Significance
Nurr1 is widely expressed in the central nervous system and plays important roles in the progression of a variety of central nervous system-related diseases, and it is a potential therapeutic target for Parkinson’s disease. Besides acting as a monomer, Nurr1 can also regulate transcription as a heterodimer with retinoid X receptor α (RXRα) and activate transcription in response to RXRα ligands. Here we reported a crystal structure of monomeric multi-domain Nurr1 bound to NBRE and an integrative model of Nurr1-RXRα heterodimer. We found that Nurr1 exhibited structure flexibility, in both monomeric and heterodimeric states, which can be modulated by the binding of DNA and/or RXRα agonist. The allosteric signaling of Nurr1 plays vital roles in its constitutive transcriptional activation.
Keywords: Nurr1, Nurr1-RXRα heterodimer, integrative analysis
Abstract
Orphan nuclear receptor Nurr1 plays important roles in the progression of various diseases, including Parkinson’s disease, neuroinflammation, Alzheimer’s disease, and multiple sclerosis. It can recognize DNA as a monomer or heterodimer with retinoid X receptor α (RXRα). But the molecular mechanism of its transcriptional activity regulation is still largely unknown. Here we obtained a crystal structure of monomer Nurr1 (DNA- and ligand-binding domains, DBD and LBD) bound to NGFI-B response element. The structure exhibited two different forms with distinct DBD orientations, unveiling the conformational flexibility of nuclear receptor monomer. We then generated an integrative model of Nurr1-RXRα heterodimer. In the context of heterodimer, the structural flexibility of Nurr1 would contribute to its transcriptional activity modulation. We demonstrated that the DNA sequence may specifically modulate the transcriptional activity of Nurr1 in the absence of RXRα agonist, but the modulation can be superseded when the agonist binds to RXRα. Together, we propose a set of signaling pathways for the constitutive transcriptional activation of Nurr1 and provide molecular mechanisms for therapeutic discovery targeting Nurr1 and Nurr1-RXRα heterodimer.
Nuclear hormone receptors are a family of transcription factors that regulate gene expression and thereby participate in the modulation of various physiological functions such as central nervous system development, homeostasis, reproduction, differentiation, metabolism, steroid production, cell differentiation, and lipid metabolism. Structurally, nuclear receptors contain the following conserved regions: N-terminal domain, a ligand-independent transcription activation region (AF-1) variable both in size and in sequence; a conserved DNA-binding domain (DBD) consisting of two zinc fingers; a hinge that is the most flexible and diverse region in nuclear receptors, mainly playing a role as the domain linker and potentially participating in the formation of nuclear receptor dimers; and a ligand-binding domain (LBD) consisting of 12 α-helices that fold into a three-layered sandwich shape with a conserved AF-2 activation region (Helix 12) (1).
Nurr1 (also known as NR4A2 or NOT) belongs to the NR4A nuclear receptor subfamily and is widely expressed in the central nervous system, especially in the substantia nigra, ventral tegmental area, and the top edge of the brain (2, 3). It is also expressed in areas such as the olfactory bulb, hippocampus, temporal cortex, hypothalamus, cerebellum, posterior hypothalamus, and cerebellar nucleus (4). Nurr1 plays important roles in the progression of a variety of central nervous system-related diseases, including neuroinflammation, Parkinson's disease (PD) (5), Alzheimer's disease (AD) (6), multiple sclerosis (MS) (7), depression (8), and Schizophrenia (9, 10). The crystal structure of Nurr1-LBD showed that, due to the presence of bulky hydrophobic side-chain residues, Nurr1 lacks the classical feature of ligand-binding pocket (LBP) and was defined as the first ligand-independent nuclear receptor (11). However, further solution analyses of Nurr1-LBD suggested that its LBP is dynamic and potentially solvent accessible (12). And a few small molecules, such as PGE1, PGA1 (13), DHI(14), and compounds like AQ and CQ (15), were identified to bind to Nurr1-LBD and enhance its transcriptional activity. These studies provided potential avenues for therapeutic interventions for diseases related to Nurr1.
Nurr1 can bind to NBRE (nerve growth factor-inducible-β-binding response element, 5′-AAAGGTCA-3′) as a monomer (16) and bind to Nur-responsive element as a homodimer (17). Furthermore, Nurr1 can form a heterodimer with retinoid X receptor α (RXRα), a receptor for 9-cis retinoic acid (18). The heterodimer can bind to NBRE, DR5 (18), or IR5 (19) motif and can be efficiently activated by RXRα agonists, such as docosahexaenoic acid, Bexarotene, LG100268, and XCT0139508, to facilitate Nurr1 functions. These RXRα compounds have been demonstrated to protect DA neurons through interaction with Nurr1-RXRα heterodimer (20–22). In PD models, a synthetic RXRα ligand BRF110 showed both neuroprotective and symptomatic effects that depend on Nurr1 expression (23). But the molecular mechanism of the signal transduction in Nurr1-RXRα is still unknown. Due to the crucial rule of Nurr1 in central nervous system-related diseases, both Nurr1 and Nurr1-RXRα are potential drug targets for treatment of these diseases.
To understand the structural basis for transcription activation by Nurr1 monomer and Nurr1-RXRα heterodimer, we attempted to crystallize the multi-domain Nurr1 or Nurr1-RXRα binding to different response elements. We obtained the crystal structure of Nurr1 (DBD-LBD) monomer in complex with NBRE. For the first time, a crystal structure of monomeric multi-domain nuclear receptor binding to DNA has been solved. We also constructed and validated an integrative structural model for multi-domain Nurr1-RXRα heterodimer binding to a IR5 motif. These studies provide structural models for the different domain organization of Nurr1 monomer and heterodimer and further uncover the mechanistic insights into their transcriptional activity regulation.
