Abstract
The monofunctional penicillin-binding dd-peptidases and penicillin-hydrolyzing serine β-lactamases diverged from a common ancestor by the acquisition of structural changes in the polypeptide chain while retaining the same folding, three-motif amino acid sequence signature, serine-assisted catalytic mechanism, and active-site topology. Fusion events gave rise to multimodular penicillin-binding proteins (PBPs). The acyl serine transferase penicillin-binding (PB) module possesses the three active-site defining motifs of the superfamily; it is linked to the carboxy end of a non-penicillin-binding (n-PB) module through a conserved fusion site; the two modules form a single polypeptide chain which folds on the exterior of the plasma membrane and is anchored by a transmembrane spanner; and the full-size PBPs cluster into two classes, A and B. In the class A PBPs, the n-PB modules are a continuum of diverging sequences; they possess a five-motif amino acid sequence signature, and conserved dicarboxylic amino acid residues are probably elements of the glycosyl transferase catalytic center. The PB modules fall into five subclasses: A1 and A2 in gram-negative bacteria and A3, A4, and A5 in gram-positive bacteria. The full-size class A PBPs combine the required enzymatic activities for peptidoglycan assembly from lipid-transported disaccharide-peptide units and almost certainly prescribe different, PB-module specific traits in peptidoglycan cross-linking. In the class B PBPs, the PB and n-PB modules cluster in a concerted manner. A PB module of subclass B2 or B3 is linked to an n-PB module of subclass B2 or B3 in gram-negative bacteria, and a PB module of subclass B1, B4, or B5 is linked to an n-PB module of subclass B1, B4, or B5 in gram-positive bacteria. Class B PBPs are involved in cell morphogenesis. The three motifs borne by the n-PB modules are probably sites for module-module interaction and the polypeptide stretches which extend between motifs 1 and 2 are sites for protein-protein interaction. The full-size class B PBPs are an assortment of orthologs and paralogs, which prescribe traits as complex as wall expansion and septum formation. PBPs of subclass B1 are unique to gram-positive bacteria. They are not essential, but they represent an important mechanism of resistance to penicillin among the enterococci and staphylococci. Natural evolution and PBP- and β-lactamase-mediated resistance show that the ability of the catalytic centers to adapt their properties to new situations is limitless. Studies of the reaction pathways by using the methods of quantum chemistry suggest that resistance to penicillin is a road of no return.
Trypsin, chymotrypsin, and other acyl serine transferases catalyze the transfer of the electrophilic group R1—CO of ester, thiolester, and amide (peptide) carbonyl donors R1—CO—X—R2 to an acceptor, HY, via the formation of a serine-ester-linked acyl (R1—CO—) enzyme intermediate. X denotes an oxygen atom, a sulfur atom, or an NH group. R1 and R2 denote the substituents of the scissile CO—X bond. When HY is H2O, the carbonyl donor is hydrolyzed and the product of enzyme deacylation is R1—COOH. When HY is an amino compound, NH2—R3, the carbonyl donor is transpeptidated and the product of enzyme deacylation is R1—CO—NH—R3.
Specialized acyl serine transferases are involved in the assembly and metabolism of the bacterial cell wall peptidoglycan. They have in common the ability to catalyze the rupture of the β-lactam amide bond of penicillin and the formation of a serine ester-linked penicilloyl enzyme. However, this intermediate is almost completely inert, the enzyme catalytic center turns over very slowly, once or less per hour, and the inactivated acyl serine transferases are easily detectable as penicillin-binding proteins (PBPs).
The production of β-lactamases of classes A, C, and D is a remarkable defensive mechanism that bacteria have developed to protect their wall peptidoglycan-synthesizing machinery against the toxic effect of penicillin. The serine β-lactamases hydrolyze penicillin into penicilloate via the formation of a serine ester-linked penicilloyl enzyme that is hydrolytically labile. On good β-lactam substrates, β-lactamases can turn over 1,000 times or more per second.
The PBPs and serine β-lactamases have been discussed in recent reviews (22, 24–28, 38, 39, 48, 49). This article focuses on questions that biochemists still strive to answer concerning the multimodular PBPs, which, globally, are the lethal targets of penicillin in susceptible bacteria. To apprehend the problem, we shall first position the multimodular PBPs within the penicilloyl serine transferases superfamily (24).
THE PENICILLOYL SERINE TRANSFERASES: A SUPERFAMILY OF MULTIPLE PERSONALITIES
The bacterial cell wall peptidoglycan (23) is a covalently closed, net-like polymer in which glycan strands are cross-linked by peptides (Fig. 1A). The glycan portion is made up of alternating β-1,4-linked units of N-acetylglucosamine and N-acetylmuramic acid arranged in linear chains. The carboxyl groups of the N-acetylmuramic acid residues are involved in amide linkages to terminal l-alanine residues of the peptide units l-alanyl-γ-d-glutamyl-l-diaminoacyl-d-alanine. Also, neighboring peptide-substituted glycan strands are cross-linked by peptide bridges which extend from the carboxyl group of the terminal d-alanine of one peptide unit to the side chain amino group of the diamino acid residue of another peptide unit.
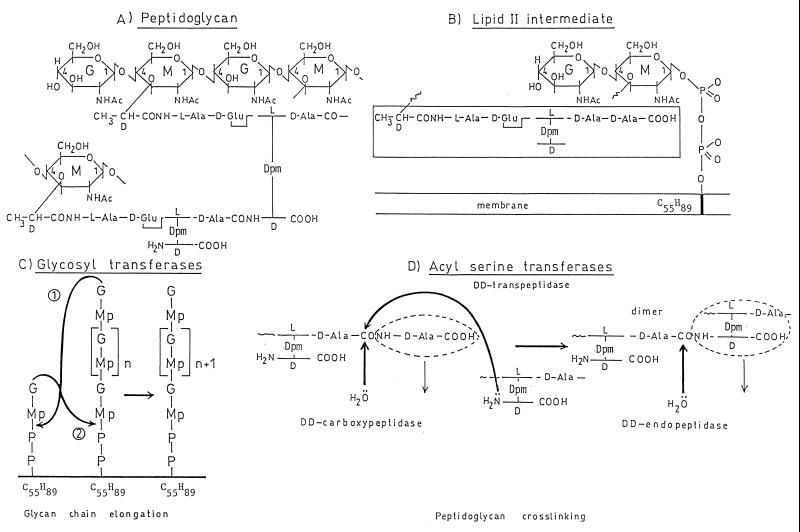
FIG. 1.
(A and B) Structure of the wall peptidoglycan of E. coli (A) and assembly from the lipid II intermediate (B). G, N-acetylglucosamine; M, N-acetylmuramic acid; Dpm, meso-diaminopimelic acid; transmembrane bar, C55H89 isoprenoid alcohol carrier; Mp, N-acetylmuramyl pentapeptide. (C) Reaction 1, glycan chain elongation at the nonreducing end of the chain; reaction 2, glycan chain elongation at the reducing end of the chain. (D) Overall reactions catalyzed by the dd-transpeptidases, dd-carboxypeptidases, and dd-endopeptidases.
The nature of the diamino acid residue of the peptide units, the composition and length of the peptide bridges, and the extent of peptidoglycan cross-linking vary according to the bacterial species. In Escherichia coli, the diaminoacid residue is meso-diaminopimelic acid, the glycan chains are substituted by peptide monomers and cross-linked peptide dimers, most of the interpeptide bridges are direct d-alanyl-(d)-meso-diaminopimelic acid bonds, and peptide oligomers larger than dimers have not been identified.
The immediate precursor of the wall peptidoglycan is lipid II (Fig. 1B). A disaccharide peptide is linked to a C55H89 undecaprenyl lipid carrier via a pyrophosphate bridge involving C1 of N-acetylmuramic acid, and the peptide borne by N-acetylmuramic acid is a pentapeptide which terminates with the sequence d-alanyl-d-alanine. From this precursor, the assembly of lipid-transported disaccharide pentapeptide units into polymeric peptidoglycan requires two enzymatic activities, a glycosyl transferase and an acyl serine transferase (66).
The glycosyl transferase-catalyzed transglycosylation proceeds through displacement of the pyrophosphate of lipid II by the 4-hydroxyl group of N-acetylglucosamine of the growing glycan chain (reaction 1 in Fig. 1C) or displacement of the pyrophosphate of the growing glycan chain by the 4-hydroxyl group of N-acetylglucosamine of lipid II (reaction 2 in Fig. 1C). This latter mechanism has been observed in Bacillus licheniformis and Micrococcus luteus (68, 69). In Escherichia coli, the absence of nascent peptidoglycan has been established, implying that its binding to preexisting peptidoglycan is concomitant with its synthesis.
The acyl serine transferase (transpeptidase)-catalyzed peptidoglycan cross-linking is made at the expense of the d-alanyl-d-alanine bond of the pentapeptide units (Fig. 1D). The reaction proceeds via the formation of a serine ester-linked peptidyl (-l-alanyl-γ-d-glutamyl-l-diaminoacyl-d-alanyl) enzyme with the concomitant release of the carboxy-terminal d-alanine of the pentapeptide, and it is achieved by the transfer of the peptidyl moiety to the side chain amino group of the diamino acid residue of another peptide. Because the reaction involves breaking a d-alanyl-d-alanine bond, the transferase is classified as a dd-transpeptidase.
PBP1a and PBP1b of E. coli each catalyze the conversion of lipid II into peptidoglycan in in vitro assays (50). These two PBPs are the prototypes of bifunctional PBPs which combine in a single polypeptide chain the required transglycosylase and dd-transpeptidase activities. Essentially, a noncleavable signal peptide which functions as a transmembrane anchor is fused to the amino end of a transglycosylase non-penicillin-binding (n-PB) module, which itself is fused to the amino end of an acyl serine transferase (dd-transpeptidase) penicillin-binding (PB) module. The two catalytic modules form a single polypeptide chain that folds on the exterior of the plasma membrane.
