Abstract
A physical map, EcoMap10, of the now completely sequenced Escherichia coli chromosome is presented. Calculated genomic positions for the eight restriction enzymes BamHI, HindIII, EcoRI, EcoRV, BglI, KpnI, PstI, and PvuII are depicted. Both sequenced and unsequenced Kohara/Isono miniset clones are aligned to this calculated restriction map. DNA sequence searches identify the precise locations of insertion sequence elements and repetitive extragenic palindrome clusters. EcoGene10, a revised set of genes and functionally uncharacterized open reading frames (ORFs), is also depicted on EcoMap10. The complete set of unnamed ORFs in EcoGene10 are assigned provisional names beginning with the letter “y” by using a systematic nomenclature.
EcoMap10, the physical map of edition 10 of the Escherichia coli K-12 linkage map, is a map of restriction sites and genomic positions of a set of bacteriophage lambda clones and includes a graphic representation of EcoGene10, a refined annotation of the Escherichia coli genome sequence. The previous version, EcoMap7, was published as part of edition 9 of the Escherichia coli K-12 linkage map (4). A brief description of EcoMap10 is provided here, and a more detailed description of EcoMap10 and the EcoGene10 data set will be published separately (16). The most significant change in EcoMap construction is that it is now based upon the complete genome sequence of E. coli K-12 strain MG1655 version M52 (4,639,221 bp) as determined by Blattner et al. (5). EcoMap10 features, including the predicted restriction sites, Kohara clone alignments, protein coding regions, gene and open reading frame (ORF) designations, insertion sequence (IS) elements, and repetitive extragenic palindrome (REP) clusters, have all been derived by using version M52 of the MG1655 DNA sequence (GenBank/EMBL/DDBJ accession no., U00096). The tables and references in the traditional map of edition 10 of the Escherichia coli K-12 linkage map (3) also apply to the genes displayed in the physical map.
ECOMAP10
Restriction Enzyme Recognition Sites
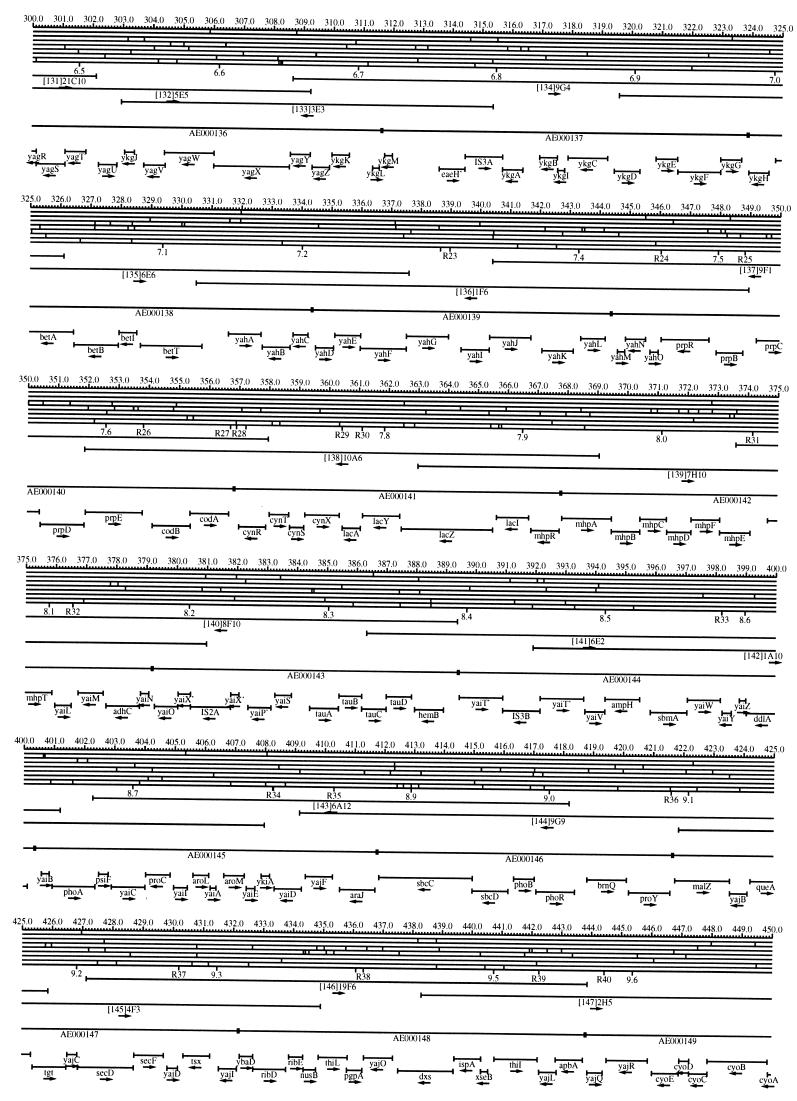
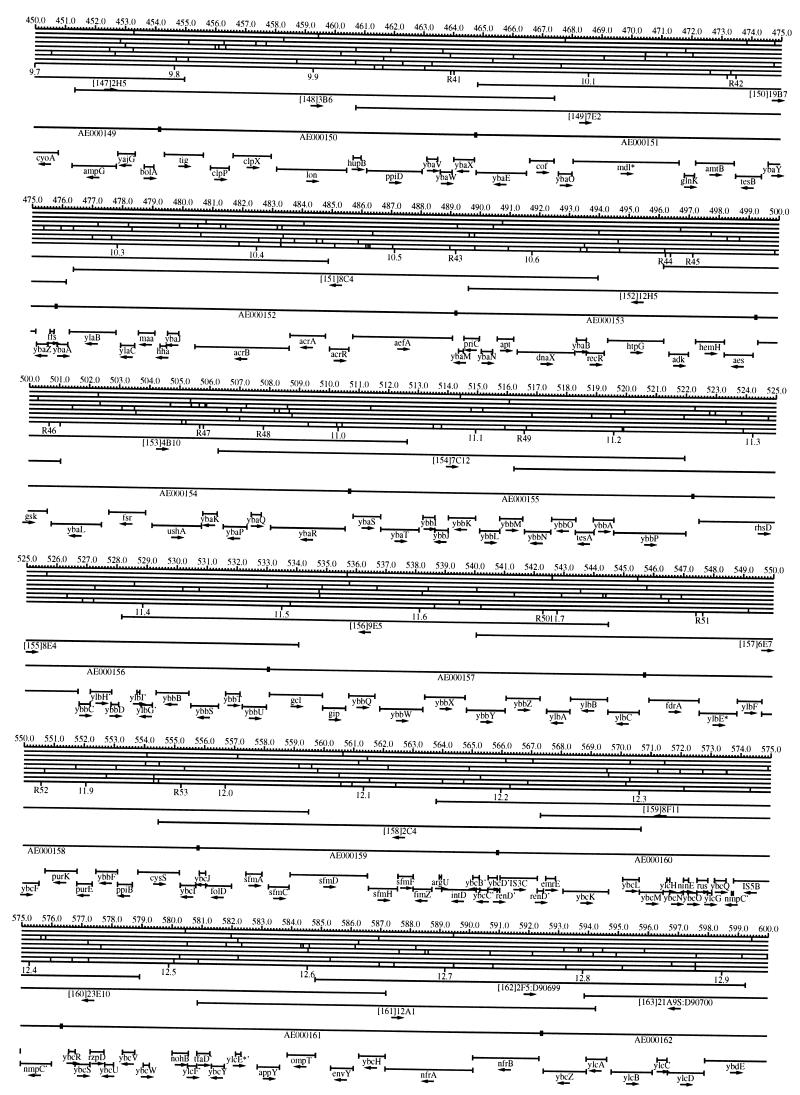
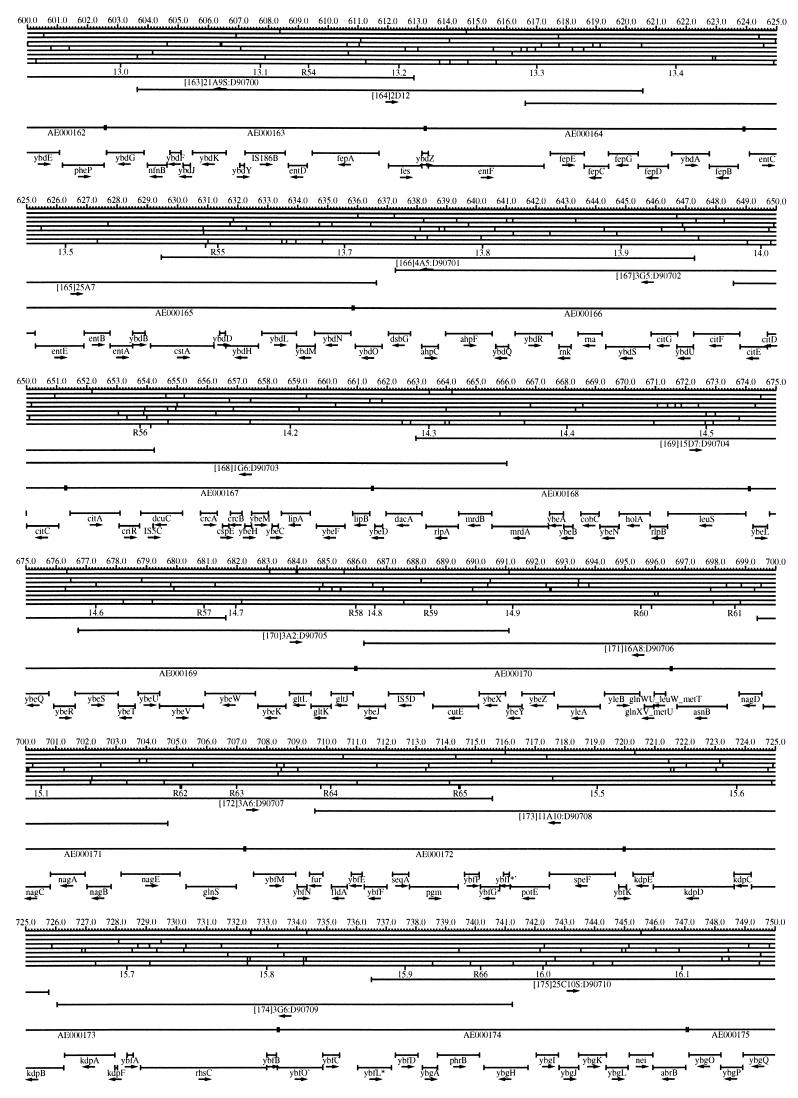
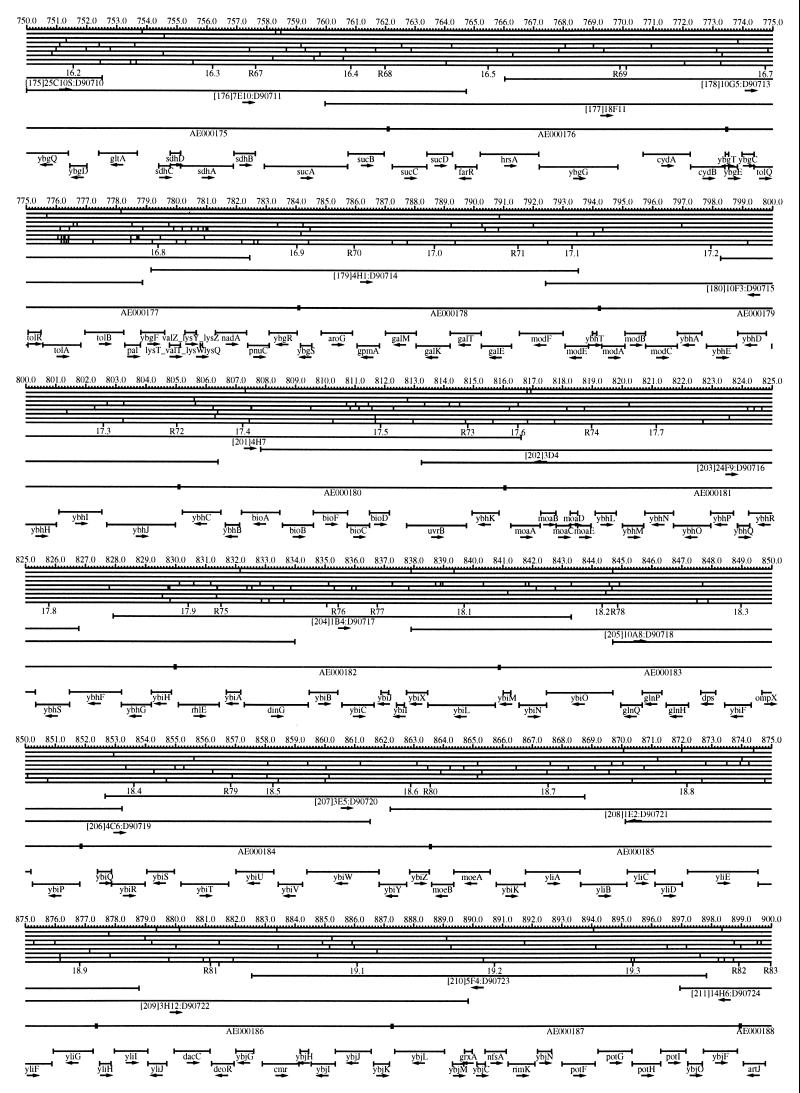
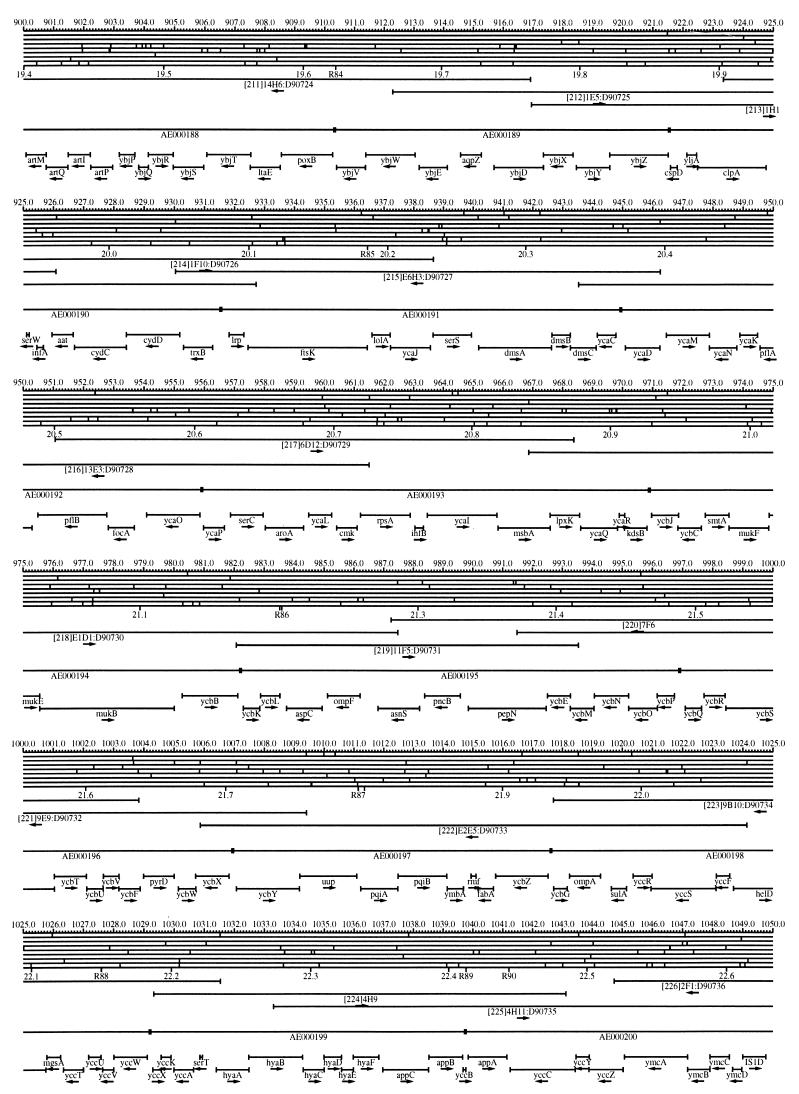
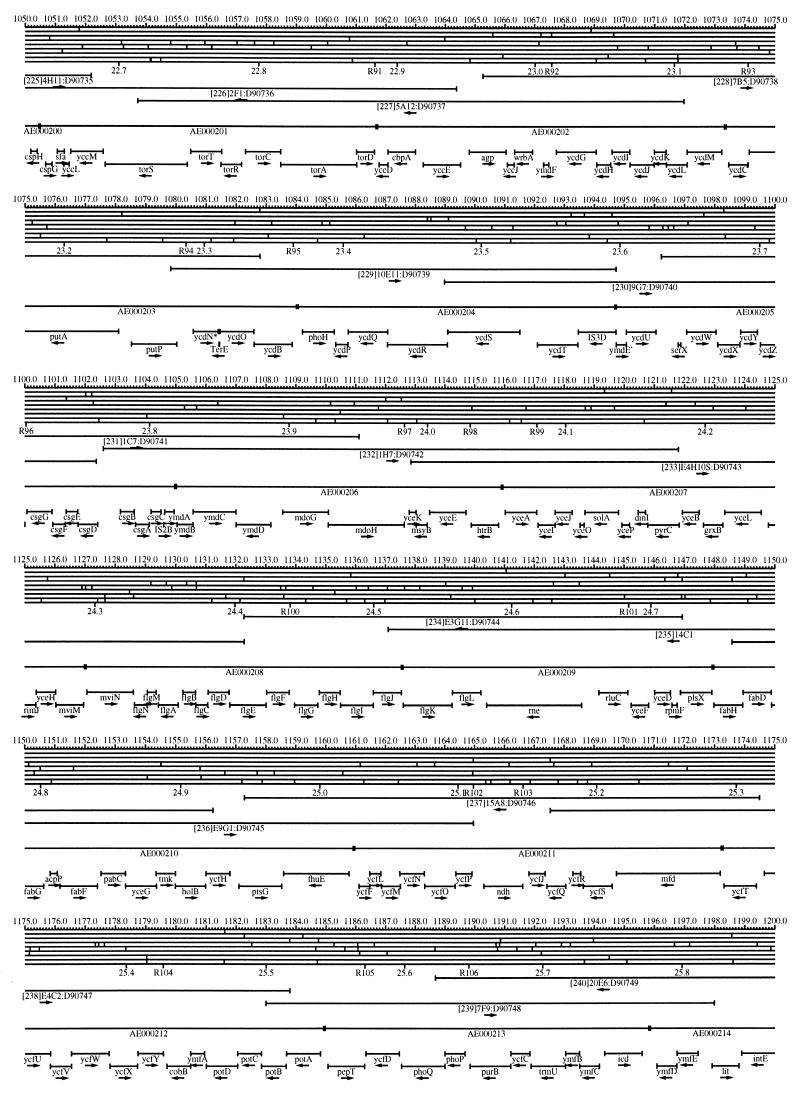
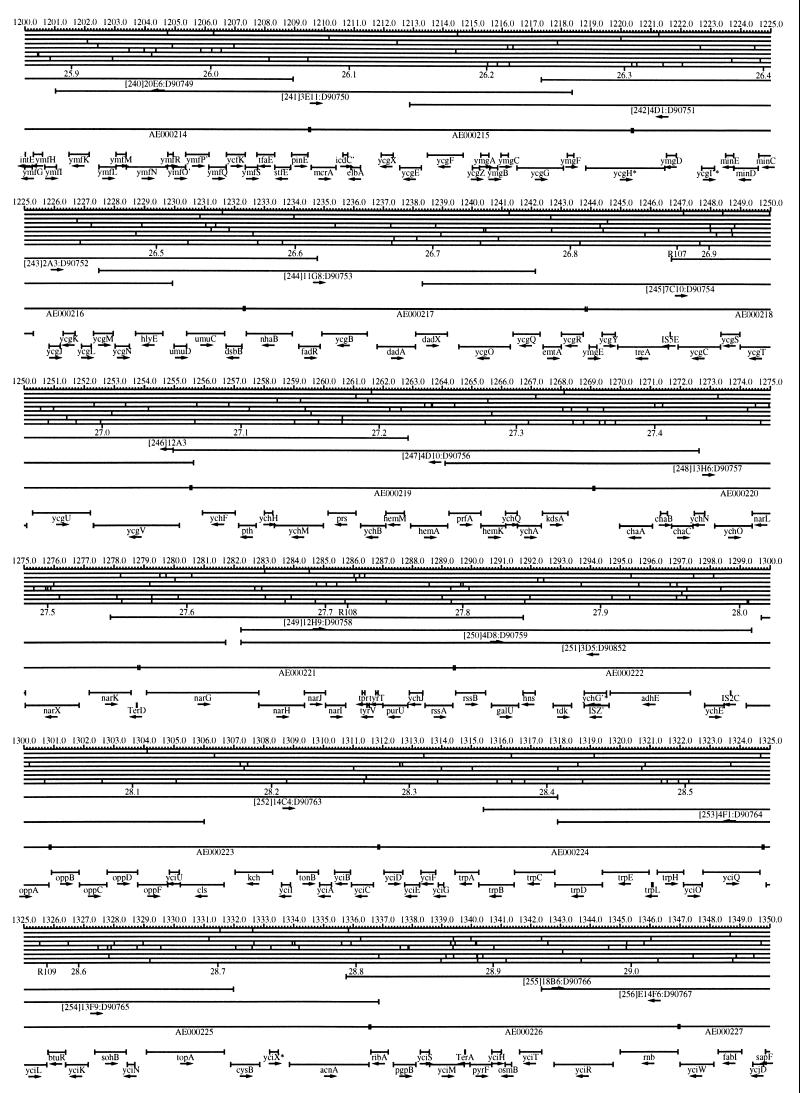
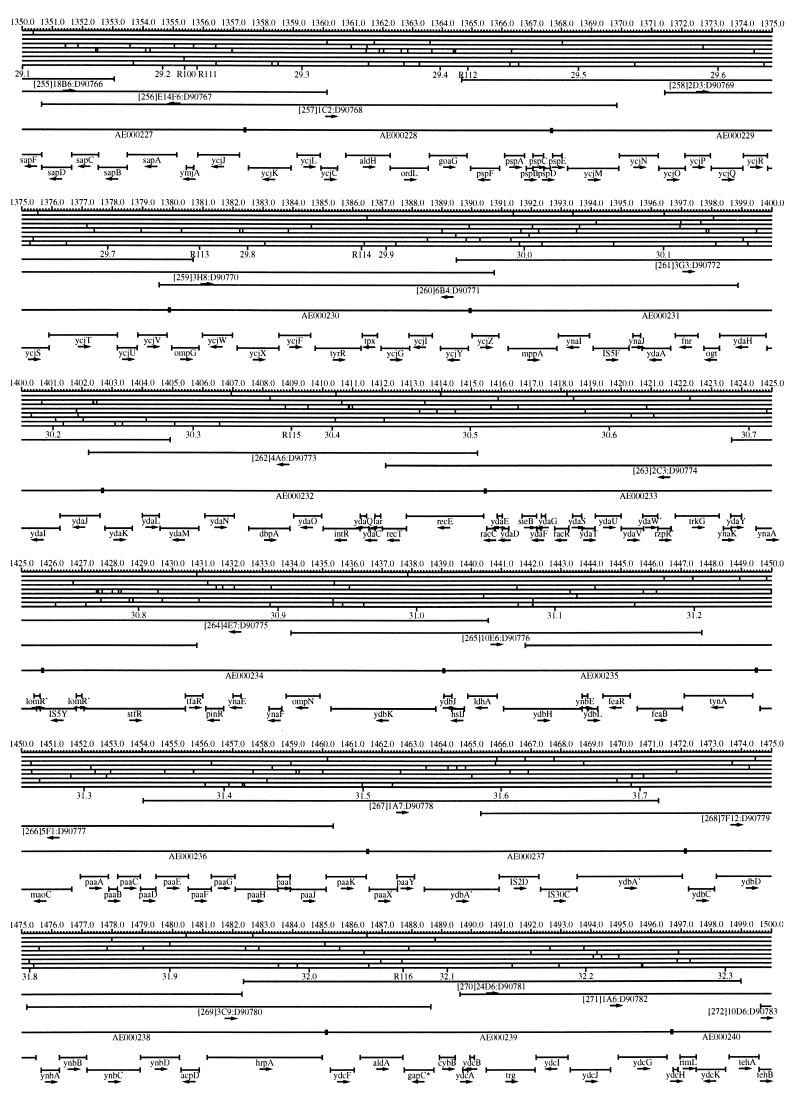
