Abstract
Introduction:
Identifying genetic factors that influence HIV-pathogenesis is critical for understanding disease pathways. Previous studies have suggested a role for the human gene ten-eleven methylcytosine dioxygenase 2 (TET2) in modulating HIV-pathogenesis.
Methods:
We assessed whether genetic variation in TET2 was associated with markers of HIV-pathogenesis using both gene level and single nucleotide polymorphism (SNP) level association in 8512 HIV-positive persons across five clinical trial cohorts.
Results:
Variation at both the gene and SNP-level of TET2 was found to be associated with levels of HIV viral load (HIV-VL) consistently in the two cohorts that recruited antiretroviral-naïve participants. The SNPs occurred in two clusters of high linkage disequilibrium (LD), one associated with high HIV-VL and the other low HIV-VL, and were predominantly found in Black participants.
Conclusion:
Genetic variation in TET2 was associated with HIV-VL in two large antiretroviral therapy (ART)-naive clinical trial cohorts. The role of TET2 in HIV-pathogenesis warrants further investigation.
Keywords: HIV, ten-eleven methylcytosine dioxygenase 2, HIV viral load, genetics, transcriptional regulation
Introduction
HIV-1 pathogenesis is a multifaceted interaction between host and virus that results in varying degrees of immune-deficiency, high levels of HIV viral load (HIV-VL), chronic inflammation, and coagulopathy. In previous studies, variations in all aspects of these responses have been linked with increased risk of adverse clinical outcomes in people with HIV (PWH) [1–11]. A number of recent studies have linked variation in the host genome with a variety of markers of HIV progression and disease [12–18]. These genetic association studies present a unique opportunity to explore potential contributors to HIV progression and disease by studying the impact of genetic variation in known or suspected host HIV enhancing or resistance factors on relevant biomarkers.
In this study, we focus on Ten-eleven methylcytosine dioxygenase 2 (TET2). TET2 encodes a protein that is involved in locus-specific DNA de-methylation of previously methylated and silenced genes through the oxidation of 5-methylcytosine (5mC) [19,20]. Loss of TET2 function is implicated in the development of both haematological malignancies and solid tumours in people without HIV (reviewed in [21]). However, there is a developing literature associating this gene with viral replication, and in particular retroviral replication, as well as viral associated malignancy. TET2 function has been linked to both Epstein-Barr virus (EBV) and Human T-cell leukaemia virus type-1 (HTLV-1) induced malignancy [22–28], while TET2 also appears to be critical for the regulation of endogenous retroviral elements [29–31]. In the context of HIV, recent work has suggested that the HIV-protein VPR selectively degrades TET2, thereby enhancing IL-6 and IFITM3 and promoting viral replication in monocyte-macrophage lineage cells [32,33], while mutations in TET2 are relatively common in HIV positive persons with clonal haematopoiesis [34]. Additionally, a key upstream regulator of TET2 (IDH1) has previously been implicated in HIV acquisition in molecular work and clinical cohorts [35]. Finally, from our own recent study, we observed associations between singal nucleotide polymorphisms (SNPs) in the TET2 gene with HIV-VL (via GWAS). Although the SNPs of interest were below the 5% minor allele frequency (MAF) cut-off used in that study [36], this signal, combined with the literature associating TET2 function with retroviral replication and viral malignancy, encouraged us to investigate this gene and its upstream regulators further.
We therefore hypothesized that TET2 is a critical regulator of HIV-pathogenesis, and that genetic variation in this gene and upstream regulators will impact gene function and therefore the specific pathogenic process it is involved in. However, as it was not abundantly clear from the molecular data how TET2 was involved in HIV-pathogenesis (both inflammatory and viral load mechanisms were proposed [32,33]), we sought to perform an exploratory targeted genetic association study to determine what, if any, role TET2 has in HIV-pathogenesis. Biomarkers tested include markers of disease progression and immune dysfunction (HIV-VL, CD4+ T-cell count, CD8+ T-cell count and CD4/CD8 ratio) as well as markers of inflammation and coagulopathy (interleukin 6 [IL-6], C-reactive protein [CRP], D-dimer). We conducted this targeted genetic association study on TET2 and two regulators of TET2 function – IDH1 and IDH2 [37]. Associations were assessed at both the gene and SNP-level using five independent trials from the International Network for Strategic Initiatives in Global HIV Trials (INSIGHT) network.
Materials and methods
Study population
Our study population included HIV-positive adults from five clinical trials: START (NCT00867048) [40], SMART [41] (NCT00027352), ESPRIT [42] (NCT00004978), STALWART [43] (NCT00110812) and FIRST [44] (NCT00000922). The trials were approved by the institutional review board or ethics committee at each contributing centre. Participants analysed in this study were those who provided written informed consent for the trials and for genetic analyses. This was only a subset of the main trial and therefore the number of participants analysed in this study is less than presented in the main trial report. Details on the individual trials can be found in the primary papers and supplemental methods, Supplemental Digital Content.
Genotyping and quality control
Participants were genotyped using a custom content Affymetrix Axiom SNP array, consisting of 770 558 probes, enriched with markers related to immune dysfunction. The Ensembl Gene database, assembly hg19/GRCh37, was used to annotate genes within a 5 kb window of each variant. Genotyping was performed at Advanced BioMedical Laboratories, using standardized quality assurance procedures. As part of SNP-level QC, probes were removed if any of the following applied: probe was duplicate, multiallelic or nonautosomal, missingness >0.03, reproducibility less than 0.90, MAF <1% or deviation from the Hardy–Weinberg equilibrium with P < 1 × 10–6. At sample-level QC, participants were excluded if there was a sex mismatch, duplication or missingness. In order to capture potentially relevant TET2 phenotypes present in related individuals, participants were not excluded due to cryptic relatedness or outlying heterozygosity. After sample and SNP QC SNPs that mapped to TET2 as well as IDH1 and IDH2, (±5 kb) were selected for further analysis. These genes included TET2 as well as IDH1 and IDH2, two regulators of TET2 function [37].
