Abstract
Bradysia odoriphaga (Diptera: Sciaridae) is a major insect pest of seven plant families including 30 commercial crops in Asia. The long-term use of chemical pesticides leads to problems such as insect resistance, environmental issues, and food contamination. Against this background, a novel pest control method should be developed. In insects, odorant-binding proteins (OBPs) transport odor molecules, including pheromones and plant volatiles, to olfactory receptors. Here, we expressed and characterized the recombinant B. odoriphaga OBP BodoOBP10, observing that it could bind the sulfur-containing compounds diallyl disulfide and methyl allyl disulfide with Ki values of 8.01 μM and 7.00 μM, respectively. Homology modeling showed that the BodoOBP10 3D structure was similar to that of a typical OBP. Both diallyl disulfide and methyl allyl disulfide bound to the same site on BodoOBP10, mediated by interactions with six hydrophobic residues Met70, Ile75, Thr89, Met90, Leu93, and Leu94, and one aromatic residue, Phe143. Furthermore, silencing BodoOBP10 expression via RNAi significantly reduced the electroantennogram (EAG) response to diallyl disulfide and methyl allyl disulfide. These findings suggest that BodoOBP10 should be involved in the recognition and localization of host plants.
Keywords: Bradysia odoriphaga, odorant-binding protein 10, ligand binding assays, RNAi, electroantennogram
The gnat Bradysia odoriphaga (Diptera: Sciaridae) is an agricultural pest that is widely distributed in China and other Asian countries (Yang et al. 2015). It affects over seven plant families, including 30 commercial crops such as Chinese chives, onion, ginger, and other Liliaceae crops (Li et al. 2015). The application of chemical insecticides is currently the main means of control (Yang et al. 2019). However, long-term use of chemicals can result in insect resistance, serious environmental pollution, and the contamination of food, adversely affecting both ecosystem balance and human health (Wang et al. 2011). Thus, novel and alternative management methods are needed for the control of B. odoriphaga.
Pheromones and plant volatiles play central roles in insect behavior, especially the regulation of insect–host plant recognition, mate identification, and oviposition (Pelosi et al. 2006, Field et al. 2000, Hansson 2011, Leal 2013, Pelosi et al. 2014). For example, the volatiles of geraniol can lure Maruca vitrata (Feng et al. 2017) and Plutella xylostella (Mu 2010) and stimulate female oviposition. Volatile sulfur compounds are the main chemical signals used by Delia antiqua in seeking host plants and oviposition sites (Harris 1988), and (Z)-3-dodecen-1-yl (E)-2-butenoate, secreted by female Cylas formicarius, attracts males (Hua et al. 2021). We have previously observed that volatile compounds from Chinese chives modulated the behavior of female B. odoriphaga, stimulating oviposition (Yang et al. 2019). However, the mechanism underlying the relationship between B. odoriphaga and host plant volatiles has not yet been determined.
Insects use their antennae to monitor chemical signals such as environmental odors and pheromones (Sato 2008, Hansson 2011). Olfaction plays a critical role in a variety of insect activities, including the selection of habitats, mates, and oviposition sites and the evasion of predators (Field et al. 2000, Gadenne et al. 2016). The olfactory system contains odorant-binding proteins (OBPs), small soluble proteins located in the sensillum lymph of the antennae (Field et al. 2000, Wang et al. 2019). The OBPs bind and transport (or termination) odor molecules to the olfactory receptors (ORs) through the sensillum lymph, thereby participating in regulating insect behavior (Leal 2013, Fan et al. 2011, Schymura et al. 2010, Jacquin-Joly 2014, Sims et al. 2022). Therefore, the interaction between OBPs and external odor molecules represents the initial step in insects’ recognition of chemical signals in the environment (Vogt et al. 1991, Hua et al. 2021).
Previous studies have shown that OBPs participate in different physiological functions as they are expressed in various tissues/organs and interact with specific chemicals (Brito et al. 2016, Pelosi et al. 2018, Rihani et al. 2021). For example, OBP3, OBP6, and OBP10 of Chrysopa pallens are expressed in the antennae and bind to 2-hexyl-1-decanol and (E)-beta-farnesene (Li et al. 2017). In Apolygus lucorum OBP22 is located mostly in the antennae where it assists in the recognition of terpenoids (Liu et al. 2019), whereas OBP14 of Adelphocoris lineolatus is mainly expressed in the heads of adults where it binds to nerolidol, β-ionone, and trans-farnesol (Sun et al. 2019). In addition, OBP10 of Apis cerana cerana is expressed in the venom gland, and knockdown of the gene increased the expression of several stress-related genes (Guo et al. 2020) while OBP49a of Drosophila melanogaster is found largely in the taste organs where it detects bitter compounds in food sources (Jeong et al. 2013). These findings suggest that OBPs may be useful targets for novel pest control strategies. For example, knockdown of C. formicarius OBP1-3 significantly altered insect odor attraction(Hua et al. 2021) and, as OBP10 of Helicoverpa armigera can bind to 1-dodecene, the compound has been used as a pest repellent (Sun et al. 2012). In B. odoriphaga, 49 OBP genes were identified by Zhao et al (Zhao et al. 2018), however, to date, relatively few OBP functions have been elucidated in B. odoriphaga although it has been observed that BodoOBP1 and BodoOBP2 (Tang et al. 2019), BodoOBP5 (Yang et al. 2021a), and BodoOBP8 (Yang et al. 2021b) can bind host plant volatiles, implying that OBPs in this species are involved in host plant selection and recognition. Our previous study showed that BodoOBP10 was highly expressed in the antennae both male and female, and this result was consisted with Zhao et al (2018). Antennae as an important olfactory organ of insects, the abundant expression of BodoOBP10 in antennae suggests that it is involved in the olfactory function of insects.
To elucidate the olfactory function of OBP10, we conducted the following investigations: 1) analysis of the BodoOBP10 sequence; 2) investigation of BodoOBP10 expression in different tissues and developmental stages; 3) assessment of the binding affinity of the recombinant BodoOBP10 protein; 4) prediction of the recombinant BodoOBP10 binding sites for ligands associated with host plant orientation and seeking behavior; 5) silencing of BodoOBP10 and measurement of the electroantennogram (EAG) responses of B. odoriphaga to two sulfur compounds.
