Abstract
N′-Acetyl-2-cyanoacetohydrazide (H2L1) and 2-cyano-N-(6-ethoxybenzo thiazol-2-yl) acetamide (HL2) ligands were used to synthesize [Cr(OAc)(H2L1)(HL2)]·2(OAc) and [Mn(H2L1)(HL2)]·Cl2·2H2O as mixed ligand complexes. All new compounds were analyzed by analytical, spectral, and computational techniques to elucidate their chemical formulae. The bidentate nature was suggested for each coordinating ligand via ON donors. The electronic transitions recorded are attributing to 4A2g(F) → 4T2g(F)(υ2) and 4A2g(F) → 4T1g(F)(υ3) types in the octahedral Cr(III) complex, while 6A1 → 4T2(G) and 6A1 → 4T1(G) transitions are attributing to the tetrahedral Mn(II) complex. These complexes were optimized by the density functional theory method to verify the bonding mode which was suggested via N(3), O(8), N(9), and N(10) donors from the mixed-ligands. Hirshfeld crystal models were demonstrated for the two ligands to indicate the distance between the functional groups within the two ligands and supporting the exclusion of self-interaction in between. Finally, the biological activity of the two mixed ligand complexes was tested by in silico ways as well as in vitro ways for confirmation. Three advanced programs were applied to measure the magnitude of biological efficiency of the two complexes toward kinase enzyme (3nzs) and breast cancer proliferation (3hy3). All in silico data suggest the superiority of the Mn(II) complex. Moreover, the in vitro assays for the two complexes that measure their antioxidant and cytotoxic activity support the distinguished activity of the Mn(II) complex.
1. Introduction
Essential bioactive compounds are mainly derived from heterocyclic compounds that include NSO donors which basically act as sensors in coordination chemistry.1,2 Among such compounds are the thiazole derivatives which could promote a distinct biological activity and also enrich the coordination field by changeable binding modes.3,4 Thiazoles are considered a powerful nucleus that contribute in designing drugs due to their extensive therapeutic efficiency.5 Diverse coordinating compounds were obtained from thiazoles and exhibited different biological effectiveness toward free radicals, general pain, hypertension, cancer, allergy, inflammation, malaria, bacteria, and fungi.6,7 The interest of mixed ligand complexes was shinned recently to enrich the coordination chemistry by new ideas, binding modes, and diverse applications.8 Mixed ligand complexes were derived from acid (phthalic, oxalic, or succinic) which mixed with heterocyclic bases to prepare Zn(II) and Cd(II) mixed-ligand complexes. Such complexes were investigated to characterize their tetrahedral geometry and then their antimicrobial activity was evaluated versus different bacteria.9 Quercetin and diimine ligands were mixed to isolate some bivalent metal ion complexes. These complexes were screened versus cancer cell lines and the mixed ligand Cu(II) complex showed the most toxic activity toward breast cancer cell.10 4-Aminoantipyrine (4-AAP) and imine derivative (H2L) were added sequential to copper, nickel, and manganese ions to prepared mixed ligand complexes. The analysis suggested that the complexes were formed with 2:1:2 molar ratio of metal/H2L/4-AAP, respectively. The antimicrobial activity of these complexes was investigated against different bacteria and fungi.11 Antiviral drugs were explored from tri-substituted thiazoles through investigation against common viruses.12,13 Octahedral complexes were formed from Mn(II), Fe(III), and Cr(III) ions with the Schiff base ligand. The electrolytic nature of the Mn(II) complex was recorded, while the non-electrolytic nature was appeared with Fe(III) and Cr(III) complexes. All compounds were screened versus bacteria and fungi to evaluate their biological influence.14 Schiff base ligand of the thiazole derivative was derived and used for coordination with Mn(III), Fe(III), Cr(III), Zr(IV), and VO(IV) ions. The complexes were elucidated by various tools and then their antibacterial activities15 were tested. The octahedral and square-pyramidal geometries were suggested for Co(II) and Cr(III) complexes, respectively. These complexes showed significant antibacterial data toward different bacteria.16
Proceeding from previous studies, we intended to prepare N′-acetyl-2-cyanoacetohydrazide and 2-cyano-N-(6-ethoxybenzo thiazol-2-yl) acetamide ligands to be used for preparing mixed ligand complexes derived from Cr(III) and Mn(II) ions. The targeted ligands have the same nature that is the cause behind their selection for this preparation to exclude their self-interaction. All new compounds were investigated by different available techniques which varied between analytical, spectral, and computational methods. The biological activity was predicated from a quantum point of view (in silico) via estimating pharmacokinetics, drug-likeness, and molecular docking studies. Then, the antioxidant activity and toxicity of the new complexes were investigated to evaluate the biological activity of the two mixed ligand complexes, practically.
2. Experimental Work
2.1. Reagents and Synthesis of Organic Compounds
The chemicals used to synthesize cyanoacetohydrazide derivative were 2-cyanoacetohydrazide and acetic anhydride, whereas those used to synthesize thiazole derivative were 2-amino-6-ethoxybenzothiazole and 3,5-dimethyl-1-cyanoacetyl. The CrCl3·6H2O and MnCl2 are the salts used to synthesize the two mixed ligand complexes, and NaOAc was used to rise the pH of the medium slightly. The ethanol (EtOH, 95%), dioxane (99.5%), and dimethyl sulfoxide (DMSO, 99%) were the solvents used either for synthesis or for analysis. All the reported chemicals are extra pure and purchased from Sigma-Aldrich then used as it is without any preliminary treatment.
2.1.1. Synthesis of N′-Acetyl-2-cyanoacetohydrazide (1) (H2L1)
A suspension of 2-cyanoacetohydrazide (5.0 g, 0.05 mol) in 50 mL of acetic anhydride was kept at 60–70 °C for 10 h. After standing overnight, a white precipitate was formed. It was collected by filtration and washed with plenty of water to give 3.50 g of N′-acetyl-2-cyanoacetohydrazide (1) (Scheme 1).
Scheme 1. Synthesis N′-Acetyl-2-cyanoacetohydrazide (1, H2L1) and 2-Cyano-N-(6-ethoxybenzo thiazol-2-yl) Acetamide (2, HL2) Ligands.
White solid, yield = 50%, mp = 175–176 °C, lit. mp = 176–178 °C.17 IR (KBr): 3273 (N–H), 3209 (N–H), 2250 (C≡N), 1620 (C=O) cm–1. 1H NMR (CDCl3): δ/ppm = 1.85 (s, 3H, CH3), 3.78 (s, 2H, CH2), 9.80 (s, 1H, NH), 9.98 (s, 1H, NH) (Figure S1).
2.1.2. Synthesis of 2-Cyano-N-(6-ethoxybenzothiazol-2-yl)acetamide (2) (HL2)
A mixture of 2-amino-6-ethoxybenzothiazole (1.94 g, 0.01 mol) and 3,5-dimethyl-1-cyanoacetyl pyrazole (1.64 g, 0.01 mol) was refluxed for 4 h in 20 mL of dioxane. The precipitate that formed after cooling of the reaction mixture was filtered off, dried, and recrystallized from ethanol (Scheme 1).
Beige powder, yield = 84%. mp = 218–219 °C, lit. mp = 214–216 °C.18 IR (νmax/cm–1): 3329 (N–H), 2222 (C≡N), 1676 (C=O). 1H NMR (DMSO-d6): δ/ppm = 1.35 (t, 3H, CH3), 3.80 (s, 2H, CH2), 4.06 (q, 2H, OCH2), 7.05–7.65 (m, 3H, Ar–H), 11.49 (s, 1H, NH) (Figure S1).
2.2. Synthesis of Mixed Ligand Complexes
The Cr(III) and Mn(II) complexes were prepared by dissolving 5 mmol (1.33 g) CrCl3·6H2O and 5 mmol (0.629 g) of MnCl2 salt in adequate amount of EtOH (10 mL), respectively. Then, equi-molar amounts of the N′-acetyl-2-cyanoacetohydrazide (H2L1) ligand (5 mmol, 0.706 g) and 2-cyano-N-(6-ethoxybenzothiazol-2-yl) acetamide (2) (HL2) ligand (5 mmol, 1.306 g) were dissolved separately in 10 mL of EtOH by heating. The solutions of the two ligands were added drop wise sequentially at the same time to each metal salt solution. This addition will give equal chance for the two ligands in coordination to the metal ions. Each reaction mixture was refluxed over night to complete the reaction, while the solid precipitates were isolated after adding NaOAc salt (in H2O), 0.5 g in Mn(II) complex and 1.5 g in Cr(III) complex.
2.3. Equipment
Most of instruments used for analysis were depicted in the Supporting Information as images for simplicity and each devise properties were typed below its image (Scheme S1). Additionally, the measurement conditions of each technique will be reported in the discussion part, individually. The 1H NMR spectra of the two ligands were registered by Bruker WP (500 MHz) in DMSO-d6 as the solvent and tetramethylsilane as an internal reference. The conductance of the complexes was determined for 1 mmol in DMSO solvent using JENWAY model 4070 Conductance Bridge. Also, the magnetic moments were determined via estimating the magnetic susceptibility at room temperature by using Johnson Matthey Magnetic Susceptibility Balance. The Cr(III) and Mn(II) contents were measured by complexometric titration by using the standard method.19 On the other hand, the absence or presence of chloride in the two complexes was detected qualitatively by AgNO3 after complete digestion of an adequate amount of the complex in conc. HNO3.19 There is no precipitate observed with the Cr(III) complex, while a white precipitate appeared with the Mn(II) complex which quantitatively evaluated gravimetrically to calculate the chloride percentage in the complex. The absence of a chloride ion in the Cr(III) complex is acceptable especially in presence of sufficient amount of NaOAc salt (1.5 g) that facilitates the coordination of the acetate group instead of chloride.
