Significance
Candidate phyla radiation (CPR) is a large clade of bacterial lineages that account for approximately one quarter of microbial diversity. Knowledge about their physiology and obligate (epi)symbioitc lifestyle is rare due to a lack of cultivation. We developed a gene-targeted method that brought about the cultivation of a novel CPR bacterium, designated Saccharibacteria TM7i, with its basibiont from an enriched sample of an insect. We showed its growth dynamics using super-resolution imaging and revealed the essential role of type IV pili in enabling motility and adhesion to the host during episymbiosis. A four-stage lifecycle is proposed for the novel CPR bacteria. Our findings will undoubtedly contribute to the cultivation and understanding of these intriguing and enigmatic microbes.
Keywords: candidate phyla radiation, Saccharibacteria, TM7, episymbiosis, type IV pili
Abstract
Recent characterization of the obligate episymbiont Saccharibacteria (TM7) belonging to the candidate phyla radiation (CPR) has expanded the extent of microbial diversity. However, the episymbiotic lifestyle of TM7 is still underexploited due to the deficiency of cultivated representatives. Here, we describe gene-targeted TM7 cultivation guided by repurposing epicPCR (emulsion, paired isolation, and concatenation PCR) to capture in situ TM7‒host associations. Using this method, we obtained a novel Saccharibacteria isolate TM7i and its host Leucobacter aridicollis J1 from Cicadae Periostracum, the castoff shell of cicada. Genomic analyses and microscopic characterizations revealed that TM7i could bind to J1 through twitching-like motility mediated by type IV pili (T4P). We further showed that the inhibition of T4P extrusion suppressed the motility and host adherence of TM7i, resulting in its reduced growth. However, the inactivation of T4P had little effect on the growth of TM7i that had already adhered to J1, suggesting the essential role of T4P in host recognition by TM7i. By capturing CPR‒host association and elaborating the T4P-dependent episymbiotic association mechanism, our studies shed light on the distinct yet widespread lifestyle of CPR bacteria.
Candidate phyla radiation (CPR) comprises large evolutionary lineages of ultrasmall bacteria lacking isolated representatives, accounting for over 26% of biodiversity in the bacterial domain (1–3). Metagenome-assembled genomes of CPR highlight major deficiencies of essential metabolic pathways (3–5), indicating that most of these enigmatic bacteria live as obligate symbionts. Cultivating CPR symbionts from various habitats to facilitate further functional analyses is one of the biggest challenges for unveiling CPR’s ecological and evolutionary roles (6). Antibiotic selection and filtrate reinfection have led to the cultivation of the first phylotype of CPR belonging to Candidatus Saccharibacteria (TM7) from the human oral cavity (7). However, the isolation process is contingent and requires much labor to screen all possible hosts. Targeted cultivation was realized by fluorescence-activated cell sorting of CPR epibionts that can be immunolabeled (8). However, the obtained isolates cannot represent the in situ interaction of CPR‒host associations, especially for low-abundance CPR groups. Furthermore, apart from limited oral TM7 isolates, reports on the cultivation of environmental CPR are still rare, which hinders the understanding of CPR’s ecological and environmental impacts.
CPR bacteria rely on intriguing mechanisms to support their parasitic lifestyles (9, 10). For example, the lifecycle of TM7 requires close contact with host cell surfaces (7, 11–13), which necessitates potential appendages for bacterial motility and host adherence. A unique insight informed by genomics is that many CPR bacteria have type IV pili (T4P) that presumably confer motility and adhesion to CPR for moving toward the host and achieving parasitic association (4, 9, 11, 14). Yet, genomic annotations for other filament appendages in the extremely streamlined genomes of CPR bacteria are rare (3). Generally, T4P are ubiquitous, retractable, and multifunctional hair-like appendages of many bacteria and archaea (15, 16) involved in different processes, including locomotion, adherence to host cells, virulence, and DNA uptake (16–19). Moreover, electron microscopy revealed pili-like appendages on the cell surface of TM7 (11) and other CPR and DPANN (an acronym of the names of the first included archaeal phyla, Candidatus Diapherotrites, Candidatus Parvarchaeota, Candidatus Aenigmarchaeota, Nanoarchaeota, and Candidatus Nanohaloarchaeota) groups (9, 14). However, current studies are insufficient to address either the physical existence or biological function of T4P in CPR bacteria. The role of T4P in supporting the parasitic lifestyle of CPR is still unclear.
EpicPCR (emulsion, paired isolation, and concatenation PCR) was initially developed for the paired association of functional genes with phylogenetic markers (20). Here, we developed a targeted cultivation approach by repurposing epicPCR to resolve in situ CPR‒host episymbiotic associations. The epicPCR-revealed host spectrum provides vital clues for the isolation of an aerobic CPR phylotype, Saccharibacteria TM7i, with its host Leucobacter aridicollis J1 from Cicadae Periostracum (CP), the castoff shell of cicada. We then carried out morphological and physiological profiling using the TM7i‒J1 symbiont. As a result, we revealed that the T4P-powered motility of TM7i facilitates active approaching and binding with J1 to start asymmetric division, elucidating the enigmatic lifestyle of the first insect-associated TM7 phylotype as a representative of epibiotic CPR in the environment.
Results
EpicPCR-Directed TM7 Symbiont Isolation.
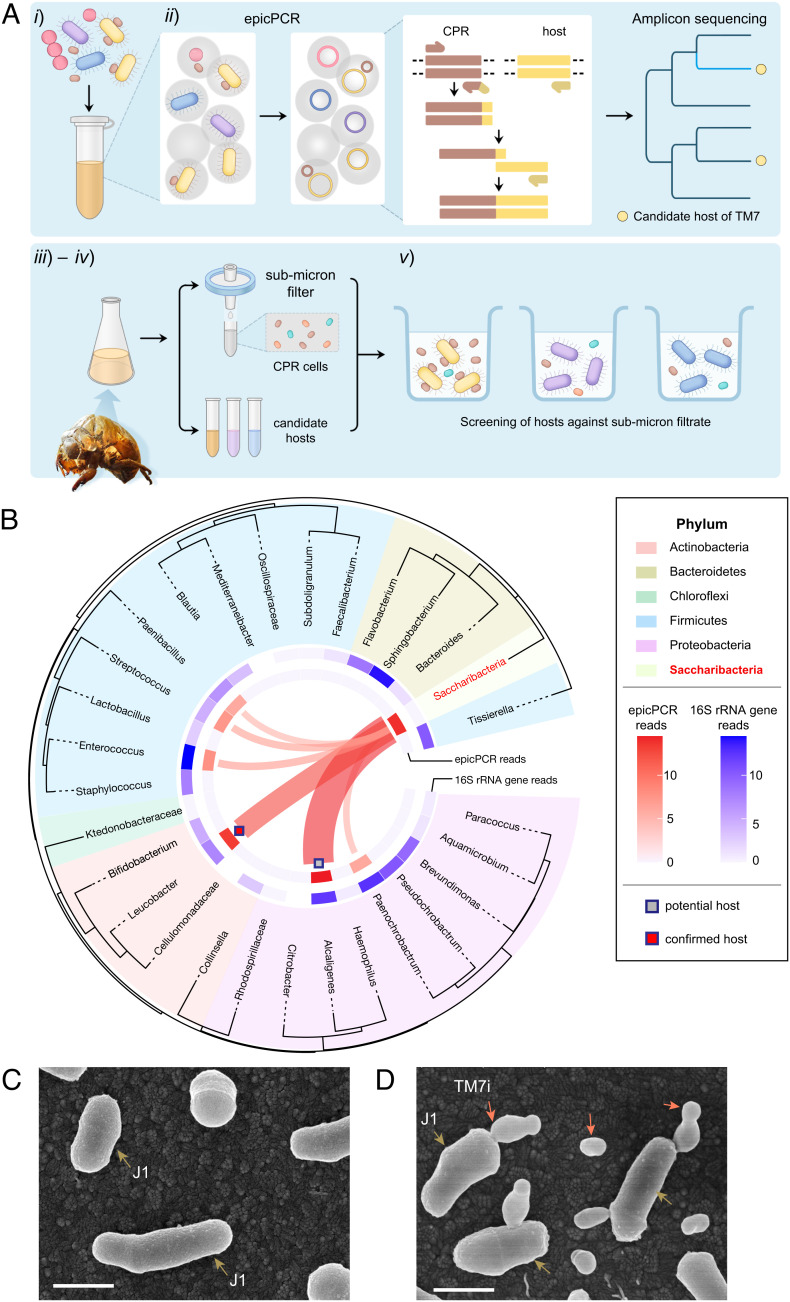
We developed an epicPCR-directed cultivation approach to improve the efficiency of epibiotic CPR isolation. The workflow is as follows (Fig. 1A): i) preparation of a microbial suspension of the microbial community containing CPR bacteria; ii) detection of epibiotic association in the community using epicPCR; iii) selective isolation of epicPCR-informed candidate hosts, which helps us to narrow down candidate hosts in complex microbial communities; iv) screening the candidate hosts against submicron filtrate containing planktonic TM7 cells from the sample according to their ultrasmall sizes (4, 7); and v) identification and recovery of CPR‒host symbionts by serial passage and PCR validation. Compared with the traditional approaches for isolating CPR bacteria, this workflow helped us to narrow down candidate hosts in complex samples and reduce the uncertainty and labor during isolation.
