) recruitment of the target DNA; cleavage and strand transfer of the transposon ends into the target; and processing of the strand transfer complex to a final product.
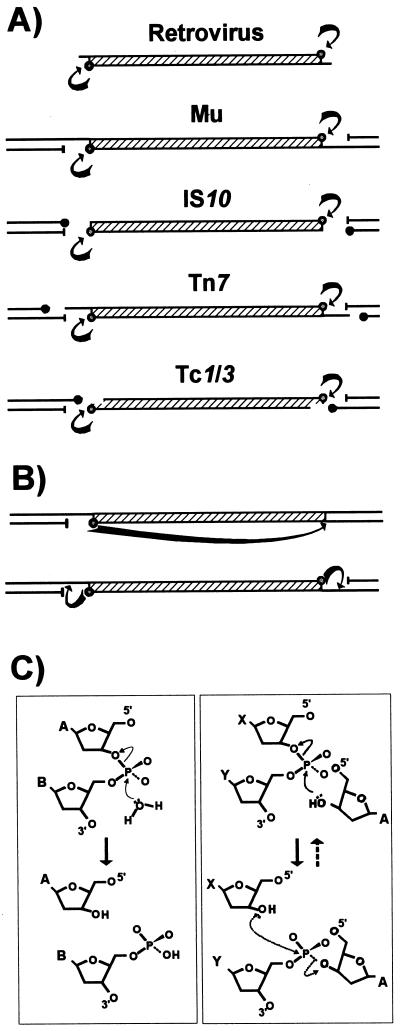
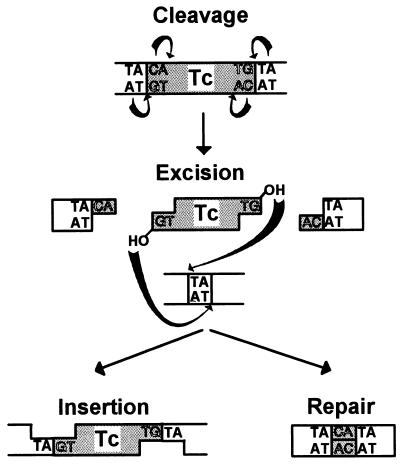
A rather surprising finding has been that the chemistry of cleavage and strand transfer is very similar, if not identical, in most of the limited collection of transposable elements analyzed in detail to date. These include retroviruses and the eukaryote Tc and mariner elements as well as bacteriophage Mu, IS10, Tn7, and IS911. The Tpase catalyzes cleavage at the 3′ ends of the element by an attacking nucleophile (generally H2O) to expose a free 3′OH group (Fig. 2A and C, left panel). This hydroxyl in turn acts as a nucleophile in the attack of a 5′-phosphate group in the target DNA in a single-step transesterification reaction (Fig. 2C, right panel). A concerted transfer of both transposon ends to the target site while maintaining the correct strand polarity results in joining of each transposon strand to opposite target strands and leaves a 3′OH group on the cleaved target strand. Under certain conditions, the enzyme is also capable of “disintegrating” the transposon end by catalyzing the attack of the 3′ target OH group on the new transposon-target junction (Fig. 2C, right panel) (59, 270, 352). The reaction(s) does not require an external energy source and does not appear to involve a covalently linked enzyme-substrate intermediate, as do certain site-specific recombination reactions (134). Furthermore, it is worth underlining that since it is the donor strand itself which performs the cleavage-ligation step in the target DNA, no cleaved target molecule is detected in the absence of strand transfer.
FIG. 2.
Different types of Tpase-mediated cleavage at transposon ends. (A) Transposons are represented by hatched boxes, and flanking donor DNA is represented by black lines. The arrows indicate Tpase-mediated cleavages at the 3′ ends of each element which give rise to active 3′OH groups (open circles) and 5′-phosphate groups (—|). Solid circles indicate 3′OH groups generated in flanking donor DNA. (B) Intramolecular strand transfer events which generate a single circularized transposon strand (top) or terminal hairpins (bottom). (C) Chemistry of the cleavage and strand transfer events. The left panel shows nucleophilic attack by a water molecule on the transposon phosphate backbone. The nucleotide shown as base A represents the terminal 3′ base of the transposon, and that marked B represent the neighboring 5′ nucleotide of the vector backbone DNA. Initial attack generates a 3′OH group on the transposon end. The right panel shows a strand transfer event. The 3′OH group at the transposon end acts as a nucleophile in the attack of the target phosphodiester backbone (bases X and Y), joining the 3′ transposon end to a 5′ target end and creating a 3′OH group on the neighboring target base (X). Also shown in this panel as dashed arrows is the disintegration reaction, in which the 3′OH of the target (X) attacks the newly created phosphodiester bond between the transposon (A) and target (Y) to regenerate the original phosphodiester bond between X and Y.
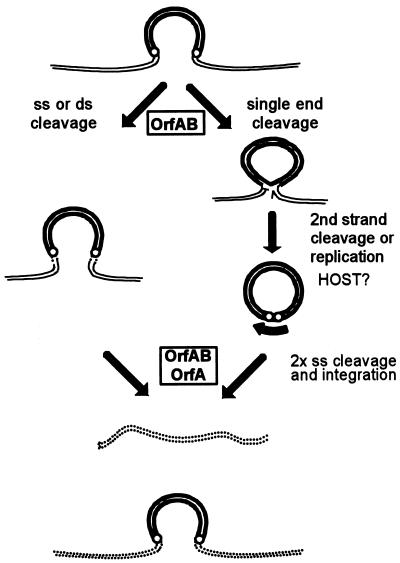
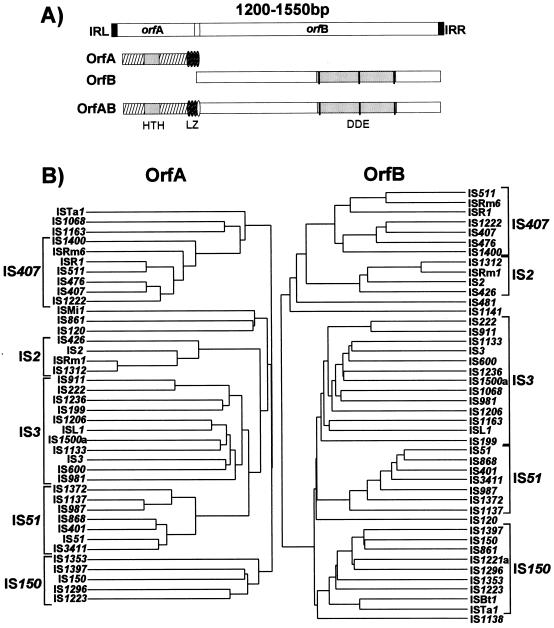
Differences in the location of the target phosphodiester bond in the initial strand transfer reaction can lead to interesting variations in the overall transposition pathway. For the IS3 family members IS911 and IS2, a frequent product is a molecule in which only one transposon DNA strand is circularized (Fig. 2B, top) (see “IS3 family, IS911” below). This results from a free 3′OH group generated at one transposon end by the Tpase with the opposite end as a target. These molecules appear to be processed into transposon circles by “resolving” the complementary strand, and the circles can then undergo integration (see Fig. 8).
FIG. 8.
Transposition pathways. Two possible pathways for transposition of IS3 family members are shown. Transposon DNA is represented by heavy double lines, donor backbone DNA is represented by fine double lines, and target DNA is represented by a double dotted line. The ends of the transposon are represented by small open circles. The left-hand pathway represents transposon excision as a linear molecule by double-strand cleavage at each end followed by strand transfer into the target molecule. It does not entail the formation of an active junction. The right-hand pathway shows passage via a single circularized strand (figure-eight) mediated by OrfAB. Formation of a circularized transposon from this intermediate is thought to require a host factor. Insertion requires both OrfAB and OrfA. The 3′OH revealed on the donor backbone is shown as a half arrow. The heavy curved arrow indicates the strong pjunc promoter created by the abutted terminal IRs on circularization.
Another variation which could in principle occur in transposition reactions is one in which the exposed 3′OH group itself cleaves the complementary strand to generate a double-strand break. This would generate hairpin structures (Fig. 2B, bottom) and result in excision of the element from its donor site. This pathway has been adopted in V(D)J recombination (346), in which an intervening segment of DNA between two coding sequences must be eliminated. Deletion is accomplished by introduction of a single-strand nick at each boundary between coding and noncoding DNA to generate an exposed a 3′OH on the coding boundary which attacks the complementary strand. The two resulting hairpin structures are then joined and assembled into a new coding joint (277). Recently, this type of hairpin structure has been detected with IS10 (175a).
DDE motif.
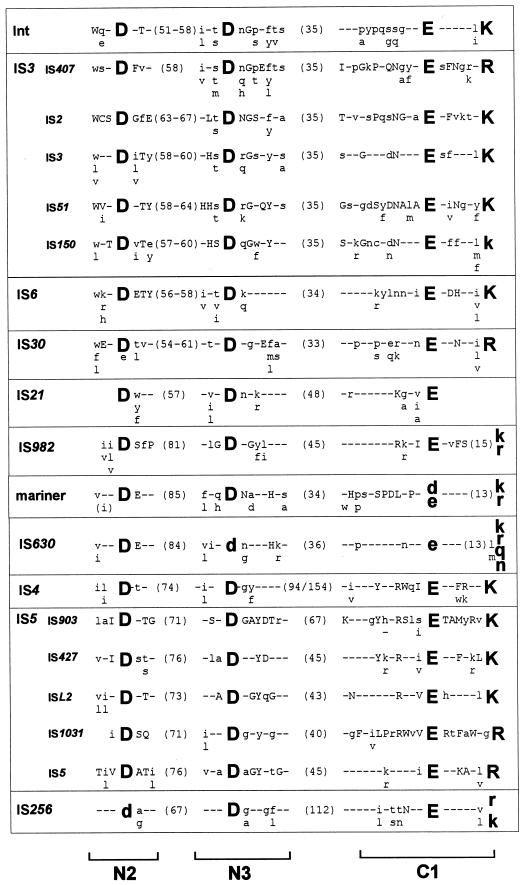
Over the last few years, it has become clear that many of the enzymes involved in the reactions described above are related and, moreover, are part of a larger family of phosphoryltransferases which also includes RNase H and the RuvC Holliday resolvase. An acidic amino acid triad present in all these enzymes is intimately involved in catalysis, and its role is presumably in coordinating divalent metal cations (in particular Mg2+) implicated in assisting the various nucleophilic attacking groups during the course of the reaction. For many ISs (the IS3 and IS6 families) and the retroviral integrases, this triad is known as the DD(35)E motif and is highly conserved, together with several additional residues (97, 172, 188) (Fig. 3) which include a K or R residue approximately 7 amino acids downstream from the E residue (87, 165, 266). In retroviruses, this motif interacts with the terminal base pairs of the element, presumably contributing to correct positioning of the transposon end in the active site (165). It is remarkable that such a motif can be found in many of the IS families defined here (Fig. 3). Although this conservation in the primary sequence is lower in certain of the other groups of elements and not all families have been explored in sufficient detail to ensure that the alignments shown in Fig. 3 are biologically relevant, mutagenic studies with some of these elements (e.g., the Mu, Tn7, IS10, and Tc1 and Tc3 Tpases) clearly underline the importance of these residues. Moreover, structural analysis has shown the presence of a related constellation of acidic amino acids arranged in a similar three-dimensional manner for retroviral integrases, bacteriophage Mu Tpase, RNase H, and RuvC (289, 290).
FIG. 3.
DDE consensus of different families. The alignments are derived from the groups presented in Table 1. Amino acids forming part of the conserved motif are shown as large bold letters. Capital letters indicate conservation within a family, and lowercase letters indicate that the particular amino acid is predominant. The numbers in parentheses show the distance in amino acids between the amino acids of the conserved motif. The retroviral integrase alignment is based on reference 266. The IS3 family is divided into the subgroups IS407, IS2, IS3, IS51, and IS150, as shown in Fig. 7B. The overall alignment (not shown) is essentially that obtained in reference 266. For IS21, see also reference 129; for mariner, see also references 87 and 295; for IS630, see also reference 87; for IS4 and IS5, see also reference 288. The IS5 family is divided into subgroups IS903, IS427, ISL3, IS1031, and IS5, as shown in Fig. 10. For IS256, see references 63 and 260. N2, N3, and C1 are regions defined in the IS4 transposon family (288).
Other chemistries?
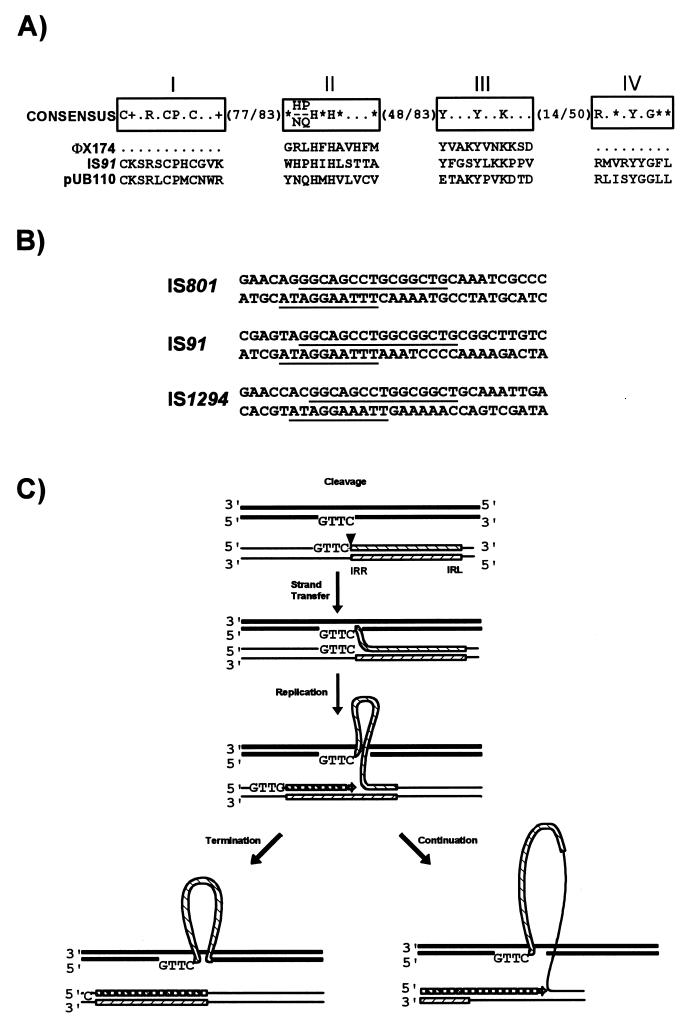
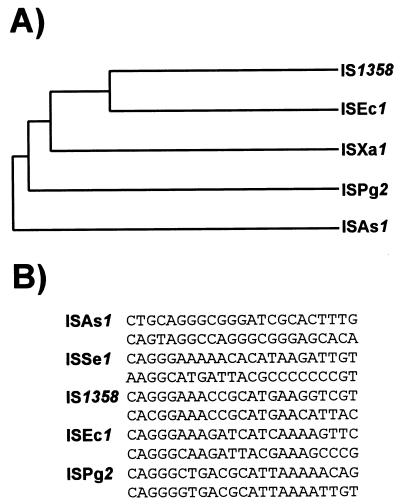
Variations and exceptions to this unifying mechanism will certainly emerge. Not all ISs exhibit a well-defined DDE triad. For example, the Tpases of one group of elements, the IS91 family, show significant similarities to enzymes associated with replicons which use a rolling-circle replication mechanism (see Fig. 15A). Indeed, evidence (232) suggests that IS91 has adopted a rolling-circle transposition mechanism similar to that proposed by Galas and Chandler (106). In addition, members of the IS110 family appear to encode a novel type of site-specific recombinase (205), while the IS1 Tpase shows limited similarity to phage λ integrase (313), and active sites for the IS66, IS200/IS605, IS1380, ISAs1, and ISL3 families (see Table 2) have yet to be defined.
FIG. 15.
IS91 family. (A) Comparison of the primary Tpase sequence with related single-strand replicases. The four conserved regions are boxed and labelled I to IV. They are separated by various numbers of nonconserved amino acids as indicated. In addition to the standard one-letter amino acid code, + and ∗ represent basic and hydrophobic amino acids, respectively. IS91 is compared to bacteriophage φX174 and plasmid pUB110 replication proteins. (B) Transposon ends. Highly conserved sequences within the termini are underlined. The upper sequence in each pair represents the left end, and the lower sequence represents the right end. (C) Proposed rolling-circle mechanism for IS91 transposition. IS91 is shown as a hatched box with left and right termini, vector DNA is shown as a fine line, and target DNA is shown as a heavy line. Initial cleavage (vertical arrowhead) occurs at IRR and is followed by strand transfer to the conserved target sequence. Replication of the displaced strand in the donor DNA then takes place with priming from the liberated 3′ donor end. The left-hand pathway shows the result of correct cleavage and termination at the right extremity of the element. The right-hand pathway shows the result of progression through the termination signal and continuation into neighboring DNA of the donor molecule.
Transposition Reactions and Different Types of Gene Rearrangement
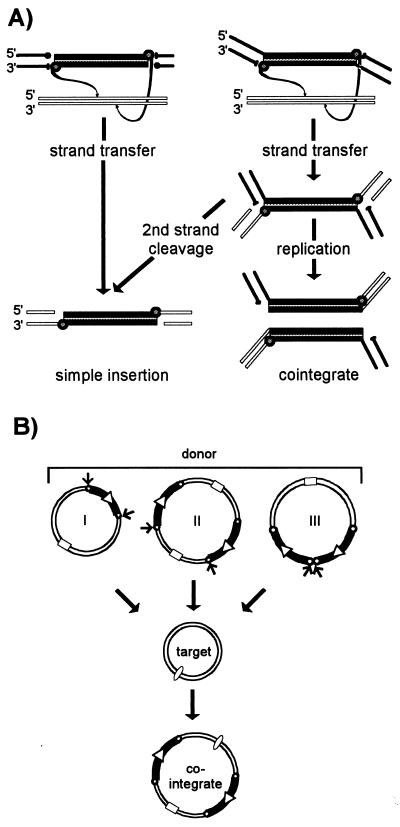
While initiation of a transposition reaction proceeds via transfer of the 3′ end of the transposon, the outcome of the reaction is governed by cleavage of the 5′ end of the element (Fig. 2A). If cleavage at the 5′ end occurs concurrently with cleavage at the 3′ end, the transposon is physically separated from its donor molecule. Strand transfer to a target then results in direct insertion of the element (Fig. 4A, left). If 5′-strand cleavage occurs only after 3′-strand transfer, the donor and target molecules become covalently linked (Fig. 4A, right). Subsequent 5′ strand cleavage will separate the element from the donor backbone and will also result in a direct insertion. On the other hand, while 3′-strand transfer joins the transposon and target, it leaves a 3′OH in the target DNA at the junction. This can act as a primer for replication of the element and generate cointegrates where donor and target molecules are separated by a single transposon copy at each junction (Fig. 4A, right, and B, structure I). It is important to note that cointegrates identical to those produced by replicative transposition can be produced by a nonreplicative process either from a plasmid dimer (structure II) (26, 210) or from tandemly repeated copies of an IS element (structure III). The 5′ cleavage can vary from element to element (Fig. 2A). In retroviruses, only the 3′ cleavage occurs, removing 2 bp from the end of the double-strand DNA viral copy. Since no donor backbone is attached to the viral DNA, direct insertion can ensue. Bacteriophage Mu similarly undergoes only 3′ cleavage, the donor backbone remains attached, and cointegrate molecules result if replication occurs (Fig. 2A and 4A, right). For IS10 and Tn7, both 3′ and 5′ cleavages occur and both elements undergo simple insertion. Double-strand cleavage at the ends of IS10 is flush and is promoted by the single Tpase protein. Double-strand cleavage at the ends of Tn7 leaves a 5′ 3-bp overhang and involves two proteins which cleave the 3′ (TnsB) and 5′ (TnsA) strands (67). Inactivation of the catalytic domain of TnsA prevents 5′-strand cleavage and results in the formation of branched-strand transfer intermediates in vitro and the production of cointegrates in vivo (231). Double-strand cleavage has also been demonstrated for the eukaryotic Tc1/3 and P elements. However, whereas cleavage occurs precisely at the 3′ end, cleavage at the 5′ ends occurs 2 bases within the element in Tc1 and Tc3 (348) and 17 bases within the element in P (20).
FIG. 4.
Simple insertions and cointegrate formation. (A) Strand transfer and replication leading to simple insertions and cointegrates. The IS DNA is shown as a shaded cylinder. Liberated transposon 3′OH groups are shown as small shaded circles, and those of the donor backbone (bold lines) are shown as filled circles. The 5′ phosphates are indicated by bars. Strand polarity is indicated. Target DNA is shown as open boxes. The left panel shows an example of an IS which undergoes double-strand cleavage prior to strand transfer. The right panel shows an element which undergoes single-strand cleavage at its ends. After strand transfer, this can evolve into a cointegrate molecule by replication or a simple insertion by second-strand cleavage. (B) Replicative and nonreplicative transposition as mechanisms leading to cointegrates. Three “cointegrate” pathways are illustrated: (I) by replicative transposition, (II) by simple insertion from a dimeric form of the donor molecule, and (III) by simple insertion from a donor carrying tandem copies of the transposable element. Transposon DNA is indicated by a heavy line, and the terminal repeats are indicated by small open circles. The relative orientation is indicated by an open arrowhead. Square and oval symbols represent compatible origins of replication and are included to visually distinguish the different replicons. Arrows show which transposon ends are involved in each reaction.
The spectrum of possible DNA rearrangements is probably even larger. A suggestion that certain Tpases may be capable of generating synapses between two ends on different molecules was originally proposed based on the results of a genetic analysis of Tn5 (210) and has more recently been demonstrated for IS10 in vitro (54). Similar behavior as well as the capacity to act on directly repeated IS ends has recently been suggested for the IS1 Tpase in vivo (198). These types of event obviously extend the spectrum of possible DNA rearrangements.
Transposition Immunity
One important property of some transposable elements is that of transposition immunity, in which a target molecule already carrying a copy of an element exhibits a significantly reduced affinity for insertion of a second copy. At present, this phenomenon appears to be limited to the more complex transposons, bacteriophage Mu and Tn7, as well as to members of the Tn3 family. To our knowledge no insertion sequences have yet been clearly demonstrated to adopt this strategy although some evidence concerning IS21 suggests that this element may show immunity (73, 128a). A priori, this behavior would be inappropriate for elements involved in the formation of compound transposons.
Although perhaps not immediately relevant to insertion sequences per se, immunity seems a sufficiently important phenomenon in the field of transposition to merit a short overview. For bacteriophage Mu, transposition immunity is displayed by target DNA carrying Mu end sequences and is transmitted by the MuB protein. MuB plays a key role in target capture and strand transfer by binding DNA in a nonspecific manner, providing a preferential target for the MuA Tpase complexed with Mu ends, and stimulating Tpase activity (199). MuB displays an ATPase activity which is stimulated both by DNA and MuA (5). ATP, but not ATP hydrolysis, is necessary for MuB binding and for strand transfer (3). Interaction of MuB with MuA (bound to the immune target) provokes ATP hydrolysis with subsequent release of MuB and consequent reduction in the attractiveness of the DNA molecule as a target (4). This mechanism serves to redistribute MuB preferentially to DNA molecules which do not contain a MuA binding site.
A similar mechanism has been proposed for transposon Tn7 (17), where the presence of the right end of Tn7 renders the target immune (12). Here the Tpase is composed of two Tn7 proteins, TnsA and TnsB. It acts in conjunction with TnsC which, like MuB, is a nonspecific DNA binding protein with ATPase activity (67).
Although transposition immunity of Tn3 and the related Tn1000 (γδ) is less well understood, it is known to require the presence of the 38-bp terminal IR on the immune target (15, 364). A major difference between Tn3 and the phage Mu and Tn7 systems is that only a single protein, the Tn3 Tpase (TnpA), appears to be involved. As in these other two systems, immunity is mediated by Tpase binding to this end (9, 252, 364). Indeed, IHF, which stimulates Tpase binding to the IRs of Tn1000 (362), also increases immunity (363).
Target Specificity
Where appropriate, insertion patterns of ISs are described in the sections dealing with the individual elements. Insertion specificity has also been treated in detail in a recent review (68). It is perhaps worthwhile, however, to summarize some of the more general issues concerning this aspect of transposition.
Target site selection differs significantly from element to element. Sequence-specific insertion is exhibited to some degree by several elements and varies considerably in stringency. It is strict in the case of one of the two Tn7 transposition pathways, where insertion occurs exclusively with high efficiency into a unique chromosomal site (attTn7) (67), and for IS91, which requires a GAAC/CAAG target sequence (233). Insertion sites are less strict but nevertheless are sequence specific for members of the IS630 and mariner/Tc families, which both require a TA dinucleotide in the target; for IS10, which prefers (but is not restricted to) the symmetric 5′-NGCTNAGCN-3′ heptanucleotide; for IS231, which shows a preference for 5′-GGG(N)5CCC-3′ (133); and for bacteriophage Mu, which shows a preference for 5′-NYG/CRN-3′ (240). For both IS10 and the Tc1/3 elements, sequences immediately adjacent to the consensus also influence the target choice (23, 261). A demonstration that IS10 Tpase directly influences the target choice has been obtained by isolation of specific Tpase mutants which exhibit distinct alterations in target choice (22). Other elements exhibit regional preferences: for example GC- or AT-rich DNA segments (IS186 [312] and IS1 [105, 236, 375], respectively). Such regional specificity could reflect more global parameters such as local DNA structure. Indeed, the degree of supercoiling (IS50 [212]), bent DNA (retroviruses [247] and IS231 [133]), replication (Tn7 [368] and IS102 [30]), transcription (IS102 [31] and Tn5/Tn10 [50]), and protein-mediated targeting to or exclusion from transcriptional control regions (Mu [354] and yeast Ty1 [86]) have all been evoked as parameters which influence target choice. The nature of the target, e.g., whether it is a plasmid or chromosome, can also play a significant role (183). Target immunity can clearly be an additional factor.
Although much information on target specificity has been obtained by analyzing individual insertions, a more powerful approach is the use of population-based methods. Such methods provide a picture which is statistically more significant. They have been applied in the analysis of retroviral integration in vitro (247, 273), in the analysis of bacteriophage Mu insertion both in vitro (240) and in vivo (354), and in the investigation of IS1-mediated adjacent deletions in vivo (342). For retroviruses, this approach has revealed a preference for the exposed face of the nucleosome DNA helix and exclusion by DNA-bound regulatory proteins. For phage Mu, it has permitted definition of the target consensus in vitro and has allowed analysis of the effect of binding of various gene regulatory proteins on insertion in vivo.
Another phenomenon which may reflect insertion site specificity is the interdigitation of various intact or partial IS elements which has been noted repeatedly in the literature. Many of these observations are anecdotal and may reflect the scars of consecutive but isolated transposition events resulting from selection for acquisition (or loss) of accessory genes (see “Sources of bacterial insertion sequences” above). Some indication of the statistical significance of this is expected to emerge from the many bacterial genome-sequencing projects under way. On the other hand, several ISs exhibit a true preference for insertion into other elements. A preferred target for IS231 is the terminal 38 bp of the transposon Tn4430, which includes both the sequence-specific and conformational components described above (133), while IS21 has been reported to show a preference for insertion close to the end of a second copy of the element located in the target plasmid (280). In this latter case, the site-specific DNA binding properties of the Tpase are presumably implicated. At the mechanistic level, this phenomenon might be related to the capacity of IS10 Tpase to form synaptic complexes with IS10 ends located on separate DNA molecules (54).
Population Dynamics and Horizontal Transfer
The distribution of many insertion sequences within and between various bacterial species has often been investigated as part of the initial characterization of a new element, usually by simple Southern hybridization. Although useful in typing strains, much of the data remains purely descriptive. Few systematic attempts have been made to determine the dynamics of insertion sequences within bacterial populations in a controlled manner.
Hartl and colleagues (131, 302) have determined the distribution of IS1, IS2, IS3, IS4, IS5, IS30, and IS103 in a heterogeneous collection of E. coli strains (ECOR collection). By fitting this data to a number of models, they concluded that these elements could be classified into three groups by the apparent strength of regulation: IS1 and IS5 (weakly regulated); IS2, IS4, and IS30 (moderately regulated); and IS3 (strongly regulated).
Based on an initial observation that bacteriophage P1 appeared to accumulate mutations due to insertion sequences when the host strain was stored in agar stabs (11), Arber and colleagues undertook a study of the changes in distribution of eight ISs (IS1, IS2, IS3, IS4, IS5, IS30, IS150, and IS186) from cultures of 118 individual clones isolated from a single 30-year-old stab of the well-characterized E. coli K-12 strain W3110 (250). The degree of variation in copy number was found to differ from element to element. When the number of each IS was counted, significant variation was noted in particular for IS5 but also for IS2, IS3, and IS30. Lower variation was observed for IS1, IS4, IS150, and IS186. These variations in copy number were roughly correlated with the number of different patterns of hybridization obtained by extensive Southern blot analysis. For IS30, the data showed that copy number diversity increased in clones which had generated a particular restriction fragment carrying a tandem dimer of the element, a configuration which results in high transposition levels (see “IS30 family” below).
Although given elements common to both these studies appear to display differences in their copy number diversity, it seems inherently unlikely that this could reflect a real difference in the behavior of a specific IS in the two sets of studies. Rather, it may occur because the E. coli W3110 strain used by Naas et al. (250) was initially homogeneous whereas members of the ECOR collection (131, 302) have presumably undergone very different selective pressures.
Horizontal transfer of ISs in nature would not be surprising in view of the number and variety of autonomous extrachromosomal elements such as bacteriophages and plasmids which can serve as vectors, particularly promiscuous plasmids with wide host ranges. Several serendipitous observations, such as the isolation of identical IS6 family members from Mycobacterium fortuitum and Flavobacterium (Arthrobacter) sp. (IS6100 [170]), clearly support the idea that horizontal transfer occurs in nature.
Some information has been obtained concerning the evolution of certain insertion sequences within the enterobacteria. Analysis of the nucleotide sequences of IS1, IS3, and IS30 from the ECOR collection and from other related enteric bacteria showed that each type of IS was highly conserved within E. coli (200). Since the degree of sequence divergence of several chromosomal genes within these clonal lineages was significantly higher, it was concluded that the ISs had a high turnover and rapid movement. Moreover, strains carrying one type of insertion element also tended to carry other types. This observation is consistent with the idea that multiple insertion sequences can be delivered by a single vector, for example a transmissible plasmid or phage (302). The homologs of these ISs carried by other species of enteric bacteria were divergent from the E. coli elements. This suggested a lower rate of transmission between species. Finally, the presence of mosaic variants of both IS1 and of IS3 in certain enteric species led to the conclusion that horizontal transmission (accompanied by recombination) had indeed occurred. Other studies have also compared the differences in the degree of nucleotide sequence variation of ISs with that of chromosomal genes. For IS1 and IS200 elements in natural populations of E. coli and Salmonella typhimurium, the results suggested that IS200 has a significantly lower frequency of horizontal transfer than does IS1 (37).
Data consistent with horizontal transfer are also emerging from studies with nonenteric bacteria. In one study, 17 isoforms of the ISS1 sequence, isolated largely from bacteria which occupy another complex ecological niche, milk and cheese, were compared. They were determined to fall into three defined subgroups. Not only were nearly identical copies of these IS6 family members isolated from distantly related Streptococcus thermophilus and Lactococcus lactis strains, but also mosaic copies were detected (40). Moreover, nearly identical IS6 family members have also been found in E. coli, Proteus vulgaris, and Pasteurella piscicida (178).
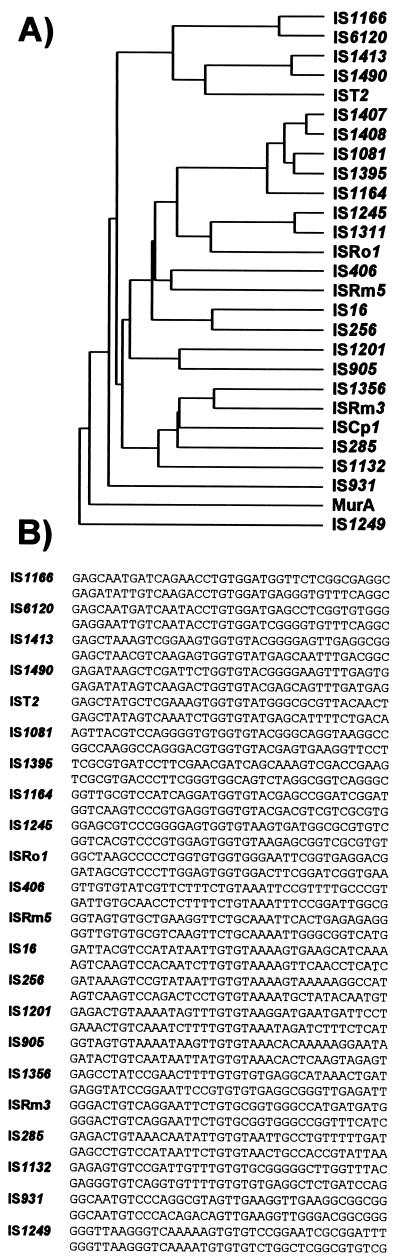
For members of the IS256 family, a phylogenetic tree of eight members was found to differ significantly from that of their host bacteria (123). Another study indicated that 10 members of the family isolated from actinomycetes formed a distinct group. While they exhibited a similar phylogenetic tree to their hosts (based on 16S RNA and superoxide dismutase genes) and most showed divergence similar to that of the 16S RNA and superoxide dismutase genes, IS1512 and IS1511 isolated from Mycobacterium gordonae showed significantly higher divergence (suggesting a higher mobility) and were more closely related to an element isolated from the Rhodococcaceae (260).