2.4. Optimizing the Structures
Well-known molecular dynamics simulation program (Gaussian 09)20 is analogous to the single crystal X-ray capacity and has been certified for structural type verification. The valence double zeta polarizing basis set (6-31G*) and Becke3–Lee–Yang–Parr (B3LYP)21 have been added to the DFT/B3LYP method applied. To the most common split-valence double-zeta plus polarization, this balanced basis set correlates. This term indicates that the valence is determined by two Gaussian orbitals, whereas the core-orbital is specified by a contract of six. The star (*) denotes polarization operations on non-hydrogen atoms. Gauss-View and Gauss-Sum 2.222 software were used to evaluate the generated computational files (log and chk) in order to retrieve all physical properties. On the other side, the fchk file was produced from chk file that was altered in the Gauss-prog panel.
2.5. In Silico Assessments
To put an accurate prediction for the biological behavior of the complexes under study before practical applications, the molecular docking study must be taken in consideration. Consequently, we selected the most advanced techniques for this purpose as the Pharmit link, Molecular operating environmental software (MOE, Ver 2018), and Swiss ADME link for confirmation. Two proteins were selected for this simulation from PDB co-crystal files as 3nzs and 3hy3 (Scheme S2), which were characterized based on the protein database. These proteins belong to the kinase enzyme (3nzs) which is essential for cell activities and breast cancer proliferation (3hy3). As is known, inhibiting such essential proteins predicates the success of the complex in controlling cancer cells.
2.5.1. Pharmacophore Query
Pharmit link (http://pharmit.csb.edu) was the primary docking tool implemented in this study regarding the two complexes toward the selected proteins. Pharmacophore was run according to the ligand-based and grid-based model23 to evaluate the interaction availability with protein receptors and searching on drug analogues. Each protein was applied as receptors that interacted with the complexes after adjusting them (mol2 file) to be suitable for this simulation software. The Pharmit search was run onto MolPort and Zn libraries to search on drug analogues.24 Moreover, the number and types of H-bonding formed with protein receptors were determined.
2.5.2. Docking Strategy (MOE)
MOE was the most advanced docking tool that offers all features of intercalation between the tested complex and pockets or receptors of proteins (3nzs and 3hy3). Either complexes or proteins must be oriented before starting each docking operation.25 The oriented compounds were saved as MDB in a new database which is ready for the docking process. Also, each protein was oriented via, selecting receptors, adding H-atoms, fixing potential energy, searching on interacting sites after that the docking will run after inserting the MDB file. The amino acid residues and dummies in protein were adjusted according to the MMFF-force field.26 The most stable docking pose was obtained among 30 poses formed during the trail. These poses are regulated by London dG-scoring function which improved two times by triangle-matcher.27 The length of each H-bonding was determined and the effective one does not exceed than 3.5 Å. To rank the inhibition efficiency of the tested complexes toward the two proteins, the scoring value of each docking process must be estimated according to known relation.28 The 2D and 3D patterns for docking operations were obtained to investigate all interaction features and shed light on the powerful inhibition.
2.5.3. ADME Parameters
Swiss link depends on the absorption, distribution, metabolism, and extraction (ADME) of small molecules to assess their physicochemical nature, pharmacokinetics, and drug-likeness properties.29 Here, we are just dealing with the two mixed ligand complexes as the targets of this study to examine their bioavailability score. The hypothetical polarity of surface area (TPSA), solubility (log S), unsaturation ratio depending on sp3 hybridized carbon, and flexible bonds were used in the viability radar calculation to assess drug-likeness. Other essential proteins that support organisms and tissues, the solubility in aqueous solution, and the partition coefficient between n-octanol and water (log Po/w) were also detected and recognized.30
2.6. In Vitro Assessments
The antioxidant activity of the two mixed ligand complexes were evaluated by the 2,2″-diphenyl-1-picrylhydrazyl (DPPH) method. The details of the work applied are displayed in the Supporting Information (Part A). Furthermore, the biological test was extended to measure the cytotoxic activity of these complexes toward liver carcinoma (HepG2), prostate cancer (PC3), and breast cancer (MCF-7) by applying (4,5-methyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide as known by the MTT assay. The details of applied methodology are displayed in the Supporting Information also (Part B) to minimize the similarity in the text.
3. Results and Discussion
3.1. General Properties of New Synthesizes
The N′-acetyl-2-cyanoacetohydrazide (H2L1) and 2-cyano-N-(6-ethoxybenzothiazol-2-yl) acetamide (2) (HL2) ligands were analyzed to estimate their CHN percentages (Table 1). Subsequently, the two mixed ligand complexes were also analyzed to propose their molecular formulae according to elemental percentages. The most fitted molar ratio of the two complexes was 1:1:1 that belongs to metal/H2L1/HL2, respectively. The acetate and chloride anions are ionically attached with Cr(III) and Mn(II) complexes, respectively. This is depending on their molar conductivity values that determined for 1 mmol in DMSO solvent, which detects the conducting property of the two complexes (Table 1).31
Table 1. Analytical and Physical Properties of New Synthesizes.
| % found (calcd.) |
||||||||
|---|---|---|---|---|---|---|---|---|
| compound empirical formula F.W. found (calculate) | color | mp (°C) | yield (%) | C | H | N | M | Λma |
| (1) H2L1, (C5H7N3O2); (141.13) | white | 175–176, lit. mp = 176–178 | 50 | 42.30 (42.55) | 4.83 (5.00) | 29.69 (29.77) | ||
| (2) HL2, (C12H11N3O2S); (261.3) | beige powder | 218–219, lit. mp = 214–216 | 84 | 55.03 (55.16) | 4.50 (4.24) | 15.88 (16.08) | ||
| (3) [Cr(OAc)(H2L1)(HL2)]·2(OAc); (C23H27CrN6O10S);631.24 (631.56) | green | >300 | 71 | 43.59 (43.74) | 4.16 (4.31) | 13.10 (13.31) | 8.20 (8.23) | 78.91 |
| (4) [Mn(H2L1) (HL2)]·Cl2·2H2O; (C17H22Cl2MnN6O6S);530.50(564.30) | yellowish green | >300 | 62 | 36.02 (36.18) | 3.74 (3.93) | 14.90 (14.89) | 9.77 (9.74) | 98.67 |
In DMSO (ohm–1 cm2 mol–1).
The interpretation of mixed ligand complexes needs special verifications that start from the two ligands themselves. The chance of interaction between the ligands was excluded depending on their similar nucleophilic nature produced either by the methylene group, N–H functions, or heteroatom centers. Also, the amidic nature of N–H functions prevents their chemical reactions with the carbonyl and/or nitrile group. These factors direct the ligands to form metal complexes without self-interaction between them.
3.2. IR Spectra
KBr-discs were prepared for the two ligands (H2L1 and HL2) to examine the changes happened under influence of IR-radiation on the functional groups. The H2L1 ligand showed vibrations of (N–H), (C≡N), and (C=O) at 3273, 3209; 2250, and 1620 cm–1, respectively. Additionally, the HL2 ligand showed vibrations of (N–H), (C≡N), (C=O), and (C=N) at 3329, 2222, 1676, and 1521 cm–1, respectively (Table 2). Comparing the spectra of Cr(III) and Mn(II) complexes (Figure S2) with that of the ligands offers an information about the binding mode inside the complexes. Significant changes were observed for υ(C=O)2 and υ(N3H) vibrations in the H2L1 ligand, while the HL2 ligand exhibited changes of υ(C=O)* and υ(C=N) vibrations due to their coordination with the metal ions. Each ligand behaved as a neutral bidentate toward the metal ions. The coordinating acetate group with the Cr(III) complex was suggested based on the appearance of υas(OAc) and υs(OAc) bands at 1457 and 1373 cm–1, respectively.32 The separation between two vibrations (Δυ = υas – υs) indicates the bidentate nature of the contributing acetate group.33 The crystal water molecules attached with the Mn(II) complex was suggested based on the noticeable broadness around 3400 cm–1 which is later supported by thermogravimetric analysis (TGA). Finally, the new bands observed at 520; 525 and 575; 643 cm–1 may be assigned to υ(M–N) and υ(M–O) vibrations in Cr(III) and Mn(II) complexes, respectively.