Materials and Methods
Insects
All larvae of B. odoriphaga were reared on Chinese chives under controlled laboratory conditions, specifically, temperature of 26 ± 1°C, 70% RH, and a photoperiod of 14:10 (L:D) h. After pupation, the emergent adults were collected in insect cages. To obtain virgins and mated females, the pupae were reared singly and paired after emergence.
Chemicals
Thirty-two volatile compounds from Chinese chives (Yang et al. 2019) and n-heptadecane from the abdomens of B. odoriphaga adult females (Li et al. 2008) were chosen as the test ligands; these are shown in Supp Table S2 (online only). The purity of the ligands exceeded 95%. N-phenyl-1-naphthylamine (1-NPN) was chosen as the fluorescent competitive assay reporter.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from different developmental stages, including eggs (n = 30), larvae (n = 10), and pupae (n = 10), different sexes, namely, female (n = 10) and male (n = 10), and different tissues, including heads (n = 30), female antennae (n = 500), male antennae (n = 500), abdomens (n = 10), and a mixture of thoraces, legs, and wings (n = 10) using TRIzol (Invitrogen), according to the provided instructions. The RNA was reverse-transcribed to cDNA using the PrimeScript II 1st Strand cDNA Synthesis Kit (TIANGEN, Beijing, China).
Cloning, Sequencing, and Phylogeny of BodoOBP10
The primers used for OBP10 RT-PCR are shown in Supp Table S1 (online only). As described in a previous method (Yang et al. 2021a), the purified PCR product was linked to the cloning vector pEASY-Blunt 3 (TransGen Biotech, China), transformed into Trans T1 cells, and sequenced. The ORFs were evaluated by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), the molecular weight by the ExPASy tool ‘ProtParam’ (http://smart.emblheidelberg.de/), and the SignalP version 5.1 server(http://www.cbs.dtu.dk/services/SignalP/) was used for signal peptide prediction. Theoretical pI values were also determined, and phylogenetic trees were constructed using MEGA 6.0 software (Tamura et al. 2013).
RT-qPCR
The RT-qPCR primers are shown in Supp Table S1 (online only). The RPL18 and RPS15 genes, as well as EF1 and ACT, were used as references for developmental stages and tissues, respectively (Yang et al. 2021a). The qRT-PCR conditions were 94°C for 30 s, followed by 40 cycles of 94°C for 5 s, 55°C for 15 s, and 72°C for 10 s. Relative expression was analyzed using the threshold cycle number (CT) and the 2−△△Ct method and the mRNA expression values were normalized to the reference genes (Pfaffl 2001). Three biological replicates were used for all assays. One-way analysis of variance (ANOVA) was used for analyzing differences in expression patterns using the LSD method. SPSS 20.0 (IBM Corp., Armonk, NY) was used for statistical analysis and the significance level was P < 0.05.
Expression and Purification of Recombinant BodoOBP10
As described in our previous study (Yang et al. 2021a), the PCR products and expression vector pBM30 were linked and expressed in E. coli BL21 (DE3) cells. Positive BodoOBP10/PBM30 clones were selected using kanamycin. When the OD value of the culture approached 0.6–0.8, 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) (0.5 mM) was added for induction, and the culture was shaken for 12 h at 28°C. The bacteria were collected by centrifugation, after sonication, the target protein BodoOBP10 was mainly expressed in the inclusion bodies. And then 50 mM Tris buffer (pH 6.8) containing 0.2% Triton X-100 and 6 M guanidine hydrochloride were used to deal with the insoluble inclusion body, respectively. Ni ion affinity chromatography (GE Healthcare) (GE-Healthcare, USA) was used for protein purification. The His tag on the protein was detached with recombinant enterokinase (Novagen, Beijing, China). The quality of the samples was assessed using SDS-PAGE and protein concentrations were measured using a BCA Protein Assay Kit (Beyotime, Shanghai, China).
Fluorescence Binding
Competitive fluorescence binding was used to analyze binding between various ligands and BodoOBP10. The recombinant BodoOBP10 was dissolved in Tris–HCl (20 mM, pH 7.4) and diluted into 2 µM stock solution. The ligands and 1-NPN were dissolved in methanol and diluted into 1mM stock solutions for next experiments. The excitation spectrum of the 1-NPN/BodoOBP10 mixture was 337 nm, with an emission spectrum between 350 and 500 nm, using a 10-nm slit width. The methods of competitive fluorescence binding were according to our previously research(Yang et al. 2021a) without modification. The competitive dissociation constants were determined from the IC50 values (the initial fluorescence intensity decreased by 50% with ligand concentration) using the formula: Ki = [IC50]/(1 + [1−NPN]/K1−NPN); where [1−NPN] and K1−NPN represent the concentration of free 1−NPN and the dissociation constant of the protein/1−NPN complex, respectively (Campanacci et al. 2003, Wei et al. 2008).
Modeling of 3D Structure of BodoOBP10
The SwissModel online platform (SWISS-MODEL (expasy.org))was used for modeling the structure of BodoOBP10. Anopheles gambiae OBP20 (AgamOBP20; 4F7F) was used as the template; the sequence identity was 43.7% between AgamOBP20 and BodoOBP10. Ligand binding sites were predicted using SYBIL 7.3. Ligand molecular conformations were predicted in Sketch mode and optimized using Tripos force fields and Gasteiger-Hückel charges. Molecular docking was performed using Surflex-Dock in SYBYL 7.3. Modeling of the BodoOBP protein and ligands was done on a Silicon Graphics (SGI) Fuel Workstation (Silicon Graphics International Corp., CA).
RNAi Experiments and EAG Assay With Volatiles
The primers for dsBodoOBP10 and dsGFP containing a T7 promoter sequence are listed in Supp Table S1 (online only). All dsRNAs were synthesized using the T7 Ribomax TM Express RNAi System (Promega, Madison, WI). RNA quality and concentrations were assessed by agarose electrophoresis and spectrophotometry (NanoDrop, Thermo Fisher, Waltham, MA), respectively.
The RNAi method was used for the injection of dsRNA into the pupae of B. odoriphaga as previously described (Yang et al. 2021a, Zhang et al. 2021) with slight modifications. Briefly, 30 nL of 6 μg/μL dsRNA (either dsOBP10 or dsGFP) were injected into one-day-old pupal abdomens using a Nanoject (Nanoliter 2000). RNA was extracted from the adults that emerged 12 h later, and the effectiveness of the RNAi was assessed by RT-qPCR. Each treatment was repeated three times.
