Abstract
Energy metabolism by bacteria is well understood from the chemiosmotic viewpoint. We know that bacteria extrude protons across the plasma membrane, establishing an electrochemical potential that provides the driving force for various kinds of physiological work. Among these are the uptake of sugars, amino acids, and other nutrients with the aid of secondary porters and the regulation of the cytoplasmic pH and of the cytoplasmic concentration of potassium and other ions. Bacteria live in diverse habitats and are often exposed to severe conditions. In some circumstances, a proton circulation cannot satisfy their requirements and must be supplemented with a complement of primary transport systems. This review is concerned with cation transport in the fermentative streptococci, particularly Enterococcus hirae. Streptococci lack respiratory chains, relying on glycolysis or arginine fermentation for the production of ATP. One of the major findings with E. hirae and other streptococci is that ATP plays a much more important role in transmembrane transport than it does in nonfermentative organisms, probably due to the inability of this organism to generate a large proton potential. The movements of cations in streptococci illustrate the interplay between a variety of primary and secondary modes of transport.
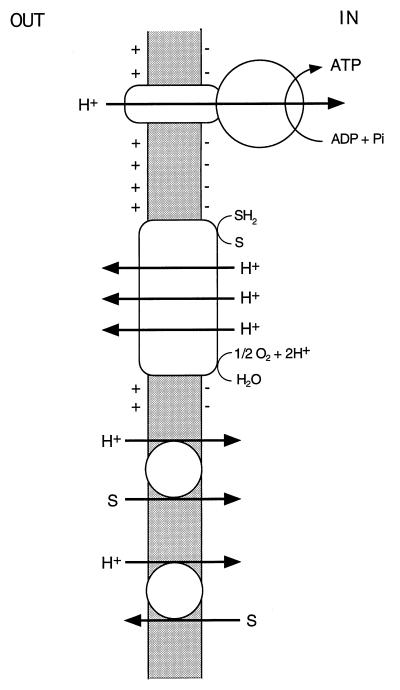
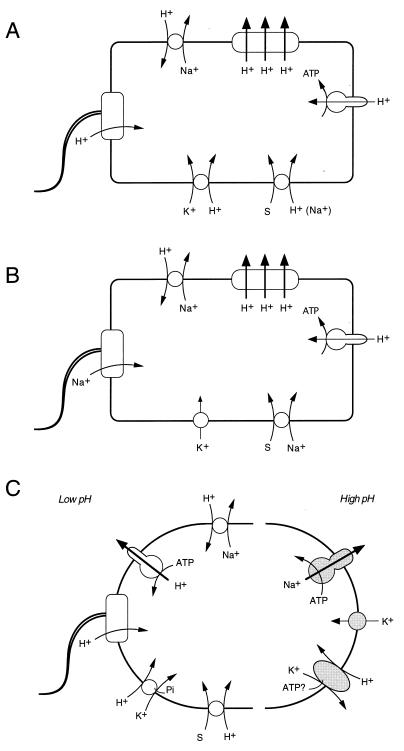
It is well established that bacteria conserve and transduce metabolic energy by means of an electrochemical gradient of hydrogen ions across the cytoplasmic membrane (ΔμH+), in accordance with the chemiosmotic theory of Peter Mitchell (168–171). According to this theory, extrusion of protons via one primary transport system or another establishes a proton potential. A primary transport system, or primary pump, is defined as active transport directly linked to a metabolic reaction; examples include electron transport by a redox chain, a proton-translocating ATPase (Fig. 1), and a light-driven reaction such as the photosynthetic reaction center and bacteriorhodopsin (72–74). The electrochemical gradient of protons ΔμH+ (proton potential, Δp) across the plasma membrane is the sum of two components, an electrical potential (ΔΨ, interior negative) and a pH gradient (ΔpH, interior alkaline). The relationship of these parameters is described by Δp = ΔΨ − ZΔpH, where ΔpH is the difference between the pH of the bulk medium and that of the cytosol and the factor Z is 2.303RT/F and is 59 mV at 25°C. The proton potential (proton motive force) can then be used by the cells to drive proton-linked energy-consuming processes. Most important, it is employed in the synthesis of ATP from ADP and inorganic phosphate by the FOF1-ATP synthase and in active transport by secondary transport systems which are not associated with a concurrent chemical reaction. Porters perform osmotic work by coupling the flux of one solute to that of another, for example protons. The linkage of coupled fluxes with the same direction in space is called symport, and the linkage of those with the opposite direction is called antiport (Fig. 1). Exergonic and endergonic reactions are thus coupled via the circulation of protons across the membrane (74, 78).
FIG. 1.
Chemiosmotic energy coupling. Electrogenic proton extrusion by the respiratory chain generates an electrochemical gradient of protons ΔμH+ (proton potential), composed of a pH gradient (inside alkaline) and a membrane potential (inside negative). Proton flow into the cytoplasm via FOF1-ATP synthase energizes formation of ATP from ADP and inorganic phosphate (Pi) and, via cotransport systems, drives active uptake (symport) or extrusion (antiport) of various substrates (S).
The maintenance of a constant internal ion composition is indispensable to all living cells. Bacteria tend to maintain the cytoplasmic pH within a narrow range and to establish gradients of K+ and Na+ ions between their cytoplasm and the surrounding medium such that the cytoplasmic K+ concentration is higher than and the Na+ concentration is lower than that of the environment. It is accepted that secondary transport systems coupled to protons mediate the movements of K+ and Na+ ions. Proton movement across the membrane is the primary event not only for energy metabolism but also for performing this homeostatic work.
Microorganisms living in aquatic habitats are directly exposed to the outside world through a cell surface layer. Their habitats commonly encompass a wide range of physical conditions: oxygen, pH, salinity, temperature, light, etc. Bacteria that cannot cope with and survive in severe environments by depending on their H+-linked machinery alone have evolved a variety of ancillary energy conversion mechanisms. It is now recognized that Na+ ions supplement the role of protons in energy transduction across the bacterial membrane (154, 228). We know of diverse sodium pumps, such as (i) Na+-translocating membrane-bound decarboxylases in Klebsiella pneumoniae, Salmonella typhimurium, Veillonella alcalescens, Propionigenium modestum, etc. (45); (ii) Na+-translocating NADH oxidoreductase in various marine bacteria such as Vibrio alginolyticus (253); and (iii) the Na+-translocating ATPase in Enterococcus hirae, which is one of the topics of this review. All these generate an electrochemical gradient of sodium ions, which is used by the cells to drive secondary Na+-linked processes such as solute transport (122, 191, 254) and flagellar rotation (96, 101). In P. modestum, the Na+ gradient generated by the decarboxylation of organic acids is used for ATP synthesis by the Na+-ATPase (94, 155, 156). Alkaliphilic bacteria use an Na+ gradient as the driving force for solute transport and flagellar rotation at high pH (96, 101, 151, 152), when the proton concentration is too low. On the other hand, halorhodopsin functions as an electrogenic chloride pump in Halobacterium halobium, generating a membrane potential (221). These specialized energy-transducing systems are again very important for ion homeostasis in particular cases.
This article centers on cation transport in streptococci. The genus Streptococcus (70) is composed of gram-positive bacteria which occur as parasitic organisms in a wide variety of human, animal, and plant habitats (31). Streptococci are important in the dairy industry, as pathogens of animals and humans, and for their role in dental caries. Most are facultatively anaerobic, but some require additional carbon dioxide for growth and some are strict anaerobes. The metabolism of streptococci is fermentative, but nutritional requirements are complex and variable. The fundamental routes of energy metabolism run as follows. Glucose is taken up and phosphorylated to glucose-6-phosphate via the phosphoenolpyruvate-dependent phosphotransferase system (259); it is subsequently converted to pyruvate and finally to lactic acid by the glycolytic pathway. In these bacteria, which lack a respiratory chain (Fig. 1), ATP produced by substrate-level phosphorylation is hydrolyzed by the FOF1-ATPase with accompanying translocation of protons; the resulting proton potential is utilized for various proton-coupled transport reactions. It is noteworthy that among streptococci (Table 1), enterococci are particularly tolerant to external stresses including high temperature, high salt concentration, alkaline pH, and the presence of bile salts (70, 160). Lactococcus lactis is also known to be moderately tolerant to these factors. Facing harsh conditions such as high salinity and alkaline pH, enterococci and probably also L. lactis have evolved special energy conservation mechanisms for cation transport and homeostasis, which other streptococci may not have.
TABLE 1.
Growth characteristics of some species of Streptococcus
| Strain | Growth at:
|
||||
|---|---|---|---|---|---|
| 10°C | 45°C | 6.5% NaCl | pH 9.6 | 40% bile | |
| Pyogenic | |||||
| S. pyogenes | − | − | − | − | − |
| S. pneumoniae | − | − | − | − | − |
| Oral | |||||
| S. sanguis | − | − | − | ||
| S. mutans | − | − | − | ||
| Enterococci | |||||
| E. hirae | + | + | + | + | + |
| Lactic | |||||
| L. lactis | + | − | − | − | + |
| Other | |||||
| S. thermophilus | − | + | − | − | − |
E. hirae (formerly Streptococcus faecalis), which is found in the intestine of higher animals, proved to be a useful system for unraveling the energetics of active transport, particularly the role of the FOF1-ATPase in chemiosmotic energy transduction (79, 80, 84–86; for reviews, see references 71 to 73). First, E. hirae, like other streptococci, lacks respiratory chains. It can generate a proton potential only by the hydrolysis of ATP, effected by the proton-translocating FOF1-ATPase. Second, its simple metabolic pathways allow the precise calculation of ATP yields from the few compounds that it can metabolize, such as glucose and arginine. Third, the cells are easily depleted of energy, because the organism does not make energy reserve polymers. Finally, like other gram-positive organisms, it is sensitive to ionophores and inhibitors that act on the cell membrane. Because of these advantages, research related to inorganic cation transport processes has been carried out primarily with enterococci, although work on other species such as L. lactis (formerly Streptococcus lactis and Streptococcus cremoris) has recently begun to be published. In the past several years, a number of genes encoding transport proteins for cations have been isolated from streptococci, allowing characterization of these transport systems at the molecular level. One of the major findings with E. hirae and other streptococci is that ATP plays a much more important role in membrane transport than it does in nonfermentative organisms; streptococci are unable to generate a large proton potential because they lack respiratory chains (77). Streptococci cope with their limited proton potential by expressing a variety of primary transport systems. In this article, I review chiefly the recent developments in cation transport by E. hirae and supplement this information with what is known of other streptococci.
CATION TRANSPORT SYSTEMS
Proton ATPase
The FOF1-ATPase is widely distributed in bacterial cell membranes. At the level of quaternary structure, the bacterial FOF1-ATPase is essentially identical to the ATPases of the inner membrane of mitochondria and of photosynthetic organelles in eukaryotic cells (36, 57, 105, 222). Its importance in membrane bioenergetics continues to be emphasized, although its universal distribution in bacterial membranes has been disproven; phylogenetically related proton ATPases of the vacuolar type replace FOF1-ATPase in archaebacteria (217) and in one eubacterium (268) (described below). All FOF1-ATPases are considered to be reversible, but physiologically the enzyme operates mainly or solely in one direction or the other. In mitochondria and respiring bacteria, the enzyme functions as an ATP synthase, mediating oxidative phosphorylation energized by the electrochemical gradient of protons (proton potential) via the respiratory chain. More than two decades ago, it was reported that some streptococci synthesize a cytochrome-like respiratory chain when the medium is supplemented with haematin (212); formation of ATP from ADP and inorganic phosphate, coupled to NADH oxidation, in cell extracts was also observed (33, 202). However, in most cases the streptococcal FOF1-ATPase does not function as ATP synthase, because of the lack of a functional H+-linked electron transport system (71, 75). In this organism, the FOF1-ATPase functions as a hydrolase and proton movements coupled to ATP hydrolysis are used for the generation of the proton potential (42, 84).
The E. hirae enzyme is of special interest because it was the first of the bacterial membrane ATPases to be discovered (9). The physiological role of streptococcal FOF1-ATPase is to alkalinize the cytoplasmic pH in the acidic pH range and to establish a proton potential as the driving force for a variety of secondary H+-linked transport systems and for H+-linked flagellar rotation in motile streptococci (165).
Molecular structure and genes.
As early as 1960, Abrams et al. (9) detected an ATP hydrolytic activity in E. hirae membranes. The enzyme purified from the membranes (4, 219) was initially reported to consist of 12 subunits (220). However, further biochemical examination of this enzyme established that E. hirae H+-ATPase belongs to the FOF1-ATPase synthase complex found in oxidative phosphorylation membranes (10–12, 158). The enzyme consists of two parts: F1, the catalytic moiety, contains five subunits (α, β, γ, δ, and ɛ), and the FO membrane sector contains three subunits (a, b and c). A subunit stoichiometry for the F1 moiety (α:β:γ:δ:ɛ = 3:3:1:1:1), which is very similar in all FOF1-ATPase complexes of bacteria, is likely (3, 10–12, 158). The stoichiometry of the FO subunits is uncertain. The F1-ATPases in S. mutans, S. sanguis (244), and L. lactis subsp. cremoris (211) have been purified. All these purified ATPases are five-subunit enzymes with molecular sizes corresponding to those of other F1-ATPases. Final proof that streptococcal H+-ATPase is an FOF1-ATPase was provided by obtaining the amino acid sequences of all these subunit molecules. First, a Russian group has chemically determined the amino acid sequence of the N,N′-dicyclohexylcarbodiimide (DCCD)-binding 7-kDa proteolipid extracted with chloroform-methanol from plasma membranes of E. hirae (148). This proteolipid, consisting of 71 amino acid residues, was considered to be the c subunit of the H+-ATPase because of the high degree of homology to the corresponding c subunit of other bacterial FOF1-ATPases. Inactivation of H+-ATPase activity by DCCD is due to the covalent modification of Glu54 in the second membrane segment of this polypeptide chain.
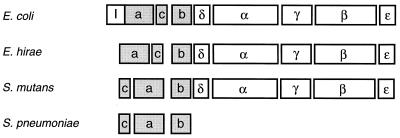
An operon encoding the FOF1-ATPase of the same strain of E. hirae was cloned by using antiserum raised against purified F1-ATPase and sequenced (Fig. 2) (223); a proteolipid was found to be the product of the c subunit gene. The order of the genes encoding eight subunits of the H+-ATPase of E. hirae is the same as that in the unc(atp) operon of Escherichia coli (Fig. 2) (59). Subsequently, the nucleotide sequence of the H+-ATPase operon of S. mutans (229) was determined, along with that of the genes encoding three subunits of the FO portion: a, b, and c, which are a part of the atp gene cluster in S. pneumoniae FOF1-ATPase (55). The deduced amino acid sequences of all these subunits clearly revealed a high similarity to those of FOF1-ATPase synthase subunits in oxidative bacteria. The E. coli unc(atp) operon contains nine genes, which encode, in order, an operator-proximal protein of unknown function, UncI (63), followed by the eight FOF1 subunits. The FOF1-ATPase operons in E. hirae and S. mutans contained no uncI gene homolog but were preceded by a relatively long (about 240-bp) intergenic space having several palindromic structures (223, 229). The expectation that this stretch of nucleotides is involved in the regulation of transcription of the operon by changes in the internal pH has not been fulfilled so far (145). In S. mutans (229) and S. pneumoniae (55), the order of the genes encoding the c and a subunits was reversed from those of E. coli and E. hirae (Fig. 2); the meaning of the reversal is unknown.
FIG. 2.
Gene organization of the proton ATP synthase/ATPase in some bacteria. Boxes indicate the open reading frames of the operons of E. coli (59), E. hirae (223), S. mutans (229), and a portion of the S. pneumoniae operon (55); the letters in boxes represent subunit names for comparison. The genes encoding hydrophobic subunits are shaded.
Enzymatic properties.
The basic catalytic properties of the streptococcal H+-ATPase were worked out first for E. hirae (for reviews, see references 7 and 8) and recently for other streptococci. Each one of these streptococcal enzymes has special features. It is well known that azide is a specific inhibitor of F1-ATPase from various sources (60). The ATP hydrolytic activity of F1-ATPase from S. mutans and L. lactis subsp. cremoris is inhibited by azide (211, 244). By contrast, the E. hirae enzyme is insensitive to azide (3). Although the sensitivity of F1 to azide is not understood at the structural level (60), this feature is convenient to discriminate enterococci from many other streptococci and is exploited in the SF medium marketed by Difco.
S. pneumoniae H+-ATPase is uniquely sensitive to amino alcohol antimalarial reagents in the erythro configuration, such as optochin, quinine, and quinidine (55, 175). These compounds and related ones specifically inhibit the membrane-bound ATPase activity. There is good correspondence between the optochin (quinine)-sensitive and optochin (quinine)-resistant strains with respect to growth and membrane ATPase. The protein responsible for the optochin (quinine)-sensitive phenotype of S. pneumoniae has been identified as the proteolipid c subunit (66 amino acid residues) of the FOF1-ATPase (55). The optochin (quinine)-resistant isolates arose by point mutations in the atpC gene encoding the c subunit (Fig. 2) and produced different single-amino-acid changes: Gly20Ala, Met23Ile, Val48Leu, or Ala49Thr (175). These four residues would be closely juxtaposed within the membrane bilayer when the c subunit is folded and would be associated with the a and b subunits to form the FO complex (62). It is speculated that the interaction of these reagents with the c subunit causes a conformational change in FO, hindering the proper presentation of the H+ translocation pathway (57). The FOF1-ATPase of a dental plaque bacterium, S. mutans, is highly sensitive to fluoride (245). Isolated F1-ATPase of S. mutans was less sensitive to fluoride than was the FOF1-ATPase holoenzyme, suggesting that the FO moiety is involved in fluoride inhibition.
Finally, the pH profile of the FOF1-ATPase is of particular importance to the physiology of streptococci; the activity has an optimum pH around 6.0 to 6.5 in E. hirae (144, 147) and S. mutans (24). On the other hand, the optimal pHs of ATPase from S. sanguis and S. salivarius are about 7.5 and 7.0, respectively (242). This difference is closely related to the physiological function of the streptococcal FOF1-ATPase as the generator of the proton potential and as regulator of the cytoplasmic pH, probably reflecting the acid tolerance of the former streptococci. The case of E. hirae FOF1-ATPase is discussed below. In contrast, the FOF1-ATPase complex of other oxidative organisms generally has a pH optimum above 7.5 (7); the pH optimum of purified E. coli F1 is around 9.5 (52).
Mechanism.
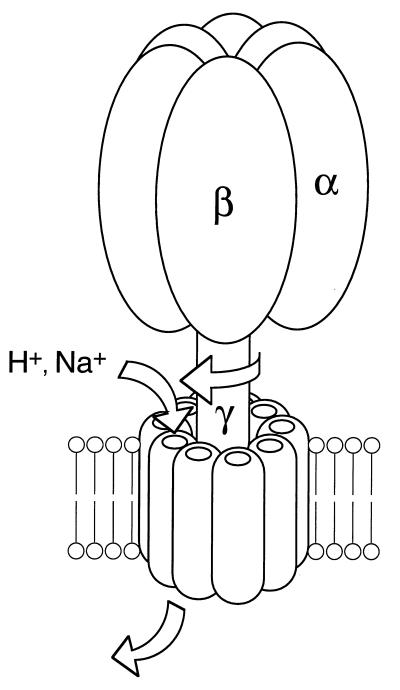
The streptococcal H+-ATPase, like other ATP synthases, is experimentally reversible; enterococcal H+-ATPase (260) and L. lactis H+-ATPase (161–163) can synthesize ATP when a large proton potential (−200 mV) is artificially imposed. Because of the lack of an energy-producing proton pump, such as an electron transport system, these enzymes do not synthesize ATP under physiological conditions. Ever since Mitchell proposed the chemiosmotic concept of energy coupling (168, 169), there has been vigorous debate over the linkage between proton flow and the ATPase reaction (Fig. 1). The molecular mechanism of FOF1-ATP synthase is not at issue in this review. However, it is worthwhile mentioning just briefly the recent fascinating progress in studies on the mechanism of the FOF1-ATPase synthase with its three catalytic sites in the F1 portion (Fig. 3) (29). A widely accepted model for energy coupling by the ATP synthase, called the binding change mechanism, has two features: (i) the major energy-requiring step is not the synthesis of ATP at the catalytic site but, rather, its release from that site, and (ii) tight binding of substrates and the release of product occur simultaneously at separate but interacting sites. It is evident from the negative cooperativity of sequential nucleotide binding that the three catalytic sites are strongly coupled. These catalytic sites may cycle in concert through the reaction. The speculation is that the required binding change is coupled to proton transport by rotation of a complex of subunits extending thorough FOF1: “rotation catalysis” (Fig. 3) (30). Based on the high-resolution structure of bovine F1 (2), which verifies that each catalytic site resides in a suitably different environment, this notion was strongly supported in biochemical (49) and spectroscopic (216) studies by independent groups. Very recently, Yoshida’s group directly visualized rotation of the F1 molecule (183); attachment of a fluorescent actin filament to the γ subunit served as a marker, which enabled them to observe this motion directly. Thus, the FOF1-ATPase is a complex that may be considered a machine, and this is best emphasized by explicit analogy to electrical motors and chemical engines; the next exciting discovery will be in the FO sector, i.e., how rotation within the FO subunit is coupled to H+ flow.
FIG. 3.
Rotating model of the FOF1-ATPase. Three αβ pairs of F1 generate three different nucleotide binding catalytic sites. Rotation of the central shaft, a composite of the γ subunit with others in FOF1, is coupled to the opening and closing of the catalytic sites. A flux of H+ (or Na+) drives the rotation of the shaft, but rotation permits the ion to flow. This model also applies to vacuolar ATPase.
Physiological work.
