Significance
High frequencies of mutations in the spliceosomal gene SF3B1 occur in many cancers. Understanding the mechanism by which these mutations affect RNA splicing is important for development of therapeutic interventions. SF3B1 mutations have been shown to disrupt the interaction of SF3B1 with another splicing factor, SUGP1, leading to splicing errors that are of cancer relevance. However, what underlies SUGP1-mediated splicing misregulation remains unknown. Here, using biochemical and splicing assays as well as structural analysis, we identified the RNA helicase DHX15 as the essential SUGP1-associated helicase and characterized in detail its critical interaction with SUGP1. Our findings thus define an SF3B1/SUGP1/DHX15 axis important for accurate splicing and provide additional insight into how mutant SF3B1 misregulates splicing in cancer.
Keywords: spliceosome, G-patch, helicase, myelodysplastic syndromes, leukemia
Abstract
SF3B1 is the most frequently mutated spliceosomal gene in cancer. Several hotspot mutations are known to disrupt the interaction of SF3B1 with another splicing factor, SUGP1, resulting in the RNA missplicing that characterizes mutant SF3B1 cancers. Properties of SUGP1, especially the presence of a G-patch motif, a structure known to function by activating DEAH-box RNA helicases, suggest the requirement of such an enzyme in SUGP1 function in splicing. However, the identity of this putative helicase has remained an important unanswered question. Here, using a variety of protein–protein interaction assays, we identify DHX15 as the critical helicase. We further show that depletion of DHX15 or expression of any of several DHX15 mutants, including one implicated in acute myeloid leukemia, partially recapitulates the splicing defects of mutant SF3B1. Moreover, a DHX15-SUGP1 G-patch fusion protein is able to incorporate into the spliceosome to rescue the splicing defects of mutant SF3B1. We also present the crystal structure of the human DHX15-SUGP1 G-patch complex, which reveals the molecular basis of their direct interaction. Our data thus demonstrate that DHX15 is the RNA helicase that functions with SUGP1 and additionally provide important insight into how mutant SF3B1 disrupts splicing in cancer.
High-throughput sequencing of many cancer genomes in recent years revealed unexpectedly high frequencies of mutations in genes encoding splicing factors (1, 2), indicating a causal role of splicing misregulation in tumorigenesis. Among the splicing genes, splicing factor 3b subunit 1 (SF3B1) is the most frequently mutated. Recurrent mutations in SF3B1 have been found in a variety of hematological malignancies and solid tumors, e.g., at a frequency of approximately 30% in myelodysplastic syndromes (MDS) (3, 4), up to 83% in the subtypes of MDS with ring sideroblasts (2, 4), up to 15% in chronic lymphocytic leukemia (5, 6), 42% in mucosal melanoma (7), 19% in uveal melanoma (8), and lower frequencies in pancreatic and breast cancers (9, 10).
Several studies have examined the mechanism by which SF3B1 cancer mutations affect splicing. It is now clear that SF3B1 mutants induce use of cryptic 3′ splice sites (ss) typically located ~10 to 30 nt upstream of the associated canonical 3′ss and that they do so by inducing recognition of alternative upstream branchsites during splicing (11–14). More than 160 misspliced cryptic 3′ss were identified in RNA samples from MDS patients with SF3B1 mutations (14). Mutant SF3B1-induced missplicing can lead to severe functional consequences. For example, mutant SF3B1 induces use of a cryptic 3′ss in transcripts encoding the kinase MAP3K7 (mitogen-activated protein kinase kinase kinase 7), and the resulting reduced levels of the kinase play a significant role in development of the severe anemia often observed in MDS patients (15). It has also been shown that mutant SF3B1 induces inclusion of a poison exon in the tumor suppressor gene bromodomain containing 9 (BRD9), promoting melanomagenesis (16).
It remains unclear how mutant SF3B1 induces recognition of alternative upstream branchsites. SF3B1 is a core subunit of the SF3B complex, which associates with U2 snRNP (small nuclear ribonucleoprotein), an essential component of the spliceosome that is critical for branchsite recognition (17). Multiple interactions are involved in branchsite recognition during early-stage spliceosome assembly. Splicing factor 1 (SF1) initially binds to the branchsite, cooperatively with the heterodimer U2 small nuclear RNA auxiliary factor (U2AF), whose two subunits U2AF1 and U2AF2 bind to the 3′ss and the adjacent polypyrimidine tract, respectively. The SF3B1-containing U2 snRNP is then recruited to the precursor mRNA (pre-mRNA), and SF1 is displaced from the branchsite with adenosine triphosphate (ATP) hydrolysis, allowing U2 small nuclear RNA (snRNA) to recognize the branchsite via base pairing (17).
Almost all of the numerous SF3B1 mutations are heterozygous missense mutations at specific residues located in the huntingtin, elongation factor 3, the A subunit of protein phosphatase 2A, and target of rapamycin 1 (HEAT) repeat domain, which provides a major scaffold for protein–protein interactions (18). The great majority of the mutated residues are clustered in HEAT repeats H4–H7 (18), suggesting a common functional mechanism for these mutations. A small number of mutations, however, are scattered in other HEAT repeats, and these may influence splicing by different mechanism(s). We previously showed that several SF3B1 mutations, including the most common hotspot mutation, K700E, disrupt the interaction of SF3B1 with another splicing factor, SURP and G-patch domain containing 1 (SUGP1), during spliceosome assembly, leading to use of alternative upstream branchsites and cryptic 3′ss during splicing (14). We also showed that mutation of the G-patch motif (G-patch) of SUGP1 recapitulated the splicing defects of mutant SF3B1 when the mutant SUGP1 was expressed in wild-type (WT) SF3B1 cells (14). The importance of the SUGP1 G-patch was further highlighted by our findings that naturally occurring SUGP1 missense mutations in cancers that recapitulate mutant SF3B1-specific missplicing all flank the G-patch (19). Because G-patch-containing proteins are well known to activate DEAH-box RNA helicases (20), we suggested that a DEAH helicase is involved in SUGP1 function in branchsite recognition. However, whether this was in fact the case and, if so, the identity of the RNA helicase remain unknown.
Given the importance of the putative RNA helicase in our understanding of SF3B1/SUGP1 function in splicing, as well as the mechanism of SF3B1 mutations in cancer, we here describe experiments that not only identify the RNA helicase but also provide biochemical and structural insights into its role in SUGP1-mediated splicing. We first employed affinity purification assays followed by mass spectrometry to reveal that DEAH-box helicase 15 (DHX15) specifically associates with the SUGP1 G-patch. We then confirmed that DHX15 interacts with SUGP1 both in vitro and in the spliceosome. We further showed that knockdown of DHX15 or expression of several DHX15 mutants, including the recurrent R222G mutation found in acute myeloid leukemia (AML) (21), partially recapitulates the splicing defects of mutant SF3B1. We also demonstrated that a DHX15-SUGP1 G-patch fusion protein is able to rescue the splicing defects of mutant SF3B1. Lastly, we determined the crystal structure of the human DHX15-SUGP1 G-patch complex, which provides detailed insights into their direct interaction. Thus, our study identifies DHX15 as the RNA helicase involved in SF3B1/SUGP1-mediated splicing and enhances our understanding of how mutant SF3B1 misregulates RNA splicing in cancer.
Results
DHX15 Specifically Associates with the SUGP1 G-Patch.
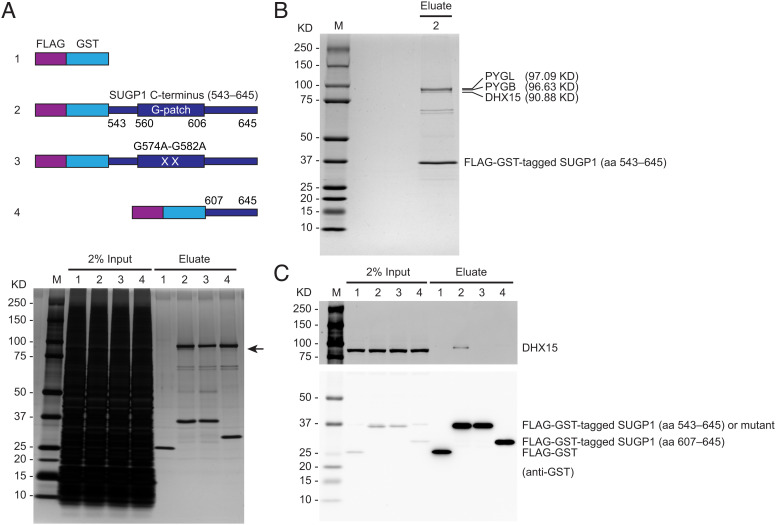
As mentioned in the Introduction, our previous study suggested the involvement of a DEAH-box helicase in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer (14). To identify the putative helicase, we employed affinity purification to isolate proteins that are associated with the SUGP1 G-patch. To this end, we made a plasmid construct encoding a SUGP1 derivative consisting of two tandem affinity tags, FLAG (a.k.a. DYKDDDDK) and GST, fused to the C terminus of SUGP1 (aa 543–645), which includes the G-patch (Fig. 1A). We also made three control plasmid constructs, specifically one encoding a mutant version of the above construct in which the two most-conserved Gly residues of the G-patch were changed to Ala residues (G574A-G582A), another encoding a portion of the SUGP1 C terminus (aa 607–645) that lacks the G-patch, and one encoding only the two affinity tags (Fig. 1A). After transfecting each of these plasmids into HEK293T cells to express the affinity-tagged proteins, we harvested the cells, prepared whole-cell extracts, and used them to perform two rounds of affinity purification, with anti-DYKDDDDK antibody and Glutathione Sepharose beads sequentially. By resolving the affinity-purified proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by silver staining, we observed several protein bands associated with the C terminus of SUGP1 (aa 543–645). One protein at an apparent molecular weight of ~91-KD associated specifically with SUGP1 (aa 543–645) but not with its G-patch mutant version or the truncated SUGP1 (aa 607–645) lacking the G-patch (Fig. 1A).