The antenna response to the diallyl disulfide and methyl allyl disulfide sulfur compounds (at 10 μg/μL) was assessed using electroantennogram (EAG) recordings, as described previously (Yang et al. 2019). Briefly, 10-μl volumes of diallyl disulfide or methyl allyl disulfide solutions were applied to filter paper strips. After 10 s, the strips were placed in glass Pasteur pipettes. The control was a filter paper strip with 10 μl of hexane; the control was tested before and after the experimental solutions. The antennae of the adult females that had emerged at 12 h treated with dsOBP10 or dsGFP were rapidly detached. The antennal root was connected to the reference glass electrode, with the tip connected to the recording electrode. Ten replicates were used for each treatment. The results were analyzed using one-way ANOVA with Tukey’s test (P < 0.05), and all results were represented by means ± SE.
Results
BodoOBP10 Identification and Phylogeny
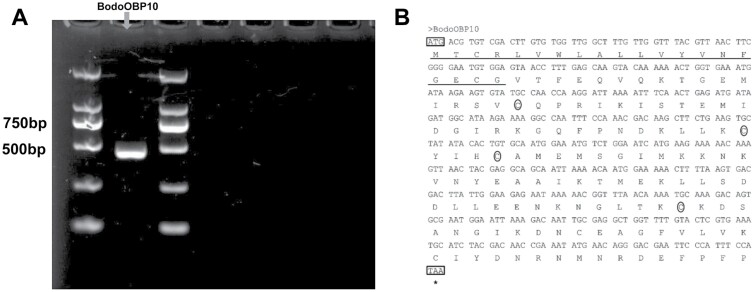
The full-length cDNA encoding BodoOBP10 (GenBank: AWC08421.1) was verified by RT-PCR (Fig. 1A). The Open Reading Frame (ORF) consisted of 435 bp encoding 144 residues, with a 19-residue at the N-terminus (Fig. 1B; Table 1). This result is consistent with the size of BodoOBP10 in Fig. 1A. The BodoOBP10 belongs to the minus-C OBP family that contains four conserved cysteines (Fig. 1B), with a predicted molecular weight (MW) of 16.48 kDa and a pI of 8.08 (Table 1). In the phylogenetic analysis, BodoOBP10 clustered with AgamOBP20, AaegOBP55, AgamOBP6, and AaegOBP27 on the same branch, indicating close relationships (Supp Fig. S1 [online only]).
Fig. 1.
(A) Amplification of full open reading frames (ORF) of BodoOBP10. The gene of BodoOBP10 is indicated in red, the molecular marker is marked on both sides of BodoOBP10. (B) Sequences alignment of BodoOBP10 of Nucleotide and deduced amino acid. The initiation and termination condons are marked in black boxes; the predicted signal peptides at the N-terminus are underlined in red; the four conserved cysteines are signed by red circle.
Table 1.
List of OBP10 genes in B. odoriphaga
| Gene | Acc. no | Length of ORF | Amino acid length | Signal peptide | Full ORF | Isoelectric point pI | Mw (kDa) |
|---|---|---|---|---|---|---|---|
| OBP10 | AWC08421.1 | 435 | 144 | 1–19 | Yes | 8.08 | 16.48 |
Spatial Profiling of BodoOBP10 Protein
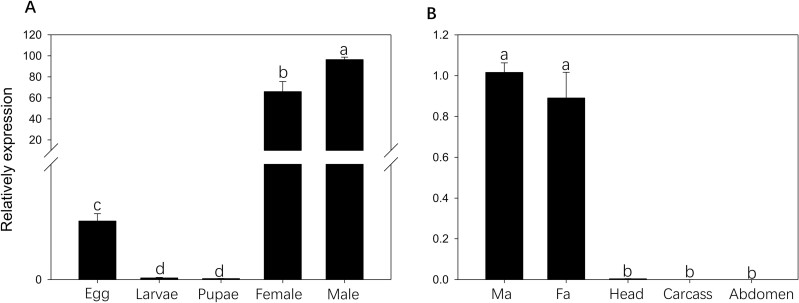
BodoOBP10 expression in various developmental stages and tissues was examined by qRT-PCR. Expression was highest at the adult stage, especially in adult males, followed by adult females (Fig. 2A). In terms of tissue expression, BodoOBP was found to be expressed specifically in antennae of both sexes, with the highest levels in male antennae. These findings were consistent with those of Zhao et al. (2018), suggesting that BodoOBP10 is related to antenna functions in B. odoriphaga (Fig. 2B).
Fig. 2.
Gene expression profiling of BodoOBP10. (A) RT-qPCR analysis in different developmental stages: Eggs; Larvae; Pupae; Female (newly emerged and virgin); Male (newly emerged and virgin). (B) RT-qPCR analysis in different tissues: FA: Female antenna; MA: Male antenna; Head; Abdomen; Carcass: leg + wing + thorax. The expression levels were estimated using the 2−ΔΔCt method. The expression level in eggs was used as a standard to compare expression levels among developmental stages, and the expression level in male antennae was used as a standard to compare expression levels among tissues. One-way analysis of variance (ANOVA) was used for analyzing differences in expression patterns using the LSD method. Data are normally distributed. SPSS 20.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis and the values are means + SE; means with different letters are significantly different (P < 0.05).
BodoOBP10 Expression and Purification
BodoOBP10 was successfully expressed in E. coli. SDS-PAGE (Supp Fig. S2 [online only]). The SDS-PAGE analysis showed that the recombinant protein BodoOBP10 had a size slightly greater than 15 kDa, which is consistent with the predicted molecular weight of 16.48 kDa (Table 1).
