Abstract
Paracoccus denitrificans and its near relative Paracoccus versutus (formerly known as Thiobacilllus versutus) have been attracting increasing attention because the aerobic respiratory system of P. denitrificans has long been regarded as a model for that of the mitochondrion, with which there are many components (e.g., cytochrome aa3 oxidase) in common. Members of the genus exhibit a great range of metabolic flexibility, particularly with respect to processes involving respiration. Prominent examples of flexibility are the use in denitrification of nitrate, nitrite, nitrous oxide, and nitric oxide as alternative electron acceptors to oxygen and the ability to use C1 compounds (e.g., methanol and methylamine) as electron donors to the respiratory chains. The proteins required for these respiratory processes are not constitutive, and the underlying complex regulatory systems that regulate their expression are beginning to be unraveled. There has been uncertainty about whether transcription in a member of the alpha-3 Proteobacteria such as P. denitrificans involves a conventional ς70-type RNA polymerase, especially since canonical −35 and −10 DNA binding sites have not been readily identified. In this review, we argue that many genes, in particular those encoding constitutive proteins, may be under the control of a ς70 RNA polymerase very closely related to that of Rhodobacter capsulatus. While the main focus is on the structure and regulation of genes coding for products involved in respiratory processes in Paracoccus, the current state of knowledge of the components of such respiratory pathways, and their biogenesis, is also reviewed.
The genus Paracoccus is one of the most distantly related of the Proteobacteria to Escherichia coli (178) as judged by 16S rRNA sequence. For many years, the sole representative of the genus was Paracoccus denitrificans, first isolated in 1908 by Beijerinck (13) as Micrococcus denitrificans. The original selection of this species was based on its ability to convert nitrate into molecular nitrogen. Improved molecular phylogenetics have led to the inclusion of Thiobacillus versutus (as Paracoccus versutus [145]) and Thiosphaera pantotropha (101, 178, 233) into the genus and to the addition of P. kocurii (203), P. alcaliphilus (301), P. aminophilus (300), P. aminovorans (300), P. thiocyanatus (145), and P. solventivorans (264). More recently, two other species have been characterized by using 16S rRNA (P. marcusii [112] and P. alkenifer [170]), but no other properties of these species have been published.
These newer species were isolated by using a range of organic and inorganic compounds, including acetone (P. solventivorans), dimethylformamide (P. aminovorans and P. aminophilus) and thiocyanate (P. thiocyanatus), as growth substrates. Recently, it has been shown that some strains of P. denitrificans can use carbon disulfide (139, 233). These properties raise the possibility of using Paracoccus species for bioremediation, particularly since most species in the genus can use nitrate and its reduction products as an alternative electron acceptor to oxygen during anaerobic respiratory growth (except P. aminovorans, P. aminophilus, and P. alcaliphilus [145]). Unifying characteristics of the species include an obligately respiratory mode of growth and the use of ribulose bisphosphate carboxylase/oxygenase to fix carbon during methylotrophic or chemolithotrophic growth. All these organisms are characterized by a high genomic guanine-plus-cytosine (G+C) content (63.8 to 70.2% [145]).
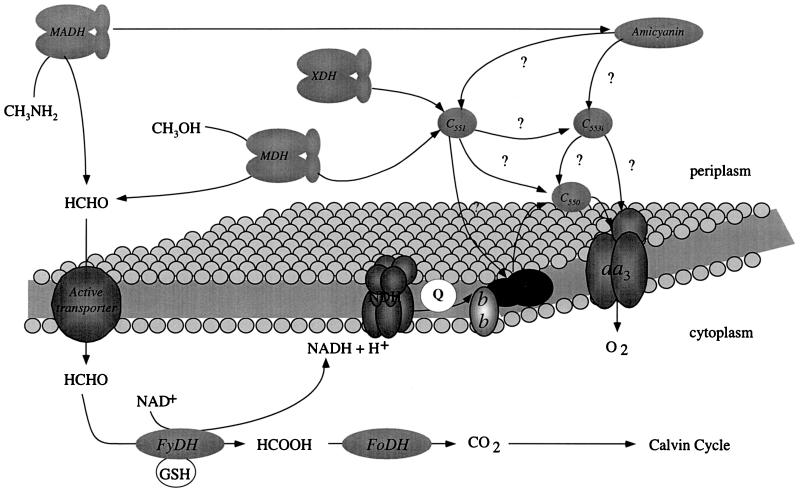
The electron transport chain used for aerobic growth by P. denitrificans has long been used as a model for the mitochondrial electron transport chain (137, 280), since it possesses a full complement of proteins with counterparts in mitochondria: electron transport flavoproteins, NADH-ubiquinone oxidoreductase, bc1 complex, c-type cytochromes, and an aa3-type terminal cytochrome oxidase (Fig. 1). This is in contrast to the usual bacterial model organism, Escherichia coli, which does not possess some of these complexes. Branches of the “conventional” electron transport chain (75a) allow the obligately respiratory members of Paracoccus to grow under different oxygen concentrations, to use N-oxides as alternative electron acceptors, and to use a variety of carbon sources, including amines and alcohols (Fig. 1). The interest in P. denitrificans electron transport has led to the striking achievement of the determination of the crystal structure of the terminal aa3-type cytochrome c oxidase (136). Other redox proteins isolated and structurally characterized from species within the genus include methylamine dehydrogenase, amicyanin, cytochrome c551 (45, 46, 73), cytochrome c550 (19), pseudoazurin (321), electron transfer flavoprotein (245), and cytochrome cd1 nitrite reductase (6, 87).
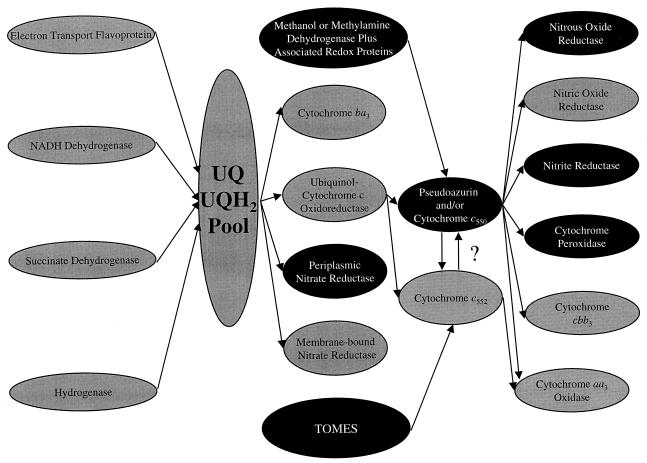
FIG. 1.
Branched electron transport chains of Paracoccus species. Enzyme complexes colored black indicate a periplasmic location. Electron transfer between cytochromes c552 and c550 has not been demonstrated experimentally but is possible, given the redox potential of the proteins. The exact nature of the roles of cytochrome c550 and pseudoazurin is currently being studied. UQ = ubiquinone, UQH2 = reduced ubiquinone. TOMES, thiosulfate oxidation multienzyme system.
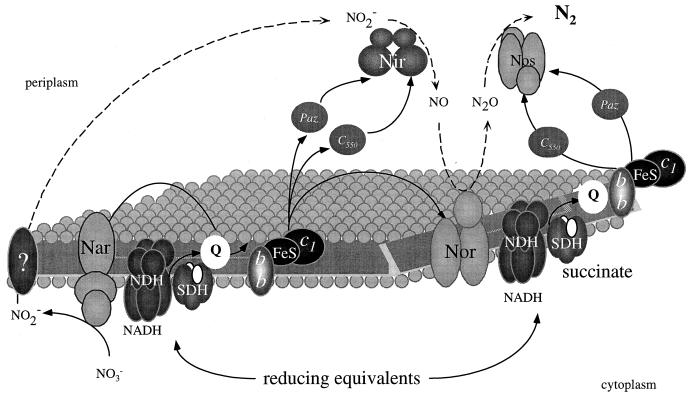
Organisms such as Paracoccus species often have to face large fluctuations in the free-oxygen concentration. The adaptive responses of P. denitrificans to changing environmental conditions sometimes resemble that of E. coli, and parts of the signal transduction cascades appear to be common between these organisms. A typical example concerns the switch from aerobic to nitrate respiration. Optimal synthesis of nitrate reductase in both organisms requires a coordinated reaction to two different types of environmental trigger: the absence of oxygen and the presence of nitrate. The molecular basis for this type of regulation in E. coli is now well understood (165, 299). To date, two proteins similar to the fumarate/nitrate respiration (FNR) regulatory protein family (important in E. coli for the sensing of oxygen and the induction and repression of several operons) have been discovered in P. denitrificans. However, many other types of regulation must occur in order to account for the diversity of electron transport shown in Fig. 1.
The derivation of the strains of P. denitrificans from the first strain isolated by Beijerinck has been well reviewed by Goodhew et al. (101), who found that the type strain (ATCC 17741) was a direct subculture of the original strain isolated by Beijerinck. Relatively few new isolates of P. denitrificans have been found, and differences between these strains of P. denitrificans have been described (101, 199, 312). However, most of the molecular biology has been performed on a single strain, P. denitrificans Pd1222. The strains used and the various loci discussed in this review are listed in Table 1. The position of the strain initially named Thiosphaera pantotropha (247) has recently been the subject of some controversy. Although Ludwig et al. (178) reclassified T. pantotropha as P. denitrificans, there is doubt (233) about this reclassification (101, 139, 281). The strain of P. denitrificans used by Ludwig et al. itself appears to be not entirely a typical P. denitrificans strain as judged by analysis of either c-type cytochromes (101) or methyl fatty acids (5). More recently, an extensive survey of various P. denitrificans strains, based on a 16S rRNA analysis, has been undertaken (233). The outcome is a proposal to name Thiosphaera pantotropha as Paracoccus pantotrophus, a species to which several strains of P. denitrificans held in culture collections for many years may be transferred. These changes in nomenclature are likely to cause confusion. It is important to note that since 1993 some research groups have continued to use the name Thiosphaera pantotropha while others have adopted P. denitrificans GB-17. A careful reading of the literature is needed to identify which strain of P. denitrificans has been used in a particular study. At the time of writing, the proposal to revive Thiosphaera pantotropha under the name Paracoccus pantotrophus seems destined for acceptance (233); we have used the name P. denitrificans GB-17 in this review. It has been our experience that apart from the necessary considerations that must be taken into account as far as antibiotic resistance are concerned, molecular genetic techniques described for P. denitrificans may equally be applied to P. denitrificans GB-17 (P. pantotrophus). The commonly used P. denitrificans Pd1222 is intrinsically resistant to spectinomycin but sensitive to streptomycin, whereas the reverse is true for P. denitrificans GB-17 (P. pantrophus). The latter is resistant to lead and arsenic, but other P. denitrificans strains are not. This review focuses mainly on P. denitrificans, the species on which the majority of molecular biological work has been performed: where necessary, we distinguish between strains of P. denitrificans, but note that the species designated P. denitrificans GB-17 also has the strain numbers LMD 92.62 and LMD 82.5 in the literature. We would refer the reader to the forthcoming article by Rainey et al. (233) for more information.
TABLE 1.
Isolated and characterized Paracoccus loci
| Strain | Gene cluster | Accession no. | Known functions | Reference(s) |
|---|---|---|---|---|
| P. denitrificans 71.11 | adk | U64203 | ATP regeneration: adenylate kinase (adk) | 66 |
| P. denitrificans Pd1222a | hemA | U12508 | Cytochrome c biosynthesis: 5-ALA synthase (hemA) | 214 |
| P. denitrificans Pd1222 | secF-hisH-ccmABCDG-ORF36-ORF117 | Z71971 | Cytochrome c biosynthesis: secretory apparatus protein (secF), histidinol phosphate transaminase (hisH) | 215 |
| P. denitrificans Pd1222 | ccmF-ccmH | AF023247 | Cytochrome c biosynthesis | 216 |
| P. denitrificans LMD 92.63b | narGHJI | Z26255Z37158 | Denitrification: membrane-bound nitrate reductase α subunit (narG), β subunit (narH), γ subunit (narI) | 23 |
| P. denitrificans Pd1222 | nirISECFD | U05002 | Denitrification: nitrite reductase transcriptional regulator (nirI), structural gene (nirS), S-adenosyl-l-methionine uroporphyrinogen methyltransferase (nirE), small c-type cytochrome (nirC) | 59 |
| P. denitrificans LMD 92.63b | nirSE | U75413 | Denitrification: nitrite reductase structural gene (nirS) | 5, 6 |
| P. denitrificans Pd1222 | nirIX | AJ001308 | Denitrification: regulation of nir gene expression | 307 |
| P. denitrificans Pd1222 | norCBQDEF | U28078 | Denitrification: nitric oxide reductase large subunit (norB), small subunit (norC) | 60 |
| P. denitrificans NCIMB 8944 | nosRZD | X74792 | Denitrification: nitrous oxide reductase transcriptional regulator (nosR), structural gene (nosZ) | 125 |
| P. denitrificansc | nosZ | AF016058 | Denitrification: nitrous oxide reductase structural gene (nosZ) | 255 |
| P. denitrificans Pd1222 | nnr | U17435 | Denitrification: FNR-like transcriptional activator (nnr) | 309 |
| P. denitrificans LMD 92.63b | pazS | Z73141 | Denitrification: pseudoazurin structural protein (pazS) | 168 |
| P. denitrificans Pd1222 | cycA-ctaDII | Y07533 | Electron transport: cytochrome c550 (cycA), iso-cytochrome c oxidase subunit II (ctaDII) | 235 |
| P. denitrificans Pd1222 | cycA-ctaDII | M27304 | Electron transport: cytochrome c550 (cycA), iso-cytochrome c oxidase subunit II (ctaDII) | 315 |
| P. versutus ATCC 25364 | ORF1-cycA-ctaDII | X62808S37058 | Electron transport: cytochrome c550 (cycA), iso-cytochrome c oxidase subunit II (ctaDII) | 296, 297 |
| P. denitrificans Pd1235 | cycM | X70367 | Electron transport: cytochrome c552 (cycM) | 294 |
| P. denitrificans Pd1235 | qoxABCD | X78196 | Electron transport: cytochrome ba3 quinol oxidase subunit II (qoxA), subunit I (qoxB), subunit III (qoxC), subunit IV (qoxD) | 244 |
| P. denitrificans ATCC 13543 | fbcFBC | M17522 | Electron transport: cytochrome bc1 complex iron sulfur subunit (fbcF), cytochrome b subunit (fbcB), cytochrome c subunit (fbcC) | 162 |
| P. denitrificans Pd1222 | ctaDI | X05829 | Electron transport: cytochrome oxidase subunit I (ctaDI) | 235 |
| P. denitrificans PD 1235 | ctaC | X05934 | Electron transport: cytochrome oxidase subunits II (ctaC) | 277 |
| P. denitrificans Pd1222 | ctaC-ctaB-ORF1-ctaG-ctaE | X05828 | Electron transport: cytochrome oxidase subunits II (ctaC) and III (ctaE) | 234 |
| P. denitrificans S1657c | ctaE | X04406 | Electron transport: cytochrome oxidase subunit III (ctaE) | 251 |
| P. denitrificans Pd1222 | ctaH | Y08372 | Electron transport: cytochrome c oxidase subunit IV (ctaH) | 325 |
| P. denitrificansc | phaAB | D49362 | Energy storage: acetoacetyl-CoA reductase (phaA), β-ketothiolase (phaB) | 336 |
| P. denitrificansc | phaC | D43764 | Energy storage: poly (3-hydroxyalkanoate) synthase (phaC) | 298 |
| P. denitrificans Pd1222 | pta-ORF3-ORF4-ORF1-ORF2-ORF5 | U08864 | Insertion sequence IS1248: phosphate acetyltransferase (pta) | 311 |
| P. denitrificans Pd1222 | ORF3-ORF4-ORF1-ORF2-ORF5 | U08856 | Insertion sequence IS1248b | 312 |
| P. denitrificans Pd1222 | ORF1-ORF2-flhA-clpP-ORF3-fghA-xoxF-cycB-xoxJI-ORF4 | U34346 | C1 metabolism: NAD-GSH-dependent formaldehyde dehydrogenase (flhA), S-formylglutathione hydrolase (fghA), PQQ-dependent dehydrogenase large subunit (xoxF), cytochrome c553i (cycB) | 116, 117, 237, 239, 240 |
| P. denitrificans Pd1222 | ORF1-flhS-ORF2-flhR-abcABC-pqqE | AJ223460 | C1 metabolism: transcriptional activator proteins (flhS, flhR), ABC transporter-type proteins (abcA, abcB, abcC), coenzyme PQQ synthesis protein E (pqqE) | 241 |
| P. denitrificans Pd1222 | mxaF | M17339 | Methanol oxidation: methanol dehydrogenase large subunit (mxaF) | 114 |
| P. denitrificans Pd1222 | mxaGIJ | M57684 | Methanol oxidation: cytochrome c551i (mxaG), methanol dehydrogenase β subunit (mxaI) | 314 |
| P. denitrificans Pd1222 | ORF1-cycB-ORF2 | M75583 | Methanol oxidation: cytochrome c553i (cycB) | 239 |
| P. denitrificans Pd1222 | mxaZYX | M92421 | Methanol oxidation: two-component regulatory proteins (mxaX, mxaY) | 118 |
| P. denitrificans Pd1222 | ORF1-mxaACKLD-ORF2-ORF3 | AJ000884 | Methanol oxidation | 242 |
| P. versutus ATCC 25364 | mauB-ORF1 | L08575 | Methylamine oxidation: methylamine dehydrogenase α subunit (mauB) | 130 |
| P. denitrificans Pd1222 | mauB-ami | X55665 | Methylamine oxidation: methylamine dehydrogenase α subunit (mauB), amicyanin (ami) | 317 |
| P. versutus ATCC 25364 | ORF1-mauB-ami | M58001 | Methylamine oxidation: methylamine dehydrogenase α subunit (mauB), amicyanin (ami) | 172, 297 |
| P. denitrificansc | mauDA | M90098 | Methylamine oxidation: methylamine dehydrogenase small subunits (mauA, mauD) | 49 |
| P. denitrificansc | mauFBE | M90099 | Methylamine oxidation: methylamine dehydrogenase small subunits (mauF, mauB, mauE) | 48 |
| P. denitrificans Pd1222 | ORF1-mauRFB | U12464 | Methylamine oxidation: LysR-type transcriptional activator (mauR), methylamine dehydrogenase large subunit (mauB) | 313 |
| P. denitrificans Pd1222 | mauED | X98581 | Methylamine oxidation | 303, 304 |
| P. versutus ATCC 25364 | mauED | L36951 | Methylamine oxidation | 130, 297 |
| P. versutus ATCC 25364 | ORF5-mauF | L36952 | Methylamine oxidation | 129 |
| P. versutus ATCC 25364 | mauG | L36953 | Methylamine oxidation | 130, 297 |
| P. denitrificans Pd1222 | mauJGMN | U15028 | Methylamine oxidation: ferredoxin-like proteins (mauM, mauN), cytochrome c peroxidase-like protein (mauG) | 304 |
| P. denitrificans NDH-1c | nqo1 | M64432, J05331 | NADH dehydrogenase: 50-kDa subunit (nqo1) | 332 |
| P. denitrificans NDH-1c | nqo2 | M74171, J05337 | NADH dehydrogenase: 25-kDa subunit (nqo2) | 331 |
| P. denitrificans NDH-1c | URF3-nqo3 | M84572 | NADH dehydrogenase: 66-kDa subunit (nqo3) | 334 |
| P. denitrificans NDH-1c | uvrA-nqo7-nqo6-nqo5-nqo4 | M93015 | NADH dehydrogenase: nd3 subunit (nqo7), ndhK subunit (nqo6), 25-kDa subunit (nqo5), 48-kDa subunit (nqo4); DNA repair protein (uvrA) | 333 |
| P. denitrificans NDH-1c | URF4-nqo8nqo9-URF5-URF6-nqo10-nqo11-nqo12-nqo14-birA | L02354, L01096 | NADH dehydrogenase: subunit VIII (nqo8), subunit IX (nqo9), subunit X (nqo10), subunit XI (nqo11), subunit XII (nqo12), subunit XIV (nqo14); biotin (acetyl-CoA carboxylase) ligase (birA) | 335 |
| P. denitrificans LMD 92.63b | dctM-napEDABC | Z36773 | Nitrate reduction: periplasmic nitrate reductase molybdenum-iron-sulfur-heme subunit (napA), diheme subunit (napB); Membrane transport: integral membrane subunit of a dicarboxylate transporter (dctM) | 24, 25, 35 |
| P. versutus | ORF1-ORF2 | U42228 | Plasmid pTAV203 | 9 |
| P. versutus | repABC | U60522 | Plasmid pTAV203: replication proteins | 8 |
| Paracoccus sp. strain 164 | ori | AF020624 | Plasmid pTM164: origin of replication | 266 |
| P. denitrificans 71.11c | secY | U64202 | Protein translocation: preprotein translocase (secY) | 228 |
| P. denitrificans Pd1222 | ORF1-fnrN-ORF278-ccoNOQPGH | U34353 | Regulation of anaerobic growth: transcriptional regulator (fnrP) | 63 |
| Electron transport: alternative oxidase cytochrome cbb3 monoheme subunit (ccoN), diheme subunit (ccoQ) | ||||
| P. denitrificansc | rrfA | X01501 | Ribosome assembly: 5S rRNA (rrfA) | 181 |
| Paracoccus sp. strain Y4 | rrnA | AB012914 | Ribosome assembly: 16S rRNA (rrnA) | 111 |
| P. alcaliphilus JCM 7364 | rrnA | D32238 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. alkenifer | rrnA | Y13827 | Ribosome assembly: 16S rRNA (rrnA) | 170 |
| P. aminophilus JCM 7686 | rrnA | D32239 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. aminovorans JCM 7685 | rrnA | D32240 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. denitrificans IAM 12479 | rrnA | D13480 | Ribosome assembly: 16S rRNA (rrnA) | 146, 147 |
| P. denitrificans LMG 4218b | rrnA | X69159 | Ribosome assembly: 16S rRNA (rrnA) | 178 |
| P. kocurii JCM 7684 | rrnA | D32241 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. marcusii | rrnA | Y12703 | Ribosome assembly: 16S rRNA (rrnA) | 112 |
| P. solventivorans DSM 6637 | rrnA | Y07705 | Ribosome assembly: 16S rRNA (rrnA) | 264 |
| P. solventivorans | rrnA | Y13826 | Ribosome assembly: 16S rRNA (rrnA) | 170 |
| P. thiocyanatus THIO11 | rrnA | D32242 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. versutus IAM 12814 | rrnA | D32243 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. versutus IAM 12815 | rrnA | D32244 | Ribosome assembly: 16S rRNA (rrnA) | 145 |
| P. denitrificans KS1b | rrnA | U58015 | Ribosome assembly: 16S rRNA (rrnA) | 139 |
| P. denitrificans KS2b | rrnA | U58016 | Ribosome assembly: 16S rRNA (rrnA) | 139 |
| P. denitrificans KL1b | rrnA | U58017 | Ribosome assembly: 16S rRNA (rrnA) | 139 |
| P. denitrificans LMG 4218b | rrlA | X87287 | Ribosome assembly: 23S rRNA (rrlA) | 179 |
| P. denitrificans 71.11c | rpsM | U64204 | Ribosome assembly: ribosomal protein S13 (rpsM) | 228 |
| P. denitrificans ATCC 13543c | sdhCDAB | U31902 | Succinate oxidation: succinate dehydrogenase b-type cytochrome subunit (sdhC), hydrophobic membrane anchor (sdhD), flavoprotein subunit (sdhA), iron-sulfur protein subunit (sdhB) | 69 |
| P. denitrificans LMD 82.5b | soxABCDEF | X79242 | Sulfur oxidation: protein B (soxB), sulfite oxidase (soxC), cytochrome (soxD), cytochrome (soxE) | 326, 327 |
| P. denitrificans Pd1222 | porG | Y09451 | Membrane proteins: porin structural protein (porG) | 254 |
| P. denitrificans Pd1222 | recA | U59631 | DNA recombination | 75 |
| P. denitrificans IFO 12442c | tyrB | Y08272 | Amino acid biosynthesis: aromatic amino acid transferase (tyrB) | 207 |
P. denitrificans Pd1222 is a derivative of DSM 413.
Now proposed to be P. pantotrophus (233).
Classification as P. denitrificans or P. pantotrophus not yet determined.
The genus Paracoccus is member of a part of the alpha Proteobacteria known as the Rhodobacter group. Paracoccus is closely related to the physiologically well-studied photosynthetic species Rhodobacter sphaeroides and Rhodobacter capsulatus, but species of Rhodovulum (147), Sagittula (100), Amaricoccus (184), Octadecobacter (103), Roseobacteria (78), and Tetracoccus (27) are also members of the group.
The molecular biology of the genus has developed considerably since de Vries et al. (68) first obtained a mutant of P. denitrificans that was amenable to genetic techniques. The review by Steinrücke and Ludwig (277) considered a number of aspects of the molecular biology of P. denitrificans, including a proposed promoter structure (unique to the genus) and related aspects of gene regulation. A considerable amount of new information has become available, which in part does not confirm the earlier proposals regarding promoter structure and otherwise is of general interest in the context of bacterial respiration, for which P. denitrificans is a model organism. Thus, a new review is timely. We consider the molecular genetics of the commonly used strains of P. denitrificans and, to a lesser extent P. versutus, the organisms on which the majority of structural, biochemical, and genetic work has been done.
(The sequences referred to in this review and their annotations are from GenBank 106.0 (released March 1998) and EMBL 54.0 (released March 1998), plus their cumulative updates until 1 May 1998, held at the Oxford University Molecular Biology Data Centre, Sir William Dunn School of Pathology, University of Oxford, South Parks Road, Oxford OX1 3RE, United Kingdom.)
GENETIC COMPOSITION OF PARACOCCUS
Relatively little work has been done on Paracoccus species to determine the overall genetic makeup of these bacteria, apart from their relatively high G+C content, which has been determined during classification studies (see, e.g., reference 199). However, since 82 loci have been sequenced (listed in Table 1), it is now possible to derive valuable information about the genomics of the genus.
The G+C content of the fragments of the P. denitrificans genome that have been sequenced to date is 65.63%. This compares well with the published genomic G+C content of 66.5% (145). The difference most probably arises because sequences submitted to the databases are biased toward coding regions of DNA. The total amount of P. denitrificans DNA sequenced (by 31 December 1997) is 166,864 bp, which can be estimated to be about 4% of the total genome (Table 2). This DNA contains 162 open reading frames (ORFs), of which 129 have had a function assigned to them, either by biochemical demonstration or by inference from closely related genes from other organisms. A compilation of this information for P. versutus is presented in Table 2.
TABLE 2.
Information for the ORFs sequenced from P. denitrificans and P. versutus
| Characteristic | P. denitrificans | P. versutus |
|---|---|---|
| % G+C from sequence | 65.63 | 64.90 |
| % G+C (experimental)a | 66.5 | 66.8 |
| Total DNA sequencedb | 166,864 | 16,092 |
| % of genome sequencedc | 4.17 | 0.81 |
| No. of ORFs identified | 162 | 19 |
| No. of ORFs (defined function)d | 129 | 13 |
| Total length of ORFs (bp)e | 115,489 | 11,718 |
Experimental data taken from reference 145.
The total DNA sequenced includes all known strains of P. denitrificans, including strain GB-17.
The total length of both genomes is taken to be 4,000 kbp, the size estimated for P. denitrificans by pulsed-field gel electrophoresis (323). The duplicate sequencing of the cycA (cytochrome c550) locus has been taken into account, but all other sequences are taken to be unique and nonoverlapping.
“Defined function” is taken to mean either that the gene product has had its biochemical effect identified experimentally or that the gene product has significant identity to a protein from another organism.
The shorter total length of the ORFs compared to the total DNA sequenced does not reflect large intergenic regions in Paracoccus; rather, it reflects the fact that sequencing targeted to specific genes leaves regions to the 3′ and 5′ of the loci that are not of sufficient length to have the definition “ORF” applied to them.
