Abstract
During the past decade the pesticidal bacterium Bacillus thuringiensis has been the subject of intensive research. These efforts have yielded considerable data about the complex relationships between the structure, mechanism of action, and genetics of the organism’s pesticidal crystal proteins, and a coherent picture of these relationships is beginning to emerge. Other studies have focused on the ecological role of the B. thuringiensis crystal proteins, their performance in agricultural and other natural settings, and the evolution of resistance mechanisms in target pests. Armed with this knowledge base and with the tools of modern biotechnology, researchers are now reporting promising results in engineering more-useful toxins and formulations, in creating transgenic plants that express pesticidal activity, and in constructing integrated management strategies to insure that these products are utilized with maximum efficiency and benefit.
GENERAL CHARACTERISTICS
The leading biorational pesticide, Bacillus thuringiensis, is a ubiquitous gram-positive, spore-forming bacterium that forms a parasporal crystal during the stationary phase of its growth cycle. B. thuringiensis was initially characterized as an insect pathogen, and its insecticidal activity was attributed largely or completely (depending on the insect) to the parasporal crystals. This observation led to the development of bioinsecticides based on B. thuringiensis for the control of certain insect species among the orders Lepidoptera, Diptera, and Coleoptera (for a review, see reference 33). There are more recent reports of B. thuringiensis isolates active against other insect orders (Hymenoptera, Homoptera, Orthoptera, and Mallophaga) and against nematodes, mites, and protozoa (109, 110). B. thuringiensis is already a useful alternative or supplement to synthetic chemical pesticide application in commercial agriculture, forest management, and mosquito control. It is also a key source of genes for transgenic expression to provide pest resistance in plants.
In 1989, Höfte and Whiteley reviewed the known cry genes and proposed a systematic nomenclature for them (164). Since then, the number of sequenced crystal protein genes (encoding Cry and Cyt proteins) has grown from 14 to well more than 100. In our accompanying work (79), we propose a revised nomenclature to accommodate this wealth of new sequence data. The present work reviews the extensive progress during the past decade in determining the gene expression, structure, and mechanism of action for these classes of proteins. The proposed revised nomenclature will be used throughout.
ECOLOGY AND PREVALENCE
B. thuringiensis seems to be indigenous to many environments (36, 65, 255). Strains have been isolated worldwide from many habitats, including soil (59, 88, 154, 255, 354), insects (59), stored-product dust (54, 65, 87, 267), and deciduous and coniferous leaves (175, 354). Isolation typically involves heat treatment to select for spores, sometimes with an acetate enrichment step (382) or antibiotic selection (89). The diversity in flagellar H-antigen agglutination reactions is one indication of the enormous genetic diversity among B. thuringiensis isolates. The Pasteur Institute has catalogued 55 different flagellar serotypes and eight nonflagellated biotypes (202, 205).
There is considerable evidence that B. thuringiensis and Bacillus cereus should be considered a single species. Classical biochemical and morphological methods of classifying bacteria have consistently failed to distinguish B. thuringiensis from B. cereus (31, 139, 177, 229, 305). Modern molecular methods—including chromosomal DNA hybridization (179), phospholipid and fatty acid analysis (40, 178), 16S rRNA sequence comparison (20, 318), amplified fragment length polymorphism analysis (181), and genomic restriction digest analysis (56, 57)—likewise support the single-species hypothesis. An attempt to distinguish B. thuringiensis isolates from B. cereus by analysis of a 16S rRNA variable region largely failed, yielding as many false positives and negatives as accurate identifications (373). The production of the parasporal crystal, the defining quality of B. thuringiensis, is too narrow a criterion for taxonomic purposes (237). Indeed, some B. cereus strains hybridize to cry1A-specific probes (56). Although we will employ the official nomenclature with two species names for these organisms, it is perhaps best to think of them as members of B. cereus sensu lato.
The remarkable diversity of B. thuringiensis strains and toxins is due at least in part to a high degree of genetic plasticity. Most B. thuringiensis toxin genes appear to reside on plasmids (138), often as parts of composite structures that include mobile genetic elements (195, 218). Many cry gene-containing plasmids appear to be conjugative in nature (137).
B. thuringiensis has developed a fascinating array of molecular mechanisms to produce large amounts of pesticidal toxins during the stationary phase of growth (8, 30). One can only speculate about the ecological value to the bacterium of using several cry gene expression systems. However, coexpression of multiple toxins is likely to increase the host range of a given strain or of a population exchanging toxin genes. One report has suggested plasmid transfer between different B. thuringiensis strains during growth within an insect (170). We are not aware of any critical experiments directed towards understanding bacterial toxin gene expression within the gut of a susceptible pest.
Persistence of B. thuringiensis spores in the laboratory, greenhouse, and field or forest environment has been reasonably well studied (299, 403, 405). B. thuringiensis spores can survive for several years after spray applications (6), although rapid declines in population and toxicity have been noted. Methods of detection have generally been limited to spore counts.
Meadows (266) has analyzed three prevailing hypothetical niches of B. thuringiensis in the environment: as an entomopathogen, as a phylloplane inhabitant, and as a soil microorganism. Available data are still insufficient to choose among these and other possibilities, although B. thuringiensis seems to have been more readily isolated from insect cadavers or stored-product dusts than from soil (36, 65). It is also noteworthy that B. thuringiensis and B. cereus are able to multiply in the insect hemocoel and to provoke septicemia (156, 157, 358). Early work recognized the presence of a number of extracellular compounds that might contribute to virulence, including phospholipases (434), other heat-labile toxin activities (reviewed in reference 332), and β-exotoxins (221). More recent characterization has shown that proteases (232), chitinases (356), and the secreted vegetative insecticidal proteins (VIPs) (108) (see below) may contribute to virulence. B. cereus and B. thuringiensis also produce antibiotic compounds that have antifungal activity (357); one of these products can act to synergize crystal protein-induced intoxication of certain lepidopterans (253). The Cry toxins are, therefore, the most prominent of a number of virulence factors allowing the development of the bacteria in dead or weakened insect larvae. Such data are at least suggestive that many strains of B. thuringiensis and some strains of B. cereus can be regarded as opportunistic insect pathogens. A more thorough understanding of the true ecological roles of B. thuringiensis would be of great importance, both for improving the reliability of risk assessment and for developing efficient methods for isolating novel B. thuringiensis strains containing useful δ-endotoxin genes.
A number of pesticidal proteins unrelated to the Cry proteins are produced by some strains of B. thuringiensis during vegetative growth (108, 401). These VIPs do not form parasporal crystal proteins and are apparently secreted from the cell. The VIPs are presently excluded from the Cry protein nomenclature because they are not crystal-forming proteins. The term VIP is a misnomer in the sense that some B. thuringiensis Cry proteins are also produced during vegetative growth as well as during the stationary and sporulation phases, most notably Cry3Aa (see “cry gene expression”). The location of the vip genes in the B. thuringiensis genome has not been reported, although it would not be surprising to find them residing on large plasmids that encode cry genes.
The vip1A gene encodes a 100-kDa protein that is apparently processed from its N terminus to yield an ∼80-kDa protein upon secretion. The 80-kDa Vip1A protein is reported to be toxic to western corn rootworm larvae in conjunction with the Vip2A protein, whose coding region is located immediately upstream (401). Interestingly, Vip1A shows sequence similarity to the protective antigen of the tripartite Bacillus anthracis toxin (298).
The vip3A gene encodes an 88-kDa protein that is produced during vegetative growth but is not processed upon secretion. Genes encoding Vip3A-type proteins appear to be common among strains of B. thuringiensis and B. cereus (108). This protein is reported to exhibit toxicity towards a wide variety of lepidopteran insect pests, including Agrotis ipsilon, Spodoptera frugiperda, Spodoptera exigua, and Helicoverpa zea (108). When fed to susceptible insects at lethal concentrations, Vip3A causes gut paralysis and lysis of midgut epithelial cells: the physical manifestations of Vip3A intoxication resemble those of the Cry proteins (431).
GENETICS AND MOLECULAR BIOLOGY
The B. thuringiensis Genome
B. thuringiensis strains have a genome size of 2.4 to 5.7 million bp (56). Physical maps have been constructed for two B. thuringiensis strains (57, 58). Comparison with B. cereus chromosomal maps suggests that all of these chromosomes have a similar organization in the half near the replication origin while displaying greater variability in the terminal half (57). Most B. thuringiensis isolates have several extrachromosomal elements, some of them circular and others linear (56). It has long been recognized that the proteins comprising the parasporal crystal are generally encoded by large plasmids (138). Sequences hybridizing to cry gene probes occur commonly among B. thuringiensis chromosomes as well (58), although it is unclear to what degree these chromosomal homologs contribute to production of the crystal.
The Transposable Elements of B. thuringiensis
The B. thuringiensis species harbors a large variety of transposable elements, including insertion sequences and transposons. The general characteristics of these elements have been extensively reviewed by Mahillon et al. (248). Here, the B. thuringiensis transposable elements are described with regard to their structural association with the cry genes.
The first studies on the structural organization of the cry1A gene environment showed that genes of this type were flanked by two sets of inverted repeated sequences (195, 218). Nucleotide sequence analysis revealed that these repetitive elements were insertion sequences that have been designated IS231 and IS232 (219, 237). IS231 belongs to the IS4 family of insertion sequences (315), and IS232 belongs to the IS21 family of insertion sequences (268). Because these elements can transpose (152, 268), it is likely that they provide mobility for the cry genes with which they form typical composite transposons. However, this hypothesis has not been tested experimentally.
Several IS231 variants have been isolated from various B. thuringiensis strains (249, 314, 316) and have been detected in representative strains from well more than half of the known B. thuringiensis serovars (212). In B. thuringiensis subsp. israelensis, an IS231 element (IS231W) is adjacent to the cry11Aa gene (4, 316). Although IS231 elements are frequently associated with cry genes, IS231-related DNA sequences have also been found in strains of B. cereus (190, 212) and Bacillus mycoides (212). In contrast, IS232 has a much smaller range among the organisms surveyed so far, appearing in only 7 of 61 B. thuringiensis serovars (212).
The cry4A gene of the israelensis subspecies is flanked by two repeated sequences in opposite orientations (45). These sequences, designated IS240, display features characteristic of insertion sequences (83). The IS240 transposase is homologous to those of the insertion sequences belonging to the IS6 family. IS240 is widely distributed in B. thuringiensis and is invariably present in known dipteran-active strains (319). Related sequences have also been detected in B. mycoides and B. cereus (212). An IS240 variant has been found upstream of the cry11B gene in the B. thuringiensis subsp. jegathesan (86) and from a plasmid of the dipteran-active strain B. thuringiensis subsp. fukuokaensis (103).
Insertion sequences have been found upstream of the cry1Ca gene (351) and downstream of a cryptic cry2Ab gene (160). These elements encode putative transposases that have significant similarities with the transposase of the IS150 element from Escherichia coli. These potential transposable elements of B. thuringiensis consequently belong to the IS3 family of insertion sequences.
The first transposable element identified in the genus Bacillus was isolated from B. thuringiensis following its spontaneous insertion into a conjugative plasmid transferred from Enterococcus faecalis (217). The genetic and structural characteristics of this transposable element fulfilled the criteria of a Tn element, and it was designated Tn4430 (216). Its transposase is homologous to those of the Tn3 family. In contrast to Tn3, however, the site-specific recombinase that mediates Tn4430 cointegrate resolution is not a resolvase but an integrase (247). Tn4430 is frequently found in the vicinity of genes of the cry1A type in various lepidopteran-active strains (196, 218, 328). However, Tn4430-like sequences have also been detected in several strains of B. cereus (56).
A transposable element designated Tn5401 was isolated from a coleopteran-active B. thuringiensis strain following its spontaneous insertion into a recombinant plasmid (27). Although nucleotide sequence analysis indicates that the structural organization of Tn5401 is similar to that of Tn4430, the transposases and the site-specific recombinases of these transposons are only distantly related (27). Tn4430 and Tn5401 are not known to coexist in any B. thuringiensis strain (27). In B. thuringiensis subsp. tenebrionis, Tn5401 is located just downstream of the cry3Aa gene (3). It is noteworthy that Tn5401 has been successfully used to construct a transposon insertion library in B. thuringiensis (251).
Two open reading frames encoding polypeptides homologous to the transposase and to the resolvase of the Tn3 family of transposons have been identified upstream of the cry16A gene found in Clostridium bifermentans (23, 82). This observation suggests that a Tn element is structurally associated with this cry gene.
Regarding the role of the transposable elements in B. thuringiensis, it is postulated that they are involved in the amplification of the cry genes in the bacterial cell, but this hypothesis has not been clearly tested. A second possible role is one of mediating the transfer of plasmids by a conduction process involving the formation of cointegrate structures between self-conjugative plasmids and chromosomal DNA or nonconjugative plasmids. Indeed, conjugation experiments suggest that Tn4430 mediates the transfer of nonconjugative plasmids by a conduction process (147). Thus, a major adaptive function for these transposable elements may be the horizontal dissemination of genetic material, including cry genes, within the B. cereus-B. thuringiensis species.
cry Gene Expression
A common characteristic of the cry genes is their expression during the stationary phase. Their products generally accumulate in the mother cell compartment to form a crystal inclusion that can account for 20 to 30% of the dry weight of the sporulated cells. The very high level of crystal protein synthesis in B. thuringiensis and its coordination with the stationary phase are controlled by a variety of mechanisms occurring at the transcriptional, posttranscriptional, and posttranslational levels. Agaisse and Lereclus (8) and Baum and Malvar (30) have recently reviewed the regulation of cry gene expression in detail. We present here a broad outline of these regulatory mechanisms.
Transcriptional Mechanisms
The cry genes have long been considered typical examples of sporulation-specific genes. However, recent studies on the expression of the cry3Aa gene have revealed that this assumption is not always valid. It is therefore necessary to distinguish, among the cry genes expressed during the stationary phase, those that are dependent on sporulation from those that are not.
Sporulation-dependent cry Gene Expression.
Extensive studies of the sporulation of B. subtilis have provided detailed information on the complex mechanisms that temporally and spatially control this differentiation process (for reviews, see references 104 and 231). At the transcriptional level, the development of sporulation is controlled by the successive activation of sigma factors, which bind the core RNA polymerase to direct the transcription from sporulation-specific promoters (275). These factors are the primary sigma factor of vegetative cells, ςA, and five factors called ςH, ςF, ςE, ςG, and ςK, which appear in that order in a temporally regulated fashion during development. The ςA and ςH factors are active in the predivisional cell, ςE and ςK are active in the mother cell, and ςF and ςG are active in the forespore.
The cry1Aa gene is a typical example of a sporulation-dependent cry gene expressed only in the mother cell compartment of B. thuringiensis. Two transcription start sites have been mapped (BtI and BtII), defining two overlapping, sequentially activated promoters (417). BtI is active between about T2 and T6 of sporulation and BtII is active from about T5 onwards (where Tn is n hours after the end of the exponential phase). Brown and Whiteley (52, 53) isolated two sigma factors, ς35 and ς28, that specifically direct transcription of cry1Aa from BtI and BtII, respectively. In vitro transcription experiments have also indicated that at least two other cry genes (cry1Ba and cry2Aa) contain either BtI alone or BtI with BtII (52).
The genes encoding ς35 and ς28 have been cloned and sequenced (1). Their deduced amino acid sequences show 88 and 85% identity with ςE and ςK of B. subtilis, respectively. B. thuringiensis ςE and ςK mutants were constructed, and cry1Aa gene expression was analyzed in these mutants (48). The results indicated that these two sigma factors regulated expression of a cry1Aa′-′lacZ transcriptional fusion in vivo. The ςK mutant produced about 50% less β-galactosidase than the wild-type strain, whereas no β-galactosidase synthesis was obtained in the ςE mutant. The latter result was anticipated, because ςE controls ςK synthesis.
Several cry gene promoters have been identified, and their sequences have been previously determined (50, 51, 94, 428, 430). Consensus sequences for promoters recognized by B. thuringiensis RNA polymerase containing ςE or ςK have been deduced from alignment of the promoter regions of these genes (8, 30). The results are that, in addition to the transcription of cry1Aa, cry1Ba, and cry2Aa, the transcription of many other cry genes (e.g., cry4Aa, cry4Ba, cry11Aa, cry15Aa, etc.) is likely to be ςE- or ςK-dependent. Analysis of cry4Aa, cry4Ba, and cry11Aa gene fusions in a B. thuringiensis sigE mutant confirms that SigE is required for their expression during sporulation (304). In addition, from a genetic analysis of B. subtilis, Yoshisue et al. (430) reported that the expression of cry4B is reduced in a spoIIID mutant strain, thus suggesting that SpoIIID, a DNA-binding protein, positively regulates the SigE-dependent transcription of cry4B. The cry18Aa gene isolated from Bacillus popilliae is successively transcribed by ςE and ςK forms of RNA polymerase from a single promoter during sporulation (433).
The expression of all these cry genes is therefore considered to be sporulation dependent. However, low-level transcription of the cry4Aa, cry4Ba, and cry11Aa genes in B. thuringiensis has been detected during the transition phase, beginning at about T−2 and lasting until the onset of sporulation (304, 429). This expression may be due to the ςH RNA polymerase, and it is suggested that Spo0A represses this weak expression, specific to the transition phase, when the cells enter the sporulation phase (304).
Sporulation-independent cry gene expression.
The cry3Aa gene, isolated from the coleopteran-active B. thuringiensis var. tenebrionis, was found to be expressed during vegetative growth, although at a lesser extent than during the stationary phase (95, 252, 339). Analysis of lacZ transcriptional fusions and primer extension experiments indicates that the cry3Aa promoter is weakly but significantly expressed during vegetative growth, is activated from the end of exponential growth until stage II of sporulation (about T3), and remains active until stage IV of sporulation (about T7) (10, 324). The cry3Aa promoter, although located unusually far upstream of the start codon (position −558), resembles promoters recognized by the primary sigma factor of vegetative cells, ςA (10). A similar promoter was found 542 bp upstream of the start codon of the cry3Bb gene (30). The expression of cry3Aa is not dependent on sporulation-specific sigma factors either in B. subtilis (7) or in B. thuringiensis (324). Moreover, cry3Aa expression is increased and prolonged in mutant strains unable to initiate sporulation (7, 213, 251, 324). The results indicate that cry3Aa expression is activated by a non-sporulation-dependent mechanism arising during the transition from exponential growth to the stationary phase. The positive effect of mutations preventing the initiation of sporulation suggests that there is an event during sporulation (e.g., the disappearance of ςA in the mother cell) that turns off cry3Aa expression (7, 324).
Posttranscriptional Mechanisms
The stability of mRNA is an important contributor to the high level of toxin production in B. thuringiensis. The half-life of cry mRNA, about 10 min, is at least fivefold greater than the half-life of an average bacterial mRNA (135).
Wong and Chang showed that the putative transcriptional terminator of the cry1Aa gene (a stem-loop structure) acts as a positive retroregulator (416). The fusion of a DNA fragment carrying this terminator with the 3′ end of heterologous genes increases the half-life of their transcripts two- to threefold, which in turn increases the expression of their gene products. It has been demonstrated in other systems that the processive activities of 3′-5′ exoribonucleases are impeded by 3′ stem-loop structures (for a review, see reference 279). It is likely, then, that the cry1Aa transcriptional terminator increases the cry mRNA stability by protecting it from exonucleolytic degradation from the 3′ end. Similar terminator sequences, potentially able to form stable stem-loop structures, are found downstream from various cry genes and may contribute to their high-level expression by stabilizing the transcripts. However, alternative processes could determine the rate of mRNA degradation, and the direct involvement of these sequences on mRNA stability has not been tested by deleting them from a cry gene and measuring stability of the message.
Between the cry3Aa promoter, located from positions −560 to −600, and the translational start codon is a region involved at a posttranscriptional level with the accumulation of cry3Aa mRNA as a stable transcript with a 5′ end corresponding to nucleotide position −129 (10). Deletion of 60 bp extending from nucleotide positions −189 to −129 has no detectable effect on the expression level or on the position of the 5′ end of the transcript (10). It is likely, then, that the initial transcript, begun hundreds of bases upstream, is processed posttranscriptionally.
Insertion of the cry3Aa 5′ untranslated region (extending from nucleotides −129 to −12) between the B. subtilis xylA promoter and a lacZ reporter gene increases about 10-fold both the stability of the lacZ fusion mRNA and the production of β-galactosidase (9). Deletion and mutation analysis indicate that the sequence required for the stabilizing effect is a perfect Shine-Dalgarno sequence (GAAAGGAGG) mapping at a position between −125 and −117; this sequence has been designated STAB-SD (9). The stability of the cry3Aa mRNA could result from an interaction between the 3′ end of 16S rRNA and STAB-SD. The binding of a 30S ribosomal subunit to this sequence may protect the mRNA against 5′-3′ ribonuclease activity, resulting in a stable transcript with a 5′ end at nucleotide position −129 (i.e., the limit of 30S subunit protection). Potential STAB-SD sequences are also present in similar positions upstream of the cry3Ba, cry3Bb, and cry3Ca genes (96, 200).
Posttranslational Mechanisms
The Cry proteins generally form crystalline inclusions in the mother cell compartment. Depending on their protoxin composition, the crystals have various forms: bipyramidal (Cry1), cuboidal (Cry2), flat rectangular (Cry3A), irregular (Cry3B), spherical (Cry4A and Cry4B), and rhomboidal (Cry11A). This ability of the protoxins to crystallize may decrease their susceptibility to premature proteolytic degradation. However, the crystals have to be solubilized rapidly and efficiently in the gut of insect larvae to become biologically active. The structure and the solubility characteristics of a crystal presumably depend on such factors as the secondary structure of the protoxin, the energy of the disulfide bonds, and the presence of additional B. thuringiensis-specific components.
Studies have shown that several cry1 genes cloned in E. coli (129) or B. subtilis (344) were able to direct the synthesis of biologically active inclusions, suggesting that the 130- to 140-kDa Cry1 protoxins can spontaneously form crystals. It is generally assumed that the cysteine-rich C-terminal half of the Cry1 protoxins contributes to crystal structure through the formation of disulfide bonds (39). A similar mechanism of protein self-assembly may be responsible for the crystal formation of other 130- to 140-kDa protoxins (e.g., Cry4, Cry5, and Cry7). The cysteine-rich C-terminal region is absent from the 73-kDa Cry3A protoxins. This protein forms a flat, rectangular crystal inclusion in which the polypeptides do not appear to be linked by disulfide bridges (35). Because this protein is able to form identical crystals in both B. thuringiensis and B. subtilis, it is possible that specific host factors are not required for the protein assembly. Analysis of the three-dimensional structure of the Cry3A toxin revealed the presence of four intermolecular salt bridges, which might participate in the formation of the crystal inclusion (222).
Various studies performed with E. coli and B. thuringiensis have demonstrated that crystallization of Cry2A (71 kDa) and Cyt1A (27 kDa) requires the presence of accessory proteins (for recent reviews, see references 8 and 30). These proteins may act at a posttranslational level to stabilize the nascent protoxin molecule and to facilitate crystallization. However, the precise mechanism of their role in crystal formation has not been determined.
Kostichka et al. (192) have reported that a Cry1Ia toxin could be found in the supernatant of B. thuringiensis cultures as a processed polypeptide of 60 kDa. The authors hypothesize that Cry1Ia is an exported protein and therefore interacts with the cellular protein export machinery. Such a characteristic, together with the fact that this toxin is synthesized early in sporulation (192), may have implications for the significance of these toxins in the ecology of B. thuringiensis. Similarly, the Cry16Aa toxin of C. bifermentans seems to be secreted during sporulation (23).
TOXIN STRUCTURE
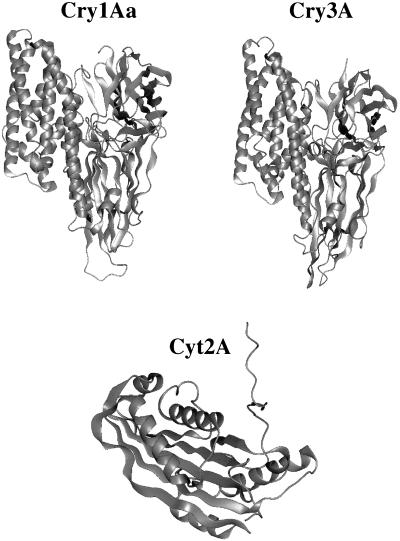
To date, the structures of three crystal proteins—Cry3A (222), Cry1Aa (148), and Cyt2A (223)—have been solved by X-ray crystallography. An analysis in the accompanying review demonstrates that Cry3A and Cry1Aa show about 36% amino acid sequence identity (79). This similarity is reflected in their three-dimensional structures; the corresponding domains can virtually be superimposed. Cyt2A, however, shows less than 20% amino acid sequence identity with Cry1Aa and Cry3A, and a similar alignment score would be obtained if the Cyt2A sequence were randomized. Not surprisingly, the Cyt2A structure is radically different from the other two structures. The structures of Cry1Aa, Cry3A, and Cyt2A are compared in Fig. 1.
FIG. 1.
Three-dimensional structures of Cry1A, Cry3A, and Cyt2A.
The Cyt toxins, unlike the Cry δ-endotoxins, are able to lyse a wide range of cell types in vitro (164). Cyt2A consists of a single domain in which two outer layers of alpha-helix wrap around a mixed beta-sheet. Cyt1A is believed to have a similar structure.
Cry3A and Cry1Aa, in contrast to Cyt2A, both possess three domains. Domain I consists of a bundle of seven antiparallel α-helices in which helix 5 is encircled by the remaining helices. Domain II consists of three antiparallel β-sheets joined in a typical “Greek key” topology, arranged in a so-called β-prism fold (330, 343). Domain III consists of two twisted, antiparallel β-sheets forming a β-sandwich with a “jelly roll” topology.
Structural and Sequence Similarities among Toxins
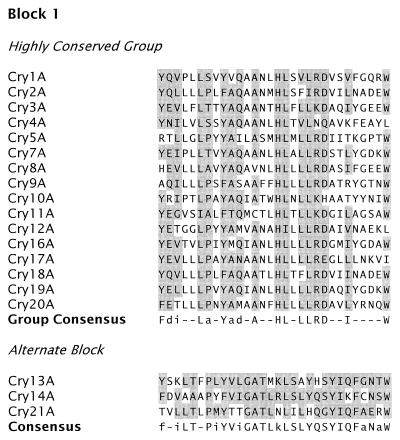
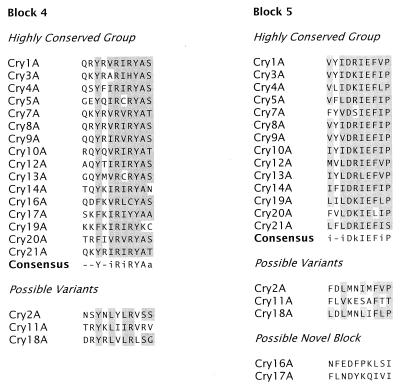
Höfte and Whiteley (164) drew attention to the five blocks of amino acids conserved among most of the Cry toxins then known. Complete amino acid sequence alignment of the Cry proteins in our data set reveals the same five tracts, or conserved blocks, in most of them (Fig. 2 and 3). Comparison of the carboxyl-terminal halves of sequences with more than 1,000 residues suggests the presence of three additional blocks lying outside the active toxic core.
FIG. 2.
Amino acid sequence blocks conserved among Cry proteins. For each block, the consensus sequence denotes the positions at which at least 75% of the aligned proteins in the group have an identical or conserved amino acid (indicated by shading). An uppercase letter within the consensus sequence indicates that at least 75% of the residues at that position are identical, while a lowercase letter indicates that at least 75% of the residues are conserved. Conserved amino acids are those that fall into the following groups: a (A, G, S, T, or P); d (D, E, N, or Q); f (F, W, or Y); i (I, L, M, or V); and k (K or R). Highly conserved sequences conform to the consensus sequence at 75% or more of its positions. Variant sequences conform to the consensus sequence of the highly conserved group at 50 to 75% of the positions. Alternate blocks are derived from groups of proteins having a consensus sequence over that sequence block that differs from the corresponding highly conserved sequence at more than half of its positions. Novel sequences have no discernible homology to a conserved block that occupies the same relative position within sequences in the conserved group.
FIG. 3.
Positions of conserved blocks among Cry proteins. The cartoon shows the sequence arrangement for each holotype toxin (e.g., Cry1Aa1) having at least one of the conserved blocks defined in the legend to Fig. 2. Sequence blocks are shown as dark gray, light gray, or white to indicate high, moderate, or low degrees of homology, respectively, to the consensus sequence for each conserved block. Variant (var) alternate (alt) are as defined in the legend to Fig. 2. The lengths of each protein and the conserved blocks within them are drawn to scale.
