Significance
Despite iterative associations between common gene variants at zinc finger protein 1 (JAZF1) and type 2 diabetes (T2D) as well as height, whether variants in JAZF1 cause these traits has not been established. We show that Jazf1 deletion leads to early growth retardation and late onset insulin resistance in Jazf1 knockout (KO) mice by regulating insulin-like growth factor 1 (IGF-1). Aging and high-fat diet increase the susceptibility of KO mice to T2D, as shown by increased fat mass and insulin resistance. Growth hormone-induced IGF-1 expression is inhibited by JAZF1 knockdown. Therefore, this study provides a functional basis in vivo by which genetic alteration in a single gene, JAZF1, affects stature and T2D.
Keywords: type 2 diabetes, insulin like growth factor 1, height, hnf4a, jazf1
Abstract
Single-nucleotide polymorphisms in the human juxtaposed with another zinc finger protein 1 (JAZF1) gene have repeatedly been associated with both type 2 diabetes (T2D) and height in multiple genome-wide association studies (GWAS); however, the mechanism by which JAZF1 causes these traits is not yet known. To investigate the possible functional role of JAZF1 in growth and glucose metabolism in vivo, we generated Jazf1 knockout (KO) mice and examined body composition and insulin sensitivity both in young and adult mice by using 1H-nuclear magnetic resonance and hyperinsulinemic-euglycemic clamp techniques. Plasma concentrations of insulin-like growth factor 1 (IGF-1) were reduced in both young and adult Jazf1 KO mice, and young Jazf1 KO mice were shorter in stature than age-matched wild-type mice. Young Jazf1 KO mice manifested reduced fat mass, whereas adult Jazf1 KO mice manifested increased fat mass and reductions in lean body mass associated with increased plasma growth hormone (GH) concentrations. Adult Jazf1 KO manifested muscle insulin resistance that was further exacerbated by high-fat diet feeding. Gene set enrichment analysis in Jazf1 KO liver identified the hepatocyte hepatic nuclear factor 4 alpha (HNF4α), which was decreased in Jazf1 KO liver and in JAZF1 knockdown cells. Moreover, GH-induced IGF-1 expression was inhibited by JAZF1 knockdown in human hepatocytes. Taken together these results demonstrate that reduction of JAZF1 leads to early growth retardation and late onset insulin resistance in vivo which may be mediated through alterations in the GH-IGF-1 axis and HNF4α.
Over the past two decades, genome-wide association studies (GWAS) have implicated numerous loci and single-nucleotide polymorphisms (SNPs) with type 2 diabetes (T2D) and obesity (1). However, the progression of our understanding from statistical genetic associations to the underlying cellular and molecular mechanisms accounting for disease risk is relatively uncommon, and a mechanistic understanding has only been advanced in <100 out of the 3,825 associations reported (2). The location of many SNPs adds to the challenge of discovering the underlying mechanisms. Over 80% of the SNPs identified by GWAS are in intergenic or intronic regions, and many of these genetic risk regions are enriched for histone modifications (3), suggesting they act as regulatory sites that function in particular cell types and/or in response to environmental factors. Due to the epigenetic complexity of gene regulation (4), investigation of pathophysiological function of these SNPs should be studied in in vivo with the relevant environmental challenges, such as diet and aging in genetically engineered mice.
In 2008, GWAS surveying hundreds of thousands of SNPs distributed throughout the human genome detected an association of a SNP (rs864745) in the first intron of the juxtaposed with another zinc finger protein 1 (JAZF1, also referred to as TIP27 and ZNF802) gene and individuals with T2D (5–7). Additionally, separate SNPs located in the first intron of JAZF1 and physically closely linked to that correlated with T2D were found to be associated with height (rs849140 and rs1635852) (8, 9) and low birth weight (rs864745) (10), suggesting a functional connection between JAZF1 and both growth and T2D. The rs864745 loci are well-replicated in multiple GWAS in different ethnic groups (111 citations for the loci at https://www.ncbi.nlm.nih.gov/snp/rs864745#publications) and show common association as the variants detection rate in individuals who fulfill the diagnostic criteria can reach ∼47% as shown in Allele Frequency Aggregator (ALFA) project data (90956/192394, https://www.ncbi.nlm.nih.gov/snp/rs864745#frequency_tab). Furthermore, the SNP showed nominally significant association with T2D in African-American cohort (11) and actually associated with mRNA expression in their muscle (12).
JAZF1 contains three putative C2H2-type zinc finger motifs, and thus it has been suggested that JAZF1 might be a transcription factor and/or regulate other protein–protein or protein–DNA interactions (13, 14). JAZF1 is ubiquitously expressed in several tissues, including liver, placenta, colon, prostate and testis, and highly conserved (98%) between mouse and human (13). However, little is known about how these properties relate to function of JAZF1 or the association with diabetes and height. Since the first identification of JAZF1 variants in GWAS, several studies have attempted to establish a causal relationship between JAZF1 mutations and T2D. Metabolic phenotyping of transgenic Jazf1 mice showed decreased body weight and increased insulin signaling (15). A recent study generated a pancreatic β cell-specific Jazf1 knockout (KO) mouse, which had no discernible phenotype under basal conditions and decreases in glucose-stimulated insulin secretion (GSIS) under hyperglycemic challenged conditions (16). In contrast, others reported that mice heterozygous for a loss of function mutation had impaired insulin secretion (17). Although these studies suggest an association between JAZF1 and T2D, the causal relationship between JAZF1 and both T2D and height is still debated. To date, there have been no reports of global Jazf1 KO mice, in which broad effects on tissues other than the pancreas might be assessed and the pathophysiological phenotypes investigated during aging.
Results
The Jazf1 KO Mice are Lean and Short at the Young Age, but Show Flipped Phenotypes Later.
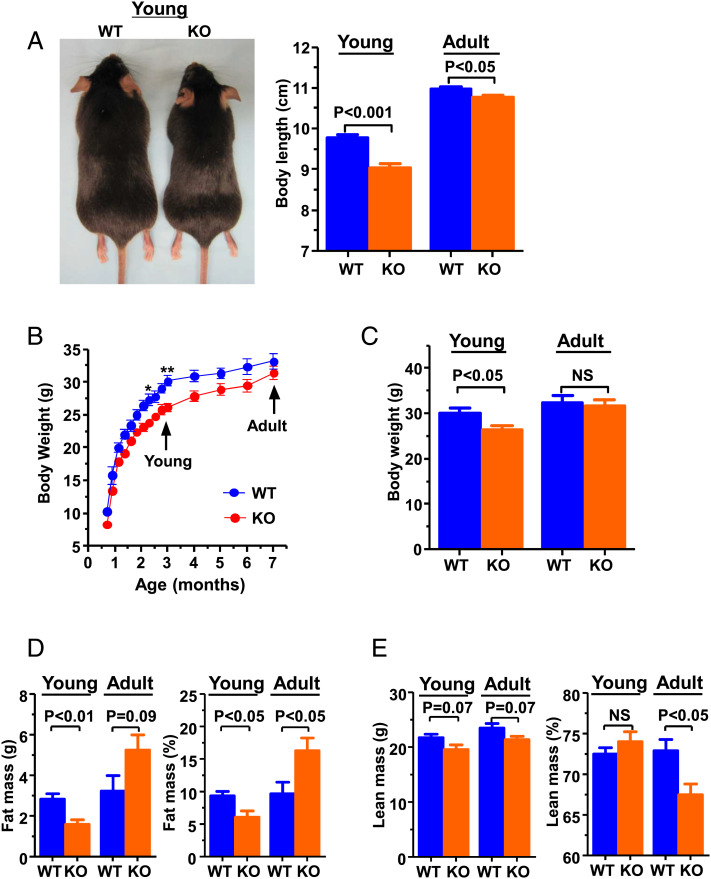
To investigate the possible role of JAZF1 in growth and glucose metabolism, two lines of Jazf1 KO mice were generated. In the first, exon 4 of Jazf1 was replaced by neomycin resistance gene cassette through homologous recombination (SI Appendix, Fig. S1A), as confirmed by southern blot and PCR analysis (SI Appendix, Fig. S1 B and C). The growth pattern and body composition of these mice were evaluated by 1H-nuclear magnetic resonance up to 7-mo-old age compared to age-matched wild-type (WT) mice. The Jazf1 KO mice were found to show early growth retardation at 3-mo-old (Fig. 1 A and B), reflected by ∼7% decrease in body length (Fig. 1A) and ∼15% reduction in body weight (Fig. 1C), relative to the WT mice. During adulthood (Adult, ∼7-mo-old), the Jazf1 KO mice rapidly caught up to WT mice in body weight (Fig. 1B), and this added weight was primarily due to increases in body fat accumulation (Fig. 1D), while lean mass remained lower (Fig. 1E). There were mild differences (less than ∼2%) in body length (Fig. 1A, Right) between WT and KO mice at ∼7-mo-old, suggest that growth retardation of Jazf1 KO mice occurred predominantly in young mice. We also analyzed a second Jazf1 KO line that contained a frameshift-inducing 8 bp deletion in the first exon of Jazf1 as described in a previous paper (18). The Jazf1 KO mice with the exon 1 deletion were also born healthy and exhibited a body weight growth pattern at the young age (SI Appendix, Fig. S1D) similar to that of the exon 4 deleted mice (Fig. 1B).
