Significance
Bacteria use the cell-to-cell communication process called quorum sensing to orchestrate group behaviors. Quorum sensing relies on extracellular molecules called autoinducers. Bacteria-infecting viruses (phages) can possess homologs of bacterial quorum-sensing receptors that detect autoinducers to control lysis–lysogeny transitions. We show that a phage LuxR-type quorum-sensing receptor is activated by the autoinducer produced by its host bacterium and is inhibited by noncognate autoinducers made by bacteria that naturally coexist with the phage’s host and by a synthetic quorum-sensing inhibitor. Our findings demonstrate that microbial community composition, mediated through quorum-sensing communication, influences phage lysis–lysogeny transitions. These results deepen the understanding of host–phage interactions in communities and could inspire new phage-specific quorum-sensing interventions.
Keywords: phage, quorum sensing, LuxR
Abstract
Viruses that infect bacteria, called phages, shape the composition of bacterial communities and are important drivers of bacterial evolution. We recently showed that temperate phages, when residing in bacteria (i.e., prophages), are capable of manipulating the bacterial cell-to-cell communication process called quorum sensing (QS). QS relies on the production, release, and population-wide detection of signaling molecules called autoinducers (AI). Gram-negative bacteria commonly employ N-acyl homoserine lactones (HSL) as AIs that are detected by LuxR-type QS receptors. Phage ARM81ld is a prophage of the aquatic bacterium Aeromonas sp. ARM81, and it encodes a homolog of a bacterial LuxR, called LuxRARM81ld. LuxRARM81ld detects host Aeromonas-produced C4-HSL, and in response, activates the phage lytic program, triggering death of its host and release of viral particles. Here, we show that phage LuxRARM81ld activity is modulated by noncognate HSL ligands and by a synthetic small molecule inhibitor. We determine that HSLs with acyl chain lengths equal to or longer than C8 antagonize LuxRARM81ld. For example, the C8-HSL AI produced by Vibrio fischeri that coexists with Aeromonads in aquatic environments, binds to and inhibits LuxRARM81ld, and consequently, protects the host from lysis. Coculture of V. fischeri with the Aeromonas sp. ARM81 lysogen suppresses phage ARM81ld virion production. We propose that the cell density and species composition of the bacterial community could determine outcomes in bacterial-phage partnerships.
Bacteria communicate and orchestrate collective behaviors using a process called quorum sensing (QS) (1). QS relies on the production, release, accumulation, and group-wide detection of molecules called autoinducers (AI). Bacteria commonly live in environments containing multiple bacterial species, and thus, different blends of QS AIs can be present. Homoserine lactones (HSL) represent a common class of QS AIs produced and detected by Gram-negative bacteria. HSL AIs possess different modifications at the C3 position and they harbor variable acyl chain lengths. LuxR-type and LuxN-type QS receptors detect HSL AIs (1). Some of these QS receptors display strict specificity for a cognate HSL AI, while others are promiscuous in HSL ligand detection (2–8). For instance, Vibrio harveyi LuxN is exclusively activated by its partner 3OHC4-HSL ligand and noncognate HSLs possessing longer acyl tails act as competitive antagonists (7). Such antagonism is thought to be a mechanism QS bacteria use to monitor and react to the presence of competing bacterial species. Specifically, species whose QS receptors are antagonized by noncognate AIs repress their QS outputs when the noncognate compounds are present, thereby avoiding leakage of QS-controlled public goods to competitors (7, 9).
Beyond QS driving interactions within and between bacterial species, we recently discovered that linear plasmid-like phages can encode LuxR-type QS receptors that detect the HSL AI produced by the bacterial host. For example, Aeromonas sp. ARM81 possesses a prophage, called ARM81ld, that encodes luxRARM81ld. LuxRARM81ld binds to and is solubilized by C4-HSL, the AI made by its Aeromonas sp. ARM81 host (10, 11). Together with a partner XREARM81ld DNA-binding protein, the LuxRARM81ld-C4-HSL complex activates transcription of a counter-oriented gene encoding a small ORF (smORFARM81ld) (10). Production of smORFARM81ld launches the phage ARM81ld lytic program, which causes host-cell lysis (10). Thus, monitoring its host’s QS status, via C4-HSL, allows phage ARM81ld to transition from its lysogenic to its lytic lifestyle and to disseminate at high host-cell density, presumably a condition that maximizes the probability of subsequent successful infection.
The finding that noncognate HSLs are inhibitory to some bacterial LuxR-type and LuxN-type receptors is intriguing because it enables bacteria to take a census of and react to nonkin bacteria in the vicinity. Whether phages that possess QS receptors also detect and respond differently to non-host-produced AIs is unknown. Here, we assess the effects of noncognate AIs on lifestyle choices made by phage ARM81ld. We demonstrate that microbial community composition, mediated through the different AIs produced, has a dramatic influence on phage ARM81ld lysis–lysogeny transitions. These results have potentially far-reaching implications for how we understand host–phage interactions in complex communities and could lead to the development of new classes of QS-targeted interventions that are phage- rather than bacteria-specific.