To allow the bacterial cell to grow and divide, morphogenetic networks channel peptidoglycan assembly into wall expansion and septum formation in a cell-cycle-dependent fashion. Central to these networks are PBPs which are similar in their modular design to the bienzymatic (transglycosylase-acyl serine transferase) PBPs. However, the n-PB module is not a transglycosylase (1). E. coli PBP2 and PBP3 are involved in cell shape maintenance and cell division, respectively (53). They are the prototypes of bifunctional PBPs which combine in a single polypeptide chain a morphogenetic determinant n-PB module and an acyl serine transferase PB module.
Throughout the bacterial cell cycle, the wall peptidoglycan undergoes constant chemical changes that do not impair the tensile strength of the polymer (34). Monofunctional serine dd-carboxypeptidases/PBPs hydrolyze d-alanyl-d-alanine bonds (Fig. 1D). They control the extent of peptidoglycan cross-linking by limiting the number of pentapeptide units available for transpeptidation. Monofunctional serine dd-carboxypeptidases/PBPs also hydrolyze peptidoglycan interpeptide bonds, which in some bacteria extend between two d-centers in the α-position to a free carboxylate, for example a d-alanyl-(d)-meso-diaminopimelic acid bond (Fig. 1D). E. coli PBP4 through PBP7 are dd-carboxy/endopeptidases. Loss of these PBPs is tolerated (73). Their functions and those of many other peptidoglycan hydrolases presumably include a role in the recycling of old peptidoglycan, a role as zipper during cell division, and a role as space maker for the insertion of new peptidoglycan material.
The serine β-lactamases, the monofunctional PBPs, and the PB modules of the multimodular PBPs fulfill different functions, and similarity in their amino acid sequences is, globally, almost nonexistent. However, they operate on R1—CO—X—R2 carbonyl donors by the same proton abstraction-donation mechanism, and their catalytic centers have a remarkably well-conserved topology.
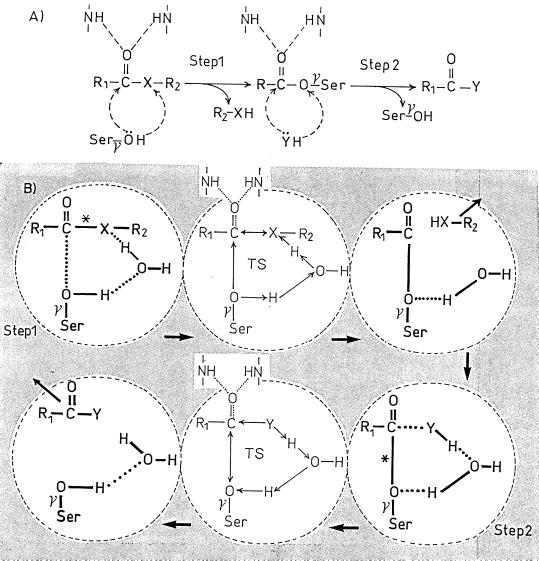
In step 1 of the catalyzed reaction (Fig. 2A), the proton of the serine γOH is abstracted, the activated Öγ attacks the carboxyl carbon atom of the CO—X bond, and the abstracted proton is back-donated to the adjacent X atom, resulting in the formation of the serine-ester-linked acyl enzyme. In step 2 (for the reaction to reach completion), the proton of the acceptor HY (an amino group or water) is abstracted, the activated Ÿ attacks the carbonyl carbon atom of the CO—O ester bond of the acyl enzyme and the abstracted proton is back-donated to the Oγ atom of the serine residue.
FIG. 2.
Acyl serine transferase-catalyzed reaction on R1—CO—X—R2 carbonyl donors. (A) Overall reaction. (B) Role of a water molecule as proton transmitter. X is O, S, or NH (in penicillin, the CO—N bond is endocyclic). HY is an acceptor, i.e., water or an amino compound. The carbonyl of the donor and that of the acyl enzyme are polarized by NH groups of the enzyme polypeptide backbone. TS, transition state; ∗, scissile bond of the carbonyl donor (step 1) and scissile bond of the acyl enzyme (step 2). The shaded area symbolizes the active-site environment.
The catalytic centers that perform this double proton shuttle are defined by three amino acid groupings, referred to as motifs. These motifs occur in the same order and with roughly the same spacing along the polypeptide chains, defining a common amino acid sequence signature (Fig. 3). Polypeptide folding brings the three motifs close to each other, forming a cavity at the boundary between an all-α domain and an α/β domain, which itself consists of a five-stranded β-sheet protected by α-helices (38).
FIG. 3.
Amino acid sequence signature of the penicilloyl serine transferases superfamily. S. K15, Streptomyces strain K15; S. R61, Streptomyces strain R61; Eco, E. coli; Bsu, B. subtilis; Spn, S. pneumoniae; Eclo, E. cloacae; Tgase, transglycosylase; Tpase, transpeptidase. PBPs and β-lactamases marked by an asterisk are of known three-dimensional structure (at 3.5-Å resolution only for Spn PBP2x). Motifs 1, 2, and 3 are at the top, and secondary structures are at the bottom. The monofunctional PBPs and β-lactamases are synthesized with a cleavable signal peptide. The R61 PBP and the β-lactamases are secreted; E. coli PBP4 is, somehow, loosely attached to the membrane; the Streptomyces strain K15 PBP is membrane associated via an internal hydrophobic segment; E. coli PBP5 and probably B. subtilis PBP5 are membrane associated via a carboxy-terminal amphiphilic helix. The multimodular PBPs are synthesized with a noncleavable signal peptide that serves as membrane anchor.
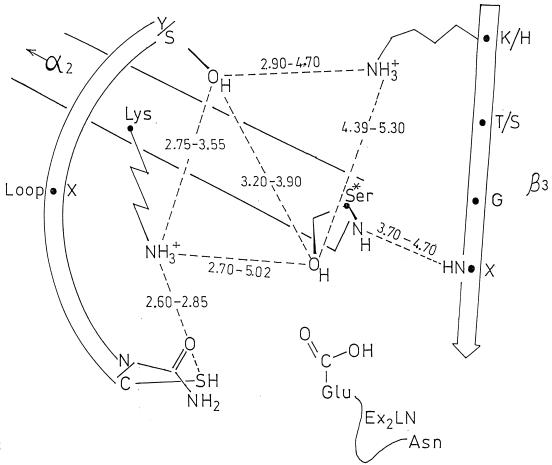
As shown in Fig. 4, motif 1, Sx2K (where S is the essential serine residue and x is a variable amino acid residue), is at the amino end of helix α2 of the all-α domain and occupies a central position in the catalytic center. Motif 2, [S/Y]x[N/C], is on a loop connecting two α-helices of the all-α domain and defines one side of the catalytic center. Motif 3, [K,H][T,S]G, is on strand β3 of the β-sheet and defines the other side of the catalytic center. Although secondary structures vary in number, size, and orientation, the spanning distances between most of the heavy atoms of the side chains of the active-site-defining motifs 1, 2, and 3 differ by less than 1 Å (Fig. 4). Variations are observed at the top and bottom of the cavities. At this latter position, the side chain of the glutamic acid residue of motif Ex2LN, found only in the class A β-lactamases, points toward the inside of the catalytic center.
FIG. 4.
Schematic representation of the catalytic center of the penicilloyl serine transferases. Average distances in angstroms between heavy atoms (O-O, O-N, S-N, and N-N) of side chains of the active-site-defining motif 1 (on helix α2), motif 2 (on a loop), and motif 3 (on strand β3) are given. The values derived from X-ray data apply to the proteins marked by an asterisk in Fig. 3. Motif 1 is, invariably, Sx2K. Motif 2, SxN, is replaced by SxC in Streptomyces strain K15 PBP and by YxN in Streptomyces strain R61 PBP and class C β-lactamases. Motif 3, K[T/S]G, is replaced by HTG in Streptomyces strain R61 PBP.
This background of structural similarity and catalytic diversity illustrates the concept that evolution often obscures the function. In the absence of direct biochemical data, the function of a penicilloyl serine transferase cannot be identified on the basis of amino acid sequence or even three-dimensional structure comparisons.
HIERARCHICAL ANALYSIS OF MULTIMODULAR PBPS
By limiting our scrutiny to the small number of available sequences, it was proposed in 1991 (24) that, depending on the motifs borne by the n-PB modules, the multimodular PBPs fall into class A (whose prototypes are the E. coli bienzymatic PBP1a and PBP1b) and class B (whose prototypes are the E. coli cell cycle PBP2 and PBP3). In recent years, the databases have expanded considerably and several bacterial genomes have been sequenced (5, 12, 20, 21, 36, 41, 65). These advances allow for better standards in screening similarity and for improvement of our ability to apprehend the structure-function relationships and, perhaps, to manipulate the functions of the PBPs in wall peptidoglycan assembly and cell morphogenesis.
Sixty-three multimodular PBPs have been analyzed. They are listed in Table 1, with their identifier codes and accession numbers. Some PBPs have been characterized biochemically, and others have been identified only on the basis of genome sequence data. This applies, in particular, to the PBPs of the spiral-shaped Helicobacter pylori (the causative agent of peptic ulcer disease) and Borrelia burgdorferi (the causative agent of Lyme disease), the filamentous Aquifex aeolicus, and the cyanobacterium Synechocystis sp. strain PCC863. A. aeolicus is a member of the most deeply branching family within the bacterial domain and is one of the most thermophilic bacteria known, with growth temperature maxima near 95°C. Synechocystis sp. strain PCC863 carries genes for oxygenic photosynthesis and is also a very deeply branching family member. Chloroplasts are believed to have evolved from cyanobacterial ancestors which developed an endosymbiotic relationship with a eukaryotic host cell.