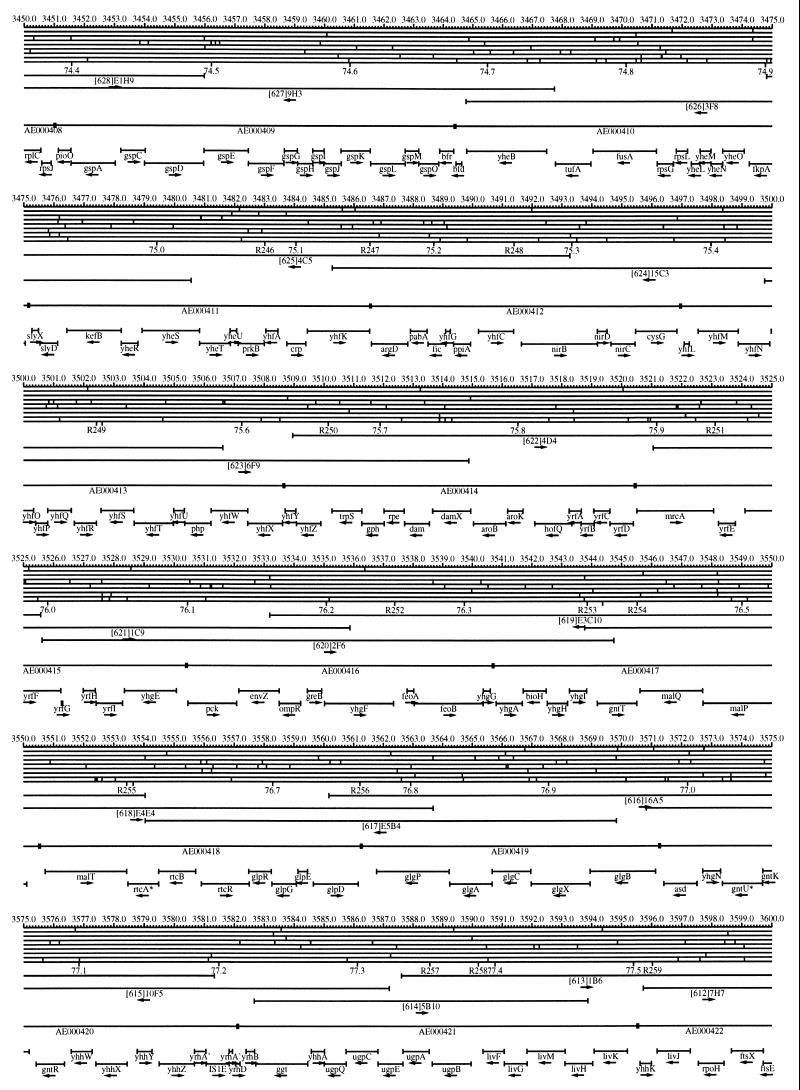
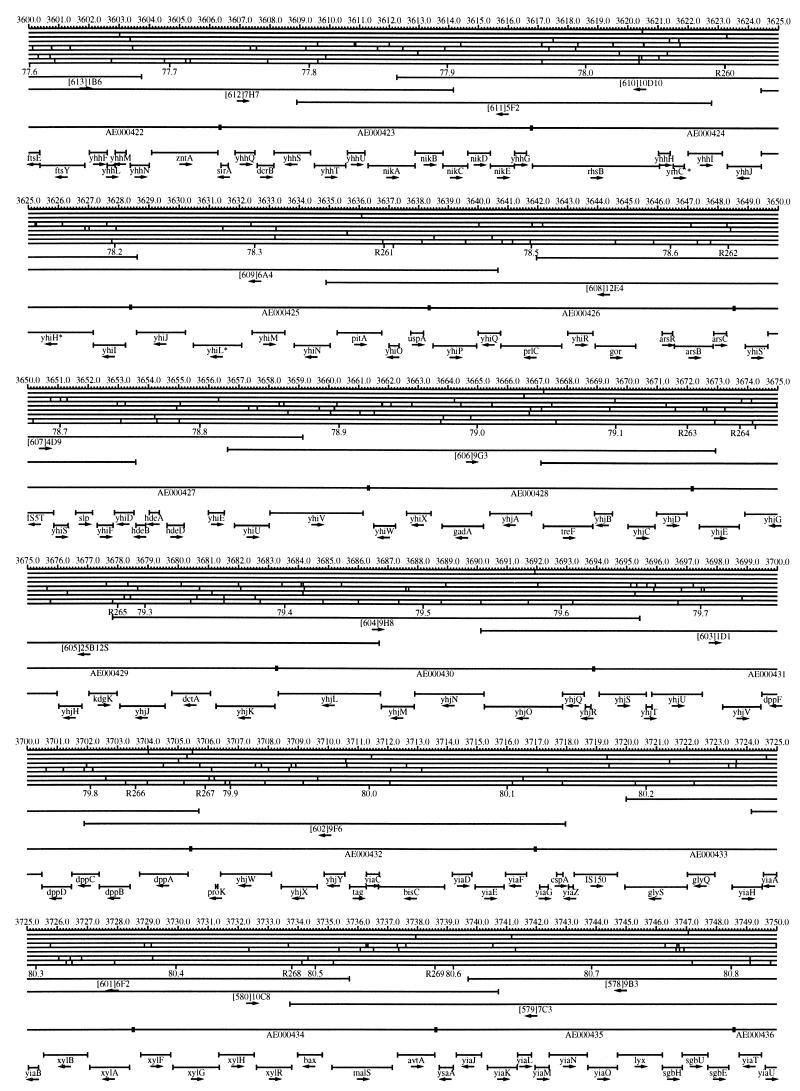
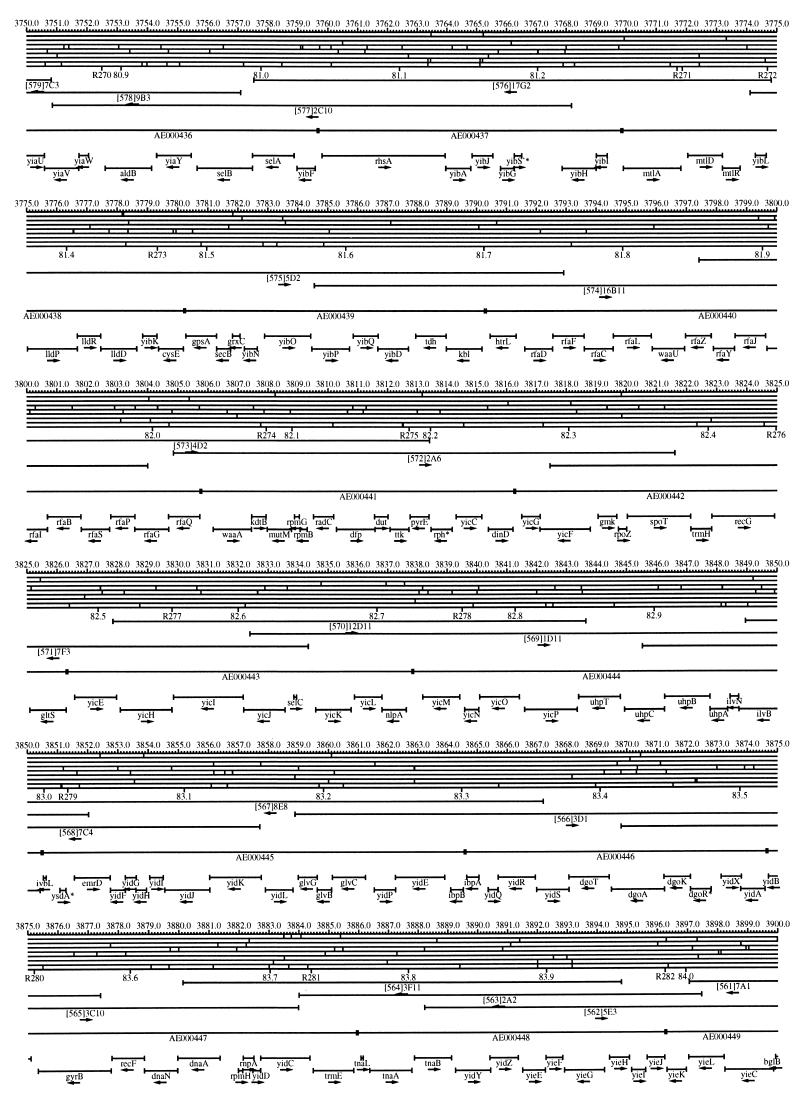
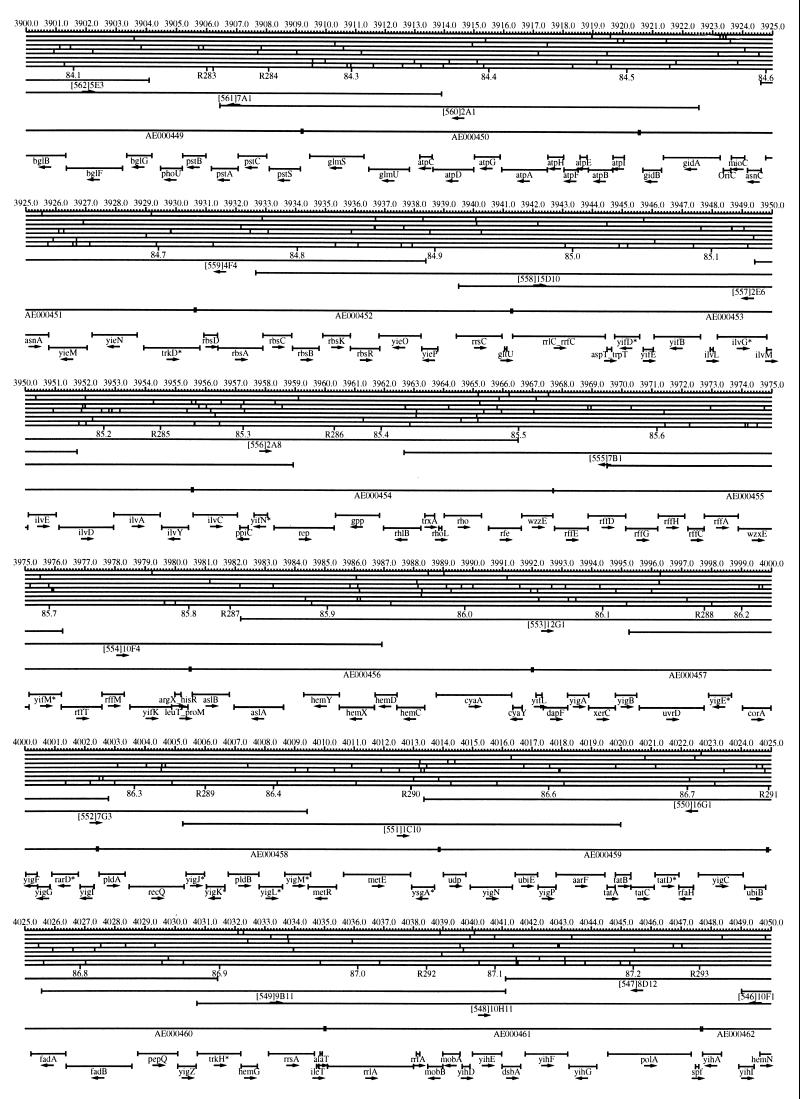
The recognition sites for the eight restriction enzymes used to create the whole genome restriction map of Kohara et al. (10) are predicted from the genomic DNA sequence. These 6-bp recognition sites are mapped at the position of their first base pair. Although any set of restriction sites can now be used to create a restriction map of the entire chromosome, this set of sites was used in order to retain continuity with the original Kohara/Isono genomic restriction map and previous EcoMap versions. This set of commonly used enzymes provides a convenient pattern of restriction sites and includes a wide range in the number of predicted recognition sites in the MG1655 genome: BamHI, 495; KpnI, 516; HindIII, 556; EcoRI, 645; PstI, 958; PvuII, 1,778; BglI, 1,919; and EcoRV, 2,040. The expected number of 6-bp restriction enzyme recognition sites in a randomly generated DNA sequence of this length and composition would be 1,133. The mean number of predicted sites for this set of eight enzymes is 1,113.