Results
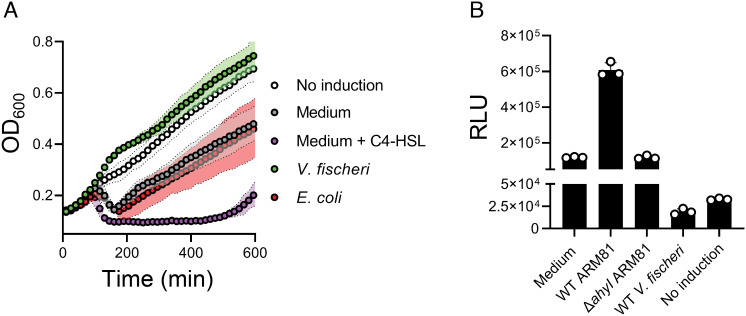
Aeromonads are known to exist in mixed microbial consortia with other QS bacteria, particularly marine Vibrios. Vibrio fischeri is one such well-studied QS bacterium. It produces 3OC6-HSL and C8-HSL (12, 13), two AIs that have longer acyl tails than the C4-HSL AI to which phage ARM81ld responds. We verified that C4-HSL is the product of the Aeromonas sp. ARM81 AhyI AI synthase using an established bioassay (SI Appendix, Fig. S1A) (10, 14). To explore the effects of signaling molecules that Aeromonas sp. ARM81 encounters in communities but that it itself does not produce on phage ARM81ld activity, we constructed an Aeromonas sp. ARM81 lysogen that was incapable of producing C4-HSL to eliminate any phage activity that occurs in response to the endogenously produced AI. We used this strain (designated ΔahyI Aeromonas sp. ARM81) in all of our assays. We introduced anhydrotetracyline (aTc)-inducible xreARM81ld-luxRARM81ld on a plasmid into ΔahyI Aeromonas sp. ARM81. We induced production of XREARM81ld-LuxRARM81ld and administered cell-free culture fluids collected from wild-type (WT) V. fischeri. As a control, we administered cell-free fluids from WT Escherichia coli, which does not produce HSL AIs. Important for our strategy is that we used a concentration of the aTc inducer sufficient to drive an intermediate level of phage-directed host-cell lysis in the absence of exogenous ligand, thus enabling us to detect increased or decreased cell death (Fig. 1A). Strikingly, cell-free culture fluids from WT V. fischeri completely suppressed cell lysis (Fig. 1A). By contrast, cell-free culture fluids from WT E. coli did not affect cell lysis relative to the medium alone (Fig. 1A).
Fig. 1.
V. fischeri cell-free culture fluids inhibit XREARM81ld-LuxRARM81ld transcriptional activity. (A) Growth of ΔahyI Aeromonas sp. ARM81 carrying aTc-inducible xreARM81ld-luxRARM81ld in a medium lacking aTc (white; No induction), a medium containing 0.1 ng mL−1 aTc supplemented with the medium alone (gray), the medium and 20 μM C4-HSL (purple), cell-free culture fluids from WT V. fischeri (3OC6-HSL+ and C8-HSL+; green), or cell-free culture fluids from E. coli (a non-HSL producer; red). (B) PsmORFARM81ld-lux expression from E. coli carrying aTc-inducible xreARM81ld-luxRARM81ld grown in a medium containing 50 ng mL−1 aTc supplemented with the medium alone, cell-free culture fluids from WT Aeromonas sp. ARM81 (C4-HSL+), cell-free culture fluids from ΔahyI Aeromonas sp. ARM81 (C4-HSL-), cell-free culture fluids from WT V. fischeri (3OC6-HSL+ and C8-HSL+), or in a medium lacking aTc (No induction). RLU denotes relative light units. Data are represented as mean ± SD with n = 3 biological replicates.
Given that XREARM81ld-LuxRARM81ld-mediated transcription of smORFARM81ld drives host-cell lysis by phage ARM81ld (10), we hypothesized that the inhibitory effect of V. fischeri culture fluids occurred through suppression of XREARM81ld-LuxRARM81ld transcriptional activity. To explore this possibility, we used recombinant E. coli harboring a PsmORFARM81ld-lux transcriptional reporter and aTc-inducible xreARM81ld-luxRARM81ld, thus excluding all other Aeromonas sp. ARM81 host and phage components from the system. Consistent with our understanding that C4-HSL activates the XREARM81ld-LuxRARM81ld pathway and occurs at concentrations relevant to that produced by Aeromonas sp. ARM81 in nature, administration of cell-free culture fluids from WT Aeromonas sp. ARM81 increased PsmORFARM81ld-lux light production fivefold over the medium alone, whereas cell-free culture fluids from ΔahyI Aeromonas sp. ARM81 had no effect (Fig. 1B). Importantly, cell-free culture fluids from WT V. fischeri inhibited light production sixfold, indeed to levels below that when expression of xreARM81ld-luxRARM81ld was not induced (Fig. 1B). Thus, V. fischeri culture fluids harbor a factor(s) that prevents phage-mediated cell lysis by inhibiting the phage-encoded XREARM81ld-LuxRARM81ld pathway.