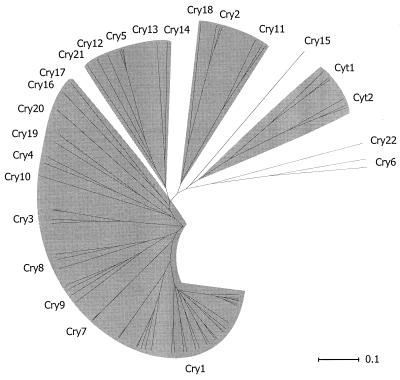
Figure 4 shows an unrooted phylogenetic tree, constructed by an unweighted pair-group method using arithmetic averages algorithm from the multiply aligned Cry and Cyt protein sequences. Five sequence similarity groups are apparent, together with a single outlying sequence (Cry15). The conserved blocks are distributed in a fashion consistent with these similarity groups. The group consisting of Cry1, Cry3, Cry4, Cry7 to Cry10, Cry16, Cry17, Cry19, and Cry20 contains all five of the core blocks. A second group consisting of Cry5, Cry12 to Cry14, and Cry21 contains recognizable homologs of blocks 1, 2, 4, and 5. Block 1 shows more variability within this second group of sequences than within the first. The proteins within this second subgroup also possess a block 2 variant; block 2 sequences show greater sequence similarity within the two groups than between them (Fig. 2). Block 3 is completely absent from this second group of Cry proteins; an unrelated sequence, highly conserved within the second subgroup but absent from the first, lies between blocks 2 and 4. For both groups, when a protein possesses the C-terminal extension, blocks 6, 7, and 8 are invariably present (Fig. 2). Members of a third sequence similarity group, composed of Cry2, Cry11, and Cry18, possess block 1 and a truncated variant of the block 2 core (Fig. 2) but lack convincing homologs of the other conserved blocks (215). An alternating arginine tract not otherwise homologous to block 4 is found near the C terminus of Cry11 and Cry18. A weak homolog of block 5 may also be present among the proteins in this group, but its significance, if any, is uncertain (Fig. 2). The other proteins in the data set—Cyt1, Cyt2, Cry6, Cry15, and Cry22—have no recognizable homologs to the conserved blocks seen in the three groups noted above.
FIG. 4.
Sequence similarity groups found among Cry and Cyt proteins. Sequences were aligned by using CLUSTAL W and a phylogenetic tree was constructed by NEIGHBOR as described in the accompanying work (79). The tree was visualized as a radial phylogram by using the TREEVIEW application. The proposed similarity groups are indicated by shading.
The conservation of blocks 1 through 5 is at least consistent with the notion that the proteins within the first subgroup, which includes Cry1 and Cry3, might adopt a similar three-domain tertiary structure. It is possible, too, that the second subgroup—Cry5, Cry12 to Cry14, and Cry21—could possess a variation of the same structural theme. The degree of sequence similarity found in the Cry2, Cry11, and Cry18 group of proteins suggests that a fold similar to that in domain I of Cry3A may be present. Indeed, the crystal structure of Cry2Aa, which has been solved but not yet published (423a), confirms this prediction. Somewhat more surprisingly, Cry2A also possesses second and third domains strikingly similar to those of Cry3A, despite the apparent absence of primary sequence homology between the two proteins over this region.
Block 1 encompasses helix 5 of domain I. As mentioned below (see “Structure-Function Interpretations”), this helix has been implicated in pore formation, a role that might explain its highly conserved nature. The central location of helix 5 within domain I also suggests an essential role in maintaining the structural integrity of the helical bundle.
Block 2 includes helix 7 of domain I and the first β-strand of domain II. These two structures comprise the region of contact between the two domains. There are three structurally equivalent salt bridges present between domain I and domain II in Cry1Aa and Cry3A (148); the residues involved lie within block 2. These interactions could be important if domain I changes its orientation relative to the rest of the molecule upon binding of the toxin to its receptor. Alternatively, the salt bridges could be responsible for maintaining the protein in a globular form during solubilization and activation.
Blocks 3, 4, and 5 each lie on one of the three buried strands within domain III. Block 3 contains the last β-strand of domain II, a structure involved in interactions between domains I and III. The central two arginines of block 4 may be involved in intermolecular salt bridges affecting crystal or oligomeric aggregation (148, 222). As Grochulski et al. have noted, however, the first and last arginines are solvent exposed (148). These residues have been implicated in channel function (68, 336, 414).
An alternative way of looking at protein families is to examine the relatedness of structural or functional segments independently (47, 378). This type of analysis helped show a correlation between domain II sequence features shared by distantly related toxins and the cross-resistance profile of a diamondback moth mutant (369).
Structure-Function Interpretations
The long hydrophobic and amphipathic helices of domain I suggest that this domain might be responsible for the formation of lytic pores in the intestinal epithelium of the target organism, one of the proposed mechanisms of Cry toxin activity (see “Mechanism of action”). Domain I bears many striking similarities to the pore-forming or membrane-translocating domains of several other bacterial protein toxins, including colicin A, diphtheria toxin, and—to a lesser extent—Pseudomonas exotoxin A (287). The pore-forming domain of colicin A consists of two central alpha-helices (α8 and α9) surrounded by eight antiparallel alpha-helices (288). Pore formation is believed to involve insertion of the hydrophobic α8-α9 helical hairpin into the membrane (101, 220). Similarly, diphtheria toxin is believed to enter the membrane via a hydrophobic helical hairpin following a pH-induced change in conformation (432). By analogy to these mechanisms, an “umbrella” model has been proposed, in which the Cry proteins also contain a hydrophobic helical hairpin (α4-α5) that initiates pore formation (222). Schwartz et al. (334) created disulfide bonds within domain I and between domains I and II in order to restrict intramolecular movements. Their results are consistent with the model described above in which helices 4 and 5 insert into the membrane while the rest of domain I flattens out on the membrane surface in an umbrella-like molten globule state. However, the lack of protein structural analysis in this work leaves open the possibility that the disulfide bonds blocked the ability of these mutant proteins to penetrate the membrane.
Similarly, little can be surmised as to the final structure of the lytic pore; a structure involving amphipathic helices (with the hydrophilic faces forming the lumen of the pore) seems the most probable. Given, however, that most domain I helices are largely amphipathic and theoretically long enough to span a membrane, little can be concluded. Even helix 2, which is split by a short nonhelical stretch, could traverse a membrane as part of a channel. Comparison of the Cry3A domain I helices with other known classes of amphipathic helices suggests that many of the helices (in particular α1, α5, and α6) show features characteristic of lytic peptides (378).
In contrast, Hodgman and Ellar (159) have proposed a “penknife” model for pore formation. In this model, based on the similarly named proposal for colicin A insertion (159), the strongly hydrophobic helices α5 and α6, which are joined by a loop at the top of the structure, open in a penknife fashion and insert into the membrane. The remainder of the molecule would remain at the membrane surface or on the receptor. Both the umbrella and penknife models are reviewed and illustrated by Knowles (185).
The surface-exposed loops at the apices of the three β-sheets of domain II, because they show similarities to immunoglobin antigen-binding sites, were initially put forward as candidates for involvement in receptor binding. Site-directed mutagenesis and segment swapping experiments, as described under “Mechanism of action,” have provided evidence in support of this model. It is interesting to note that domain II has a fold similar to that of the plant lectin jacalin (330). Jacalin is known to bind carbohydrates via the exposed loops at the apex of its β-prism fold, whereas at least one Cry protein (Cry1Ac) is believed to recognize carbohydrate moieties on its receptor (188).
The β-sandwich structure of domain III could play a number of key roles in the biochemistry of the toxin molecule. Li et al. (222) suggest that domain III functions in maintaining the structural integrity of the toxin molecule, perhaps by protecting it from proteolysis within the gut of the target organism—but of course all three domains would have to share this characteristic. From studies in other systems where toxin-receptor interaction leads to pore formation, it is known that β-strand structures can participate in receptor binding (11, 71), membrane penetration (283), and ion channel function (241, 242, 427). None of these roles has been ruled out for domain III of Cry proteins; indeed, there is at least some evidence suggesting a role for domain III in receptor binding in certain systems (see “Mechanism of action” below).
Although solving the structure of one of the Cyt toxins has not really clarified their toxic mechanism, the predominantly β-sheet structure of Cyt2A suggests a pore based on a β-barrel (223). Three of the strands are sufficiently long to span the hydrophobic core of the membrane, and the sheet formed by them shows an amphiphilic or hydrophobic character. Theoretically the number of monomers required to form a barrel of sufficient size would be four to six. Various laboratories (75, 243, 244) have observed that Cyt1A (which is believed to have a common structure with Cyt2A) aggregates on the surface of the target cell but not in solution prior to binding to the cell surface. Using synthetic peptides, Gazit et al. (125) provided further evidence that the Cyt1A toxin self-assembles within the membrane and also identified two α-helices (A and C) that appeared to be involved in both membrane interaction and intermolecular assembly. Mathematical modeling hypothesized that Cyt1A exists as a 12-toxin oligomer (243). No receptor-binding motif could be identified in the Cyt2A structure, although the use of monoclonal antibodies has identified a putative cell binding region on Cyt1A (76). Using a number of different biophysical techniques, Butko et al. (55) have also studied the interaction of Cyt1A with lipid membranes. They observed a considerable loosening of the tertiary structure of the toxin upon lipid binding but could find no evidence that the toxin actually enters the membrane. The authors suggest that Cyt1A exerts its effect via a general, detergent-like perturbation of the membrane.
MECHANISM OF ACTION
General Features
The mechanism of action of the B. thuringiensis Cry proteins involves solubilization of the crystal in the insect midgut, proteolytic processing of the protoxin by midgut proteases, binding of the Cry toxin to midgut receptors, and insertion of the toxin into the apical membrane to create ion channels or pores. Crystals are comprised of protoxins. For the protoxins to become active, a susceptible insect must eat them. For most lepidopterans, protoxins are solubilized under the alkaline conditions of the insect midgut (162). Differences in the extent of solubilization sometimes explain differences in the degree of toxicity among Cry proteins (18, 98). A reduction in solubility is speculated to be one potential mechanism for insect resistance (265). For at least one protein, Cry3A, nicking by chymotrypsin-like enzymes in the midgut may be necessary for solubilization (60).
After solubilization, many protoxins must be processed by insect midgut proteases (203, 379) to become activated toxins. The major proteases of the lepidopteran insect midgut are trypsin-like (204, 270) or chymotrypsin-like (174, 280, 297). The Cry1A protoxins are digested to a 65-kDa toxin protein in a processive manner starting at the C terminus and proceeding toward the 55- to 65-kDa toxic core (69, 73). The carboxy-terminal end of the protoxin, which initially appears to be wound around the toxin in an escargot-like manner, is clipped off processively in 10-kDa sections during processing of the protoxin (74). An interesting and unexpected finding is that DNA is intimately associated with the crystal and appears to play a role in proteolytic processing (38, 76a). The mature Cry1A toxin is cleaved at R28 at the amino-terminal end (277); Cry1Ac, at least, is cleaved at K623 on the carboxy-terminal end (37). Two stages of processing have been detected for Cry1Ia with trypsin or Ostrinia nubilalis midgut proteases: a fully toxic intermediate, with an N terminus at protoxin residue 45 and a C terminus at residue 655 or 659, is further processed to a partially toxic core, with an N terminus clipped to residue 156 (340).
Activated Cry toxins have two known functions, receptor binding and ion channel activity. The activated toxin binds readily to specific receptors on the apical brush border of the midgut microvillae of susceptible insects (161–163). Binding is a two-stage process involving reversible (161, 162) and irreversible (166, 307, 395) steps. The latter steps may involve a tight binding between the toxin and receptor, insertion of the toxin into the apical membrane, or both. It has been generally assumed that irreversible binding is exclusively associated with membrane insertion (166, 307, 395). Certainly the recent report that truncated Cry1Ab molecules containing only domains II and III can still bind to midgut receptors, but only reversibly, supports the notion that irreversible binding requires the insertion of domain I (116). Yet at least some published data is consistent with the notion of tight binding to purified receptors. Tight binding of Cry1Aa and Cry1Ab to purified Manduca sexta aminopeptidase N (APN) has been observed (256), and Cry1Ac may also show some degree of irreversible binding to M. sexta APN. There are likewise indications of irreversible binding for Cry1Ac to purified Lymantria dispar APN (172, 389). Finally, Vadlamudi et al. (385) calculated similar binding constants when toxin bound to brush border membrane vesicles (BBMV) and to nitrocellulose-immobilized receptor (i.e., a ligand blot).
In M. sexta, the Cry1Ab receptor is believed to be a cadherin-like 210-kDa membrane protein (119, 180, 385), while the Cry1Ac and Cry1C receptors have been identified as APN proteins with molecular masses of 120 and 106 kDa, respectively (183, 234, 329). Incorporation of purified 120-kDa APN into planar lipid bilayers catalyzed channel formation by Cry1Aa, Cry1Ac, and Cry1C (335). These receptor assignments can be difficult to reconcile with some ligand blot binding data, however (90, 208). There is also some evidence that domain II from either Cry1Ab or Cry1Ac can promote binding to the larger protein, while domain III of Cry1Ac promotes binding to the presumed APN (91). Alkaline phosphatase has also been proposed to be a Cry1Ac receptor (329). The recent cloning of the putative 210-kDa (386) and 120-kDa (184) Cry1Ac receptors opens exciting possibilities for studies on toxin-receptor interactions. In Heliothis virescens, three aminopeptidases bound to Cry1Ac on toxin affinity columns. One of them, a 170-kDa APN, bound Cry1Aa, Cry1Ab, and Cry1Ac, but not Cry1C or Cry1E. N-Acetylgalactosamine inhibited the binding of Cry1Ac but not that of Cry1Aa or Cry1Ab. The three Cry1A toxins each recognized a high-affinity and a low-affinity binding site on this 170-kDa APN (235). In gypsy moth (L. dispar), the Cry1Ac receptor also seems to be APN, while Cry1Aa and Cry1Ab bind to a 210-kDa brush border membrane vesicle (BBMV) protein (388, 389). In Plutella xylostella (236) and Bombyx mori (425) as well, APN appears to function as a Cry1Ac binding protein. An M. sexta gene encoding a Cry1Ab-binding APN has also been cloned, as has its P. xylostella homolog (92).
Insertion into the apical membrane of the columnar epithelial cells follows the initial receptor-mediated binding, rendering the toxin insensitive to proteases and monoclonal antibodies (415) and inducing ion channels or nonspecific pores in the target membrane. In vitro electrophysiological studies of voltage-clamping of lipid bilayers (338, 348) and sections of whole insect midguts (67, 68, 153, 225, 307) support the functional role of the toxin in pore or ion channel formation. The nature of the ion channel or pore-forming activity of Cry toxins in the insect is still controversial. It is alternatively described as a large lytic pore that is not specific for particular ions (see reference 187 and “Structure-function interpretations”) or as an ion-specific channel that disrupts the membrane potential but does not necessarily lyse midgut epithelial cells (see below).
Several recent reviews have considered the mechanism or mode of action of Cry toxins (126, 134, 158, 185, 186, 378, 412, 424). Some of these reviews have presented models for the mode of action. The present review considers the newest primary data on receptor binding and ion channel activity and critically evaluates the extant models.
General Receptor Binding and Kinetic Considerations
Soon after methods were developed for preparing insect BBMV (411), BBMV became the subjects of toxin binding studies (323, 413). Several groups were able to correlate a toxin’s insect specificity with its affinity for specific receptors on BBMV of susceptible insects (162, 163, 395). In vivo experiments have also confirmed that Cry proteins bind to microvillae in the midgut (49, 93, 426).
A set of in vitro-constructed reciprocal recombinants between Cry1Aa and Cry1Ac (130, 131) provided evidence that insect specificity was localized in the central domain of the toxin for some insects (B. mori and Trichoplusia ni) and the central and C-terminal domains for others (H. virescens). Visser et al. (397) reviewed the use of domain substitutions to locate specificity regions. Van Rie et al. (395) demonstrated that receptor binding correlated with insect specificity, and Lee et al. (209) demonstrated that the specificity and binding domains were colinear for Cry1Aa against B. mori. Examination of the crystal structure of Cry3A (222) suggested a physical basis for receptor binding (see “Toxin structure,” above) by the loops of domain II. This suggestion has now been substantiated by site-directed mutagenesis.
Early work by Hoffman et al. (162), Van Rie et al. (395), and others employed competition binding studies to demonstrate a correlation between toxin affinity and insecticidal activity. In a paradoxical finding, however, Wolfersberger (413) observed that Cry1Ab was more active than Cry1Ac against gypsy moth larvae, despite exhibiting a relatively weaker binding affinity. Other examples of this phenomenon—a lack of correlation between receptor binding affinity and insecticidal activity—are now known (123, 327, 395). Liang et al. (224) evaluated binding affinity and dissociation (both reversible and irreversible binding) of Cry1Aa, Cry1Ab, and Cry1Ac with gypsy moth BBMV. While they confirmed that the affinity of Cry1Ab was not directly related to toxin activity, they did observe a direct correlation between the irreversible binding rate and toxicity. Ihara et al. had earlier stressed the importance of considering irreversible binding in explaining the difference in toxicity of Cry1Aa and Cry1Ab to B. mori (166).
Prior to the work of Liang et al. (224), kinetic analysis of Cry toxin-receptor binding relied on the Hill (161) or Scatchard (395) equations that assume a strictly reversible binding:
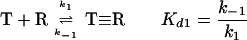 |
1 |
where T is a Cry toxin, R is a receptor for this toxin, T≡R is a toxin that is reversibly bound to the receptor, Kd1 is the dissociation constant k1 is the on rate, and k−1 is the off rate.
In reality, the toxin becomes irreversibly associated with the apical membrane by insertion (415), giving the following kinetic diagram (224) (including two models for the inserted state of the toxin):
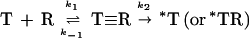 |
2 |
where T, R, and T≡R are as described for equation 1; *T is an irreversibly bound toxin, presumably inserted into the membrane but not associated with a receptor; and *TR is an irreversibly bound toxin which is still associated with a receptor.
Given the irreversible rate component k2, the reaction cannot reach equilibrium; as the toxin-receptor complex is formed, it is drained away by insertion. Therefore, competition or binding experiments under conditions where insertion can take place (equation 2) do not yield true Kd values (224). Since equilibrium conditions are not obtained, equation 2 should not be considered any more valid for calculation of a classical dissociation constant, Kd, than equation 1. Alternate values, such as the 50% inhibitory concentration (224, 257) or Kcom, the so-called competition constant (206, 208, 308, 422), have been used for Kd under these conditions. Under some conditions insertion should not occur, i.e., ligand blotting of 125I-labeled Cry1Ac to purified gypsy moth 120-kDa receptor (207) or binding of unlabeled Cry1Ac to purified M. sexta 120-kDa receptor fixed to dextran surfaces in surface plasmon resonance analysis (256). In both cases, the calculated Kd was 100 times that obtained with BBMV, suggesting that the effect of k2 upon the reversible reaction is considerable. In contrast, competition binding of Cry1Ab to the 210-kDa receptor on a ligand blot differed little from calculated competition binding to M. sexta BBMV (385) or to the cloned 210-kDa receptor expressed in human embryonic 293 cells (386) (708 pM, 1,000 pM, and 1,015 pM, respectively). It may be that the rate of insertion, k2, is negligible for the 210-kDa receptor, perhaps due to either extremely tight binding to this receptor or a failure to insert.
Role of Domain II Loop Regions
The prediction that domain II is involved in receptor binding (131, 222) has led to extensive substitution of loop residues in this domain in Cry3A, Cry1A, and Cry1C by mutagenesis (Fig. 5). Data on the effects of mutations in sequences encoding domain II loop regions of selected Cry toxins are summarized in Table 1. Perusal of these data indicates that mutations may have either a negative or positive effect on binding and toxicity and that mutations in different loop regions, sometimes involving the same type of amino acid residue, can have a different effect on binding. Minor changes in binding usually do not have a major effect on toxicity, but a major positive or negative effect has a corresponding positive or negative effect on toxicity. Furthermore, either binding affinity (as measured by competition binding) or irreversible binding may effect toxicity, and for a few mutant proteins one of these parameters may be positive (increased affinity) while the other may be negative (increased dissociation), with an overall negative effect on toxicity. It is apparent that the same mutation in a toxin can have quite different results on different insects. A more complete description of domain II loop mutations is given in a recent review (311).
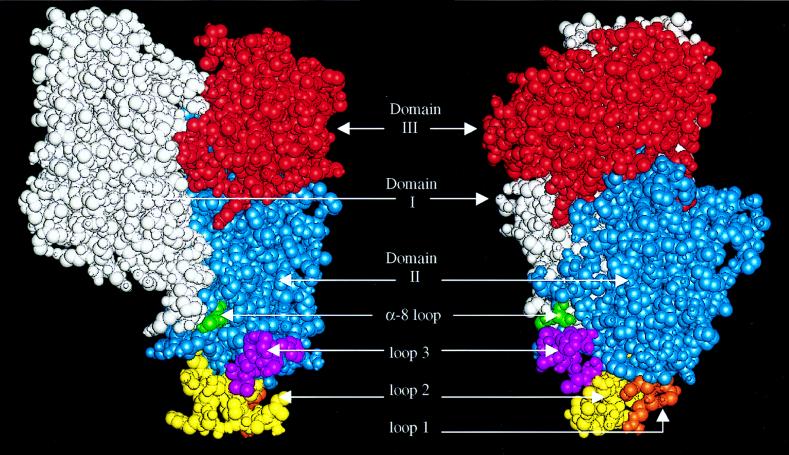
FIG. 5.
Predicted three-dimensional structure of Cry1Ab highlighting the domain II residues shown by mutagenesis to be involved in receptor binding. Domains I (white), II (blue), and III (red) and portions of loops 1 (orange), 2 (yellow), and 3 (violet) and the α8 loop (green) are shown as space-filling molecular structures in the standard presentation (left) and rotated 90° (right).
TABLE 1.
Effects of mutations in and around domain II loops of selected Cry toxins
| Gene | Loop | Sequence | Mutation | Residue(s) | Effect on bindinga,b
|
Toxicity (fold)b | Insect | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Competition | Irreversible | ||||||||
| cry1Aa | 1c | FNY | AAA | 313–315 | None | −2× | Lower | B. mori | 197 |
| cry1Aa | 2 | LYRRIIL | Δd | 365–371 | −10× | ND | −1,000 | B. mori | 233 |
| cry1Aa | 2 | LYRRIIL | AAAAAAA | 365–371 | −10× | ND | −1,000 | B. mori | 233 |
| cry1Abe | α8 | AL | GS | 282–283 | None | None | None | M. sexta | 211 |
| cry1Ab | α8 | AL | GS | 282–283 | +10× | ND | +7 | L. dispar | 211 |
| cry1Ab | 2 | RRP | AAA | 368–370 | No binding | ND | −667 | M. sexta | 309 |
| cry1Ab | 2 | RRP | AAA | 368–370 | No binding | ND | −36 | H. virescens | 309 |
| cry1Ab | 2 | PFNIGI | Δ | 370–375 | None | −30% | −600 | M. sexta | 307 |
| cry1Ab | 2 | PFNIGI | Δ | 370–375 | None | −30% | −54.6 | H. virescens | 309 |
| cry1Ab | 2 | F | A | 371 | −2× | −35% | −600 | M. sexta | 307 |
| cry1Ab | 2 | F | A | 371 | +0.7× | ND | +1.6 | H. virescens | 309 |
| cry1Ab | 2 | F | C | 371 | None | −35% | −600 | M. sexta | 309 |
| cry1Ab | 2 | F | V | 371 | None | −30% | −400 | M. sexta | 309 |
| cry1Ab | 2 | F | S | 371 | None | −20% | −40 | M. sexta | 309 |
| cry1Ab | 2 | F | L | 371 | None | −10% | −10 | M. sexta | 309 |
| cry1Ab | 2 | F | Y | 371 | None | −5% | −6 | M. sexta | 309 |
| cry1Ab | 2 | F | W | 371 | None | None | None | M. sexta | 309 |
| cry1Ab | 2 | N | A | 372 | −2× | −20% | −2 | M. sexta | 309 |
| cry1Ab | 2 | N | A | 372 | −1.3× | ND | −1.6 | H. virescens | 309 |
| cry1Ab | 2 | N | A | 372 | +4.4× | None | +8.5 | L. dispar | 308 |
| cry1Ab | 2 | N | G | 372 | +4.4× | None | +8.5 | L. dispar | 308 |
| cry1Ab | 2 | G | A | 374 | −2× | −20× | −348 | M. sexta | 307 |
| cry1Ab | 2 | G | A | 374 | −5.7× | ND | −8.7 | H. virescens | 309 |
| cry1Ab | 2 | I | A | 375 | None | −5% | −2.4 | M. sexta | 307 |
| cry1Ab | 2 | I | A | 375 | −3.6× | ND | −4.9 | H. virescens | 309 |
| cry1Ab | 3 | S | A | 438 | −1.5× | None | −4.7 | M. sexta | 310 |
| cry1Ab | 3 | G | A | 439 | −11.7× | None | −103 | M. sexta | 310 |
| cry1Ab | 3 | F | A | 440 | −8.9× | None | −19.6 | M. sexta | 310 |
| cry1Ab | 3 | S | A | 441 | None | None | None | M. sexta | 310 |
| cry1Ab | 3 | N | A | 442 | −1.6× | None | −3.8 | M. sexta | 310 |
| cry1Ab | 3 | S | A | 443 | −1.5× | None | −4.5 | M. sexta | 310 |
| cry1Ac | 1 | GYY | VSF | 312–314 | −0.7× | −7.7 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | VYF | 312–314 | −1.8× | −6.2 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | VSY | 312–314 | +4.0× | −1.2 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | GYS | 312–314 | +2.6× | −1.2 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | GYF | 312–314 | +3.4× | −1.1 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | AYY | 312–314 | +2.2× | −1.1 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | ASY | 312–314 | −2.2× | −1.0 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | GSY | 312–314 | −1.4× | −1.0 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | GFS | 312–314 | −1.2× | −1.0 | M. sexta | 349 | |
| cry1Ac | 1 | GYY | GFF | 312–314 | None | None | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | YRIP | 367–370 | −10.8× | −7.4 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | YKKA | 367–370 | −7.7× | −2.2 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | SKRP | 367–370 | −3.5× | −2.4 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | FIRP | 367–370 | −1.2× | −6.5 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | YTRP | 367–370 | −1.1× | −5.8 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | YRRA | 367–370 | −1.1× | −2.1 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | FKRA | 367–370 | None | None | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | YRKP | 367–370 | None | +1.8 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | FKRA | 367–370 | None | −1.3 | M. sexta | 349 | |
| cry1Ac | 2 | YRRP | FKRA | 367–370 | None | +2.0 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SDFS | 438–441 | −14.8× | >1,000 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | IVFS | 438–441 | −14.3× | >1,000 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SVFI | 439–441 | −8.2× | >1,000 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SVFS | 440–441 | −12.9× | −33.3 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SAFS | 438–441 | −12.3× | −33.3 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | TASS | 439–441 | −4.8× | −33.3 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SAYS | 440–441 | −3.5× | −33.3 | M. sexta | 349 | |
| cry1Ac | 3 | SGFS | SASS | 439–441 | −5.7× | −11.1 | M. sexta | 349 | |
| Competition | Irreversible | ||||||||
| cry1Ac | 3 | SGFS | NGYI | 440–441 | −2.8× | None | −11.1 | M. sexta | 349 |
| cry1Ac | 3 | SGFS | IGFI | 438–441 | −3.4× | None | −6.3 | M. sexta | 349 |
| cry1Ac | 3 | SGFS | TGYS | 439–441 | −1.9× | None | −2.6 | M. sexta | 349 |
| cry1Ac | 3 | SGFSS | IGFS | 440–441 | None | None | −2.6 | M. sexta | 349 |
| cry1Ac | 3 | SGFS | SGSS | 438–441 | None | None | −1.0 | M. sexta | 349 |
| cry1Ac | 3 | SGFS | SGFT | 439–441 | +9.6× | None | None | M. sexta | 349 |
| cry1Ac | 3 | SGFS | SGYS | 440–441 | −1.5× | None | None | M. sexta | 349 |
| cry3A | 1 | YYGND | AAAAA | 350–354 | −9× | ND | None | T. molitor | 422 |
| cry3A | 2 | PS | AA | 412–413 | None | ND | None | T. molitor | 422 |
| cry3A | 3 | MQGSRG | AAAAAA | 481–486 | −4× | +20% | +2.4 | T. molitor | 422 |
Competition results are given as fold values, while irreversible binding results are given as fold values or percentages.
Values with minus signs are decreases, while values with plus signs are increases.
Sequences around, not in, loop 1.
Δ, deletion of sequences.
Cry1Ab and Cry1Ac loops predicted from alignment with Cry1Aa.
In summary, the binding picture for domain II is complex. Results clearly suggest that all of the loops of domain II can participate in receptor binding, although perhaps not all at the same time for a given insect or receptor. Different toxins may have the same amino acid sequence in the loops of domain II (e.g., Cry1Ab and Cry1Ac) yet bind to different receptors, at least on ligand blots. The available data seem to show an intriguing similarity between the receptor binding loops of domain II and other known protein-protein epitopes; i.e., a hydrophobic residue capable of tight binding to the receptor is surrounded by hydrophobic or charged residues. Similar interactions have been noted in several other systems (for a general review, see reference 300). A striking demonstration of the importance of a hydrophobic residue in irreversible binding was a series of mutations in F371 of Cry1Ab loop 2 to residues of lower hydrophobicity. This reduction in hydrophobicity was correlated with the gradient of reduced irreversible binding and toxicity (309).