Kohara/Isono Miniset Clones
The Kohara/Isono miniset is a widely used collection of ordered E. coli bacteriophage lambda clones derived from strain E. coli K-12 W3110 (10). Four hundred and seventy-three of the original 476 miniset clones have been aligned to EcoMap10. Seven of the clones were split into two portions labeled A and B because they crossed the 0-min point, the IN(rrnD-rrnE)1 inversion endpoints, or the sites of a duplication and translocation of the tdc region specific to the Kohara/Isono version of W3110, as previously described (4, 11, 14, 17, 19, 21). One hundred and eighty-six of the clones are present in GenBank/EMBL/DDBJ as individual sequence entries, and these clones are precisely aligned to the genomic DNA sequence since their chromosomal DNA inserts have been sequenced (1, 9, 13, 22). The remaining clones were positioned by using the gel electrophoresis-derived restriction enzyme map of Kohara et al. (10) as previously described (14, 17). These clones are referred to as “unsequenced” because there are no individual GenBank/EMBL/DDBJ records available for them, even though many of them may in fact have already been sequenced. When additional information about the remaining clones becomes available, this information will be incorporated into the EcoMap alignments. Most of the miniset clones are Sau3A partial restriction fragments cloned into the BamHI site of lambda EMBL4, and no attempt was made to align the ends of the unsequenced clones to specific Sau3A sites in the genomic sequence. Twenty-four of the miniset clones depicted in EcoMap10 are EcoRI partial fragments cloned into the EcoRI site of lambda 2001, identified by clone names that begin with the designations E1 to E25. Fourteen of these have GenBank/EMBL/DDBJ entries and have terminal EcoRI restriction sites in the database entries that are all aligned to EcoRI sites in the genomic DNA sequence. The alignments of the 10 unsequenced EcoRI clones were manually adjusted so that their ends align to EcoRI restriction sites in the genomic sequence. The orientations depicted for the Kohara/Isono clone inserts indicate that the right arm of lambda is to the right of the insert’s restriction map as depicted in EcoMap10 (positive orientation, rightward arrow) or to the left (negative orientation, leftward arrow).