Fig. 1.
Growth pattern and body composition of WT and Jazf1 KO mice. (A) Representative pictures for WT and KO mice at 3 mo of age and body length measured from nose to rump (n = 5–6 for each group). (B) Body weight growing curves for weaning to 7-mo-old (n = 8 for each group) and (C) average body weight for WT and Jazf1 KO mice. (D) Body fat and (E) lean mass composition by 1H-NMR in WT and Jazf1 KO mice (n = 5 for each group in panel C to E). Young: animals age at ∼3-mo-old. Adult: animals age at ∼7-mo-old. All data are mean ± SEM. *P < 0.05 and **P < 0.01 by two-way ANOVA with post hoc analysis. Statistical numbers are by two-tailed Student's t test.
The Jazf1 KO Mice Show a Level of Low Plasma Insulin-Like Growth Factor 1.
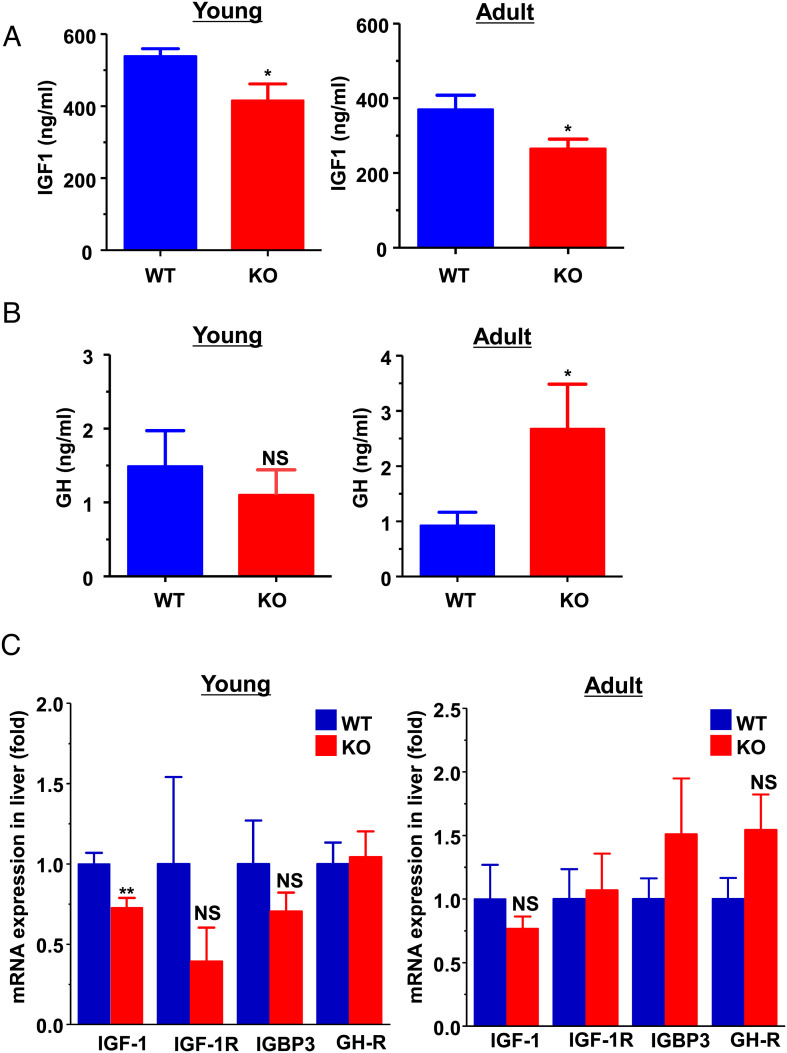
The growth hormone (GH)/insulin-like growth factor 1 (IGF-1) axis is intimately involved in the integration of a multitude of signals that regulate systemic growth and metabolism throughout fetal and postnatal development (19, 20). The postnatal growth-promoting effects of GH are mediated primarily through regulating expression of IGF-1, which is produced primarily by the liver as an endocrine hormone (21, 22). Thus, we measured circulating concentrations and hepatic expression of IGF-1 and GH in young (∼3 mo-old) and adult (∼7 mo-old) animals after overnight fast. Consistent with the decreased body length in young KO mice (Fig. 1A) and lean body mass in adult KO mice (Fig. 1E), the circulating total IGF-1 concentration (Fig. 2A and SI Appendix, Fig. S1E) and hepatic IGF-1 mRNA expression (Fig. 2C) were significantly decreased in the young KO mice. There were no significant differences in plasma GH concentration (Fig. 2B), hepatic mRNA expressions of GH receptor (GH-R), IGF-1 receptor (IGF-1R), and IGF-binding protein 3 between the genotypes (Fig. 2C) in young mice. In adult mice, the plasma GH concentrations were significantly increased (Fig. 2B), while the total plasma IGF-1 concentration remained low in the adult KO mice (Fig. 2A). The findings are consistent with the previous studies demonstrating that IGF-1 is produced primarily in the liver under the direct influence of GH and has a negative feedback on GH release (23, 24). These data suggest that a compensatory increase of GH production may occur in KO mice during aging, and that the deletion of Jazf1 may primarily act through a defect in hepatic IGF-1 production.
Fig. 2.
Serum IGF-1 and growth hormone concentration and hepatic mRNA expression in WT and Jazf1 KO mice. Animals were fed on regular chow and overnight fasted before sampling. (A) Plasma IGF-1 concentration and (B) growth hormone concentration in WT and Jazf1 KO mice. (C) Relative mRNA expression in the liver from WT and Jazf1 KO mice. (n = 5–6 for Young groups, n = 11–12 for Adult groups) All data are mean ± SEM. *P < 0.05 and **P < 0.01 by two-tailed Student's t test, compared to WT mice.
Glucose Stimulated Insulin Secretion is Not Altered in Both Young and Adult Jazf1 KO Mice.
JAZF1 has been reported as a transcriptional repressor of testicular receptor 4 (TR4, also referred to as TAK1 or NR2C2) (13), which can play a role in tumor suppression or cancer promotion (25, 26) and metabolism (27, 28). Tr4 KO mice showed low IGF-1 serum concentrations and early growth retardation (27), similar to what we observed in Jazf1 KO mice. A recent study showed decreased GSIS in beta cell-specific Jazf1 KO mice under hyperglycemic condition (16) and in beta cells differentiated from Jazf1 KO (17). Thus, we tested whether TR4 expression or insulin secretion was affected in our animals. However, we found no significant alterations in hepatic Tr4 expression in both young and adult KO mice compared to WT mice (SI Appendix, Fig. S2A). Basal plasma insulin concentrations and GSIS, assessed during intraperitoneal glucose tolerance test (IPGTT), were not altered in either the young (SI Appendix, Fig. S2C and Fig. 3E) or adult (SI Appendix, Fig. S4F) Jazf1 KO mice. Furthermore, there were no appreciable differences between WT and Jazf1 KO mice for cerebellum size, abnormal maternal behavior, or postnatal pup survival that have been reported with Tr4 KO mice (27, 28). These data suggest that neither insulin secretion nor Tr4 expression is associated with the growth phenotypes observed in Jazf1 KO mice. Thus, it is unlikely that TR4 plays a role in the early growth retardation phenotype in the Jazf1 KO mice. Further, if Tr4 expression was actually increased, we would have expected young Jazf1 KO mice to be taller, not shorter.