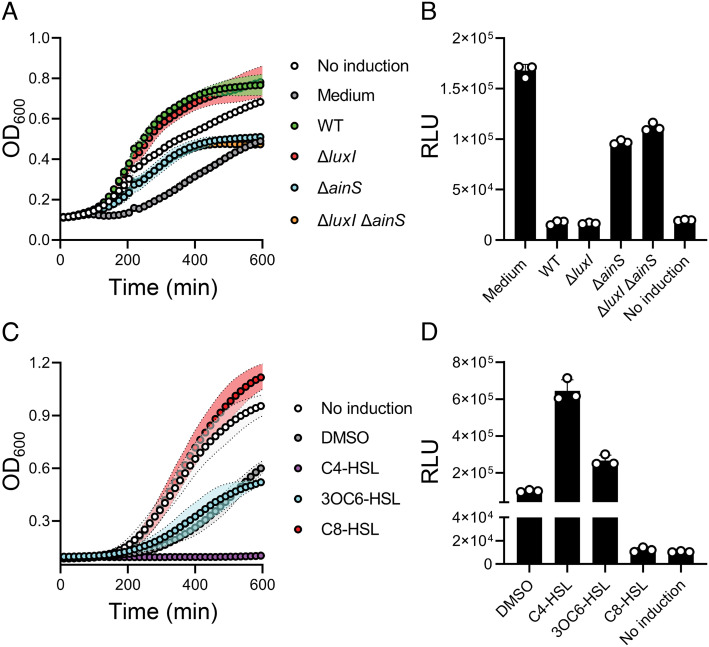
As noted, V. fischeri makes two HSL AIs, 3OC6-HSL and C8-HSL. To test whether the inhibition of Aeromonas sp. ARM81 lysis shown in Fig. 1A is due to one or both of these AIs, we administered cell-free culture fluids harvested from ΔluxI V. fischeri, which makes no 3OC6-HSL, ΔainS V. fischeri which makes no C8-HSL, and ΔluxI ΔainS V. fischeri which makes neither AI to the Aeromonas sp. ARM81 lysogen (15–17). Identical to the case of WT V. fischeri cell-free culture fluids, addition of cell-free culture fluids from ΔluxI V. fischeri inhibited Aeromonas sp. ARM81 lysis. By contrast, cell-free culture fluids from ΔainS or ΔluxI ΔainS V. fischeri only drove basal-level lysis of Aeromonas sp. ARM81, i.e., to the same level as when the medium alone was added (Fig. 2A). Consistent with this result, WT and ΔluxI culture fluids decreased PsmORFARM81ld-lux output 10-fold, while ΔainS and ΔluxI ΔainS culture fluids had less than a twofold effect (Fig. 2B). These findings suggest that the V. fischeri AIs, primarily C8-HSL, antagonize LuxRARM81ld. Indeed, administration of synthetic C8-HSL to the Aeromonas sp. ARM81 lysogen inhibited cell lysis and decreased reporter output eightfold (Fig. 2 C and D, respectively). By comparison, synthetic 3OC6-HSL had no effect on lysis and a modest activating effect (2.5-fold) on PsmORFARM81ld expression (Fig. 2 C and D, respectively). Maximum cell lysis and maximum activation of the reporter by C4-HSL are shown as controls (Fig. 2 C and D, respectively). Likely, C8-HSL is a more potent antagonist than 3OC6-HSL is an agonist of LuxRARM81ld. Thus, C8-HSL is the V. fischeri AI that prevents the induction of the Aeromonas sp. ARM81 prophage.
Fig. 2.