INSERTION SEQUENCE FAMILIES
Occurrence, Variety, and Systematics
Our current database contains 500 ISs isolated from 73 genera representing 159 bacterial species of both eubacteria and the archaea (Table 1). This list does not include ISs identified from the various genome-sequencing projects. It is important to note that the majority of these have not been tested for transposition activity and some may therefore carry mutations which render them inactive. In some instances, the alignments of translation products clearly indicated the occurrence of translation termination codons or the presence of one or more frame changes in the published sequence. It is possible that at least some of these are the result of sequencing errors. In many cases there is also some ambiguity concerning the exact tip of the IS and the number of directly repeated target base pairs generated. Where appropriate, these are noted in the text and the accompanying tables. In spite of the limitations inherent in the available data, we have been able to include 443 members of the collection in 17 families based on combinations of the following criteria: (i) similarities in genetic organization (arrangement of open reading frames); (ii) marked identities or similarities in their Tpases (common domains or motifs); (iii) similar features of their ends (terminal IRs); and (iv) fate of the nucleotide sequence of their target sites (generation of a direct target duplication of determined length). The general features of these families are shown in Table 2 and are presented in greater detail below. It should be noted that two families show some similarities to eukaryotic mobile elements: IS630 is related to the widespread Tc/mariner group (87), whereas IS256 is very distantly related to the plant transposon MuDR (91). We underline here that the classification scheme described below is not rigid and is provided only as a framework. Some families are more coherent and better established than others, and there are numerous uncertainties in several of the attributions of family status.
TABLE 1.
Database used
| Name | Synonym(s) | Isoform | Family | Group | Origin | Accession no. | Length (bp) | IRa | DRb |
|---|---|---|---|---|---|---|---|---|---|
| IS1A | IS1E, IS1K | IS1 | Escherichia coli W3110 | X52534 | 768 | 18/23 | 9 | ||
| IS1B | IS1C | IS1A | IS1 | Escherichia coli W3110 | X17345 | 768 | 18/23 | 8, 9, 10, 14 | |
| IS1D | IS1A | IS1 | Escherichia coli W3110 | X52536 | 768 | 18/23 | 9 | ||
| IS1F | IS1X1d | IS1 | Escherichia coli W3110 | X52538 | 768 | 20/23 | NDc | ||
| IS1G | IS1R | IS1 | Escherichia coli C600 | J01730 [V]e | 768 | 17/23 | 8 | ||
| IS1H | IS1 | Escherichia coli ECOR50 | U15127 | 764 | 21/23 | ND | |||
| IS1N | NuXi | IS1 | Shigella dysenteriae | J01737 | 766 | 20/23 | ND | ||
| IS1R | IS1A | IS1 | Escherichia coli (pR100) | J01730 | 768 | 18/23 | ND | ||
| IS1S | IS1A | IS1 | Shigella sonnei | M37615 | 768 | 18/23 | ND | ||
| IS1SD | IS1A | IS1 | Shigella dysenteriae | J01731 | 768 | 16/23 | ND | ||
| IS1X1 | IS1 | Shigella flexneri | M37616 | 768 | 20/23 | ND | |||
| IS1X2 | IS1X1 | IS1 | Escherichia vulneris ATCC 29943 | Z11605 | 768 | 18/23 | ND | ||
| IS1X3 | IS1 | Escherichia fergusonii ATCC 35469 | Z11603 [P]f | 694 | ND | ND | |||
| IS1X4 | IS1X2 | IS1 | Escherichia hermannii ATCC 33652 | Z11604 | 768 | 20/23 | ND | ||
| IS2 | IS3 | IS2 | Escherichia coli K-12 | M18426 | 1,331 | 32/41 | 5 | ||
| IS3 | IS3 | IS3 | Escherichia coli K-12 | X02311 | 1,258 | 29/40 | 3, 4 | ||
| IS3E | IS3 | IS3 | IS3 | IS3 | Escherichia coli ATCC 35382 | Z11606 | >1,181 | ND | ND |
| IS3F | IS3 | IS3 | IS3 | IS3 | Escherichia fergusonii ATCC 35469 | Z11607 | >1,184 | ND | ND |
| IS3G | IS3 | IS3 | IS3 | IS3 | Escherichia fergusonii ATCC 35471 | Z11608 | >1,184 | ND | ND |
| IS3H | IS3 | IS3 | IS3 | IS3 | Shigella dysenteriae ATCC 13313 | Z11609 | >1,187 | ND | ND |
| IS4 | IS4 | Escherichia coli K-12 | J01733 | 1,426 | 16/18 | 11, 12, 13 | |||
| IS4Sa | IS4 | Synechocystis sp. strain PCC6803 | U38915 | 1,303 | 15/19 | ND | |||
| IS5 | IS5 | IS5 | Escherichia coli K-12 (Lambda KH100) | J01735 | 1,195 | 15/16 | 4 | ||
| IS5D | IS5-Delta | IS5 | IS5 | IS5 | Escherichia coli DH5-alpha | X13668 | 1,283 | 15/16 | 4 |
| IS5Sa | IS5 | IS1031 | Synechocystis sp. strain PCC6803 | U38799 | 871 | 15/17 | 3 | ||
| IS5Sb | IS5Sc | IS5Sa | IS5 | IS1031 | Synechocystis sp. strain PCC6803 | U38915 | 871 | 15/17 | 3 |
| IS10L | IS10R | IS4 | Salmonella typhimurium (Tn10) | J01829 [V] | 1,329 | 17/22 | |||
| IS10R | IS10 | IS4 | Salmonella typhimurium (Tn10) | J01829 | 1,329 | 17/22 | 9 | ||
| IS15 | IS15L, IS15R, IS1522 | IS26 | IS6 | Salmonella panama LA46 (Tn1525 from pIP112) | M12900 | 1,648 | 14 | 8 | |
| IS15DI | IS15-DeltaI | IS26 | IS6 | Salmonella panama LA46 (Tn1525 from pIP112) | M12900 | 820 | 14 | 8 | |
| IS15DII | IS15-DeltaII | IS26 | IS6 | Salmonella panama LA46 (Tn1525 from pIP112) | M12900 [V] | 820 | 14 | ND | |
| IS15DIII | IS26 | IS6 | Campylobacter sp. strain BM2196 | M12900 [V] [P] | >667 | ND | ND | ||
| IS15DIV | IS26 | IS6 | Salmonella typhimurium (pBP11) | X13616 | 820 | 14 | ND | ||
| IS16 | IS256 | Enterococcus faecalis BM4281(Tn1547) | U35366 | 1,466 | 28/42 | 8 | |||
| IS21 | IS8 | IS21 | Pseudomonas aeruginosa PAO25 (pR68.45) | X14793 | 2,131 | 30/42 | 4, (5) | ||
| IS22 | ISNCYg | Pseudomonas aeruginosa PAO (pFP110) | ND | 7,300 | ND | ND | |||
| IS26 | IS6, IS26L, IS26R, IS46, IS140, IS160, IS176 | IS6 | Proteus vulgaris UR-75 (Tn2680 from pRts1) | X00011 | 820 | 14 | 8 | ||
| IS30 | IS121, Tn2700, Tn2702 | IS30 | Escherichia coli K-12 (Tn2671 from pNR1-Basel) | X00792 | 1,221 | 23/26 | 2 | ||
| IS30D | IS30 | IS30 | IS30 | Escherichia coli K-12 | X62680 | 1,221 | 23/26 | 2 | |
| IS30H | IS30 | IS30 | Escherichia hermannii ATCC 33652 | Z11753 | 1,221 | 23/26 | ND | ||
| IS50L | IS50R | IS4 | Escherichia coli (Tn5) | U15572 | 1,534 | 8/9 | |||
| IS50R | IS50 | IS4 | Escherichia coli DB729 (Tn5 from pJR67) | U15573 | 1,534 | 8/9 | 8, 9, 10 | ||
| IS51 | IS3 | IS51 | Pseudomonas syringae pv. savastanoi TK2009-5 (pIAA1) | M14365 | 1,311 | 26 | 3 | ||
| IS52 | IS5 | IS5 | Pseudomonas syringae pv. savastanoi PB205-1L (pIAA2) | M14366 | 1,209 | 9/10 | 4 | ||
| IS53 | IS21 | Pseudomonas syringae pv. savastanoi PB213 (pIAA2) | M83932 | 2,555 | 24/27 | 8 | |||
| IS53K | IS21 | Pseudomonas oleovorans TF4-1L (OCT) | J04618 [P] | >316 | ND | ND | |||
| IS60 | ISNCY | Agrobacterium tumefaciens LBA4060 (pAL108) | ND | 1,200 | ND | ND | |||
| IS66 | IS66t, IS66v2 | IS66 | Agrobacterium tumefaciens A66 (pTiA66) | M10204 | 2,548 | 18/20 | 8 | ||
| IS66-1 | IS66v1 | IS66 | IS66 | Agrobacterium tumefaciens (pTi15955) | ND | 2,556 | 18/20 | 8 | |
| IS70 | ISNCY | Proteus mirabilis (pR772) | ND | 3,700 | ND | ND | |||
| IS71 | IS66 | Agrobacterium tumefaciens (pTi15955) | ND | 2,386 | 9/11 | 8 | |||
| IS91 | IS91 | Escherichia coli EC185 (pSU233) | X17114 | 1,830 | 0 | 0 | |||
| IS91B | IS91 | IS91 | Escherichia coli G7 (pRI8801) | X77671 | >549 | ND | ND | ||
| IS92L | IS91 | IS91 | Escherichia coli SU100 (pHly152) | ND | 3,100 | ND | ND | ||
| IS92R | IS91 | IS91 | Escherichia coli SU100 (pHly152) | ND | 2,150 | ND | ND | ||
| IS100 | IS21 | Yersinia pestis 106 Otten | Z32853 | 1,953 | 20/28 | 5 | |||
| IS100kyp | IS100 | IS21 | Yersinia pseudotuberculosis (pKYP1) | U59875 | 1,954 | 20/28 | ND | ||
| IS100L | IS100 | IS21 | Yersinia pestis EV76 (pLcr) | X78302 | 1,950 | 20/28 | 5 | ||
| IS100X | IS100 | IS21 | Yersinia pestis EV7651F | L19030 | 1,924 | 20/28 | ND | ||
| IS102 | IS903 | IS5 | IS903 | Escherichia coli (pSC101) | J01728 | 1,057 | 18 | 9 | |
| IS103 | IS150 | IS3 | IS150 | Escherichia coli K-12 x342 | X07037 [V] | 1,443 | 22/31 | 4 | |
| IS110 | IS110 | Streptomyces coelicolor A3(2) | Y00434 | 1,558 | 0 | (0) | |||
| IS112 | IS5 | ISL2 | Streptomyces albus G J1147 | X56644 | 883 | 16/20 | 2 | ||
| IS116 | IS110 | Streptomyces clavuligerus NRRL3585 | M31716 | 1,421 | 0 | 0 | |||
| IS117 | A3(2) minicircle | IS110 | Streptomyces coelicolor A3(2) M130 | X15942 | 2,527 | 0 | 0 | ||
| IS120 | IS3 | IS150 | Clostridium thermocellum | ND | 1,447 | 10/12 | 3 | ||
| IS150 | IS3 | IS150 | Escherichia coli K-12 431 | X07037 | 1,443 | 22/31 | 3 | ||
| IS161 | ISNCY | Escherichia coli (Tn2424) | ND | 1,700 | ND | ND | |||
| IS186A | IS186B | IS4 | Escherichia coli RR1 | M11300 | 1,341 | 23 | 10 | ||
| IS186B | IS186-357, IS186-409 | IS4 | Escherichia coli RR1 | X03123 | 1,338 | 23 | (8), 10, 11 | ||
| IS199 | IS3 | IS150 | Streptococcus mutans V403 | L23843 | 1,220 | 31/43 | 3 | ||
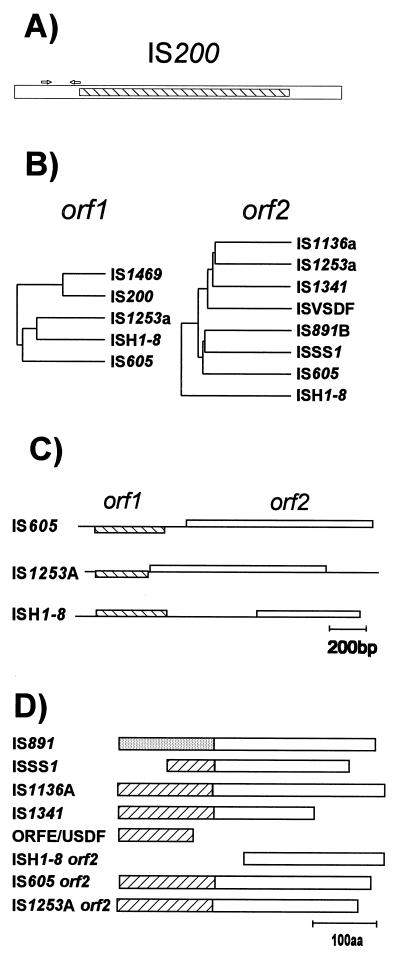
| IS200 | IS605 | IS200 | Salmonella typhimurium TR6238 | X56834 | 707 | 0 | 0 (2) | ||
| IS200A | IS200 | IS200 | IS605 | IS200 | Salmonella typhimurium LT2 | X56834 [V] [P] | 700 | 0 | 0 (2) |
| IS200B | IS200 | IS605 | IS200 | Escherichia coli ECOR8 | L25844 [P] | >601 | ND | ND | |
| IS200C | IS200 | IS200B | IS605 | IS200 | Escherichia coli ECOR51 | L25845 [P] | >601 | ND | ND |
| IS200D | IS200B | IS605 | IS200 | Escherichia coli ECOR63 | L25846 [P] | >601 | ND | ND | |
| IS200E | IS200 | IS200B | IS605 | IS200 | Escherichia coli ECOR72 | L25847 [P] | >601 | ND | ND |
| IS200F | IS200 | IS200 | IS605 | IS200 | Salmonella typhimurium SARA17 | Z54217 | 709 | 0 | ND |
| IS200G | IS605 | IS200 | Yersinia pestis 6/69M | U22457 | 707 | 0 | ND | ||
| IS200H | IS200-SAO | IS200 | IS605 | IS200 | Salmonella abortusovis SS44 | Y08755 | 708 | 0 | ND |
| IS200P | IS200-VP | IS200 | IS605 | IS200 | Salmonella typhimurium LT2 (pSLT) | Y09564 | 708 | 0 | 1 |
| IS200S | IS605 | IS200 | Streptococcus pneumoniae type 23 | L20670 [P] | >651 | 0 | ND | ||
| IS204 | ISL3 | Nocardia asteroides YP21 | U10634 | 1,453 | 19/23 | 8 | |||
| IS222 | IS3 | IS3 | Pseudomonas aeruginosa (phage D3c) | U00100 | 1,232 | 20/23 | ND | ||
| IS231A | IR1, IR1750′ | IS4 | Bacillus thuringiensis subsp. thuringiensis berliner 1715 (p65kb) | X03397 | 1,656 | 20 | 10, 11, 12 | ||
| IS231B | IR1, IR1750* | IS231A | IS4 | Bacillus thuringensis subsp. thuringiensis berliner 1715 (p65kb) | M16158 | 1,643 | 7/20 | ND | |
| IS231C | IR1, IR1750 | IS4 | Bacillus thuringiensis subsp. thuringiensis berliner 1715 (p65kb) | M16159 | 1,656 | 19/20 | 11 | ||
| IS231D | IS4 | Bacillus thuringiensis subsp. finitimus | X63383 | 1,657 | 17/20 | ND | |||
| IS231E | IS4 | Bacillus thuringiensis subsp. finitimus | X63384 | 1,614 | 0/20 | ND | |||
| IS231F | IS4 | Bacillus thuringiensis subsp. israelensis (p112kb) | X63385 | 1,655 | 19/20 | 12 | |||
| IS231G | IS4 | Bacillus thuringiensis subsp. darmstadiensis 73-E-10-2 | M93054 | 1,649 | 20 | ND | |||
| IS231H | IS4 | Bacillus thuringiensis subsp. darmstadiensis 73-E-10-2 | M93054 [P] | >817 | ND | ND | |||
| IS231V | IS4 | Bacillus thuringiensis subsp. israelensis (p112kb) | M86926 [V] | 1,964 | 21/22 | ND | |||
| IS231W | IS231V | IS4 | Bacillus thuringiensis subsp. israelensis (p112kb) | M86926 | 1,964 | 21/22 | ND | ||
| IS232A | IR2, IS232, IR2150 | IS21 | Bacillus thuringiensis subsp. thuringiensis berliner 1715 (p65kb) | M38370 | 2,184 | 48/67 | ND | ||
| IS232B | IR2, IS232, IR2150 | IS232A | IS21 | Bacillus thuringiensis subsp. kurstaki HD73 (p75kb) | M77344 [P] | 2,200 | 28/37 | ND | |
| IS232C | IR2, IS232, IR2150 | IS232A | IS21 | Bacillus thuringiensis subsp. kurstaki HD73 (p75kb) | ND | 2,200 | 28/37 | ND | |
| IS233A | IS982 | Bacillus thuringiensis subsp. galleriae T05001 | ND | 1,028 | ND | 8 | |||
| IS240A | IRA | IS6 | Bacillus thuringiensis subsp. israelensis (p112kb) | M23740 | 861 | 16/17 | ND | ||
| IS240B | IRB | IS240A | IS6 | Bacillus thuringiensis subsp. israelensis (p112kb) | M23741 | 861 | 16/17 | ND | |
| IS240C | IS6 | Bacillus cereus CER484 | ND | 817 | 16/17 | ND | |||
| IS240F | IS6 | Bacillus thuringiensis subsp. fukuokaensis 84I113 (p197kb) | Y09946 | 806 | 16/17 | ND | |||
| IS256 | IS256E, IS256L, IS256R | IS256 | Staphylococcus aureus (Tn4001 from pSK1) | M18086 | 1,324 | 17/26 | 8 | ||
| IS257-1 | IS257/1 | IS257R1 | IS6 | Staphylococcus aureus (pSH6) | X53952 | 791 | 21/26 | ND | |
| IS257-2 | IS257/2 | IS257R1 | IS6 | Staphylococcus aureus (pSH6) | X53951 | 790 | 20/26 | ND | |
| IS257-3 | IS257/3 | IS257R1 | IS6 | Staphylococcus aureus (pSH6) | X53951 | 789 | 23/27 | ND | |
| IS257R1 | IS6 | Staphylococcus aureus (Tn4003 from pSK1) | X13290 | 790 | 18/20 | 8 | |||
| IS257R2 | IS257L | IS257R1 | IS6 | Staphylococcus aureus (Tn4003 from pSK1) | X13290 | 789 | 23/27 | 8 | |
| IS281 | ISNCY | Streptomyces lividans 803 (phage C43) | ND | 1,400 | ND | ND | |||
| IS285 | IS256 | Yersinia pestis 358 (pLcr) | X78303 | 1,318 | 22/29 | 8 | |||
| IS292 | IS66 | Agrobacterium sp. strain X88-292 (pTi292) | L29283 | 2,496 | 19/21 | 8 | |||
| IS298 | ISNCY | Caulobacter crescentus SC298 | ND | 800 | 12/16 | 4 | |||
| IS401 | IS3 | IS51 | Burkholderia cepacia ATCC 17616 (pTGL6) | L09108 | 1,316 | 23/26 | 3 | ||
| IS402 | IS5 | IS427 | Burkholderia cepacia ATCC 17616 (pTGL6) | M33173 | 914 | 16/17 | 3 | ||
| IS403 | ISNCY | Burkholderia cepacia 249 | ND | 800 | ND | ND | |||
| IS404 | ISNCY | Burkholderia cepacia 249 | ND | 1,100 | ND | ND | |||
| IS405 | ISNCY | Burkholderia cepacia ATCC 17616 (pTGL1) | ND | 1,500 | ND | ND | |||
| IS406 | IS256 | Burkholderia cepacia ATCC 17616 | M83145 | 1,367 | 22/27 | 8 | |||
| IS407 | IS3 | IS407 | Burkholderia cepacia ATCC 17616 | M82980 | 1,236 | 31/49 | 4 | ||
| IS408 | IS21 | Burkholderia cepacia ATCC 17616 (pTGL1) | L09108 [P] | >2,530 | 40/48 | 8 | |||
| IS411 | ISNCY | Burkholderia cepacia ATCC 17616 (pTGL1) | ND | 2,000 | ND | ND | |||
| IS415 | ISNCY | Burkholderia cepacia ATCC 17616 | ND | 2,100 | ND | ND | |||
| IS421 | IS186 | IS4 | Escherichia coli JA221 | Y07501 | 1,342 | 23 | 11 | ||
| IS426 | IS136, ISAT1 | IS3 | IS2 | Agrobacterium tumefaciens A208 (pTiT37) | X56562 | 1,319 | 24/33 | 5 | |
| IS427 | IS5 | IS427 | Agrobacterium tumefaciens T37 (pTiT37) | M55562 | 1,271 | 13/16 | 2 | ||
| IS431L | IS257R1 | IS6 | Staphylococcus aureus (pI524) | M18437 | 788 | 19/22 | ND | ||
| IS431mec | IS257R1 | IS6 | Staphylococcus aureus BB270 | X53818 | 790 | 18/20 | ND | ||
| IS431R | IS257R1 | IS6 | Staphylococcus aureus (pI524) | M18437 [V] | 790 | 17/20 | ND | ||
| IS466 | IS6 | Streptomyces coelicolor K605 (pSCP1) | S70701, S70702 [P] | 1,628 | 32/37 | ND | |||
| IS476 | IS3 | IS407 | Xanthomonas campestris pv. vesicatoria 81-23 race 2 | M28557 | 1,226 | 22/27 | 4 | ||
| IS481 | IS3 | IS481 | Bordetella pertussis Tohama | M22031 | 1,043 | 22/23 | 6 | ||
| IS481G | IS481 | IS3 | IS481 | Bordetella pertussis NCTC 10908 | M22031 [V] | 1,039 | 22/23 | 6 | |
| IS481P | ISPARK | IS481 | IS3 | IS481 | Bordetella pertussis 18-323 | M28220 [V] [P] | >525 | 0/23 | ND |
| IS481v1 | RS1 | IS481 | IS3 | IS481 | Bordetella pertussis BPH30 | M28220 | 1,043 | 20/23 | 6 |
| IS481v2 | RS2 | IS481 | IS3 | IS481 | Bordetella pertussis Wellcome28 | M28220 [V] | 1,043 | 20/23 | 6 |
| IS492 | IS110 | Pseudomonas atlantica | M24471 | 1,202 | 0 | 5 | |||
| IS493 | IS5 | ISL2 | Streptomyces lividans CT2 | M28508 | 1,643 | 21/24 | 3 | ||
| IS511 | IS125 | IS3 | IS407 | Caulobacter crescentus CB15 | U39501 | 1,266 | 34/41 | 4 | |
| IS600 | IS3 | IS3 | Shigella sonnei | X05952 | 1,264 | 19/27 | 3 | ||
| IS602L | IS903 | IS5 | IS903 | Capnocytophaga ochraceus (Tn602 from pGD10) | M20309 [P] | 1,057 | 18 | 9 | |
| IS602R | IS903 | IS5 | IS903 | Capnocytophaga ochraceus (Tn602 from pGD10) | M20309 [P] | 1,057 | 18 | 9 | |
| IS605 | IShp, RS2 | IS605 | Helicobacter pylori NCTC 11638 | ND | 1,904 | 0 | ND | ||
| IS629 | IS3411 | IS3 | IS51 | Shigella sonnei | X51586 | 1,310 | 23/25 | ND | |
| IS630 | Tn4730, Tn4731, Tn4733 | IS630 | Shigella sonnei | X05955 | 1,153 | 21/29 | 2 | ||
| IS640 | IS21 | IS21 | Shigella sonnei | X05956 | >1,085 | ND | ND | ||
| IS701 | IS4 | Calothrix sp. strain PCC7601 | X60383 | 1,389 | 22/25 | 4 | |||
| IS702 | IS5 | ISL2 | Calothrix sp. strain PCC7601 | X60384 | 1,098 | 33/40 | 3 | ||
| IS711 | IS5 | IS427 | Brucella ovis ATCC25840 | M94960 | 842 | 20/25 | 2 | ||
| IS801 | RS-I | IS91 | Pseudomonas syringae pv. phaseolicola LR781 (pMMC7105) | X57269 | 1,512 | 0 | 0 | ||
| IS802 | RS-II | ISNCY | Pseudomonas syringae pv. phaseolicola LR781 (pMMC7105) | ND | <2,600 | ND | ND | ||
| IS803 | RS-III | ISNCY | Pseudomonas syringae pv. phaseolicola LR781 (pMMC7105) | ND | ND | ND | ND | ||
| IS861 | IS3 | IS150 | Streptococcus agalactiae COH-I | M22449 | 1,442 | 22/26 | 3 | ||
| IS866 | IST-1 | IS66 | Agrobacterium tumefaciens (pTiTm4) | M25805 | 2,716 | 24/27 | 8 | ||
| IS867 | IS867a, IST-2, IS867b, IST-3 | IS66 | Agrobacterium tumefaciens (pTiTm4) | M63056 [P] | 2,700 | ND | ND | ||
| IS868 | IS3 | IS51 | Agrobacterium tumefaciens AB3 (pTiAB3) | X55075 | 1,321 | 25/26 | ND | ||
| IS869 | IS5 | IS427 | Agrobacterium tumefaciens (pTiAB3) | X53945 | 849 | 14/16 | 2 | ||
| IS869-1 | IS5 | IS427 | Agrobacterium tumefaciens (pTi15955) | ND | 849 | 4 | 2 | ||
| IS870 | IS870.1, IS870.2, IS870.3 | IS630 | Agrobacterium vitis 2657 (pTiAg57) | Z18270 | 1,146 | 10 | 2 | ||
| IS891 | IS605 | Anabaena sp. strain M-131 | M24855 | 1,352 | 0 | 0 | |||
| IS892 | ISNCY | Anabaena sp. strain PCC7120 | M64297 | 1,675 | 21/24 | 8 | |||
| IS893 | ISNCY | Anabaena strain PCC7120 | ND | 1,200 | ND | ND | |||
| IS894 | ISNCY | Anabaena sp. strain PCC7120 | ND | 1,900 | ND | ND | |||
| IS895 | mys | IS630 | Anabaena sp. strain PCC7120 | M67475 | 1,192 | 26/28 | 2 | ||
| IS897 | ISNCY | Anabaena sp. strain PCC7120 | ND | 1,500 | ND | ND | |||
| IS898 | ISNCY | Anabaena sp. strain PCC7120 | ND | 1,000 | ND | ND | |||
| IS900 | IS110 | Mycobacterium paratuberculosis | X16293 | 1,451 | 0 | 0 | |||
| IS901 | IS110 | Mycobacterium avium FP8589 | X59272 | 1,472 | 0 | 0 | |||
| IS902 | IS901 | IS110 | Mycobacterium avium subsp. silvaticum 0016 | X58030 | 1,470 | 0 | 0 | ||
| IS903 | IS903L, IS903R, Tn55, Tn601 | IS5 | IS903 | Escherichia coli (Tn903 from pR6-5) | J01839 | 1,057 | 18 | 9 | |
| IS903B | IS903.B | IS903 | IS5 | IS903 | Escherichia coli (Tn2680 from pRts1) | X02527 | 1,057 | 18 | 9 |
| IS904 | IS1068 | IS3 | IS3 | Lactococcus lactis subsp. lactis FI5876 | M27276 | 1,241 | 31/39 | (4) | |
| IS905A | IS905 | IS256 | Lactococcus lactis subsp. cremoris FI7304 | L20851 | 1,313 | 20/28 | 8 | ||
| IS905B | IS905 | IS905A | IS256 | Lactococcus lactis subsp. cremoris MG1614 | L20851 [V] | 1,313 | 20/28 | 8 | |
| IS911 | ISSHO1 | IS3 | IS3 | Shigella dysenteriae ATCC 11456 | X17613 | 1,250 | 25/36 | 3, 4 | |
| IS931 | RS1100, IS933 | IS256 | Burkholderia cepacia AC1100 | M25495 | 1,477 | 38/39 | 8 | ||
| IS932 | ISNCY | Burkholderia cepacia AC1100 | ND | 3,400 | ND | ND | |||
| IS942 | IS4 | Bacteroides fragilis TAL3636 | J03326 | 1,598 | 13/15 | 8 | |||
| IS946M | IS946 | ISS1S | IS6 | Lactococcus lactis subsp. lactis T-EK1 (pTR2030) | ND | 800 | ND | 8 | |
| IS946V | IS946 | ISS1S | IS6 | Lactococcus lactis subsp. lactis T-EK1 (pTR2030) | M33868 | 808 | 18 | 8 | |
| IS981 | IS3 | IS3 | Lactococcus lactis subsp. lactis LM0230 | M33933 | 1,224 | 18/28 | 3 | ||
| IS982 | IS982A | IS982 | Lactococcus lactis subsp. cremoris SK11 (pSK11L) | L34754 | 999 | 19/20 | ND | ||
| IS982B | IS982 | IS982 | IS982 | Lactococcus lactis bv. diacetylactis CRL264 (pCIT264) | S77101 | 999 | 19/20 | ND | |
| IS982C | IS982 | IS982 | Lactococcus lactis subsp. cremoris NIZOB40 (pNZ4000) | U93364 | 999 | 19/20 | ND | ||
| IS986 | IS6110 | IS3 | IS51 | Mycobacterium tuberculosis | X52471 | 1,354 | 25/28 | ND | |
| IS987 | IS6110 | IS3 | IS51 | Mycobacterium bovis BCG44 | X57835 | 1,355 | 25/28 | 3 | |
| IS1000A | IS110 | Thermus thermophilus HB8 | M33159 | 1,196 | 5/6 | 0 | |||
| IS1000B | IS1000A | IS110 | Thermus thermophilus HB8 | M33159 [V] | 1,196 | 5/6 | 0 | ||
| IS1001 | IS1001A, IS1001B, IS1001C | ISL3 | Bordetella parapertussis B24 | X66858 | 1,306 | 21/26 | ND | ||
| IS1004 | IS605 | IS200 | Vibrio cholerae O1 | Z67733 | 628 | ND | ND | ||
| IS1016C | ORF3/frpC, IS1016N | ISNCY | Neisseria meningitidis FAM20 | L06299 | 713 | 19 | ND | ||
| IS1016V1 | IS1016V6 | ISNCY | Haemophilus influenzae RM118 | X58173 | 711 | 19 | ND | ||
| IS1016V2 | IS1016V6 | ISNCY | Haemophilus influenzae RM153 | X58174 | 711 | 19 | ND | ||
| IS1016V3 | IS1016V6 | ISNCY | Haemophilus influenzae RM926 | X58175 | 711 | 18/19 | ND | ||
| IS1016V4 | IS1016V6 | ISNCY | Haemophilus influenzae RM926 | X58176 | 673 | 19/0 | ND | ||
| IS1016V5 | IS1016V6 | ISNCY | Haemophilus influenzae RM7004 | X58177 | 242 | 19/0 | ND | ||
| IS1016V6 | ISNCY | Haemophilus influenzae bg. aegyptius BPF | X59756 | 712 | 19 | ND | |||
| IS1031A | IS1031B | IS5 | IS1031 | Acetobacter xylinum ATCC 23769 | M80805 | 930 | 22/24 | 3 | |
| IS1031C | IS5 | IS1031 | Acetobacter xylinum ATCC 23769 | M98777 | 930 | 23/26 | 3 | ||
| IS1031D | IS1031C | IS5 | IS1031 | Acetobacter xylinum ATCC 23769 | M98778 | 930 | 23/26 | 3 | |
| IS1032 | IS5 | IS1031 | Acetobacter xylinum ATCC 23770 | M80805 | 916 | 9/15 | 3 | ||
| IS1051 | IS5 | IS5 | Xanthomonas campestris pv. dieffenbachiae 11044 | X70380 | 1,158 | 13/15 | ND | ||
| IS1066 | IS630 | Pseudomonas sp. strain P51 (Tn5280 from pP51) | M61114 | 1,137 | 10 | 2 | |||
| IS1067 | IS1066 | IS630 | Pseudomonas sp. strain P51 (Tn5280 from pP51) | M61114 [V] | 1,137 | 10 | 2 | ||
| IS1068 | IS904N | IS3 | IS3 | Lactococcus lactis subsp. lactis NIZOR5 (Tn5276) | X52273 | 1,245 | 31/39 | ND | |
| IS1069 | IS1068 | IS3 | IS3 | Lactococcus lactis subsp. lactis NIZO22186 | X78469 | 1,245 | 31/39 | ND | |
| IS1070 | IS30 | Leuconostoc lactis NZ6009 (pNZ63) | U17353 | 1,027 | 24/28 | 0 (?) | |||
| IS1076 | IS1076L, IS1076R, Tn5286 | IS1068 | IS3 | IS3 | Lactococcus lactis subsp. lactis CNRZ270 (pUCL22) | X53013 | 1,296 | 31/39 | 3 |
| IS1081 | IS256 | Mycobacterium bovis TMC410 | X61270 | 1,435 | 19/26 | 8 | |||
| IS1086 | IS30 | Ralstonia eutropha AE104 | X58441 | 1,106 | 22/28 | 3 | |||
| IS1096 | ISL3 | Mycobacterium smegmatis ATCC 607 | M76495 | 2,259 | 24/26 | 8 | |||
| IS1106 | IS5 | IS5 | Neisseria meningitidis B15 | Z11857 | 1,137 | 35/36 | ND | ||
| IS1110 | IS110 | Mycobacterium avium LR541 | Z23003 | 1,462 | 0 | 0 | |||
| IS1111A | IS110 | Coxiella burnetii NineMile7 | M80806 | 1,450 | 7 | ND | |||
| IS1111B | IS1111A | IS110 | Coxiella burnetii NineMile7 | M80806 [V] | 1,458 | 7 | ND | ||
| IS1111C | IS1111A | IS110 | Coxiella burnetii NineMile7 | M80806 [V] | 1,452 | 7 | ND | ||
| IS1112 | ISXoo1 | IS30 | Xanthomonas oryzae pv. oryzae PXO86Rif | ND | 1,055 | 22/25 | 3 | ||
| IS1112a | IS203a | IS1112 | IS30 | Xanthomonas oryzae pv. oryzae PXO96Rif | ND | 1,055 | 23/25 | 3 | |
| IS1112b | IS203b, IS203c | IS1112 | IS30 | Xanthomonas oryzae pv. oryzae PX112 | ND | 1,055 | 21/25 | 3 | |
| IS1113 | TNX1 | ISNCY | Xanthomonas oryzae pv. oryzae | ND | 1,050 | ND | ND | ||
| IS1126 | IS5 | IS5 | Porphyromonas gingivalis W83 | X77924 | 1,336 | 12 | 5 | ||
| IS1131 | ISC | IS66 | Agrobacterium tumefaciens PO22 (pTi) | M82888 | 2,773 | 12 | 8 | ||
| IS1132 | DI-2 | IS256 | Corynebacterium diphtheriae Belfanti 1030 | A07012 | 1,441 | 21/28 | 8 | ||
| IS1133 | IS3 | IS3 | Erwinia amylovora CA11 (Tn5393 from pEa34) | Z12167 | 1,232 | 24/28 | 3 | ||
| IS1136A | IS605 | Saccharopolyspora erythraea NRRL 2338 | L07626 | 1,406 | 7/13 | (0) | |||
| IS1136B | IS1136A | IS605 | Saccharopolyspora erythraea NRRL 2338 | L07627, L07628 [P] | 1,400 | 8/14 | ND | ||
| IS1136C | IS605 | Saccharopolyspora erythraea NRRL 2338 | L07629, L07630 [P] | 1,400 | ND | ND | |||
| IS1136D | IS605 | Saccharopolyspora erythraea NRRL 2338 | L07631, L07632 [P] | 1,400 | ND | ND | |||
| IS1137 | IS3 | IS51 | Mycobacterium smegmatis | X70913 | 1,361 | 22/24 | 3 | ||
| IS1138 | IS3 | IS150 | Mycoplasma pulmonis KD735-26 | Z16416 | 1,288 | 18 | 3 | ||
| IS1138B | IS1138 | IS3 | IS150 | Mycoplasma pulmonis KD735-26 | Z16416 [V] | 1,288 | 18 | 3 | |
| IS1139 | IS1161 | IS30 | Streptococcus salivarius ATCC 25975 | Z17279 | 1,162 | 20/27 | 0 (?) | ||
| IS1141 | IS3 | IS3 | Mycobacterium intracellulare | L10239 | 1,588 | 18/23 | ND | ||
| IS1151 | IS4 | Clostridium perfringens NCTC 2062 | Z18246 | 1,689 | 21 | ND | |||
| IS1161 | IS30 | Streptococcus salivarius ATCC 25975 | L07794 | 1,165 | 22/28 | 0 (?) | |||
| IS1162 | IS21 | Pseudomonas fluorescens ST (pEG) | X79443 | 2,634 | 37/50 | ND | |||
| IS1163 | IS3 | IS3 | Lactobacillus sake L45 | X75164 | 1,180 | 32/48 | 3 | ||
| IS1164 | IS256 | Rhodococcus rhodochrous J1 | D67027 | 1,430 | 18/27 | 8 | |||