Generation of the proton potential is one of the major functions of the streptococcal FOF1-ATPase; this is subsequently utilized by means of secondary H+-coupled transport systems, in accord with the chemiosmotic viewpoint (71–75). The magnitude of the proton potential is affected by several factors: the proton conductance of the cell membrane and the permeability of the membrane to charged molecules and to ions. Furthermore, it is also affected by cellular activities related to growth. L. lactis cells glycolyzing in a buffer at pH 6 maintained a proton potential of −160 mV, while that of growing organisms was only −140 mV (128). The difference possibly results from the partial consumption of the proton potential by various H+-coupled transport systems. The maximum size of the proton potential generated in enterococci is almost −130 to −150 mV at acidic pH (144). With regard to the uptake of metabolites, if we assume symport of a metabolite with one proton, a proton potential of −180 mV would suffice to sustain a concentration gradient of 103 while −240 mV would support a gradient of 104 (214); physiological gradients of transport substrates fall within this range. On the other hand, at alkaline pH, the H+-ATPase activity is minimal and the proton potential is too low for adequate accumulation of solutes by H+-linked porters (110). This leads one to expect the evolution of various primary transport pumps to supplement the H+-linked secondary ones.
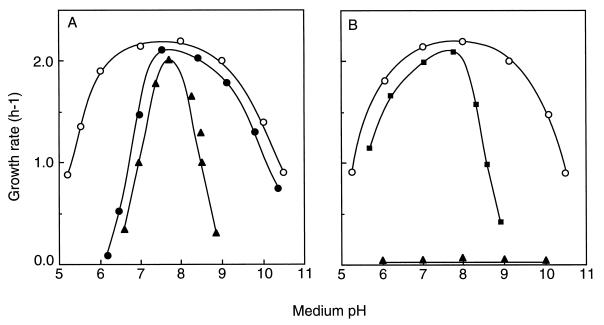
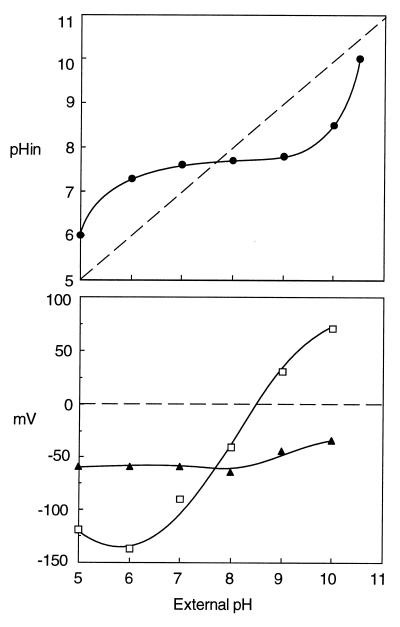
Streptococci grow over a relatively wide range of pH; the pH profile of enterococci is particularly broad, ranging from pH 5 to 11 (Fig. 4). Around an external pH from 6 to 9, where streptococci grow at their optimal rate, the cytoplasmic pH is estimated to be between 7.5 and 8.0 (145). When streptococci are grown to the stationary phase, the pH of the growth medium falls as low as 4.5, due to acid production by glycolysis. The cytoplasmic pH of such cells was about 5.0, but glycolysis recovered upon neutralization of the cytoplasm (270), suggesting that homeostasis of the cytoplasmic pH is essential to the physiology of streptococci. An elegant experiment indicating the importance of pH homeostasis in streptococci was performed by Harold and Van Brunt (82). They demonstrated that under certain conditions E. hirae can grow in the presence of an ionophore. In the presence of an ionophore such as gramicidin D, protons move freely across the membrane and hence any pH gradient collapses; the cytoplasmic pH of these cells is proportional to the pH of the medium. In high-K+ complex medium containing this ionophore, the growth of E. hirae is limited to a narrow pH range, around 7.8 (Fig. 4A). The cell stop growing at pH 6 or 9 in the presence of the ionophore. Regulation of the cytoplasmic pH at acidic or alkaline external pH is thus essential for optimal growth of E. hirae. Harold and coworkers (79, 80, 84–86) clearly demonstrated that expulsion of protons via the H+-ATPase results in cytoplasmic alkalinization. The idea that the H+-ATPase is the instrument for alkalinization at acidic external pH was validated by genetics. E. hirae mutants, defective in the alkalinization of the cytoplasmic pH at acidic pH, were isolated as acid-sensitive mutants which grew at a pH around 7.5 but failed to grow at acid pHs below 6.0 (143).
FIG. 4.
Growth rates of E. hirae strains as a function of medium pH. Cells were grown in medium KTY (high K+, low Na+) (A) and medium NaTY (high Na+, low K+) (B). Symbols: ○, strain 9790 (wild type); •, strain AS25 (proton ATPase mutant); ■, strain Nak1 (sodium ATPase mutant); ▴, strain 9790 in the presence of gramicidin D.
Mutant AS25, one such acid-sensitive mutant (Table 2), grew at acidic external pH just as the wild type does in the presence of gramicidin D (143) (Fig. 4A). AS25 growing at acid pH failed to alkalinize its cytoplasm, whose pH was proportional to that of the medium; this is how the wild type behaves in the presence of gramicidin D (140). In alkaline medium, by contrast, AS25 grew normally (Fig. 4A). The cytoplasmic pH of the wild-type strain at pH 9 to 10 was acidified to 8.2 to 8.7 (109). The cytoplasmic pH of this mutant at alkaline pH was also acidified to a value identical to that of the wild type. Mutant AS25 is defective only in the regulatory system for cytoplasmic alkalinization at acidic pH; the separate machinery for pH regulation at alkaline external pH will be described below. A correlation between the H+-ATPase and alkalinization of the cytoplasmic pH was obtained by showing that activities for both ATP hydrolysis and proton extrusion are very low in mutant AS25 (143, 144). The idea that the mutant may be unable to maintain pH homeostasis as a result of an increase in membrane conductance to protons has been disproved. Mutants lacking the H+-ATPase have not been isolated from other streptococci, but alkalinization of the cytoplasmic pH in S. mutans was inhibited by DCCD, a well-known inhibitor of that enzyme (42). Evidently, streptococci generally rely on the H+-ATPase for pH homeostasis in acidic media.
TABLE 2.
Mutants of E. hirae deficient in ion transporta
| Strain | Lesion | Reference |
|---|---|---|
| 9790 | None (parent strain) | |
| AS25 | Defective FOF1-ATPase | 143 |
| 7683 | Defective sodium extrusion (probable double mutation of Na+-ATPase and Na+/H+ antiporter) | 83 |
| Nak1 | Defective vacuolar Na+-ATPase (probable nonsense mutation of ntpA gene) | 116 |
| WD4 | Disrupted napA gene | 263 |
| JEM2 | Disrupted ntpJ gene | 176 |
| 576B | Defective KtrI (change in cation specificity) | 76 |
| 325B | Defective K+ retention | 87 |
This table lists only mutants referred to in the text.
In 1971, Abrams and Smith (6) showed that the E. hirae H+-ATPase level increased when cells were grown on medium containing a limiting level of K+ ions. Subsequently, an increase in the H+-ATPase level of the membranes was observed, both when the cytoplasmic pH was lowered by the addition of gramicidin D and when cells were grown on an acid medium (5, 147). The amount of H+-ATPase increased four- to fivefold upon the addition of gramicidin D. When E. hirae cells were transferred to acidic medium, the cytoplasmic pH was alkalinized as the amount of H+-ATPase increased (145). When the increase in the H+-ATPase was blocked by chloramphenicol, the ability of the cell to maintain an alkaline cytoplasmic pH was impaired. These results suggest that a change in the amount of the H+-ATPase is also important for cytoplasmic pH alkalinization (246). An increase in the amount of H+-ATPase when the cells were grown at the lowest pH permitting growth was also observed in S. mutans and S. bovis (172).
One would expect the increase in membrane ATPase to be the result of enhanced synthesis (145), but this is not the case. First, the amount of mRNA for the H+-ATPase operon was only slightly changed under the culture conditions that influence the enzyme level in both E. hirae and S. mutans (229). Second, Western blotting experiments revealed that the amount of each ATPase subunit in cell lysates remained nearly constant while the amount of membrane-bound, functional ATPase increased; apparently, not all the cell enzyme is necessarily assembled into the membranes. On the other hand, the elevated H+-ATPase level at acidic pH decreased when the medium was brought to alkaline pH. Abrams and Jensen (5) proposed selective degradation of the α subunit of the H+-ATPase at an alkaline pH; degradation of the subunit may decrease the level of functional enzyme. Thus, the regulatory step for the functional H+-ATPase level is probably at the posttranscriptional level. It may be speculated that the amount of functional H+-ATPase in the membrane is regulated at multiple levels: synthesis, assembly, and turnover (142). We conclude that changes in the amount of functional H+-ATPase are essential to the regulation of cytoplasmic pH in acidic media (141), but the mechanism is not simple and is still unclear.
Sodium Transport Systems
Bacteria actively extrude sodium ions and maintain the electrochemical concentration gradient of sodium directed inward. The significance of the sodium gradient in bacteria is well known (73, 74, 154, 228); the sodium current is frequently linked with cotransport systems (191) and can serve as the driving force for flagellar rotation (101). In marine bacteria, a variety of transport systems are linked to Na+ rather than H+ (122, 254); in this case, the sodium circulation is primary rather than supplemental to the proton circulation. The mechanism of sodium extrusion is generally thought to be secondary antiport of sodium ions for protons, energized by the proton potential, just as Mitchell envisaged (71, 72, 169, 171). However, the activity of the Na+/H+ antiporter is supplemented by a variety of primary transport systems energized by ATP hydrolysis, redox potential, or decarboxylation. Sodium transport in streptococci has been extensively studied in E. hirae (241). In this bacterium, the investigation of sodium transport grew out of Mitchell’s antiport hypothesis but now illustrates the interplay between the primary and secondary modes of energy-linked transport (77).
Na+/H+ antiporter.
Harold and his colleagues contributed fundamental information on Na+ transport in E. hirae. Early studies on Na+ transport in E. hirae were interpreted as providing support for a Na+/H+ antiporter driven by the proton potential. Sodium extrusion from the cells against the Na+ gradient was blocked by DCCD, an inhibitor of the proton-translocating FOF1-ATPase, suggesting that sodium efflux requires a proton potential. Furthermore, H+ influx accompanying Na+ efflux was observed in alkalinized Na+-loaded cells metabolizing in Na+-free buffer. In this experiment, the driving force for Na+ efflux was presumably the Na+ gradient directed outward, since DCCD prevented establishment of the proton potential (80). These findings provided the first confirmation of the proposal that bacteria contain an Na+/H+ antiporter (264). For some years, this antiporter activity was held to be an artifact connected with the newly discovered Na+-translocating ATPase (for details, see references 88 and 114). Briefly, the hypothesis was that the Na+ pump of E. hirae catalyzes an ATP-driven exchange of H+ for Na+. The antiporter activity, visible only in membrane vesicles or in a mutant deficient in the Na+ pump, was considered to be the antiporter moiety of a modular ATP-driven exchanger of Na+ for H+ (83, 89, 91). This interpretation, however, proved incorrect. In the wild-type strain cultured on Na+-limited medium, in which the Na+-inducible Na+-ATPase level was minimal, the Na+/H+ antiport activity was clearly observed (108). In response to an artificially imposed pH gradient (with exterior acid), energy-depleted cells exhibited a transient sodium extrusion which was unaffected by treatments that dissipated the membrane potential but was blocked by proton conductors. One must conclude that E. hirae has two separate Na+ extrusion systems: an Na+-ATPase and an Na+/H antiporter.
Mutant 7683, which is totally negative in sodium-extruding activity, was unable to grow in high-Na+ medium (83). The E. hirae Na+/H+ antiporter gene, napA, was cloned by screening for the recovery of growth of this mutant at high Na+. This gene encodes a 42-kDa hydrophobic polypeptide having 12 putative membrane-spanning regions, like most other secondary porters (263). Everted vesicles of strain WD4, in which the napA gene is disrupted, did not show uptake of 22Na+ in response to a proton potential. When the napA gene was expressed in E. coli, NapA-dependent, Na+-coupled H+ flux was observed (240). The amino acid residues conserved in various Na+-linked secondary transporters were also found in the amino acid sequence of the NapA protein (191), although their significance in the molecular mechanism is unclear. However, from the standpoint of the evolution of cation porters, it is noteworthy that the overall sequence of NapA was highly similar to that of E. coli KefC K+ efflux protein rather than to that of the NhaA Na+/H+ antiporter of E. coli (210). NapA recognizes Li+ as well as Na+ as the substrate.
It has been considered that the Na+/H+ antiporter may be constitutive, because full Na+/H+ antiporter activity was observed even in Na+-limited media (less than 1 mM Na+). However, Solioz’s group now suggests that the amount of the NapA protein is regulated at the transcriptional level, responding to the concentration of Na+ as well as Li+ in the medium (238). Although no one doubts the ubiquitous distribution of Na+/H+ antiporters in bacteria, it has not so far been reported in other streptococci. E. coli contains three Na+/H+ antiport systems, NhaA, NhaB, and ChaA; they differ from one another in activity, optimal pH, and inducibility (192). We also have no information about multiplicity of Na+/H+ antiporters in streptococci.
Na+-ATPase.
(i) Discovery.
Although most of the early data on Na+ extrusion by E. hirae cells fit the sodium/proton antiport model, there was one important observation which could not be easily explained by this model: net Na+ movement and 22Na+-Na+ exchange were seen only in cells capable of generating ATP (80). The search for an answer to this question led to the discovery of the sodium-translocating ATPase in E. hirae. Sodium extrusion against a concentration gradient, under conditions such that the proton potential has been totally dissipated by the presence of DCCD or protonophores and valinomycin, was readily induced by the addition of glucose or arginine. Since the energy donor common to the metabolism of glucose and arginine is ATP, Na+ extrusion was attributed to an ATP-driven Na+ pump (89). ATP-driven 22Na+ uptake and Na+-stimulated ATP hydrolysis were observed in everted membrane vesicles in the presence of DCCD and the ionophores (90). No Na+-pumping activity was detected in vesicles of mutant 7683 (83); this mutant is now considered to be a double mutant defective in both Na+-ATPase and the Na+/H+ antiporter (114, 232). All these results suggest that a Na+-translocating ATPase exists in the cell membrane of E. hirae, which was the first bacterium in which a Na+-ATPase was discovered. Na+ movements unconnected to a proton potential have recently been reported in S. bovis (241); an Na+-ATPase is likely to be responsible.
(ii) The sodium pump is a V-ATPase.
Ion-motive ATPases are divided into two categories: one which forms phosphorylated intermediates (E-P enzyme; P-ATPase) and the other which does not. P-ATPases are exemplified by the Na+,K+-ATPase, H+,K+-ATPase and Ca2+-ATPase of higher organisms and by a variety of ion-translocating ATPases in bacteria (194, 195). The ATPases which do not form E-P intermediates are now divided into two types: FOF1-ATPase (F-ATPase) and vacuolar ATPase (V-ATPase). F-ATPase functions as an ATP synthase in oxidative bacterial membranes and as the proton pump in the membranes of fermentative bacteria such as streptococci. On the other hand, the V-ATPase is known as the proton pump of acidic organelles, such as the vacuoles of fungi and plants and various endosomes of animal cells (58, 180, 181). Archaebacteria contain a V-ATPase, which is believed to mediate ATP synthesis (217). Both ATPases are quite similar multisubunit enzymes consisting of a hydrophilic catalytic portion (F1 and V1, respectively) and a membrane-embedded portion (FO and VO). The proteolipid of the membrane sector, which contains a DCCD-reactive acidic amino acid residue, is thought to be the pathway by which protons cross the membrane. However, a stretch of the sequence (about 90 amino acid residues) is commonly conserved in the N-terminal region of the V-ATPase A subunit but is not found in the sequence of the β subunit of E. coli F-ATPase. The size of the eukaryotic V-ATPase proteolipid is generally 16 to 17 kDa and is thought to have arisen by tandem duplication of the 7- to 8-kDa c subunit gene of the F-ATPase (164). The high homology between the amino acid sequences of several major subunits of these ATPases suggests a close evolutionary relationship between them (66, 164, 181, 182). The V-ATPase has been called a “big sister” of the F-ATPase (14).
Although the “modular pump model” has been excluded, we expected the E. hirae Na+-ATPase to be distinct from other ion-motive ATPases, because of its resistance to both vanadate, an inhibitor of P-ATPase, and DCCD, an inhibitor of F- and V-ATPases. An antibody to purified E. hirae F1-ATPase did not inhibit the Na+-ATPase (114, 142). A decade after the discovery of the Na+-ATPase, attempts at purification (118) showed that the enzyme is complex, containing both peripheral and membrane-embedded subunits. Everted vesicles were treated with EDTA for the purpose of peeling off the unrelated peripheral proteins from the membranes, especially F1-ATPase. Contrary to expectations, Na+-stimulated ATP hydrolytic activity of the membranes was easily removed by this treatment. However, Na+-ATPase activity was fully restored by addition of an excess of Mg2+ to this EDTA-treated membrane suspension. The component essential for the activity of this enzyme was released and detected by polyacrylamide gel electrophoresis (Fig. 5). By appropriate staining procedures, we were able to detect an ATP-hydrolyzing protein just below the F1 moiety of F-ATPase, which was also released. Identification of this protein as the Na+-ATPase component rests on the following observations. First, this protein was not observed in EDTA extracts from the Na+-ATPase-negative mutant Nak1 (116). It is now known that this mutant is unable to produce one of the major subunits (subunit A, 65 kDa) of Na+-ATPase; therefore, the catalytic V1 moiety is not assembled. Second, we know that the Na+-ATPase is not constitutive; the amount of Na+-ATPase in the membranes increases in cells cultured in high-Na+ medium (112, 114, 136). The amount of this protein in the EDTA extract altered in parallel with the activity of the Na+-ATPase. The ATPase was subsequently purified (119); its molecular mass was about 400 kDa, consisting of 65-, 56-, and 29-kDa polypeptides with a probable stoichiometry of 3:3:1. Rabbit antiserum against purified V1-ATPase did inhibit the Na+-stimulated ATP hydrolytic activity of the membranes.
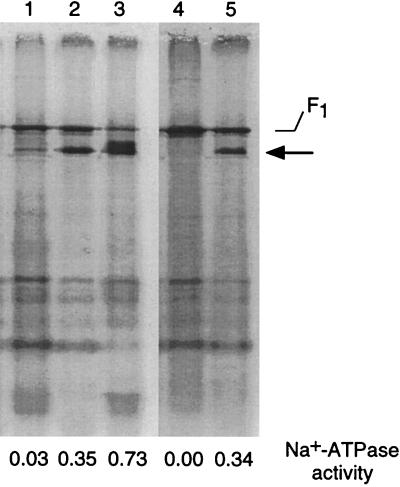
FIG. 5.
Polyacrylamide gel electrophoresis of EDTA extracts of E. hirae membranes. EDTA extracts were prepared from membrane vesicles of E. hirae strains cultured in various media. The Na+-ATPase activities of each of the vesicles are shown underneath. Lanes: 1, 9790 (wild type) in low Na+; 2, 9790 in high Na+; 3, 9790 in high Na+ at high pH; 4, Nak1 (Na+-ATPase mutant) in high Na+; 5, revertant of Nak1 in high Na+. Reprinted from reference 117 with permission of the publisher.
Several early observations suggested that the E. hirae Na+-ATPase belongs to the class of vacuolar ATPases (117). (i) It is sensitive to V-ATPase inhibitors such as nitrate and N-ethylmaleimide (NEM), (ii) antiserum against two major subunits of animal V-ATPase cross-reacted with those of the ATPase, and (iii) the N-terminal amino acid sequences of these 65- and 56-kDa subunits of purified enzyme are similar to those of two major subunits of other V-ATPases. Direct evidence that the E. hirae Na+-ATPase is a V-ATPase was obtained by cloning the genes. The ntp operon, ntpFIKECGAB D(H)J, encoding this enzyme (125, 248, 249) was cloned by use of a probe consisting of a PCR-amplified DNA fragment corresponding to the N-terminal part of the 65-kDa subunit (123) (Fig. 6A). The 65-, 56-, and 29-kDa subunits of the purified ATPase were identified as the ntpA, ntpB, and ntpD gene products, respectively. Furthermore, extensive gene disruption experiments (reference 232 and our unpublished data) indicate that all these ntp genes, except for the ntpH and ntpJ genes, are required for expression of the Na+-ATPase. The ntpJ gene encodes a component of the K+ transport system (KtrII system) (176). The status of the minigene ntpH (183 bp) as an open reading frame is in doubt. The Na+-ATPase was very recently purified from the membranes of cells in which the amount of Na+-ATPase was amplified by introducing the ntp operon (177) (Fig. 6B). Purified Na+-ATPase consists of nine polypeptides, all of which were identified as ntp gene products, (the ntpH and ntpJ products were not present). Proteoliposomes reconstituted with purified ATPase showed ATP-driven 22Na+ transport which was accelerated by valinomycin and protonophore but blocked by monensin; it seems to be electrogenic (Fig. 7). We conclude that E. hirae Na+-ATPase is an electrogenic sodium pump consisting of nine Ntp proteins (120).
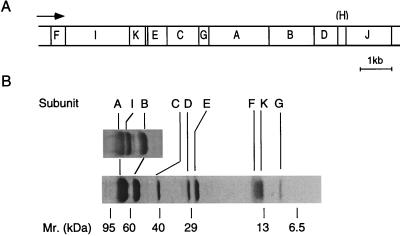
FIG. 6.
Organization of the ntp operon and subunit structure of the E. hirae Na+-ATPase. (A) Gene organization. The arrow indicates transcriptional direction. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of ATPase at different concentrations of gel. Reprinted from reference 177 with permission of the publisher.
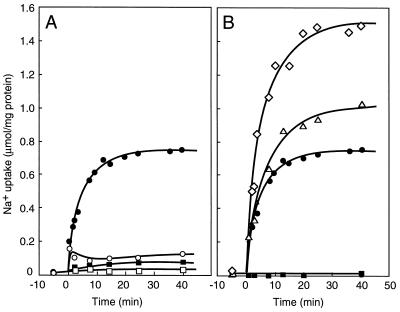
FIG. 7.
Na+ uptake into proteoliposomes reconstituted with Na+-ATPase. Uptake was started by the addition of 5 mM ATP at 0 min. Inhibitors or ionophores were added at −10 min. (A) Symbols: □, no ATP; •, ATP; ○, ATP plus 50 mM KNO3; ■, ATP plus 50 μM destruxin B. (B) Symbols: •, ATP; ◊, ATP plus 25 μM valinomycin; ▵, ATP plus 25 μM CCCP; ■, ATP plus 25 μM monensin. Reprinted from reference 177 with permission of the publisher.