Within the ORFs from P. denitrificans identified so far, there is a bias at the third codon to guanine or cytosine. These nucleotides occur in the third position in 84.03% of codons. The GC bias is also reflected in the frequency at the first (61.13%) but not the second (52.73%) position (Table 3). This codon usage is slightly different from that previously reported (277) because of the larger number of ORFs considered in this study. The additional ORFs included here also contain very rare codons (CTA and TTA for leucine for example), so that all combinations are represented. Hence, the tRNA composition of P. denitrificans cannot be deduced confidently from codon usage. A similar codon bias is also seen in P. versutus (data not shown), but guanine or cytosine occurs in the first position in 67.13% of codons. The codon usage tables used in this review (Genetics Computer Group format) are available on request from the authors for both P. denitrificans and P. versutus.
TABLE 3.
Codon usage in ORFs of P. denitrificansa
| Amino acid | Codon | No. | No./1,000 | Fraction | Amino acid | Codon | No. | No./1,000 | Fraction | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gly | GGG | 155 | 12.44 | 0.16 | Trp | TGG | 320 | 25.69 | 1.00 | |
| Gly | GGA | 50 | 4.01 | 0.05 | End | TGA | 96 | 7.71 | 0.87 | |
| Gly | GGT | 56 | 4.50 | 0.06 | Cys | TGT | 26 | 2.09 | 0.12 | |
| Gly | GGC | 731 | 58.68 | 0.74 | Cys | TGC | 198 | 15.89 | 0.88 | |
| Glu | GAG | 351 | 28.18 | 0.61 | End | TAG | 7 | 0.56 | 0.06 | |
| Glu | GAA | 224 | 17.98 | 0.39 | End | TAA | 7 | 0.56 | 0.06 | |
| Asp | GAT | 126 | 10.11 | 0.23 | Tyr | TAT | 178 | 14.29 | 0.57 | |
| Asp | GAC | 425 | 34.12 | 0.77 | Tyr | TAC | 134 | 10.76 | 0.43 | |
| Val | GTG | 344 | 27.61 | 0.46 | Leu | TTG | 78 | 6.26 | 0.07 | |
| Val | GTA | 11 | 0.88 | 0.01 | Leu | TTA | 4 | 0.32 | <0.01 | |
| Val | GTT | 36 | 2.89 | 0.05 | Phe | TTT | 53 | 4.25 | 0.11 | |
| Val | GTC | 361 | 28.98 | 0.48 | Phe | TTC | 419 | 33.64 | 0.89 | |
| Ala | GCG | 608 | 48.81 | 0.39 | Ser | TCG | 349 | 28.02 | 0.45 | |
| Ala | GCA | 123 | 9.87 | 0.08 | Ser | TCA | 65 | 5.22 | 0.08 | |
| Ala | GCT | 80 | 6.42 | 0.05 | Ser | TCT | 44 | 3.53 | 0.06 | |
| Ala | GCC | 745 | 59.81 | 0.48 | Ser | TCC | 130 | 10.44 | 0.17 | |
| Arg | AGG | 142 | 11.40 | 0.14 | Arg | CGG | 242 | 19.43 | 0.24 | |
| Arg | AGA | 57 | 4.58 | 0.06 | Arg | CGA | 55 | 4.42 | 0.05 | |
| Ser | AGT | 21 | 1.69 | 0.03 | Arg | CGT | 30 | 2.41 | 0.03 | |
| Ser | AGC | 172 | 13.81 | 0.22 | Arg | CGC | 497 | 39.90 | 0.49 | |
| Lys | AAG | 279 | 22.40 | 0.81 | Gln | CAG | 253 | 20.31 | 0.81 | |
| Lys | AAA | 64 | 5.14 | 0.19 | Gln | CAA | 60 | 4.82 | 0.19 | |
| Asn | AAT | 53 | 4.25 | 0.19 | His | CAT | 118 | 9.47 | 0.55 | |
| Asn | AAC | 229 | 18.38 | 0.81 | His | CAC | 95 | 7.63 | 0.45 | |
| Met | ATG | 401 | 32.19 | 1.00 | Leu | CTG | 790 | 63.42 | 0.74 | |
| Ile | ATA | 14 | 1.12 | 0.02 | Leu | CTA | 4 | 0.32 | <0.01 | |
| Ile | ATT | 48 | 3.85 | 0.08 | Leu | CTT | 87 | 6.98 | 0.08 | |
| Ile | ATC | 533 | 42.79 | 0.90 | Leu | CTC | 103 | 8.27 | 0.10 | |
| Thr | ACG | 230 | 18.46 | 0.32 | Pro | CCG | 458 | 36.77 | 0.54 | |
| Thr | ACA | 54 | 4.33 | 0.07 | Pro | CCA | 64 | 5.14 | 0.07 | |
| Thr | ACT | 21 | 1.69 | 0.03 | Pro | CCT | 61 | 4.90 | 0.07 | |
| Thr | ACC | 416 | 33.39 | 0.58 | Pro | CCC | 272 | 21.84 | 0.32 |
Including P. denitrificans GB-17.
Megaplasmids and Genomic Structure
Bacterial genomes are generally believed to be a single circular DNA molecule, with the model being E. coli. The alpha subgroup of the Proteobacteria, a division which includes Paracoccus, contains notable exceptions to this: Rhodobacter sphaeroides has two circular chromosomes (282), and Rhizobium meliloti has three (267). Agrobacterium tumefaciens C58 also has two chromosomes, but one is linear and the other is circular (2). The reason why these bacteria have multiple replicons is obscure: although some genes are duplicated (for example, the two copies of the carbon dioxide fixation genes in R. sphaeroides on separate chromosomes [93]), genes forming enzymes for a complete pathway are scattered over all the replicons (see http://capsulapedia.uchicago.edu for emerging results on R. capsulatus). P. denitrificans has also proved to have an unusual genomic structure.
When chromosomal DNA from P. denitrificans Pd1222 was separated by pulsed-field gel electrophoresis, it became apparent that the genome consists of three distinct DNA molecules of 1.83, 1.16, and 0.67 Mbp (323), designated molecules I, II and III, respectively. The behavior of the molecules under various electrophoretic conditions suggested that at least the two smaller ones were linear. To determine if the molecules were large plasmids conferring specific properties to P. denitrificans, probes to respiratory genes were used to gain an insight into gene distribution. Genes coding for the aa3-type oxidase were spread between molecules I and II, while ubiquinol oxidase genes were found on molecule III. The genes encoding specific pathways, if transcribed from separate loci, appeared to be randomly distributed: for example, a methanol oxidation gene (mxa) was found on molecule I but the cytochrome c550 structural gene (cycA) and the S-formylglutathione hydrolase gene (fghA) were found on molecule II. It thus seems likely that these three replicons comprise the P. denitrificans genome and will not be replicated independently of one another (323). However, the presence or absence of rRNA genes was not investigated, and so it was not possible to say which, if any, of these molecules were true chromosomes.
The composition of the genomes of other strains and species of Paracoccus varies. P. denitrificans GB-17 and DSM 65 both possess four DNA molecules of 2.2, 1.5, 0.71, and 0.5 Mbp (323) and are proposed P. pantotrophus strains (233). The electrophoretic characteristics of the 0.71-Mbp molecule indicate that this molecule is in closed-circular rather than linear form. Additionally, a much smaller molecule of less than 1 Mbp was seen in some preparations (323). A plasmid (pTAV1) of 107 kbp has been isolated from P. versutus and has been used to construct minireplicons (8, 9). P. versutus cured of the plasmid retained wild-type growth characteristics, except with respect to cesium and barium resistance (8). A second linear replicon (pTAV2) has also been found in P. versutus (201).
Restriction and Modification Systems and the SOS Response
The possession of an efficient means of ameliorating the effects of the introduction of foreign DNA into a cell is an important trait for a microorganism living in environments where mixed cultures occur. However, when these bacteria are transferred from their environment to the laboratory, DNA restriction and modification systems present a problem to the molecular geneticist. Studies of regulation in P. denitrificans NCIMB 8944 (traditionally used for biochemical studies) were hampered by the lack of a mutant suitable for the maintenance of plasmids without significant recombination into the genome. Stable inheritance of extrachromosomal material does, however, occur in P. versutus, as well as in P. denitrificans GB-17. Furthermore, the type culture of P. denitrificans (ATCC 17741) will maintain plasmids in the wild-type form of the strain (143).
An undefined P. denitrificans N-methyl-N′-nitro-N-nitrosoguanidine chemical mutant (Pd1222) which had a recombination-minus phenotype and an enhanced frequency of conjugation was isolated from DSM 413 (68). The useful property of resistance to rifampin was subsequently introduced, and this antibiotic resistance can be used to select against E. coli strains present in bi- or triparental mating experiments. However, the apparatus for recombination of plasmid DNA with the genome still remained in this mutant, and to manufacture a truly recombinant-deficient organism, Fernandez de Henestrosa et al. (75) isolated and mutated the recA gene of P. denitrificans Pd1222. This new derivative should prove valuable in future work.
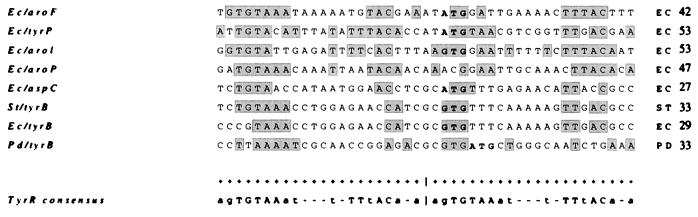
Despite the high identity of recA proteins within the Proteobacteria (144) and the high identity within the coding regions (P. denitrificans recA is 88.6% identical to the Rhodobacter sphaeroides gene and 64.3% identical to that from E. coli [75]), regulation of P. denitrificans recA differed not only from that of E. coli but also from that of the phylogenetic near neighbor R. sphaeroides. No LexA binding site could be seen in the putative promoter region of P. denitrificans recA, but the use of a plasmid containing the promoter translationally fused to a reporter suggested that conditions for repression and activation of the gene in P. denitrificans were similar to those required by E. coli. Further evidence for differences in the details of control of the recA gene in P. denitrificans were obtained when the reporter gene was fused to recA promoters of Rhizobium etli, R. sphaeroides, and R. capsulatus (75). When these fusions were introduced into P. denitrificans, the reporter was induced (on the addition of mitomycin C, which induces the SOS response) only from the Rhizobium etli promoter. This was not the expected result in view of the closer phylogenetic relationship of Paracoccus and Rhodobacter than of Paracoccus and Rhizobium. Examination of the promoter sequences revealed little similarity between the Rhodobacter promoters and that of Paracoccus, but the Rhizobium etli promoter contained a similar region of dyad symmetry (5′-TTGN10CAA-3′ in P. denitrificans and in R. etli, N = 11). Interruption of this inverted repeat in Rhizobium etli led to inactivation of the recA promoter (284). It would thus appear that P. denitrificans possesses a recA system more like that found in the rhizobia than in Rhodobacter species.
rRNA Genes
rRNA functions in the assembly of the ribosome but has assumed new significance with the realization that it can be regarded as a molecular clock (204, 329). The 5S, 16S, and 23S genes of P. denitrificans have been sequenced (Table 1), as have the 16S genes from all of the other species of Paracoccus. Unfortunately, due to the use of thermal polymerase amplification involving primers to conserved sequences within the genes, little information can be obtained about the promoters, which would be expected to be of the ς70 RNA polymerase (RNAP) type. The derivation of a consensus ribosome binding site from the 16S rRNA sequence has been discussed previously (277).
The transcript from the 23S gene is unusual in that it seems to be unstable in some preparations when isolated with total RNA from P. denitrificans GB-17 (P. pantotrophus), appearing on formamide-agarose gels as two smaller molecules (one the same size as 16S rRNA) cleaved at a distinct site (252). This phenomenon has also been noted in R. capsulatus (343). Since the integrity of the 16S rRNA transcript is often used as an indicator of the state of degradation of RNA, this may give a misleading result when considering the quality of a P. denitrificans GB-17 total RNA preparation. The instability of the 23S rRNA might indicate the presence of an intervening sequence (usually originating from an insertion sequence or other mobile genetic element, appearing as inverted repeats and/or an ORF[s] in the middle of some rRNA genes). Such intervening sequences have been found in several bacteria, including Salmonella typhimurium, and result in no apparent intact 23S rRNA in the cell (105).
Insertion Sequences
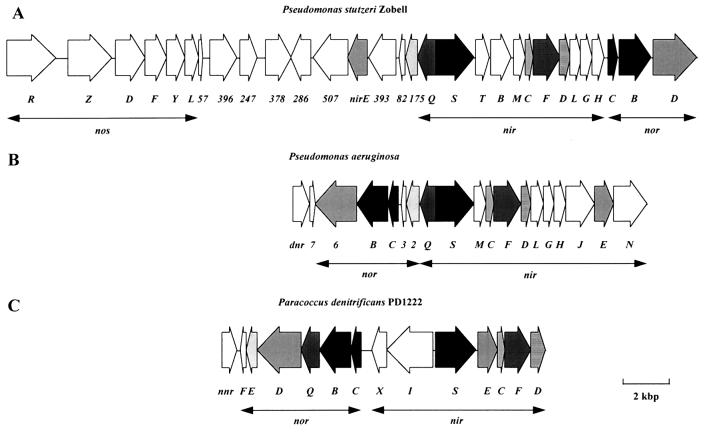
Bacterial insertion sequence (IS) elements are small, discrete elements of DNA that are integrated into the host genome or, more frequently, into naturally occurring plasmids in bacteria. The coding capacity of these elements is often limited to the synthesis of transposase, the protein which drives the transpositional event and allows the element to jump along the host DNA. The IS element IS1248, which was characterized in P. denitrificans, belongs to a larger family of elements that are found in strains belonging to different clusters of gram-positive as well as gram-negative bacteria. This family includes IS869 and IS427 of Agrobacterium tumefaciens (67, 224), IS402 of Pseudomonas cepacia (77), ISmyco of Mycobacterium tuberculosis (183), IS1106 of Neisseria meningitidis (156), Tn4811 of Streptomyces lividans (44), ISRm4 and a similar element from Rhizobium meliloti (84, 227, 268), and IS1031 of Acetobacter xylinum (50). Trapping of IS1248 occurred during plasmid transfer experiments with derivatives of suicide vector pRVS3, which appeared to be integrated into the genome via IS1248-mediated cointegrate formation (311). The finding that the vector was flanked by identical copies of the transposed IS element as well as of the target site, 5′-CTAG-3′, even suggested that integration had occurred via replicative transposition, an event which is preceded by a staggered cleavage of the IS target site, resulting in duplication of it. IS1248 is 830 bp long and has 13-bp imperfect inverted repeats at the borders. Two of the five ORFs identified in IS1248 correspond to counterparts from the other members of this IS family. Since these putative genes have the potential to encode proteins that are hydrophilic overall and have relatively high isoelectric points, they might be the candidates for the transposase function. Two sequences are found in the inverted repeats of IS1248, which have been suggested to be involved in the transpositional event. The first sequence, 5′-GANNNNTTGAT-3′, resembles the binding site for the integration host factor, which is involved in stimulation of transposition of a number of IS elements (90). The second sequence, 5′-GNNTCATAA-3′, is identical to that found in related elements and may be a recognition site for their transposases. IS1248 is present in multiple (four to six) copies in the genome of many strains of Paracoccus (312), and the pattern of IS1248-hybridizing fragments appeared to be different in P. denitrificans Pd1222 and P. denitrificans GB-17 (P. pantotrophus). IS1248 is not present in P. versutus, suggesting that it invaded P. denitrificans after these two species had branched from a common ancestor. This suggestion would support the idea of horizontal gene transfer (312).
Apart from the IS1248-mediated integration mechanism, P. denitrificans has a second mechanism involved in the integration of heterologous DNA into its genome (312). The result of the latter type of integration is different from that observed for IS1248, in that the integrated DNA is not flanked by two identical sequences. Furthermore, the DNA sequences of the donor backbone and the target DNA at the integration site were found to be similar and to resemble the res site found in transposons belonging to the Tn3 family (90, 152). These res sites are an essential part of the transposon-mediated site-specific recombination system involved in cointegrate resolution. At least two copies of this integrative element are present in the genome of P. denitrificans (312).
Regulation of Transcription in Paracoccus
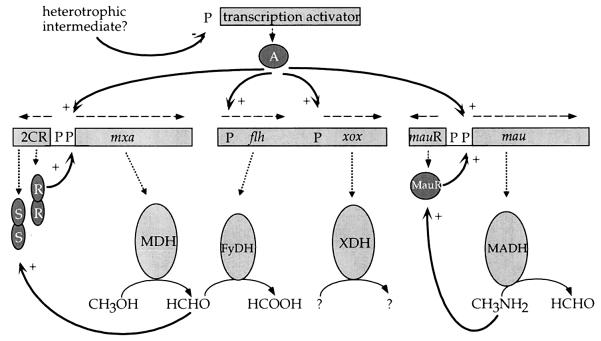
When considering how and when a particular gene from Paracoccus is transcribed, researchers find themselves in an unusual position. It is possible to define transcript start sites and some regulatory protein binding sites (such as FNR-type proteins [310]) but not to determine where RNAP might bind or even which type of RNAP is effective. Consensus sequences that have been proposed previously (277) for Paracoccus are, as discussed below, unsuitable. In the absence of any direct biochemical or genetic evidence for the presence of an RNAP of the ς70 type in Paracoccus, it is difficult to define clearly the elements of promoters from this genus that may be involved in transcription. The promoter regions that have been sequenced rarely contain the typical −10 or −35 motifs, and workers studying Paracoccus frequently note that its promoters rarely function in aerobically grown E. coli.
An obligately respiratory organism such as P. denitrificans achieves metabolic flexibility by having many alternative electron transport chains. The bacterium must have some overall control of these branched electron transport pathways it possesses: in many cases, the concentration of more than one respiratory enzyme is either elevated or diminished under a particular growth condition, suggesting that a single regulatory protein has pleiotropic control over the expression of their allocated genes. Comparison of the promoter regions in front of the known respiratory genes and gene clusters revealed a number of sequences with a minimum of 8 bases conserved in two or more of the putative promoter regions. Palindromic sequences, which may be probable candidates for binding transcriptional activators or DNA binding proteins, can be selected from these conserved sequences. A list of these sequences is presented in Table 4. Whether these sequences indeed act as regulatory elements is speculative at the moment, but these palindromes do not resemble those noted in R. capsulatus (5′-GTGTAART-N6-TTACAC-3′ [1]), nor, in most cases, do they conform to the consensus sequence for E. coli transcriptional regulatory factors (5′-TGTGT-N6–10-ACACA-3′ [95]).
TABLE 4.
Palindromic sequences present in regions upstream of respiratory genes
| Palindrome (consensus) | Sequence | Promoter | Distance relative to:
|
|
|---|---|---|---|---|
| Transcriptional starta | Translational start | |||
| ACG...CGT | ACGG.CCGT | fbc | −22 | 52 |
| ACG...CGT | etf | NNb | 90 | |
| T.GCA.....TGC.A | T.GCAGC.GCTGC.A | nqo | NN | 125 |
| TCGCA.....TGCGA | ccoN | NN | 45 | |
| TCGCA.....TGCCC | nap | NN | 11 | |
| T.GCAA...TTGC.G | ccoN | NN | 192 | |
| TCGC...GCGA | TCGCA.TGCGA | ctaDII | NN | 82 |
| TCGCC.TGCGA | cycA | 81 | 133 | |
| TCGCA.GGCGA | qox | 44 | 98 | |
| TCGCT.ACCGA | qox | −34 | 20 | |
| TGCC.GGCA | TGCT.GGCA | ccoG | NN | 79 |
| TGCC.GGCA | ccoG | NN | 174 | |
| TGCC.GGCA | qox | 129 | 183 | |
| CCTGC.....GCAGG | CCTGCCG.CGGCATG | ctaC | NN | 25 |
| CCTGC.....GCAGG | ccoN | NN | 207 | |
| ATC....GAT | ATCCCGGTAT | ctaDI | NN | 42 |
| ATCCCGAGAT | ccoG | NN | 272 | |
| ATTCCGGGAT | ccoN | NN | 151 | |
| ATCG..CGAT | nap | NN | 294 | |
| ATC.GC.GAT | cycM | NN | 5 | |
| ATC.GC.GAT | sdh | −5 | 128 | |
| TGC......GCA | TGCG....CGCA | nap | NN | 124 |
| TGCG....CGCA | ccoG | NN | 46 | |
| TGC.TTAG.GCA | fbc | 126 | 200 | |
| TGC.A..T.GCA | qox | 5 | 59 | |
Minus sign indicates that it is located downstream.
NN, not known.
Promoter structure in Paracoccus and the Rhodobacter group of the alpha Proteobacteria.
There has been no further review of Paracoccus promoter sequences since Steinrücke and Ludwig (277) deduced a consensus sequence (5′-TCGGGGN-N(18 ± 2)-GATNGS-3′) based on promoters from Paracoccus, Rhodobacter, and Bradyrhizobium. Surprisingly, little attention has been paid in general to promoters in the alpha-Proteobacteria, the division of the Eubacteria to which Paracoccus belongs. Although alternative polymerases (e.g., RpoN [37] of R. capsulatus) have been isolated, purified, and characterized, work on the binding of housekeeping holopolymerase to constitutively induced promoters is just beginning (54, 180), with Rhodobacter as the model organism. However, alignment of Paracoccus and Rhodobacter promoters (Table 5) indicates that most constitutive promoters have some sequences in common.
TABLE 5.
Promoters from the Rhodobacter group of the alpha Proteobacteria
| Strain/promotera | Promoter regionb | Reference(s) |
|---|---|---|
| Ec/consensusa | TTGACA N15–19 TATAAT | 120 |
| Group A | ||
| Rc/bchC | ATCAAATTGACAGTCGGGCGTGTAAGTTCAATGATACACACAGG | 54 |
| Rs/groE | CCCCCGTTGACAGGTGCCGGACGCTCTCATATCTCTCGCGTCG | 166 |
| Pd/qoxA* | TTGACCTAGATCAAGGTAATTCACCCCGCCATGTTGC | 346 |
| Rc/radC | GAGATCTTGACGAGGCGCAGCCGGTTGCCGTCTCTTCCGACC | 197 |
| Rs/trxA | GACAACTTGACGCGGCAGGGGGCCATCCTTACTTTCCAAGCA | 222 |
| Rc/recA | ATTTGATTGCAATGTTCCGCAATTGTACTTCATACCATGAGAA | 76 |
| Rc/cpeA | ATGCAATTGGAATTTATCAAAATCGCCTTCTAGTTTCAATCGC | 81 |
| Rc/puc | TTACACTTGATCGCCGACACTTGGGCTCCCATAGTGCGTCTCA | 54 |
| Rc/fdxA | CTGCTCTTGATTGATCGCCCCCGGAGGGCTAGGACATCCC | 72, 248 |
| Rc/atpH | TGGTTGCGAGGGTCTTGATGCTCTGCTAGACGCAACCCCCGA | 31 |
| Rs/hupS | TTCTCATTGGCGGAAACCGGCCCAACCATGAGAATTCCACTCTA | 288 |
| Rs/coxII | TTTTCCTTGTCGTCGCCAAAACAATATGGTCTCAATCGGTTCA | 79 |
| Rc/lepB | TTGACTTGCCCCCCGCATCGGCCAAGGAAGACAAGGTTTCAAC | 154 |
| Rc/dnaK | CAGTTCTTGCAGGGCTATTTTCCCCTCCTTATATACGCCGG | 198 |
| Rs/rrnB | ATCCGCTTGCGCCCGGGGCCGTCTGCTCCTAGAAACCGCTTC | 71 |
| Rs/rrnA | TTCCTCTTGCGGGTTTTTTTGCGGTTCCCTAGATAGCGCCTC | 71 |
| Rs/rrnC | TTCCTCTTGCGGGTTTTTTTGCGGTTCCCTAGATAGCGCCTC | 71 |
| Rc/rpoD | TTTGATTCGCCCCTGTGATTCGCCGCCGTGATTCGATAAAAC | 223 |
| Group B | ||
| Pd/sdhC | ATGTGATCACAGCTGCCGTTTGCGTGATCACAAAAATGGAAA | 69 |
| Rc/porA | GGCCGCTTCGGTCCCGTTTTCGTTGCAATATGAGGCGTGG | 289 |
| Rs/cycF | TTCCAGTATGTCTGTCGCGACCGGCGGGCTAGATTTCCGGGA | 79, 80 |
| Rs/norC | CCCGCAACCTCTCCGGCGCCGGCGCGGGCTAGAGGAGGTCCA | 7 |
| Pd/ctaC* | GATAGGTATGGCTTGCCGCCGGGGTAAGATATGGTTCTGGTG | 277 |
| Pd/fbc* | CCGCTGGACTGACGGGGATTTGATCGCTAGAACCGCGC | 277 |
| Pd/porG* | ATGCGGGGGGGAAAGCGCGACACCACTTGCATACCCCAAAC | 254 |
| Rc/pufG | CTCTGGACCGGATCGTGTCGCAACACCCGGTTCTGACACGGA | 11 |
| Rc/pufQ | CGCGCGACGGGCACCCCCTTCATGGTTACATGGGTA | 54 |
| Rs/chrR1 | GCCTGATCCAGACTGGCCCGGCCGCCGTAAGAAGGACGTTA | 257 |
| Rs/chrR2 | GCGGATGCAAGCCGGGGGAGGGTTTCCTATCTTCACCTCCGG | 257 |
| Rc/cycA | ATATGCGACCTTTTGCCTTGTGGTAAAGCGTGGACCG | 54, 70 |
| Pd/pazS | TTCTCAAGCCGAATGCCCTGTACCGGACCTAAACCAGGCACG | 168 |
| Group C | ||
| Pd/cycA1 | CCTGTATTCTGCCCGCTTGGCACATGATAGCCCTGTCAATCG | 279 |
| Pd/cycA2 | TCTGCCCGCTTGGCACATGATAGCCCTGTCAATCGGGAAGCG | 279 |
| Rc/fruB | TTTCGCGCGCGAACTCTGCCCCATGGGCGATGGCCG | 54 |
| Rc/xdhA | TTCAACGCGCCGCGGGCCGAAATCCTTGCCAGCGCCAAAGCC | 167 |
| Rs/crtA | GGGCGGACATTAGTCGCGAAGTCGCACCGCCCTCAAGCCGCA | 164 |
| Rs/ctaD | GGAACAAGACCGGCGTCTACAGATATCCGGGAGATGCTGGCA | 79 |
| Rs/cycA | CCGGAACGCGCGGCCCGCAGTAGTGATTGTGTGCCGGCGGCA | 70 |
| Rs/fbcA | GCGCCGCAAGATCGAGCCCGACCCGCGCGAGCCGCGCTACCT | 89 |
| Rs/nirK | CGCAAACTCCGGCCTCTCCAGAGGATCTACCGATCGGGTCAT | 7 |
| Rs/pucT | GAATCTGTCAGCGCAATGTGACACCCATAATGCGAGCCGGGG | 94 |
| Rs/pufG | ATCCGCCGCGCGACGGGCACCCCCTTCATGGGTTACATGGGT | 151 |
All promoters have had their transcript start sites (the last base in each sequence) determined by primer extension or S1 nuclease (∗) assays. Group A consists of promoters possessing either or both of the −10 and −35 hexamers and are ranked according to the identity of the −35 region to that of the E. coli consensus. Group B consists of promoters with a −10 region 5 to 9 bp from the transcript start site but no −35 region. The promoters in group C have RNAP binding sites that cannot be identified easily, but this does not necessarily disqualify them from ς70 RNA polymerase dependency. R. capsulatus glnB, nifA1, and nifA2 promoters are not included because they have been shown to be RpoN-dependent (82). Pd, Paracoccus denitrificans; Rc, Rhodobacter sphaeroides; Rs, Rhodobacter capsulatus; Ec, E. coli.
Underlined hexamers have 50% or more identity to the appropriate E. coli consensus.