Fluorescence binding Assay of BodoOBP10
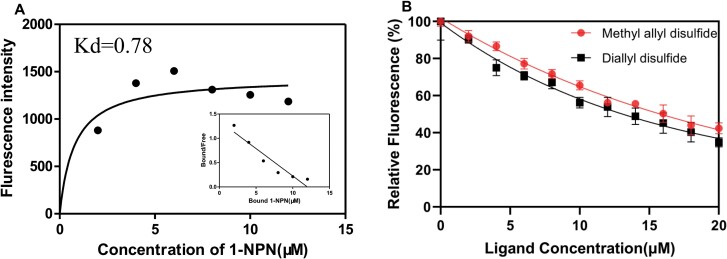
The fluorescence reporter, 1-NPN, was used to assess binding between BodoOBP10 and the different ligands. BodoOBP10 showed high-affinity binding to 1-NPN, with a dissociation constant (Ki) of 0.78 μM (Fig. 3A). The competitive fluorescence binding graphs indicated that the [BodoOBP10/1-NPN] mixture bound the two sulfur compounds diallyl disulfide and methyl allyl disulfide, with Ki values of 8.01 and 7.00 μM, respectively (Fig. 3B; Supp Table S3 [online only]). However, the [BodoOBP10/1-NPN] mixture only bound weakly to other compounds (Supp Table S3 [online only]). These results indicate that BodoOBP10 is a functional protein allowing B. odoriphaga to recognize sulfur-containing volatiles, with strong affinity for diallyl disulfide and methyl allyl disulfide.
Fig. 3.
Competitive binding curves of BodoOBP10 with ligands. (A) Binding curves for 1-NPN and Scatchard plots; (B) Competitive binding curves of two sulfur compounds with BodoOBP10. (Round) Methyl allyl disulfide; (Square) Diallyl disulfide. Each treatment was repeated three times and the values represents the means.
3D Modeling and Molecular Docking of BodoOBP10
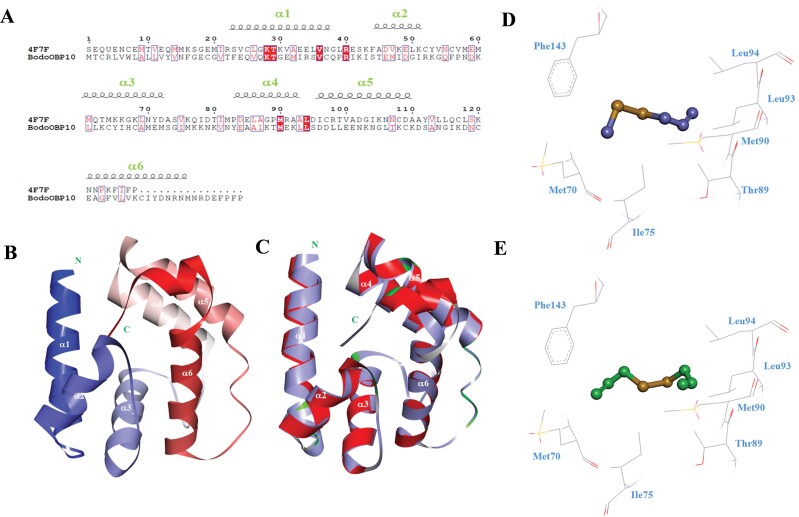
As shown in Fig. 4A, the crystal structure of AgamOBP20 (4F7F) was used as the modeling template; the sequence identity between AgamOBP20 and BodoOBP10 was 43.7%. The modeled structure of BodoOBP10 was mostly helical and showed that the six α-helices of BodoOBP10 were located between Thr22-Gln38 (α1), Ser44-Ile51 (α2), Leu61-Met72 (α3), Tyr83-Leu93 (α4), Asp96-Lys110 (α5), and Glu121-Met127 (α6) (Fig. 4A). The structure also included four conserved cysteine residues, forming two disulfide bonds between Cys37-Cys68 and Cys109-Cys129, to further stabilize the whole structure (Fig. 4B). The structures of BodoOBP10 and the template AgamOBP20 aligned well, and the values of GMQE was 0.7 (Fig. 4C).
Fig. 4.
3D structural modeling and molecular docking of BodoOBP10. (A) The sequence alignment between the template protein AgamOBP20 and the target protein BodoOBP10; the black coil represents the α-helix; (B) The 3D model of the target protein BodoOBP10 based on the crystal structure of the template protein of AgamOBP20; the position of α-helix is marked by white; (C) The alignment plot of the target protein BodoOBP10 (red) and the template protein AgamOBP20 (purple, ID: 4F7F). Predicted binding mode and key residues between the target protein BodoOBP10 and methyl allyl disulfide (purple) (D), diallyl disulfide (green) (E). Residues are marked in blue.
Molecular docking was then used to analyze the key residues involved in BodoOBP-ligand binding (Fig. 4). As shown by fluorescence binding, BodoOBP10 bound with high affinity (Ki < 10.00 μM) to both diallyl disulfide and methyl allyl disulfide. Thus, two disulfide compounds were selected as the ligands and used for molecular docking with BodoOBP10 protein. The results showed both compounds bound to almost the same site on BodoOBP10 due to their similarities in 3D structure and binding conformation; this was especially evident with the disulfide bonds (Fig. 4). In addition, the result of molecular docking also indicated that the existence of van der Waals interaction between ligand and protein (Fig. 4). Both ligands were surrounded by some same hydrophobic residues, namely, Met70, Ile75, Thr89, Met90, Leu93, and Leu94, and one aromatic residue, Phe143 (Fig. 4D and E).
Functional Analysis of BodoOBP10
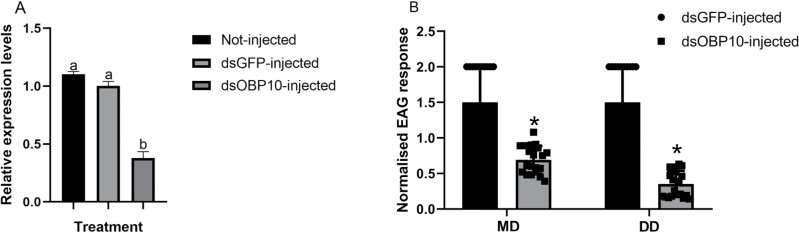
The function of BodoOBP10 was investigated using RNAi.After injection with dsOBP10, dsGFP, or no injection, the BodoOBP10 expression level was significant reduced compared with the control (no injection or injected with dsGFP) (P < 0.05) (Fig. 5A). In the EAG assays, compared with the control (dsGFP-injected), the EAG responses in dsOBP10-injected insects were significantly reduced in response to methyl allyl disulfide (t = 9.044, df = 18, P < 0.001) and diallyl disulfide (t = 11.374, df = 18, P < 0.001) (Fig. 5B).
Fig. 5.