Statistical analyses
Association of TET2 pathway genetic variation with markers of HIV-pathogenesis
Gene level associations were estimated using the SKAT-O method [45–47]. To control for population stratification, we calculated eigenvectors of the genetic data using EIGENSTRAT [48]. Eigenvectors were calculated independently for each trial cohort. Eigenvectors were calculated using all SNPs (not just TET2 pathway SNPs) that passed QC and were present above 5% MAF. The first four eigenvectors and sex were included as explanatory variables in the SKAT-O model.
We investigated associations of TET2 SNPs with study entry measurements of HIV-VL, IL-6, hs-CRP, D-dimer, CD4+ T-cell count, CD8+ T-cell count, CD4/CD8 ratio and CD4 nadir, using separate linear regression models for each biomarker (response) and SNP (predictor) combination. Associations were assessed using an additive genetic model. Associations with HIV-VL were only assessed in ART-naive cohorts (START, FIRST and STALWART). In order to ensure an approximately normal distribution of the biomarkers, HIV-VL underwent log10 transformation, IL-6, hs-CRP and D-dimer underwent log2-transformation, whereas CD4+ T-cell count, CD8+ T-cell count, CD4/CD8 ratio and CD4 nadir were not transformed. The first four eigenvectors and sex were included as explanatory variables in the regression models. Associations were estimated separately in each of the five cohorts. To control for inflation of Type I error due to multiple testing, we used the Benjamini–Hochberg FDR method to limit the false discovery rate to at least 5% for each biomarker and cohort [49].
We conducted several sensitivity analyses on the SNPs that were associated with HIV-VL in the START and FIRST studies. These are described in supplemental methods, Supplemental Digital Content.
To assess whether alternate alleles were more frequent among Black compared with non-Black participants, we used a two-proportions z-test. To determine whether SNPs occurred in clusters of high linkage disequilibrium (LD), we ranked the SNPs associated with HIV-VL in both START and FIRST by effect size and performed pairwise LD analyses using LDheatmap [50]. The LDheatmaps were made independently for the START and FIRST cohorts and included all participants from the respective cohorts.
All statistical analyses were performed using R version 3.5.1 [51]. Figures were generated using R or GraphPad Prism.
Results
Characteristics of participants in the INSIGHT genetic cohorts
A total of 8512 participants from the five cohorts were included in this study. Baseline characteristics are summarized in Table 1 and distributions of the biomarkers tested are shown in Figures 1–5, Supplemental Digital Content. Participant characteristics varied substantially between studies. The START (early HIV infection with preserved immune parameters) and FIRST (advanced untreated HIV infection) trials were the only studies that enrolled exclusively antiretroviral therapy (ART)-naive participants. Participants were predominantly White, with a large percentage of Black participants in the START, SMART and FIRST studies, 23, 38, and 57%, respectively. SMART and FIRST enrolled predominantly in the United States, whereas the other studies had wider enrolment profiles (including Europe, Asia, Africa, Australia, South America). The SCREE plots for the eigenvectors calculated to control for this population stratification are presented in Figure 6, Supplemental Digital Content.
Table 1.
Study entry demographics for the five cohorts.
| START | FIRST | ESPRIT | SMART | STALWART | |
| Participants (n = ) | 2546 | 544 | 2891 | 2283 | 244 |
| Age (years): median (IQR) | 36 (29, 45) | 38 (32, 44) | 40 (34, 46) | 44 (38, 50) | 36 (30, 44) |
| Female (%) | 20.1 | 20.2 | 18 | 26 | 18 |
| Race | |||||
| Asian (%) | 1 | <1 | 12 | 1 | 27 |
| Black (%) | 23 | 57 | 9 | 38 | 6 |
| White/other (%) | 76 | 43 | 79 | 60 | 68 |
| CD4+ cell count (cells/μl): median (IQR) | 651 (585, 759) | 181 (43, 345) | 451 (368, 582) | 572 (455, 773) | 419 (358, 516) |
| HIV RNA level (log10 copies/ml): median (IQR) | 4.17 (3.54, 4.66) | 5.09 (4.53, 5.54) | 1.70 (1.70, 2.60) | 2.60 (1.70, 3.45) | 4.38 (3.86, 4.79) |
| CD8+ T-cell count (cells/μl): median (IQR) | 1062 (790, 1431) | n/a | n/a | n/a | n/a |
| Nadir CD4+ T-cell count (cells/μl): median (IQR) | 545 (473, 642) | n/a | n/a | n/a | n/a |
| CD4/CD8 ratio: median (IQR) | 0.62 (0.46, 0.84) | n/a | n/a | n/a | n/a |
| IL-6 (pg/ml): median (IQR) | 1.47 (1.02, 2.21) | n/a | 1.90 (1.30, 2.80) | 1.91 (1.19, 3.23) | n/a |
| CRP (μg/ml): median (IQR) | 1.82 (0.77, 4.15) | n/a | 1.49 (0.69, 3.21) | 1.79 (0.76, 4.51) | n/a |
| D-dimer (μg/ml): median (IQR) | 0.31 (0.22, 0.47) | n/a | 0.26 (0.18, 0.37) | 0.22 (0.13, 0.40) | n/a |
| On ART at study entry (%) | 0 | 0 | 100 | 79.5 | 0 |
| ART-naive (%) | 100 | 100 | 0 | 6.0 | 78 |
| Geographical region a | |||||
| U.S. (%) | 18 | 100 | 25 | 80.4 | 10 |
| Europe/Australia/Israel (%) | 49 | 0 | 49 | 11.2 | 34 |
| South America/Mexico (%) | 20 | 0 | 14 | 5.5 | 30 |
| Asia (%) | 0 | 0 | 11 | 0.4 | 25 |
| Africa (%) | 13 | 0 | 1 | 2.5 | <1% |
CRP, C-reactive protein; IL-6, interleukin 6; IQR, interquartile range.