Fig. 1.
DHX15 specifically associates with the SUGP1 G-patch. (A, Upper) Schematic representation of plasmid constructs (1 to 4) expressing FLAG-GST tandem tags, FLAG-GST-tagged SUGP1 (aa 543–645), its G574A-G582A mutant, and FLAG-GST-tagged SUGP1 (aa 607–645), respectively. (Lower) Each of the four constructs was transfected into HEK293T cells, followed by small-scale affinity purification using FLAG and GST tags. The purified proteins in eluates were resolved by SDS-PAGE, followed by silver staining. (B) HEK293T cells were transfected with construct 2 in (A), followed by large-scale affinity purification using FLAG and GST tags. The purified proteins were resolved by SDS-PAGE, followed by staining with QC Colloidal Coomassie Stain (Bio-Rad). (C) Aliquots of the purified proteins in (A) were resolved by SDS-PAGE, followed by western blotting. In all panels: M, Precision Plus Protein marker (Bio-Rad).
To identify the above 91-KD protein, we performed a large-scale purification using the plasmid expressing the affinity-tagged SUGP1 (aa 543–645). After resolving the affinity-purified proteins by SDS-PAGE followed by colloidal Coomassie staining (Fig. 1B), we cut out the relevant gel region (~85 to 100 KD) and identified the proteins in this gel section by mass spectrometry. We identified a total of 25 proteins, each with at least one unique peptide (Dataset S1). Because the great majority of these proteins were of very low abundance (no more than three unique peptides each), we focused on the three most abundant ones that have at least 38 unique peptides each and whose molecular weights match the sizes of the protein bands in the gel (Fig. 1B). Based on the molecular weights of these three proteins, the 91-KD protein corresponds to the DEAH helicase DHX15 (Fig. 1B). By using a DHX15-specific antibody to immunoblot aliquots of the affinity-purified proteins (same as those shown in Fig. 1A), we confirmed that the protein specifically associated with SUGP1 (aa 543–645) was indeed DHX15 (Fig. 1C). The other two proteins, PYGL and PYGB (glycogen phosphorylase, liver and brain isoforms), appeared in the silver-stained gel to be associated with both the G-patch mutant SUGP1 (aa 543–645) and the truncated SUGP1 (aa 607–645) lacking the G-patch (Fig. 1A). This result suggests that these two proteins associate with other parts of the SUGP1 C terminus rather than with the G-patch. Furthermore, PYGL and PYGB were not among the proteins we previously identified in SF3B1-associated spliceosomal complexes (14), and neither of them has an obvious function related to RNA splicing. We reasoned that it is unlikely that PYGL and PYGB are involved in SUGP1 G-patch-regulated splicing, and therefore in subsequent experiments, we focused on DHX15 only.
DHX15 Interacts with SUGP1 both In Vitro and in the Spliceosome.
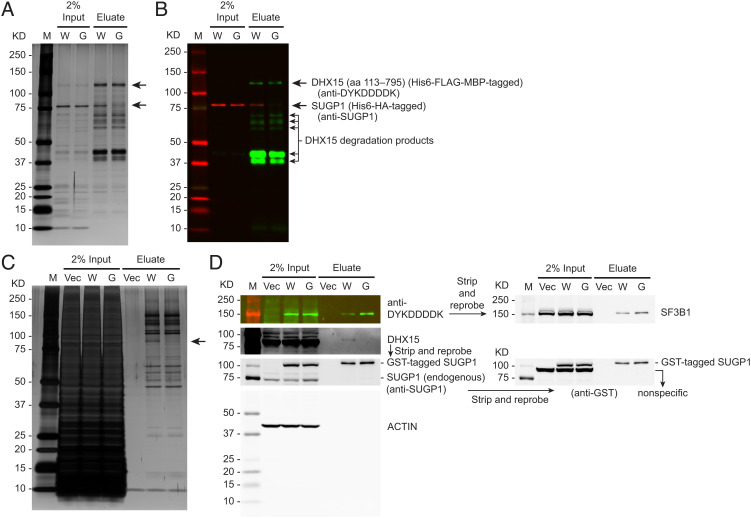
Next, we wanted to determine whether DHX15 interacts with SUGP1 directly. To this end, we purified SUGP1 (full length) and DHX15 (aa 113–795), which lacks the unstructured N-terminal region (22, 23), with affinity tags from Escherichia coli, and then performed an in vitro interaction assay by co-immunoprecipitation (CoIP). We used a His6 (hexahistidine) tag as the purification tag for both proteins. We added an MBP (maltose-binding protein) tag to the N terminus of DHX15 (aa 113–795) because it is known to improve recombinant protein solubility (24, 25). We also added a FLAG tag to DHX15 (aa 113–795) for CoIP purposes. By incubating the purified SUGP1 (full length) and DHX15 (aa 113–795) proteins together followed by CoIP with anti-DYKDDDDK (FLAG) antibody, we found that DHX15 (aa 113–795) bound to WT SUGP1 in vitro, but not to G-patch mutant SUGP1 (Fig. 2 A and B). We note that DHX15 was challenging to purify, giving rise to some degradation products as detected by western blotting (Fig. 2B). This observation does not affect our conclusion that the two proteins interact directly.
Fig. 2.
DHX15 interacts with SUGP1 both in vitro and in the spliceosome. (A and B) Purified His6-HA (hemagglutinin)-tagged SUGP1 WT (W) and its G574A-G582A mutant (G) were co-immunoprecipitated with purified His6-FLAG-MBP-tagged DHX15 (aa 113–795), followed by SDS-PAGE and silver staining (A) or western blotting (B). (C and D) HEK293T cells were transfected with empty vector plasmid (Vec) or a mixture of two plasmids co-expressing His6-FLAG-tagged SF3B1 with GST-tagged SUGP1 WT (W) or with its G574A-G582A mutant (G), followed by small-scale affinity purification using FLAG and GST tags. The purified proteins were resolved by SDS-PAGE, followed by silver staining (C) or western blotting (D). In all panels: M, Precision Plus Protein marker (Bio-Rad).
We next asked whether DHX15 interacts with SUGP1 in the spliceosome. Because the spliceosome is a large protein-RNA complex that rapidly changes its composition and structure at different stages of the splicing reaction (17), and because we did not know whether SUGP1 was in a stage-specific spliceosomal complex or a transitional complex, we purified all spliceosomal complexes from cell extracts using affinity-tagged SF3B1 (a major component of the spliceosome). We isolated SUGP1-containing spliceosomes using two rounds of affinity purification following co-expression of affinity-tagged SF3B1 and SUGP1 (WT or G-patch mutant) proteins exogenously in HEK293T cells. The first round involved pull-down of spliceosomal complexes using a FLAG tag attached to the N terminus of SF3B1 followed by a second round involving pull-down of WT or G-patch mutant SUGP1-containing spliceosomes using a GST tag attached to the N terminus of the SUGP1 derivatives. By resolving the affinity-purified spliceosomal complexes by SDS-PAGE followed by silver staining, we observed that only one protein band (~91-KD; the expected size of DHX15) was obviously missing (or greatly reduced) from the G-patch mutant SUGP1-containing spliceosomes compared to the WT SUGP1-containing spliceosomes (Fig. 2C). Western blotting with a DHX15-specific antibody confirmed that the missing protein was DHX15 (Fig. 2D). These results suggest that it is solely DHX15 (i.e., no other potentially redundant helicase) that interacts with the SUGP1 G-patch in the spliceosome and that the incorporation of SUGP1 with a mutant G-patch can prevent DHX15 recruitment.
Knockdown of DHX15 or Expression of Several DHX15 Mutants Partially Recapitulates the Splicing Defects of Mutant SF3B1.
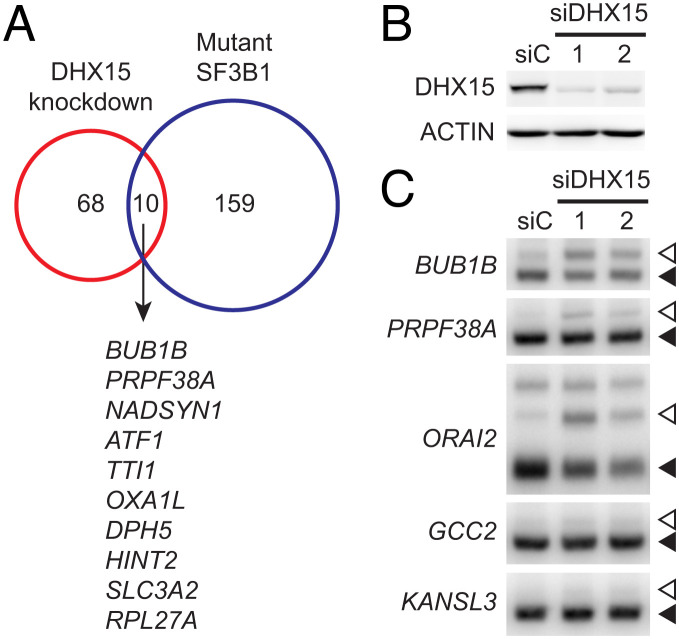
The next important question is whether DHX15 is involved in SUGP1-mediated RNA splicing. Cancer-associated SF3B1 mutations induce aberrant splicing of cryptic 3′ss by disrupting the interaction of SF3B1 with SUGP1 and loss of SUGP1 accounts for the splicing defects (14). We therefore asked whether depletion of DHX15 also induces missplicing of cryptic 3′ss. First, we analyzed publicly available RNA-sequencing (RNA-seq) data from HeLa cells with or without DHX15 knockdown (26). By using the same computational method we used in our previous study to identify mutant SF3B1-specific cryptic 3′ss (but with slightly less stringent threshold parameters that are necessary due to the smaller sample size) (14), we identified 78 cryptic 3′ss that were misspliced upon DHX15 knockdown (Dataset S2), 10 (i.e., 13%) of which overlapped with those misspliced by mutant SF3B1 (Fig. 3A; we discuss below why these numbers might be low). Next, using two independent small interfering RNAs (siRNAs) targeting DHX15, we knocked down DHX15 in HEK293T cells (knockdown efficiency shown in Fig. 3B) and performed 32P RT-PCR with the purified RNAs to detect 3′ss missplicing. By examining the top two overlapping cryptic 3′ss (in BUB1B and PRPF38A), as well as three of the top target cryptic 3′ss of mutant SF3B1 (in ORAI2, GCC2, and KANSL3) that we previously validated (14), we found that DHX15 knockdown also partially recapitulated cryptic 3′ss missplicing, e.g., of BUB1B, PRPF38A, and ORAI2, but not of GCC2 or KANSL3 pre-mRNA (Fig. 3C).