RNAi and electroantennograms (EAG) assays of BodoOBP10. (A) Effect of RNAi treatment on the transcript levels of BodoOBP10. (B) EAG responses of B. odoriphaga after RNAi. MD: Methyl allyl disulfide; DD: Diallyl disulfide. One-way analysis of variance (ANOVA) was used for analyzing differences in expression patterns using the LSD method. Data are normally distributed. SPSS 20.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis and the values are means + SE; means with different letters are significantly different (P < 0.05).
Discussion
OBPs have an important function in the olfactory systems of insects. They transport chemical cues such as pheromones and volatile compounds released from host plants to the ORs on olfactory neurons, ultimately giving rise to behavioral responses (Brito et al. 2016, Leal 2013, Yao et al. 2021, Zhang et al. 2022, Sims et al. 2022, Rihani et al. 2021). Therefore, insect OBPs may be effective molecular targets for pest control.
In order to understand the evolutionary relationship between OBP10 and other diptera insects, we constructed a phylogenetic tree using dipteran OBPs sequences. The result showed that BodoOBP10 was located in a single clade of the phylogenetic tree, and clustered with AgamOBP20, AaegOBP55, AgamOBP6, and AaegOBP27 on the same branch (Supp Fig. S1 [online only]), indicating that these OBPs have close relationships and may have came from common ancestral protein.
Here, we analyzed the expression of BodoOBP10 showing that the gene was expressed largely in the adult insect stages. Expression was high in the antennae of female and male adults, with higher levels in males (Fig. 2A and B). These findings are consistent with those of a previous transcriptomic study of B. odoriphaga (Zhao et al. 2018). Insect OBPs have been found in many organs, suggesting that they are involved in specific physiological functions (Brito et al. 2016, Pelosi et al. 2018, Li et al. 2022, Sims et al. 2022, Rihani et al. 2021). The abundant expression of OBPs genes in insect antennae suggests that it is mainly involved in the olfactory function of insects(Yang et al. 2016, Pelosi et al. 2018). Other studies have also shown high expression of OBPs in male antennae, suggesting involvement in courtship (Liu et al. 2022, Chen et al. 2018a, b). Hence, we hypothesize that OBP10 is likely involved in courtship as well as olfactory function of B. odoriphaga of B. odoriphaga.
Competitive fluorescence assays are frequently used for studying binding between olfaction-associated proteins and odorant compounds, such as pheromones and plant volatiles, and have been used in a variety of insect species, including Holotrichia oblita (Yin et al. 2019), C. pallens (Li et al. 2017), Sogatella furcifera (He and He 2014, Chen et al. 2018b), Diaphorina citri (Zhang et al. 2020, Wang et al. 2020, Liu et al. 2021), and Athetis lepigone (Yao et al. 2021). Here, we choose the extract of abdomen of B. odoriphaga n-heptadecane and 32 host plant volatiles to investigate binding with BodoOBP10. BodoOBP10 was found to bind two sulfur-containing plant volatiles with high affinity, namely, diallyl disulfide (Ki = 8.01 μM) and methyl allyl disulfide (Ki = 7.00 μM), with minimal binding to n-heptadecane (Ki > 50μM). n-Heptadecane was extracted from the abdomens of adult females and can lead to behavioral responses in male B. odoriphaga (Li et al. 2008). These results suggest that n-heptadecane was mainly recognized by other OBPs but not by OBP10. Additionally, the two sulfur-containing compounds diallyl disulfide and methyl allyl disulfide not only could stimulate the EAG responses of B. odoriphaga adults but could also stimulate female oviposition (Yang et al. 2019). Thus, these results suggest that BodoOBP10 is involved in the detection of host plant volatiles and mating sites.
To further identify the residues involved in the binding of diallyl disulfide and methyl allyl disulfide to BodoOBP10, we used 3D modeling and molecular docking. Based on the secondary structure and 3D model, BodoOBP10 contains four conserved cysteines that formed two disulfide bonds (Cys37-Cys68 and Cys109-Cys129) indicating that it may be a minus-C OBP type. These result are consistent with those previously reported, including Bactrocera minax OBP9(Yao et al. 2021), Harmonia axyridis OBP1 and OBP3 (Qu et al, 2021). In addition, the results of molecular docking showed that six hydrophobic residues, Met70, Ile75, Thr89, Met90, Leu93, and Leu94, formed a hydrophobic binding pocket, contributing to binding. Similar hydrophobic pockets involved in binding have been reported for insect OBPs; e.g., Arg107 in D. citri OBP7 (Liu et al. 2021), Tyr77, Ile41, Ala116, Ala113, Lys38, Gln43, and Ile114 in Aphid Sitobion OBP9 (Ullah et al. 2020), Trp50, Leu67, and Phe113 in Nilaparvata lugens OBP8 (He et al. 2019), Lys123 in Helicoverpa armigera OBP7 (Dong et al. 2017), Leu99, Leu103, Ala143, Tyr107, Phe142, and Trp144 in B. odoriphaga OBP5 (Yang et al. 2021a), and Ile96, Ile103, Ala107, and Leu111 in B. odoriphaga OBP8 (Yang et al. 2021b). This suggests that the residues Met70, Ile75, Thr89, Met90, Leu93, Leu94, and Phe143 are good candidates of binding sites for recognizing and binding of diallyl disulfide and methyl allyl disulfide by BodoOBP10. However, the functions of these residues need further experiments to confirm.
RNAi was used to verify the role of BodoOBP10 in B. odoriphaga chemoreception. Compared with the control (dsGFP-injected), the EAG response of dsOBP10-injected insects to both methyl allyl disulfide and diallyl disulfide was significantly reduced. This finding is consistent with previous reports where, for example, knockdown of OBP2 in Aphis gossypii significantly lowered EAG responses to host plant volatiles (Rebijith et al. 2016). In A. lineolatus, silencing of OBP4also reduced EAG responses to six semiochemicals (Zhang et al. 2017) while silencing of OBP7 in D. citri lowered the EAG activities of adults towards four volatile compounds (Liu et al. 2021). Interestingly, our previous investigation of EAG and behavioral responses showed that the antennae of adult B. odoriphaga also exhibited a significant preference for the sulfur-containing compounds methyl allyl disulfide and diallyl disulfide (Yang et al. 2019). Therefore, we believe that diallyl disulfide and methyl allyl disulfide can be recognized by BodoOBP10, and BodoOBP10 is involved in the host plant recognition of B. odoriphaga.