Results
Crystal Structure of Nurr1 DBD-LBD Binding to NBRE.
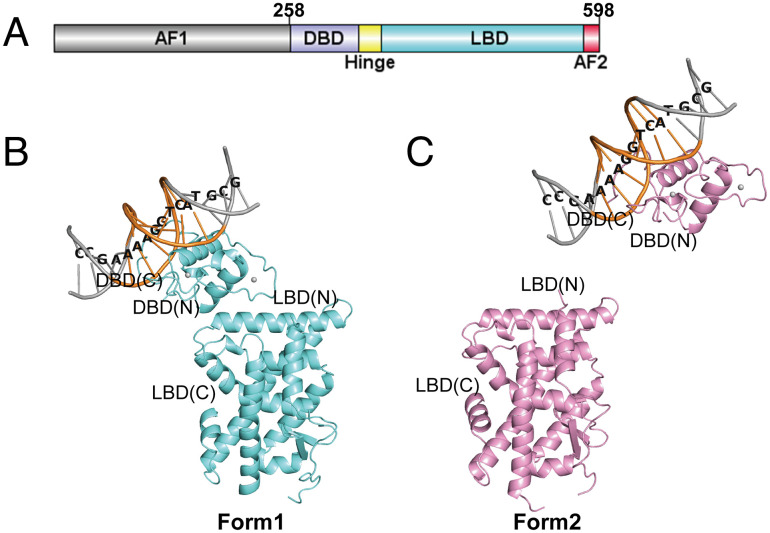
We constructed a multi-domain Nurr1 plasmid including DBD, hinge region, and LBD, named as Nurr1258 (Fig. 1A). We determined the crystal structure of Nurr1258 monomer binding to NGFI-B response element (CCGAAAAGGTCATGCG) to 3.1 Å resolution (SI Appendix, Table S1). The asymmetric unit of the crystal contains four molecules of Nurr1258 with interpretable electron density for DBD, LBD, and the DNA. The region between DBD and LBD, from residue 347 to 360, is invisible in the electron density map and therefore not modeled in the final structure. The DBD and LBD can be organized into two different settings, herein defined as Forms 1 and 2. In Form 1 (chains A and B) (Fig. 1B), the DBD anchors on the loop between Helix 8 and Helix 9 in LBD. In this form, the Helix 2 of DBD and Helix 9 of LBD are involved in the inter-domain interface, whose buried surface areas are 259 and 283 Å2 in chains A and B, respectively. In Form 2 (chains C and D), the DBD and LBD are further away from each other (Fig. 1C).
Fig. 1.
Crystal structure of Nurr1258-NBRE (PDB: 7WNH). (A) Domain structure of Nurr1. Overall structure of Nurr1258-NBRE in two forms: Form 1 (B, aquamarine) and Form 2 (C, pink). NRBE binding motif was highlight by orange and the DNA sequence was labeled. The N and C termini of DBD and LBD were labeled.
The overall structures of DBD in four molecules are very similar to each other with a root-mean-square deviation (RMSD) value of 0.10 Å for Cα atoms. The DBD closely resembles the rat NGFI-B (rNGFI-B, PDB ID: 1CIT (24)) binding to the same element, with a RMSD of 0.43 Å. Residues E281, K284, K288, and R289 on Helix 1 of DBD are involved in binding to the major groove of DNA, in a similar manner as those in rNGFI-B. However, compared to rNGFI-B, the DNA chains binding to the C terminus of DBD untwist slightly in the structure of Nurr1258-NBRE, which is similar to the structure of Nurr1-DBD binding with inverted repeat element (PDB ID: 6L6Q (17)) (SI Appendix, Fig. S1A).
Interestingly, structure superposition of the four molecules of Nurr1258 LBD shows conformation changes around Helix 10/11 and Helix 12, which can be grouped into two forms. Group I, including chains B and D, is almost identical to the LBD structure reported previously in apo-form (11). In group II (chains A and C), the loop between Helix 9 and Helix 10/11, also referred as Ω loop, and the N-terminal region of Helix 10/11 exhibits a higher degree of flexibility, resulting in a slight bending in the middle of Helix 10/11 where P560 is shifted about 3.3 Å. Additionally, Helix 12 swings inward about 15.2°, which is firstly observed in reported Nurr1-LBD structures (SI Appendix, Fig. S1B).
Solution Studies Using XL-MS and Small-Angle X-ray Scattering (SAXS) Confirm the Orientation Variability between DBD and LBD in Nurr1 Monomer.