The V. fischeri C8-HSL AI antagonizes LuxRARM81ld. (A) Growth of ΔahyI Aeromonas sp. ARM81 carrying aTc-inducible xreARM81ld-luxRARM81ld in a medium lacking aTc (white; No induction), a medium containing aTc supplemented with the medium alone (gray), cell-free culture fluids from WT (3OC6-HSL+ and C8-HSL+; green), ΔluxI (3OC6-HSL− and C8-HSL+; red), ΔainS (3OC6-HSL+ and C8-HSL−; cyan), or ΔluxI ΔainS (3OC6-HSL− and C8-HSL−; orange) V. fischeri. (B) PsmORFARM81ld-lux expression from E. coli carrying aTc-inducible xreARM81ld-luxRARM81ld grown in a medium containing aTc supplemented with the medium alone, cell-free culture fluids from WT (3OC6-HSL+ and C8-HSL+) V. fischeri ΔluxI (3OC6-HSL− and C8-HSL+) V. fischeri ΔainS (3OC6-HSL+ and C8-HSL−) V. fischeri ΔluxI ΔainS (3OC6-HSL− and C8-HSL−) V. fischeri, or in a medium lacking aTc (No induction). (C) Growth of ΔahyI Aeromonas sp. ARM81 carrying aTc-inducible xreARM81ld-luxRARM81ld in a medium lacking aTc (white; No induction), the medium containing aTc supplemented with DMSO (gray), C4-HSL (purple), 3OC6-HSL (cyan) or C8-HSL (red). All HSLs were supplied at 20 µM. (D) PsmORFARM81ld-lux expression from E. coli carrying aTc-inducible xreARM81ld-luxRARM81ld grown in a medium containing aTc supplemented with DMSO, C4-HSL, 3OC6-HSL, C8-HSL, or in a medium lacking aTc (No induction). HSL concentrations as in (C). Data are represented as mean ± SD with n = 3 biological replicates. RLU as in Fig. 1B (B, D). aTc; 0.1 ng mL−1 (A, C), 50 ng mL−1 (B) and 25 ng mL−1 (D).
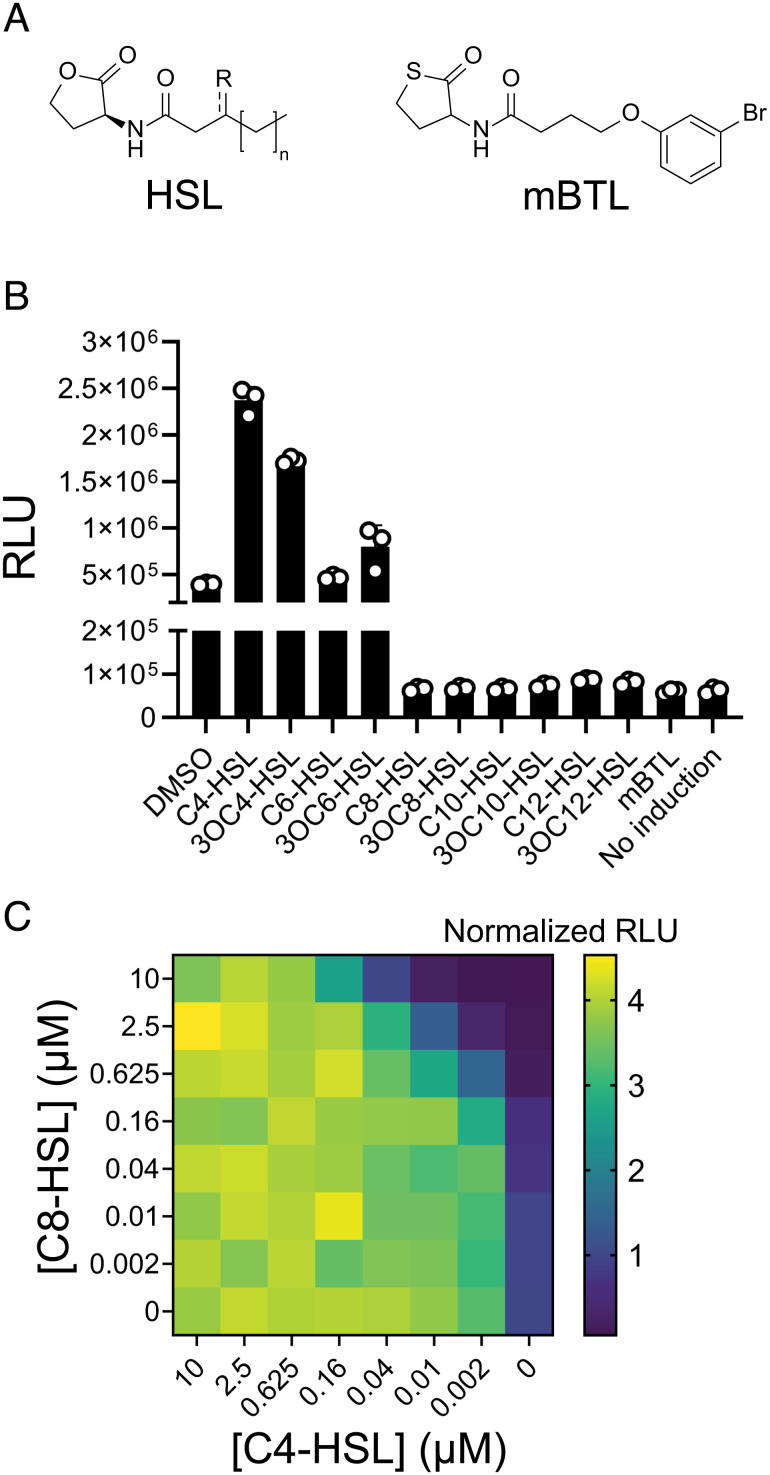
Despite our finding that C4-HSL promotes and C8-HSL prevents XREARM81ld-LuxRARM81ld-driven host-cell lysis, both HSLs solubilize LuxRARM81ld (SI Appendix, Fig. S1B) (11). We thus wondered what features of HSL ligands distinguish inhibition from activation of LuxRARM81ld. To probe this question, we administered a panel of synthetic HSLs to the E. coli PsmORFARM81ld-lux reporter (Fig. 3 A and B). Light output increased in the presence of C4-HSL, 3OC4-HSL, and 3OC6-HSL (Fig. 3B). C6-HSL had no effect (Fig. 3B). Conversely, HSLs with chain lengths of C8 or longer reduced PsmORFARM81d expression fivefold to sevenfold relative to the basal activity generated by the presence of XREARM81ld and LuxRARM81ld (Fig. 3B). We also assayed the compound meta-bromo-thiolactone (mBTL, Fig. 3A), a synthetic inhibitor of LuxR-driven QS (18). Similar to the longer acyl chain HSL AIs, mBTL inhibited PsmORFARM81ld-lux activity 6.5-fold (Fig. 3B). Finally, simultaneous administration of the C4-HSL agonist and the C8-HSL antagonist revealed that LuxRARM81ld is highly sensitive to and prefers C4-HSL, but C8-HSL can compete for binding when provided at 60- to 250-fold higher concentrations (Fig. 3C). This result is consistent with the finding that C4-HSL solubilizes LuxRARM81ld more effectively than C8-HSL (SI Appendix, Fig. S1B) (11).