Country/region of residence.
SNP quality control and minor allele frequency filtering
Eight hundred and eighty-eight SNPs spanning the TET2 (796), IDH1 (7) and IDH2 (85) genes were present in the GeneCHIP array. After SNP quality control (QC) and cohort specific MAF filtering, a total of 292 SNPs were included in START genetic association analyses, 345 in FIRST, 317 in SMART, 262 in ESPRIT and 276 in STALWART.
Association of genetic variation in the ten-eleven methylcytosine dioxygenase 2 pathway with markers of HIV-pathogenesis
First, the associations between genes of the TET2 pathway and markers of interest were assessed using the SKAT-O method. TET2 was associated with levels of HIV-VL at study entry in the two trials that included only ART-naive participants, FIRST (P = 0.0005) and START (P = 0.0001), but not any of the other cohorts (Table 2). TET2 was also associated with CD4+ T-cell count in the START study (P = 0.016) (Table 2). TET2 was not associated with any other marker in any of the five studies. SKAT-O did not reveal any significant associations with either IDH1 or IDH2 (data not shown).
Table 2.
SKAT-O associations between TET2 and markers of HIV-pathogenesis in each of the five cohorts.
| Outcome | START | FIRST | ESPRIT | SMART | STALWART |
| CD4+ T-cell count | 0.023 | 0.868 | 0.126 | 0.391 | 0.604 |
| CD4/CD8 ratio | 0.843 | N/A | N/A | N/A | N/A |
| CD4 nadir | 0.430 | N/A | N/A | N/A | N/A |
| CD8+ T-cell count | 1.000 | N/A | N/A | N/A | N/A |
| HIV-VL | 0.000152 | 0.00067 | Not assesseda | Not assesseda | 0.458 |
| IL-6 | 0.062 | N/A | 0.071 | 0.431 | N/A |
| CRP | 0.806 | N/A | 0.059 | 0.470 | N/A |
| D-dimer | 0.905 | N/A | 0.244 | 0.791 | N/A |
CRP, C-reactive protein; IL-6, interleukin 6; TET2, ten-eleven methylcytosine dioxygenase 2; VL, viral load.
As ESPRIT and SMART did not recruit ART-naive individuals, HIV-VL was not assessed in these cohorts.
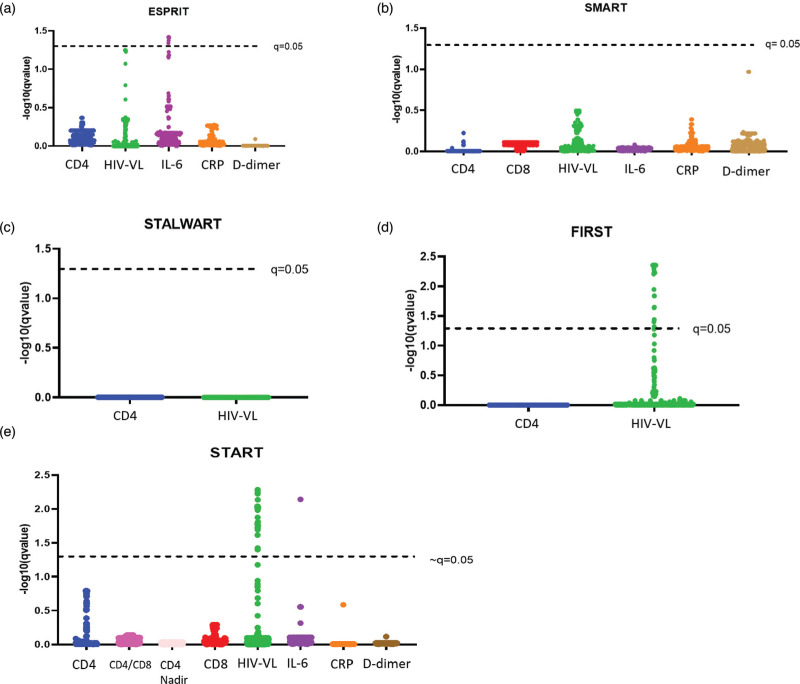
Next, we estimated associations of individual SNPs with each of the biomarkers, separately for each of the five cohorts. In this analysis, 15 SNPs were significantly (q < 0.05) associated with HIV-VL in both the START and FIRST cohorts (Fig. 1d, e; Table 3). Additionally, we found 13 and 8 variants, respectively, that were associated with HIV-VL in either START or FIRST, but not in both studies. The SNPs that were significantly associated with HIV-VL in only one of the two studies showed a similar effect size and direction in the other study. All SNPs associated with HIV-VL were located within the TET2 gene, except for a single SNP located in IDH1 (rs12694101). SNPs that were associated with baseline HIV-VL in the START study were also associated with longitudinal VL levels while participants remained off ART (Figure 7, Supplemental Digital Content). The alternate allele of the SNPs associated with HIV-VL were predominantly found in PWH of Black race (Tables 2–5, Supplemental Digital Content) across a variety of geographic regions of enrolment (Tables 2 and 3, Supplemental Digital Content).