Few genes from Paracoccus have had their transcription start sites determined, an essential step in defining the exact position of possible promoter sequences. The start sites for qoxA (quinol oxidase [346]), sdhC (succinate dehydrogenase [69]), ctaC (cytochrome oxidase [277]), fbc (bc1 complex [277]), porG (porin [254]), pazS (pseudoazurin [168]), and cycA (cytochrome c550 [279]) have been published, and determination of the nir genes is in progress (5). Of these, one would expect the promoters for succinate dehydrogenase, cytochrome oxidase, bc1 complex, quinol oxidase, and porin to be under the control of a housekeeping polymerase, in that they are constitutively produced. They all have an unusual base usage biased towards A and T in the 40 bp upstream of the transcript start site (41 to 60% G+C, compared with 66.5% G+C for the whole genome), the expected promoter location. A · T base pairing in the −10 (TATAAT) and −35 (TTGACA) regions of the model E. coli ς70-dependent promoter is thought to contribute to local melting of the double helix, allowing holo RNAP to function (reference 36 and references therein).
Since comparatively few Paracoccus promoters have been thoroughly characterized, little can be deduced if they are considered in isolation. However, if the reasonable assumption is made that transcription in closely related bacteria will be very similar, results obtained within the Rhodobacter group (essentially R. capsulatus and R. sphaeroides as well as P. denitrificans) can also be considered. Although this allows the study of 45 promoters, the majority of these are from genes which are considered to be highly regulated. Data for more promoters from genes with housekeeping functions would provide more information for derivation of a consensus for the equivalent of the ς70 RNAP binding site. However, alignment of the promoters with respect to their transcript start sites does reveal sequences in some promoters that resemble those of the E. coli ς70 consensus sequence.
Although studies of mutants with site-directed mutations of the bch operon of R. capsulatus (180) indicated that transcription was dependent on bases at −10 and −35, no promoter yet characterized from Rhodobacter or Paracoccus has sequences (Table 5) that conform exactly to the canonical E. coli ς70 motif (5′-TTGACA-N(15–19)-TATAAT-N(5–9)-3′, first derived by Hawley and McClure [120]). In vitro and in vivo studies of five R. capsulatus ς70-dependent promoters (54) have led to a loose definition of the likely housekeeping holopolymerase binding sites: the −35 hexamer was found to be TTGACN, and the −10 motif was of such variable composition that “AT rich” was a sufficient description (54). Examination of the R. sphaeroides rrn operon promoters again revealed −35 regions resembling those of E. coli (Table 5) but found less similarity in the −10 region (70): it could have been expected that the rrn operons would have strong promoters conforming to the consensus ς70 for the genus.
Alignment of the promoters from the Rhodobacter group still gives no clear picture of a consensus sequence (Table 5). The promoters fall into three groups: those with a sequence at −35 with 50% or more identity to the E. coli consensus; those with a sequence at −10 with 50% or more identity; and those that have polymerase binding sites which are not easily identifiable from sequence data alone. Most of the Paracoccus promoters fall into one of the first two categories, but, surprisingly, neither of the cycA promoters (279) can be included in these groups. Although the influence of ς70 cannot be ruled out completely, since footprinting studies have not been performed, it has been demonstrated that cycA is transcribed under all the conditions tested (279, 315), including aerobic or anaerobic growth on succinate and aerobic growth on methanol or methylamine. Given the results obtained with R. capsulatus purified ς70 (54), it seems likely that qoxA is transcribed with the aid of this sigma factor, since the hexamer TTGACC appears 35 bp upstream of the transcription start site (Table 5). However, the definite assignment as “ς70 dependent” to the Paracoccus promoters that have a −10 hexamer alone is questionable. The constitutive expression of succinate dehydrogenase, cytochrome oxidase, the bc1 complex, and porin has been experimentally determined, but the variation in composition and the position of candidate −10 hexamers show that the sequence results are only a very preliminary indication.
Considering all the promoters aligned in Table 5, the shortcomings of a sequence-based approach are illustrated. For example, the R. sphaeroides ctaD (cytochrome oxidase) gene has no clear −10 or −35 sequences, yet P. denitrificans ctaC has a −10 hexamer 67% identical to that of the E. coli consensus. However, the alignment does show that TTG in the −35 hexamer is a common characteristic of Rhodobacter group promoters. Because it is so difficult to identify candidate promoter regions, we suggest that definition of polymerase binding sites should be attempted only if the promoter/operator has been mapped by either S1 nuclease protection assay or primer extension experiments.
Even though it is not yet possible to derive a consensus sequence for a Paracoccus promoter, there are indications that a ς70-type RNAP is present in the cell. The best evidence for this in P. denitrificans comes from studies with a broad-host-range vector containing the promoter of bacteriophage T4 gene 32 fused to xylE (85). After construction of the vector in E. coli, the plasmid was introduced by conjugation into Agrobacterium, Erwinia, Xanthomonas, Pseudomonas, and Paracoccus species and the transcription of the fusion was studied in comparison with that in E. coli. The transcription start site of the plasmid-borne fusion was identical in all the genera studied and was found to be downstream of hexamers resembling a ς70 promoter. In addition, the resulting mRNA molecules were subject to posttranscriptional modification and were processed in a similar manner. Although it could be argued, in the absence of ς70 footprinting studies in each strain, that the transcript arose from fortuitous promoters, the exact coincidence of the transcript start sites is indicative of a common core polymerase subunit.
Although the promoters in the Rhodobacter group appear to be anomalous compared to the canonical ς70 promoter, recent work (106) has suggested that all the eubacteria have a very similar principal sigma factor. Examination of the variation in sigma factor protein sequences suggests that all housekeeping polymerase-dependent promoters have DNA sequences at −10 and −35 which bear some resemblance to the classical binding sites proposed by Hawley and McClure (120). The differences exhibited by the promoters in Table 5 could be explained by the interaction of polymerases with other unknown protein factors, rather than some intrinsic difference in the holopolymerase itself. The requirement for other factors may go some way to explaining the inability of E. coli to initiate transcription from the Paracoccus promoters tested so far.
Termination of transcription.
To date, no direct experimental evidence exists for any termination event in Paracoccus. However, mRNA analysis and other indirect evidence suggests that Paracoccus possesses both factor-dependent and factor-independent pathways for termination of transcription. A truncated form of the Rho-dependent terminator gene (rho′) of R. sphaeroides 2.4.1 was lethal in the wild-type organism but partially interfered with the transcription termination machinery of E. coli. When rho′ was introduced into P. denitrificans ATCC 17741, the construct was also found to be toxic (99). This suggests not only that a Rho-like system exists in the genus Paracoccus but also that the mechanism of termination is the same for R. sphaeroides and perhaps that the structure of Rho in these organisms is similar as well.
Some of the stem-loop structures found downstream of genes such as cycH (involved in cytochrome c biogenesis [215]) conform to the classical factor-independent structure (a stem-loop followed at the 3′ end by an AT-rich region). The run of A or T is particularly noticeable against the GC-rich Paracoccus DNA, but it includes some guanine and cytosine residues. A similar factor-independent termination site can be proposed when considering the region between nirS (cytochrome cd1 nitrite reductase) and nirE (a putative methyl transferase) of P. denitrificans (59). Studies involving Northern blotting indicate that this terminator is functional during denitrifying growth in P. denitrificans (252).
Many transcriptional terminators have been deduced from putative stem-loop structures in DNA sequences. Stem-loops with highly negative Gibbs free energy can be found within one of the methanol oxidation operons (mxaFJGIR). However, these structures may equally confer stability to mRNA, forming the sort of stem-loop structures found in the puf operon of R. capsulatus (121).
In summary, the genus Paracoccus contains bacteria with multiple replicons. Genes and insertion sequences are distributed among these replicons, and to date no megaplasmid has been assigned a specific function. Despite a growing amount of sequence data and analysis, no obvious consensus promoter sequence is immediately apparent by simple inspection. This has led to the idea that P. denitrificans, along with the members of the alpha Proteobacteria, has very different promoter regions from those in E. coli. However, careful analysis of a variety of available data suggests that at least for some of the genes there is similarity to typical ς70 hexameric sequences. It also seems probable that there are novel regulation mechanisms (e.g., involving different regulators or sigma factors) yet to be discovered in P. denitrificans and its relatives.
REGULATION OF RESPIRATORY GENES
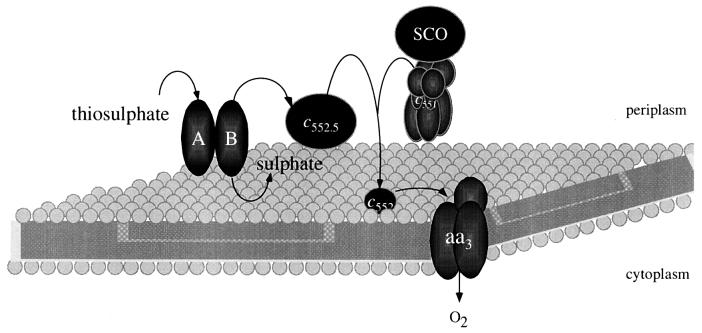
A number of respiratory systems and their underlying biosynthesis genes from Paracoccus have been characterized by molecular biological methods. The regulation of these genes has some features in common with the regulation of the genes in the more intensively studied organisms such as E. coli. The loci sequenced, their accession numbers, and the strain of origin are listed in Table 1. The respiratory pathways of Paracoccus are dependent on many metalloproteins, the best characterized of which are c-type cytochromes (Table 6). Although the biochemical mechanisms by which these proteins are synthesized are only just becoming understood, P. denitrificans has proved to be a good model organism for these studies, producing c-type cytochromes aerobically as well as under oxygen limitation.
TABLE 6.
Well-characterized Paracoccus c-type cytochromes
| Cytochrome c | Mol mass (kDa) | No. of c heme centers | Gene | Role or probable role |
|---|---|---|---|---|
| Cytochrome c550 | 14 | 1 | cycA | General periplasmic electron carrier |
| Cytochrome c1 | 45a | 1 | fbcC | Component of bc1 complex |
| Cytochrome c552 | 22 | 1 | cycM | Electron donor to aa3-type cytochrome oxidase |
| Cytochrome c551i | 18 | 1 | mxaG | Electron acceptor for MDH |
| Cytochrome c553i | 23 | 1 | cycB | Putative electron carrier during methylotrophic growth |
| Cytochrome c551 (putative) | 40 | 2 | soxD | Electron transport during growth on thiosulfate |
| Cytochrome c′ | 12 | 1 | Unknown | |
| Cytochrome c peroxidase | 42 | 2 | Removal of hydrogen peroxide from the periplasm | |
| Cytochrome cd1 | 65 | 1 | nirS | Nitrite reductase |
| CcoO | 30 | 1 | ccoO | Component of cbb3-type cytochrome oxidase |
| CcoP | 45 | 2 | ccoP | Component of cbb3-type cytochrome oxidase |
| MauG | 40 | 2 | mauG | Synthesis of prosthetic group of MADH |
| NapB | 15 | 2 | napB | Component of periplasmic nitrate reductase |
| NapC | 27 | 4 | napC | NirT homologue, electron donor to periplasmic nitrate reductase |
| NirC | 10 | 1 | nirC | Unknown |
| NorC | 14 | 1 | norC | Component of nitric oxide reductase |
| SoxE | 26 | 1 | soxE | Electron transport during growth on thiosulfate |
Cytochrome c1 migrates with an apparent molecular mass of 60 to 68 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Cytochrome c Biogenesis
Cytochromes c are distinguished from cytochromes of other classes by covalent attachment of the heme moiety to the cytochrome polypeptide via thioether links between the two protoporphyrin IX vinyl groups and the thiol groups of two cysteine residues in the conserved motif Cys-X-Y-Cys-His. The process of c-type cytochrome biosynthesis thus includes posttranslational modification of the apocytochrome polypeptide. A number of lines of evidence indicate that in gram-negative bacteria this process takes place in the periplasm, although this has yet to be rigorously demonstrated experimentally. Eight genes required for c-type cytochrome maturation have now been identified in P. denitrificans; all are clearly homologous to genes found in a number of other gram-negative bacteria including Bradyrhizobium japonicum, R. capsulatus, and E. coli (for comprehensive reviews, see references 218 and 285). The organization of the known P. denitrificans c-type cytochrome biosynthetic genes resembles that in R. capsulatus, in that they are distributed over at least three loci, but differs from that in E. coli, in which genes are clustered at a single locus (ccmABCDEFGH), and in the Rhizobiaceae, in which the genes are present at two loci (cycHJKL and cycVWZXY). Southern blotting of a cosmid library suggests that the three loci are separated by at least 20 kbp in the P. denitrificans genome (200). The E. coli nomenclature has been adopted for the P. denitrificans genes; an exception is cycH, which has no clear equivalent in E. coli (although it exhibits some similarity to the C-terminal region of ccmH). No gene corresponding to ccmE/cycJ has been identified in P. denitrificans but has for R. capsulatus (see http://capsulapedia.uchicago.edu).
ccmA, ccmB, ccmC, ccmD, and ccmG.
ccmA, ccmB, and ccmC appear to encode the components of a membrane transporter of the ABC (ATP-binding cassette) superfamily. The corresponding hypothetical transporters in B. japonicum and R. capsulatus have been suggested to translocate heme or apocytochromes to the periplasm; however, sequence analysis indicates no similarity between CcmB and CcmC (or their homologues) and the membrane-integral components of transporters mediating the uptake of heme or other iron complexes. Supplementation of growth media with heme did not stimulate c-type cytochrome formation in mutants disrupted in ccmA or ccmB, although it elevated the levels of soluble hemoproteins and membrane-bound cytochromes b, suggesting that exogenous heme can traverse both outer and inner membranes in P. denitrificans. Expression of an apocytochrome c550-alkaline phosphatase fusion protein and of apocytochrome cd1 was unaffected in a ccmB::Tn5 mutant. These results suggest that the substrate for the putative CcmABC transporter may be neither heme nor c-type apocytochromes (217).
CcmD is predicted to comprise a single membrane-spanning α-helix and a small (about 30 amino acids) cytoplasmically oriented hydrophilic domain. Its function is obscure. The ccmG gene encodes a soluble periplasmic thioredoxin-like protein; disruption of ccmG (P. denitrificans DP307; ccmG::Ω) not only abolished c-type cytochrome biogenesis but also almost completely eliminated assembly of the aa3-type cytochrome oxidase and rendered the mutant strain incapable of growth on rich media such as Luria-Bertani medium (215). Dithiothreitol promoted the growth of DP307 on rich media and substantially restored assembly of the aa3-type cytochrome oxidase, although it did not restore c-type cytochrome biogenesis. Assembly of the disulfide-bridged proteins methanol dehydrogenase and E. coli alkaline phosphatase was unaffected in DP307. CcmG is proposed to act in vivo to reduce disulfide bonds in certain protein substrates including c-type cytochrome polypeptides and/or polypeptides involved in c-type cytochrome biogenesis (216).
cycH.
Disruption of cycH (ccmI has also been suggested as a suitable name [218]) results in loss of soluble c-type cytochromes, but low levels of membrane cytochromes c (estimated at 5 to 10% of wild-type levels) remain. Thus, CycH is not absolutely required for c-type cytochrome assembly in P. denitrificans, but it clearly increases the efficiency of the process manyfold (215). Analysis of a cycH-lacZ fusion indicates that it is expressed during aerobic growth but is induced fourfold under anaerobic growth conditions and that this induction is mediated by the transcriptional activator FnrP but not by the closely related protein Nnr (210). FnrP and Nnr are discussed further in the context of the regulation of denitrification (see below).
ccmF and ccmH.
The P. denitrificans ccmF and ccmH have recently been established (225). CcmF is predicted to be a membrane-integral protein with 11 or more membrane-spanning α-helices, and, as such, it is potentially a transporter; however, supplementation of growth media with heme did not stimulate c-type cytochrome formation in a mutant disrupted in ccmF. CcmH has a Cys-X-X-Cys motif and thus may be a protein-disulfide oxidoreductase, but a ccmH mutant has yet to be constructed and characterized.
hemA.
While not sensu stricto a c-type cytochrome biogenesis gene, the P. denitrificans hemA gene (coding for 5-aminolevulinic acid [5-ALA] synthase) was identified during screening for mutants defective in c-type cytochrome assembly. A transposon mutant in which Tn5::phoA had integrated in the hemA promoter region, reducing but not eliminating hemA expression, was obtained. This had the effect of reducing the levels of a- and b-type cytochromes and membrane-bound c-type cytochromes in the mutant strain to about 50% of those in Pd1222 and virtually eliminating the formation of soluble periplasmic cytochromes c. Disruption of the hemA structural gene led to 5-ALA auxotrophy, indicating that P. denitrificans, like R. capsulatus but unlike R. sphaeroides (127, 195), possesses only one 5-ALA synthase (confirmed by Southern blotting) and that no 5-ALA synthase-independent route of 5-ALA synthesis exists in P. denitrificans (214).
Genes of Oxygen Respiration
All species of Paracoccus are obligately respiratory and have no pathways that allow them a fermentative mode of growth. Although P. denitrificans is noted for the similarity of its aerobic electron transport chain to that of mitochondria, it differs from them in that it uses alternative terminal oxidases depending on the aerobic state of the immediate environment. The genes of oxygen respiration must be regulated in response to oxygen, up to the point of anaerobiosis. In this scenario, nitrate (if present in sufficient concentration) may act as an alternative electron acceptor to oxygen.
NADH-ubiquinone oxidoreductase.
The NADH-ubiquinone oxidoreductase holoenzyme from P. denitrificans is thought to contain at least 14 subunits, whereas that from mitochondria is considerably more complex, with 28 additional subunits (126, 258). Despite the difference in subunit composition, the function of the two enzymes is the same and there is considerable protein sequence homology between equivalent subunits (331, 332). Therefore, the Paracoccus proteins are named after their mitochondrial counterparts (Nqo1, Nqo2 etc.). The genes coding for these subunits are found in an operon between an ORF possibly coding for the Paracoccus UvrA (a DNA repair enzyme), and another gene (ORF240) similar to birA (biotin [acetyl coenzyme A (CoA) carboxylase] ligase). P. denitrificans UvrA has 74 and 71% identity to the equivalent E. coli (131) and Micrococcus luteus (263) proteins, respectively, while P. denitrificans BirA is 31% identical to the equivalent E. coli protein. The proposal that a bacterium such as Paracoccus is the forerunner of the eukaryotic mitochondrion (137) receives little support from the gene order of this nqo operon: the arrangement of the genes is more similar to that of chloroplasts (e.g., liverwort [202]) than to that of the bovine mitochondrion.
The region upstream of the initiation codon of nqo7 (the first subunit to be transcribed), is rich in long inverted repeats. However, there has been no attempt to define the base pair composition of a promoter. Termination appears to be Rho independent, probably occurring at a potential stem-loop structure between nqo14 and birA. A second inverted repeat is found within the coding region of birA, but it is more likely that this serves to stabilize birA mRNA than that it is a duplicated terminator.
Although 14 subunits of the NADH-ubiquinone oxidoreductase have had genes assigned to them (331–335), three ORFs within the operon are still of unknown function. N-terminal sequencing of the subunits of the holoenzyme has not revealed sequences similar to those of the putative products of the translated ORFs, and no homology to these unknown proteins can be found in the databases. Steinrücke and Ludwig (277) note that URF2 is similar to ctaG of the P. denitrificans cytochrome oxidase operon (275, 331, 332, 334).
Succinate dehydrogenase.
Succinate dehydrogenase of P. denitrificans has been purified (226) and shown to have four subunits, and the genes for these subunits appear to be in an operon (69). The enzyme contains covalently bound flavin, iron-sulfur centers, and cytochrome b, thus showing considerable amino acid sequence similarity to its mitochondrial counterpart. The promoter of the P. denitrificans sdhCDAB operon has been characterized (69), but sequence analysis alone does not provide much information on this constitutively expressed cluster (160, 226). The presence of ς70 hexamers at −10 with respect to the transcription start site (Table 5) suggests that the genes are transcribed with a ς70-like RNA polymerase.
The cytochrome bc1 complex.
After an early report on the purification and characterization of cytochrome c1 from P. denitrificans as a polypeptide with an unusually high molecular mass (177), the bc1 complex was isolated initially as a “supercomplex” along with cytochrome c oxidase and a membrane-bound cytochrome c552, yielding high quinol-oxidizing activity (26). Subsequently, its subunit composition was confirmed unequivocally (340), showing that only the three subunits carrying redox centers make up a complex that is also fully competent in free energy transduction (341). The cloning of the corresponding genes (162) revealed an operon structure, fbcFBC, coding for the Rieske FeS subunit, the cytochrome b, and the cytochrome c1 subunit. The latter is unique in having an additional N-terminal domain of around 150 amino acids (compared to the eukaryotic mitochondrial proteins, explaining the higher molecular weight of the P. denitrificans protein), with a characteristic composition (40% alanine, 38% acidic residues, and no basic residues [162]); its function is still not understood. While cytochrome b shows an amazingly high degree of sequence identity to other bacterial and mitochondrial subunits (see references 155, 285, and 290 for reviews), the existence of an additional transmembrane helix in the N terminus of the protein has been suggested (153) on the basis of using monoclonal antibody fragments in conjunction with electron microscopy. Gene and operon deletion studies, as well as expression of the fbc operon from a multicopy plasmid (92), resulted in a considerable overexpression of the complex in the homologous host. Once again, an E. coli ς70-like −10 region (TAGAAC; Table 5) can be found in the promoter region.
Cytochrome aa3.
P. denitrificans can use several terminal electron acceptors during aerobic growth, the best characterized of which is the aa3-type cytochrome c oxidase (cytochrome aa3). The biochemistry of this multisubunit enzyme has been reviewed extensively (62, 88, 109, 206, 291). The structure of the holoenzyme has been determined to a resolution of 2.8 Å by X-ray crystallography (136).
The cytochrome oxidase of P. denitrificans is a four-subunit enzyme (136). Three of these subunits have eukaryotic equivalents encoded mitochondrially, but the mitochondrial holoenzyme has an additional 10 subunits which are coded for in the nucleus (38, 291). Initially, only two subunits could be isolated from P. denitrificans (176). By use of labeled oligonucleotides, designed to hybridize to the conserved regions of the mitochondrially encoded subunit 3 of aa3, Raitio et al. (234) demonstrated that P. denitrificans possessed an equivalent polypeptide. Subunit I contains a heme a and a heme a3 prosthetic group, which, together with CuB, forms the active site. Subunit I is transcribed from the ctaDII gene, which was cloned independently by Raitio et al. (235) and van Spanning et al. (315). The product of this gene is a constituent of the aa3 complex under physiological conditions, although an isogene (ctaDI) exists. The isogenes have nearly 90% identity (235) and are found in different loci on the P. denitrificans genome. The ctaDI gene can be expressed in P. denitrificans only if it is maintained on a plasmid, possibly due to a gene dosage or repressor titration effect.
The genes coding for cytochrome oxidase subunits II and III (208, 234, 278) are found in an operon, ctaCB-ORF1-ctaGE. CtaC (subunit II) contains a copper A center, which is the entry point of electrons to the aa3 holoenzyme (123), although alternative electron pathways have been proposed. The ctaE gene codes for subunit III, an integral membrane protein of unknown function (108). The two remaining cta genes in the operon code for enzymes involved in posttranslational processing of subunit I (276, 278). CtaB catalyzes the conversion of heme b to heme o (91). The insertion of heme has been postulated to be mediated by CtaG (276). The function of the ORF found in the operon is currently unknown. The promoter region of the ctaCB-ORF1-ctaGE operon has been examined in some detail. The transcription start site was mapped with S1 nuclease (278) and was found to be 34 bp upstream of the translation initiation codon. Although there is a clearly discernible −10 region (that might indicate binding of a ς70 RNA polymerase), when the operon was expressed in E. coli, it was thought that initiation of transcription originated in the plasmid vector used, rather than from the native promoter (278).
Cytochrome oxidase was considered for many years to be composed of three subunits, although Haltia had copurified a small polypeptide with the oxidase (110). Determination of the crystal structure of the holoenzyme revealed that this polypeptide was a fourth subunit (136, 205), consisting of a cytoplasmic and a transmembrane domain. The peptide sequence enabled recovery of the gene (ctaH) from a P. denitrificans genomic library (325). Deletion of the ctaH gene did not seem to have any effect on the assembly of the other three subunits or their prosthetic groups or on the in vitro activity (325). Despite a similar spatial location (although no homology) to subunit IV of quinol oxidases, no role could be assigned to this polypeptide, and it has been speculated that it is an evolutionary remnant (325).
The ctaH gene is transcribed from its own promoter at a locus removed from ctaDI, ctaDII, and ctaCB-ORF1-ctaGE. Both the ctaCB-ORF1-ctaGE and ctaDII loci have upstream regions rich in inverted repeats, but to date it has not been possible to relate this to regulation. Since P. denitrificans uses three different terminal oxidases, maximally expressed under different oxygen concentrations (Fig. 2), there must be fairly stringent control of expression of terminal electron transfer complexes. The region upstream of ctaH has some elements in common with the ctaC promoter (325), but no clear picture has yet emerged of the elements responsible for the coregulation of the four cta loci.
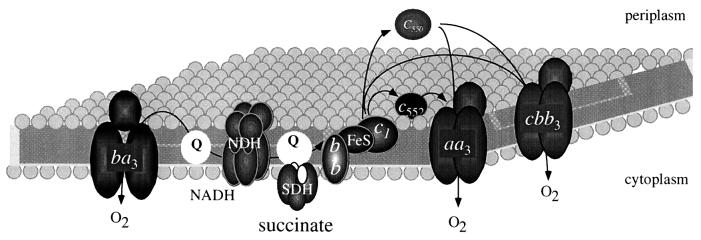
FIG. 2.
Branched electron transport pathway of P. denitrificans under various conditions of oxygen limitation. FeS, iron-sulfur center; NDH, NADH-ubiquinone oxidoreductase; SDH, succinate dehydrogenase; Q, ubiquinone pool.
The cbb3-type oxidase.
Bacteria that rely entirely on respiration for the liberation of free energy are challenged under near-anoxic conditions since the terminal oxidases of the aa3 and bo3 or ba3 type are unable to function at exceptionally low oxygen concentrations. One of the strategies for survival under these conditions is the recruitment of a cbb3-type oxidase, which has a relatively high affinity for oxygen. This type of oxidase was first encountered in endosymbiotic rhizobia, which use it during nitrogen fixation in the root nodule (142, 182, 230, 231). The finding that its derived Km value for oxygen is 7 nM may explain why the cbb3-type oxidase supports the growth of the bacteroids in these nodules, where the free-oxygen concentration is only 3 to 22 nM (230).
The oxidase has three subunits, which are encoded by the ccoNOQP gene cluster: CcoN, CcoO, and CcoP (63). The last two subunits are membrane-bound mono- and diheme c-type cytochromes, respectively, which may function as electron entry sites. CcoN is the catalytic subunit I with two hemes b and a copper ion. The architecture and metal-heme arrangement are similar to those of subunits I of the other members of the heme copper oxidases and to that of subunit I of nitric oxide reductase (42, 302). The role of CcoQ is not yet clear.
Apart from P. denitrificans, other bacteria are able to synthesize an oxidase of the cbb3 type (63, 286). The importance of this oxidase for P. denitrificans has been stressed by the observations that a mutant lacking the ccoN gene was unable to grow under microaerobic conditions (61). Detailed studies on the properties of the cbb3-type oxidase of P. denitrificans have been facilitated by the construction of a mutant strain that lacks the other types of oxidase. Proton translocation measurements of this mutant showed that the oxidase has the capacity to pump protons (63). Recent analyses of growth efficiencies of the wild-type and a set of double oxidase mutant strains, as determined from chemo- and auxostat experiments, have suggested that the cbb3-type oxidase transduces free energy as efficiently as the aa3-type oxidase does (306). However, amino acid residues implicated in the pathway of pumped protons and chemical protons, which are conserved in aa3-type cytochrome c and bo3-type quinol oxidases (128, 136), are not present at the corresponding positions of the cbb3-type oxidases (63, 302). It may thus be that the proton channel in the cbb3-type oxidases is composed of residues different from those of the other types or that the mechanism of proton translocation is not yet understood properly.
Expression of the cco gene cluster in P. denitrificans is under control of the FnrP protein as judged by the presence of an FnrP binding site, TTGAC-N4-ATCAA, in the ccoN promoter region and by the finding that FnrP mutants showed a decreased expression of this type of oxidase (310). This conclusion is in agreement with the observation that synthesis of the cbb3-type oxidase increases with decreasing oxygen concentrations (32).
Quinol oxidase.