Fig. 3.
Knockdown of DHX15 partially recapitulates the splicing defects of mutant SF3B1. (A) Venn diagram showing the overlap between the cryptic 3′ss misregulated upon DHX15 knockdown and those misspliced by mutant SF3B1 (14). The genes harboring the overlapping cryptic 3′ss are sorted by difference in cryptic 3′ss usage between DHX15 knockdown and control. (B) HEK293T cells were transfected with a negative control siRNA (siC) or one of two independent siRNAs targeting DHX15, followed by western blotting. (C) 32P RT-PCR products of the cryptic 3′ss (open arrowheads) and canonical 3′ss (solid arrowheads) of the indicated genes in HEK293T cells in (B).
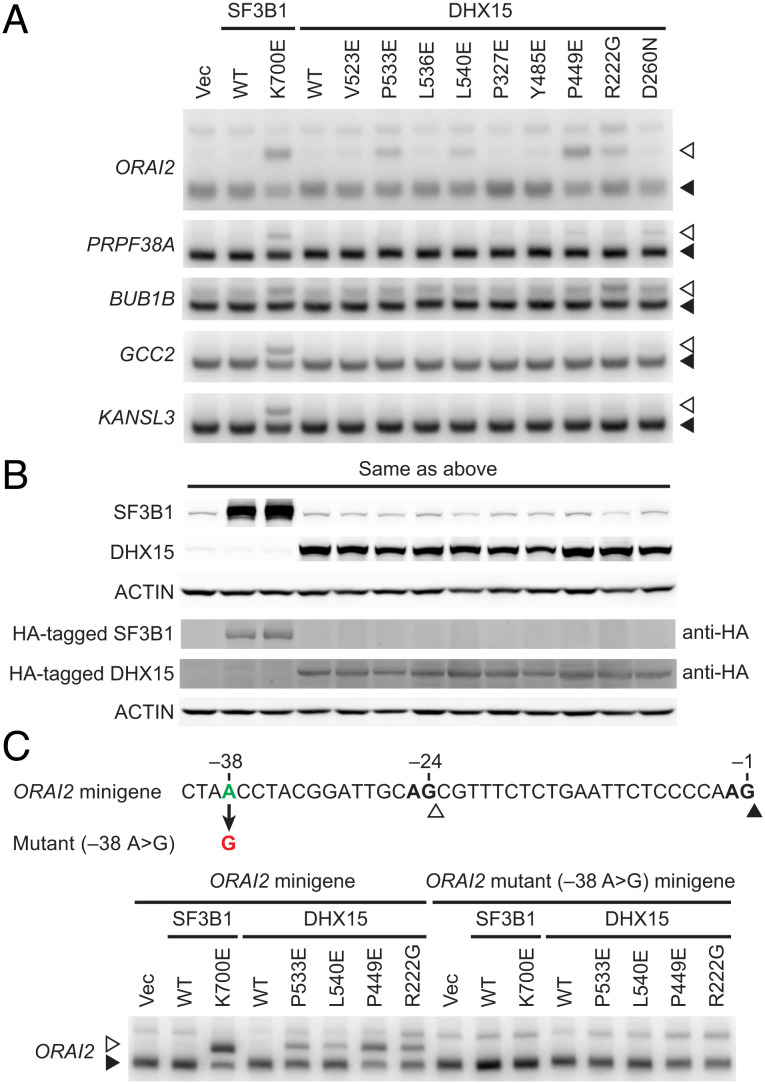
We previously showed that expression of SUGP1 derivatives with a mutant G-patch or containing cancer mutations partially recapitulated mutant SF3B1-induced missplicing (14, 19). We therefore asked whether expression of DHX15 mutants also induces cryptic 3′ss missplicing. To this end, we made plasmid constructs expressing different DHX15 mutant derivatives. These included six with single amino acid changes known to be important for interaction with the G-patch of NKRF (a ribosome biogenesis factor whose G-patch interacts with DHX15) (23), one with a single residue change at the β-turn near the G-patch binding region (P449E), one containing the R222G mutation found in AML (21), and finally one with a mutation in the DEAH-box (D260N). We then co-expressed each of the DHX15 mutants with FLAG-GST tandem tagged SUGP1 (aa 543–645) in HEK293T cells, followed by two rounds of affinity purification of the tagged SUGP1 derivative with anti-DYKDDDDK antibody and Glutathione Sepharose beads sequentially. The results confirmed that the six mutations important for NKRF G-patch binding and the mutation at the β-turn near the binding region all disrupted interaction of DHX15 with the SUGP1 G-patch (SI Appendix, Fig. S1). The D260N mutation also reduced interaction (but to a lesser degree), whereas the AML-associated R222G mutation did not affect interaction (SI Appendix, Fig. S1). To examine whether the DHX15 mutants affect cryptic 3′ss splicing, we expressed each of the mutants by themselves in HEK293T cells and performed 32P RT-PCR with RNAs purified 48 h following transfection. We found that expression of some (but not all) of the DHX15 mutants, including importantly the R222G AML mutant, partially recapitulated missplicing of mutant SF3B1-specific cryptic 3′ss, e.g., in the ORAI2 pre-mRNA (Fig. 4 A and B).
Fig. 4.
Expression of several DHX15 mutants partially recapitulates the splicing defects of mutant SF3B1. (A) HEK293T cells were transfected with Vec or expression plasmid for HA-tagged SF3B1 (WT or K700E) or HA-tagged DHX15 (WT or one of the mutants as indicated), followed by 32P RT-PCR of the cryptic 3′ss (open arrowheads) and canonical 3′ss (solid arrowheads) of the indicated genes. (B) Western blotting of protein extracts from HEK293T cells in (A). anti-HA, rabbit polyclonal anti-HA tag antibody. (C) The ORAI2 minigene or its mutant (–38 A > G) minigene was co-transfected with Vec or expression plasmid for HA-tagged SF3B1 (WT or K700E) or HA-tagged DHX15 (WT or one of the mutants as indicated), followed by 32P RT-PCR of the cryptic 3′ss (open arrowheads) and canonical 3′ss (solid arrowheads) from the ORAI2 minigenes.
It is well known that mutant SF3B1 induces missplicing of cryptic 3′ss by promoting usage of upstream branch points (11, 12, 14). To test whether the DHX15 mutants also induce use of an upstream branch point, we performed splicing assays using the WT ORAI2 minigene and its mutant derivative with the upstream branch point mutated (A to G) that we generated in our previous study (14). We co-transfected each of four DHX15 mutant constructs with either WT or mutant ORAI2 minigene in HEK293T cells and then performed 32P RT-PCR with the isolated RNAs. Results showed that all four DHX15 mutants induced use of the cryptic 3′ss in the WT ORAI2 minigene, just as K700E SF3B1 does, while the branch point mutation in the mutant ORAI2 minigene abolished the abilities of these DHX15 mutants (as well as K700E SF3B1) to induce use of the cryptic 3′ss (Fig. 4C). These results suggest that DHX15 mutants not only partially recapitulate the missplicing of cryptic 3′ss, but also require an upstream branch point for missplicing (a key feature of mutant SF3B1).
DHX15-SUGP1 G-Patch Fusion Protein Incorporates into the Spliceosome and Rescues the Splicing Defects of Mutant SF3B1.