TABLE 1.
Code, accession number, class, and size for multimodular PBPs
| Sourcea | PBP code | Data bankb | Accession no. | Subclassc | Residue no. |
|---|---|---|---|---|---|
| Gram-positive bacteria | |||||
| +Bacillus subtilis | Bsu1 | S | P39793 | A3 | 914 |
| Bsu2c | S | P38050 | A4 | 714 | |
| Bsu4 | S | P40750 | A5 | 624 | |
| Bsu3 | S | P42971 | B1 | 668 | |
| BsuVD | S | Q03524 | B3 | 645 | |
| Bsu2b | S | Q07868 | B4 | 716 | |
| Bsu2a | S | P54488 | B5 | 716 | |
| Enterococcus faecalis | Efas5 | E | X78425 | B1 | 679 |
| Enterococcus faecium | Efam5 | E | X84859 | B1 | 673 |
| EfamC | E | U94707 | B4 | 742 | |
| Enterococcus hirae | Ehi3r | P | A36903 | B1 | 678 |
| Ehi5 | E | X62280 | B1 | 678 | |
| Ehi3s | E | Y13922 | B4 | 730 | |
| Staphylococcus aureus | Sau2 | P | S43693 | A3 | 716 |
| Sau2a | P | JQ0773 | B1 | 668 | |
| SauB | E | U94706 | B4 | 646 | |
| Staphylococcus sciuri | Ssc2 | E | Y09223 | B1 | 666 |
| Streptococcus oralis | Sor1a | S | Q00573 | A3 | 637 |
| Streptococcus pneumoniae | Spn1a | S | Q04707 | A3 | 719 |
| Spn2a | E | AJ002292 | A4 | 731 | |
| Spn1b | E | AJ002291 | A5 | 821 | |
| Spn2x | S | P14677 | B4 | 750 | |
| Spn2b | S | P10524 | B5 | 679 | |
| Streptococcus pyogenes | Spy263 | O | contig 263 | A3 | 721 |
| Spy250 | O | contig 250 | A4 | 778 | |
| Spy286 | O | contig 286 | A5 | 723 | |
| Spy290 | O | contig 290 | B4 | 752 | |
| Streptococcus thermophilus | Sth2b | E | U58210 | B5 | 704 |
| Actinomycetales | |||||
| Mycobacterium leprae | Mle1* | E | L39923 | A | 686 |
| Mle1 | E | L01263 | A | 821 | |
| Mycobacterium tuberculosis | Mtu1 | E | Z80775 | A | 665 |
| Streptomyces clavuligerus | Sclpcbr | E | U56256 | A:B | 551 |
| Streptomyces coelicolor | Scoa3 | E | Y14206 | B:B2 | 770 |
| Gram-negative bacteria | |||||
| Citrobacter freundii | Cfr1b | P | S57580 | A2 | 846 |
| +Escherichia coli | Eco1a | S | P02918 | A1 | 850 |
| Eco1b | S | P02919 | A2 | 844 | |
| Eco1c | E | U88571 | A | 770 | |
| Eco2 | S | P08150 | B2 | 633 | |
| Eco3 | S | P04286 | B3 | 588 | |
| +Haemophilus influenzae | Hin1a | S | P31776 | A1 | 853 |
| Hin1b | S | P45345 | A2 | 781 | |
| Hin2 | P | C64044 | B2 | 651 | |
| Hin3 | P | G64184 | B3 | 610 | |
| Neisseria gonorrhoeae | Ngo1 | E | U72876 | A1 | 798 |
| Ngo2 | P | S49090 | B3 | 582 | |
| Neisseria meningitidis | Nme1 | E | U80933 | A1 | 798 |
| Nme2 | E | X59624 | B3 | 584 | |
| Pseudomonas aeruginosa | Pae1a | E | U73780 | A1 | 822 |
| Pae3 | P | S54872 | B3 | 579 | |
| Pae3a | E | X95517 | B3 | 565 | |
| Spiral-shaped and filamentous gram-negative related bacteria | |||||
| +Helicobacter pylori | Hpy1a | E | AE000573 | A2 | 659 |
| Hpy2 | E | AE000654 | B:B2 | 588 | |
| Hpy3 | T | HP1556 | B:B3 | 615 | |
| +Borrelia burgdorferi | Bbu3 | G | AE001173 | A1 | 932 |
| Bbu2 | G | AE001171 | B:B2 | 599 | |
| Bbu1 | G | AE001125 | B3 | 629 | |
| +Aquifex aeolicus | Aaemrca | G | AE000699 | A1 | 726 |
| AaeA1 | G | AE000728 | B2 | 595 | |
| AaeA2 | G | AE000695 | B3 | 578 | |
| Cyanobacteria | |||||
| +Synechocystis strain PCC6803 | Syn1a | K | sll0002 | A1 | 885 |
| Synmrca | K | sll1434 | A1 | 650 | |
| Synmrcb | K | slr1710 | A1 | 749 | |
| Syn3 | K | sll1833 | B3 | 607 |
+ denotes that the genome has been sequenced.
E, EMBL DNA data bank; G, GenBank; K, Kazusa DNA Research Institute data bank at http://www.kazusa.or.jap; O, Oklahoma University DNA data bank at http://www.genome.ou.edu; P, PIR data bank; S, Swiss-Prot data bank; T, TIGR Microbial data bank at http://www.tigr.org.
Letters without suffixes indicate that the PBP does not belong to a defined subclass. A:B indicates that the n-PB and PB modules of the PBP are of classes A and B, respectively. B:B2 and B:B3 indicate that the n-PB module of the PBP is an outlier of class B.
Classically, the relationships between genes from different genomes are represented as a system of orthologs and paralogs. Ortholog genes in different organisms have evolved from a common ancestor, and the encoded ortholog proteins normally retain the same function. Paralog genes are related by duplication within a genome, and paralog proteins normally evolve new functions. Often, the picture is more complex because genes may have been acquired by horizontal transfer, so that paralogs may have arisen from orthologs and vice versa.
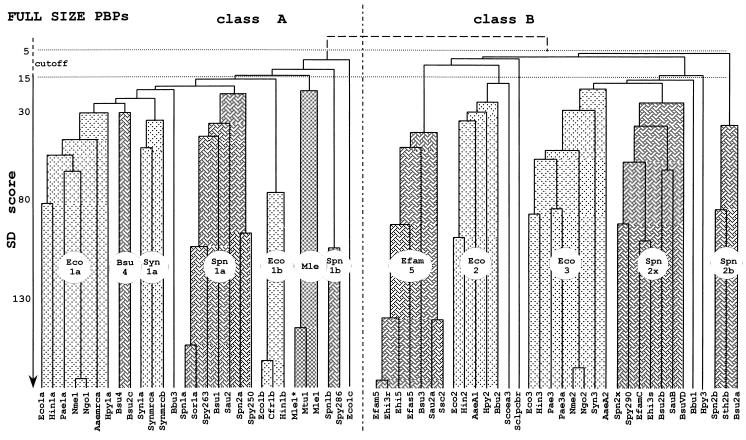
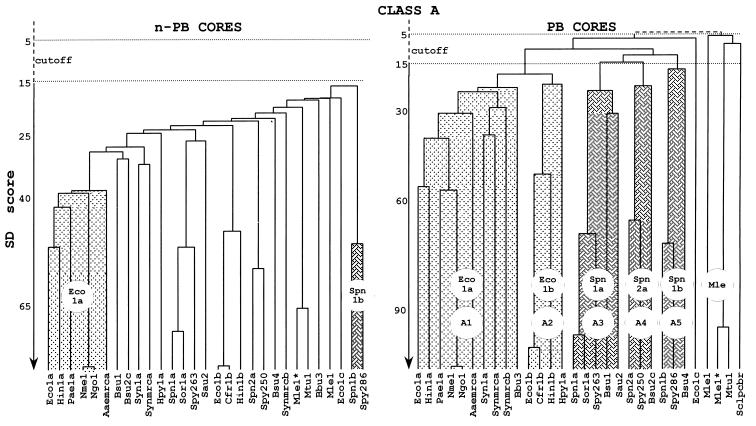
To shed light on the structure-function and evolutionary relationships of the multimodular PBPs and the differential roles that they play in wall peptidoglycan assembly, the amino acid sequences of the 63 PBPs have been analyzed by hierarchically combining pairwise comparisons of two sequences, either a sequence and a preexisting alignment or two preexisting alignments (2, 45). By using PileUp from the Wisconsin package, the full-size PBPs generate the dendrograms shown in Fig. 5. The PBP identifier codes are at the bottom, the sequences joined toward the bottom have greater similarity than the sequences joined further to the top, and the vertical axis is roughly calibrated in similarity scores calculated for selected pairs of sequences.
FIG. 5.
Hierarchical analysis of full-size, multimodular PBPs. PBP codes are given in Table 1. Each cluster (or subclass) is identified by a prototypic PBP. For SD score values and more details, see the text.
A score is the standard deviation value above that expected from a run of 100 randomized pairs of sequences with the same amino acid composition as the two sequences under comparison. The score calculated for a pair of identical sequences varies depending on the length and, to some extent, the composition of the polypeptide chain. Thus, the scores calculated for Bacillus subtilis PBP1a (914 amino acid residues) and Pseudomonas aeruginosa PBP3a (565 amino acid residues) are 173 and 145, respectively. In spite of these limitations, dendrograms calculated for proteins with different molecular masses give a reasonably accurate picture of the hierarchical relationship. Scores larger than 15 express statistically significant similarity, scores smaller than 5 express lack of similarity, and scores between 15 and 5 define a cutoff region.