Not included above is a discussion of work on two putative surface loops of domain II of Cry1C (loop 1, 317GRNF320, and loop 2, 374QPWP377) (350). This study did not evaluate the effect of mutational alteration of loop residues on binding, but examined cytotoxicity with cultured Spodoptera Sf9 cells and toxicity with Aedes aegypti larvae. The results indicated that specificity differences for Cry1C between Sf9 cells and A. aegypti larvae could be changed radically by single point mutations in the loops. For example, an R-to-I mutation at position 318 (R318I) abolished mosquitocidal activity but retained 80% cytotoxicity to Sf9 cells. Likewise, several mutations caused a loss of mosquitocidal activity with only a marginal loss of cytolytic activity against Sf9 cells. Substitutions that altered the charge, such as Q374E, completely abolished activity against both cells and mosquito larvae.
Role of Domain III in Receptor Binding
Domain III has also been implicated in receptor binding. As mentioned above, several groups (130, 331) have suggested a role for domain III of Cry1Ac in H. virescens specificity. Masson et al. (258) extended the suggestion to include CF-1 cells. Aronson et al. (19) mutated a hypervariable region of domain III (residues 500 to 509) of Cry1Ac. Mutations S503A and S504A resulted in lower toxicity to M. sexta, with a corresponding decrease in binding to BBMV proteins on ligand blots. Lee et al. (211) analyzed homolog scanning mutants that exchanged domain III between Cry1Aa and Cry1Ac. Hybrid proteins containing the Cry1Aa domain III bound a 210-kDa receptor while hybrid proteins containing the Cry1Ac domain III bound a 120-kDa receptor in gypsy moth. Domain switching experiments have also suggested a role for Cry1Ab domain III in binding to S. exigua (90). Finally, there is one report suggesting a biotin-binding activity for domain III (99), although a role for this activity in receptor binding has not been demonstrated directly.
Membrane Insertion
Mutations in domain I have been shown to affect the ability of the toxin to dissociate from the binding complex. Wu and Aronson (419) created several mutations in domain I of Cry1Ac. The A92D and R93G mutations (at the base of α3) dramatically reduced toxicity to M. sexta. A loss of toxicity by the A92D mutation was also observed in Cry1Aa and Cry1Ab. A series of substitution residues at the 92 and 93 positions revealed that at position 92 only a negatively charged residue caused a loss of toxicity. Any substitution of R93 except the positively charged Lys caused a loss of toxicity. The authors concluded that a positively charged surface is important for toxicity. Chen et al. (67) repeated the mutation at the A92 position in Cry1Ab with A92E. In agreement with Wu and Aronson’s result (419), toxicity was almost completely lost. Although competition binding of the mutant toxin to M. sexta was not affected, irreversible binding was severely disrupted. Chen et al. (67) further demonstrated that Y153 mutations (at the loop between the bottoms of α4 and α5, on the same surface as A92E) introducing a negative charge had a negative effect on membrane insertion.
In summary, binding studies reveal three types of mutants. Certain mutations in domain II (A mutants) affect competition but not dissociation. Examples are Cry1Ab 368RRP370 (309) and Cry1Ab loop 3 mutations F440A and G439A (310). Certain other mutations in domain II (B mutants) affect dissociation but not competition. Examples are Cry1Ab F371A (and most other substitutions except Trp) and G439A (307). In domain I, certain mutations (C mutants) affect insertion of toxin into the membrane. The distinction between B and C mutants may be arbitrary; it assumes different functions for domains I and II, a point still lacking definitive proof. Examples of C mutants are Cry1Ac A92D or R93G (419) and Cry1Ab A92E or Y153D (67). In the above cases, all of these effects were observed in the same toxin (Cry1Ab) and insect (M. sexta) system. Cry3A loop 3 mutants have also been described in which effects on both competition and dissociation were observed (422).
Masson et al. (256) describe differences in off rates for two Cry1Ac toxins that differ in three residues: L366F, F439S, and a deletion of D442. While these differences might be due to other causes, it is interesting that position 366 and positions 439 to 442 occur in loops 2 and 3, respectively. Wells (402) describes human growth hormone mutants in which alanine substitution of positively charged residues affects on rates, and other alanine-scanning mutants in large hydrophobic residues affect off rates. A similar pattern is observed in the Cry toxin mutations of the receptor binding loops. Positive residues may be involved in long-range orientation of the toxin to the receptor, affecting the on rate. In some cases, large hydrophobic residues were involved in tight binding, and their mutants affected the off rate; in other cases, mutations in large hydrophobic residues affected competition binding (that is, on rates).
Ion Channel Activity
The ion channel activity of Cry toxins has been explored by a wide variety of techniques. The toxin has been studied with complete proteins, with domain I in isolation, with synthetic peptides mimicking particular α-helices, and with mutants that disrupt ion channel function.
Considerable work has been reported on the effects of Cry toxins on insect tissue culture cells. Work with CF-1 cells has led to the colloidal osmotic lysis model for the cytolytic activity of Cry toxins (187). This model proposes that an influx of water, along with ions, results in cell swelling and eventually lysis. When exposed to microgram amounts of activated toxin, cells leaked a variety of electrolytes tested, including CrO42−, uridine, and Rb+. Under these conditions, then, Cry toxins form a nonspecific pore. Wolfersberger (412) lists the problems that arise from experiments with established cell cultures. The cells are normally maintained at a pH of 6.8—not the basic pH found in the lumen of many insect midguts. They lack normal midgut receptors (161) and do not respond as specifically to toxins as does the whole insect (410). They are tolerant to nearly 1,000-fold-greater levels of toxin than insects under physiological conditions (187). From experiments on tissue culture cells it is clear, however, that Cry toxins have a fairly general capacity to insert into membranes and form large, nonspecific pores under certain conditions, including high-toxin concentrations, long incubation times, and relatively low pHs.
Several techniques have been employed to study the ion channel activity of the B. thuringiensis Cry proteins. Harvey and Wolfersberger (153) used electrophysiological analysis of sections of whole midgut of M. sexta to measure short circuit current inhibition (ISC). The mechanism of ISC is explained in the excellent review by Wolfersberger (412). Results of recent studies (67, 68), using nanomolar concentrations of toxin, have supported the validity of the voltage clamping technique as an assessment of Cry toxin activity correlating well with bioassays.
Several groups have examined Cry toxin ion channel activity in planar lipid bilayer (PLB) systems. Slatin et al. (348) examined Cry1Ac and Cry3A in PLB membranes of various compositions and found that toxins formed cation-selective channels. Cry1Ac ion channels exhibited multiple opening and closing states (indicating more than one single-channel conductance level or cooperative gating). Cry1Ac channels were commonly 600 pS in size (in 300 mM KCl), while Cry3A formed larger channels of 4,000 pS. Channels did not form at pH 7 but did form at pH 9.7.
In a pivotal paper on Cry protein ion channel activity, Schwartz et al. (338) reported a pH effect on the type and size of ion channels made by Cry1C in PLBs. Under alkaline conditions (pH 9.5), cationic channels of 100 to 200 pS were formed, exhibiting multiple conductance states. Under acidic conditions (pH 6.0), anionic channels of different sizes (8 to 120 pS) were observed. These channels were inhibited by zinc added to the cis chamber, but not to the trans chamber, indicating directionality of the channel. The authors note that behavior of the toxins at pH 6 is similar to that recorded in native membranes of cultured insect cells (grown at pH 6.3) (337). This observation may clarify the nonselectivity of Cry proteins on cultured insect cells (187). The physical basis of pH-dependent selectivity may be related to the observation that α-helical content, as measured by circular dichroism, changes radically with pH (72, 111, 189). It is speculated that pH can alter the pitch or arrangement of the α-helices of domain I and change the nature of the ion channel. In general, the role of pH in ion specificity is thought to be by titration of charged amino acids lining the aqueous pore, but pH changes on Cry channels have global effects on ion specificity and pore size.
Channel formation in PLBs has also been observed with N-terminal fragments (essentially domain I) of Cry1Ac (399) and Cry3Bb (398), and with α5 helix peptides of Cry1Ac (80) and Cry3A (127, 128). The α7 helix alone did not form channels, but in the presence of the α5 helix it assembled and penetrated membranes better than did α5 complexes alone (126). Channels formed by the α5 helix, unlike those formed by full-length toxins, are small (60 pS) and hemolytic (127) and prefer acidic phospholipid vesicles (80, 127). The channels formed with Cry1Ac N-terminal fragments differed from those formed by whole toxins in having only a single conductance state, being less cation selective, and showing no toxicity to whole insects. They did, however, have similar conductance levels (200 to 600 pS). They also exhibited twice the Rb+ efflux from phospholipid vesicles as did full-length toxins (399). In contrast, N-terminal fragments of Cry3Bb were quantitatively similar to the full-length toxin, but exhibited less Rb+ efflux than full-length toxins with phospholipid vesicles. In summary, these results show qualitative support for the model that domain I constitutes, or at least participates in, the ion channel.
Domain III has also been reported to play a role in ion channel activity. Chen et al. (68) analyzed an alternating arginine region in β-sheet 17 (conserved block 4), a sequence superficially similar to the positively charged face on the S-4 helix in classical ion channels. While alteration of the central arginines caused structural alterations in Cry1Aa, conservative substitutions of the outermost arginines were stable and led to reduction of activity, as measured by bioassays and by voltage clamping of M. sexta midgut sections. These altered toxins were also examined by the BBMV permeability-light scattering assay (414) and in lipid bilayers for conductance (336). Both methods detect an alteration of ion channel activity caused by these conservative alterations in this β-sheet of domain III.
Reconstitution systems involving BBMV fused with lipid bilayers have been recently reported from two laboratories. Martin and Wolfersberger (254) measured Cry1Ac channels in PLBs that were fused with M. sexta BBMV. The addition of 1.5 nM of toxin resulted in very large channels (>260 nS) at pH 9.6. The smallest toxin-dependent increase in conductance was 13 nS, which may represent a single membrane pore. Thus, these channels were capable of very large changes in conductance state (in 13-nS increments) but were never observed to close. Channel behavior was also pH dependent. At pH 8.8, smaller channels of 2 to 3 nS were observed. The authors concluded that pores of the largest size would be 2.2 nm in diameter (more than twice the diameter previously measured in bilayers), and that such differences in properties favor active involvement of BBMV proteins in the pore formation. More recently Carroll and Ellar (62) measured the size changes of M. sexta BBMV in an environment of high osmotic pressure and high Cry1Ac concentrations. The rate of Cry1Ac-induced swelling varied with the radius of the solutes used, allowing for an estimate of Cry1Ac pore size. Under these conditions, large pores were formed (2.4 nm at pH 8.7 and 2.6 nm at pH 9.8).
Lorence et al. (230) also have reported intrinsic ion channels in S. frugiperda BBMV. These cationic channels were small (31, 47, and 76 pS), of low selectivity (permeability relative to K+ is >80% for Na+, Li+, Cs+, Rb+, and NH4+), and were inhibited by standard channel blockers. The addition of Cry1C or Cry1D toxin resulted in large cationic channels of 50, 106, and 360 pS that showed greater K+ selectivity but were not exclusively K+ channels. The Cry1D channels formed in whole S. frugiperda BBMV were reported to be blocked by Ba+ and Ca2+ and less so by triethanolamine, in agreement with an earlier report on the blocking of inhibition of ISC on M. sexta midguts (77). These experiments were performed at pH 9.0; no anionic channels were observed under these conditions. The latter result differs from light scattering results from M. sexta BBMV with Cry1Ac at pH 7.5 (61). Interestingly, while the insecticidal activity against first-instar S. frugiperda for Cry1C was greater than that for Cry1D, the channel-forming activities for Cry1C and Cry1D on BBMV taken from second-instar larvae were equal and that for Cry1C was less than that for Cry1D on BBMV from fifth-instar larvae. Clearly the fused BBMV-lipid bilayer studies raise interesting questions and open new avenues for understanding Cry toxin action.
Mutants with Enhanced Activity
A primary goal of protein engineering of the Cry proteins is to create better pesticides through rational design. A few examples of this effort are now starting to appear. A mutation (H168R) in helix α5 of Cry1Ac, domain I, caused a twofold increase in toxicity against M. sexta (419). Further characterization of this mutant (165) revealed that the increased toxicity was correlated with the rate of irreversible binding (kobs). Jellis et al. (171) have also described multiple mutations in domain I that increased toxicity; however, the mechanism of action of these mutants has not been addressed. An R204A mutation in domain I of Cry4B resulted in a threefold increase in activity against mosquitoes, perhaps by removing a site of proteolytic instability (16).
Several mutations in domain II have led to increased toxicity. Loop 3 (481MQGSRG486) of domain II Cry3A was mutated to alanines, and a 2.4-fold increase in toxicity against Tenebrio molitor was observed (422). An increase in irreversible binding was correlated with this increase in toxicity. Other mutations in loop 1 of Cry3A have significantly improved toxicity against T. molitor (11.4-fold); Chrysomela scripta, cottonwood leaf beetle (2.5-fold); and Leptinotarsa decemlineata, Colorado potato beetle (1.9-fold) (423). An increase in irreversible binding was correlated with the increase in toxicity for these mutants as well. In Cry1Ab, a combination of mutations in the α8 loop and loop 2 resulted in a 32-fold increase in toxicity to L. dispar over the background gene product and a 4-fold improvement over the previously best-known gene product (Cry1Aa) (308). The mechanism of increase in toxicity is correlated to improvement in initial binding affinity in this case.
In summary, the B. thuringiensis Cry protein behaves as a bona fide ion channel in lipid bilayers and in the midgut epithelium. As such it represents one of the few ion channels that has a known structure. The contradictory results and confusion concerning the selectivity and size of the pore may be due to the range of experimental conditions employed but more importantly may reflect the adaptability of the toxin to different physiological conditions which exist in its functional environments. In the alkaline midgut, the toxin may function as a cation channel (338), taking advantage of the large K+ gradient that exists in some insect midgut environments. As the pH falls due to cell lysis or leakage, the toxin may function as an anion channel (338), further wounding the epithelial cells. In large amounts, the Cry protein may form very large leakage pores, resulting in cell lysis and disruption of the midgut epithelium. Continued intensive research effort, now under way, will clarify the mechanism of action of the Cry proteins.
Effect of Synergistic Interactions on Toxin Potency
B. thuringiensis subsp. israelensis.
Wu and Chang (420) were the first to observe that when protein fractions from the purified inclusion body of B. thuringiensis subsp. israelensis were mixed and assayed against A. aegypti larvae, the activity of some combinations was greater than would have been expected from the activity of the individual fractions. Other reports followed, confirming synergistic interactions among various toxins of B. thuringiensis subsp. israelensis (15, 64, 70, 78, 85, 303, 421). In evaluating these studies, it is difficult to establish the precise contribution of each toxin (either alone or in combination) towards the overall toxicity of the inclusion. Part of the problem is the large variation in reported toxicities for individual toxins, probably due to differences in experimental conditions. Complicating factors include host-dependent differences in the size, quality, and solubility of crystals among the various expression systems used (15); differences in presenting the proteins to the larvae (soluble or reprecipitated form); variation in bioassay conditions, including larval age and diet; and natural variation in insect populations (317).
A recent study (78) attempted to overcome these problems by assaying the toxins under constant experimental conditions. From these data, it can be deduced that the order of relative activities of the individual toxins against A. aegypti larvae (based on the 50% lethal concentration [LC50]) is (from greatest to least) Cry11A, Cry4B, Cry4A, and Cyt1A. Synergistic interactions were demonstrated with all combinations of toxins used, although the extent of this interaction was dependent on the combination. No combination, however, was as active as was the native B. thuringiensis var. israelensis inclusion. There might be additional factors important for toxicity associated with the native crystal. It is also possible that native crystals might be ingested or solubilized more efficiently than those from the recombinant strains are. Additionally, the presentation of all four toxins in a single crystal might be more efficient than a mixture of four inclusions.
In an alternative approach to study the relative contributions of the B. thuringiensis var. israelensis toxins to the overall toxicity, strains have been made in which either the cry11A gene or the cyt1A gene were genetically inactivated. The effect of inactivating cry11A (301) was to halve the toxicity of the resulting strain to A. aegypti larvae. In contrast, inactivating the cyt1A gene (84) produced a strain with similar toxicity to the native strain, suggesting that Cyt1A was not essential for mosquitocidal activity. In interpreting those results, however, one should keep in mind the relative activities of the individual toxins (78). If the crystals produced by the cyt1A null mutant contain relatively greater proportions of the more active toxins than those found in wild-type crystals, one would expect the mutant strain to be considerably more toxic than the wild-type strain. The fact that the presence of Cyt1A in crystals does not dilute their potency suggests that this protein is indeed an important component of the B. thuringiensis subsp. israelensis mosquitocidal arsenal. As such, Cyt1A may provide a redundant set of synergistic interactions.
Little is known about the mechanism of this synergistic interaction. A comparison of the dose-response curves for the individual B. thuringiensis subsp. israelensis toxins (78) shows a clear difference between Cyt1A and the Cry toxins. Thus, Cyt1A may act in a different way than the Cry toxins. Cyt1A has a completely different structure than the Cry toxins (223) and appears to interact with a different type of receptor (375). Ravoahangimalala and Charles (312) found that Cyt1A, when added alone to midgut tissue sections of Anopheles gambiae, bound to the microvilli of all midgut and anterior stomach cells (with the exception of the peritrophic membrane-secreting cardia cells). In contrast, the Cry toxins bound only weakly to anterior stomach cells. When the complete set of B. thuringiensis subsp. israelensis toxins were added to insects in vivo, Cyt1A was not found to be bound to the anterior stomach cells (313). Although this negative result could have been an artifact, it might also represent a strong association between the Cry and Cyt toxins that could form the basis of a synergistic interaction. An additional consequence of this synergism is discussed under “Resistance Management” below.
Much of the work discussed above was concerned with activity against A. aegypti larvae. Synergism has also been established between different toxin combinations against both Culex pipiens and Anopheles stephensi (85, 303).
Other B. thuringiensis strains.
Synergistic interactions between toxins other than those from B. thuringiensis subsp. israelensis were reported in 1991 by van Frankenhuyzen et al. (393). Interactions were observed between the individual Cry1 toxins of HD-1 against a number of forest-defoliating insects. The data presented in that report (393) were later reevaluated by Tabashnik (363), who applied a more rigorous mathematical treatment to the toxicity data and concluded that synergism could not, in fact, be satisfactorily demonstrated. Recently, however, synergism has been observed between Cry1 proteins. The relative toxicities of Cry1Aa, Cry1Ab, and Cry1Ac against L. dispar and B. mori were investigated in force-feeding experiments (207). While synergism was observed between Cry1Aa and Cry1Ac for L. dispar by using the mathematical approach of Tabashnik (363), an antagonistic effect was exhibited between Cry1Aa and Cry1Ab. No synergistic effect on B. mori was observed with any toxin combination. The authors also noted that synergistic interactions were observed both in the bioassay and in ISC. The authors speculated that the pores formed by different toxins act in a cooperative way or that a more efficient pore is formed from a hetero-oligomer of different toxins. The presence of certain toxins might enhance the activity of another by preventing nonproductive binding. Whatever the actual mechanism, it is clear that the interaction is insect specific, a fact that may reflect differences in receptor affinities for each toxin.
In addition to synergistic interactions between different toxins, similar potentiating effects on toxicity have been observed between certain toxins and spores (85, 100, 173, 271, 273, 372) and also between toxins and other bacteria (100). In each case, septicemia caused by the spores or bacteria infecting the insect, whose midgut has become ulcerated as a result of the toxin, is believed to be the cause of this observed synergism. In addition, the presence of the B. thuringiensis spore with the Cry proteins may even reduce the likelihood of insect resistance development in some instances (272).
BIOTECHNOLOGY OF B. THURINGIENSIS
Application of Cry Proteins for Pest Control and Plant Protection
B. thuringiensis is now the most widely used biologically produced pest control agent. In 1995, worldwide sales of B. thuringiensis were projected at $90 million (353), representing about 2% of the total global insecticide market (199). Rowe et al. (322) reported that the annual worldwide distribution of B. thuringiensis amounts to 2.3 × 106 kg. As of early 1998, there were nearly 200 registered B. thuringiensis products in the United States (Table 2) (381). While the use of biological pesticides in agriculture remains significantly behind that of synthetic chemical pesticides, several environmental and safety considerations favor the future development of B. thuringiensis. Cry proteins that have been studied thus far are not pathogenic to mammals, birds, amphibians, or reptiles, but are very specific to the groups of insects and invertebrate pests against which they have activity. Cry-based pesticides generally have low costs for development and registration. B. thuringiensis subsp. israelensis, for example, had a development cost estimated at 1/40 that of a comparable novel synthetic chemical pesticide (32). Finally, the mode of action for the Cry proteins differs completely from the modes of action of known synthetic chemical pesticides, making Cry proteins key components of integrated pest management strategies aimed at preserving natural enemies of pests and managing insect resistance.
TABLE 2.
Microbial pesticides registered by the U.S. Environmental Protection Agency as of 1997
| Agent | Active ingredient(s) | Crop | Yr registered | No. of products | Target pest |
|---|---|---|---|---|---|
| Bacterium | B. popilliae, B. lentimorbus | 1948 | 2 | Japanese beetle larva | |
| B. thuringiensis subsp. kurstaki | 1961 | 127 | Lepidopteran larva | ||
| B. thuringiensis subsp. israelensis | 1981 | 26 | Dipteran larva | ||
| B. thuringiensis subsp. Berliner | 1984 | 1 | Lepidopteran larva | ||
| B. thuringiensis subsp. tenebrionis | 1988 | 6 | Coleopteran larva | ||
| B. thuringiensis subsp. kurstaki EG2348 | 1989 | 4 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki EG2424 | 1989 | 1 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki EG2371 | 1990 | 3 | Lepidopteran larva | ||
| B. sphaericus | 1991 | 1 | Dipteran larva | ||
| B. thuringiensis subsp. aizawai GC-91 | 1992 | 2 | Lepidopteran larva | ||
| B. thuringiensis subsp. aizawai | 1992 | 2 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki BMP123 | 1993 | 5 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki EG7673 | 1995 | 2 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki EG7673 | 1995 | 2 | Colorado potato beetle | ||
| B. thuringiensis subsp. kurstaki EG7841 | 1996 | 1 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki EG7826 | 1996 | 3 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki M200 | 1996 | 1 | Lepidopteran larva | ||
| Nonviable microbial pesticide | |||||
| B. thuringiensis subsp. kurstaki delta-endotoxin in killed P. fluorescens | 1991 | 2 | Lepidopteran larva | ||
| B. thuringiensis subsp. san diego delta-endotoxin in killed P. fluorescens | 1991 | 1 | Coleopteran larva | ||
| B. thuringiensis Cry1Ac and Cry1C delta-endotoxin in killed P. fluorescens | 1995 | 1 | Lepidopteran larva | ||
| B. thuringiensis subsp. kurstaki Cry1C delta-endotoxin in killed P. fluorescens | 1996 | 1 | Lepidopteran larva | ||
| Plant pesticide | B. thuringiensis Cry3A delta-endotoxin | Potato | 1995 | 1 | Colorado potato beetle |
| B. thuringiensis Cry1Ab delta-endotoxin | Corn | 1995 | 2 | Lepidopteran larva | |
| B. thuringiensis Cry1Ac delta-endotoxin | Cotton | 1995 | 1 | Lepidopteran larva | |
| B. thuringiensis Cry1Ab delta-endotoxin | Corn | 1996 | 2 | Lepidopteran larva | |
| B. thuringiensis subsp. kurstaki delta-endotoxin from HD-1-derived plasmid vector pZ01502 | Corn | 1996 | 2 | Lepidopteran larva | |
| B. thuringiensis subsp. kurstaki Cry1Ac delta-endotoxin | Corn | 1997 | 1 | Lepidopteran larva |
Forestry
The transfer of emphasis to environmentally friendly pesticides that have minimal effects on natural enemies of Lepidoptera (14) has already begun in the forests of the United States, where B. thuringiensis has become the major pesticide used against the gypsy moth (239). B. thuringiensis products for the forest industry have been based primarily on B. thuringiensis HD-1 subsp. kurstaki (102), which produces Cry1Aa, Cry1Ab, Cry1Ac, and Cry2Aa toxins. The gypsy moth is by no means the only forest pest that can be controlled successfully with B. thuringiensis (392, 393). Currently targeted pests include the spruce budworm (Canada), the nun moth (Poland), the Asian gypsy moth (United States, Canada, and the Far East), the pine processionary moth (Spain and France), and the European pine shoot moth (South America) (46).
Control of Mosquitoes and Blackflies
Since its discovery in 1977 (136), B. thuringiensis subsp. israelensis has proved to be one of the most effective and potent biological pesticides (for reviews, see references 32 and 81). Its discovery came at an auspicious moment because of the mounting resistance of mosquitoes and blackflies to synthetic chemical pesticides. Five B. thuringiensis subsp. israelensis cry and cyt genes encode dipteran-active toxins: cry4A, cry4B, cry10A, cry11A, and cyt1A (cytolysin). In addition, the Cyt1A cytolysin may synergize the activity of other Cry toxins (see “Effect of synergistic interactions on toxin potency”). These five genes are all found on a large plasmid of about 72 MDa that can be transferred to other B. thuringiensis strains by a conjugation-like process (137). Interestingly, this same set of toxins has also been discovered in isolates from several other B. thuringiensis serotypes (286), suggesting that the conjugation process analyzed in the laboratory may have environmental significance for horizontal transfer of cry genes among B. thuringiensis populations.
Given the severe impact of mosquito- and blackfly-borne human diseases, there is considerable interest in identifying additional dipteran-active toxins. Mosquitocidal activity has been reported for Cry2Aa (408), Cry1Ab (150, 151), and Cry1Ca (352). The cytolytic Cyt1A and Cyt2A crystal proteins also show some degree of dipteran specificity in vivo (191). New mosquitocidal cry genes have also been recently reported (e.g., cry11B and cry16A [85]), as well as several new isolates containing uncharacterized cry genes with mosquitocidal activity (289, 306). A surprising source of additional Cry-related mosquitocidal proteins is the bacterium C. bifermentans subsp. malaysia (23, 82), the toxins of which we have designated Cry17A, Cry18A, and Cry19A in the accompanying paper (79).
Developing New Cry Biopesticides Based on B. thuringiensis
B. thuringiensis has evolved to produce large quantities of crystal proteins (for reviews, see references 8 and 30), making it a logical host for developing improved Cry biopesticides. Natural isolates of B. thuringiensis can produce several different crystal proteins, each of which may exhibit different, perhaps even undesirable, target specificity (164, 199). On the other hand, certain combinations of Cry proteins have been shown to exhibit synergistic effects (64, 78, 207, 303, 421). Accordingly, genetic manipulation of B. thuringiensis—to create combinations of genes more useful for a given purpose than those known to occur in natural isolates—may be desirable.
A conjugation-like system has been used to transfer Cry-encoding plasmids from one strain to another (137), but most cry genes are not readily transmissible by this process. Nevertheless, a number of transconjugant and naturally occurring strains producing Cry proteins distinct from those of B. thuringiensis HD-1 subsp. kurstaki, including strains of B. thuringiensis subsp. aizawai and B. thuringiensis subsp. morrisoni, have been registered with the U.S. Environmental Protection Agency.
A breakthrough development for engineering B. thuringiensis and B. cereus came in 1989 when several groups independently applied electroporation technology to transform vegetative cells with plasmid DNA (34, 42, 214, 246, 259, 333). These protocols differed in cell preparation methods, buffer components, and electric pulse parameters, but each could achieve frequencies of 102 to 105 transformants per μg of plasmid DNA with a wide variety of hosts and vectors. Macaluso and Mettus (238) added the important observation that some B. thuringiensis strains restrict methylated DNA. Plasmid DNA isolated from Bacillus megaterium or Dcm− strains of E. coli transformed B. thuringiensis with much higher frequencies than did DNA isolated from B. subtilis or Dcm+ strains of E. coli. Their data also provided evidence that several restriction systems exist within the B. thuringiensis species. The use of unmethylated DNA with the Macaluso and Mettus protocol allows transformation frequencies as high as 3 × 106 to be achieved.
A variety of shuttle vectors, some employing B. thuringiensis plasmid replicons (17, 28, 63, 122), has been used to introduce cloned cry genes into B. thuringiensis (124). Alternatively, integrational vectors have been used to insert cry genes by homologous recombination into resident plasmids (2, 219) or the chromosome (176). Plasmid vector systems employing B. thuringiensis site-specific recombination systems have been developed to construct recombinant B. thuringiensis strains for new bioinsecticide products (26, 29, 325, 326).