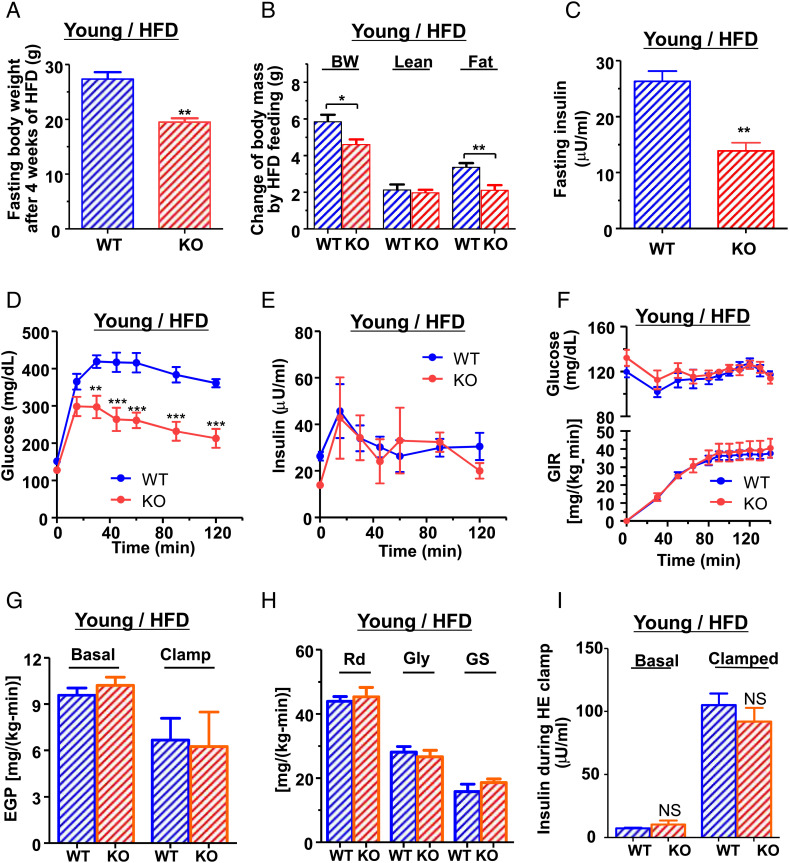
Fig. 3.
Diet-induced obesity and insulin resistance in young animals. Young KO mice were protected from diet-induced obesity without improvements in insulin sensitivity. (A) Fasting body weight of WT and KO mice after 4 wk of HFD feeding at 3 mo of age. (B) Changes of body weight (BW), lean mass, and fat mass during HFD feeding. (C) Fasting plasma insulin concentration after HFD feeding at 3 mo of age. (n = 5–6 for each group) (D) Plasma glucose and (E) insulin concentration during IPGTT at ∼3 mo after 4 wk of HFD feeding. (F–I) Hyperinsulinemic-euglycemic clamp study after 4 wk HFD feeding at the age ∼3 mo. (n = 6–8 for each group) (F) Plasma glucose concentration and glucose infusion rate (GIR). (G) Basal and insulin suppressed endogenous glucose production (EGP). (H) Insulin-stimulated whole-body glucose uptake (Rd) and glycolysis (Gly) and glycogen synthesis (GS). (I) Plasma insulin concentration during the clamp study. All data are mean ± SEM. *P < 0.05 and **P < 0.01 by two-tailed Student's t test for panel A–C and by two-way ANOVA for panel D–F.
In Young Mice, Jazf1 Deletion Protects Diet-Induced Obesity but not Insulin Resistance.
We next examined whether the disruption of Jazf1 might cause insulin resistance (29, 30), which is an early hallmark in the development of T2D. Glucose tolerance and GSIS were assessed by IPGTTs in both young and adult mice fed either a regular chow (RC) or high-fat diet (HFD). Between the group of young KO and WT mice fed RC, there were no differences in plasma glucose and insulin concentrations during IPGTT (SI Appendix, Fig. S2 B and C), suggesting identical glucose tolerance and GSIS in this group. However, when placed on a HFD for 4 wk, marked differences emerged between young WT and KO mice. The diet-induced obesity was dramatically prevented in Jazf1 KO mice as shown by markedly decreased fasting body weight (Fig. 3A) and the gain of body fat content during HFD feeding (Fig. 3B). Furthermore, during IPGTT, fasting plasma insulin concentration (Fig. 3C) and plasma glucose concentrations (Fig. 3D) were lower in KO mice with identical GSIS levels (Fig. 3E). These data suggest that Jazf1 deletion protects HFD-induced obesity and improves glucose tolerance in young mice.
However, it is possible that the marked reduction in body mass of Jazf1 KO mice could have introduced a significant confounder in the IPGTT results since the glucose load is scaled to total body weight (31). To definitively examine whole-body and tissue-specific rates of insulin-stimulated glucose metabolism, we next performed hyperinsulinemic-euglycemic (HE) clamp studies combined with glucose isotopes to assess basal and insulin-stimulated rates of glucose metabolism in liver and skeletal muscle. Surprisingly, despite ∼30% reduction in body weight in the KO mice, whole-body insulin sensitivity as reflected by glucose infusion rate (GIR) required to maintain euglycemia during the HE clamp was identical between two groups (Fig. 3F). There were no differences in hepatic insulin sensitivity between the WT and Jazf1 KO mice, determined by the suppression of endogenous glucose production (EGP) during the HE clamp (Fig. 3G) as well as insulin-stimulated whole-body glucose disposal (Rd), glycolysis, and glycogen synthesis (Fig. 3H). Plasma insulin concentrations after HE clamp study were similar between WT and Jazf1 KO mice (Fig. 3I). These data suggest that in young mice, Jazf1 deletion primarily decreases adiposity on a HFD without altering insulin sensitivity.
In Adult Mice, Jazf1 Deletion Causes Muscle Insulin Resistance.
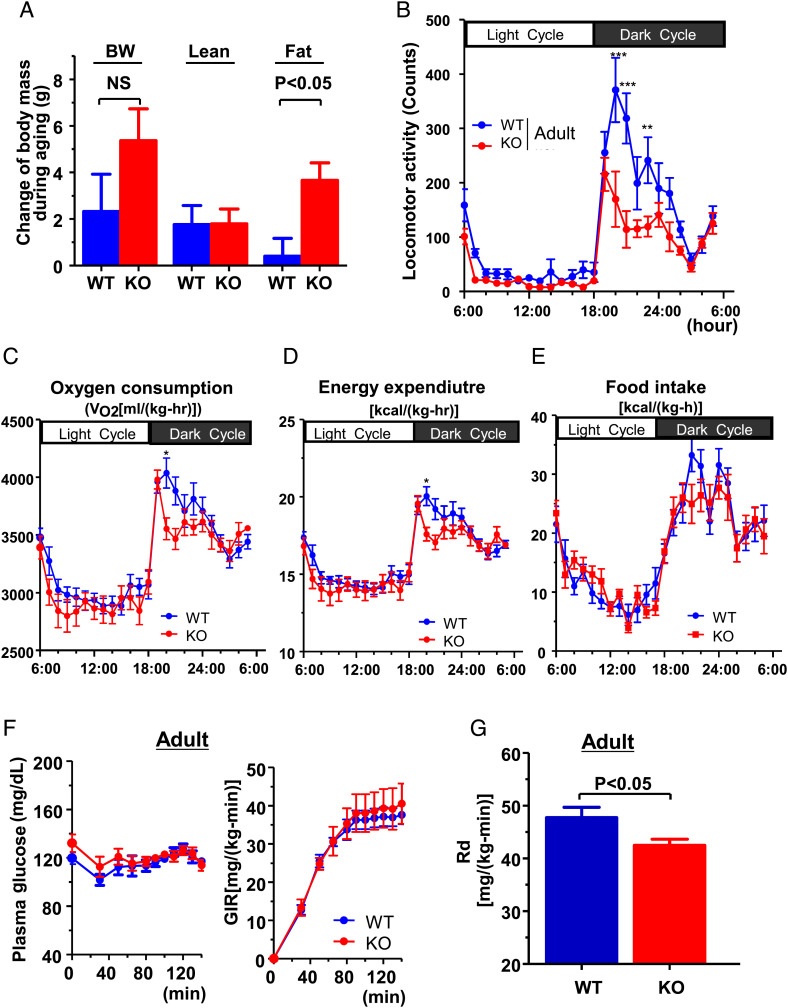
Human subjects with a polymorphism in the promoter of the IGF-1 gene typically present with low IGF-1 concentration, short stature (32, 33) and increased risk of T2D in middle age (55∼65 y old) (32). Thus, we further tested the impact of Jazf1 deletion in combination with aging. As described, Jazf1 KO mice caught up to WT mice in body weight and body length during aging, and body weight was not significantly different between genotypes at the age of 7 mos (Fig. 1B). With aging, the increased body weight in KO mice was mostly accounted for by fat mass (Fig. 4A), resulting in lower lean mass percentage in the KO mice compared to WT mice (Fig. 1E). Consistent with the decreased lean mass and increased fat mass, spontaneous locomotor activity was lower in KO mice during dark cycle (Fig. 4B). These differences may account for observed differences in energy balance. Adult KO mice have decreased whole-body oxygen consumption (Fig. 4C) and energy expenditure (Fig. 4D) without differences in food intake (Fig. 4E). The HE clamp study found that there was a slight but significant decrease in insulin-stimulated glucose disposal (Rd) in KO mice (Fig. 4G), indicating muscle insulin resistance. However, GIR required to maintain euglycemia (∼120 mg/dl) (Fig. 4F), hepatic insulin sensitivity (SI Appendix, Fig. S3A), and plasma insulin concentrations (SI Appendix, Fig. S3B) was not significantly different between two groups. Taken together, these data indicate that the Jazf1 deletion decreases circulating IGF-1 concentration, lean mass, and locomotor activity accounting for a decrease in whole-body energy expenditure, which in turn may predispose the Jazf1 KO mice to accumulation of body fat and muscle insulin resistance with age even under RC fed conditions.