Fig. 3.
Noncognate HSL AIs with chain lengths of C8 or longer, and the synthetic compound mBTL, inhibit LuxRARM81ld activity. (A) General structure of an HSL AI (R = O or H; n = 0, 2, 4, 6, or 8) and the structure of the synthetic compound mBTL. (B) PsmORFARM81ld-lux expression from E. coli carrying aTc-inducible xreARM81ld-luxRARM81ld grown in a medium containing aTc supplemented with DMSO or the indicated compounds or in a medium lacking aTc (No induction). HSL concentrations as in Fig. 2C. (C) PsmORFARM81ld-lux expression from E. coli carrying aTc-inducible xreARM81ld-luxRARM81ld grown in a medium containing aTc and the indicated concentrations of C4-HSL and C8-HSL. Data are shown as a heatmap. Normalized RLU refers to the RLU of each sample relative to the RLU of the sample administered DMSO only, which was set to 1.0. Data are represented as mean ± SD with n = 3 biological replicates (B) or as mean with n = 2 biological replicates (C). RLU as in Fig. 1B (B, C). aTc; 25 ng mL−1 (B, C).
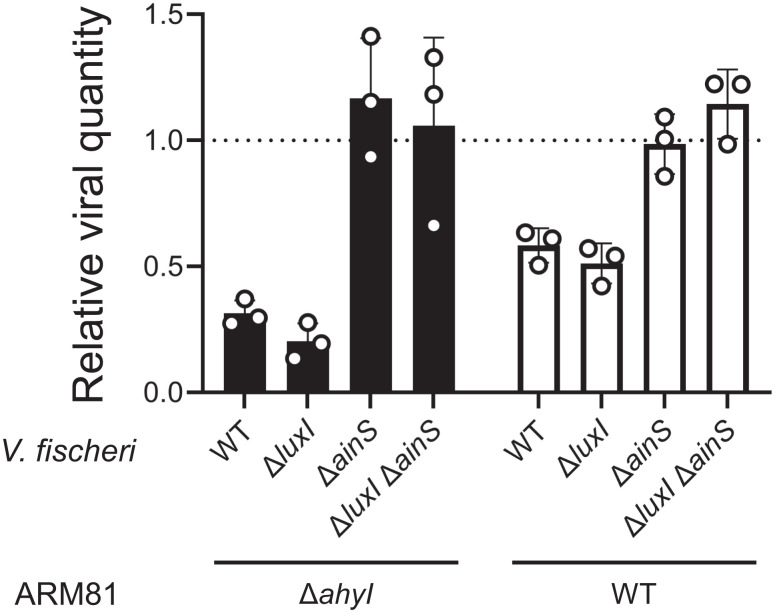
Our above results imply that in mixed-species communities, whether the Aeromonas sp. ARM81 lysogen is killed by or protected from prophage induction could depend on whether other species in the vicinal community are QS-proficient bacteria or not, and if the former, on what particular HSLs they produce. To garner evidence for this notion, we grew the ΔahyI Aeromonas sp. ARM81 lysogen harboring inducible xreARM81ld-luxRARM81ld, alone or in combination with either WT, ΔluxI, ΔainS, or ΔluxI ΔainS V. fischeri. Quantitation of the ARM81ld phage-to-host ratio revealed that the viral load was approximately fourfold lower when ΔahyI Aeromonas sp. ARM81 was grown in coculture with V. fischeri that produce C8-HSL (WT and ΔluxI V. fischeri) than when ΔahyI Aeromonas sp. ARM81 was grown in monoculture or in coculture with V. fischeri strains that lacked the ability to produce C8-HSL (ΔainS or ΔluxI ΔainS V. fischeri) (Fig. 4, black bars). A similar trend but, not surprisingly, with a reduced effect occurred when the WT Aeromonas sp. ARM81 lysogen that produces endogenous C4-HSL was used (Fig. 4, white bars). Together, these results indicate that, under the conditions tested, the presence of V. fischeri suppresses induction of phage ARM81ld and diminishes release of phage particles, including from the C4-HSL producing (WT) Aeromonas sp. ARM81 lysogen. The inhibitory effect relies on V. fischeri production of C8-HSL and operates by C8-HSL antagonism of the phage-encoded QS receptor in the neighboring Aeromonas sp. ARM81 lysogen. Regarding consequences to V. fischeri, the other participant in our experiments, while not tested here, earlier reports suggest that Aeromonas-produced C4-HSL does not alter the V. fischeri QS output (19, 20).