Homologous recombination has been used to create null mutants in vivo. Applications of this technique have included disruptions of cry and cyt genes to assess their contribution to pesticidal activity (85, 301) and inactivation of protease production genes to increase crystal production and stability (97, 370). Recent progress in understanding cry gene expression has allowed the construction of asporogenous B. thuringiensis strains that nevertheless produce crystals; these crystals remain encapsulated in the mother cell compartment (48, 213). Much remains unclear about the fate of naked Cry toxins in the environment, although they appear to be quite sensitive to degradation by natural soil microbes (404). It is a plausible but untested hypothesis that encapsulation within the mother cell can improve toxin persistence in sprayed applications.
Alternative Delivery Systems for Cry Proteins
Crystal genes were introduced into E. coli, B. subtilis, B. megaterium, and Pseudomonas fluorescens long before there was an efficient transformation system available for B. thuringiensis (for a review, see reference 124). Fermentations of recombinant pseudomonads have been used to produce concentrated aqueous biopesticide formulations consisting of Cry inclusions encapsulated in dead cells. These encapsulated forms of the Cry proteins have been reported to show improved persistence in the environment (121). Fermentations of pseudomonads producing different Cry proteins can be combined in a single formulation to expand the range of target insects controlled. The production or activity of certain Cry proteins in P. fluorescens has been improved by the use of chimeric cry genes containing a substantial portion of the Cry1Ab carboxyl-terminal region (376, 377). It is anticipated that engineered forms of the Cry proteins showing improved potency or yield, regardless of their host, will make Cry biopesticides a more attractive and practical alternative to synthetic chemical control agents.
The primary rationale for using live endophytic or epiphytic bacteria as hosts is to prolong the persistence of Cry proteins in the field by using a host that can propagate itself at the site of feeding and continue to produce crystal protein. The cry1Ac gene, for example, has been introduced into the endophytic bacterium Clavibacter xyli on an integrative plasmid (201), and the resulting recombinant strain has been used to inoculate corn for the control of European corn borer infestation (380). Endophytic isolates of B. cereus have been used as hosts for the cry2Aa gene (245), and a B. megaterium isolate that persists in the phyllosphere (43) has been used as a host for cry1A genes. Similarly, cry genes have been transferred into other plant colonizers, including Azospirillum spp., Rhizobium leguminosarum, Pseudomonas cepacia, and P. fluorescens (281, 282, 347, 361, 384). Alternative delivery systems have also been sought for the dipteran-active toxins of B. thuringiensis subsp. israelensis to increase their persistence in the aquatic feeding zone. Such hosts include Bacillus sphaericus (22, 302), Caulobacter crescentus (374), and the cyanobacteria Agmenellum quadruplicatum (359) and Synechococcus spp. (355).
Expression of B. thuringiensis cry Genes in Plants
Several cry genes have been introduced into plants, starting with tobacco (24, 387) and now including many major crop species (5, 120, 193, 278, 294, 296, 391). Because this subject has been well reviewed in recent years (107, 290), we will limit our discussion to a few important points.
When unmodified crystal protein genes are fused with expression signals used in the plant nucleus, protein production is quite poor compared to that of similar transcription units containing typical plant marker genes (390). Nucleus-directed expression of full-length unmodified genes has been reported for some plants (114, 115). However, truncation of the unmodified genes to synthesize only the toxic portion of the protein typically results in much improved, but still comparatively low, expression (24, 114, 387).
The relatively A+T-rich Bacillus DNA contains a number of sequences that could provide signals deleterious to gene expression in plants, such as splice sites, poly(A) addition sites, ATTTA sequences, mRNA degradation signals, and transcription termination sites, as well as a codon usage biased away from that used in plants. When the Bacillus sequences are extensively modified, with synonymous codons to reduce or eliminate the potentially deleterious sequences and generate a codon bias more like that of a plant, expression improves dramatically (5, 120, 193, 294, 296). In some cases, less extensive changes in the coding region have also led to fairly dramatic increases in expression (295, 390, 391). The study of van Aarssen et al. (390) is noteworthy in that it points to fortuitous splicing signals in the Bacillus coding region as being a significant barrier to expression of cry1Ab in plants. In contrast to expression from the nucleus, an unmodified cry1Ac gene was expressed at very high levels in the chloroplasts of tobacco (260).
The year 1996 marked a milestone in agricultural biotechnology: for the first time, varieties of potato, cotton, and corn containing modified cry genes were sold to growers. The production of Cry proteins in planta can offer several benefits. Because the toxins are produced continuously and apparently persist for some time in plant tissue (345, 346), fewer applications of other insecticides are needed, reducing field management costs. Like B. thuringiensis-based biopesticides, such “enhanced seed systems” are less harmful to the environment than synthetic chemical insecticides and typically do not affect beneficial (e.g., predatory and parasitic) insects. The plant delivery system also expands the range of pests targeted for control with Cry proteins, including sucking and boring insects, root-dwelling insects, and nematodes.
In addition to concerns regarding the development of natural resistance towards the B. thuringiensis toxins, the impact of gene flow to wild relatives needs to be assessed. Preliminary experiments documented the possibility of cross hybridization among members of the family Brassicaceae and an increased survivorship of Brassica napus with a B. thuringiensis transgene under certain conditions (360). From these data it could be inferred that transgenic B. napus may transfer its insecticidal B. thuringiensis gene into wild relatives (360). However, analysis with respect to the stable inheritance and expression of the insect-resistant phenotype in the offspring of any such hybrids is needed to determine the likelihood and impact of such a transfer.
Insect Resistance to B. thuringiensis Toxins
Laboratory-selected strains.
Over 500 species of insects have become resistant to one or multiple synthetic chemical insecticides (132). In the past it was hoped that insects would not develop resistance to B. thuringiensis toxins, since B. thuringiensis and insects have coevolved. Starting in the mid-1980s, however, a number of insect populations of several different species with different levels of resistance to B. thuringiensis crystal proteins were obtained by laboratory selection experiments, using either laboratory-adapted insects or insects collected from wild populations (112, 364). The degree of resistance observed in an insect population is typically expressed as the resistance ratio (number of LC50-resistant insects/number of LC50-sensitive insects), and while resistance ratios determined by different types of bioassay are correlated, they are known to give different values (293), so that some care is required in comparing results. Examples of laboratory-selected insects resistant to individual Cry toxins include the Indianmeal moth (Plodia interpunctella) (262), the almond moth (Cadra cautella) (263), the Colorado potato beetle (Leptinotarsa decemlineata) (406), the cottonwood leaf beetle (C. scripta) (25), the cabbage looper (T. ni) (106), the cotton leafworm (Spodoptera littoralis) (276), the beet armyworm (S. exigua) (272), the tobacco budworm (H. virescens) (145, 210, 362), the European corn borer (O. nubilalis) (41), and the mosquito Culex quinquefasciatus (133). Instances of resistance discussed in the text below are summarized in Table 3.
TABLE 3.
Selected list of insect species and strains with resistance to Cry toxins
| Species | Location | Environment | Generation (n)a | Selective agent(s)b | Resistancec | Resistance mechanism(s)e | Reference(s) |
|---|---|---|---|---|---|---|---|
| P. interpunctella | Oklahoma | Laboratory | 13 | Dipel | Dipel (100) | 261 | |
| 36 | Dipel (>250) | 263 | |||||
| Dipel (>30) | 396 | ||||||
| Cry1Ab (866)d | Reduced binding | 396 | |||||
| P. interpunctella | Kansas | Laboratory | B. thuringiensis subsp. entomocidus HD-198 | Cry1Ab (13) | 284, 285 | ||
| Cry1Ac (128) | Reduced protoxin activation | 284, 285 | |||||
| Cry1Ca (6) | 284, 285 | ||||||
| P. interpunctella | Kansas | Laboratory | B. thuringiensis subsp., aizawai | Cry1Aa (17) | 264 | ||
| Cry1Ab (226) | 264 | ||||||
| Cry1Ac (789) | Reduced binding | 243, 264 | |||||
| Cry1Ba (44) | 264 | ||||||
| Cry1Ca (19) | 264 | ||||||
| Cry2A (24) | 264 | ||||||
| P. xylostella | Philippines | Field | Dipel | Cry1Ab (>200) | Reduced binding | 113 | |
| Cry1Ba (2) | 113 | ||||||
| Cry1Ca (0.5) | 113 | ||||||
| P. xylostella | Philippines | Field | Dipel | Cry1Aa (1.3) | 21 | ||
| Cry1Ab (236) | Reduced binding | 21, 368 | |||||
| Cry1Ac (1) | 21 | ||||||
| P. xylostella | Hawaii | Field/laboratory | B. thuringiensis, Dipel | Cry1Ac (>59) | Reduced binding | 365 | |
| Dipel (>130) | 365 | ||||||
| Cry1Aa (>100) | 369 | ||||||
| Cry1Ab (>100) | 369 | ||||||
| Cry1Ac (>100) | 369 | ||||||
| Cry1Ba (3) | 369 | ||||||
| Cry1Bb (6) | 369 | ||||||
| Cry1Ca (2) | 369 | ||||||
| Cry1Da (3) | 369 | ||||||
| Cry1Fa (>100) | 369 | ||||||
| Cry1Ia (3) | 369 | ||||||
| Cry1Ja (>140) | 369 | ||||||
| Cry2A (6) | 369 | ||||||
| P. xylostella | Hawaii | Field/laboratory | B. thuringiensis, B. thuringiensis subsp. aizawai, Dipel | Cry1C (22) | 228 | ||
| B. thuringiensis subsp. kurstaki HD-1 (134) | 228 | ||||||
| B. thuringiensis subsp. aizawai (3) | 228 | ||||||
| P. xylostella | Florida | Field | B. thuringiensis | Javelin (1,640) | 341 | ||
| Dipel (22) | 341 | ||||||
| Cry1Aa (>200) | 372 | ||||||
| Cry1Ab (>200) | Reduced binding | 372 | |||||
| Cry1Ac (>200) | 372 | ||||||
| Cry1Ba (2.5) | 372 | ||||||
| Cry1Ca (3.4) | 372 | ||||||
| Cry1Da (1) | 372 | ||||||
| P. xylostella | Pennsylvania | Field/laboratory | B. thuringiensis subsp. kurstaki | Cry1Aa (high) | 368 | ||
| Cry1Ab (high) | Reduced binding | 368 | |||||
| Cry1Ac (high) | 368 | ||||||
| Cry1Ca (none) | 368 | ||||||
| Cry1Fa (high) | 368 | ||||||
| Cry1Ja (high) | 368 | ||||||
| H. virescens | North Carolina | Laboratory | Cry1A(c) | Cry1Aa | Reduced binding | 210 | |
| Cry1Ab (2,300) | No altered binding | 144, 210 | |||||
| Cry1Ac (10,000) | No altered binding | 144, 210 | |||||
| Cry1Fa (3,700) | 144 | ||||||
| Cry2A (25) | 144 | ||||||
| H. virescens | North Carolina | Laboratory | Cry1A(b), Dipel | Cry1Ab (71) | Slightly altered binding | 240 | |
| Cry1Ac (16) | Slightly altered binding | 240 | |||||
| Dipel (57) | 240 | ||||||
| H. virescens | North Carolina | Laboratory | 17 | Cry1A(c) | Cry1Ab (13) | Decreased protoxin activation; | 117, 145 |
| Increased toxin degradation | |||||||
| 17 | Cry1Ac (50) | No altered binding | 145 | ||||
| 17 | Cry2A (53) | 145 | |||||
| S. exigua | Alabama | Laboratory | 10 | B. thuringiensis subsp. kurstaki HD-1 | B. thuringiensis subsp. kurstaki HD-1 (1) | 272 | |
| Laboratory | 24 | Cry1Ca | Cry1Ca (100) | Reduced total binding; increased nonspecific binding | 272 | ||
| 20 | Cry1Ab (20)d | 272 | |||||
| 34 | Cry2A (73) | 272 | |||||
| 34 | Cry9Ca (12) | 272 |
When available, the number of generations of selection at which the insects were tested is given.
Some commercial formulations of B. thuringiensis were used; Dipel and Javelin are tradenames of formulations of B. thuringiensis subsp. kurstaki.
Given are the agent to which resistance was developed and (in parentheses) the resistance ratio (LC50 or 50% lethal dose LC50 [LD50] of resistant strain divided by LC50 [or LD50] of susceptible control strain).
Estimated ratio.
When available, the mechanism(s) of resistance to the particular toxin is given. In the case of binding, only the results of binding experiments to native BBMV or to tissue sections are given.
In 1985, McGaughey (262) reported that Indianmeal moth populations from grain storage bins that had been treated for 1 to 5 months with a B. thuringiensis subsp. kurstaki formulation had a small but significant increase in LC50s relative to populations in untreated bins. Laboratory experiments with colonies collected from treated bins demonstrated measurable increases in resistance after only two generations of selection. After 15 generations of selection, insects from the treated colony showed LC50s nearly 100-fold greater than those shown by control colonies. The resistance trait proved to be recessive. When selection was removed before resistance became fixed, resistance levels decreased (263). A later study determined that resistance was correlated with a 50-fold decrease in binding affinity of a receptor for the Cry1Ab protein, one of the toxins in the B. thuringiensis formulation used for selection (396). In contrast, this Cry1Ab-resistant population showed an increased susceptibility to Cry1Ca, a protein not present in the selective formulation, and a corresponding increase in binding sites on the midgut for the Cry1Ca protein.
Several additional colonies of P. interpunctella were selected for resistance to B. thuringiensis strains having, in some cases, toxin compositions different from the one described above (264). The LC50s for several toxins were determined for each colony. While resistance ratios for Cry1Ac and Cry1Ab were most dramatic (24 to >2,000), resistance ratios of >10 were also found for Cry1Aa, Cry1Ba, Cry1Ca, and Cry2Aa in some of the colonies. A high level of resistance to Cry1Ac in three of the colonies was noteworthy, because the selective B. thuringiensis strains were reported not to produce that toxin. The toxin binding characteristics of Cry1Ac to BBMV proteins and tissue sections of several of these colonies have been studied (274). Binding to an 80-kDa BBMV protein appeared unaltered in ligand blots using BBMV from sensitive and several resistant insect colonies. By contrast, the binding of fluorescein isothiocyanate-labeled Cry1Ac toxin to midgut cells from insects selected with Dipel or HD-133 was much reduced compared to results with sensitive insects. For a P. interpunctella colony under selection with B. thuringiensis subsp. entomocidus HD-198, resistance to Cry1Ac was correlated with reduced in vitro activation of Cry1Ac protoxin by midgut extracts from resistant larvae (285). Examination of midgut enzymes in protease activity blots revealed that one of the two major trypsin-like proteases found in P. interpunctella was missing in the mutant. A similar result was also observed for a colony resistant to B. thuringiensis subsp. aizawai HD-133. In genetic crosses, the protease-deficient and Cry1Ac-resistant phenotypes cosegregated as a recessive trait (284).
Colonies of H. virescens with different levels of resistance and different resistance mechanisms have also been obtained in selection experiments with B. thuringiensis strains and proteins. In an H. virescens population selected on Cry1Ab protoxin expressed by an engineered P. fluorescens strain, resistance to Cry1Ab increased to 20-fold after seven generations. Resistance further increased to 71-fold after four additional generations of selection with Dipel, a formulated B. thuringiensis product containing several crystal proteins, including Cry1Ab (362). The toxin showed a lower binding affinity to a higher number of binding sites within the insect gut, but the change in binding characteristics was considered insufficient to explain the resistance (240).
Selection of another H. virescens population with Cry1Ac protoxin as produced by a natural B. thuringiensis strain resulted in a 50-fold resistance to Cry1Ac, a 13-fold resistance to Cry1Ab, and a 53-fold resistance to Cry2Aa (145). Larvae from this population could not survive on transgenic tobacco plants with moderate (0.01%) levels of Cry1Ab (194). Altered toxin binding was not implicated as a factor in resistance, an observation that again suggests the existence of multiple resistance mechanisms.
Very high levels of resistance to Cry1Ac (over 10,000-fold) and to Cry1Ab (more than 2,000-fold) were obtained in H. virescens by selection with Cry1Ac (144). The H. virescens colony was highly cross-resistant to Cry1Aa and Cry1Fa but displayed minimal resistance to Cry1Ba and Cry1Ca. A recent study (146) showing that Cry1Fa and Cry1Ab compete for the same receptor, at least in P. xylostella, provides a plausible explanation for this observation. Larvae of this resistant H. virescens strain survived significantly better than susceptible larvae (144) on transgenic tobacco plants reported to produce levels of Cry1Ab up to 0.007% of soluble protein (400). Surprisingly, the binding of Cry1Ac (the selective toxin) and Cry1Ab was unchanged while the binding of Cry1Aa was dramatically reduced (210). It had already been demonstrated that Cry1Ac also binds to the Cry1Aa binding site in H. virescens (395). Consequently, it was proposed that the altered Cry1Aa binding site caused resistance to all three Cry1A toxins and that the additional binding sites recognized by Cry1Ab and Cry1Ac might not be involved in toxicity (210). The allele conferring most of the resistance phenotype of this strain has been mapped to a 10-centimorgan region on linkage group 9 of H. virescens at a locus termed BTR4 (155). The initial frequency of this resistance allele in wild H. virescens populations in the Southeastern United States was estimated to be between 1 in 500 and 1 in 667 (143), which is consistent with estimates based on initial populations of insects used in selection experiments (1 in 200 to 1 in 2,000) (142).
Selection experiments using Cry1Ca have generated resistant strains of Spodoptera species. An S. littoralis colony with >500-fold resistance was obtained (276). These insects were cross-resistant to Cry1Da (7-fold) and Cry1Ea (34-fold). However, their susceptibility to Cry1Fa was unchanged, consistent with the observation that Cry1Fa and Cry1Ca compete for different receptors, at least in P. xylostella (146). An analysis of the inheritance of resistance in this S. littoralis strain indicates it is partially recessive and probably multifactorial (66). Moar et al. (272) developed an S. exigua strain resistant to Cry1Ca toxin. The basis of resistance could not be entirely explained by changes in toxin binding characteristics. This insect strain was cross-resistant to Cry1Ab, Cry2Aa, Cry9C, and a Cry1Ea-Cry1Ca hybrid protein (44).
Given the multiple steps in processing the crystal to an active toxin (see “Mechanism of Action”), it is not surprising that insect populations might develop various means of resisting intoxication. It is important, however, to keep in mind that selection in the laboratory may be very different from selection that occurs in the field. Insect populations maintained in the laboratory presumably have a considerably lower level of genetic diversity than field populations. Several laboratory experiments to select for B. thuringiensis resistance in diamondback moths failed, although the diamondback moth is the only known insect reported so far to have developed resistance to B. thuringiensis in the field. It is possible that the genetic diversity of the starting populations was too narrow and thus did not include resistance alleles. In the laboratory, insect populations are genetically isolated; dilution of resistance by mating with susceptible insects, as observed in field populations, is excluded. In addition, the natural environment may contain factors affecting the viability or fecundity of resistant insects, factors excluded from the controlled environment of the laboratory. Resistance mechanisms can be associated with certain fitness costs that can be deleterious under natural conditions (383). Natural enemies, such as predators and parasites, can influence the development of resistance to B. thuringiensis by preferring either the intoxicated, susceptible or the healthy, resistant insects. In the former case, one would expect an increase in resistance development, while in the latter, natural enemies can help to retard resistance development to B. thuringiensis. Nevertheless, selection experiments in the laboratory are valuable because they reveal possible resistance mechanisms and make genetic studies of resistance possible.
Field-selected strains.
The first case of field-selected resistance to B. thuringiensis was reported from Hawaii, where populations of diamondback moth (P. xylostella) showed different levels of susceptibility to a formulated B. thuringiensis product (Dipel). Populations from heavily treated areas proved more resistant than populations treated at lower levels, with the highest level of resistance at 30-fold (365). Laboratory selection rapidly increased resistance to >1,000-fold (366). A study of the resistance mechanism showed a reduced binding of the Cry1Ac protein to gut BBMV (365). However, immunohistochemical (105) and surface plasmon resonance (257) analyses demonstrated the presence of at least some receptor molecules on the midgut of this resistant insect strain. The resistance trait is conferred largely by a single autosomal recessive locus (367, 368). This “Hawaii” resistance allele simultaneously confers cross-resistance to Cry1Aa, Cry1Ab, Cry1Ac, Cry1Fa, and Cry1Ja but not to Cry1Ba, Cry1Bb, Cry1Ca, Cry1Da, Cry1Ia, or Cry2Aa (369). At least one Cry1A-resistant diamondback moth strain has been shown to be very susceptible to Cry9C (198). The toxins in the cross-resistance group have significant amino acid sequence similarity in domain II, a region believed to be important for receptor binding in many systems (see “Mechanism of Action”). Furthermore, Cry1Aa, Cry1Ac (21), and Cry1F (146), but not Cry1B or Cry1C (113), compete for the Cry1Ab binding site in P. xylostella, observations that clearly correspond to the cross-resistance data. A phenotypically similar resistant strain collected in Pennsylvania carries a resistance allele at the same multitoxin resistance locus (368).
A P. xylostella strain collected in Florida showed very high resistance to a B. thuringiensis subsp. kurstaki formulation and low-level resistance to B. thuringiensis subsp. aizawai (341). The strain has been estimated to have >200-fold resistance to Cry1Aa, Cry1Ab, and Cry1Ac and 60-fold resistance to the HD-1 spore but near wild-type sensitivity to Cry1B, Cry1C, and Cry1D. Binding of Cry1Ab, but not Cry1B, was reduced with midgut tissue sections and native BBMV prepared from the resistant strain (372). The existence of a single-locus resistance allele with autosomal, incompletely recessive inheritance best fits the genetic data for B. thuringiensis var. kurstaki resistance in this strain (371). A simple and plausible explanation is that the multitoxin resistance locus altered in the Hawaii and Pennsylvania strains is also affected in the Florida population, but this possibility has not been tested. The resistance phenotype was not associated with any fitness costs and, after an initial decrease in resistance during the first three generations, remained stable at a high level even in the absence of selection (371). Diamondback moth populations with a similar resistance phenotype—high-level resistance to B. thuringiensis subsp. kurstaki formulations and low-level resistance to B. thuringiensis subsp. aizawai—have also been isolated in Indonesia (341), Malaysia (167), Central America (292), and several states within the continental United States (341). Data are insufficient, however, to compare these strains to the resistant Hawaii, Pennsylvania, or Florida populations in stability, inheritance, or mechanism of resistance.
A field population of diamondback moths from the Philippines showed partial resistance to Cry1Aa, Cry1Ab, and Cry1Ac, but full sensitivity to Cry1C, Cry1F, and Cry1J (368). Binding to resistant-strain BBMV was reduced for Cry1Ab but apparently unaffected for Cry1Aa, Cry1Ac, or Cry1C. Interestingly, the Cry1Ab single-resistance phenotype appeared to be due to an autosomal, recessive mutation at the multitoxin resistance locus implicated in the resistant Hawaii and Pennsylvania strains, although the Philippines allele conferred no cross-resistance. Inheritance of resistances to Cry1Aa and Cry1Ac was expressed in an autosomal dominant and semidominant fashion, respectively, at the test dose employed (368). Cry1Ab binding was also implicated in the resistance mechanism of a strain isolated earlier from the same region of the Philippines (49, 113), although the cross-resistance phenotypes and inheritance patterns of this earlier isolate were not rigorously analyzed.
Resistance to B. thuringiensis subsp. kurstaki products and resulting failure in diamondback moth control has resulted in extensive use of B. thuringiensis subsp. aizawai-based insecticides in certain locations. Insects in two colonies from Hawaii have up to a 20-fold resistance to Cry1Ca compared to several other colonies, including one obtained earlier from the same location, as well as moderately high resistance to Cry1Ab and kurstaki subspecies-based formulations (228). Following additional selection in the laboratory, Cry1Ca resistance increased to 60-fold over control levels. The Cry1C resistance trait was shown to segregate independently from the Cry1Ab resistance determinant, behaving as an additive autosomal trait, appearing recessive at high test doses of toxin and dominant at low test doses (227).
A Malaysian strain simultaneously highly resistant to the kurstaki subspecies and the aizawai subspecies was apparently mutated in several loci (418). A Cry1Ab resistance allele, associated with reduced binding to BBMV receptors, was partially responsible for resistance to both subspecies. In contrast, binding of Cry1Aa, Cry1Ac, and Cry1C showed no gross alterations compared with BBMV from the sensitive strain. Genetic determinants responsible for subspecies kurstaki-specific and subspecies aizawai-specific resistance segregated separately from each other and from the Cry1Ab resistance allele in genetic experiments (418).
These studies suggest that a single locus, perhaps encoding a common receptor for many of the Cry1A toxins, can mutate to multitoxin resistance in P. xylostella. A different type of mutation at the same locus might alter the binding site for Cry1Ab, while leaving binding sites for other toxins on the same receptor unaffected. Unlinked loci affecting other events in toxicity, either before or after the binding step, can mutate to provide specific resistance to other Cry toxins. Additional studies along the lines of that conducted by Tabashnik et al. (368), using other resistant strains, are urgently needed to clarify the genetic and mechanistic picture.
It is clear, however, that the case history of P. xylostella presents a cautionary tale for the use of B. thuringiensis and its toxins in agriculture. After less than 2 decades of intensive subspecies kurstaki use in crucifer agriculture, resistant insects have evolved in numerous geographically isolated regions of the world, and subspecies aizawai resistance is beginning to appear even more rapidly. Injudicious use of Cry toxins could rapidly render them ineffective against other major crop pests, squandering a precious resource at a time when synthetic organic pesticides are already increasingly ineffective. Various alleles showing cross-resistance, dominant inheritance, or stability in the absence of selection have been detected in resistant field lines of P. xylostella, phenomena with far-reaching implications for resistance management. These observations underscore a critical need for increased emphasis and funding on an international scale for all aspects of Cry toxin research.
Resistance Management
Resistance management strategies try to prevent or diminish the selection of the rare individuals carrying resistance genes and hence to keep the frequency of resistance genes sufficiently low for insect control. Strategy development generally relies heavily on theoretical assumptions and on computer models simulating insect population growth under various conditions (12, 141, 168, 250, 320, 321, 364). Proposed strategies include the use of multiple toxins (stacking or pyramiding), crop rotation, high or ultrahigh dosages, and spatial or temporal refugia (265, 364). Only recently have some of the proposed tactics been experimentally evaluated on a small scale (342). Retrospective analysis of resistance development does support the use of refugia (364). It is clear that the real value of the different proposed tactics can only be tested in larger-scale field trials.
It is expected that each pest-crop complex may require a specific implementation of certain resistance management strategies that may have to address the use of both B. thuringiensis sprays and transgenic crops. Experience with transgenic crops expressing cry genes grown under different agronomic conditions is essential to define the requirements of resistance management. It is equally important to design a resistance management strategy acceptable to everyone involved: technology suppliers, seed companies, extension workers, crop consultants, regulators, and, most of all, growers (182).
In transgenic plants, selection pressure could be reduced by restricting the expression of the crystal protein genes to certain tissues of the crop (those most susceptible to pest damage) so that only certain parts of the plant are fully protected, the remainder providing a form of spatial refuge (but see the concerns raised in reference 250). It has been proposed that cotton lines in which cry gene expression is limited to the young bolls may not suffer dramatic yield loss from Heliothis larvae feeding on other plant structures, since cotton plants can compensate for a high degree of pest damage (140). Crystal protein gene expression could be triggered by the feeding of the insect itself in a transgenic plant, with resident cry genes controlled by wound-inducible promoters (291). If plants were to express B. thuringiensis toxin only in response to specific damage thresholds, it might provide a mechanism to diminish toxin exposure to insects. Alternatively, toxin expression could be induced by the application of a chemical (409). In this way, a farmer would have the option to have Cry toxin present in the crops only when insect densities exceed an economic threshold.
Another management option is the rotation of plants or sprays of a particular B. thuringiensis toxin with those having another toxin type that binds to a different receptor. This strategy has potential value when a fitness cost is associated with resistance. Such fitness costs have been reported in P. xylostella lines, in which resistant males have lower mating success than their nonresistant competitors (149). Insects resistant to one Cry toxin type would be at a disadvantage during the next growth season when a different toxin type is used, resulting in a decrease of the frequency of the corresponding resistance gene. Ideally, reversion to susceptibility for this Cry toxin type should occur within the growth season. Tabashnik et al. (365) noticed that revertant diamondback moth populations responded rapidly to reselection and susceptibility was not fully restored.
If transgenic plants can express a cry gene at doses high enough to kill even homozygous resistant insects, that crop will become a nonhost. While such an ultrahigh dose might be impractical with a sprayable product due to high cost, incomplete coverage, toxin breakdown, and plant growth, it may be possible with toxin-engineered plants, taking into account the currently attainable levels of Cry expression in planta (169). For example, a Colorado potato beetle population 100-fold resistant to a Cry3A-containing B. thuringiensis spray could not survive on potato plants expressing the same protein (13, 407). It remains to be seen if a combination of toxins with ultrahigh expression can overcome all homozygous resistance alleles, changing the crops into nonhost plants.