| IS1165 | ISL3 | Leuconostoc mesenteroides subsp. cremoris DB1165 (p30kb) | X62617 | 1,553 | 20/39 | ND | |||
| IS1166 | IS256 | Rhodococcus sp. strain IGTS8 (pSOX) | U08850 | 1,469 | 19/25 | ND | |||
| IS1167 | ISL3 | Streptococcus pneumoniae CP1200 | M36180 | 1,435 | 17/24 | 8 (?) | |||
| IS1167L | IS1167 | ISL3 | Streptococcus pneumoniae 13868 | Z83335 | 1,484 | 18/24 | ND | ||
| IS1167R | IS1167 | ISL3 | Streptococcus pneumoniae 13868 | Z83335 | 1,464 | 20/24 | ND | ||
| IS1168 | IS5 | IS5 | Bacteroides vulgatus BV17 (pIP417) | X71444 | 1,320 | 13/17 | ND | ||
| IS1168F | IS1168 | IS1168 | IS5 | IS5 | Bacteroides fragilis BF8 | X71443 [P] | >777 | ND | ND |
| IS1169 | IS5 | IS5 | Bacteroides fragilis BF-F238 (pIP421) | X76949 | 1,317 | 16/20 | ND | ||
| IS1170 | IS4 | Bacteroides thetaiotaomicron BT13 (pIP419) | X76948 | 1,604 | 12/15 | ND | |||
| IS1181 | ISL3 | Staphylococcus aureus BM3121 | L14544 | 1,512 | 17/23 | 8 | |||
| IS1182 | ISNCY | IS1182 | Staphylococcus aureus BM3121 | L43082 | 1,864 | 14/16 | ND | ||
| IS1186 | IS1168 | IS5 | IS5 | Bacteroides fragilis BFr81R | X72301 | 1,300 | 13/17 | 4 | |
| IS1191 | IS905A | IS256 | Streptococcus thermophilus CNRZ368 | X71808 | 1,313 | 20/28 | 8 | ||
| IS1193D | IS1193 | ISL3 | Streptococcus thermophilus CNRZ368 | Y13713 | 1,411 | 17/24 | 8 | ||
| IS1194 | IS5 | IS5 | Streptococcus thermophilus CNRZ368 | Y13626 | 1,200 | 15/16 | ND | ||
| IS1201 | IS256 | Lactobacillus helveticus CNRZ1094 (p34kb) | L26311 | 1,387 | 20/24 | 8 | |||
| IS1202 | ISNCY | Streptococcus pneumoniae SSZ serotype 19F | U04047 | 1,747 | 18/23 | 23 (?) | |||
| IS1203 | IS3411 | IS3 | IS51 | Escherichia coli O111:H− PH | U06468 | 1,310 | 22/25 | ND | |
| IS1203E | IS3411 | IS3 | IS51 | Escherichia coli O157:H7 EDL933 (pO157) | X97542 | 1,032 | ND | ND | |
| IS1206 | ISaB1 | IS3 | IS3 | Corynebacterium glutamicum Bl15 | X69104 | 1,290 | 19/26 | 3 | |
| IS1207 | ISbB1 | IS31831 | ISL3 | Corynebacterium glutamicum Bl15 | ND | 1,450 | 24 | 8 | |
| IS1216 | IS6 | Enterococcus hirae S185 | X81654 | 809 | 19 | ND | |||
| IS1216E | IS1216 | IS6 | Enterococcus faecium HM1073 | U49512 | 808 | 24 | ND | ||
| IS1216V | IS1216 | IS6 | Enterococcus sp. (pHKK701) | L38972 | 809 | 18 | ND | ||
| IS1221A | G1135.2 | IS3 | IS150 | Mycoplasma hyorhinis GDL-1 | U01217 | 1,513 | 23/28 | ||
| IS1121B | IS1221 | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis GDL-1 | L33925 | 1,519 | 22/28 | ND |
| IS1221C | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis GDL-1 | L33927 | 1,518 | 23/28 | ND | |
| IS1221D | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis GDL-1 | L33928 | >813 | ND | ND | |
| IS1221E | IS1221A | IS3 | IS150 | Mycoplasma hyopneumoniae J | L33924 | 1,511 | 15/19 | ND | |
| IS1221F | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis | L33926 | 1,509 | 22/28 | ND | |
| IS1221G | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis SK76 | X62936 | 1,527 | 16/26 | 4 (?) | |
| IS1221H | ISJ1251 | IS1221A | IS3 | IS150 | Mycoplasma hyopneumoniae J | X17372 [V] | 1,557 | 15/20 | ND |
| IS1221I | ISJ1252 | IS1221A | IS3 | IS150 | Mycoplasma hyopneumoniae J | X17372 | 1,554 | 21/27 | ND |
| IS1221J | IS1221A | IS3 | IS150 | Mycoplasma hyorhinis MM96 | M37339 [P] | >773 | ND | ND | |
| IS1222 | IS3 | IS407 | Enterobacter agglomerans 339 (pEA9) | X78052 | 1,221 | 22/25 | ND | ||
| IS1223 | IS3 | IS150 | Lactobacillus johnsonii ATCC 11506 | U09558 | 1,492 | 21/26 | 3 | ||
| IS1236 | IS3 | IS3 | Acinetobacter calcoaceticus ADP1 | U03772 | 1,237 | 30/39 | 3 | ||
| IS1237 | IS5 | IS427 | Clavibacter xyli subsp. cynodontis | X75973 | 899 | 19 | 3 | ||
| IS1239 | IS30 | Streptococcus pyogenes MGAS1898 (M15) | U11799 | 1,080 | 7/15 | ND | |||
| IS1245 | IS256 | Mycobacterium avium | L33879 | 1,414 | 31/40 | ND | |||
| IS1246 | IS5 | IS5 | Pseudomonas putida mt-2 (pWWO) | L11583 | 1,275 | 15/16 | ND | ||
| IS1247 | IS1380 | Xanthobacter autotrophicus GJ10 | X84038 | 1,672 | 12/16 | 4 | |||
| IS1248A | IS1248 | IS5 | IS427 | Paracoccus denitrificans PdX13 | U08864 | 832 | 13/14 | 2 | |
| IS1248B | IS1248 | IS1248A | IS5 | IS427 | Paracoccus denitrificans PdX22 | U08856 | 832 | 13/14 | 2 |
| IS1249 | IS256 | Corynebacterium xerosis M82B (pTP10) | U21300 | 1,385 | 27/31 | 8 | |||
| IS1251 | ISL3 | Enterococcus faecium GUC | L34675 | 1,496 | 16/24 | ND | |||
| IS1252 | IS30 | Enterococcus sp. (pHKK701) | L38972 | 1,065 | 21/28 | ND | |||
| IS1253A | IS1253 | IS605 | Dichelobacter nodosus AC3577 (pJIR896) | U34772 | 1,689 | 0 | ND | ||
| IS1253B | IS1253 | IS1253A | IS605 | Dichelobacter nodosus A198 | U34771 | 1,698 | 0 | ND | |
| IS1272 | ISNCY | IS1182 | Staphylococcus haemolyticus Y176 | U35635 | 1,935 | 15/16 | ND | ||
| IS1294 | IS91 | Escherichia coli (pUB2380) | X82430 | 1,689 | 0 | 0 | |||
| IS1295 | IS256 | Rhodococcus sp. strain IGTS8 (pSOX) | U08850 | 1,455 | 23/29 | ND | |||
| IS1296 | IS3 | IS150 | Mycoplasma mycoides subsp. mycoides L2 | X84021 | 1,485 | ND | ND | ||
| IS1297 | ISS1N | IS6 | Leuconostoc mesenteroides subsp. dextranicum NZDRI2218 | U59101 | 808 | 17/18 | ND | ||
| IS1301 | IS5 | IS427 | Neisseria meningitidis B1940 | Z49092 | 844 | 15/17 | 2 | ||
| IS1311 | IS256 | Mycobacterium avium IMM147460 | U16276 | >1,317 | 15 | ND | |||
| IS1312 | IS3 | IS2 | Agrobacterium tumefaciens Bo542 (pTiBo542) | U19148 | 1,317 | 17/23 | 5 | ||
| IS1313 | IS66 | Agrobacterium tumefaciens Bo542 (pTiBo542) | U19149 | 2,547 | 20/24 | 8 | |||
| IS1326 | IS21 | Pseudomonas aeruginosa (In0 from pVS1) | U38187 | 2,470 | 22/26 | ND | |||
| IS1327 | IS1327L, IS1327R | IS6 | Erwinia herbicola pv. gypsophilae Eh824-1 (pPATH) | X87144 | 810 | 15/16 | ND | ||
| IS1328 | RS.2 | IS110 | Yersinia enterocolitica WA-314 | Z48244 | 1,355 | 0 | 0 | ||
| IS1341 | IS605 | Thermophilic bacterium PS3 | D38778 | 1,353 | 10/11 | ND | |||
| IS1353 | IS3 | IS150 | Pseudomonas aeruginosa Tn21::In2 | U40482 | 1,613 | 20/24 | ND | ||
| IS1356 | IS256 | Burkholderia cepacia | U44828 | 1,354 | 14/24 | 9 | |||
| IS1358 | VcIs1, rfbQRS | ISAs1 | Vibrio cholerae O139 Bengal | U24571 | 1,326 | 16/17 | ND | ||
| IS1372 | IS3 | IS51 | Streptomyces lividans ZX7 | U50076 | 1,304 | 22/27 | 3 | ||
| IS1373 | IS5 | ISL2 | Streptomyces lividans 66 1326.32 | U05249 | 817 | 9/15 | 2 | ||
| IS1380A | IS1380 | Acetobacter pasteurianus NC11380 | D90424 | 1,665 | 13/15 | 4 | |||
| IS1380B | IS1380A | IS1380 | Acetobacter pasteurianus NC11380 | D90424 [V] | 1,665 | 12/15 | 4 | ||
| IS1381A | IS5 | ISL2 | Streptococcus pneumoniae R6 | Z77725 | 846 | 18/20 | ND | ||
| IS1381B | IS1381A | IS5 | ISL2 | Streptococcus pneumoniae R6 | Z77726 | 865 | 18/20 | ND | |
| IS1381C | IS1381A | IS5 | ISL2 | Streptococcus pneumoniae R6 | Z77727 [P] | >823 | ND | ND | |
| IS1381D | IS5 | ISL2 | Streptococcus mitis NCTC 12261 | Z82003 [P] | >823 | ND | ND | ||
| IS1395 | IS256 | Mycobacterium xenopi | U35051 | >1,323 | 14/15 | ND | |||
| IS1397 | IS3 | IS150 | Escherichia coli EPEC25 | X92970 | 1,432 | 20/25 | 3, 4 | ||
| IS1400 | RS.1 | IS3 | IS407 | Yersinia enterocolitica Ye 8081 | X94452 | 1,207 | 30/39 | 4 | |
| IS1407 | IS256 | Mycobacterium celatum | X97307 | >1,399 | ND | ND | |||
| IS1408 | IS256 | Mycobacterium branderi | U62766 | >1,325 | ND | ND | |||
| IS1412 | IS1380 | Sphingomonas sp. strain CF06 | U50714 | 1,656 | 12/18 | ND | |||
| IS1413 | IS256 | Burkholderia cepacia AC1100 | U58191 | 1,429 | 24/32 | 9 | |||
| IS1415 | IS21 | Rhodococcus erythropolis NI86/21 | AF002247 | 2,580 | 24/36 | 5, 6 | |||
| IS1452 | IS4 | Acetobacter pasteurianus NCI 1452 | D63923 | 1,411 | 21 | 4 | |||
| IS1469 | IS200 | IS605 | IS200 | Clostridium perfringens NCTC 8239 | X71844 | 717 | 0 | ND | |
| IS1470 | IS30 | Clostridium perfringens NCTC 8239 | X71844 | 1,212 | 22/27 | ND | |||
| IS1471 | IS630 | Burkholderia cepacia 2a (pIJB1) | U67938 | 1,113 | 18/25 | 2 | |||
| IS1476 | ISL3 | Enterococcus faecium | U63997 | 1,500 | 18/27 | ND | |||
| IS1490 | IS256 | Burkholderia cepacia AC1100 | U80795 | 1,433 | 19/25 | 8 | |||
| IS1500A | IS3 | IS3 | Leptospira interrogans sv. icterohaemorrhagiae Verdun | U13012 | 1,235 | 24/31 | ND | ||
| IS1500B | IS1500A | IS3 | IS3 | Leptospira interrogans sv. icterohaemorrhagiae RZ11 | U13013 | 1,235 | 24/31 | ND | |
| IS1511 | IS256 | Mycobacterium gordonae ATCC 14470 | U95315 | 1,142 | ND | ND | |||
| IS1512 | IS256 | Mycobacterium gordonae 960592 | U95314 | 1,428 | 18/24 | ND | |||
| IS1533 | IS110 | Leptospira borgpetersenii sv. hardjo type hardjo-bovis RZ33 | M82880 | 1,460 | 13/15 | ND | |||
| IS1533W | IS1533 | IS110 | Leptospira borgpetersenii sv. hardjo type hardjo-bovis | M57713 | 1,452 | 13/15 | ND | ||
| IS1549 | IS4 | Mycobacterium smegmatis LR222 | ND | 1,634 | 11 | ND | |||
| IS1936 | IS1936L, IS1936R | IS6 | Salmonella wien 20/70 (Tn1935 from pZM3) | ND | 800 | ND | ND | ||
| IS3411 | IS3411L, IS3411R | IS629 | IS3 | IS51 | Escherichia coli (Tn3411 from pOH3001) | M19532 | 1,310 | 24/27 | 3 |
| IS4321L | IS110 | Enterobacter aerogenes (pR751) | U67194 | 1,347 | 0 | ND | |||
| IS4321R | IS4321L | IS110 | Enterobacter aerogenes (pR751) | U67194 | 1,326 | 0 | ND | ||
| IS4351 | IS4400, IS4351L, IS4351R | IS30 | Bacteroides fragilis V479-1 (Tn4351 from pBF4) | M17124 | 1,155 | 20/25 | 3 | ||
| IS4502 | Tn4502 | IS256 | Caedibacter taeniospiralis 116 (pKAP) | U60645 | 1,412 | 22/26 | ND | ||
| IS4521L | LTR | IS4521R | IS3 | IS2 | Escherichia coli DH1 (Tn4521) | M17617 | 722 | ND | ND |
| IS4521R | RTR | IS3 | IS2 | Escherichia coli DH1 (Tn4521) | M17618 | 1,226 | 11 | ND | |
| IS4811 | Tn4811, IS456 | IS5 | IS1031 | Streptomyces lividans TK64 | Z11519 | 5,396 | 9/11 | 3 | |
| IS5376 | IS21 | Bacillus stearothermophilus CU21 | X67861 | 2,107 | 39/50 | 5 | |||
| IS5377 | IS4 | Bacillus stearothermophilus CU21 | X67862 | 1,249 | 18/20 | 9 | |||
| IS5469 | Tn5469 | IS6 | Fremyella diplosiphon Fd33R1 | U33002 | 4,894 | 22/25 | ND | ||
| IS6100 | IS6100L, IS6100R, Tn610 | IS6 | Mycobacterium fortuitum FC1 (Tn610) | X53635 | 880 | 14 | ND | ||
| IS6110 | IS3 | IS51 | Mycobacterium tuberculosis H37Rv | X17348 | 1,354 | 25/28 | 3, 4 | ||
| IS6120 | IS256 | Mycobacterium smegmatis mc2155 | M69182 | 1,486 | 21/24 | 9 | |||
| IS6501 | IS711 | IS5 | IS427 | Brucella ovis ATCC 25840 | X71024 | 838 | 20/25 | 2 | |
| IS6770 | ISEF1 | IS30 | Enterococcus faecalis CV34 (pLRM1) | L28754 | 1,065 | 22/28 | ND | ||
| IS8402 | IS4 | Synechocystis sp. BO8402 | L76080 | 1,322 | 19/22 | 8 | |||
| IS12528 | IS5 | IS1031 | Gluconobacter suboxydans IFO12528 | D86632 | 905 | 22/26 | 3 | ||
| IS13869 | IS-Bl | ISL3 | Brevibacterium lactofermentum ATCC 13869 | Z66534 | 1,437 | 33/36 | 8 | ||
| IS31831 | IS-Cg | ISL3 | Corynebacterium glutamicum ATCC 31831 | D17429 | 1,453 | 19/24 | 8 | ||
| ISAa1 | IS605 | IS200 | Actinobacillus (Haemophilus) actinomycetemcomitans FDCY4 | D64078 | 718 | 0 | ND | ||
| ISAe1 | ISAE1 | ISL3 | Ralstonia eutropha H1-4 | M86608 | 1,313 | 18/24 | 8 | ||
| ISAr1 | Riat1 | IS630 | Agrobacterium rhizogenes HR1 | K03313 | 1,146 | 11 | 2 | ||
| ISAs1 | ISAS1 | ISAs1 | Aeromonas salmonicida A450-ceg6 | L27156 | 1,223 | 20/22 | 8 | ||
| ISAs2 | ISAS2 | IS30 | Aeromonas salmonicida A449-ceg4 | L27157 | 1,084 | 24/29 | 3 | ||
| ISAsp1 | ISL3 | Anabaena sp. strain PCC7120 | U13767 | 1,419 | 19/22 | ND | |||
| ISBf1 | IS21 | Bacteroides fragilis RBF103 | U05888 | 2,787 | 6/8 | ND | |||
| ISBf2 | IS21 | Bacteroides fragilis RBF49 | U05886 | >355 | ND | ND | |||
| ISBj1 | HRS1, HRS1a | IS1380 | Bradyrhizobium japonicum serocluster 123 USDA424 | L09226 | 2,071 | 6/7 | 4 | ||
| ISBs1 | RSBst-alpha | IS982 | Bacillus stearothermophilus CU21 | Z21626 | 996 | 18/21 | ND | ||
| ISBs2 | RSBst-beta | IS630 | Bacillus stearothermophilus CU21 | Z21626 [P] | >660 | ND | ND | ||
| ISBt1 | orfX | IS3 | IS150 | Bacillus thuringiensis subsp. aizawai HD229 | L29100 | 999 | 15/17 | ND | |
| ISBt2 | IS3 | IS150 | Bacillus thuringiensis YBT-226 | S68409 [P] | >187 | ND | ND | ||
| ISC1217 | ISNCY | Sulfolobus solfataricus DSM1616 | X70335 | 1,147 | 10/13 | 6 | |||
| ISCm1 | RS | ISNCY | Clavibacter michiganense subsp. sepedonicum Cs3R (pCS1) | ND | 1,300 | ND | ND | ||
| ISCp1 | ORF2-4 | IS256 | Clostridium perfringens NCTC 2062 | X60694 [P] | >1,128 | ND | ND | ||
| ISCx1 | ISL3 | Corynebacterium xerosis M82B (pTP10) | U21300 [P] | >536 | ND | ND | |||
| ISEc1 | H-rptB, RhsB | ISAs1 | Escherichia coli K-12 | L02370 | 1,291 | 14/18 | ND | ||
| ISEc2 | H-rptC1, RhsC | ISEc1 | ISAs1 | Escherichia coli K-12 | L02373 | 380 | ND | ND | |
| ISEc3 | H-rptC2, RhsC | ISEc1 | ISAs1 | Escherichia coli K-12 | L02373 | 1,290 | 16/18 | ND | |
| ISEc4 | H-rptC3, RhsC | ISEc1 | ISAs1 | Escherichia coli K-12 | L02373 | 954 | ND | ND | |
| ISEc5 | H-rptE, RhsE | ISEc1 | ISAs1 | Escherichia coli K-12 | L02372 | 1,291 | 14/19 | ND | |
| ISEc6 | H-rpt, min.5 | ISEc1 | ISAs1 | Escherichia coli K-12 | D83536 | 291 | ND | ND | |
| ISEc7 | H-rpt, RhsF | ISEc1 | ISAs1 | Escherichia coli ECOR 50 | U15125, U15126 [P] | >276 | 18/25 | ND | |
| ISFn1 | IS91 | Fusobacterium necrophorum biotype AB FnS1 | X68876 [P] | >712 | ND | ND | |||
| ISH1 | IS5 | ISH1 | Halobacterium salinarium S9 | J01725 | 1,118 | 8/9 | 8 | ||
| ISH1-8 | ISH1.8 | IS605 | Halobacterium salinarium (phage H1) | X00805 | 1,887 | 0 | (0) | ||
| ISH2 | IS4 | Halobacterium salinarium S9 | J01726 | 521 | 19 | 10, 11, 12, 20 | |||
| ISH3A | SH3 | ISH27-1 | IS4 | Halobacterium salinarium R1 | L19296 [P] | 1,378 | 15/16 | 5 | |
| ISH3B | ISH27-1 | IS4 | Halobacterium salinarium NRC-1 (pNRC100) | ND | 1,400 | 15/16 | ND | ||
| ISH4 | ISH27-2 | IS4 | Halobacterium salinarium SD104 | ND | 1,375 | 15/16 | ND | ||
| ISH6 | ISH27-1 | IS4 | Halobacterium salinarium NRC-1 (pNRC100) | ND | 1,400 | 15/16 | ND | ||
| ISH8 | ISH26 | IS4 | Halobacterium salinarium SD108 | M58557 [P] | 1,395 | 16/18 | ND | ||
| ISH11 | IS5 | IS427 | Halobacterium salinarium R1 | X54752 | 1,068 | 15 | (7) | ||
| ISH23 | ISH50 | ISNCY | Halobacterium salinarium PHH1 (pHH1) | ND | 1,000 | 23/29 | 9 | ||
| ISH24 | ISNCY | Halobacterium salinarium PHH1 (pHH1) | ND | 3,000 | 10 | 7 | |||
| ISH26 | IS4 | Halobacterium salinarium PHH1 (pHH1) | X04832 | 1,384 | 19 | 11 | |||
| ISH26-1 | ISH26 | IS4 | Halobacterium salinarium | X04832 | 1,705 | 16 | 11 | ||
| ISH27-1 | ISH61 | IS4 | Halobacterium salinarium PHH1 (pHH1) | X54432 | 1,398 | 16 | 5 | ||
| ISH27-2 | ISH61 | IS4 | Halobacterium salinarium PHH1 (pHH1) | X54433 | 1,389 | 16 | 5 | ||
| ISH27-3 | ISH61 | ISH27-2 | IS4 | Halobacterium salinarium | X54434 | 1,389 | 16 | 5 | |
| ISH28 | IS5 | ISH1 | Halobacterium salinarium PHH1 (pHH1) | X59158 | 932 | 16 | 8 | ||
| ISH50 | ISH4 | ISNCY | Halobacterium salinarium R1 (pHH1) | X01584 | 996 | 23/29 | 8 | ||
| ISH51-1 | ISH51L | IS4 | Haloferax volcanii | X04389 | 1,370 | 15/16 | 5 | ||
| ISH51-2 | ISH51R | IS4 | Haloferax volcanii | X04389 | 1,379 | 15/16 | 5 | ||
| ISH51-3 | IS4 | Haloferax volcanii DS2 | ND | 1,400 | ND | ND | |||
| ISHs1 | ISHS1 | ISNCY | Halobacterium salinarium S1 | M38315 | 1,449 | 24/27 | 8 | ||
| ISIMb1 | IS110 | Moraxella bovis EPP63 | M32345 | >968 | ND | ND | |||
| ISIMl1 | ISIMb1 | IS110 | Moraxella lacunata ATCC 17956 | M34367 | >969 | ND | ND | ||
| ISJp4 | ISJP4 | IS5 | IS427 | Ralstonia eutropha JMP134 (pJP4) | U16782 | 915 | 18/19 | 3 | |
| ISL1 | IS3 | IS3 | Lactobacillus casei S-1 (phage FSV-B) | X02734 | 1,257 | 23/40 | 3 | ||
| ISL2 | IS5 | ISL2 | Lactobacillus helveticus LH28 | X77332 | 858 | 16 | 3 | ||
| ISL2A | ISL2 | ISL2 | IS5 | ISL2 | Lactobacillus helveticus LH27 | X77332 [V] | 858 | 16 | 3 |
| ISL3 | ISL3 | Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 | X79114 | 1,494 | 27/38 | 8 | |||
| ISLd1 | ISRLdTAL11451 | ISNCY | Leucaena diversifolia (Rhizobium sp.) TAL1145 | ND | 2,500 | ND | ND | ||
| ISLh1 | IS982 | Lactobacillus helveticus ATCC 15009 (pLH3) | X81979 | 962 | 32/35 | ND | |||
| ISLll1 | IS982 | IS982 | Lactococcus lactis subsp. lactis ME2 (pTN20) | M95956 [P] | >244 | ND | ND | ||
| ISLtaq1 | IS3 | IS150 | Thermus aquaticus ATCC 25104 | D14009 | 1,187 | 7 | ND | ||
| ISM1 | ISNCY | Methanobrevibacter smithii | X02587 | 1,381 | 33/34 | 8 | |||
| ISMi1 | ISMINC | IS3 | IS150 | Mycoplasma incognitus | M34841 | 1,405 | 22/29 | 3 | |
| ISMk1 | ISNCY | Mycobacterium kansasii | L11041 | 947 | ND | ND | |||
| ISMt1 | ISTUB1 | IS5 | IS427 | Mycobacterium tuberculosis H37Rv | X65618 | 969 | 16/17 | 4 | |
| ISMt2 | IS21 | Mycobacterium tuberculosis H37Rv | Z77165 | >2,200 | ND | ND | |||
| ISMt3 | IS21 | Mycobacterium tuberculosis H37Rv | Z83858 | 2,213 | 40/50 | 5 | |||
| ISNg1 | ISNGB | IS3 | IS150 | Neisseria gonorrhoeae 5019 | M19676 | 1,446 | 5/6 | ND | |
| ISPa1 | IS-PA-1 | ISNCY | Pseudomonas aeruginosa PA103 | M27175 | 1,035 | 0 | 5 | ||
| ISPa2 | IS-PA-2 | ISNCY | Pseudomonas aeruginosa Ps388 | M27186 | 785 | 0 | 5 | ||
| ISPa4 | IS-PA-4 | ISNCY | Pseudomonas aeruginosa PAO-muc | U16785 | 2,564 | 0 | ND | ||
| ISPa5 | IS-PA-5 | ISNCY | Pseudomonas aeruginosa PAO-muc | U16785 | 965 | 0 | ND | ||
| ISPa6 | IS-PA-6 | ISNCY | Pseudomonas aeruginosa PAO-muc | U16784 | 1,329 | 0 | ND | ||
| ISPg2 | PGIS2 | ISAs1 | Porphyromonas gingivalis A7436 | U77885 | 1,207 | 18/19 | 10 | ||
| ISPp1 | IS30 | Pedicococcus pentosaceus PPE1.0 | Z32771 | 1,039 | 21/23 | ND | |||
| ISPp2 | ISL3 | Pseudomonas putida ML2 | U25434 | >1,280 | ND | ND | |||
| ISpRm1 | ISpRm20111, ISpRmUSDA102 4-2 | ISRm1 | IS3 | IS2 | Rhizobium meliloti 2011 | ND | 1,400 | ND | ND |
| ISpRm3 | ISpRm22012-1, ISpRm22013-1 | ISNCY | Rhizobium meliloti 220-12 | ND | 750 | ND | ND | ||
| ISPs1 | IS5 | IS427 | Pseudomonas syringae pv. savastonoi | M11035 [P] | >203 | ND | ND | ||
| ISPsp1 | IS5 | IS5 | Pseudomonas sp. strain EST1001 (pEST1226) | M57500 | 1,188 | 9/12 | (4) | ||
| ISPsp2 | Tn401 | ISL3 | Pseudomonas sp. strain EST1001 (pEST1226) | M57500 | 1,416 | 23/24 | ND | ||
| ISR1 | IS3 | IS407 | Rhizobium lupini class IV (pRP4) | X06616 | 1,259 | 12/13 | 4 | ||
| ISRf1 | IS630 | Rhizobium fredii USDA 257 | M73698 | 1,136 | 10/17 | 2 | |||
| ISRhf1 | IS-Rf | IS3 | IS3 | Rhodococcus fascians DSM 20131 | ND | 1,300 | 15/19 | 3 | |
| ISRj1 | RSalpha, RSRj-alpha-9 | IS630 | Bradyrhizobium japonicum USDA 110 | X02581 | 1,122 | 6/8 | 2 | ||
| ISRj2 | RSbeta, RSRj-beta | ISNCY | Bradyrhizobium japonicum USDA 110 | ND | 950 | ND | ND | ||
| ISRj3 | RSgamma | ISNCY | Bradyrhizobium japonicum USDA 110 | ND | 1,000 | ND | ND | ||
| ISRj4 | RSdelta | ISNCY | Bradyrhizobium japonicum USDA 110 | ND | 1,000 | ND | ND | ||
| ISRj5 | RSepsilon | ISNCY | Bradyrhizobium japonicum USDA 110 | ND | 1,000 | ND | ND | ||
| ISRj6 | RSzêta | ISNCY | Bradyrhizobium japonicum USDA 110 | ND | 2,000 | ND | ND | ||
| ISRl1 | IS66 | Rhizobium leguminosarum bv. viciae 897 | L19650 | 2,495 | 14/15 | ND | |||
| ISRl2 | ISRlF7-2 | IS5 | IS1031 | Rhizobium leguminosarum bv. viciae MSDJ4184 | Z37965 | 932 | 14/17 | 3 | |
| ISRle39A | IS6 | Rhizobium leguminosarum VF39SM | X99520 | 890 | 18/22 | ND | |||
| ISRle39B | ISRle39A | IS6 | Rhizobium leguminosarum VF39SM | X99520 | 890 | 17/22 | ND | ||
| ISRm1 | IS3 | IS2 | Rhizobium meliloti 1021 (pRmeSU47a) | X56563 | 1,319 | 25/32 | 5 | ||
| ISRm2 | IS66 | Rhizobium meliloti 41 (pRme41a) | M21471, M21472, M15786, M15785 [P] | 2,700 | 24/25 | 8 | |||
| ISRm3 | ISRm-3 | IS256 | Rhizobium meliloti 102F70 | M60971 | 1,298 | 30 | 8, 9 | ||
| ISRm3G | ISRm3 | IS256 | Rhizobium meliloti GR4 (pRmeGR4b) | X63715 | 1,298 | 30 | 9 | ||
| ISRm4 | IS5 | IS1031 | Rhizobium meliloti GR4 | X65471 | 933 | 12/17 | 3 | ||
| ISRm5 | IS256 | Rhizobium meliloti IZ450 | U08627 | 1,340 | 22/27 | 8 | |||
| ISRm6 | IS3 | IS407 | Rhizobium meliloti GR4 (pRmeGR4b) | X95567 | 1,261 | 21/26 | 4 | ||
| ISRm7 | IS3 | IS407 | Rhizobium meliloti (pRmJT5) | L76274 | >912 | ND | ND | ||
| ISRm8 | IS66 | Rhizobium meliloti 41 (pSym-a) | L08667 | >1,235 | ND | ND | |||
| ISRm2011-2 | ISp-Rm2011-2 | IS630 | Rhizobium meliloti 2011 | U22370 | 1,053 | 16/19 | 2 | ||
| ISRo1 | IS256 | Rhodococcus opacus MR11 (pHG201) | U70364 | 1,474 | 15/23 | 8 | |||
| ISRsp1 | IS66 | Rhizobium sp. strain NGR234 (pSym) | X74068 | >2,200 | ND | ND | |||
| ISRsp2 | IS21 | Rhizobium sp. strain NGR234 (pNGR234a) | AE000081 | 2,623 | 12/16 | ND | |||
| ISS1A | ISS1S | IS6 | Lactococcus lactis subsp. lactis IL964 (pIL105) | L35176 | 807 | 18 | ND | ||
| ISS1B | ISS1S | IS6 | Lactococcus lactis subsp. lactis bv. diacetylactic B-1 (pTL02) | D63820 | 808 | 18 | ND | ||
| ISS1CH | ISS1N | IS6 | Lactococcus lactis subsp. lactis CNRZ270 | X60456 | 810 | 18 | ND | ||
| ISS1D | ISS1/pTD1 | ISS1N | IS6 | Lactococcus lactis subsp. lactis W67 (pTD1) | X62737 | 808 | 17/18 | ND | |
| ISS1E | ISS1N | IS6 | Lactococcus lactis MG1820 | X60734 | 808 | 15/18 | ND | ||
| ISS1M | ISS1N | IS6 | Lactococcus lactis subsp. cremoris SK11 (pSK111) | X59946 | 808 | 16/18 | ND | ||
| ISS1N | ORF-N1 | IS6 | Lactococcus lactis subsp. cremoris SK11 (pSK111) | M37395 | 808 | 18 | ND | ||
| ISS1RS | RS1, RS2 | ISS1S | IS6 | Lactococcus lactis subsp. lactis CNRZ270 (pUCL22) | X60594 | 808 | 18 | 8 | |
| ISS1S | SK7 | IS6 | Lactococcus lactis subsp. lactis ML3 (pSK08) | M18294 | 808 | 18 | 8 | ||
| ISS1T | SK13 | ISS1S | IS6 | Lactococcus lactis subsp. lactis ML3 (pSK08) | Z14022 | 808 | 18 | 8 | |
| ISS1W | ORF-W2 | IS6 | Lactococcus lactis subsp. cremoris Wg2 (pWV05) | M37396 | 809 | 17/18 | ND | ||
| ISS1X | ISS1S | IS6 | Lactococcus lactis subsp. lactis IL416 | M77708 | 808 | 18 | ND | ||
| ISS1Y | IS1216 | IS6 | Enterococcus faecalis CH19 | ND | >448 | ND | ND | ||
| ISS1Z | ISS1S | IS6 | Lactococcus lactis subsp. lactis OZS1 (pOZS550) | M85192 | 808 | 18 | ND | ||
| ISSc1 | IS30 | Spiroplasma citri (phage SpV1-R8A2B) | X51344 | ND | 25/31 | ND | |||
| ISSd1 | ISS1 | IS600 | IS3 | IS3 | Shigella dysenteriae 1-60R | M21947 | 1,266 | 19/27 | ND |
| ISSe1 | H-rpt | ISAs1 | Salmonella enterica D2 sv. Strasbourg M388 | U04165 | 1,284 | 5/9 | ND | ||
| ISSf1 | IS5 | IS1031 | Shigella flexneri (pR387) | X07848 [P] | >257 | ND | ND | ||
| ISSg1 | ISL3 | Streptococcus gordonii M5 | ND | 1,186 | 17/24 | ND | |||
| ISSm1 | IS1247 | IS1380 | Serratia marcescens 82041944 | M97172 [P] | >654 | ND | ND | ||
| ISSs1 | IS605 | Synechococcus sp. PCC7002 | M93569 | >1,100 | ND | ND | |||
| IST2 | IS256 | Thiobacillus ferroxidans ATCC 19859 | J03859 | 1,408 | 22/28 | 9 | |||
| IST3091 | IS3091 | IS30 | Thiobacillus ferroxidans tfi-91 (pTF191) | U32113 | 1,077 | >14/19 | ND | ||
| ISTcSa | IS630 | Synechocystis sp. strain PCC6803 | U38915 | 947 | 20/24 | 2 | |||
| ISV1L | ISVm5-2 | IS5 | IS903 | Vibrio parahaemoliticus WP1 | M64126 | 1,046 | 15/18 | ND | |
| ISV3L | IS5 | IS903 | Vibrio parahaemoliticus AQ3776 | M64121 | 1,054 | 19 | ND | ||
| ISV4R | IS5 | IS903 | Vibrio parahaemoliticus AQ3776 | M64124 | 1,043 | 17/19 | ND | ||
| ISV5L | ISV3L | IS5 | IS903 | Vibrio parahaemoliticus AQ3860 | M64122 | 1,058 | 19 | ND | |
| ISV5R | ISV3L | IS5 | IS903 | Vibrio parahaemoliticus AQ3860 | M64123 | 1,053 | 19 | ND | |
| ISVa1 | ISV-A1 | IS5 | IS903 | Vibrio anguillarum 775 (pJM1) | L40497 | 1,041 | 27/39 | 9 | |
| ISVa2 | ISV-A2 | IS5 | IS903 | Vibrio anguillarum 775 (pJM1) | L40498 | 1,054 | 21/25 | 9 | |
| ISVm2-4 | ISVM2-4 | ISVm5-2 | IS5 | IS903 | Vibrio mimicus 6 | M64131 | 964 | 18 | ND |
| ISVm5-2 | ISVM5-2 | IS5 | IS903 | Vibrio mimicus 6 | M64129 | 1,044 | 17/18 | ND | |
| ISVm7-2 | ISVM7-2 | IS5 | IS903 | Vibrio mimicus 6 | M64130 | 748 | 18 | ND | |
| ISVmL | ISVML | ISVm5-2 | IS5 | IS903 | Vibrio mimicus 6 | M64127 | 1,031 | 17/18 | ND |
| ISVmR | ISVMR | ISVm5-2 | IS5 | IS903 | Vibrio mimicus 6 | M64128 | 1,031 | 15/18 | ND |
| ISVNR | ISV4R | IS5 | IS903 | Vibrio cholerae 91 | M64125 | 881 | 16/19 | ND | |
| ISWz1 | IS91 | Weeksella zoohelcum | U14952 | >1,219 | ND | ND | |||
| ISXa1 | ISAs1 | Xanthobacter autotrophicus GJ10 | M26950 [P] | >947 | ND | ND | |||
| ISXc1 | ISXcB100-1 | ISNCY | Xanthomonas campestris B100 | ND | 1,300 | ND | ND | ||
| ISXc2 | ISXcB100-2 | ISNCY | Xanthomonas campestris B100 | ND | 1,700 | ND | ND | ||
| ISXc4 | ISNCY | Xanthomonas campestris pv. citri XW45 (pXW45N) | ND | 5,500 | ND | ND | |||
| ISXc5 | ISNCY | ISXc4 | Xanthomonas campestris pv. citri XW45 (pXW45J) | ND | 6,900 | ND | ND | ||
| ISYe | IS3 | IS150 | Yersinia enterocolitica 8081c | M29945 [P] | 884 | ND | ND |
IR indicates the length(s) of the terminal IR(s) in base pairs. A unique number refers to two IRs with the same length.