The deduced amino acid sequences of these ntp gene products revealed a striking similarity between the Ntp subunits and those of the eukaryotic V-ATPase, especially those from yeast (Table 3). Three major subunits, the 66-kDa NtpA and 51-kDa NtpB subunits of the V1 portion and the 16-kDa NtpK (proteolipid) subunit, are highly similar to those of the V-ATPases. A stretch of the sequence commonly conserved in the N-terminal region of the Walker A motif of V-ATPase A subunit was also found in the sequence from residues 122 to 208 of NtpA. Amino acid clusters such as the motif GlyXXXXGlyLys(Thr/Ser), which are specifically conserved in the sequences of nucleotide binding proteins (262), were all conserved in the sequence of the NtpA subunit; a similar sequence, GlyXXXGlyLysThr, is found in the NtpB subunit. The NtpK proteolipid subunit, which is the putative ion-translocating pathway of the VO moiety, has a DCCD-reactive glutamic acid residue (Glu139) in its fourth membrane-spanning domain, probably derived by tandem duplication of the c subunit of F-ATPase (164). Other Na+-ATPase subunits are moderately similar to the corresponding ones of yeast V-ATPase. The similarity of NtpC and NtpI to the corresponding yeast V-ATPase subunit was less prominent, but some amino acid clusters conserved among corresponding subunits in eukaryotic vacuolar ATPases are also conserved in the bacterial sequences. E. hirae Na+-ATPase clearly belongs to the V-ATPase family. This bacterium has both an F-ATPase and a V-ATPase, functionally coexisting in the same cell membrane (Fig. 5).
TABLE 3.
Similarities between E. hirae Na+-ATPase subunits and S. cerevisiae proteinsa
| Na+-ATPase subunit (kDa) | S. cerevisiae protein (kDa) | Identity (similarity) (%) |
|---|---|---|
| A (66) | Vma1p (69) | 50 |
| B (51) | Vma2p (60) | 53 |
| C (38) | Vma6p (36) | 15 (36) |
| D (27) | Vma8p (32) | 23 (48) |
| E (23) | Vma4p (27) | 22 (39) |
| F (14) | Vma10p (13) | 29 (46) |
| G (11) | Vma7p (14) | 26 (52) |
| H (7) | ||
| I (76) | Vph1p (100) | 16 (37) |
| J (49) | Trk2p (101) | 27 (54) |
| K (16) | Vma3p (17) | 25 (51) |
Reproduced from reference 249 with permission of the publisher.
(iii) Catalytic properties.
The activity of E. hirae Na+-ATPase is maximal at pH 8.5 to 9.0 but not detectable at pH 6.0; the pH profile of ATPase activity fits its importance to the physiology of this bacterium at alkaline pH, as described below. This ATPase is stimulated by Na+ or Li+ ions but not by K+ and Ca2+ ions. The ATP hydrolytic activity of the purified VOV1 enzyme absolutely requires Na+; the kinetics of ATP hydrolysis showed at least two different affinities for Na+ (Km values of 20 and 4 mM), and probably one more. These different Na+ affinities of the ATPase are definitely linked to its mechanism, but their meaning remains unsolved. The affinity of this enzyme for ATP is 0.5 mM for both the purified enzyme and the membrane-bound form (115, 177).
E. hirae Na+-ATPase was initially proposed to be insensitive to DCCD (89–91). However, it is now clear that the activity of Na+-ATPase is specifically inhibited by this reagent (177); a glutamic acid residue (Glu139) in the NtpK proteolipid of the complex is probably covalently modified by DCCD. Why did the activity of the enzyme appear to be insensitive to DCCD? The trick is the fact that the reaction of DCCD with the Na+-ATPase is blocked by the coupling ions Na+ and Li+ (138). Na+ and Li+ ions compete with the attack of DCCD. In all previous experiments in which DCCD-insensitive 22Na+ extrusion activity was observed, this reagent, blocking the generation of the proton potential by the H+-ATPase, was added after the cells or vesicles had been loaded with sodium ions. No proton potential-independent Na+ extrusion was observed when DCCD was added before Na+ loading. It was just an unexpected bit of luck in the history of Na+-ATPase in enterococci.
The macrolide antibiotic bafilomycin A1 is a well-known inhibitor of V-ATPases (28) and probably attacks the VO portion of the eukaryotic enzymes (271). However, the effect of this antibiotic on bacterial V-ATPase was insignificant. Concanamycin A, another macrolide antibiotic, did not inhibit E. hirae Na+-ATPase when used at 5 μM. Recently it was shown that two proteolipid subunits, Vma11p and Vma16p, are also essential for the activity and assembly of yeast V-ATPase (95). More than 13 subunits make up yeast V-ATPase, and the same is probably true for Caenorhabditis elegans (190). The structures of bacterial V-ATPase and eukaryotic V-ATPase are homologous, but their fine structures are distinct, probably resulting in their differential sensitivity to antibiotics. Destruxin B, a peptide antibiotic which may attack the V1 catalytic portion of V-ATPase (179), is as effective with E. hirae Na+-ATPase as with the eukaryotic enzyme. The effect of nitrate on eukaryotic V-ATPase is attributed to its chaotropic property; the V1 catalytic moiety is detached and dissociated from the VO membrane sector (173, 207, 258). E. hirae Na+-ATPase and archaebacterial V-ATPase (1) are also inhibited by nitrate, but inhibition occurs at a much lower concentration and is probably not chaotropic. E. hirae Na+-ATPase is inhibited by NEM, but the Ki value (0.15 mM) is higher than the micromolar amounts required by eukaryotic V-ATPase. It has been proposed that the cysteine residue in the Walker A motif (GlyXXXCysGlyLysThr), commonly conserved in the A subunit of eukaryotic V-ATPase, is attacked by sulfhydryl reagents (54). The corresponding amino acid residue in E. hirae Na+-ATPase is alanine (Ala236) of the NtpA subunit. Replacement of the Ala236 by cysteine greatly enhanced the sensitivity of the Na+-ATPase to NEM (247).
(iv) Mechanism.
There must be common principles in the energy-transducing machinery of V-ATPase and F-ATPase molecules. It is likely that in both cases, energy transfer from ATP hydrolysis calls for three catalytic sites on the V1 moiety and an ion-translocating pathway through the VO proteolipid (133). The rotation catalysis mechanism, experimentally verified for the F-ATPase, is probably applicable to the V-ATPase (Fig. 3). Which subunit of the V-ATPase does rotate, as does the γ subunit of F-ATPase? The most likely candidate is the D subunit, even though its sequence similarity to γ is minimal. The reason is that the D subunit makes up the core of the V1 complex, together with A and B subunits, in purified V1 (119, 268).
ATP hydrolysis by the isolated V1 moiety was not stimulated by Na+ (118). Direct interaction between the V1 and VO moieties is a prerequisite for Na+-stimulated ATPase activity, suggesting that the binding site for Na+ is in the VO moiety. As described above, the reaction of DCCD with the 16-kDa proteolipid is blocked by Na+ or Li+ (138). It has been reported that certain subunits can be exchanged between the H+-ATPase of E. coli and the Na+-ATPase of P. modestum; a hybrid of E. coli F1 and P. modestum FO translocates Na+ (106, 157). Furthermore, certain amino acid replacements in the c subunit of P. modestum Na+-ATPase altered its ion selectivity (107). It is therefore speculated that the determinants for the ion selectivity of F-ATPase and V-ATPase reside in the amino acid residues of the proteolipid subunit (138, 204).
(v) Linkage with K+ movement.
Before finding the structural information on the Na+-ATPase by gene cloning, Harold and I suggested that this ATPase may be a Na+(K+)-ATPase (112). Potassium uptake via the KtrII K+ transport system is somehow linked to the Na+-ATPase, which, in intact cells, expels Na+ ions by exchange for K+. Moreover, the ntpJ gene, which is known to encode the K+ uptake capacity of the transport system, is cotranscribed with other subunits of the Na+-ATPase (Fig. 6A) (178). However, the NtpJ protein is separable from the Na+-ATPase complex by centrifugation of the solubilized fraction (177). Moreover, strain JEM2, in which the ntpJ gene has been disrupted (and which lacked K+ uptake), still contained normal Na+-ATPase (176). It therefore appears that the KtrII K+ uptake system is not mechanically linked with the Na+-ATPase complex; the K+/Na+ exchange model of this enzyme should be considered withdrawn.
(vi) Inducibility.
In 1984, Kinoshita et al. (136) reported that the Na+-ATPase level of E. hirae is not constant. They measured the Na+-stimulated ATPase in mutant AS25, deficient in the H+-ATPase, and observed much higher levels of Na+-ATPase in this mutant than those found by Heefner and Harold (90) in the wild type. Moreover, both the rate of sodium transport by intact cells and the activity of Na+-ATPase in vesicles were altered by the culture conditions. Mutant cells grown in high-Na+ (0.12 M) media exhibited high activities, while those grown in low-Na+ (5 to 10 mM) media exhibited much lower enzymatic and transport activities. Sodium transoort and Na+-ATPase activity in the wild-type strain were much lower than those in the mutant strain. When the wild-type cells were grown in the presence of a protonophore, carbonyl cyanide m-chlorophenylhydrazone (CCCP), both Na+ transport and Na+-ATPase activity were elevated. Furthermore, when the wild-type cells were grown at alkaline pH, both activities increased significantly (115). The sodium ATPase is thus induced when cells are grown on media rich in sodium, particularly under conditions that limit the generation of a proton potential, indicating that an increase in the cytoplasmic sodium level serves as the signal. Western blotting and Northern blotting experiments revealed substantial correlation between the amount of this enzyme and expression of the ntp operon (178). Even when the cells were grown on low-Na+ medium, the ionophores monensin and gramicidin D, which render the membrane permeable to Na+, significantly increased the amounts of Na+-ATPase and of the mRNAs for the operon (178). All these data are explained by the hypothesis that the Na+-ATPase is induced at the transcriptional level by an increase in the cytoplasmic Na+ concentration. In E. coli, transcriptional regulation of the nhaA Na+/H+ antiporter gene, responding to Na+ and Li+ ions (193), is achieved by the NhaR regulatory protein, which is a member of the LysR protein family (203). The next step for understanding the regulation of the ntp operon, in concert with that of the napA antiporter gene (238), is to discover the system that senses the Na+ concentration.
(vii) Comparison with other Na+-ATPases.
Whether a vacuolar-type Na+-ATPase occurs in other streptococci is not clear. Based on their tolerance to salinity and alkaline pH, some related streptococci such as L. lactis and S. bovis appear to possess a Na+-ATPase, although it may be less prominent. We have already found that everted membrane vesicles of L. lactis contain a Na+-stimulated ATPase; reactions with antibodies directed against components of the E. hirae enzyme suggest the presence of homologs to NtpA and NtpB (124). An Na+-translocating ATPase, recently discovered in the thermophilic Clostridium fervidus (237), is very likely to be of the vacuolar type as judged by the enzymatic and biochemical properties of the purified enzyme (97). Furthermore, the enzymatic features of an Na+-stimulated-ATPase observed in Mycoplasma mycoides (a parasitic glycolytic organism) are virtually identical to those of the E. hirae enzyme (25), and an Na+-stimulated ATPase from Acholeplasma laidlawii seems to be of the V type as judged by its subunit composition (104, 159).
On the other hand, the electrogenic Na+-ATPases found in P. modestum, Acetobacterium woodii (208), Methanosarcina mazei (23) and other organisms belong to the F-ATPase class. It is proposed that these ATPases perform ATP synthesis, energized by the Na+ gradient generated by a metabolic sodium pump. The P. modestum Na+-ATPase has contributed powerfully to analysis of the mechanism of F-ATPases (107, 138, 155–157). To date, no P-type Na+-ATPase has been found in bacteria.
(viii) Distribution and evolution of V-ATPase.
In the prokaryotic world, V-ATPases were first found in archaebacteria but are now known to be distributed among eubacteria such as E. hirae and Thermus thermophilus (268). Sequencing and characterization of the genes encoding the H+-translocating V-ATPases of other bacteria (266) suggest that all these enzymes consist of nine subunits, identical to the number for the E. hirae Na+-ATPase. We have found that nine open reading frames in the genome of Methanococcus jannaschii (35) correspond to individual ntp genes for the E. hirae Na+-ATPase subunits (248, 249), although the ion specificity of this putative V-ATPase is unknown. In these V-ATPase clusters, the order of the subunit genes is almost always the same. The Sulfolobus acidocaldarius V-ATPase operon is slightly different; the operon consists of only six genes (43). Since the whole subunit structure of Sulfolobus ATPase is unsettled, unidentified subunits may be encoded on separate genes.
E. hirae is the organism that has clearly proved that functional F- and V-ATPases can coexist on the same plasma membrane; there is no particular similarity between the subunits of these ATPases found in the same organism. Coexistence of V- and F-ATPases in a single bacterial cell is no longer exceptional. Both ATPases are also present in Methanosarcina mazei (23, 266); in this bacterium, an F-ATPase transports Na+ and a V-ATPase transports H+. Recent analyses by PCR have suggested that various eubacteria containing F-ATPase also possess genes similar to those for V-ATPase and that the archaebacterium Methanosarcina barkeri, in which V-ATPase functions, possesses an F-ATPase-like gene (243). It will be interesting to learn whether functional ATPases are encoded by these genes.
V-ATPase and F-ATPase exhibit similar structural features, and they are evolutionarily distantly related (66, 164, 181, 182); the A and B subunits of V-ATPase and the α and β subunits of F-ATPase are similar to each other, and the 16- to 17-kDa proteolipids of the eukaryotic V-ATPase and E. hirae Na+-ATPase are likely to be tandem duplications of the c subunit gene of the F-ATPase (164). Phylogenetic analysis of the homologous ATPase subunits sustains rigorous argument about ATPase evolution, but no firm conclusions have been reached (26, 92). However, it is believed that V-ATPase diverged from F-ATPase relatively early in evolution (182). It seems to me that the development of V-ATPase occurred between the archaeal and eukaryotic domains. Following extensive divergence of V-ATPases from F-ATPases, the archaea diverged from the “proeukaryotes.” Only in the proeukaryotic lineage, prior to the diversification of eukaryotes, did the tandem intragenic duplication of the c subunit occur. These genes may subsequently have been acquired by bacteria as a result of horizontal transmission of the ATPase genes. Among them, some species may have lost the gene for one of these ATPases, or at least the capability of its expression. A few organisms have retained the capability of expressing the genes for both ATPases (26, 92); one of these ATPases apparently underwent alteration of cation specificities, i.e., Na+ instead of H+, presumably in the course of adaptation to the environment.
The archaebacterial V-ATPase proteolipids so far reported clearly did not result from duplications of the c subunit gene of the F-ATPase (44). They are smaller than eukaryotic proteolipids. The significance of the proteolipid size in archaebacterial ATPase is obscure, but it is speculated that the F-ATPase proteolipid-like size may be relevant to the role of archaebacterial ATPase as an ATP synthase, not a proton pump (217).
Pathway of Na+ entry.
At least one route for Na+ entry must exist, since a Na+ circulation is important for the growth of streptococci. The nature of this pathway is still unknown. The limited data at hand suggest that the Na+/H+ antiporter mentioned above may allow Na+ ions to enter cells whose membrane potential has been collapsed (108, 114). A different pathway was observed by Heefner and Harold (89), who showed that when a membrane potential (inside negative) was imposed by K+ efflux, the cells took up Na+ ions by exchange for K+. The process involved has low affinity but high capacity (Km > 20 mM; Vmax > 50 nmol/min/mg of cells) and apparently responds to both the concentration gradient and the electrical potential. Because of its low affinity, we suspect that this pathway is relatively nonspecific and reflects some kind of leakage down the electrochemical potential gradient.
Physiological work.
Like all other living cells, streptococci exclude Na+ ions and accumulate K+ ions. There is no simple answer to the question why cells expend energy on generating this ion gradient. Generation of a sodium gradient is an essential aspect of any sodium circulation. The reason for the obligatory role of sodium circulation in streptococci is not evident, except for the accumulation of K+ at alkaline pH, which is somehow linked to Na+ extrusion. Na+-dependent secondary transport has been reported only for S. bovis (215). There is also a body of evidence to suggest that a high Na+ concentration in the cytoplasm is generally inhibitory to cell physiology, although specific targets have not been identified (83). On the other hand, it has long been known that K+ ions are the major cations specifically required for protein synthesis.
E. hirae 7683, a mutant totally defective in sodium extrusion, was unable to grow at any pH in K+-limited, high-Na+ medium; by contrast, in Na+-limited, high-K+ medium, the mutant grew nearly as well as the wild-type strain did (83). Although the activities of the H+-ATPase and of the major K+ uptake system, KtrI, were normal, sodium-loaded cells of the mutant failed to accumulate K+; all the intracellular anions were occupied by Na+ ions, which could not be expelled. However, growth of the mutant cells resumed when the K+/Na+ ratio of the medium was increased; under those conditions, the intracellular K+/Na+ ratio rose concomitantly. The wild-type strain behaved likewise in Na+-rich medium in the presence of gramicidin D (Fig. 4); here again, the effect of the ionophore was a function of the K+/Na+ ratio in the medium. It is clear that one substantial role of sodium extrusion is to make room for K+ ions. This task is very simple but important in homeostasis.
In high-Na+ medium, a mutant lacking the Na+-ATPase activity did not grow at alkaline pH; growth at acidic pH was normal (Fig. 4B). The Na+-ATPase is indispensable for Na+ extrusion at alkaline pH, but at acidic pH the Na+/H+ antiporter compensates for its absence. On the other hand, the pH profile of growth of the napA-disrupted strain WD4 in high-Na+ medium did not differ from that of growth of the parent strain (134). The NapA function (Na+/H+ antiport) is dispensable for the physiology of E. hirae, even in high-Na+ medium, at any pH. Regulation of the Na+-ATPase level compensates for any deficiency in NapA, even at acidic pH (134).
It has been suggested that the Na+/H+ antiporter plays an important role, not only in sodium extrusion but also in cytoplasmic acidification at alkaline pH (152, 192). The effect depends on proton influx via the Na+/H+ antiporter in response to the membrane potential. However, this proposal cannot apply to E. hirae, since the napA mutant grows well at alkaline pH when the membrane potential is minimal. Cytoplasmic acidification in E. hirae depends on another system, a K+/H+ antiporter pump.
Potassium Transport Systems
Potassium is the major intracellular cation for all living cells from animals to microorganisms. Intracellular K+ concentrations are kept in the range of 0.1 to 0.5 M in E. coli and Salmonella typhimurium and 0.4 to 0.6 M in E. hirae; the K+ concentrations remain high even when extracellular concentrations drop to starvation levels. Maintaining the intracellular K+ level requires several parallel potassium transport systems (17). Investigation of bacterial K+ transport is most advanced in E. coli and is founded on powerful genetics and molecular biology. A number of separate K+ transport systems are thought to occur in E. coli (51); the two three-component TrkG and TrkH systems and the one-component Kup system are constitutively expressed, while the three-component Kdp-ATPase system is derepressed under conditions of K+ starvation or osmotic upshock (153). In addition, there are two transport pathways for K+ efflux in E. coli: KefB and KefC (20, 56). These efflux systems are regulated by glutathione; whether they operate as porters or channels is currently being examined.
E. hirae possesses two K+ uptake systems, KtrI and KtrII, and one extrusion system, here designated Kep (K+ extrusion pump). KtrI appears similar to the Trk system of E. coli, but the other two systems show unique features not seen in other bacteria. The molecular characterization of these K+ transport systems in enterococci has just been initiated.
KtrI.
The KtrI system is the major potassium uptake pathway under most conditions; it has a pH optimum near 7 and an apparent Km of about 0.2 mM, and it attains a maximal rate of about 70 nmol of K+ min−1 mg of cells−1. This rate is comparable to the rate of glycolysis (100 to 150 nmol of lactate min−1 mg of cells−1), and under certain conditions cells do take up K+ at a rate that approaches their overall metabolic rate. KtrI also transports Rb+ with kinetics similar to those of K+, and it appears to be constitutive. An early observation that the rates of K+ and Rb+ uptake via this system increased in E. hirae grown on K+-deficient medium may reflect an increased activity of the F-type H+-ATPase in such a medium, as described above (6). There is no clear evidence for activation of KtrI activity by a change in turgor pressure or in the osmotic pressure of the medium.
The nature of energy coupling in this system was addressed by Bakker and Harold (18). KtrI can establish a potassium concentration gradient, [K+]in/[K+]out, of 5 × 104 or even 1 × 105. The membrane potential is at most −150 mV and is often much lower under physiological conditions, suggesting that the cellular K+ pool cannot simply be in electrochemical equilibrium with the membrane potential.
That KtrI has a requirement for metabolic energy was inferred from experiments in which a membrane potential was imposed artificially by shifting the pH in the presence of a protonophore. DCCD was present in these experiments to block the H+-ATPase and thus prevent the generation of a proton potential by ATP hydrolysis. When a membrane potential alone was imposed, K+ uptake was insignificant. There was somewhat greater K+ uptake when glucose was present and the protonophore was omitted. A high rate of K+ uptake was induced when both glucose and a protonophore were present. The requirement for glucose shows that some metabolic product, possibly ATP, is required for KtrI. Thus, K+ uptake by KtrI requires the cells to generate both ATP and a membrane potential. Since potassium uptake depolarizes the cells, it is probably accompanied by the net influx of a positive charge, possibly protons. From the mechanistic viewpoint, KtrI might be either a pump or a porter. Which mechanism fits this system? If it is a primary pump, such as an ATP-driven K+ pump, it is likely to be a K+-ATPase. Considering the Vmax of KtrI activity, a K+-stimulated ATPase activity should be readily observable in membrane vesicles when the H+-ATPase is blocked. Repeated efforts to find such a K+-stimulated ATPase have not been fruitful. A comment is in order here concerning a vanadate-sensitive ATPase that was purified from E. hirae cell membranes by Solioz’s group (100). The entire subject of K+ transport in E. hirae was confused for a time by reports claiming that this enzyme is a K+-transport ATPase, probably KtrI. This claim has now been withdrawn (15) for lack of supporting evidence.
