Significance
Microbial exposure in early life can have long-term consequences on health. However, our understanding of how the microbial environment shapes the offspring’s immune system is incomplete. In this report, we found that the proportion and function of fetal-derived CD8+ T cells are altered by the early microbial environment. These changes persist into adulthood and affect immune susceptibility to intracellular pathogens. Our findings provide key insights into how microbial exposure leads to individual variation in immune responsiveness, suggesting that we may be able to predict infection outcomes and disease risk based on the ratio of fetal- to adult-derived CD8+ T cells present in the adult immune system.
Keywords: immune training, CD8+ T cells, pet-shop dirty mouse model, developmental layering, developmental origins of adult health and disease
Abstract
Microbial exposure during development can elicit long-lasting effects on the health of an individual. However, how microbial exposure in early life leads to permanent changes in the immune system is unknown. Here, we show that the microbial environment alters the set point for immune susceptibility by altering the developmental architecture of the CD8+ T cell compartment. In particular, early microbial exposure results in the preferential expansion of highly responsive fetal-derived CD8+ T cells that persist into adulthood and provide the host with enhanced immune protection against intracellular pathogens. Interestingly, microbial education of fetal-derived CD8+ T cells occurs during thymic development rather than in the periphery and involves the acquisition of a more effector-like epigenetic program. Collectively, our results provide a conceptual framework for understanding how microbial colonization in early life leads to lifelong changes in the immune system.
Microbial exposure in early life can permanently program an individual’s immune system and alter lifelong disease risk. For example, children exposed to farm environments are less likely to develop asthma, whereas antibiotic use in early life is associated with an increased risk of developing inflammatory bowel disease and diabetes (1–4). This phenomenon, often referred to as “developmental programming”, is founded on the idea that various organ systems adapt a phenotype that is best suited for the environment during early development (5). However, most of these studies are based on epidemiological associations, and the underlying mechanisms remain undefined. While previous reports have highlighted the aspects of immune development that can be altered by the early microbial environment (e.g., thymic output, lymph node growth, and epigenetic modifications) (6–10), one key variable left unexamined is the developmental layering of the immune system (11–13).
The development of the immune system was long thought to progress in a linear manner from fetal life to adulthood. Now, numerous studies have established that the immune system is stratified into layers of distinct immune cells, which develop sequentially from unique waves of hematopoietic stem cells (HSCs) (14–18). In general, fetal HSCs produce a layer of fast-acting lymphocytes (e.g., neonatal CD8+ T cells, B1 B cells, and gamma/delta T cells) that persist into adulthood and represent the early effectors, while adult HSCs produce a layer of slower-acting lymphocytes (e.g., adult CD8+ T cells and B2 B cells) that preferentially contribute to the memory population (19–21). These fetal and adult developmental layers of the CD8+ T cell pool exhibit unique roles during infection (15, 22). Interestingly, the developmental switch between fetal and adult lymphopoiesis occurs at the same time the host is transitioning from the protected environment of the uterus to the antigen-rich environment of the outside world (23–25). An important and unanswered question is whether the early microbial environment alters lifelong disease risk by modulating the layering of immune cells during ontogeny.
In this report, we used a fate mapping “timestamp” approach to compare immune development in standard laboratory mice raised in a “clean” (specific pathogen-free) facility or a “dirty” (pet shop-exposed) environment. Our timestamp approach allows us to permanently label cells produced at different ages and track their contribution to immune responses in later life. Using this approach, we describe how early microbial exposure in dirty pet-shop mice alters the developmental layers of the CD8+ T cell compartment and host response to infection in adulthood.
Results
Early Microbial Exposure Enhances CD8+ T Cell-Mediated Immune Protection in Adulthood.