Table 2. Significant IR Bands for the Ligands and Their Mixed-Ligand Complexesa.
| compound | υ(CN) | υ(NH) | υ(C=O)1 | υ(C=O)2 | υ(C=O)* | υ(C=N) | δ(NH) | υas(OAc); υs(OAc) | υ(M–O) | υ(M–N) |
|---|---|---|---|---|---|---|---|---|---|---|
| (1) H2L1 | 2250 | 3270 | 1620 | 1620 | ||||||
| 3209 | ||||||||||
| (2) HL2 | 2222 | 3329 | 1676 | 1521 | ||||||
| (3) [Cr(OAc) (H2L1)(HL2)]·2(OAc) | 2250 | 3399 | 1620 | 1610 | 1670 | 1547 | 1457 | 1457 | 575 | 520 |
| 3150 | 1373 | |||||||||
| (4) [Mn(H2L1)(HL2)]·Cl2·2H2O | 2250 | B. C. at 3448 | 1642 | Sh, 1600 | 1681 | 1563 | 1412 | 643 | 525 |
B. C. Board centered for υ(OH) of water + υ(NH); Sh Shoulder.
3.3. UV–Vis Spectra and Magnetic Susceptibility
To obtain electronic transitions inside a chemical compound, a sample of it must be subjected to UV–vis radiations. This sample may be as emulsion in nujol or dissolved either in DMSO or DMF for the trace amount. Here, the two complexes were dissolved in DMSO solvent on cold and then scanned by radiations in instrument (200–1000 nm range).34 The ligand field transitions (d–d transitions), charge transfer, and intra-ligand transitions were recorded (Figure S3) to investigate the geometry of coordination of each complex. All observable transitions were extracted (nm) then changed to their wavenumber values (cm–1, Table 3). The [Cr(OAc)(H2L1)(HL2)]·2(OAc) complex showed 4A2g(F) → 4T2g(F)(υ2) and 4A2g(F) → 4T1g(F) (υ3) transitions at 11,299 and 16,835 cm–1, respectively.35 These transitions are attributing to transitions within the octahedral geometry of the d3 system. Furthermore, the [Mn(H2L1) (HL2)]·Cl2·2H2O complex showed 6A1 → 4T2(G)(υ2) and 6A1 → 4T1(G)(υ1) transitions at 20,321 and 14,281 cm–1, respectively.36 These weak transitions assign to that of tetrahedral geometry of high spin d5 system. On the other hand, the ligand field parameters as nephelauxetic (β), Racah (B), and crystal field stabilization energy (Δ = 10Dq) were calculated for the two complexes (Table 3). The calculated values close to that known for octahedral Cr(III) and tetrahedral Mn(II) complexes34−36 which formed from strong covalent bonds with coordinating ligands. The B parameter of the Cr(III) complex was calculated by using the following equation; υ3 = 15B + 30Dq – υ2, where υ1 = 10Dq so υ3 = 15B + 3υ1 – υ2 then after calculating B we could calculate Δ value from this relation Δo/B = 24. The β value can be calculated from this relation β = B (complex)/Bo(free ion) by knowing Bo of Cr(III) which equals 1030 cm–1. Meanwhile, the parameters of the Mn(II) complex was calculated from the ratio of υ2(E2)/υ1(E1) which used to determine the E2/B ratio from the energy table and then B value can be estimated. Consequently, the ratio of Δo/B = 11 was used to calculate Δo and the β was calculated by knowing the Bo value (free ion = 960 cm–1).34 The magnetic moment (μeff) values of Cr(III) and Mn(II) complexes were determined at room temperature and their values are 4.63 and 4.69 BM, respectively. Such values confirming the geometries suggested for weak field ligands with high spin states (Scheme 2).
Table 3. Magnetic Susceptibility, Transition Bands of Electrons, and Ligand Field Parameters.
| compound | μeff. (BM) | ligand field bands (cm–1) ; assignments | intra-ligand transitions ; charge transfer bands (cm–1) | Dq (cm–1) | B (cm–1) | β | geometry |
|---|---|---|---|---|---|---|---|
| [Cr(OAc)(H2L1) (HL2)]·2(OAc) | 4.63 | 16,835; 4A2g(F) → 4T1g(F)(υ3) | 44,400; 38,168 | 1129.9 | 607.14 | 0.661 | octahedral |
| 11,299; 4A2g(F) → 4T2g(F)(υ2) | 30,395; 23,148 | ||||||
| [Mn(H2L1)(HL2)]·Cl2·2H2O | 4.69 | 20,321; 6A1 → 4T2(G) | 44, 743; 36,496 | 654.54 | 595.04 | 0.692 | tetrahedral |
| 14,281; 6A1 → 4T1(G) | 34,602; 27,789 |
Scheme 2. Structures of Two Mixed-Ligand Complexes.
3.4. Thermogravimetric Analysis
TGA technique aims to elucidate the hydrating nature of the Mn(II) complex that matches the presence of two water molecules and, on the other hand, confirms the dehydrating nature of the Cr(III) complex. The process was performed on solid samples under nitrogen at a constant rate of heating (10 °C min–1) and on a broad range of temperatures (20–800 °C, Figure S4). The decomposition curve of the Cr(III) complex was limited on two steps that were started at 185 and ended at 675 °C (Table S1). These two steps reflect the complete distortion for the complex and the organic ligands were divided to small moieties that easily evaporated leaving carbon atoms polluted the chromium metal. On the other side, the decomposition curve of the Mn(II) complex was extended to four steps that started at low temperature (25 °C) and ended at 742 °C (Table S1). The first step recorded the dehydration of the complex by losing 2H2O by 6.56 (calcd. 6.38%) mass loss. Meanwhile, the following steps are capable for complete distortion for the complex and leaving residues from carbon atoms that polluted the MnO molecule. The lower thermal stability of the [Mn(H2L1)(HL2)]·Cl2·2H2O complex verifies the presence of hydrated water molecules that suggested previously.
3.5. Mass Spectroscopy
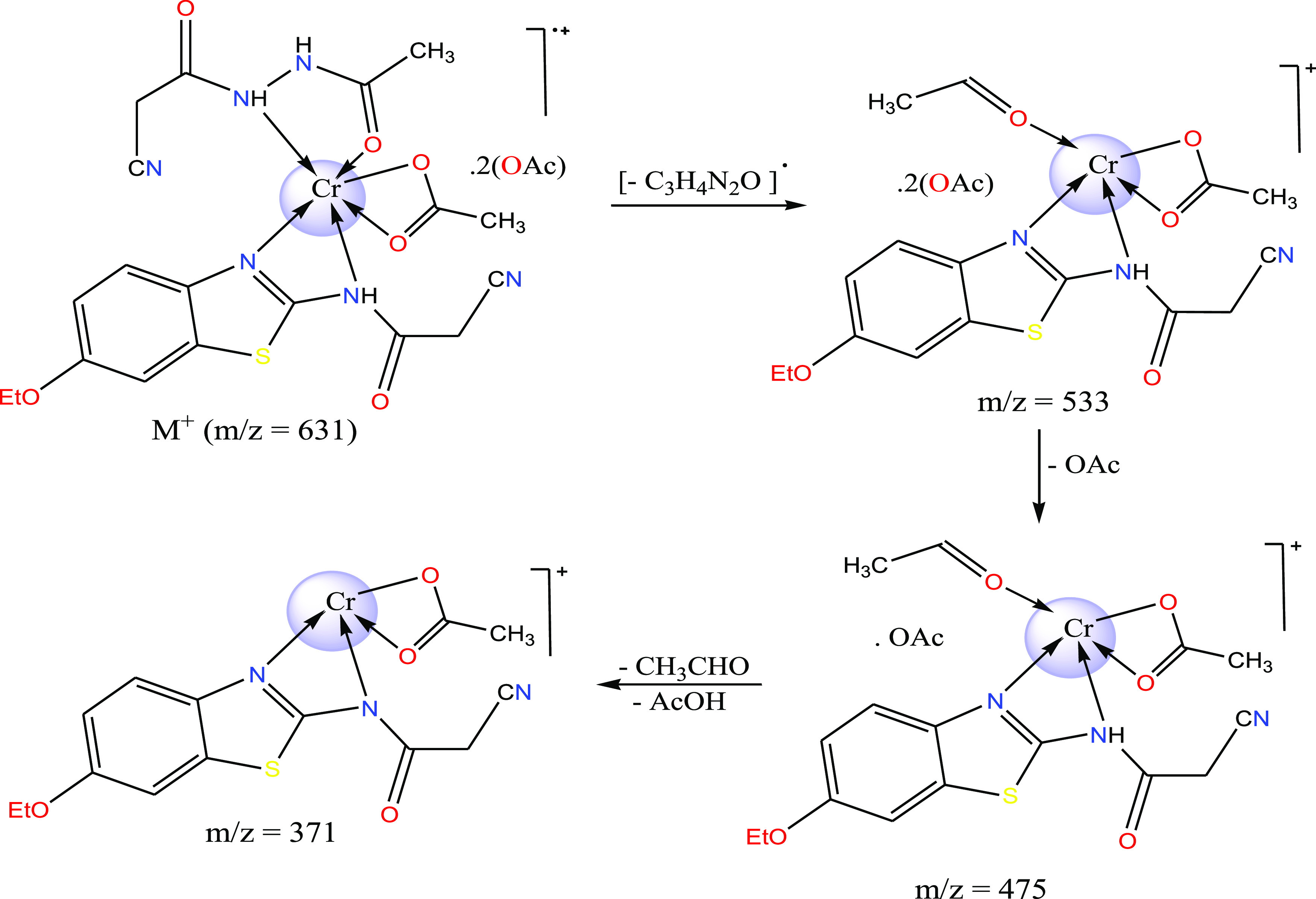
The importance of mass spectral analysis is growing in failure of isolating single crystal for the compound targeted for analysis. The two mixed-ligand complexes were subjected to an accelerated beam of electrons at 70 eV potential difference, 40 °C/min heating rate and over mass/charge range from 50 to 800. The successive fragmentation peaks were recorded on the displayed patterns (Figure S5) to elucidate the chemical formulae of Cr(III) and Mn(II) complexes. The molecular ion peaks appeared in the patterns were 631.24(calcd. 631.56) and 530.50 (calcd. 564.30) for the two complexes, respectively. The ion peak of Cr(III) complex formula could being assign to [M+] which confirms the suggested formula of the complex. Meanwhile, the ion peak of Mn(II) complex formula could being assigned to [M+ + 2] also but after losing the two water molecules as usually happened in some cases.37 This initial decomposition of water molecules is known to occur in the vaporization stage before recoding the formula peak.37 Furthermore, the fragmentation stages in the two complexes yield chromium or manganese isotopes as appeared around m/z = 54 or 55, respectively. Finally, the suitable fragmentation strategy was suggested for the Cr(III) complex, as depicted in Scheme 3.
Scheme 3. Fragmentation Scheme of the Cr(III) Complex.