Bakker and Harold (18) favor the hypothesis that KtrI is a secondary porter that mediates symport of K+ with a proton(s) and is regulated by phosphorylation (or by some other ATP-dependent covalent modification). By the trick of coupling the uptake of K+ to that of a proton, the driving force is greatly increased, and a potassium concentration gradient of 104 can be compatible with a membrane potential of −120 mV. Regulation of KtrI by ATP would serve the physiological purpose of inhibiting rapid K+ loss in energy-depleted cells. The energetics of KtrI are similar to those of the Trk system of E. coli and of potassium transport in Bacillus acidocaldarius (167). All these K+ transport systems show high capacity of K+ accumulation, which requires both ATP and the membrane potential. The TrkA subunit, a peripheral membrane protein of the E. coli Trk system, resembles NAD+-dependent dehydrogenase (218); Schlösser et al. speculate that the ATP requirement is secondary via NAD binding to the TrkA protein (218).
A unique E. hirae mutant with an alteration in this system is strain 576B, in which the competitive inhibition of K+ uptake by Na+ is markedly increased (76). This mutation appears to reduce the specificity of the cation binding site, especially at pH 7.5, and as a result a high concentration of Na+ in the medium inhibits K+ uptake and growth.
KtrII (NtpJ).
A second uptake system for K+ in E. hirae, one that is not dependent on the proton potential, was discovered by Kobayashi (139) during studies of mutant AS25, which is defective in the H+-ATPase and the generation of the proton potential. The small proton potential in this mutant should not support high activity of KtrI, yet the cells did accumulate K+ 5,000-fold. This proton potential-independent uptake system, KtrII, has a pH optimum around 9, does not transport Rb+ very well, and has a Km for K+ of 0.5 mM and a Vmax of about 20 nmol of K+ min−1 mg of cell−1. KtrII activity is inducible, but it does not respond to K+ deprivation, nor is it repressed by excess K+; KtrII is synthesized in response to the need of the cell to expel Na+, because its activity is induced to a high level when mutant AS25 is grown in sodium-rich media. In the wild type, high levels of KtrII are induced when the cells are grown in sodium-rich media supplemented with a protonophore. Thus, the behavior of KtrII appears to be quite unlike that of Kdp, the K+-translocating ATPase of E. coli. KtrII cannot scavenge trace amounts of K+ from the medium, but it does permit the cells to grow under conditions that render KtrI inoperative.
In a study of the energetics of KtrII, Harold and I proposed the simplest mechanism, a direct exchange of Na+ for K+ ions by the Na+-ATPase (112). K+ uptake via KtrII is strictly coupled to Na+ extrusion by the Na+-ATPase, although we observed little or no stimulation of the ATPase activity by K+ ions or by Na+ and K+ together (for details, see reference 114). Furthermore, uptake of K+ and Rb+ was examined in Na+-loaded cells of the wild-type strain, grown either on Na+-limited medium or on high-Na+ (more than 500 mM) medium to induce the Na+-ATPase (112, 136); DCCD was absent from the assay buffer. Based on the cation specificities of KtrI and KtrII as described above, we regard Rb+ uptake as a measure of KtrI and K+-specific uptake as a measure of KtrII. The Na+/H+ antiporter can operate under these experimental conditions, and Na+ extrusion via the Na+-ATPase should not be rate limiting for the accumulation by either system. Rb+ uptake by KtrI was unaffected by the growth conditions, but K+-specific uptake activity by KtrII obviously increased in cells grown on high-Na+ medium in which the Na+-ATPase was highly induced (110), suggesting that induction of KtrII as well as the Na+-ATPase responds to the sodium content of the medium.
By sequencing the Na+-ATPase operon, we found an interesting cistron at the tail, designated ntpJ (Fig. 6A). The ntpJ gene encodes a putative 49-kDa hydrophobic protein with at least 10 membrane-spanning segments, which does not resemble any other V-ATPase subunit (249). The NtpJ protein resembles various K+ transporter proteins such as Trk1p and Trk2p of yeast and TrkG and TrkH of the E. coli Trk system (17). In the ntpJ-disrupted strain JEM2, there was no K+ uptake via KtrII (Fig. 8) but KtrI was operative. Thus, it is obvious that the ntpJ gene encodes a component of the KtrII system; coinducibility of KtrII with the Na+-ATPase was thus proven at the gene level. However, KtrII is not intimately linked to the Na+-ATPase, since the assembly and activity of the latter in mutant JEM2 were normal (176). We considered the possibility that KtrII (NtpJ) represents a Na+/K+ symporter. However, this seems unlikely since the K+ gradient established by KtrII is far greater than the sodium gradient established by the Na+-ATPase under our experimental conditions. It is also uncertain whether KtrII is wholly encoded by the ntpJ gene or requires additional components; both the composition and the mechanism of KtrII remain mysterious. The ntpJ-disrupted strain JEM2, in which KtrII was defective, did not grow at alkaline pH in K+-limited (less than 1 mM K+) medium but grew normally at pH 7. The KtrII system evidently serves to accumulate K+ under conditions of K+ limitation and pH such that KtrI is inoperative. Thus, the ntp operon is an interesting one, since it contains genes that encode cation transport systems to cope with environments rich in sodium and at alkaline pH.
FIG. 8.
KtrII K+ uptake activities of E. hirae strains. Movements of K+ (•, ○) and Na+ (■, □) at pH 9.0 were assayed with Na+-loaded cells in the presence of protonophore; the reaction was initiated by the addition of 1 mM KCl at 10 min as indicated by the arrows. Solid symbols, 10 mM glucose added; open symbols, no glucose. (A) 9790 (parent). (B) Nak1 (sodium ATPase mutant). (C) JEM2 (mutant in which the ntpJ gene is disrupted by insertion of erythromycin resistance gene). Reprinted from reference 176 with permission of the publisher.
Potassium extrusion.
K+ efflux in streptococci and its relation to K+ uptake are not yet fully understood. NEM-sensitive downhill K+ extrusion, analogous to the Kef system of E. coli, has been observed in several bacteria but not in streptococci or enterococci (46). Passive K+ efflux seen in the presence of uncouplers requires ATP (or some other energy metabolite) (18), but whether such efflux occurs through KtrI (a putative ATP-regulated K+/H+ symporter) or by some other pathways is not known. The 325B mutant, originally isolated as a strain that requires elevated concentrations of K+ for growth, is defective in K+ retention (87). The normal kinetics of K+ influx in this mutant suggest that the lesion affects a separate system, probably one that mediates K+ efflux.
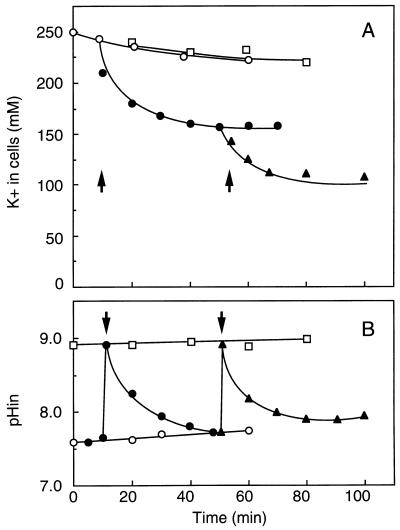
A unique K+ extrusion system, Kep, has been discovered in E. hirae (113): K+ extrusion occurred only when the cytoplasmic pH was alkaline, and it required the generation of ATP (or a related metabolite). As shown in Fig. 9, glycolyzing cells suspended in an alkaline medium extruded K+ ions, even against a K+ concentration gradient, provided that the medium contained a weak permeant base (e.g., diethanolamine or methylamine). The amine renders the cytoplasmic pH alkaline; when conditions were arranged so as to keep the cytoplasm neutral, no K+ extrusion was seen. K+ extrusion required the presence of either glucose or arginine and was unaffected by protonophores or by inhibitors of the H+-ATPase. When the medium contained radioactive methylamine, the cells accumulated the base to an extent stoichiometrically equivalent to the amount of K+ lost. Concomitantly, the cytoplasmic pH fell from 8.8 to 7.6, at which point K+ extrusion ceased: this system is strictly regulated by the cytoplasmic pH. It is hard to distinguish K+/ammonium (methylammonium) antiport from K+/H+ antiport. However, the following observations point to K+/H+ antiport: (i) K+/amine (ammonium) exchange occurs only at alkaline pH, as expected if the unprotonated form of amine was the species crossing the membrane; (ii) a high concentration of amine is required, suggesting that the amine serves to dissipate the pH gradient (inside acidic) at alkaline pH; and, most importantly, (iii) radioactive methylamine uptake coupled to K+ extrusion does not display saturation kinetics even at extracellular concentrations as high as 50 mM. Thus, in E. hirae, an ATP-driven transport system that expels K+ by exchange for H+ takes part in the circulation of these ions at an alkaline cytoplasmic pH. The FOF1-ATPase may not be the only proton pump in this bacterium.
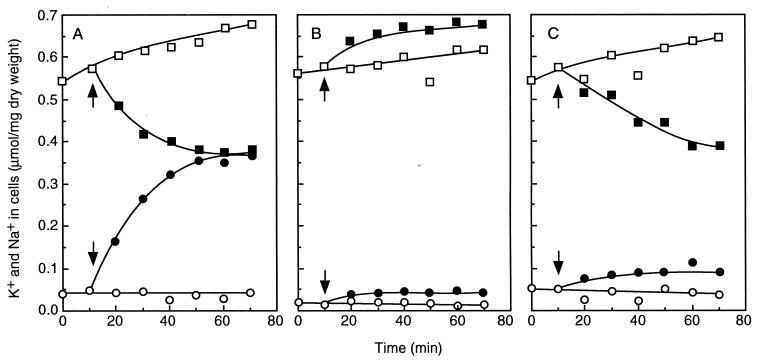
FIG. 9.
Effect of diethanolamine on uphill K+ extrusion and the internal pH of E. hirae. The K+ movement (A) and the internal pH change (B) of the cells at pH 9.0 were monitored in a high-K+ buffer (250 mM K+). Solid symbols, glucose added; open symbols, no glucose. The reaction was initiated by addition of diethanolamine as indicated by the arrows. Symbols: ○, no addition; •, 20 mM diethanolamine; ▴, 50 mM diethanolamine; □, control (in a buffer containing 50 mM diethanolamine but not glucose). Reprinted from reference 113 with permission of the publisher.
Additional potassium transport systems.
Another active-transport system with low affinity and high capacity participates in K+ accumulation at alkaline pH. The KtrII-defective strain JEM2, in which the ntpJ gene is disrupted, did not grow at alkaline pH in K+-limited medium but grew well in the presence of 5 to 10 mM K+. Even though KtrII is inoperative, about a 20-fold concentration gradient of K+ is generated in this strain at alkaline pH (176). This system also recognizes Rb+. A novel mutant, distinct from KtrII, defective in K+ accumulation at alkaline pH has been isolated. The level of K+ accumulation in this strain is about half that in the wild type. The characteristics of this K+(Rb+) transport system, which apparently responds to the membrane potential, are under investigation (135). Potassium transport has not been extensively studied in other streptococci, but there is a report of Na+-dependent uptake of thallous ions (an analog of K+) by L. lactis (127).
Physiological work.
One of the major roles of K+ is charge neutralization of cellular anions. One might think that other cations, such as Na+ or Mg2+, could neutralize cellular anions equally well, but in fact cells loaded with Na+ glycolyze and transport substrate normally but do not grow (83). The choice of K+ over other cations as the main intracellular cation must have some deep evolutionary reasons, which have never been quite resolved. It is generally agreed that K+ ions make a more “compatible” solute than Na+ ions, in the sense that the former are less destructive to cellular macromolecules and their associated water shells (78, 265). Indeed, Rb+ can completely replace K+ in growing cells of E. hirae. Some bacterial enzymes are activated by K+, but all bacteria require K+ for protein synthesis. A high intracellular K+ concentration may be required for polysome stability (50). Another important role of K+ transport is the regulation of the cytoplasmic pH, which is discussed below. In E. coli, turgor pressure is maintained in part by regulating intracellular K+ (17, 51), and the same may be true for plant cells, but it does not appear to be the case in enterococci. In E. hirae, for instance, K+ is the major cellular osmolite, but there is no evidence that K+ transport is altered by changes in cell turgor. Finally, the relatively massive K+ gradient directed outward (and the Na+ gradient directed inward) constitute a kind of energy storage that can, if necessary, drive motility and transport (228). This option may well be an extra benefit that makes the difference under some conditions.
Calcium Transport Systems
Calcium extrusion.
Calcium transport was first investigated in E. hirae. Using both intact cells and everted membrane vesicles, Kobayashi et al. demonstrated that E. hirae extrudes Ca2+ by a primary active transport system (146). Calcium is readily expelled from the cells in the absence of a detectable proton potential, establishing a gradient of 30:1 (out/in). In everted membrane vesicles, ATP-driven Ca2+ accumulation was observed in the absence of a proton potential. Autologous 45Ca2+/Ca2+ exchange is demonstrable in both intact cells and vesicles, and it requires ATP. The Km for the uptake activity by the vesicles varies with the pH: at the optimal pH of 7.0, the Km is 0.1 mM, and at pH 6.0 it increases to 1.0 mM. Ca2+ transport is not significantly inhibited by ruthenium red, NEM, or lanthanum. The effect of ionophores on Ca2+ transport suggests that the process is electroneutral; probably two H+ ions are exchanged for each Ca2+ ion. Ca2+ transport, linked to ATP but not the proton potential, was also observed in L. lactis and S. sanguis (13, 98, 213). In all these streptococci, ATP-driven 45Ca2+ uptake is inhibited by vanadate, an inhibitor of the P-ATPase, suggesting that transport is mediated by a Ca2+-translocating ATPase. However, Ca2+-stimulated ATP hydrolysis by the membranes has not been detected.
In contrast, Ca2+ efflux in L. lactis subsp. cremoris is mediated by a secondary transport system catalyzing the exchange of calcium ions and protons; collapse of the proton potential by protonophores or inhibition of the H+-ATPase by DCCD strongly inhibits Ca2+ extrusion (47). Further studies of Ca2+ efflux at different external pH values, in the presence of either valinomycin or nigericin, suggested that Ca2+ efflux from intact cells is an electrogenic process. Uptake of Ca2+ in response to a proton potential was demonstrated by the use of membrane vesicles fused to proteoliposomes containing the light-driven proton pump bacteriorhodopsin. It has been reported that 45Ca2+ efflux from energy-depleted S. pneumoniae cells could be induced by an imposed Na+ gradient; the authors speculate that Na+/Ca2+ antiport is responsible (255, 257). This efflux was inhibited by the amiloride derivative 2′,4′-dimethylbenzamil (DMB). Very little is known about the molecular features of Ca2+ transport systems in bacteria. In E. coli, the chaA gene was regarded as the putative gene for a Ca2+/H+ antiporter (103); however, it now appears that the ChaA protein functions physiologically as a Na+/H+ antiporter, recognizing sodium rather than calcium as its substrate (189).
Calcium influx.
No Ca2+-specific uptake system has been found in E. hirae. When a membrane potential, interior negative, was present, 45Ca2+ uptake was proportional to the external Ca2+ concentration over a range of 0.1 to 10 mM (146). This potential-driven Ca2+ uptake probably occurs via a nonspecific leak pathway. On the other hand, an electrogenic 45Ca2+ influx in ATP-depleted S. pneumoniae is sensitive to the amiloride derivative DMB at 4 μM (255, 257); in this case, Ca2+ movement may be mediated by a specific porter.
Physiological work.
Calcium ions have often been designated a second messenger in eukaryotes (37). In prokaryotes, Ca2+ is also involved in a variety of cellular processes: proteolysis in sporulation of Bacillus subtilis (188), chemotaxis in E. coli (252), etc. Ca2+ ions activate autophosphorylation of the heat shock protein DnaK (38) and of EnvZ (206). In S. pneumoniae, about 0.15 mM calcium is required for cell growth (255, 256). When Ca2+ transport was inhibited by DMB at high concentrations of external Ca2+ (1 mM), the growth of S. pneumoniae ceased. Homeostasis of the internal calcium ion level should be important for bacterial physiology. Calcium transport systems are expected to do this work, although the details of their role are not obvious. Furthermore, high concentrations of Ca2+ ions induce competence of S. pneumoniae for genetic transformation in exponentially growing cultures and lysis in early stationary phase. It is also proposed that DNA uptake at competence constitutes a homeostatic response to elevated Ca2+ levels (255). Competence induction and autolysis are prevented by inhibition of Ca2+ transport with the amiloride derivative. On the other hand, E. hirae grows well in a defined medium in which the free Ca2+ concentration is less than 10−8 to 10−9 M, and no physiological role of calcium has ever been proposed.
Transport Systems for Other Cations
The uptake of magnesium, a major cation in cellular physiology, has been examined in S. typhimurium and other bacteria (230, 231, 250) but not in streptococci. Trace cations are universally required as cofactors in cellular metabolism, but their uptake has not been well studied in streptococci; a few papers have suggested the existence of transport systems specific for Mn2+ (22) and Fe3+ (53). Recently, a systematic study of copper transport by E. hirae was begun by Solioz and his colleagues.
Copper ATPase.
Copper is an essential heavy metal ion that serves as a cofactor for many redox enzymes, such as cytochrome c oxidase, superoxide dismutase, lysyl oxidase, and dopamine β-hydroxylase. On the other hand, copper can be very toxic to both prokaryotic and eukaryotic cells through formation of free radicals. Homeostatic mechanisms have evolved to strictly balance the internal copper concentration. Regulation of the cytoplasmic copper level appears to involve influx and efflux pathways in addition to the chemical modification of copper in the cytoplasm (32). Genes involved in the control of the cytoplasmic copper level have been identified in several bacteria (225, 227). E. coli, for example, has a number of chromosomal genes, as well as a plasmid-borne one, involved in copper resistance (226). These genes have already been sequenced, but their functions remain unclear (224). On the other hand, two kinds of copper transport ATPases have been reported as the machinery for copper homeostasis of E. hirae, one directed inward and the other directed outward (234, 236).
(i) Structure.
The copper ATPase in E. hirae was discovered by accident. Solioz and coworkers (187) were attempting to clone the gene encoding a vanadate-sensitive ATPase (100), which they had tentatively identified as the KtrI K+ transport system, by use of an antibody raised against a sample of purified ATPase (235). Since this ATPase fraction was a relatively crude preparation, they inadvertently cloned a cop operon that does not code for KtrI but is involved in conferring copper homeostasis on E. hirae (186) (Fig. 10). The cop operon consists of four genes, copYZAB (186, 239). The copY (145 amino acid residues) and copZ (69 amino acid residues) genes encode hydrophilic proteins involved in the regulation of this operon; copA and copB encode ATPases with 727 and 745 amino acid residues, respectively. CopA and CopB exhibit significant sequence similarity and are also similar to other P-ATPases. Most prominently, they resemble the human Menkes gene product (261), a copper-ATPase, with 43% similarity to CopA and 33% similarity to CopB. The sequence AspLysThrGlyThr, considered to be the phosphorylation site in all known P-ATPases, is conserved in both CopA and CopB; the aspartic acid residue that forms the acyl phosphate intermediate characteristic of these enzymes is also found (267). While CopA and CopB display extensive overall sequence similarity, their N-terminal sequences are different. The sequence GlyXCysXXCys in the N-terminal region of CopA is similar to those of various metal binding proteins, such as MerA, the mercuric reductase of Thiobacillus ferrooxidans (102), CadA, the cadmium extrusion ATPase of Staphylococcus aureus (227), and the Menkes copper ATPase. The N terminus of CopB contains three repeats of the consensus sequence MetXHisXXMetSerGlyMet, which is very similar to repeats present in the Pseudomonas syringae pv. tomato CopA protein (166), known to be a periplasmic copper binding protein (40). These conserved sequences in CopA and CopB appear to represent the binding sites for heavy metal ions, responsible for its metal specificity.
FIG. 10.
Schematic representation of the E. hirae copper ATPase (cop) operon. Boxes indicate open reading frames of the cop operon, and letters represent the gene names: copA and copB encode P-type ATPases for uptake and extrusion of copper, respectively; copY and copX (shaded) encode regulator proteins sensing copper. The black box represents the promoter/operator region.
Copper transport by intact cells has yet to be investigated, and information about the biochemical properties and hydrolytic activities of these ATPases is limited. However, the roles of CopA and CopB in copper homeostasis are clear from the copper sensitivity of strains in which the cop genes were disrupted; the copA mutant, as well as the wild type, grew in high concentrations of copper, but the copB mutant did not. The copA mutant ceased to grow when the external metal ion level was decreased by complex formation with 8-hydroxyquinoline, conditions that do not affect the growth of the wild type (186). It is suggested that copA codes for copper uptake and copB codes for copper extrusion. ATP-driven copper uptake by CopB in everted membrane vesicles has been observed (233).
Although copper ATPases have not been studied in other streptococci, they are widely distributed among microorganisms. Genes for copper uptake (ctaA) and copper efflux (pacS) have been identified in the cyanobacterium Synechococcus sp. strain PCC7942 (126, 196). hpCopA of Helicobacter pylori (64) and ccc2 of Saccharomyces cerevisiae (269) are postulated to encode a copper-pumping ATPase. Most notably, genes for two copper ATPases have been identified in humans (34, 261); mutations in these genes cause the lethal diseases Menkes’ syndrome and Wilson’s disease. Their roles in disease have been predicted largely on the basis of bacterial homologs, and information about bacterial enzymes will contribute insight into the mechanisms of copper ATPases.
(ii) Physiological work.
Although it has not been established that copper is indispensable for the growth of E. hirae, copper probably plays a role as cofactor for some metabolic enzymes as seen in other bacteria. The growth requirement of microorganisms for copper is satisfied by less than 10 μM copper (39), which is not toxic to E. hirae and does not induce the cop operon. The cytoplasmic copper concentration may therefore be in the micromolar range, and a Km of 1 μM for copper transport by CopB (233) appears reasonable. It is interesting that the CopB ATPase transports 64Cu+ in membrane vesicles. 110mAg+ was also transported by CopB ATPase at a higher rate and lower affinity, suggesting that CopB recognizes these metals in the monovalent form. Cytoplasmic copper occurs in the monovalent form, either due to the reducing environment of the cytoplasm or through the action of reductases. From the finding that CopB ATPase transports silver (Ag+) in membrane vesicles, it is likely that CopB plays a role in the detoxification of silver. Energy-dependent efflux of silver ions is lost in the copB-disrupted strain (239). However, the growth of the copB mutant as well as the wild type was inhibited by 5 μM Ag+, suggesting a minimal contribution of CopB to silver detoxification in vivo. By contrast, the copA-disrupted strain tolerates considerably higher silver concentrations than the wild-type strain does (186). Silver may be accumulated by the cells via CopA, which has a broad substrate spectrum for several metal ions.