Fig. 4.
Measurement of energy balance and insulin resistance in adult mice fed regular chow. (A) Changes of body weight (BW), lean mass, and fat mass during aging between young and adult WT and KO mice on regular chow (n = 5 for each group). Time course metabolic patterns obtained during 72 h of analysis for (B) locomotor activity, (C) VO2, (D) energy expenditure, and (E) food intake (n = 8 for each group in panel B–E). (F) Plasma glucose and glucose infusion rate (GIR) and (G) insulin-stimulated whole-body glucose uptake (Rd) during hyperinsulinemic-euglycemic clamp study on regular chow at the age of 7 mo. (n = 5–6 for each group in panel F and G). All data are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by two-way ANOVA with post hoc analysis. Statistical numbers are by two-tailed Student's t test.
In Adult Mice, Muscle Insulin Resistance is Further Exacerbated by HFD Challenging.
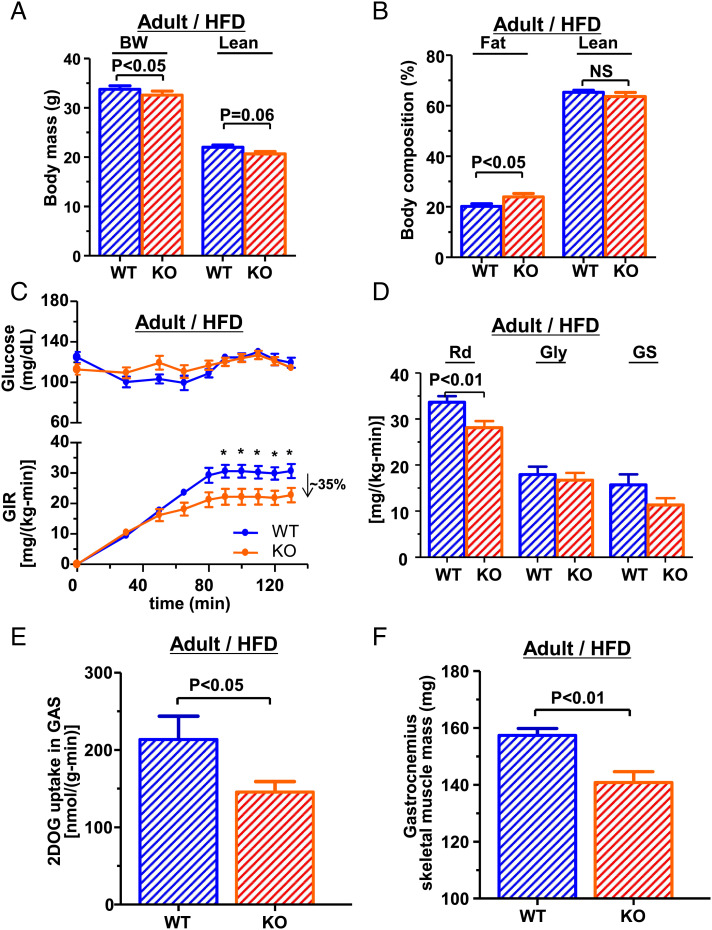
We next examined whether this metabolic phenotype could be further exacerbated with a HFD in adult mice. Following 4 wk of HFD feeding, the difference in body weight and body composition of adult mice on RC (Fig. 1 C–E) remained similar between genotypes. Though the Jazf1 KO mice were lighter (Fig. 5A), they had a higher body fat percentage, compared to WT mice (Fig. 5B). The HE clamp study found that the GIR required to maintain euglycemia was ∼35% lower in Jazf1 KO mice (Fig. 5C), attributable to a ∼20% reduction in insulin-stimulated Rd (Fig. 5D), and decreased insulin-stimulated skeletal muscle 2-deoxyglucose uptake rate (Fig. 5E). Gastrocnemius skeletal muscle mass was significantly decreased in Jazf1 KO mice (Fig. 5F). Interestingly, there were slight decreases in basal rates of EGP (SI Appendix, Fig. S4A) in the adult KO mice after HFD feeding, but neither fasting plasma glucose concentration (Fig. 5C) nor the hepatic insulin sensitivity, determined by suppression of EGP during the HE clamp study (SI Appendix, Fig. S4 A and B), was altered in Jazf1 KO mice compared to age-matched WT mice. Plasma insulin concentration was not different between genotypes (SI Appendix, Fig. S4C). There was a non-significant trend for increases in intramuscular triglyceride accumulation in Jazf1 KO mice after HFD (SI Appendix, Fig. S4D). This may be a consequence of elevated GH in the adult KO mice (Fig. 2B), as GH has been known to increase fatty acids release from adipose tissue and insulin resistance (34). In contrast, this subtle difference in insulin action was not evident following an IPGTT with similar plasma glucose and insulin concentration curves (SI Appendix, Fig. S4 E and F). Together, these data indicating the presence of muscle insulin resistance in the KO mice mainly accounted for whole-body insulin resistance combined with decreased in muscle mass. Furthermore, these data indicate that muscle insulin resistance was exacerbated by HFD feeding in the adult KO mice, suggesting that loss of Jazf1 increased the susceptibility of these mice to diet-induced insulin muscle resistance by aging and diet.
Fig. 5.
Body composition and insulin resistance in adult mice fed high-fat diet. Hyperinsulinemic-euglycemic clamp study after 4 wk high-fat diet (HFD) feeding at the age ∼7 mo (n = 9 for each group). (A) Body weight and lean body mass, (B) percentage of body fat and lean mass in in WT and Jazf1 KO mice fed HFD. (C) Plasma glucose concentration and glucose infusion rate (GIR) during hyperinsulinemic-euglycemic clamp study in adult WT and KO mice fed high-fat diet. (D) Insulin-stimulated whole-body glucose uptake (Rd), glycolysis (Gly), and glycogen synthesis (GS) during hyperinsulinemic-euglycemic clamp study. (E) Insulin-stimulated 2-DOG uptakes in gastrocnemius (GAS) muscle during the clamp study. (F) GAS muscle mass (n = 9 for each group). All data are mean ± SEM. *P < 0.05 by two-way ANOVA with post hoc analysis. Statistical numbers are by two-tailed Student's t test.
Gene Expression Sets Related to GH Response, Glucose/Lipid Metabolism, and Hepatic Nuclear Factor 4 Alpha Pathways are Enriched in the Adult Jazf1 KO Liver.