The results of this analysis (Fig. 5) are consistent with those described recently (13, 49, 55). The 63 full-size PBPs fall into two unbridgeable classes, A and B (score, <<5). Class A comprises 29 PBPs. The PBPs of gram-negative bacteria fall into three clusters (whose prototypes are E. coli PBP1a, Synechocystis PBP1a, and E. coli PBP1b), the PBPs of gram-positive bacteria fall into three clusters (whose prototypes are B. subtilis PBP4, Streptococcus pneumoniae PBP1a, and S. pneumoniae PBP1b), and the mycobacterial PBPs form one distinct cluster. Class B comprises 34 PBPs. The PBPs of gram-negative bacteria fall into two clusters (whose prototypes are E. coli PBP2 and E. coli PBP3), and the PBPs of gram-positive bacteria fall into three clusters (whose prototypes are Enterococcus faecium PBP5, S. pneumoniae PBP2x, and S. pneumoniae PBP2b). Classes A and B contain PBP outliers, which fall outside the clusters.
CORE-BASED CLUSTERING OF MULTIMODULAR PBPS
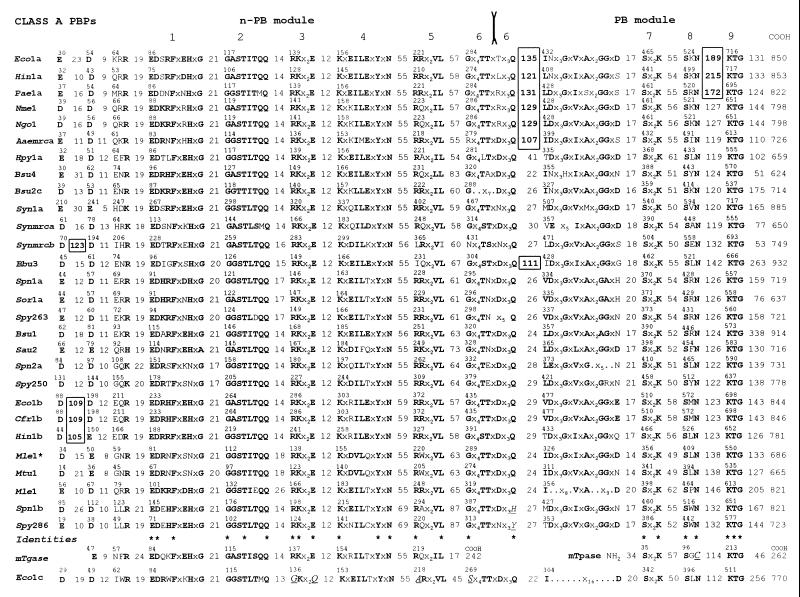
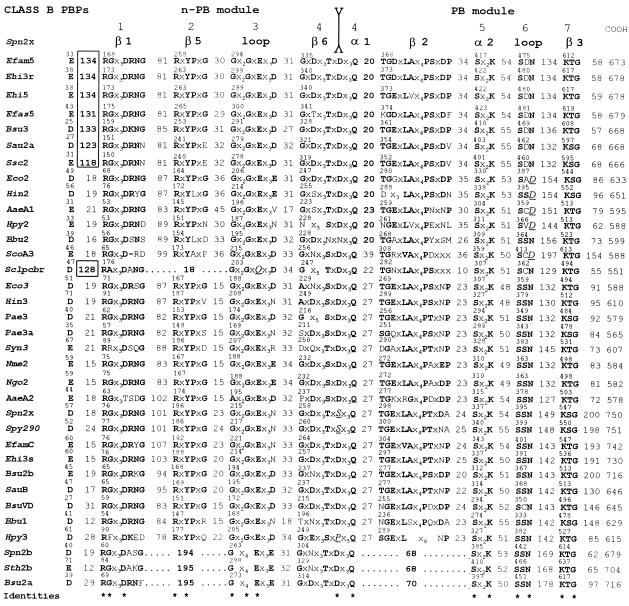
To identify features that may be responsible for the clustering shown in Fig. 5, the amino acid sequences of the 63 full-size PBPs have been aligned in a way that highlights the class-specific motifs. In Fig. 6 and 7, the sequences for class A and class B PBPs, respectively, are presented in the same order from top to bottom as in the dendrogram of Fig. 5 from left to right. The sequences each start at the first dicarboxylic acid, D or E, immediately downstream from the hydrophobic sequence assumed to function as membrane anchor. The conserved motifs that contain identities are numbered at the top of the figures. Equivalent amino acid residues (A and G; K, R, and H; D and E; I, L, and V) are in boldface type. The intermotif distances are given as the number of amino acid residues. Since it is known that motif 4 of S. pneumoniae class B PBP2x is the fusion site between strand β6 at the carboxy end of the n-PB module and helix α1 at the amino end of the PB module (57), motif 4 of all class B PBPs and the equivalent motif 6 of all class A PBPs are assumed to represent the intermodule junctions.
FIG. 6.
Amino acid sequence analysis of full-size, multimodular class A PBPs. Conserved motifs, intermodule junction sites, inserts, and amino- and carboxy-terminal extensions are shown. mTgase, putative monofunctional transglycosylase of E. coli (Swiss-Prot accession no. P46022). mTpase, monofunctional dd-transpeptidase/PBP of Streptomyces strain K15 (Swiss-Prot accession no. P39042); Mle1∗, high-affinity, thermolabile class A PBP of M. leprae (43). Asterisks at the bottom of the figure highlight identities defining the amino acid sequence signatures of the modules. Amino acid residues that do not obey the consensus are in underlined italics. For more details, see the text.
FIG. 7.
Amino acid sequence analysis of full-size, multimodular class B PBPs. Conserved motifs, intermodule junction sites, inserts, and carboxy-terminal extensions are shown. Secondary structures of the S. pneumoniae PBP2x are shown at the top. Identities (asterisks) define the amino acid sequence signatures of the modules. Amino acid residues that do not obey the consensus are in underlined italics. For more details, see the text.
The picture which emerges from these alignments is one of distinctive motifs in the n-PB modules of class A PBPs versus class B PBPs, of conserved penicilloyl serine transferase motifs in the PB modules of both class A and class B PBPs, and of adducts occurring at various places along the polypeptide chains.
Cytosolic tails are present when the sequence between the amino end of the protein and the first dicarboxylic acid, D or E, is more than about 60 to 70 amino acid residues long. Carboxy-terminal extensions are present when the sequence between motif K[T/S]G and the carboxy end of the protein is more than about 60 to 70 amino acid residues long. The carboxy-terminal extensions vary widely in size. Residues 780 to 844 in E. coli PBP1b are dispensable (37), but residues 762 to 780 at the carboxy end of the PB module are not (42). Internal inserts (boxed in Fig. 6 and 7) are also present. They occur between the membrane anchor and motif 1 in class A and class B PBPs, downstream from the junction site in class A PBPs, and between motifs 8 and 9 of the PB modules of class A PBPs.
As a result of these alignments, the cores of the n-PB modules can be defined as the sequences extending from the amino end of motif 1 to the carboxy end of motif 6 (class A) or motif 4 (class B). Likewise, the cores of the PB modules can be defined as the sequences starting 60 amino acid residues upstream from motif Sx2K and terminating 70 amino acid residues downstream from motif KTG or at the carboxy end of PBPs which have no carboxy-terminal extensions. On this basis, the hierarchical analysis of the cores of the n-PB modules (n-PB cores) and the cores of the associated PB modules (PB cores) leads to important observations.
The n-PB cores which bear the distinctive class-specific motifs fall into two unbridgeable groups, class A and class B (scores, ≤5), as expected. Contrary to expectations, the PB cores which all bear the penicilloyl serine transferases motifs also fall in the same unbridgeable classes A and B (scores, ≤5).
As shown in Fig. 8, the n-PB cores of the 29 class A PBPs form a continuum of diverging sequences which, from the E. coli PBP1a cluster to the S. pneumoniae PBP1b cluster, are related by scores larger than 15. In contrast, the associated PB cores of PBPs from gram-negative bacteria fall into two subclasses, A1 (whose prototype is E. coli PBP1a) and A2 (whose prototype is E. coli PBP1b), which form a supercluster; and those of PBPs from gram-positive bacteria fall into three subclasses, A3 (whose prototype is S. pneumoniae PBP1a), A4 (whose prototype is S. pneumoniae PBP2a), and A5 (whose prototype is S. pneumoniae PBP1b), which also form a supercluster. It should also be noted that the PB cores of A. aeolicus PBP mrca, Synechocystis PBPs 1a, mrca, and mrcb, and B. burgdorferi PBP3 belong to subclass A1; the PB core of H. pylori PBP1a belongs to subclass A2; the PB cores of Mycobacterium leprae PBP1* and M. tuberculosis PBP1 form a distinct cluster; and the PB cores of E. coli PBP1c, M. leprae PBP1, and Streptomyces clavuligerus PBP pcbr are outliers. M. leprae PBP1 is thermostable and binds penicillin with a very low affinity (3). In contrast, M. leprae PBP1* is thermolabile and binds penicillin with a high affinity (43).
FIG. 8.
Hierarchical analysis of the n-PB cores and PB cores of the multimodular class A PBPs. PBP codes are given in Table 1.