Caution must be taken if EcoMap10 is used as the source of a restriction map for the Kohara/Isono miniset clone since the miniset was derived from W3110. In addition to the rare occurrence of DNA sequence errors and strain-specific DNA sequence polymorphisms that might lead to minor restriction map differences, there are major differences due to genome rearrangements (noted above) and the W3110-specific IS elements (see below) (reviewed in reference 6). Solutions to this problem include using the DNA sequence database entries for the sequenced clone subset or using the original Kohara/Isono W3110 restriction map (10) for the unsequenced subset of clones. In either case, the experimental verification of critical restriction sites is recommended.
IS Elements and REP Clusters
IS and REP (also called PU) elements are repeated DNA sequences and major extragenic features of the E. coli chromosome (2, 6). The positions of the IS elements present in MG1655 are determined by searching the complete genomic MG1655 DNA sequence with representative IS family member sequences. The positions of the W3110-specific IS element insertion points are determined from the sequenced W3110 clones whenever possible or estimated from the physical mapping data as previously described (14). The orientations of the IS elements indicate the direction of transcription of the transposase gene, as previously described (4, 6). The IS5 family element orientations were depicted incorrectly in EcoMap7 (4), and this error has been corrected in EcoMap10. Three putative IS-related sequences of unknown origin were identified and are temporarily designated ISX (2793.3 kb), ISY (2714.1 kb), and ISZ (1293.8 kb). The IS-encoded genes are not considered E. coli genes in EcoGene, and it is the full length of the IS element that is represented, not the coding regions contained within them.