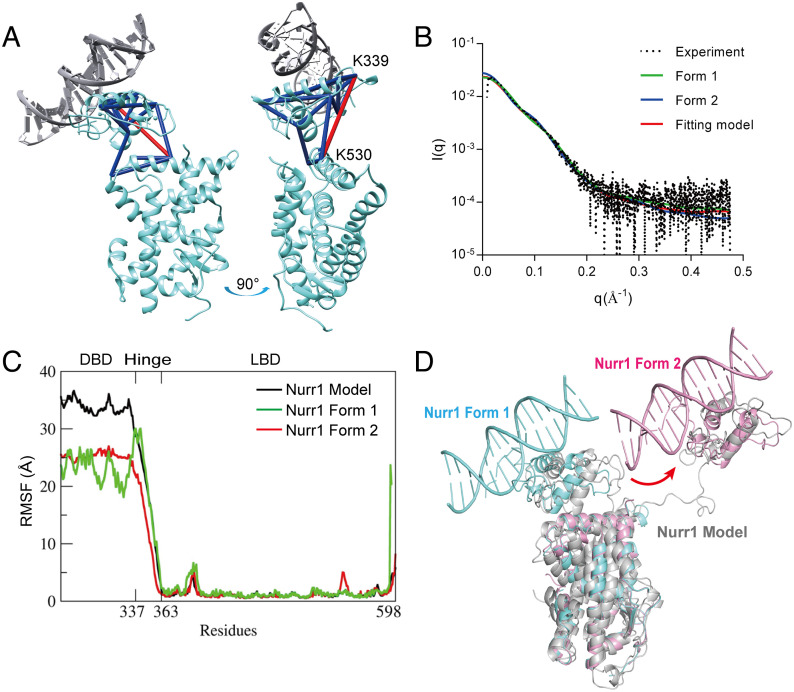
To confirm the existence of conformations observed in the crystal structure, we performed chemical cross-linking mass spectrometry (XL-MS) experiment for Nurr1258-NBRE using bis(sulfosuccinimidyl) suberate (BS3) cross-linker and SAXS experiment. BS3 linker has a 11.4 Å long arm when fully extended, thus allowing the two Cα atoms of cross-linked lysine residues to be up to ∼24 Å apart in theory. However, the linker flexibility and structure dynamics in solution can lead to a larger range of observed inter-residue distances for the covalently linked lysine residues. XL-MS studies on proteins of known structure frequently report cross-links that exceed the theoretical distance. Therefore, a distance constraint of 26–30 Å between Cα atoms for BS3 cross-linked lysine residues is also appropriate (25). In this study, we obtained 15 high-confidence (confidence score >10, calculated by pLink2 (26)) BS3 cross-linked peptides. We set the upper limit of distance constraint for the cross-linked lysine–lysine (Cα-Cα) as 30 Å to validate the structure. Fourteen out of the 15 cross-linked peptides were satisfied with the Form 1 model in the crystal structure. The remaining one cross-linker connected K339 and K530 in this model, with a Cα–Cα distance of 33.7 Å in the crystal structure, which may result from the conformational flexibility of the protein in solution (Fig. 2A). On the other hand, the distance between DBD and LBD in Form 2 is too long to be captured in the BS3 cross-linking. Additionally, we performed XL-MS for Nurr1258 without DNA binding and mapped the cross-linked lysine on Form 1. The results showed that 31% of high-confidence cross-links were not consistent with the structure, indicating significant conformation change for Nurr1 upon binding to DNA (SI Appendix, Fig. S2).
Fig. 2.
Analysis of the two forms observed in the crystal structure using XL-MS, SAXS, and MD simulation. (A) High-confidence cross-links of Nurr1-NBRE were labeled on Form 1. The cross-links consistent with the structure were colored in blue. Only one cross-link not consistent with the structure was labeled and colored in red. (B) Curve-scattering patterns computed by CRYSOL for SAXS data. (C) RMSF analysis from the three MD calculations (with different starting models) for Nurr1 DBD-LBD monomer. The x-axis represents residue number (from DBD to LBD), and the y-axis is the fluctuation of the corresponding residues in Å. Most of the residues move within 2–5 Å range, and the DBD and hinge region exhibit larger atomic fluctuations than the rest of the structure. (D) Models identified in the trajectories (in grey, MD calculation starting from the Robetta model) were overlapped with Nurr1 Form 1 (aquamarine) and Form 2 (pink).
Furthermore, we used SAXS to analyze the two forms of Nurr1-NBRE in solution (Fig. 2B and SI Appendix, Table S2). The calculated molecular weight from the SAXS data (about 42.8 kD) is consistent with the theoretical value ~43 kD. A high NSD value suggests that the complex had multiple conformations in solution. Using OLIGOMER (27), we identified the volume fractions of Form 1 and Form 2 as 0.61 and 0.39, respectively. And the χ2 value of the fitting model is 1.64, calculated by CRYSOL (28) (Fig. 2B).
Crystal structures of multi-domain protein or protein complex represent snapshots of how molecules organize or pack in the crystal lattice. In this monomeric structure of Nurr1 (DBD-LBD) binding with DNA, we unexpectedly observed two forms of domain organization. Together with XL-MS and SAXS data, our results clearly show the conformation variation between DBD and LBD when the monomeric Nurr1 binds to NBRE.
MD Calculation Demonstrates the Flexibility of Hinge Region of Nurr1.
We then conducted MD simulations for the Nurr1 DBD-LBD model in explicit water to investigate its structure dynamics. The initial model was predicted by Robetta online Website (https://robetta.bakerlab.org) using cross-linking restraints. MD trajectories analysis showed that the hinge region of Nurr1 is very flexible, and thus the relative positions of DBD and LBD have a lot of spatial possibilities (Fig. 2C). Models resembling the two forms of Nurr1 in the crystal structure can be identified in the trajectories (Fig. 2D). We also used the two models from the crystal structure as the starting points to conduct the simulation. Interestingly, starting from the Form 1 structure, we can obtain models that have a similar relative orientation of DBD and LBD as in Form 2 from the MD trajectory (SI Appendix, Fig. S3). Taken together, MD results confirm that Nurr1 exhibits great flexibility, mostly in the hinge region, when it exists alone.
Nurr1-RXRα Heterodimer Binds with IR5.