Conclusion
In conclusion, BodoOBP10 is mainly expressed in the antennae of both female and male insects, with higher levels in males, and specifically binds two sulfur-containing compounds, methyl allyl disulfide and diallyl disulfide. Modeling and docking studies showed that the BodoOBP10 structure is similar to that of a classic OBP and that six hydrophobic residues, Met70, Ile75, Thr89, Met90, Leu93, and Leu94, together with Phe143, on BodoOBP10 are candidates of binding sites with the two ligands. Furthermore, silencing OBP10 reduced EAG responses to both methyl allyl disulfide and diallyl disulfide. Therefore, our findings indicate that BodoOBP10 plays a vital role in olfactory perception including host plant recognition and location in B. odoriphaga and suggest that it may be a useful target for controlling B. odoriphaga in crops.
Supplementary Material
Acknowledgments
The research of Y.-T.Y., J.-Q.Z., and F.W., was funded and carried out by Zhang Youjun’s Laboratory, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China. Finally, we thank the reviewers for their careful reading of our manuscript, and helpful and constructive suggestions both in English and data presentation. This work was funded by the National Natural Science Foundation of China (32102202) to Y.-T.Y., and China Agriculture Research System (CARS-24-C-02) to Y.-J.Z.
Contributor Information
Jiaqi Zhu, Hubei Engineering Technology Center for Pest Forewarning and Management, Institute of Insect Sciences, College of Agriculture, Yangtze University, Jingzhou 434000, Hubei, China.
Fu Wang, Hubei Engineering Technology Center for Pest Forewarning and Management, Institute of Insect Sciences, College of Agriculture, Yangtze University, Jingzhou 434000, Hubei, China.
Youjun Zhang, Department of Plant Protection, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China.
Yuting Yang, Hubei Engineering Technology Center for Pest Forewarning and Management, Institute of Insect Sciences, College of Agriculture, Yangtze University, Jingzhou 434000, Hubei, China.
Dengke Hua, Institute of Agricultural Quality Standards and Testing Technology Research, Hubei Academy of Agricultural Sciences, Hubei Key Laboratory of Nutritional Quality and Safety of Agro Products, Wuhan 430064, Hubei, China.
Author Contributions
Conceptualization: Yuting Yang, Dengke Hua, Youjun Zhang; Methodology: Jiaqi Zhu, Yuting Yang, Fu Wang; Formal analysis and investigation: Yuting Yang, Jiaqi Zhu; Writing - original draft preparation: Jiaqi Zhu, Yuting Yang; Writing - review and editing: Yuting Yang, Dengke Hua; Supervision: Yuting Yang, Dengke Hua.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References Cited
- Brito, N. F., Moreira M. F., and Melo A. C. A... 2016. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 95: 51–65. doi: 10.1016/j.jinsphys.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Campanacci, V., Lartigue A., Hallberg B. M., Jones T. A., Giudici-Orticoni M. T., Tegoni M., and Cambillau C... 2003. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Natl. Acad. Sci. U.S.A. 100: 5069–5074. doi: 10.1073/pnas.0836654100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. L., Li G. W., Xu X. L., and Wu J. X... 2018a. Molecular and functional characterization of odorant binding protein 7 from the oriental fruit moth Grapholita molesta (Busck) (Lepidoptera: Tortricidae). Front. Physiol. 9: 1762. doi: 10.3389/fphys.2018.01762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. L., Pan Y. F., Ma Y. F., Wang J., He M., and He P... 2018b. Binding affinity characterization of an antennae-enriched chemosensory protein from the white-backed planthopper, Sogatella furcifera (Horvath), with host plant volatiles. Pest Bioch. Physiol. 152: 1–7. [DOI] [PubMed] [Google Scholar]
- Dong, K., Sun L., Liu J. T., Liu J. T., Gu S. H., Zhou J. J., Yang R. N., Dhiloo K. H., Gao X. W., Guo Y.Y., and Zhang Y. J... 2017. RNAi-induced electrophysiological and behavioral changes reveal two pheromone binding proteins of Helicoverpa armigera involved in the perception of the main sex pheromone component Z11-16:Ald. J. Chem. Ecol. 43: 207–214. [DOI] [PubMed] [Google Scholar]
- Field, L. M., Pickett J. A., and Wadhams L. J... 2000. Molecular studies in insect olfaction. Insect. Mol. Biol. 9: 545–551. doi: 10.1046/j.1365-2583.2000.00221.x [DOI] [PubMed] [Google Scholar]
- Feng, B., Qian K., and Du Y. J... 2017. Floral volatiles from Vigna unguiculata are olfactory and gustatory stimulants for oviposition by the Bean Pod Borer Moth Maruca vitrata. Insect. 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Francis F., Liu Y., Chen J. L., and Cheng D. F... 2011. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 10: 3056–3069. doi: 10.4238/2011.December.8.2 [DOI] [PubMed] [Google Scholar]
- Gadenne, C., Barrozo R. B., and Anton S... 2016. Plasticity in insect olfaction: to smell or not to smell? Annu. Rev. Entomol. 61: 317–333. doi: 10.1146/annurev-ento-010715-023523 [DOI] [PubMed] [Google Scholar]
- Guo, D., Hao C., Cui X. P., Wang Y., Liu Z. G., Xu B. H., and Guo X. Q... 2020. Molecular and functional characterization of the novel odorant-binding protein gene AccOBP10 from Apis cerana cerana. J. Bioch. 169: 215–225. [DOI] [PubMed] [Google Scholar]
- Hansson, B. S., and Stensmyr M. C... 2011. Evolution of insect olfaction. Neuron. 72: 698–711. doi: 10.1016/j.neuron.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Harris, M. O., and Miller J. R... 1988. Host-acceptance behavior in an herbivorous fly, Delia antiqua. J. Insect Physiol. 3: 179–190. [Google Scholar]
- Hua, J. F., Pan C., Huang Y. M., Li Y. Q., Li H. F., Wu C. R., Chen T. Y., Ma D. F., and Li Z. Y... 2021. Functional characteristic analysis of three odorant-binding proteins from the sweet potato weevil (Cylas formicarius) in the perception of sex pheromones and host plant volatiles. Pest Manag. Sci. 77: 300–312. [DOI] [PubMed] [Google Scholar]