Fig. 1.
Gene-targeted episymbiont cultivation leads to the isolation of the first insect-associated Saccharibacteria (TM7) phylotype. (A) The workflow of gene-targeted TM7 symbiont isolation include five steps: (i) prepare microbial suspension, (ii) profile the TM7‒host association using epicPCR (emulsion, paired isolation, and concatenation PCR), (iii) selectively cultivate the candidate hosts, (iv) filtrate reinfection, and (v) identification and recovery of TM7‒host symbionts. (B) EpicPCR-informed TM7‒host associations in the enrichment community of Cicadae Periostracum. We built the phylogenetic tree shown in the outer circle with 16S rRNA gene sequences. Taxon groups are depicted as colored segments. The relative abundance of 16S rRNA amplicon sequencing and epicPCR-informed associations are annotated in the inner blue and red rings, respectively. Ribbons connecting to TM7 indicate potential TM7‒host associations. (C and D) The information obtained through epicPCR led to the cultivation of Saccharibacteria TM7i with the microbial host L. aridicollis J1, as shown in the scanning electron microscopy (SEM) images. (Scale bar, 500 nm.) (C) Pure culture of actinobacterial host J1. Brown arrows indicate J1 cells. (D) TM7i‒J1 symbiont. Red arrows indicate either planktonic or host-adhered TM7i cells.
The epicPCR experiment began with the emulsification of a bacterial suspension containing CPR symbionts into picoliter water-in-oil polyacrylamide droplets via stochastic encapsulation of single cells, followed by cell lysis and concatenation PCR to generate amplicons linking sequences of CPR with their hosts (Fig. 1A). The PCR products were extracted and sequenced to disclose the epibiotic associations of CPR bacteria with candidate hosts. We validated the performance of epicPCR-directed TM7‒host association detection by mock TM7‒host episymbiont samples containing Escherichia coli cells transformed with a TM7-specific plasmid (SI Appendix, Supplementary Note and Fig. S1).
Next, a microbial enrichment from CP was chosen for the evaluation of epicPCR-directed cultivation of TM7 symbionts (SI Appendix, Fig. S2). The existence of TM7 in CP samples was detected by accident when we attempted to assess microbial contamination in various traditional Chinese medicines. The amplicon sequencing of 16S rRNA genes showed that TM7 and Actinobacteria accounted for 3.10% and 26.36% of the total reads in the microbial community of the original sample, respectively (SI Appendix, Fig. S2A). In contrast, no reads belonging to the TM7 phylum were observed in enriched samples from cicada larvae (CL). Moreover, TM7 persisted during serial passages of the enrichment supplemented with streptomycin at concentrations from 50 μg/mL to 200 μg/mL (SI Appendix, Fig. S2 B and C). Consequently, we performed epicPCR experiments to assess TM7‒host epibiotic associations in the enrichment, indicating high-degree associations of TM7 with Alcaligenes and Leucobacter (Fig. 1B). Next, instead of exhausting efforts to recover all microbial species, we selectively isolated Alcaligenes sp. and L. aridicollis J1. The isolation of candidate hosts was confirmed by Sanger Sequencing of PCR-amplified 16S rRNA Gene. Then, we incubated them with the 0.22-μm filtrate of the enrichment separately. TM7 was amplified with J1 and preserved in broth and agar culture during multiple passages (SI Appendix, Fig. S2D) but could not grow with the Alcaligenes isolate. Subsequently, the scanning electron microscopy (SEM) images showed the interaction of J1 (in pure culture, Fig. 1C) and TM7 cells with submicron size (in coculture with J1, Fig. 1D). The obtained TM7‒J1 symbiont was also validated by PCR detection of multiple passages using TM7 specific primers (SI Appendix, Table S3). Further experiments confirmed the stable coculture of a novel insect-associated TM7 strain designated TM7i. The 16S rRNA gene of TM7i shared 93.9% similarity with that of TM7x (SI Appendix, Figs. S3 and S4), which suggests that it is the first cultivated member of a novel genus-level TM7 lineage. Therefore, we proposed it as a new species of a new genus, with the nomenclature Leucosynbacter cicadicola gen. nov., sp. nov. given. Compared with previous CPR isolates that triggered reduced growth (13) or lysis of host cells (11, 14), TM7i had minor impacts on the growth and cell size of J1 under an aerobic condition (SI Appendix, Fig. S2 E and F).
Morphological Characterization Highlights TM7i’s Pili-Like Appendages.
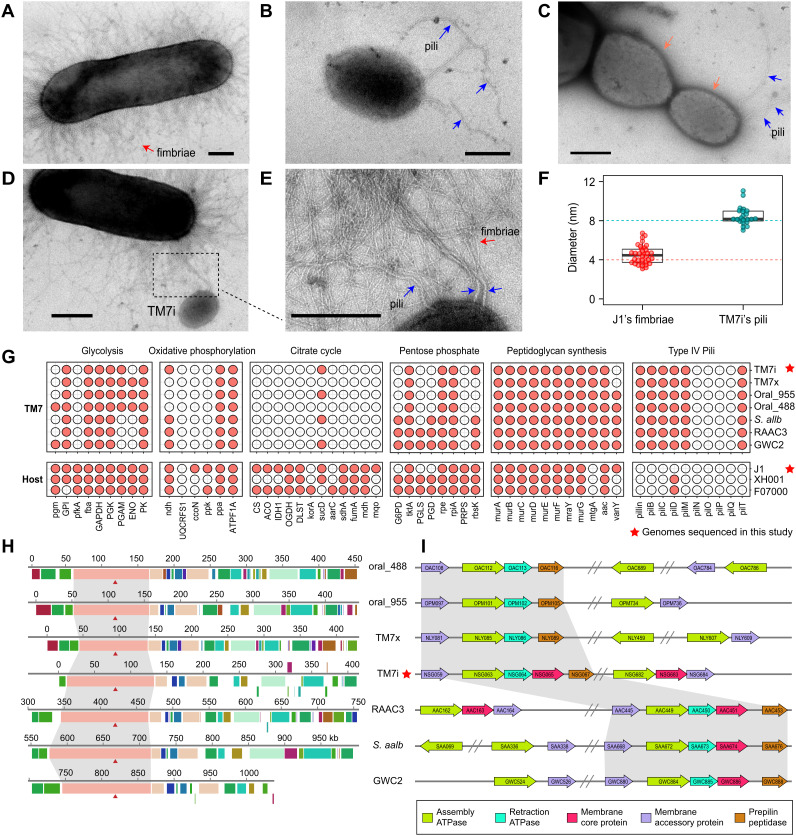
Next, we used transmission electron microscopy (TEM) to capture the morphological features of the freshly cultivated TM7i‒J1 symbiont. TEM showed that J1 is covered with thin and short fimbriae with an average diameter of approximately 4 nm (Fig. 2A). The genomic and transcriptomic profiles of J1 also indicate that it harbors a fimbriae expression system (SI Appendix, Table S1). In contrast, planktonic TM7i cells have an extremely small size of 200‒300 nm with a few pili-like appendages with a diameter of ~8 nm (Fig. 2 B and F). TM7 can adhere to a J1 cell and generate progeny cells by asymmetric division (Fig. 2C). Interestingly, we also observed pilus formation on the distal end of progeny TM7i cells, indicating that pili may facilitate the dispersal of progeny cells for recurrent host infection. We can distinguish pili-like appendages of TM7i from J1 fimbriae based on longer lengths and thicker diameters (Fig. 2F). The diameter of TM7i’s pili-like appendages is consistent with the typical size of bacterial T4P (16). Our TEM images also captured the interaction of planktonic TM7i cells with J1 cells via TM7i’s pili-like appendages (Fig. 2 D and E and SI Appendix, Fig. S5).