DR indicates the number of target base pairs duplicated on insertion. Numbers in parentheses are still uncertain.
ND, not determined.
Elements which are underlined are those to which we have given a (new) name.
[V] indicates a variant of the proposed accession number.
[P] indicates a partial DNA sequence.
ISNCY refers to unclassified IS elements.
Sequences of the (putative) Tpases were aligned by PileUp software (Wisconsin package version 9.1; Genetics Computer Group, Madison, Wis.) to generate a table of pairwise distances (Distances software, Genetics Computer Group package). These data were converted into dendrograms of relatedness (relationships) by using Growtree software by the UPGMA method (unweighted pair group method using arithmetic averages). We emphasize that we have not explored these in detail and that they do not necessarily represent phylogenetic relationships between the different elements.
Figure 5 shows the distribution of insertion sequences between families. In this global analysis, we have distinguished IS isoforms, of which there are 158. We define these as elements which show a divergence of less than 5% in the amino acid sequence of their potential proteins. When reading frames are obviously interrupted by mutation or errors in sequence determination, a second rule adopted has been a divergence in the nucleic acid sequence of less than 10%. This rather arbitrary choice is for convenience only. The IS3 (50 members excluding isoforms) and IS5 (47 members) families are clearly the most preponderant, followed by IS256 (29 members) and IS4 (28 members). Together, these represent nearly 50% of the 337 distinct (non-isoform) database entries. The nucleotide sequences of 33 ISs were not available and a further 21 ISs did not fall into the established families.
FIG. 5.
IS distribution among different families. The figure shows the number distribution of the entire IS database into the various IS families. The numbers of isoforms are indicated as the open boxes, and the distinct individual members are shown as shaded boxes. NCY, not classified. ND, nucleotide sequence not determined.
The distribution of ISs among different bacterial genera and groups (146a) is presented in Tables 3 and 4 (including isoforms). What has become increasingly clear since the publication of Mobile DNA in 1989 (29) is that members of most individual families can be distributed across many different eubacterial and archaebacterial genera (Table 3). So far, two exceptions are IS1, which appears to be limited to the enterobacteria, and IS66, which is restricted to bacteria of the rhizosphere. Not unexpectedly, the groups of bacteria in which the largest number of elements have been documented are also those which have received the most attention (Table 4). This implies that a wealth of elements remain to be discovered in the less well characterized groups.
TABLE 3.
IS family distribution among bacterial genera
| Genus | IS1 | IS3 | IS4 | IS5 | IS6 | IS21 | IS30 | IS66 | IS91 | IS110 | IS256 | IS605 | IS630 | IS982 | IS1380 | ISAs1 | ISL3 | NCYa | NDa | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetobacter | 1 | 4 | 2 | 7 | ||||||||||||||||
| Acinetobacter | 1 | 1 | ||||||||||||||||||
| Actinobacillus | 1 | 1 | ||||||||||||||||||
| Aeromonas | 1 | 1 | 2 | |||||||||||||||||
| Agrobacterium | 3 | 3 | 8 | 2 | 1 | 17 | ||||||||||||||
| Anabaena | 1 | 1 | 1 | 1 | 4 | 8 | ||||||||||||||
| Bacillus | 2 | 11 | 4 | 4 | 1 | 2 | 24 | |||||||||||||
| Bacteroides | 2 | 4 | 2 | 1 | 9 | |||||||||||||||
| Bordetella | 5 | 1 | 6 | |||||||||||||||||
| Bradyrhizobium | 1 | 1 | 5 | 7 | ||||||||||||||||
| Brevibacterium | 1 | 1 | ||||||||||||||||||
| Brucella | 2 | 2 | ||||||||||||||||||
| Burkholderia | 2 | 1 | 1 | 5 | 1 | 6 | 16 | |||||||||||||
| Caedibacter | 1 | 1 | ||||||||||||||||||
| Calothrix | 1 | 1 | 2 | |||||||||||||||||
| Campylobacter | 1 | 1 | ||||||||||||||||||
| Capnocytophaga | 2 | 2 | ||||||||||||||||||
| Caulobacter | 1 | 1 | 2 | |||||||||||||||||
| Clavibacter | 1 | 1 | 2 | |||||||||||||||||
| Clostridium | 1 | 1 | 1 | 1 | 1 | 5 | ||||||||||||||
| Corynebacterium | 1 | 2 | 3 | 6 | ||||||||||||||||
| Coxiella | 3 | 3 | ||||||||||||||||||
| Dichelobacter | 2 | 2 | ||||||||||||||||||
| Enterobacter | 1 | 2 | 3 | |||||||||||||||||
| Enterococcus | 4 | 2 | 1 | 2 | 9 | |||||||||||||||
| Erwinia | 1 | 1 | 2 | |||||||||||||||||
| Escherichia | 10 | 13 | 6 | 5 | 3 | 5 | 4 | 7 | 1 | 54 | ||||||||||
| Fremyella | 1 | 1 | ||||||||||||||||||
| Fusobacterium | 1 | 1 | ||||||||||||||||||
| Gluconobacter | 1 | 1 | ||||||||||||||||||
| Haemophilus | 6 | 6 | ||||||||||||||||||
| Halobacterium | 11 | 3 | 1 | 2 | 2 | 19 | ||||||||||||||
| Haloferax | 3 | 3 | ||||||||||||||||||
| Helicobacter | 1 | 1 | ||||||||||||||||||
| Lactobacillus | 3 | 2 | 1 | 1 | 1 | 8 | ||||||||||||||
| Lactococcus | 5 | 15 | 2 | 4 | 26 | |||||||||||||||
| Leptospira | 2 | 2 | 4 | |||||||||||||||||
| Leucaena | 1 | 1 | ||||||||||||||||||
| Leuconostoc | 1 | 1 | 1 | 3 | ||||||||||||||||
| Methanobrevibacter | 1 | 1 | ||||||||||||||||||
| Moraxella | 2 | 2 | ||||||||||||||||||
| Mycobacterium | 5 | 1 | 1 | 1 | 2 | 4 | 9 | 1 | 1 | 25 | ||||||||||
| Mycoplasma | 14 | 14 | ||||||||||||||||||
| Neisseria | 1 | 2 | 1 | 4 | ||||||||||||||||
| Nocardia | 1 | 1 | ||||||||||||||||||
| Paracoccus | 2 | 2 | ||||||||||||||||||
| Pediococcus | 1 | 1 | ||||||||||||||||||
| Porphyromonas | 1 | 1 | 2 | |||||||||||||||||
| Proteus | 1 | 1 | 2 | |||||||||||||||||
| Pseudomonas | 3 | 4 | 5 | 1 | 1 | 2 | 2 | 5 | 3 | 26 | ||||||||||
| Ralstonia | 1 | 1 | 1 | 3 | ||||||||||||||||
| Rhizobium | 5 | 2 | 2 | 1 | 4 | 3 | 2 | 1 | 20 | |||||||||||
| Rhodococcus | 1 | 1 | 4 | 6 | ||||||||||||||||
| Saccharopolyspora | 4 | 4 | ||||||||||||||||||
| Salmonella | 2 | 5 | 5 | 1 | 13 | |||||||||||||||
| Serratia | 1 | 1 | ||||||||||||||||||
| Shigella | 4 | 5 | 1 | 1 | 1 | 12 | ||||||||||||||
| Sphingomonas | 1 | 1 | ||||||||||||||||||
| Spiroplasma | 1 | 1 | ||||||||||||||||||
| Staphylococcus | 8 | 1 | 1 | 2 | 12 | |||||||||||||||
| Streptococcus | 2 | 5 | 3 | 1 | 1 | 5 | 1 | 18 | ||||||||||||
| Streptomyces | 1 | 4 | 1 | 3 | 1 | 10 | ||||||||||||||
| Sulfolobus | 1 | 1 | ||||||||||||||||||
| Synechococcus | 1 | 1 | ||||||||||||||||||
| Synechocystis | 2 | 2 | 1 | 5 | ||||||||||||||||
| Thermophilic | 1 | 1 | ||||||||||||||||||
| Thermus | 1 | 2 | 3 | |||||||||||||||||
| Thiobacillus | 1 | 1 | 2 | |||||||||||||||||
| Vibrio | 13 | 1 | 1 | 15 | ||||||||||||||||
| Weeksella | 1 | 1 | ||||||||||||||||||
| Xanthobacter | 1 | 1 | 2 | |||||||||||||||||
| Xanthomonas | 1 | 1 | 3 | 5 | 10 | |||||||||||||||
| Yersinia | 2 | 4 | 1 | 1 | 1 | 9 | ||||||||||||||
| Total | 14 | 82 | 41 | 68 | 45 | 21 | 19 | 12 | 8 | 20 | 33 | 25 | 12 | 7 | 6 | 12 | 21 | 21 | 33 | 500 |
NCY, not classified; ND, sequence not determined.
TABLE 4.
IS distribution among bacterial groups
| Bacterial group | IS1 | IS3 | IS4 | IS5 | IS6 | IS21 | IS30 | IS66 | IS91 | IS110 | IS256 | IS605 | IS630 | IS982 | IS1380 | ISAs1 | ISL3 | NCYa | NDa | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spirochetes | 2 | 2 | 4 | |||||||||||||||||
| Vibrioid gram negative | 1 | 1 | 2 | |||||||||||||||||
| Curved gram negative | 0 | |||||||||||||||||||
| Gram-negative rods and cocci | 22 | 1 | 23 | 2 | 7 | 4 | 12 | 2 | 5 | 9 | 8 | 5 | 1 | 4 | 6 | 22 | 133 | |||
| Facultatively anaerobic gram negative | 14 | 22 | 8 | 19 | 7 | 5 | 4 | 5 | 3 | 1 | 12 | 1 | 1 | 10 | 6 | 2 | 120 | |||
| Anaerobic gram negative | 2 | 5 | 2 | 1 | 1 | 2 | 1 | 14 | ||||||||||||
| SO4 or S reducing | 0 | |||||||||||||||||||
| Anaerobic gram-negative cocci | 0 | |||||||||||||||||||
| Rickettsiae and Chlamydiae | 3 | 3 | ||||||||||||||||||
| Anoxygenic phototrophs | 0 | |||||||||||||||||||
| Oxygenic phototrophs | 3 | 3 | 1 | 2 | 2 | 1 | 1 | 4 | 17 | |||||||||||
| Aerobic chemolithotrophs | 1 | 1 | 2 | |||||||||||||||||
| Budding bacteria | 1 | 1 | 2 | |||||||||||||||||
| Sheathed bacteria | 0 | |||||||||||||||||||
| Gliding bacteria | 2 | 2 | ||||||||||||||||||
| Myxobacteria | 0 | |||||||||||||||||||
| Gram-positive cocci | 7 | 5 | 28 | 7 | 5 | 1 | 4 | 9 | 3 | 69 | ||||||||||
| Sporeforming gram positive | 3 | 12 | 4 | 4 | 1 | 1 | 1 | 1 | 2 | 29 | ||||||||||
| Regular nonsporeforming gram positive | 3 | 2 | 1 | 1 | 1 | 8 | ||||||||||||||
| Irregular nonsporeforming Gram positive | 1 | 1 | 2 | 4 | 1 | 9 | ||||||||||||||
| Mycobacteria | 5 | 1 | 1 | 1 | 2 | 4 | 9 | 1 | 1 | 25 | ||||||||||
| Nocardioform actinomycetes | 1 | 1 | 4 | 4 | 1 | 11 | ||||||||||||||
| Multilocular actinomycetes | 0 | |||||||||||||||||||
| Actinoplanetes | 0 | |||||||||||||||||||
| Streptomyces | 1 | 4 | 1 | 3 | 1 | 10 | ||||||||||||||
| Maduromycetes | 0 | |||||||||||||||||||
| Thermonosporas | 0 | |||||||||||||||||||
| Thermoactinomycetes | 0 | |||||||||||||||||||
| Other actinomycetes | 0 | |||||||||||||||||||
| Mycoplasmas | 14 | 1 | 15 | |||||||||||||||||
| Methanogens | 1 | 1 | ||||||||||||||||||
| Archeal SO4 reducers | 0 | |||||||||||||||||||
| Halobacteria | 14 | 3 | 1 | 2 | 2 | 22 | ||||||||||||||
| Cell-wall-less archaebacteria | 0 | |||||||||||||||||||
| Hyperthermophilic | 1 | 1 | 2 | |||||||||||||||||
| Total | 14 | 82 | 41 | 68 | 45 | 21 | 19 | 12 | 8 | 20 | 33 | 25 | 12 | 7 | 6 | 12 | 21 | 21 | 33 | 500 |
NCY, not classified; ND, sequence not determined.
In the following, we describe first the essential features of each IS family and subsequently the current state of knowledge about the control of gene expression and transposition mechanism of the better-studied IS sequences within each family.
IS1 Family
IS1 was one of the first bacterial insertion sequences to be isolated and characterized (100, 144). The original examples were obtained from an F′lac-proB plasmid (IS1K [167]) and the multiple drug resistance plasmid R100 (IS1R [254]). The nucleotide sequences of 17 variants of this IS from Escherichia and Shigella species have been determined (200, 343, 379). Of these, three have proved to be duplicates and one is only partially complete. Nine of the others exhibit sequence divergence of between 0.52 and 10% at the nucleic acid level. These have been called IS1 isoforms. Two examples, IS1N and IS1H, are significantly different from the others (45 to 47% divergence in nucleotide sequence; 55 to 58% divergence at the protein level) but similar to each other (14 to 19% divergence at the protein level) and might be considered distinct members of the family. Except for IS1K(A) and IS1R(G), transposition of these elements has not been directly demonstrated experimentally in a controlled way but is implied from the isolation of mutants with spontaneous mutations in various genes. IS1 is a component of several compound transposons such as Tn9 (219) and Tn1681 (318), where it is present in direct or inverted orientation flanking a chloramphenicol acetyltransferase and heat-stable toxin gene, respectively.
Integration of IS1 is accompanied by duplication of generally 9 bp in the target DNA at the site of insertion (48, 117). Direct target repeats of 8, 10, and 14 bp (154, 220, 312) have also been observed. The frequency of their appearance is increased by mutations within the Tpase gene (220). The element exhibits a strong preference for insertion into AT-rich target regions (105, 236, 375).
IS1 (Fig. 6) is one of the smallest “autonomous” bacterial insertion sequences isolated so far. It is 768 bp long, includes two approximately 23-bp imperfect inverted repeats (IRL and IRR) located at its ends (167, 254), and two partly overlapping open reading frames (insA and insB′) located in the 0 and −1 relative translational phases, respectively (Fig. 6A). The integrity of these frames is essential for transposition (164, 222).
FIG. 6.
Organization of IS1. (A) Structure of IS1. Left (IRL) and right (IRR) terminal IRs are shown as solid boxes. The relative positions of the insA and insB′ reading frames, together with their overlap region, are shown within the open box representing IS1. The IS1 promoter pIRL, partially located in IRL, is indicated by a small arrow. IHF binding sites, located partially within each terminal IR, are shown as small open boxes. The InsA protein is represented as a hatched box beneath. The InsA and InsB′ components of the InsAB′ frameshift product are shown as hatched and stippled boxes, respectively. Arrows indicate the probable region of action of InsA and InsAB′ proteins. The effect of InsA and InsAB′ on transposition is shown above. (B) RNA and protein sequence in the crossover region between the two open reading frames. Codons shown above the RNA sequence show the product of direct translational readout. Those below show the product of a −1 translational frameshift. The heptanucleotide A6C frameshift sequence involved in production of InsAB′ from the wild-type IS1 coding sequence is indicated in boldface type, as is the UAA termination codon for InsA.
A transcript is initiated from a promoter, pIRL, partially located in IRL (367) and is translated to give two products: InsA and InsAB′. The first and more abundant protein is encoded by the insA frame alone. The small, basic InsA protein binds specifically to the IRs (376, 377) and represses transcription from pIRL (221, 378). It also appears to inhibit transposition, probably by competing with the Tpase for binding to the ends of the element (95). The second protein, InsAB′, is the Tpase of the element. Its production results from a programmed translational frameshift between the insA and insB′ frames which occurs at a frequency of approximately 1%. The site of frameshifting is an A6C motif located at the 3′ end of the upstream insA frame (95, 310) (Fig. 6B). Natural transposition of IS1 occurs at a relatively low frequency (approximately 10−7 in a standard mating assay). Insertion of an additional A residue within the A6C motif to yield A7C or replacement of the motif with GA2GA3C fuses the two reading frames, leading to constitutive production of the Tpase while eliminating the production of InsA. This results in levels of transposition of between 0.1 and 1% in vivo (94, 95). Indirect evidence has been presented suggesting that a translational restart within the insA frame gives rise to an InsAB′ protein with an N-terminal deletion. It has been proposed that this protein is the true Tpase (229, 230). No independent product of the downstream frame, insB′, alone has been detected. Transposition activity appears to depend on the InsAB′/InsA ratio (95). Since this is relatively insensitive to the intensity of transcription, this arrangement ensures that IS1 is not activated by high levels of impinging transcription following insertion into highly expressed genes.
An additional control of Tpase expression may be exercised at the level of transcription termination. Early studies on the organization of IS1 identified a region at the end of the insA gene which behaves as a Rho-dependent transcription terminator (151, 272). Premature transcription termination would therefore result in the production of an mRNA lacking the insB frame. The role of this sequence in the control of IS1 transposition remains to be determined.
IS1 generates both simple insertions and replicon fusions (cointegrates) which are composed of two copies of the insertion sequence in DR, one at each junction between the target and donor replicons. The occurrence of stable cointegrates as transposition end products led to the suggestion that transposition of IS1 can proceed in a replicative manner (106) while simple insertions may occur without replication (34). Thus, IS1 may be capable of both replicative and conservative transposition. More convincing evidence in support of a duplicative transposition pathway has recently been obtained by analyzing the products of intramolecular transposition (342). In addition, these and other studies (307) detected excised circular copies of the IS1-derived transposon, and it was suggested that, as in the case of IS911 (see “IS3 family” below), such forms may integrate into a target molecule to give rise to simple insertions.
High levels of InsAB′ in the presence of suitable IS1 ends induce the host SOS response, possibly reflecting endonucleolytic activity of the IS1 Tpase (198). By using this in vivo assay system, originally developed for screening mutants of the IS10 Tpase (see “IS4 family” below), it was possible to show that for relatively short artificial derivatives of IS1, the level of response depends in a periodic manner on the distance between the ends. The periodicity was found to be about 10 to 11 bp and was also reflected in the transposition activity, suggesting a requirement for correct helical positioning of both ends. Two directly repeated ends were also capable of eliciting the SOS response, although they were not capable of giving productive transposition.
Inspection of the amino acid sequence of InsAB′ indicates the presence of an amino acid triad, H200, R203, and Y231, reminiscent of the catalytic site of integrases of the phage λ family (2, 13) in the C-terminal end of the protein. Derivatives with the H200L, R203L, or Y231F mutation are inactive in several assays whereas an H200Q mutation retains a low level of activity. The relative activities of these mutants parallel those of similar mutants with mutations in the Flp recombinase (313). At present it is difficult to assess the relevance of these similarities to the transposition mechanism. It should be noted that in IS1N (253) and IS1H (379) the H200 residue is replaced with R but neither of these IS1 derivatives has been demonstrated to be capable of autonomous transposition.
IS3 Family
General features.
Over 80 members of the IS3 family have been found so far in 40 bacterial species. Of these, 32 proved to be isoforms or duplicate isolates and 11 were only partial sequences. The G+C content of members of the family ranges from 70% in the mycobacterial examples to 25% in those isolated from Mycoplasma species. In spite of this enormous variation, they are strikingly similar in many respects and form an extremely coherent and highly related family. Most show a similar G+C content to their host organism. Several family members are part of compound transposons. These include IS3411 flanking genes for citrate utilization (158); IS4521, which flanks a heat-stable enterotoxin gene in enterotoxinogenic E. coli (148); and IS1706 (149), which flanks genes of the Clp protease/chaperone family.
Members are characterized by lengths of between 1,200 and 1,550 bp, with one exception (IS481 [1,045 bp]), and inverted terminal repeats in the range of 20 to 40 bp. These repeats are variable but clearly related (Fig. 7C). The majority of the elements terminate with 5′-TG-----CA-3′ and present an internal block of G and C residues of variable length. IS3 family members generally have two consecutive and partially overlapping reading frames, orfA and orfB, in the relative translational reading phases 0 and −1, respectively (Fig. 7A). It has been demonstrated in at least three cases (IS150 [351], IS3 [308], and IS911 [267]) that in addition to the product of the upstream frame, OrfA, a fusion protein, OrfAB, is generated by programmed translational frameshifting (55). However, in contrast to IS1, the product of the downstream frame, OrfB, is also detected. The frequency of frameshifting varies from element to element. It is approximately 50% for IS150 (351) and only 15% for IS911 (267).
FIG. 7.
IS3 family. (A) General organization of IS3 family members. The solid boxes indicate the left (IRL) and right (IRR) terminal IRs. Transcription probably occurs from a weak promoter located partially in IRL. The two consecutive overlapping open reading frames are indicated (orfA and orfB) and are arranged in reading phases 0 and −1 respectively. The products of these frames are shown below. OrfA and OrfB are shown as hatched and open boxes, respectively. The position of a potential helix-turn-helix motif (HTH) is shown as a stippled box in OrfA, and the DDE catalytic domain is shown as a stippled box in OrfB. A potential leucine zipper (LZ) at the C-terminal end of OrfA and extending into OrfAB is also indicated. Each leucine heptad is indicated by an oval. Those present in the OrfA domain are cross-hatched, whereas that deriving from the frameshifted product is open. (B) Dendrogram based on the alignment of the amino acid sequences of predicted OrfA proteins from 40 different elements (left) and 44 predicted OrfB frames (right). The major groups are indicated by brackets. (C) Nucleotide sequences of the terminal IRs of two representative elements of each subgroup, together with some of the elements which do not clearly form part of these groups.
Several members exhibit an organization which does not apparently conform to the generic IS3 member. In IS120, for example, the relationship between the reading phases of the upstream and downstream open reading frames appears to be +1 rather than −1, while in ISNg1 and ISYe1 the characteristic motifs of OrfB (see below) are distributed between reading phases. Other members, such as IS1076, IS1138, IS1221, and IS1141, exhibit only one long open reading frame. Although these may be true variants, it cannot at present be ruled out that the variations are simply due to errors in sequence determination. IS481 is significantly shorter (1,045 bp) than the others and carries only one open reading frame. Several individual examples of this element have been isolated. It is included in this family since it shows strong similarities in its putative Tpase sequence.
Family members from Mycoplasma species merit special attention. Not only does the host use a nonuniversal genetic code in which the opal termination codon TGA directs the insertion of tryptophan (257), but also their genomes are among the smallest bacterial genomes known and are extremely AT rich. To date, four different IS3 family members have been observed in Mycoplasma and the nucleotide sequence of 14 individual examples has been determined. Of these, only IS1138 and IS1138b have been directly demonstrated to undergo autonomous transposition (33). All exhibit similarly high A+T contents, and this unusual base composition could lead to difficulties in sequence determination. It is remarkable that typical IS3 family characters have been maintained in such an “extreme” genetic environment. Nine of these are closely related and form a group of isoelements which have been called IS1221. As indicated above, one of these carries a single long reading frame (representing orfA plus orfB) instead of two consecutive overlapping frames. The others each carry insertions or deletions which destroy the equivalent of orfA, orfB, or both. Expression studies with E. coli indicate that a protein, equivalent to OrfAB, is indeed produced from the long open reading frame of IS1221. Interestingly, it appears that a second truncated protein, equivalent to OrfA, may be generated from orfAB by translational frameshifting, representing an inverted expression pattern to the majority of the family members (382). Although this appears not to be a general rule for IS3 family members originating from Mycoplasma hosts, the presence of a similar single-frame arrangement in a second member, IS1138, indicates that it might not be rare. Because of the extremely high A+T content of these elements, many potential frameshift windows of the A6(G/C) or A7 type are expected to occur. Only direct experiments will therefore be able to determine which, if any, of these sequences are used to generate the Tpase or, conversely, an OrfA-like protein.
Extensive alignment studies of the predicted OrfA and OrfB amino acid sequences between themselves and with those of other transposable elements (87, 97, 172, 176, 288) have provided insights into the structure-function relationships of the proteins. The predicted primary amino acid sequences of the various OrfA proteins from different members of the family exhibit a relatively strong α-helix–turn–α-helix motif (Fig. 7A), suggesting that they might provide sequence-specific binding to the terminal IRs of their particular IS (304). The OrfB products, on the other hand, carry a DD(35)E motif and have additional identities to retroviral integrases and various other Tpases (Fig. 3) (266). These include two amino acids located 4 and 7 residues downstream from the glutamate residue. Interestingly, many members carry a putative leucine zipper located at the end of OrfA (and sometimes extending into the OrfB region of the OrfAB protein) (41, 344, 382). Studies with IS911 and IS2 strongly suggest that this represents one domain of multimerization of the proteins (137a, 138, 139, 204).