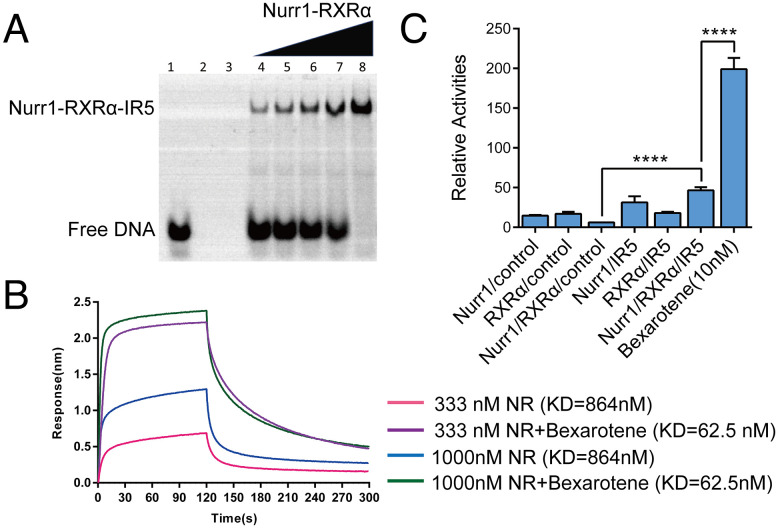
Using high-throughput SELEX and ChIP sequencing, Arttu Jolma et al. revealed that Nurr1-RXRα heterodimer can bind to a IR5 motif that consists of two inverted half-sites spaced by five nucleotides (19). We constructed a plasmid that contains DBD, hinge region, and LBD of both Nurr1 and RXRα, and successfully obtained the heterodimer protein named Nurr1258-RXRα98. We carried out electrophoretic mobility shift assay (EMSA) to confirm the binding ability of IR5 to Nurr1258-RXRα98. Purified Nurr1258-RXRα98 heterodimer was incubated with IR5-cy3 in the presence of RXRα agonist Bexarotene. Samples were detected on a native polyacrylamide gel. As shown in Fig. 3A, Nurr1258-RXRα98 can form a complex with IR5. Moreover, we used biolayer interferometry (BLI) to probe the binding affinity of IR5 to Nurr1258-RXRα98. As shown in Fig. 3B, the binding affinity of IR5 to Nurr1258-RXRα98 heterodimer is 864 nM in the absence of Bexarotene, and it is 62.5 nM when Bexarotene is added.
Fig. 3.
Nurr1-RXRα heterodimer binding with IR5 (A) Binding of Nurr1258-RXRα98 with IR5 was determined by EMSA, lane 1 shows free IR5 without protein, lane 2 shows free Nurr1258-RXRα98 without IR5, lane 3 is empty control, lane 4–8 show the IR5:Nurr1258-RXRα98 in molar ratio of 1:0.5, 1:0.75, 1:1, 1:1.25, 1:2, respectively. (B) Binding affinity of IR5 on Nurr1258-RXRα98 (NR) measured by BLI. (C) Luciferase reporter assays testing the transcriptional ability of Nurr1258-RXRα98 to IR5 in the absence and presence of Bexarotene. The control is pGL3-promoter vector.
We then performed luciferase reporter assay to assess the transcription activity of Nurr1-RXRα heterodimer on IR5. Full-length Nurr1 and RXRα were co-transfected in HEK293T along with an IR5-luciferase reporter construct in the absence or presence of Bexarotene. Nurr1 and RXRα could co-express in cells and recognize IR5 efficiently. Bexarotene significantly increased the transcription activity of Nurr1-RXRα on IR5 (EC50 = 50 nM) (Fig. 3C and SI Appendix, Fig. S4). Collectively, Nurr1 can form a heterodimer with RXRα and specifically recognize IR5 response element and RXRα agonist Bexarotene can efficiently enhance the binding affinity and transcription activity of the heterodimer on IR5.
Integrative Modeling of Nurr1-RXRα Heterodimer When Binding with IR5.
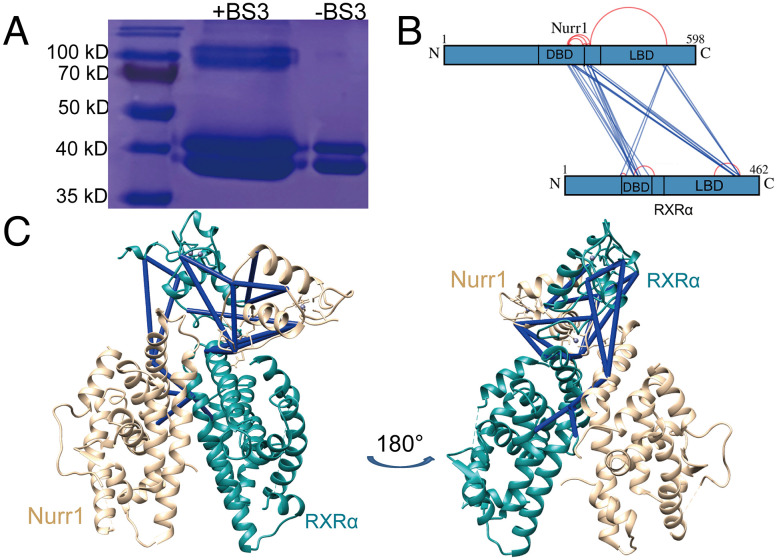
To obtain intra- and intermolecular distance information for Nurr1258-RXRα98-IR5 complex, we performed cross-linking mass spectrometry (XL-MS) experiments using two different types of cross-linkers, bis(sulfosuccinimidyl) suberate (BS3) and EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride). The analysis of cross-linked samples via SDS–PAGE revealed a few bands at about 80–110 kD, which may result from the cross-linking. We took the major heterodimer band at about 80 kD for further analysis via liquid chromatography/tandem MS after in-gel trypsin digestion (Fig. 4A). We obtained 35 high-confidence cross-linked peptides (confidence score > 10, calculated by pLink2), 12 of them were intramolecular cross-linked peptides and 23 of them were intermolecular cross-linked peptides. There was no intramolecular cross-linked peptide between DBD and LBD, neither in Nurr1258 nor in RXRα98. However, among the intermolecular cross-linked peptides, four were found to cross-link Nurr1258 DBD and RXRα98 LBD, and two were found to cross-link Nurr1258 LBD and RXRα98 DBD (Fig. 4B). We obtained similar results in EDC XL-MS experiment (SI Appendix, Fig. S5). Atomic models of Nurr1258-RXRα98 were then generated by using DISVIS (29) and HADDOCK (30), combined with the distance information provided by XL-MS. The final model is in excellent agreement with all high-confidence cross-linked data (Fig. 4C).