REP elements have been postulated to have a variety of RNA- and DNA-related functions, but the stabilization of mRNA is the only firmly established function (2). The positions of individual REP elements were determined by a variety of pattern searches, as will be described elsewhere (16). This approach identified nearly all previously reported REP elements (2) and was used to locate new REP elements. The few REP elements identified earlier that were missed by this approach were annotated manually. Individual REP elements occur in intergenic REP clusters, also called bacterial interspersed mosaic elements (BIMEs) containing from 1 to 12 REP elements interspersed with other small conserved sequences (2, 8). Three hundred and fifty-five REP clusters (BIMEs) containing a total of 697 individual REP elements were identified. Particular attention was given to the detection of a class of REP-like putative bidirectional transcription terminators referred to as PU* or Y* (2, 7). A total of 108 individual Y* elements are included in the REP tabulation (16). Y* elements can also be found as subsequences of a number of other REP elements, but these overlapping Y* elements are not counted separately in the REP tabulation. The serially numbered REP clusters (BIMEs) identified in the MG1655 genome sequence are denoted R1 to R355 directly under the restriction map portion of EcoMap10 along with the minute position labels.
Genes and ORFs
A detailed description of the annotation of genes and functionally uncharacterized ORFs in EcoGene10 is presented in a separate publication (16). The entire genome sequence annotation of protein coding regions has been reviewed, and revisions have been made to approximately 15% of them. The most frequent revisions were the choice of an alternative translation start site, although sequences encoding small proteins were added and deleted from the set of coding regions as well. These two areas were acknowledged as difficult aspects of protein coding region annotation (5), and the EcoGene annotation should be thought of as one view of the E. coli K-12 genome. Producing a set of predicted protein sequences as