3.6. X-ray Analyses Either via Energy-Dispersive or Diffraction
Energy-dispersive X-ray analysis confirms the coordination process itself via displaying singlet-set of peaks attributed to the Cr(III) or Mn(II) element (Figure S6) in the complex. This analysis depends on accelerated X-ray photons to penetrate the inner shells (K- or L-shell). These photons excite the electrons then leave positive holes which easily attract another electrons from higher levels (L- or M-shell). This relaxation is attached with emitted radiation which draws the pattern peaks of K-series (Ka1, Ka2, or Kb) or L-series. Such peaks characterize the elements according to the unique electronic configuration of each element, while L-series is the most significant.38 Also, the wt/at % was determined regarding to the indicator element (heavier element as the metal).
X-ray diffraction (XRD) technique could identify the magnitude of crystallinity of the tested complexes as well as determines the crystal properties of crystalline one. X-rays were used over 10° < 2θ < 80° range after their emission from the tube containing stimulated electrons that bombarding copper target (Cu/Kα). The obtained patterns (Figures 1 and S7) that recorded by the diffracted radiations39 were used to calculate the crystal parameters for the two complexes. Deby–Scherrer [β = 0.94 λ/(S cos θ)] and Bragg (nλ = 2d sin θ) equations were applied according to full width at half-maximum (FWHM) to estimate the crystal parameters as follows: diffraction angle of sharp peak (2θ°), d-spacing (Å), FWHM (β), intensity (I), and the size (S, Å).40 It is easy to show the extent of crystallinity of the Mn(II) complex based on the sharp peaks appeared in its pattern. Meanwhile, the pattern of the Cr(III) complex reflects its lower crystallinity, although we could calculate its crystal parameters also. The calculated crystal parameters of the Mn(II) complex were 11.277, 7.84017, 0.2667, 18.0, and 5.4563°, respectively, whereas the parameters calculated for the Cr(III) complex were 25.12, 3.54207, 0.6444, 6.23, and 5.1161°, respectively. The sizes calculated for Mn(II) and Cr(III) complexes are 54.563 and 51.161 nm which fall in the nanometer range perfectly. This feature is strongly preferable for various applications among that the biological activity which is already applied in this research.
Figure 1.
XRD scanning for the Mn(II) mixed-ligand complex.
3.7. Computational Study
3.7.1. Optimizing the Structures
Under standard conditions, Gaussian 09 program produced three files that visualized on program screen to estimate quantum calculations. Such features are the optimized structure (Figure S8), computational parameters (Table 4), 3D-maps for frontier orbitals (Figure 2), and iso-surface with array plots (Figure S9). Investigating the extracted parameters and maps leads to aggregate the following information.
Table 4. Essential Log-File Parameters for Optimized Compounds.
| the compound | bond lengths (Å) | bond angles (°) | Mullikan’s charges |
|---|---|---|---|
| H2L1 | C(6)=O(8) 1.20901 | C(7)–C(6)–O(8) 122.85 | N(3) = −0.185547 |
| HOMO = −0.24167 eV | C(2)–N(3) 1.36986 | C(7)–C(6)–N(5) 119.55 | O(8) = −0.418200 |
| LUMO = −0.01739 eV | N(3)–N(5) 1.35435 | C(6)–N(5)–N(3) 121.48 | O(4) = −0.405864 |
| ΔE = 0.22428 eV | N(5) = −0.102243 | ||
| dipole moment = 4.112 debye | |||
| formation energy = −509.246 au | |||
| HL2 | N(10)–C(11) 1.36662 | C(11)–N(10)–C(8) 121.16 | N(9) = −0.422790 |
| HOMO = −0.21884 eV | N(9)=C(8) 1.30832 | C(8)–N(9)–C(5) 121.31 | N(10) = −0.330873 |
| LUMO = −0.05173 eV | C(11)=O(12) 1.20788 | N(9)–C(8)–N(10) 118.27 | O(12) = −0.330617 |
| ΔE = 0.16711 eV | |||
| dipole moment = 7.403 debye | |||
| formation energy = −1176.498 au | |||
| [Cr(OAc)(H2L1)(HL2)]·2(OAc) | C(6)=O(8) 1.21878 | C(7)–C(6)–N(5) 114.62 | N(3) = −0.001477 |
| HOMO = −0.20625 eV | C(2)–N(3) 1.38129 | N(5)–N(3)–C(2) 118.34 | O(8) = −0.000490 |
| LUMO = −0.12457 eV | C(25)=N(26) 1.30757 | C(22)–N(26)–C(25) 111.35 | N(26) = −0.000524 |
| ΔE = 0.08168 eV | C(25)–N(27) 1.34750 | C(25)–N(27)–C(28) 131.33 | N(27) = −0.000030 |
| dipole moment = 10.592 debye | N(3)–Cr(47)–O(8) 84.82 | ||
| formation energy = −2958.939 au | |||
| [Mn(H2L1)(HL2)]·Cl2·2H2O | C(6)=O(8) 1.21845 | C(7)–C(6)–N(5) 114.81 | N(3) = −0.153868 |
| HOMO = −0.17602 eV | C(2)–N(3) 1.36985 | N(5)–N(3)–C(2) 119.69 | O(8) = −0.456864 |
| LUMO = −0.10068 eV | C(25)=N(26) 1.30780 | C(22)–N(26)–C(25) 111.51 | N(26) = −0.596077 |
| ΔE = 0.07534 eV | C(25)–N(27) 1.34740 | C(25)–N(27)–C(28) 131.45 | N(27) = −0.450556 |
| dipole moment = 8.798 debye | N(3)–Mn(47)–O(8) 105.67 | ||
| formation energy = −2825.216 au |
Figure 2.
HOMO and LUMO images for the two ligands (H2L1 and HL2) and their mixed-ligand complexes.
3.7.1.1. General Properties
The two ligands (H2L1 and HL2) are characterized by their high nucleophilicity according to the atomic charges displayed in Table 4. In particular, the calculated charges of N(3) and O(8) donors in the H2L1 ligand, as well as the N(9) and N(10) donors in the HL2 ligand, reflect their comparable nucleophilic feature. These charges are severely minimized in the Cr(III) complex, while they improved in Mn(II) complex due to Mn → ligand charge transfer through π-back donation.41,42 On the other hand, the optimized structures (Figure S7) reflect the best orientation of the donors (N3; O8 and N9; N10) that coordinated from the two mixed-ligands used. This supports their facilitated coordination toward both Cr(III) and Mn(II) ions without twisting or bond strain for their functional groups. This is observed easily in the bond lengths (Å) of the functional groups that selected for comparison in Table 4. A noticeable elongation of the coordinating functional groups appeared in normal range due to complexation without any change in hybridization of the atoms. Some bond angles were estimated for the two free ligands as well as the two mixed ligand complexes (Table 4). The bond angles in the two ligands indicate the regularity of trigonal planer geometry (∼120°) of the atoms. While in the two complexes, these angles appeared shifted from such regularity particularly that include the coordinating functional groups. In addition, the bond angles centered by the Cr(III) ion seem close to 90°, while that centered by the Mn(II) ion seem close to 109°. These angles supported the octahedral and tetrahedral geometries of the two complexes, respectively.43,44
The energy values of highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) levels were estimated for all compounds (Table 4) to determine the band gap in between (ΔE = ELUMO – EHOMO). The calculated values detect the reduction of band gap in the two complexes which clarify the impact of metal ions on the electronic transitions inside chemical systems. The electronic transitions inside the complexes are extremely facilitated based on reduced ΔE values. The dipole moment values detect the reduced polarity of the ligands as well as the Mn(II) complex which expect their lipophilicity that leads to effective biological activity. Also, the formation energy values (au) detect the higher stability of the two complexes.45
3.7.1.2. 3D-Maps
The HOMO and LUMO images were constructed over the contours of the optimized structures. Such orbitals appeared covering most functional groups in the two free ligands, which indicates that the first electronic transition is confined between coordinating functional groups,43 whereas the shape of these orbitals were significantly changed in the two complexes. The HOMO levels seem concentrated on the H2L1 ligand but the LUMO levels appeared concentrated around the central atom (Figure 2).
Furthermore, array plot shapes of the iso-surface were developed to assess the unsaturation degree of the compound surface (Figure S9). This kind of image was made after measuring the electron density at various locations on the surface grid and correlating it. The 2D-array plot divides the compound onto sections, with the inner contour denoted by the thin yellow lines around the edge. On the other side, the red lines denote the exterior contour limit. The two ligands and their Mn(II) complex exhibited in the images as an elongation of exterior counters. The capability of the ligands and their Mn(II) complex to absorb electrons from their environment is essentially represented by this property. As it would if it interacted with amino acids in biological systems, this might lead to their interaction with additional functional groups. Therefore, the effective biological activity may be expected from the Mn(II) complex.
3.7.2. Hirshfeld Study
This study was executed by crystal explorer software which is capable to build the crystal form of the chemical compound to identify significant properties among that the strength of the crystal packing.46 Here, we considered only H2L1 and HL2 ligands to assess the probability of contact in between which is not preferable to happen during coordination. Each ligand was optimized on VESTA package47 to be in a suitable format for the explorer program, and then, the optimized file was imported on the crystal program to run. The 3D-models (Figure 3) were obtained by using dnorm (A) which depends on normalized contact distance and shape index (B) to identify the contact strength between crystals. Red, white, and blue colors were the indicators for short, moderate, and long length (H-bonding) between crystals in the compound packing style. These lengths recorded reflect strong, moderate, and weak contact between crystals based on van der Waals radii.47 As observed in Figure 3, the red spots in the two ligands were centered by oxygen atoms. This clarifies the probability of closeness of the carbonyl groups during their reaction with the metal ions. Therefore, these groups have the same nature and then the probability of self-reaction in between is completely excluded as reported previously (Section 3.1). Furthermore, the 2D-models (Figure S10) were constructed to identify the contributing atoms in surface contact. As appeared, the nitrogen and oxygen atoms were the contributing in the surface contact, while the length percentage of N–H in crystal packing is 21.0% in H2L1 and 17.8% in HL2. This longer length detects their farther presence during their coordination with the Cr(III) or Mn(II) ion.48 Also, the length percentage of O–H in crystal packing is 11.1 in H2L1 and 0.1% in HL2. This detects the closeness of two identical functional groups during the coordination process, which leads to a safe mixed-ligand complexation process.