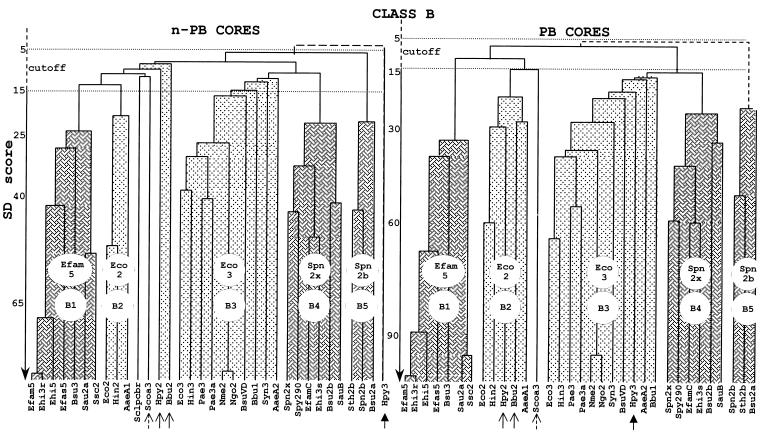
As shown in Fig. 9, the n-PB and PB cores of class B PBPs from gram-positive bacteria fall into three distinct subclasses, B1 (whose prototype is Enterococcus faecium PBP5), B4 (whose prototype is S. pneumoniae PBP2x), and B5 (whose prototype is S. pneumoniae PBP2b). Remarkably, an n-PB core of subclass B1, B4, or B5 is linked to a PB core of subclass B1, B4, or B5, respectively. As also shown in Fig. 9, the n-PB and PB cores of class B PBPs from gram-negative bacteria fall into two distinct subclasses, B2 (whose prototype is E. coli PBP2) and B3 (whose prototype is E. coli PBP3). Almost invariably, an n-PB core of subclass B2 or B3 is linked to a PB core of subclass B2 or B3, respectively. It should be noted that the n-PB cores of H. pylori PBP2, B. burgdorferi PBP2, and Streptomyces coelicolor PBP3a are outliers and that the corresponding associated PB cores belong to subclass B2 (thin and broken arrows in Fig. 9). Likewise, the n-PB core of H. pylori PBP3 is an outlier and the associated PB core belongs to subclass B3 (thick arrow in Fig. 9).
FIG. 9.
Hierarchical analysis of the n-PB cores and PB cores of the multimodular class B PBPs. PBP codes are given in Table 1.
Streptomyces clavuligerus PBP pcbr is the only wild card which really breaks the pattern. The full-size PBP is of class B, the n-PB core is of class B, and the PB core is of class A. This PBP, whose gene is located downstream from the isopenicillin synthase-encoding gene in the cephamycin cluster, is probably responsible for the penicillin resistance of this organism (56).
The picture which results from the above analysis is almost certainly far short of being complete. As the structure databases continue to expand, existing subclasses will become more populated and new subclasses will be identified. Moreover, PBPs of the spiral-shaped H. pylori and B. burgdorferi and that of Streptomyces coelicolor suggest that mixed subclasses of PBPs may exist. Unrelated n-PB modules may be linked to PB modules belonging to a same subclass.
Adducts (Fig. 6 and 7; Table 2) allow the core-based clustering to be refined. Three PBPs of subclass A2 (Eco1b, Cfr1b, and Hin1b) have structurally related inserts downstream from the membrane anchor (>30% identity [Table 2]). The PBPs of subclass B1 (Efam5, Ehi3r, Ehi5, Efas5, Bsu3, Sau2a, and Ssc2) also have inserts at this position (Table 2). The enterococcal and Bacillus inserts are structurally related (>23% identity), the two staphylococcal inserts have 71% identity, and the two groups of inserts are distinctly related to each other (17% identity). Three PBPs of subclass A1 (Eco1a, Hin1a, and Pae1a) have inserts between motif 8 and motif 9 of the PB module which are structurally related (>30% identity [Table 2]). Six PBPs of subclass A1 (Eco1a, Hin1a, Pae1a, Nme1/Ngo1, Aaemrca, and Bbu3) have inserts located downstream from the intermodule junction (Table 2). The E. coli, Haemophilus influenzae, P. aeruginosa, and Neisseria inserts are structurally related (>25% identity).
TABLE 2.
Pairwise comparison of the amino acid sequences of inserts of multimodular PBPs
| Subclass | PBPa | Similarity (%) |
|---|---|---|
| A2 | Eco1b D88-D198/Cfr1b D88–D198 | 91 |
| /Hin1b D44–E150 | 32 | |
| B1 | Efam5 E33-R168/Ehi3r E38–R173 | 97 |
| /Ehi5 E38–R173 | 65 | |
| /Efas5 E43–R175 | 35 | |
| /Bsu3 D25–R159 | 27 | |
| Ehi5 E38–R173/Efas5 E43–R175 | 37 | |
| /Bsu3 D25–R159 | 28 | |
| Efas5 E43–R175/Bsu3 D25–R159 | 23 | |
| Sau2a D27–R151/Ssc2 E31–R150 | 71 | |
| /Efam5 E33–R168 | 17 | |
| /Ehi3r E38–R173 | 17 | |
| /Ehi5 E38–R173 | 17 | |
| /Efas5 E43–R175 | 17 | |
| /Bsu3 D25–R159 | 18 | |
| A1 | Eco1a N526–K716/Hin1a N501–K717 | 48 |
| /Pae1a N522–K695 | 31 | |
| Hin1a N501–K717/Pae1a N522–K695 | 31 | |
| A1 | Nme1/Ngo1 Q298–L428/Eco1a Q296–I432 | 25 |
| /Hin1a Q286–L408 | 26 | |
| /Pae1a Q296–L428 | 26 | |
| /Aaemrca Q291–I399 | 21 | |
| /Bbu3 Q316–I428 | 6 | |
| Eco1a Q296–I432/Hin1a Q286–L408 | 45 | |
| /Pae1a Q296–L428 | 40 | |
| /Aaemrca Q291–I399 | 16 | |
| /Bbu3 Q316–I428 | 4 | |
| Hin1a Q286–L408/Pae1a Q296–L428 | 32 | |
| /Aaemrca Q291–I399 | 14 | |
| /Bbu3 Q316–I428 | 9 | |
| Pae1a Q296–L428/Aaemrca Q291–I399 | 18 | |
| /Bbu3 Q316–I428 | 10 | |
| Aaemrca Q291–I399/Bbu3 Q316–I428 | 15 |
The PBP codes are given in Table 1. PBPs Nme1 and Ngo1 have 98% identity.
Similarity is expressed as percent identity.
EVOLUTION OF MULTIMODULAR PBPS
The multimodular PBPs are an exemplary model of molecular evolution that gave rise to two classes of PBPs and to a prolific expansion of subclasses by fusion among several polypeptide chains (membrane anchor, n-PB core, PB core), acquisition of adducts, adaptive radiation, and speciation. Adaptive radiation is a term applied to the spread of species of common ancestry into different niches (70). It is extended in the present context to the spread of PBPs into the gram-negative bacteria, gram-positive bacteria, and other bacterial groups.
These structural changes occurred in combination with conservation of the core-specific amino acid sequence signatures. They resulted in the making of fully integrated polypeptide hybrids. These hybrids combine the main properties of the parental chains and are endowed with new properties because of noncovalent interactions between the constitutive modules.
Polypeptide folding and module swapping illustrate this notion. All the attempts made to produce the PB modules of E. coli PBP1b of subclass A2 (unpublished data), E. coli PBP3 of subclass B3 (29), Enterococcus hirae PBP5 of subclass B1 (51), and Staphylococcus aureus PBP2a of the same subclass B1 (71) as independent stable, penicillin-binding entities (using various expression-secretion vectors) failed. In contrast to the monofunctional PBPs, which are autonomous folding entities, the PB modules of multimodular PBPs have lost the ability to fold by themselves. They require the assistance of the associated n-PB modules, a property which implies precise and specific module-module interactions. While correct folding (in terms of penicillin binding) of the associated n-PB and PB modules is independent of the transporting signal peptide sequence, the in vivo activity of E. coli PBP3 is membrane anchor module dependent, indicating that the membrane anchor and its cytosolic tail have more sophisticated functions than that of a simple anchoring device (29, 31).
A plausible interpretation of the core-based clustering of the class A PBPs (Fig. 8) is that the gram-negative PBPs of subclasses A1 and A2 are paralogs (i.e., performing different functions); the gram-positive PBPs of subclasses A3, A4, and A5 are also paralogs; and the gram-negative PBPs of subclass A1 or A2 and the gram-positive PBPs of subclass A3, A4, or A5 may be orthologs (i.e., performing similar functions in these two groups of bacteria). Likewise, the clustering of the class B PBPs (Fig. 9) suggest that the gram-negative PBPs of subclasses B2 and B3 are paralogs; the gram-positive PBPs of subclasses B1, B4, and B5 are also paralogs; and the gram-negative PBPs of subclass B2 or B3 and the gram-positive PBPs of subclass B4 or B5 may be orthologs. As discussed below, the gram-positive PBPs of subclass B1 have no equivalents in the gram-negative bacteria.
The ensuing sections confront this scheme with facts. The associated n-PB and PB cores comprise about 500 to 550 amino acid residues, whereas the full-size PBPs are polypeptide chains 600 to 900 amino acid residues long. Because the inserts and extensions are not taken into consideration, an important proportion of the information may be lost in some cases. Moreover, the biochemistry audit of the multimodular PBPs is far short of reality. As stated above, attributing a function to a protein in the absence of direct biochemical data is a dangerous exercise.
FUNCTIONS OF MULTIMODULAR CLASS A PBPS
E. coli PBP1a of subclass A1 and E. coli PBP1b of subclass A2 are the only class A PBPs which have been identified biochemically as bifunctional transglycosylase (n-PB module)-transpeptidase (PB module) entities. However, the class A PBPs have two remarkable features. The n-PB cores have an extended amino acid sequence signature in the form of six conserved motifs (Fig. 6), and they underwent steady divergence without marked adaptive radiation (Fig. 8), suggesting conserved functionability. In contrast, the PB cores, while retaining the penicilloyl serine transferase signature (Fig. 6), have evolved in two gram-negative PBPs of subclasses A1 and A2 that form a supercluster and three gram-positive PBPs of subclasses A3, A4, and A5 that also form a supercluster (Fig. 8), suggesting differential functionability. On the basis of this conjecture, plausible suggestions can be made.