accurately as possible was the goal of the reannotation effort, but experimental verification is the only way to establish the coding regions definitively. Published experimental data was used to establish gene intervals as much as possible. Anyone wishing to communicate additional prepublication information directly is encouraged to do so, especially if he or she has no objection to the information being made publicly available in the EcoGene and SWISS-PROT databases as a personal communication. The E. coli genome sequence annotation refinement has been a close collaboration with the curator of the SWISS-PROT database, Amos Bairoch.
Partial or frameshifted ORFs and genes are marked in Fig. 1 (see the figure legend). In most cases, but not all, the presence of a frameshift or deletion is based on sequence analysis alone and thus should be considered a prediction. It is not known if any particular putative frameshift or deletion is the result of a DNA sequencing error, a cloning artifact, an adaptation to the laboratory environment, natural evolutionary pressure, or pseudogene formation. Errors introduced during the reannotation process are also possible, and everyone is encouraged to contact this author or SWISS-PROT if he or she thinks an error has been made; we will take appropriate steps to update our databases. These sequence-based frameshift predictions should assist in the experimental determination of the source of the frameshifts.
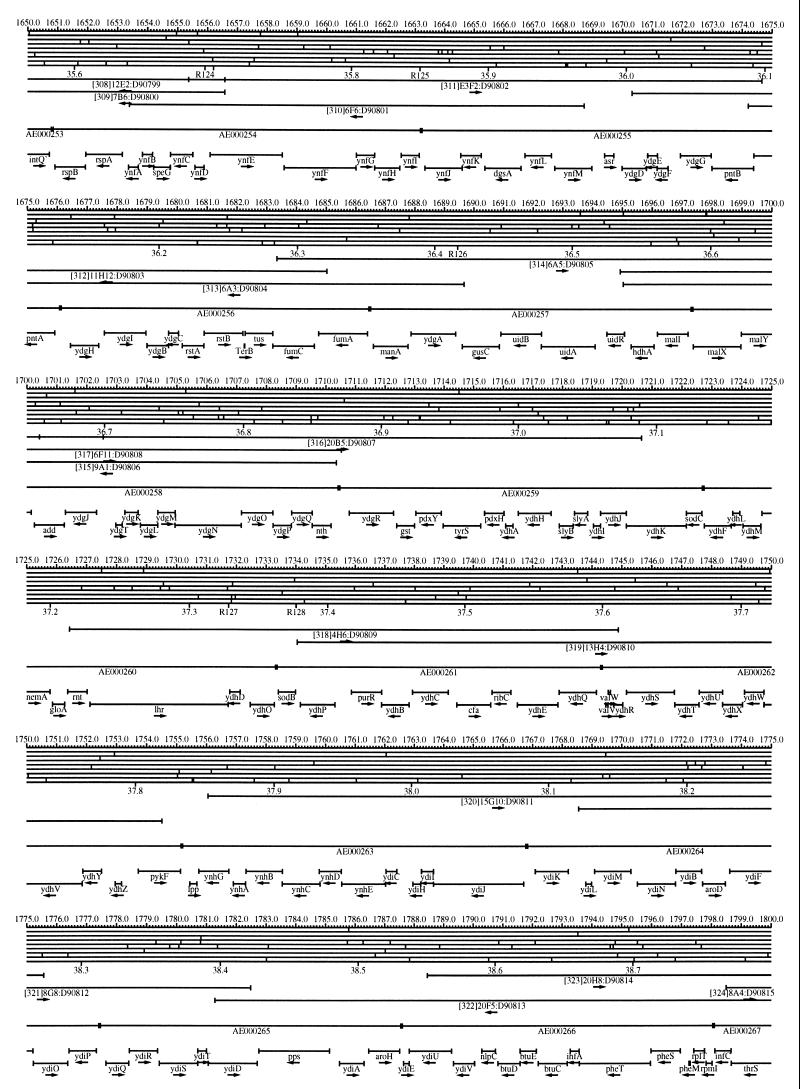
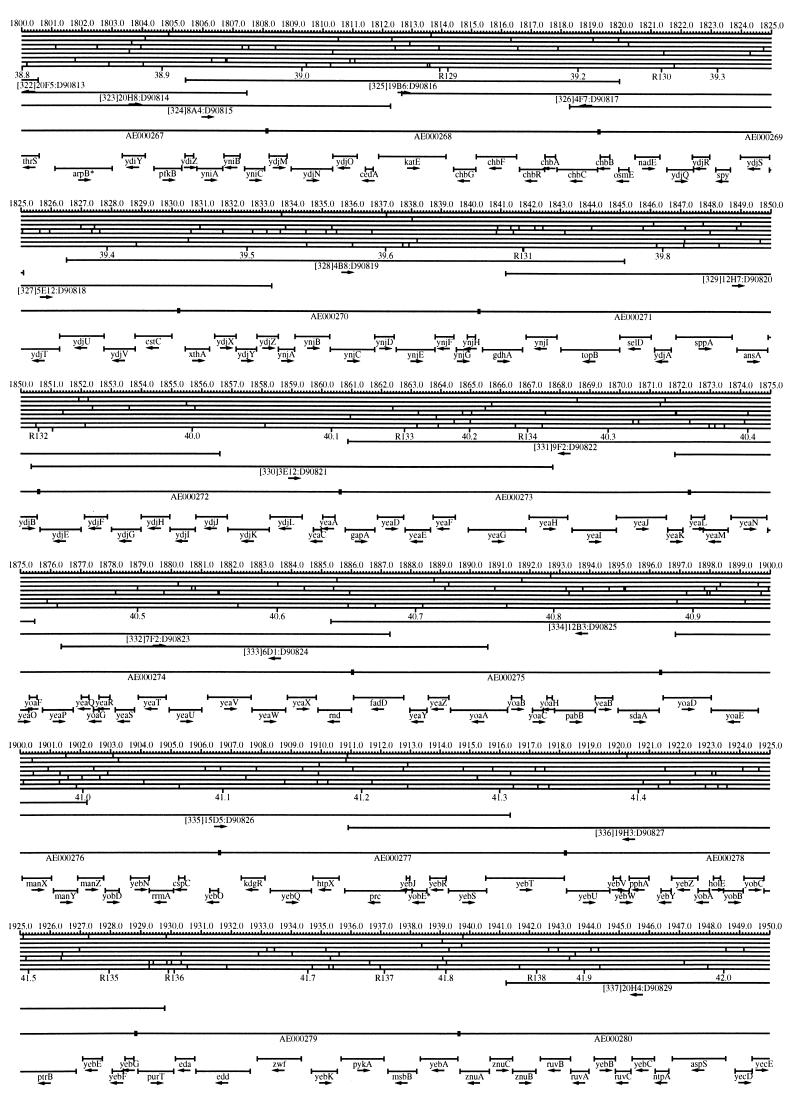
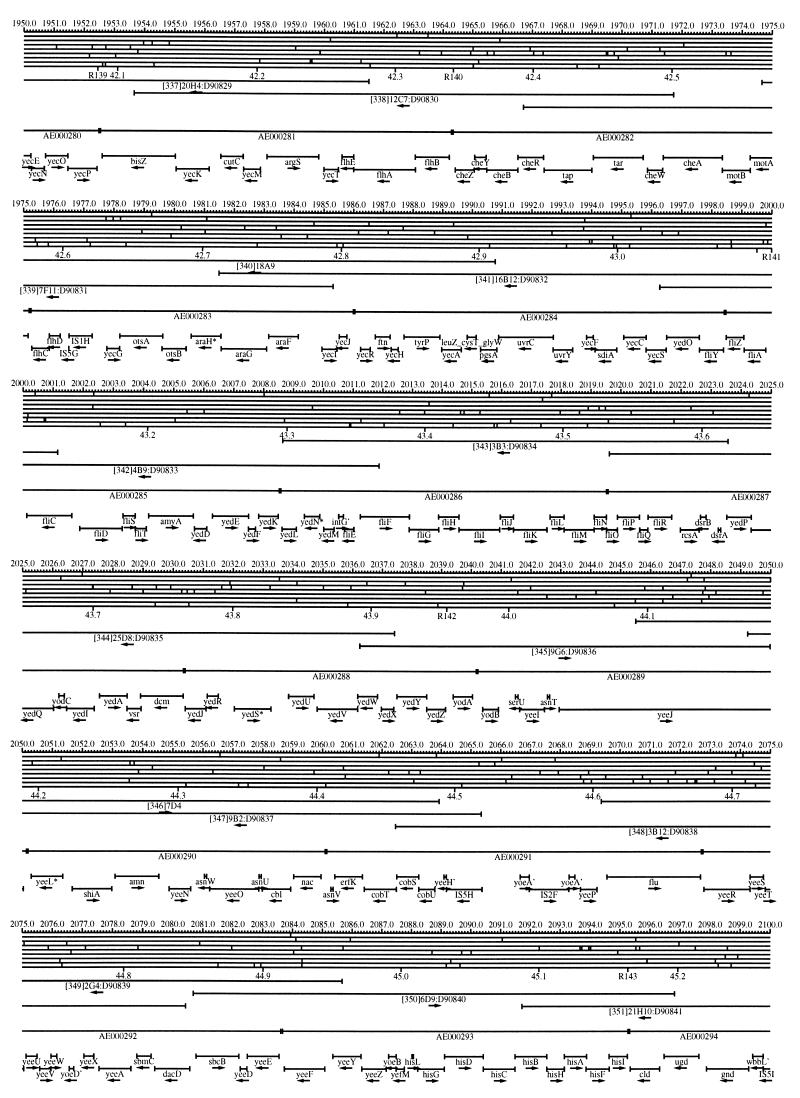
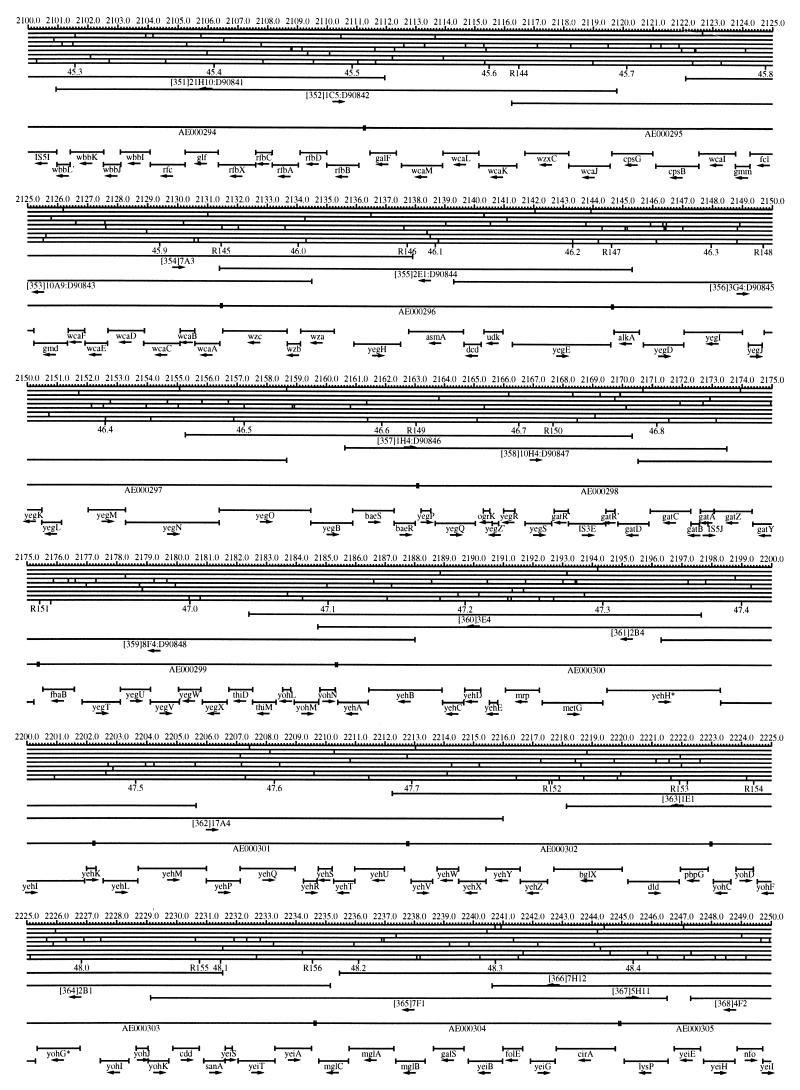
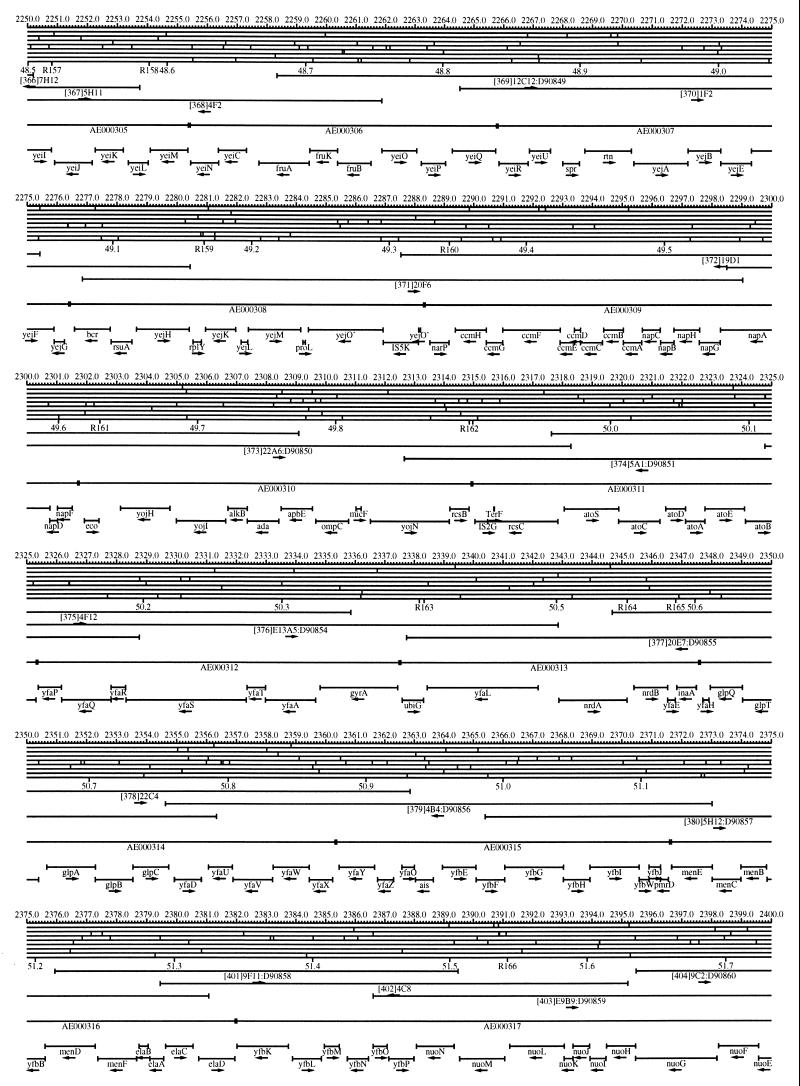
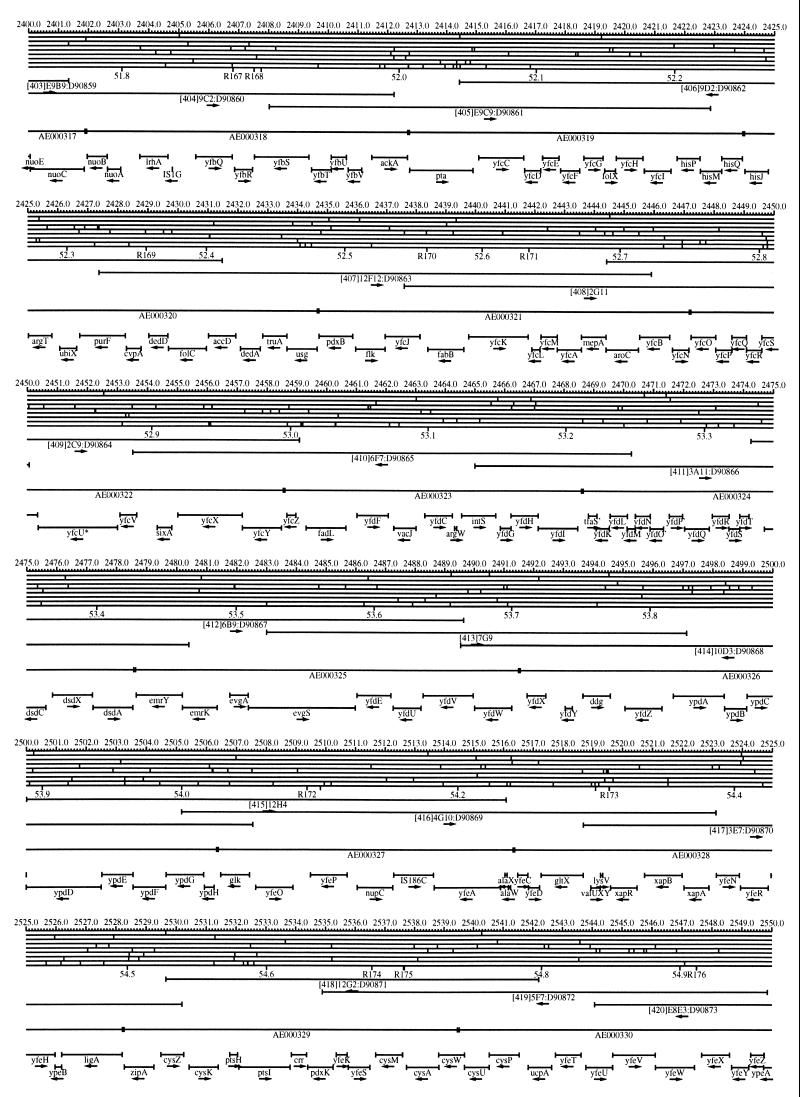
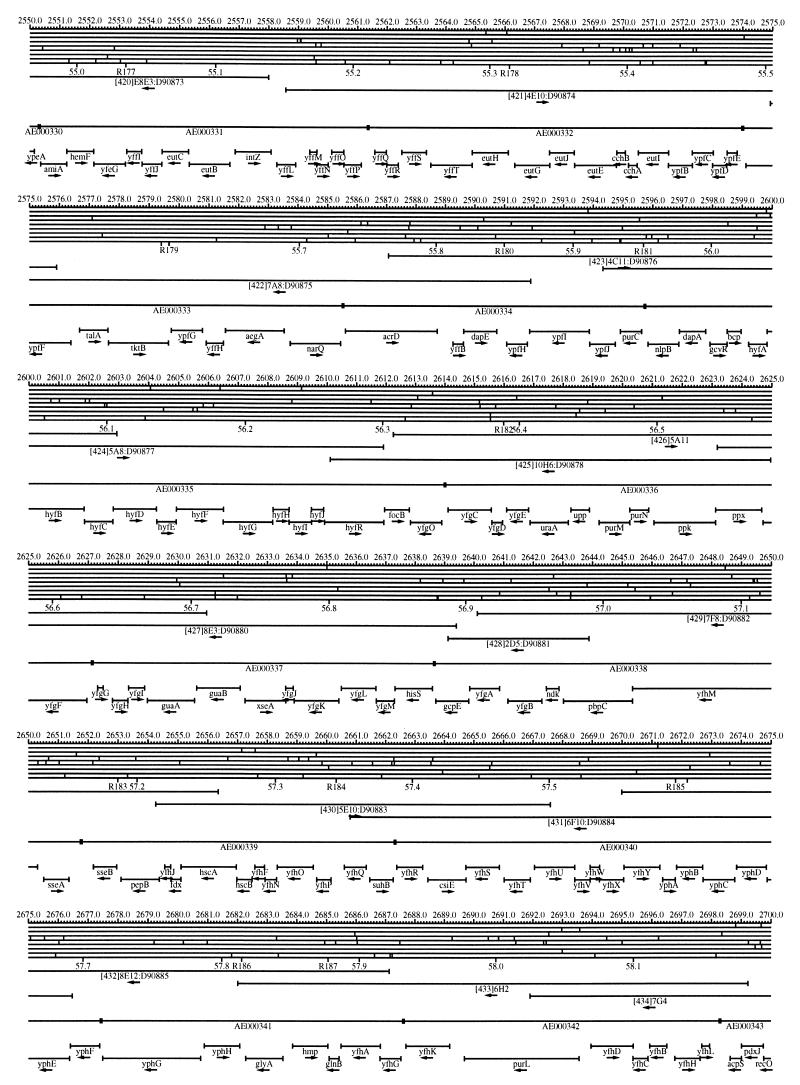
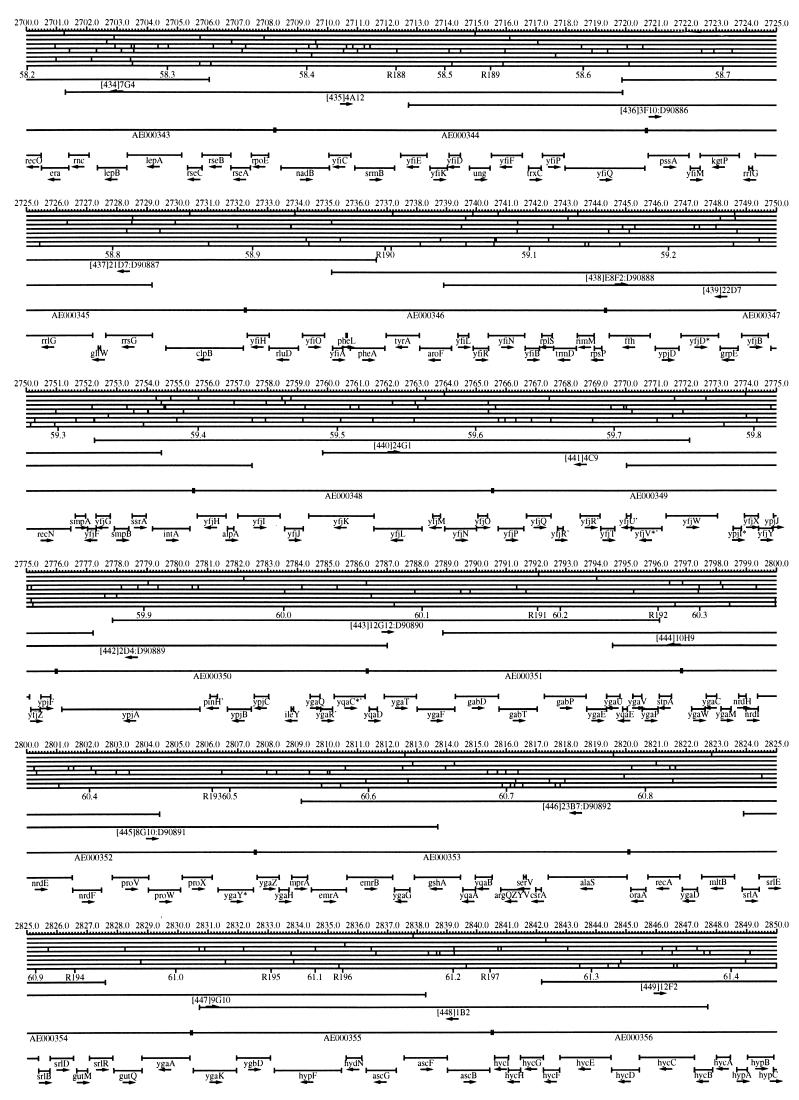
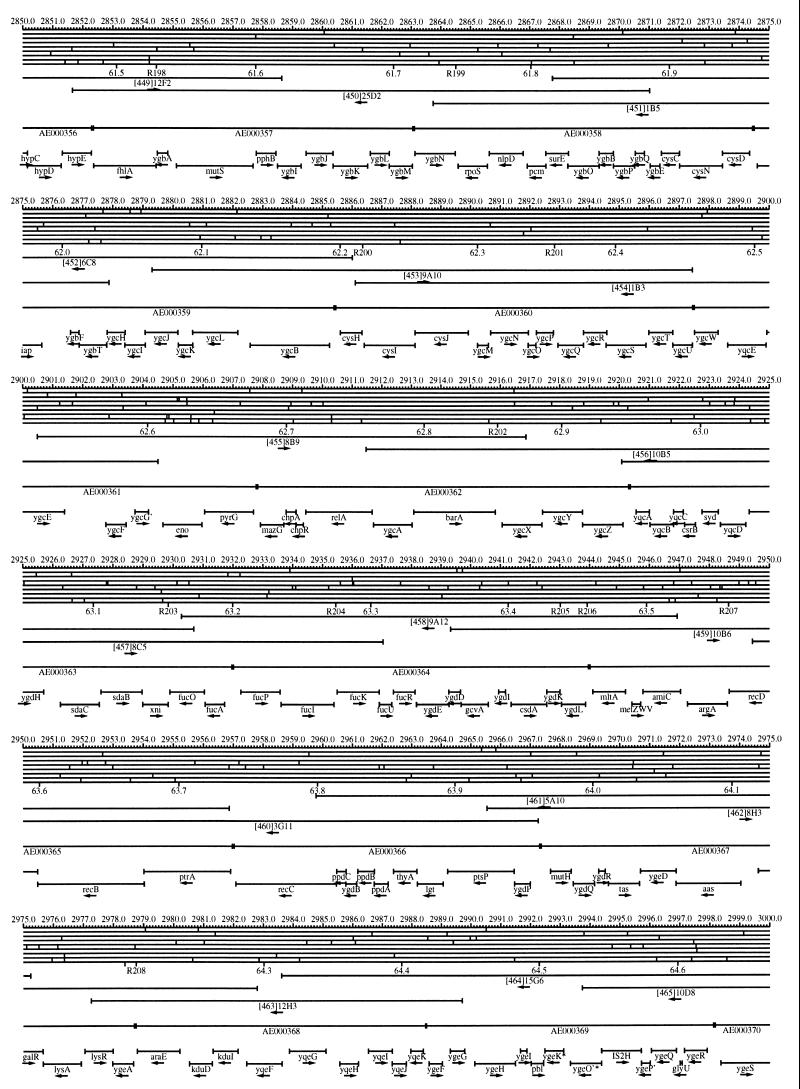
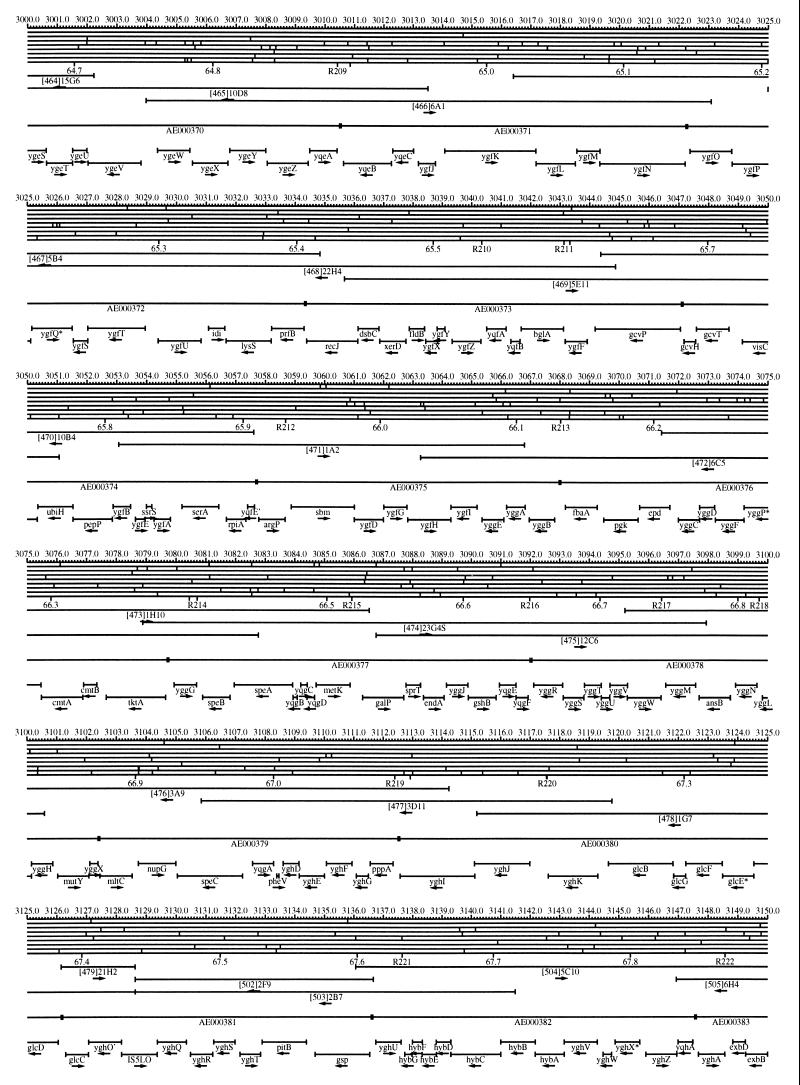
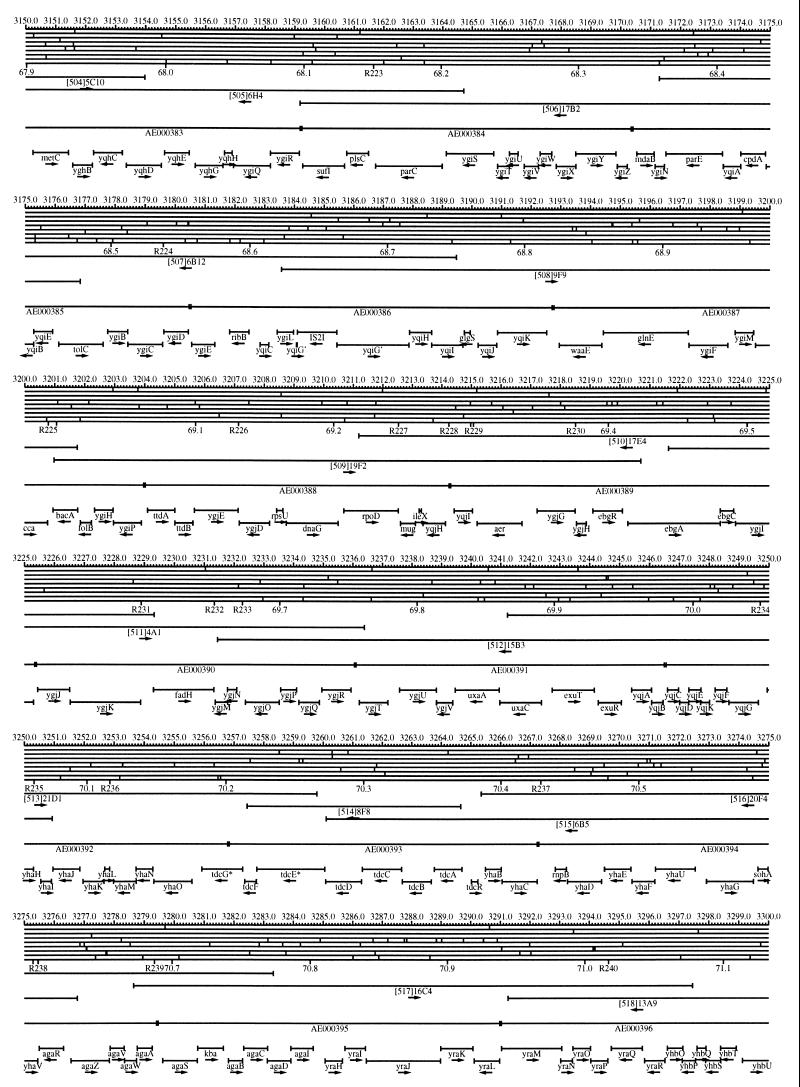
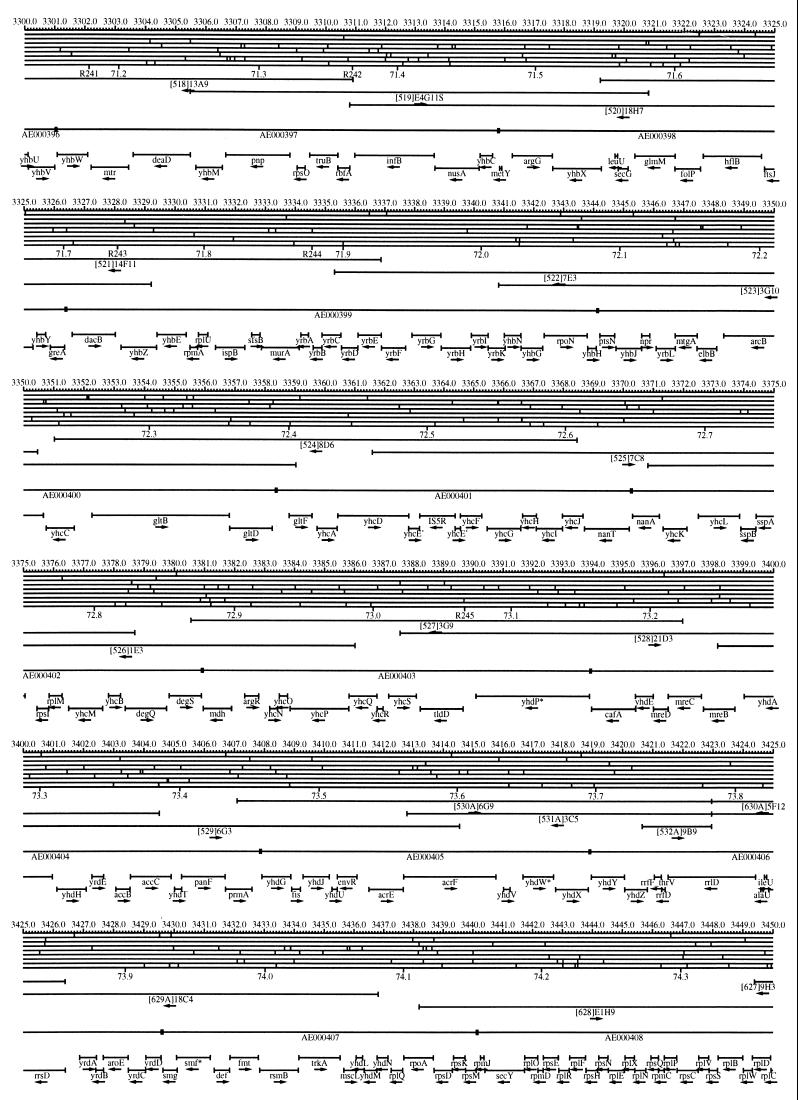
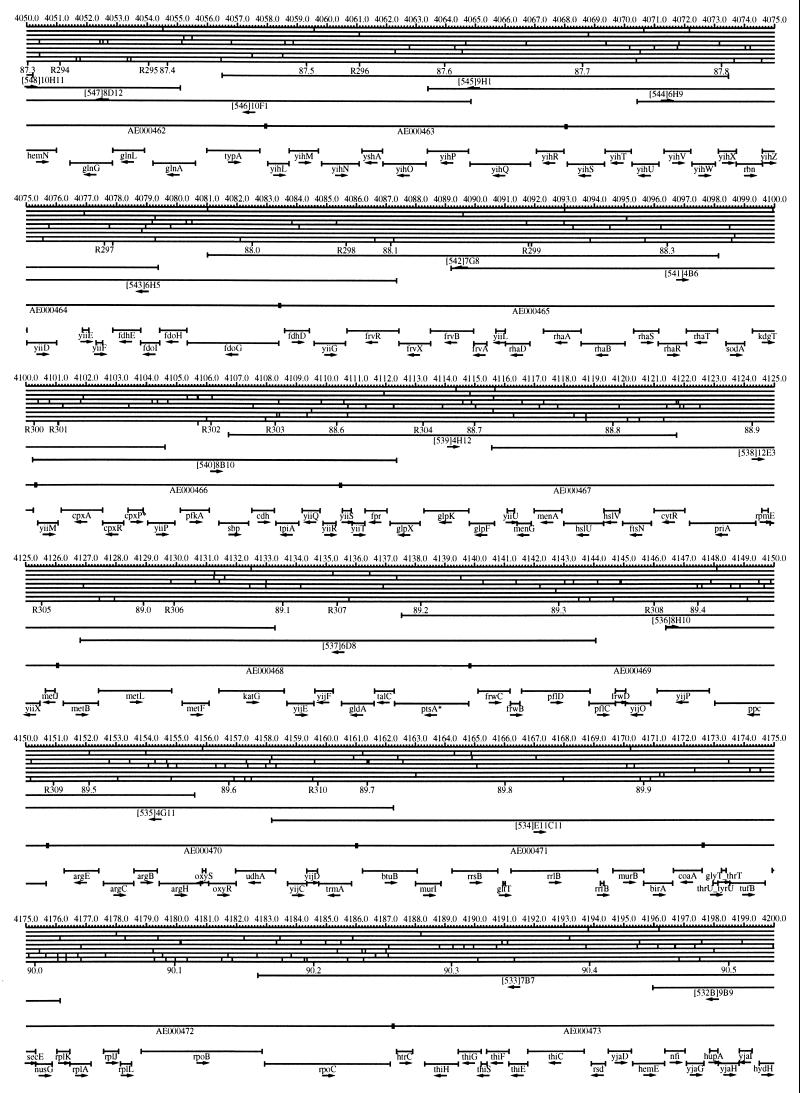
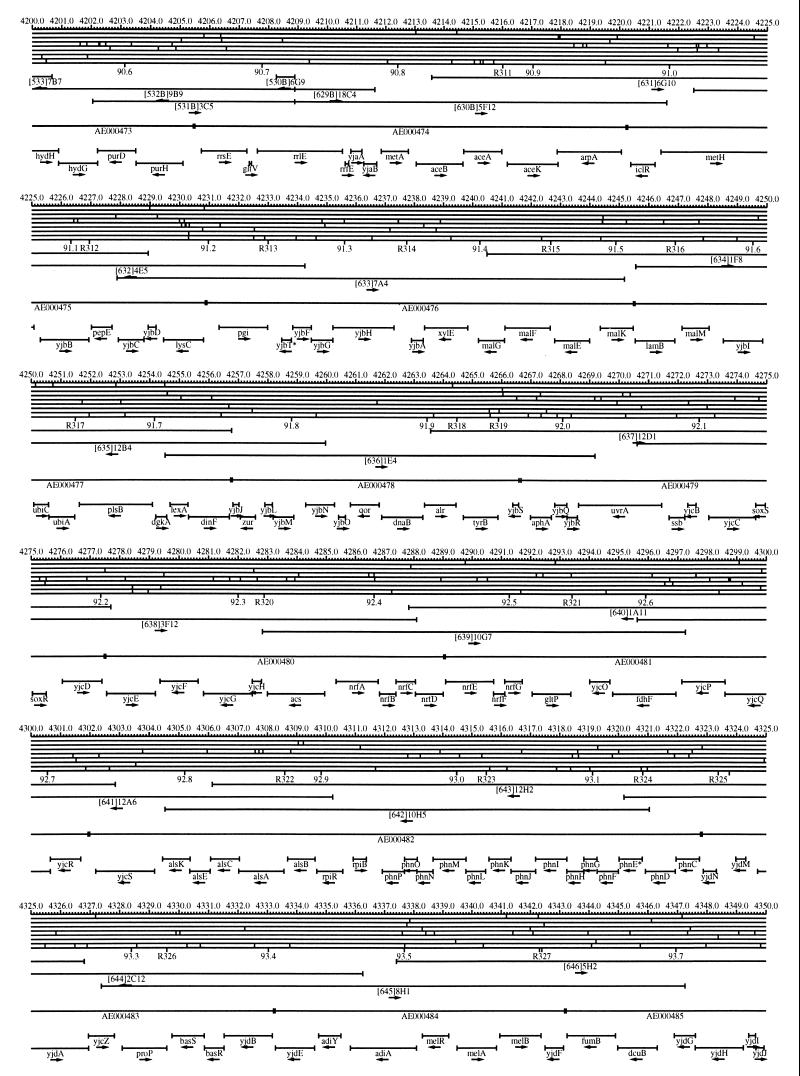
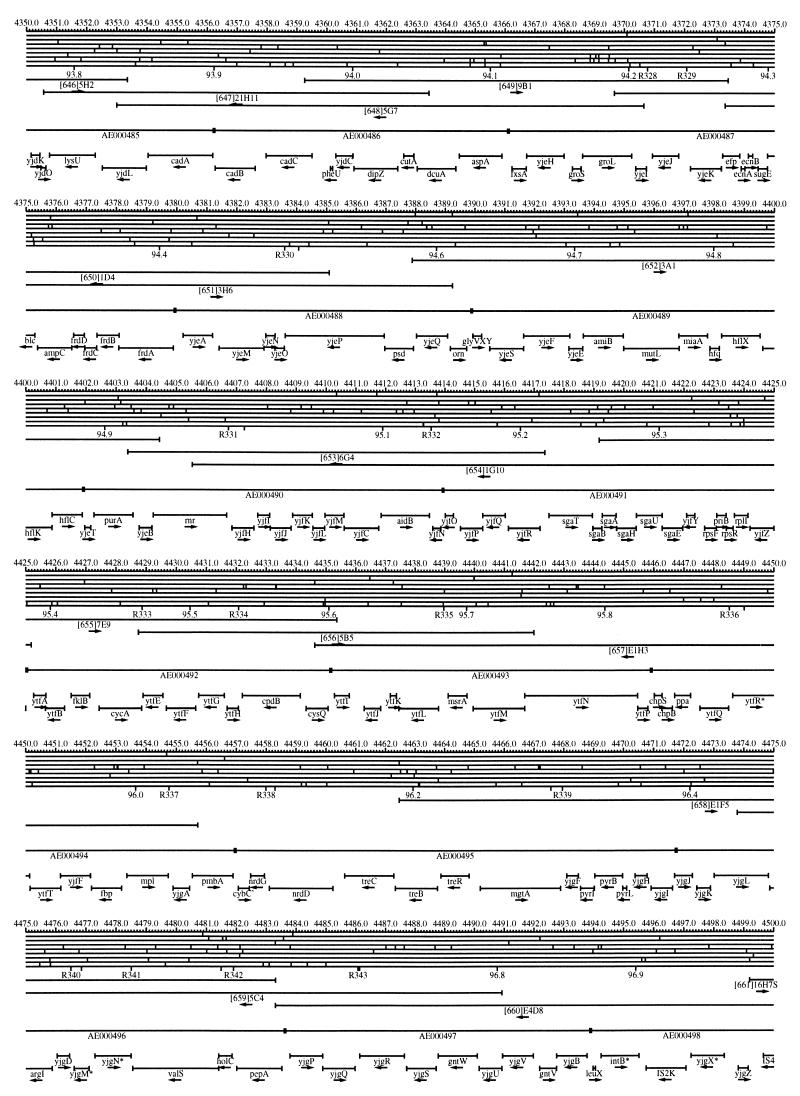
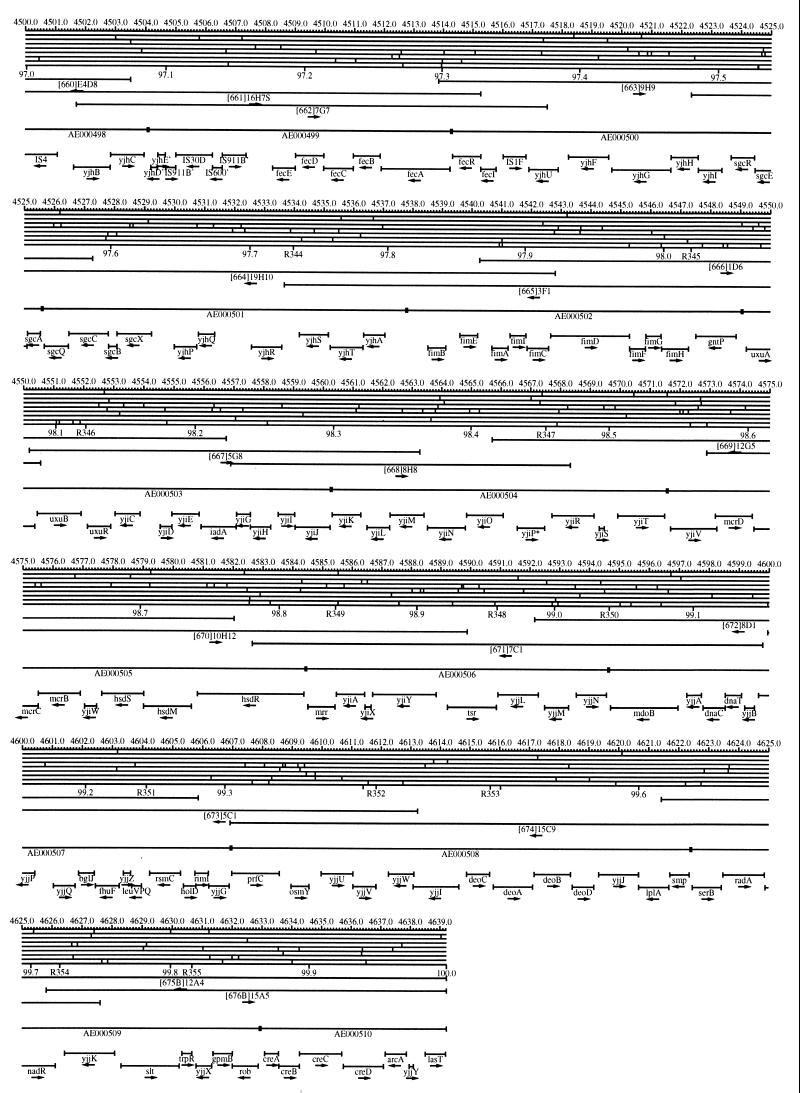
FIG. 1.