Figure 3.
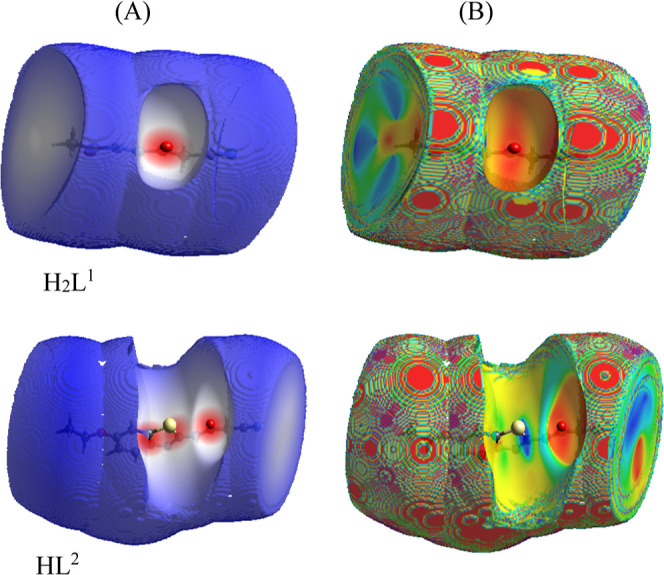
dnorm (A) and shape index (B) of H2L1 and HL2 ligands.
3.8. Biological Investigation
In this investigation we aimed to evaluate the extent of reactivity of the two mixed ligand complexes via computational methods which followed by practical methods belong to antioxidant and cytotoxicity tests.
3.8.1. In Silico Assays
This computation was used to visualize the inhibition feature of the two complexes before practical tests. Three advanced programs (A–C) were used for this in silico assays to measure their magnitude of biological efficiency toward kinase enzyme (3nzs) which is essential for cell activities and breast cancer proliferation (3hy3).
3.8.1.1. Pharmacophore Profile
Pharmit server was used to put an introductory view about the interaction between the two complexes and the selected proteins (3nzs and 3hy3).49,50 The pharmacophore profiles were presented (Figure 4) for investigation. As seen, the structure of the Cr(III) complex is significantly farther of both proteins. This predicates the lower activity of such a complex due to the absence of any interaction with protein pockets. On the other side, the structure of the Mn(II) complex is relatively interacted with 3hy3, while it is completely penetrating the pockets of the 3nzs protein. This expects a significant reactivity toward kinase enzyme but as an excellent inhibitor for breast cancer cell growth.51 The H-bonding appeared with the Mn(II) complex and 3nzs receptors were H-acceptor (acc = 9) and H-donor (don = 3). Finally, searching at MolPort and Zn libraries detects that there is no analogue for the Mn(II) complex among the drugs already found.
Figure 4.
Pharmit search for interaction of mixed-ligand complexes with 3nzs and 3hy3 proteins.
3.8.1.2. Molecular Docking (MOE)
MOE software able to give all interaction details in between the complexes and proteins. This is more advanced than the previous tool; it gives a view nearer to the true behavior in living cells.52 The docking parameters (Table 5) as well as the 2D or 3D shapes (Figures S11 and S12) of interaction were exported and displayed for studying. The following remarks were aggregated to rank the inhibition activity of the Cr(III) or Mn(II) complex toward 3nzs and 3hy3 proteins;
-
(i)
The scoring value of energy (S) was calculated and represents the following inhibition order: Mn(II) complex-3nzs > Mn(II) complex-3hy3 > Cr(III) complex -3nzs. The best inhibition was expected from the Mn(II) complex toward the two proteins especially with that of breast cancer cells (3nzs).53 The interaction paths are mostly true due to the formation of natural H-bonding (≤3.5 Å), except the interaction path of Cr(III) complex toward 3hy3 protein.
-
(ii)
Regarding the two proteins, polar (pink circle) and basic (blue circle) residues were the basically contributors in interaction with the Mn(II) complex (Figures S11 and S12). Such an interaction was carried out by the side chain acceptor and backbone acceptor types mainly via H-acceptor but the H-donor in one case only.
-
(iii)
The interacting sites in the Mn(II) complex were O4, O24, N11, and N22, while those of the Cr(III) complex were N3 and N38. The amino acid residues that interacted with the Mn(II) complex were GLU 880(A) and ARG 849(A) from 3nzs, while the residues from 3hy3 were SER 7(A), ARG 14(A), GLY 147(A), and LYS 10(A). On the other hand, the residues from 3nzs protein were SER 236(A) and ASP 239(A).54
-
(iv)
The ligand exposure surface in docking complexes is significantly lower regarding the docking of the Mn(II) complex, while this surface is significantly broad regarding the docking of the Cr(III) complex. This points to the saturated surface of the Mn(II) complex by interaction which reached to the maximum level. The formation energy values of docking complexes belong to the Mn(II) complex were reached to −16.4 kcal/mol with 3nzs protein and −12.1 kcal/mol with 3hy3 protein. These values signify the stability of its docking complexes, while the values belong to the Cr(III) complex docking are very low.54 Finally, this investigation expects the superiority of the Mn(II) complex in biological field especially in cytotoxicity.
Table 5. Interaction Parameters for Docking Results of Two Mixed-Ligand Complexes toward Two Functional Proteins.
| compounds | proteins | ligand | receptor | interaction | distance (Å) | E (kcal/mol) | S (energy score) |
|---|---|---|---|---|---|---|---|
| Cr(III) complex | 3hy3 | N36 | OD1 ASP 189 (A) | H-donor | 3.90 | –0.7 | –5.775 |
| 3nzs | N38 | OG SER 236 (A) | H-donor | 3.09 | –5.7 | –6.1060 | |
| N3 | OD2 ASP 239 (A) | ionic | 3.19 | –3.3 | |||
| Mn(II) complex | 3hy3 | O4 | OG SER 7 (A) | H-acceptor | 297 | –1.4 | –6.1243 |
| N11 | NH2 ARG 14 (A) | H-acceptor | 3.43 | –12.0 | |||
| N11 | N GLY 147 (A) | H-acceptor | 3.09 | –4.2 | |||
| O24 | NZ LYS 10 (A) | H-acceptor | 3.02 | –4.0 | |||
| 3nzs | N22 | OE1 GLU 880 (A) | H-donor | 3.04 | –16.4 | –6.9622 | |
| S18 | NH2 ARG 849 (A) | H-acceptor | 3.54 | –1.6 |
3.8.1.3. ADME Parameters
Swiss link was implemented to estimate ADME parameters which produce intensive insights into pharmacokinetics and drug-likeness score of the tested complexes (Table 6). Additionally, the physicochemical properties of the two complexes such as polarity (TPSA), molar reactivity, and rotatable bonds were calculated. The lipophilicity of the two complexes was assessed based on XLOGP3, Log S, and Log Po/w parameters. Pharmacokinetics of the two complexes were tested via Log Kp (permeability of skin), P-glycoprotein (P-gp) substrate, GI absorption, BBB permeation, and cytochromes P450 (CYP) parameters. Meanwhile, the drug-likeness property was evaluated from the bioavailability radar which showed six parameters over the axis. These parameters are XLOGP3, polarization, unsaturation, size, flexibility, and solubility (Table 6). The pink area showed the optimum range for the six indexes, so the drug-likeness depends on the range of such area for each tested compound regarding these indexes.55 The radar of the Cr(III) complex exhibited XLOGP3 (1.42), saturation limit (sp3 ≥ 0.25), size (small), polarity (203.58), and flexibility (6) parameters that detect its reduced drug-like properties. Meanwhile, the parameters of the Mn(II) complex reflect its elevated drug-like features depending on its lower lipophilicity and polarity values. Furthermore, the solubility indices (Log Po/w and Log S) were also estimated for the two complexes (Table 6). The partition coefficient (Log Po/w) of the compounds in n-octane/water solvent as well as the Log S parameter detects their ability for water solubility. Consequently, the reduced values recorded with the Mn(II) complex support its lipophilicity that elevates its biological activity.
Table 6. ADME Parameters Estimated for the Two Mixed Ligand Complexes.
Regarding pharmacokinetic parameters, we investigated the behavior of the two complexes against P-gp and cytochrome P450 (CYP) enzymes. As known, the excretion of the P-gp protein in tumor cells resists the action of drugs, while on the opposite side, this protein protects the nerve cells from xenobiotics. Regarding cytochromes (CYP), the behavior of these complexes was examined toward collection of iso-enzymes such as CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 which are essential to eliminate drug residues through metabolism.56 These complexes are the substrates of P-gp which indicates their rule in protecting the cell organisms or tissues. Also, the two complexes inhibit only one iso-enzyme (CYP2C19) among the five CYP. Consequently, the metabolism of drug residues is not highly affected and there is no significant drug accumulation in living cells. The skin permeation ability was evaluated by log Kp (cm/s) values which are determined from the relation between lipophilicity and the size of the tested complex. The smaller the negative value as in the Mn(II) complex (−8.18 cm/s), the higher the skin permeability feature.