Fig. 4.
V. fischeri that produces C8-HSL prevents phage ARM81ld-driven viral production in coculture with the ΔahyI and WT ARM81 lysogens. Detection of phage ARM81ld obtained from cultures of ΔahyI Aeromonas sp. ARM81 (black bars) or WT Aeromonas sp. ARM81 (white bars) carrying aTc-inducible xreARM81ld-luxRARM81ld that were grown in coculture with the indicated V. fischeri strains. Relative viral quantity is the amount of phage ARM81ld DNA in a sample compared with the amount of Aeromonas sp. ARM81 host DNA. Data are represented as mean ± SD with n = 3 biological replicates and n = 3 technical replicates. aTc; 0.1 ng mL−1.
Discussion
Here, we show that the outcome of the phage ARM81ld lysis–lysogeny transition can be altered by other bacterial species in the community that engage in QS and produce noncognate HSL AIs. Our findings suggest that phage ARM81ld monitors its host’s QS status and also the cell density and species composition of the vicinal community. The information it garners exists in the form of QS chemical cues, and it integrates that information into its lysis–lysogeny decision-making mechanism. We propose that, in communities in which multiple bacterial species and phages coexist, detection of a variety of HSL AIs could benefit the phage or the host, and which entity receives the benefit likely depends on the particular circumstances. First, regarding a possible benefit to the phage: Antagonism of LuxRARM81ld by noncognate HSL AIs could prevent premature launch of the phage ARM81ld lytic cascade, and release of viral particles under conditions where Aeromonads make up only a minority of a mixed-species community. Because the phage ARM81ld host range is likely limited to Aeromonads, this mechanism could prevent phage ARM81ld from launching its lytic cycle when the likelihood of released virions encountering suitable bacteria to infect is low. Alternatively, regarding a possible benefit to the Aeromonas host: The production of noncognate AIs by other members of the vicinal bacterial community could suppress QS-mediated induction of the Aeromonas sp. ARM81 lysogen, curb release of phage ARM81ld virions, and thereby protect existing Aeromonads harboring prophages from killing as well as protect neighboring susceptible Aeromonads from infection. While it remains to be tested, the possibility exists that Vibrios receive benefits when lysis of Aeromonas is prevented. As examples, Aeromonads could produce public goods that Vibrios can exploit, or possibly, stable microbial communities require Aeromonads to be present in sufficient numbers.
Beyond exploring the effects of noncognate AIs on phages in bacterial communities, we demonstrated that the synthetic mBTL compound antagonizes XREARM81ld-LuxRARM81ld transcriptional activity, and in doing so, prevents lysis of Aeromonas sp. ARM81. This finding suggests that synthetic molecules designed against bacterial QS systems may have significant and unintended consequences on prophages and other mobile genetic elements that may not be present in all isolates. As we continue to uncover diverse and unexpected roles phages play in biology, the ability to develop small molecules to manipulate phage-specific rather than bacteria-specific activities may be useful. Discovering and characterizing new phage regulatory systems, like that of phage ARM81ld, could be an important step for consideration in advancing this goal.
Materials & Methods
Bacterial Strains and Growth Conditions.