Fig. 2.
Microscopy observation and comparative genomics analyses. (A) Transmission electron microscopy (TEM) of a J1 cell with fimbriae. (B) TEM of a planktonic TM7i cell with pili-like appendages. Blue arrows indicate pili. (C) The asymmetric division of TM7i adhered to J1. Divided TM7i cells are marked with red arrows. Blue arrows indicate pili on the distal pole of the progeny cell. (D and E) TM7i pili intertwined with J1 fimbriae. The blue arrows in the zoom-in view indicate TM7i T4P. (The scale bar is 200 nm.) (F) Diameter comparison of TM7i pili and J1 fimbriae (for fimbriae, n = 22; for pili, n = 34). (G) Profile of the presence or absence of metabolic or biosynthetic potentials (columns) for seven selected TM7 bacteria and three representative hosts. The type IV pili (T4P) gene cluster is highly conserved in all TM7 genomes, including TM7i. The red star indicates genomes sequenced in this study. (H) Locally collinear blocks conserved among TM7 genomes. Red triangles indicate pilT, which encodes an ATPase for pili retraction. (I) The annotation of T4P-related genes throughout the TM7 genomes. The direction of the arrow indicates the gene direction. Double slashes indicate intervals between gene clusters. The region of genes was marked by the number of open reading frames (ORFs) confirmed in both the KEGG and NCBI databases.
Widespread Conservation of T4P in TM7 Genomes.
We then carried out multigenome analysis using genomes of TM7i and J1 obtained in this study, together with representative TM7 and host genomes in the NCBI database (see Materials and Methods for details). Whole-genome profiling of seven complete TM7 genomes indicated deeply reduced genomic repertoires with deficiencies in essential metabolic pathways, including glycolysis, oxidative phosphorylation, and pentose phosphate pathways, indicating energy and nutrition dependencies on their hosts. However, the peptidoglycan biosynthesis pathway and T4P-assembly gene clusters were highly conserved in all analyzed TM7 genomes (Fig. 2G). The transcriptomic profiling of TM7i also shows that all T4P-related genes have a consistent level of expression (SI Appendix, Table S2). The T4P gene cluster contains pilin as the fimbrial protein; pilB and pilT genes encode T4P elongation and retraction ATPases, while pilC and pilM genes encode the membrane core protein and the accessory protein, respectively. These proteins underline the molecular capacity for TM7 bacteria to assemble or retract type IV pili (16). Multiple genomic alignments of TM7 genomes showed that T4P-related genes occurred in one of the major locally collinear blocks (Fig. 2H). We also found a highly conserved orientation of the T4P gene cluster in selected TM7 genomes (Fig. 2I).
T4P Enables the Twitching Motility of TM7i.
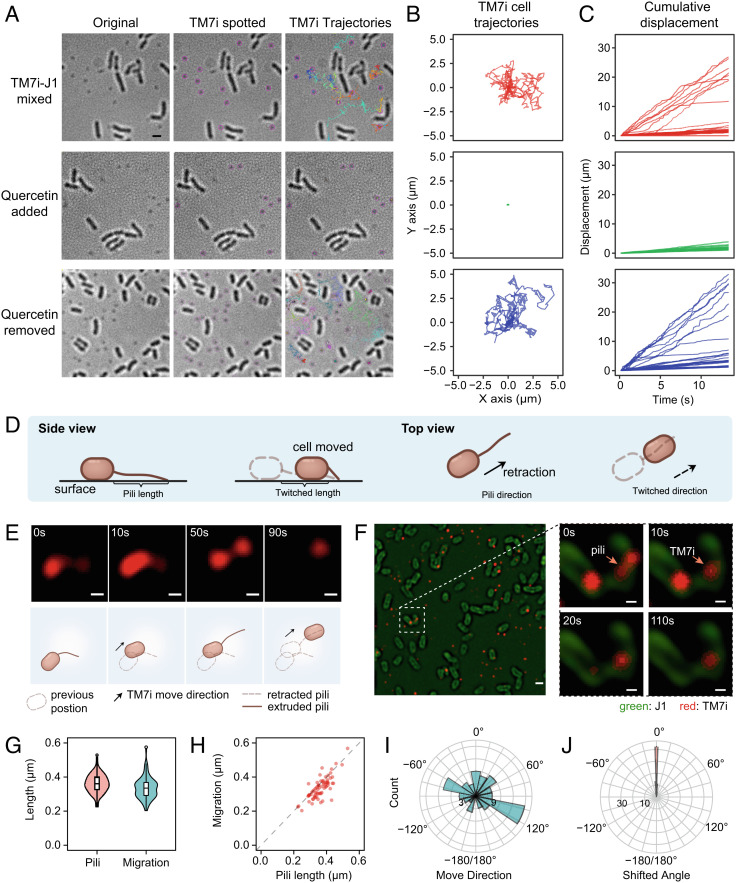
To further confirm that T4P drives TM7i motility, we tested whether the motility can be inhibited by quercetin, a PilB inhibitor that prevents T4P extrusion (21). (Fig. 3 A‒C). We constrained the bacterial mixture in the thin layer formed by a gel pad and coverslip and used bright field microscopy to observe the dynamics of the TM7i‒J1 interaction. In all microscopic motility assays, J1 cells were immotile (Movie S1). Real-time imaging showed that planktonic TM7i cells enriched in the filtrate rarely moved, except when J1 cells were added (Fig. 3A and Movie S1). When TM7i was mixed with J1, 26% of TM7i cells migrated significantly with cumulative displacement > 5 μm within 30 s (n = 30), and the fastest average migration rate reached 2.02 μm/s (Fig. 3 B and C and Movie S1). As expected, the addition of quercetin strongly reduced the motility of TM7i cells at a concentration of 200 μM in vivo (Fig. 3 E and F). Furthermore, the inhibited motility of TM7i was restored upon quercetin removal, proving that quercetin inhibition of pilB is competitive (21).
Fig. 3.