The level of CopB ATPase, the last gene product of the cop operon (Fig. 10), is enhanced by an increase in the copper concentration of the growth medium and also in response to Ag+ or Cd2+ (187). On the other hand, an increase in the level of the CopB protein was also observed when o-phenanthroline or 8-hydroxyquinoline was added to chelate heavy metal ions in media. It thus appears that either low or high concentrations of copper ions lead to the induction of both CopA and CopB ATPases (185). The simultaneous stimulation of both cop genes in the regulation of intracellular copper is quite mystifying and requires clarification.
The CopY and CopZ proteins participate in the control of the levels of the CopA and CopB proteins (239). Both proteins contain metal-bonding motifs and exhibit significant sequence similarity to other known regulatory proteins. In the copY-disrupted strain, constitutive overexpression of CopA and CopB occurred, resulting in a copper-dependent phenotype. In contrast, the expression of both copA and copB genes was suppressed by disruption of the copZ gene, rendering the cells sensitive to copper. Both null mutants could be complemented by a plasmid harboring the copY and copZ genes. Based on these results, it is proposed that copY and copZ encode trans-acting regulatory proteins, required for expression of the cop operon and dependent upon the copper level. CopY apparently acts as a copper-induced repressor and CopZ as an activator (184). CopY has been shown to interact with the operator, probably as a homodimer, and the critical nucleotides have been identified.
ENERGY TRANSDUCTION
ATP Production
Sugar metabolism.
Streptococci, lacking a respiratory chain, metabolize carbohydrate without net production of reducing equivalents. A case in point is the degradation of glucose to lactic acid, resulting in the production of 2 mol of ATP per mol of glucose metabolized. Some streptococci, however, have a limited capacity to utilize exogenous electron acceptors for the degradation of substrates. Carbohydrate metabolism in L. lactis and S. thermophilus is affected by the presence of oxygen. Under aerobic conditions, L. lactis converts lactose almost quantitatively to acetic acid plus carbon dioxide, thereby producing an excess of NADH (197). These streptococci contain flavin-type NADH oxidases and NADH peroxidases, which obviates the need to reduce pyruvate to lactate or to reduce acetyl coenzyme A to ethanol. By forming acetic acid instead of lactic acid and using an oxidase to reoxidize the NADH produced, additional ATP can be formed by substrate-level phosphorylation. The metabolic end products of glucose metabolism in some other streptococci are also affected by culture conditions; the amounts of acetate and ethanol as end products increased anaerobically at alkaline pH. The glycolytic metabolism of enterococci at alkaline pH is not the same as at acidic pH, switching to a more effective production of ATP in the presence of high concentrations of carbon dioxide (67, 68). The metabolic pattern is also altered under anaerobic conditions: if fumarate reductase is present, carbohydrate metabolism by streptococci may also yield acetic acid as an end product and thus result in extra ATP. In this scheme, electrons are transferred from NADH dehydrogenase to fumarate reductase. Although these components are present in L. lactis (93), the participation of this electron transfer chain in the oxidation of NADH under anaerobic conditions has not been demonstrated.
The initial event in the metabolism of sugars involves their translocation across the cytoplasmic membrane. Three kinds of transport systems are known to translocate carbohydrates: (i) phosphoenolypruvate-dependent sugar phosphotransferases; (ii) sugar transport ATPases, members of the ATP binding cassette transport family; and (iii) ion-linked sugar transport and sugar exchange mechanisms. Different transport mechanisms for a wide variety of sugars are observed in streptococci.
Deiminase pathway.
In addition to ATP production by the glycolytic pathway, some streptococci generate ATP by metabolism of arginine or agmatine via the corresponding deiminase pathways (41). In the initial step of these reactions, arginine or agmatine is taken up via an antiporter which catalyzes a one-for-one exchange of arginine for ornithine (198) or of agmatine for putrescine (48). No additional energy is required for translocation of these solutes. Consequently, the net yield of metabolic energy from arginine and agmatine is 1 mol of ATP per mol of substrate metabolized.
Generation of the Proton Potential
Machinery.
The generation of the proton potential in streptococci is clearly the function of the F-type, H+-ATPase; the mutant defective in H+-ATPase did not generate it. Some time ago, it was reported that certain streptococci have a cytochrome-like respiratory chain (212), but nobody has written of it since. On the other hand, certain combinations of secondary transport and metabolism may contribute to the proton potential. Lactic acid bacteria produce large quantities of cytoplasmic fermentation products, building up a considerable concentration gradient directed outward. Lactic acid and other metabolic end products commonly leave the cell by way of a transport carrier, together with one or more protons. When the gradient is large enough, it can contribute significantly to the total proton potential, conserving metabolic energy by transformation of a product gradient into a proton potential. In some streptococci, the lactate porter appears to be electrogenic, with a stoichiometry of 2H+ per lactate (251). Net efflux of lactate plus proteins contributes to the proton gradient and thus participates in the conversion of metabolic energy into that of an ion gradient. Another mechanism for the generation of a proton potential, without participation of any proton pump, has been proposed for L. lactis (201). In this pathway, l-malate taken up by the cells is decarboxylated by malolactic enzyme to yield l-lactic acid and carbon dioxide, after which the products leave the cells. Monoanionic l-malate is taken up either by exchange for l-lactic acid or by uniport; in consequence, a negative charge is translocated from the medium into the cytoplasm, generating a membrane potential inside negative. Since charge compensation in the decarboxylation reaction requires the consumption of a proton, the cytoplasm is alkalinized relative to the outside medium and a pH gradient is generated. Thus, the free energy of decarboxylation is converted into a pH gradient simply by compartmentalization of the metabolites. It has been shown for L. lactis that l-malate utilization results in the formation of a proton potential of about −175 mV, which is high enough to drive ATP synthesis via the F-ATPase. Whether the ion gradient thus generated actually does synthesize ATP via the F-ATPase is not known. In any event, the combined action of an electrogenic transport process (uptake of substrate and excretion of product) together with the decarboxylation reaction allows a significant portion of the free energy to be conserved in the form of ion gradient.
Magnitude of the proton potential.
In microorganisms of sufficient size, especially the fungus Neurospora and the cells of many algae, the internal pH and the electrical potential can be measured directly by means of microelectrodes. Bacteria are too small for this procedure, and so indirect approaches must be used; these usually involve dyes that respond to pH or electrical potential. Most of the measurements presently available are based on the principle that any ion that moves passively across the membrane should be distributed between the cytoplasm and medium in accordance with the Nernst equation. The methods used are reasonably well standardized and have been reviewed in detail (27, 129). The membrane potential is usually measured by the accumulation of radioactive lipid-soluble ions, a cation when the potential is inside negative and an anion when it is inside positive; the pH gradient is calculated from the uptake of lipid-soluble weak acids or bases, e.g., benzoate or acetylsalicylic acid (aspirin) when the internal pH is more alkaline than that of the medium and methylamine or benzylamine when it is more acidic. Nuclear magnetic resonance spectroscopy provides the most accurate measurement of the internal pH presently available. In all cases, corrections have to be applied to the data, which limit the accuracy of the determination. The method of pH measurement in bacteria and its limitations have been reviewed (27, 129).
In aerobic bacteria such as E. coli, the proton potential generated by the respiratory chain poises the equilibrium of the ATPase reaction in oxidative phosphorylation. The proton potential is estimated to be at most −200 mV by the methods mentioned above. There is no doubt that this value is substantially lower than the thermodynamic maximum, for reasons related to the flux of other ions; for example, its magnitude is affected by the concentrations of both K+ and Na+ (130). However, assuming that the FOF1-ATPase translocates three protons per reaction cycle (3H+/ATP), a proton potential of −200 mV would support the observed phosphorylation potential, ΔGp, of −58 kJ/mol. It is of general significance that the proton potential of fermentative bacteria is distinctly lower than that of respiring cells. When growing cells of Staphylococcus aureus were compared under aerobic and anaerobic conditions, the proton potentials at pH 6 were −250 and −160 mV, respectively (128). Observed values for respiring cells are entirely consistent with expectations. Therefore, the difference probably reflects the fact that aerobic cells extrude protons by redox reactions while anaerobic ones rely primarily on the hydrolysis of ATP by the FOF1-ATPase. In enterococci and streptococci, the magnitude of the proton potential is much smaller, ranging from −120 to −150 mV in all these cases (144). These bacteria do not need the highest “threshold” magnitude of proton potential for physiological work, because proton flow is not required for ATP formation.
Figure 11 displays the magnitude of the proton potential of E. hirae as a function of external pH from 5 to 10. Cells were grown in high-K+, low-Na+ medium (120 mM K+, less than 20 mM Na+). The proton potential is maximal at a medium pH of 6 to 7, reflecting the optimal pH of the H+-ATPase activity, but declines as the external pH is made more alkaline. At pH values around 9, the proton potential is nearly zero, and beyond that it is reversed. Low values of the proton potential at an alkaline pH have also been observed in L. lactis and L. lactis subsp. cremoris (132). Thus, chemiosmotic energy transduction cannot operate in streptococci at alkaline pH.
FIG. 11.
Internal pH, membrane potential, and proton potential of E. hirae at various external pHs. E. hirae was grown in KTY (high K+, low Na+) medium. Symbols: •, internal pH; ▴, membrane potential; □, proton potential.
The same is true of zygotes and the large marine alga Pelvetia, in which the proton potential across the plasma membrane varies dramatically with the external pH (65). The internal pH and the electric potential were directly measured with microelectrodes (65, 150). In this organism, the relatively constant cytoplasmic pH (from 7.4 to 7.5) and membrane potential (−50 mV) induce a decrease in the proton potential at alkaline pH. At the crossover point at an external pH of 7.5, the direction of the proton gradient was reversed.
E. hirae copes with the decrease in the proton potential at alkaline pH, with the help of a number of primary ATP-driven pumps for K+ and Na+ and perhaps for other metabolites.
Utilization of the Proton Potential
Transport systems for most amino acids are coupled to H+ (197). In L. lactis, branched-chain aliphatic amino acids (Leu, Ile, and Val), neutral amino acids (Ala and Gly), aliphatic amino acids with a hydroxyl side chain (Ser and Thr), and aromatic amino acids (Phe, Tyr, and Trp) are transported by separate H+-linked mechanisms. In E. hirae (16) and Streptococcus pyogenes (209), the neutral amino acids, glycine, alanine, serine and threonine, are transported by a common H+-linked system. Proton-coupled amino acid transport has also been proposed for S. thermophilus, S. agalactiae, and S. pneumoniae, usually on the basis of inhibitor effects (199). By contrast, ATP-driven uptake accounts for the accumulation of glutamic acid, glutamine, proline, and asparagine in several organisms (81, 200); all these amino acids serve as intracellular osmolytes, involved in the regulation of turgor. In L. lactis, di- and tripeptides are taken up by a pH gradient-driven transport system (69). Proton-linked cation transport is exemplified by the Na+/H+ antiporter and possibly by KtrI in E. hirae and by the Ca2+/H+ antiporter in L. lactis subsp. cremoris. Protons also serve as the coupling ions for flagellar rotation (165); motility was observed at acidic pH but not at alkaline pH, reflecting the size of proton potential at different pHs.
Generation and Utilization of the Sodium Gradient
There are very few examples of sodium-dependent secondary-transport systems in streptococci. Sodium-dependent uptake of amino acids has been reported in S. bovis (215), and sodium-dependent uptake of K+ (Tl+) has been reported in L. lactis (127). There is no substantial evidence for any essential functions of this kind in enterococci. In E. hirae, an Na+ electrochemical gradient of as much as −110 mV can be generated under certain conditions, such as alkaline pH (114, 115). I doubt that organisms fail to utilize such a powerful energy source in their metabolic economy. The mechanism of KtrII, the NtpJ-dependent K+ uptake system, is still under investigation, but coupled movement of Na+ and K+ is very likely (115, 176). E. hirae shows no Na+ requirement for growth at neutral pH, if one ignores the contamination level of less than 1 mM Na+. At alkaline pH and with limited K+, the growth of E. hirae requires the presence of sodium ions in the medium (115). Further investigation of the Na+-coupled secondary systems is required to clarify the significance of the Na+ circulation (228) in streptococci.
ROLE OF CATION TRANSPORT IN HOMEOSTASIS
Regulation of the Cytoplasmic pH
We should note that because of their small size, bacterial cells contain few free protons. Taking the volume of a bacterial cell to be 10−15 liters, the numbers of free protons is calculated to be 6 at pH 6 and less than 1 at pH 8 or 9. However, bacteria do have a vast number of bound protons, and equilibrium between free and bound protons should be attained in the cytoplasm. Protons are available even at alkaline pHs above 9, even though there are no free protons at any given moment.
Regulation of the cytoplasmic pH is essential for the survival of bacteria, which must be able to accommodate variations of the environmental pH, metabolic reactions that produce or consume internal protons, and the movement of acids and bases across the plasma membrane. Cellular activities demand that the cytoplasmic pH be kept within relatively narrow limits. Homeostasis of cytoplasmic pH in bacteria was first demonstrated in streptococci by Harold et al. (84). Subsequently, there have been reports dealing with other bacteria (for reviews, see references 27 and 73). Of interest is the report that the internal pH of membrane vesicles prepared from E. coli remains constant even though such vesicles have no cytoplasmic metabolic activity (205). Over the years, various mechanisms have been proposed to explain how cells maintain a constant cytoplasmic pH. Half a century ago, Gale (61) proposed that amino acid decarboxylase is induced at alkaline pH and deaminase is induced at acidic pH. In Thermoplasma acidophila, pH regulation may depend on a Donnan potential and a plasma membrane permeable to cations (99). However, the common opinion today holds that the cytoplasmic pH in bacteria is regulated mainly by H+-linked transport systems located in membranes (for reviews, see references 27, 73, 151, and 152). In E. coli, the respiratory proton pump and H+-linked transport systems are clearly involved in pH homeostasis. Acidic and alkaline external pHs call for separate and distinct mechanisms to regulate the cytoplasmic pH. In E. hirae, the H+-ATPase plays a role in pH homeostasis at acidic external pH whereas an independent machinery operates at alkaline pH when the magnitude of proton potential is minimal.
Acidic pH.
At acidic pH (external pH below 7), proton expulsion by the FOF1-ATPase alkalinizes the cytoplasmic pH and contributes to its control but is not sufficient by itself. Proton extrusion by the H+-ATPase is electrogenic, and hence a membrane potential is generated (79, 80, 84). Because the membrane capacitance is low (about 1 μF cm−2), extrusion of a substantial quantity of protons requires dissipation of the membrane potential. Cytoplasmic alkalinization is accompanied by the electrogenic accumulation of potassium ions in E. hirae (19) and L. lactis (131, 132). The cytoplasm can also be alkalinized in the presence of potassium ions plus valinomycin, an ionophore for K+, or of dimethyldibenzyl ammonium ion (DDA+), a synthetic organic cation (79, 131). Cytoplasmic alkalinization can even be achieved by the accumulation of sodium ions in mutant 7683. Since this mutant is totally defective in Na+ extrusion, sodium ions probably accumulate via a leak pathway in response to the membrane potential (83). To keep the cytoplasmic pH constant, the activity responsible for cytoplasmic alkalinization must be high when the cytoplasm is acidic but must diminish as the cytoplasmic pH is raised. The mechanism appears to be simple. It is attributed to a feature of the H+-ATPase, which has an optimal pH at 6.5; the activity is very low at an alkaline pH (132, 145). The amount of functional H+-ATPase in membranes varies in response to the cytoplasmic pH, as described above. Cytoplasmic pH at acidic external pH is thus kept constant by changes in both the amount and activity of the H+-ATPase (141). Since the cytoplasmic pH of mutant AS25 deficient in the H+-ATPase was not excessively acidic at pH 7 (143), the cytoplasmic pH at this external pH is not a steady state of concurrent alkalinizing and acidifying mechanisms.
E. hirae maintains a basal level of H+-ATPase even at alkaline pH (145), even though the H+-ATPase does not play a role in pH regulation under these conditions (Fig. 4A). The function of the H+-ATPase under these conditions is not clear; it may be required to generate a membrane potential, albeit small, or allow rapid resumption of growth if the pH should shift back from alkaline to acid.
Alkaline pH.
Nigericin is an ionophore that exchanges K+ for H+. Its effect on the growth of E. hirae depends on the pH: in high-K+ medium, growth was unaffected at pH 7 (82) but was blocked at pH 9.6 (109). This appears to reflect the regulation of cytoplasmic pH. Cells growing at alkaline pH (pH 9.0 to 9.5) acidify their cytoplasm to pH 7.8 to 8.0 (Fig. 11). In the presence of nigericin, the cytoplasmic pH is identical to the external pH, and pH 9.6 is inhibitory. When the pH of the medium was brought back to 7.0, growth of the cells resumed. Cytoplasmic acidification appears to be required for the growth of E. hirae in alkaline medium.
In several bacteria, acidification of the cytoplasmic pH has been attributed to secondary porters that exchange K+ for H+ or Na+ for H+. This cannot be true for E. hirae. Even in medium containing more than 200 mM K+, where the ratio of internal to external K+ is close to 1, this organism acidifies the cytoplasm and grows well at alkaline pH. Influx of protons via a secondary K+/H+ antiporter can be excluded, unless the stoichiometry of H+/K+ is extraordinarily large. Acidification by proton influx via the Na+/H+ antiporter can also be excluded; even assuming a stoichiometry of H+/Na+ greater than 1 (152), such that antiport can be energized by the membrane potential, the membrane potential in alkaline medium is too small. As shown in Fig. 4A, the growth of AS25, the mutant defective in H+-ATPase, was normal at alkaline pH. Cytoplasmic acidification of this bacterium at alkaline pH calls for a new mechanism, probably the energy-linked K+/H+ antiport system designated Kep. A physiological role for K+/H+ antiport in pH regulation at alkaline external pH was suggested by the observation that high concentrations of ammonium ions did not inhibit the growth of E. hirae at alkaline pH (121). When the cytoplasm was made alkaline with ammonia, recovery of the normal intracellular pH of 8.0 was accompanied by K+ efflux (121). Mutants defective in Kep activity were isolated as follows. In the first step, mutants that grew well when the pH of the medium was lower than 8.0 but failed to grow at pH 9 were selected. These mutants were then screened, one by one, to identify those that had a defect in uphill K+ extrusion. All these Kep-defective mutants failed to acidify the cytoplasm in alkaline media (111). Molecular characterization of this new system is under way. Interestingly, the Kep mutants grew normally at pH near 7, indicating that the Kep K+ extrusion system is dispensable at acidic pH. In sum, the H+-ATPase is required for pH regulation in acidic media and the Kep system is required for pH regulation in alkaline ones. In medium near neutrality, neither proton-translocating pathway is required (82).
The role of the H+-ATPase in cytoplasmic pH regulation at acidic external pH has also been demonstrated in S. mutans (229). However, acidification of the cytoplasmic pH at alkaline external pH has not been reported for other streptococci, possibly reflecting the intolerance of other streptococci to highly alkaline pH (Table 1).
There has been some argument about the estimation of the cytoplasmic pH at alkaline external pH. Mugikura et al. (174) criticized the measurement of cytoplasmic pH by the use of membrane-permeable amines and proposed that neither E. coli nor enterococci acidify the cytoplasm when growing at alkaline pH. They claim that alkalinization at acidic pH is the sole mechanism of cytoplasmic pH control. The estimation of the internal pH by the use of permeable probes has always been controversial (27, 129), and we could not completely disprove the critique of its “accuracy.” However, supposing that the cytoplasmic pH of enterococci growing at alkaline external pH were the same as that of the medium, we would also have to postulate that the intracellular metabolism is uncommonly indifferent to alkalinity. This seems implausible, since streptococci can grow at pHs as high as 11 (Table 1). Regulation of the cytoplasmic pH by acidification at alkaline pH environment makes physiological sense, as described above. Incidentally, acidification of the cytoplasmic pH and generation of a reverse pH potential at alkaline pH have also been reported in a marine alga by measurements with microelectrodes (65).
Although the cytoplasmic pH is influenced by cellular metabolism and by the operation of transport systems, its regulation is thus explained mainly by chemiosmotic movements of protons through several cation transport systems. Evolution of special transport systems may be the simplest and most effective solution to the problem of regulating the cytoplasmic pH.
Circulations of K+ and Na+ and Adaptation to the Environment
The distribution of various transport systems for H+, K+, and Na+ ions described in this article is fundamental to streptococcal physiology; by these transport systems, streptococci enlarge their natural habitat, as illustrated in Fig. 12C. Cells of E. hirae flourish in K+-deficient media over a broad range of environmental pHs (from 5 to 11), even in the presence of high NaCl concentrations. At acidic external pH, the proton potential generated by the H+-ATPase, although smaller than that of aerobes, suffices to drive K+ accumulation by KtrI (K+/H+ symporter), sodium expulsion by the Na+/H+ antiporter, flagellar motion, and other proton-linked transport systems. The cytoplasmic pH is alkalinized to 7.5 to 8.0 for optimal growth by the interplay of electrogenic H+ expulsion via the H+-ATPase and electrogenic K+ influx via KtrI, but as the cytoplasmic pH rises above the pH optimum of the F-ATPase (around pH 6.5), proton translocation by the ATPase will diminish.
FIG. 12.