Initially it was speculated that quinol oxidase activity in Paracoccus could be ascribed to cytochrome o (51), based mainly on spectroscopic data. Less than a decade ago, more detailed studies indicated the presence of a proton-pumping quinol oxidase in whole cells (232). The complex thought to be responsible was purified shortly afterward from membranes of P. denitrificans (175). Several characteristics of this complex, such as its function and subunit pattern, suggested some similarities to the bo3 quinol oxidase studied extensively in E. coli (reference 47 and references therein). This was substantiated by sequencing of the genes for the Paracoccus quinol oxidase (62).
The Paracoccus quinol oxidase is a member of the superfamily of heme copper oxidases (250). Six conserved histidine residues, which may act as ligands to the two heme components and one copper atom, were identified in the largest of the genes coding for subunit I (62). The four genes of the qox locus are arranged in an operon-like fashion (244), and they all show a remarkable degree of identity when translated not only to the four subunits of their E. coli counterparts but also to other oxidases.
Detailed biochemical and spectroscopic studies have shown that the quinol oxidase of P. denitrificans carries one heme b, one heme a, and one copper atom as redox-active groups in subunit I, QoxB. The last two constitute the binuclear center of oxygen reduction (345, 346). The observation that only one copper atom is present per enzyme complex reflects the fact that quinol oxidases do not possess a binuclear CuA center in their homologue of subunit II. The ba3 heme composition of this oxidase is essential for maintaining its catalytic competence, as was seen when an inactive quinol oxidase variant was isolated from a Paracoccus strain unable to synthesize heme a (346). Nevertheless, there are conflicting reports on the heme composition of the quinol oxidase, based on heme analysis of membranes from an aa3-type cytochrome oxidase mutant (62, 307).
Recently, two-dimensional crystals of the bo3-type quinol oxidase of E. coli were analyzed by cryoelectron microscopy (98). Although the resolution of about 6 Å is too low to identify individual residues, it definitely shows a transmembrane helix and subunit arrangement similar to the structure of the aa3-type cytochrome oxidase of P. denitrificans (136), apart from differences in the nonhomologous subunits IV. Given the close relationship of the P. denitrificans quinol oxidase to its E. coli homologue, a similar spatial structure can be inferred from these results. Another indication of their structural similarity is the retention of some enzymatic activity (about 20% both in membranes and in the isolated complex compared to the native P. denitrificans quinol oxidase) after replacement of the qoxA gene coding for subunit II of the P. denitrificans enzyme by the equivalent gene, cyoA, from the bo3-type quinol oxidase of E. coli (324). It is not yet clear whether the chimeric quinol oxidase retains the ability to translocate one proton across the membrane per electron transferred to oxygen, as do the parent oxidases.
The qox promoter region contains both an FNR box (34 bp upstream of the transcript start site [Table 5]) and a sequence (TTGACC) very close to the E. coli ς70 −35 hexamer (TTGACA). The presence of a binding site for FnrP (310) in the promoter region of the qox operon encoding the quinol oxidase might indicate regulation of transcription in response to the level of oxygen or changes in the redox potential. However, no significant decrease in expression was observed with a synthetic promoter/reporter gene construct when reporter levels were measured under aerobic growth conditions compared to those measured under anaerobic growth conditions (346). Surprisingly, the same study revealed the positive influence of nitrate and nitrite (added to the growth medium) on the expression of quinol oxidase under aerobic conditions. Additional regulatory studies are needed before the role and importance of this oxidase for the metabolic flexibility and growth of Paracoccus can be assessed.
Cytochrome c550.
Cytochrome c550 is believed to function as an electron donor in several respiratory pathways including those for denitrification (30, 185), methanol oxidation and methylamine oxidation (57, 62, 74). The P. denitrificans locus coding for cytochrome c550 is found just upstream of ctaDII (235, 315), separated by a putative Rho-dependent terminator. A similar gene order is found in the P. versutus locus, and the two species have 89% DNA identity over comparable regions. Truncated versions of the cycA-ctaDII locus from P. versutus fused to a reporter and transferred to E. coli showed that the two genes are separated by a strong terminator. No activity from the P. versutus ctaDII promoter region could be demonstrated in E. coli (296).
The gene coding for cytochrome c550 (cycA) is subject to extensive regulation, since the expression is raised above a basal level in response to the induction of various respiratory pathways (209, 211, 275). Dual transcription start sites (separated by only 8 bp) for the gene have been determined by primer extension studies (279), but it still remains unclear which elements in the putative promoter regions are responsible for the regulation of the gene. Furthermore, examination of promoters by the authors of this review revealed no clear −10 or −35 sequences in the appropriate positions upstream of either transcription start site proposed (Table 5).
Cytochrome c552.
Among the many c-type cytochromes found in P. denitrificans (Table 6), a membrane-bound polypeptide, cytochrome c552 (CycM), is the most likely mediator between the cytochrome bc1 complex and the heme aa3 cytochrome c oxidase, as suggested by a number of arguments. (i) Under certain solubilization conditions, a complex of this cytochrome either with the oxidase or with oxidase and the bc1 complex has been isolated from membranes (26, 109); a high rate of electron transfer points at its efficient role as a redox link. (ii) Specific antibodies obtained against the purified protein (294) inhibited electron transport in membranes between NADH and oxygen but had no effect when partial reactions were assayed with mitochondrial cytochrome c. (iii) Cloning of the gene encoding cytochrome c552 (294) showed a tripartite structure of the polypeptide with an N-terminal membrane anchor; membranes isolated from gene deletion mutants (293) were blocked in their electron transport from NADH to oxygen via complexes I, III, and IV, and this inhibition could be partially overcome by the addition of mitochondrial cytochrome c.
Electron transport flavoprotein.
Electron transport flavoproteins (ETF) of bacteria transfer electrons between flavoprotein dehydrogenases and the respiratory chain. The electron acceptor of ETF is presumably ubiquinone in P. denitrificans. Although the mitochondrial ETF accepts electrons from a wide variety of dehydrogenases, the P. denitrificans protein shows a more restricted substrate range. ETF has been isolated in abundance from trimethylamine-grown cells but has also be shown to transfer electrons from glutyryl coenzyme A (CoA) dehydrogenase (135). However, it does not accept electrons from periplasmic dehydrogenases such as methanol dehydrogenase and methylamine dehydrogenase (56, 273).
The genes encoding the two subunits of the heterodimeric protein were not given a gene designation and will be called etfS and etfL (for the small- and large-subunit genes, respectively, of this heterodimer) for the purposes of this review. These two genes are found in a short operon (12), much like the orthologous gene cluster fixAB of Azorhizobium caulonidulans (4). No gene coding for ETF-ubiquinone oxidoreductase has yet been discovered in Paracoccus. The etfSL locus is characterized by an abundance of inverted repeats upstream and downstream of the structural genes, and in P. denitrificans, these repeats probably prevent expression of the genes in heterologous systems (12).
Interest in P. denitrificans ETF centers on the similarity of the protein to the mammalian mitochondrial equivalent. Some human diseases are associated with mutations in ETF, and although P. denitrificans ETF differs considerably in substrate range, the potential for overexpression has made it an attractive model. The differences between the human and Paracoccus ETFs have been overcome to some extent by the production of chimeric proteins: these and the wild-type proteins have been the subject of X-ray crystallography studies, yielding structures to a resolution of 2.5 Å (245).
Respiratory Denitrification Genes
Denitrification proceeds via nitrogen oxide intermediates (Fig. 3). A membrane-bound nitrate reductase with its active site facing the cytoplasm yields nitrite, which is converted to nitric oxide (NO) by a periplasmic reductase. The NO is further reduced by a periplasmic membrane-bound nitric oxide reductase, and molecular nitrogen is generated by a soluble periplasmic nitrous oxide reductase. This reaction pathway is also seen in other bacteria, including Pseudomonas aeruginosa, Pseudomonas stutzeri, and R. sphaeroides (forma denitrificans), and parts of the pathway are found in fungi (157). The bacterial enzymes of denitrification are essentially similar in all organisms, except that there are two types of nitrite reductase: a copper-containing protein and a cytochrome containing c and d1 hemes. The two types of nitrite reductase have yet to be found together in a single bacterial species (342). P. denitrificans has the latter. The enzymology of denitrification has been reviewed thoroughly (22, 347, 348, 350).
FIG. 3.
Denitrification pathway of P. denitrificans, with succinate as the carbon and energy source. FeS, iron-sulfur center; NDH, NADH-ubiquinone oxidoreductase; SDH, succinate dehydrogenase; Nar, nitrate reductase; Nor, nitric oxide reductase; Nir, nitrite reductase; Nos, nitrous oxide reductase; Q, ubiquinone pool; ?, unknown transporter. The exact electron pathway(s) through the c-type cytochromes and pseudoazurin is not known.
Organization of denitrification genes.
In P. denitrificans, nir genes (associated with nitrite reduction) and nor genes (associated with nitric oxide reduction) are clustered on a 17.7-kbp fragment, essentially transcribed divergently from one another (Fig. 4). There seems to be little variation in genetic organization between the strains of P. denitrificans studied (252). In Pseudomonas stutzeri, the other organism for which the genetics of denitrification have been extensively studied, the nos genes (nitrous oxide reductase associated) are also found in the same 30-kbp region of the genome as the nir and nor genes (34, 97, 347, 348, 350). The order of the genes within the nir, nor, and nos clusters varies among organisms, but the structural genes (nirS and norCB) are transcribed from the 5′ end of an operon (34, 59, 252). It is probable that other nir, nos, and perhaps nor genes remain undiscovered in P. denitrificans, given that the Pseudomonas stutzeri cluster contains at least five genes so far unidentified in Paracoccus (348). It is not yet known whether the nitrate reductase (nar) genes map to the same region of the chromosome as the nir, nor, and nos genes in any of the denitrifying bacteria.
FIG. 4.
Denitrification gene clusters of Pseudomonas stutzeri (A), Pseudomonas aeruginosa (B), and P. denitrificans Pd1222 (C). Reproduced with permission from reference 251a.
Nitrate reductases.
Two nitrate reductases, a membrane-bound enzyme induced during oxygen limitation and nitrate availability and a constitutively expressed periplasmic reductase, can be active during denitrification (16). Insertion of Tn5 into a structural gene of the membrane-bound nitrate reductase of P. denitrificans resulted in anaerobic overexpression of periplasmic nitrate reductase activity (15).
(i) Membrane-bound nitrate reductase.
The nitrate reductase induced during denitrification is of a similar type to that found in E. coli. It is a three-subunit protein, with the transmembrane γ subunit anchoring the cytoplasmically exposed α and β subunits to the membrane. Deletion of the γ subunit stops nitrate reductase activity in whole cells, but the activity of the remaining αβ dimer can still be detected in a nonphysiological assay with methyl viologen as an electron donor (16). To date, only the 5′ region of narG (the α subunit), plus narH (the β subunit), narJ (unknown function), and narI (the γ subunit) have been sequenced (23), and so it can be assumed only that the genes form an operon much like that found in E. coli.
(ii) Periplasmic nitrate reductase.
The periplasmic nitrate reductase is a molybdopterin protein containing both heme and nonheme iron (18, 25, 35, 259). The structural and biosynthetic peptides required for construction of the holoenzyme are coded for by the napEDABC operon (24). The large and small subunits are coded for by napA and napB, respectively. Sequence analysis shows that the large subunit contains the molybdenum center and an iron-sulfur center whereas the smaller subunit has two heme binding sites. The exact functions of the cytoplasmic NapD and membrane-bound NapE are currently under investigation, but it is thought that NapC is a tetraheme c-type cytochrome (247a) and the electron donor to the holoreductase (24); evidence for this role has been presented for R. sphaeroides NapC (243). It has similarity to the tetraheme cytochrome NirT (implicated in denitrification in Pseudomonas stutzeri [97]).
The induction of the nap gene cluster does not follow that of denitrification. Although the proteins are expressed during anaerobic growth, maximal expression is found during aerobic growth on substrates such as butyrate (260). Examination of the region upstream of napE reveals no easily distinguishable binding sites for regulatory elements.
Nitrite reductase.
The reduction of nitrite to nitric oxide is the first committed step of the denitrification process. The NO produced can be used as a substrate only by nitric oxide reductase and must be quickly removed from the cell because it is toxic at quite low concentrations. The physiological electron donors to nitrite reductase in P. denitrificans are cytochrome c550 (CycA) and the copper-containing pseudoazurin, PazS. No confusion of azurin with pseudoazurin should occur: they are connected merely by both being copper proteins (168, 189, 192, 321). The existence of two donors with two differing metal centers probably allows the organism to maintain the electron transport chain under different metal ion-limited conditions.
Nitrite reductase has been the subject of study for a number of years, and the protein was first purified by Newton (196). The enzyme has an alternative function as a cytochrome oxidase, but this is probably nonphysiological. The enzyme has two hemes as prosthetic groups, a normal c-type heme and a unique d1 heme. Both of these hemes are added to the reductase in the periplasm or during the preprotein’s translocation to the periplasm (211, 212). To date, the d1 heme has been found only in other bacterial cytochrome nitrite reductases. A crystal structure (87) reveals that the two hemes lie in distinct domains: one domain somewhat resembles the class I c-type cytochromes such as c550, with the c-type heme covalently attached to the N terminus of the domain; the second domain contains the d1 heme noncovalently held in a complex β-sheet structure. The structure of the d1 domain can be regarded as an eight-bladed propeller (87), similar to the structure of methanol dehydrogenase (6) but with a d1 heme active site held in the center.
(i) Biosynthesis of nitrite reductase.
The structural gene for cytochrome cd1 nitrite reductase (nirS) was sequenced by de Boer et al. (59). The structural gene maps close to those of nitric oxide reductase but is transcribed in the opposite direction (Fig. 4). Immediately upstream of nirS lie two ORFs (nirI and nirX), which, by interruption mutagenesis, are found to be essential for nitrite reduction. Neither NirI nor NirX is thought to have any function in the biosynthesis of nitrite reductase, but they both influence the transcription of the gene (307). Downstream of nirS lie four other ORFs, also essential for nitrite reduction, nirE, nirC, nirF, and nirD (59), which may be transcribed from a promoter in the nirS-nirE intergenic region (252). Interruption of these genes by introduction of a kanamycin cassette does not have any effect on the transcription of the structural gene or on the insertion of c-type heme into the polypeptide (59), but no d1 heme appears to be made in these mutants. NirE resembles, by homology, CysG of E. coli, a uroporphyrinogen methyltransferase implicated in the synthesis of the sulfite reductase prosthetic group, siroheme (53). It thus seems probable that NirE is involved in the addition of methyl groups to a precursor to form d1 heme, but since the biosynthetic pathway for this prosthetic group remains uncharacterized, it is not possible to determine exactly how NirE acts. NirC codes for a low-molecular-weight class I c-type cytochrome, which plays an undefined part in the formation of d1 heme. This cytochrome has not been seen in heme-stained extracts of P. denitrificans (252).
nirF codes for a protein with some degree of identity to the d1 domain of the nitrite reductase structural gene. Sequence identity of NirF to the NirS d1 domain may support the postulate that NirF is similar to this domain (97). Although NirF has aspartate residues in a similar spacing to NirS and other propeller structures (a motif that may be common to many propeller folds [6]), no clear structural similarity can be predicted (6). It has been suggested that NirF may be acting as a scaffold on which the d1 heme is assembled prior to insertion into nitrite reductase (97), but the periplasmic location of the protein and the difference in deduced structure argue against this theorem (6). The last gene in the cluster so far identified is nirD, an ORF of unknown function.
Nitric oxide reductase.
For some time it was a matter of debate whether nitric oxide was really an intermediate in denitrification, but the matter has been resolved by the finding of a separate nitric oxide reductase complex (39, 40), the trapping of NO during denitrification (39, 102, 161), and, ultimately, the interruption of the genes encoding the nitric oxide reductase proteins (34, 60), yielding a mutant that is unable to grow anaerobically.
The nitric oxide reductase of P. denitrificans has been purified (40, 96) as a two-subunit protein. The larger subunit binds heme b, while the smaller one binds heme c. The complex has also been shown to contain nonheme iron (96, 122). The smaller subunit is anchored to the membrane via a single α-helix, with the c-heme domain acting as a soluble periplasmic polypeptide. The large subunit has 12 membrane-spanning helices, anchoring all of the protein within the membrane except for the N and C termini. Despite the similarity to heme copper oxidases (249, 302), which can act as primary proton pumps (62, 63, 219, 236, 250), nitric oxide reductase has no proton translocation activity (17, 39, 261).
The genes associated with nitric oxide reductase are found in a cluster upstream of the nir locus (60). A small ORF, norC, codes for the c-heme subunit, and norB codes for the larger b-heme subunit. The norCB genes appear to be cotranscribed with four other genes that are essential for the maturation of nitric oxide reductase (norQDEF), although their exact function remains to be determined. NorQ contains two putative nucleotide binding motifs and has been found in other denitrifiers including Pseudomonas stutzeri (351) and R. sphaeroides (7). NorQ has been implicated in the formation of nitrite reductase in Pseudomonas stutzeri (141). The gene product of norD appears to be cytoplasmically located like NorQ, suggesting that both influence transcription rather than protein maturation. A knockout of Pseudomonas stutzeri norQ affects nirS and norCB activities; a knockout of Pseudomonas stutzeri nirQ also affects nirS and norCB. The norQ knockout has the same effect in P. denitrificans, but as yet nirQ has not been isolated. The genes norE and norF have no known function. NorF of P. denitrificans has no known orthologues, but NorE is similar to cytochrome oxidase subunit III (60). Knockouts of norE and norF affect only nitric oxide reductase activity (60). Several experiments suggest that the nitrite reductase and nitric oxide reductase are in some way linked, perhaps as a multienzyme complex. Both de Boer et al. (59) and Jungst et al. (140) describe a lowering of nitric oxide reductase activity on insertional mutagenesis of nirS, despite there being no obvious transcriptional link between nirS and norCB in either Paracoccus or Pseudomonas (Fig. 4).
Nitrous oxide reductase.
The formation of molecular nitrogen from nitrous oxide is carried out in P. denitrificans by a periplasmic copper nitrous oxide reductase (21). The redox centers of this protein are of a copper A type and an unusual copper Z type (reference 350 and references therein). The nosZ gene coding for the structural protein of nitrous oxide reductase has been sequenced, along with part of an upstream ORF cotranscribed with it, named nosR (125).
NosR of P. denitrificans is the counterpart of the Pseudomonas stutzeri NosR protein (55). Protein sequence analysis indicates that NosR is a membrane protein with relatively large soluble domains facing the periplasm and cytoplasm. The cytoplasmic domain harbors two cysteine signatures indicative of the presence of [4Fe-4S] clusters. NosR has a high degree of homology to NirI (307). Other structurally related proteins are NapH, MauN, and CcoG (21, 22). Disruption of the nosR gene from Pseudomonas stutzeri yielded a mutant unable to transcribe the nos gene cluster (55). It was suggested that NosR itself was the transcriptional activator based on the identification of a helix-turn-helix-like DNA binding motif. Other examples of transcriptional regulators that may bind both the membrane and DNA have been found in other organisms, including ToxR (which regulates cholera toxin expression in Vibrio cholerae [187]).
Pseudoazurin.
Despite the roles in electron transport assigned to CycA (cytochrome c550), a knockout of its gene (315, 316) did not lead to cessation of anaerobic or methylotrophic growth. This does not necessarily contradict the postulate that cytochrome c550 is involved in these processes (Fig. 1–3, 5), but it suggests that the role of this cytochrome can be duplicated by another protein; the blue copper protein pseudoazurin (PazS) has been proposed as a candidate (132, 189, 192, 193) for a substitute in denitrification. A cycA pazS double deletion is currently under construction.
FIG. 5.
Electron transport pathway of P. denitrificans during growth on methylamine and methanol. FyDH, formaldehyde dehydrogenase; FoDH, formate dehydrogenase; FeS, iron-sulfur center; NDH, NADH-ubiquinone oxidoreductase; GSH, glutathione. The exact electron pathway(s) through the c-type cytochromes is not known.
The pazS gene may be under the control of an Fnr-like protein, as indicated by the presence of an FNR box 42.5 bp upstream of the transcript start site (168), but there is no experimental evidence whether this putative transcriptional activator is FnrP or Nnr (see below). There is also a −10 hexamer (TAAACC) with low similarity to that of E. coli (TATAAT [Table 5]). In addition, the promoter/operator region of pasZ contains some apparently anomalous motifs. An apparent NtrA box can be found in the promoter region, but it is centered at −82 bp with respect to the transcript start site determined during growth on nitrate. Since ς54-controlled genes usually have the NtrA box from −24 to −11, this may be either the binding site for a second promoter or coincidental (253). Another puzzling nucleotide structure is found just upstream of the transcript start site. In this case, a palindrome is formed in the 5′ end of the mRNA. This might be expected to increase mRNA stability, but it appears to inhibit isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent expression in E. coli (168). Its removal leads to a twofold elevation in the yield of the protein when expressed heterologously, a finding previously observed with the Ralstonia (Alcaligenes) eutrophus S-6 pseudoazurin (337).
Regulation of denitrification and integration with oxygen respiration.
Oxygen concentration can be sensed by a protein designated FNR (Fumarate Nitrate Reduction), which in E. coli activates the transcription of the genes involved in anaerobic metabolism (107, 269, 271, 299). FNR has four important functional domains. First, the N-terminal domain contains three cysteine residues. Together with a fourth cysteine residue located in the central domain, they may bind a redox-sensitive [4Fe-4S] cluster (10, 107, 150). Second, a sequence of residues in the central domain of FNR forms a loop-like structure that contacts RNA polymerase (322). Third, FNR contains an α-helical domain surrounding Asp154, which is probably involved in dimerization (10, 262). Finally, at the C-terminal end it contains a helix-turn-helix DNA binding motif, which recognizes the DNA target site with the consensus sequence TTGAT-N4-ATCAA (the FNR box). Two invariant amino acid residues (Glu209 and Ser212) in this region are essential for binding to the FNR box (269, 270). Anaerobic transcription activation by FNR depends on the presence of the redox-sensitive [4Fe-4S] cluster. Since this cluster is rapidly destroyed upon exposure to oxygen (104, 150, 265), increasing activation of FNR may be expected at decreasing oxygen concentrations. In its active form, FNR may dimerize with a partner molecule and bind strongly to the FNR box. Usually, this box is located directly upstream of the RNA polymerase binding site at the −35, −10 promoter consensus sequences. Such an organization of binding sites allows physical contact between the FNR dimer and RNA polymerase. Once this contact occurs, the transcription of a number of target genes essential for anaerobic metabolism gets underway.
Although expression of the E. coli nar operon requires active FNR, optimal expression of this locus is achieved only when the nitrate-sensing regulatory device is activated too. This device is composed of two proteins, a protein histidine kinase (NarX) and a DNA binding response regulator (NarL), a set of proteins that belongs to the family of two-component regulators. Since the behavior of P. denitrificans with respect to the on-off set of nitrate reductase synthesis is similar to that of E. coli, one may expect that both organisms will make use of similar protein-based signal transduction circuits. Partial evidence for this suggestion has recently been obtained from the identification of the P. denitrificans counterpart of the E. coli fnr gene, designated fnrP. Its product, FnrP, resembles FNR to a large extent, and all four functional domains that are found in FNR are also present in FnrP. The fnrP gene was identified close to the ccoNOQP locus encoding the cbb3-type cytochrome c oxidase. A mutation in fnrP had pleiotropic effects during growth at oxygen depletion. The mutant was unable to express membrane-bound nitrate reductase (the product of the narGHJI operon) and cytochrome c peroxidase (encoded by the ccp gene [307]), whereas it had attenuated levels of the cbb3-type cytochrome oxidase and increased levels of the quinol oxidase (encoded by the qoxABCD locus). Target sites of FnrP, 5′-TTGAC-N4-ATCAA-3′, which resemble the E. coli FNR box were identified in the promoter regions of the cco, qox, and fnrP genes and operons (those of the ccp gene and the nar operon have yet to be sequenced). In the promoter region of the qox locus, this box appears to be located within the RNA polymerase binding site rather than upstream of it, which may explain the observed repression of transcription of the qox locus by FnrP. The presence of such a box in the fnrP promoter suggests that its expression is autoregulated, as is the case for fnr from E. coli. The expression of two other gene clusters that are important for anaerobic growth with nitrate, the nir and nor gene clusters encoding nitrite reductase and nitric oxide reductase, respectively, turned out to be under the control of NNR, which is a second FNR homologue found in P. denitrificans (309). NNR shares all but one of the features of FNR homologues: it does not contain the redox sensitive [4Fe-4S] cluster. The nnr gene is located close to the locus that encompasses the nir and nor gene clusters. A mutation in the nnr gene resulted in an inability to synthesize nitrite reductase, while the expression of nitric oxide reduction was decreased to about one-quarter of the wild-type level. The expression of other gene clusters (ccoNOQP, qoxABCD, narGHJI, and ccp) was not affected by the nnr mutation. DNA binding sites for NNR, which are virtually identical to the FnrP binding site, were found in the promoter regions of both gene clusters. Despite this degree of identity, NNR and FnrP were unable to take over each other’s tasks in gene regulation during denitrification. Apparently, expression of each set of target genes requires additional transcription factors that are dedicated to either NNR- or FnrP-mediated control. Alignment studies of a number of FNR homologues of different bacterial origins have led to the suggestion that this family of regulators may be divided into three more or less discrete groups, each with proteins harboring specific signatures of invariant amino acid residues within their functional domains. Such a specificity in a resulting RNAP contact site may thus reflect specificity of proteins from each group for a particular type of sigma factor. Sigma factors are proteins that bind reversibly to the catalytically active core RNAP and are required for transcription initiation. Since NNR and FnrP fall into two different groups, this could mean that the dedicated transcription factors suggested for NNR and FnrP are specific sigma factors. In many bacterial species, there are sigma factors other than the primary ones; the latter are ς70 homologues that are involved in transcription of the majority of housekeeping genes. Alternative sigma factors often control the transcription of coordinately regulated sets of genes from promoter sequences that are different from those recognized by the primary sigma factor. Examples of these alternative sigma factors are RpoN (control of genes involved in nitrogen metabolism), RpoH (control of heat shock genes), and RpoS (control of gene expression at the stationary phase of growth). Examination of the upstream regions of the genes that are regulated by NNR (nirS, nirI, and norCB) reveal no obvious ς70 RNAP binding sites; instead, other common motifs that might have some function in transcription have emerged (5).
The nature of the signals sensed by FnrP and NNR is still not fully established. FnrP closely resembles E. coli FNR in that it contains the cysteine signature for ligating a redox-sensitive [4Fe-4S] cluster. In E. coli, the concentration of molecular oxygen is the most important factor involved in modulation of FNR activity. In vitro experiments showed that the [4Fe-4S] cluster is rapidly destroyed by molecular oxygen, resulting in inactive FNR (165). FnrP of P. denitrificans, however, is still capable of activating the transcription of its target genes at aerobic growth conditions. Under these conditions, FnrP activation appears to increase at increasing reduction levels of the components of the respiratory network. These observations suggest that the [4Fe-4S] cluster of FnrP requires reduction for its full activation and that it is less sensitive to molecular oxygen than is the E. coli FNR protein. It has been shown that the latter protein may acquire an enhanced stability toward oxygen when the Asp residue at position 154 is changed into an Ala residue. Interestingly, there is an Ala residue in P. denitrificans FnrP at the position corresponding to Asp154 of the E. coli FNR protein.
Despite the diversity of promoters, comparison of the anaeroboxes allows the deduction of some Paracoccus promoter features. To date only two NNR/FNR-regulated promoters have had their transcription start sites determined. The genes for pseudoazurin (pazS [168]) and NNR-controlled nitrite reductase (nirS [252]) have anaeroboxes. In both cases, the FNR-like box is centered at a position 41.5 bases upstream of the transcript start site.
The closely related bacterium R. sphaeroides also has an NNR-like (NnrR) system, nirK, controlling a copper-containing nitrite reductase (287). Again, this system works in parallel with a conventional FNR-type protein. By using lacZ fusions to nirK and organic compounds from which NO was generated in the medium of R. sphaeroides, it was shown that NO was the signal to which the R. sphaeroides Nnr was responding (163). It is postulated that nirK is switched on for most of the time, albeit at very low levels. In the presence of nitrite (from nitrate reductases), a small quantity of NO can be produced. NO is argued to be a biologically very active signal molecule by analogy to eukoryotic NO-responsive systems and because of the low levels of exogenous NO required to stimulate R. sphaeroides nirK fusions (163). NO switches on NnrR, which in turns stimulates transcription of nirK, so that more NO is produced. The transcription of norCB is partially dependent on the same NnrP protein; therefore, NO does not accumulate to toxic levels (7, 154).