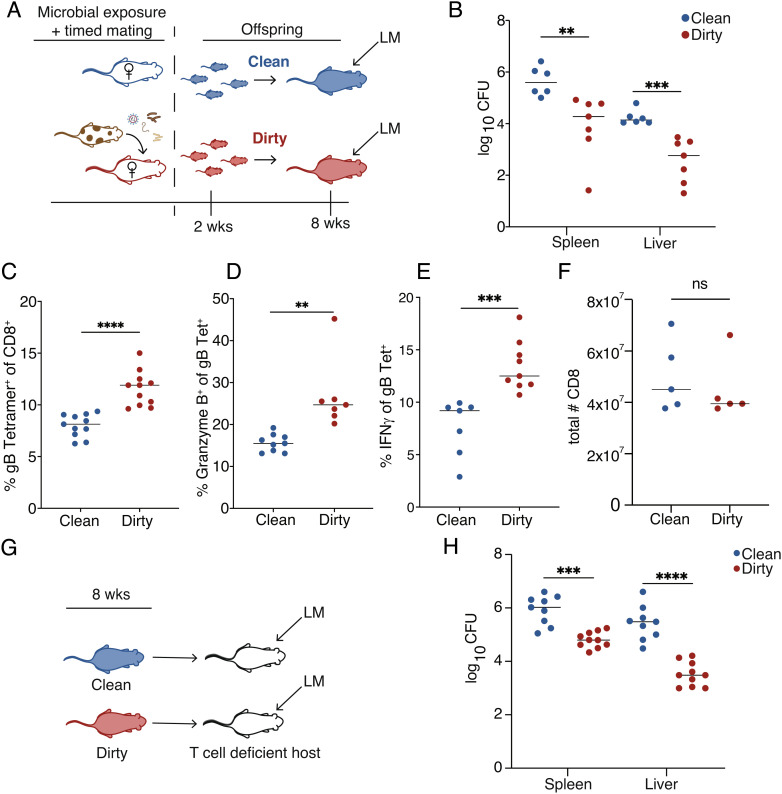
To examine the long-term impact of early microbial exposure on immune susceptibility, we modified the pet-shop mouse model (26–28) to provide microbial exposure beginning in early gestation (Fig. 1A). The pups raised in a dirty environment were born to mothers that were uniformly exposed to materials from pet-shop mice prior to mating (SI Appendix, Table S1 and Fig. S1). The pups raised in a clean environment correspond to mice raised in a standard (specific pathogen-free) lab facility. First, we challenged the offspring with Listeria monocytogenes (LM), a pathogen that requires CD8+ T cells for clearance. We found that mice raised in a dirty environment exhibited an enhanced ability to eliminate LM (Fig. 1B) and mounted a more robust antigen-specific CD8+ T cell response (Fig. 1 C–E), despite having similar numbers of CD8+ T cells before infection (Fig. 1F). We considered the possibility that the dirty environment primed CD8+ T cells to respond more quickly. However, CD8+ T cell adoptive transfer experiments showed that even when placed in the same environment, dirty cells were more protective than their clean counterparts (Fig. 1 F and G). Collectively, these data demonstrate that microbial exposure induces changes to the CD8+ T cell compartment that enhance its ability to protect the host against infection.
Fig. 1.
Early microbial exposure enhances immunity in adulthood. (A) Approach to generate dirty mice: Female laboratory mice were exposed to fecal and cage contents of pet-shop mice for 4 wks. Their offspring were maintained in a dirty environment. (B) Bacterial load at 3 d post infection (dpi) in 8-wk-old mice that were directly infected with 1 × 104 CFU of WT LM-gB. (C) Percent of CD8+ T cells that are positive for the Kb:gB498–505 tetramer (D) Granzyme B and (E) interferon-gamma at 5 dpi. (F) Total number of splenic CD8+ T cells. (G) Approach to transfer purified CD8s into T cell-deficient hosts. (H) Bacterial load at 3 dpi from TCRα KO recipients that received 5 × 106 purified CD8s from clean or dirty 8-wk-old donor mice and subsequently infected with LM-gB. Data for B are pooled from two independent experiments (n = 3–4 mice/group) and are ±SEM. Data from C–E are pooled from two independent experiments (n = 4–5 mice/group) and are ±SEM. Data for G are pooled from two independent experiments (n = 4–5 mice/group) and are mean ± SEM. Statistical significance was determined by Student’s t test (ns = not significant, **P < 0.005, ***P < 0.0005, ***P < 0.0005, ****P < 0.00005).
Early Microbial Exposure Leads to an Accumulation of Fetal-Derived CD8+ T Cells in Adulthood.
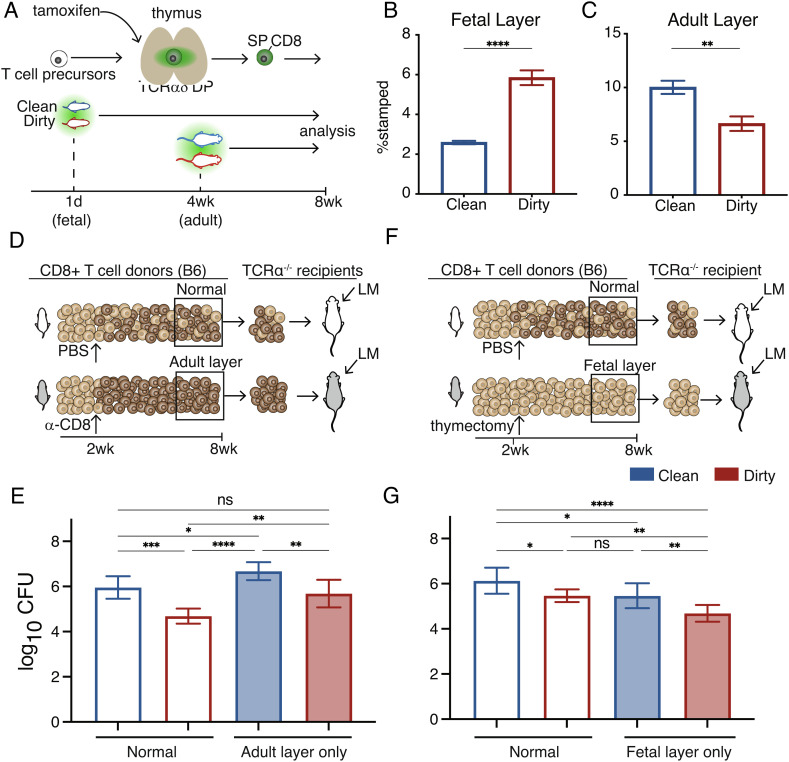
We next sought to understand how a more microbially diverse environment in early life leads to a more protective CD8+ T cell compartment in adulthood. On the one hand, the microbial environment may enhance the “average phenotype” or function of all CD8+ T cells in the starting pool in a similar manner (the “uniform” model). Alternatively, microbes could alter immune responsiveness by changing the proportion of cells that are derived from different developmental periods (the “layered” model). To distinguish between the uniform and layered models of microbial education, we first determined whether the developmental layering of CD8+ T cells was altered in dirty mice. To do this, we used fate-mapping timestamp mice (see Methods) to permanently mark or timestamp a wave of cells at discrete times of thymic development. We timestamped dirty and clean pups at birth to mark fetal-derived cells (29) and at 28 d to mark adult-derived cells (Fig. 2A) (24). At 8 wk of age, we found a higher proportion of fetal-derived CD8+ T cells and a lower proportion of adult-derived CD8+ T cells present in dirty mice (Fig. 2 B and C). This pattern persisted across multiple stages of development (SI Appendix, Fig. S2). Importantly, cells with different developmental origins had an antigen-inexperienced (CD49dlo) phenotype (30–32), indicating that the increased proportion of fetal-derived CD8+ T cells in dirty mice was not due to an expansion of antigen-specific cells but rather to changes in the developmental layering of the naive CD8+ T cell compartment (SI Appendix, Fig. S3). We also found that delaying microbial exposure to adulthood did not lead to an increase of CD8+ T cells in the fetal layer (SI Appendix, Fig. S4), indicating that there is a critical window of opportunity in early life when the developmental layers of the CD8+ T cell compartment can be altered.
Fig. 2.
Expansion of the fetal layer drives enhanced protection in dirty mice. (A) Approach to timestamp the fetal and adult layers in clean and dirty mice. (B) The proportion of fetal- and (C) adult-derived CD8+ T cells in 8-wk-old clean and dirty mice. (D) Schematic showing the approach to deplete the fetal layer. (E) Pathogen burden in clean and dirty mice lacking the fetal layer of CD8+ T cells. (F) Schematic showing the approach to prevent the adult layer from forming. (G) Pathogen burden in clean and dirty mice lacking the adult layer of CD8+ T cells. Data for B and C are ±SEM and are pooled from two independent experiments (n = 3–4 mice/group). Data for E and G are mean ± SEM and are pooled from two independent experiments (n = 3–4 mice/group). Statistical significance for B and C was determined by Student’s t test. Significance for E and F was determined by a one-way ANOVA with a Tukey multiple comparison posttest (ns = not significant, *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005).
Adjusting the Developmental Layers in Clean and Dirty Mice Normalizes Immune Susceptibility.
The timestamping experiments suggest that dirty mice mount a more protective CD8+ T cell response because they are composed of a larger proportion of fast-acting, fetal-derived cells prior to infection. However, we wanted to know whether fetal-derived CD8+ T cells provide enhanced protection against intracellular pathogens in adulthood and whether the observed changes in the proportions of fetal- and adult-derived CD8+ T cells in dirty mice were functionally significant. To address these questions, we designed experiments to see if we could normalize susceptibility in clean and dirty mice by manipulating the developmental layers in the CD8+ T cell compartment. Our approach involved reconstituting T cell-deficient recipient mice with clean or dirty donor CD8+ T cells that lack either the fetal or adult layer. To ablate the fetal layer, we administered a CD8α-depleting antibody to clean or dirty mice at 2 wk of age (SI Appendix, Fig. S5). Once the mice reached 8 wk of age, we transferred an equal number of purified CD8+ T cells from depleted donor mice into T cell-deficient recipients and infected the recipients with LM (Fig. 2D). Importantly, recipient mice that received cells from dirty mice lacking the fetal layer had a pathogen load comparable to clean mice with a normal CD8+ T cell compartment (Fig. 2E). To test whether immune susceptibility was changed in mice lacking the adult layer of CD8+ T cells, we performed thymectomies on clean and dirty mice at 2.5–3 wk of age to prevent the adult layer from forming (Fig. 2F). When the mice were 8 wk old, we again transferred an equal number of donor CD8+ T cells into the T cell-deficient recipients and infected the mice with LM. Notably, the recipient mice that received cells from clean mice with only the fetal layer had a similar pathogen load compared to recipient mice that received cells from dirty mice (Fig. 2G). Together, these results establish the fetal layer as essential for enhanced immunity in dirty mice.
Early Microbial Exposure Leads to Enhanced Responsiveness of Fetal-Derived CD8+ T Cells.