- He, M., and He P... 2014. Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the white-backed planthopper Sogatella furcifera. Comp. Biochem. Physiol. B Biochem. Mol. Bio. 174: 1–8. [DOI] [PubMed] [Google Scholar]
- He, P., Durand N., and Dong S. L... 2019. Insect olfactory proteins from gene identification to functional characterization. Front. Physiol. 10: 1313. doi: 10.3389/fphys.2019.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquin-Joly, E., and Merlin C... 2014. Insect olfactory receptors: Contributions of molecular biology to chemical ecology. J. Chem. Ecol. 30: 2359–2397. [DOI] [PubMed] [Google Scholar]
- Jeong, Y. T., Shim J., Oh S. R., Yoon H. I., Kim C. H., Moon S. J., and Montell C... 2013. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 79: 725–737. doi: 10.1016/j.neuron.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. X., Yang Y. T., Xie W., Wu Q. J., Xu B. Y., Wang S. L., Wang S. J., and Zhang Y. J... 2015. Effects of temperature on the age-stage, two-sex life table of Bradysia odoriphaga (Diptera: Sciaridae). J. Econ. Entomol. 108: 126–134. [DOI] [PubMed] [Google Scholar]
- Leal, W. S. 2013. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58: 373–391. doi: 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- Li, Z. Q., Zhang S., Cai X. M., Luo J. Y., Dong S. L., Cui J. J., and Chen Z. M... 2017. Three odorant binding proteins may regulate the behavioural response of Chrysopa pallens to plant volatiles and the aphid alarm pheromone (E)-beta-farnesene. Insect. Mol. Biol. 26: 255–265. doi: 10.1111/imb.12295 [DOI] [PubMed] [Google Scholar]
- Liu, H., Duan H., Wang Q., Xiao Y., Wang Q., Xiao Q., Sun L., and Zhang Y. J... 2019. Key amino residues determining binding activities of the odorant binding protein AlucOBP22 to two host plant terpenoids of Apolygus lucorum. J. Agric. Food Chem. 67: 5949–5956. [DOI] [PubMed] [Google Scholar]
- Li, H. J., He X. K., Zeng A. J., Liu Y. J., and Jiang S. R... 2008. Evidence of a female sex pheromone in Bradysia odoriphaga (Diptera: Sciaridae). Can. Entomol. 140: 324–326. doi: 10.4039/n08-005 [DOI] [Google Scholar]
- Li, L. L., Huang J. R., Xu J. W., Yao W. C., Yang H. H., Shao L., Zhang H. R., Zhu Y., Dewer X. Y., and Zhang Y. N... 2022. Ligand-binding properties of odorant-binding protein 6 in Athetis lepigone to sex pheromones and maize volatiles. Pest Manag. Sci. 78: 52–62. [DOI] [PubMed] [Google Scholar]
- Liu, H., Wang C., Qiu C. L., Shi J. H., Sun Z., Hu X. J., Liu L., and Wang M. Q... 2022. A salivary odorant binding protein mediates Nilaparvata lugens feeding and host plant phytohormone suppression. Inter. J. Mol. Sci. 22: 4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. Q., Jiang H. B., Fan J. Y., Liu T. Y., Meng L. W., Liu Y., Yu H. Z., Dou W., and Wang J. J... 2021. An odorant-binding protein of Asian citrus psyllid, Diaphorina citri, participates in the response of host plant volatiles. Pest Manag. Sci. 77: 3068–3079. doi: 10.1002/ps.6352 [DOI] [PubMed] [Google Scholar]
- Mu, L. L., and Li G. Q... 2010. Influence of methanol-treatedRaphanus sativuson Plutella xylostellaandPhyllotreta striolata. Acta Phytophy. Sin. 37: 575–576. [Google Scholar]
- Pelosi, P., Zhou J. J., Ban L. P., and Calvello M... 2006. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63: 1658–1676. doi: 10.1007/s00018-005-5607-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P., Iovinella I., Felicioli A., and Dani F. R... 2014. Soluble proteins of chemical communication: an overview across arthropods. Front. Physiol. 5: 320. doi: 10.3389/fphys.2014.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P., Iovinella I., Zhu J., Wang G., and Dani F. R... 2018. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93: 184–200. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: 45e2002–45e2045. doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, C., Wang R., Che W. N., Li F. Q., Zhao H. P., Wei Y. Y., Luo C., and Xue M... 2021. Identification and tissue distribution of odorant binding protein genes in Harmonia axyridis (Coleoptera: Coccinellidae). J. Integr. Agric. 20: 2204–2213. [Google Scholar]
- Rihani, K., Ferveur J. F., and Briand L... 2021. The 40-year mystery of insect odorant-binding proteins. Biomolecules. 11: 509. doi: 10.3390/biom11040509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebijith, K. B., Asokan R., Hande H. R., Krishna K. N. K., Krishna V., Vinutha J., and Bakthavatsalam N... 2016. RNA interference of odorant-binding protein 2 (OBP2) of the cotton aphid, Aphis gossypii (glover), resulted in altered electrophysiological responses. Appl. Biochem. Biotech. 178: 251–266. [DOI] [PubMed] [Google Scholar]
- Sims, C., Birkett M. A., and Withall D. M... 2022. Enantiomeric discrimination in insects: the role of OBPs and ORs. Insects. 13: 368. doi: 10.3390/insects13040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., and Touhara K... 2008. Insect olfaction: receptors, signal transduction, and behavior. Results Probl. Cell Death Differ. 47: 121–138. 10.1007/400_2008_10 [DOI] [PubMed] [Google Scholar]
- Schymura, D., Forstner M., Schultze A., Kröber T., Swevers L., Latrou K., and Jürgen K. J... 2010. Antennal expression pattern of two olfactory receptors and an odorant binding protein implicated in host odor detection by the malaria vector Anopheles gambiae. Int. J. Biol. Sci. 6: 614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., Li Y., Zhang Z. D., Guo H. W., Xiao Q., Wang Q., and Zhang Y. J... 2019. Expression patterns and ligand binding characterization of Plus-C odorant-binding protein 14 from Adelphocoris lineolatus(Goeze). Comp. Biochem. Physiol. Part B. 227: 75–82. [DOI] [PubMed] [Google Scholar]