Genomically conserved T4P enabled motility of TM7i cells. (A) Inhibition of TM7i motility in vivo using T4P inhibitor quercetin. TM7i exhibited high motility in symbiosis with J1 (Top row); quercetin (200 μM) reduced TM7i motility (Middle row); the motility of TM7i cells was recovered after quercetin removal (Bottom row). Original bright field images (Left), TM7i cells spotted overlay (Middle), and TM7i trajectories overlay (Right) within a 30-s time-lapse recording are shown. Identified TM7i cells are marked with purple circles in the Middle panel. Trajectories of motile TM7i cells are marked with different colors. (B) Trajectory plot of individual cells corresponding to Panel (A). For each group, 30 trajectories were acquired randomly and plotted from three independent experiments. (C) Cumulative displacement against time for the three groups. (D) Schematic of T4P-mediated movement of bacterial cells. (E) Structured illumination microscopy (SIM) imaging of pili-mediated motility of TM7i labeled with NHS ester of Alexa Fluor 555. The schematics depict the interpretive cell layout. (Scale bar, 200 nm.) (F) SIM images show that TM7i infects J1 by pili adhesion (also see Movie S2). J1 and TM7i cells were separately labeled and freshly mixed for live SIM imaging. J1 cells were labeled with NHS ester of Alexa Fluor 488. Images are indicative of three independent experiments. (Scale bar in left, 500 nm; scale bars for zoom-in views, 200 nm.) (G) Box plot of the extruded T4P length and the consequent cell migration. (H) The consistency between the extruded T4P length and the resultant cell migration. (I) The directions of TM7i movements (n = 73). (J) The shifting angle between T4P extrusion and the resultant movement.
Twitching motility occurs in many bacteria to facilitate and maintain surface colonization and host infection. Typically, it involves the extrusion, tethering, and retraction of T4P that allows bacterial translocation over surfaces (Fig. 3D). We performed nanometer-resolution fluorescence imaging of TM7 symbionts using structured illumination microscopy (SIM) to observe TM7’s pili in motion. First, TM7i cells were separated from coculture by 0.45 μm filtration and labeled with Alexa Fluor 555 NHS ester probes (red fluorescence) (22). Then, we brought TM7i cells into contact with J1 cells labeled with Alexa Fluor 488 NHS ester probes (green fluorescence) surrounded by a wetting layer between an agar pad and a cover glass for multicolor SIM imaging at a frame rate of 0.1 fps. We observed that TM7i cells actively extruded or retracted their pili, triggering the dynamic translocation of TM7i cells (Fig. 3E and Movie S2). Interestingly, some TM7i cells moved toward J1 cells and became sessile on J1 cells (Fig. 3F and Movie S2). The surface-tethered pili became straightened and had an average length of 0.361 ± 0.056 μm (mean ± SD, n = 73). TM7i cells moved in the direction of the surface-tethered pili with an average twitching distance of 0.335 ± 0.063 μm, slightly shorter than the pili length (Fig. 3G). The twitching motility precisely aligned with the length and direction of straightened pili extrusions that were surface-tethered, with 91% of events having a shifting angle of less than 5° (Fig. 3 H‒J). Thus, this pili-driven motility is highly consistent with typical T4P-dependent twitching motility (Fig. 3D). Since TM7i has a streamlined genome without any other filamentous appendages identified, we thus provide strong evidence for TM7i’s twitching motility enabled by T4P.
T4P-Dependent Motility Affects TM7i Proliferation.
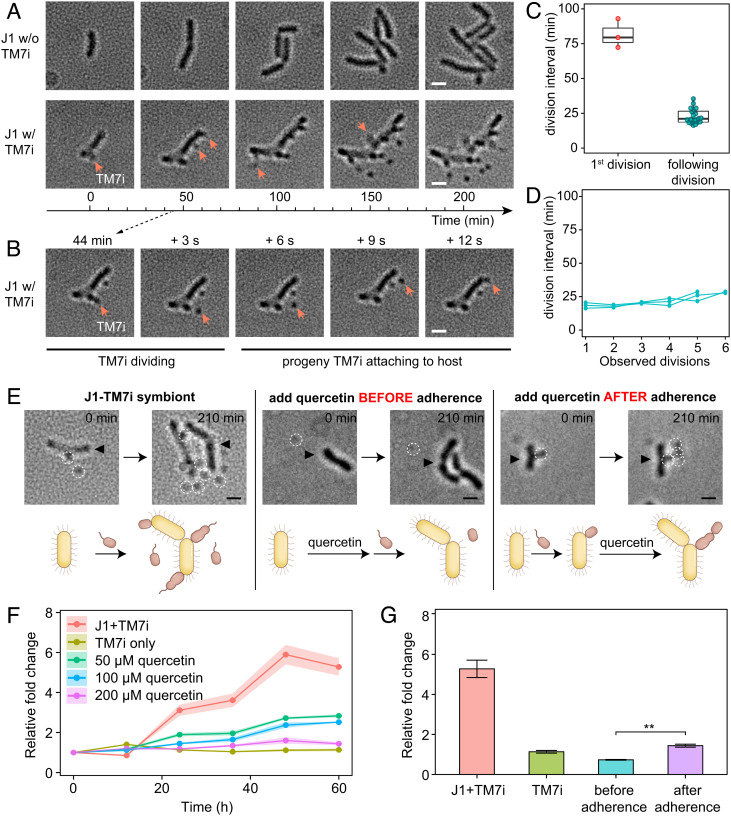
To investigate the effect of T4P-dependent twitching motility on the parasitic lifestyle of TM7i, we recorded the proliferation of TM7i‒J1 symbionts via long-term microscopy observations. The results revealed the continuous propagation of progeny TM7i cells by host-adhered TM7i cells that exhibited highly active motility surrounding the host. (Fig. 4A and Movie S3). In some cases, newly generated progeny TM7i cells rapidly translocated and adhered to J1 cells within 12 s (Fig. 4B). New J1-adhered TM7i cells required 86.2 ± 7.4 min to grow in size before asymmetric division to produce the first progeny cell in a budding-like manner, followed by continuous propagation of progeny cells with an average division interval of 22.9 ± 5.3 min (Fig. 4 C and D). Compared to its tiny size, the surprisingly high motility of TM7i may help TM7i cells search for and infect new host cells, which might be essential for the survival of TM7i with major metabolic and biosynthetic deficiencies.
Fig. 4.
T4P-dependent motility affects TM7i proliferation. (A) Long-term time-lapse imaging visualizes the propagation of the TM7i‒J1 symbiont (also see Movie S3). Red arrows indicate newly divided TM7i cells. (The scale bar is 500 nm.) (B) Propagation of a progeny TM7i cell to another localization site within 12 s. (C) The average division intervals differ for the first and the following divisions. (D) Continuous intervals of TM7i division after the first division. (E) T4P inhibition with quercetin blocks twitching motility and host infection. Compared with TM7‒J1 coculture, quercetin inhibited the twitching motility of planktonic cells and halted host infection. However, quercetin did not suppress the propagation of progeny cells for host-adhered TM7i cells. White dashed circles indicate TM7i cells, and black triangles indicate J1 cells. (The scale bar is 500 nm.) (F) Reduced TM7i growth under quercetin treatments compared with the no quercetin control. Log2-fold change values were generated by real-time PCR of the TM7i-specific 16S rRNA gene. (G) Timing of adding inhibitor affects growth rate; quercetin added before adherence shows an even slower TM7i growth (**P < 0.01).
To examine whether the inhibition of T4P-dependent twitching motility affects the proliferation of TM7i, we performed T4P inhibition assays by adding 200 μM quercetin before and after host adherence in parallel, using the no quercetin group as the control (Fig. 4E and Movie S3). As expected, the addition of quercetin prior to the adherence suppressed the motility and growth of TM7i cells, as quercetin suppressed the assembly and extrusion of T4P. However, when quercetin was added after preincubation of TM7i with J1, the host-adhered TM7i cells continued to generate progeny cells (Movie S3), which indicated the inhibition of T4P had weak effects on cell growth and division. However, the newly divided daughter cells did not undergo significant motion, failing to further disperse and infect other host cells. Next, we generated the calibration curve based on serial dilution using quantitative PCR (SI Appendix, Fig. S6A) and obtained the relative growth change of TM7i in TM7i‒J1 broth cultures with 50 to 200 μM quercetin compared to the no quercetin control. We found that quercetin dose-dependently lowered the growth of TM7i during symbiosis with J1 at the population level (Fig. 4F). At all tested quercetin concentrations, the growth of L. aridicollis J1 was not significantly affected (SI Appendix, Fig. S6B). Moreover, the timing of quercetin addition affected the proliferation of TM7i cells, as quercetin added before host adherence resulted in an even slower growth rate compared with that observed when quercetin was added after adherence (Fig. 4G). Our results suggested that TM7i’s T4P plays an essential role in recognizing and adhering to J1 hosts.