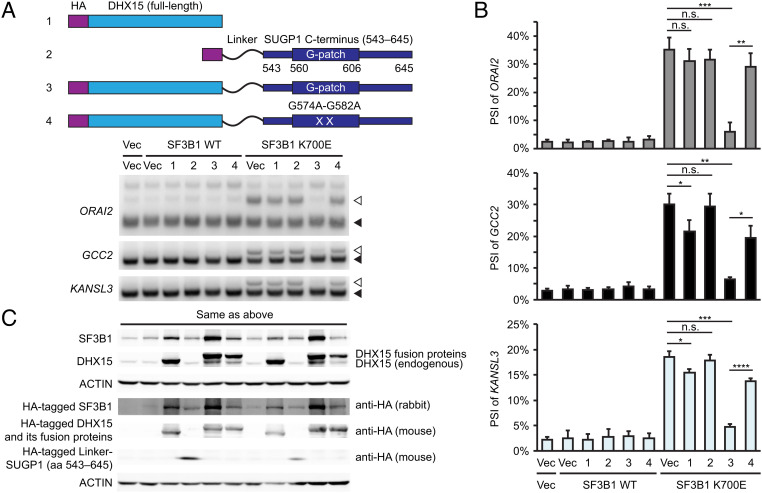
We next investigated whether high levels of DHX15 could rescue splicing defects caused by mutant SF3B1. Indeed, we showed previously that SUGP1 overexpression partially rescued K700E SF3B1-induced missplicing (14). However, similar experiments overexpressing DHX15 revealed only slight rescue of the splicing defects in GCC2 and KANSL3, but not in ORAI2 (Fig. 5 A–C, construct 1 compared to the vector control). This inefficient rescue may reflect the absence of SUGP1, specifically of the G-patch required to activate DHX15. To investigate this possibility, we made a fusion protein construct by fusing DHX15 with SUGP1 (aa 543–645, including the G-patch and its surrounding regions) separated by a flexible linker (GGGGSGGGGSGGGGS) to allow certain plasticity for protein folding (27) (SI Appendix, Fig. S2A). Our hypothesis was that the SUGP1 G-patch will constitutively activate DHX15 so that the fusion protein is able to rescue splicing defects of mutant SF3B1 even when SUGP1 itself is absent from the mutant SF3B1 spliceosome.
Fig. 5.
DHX15-SUGP1 G-patch fusion protein rescues the splicing defects of mutant SF3B1. (A, Upper) Schematic representation of plasmid constructs (1 to 4) expressing HA-tagged DHX15, HA-tagged GGGGSGGGGSGGGGS (Linker)-SUGP1 (aa 543–645), HA-tagged DHX15-SUGP1 (aa 543–645) fusion protein, and its G574A-G582A mutant, respectively. (Lower) Vec or expression plasmid for HA-tagged SF3B1 (WT or K700E) was co-transfected with Vec or one of the four constructs (as indicated) in HEK293T cells, followed by 32P RT-PCR of the cryptic 3′ss (open arrowheads) and canonical 3′ss (solid arrowheads) of the indicated genes. (B) Quantification of the 32P RT-PCR products in (A). PSI (percent spliced in) of the cryptic 3′ss of the indicated genes is shown. Error bars represent SDs of the means (n = 3; three independent experiments). n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (unpaired, two-tailed, and unequal variance t test). (C) Western blotting of protein extracts from HEK293T cells in (A).
To test this hypothesis, we first examined whether the DHX15-SUGP1 G-patch fusion protein is able to incorporate into the spliceosome. We co-expressed the fusion protein with either WT or K700E mutant SF3B1 in HEK293T cells and then performed two rounds of affinity purification of the spliceosomes by using tandem tags (FLAG and His6) attached to the N terminus of SF3B1. By resolving the affinity-purified spliceosomes by SDS-PAGE followed by either silver staining or western blotting, we found that the DHX15-SUGP1 G-patch fusion protein was indeed incorporated into both WT and K700E SF3B1 spliceosomes (SI Appendix, Fig. S2 A and B). Notably, a derivative of the fusion protein with a mutant G-patch was also incorporated into the spliceosome (SI Appendix, Fig. S2 A and B). To examine whether either fusion protein was able to rescue the missplicing of cryptic 3′ss by mutant SF3B1, we co-expressed them with either WT or K700E mutant SF3B1 in HEK293T cells and performed 32P RT-PCR with the purified RNAs. Strikingly, whereas a derivative consisting of only the SUGP1 G-patch-containing region with the linker failed to rescue any missplicing (Fig. 5 A–C, construct 2), the DHX15-SUGP1 G-patch fusion protein almost completely rescued the splicing defects induced by mutant SF3B1 (Fig. 5 A–C, construct 3). Notably, rescue was abolished (in ORAI2) or significantly reduced (in GCC2 and KANSL3) by the mutant G-patch (Fig. 5 A–C, construct 4).
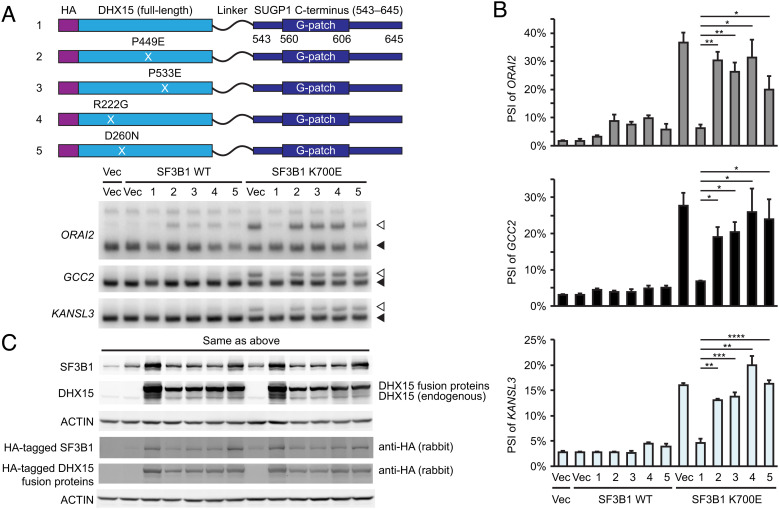
Given the highly efficient rescue of K700E SF3B1-induced missplicing achieved by the DHX15-SUGP1 G-patch fusion protein, we next asked whether rescue activity would be affected by any of the DHX15 mutations analyzed above (see Fig. 4). To this end, we introduced each of the four mutations, including the R222G AML-associated mutation, into the DHX15 portion of the fusion protein, expressed the proteins along with WT or K700E SF3B1 in HEK293T cells, and analyzed purified RNAs by 32P RT-PCR as above. Strikingly, all four DHX15 mutations, including R222G, either completely or partially abolished the abilities of the fusion proteins to rescue the splicing defects of mutant SF3B1 (Fig. 6 A–C).
Fig. 6.
Mutations in the DHX15-SUGP1 G-patch fusion protein abolish the rescue effect. (A, Upper) Schematic representation of plasmid constructs (1 to 5) expressing HA-tagged DHX15-SUGP1 (aa 543–645) fusion protein and its mutants P449E, P533E, R222G, and D260N, respectively. (Lower) Vec or expression plasmid for HA-tagged SF3B1 (WT or K700E) was co-transfected with Vec or one of the five constructs (as indicated) in HEK293T cells, followed by 32P RT-PCR of the cryptic 3′ss (open arrowheads) and canonical 3′ss (solid arrowheads) of the indicated genes. (B) Quantification of the 32P RT-PCR products in (A). PSI of the cryptic 3′ss of the indicated genes is shown. Error bars represent SDs of the means (n = 3; three independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (unpaired, two-tailed, and unequal variance t test). (C) Western blotting of protein extracts from HEK293T cells in (A).
Structure of the DHX15-SUGP1 G-Patch Complex.
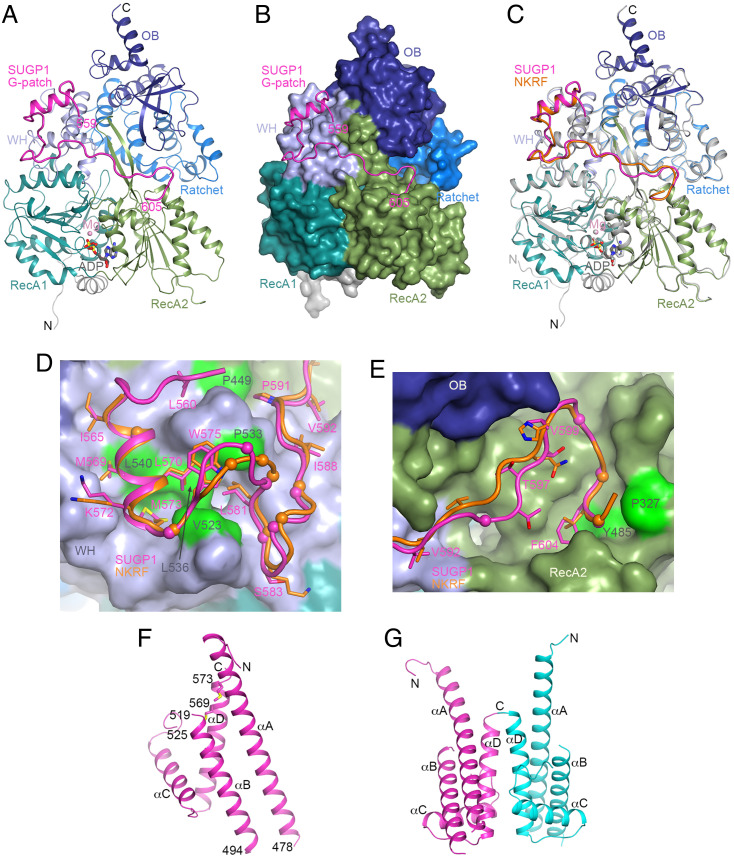
To gain detailed insights into the interaction between DHX15 and the SUGP1 G-patch, we determined the crystal structure of their complex at 1.8 Å resolution (Fig. 7A and SI Appendix, Table S1). SUGP1 residues 559–605, containing essentially the entire G-patch, are included in the atomic model. The G-patch has a continuous electron density map for the backbone except residues Lys594 and Gly595, which have no direct contacts with DHX15 and are likely more flexible. Approximately 1,800 Å2 of the surface area of the G-patch is buried in the interface with DHX15. The small N-terminal helix (brace helix, residues 565–573) and the following segment (residues 574–592) have extensive interactions with the DHX15 winged helix (WH) domain, while the C-terminal brace loop (residues 600–605) interacts with the DHX15 RecA2 domain (Fig. 7B).