EcoMap10, a DNA sequence-derived map depicting restriction sites, Kohara/Isono clones, genes, ORFs, IS elements, and REP clusters of the E. coli K-12 chromosome. The derivation of this map from the complete genome sequence of E. coli K-12 strain MG1655 is briefly described in the text. The map depicts sites for eight restriction enzymes (top line to bottom line: BamHI, HindIII, EcoRI, EcoRV, BglI, KpnI, PstI, and PvuII). Above the restriction map are position coordinates in kilobases; immediately below the map are minute coordinates (in 0.1-min increments). Also immediately below the map are the designations R1 to R355 referring to the 355 serially numbered REP clusters, placed at the genomic position of the base pair at their left ends. Some minute designations were omitted as they overlapped with the REP serial numbers, but the tick marks for these unlabeled 0.1-minute positions are present, and their values can be easily determined from the flanking minute values. The first set of spanning lines below the map represent the genomic positions and clone insert orientations of the Kohara miniset clones. Those Kohara miniset clone W3110 chromosomal DNA inserts that have been completely sequenced are additionally labeled with their GenBank/EMBL/DDBJ accession numbers, D90699 to D90892 (1, 9, 13, 22). The second set of spanning lines, labeled with database accession numbers AE000111 to AE000510, represent the locations of the GenBank/EMBL/DDBJ complete-genome MG1655 sequence entries of Blattner et al. (5). The third set of spanning lines depict the positions and orientations of the genes, ORFs, and IS elements that constitute EcoGene10. An asterisk following a gene or ORF name indicates that a frameshift or in-frame stop codon that prevents the EcoGene10 representation of the coding region from being translated is present in the genome sequence. A prime indicates a partial EcoGene entry, i.e., a deletion or IS element insertion is predicted to have disrupted the ancestral complete gene, ORF, or IS element. This figure was created by using the PrintMap Postscript drawing program, which implements the Plasmid Description Language developed by Craig Werner (18).
The traditional and physical maps of edition 10 of the Escherichia coli K-12 linkage map are closely correlated. When there is a choice of several synonyms to use as the primary gene name, the physical map uses the same primary gene name as the traditional map. The primary names of genes not yet in the E. coli Genetic Stock Center (CGSC) database are considered provisional primary gene names. When choosing names for genes that are being functionally characterized for the first time, gene names already present in the database at the CGSC or the EcoGene database should be avoided. Guidance on naming and renaming genes is given in the paper containing the traditional map (3).
For the cases in which no standard-format gene name was assigned to a functionally uncharacterized gene or ORF, a systematic ORF nomenclature, the “y” naming system, was used to generate a provisional name (4, 15, 20). The first three letters of a “y” name are based on the map position of an ORF at the time the name was assigned. Similar to the “z” naming system for transposon insertions, ya[a to j]A to Z designates ORFs in the 0- to 10-min region of the chromosome, yb[a to j]A to Z designates ORFs in the 11- to 20-min region, and so on. The fourth letters (A to Z) can be assigned in any order within the 1-min interval. If all 26 names in any 1-min interval are exhausted, a new second letter is assigned to generate another 26 possibilities; additional ORFs after yaaZ would be ykaA, ykaB, and so on; additional ORFs after ybaZ would be ylaA, ylaB, and so on. The “y” names are not reused if a “y” ORF is given a new gene name or if an ORF becomes defunct, e.g., if a frameshift correction fuses two adjacent ORFs. Map locations provide a convenient and systematic method for naming ORFs, and the “y” names can guide one to an approximate map position. However, to avoid unnecessary renaming the “y” name of an ORF is not changed if a map revision moves it into an adjacent minute interval. The “y” names are now assigned to all the functionally uncharacterized, unnamed ORFs in EcoGene10. Once a new function is established for an E. coli gene, the provisional “y” name should be abandoned and a new gene name should be chosen.
Information concerning the availability of the EcoMap10 and EcoGene10 electronic datasets in various formats, including the Colibri database management program (12), can be obtained at http://cesspit.med.miami.edu. Additional information about the genes and ORFs in EcoGene10 is contained in SWISS-PROT records (http://www.expasy.ch/sprot) that can be accessed by using the names that are depicted on EcoMap10 and that are indexed in a master file (http://www.expasy.ch/cgi-bin/lists?ecoli.txt.
ACKNOWLEDGMENTS
This work was supported by funds made available to K.E.R. from a Lucille P. Markey Charitable Trust grant to the Department of Biochemistry and Molecular Biology at the University of Miami School of Medicine.
I am especially indebted to Amos Bairoch for his dedication to E. coli, his enthusiastic support of EcoGene, and for the many gene discoveries, literature citations, and protein sequence refinements that he has shared with me since the beginning of the EcoGene project. I thank Yuji Kohara and Katsumi Isono for providing the miniset of E. coli lambda clones, for allowing me to freely redistribute them, and for providing the original individual restriction maps of each miniset clone in electronic format. My collaboration with Mary Berlyn of the CGSC has been an essential component of the EcoMap/EcoGene project, and I am grateful for her patience and kindness throughout our data sharing and map coordination effort. I would also like to thank and acknowledge Gabrielle Redfern, Yuhong Zuo, Webb Miller, Karl Sirotkin, Craig Werner, Gerald Bouffard, Bobby Baum, Mark Borodovsky, Nir Hus, Rick Mitchell, Valerie Wasinger, Peter Maxwell, Ian Humphery-Smith, Ivan Moszer, and Antoine Danchin variously for general assistance, programming support, and helpful comments as well as for their continuing friendship. I acknowledge this work as a being derived from the many scientific contributions of the entire E. coli research community and extend my sincere gratitude to this community for their contributions. I gratefully acknowledge Fred Blattner and his colleagues for the complete MG1655 sequence and all the members of the Japanese research consortium who participated in the sequencing of the W3110 genome.
REFERENCES
- 1.Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kasai H, Kashimoto K, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasunddaram S, Tagami H, Takeda J, Takemoto K, Takeuchi Y, Wada C, Yamamoto Y, Horiuchi T. A 570-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 28.0-40.1 min region on the linkage map. DNA Res. 1996;3:363–377. doi: 10.1093/dnares/3.6.363. [DOI] [PubMed] [Google Scholar]
- 2.Bachellier S, Gilson E, Hofnung M, Hill C W. Repeated sequences. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2012–2040. [Google Scholar]
- 3.Berlyn M B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlyn M B, Low K B, Rudd K E. Integrated linkage map of Escherichia coli K-12, edition 9. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1715–1902. [Google Scholar]
- 5.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Deonier R C. Native insertion sequence elements: locations, distributions, and sequence relationships. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2000–2011. [Google Scholar]
- 7.Gilson E, Rousset J P, Clement J M, Hofnung M. A subfamily of E. coli palindromic units implicated in transcription termination? Ann Inst Pasteur Microbiol. 1986;137B:259–270. doi: 10.1016/s0769-2609(86)80116-8. [DOI] [PubMed] [Google Scholar]
- 8.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. The BIME family of bacterial highly repetitive sequences. Res Microbiol. 1991;142:217–222. doi: 10.1016/0923-2508(91)90033-7. [DOI] [PubMed] [Google Scholar]
- 9.Itoh T, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Kasai H, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Sato N, Sampei G, Seki Y, Sivasunddaram S, Tagami H, Takeda J, Takemoto K, Wada C, Yamamoto Y, Horiuchi T. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1-50.0 min region on the linkage map. DNA Res. 1996;3:379–392. doi: 10.1093/dnares/3.6.379. [DOI] [PubMed] [Google Scholar]
- 10.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 11.Komine Y, Inokuchi H. Precise mapping of the rnpB gene encoding the RNA component of RNase P in Escherichia coli K-12. J Bacteriol. 1991;173:1813–1816. doi: 10.1128/jb.173.5.1813-1816.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medigue C, Viari A, Henaut A, Danchin A. Colibri: a functional data base for the Escherichia coli genome. Microbiol Rev. 1993;57:623–654. doi: 10.1128/mr.57.3.623-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshima T, Aiba H, Baba T, Fujita K, Hayashi K, Honjo A, Ikemoto K, Inada T, Itoh T, Kajihara M, Kanai K, Kashimoto K, Kimura S, Kitagawa M, Makino K, Masuda S, Miki T, Mizobuchi K, Mori H, Motomura K, Nakamura Y, Nashimoto H, Nishio Y, Saito N, Sampei G, Seki Y, Tagami H, Takemoto K, Wada C, Yamamoto Y, Yano M, Horiuchi T. A 718-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 12.7-28.0 min region on the linkage map. DNA Res. 1996;3:137–155. doi: 10.1093/dnares/3.3.137. [DOI] [PubMed] [Google Scholar]
- 14.Rudd K E. Alignment of E. coli DNA sequences to a revised, integrated genomic restriction map. In: Miller J, editor. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 2.3–2.4.3. [Google Scholar]
- 15.Rudd K E. Maps, genes, sequences, and computers: an Escherichia coli case study. ASM News. 1993;59:335–341. [Google Scholar]
- 16.Rudd, K. E., M. K. B. Berlyn, Y. Zuo, A. Danchin, I. Moszer, N. Hus, R. Mitchell, and A. Bairoch. Submitted for publication.
- 17.Rudd K E, Miller W, Ostell J, Benson D A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990;18:313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudd K E, Miller W, Werner C, Ostell J, Tolstoshev C, Satterfield S G. Mapping sequenced E. coli genes by computer: software, strategies and examples. Nucleic Acids Res. 1991;19:637–647. doi: 10.1093/nar/19.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweizer H P, Datta P. Physical map location of the tdc operon of Escherichia coli. J Bacteriol. 1990;172:2825. doi: 10.1128/jb.172.6.2825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart A. Genetic nomenclature guide including information on genomic databases. New York, N.Y: Elsevier; 1995. [Google Scholar]
- 21.Umeda M, Ohtsubo E. Mapping of insertion element IS5 in the Escherichia coli K-12 chromosome. Chromosomal rearrangements mediated by IS5. J Mol Biol. 1990;213:229–237. doi: 10.1016/S0022-2836(05)80186-X. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto Y, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kimura S, Kitagawa M, Makino K, Miki T, Mitsuhashi N, Mizobuchi K, Mori H, Nakade S, Nakamura Y, Nashimoto H, Oshima T, Oyama S, Saito N, Sampei G, Satoh Y, Sivasundaram S, Tagami H, Takahashi H, Takeda J, Takemoto K, Uehara K, Wada C, Yamagata S, Horiuchi T. Construction of a contiguous 874-kb sequence of the Escherichia coli-K12 genome corresponding to 50.0-68.8 min on the linkage map and analysis of its sequence features. DNA Res. 1997;4:91–113. doi: 10.1093/dnares/4.2.91. [DOI] [PubMed] [Google Scholar]