Size-Differentiated Lifecycle of TM7i with Its Host.
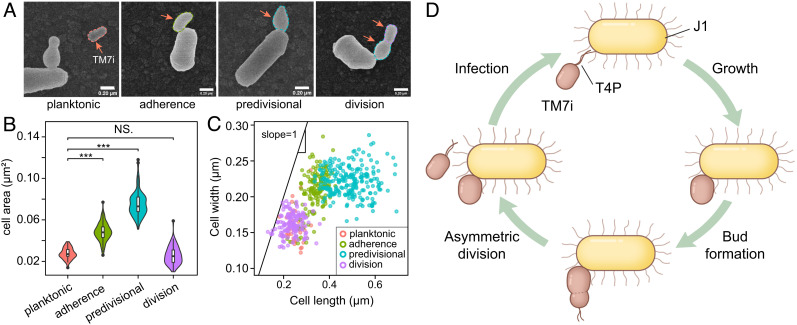
To determine the growth patterns of TM7i, we quantitatively characterized TM7i cells in symbiosis with J1 using SEM images (Fig. 5 A‒C). The results suggest that TM7i cells have a size-differentiated lifecycle consisting of four stages (Fig. 5D). i) In the planktonic (free-floating) stage, TM7i cells exhibit their smallest average widths of 160 ± 24 nm and lengths of 250 ± 31 nm (mean ± SD, n = 41). ii) In the adherence stage, planktonic TM7i cells adhere to J1 cells and become sessile. The cell size gradually increases, with an average length increase to 337 ± 41 nm and a cell area average increase to 0.049 ± 0.008 μm2 (mean ± SD, n = 143). iii) In the predivision stage, the adhered TM7i cells generate budding-like progeny cells on their distal ends, followed by a gradual expansion of progeny cell sizes. During this stage, the TM7i length reached 693 nm and had an average length of 489 ± 78 nm (mean ± SD, n = 213). After TM7i adhered to their host, the length and width of sessile TM7i cells began to grow synchronously with a slope close to 1, while TM7i expanded mainly along its length at the budding-like stage (Fig. 5C). iv) In the division stage, clear dividing sections were observed between the sessile cell and progeny cells, followed by the release of the progeny cell by asymmetric division.
Fig. 5.
TM7i’s four-stage lifecycle. (A) Microscopic analysis of the TM7i‒J1 symbiont showed that TM7i has four growth stages: planktonic, adherence, predivisional, and division. Images are indicative of three independent experiments. (Scale bar, 200 nm.) (B) Violin plot of cell sizes quantitatively assessed for the four aforementioned cell types (***P < 0.001; NS.P = 0.22). (C) Scatter plot of TM7i cell sizes classified according to the four cell types. We hypothesize that after infection, TM7i cells expand their lengths and widths synchronously to the adherence stage. This was followed by the expansion of the cell length to the predivisional stage. (D) Scheme of the epibiotic lifecycle of TM7i. Proliferation occurred during adherence through budding-like asymmetric division, generating a smaller progeny cell. Progeny cells approach host cells by motility mechanisms, which is enabled by T4P.
Upon the contacting interface of the TM7i‒J1 symbiont, a tube-like structure can be seen in some SEM images (SI Appendix, Fig. S7A), which may help mass exchange between the host cell and the parasitic TM7i cell. Progeny TM7i cells exhibited similar sizes as free TM7i cells (n = 126), with average lengths of 238 ± 41 nm (Fig. 5 B and C and SI Appendix, Fig. S7 B and C). In most cases, a J1 cell can be infected by one or two TM7i cells. Regarding the position of the infecting TM7i cells, SEM imaging showed that TM7i cells preferentially adhered to the cell poles of J1 (SI Appendix, Fig. S7 C and D), in agreement with bright-field microscopy data (Fig. 4 and Movie S3). Moreover, T4P discretely formed on the distal pole of progeny cells, which may aid their dispersal (Fig. 2C). The quantitative analysis indicates that TM7i relies on J1 for proliferation and has an asymmetric division lifecycle with strict control of cell sizes and shapes (Fig. 5D).
Discussion
CPR bacteria comprise enormous uncultivated superphyla that contribute to the recent expansion of the tree of life (6), representing a massive part of microbial “dark matter” with significant evolutionary and ecological impacts (3, 4, 23). However, despite CPR’s prevalence uncovered by metagenome-enabled phylogenomic analyses, the biological perception of CPR is very limited, as most of them are uncultured. Specifically, TM7 is a ubiquitous CPR group widely discovered in soils, sediments, wastewater, mammals, and humans (the gastrointestinal tract and oral cavity) (1, 3, 24, 25). Instead, the occurrence of TM7 in insects has rarely been reported and studied. In this study, we discovered a high abundance of TM7 in the microbiota of CP, while no TM7 was detected in CL. This result suggests that TM7 bacteria only exist in the cicada microbiota at specific lifecycle stages or body parts, indicating the existence of unknown equilibrium and interactions among cicada, Actinobacteria, and TM7. In particular, TM7i isolated in this study provides a unique phylotype that can grow aerobically for further physiological investigations. Insects are the most dominant animal class on Earth (26, 27). Close symbiotic relationships between insects and microbes have been discovered that contribute to digestion, development, and pathogen resistance (28). Our study proves that insects are a rich reservoir of CPR‒host symbionts and indicates enigmatic roles CPR bacteria may play in the cicada lifecycle.
CPR bacteria have been suggested as obligate symbionts because of genomic evidence and the lack of cultured representatives (7, 9). Notably, the isolation of CPR bacteria remains a challenge partly because of the inability to reconstruct in situ CPR‒host associations using traditional amplicon or metagenomic sequencing. As we noticed, epicPCR has been recently used to detect spatial associations between species via single-cell coisolation in picoliter emulsions, as demonstrated by the revelation of in situ phage‒bacteria associations (20). Here, we made unique use of epicPCR to detect episymbiotic associations of CPR with their hosts, providing direct guidance and facilitating the successful coisolation of obligate episymbiotic TM7i cells with their host. Likewise, epicPCR can detect microbe‒microbe associations in a high-throughput manner. Apart from obligate microbial epibionts in CPR, we may also use epicPCR to capture host associations of DPANN (29) and other epibiotic or endobiotic associations of free-living bacteria. In future work, we will further evaluate the full advantage of epicPCR in epibiotic association screening by designing epicPCR systems for different CPR or DPANN groups and performing large-scale targeted cultivation to recover more isolates from various habitats.