NO is also a regulator of nitrite reductase in P. denitrificans (307); the oxygen tension of the cell is not the sole modulator of transcription and hence of the activity of the denitrification enzymes. This may give some credence to the suggestion that P. denitrificans LMD 82.5 can denitrify aerobically (14, 29, 39, 41, 58, 171, 246, 247). The presence of nitrate could indirectly act as a stimulus, since nitrate would be converted to nitrite by nitrate reductase(s) and NO would be generated by nitrite reductase, which is present at low levels in P. denitrificans even under aerobic conditions (Northern blotting experiments revealed a low level of transcription of the nirS gene) (252). Experiments are under way in several laboratories to find strategies to characterize completely the signal transduction pathway of the denitrification cluster.
Genes for Autotrophy
When P. denitrificans experiences a shortage of heterotrophic substrates, it can switch to chemoautotrophic growth, during which hydrogen or thiosulfate is oxidized (86, 138, 327). The carbon supply is provided by the fixation of atmospheric carbon dioxide via the Calvin cycle under these growth conditions. The bacterium is also able to grow on C1 substrates such as methanol or methylamine (reviewed in reference 119), and these processes are also considered autotrophic, since these carbon sources are completely oxidized before assimilation (318).
Methanol dehydrogenase.
Not all Paracoccus species can utilize methanol. P. denitrificans grows well on this substrate, but the closely related P. denitrificans GB-17 is unable to use methanol as a sole source of carbon and energy. The latter strain does not carry the full complement of methanol oxidation genes, but mutagenic events (191) can restore the methanol dehydrogenase (MDH) activity specifically by the extension of the substrate range of an alcohol dehydrogenase (238).
In the presence of methanol as the sole source of energy, P. denitrificans induces the expression of the mxa gene cluster, encoding MDH and its dedicated electron acceptor cytochrome c551i (119, 316). Both proteins are located in the periplasm, and in subsequent redox steps they connect the oxidation of methanol to the reduction of cytochromes c550 and c552. Methanol-oxidizing systems are found in a wide variety of methylotrophic bacteria (3). Apart from P. denitrificans, much of the current knowledge about the physiology and molecular genetics of this system has been obtained from studies on the facultative methylotrophic bacteria Methylobacterium extorquens AM1, Methylobacterium organophilum XX, and Methylobacterium organophilum DSM 760 (reviewed in reference 169). These studies revealed that the synthesis of a fully active methanol-oxidizing pathway requires a total of at least 32 genes, among which are the pqq genes involved in cofactor biosynthesis and a comprehensive set of so-called mxa, mxb, mxc, and mxd genes, some of which encode the structural enzymes, others of which encode enzymes involved in calcium insertion, and yet others of which encode proteins involved in regulation of gene expression and protein activity (169).
Methylamine dehydrogenase.
Growth on methylamine is accompanied by the appearance in the periplasm of methylamine dehydrogenase (MADH) and its electron acceptor, the blue copper protein amicyanin, both of which are encoded by the mau gene cluster (48, 49, 304, 313). Electrons derived from the oxidation of methylamine are transferred via amicyanin to the electron transport network at the level of cytochrome c (64, 133, 295, 314). The mau gene cluster of P. denitrificans consists of 11 genes, 10 of which encode the structural proteins or proteins involved in cofactor biosynthesis and are transcribed in one direction whereas the 11th regulatory gene (mauR) is located upstream and is divergently transcribed (48, 304, 313).
In addition to the proteins expressed specifically during and involved in the oxidation of methanol and methylamine, a cytochrome c553i is expressed during growth on these C1 substrates (33, 134). The gene encoding this cytochrome (cycB) has been cloned and shown to be part of the gene cluster designated xox (239). Surprisingly, the products of the genes located upstream and downstream of cycB (xoxF and xoxJ, respectively) had a large degree of identity to the α subunit of MDH and MxaJ (a protein of unknown function), respectively. The role of this putative oxidizing branch still remains obscure, since mutational analysis of this gene cluster did not reveal a clear function for the proteins (240).
Formaldehyde dehydrogenase.
The oxidation product of both methanol and methylamine is formaldehyde, which is translocated across the membrane to the cytoplasm. This transport process has been suggested to involve a specific formaldehyde carrier (159). In the cytoplasm, formaldehyde is further oxidized to formate via two consecutive reactions. In the first reaction, a NAD-linked glutathione-dependent formaldehyde dehydrogenase (FlhA) catalyzes the oxidation of formaldehyde into S-formylglutathione (305). In the second reaction, the latter molecule is converted into formate by S-formylglutathione hydrolase, which is a human esterase D homologue (117). The flhA gene cluster of P. denitrificans encoding the two enzymes has recently been identified upstream of the xox gene cluster (240). Finally, formate is oxidized to carbon dioxide predominantly by a NAD-dependent formate dehydrogenase (3). It has been reported, however, that P. denitrificans can synthesize isoenzymes of the formaldehyde and formate dehydrogenases that are NAD independent (113). A schematic view of the respiratory network that operates during C1 metabolism is presented in Fig. 5.
Regulation of the metabolism of C1 compounds.
Expression of the components of the methanol- and methylamine-oxidizing branches is subject to complex regulatory cascades that have to orchestrate the optimal composition of the respiratory network under these growth conditions to maintain respiratory electron flows at competitive rates on the one hand but prevent the accumulation of the toxic intermediate formaldehyde on the other hand. Moreover, part of these signal transduction systems coordinate gene expression according to an energetic hierarchy, ensuring that the C1-oxidizing branches are expressed only when the cell is unable to metabolize more energetically favorable substrates (119). This view is corroborated by extensive physiological and biochemical studies on this type of metabolism. MADH and amicyanin are synthesized only when the cell uses methylamine (and to a lesser extent other alkylamines) as the sole source of energy (118, 213, 317). MDH and XDH and their dedicated cytochromes are expressed not only during growth on methanol but also during growth on methylamine or choline. Since the oxidation of all these carbon sources yields formaldehyde, it has been postulated that the latter molecule is an important trigger in the regulation of expression of these gene clusters (113). The synthesis of all the enzymes involved in the consecutive steps of methanol or methylamine oxidation is blocked if the cells can metabolize multicarbon substrates in addition to these C1 growth substrates (118, 213, 313). Growth during C1 metabolism also affects the regulation of expression of the other components of the branches, cytochrome c550 and the cytochrome c oxidases. The amount of these components increases two- to fivefold under these conditions compared to that obtained during heterotrophic growth (279, 313). Apparently, this increased supply meets the demands of increased velocities of electron transfer within this part of the respiratory network. To date, a number of different regulatory proteins that control the expression of the flh, xox, mxa, and mau gene clusters (115), as well that of the cco and qox gene clusters, have been identified (308–310).
Expression of the mxa genes is controlled by the products of the mxaXYZ genes, which are located upstream of the structural mxa gene cluster (118). MxaY and MxaX are a protein histidine kinase and a DNA binding response regulator, respectively, and form a set of proteins that belongs to the family of two-component regulators. MxaZ has no significant homology to any protein in the databases. As suggested above, the effector molecule for the sensor domain of MxaY is probably formaldehyde. The general principle for signal transduction via two-component regulatory systems is that binding of the effector molecule triggers the autophosphorylation of the kinase, after which the phosphate group is transferred to the response regulator. Once phosphorylated, the latter may bind to the promoter region of its target gene in order to facilitate transcription by the RNAP (220). Surprisingly, however, MxaY was shown to be dispensable in this cascade, suggesting that an alternative kinase takes over its role (339). The role of MxaZ in this cascade is still unclear. The MxaYX regulatory system appears to be specifically dedicated to the regulation of expression of the mxa genes; mxaX mutants showed an unimpaired expression of the mau, flhA, and xox genes (116, 118). A two-component system, MxcQE, with a function similar to that of MxaYX has now also been identified in Methylobacterium organophilum XX. The regulatory gene cluster is, however, separated by more than 40 kbp from its target, the mxa locus (330).
The methylamine gene cluster of P. denitrificans is regulated by MauR, which is a transcription activator that belongs to the family of one-component LysR-type regulators (313). The spatial organization of the functional domains involved in DNA binding, signal perception, and dimerization in the members of this family is conserved (256). Usually, the molecule that is oxidized by the proteins encoded by the target locus of this type of gene activator also serves as the trigger molecule for activation of the latter. This finding would be in agreement with the suggestion that methylamine is the molecule that triggers the expression of the mau gene cluster (313). The current view with respect to the mode of action of LysR-type regulators is that signal perception induces dimerization of the transcription activator, which is followed by binding to a target site upstream of the promoter. As a result, the dimer may come into contact with RNAP, after which transcription gets under way. In agreement with this view was the observation that MauR binds to the mau promoter region in a gel shift assay (65). The target site for LysR-type activators is usually a conserved T-N11-A motif that harbors small palindromic sequences at its ends. Such a motif, however, has not been recognized in the mau promoter region, indicating that the target site of MauR is slightly different from the conserved one. The target site for Pseudomonas putida CatR, another member of the LysR-type activators, is G-N11-A (221), suggesting that small deviations of the target sequence do occur. Promoter probe studies have further shown that P. denitrificans mau gene expression increases almost 1,600-fold during growth on methylamine compared to that during heterotrophic growth. This increase was not observed in the P. denitrificans MauR mutant, and this result confirms that MauR is a regulator of methylamine oxidation (65). The expression of the mauR gene itself is more or less constant, independent of the growth conditions and of MauR itself. Apparently, its expression is not autoregulated, a feature which is different from that of most other lysR-type genes. A counterpart of mauR has been described in P. versutus (130). Analysis of the corresponding region upstream of the mau gene cluster from Methylobacterium extorquens AM1 did not reveal a mauR-like gene (48).
Apart from their dedicated activators, the regulation of both mxa and mau gene clusters is subject to some kind of hierarchical control that suppresses transcription during heterotrophic growth. Indeed, it has been shown that the expression of the mau genes is virtually blocked when the bacterium grows in the simultaneous presence of succinate and methylamine (213, 313). Further, both MADH and MDH are absent once succinate is added to the growth medium in addition to their corresponding substrates (119). The view that a global transcription activator is responsible for these phenomena has recently been corroborated by the analysis of a mutant disturbed in many aspects of C1 metabolism (115). This mutant was unable to grow on methanol, methylamine, or choline. Moreover, the expression of the flhA, mxa, mau, and xox genes was completely blocked. These pleiotropic effects were explained by assuming that the mutated gene in this strain encodes a regulator that affects the expression of all these C1 genes and gene clusters. Apparently, this regulator is not a repressor protein, since one would expect constitutive expression rather than repression in the knockout situation. Therefore, it has been suggested that this protein acts as a second activator in synergism with either of the two dedicated activators of mxa and mau gene expression. If this view is correct, one may expect that the presence and/or activity of this second activator would be suppressed during heterotrophic growth. The identification of a gene coding for this second activator has recently been facilitated by the isolation of a genomic locus that fully complements the mutation (116). A scheme of the regulatory pathways that control gene expression during C1 metabolism is shown in Fig. 6.
FIG. 6.
Regulation of gene expression during C1 metabolism in P. denitrificans. P, promoter; FyDH, formaldehyde dehydrogenase; XDH, xox gene products; Ami, amicyanin; SS, unknown signal sensor; RR, unknown response regulator; 2CR, two-component regulatory genes.
Sulfur oxidation.
The periplasmic enzymes specifically involved in the use of thiosulfate by P. versutus (enzyme A, enzyme B, cytochrome c552.5, sulfite:cytochrome c oxidoreductase, cytochrome c551, and cytochrome c552) (Fig. 7) have also been called the thiosulfate-oxidizing multienzyme system (TOMES) (149). This system allows P. versutus to grow autotrophically with compounds such as thiosulfate or sulfite as the sole source of energy, and the system has been biochemically characterized (references 148 and 149 and references therein). P. denitrificans can also use thiosulfate as an energy source for autotrophic growth (86, 319), and P. denitrificans GB-17 (P. pantotrophus) was originally isolated as a facultative chemolithotroph (247). Strain GB-17 was found to be the most amenable strain for Tn5 mutagenesis and subsequent selection for sulfur oxidation (Sox) mutants (43).
FIG. 7.
Thiosulfate-oxidizing multienzyme system. The figure is derived from the scheme devised by Kelly (140) for P. versutus. SCO, sulfite:cytochrome c oxidoreductase; A, enzyme A (SoxA); B, enzyme B (SoxB).
One strain deficient in thiosulfate oxidation (43) was found to be missing one of the megaplasmids (that of 450 kbp) characteristic of P. denitrificans GB-17 strains (43, 190, 312). Sulfur oxidation cannot be definitively mapped exactly to this molecule, since the smaller (110-kbp) megaplasmid was also reduced in size. Another of the Sox mutants of P. denitrificans with a full complement of megaplasmids (TP19) was used to isolate a locus coding for some of the TOMES proteins (188). The 13-kbp region isolated hybridized with identical bands of EcoRI-restricted genomic DNA of type strains of P. denitrificans, but P. versutus showed a different hybridization pattern. Although the sox locus hybridized to R. capsulatus DNA, other thiosulfate-oxidizing bacteria, such as members of the “true” thiobacilli, did not appear to contain homologous sequences. The region in which the Tn5 had interrupted sox expression was named soxB (326); it coded for a periplasmic protein with similarity to the protein sequence of a sulfur oxidation enzyme purified from P. versutus. This enzyme (protein B) has been shown to play an essential part in thiosulfate oxidation (173).
Further sequencing of the sox locus (327) reveals further ORFs, possibly transcriptionally linked to soxB. The ORF named soxC has high identity to the nucleus-encoded sulfite oxidases of mammalian cells. Despite the differences in conserved residues between SoxC and other molybdopterin-containing proteins (327), the presence of an unusually long periplasmic targeting sequence containing a double arginine (20, 349) suggests that P. denitrificans GB-17 SoxC is a molybdoprotein. The activity of SoxC as sulfite dehydrogenase was confirmed by the absence of activity in a soxC mutant (327). Of the remaining ORFs, soxD and soxE code for c-type cytochromes (perhaps corresponding to c551 and c552.5, respectively) whereas soxF codes for a protein with high homology to the flavin domain of Chromatium vinosum flavocytochrome c. The role of this flavoprotein has yet to be integrated into the TOMES scheme proposed by Kelly et al. (149).
To summarize this section on the regulation of respiratory genes, it has emerged that the control of the changeover by P. denitrificans for aerobic growth involves an unanticipated complexity. Particularly intriguing is the existence of the Nnr protein as well as FnrP, when considered in the context that they can each differentiate between apparently identical DNA binding sequences. Hence, FnrP seemingly cannot substitute for Nnr and vice versa. The activation of Nnr by NO is also a remarkable finding. A related complexity applies to the regulation of genes for C1 metabolism. Here again, a toxic intermediate, in this case formaldehyde, appears to be the signalling molecule.
OTHER NONRESPIRATORY SYSTEMS
Members of the genus Paracoccus have not been studied solely with respect to their electron transport chains and unusual modes of metabolism. P. denitrificans, in particular, has proved to be a source of proteins (and their corresponding genes) with properties differing slightly from their better-known E. coli counterparts. In the remaining part of the review, a survey of these genes is presented. The only loci that are not discussed below are those of ribosomal protein S13, preprotein translocase (228), regulators of C1 metabolism (241), and additional methanol oxidation genes (242). Although these loci have been sequenced and deposited in the database, there is no complementary biochemical information available.
Poly-β-Hydroxybutyrate Synthesis
P. denitrificans can produce a variety of polyhydroxyalkanoates, presumably as a reserve of carbon and energy (338). The homopolyester poly(3-hydroxybutyrate) is the predominant product during growth on methanol, but if n-pentanol is used, poly(3-hydroxyvalerate) is produced. The copolyester poly(3-hydroxybutyrate-co-3-hydroxyvalerate) is produced during growth on n-propanol (338). The genes responsible for this process (phA [β-ketothiolase], phaB [acetoacetyl coenzyme A reductase], and phaC [poly(3-hydroxyalkanoate) synthase]) have been identified in several bacteria, notably the closely related R. sphaeroides (274). The genes phaA, phaB (336), and phaC (298) have been identified in P. denitrificans. PhaA is 56% identical to PhaA of R. sphaeroides, and the 1,902-bp gene is possibly transcribed from its own promoter in E. coli (298), as judged from in vivo lacZ fusion studies. No detailed study of the promoter region has been made. Certainly, there is no obvious sequence corresponding to the E. coli ς70 consensus, but the gene is followed by a Rho-independent terminator-like sequence. Reintroduction of the gene into P. denitrificans on a broad-host-range plasmid leads to a twofold increase in the amount of polyhydroxyalkanoate synthesized (298).
Aromatic Amino Acid Transferase
The gene tyrB codes for a multifunctional enzyme, aromatic amino acid transferase, involved in transamination reactions between amino acids (phenylalanine or tyrosine) and 2-oxoglutarate. In E. coli, the action of TyrB is complemented by a very similar enzyme, AspC, which transfers amino groups between aspartate and 2-oxoglutarate (83). The tyrB gene is subject to product repression in E. coli, with the concentration of tyrosine mediating expression of the gene via the TyrR repressor (reference 229 and references therein).
The DNA encoding TyrB from P. denitrificans has been sequenced, and the primary sequence of the protein showed high similarity to that of the E. coli protein (207). The restricted substrate specificity of the P. denitrificans enzyme implies that this protein can be better used as a model for a dual-substrate recognition than can the more catholic E. coli enzyme. Although active P. denitrificans TyrB can be expressed in appropriate mutants of E. coli, little interest has been shown in the regulation of tyrB in P. denitrificans. However, the putative P. denitrificans tyrB promoter/operator region has many elements in common with its enterobacterial counterparts. Sequences with significant identity to E. coli TyrR boxes (33% compared to the TyrR box consensus) can be found in a similar position relative to the tyrB promoters of E. coli (83) and Salmonella typhimurium (194) (Fig. 8). The TyrR boxes in the tyrB genes of E. coli and S. typhimurium have similarly low identity (27 and 33%, respectively) to the consensus. The optimal alignment is found if GTG is considered to be the start codon, rather than the ATG used in vivo (207).
FIG. 8.
Comparison of TyrR-dependent promoters in E. coli (Ec) and S. typhimurium (St) with the tyrB promoter region of P. denitrificans (Pd). The number at the end of the sequence denotes the percent identity of the TyrR box to the TyrR consensus.
The tyrB gene was characterized from an earlier clone of P. denitrificans aromatic amino acid transferase locus, isolated by Tagaki et al. (283). This sequence, although not yet in the databases, has a large region (>1,000 bp) of upstream DNA, which contains an ORF transcribed in the same direction as tyrB (5). This ORF is most likely to be the gene encoding thiosulfate sulfur transferase (see below). Transcriptional readthrough from the putative sulfur transferase gene to tyrB is probably prevented by the rho-independent-like termination sequence found between the two genes (5).
Porin
Porin from P. denitrificans was first described by Zalman and Nikaido (344) as a dimer, but subsequent experiments (320) and its eventual crystallization (124) showed that porin is trimeric. The porin gene (porG) has had its transcript start site determined by S1 nuclease mapping (Table 5). Although an E. coli ς70-like hexamer can be seen at the −10 region, the presence of a −35 hexamer is not clear. The 3′ region of the porG locus includes a terminator-like stem-loop structure (254).
Adenylate Kinase
The adenylate kinase of P. denitrificans is a member of the nucleotide phosphate kinase family (272) and plays a role in phosphate group turnover, allowing the formation of ATP from ADP. Sequencing of the adk gene revealed that the protein from P. denitrificans is unusual in that it is more similar to the adenylate kinases from gram-positive bacteria than to those of its nearer relations, the gram-negative bacteria (66). Adenylate kinase from P. denitrificans is thought to contain zinc and iron, although the exact binding site of these ions has yet to be determined (66).
Thiosulfate Sulfur Transferase (Rhodanese)
A protein translated from DNA sequenced by Tagaki et al. (283), upstream of tyrB (see above), showed high homology to the thiosulfate sulfur transferases (rhodanese) of E. coli, (57% identity) and to a lesser extent to the mammalian isoenzymes from cows (41% identity) and humans (36% identity). The identities were deduced after the published ORF had frameshifts resolved to maximize identity (5). The function of this protein in either eubacterial or mammalian cells is not clear, but it has been implicated in the cysteine and methionine catabolic pathways and also in the detoxification of cyanide. ORFs similar to those coding for rhodanese have been found in all the eubacterial genomes sequenced so far. Although rhodanese is periplasmic in P. versutus (174), the signal sequence in the P. denitrificans protein is not clear.
It is difficult from the homology alone to propose a function for the rhodanese ORF. The E. coli genome (28) contains three rhodanese-like ORFs. Despite the name, this class of protein is not implicated in the oxidation of thiosulfate in P. versutus (174). Further upstream of the putative rhodanese gene is another ORF, probably coding for the polypeptide named “small protein b” (sprB [186]) in the E. coli, Neisseria meningitidis, Haemophilus influenzae, and Enterococcus faecalis genome-sequencing projects.
CONCLUDING REMARKS
The endowment of considerable metabolic versatility, especially with regard to respiration, requires P. denitrificans to have mechanisms for selecting which set or sets of respiratory genes to express. One prominent example is the switch from aerobic respiration with oxygen as a terminal electron acceptor to anaerobic respiration with an alternative terminal electron acceptor. In the latter situation, (i.e., denitrification), reductases for nitrate, nitrite, nitric oxide, or nitrous oxide have to be expressed along with any genes necessary for their biogenesis. It might have been expected, contrary to the experimental results, that a single global regulator would control denitrification because P. denitrificans and other denitrifying members of the genus Paracoccus have only two choices to sustain ATP production, aerobic or anaerobic respiration, there being no mode of fermentation.
The preference for aerobic respiration can be rationalized on the basis that passage of electrons to any oxidase of the respiratory system probably leads to a higher stoichiometry of proton translocation, and hence of ATP synthesis, than does electron transport to any of the reductases associated with the denitrification mode of respiration. It is emerging that the switch from aerobic to anaerobic respiration is controlled by a number of interacting factors rather than a single global factor. Clearly, there is a complex regulatory network. As the dissolved-oxygen tension declines, the transcriptional regulator FnrP is activated, thus stimulating the expression of the high-affinity oxidase (cytochrome cbb3) as well as the respiratory nitrate reductase. It is likely, though not yet proven, that the expression of the latter will be further stimulated by the availability of nitrate, sensed by a NarL/NarX system analogous to that found in E. coli. However, the reductases for nitrite, nitric oxide, or nitrous oxide are not controlled, at least directly, by FnrP. Under natural conditions, it is possible that P. denitrificans encounters nitrous oxide alone, in which case switching on the entire denitrification pathway from the nitrate reductase onward would be wasteful. Nitrite and nitric oxide reductases must be coexpressed to stop the accumulation of toxic nitric oxide. They require for their expression the active form of Nnr, a second FNR-like transcription factor whose activation mechanism is at present unclear, despite the sequence similarity to FNR both in protein sequence and in the DNA binding sites upstream of the genes that the two proteins control. The evidence for the mechanism of action of Nnr suggests that the controlling signal is nitric oxide, in which case we are confronted with the relatively unusual situation in which the product of the nitrite reductase reaction activates expression of the enzyme responsible for its formation.
The denitrification system of respiratory proteins certainly has yet to reveal all the secrets of its regulatory mechanisms. For example, NirI, a protein of an unknown function, plays an undefined role in mechanisms of control for the expression of nitrous oxide reductase, which appears to be independent of both the FnrP and Nnr regulons. The reason for this complexity is not clear. To an extent, it may reflect the fact that the complete denitrification pathway comprises a set of proteins which individually or in some combination are also found in nondenitrifying bacteria (348). Thus, for example, nitrate reductase, with a similar gene organization and likely regulation to P. denitrificans, is found in E. coli. The occurrence of nitrite reductase genes and nitric oxide reductase genes in a cluster along with DNA coding for their regulatory factors on the genome of P. denitrificans could mean that these proteins were utilized by the bacterium later in its evolution, after nitrate reductase, and that the regulation of the complete denitrification pathway is overlaid on other existing sensory mechanisms.
The elucidation of the complex regulatory network that controls denitrification also requires identification of the types of RNAP that are involved. Our scrutiny of promoters in the alpha Proteobacteria shows that the previous consensus proposed for a ς70 polymerase binding site (5′-TCGGGN- N(18 ± 2)-GATNGS-3′) (277), a binding site that is supposedly unique to Rhodobacter and Paracoccus, is not sufficient to define housekeeping promoters. Our analysis shows that Paracoccus and the alpha Proteobacteria have RNAP binding sites with some resemblance to the canonical E. coli sequence (5′-TTGACA- N(17 ± 1)-TATAAT-3′) (120), even though the data set is biased toward respiratory genes. The occurrence of canonical ς70 promoters is also indicated by the observation that counterparts of other E. coli DNA binding proteins recognize sequences similar to those described for E. coli. Clear examples are the binding sites for TyrR and FnrP in P. denitrificans. However, the definition of the fine structure of Paracoccus promoters requires the acquisition of more data, particularly from genes involved in the housekeeping functions of Paracoccus. However, preliminary results from the sequences of several promoter regions indicate that other sigma factors are involved in the transcription of denitrification genes. The discrimination between FnrP and Nnr activation could be on this basis.
Whereas the switch from aerobic respiration to denitrification involves a choice of electron acceptor, a choice of electron donor can also be made by P. denitrificans. Thus, if the bacterium encounters both a heterotrophic substrate (such as glucose, succinate or mannitol) and a C1 substrate, like methanol or methylamine, its strategy is such that it will first metabolize the heterotrophic substrate and then metabolize the C1 substrate. This strategy is sensible in that not only is the growth yield on heterotrophic substrates higher than that on C1 substrates but also growth on the latter will present the cell with the toxic intermediate formaldehyde. The ability to choose the potential electron source is achieved by a very complex signal transduction network operating at the level of transcription. The current view envisages that all of the genes required for C1 metabolism are silent during heterotrophic growth because a global transcriptional activator required for the transcription of all of them either is not synthesized or is inactive under these growth conditions. The concentration and/or activity of the global activator increases during depletion of the heterotrophic substrate and prepares the cell to switch metabolism. A dedicated signal sensor of a two-component regulatory system is activated by detecting formaldehyde, which is the reaction product of methanol oxidation. The partner response regulator in turn acts synergistically with the global activator, and the mxa operon, encoding the enzymes required for methanol metabolism, is transcribed. In a similar sequence of events, enzymes required for methylamine metabolism are expressed. Signal transduction in this case, however, is mediated by a one-component transcriptional activator, MauR, which induces the mau operon encoding methylamine dehydrogenase upon binding of methylamine.
This survey of the molecular biology of Paracoccus has revealed that genes encoding respiratory proteins are scattered over both the replicons. It would appear that there is no need for a proximal location for these genes, a situation somewhat different from that found in clusters of genes encoding more defined metabolic pathways, such as the mxa, mau, nir, nor, and nos genes. These genomic loci encompass the genes encoding not only the structural protein but also the regulators of transcription, together with proteins required for activation, cofactor processing, and transport. In some instances, an electron transport donor or acceptor protein is also found on contiguous DNA (e.g., the mxa system). A coherent expression of all these proteins allows an efficient and rapid adaptation.
ACKNOWLEDGMENTS
We acknowledge Dr. D. Richardson (University of East Anglia), N. F. W. Saunders (University of Oxford), and N. Harms (Vrije Universiteit Amsterdam) for useful discussions. We also thank N. F. W. Saunders for permission to use Fig. 6. We thank those who allowed us access to unpublished or submitted material.
R.S. acknowledges support by the Netherlands Foundation for Chemical Research, and the Netherlands Organization for Scientific Research. O.-M.H.R. thanks the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie for support through grants to B.L. S.C.B. was supported by BBSRC grant GR/J36457 to S.J.F.