The IS3 family can be divided into several subgroups (Table 2) defined by deep branching in the alignment of the various OrfB sequences (Fig. 7B). We have designated these the IS2 and IS407 subgroups (which appear closely related) and the IS3, IS51, and IS150 groups. One feature which lends biological credence to these subgroups is that they also clearly appear clustered (with some exceptions) in the results of the alignments with the upstream OrfA protein. Moreover, there is a strong correlation between the members of each group and the number of base pairs of target DNA duplicated on insertion: for elements in the IS2 subgroup, insertion invariably leads to a 5-bp direct target repeat; for the IS407 subgroup, a 4-bp repeat is observed; while for the other groups, a 3-bp repeat is obtained. In the latter cases, some of the elements, e.g., IS911, occasionally generate 4-bp repeats. This clustering is also exhibited to some extent in the nucleotide sequence of the terminal IRs (Fig. 7C) and is particularly marked in the IS2, IS51, and IS407 subgroups. It can also be observed in the primary sequence details of the putative leucine zipper (data not shown). IS481 appears to be relatively separated from the major groups. This is also reflected by the fact that it is the only known example which appears to generate a 6-bp direct target repeat (consensus NCTAGN) (325a).
Several members carry GATC methylation sites within 50 bp of their ends, which have been shown in one case, IS3, to modulate transposition activity (320). However, this is not a general characteristic of the family, nor is it restricted to any particular subgroup.
Little is known about the insertion specificity of members of the family. IS2 exhibits a preference for a region of bacteriophage P1, but the basis of this preference is at present unknown (311). Both IS911 (269) and IS150 (361) have been found next to sequences which resemble their IRs, and IS1397 is invariably located within intergenic repeated sequences in E. coli (bacterial interspersed mosaic elements [16]).
Finally, an element isolated from the ECOR collection of E. coli and closely related to IS3411 carries a group II intron (98). The implication of this for the regulation transposition of this element has not been investigated.
IS911.
One of the best-characterized members of the IS3 family is IS911. OrfA and OrfAB bind specifically to the terminal IRs (139, 264). Constitutive production of OrfAB from a contiguous orfAB frame (generated by mutation and eliminating production of OrfA) leads to only a modest increase in intermolecular transposition activity. In contrast, high production of OrfAB in this way either in cis or in trans stimulates the production of excised circular transposon copies, whose formation is best explained by a single-ended attack by one IS extremity on the other (268). This is thought to occur in a two-step process (Fig. 8) in which one end of the transposon is cleaved to generate a free 3′OH, which, in turn, is used as a nucleophile to attack the opposite end. This results in the circularization of a single transposon strand visualized as a figure-eight molecule in which the transposon ends are joined by a single-strand bridge (Fig. 8) (265). It leaves a 3′OH group on the vector backbone at the point of insertion. Kinetic studies suggest that the figure-eight species is processed into transposon circles.
The figure-eight recombination reaction has been reproduced by using a cell-free system, but no transposon circles, excised linear transposon species, or indeed any product resulting from double-strand cleavage at the transposon ends was detected in the in vitro system. It is not known how the figure-eight form might be processed into transposon circles, although host factors which promote either replication or second-strand recombination are thought to be involved (270). By using a purified figure-eight substrate, the cell-free system was found to support a reaction equivalent to the disintegration characteristic of retroviral integrases (59) (see “Reaction mechanisms” above). Moreover, this activity is also exhibited by the OrfB region of the protein alone, demonstrating that the DD(35)E domain carries out catalysis (270).
Although it is not yet clear whether transposon circles are “natural” transposition intermediates, they are efficient Tpase substrates for intermolecular transposition in vivo (338; also see reference 269). Simultaneous high-level expression of OrfA with the OrfAB protein greatly reduces or eliminates the formation of excised transposon circles (and the figure-eight species) and stimulates intermolecular transposition. It also stimulates intramolecular transposition of a plasmid carrying a cloned circle junction. The development of an intermolecular transposition system in vitro (339) has demonstrated highly efficient integration of transposon circles in a reaction which requires both OrfAB and OrfA. Integration does not require a supercoiled donor molecule and is optimal when the abutted IRs are separated by 3 bp, as occurs in the circle junction. More recently, linear derivatives of IS911 have been observed in vivo. They appear to be derived from the transposon circle rather than resulting from direct excision from the donor plasmid molecule. Moreover, while they undergo integration in vitro, the efficiency of this reaction is significantly reduced compared to that of the transposon circles (337a).
One striking feature of transposon circularization is that it creates a strong promoter at the circle junction in which IRR contributes a −35 hexamer and IRL contributes a −10 hexamer (294). This has suggested a novel mechanism for autoregulation of transposition (Fig. 8, right-hand pathway). Transposon circles are proposed to be generated at low frequencies by a combination of low Tpase levels (from the weak endogenous promoter) and host functions (to ensure processing of the second strand). Once formed, the junction promoter ensures high levels of Tpase which is capable of binding to the abutted ends, introducing two single-strand breaks (one at each end) generating an opened transposon molecule. Transfer of both ends to a suitable target completes the transposition cycle. Insertion results in the destruction of the efficient junction promoter. This model does not require double-strand cleavage and therefore takes into account the observation that the only activity detected for the OrfAB protein is cleavage and transfer of a single DNA strand. It also ensures that a suitable substrate is present before high levels of Tpase are produced. The results do not rule out alternative pathways involving simple excision (Fig. 8, left-hand pathway).
Other members.
Several other members of this family are also being analyzed in detail. These include IS2, IS3, and IS150. All three have been shown to generate circles when supplied with high levels of the fused-frame Tpase (208, 308, 361).
IS3 also generates adjacent deletions (308) but, unlike IS911, appears to undergo excision from the donor molecule as a linear form following a staggered double-strand break at each end. These forms have a 3-base 5′ overhang and may be an alternative type of transposition intermediate (309) (Fig. 8, left-hand pathway). Such forms may be equivalent to the linear IS911 species derived from transposon circles. In addition, IS3-derivative transposons in which two abutted ends have been engineered undergo high levels of transposition (320). Insertion of IS3 creates generally 3-bp and sometimes 4-bp direct target repeats. It is significant that plasmids in which the IRs are separated by 4 bp are more active than those in which they are separated by 8 bp. In these studies the authors were unable to engineer derivatives with two complete tandem IS3 elements. This may be the result of the formation of a strong hybrid promoter which, as described for IS911 and other ISs (see above), drives high levels of Tpase expression. This configuration of ends is equivalent to that found at the circle junction and suggests that abutted ends of IS3 are also efficient substrates in transposition.
IS2 generates direct target duplications of 5 bp on insertion (111), although transposon circles generated with this element carry only a single base pair separating IRL and IRR (208). Moreover, while IS2 carries a conserved terminal 5′-CA-3′ at its right end, the left end terminates with 5′-TG-3′. Further analysis of IS2 circles has demonstrated that the atypical IRL does not act as a strand donor but uniquely as a target in the circularization reaction. Functional studies indicate that the product of the upstream orfA may inhibit transposition (147). It has been shown to bind specifically to IRL at a sequence which overlaps the −10 hexamer of the resident Tpase promoter and to repress expression of OrfA. It does not appear to bind IRR (note that in the original article the authors reverse the standard definition of IRL and IRR [147]). Several other elements also exhibit small IR sequences which flank the −10 hexamer of the putative resident Tpase promoter. IS2-derivative transposons in which two abutted ends have been engineered also undergo high levels of transposition (208, 330), and, like IS911, the circle junction of IS2 also constitutes a strong promoter capable of driving Tpase expression. Several (but not all) IS3 family elements may also carry similarly located potential −35 and −10 sequences within their IRs.
IS4 Family
General features.
Two subgroups were originally included in the IS4 family (288). However, we have decided to separate the IS4 and IS5 subgroups into families in their own right. The IS4 family is quite heterogeneous. There are 41 members, including 13 isoforms. It is difficult to justify division of this family into well-defined subgroups based on the relationships between their Tpases, although one major grouping (IS231A through IS4 [Fig. 9A]) shows relatively closely related IRs (Fig. 9B) and tends to generate target repeats of between 10 and 13 bp (Table 2). Many, such as IS10 and IS50, are involved in compound transposons (29). A single open reading frame spans most of the element. A previous alignment of these Tpases (288) revealed several regions of amino acid conservation: N1, N2, N3, and C1 (226), which encompass the D (N2), D (N3), and E (C1) regions of typical DDE motifs, respectively (Fig. 3). The distance between N3 and C1 in members of the IS4 family is about 100 amino acids, compared with 33 to 40 amino acids for Tpases with the canonical DDE motif. In the case of IS10 (see below), the importance of these regions has received experimental support. Several members carry GATC methylation sites, which, for both IS10 and IS50, have been shown to play a modulating role in transposition activity (see below).
FIG. 9.
IS4 family. (A) Dendrogram based on alignments of the putative Tpases. (B) Terminal IRs of selected members. (C) Schematic representation of IS10 and IS50. The terminal IRL (OE) and IRR (IE) are shown as solid boxes. Dam methylation sites (∗) are also shown. For IS10, the Tpase promoter, pIN, and the antiRNA promoter, pOUT, are indicated as horizontal arrows. A mechanistically important IHF site is indicated by an open box next to IRL. The Tpase is represented underneath. Stippled boxes indicate the positions of the consensus sequence within members of the IS4 family (from positions 93 to 132, 157 to 187, and 266 to 326). I and II indicate patch I and patch II, respectively, as defined by mutagenesis. The vertical arrow indicates a protease-sensitive site. For IS50, the promoters for Tpase and inhibitor protein, p1 and p2, respectively, are indicated as horizontal arrows. DnaA and Fis binding sites, located close to the left and right ends, respectively, are indicated by open boxes.
IS10 and IS50 (Fig. 9C) are certainly the best-characterized members of this group. Both transpose by a “cut-and-paste” mechanism, as does IS231A (205a). The mechanism of transposition of other members of this group is at present unknown.
IS10.
IS10 (Fig. 9C) forms part of the composite tetracycline resistance transposon Tn10. Since it has been recently described in a comprehensive review (179), only a brief summary is included here. Insertion results in a 9-bp target duplication and occurs in a relatively well defined target sequence with the symmetric 6-bp consensus: 5′-NGCTNAGCN-3′. It carries 22-bp terminal IRs, called outside (OE = IRL) and inside (IE = IRR) ends, describing their relative position in Tn10, and exhibits an elaborate ensemble of mechanisms to control its activity. The Tpase of IS10 is expressed from a single long reading frame. Expression is protected from activation by external transcription by an IR sequence located close to the left end (see “Sequestration of translation initiation signals” above). The activity of the Tpase promoter (pIN) is modulated by the presence of a site for the Dam methylase located within the −10 hexamer. Methylation reduces pIN activity, but after replication, this site becomes transiently hemimethylated and the promoter strength is concomitantly increased. A second level of control is exercised by a small antisense RNA molecule transcribed from a second, outwardly directed, promoter located proximal to IRL (pOUT). This RNA is complementary to the region of Tpase mRNA carrying the ribosome binding site, acts in trans, and interacts with the mRNA to inhibit ribosome binding.
Dam methylation also exerts control at another level. In addition to the Dam site located in IRL, another site is localized in IRR. When these sites are methylated, the activity of the ends in transposition is reduced, whereas when they are hemimethylated, the transposition activity is increased. With this arrangement, both Tpase expression and transposition activity are coupled to replication of the donor molecule. Since transposition of IS10 is nonreplicative, this ensures that passive replication of the IS occurs before transposition takes place. A binding site for the host IHF protein is also located at the left end of IS10, outside IRL. IHF plays a subtle role in IS10 transposition in vivo by influencing the nature of transposition products (314).
Overproduction of Tpase in vivo in the presence of a suitable IS10 derivative transposon leads to the appearance of a variety of transposon forms. Among these are excised transposon circles (243). These are not equivalent to those observed for IS3 family members (see “IS3 family” above) and have proved to be rearranged circular products of transposition (179, 244). In addition, linear transposon species which are retained in a circular configuration by Tpase bound at the ends are also observed. IS10 transposition involves double-strand cleavage at the ends of the IS, thus separating it from the donor backbone. An in vitro IS10 transposition reaction has been developed by using a circular plasmid donor substrate (53). This generates an array of intermediates and products, some of which are identical to those found in vivo. However, additional species can be detected. These include the results of single-ended transpositions (lariat structures), reactions involving synapsis of directly repeated IRs, and even reactions between IRs on separate molecules (54). A simpler assay employs a linear double-stranded oligonucleotide substrate including the “outside” end of IS10, which carries the IHF binding site. This assay has allowed the isolation and analysis of a synaptic complex between two copies of the substrate DNA fragment, Tpase, and IHF. IHF is essential for Tpase binding and for the formation of synaptic complexes (299). By using this system, the two single-strand cleavages which take place at each of the transposon ends have been shown to occur in a specific order: the transferred strand is the first to be cleaved (38). More recently, data have been obtained suggesting that cleavage of the second strand at each end is promoted by nucleophilic attack with the liberated 3′OH generated in cleavage of the first strand, giving rise to hairpin structures at both ends (175a).
As indicated above (see “Terminal inverted repeats”), the ends of IS10 can be divided into an internal Tpase binding domain and a terminal domain intimately involved in cleavage and strand transfer. Tpase function has also been probed by the isolation of Tpase mutants. Several ingenious screening procedures have been devised. One of these was a scheme for isolating derivatives with altered target specificity (ATS). A second exploited the capacity of IS10 Tpase to induce the host SOS response in the presence of suitably oriented IS10 ends. Mutants presumably defective in strand transfer but retaining the ability to generate an SOS response have been obtained. Most fall within two regions, called patch I and patch II (Fig. 9C) (137), corresponding to the N3 and C1 regions of the IS4 group Tpases (Fig. 3) (179, 226, 288). Moreover, intragenic suppressors of the SOS and ATS mutations, derivatives which suppress the effect of mutations within the IS10 ends, and mutations giving a hypernicking phenotype have also been localized in these regions (see reference 179 for a summary). Partial proteolysis of the Tpase generates an N-terminal domain containing patch I and a C-terminal domain containing part of patch II, which are connected in the complete protein by a short proteolytically sensitive linker region. This region also carries part of patch II. Interestingly, Tpase activity can be reconstituted with a mixture of the two proteolytic fragments (192). Recently, evidence has been presented suggesting that a single active site in the enzyme is used for 3′ and 5′ cleavage and for strand transfer (39).
IS50.
IS50 forms part of transposon Tn5 and, like IS10, has been extensively studied (reviewed in references 27 and 286). It differs significantly from IS10 in its organization and control (Fig. 9C). It inserts without notable target sequence specificity and carries small imperfect terminal IRs. The terminal 19-bp sequence is critical for transposition. Like IS10, IS50 has adopted an elaborate series of mechanisms to control its activity. It encodes two proteins, the Tpase, called Tnp, expressed from a single long reading frame, and an inhibitor protein, Inh, translated in the same frame as Tnp but using an alternative initiation codon and lacking the N-terminal 55 amino acids. These proteins appear to be expressed from separate (and possibly competing) promoters, P1 and P2, respectively (Fig. 9C), which are located downstream from the terminal IR (182). As in the case of IS10, Tpase expression is protected from activation by external transcription by an inverted repeat sequence located close to the left end. Moreover, the presence of Dam methylation sites within the Tpase promoter, T1, renders Tnp expression sensitive to methylation while having little effect on the activity of the Inh promoter T2.
The ends of IS50, known as OE (=IRL) and IE (=IRR), carry a series of sites which modulate Tnp expression and transposition activity. Extensive mutagenesis has shown that these ends conform to the general two-domain organization with a protein binding domain and a terminal domain (166). Tnp specifically binds both 19-bp end sequences. OE includes a binding site for the host DnaA protein, and transposition activity is reduced in a dnaA host (371). IE is somewhat more complex. It carries a binding site for the host protein, Fis, and three consecutive Dam methylation sites. Transposition activity is reduced by the presence of the Fis site (359) and by methylation of IE (369). The effect of methylation has been directly attributed to interference with Tnp binding (286).
Some analysis of Tpase organization and function has been undertaken. While Tnp specifically binds OE and IE, Inh does not (75). Moreover, the introduction of missense mutations or deletion of the 11 N-terminal amino acids destroys DNA binding activity (357). This has suggested that the DNA binding domain lies at least partially within the N-terminal 55 amino acids of Tnp. Interestingly, deletion of the C-terminal 100 amino acids increases the specific affinity of the protein for OE, suggesting that binding activity may be masked by the C-terminal part of the protein.
It is thought that the inhibitory action of Inh involves the formation of (inactive) heteromultimers between Inh and Tnp. Inh appears to be able to act in trans, whereas, like many Tpases, Tnp shows preferential action in cis.
More recently, an in vitro transposition system has been developed. This has clearly demonstrated that, like IS10 Tpase, IS50 Tpase is capable of introducing double-strand cuts at the ends of the element separating it from the donor plasmid backbone (116a).
IS231.
IS231A was isolated from Bacillus thuringiensis, in which it flanked a δ-endotoxin crystal protein gene (225), and it has been proposed that it forms part of a composite transposon. IS231A is active in E. coli. Ten other isoforms have been isolated. Sequence analysis of the terminal IRs of these elements suggests that they conform to the two-domain structure (Fig. 1) with conservation of bp 1 to 3 and 6 to 12. Like IS10 and IS50, a potential ribosome binding site for the Tpase gene would be sequestered in a secondary structure in transcripts originating outside the element. Although most examples carry a single open reading frame, Tpase expression from two elements (IS231V and IS231W) may occur by a +1 and +2 frameshift, respectively, but this has yet to be confirmed. Little is known about the transposition mechanism of this element, although it exhibits strong target specificity and recent evidence suggests that it transposes by a nonreplicative cut-and-paste mechanism (205a).
IS5 Family
General features.
The IS5 family is a relatively heterogeneous group composed of 47 distinct members and 21 isoelements. They include sequences from both eubacteria and the archaea. The majority carry only a single open reading frame. Their lengths range from 850 bp (IS869) to 1,643 bp (IS493). The latter carries a second open reading frame upstream of the “Tpase” frame which is inessential for transposition (18). Similarly, IS4811 (Tn4811 [57]), which is greater than 5 kb, clearly contains other, as yet uncharacterized genes. The major feature which defines this group is the similarity between their putative Tpases (288). In particular, this includes the N2, N3, and C1 domains carried by the IS4 group (Fig. 3). However, Tpases of the IS5 family exhibit a spacing between the N3 and C1 domains of approximately 40 residues, a distance more consistent with the canonical DDE motif. This group can be divided into six subgroups according to the depth of branching between them (Fig. 10A). The division is supported by similarities in the terminal IRs (Fig. 10B), by correlation with the length of the target duplication, and, to a lesser extent, by the typical length of the entire IS (Table 2). It is important to note that most members of the IS427 subgroup exhibit two partially overlapping open reading frames in a configuration in which the downstream frame is in phase −1 compared to the upstream frame and that most have a potential −1 frameshift window. Several members of the family are associated with compound transposons. These include IS903 and IS602, which form part of the kanamycin resistance transposons Tn903 (118) and Tn602 (326), respectively, and ISVa1/ISVa2, which form part of a transposon carrying iron transport genes (336).
FIG. 10.
IS5 family. (A) Dendrogram based on Tpase alignments, showing the division of the family into various subgroups. (B) Terminal IRs of two members of each subgroup, together with those of several elements which fall outside these groups.
Several members exhibit GATC sites within their terminal 50 bp. This includes all members of the IS903 subgroup and about 50% of the members of the IS1031 and IS427 subgroups. IS903 transposition activity is modulated by Dam in vivo (cited in reference 292).
For two subgroups, IS5 and IS427, and for two members of the ISL2 group (IS112 and IS1373), a preferred target sequence, YTAR (often CTAG), is observed in which either all four base pairs or the central TA is duplicated on insertion. It is important to underline that in many cases, the sequence of the original target site before insertion is not available. This can introduce ambiguities not only in estimating the number of duplicated target base pairs but also in defining the IRs. It is particularly important in several cases where the target repeat is symmetrical (e.g., CTAG) and where it is impossible to distinguish whether the element duplicates 2 or 4 bp and therefore to determine the exact ends of the element. Alignment of the ends of these elements in subgroups has permitted a number of ambiguities to be resolved. Members of the ISL2 group which generate 3-bp DRs exhibit a preference for ANT, while those from the IS1031 group (which generate exclusively a 3-bp DR) exhibit a preference for insertion sites with the sequence TNA. Neither the small ISH1 group (8-bp DRs) nor the IS903 group (9-bp DRs) exhibits marked target specificity.
Only two of these elements, IS5 and IS903, have received significant attention.
IS5.
In spite of the importance of IS5 in generating mutations, the published work concerning this element is largely directed to an understanding of its coding capacity and expression properties. IS5 carries one large open reading frame ins5A, which spans the entire element and is essential for transposition (303), and two small open reading frames, ins5B and ins5C, whose relevance to transposition remains to be demonstrated. Nothing is known about the transposition mechanism of this element.
IS903.
The ends of IS903 carry IRs of 18 bp which exhibit the typical two-domain organization (Fig. 1) (82). Transposase has been shown to bind specifically to the ends by using a region located in the amino-terminal portion of the protein (79, 332). In addition, recent results have revealed a region possibly involved in the formation of higher-order multimers and has pinpointed residues probably involved in catalysis (332). Little is known about its transposition mechanism, although an elegant genetic analysis provided strong evidence that IS903 is not only capable of undergoing direct insertion but can also generate adjacent deletions in a duplicative manner (356).
IS6 Family
The IS6 family was named after the directly repeated insertion sequences in transposon Tn6 (28). Historically, several of these elements were given individual IS numbers and have subsequently been determined to be identical. Many have been found as part of compound transposons. The IS6 family of elements is composed of 45 members, 30 of which are isocopies. Several distinct family members have been shown to transpose. The putative Tpases are very closely related and show identity levels ranging from 40 to 94%. They generally range in length from 789 bp (IS257) to 880 bp (IS6100). However, one isolate, IS15, corresponds to an insertion of one iso-IS6 (IS15Δ) into another (341). Although it has been included in the family based on similarities of its terminal IRs (Fig. 11B), IS466 from Streptomyces coelicolor is 1,628 bp long and the Tpase open reading frame has yet to be determined. All carry short related (15 to 20 bp) terminal IRs and generally create 8-bp direct target repeats. No marked target selectivity has been observed (241). Interestingly, sequences identical to the Mycobacterium fortuitum IS6100 have recently been identified as part of a plasmid-associated catabolic transposon carrying genes for nylon degradation in Arthrobacter sp. (170), from the Pseudomonas aeruginosa R1003 plasmid (132), and within the Xanthomonas campestris transposon Tn5393b (328). This suggests a relatively recent horizontal dissemination.
FIG. 11.
IS6 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs. (C) Transposition mechanism. A target plasmid is distinguished by an open oval representing the origin of replication. The transposon carried by the donor plasmid is composed of two copies of the IS (heavy double lines terminated by small circles) in direct relative orientation (indicated by the open arrowhead) flanking an interstitial DNA segment (shown as a zigzag). The donor plasmid is distinguished by an open rectangle representing its origin of replication. Tpase-mediated replicon fusion of the two molecules generates a third copy of the IS in the same orientation as the original pair (open arrowhead). Homologous recombination, using the recA system, between any two copies can in principle occur. This will either regenerate the donor plasmid, leaving a single IS copy in the target, delete the transposon, or transfer the transposon to the target (as shown), leaving a single copy of the IS in the donor molecule.
In IS26, an open reading frame is transcribed from a promoter located within the first 82 bp of the left end. It stretches across almost the entire IS and is required for transposition activity (241), and the predicted amino acid sequence of the corresponding protein exhibits a strong DDE motif (Fig. 3). Translation products of this frame have been demonstrated for IS240 (76) and IS26 (350a). Little is known about the control of Tpase expression, although recent evidence obtained with IS6100 in Streptomyces lividans (317) and IS26 in E. coli (350a) indicates that the transposition activity is significantly increased when the element is placed downstream from a strong promoter.
Where analyzed, members of the IS6 family give rise exclusively to replicon fusions (cointegrates) in which the donor and target replicons are separated by two directly repeated IS copies (e.g., IS15Δ, IS26, IS257, and IS1936). Transposition of these elements is therefore presumably accompanied by replication. Following cointegration, a resolution step is required to separate donor and target replicons and to transfer the transposon to the target replicon. Recombination between directly repeated ISs necessary for this separation occurs by homologous recombination and requires a recombination-proficient host (Fig. 11C).
An interesting similarity has been observed to a 4,904-bp element isolated from the filamentous cyanobacterium Fremyella diplosiphon (Tn5469 [IS5469] [168]). This relatively long element carries three consecutive open reading frames. The protein encoded by the first and longest frame, TnpA, is 909 residues long, compared to the IS6 family Tpases of between 220 and 260 residues. While TnpA contains an N-terminal region which exhibits significant conservation with respect to the IS6 Tpases (27 of 80 conserved residues over approximately 220 residues) (Fig. 11A), this region does not carry the DDE motif. On the other hand, a clear DDE motif resembling the Tpases of Tn5090 (Klebsiella aerogenes plasmid R751 [276]) and of the relatively complex transposon Tn552 (Staphylococcus aureus [298]) is located in the C-terminal region. Moreover, the ends of Tn5469 exhibit a strong similarity to those of the IS6 family members. This raises the possibility that recognition of the ends is determined by the IS6-like N-terminal region while catalysis is governed by a Tn552-related recombinase.
IS21 Family
General features.
There are 15 distinct members of the IS21 family (but only 11 for which the entire nucleotide sequence has been established [Fig. 12B]) together with 6 iso-ISs. They carry related terminal IRs whose lengths may vary between 11 bp (IS21) and 50 bp (IS5376) and generally terminate in the dinucleotide 5′-CA-3′. Several members, but not IS21 itself, carry multiple repeated sequences at their ends which include part of the terminal IRs (Fig. 12A and C) and which may represent Tpase binding sites. Insertion of these elements results in a direct target repeat of 4 bp or, more frequently, 5 bp, while two members (IS53 and IS408) may generate 8 bp. They exhibit two consecutive open reading frames: a long upstream frame designated istA and a shorter downstream frame designated istB (Fig. 12A). The putative IstA and IstB proteins carry several blocks of highly conserved residues. Overall identities range from 10 to 59% for IstA and from 25 to 67% for IstB. The istB frame may be located in a relative reading phase of −1 (e.g., IS21 and IS5376) or +1 (e.g., IS232 and IS1326) compared to istA. It can be slightly separated from istA (17 bp for IS408) or can overlap for 1 bp (IS21) or for several base pairs (IS232, IS5376, and IS1326); it is generally preceded by a potential ribosome binding site. The arrangement of the two reading frames suggests that translational coupling may occur (Fig. 12A). The istA reading frame carries a motif related to the widespread integrase DDE motif but lacking the conserved K/R residue (Fig. 3) and a potential helix-turn-helix motif, while the istB frame carries a relatively well conserved potential nucleoside triphosphate binding domain (319). Sequence alignments of IstA and of IstB gives very similar relationships (Fig. 12B).
FIG. 12.
IS21 family. (A) General organization. The terminal IRL and IRR are shown as solid boxes. The position of the istA and istB reading frames is also shown. The horizontal lines below show the relative positions of the multiply repeated elements whose sequences are presented in panel C. IstA (hatched box) together with the potential DDE motif (stippled box) and IstB (open box) are indicated below. The possibility of translational coupling between the two reading frames is indicated. (B) Dendrogram derived from alignment of the IstA and IstB gene products. (C) Nucleotide sequences of the multiple terminal repeats, together with their coordinates. CS, complementary strand. L1, L2, and L3, and R1 and R2, indicate internal repeated sequences at the left and right ends, respectively.
A partial sequence lacking the right end is available for one member, IS408. For another member, IS53, 45% identity of the potential istA frame with that of IS1326 can be obtained by the introduction of several frameshifts. An additional sequence, IS640 from Shigella sonnei, is a truncated iso-IS21. It carries an istA reading frame which is nearly identical to that of IS21 but lacks istB and the right terminal IR. The “intact” members have lengths of between 2 and 2.5 kb and are therefore among the largest bacterial IS elements.
IS21.
IS21 has been studied in some detail. It was discovered in Pseudomonas aeruginosa as a constituent of R68. This conjugative plasmid carries a single copy of IS21 and is inefficient in chromosome transfer. A derivative of R68, R68.45, exhibiting a marked increase in the transfer of chromosomal genes, was isolated (291) and proved to carry two directly repeated copies of IS21 separated by 3 bp (283). Tandem duplication is presumably favored by the marked tendency of IS21 to insert in, or close to, an IS21 end but may also occur by unequal crossing over during replication of the donor plasmid or, as suggested for IS30, whose transposition mechanism shows some similarity to that of IS21 (see “IS30 family” below), by dimerization of the donor plasmid (Fig. 4B, structure II) and deletion of DNA between the two IS copies (structure III). The tandemly repeated copies of IS21 promote insertion of the entire plasmid in a transposition event involving the abutted terminal IRs. Integration results in loss of the 3 bp located between the two ends in the parental R68.45 plasmid. The resulting replicon fusion thus has the appearance of a true cointegrate (i.e., with a single directly repeated copy of the IS at each junction), although transposition does not involve duplication of the insertion sequence during the insertion event. Both IstA and IstB are required for integration (281), and production of a second IstA derivative (from an alternative translational initiation codon eight codons downstream of that of IstA itself) greatly increases the efficiency of replicon fusions (282) for reasons that are at present unknown. Like retroviruses and several other transposable elements, IS21 terminates with 5′-CA-3′. Mutation of the terminal A abolishes replicon fusion (129).
The duplication of IS21 in R68.45 not only creates an efficient integrative donor structure by providing two closely spaced and correctly oriented IRs but also generates a strong promoter in which a −35 box located in the IRR of the upstream element is combined with a −10 box located in the abutted IRL of the downstream copy. This directs high levels of production of the IS21 transposition proteins (283) in a similar manner to the junction promoter created by circularization of IS911 and IS2. Insertion of IS21 downstream of a strong promoter results in the efficient expression of transposition proteins. Thus, IS21 appears not to have adopted a mechanism which “protects” it against high levels of impinging transcription. It is presumably the necessity for a transposition substrate carrying two abutted ends and the low frequency of formation of such molecules which acts as a control on transposition activity. More recently, a low level of circular IS21 copies has been detected in preparations of IS21-carrying plasmid DNA (302a), raising the possibility that IS21, like at least some of the IS3 family members, also transposes via a circular intermediate.
It has been demonstrated in vitro, using E. coli cell extracts enriched for IstA, that this protein is responsible for the 3′-end cleavage of IS21 (281). A substrate carrying two abutted IS21 ends separated by 3 bp is cleaved specifically at both 3′ ends in an IstA-dependent way.
IS30 Family
General features.
The IS30 family comprises 15 distinct complete members whose nucleotide sequence has been determined and 4 isoelements (Fig. 13A). Their lengths vary between 1,027 bp (IS1070) and 1,221 bp (IS30) with a single open reading frame of between 293 and 383 codons, which spans almost the entire length. The termination codon is generally very close to the right end. Alignment of the putative Tpases reveals a well-conserved DD(33)E motif. An open reading frame from a Spiroplasma citri phage (φSc1) is distantly related to the family. The terminal IRs are in the range of 20 to 30 bp and exhibit significant homologies (Fig. 13B). It should be noted that the tips of the elements, as defined by the various authors, show significant sequence variation. Where it has been determined, 2- or 3-bp DRs are generated on insertion.
FIG. 13.
IS30 family. (A) Dendrogram based on Tpase alignments. φSc1 is a homologous reading frame detected in a Spiroplasma citri bacteriophage. (B) Terminal IRs.
IS30.
Of the different members of this family, IS30 is the best characterized. It shows pronounced insertion specificity and generates DRs of 2 bp (51). It terminates with the dinucleotide 5′-CA-3′ (72), although many other members of the family do not. The long open reading frame is preceded by a relatively weak promoter (p30A) capable of driving Tpase expression with its −35 hexamer within IRL (71) and contains a weak internal transcription terminator. Premature termination of transcription at this site could lead to the formation of Tpase molecules truncated at their C-terminal end. Since the specific DNA binding domain which recognizes the terminal IRs is located in the N-terminal one-third of the protein (322), the potential truncated derivatives may lack the catalytic domain and regulate transposition activity by competition or interaction with a full-length Tpase. This arrangement may play the same role at the transcriptional level as does frameshifting at the translational level for certain other elements. A second weak promoter (p30C) located on the opposite strand upstream of a short open reading frame has been detected (71), and it has recently been shown that a small (150-nucleotide) transcript driven from this promoter and contained entirely within the Tpase transcript acts as an antisense control by reducing translation of the Tpase mRNA (14).
As for IS21 and IS911, the assembly of abutted IRL and IRR copies creates a strong promoter. In a tandem dimer of the element, this may result in increased expression of the Tpase from the downstream copy (70). The IRL-IRR junction is also recombinationally unstable. It promotes transposition reactions at high frequency in vivo (255), presumably in a similar way to the equivalent junction of IS21 described above. The activity of the junction appears maximal when the spacer between the abutted ends is 2 bp (the length of target duplications introduced when IS30 inserts). Lower activities are obtained with 1- and 3-bp spacers, while little or no activity is observed for 0 or >3 bp (249a).
IS66 Family
The IS66 family is composed of 12 members limited to agrobacteria and rhizobia. These include one isoelement and several elements for which only partial sequence data have been obtained. Members of this family are between 2.49 and 2.73 kb long and have very similar terminal IRs of between 15 and 27 bp (Fig. 14B). These elements are flanked by 8-bp direct target repeats. Each contains a number of open reading frames which varies among elements due to slight variations in the nucleotide sequences (either real or due to sequencing errors). We have used IS866 as a reference since it appears to carry the most representative set of open reading frames (Fig. 14A). However, we underline the possibility that errors in this interpretation may well exist. We have defined here five open reading frames common to IS66, IS292, and IS866. While having the expected length and terminal IRs, IS1313 carries only a single reading frame with similarity to those of IS66 family members (open reading frame 2). An interesting observation concerning this family is the presence of a group II intron signature in open reading frame 5 (180), although its significance is at present unknown. No studies have addressed the transposition of IS66 family members.