Circulation of H+, Na+, and K+ in bacteria. (A) E. coli. The proton current is generated by the electrogenic extrusion of protons through the respiratory chain. Protons return to the cytosol by one primary system, FOF1-ATP synthase, and several secondary systems. Among these are Trk (K+ uptake), Nha Na+/H+ antiporters (Na+ extrusion), the flagellar motor, and various proton-linked metabolite porters. Some porters are coupled to the Na+ circulation; the K+ transport systems Kup and Kef are not shown. (B) Alkaliphilic bacteria. Ion movements are similar to those of E. coli. However, flagellar motion and many secondary-transport systems are energized by Na+ circulation; K+ transport has been little studied. (C) E. hirae. At acidic pH, the proton current is generated by the FOF1-ATPase and completed by several secondary pathways that perform useful work: KtrI (K+ uptake), NapA (Na+ extrusion), the flagellar motor, and various proton-linked transport systems. At alkaline pH, there is no obvious proton circulation. Ion transport relies on ATP-driven systems including the vacuolar Na+-ATPase, Kep (K+/H+ exchange), and K+ uptake via KtrII.
High sodium concentrations pose a problem for the cells. Sodium ions leak into the cytoplasm, collapsing the membrane potential, as reported for L. lactis (21). Under these conditions, electrogenic H+ expulsion by the H+-ATPase can be accelerated. However, since the activity of the H+-ATPase itself is limited by the internal pH, a decrease in the membrane potential cannot be totally compensated. The internal pH remains constant, the membrane potential decreases, and the proton potential is limited. Na+ extrusion via the antiporter is insufficient to suppress the increase in cytoplasmic Na+, and K+ uptake via KtrI, which depends on the proton potential, comes to a halt. For all these reasons, continued growth of the cells depends on the induction of the Na+-ATPase together with the KtrII K+ uptake system; the ntp operon is responsive to an increase in the intracellular Na+ (178). These two transport systems were even expressed in wild-type E. hirae cultured at acidic pH in high Na+ medium (110, 114, 115).
Expression of the napA Na+/H+ antiporter gene is also stimulated in high-Na+ medium (238). Even mutants in which the Na+-ATPase is defective grew normally in high-Na+ medium at acidic pH (116). Mutants defective in the napA gene, which lack the Na+/H+ antiporter, also grew normally in high-Na+ medium at acidic pH, presumably thanks to the induction of the Na+-ATPase (134). Thus, these two sodium extrusion systems cope cooperatively with a decrease in the proton potential by high Na+ ions.
Growth at alkaline pH poses an even graver problem than does excess Na+. When the external pH rises above 8, the proton potential is drastically decreased. Acidification of the cytoplasmic pH to pH 8.2 or less is indispensable for cell physiology; with the influx of protons, the proton potential across the cell membrane goes to zero and in some cases becomes positive. Proton-linked transport systems, such as KtrI and the flagellar motor, cease to function: the Na+/H+ antiporter may reverse direction, letting Na+ flow inward. For the cells to regulate their cytoplasmic pH, they must accumulate K+ and use K+/H+ antiport to acidify the cytoplasm. Under laboratory culture conditions in a K+-rich medium, passive K+ uptake may suffice. However, when the pH is alkaline and the K+ concentration is limited as well, active uptake of K+ is required; that is the role of KtrII. Cell growth at alkaline pH depends on expression of the ntp operon; even in low-Na+ medium, the Na+-ATPase is amplified when the pH is high. Under certain conditions, growth of E. hirae at alkaline pH requires the inclusion of Na+ ions in the medium (115). Mutants deficient in the Na+-ATPase are easily selected by their inability to grow at alkaline pH (116). When the K+ content of the medium is low (2 mM or less) and the pH is alkaline, KtrII provides the only pathway for the efficient uptake of K+ (176). In the presence of more K+ (10 mM or more), another K+ transport system comes into play; this may depend on the membrane potential but has not been studied (135, 176).
Most of the other streptococci do not tolerate extremely alkaline pH, where these cation pumps are important for cell physiology. Information on cation transport systems in other streptococci is limited, but the circulations of K+ and Na+ are probably linked with proton current.
Is Cation Circulation Obligatory for Growth?
With the proton circulation playing so many roles in the bacterial energy economy, one might expect it to be an obligatory feature. This is not always the case. More than two decades ago, Harold and Van Brunt (82) showed that enterococci can grow in the presence of the powerful cation conductor gramicidin D, which makes the cell membrane permeable to all small monovalent cations including H+, Na+, and K+. In the presence of gramicidin D, the electrochemical potentials for all these ions go to zero, although cations still accumulate nonselectively in the cells due to their content of fixed anionic substances, such as nucleic acids. It follows that the circulation of H+, Na+, and K+ is dispensable for producing the architecture of the bacterial cell. However, growth in the presence of gramicidin D requires particular nutritional circumstances. The cells require a medium of high K+ concentration and low Na+ concentration; the cytoplasmic pH must not be far from 7; and high concentrations of extracellular nutrients such as amino acids are needed. In other words, the maintenance of a high cytoplasmic K+ level and of a slightly alkaline cytoplasmic pH would be essential to bacterial growth; the key functions of these properties are not particularly clear. The effect of ionophores on cell growth has not been extended to other streptococci. The effect of a protonophore, CCCP, on the growth of E. coli supports the conclusion that bacteria can grow in the absence of the proton potential when glucose is used as an energy source (137, 174). It is not clear how cells in the presence of CCCP secrete proteins into the periplasm, which depends on the proton potential (at least in E. coli). A variety of drug resistance systems are distributed in bacteria (149); some kinds of drug extrusion pumps have been found in L. lactis. The effectiveness of various drugs on growing cells must be considered with care, although there is so far no discrepancy in the interpretation of the effect of ionophores on enterococci.
All these things probably tell us something fundamental about cation transport. Cells require the proton/cation circulation to keep their internal environment constant and different from the external one. In the laboratory, we can arrange matters so that this function is dispensable; however, in the natural world it is essential. A diversity of cation transport systems allows bacteria to maintain homeostasis in the face of a changing external environment and affords them a broader habitat.
CONCLUDING REMARKS
The essence of homeostasis in bacteria has been quite elegantly summarized by Harold in his book The Vital Force: a Study of Bioenergetics (74). “The internal environment of microorganisms, like that of other living cells, generally differs substantially from the external one in chemical composition and in physical properties. It also remains remarkably constant in the face of external fluctuations. Bacteria, unlike higher organisms, can often survive gross changes in cytosolic composition, but they reestablish the normal one before growth resumes. The reasons are simple enough. The cytoplasmic pH should not be too far from neutrality, which is optimal for most metabolism. K+ is always the predominant cytoplasmic cation, while the more abundant Na+ is excluded, although why that is so is not altogether clear. Furthermore, a number of physiological parameters must be subject to regulatory mechanisms that counter perturbations and maintain optimal values. These include the proton potential, the cytosolic pH, the phosphorylation potential of ATP, the adenine nucleotide energy change, the redox potential, and turgor pressure which supplies the driving force for wall expansion. Homeostasis is obviously an important use of biological energy, and it illustrates the interplay of the proton circulation with other modes of energy generation.”
There should be a relationship between the ion species of the energy transfer reactions found in any given organism and its habitat or metabolic economy (Fig. 12). In aerobic bacteria such as E. coli, which establish a large proton potential by the respiratory chain, H+ serves as the coupling ion for membrane energy transduction. K+ uptake by the Trk system, Na+ extrusion by the Nha systems, and a variety of nutrient carriers are linked to H+ (Fig. 12A). In alkaliphilic and halphilic aerobes, Na+ serves as a second important coupling ion for energy transduction (74, 228); the choice of Na+ in addition to H+ as the coupling ion is sensible in their environment. The Na+ gradient, generated by a combination of H+-translocating electron transport and Na+/H+ antiporter or Na+-translocating electron transport in some marine bacteria (253), is used as the driving force for various kinds of nutrient uptake and for flagellar motion (Fig. 12B). A gradient of H+ or Na+ is thus utilized as the driving force for various secondary porters in most bacteria.
The same is true in enterococci and in other streptococci growing at a pH that is either neutral or acidic (Fig. 12C). However, growing streptococci maintain a significantly lower proton potential than do growing anaerobic bacteria. Some streptococci, such as enterococci, can grow at alkaline pH where the proton potential is negligible. A number of ATP-linked cation transport systems, including the Kep K+ transport system and the Na+-translocating vacuolar ATPase, unique to streptococci, surely contribute to this ability. When growing at alkaline pH, streptococci also generate a sodium electrochemical potential large enough to power secondary systems; whether they use it for that purpose is not clear. The information available is limited, and it seems premature to draw conclusions about the role of Na+ as a general coupling ion. The functions of the Na+ circulation will become clear when more is known of the distribution of sodium-linked transport systems among streptococci.
The molecular analysis of membrane transport proteins in streptococci is just beginning, with some progress being made on the genetic level. When the genome sequence of streptococci is fully determined, we shall be able to look at the evolution of cation transport systems discovered in this bacterium and perhaps find how the various primary and secondary systems are distributed and why.
ACKNOWLEDGMENTS
I first acknowledge the remarkable contribution of F. M. Harold and his colleagues, who pioneered the study of ion transport in streptococci and linked it to Peter Mitchell’s insights into chemiosmotic energy transduction. I am personally grateful to F.M.H. for his advice and support and for his help in preparing this review for publication. I thank H. Tokuda and R. Hirata for critical reading of the manuscript.
The work in my laboratory described in this review was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Culture and Sports, Japan, and also by the Hamaguchi Biochemistry Foundation, the Salt Science Research Foundation and the Takeda Science Foundation.
REFERENCES
- 1.Abe T, Ihara K, Mukohata Y. Abstracts of the 17th Annual Meeting of the Japan Bioenergetics Group. 1991. NO3− inhibits the A-type ATPase, but not the ATP synthase in Halobacteria; pp. 78–79. [Google Scholar]
- 2.Abrahams J P, Leslie A G, Lutter R, Walker J E. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 3.Abrams A. The proton-translocating membrane ATPase (F1F0) in Streptococcus faecalis. In: Martonosi A N, editor. Enzymes in biological membranes. 2nd ed. Vol. 4. New York, N.Y: Plenum Press; 1985. pp. 177–193. [Google Scholar]
- 4.Abrams A, Baron C. The isolation and subunit structures of streptococcal membrane adenosine triphosphatase. Biochemistry. 1967;6:225–229. doi: 10.1021/bi00853a035. [DOI] [PubMed] [Google Scholar]
- 5.Abrams A, Jensen C. Altered expression of the H+ ATPase in Streptococcus faecalis membranes. Biochem Biophys Res Commun. 1984;122:151–157. doi: 10.1016/0006-291x(84)90452-2. [DOI] [PubMed] [Google Scholar]
- 6.Abrams A, Smith J B. Increased membrane ATPase and K+ transport rates in Streptococcus faecalis induced by K+ restriction during growth. Biochem Biophys Res Commun. 1971;44:1488–1495. doi: 10.1016/s0006-291x(71)80254-1. [DOI] [PubMed] [Google Scholar]
- 7.Abrams A, Smith J B. Bacterial membrane ATPase. Enzymes. 1974;10:395–429. [Google Scholar]
- 8.Abrams A, Leimgruber R M. The N,N′-dicyclohexylcarbodiimide-sensitive ATPase in Streptococcus faecalis membranes. In: Martonosi A N, editor. Membranes and transport. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 465–471. [Google Scholar]
- 9.Abrams A, McNamara P, Johnson F B. Adenosine triphosphatase in isolated bacterial cell membranes. J Biol Chem. 1960;235:3659–3662. [PubMed] [Google Scholar]
- 10.Abrams A, Jensen C, Morris D. Studies of substructure and tightly bound nucleotide in bacterial membrane ATPase. J Supramol Struct. 1975;3:261–274. doi: 10.1002/jss.400030309. [DOI] [PubMed] [Google Scholar]
- 11.Abrams A, Jensen C, Morris D. Role of Mg2+ ions in the subunit structure and membrane binding properties of bacterial energy transducing ATPase. Biochem Biophys Res Commun. 1976;69:804–811. doi: 10.1016/0006-291x(76)90946-3. [DOI] [PubMed] [Google Scholar]
- 12.Abrams A, Morris D, Jensen C. Chymotryptic conversion of bacterial membrane ATPase to an active form with modified α chains and defective membrane binding properties. Biochemistry. 1976;15:5560–5566. doi: 10.1021/bi00670a021. [DOI] [PubMed] [Google Scholar]
- 13.Ambudkar S V, Lynn A R, Maloney P C, Rosen B P. Reconstitution of ATP-dependent calcium transport from streptococci. J Biol Chem. 1986;261:15596–15600. [PubMed] [Google Scholar]
- 14.Anraku Y. Structure and function of the yeast vacuolar membrane H+-ATPase. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Science B.V.; 1996. pp. 93–109. [Google Scholar]
- 15.Apell H J, Solioz M. Electrogenic transport by the Enterococcus hirae ATPase. Biochim Biophys Acta. 1990;1017:221–228. doi: 10.1016/0005-2728(90)90188-a. [DOI] [PubMed] [Google Scholar]
- 16.Asghar S S, Levin E, Harold F M. Accumulation of neutral amino acids by Streptococcus faecalis. Energy coupling by a proton-motive force. J Biol Chem. 1973;248:5225–5233. [PubMed] [Google Scholar]
- 17.Bakker E P. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press, Inc.; 1993. [Google Scholar]
- 18.Bakker E P, Harold F M. Energy coupling to potassium transport in Streptococcus faecalis: interplay of ATP and the protonmotive force. J Biol Chem. 1980;255:433–440. [PubMed] [Google Scholar]
- 19.Bakker E P, Mangerich W E. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J Bacteriol. 1981;147:820–826. doi: 10.1128/jb.147.3.820-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakker E P, Booth I R, Dinnbier U, Epstein W, Gajewski A. Evidence for multiple K+ export systems in Escherichia coli. J Bacteriol. 1987;169:3743–3749. doi: 10.1128/jb.169.8.3743-3749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barker S L, Kashket E R. Effects of sodium ions on the electrical and pH gradients across the membrane of Streptococcus lactis cells. J Supramol Struct. 1977;6:383–388. doi: 10.1002/jss.400060311. [DOI] [PubMed] [Google Scholar]
- 22.Bauer P D, Trapp C, Drake D, Taylor K G, Doyle R J. Acquisition of manganous ions by mutans group streptococci. J Bacteriol. 1993;175:819–825. doi: 10.1128/jb.175.3.819-825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becher B, Müller V. ΔμNa+ drives the synthesis of ATP via an ΔμNa+-translocating F1F0-ATP synthase in membrane vesicles of the archaeon Methanosarcina mazei Gö1. J Bacteriol. 1994;176:2543–2550. doi: 10.1128/jb.176.9.2543-2550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender G R, Sutton S V, Marquis R E. Acid tolerance, proton permeabilities and membrane ATPases of oral streptococci. Infect Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benyoucef M, Rigaud J L, Leblanc G. Cation transport mechanisms in Mycoplasma mycoides var. Capri cells. The nature of the link between K+ and Na+ transport. Biochem J. 1982;208:539–547. doi: 10.1042/bj2080539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair A, Ngo L, Park J, Paulsen I T, Saier M H., Jr Phylogenetic analyses of the homologous transmembrane channel-forming proteins of the F0F1-ATPases of bacteria, chloroplasts and mitochondria. Microbiology. 1996;142:17–32. doi: 10.1099/13500872-142-1-17. [DOI] [PubMed] [Google Scholar]
- 27.Booth I R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman E J, Siebers A, Altendorf K. Bafilomycin: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer P D. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 30.Boyer P D, Kohlbrener W E. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 31.Bridge P D, Sneath P H A. Numerical taxonomy of Streptococcus. J Gen Microbiol. 1983;129:565–597. doi: 10.1099/00221287-129-3-565. [DOI] [PubMed] [Google Scholar]
- 32.Brown N L, Camakaris J, Lee B T, Williams T, Morby A P, Parkhill J, Rouch D A. Bacterial resistances to mercury and copper. J Cell Biochem. 1991;46:106–114. doi: 10.1002/jcb.240460204. [DOI] [PubMed] [Google Scholar]
- 33.Bryan-Jones D B, Whittenbury R. Haematin-dependent oxidative phosphorylation in Streptococcus faecalis. J Gen Microbiol. 1969;58:247–260. doi: 10.1099/00221287-58-2-247. [DOI] [PubMed] [Google Scholar]
- 34.Bull P C, Thomas G R, Rommens J M, Forbes J R, Cox D W. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 35.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 36.Capaldi R A, Aggeler R, Turina P, Wilkens S. Coupling between catalytic sites and the proton channel in F1F0-type ATPases. Trends Biochem Sci. 1994;19:284–289. doi: 10.1016/0968-0004(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 37.Carafoli E. Intracellular calcium homeostasis. Annu Rev Biochem. 1987;56:395–433. doi: 10.1146/annurev.bi.56.070187.002143. [DOI] [PubMed] [Google Scholar]
- 38.Cegielska A, Georgopoulos C. Functional domains of the Escherichia coli dnaK heat shock protein as revealed by mutational analysis. J Biol Chem. 1989;264:21122–21130. [PubMed] [Google Scholar]
- 39.Cervantes C, Gutierrez-Corona F. Copper resistance mechanisms in bacteria and fungi. FEMS Microbiol Rev. 1994;14:121–137. doi: 10.1111/j.1574-6976.1994.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 40.Cha J S, Cooksey D A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dashper S G, Reynolds E C. pH regulation by Streptococcus mutans. J Dent Res. 1992;71:1159–1165. doi: 10.1177/00220345920710050601. [DOI] [PubMed] [Google Scholar]
- 43.Denda K, Konishi J, Hajiro K, Oshima T, Date T, Yoshida M. Structure of an ATPase operon of an acidothermophilic archaebacterium, Sulfolobus acidocaldarius. J Biol Chem. 1990;265:21509–21513. [PubMed] [Google Scholar]
- 44.Denda K, Konishi J, Oshima T, Date T, Yoshida M. A gene encoding the proteolipid subunit of Sulfolobus acidocaldarius ATPase complex. J Biol Chem. 1989;264:7119–7121. [PubMed] [Google Scholar]