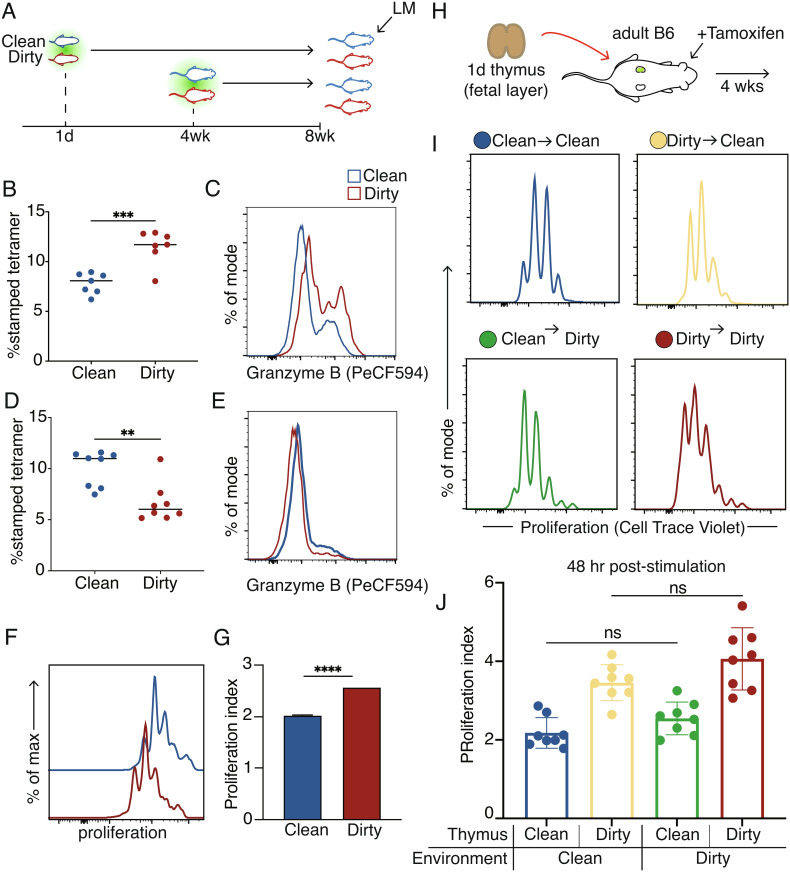
One of the most intriguing findings was that recipient mice receiving fetal-derived cells from dirty mice had the lowest pathogen load (Fig. 2G), suggesting that fetal cells from dirty mice may be even more protective than their counterparts from clean mice. To examine the impact of microbial exposure on the behavior of fetal- and adult-derived CD8+ T cells, we compared the function and phenotype of the developmental layers in clean and dirty mice after direct infection with LM (Fig. 3A). Interestingly, fetal-derived cells from dirty mice mounted an enhanced response and produced more of the effector protein granzyme B compared to their clean counterparts (Fig. 3 B and C). In contrast, adult-derived cells from dirty mice contributed less to the response while producing lower levels of granzyme B (Fig. 3 D and E), most likely due to increased competition from fetal-derived CD8+ T cells.
Fig. 3.
Fetal-derived cells in the dirty environment are more responsive to stimulation. (A) Approach to infect clean or dirty timestamp mice with Listeria. (B) The proportion of antigen-specific fetal-derived cells at 5 d post-LM infection. (C) Representative histogram showing Granzyme B production after infection in fetal-derived cells. (D) The proportion of antigen-specific adult-derived CD8+ T cells at 5 d post-LM infection. (E) Representative histogram showing Granzyme B production in adult-derived cells. (F) Representative histograms of proliferation peaks after aCD3/28 activation. (G) Division index of clean or dirty fetal-derived cells. (H) Experimental approach for kidney capsule thymic transplant surgeries. (I) Representative proliferation peaks 48 h after in vitro activation with aCD3/CD28. (J) Division index of fetal-derived cells after aCD3/28 stimulation. Data for B and D are ±SEM and are pooled from two independent experiments (n = 3–4 mice/group). Statistical significance for B and D was determined by Student’s t test. Statistical significance for J was determined by a one-way ANOVA with a Tukey multiple comparison posttest (ns = not significant, **P < 0.005, ***P < 0.0005).
To test whether the behavior of fetal-derived CD8+ T cells in dirty mice is pathogen specific, we challenged our timestamp mice with Vaccinia virus, a large DNA virus. Even though Listeria and Vaccinia infect distinct cell types, elicit different cytokines, and exhibit altered antigen kinetics, a more robust response by fetal CD8+ T cells in dirty mice was observed in both infectious scenarios (SI Appendix, Fig. S6). These data demonstrate that our results are not pathogen dependent and that the microbial environment preferentially enhances the functions of fetal-derived CD8+ T cells in response to both intracellular bacteria and viruses.