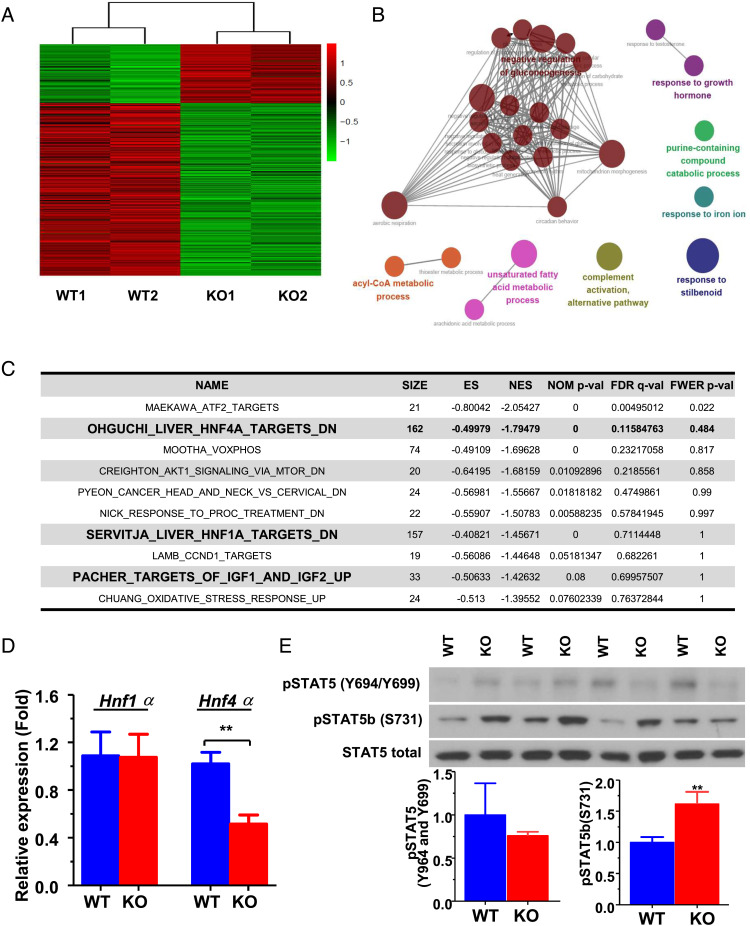
Decreased serum IGF-1 concentration was a common finding in both young and adult in Jazf1 KO mice, and hints at an underlying mechanism of action by which loss of JAZF1 can lead to metabolic diseases. We performed an RNA-Seq transcriptomic analysis to further probe the association between JAZF1 and IGF-1 using RNA extracted from liver of adult WT and Jazf1 KO mice fed RC. Differentially expressed genes (DEGs) were determined by false discovery rate threshold (FDR) <0.05 and genes with fold change greater than 2. As shown in the heatmap visualizing all the DEGs (Fig. 6A and SI Appendix, Table S1), Jazf1 KO mice have a distinct expression pattern compared to the WT mice. When gene sets from gene ontology (GO) biological processes were compared to these DEGs, the top 27 up-regulated pathways identified in Jazf1 KO mice were pathways relevant to GH response, gluconeogenesis, acyl-CoA metabolism, and unsaturated fatty acid metabolism (Fig. 6B and SI Appendix, Table S2). Analysis using chemical and genetic perturbation (CGP) gene sets showed that the liver transcriptomic profile of Jazf1 KO mice is similar to those of liver-specific hepatic nuclear factor 4 alpha (Hnf4α) KO mice, as the gene set of “OHGUCHI_LIVER HNF4A_TARGETS_DN” ranked second on the list of enriched CGP gene sets (Fig. 6C). Also, among the top ten list of CGP gene sets included “SERVITJA_LIVER_HNF1A_TARGETS_DN” and “PACHER_ TARGETS OF_IGF1 AND_IGF2_UP” sets (Fig. 5C). These results indicate that gene sets related to GH response, glucose/lipid metabolism, and HNF4α pathways are highly affected by Jazf1 deletion and likely associated with the low circulating IGF-1 phenotype found in Jazf1 KO mice. Indeed, there was a dramatic decrease in the mRNA expression of the Hnf4α gene by ∼60% in Jazf1 KO liver, compared to WT mice (Fig. 5D).
Fig. 6.
Transcriptomic analysis in the liver from adult WT and Jazf1 KO mice fed regular chow. (A) Heatmap for the differentially expressed genes between WT and KO mouse liver. (B) Enrichment map: commonality of positive enrichments in the gene ontology (GO) gene sets after GSEA assessment using the 108 up-regulated genes in Jazf1 KO mouse liver. (C) The list of top ten enriched CGP gene sets in Jazf1 KO liver with enrichment score (ES), the nominal P-value (NOM P-val), and false discovery rate (FDR Q-val). (D) The mRNA expression of Hnf1α and Hnf4α in the liver from WT and Jazf1 KO mice fed regular chow. (E) Phosphorylation of STAT5B protein in the liver from adult WT and KO mice fed regular chow. N = 4–5 for each group. All data are mean ± SEM for panel D and E. *P < 0.05, **P < 0.01 by two-tailed Student's t test.
Compensatory Increased GH Response Signaling Is Present in the Adult Jazf1 KO Liver.
HNF4α (also referred to as nuclear receptor subfamily 2, group A, member 1) and the protein signal transducers and activators of transcription 5b (STAT5B) exhibit bidirectional crosstalk that may enhance HNF4α-dependent gene transcription but inhibit STAT5B activity via the inhibitory effects of HNF4α on JAK2 phosphorylation. A consequence may be inhibition of STAT5B signaling initiated by the GH-R at the cell surface (35). Consistent with the increases in the circulating GH levels (Fig. 2B) and trend of increases in hepatic GH-R mRNA expression in adult Jazf1 KO mice, we found ∼60% increase in the phosphorylation of STAT5B on serine 731 in the liver of adult KO mice, compared to WT mice (Fig. 6E). These data demonstrated that GH signaling in Jazf1 KO mice is more activated through the step of STAT5B phosphorylation in adult mice and suggest a possible mechanism by which Jazf1 KO increased the GH response. This finding may explain the patterns of catch-up growth and the late onset of insulin resistance in Jazf1 KO mice during aging, as sustained GH-induced lipolysis causally linked to insulin resistance (34, 36) and the impairment of the metabolic action of insulin in patients is well known with GH-producing pituitary tumors (37).
JAZF1 Plays a Role in GH-Induced IGF-1 Expression in Human Hepatocytes.
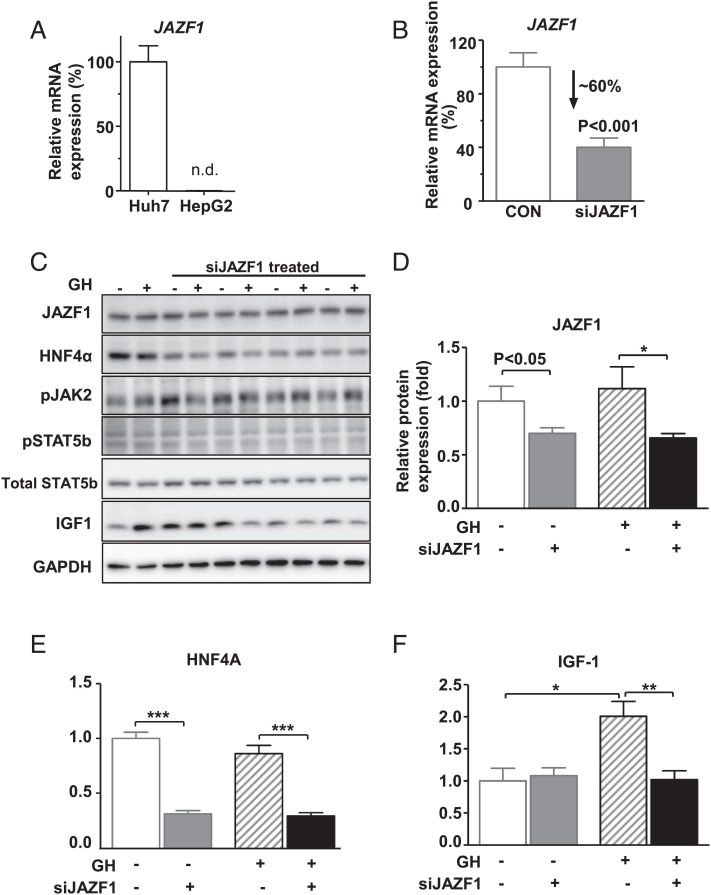
To further test whether JAZF1 directly alters IGF-1 transcription in the GH-IGF-1 axis pathway, we examined GH-induced production of IGF-1 by using JAZF1-specific siRNA (siJAZF1) transfection in cultured cells. Among the human hepatoma cell lines, the mRNA expression of JAZF1 was found to be abundant in the Huh7 cells, but not detectable in HepG2 cells (Fig. 7A). After JAZF1-specific siRNA (siJAZF1) transfection of Huh7 cells, there were ∼60% and ∼40% reductions in the mRNA (Fig. 7B) and protein levels of JAZF1 (Fig. 7 C and D), respectively. Deletion of JAZF1 dramatically decreased HNF4A protein expression in Huh7 cells (Fig. 7E), consistent with the results of animal experiments in which we observed a decrease in Hnf4α expression in Jazf1 KO mice (Fig. 6D). Moreover, GH-induced IGF-1 expression was almost completely blocked by siJAZF1 treatment (Fig. 7F). These data suggest that JAZF1 is required for hepatic IGF-1 production, especially under GH-stimulated conditions.
Fig. 7.