Dicarboxylic amino acid residues E and D of motif 1 and E of motif 3 present in all class A PBPs (Fig. 6) are almost certainly important components of the transglycosylase catalytic center of the n-PB modules. In this respect, E. coli PBP1c looks like a class A PBP mutant. The changes R→G and E→Q in motif 3 and R→A in motif 5 suggest that the n-PB module of this PBP may not be a transglycosylase.
Peptidoglycan cross-linking by the acyl serine transferase-PB module of the class A PBPs is more complex than expected. PBP1a of subclass A1 and PBP1b of subclass A2 substitute for each other in E. coli (37). Loss of either PBP1a or PBP1b is tolerated, but loss of the two class a PBPs is fatal. Under these extreme conditions, PBP1c cannot rescue E. coli from death.
PBP1 of subclass A3, PBP2c of subclass A4, and PBP4 of subclass A5 also substitute for each other in B. subtilis (59). Loss of PBP1 is associated with slight growth and cell morphology defects, these defects are exacerbated by the additional loss of PBP4 but not that of PBP2c, and loss of all three PBPs slows growth even further but is not fatal. Under these extreme conditions (and as discussed in the next section), B. subtilis PBP3 of subclass B1 might take over the functions of the PBPs of subclasses A3, A4, and A5.
Although performing the same transglycosylase-transpeptidase activities, PBP1a and PBP1b in E. coli and, likewise, PBP1, PBP2c, and PBP4 in B. subtilis are probably paralogs, prescribing distinct, subtle traits in peptidoglycan cross-linking in these two organisms that are difficult to detect.
FUNCTIONS OF MULTIMODULAR CLASS B PBPS
The class B PBPs evolved differently (Fig. 9). The n-PB cores and PB cores underwent adaptive radiation in such a concerted manner that in the gram-negative bacteria an n-PB core of subclass B2 or B3 is linked, almost invariably, to a PB core of subclass B2 or B3, respectively, and in the gram-positive bacteria an n-PB core of subclass B1, B4, or B5 is linked to a PB core of subclass B1, B4, or B5, respectively.
Class B PBPs of Gram-Negative Bacteria
E. coli PBP2 of subclass B2 and E. coli PBP3 of subclass B3 cannot substitute for each other. They are paralogs. E. coli PBP3 of subclass B3 and a set of non-PBPs work together to initiate septum formation (53, 67). In recent years, the catalogue of these non-PBPs has grown considerably. The genes encoding PBP3 and six other cell cycle proteins reside in the division and cell wall (dcw) cluster at the 2-min region of the chromosome (67). PBP3, FtsQ (10), and FtsL, a protein bearing a putative leucine zipper motif (30), are membrane bound, with the bulk of the polypeptide chains exposed on the outer face of the plasma membrane. FtsW is an integral membrane protein with loops exposed on both faces of the membrane (6). FtsA, an isolog of the DnaK-actin family of ATPases, is cytosolic when phosphorylated and is membrane associated when unphosphorylated (63). MraW, a protein bearing a putative S-adenosylmethionine-binding motif (9), and FtsZ, a GTPase similar to tubulin (47), are cytosolic. FtsZ is of known three-dimensional structure (46, 54). It functions as a cytoskeletal element mediating the invagination of the septum.
E. coli PBP2 of subclass B2, RodA (an integral membrane protein similar to FtsW), and the monofunctional dd-carboxypeptidase PBP5, whose genes are located at the 14-min region of the chromosome, are involved in wall expansion and shape maintenance (35, 61). PBP2, RodA, and ribosomal activities are coordinated by a chain of interacting elements, one of which is regulated by the nucleotide guanosine 5′-diphosphate 3′-diphosphate ppGpp.
E. coli PBP2 is not required for septum synthesis. Loss of PBP2 results in a block of cell division; however, in the absence of PBP2, cell division and viability are restored by increasing the pool of ppGpp or the level of FtsQAZ, showing that the cell septation and cell shape networks are interconnected (35). A fascinating story is now beginning to unfold involving the description of these networks in terms of interaction between the constitutive components. In spite of these advances, the questions of how these proteins work together and how the bacterial cell choreographs the interplay remain before us.
Class B PBPs of Gram-Positive Bacteria
The gram-positive bacteria possess two class B PBPs of subclasses B4 and B5, and some of them possess one additional PBP of subclass B1 (Fig. 9). The gram-positive PBPs of subclass B4 and the gram-negative PBPs of subclass B3 are almost certainly orthologs involved in cell division. Inactivation of Enterococcus hirae PBP3s of subclass B4 results in a block of septum formation (11), and a 10-kb segment of the E. hirae chromosome that contains the PBP3s-encoding gene also contains genes that code for proteins similar to MraW, FtsL, FtsQ, FtsW, FtsA, and FtsZ (18), a situation comparable to that of the E. coli dcw cluster. B. subtilis has a dcw cluster with a similar organization (7). However, this dcw cluster contains two PBP-encoding genes. PBP2b, which is involved in the metabolism of the vegetative cells, belongs to the gram-positive PBPs of subclass B4. PBPVD, which is involved in sporulation, belongs to the gram-negative PBPs of subclass B3.
The gram-positive PBPs of subclass B5 are of unknown function, but they are important. In Streptococcus pneumoniae, PBP2b of subclass B5 and PBP2x of subclass B4 cannot substitute for each other (40). They are paralogs. PBP2b and PBP2x are the first PBPs to be affected during selection of laboratory mutants with a reduced affinity for cefotaxime and piperacillin, respectively, and the modified PBP2b and PBP2x each confer resistance upon transformation (32).
The gram-positive PBPs of subclass B1 are endowed with unique properties. They represent an important mechanism of resistance to penicillin. The enterococcal and staphylococcal PBPs of subclass B1 have a very low affinity for the drug. They allow the strains that (over)produce them to grow in the presence of penicillin at concentrations sufficient to inactivate the PB modules of all the other multimodular PBPs of class A and class B (4, 19, 58, 60). To all appearances, the low-affinity PBPs of subclass B1 (including, perhaps, B. subtilis PBP3) can perform the basic functions required for wall peptidoglycan assembly in a cell-cycle-dependent fashion in conjunction, presumably, with the transglycosylase n-PB module of class A PBPs or with monofunctional transglycosylases (64).
Class B PBPs in Cell Morphogenesis
The question of how the class B PBPs function in vivo is left open. However, the unprecedented amino acid sequence signature of their n-PB modules (Fig. 7) and the three-dimensional structure of S. pneumoniae PBP2x of subclass B4 (57) are worthy of reflection.
The n-PB module of PBP2x is shaped like a pair of sugar tongs whose head fits in a noncatalytic groove of the PB module. Remarkably, motifs 1 to 4 of the n-PB module are located in the head of the “sugar tongs” in interaction with the PB module, and the 101-amino-acid residue polypeptide stretch that extends between motifs 1 and 2 is well exposed at the surface of the protein. Other class B PBPs probably adopt the same basic folded structure but with subclass- and species-specific variations. Key elements of the amino acid sequence-folding information for E. coli PBP3 of subclass B3 reside in the sequence G57 to W110, which contains motif 1, and in motif 3. Alterations of E193 of motif 3 results in the production of misfolded, unstable protein mutants that are rapidly degraded (29).
S. pneumoniae PBP2x and E. coli PBP3 each catalyze peptide bond formation from properly structured thiolester carbonyl donors and amino acceptors (1). Based on this observation, the acyl serine transferase-PB module of the class B PBPs probably prescribes species-specific traits related to peptidoglycan cross-linking, and this activity might be regulated by the associated n-PB module itself in interaction with other components of the morphogenetic networks. These speculations illustrate how little we know of the biochemistry of the class B PBPs.
AN AMAZING PANOPLY OF MULTIMODULAR PBPS
As shown in Table 1, B. subtilis possesses one PBP each in subclasses A3, A4, and A5 and one PBP each in subclasses B1, B3, B4, and B5 (one of them is involved in sporulation). S. pneumoniae possesses one PBP each in subclasses A3, A4, and A5 and one PBP each in subclasses B4 and B5. E. coli and Haemophilus influenzae possess one PBP each in subclasses A1 and A2 (not counting E. coli PBP1c) and one PBP each in subclasses B2 and B3. A. aeolicus possesses one PBP of subclass A1 and one PBP each in subclasses B2 and B3. Synechocystis strain PCC6803 possesses three PBPs of subclass A1 and one PBP of subclass B3. Helicobacter pylori possesses one PBP of subclass A2 and two mixed class B PBPs whose PB cores belong to subclasses B2 and B3, respectively.
The differentiation of multimodular PBPs into classes A and B is almost certainly an ancient evolutionary event. The question of which properties of the wall peptidoglycan assembly machinery determine the variable, species-specific assortment of multimodular PBPs is left open.
PENICILLIN-ORIENTED EVOLUTION
Diverging evolution gave rise to many different groups, classes, and subclasses of penicilloyl serine transferases. The use of β-lactam antibiotics functions to fuel the emergence of β-lactamases with widely varying spectra against β-lactam antibiotics. In recent years, more than 50 variants of the class A TEM β-lactamase (8) and many variants of the class A SHV β-lactamase have been identified in clinical isolates. They each arose by alteration of a limited number of amino acid residues in the corresponding wild-type enzymes. Another result of this directional evolution is the emergence and the spread of multimodular PBPs with a low or decreased affinity for the drug among important bacterial pathogens.