We considered the possibility that the enhanced responsiveness of fetal-derived cells in dirty mice was due to cell-intrinsic differences. To test this, we compared the ability of cells from clean and dirty mice to proliferate after in vitro stimulation via the T cell receptor (TCR). Fetal-derived cells from dirty mice turned out to be more proliferative than their clean counterparts (Fig. 3 F and G). We also considered whether this enhanced ability of dirty fetal-derived cells to proliferate could relate to differences in their starting phenotype. Indeed, we found that fetal-derived CD8+ T cells from dirty mice have the highest proportion of virtual memory (VM) cells, a subset of antigen-inexperienced cells that are functionally similar to memory cells, which could explain their more proliferative response after stimulation (SI Appendix, Fig. S7 A and B) (33, 34). However, even after controlling for their initial phenotype status, we found that dirty fetal-derived CD8+ T cells still underwent division at higher rates in the steady state and in response to TCR stimulation (SI Appendix, Fig. S8 C–E). This indicates that the microbial environment programs the fetal layer of CD8+ T cells to be highly responsive to antigenic stimulation at the individual cell level.
Microbial Education Occurs During Thymic Development.
An important question is when does the microbial environment “instruct” fetal-derived CD8+ T cells to behave differently? The simplest explanation is that fetal-derived CD8+ T cells are programmed differently in clean and dirty mice by environmental factors in the periphery. However, recent studies have suggested that “training” of immune cells can occur during the early stages of their development (35–37), raising the possibility that the microbial education of CD8+ T cells takes place in progenitors during thymic development. To differentiate between these possibilities, we took newborn timestamp thymii from clean or dirty donor mice and transplanted them under the kidney capsule of clean or dirty adult recipients (Fig. 3H). We administered tamoxifen immediately after thymic transplantation to mark a wave of fetal-derived thymic precursors and tracked their behavior after maturing in either a clean or dirty peripheral environment. Interestingly, we observed a preferential accumulation of fetal-derived CD8+ T cells from dirty mice that was largely independent of the microbial status of the peripheral environment (SI Appendix, Fig S8 A and B). In addition, the fetal-derived CD8+ T cells from dirty mice exhibited an enhanced ability to proliferate after stimulation, regardless of whether they were matured in a clean or dirty environment (Fig. 3 I and J). We also considered whether antigen experience may be driving the enhanced proliferation of fetal-derived CD8+ T cells in dirty mice. However, the expression levels of CD49d were comparable in clean and dirty CD8+ T cells matured in the same environment (SI Appendix, Fig. S8C). Collectively, these data suggest that microbial programming of fetal-derived CD8+ T cells occurs in the progenitors and is retained in the mature cells after they have exited the thymus.
Fetal-Derived CD8+ T Cells Are Trained by the Microbial Environment.
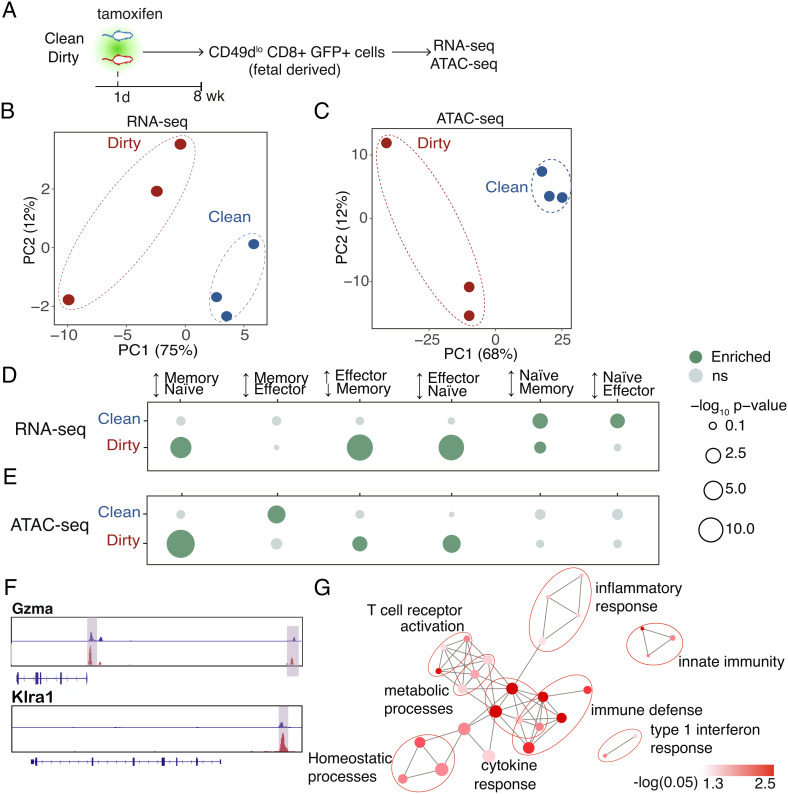
Lastly, we sought to understand the regulatory basis of how the microbial environment enhances the ability of peripheral fetal-derived CD8+ T cells to respond to infection. We previously reported that fetal-derived CD8+ T cells respond more rapidly to infection because they have increased chromatin accessibility at regulatory sites for effector genes before stimulation (22). However, the enhanced immune functions in dirty mice suggest that fetal-derived CD8+ T cells may undergo additional programming when developed in a dirty environment. To address this possibility, we compared the transcriptomes (RNA-seq) and chromatin landscapes (ATAC-seq) of fetal-derived CD8+ T cells from clean and dirty mice at 8 wk of age (Fig. 4A). For these studies, we sorted fetal-derived CD8+ T cells with a CD49dlo phenotype to focus on antigen-inexperienced cells (30–32) and generated fully matched RNA- and ATAC-seq data sets.