A very attractive resistance management tactic is the combination of a high-dose strategy with the use of refugia (toxin-free areas). The principle is to express Cry toxins at such a dose that nearly all heterozygotic carriers of resistance alleles will be killed. Survivors would most likely mate with the sensitive insects harbored in the nearby refuge. Consequently, a population of homozygous resistant insects would be unlikely to emerge. B. thuringiensis resistance is in fact a recessive trait in at least some insect species (364); with the high levels of expression now attainable in planta (e.g., a dose 50-fold higher than the LC50) (193), and with essentially complete foliar coverage, it may be reasonable to attain nearly total killing of heterozygotes. Indeed, Metz et al. (269) demonstrated that F1 larvae from a cross between a susceptible laboratory P. xylostella colony and a field-resistant colony did not survive on transgenic broccoli expressing Cry1Ac (341). It has been reported that the inclusion of refuge plants in cages with transgenic broccoli plants resulted in slower evolution of resistance in populations of P. xylostella (342). Supporting evidence also comes from selection experiments using B. thuringiensis subsp. aizawai and a diamondback moth population that had evolved resistance to Cry1Ab and Cry1Ca in the field. In these studies, a 10% refuge delayed resistance over a nine-generation test (226). Depending on the crop, refugia may be naturally present or may need to be created by the planting of nontransgenic plots. Refugia should be uncontaminated, and there should be random mating between resistant and nonresistant insects (141). Refugia that are temporally and spatially contiguous with the transgenic crop could fulfill these requirements (118). See the work of Gould (142) for a broader discussion from a perspective of population dynamics and evolution.
A specific planting strategy that has been recommended to reduce selection is the use of seed mixtures of toxin-expressing and toxin-free plants to provide prepackaged refugia. The seed mix strategy, still controversial, would probably only be effective for insect species whose larvae move very little between plants (250, 364) or whose adults acquire a mate visually over a short distance (320).
Another valuable option for resistance management, in combination with the use of refugia, is the expression of multiple Cry proteins in crops or incorporation of multiple proteins in B. thuringiensis sprays, provided these toxins have different modes of action (321) with respect to the insect’s mechanism of resistance. Cry toxins that recognize different receptors in the same target species could be deployed in this strategy, since they are less prone to cross-resistance. As noted above, diamondback moth populations resistant to field applications of Cry1A-containing B. thuringiensis formulations showed minimal cross-resistance to other crystal proteins such as Cry1Ba, Cry1Bb, Cry1Ca, Cry1Da, Cry1Ia, Cry2A, and Cry9Ca, while they were cross-resistant to Cry1Fa and Cry1Ja (198, 365, 369, 372). There are several other insect species in which Cry toxins with different receptor specificities are known (93, 105, 113, 163, 198, 394, 395). For many insect species, multiple Cry1A proteins would not be an appropriate choice, since some of these proteins share binding sites with one another (94, 106, 395, 413) and even with other toxins of the Cry1 class (97). Yet for other insects, Cry1A proteins have been shown, at least on ligand blots, to recognize different binding proteins (211, 385, 386, 388). Additionally, B. thuringiensis Cry toxins could be combined with other insecticidal proteins. The multiple-attack strategy assumes that within a population, if insects homozygous for one resistance gene are rare, then insects homozygous for multiple resistance genes are extremely rare. Crops or sprays deploying multiple toxins would still control even insects homozygous for one or two resistance genes yet heterozygous for another gene. A critical condition for the success of this strategy is that each of the insecticides on its own should have high mortality for susceptible homozygotes (321). An example is O. nubilalis, in which Cry1Ab and Cry1Ba, both highly active, bind to different receptors (94). A strong argument for the utility of multiple-gene pyramiding is found in the recent results of Georghiou and Wirth (133). Their field-collected C. quinquefasciatus populations readily developed resistance in the laboratory to a single B. thuringiensis subsp. israelensis toxin (Cry11A) but remained remarkably sensitive when selection was with the full complement of toxins from this variety.
Due to the urgent need for a more complete understanding of the parameters of effective resistance management, companies developing B. thuringiensis biopesticidal sprays and transgenic plants formed the B. thuringiensis Management Working Group in 1988 to promote research on the judicious use of B. thuringiensis products. It is hoped that an increased understanding of the complex interplay among Cry toxins, their bacterial hosts, their target organisms, and the ecosystems they share will allow for the long-term, effective use of Cry toxins for pest management.
ACKNOWLEDGMENTS
The Bacillus Genetic Stock Center is supported by National Science Foundation grant DBI-9319712 and by industrial sponsorships.
We thank Oscar Alzate for preparing the figure showing three-dimensional structures of Cry proteins. We also thank two anonymous reviewers for their unusually thorough analyses of our manuscript.
ADDENDUM IN PROOF
After this review was accepted for publication, an analysis of the effect of ligand blot conditions on the binding of Cry1A toxins to the cadherin-like 210-kDA receptor from M. sexta was published (T. P. Keaton, B. R. Francis, W. S. A. Maaty, and L. A. Bulla, Jr., Appl. Environ. Microbiol. 64:2158–2165, 1998). Under a variety of conditions, this cadherin-like protein bound not only Cry1Ab but also Cry1Aa and Cry1Ac, suggesting that it is an important receptor for all three Cry1A proteins.
REFERENCES
- 1.Adams L F, Brown K L, Whiteley H R. Molecular cloning and characterization of two genes encoding sigma factors that direct transcription from a Bacillus thuringiensis crystal protein gene promoter. J Bacteriol. 1991;173:3846–3854. doi: 10.1128/jb.173.12.3846-3854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams L F, Mathewes S, O’Hara P, Petersen A, Gürtler H. Elucidation of the mechanism of CryIIIA overproduction in a mutagenized strain of Bacillus thuringiensis var. tenebrionis. Mol Microbiol. 1994;14:381–389. doi: 10.1111/j.1365-2958.1994.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 3.Adams L F, Thomas M D. Presented at the Seventh International Conference on Bacillus. Paris, France: Institut Pasteur; 1993. [Google Scholar]
- 4.Adams L F, Visick J E, Whiteley H R. A 20-kilodalton protein is required for efficient production of the Bacillus thuringiensis subsp. israelensis 27-kilodalton crystal protein in Escherichia coli. J Bacteriol. 1989;171:521–530. doi: 10.1128/jb.171.1.521-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adang M J, Brody M S, Cardineau G, Eagan N, Roush R T, Shewmaker C K, Jones A, Oakes J V, McBride K E. The reconstruction and expression of a Bacillus thuringiensis cryIIIA gene in protoplasts and potato plants. Plant Mol Biol. 1993;21:1131–1145. doi: 10.1007/BF00023609. [DOI] [PubMed] [Google Scholar]
- 6.Addison J A. Persistence and non-target effects of Bacillus thuringiensis in soil: a review. Can J For Res. 1993;23:2329–2342. [Google Scholar]
- 7.Agaisse H, Lereclus D. Expression in Bacillus subtilis of the Bacillus thuringiensis cryIIIA toxin gene is not dependent on a sporulation-specific sigma factor and is increased in a spo0A mutant. J Bacteriol. 1994;176:4734–4741. doi: 10.1128/jb.176.15.4734-4741.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agaisse H, Lereclus D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agaisse H, Lereclus D. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol Microbiol. 1996;20:633–643. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- 10.Agaisse H, Lereclus D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol. 1994;13:97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 11.Allured V S, Collier R J, Carroll S F, McKay D B. Structure of exotoxin A of Pseudomonas aeruginosa at 3.0-Ångstrom resolution. Proc Natl Acad Sci USA. 1986;83:1320–1324. doi: 10.1073/pnas.83.5.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alstad D N, Andow D A. Managing the evolution of insect resistance to transgenic plants. Science. 1995;268:1894–1896. doi: 10.1126/science.268.5219.1894. [DOI] [PubMed] [Google Scholar]
- 13.Altre J A, Grafius E J, Whalon M E. Feeding behavior of CryIIIA-resistant and susceptible Colorado potato beetle (Coleoptera: Chrysomelidae) larvae on Bacillus thuringiensis tenebrionis-transgenic CryIIIA-treated and untreated potato foliage. J Econ Entomol. 1996;89:311–317. [Google Scholar]
- 14.Andreadis T G, Dubois N R, Moore R E B, Anderson J F, Lewis F B. Single applications of high concentrations of Bacillus thuringiensis for control of gypsy moth (Lepidoptera: Lymantriidae) populations and their impact on parasitism and disease. J Econ Entomol. 1983;76:1417–1422. [Google Scholar]
- 15.Angsuthanasombat C, Crickmore N, Ellar D J. Comparison of Bacillus thuringiensis subsp. israelensis CryIVA and CryIVB cloned toxins reveals synergism in vivo. FEMS Microbiol Lett. 1992;94:63–68. doi: 10.1016/0378-1097(92)90584-b. [DOI] [PubMed] [Google Scholar]
- 16.Angsuthanasombat C, Crickmore N, Ellar D J. Effects on toxicity of eliminating a cleavage site in a predicted interhelical loop in Bacillus thuringiensis CryIVB delta endotoxin. FEMS Microbiol Lett. 1993;111:255–261. doi: 10.1111/j.1574-6968.1993.tb06395.x. [DOI] [PubMed] [Google Scholar]
- 17.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 18.Aronson A I, Han E-S, McGaughey W, Johnson D. The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol. 1991;57:981–986. doi: 10.1128/aem.57.4.981-986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson A I, Wu D, Zhang C. Mutagenesis of specificity and toxicity regions of a Bacillus thuringiensis protoxin gene. J Bacteriol. 1995;177:4059–4065. doi: 10.1128/jb.177.14.4059-4065.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ash C, Farrow J A E, Darsh M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 21.Ballester V, Escriche B, Mensua J L, Riethmacher G W, Ferré J. Lack of cross-resistance to other Bacillus thuringiensis crystal proteins in a population of Plutella xylostella highly resistant to CryIA(b) Biocontrol Technol. 1994;4:437–443. [Google Scholar]
- 22.Bar E, Lieman-Hurwitz J, Rahamim E, Keynan A, Sandler N. Cloning and expression of Bacillus thuringiensis israelensis δ-endotoxin in B. sphaericus. J Invertebr Pathol. 1991;57:149–158. doi: 10.1016/0022-2011(91)90110-c. [DOI] [PubMed] [Google Scholar]
- 23.Barloy F, Delécluse A, Nicolas L, Lecadet M-M. Cloning and expression of the first anaerobic toxin gene from Clostridium bifermentans subsp. malaysia, encoding a new mosquitocidal protein with homologies to Bacillus thuringiensis delta-endotoxins. J Bacteriol. 1996;178:3099–3105. doi: 10.1128/jb.178.11.3099-3105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton K A, Whiteley H R, Yang N-S. Bacillus thuringiensis δ-endotoxin expressed in transgenic Nicotiana tabacum provides resistance to lepidopteran insects. Plant Physiol. 1987;85:1103–1109. doi: 10.1104/pp.85.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer L S, Koller C N, Miller D L, Hollingworth R M. Presented at the XXVIIth Annual Meeting of the Society for Invertebrate Pathology, Montpellier, France. 1994. [Google Scholar]
- 26.Baum, J. A. August 1995. U.S. patent 5,441,884.
- 27.Baum J A. Tn5401, a new class II transposable element from Bacillus thuringiensis. J Bacteriol. 1994;176:2835–2845. doi: 10.1128/jb.176.10.2835-2845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baum J A, Coyle D M, Gilbert M P, Jany C S, Gawron-Burke C. Novel cloning vectors for Bacillus thuringiensis. Appl Environ Microbiol. 1990;56:3420–3428. doi: 10.1128/aem.56.11.3420-3428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baum J A, Kakefuda M, Gawron-Burke C. Engineering Bacillus thuringiensis bioinsecticides with an indigenous site-specific recombination system. Appl Environ Microbiol. 1996;62:4367–4373. doi: 10.1128/aem.62.12.4367-4373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baum J A, Malvar T. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol. 1995;18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x. [DOI] [PubMed] [Google Scholar]
- 31.Baumann L, Okamoto K, Unterman B M, Lynch M J, Baumann P. Phenotypic characterization of Bacillus thuringiensis and Bacillus cereus. J Invertebr Pathol. 1984;44:329–341. [Google Scholar]
- 32.Becker N, Margalit J. Use of Bacillus thuringiensis israelensis against mosquitoes and blackflies. In: Entwistle P F, Cory P F, Bailey M J, Higgs S, editors. Bacillus thuringiensis, an environmental biopesticide: theory and practice. J. New York, N.Y: Wiley & Sons; 1993. pp. 145–170. [Google Scholar]
- 33.Beegle C C, Yamamoto T. History of Bacillus thuringiensis Berliner research and development. Can Entomol. 1992;124:587–616. [Google Scholar]
- 34.Belliveau B H, Trevors J H. Transformation of Bacillus cereus vegetative cells by electroporation. Appl Environ Microbiol. 1989;55:1649–1652. doi: 10.1128/aem.55.6.1649-1652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernhard K. Studies on the delta-endotoxin of Bacillus thuringiensis var. tenebrionis. FEMS Microbiol Lett. 1986;33:261–265. [Google Scholar]
- 36.Bernhard K, Jarrett P, Meadows M, Butt J, Ellis D J, Roberts G M, Pauli S, Rodgers P, Burges H D. Natural isolates of Bacillus thuringiensis: worldwide distribution, characterization, and activity against insect pests. J Invertebr Pathol. 1997;70:59–68. [Google Scholar]
- 37.Bietlot H P, Carey P R, Pozsgay M, Kaplan H. Isolation of carboxyl-terminal peptides from proteins by diagonal electrophoresis: application to the entomocidal toxin from Bacillus thuringiensis. Anal Biochem. 1989;181:212–215. doi: 10.1016/0003-2697(89)90231-5. [DOI] [PubMed] [Google Scholar]
- 38.Bietlot H P, Schernthaner J P, Milne R E, Clairmont F R, Bhella R S, Kaplan H. Evidence that the CryIA crystal protein from Bacillus thuringiensis is associated with DNA. J Biol Chem. 1993;268:8240–8245. [PubMed] [Google Scholar]
- 39.Bietlot H P, Vishnubhatla I, Carey P R, Pozsgay M, Kaplan H. Characterization of the cysteine residues and disulfide linkages in the protein crystal of Bacillus thuringiensis. Biochem J. 1990;267:309–316. doi: 10.1042/bj2670309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Black G E, Snyder A P, Heroux K S. Chemotaxonomic differentiation between the Bacillus cereus group and Bacillus subtilis by phospholipid extracts analyzed with electrospray ionization tandem mass spectrometry. J Microbiol Methods. 1997;28:187–199. [Google Scholar]
- 41.Bolin P C, Hutchison W D, Andow D A. Presented at the XXVIIIth Annual Meeting of the Society for Invertebrate Pathology. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 42.Bone E J, Ellar D J. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett. 1989;58:171–178. doi: 10.1016/0378-1097(89)90033-5. [DOI] [PubMed] [Google Scholar]
- 43.Bora R S, Murty M G, Shenbagarathai R, Sekar V. Introduction of a lepidopteran-specific insecticidal crystal protein gene of Bacillus thuringiensis subsp. kurstaki by conjugal transfer into a Bacillus megaterium strain that persists in the cotton phyllosphere. Appl Environ Microbiol. 1994;60:214–222. doi: 10.1128/aem.60.1.214-222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosch D, Schipper B, van der Kleij H, de Maagd R A, Stiekema W J. Recombinant Bacillus thuringiensis crystal proteins with new properties: possibilities for resistance management. Bio/Technology. 1994;12:915–918. doi: 10.1038/nbt0994-915. [DOI] [PubMed] [Google Scholar]
- 45.Bourgouin C, Delécluse A, Ribier J, Klier A, Rapoport G. A Bacillus thuringiensis subsp. israelensis gene encoding a 125-kilodalton larvicidal polypeptide is associated with inverted repeat sequences. J Bacteriol. 1988;170:3575–3583. doi: 10.1128/jb.170.8.3575-3583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowen, T. 1996. Unpublished results.
- 47.Bravo A. Phylogenetic relationships of Bacillus thuringiensis δ-endotoxin family proteins and their functional domains. J Bacteriol. 1997;179:2793–2801. doi: 10.1128/jb.179.9.2793-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bravo A, Agaisse H, Salamitou S, Lereclus D. Analysis of cryIAa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol Gen Genet. 1996;250:734–741. doi: 10.1007/BF02172985. [DOI] [PubMed] [Google Scholar]
- 49.Bravo A, Jansens S, Peferoen M. Immunocytochemical localization of Bacillus thuringiensis insecticidal crystal proteins in intoxicated insects. J Invertebr Pathol. 1992;60:237–246. [Google Scholar]
- 50.Brizzard B L, Schnepf H E, Kronstad J W. Expression of the cryIB crystal protein gene of Bacillus thuringiensis. Mol Gen Genet. 1991;231:59–64. doi: 10.1007/BF00293822. [DOI] [PubMed] [Google Scholar]
- 51.Brown K L. Transcriptional regulation of the Bacillus thuringiensis subsp. thompsoni crystal protein gene operon. J Bacteriol. 1993;175:7951–7957. doi: 10.1128/jb.175.24.7951-7957.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown K L, Whiteley H R. Isolation of a Bacillus thuringiensis RNA polymerase capable of transcribing crystal protein genes. Proc Natl Acad Sci USA. 1988;85:4166–4170. doi: 10.1073/pnas.85.12.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown K L, Whiteley H R. Isolation of the second Bacillus thuringiensis RNA polymerase that transcribes from a crystal protein gene promoter. J Bacteriol. 1990;172:6682–6688. doi: 10.1128/jb.172.12.6682-6688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burges H D, Hurst J A. Ecology of Bacillus thuringiensis in storage moths. J Invertebr Pathol. 1977;30:131–139. [Google Scholar]
- 55.Butko P, Huang F, Pusztai-Carey M, Surewicz W K K. Interaction of the δ-endotoxin CytA from Bacillus thuringiensis var. israelensis with lipid membranes. Biochemistry. 1997;36:12862–12868. doi: 10.1021/bi9702389. [DOI] [PubMed] [Google Scholar]
- 56.Carlson C R, Caugant D A, Kolstø A-B. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl Environ Microbiol. 1994;60:1719–1725. doi: 10.1128/aem.60.6.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlson C R, Johansen T, Lecadet M-M, Kolstø A-B. Genomic organization of the entomopathogenic bacterium Bacillus thuringiensis subsp. berliner 1715. Microbiology. 1996;142:1625–1634. [Google Scholar]
- 58.Carlson C R, Kolstø A-B. A complete physical map of a Bacillus thuringiensis chromosome. J Bacteriol. 1993;175:1053–1060. doi: 10.1128/jb.175.4.1053-1060.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carozzi N B, Kramer V C, Warren G W, Evola S, Koziel M G. Prediction of insecticidal activity of Bacillus thuringiensis strains by polymerase chain reaction product profiles. Appl Environ Microbiol. 1991;57:3057–3061. doi: 10.1128/aem.57.11.3057-3061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carroll J, Convents D, Van Damme J, Boets A, Van Rie J, Ellar D J. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A delta-endotoxin may facilitate its coleopteran toxicity. J Invertebr Pathol. 1997;70:41–49. doi: 10.1006/jipa.1997.4656. [DOI] [PubMed] [Google Scholar]
- 61.Carroll J, Ellar D J. An analysis of Bacillus thuringiensis δ-endotoxin action on insect-midgut-membrane permeability using a light-scattering assay. Eur J Biochem. 1993;214:771–778. doi: 10.1111/j.1432-1033.1993.tb17979.x. [DOI] [PubMed] [Google Scholar]
- 62.Carroll J, Ellar D J. Analysis of the large aqueous pores produced by a Bacillus thuringiensis protein insecticide in Manduca sexta midgut-brush-border-membrane vesicles. Eur J Biochem. 1997;245:797–804. doi: 10.1111/j.1432-1033.1997.00797.x. [DOI] [PubMed] [Google Scholar]
- 63.Chak K-F, Tseng M-Y, Yamamoto T. Expression of the crystal protein gene under the control of the α-amylase promoter in Bacillus thuringiensis strains. Appl Environ Microbiol. 1994;60:2304–2310. doi: 10.1128/aem.60.7.2304-2310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang C, Yu Y-M, Dai S-M, Law S K, Gill S S. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–821. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaufaux J, Marchal M, Gilois N, Jehanno I, Buisson C. Investigation of natural strains of Bacillus thuringiensis in different biotopes throughout the world. Can J Microbiol. 1997;43:337–343. [Google Scholar]
- 66.Chaufaux J, Muller-Cohn J, Buisson C, Sanchis V, Lereclus D, Pastuer N. Inheritance of resistance to the Bacillus thuringiensis CryIC toxin in Spodoptera littoralis (Lepidoptera: Noctuidae) J Econ Entomol. 1997;90:873–878. [Google Scholar]
- 67.Chen X J, Curtiss A, Alcantara E, Dean D H. Mutations in domain I of Bacillus thuringiensis δ-endotoxin CryIAb reduce the irreversible binding of toxin to Manduca sexta brush border membrane vesicles. J Biol Chem. 1995;270:6412–6419. doi: 10.1074/jbc.270.11.6412. [DOI] [PubMed] [Google Scholar]
- 68.Chen X J, Lee M K, Dean D H. Site-directed mutations in a highly conserved region of Bacillus thuringiensis δ-endotoxin affect inhibition of short circuit current across Bombyx mori midguts. Proc Natl Acad Sci USA. 1993;90:9041–9045. doi: 10.1073/pnas.90.19.9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chestukhina G G, Kostina L I, Mikhailova A L, Tyurin S A, Klepikova F S, Stepanov V M. The main features of Bacillus thuringiensis delta-endotoxin molecular structure. Arch Microbiol. 1982;132:159–162. [Google Scholar]
- 70.Chilcott C N, Ellar D J. Comparative study of Bacillus thuringiensis var. israelensis crystal proteins in vivo and in vitro. J Gen Microbiol. 1988;134:2551–2558. doi: 10.1099/00221287-134-9-2551. [DOI] [PubMed] [Google Scholar]
- 71.Choe S, Bennett M J, Fujii G, Curmi P M G, Kantardjieff K A, Collier R J, Eisenberg D. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- 72.Choma C T, Kaplan H. Folding and unfolding of the protoxin from Bacillus thuringiensis: evidence that the toxic moiety is present in an active conformation. Biochemistry. 1990;29:10971–10977. doi: 10.1021/bi00501a015. [DOI] [PubMed] [Google Scholar]
- 73.Choma C T, Surewicz W K, Carey P R, Pozsgay M, Kaplan H. Secondary structure of the entomocidal toxin from Bacillus thuringiensis subsp. kurstaki HD-73. J Protein Chem. 1990;9:87–94. doi: 10.1007/BF01024989. [DOI] [PubMed] [Google Scholar]
- 74.Choma C T, Surewicz W K, Kaplan H. The toxic moiety of the Bacillus thuringiensis protoxin undergoes a conformational change upon activation. Biochem Biophys Res Commun. 1991;179:933–938. doi: 10.1016/0006-291x(91)91908-u. [DOI] [PubMed] [Google Scholar]
- 75.Chow E, Singh G J P, Gill S S. Binding and aggregation of the 25-kilodalton toxin of Bacillus thuringiensis subsp. israelensis to cell membranes and alteration by monoclonal antibodies and amino acid modifiers. Appl Environ Microbiol. 1989;55:2779–2788. doi: 10.1128/aem.55.11.2779-2788.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chow E, Singh G J P, Gill S S. Binding and aggregation of the 25-kilodalton toxin of Bacillus thuringiensis subsp. israelensis to cell membranes and alteration by monoclonal antibodies and amino acid modifiers. Appl Environ Microbiol. 1995;55:2779–2788. doi: 10.1128/aem.55.11.2779-2788.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76a.Clairmont F R, Milne R E, Pham V T, Carrière M B, Kaplan H. Role of DNA in the activation of the Cry1A insecticidal crystal protein from Bacillus thuringiensis. J Biol Chem. 1998;273:9292–9296. doi: 10.1074/jbc.273.15.9292. [DOI] [PubMed] [Google Scholar]
- 77.Crawford D N, Harvey W R. Barium and calcium block Bacillus thuringiensis subspecies kurstaki δ-endotoxins inhibition of potassium current across isolated midgut of larval Manduca sexta. J Exp Biol. 1988;137:277–286. doi: 10.1242/jeb.137.1.277. [DOI] [PubMed] [Google Scholar]
- 78.Crickmore N, Bone E J, Williams J A, Ellar D J. Contribution of the individual components of the δ-endotoxin crystal to the mosquitocidal activity of Bacillus thuringiensis subsp. israelensis. FEMS Microbiol Lett. 1995;131:249–254. [Google Scholar]
- 79.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cummings C E, Armstrong G, Hodgman T C, Ellar D J. Structural and functional studies of a synthetic peptide mimicking a proposed membrane inserting region of a Bacillus thuringiensis δ-endotoxin. Mol Membr Biol. 1994;11:87–92. doi: 10.3109/09687689409162225. [DOI] [PubMed] [Google Scholar]
- 81.de Barjac H, Sutherland D J. Bacterial control of mosquitoes and blackflies. New Brunswick, N.J: Rutgers University Press; 1990. [Google Scholar]
- 82.Delécluse, A. Unpublished results.