Enterococci, although related to streptococci, are 10- to 100-fold more resistant to penicillin because they produce one or two low-affinity PBPs of subclass B1. Enterococcus faecium is the most resistant species, and new populations of clinical isolates have appeared in different countries (44, 74). The gene encoding the low-affinity PBP5 is chromosomal in E. faecium and E. hirae R40. In contrast, E. hirae S185R (a derivative of a swine isolate) possesses a large plasmid that carries several copies of the gene encoding the low-affinity PBP3r (preceded by a psr-like negative regulatory gene), several IS1216 insertion sequences, and the determinants of resistance to streptomycin and erythromycin (60). These elements form a transposon-like structure, a situation eminently favorable for the horizontal spread of resistance to these antibiotics. Because pbp3r and psr3r are on a 1.3-kb DNA segment whose sequence is 99% homologous to that present in the E. faecium D63R chromosome, a DNA fragment might have been excised from E. faecium and inserted in a plasmid of E. hirae or in a plasmid of E. faecium which was then transferred to E. hirae.
In staphylococci, acquired resistance is also caused by the acquisition of a low-affinity PBP of subclass B1, known as PBP2a or PBP2′. The encoding mecA is chromosomal, and its environment is somewhat similar to that of the low-affinity PBP-encoding genes in streptococci (4). IS257, which is similar to IS1216, is probably involved in the integration of mecA in the chromosome of MRSA (methicillin-resistant S. aureus) strains, in the amplification phenomena observed in highly resistant staphylococcal mutants, in the evolution of the mec locus, and, perhaps, in the clustering of additional antibiotic resistance determinants. Strains close to the squirrel S. sciuri might be the sources of mecA in human staphylococcal pathogens (72).
In Streptococcus pneumoniae, N. meningitidis, and N. gonorrhoeae, acquired resistance is caused by the acquisition of altered forms of wild-type class A and class B PBPs. These PBPs of decreased affinity for the drug are the products of mosaic genes in which sensitive sequence blocks are replaced by homologous, resistant sequence blocks from related species by recombinational events (17). Of particular concern is this type of resistance in pneumococci, in which it is increasing worldwide. Mosaic structures have been described for the S. pneumoniae genes that encode PBP1a, PBP2a, and PBP1b of subclasses A3, A4, and A5, PBP2x of subclass B4, and PBP2b of subclass B5 (33). More than 20 variants of PBP2x have been identified in clinical isolates (32). Resistant blocks diverge from the sensitive ones by about 20% in nucleotides, and the mosaic PBPs diverge from the sensitive ones by about 15% in amino acid residues. The origin of the foreign DNA sequences is unknown, but two to four different sources may be involved (32).
TUNING UP THE CATALYTIC TWO-STROKE ENGINE
Diverging evolution, penicillin-oriented evolution, and site-directed mutagenesis (which allows protein mutants derived from extant genes to be produced through a mode unexplored, or explored but not retained, by natural selection) each show that the ability of the serine β-lactamases and the PB modules of the multimodular PBPs to adapt their catalytic properties to new situations is limitless. As a corollary, the ability of the proton abstraction-donation machinery of the acyl serine transferase catalytic center (Fig. 2A) to acquire new properties in response to structural changes in the enzyme polypeptide backbone is also limitless.
Michaelis complexes are formed by the noncovalent binding of carbonyl donors to the enzyme catalytic center. Dense hydrogen-bonding networks, comparable to three-dimensional cobwebs, connect the side chains and functional groupings of the ligand, the side chains of amino acid residues of the active-site-defining motifs, and several water molecules, one of which is hydrogen bonded to the γOH of the essential serine residue of motif Sx2K.
Formation of the Michaelis complex launches catalysis. At this level of the investigation (the angstrom level), the methods of quantum chemistry only highlight the underlying mechanism. As derived from theoretical studies first carried out on chymotrypsin (14–16), the simplest model of the engine that performs the required proton shuttle for catalysis is a six-membered ring in which a water molecule bridges the hydrogen atom of the serine γOH and the X atom of the scissile CO—X bond of the carbonyl donor (Fig. 2B).
Assuming that the engine which is created upon binding of the ligand to the catalytic center is in perfect tune, two backbone NH groups polarize the carbonyl of the CO—X bond, the proton of the serine γOH is transmitted to the X atom via the water molecule, and the serine residue is acylated (step 1). In turn, the serine-ester-linked acyl enzyme adopts the required conformation for the creation of a six-membered ring which, upon entry of the acceptor HY molecule, achieves enzyme deacylation (step 2). In the penicilloyl serine transferases, the backbone NH groups belong to the active-site serine itself and to the amino acid residue immediately downstream from motif [K/H][T/S]G on strand β3.
Admittedly, the efficacy with which the two-stroke engine overcomes the free energy barrier of the reactions via the transition states depends on many parameters. These parameters include the freedom of the catalytic water molecule itself, the ease with which the donor and the acceptor molecules can undergo deformation, and the ease with which the enzyme polypeptide backbone can undergo relaxation. Moreover, the six-membered ring model is an oversimplification. Additional water molecules, the imidazole moiety of the histidine residue of the catalytic triad of chymotrypsin, the ɛ amino group of the conserved lysine residue of motif Sx2K of the penicilloyl serine transferases, the γ carboxylate of the glutamic acid residue of motif Ex2LN of class A β-lactamases, and side chains of other amino acid residues at the boundary of the catalytic center shape and orient the two-stroke engine and/or are involved in proton transmission.
To understand the effects of these subtle changes, potential energy hypersurfaces (the dimensionality of which is 3 N − 6, where N is the number of atoms of the system) can be explored. They describe faithfully the geometric rearrangements and the electronic redistributions that the interacting partners, the enzyme catalytic center and the ligands, undergo along the reaction pathway (28). Minute structural changes in the enzyme and/or the ligand with which it is reacting may alter the entire hydrogen-bonding conformation of the Michaelis complex. As a corollary, the characteristics of the potential energy hypersurfaces and the reaction pathways are both ligand and enzyme specific.
One conclusion of these studies is that mutations that affect the polypeptide chain of a β-lactamase or a multimodular PBP may result in the creation of a two-stroke engine of increased catalyzed hydrolysis of a β-lactam antibiotic by the β-lactamase or of decreased affinity of the PBP for the drug (without alteration of its physiological function). In the context of penicillin-oriented evolution, these mutations are selected and the encoded β-lactamase and PBP mutants are maintained. There are no signs indicating that the resistant bacteria which produce these protein mutants suffer from decreased fitness and could not compete with the sensitive ones in a penicillin-free world. The absence of penicillin is not a selective pressure. Resistance is probably a road of no return (52, 62).
CONCLUSIONS AND FUTURE DIRECTIONS
The simplest conceivable event at the level of the gene can result in the emergence of penicillin (and other antibiotics) resistance determinants. The bacterial world behaves as an enormous organism whose cells exchange their genes with great ease, and the opportunities for the exchange of genetic material in nature are considerable. The antibiotics are societal drugs: a resistance gene which appears somewhere can spread far and fast. The use of current antibiotics in ways that would prevent the worldwide prevalence of resistant bacterial strains is a peremptory necessity.
Direct approaches to the drug resistance problem would be to design new antibiotics that bacteria have never seen before. One approach rests upon the knowledge of the structure and functioning of old targets at the most fundamental level. Multimodular PBPs which are assigned to different functional classes and subclasses and enzymes involved in the synthesis of lipid II are in front of the stage. Another approach rests upon the identification of new targets. The non-PB cell cycle proteins of the morphogenetic networks are good places to look. The ultimate goal, however, will be never reached, due to the evolutionary characteristics of the biological systems.
ACKNOWLEDGMENTS
This work was supported in part by the Belgian programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Services fédéraux des affaires scientifiques, techniques et culturelles (PAI no. P4/03) and the Fonds de la Recherche Fondamentale Collective (contract 2.4534.95). C.G. is Chercheur qualifié of the Fonds National de la Recherche Scientifique, Brussels, Belgium.
We thank Martine Nguyen-Distèche, Claudine Fraipont, Jacques Coyette, and Georges Dive, Centre d’Ingénierie des Protéines, Regine Hakenbeck (Kaiserslautern University, Germany), Jean van Heijenoort (CNRS, Orsay, France), and two anonymous referees for their authoritative comments and suggestions.