FIG. 14.
IS66 family. (A) Organization of IS866. A “best-guess” diagram of the open reading frames is shown. All are transcribed from left to right. The difference in shading is simply to facilitate their distinction. (B) Terminal IRs.
IS91 Family
General features.
The IS91 family is composed of eight members, of which one, ISFn1, has been only partially determined (145) and three are isoforms. Two additional members, IS92L and R form part of a composite transposon. They exhibit homology to IS91 as judged by Southern hybridization but appear to contain various large deletions and substitutions (374). Another member, IS1294, was discovered on plasmid pUB2380 (64). Comparison with other members of the family suggests that its Tpase could be extended to 389 amino acids rather than the 312 amino acids. This modification would increase to 62% the identity to the Tpase of IS801, another member of the family. The putative IS91 Tpase exhibits 36% identity to that of IS801. One notable feature of several members is that while their 3′ ends are relatively constant, there is significant variation in their lengths at the 5′ end, excluding a well-conserved terminal 20 to 25 bp. This may be related to their particular transposition mechanism, discussed below.
IS91 carries short (7- to 8-bp) imperfect terminal IRs, inserts into a rather specific tetranucleotide sequence (GAAC and CAAG), and does not generate direct target repeats on insertion (233). An alignment of the ends of IS91 with those of IS801 and IS1294 shows highly conserved regions at both the left and right ends (Fig. 15B) and suggests that they also use the same target sequence (235). They carry a long open reading frame whose potential products have significant similarities and are related to a family of replication proteins of bacteriophages and small gram-positive plasmids which propagate by a rolling-circle replication mechanism (155, 234). The similarity is localised to a consensus of four motifs (Fig. 15A). These Tpases are more closely related to the φX174 gpA protein than to the plasmid Rep protein family, since they carry two tyrosine residues in motif III. In φX174, they are directly involved in catalyzing strand cleavage and transfer (136).
IS91.
In addition to suggestions derived from protein homologies, experimental evidence has been obtained which strongly suggests that IS91 transposition occurs by a polarized rolling-circle replicative mechanism (Fig. 15C) (232) similar to that proposed by Galas and Chandler (106). This conclusion is based on the observations that 81 bp at the right end of the element, including the terminal IRR, is sufficient for transposition and that the terminal IRL is dispensable. The target sequence GAAC or CAAG abutting IRR is also essential for further transposition. The products of “one-ended” transposition (in the absence of IRL) occur at high frequency, and all carry a constant end defined by IRR and a variable end defined by a copy of the target consensus located in the donor plasmid. Moreover, there is a high level of multiple tandem insertions, as though the “transposition-replication” apparatus recognizes the donor “target” consensus inefficiently and continues a second round. In the model proposed by de la Cruz and collaborators (232) and illustrated in Fig. 15C, donor strand cleavage results in a covalent complex between the 5′ IRR end and Tpase. This is followed by single-strand transfer into the target DNA at a site containing a consensus tetranucleotide. The attached single strand of the IS is displaced by replication in the donor molecule. Termination is triggered when the complex reaches either the 3′ IRL end or a tetranucleotide consensus sequence in the donor. The model does not directly address second-strand synthesis of the transferred IS, which could occur simply by replication of the recipient replicon. A second member of this family, IS1294, appears to promote natural one-ended transposition (64).
IS110 Family
The IS110 family is somewhat disparate. It contains 20 members (of which 7 are isoforms) (Fig. 16) varying between 1,136 and 1,558 bp, with most clustered in the 1,450-bp range. All family members have no or very small (<7-bp) IRs and do not generally create direct target repeats (IS492 might generate 5-bp repeats [19]). Little overall similarity can be detected between the ends. A single long, relatively well conserved reading frame is present and shows some groups of identity within the N- and C-terminal portions. These elements share a highly conserved tetrad motif with reverse transcriptase (142), whose significance is at present unknown. Unlike IS91-related elements, no obvious rolling-circle replication signature is present in the predicted protein. The inclusion in this family of two additional elements, the Streptomyces coelicolor minicircle IS117 (140) and a Moraxella bovis (and M. lacunata) DNA inversion system determining pilus synthesis (Piv), is based on significant similarities in their putative recombinase proteins (205). It should be noted that this family of recombinases is not related to the site-specific recombinases of the λ integrase or resolvase/invertase families.
FIG. 16.
IS110 family. Only the dendrogram based on Tpase alignments is shown.
Little information is available concerning the mechanism of transposition of these elements. However, S. coelicolor IS117 was initially demonstrated in a circular form and integrates at a frequency 2 orders of magnitude higher than when cloned as a linear copy (141). In recent studies with IS492 (259a) and IS900 (231a), DNA fragments carrying abutted IS ends have been detected in vivo. Their appearance is dependent on an intact Tpase gene, and their nucleotide sequence is consistent with the formation of a circular form of the element (although it is also consistent with the presence of a head-to-tail dimer). Moreover, the junction of IS492, like several other ISs, appears to form a strong promoter in E. coli (89b). For both IS117 (141) and IS900 (231a), the target sequences exhibit some similarities to the circle junction, suggesting that insertion may occur by a site-specific recombination.
Finally, although little data is available concerning enzymatic activities of the putative Tpases of this family of elements, IS900 Tpase has been detected by immunological methods in the Mycobacterium paratuberculosis host (335) and IS492 Tpase has recently been purified and exhibits DNA cleavage activity specific for the ends of the element (259a).
IS200/IS605 Group
Although IS200 has been described as an autonomous element, it is often associated with other open reading frames in the form of a composite. This was first observed with the IS605 element from Helicobacter pylori. In an attempt to group these elements to facilitate searches in the database, we have elected to refer the ensemble as the IS605 group or family.
General features of IS200.
IS200 was originally detected in Salmonella typhimurium (194), but related elements have since been found in a variety of bacterial species including certain Shigella (112), Clostridium (44), and Streptococcus pneumoniae (60a). To date, 16 members of the family and 9 isoelements have been found. The most recent sequence determination (32) indicates that it is 707 bp long (and not 708 bp as previously described [113]), with a single long open reading frame preceded by several sequences capable of forming hairpin structures (Fig. 17A). One of these, which is located within 20 bp of the end, is thought to act as a transcriptional terminator and to prevent impinging transcription from activating the element (195), while another may sequester the ribosome binding site of the Tpase gene (32). The predicted product of its single open reading frame exhibits some similarities to that of open reading frame 1 (orf1) of Halobacterium ISH1-8. IS200 does not carry terminal IRs. Despite much effort, few cases of IS200 transposition or other forms of rearrangement have been documented (127).
FIG. 17.
IS200 complex. (A) Organization of IS200. Short IRs (open arrows) are shown at the left end, and the relative position of the potential open reading frame (hatched box) is indicated. (B) Dendrogram of IS200 family Tpases, orf1 (left) and the associated orf2 reading frames (right). (C) Relative localization of orf1 and orf2 in selected examples. The convention for the orientation of each reading frame is that frames shown above the line are transcribed to the right while those below the line are transcribed to the left. (D) Relationship between various examples of orf2 and other IS elements. aa, amino acids.
IS200 as a complex: the IS605 family?
In the few examples of IS200 insertions in the S. typhimurium genome where the sequence of the unoccupied target site is also known, it seems clear that transposition resulted from a simple insertion of IS200 alone (32, 47, 256). On the other hand, in a significant number of IS200 isolates from other species, most of which were defined during sequence analysis of virulence determinants in the respective hosts, the Tpase open reading frame (orf1 in Fig. 17B) has been found close to open reading frames associated with other presumed IS elements (orf2 in Fig. 17B). This may simply reflect insertion preferences of one or another of the ISs. However, in Helicobacter pylori, an IS200-like Tpase, orf1, is located next to (and is transcribed in a divergent manner from) a second reading frame, orf2, which is related to the putative Tpase of IS1341. This unit, called IS605, is found repeated several times in the genome, and the two reading frames are always associated (25a, 337). A similar arrangement is found in IS1253A from Dichelobacter nodosus (36), where orf2 is related to the putative Tpase of IS1136. In the Halobacterium element ISH1-8, orf2 is related to IS891, although in this case both open reading frames are transcribed in the same direction (Fig. 17C). In most isolates describing the individual IS elements (IS891, IS1136, etc.), as well as for the majority of “individual” IS200 derivatives identified, insufficient flanking-nucleotide sequence information is available to determine whether similar associations of open reading frames occur. It should be noted that for convenience, the entire complex is referred to as the IS605 family in Table 1.
Another interesting taxonomic characteristic is that the different orf2 frames have N-terminal and C-terminal domains which are interrelated, albeit distantly (Fig. 17D) (36), and could themselves be considered a group. As illustrated in Fig. 17D, the N-terminal portions of the single open reading frames of ISSs1, IS1136A, and IS1341 and the N-terminal ends of orf2 of IS605 and IS1253A (as well as an entire open reading frame, orfE, located on a Salmonella virulence plasmid, [125]) have similarities, as do all the C-terminal domains (including that of IS891) and the entire orf2 of ISH1-8. These relationships require further exploration.
IS256 Family
A total of 33 members of the IS256 family, including 2 isoforms, have been identified from a wide range of organisms. They are 1,298 bp (ISRm3) to 1,486 bp (IS6120) long. IS256 was originally isolated as a component of the compound transposon Tn4001 (218). Members carry related terminal IRs of between 24 and 41 bp (Fig. 18B), and most generate 8-bp direct target repeats but some 9-bp duplications have been observed. Interestingly, they also appear to carry internal IRs close to the ends (123, 260). A single long open reading frame carrying a potential DDE motif with a spacing of 112 residues between the second D and E residues together with a correctly placed K/R residue (Fig. 3) is present in most members, although the closely related IST2 (Thiobacillus ferrooxidans) and IS6120 (Mycobacterium smegmatis) appear to encode two open reading frames and may represent members of a distinct subgroup (124) (Fig. 18A). However, the intervening putative amino acid sequence contains residues conserved in the single open reading of other members of the family. For example, in IST2, a single change of phase extends the N-terminal region of the longer open reading frame by 140 amino acids, which contains additional identities to other members of the family. It has been suggested that translational frameshifting may occur to generate a single transframe Tpase (124). This subgroup also includes most cases of 9-bp target repeats. For IS285, which has similarities to ISRm3, optimization of the alignment between the two Tpases leads us to propose the presence of a single open reading frame. Three frame changes are sufficient to generate the single reading frame, which then shows extensive identity (44%) to ISRm3. The actual configuration of the open reading frames in these cases requires experimental clarification. The overall similarity between the majority of members of the family (Fig. 18) is between 22 and 48%. IS931 forms an outgroup, while IS1249 shows similarity in a subdomain of the putative Tpase and also has somewhat related terminal IRs. The other members form three deep branching groups, each showing between 30 and 40% similarity. No striking statistical conservation in target sequences can be deduced from the limited number of insertions analyzed, and little is known about the mechanism of transposition of this family.
FIG. 18.
IS256 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
An interesting feature of this family is that it may also contain eukaryote homologs. The putative MurA gene product of the autonomous mutator element of Zea mays, MuDR (see “MuDR” below) exhibits some weak similarities to the Tpases of this group (91). MuDR generates 9-bp target repeats.
IS630 Family
The IS630 family is composed of 12 members, of which 1 is an isoform and 1 has been only partially sequenced. They are interesting from several points of view, not the least being significant similarities to eukaryote “insertion sequences,” such as Tc1 and Tc3 of the nematode Caenorhabditis elegans and the mariner family (366) (Fig. 3). They are between 1,100 and 1,200 bp long and show a high target specificity. Since the conserved target sequence appears to be duplicated at each end of the element, it is formally possible that one or other copy is an integral part of a terminal IR of the IS. This arrangement therefore introduces an ambiguity not only in determining the exact tips of the IS but also in assessing the number of duplicated target base pairs (see “IS5 family” above).
Detailed studies concerning the target DNA sequence have been carried out for IS630 (333). The target sequence was determined before and after insertion, and the results suggested that insertion generated a duplication of an invariant target TA dinucleotide. That this dinucleotide does not form an integral part of the IS was investigated by site-specific mutagenesis of the transposon donor to eliminate the terminal TA. Transposition of the IS from the mutated donor molecule resulted in insertions which all exhibited a flanking TA DR. This clearly demonstrates that insertion results in the duplication of the central TA dinucleotide (333). Further analysis demonstrated that IS630 exhibits a strong preference for a 5′-CTAG-3′ target sequence (334). Moreover, point mutation of the CTAG target sites reduced or eliminated their attractiveness as insertion hot spots.
None of the other insertion sequences of this family have been analyzed in sufficient detail to determine unambiguously the exact tips of the element and the number of target base pairs duplicated. All known insertions of these elements are consistent with a TA duplication (Fig. 19B). For two insertions of the Agrobacterium vitis element IS870, it was possible to determine the target sequence prior to insertion. This sequence was found to carry a single CTAG copy (103), an observation which remains consistent with a simple TA dinucleotide target duplication. For coherence, we have therefore assumed that all members of this family generate an identical (TA) target duplication and have therefore defined the tips of the elements accordingly (Fig. 19B).
FIG. 19.
IS630 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
While IS895 appears to contain two consecutive open reading frames (7), the other members of the family all carry a single long reading frame. Three elements (IS870, ISAr1, and ISRf1) show more than 70% identity, two (IS1066 and ISRj1) show about 50% identity to each other and 40% to the other members, and two show less than 20% identity to each other and to the other members. It is interesting that the three elements most closely related on the basis of Tpase similarities (IS870, ISRf1, and ISAr1) appear not to carry translation termination codons for the Tpase gene. However, insertion into the specific target site, CTAG, with concomitant duplication (of either 2 or 4 bp) generates a TAG termination codon in phase with the Tpase gene. The influence of this arrangement on the transposition of the IS elements has yet to be determined.
Very little is known about the transposition properties of these elements. In IS630, while direct insertions can be observed at reasonable frequencies, no cointegrates could be detected (333).
IS982 Family
The IS982 family is composed of four members and two isoelements, for which the entire nucleotide sequence is known (Fig. 20A). It is restricted at present to gram-positive bacteria. The family members are between 962 and 1,026 bp long and carry similar terminal IRs of between 18 and 35 bp (Fig. 20B). Their termini are closely related and have been defined by comparison of different individual copies (Fig. 20). As for several other IS elements, IS982B from Lactococcus lactis has been shown to provide a −35 hexamer in its right terminal IR which is capable of forming a hybrid promoter with a resident −10 sequence located at the target site (214). Family members contain a single open reading frame capable of encoding a protein of between 271 and 296 amino acids; the proteins exhibit between 33 and 44% amino acid identity among the family members. A possible DDE motif is also apparent (Fig. 3) but lacks the conserved K/R residue. Little is known about the transposition of these elements.
FIG. 20.
IS982 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
IS1380 Family
Six members of the IS1380 family (345) have been identified, of which two are isoforms (Fig. 21A). They are between 1,665 bp (IS1380) and 2,071 bp (ISBj1) long, and each carries a single long open reading frame. IS1380 is present in extremely high copy number (approximately 100) in its host, Acetobacter pasteurianus (331). For ISBj1, a simple phase change around coordinate 1365 results in the extension of the potential Tpase open reading frame from 328 to 454 amino acids (similar to that of the other group members) and significantly increases the quality of the alignment. The family members exhibit between 30 and 50% amino acid identity in the potential Tpase. In addition, ISBj1 carries a 112-bp duplication in its right end. The terminal IRs are related (Fig. 21B) and are approximately 15 bp long. Insertion appears to generate a target duplication of 4 bp. Note that the external sequence of ISBj1 is also consistent with a 4-bp duplication.
FIG. 21.
IS1380 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
ISAs1 Family
The ISAs1 family is composed of 12 members, of which 6 are isoforms, restricted at present to gram-negative bacteria. They are 1,207 bp (ISPg1) to 1,326 bp (IS1358) long and generally carry terminal IRs of between 14 and 22 bp (Fig. 22) (although several of the IRs remain to be defined). A single open reading frame of between 294 and 376 amino acids occupies almost the entire length and shows between 26 and 50% identity. The putative Tpase of IS1358 has been visualized with a phage T7 promoter-driven derivative (327), and that of ISAs1 has been detected in E. coli minicells (126).
FIG. 22.
ISAs1 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
The family also includes the “H-repeats.” These form part of several so-called Rhs (rearrangement hot spot) elements (143, 379). E. coli K-12 contains five large repetitions of this type (RhsA to RhsE, scattered around the chromosome with lengths of between 3.7 and 9.6 kb). They represent nearly 1% of the chromosome and provide homology for RecA-dependent rearrangements. These elements are present in many but not all wild-type isolates of E. coli. The most prominent Rhs component is a giant core open reading frame, whose features are suggestive of a cell wall surface ligand binding protein. Each Rhs element also contains another repeated sequence, the H-rpt element (Hinc repeat), displaying features of typical insertion sequences, although no transposition activity has yet been detected. For the sake of clarity, the H-rpt DNA sequences B (RhsB), C1 to C3 (RhsC), E (RhsE), and min.5 (381), as well as H-rptF (379), have been renamed ISEc1 to ISEc7, respectively (Table 1). It is interesting that other members of the family (IS1358, ISAs1, and ISSe1) have been found associated with cell surface component genes in their respective hosts. Little is known about the transposition properties of this family of elements. However, recent experiments with the Vibrio cholerae element IS1358 have demonstrated that insertion generates 10-bp DRs and that in E. coli, it undergoes simple insertion into a target plasmid, pOX38 (89a).
ISL3 Family
The ISL3 family is presently composed of 21 known members, of which 3 are isoforms. Except for ISCx1, for which only a partial sequence is available, and IS1069, which is significantly larger, they range in size from 1,186 bp (ISSg1) to 1,553 bp (IS1165). They carry IRs of between 15 and 39 bp, which are closely related (Fig. 23) and generate a direct target repeat of 8 bp. One long open reading frame is present which encodes potential proteins of roughly 400 to 440 amino acids, showing good alignment, particularly in the C-terminal half. IS1096 harbors two open reading frames. The upstream open reading frame exhibits similarities to the ISL3 family Tpases. The second, tnpR, is related to open reading frames from Agrobacterium rhizogenes and Rhizobium plasmids. In IS1167, the single reading frame appears to be distributed between two consecutive open reading frames with a potential for translational coupling suggested by overlapping initiation and termination codons (ATGA). For analysis purposes, we have considered only one fused frame. The elements fall into three deeply branching groups with about 35, 30, and 25 to 60% identities, respectively (Fig. 23). Small sequences (130 to 340 bp) related to the IS1167 IRs have been detected in Streptococcus sanguis, S. pneumoniae, and S. agalactiae (8, 383). No obvious target sequence specificity has yet been observed, although there is some suggestion that there may be a preference for AT-rich regions.
FIG. 23.
ISL3 family. (A) Dendrogram based on Tpase alignments. (B) Terminal IRs.
Transposition of most of these elements has been demonstrated, but no analysis of their transposition mechanism has appeared. In one case, IS31831, derivatives of which have been used in mutagenesis, the majority of insertions were found to be simple and only rare clones carried accompanying vector DNA (350).
Unclassified Elements
Of the 500 ISs in the database, 54 remain to be classified. These are either elements whose nucleotide sequence is entirely unknown or with limited sequence information insufficient for allocation (33 examples) or elements whose entire nucleotide sequence is known but which show no marked relationships with more than one other element (21 ISs of which 5 are isoforms).
EUKARYOTIC INSERTION SEQUENCES
In addition to the large number of bacterial IS elements, a significant number of eukaryotic elements have been documented. The largest class described, and arguably the best characterized, is the mariner/Tc family, originally observed in Drosophila mauritiana (161) and C. elegans (92). These elements are structurally the closest to bacterial ISs. A second type of element which has also received extensive attention is the P element of D. melanogaster. A distinguishing character of this element is that Tpase binds to a subterminal region rather than to the terminal IRs. Binding of Tpase to subterminal motifs rather than to the terminal IRs is also a characteristic of two other groups of eukaryotic elements whose lengths exceed 4 kb: the CACTA family (restricted at present to plants), and the Ac family (with members in both plants and insects). Two characteristics of several eukaryotic elements which were thought to distinguish them from elements in eubacteria and archaebacteria are the presence of large numbers of defective copies and the facility with which an intact copy in the same genome can provide functions in trans for their mobilization (for reviews, see references 25, 93, and 190). However, studies of certain archaebacterial elements (146) and the accumulating data from bacterial genome projects suggest that multiple copies of inactive elements may not be limited to eukaryotes.
mariner/Tc Family
The presence of such elements has been extensively examined primarily in insect species (294), although they are found in many phyla including vertebrates, arthropods, ciliates, and fungi (reference 261 and references therein). The elements have been detected by using a specific PCR probe which exploits the presence of a highly conserved Tpase sequence.
The mariner family includes the Tc elements originally detected in C. elegans (92). These have been the subject of extensive analysis. Alignment of their Tpases (87) and their target specificity suggest that they are related to the IS630 family of bacterial elements. Tc1, Tc3, and mariner show significant similarities (Fig. 3). Moreover, the insertion specificity of both Tc1 and Tc3 is strict. Analysis of 204 Tc1 and 166 Tc3 insertions indicated that all occurred into a TA dinucleotide which was duplicated during the insertion event (349), a target choice which is identical to that of IS630.
Both Tc1 and the related Tc3 element carry a single transposase gene, tc1A and tc3A, respectively, each interrupted by one intron at a different position. Tc1 is 1,610 bp long and carries terminal IR sequences of 54 bp, while Tc3 is 2,335 bp long with 462-bp IRs. The reason for this large difference in IR length is unknown. The IR sequences are recognized and bound by their cognate Tpases in vitro (62, 353). This is consistent with the limited conservation of nucleotides between the IRs of each element. Moreover, it has been shown that Tpase induction in vivo from a heat shock promoter leads to high levels of transposition of the cognate element (347, 353). Tc1A and Tc3A also carry the DD(35)E motif (87) (Fig. 3). Point mutations in these residues render the Tc3 enzyme inactive in vivo (348). As in several bacterial Tpases, a sequence-specific DNA binding domain is located at the N-terminal end of both Tpases. It is followed by a region which overlaps the DD(35)E region and binds DNA nonspecifically. Interestingly, the N-terminal domain of Tc1A shows similarities and identities over a 40-amino-acid stretch to the N-terminal region of the IS30 Tpase, which is itself sufficient for binding to IS30 IRs. Also in the case of Tc1A, band shift assays, footprinting studies, and methylation interference studies have demonstrated a binding site between nucleotides 5 and 26 of the IR. Nucleotides 1 to 4 are not necessary for binding (353). This observation is consistent with the notion that the Tc1 IR sequences are organized into a two-domain structure similar to that of several bacterial elements (Fig. 1). The binding site can be further subdivided into two distinct regions: one closer to the tip, which is contacted by the specific N-terminal Tpase domain, and one which is contacted by the nonspecific DNA binding domain (352). A leucine zipper motif has also been detected in the Tpases of certain members of this group (160). As in the IS3 family, this may represent a multimerization domain, although recent structural studies with a truncated transposase derivative indicated that this region did not assume a coiled-coil motif (90).
Purified Tc1A has been demonstrated to have both endonuclease and phosphoryltransferase activities in vitro (Fig. 24). The major endonucleolytic activity detected was at the 5′ ends rather than, as might have been expected based on the activities of other Tpases (352), at the 3′ ends of the element. Moreover, cleavage was shown to occur specifically at the phosphodiester bond between the second and third nucleotide within the transposon. Phosphotransferase (strand transfer) activity was also observed in a “disintegration” assay.
FIG. 24.
Tc and mariner elements. Tpase-mediated cleavage at the terminal 3′ phosphodiester bond and at 2 nucleotides within the 5′ ends is indicated by curved arrows. Excision of the element (grey box) leaves two 2-nucleotide 3′ extensions in the vector backbone (open flanking box). Insertion is thought to occur by simple nucleophilic attack by the free 3′OH groups of a specific TA target dinucleotide, as shown by the long curved arrows. Repair of the donor backbone will leave a 2-bp insertion (footprint) compared to the original target sequence. This must proceed via the formation of a mismatch joint presumably followed by repair or replication to resolve this mismatch.
Transposon circles have been detected in vivo (92, 275, 296), but their role as possible intermediates in transposition is not entirely clear. Tc1 and Tc3 appear to transpose by a cut-and-paste mechanism. Induction of Tc3A in vivo results in excision of Tc3 as a linear form. One interesting characteristic of these molecules is that while the cleavage at the 3′ ends of the element occurs precisely between the transposon and its flanking sequences, 5′ cleavage occurs 2 nucleotides within the transposon in a manner similar to that observed in vitro (348). The result of these cleavages is that 2 nucleotides of each transposon end remain behind in the donor site, leaving a “footprint”: TACATA (where the flanking TA dinucleotide is that generated by duplication during the original integration) (Fig. 24). It has been noted by Plasterk (261) that a related footprint observed for Mos1 (TACCATA) suggests that excision of these elements occurs with a 3-nucleotide stagger.
Recently, systems which support Tc1 and mariner transposition in vitro have been described (196, 353). In both cases, it has been demonstrated that the Tpase is the only protein required for transposition. For Tc1, double-strand cleavage was observed not only at each end of an element carried on a supercoiled plasmid donor molecule but also on molecules carrying a single end. Excision, but not cleavage of a single end, appeared somewhat reduced when the donor plasmid was linearized suggesting that supercoiling stimulates the two-ended reactions. The cleaved ends were shown to carry a 2-base 3′ overhang, as shown in Fig. 24. Intermolecular transposition was demonstrated by using a genetic assay, and the resulting products showed a similar target specificity to those analyzed in vivo. For mariner (Himar1 from the hornfly, Haematobia irritans), the purified Tpase was demonstrated to generate a footprint in the terminal IR and to promote double-strand cleavage similar to that found with Tc1 but with a 3-base 3′ overhang (196).
P Element
The P element of D. melanogaster was discovered by its role in the syndrome known as hybrid dysgenesis (see, for example, reference 177). This syndrome is characterized by a loss of fertility in a cross between males which carry an autonomous P element and females which do not; it is due to high level P transposition in germ line cells. Tpase expression requires correct splicing of an mRNA which contains several introns. In particular, removal of one intron (intron 2-3) occurs only in the germ line, limiting transposition activity to these cells (see reference 93 for review). The element has been used extensively in mutagenesis of its host organism by using specifically tailored P derivatives. Like Tc1, Tc3, and mariner, it transposes by a cut-and-paste mechanism, and together with these, it is one of the best-characterized eukaryotic elements. It is 2.9 kb long, generates 8-bp target repeats on insertion, and specifies an 87-kDa Tpase produced from a single open reading frame which carries three introns. Transposition requires approximately 150 bp at each end, including 31-bp terminal IRs (248). Tpase does not bind to these, however, but instead recognizes neighboring internal sites (173). The terminal IRs are bound by a host protein which has been reported to show homology to KU (174), a protein also implicated in double-strand break repair and V(D)J recombination (297). Transposase binding to the “left” end represses Tpase expression (174). In addition, two types of repressor molecule have been detected, presumably encoded by defective (nonautonomous) elements (115). The first is an approximately 66-kDa derivative truncated at the C-terminal end of the Tpase. The second, KP, is a 207-amino-acid protein produced from an internally deleted element. KP forms dimers and, unlike the full-length Tpase, binds to multiple sites at the ends of P (202). A DNA binding domain has been localized to the N-terminal 98 amino acids. A leucine zipper motif has also been detected and has been shown to be important for KP activity in vivo (10) and to be necessary for dimerization in vitro (202).
In addition to Tpase binding, two other characteristics distinguish P from the elements discussed above. Studies with an in vitro system have shown that the Tpase requires GTP as a cofactor, although nonhydrolyzable analogs are also effective (175). Interestingly, a single-amino-acid change has recently been shown to switch cofactor specificity from GTP to XTP in vivo (246). The second characteristic concerns the nature of the double-strand Tpase-induced cleavages at each end. While 3′ cleavage occurs at the terminal nucleotide, 5′ cleavage occurs 17 bp within the element (20). This position, within the terminal IRs, is directly adjacent to the host factor binding site. The in vitro system has also revealed that both the left and right ends are required for cleavage activity, suggesting that prior formation of a synaptic complex is required.
Other Families
CACTA family.
The CACTA family (114) is named according to the nucleotide sequence of the highly conserved terminal IRs of its members. This group is currently restricted to plants and includes En/Spm and some Tam elements (Tam1, Tam2, Tam4, Tam7, Tam8, Tam9). Full-length members are large (>4 kb), carry nearly identical terminal IRs of 13 bp, and generate target duplications of 3 bp. Transposition requires both the left ∼180 bp and the right ∼300 bp. The En element produces two proteins by relatively complex differential splicing: TNPA (67 kDa) and TNPD (131 kDa). Both are required for transposition. TNPA binds a reiterated subterminal 12-bp motif but not the terminal IRs. The equivalents of TNPA from different members of this group do not appear to exhibit significant identities in primary sequence. This would be consistent with a role in binding to the element-specific reiterative repeats. Both DNA binding and multimerization domains have been localized for TNPA of En (340). On the other hand, the second and larger protein species shows significant identity (45% between the TNP2 proteins of Tam1 and TNPD), and for this reason it has been proposed that it binds the related terminal IRs of the elements and catalyzes cleavage.
Ac.
The organization of the Ac element (190) of maize is similar to that of Tam 3, P, and hobo. It is 4,565 bp long and carries 11-bp imperfect terminal IRs and numerous reiterated subterminal 6-bp motifs. Insertion results in an 8-bp target duplication. A single open reading frame with four introns specifies a 112-kDa Tpase. Approximately 240 bp is required at each end for efficient transposition, and the frequency decreases as this is reduced to ∼100 bp. Both left and right ends are required. Like those of P and En, the Tpase binds to the subterminal repeats, probably in a cooperative manner using a domain located in the C-terminal end of the protein. It binds with significantly lower affinity to the terminal IRs by using a domain located in the N-terminal end (21). The C-terminal 600-amino-acid sequence shows homology to the putative Tpases of Tam3 and hobo, family members from plant and insect species, respectively.
MuDR.
The MuDR transposon (25), the mutator element of maize, is highly active and has been used extensively to tag various maize mutants. The autonomous element is approximately 5 kb long, carries ∼200-bp terminal IRs, and generates 9-bp target repeats on insertion. It contains two open reading frames transcribed in a convergent manner from promoters located in the terminal IRs and capable of specifying proteins of 207 (MURB) and 823 (MURA) amino acids. MURA is required for transposition and shows some similarities to the Tpases of the IS256 family of elements (91). It is also capable of binding specifically to the terminal IRs of MuDR (24). Little is known about transposition of this family of elements at the molecular level.
CONCLUSIONS AND PERSPECTIVES
One of the major objectives of this review was to assemble available information concerning bacterial insertion sequences into a coherent framework. This has proved a daunting task. The various families defined here provide a starting point for the classification of these relatively simple transposable elements. It is clear, however, that this scheme will have to be updated and extended as additional ISs are isolated and characterized. Further examples are beginning to accumulate at increasing rates as a result of efforts in small-genome sequencing. The recently published nucleotide sequence of the Rhizobium sp. strain NGR234 megaplasmid, pNGR234a (536 kbp) (104), for example, alone includes between 30 and 40 potential ISs, and a large number have been identified in other genomes such as that of Mycobacterium tuberculosis (60b). The amount of information which is presented here represents only a fraction of that included in our IS database. The entire database is available at http://pc4.sisc.ucl.ac.be/is.html with appropriate links to the international sequence databases.
Although we have provided dendrograms for many of the families described here, these do not imply any particular phylogenetic relationship. The collection of sequences in this database will provide a useful starting point for phylogenetic analyses.
Structuring the known ISs into 17 families underscores the immense diversity of these elements. However, one characteristic which encompasses many of the IS families, including the majority of known IS elements, is a real or potential DDE motif. This has been demonstrated to be mechanistically important in retroviral, phage Mu, Tn7, IS10, and Tc1 and Tc3 transposition. It will therefore be of considerable interest to confirm the relevance of this catalytic triad in at least some additional families and to determine the spatial relationship of the residues within the three-dimensional structure. On the other hand, although the DDE motif appears to be a common theme in many of the elements described here, implying a shared chemistry, the detailed ways in which elements of each family assemble the individual cleavage and strand transfer reactions can vary and result in important differences in overall mechanism. Thus, certain elements with DDE chemistry generate replicon fusions exclusively (Mu and IS6) while others are excised from their donor molecule before forming a synaptic complex with the target molecule (IS10, IS50, and IS911) and yet others appear to require the presence of a target site prior to donor strand cleavage (Tn7). Details of second-strand cleavage also vary. Further studies of selected members of the DDE superfamily will undoubtedly uncover additional combinations of reaction steps and extend the known range of DNA transactions which contribute to genome plasticity.
Finally, while members of the DDE family represent the majority of known IS elements, a significant fraction do not. These families include IS1, IS91, and IS110 (which are at present being actively analyzed) and IS1380 and ISAs1 (which to our knowledge are not). From a mechanistic and evolutionary point of view, it is important to understand how these elements may be related to bacteriophage λ integration (IS1), single-strand phage and plasmid replication (IS91), and site-specific inversion systems (IS110).