GH-stimulated IGF-1 response after JAZF1 knockdown in Huh7 human hepatocytes. (A) mRNA expression of JAZF1 in Huh7 and HepG2 cells. (B) mRNA expression of JAZF1 after JAZF1-specific siRNA (siJAZF1) treatment in Huh7 cells. (C) Western blot for HNF4α, JAK-STAT-IGF-1 protein in control, and siJAZF1 cells after GH treatment ((50 ng/ml, 1 h), siRNA 30 pmol, and P-JAK2 (T1007, T1008) P-STAT5b (s731)). Normalized western blot data for (D) JAZF1, (E) HNF4α, and (F) IGF-1 protein expression in control and siJAZF1 cells after GH treatment. N = 3–4 for each group. All data are mean ± SEM. *P < 0.05 by one-way ANOVA with post hoc analysis. Statistical numbers are by two-tailed Student's t test. n.d., non-detected.
Discussion
GWAS have proven successful in identifying genetic associations with complex traits. However, while these GWAS approaches have discovered statistically robust associations between many DNA sequence variants and traits (38, 39) including T2D (5, 40, 41), the predictive value of any single variant for assessing an individual’s disease risk has generally been disappointing as each of the identified genes has a relatively small effect on disease risk (40, 41). The link is further complicated when considering traits with significant heterogeneity (like insulin resistance and T2D) that are also influenced by environmental factors, such as aging and diet. Thus, understanding the causal relationship between a genetic marker and a trait remains a challenge, especially for genetically and environmentally complex traits like height, weight, and T2D. The present study is the first of which we are aware to propose a mechanism by which JAZF1 impacts on growth and insulin resistance through the GH-IGF-1 axis and HNF4α. Deletion of the Jazf1 in mice leads to early growth retardation, which was accounted by reduced plasma IGF-1 levels, and in adulthood to decreased muscle mass, increased GH level, fat mass, and insulin resistance. The loss of JAZF1 impacts GH-mediated activation of HNF4A and consequently impedes increases in IGF-1. The Jazf1 KO mice have a decrease in muscle mass and “short stature.” In young mice and on a low-fat diet, the effects on glucose metabolism are minimal. But, with age and a HFD, the decrease in muscle mass contributes to greater insulin resistance, and these data provide insight into the interplay between mutations in JAZF1 with aging and diet in the development of insulin resistance.
To further address if there are any potential relevance of our results to other SNPs associated with T2D and height, we have compared the DEGs with previously reported genes associated with T2D (665 genes) and height (1,404 genes) selected from the GWAS catalog DB (https://www.ebi.ac.uk/gwas/docs/file-downloads). Two genes (Bcl6 and Serpine2) reported in height are increased in Jazf1 KO liver. Interestingly, it has been reported that liver-specific KO of Bcl6 (B cell lymphoma 6) suppresses progression of non-alcoholic steatohepatitis in mice (42, 43). Serpine2 (Serpin Family E Member 2) has been also reported to have a role in cardiac fibrosis as knockdown of serpine2 using RNAi-lentivirus attenuates cardiac fibrosis in a mouse model (44). Taken together, these results suggest a potential relevance of other SNPs to T2D and height in the Jazf1 KO mice and will be interesting to pursue in future studies.
Our data suggest that the decrease in circulating IGF-1 concentration is the key link between genetic polymorphisms in JAZF1 and differences observed in human phenotypes. Several studies have investigated the effect of IGF-1 on growth and its relation to T2D. In humans, approximately 10% of all children born small for gestational age (SGA) continue to have a short stature throughout childhood and adolescence (45), and it has been considered that disturbances in the GH-IGF-1 axis might underlie this reduction in height (46). IGF-1R deficiency causes intrauterine growth retardation or postnatal growth retardation (47). Subjects with a variant in the IGF-1 gene typically present with low IGF-1 concentration and short stature (32, 33) and increased risk of T2D in middle age (55–65 y old) (32). Large longitudinal studies, including the National Health and Nutrition Examination Survey III, reported a higher risk of insulin resistance, metabolic syndrome, and T2D in subjects with low serum IGF-1 concentrations or low IGF-1/IGF-1 binding protein ratios (48–51). Animal models also showed that a deletion of hepatic IGF-1 production, resulting in an 80% reduction in plasma IGF-1 levels, led to hyperinsulinemia and abnormal glucose clearance (52) and muscle insulin resistance (53). Our data are consistent with these previous studies indicating important roles of IGF-1 in growth and glucose metabolism and provide a potential explanation for the clinical association between T2D and both SGA (54) and height (32) as well as for the effects on subjects with certain JAZF1 variants (5, 8–10).
In conclusion, we propose a novel mechanism connecting genetic polymorphisms in the gene JAZF1, to decreased IGF-1 production, growth and insulin resistance, and envision three ways that JAZF1 might directly be associated with pathogenesis of T2D: i) Low IGF-1 levels intimately participate in muscle mass and locomotor activity, and/or ii) increased fat mass by aging, and/or iii) inactivating HNF4α transcriptional activity which might give rise to the maturity-onset diabetes of the young type 1 (MODY1)(55). This study also provides insight into the interplay between genetic variations in JAZF1 with aging and diet in the development of insulin resistance. Regardless of the exact mechanism, based on many different GWAS studies (5–10, 56), JAZF1 appears to affect two traits by regulating IGF-1, and therefore, this study provides a functional basis in vivo by which genetic alterations in a single gene, JAZF1, affect multiple traits, including stature and T2D.
Materials and Methods
Study Design.
To investigate the possible role of JAZF1 in growth and glucose metabolism, two lines of Jazf1 KO mice were generated in which different parts of the Jazf1 gene were deleted, and analyses were performed on these mice measuring body composition, whole-body energy balance, glucose tolerance, and insulin sensitivity (57, 58) on both RC and HFD-fed condition compared to age-matched littermate WT mice. To investigate the underlying mechanism related to IGF-1 axis, a transcriptomic analysis and cell culture studies were performed.
Generation of Jazf1 KO Mice.
DNA KO vector (pKO915) carrying two pieces of DNA fragments of murine Jazf1 flanking the two LoxP sites was inserted into mouse enrichment score (ES) cells. Through homologous recombination, the entire sequence of Jazf1 exon4 was replaced with LoxP-Neomycine cassette (SI Appendix, Fig. S1A). After selection for neomycin resistance, ES cells were injected into blastocysts that were subsequently introduced into the uteri of pseudopregnant female mice. Resulting chimeric mice are bred to produce heterozygotes, which are genotyped using southern blot analysis as previously described (56) (SI Appendix, Fig. S1B). Confirmed heterozygotes were bred to produce homozygotes. PCR to evaluate the status of the Jazf1 gene was carried out using primers JAZF1-205F: CAGCCAACCTATGTTGCCCTGAGTTAC and JAZF1-329R: GACACTGAGCTGGACAGAGTCAGCG (59) (SI Appendix, Fig. S1C). Animals were further backcrossed at least ten generations onto C57BL/6 in the Yale animal facility and used for all analyses presented here. Male mice were used to avoid confounding effects of estrous cycles. The ∼3-mo-old (young) and ∼6 to 8-mo-old (adult) male Jazf1 KO mice and their age-matched littermates were used as controls (WT). To minimize environmental differences, mice were individually housed for at least 2 wk before each experiment. They were fed ad libitum RC diet (17% calories from fat, TD2018S, Harlan Teklad) or HFD (60% calories from fat, D12492; Research Diets) and housed under controlled conditions of temperature (23°C) and lighting (12-h light/12-h dark) with free access to water. The study was conducted at the NIH-Yale Mouse Metabolic Phenotyping Center. Mice with CRISPR/Cas9-induced 8 bp frameshift deletion within the protein-coding sequence in exon 1 of Jazf1 were generated as described in previous paper (18) and studied in different animal facility at Vanderbilt University. All procedures were approved by the Vanderbilt or Yale University Animal Care and Use Committee.
Body Composition and Metabolic Study.
Body length was measured from nose to rump (tail insertion) at 3 mo old and 7 mo after anesthesia with isoflurane. Fat and lean body masses were assessed by 1H-nuclear magnetic resonance spectroscopy (Bruker BioSpin) on WT and Jazf1 KO mice without anesthesia and expressed as percentages of total body weight. A comprehensive animal metabolic monitoring system (CLAMS; Columbus Instruments) was used to evaluate activity, food consumption, and energy expenditure for body weight and fat matched animals. Energy expenditure and food intake data were normalized with respect to body weight. Energy expenditure and respiratory quotient (RQ) were calculated from the gas exchange data as follows: energy expenditure = (3.815 + 1.232 × RQ) × VO2, where RQ is the ratio of VCO2 to VO2, which changes depending on the energy source the animal is using. Activity was measured on x and z axes using infrared beams to count the number of beam breaks during the specified measurement period. Feeding was measured by recording the difference in the scale measurement of the center feeder from one-time point to another.
HE Clamp Study.