REFERENCES
- 1.Adam M, Fraipont C, Rhazi N, Nguyen-Distèche M, Lakaye B, Frère J M, Devreese B, Van Beeumen J, van Heijenoort Y, van Heijenoort J, Ghuysen J M. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyses peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from lipid II intermediate. J Bacteriol. 1997;179:6005–6009. doi: 10.1128/jb.179.19.6005-6009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton G J. Protein multiple sequence alignment and flexible pattern matching. Methods Enzymol. 1990;183:403–428. doi: 10.1016/0076-6879(90)83027-7. [DOI] [PubMed] [Google Scholar]
- 3.Basu J, Mahapatra S, Kundu M, Mukhopadhyay S, Nguyen-Distèche M, Dubois P, Joris B, Van Beeumen J, Cole S T, Chakrabarti P, Ghuysen J M. Identification and overexpression in Escherichia coli of a Mycobacterium leprae gene, pon1, encoding high-molecular-mass class A penicillin-binding protein PBP1. J Bacteriol. 1996;178:1707–1711. doi: 10.1128/jb.178.6.1707-1711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bächi B. Expression of resistance to methicillin. Trends Microbiol. 1994;2:389–393. doi: 10.1016/0966-842x(94)90617-3. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, et al. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan C E, Henriques A O, Piggot P S. Cell wall changes during bacterial endospore formation. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 167–183. [Google Scholar]
- 8.Bush K, Jacoby G. Nomenclature of TEM β-lactamases. J Antimicrob Chemother. 1997;39:1–3. doi: 10.1093/jac/39.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Carrión M, Gómez M J, Ayala J A. Molecular analysis of the gene mraW at the dcw cluster of Escherichia coli. Workshop on Structure, Function and Controls in Microbial Division. Ist Juan March Estud Investig. 1995;42:17–18. [Google Scholar]
- 10.Carson M J, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutants. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyette J, Somzé A, Briquet J J, Ghuysen J M, Fontana R. Function of penicillin-binding protein 3 in Streptococcus faecium. In: Hakenbeck R, Höltje J V, Labischinski H, editors. The target of penicillin. Berlin, Germany: Walter de Gruyter & Co.; 1983. pp. 523–530. [Google Scholar]
- 12.Deckert G, et al. The complete genome of the hyperthermophilic bacteria Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.DiBernardino M, Dijkstra A, Stüber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesizing enzymes. Overexpression and determination of the glycan-polymerizing activity. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 14.Dive G, Dehareng D, Ghosez L. Catalytic reaction pathways approached by quantum chemistry: a challenge. Cell Mol Life Sci. 1998;54:378–382. [Google Scholar]
- 15.Dive G, Dehareng D, Ghuysen J M. A detailed study of a molecule into a molecule: the N-acetyl-l-tryptophanamide in active site model of the α-chymotrypsin. J Am Chem Soc. 1994;116:2548–2556. [Google Scholar]
- 16.Dive G, Dehareng D, Peeters D. Proposition for the acylation mechanism of serine proteases: a one-step process? Int J Quantum Chem. 1996;58:85–107. [Google Scholar]
- 17.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin-binding protein-mediated resistance to β-lactam antibiotics. Trends Microbiol. 1994;2:361–366. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 18.Duez, C., I. Thamm, F. Sapunaric, J. Coyette, and J. M. Ghuysen. The division and cell wall gene cluster of Enterococcus hirae S185. DNA Sequence, in press. [DOI] [PubMed]
- 19.El Kharroubi A, Jacques P, Piras G, Coyette J, Van Beeumen J, Ghuysen J M. The Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant Staphylococcus aureus penicillin-binding protein 2′ are homologs. Biochem J. 1991;280:463–469. [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Frazer C M, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1998;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 22.Frère J M. β-Lactamases and bacterial resistance to antibiotics. Mol Microbiol. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 23.Ghuysen J M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 24.Ghuysen J M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 25.Ghuysen J M. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 26.Ghuysen J M. Penicillin-binding proteins. Wall peptidoglycan assembly and resistance to penicillin: facts, doubts and hopes. Int J Antimicrob Agents. 1997;8:45–60. doi: 10.1016/s0924-8579(96)00358-5. [DOI] [PubMed] [Google Scholar]
- 27.Ghuysen J M, Charlier P, Coyette J, Duez C, Fonzé E, Fraipont C, Goffin C, Joris B, Nguyen-Distèche M. Penicillin and beyond: evolution, protein fold, multimodular polypeptides, and multiprotein complexes. Microb Drug Resist. 1996;2:163–175. doi: 10.1089/mdr.1996.2.163. [DOI] [PubMed] [Google Scholar]
- 28.Ghuysen J M, Dive G. Biochemistry of the penicilloyl serine transferases. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 103–130. [Google Scholar]
- 29.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J M. The non-penicillin-binding module of the tripartite penicillin binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzman L M, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 31.Guzman L M, Weiss D S, Beckwith J. Domain swapping analysis of FtsI, FtsL, and FtsQ: bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakenbeck R. Target-mediated resistance to β-lactam antibiotics. Biochem Pharmacol. 1995;50:1121–1127. doi: 10.1016/0006-2952(95)00158-v. [DOI] [PubMed] [Google Scholar]
- 33.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höltje J V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joseleau-Petit D, Thévenet D, D’Ari R. ppGpp concentration, growth without PBP2 activity, and growth-rate control in Escherichia coli. Mol Microbiol. 1994;13:911–917. doi: 10.1111/j.1365-2958.1994.tb00482.x. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko T, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 37.Kato J I, Hideho S, Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- 38.Kelly J A, Kuzin A P, Charlier P, Fonzé E. X-ray studies of enzymes that interact with penicillins. Cell Mol Life Sci. 1991;54:353–358. doi: 10.1007/s000180050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krauss J, van der Linden M, Grebe T, Hakenbeck R. Penicillin-binding proteins 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb Drug Resist. 1996;2:183–186. doi: 10.1089/mdr.1996.2.183. [DOI] [PubMed] [Google Scholar]
- 41.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 42.Lefèvre F, Rémy M H, Masson J M. Topographical and functional investigation of Escherichia coli penicillin-binding protein 1b by alanine stretch scanning mutagenesis. J Bacteriol. 1997;179:4761–4767. doi: 10.1128/jb.179.15.4761-4767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lepage S, Dubois P, Ghosh T, Joris B, Mahapatra S, Kundu M, Basu J, Chakrabarti P, Cole S T, Nguyen-Distèche M, Ghuysen J M. Dual multimodular class A penicillin-binding proteins in Mycobacterium leprae. J Bacteriol. 1997;179:4627–4630. doi: 10.1128/jb.179.14.4627-4630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligozzi M, Pittaluga F, Fontana R. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1996;40:354–357. doi: 10.1128/aac.40.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livingstone G D, Barton G J. Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol. 1996;266:497–512. doi: 10.1016/s0076-6879(96)66031-5. [DOI] [PubMed] [Google Scholar]
- 46.Löwe J, Amos L A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 47.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 48.Matagne A, Lamotte-Brasseur J, Frère J M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuhashi M. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanisms of their regulation. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 55–72. [Google Scholar]
- 51.Mollerach M, Partoune P, Coyette J, Ghuysen J M. Importance of the E46-D160 polypeptide segment of the non-penicillin-binding module for the stability of the low-affinity, multimodular class B penicillin-binding protein 5 of Enterococcus hirae. J Bacteriol. 1996;178:1774–1775. doi: 10.1128/jb.178.6.1774-1775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morell V. Antibiotic resistance: road of no return. Science. 1997;278:575–576. doi: 10.1126/science.278.5338.575b. [DOI] [PubMed] [Google Scholar]
- 53.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nogales E, Downing K H, Amos L A, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 55.Paik J, Jendrossek D, Hakenbeck R. A putative monofunctional glycosyltransferase is expressed in Ralstonia eutropha. J Bacteriol. 1997;179:4061–4065. doi: 10.1128/jb.179.12.4061-4065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paradkar A S, Aidoo K A, Wong A, Jensen S E. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pares S, Mouz N, Pétillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 58.Piras G, Raze D, El Kharroubi A, Hastir D, Englebert S, Coyette J, Ghuysen J M. Cloning and sequencing of the low affinity penicillin-binding protein 3r-encoding gene of Enterococcus hirae S185. Modular design and structural organization of the protein. J Bacteriol. 1993;175:2844–2852. doi: 10.1128/jb.175.10.2844-2852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raze D, Dardenne O, Hallut S, Martinez-Bueno M, Coyette J, Ghuysen J M. The low-affinity penicillin-binding protein 3R-encoding gene of Enterococcus hirae S185R is borne on a plasmid carrying other antibiotic resistance determinants. Antimicrob Agents Chemother. 1998;42:534–539. doi: 10.1128/aac.42.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddy P S, Raghavan A, Chatterjee D. Evidence of a ppGpp-binding site in Escherichia coli RNA polymerase: proximity relationship with the rifampicin-binding domain. Mol Microbiol. 1995;15:255–265. doi: 10.1111/j.1365-2958.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- 62.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez M, Valencia A, Ferrandiz M J, Sander C, Vicente M. Correlations between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 1994;13:4919–4925. doi: 10.1002/j.1460-2075.1994.tb06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spratt B G, Zhou J, Taylor M, Merrick M J. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;19:639–640. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 65.Tomb J F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 66.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 67.Vicente M, Errington J. Structure, function and controls in microbial division. Mol Microbiol. 1996;20:1–7. doi: 10.1111/j.1365-2958.1996.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 68.Ward J B, Perkins H R. The direction of glycan synthesis in a bacterial peptidoglycan. Biochem J. 1973;135:721–728. doi: 10.1042/bj1350721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weston A, Ward J B, Perkins H R. Biosynthesis of peptidoglycan in wall plus membrane preparations from Micrococcus luteus: direction of chain extension, length of chains and effect of penicillin on cross-linking. J Gen Microbiol. 1977;99:171–181. [Google Scholar]
- 70.Wilson E O. The diversity of life. W. W. New York, N.Y: Norton & Co.; 1993. [Google Scholar]
- 71.Wu E C Y, Alborn Jr W E, Flokowitsch J E, Hoskins J, Unal S, Blaszczak L C, Preston D A, Skatrud P L. Site-directed mutagenesis of the mecA gene from methicillin-resistant strains of Staphylococcus aureus. J Bacteriol. 1994;176:443–449. doi: 10.1128/jb.176.2.443-449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the origin of an antibiotic-resistance gene: methicillin-susceptible strains of the animal species Staphylococcus sciuri carry as a native gene a homologue of the Staphylococcus aureus methicillin resistance determinant mecA. Microb Drug Resist. 1996;2:435–441. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 73.Young K D, Denome S A, Elf P K, Henderson T A. The Bacterial Cell Cycle Workshop in Chorin, Germany 13 to 17 September 1997. 1997. Use of a comprehensive set of PBP mutants to investigate peptidoglycan synthesis in Escherichia coli; p. 41. [Google Scholar]
- 74.Zorzi W, Zhou X Y, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol. 1996;178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]