Fig. 1.
Associations between SNPs and markers of HIV pathogenesis.
Q-values were −log10 transformed and plotted on the y-axis; higher values denote smaller q-values, and stronger evidence for the presence of an association. SNPs are grouped by associations to a specific marker of interest and plotted on the x-axis. Each dot represents a single SNP. Associations are considered significant with q-value <0.05.
Table 3.
Results and description of SNPs that were observed to be associated with HIV-VL in the START and FIRST cohorts.
| dbSNP RS ID | Gene (±5 kb) | Chr | Physical position | Strand | Ref allele | Alt allele | START estimatea | START P-value | START q-value | START rank | FIRST estimatea | FIRST P-value | FIRST q-value | FIRST rank |
| rs77936891 | TET2 | 4 | 106122657 | + | A | G | −0.264 | 0.000025 | 0.0052 | 1 | −0.1869 | 0.028 | 0.2587 | 37 |
| rs75555015 | TET2 | 4 | 106151781 | + | A | G | −0.2839 | 0.000036 | 0.0052 | 2 | −0.1838 | 0.059 | 0.4421 | 45 |
| rs115930414 | TET2 | 4 | 106173177 | + | G | T | −0.3001 | 0.000059 | 0.0057 | 3 | −0.191 | 0.057 | 0.4421 | 44 |
| rs78305976 | TET2 | 4 | 106095219 | + | T | A | −0.2705 | 0.000082 | 0.0059 | 4 | −0.1934 | 0.023 | 0.2386 | 33 |
| rs139100435 | TET2 | 4 | 106142080 | + | CTT | − | 0.3156 | 0.00012 | 0.0073 | 5 | 0.5107 | 0.00031 | 0.0072 | 15 |
| rs80014899 | TET2 | 4 | 106141445 | + | A | G | −0.2453 | 0.00021 | 0.0092 | 6 | −0.1728 | 0.040 | 0.3282 | 42 |
| rs72955180 | TET2 | 4 | 106133750 | + | G | T | 0.4022 | 0.00024 | 0.0092 | 7 | N/A | N/A | N/A | N/A |
| rs72950501 | TET2 | 4 | 106197936 | + | C | T | 0.3072 | 0.00028 | 0.0092 | 8 | 0.5511 | 0.000081 | 0.0058 | 4 |
| rs76644731 | TET2 | 4 | 106132312 | + | A | G | −0.2451 | 0.00028 | 0.0092 | 9 | −0.1894 | 0.026 | 0.2531 | 35 |
| rs72955179 | TET2 | 4 | 106133471 | + | G | A | 0.3022 | 0.00034 | 0.0096 | 10 | 0.5107 | 0.00031 | 0.0072 | 14 |
| rs72955193 | TET2 | 4 | 106145468 | + | A | G | 0.3781 | 0.00039 | 0.0096 | 11 | 0.5505 | 0.00017 | 0.0058 | 9 |
| rs17035325 | TET2 | 4 | 106110796 | + | T | C | −0.2394 | 0.00040 | 0.0096 | 12 | −0.1761 | 0.036 | 0.305 | 41 |
| rs77738124 | TET2 | 4 | 106173540 | + | T | C | −0.2367 | 0.00044 | 0.0096 | 13 | −0.1561 | 0.059 | 0.4421 | 46 |
| rs59519484 | TET2 | 4 | 106190732 | + | C | A | 0.2992 | 0.00046 | 0.0096 | 14 | 0.5141 | 0.00036 | 0.0077 | 16 |
| rs142786189 | TET2 | 4 | 106162216 | + | T | − | −0.2393 | 0.00053 | 0.0104 | 15 | −0.1902 | 0.027 | 0.2587 | 36 |
| rs78763791 | LOC101929491;TET2 | 4 | 106065102 | + | C | A | −0.2107 | 0.00073 | 0.0133 | 16 | −0.2485 | 0.0025 | 0.0371 | 23 |
| rs72963014 | TET2 | 4 | 106168623 | + | C | G | 0.348 | 0.00091 | 0.0155 | 17 | 0.5336 | 0.00022 | 0.0062 | 12 |
| rs72963032 | TET2 | 4 | 106182446 | + | A | G | 0.3407 | 0.0010 | 0.0164 | 18 | 0.5451 | 0.00011 | 0.0058 | 6 |
| rs59479204 | TET2 | 4 | 106160495 | + | A | G | 0.3411 | 0.0011 | 0.0164 | 19 | 0.5458 | 0.00018 | 0.0058 | 11 |
| rs17035308 | LOC101929491;TET2 | 4 | 106064349 | + | G | C | −0.213 | 0.0012 | 0.0169 | 20 | −0.2392 | 0.0038 | 0.0521 | 25 |
| rs58322634 | TET2 | 4 | 106166139 | + | A | G | 0.2467 | 0.0013 | 0.0176 | 21 | 0.5561 | 0.000040 | 0.0058 | 1 |
| rs137883243 | TET2 | 4 | 106184236 | + | ATATAAA | − | −0.2338 | 0.0013 | 0.0178 | 22 | −0.2156 | 0.026 | 0.2531 | 34 |
| rs72961199 | TET2 | 4 | 106159386 | + | G | A | 0.3389 | 0.0015 | 0.0189 | 23 | 0.554 | 0.00015 | 0.0058 | 7 |