Seven days prior to the HE clamp studies, indwelling catheters were placed into the right internal jugular vein extending to the right atrium. After an overnight fast, [3-3H]-glucose (HPLC purified; PerkinElmer) was infused at a rate of 0.05 μCi/min for 2 h to assess the basal glucose turnover, and a HE clamp was conducted for 140 min with a primed/continuous infusion of human insulin (21 mU/kg prime over 3 min, 3 mU/(kg-min) infusion for RC group; 31.5 mU/kg prime over 3 min, 4.5 mU/(kg-min) infusion for HFD group; Novo Nordisk) as described previously (30, 57). During the clamp, plasma glucose was maintained at basal concentrations (∼120 mg/dl). Rates of basal and insulin-stimulated whole-body glucose fluxes and tissue glucose uptake were determined by bolus injection of 2-deoxy-D-[1-14C] glucose (2-DOG, PerkinElmer).
IPGTT.
Mice were maintained on RC or HFD for 4 wk, and the glucose stimulated insulin secretion was analyzed under the age-matched condition. Body weight was significantly lower in KO mice. After an overnight fast, mice were weighed and received a bolus intraperitoneal injection of glucose (1 g/kg). Blood samples were obtained from a tail vein at baseline (0) and 15, 30, 45, 60, 90, and 120 min after glucose challenge for determination of plasma glucose and insulin concentrations.
Plasma Parameters.
Blood samples were taken from the tail vein after 16 h overnight fast in Jazf1 KO and WT mice. Plasma IGF-1 and GH levels were measured using ELISA kits (22-IG1MS-E01, ALPCO, NH, USA and EZRMGH-45 K, Millipore, MA, USA), according to the manufacturer’s instructions. Plasma insulin levels were measured by radioimmunoassay (Linco). Plasma glucose measured by glucose analyzer (YSI).
RNA-Seq and Gene Set Enrichment Analysis (GSEA).
To access the potential pathway related to IGF-1 in Jazf1 KO mice, RNA sequencing (RNA-Seq) data in WT and Jazf1 KO mice liver were analyzed with open-source software v4.1.0 (https://software.broadinstitute.org/gsea/index.jsp) to perform a GSEA of DEGs using the GO biological process and CGPs gene sets. P-values were calculated by permuting the data 1,000 times to find enriched gene sets. The GSEA software produces a nominal P-value and a false discovery rate (FDR; Q-value). Gene sets that were up-regulated or down-regulated with a Q-value < 0.05 were considered significant.
Cell Culture.
The human hepatocellular carcinoma cell line Huh7 was cultured in Dulbecco's Modified Eagle's Medium containing 25 mM glucose, 10% heat-inactivated fetal bovine serum, and 10 μg/ml streptomycin and 10 U/ml penicillin (Welgene, Deagu, Korea). The cells were maintained at 37°C in an atmosphere of humidified air with 5% CO2 in a cell culture incubator. Huh7 cells were seeded in 150 mm tissue culture-treated polystyrene plates (SPL Life Sciences, Pocheon, Korea), grown to ∼80% confluence over 2 d before use or additional passaging.
siRNA Transfection and GH Treatment.
siRNAs were designed and synthesized by Bioneer using human JAZF1 siRNA sequences 5′-CGAAGCUCUCGCUGACUCU-3′ and 5′-UCUCGAUGUAGCACCAUGA-3′. For transfection, Huh7 cells were seeded into six-well plate at 1 × 105 cells and incubated for overnight. siRNA transfection was carried out following the instruction of Lipofectamine RNAiMAX transfection reagent (Invitrogen, CA, USA). After 24 h, the media were replaced with serum-free DMEM containing 25 mM glucose for 3 h, and 50 ng/ml human GH (Sigma, CA, USA) dissolving in sterile PBS with 0.1% human albumin serum (Sigma, CA, USA) was added to the media. After incubation for 1 h, cells were harvested for western blot analysis of IGF-1, HNF4A, and JAK2/STAT5 signaling.
RT-PCR.
Tissue samples were obtained from mice maintained on RC or HFD for 6 wk fasted overnight and euthanized by isoflurane. Total RNA was isolated using Trizol by manufacturer instructions (Invitrogen) from snap frozen liver tissue and harvested cells, and cDNA was reverse transcribed from 1 µg total RNA. Quantitative real-time PCR was performed using Mx7500 multiplex PCR system with SYBRTM Green PCR core reagents (Applied Biosystems, Carlsbad, CA, USA). Relative quantification of gene expression was performed using the standard curve method with beta actin as the housekeeping gene. The list and sequences of PCR primers used are described provided in SI Appendix, Table S3.
Tissue Lipid Measurement.
Tissue triglyceride was extracted using the method of Bligh and Dyer (60) and measured using a DCL triglyceride reagent (Diagnostic Chemicals Ltd, Oxford, Connecticut, USA).
Western Blotting.
Snap frozen liver and harvested cells were homogenized in lysis buffer containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 250 mM NaCl, 0.1% NP40, 5 mM EDTA, phosphatase inhibitor, and complete proteinase inhibitor (Roche Molecular Biochemicals, Indianapolis, Indiana, USA). After centrifugation at 10,000 rpm for 15 min, the supernatants were resolved by SDS-PAGE and transferred to Immobilon-P Polyvinylidene difluoride (PVDF) membranes and immunoblotted. Immunoblots were quantitated from multiple exposures using ImageJ (NIH). The antibodies used for western blotting were as follows: JAZF1 (sc-376503, Santa cruz), human IGF-1 (ab5972, Abcam), phospho-STAT5b (Ser731) (Ab52211, Abcam), phosphor-STAT5b (Tyr699) (05-495, Sigma), STAT5b (ab178941, Abcam, Cambridge, MA, USA), HNF4A (MA1-199, Invitrogen), phospho-JAK2 (Tyr1007) (44-426G, Invitrogen), and GAPDH (5174, Cell signaling).
Statistics.
Values are expressed as mean ± SEM. The significance of the differences in mean values among two groups was evaluated by two-tailed Student's t tests. More than three groups and multiple time point groups were evaluated by ANOVA followed by post hoc analysis using the Bonferroni's multiple comparison test. P-values less than 0.05 were considered significant.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Andreas L. Birkenfeld, Francois R. Jornayvaz, Shoichi Kanda, Michael J. Jurczak, Joao Camporez, David Frederick, Mario Kahn, Blas Guigni, Jinglan Wang, Yanna Kosover, and Aida Groszmann at Yale University and Sung-Yup Cho at Seoul National University for their helpful communication and excellent technical supports in these studies. These studies were support by grants from the National Institutes of Health/NIDDK (R01 DK116774, R01 DK119968, R01 DK113984, R01 DK085087-01, P30 DK045735) and a VA Merit award (I01 BX000901) and National Research Foundation of Korea (NRF) (NRF-2020R1A2B5B01002789 and NRF-2018M3A9F3056405).
Author contributions
H.-Y.L., V.T.S., M.A.M., J.S., and G.I.S. designed research; H.-Y.L., H.R.J., H.L., V.T.S., K.D.D., A.B.O., M.A.M., and J.S. performed research; M.A.M. and J.S. contributed new reagents/analytic tools; H.-Y.L., H.R.J., H.L., V.T.S., K.D.D., A.B.O., M.A.M., J.S., and G.I.S. analyzed data; and H.-Y.L., V.T.S., M.A.M., J.S., and G.I.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: D.A., Broad Institute of Harvard and MIT; and P.F., Imperial College London.
Contributor Information
Mark A. Magnuson, Email: mark.magnuson@vanderbilt.edu.
Jeffrey Sklar, Email: jeffrey.sklar@yale.edu.