E. coli strains were grown with aeration in Luria–Bertani (LB-Miller, BD-Difco) broth. Aeromonas sp. ARM81 and V. fischeri strains were grown in LB with 3% NaCl. All strains were grown at 30 °C. Strains used in the study are listed in SI Appendix, Table S1. Unless otherwise noted, the following antibiotics and concentrations were used: 100 μg mL−1 ampicillin (Amp, Sigma), 50 μg mL−1 kanamycin (Kan, GoldBio), and 5 μg mL−1 chloramphenicol (Cm, Sigma). Inducers were used as follows: E. coli: 200 μM isopropyl beta-D-1-thiogalactopyranoside (IPTG, GoldBio), 0.1% L-arabinose (Sigma), and 50 ng mL−1 or 25 ng mL−1 anhydrotetracycline (aTc, Clontech) and Aeromonas sp. ARM81: 0.1 ng mL−1 aTc. HSL AIs were supplied at a final concentration of 20 μM, unless otherwise indicated.
Cloning Techniques.
All primers and dsDNA (gene blocks) used for plasmid construction and qPCR, listed in SI Appendix, Table S2, were obtained from Integrated DNA Technologies. Gibson assembly, and traditional cloning methods were employed for all constructions. PCR with iProof was used to generate insert and backbone DNA. Gibson assembly relied on the HiFi DNA assembly mix (NEB). All enzymes used in cloning were obtained from NEB. Plasmids used in this study are listed in SI Appendix, Table S3. Transfer of plasmids into Aeromonas sp. ARM81 was carried out by conjugation followed by selective plating on LB supplemented with Kan and Cm.
Lysis and Reporter Assays.
For ΔahyI Aeromonas sp. ARM81 growth and lysis assays, overnight cultures were back-diluted 1:50 with fresh medium and appropriate antibiotics before being dispensed into 96-well plates (Corning Costar 3904). Cultures were grown in the plates for 60 min prior to administration of aTc, cell-free culture fluids, HSLs, or mBTL. E. coli reporter assays were carried out as above with the following modifications: Overnight cultures were back-diluted 1:100 with fresh medium and appropriate antibiotics, dispensed into 96-well plates, and immediately supplied aTc, cell-free culture fluids, or HSLs. In all cases, cell-free culture fluids were administered at 30% (w/v), and plate wells that did not receive treatment received equal volumes of the growth medium or DMSO, as specified. To make cell-free culture fluids, overnight cultures of V. fischeri, Aeromonas sp. ARM81, and E. coli strains were grown in LB + 3% NaCl, and cells were removed by centrifugation. The clarified supernatants were collected and filtered through 0.22-μM filters (Corning SpinX). A BioTek Synergy Neo2 multimode reader was used to measure OD600 and bioluminescence. Relative light units (RLU) were calculated by dividing the bioluminescence readings by the OD600 reading at that time.
Total Protein and In-Gel HALO Detection to Assess Phage LuxRARM81ld Solubility.
Overnight cultures of E. coli T7 Express lysY/Iq carrying the plasmid with the LuxRARM81ld-HALO-HIS fusion were diluted 1:200 in 15 mL medium and grown at 37 °C to OD600 ~ 0.5. Subsequently, 200 µM IPTG was added to each culture before it was divided into 3 equal volumes, and the aliquots received 75 µM C4-HSL, 75 µM C8-HSL, or an equivalent volume of DMSO. The cultures were returned to growth at 37 °C for an additional 3 h prior to cell collection by centrifugation. Pellets were stored at −80 °C prior to processing. Cell pellets were resuspended in a lysis buffer containing BugBuster, benzonase, and 1 µM HALO-Alexa660 (excitation/emission: 663/690 nm) and incubated at room temperature for 15 min. The resulting whole-cell lysates were loaded onto 4 to 20% SDS-PAGE stain-free gels, which were imaged using an ImageQuant LAS 4,000 imager under the Cy5 setting for HALO-Alexa660 before being exposed to UV-light for 7 min and reimaged under the EtBr setting for total protein. Exposure times never exceeded 30 s.
qPCR Measurement of Relative Phage ARM81ld Viral Load from Cocultures.
Triplicate colonies of WT and ΔahyI Aeromonas sp. ARM81 and V. fischeri strains were each resuspended in 1 mL fresh growth medium and incubated at 30 °C until the cultures reached OD600 ~0.5. Cultures were back-diluted 1:100 into a fresh growth medium and combined at a 1:5 ratio of Aeromonas sp. ARM81:V. fischeri. The Aeromonas sp. ARM81 monoculture control was prepared in parallel by dilution of the Aeromonas sp. ARM81 culture 1:5 in the growth medium. The mono- and cocultures were dispensed into a 96-well plate and incubated at 30 °C with shaking for ~10 h, at which point 10-uL aliquots were collected, heated to 95 °C for 10 min, and diluted 1:1,000 in water. SYBR Green mix (Quanta) and the Applied Biosystems QuantStudio 6 Flex Real-Time PCR detection system (Thermo) were used for real-time PCR. Data were processed and analyzed (Pfaffl method) by comparing the relative amplification within samples from reactions using an ARM81ld phage-specific primer pair (targeting cIARM81ld, SI Appendix, Table S3) to that from reactions using an Aeromonas sp. ARM81 host-specific primer pair (targeting rpoB, SI Appendix, Table S3). The relative phage ARM81ld viral load was determined by dividing the ARM81 phage-to-host amplification ratio from each coculture condition by that of the Aeromonas sp. ARM81 monoculture.