ACKNOWLEDGMENTS
Special thanks are due to R. Reszöhazy for his help in an early stage of this review. We acknowledge the cooperation of many colleagues in sharing unpublished results before publication, A. Gaeckle and F. Rodriguez for designing and installing the Web site, M. Boschet for taming the bibliography, and J. Doris for work on some of the Tpase alignments. R. Alazard, Y. Chen, G. Duval-Valentin, L. Haren, C. Léonard, C. Normand, P. Polard, B. Ton-Hoang, M.-C. Serre, C. Turlan, and D. Zerbib provided invaluable suggestions during the course of this work. M. Bétermier, F. de la Cruz, I. Goryshin, D. Haas, D. Haniford, J. McFadden, T. Naas, R. Plasterk, W. Reznikoff, P. Sharpe, and A. Toussaint provided valuable input for various parts of the manuscript.
Work in the Chandler laboratory was supported by the CNRS and grants from the Région Midi-Pyrenées, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer (leg Petijean), the Association National de Recherche Contre le SIDA, and an EU network grant in the HCM program. Work in the Mahillon laboratory was supported by the FNRS and the Université catholique de Louvain.
REFERENCES
- 1.Reference deleted.
- 2.Abremski K E, Hoess R H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992;5:87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Adzuma K, Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of Mu B protein. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- 4.Adzuma K, Mizuuchi K. Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989;57:41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- 5.Adzuma K, Mizuuchi K. Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage Mu. Involvement of protein oligomerization in the ATPase cycle. J Biol Chem. 1991;266:6159–6167. [PubMed] [Google Scholar]
- 6.Ahmed A. Evidence for replicative transposition of Tn5 and Tn9. J Mol Biol. 1986;191:75–84. doi: 10.1016/0022-2836(86)90423-7. [DOI] [PubMed] [Google Scholar]
- 7.Alam J, Vrba J M, Cai Y, Martin J A, Weislo L J, Curtis S E. Characterization of the IS895 family of insertion sequences from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1991;173:5778–5783. doi: 10.1128/jb.173.18.5778-5783.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alloing G, Trombe M C, Claverys J P. The ami locus of the gram-positive bacterium Streptococcus pneumoniae is similar to binding protein-dependent transport operons of gram-negative bacteria. Mol Microbiol. 1990;4:633–644. doi: 10.1111/j.1365-2958.1990.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 9.Amemura J, Ichikawa H, Ohtsubo E. Tn3 transposition immunity is conferred by the transposase-binding domain in the terminal inverted-repeat sequence of Tn3. Gene. 1990;88:21–24. doi: 10.1016/0378-1119(90)90055-v. [DOI] [PubMed] [Google Scholar]
- 10.Andrews J D, Gloor G B. A role for the KP leucine zipper in regulating P element transposition in Drosophila melanogaster. Genetics. 1995;141:587–594. doi: 10.1093/genetics/141.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arber W, Iida S, Jutte H, Caspers P, Meyer J, Hanni C. Rearrangements of genetic material in Escherichia coli as observed on the bacteriophage P1 plasmid. Cold Spring Harbor Symp Quant Biol. 1979;43(2):1197–1208. doi: 10.1101/sqb.1979.043.01.136. [DOI] [PubMed] [Google Scholar]
- 12.Arciszewska L K, Drake D, Craig N L. Transposon Tn7. cis-Acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 13.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson L S. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arini A, Keller M P, Arber W. An antisense RNA in IS30 regulates the translational expression of the transposase. Biol Chem. 1997;378:1421–1431. doi: 10.1515/bchm.1997.378.12.1421. [DOI] [PubMed] [Google Scholar]
- 15.Arthur A, Nimmo E, Hettle S, Sherratt D. Transposition and transposition immunity of transposon Tn3 derivatives having different ends. EMBO J. 1984;3:1723–1729. doi: 10.1002/j.1460-2075.1984.tb02038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bachellier S, Clement J M, Hofnung M, Gilson E. Bacterial interspersed mosaic elements (BIMEs) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics. 1997;145:551–562. doi: 10.1093/genetics/145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bainton R J, Kubo K M, Feng J N, Craig N L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 18.Baltz R H, Hahn D R, McHenney M A, Solenberg P J. Transposition of Tn5096 and related transposons in Streptomyces species. Gene. 1992;115:61–65. doi: 10.1016/0378-1119(92)90541-v. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett D H, Silverman M. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J Bacteriol. 1989;171:1763–1766. doi: 10.1128/jb.171.3.1763-1766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beall E L, Rio D C. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 1997;11:2137–2151. doi: 10.1101/gad.11.16.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker H A, Kunze R. Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol Gen Genet. 1997;254:219–230. doi: 10.1007/s004380050410. [DOI] [PubMed] [Google Scholar]
- 22.Bender J, Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992;11:741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci USA. 1992;89:7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benito M I, Walbot V. Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol Cell Biol. 1997;17:5165–5175. doi: 10.1128/mcb.17.9.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennetzen J L. The Mutator transposable element system of maize. Curr Top Microbiol Immunol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- 25a.Berg, D. Personal communication.
- 26.Berg D E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci USA. 1983;80:792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg D E. Transposon Tn5. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 185–210. [Google Scholar]
- 28.Berg D E, Davies J, Allet B, Rochaix J D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci USA. 1975;72:3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. [Google Scholar]
- 30.Bernardi F, Bernardi A. Role of replication in IS102-mediated deletion formation. Mol Gen Genet. 1987;209:453–457. doi: 10.1007/BF00331149. [DOI] [PubMed] [Google Scholar]
- 31.Bernardi F, Bernardi A. Transcription of the target is required for IS102 mediated deletions. Mol Gen Genet. 1988;212:265–270. doi: 10.1007/BF00334695. [DOI] [PubMed] [Google Scholar]
- 32.Beuzon C R, Casadesus J. Conserved structure of IS200 elements in Salmonella. Nucleic Acids Res. 1997;25:1355–1361. doi: 10.1093/nar/25.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhugra B, Dybvig K. Identification and characterization of IS1138, a transposable element from Mycoplasma pulmonis that belongs to the IS3 family. Mol Microbiol. 1993;7:577–584. doi: 10.1111/j.1365-2958.1993.tb01148.x. [DOI] [PubMed] [Google Scholar]
- 34.Biel S W, Berg D E. Mechanism of IS1 transposition in E. coli: choice between simple insertion and cointegration. Genetics. 1984;108:319–330. doi: 10.1093/genetics/108.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billington S J, Sinistaj M, Cheetham B F, Ayres A, Moses E K, Katz M E, Rood J I. Identification of a native Dichelobacter nodosus plasmid and implications for the evolution of the vap regions. Gene. 1996;172:111–116. doi: 10.1016/0378-1119(96)00032-7. [DOI] [PubMed] [Google Scholar]
- 37.Bisercic M, Ochman H. The ancestry of insertion sequences common to Escherichia coli and Salmonella typhimurium. J Bacteriol. 1993;175:7863–7868. doi: 10.1128/jb.175.24.7863-7868.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolland S, Kleckner N. The two single-strand cleavages at each end of Tn10 occur in a specific order during transposition. Proc Natl Acad Sci USA. 1995;92:7814–7818. doi: 10.1073/pnas.92.17.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 40.Bourgoin F, Guedon G, Pebay M, Roussel Y, Panis C, Decaris B. Characterization of a mosaic ISS1 element and evidence for the recent horizontal transfer of two different types of ISS1 between Streptococcus thermophilus and Lactococcus lactis. Gene. 1996;178:15–23. doi: 10.1016/0378-1119(96)00316-2. [DOI] [PubMed] [Google Scholar]
- 41.Boursaux-Eude C, Saint Girons I, Zuerner R. IS1500, an IS3-like element from Leptospira interrogans. Microbiology. 1995;141:2165–2173. doi: 10.1099/13500872-141-9-2165. [DOI] [PubMed] [Google Scholar]
- 42.Bouziane M, Cherny D I, Mouscadet J F, Auclair C. Alternate strand DNA triple helix-mediated inhibition of HIV-1 U5 long terminal repeat integration in vitro. J Biol Chem. 1996;271:10359–10364. doi: 10.1074/jbc.271.17.10359. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Brynestad S, Iwanejko L A, Stewart G S, Granum P E. A complex array of Hpr consensus DNA recognition sequences proximal to the enterotoxin gene in Clostridium perfringens type A. Microbiology. 1994;140:97–104. doi: 10.1099/13500872-140-1-97. [DOI] [PubMed] [Google Scholar]
- 45.Brynestad S, Synstad B, Granum P E. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology. 1997;143:2109–2115. doi: 10.1099/00221287-143-7-2109. [DOI] [PubMed] [Google Scholar]
- 46.Bukhari A I, Shapiro J A, Adhya S L. DNA insertion elements, plasmids, and episomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1977. [Google Scholar]
- 47.Burnens A P, Stanley J, Sack R, Hunziker P, Brodard I, Nicolet J. The flagellin N-methylase gene fliB and an adjacent serovar-specific IS200 element in Salmonella typhimurium. Microbiology. 1997;143:1539–1547. doi: 10.1099/00221287-143-5-1539. [DOI] [PubMed] [Google Scholar]
- 48.Calos M P, Johnsrud L, Miller J H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978;13:411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- 49.Campbell Wyndham R, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 50.Casadesus J, Roth J R. Transcriptional occlusion of transposon targets. Mol Gen Genet. 1989;216:204–209. doi: 10.1007/BF00334357. [DOI] [PubMed] [Google Scholar]
- 51.Caspers P, Dalrymple B, Iida S, Arber W. IS30, a new insertion sequence of Escherichia coli K12. Mol Gen Genet. 1984;196:68–73. doi: 10.1007/BF00334094. [DOI] [PubMed] [Google Scholar]
- 52.Chaconas G, Lavoie B D, Watson M A. DNA transposition: jumping gene machine, some assembly required. Curr Biol. 1996;6:817–820. doi: 10.1016/s0960-9822(02)00603-6. [DOI] [PubMed] [Google Scholar]
- 53.Chalmers R M, Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 54.Chalmers R M, Kleckner N. IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 1996;15:5112–5122. [PMC free article] [PubMed] [Google Scholar]
- 55.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 56.Charlier D, Piette J, Glansdorff N. IS3 can function as a mobile promoter in E. coli. Nucleic Acids Res. 1982;10:5935–5948. doi: 10.1093/nar/10.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C W, Yu T W, Chung H M, Chou C F. Discovery and characterization of a new transposable element, Tn4811, in Streptomyces lividans 66. J Bacteriol. 1992;174:7762–7769. doi: 10.1128/jb.174.23.7762-7769.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Chen, Y., and J. Mahillon. Unpublished data.
- 58.Cheng X, Nicolet J, Poumarat F, Regalla J, Thiaucourt F, Frey J. Insertion element IS1296 in Mycoplasma mycoides subsp. mycoides small colony identifies a European clonal line distinct from African and Australian strains. Microbiology. 1996;141:3221–3228. doi: 10.1099/13500872-141-12-3221. [DOI] [PubMed] [Google Scholar]
- 59.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 60.Ciampi M S, Schmid M B, Roth J R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci USA. 1982;79:5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60a.Claverys, J. P. Personal communication.
- 60b.Cole S T, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 61.Collins C M, Gutman D M. Insertional inactivation of an Escherichia coli urease gene by IS3411. J Bacteriol. 1992;174:883–888. doi: 10.1128/jb.174.3.883-888.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colloms S D, van Luenen H G, Plasterk R H. DNA binding activities of the Caenorhabditis elegans Tc3 transposase. Nucleic Acids Res. 1994;22:5548–5554. doi: 10.1093/nar/22.25.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colonna B, Bernardini M, Micheli G, Maimone F, Nicoletti M, Casalino M. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936-kanamycin resistance element. Plasmid. 1988;20:221–231. doi: 10.1016/0147-619x(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 64.Comanducci A, Dodd H M, Bennett P M. pUB2380: an R plasmid encoding a unique, natural one-ended transposition system. In: Butler L O, Harwood C, Moseley B E B, editors. Genetic transformation and expression. Andover, England: Intercept; 1989. pp. 305–311. [Google Scholar]
- 65.Cornillot E, Saint-Joanis B, Daube G, Katayama S, Granum P E, Canard B, Cole S T. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol Microbiol. 1995;15:639–647. doi: 10.1111/j.1365-2958.1995.tb02373.x. [DOI] [PubMed] [Google Scholar]
- 66.Craig N L. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 67.Craig N L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 68.Craig N L. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 69.Craigie R, Mizuuchi M, Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984;39:387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- 70.Dalrymple B. Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. Mol Gen Genet. 1987;207:413–420. doi: 10.1007/BF00331609. [DOI] [PubMed] [Google Scholar]
- 71.Dalrymple B, Arber W. Promotion of RNA transcription on the insertion element IS30 of E. coli K12. EMBO J. 1985;4:2687–2693. doi: 10.1002/j.1460-2075.1985.tb03988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dalrymple B, Caspers P, Arber W. Nucleotide sequence of the prokaryotic mobile genetic element IS30. EMBO J. 1984;3:2145–2149. doi: 10.1002/j.1460-2075.1984.tb02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danilevich V N, Kostiuchenko D A. Immunity to repeated transposition of the insertion sequence IS21. Mol Biol (Moscow) 1985;19:1242–1250. [PubMed] [Google Scholar]
- 74.Davis M A, Simons R W, Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985;43:379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- 75.de la Cruz N B, Weinreich M D, Wiegand T W, Krebs M P, Reznikoff W S. Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. J Bacteriol. 1993;175:6932–6938. doi: 10.1128/jb.175.21.6932-6938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delécluse A, Bourgouin C, Klier A, Rapoport G. Nucleotide sequence and characterization of a new insertion element, IS240, from Bacillus thuringiensis israelensis. Plasmid. 1989;21:71–78. doi: 10.1016/0147-619x(89)90088-7. [DOI] [PubMed] [Google Scholar]
- 77.De Meirsman C, Van Soom C, Verreth C, Van Gool A, Vanderleyden J. Nucleotide sequence analysis of IS427 and its target sites in Agrobacterium tumefaciens T37. Plasmid. 1990;24:227–234. doi: 10.1016/0147-619x(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 78.Demuth D R, Duan Y, Jenkinson H F, McNab R, Gil S, Lamont R J. Interruption of the Streptococcus gordonii M5 sspA/sspB intergenic region by an insertion sequence related to IS1167 of Streptococcus pneumoniae. Microbiology. 1997;143:2047–2055. doi: 10.1099/00221287-143-6-2047. [DOI] [PubMed] [Google Scholar]
- 79.Derbyshire K M, Grindley N D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992;11:3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Derbyshire K M, Grindley N D. Cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. Mol Microbiol. 1996;21:1261–1272. doi: 10.1111/j.1365-2958.1996.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 81.Reference deleted.
- 82.Derbyshire K M, Hwang L, Grindley N D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Derbyshire K M, Kramer M, Grindley N D. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc Natl Acad Sci USA. 1990;87:4048–4052. doi: 10.1073/pnas.87.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reference deleted.
- 85.DeShazer D, Wood G E, Friedman R L. Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. Mol Microbiol. 1994;14:123–130. doi: 10.1111/j.1365-2958.1994.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 86.Devine S E, Boeke J D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 86a.Di Gioia D, Peel M, Fava F, Wyndham R C. Structures of homologous composite transposons carrying cbaABC genes from Europe and North America. Appl Environ Microbiol. 1998;64:1940–1946. doi: 10.1128/aem.64.5.1940-1946.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doak T G, Doerder F P, Jahn C L, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dodson K W, Berg D E. Factors affecting transposition activity of IS50 and Tn5 ends. Gene. 1989;76:207–213. doi: 10.1016/0378-1119(89)90161-3. [DOI] [PubMed] [Google Scholar]
- 89.Doolittle W F, Kirkwood T B, Dempster M A. Selfish DNAs with self-restraint. Nature. 1984;307:501–502. doi: 10.1038/307501b0. [DOI] [PubMed] [Google Scholar]
- 89a.Dumontier, P. Trieu-Cuot, and P. Berche. Personal communication.
- 89b.Duval-Valentin, G., and M. Chandler. Unpublished data.
- 90.Eijkelenboom A P, van den Ent F M, Vos A, Doreleijers J F, Hard K, Tullius T D, Plasterk R H, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 91.Eisen J A, Benito M I, Walbot V. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Emmons S W, Yesner L, Ruan K S, Katzenberg D. Evidence for a transposon in Caenorhabditis elegans. Cell. 1983;32:55–65. doi: 10.1016/0092-8674(83)90496-8. [DOI] [PubMed] [Google Scholar]
- 93.Engels W R. P elements in Drosophila. Curr Top Microbiol Immunol. 1996;204:103–123. doi: 10.1007/978-3-642-79795-8_5. [DOI] [PubMed] [Google Scholar]
- 94.Escoubas J M, Lane D, Chandler M. Is the IS1 transposase, InsAB′, the only IS1-encoded protein required for efficient transposition? J Bacteriol. 1994;176:5864–5867. doi: 10.1128/jb.176.18.5864-5867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Escoubas J M, Prere M F, Fayet O, Salvignol I, Galas D, Zerbib D, Chandler M. Translational control of transposition activity of the bacterial insertion sequence IS1. EMBO J. 1991;10:705–712. doi: 10.1002/j.1460-2075.1991.tb08000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fayet O, Ramond P, Polard P, Prere M F, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- 98.Ferat J L, Le Gouar M, Michel F. Multiple group II self-splicing introns in mobile DNA from Escherichia coli. C R Acad Sci Ser III. 1994;317:141–148. [PubMed] [Google Scholar]
- 99.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 100.Fiandt M, Szybalski W, Malamy M H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119:223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- 101.Filippov A A, Oleinikov P N, Drozdov A V, Protsenko O A. The role of IS-elements of Yersinia pestis (Lehmann, Neumann) in the emergence of calcium-independent mutations. Genetika. 1990;26:1740–1748. [PubMed] [Google Scholar]
- 102.Finnegan D J. Transposable elements: how non-LTR retrotransposons do it. Curr Biol. 1997;7:R245–R248. doi: 10.1016/s0960-9822(06)00112-6. [DOI] [PubMed] [Google Scholar]
- 103.Fournier P, Paulus F, Otten L. IS870 requires a 5′-CTAG-3′ target sequence to generate the stop codon for its large ORF1. J Bacteriol. 1993;175:3151–3160. doi: 10.1128/jb.175.10.3151-3160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 105.Galas D J, Calos M P, Miller J H. Sequence analysis of Tn9 insertions in the lacZ gene. J Mol Biol. 1980;144:19–41. doi: 10.1016/0022-2836(80)90213-2. [DOI] [PubMed] [Google Scholar]
- 106.Galas D J, Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982;154:245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- 107.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 108.Gamas P, Galas D, Chandler M. DNA sequence at the end of IS1 required for transposition. Nature. 1985;317:458–460. doi: 10.1038/317458a0. [DOI] [PubMed] [Google Scholar]
- 109.Garcia M I, Labigne A, Le Bouguenec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol. 1994;176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 111.Ghosal D, Sommer H, Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979;6:1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gibert I, Barbe J, Casadesus J. Distribution of insertion sequence IS200 in Salmonella and Shigella. J Gen Microbiol. 1990;136:2555–2560. doi: 10.1099/00221287-136-12-2555. [DOI] [PubMed] [Google Scholar]
- 113.Gibert I, Carroll K, Hillyard D R, Barbe J, Casadesus J. IS200 is not a member of the IS600 family of insertion sequences. Nucleic Acids Res. 1991;19:1343. doi: 10.1093/nar/19.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gierl A. The En/Spm transposable element of maize. Curr Top Microbiol Immunol. 1996;204:145–159. doi: 10.1007/978-3-642-79795-8_7. [DOI] [PubMed] [Google Scholar]
- 115.Gloor G B, Preston C R, Johnson-Schlitz D M, Nassif N A, Phillis R W, Benz W K, Robertson H M, Engels W R. Type I repressors of P element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goosen N, van de Putte P. Role of ner protein in bacteriophage Mu transposition. J Bacteriol. 1986;167:503–507. doi: 10.1128/jb.167.2.503-507.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116a.Goryshin I Y, Reznikoff W S. Tn5 in vitro transposition. J Biol Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 117.Grindley N D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978;13:419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- 118.Grindley N D, Joyce C M. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc Natl Acad Sci USA. 1980;77:7176–7180. doi: 10.1073/pnas.77.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reference deleted.
- 120.Grindley N D, Joyce C M. Analysis of the structure and function of the kanamycin-resistance transposon Tn903. Cold Spring Harbor Symp Quant Biol. 1981;45(1):125–133. doi: 10.1101/sqb.1981.045.01.021. [DOI] [PubMed] [Google Scholar]
- 121.Grindley N D, Leschziner A E. DNA transposition: from a black box to a color monitor. Cell. 1995;83:1063–1066. doi: 10.1016/0092-8674(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 122.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 123.Guedon G, Bourgoin F, Pebay M, Roussel Y, Colmin C, Simonet J M, Decaris B. Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol Microbiol. 1995;16:69–78. doi: 10.1111/j.1365-2958.1995.tb02392.x. [DOI] [PubMed] [Google Scholar]
- 124.Guilhot C, Gicquel B, Davies J, Martin C. Isolation and analysis of IS6120, a new insertion sequence from Mycobacterium smegmatis. Mol Microbiol. 1992;6:107–113. doi: 10.1111/j.1365-2958.1992.tb00842.x. [DOI] [PubMed] [Google Scholar]
- 125.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis and DNA sequence of a 7.8-kb virulence region of the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 126.Gustafson C E, Chu S, Trust T J. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 127.Haack K R. The activity of IS200 in Salmonella typhimurium. PhD thesis. Salt Lake City: University of Utah; 1995. [Google Scholar]
- 128.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128a.Haas, D. Personal communication.
- 129.Haas D, Berger B, Schmid S, Seitz T, Reimmann C. Insertion sequence IS21: Related insertion sequence elements, transpositional mechanisms, and application to linker insertion mutagenesis. In: Nakazawa T, editor. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 238–249. [Google Scholar]
- 130.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 131.Hall B G, Parker L L, Betts P W, DuBose R F, Sawyer S A, Hartl D L. IS103, a new insertion element in Escherichia coli: characterization and distribution in natural populations. Genetics. 1989;121:423–431. doi: 10.1093/genetics/121.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hall R M, Brown H J, Brookes D E, Stokes H W. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hallet B, Rezsöhazy R, Mahillon J, Delcour J. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol Microbiol. 1994;14:131–139. doi: 10.1111/j.1365-2958.1994.tb01273.x. [DOI] [PubMed] [Google Scholar]
- 134.Hallet B, Sherratt D J. Transposition and site-specific recombination: adapting DNA cut- and- paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 135.Hammerschmidt S, Hilse R, van Putten J P, Gerardy-Schahn R, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 136.Hanai R, Wang J C. The mechanism of sequence-specific DNA cleavage and strand transfer by phi X174 gene A* protein. J Biol Chem. 1993;268:23830–23836. [PubMed] [Google Scholar]
- 137.Haniford D B, Chelouche A R, Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989;59:385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- 137a.Haren, L., and M. Chandler. Unpublished results.
- 138.Haren L, Betermier M, Polard P, Chandler M. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol Microbiol. 1997;25:531–540. doi: 10.1046/j.1365-2958.1997.4951854.x. [DOI] [PubMed] [Google Scholar]
- 139.Haren, L., P. Polard, B. Ton-Hoang, and M. Chandler. J. Mol. Biol., in press. [DOI] [PubMed]
- 140.Henderson D J, Brolle D F, Kieser T, Melton R E, Hopwood D A. Transposition of IS117 (the Streptomyces coelicolor A 3 (2) mini-circle) to and from a cloned target site and into secondary chromosomal sites. Mol Gen Genet. 1990;224:65–71. doi: 10.1007/BF00259452. [DOI] [PubMed] [Google Scholar]
- 141.Henderson D J, Lydiate D J, Hopwood D A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2) Mol Microbiol. 1989;3:1307–1318. doi: 10.1111/j.1365-2958.1989.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 142.Hernandez Perez M, Fomukong N G, Hellyer T, Brown I N, Dale J W. Characterization of IS1110, a highly mobile genetic element from Mycobacterium avium. Mol Microbiol. 1994;12:717–724. doi: 10.1111/j.1365-2958.1994.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 143.Hill C W, Sandt C H, Vlazny D A. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol Microbiol. 1994;12:865–871. doi: 10.1111/j.1365-2958.1994.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 144.Hirsch H J, Starlinger P, Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119:191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- 145.Hodgson A L, Nicholson L A, Doran T J, Corner L A. Restriction fragment length polymorphism analysis of Fusobacterium necrophorum using a novel repeat DNA sequence and a 16S rRNA gene probe. FEMS Microbiol Lett. 1993;107:205–210. doi: 10.1111/j.1574-6968.1993.tb06031.x. [DOI] [PubMed] [Google Scholar]
- 146.Hofman J D, Schalkwyk L C, Doolittle W F. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium, Halobacterium volcanii. Nucleic Acids Res. 1986;14:6983–7000. doi: 10.1093/nar/14.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146a.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 147.Hu S T, Hwang J H, Lee L C, Lee C H, Li P L, Hsieh Y C. Functional analysis of the 14 kDa protein of insertion sequence 2. J Mol Biol. 1994;236:503–513. doi: 10.1006/jmbi.1994.1161. [DOI] [PubMed] [Google Scholar]
- 148.Hu S T, Lee C H. Characterization of the transposon carrying the STII gene of enterotoxigenic Escherichia coli. Mol Gen Genet. 1988;214:490–495. doi: 10.1007/BF00330485. [DOI] [PubMed] [Google Scholar]
- 149.Huang D C, Novel M, Novel G. A transposon-like element on the lactose plasmid of Lactococcus lactis subsp. lactis Z270. FEMS Microbiol Lett. 1991;61:101–106. doi: 10.1016/0378-1097(91)90021-2. [DOI] [PubMed] [Google Scholar]
- 150.Hubner A, Hendrickson W. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2,4,5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J Bacteriol. 1997;179:2717–2723. doi: 10.1128/jb.179.8.2717-2723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hubner P, Iida S, Arber W. A transcriptional terminator sequence in the prokaryotic transposable element IS1. Mol Gen Genet. 1987;206:485–490. doi: 10.1007/BF00428889. [DOI] [PubMed] [Google Scholar]
- 152.Huisman O, Errada P R, Signon L, Kleckner N. Mutational analysis of IS10’s outside end. EMBO J. 1989;8:2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ichikawa H, Ikeda K, Amemura J, Ohtsubo E. Two domains in the terminal inverted-repeat sequence of transposon Tn3. Gene. 1990;86:11–17. doi: 10.1016/0378-1119(90)90108-4. [DOI] [PubMed] [Google Scholar]
- 154.Iida S, Hiestand-Nauer R, Arber W. Transposable element IS1 intrinsically generates target duplications of variable length. Proc Natl Acad Sci USA. 1985;82:839–843. doi: 10.1073/pnas.82.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Isberg R R, Syvanen M. Replicon fusions promoted by the inverted repeats of Tn5. The right repeat is an insertion sequence. J Mol Biol. 1981;150:15–32. doi: 10.1016/0022-2836(81)90322-3. [DOI] [PubMed] [Google Scholar]
- 157.Isberg R R, Syvanen M. DNA gyrase is a host factor required for transposition of Tn5. Cell. 1982;30:9–18. doi: 10.1016/0092-8674(82)90006-x. [DOI] [PubMed] [Google Scholar]
- 158.Ishiguro N, Sato G. Spontaneous deletion of citrate-utilizing ability promoted by insertion sequences. J Bacteriol. 1984;160:642–650. doi: 10.1128/jb.160.2.642-650.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ishiguro N, Sato G, Sasakawa C, Danbara H, Yoshikawa M. Identification of citrate utilization transposon Tn3411 from a naturally occurring citrate utilization plasmid. J Bacteriol. 1982;149:961–968. doi: 10.1128/jb.149.3.961-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ivics Z, Izsvak Z, Minter A, Hackett P B. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jacobson J W, Medhora M M, Hartl D L. Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci USA. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jain C, Kleckner N. Preferential cis action of IS10 transposase depends upon its mode of synthesis. Mol Microbiol. 1993;9:249–260. doi: 10.1111/j.1365-2958.1993.tb01687.x. [DOI] [PubMed] [Google Scholar]
- 163.Reference deleted.
- 164.Jakowec M, Prentki P, Chandler M, Galas D J. Mutational analysis of the open reading frames in the transposable element IS1. Genetics. 1988;120:47–55. doi: 10.1093/genetics/120.1.47. . (Erratum, 121:393, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Johnson R C, Reznikoff W S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983;304:280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- 167.Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979;169:213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- 168.Kahn K, Schaefer M R. Characterization of transposon Tn5469 from the cyanobacterium Fremyella diplosiphon. J Bacteriol. 1995;177:7026–7032. doi: 10.1128/jb.177.24.7026-7032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kanazawa H, Kiyasu T, Noumi T, Futai M, Yamaguchi K. Insertions of transposable elements in the promoter proximal region of the gene cluster for Escherichia coli H+-ATPase: 8 base pair repeat generated by insertion of IS1. Mol Gen Genet. 1984;194:179–187. doi: 10.1007/BF00383514. [DOI] [PubMed] [Google Scholar]
- 170.Kato K, Ohtsuki K, Mitsuda H, Yomo T, Negoro S, Urabe I. Insertion sequence IS6100 on plasmid pOAD2, which degrades nylon oligomers. J Bacteriol. 1994;176:1197–1200. doi: 10.1128/jb.176.4.1197-1200.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 172.Katzman M, Mack J P, Skalka A M, Leis J. A covalent complex between retroviral integrase and nicked substrate DNA. Proc Natl Acad Sci USA. 1991;88:4695–4699. doi: 10.1073/pnas.88.11.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kaufman P D, Doll R F, Rio D C. Drosophila P element transposase recognizes internal P element DNA sequences. Cell. 1989;59:359–371. doi: 10.1016/0092-8674(89)90297-3. [DOI] [PubMed] [Google Scholar]
- 174.Kaufman P D, Rio D C. Drosophila P-element transposase is a transcriptional repressor in vitro. Proc Natl Acad Sci USA. 1991;88:2613–2617. doi: 10.1073/pnas.88.7.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kaufman P D, Rio D C. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992;69:27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- 175a.Kennedy, A., A. Guhathakurta, N. Kleckner, and D. Haniford. Personal communication.
- 176.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. . (Erratum, 19:1358.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kidwell M G. Hybrid dysgenesis in Drosophila melanogaster: partial sterility associated with embryo lethality in the P-M system. Genet Res. 1984;44:11–28. doi: 10.1017/s0016672300026215. [DOI] [PubMed] [Google Scholar]
- 178.Kim E H, Aoki T. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1994;38:31–38. doi: 10.1111/j.1348-0421.1994.tb01741.x. [DOI] [PubMed] [Google Scholar]
- 179.Kleckner N, Chalmers R M, Kwon D, Sakai J, Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanisms and regulation in vivo and in vitro. In: Saedler H, Gierl A, editors. Transposable elements. Heidelberg, Germany: Springer-Verlag KG; 1996. pp. 49–82. [DOI] [PubMed] [Google Scholar]
- 180.Knoop V, Brennicke A. Evidence for a group II intron in Escherichia coli inserted into a highly conserved reading frame associated with mobile DNA sequences. Nucleic Acids Res. 1994;22:1167–1171. doi: 10.1093/nar/22.7.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Koonin E V, Ilyina T V. Computer-assisted dissection of rolling circle DNA replication. Biosystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 182.Krebs M P, Reznikoff W S. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 183.Kretschmer P J, Cohen S N. Selected translocation of plasmid genes: frequency and regional specificity of translocation of the Tn3 element. J Bacteriol. 1977;130:888–899. doi: 10.1128/jb.130.2.888-899.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kruklitis R, Welty D J, Nakai H. ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 185.Kuan C T, Liu S K, Tessman I. Excision and transposition of Tn5 as an SOS activity in Escherichia coli. Genetics. 1991;128:45–57. doi: 10.1093/genetics/128.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kuan C T, Tessman I. LexA protein of Escherichia coli represses expression of the Tn5 transposase gene. J Bacteriol. 1991;173:6406–6410. doi: 10.1128/jb.173.20.6406-6410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kuan C T, Tessman I. Further evidence that transposition of Tn5 in Escherichia coli is strongly enhanced by constitutively activated RecA proteins. J Bacteriol. 1992;174:6872–6877. doi: 10.1128/jb.174.21.6872-6877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 190.Kunze R. The maize transposable element activator (Ac) Curr Top Microbiol Immunol. 1996;204:161–194. doi: 10.1007/978-3-642-79795-8_8. [DOI] [PubMed] [Google Scholar]
- 191.Kunze Z M, Wall S, Appelberg R, Silva M T, Portaels F, McFadden J J. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol Microbiol. 1991;5:2265–2272. doi: 10.1111/j.1365-2958.1991.tb02157.x. [DOI] [PubMed] [Google Scholar]
- 192.Kwon D, Chalmers R M, Kleckner N. Structural domains of IS10 transposase and reconstitution of transposition activity from proteolytic fragments lacking an interdomain linker. Proc Natl Acad Sci USA. 1995;92:8234–8238. doi: 10.1073/pnas.92.18.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Toussaint A. Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp protease. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 194.Lam S, Roth J R. Genetic mapping of IS200 copies in Salmonella typhimurium strain LT2. Genetics. 1983;105:801–811. doi: 10.1093/genetics/105.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lam S, Roth J R. IS200: a Salmonella-specific insertion sequence. Cell. 1983;34:951–960. doi: 10.1016/0092-8674(83)90552-4. [DOI] [PubMed] [Google Scholar]
- 196.Lampe D J, Churchill M E, Robertson H M. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. . (Erratum, 16:4153, 1997.) [PMC free article] [PubMed] [Google Scholar]
- 197.Reference deleted.