Fig. 4.
Dirty fetal-derived cells lose their naive programming. (A) Schematic of sorting strategy. (B) Principal component analysis of genome-wide sequencing from RNA- and (C) ATAC-sequencing data sets. (D) Enrichment analysis for various CD8+ T cell states in genes with increased expression and/or (E) increased accessibility in clean and dirty cells. (F) Genome browser views of a dirty-upregulated gene and a dirty-poised gene. (G) Enrichment analysis for GO “biological process” terms in dirty-poised genes. Pathways are connected if they share 30% or more genes. Node sizes denote gene set sizes. Statistical significance of differential expression and differential accessibility was determined by the Wald test (D) and quasilikelihood F test (E) with Benjamini–Hochberg correction (P < 0.05). Corrected P-values for enrichment or depletion denoted by dot size and color. Enrichment values in D and F were determined by a hypergeometric test followed by Benjamini–Hochberg correction. (P < 0.05).
Consistent with their unique behavior during infection, fetal-derived cells from dirty mice exhibited distinct transcriptomic and epigenomic profiles (Fig. 4 B and C). To understand the nature of these differences, we compared the profiles of upregulated genes in clean and dirty cells to establish gene sets that reflect different CD8+ T cell states. Dirty fetal-derived cells exhibited increased expression and accessibility for genes corresponding to effector or memory cells (Fig. 4 D–F). Since fetal-derived cells have higher proportions of VM cells, we also compared the gene expression profiles and chromatin landscapes of phenotype-matched cells from clean and dirty mice and found that the distinct loss of the naive state in dirty mice persisted in phenotypically similar populations (SI Appendix, Fig. S9). Gene ontology (GO) analysis of the poised genes, which are not differentially expressed but have altered accessibility in the corresponding promoter and enhancer regions, revealed that the epigenome of dirty fetal-derived cells was more associated with immune defense (innate immunity, inflammatory response, and TCR activation) and cytokine response (type I IFN, cytokine response), while their clean counterparts overlapped with more general developmental processes (Fig. 4G and SI Appendix, Fig. S10). Overall, these data suggest that mice raised in a dirty environment mount stronger responses to infection because their T cells are epigenetically “primed” for stimulation.
Discussion
In this study, we offer a conceptual framework for understanding how the microbial environment alters immune susceptibility in adulthood. By tracking T cells produced at different stages of life, we uncovered a link between microbial exposure, developmental layering, and immune susceptibility. In response to increased microbial exposure, the developing immune system adapts by expanding the “fast-acting” fetal layer of CD8+ T cells, allowing the host to mount a more targeted response to early infection in adulthood. When microbial exposure is reduced, the slower-acting adult layer of cells preferentially accumulates, leaving the host more susceptible to intracellular pathogens. These findings offer insights into how variation in microbial exposure in early life can program lifelong intraindividual variation in the immune system and may allow us to predict infection outcomes and disease risk based on the ratio of fetal- to adult-derived CD8+ T cells present in adults.
Our results are significant because there is a prevailing notion that the CD8+ T cell compartment is a homogenous pool composed of a single lineage of cells without a structure. Although we previously showed that the CD8+ T cell compartment is composed of CD8+ T cells with distinct developmental origins (22, 38), an important and unanswered question was whether the number of fetal- to adult-derived CD8+ T cells present in the starting population corresponded to changes in susceptibility to infection. Here, we extend our earlier work by showing that (i) developmental layering of the CD8+ T cell compartment does indeed have a direct effect on how the host responds to intracellular pathogens, and (ii) this layering can be shaped by the microbial environment.
Perhaps one of the most interesting findings is that the developmental layering of the CD8+ T cell compartment can be altered by changes in microbial exposure in early life but not in adulthood. This finding is consistent with earlier studies, suggesting that there is a “window of opportunity” in early life in which the immune system can be programmed by environmental factors (23, 39, 40). However, with progressing age, a loss of plasticity occurs and the window of opportunity closes. We propose that the window of opportunity exists in the immune system during perinatal life because functionally distinct immune cells derived from unique HSCs are present. It is these cells that are most affected by the microbial environment.
Finally, our findings offer a perspective on immune training, which currently focuses on understanding how innate immune cells, such as natural killer cells and macrophages, retain the memory of past infections (41). The trained immune phenotype in innate cells is associated with epigenetic reprogramming, allowing them to respond more strongly to subsequent infections. We found that fetal-derived CD8+ T cells in dirty mice similarly exhibit enhanced functions related to increased accessibility at effector genes, indicating that immune training is not limited to innate immune cells and that CD8+ T cells are also programmed by the microbial environment. In the future, it will be important to examine how other environmental factors, such as stress, diet, and toxins alter the developmental layering and programming of CD8+ T cells. Knowledge gained from these studies will broaden our fundamental understanding of immune ontogeny and cell-mediated immunity.