- 83.Delécluse A, Bourgouin C, Klier A, Rapoport G. Nucleotide sequence and characterization of a new insertion element, IS240, from Bacillus thuringiensis israelensis. Plasmid. 1989;21:71–78. doi: 10.1016/0147-619x(89)90088-7. [DOI] [PubMed] [Google Scholar]
- 84.Delécluse A, Charles J-F, Klier A, Rapoport G. Deletion by in vivo recombination shows that the 28-kilodalton cytolytic polypeptide from Bacillus thuringiensis subsp. israelensis is not essential for mosquitocidal activity. J Bacteriol. 1991;173:3374–3381. doi: 10.1128/jb.173.11.3374-3381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delécluse A, Poncet S, Klier A, Rapoport G. Expression of cryIVA and cryIVB genes, independently or in combination, in a crystal-negative strain of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1993;59:3922–3927. doi: 10.1128/aem.59.11.3922-3927.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delécluse A, Rosso M-L, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delucca A J, II, Palmgren M S, de Barjac H. A new serovar of Bacillus thuringiensis from grain dust: Bacillus thuringiensis serovar colmeri (serovar 21) J Invertebr Pathol. 1984;43:437–438. [Google Scholar]
- 88.Delucca A J, II, Simonson J, Larson A. Two new serovars of Bacillus thuringiensis: serovars dakota and indiana (serovars 15 and 16) J Invertebr Pathol. 1979;34:323–324. [Google Scholar]
- 89.Delucca A J, II, Simonson J G, Larson A D. Bacillus thuringiensis distribution in soils of the United States. Can J Microbiol. 1981;27:865–870. doi: 10.1139/m81-137. [DOI] [PubMed] [Google Scholar]
- 90.de Maagd R A, Kwa M S G, van der Klei H, Yamamoto T, Schipper B, Vlak J M, Stiekema W J, Bosch D. Domain III substitution in Bacillus thuringiensis CryIA(b) results in superior toxicity for Spodoptera exigua and altered membrane protein recognition. Appl Environ Microbiol. 1996;62:1537–1543. doi: 10.1128/aem.62.5.1537-1543.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Maagd R A, van der Klei H, Bakker P L, Stiekema W J, Bosch D. Different domains of Bacillus thuringiensis δ-endotoxins can bind to insect midgut membrane proteins on ligand blots. Appl Environ Microbiol. 1996;62:2753–2757. doi: 10.1128/aem.62.8.2753-2757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Denolf P, Hendrickx K, Van Damme J, Jansens S, Peferoen M, Degheele D, Van Rie J. Cloning and characterization of Manduca sexta and Plutella xylostella midgut aminopeptidase N enzymes related to Bacillus thuringiensis toxin-binding proteins. Eur J Biochem. 1997;248:748–761. doi: 10.1111/j.1432-1033.1997.t01-1-00748.x. [DOI] [PubMed] [Google Scholar]
- 93.Denolf P, Jansens S, Peferoen M, Degheele D, Van Rie J. Two different Bacillus thuringiensis delta-endotoxin receptors in the midgut brush border membrane of the European corn borer, Ostrinia nubilalis (Hübner) (Lepidoptera: Pyralidae) Appl Environ Microbiol. 1993;59:1828–1837. doi: 10.1128/aem.59.6.1828-1837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dervyn E, Poncet S, Klier A, Rapoport G. Transcriptional regulation of the cryIVD gene operon from Bacillus thuringiensis subsp. israelensis. J Bacteriol. 1995;177:2283–2291. doi: 10.1128/jb.177.9.2283-2291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.De Souza M T, Lecadet M-M, Lereclus D. Full expression of the cryIIIA toxin gene of Bacillus thuringiensis requires a distant upstream DNA sequence affecting transcription. J Bacteriol. 1993;175:2952–2960. doi: 10.1128/jb.175.10.2952-2960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donovan W P, Rupar M J, Slaney A C, Malvar T, Gawron-Burke M C, Johnson T B. Characterization of two genes encoding Bacillus thuringiensis insecticidal crystal proteins toxic to Coleoptera species. Appl Environ Microbiol. 1992;58:3921–3927. doi: 10.1128/aem.58.12.3921-3927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Donovan W P, Tan Y, Slaney A C. Cloning of the nprA gene for neutral protease A of Bacillus thuringiensis and effect of in vivo deletion of nprA on insecticidal crystal protein. Appl Environ Microbiol. 1997;63:2311–2317. doi: 10.1128/aem.63.6.2311-2317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Du C, Martin P A W, Nickerson K W. Comparison of disulfide contents and solubility at alkaline pH of insecticidal and noninsecticidal Bacillus thuringiensis protein crystals. Appl Environ Microbiol. 1994;60:3847–3853. doi: 10.1128/aem.60.10.3847-3853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Du C, Nickerson K W. The Bacillus thuringiensis insecticidal toxin binds biotin-containing proteins. Appl Environ Microbiol. 1996;62:2932–2939. doi: 10.1128/aem.62.8.2932-2939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dubois N R, Dean D H. Synergism between Cry1A insecticidal crystal proteins and spores of Bacillus thuringiensis, other bacterial spores, and vegetative cells against Lymantria dispar (Lepidoptera: Lymantriidae) larvae. Environ Entomol. 1995;24:1741–1747. [Google Scholar]
- 101.Duche D, Parker M W, González-Mañas J-M, Pattus F, Baty D. Uncoupled steps of the colicin A pore formation demonstrated by disulphide bond engineering. J Biol Chem. 1994;269:6332–6339. [PubMed] [Google Scholar]
- 102.Dulmage H T. Insecticidal activity of HD-1, a new isolate of Bacillus thuringiensis var. alesti. J Invertebr Pathol. 1970;15:232–239. [Google Scholar]
- 103.Dunn M G, Ellar D J. Identification of two sequence elements associated with the gene encoding the 24-kDa crystalline component in Bacillus thuringiensis ssp. fukuokaensis: an example of transposable element archaeology. Plasmid. 1997;37:205–215. doi: 10.1006/plas.1997.1283. [DOI] [PubMed] [Google Scholar]
- 104.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Escriche B, Tabashnik B, Finson N, Ferré J. Immunohistochemical detection of binding of CryIA crystal proteins of Bacillus thuringiensis in highly resistant strains of Plutella xylostella (L.) from Hawaii. Biochem Biophys Res Commun. 1995;212:388–395. doi: 10.1006/bbrc.1995.1982. [DOI] [PubMed] [Google Scholar]
- 106.Estada U, Ferré J. Binding of insecticidal crystal proteins of Bacillus thuringiensis to the midgut brush border of the cabbage looper, Trichoplusia ni (Hübner) (Lepidoptera: Noctuidae), and selection for resistance to one of the crystal proteins. Appl Environ Microbiol. 1994;60:3840–3846. doi: 10.1128/aem.60.10.3840-3846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Estruch J J, Carozzi N B, Desai N, Duck N B, Warren G W, Koziel M G. Transgenic plants: an emerging approach to pest control. Nat Biotechnol. 1997;15:137–141. doi: 10.1038/nbt0297-137. [DOI] [PubMed] [Google Scholar]
- 108.Estruch J J, Warren G W, Mullins M A, Nye G J, Craig J A, Koziel M G. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc Natl Acad Sci USA. 1996;93:5389–5394. doi: 10.1073/pnas.93.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feitelson J S. The Bacillus thuringiensis family tree. In: Kim L, editor. Advanced engineered pesticides. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 63–71. [Google Scholar]
- 110.Feitelson J S, Payne J, Kim L. Bacillus thuringiensis: insects and beyond. Bio/Technology. 1992;10:271–275. [Google Scholar]
- 111.Feng Q, Becktel W J. pH induced conformational transitions of CryIA(a), CryIA(c) and CryIIIA δ-endotoxins in Bacillus thuringiensis. Biochemistry. 1994;33:8521–8526. doi: 10.1021/bi00194a017. [DOI] [PubMed] [Google Scholar]
- 112.Ferré J, Escriche B, Bel Y, Van Rie J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis insecticidal crystal proteins. FEMS Microbiol Lett. 1995;132:1–7. [Google Scholar]
- 113.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fischhoff D A, Bowdisch K S, Perlak F J, Marrone P G, McCormick S H, Niedermeyer J G, Dean D A, Kusano-Kretzmer K, Mayer E J, Rochester D E, Rogers S G, Fraley R T. Insect tolerant transgenic tomato plants. Bio/Technology. 1987;5:807–813. [Google Scholar]
- 115.Fischhoff, D. A., and S. G. Rogers. 1994. U.S. patent 5,349,124.
- 116.Flores H, Soberon X, Sanchez J, Bravo A. Isolated domain II and III from the Bacillus thuringiensis Cry1 Ab delta-endotoxin binds to lepidopteran midgut membranes. FEBS Lett. 1997;414:313–318. doi: 10.1016/s0014-5793(97)01015-6. [DOI] [PubMed] [Google Scholar]
- 117.Forcada C, Alcacer E, Garcera M D, Martinez R. Differences in the midgut proteolytic activity of two Heliothis virescens strains, one susceptible and one resistant to Bacillus thuringiensis toxins. Arch Insect Biochem Physiol. 1996;31:257–272. [Google Scholar]
- 118.Forrester N W. Resistance management options for conventional Bacillus thuringiensis and transgenic plants in Australian summer field crops. Biocontrol Sci Technol. 1994;4:549–553. [Google Scholar]
- 119.Francis B R, Bulla L A., Jr Further characterization of BT-R1, the cadherin-like receptor for Cry1Ab toxin in tobacco hornworm (Manduca sexta) midguts. Insect Biochem Mol Biol. 1997;27:541–550. doi: 10.1016/s0965-1748(97)00029-5. [DOI] [PubMed] [Google Scholar]
- 120.Fujimoto H, Itoh K, Yamamoto M, Kyozuka J, Shimamoto K. Insect resistant rice generated by introduction of a modified δ-endotoxin gene of Bacillus thuringiensis. Bio/Technology. 1993;11:1151–1155. doi: 10.1038/nbt1093-1151. [DOI] [PubMed] [Google Scholar]
- 121.Gaertner F H, Quick T C, Thompson M A. CellCap: an encapsulation system for insecticidal biotoxin proteins. In: Kim L, editor. Advanced engineered pesticides. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 73–83. [Google Scholar]
- 122.Gamel P H, Piot J C. Characterization and properties of a novel plasmid vector for Bacillus thuringiensis displaying compatibility with host plasmids. Gene. 1992;120:17–26. doi: 10.1016/0378-1119(92)90004-9. [DOI] [PubMed] [Google Scholar]
- 123.Garczynski S F, Crim J W, Adang M J. Identification of putative insect brush border membrane-binding molecules specific to Bacillus thuringiensis δ-endotoxin by protein blot analysis. Appl Environ Microbiol. 1991;57:2816–2820. doi: 10.1128/aem.57.10.2816-2820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gawron-Burke C, Baum J A. Genetic manipulation of Bacillus thuringiensis insecticidal crystal protein genes in bacteria. In: Setlow J K, editor. Genetic engineering: principles and methods. Vol. 13. New York, N.Y: Plenum Press; 1991. pp. 237–263. [DOI] [PubMed] [Google Scholar]
- 125.Gazit E, Burshtein N, Ellar D J, Sawyer T, Shai Y. Bacillus thuringiensis cytolytic toxin associates specifically with its synthetic helices A and C in the membrane bound state. Implications for the assembly of oligomeric transmembrane pores. Biochemistry. 1997;36:15546–15554. doi: 10.1021/bi9707584. [DOI] [PubMed] [Google Scholar]
- 126.Gazit E, Shai Y. The assembly and organization of the α5 and α7 helices from the pore-forming domain of Bacillus thuringiensis δ-endotoxin. J Biol Chem. 1995;270:2571–2578. doi: 10.1074/jbc.270.6.2571. [DOI] [PubMed] [Google Scholar]
- 127.Gazit E, Shai Y. Structural and functional characterization of the α5 segment of Bacillus thuringiensis δ-endotoxin. Biochemistry. 1993;32:3429–3436. doi: 10.1021/bi00064a029. [DOI] [PubMed] [Google Scholar]
- 128.Gazit E, Shai Y. Structural characterization, membrane interaction, and specific assembly within phospholipid membranes of hydrophobic segments from Bacillus thuringiensis var. israelensis cytolytic toxin. Biochemistry. 1993;32:12363–12371. doi: 10.1021/bi00097a013. [DOI] [PubMed] [Google Scholar]
- 129.Ge A Z, Pfister R M, Dean D H. Hyperexpression of a Bacillus thuringiensis delta-endotoxin-encoding gene in Escherichia coli: properties of the product. Gene. 1990;93:49–54. doi: 10.1016/0378-1119(90)90134-d. [DOI] [PubMed] [Google Scholar]
- 130.Ge A Z, Rivers D, Milne R, Dean D H. Functional domains of Bacillus thuringiensis insecticidal crystal proteins: refinement of Heliothis virescens and Trichoplusia ni specificity domains on CryIA(c) J Biol Chem. 1991;266:17954–17958. [PubMed] [Google Scholar]
- 131.Ge A Z, Shivarova N I, Dean D H. Location of the Bombyx mori specificity domain on a Bacillus thuringiensis δ-endotoxin protein. Proc Natl Acad Sci USA. 1989;86:4037–4041. doi: 10.1073/pnas.86.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Georghiou G P, Lagunes-Tejeda A. The occurrence of resistance to pesticides in arthropods. Rome, Italy: Food and Agriculture Organization of the United Nations; 1991. [Google Scholar]
- 133.Georghiou G P, Wirth M C. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae) Appl Environ Microbiol. 1997;63:1095–1101. doi: 10.1128/aem.63.3.1095-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gill S S, Cowles E A, Pietrantonio P V. The mode of action of Bacillus thuringiensis endotoxins. Annu Rev Entomol. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- 135.Glatron M F, Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 136.Goldberg L J, Margalit J. A bacterial spore demonstrating rapid larvicidal activity against Anopheles sergentii, Uranotaenia unguiculata, Culex univitattus, Aedes aegypti, and Culex pipiens. Mosquito News. 1977;37:355–358. [Google Scholar]
- 137.González J M, Jr, Brown B J, Carlton B C. Transfer of Bacillus thuringiensis plasmids coding for δ-endotoxin among strains of B. thuringiensis and B. cereus. Proc Natl Acad Sci USA. 1982;79:6951–6955. doi: 10.1073/pnas.79.22.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.González J M, Jr, Dulmage H T, Carlton B C. Correlation between specific plasmids and δ-endotoxin production in Bacillus thuringiensis. Plasmid. 1981;5:351–365. doi: 10.1016/0147-619x(81)90010-x. [DOI] [PubMed] [Google Scholar]
- 139.Gordon R E, Haynes W C, Pang C H-N. The genus Bacillus. U.S. Washington, D.C: Department of Agriculture; 1973. [Google Scholar]
- 140.Gould F. Evolutionary biology and genetically engineered crops. BioScience. 1988;38:26–33. [Google Scholar]
- 141.Gould F. Potential and problems with high-dose strategies for pesticidal engineered crops. Biocontrol Sci Technol. 1994;4:451–461. [Google Scholar]
- 142.Gould F. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu Rev Entomol. 1998;43:701–726. doi: 10.1146/annurev.ento.43.1.701. [DOI] [PubMed] [Google Scholar]
- 143.Gould F, Anderson A, Jones A, Sumerford D, Heckel D G, Lopez J, Micinski S, Leonard R, Laster M. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. Proc Natl Acad Sci USA. 1997;94:3519–3523. doi: 10.1073/pnas.94.8.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gould F, Anderson A, Reynolds A, Bumgarner L, Moar W. Selection and genetic analysis of a Heliothis virescens (Lepidoptera: Noctuidae) strain with high levels of resistance to Bacillus thuringiensis toxins. J Econ Entomol. 1995;88:1545–1559. [Google Scholar]
- 145.Gould F, Martinez-Ramirez A, Anderson A, Ferré J, Silva F J, Moar W J. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc Natl Acad Sci USA. 1992;89:7986–7990. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Granero F, Ballester V, Ferré J. Bacillus thuringiensis crystal proteins Cry1Ab and Cry1Fa share a high affinity binding site in Plutella xylostella (L.) Biochem Biophys Res Commun. 1996;224:779–783. doi: 10.1006/bbrc.1996.1099. [DOI] [PubMed] [Google Scholar]
- 147.Green B D, Battisti L, Thorne C B. Involvement of Tn4430 in transfer of Bacillus anthracis plasmids mediated by Bacillus thuringiensis plasmid pXO12. J Bacteriol. 1989;171:104–113. doi: 10.1128/jb.171.1.104-113.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz J-L, Brousseau R, Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J Mol Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 149.Groeters F R, Tabashnik B E, Finson N, Johnson M W. Resistance to Bacillus thuringiensis affects mating success of the diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1993;86:1035–1039. [Google Scholar]
- 150.Haider M Z, Ellar D J. Analysis of the molecular basis of insecticidal specificity of Bacillus thuringiensis crystal δ-endotoxin. Biochem J. 1987;248:197–201. doi: 10.1042/bj2480197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haider M Z, Knowles B H, Ellar D J. Specificity of Bacillus thuringiensis var. colmeri insecticidal δ-endotoxin is determined by differential proteolytic processing of the protoxin by larval gut proteases. Eur J Biochem. 1986;156:531–540. doi: 10.1111/j.1432-1033.1986.tb09612.x. [DOI] [PubMed] [Google Scholar]
- 152.Hallet B, Rezsöhazy R, Delcour J. IS231A from Bacillus thuringiensis is functional in Escherichia coli: transposition and insertion specificity. J Bacteriol. 1991;173:4526–4529. doi: 10.1128/jb.173.14.4526-4529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Harvey W R, Wolfersberger M G. Mechanism of inhibition of active potassium transport in isolated midgut of Manduca sexta by Bacillus thuringiensis endotoxin. J Exp Biol. 1979;83:293–304. doi: 10.1242/jeb.83.1.293. [DOI] [PubMed] [Google Scholar]
- 154.Hastowo S, Lay B W, Ohba M. Naturally occurring Bacillus thuringiensis in Indonesia. J Appl Bacteriol. 1992;73:108–113. [Google Scholar]
- 155.Heckel D G, Gahan L C, Gould F, Anderson A. Identification of a linkage group with a major effect on resistance to Bacillus thuringiensis Cry1Ac endotoxin in the tobacco budworm (Lepidoptera: Noctuidae) J Econ Entomol. 1997;90:75–86. [Google Scholar]
- 156.Heierson A, Sidén I, Kivaisi A, Boman H G. Bacteriophage-resistant mutants of Bacillus thuringiensis with decreased virulence in pupae of Hyalophora cecropia. J Bacteriol. 1986;167:18–24. doi: 10.1128/jb.167.1.18-24.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Heimpel A M. Investigations of the mode of action of strains of Bacillus cereus Fr. and Fr. pathogenic for the larch sawfly, Pristiphor erichsonii (Htg.) Can J Zool. 1955;33:311–326. doi: 10.1139/m57-059. [DOI] [PubMed] [Google Scholar]
- 158.Himeno M, Ihara H. Molecular action of insecticides on ion channels. Washington, D.C: American Chemical Society; 1995. Mode of action of δ-endotoxin from Bacillus thuringiensis var. aizawai. In J. M. Clark (ed.) [Google Scholar]
- 159.Hodgman T C, Ellar D J. Models for the structure and function of the Bacillus thuringiensis δ-endotoxins determined by compilational analysis. DNA Seq. 1990;1:97–106. doi: 10.3109/10425179009016037. [DOI] [PubMed] [Google Scholar]
- 160.Hodgman T C, Ziniu Y, Shen J, Ellar D J. Identification of a cryptic gene associated with an insertion sequence not previously identified in Bacillus thuringiensis. FEMS Microbiol Lett. 1993;114:23–29. doi: 10.1016/0378-1097(93)90136-p. [DOI] [PubMed] [Google Scholar]
- 161.Hofmann C, Lüthy P. Binding and activity of Bacillus thuringiensis delta-endotoxin to invertebrate cells. Arch Microbiol. 1986;146:7–11. doi: 10.1007/BF00690150. [DOI] [PubMed] [Google Scholar]
- 162.Hofmann C, Lüthy P, Hütter R, Pliska V. Binding of the delta-endotoxin from Bacillus thuringiensis to brush-border membrane vesicles of the cabbage butterfly (Pieris brassicae) Eur J Biochem. 1988;173:85–91. doi: 10.1111/j.1432-1033.1988.tb13970.x. [DOI] [PubMed] [Google Scholar]
- 163.Hofmann C, Vanderbruggen H, Höfte H, Van Rie J, Jansens S, Van Mellaert H. Specificity of Bacillus thuringiensis δ-endotoxins is correlated with the presence of high-affinity binding sites in the brush border membrane of target insect midguts. Proc Natl Acad Sci USA. 1988;85:7844–7848. doi: 10.1073/pnas.85.21.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hussain, S.-R. A., O. Alzate, J. A. Cotrill, A. I. Aronson, and D. H. Dean. 1996. Unpublished observation.
- 166.Ihara H, Kuroda E, Wadano A, Himeno M. Specific toxicity of δ-endotoxins from Bacillus thuringiensis to Bombyx mori. Biosci Biotechnol Biochem. 1993;57:200–204. doi: 10.1271/bbb.57.200. [DOI] [PubMed] [Google Scholar]
- 167.Iqbal M, Verkerk R H J, Furlong M J, Rahman S A, Wright D J. Evidence for resistance to Bacillus thuringiensis (Bt) subsp. kurstaki HD-1, Bt subsp. aizawai, and avamectin in field populations of Plutella xylostella from Malaysia. Pestic Sci. 1996;48:89–97. [Google Scholar]
- 168.Ives A R, Alstad D N, Andow D A. Evolution of insect resistance to Bacillus thuringiensis-transformed plants. Science. 1996;273:1412–1413. doi: 10.1126/science.273.5280.1413. [DOI] [PubMed] [Google Scholar]
- 169.Jansens S, Van Vliet A, Dickburt C, Buysse L, Piens C, Saey B, De Wulf A, Gossele V, Paez A, Goebel E, Peferoen M. Transgenic corn expressing a Cry9C insecticidal protein from Bacillus thuringiensis protected from European corn borer damage. Crop Sci. 1997;37:1616–1624. [Google Scholar]
- 170.Jarrett P, Stephenson M. Plasmid transfer between strains of Bacillus thuringiensis infecting Galleria mellonella and Spodoptera littoralis. Appl Environ Microbiol. 1990;56:1608–1614. doi: 10.1128/aem.56.6.1608-1614.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jellis C, Bassand D, Beerman N, Dennis C, Farrell K, Piot J C, Rusche J, Carson H, Witt D. Molecular biology of Bacillus thuringiensis and potential benefits to agriculture. Isr J Entomol. 1989;23:189–199. [Google Scholar]
- 172.Jenkins, J., O. Alzate, and D. H. Dean. 1998. Unpublished observation.
- 173.Johnson D E, McGaughey W H. Contribution of Bacillus thuringiensis spores to toxicity of purified Cry proteins towards Indianmeal moth larvae. Curr Microbiol. 1996;33:54–59. doi: 10.1007/s002849900074. [DOI] [PubMed] [Google Scholar]
- 174.Johnston K A, Lee M J, Brough C, Hilder V A, Gatehouse A M R, Gatehouse J A. Protease activities in the larval midgut of Heliothis virescens: Evidence for trypsin and chymotrypsin-like enzymes. Insect Biochem Mol Biol. 1995;25:375–383. [Google Scholar]
- 175.Kaelin P, Morel P, Gadani F. Isolation of Bacillus thuringiensis from stored tobacco and Lasioderma serricorne (F.) Appl Environ Microbiol. 1994;60:19–25. doi: 10.1128/aem.60.1.19-25.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kalman S, Kiehne K L, Cooper N, Reynoso M S, Yamamoto T. Enhanced production of insecticidal proteins in Bacillus thuringiensis strains carrying an additional crystal protein gene in their chromosomes. Appl Environ Microbiol. 1995;61:3063–3068. doi: 10.1128/aem.61.8.3063-3068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kämpfer P. Application of miniaturized physiological tests in numerical classification and identification of some Bacilli. J Gen Appl Microbiol. 1991;37:225–247. [Google Scholar]
- 178.Kämpfer P. Limits and possibilities of total fatty acid analysis for classification of Bacillus species. Syst Appl Microbiol. 1994;17:86–98. [Google Scholar]
- 179.Kaneko T, Nozaki R, Aizawa K. Deoxyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Microbiol Immunol. 1978;22:639–641. doi: 10.1111/j.1348-0421.1978.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 180.Keeton T P, Bulla L A., Jr Ligand specificity and affinity of BT-R1, the Bacillus thuringiensis Cry1A toxin receptor from Manduca sexta, expressed in mammalian and insect cell cultures. Appl Environ Microbiol. 1997;63:3419–3425. doi: 10.1128/aem.63.9.3419-3425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kennedy G G, Whalon M E. Managing pest resistance to Bacillus thuringiensis endotoxins: constraints and incentives to implementation. J Econ Entomol. 1995;88:454–460. [Google Scholar]
- 183.Knight P J K, Crickmore N, Ellar D J. The receptor for Bacillus thuringiensis CryIA(c) delta-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol Microbiol. 1994;11:429–436. doi: 10.1111/j.1365-2958.1994.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Knight P J K, Knowles B H, Ellar D J. Molecular cloning of an insect aminopeptidase N that serves as a receptor for Bacillus thuringiensis CryIA(c) toxin. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]
- 185.Knowles B H. Mechanism of action of Bacillus thuringiensis insecticidal δ-endotoxins. Adv Insect Physiol. 1994;24:275–308. [Google Scholar]
- 186.Knowles B H, Dow J A T. The crystal δ-endotoxins of Bacillus thuringiensis: models for their mechanism of action on the insect gut. Bioessays. 1993;15:469–476. [Google Scholar]
- 187.Knowles B H, Ellar D J. Colloid-osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis δ-endotoxins with different insect specificity. Biochim Biophys Acta. 1987;924:509–518. [Google Scholar]
- 188.Knowles B H, Thomas W E, Ellar D J. Lectin-like binding of Bacillus thuringiensis var. kurstaki lipidopteran-specific toxin is an initial step in insecticidal action. FEBS Lett. 1984;168:197–202. doi: 10.1016/0014-5793(84)80245-8. [DOI] [PubMed] [Google Scholar]
- 189.Koller C N, Bauer L S, Hollingworth R M. Characterization of the pH-mediated solubility of Bacillus thuringiensis var. san diego native δ-endotoxin crystals. Biochem Biophys Res Commun. 1992;184:692–699. doi: 10.1016/0006-291x(92)90645-2. [DOI] [PubMed] [Google Scholar]
- 190.Kolstø, A.-B. Personal communication.
- 191.Koni P A, Ellar D J. Biochemical characterization of Bacillus thuringiensis cytolytic δ-endotoxins. Microbiology. 1994;140:1869–1880. doi: 10.1099/13500872-140-8-1869. [DOI] [PubMed] [Google Scholar]
- 192.Kostichka K, Warren G W, Mullins M, Mullins A D, Craig J A, Koziel M G, Estruch J J. Cloning of a cryV-type insecticidal protein gene from Bacillus thuringiensis: the cryV-encoded protein is expressed early in stationary phase. J Bacteriol. 1996;178:2141–2144. doi: 10.1128/jb.178.7.2141-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Koziel M G, Beland G L, Bowman C, Carozzi N B, Crenshaw R, Crossland L, Dawson J, Desai N, Hill M, Kadwell S, Launis K, Lewis K, Maddox D, McPherson K, Meghji M R, Merlin E, Rhodes R, Warren G W, Wright M, Evola S V. Field performance of elite transgenic maize plants expressing an insecticidal protein derived from Bacillus thuringiensis. Bio/Technology. 1993;11:194–200. [Google Scholar]
- 194.Koziel M G, Carozzi N B, Currier T C, Warren G W, Evola S V. The insecticidal crystal proteins of Bacillus thuringiensis: past, present and future uses. Biotech Genet Eng Rev. 1993;11:171–228. [Google Scholar]
- 195.Kronstad J W, Whiteley H R. Inverted repeat sequences flank a Bacillus thuringiensis crystal protein gene. J Bacteriol. 1984;160:95–102. doi: 10.1128/jb.160.1.95-102.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kronstad J W, Whiteley H R. Three classes of homologous Bacillus thuringiensis crystal-protein genes. Gene. 1986;43:29–40. doi: 10.1016/0378-1119(86)90005-3. [DOI] [PubMed] [Google Scholar]
- 197.Kwak I-S, Lu H, Dean D H. Exploration of receptor binding of Bacillus thuringiensis toxins. Mem Inst Oswaldo Cruz. 1995;90:75–79. doi: 10.1590/s0074-02761995000100017. [DOI] [PubMed] [Google Scholar]
- 198.Lambert B, Buysse L, Decock C, Jansens S, Piens C, Saey B, Seurinck J, Van Audenhove K, Van Rie J, Van Vliet A, Peferoen M. A Bacillus thuringiensis insecticidal protein with a high activity against members of the family Noctuidae. Appl Environ Microbiol. 1996;62:80–86. doi: 10.1128/aem.62.1.80-86.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lambert B, Peferoen M. Insecticidal promise of Bacillus thuringiensis. Facts and mysteries about a successful biopesticide. BioScience. 1992;42:112–122. [Google Scholar]
- 200.Lambert B, Theunis W, Agouda R, Van Audenhove K, C. D, Jansens S, Seurinck J, Peferoen M. Nucleotide sequence of gene cryIIID encoding a novel coleopteran-active crystal protein from strain BTI109P of Bacillus thuringiensis subsp. kurstaki. Gene. 1992;110:131–132. doi: 10.1016/0378-1119(92)90457-z. [DOI] [PubMed] [Google Scholar]
- 201.Lampel J S, Canter G L, Dimock M B, Kelly J L, Anderson J J, Uratani B B, Foulke J S, Jr, Turner J T. Integrative cloning, expression, and stability of the cryIA(c) gene from Bacillus thuringiensis subsp. kurstaki in a recombinant strain of Clavibacter xyli subsp. cynodontis. Appl Environ Microbiol. 1994;60:501–508. doi: 10.1128/aem.60.2.501-508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lecadet, M.-M. 1997. Personal communication.