REFERENCES
- 1.Alberti M, Burke D H, Hearst J E. Structure and sequence of the photosynthetic gene cluster. In: Blakenship R E, Madigan M T, Bauer C E, editors. Advances in photosynthesis, anoxygenic photosynthetic bacteria. Vol. 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1083–1106. [Google Scholar]
- 2.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;37:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- 4.Arigoni F, Kaminski P A, Hennecke H, Elmerich C. Nucleotide sequence of the fixABC region of Azorhizobium caulinodans ORS571: similarity of the fixB product with eukaryotic flavoproteins, characterization of fixX, and identification of nifW. Mol Gen Genet. 1991;225:514–520. doi: 10.1007/BF00261695. [DOI] [PubMed] [Google Scholar]
- 5.Baker, S. C., N. F. W. Saunders, and S. J. Ferguson. Unpublished results.
- 6.Baker S C, Saunders N F W, Willis A C, Ferguson S J, Hajdu J, Fulop V. Cytochrome cd1 structure: unusual haem environments in a nitrite reductase and analysis of factors contributing to beta-propeller folds. J Mol Biol. 1997;269:440–455. doi: 10.1006/jmbi.1997.1070. [DOI] [PubMed] [Google Scholar]
- 7.Bartnikas T B, Tosques I E, Laratta W P, Shi J, Shapleigh J P. Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:3534–3540. doi: 10.1128/jb.179.11.3534-3540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartosik D, Baj J, Plasota M, Piechucka E, Wlodarczyk M. Analysis of Thiobacillus versutus pTAV1 plasmid functions. Acta Microbiol Pol. 1993;42:97–100. [Google Scholar]
- 9.Bartosik D, Baj J, Wlodarczyk M. Construction and preliminary characterisation of mini-derivatives of a large (107-kb) cryptic plasmid of the sulfur bacterium Thiobacillus versutus. FEMS Microbiol Lett. 1995;129:169–174. [Google Scholar]
- 10.Bates D M, Lazazzera B A, Kiley P J. Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J Bacteriol. 1995;177:3972–3978. doi: 10.1128/jb.177.14.3972-3978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer C E, Young D A, Marrs B L. Analysis of the Rhodobacter capsulatus puf operon. Location of the oxygen-regulated promoter region and the identification of an additional puf-encoded gene. J Biol Chem. 1988;263:4820–4827. [PubMed] [Google Scholar]
- 12.Bedzyk L A, Escudero K W, Gill R E, Griffin K J, Frerman F E. Cloning, sequencing, and expression of the genes encoding subunits of Paracoccus denitrificans electron transfer flavoprotein. J Biol Chem. 1993;268:20211–20217. [PubMed] [Google Scholar]
- 13.Beijerinck M. Bildung und Verbrauch von Stickoxydul durch Bakterien. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt II. 1910;25:30–63. [Google Scholar]
- 14.Bell L C, Ferguson S J. Nitric and nitrous oxide reductases are active under aerobic conditions in cells of Thiosphaera pantotropha. Biochem J. 1991;273:423–427. doi: 10.1042/bj2730423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell L C, Page M D, Berks B C, Richardson D J, Ferguson S J. Insertion of transposon Tn5 into a structural gene of the membrane-bound nitrate reductase of Thiosphaera pantotropha results in anaerobic overexpression of periplasmic nitrate reductase activity. J Gen Microbiol. 1993;139:3205–3214. doi: 10.1099/00221287-139-12-3205. [DOI] [PubMed] [Google Scholar]
- 16.Bell L C, Richardson D J, Ferguson S J. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. The periplasmic enzyme catalyzes the first step in aerobic denitrification. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- 17.Bell L C, Richardson D J, Ferguson S J. Identification of nitric oxide reductase activity in Rhodobacter capsulatus: the electron transport pathway can either use or bypass both cytochrome c2 and the cytochrome bc1 complex. J Gen Microbiol. 1992;138:437–443. doi: 10.1099/00221287-138-3-437. [DOI] [PubMed] [Google Scholar]
- 18.Bennett B, Charnock J M, Sears H J, Berks B C, Thomson A J, Ferguson S J, Garner C D, Richardson D J. Structural investigation of the molybdenum site of the periplasmic nitrate reductase from Thiosphaera pantotropha by X-ray absorption spectroscopy. Biochem J. 1996;317:557–563. doi: 10.1042/bj3170557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benning M M, Meyer T E, Holden H M. X-ray structure of the cytochrome c2 isolated from Paracoccus denitrificans refined to 1.7-A resolution. Arch Biochem Biophys. 1994;310:460–466. doi: 10.1006/abbi.1994.1193. [DOI] [PubMed] [Google Scholar]
- 20.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 21.Berks B C, Baratta D, Richardson J, Ferguson S J. Purification and characterization of a nitrous oxide reductase from Thiosphaera pantotropha. Implications for the mechanism of aerobic nitrous oxide reduction. Eur J Biochem. 1993;212:467–476. doi: 10.1111/j.1432-1033.1993.tb17683.x. [DOI] [PubMed] [Google Scholar]
- 22.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyze the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 23.Berks B C, Page M D, Richardson D J, Reilly A, Cavill A, Outen F, Ferguson S J. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol Microbiol. 1995;15:319–331. doi: 10.1111/j.1365-2958.1995.tb02246.x. [DOI] [PubMed] [Google Scholar]
- 24.Berks B C, Richardson D J, Reilly A, Willis A C, Ferguson S J. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem J. 1995;309:983–992. doi: 10.1042/bj3090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berks B C, Richardson D J, Robinson C, Reilly A, Aplin R T, Ferguson S J. Purification and characterization of the periplasmic nitrate reductase from Thiosphaera pantotropha. Eur J Biochem. 1994;220:117–124. doi: 10.1111/j.1432-1033.1994.tb18605.x. [DOI] [PubMed] [Google Scholar]
- 26.Berry E A, Trumpower B L. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 27.Blackall L L, Rossetti S, Christensson C, Cunningham M, Hartman P, Hugenholtz P, Tandoi V. The characterization and description of representatives of ‘G’ bacteria from activated sludge plants. Lett Appl Microbiol. 1997;25:63–69. doi: 10.1046/j.1472-765x.1997.00176.x. [DOI] [PubMed] [Google Scholar]
- 28.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 29.Bonin P, Gilewicz M. A direct demonstration of “co-respiration” of oxygen and nitrogen oxides by Pseudomonas nautica: some spectral and kinetic properties of the respiratory components. FEMS Microbiol Lett. 1991;80:183–188. [Google Scholar]
- 30.Boogerd F C, van Verseveld H W, Stouthamer A H. Electron transport to nitrous oxide in Paracoccus denitrificans. FEBS Lett. 1980;113:279–284. doi: 10.1016/0014-5793(80)80609-0. [DOI] [PubMed] [Google Scholar]
- 31.Borghese R, Crimi M, Fava L, Melandri B A. The ATP synthase atpHAGDC (F-1) operon from Rhodobacter capsulatus. J Bacteriol. 1998;180:416–421. doi: 10.1128/jb.180.2.416-421.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosma G. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1989. [Google Scholar]
- 33.Bosma G, Braster M, Stouthamer A H, van Verseveld H W. Subfractionation and characterization of soluble c-type cytochromes from Paracoccus denitrificans cultured under various limiting conditions in the chemostat. Eur J Biochem. 1987;165:665–670. doi: 10.1111/j.1432-1033.1987.tb11492.x. [DOI] [PubMed] [Google Scholar]
- 34.Braun C, Zumft W G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J Bacteriol. 1992;174:2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breton J, Berks B C, Reilly A, Thomson A J, Ferguson S J, Richardson D J. Characterization of the paramagnetic iron-containing redox centres of Thiosphaera pantotropha periplasmic nitrate reductase. FEBS Lett. 1994;345:76–80. doi: 10.1016/0014-5793(94)00445-5. [DOI] [PubMed] [Google Scholar]
- 36.Burns H D, Belyaeva T A, Busby S J W, Minchin S D. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem J. 1996;317:305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannon W, Missailidis S, Austin S, Moore M, Drake A, Buck M. Purification and activities of the Rhodobacter capsulatus RpoN (sigma(N)) protein. Mol Microbiol. 1996;21:233–245. doi: 10.1046/j.1365-2958.1996.6181334.x. [DOI] [PubMed] [Google Scholar]
- 38.Capaldi R A. Structure and function of cytochrome c oxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- 39.Carr G J, Ferguson S J. Nitric oxide formed by nitrite reductase of Paracoccus denitrificans is sufficiently stable to inhibit cytochrome oxidase activity and is reduced by its reductase under aerobic conditions. Biochim Biophys Acta. 1990;1017:57–62. doi: 10.1016/0005-2728(90)90178-7. [DOI] [PubMed] [Google Scholar]
- 40.Carr G J, Page M D, Ferguson S J. The energy conserving nitric oxide reductase system in Paracoccus denitrificans: distinction from the nitrite reductase that catalyses synthesis of nitric oxide and evidence from trapping experiments for nitric oxide as a free intermediate during denitrification. Eur J Biochem. 1989;179:683–692. doi: 10.1111/j.1432-1033.1989.tb14601.x. [DOI] [PubMed] [Google Scholar]
- 41.Carter J P, Hsiao Y H, Spiro S, Richardson D J. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl Environ Microbiol. 1995;61:2852–2858. doi: 10.1128/aem.61.8.2852-2858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castresana J, Lubben M, Saraste M, Higgins D G. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandra T S, Friedrich C G. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J Bacteriol. 1986;166:446–452. doi: 10.1128/jb.166.2.446-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C W, Yu T W, Chung H M, Chou C F. Discovery and characterization of a new transposable element, Tn4811, in Streptomyces lividans 66. J Bacteriol. 1992;174:7762–7769. doi: 10.1128/jb.174.23.7762-7769.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Durley R C E, Mathews F S, Davidson V L. Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i. Science. 1994;264:86–90. doi: 10.1126/science.8140419. [DOI] [PubMed] [Google Scholar]
- 46.Chen L, Mathews F S, Davidson V L, Huizinga E G, Vellieux F M, Hol W G. Three-dimensional structure of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans determined by molecular replacement at 2.8 A resolution. Proteins. 1992;14:288–299. doi: 10.1002/prot.340140214. [DOI] [PubMed] [Google Scholar]
- 47.Chepuri V, Lemieux L, Au D C-T, Gennis R B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990;265:11185–11192. [PubMed] [Google Scholar]
- 48.Chistoserdov A Y, Boyd J, Mathews F S, Lidstrom M E. The genetic organization of the mau gene cluster of the facultative autotroph Paracoccus denitrificans. Biochem Biophys Res Commun. 1992;184:1181–1189. doi: 10.1016/s0006-291x(05)80007-5. [DOI] [PubMed] [Google Scholar]
- 49.Chistoserdov A Y, Chistoserdova L V, McIntire W S, Lidstrom M E. Genetic organization of the mau gene cluster in Methylobacterium extorquens AM1: complete nucleotide sequence and generation and characteristics of mau mutants. J Bacteriol. 1994;176:4052–4065. doi: 10.1128/jb.176.13.4052-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coucheron D H. A family of IS1031 elements in the genome of Acetobacter xylinum: nucleotide sequences and strain distribution. Mol Microbiol. 1993;9:211–218. doi: 10.1111/j.1365-2958.1993.tb01682.x. [DOI] [PubMed] [Google Scholar]
- 51.Cox J C, Ingledew W J, Haddock B A, Lawford H G. The variable cytochrome content of Paracoccus denitrificans grown aerobically under different conditions. FEBS Lett. 1978;93:261–265. doi: 10.1016/0014-5793(78)81117-x. [DOI] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Crouzet J, Cameron B, Cauchois L, Rigault S, Rouyez M C, Blanche F, Thibaut D, Debussche L. Genetic and sequence analysis of an 8.7-kilobase Pseudomonas denitrificans fragment carrying eight genes involved in transformation of precorrin 2 to cobyrinic acid. J Bacteriol. 1990;172:5980–5990. doi: 10.1128/jb.172.10.5980-5990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cullen P J, Kaufman K J, Bowman W C, Kranz R G. Characterization of the Rhodobacter capsulatus housekeeping RNA polymerase. J Biol Chem. 1997;272:27266–27273. doi: 10.1074/jbc.272.43.27266. [DOI] [PubMed] [Google Scholar]
- 55.Cuypers H, Viebrock-Sambale A, Zumft W G. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J Bacteriol. 1992;174:5332–5339. doi: 10.1128/jb.174.16.5332-5339.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidson V L, Husain M, Neher J W. Electron transfer flavoprotein from Methylophilus methylotrophus: properties, comparison with other electron transfer flavoproteins, and regulation of expression by carbon source. J Bacteriol. 1986;166:812–817. doi: 10.1128/jb.166.3.812-817.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson V L, Kumar M A. Cytochrome c550 mediates electron transfer from inducible periplasmic c-type cytochromes to the cytoplasmic membrane of Paracoccus denitrificans. FEBS Lett. 1989;245:271–273. doi: 10.1016/0014-5793(89)80235-2. [DOI] [PubMed] [Google Scholar]
- 58.Davies K J, Lloyd D, Boddy L. The effect of oxygen on denitrification in Paracoccus denitrificans and Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:2445–2451. doi: 10.1099/00221287-135-9-2445. [DOI] [PubMed] [Google Scholar]
- 59.de Boer A P N, Reijnders W N M, Kuenen J G, Stouthamer A H, van Spanning R J M. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction in Paracoccus denitrificans. Antonie Leeuwenhoek. 1994;66:111–127. doi: 10.1007/BF00871635. [DOI] [PubMed] [Google Scholar]
- 60.de Boer A P N, van der Oost J, Reijnders W N M, Westerhoff H V, Stouthamer A H, van Spanning R J M. Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur J Biochem. 1996;242:592–600. doi: 10.1111/j.1432-1033.1996.0592r.x. [DOI] [PubMed] [Google Scholar]
- 61.de Gier J W L. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1995. [Google Scholar]
- 62.de Gier J W L, Luebben M, Reijnders W N M, Tipker C A, Slotboom D J, van Spanning R J M, Stouthamer A H, van der Oost J. Terminal oxidases of Paracoccus denitrificans. Mol Microbiol. 1994;13:183–196. doi: 10.1111/j.1365-2958.1994.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 63.de Gier J W L, Schepper M, Reijnders W N M, Vandyck S J, Slotboom D J, Warne A, Saraste M, Krab K, Finel M, Stouthamer A H, van Spanning R J M, van der Oost J. Structural and functional analysis of aa3-type and cbb3-type cytochrome c oxidases of Paracoccus denitrificans reveals significant differences in proton-pump design. Mol Microbiol. 1996;20:1247–1260. doi: 10.1111/j.1365-2958.1996.tb02644.x. [DOI] [PubMed] [Google Scholar]
- 64.de Gier J W L, van der Oost J, Harms N, Stouthamer A H, van Spanning R J M. The oxidation of methylamine in Paracoccus denitrificans. Eur J Biochem. 1995;229:148–154. doi: 10.1111/j.1432-1033.1995.0148l.x. [DOI] [PubMed] [Google Scholar]
- 65.Delorme C, Huisman T T, Reijnders W N M, Chan Y L, Harms N, Stouthamer A H, van Spanning R J M. Expression of the mau gene cluster of Paracoccus denitrificans is controlled by MauR and a second transcription regulator. Microbiology. 1997;143:793–801. doi: 10.1099/00221287-143-3-793. [DOI] [PubMed] [Google Scholar]
- 66.Deligiannakis Y, Boussac A, Botin H, Perrier V, Barzu O, Giles A M. A new non-heme iron environment in Paracoccus denitrificans adenylate kinase studied by electron paramagnetic resonance and electron spin echo envelope modulation spectroscopy. Biochemistry. 1997;361:9446–9452. doi: 10.1021/bi970021e. [DOI] [PubMed] [Google Scholar]
- 67.de Miersman C, van Soom C, Verreth C, van Gool A, Vanderleyden J. Nucleotide sequence of IS427 and its target sites in Agrobacterium tumefaciens T37. Plasmid. 1990;24:227–234. doi: 10.1016/0147-619x(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 68.de Vries G E, Harms N, Hoogendijk J, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a (nGATCn)-DNA-modifying property. Arch Microbiol. 1989;152:52–57. [Google Scholar]
- 69.Dickins, M. A., T. Dhawan, R. P. Gunsalus, I. Schroeder, and G. Cecchini. Cloning, sequencing and expression of the succinate-ubiquinone oxidoreductase (sdhCDAB) operon from Paracoccus denitrificans. Unpublished results. Accession no. U31902.
- 70.Donohue T J, McEwan A G, Kaplan S. Cloning, DNA sequence, and expression of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1986;168:962–972. doi: 10.1128/jb.168.2.962-972.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dryden S C, Kaplan S. Identification of cis acting regulatory regions upstream of the rRNA operons of Rhodobacter sphaeroides. J Bacteriol. 1993;175:6392–6402. doi: 10.1128/jb.175.20.6392-6402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duport C, Jouanneau Y, Vignais P M. Transcriptional analysis and promoter mapping of the fdxA gene which encodes the 7Fe ferredoxin (FdII) of Rhodobacter capsulatus. Mol Gen Genet. 1992;231:323–328. doi: 10.1007/BF00279806. [DOI] [PubMed] [Google Scholar]
- 73.Durley R, Chen L, Lim L W, Mathews F S, Davidson V L. Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution. Protein Sci. 1993;2:739–752. doi: 10.1002/pro.5560020506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73a.Ferguson S J. The Paracoccus denitrificans electron transport system: aspects of organization, structures and biogenesis. NATO ASI Ser. 1998;c512:77–88. [Google Scholar]
- 74.Ferguson S J, Jackson J B, McEwan A G. Anaerobic respiration in the Rhodospirillaceae: characterisation of pathways and evaluation of roles in redox balancing during photosynthesis. FEMS Microbiol Rev. 1987;46:117–143. [Google Scholar]
- 75.Fernandez de Henestrosa A R, del Rey A, Tarraga R, Barbé J. Cloning and characterization of the recA gene of Paracoccus denitrificans and construction of a recA-deficient mutant. FEMS Microbiol Lett. 1997;147:209–213. doi: 10.1111/j.1574-6968.1997.tb10243.x. [DOI] [PubMed] [Google Scholar]
- 76.Fernandez de Henestrosa A R, Labazi M, Lopez M M, Barbé J. Functional analysis of the recA promoter of Rhodobacter capsulatus. Mol Gen Genet. 1997;255:487–494. doi: 10.1007/s004380050521. [DOI] [PubMed] [Google Scholar]
- 77.Ferrante A A, Lessie T G. Nucleotide sequence of IS402 from Pseudomonas cepacia. Gene. 1991;102:143–144. doi: 10.1016/0378-1119(91)90555-p. [DOI] [PubMed] [Google Scholar]
- 78.Feurst J A, Hawkins J A, Holmes A, Sly L I, Moore C, Stackebrandt E. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from freshwater. Int J Syst Bacteriol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 79.Flory J E, Donohue T J. Transcriptional control of several aerobically induced cytochrome structural genes in Rhodobacter sphaeroides. Microbiology. 1997;143:3101–3110. doi: 10.1099/00221287-143-10-3101. [DOI] [PubMed] [Google Scholar]
- 80.Flory J E, Donohue T J. Organization and expression of the Rhodobacter sphaeroides cycFG operon. J Bacteriol. 1995;177:4311–4320. doi: 10.1128/jb.177.15.4311-4320.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forkl H, Drews G, Tadros M H. Promoter analysis of the catalase-peroxidase gene (cpeA) from Rhodobacter capsulatus. FEMS Microbiol Lett. 1996;137:169–174. doi: 10.1111/j.1574-6968.1996.tb08101.x. [DOI] [PubMed] [Google Scholar]
- 82.Foster-Hartnett D, Kranz R G. The Rhodobacter capsulatus glnB gene is regulated by NtrC at tandem rpoN-independent promoters. J Bacteriol. 1994;176:5171–5176. doi: 10.1128/jb.176.16.5171-5176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reference deleted.
- 84.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 85.Frey J, Mudd E A, Krisch H M. A bacteriophage T4 expression cassette that functions efficiently in a wide range of gram-negative bacteria. Gene. 1988;62:237–247. doi: 10.1016/0378-1119(88)90562-8. [DOI] [PubMed] [Google Scholar]
- 86.Friedrich C G, Mitrenga G. Oxidation of thiosulfate by Paracoccus denitrificans and other hydrogen bacteria. FEMS Microbiol Lett. 1981;10:209–212. [Google Scholar]
- 87.Fülöp V, Moir J W, Ferguson S J, Hajdù J. The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell. 1995;81:369–377. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- 88.Gabel C, Bittinger M A, Maier R J. Cytochrome aa3 gene regulation in members of the family Rhizobiaceae: comparison of copper and oxygen effects in Bradyrhizobium japonicum and Rhizobium tropici. Appl Environ Microbiol. 1994;60:141–148. doi: 10.1128/aem.60.1.141-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gabellini N, Sebald W. Nucleotide sequence and transcription of the fbc operon from Rhodopseudomonas sphaeroides. Evaluation of the deduced amino acid sequences of the FeS protein, cytochrome b and cytochrome c1. Eur J Biochem. 1986;154:569–579. doi: 10.1111/j.1432-1033.1986.tb09437.x. [DOI] [PubMed] [Google Scholar]
- 90.Galas D J, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 91.Garcià-Horsman J A, Berry E, Shapleigh J P, Alben J O, Gennis G B. A novel cytochrome c oxidase from Rhodobacter sphaeroides that lacks CuA. Biochemistry. 1994;33:3113–3119. doi: 10.1021/bi00176a046. [DOI] [PubMed] [Google Scholar]
- 92.Gerhus E, Steinrucke P, Ludwig B. Paracoccus denitrificans cytochrome c1 gene replacement mutants. J Bacteriol. 1990;172:2392–2400. doi: 10.1128/jb.172.5.2392-2400.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gibson J L, Tabita F R. Localization and mapping of CO2 fixation genes within two gene clusters in Rhodobacter sphaeroides. J Bacteriol. 1988;170:2153–2158. doi: 10.1128/jb.170.5.2153-2158.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gibson L C, McGlynn P, Chaudhri M, Hunter C N. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. II. Analysis of a region of the genome encoding hemF and the puc operon. Mol Microbiol. 1992;6:3171–3186. doi: 10.1111/j.1365-2958.1992.tb01773.x. [DOI] [PubMed] [Google Scholar]
- 95.Gicquel-Sanzey B, Cossart P. Homologies between different prokaryotic DNA-binding regulatory proteins and between their sites of action. EMBO J. 1982;1:591–595. doi: 10.1002/j.1460-2075.1982.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Girsch P, de Vries S. Purification and initial kinetic and spectroscopic characterization of NO reductase from Paracoccus denitrificans. Biochim Biophys Acta. 1997;1318:202–216. doi: 10.1016/s0005-2728(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 97.Glockner A B, Zumft W G. Sequence analysis of an internal 9.72 kb segment from the 30 kb denitrification gene cluster of Pseudomonas stutzeri. Biochim Biophys Acta. 1996;1277:6–12. doi: 10.1016/s0005-2728(96)00108-9. [DOI] [PubMed] [Google Scholar]
- 98.Gohlke U, Warne A, Saraste M. Projection structure of the cytochrome bo ubiquinol oxidase from Escherichia coli at 6 Å resolution. EMBO J. 1997;16:1181–1188. doi: 10.1093/emboj/16.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gomelsky M, Kaplan S. The Rhodobacter sphaeroides 2.4.1 rho gene: expression and genetic analysis of structure and function. J Bacteriol. 1996;178:1946–1954. doi: 10.1128/jb.178.7.1946-1954.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gonzalez J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goodhew C F, Pettigrew G W, Devreese B, Vanbeeumen J, Van Spanning R J M, Baker S C, Saunders N, Ferguson S J, Thompson I P. The cytochromes c550 of Paracoccus denitrificans and Thiosphaera pantotropha: a need for re-evaluation of the history of Paracoccus cultures. FEMS Microbiol Lett. 1996;137:95–101. [Google Scholar]
- 102.Goretski J, Hollocher T C. Trapping of nitric oxide produced during denitrification by extracellular hemoglobin. J Biol Chem. 1988;263:2316–2323. [PubMed] [Google Scholar]
- 103.Gosink, J. J. 1998. Taxonomy, biogeography, and evolution of polar gas vacuolate bacteria. Unpublished results. Accession no. U73725.
- 104.Green J, Bennett B, Jordan P, Ralph E T, Thomson A J, Guest J R. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem J. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gregory S, O’Connor M, Dahlberg A. Functional Escherichia coli 23S rRNAs containing processed and unprocessed intervening sequences from Salmonella typhimurium. Nucleic Acids Res. 1996;24:4918–4923. doi: 10.1093/nar/24.24.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gruber T M, Bryant D A. Molecular systematic studies of Eubacteria, using ς70-type sigma factors of group 1 and group 2. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gunsalus R P, Park S J. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 108.Haltia T, Finel M, Harms N, Nakari T, Raitio M, Wikstrom M, Saraste M. Deletion of the gene for subunit III leads to defective assembly of bacterial cytochrome oxidase. EMBO J. 1989;8:3571–3579. doi: 10.1002/j.1460-2075.1989.tb08529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haltia T, Puustinen A, Finel M. The Paracoccus denitrificans aa3 has a third subunit. Eur J Biochem. 1988;172:543–546. doi: 10.1111/j.1432-1033.1988.tb13923.x. [DOI] [PubMed] [Google Scholar]
- 110.Haltia T, Semo N, Arrondo J L R, Goin F M, Freire E. Thermodynamic and structural stability of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 1994;33:9731–9741. doi: 10.1021/bi00198a044. [DOI] [PubMed] [Google Scholar]
- 111.Hamada, T. 1998. Use of gyrB gene analysis to investigate phylogeny of marine bacteria. Unpublished results. Accession no. AB012914.
- 112.Harker M, Hirschberg J, Oren A. Paracoccus marcusii sp. nov., an orange gram-negative coccus. Int J Syst Bacteriol. 1998;48:543–548. doi: 10.1099/00207713-48-2-543. [DOI] [PubMed] [Google Scholar]
- 113.Harms N. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1988. [Google Scholar]
- 114.Harms N, de Vries G E, Maurer K, Hoogendijk J, Stouthamer A H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987;169:3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harms N, de-Vries G E, Maurer K, Veltkamp E, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with defects in the metabolism of one-carbon compounds. J Bacteriol. 1985;164:1064–1070. doi: 10.1128/jb.164.3.1064-1070.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harms N, Ras J, Koning S, Reijnders W M N, Stouthamer A H, van Spanning R J M. Genetics of C1 metabolism regulation in Paracoccus denitrificans. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Delft, The Netherlands: Kluwer Academic Publishers; 1996. pp. 126–132. [Google Scholar]
- 117.Harms N, Ras J, Reijnders W N, van Spanning R J M, Stouthamer A H. S-Formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification. J Bacteriol. 1996;178:6296–6299. doi: 10.1128/jb.178.21.6296-6299.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Harms N, Reijnders W N M, Anazawa H, van der Palen C J, van Spanning R J M, Oltmann L F, Stouthamer A H. Identification of a two-component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993;8:457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 119.Harms N, van Spanning R J M. C1 metabolism in Paracoccus denitrificans: genetics of Paracoccus denitrificans. J Bioenerg Biomembr. 1991;23:187–210. doi: 10.1007/BF00762217. [DOI] [PubMed] [Google Scholar]
- 120.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heck C, Rothfuchs R, Jđger A, Rauhut R, Klug G. Effect of the pufQ-pufB intercistronic region on puf mRNA stability in Rhodobacter capsulatus. Mol Microbiol. 1996;20:1165–1178. doi: 10.1111/j.1365-2958.1996.tb02637.x. [DOI] [PubMed] [Google Scholar]
- 122.Heiss B, Frunzke K, Zumft W G. Formation of the N-N bond from nitric oxide by a membrane-bound cytochrome bc complex of nitrate-respiring (denitrifying) Pseudomonas stutzeri. J Bacteriol. 1989;171:3288–3297. doi: 10.1128/jb.171.6.3288-3297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hill B C. The sequence of electron carriers in the reaction of cytochrome c with oxygen. J Bioenerg Biomembr. 1993;25:115–120. doi: 10.1007/BF00762853. [DOI] [PubMed] [Google Scholar]
- 124.Hirsch A, Breed J, Saxena K, Richter O-H M, Ludwig B, Diedrichs K, Welte W. The structural porin from Paracoccus denitrificans at 3.1 Å resolution. FEBS Lett. 1997;404:208–210. doi: 10.1016/s0014-5793(97)00131-2. [DOI] [PubMed] [Google Scholar]
- 125.Hoeren F U, Berks B C, Ferguson S J, McCarthy J E G. Sequence and expression of the gene encoding the respiratory nitrous-oxide reductase from Paracoccus denitrificans: new and conserved structural and regulatory motifs. Eur J Biochem. 1993;218:49–57. doi: 10.1111/j.1432-1033.1993.tb18350.x. [DOI] [PubMed] [Google Scholar]
- 126.Hofhaus G, Weiss H, Leonard K. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (complex I) J Mol Biol. 1991;221:1027–1044. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 127.Hornberger U, Liebetanz R, Tichy H-V, Drews G. Cloning and sequencing of the hemA gene of Rhodobacter capsulatus and isolation of a 5-aminolevulinic acid-dependent mutant strain. Mol Gen Genet. 1990;221:371–378. doi: 10.1007/BF00259402. [DOI] [PubMed] [Google Scholar]
- 128.Hosler P, Ferguson-Miller S, Calhoun M W, Thomas J W, Hill J, Lemieux L, Ma J X, Georgiou C, Fetter J, Shapleigh J, Tecklenburg M M J, Babcock G T, Gennis R B. Insight into the active-site structure and function of cytochrome oxidase by analysis of site-directed mutants of bacterial cytochrome-aa3 and cytochrome-bo. J Bioenerg Biomembr. 1993;25:121–136. doi: 10.1007/BF00762854. [DOI] [PubMed] [Google Scholar]
- 129.Huitema, F., J. A. Duine, and G. W. Canters. Sequence of the genes surrounding the genes encoding methylamine dehydrogenase from Thiobacillus versutus and influence of those genes on expression of the MADH-subunits in Escherichia coli. Unpublished results. Accession no. L36951, L36952, and L36953.