Quantitation and Statistical Analyses.
Software used to acquire and analyze data generated in this study consisted of: GraphPad Prism9 for analysis of growth- and reporter-based experiments; Gen5 for collection of growth- and reporter-based data; SnapGene v6 for primer design; QuantStudio for qPCR quantitation; and FIJI for image analyses. Data are presented as mean ± SD. The numbers of independent biological replicates for each experiment are indicated in the figure legends.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank all members of the Bassler lab for insightful discussions. This work was supported by the HHMI, NSF grant MCB-2043238, and the NIH grant R37GM065859 to B.L.B. J.E.S. is a HHMI Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. O.P.D. was supported by the NIGMS T32GM007388 grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Author contributions
J.E.S., O.P.D., and B.L.B. designed research; J.E.S. and O.P.D. performed research; J.E.S. and O.P.D. contributed new reagents/analytic tools; J.E.S., O.P.D., and B.L.B. analyzed data; and J.E.S., O.P.D., and B.L.B. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Reviewers: L.A.M., The Rockefeller University; and R.S., Weizmann Institute of Science.
Data, Materials, and Software Availability
All growth data, reporter data, and unprocessed gels presented in each panel of this study are provided in Dataset S1 and available on Zenodo (21).
Supporting Information
References
- 1.Papenfort K., Bassler B. L., Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. M., SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galloway W. R. J. D., Hodgkinson J. T., Bowden S. D., Welch M., Spring D. R., Quorum sensing in gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 111, 28–67 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Nguyen Y., et al. , Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio 6, e02429-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawver L. A., Jung S. A., Ng W. L., Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCready A. R., Paczkowski J. E., Henke B. R., Bassler B. L., Structural determinants driving homoserine lactone ligand selection in the Pseudomonas aeruginosa LasR quorum-sensing receptor. Proc. Natl. Acad. Sci. U.S.A. 116, 245–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ke X., Miller L. C., Bassler B. L., Determinants governing ligand specificity of the Vibrio harveyi LuxN quorum-sensing receptor. Mol. Microbiol. 95, 127–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdt J. P., et al. , Chemical interrogation of LuxR-type quorum sensing receptors reveals new insights into receptor selectivity and the potential for interspecies bacterial signaling. ACS Chem. Biol. 12, 2457–2464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandoz K. M., Mitzimberg S. M., Schuster M., From the cover: Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104, 15876 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silpe J. E., Duddy O. P., Hussain F. A., Forsberg K. J., Bassler B. L., Small protein modules dictate prophage fates during polylysogeny. bioRxiv[Preprint]. 10.1101/2022.09.16.508337 (2022). [DOI] [PMC free article] [PubMed]
- 11.Silpe J. E., Bassler B. L., Phage-encoded LuxR-type receptors responsive to host-produced bacterial quorum-sensing autoinducers. mBio 10, e00638-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo A., Blough N. V., Dunlap P. V., Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 176, 7558 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberhard A., et al. , Structural identification of autoinducer of photobacterium fischeri luciferase. Biochemistry 20, 2444–2449 (1981). [DOI] [PubMed] [Google Scholar]
- 14.Paczkowski J. E., et al. , Flavonoids suppress pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 292, 4064–4076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupp C., Ruby E. G., Vibrio fischeri LuxS and AinS: Comparative study of two signal synthases. J. Bacteriol. 186, 3873 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engebrecht J. A., Silverman M., Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4154 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilson L., Kuo A., Dunlap P. V., AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177, 6946 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Loughlin C. T., et al. , A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 110, 17981–17986 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins A. C., Arnold F. H., Stuermer R., Hauer B., Leadbetter J. R., Directed evolution of Vibrio fischeri LuxR for improved response to butanoyl-homoserine lactone. Appl. Environ. Microbiol. 73, 5775–5781 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimbrough J. H., E. V. Stabb, Substrate specificity and function of the pheromone receptor AinR in Vibrio fischeri ES114. J. Bacteriol. 195, 5223–5232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silpe Justin E., Duddy Olivia P., Bassler Bonnie L., Data for Natural and synthetic small molecule inhibitors of a phage-encoded quorumsensing receptor affect phage-host dynamics in mixed bacterial communities. Zenodo. 10.5281/zenodo.7209272. Deposited: October 15, 20226 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All growth data, reporter data, and unprocessed gels presented in each panel of this study are provided in Dataset S1 and available on Zenodo (21).