- 198.Lane D, Cavaille J, Chandler M. Induction of the SOS response by IS1 transposase. J Mol Biol. 1994;242:339–350. doi: 10.1006/jmbi.1994.1585. [DOI] [PubMed] [Google Scholar]
- 199.Lavoie B D, Chaconas G. Transposition of phage Mu DNA. Curr Top Microbiol Immunol. 1996;204:83–102. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- 200.Lawrence J G, Ochman H, Hartl D L. The evolution of insertion sequences within enteric bacteria. Genetics. 1992;131:9–20. doi: 10.1093/genetics/131.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lederberg E M. Plasmid reference center registry of transposon (Tn) allocations through July 1981. Gene. 1981;16:59–61. doi: 10.1016/0378-1119(81)90060-3. [DOI] [PubMed] [Google Scholar]
- 202.Lee C C, Mul Y M, Rio D C. The Drosophila P-element KP repressor protein dimerizes and interacts with multiple sites on P-element DNA. Mol Cell Biol. 1996;16:5616–5622. doi: 10.1128/mcb.16.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Leelaporn A, Firth N, Byrne M E, Roper E, Skurray R A. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother. 1994;38:2238–2244. doi: 10.1128/aac.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lei G S, Hu S T. Functional domains of the InsA protein of IS2. J Bacteriol. 1997;179:6238–6243. doi: 10.1128/jb.179.20.6238-6243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lenich A G, Glasgow A C. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J Bacteriol. 1994;176:4160–4164. doi: 10.1128/jb.176.13.4160-4164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205a.Léonard, C., and J. Mahillon. IS231A transposition: conservative versus replicative pathway. Res. Microbiol., in press. [DOI] [PubMed]
- 206.Levchenko I, Luo L, Baker T A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 207.Levchenko I, Yamauchi M, Baker T A. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 208.Lewis L A, Grindley N D. Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol. 1997;25:517–529. doi: 10.1046/j.1365-2958.1997.4871848.x. [DOI] [PubMed] [Google Scholar]
- 209.Reference deleted.
- 210.Lichens-Park A, Syvanen M. Cointegrate formation by IS50 requires multiple donor molecules. Mol Gen Genet. 1988;211:244–251. doi: 10.1007/BF00330600. [DOI] [PubMed] [Google Scholar]
- 211.Reference deleted.
- 212.Lodge J K, Berg D E. Mutations that affect Tn5 insertion into pBR322: importance of local DNA supercoiling. J Bacteriol. 1990;172:5956–5960. doi: 10.1128/jb.172.10.5956-5960.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lohe A R, Sullivan D T, Hartl D L. Subunit interactions in the mariner transposase. Genetics. 1996;144:1087–1095. doi: 10.1093/genetics/144.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lopez de Felipe F, Magni C, de Mendoza D, Lopez P. Transcriptional activation of the citrate permease P gene of Lactococcus lactis biovar diacetylactis by an insertion sequence-like element present in plasmid pCIT264. Mol Gen Genet. 1996;250:428–436. doi: 10.1007/BF02174031. [DOI] [PubMed] [Google Scholar]
- 215.Louarn J M, Bouche J P, Legendre F, Louarn J, Patte J. Characterization and properties of very large inversions of the E. coli chromosome along the origin-to-terminus axis. Mol Gen Genet. 1985;201:467–476. doi: 10.1007/BF00331341. [DOI] [PubMed] [Google Scholar]
- 215a.Low K B. Hfr strains of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2402–2412. [Google Scholar]
- 216.Lundblad V, Taylor A F, Smith G R, Kleckner N. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. Proc Natl Acad Sci USA. 1984;81:824–828. doi: 10.1073/pnas.81.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Luthi K, Moser M, Ryser J, Weber H. Evidence for a role of translational frameshifting in the expression of transposition activity of the bacterial insertion element IS1. Gene. 1990;88:15–20. doi: 10.1016/0378-1119(90)90054-u. [DOI] [PubMed] [Google Scholar]
- 218.Lyon B R, Gillespie M T, Skurray R A. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J Gen Microbiol. 1987;133:3031–3038. doi: 10.1099/00221287-133-11-3031. [DOI] [PubMed] [Google Scholar]
- 219.MacHattie L A, Jackowski J B. Physical structure and deletion effects of the chloramphenicol resistance element Tn9 in phage lambda. In: Bukhari A I, Shapiro J A, Adhya S L, editors. DNA insertion elements, plasmids, and episomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1977. pp. 219–228. [Google Scholar]
- 220.Machida C, Machida Y. Base substitutions in transposable element IS1 cause DNA duplication of variable length at the target site for plasmid co-integration. EMBO J. 1987;6:1799–1803. doi: 10.1002/j.1460-2075.1987.tb02433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Machida C, Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989;208:567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- 222.Machida Y, Machida C, Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984;177:229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- 223.Machida Y, Machida C, Ohtsubo H, Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci USA. 1982;79:277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Maekawa T, Amemura-Maekawa J, Ohtsubo E. DNA binding domains in Tn3 transposase. Mol Gen Genet. 1993;236:267–274. doi: 10.1007/BF00277122. [DOI] [PubMed] [Google Scholar]
- 224a.Maekawa T, Yanagihara K, Ohtsubo E. Specific nicking at the 3′ ends of the terminal inverted repeat sequences in transposon Tn3 by transposase and an E. coli protein ACP. Genes Cells. 1996;1:1017–1030. doi: 10.1046/j.1365-2443.1996.d01-221.x. [DOI] [PubMed] [Google Scholar]
- 225.Mahillon J, Rezsöhazy R, Hallet B, Delcour J. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica. 1994;93:13–26. doi: 10.1007/BF01435236. [DOI] [PubMed] [Google Scholar]
- 226.Mahillon J, Seurinck J, van Rompuy L, Delcour J, Zabeau M. Nucleotide sequence and structural organization of an insertion sequence element (IS231) from Bacillus thuringiensis strain berliner 1715. EMBO J. 1985;4:3895–3899. doi: 10.1002/j.1460-2075.1985.tb04163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Makris J C, Nordmann P L, Reznikoff W S. Mutational analysis of insertion sequence 50 (IS50) and transposon 5 (Tn5) ends. Proc Natl Acad Sci USA. 1988;85:2224–2228. doi: 10.1073/pnas.85.7.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mariani F, Piccolella E, Colizzi V, Rappuoli R, Gross R. Characterization of an IS-like element from Mycobacterium tuberculosis. J Gen Microbiol. 1993;139:1767–1772. doi: 10.1099/00221287-139-8-1767. [DOI] [PubMed] [Google Scholar]
- 229.Matsutani S. Genetic evidence for IS1 transposition regulated by InsA and the delta InsA-B′-InsB species, which is generated by translation from two alternative internal initiation sites and frameshifting. J Mol Biol. 1994;240:52–65. doi: 10.1006/jmbi.1994.1417. [DOI] [PubMed] [Google Scholar]
- 230.Matsutani S. Genetic analyses of the interactions of the IS1-encoded proteins with the left end of IS1 and its insertion hotspot. J Mol Biol. 1997;267:548–560. doi: 10.1006/jmbi.1996.0894. [DOI] [PubMed] [Google Scholar]
- 231.May E W, Craig N L. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- 231a.McFadden, J. Personal communication.
- 232.Mendiola M V, Bernales I, de la Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mendiola M V, de la Cruz F. Specificity of insertion of IS91, an insertion sequence present in alpha-haemolysin plasmids of Escherichia coli. Mol Microbiol. 1989;3:979–984. doi: 10.1111/j.1365-2958.1989.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 234.Mendiola M V, de la Cruz F. IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res. 1992;20:3521–3521. doi: 10.1093/nar/20.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mendiola M V, Jubete Y, de la Cruz F. DNA sequence of IS91 and identification of the transposase gene. J Bacteriol. 1992;174:1345–1351. doi: 10.1128/jb.174.4.1345-1351.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Meyer J, Iida S, Arber W. Does the insertion element IS1 transpose preferentially into A+T-rich DNA segments? Mol Gen Genet. 1980;178:471–473. doi: 10.1007/BF00270502. [DOI] [PubMed] [Google Scholar]
- 237.Mhammedi-Alaoui A, Pato M, Gama M J, Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 238.Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992;267:21273–21276. [PubMed] [Google Scholar]
- 239.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 240.Mizuuchi M, Mizuuchi K. Target site selection in transposition of phage Mu. Cold Spring Harbor Symp Quant Biol. 1993;58:515–523. doi: 10.1101/sqb.1993.058.01.058. [DOI] [PubMed] [Google Scholar]
- 241.Mollet B, Iida S, Arber W. Gene organization and target specificity of the prokaryotic mobile genetic element IS26. Mol Gen Genet. 1985;201:198–203. doi: 10.1007/BF00425660. [DOI] [PubMed] [Google Scholar]
- 242.Moore S P, Powers M, Garfinkel D J. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morisato D, Kleckner N. Transposase promotes double strand breaks and single strand joints at Tn10 termini in vivo. Cell. 1984;39:181–190. doi: 10.1016/0092-8674(84)90204-6. [DOI] [PubMed] [Google Scholar]
- 244.Morisato D, Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987;51:101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 245.Morisato D, Way J C, Kim H J, Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983;32:799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- 246.Mul Y M, Rio D C. Reprogramming the purine nucleotide cofactor requirement of Drosophila P element transposase in vivo. EMBO J. 1997;16:4441–4447. doi: 10.1093/emboj/16.14.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Muller H P, Varmus H E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4704–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mullins M C, Rio D C, Rubin G M. cis-acting DNA sequence requirements for P-element transposition. Genes Dev. 1989;3:729–738. doi: 10.1101/gad.3.5.729. [DOI] [PubMed] [Google Scholar]
- 249.Murphy E. Transposable elements in gram-positive bacteria. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 269–288. [Google Scholar]
- 249a.Naas, T., and W. Arber. Personal communication.
- 250.Naas T, Blot M, Fitch W M, Arber W. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics. 1994;136:721–730. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Nakatsu C, Ng J, Singh R, Straus N, Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci USA. 1991;88:8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nissley D V, Lindh F, Fennewald M A. Mutations in the inverted repeats of Tn3 affect binding of transposase and transposition immunity. J Mol Biol. 1991;218:335–347. doi: 10.1016/0022-2836(91)90716-j. [DOI] [PubMed] [Google Scholar]
- 253.Ohtsubo H, Nyman K, Doroszkiewicz W, Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981;292:640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- 254.Ohtsubo H, Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci USA. 1978;75:615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Olasz F, Stalder R, Arber W. Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. Mol Gen Genet. 1993;239:177–187. doi: 10.1007/BF00281616. [DOI] [PubMed] [Google Scholar]
- 256.O’Reilly C, Black G W, Laffey R, McConnell D J. Molecular analysis of an IS200 insertion in the gpt gene of Salmonella typhimurium LT2. J Bacteriol. 1990;172:6599–6601. doi: 10.1128/jb.172.11.6599-6601.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Osawa S, Jukes T H, Watanabe K, Muto A. Recent evidence for evolution of the genetic code. Microbiol Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Otten L, Canaday J, Gerard J C, Fournier P, Crouzet P, Paulus F. Evolution of agrobacteria and their Ti plasmids—a review. Mol Plant-Microbe Interact. 1992;5:279–287. doi: 10.1094/mpmi-5-279. [DOI] [PubMed] [Google Scholar]
- 259.Pato M L, Banerjee M. The Mu strong gyrase-binding site promotes efficient synapsis of the prophage termini. Mol Microbiol. 1996;22:283–292. doi: 10.1046/j.1365-2958.1996.00115.x. [DOI] [PubMed] [Google Scholar]
- 259a.Perkins-Balding, D., and A. C. Glasgow. Personal communication.
- 260.Picardeau M, Bull T J, Vincent V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol Lett. 1997;154:95–102. doi: 10.1111/j.1574-6968.1997.tb12629.x. [DOI] [PubMed] [Google Scholar]
- 261.Plasterk R H. The Tc1/mariner transposon family. Curr Top Microbiol Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 262.Podglajen I, Breuil J, Collatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1994;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 263.Podladchikova O N, Dikhanov G G, Rakin A V, Heesemann J. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol Lett. 1994;121:269–274. doi: 10.1111/j.1574-6968.1994.tb07111.x. [DOI] [PubMed] [Google Scholar]
- 264.Polard P. Etude de l’elément transposable bacterien IS911: mise en evidence et charactérisation des facteurs necessaires a sa transposition. Ph.D. thesis. Toulouse, France: Université Paul Sabatier; 1993. [Google Scholar]
- 265.Polard P, Chandler M. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 1995;9:2846–2858. doi: 10.1101/gad.9.22.2846. [DOI] [PubMed] [Google Scholar]
- 266.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 267.Polard P, Prère M F, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- 268.Polard P, Prère M F, Fayet O, Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992;11:5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Polard P, Seroude L, Fayet O, Prère M F, Chandler M. One-ended insertion of IS911. J Bacteriol. 1994;176:1192–1196. doi: 10.1128/jb.176.4.1192-1196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Polard P, Ton-Hoang B, Haren L, Betermier M, Walczak R, Chandler M. IS911-mediated transpositional recombination in vitro. J Mol Biol. 1996;264:68–81. doi: 10.1006/jmbi.1996.0624. [DOI] [PubMed] [Google Scholar]
- 271.Prentki P, Pham M H, Gamas P, Chandler M, Galas D J. Artificial transposable elements in the study of the ends of IS1. Gene. 1987;61:91–101. doi: 10.1016/0378-1119(87)90368-4. [DOI] [PubMed] [Google Scholar]
- 272.Prentki P, Teter B, Chandler M, Galas D J. Functional promoters created by the insertion of transposable element IS1. J Mol Biol. 1986;191:383–393. doi: 10.1016/0022-2836(86)90134-8. [DOI] [PubMed] [Google Scholar]
- 273.Pryciak P M, Varmus H E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 274.Radice A D, Bugaj B, Fitch D H, Emmons S W. Widespread occurrence of the Tc1 transposon family: Tc1-like transposons from teleost fish. Mol Gen Genet. 1994;244:606–612. doi: 10.1007/BF00282750. [DOI] [PubMed] [Google Scholar]
- 275.Radice A D, Emmons S W. Extrachromosomal circular copies of the transposon Tc1. Nucleic Acids Res. 1993;21:2663–2667. doi: 10.1093/nar/21.11.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Radström P, Skold O, Swedberg G, Flensburg J, Roy P H, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ramsden D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 278.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 279.Reif H J, Saedler H. IS1 is Involved in Deletion Formation in the gal region of E. coli K12. Mol Gen Genet. 1974;137:17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- 280.Reimmann C, Haas D. Mode of replicon fusion mediated by the duplicated insertion sequence IS21 in Escherichia coli. Genetics. 1987;115:619–625. doi: 10.1093/genetics/115.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Reimmann C, Haas D. The istA gene of insertion sequence IS21 is essential for cleavage at the inner 3′ ends of tandemly repeated IS21 elements in vitro. EMBO J. 1990;9:4055–4063. doi: 10.1002/j.1460-2075.1990.tb07627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Reimmann C, Haas D. Mobilization of chromosomes and nonconjugative plasmids by cointegrative mechanisms. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 137–188. [Google Scholar]
- 283.Reimmann C, Moore R, Little S, Savioz A, Willetts N S, Haas D. Genetic structure, function and regulation of the transposable element IS21. Mol Gen Genet. 1989;215:416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- 284.Reynolds A E, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 285.Reynolds A E, Mahadevan S, LeGrice S F, Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 286.Reznikoff W S. The Tn5 transposon. Annu Rev Microbiol. 1993;47:945–963. doi: 10.1146/annurev.mi.47.100193.004501. [DOI] [PubMed] [Google Scholar]
- 287.Reznikoff W S, Jilk R, Krebs M P, Makris J C, Nordmann P L, Weinreich M, Wiegand T. Tn5 lacZ translation fusion element: isolation and analysis of transposition mutants. Methods Enzymol. 1993;217:312–322. doi: 10.1016/0076-6879(93)17072-d. [DOI] [PubMed] [Google Scholar]
- 288.Rezsöhazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 289.Rice P, Craigie R, Davies D R. Retroviral integrases and their cousins. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 290.Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 291.Riess G, Holloway B W, Puhler A. R68.45, a plasmid with chromosome mobilizing ability (Cma) carries a tandem duplication. Genet Res. 1980;36:99–109. doi: 10.1017/s0016672300019704. [DOI] [PubMed] [Google Scholar]
- 292.Roberts D, Hoopes B C, McClure W R, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 293.Reference deleted.
- 294.Robertson H M. The mariner transposable element is widespread in insects. Nature. 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 295.Robertson H M, Lampe D J. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol Biol Evol. 1995;12:850–862. doi: 10.1093/oxfordjournals.molbev.a040262. [DOI] [PubMed] [Google Scholar]
- 296.Rose A M, Snutch T P. Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans. Nature. 1984;311:485–486. doi: 10.1038/311485a0. [DOI] [PubMed] [Google Scholar]
- 297.Roth D B, Lindahl T, Gellert M. Repair and recombination. How to make ends meet. Curr Biol. 1995;5:496–499. doi: 10.1016/s0960-9822(95)00101-1. [DOI] [PubMed] [Google Scholar]
- 298.Rowland S J, Dyke K G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 299.Sakai J, Chalmers R M, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Sasakawa C, Uno Y, Yoshikawa M. The requirement for both DNA polymerase and 5′ to 3′ exonuclease activities of DNA polymerase I during Tn5 transposition. Mol Gen Genet. 1981;182:19–24. doi: 10.1007/BF00422761. [DOI] [PubMed] [Google Scholar]
- 301.Savic D J, Romac S P, Ehrlich S D. Inversion in the lactose region of Escherichia coli K-12: inversion termini map within IS3 elements alpha 3 beta 3 and beta 5 alpha 5. J Bacteriol. 1983;155:943–946. doi: 10.1128/jb.155.2.943-946.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sawyer S A, Dykhuizen D E, DuBose R F, Green L, Mutangadura-Mhlanga T, Wolczyk D F, Hartl D L. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987;115:51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302a.Schmid, S., and D. Haas. Personal communication.
- 303.Schnetz K, Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schwartz E, Kroger M, Rak B. IS150: distribution, nucleotide sequence and phylogenetic relationships of a new E. coli insertion element. Nucleic Acids Res. 1988;16:6789–6802. doi: 10.1093/nar/16.14.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Scordilis G E, Ree H, Lessie T G. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J Bacteriol. 1987;169:8–13. doi: 10.1128/jb.169.1.8-13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 307.Sekine Y, Eisaki N, Kobayashi K, Ohtsubo E. Isolation and characterization of IS1 circles. Gene. 1997;191:183–190. doi: 10.1016/s0378-1119(97)00057-7. [DOI] [PubMed] [Google Scholar]
- 308.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 309.Sekine Y, Eisaki N, Ohtsubo E. Identification and characterization of the linear IS3 molecules generated by staggered breaks. J Biol Chem. 1996;271:197–202. doi: 10.1074/jbc.271.1.197. [DOI] [PubMed] [Google Scholar]
- 310.Sekine Y, Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci USA. 1989;86:4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Sengstag C, Arber W. A cloned DNA fragment from bacteriophage P1 enhances IS2 insertion. Mol Gen Genet. 1987;206:344–351. doi: 10.1007/BF00333593. [DOI] [PubMed] [Google Scholar]
- 312.Sengstag C, Iida S, Hiestand-Nauer R, Arber W. Terminal inverted repeats of prokaryotic transposable element IS186 which can generate duplications of variable length at an identical target sequence. Gene. 1986;49:153–156. doi: 10.1016/0378-1119(86)90395-1. [DOI] [PubMed] [Google Scholar]
- 313.Serre M C, Turlan C, Bortolin M, Chandler M. Mutagenesis of the IS1 transposase: importance of a His-Arg-Tyr triad for activity. J Bacteriol. 1995;177:5070–5077. doi: 10.1128/jb.177.17.5070-5077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313a.Sharpe, P., and N. Craig. Personal communication.
- 314.Signon L, Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 315.Skalka A M. Retroviral DNA integration: lessons for transposon shuffling. Gene. 1993;135:175–182. doi: 10.1016/0378-1119(93)90063-9. [DOI] [PubMed] [Google Scholar]
- 316.Small P M, van Embden J D A. Molecular epidemiology of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 569–582. [Google Scholar]
- 317.Smith B, Dyson P. Inducible transposition in Streptomyces lividans of insertion sequence IS6100 from Mycobacterium fortuitum. Mol Microbiol. 1995;18:933–941. doi: 10.1111/j.1365-2958.1995.18050933.x. [DOI] [PubMed] [Google Scholar]
- 318.So M, Heffron F, McCarthy B J. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979;277:453–456. doi: 10.1038/277453a0. [DOI] [PubMed] [Google Scholar]
- 319.Solinas F, Marconi A M, Ruzzi M, Zennaro E. Characterization and sequence of a novel insertion sequence, IS1162, from Pseudomonas fluorescens. Gene. 1995;155:77–82. doi: 10.1016/0378-1119(94)00922-f. [DOI] [PubMed] [Google Scholar]
- 320.Spielmann-Ryser J, Moser M, Kast P, Weber H. Factors determining the frequency of plasmid cointegrate formation mediated by insertion sequence IS3 from Escherichia coli. Mol Gen Genet. 1991;226:441–448. doi: 10.1007/BF00260657. [DOI] [PubMed] [Google Scholar]
- 321.Reference deleted.
- 322.Stalder R, Caspers P, Olasz F, Arber W. The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. J Biol Chem. 1990;265:3757–3762. [PubMed] [Google Scholar]
- 323.Stanley J, Baquar N, Threlfall E J. Genotypes and phylogenetic relationships of Salmonella typhimurium are defined by molecular fingerprinting of IS200 and 16S rrn loci. J Gen Microbiol. 1993;139:1133–1140. doi: 10.1099/00221287-139-6-1133. [DOI] [PubMed] [Google Scholar]
- 324.Stark W M, Boocock M R, Sherratt D J. Site-specific recombination by Tn3 resolvase. Trends Genet. 1989;5:304–309. doi: 10.1016/0168-9525(89)90113-3. [DOI] [PubMed] [Google Scholar]
- 325.Sternglanz R, DiNardo S, Voelkel K A, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang J C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325a.Stibitz, S. Personal communication.
- 326.Stibitz S, Davies J E. Tn602: a naturally occurring relative of Tn903 with direct repeats. Plasmid. 1987;17:202–209. doi: 10.1016/0147-619x(87)90028-x. [DOI] [PubMed] [Google Scholar]
- 327.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Sundin G W, Bender C L. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl Environ Microbiol. 1995;61:2891–2897. doi: 10.1128/aem.61.8.2891-2897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Syvanen M, Hopkins J D, Clements M. A new class of mutants in DNA polymerase I that affects gene transposition. J Mol Biol. 1982;158:203–212. doi: 10.1016/0022-2836(82)90429-6. [DOI] [PubMed] [Google Scholar]
- 330.Szeverenyi I, Bodoky T, Olasz F. Isolation, characterization and transposition of an (IS2)2 intermediate. Mol Gen Genet. 1996;251:281–289. doi: 10.1007/BF02172518. [DOI] [PubMed] [Google Scholar]
- 331.Takemura H, Horinouchi S, Beppu T. Novel insertion sequence IS1380 from Acetobacter pasteurianus is involved in loss of ethanol-oxidizing ability. J Bacteriol. 1991;173:7070–7076. doi: 10.1128/jb.173.22.7070-7076.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Tavakoli N P, DeVost J, Derbyshire K M. Defining functional regions of the IS903 transposase. J Mol Biol. 1997;274:491–504. doi: 10.1006/jmbi.1997.1410. [DOI] [PubMed] [Google Scholar]
- 333.Tenzen T, Matsutani S, Ohtsubo E. Site-specific transposition of insertion sequence IS630. J Bacteriol. 1990;172:3830–3836. doi: 10.1128/jb.172.7.3830-3836.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Tenzen T, Ohtsubo E. Preferential transposition of an IS630-associated composite transposon to TA in the 5′-CTAG-3′ sequence. J Bacteriol. 1991;173:6207–6212. doi: 10.1128/jb.173.19.6207-6212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Tizard M L, Moss M T, Sanderson J D, Austen B M, Hermon-Taylor J. p43, the protein product of the atypical insertion sequence IS900, is expressed in Mycobacterium paratuberculosis. J Gen Microbiol. 1992;138:1729–1736. doi: 10.1099/00221287-138-8-1729. [DOI] [PubMed] [Google Scholar]
- 336.Tolmasky M E, Crosa J H. Iron transport genes of the pJM1-mediated iron uptake system of Vibrio anguillarum are included in a transposonlike structure. Plasmid. 1995;33:180–190. doi: 10.1006/plas.1995.1019. [DOI] [PubMed] [Google Scholar]
- 337.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 337a.Ton-Hoang, B., and M. Chandler. Unpublished results.
- 338.Ton-Hoang B, Betermier M, Polard P, Chandler M. Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J. 1997;16:3357–3371. doi: 10.1093/emboj/16.11.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ton-Hoang B, Polard P, Chandler M. Efficient transposition of IS911 circles in vitro. EMBO J. 1998;17:1169–1181. doi: 10.1093/emboj/17.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Trentmann S M, Saedler H, Gierl A. The transposable element En/Spm-encoded TNPA protein contains a DNA binding and a dimerization domain. Mol Gen Genet. 1993;238:201–208. doi: 10.1007/BF00279548. [DOI] [PubMed] [Google Scholar]
- 341.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the transposable element IS15. Gene. 1984;30:113–120. doi: 10.1016/0378-1119(84)90111-2. [DOI] [PubMed] [Google Scholar]
- 342.Turlan C, Chandler M. IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J. 1995;14:5410–5421. doi: 10.1002/j.1460-2075.1995.tb00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Umeda M, Ohtsubo E. Four types of IS1 with differences in nucleotide sequence reside in the Escherichia coli K-12 chromosome. Gene. 1991;98:1–5. doi: 10.1016/0378-1119(91)90096-t. [DOI] [PubMed] [Google Scholar]
- 344.Utsumi R, Ikeda M, Horie T, Yamamoto M, Ichihara A, Taniguchi Y, Hashimoto R, Tanabe H, Obata K, Noda M. Isolation and characterization of the IS3-like element from Thermus aquaticus. Biosci Biotechnol Biochem. 1995;59:1707–1711. doi: 10.1271/bbb.59.1707. [DOI] [PubMed] [Google Scholar]
- 345.van der Ploeg J, Willemsen M, van Hall G, Janssen D B. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol. 1995;177:1348–1356. doi: 10.1128/jb.177.5.1348-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 347.van Luenen H G, Colloms S D, Plasterk R H. Mobilization of quiet, endogenous Tc3 transposons of Caenorhabditis elegans by forced expression of Tc3 transposase. EMBO J. 1993;12:2513–2520. doi: 10.1002/j.1460-2075.1993.tb05906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.van Luenen H G, Colloms S D, Plasterk R H. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994;7984:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 349.van Luenen H G, Plasterk R H. Target site choice of the related transposable elements Tc1 and Tc3 of Caenorhabditis elegans. Nucleic Acids Res. 1994;22:262–269. doi: 10.1093/nar/22.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Vertes A A, Inui M, Kobayashi M, Kurusu Y, Yukawa H. Isolation and characterization of IS31831, a transposable element from Corynebacterium glutamicum. Mol Microbiol. 1994;11:739–746. doi: 10.1111/j.1365-2958.1994.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 350a.Vögele, K. Personal communication.
- 351.Vögele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Vos J C, Plasterk R H. Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 1994;13:6125–6132. doi: 10.1002/j.1460-2075.1994.tb06959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Vos J C, van Luenen H G, Plasterk R H. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 1993;7:1244–1253. doi: 10.1101/gad.7.7a.1244. [DOI] [PubMed] [Google Scholar]
- 354.Wang X, Higgins N P. ‘Muprints’ of the lac operon demonstrate physiological control over the randomness of in vivo transposition. Mol Microbiol. 1994;12:665–677. doi: 10.1111/j.1365-2958.1994.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 355.Reference deleted.
- 356.Weinert T A, Schaus N A, Grindley N D. Insertion sequence duplication in transpositional recombination. Science. 1983;222:755–765. doi: 10.1126/science.6314502. [DOI] [PubMed] [Google Scholar]
- 357.Weinreich M D, Mahnke-Braam L, Reznikoff W S. A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. J Mol Biol. 1994;241:166–177. doi: 10.1006/jmbi.1994.1486. [DOI] [PubMed] [Google Scholar]
- 358.Weinreich M D, Makris J C, Reznikoff W S. Induction of the SOS response in Escherichia coli inhibits Tn5 and IS50 transposition. J Bacteriol. 1991;173:6910–6918. doi: 10.1128/jb.173.21.6910-6918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Weinreich M D, Reznikoff W S. Fis plays a role in Tn5 and IS50 transposition. J Bacteriol. 1992;174:4530–4537. doi: 10.1128/jb.174.14.4530-4537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Welty D J, Jones J M, Nakai H. Communication of ClpXP protease hypersensitivity to bacteriophage Mu repressor isoforms. J Mol Biol. 1997;272:31–41. doi: 10.1006/jmbi.1997.1193. [DOI] [PubMed] [Google Scholar]
- 361.Welz C. Functionelle analyse des Bakteriellen Insertionelements IS150. Ph.D. thesis. Freiburg, Germany: Albert-Ludwigs-Universität Freiburg; 1993. [Google Scholar]
- 362.Wiater L A, Grindley N D. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988;7:1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wiater L A, Grindley N D. Integration host factor increases the transpositional immunity conferred by gamma delta ends. J Bacteriol. 1990;172:4951–4958. doi: 10.1128/jb.172.9.4951-4958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Wiater L A, Grindley N D. Uncoupling of transpositional immunity from gamma delta transposition by a mutation at the end of gamma delta. J Bacteriol. 1990;172:4959–4963. doi: 10.1128/jb.172.9.4959-4963.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Wiegand T W, Reznikoff W S. Interaction of Tn5 transposase with the transposon termini. J Mol Biol. 1994;235:486–495. doi: 10.1006/jmbi.1994.1008. [DOI] [PubMed] [Google Scholar]
- 366.Williams K, Doak T G, Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993;12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wishart W L, Machida C, Ohtsubo H, Ohtsubo E. Escherichia coli RNA polymerase binding sites and transcription initiation sites in the transposon Tn3. Gene. 1983;24:99–113. doi: 10.1016/0378-1119(83)90135-x. [DOI] [PubMed] [Google Scholar]
- 368.Wolkow C A, DeBoy R T, Craig N L. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 369.Yin J C, Krebs M P, Reznikoff W S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988;199:35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- 370.Reference deleted.
- 371.Yin J C, Reznikoff W S. dnaA, an essential host gene, and Tn5 transposition. J Bacteriol. 1987;169:4637–4645. doi: 10.1128/jb.169.10.4637-4645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Youngman P. Transposons and their applications. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 585–596. [Google Scholar]
- 373.Yurieva O, Nikiforov V. Catalytic center quest: comparison of transposases belonging to the Tn3 family reveals an invariant triad of acidic amino acid residues. Biochem Mol Biol Int. 1996;38:15–20. [PubMed] [Google Scholar]
- 374.Zabala J C, Garcia-Lobo J M, Diaz-Aroca E, de la Cruz F, Ortiz J M. Escherichia coli alpha-haemolysin synthesis and export genes are flanked by a direct repetition of IS91-like elements. Mol Gen Genet. 1984;197:90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]
- 374a.Zablweska, B., et al. Unpublished data.
- 374b.Zerbib, D., and M. Chandler. Unpublished data.
- 375.Zerbib D, Gamas P, Chandler M, Prentki P, Bass S, Galas D. Specificity of insertion of IS1. J Mol Biol. 1985;185:517–524. doi: 10.1016/0022-2836(85)90068-3. [DOI] [PubMed] [Google Scholar]
- 376.Zerbib D, Jakowec M, Prentki P, Galas D J, Chandler M. Expression of proteins essential for IS1 transposition: specific binding of InsA to the ends of IS1. EMBO J. 1987;6:3163–3169. doi: 10.1002/j.1460-2075.1987.tb02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zerbib D, Polard P, Escoubas J M, Galas D, Chandler M. The regulatory role of the IS1-encoded InsA protein in transposition. Mol Microbiol. 1990;4:471–477. doi: 10.1111/j.1365-2958.1990.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 378.Zerbib D, Prentki P, Gamas P, Freund E, Galas D J, Chandler M. Functional organization of the ends of IS1: specific binding site for an IS1-encoded protein. Mol Microbiol. 1990;4:1477–1486. [PubMed] [Google Scholar]
- 379.Zhao S, Hill C W. Reshuffling of Rhs components to create a new element. J Bacteriol. 1995;177:1393–1398. doi: 10.1128/jb.177.5.1393-1398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Reference deleted.
- 381.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Zheng J, McIntosh M A. Characterization of IS1221 from Mycoplasma hyorhinis: expression of its putative transposase in Escherichia coli incorporates a ribosomal frameshift mechanism. Mol Microbiol. 1995;16:669–685. doi: 10.1111/j.1365-2958.1995.tb02429.x. [DOI] [PubMed] [Google Scholar]
- 383.Zhou L, Hui F M, Morrison D A. Characterization of IS1167, a new insertion sequence in Streptococcus pneumoniae. Plasmid. 1995;33:127–138. doi: 10.1006/plas.1995.1014. [DOI] [PubMed] [Google Scholar]