Gerald I. Shulman, Email: gerald.shulman@yale.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.D. I. G., Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46, 234–244 (2014), 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallagher M. D., Chen-Plotkin A. S., The post-GWAS era: From association to function. Am. J. Hum. Genet. 102, 717–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012), 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Bailey S. D., Lupien M., Laying a solid foundation for Manhattan–’setting the functional basis for the post-GWAS era’. Trends Genet. 30, 140–149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeggini E., et al. , Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees S. D., et al. , Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia 54, 1368–1374 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Zhou D. Z., et al. , Variations in/nearby genes coding for JAZF1, TSPAN8/LGR5 and HHEX-IDE and risk of type 2 diabetes in Han Chinese. J. Hum. Genet. 55, 810–815 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Johansson A., et al. , Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 18, 373–380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soranzo N., et al. , Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 5, e1000445 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulizzi N., et al. , Interaction between prenatal growth and high-risk genotypes in the development of type 2 diabetes. Diabetologia 52, 825–829 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Langberg K. A., et al. , Single nucleotide polymorphisms in JAZF1 and BCL11A gene are nominally associated with type 2 diabetes in African-American families from the GENNID study. J. Hum. Genet. 57, 57–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma N. K., Langberg K. A., Mondal A. K., Elbein S. C., Das S. K., Type 2 diabetes (T2D) associated polymorphisms regulate expression of adjacent transcripts in transformed lymphocytes, adipose, and muscle from Caucasian and African-American subjects. J. Clin. Endocrinol. Metab. 96, E394–E403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima T., Fujino S., Nakanishi G., Kim Y. S., Jetten A. M., TIP27: A novel repressor of the nuclear orphan receptor TAK1/TR4. Nucleic. Acids. Res. 32, 4194–4204 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., et al. , The role of JAZF1 on lipid metabolism and related genes in vitro. Metab. Clin. Exp. 60, 523–530 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Zhou M., et al. , Effect of central JAZF1 on glucose production is regulated by the PI3K-Akt-AMPK pathway. FASEB J. 34, 7058–7074 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Kobiita A., et al. , The diabetes gene JAZF1 is essential for the homeostatic control of ribosome biogenesis and function in metabolic stress. Cell Rep. 32, 107846 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Park S. J., et al. , Jazf1 acts as a regulator of insulin-producing beta-cell differentiation in induced pluripotent stem cells and glucose homeostasis in mice. FEBS J. 288, 4412–4427 (2021), 10.1111/febs.15751. [DOI] [PubMed] [Google Scholar]
- 18.Osipovich A. B., et al. , A developmental lineage-based gene co-expression network for mouse pancreatic beta-cells reveals a role for Zfp800 in pancreas development. Development 148, dev196964 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dattani M., Preece M., Growth hormone deficiency and related disorders: Insights into causation, diagnosis, and treatment. Lancet 363, 1977–1987 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Bermejo A., Buckway C. K., Rosenfeld R. G., Genetic defects of the growth hormone-insulin-like growth factor axis. Trends Endocrinol. Metab. 11, 39–49 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Baker J., Liu J. P., Robertson E. J., Efstratiadis A., Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75, 73–82 (1993). [PubMed] [Google Scholar]
- 22.Liu J. P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A., Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72 (1993). [PubMed] [Google Scholar]
- 23.Berelowitz M., et al. , Somatomedin-C mediates growth hormone negative feedback by effects on both the hypothalamus and the pituitary. Science 212, 1279–1281 (1981). [DOI] [PubMed] [Google Scholar]
- 24.Tannenbaum G. S., Guyda H. J., Posner B. I., Insulin-like growth factors: A role in growth hormone negative feedback and body weight regulation via brain. Science 220, 77–79 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Sung Y., et al. , Jazf1 promotes prostate cancer progression by activating JNK/Slug. Oncotarget 9, 755–765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang L., et al. , JAZF1 suppresses papillary thyroid carcinoma cell proliferation and facilitates apoptosis via regulating TAK1/NF-kappaB pathways. Onco Targets Ther. 12, 10501–10514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins L. L., et al. , Growth retardation and abnormal maternal behavior in mice lacking testicular orphan nuclear receptor 4. Proc. Natl. Acad. Sci. U.S.A. 101, 15058–15063 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y. S., et al. , Altered cerebellar development in nuclear receptor TAK1/ TR4 null mice is associated with deficits in GLAST(+) glia, alterations in social behavior, motor learning, startle reactivity, and microglia. Cerebellum 9, 310–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayala J. E., et al. , Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis. Model Mech. 3, 525–534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H. Y., et al. , Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 12, 668–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faerch K., et al. , Impact of glucose tolerance status, sex, and body size on glucose absorption patterns during OGTTs. Diabetes Care 36, 3691–3697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaessen N., et al. , A polymorphism in the gene for IGF-I: Functional properties and risk for type 2 diabetes and myocardial infarction. Diabetes 50, 637–642 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Rietveld I., et al. , A polymorphic CA repeat in the IGF-I gene is associated with gender-specific differences in body height, but has no effect on the secular trend in body height. Clin. Endocrinol. (Oxf.) 61, 195–203 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Kopchick J. J., Berryman D. E., Puri V., Lee K. Y., Jorgensen J. O. L., The effects of growth hormone on adipose tissue: Old observations, new mechanisms. Nat. Rev. Endocrinol. 16, 135–146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S. H., Wiwi C. A., Waxman D. J., Signalling cross-talk between hepatocyte nuclear factor 4alpha and growth-hormone-activated STAT5b. Biochem. J. 397, 159–168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma V. M., et al. , Growth hormone acts along the PPARgamma-FSP27 axis to stimulate lipolysis in human adipocytes. Am. J. Physiol. Endocrinol. Metab. 316, E34–E42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizza R. A., Mandarino L. J., Gerich J. E., Effects of growth hormone on insulin action in man. Mechanisms of insulin resistance, impaired suppression of glucose production, and impaired stimulation of glucose utilization. Diabetes 31, 663–669 (1982). [DOI] [PubMed] [Google Scholar]
- 38.Manolio T. A., Brooks L. D., Collins F. S., A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manolio T. A., Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363, 166–176 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Zeggini E., et al. , Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voight B. F., et al. , Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 42, 579–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chikada H., Ida K., Nishikawa Y., Inagaki Y., Kamiya A., Liver-specific knockout of B cell lymphoma 6 suppresses progression of non-alcoholic steatohepatitis in mice. Sci. Rep. 10, 9704 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., et al. , B-cell lymphoma 6 alleviates nonalcoholic fatty liver disease in mice through suppression of fatty acid transporter CD36. Cell Death Dis. 13, 359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., et al. , Overexpression of Serpine2/protease nexin-1 contribute to pathological cardiac fibrosis via increasing collagen deposition. Sci. Rep. 6, 37635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hokken-Koelega A. C., et al. , Children born small for gestational age: Do they catch up? Pediatr. Res. 38, 267–271 (1995). [DOI] [PubMed] [Google Scholar]
- 46.de Waal W. J., Hokken-Koelega A. C., Stijnen T., de Muinck Keizer-Schrama S. M., Drop S. L., Endogenous and stimulated GH secretion, urinary gh excretion, and plasma IGF-I and IGF-II levels in prepubertal children with short stature after intrauterine growth retardation. The Dutch Working Group on Growth Hormone. Clin. Endocrinol. (Oxf) 41, 621–630 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Abuzzahab M. J., et al. , IGF-I receptor mutations resulting in intrauterine and postnatal growth retardation. N. Engl. J. Med. 349, 2211–2222 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Sandhu M. S., et al. , Circulating concentrations of insulin-like growth factor-I and development of glucose intolerance: A prospective observational study. Lancet 359, 1740–1745 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Sierra-Johnson J., et al. , IGF-I/IGFBP-3 ratio: A mechanistic insight into the metabolic syndrome. Clin. Sci. (Lond.) 116, 507–512 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Colao A., et al. , Relationships between serum IGF1 levels, blood pressure, and glucose tolerance: An observational, exploratory study in 404 subjects. Eur. J. Endocrinol. 159, 389–397 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Sesti G., et al. , Plasma concentration of IGF-I is independently associated with insulin sensitivity in subjects with different degrees of glucose tolerance. Diabetes Care 28, 120–125 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Sjogren K., et al. , Liver-derived IGF-I is of importance for normal carbohydrate and lipid metabolism. Diabetes 50, 1539–1545 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Yakar S., et al. , Liver-specific igf-1 gene deletion leads to muscle insulin insensitivity. Diabetes 50, 1110–1118 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Hofman P. L., et al. , Premature birth and later insulin resistance. N. Engl. J. Med. 351, 2179–2186 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Warncke K., et al. , Frequency and characteristics of MODY 1 (HNF4A mutation) and MODY 5 (HNF1B mutation): Analysis from the DPV database. J Clin Endocrinol Metab. 104, 845–855 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Lyssenko V., et al. , Clinical risk factors, DNA variants, and the development of type 2 diabetes. N. Engl. J. Med. 359, 2220–2232 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Lee H. Y., et al. , Apolipoprotein CIII overexpressing mice are predisposed to diet-induced hepatic steatosis and hepatic insulin resistance. Hepatology 54, 1650–1660 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H. Y., Jeong K. H., Choi C. S.; International Mouse Phenotyping Consortium, In-depth metabolic phenotyping of genetically engineered mouse models in obesity and diabetes. Mamm. Genome 25, 508–521 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Li H., et al. , Effects of rearrangement and allelic exclusion of JJAZ1/SUZ12 on cell proliferation and survival. Proc. Natl. Acad. Sci. U.S.A. 104, 20001–20006 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bligh E. G., Dyer W. J., A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.