Fig. 7.
Crystal structure of the DHX15-SUGP1 G-patch complex. (A) Schematic drawing of the structure of the human DHX15-SUGP1 G-patch complex. The domains in DHX15 are given different colors and labeled. The SUGP1 G-patch is in magenta. The bound adenosine diphosphate (ADP) molecule is shown as a stick model (gray for carbon atoms) and Mg2+ ion as a sphere (pink). WH, winged helix; OB, oligonucleotide/oligosaccharide binding. (B) The structure of DHX15 is shown as a molecular surface, colored by domains. The SUGP1 G-patch interacts with the WH and RecA2 domains. (C) Overlay of the structures of human DHX15-SUGP1 G-patch complex (in color) and DHX15-NKRF G-patch (DHX15 in gray and NKRF in orange). (D) Interaction of the N-terminal part of the SUGP1 G-patch with the DHX15 WH domain. DHX15 is shown as a surface, and side chains in the SUGP1 G-patch (in magenta) making large contributions to the interface are shown as sticks. The NKRF G-patch is shown for comparison (orange). The Cα atoms of glycine residues are indicated by spheres. DHX15 residues that were selected for mutation (SI Appendix, Fig. S1) are colored in green and labeled in dark gray. (E) Interaction of the C-terminal part of the SUGP1 G-patch with the DHX15 RecA2 domain. (F) Crystal structure of the C-terminal region of SUGP1 (aa 433–577). The four helices are labeled, and the side chains of Met569 and Met573 are shown as sticks. (G) A dimer of the C-terminal region of SUGP1 in the crystal. The second monomer is shown in cyan, and the twofold axis of the dimer is vertical. Conformational differences for helix αA in the two monomers are visible. The structure figures are produced with PyMOL (www.pymol.org).
The overall structure of the DHX15-SUGP1 G-patch complex is similar to that of the DHX15-NKRF G-patch complex (Fig. 7C) (23) and related structures (20, 28, 29). The rms distance (r.m.s.d) is 0.34 Å for 673 equivalent Cα atoms of DHX15 in the SUGP1 and NKRF complexes. For the G-patch, five additional residues (559–563) at the N terminus are observed in the SUGP1 complex as compared to the NKRF complex, with Leu560 buried in the interface with DHX15 (Fig. 7D). This segment is also partially stabilized by crystal packing. The brace helix and the following segment have similar interactions with the DHX15 WH domain in the two structures (Fig. 7D). A conformational difference is observed for SUGP1 residues 576–580, due to the insertion of a residue in this segment. Similarly, the insertion of a residue in the 593–598 segment in SUGP1 is reflected by conformational differences with NKRF, although the following brace loop has a similar conformation and interaction with the DHX15 RecA2 domain (Fig. 7E). This insertion also contributes to the disordering of residues Lys594 and Gly595, as described earlier.
Consistent with the structural information, the mutated DHX15 residues that disrupted interactions with SUGP1 and in some cases affected splicing (SI Appendix, Fig. S1) are all located in the interface with the SUGP1 G-patch (Fig. 7 D and E and SI Appendix, Fig. S3 A and B). Especially notable, Tyr485 of DHX15 is π-stacked with Phe604 in the G-patch (SI Appendix, Fig. S3B). However, the Arg222 residue of DHX15 is located in a β-reverse turn in the RecA1 domain and makes no contact with the SUGP1 G-patch (SI Appendix, Fig. S3C), consistent with our CoIP data that the AML-associated R222G mutation does not affect the interaction of DHX15 with the SUGP1 G-patch (SI Appendix, Fig. S1).
Many of the glycine residues in the SUGP1 G-patch are conserved in NKRF and assume similar conformations in the two structures (Fig. 7 D and E). Some of the Gly residues are located at sharp, left-handed turns in the backbone of the G-patch (e.g., 574, 578, 585, and 601), while others are packed close against DHX15 (566, 582, 587, 603) or within the G-patch (580). Therefore, the glycine residues are important for the conformation of the G-patch and/or the tight contact with DHX15.
Structure of the SUGP1 C-Terminal Region.
To our knowledge, the only structures of G-patches previously reported have been in complex with their interacting DEAH helicases. We attempted to determine the structure of the SUGP1 C-terminal region (including the G-patch) on its own. We obtained a 2.4 Å resolution crystal structure for residues 433–577 of SUGP1 (Fig. 7F and SI Appendix, Table S1), which includes the N-terminal region of the G-patch. The structure contains four helices (αA to αD), with αA having ten turns (439–476). The loops connecting the helices are highly flexible, with most of the residues connecting αA and αB (479–493) being disordered. There are two molecules of SUGP1 in the asymmetric unit of the crystal, with generally similar structures, although some differences can be seen in the relative positions of αA. The two molecules form a dimer, mediated primarily by helix αD (552–573) (Fig. 7G). This dimer is likely a crystal-packing artifact, as the protein sample is a monomer in solution based on gel filtration chromatography.
The N-terminal region of the G-patch (residues 565–573) forms a short helix (brace helix) in the DHX15 complex (Fig. 7A), while it is a part of the much longer helix αD (residues 552–573) in SUGP1 alone (Fig. 7G). In addition, residues Met569 and Met573 are in the interface between SUGP1 and DHX15 in the complex (Fig. 7D), while these two residues are in the hydrophobic core of the four helices in SUGP1 alone (Fig. 7G). There are thus extensive differences between the crystal structures of the SUGP1 G-patch alone and the complex with DHX15.
Discussion
Our previous study showed that cancer-associated SF3B1 mutations misregulate splicing by disrupting the interaction of SF3B1 with SUGP1 (14). We further provided evidence that missplicing could be recapitulated by expression of a G-patch mutant SUGP1, implicating an unknown DEAH-box RNA helicase in the process. Here we present biochemical and structural evidence that the SUGP1 G-patch indeed binds a DEAH helicase with high specificity. This helicase is DHX15, which itself harbors a cancer-associated mutation that we have shown partially recapitulates related missplicing. Below we discuss the importance and significance of these findings, as well as how they extend our understanding of splicing dysregulation in cancer.
DHX15 and its yeast homolog Prp43 have been known to function in multiple cellular processes, including pre-mRNA splicing and ribosomal RNA biogenesis (20). In different processes, DHX15/Prp43 proteins are activated by different G-patch-containing cofactors to fulfill their different functions (including promoting disassembly of RNA-protein complexes). For example, Prp43 is activated by G-patch factor Ntr1 to disassemble intron–lariat spliceosomes (ILS) at the late stage of splicing (30), and in humans, DHX15 is activated by G-patch factor TFIP11 to facilitate disassembly of the post-splicing 40S lariat–intron complex (31). During ribosome biogenesis, DHX15 is activated by G-patch factor NKRF to promote pre-rRNA cleavage at the A′ site (32). In all these cases, binding to a G-patch factor is a common mechanism of activating DHX15/Prp43. Here we identify SUGP1 as another G-patch-containing factor that activates DHX15 and further provide detailed structural insights into the interaction between DHX15 and the SUGP1 G-patch. The DHX15-SUGP1 G-patch complex structure is highly similar to that of the DHX15-NKRF G-patch complex previously published (23). Therefore, we envision that the SUGP1 G-patch activates DHX15 in the same way as the NKRF G-patch does, i.e., the G-patch tethers together the WH and RecA2 domains of DHX15, leading to a conformational change in DHX15 that facilitates its stable binding to RNA substrates and concomitantly stimulates its ATPase and RNA unwinding activities (23).
How is DHX15 involved in SUGP1 G-patch regulated splicing, and what is its role in altered branchsite selection by mutant SF3B1 in cancer? Based on our data presented here and data in previously published studies, we suggest two possible models. Mutant SF3B1-regulated introns harbor two potential branchsites (one canonical and the other typically upstream) and two possible 3′ss (one canonical and the other cryptic) (11, 12, 14). WT SF3B1-containing spliceosomes use the canonical branchsite and 3′ss, whereas mutant SF3B1 spliceosomes recognize the upstream branchsite and cryptic 3′ss. At the early stage of splicing, the canonical branchsite is bound by SF1 with the help of U2AF (the U2AF2/U2AF1 heterodimer), which recognizes and anchors to the canonical 3′ss (33, 34). Accordingly, the canonical branchsite is generally located close to the canonical 3′ss within reach of SF1/U2AF. During U2 snRNP-branchsite recognition, SF1 has to be displaced from the branchsite, which requires ATP hydrolysis (17).
In one model to explain the involvement of SUGP1/DHX15 in branchsite recognition, we suggest that DHX15 promotes recognition of canonical branchsites. SUGP1 is first recruited to U2 snRNP by its interaction with SF3B1. Because SUGP1 has two SURP domains and one U2AF2-binding site (14, 35), and SURP domains are known to interact with SF1 (36), SUGP1 may localize U2 snRNP to the vicinity of the canonical branchsite by its interactions with SF1 and U2AF2. The SUGP1 G-patch then activates DHX15 to displace SF1 from the canonical branchsite, possibly by a mechanism of helicase tracking on the pre-mRNA in an ATP-dependent fashion (37). The exposed canonical branchsite is then recognized by U2 snRNA via base pairing. When SF3B1 is mutated in cancer, its interaction with SUGP1 is disrupted, leading to loss of SUGP1 during spliceosome assembly (14). In the absence of SUGP1, DHX15 is not activated and SF1 is not efficiently displaced from the canonical branchsite, blocking U2 snRNP recognition. The mutant SF3B1 spliceosome then has to recognize an upstream branchsite, possibly through branchsite scanning by p14 (38). If there is an appropriately spaced cryptic 3′ss, then that 3′ss is used and missplicing occurs. In cases where a cryptic 3′ss is absent, splicing may still proceed using the canonical 3′ss but at a reduced rate (39–41).