- Sun, Y. F., De F., Biasio H. L., Qiao I., Iovinella S. X., Yang Y., Ling L., Riviello D., Battaglia, P.Falabella, X. L. Yang, and Pelosi P... 2012. Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS One. 7: e32759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, B. W., Tai S. L., Dai W., and Zhang C. N... 2019. Expression and functional analysis of two odorant-binding proteins from Bradysia odoriphaga (Diptera: Sciaridae). J. Agric. Food Chem. 67: 3565–3574. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Stecher G., Peterson D., Filipski A., and Kumar S... 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, R. M. K., Quershi S. R., Adeel M. M., Abdelnabby H., Waris M. I., Duan S. G., and Wang M. Q... 2020. An odorant binding protein (SaveOBP9) involved in chemoreception of the wheat aphid Sitobion avenae. Inter. J. Mol. Sci. 21: 8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, R. G., Prestwich G. D., and Lerner M. R... 1991. Odorant-binding-protein sub-families associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 22: 74–84. doi: 10.1002/neu.480220108 [DOI] [PubMed] [Google Scholar]
- Wang, W. J., Zhang T., Chen J. M., and Chen Z. D... 2011. Present situation and control technology of pesticide residue in Chinese Chives. Shandong Agric. Sci. 10: 82–84. In Chinese. [Google Scholar]
- Wang, Q., Zhou J. J., Liu J. T., Huang G. Z., Xu W. Y., Zhang Q., Chen J. L., Zhang Y. J., Li X. C., and Gu S. H... 2019. Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids. Insect. Mol. Biol. 28: 1–22. doi: 10.1111/imb.12513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Brandazza A., and Pelosi P... 2008. Binding of polycyclic aromatic hydrocarbons to mutants of odorant-binding protein: a first step towards biosensors for environmental monitoring. Biochim. Biophys. Acta (BBA) Proteins Proteomics. 1784: 666–671. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Gao C., Liu J., Zhou W., and Zeng X... 2020. Host plant odors and their recognition by the odorant-binding proteins of Diaphorinacitri Kuwayama (Hemiptera: Psyllidae). Pest Manag. Sci. 76: 2453–2464. doi: 10.1002/ps.5786 [DOI] [PubMed] [Google Scholar]
- Yang, Y. T., Li W. X., Xie W., Wu Q. J., Xu B. Y., Wang S. L., Li C. R., and Zhang Y. J... 2015. Development of Bradysia odoriphaga (Diptera: Sciaridae) as affected by humidity: an age-stage, two-sex, life-table study. Appl. Entomol. Zool. 50: 3–10. [Google Scholar]
- Yang, K., Yan L., Niu D. J., Wei D., Li F., Wang G. R., and Dong S. L... 2016. Identification of novel odorant binding protein genes and functional characterization of OBP8 in Chilo suppressalis (Walker). Gene. 591: 425–432. doi: 10.1016/j.gene.2016.06.052. [DOI] [PubMed] [Google Scholar]
- Yang, Y. T., Su Q., Shi L. L., Chen G., Zeng Y., Shi C. H., and Zhang Y. J... 2019. Electrophysiological and behavioural responses of Bradysia odoriphaga Yang et Zhang (Diptera: Sciaridae) to volatiles from its host plant, Chinese chives (Allium tuberosum Rottler). J. Econ. Entomol. 112: 1638–1644. [DOI] [PubMed] [Google Scholar]
- Yang, Y. T., Hua D. K., Shi C. H., Xie W., and Zhang Y. J... 2021a. Molecular and binding characteristic of OBP5 of Bradysia odoriphaga (Diptera: Sciaridae). J. Econ. Entomol. 114: 1509–1516. [DOI] [PubMed] [Google Scholar]
- Yang, Y. T., Luo L., Tian L. X., Zhao C. W., Niu H., Hu Y. F., Shi C. H., Xie W., and Zhang Y. J... 2021b. Function and characterization analysis of BodoOBP8 from Bradysia odoriphaga (Diptera: Sciaridae) in the recognition of plant volatiles and sex pheromones. Insects. 12: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. X., Zhao M. M., Zhong L., Li Y., Li D. Z., Deng Z. N., and Ma X. F... 2021. Characterization of the binding ability of the odorant binding protein BminOBP9 of Bactrocera minax to citrus volatiles. Pest Manag. Sci. 77: 1214–1225. [DOI] [PubMed] [Google Scholar]
- Yin, J., Wang C., Fang C., Zhang S., Cao Y., Li K. B., and Lea W. S... 2019. Functional characterization of odorant-binding proteins from the scarab beetle Holotrichia oblita based on semiochemical-induced expression alteration and gene silencing. Insect Biochem. Mol. Biol. 104: 11–19. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Ding J., Zhang Z., Liu F., Zhou C., and Mu W... 2018. Sex- and tissue-specific expression profiles of odorant binding protein and chemosensory protein genes in Bradysia odoriphaga (Diptera: Sciaridae). Front. Physiol. 9: 107. doi: 10.3389/fphys.2018.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. N., Tang B. W., Zhou T. L., Yu X. T., Hu M. F., and Dai W... 2021. Involvement of chemosensory protein BodoCSP1 in perception of host plant volatiles in Bradysia odoriphaga. J. Agric. Food Chem. 69: 10797–10806. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. K., Li Z. B., Chen D. K., Wu S. Y., Wang H. H., Li Y. L., and Lei Z. R... 2022. The molecular identification, odor binding characterization, and immunolocalization of odorant-binding proteins inLiriomyza trifolii. Pest Bioch. Physiol. 181: 105016. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Chen J. L., Lin J. H., Lin J. T., and Wu Z. Z... 2020. Odorant-binding proteins and chemosensory proteins potentially involved in host plant recognition in the Asian citrus psyllid Diaphorina citri. Pest Manag. Sci. 76: 2609–2618. doi: 10.1002/ps.5799 [DOI] [PubMed] [Google Scholar]
- Zhang, X. Y., Zhu X. Q., Gu S. H., Zhou Y. L., Wang S. Y., Zhang Y. J., and Guo Y. Y... 2017. Silencing of odorant binding protein gene AlinOBP4 by RNAi induces declining electrophysiological responses of Adelphocoris lineolatus to six semiochemicals. Insect Sci. 24: 789–797. doi: 10.1111/1744-7917.12365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.