| rs60382101 | TET2 | 4 | 106160161 | + | A | G | 0.3372 | 0.0016 | 0.0192 | 24 | 0.554 | 0.00015 | 0.0058 | 8 |
| rs72963038 | TET2 | 4 | 106189156 | + | C | A | 0.3282 | 0.0017 | 0.0203 | 25 | 0.5731 | 0.000053 | 0.0058 | 2 |
| rs79305653 | TET2 | 4 | 106200234 | + | T | C | −0.2254 | 0.0022 | 0.0242 | 26 | −0.1809 | 0.062 | 0.4476 | 48 |
| rs72952305 | TET2 | 4 | 106201653 | + | T | G | 0.314 | 0.0035 | 0.0377 | 27 | 0.544 | 0.00026 | 0.0069 | 13 |
| rs72963036 | TET2 | 4 | 106188548 | + | A | T | 0.3011 | 0.0038 | 0.0397 | 28 | 0.577 | 0.00010 | 0.0058 | 5 |
| rs72963031 | TET2 | 4 | 106182132 | + | A | T | 0.2422 | 0.014 | 0.1276 | 31 | 0.4754 | 0.00068 | 0.0138 | 17 |
| rs60786079 | TET2 | 4 | 106197750 | + | G | A | 0.1663 | 0.015 | 0.1402 | 32 | 0.5261 | 0.000061 | 0.0058 | 3 |
| rs72963046 | TET2 | 4 | 106194419 | + | C | A | 0.2547 | 0.019 | 0.1601 | 34 | 0.5501 | 0.00017 | 0.0058 | 10 |
| rs6811468 | TET2 | 4 | 106082473 | + | A | G | 0.1042 | 0.19 | 0.7998 | 69 | 0.3239 | 0.0024 | 0.0371 | 22 |
| rs12694101 | IDH1; PIKFYVE | 2 | 209128892 | + | G | A | 0.0096 | 0.77 | 0.9674 | 231 | −0.1913 | 0.0014 | 0.0233 | 20 |
| rs72963007 | TET2 | 4 | 106164723 | + | G | A | N/A | N/A | N/A | N/A | 0.5476 | 0.00093 | 0.0169 | 19 |
| rs72961197 | TET2 | 4 | 106158738 | + | G | A | N/A | N/A | N/A | N/A | 0.5476 | 0.00093 | 0.0169 | 18 |
| rs72955158 | TET2 | 4 | 106110032 | + | A | G | N/A | N/A | N/A | N/A | 0.4082 | 0.0018 | 0.0302 | 21 |
N/A indicates that a particular SNP did not pass QC or the MAF cut-off for that particular study and was therefore not included in the GLM.
MAF, minor allele frequency; QC, quality control; TET2, ten-eleven methylcytosine dioxygenase 2; VL, viral load.
∗SNPs were ordered by P-value (low to high) with rank = 1 for the SNP with the lowest P-value.
Estimates are presented for the alternate allele.
The results of the sensitivity analyses for SNPs significantly associated with HIV-VL in START and FIRST were consistent with the main models (Tables 6 and 7, Supplemental Digital Content and Figures 8–10, Supplemental Digital Content). For the subset analyses, 578 and 309 participants were assessed in the Black only analysis, for START and FIRST, respectively, 2444 participants were assessed in the analysis that excluded participants exhibiting cryptic and outlying heterozygosity, from the START study only. 2365 participants were assessed in the analysis that excluded recently infected participants, from the START study only.
We also identified six SNPs that were associated with IL-6 levels at study entry in the ESPRIT cohort (Fig. 1a) and one in the START cohort (Fig. 1e). However, these SNPs were not the same across the different cohorts (Table 1, Supplemental Digital Content). Associations with other markers of interest were not observed in any of the cohorts.
Linkage disequilibrium of SNPs associated with HIV-viral load
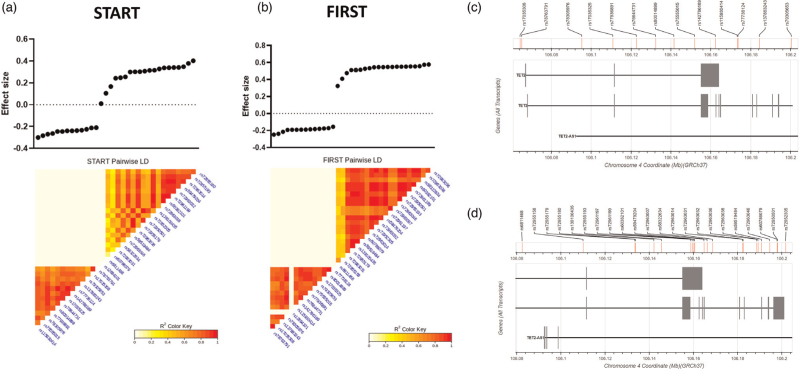
Among the 36 SNPs associated with HIV-VL at study entry in the START and FIRST cohorts, 14 were associated with lower viral load, while 22 were associated with a higher HIV-VL. These SNPs form two clusters of high linkage disequilibrium (LD), one containing the SNPs that were associated with high HIV-VL, the other all SNPs associated with low HIV-VL (Fig. 2).
Fig. 2.
Pairwise linkage disequilibrium (LD) analysis of SNPs significantly associated with VL in the START (a) and/or FIRST (b) cohorts.