Fig. 4.
Calculation of the Nurr1-RXRα heterodimer model combined with XL-MS data. (A) SDS–PAGE analysis of Nurr1258-RXRα98-IR5 with and without cross-linker (BS3). (B) Map of high-confidence BS3 cross-linked residues shown schematically on the Nurr1 and RXRα sequence, dark blue lines connected the intermolecular cross-linked peptides, red arc connected the intramolecular cross-linked peptides. (C) High-confidence cross-linked were labeled on top of Nurr1258-RXRα98 model (Nurr1, wheat; RXRα, teal), dark blue for distance satisfied cross-links.
Hydrogen–Deuterium Exchange Mass Spectrometry (HDX-MS) Experiments Confirm the Inter-Domain Interface in Nurr1-RXRα Heterodimer.
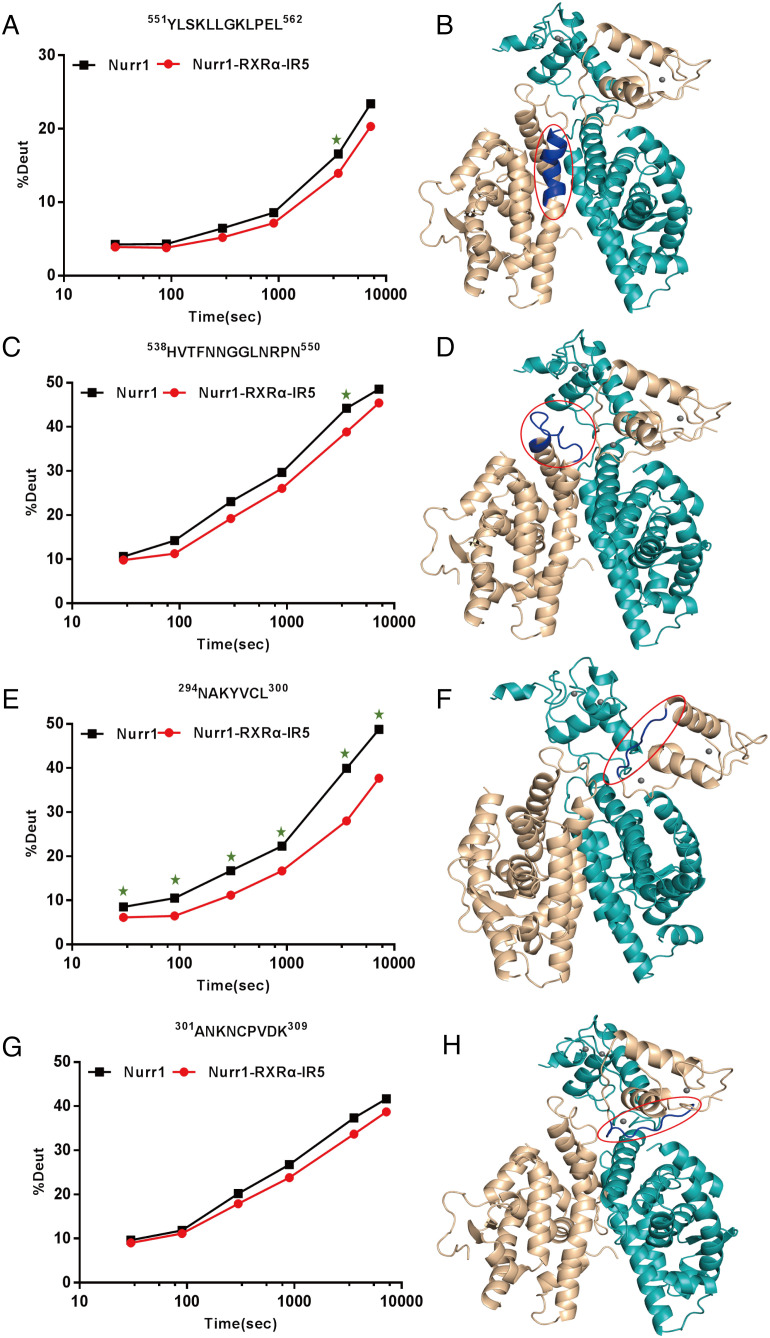
Nurr1258-RXRα98 model showed an X-cross shape (Fig. 4C). The two LBDs utilize the classical dimerization sites (Helix 9-10, Loop 8-9, and Helix 7) (31), which agrees with previous study that Nurr1 I-box (K554-L562) is very important for Nurr1-RXRa dimerization (32). To probe changes in protein structures between Nurr1258 monomer and Nurr1258-RXRα98-IR5, we performed HDX-MS analysis. Decreases in deuterium uptake were observed in the I-box 551YLSKLLGKLPEL562 of Nurr1258 in Nurr1258-RXRα98-IR5, compared with Nurr1258 alone (Fig. 5 A and B). Additionally, it is noted that a single patch (on Helix 9 and Helix 10, also referred to as Ω loop) on the LBDs of distinct subunit partners of RXRα tends to participate in domain interaction between LBD and DBD (33). In our model, this patch (538HVTFNNGGLNRPN550), positioning at the Nurr1258-LBD and RXRα98-DBD interface, exhibited decrease in deuterium uptake (Fig. 5 C and D). This suggested that dynamic allostery of this patch participates in the signal transmission between Nurr1 and RXRα.