Moreover, the absorption of these complexes in the gastrointestinal tract (HIA) and barrier of brain (BBB) was evaluated through BOILED-Egg images (Table 6). The BOILED-Egg images refer to the relation between WLOGP and TPSA. The absorption in the gastrointestinal tract was indicated from the white zone, while the absorption in the brain was indicated from the yellow region (yolk).56 The blue point appeared in the image of the Mn(II) complex supports its function as the substrate of P-gp (PGP+) but this complex did not absorb in the gastrointestinal tract or brain barrier. Meanwhile, the Cr(III) complex did not have any indication about this property.
To expect the pathways of the Mn(II) complex as the most active one inside living cells, Swiss-Target Prediction tool was run to reach this target. The resulting figure (Figure 5) was displayed for clarification. The Mn(II) complex could interact with 11-β-hydroxysteroid dehydrogenase enzyme 26.7%, cyclooxygenase-2 oxidoreductase by 6.7%, glycine transporter 1 (by homology) electrochemical transporter by 13.3%, and acetylcholinesterase hydrolase by 13.3% also.
Figure 5.

Predicated biological targets of the Mn(II) complex inside living cells.
3.8.2. In Vitro Assays
This part aimed to evaluate the biological activity of the two mixed ligand complexes based on in silico results which expect their biological efficiency, in particular to Mn(II) complex. Here, we focused on testing their antioxidant and anticancer activities to shed a light on the best therapeutic behavior.
3.8.2.1. Antioxidant Activity
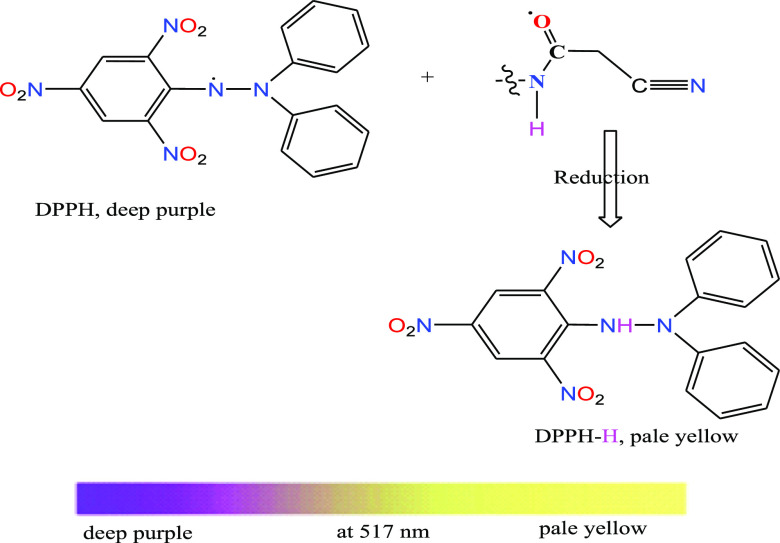
We know the dangerous role of free radicals inside human cells, so controlling the free radicals is a precious target of researchers. The free radicals (DPPH•) generated from the DPPH sensor in a polar solvent could be overcome by the two tested complexes which may take a hypothetical symbol of AH. The reaction between the antioxidant compound (AH) and the sensor (DPPH) must follow this reaction; DPPH• + AH → DPPH-H + A•.38 Therefore, the complex release hydrogen radicals have an antioxidant behavior toward DPPH• and then the concentration of such free radical will decrease leading to low absorption at 517 nm.57 The release of hydrogen radicals was expected from the amide group in the coordinating ligands (Scheme 4) depending on the liability of NH hydrogen due to its neighboring with the carbonyl group. The IC50 values (mg/mL, Table 7) were calculated for the two complexes compared to the reference (ascorbic acid). The antioxidant behavior of the Mn(II) complex is highly significant.
Scheme 4. Antioxidant Mechanism which May be Happened from the Complexes.
Table 7. DPPH Inhibition of New Complexes.
| acrchemical compounds | DPPH IC50 (mg/mL) |
|---|---|
| ascorbic acid | 5.39 |
| [Cr(OAc)(H2L1)(HL2)]·2(OAc) | 13.21 |
| [Mn(H2L1)(HL2)]·Cl2·2H2O | 9.14 |
3.8.2.2. Cytotoxicity
After treating with the novel complexes in comparison to 5-fluorouracil (5-Fu), the MTT technique was used to measure the cell viability of liver carcinoma (HepG2), prostate cancer (PC3), and breast cancer cells (MCF-7). Table 8 lists the results for the IC50 (μg/mL), and the Mn(II) complex demonstrated the greatest toxicity for breast cancer cells (MCF-7). As a result, silico and vitro studies predicted the potential biological activity of the Mn(II) complex. This activity may be due to the presence of the thiazole ring, C=N, C=O, and NH groups that improved the penetration capability of the tested complexes within the cell membrane. Moreover, Tweedy’s theory of chelation reflects a light on the role of metal ion on the biological activity of the complex which varied the activity from the complex to another for the same ligand. The charge of the metal ion after coordination was reduced to extent permit the miscibility of such complex in cell lipids. The charge of Mn(II) ion was highly reduced compared to the Cr(III) charge which already higher than the Mn(II) ion.58
Table 8. Antitumor Activity of the Two Mixed-Ligand Complexes against Three Cancer Cell Lines.
| in vitro
cytotoxicity IC50 (μg/mL)a |
|||
|---|---|---|---|
| compounds | HePG2 | PC3 | MCF-7 |
| [Cr(OAc)(H2L1)(HL2)]·2(OAc) | 26.28 ± 1.1 | 16.64 ± 9.0 | 13.36 ± 0.8 |
| [Mn(H2L1)(HL2)]·Cl2·2H2O | 15.68 ± 1.6 | 12.44 ± 1.2 | 9.87 ± 1.0 |
| 5-FUb | 7.91 ± 0.3 | 8.22 ± 0.2 | 5.60 ± 0.2 |
IC50 (μg/mL): 1–10 (very strong), 11–20 (strong), 21–50 (moderate), 51–100 (weak), and above 100 (non-cytotoxic).
5-FU = 5-fluorouracil is the reference drug for anticancer tests.
4. Conclusions
Two mixed ligand complexes were prepared from Cr(III) and Mn(II) ions. The molar ratios of the two complexes appeared the same by the 1M/1ligand1/1ligand2 ratio. All compounds were characterized by available analytical and spectral techniques to know their molecular and structural formulae. The electronic transitions and magnetic moment values point to the octahedral geometry of Cr(III) complex, whereas to the tetrahedral one of the Mn(II) complex. All structures were optimized to confirm the mode of bonding for the two ligands inside the coordination sphere. Moreover, the biological activity was extended onto two pathways, the first interested in silico and the second interested in vitro to strengthen the application part. In silico was performed by pharmacophore search, MOE docking, and Swiss ADME link. All extracted parameters or figures signify the superiority of the Mn(II) complex in biological applications. This assumption was confirmed lately from in vitro which investigated the antioxidant and antitumor activity of the two complexes. Also, the calculated IC50 values supported the distinguished biological behavior of Mn(II) complex toward DPPH radical and breast cancer cell line (MCF-7).
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by grant code: (22UQU4350527DSR03).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c07407.
Outlines of methodology of antioxidant activity and cytotoxicity, analytical tools utilized for analysis of new mixed-ligand complexes, PDB files of treated pathogen cell proteins, spectral images, TGA curves, EDX patterns, XRD patterns, iso-surface with array plots, 2D-fingerprint of the atoms, 2D and 3D interaction patterns, and TGA data (PDF)
The authors declare no competing financial interest.
Notes
The author(s) agree to publish the article under the Creative Commons Attribution License. All data generated or analyzed during this study are included in this published article.