- 45.Dimroth P. Sodium ion transport decarboxylases and other aspects of sodium ion cycling in bacteria. Microbiol Rev. 1987;51:320–340. doi: 10.1128/mr.51.3.320-340.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Douglas R M, Roberts J A, Munro A W, Ritchie G Y, Lamb A J, Booth I R. The distribution of homologues of the Escherichia coli KefC K+-efflux system in other bacterial species. J Gen Microbiol. 1991;137:1999–2005. doi: 10.1099/00221287-137-8-1999. [DOI] [PubMed] [Google Scholar]
- 47.Driessen A J, Konings W N. Calcium transport in membrane vesicles of Streptococcus cremoris. Eur J Biochem. 1986;159:149–155. doi: 10.1111/j.1432-1033.1986.tb09845.x. [DOI] [PubMed] [Google Scholar]
- 48.Driessen A J, Smid E J, Konings W N. Transport of diamines by Enterococcus faecalis is mediated by an agmatine-putrescine antiporter. J Bacteriol. 1988;170:4522–4527. doi: 10.1128/jb.170.10.4522-4527.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ennis H L, Sells B H. Breakdown and re-formation of polysomes in Escherichia coli during inhibition of protein synthesis. Biochim Biophys Acta. 1968;161:503–508. doi: 10.1016/0005-2787(68)90126-3. [DOI] [PubMed] [Google Scholar]
- 51.Epstein W, Burrman E, McLaggan D, Naprstek J. Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem Soc Trans. 1993;21:1006–1010. doi: 10.1042/bst0211006. [DOI] [PubMed] [Google Scholar]
- 52.Evans D J., Jr Membrane adenosine triphosphatase of Escherichia coli: activation by calcium ion and inhibition by monovalent cations. J Bacteriol. 1969;100:914–922. doi: 10.1128/jb.100.2.914-922.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans S L, Arceneaux J E, Byers B R, Martin M E, Aranha H. Ferrous iron transport in Streptococcus mutans. J Bacteriol. 1986;168:1096–1099. doi: 10.1128/jb.168.3.1096-1099.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng Y, Forgac M. Cysteine 254 of the 73-kDa A subunit is responsible for inhibition of the coated vesicle H+-ATPase upon modification by sulfhydryl reagents. J Biol Chem. 1992;267:5817–5822. [PubMed] [Google Scholar]
- 55.Fenoll A, Muñoz R, García E, de la Campa A G. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the F0 complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol Microbiol. 1994;12:587–598. doi: 10.1111/j.1365-2958.1994.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 56.Ferguson G P, Munro A W, Douglas R M, McLaggan D, Booth I R. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol Microbiol. 1993;9:1297–1303. doi: 10.1111/j.1365-2958.1993.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 57.Fillingame R H. Molecular mechanics of ATP synthesis by F1F0-type H+-translocating ATP synthases. In: Krulwich T A, editor. The bacteria: a treatise on structure and function. New York, N.Y: Academic Press, Inc.; 1990. pp. 345–391. [Google Scholar]
- 58.Forgac M. Structure and function of vacuolar class of ATP-driven proton pumps. Physiol Rev. 1989;69:765–796. doi: 10.1152/physrev.1989.69.3.765. [DOI] [PubMed] [Google Scholar]
- 59.Futai M, Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): biochemical and molecular biological approaches. Microbiol Rev. 1983;47:285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Futai M, Noumi T, Maeda M. ATP synthase (H+-ATPase): results by combined biochemical and molecular biological approaches. Annu Rev Biochem. 1989;58:111–136. doi: 10.1146/annurev.bi.58.070189.000551. [DOI] [PubMed] [Google Scholar]
- 61.Gale E F. Factors influencing the enzymic activities of bacteria. Bacteriol Rev. 1943;7:139–173. doi: 10.1128/br.7.3.139-173.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garvin M E, Fillingame R H. Helical structure and folding of subunit c of F1F0 ATP synthase: 1H NMR resonance assignments and NOE analysis. Biochemistry. 1993;32:12167–12177. doi: 10.1021/bi00096a029. [DOI] [PubMed] [Google Scholar]
- 63.Gay N J. Construction and characterization of an Escherichia coli strain with a uncI mutation. J Bacteriol. 1984;158:820–825. doi: 10.1128/jb.158.3.820-825.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge Z, Hiratsuka K, Taylor D E. Nucleotide sequence and mutational analysis indicate that two Helicobacter pylori genes encode a P-type ATPase and a cation-binding protein associated with copper transport. Mol Microbiol. 1995;15:97–106. doi: 10.1111/j.1365-2958.1995.tb02224.x. [DOI] [PubMed] [Google Scholar]
- 65.Gibbon B, Kropf D L. Intracellular pH and its regulation in Pelvetia zygotes. Dev Biol. 1993;157:259–268. doi: 10.1006/dbio.1993.1130. [DOI] [PubMed] [Google Scholar]
- 66.Gogarten J P, Kibak H, Dittrich P, Taiz L, Bowman E J, Bowman B J, Manolson M F, Poole R J, Date T, Oshima T, Yoshida M. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Graham A F, Lund B M. The effect of alkaline pH on growth and metabolic products of a motile, yellow-pigmented Streptococcus sp. J Gen Microbiol. 1983;129:2429–2435. [Google Scholar]
- 68.Gunsalus I C, Niven C F. The effect of pH on the lactic acid fermentation. J Biol Chem. 1942;145:131–136. [Google Scholar]
- 69.Hagting A, Kunji E R, Leenhouts K J, Poolman B, Konings W N. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem. 1994;269:11391–11399. [PubMed] [Google Scholar]
- 70.Hardie J M. Genus Streptococcus. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1043–1071. [Google Scholar]
- 71.Harold F M. Conservation and transformation of energy by bacterial membranes. Bacteriol Rev. 1972;36:172–230. doi: 10.1128/br.36.2.172-230.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harold F M. Membrane and energy transduction in bacteria. Curr Top Bioenerg. 1977;6:84–151. [Google Scholar]
- 73.Harold F M. Ion currents and physiological functions in microorganisms. Annu Rev Microbiol. 1977;31:181–203. doi: 10.1146/annurev.mi.31.100177.001145. [DOI] [PubMed] [Google Scholar]
- 74.Harold F M. The vital force: a study of bioenergetics. W. H. New York, N.Y: Freeman & Co.; 1986. [Google Scholar]
- 75.Harold F M, Altendorf K. Cation transport in bacteria: K+, Na+, and H+ Curr Top Membr Transp. 1974;5:1–50. [Google Scholar]
- 76.Harold F M, Baarda J R. Inhibition of potassium transport by sodium in a mutant of Streptococcus faecalis. Biochemistry. 1967;6:3107–3110. doi: 10.1021/bi00862a018. [DOI] [PubMed] [Google Scholar]
- 77.Harold F M, Kakinuma Y. Primary and secondary transport of cations in bacteria. Ann N Y Acad Sci. 1985;456:375–383. doi: 10.1111/j.1749-6632.1985.tb14888.x. [DOI] [PubMed] [Google Scholar]
- 78.Harold F M, Maloney P. Energy transduction by ion currents. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 283–306. [Google Scholar]
- 79.Harold F M, Papineau D. Cation transport and electrogenesis in Streptococcus faecalis. I. The membrane-potential. J Membr Biol. 1972;8:27–44. doi: 10.1007/BF01868093. [DOI] [PubMed] [Google Scholar]
- 80.Harold F M, Papineau D. Cation transport and electrogenesis in Streptococcus faecalis. II. Proton and sodium extrusion. J Membr Biol. 1972;8:45–62. doi: 10.1007/BF01868094. [DOI] [PubMed] [Google Scholar]
- 81.Harold F M, Spitz E. Accumulation of arsenate, phosphate and aspartate by Streptococcus faecalis. J Bacteriol. 1975;122:266–277. doi: 10.1128/jb.122.1.266-277.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harold F M, Van Brunt J. Circulation of H+ and K+ across the plasma membrane is not obligatory for bacterial growth. Science. 1977;197:372–373. doi: 10.1126/science.69317. [DOI] [PubMed] [Google Scholar]
- 83.Harold F M, Baarda J R, Pavlasova E. Extrusion of sodium and hydrogen ions as the primary process in potassium ion accumulation by Streptococcus faecalis. J Bacteriol. 1970;101:152–159. doi: 10.1128/jb.101.1.152-159.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harold F M, Pavlasova E, Baarda J R. A transmembrane pH gradient in Streptococcus faecalis: origin, and dissipation by proton conductors and N,N′-dicyclohexylcarbodimide. Biochim Biophys Acta. 1970;196:235–244. doi: 10.1016/0005-2736(70)90011-8. [DOI] [PubMed] [Google Scholar]
- 85.Harold F M, Baarda J R, Baron C, Abrams A. Dio 9 and chlorhexidine: inhibitors of membrane-bound ATPase and of cation transport in Streptococcus faecalis. Biochim Biophys Acta. 1969;183:129–136. doi: 10.1016/0005-2736(69)90136-9. [DOI] [PubMed] [Google Scholar]
- 86.Harold F M, Baarda J R, Baron C, Abrams A. Inhibition of membrane-bound adenosine triphosphatase and of cation transport in Streptococcus faecalis by N,N′-dicyclohexylcarbodiimide. J Biol Chem. 1969;244:2261–2268. [PubMed] [Google Scholar]
- 87.Harold F M, Harold R L, Baarda J R, Abrams A. A genetic defect in retention of potassium by Streptococcus faecalis. Biochemistry. 1967;6:1777–1784. doi: 10.1021/bi00858a028. [DOI] [PubMed] [Google Scholar]
- 88.Heefner D L. Transport of H+, K+, Na+ and Ca++ in Streptococcus. Mol Cell Biochem. 1982;44:81–106. doi: 10.1007/BF00226893. [DOI] [PubMed] [Google Scholar]
- 89.Heefner D L, Harold F M. ATP-linked sodium transport in Streptococcus faecalis. I. The sodium circulation. J Biol Chem. 1980;255:11396–11402. [PubMed] [Google Scholar]
- 90.Heefner D L, Harold F M. ATP-driven sodium pump in Streptococcus faecalis. Proc Natl Acad Sci USA. 1982;79:2798–2802. doi: 10.1073/pnas.79.9.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heefner D L, Kobayashi H, Harold F M. ATP-linked sodium transport in Streptococcus faecalis. II. Energy coupling in everted membrane vesicles. J Biol Chem. 1980;255:11403–11407. [PubMed] [Google Scholar]
- 92.Hilario E, Gogarten J P. Horizontal transfer of ATPase genes-the tree of life becomes a net of life. Biosystems. 1993;31:111–119. doi: 10.1016/0303-2647(93)90038-e. [DOI] [PubMed] [Google Scholar]
- 93.Hillier A J, Jericho R E, Green S M, Jago G R. Properties and function of fumarate reductase (NADH) in Streptococcus lactis. Aust J Biol Sci. 1979;32:625–635. doi: 10.1071/bi9790625. [DOI] [PubMed] [Google Scholar]
- 94.Hilpert W, Schink B, Dimroth P. Life by a new decarboxylation-dependent energy conservation mechanism with Na+ as coupling ion. EMBO J. 1984;3:1665–1670. doi: 10.1002/j.1460-2075.1984.tb02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hirata R, Graham L A, Takatsuki A, Stevens T H, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 96.Hirota N, Imae Y. Na+-driven flagellar motors of alkalophilic Bacillus strain YN-1. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 97.Honer zu Bentrup K, Ubbink-Kok T, Lolkema J S, Konings W N. An Na+-pumping V1V0-ATPase complex in the thermophilic bacterium Clostridium fervidus. J Bacteriol. 1997;179:1274–1279. doi: 10.1128/jb.179.4.1274-1279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Houng H S, Lynn A R, Rosen B P. ATP-driven calcium transport in membrane vesicles of Streptococcus sanguis. J Bacteriol. 1986;168:1040–1044. doi: 10.1128/jb.168.2.1040-1044.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsung J C, Haug A. Intracellular pH of Thermoplasma acidophila. Biochim Biophys Acta. 1975;389:477–482. doi: 10.1016/0005-2736(75)90158-3. [DOI] [PubMed] [Google Scholar]
- 100.Hugentobler G, Heid I, Solioz M. Purification of a putative K+-ATPase from Streptococcus faecalis. J Biol Chem. 1983;258:7611–7617. [PubMed] [Google Scholar]
- 101.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 102.Inoue C, Sugawara K, Shiratori T, Kusano T, Kitagawa Y. Nucleotide sequence of the Thiobacillus ferroxidans chromosomal gene encoding mercuric reductase. Gene. 1989;84:47–54. doi: 10.1016/0378-1119(89)90138-8. [DOI] [PubMed] [Google Scholar]
- 103.Ivey D M, Guffanti A A, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich T A. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- 104.Jinks D C, Silvius J R, McElhaney R N. Physiological role and membrane lipid modulation of the membrane-bound (Mg2+, Na+)-adenosine triphosphatase activity in Acholeplasma laidlawii. J Bacteriol. 1978;136:1027–1036. doi: 10.1128/jb.136.3.1027-1036.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kagawa Y. Proton motive ATP synthase. In: Ernster L, editor. Bioenergetics. Amsterdam, The Netherlands: Elsevier Science Publishers BV; 1984. pp. 149–186. [Google Scholar]
- 106.Kaim G, Dimroth P. Formation of a functionally active sodium-translocating hybrid F1F0 ATPase in Escherichia coli by homologous recombination. Eur J Biochem. 1993;218:937–944. doi: 10.1111/j.1432-1033.1993.tb18450.x. [DOI] [PubMed] [Google Scholar]
- 107.Kaim G, Dimroth P. A double mutation in subunit c of the Na+-specific F1F0-ATPase of Propionigenium modestum results in a switch from Na+ to H+-coupled ATP synthesis in the Escherichia coli host cells. J Mol Biol. 1995;253:726–738. doi: 10.1006/jmbi.1995.0586. [DOI] [PubMed] [Google Scholar]
- 108.Kakinuma Y. Sodium/proton antiporter in Streptococcus faecalis. J Bacteriol. 1987;169:3886–3890. doi: 10.1128/jb.169.9.3886-3890.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kakinuma Y. Lowering of cytoplasmic pH is essential for growth of Streptococcus faecalis at high pH. J Bacteriol. 1987;169:4403–4405. doi: 10.1128/jb.169.9.4403-4405.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kakinuma Y. K+ transport in Enterococcus hirae. In: Bakker E P, editor. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 277–290. [Google Scholar]
- 111.Kakinuma, Y. Unpublished data.
- 112.Kakinuma Y, Harold F M. ATP-driven exchange of Na+ and K+ ions by Streptococcus faecalis. J Biol Chem. 1985;260:2086–2091. [PubMed] [Google Scholar]
- 113.Kakinuma Y, Igarashi K. Active potassium extrusion regulated by intracellular pH in Streptococcus faecalis. J Biol Chem. 1988;263:14166–14170. [PubMed] [Google Scholar]
- 114.Kakinuma Y, Igarashi K. Sodium-translocating adenosine triphosphatase in Streptococcus faecalis. J Bioenerg Biomembr. 1989;21:679–692. doi: 10.1007/BF00762686. [DOI] [PubMed] [Google Scholar]
- 115.Kakinuma Y, Igarashi K. Amplification of the Na+-ATPase of Streptococcus faecalis at alkaline pH. FEBS Lett. 1990;261:135–139. doi: 10.1016/0014-5793(90)80654-2. [DOI] [PubMed] [Google Scholar]
- 116.Kakinuma Y, Igarashi K. Mutants of Streptococcus faecalis sensitive to alkaline pH lack Na+-ATPase. J Bacteriol. 1990;172:1732–1735. doi: 10.1128/jb.172.4.1732-1735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kakinuma Y, Igarashi K. Some features of the Streptococcus faecalis Na+-ATPase resemble those of the vacuolar-type ATPases. FEBS Lett. 1990;271:97–101. doi: 10.1016/0014-5793(90)80381-r. [DOI] [PubMed] [Google Scholar]
- 118.Kakinuma Y, Igarashi K. Release of the component of Streptococcus faecalis Na+-ATPase from the membranes. FEBS Lett. 1990;271:102–105. doi: 10.1016/0014-5793(90)80382-s. [DOI] [PubMed] [Google Scholar]
- 119.Kakinuma Y, Igarashi K. Purification and characterization of the catalytic moiety of vacuolar-type Na+-ATPase from Enterococcus hirae. J Biochem. 1994;116:1302–1308. doi: 10.1093/oxfordjournals.jbchem.a124679. [DOI] [PubMed] [Google Scholar]
- 120.Kakinuma Y, Igarashi K. Electrogenic Na+ transport by Enterococcus hirae Na+-ATPase. FEBS Lett. 1995;359:255–258. doi: 10.1016/0014-5793(95)00056-f. [DOI] [PubMed] [Google Scholar]
- 121.Kakinuma Y, Igarashi K. Potassium/proton antiport system of Enterococcus hirae growing at high pH. J Bacteriol. 1995;177:2227–2229. doi: 10.1128/jb.177.8.2227-2229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kakinuma Y, Unemoto T. Sucrose uptake is driven by the Na+ electrochemical potential in the marine bacterium Vibrio alginolyticus. J Bacteriol. 1985;163:1293–1295. doi: 10.1128/jb.163.3.1293-1295.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kakinuma Y, Igarashi K, Konishi K, Yamato I. Primary structure of the α subunit of vacuolar-type Na+-ATPase in Enterococcus hirae: amplification of an 1,000 bp fragment by polymerase chain reaction. FEBS Lett. 1991;292:64–68. doi: 10.1016/0014-5793(91)80835-q. [DOI] [PubMed] [Google Scholar]
- 124.Kakinuma, Y., M. Ikegami, M. Kawano, and K. Igarashi. Unpublished data.
- 125.Kakinuma Y, Kakinuma S, Takase K, Konishi K, Igarashi K, Yamato I. A gene encoding the 16-kDa proteolipid subunit of Enterococcus hirae Na+-ATPase complex. Biochem Biophys Res Commun. 1993;195:1063–1069. doi: 10.1006/bbrc.1993.2152. [DOI] [PubMed] [Google Scholar]
- 126.Kanamaru K, Kashiwagi S, Mizuno T. A copper-transporting P-type ATPase found in the thylakoid membrane of the cyanobacterium Synechococcus species PCC7942. Mol Microbiol. 1994;13:369–377. doi: 10.1111/j.1365-2958.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 127.Kashket E R. Active transport of thallous ions by Streptococcus lactis. J Biol Chem. 1979;254:8129–8131. [PubMed] [Google Scholar]
- 128.Kashket E R. Proton motive force in growing Streptococcus lactis and Staphylococcus aureus cells under aerobic and anaerobic conditions. J Bacteriol. 1981;146:369–376. doi: 10.1128/jb.146.1.369-376.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kashket E R. The proton motive force in bacteria: a critical assessment of methods. Annu Rev Microbiol. 1985;39:219–242. doi: 10.1146/annurev.mi.39.100185.001251. [DOI] [PubMed] [Google Scholar]
- 130.Kashket E R. Effects of K+ and Na+ on the proton motive force of respiring Escherichia coli at alkaline pH. J Bacteriol. 1985;163:423–429. doi: 10.1128/jb.163.2.423-429.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kashket E R, Barker S L. Effects of potassium ions on the electrical and pH gradients across the membrane of Streptococcus lactis cells. J Bacteriol. 1977;130:1017–1023. doi: 10.1128/jb.130.3.1017-1023.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kashket E R, Blanchard A G, Metzger W C. Proton motive force during growth of Streptococcus lactis cells. J Bacteriol. 1980;143:128–134. doi: 10.1128/jb.143.1.128-134.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaso V N, Boyer P D. Vacuolar ATPases, like F1,F0-ATPases, show a strong dependence of the reaction velocity on the binding of more than one ATP per enzyme. Proc Natl Acad Sci USA. 1989;86:8708–8711. doi: 10.1073/pnas.86.22.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kawano, M., K. Igarashi, and Y. Kakinuma. Submitted for publication.
- 135.Kawano, M., K. Igarashi, and Y. Kakinuma. Unpublished data.
- 136.Kinoshita N, Unemoto T, Kobayashi H. Sodium-stimulated ATPase in Streptococcus faecalis. J Bacteriol. 1984;158:844–848. doi: 10.1128/jb.158.3.844-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kinoshita N, Unemoto T, Kobayashi H. Proton motive force is not obligatory for growth of Escherichia coli. J Bacteriol. 1984;160:1071–1077. doi: 10.1128/jb.160.3.1074-1077.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kluge C, Dimroth P. Specific protection by Na+ or Li+ of the F1F0-ATPase of Propionigenium modestum from the reaction with dicyclohexylcarbodiimide. J Biol Chem. 1993;268:14557–14560. [PubMed] [Google Scholar]
- 139.Kobayashi H. Second system for potassium transport in Streptococcus faecalis. J Bacteriol. 1982;150:506–511. doi: 10.1128/jb.150.2.506-511.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kobayashi H. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J Biol Chem. 1985;260:72–76. [PubMed] [Google Scholar]
- 141.Kobayashi H. Regulation of cytoplasmic pH in streptococci. In: Reizer J, Peterkofsky A, editors. Sugar transport and metabolism in gram-positive bacteria. Chichester, England: Ellis Horwood Ltd.; 1987. pp. 255–269. [Google Scholar]
- 142.Kobayashi, H. Personal communication.