- 130.Huitema F, van Beeumen J, van Driessche G, Duine J A, Canters G W. Cloning and sequencing of the gene coding for the large subunit of methylamine dehydrogenase from Thiobacillus versutus. J Bacteriol. 1993;175:6254–6259. doi: 10.1128/jb.175.19.6254-6259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Husain I, van Houten B, Thomas D C, Sancar A. Sequences of Escherichia coli uvrA gene and protein reveal two potential ATP binding sites. J Biol Chem. 1986;261:4895–4901. [PubMed] [Google Scholar]
- 132.Husain M, Davidson V L. An inducible periplasmic blue copper protein from Paracoccus denitrificans: purification, properties, and physiological role. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 133.Husain M, Davidson V L. Characterization of two inducible periplasmic c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986;261:8577–8580. [PubMed] [Google Scholar]
- 134.Husain M, Davidson V L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987;169:1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Husain M, Steenkamp D J. Partial purification and characterization of glutaryl-coenzyme A dehydrogenase, electron transfer flavoprotein, and electron transfer flavoprotein-Q oxidoreductase from Paracoccus denitrificans. J Bacteriol. 1985;163:709–715. doi: 10.1128/jb.163.2.709-715.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 137.John P, Whatley F R. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature. 1975;254:495–498. doi: 10.1038/254495a0. [DOI] [PubMed] [Google Scholar]
- 138.John P, Whatley F R. The bioenergetics of Paracoccus denitrificans. Biochim Biophys Acta. 1977;463:129–153. doi: 10.1016/0304-4173(77)90006-4. [DOI] [PubMed] [Google Scholar]
- 139.Jordan S L, McDonald I R, Kraczkiewicz Dowjat A J, Kelly D P, Rainey F A, Murrell J C, Wood A P. Autotrophic growth on carbon disulfide is a property of novel strains of Paracoccus denitrificans. Arch Microbiol. 1997;168:225–236. doi: 10.1007/s002030050492. [DOI] [PubMed] [Google Scholar]
- 140.Jungst A, Braun C, Zumft W G. Close linkage in Pseudomonas stutzeri of the structural genes for respiratory nitrite reductase and nitrous oxide reductase, and other essential genes for denitrification. Mol Gen Genet. 1991;225:241–248. doi: 10.1007/BF00269855. [DOI] [PubMed] [Google Scholar]
- 141.Jungst A, Zumft W G. Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 1992;314:308–314. doi: 10.1016/0014-5793(92)81495-8. [DOI] [PubMed] [Google Scholar]
- 142.Kahn D, Batut J, Daveran M-L, Fourment J. Structure and regulation of the fixNOQP operon from Rhizobium meliloti. In: Palacios R, Mora J, Newton W E, editors. New horizons in nitrogen fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. p. 474. [Google Scholar]
- 143.Kaplan, S. Personal communication.
- 144.Karlin S, Brocchieri L. Evolutionary conservation of recA genes in relation to protein structure and function. J Bacteriol. 1996;178:1881–1894. doi: 10.1128/jb.178.7.1881-1894.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Katayama Y, Hiraishi A, Kuraishi H. Paracoccus thiocyanatus sp. nov., a new species of thiocyanate-utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus. Microbiology. 1995;141:1469–1477. doi: 10.1099/13500872-141-6-1469. [DOI] [PubMed] [Google Scholar]
- 146.Kawasaki H, Hoshino Y, Hirata A, Yamasato K. Is intracytoplasmic membrane structure a generic criterion? It does not coincide with phylogenetic interrelationships among phototrophic purple nonsulfur bacteria. Arch Microbiol. 1993;160:358–362. doi: 10.1007/BF00252221. [DOI] [PubMed] [Google Scholar]
- 147.Kawasaki H, Hoshino Y, Yamasato K. Phylogenetic diversity of phototrophic purple non-sulfur bacteria in the Proteobacteria alpha group. FEMS Microbiol Lett. 1993;112:61–66. [Google Scholar]
- 148.Kelly D P. Oxidation of sulphur compounds. Symp Soc Gen Microbiol. 1988;42:65–98. [Google Scholar]
- 149.Kelly D P, Shergill J K, Lu W-P, Wood A P. Oxidative metabolism of inorganic sulphur compounds by bacteria. Antonie Leeuwenhoek. 1997;71:95–108. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 150.Khoroshilova N, Beinert H, Kiley P J. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci USA. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kiley P J, Donohue T J, Havelka W A, Kaplan S. DNA sequence and in vitro expression of the B875 light-harvesting polypeptides of Rhodobacter sphaeroides. J Bacteriol. 1987;169:742–750. doi: 10.1128/jb.169.2.742-750.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kitts P A, Symington L S, Dyson P, Sherratt D J. Transposon-encoded site specific recombination: nature of the Tn3 DNA sequences which constitute the recombination site res. EMBO J. 1983;2:1055–1060. doi: 10.1002/j.1460-2075.1983.tb01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kleymann G, Iwata S, Weismuller K H, Haase W, Michel H. Immunoelectron microscopy and epitope mapping with monoclonal antibodies suggest the existence of an additional N-terminal transmembrane helix in the cytochrome b subunit of bacterial ubiquinol:cytochrome c oxidoreductases. Eur J Biochem. 1995;230:359–363. doi: 10.1111/j.1432-1033.1995.tb20571.x. [DOI] [PubMed] [Google Scholar]
- 154.Klug G, Jager A, Heck C, Rauhut R. Identification, sequence analysis, and expression of the lepB gene for a leader peptidase in Rhodobacter capsulatus. Mol Gen Genet. 1997;253:666–673. doi: 10.1007/s004380050370. [DOI] [PubMed] [Google Scholar]
- 155.Knaff D B. The cytochrome bc1 complexes of photosynthetic purple bacteria. Photosynth Res. 1993;35:117–133. doi: 10.1007/BF00014743. [DOI] [PubMed] [Google Scholar]
- 156.Knight A I, Ni H, Cartwright K A V, McFadden J J. Identification and characterization of a novel insertion sequence, IS1106, downstream of the porA gene in B15 Neisseria meningitidis. Mol Microbiol. 1992;6:1565–1573. doi: 10.1111/j.1365-2958.1992.tb00878.x. [DOI] [PubMed] [Google Scholar]
- 157.Kobayashi M, Matsuo Y, Takimoto A, Suzuki S, Maruo F, Shoun H. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J Biol Chem. 1996;271:16263–16267. doi: 10.1074/jbc.271.27.16263. [DOI] [PubMed] [Google Scholar]
- 158.Reference deleted.
- 159.Kostler M, Kleiner D. Assimilation of methylamine by Paracoccus denitrificans involves formaldehyde transport by a specific carrier. FEMS Microbiol Lett. 1989;53:1–4. doi: 10.1016/0378-1097(89)90356-x. [DOI] [PubMed] [Google Scholar]
- 160.Kucera I, Kozak L, Dadak V. Is the ubiquinone pool in the respiratory chain of the bacterium Paracoccus denitrificans really unhomogeneous? Arch Biochem Biophys. 1987;253:199–204. doi: 10.1016/0003-9861(87)90652-7. [DOI] [PubMed] [Google Scholar]
- 161.Kucera I. The release of nitric oxide from denitrifying cells of Paracoccus denitrificans by an uncoupler is the basis of a new oscillation. FEBS Lett. 1989;249:56–58. [Google Scholar]
- 162.Kurowski B, Ludwig B. The genes of the Paracoccus denitrificans bc1 complex. Nucleotide sequence and homologies between bacterial and mitochondrial subunits. J Biol Chem. 1987;262:13805–13811. [PubMed] [Google Scholar]
- 163.Kwiatkowski A V, Shapleigh J P. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Biol Chem. 1996;271:24382–24388. doi: 10.1074/jbc.271.40.24382. [DOI] [PubMed] [Google Scholar]
- 164.Lang H P, Cogdell R J, Takaichi S, Hunter C N. Complete DNA sequence, specific Tn5 insertion map, and gene assignment of the carotenoid biosynthesis pathway of Rhodobacter sphaeroides. J Bacteriol. 1995;177:2064–2073. doi: 10.1128/jb.177.8.2064-2073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 166.Lee W T, Terlesky K C, Tabita F R. Cloning and characterization of two groESL operons of Rhodobacter sphaeroides: transcriptional regulation of the heat-induced groESL operon. J Bacteriol. 1997;179:487–495. doi: 10.1128/jb.179.2.487-495.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Leimkuhler S, Kern M, Solomon P S, McEwan A G, Schwarz G, Mendel R R, Klipp W. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol Microbiol. 1998;27:853–869. doi: 10.1046/j.1365-2958.1998.00733.x. [DOI] [PubMed] [Google Scholar]
- 168.Leung Y C, Chan C, Reader J S, Willis A C, van Spanning R J M, Ferguson S J, Radford S E. The pseudoazurin gene from Thiosphaera pantotropha: analysis of upstream putative regulatory sequences and overexpression in Escherichia coli. Biochem J. 1997;321:699–705. doi: 10.1042/bj3210699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lidstrom M E, Anthony C, Biville F, Gasser F, Goodwin P, Hanson R S, Harms N. New unified nomenclature for genes involved in the oxidation of methanol in Gram-negative bacteria. FEMS Microbiol Lett. 1994;117:103–106. doi: 10.1111/j.1574-6968.1994.tb06749.x. [DOI] [PubMed] [Google Scholar]
- 170.Lipski A, Reichert K, Reuter B, Sproer C, Altendorf K. Identification of bacterial isolates from biofilters as Paracoccus alkenifer sp. nov. and Paracoccus solventivorans with emended description of Paracoccus solventivorans. Int J Syst Bacteriol. 1998;48:529–536. doi: 10.1099/00207713-48-2-529. [DOI] [PubMed] [Google Scholar]
- 171.Lloyd D, Boddy L, Davies K J P. Persistence of bacterial denitrification capacity under aerobic conditions: the rule rather than the exception. FEMS Microbiol Ecol. 1987;45:103–106. [Google Scholar]
- 172.Lommen A, Wijmenga S, Hilbers C W, Canters G W. Assignment of the 600-MHz 1H-NMR spectrum of amicyanin from Thiobacillus versutus by two-dimensional NMR methods provides information on secondary structure. Eur J Biochem. 1991;201:695–702. doi: 10.1111/j.1432-1033.1991.tb16330.x. [DOI] [PubMed] [Google Scholar]
- 173.Lu W P, Kelly D P. Purification and some properties of two principle enzymes of the thiosulphate-oxidising multi-enzyme system from Thiobacillus A2. J Gen Microbiol. 1983;129:3549–3564. [Google Scholar]
- 174.Lu W P, Kelly D P. Rhodanese: an enzyme not necessary for thiosulphate oxidation by Thiobacillus A2. FEMS Microbiol Lett. 1983;18:289–292. [Google Scholar]
- 175.Ludwig B. Terminal oxidases in Paracoccus denitrificans. Biochim Biophys Acta. 1992;1101:195–197. [PubMed] [Google Scholar]
- 176.Ludwig B, Schatz G. A two-subunit cytochrome c oxidase (cytochrome aa3) from Paracoccus denitrificans. Proc Natl Acad Sci USA. 1980;77:196–200. doi: 10.1073/pnas.77.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ludwig B, Suda K, Cerletti N. Cytochrome c1 from Paracoccus denitrificans. Eur J Biochem. 1983;137:597–602. doi: 10.1111/j.1432-1033.1983.tb07867.x. [DOI] [PubMed] [Google Scholar]
- 178.Ludwig W, Mittenhuber G, Friedrich C G. Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int J Syst Bacteriol. 1993;43:363–367. doi: 10.1099/00207713-43-2-363. [DOI] [PubMed] [Google Scholar]
- 179.Ludwig W, Rossello-Mora R, Aznar R, Klugbauer S, Spring S, Reetz K, Beimfohr C, Brockmann E, Kirchhof G, Dorn S, Bachleitner M, Klugbauer N, Springer N, Lane D, Nietupsky R, Weizenegger M, Schleifer K H. Comparative sequence analysis of 23S rRNA from proteobacteria. Syst Appl Microbiol. 1995;18:164–188. [Google Scholar]
- 180.Ma D, Cook D N, O’Brien D A, Hearst J E. Analysis of the promoter and regulatory sequences of an oxygen-regulated bch operon in Rhodobacter capsulatus by site-directed mutagenesis. J Bacteriol. 1993;175:2037–2045. doi: 10.1128/jb.175.7.2037-2045.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.MacKay R M, Salgado D, Bonen L, Stackebrandt E, Doolittle W F. The 5S ribosomal RNAs of Paracoccus denitrificans and Prochloron. Nucleic Acids Res. 1982;10:2963–2970. doi: 10.1093/nar/10.9.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mandon K, Kaminski P A, Elmerich C. Functional analysis of the fixNOQP region of Azorhizobium caulinodans. J Bacteriol. 1994;176:2560–2568. doi: 10.1128/jb.176.9.2560-2568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mariani F, Piccolella E, Colizzi V, Rappuoli R, Gross R. Characterization of an IS-like element from Mycobacterium tuberculosis. J Gen Microbiol. 1993;139:1767–1772. doi: 10.1099/00221287-139-8-1767. [DOI] [PubMed] [Google Scholar]
- 184.Maszenan A M, Seviour R J, Patel B K, Rees G N, McDougall B M. Amaricoccus gen. nov., a gram-negative coccus occurring in regular packages or tertrads, isolated from activated sludges biomass, and descriptions of Amaricoccus veronensis sp. nov., Amaricoccus tamworthensis sp. nov., and Amaricoccus kaplicencis sp. nov. Int J Syst Bacteriol. 1997;47:727–734. doi: 10.1099/00207713-47-3-727. [DOI] [PubMed] [Google Scholar]
- 185.Mat’chova I, Kucera I. Evidence for the role of soluble cytochrome c in the dissimilatory reduction of nitrite and nitrous oxide by cells of Paracoccus denitrificans. Biochim Biophys Acta. 1991;1058:256–260. doi: 10.1016/s0005-2728(05)80245-2. [DOI] [PubMed] [Google Scholar]
- 186.Miczak A, Chauhan A K, Apirion D. Two new genes located between 2758 and 2761 kilobase pairs on the Escherichia coli genome. J Bacteriol. 1991;173:3271–3272. doi: 10.1128/jb.173.11.3271-3272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 188.Mittenhuber G, Sonomoto K, Egert M, Friedrich C G. Identification of the DNA region responsible for sulfur-oxidizing ability of Thiosphaera pantotropha. J Bacteriol. 1991;173:7340–7344. doi: 10.1128/jb.173.22.7340-7344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Moir J W B, Baratta D, Richardson D J, Ferguson S J. The purification of a cd1-type nitrite reductase from, and the absence of a copper-type nitrite reductase from, the aerobic denitrifier Thiosphaera pantotropha: the role of pseudoazurin as an electron donor. Eur J Biochem. 1993;212:377–385. doi: 10.1111/j.1432-1033.1993.tb17672.x. [DOI] [PubMed] [Google Scholar]
- 190.Moir J W B. D.Phil. thesis. Oxford, United Kingdom: University of Oxford; 1993. [Google Scholar]
- 191.Moir J W B, Ferguson S J. Spontaneous mutation of Thiosphaera pantotropha enabling growth on methanol correlates with synthesis of a 26 kDa c-type cytochrome. FEMS Microbiol Lett. 1993;113:321–326. doi: 10.1111/j.1574-6968.1993.tb06534.x. [DOI] [PubMed] [Google Scholar]
- 192.Moir J W B, Ferguson S J. Properties of a Paracoccus denitrificans mutant deleted in cytochrome c550 indicate that a copper protein can substitute for this cytochrome in electron transport to nitrite, nitric oxide and nitrous oxide. Microbiology. 1994;140:389–397. [Google Scholar]
- 193.Moir J W B, Richardson D J, Ferguson S J. The expression of redox proteins of denitrification in Thiosphaera pantotropha grown with oxygen, nitrate, and nitrous oxide as electron acceptors. Arch Microbiol. 1995;164:43–49. [Google Scholar]
- 194.Nakai Y, Hayashi H, Kagamiyama H. Cloning and characterization of the tyrB gene from Salmonella typhimurium. Biochim Biophys Acta. 1996;1308:189–192. doi: 10.1016/0167-4781(96)00113-3. [DOI] [PubMed] [Google Scholar]
- 195.Neidle E L, Kaplan S. 5-Aminolevulinic acid availability and control of spectral complex formation in hemA and hemT mutants of Rhodobacter sphaeroides. J Bacteriol. 1993;175:2304–2313. doi: 10.1128/jb.175.8.2304-2313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Newton N. The two-haem nitrite reductase of Micrococcus denitrificans. Biochim Biophys Acta. 1969;185:316–331. doi: 10.1016/0005-2744(69)90425-2. [DOI] [PubMed] [Google Scholar]
- 197.Nickel, C. M. Rhodobacter capsulatus radC. Unpublished results. Accession no. U74017.
- 198.Nickel C M, Vandekerckhove J, Beyer P, Tadros M H. Molecular analysis of the Rhodobacter capsulatus chaperone dnaKJ operon: purification and characterization of DnaK. Gene. 1997;192:251–259. doi: 10.1016/s0378-1119(97)00085-1. [DOI] [PubMed] [Google Scholar]
- 199.Nokhal T H, Schlegel H G. Taxonomic study of Paracoccus denitrificans. Int J Syst Bacteriol. 1983;33:26–37. [Google Scholar]
- 200.Norris, H. A. C., and S. J. Ferguson. Unpublished data.
- 201.Nowicka B, Meler A, Wlodarczyk M, Wolska K I. Several lines of evidence for the linearity of Thiobacillus versutus extrachromosomal pTAV2 DNA. Acta Microbiol Pol. 1990;39:205–210. [Google Scholar]
- 202.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 203.Ohara M, Katayama Y, Tsuzaki M, Nakamoto S, Kuraishi H. Paracoccus kocurii sp. nov., a tetramethylammonium-assimilating bacterium. Int J Syst Bacteriol. 1990;40:292–296. doi: 10.1099/00207713-40-3-292. [DOI] [PubMed] [Google Scholar]
- 204.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ostermeier C, Iwata S, Ludwig B, Michel H. Fv fragment-mediated crystallization of the membrane protein bacterial cytochrome c oxidase. Nat Struct Biol. 1995;10:842–846. doi: 10.1038/nsb1095-842. [DOI] [PubMed] [Google Scholar]
- 206.Ostermeier C, Iwata S, Michel H. Cytochrome c oxidase. Curr Opin Struct Biol. 1996;6:460–466. doi: 10.1016/s0959-440x(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 207.Oue S, Okamoto A, Nakai Y, Nakahira M, Shibatani T, Hayashi H, Kagamiyama H. Paracoccus denitrificans aromatic amino acid aminotransferase: a model enzyme for the study of dual substrate recognition mechanism. J Biochem. 1997;121:161–171. doi: 10.1093/oxfordjournals.jbchem.a021561. [DOI] [PubMed] [Google Scholar]
- 208.Paetow B, Panskus G, Ludwig B. Cloning of Paracoccus cytochrome c oxidase subunit II. J Inorg Biochem. 1985;23:183–186. doi: 10.1016/0162-0134(85)85024-8. [DOI] [PubMed] [Google Scholar]
- 209.Page M D, Carr G, Bell L C, Ferguson S J. Structure, control and assembly of a bacterial electron transport system as exemplified by Paracoccus denitrificans. Biochem Soc Trans. 1989;17:991–993. doi: 10.1042/bst0170991. [DOI] [PubMed] [Google Scholar]
- 210.Page, M. D., and S. J. Ferguson. Unpublished results.
- 211.Page M D, Ferguson S J. Apo forms of cytochrome c550 and cytochrome cd1 are translocated to the periplasm of Paracoccus denitrificans in the absence of haem incorporation caused by either mutation or inhibition of haem synthesis. Mol Microbiol. 1990;4:1181–1182. doi: 10.1111/j.1365-2958.1990.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 212.Page M D, Ferguson S J. A bacterial c-type cytochrome can be translocated to the periplasm as an apo form; the biosynthesis of cytochrome cd1 (nitrite reductase) from Paracoccus denitrificans. Mol Microbiol. 1990;3:653–661. doi: 10.1111/j.1365-2958.1989.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 213.Page M D, Ferguson S J. Mutants of Methylobacterium extorquens and Paracoccus denitrificans deficient in c-type cytochrome biogenesis synthesise the methylamine-dehydrogenase polypeptides but cannot assemble the tryptophan-tryptophylquinone group. Eur J Biochem. 1993;218:711–717. doi: 10.1111/j.1432-1033.1993.tb18425.x. [DOI] [PubMed] [Google Scholar]
- 214.Page M D, Ferguson S J. Differential reduction in periplasmic and membrane-bound c-type cytochromes in a Paracoccus denitrificans mutant partially deficient in 5-aminolevulinate synthase. J Bacteriol. 1994;176:5919–5928. doi: 10.1128/jb.176.19.5919-5928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Page M D, Ferguson S J. Cloning and sequence analysis of cycH gene from Paracoccus denitrificans: the cycH gene product is required for assembly of all c-type cytochromes, including cytochrome c1. Mol Microbiol. 1995;15:307–318. doi: 10.1111/j.1365-2958.1995.tb02245.x. [DOI] [PubMed] [Google Scholar]
- 216.Page M D, Ferguson S J. Paracoccus denitrificans CcmG is a periplasmic protein-disulphide oxidoreductase required for c- and aa3-type cytochrome biogenesis: evidence for a reductase role in vivo. Mol Microbiol. 1997;24:977–990. doi: 10.1046/j.1365-2958.1997.4061775.x. [DOI] [PubMed] [Google Scholar]
- 217.Page M D, Pearce D A, Norris H A C, Ferguson S J. The Paracoccus denitrificans ccmA, B and C genes: cloning and sequencing, and analysis of the potential of their products to form a haem or apo-c-type cytochrome transporter. Microbiology. 1997;143:563–576. doi: 10.1099/00221287-143-2-563. [DOI] [PubMed] [Google Scholar]
- 218.Page M D, Sambongi Y, Ferguson S J. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- 219.Papa S, Lorusson M, Capitanio N. Mechanistic and phenomenological features of proton pumps in the respiratory chain of mitochondria. J Bioenerg Biomembr. 1994;26:609–618. doi: 10.1007/BF00831535. [DOI] [PubMed] [Google Scholar]
- 220.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 221.Parsek M R, Ye R W, Pun P, Chakrabarty A M. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region: catBC promoter activation by CatR. J Biol Chem. 1994;269:11279–11284. [PubMed] [Google Scholar]
- 222.Pasternak C, Assemat K, Breton A M, Clement-Metral J D, Klug G. Expression of the thioredoxin gene (trxA) in Rhodobacter sphaeroides is regulated by oxygen. Mol Gen Genet. 1996;250:189–196. doi: 10.1007/BF02174178. [DOI] [PubMed] [Google Scholar]
- 223.Pasternak C, Chen W, Heck C, Klug G. Cloning, nucleotide sequence and characterization of the rpoD gene encoding the primary sigma factor of Rhodobacter capsulatus. Gene. 1996;176:177–184. doi: 10.1016/0378-1119(96)00243-0. [DOI] [PubMed] [Google Scholar]
- 224.Paulus F, Canaday J, Vincent F, Bonnard G, Kares C, Otten L. Sequence of the iaa and ipt region of different Agrobacterium tumefaciens biotype III octopine strains: reconstruction of octopine Ti plasmid evolution. Plant Mol Biol. 1991;16:601–614. doi: 10.1007/BF00023425. [DOI] [PubMed] [Google Scholar]
- 225.Pearce D A, Page M D, Norris H A C, Tomlinson E, Ferguson S J. Identification of the Paracoccus denitrificans ccmF and ccmH genes: disruption of ccmF, coding for a putative transporter, results in formation of an unstable apo cytochrome c and deficiency in siderophore production. Microbiology. 1998;144:467–477. doi: 10.1099/00221287-144-2-467. [DOI] [PubMed] [Google Scholar]
- 226.Pennoyer J D, Ohnishi T, Trumpower B L. Purification and properties of succinate-ubiquinone oxidoreductase complex from Paracoccus denitrificans. Biochim Biophys Acta. 1988;935:195–207. doi: 10.1016/0005-2728(88)90216-2. [DOI] [PubMed] [Google Scholar]
- 227.Perret, X., R. Fellay, A. J. Bjourson, J. E. Cooper, S. Brenner, and W. J. Broughton. Unpublished results. Accession no. U00090.
- 228.Perrier, V., A. Boussac, O. Meier, O. Barzu, and A.-M. Gilles. Adenylate kinase from P. denitrificans, an iron and zinc binding protein, catalyzes phosphorylation of AMP and reduction of cytochrome c. Unpublished results. Accession no. U64202, U64203, and U64204.