- 203.Lecadet M-M, Dedonder R. Enzymatic hydrolysis of the crystals of Bacillus thuringiensis by the proteases of Pieris brassicae. I. Preparation and fractionation of the lysates. J Invertebr Pathol. 1967;9:310–321. [Google Scholar]
- 204.Lecadet M-M, Dedonder R. Les protéases de Pieris brassicae. I. Purification et propriétés. Bull Soc Chim Biol. 1966;48:631–660. [PubMed] [Google Scholar]
- 205.Lecadet M-M, Frachon E. Presented at the XXVIIth Annual Meeting of the Society for Invertebrate Pathology, Montpellier, France. 1994. [Google Scholar]
- 206.Lee M K, Aguda R M, Cohen M B, Gould F L, Dean D H. Determination of binding of Bacillus thuringiensis δ-endotoxin receptors to rice stem borer midguts. Appl Environ Microbiol. 1997;63:1453–1459. doi: 10.1128/aem.63.4.1453-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lee M K, Curtiss A, Alcantara E, Dean D H. Synergistic effect of the Bacillus thuringiensis toxins CryIAa and CryIAc on the gypsy moth, Lymantria dispar. Appl Environ Microbiol. 1996;62:583–586. doi: 10.1128/aem.62.2.583-586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lee M K, Dean D H. Inconsistencies in determining Bacillus thuringiensis toxin binding sites relationship by comparing competition assays with ligand blotting. Biochem Biophys Res Commun. 1996;220:575–580. doi: 10.1006/bbrc.1996.0445. [DOI] [PubMed] [Google Scholar]
- 209.Lee M K, Milne R E, Ge A Z, Dean D H. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis δ-endotoxin. J Biol Chem. 1992;267:3115–3121. [PubMed] [Google Scholar]
- 210.Lee M K, Rajamohan F, Gould F, Dean D H. Resistance to Bacillus thuringiensis CryIA δ-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration. Appl Environ Microbiol. 1995;61:3836–3842. doi: 10.1128/aem.61.11.3836-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lee M K, Young B A, Dean D H. Domain III exchanges of Bacillus thuringiensis CryIA toxins affect binding to different gypsy moth midgut receptors. Biochem Biophys Res Commun. 1995;216:306–312. doi: 10.1006/bbrc.1995.2625. [DOI] [PubMed] [Google Scholar]
- 212.Léonard C, Chen Y, Mahillon J. Diversity and different distribution of IS231, IS232 and IS240 among Bacillus cereus, Bacillus thuringiensis and Bacillus mycoides. Microbiology. 1997;143:2537–2547. doi: 10.1099/00221287-143-8-2537. [DOI] [PubMed] [Google Scholar]
- 213.Lereclus D, Agaisse H, Gominet M, Chaufaux J. Overproduction of encapsulated insecticidal crystal proteins in a Bacillus thuringiensis sp0A mutant. Bio/Technology. 1995;13:67–71. doi: 10.1038/nbt0195-67. [DOI] [PubMed] [Google Scholar]
- 214.Lereclus D, Arantès O, Chaufaux J, Lecadet M-M. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;60:211–218. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 215.Lereclus D, Bourgouin C, Lecadet M-M, Klier A, Rapoport G. Role, structure, and molecular organization of the genes coding for parasporal δ-endotoxins of Bacillus thuringiensis. In I. Smith, R. A. Slepecky, and P. Setlow (ed.), Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. Washington, D.C: American Society for Microbiology; 1989. [Google Scholar]
- 216.Lereclus D, Mahillon J, Menou G, Lecadet M-M. Identification of Tn4430, a transposon of Bacillus thuringiensis functional in Escherichia coli. Mol Gen Genet. 1986;204:52–57. doi: 10.1007/BF00330186. [DOI] [PubMed] [Google Scholar]
- 217.Lereclus D, Menou G, Lecadet M-M. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAMβ1. Mol Gen Genet. 1983;191:307–313. doi: 10.1007/BF00334831. [DOI] [PubMed] [Google Scholar]
- 218.Lereclus D, Ribier J, Klier A, Menou G, Lecadet M-M. A transposon-like structure related to the δ-endotoxin gene of Bacillus thuringiensis. EMBO J. 1984;3:2561–2567. doi: 10.1002/j.1460-2075.1984.tb02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lereclus D, Vallade M, Chaufaux J, Arantes O, Rambaud S. Expansion of the insecticidal host range of Bacillus thuringiensis by in vivo genetic recombination. Bio/Technology. 1992;10:418–421. doi: 10.1038/nbt0492-418. [DOI] [PubMed] [Google Scholar]
- 220.Lesieru C, Vecsey-Semjen B, Abrami L, Fivaz M, Van Der Goot F G. Membrane insertion: the strategies of toxins. Mol Membr Biol. 1997;14:45–64. doi: 10.3109/09687689709068435. [DOI] [PubMed] [Google Scholar]
- 221.Levinson B L. High-performance liquid chromatography analysis of two beta-exotoxins produced by some Bacillus thuringiensis strains. In: Hickle L A, Fitch W L, editors. Analytical chemistry of Bacillus thuringiensis. Washington, D.C: American Chemical Society; 1990. pp. 115–136. [Google Scholar]
- 222.Li J, Carroll J, Ellar D J. Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 223.Li J, Koni P A, Ellar D J. Structure of the mosquitocidal δ-endotoxin CytB from Bacillus thuringiensis sp. kyushuensis and implications for membrane pore formation. J Mol Biol. 1996;257:129–152. doi: 10.1006/jmbi.1996.0152. [DOI] [PubMed] [Google Scholar]
- 224.Liang Y, Patel S S, Dean D H. Irreversible binding kinetics of Bacillus thuringiensis CryIA δ-endotoxins to gypsy moth brush border membrane vesicles is directly correlated to toxicity. J Biol Chem. 1995;270:24719–24724. doi: 10.1074/jbc.270.42.24719. [DOI] [PubMed] [Google Scholar]
- 225.Liebig B, Stetson D L, Dean D H. Quantification of the effect of Bacillus thuringiensis toxins on short-circuit current in the midgut of Bombyx mori. J Insect Physiol. 1995;41:17–22. [Google Scholar]
- 226.Liu Y-B, Tabashnik B E. Experimental evidence that refuges delay insect adaptation to Bacillus thuringiensis. Proc R Soc Lond Ser B. 1997;264:605–610. [Google Scholar]
- 227.Liu Y-B, Tabashnik B E. Inheritance of resistance to the Bacillus thuringiensis toxin Cry1C in the diamondback moth. Appl Environ Microbiol. 1997;63:2218–2223. doi: 10.1128/aem.63.6.2218-2223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu Y-B, Tabashnik B E, Pusztai-Carey M. Field-evolved resistance to Bacillus thuringiensis toxin CryIC in diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1996;89:798–804. [Google Scholar]
- 229.Logan N A, Berkley R C W. Identification of Bacillus strains using the API system. J Gen Microbiol. 1984;130:1871–1882. doi: 10.1099/00221287-130-7-1871. [DOI] [PubMed] [Google Scholar]
- 230.Lorence A, Darszon A, Diaz C, Liévano A, Quintero R, Bravo A. δ-endotoxins induce cation channels in Spodoptera frugiperda brush border membranes in suspension and in planar lipid bilayers. FEBS Lett. 1995;360:217–222. doi: 10.1016/0014-5793(95)00092-n. [DOI] [PubMed] [Google Scholar]
- 231.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 232.Lövgren A, Zhang M-Y, Engström A, Dalhammar G, Landén R. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol Microbiol. 1990;4:2137–2146. doi: 10.1111/j.1365-2958.1990.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 233.Lu H, Rajamohan F, Dean D H. Identification of amino acid residues of Bacillus thuringiensis δ-endotoxin CryIAa associated with membrane binding and toxicity to Bombyx mori. J Bacteriol. 1994;176:5554–5559. doi: 10.1128/jb.176.17.5554-5559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Luo K, Lu Y-J, Adang M J. A 106 kDa form of aminopeptidase is a receptor for Bacillus thuringiensis CryIC delta-endotoxin in the brush border membrane of Manduca sexta. Insect Biochem Mol Biol. 1996;26:783–791. [Google Scholar]
- 235.Luo K, Sangadala S, Masson L, Mazza A, Brousseau R, Adang M J. The Heliothis virescens 170 kDa aminopeptidase functions as “receptor A” by mediating specific Bacillus thuringiensis Cry1A δ-endotoxin binding and pore formation. Insect Biochem Mol Biol. 1997;27:735–743. doi: 10.1016/s0965-1748(97)00052-0. [DOI] [PubMed] [Google Scholar]
- 236.Luo K, Tabashnik B E, Adang M J. Binding of Bacillus thuringiensis Cry1Ac toxin to aminopeptidase in susceptible and resistant diamondback moths (Plutella xylostella) Appl Environ Microbiol. 1997;63:1024–1027. doi: 10.1128/aem.63.3.1024-1027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lysenko O. Bacillus thuringiensis: evolution of a taxonomic conception. J Invertebr Pathol. 1983;42:295–298. [Google Scholar]
- 238.Macaluso A, Mettus A M. Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. J Bacteriol. 1991;173:1353–1356. doi: 10.1128/jb.173.3.1353-1356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Machesky H. USDA forest service gypsy moth aerial suppression/eradication projects. Gypsy Moth News. 1989;20:2–3. [Google Scholar]
- 240.MacIntosh S C, Stone T B, Jokerst R S, Fuchs R L. Binding of Bacillus thuringiensis proteins to a laboratory-selected line of Heliothis virescens. Proc Natl Acad Sci USA. 1991;88:8930–8933. doi: 10.1073/pnas.88.20.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.MacKinnon R, Miller C. Mutant potassium channels with altered binding of charybdotoxin, a pore-blocking peptide inhibitor. Science. 1989;245:1382–1385. doi: 10.1126/science.2476850. [DOI] [PubMed] [Google Scholar]
- 242.MacKinnon R, Yellen G. Mutations affecting TEA blockade and ion permeation in voltage-activated K+ channels. Science. 1990;250:276–279. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- 243.Maddrell S H P, Lane N J, Harrison J B, Overton J A, Moreton R B. The initial stages in the action of an insecticidal δ-endotoxin of Bacillus thuringiensis var. israelensis on the epithelial cells of the Malpighian tubules of the insect, Rhodnius prolixus. J Cell Sci. 1988;90:131–144. doi: 10.1242/jcs.90.1.131. [DOI] [PubMed] [Google Scholar]
- 244.Maddrell S H P, Overton J A, Ellar D J, Knowles B H. Action of activated 27 000 Mr toxin from Bacillus thuringiensis var. israelensis on Malpighian tubules of the insect, Rhodnius prolixus. J Cell Sci. 1989;94:601–608. doi: 10.1242/jcs.94.3.601. [DOI] [PubMed] [Google Scholar]
- 245.Mahaffee W F, Moar W J, Kloepper J W. Bacterial endophytes genetically engineered to express the CryIIA δ-endotoxin from Bacillus thuringiensis subsp. kurstaki. In: Ryder M H, Stevens P M, Bowen G D, editors. Improving plant productivity with rhizosphere bacteria. East Melbourne, Victoria, Australia: CSIRO Publications; 1994. pp. 245–246. [Google Scholar]
- 246.Mahillon J, Chungjatupornchai W, Decock J, Dierickx S, Michiels F, Peferoen M, Joos H. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett. 1989;60:205–210. [Google Scholar]
- 247.Mahillon J, Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988;7:1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mahillon J, Rezsöhazy R, Hallet B, Delcour J. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica. 1994;93:13–26. doi: 10.1007/BF01435236. [DOI] [PubMed] [Google Scholar]
- 249.Mahillon J, Seurinck J, Delcour J, Zabeau M. Cloning and nucleotide sequence of different iso-IS231 elements and their structural association with the Tn4430 transposon in Bacillus thuringiensis. Gene. 1987;51:187–196. doi: 10.1016/0378-1119(87)90307-6. [DOI] [PubMed] [Google Scholar]
- 250.Mallet J, Porter P. Preventing insect adaptation to insect-resistant crops: are seed mixtures or refugia the best strategy? Proc R Soc Lond Ser B. 1992;250:165–169. [Google Scholar]
- 251.Malvar T, Baum J A. Tn5401 disruption of the spo0F gene, identified by direct chromosomal sequencing, results in CryIIIA overproduction in Bacillus thuringiensis. J Bacteriol. 1994;176:4750–4753. doi: 10.1128/jb.176.15.4750-4753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Malvar T, Gawron-Burke C, Baum J A. Overexpression of Bacillus thuringiensis HknA, a histidine protein kinase homolog, bypasses early Spo− mutations that result in CryIIIA overproduction. J Bacteriol. 1994;176:4742–4749. doi: 10.1128/jb.176.15.4742-4749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Manker, D. C., W. D. Lidster, R. L. Starnes, and S. C. MacIntosh. May 1994. World Intellectual Property Organization patent WO 94/09630.
- 254.Martin F G, Wolfersberger M G. Bacillus thuringiensis δ-endotoxin and larval Manduca sexta midgut brush-border membrane vesicles act synergistically to cause very large increases in the conductance of planar lipid bilayers. J Exp Biol. 1995;198:91–96. doi: 10.1242/jeb.198.1.91. [DOI] [PubMed] [Google Scholar]
- 255.Martin P A W, Travers R S. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl Environ Microbiol. 1989;55:2437–2442. doi: 10.1128/aem.55.10.2437-2442.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Masson L, Lu Y-J, Mazza A, Brousseau R, Adang M J. The CryIA(c) receptor purified from Manduca sexta displays multiple specificities. J Biol Chem. 1995;270:20309–20315. doi: 10.1074/jbc.270.35.20309. [DOI] [PubMed] [Google Scholar]
- 257.Masson L, Mazza A, Brousseau R, Tabashnik B. Kinetics of Bacillus thuringiensis toxin binding with brush border membrane vesicles from susceptible and resistant larvae of Plutella xylostella. J Biol Chem. 1995;270:11887–11896. doi: 10.1074/jbc.270.20.11887. [DOI] [PubMed] [Google Scholar]
- 258.Masson L, Mazza A, Gringorten L, Baines D, Aneliunas V, Brousseau R. Specificity domain localization of Bacillus thuringiensis insecticidal toxins is highly dependent on the bioassay system. Mol Microbiol. 1994;14:851–860. doi: 10.1111/j.1365-2958.1994.tb01321.x. [DOI] [PubMed] [Google Scholar]
- 259.Masson L, Préfontaine G, Brousseau R. Transformation of Bacillus thuringiensis vegetative cells by electroporation. FEMS Microbiol Lett. 1989;60:273–278. doi: 10.1016/0378-1097(89)90409-6. [DOI] [PubMed] [Google Scholar]
- 260.McBride K E, Svab Z, Schaaf D J, Hogan P S, Stalker D M, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of insecticidal crystal protein in tobacco. Bio/Technology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 261.McGaughey W H. Evaluation of Bacillus thuringiensis for controlling Indianmeal moths (Lepidoptera: Pyralidae) in farm grain bins and elevator silos. J Econ Entomol. 1985;78:1089–1094. [Google Scholar]
- 262.McGaughey W H. Insect resistance to the biological insecticide Bacillus thuringiensis. Science. 1985;229:193–195. doi: 10.1126/science.229.4709.193. [DOI] [PubMed] [Google Scholar]
- 263.McGaughey W H, Beeman R W. Resistance to Bacillus thuringiensis in colonies of Indianmeal moth and almond moth (Lepidoptera: Pyralidae) J Econ Entomol. 1988;81:28–33. [Google Scholar]
- 264.McGaughey W H, Johnson D E. Influence of crystal protein composition of Bacillus thuringiensis strains on cross-resistance in Indianmeal moths (Lepidoptera: Pyralidae) J Econ Entomol. 1994;87:535–540. [Google Scholar]
- 265.McGaughey W H, Whalon M E. Managing insect resistance to Bacillus thuringiensis toxins. Science. 1992;258:1451–1455. doi: 10.1126/science.258.5087.1451. [DOI] [PubMed] [Google Scholar]
- 266.Meadows M P. Bacillus thuringiensis in the environment: ecology and risk assessment. In: Entwistle P F, Cory J S, Bailey M J, Higgs S, editors. Bacillus thuringiensis, an environmental biopesticide: theory and practice. New York, N.Y: Wiley; 1993. pp. 193–220. [Google Scholar]
- 267.Meadows M P, Ellis D J, Butt J, Jarrett P, Burges H D. Distribution, frequency, and diversity of Bacillus thuringiensis in an animal feed mill. Appl Environ Microbiol. 1992;58:1344–1350. doi: 10.1128/aem.58.4.1344-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Menou G, Mahillon J, Lecadet M-M, Lereclus D. Structural and genetic organization of IS232, a new insertion sequence of Bacillus thuringiensis. J Bacteriol. 1990;172:6689–6696. doi: 10.1128/jb.172.12.6689-6696.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Metz T D, Roush R T, Tang J D, Shelton A M, Earle E D. Transgenic broccoli expressing a Bacillus thuringiensis insecticidal crystal protein: implications for pest management strategies. Mol Breed. 1995;1:309–317. [Google Scholar]
- 270.Milne R, Kaplan H. Purification and characterization of a trypsin-like digestive enzyme from spruce budworm (Choristoneura fumiferana) responsible for the activation of δ-endotoxin from Bacillus thuringiensis. Insect Biochem Mol Biol. 1993;23:663–673. doi: 10.1016/0965-1748(93)90040-y. [DOI] [PubMed] [Google Scholar]
- 271.Miyasono M, Inagaki S, Yamamoto M, Ohba K, Ishiguro T, Takeda R, Hayashi Y. Enhancement of δ-endotoxin activity by toxin-free spore of Bacillus thuringiensis against the Diamondback Moth, Plutella xylostella. J Invertebr Pathol. 1994;63:111–112. [Google Scholar]
- 272.Moar W J, Pusztai-Carey M, van Faassen H, Bosch D, Frutos R, Rang C, Luo K, Adang M J. Development of Bacillus thuringiensis CryIC resistance by Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae) Appl Environ Microbiol. 1995;61:2086–2092. doi: 10.1128/aem.61.6.2086-2092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Moar W J, Trumble J T, Federici B A. Comparative toxicity of spores and crystals from the NRD-12 and HD-1 strains of Bacillus thuringiensis subsp. kurstaki to neonate beet armyworm (Lepidoptera: Noctuidae) J Econ Entomol. 1989;82:1593–1603. [Google Scholar]
- 274.Mohammed S I, Johnson D E, Aronson A I. Altered binding of the Cry1Ac toxin to larval membranes but not to the toxin-binding protein in Plodia interpunctella selected for resistance to different Bacillus thuringiensis isolates. Appl Environ Microbiol. 1996;62:4168–4173. doi: 10.1128/aem.62.11.4168-4173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Moran C P. RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 276.Müller-Cohn J, Chaufaux J, Buisson C, Gilois N, Sanchis V, Lereclus D. Spodoptera littoralis (Lepidoptera: Noctuidae) resistance to CryIC and cross-resistance to other Bacillus thuringiensis crystal toxins. J Econ Entomol. 1996;89:791–797. [Google Scholar]
- 277.Nagamatsu Y, Itai Y, Hatanaka C, Funatsu G, Hayashi K. A toxic fragment from the entomocidal crystal protein of Bacillus thuringiensis. Agric Biol Chem. 1984;48:611–619. [Google Scholar]
- 278.Nayak P, Basu D, Das S, Basu A, Ghosh D, Ramakrishnan N A, Ghosh M, Sen S K. Transgenic elite indica rice plants expressing CryIAc delta-endotoxin of Bacillus thuringiensis are resistant against yellow stem borer (Scirpophaga incertulas) Proc Natl Acad Sci USA. 1997;94:2111–2116. doi: 10.1073/pnas.94.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Nierlich D P, Murakawa G J. The decay of bacterial messenger RNA. Prog Nucl Acid Res Mol Biol. 1996;52:153–216. doi: 10.1016/s0079-6603(08)60967-8. [DOI] [PubMed] [Google Scholar]
- 280.Novillo C, Castañera P, Ortego F. Characterization and distribution of chymotrypsin-like and other digestive proteases in Colorado potato beetle larvae. Arch Insect Biochem Physiol. 1997;36:181–201. [Google Scholar]
- 281.Obukowicz M G, Perlak F J, Kusano-Kretzmer K, Mayer E J, Bolten S L, Watrud L S. Tn5-mediated integration of the delta-endotoxin gene from Bacillus thuringiensis into the chromosome of root-colonizing pseudomonads. J Bacteriol. 1986;168:982–989. doi: 10.1128/jb.168.2.982-989.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Obukowicz M G, Perlak F J, Kusano-Kretzmer K, Mayer E J, Watrud L S. Integration of the delta endotoxin gene of Bacillus thuringiensis into the chromosome of root-colonizing strains of pseudomonads using Tn5. Gene. 1986;45:327–331. doi: 10.1016/0378-1119(86)90031-4. [DOI] [PubMed] [Google Scholar]
- 283.Ojcius D M, Young J D E. Cytolytic pore-forming proteins and peptides—is there a common structural motif? Trends Biochem Sci. 1991;16:225–229. doi: 10.1016/0968-0004(91)90090-i. [DOI] [PubMed] [Google Scholar]
- 284.Oppert B, Kramer K J, Beeman R W, Johnson D, Mcgaughey W H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J Biol Chem. 1997;272:23473–23476. doi: 10.1074/jbc.272.38.23473. [DOI] [PubMed] [Google Scholar]
- 285.Oppert B, Kramer K J, Johnson D E, MacIntosh S C, McGaughey W H. Altered protoxin activation by midgut enzymes from a Bacillus thuringiensis resistant strain of Plodia interpunctella. Biochem Biophys Res Commun. 1994;198:940–947. doi: 10.1006/bbrc.1994.1134. [DOI] [PubMed] [Google Scholar]
- 286.Padua L E, Ohba M, Aizawa K. The isolates of Bacillus thuringiensis serotype 10 with a highly preferential toxicity to mosquito larvae. J Invertebr Pathol. 1980;36:180–186. doi: 10.1016/0022-2011(80)90022-1. [DOI] [PubMed] [Google Scholar]
- 287.Parker M W, Pattus F. Rendering a membrane protein soluble in water: a common packing motif in bacterial protein toxins. Trends Biochem Sci. 1993;18:391–395. doi: 10.1016/0968-0004(93)90096-6. [DOI] [PubMed] [Google Scholar]
- 288.Parker M W, Postma J P M, Pattus F, Tucker A D, Tsernoglou D. Refined structure of pore-forming domain of colicin A at 2.4 Å resolution. J Mol Biol. 1992;224:639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 289.Payne, J., K. A. Uyeda, C. J. Stalder, and T. E. Michaels. March 1994. U. S. patient 5,298,245.
- 290.Peferoen M. Progress and prospects for field use of Bt genes in crops. Trends Biotechnol. 1997;15:173–177. [Google Scholar]
- 291.Peferoen M, Jansens S, Reynaerts A, Leemans J. In: Molecular and cellular biology of the potato. Vayda M E, Park W C, editors. Wallingford, United Kingdom: CAB International; 1990. p. 193. [Google Scholar]
- 292.Perez C J, Shelton A M. Resistance of Plutella xylostella (Lepidoptera: Plutellidae) to Bacillus thuringiensis Berliner in Central America. J Econ Entomol. 1997;90:87–93. [Google Scholar]
- 293.Perez C J, Tang J D, Shelton A M. Comparison of leaf-dip and diet bioassays for monitoring Bacillus thuringiensis resistance in field populations of diamondback moth (Lepidoptera:Plutelliidae) J Econ Entomol. 1997;90:94–101. [Google Scholar]
- 294.Perlak F J, Deaton R W, Armstrong T A, Fuchs R L, Sims S R, Greenplate J T, Fischhoff D A. Insect resistant cotton plants. Bio/Technology. 1990;8:939–943. doi: 10.1038/nbt1090-939. [DOI] [PubMed] [Google Scholar]
- 295.Perlak F J, Fuchs R L, Dean D A, McPherson S L, Fischhoff D A. Modification of the coding sequence enhances plant expression of insect control protein genes. Proc Natl Acad Sci USA. 1991;88:3324–3328. doi: 10.1073/pnas.88.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Perlak F J, Stone T B, Muskopf Y M, Petersen L J, Parker G B, McPherson S A, Wyman J, Love S, Reed G, Biever D, Fischhoff D A. Genetically improved potatoes: protection from damage by Colorado potato beetles. Plant Mol Biol. 1993;22:313–321. doi: 10.1007/BF00014938. [DOI] [PubMed] [Google Scholar]
- 297.Peterson A M, Fernando G J P, Wells M A. Purification, characterization and cDNA sequence of an alkaline chymotrypsin from the midgut of Manduca sexta. Insect Biochem Mol Biol. 1995;25:765–774. doi: 10.1016/0965-1748(94)00092-v. [DOI] [PubMed] [Google Scholar]
- 298.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 299.Petras S F, Casida L E., Jr Survival of Bacillus thuringiensis spores in soil. Appl Environ Microbiol. 1985;50:1496–1501. doi: 10.1128/aem.50.6.1496-1501.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Phizicky E M, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Poncet S, Anello G, Delécluse A, Klier A, Rapoport G. Role of the CryIVD polypeptide in the overall toxicity of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 1993;59:3928–3930. doi: 10.1128/aem.59.11.3928-3930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Poncet S, Delécluse A, Anello G, Klier A, Rapoport G. Transfer and expression of the cryIVB and cryIVD genes of Bacillus thuringiensis subsp. israelensis in Bacillus sphaericus 2297. FEMS Microbiol Lett. 1994;117:91–96. [Google Scholar]
- 303.Poncet S, Delécluse A, Klier A, Rapoport G. Evaluation of synergistic interactions between the CryIVA, CryIVB and CryIVD toxic components of B. thuringiensis subsp. israelensis crystals. J Invertebr Pathol. 1995;66:131–135. [Google Scholar]
- 304.Poncet S, Dervyn E, Klier A, Rapoport G. Spo0A represses transcription of the cry toxin genes in Bacillus thuringiensis. Microbiology. 1997;143:2743–2751. doi: 10.1099/00221287-143-8-2743. [DOI] [PubMed] [Google Scholar]
- 305.Priest F G, Goodfellow M, Todd C. A numerical classification of the genus Bacillus. J Gen Microbiol. 1988;134:1847–1882. doi: 10.1099/00221287-134-7-1847. [DOI] [PubMed] [Google Scholar]
- 306.Ragni A, Thiéry I, Delécluse A. Characterization of six highly mosquitocidal Bacillus thuringiensis strains that do not belong to the H-14 serotype. Curr Microbiol. 1996;32:48–54. doi: 10.1007/s002849900009. [DOI] [PubMed] [Google Scholar]
- 307.Rajamohan F, Alcantara E, Lee M K, Chen X J, Curtiss A, Dean D H. Single amino acid changes in domain II of Bacillus thuringiensis CryIAb δ-endotoxin affect irreversible binding to Manduca sexta midgut membrane vesicles. J Bacteriol. 1995;177:2276–2282. doi: 10.1128/jb.177.9.2276-2282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Rajamohan F, Alzate O, Cotrill J A, Curtiss A, Dean D H. Protein engineering of Bacillus thuringiensis δ-endotoxin: mutations at domain II of Cry1Ab enhance receptor affinity and toxicity towards gypsy moth larvae. Proc Natl Acad Sci USA. 1996;93:14338–14343. doi: 10.1073/pnas.93.25.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rajamohan F, Cotrill J A, Gould F, Dean D H. Role of domain II, loop 2 residues of Bacillus thuringiensis CryIAb δ-endotoxin in reversible and irreversible binding to Manduca sexta and Heliothis virescens. J Biol Chem. 1996;271:2390–2396. doi: 10.1074/jbc.271.5.2390. [DOI] [PubMed] [Google Scholar]
- 310.Rajamohan F, Hussain S-R A, Cotrill J A, Gould F, Dean D H. Mutations in domain II, loop 3 of Bacillus thuringiensis CryIAa and CryIAb δ-endotoxins suggest loop 3 is involved in initial binding to lepidopteran midguts. J Biol Chem. 1996;271:25220–25226. doi: 10.1074/jbc.271.41.25220. [DOI] [PubMed] [Google Scholar]
- 311.Rajamohan F, Lee M K, Dean D H. Bacillus thuringiensis insecticidal proteins: molecular mode of action. Progress in Nucleic Acid Research and Molecular Biology. Vol. 60. New York, N.Y: Academic Press; 1998. [DOI] [PubMed] [Google Scholar]
- 312.Ravoahangimalala O, Charles J-F. In vitro binding of Bacillus thuringiensis var. israelensis individual toxins to midgut cells of Anopheles gambiae larvae (Diptera: Culicidae) FEBS Lett. 1995;362:111–115. doi: 10.1016/0014-5793(95)00220-4. [DOI] [PubMed] [Google Scholar]
- 313.Ravoahangimalala O, Charles J F, Schoeller-Raccaud J. Immunological localization of Bacillus thuringiensis serovar israelensis toxins in midgut cells of intoxicated Anopheles gambiae larvae Diptera: Culicidae. Res Microbiol. 1993;144:271–278. doi: 10.1016/0923-2508(93)90011-p. [DOI] [PubMed] [Google Scholar]
- 314.Rezsöhazy R, Hallet B, Delcour J. IS231D, E and F, three new insertion sequences in Bacillus thuringiensis: extension of the IS231 family. Mol Microbiol. 1992;6:1959–1967. doi: 10.1111/j.1365-2958.1992.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 315.Rezsöhazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 316.Rezsöhazy R, Hallet B, Mahillon J, Delcour J. IS231V and W from Bacillus thuringiensis subsp. israelensis, two distant members of the IS231 family of insertion sequences. Plasmid. 1993;30:141–149. doi: 10.1006/plas.1993.1041. [DOI] [PubMed] [Google Scholar]
- 317.Robertson J L, Preisler H K, Ng S S, Hickle L A, Gelernter W D. Natural variation: a complicating factor in bioassays with chemical and microbial pesticides. J Econ Entomol. 1995;88:1–10. [Google Scholar]
- 318.Rössler D, Ludwig W, Schleifer K H, Lin C, McGill T J, Wisotzkey J D, Jurtshuk P, Jr, Fox G E. Phylogenetic diversity in the genus Bacillus as seen by 16S rRNA sequencing studies. Syst Appl Microbiol. 1991;14:266–269. doi: 10.1016/S0723-2020(11)80379-6. [DOI] [PubMed] [Google Scholar]
- 319.Rosso M-L, Delécluse A. Distribution of the insertion element IS240 among Bacillus thuringiensis strains. Curr Microbiol. 1997;34:348–353. doi: 10.1007/s002849900194. [DOI] [PubMed] [Google Scholar]
- 320.Roush R T. Can we slow adaptation by pests to insect transgenic crops? In: Persley G J, editor. Biotechnology and integrated pest management. Wallingford, United Kingdom: CAB International; 1996. pp. 242–263. [Google Scholar]
- 321.Roush R T. Managing pests and their resistance to Bacillus thuringiensis: can transgenic crops be better than sprays? Biocontrol Sci Technol. 1994;4:501–516. [Google Scholar]
- 322.Rowe G E, Margaritis A, Dulmage H T. Bioprocess developments in the production of bioinsecticides by Bacillus thuringiensis. Crit Rev Biotechnol. 1987;6:87–127. [Google Scholar]
- 323.Sacchi V F, Parenti P, Hanozet G M, Giordana B, Lüthy P, Wolfersberger M G. Bacillus thuringiensis toxin inhibits K+-gradient-dependent amino acid transport across the brush border membrane of Pieris brassicae midgut cells. FEBS Lett. 1986;204:213–218. [Google Scholar]
- 324.Salamitou S, Agaisse H, Bravo A, Lereclus D. Genetic analysis of cryIIIA gene expression in Bacillus thuringiensis. Microbiology. 1996;142:2049–2055. doi: 10.1099/13500872-142-8-2049. [DOI] [PubMed] [Google Scholar]
- 325.Sanchis V, Agaisse H, Chaufaux J, Lereclus D. Construction of new insecticidal Bacillus thuringiensis recombinant strains by using the sporulation non-dependent expression system of cryIIIA and a site specific recombination vector. J Biotechnol. 1996;48:81–96. doi: 10.1016/0168-1656(96)01404-6. [DOI] [PubMed] [Google Scholar]
- 326.Sanchis V, Agaisse H, Chaufaux J, Lereclus D. A recombinase-mediated system for elimination of antibiotic resistance gene markers from genetically engineered Bacillus thuringiensis strains. Appl Environ Microbiol. 1997;63:779–784. doi: 10.1128/aem.63.2.779-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Sanchis V, Ellar D J. Identification and partial purification of a Bacillus thuringiensis CryIC δ-endotoxin binding protein from Spodoptera littoralis gut membranes. FEBS Lett. 1993;316:264–268. doi: 10.1016/0014-5793(93)81305-j. [DOI] [PubMed] [Google Scholar]
- 328.Sanchis V, Lereclus D, Menou G, Chaufaux J, Lecadet M-M. Multiplicity of δ-endotoxin genes with different insecticidal specificities in Bacillus thuringiensis aizawai 7.29. Mol Microbiol. 1988;2:393–404. doi: 10.1111/j.1365-2958.1988.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 329.Sangadala S, Walters F S, English L H, Adang M J. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb+-K+ efflux in vitro. J Biol Chem. 1994;269:10088–10092. [PubMed] [Google Scholar]
- 330.Sankaranarayanan R, Sekar K, Banerjee R, Sharma V, Surolia A, Vijayan M. A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a β-prism fold. Nat Struct Biol. 1996;3:596–603. doi: 10.1038/nsb0796-596. [DOI] [PubMed] [Google Scholar]
- 331.Schnepf H E, Tomczak K, Ortega J P, Whiteley H R. Specificity-determining regions of a lepidopteran-specific insecticidal protein produced by Bacillus thuringiensis. J Biol Chem. 1990;265:20923–20930. [PubMed] [Google Scholar]
- 332.Schnepf H E, Whiteley H R. Protein toxins of Bacilli. In: Hoch J A, Setlow P, editors. Molecular biology of microbial differentiation. Washington, D.C: American Society for Microbiology; 1985. pp. 209–216. [Google Scholar]
- 333.Schurter W, Geiser M, Mathé D. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned delta-endotoxin gene. Mol Gen Genet. 1989;218:177–181. doi: 10.1007/BF00330581. [DOI] [PubMed] [Google Scholar]
- 334.Schwartz J L, Juteau M, Grochulski P, Cygler M, Préfontaine G, Brousseau R, Masson L. Restriction of intramolecular movements within the CrylAa toxin molecule of Bacillus thuringiensis through disulfide bond engineering. FEBS Lett. 1997;410:397–402. doi: 10.1016/s0014-5793(97)00626-1. [DOI] [PubMed] [Google Scholar]
- 335.Schwartz J L, Lu Y J, Sohnlein P, Brousseau R, Laprade R, Masson L, Adang M J. Ion channels formed in planar lipid bilayers by Bacillus thuringiensis toxins in the presence of Manduca sexta midgut receptors. FEBS Lett. 1997;412:270–276. doi: 10.1016/s0014-5793(97)00801-6. [DOI] [PubMed] [Google Scholar]
- 336.Schwartz J L, Potvin L, Chen X J, Brousseau R, Laprade R, Dean D H. Single-site mutations in the conserved alternating-arginine region affect ionic channels formed by CryIAa, a Bacillus thuringiensis toxin. Appl Environ Microbiol. 1997;63:3978–3984. doi: 10.1128/aem.63.10.3978-3984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Schwartz J-L, Garneau L, Masson L, Brousseau R. Early response of cultured lepidopteran cells to exposure to δ-endotoxin from Bacillus thuringiensis: involvement of calcium and anionic channels. Biochim Biophys Acta. 1991;1065:250–260. doi: 10.1016/0005-2736(91)90237-3. [DOI] [PubMed] [Google Scholar]
- 338.Schwartz J-L, Garneau L, Savaria D, Masson L, Brousseau R, Rousseau E. Lepidopteran-specific crystal toxins from Bacillus thuringiensis form cation- and anion-selective channels in planar lipid bilayers. J Membr Biol. 1993;132:53–62. doi: 10.1007/BF00233051. [DOI] [PubMed] [Google Scholar]
- 339.Sekar V. The insecticidal crystal protein gene is expressed in vegetative cells of Bacillus thuringiensis var. tenebrionis. Curr Microbiol. 1988;17:347–349. [Google Scholar]
- 340.Sekar V, Held B, Tippett J, Amirhusin B, Robert P, Wang K, Wilson H M. Biochemical and molecular characterization of the insecticidal fragment of CryV. Appl Environ Microbiol. 1997;63:2798–2801. doi: 10.1128/aem.63.7.2798-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Shelton A M, Robertson J L, Tang J D, Perez C, Eigenbrode S D, Preisler H K, Wilsey W K, Cooley R J. Resistance of diamondback moth (Lepidoptera: Plutellidae) to Bacillus thuringiensis subspecies in the field. J Econ Entomol. 1993;86:697–705. [Google Scholar]
- 342.Shelton A M, Roush R T, Tang J D, Perez C J, Earle E D. Presented at the XXVIIIth Annual Meeting of the Society for Invertebrate Pathology. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 343.Shimizu T, Morikawa K. The β-prism: a new folding motif. Trends Biochem Sci. 1996;21:3–6. [PubMed] [Google Scholar]
- 344.Shivakumar A G, Gundling G J, Benson T A, Casuto D, Miller M F, Spear B B. Vegetative expression of the δ-endotoxin genes of Bacillus thuringiensis subsp. kurstaki in Bacillus subtilis. J Bacteriol. 1986;166:194–204. doi: 10.1128/jb.166.1.194-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Sims S R, Holden L R. Insect bioassay for determining soil degradation of Bacillus thuringiensis subsp. kurstaki CryIA(b) protein in corn tissue. Environ Entomol. 1996;25:659–664. [Google Scholar]
- 346.Sims S R, Ream J E. Soil inactivation of the Bacillus thuringiensis subsp. kurstaki CryIIA insecticidal protein within transgenic cotton tissue: laboratory microcosm and field studies. J Agric Food Chem. 1997;45:1502–1505. [Google Scholar]
- 347.Skøt L, Harrison S P, Nath A, Mytton L R, Clifford B C. Expression of insecticidal activity in Rhizobium containing the δ-endotoxin gene cloned from Bacillus thuringiensis subsp. tenebrionis. Plant Soil. 1990;127:285–295. [Google Scholar]
- 348.Slatin S L, Abrams C K, English L. Delta-endotoxins form cation-selective channels in planar lipid bilayers. Biochem Biophys Res Commun. 1990;169:765–772. doi: 10.1016/0006-291x(90)90397-6. [DOI] [PubMed] [Google Scholar]
- 349.Smedley D P, Ellar D J. Mutagenesis of three surface-exposed loops of a Bacillus thuringiensis insecticidal toxin reveals residues important for toxicity, receptor recognition and possibly membrane insertion. Microbiology. 1996;142:1617–1624. doi: 10.1099/13500872-142-7-1617. [DOI] [PubMed] [Google Scholar]
- 350.Smith G P, Ellar D J. Mutagenesis of two surface-exposed loops of the Bacillus thuringiensis CryIC δ-endotoxin affects insecticidal specificity. Biochem J. 1994;302:611–616. doi: 10.1042/bj3020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Smith G P, Ellar D J, Keeler S J, Seip C E. Nucleotide sequence and analysis of an insertion sequence from Bacillus thuringiensis related to IS150. Plasmid. 1994;32:10–18. doi: 10.1006/plas.1994.1039. [DOI] [PubMed] [Google Scholar]
- 352.Smith G P, Merrick J D, Bone E J, Ellar D J. Mosquitocidal activity of the CryIC δ-endotoxin from Bacillus thuringiensis subsp. aizawai. Appl Environ Microbiol. 1996;62:680–684. doi: 10.1128/aem.62.2.680-684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Smith, R. A. 1996. Personal communication.