- 143.Kobayashi H, Unemoto T. Streptococcus faecalis mutants defective in regulation of cytoplasmic pH. J Bacteriol. 1980;143:1187–1193. doi: 10.1128/jb.143.3.1187-1193.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kobayashi H, Murakami N, Unemoto T. Regulation of the cytoplasmic pH in Streptococcus faecalis. J Biol Chem. 1982;257:13246–13252. [PubMed] [Google Scholar]
- 145.Kobayashi H, Suzuki T, Unemoto T. Streptococcal cytoplasmic pH is regulated by changes in amount and activity of a proton-translocating ATPase. J Biol Chem. 1986;261:627–630. [PubMed] [Google Scholar]
- 146.Kobayashi H, Van Brunt J, Harold F M. ATP-Iinked calcium transport in cells and membrane vesicles of Streptococcus faecalis. J Biol Chem. 1978;253:2085–2092. [PubMed] [Google Scholar]
- 147.Kobayashi H, Suzuki T, Kinoshita N, Unemoto T. Amplification of the Streptococcus faecalis proton-translocating ATPase by a decrease in cytoplasmic pH. J Bacteriol. 1984;158:1157–1160. doi: 10.1128/jb.158.3.1157-1160.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kocherginskaia S A, Shakhparonov M I, Aldanova N A, Modianov N N. Primary structure of the dicyclohexylcarbodiimide-binding subunit of Streptococcus faecalis H+-ATPase. Bioorg Khim. 1983;9:746–755. [PubMed] [Google Scholar]
- 149.Konings W N, Lolkema J S, Bolhuis H, van Veen H W, Poolman B, Driessen A J. The role of transport processes in survival of lactic acid bacteria. Energy transduction and multidrug resistance. Antonie Leeuwenhoek. 1997;71:117–128. doi: 10.1023/a:1000143525601. [DOI] [PubMed] [Google Scholar]
- 150.Kropf D L. Establishment and expression of cellular polarity in fucoid zygotes. Microbiol Rev. 1992;56:316–339. doi: 10.1128/mr.56.2.316-339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Krulwich T A, Guffanti A A. Physiology of acidophilic and alkalophilic bacteria. Adv Microbiol Physiol. 1983;24:173–214. doi: 10.1016/s0065-2911(08)60386-0. [DOI] [PubMed] [Google Scholar]
- 152.Krulwich T A, Cheng J, Guffanti A A. The role of monovalent cation/proton antiporters in Na+-resistance and pH homeostasis in Bacillus: an alkaliphile versus a neutralophile. J Exp Biol. 1994;196:457–470. doi: 10.1242/jeb.196.1.457. [DOI] [PubMed] [Google Scholar]
- 153.Laimins L A, Rhoads D B, Epstein W. Osmotic control of kdp operon expression in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:464–468. doi: 10.1073/pnas.78.1.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lanyi J K. The role of Na+ in transport processes of bacterial membranes. Biochim Biophys Acta. 1979;559:377–397. doi: 10.1016/0304-4157(79)90011-x. [DOI] [PubMed] [Google Scholar]
- 155.Laubinger W, Dimroth P. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur J Biochem. 1987;168:475–480. doi: 10.1111/j.1432-1033.1987.tb13441.x. [DOI] [PubMed] [Google Scholar]
- 156.Laubinger W, Dimroth P. Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry. 1988;27:7531–7537. doi: 10.1021/bi00419a053. [DOI] [PubMed] [Google Scholar]
- 157.Laubinger W, Deckers-Hebestreit G, Altendorf K, Dimroth P. A hybrid adenosinetriphosphatase composed of F1 of Escherichia coli and F0 of Propionigenium modestum is a functional sodium ion pump. Biochemistry. 1990;29:5458–5463. doi: 10.1021/bi00475a008. [DOI] [PubMed] [Google Scholar]
- 158.Leimgruber R M, Jensen C, Abrams A. Purification and characterization of the membrane adenosine triphosphatase complex from the wild-type and N,N′-dicyclohexylcarbodiimide-resistant strains of Streptococcus faecalis. J Bacteriol. 1981;147:363–372. doi: 10.1128/jb.147.2.363-372.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lewis R N A H, McElhaney R N. Purification and characterization of the membrane (Na+ + Mg2+)-ATPase from Acholeplasma laidlwaii B. Biochim Biophys Acta. 1983;735:113–122. doi: 10.1016/0005-2736(83)90266-3. [DOI] [PubMed] [Google Scholar]
- 160.Ludwig W, Seewaldt E, Kilpper-Bälz R, Schleifer K H, Magrum L, Woese C, Fox G E, Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985;131:543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 161.Maloney P C. Obligatory coupling between proton entry and the synthesis of adenosine 5′-triphosphate in Streptococcus lactis. J Bacteriol. 1977;132:564–575. doi: 10.1128/jb.132.2.564-575.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maloney P C, Wilson T H. ATP synthesis driven by a protonmotive force in Streptococcus lactis. J Membr Biol. 1975;25:285–310. doi: 10.1007/BF01868580. [DOI] [PubMed] [Google Scholar]
- 163.Maloney P C, Kashket E R, Wilson T H. A protonmotive force drives ATP synthesis in bacteria. Proc Natl Acad Sci USA. 1974;71:3896–3900. doi: 10.1073/pnas.71.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mandel M, Moriyama Y, Hulmes J D, Pan Y C, Nelson H, Nelson N. cDNA sequence encoding the 16-kDa proteolipid of chromaffin granules implies gene duplication in the evolution of H+-ATPases. Proc Natl Acad Sci USA. 1988;85:5521–5524. doi: 10.1073/pnas.85.15.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Manson M, Tedesco P, Berg H, Harold F M, van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–33064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mellano M A, Cooksey D A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988;170:2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Michels M, Bakker E P. Low affinity potassium uptake system in Bacillus acidocaldarius. J Bacteriol. 1987;169:4335–4341. doi: 10.1128/jb.169.9.4335-4341.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 169.Mitchell P. Chemiosmotic coupling and energy transduction. Bodmin, England: Glynn Research; 1968. [Google Scholar]
- 170.Mitchell P. Performance and conservation of osmotic work by proton-coupled solute porter systems. J Bioenerg. 1973;4:63–91. doi: 10.1007/BF01516051. [DOI] [PubMed] [Google Scholar]
- 171.Mitchell P. Vectorial chemistry and the molecular mechanics of chemiosmotic coupling: power transmission by proticity. Biochem Soc Trans. 1976;4:399–430. doi: 10.1042/bst0040399. [DOI] [PubMed] [Google Scholar]
- 172.Miwa T, Esaki H, Umemori J, Hino T. Activity of H+-ATPase in ruminal bacteria with special reference to acid tolerance. Appl Environ Microbiol. 1997;63:2155–2158. doi: 10.1128/aem.63.6.2155-2158.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Moriyama Y, Nelson N. Cold inactivation of vacuolar-proton ATPases. J Biol Chem. 1989;265:3577–3582. [PubMed] [Google Scholar]
- 174.Mugikura S, Nishikawa M, Igarashi K, Kobayashi H. Maintenance of a neutral cytoplasmic pH is not obligatory for growth of Escherichia coli and Streptococcus faecalis at an alkaline pH. J Biochem. 1990;108:86–91. doi: 10.1093/oxfordjournals.jbchem.a123168. [DOI] [PubMed] [Google Scholar]
- 175.Muñoz R, García E, De la Campa A G. Quinine specifically inhibits the proteolipid subunit of the F0F1 H+-ATPase of Streptococcus pneumoniae. J Bacteriol. 1996;178:2455–2458. doi: 10.1128/jb.178.8.2455-2458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. The ntpJ gene in the Enterococcus hirae ntp operon encodes the component of KtrII potassium transport system functionally independent of vacuolar Na+-ATPase. J Biol Chem. 1996;271:10042–10047. doi: 10.1074/jbc.271.17.10042. [DOI] [PubMed] [Google Scholar]
- 177.Murata T, Takase K, Yamato I, Igarashi K, Kakinuma Y. Purification and reconstitution of Na+-translocating vacuolar ATPase from Enterococcus hirae. J Biol Chem. 1997;272:24885–24890. doi: 10.1074/jbc.272.40.24885. [DOI] [PubMed] [Google Scholar]
- 178.Murata T, Yamato I, Igarashi K, Kakinuma Y. Intracellular Na+ regulates the transcription of the ntp operon encoding vacuolar-type Na+-translocating ATPase in Enterococcus hirae. J Biol Chem. 1996;271:23661–23666. doi: 10.1074/jbc.271.39.23661. [DOI] [PubMed] [Google Scholar]
- 179.Muroi M, Shiragami N, Takatsuki A. Destruxin B, a specific and readily reversible inhibitor of vacuolar-type H+-translocating ATPase. Biochem Biophys Res Commun. 1994;205:1358–1365. doi: 10.1006/bbrc.1994.2815. [DOI] [PubMed] [Google Scholar]
- 180.Nelson N. Structure, function and evolution of proton ATPases. Plant Physiol. 1988;86:1–3. doi: 10.1104/pp.86.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nelson N. Evolution of organellar proton-ATPases. Biochim Biophys Acta. 1992;1100:109–124. doi: 10.1016/0005-2728(92)90072-a. [DOI] [PubMed] [Google Scholar]
- 182.Nelson N, Taiz L. The evolution of H+-ATPases. Trends Biochem Sci. 1989;14:113–116. doi: 10.1016/0968-0004(89)90134-5. [DOI] [PubMed] [Google Scholar]
- 183.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 184.Odermatt A, Solioz M. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J Biol Chem. 1995;270:4349–4354. doi: 10.1074/jbc.270.9.4349. [DOI] [PubMed] [Google Scholar]
- 185.Odermatt A, Krapf R, Solioz M. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem Biophys Res Commun. 1994;202:44–48. doi: 10.1006/bbrc.1994.1891. [DOI] [PubMed] [Google Scholar]
- 186.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 187.Odermatt A, Suter H, Krapf R, Solioz M. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann N Y Acad Sci. 1992;671:484–486. doi: 10.1111/j.1749-6632.1992.tb43836.x. [DOI] [PubMed] [Google Scholar]
- 188.O’Hara M B, Hageman J H. Energy and calcium ion dependence of proteolysis during sporulation of Bacillus subtilis cells. J Bacteriol. 1990;172:4161–4170. doi: 10.1128/jb.172.8.4161-4170.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ohyama T, Igarashi K, Kobayashi H. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J Bacteriol. 1994;176:4311–4315. doi: 10.1128/jb.176.14.4311-4315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Oka T, Yamamoto R, Futai M. Three vha genes encode proteolipids of Caenorhabditis elegans vacuolar-type ATPase. Gene structures and preferential expression in an H-shaped excretory cell and pectal cells. J Biol Chem. 1997;272:24371–24379. doi: 10.1074/jbc.272.39.24387. [DOI] [PubMed] [Google Scholar]
- 191.Padan E, Schuldiner S. Na+ transport systems in prokaryotes. In: Bakker E P, editor. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 3–24. [Google Scholar]
- 192.Padan E, Schuldiner S. Bacterial Na+/H+ antiporters—molecular biology, biochemistry and physiology. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. Amsterdam, The Netherlands: Elsevier Science BV; 1996. pp. 501–531. [Google Scholar]
- 193.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s) J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 194.Pedersen P L, Carafoli E. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem Sci. 1987;12:146–150. [Google Scholar]
- 195.Pedersen P L, Carafoli E. Ion motive ATPases. II. Energy coupling and work output. Trends Biochem Sci. 1987;12:186–189. [Google Scholar]
- 196.Phung L T, Ajlani G, Haselkorn R. P-type ATPase from the cyanobacterium Synechococcus 7942 related to the human Menkes and Wilson disease gene products. Proc Natl Acad Sci USA. 1994;91:9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Poolman B. Energy transduction in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:125–148. doi: 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 198.Poolman B, Driessen A J, Konings W N. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol. 1987;169:5597–5604. doi: 10.1128/jb.169.12.5597-5604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Poolman B, Driessen A J, Konings W N. Regulation of solute transport in streptococci by external and internal pH values. Microbiol Rev. 1987;51:498–508. doi: 10.1128/mr.51.4.498-508.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Poolman B, Smid E J, Konings W N. Kinetic properties of a phosphate-bond-driven glutamate-glutamine transport system in Streptococcus lactis and Streptococcus cremoris. J Bacteriol. 1987;169:2755–2761. doi: 10.1128/jb.169.6.2755-2761.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Poolman N, Molenaar D, Smid E J, Ubbink T, Abee T, Renault P P, Konings W N. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J Bacteriol. 1991;173:6030–6037. doi: 10.1128/jb.173.19.6030-6037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pritchard G G, Wimpenny J W T. Cytochrome formation, oxygen-induced proton extrusion and respiratory activity in Streptococcus var. zymogenes grown in the presence of haematin. J Gen Microbiol. 1978;104:15–22. doi: 10.1099/00221287-104-1-15. [DOI] [PubMed] [Google Scholar]
- 203.Rahav-Manor O, Carmel O, Karpel R, Taglicht D, Glaser G, Schuldiner S, Padan E. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J Biol Chem. 1992;267:10433–10438. [PubMed] [Google Scholar]
- 204.Rahlfs S, Müller V. Sequence of subunit c of the Na+-translocating F1F0 ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett. 1997;404:269–271. doi: 10.1016/s0014-5793(97)00088-4. [DOI] [PubMed] [Google Scholar]
- 205.Ramos S, Kaback H R. The electrochemical proton gradient in Escherichia coli membrane vesicles. Biochemistry. 1977;16:848–854. doi: 10.1021/bi00624a006. [DOI] [PubMed] [Google Scholar]
- 206.Rampersaud A, Utsumi R, Delgado J, Forst S A, Inouye M. Ca2+-enhanced phosphorylation of a chimeric protein kinase involved with bacterial signal transduction. J Biol Chem. 1991;266:7633–7637. [PubMed] [Google Scholar]
- 207.Rea P A, Griffith C J, Manolson M F, Sanders D. Irreversible inhibition of H+-ATPase of higher plant tonoplast by chaotropic anions: evidence of peripheral location of nucleotide binding subunits. Biochim Biophys Acta. 1987;904:1–12. [Google Scholar]
- 208.Reidlinger J, Müller V. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1F0-type enzyme. Eur J Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 209.Reizer J, Panos C. Transport of alpha-aminoisobutyric acid by Streptococcus pyogenes and its derived l-form. J Bacteriol. 1982;149:211–220. doi: 10.1128/jb.149.1.211-220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Reizer J, Reizer A, Saier M H., Jr The putative Na+/H+ antiporter (NapA) of Enterococcus hirae is homologous to the putative K+/H+ antiporter (KefC) of Escherichia coli. FEMS Microbiol Lett. 1992;94:161–164. doi: 10.1016/0378-1097(92)90601-j. [DOI] [PubMed] [Google Scholar]
- 211.Rimpilainen M A, Mettanen T T, Niskasaari K, Forsen R I. The F1-ATPase from Streptococcus cremoris: isolation, purification and partial characterization. Int J Biochem. 1988;20:1117–1124. doi: 10.1016/0020-711x(88)90257-1. [DOI] [PubMed] [Google Scholar]
- 212.Ritchey T W, Seeley H W., Jr Distribution of cytochrome-like respiration in Streptococci. J Gen Microbiol. 1976;93:195–203. doi: 10.1099/00221287-93-2-195. [DOI] [PubMed] [Google Scholar]
- 213.Rosen B P, Ambudkar S V, Borbolla M G, Chen C M, Houng H S, Mobley H L, Tsujibo H, Zlotnick G W. Ion extrusion systems in bacteria. Ann N Y Acad Sci. 1985;456:235–244. doi: 10.1111/j.1749-6632.1985.tb14870.x. [DOI] [PubMed] [Google Scholar]
- 214.Rottenberg H. The driving force for proton(s) metabolite cotransport in bacterial cells. FEBS Lett. 1976;66:159–163. doi: 10.1016/0014-5793(76)80493-0. [DOI] [PubMed] [Google Scholar]
- 215.Russell J B, Strobel H J, Driessen A J, Konings W N. Sodium-dependent transport of neutral amino acids by whole cells and membrane vesicles of Streptococcus bovis, a ruminal bacterium. J Bacteriol. 1988;170:3531–3536. doi: 10.1128/jb.170.8.3531-3536.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sabbert D, Engelbrecht S, Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 217.Schäfer G, Meyering-Vos M. The plasma membrane ATPase of archaebacteria: a chimeric energy converter. Ann N Y Acad Sci. 1992;671:293–309. doi: 10.1111/j.1749-6632.1992.tb43804.x. [DOI] [PubMed] [Google Scholar]
- 218.Schlösser A, Hamann A, Bossemeyer D, Schneider E, Bakker E P. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol Microbiol. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 219.Schnebli H P, Abrams A. Membrane adenosine triphosphatase from Streptococcus faecalis: preparation and homogeneity. J Biol Chem. 1970;245:1115–1121. [PubMed] [Google Scholar]
- 220.Schnebli H P, Vatter A E, Abrams A. Membrane adenosine triphosphatase from Streptococcus faecalis: molecular weight, subunit structure, and amino acid composition. J Biol Chem. 1970;245:1122–1127. [PubMed] [Google Scholar]
- 221.Schobert B, Lanyi J K. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 222.Senior A. The proton-translocating ATPase of Escherichia coli. Annu Rev Biophys Biophys Chem. 1990;19:7–41. doi: 10.1146/annurev.bb.19.060190.000255. [DOI] [PubMed] [Google Scholar]
- 223.Shibata C, Ehara T, Tomura K, Igarashi K, Kobayashi H. Gene structure of Enterococcus hirae (Streptococcus faecalis) F1F0-ATPase, which functions as a regulator of cytoplasmic pH. J Bacteriol. 1992;174:6117–6124. doi: 10.1128/jb.174.19.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Silver S. Transport of inorganic cations. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1091–1102. [Google Scholar]
- 225.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 226.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Silver S, Nucifora G, Chu L, Misra T K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989;14:76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- 228.Skulachev V P. Membrane bioenergetics. Berlin, Germany: Springer-Verlag KG; 1988. [Google Scholar]
- 229.Smith A J, Quivey R G, Jr, Faustoferri R C. Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon. Gene. 1996;183:87–96. doi: 10.1016/s0378-1119(96)00502-1. [DOI] [PubMed] [Google Scholar]
- 230.Smith D L, Maguire M E. Distribution of the CorA Mg2+ transport system in gram-negative bacteria. J Bacteriol. 1995;177:1638–1640. doi: 10.1128/jb.177.6.1638-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Snavely M D, Miller C G, Maguire M E. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- 232.Solioz M, Davies K. Operon of vacuolar-type Na+-ATPase of Enterococcus hirae. J Biol Chem. 1994;269:9453–9459. [PubMed] [Google Scholar]
- 233.Solioz M, Odermatt A. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem. 1995;270:9217–9221. doi: 10.1074/jbc.270.16.9217. [DOI] [PubMed] [Google Scholar]
- 234.Solioz M, Vulpe C. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci. 1996;21:237–241. [PubMed] [Google Scholar]
- 235.Solioz M, Mathews S, Furst P. Cloning of the K+-ATPase of Streptococcus faecalis. Structural and evolutionary implications of its homology to the KdpB-protein of Escherichia coli. J Biol Chem. 1987;262:7358–7362. [PubMed] [Google Scholar]
- 236.Solioz M, Odermatt A, Krapf R. Copper pumping ATPases: common concepts in bacteria and man. FEBS Lett. 1994;346:44–47. doi: 10.1016/0014-5793(94)00316-5. [DOI] [PubMed] [Google Scholar]
- 237.Speelmans G, Poolman B, Abee T, Konings W N. Energy transduction in the thermophilic anaerobic bacterium Clostridium fervidus is exclusively coupled to sodium ions. Proc Natl Acad Sci USA. 1993;90:7975–7979. doi: 10.1073/pnas.90.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Strausak D, Solioz M. Regulation of sodium transport in Enterococcus hirae. EBEC Short Rep. 1994;8:77. [Google Scholar]
- 239.Strausak D, Solioz M. CopY is a copper-inducible repressor of the Enterococcus hirae copper ATPases. J Biol Chem. 1997;272:8932–8936. doi: 10.1074/jbc.272.14.8932. [DOI] [PubMed] [Google Scholar]
- 240.Strausak D, Waser M, Solioz M. Functional expression of the Enterococcus hirae NaH-antiporter in Escherichia coli. J Biol Chem. 1993;268:26334–26337. [PubMed] [Google Scholar]
- 241.Strobel H J, Russell J B. Non-proton-motive-force-dependent sodium efflux from the ruminal bacterium Streptococcus bovis: bound versus free pools. Appl Environ Microbiol. 1989;55:2664–2668. doi: 10.1128/aem.55.10.2664-2668.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sturr M G, Marquis R E. Comparative acid tolerances and inhibitor sensitivities of isolated F-ATPases of oral lactic acid bacteria. Appl Environ Microbiol. 1992;58:2287–2291. doi: 10.1128/aem.58.7.2287-2291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sumi M, Sato M H, Denda K, Date T, Yoshida M. A DNA fragment homologous to F1-ATPase beta subunit was amplified from genomic DNA of Methanosarcina barkeri. Indication of an archaebacterial F-type ATPase. FEBS Lett. 1992;314:207–210. doi: 10.1016/0014-5793(92)81472-x. [DOI] [PubMed] [Google Scholar]
- 244.Sutton S V, Marquis R E. Membrane-associated and solubilized ATPases of Streptococcus mutans and Streptococcus sanguis. J Dent Res. 1987;66:1095–1098. doi: 10.1177/00220345870660060201. [DOI] [PubMed] [Google Scholar]
- 245.Sutton S V, Bender G R, Marquis R E. Fluoride inhibition of proton-translocating ATPases of oral bacteria. Infect Immun. 1987;55:2597–2603. doi: 10.1128/iai.55.11.2597-2603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Suzuki T, Kobayashi H. Regulation of the cytoplasmic pH by a proton-translocating ATPase in Streptococcus faecalis (faecium). A computer simulation. Eur J Biochem. 1989;180:467–471. doi: 10.1111/j.1432-1033.1989.tb14669.x. [DOI] [PubMed] [Google Scholar]
- 247.Takase K. Ph.D. thesis. Chiba, Japan: Science University of Tokyo; 1997. [Google Scholar]
- 248.Takase K, Yamato I, Kakinuma Y. Cloning and sequencing of the genes coding for the A and B subunits of vacuolar-type Na+-ATPase from Enterococcus hirae. J Biol Chem. 1993;268:11610–11616. [PubMed] [Google Scholar]
- 249.Takase K, Kakinuma S, Yamato I, Konishi K, Igarashi K, Kakinuma Y. Sequencing and characterization of the ntp gene cluster for Na+-translocating ATPase in Enterococcus hirae. J Biol Chem. 1994;269:11037–11044. [PubMed] [Google Scholar]
- 250.Tao T, Snavely M D, Farr S G, Maguire M E. Magnesium transport in Salmonella typhimurium mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J Bacteriol. 1995;177:2654–2662. doi: 10.1128/jb.177.10.2654-2662.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.tenBrink B, Konings W N. Electrochemical proton gradient and lactate concentration gradient in Streptococcus cremoris cells grown in batch culture. J Bacteriol. 1982;152:682–686. doi: 10.1128/jb.152.2.682-686.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tisa L S, Adler J. Calcium ions are involved in Escherichia coli chemotaxis. Proc Natl Acad Sci USA. 1992;89:11804–11808. doi: 10.1073/pnas.89.24.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tokuda H, Unemoto T. Characterization of the respiration-dependent Na+ pump in the marine bacterium Vibrio alginolyticus. J Biol Chem. 1982;257:10007–10014. [PubMed] [Google Scholar]
- 254.Tokuda H, Sugasawa M, Unemoto T. Roles of Na+ and K+ in α-aminoisobutyric acid transport by the marine bacterium Vibrio alginolyticus. J Biol Chem. 1982;257:788–794. [PubMed] [Google Scholar]
- 255.Trombe M C. Characterization of a calcium porter of Streptococcus pneumoniae involved in calcium regulation of growth and competence. J Gen Microbiol. 1993;139:433–439. doi: 10.1099/00221287-139-3-433. [DOI] [PubMed] [Google Scholar]
- 256.Trombe M C, Clave C, Manias J M. Calcium regulation of growth and differentiation in Streptococcus pneumoniae. J Gen Microbiol. 1992;138:77–84. doi: 10.1099/00221287-138-1-77. [DOI] [PubMed] [Google Scholar]
- 257.Trombe M C, Rieux V, Baille F. Mutations which alter the kinetics of calcium transport alter the regulation of competence in Streptococcus pneumoniae. J Bacteriol. 1994;176:1992–1996. doi: 10.1128/jb.176.7.1992-1996.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Uchida E, Ohsumi Y, Anraku Y. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1985;260:1090–1095. [PubMed] [Google Scholar]
- 259.Vadeboncoeur C, Pelletier M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev. 1997;19:187–207. doi: 10.1111/j.1574-6976.1997.tb00297.x. [DOI] [PubMed] [Google Scholar]
- 260.van der Drift C, Janssen D B, van Wezenbeek P M. Hydrolysis and synthesis of ATP by membrane-bound ATPase from a motile Streptococcus. Arch Microbiol. 1978;119:31–36. doi: 10.1007/BF00407924. [DOI] [PubMed] [Google Scholar]
- 261.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat Genet. 1993;3:7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- 262.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Waser M, Hess-Bienz D, Davies K, Solioz M. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J Biol Chem. 1992;267:5396–5400. [PubMed] [Google Scholar]
- 264.West I C, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wiggins P M. Role of water in some biological processes. Microbiol Rev. 1990;54:432–449. doi: 10.1128/mr.54.4.432-449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wilms R, Freiberg C, Wegerle E, Meier I, Mayer F, Müller V. Subunit structure and organization of the genes of the A1A0 ATPase from the archaeon Methanosarcina mazei Gö1. J Biol Chem. 1996;271:18843–18852. doi: 10.1074/jbc.271.31.18843. [DOI] [PubMed] [Google Scholar]
- 267.Wyler-Duda P, Solioz M. Phosphoenzyme formation by purified, reconstituted copper ATPase of Enterococcus hirae. FEBS Lett. 1996;399:143–146. doi: 10.1016/s0014-5793(96)01306-3. [DOI] [PubMed] [Google Scholar]
- 268.Yokoyama K, Oshima T, Yoshida M. Thermus thermophilus membrane-associated ATPase: indication of an eubacterial V-type ATPase. J Biol Chem. 1990;265:21946–21950. [PubMed] [Google Scholar]
- 269.Yuan D S, Stearman R, Dancis A, Dunn T, Beeler T, Klausner R D. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron. Proc Natl Acad Sci USA. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zarlengo M, Abrams A. Selective penetration of ammonia and alkylamines into Streptococcus faecalis and their effect on glycolysis. Biochim Biophys Acta. 1963;71:65–77. doi: 10.1016/0006-3002(63)90986-7. [DOI] [PubMed] [Google Scholar]
- 271.Zhang J, Feng Y, Forgac M. Proton conduction and bafilomycin binding by the V0 domain of the coated vesicle V-ATPase. J Biol Chem. 1994;269:23518–23523. [PubMed] [Google Scholar]