We affirmed that conserved T4P confers the essential motility for the epibiotic association of TM7i with its bacterial host J1, yet T4P may play a multifunctional role in the episymbiotic lifestyle. T4P is a class of pole-located extracellular filaments that exhibit twitching motility in bacterial cells (16, 17, 30). T4P existence has been proposed for the CPR domain (3, 10, 11, 31), and pili-like appendages have been observed on other CPR bacteria using electron microscopy (9, 11). However, the T4P-enabled motility of CPR has not yet been observed, and its function in CPR‒host associations has not been determined. The morphology, location, and T4P inhibition assays of this study provide early evidence for the presence and function of T4P in TM7, demonstrating its essential role in the episymbiotic lifestyle of TM7 and other CPR bacteria. Hitherto, we are unable to construct T4P-deficient mutants of TM7i by genetic modification and examine whether they exhibit lowered motility and survivability, because of the obligate episymbiotic lifestyle. In addition, whether T4P play a role in TM7‒host recognition is still unknown. Since T4P can sense surfaces and trigger retraction (32, 33), and T4P of some bacterial pathogens are involved in receptor recognition in pathogenic adhesion to human cells (34–36), TM7i may develop specialized mechanisms for selective host recognition using its T4P. Furthermore, the enrichment of DNA processing protein A (DprA) and competence protein EC (ComEC) in genomes of CPR bacteria also suggests the capacity of T4P-mediated DNA uptake in many CPR bacteria, which may serve as a potential material transfer mechanism (10). Based on the above reasons, we believe that T4P play a multifunctional role in supporting episymbiotic lifestyles of TM7 and other CPR bacteria; T4P-dependent motility may be universally crucial for the survival of CPR groups in various habitats, enabling cell translocation and adherence of planktonic CPR to their microbial hosts to initialize (epi)symbiosis (7, 9).
We also depicted the rapid and periodic budding-like lifecycle of TM7i with strict control of cell numbers, sizes, shapes, division patterns, and localization at the host cell surface (Figs. 4 and 5). The budding divisions associated with CPR were first proposed for TM7x based on fluorescence in situ hybridization (FISH) and SEM data (37). Nevertheless, our real-time observations raise the possibility that unequal divisions are widespread in the lifecycle of epibiotic TM7 bacteria. Considering the highly streamlined genome size of TM7i, its precisely regulated lifecycle is unusual and deserves further investigation. Importantly, we noted differences in the intervals between the initial and subsequent divisions. The extra time in the initial division may be attributed to the establishment of symbiosis and cell volume increment before division. However, intriguing questions remain: What mechanisms dominate the establishment of symbiosis? How can TM7i cells derive nutrients from host cells for growth and proliferation? How can TM7i cells generate progeny cells by asymmetric division? The answers to these questions may promote the understanding of intercellular decision-making in CPR‒host symbiosis and may revolutionize our view of microbial cell‒cell interactions. It is likely that the T4P-triggered epibiotic lifecycle described in this study may be widely adopted by members of the CPR with vast diversity and habitat distribution. Nevertheless, further studies are required to interpret the unique lifestyle of CPR symbionts, which will lead to a better understanding of their biological, ecological, and evolutionary importance in the biosphere.
Materials and Methods
Sample Collection and Enrichment.
CP was provided as traditional Chinese medicine from a pharmaceutical factory in Hangzhou, China. Approximately 0.5 g of CP was added to 50 mL of brain heart infusion (BHI) broth for enrichment over 2 d in a 28 °C incubation shaker with a shaking speed of 180 rpm. PCR assays with TM7-specific primers (F1-TM7-580F and 910R, SI Appendix, Table S3) were performed to determine whether TM7 was present in the enriched samples, as evaluated by visualization of products on 0.8% agarose gels by electrophoresis.
EpicPCR.
For the enrichment with TM7 detected positively by PCR during three continuous passages, epicPCR was performed as previously described (20) with modifications described below. Enriched samples were suspended in sterile water and vortexed. After SYTO9-labeling, cell densities were adjusted to 10‒20 × 106 cells per 30-μL based on fluorescence microscopic cell counting and dilution. After the sample was encapsulated into hydrogel beads, 20-μL beads were combined with 2× Phusion High-Fidelity PCR Master Mix with HF Buffer (1× final concentration; New England Biolabs, Ipswich, MA, USA), UltraPure molecular grade water (ThermoFisher, Waltham, MA, USA), F1-TM7-580F (1.0 μM final concentration), R1-F2-27F-910R (linking primer, 0.1 μM final concentration), R2-519R (1.0 μM final concentration), bovine serum albumin (0.02 mg/mL final concentration), and Tween 20 (0.2% v/v final concentration) to a final volume of 100 μL (SI Appendix, Table S3). PCR reagents were mixed with ABIL oil and emulsified by vortexing at a maximum speed.
Emulsions were loaded as 60-μL aliquots and amplified with the following conditions: denaturation at 94 °C for 5 min; 35 cycles of denaturation at 94 °C for 10 s, annealing at 54 °C for 30 s and extension at 72 °C for 30 s; and a final extension at 72 °C for 5 min; this step replicates an ~300 bp sequence from the TM7 genomes, with a universal 16S rRNA gene primer end that allowed initialization of host sequence amplification by PCR to produce an ~800 bp fused fragment.
Amplified PCR products were extracted using the StarPrep DNA Fragment Purification Kit (Genestar, Beijing, China) according to the manufacturer’s instructions. After extraction, nested PCR was performed using the fused DNA amplicons to produce an ~500 bp internal sequence with adapters for Illumina sequencing. Finally, purified amplicons were subjected to equimolar library pooling and paired-end sequencing on the MiSeq PE300 platform (Illumina, San Diego, CA, USA) according to standard protocols (Majorbio Bio-Pharm Technology, Shanghai, China).
Raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp (version 0.20.0) and merged with FLASH (version 1.2.7). Samples were distinguished according to barcodes and primers, while clean reads were further processed using QIIME2 (38). A Python script was used to separate amplicon sequences that contained both the TM7 primer and universal primer fragments. Separated sequences were then reanalyzed in QIIME2 to determine strain identities.
Bacterial Isolation and Growth.
L. aridicollis J1 was isolated in BHI broth supplemented with streptomycin (100 μg/mL) and nalidixic acid (30 μg/mL) incubated at 28 °C. An Alcaligenes sp. strain was also directly isolated from the cicada enrichment. L. aridicollis J1 and Alcaligenes sp. cultures were separately mixed with the filtrate from the enrichment sample (500 μL filtrate in 50 mL of culture broth). To evaluate whether TM7 cells can grow in the mixed cultures, PCR amplification of TM7-specific 16S rRNA genes was performed with TM7-specific primers (SI Appendix, Table S3). Agarose gel analysis and Sanger sequencing of amplicons were additionally performed to identify DNA products of PCR targeting TM7. For PCR-positive cocultures, the same assay was performed for consecutive 2-d passages to confirm the existence of a positive symbiosis.
For growth curve analysis, uninfected J1 was mixed with coculture filtrate that contained TM7i cells to test the impact of TM7i infection on J1 growth. A 2-d TM7i‒J1 coculture was filtered using a 0.22-μm filter to separate TM7i cells. Next, the J1 broth culture in the BHI medium was diluted to 1/100 from a culture with an OD600 = 0.4. Different volumes of TM7i filtrate (e.g., 0, 1, 10, and 100 μL) were then added to J1 suspension in a Honeycomb plate (Oy Growth Curves Ab Ltd, Helsinki, Finland), followed by the addition of BHI medium to reach a volume of 300 μL. The growth curves of TM7i and J1 were measured after incubating at 28 °C with shaking at 200 rpm using BioScreen (Oy Growth Curves Ab Ltd, Helsinki, Finland). Each group comprised three biological and three technical replicates.
Genome Sequencing and Assembly.
The complete genomes of TM7i and L. aridicollis J1 were sequenced using the PacBio Sequel and Illumina NovaSeq PE150 platforms. PacBio Sequel platform libraries were constructed for sequencing with an insert size of 10 kbp using the SMRTbell™ Template kit (Pacific Biosciences, Menlo Park, CA, USA). Next, a total of 1 μg of DNA per sample was used as input material for DNA sample preparations for Illumina NovaSeq library construction. Sequencing libraries were then generated using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) with index adapters added to identify sequences belonging to individual samples.