- 354.Smith R A, Couche G A. The phylloplane as a source of Bacillus thuringiensis variants. Appl Environ Microbiol. 1991;57:311–315. doi: 10.1128/aem.57.1.311-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Soltes-Rak E, Kushner D J, Williams D D, Coleman J R. Effect of promoter modification on mosquitocidal cryIVB gene expression in Synechococcus sp. strain 7942. Appl Environ Microbiol. 1993;59:2404–2410. doi: 10.1128/aem.59.8.2404-2410.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Sonngay, S., and W. Panbangred. 1997. Unpublished observation.
- 357.Stabb E V, Jacobson L M, Handelsman J. Zwittermycin A-producing strains of Bacillus cereus from diverse soils. Appl Environ Microbiol. 1994;60:4404–4412. doi: 10.1128/aem.60.12.4404-4412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Stephens J M. Disease in codling moth larvae produced by several strains of Bacillus cereus. Can J Zool. 1952;30:30–40. [Google Scholar]
- 359.Stevens S E, Jr, Murphy R C, Lamoreaux W J, Coons L B. A genetically engineered mosquitocidal cyanobacterium. J Appl Phycol. 1994;6:187–197. [Google Scholar]
- 360.Stewart C N, Jr, All J N, Raymer P L, Ramachandran S. Increased fitness of transgenic insecticidal rapeseed under insect selection pressure. Mol Ecol. 1997;6:773–779. [Google Scholar]
- 361.Stock C A, McLoughlin T J, Klein J A, Adang M J. Expression of a Bacillus thuringiensis crystal protein gene in Pseudomonas cepacia 526. Can J Microbiol. 1990;36:879–884. [Google Scholar]
- 362.Stone T B, Sims S R, Marrone P G. Selection of tobacco budworm for resistance to a genetically engineered Pseudomonas fluorescens containing the δ-endotoxin of Bacillus thuringiensis subsp. kurstaki. J Invertebr Pathol. 1989;53:228–234. [Google Scholar]
- 363.Tabashnik B E. Evaluation of synergism among Bacillus thuringiensis toxins. Appl Environ Microbiol. 1992;58:3343–3346. doi: 10.1128/aem.58.10.3343-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Tabashnik B E. Evolution of resistance to Bacillus thuringiensis. Annu Rev Entomol. 1994;39:47–79. doi: 10.1146/annurev.ento.54.110807.090518. [DOI] [PubMed] [Google Scholar]
- 365.Tabashnik B E, Finson N, Groeters F R, Moar W J, Johnson M W, Luo K, Adang M J. Reversal of resistance to Bacillus thuringiensis in Plutella xylostella. Proc Natl Acad Sci USA. 1994;91:4120–4124. doi: 10.1073/pnas.91.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Tabashnik B E, Finson N, Johnson M W, Moar W J. Resistance to toxins from Bacillus thuringiensis subsp. kurstaki causes minimal cross-resistance to B. thuringiensis subsp. aizawai in the diamondback moth (Lepidoptera: Plutellidae) Appl Environ Microbiol. 1993;59:1332–1335. doi: 10.1128/aem.59.5.1332-1335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Tabashnik B E, Liu Y-B, Finson N, Masson L, Heckel D G. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Tabashnik B E, Liu Y-B, Malvar T, Heckel D G, Masson L, Ballester V, Granero F, Ménsua J L, Ferré J. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc Natl Acad Sci USA. 1997;94:12780–12785. doi: 10.1073/pnas.94.24.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Tabashnik B E, Malvar T, Liu Y B, Finson N, Borthakur D, Shin B S, Park S H, Masson L, de Maagd R A, Bosch D. Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl Environ Microbiol. 1996;62:2839–2844. doi: 10.1128/aem.62.8.2839-2844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Tan Y, Donovan W. Abstracts of the 95th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1995. Cloning and characterization of the alkaline protease gene of Bacillus thuringiensis, abstr. Q-40; p. 406. [Google Scholar]
- 371.Tang J D, Gilboa S, Roush R T, Shelton A M. Inheritance, stability, and lack-of-fitness costs of field-selected resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae) from Florida. J Econ Entomol. 1997;90:732–741. [Google Scholar]
- 372.Tang J D, Shelton A M, Van Rie J, De Roeck S, Moar W J, Roush R T, Peferoen M. Toxicity of Bacillus thuringiensis spore and crystal protein to resistant diamondback moth (Plutella xylostella) Appl Environ Microbiol. 1996;62:564–569. doi: 10.1128/aem.62.2.564-569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Te Giffel M C, Beumer R R, Klijn N, Wagendorp A, Rombouts F M. Discrimination between Bacillus cereus and Bacillus thuringiensis using specific DNA probes based on variable regions of 16S rRNA. FEMS Microbiol Lett. 1997;146:47–51. doi: 10.1111/j.1574-6968.1997.tb10169.x. [DOI] [PubMed] [Google Scholar]
- 374.Thanabalu T, Hindley J, Brenner S, Oei C, Berry C. Expression of the mosquitocidal toxins of Bacillus sphaericus and Bacillus thuringiensis subsp. israelensis by recombinant Caulobacter crescentus, a vehicle for biological control of aquatic insect larvae. Appl Environ Microbiol. 1992;58:905–910. doi: 10.1128/aem.58.3.905-910.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Thomas W E, Ellar D J. Mechanism of action of Bacillus thuringiensis var. israelensis insecticidal δ-endotoxin. FEBS Lett. 1983;154:362–368. doi: 10.1016/0014-5793(83)80183-5. [DOI] [PubMed] [Google Scholar]
- 376.Thompson, M., and G. E. Schwab. November 1995. World Intellectual Property Organization patent WO 95/30752.
- 377.Thompson, M., G. E. Schwab, H. E. Schnepf, and B. Stockhoff. November 1995. World Intellectual Property Organization patent WO 95/30753.
- 378.Thompson M A, Schnepf H E, Feitelson J S. Structure, function, and engineering of Bacillus thuringiensis toxins. In: Setlow J K, editor. Genetic engineering: principles and methods. Vol. 17. New York, N.Y: Plenum Press; 1995. pp. 99–117. [PubMed] [Google Scholar]
- 379.Tojo A, Aizawa K. Dissolution and degradation of Bacillus thuringiensis δ-endotoxin by gut juice protease of the silkworm Bombyx mori. Appl Environ Microbiol. 1983;45:576–580. doi: 10.1128/aem.45.2.576-580.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Tomasino S F, Leister R T, Dimock M B, Beach R M, Kelly J L. Field performance of Clavibacter xyli subsp. cynodontis expressing the insecticidal crystal protein gene cry1A(c) of Bacillus thuringiensis against European corn borer in field corn. Biol Controls. 1995;5:442–448. [Google Scholar]
- 381.Torla, R. 1998. Personal communication.
- 382.Travers R S, Martin P A W, Reichelderfer C F. Selective process for efficient isolation of soil Bacillus sp. Appl Environ Microbiol. 1987;53:1263–1266. doi: 10.1128/aem.53.6.1263-1266.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Trisyono A, Whalon M E. Fitness costs of resistance to Bacillus thuringiensis in Colorado potato beetle (Coleoptera: Chrysomelidae) J Econ Entomol. 1997;90:267–271. doi: 10.1093/jee/88.1.21. [DOI] [PubMed] [Google Scholar]
- 384.Udayasuriyan V, Nakamura A, Masaki H, Uozumi T. Transfer of an insecticidal protein gene of Bacillus thuringiensis into plant-colonizing Azospirillum. World J Microbiol Biotechnol. 1995;11:163–167. doi: 10.1007/BF00704640. [DOI] [PubMed] [Google Scholar]
- 385.Vadlamudi R K, Ji T H, Bulla L A., Jr A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner. J Biol Chem. 1993;268:12334–12340. [PubMed] [Google Scholar]
- 386.Vadlamudi R K, Weber E, Ji I, Ji T H, Bulla L A., Jr Cloning and expression of a receptor for an insecticidal toxin of Bacillus thuringiensis. J Biol Chem. 1995;270:5490–5494. doi: 10.1074/jbc.270.10.5490. [DOI] [PubMed] [Google Scholar]
- 387.Vaeck M, Reynaerts A, Höfte H, Jansens S, De Beukeleer M, Dean C, Zabeau M, Van Montagu M, Leemans J. Transgenic plants protected from insect attack. Nature. 1987;328:33–37. [Google Scholar]
- 388.Valaitis A P, Lee M K, Rajamohan F, Dean D H. Brush border membrane aminopeptidase-N in the midgut of the gypsy moth serves as the receptor for the CryIA(c) δ-endotoxin of Bacillus thuringiensis. Insect Biochem Mol Biol. 1995;25:1143–1151. doi: 10.1016/0965-1748(95)00050-x. [DOI] [PubMed] [Google Scholar]
- 389.Valaitis A P, Mazza A, Brousseau R, Masson L. Interaction analyses of Bacillus thuringiensis Cry1A toxins with two aminopeptidases from gypsy moth midgut brush border membranes. Insect Biochem Mol Biol. 1997;27:529–539. [Google Scholar]
- 390.van Aarssen R, Soetaert P, Stam M, Dockx J, Gossele V, Seurinck J, Reynaerts A, Cornelissen M. cryIA(b) transcript formation in tobacco is inefficient. Plant Mol Biol. 1995;28:513–524. doi: 10.1007/BF00020398. [DOI] [PubMed] [Google Scholar]
- 391.van der Salm T, Bosch D, Honée G, Feng L, Munsterman E, Bakker P, Stiekema W J, Visser B. Insect resistance of transgenic plants that express modified Bacillus thuringiensis cryIA(b) and cryIC genes: a resistance management strategy. Plant Mol Biol. 1994;26:51–59. doi: 10.1007/BF00039519. [DOI] [PubMed] [Google Scholar]
- 392.van Frankenhuyzen K, Gringorten J L, Gauthier D, Milne R E, Masson L, Peferoen M. Toxicity of activated CryI proteins from Bacillus thuringiensis to six forest Lepidoptera and Bombyx mori. J Invertebr Pathol. 1993;62:295–301. [Google Scholar]
- 393.van Frankenhuyzen K, Gringorten J L, Milne R E, Gauthier D, Pusztai M, Brousseau R, Masson L. Specificity of activated CryIA proteins from Bacillus thuringiensis subsp. kurstaki HD-1 for defoliating forest Lepidoptera. Appl Environ Microbiol. 1991;57:1650–1655. doi: 10.1128/aem.57.6.1650-1655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Specificity of Bacillus thuringiensis δ-endotoxin: importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur J Biochem. 1989;186:239–247. doi: 10.1111/j.1432-1033.1989.tb15201.x. [DOI] [PubMed] [Google Scholar]
- 396.Van Rie J, McGaughey W H, Johnson D E, Barnett B D, Van Mellaert H. Mechanism of insect resistance to the microbial insecticide Bacillus thuringiensis. Science. 1990;247:72–74. doi: 10.1126/science.2294593. [DOI] [PubMed] [Google Scholar]
- 397.Visser B, Bosch D, Honée G. Domain-function studies of Bacillus thuringiensis crystal proteins: a genetic approach. In: Entwistle P F, Cory J S, Bailey M J, Higgs S, editors. Bacillus thuringiensis, an environmental biopesticide: theory and practice. Chichester, United Kingdom: John Wiley & Sons; 1993. pp. 71–88. [Google Scholar]
- 398.Von Tersch M A, Slatin S L, Kulesza C A, English L H. Membrane-permeabilizing activities of Bacillus thuringiensis coleopteran-active toxin CryIIIB2 and CryIIIB2 domain I peptide. Appl Environ Microbiol. 1994;60:3711–3717. doi: 10.1128/aem.60.10.3711-3717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Walters F S, Slatin S L, Kulesza C A, English L H. Ion channel activity of N-terminal fragments from CryIA(c) delta-endotoxin. Biochem Biophys Res Commun. 1993;196:921–926. doi: 10.1006/bbrc.1993.2337. [DOI] [PubMed] [Google Scholar]
- 400.Warren G W, Carozzi N B, Desai N, Koziel M G. Field evaluation of transgenic tobacco containing a Bacillus thuringiensis insecticidal protein gene. J Econ Entomol. 1992;85:1651–1659. [Google Scholar]
- 401.Warren, G. W., M. G. Koziel, M. A. Mullins, G. J. Nye, N. Desai, B. Carr, and N. K. Kostichka. September 1994. World Intellectual Property Organization patent WO 94/21795.
- 402.Wells J A. Binding in the growth hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.West A W, Burges H D, Dixon T J, Wyborn C H. Survival of Bacillus thuringiensis and Bacillus cereus spore inocula in soil: effects of pH, moisture, nutrient availability and indigenous microorganisms. Soil Biol Biochem. 1985;17:657–665. [Google Scholar]
- 404.West A W, Burges H D, White R J, Wyborn C H. Persistence of Bacillus thuringiensis parasporal crystal insecticidal activity in soil. J Invertebr Pathol. 1984;44:128–133. [Google Scholar]
- 405.West A W, Burges H D, Wyborn C H. Effect of incubation in natural and autoclaved soil upon potency and viability of Bacillus thuringiensis. J Invertebr Pathol. 1984;44:121–127. [Google Scholar]
- 406.Whalon M E, Miller D L, Hollingworth R M, Grafius E J, Miller J R. Selection of a Colorado potato beetle (Coleoptera: Chrysomelidae) strain resistant to Bacillus thuringiensis. J Econ Entomol. 1993;86:226–233. [Google Scholar]
- 407.Whalon M E, Wierenga J M. Bacillus thuringiensis resistant Colorado potato beetle and transgenic plants: some operational and ecological implications for deployment. Biocontrol Sci Technol. 1994;4:555–561. [Google Scholar]
- 408.Widner W R, Whiteley H R. Location of the dipteran specificity region in a lepidopteran-dipteran crystal protein from Bacillus thuringiensis. J Bacteriol. 1990;172:2826–2832. doi: 10.1128/jb.172.6.2826-2832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Williams S, Friedrich L, Dincher S, Carozzi N, Kessmann H, Ward E, Ryals J. Chemical regulation of Bacillus thuringiensis δ-endotoxin expression in transgenic plants. Bio/Technology. 1992;10:540–543. [Google Scholar]
- 410.Witt D P, Carson H, Hodgdon J C, Vlak J M, Peters D. Cytotoxicity of Bacillus thuringiensis δ-endotoxin to cultured CF-1 cells does not correlate with in vivo activity towards spruce budworm larvae. In: Samson R A, editor. Fundamental and applied aspects of invertebrate pathology. Wageningen, The Netherlands: Foundation of the Fourth International Colloquium of Invertebrate Pathology; 1986. pp. 3–6. [Google Scholar]
- 411.Wolfersberger M G. Enzymology of plasma membranes of insect intestinal cells. Am Zool. 1984;24:187–197. [Google Scholar]
- 412.Wolfersberger M G. Permeability of Bacillus thuringiensis CryI toxin channels. In: Clark J M, editor. Molecular action of insecticides on ion channels. Washington, D.C: American Chemical Society; 1995. [Google Scholar]
- 413.Wolfersberger M G. The toxicity of two Bacillus thuringiensis δ-endotoxins to gypsy moth larvae is inversely related to the affinity of binding sites on midgut brush border membranes for the toxins. Experientia. 1990;46:475–477. doi: 10.1007/BF01954236. [DOI] [PubMed] [Google Scholar]
- 414.Wolfersberger M G, Chen X J, Dean D H. Site-directed mutations in the third domain of Bacillus thuringiensis δ-endotoxin CryIAa affect its ability to increase the permeability of Bombyx mori midgut brush border membrane vesicles. Appl Environ Microbiol. 1996;62:279–282. doi: 10.1128/aem.62.1.279-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Wolfersberger M G, Hofmann C, Lüthy P. Interaction of Bacillus thuringiensis delta-endotoxin with membrane vesicles isolated from lepidopteran larval midgut. In: Falmagne P, Alouf J E, Fehrenbach F J, Jeljaszewicz J, Thelestam M, editors. Bacterial protein toxins. Stuttgart, Germany: Gustav Fischer Verlag; 1986. pp. 237–238. [Google Scholar]
- 416.Wong H C, Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci USA. 1986;83:3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Wong H C, Schnepf H E, Whiteley H R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983;258:1960–1967. [PubMed] [Google Scholar]
- 418.Wright D J, Iqbal M, Granero F, Ferré J. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl Environ Microbiol. 1997;63:1814–1819. doi: 10.1128/aem.63.5.1814-1819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Wu D, Aronson A I. Localized mutagenesis defines regions of the Bacillus thuringiensis δ-endotoxin involved in toxicity and specificity. J Biol Chem. 1992;267:2311–2317. [PubMed] [Google Scholar]
- 420.Wu D, Chang F N. Synergism in mosquitocidal activity of 26 and 65 kDa proteins from Bacillus thuringiensis subsp. israelensis crystal. FEBS Lett. 1985;190:232–236. [Google Scholar]
- 421.Wu D, Johnson J J, Federici B A. Synergism of mosquitocidal toxicity between CytA and CryIVD proteins using inclusions produced from cloned genes of Bacillus thuringiensis. Mol Microbiol. 1994;13:965–972. doi: 10.1111/j.1365-2958.1994.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 422.Wu S-J, Dean D H. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA δ-endotoxin. J Mol Biol. 1996;255:628–640. doi: 10.1006/jmbi.1996.0052. [DOI] [PubMed] [Google Scholar]
- 423.Wu, S.-J., C. N. Koller, D. L. Miller, L. S. Bauer, and D. H. Dean. 1996. Unpublished observation.
- 423a.Yamamoto, T. Personal communication.
- 424.Yamamoto T, Powell G K. Bacillus thuringiensis crystal proteins: recent advances in understanding its insecticidal activity. In: Kim L, editor. Advanced engineered pesticides. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 3–42. [Google Scholar]
- 425.Yaoi K, Kadotani T, Kuwana H, Shinkawa A, Takahashi T, Iwahana H, Isato R. Aminopeptidase N from Bombyx mori as a candidate for the receptor of Bacillus thuringiensis Cry1Aa toxin. Eur J Biochem. 1997;246:652–657. doi: 10.1111/j.1432-1033.1997.t01-1-00652.x. [DOI] [PubMed] [Google Scholar]
- 426.Yi S, Pang A S D, van Frankenhuyzen K. Immunocytochemical localization of Bacillus thuringiensis CryI toxins in the midguts of three forest insects and Bombyx mori. Can J Microbiol. 1996;42:634–641. [Google Scholar]
- 427.Yool A J. Block of the inactivating potassium channel by clofilium and hydroxylamine depends on the sequence of the pore region. Mol Pharmacol. 1994;46:970–976. [PubMed] [Google Scholar]
- 428.Yoshisue H, Fukada T, Yoshida K-I, Sen K, Kurosawa S-I, Sakai H, Komano T. Transcriptional regulation of Bacillus thuringiensis subsp. israelensis mosquito larvicidal crystal protein gene cryIVA. J Bacteriol. 1993;175:2750–2753. doi: 10.1128/jb.175.9.2750-2753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Yoshisue H, Ihara K, Nishimoto T, Sakai H, Komano T. Expression of the genes for insecticidal crystal proteins in Bacillus thuringiensis: cryIVA, not cryIVB, is transcribed by RNA polymerase containing Sigma H and that containing Sigma E. FEMS Microbiol Lett. 1995;127:65–72. doi: 10.1111/j.1574-6968.1995.tb07451.x. [DOI] [PubMed] [Google Scholar]
- 430.Yoshisue H, Nishimoto T, Sakai H, Komano T. Identification of a promoter for the crystal protein-encoding gene cryIVB from Bacillus thuringiensis subsp. israelensis. Gene. 1993;137:247–251. doi: 10.1016/0378-1119(93)90015-u. [DOI] [PubMed] [Google Scholar]
- 431.Yu C-G, Mullins M A, Warren G W, Koziel M G, Estruch J J. The Bacillus thuringiensis vegetative insecticidal protein Vip3A lyses midgut epithelial cells of susceptible insects. Appl Environ Microbiol. 1997;63:532–536. doi: 10.1128/aem.63.2.532-536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Zhan H, Choe S, Huynh P D, Finkelstein A, Eisenberg D, Collier R J. Dynamic transitions of the transmembrane domain of diphtheria toxin: disulfide trapping and fluorescence proximity studies. Biochemistry. 1994;33:11254–11263. doi: 10.1021/bi00203a022. [DOI] [PubMed] [Google Scholar]
- 433.Zhang, J., H. U. Schairer, W. Schnetter, D. Lereclus, and H. Agaisse.Bacillus popilliae cry18Aa operon is transcribed by Sigma E and Sigma K forms of RNA polymerase from a single initiation site. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 434.Zhang M-Y, Lövgren A, Low M G, Landén R. Characterization of an avirulent pleiotropic mutant of the insect pathogen Bacillus thuringiensis: reduced expression of flagellin and phospholipases. Infect Immun. 1993;61:4947–4954. doi: 10.1128/iai.61.12.4947-4954.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]