- 229.Pittard J. The various strategies within the TyrR regulation of Escherichia coli to modulate gene expression. Genes Cells. 1996;1:717–725. doi: 10.1111/j.1365-2443.1996.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 230.Preisig O, Anthamattan D, Hennecke H. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Preisig O, Zufferey R, Thöny-Meyer L, Appleby C A, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Puustinen A, Finel M, Virkki M, Wikstrom M. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 1989;249:163–167. doi: 10.1016/0014-5793(89)80616-7. [DOI] [PubMed] [Google Scholar]
- 233.Rainey, F. A. D. P. Kelly, E. Stackebrandt, J. Burkhardt, A. Hiraishi, Y. Katayama, and A. P. Wood. A reevaluation of the taxonomy of Paracoccus denitrificans and a proposal for the creation of Paracoccus pantotrophus comb. nov. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 234.Raitio M, Jalli T, Saraste M. Isolation and characterization of genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987;6:2825–2833. doi: 10.1002/j.1460-2075.1987.tb02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Raitio M, Pispa J M, Metso T, Saraste M. Are there isoenzymes of cytochrome c oxidase in Paracoccus denitrificans? FEBS Lett. 1990;261:431–435. doi: 10.1016/0014-5793(90)80609-m. [DOI] [PubMed] [Google Scholar]
- 236.Raitio M, Wikstrom M. An alternative cytochrome oxidase of Paracoccus denitrificans functions as a proton pump. Biochim Biophys Acta. 1994;1186:100–106. [Google Scholar]
- 237.Ras J. Ph.D. thesis. Amsterdam, The Netherlands: Vrije Universiteit; 1995. [Google Scholar]
- 238.Ras J, Hazelaar M J, Robertson L A, Kuenen J G, van Spanning R J M, Stouthamer A H, Harms N. Methanol oxidation in a spontaneous mutant of Thiosphaera pantotropha with a methanol positive phenotype is catalyzed by a dye-linked ethanol dehydrogenase. FEMS Microbiol Lett. 1995;127:159–164. [Google Scholar]
- 239.Ras J, Reijnders W N M, van Spanning R J M, Harms N, Oltmann L F, Stouthamer A H. Isolation, sequencing, and mutagenesis of the gene encoding cytochrome c553i of Paracoccus denitrificans and characterization of the mutant strain. J Bacteriol. 1991;173:6971–6979. doi: 10.1128/jb.173.21.6971-6979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ras J, van Ophem P W, Reijnders W N M, van Spanning R J M, Duine J A, Stouthamer A H, Harms N. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol. 1995;177:247–251. doi: 10.1128/jb.177.1.247-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Reijnders, W. N. M., and N. Harms. 1998. A global two-component system regulates methanol, methylamine and formaldehyde oxidation in Paracoccus denitrificans. Unpublished results. Accession no. AJ223460. [DOI] [PMC free article] [PubMed]
- 242.Reijnders, W. N. M., and N. Harms. 1998. Paracoccus denitrificans, MxaA, MxaC, MxaK, MxaL, MxaD and ORFs. Unpublished results. Accession no. AJ000884.
- 243.Reyes F, Roldan M D, Klipp W, Castillo F, Moreno-Vivian C. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductases. Mol Microbiol. 1996;19:1307–1318. doi: 10.1111/j.1365-2958.1996.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 244.Richter O-M H, Tao J S, Turba A, Ludwig B. Cytochrome ba3 functions as a quinol oxidase in Paracoccus denitrificans. Purification, cloning, and sequence comparison. J Biol Chem. 1994;269:23079–23086. [PubMed] [Google Scholar]
- 245.Roberts D L, Herrick K R, Frerman F E, Kim J J P. Crystallization and preliminary X-ray analysis of electron transfer flavoproteins from human and Paracoccus denitrificans. Protein Sci. 1995;4:1654–1657. doi: 10.1002/pro.5560040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Robertson L A, Cornelisse R, de Vos P, Hadioetomo R, Kuenen J G. Aerobic denitrification in various heterotrophic nitrifiers. Antonie Leeuwenhoek. 1989;56:289–300. doi: 10.1007/BF00443743. [DOI] [PubMed] [Google Scholar]
- 247.Robertson L A, Kuenen J G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulphur bacterium. J Gen Microbiol. 1983;129:2847–2855. [Google Scholar]
- 247a.Roldán, M. D., H. J. Sears, M. R. Cheeseman, S. J. Ferguson, A. J. Thomson, B. C. Berks, and D. J. Richardson. Spectroscopic characterization of a novel multiheme c-type cytochrome widely implicated in bacterial respiratory electron transport. J. Biol. Chem., in press. [DOI] [PubMed]
- 248.Saeki K, Suetsugi Y, Tokuda K-I, Miyatake Y, Young D A, Mars B L, Matsubara H. Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J Biol Chem. 1991;266:12889–12895. [PubMed] [Google Scholar]
- 249.Saraste M, Castresana J. Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 1994;341:1–4. doi: 10.1016/0014-5793(94)80228-9. [DOI] [PubMed] [Google Scholar]
- 250.Saraste M, Holm L, Lemieux L, Lubben M, van der Oost J. The happy family of cytochrome oxidases. Biochem Soc Trans. 1991;19:608–612. doi: 10.1042/bst0190608. [DOI] [PubMed] [Google Scholar]
- 251.Saraste M, Raitio M, Jalli T, Peramaa A. A gene in Paracoccus for subunit III of cytochrome oxidase. FEBS Lett. 1986;206:154–156. doi: 10.1016/0014-5793(86)81359-x. [DOI] [PubMed] [Google Scholar]
- 251a.Saunders N F W. Cloning, sequence analysis and studies on the expression of the nirS gene, encoding cytochrome cd1 nitrite reductase, from Thiosphaera pantotropha. D.Phil. thesis. Oxford, United Kingdom: University of Oxford; 1997. [Google Scholar]
- 252.Saunders, N. F. W., S. C. Baker, and S. J. Ferguson. 1997. Unpublished results.
- 253.Savioz A, Zimmermann A, Haas D. Pseudomonas aeruginosa promoters which contain a conserved GG-N10-GC motif but appear to be RpoN-independent. Mol Gen Genet. 1993;238:74–80. doi: 10.1007/BF00279533. [DOI] [PubMed] [Google Scholar]
- 254.Saxena K, Richter O-M H, Ludwig B, Benz R. Molecular cloning and functional characterization of the Paracoccus denitrificans porin. Eur J Biochem. 1997;245:300–306. doi: 10.1111/j.1432-1033.1997.00300.x. [DOI] [PubMed] [Google Scholar]
- 255.Scala D J, Kerkhof L J. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett. 1998;162:61–68. doi: 10.1111/j.1574-6968.1998.tb12979.x. [DOI] [PubMed] [Google Scholar]
- 256.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 257.Schilke B A, Donohue T J. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J Bacteriol. 1995;177:1929–1937. doi: 10.1128/jb.177.8.1929-1937.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Schulte U, Fecke W, Krull C, Nehls U, Schmiede A, Schneider R, Ohnishi T, Weiss H. In vivo dissection of the mitochondrial respiratory NADH: ubiquinone oxidoreductase (complex I) Biochim Biophys Acta. 1994;1187:121–124. doi: 10.1016/0005-2728(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 259.Sears H J, Bennett B, Spiro S, Thomson A J, Richardson D J. Identification of periplasmic nitrate reductase Mo(V) EPR signals in intact cells of Paracoccus denitrificans. Biochem J. 1995;310:311–314. doi: 10.1042/bj3100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sears H J, Spiro S, Richardson D J. Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology. 1997;143:3767–3774. doi: 10.1099/00221287-143-12-3767. [DOI] [PubMed] [Google Scholar]
- 261.Shapleigh J P, Payne W J. Nitric oxide-dependent proton translocation in various denitrifiers. J Bacteriol. 1985;163:837–840. doi: 10.1128/jb.163.3.837-840.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shaw D J, Rice D W, Guest J R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983;1983:241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- 263.Shiota S, Nakayama H. Micrococcus luteus homolog of the Escherichia coli uvrA gene: identification of a mutation in the UV-sensitive mutant DB7. Mol Gen Genet. 1989;217:332–340. doi: 10.1007/BF02464901. [DOI] [PubMed] [Google Scholar]
- 264.Siller H, Rainey F A, Stackebrandt E, Winter J. Isolation and characterization of a new gram-negative, acetone-degrading, nitrate-reducing bacterium from soil, Paracoccus solventivorans sp. nov. Int J Syst Bacteriol. 1996;46:1125–1130. doi: 10.1099/00207713-46-4-1125. [DOI] [PubMed] [Google Scholar]
- 265.Six S, Trageser M, Kojro E, Fahrenholz F, Unden G. Reactivity of the N-terminal cysteine residues in active and inactive forms of FNR, an O2-responsive, Fe containing transcriptional regulator of Escherichia coli. J Inorg Biochem. 1996;62:89–102. doi: 10.1016/0162-0134(95)00091-7. [DOI] [PubMed] [Google Scholar]
- 266.Sobecky, P. A., T. J. Mincer, M. C. Chang, A. Toukdarian, and D. R. Helinski. 1998. Unpublished results. Accession no. AF020624.
- 267.Sobral B W S, Honeycutt R J, Atherly A G, Mcclelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991;173:5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Soto M J, Zorzano A, Olivares J, Toro N. Sequence of ISRm4 from Rhizobium meliloti strain GR4. Gene. 1992;120:125–126. doi: 10.1016/0378-1119(92)90020-p. [DOI] [PubMed] [Google Scholar]
- 269.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 270.Spiro S, Gaston K L, Bell A I, Roberts R E, Busby S J W, Guest J R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 271.Spiro S, Guest J R. Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem Sci. 1991;16:310–314. doi: 10.1016/0968-0004(91)90125-f. [DOI] [PubMed] [Google Scholar]
- 272.Spurgin P, Tomasselli A G, Schiltz E. The amino acid sequence of adenylate kinase from Paracoccus denitrificans and its relationship to mitochondrial and microbial adenylate kinases. Eur J Biochem. 1989;179:621–628. [PubMed] [Google Scholar]
- 273.Steenkamp D J, Gallup M. The natural flavoprotein electron acceptor of trimethylamine dehydrogenase. J Biol Chem. 1978;253:4086–4089. [PubMed] [Google Scholar]
- 274.Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1993;10:347–350. doi: 10.1111/j.1574-6968.1993.tb05875.x. [DOI] [PubMed] [Google Scholar]
- 275.Steinrücke P, Gerhus E, Jetzek M, Turba A, Ludwig B. The cytochrome c reductase/oxidase respiratory pathway of Paracoccus denitrificans: genetic and functional studies. J Bioenerg Biomembr. 1991;23:227–239. doi: 10.1007/BF00762219. [DOI] [PubMed] [Google Scholar]
- 276.Steinrücke P, Gerhus E, Ludwig B. Paracoccus denitrificans mutants deleted in the gene for subunit II of cytochrome c oxidase also lack subunit I. J Biol Chem. 1991;266:7676–7681. [PubMed] [Google Scholar]
- 277.Steinrücke P, Ludwig B. Genetics of Paracoccus denitrificans. FEMS Microbiol Rev. 1993;104:83–118. doi: 10.1016/0378-1097(93)90505-v. [DOI] [PubMed] [Google Scholar]
- 278.Steinrücke P, Steffens G C, Panskus G, Buse G, Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987;167:431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- 279.Stoll R, Page M D, Sambongi Y, Ferguson S J. Cytochrome c550 expression in Paracoccus denitrificans strongly depends on growth condition: identification of promoter region for cycA by transcription start analysis. Microbiology. 1996;142:2577–2585. doi: 10.1099/00221287-142-9-2577. [DOI] [PubMed] [Google Scholar]
- 280.Stouthamer A H. Metabolic pathways in Paracoccus denitrificans and closely related bacteria in relation to the phylogeny of prokaryotes. Antonie Leeuwenhoek. 1992;61:1–33. doi: 10.1007/BF00572119. [DOI] [PubMed] [Google Scholar]
- 281.Stouthamer A H, de Boer A P N, van der Oost J, van Spanning R J M. Emerging principles of inorganic nitrogen metabolism in Paracoccus denitrificans and related bacteria. Antonie Leeuwenhoek. 1997;71:33–41. doi: 10.1023/a:1000113824961. [DOI] [PubMed] [Google Scholar]
- 282.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989;171:5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tagaki T, Taniguchi T, Yamamoto Y, Shibatani T. Molecular cloning of the l-phenylalanine transaminase gene from Paracoccus denitrificans in Escherichia coli K12. Biotechnol Appl Biochem. 1991;13:112–119. [PubMed] [Google Scholar]
- 284.Tapias A, Fernandez de Henestrosa A R, Barbé J. Characterization of the promoter of the Rhizobium etli recA gene. J Bacteriol. 1997;179:1573–1579. doi: 10.1128/jb.179.5.1573-1579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Thöny-Meyer L, Beck C, Preisig O, Hennecke H. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol Microbiol. 1994;14:705–716. doi: 10.1111/j.1365-2958.1994.tb01308.x. [DOI] [PubMed] [Google Scholar]
- 287.Tosques I E, Kwiatkowski A V, Shi J, Shapleigh J P. Characterization and regulation of the gene encoding nitrite reductase in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:1090–1095. doi: 10.1128/jb.179.4.1090-1095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Toussaint B, Daspremont R D, Delicattree I, Berchet V, Elsen S, Colbeau A, Dischert W, Lazzaroni Y, Vignais P M. The Rhodobacter capsulatus hupSLC promoter: identification of cis-regulatory elements and of trans-activating factors involved in H2 activation of hupSLC transcription. Mol Microbiol. 1997;26:927–937. doi: 10.1046/j.1365-2958.1997.6291996.x. [DOI] [PubMed] [Google Scholar]
- 289.Trieschmann M D A, Pattus F, Tadros M H. Molecular analysis of the Rhodobacter capsulatus B10 porin (porCa) gene: purification and biochemical characterization of the porin protein. Mol Gen Genet. 1996;253:253–258. doi: 10.1007/s004380050320. [DOI] [PubMed] [Google Scholar]
- 290.Trumpower B L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Trumpower B L, Gennis R B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 292.Reference deleted.
- 293.Turba A. Ph.D. thesis. Frankfurt, Germany: University of Frankfurt; 1993. [Google Scholar]
- 294.Turba A, Jetzek M, Ludwig B. Purification of Paracoccus denitrificans cytochrome c552 and sequence analysis of the gene. Eur J Biochem. 1995;231:259–265. [PubMed] [Google Scholar]
- 295.Ubbink M, Hunt N I, Hill H A O, Canters G W. Kinetics of the reduction of wild-type and mutant cytochrome c550 by methylamine dehydrogenase and amicyanin from Thiobacillus versutus. Eur J Biochem. 1994;222:561–571. doi: 10.1111/j.1432-1033.1994.tb18898.x. [DOI] [PubMed] [Google Scholar]
- 296.Ubbink M, van Beeumen J, Canters G W. Cytochrome c550 from Thiobacillus versutus: cloning, expression in Escherichia coli, and purification of the heterologous holoprotein. J Bacteriol. 1993;174:3707–3714. doi: 10.1128/jb.174.11.3707-3714.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ubbink M, van Kleef M A, Kleinjan D J, Hoitink C W, Huitema F, Beintema J J, Duine J A, Canters G W. Cloning, sequencing and expression studies of the genes encoding amicyanin and the beta-subunit of methylamine dehydrogenase from Thiobacillus versutus. Eur J Biochem. 1991;202:1003–1012. doi: 10.1111/j.1432-1033.1991.tb16462.x. [DOI] [PubMed] [Google Scholar]
- 298.Ueda S, Yabutani T, Maehara A, Yamane S. Molecular analysis of the poly(3-hydroxyalkanoate) synthase gene from a methylotrophic bacterium, Paracoccus denitrificans. J Bacteriol. 1996;178:774–779. doi: 10.1128/jb.178.3.774-779.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 300.Urakami T, Araki H, Oyangi H, Suzuki K-I, Komagata K. Paracoccus aminophilus sp. nov. and Paracoccus aminovorans sp. nov., which utilize N,N-dimethylformamide. Int J Syst Bacteriol. 1990;40:287–291. doi: 10.1099/00207713-40-3-287. [DOI] [PubMed] [Google Scholar]
- 301.Urakami T, Tamaoka J, Suzuki K-I, Komagata K. Paracoccus alcaliphilus sp. nov., an alciphilic and facultatively methylotrophic bacterium. Int J Syst Bacteriol. 1989;39:116–121. [Google Scholar]
- 302.van der Oost J, de Boer A P N, de Gier J W L, Zumft W G, Stouthamer A H, van Spanning R J M. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol Lett. 1994;121:1–9. doi: 10.1111/j.1574-6968.1994.tb07067.x. [DOI] [PubMed] [Google Scholar]
- 303.van der Palen C J N M, Reijnders W N M, de Vries S, Stouthamer A H, Duine J A, van Spanning R J M. An essential role of mauE and mauD in methylamine metabolism in Paracoccus denitrificans. Antonie Leeuwenhoek. 1997;72:219–228. doi: 10.1023/a:1000441925796. [DOI] [PubMed] [Google Scholar]
- 304.van der Palen C J N M, Slotboom D J, Jongejan L, Reijnders W N M, Harms N, Duine J A, van Spanning R J M. Mutational analysis of mau genes involved in methylamine metabolism in Paracoccus denitrificans. Eur J Biochem. 1995;230:860–871. doi: 10.1111/j.1432-1033.1995.tb20629.x. [DOI] [PubMed] [Google Scholar]
- 305.van Ophem P W, Duine J A. NAD- and co-substrate (GSH or factor)-dependent formaldehyde dehydrogenases from methylotrophic microorganisms act as a class III alcohol dehydrogenase. FEMS Microbiol Lett. 1994;116:87–93. [Google Scholar]
- 306.van Spanning, R. J. M., and H. van Verseveld. Unpublished results.
- 307.van Spanning, R. J. M. Unpublished results.
- 308.van Spanning R J M, de Boer A P N, Reijnders W N M, de Gier J W L, Delorme C O, Stouthamer A H, Westerhoff H V, Harms N, van der Oost J. Regulation of oxidative phosphorylation: The flexible respiratory network of Paracoccus denitrificans. J Bioenerg Biomembr. 1995;27:499–512. doi: 10.1007/BF02110190. [DOI] [PubMed] [Google Scholar]
- 309.van Spanning R J M, de Boer A P N, Reijnders W N M, Spiro S, Westerhoff H V, Stouthamer A H, van der Oost J. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 1995;360:151–154. doi: 10.1016/0014-5793(95)00091-m. [DOI] [PubMed] [Google Scholar]
- 310.van Spanning R J M, de Boer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, van der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 311.van Spanning R J M, de Boer A P N, Slotboom D J, Reijnders W N M, Stouthamer A H. Isolation and characterization of a novel insertion sequence element, IS1248, in Paracoccus denitrificans. Plasmid. 1995;34:11–21. doi: 10.1006/plas.1995.1029. [DOI] [PubMed] [Google Scholar]
- 312.van Spanning R J M, Reijnders W N M, Stouthamer A H. Integration of heterologous DNA into the genome of Paracoccus denitrificans is mediated by a family of IS1248-related elements and a second type of integrative recombination event. J Bacteriol. 1995;177:4772–4778. doi: 10.1128/jb.177.16.4772-4778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.van Spanning R J M, van der Palen C J N M, Slotboom D J, Reijnders W N M, Stouthamer A H, Duine J A. Expression of the mau genes involved in methylamine metabolism in Paracoccus denitrificans is under control of a LysR-type transcriptional activator. Eur J Biochem. 1994;226:201–210. doi: 10.1111/j.1432-1033.1994.tb20042.x. [DOI] [PubMed] [Google Scholar]
- 314.van Spanning R J M, Wansell C W, de Boer A P N, Hazelaar M J, Anazawa H, Harms N, Oltmann L F, Stouthamer A H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991;173:6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.van Spanning R J M, Wansell C W, Harms N, Oltmann L F, Stouthamer A H. Mutagenesis of the gene encoding cytochrome c550 of Paracoccus denitrificans and analysis of the resultant physiological effects. J Bacteriol. 1990;172:986–996. doi: 10.1128/jb.172.2.986-996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.van Spanning R J M, Wansell C W, Reijnders W N M, Harms N, Ras J, Oltmann L F, Stouthamer A H. A method for the introduction of unmarked mutations in the genome of Paracoccus denitrificans: construction of strains with multiple mutations in the genes encoding periplasmic c550, c551i, and c553i. J Bacteriol. 1991;173:6962–6970. doi: 10.1128/jb.173.21.6962-6970.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.van Spanning R J M, Wansell C W, Reijnders W N, Oltmann L F, Stouthamer A H. Mutagenesis of the gene encoding amicyanin of Paracoccus denitrificans and the resultant effect on methylamine oxidation. FEBS Lett. 1990;275:217–220. doi: 10.1016/0014-5793(90)81475-4. [DOI] [PubMed] [Google Scholar]
- 318.van Verseveld H W, Stouthamer A H. Growth yields and the efficiency of oxidative phosphorylation during autotrophic growth of Paracoccus denitrificans on methanol and formate. Arch Microbiol. 1978;118:21–26. doi: 10.1007/BF00406069. [DOI] [PubMed] [Google Scholar]
- 319.Verhoeven W. Studies on true dissimilatory nitrate reduction. V. Nitric oxide production and consumption by microorganisms. Antonie Leeuwenhoek. 1956;22:384–406. doi: 10.1007/BF02538352. [DOI] [PubMed] [Google Scholar]
- 320.Wiese A, Schröder G, Brandenberg K, Hirsch A, Welte W, Seydel V. Influence of the lipid matrix on incorporation and function of LPS-free porin from Paracoccus denitrificans. Biochim Biophys Acta. 1994;1190:231–242. doi: 10.1016/0005-2736(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 321.Williams P A, Fulop V, Leung Y C, Chan C, Moir J W B, Howlett G, Ferguson S J, Radford S E, Hajdu J. Pseudospecific docking surfaces on electron transfer proteins as illustrated by pseudoazurin, cytochrome c550 and cytochrome cd1 nitrite reductase. Nat Struct Biol. 1995;2:975–982. doi: 10.1038/nsb1195-975. [DOI] [PubMed] [Google Scholar]
- 322.Williams R, Bell A, Sims G, Busby S. The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res. 1991;19:6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Winterstein C, Ludwig B. Genes coding for respiratory complexes map on all three chromosomes of the Paracoccus denitrificans genome. Arch Microbiol. 1998;169:275–281. doi: 10.1007/s002030050572. [DOI] [PubMed] [Google Scholar]
- 324.Winterstein C, Richter O-M, Ludwig B. Chimeric quinol oxidases expressed in Paracoccus denitrificans. NATO ASI Ser. 1998;c512:259–269. [Google Scholar]
- 325.Witt H, Ludwig B. Isolation, analysis and deletion of the gene coding for subunit IV of cytochrome c oxidase in Paracoccus denitrificans. J Biol Chem. 1997;272:5514–5517. doi: 10.1074/jbc.272.9.5514. [DOI] [PubMed] [Google Scholar]
- 326.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB-17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wodara C, Kostka S, Egert M, Kelly D P, Friedrich C G. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB-17. J Bacteriol. 1994;176:6188–6191. doi: 10.1128/jb.176.20.6188-6191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Reference deleted.
- 329.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Xu H H, Janka J J, Viebahn M, Hanson R S. Nucleotide sequence of the mxcQ and mxcE genes, required for methanol dehydrogenase synthesis in Methylobacterium organophilum XX: a two-component regulatory system. Microbiology. 1995;141:2543–2551. doi: 10.1099/13500872-141-10-2543. [DOI] [PubMed] [Google Scholar]
- 331.Xu X M, Matsuno-Yagi A, Yagi T. Characterization of the 25-kilodalton subunit of the energy-transducing NADH-ubiquinone oxidoreductase of Paracoccus denitrificans: sequence similarity to the 24-kilodalton subunit of the flavoprotein fraction of mammalian complex I. Biochemistry. 1991;30:8678–8684. doi: 10.1021/bi00099a027. [DOI] [PubMed] [Google Scholar]
- 332.Xu X M, Matsuno-Yagi A, Yagi T. The NADH-binding subunit of the energy-transducing NADH-ubiquinone oxidoreductase of Paracoccus denitrificans: gene cloning and deduced primary structure. Biochemistry. 1991;30:6422–6428. doi: 10.1021/bi00240a012. [DOI] [PubMed] [Google Scholar]
- 333.Xu X M, Matsuno-Yagi A, Yagi T. Gene cluster of the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans: characterization of four structural gene products. Biochemistry. 1992;31:6925–6932. doi: 10.1021/bi00145a009. [DOI] [PubMed] [Google Scholar]
- 334.Xu X M, Matsuno-Yagi A, Yagi T. Structural features of the 66-kDa subunit of the energy-transducing NADH-ubiquinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Arch Biochem Biophys. 1992;296:40–48. doi: 10.1016/0003-9861(92)90542-5. [DOI] [PubMed] [Google Scholar]
- 335.Xu X M, Matsuno-Yagi A, Yagi T. DNA sequencing of the seven remaining structural genes of the gene cluster encoding the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry. 1993;32:968–981. doi: 10.1021/bi00054a030. [DOI] [PubMed] [Google Scholar]
- 336.Yabutani T, Maehara A, Ueda S, Yamane T. Analysis of β-thioketolase and aceto-acetyl-coA reductase genes of a methylotrophic bacterium, Paracoccus denitrificans, and their expression in Escherichia coli. FEMS Microbiol Lett. 1995;133:85–90. doi: 10.1111/j.1574-6968.1995.tb07865.x. [DOI] [PubMed] [Google Scholar]
- 337.Yamamoto K, Uozumi T, Beppu T. The blue copper protein gene of Alcaligenes faecalis S-6 directs secretion of blue copper protein from Escherichia coli cells. J Bacteriol. 1987;169:5648–5652. doi: 10.1128/jb.169.12.5648-5652.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Yamane T, Chen X-F, Uda S. Polyhroxyalkanoate synthesis from alcohols during the growth of Paracoccus denitrificans. FEMS Microbiol Lett. 1996;135:207–211. [Google Scholar]
- 339.Yang H, Reijnders W N M, van Spanning R J M, Stouthamer A H, Harms N. Expression of the structural max genes in Paracoccus denitrificans follows wild-type regulation in mutants with a deletion in mxaY, the gene encoding the signal sensor. Microbiology. 1995;141:825–830. [Google Scholar]
- 340.Yang X, Trumpower B L. Purification of a three-subunit ubiquinol-cytochrome c oxidoreductase complex from Paracoccus denitrificans. J Biol Chem. 1986;261:12282–12289. [PubMed] [Google Scholar]
- 341.Yang X, Trumpower B L. Protonmotive Q cycle pathway of electron transfer and energy transduction in the three-subunit ubiquinol-cytochrome c oxidoreductase complex of Paracoccus denitrificans. J Biol Chem. 1988;263:11962–11970. [PubMed] [Google Scholar]
- 342.Ye R W, Fries M R, Bezborodnikov S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1993;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Yu P L, Hohn B, Falk H, Drews G. Molecular cloning of the ribosomal RNA genes of the photosynthetic bacterium Rhodopseudomonas capsulata. Mol Gen Genet. 1982;188:392–398. [Google Scholar]
- 344.Zalman L S, Nikaido H. Dimeric porin from Paracoccus denitrificans. J Bacteriol. 1984;162:430–433. doi: 10.1128/jb.162.1.430-433.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zickermann I, Anemuller S, Richter O-M H, Tautu O S, Link T A, Ludwig B. Biochemical and spectroscopic properties of the four-subunit quinol oxidase (cytochrome ba3) from Paracoccus denitrificans. Biochim Biophys Acta. 1996;1277:93–102. doi: 10.1016/s0005-2728(96)00086-2. [DOI] [PubMed] [Google Scholar]
- 346.Zickermann I, Tautu O S, Link T A, Korn M, Ludwig B, Richter O-M H. Expression studies on the ba3 quinol oxidase from Paracoccus denitrificans. A bb3 variant is enzymatically inactive. Eur J Biochem. 1996;246:618–624. doi: 10.1111/j.1432-1033.1997.00618.x. [DOI] [PubMed] [Google Scholar]
- 347.Zumft W G. The biological role of nitric oxide in bacteria. Arch Microbiol. 1993;160:253–264. doi: 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]
- 348.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1998;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zumft W G, Deusch A, Lochelt S, Cuypers H, Friedrich B, Schneider B. Derived amino acid sequences of the nosZ (respiratory N2O reductase) from Alcaligenes eutrophus, Pseudomonas aeruginosa and Pseudomonas stutzeri reveal potential copper binding residues. Implications for the CuA site of N2O reductase and cytochrome-c oxidase. Eur J Biochem. 1992;208:31–40. doi: 10.1111/j.1432-1033.1992.tb17156.x. [DOI] [PubMed] [Google Scholar]
- 350.Zumft W G, Korner H. Enzyme diversity and mosaic gene organisation in denitrification. Antonie Leeuwenhoek. 1997;71:43–58. doi: 10.1023/a:1000112008026. [DOI] [PubMed] [Google Scholar]
- 351.Zumft W G, Braun C, Cuypers H. Nitric oxide reductase from Pseudomonas stutzeri. Primary structure and gene organisation of a novel bacterial cytochrome bc complex. Eur J Biochem. 1994;219:481–490. doi: 10.1111/j.1432-1033.1994.tb19962.x. [DOI] [PubMed] [Google Scholar]