Preliminary library assembly was performed using the SMRT Link software program (version v5.0.1). To ensure the accuracy of subsequent analyses, low-quality reads (<500 bp) were filtered to obtain clean data. The automatic error correction function of the SMRT portal was used, and long reads (>6,000 bp) were selected as seed sequences, while shorter reads were aligned to the seed sequences using BLASR (39). The assembly was then corrected using the variant caller module of SMRT Link, while the arrow algorithm was used to correct and count variant sites in preliminary assembly results. The final assembly was corrected with the Illumina short-read data. The corrected assembly was used as the reference sequence for BLAST searches of the Illumina data. In addition, the final assembly required minimum base mass values of 20, minimum read depth values of 4, and maximum read depth values of 1,000. Assembly cyclization was conducted based on the overlap between head and tail regions.
Phylogenetic Analysis.
The 16S rRNA gene-based phylogenetic analysis of strain TM7i and host strain J1 was performed in the ARB program (40). Full-length 16S sequences were obtained from public databases, including eHOMD (41), EzBioCloud (42), and NCBI. We aligned all 16S rRNA reads with SINA v1.2.11 (43) and the SILVA database (release 138.1). Next, we used FastTree to build the maximum-likelihood phylogenetic tree by applying the GTR+G model with 100 bootstrap replicates (44). The phylogenomic tree was reconstructed using 86 core genes from all 723 publicly available genome sequences of TM7 bacteria taken from NCBI (by Oct. 2021), including 184 genomes containing 16S rRNA gene sequences. These 86 core genes were retrieved using the Up-to-date Bacterial Core Gene tool (45). A concatenated sequence of 86 genes was used to reconstruct the FastTree-based phylogenomic tree.
Multigenome Analysis of TM7 Genomes.
Six representative TM7 genomes were downloaded from the NCBI database and annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) by BlastKOALA for bacterial metabolic pathway analysis (46). Each KEGG ortholog annotated within the genomes was classified by Enzyme Nomenclature numbers. Multiple genome alignments were performed using progressive mauve with default parameters (47). For type IV pili (T4P) gene cluster analysis, representative genomes were annotated by BlastKOALA and the NCBI database. T4P genes annotated identically from two databases were extracted and presented.
Electron Microscopy Imaging of the TM7i‒J1 Symbiont.
The TM7i‒J1 symbiont was passaged and cocultured for 24 h and prepared for electron microscopy. Ultramicrotomy was employed in sectioning the symbiont cells for observation using TEM (transmission electron microscope, JEM-1400, JEOL, Tokyo, Japan). Ultrathin sections of approximately 70 nm thickness were stained with 2% uranyl acetate and lead citrate. Negative staining was used to observe the intertwinement of pili‒fimbriae of the symbiont under TEM. The diameters of TM7i T4P and J1 fimbriae were measured based on the thickness of pili perpendicular to pili direction. SEM was performed using a HITACHI SU8018 (Hitachi, Tokyo, Japan), before which critical point drying (EM CPD300, LEICA, Wetzlar, Germany) and gold coating were employed. SEM images were analyzed using ImageJ software (NIH, USA).
Pili Labeling and Super-Resolution Fluorescence Imaging.
Fluorescence probes (Macklin, Shanghai, China) of Alexa Fluor NHS esters were used to separately label surface proteins of TM7i and J1 cells, as previously described (22). Free TM7 cells were isolated from the filtrate of a 48-h TM7i‒J1 coculture by passing it through a 0.45-μm filter (Jinteng, Tianjin, China). J1 and TM7i cells were collected by centrifugation at 7,000 × g for 10 min. The cell pellets were then washed with 1 mL of M9 medium three times and resuspended in 0.5 mL of M9 medium. Then, 40 μL of 1 mg/mL fluorescence probes and 35 μL of 1 M sodium bicarbonate were added to label the cells and the pH was adjusted to 8.0‒8.5. The samples were then lightly shaken for 60 min at room temperature in the dark.
After labeling, cells were centrifuged and washed twice with 1 mL of M9 medium and resuspended in M9 to achieve an ideal cell density for imaging. We added 2-μL cell suspensions of pure TM7i or TM7i‒J1 coculture onto Nunc™ glass-bottom dishes (ThermoFisher, Waltham, MA, USA). A gellan gum pad (0.2% w/v) of M9 medium was then placed onto the cell suspension. Fluorescence imaging was performed using a Super-Resolution Microscope System (N-SIMS, Nikon, Tokyo, Japan) with a 100× objective lens (Plan Fluo, NA 1.49, oil immersion) that can achieve 115 nm spatial resolution. Images were taken with an exposure time of 500 ms in 10-s intervals. Line lengths connecting the center of TM7i cells and pili tips were measured to measure pili lengths in the fluorescence images using ImageJ. Twitch lengths were measured based on line length between the center of movement beginning and the ends. Shifted angles were calculated that represent differences in the pili and twitch directions.
T4P Inhibition Assays.
Gellan gum pads (0.2% w/v) of BHI medium containing 200 μM quercetin (dissolved in DMSO) or an equivalent volume of DMSO were freshly made prior to experiments. TM7i cells were harvested as previously discussed and mixed with J1 cells during exponential growth (OD600 = 0.8). Then, quercetin or DMSO was added to the cell suspension and incubated for 20 min. For the quercetin-treated recovery group, the cell suspension incubated with quercetin was harvested by centrifugation at 14000 × g for 5 min and resuspended in BHI medium for a 20-min incubation. After incubation, the cell suspension was loaded under the gel pad over a glass coverslip for bright field observation. The experiment was repeated at least three times. Movies were captured at 8 fps with a 50-ms exposure time using an Eclipse Ti microscope (Nikon, Tokyo, Japan) equipped with a 100× objective lens (Plan Fluo, NA 1.45, oil immersion, Nikon, Tokyo, Japan).
For TM7i growth assay under quercetin treatments, the preparation of coculture samples has a slight difference considering whether TM7i cells were attached or not. For TM7i growth in different concentration of quercetin, 200-μL TM7i filtrate (prepared as mentioned before) were transferred to 50 mL BHI medium supplemented with 1% J1 cells during late-exponential growth (OD600 = 1.2) for a 20-min incubation at 28 °C and a shaking speed of 180 rpm. This incubation allows TM7i to attach to the host. After incubation, 50 μM, 100 μM, or 200 μM quercetin, or an equivalent volume of DMSO as a blank control was added to respective groups. Each group had three biological replicates. To inhibit T4P of TM7i cells before attachment, 200-μL TM7i filtrate was directly added to 50 mL BHI culture containing 200 μM quercetin or an equivalent volume of DMSO as the control for a 20-min treatment. After the incubation, 1% J1 was added to the culture to initiate coculture. Each group had three biological replicates. The cultured samples were subjected to quantitative PCR (qPCR) with TM7-specific primers to evaluate TM7i growth in the samples. The reactions consisted of 0.5 μL of cultured samples, 0.5 μL of each primer, 12.5 μL of 2× Ssofast Evagreen Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and water added to a final volume of 25 μL. Negative controls (J1 cultured sample or water) were used to exclude nonspecific amplification of DNA contamination. All samples and negative controls were amplified in triplicate. qPCRs were performed using Quantagene q225 (Kubo Technology, Beijing, China).
Supplementary Material
Appendix 01 (PDF)
Inhibition of TM7i T4P affects cell motility. The bacterial suspension containing pure J1 or J1 mixed with TM7i was sandwiched within a thin layer between a gel pad and a cover glass. TM7i cells were spotted and tracked to show motility. TM7i cells exhibited motility in the presence of J1 cells. The T4P inhibitor reversibly inhibited the motility of TM7i.
T4P trigger TM7i motility and interaction with its host. TM7i and J1 cells were separately labeled with red and green fluorescent NHS ester probes and sandwiched within a thin layer between a gel pad and a cover glass for dynamic structured illumination microscopy (SIM) imaging. 1) A TM7i cell dynamically extruded and retracted its pili, which is typical for T4P; 2) T4P triggered the continuous translocation of the TM7i cell on the surface; 3) A TM7i cell moved toward a J1 cell with T4P-mediated motility and became sessile on the J1 cell.