Methods
Key information is summarized below. Extended methods can be found in SI Appendix.
Mice and Microbial Exposure.
C57BL/6 mice were purchased from Charles River Laboratories. ZsGreen and TCRδCre-ERT2 mice were obtained from Jackson Laboratories. Pet-shop mice were purchased from pet shops. Generation of dirty mice occurred at an off-campus mouse facility. The microbial material from pet-shop mice was transferred to female laboratory mice through an oral gavage from the fecal material and bedding transfer over a 4-wk period. The facilities are accredited by the American Association of Accreditation of Laboratory Animal Care. All protocols regarding animal use were reviewed and approved by the Institutional Animal Care and Use Committee at Cornell University.
Adoptive Transfer and Pathogen Burden.
First, 5 × 106 purified CD8+ T cells were injected intravenously into a TCRα knockout recipient. The next day, mice were infected with 1 × 104 CFU of wild-type (WT) LM expressing the gB-8p peptide (Lm-gB), as previously described (42). At 3 d post infection (peak of bacterial growth), lysed tissue homogenate from the spleen and liver were serially plated and colony growth enumerated.
In Vitro Stimulation.
CD8+ T cells were isolated from the spleen using positive magnetic selection with CD8 microbeads (Miltenyi). Cells were labeled with Cell Trace Violet and stimulated with plate-bound anti-CD3 (2C11), then cultured with complete RPMI supplemented with 100 U/ml human IL2 and 4 μg CD28/ml (37.51). Cells were harvested at the time points indicated in the figure legends, stained for surface markers, and analyzed by flow cytometry.
Flow Cytometry.
For surface staining, cells were processed using an IC fixation kit (Invitrogen), according to the manufacturer’s protocol. Samples that underwent intracellular staining were processed using Foxp3/Transcription Factor Staining Buffer, according to the manufacturer’s instructions.
Statistical Analysis.
Error bars are represented by SEM or SD. Unless otherwise stated in the figure legends, statistical significance was determined by Student’s t test or by a 1- or 2-way ANOVA followed by an appropriate posttest, as indicated in the figure legends. Significance is denoted by the following: *P < 0.05, **P < 0.05, ***P < 0.0005, ****P < 0.00005.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the Cornell Center for Animal Resource and Education (CARE) for expert mouse breeding assistance. Cell sorting was done at Cornell University’s Flow Cytometry Facility in the Biotechnology Resource Center. This work was supported by NIH awards R01 AI142867 (to B.D.R. from the National Institute of Allergy and Infectious Disease), R01 HD107798 (to B.D.R. from the National Institute of Child Health and Human Development), F31 AI157236 (to C.T. from the National Institute of Allergy and Infectious Disease), and P50HD076210 (to A.G. from National Institute of Child Health and Human Development). M.P.D. is supported by National Health and Medical Research Council (NHMRC) (Australia) Investigator Grant (1173027).
Author contributions
C.T., M.P.D., A.G., and B.D.R. designed research; C.T., C.W.P.D., K.J.Y.M., S.P.W., J.K.G., and N.L.S. performed research; C.T., D.S.I., A.R., J.K.G., M.P.D., N.L.S., A.G., and B.D.R. analyzed data; and C.T., D.S.I., N.L.S., A.G., and B.D.R. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Darabi B., Rahmati S., HafeziAhmadi M. R., Badfar G., Azami M., The association between caesarean section and childhood asthma: An updated systematic review and meta-analysis. Allergy Asthma Clin. Immunol. 15, 62 (2019), 10.1186/s13223-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ege M. J., et al. , Prenatal exposure to a farm environment modifies atopic sensitization at birth. J. Allergy Clin. Immunol. 122, 407–412, 412 e401–404 (2008), 10.1016/j.jaci.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Kirjavainen P. V., et al. , Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat. Med. 25, 1089–1095 (2019), 10.1038/s41591-019-0469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kronman M. P., Zaoutis T. E., Haynes K., Feng R., Coffin S. E., Antibiotic exposure and IBD development among children: A population-based cohort study. Pediatrics 130, e794–803 (2012), 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateson P., et al. , Developmental plasticity and human health. Nature 430, 419–421 (2004), 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z., et al. , Peripheral lymphoid volume expansion and maintenance are controlled by gut microbiota via RALDH+ dendritic cells. Immunity 44, 330–342 (2016), 10.1016/j.immuni.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Pavert S. A., et al. , Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 508, 123–127 (2014), 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ennamorati M., et al. , Intestinal microbes influence development of thymic lymphocytes in early life. Proc. Natl. Acad. Sci. U.S.A. 117, 2570–2578 (2020), 10.1073/pnas.1915047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legoux F., et al. , Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366, 494–499 (2019), 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 10.Gury-BenAri M., et al. , The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246 e1213 (2016), 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 11.Davenport M. P., Smith N. L., Rudd B. D., Building a T cell compartment: How immune cell development shapes function. Nat. Rev. Immunol. 20, 499–506 (2020), 10.1038/s41577-020-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzenberg L. A., Herzenberg L. A., Toward a layered immune system. Cell 59, 953–954 (1989), 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]