Fig. 5.
HDX-MS experiments reveal deuterium uptake decrease in regions participated in the formation of Nurr1258-RXRα98 heterodimer. (A, C, E, G) Deuterium uptake plot for peptides (in Nurr1) compared between Nurr1258 (black) and Nurr1258-RXRα98-IR5 (red). (B, D, F, H) Peptides with decreased deuterium uptake (dark blue) were mapped to the Nurr1258-RXRα98 model (Nurr1, wheat; RXRα, teal). The green stars in A, C, E stand for significant differences in the uptake plot between two protein samples.
Meanwhile, in the integrative modeling process, we obtained a variety of models with similar LBD-LBD formation but entirely distinct DBDs locations. This indicated that the DBDs may be flexible in the dimer formation. Interestingly, the interface of the symmetrical homodimer Nurr1-DBD in NR4A2-DBD-IR5 complex mainly consists of residues 294NAKYVC300 of Nurr1-DBD (17). In our model, likewise, this region contributes to the dimerization of Nurr1-DBD and RXRα-DBD. The observed deuterium uptake decrease in this region corroborates well with our model (Fig. 5 E and F). Moreover, we observed deuterium uptake decrease of a peptide 301ANKNCPVDK309 which is at the interface between Nurr1-DBD and RXRα-LBD in the model, but there was no significant difference (calculated by HDExaminer) between the two plots, suggesting that this interface may not be stable in solution. (Fig. 5 G and H).
MD Analysis of the Nurr1258-RXRα98 Heterodimer Complex.
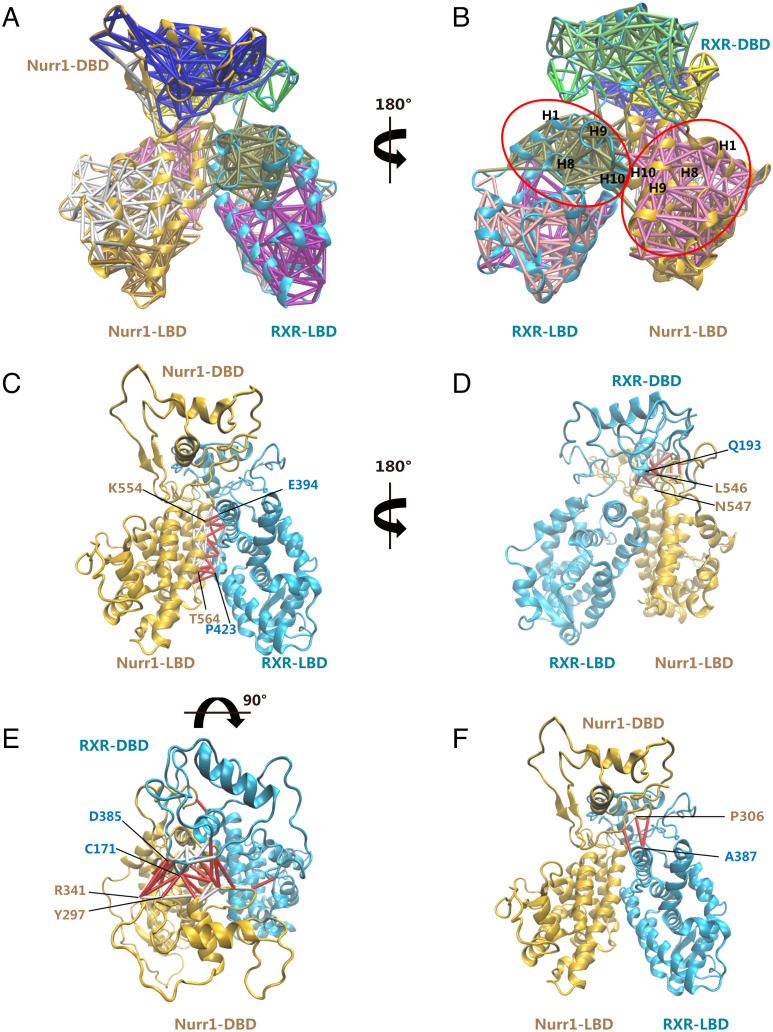
We performed MD simulations for the generated Nurr1258-RXRα98 heterodimer model to analyze the inter-domain interfaces. Then we investigated the dynamics of the residues within Nurr1258-RXRα98 domain interfaces by conducting dynamical network analysis (34). Dynamical network describes the location and extent of concerted motions in a biomolecular system and can be used to identify communication pathways between molecular components. The network consists of a set of nodes and a set of edges connecting pairs of nodes. We assigned one node to each amino acid centering at Cα atom. Then we drew edges between nodes whose residues were within a cutoff distance (4.5 Å) for at least 75% of the MD trajectory. Additionally, we identified smaller subnetworks denoted as communities, which partitioned the original network and corresponded to sets of residues that moved in concert with each other. Residues situated in the interface between pairs of communities were defined as critical nodes, carrying most of the communications between communities.