Supplementary Material
References
- a Al-Fahemi J. H.; Saad F. A.; El-Metwaly N. M.; Farghaly T. A.; ElGhalban M. G. Appl. Organomet. Chem. 2017, 31, 3787. 10.1002/aoc.3787. [DOI] [Google Scholar]; b Frija L. M. T.; Pombeiro A. J. L.; Kopylovich M. N. Coordination chemistry of thiazoles, isothiazoles and thiadiazoles. Coord. Chem. Rev. 2016, 308, 32–55. 10.1016/j.ccr.2015.10.003. [DOI] [Google Scholar]
- Althagafi I.; El-Metwaly N. M.; Elghalban M.; Farghaly T. A.; Khedr A. M. Synthesis of Pyrazolone Derivatives and Their Nanometer Ag(I) Complexes and Physicochemical, DNA Binding, Antitumor, and Theoretical Implementations. Bioinorg. Chem. Appl. 2018, 2018, 1. 10.1155/2018/2727619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Abbas E. M. H.; Farghaly T. A.; Sabour R.; Shaaban M. R.; Abdallah Z. A. Design, synthesis, cytotoxicity, and molecular docking studies of novel thiazolyl–hydrazone derivatives as histone lysine acetyl-transferase inhibitors and apoptosis inducers. Arch. Pharm. 2022, 355, e2200076 10.1002/ardp.202200076. [DOI] [PubMed] [Google Scholar]; b Mahmoud H. K.; Gomha S. M.; Farghaly T. A.; Awad H. M. Synthesis of Thiazole Linked Imidazo[2,1-b]Thiazoles as Anticancer Agents. Polycyclic Aromat. Compd. 2021, 41, 1608–1622. 10.1080/10406638.2019.1689514. [DOI] [Google Scholar]
- a Hussain R.; Iqbal S.; Shah M.; Rehman W.; Khan S.; Rasheed L.; Rahim F.; Dera A. A.; Kehili S.; Elkaeed E. B.; Awwad N. S.; Bajaber M. A.; Alahmdi M.; Alrbyawi H.; Alsaab H. O. Synthesis of Novel Benzimidazole-Based Thiazole Derivatives as Multipotent Inhibitors of α-Amylase and α-Glucosidase: In Vitro Evaluation along with Molecular Docking Study. Molecules 2022, 27, 6457. 10.3390/molecules27196457. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Farghaly T. A.; Masaret G. S.; Muhammad Z. A.; Harras M. F. Discovery of thiazole-based-chalcones and 4-hetarylthiazoles as potent anticancer agents: Synthesis, docking study and anticancer activity. Bioorg. Chem. 2020, 98, 103761. 10.1016/j.bioorg.2020.103761. [DOI] [PubMed] [Google Scholar]
- Sokolova A. S.; Yarovaya O. I.; Bormotov N. I.; Shishkina L. N.; Salakhutdinov N. F. Synthesis and antiviral activity of camphor-based 1,3-thiazolidin-4-one and thiazole derivatives as Orthopoxvirus-reproduction inhibitors. MedChemComm 2018, 9, 1746. 10.1039/c8md00347e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cascioferro S.; Parrino B.; Carbone D.; Schillaci D.; Giovannetti E.; Cirrincione G.; Diana P. Thiazoles, Their Benzofused Systems, and Thiazolidinone Derivatives: Versatile and Promising Tools to Combat Antibiotic Resistance. J. Med. Chem. 2020, 63, 7923. 10.1021/acs.jmedchem.9b01245. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zou X.; Shi P.; Feng A.; Mei M.; Li Y. Two metal complex derivatives of pyridine thiazole ligand: synthesis, characterization and biological activity. Transition Met. Chem. 2021, 46, 263. 10.1007/s11243-020-00442-4. [DOI] [Google Scholar]
- Shih S.-R.; Chu T.-Y.; Reddy G. R.; Tseng S.-N.; Chen H.-L.; Tang W.-F.; Wu M.-S.; Yeh J.-Y.; Chao Y.-S.; Hsu J. T.; Hsieh H.-P.; Horng J.-T. J. Biomed. Sci. 2010, 17, 13. 10.1186/1423-0127-17-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaker S. A.; Ali Salih H. A. M. A. Orient. J. Chem. 2010, 2, 371. [Google Scholar]
- Nesa S.; Hossain S.; Nasira S.; Uddin N.; Ashrafuzzaman M.; Habib A.; Rashid A.; Haque M. Mixed ligand complexes: Synthesis, characterization and antibacterial activity investigation. Int. J. Chem. Stud. 2020, 8, 306–312. 10.22271/chemi.2020.v8.i1d.8267. [DOI] [Google Scholar]
- Mutlu Gençkal H. M.; Erkisa M.; Alper P.; Sahin S.; Ulukaya E.; Ari F. Mixed ligand complexes of Co(II), Ni(II) and Cu(II) with quercetin and diimine ligands: synthesis, characterization, anti-cancer and anti-oxidant activity. J. Biol. Inorg Chem. 2020, 25, 161–177. 10.1007/s00775-019-01749-z. [DOI] [PubMed] [Google Scholar]
- El-Sonbati A. Z.; El-Mogazy M. A.; Nozha S. G.; Diab M. A.; Abou-Dobara M. I.; Eldesoky A. M.; Morgan Sh. M. Mixed ligand transition metal(II) complexes: Characterization, spectral, electrochemical studies, molecular docking and bacteriological application. J. Mol. Struct. 2022, 1248, 131498. 10.1016/j.molstruc.2021.131498. [DOI] [Google Scholar]
- Yan S.; Appleby T.; Larson G.; Wu J. Z.; Hamatake R.; Hong Z.; Yao N. Structure-based design of a novel thiazolone scaffold as HCV NS5B polymerase allosteric inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 5888. 10.1016/j.bmcl.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Dawood K. M.; Eldebss T. M. A.; El-Zahabi H. S. A.; Yousef M. H. Synthesis and antiviral activity of some new bis-1,3-thiazole derivatives. Eur. J. Med. Chem. 2015, 102, 266. 10.1016/j.ejmech.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Al-Wasidi A. S.; Naglah A. M.; Saad F. A.; Abdelrahman E. A. Modification of Silica Nanoparticles with 4,6-Diacetylresorcinol as a Novel Composite for the Efficient Removal of Pb(II), Cu(II), Co(II), and Ni(II) Ions from Aqueous Media. J. Inorg. Organomet. Polym. 2022, 32, 2332. 10.1007/s10904-022-02282-4. [DOI] [Google Scholar]
- Kelode S. R. Synthesis, characterization and antimicrobial activity Cr(III), Mn(III), Fe(III), VO(IV), Zr(IV) and UO2(VI) metal complexes derived from bidentate thiazole Schiff base. J. Chem. Pharm. Sci. 2013, 5, 100–103. [Google Scholar]
- Kafi-Ahmadi L.; Shirmohammadzadeh L. Synthesis of Co(II) and Cr(III) salicylidenic Schiff base complexes derived from thiourea as precursors for nano-sized Co3O4 and Cr2O3 and their catalytic, antibacterial properties. J. Nanostruct. Chem. 2017, 7, 179–190. 10.1007/s40097-017-0221-x. [DOI] [Google Scholar]
- Martin N.; Quinteiro M.; Seoane C.; Soto J. L.; Fonseca I.; Florencio F.; Sanz J. Two rings in one step: a novel 1, 2, 4-triazolo [1, 5-a] pyridone with an unusual crystal structure. J. Org. Chem. 1990, 55, 2259–2262. 10.1021/jo00294a058. [DOI] [Google Scholar]
- Abdel-Galil E.; Moawad E. B.; El-Mekabaty A.; Said G. E. Synthesis, characterization and antibacterial activity of some new thiazole and thiazolidinone derivatives containing phenyl benzoate moiety. Synth. Commun. 2018, 48, 2083–2092. 10.1080/00397911.2018.1482349. [DOI] [Google Scholar]
- Vogel A. I.Quantitative Inorganic Analysis; Longmans: London, 1989. [Google Scholar]
- Frisch M.; Trucks G.; Schlegel H.; Scuseria G.; Robb M.; Cheeseman J.; Scalmani G.; Barone V.; Mennucci B.; Petersson G.. Gaussian 09, Revision A1; Gaussian: Wallingford, CT, USA, 2009.
- Lee C.; Yang W.; Parr R. G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785. 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Dennington R.; Keith T.; Millam J.. Gauss View, Version 5; Semichem Inc.: Shawnee Mission, KS, 2009.
- Willey C. D.; Bonner J. A.. Interaction of Chemotherapy and Radiation Gunderson LLTepper JEBT-CRO; Elsevier, 2012; Vol. 65. [Google Scholar]
- Sunseri J.; Koes D. Pharmit: interactive exploration of chemical space. Nucleic Acids Res. 2016, 44, W442. 10.1093/nar/gkw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Musyoka T. M.; Kanzi A. M.; Lobb K. A.; Tastan Bishop Ö.T. Structure based docking and molecular dynamic studies of plasmodial cysteine proteases against a South African natural compound and its analogs. Sci. Rep. 2016, 6, 23690. 10.1038/srep23690. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang Q.; Zhang Y.; Wu L.; Niu S.; Song C.; Zhang Z.; Lu G.; Qiao C.; Hu Y.; Yuen K. Y.; Wang Q.; Zhou H.; Yan J.; Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894. 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-nami S. Y.; Aljuhani E.; Althagafi I.; Abumelha H. M.; Bawazeer T. M.; Al-Solimy A. M.; Al-Ahmed Z. A.; Al-Zahrani F.; El-Metwaly N. Synthesis and Characterization for New Nanometer Cu(II) Complexes, Conformational Study and Molecular Docking Approach Compatible with Promising in Vitro Screening. Arabian J. Sci. Eng. 2021, 46, 365. 10.1007/s13369-020-04814-x. [DOI] [Google Scholar]
- Refat M. S.; Bayazeed A.; Katouah H.; Shah R.; Morad M.; Abualnaja M.; Alsaigh S.; Saad F.; El-Metwaly N. In-silico studies for kinetin hormone and its alkaline earth metal ion complexes as anti-aging cosmetics; synthesis, characterization and ability for controlling collagen-inhibitors. J. Mol. Struct. 2021, 1232, 130041. 10.1016/j.molstruc.2021.130041. [DOI] [Google Scholar]
- Shah R.; Katouah H.; Sedayo A. A.; Abualnaja M.; Aljohani M. M.; Saad F.; Zaky R.; El-Metwaly N. M. Practical and computational studies on novel Schiff base complexes derived from green synthesis approach: conductometry as well as in-vitro screening supported by in-silico study. J. Mol. Liq. 2020, 319, 114116. 10.1016/j.molliq.2020.114116. [DOI] [Google Scholar]