Inhibition of TM7i T4P affects host infection. A 3-h long-term microscopic imaging of the TM7i–J1 symbiont was carried out. The bacterial suspension containing pure J1 or mixed with TM7i was sandwiched within a thin layer between a gel pad and a cover glass. TM7i cells exhibited active motility after division. The T4P inhibitor immobilized free-floating and progeny TM7i cells, preventing them from adhering to potential host cells.
Acknowledgments
This work was funded by the National Key Research & Development Program of China (2021YFC2301000, 2021YFA0717000, and 2021YFC2103300) and the National Natural Science Foundation of China (92251302, 91951103, 31970091, 21822408). We thank Songnian Hu, Linqi Wang, Yingfeng Luo, and Li Huang from the Institute of Microbiology Chinese Academy of Sciences, for insightful discussions and suggestions. We thank Chunli Li, Jingnan Liang, and Lei Su from the Institutional Center for Shared Technologies and Facilities for their help with microscopic imaging.
Author contributions
B.X. and W.D. designed research; B.X., J.W., Z.W., and D.C. performed research; B.X. and W.D. contributed new reagents/analytic tools; B.X., J.W., Y.N., B.H., and W.D. analyzed data; and B.X., J.W., Y.N., J.T., D.C., X.-L.W., and W.D. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. The genomes of TM7i and J1 have been deposited in NCBI GenBank under accession codes CP075337 and CP075339, respectively. The raw data generated from epicPCR have been deposited in NCBI Sequence Read Archive (SRA) database (BioSample Accession Number: SAMN19276902).
Supporting Information
References
- 1.Parks D. H., et al. , Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Brown C. T., et al. , Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Luef B., et al. , Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 6, 6372 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Castelle C. J., et al. , Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol. 16, 629–645 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Castelle C. J., Banfield J. F., Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 172, 1181–1197 (2018). [DOI] [PubMed] [Google Scholar]
- 7.He X., et al. , Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. U.S.A. 112, 244–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross K. L., et al. , Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He C., et al. , Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat. Microbiol. 6, 354–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meheust R., Burstein D., Castelle C. J., Banfield J. F., The distinction of CPR bacteria from other bacteria based on protein family content. Nat. Commun. 10, 4173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batinovic S., Rose J. J. A., Ratcliffe J., Seviour R. J., Petrovski S., Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat. Microbiol. 6, 703–711 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Tian J., et al. , Acquisition of the arginine deiminase system benefits epiparasitic Saccharibacteria and their host bacteria in a mammalian niche environment. Proc. Natl. Acad. Sci. U.S.A. 119, e2114909119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utter D. R., He X., Cavanaugh C. M., McLean J. S., Bor B., The saccharibacterium TM7x elicits differential responses across its host range. ISME J. 14, 3054–3067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira D., Zivanovic Y., Lopez-Archilla A. I., Iniesto M., Lopez-Garcia P., Reductive evolution and unique predatory mode in the CPR bacterium Vampirococcus lugosii. Nat. Commun. 12, 2454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufrene Y. F., Persat A., Mechanomicrobiology: How bacteria sense and respond to forces. Nat. Rev. Microbiol. 18, 227–240 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Mattick J. S., Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Eriksson J., et al. , Characterization of motility and piliation in pathogenic Neisseria. BMC Microbiol. 15, 92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellison C. K., et al. , Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 3, 773–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer S. J., et al. , Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 10, 427–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dye K. J., Vogelaar N. J., Sobrado P., Yang Z., High-throughput screen for inhibitors of the type IV pilus assembly ATPase PilB. mSphere 6, e121–e129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth R., et al. , The mode of cell wall growth in selected archaea is similar to the general mode of cell wall growth in bacteria as revealed by fluorescent dye analysis. Appl. Environ. Microbiol. 77, 1556–1562 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinke C., et al. , Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Gharechahi J., et al. , Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 15, 1108–1120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N., et al. , Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13, R42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stork N. E., How many species of insects and other terrestrial arthropods are there on Earth? Annu. Rev. Entomol. 63, 31–45 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Basset Y., Lamarre G. P. A., Toward a world that values insects. Science 364, 1230–1231 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jing T. Z., Qi F. H., Wang Z. Y., Most dominant roles of insect gut bacteria: Digestion, detoxification, or essential nutrient provision? Microbiome 8, 38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai H. D., et al. , Insight into the symbiotic lifestyle of DPANN archaea revealed by cultivation and genome analyses. Proc. Natl. Acad. Sci. U.S.A. 119, e2115449119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ottow J. C., Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 29, 79–108 (1975). [DOI] [PubMed] [Google Scholar]
- 31.McLean J. S., et al. , Acquisition and adaptation of ultra-small parasitic reduced genome bacteria to mammalian hosts. Cell Rep. 32, 107939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison C. K., et al. , Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Toole G. A., Wong G. C., Sensational biofilms: Surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Guennec L., et al. , Receptor recognition by meningococcal type IV pili relies on a specific complex N-glycan. Proc. Natl. Acad. Sci. U.S.A. 117, 2606–2612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virion Z., et al. , Sialic acid mediated mechanical activation of β2 adrenergic receptors by bacterial pili. Nat. Commun. 10, 4752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dos S. S. I., et al. , Meningococcal disease: A paradigm of type-IV pilus dependent pathogenesis. Cell Microbiol. 22, e13185 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Bor B., et al. , Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont Actinomyces. Microb. Ecol. 71, 243–255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolyen E., et al. , Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaisson M. J., Tesler G., Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): Application and theory. BMC Bioinformatics 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig W., et al. , ARB: A software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Escapa I. F., et al. , New insights into human nostril microbiome from the expanded Human Oral Microbiome Database (eHOMD): A resource for species-level identification of microbiome data from the aerodigestive tract. mSystems 3, e118–e187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon S. H., et al. , Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse E., Peplies J., Glockner F. O., SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Na S. I., et al. , UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Kanehisa M., Sato Y., Morishima K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Darling A. C., Mau B., Blattner F. R., Perna N. T., Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Inhibition of TM7i T4P affects cell motility. The bacterial suspension containing pure J1 or J1 mixed with TM7i was sandwiched within a thin layer between a gel pad and a cover glass. TM7i cells were spotted and tracked to show motility. TM7i cells exhibited motility in the presence of J1 cells. The T4P inhibitor reversibly inhibited the motility of TM7i.
T4P trigger TM7i motility and interaction with its host. TM7i and J1 cells were separately labeled with red and green fluorescent NHS ester probes and sandwiched within a thin layer between a gel pad and a cover glass for dynamic structured illumination microscopy (SIM) imaging. 1) A TM7i cell dynamically extruded and retracted its pili, which is typical for T4P; 2) T4P triggered the continuous translocation of the TM7i cell on the surface; 3) A TM7i cell moved toward a J1 cell with T4P-mediated motility and became sessile on the J1 cell.
Inhibition of TM7i T4P affects host infection. A 3-h long-term microscopic imaging of the TM7i–J1 symbiont was carried out. The bacterial suspension containing pure J1 or mixed with TM7i was sandwiched within a thin layer between a gel pad and a cover glass. TM7i cells exhibited active motility after division. The T4P inhibitor immobilized free-floating and progeny TM7i cells, preventing them from adhering to potential host cells.
Data Availability Statement
All study data are included in the article and/or SI Appendix. The genomes of TM7i and J1 have been deposited in NCBI GenBank under accession codes CP075337 and CP075339, respectively. The raw data generated from epicPCR have been deposited in NCBI Sequence Read Archive (SRA) database (BioSample Accession Number: SAMN19276902).