Fig. 6 A and B shows dynamical networks calculated from the last 600 ns of the MD trajectory, mapping onto the Nurr1-RXRα model after the final MD step. The critical nodes lie on all the inter-domain interfaces within the heterodimer, suggesting that these residues carry the majority of coupled motions between Nurr1 and RXRα (Fig. 6 C–F). Among the communities we identified, the motion of those consisting of residues in LBD (H1, H8, H9, and H10) is compactly coupled to the motion of the corresponding DBD and LBD partner. Consequently, the motion of the communities formed by residues in Nurr1 is strongly coupled to the motion of those in RXRα, via a tight network of interactions. These findings hint a potential role of the long-range communication in signal transductions between Nurr1 and RXRα.
Fig. 6.
Analysis of the inter-domain contacts in the Nurr1-RXRα Model. (A, B) Dynamical network analysis of Nurr1-RXRα MD simulations. Subnetworks are shown in different colors with corresponding edges plotted onto the last snapshot from the MD simulations. Communities consisting of residues in LBD (H1, H8, H9, and H10) were circled in red. Critical edges connecting networks across and edges connecting nodes within the same network across the (C) Nurr1-LBD: RXRα-LBD interface, (D) Nurr1-LBD: RXRα-DBD interface, (E) Nurr1-DBD: RXRα-DBD interface, and (F) Nurr1-DBD: RXRα-LBD interface are colored red and labeled by their corresponding node residues.
We further analyzed the details of edges in all inter-domain interface (SI Appendix, Table S3). Critical nodes were chosen on the four inter-domain interfaces to analyze their Cβ distance dynamics. Overall, there are numerous edges in the Nurr1-LBD: RXRα-LBD interface, suggesting a very stable interface (Fig. 6C). And the Cβ distance of residues at this interface remained very close to their starting values in the model (SI Appendix, Fig. S6A). The Cβ distance of residues at the Nurr1 LBD-RXRα DBD and Nurr1 DBD-RXRα DBD interface did not change significantly as they remained relatively stable during the MD simulation (SI Appendix, Fig. S6 B and C). However, there are only three edges and one critical-node connection at the interface of Nurr1-DBD and RXRα-LBD, demonstrating that there is no close connection in this interface during the MD simulation (Fig. 6F and SI Appendix, Fig. S6D). Additionally, we performed optimal and suboptimal path analysis between key residues in the heterodimer. I268 in RXRα-LBD is critical for ligand binding. P560 in Nurr1-LBD plays a significant role for heterodimer interface communication. K156 in RXRα-DBD and K288 in Nurr1-DBD are key residues for DNA binding. There are numerous suboptimal paths, alternative paths in the network with nearly equivalent distance, linking these residues (SI Appendix, Fig. S7). This analysis suggests that the inter-community junctions are crucial regions responsible for the allosteric communication between Nurr1 and RXRα.
Functional Validation and Putative Model for Nurr1-RXRα Binding to DNA.
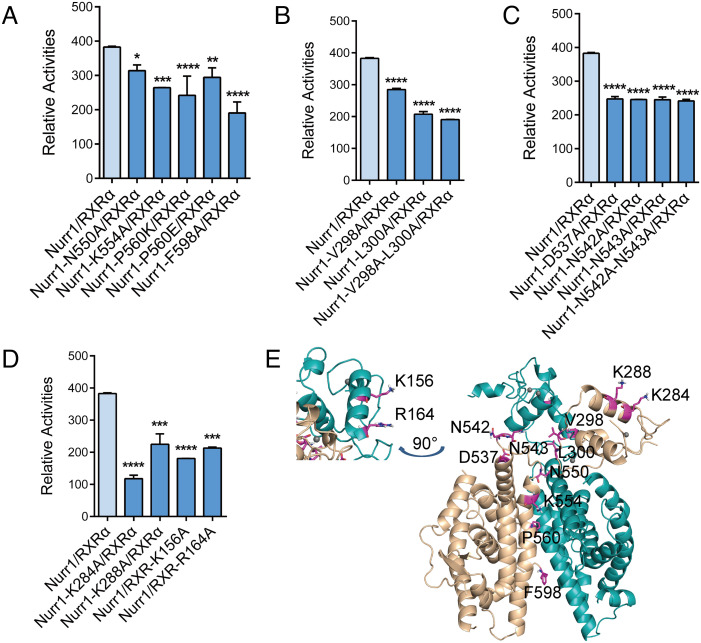
Using cell-based functional studies, we investigated the involvements of the domain-domain interfaces of Nurr1 and RXRα for the transcriptional activities on IR5. As we mentioned above, the interface of Nurr1-DBD and RXR-LBD was unstable in the complex, so the cell-based assays were performed focusing on the other interfaces in the complex. To probe the importance of Nurr1-LBD and RXRα-LBD interactions for the transcriptional activity, we constructed mutations (N550A, K554A, P560E, P560K and F598A) in Nurr1. According to our model, the mutation of N550, K554, and P560 would influence the dimerization of the two LBDs by breaking the hydrogen bond and hydrophobic interaction. F598 is sandwiched between L596 from Nurr1 and L430/K431 from RXRα and may function to stabilize the two LBDs’ interaction. The ability of these mutants to activate transcription in the context of heterodimer was examined by transfection in human H293K cells. All mutants showed a decrease in the transcriptional activity of Nurr1-RXRα on IR5 (Fig. 7 A, E). We also constructed mutations (V298A, L300A, V298A-C299-L300A) in Nurr1 that participated in the interaction between Nurr1-DBD and RXRα DBD, and mutations (D537A, N542A, N543A, N542A-N543A) in Nurr1 at the interface between Nurr1-LBD and RXRα DBD. The mutations of N542 and N543 could alter the conformation of the Ω loop of Nurr1, thereby affecting the conformation of helix 10/11 of Nurr1. The results showed that these mutants on Nurr1 could also reduce, to different degrees, the transcriptional activity (Fig. 7 B, C, and E). Taken together, these results indicated that these three interfaces in Nurr1-RXRα complex are critical for the transcriptional signal transmission.