Another possible way DXH15 may function in branchsite selection is as a quality control mechanism, by promoting dissociation of U2 snRNP that assembles on upstream cryptic branchsites. This idea stems from the recent discovery that DHX15 has a role in quality control of defective branchsite recognition in vitro (42), as well as from previously reported functions of Prp43 in yeast. It was found that DHX15 can destabilize the interaction between U2 snRNP and a minimal RNA substrate in a stalled and unproductive A-complex (42). In yeast, a constitutively active Prp43 derivative (fused to the Ntr1 G-patch; see below) can also target the U2 snRNP-intron interaction for dissociation from early spliceosome complexes (43). Therefore, under normal splicing conditions, DHX15 may promote dissociation of U2 snRNP only from the upstream branchsite (e.g., due to a potentially suboptimal conformation). When SF3B1 is mutated, SUGP1 is not recruited and hence DHX15 is not activated, allowing mutant SF3B1-containing U2 snRNP to remain associated with the suboptimal upstream branchsite. As a result, splicing proceeds, and if a cryptic 3′ss is present, generates the splicing defects seen in many mutant SF3B1 cancers.
More insight into SUGP1-DHX15 function comes from studies on Prp43 and its G-patch activator Ntr1. Prp43 together with Ntr1 and another protein, Ntr2, forms the NTR complex, which dissociates late stage ILS (44), as well as earlier stalled or defective intermediates (45). Interestingly, a Prp43-Ntr1 G-patch fusion protein can disassemble ILS as efficiently as the NTR in cell extracts, but has lost its specificity and also disrupts normal earlier spliceosomal complexes that are not targets of NTR (43). This suggests that regions in Ntr1 outside the G-patch contribute to proper targeting of activated Prp43 (43, 46). In the case of SUGP1-DHX15, we suggest the SUGP1 SURP and U2AF2-binding motifs target DHX15 to early-stage spliceosomes. This could be to position activated DHX15 either to displace SF1 in the normal course of splicing, or to disrupt aberrant splicing complexes that assemble on upstream cryptic splicing signals, depending on which if either of the two models we proposed is correct. We note that the spatial relationship between the branchsite and SF1/U2AF on the pre-mRNA would be different in the cryptic and canonical pre-spliceosomal complexes and evidence showed that cryptic branchsites lead to much slowed splicing (39–41). Therefore, it is possible that the suboptimal spatial configurations could be recognized by SUGP1 to target DHX15 to dismantle the cryptic complexes.
The above two models for DHX15 function in altered branchsite selection are speculative, and further work is required to determine which of them is correct. Nonetheless, the two models are not necessarily mutually exclusive, i.e., DHX15 may function both to promote canonical branchsite recognition and at the same time to facilitate dissociation of U2 snRNP from suboptimal cryptic branchsites.
It is important to note that DHX15 can be recruited to the spliceosome in the absence of SUGP1. In our previous study (14), our mass spectrometry data showed that DHX15 is present almost equally in both WT and mutant SF3B1-associated spliceosomal complexes, i.e., with or without SUGP1. Our data here showed that the DHX15-SUGP1 G-patch fusion protein is incorporated into both WT and mutant SF3B1 spliceosomes, again indicating that DHX15 is recruited to the spliceosome independently of SUGP1. How this occurs is not known, but it possibly involves interaction of DHX15 with DDX42 (a SF3b subunit formerly known as SF3b125) (47). The SUGP1 G-patch thus functions solely for DHX15 activation, not for DHX15 recruitment. However, expression of SUGP1 with a mutated G-patch was found not only to induce robust cryptic 3′ss usage (14), but also as shown here to prevent recruitment of DHX15 into the spliceosome. Given that SUGP1, and hence its G-patch, is not required for DHX15 recruitment, this result likely reflects a dominant negative effect of the mutant G-patch, due perhaps to an altered conformation and resultant steric hindrance that interferes with DHX15 association with the spliceosome.
We note that knockdown of DHX15 only partially recapitulated the splicing defects of mutant SF3B1. For example, in the publicly available RNA-seq data we analyzed (26), only 13% of the cryptic 3′ss misspliced upon DHX15 knockdown overlap with those regulated by mutant SF3B1. This low level of overlap may be explained in at least two ways. First, the DHX15 knockdown efficiency might not have been optimal in the RNA-seq data (knockdown efficiency was not indicated), leading to missing of some target cryptic 3′ss. For example, in our own experiments we knocked down DHX15 in HEK293T cells with a high efficiency of ~80% and detected the misspliced cryptic 3′ss of ORAI2, whereas we did not detect the ORAI2 cryptic 3′ss in the RNA-seq data. Second, as already mentioned DHX15 is involved in multiple different cellular processes and can interact with multiple other G-patch factors (20). Therefore, knockdown of DHX15 not only affects SUGP1-regulated splicing, but also affects other DHX15-dependent processes, which in turn may directly or indirectly suppress SUGP1-regulated splicing defects. For example, we did not detect cryptic 3′ss usage with GCC2 and KANSL3 upon DHX15 knockdown in our experiments (nor in the RNA-seq data). However, we did detect missplicing of both of these transcripts in our previous study following expression of SUGP1 with a mutated G-patch (14), which we showed here prevents recruitment of DHX15 (and only DHX15) to the spliceosome. In addition, we showed that the DHX15-SUGP1 G-patch fusion protein rescues splicing of both GCC2 and KANSL3. Taken together, these results explain, at least in part, the limited overlap of mutant SF3B1 and DHX15 knockdown induced missplicing, as well as confirm that DHX15 is solely responsible for SUGP1 G-patch-regulated missplicing.
Our experiments have provided insights into the mechanism by which the AML-associated R222G mutation in DHX15 affects splicing. This mutation has been found in 7% (6/85) of AML patients with RUNX1-RUNX1T1 rearrangement (21). These authors showed by CoIP that R222G reduced interaction of DHX15 with the G-patch factor TFIP11. It was therefore unexpected that our experiments revealed that R222G does not affect DHX15 interaction with the SUGP1 G-patch. This result is consistent with the DHX15-SUGP1 G-patch structure showing that R222 does not make any contact with the SUGP1 G-patch. It thus may be that the SUGP1 and TFIP11 G-patches interact differently with DHX15. Although exactly how R222G affects splicing requires further study, R222G mutant DHX15 is expected to induce cancer-driving splicing defects (especially in the RUNX1-RUNX1T1 background) and expression of this mutant in HEK293T cells recapitulated, albeit only partially, the splicing defects of mutant SF3B1, e.g., in ORAI2 transcripts. Furthermore, we showed that the R222G mutation completely abolished the ability of the DHX15-SUGP1 G-patch fusion protein to rescue SF3B1 K700E-induced cryptic 3′ss missplicing in all transcripts tested. Why we observed greater effects with this rescue assay compared to simple expression of the R222G mutant protein is not entirely clear. It may however reflect the fact that the DHX15-SUGP1 G-patch fusion protein with the R222G mutation only affects SUGP1-regulated cryptic 3′ss splicing, whereas the R222G DHX15 alone likely affects other G-patch factor-regulated processes (which may in turn suppress some of the cryptic 3′ss).
In summary, we have identified DHX15 as the RNA helicase involved in SF3B1/SUGP1-mediated splicing. The SUGP1 G-patch interacts directly and specifically with DHX15, and this interaction is required for DHX15 activation but not for recruitment to the spliceosome. Our findings shed new light on how mutant SF3B1 misregulates RNA splicing in cancer and define an SF3B1/SUGP1/DHX15 axis where mutations in any of these factors cause related defects in splicing.
Materials and Methods
Expression Plasmid Constructs.
His6-FLAG-tagged SF3B1 (WT and K700E) and HA-tagged SF3B1 (WT and K700E) were cloned in p3xFLAG-CMV-14 (Sigma-Aldrich) in our previous study (14). HA-tagged DHX15 (codon optimized by GenScript) was cloned in p3xFLAG-CMV-14 (Sigma-Aldrich) using HindIII and BamHI sites. Its mutant constructs (V523E, P533E, L536E, L540E, P327E, Y485E, P449E, R222G, and D260N) were generated by site-directed mutagenesis using overlap extension PCR (48). FLAG-GST tandem tags, FLAG-GST-tagged SUGP1 (aa 543–645), FLAG-GST-tagged SUGP1 (aa 607–645), and GST-tagged SUGP1 were cloned in p3xFLAG-CMV-14 (Sigma-Aldrich) using HindIII and BamHI sites. The G574A-G582A mutant constructs of FLAG-GST-tagged SUGP1 (aa 543–645) and GST-tagged SUGP1 were generated by site-directed mutagenesis using overlap extension PCR (48). HA-tagged DHX15-SUGP1 (aa 543–645) fusion construct and HA-tagged GGGGSGGGGSGGGGS-SUGP1 (aa 543–645) were cloned in p3xFLAG-CMV-14 (Sigma-Aldrich) using HindIII and BamHI sites. Each of the mutations (P449E, P533E, R222G, and D260N in DHX15, as well as G574A-G582A in SUGP1 G-patch) was introduced into the HA-tagged DHX15-SUGP1 (aa 543–645) fusion construct by site-directed mutagenesis using overlap extension PCR (48). In all of the above constructs, the affinity tags were added to the N termini of the proteins. In addition, two stop codons were added immediately following the last codon of each protein, and therefore, the 3xFLAG tag in the vector downstream of the stop codons was not added to the final protein product.
Affinity Purification Using Two Tags.
For affinity purification using two tags, see detailed methods in SI Appendix.