The LD shows two distinct clusters of SNPs in both studies, consisting of SNPs that are associated with lower VL (on the left), and those associated with higher VL (on the right). All TET2 SNPs that associated with higher VL in START are also associated with higher VL in FIRST. Likewise, TET2 SNPs that associated with a lower VL in START are also associated with a lower VL in FIRST. LD is measured using R2 of each SNP pair; darker colour denotes stronger associations. (c) TET2 SNPs associated with lower VL are spread out across the TET2 gene. (d) TET2 SNPs associated with higher VL tend to cluster towards the 5’ end of TET2. TET2, ten-eleven methylcytosine dioxygenase 2; VL, viral load.
Discussion
Here we report on the effect of genetic variation in TET2 and related genes on clinical markers of HIV-pathogenesis. Gene level analysis revealed significant associations between TET2 with HIV-VL in two independent ART-naive cohorts (START and FIRST), and with CD4+ T-cell count in one cohort (START). No gene level associations were observed for IDH1 or IDH2. SNP level analyses confirmed an association with HIV-VL, identifying a number of SNPs associated with HIV-VL in the START and FIRST cohorts. The association with CD4+ T-cell count was not confirmed at the SNP-level. No associations were observed with any of the other biomarkers.
Our findings at both gene and SNP level suggest that variation in the TET2 gene impacts HIV-VL in PWH not taking ART. This effect on HIV-VL appears to be driven by several SNPs that group into two distinct LD clusters, and are predominantly seen in persons of Black race. In the START cohort, we observed 13 SNPs significantly (q-value < 0.05) associated with lower HIV-VL at study entry, whereas 15 SNPs were significantly associated with a higher HIV-VL at study entry. These results were replicated using a linear mixed effects model which included follow-up HIV-VL data. All SNPs in the LD cluster associated with higher HIV-VL in START were associated with a higher HIV-VL in FIRST; whereas all SNPs in the LD cluster associated with lower HIV-VL in START were also associated with a lower HIV-VL in the FIRST study. Many of the SNPs (15/36) in the two clusters were significant across both studies, while the SNPs that were significantly associated with HIV-VL in only one of the two studies showed a similar effect size and direction in the other. Since SNP MAF and QC filtering were performed per study, some SNPs were only present in one study or the other. Amongst these discordant SNPs was rs72963007, which was significantly associated (β = 0.55, q = 0.02) with HIV-VL in the FIRST cohort but was below the MAF cut-off in the START cohort. Of the TET2 SNPs that were associated with HIV-VL in our study, only rs72963007 has been reported in the literature, where it was associated with an increased risk of adult T-cell leukaemia caused by HTLV-1 in persons of African descent [23]. Like HIV, HTLV-1 is a retrovirus, with many similar genetic elements, and this association suggests a critical role for this SNP (and TET2 more generally) in retroviral pathogenesis.
The exact mechanism through which TET2 affects HIV-VL, and why some SNPs are associated with higher HIV-VL and others with lower HIV-VL, is not clear. Prior molecular work has focused on the relationship between TET2 and HIV-pathogenesis in monocyte/macrophage cells and cell-lines [32,33], with recent work suggesting that TET2 is only downregulated in HIV-infected monocyte-derived-macrophages and not CD4+ T cells [38]. If true, this could explain the relatively small effect these SNPs have on HIV-VL, as monocyte derived macrophages represent a smaller subset of the total infected cells than their T-cell counterparts. The SNPs themselves are in high LD and are spread out across the TET2 gene (although those associated with higher HIV-VL tend to congregate in the 5’ end of TET2). Given the previous evidence associating TET2 with transcriptional regulation of endogenous retroviral elements [29–31], it may be that TET2 is playing a role in transcriptional regulation of HIV. Alternatively, previously studies have suggested two independent mechanisms for TET2 related enhancement of HIV-VL, mediated through the VPR. The first via modulation of IL-6 and the second via IFITM3 [32,33], and it is possible that these SNPs are either preventing or enhancing one of these mechanisms. Although, in our study we did not observe consistent associations between TET2 and IL-6, and further laboratory work is required to elucidate the relationship between TET2 and HIV-VL.
Our study has several limitations. First, the five cohorts have different populations. This is particularly relevant for participants from the two trials that recruited only ART-naive participants. Those in FIRST were more progressed than participants of START (as indicated by lower CD4+ T-cell count and higher HIV-VL). This difference means the HIV-VL phenotype is not directly comparable between these two cohorts, but as associations were found in both cohorts, it may be that the mechanism through which the TET2 mutations affect HIV-VL is independent of progression status. However, this also means that we may have been able to validate other signals (e.g. the association with CD4+ T-cell count and gene-level variation in the START cohort) if we were able to access cohorts with similar characteristics. Another limitation is the large number of associations explored and the less stringent multiple testing correction methodology used (compared to the Bonferroni method preferred by large GWAS’). Instead, we relied on validation of the associations across the more than one cohort to increase confidence in our observations. We believe this, combined with the totality of molecular evidence suggesting that TET2 is involved in retroviral and HIV-pathogenesis, justifies the use of the less stringent multiple testing threshold utilized. Despite this, the exploratory nature of this study means that results should be interpreted cautiously and further studies (both laboratory based and in a different clinical populations) are required to validate the results observed here. Finally, the associations for both IDH1 and IDH2 were limited by the number of SNPs from these genes in the Affymetrix array we utilized. Only seven and 85 SNPs, compared to 796 for TET2, were included for IDH1 and IDH2, respectively.
In summary, we showed that genetic variations in TET2 were associated with levels of HIV-VL in ART-naive HIV-positive persons. These results support previous work reporting a key role for TET2 in retroviral replication and pathogenesis. This is the first in-human evidence of the importance of this gene in HIV-pathogenesis. These observations warrant further investigation in additional cohorts and through molecular work. Of particularly interest is exploring the effects of these SNPs in larger populations of Black persons, since the risk allele of this SNP appears substantially more frequent in this subgroup. This group remains understudied in large genetic association studies, despite Black persons representing the vast majority of PWH globally [39].