- Alaysuy O.; Abumelha H. M.; Alsoliemy A.; Alharbi A.; Alatawi N. M.; Osman H. E. M.; Zaky R.; El-Metwaly N. M. Elucidating of new hydrazide-based complexes derived from Pd (II), Cu (II) and Cd (II) ions: studies concerning spectral, DFT, Hirshfeld-crystal, biological screening beside Swiss-ADME verification. J. Mol. Struct. 2022, 1259, 132748. 10.1016/j.molstruc.2022.132748. [DOI] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary W. J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81. 10.1016/s0010-8545(00)80009-0. [DOI] [Google Scholar]
- Abu-Dief A. M.; El-Metwaly N. M.; Alzahrani S. O.; Alkhatib F.; Abualnaja M. M.; El-Dabea T.; El-Remaily M. Synthesis and characterization of Fe (III), Pd (II) and Cu (II)-thiazole complexes; DFT, pharmacophore modeling, in-vitro assay and DNA binding studies. J. Mol. Liq. 2021, 326, 115277. 10.1016/j.molliq.2021.115277. [DOI] [Google Scholar]
- Nakamoto K.Infrared spectra of inorganic, coordination compounds; Wiley Interscience: New York, 1970; Vol. 25, p 232. [Google Scholar]
- Lever A. B. P.Inorganic Electronic Spectroscopy, 2nd ed.; Elsevier: New York, 1984. [Google Scholar]
- El-Metwaly N.; Katouah H.; Aljuhani E.; Alharbi A.; Alkhatib F.; Aljohani M.; Alzahrani S.; Alfaifi M. Y.; Khedr A. M. Synthesis and elucidation for new nanosized Cr (III)-pyrazolin complexes; crystal surface properties, antitumor simulation studies beside practical apoptotic path. J. Inorg. Organomet. Polym. 2020, 30, 4142–4154. 10.1007/s10904-020-01561-2. [DOI] [Google Scholar]
- Shaker S. A.; Khaledi H.; Cheah S.-C.; Ali H. M. New Mn(II), Ni(II), Cd(II), Pb(II) complexes with 2-methylbenzimidazole and other ligands. Synthesis, spectroscopic characterization, crystal structure, magnetic susceptibility and biological activity studies. Arabian J. Chem. 2016, 9, S1943–S1950. 10.1016/j.arabjc.2012.06.013. [DOI] [Google Scholar]
- Refat M. S.; El-Metwaly N. M. Spectral, thermal and biological studies of Mn (II) and Cu (II) complexes with two thiosemicarbazide derivatives. Spectrochim. Acta, Part A 2012, 92, 336. 10.1016/j.saa.2012.02.041. [DOI] [PubMed] [Google Scholar]
- Alkhamis K.; Alsoliemy A.; Aljohani M. M.; Alrefaei A. F.; Abumelha H. M.; Mahmoud M. H. H.; Zaky R.; El-Metwaly N. M. Conductometry of nano-sized zinc sulfate; synthesis and characterization of new hydrazone complexes: conformational and in-vitro assay. J. Mol. Liq. 2021, 340, 117167. 10.1016/j.molliq.2021.117167. [DOI] [Google Scholar]
- Cullity B. D.Elements of X–ray diffraction, 1st ed.; Addison −Wesley Inc., 1993. [Google Scholar]
- Velumani S.; Mathew X.; Sebastian P. J.; Narayandass S. K.; Mangalaraj D. Structural and optical properties of hot wall deposited CdSe thin films. Sol. Energy Mater. Sol. Cells 2003, 76, 347. 10.1016/s0927-0248(02)00287-8. [DOI] [Google Scholar]
- Alzahrani S.; Morad M.; Bayazeed A.; Aljohani M.; Alkhatib F.; Shah R.; Katouah H.; Abumelha H. M.; Althagafi I.; Zaky R.; El-Metwaly N. M. Ball milling approach to prepare new Cd(II) and Zn(II) complexes; characterization, crystal packing, cyclic voltammetry and MOE-docking agrees with biological assay. J. Mol. Struct. 2020, 1218, 128473. 10.1016/j.molstruc.2020.128473. [DOI] [Google Scholar]
- Almalki S. A.; Bawazeer T. M.; Asghar B.; Alharbi A.; Aljohani M. M.; Khalifa M. E.; El-Metwaly N. M. Synthesis and characterization of new thiazole-based Co (II) and Cu (II) complexes; therapeutic function of thiazole towards COVID-19 in comparing to current antivirals in treatment protocol. J. Mol. Struct. 2021, 1244, 130961. 10.1016/j.molstruc.2021.130961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazetis G.; James B. D. Molecular models of brown coal containing inorganic species. Org. Geochem. 2006, 37, 244. 10.1016/j.orggeochem.2005.07.006. [DOI] [Google Scholar]
- Shaaban S.; Ferjani H.; Althagafi I.; Yousef T. Crystal structure, Hirshfeld surface analysis, and DFT calculations of methyl (Z)-4-((4-((4-bromobenzyl)selanyl)phenyl)amino)-4-oxobut-2-enoate. J. Mol. Struct. 2021, 1245, 131072. 10.1016/j.molstruc.2021.131072. [DOI] [Google Scholar]
- Al-Hazmi G. A. A.; Abou-Melha K. S.; El-Metwaly N. M.; Althagafi I.; Shaaban F.; Zaki R. Green synthesis approach for Fe (III), Cu (II), Zn (II) and Ni (II)-Schiff base complexes, spectral, conformational, MOE-docking and biological studies. Appl. Organomet. Chem. 2020, 34, e5403 10.1002/aoc.5403. [DOI] [Google Scholar]
- Turner M. J.; McKinnon J. J.; Jayatilaka D.; Spackman M. A. Visualization and characterization of voids in crystalline materials. CrystEngComm 2011, 13, 1804–1813. 10.1039/c0ce00683a. [DOI] [Google Scholar]
- Momma K.; Izumi F. VESTA 3 for three-dimensional visualization of crystal, volumetric and morphology data. J. Appl. Crystallogr. 2011, 44, 1272–1276. 10.1107/s0021889811038970. [DOI] [Google Scholar]
- Katouah H.; Hameed A. M.; Alharbi A.; Alkhatib F.; Shah R.; Alzahrani S.; Zaky R.; El-Metwaly N. M. Green Synthesis Strategy for New Schiff-Base Complexes: Characterization, Conductometry, In Vitro Assay Confirmed by In Silico Approach. ChemistrySelect 2020, 5, 10256–10268. 10.1002/slct.202002388. [DOI] [Google Scholar]
- Mu P.; Karuppasamy R. Discovery of human autophagy initiation kinase ULK1 inhibitors by multi-directional in silico screening strategies. J. Recept. Signal Transduction 2019, 39, 122–133. 10.1080/10799893.2019.1638401. [DOI] [PubMed] [Google Scholar]
- Kandakatla N.; Ramakrishnan G. Ligand based pharmacophore modeling and virtual screening studies to design novel HDAC2 inhibitors. Adv. Bioinf. 2014, 2014, 812148. 10.1155/2014/812148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E. L.; Simmers C.; Knight D. A. Cobalt complexes as antiviral and antibacterial agents. Pharmaceuticals 2010, 3, 1711–1728. 10.3390/ph3061711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrecht A. S.; Urban N.; Schaefer M.; Röse A.; Kless A.; Meents J. E.; Lampert A.; Abdelrahman A.; Müller C. E.; Schmalzing G.; Hausmann R. Identification of aurintricarboxylic acid as a potent allosteric antagonist of P2X1 and P2X3 receptors. Neuropharmacology 2019, 158, 107749. 10.1016/j.neuropharm.2019.107749. [DOI] [PubMed] [Google Scholar]
- Angelusiu M. V.; Barbuceanu S. F.; Draghici C.; Almajan G. L. New Cu (II), Co (II), Ni (II) complexes with aroyl-hydrazone based ligand. Synthesis, spectroscopic characterization and in vitro antibacterial evaluation. Eur. J. Med. Chem. 2010, 45, 2055. 10.1016/j.ejmech.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Refat M. S.; Sedayo A. A.; Sayqal A.; Alharbi A.; Katouah H. A.; Abumelha H. M.; Alzahrani S.; Alkhatib F.; Althagafi I.; El-Metwaly N. Aurintricarboxylic acid and its metal ion complexes in comparative virtual screening versus Lopinavir and Hydroxychloroquine in fighting COVID-19 pandemic: Synthesis and characterization. Inorg. Chem. Commun. 2021, 126, 108472. 10.1016/j.inoche.2021.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton R. R.; Gauci R.; Junk P. C.; Lindoy L. F.; Luckay R. C.; Meehan G. V.; Price J. R.; Turner P.; Wei G. Macrocyclic ligand design. Structure–function relationships involving the interaction of pyridinyl-containing, mixed oxygen–nitrogen donor macrocycles with cobalt (II), nickel (II), copper (II), zinc (II), cadmium (II), silver (I) and lead (II). J. Chem. Soc., Dalton Trans. 2002, 10, 2185–2193. 10.1039/b201195f. [DOI] [Google Scholar]
- Alatawi N. M.; Alsharief H. H.; Alharbi A.; Alhasani M.; Attar R. M. S.; Khalifa M. E.; Abu-Dief A. M.; El-Metwaly N. M. Simulation for the behavior of new Fe (III) and Cr (III)-thiophenyl complexes towards DNA polymerase: synthesis, characterization, eukaryotic DNA and Hartree–Fock computation. Chem. Pap. 2022, 76, 3919. [Google Scholar]
- Alkhamis K.; Alkhatib F.; Alsoliemy A.; Alrefaei A. F.; Katouah H. A.; Osman H. E.; Mersal G. A.; Zaky R.; El-Metwaly N. M. Elucidation for coordination features of hydrazide ligand under influence of variable anions in bivalent transition metal salts; green synthesis, biological activity confirmed by in-silico approaches. J. Mol. Struct. 2021, 1238, 130410. 10.1016/j.molstruc.2021.130410. [DOI] [Google Scholar]
- a Abumelha H. M.; Al-Fahemi J. H.; Althagafi I.; Bayazeed A. A.; Al-Ahmed Z. A.; Khedr A. M.; El-Metwaly N. Deliberate-characterization for Ni (II)-Schiff Base complexes: promising in-vitro anticancer feature that matched MOE docking-approach. J. Inorg. Organomet. Polym. Mater. 2020, 30, 3277. 10.1007/s10904-020-01503-y. [DOI] [Google Scholar]; b Farghaly T. A.; Althagafi I.; Ibrahim M. H.; Al-Qurashi N. T.; Farooq U. Synthesis under microwaves irradiation, structure elucidation, docking study for inhibiting COVID-19 and DFT calculations of novel azoles incorporated indole moiety. J. Mol. Struct. 2021, 1244, 131263. 10.1016/j.molstruc.2021.131263. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.