Recombinant Protein Purification and In Vitro Protein–Protein Interaction.
For recombinant protein purification and in vitro protein–protein interaction, see detailed methods in SI Appendix.
Computational Identification of Cryptic 3′SS.
For computational identification of cryptic 3′ss, see detailed methods in SI Appendix.
32P RT-PCR.
32P RT-PCR was performed as described (14). Briefly, 2 μg total RNA was reverse-transcribed with 50 pmol oligo-dT primer using 0.3-μL Maxima Reverse Transcriptase (Thermo Fisher Scientific). The synthesized cDNA was then diluted 1:10 in H2O, and 1.2 μL was used as template in a 10-μL PCR reaction containing 0.6 μCi [α-32P] dCTP. PCR products were subjected to 6% non-denaturing PAGE, followed by phosphor imaging (GE Healthcare). Primers used in the PCR reactions were BUB1B forward, 5′-GAAACTTCACTTGCGGAGAACA-3′; BUB1B reverse, 5′-GCAGGAGGACTTTTATTCTTCTTTTCTG-3′; PRPF38A forward, 5′-TCCCAGAAGGCGGAGTCG-3′; and PRPF38A reverse, 5′-GTGATGACCTGGGGAC TTGG-3′. Primers used for PCR of ORAI2, GCC2, and KANSL3 were listed in our previous study (14).
Western Blotting.
Western blotting was performed as described (14). Briefly, protein samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes, followed by immunoblotting with primary and secondary antibodies. Primary antibodies were anti-DHX15 (Bethyl Laboratories, A300-389A, 1:1,000), anti-Glutathione S-transferase (GST) (Invitrogen, A5800, 1:1,500), anti-DYKDDDDK (GenScript, A00187, 1:1,000), anti-SUGP1 (Bethyl Laboratories, A304-675A-M, 1:1,000), anti-SUGP1 (Sigma-Aldrich, HPA004890, 1:1,000), anti-SF3B1 (Bethyl Laboratories, A300-996A, 1:1,000), anti-ACTIN (Sigma-Aldrich, A2066, 1:2,000), anti-HA rabbit polyclonal (Abm, G166, 1:1,000), and anti-HA mouse monoclonal (Sigma-Aldrich, H3663, 1:1,000). Secondary antibodies were Donkey anti-Rabbit IgG (LI-COR, 926-68073, 1:5,000) and Goat anti-Mouse IgG (LI-COR, 926-32210, 1:5,000). Immunofluorescence signals on the membranes were then detected using the ChemiDoc Imaging System (Bio-Rad).
Knockdown Experiments.
Knockdown experiments were performed as described (14). Briefly, two rounds of siRNA transfection were performed. In the first round, 20 pmol siRNA were first mixed with 3-μL DharmaFECT 1 reagent (Dharmacon) in Opti-MEM (Thermo Fisher Scientific). This transfection mixture was then mixed with 300,000 HEK293T cells in fresh growth medium, followed by seeding the cells in one well of a six well plate. The total volume of each well is 2 mL, so that the final concentration of siRNA is 10 nM. After 24 h of incubation, the cells were transfected again with siRNA at a final concentration of 10 nM. At 24 h post second-round transfection (i.e., 48 h post initial transfection), cells were collected for total RNA extraction and protein isolation using TRIzol (Thermo Fisher Scientific). Two independent siRNAs targeting DHX15 (siDHX15-1 and siDHX15-2) and one negative control siRNA (siC) were used, and their sequences were siDHX15-1 sense strand, 5′-GGUCUACAAUCCUCGAAUCdTdT-3′; siDHX15-1 antisense strand, 5′-GAUUCGAGGAUUGUAGACCdTdT-3′; siDHX15-2 sense strand, 5′-GGAGUUGCGAGCUUCAACAdTdT-3′; siDHX15-2 antisense strand, 5′-UGUUGAAGCUCGCAACUCCdTdT-3′; siC sense strand, 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (Shanghai GenePharma); and siC antisense strand, 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (Shanghai GenePharma).
Plasmid Expression Experiments.
HEK293T cells were seeded in six well plates with 300,000 cells per well. After 24 h of incubation, each well of cells was transfected with 2 μg of expression plasmid DNA (or a mixture of two plasmids with 1 μg each for co-expression experiments) using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 h post-transfection, cells were collected for total RNA extraction and protein isolation using TRIzol (Thermo Fisher Scientific).
Minigene Assays.
The ORAI2 minigene and its mutant (–38 A > G) minigene were cloned in pcDNA3 (Invitrogen) in our previous study (14). HEK293T cells were seeded in six well plates with 300,000 cells per well. After 24 h of incubation, each well of cells was transfected with a mixture of 100 ng minigene and 2 μg expression plasmid DNA using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 h post-transfection, total RNA was extracted from the transfected cells using TRIzol (Thermo Fisher Scientific), followed by treatment with DNase I (New England Biolabs). RT-PCR was then performed as described (14). Briefly, 2 μg DNase-treated total RNA was reverse-transcribed with 50 pmol oligo-dT primer and 0.2 pmol vector-specific reverse primer (5′-TAGAAGGCACAGTCGAGG-3′) using 0.3-μL Maxima Reverse Transcriptase (Thermo Fisher Scientific), followed by PCR containing [α-32P] dCTP with vector-specific forward primer (5′-TAATACGACTCACTATAGGGAG-3′) and ORAI2 reverse primer (5′-CTCTCCATCCCATCTCCTTG-3′). PCR products were subjected to 6% non-denaturing PAGE, followed by phosphor imaging (GE Healthcare).
Structure of the DHX15-SUGP1 G-Patch Complex.
Human DHX15 (residues 113–795) and SUGP1 G-patch (residues 543–614) were cloned into pFastBac Serious-438 MacroBac vector and co-expressed in insect cells (Trichoplusia ni, Expression Systems) at 27 °C for 48 to 72 h. DHX15 (residues 113–795) was fused with an N-terminal His6, MBP, and TEV protease cleavage site, while the SUGP1 G-patch has an N-terminal MBP followed by a TEV protease cleavage site. Harvested cell pellets were resuspended in lysis buffer containing 20 mM Tris (pH 8.0), 300 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol and supplemented with SIGMAFAST™ EDTA-free protease inhibitor cocktail (Sigma-Aldrich). Suspended cells were lysed by sonication, followed by centrifugation to remove debris. The lysate was further cleared by 0.8 μm filtration (Cytiva) before being incubated with pre-equilibrated Ni-NTA beads (Qiagen) for 1 h at 4 °C. The nickel beads were washed with lysis buffer, and then the target proteins were eluted with 20 mM Tris (pH 8.0), 300 mM NaCl, 250 mM imidazole, and 10 mM β-mercaptoethanol. The fractions containing the target proteins were pooled and diluted to reduce imidazole concentration using the gel filtration running buffer [20 mM Tris (pH 8.0), 250 mM NaCl, and 5 mM dithiothreitol (DTT)]. The affinity tag was cleaved by TEV protease at 4 °C overnight. Digested protein sample was incubated with amylose resin (New England BioLabs) for 1 h at 4 °C, and the flow through was loaded onto a pre-equilibrated HiLoad 16/600 Superdex 200 prep grade gel filtration column (Cytiva). Fractions with high absorbance at 280 nm were pooled, and the mixture was diluted to a low salt concentration using 20 mM Tris (pH 8.0) and 5 mM DTT. After binding to the HiTrap Q HP column (Cytiva), the protein complex was eluted using a gradient of NaCl increasing to 1.0 M. The protein complex was exchanged to a buffer containing 20 mM Tris (pH 8.0), 250 mM NaCl, and 5 mM DTT, concentrated to ~14.7 mg/mL, flash frozen in liquid nitrogen, and stored at –80 °C.
Crystals of human DHX15-SUGP1 G-patch (residues 543–614) complex were obtained by the hanging-drop vapor diffusion method. The protein solution is composed of 5 mg/mL of complex, 5 mM ADP, and 10 mM MgCl2. Two-μL hanging drop was set with 1:1 ratio of protein solution and reservoir solution at 21 °C. Crystals were observed under conditions containing PEG3350 and sulfates, such as ammonium sulfate, lithium sulfate, or sodium sulfate. After several rounds of optimization, football-shaped crystals more than 150 μm in length were obtained using 24 to 25% (w/v) PEG3350, 200 to 250 mM ammonium sulfate, 100 mM HEPES (pH 7.2 to 7.7). Crystals were cryo-protected with mother liquor supplemented with 20% (v/v) glycerol and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at beamline 24-ID-E (NE-CAT) of Advanced Photon Source (Argonne, USA) using a Dectris EIGER 16M detector. The diffraction images were processed using X-ray Detector Software (XDS) (49). The crystal is isomorphous to that of DHX15 in complex with NKRF G-patch (PDB entry 6SH6) (23). After structure refinement of DHX15 itself, clear electron density was observed for most of the SUGP1 G-patch, Mg-ADP, and three residues (Ser-Asn-Met) from the expression tag at the N terminus, and they were modeled into the density. The structure refinement was carried out with Python-based Hierarchical ENvironment for Integrated Xtallography (PHENIX) (50) and manual model building with Coot (Crystallographic Object-Oriented Toolkit) (51). The quality of the model was assessed using MolProbity (52). The crystallographic information is summarized in SI Appendix, Table S1.
Structures of the SUGP1 C-Terminal Region.