Acknowledgements
The authors would like to thank all participants and staff from the trials described in this study.
See N Engl J Med 2015;373:795–807 for the complete list of START investigators.
See Lancet 2006;368:2125–35 for the complete list of FIRST investigators.
See N Engl J Med 2006;355:2283–96 for the complete list of SMART investigators.
See N Engl J Med 2009;361:1548–59 for the complete list of ESPRIT investigators.
See PLoS ONE 2010; 5(2):e9334. doi:10.1371/journal.pone.0009334 for the complete list of STALWART investigators.
Funding: This study was funded by the Danish National Research Foundation (DNRF126). The clinical trials described in this study were supported by the National Institute of Health (UM1-AI06864, UM1-AI120197, 1U01-AI36780, U01-AI46957, U01-AI042170, U01-AI046362), National Institute of Allergy and Infectious Diseases, National Institutes of Health Clinical Center, National Cancer Institute, National Heart, Lung, and Blood Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute of Arthritis and Musculoskeletal and Skin Diseases, Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (France), National Health and Medical Research Council (Australia), National Research Foundation (Denmark), Bundes ministerium für Bildung und Forschung (Germany), European AIDS Treatment Network, Medical Research Council (United Kingdom), National Institute for Health Research, National Health Service (United Kingdom), and University of Minnesota. Antiretroviral drugs were donated to the central drug repository by AbbVie, Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline/ViiV Healthcare, Janssen Scientific Affairs, and Merck.
Conflicts of interest
D.D.M., C.R.M, A.G.Z., J.R., M.H. and J.D.L. were supported by the Danish National Research Foundation (DNRF126). B.G. was supported by the National Institute of Health (UM1 AI120197). A.H.B. was supported by the Lundbeckfonden (grant number R219-2016-762). L.D.D. was supported by the EACS Medical Exchange Programme and the Spanish Society of Infectious Diseases and Clinical Microbiology. M.H. participated in advisory boards for AstraZeneca, Gilead, GSK, MSD, Roche and Sobi and received speakers honoraria from Gilead and GSK. M.P. received Funding from BMS, Celgene, Gilead, Janssen, ViiV Pharmaceuticals all outside submitted work (to institution). Provision of drug and other materials for studies from Astex/Otsuka, Celgene, CSL, Emergent Biosolutions, Janssen Grifols, Takeda, Verastem, ViiV Pharmaceuticals, all outside submitted work (to institution). Advisory board/speakers panel for Celgene, Gilead, outside submitted work. For the remaining authors, no conflicts relating to the submitted work were declared.
Supplementary Material
B.G. and C.R.M. contributed equally to the study.
Supplemental digital content is available for this article.
References
- 1.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 2008; 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ledwaba L, Tavel JA, Khabo P, Maja P, Qin J, Sangweni P, et al. Pre-ART levels of inflammation and coagulation markers are strong predictors of death in a South African cohort with advanced HIV disease. PLoS One 2012; 7:e24243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wada NI, Bream JH, Martinez-Maza O, Macatangay B, Galvin SR, Margolick JB, et al. Inflammatory biomarkers and mortality risk among HIV-suppressed men: a multisite prospective cohort study. Clin Infect Dis 2016; 63:984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 2014; 210:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 2014; 210:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byakwaga H, Boum Y, 2nd, Huang Y, Muzoora C, Kembabazi A, Weiser SD. The kynurenine pathway of tryptophan catabolism, CD4+ T-cell recovery, and mortality among HIV-infected Ugandans initiating antiretroviral therapy. J Infect Dis 2014; 210:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S, Byakwaga H, Boum Y, Burdo TH, Williams KC, Lederman MM, et al. Immunologic pathways that predict mortality in HIV-infected Ugandans initiating Antiretroviral therapy. J Infect Dis 2017; 215:1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tien PC, Choi AI, Zolopa AR, Benson C, Tracy R, Scherzer R, et al. Inflammation and mortality in HIV-infected adults: analysis of the FRAM study cohort. J Acquir Immune Defic Syndr 2010; 55:316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis 2011; 203:780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudsen TB, Ertner G, Petersen J, Moller HJ, Moestrup SK, Eugen-Olsen J, et al. Plasma soluble CD163 level independently predicts all-cause mortality in HIV-1-infected individuals. J Infect Dis 2016; 214:1198–1204. [DOI] [PubMed] [Google Scholar]
- 11.Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, et al. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog 2017; 13:e1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telenti A, Goldstein DB. Genomics meets HIV-1. Nat Rev Microbiol 2006; 4:865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limou S, Zagury JF. Immunogenetics: genome-wide association of non-progressive HIV and viral load control: HLA genes and beyond. Front Immunol 2013; 4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellay J, Shianna KV, Telenti A, Goldstein DB. Host genetics and HIV-1: the final phase?. PLoS Pathog 2010; 6:e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet 2010; 26:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fellay J. Host genetics influences on HIV type-1 disease. Antivir Ther 2009; 14:731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLaren PJ, Carrington M. The impact of host genetic variation on infection with HIV-1. Nat Immunol 2015; 16:577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren PJ, Coulonges C, Bartha I, Lenz TL, Deutsch AJ, Bashirova A, et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc Natl Acad Sci USA 2015; 112:14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 2016; 30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18:517. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet 2014; 30:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh CH, Bai XT, Moles R, Ratner L, Waldmann TA, Watanabe T, et al. Mutation of epigenetic regulators TET2 and MLL3 in patients with HTLV-I-induced acute adult T-cell leukemia. Mol Cancer 2016; 15:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcais A, Waast L, Bruneau J, Hanssens K, Asnafi V, Gaulard P, et al. Adult T cell leukemia aggressiveness correlates with loss of both 5-hydroxymethylcytosine and TET2 expression. Oncotarget 2017; 8:52256–52268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu F, Wiedmer A, Martin KA, Wickramasinghe P, Kossenkov AV, Lieberman PM. Coordinate regulation of TET2 and EBNA2 controls the DNA methylation state of latent Epstein-Barr virus. J Virol 2017; 91:e00804–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao LM, Zhao S, Zhang WY, Wang M, Li HF, Lizaso A, et al. Somatic mutations in KMT2D and TET2 associated with worse prognosis in Epstein-Barr virus-associated T or natural killer-cell lymphoproliferative disorders. Cancer Biol Ther 2019; 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Namba-Fukuyo H, Funata S, Matsusaka K, Fukuyo M, Rahmutulla B, Mano Y, et al. TET2 functions as a resistance factor against DNA methylation acquisition during Epstein-Barr virus infection. Oncotarget 2016; 7:81512–81526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saillard C, Guermouche H, Derrieux C, Bruneau J, Frenzel L, Couronne L, et al. Response to 5-azacytidine in a patient with TET2-mutated angioimmunoblastic T-cell lymphoma and chronic myelomonocytic leukaemia preceded by an EBV-positive large B-cell lymphoma. Hematol Oncol 2017; 35:864–868. [DOI] [PubMed] [Google Scholar]
- 28.Wille CK, Nawandar DM, Henning AN, Ma S, Oetting KM, Lee D, et al. 5-hydroxymethylation of the EBV genome regulates the latent to lytic switch. Proc Natl Acad Sci USA 2015; 112:E7257–E7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guallar D, Bi X, Pardavila JA, Huang X, Saenz C, Shi X, et al. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Genet 2018; 50:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deniz O, de la Rica L, Cheng KCL, Spensberger D, Branco MR. SETDB1 prevents TET2-dependent activation of IAP retroelements in naive embryonic stem cells. Genome Biol 2018; 19:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Topham JT, Titmuss E, Pleasance ED, Williamson LM, Karasinska JM, Culibrk L, et al. Endogenous retrovirus transcript levels are associated with immunogenic signatures in multiple metastatic cancer types. Mol Cancer Ther 2020; 19:1889–1897. [DOI] [PubMed] [Google Scholar]
- 32.Lv L, Wang Q, Xu Y, Tsao LC, Nakagawa T, Guo H, et al. Vpr targets TET2 for Degradation by CRL4(VprBP) E3 ligase to sustain IL-6 expression and enhance HIV-1 replication. Mol Cell 2018; 70:961–970. e965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Su L. Vpr enhances HIV-1 Env processing and virion infectivity in macrophages by modulating TET2-dependent IFITM3 expression. MBio 2019; 10:e01344-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dharan NJ, Yeh P, Bloch M, Yeung MM, Baker D, Guinto J, et al. HIV is associated with an increased risk of age-related clonal hematopoiesis among older adults. Nat Med 2021; 27:1006–1011. [DOI] [PubMed] [Google Scholar]
- 35.Chinn LW, Tang M, Kessing BD, Lautenberger JA, Troyer JL, Malasky MJ, et al. Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection. J Infect Dis 2010; 202:1836–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekenberg C, Tang ME, Zucco AG, Murray DD, MacPherson CR, Hu X, et al. Single nucleotide polymorphisms in HLA alleles are associated with HIV-1 viral load in demographically diverse, ART-naive participants from the START trial. J Infect Dis 2019; 220:1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu F, Zankharia U, Vladimirova O, Yi Y, Collman RG, Lieberman PM. Epigenetic landscape of HIV-1 infection in primary human macrophage. J Virol 2022; 96:e0016222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UNAIDS. Global AIDS Update. In. unaids.org; 2019. [Google Scholar]
- 40.Group ISS, Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, et al. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 2015; 373:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Sadr WM, Lundgren J, Neaton JD, Gordin F, Abrams D, et al. Strategies for Management of Antiretroviral Therapy Study Group. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med 2006; 355:2283–2296. [DOI] [PubMed] [Google Scholar]
- 42.Grou I-ES, Committee SS, Abrams D, Levy Y, Losso MH, Babiker A, et al. Interleukin-2 therapy in patients with HIV infection. N Engl J Med 2009; 361:1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavel JA, Group ISS, Babiker A, Fox L, Gey D, Lopardo G, et al. Effects of intermittent IL-2 alone or with peri-cycle antiretroviral therapy in early HIV infection: the STALWART study. PLoS One 2010; 5:e9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacArthur RD, Novak RM, Peng G, Chen L, Xiang Y, Hullsiek KH, et al. A comparison of three highly active antiretroviral treatment strategies consisting of nonnucleoside reverse transcriptase inhibitors, protease inhibitors, or both in the presence of nucleoside reverse transcriptase inhibitors as initial therapy (CPCRA 058 FIRST Study): a long-term randomised trial. Lancet 2006; 368:2125–2135. [DOI] [PubMed] [Google Scholar]
- 45.Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am J Hum Genet 2013; 92:841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012; 13:762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 2011; 89:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38:904–909. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 1995; 57:289–300. [Google Scholar]
- 50.Shin J-H BS, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Soft; 2006;16:1–9. [Google Scholar]
- 51.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2018. Available at: https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.