- 13.Ikuta K., et al. , A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell 62, 863–874 (1990), 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 14.Mold J. E., et al. , Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 330, 1695–1699 (2010), 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., et al. , Fetal and adult progenitors give rise to unique populations of CD8+ T cells. Blood 128, 3073–3082 (2016), 10.1182/blood-2016-06-725366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tieppo P., et al. , The human fetal thymus generates invariant effector gammadelta T cells. J. Exp. Med. 217 (2020), 10.1084/jem.20190580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramond C., et al. , Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat. Immunol. 15, 27–35 (2014), 10.1038/ni.2782. [DOI] [PubMed] [Google Scholar]
- 18.Montecino-Rodriguez E., et al. , Distinct genetic networks orchestrate the emergence of specific waves of fetal and adult B-1 and B-2 development. Immunity 45, 527–539 (2016), 10.1016/j.immuni.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montecino-Rodriguez E., Dorshkind K., B-1 B cell development in the fetus and adult. Immunity 36, 13–21 (2012), 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudd B. D., Neonatal T cells: A reinterpretation. Annu. Rev. Immunol. 38, 229–247 (2020), 10.1146/annurev-immunol-091319-083608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan L., et al. , A fetal wave of human type 3 effector gammadelta cells with restricted TCR diversity persists into adulthood. Sci. Immunol. 6 (2021), 10.1126/sciimmunol.abf0125. [DOI] [PubMed] [Google Scholar]
- 22.Smith N. L., et al. , Developmental origin governs CD8(+) T cell fate decisions during infection. Cell 174, 117–130 e114 (2018), 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Jain N., The early life education of the immune system: Moms, microbes and (missed) opportunities. Gut. Microbes 12, 1824564 (2020), 10.1080/19490976.2020.1824564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim I., Saunders T. L., Morrison S. J., Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130, 470–483 (2007), 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowie M. B., et al. , Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. U.S.A. 104, 5878–5882 (2007), 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beura L. K., et al. , Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016), 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huggins M. A., et al. , Microbial exposure enhances immunity to pathogens recognized by TLR2 but increases susceptibility to cytokine storm through TLR4 sensitization. Cell Rep. 28, 1729–1743 e1725 (2019), 10.1016/j.celrep.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labuda J. C., Fong K. D., McSorley S. J., Cohousing with dirty mice increases the frequency of memory T cells and has variable effects on intracellular bacterial infection. Immunohorizons 6, 184–190 (2022), 10.4049/immunohorizons.2100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adkins B., Developmental regulation of the intrathymic T cell precursor population. J. Immunol 146, 1387–1393 (1991). [PubMed] [Google Scholar]
- 30.Haluszczak C., et al. , The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206, 435–448 (2009), 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sosinowski T., et al. , CD8alpha+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J. Immunol. 190, 1936–1947 (2013), 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu B. C., Martin B. E., Stolberg V. R., Chensue S. W., Cutting edge: Central memory CD8 T cells in aged mice are virtual memory cells. J. Immunol. 191, 5793–5796 (2013), 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White J. T., et al. , Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat. Commun. 7, 11291 (2016), 10.1038/ncomms11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J. Y., Hamilton S. E., Akue A. D., Hogquist K. A., Jameson S. C., Virtual memory CD8 T cells display unique functional properties. Proc. Natl. Acad. Sci. U.S.A. 110, 13498–13503 (2013), 10.1073/pnas.1307572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim A. I., et al. , Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science 373 (2021), 10.1126/science.abf3002. [DOI] [PubMed] [Google Scholar]
- 36.Mitroulis I., et al. , Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161 e112 (2018), 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaufmann E., et al. , BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190 e119 (2018), 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 38.Reynaldi A., et al. , Fate mapping reveals the age structure of the peripheral T cell compartment. Proc. Natl. Acad. Sci. U.S.A. 116, 3974–3981 (2019), 10.1073/pnas.1811634116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torow N., Hornef M. W., The neonatal window of opportunity: Setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 198, 557–563 (2017), 10.4049/jimmunol.1601253. [DOI] [PubMed] [Google Scholar]
- 40.Apostol A. C., Jensen K. D. C., Beaudin A. E., Training the fetal immune system through maternal inflammation-A Layered hygiene hypothesis. Front. Immunol. 11, 123 (2020), 10.3389/fimmu.2020.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netea M. G., et al. , Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020), 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith N. L., et al. , Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J. Immunol. 193, 177–184 (2014), 10.4049/jimmunol.1400553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the manuscript and/or SI Appendix.