Human SUGP1 (residues 433–577, 433–586, 433–597, 433–605, 433–611, 433–633, and 433–645) were cloned into plasmid pET28a (Novagen) with an N-terminal His6 tag. The expression plasmid was transformed into competent cells and induced using 0.4 mM isopropyl-β-thio-D-galactopyranoside when A600 is 0.8. After incubation overnight at 20 °C, the cells were harvested and resuspended in lysis buffer [20 mM Tris (pH 8.0), 500 mM NaCl, and 5% (v/v) glycerol] supplemented with 1 mM phenylmethylsulfonyl fluroride and lysed with ultrasonication. The lysate was incubated with Ni-NTA resin (Qiagen) and washed with 50 column volumes lysis buffer containing 20 mM imidazole. Bound protein was eluted with lysis buffer containing 250 mM imidazole and loaded onto a Superdex 200 column (Cytiva) equilibrated with gel filtration buffer [20 mM Tris (pH 8.0), 150 mM NaCl]. The fractions of the protein peak were collected and concentrated to 10 mg/mL for SUGP1 433–577 and set up for crystallization with the sitting-drop vapor diffusion method at 20 °C. Crystals were observed in many conditions containing polyethylene glycol 3350. The best crystals were grown in 0.2 M ammonium tartrate dibasic, pH 7.0, and 20% (w/v) polyethylene glycol 3350. Using mother liquor supplemented with 30% (v/v) glycerol as cryo-protectant, the crystals were frozen in liquid nitrogen before data collection. X-ray diffraction data were collected at 100 K at NE-CAT beamline 24-ID-C of Advanced Photon Source (APS) at Argonne National Laboratory and processed using program XDS (49).
To solve the structure, the selenium methionine protein was purified following the same protocol and crystals were obtained in 0.1 M ammonium acetate, 0.015 M magnesium acetate tetrahydrate, 0.05 M sodium cacodylate trihydrate (pH 6.5), and 10% (v/v) 2-propanol. Single-wavelength anomalous dispersion data were collected at 24-ID-C of APS. The phase was solved and improved, and model was auto-built with PHENIX (50). Structure refinement was performed using PHENIX against the native dataset to 2.4 Å resolution, and manual model building was carried out with Coot (51). The crystallographic information is summarized in SI Appendix, Table S1.
To obtain more information on the G-patch region, we examined additional samples covering residues 433–586, 433–597, 433–605, 433–611, 433–633, and 433–645. We were able to determine structures of the samples containing residues 433–586 (2.8 Å resolution, SI Appendix, Table S1) and 433–597. However, no electron density was observed for the additional residues, and the structures are essentially the same as that for residues 433–577, suggesting that these additional residues are highly flexible in SUGP1 alone. The other samples failed to crystallize or produced crystals that did not diffract.
For human SUGP1 433–586, the protein was concentrated to 20 mg/mL and crystals were grown in 0.1 M Bis-Tris (pH 7.5), 25% (w/v) polyethylene glycol 3350. Addition of 5% (v/v) polyethylene glycol 400 was later found to result in crystals with better diffraction properties. X-ray data were collected at APS beamline 24-ID-E and processed with XDS. The crystal is isomorphous to that of SUGP1 433–577 (SI Appendix, Table S1).
Quantification and Statistical Analysis.
Radioactive signals of 32P RT-PCR products were quantified using ImageQuant (Molecular Dynamics), from which PSI values of cryptic 3′ss were calculated. Error bars represent SDs of the means (n = 3; three independent experiments). Unpaired, two-tailed, and unequal variance t tests were performed using Microsoft Excel (Microsoft). A P value less than 0.05 is considered statistically significant.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Acknowledgments
We thank Yu Chun Mu for technical help in this study. The structural work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This work was supported by the National Institutes of Health (grants R35 GM118136 to J.L.M. and R35 GM118093 to L.T.), the National Natural Science Foundation of China (grant 32170565 to Z.L.), and the Chinese Academy of Sciences Hundred Talents Program (Z.L.). J.Z. and J.L.M. were supported in part by a grant from Celgene Pharmaceutical Company (currently Bristol Myers Squibb).
Author contributions
J.Z., J.H., K.X., P.X., Y.H., Z.L., L.T., and J.L.M. designed research; J.Z., J.H., K.X., P.X., and Y.H. performed research; J.Z., J.H., K.X., P.X., Y.H., Z.L., L.T., and J.L.M. analyzed data; and J.Z., J.H., K.X., Z.L., L.T., and J.L.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: D.L.B., University of California, Los Angeles; and T.W.N., Case Western.
Data, Materials, and Software Availability
The atomic coordinates for the structures have been deposited in the Protein Data Bank (PDB; https://www.wwpdb.org/) under accession numbers 8EJM (human DHX15 in complex with SUGP1 G-patch), 8GXL (human SUGP1 433–577), and 8GXM (human SUGP1 433–586). The raw mass spectrometry data and search results have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under accession number MSV000090492. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Yoshida K., Ogawa S., Splicing factor mutations and cancer. Wiley Interdiscip. Rev. RNA 5, 445–459 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Yoshida K., et al. , Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Haferlach T., et al. , Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E., et al. , Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quesada V., Ramsay A. J., Lopez-Otin C., Chronic lymphocytic leukemia with SF3B1 mutation. N. Engl. J. Med. 366, 2530 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Wang L., et al. , SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hintzsche J. D., et al. , Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res. 27, 189–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbour J. W., et al. , Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 45, 133–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biankin A. V., et al. , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis M. J., et al. , Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alsafadi S., et al. , Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darman R. B., et al. , Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015). [DOI] [PubMed] [Google Scholar]
- 13.DeBoever C., et al. , Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 11, e1004105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J., et al. , Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, e1–e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieu Y. K., et al. , SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc. Natl. Acad. Sci. U.S.A. 119, e2111703119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue D., et al. , Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl M. C., Will C. L., Lührmann R., The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Cretu C., et al. , Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Liu Z., et al. , Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation. Proc. Natl. Acad. Sci. U.S.A. 117, 10305–10312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnsack K. E., Ficner R., Bohnsack M. T., Jonas S., Regulation of DEAH-box RNA helicases by G-patch proteins. Biol. Chem. 402, 561–579 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Faber Z. J., et al. , The genomic landscape of core-binding factor acute myeloid leukemias. Nat. Genet. 48, 1551–1556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami K., Nakano K., Shimizu T., Ohto U., The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family. Acta Crystallogr. F Struct. Biol. Commun. 73, 347–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studer M. K., Ivanović L., Weber M. E., Marti S., Jonas S., Structural basis for DEAH-helicase activation by G-patch proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 7159–7170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pryor K. D., Leiting B., High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 10, 309–319 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Fox J. D., Kapust R. B., Waugh D. S., Single amino acid substitutions on the surface of Escherichia coli maltose-binding protein can have a profound impact on the solubility of fusion proteins. Protein Sci. 10, 622–630 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duchemin A., et al. , DHX15-independent roles for TFIP11 in U6 snRNA modification, U4/U6.U5 tri-snRNP assembly and pre-mRNA splicing fidelity. Nat. Commun. 12, 6648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Zaro J. L., Shen W. C., Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 65, 1357–1369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamann F., et al. , Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2. Proc. Natl. Acad. Sci. U.S.A. 117, 2948–2956 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai R., et al. , Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2. Science 371, eabe8863 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Tanaka N., Aronova A., Schwer B., Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 21, 2312–2325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto R., Kataoka N., Okawa K., Ohno M., Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 37, 891–902 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memet I., Doebele C., Sloan K. E., Bohnsack M. T., The G-patch protein NF-kappaB-repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res. 45, 5359–5374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berglund J. A., Abovich N., Rosbash M., A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12, 858–867 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruskin B., Zamore P. D., Green M. R., A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52, 207–219 (1988). [DOI] [PubMed] [Google Scholar]
- 35.Sampson N. D., Hewitt J. E., SF4 and SFRS14, two related putative splicing factors on human chromosome 19p13.11. Gene 305, 91–100 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Crisci A., et al. , Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 43, 10456–10473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jankowsky E., Bowers H., Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 34, 4181–4188 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perea W., Schroeder K. T., Bryant A. N., Greenbaum N. L., Interaction between the spliceosomal pre-mRNA branch site and U2 snRNP protein p14. Biochemistry 55, 629–632 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Noble J. C., Pan Z. Q., Prives C., Manley J. L., Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell 50, 227–236 (1987). [DOI] [PubMed] [Google Scholar]
- 40.Padgett R. A., et al. , Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc. Natl. Acad. Sci. U.S.A. 82, 8349–8353 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruskin B., Greene J. M., Green M. R., Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell 41, 833–844 (1985). [DOI] [PubMed] [Google Scholar]
- 42.Maul-Newby H. M., et al. , A model for DHX15 mediated disassembly of A-complex spliceosomes. RNA 28, 583–595 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fourmann J. B., et al. , The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. Elife 5, e15564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai R. T., et al. , Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19, 2991–3003 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandit S., Lynn B., Rymond B. C., Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. U.S.A. 103, 13700–13705 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fourmann J. B., Tauchert M. J., Ficner R., Fabrizio P., Lührmann R., Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res. 45, 4068–4080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hegele A., et al. , Dynamic protein-protein interaction wiring of the human spliceosome. Mol. Cell 45, 567–580 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R., Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989). [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Cryst. D66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. D., Coot: Model-building tools for molecular graphics. Acta Cryst. D60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Data Availability Statement
The atomic coordinates for the structures have been deposited in the Protein Data Bank (PDB; https://www.wwpdb.org/) under accession numbers 8EJM (human DHX15 in complex with SUGP1 G-patch), 8GXL (human SUGP1 433–577), and 8GXM (human SUGP1 433–586). The raw mass spectrometry data and search results have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under accession number MSV000090492. All other study data are included in the article and/or SI Appendix.