Fig. 7.
Effects of mutations at distinct interfaces in Nurr1-RXR α -IR5. Transcription activity for mutations of Nurr1 at (A) Nurr1-LBD and RXRα-LBD interface, (B) Nurr1-DBD and RXRα-DBD interface, (C) Nurr1-LBD and RXRα-DBD interface. (D) Transcription activity for mutations of Nurr1 and RXRα at the potential DNA-binding interface of Nurr1-RXRα complex. (E) The mutated residues were mapped on the Nurr1-RXRα model.
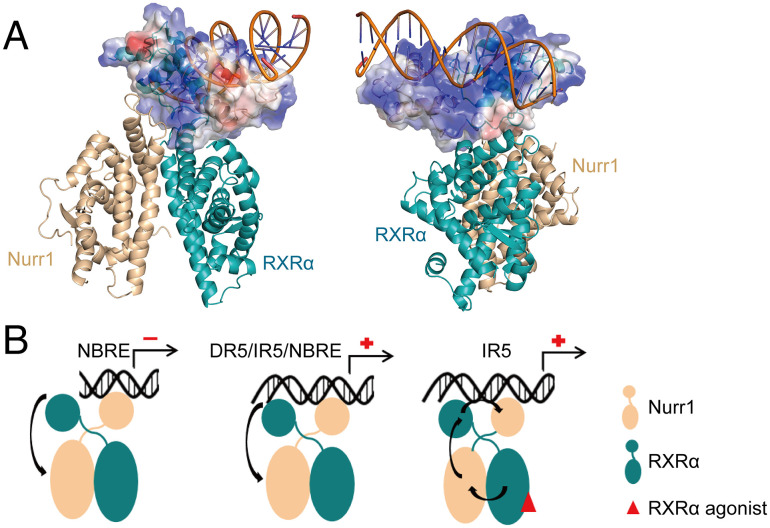
The crystal structure studies of Nurr1 DBD and RXRα DBD binding to DNA indicated that the N-terminal Helix 1 of Nurr1-DBD and RXRα-DBD predominantly interact with the major groove of DNA (17, 35). To probe the DNA binding site on the DBDs of Nurr1-RXRα, we constructed mutations (K284A/K288A in Nurr1 and K156A/R164A in RXRα) on Helix1 of Nurr1 and RXRα DBD, which were mentioned in previous studies to participate in the interaction between the DBDs and DNA. The transcriptional activity decreased for all the mutants, compared to wild-type Nurr1-RXRα (Fig. 7 D and E). These suggest that the binding interface between the Nurr1-RXRα heterodimer and IR5 must be in the first helix of the two DBDs, corroborating well with our model in which the Helix 1 of the two DBDs apparently forms a positively charged surface for DNA binding (Fig. 8A). We used Paradock (36) to dock IR5 onto the Nurr1258-RXRα98 heterodimer. The two positively charged regions can insert into the two consecutive DNA major grooves, as shown in Fig. 8A and SI Appendix, Fig. S8. However, due to the limitation of rigid docking, the orientation of the helix to the major groove may not be precisely determined. The whole complex will likely undergo conformational change when binding to IR5 and warrant further studies.
Fig. 8.
A putative model for Nurr1 binding to its response elements. (A) Nurr1258-RXRα98-IR5 model generated by Paradock, the DBDs are shown as electrostatic potential surface, blue for positive charge and red for negative charge. (B) In the absence of RXRα agonist, the transcriptional activity of Nurr1 in the Nurr1-RXRα heterodimer may depend on DNA sequence. RXRα agonist can facilitate a series of conformation allostery to transmit the transcriptional active signal. Blue for Nurr1, orange for RXRα, and red oval for RXRα agonist.
Discussion
With a hinge connecting DBD and LBD, the overall structure of nuclear receptors is generally very flexible. It is challenging to assess the allosteric communications between the nuclear receptor domains. Current understanding of the mechanistic action of nuclear receptors has mostly relied on analyses of the individual DBD or LBD. To date, only four crystal structures containing multi-domain nuclear receptors bound to DNA have been reported, and all of them are hetero-/homodimers (33, 37–39). Here we determined the crystal structure of monomeric multi-domain Nurr1 binding with NBRE. We found two forms of structures with different spatial arrangement between the DBD and LBD. Form 1 exhibits a compact formation, and Form 2 shows an expanding form. Both forms can be identified from solution studies. The structures present clear evidence for the high flexibility of nuclear receptors in the monomeric form. Many studies suggested that intramolecular DBD-LBD interactions mediate signal transduction in nuclear receptors, and ligands or coactivators can trigger the conformation allostery of LBDs to facilitate the interaction. However, there is no direct interaction between the DBD and LBD in both forms of our monomeric Nurr1 structures, even in Form 1. Our XL-MS experiments, with and without DNA, for the monomeric Nurr1 reveal the conformational changes upon binding to DNA. This suggests that the recognition and binding process of Nurr1 to its target gene may contribute to the constitutive conformation allostery of Nurr1, instead of binding to ligands or coactivators. Therefore, the conformation allostery of Nurr1 may contribute to its constitutive active ability. Our finding provides a novel molecular mechanism for the ligand/coactivator-independent activity of orphan nuclear receptors.
Given that Nurr1 as a ligand-independent nuclear receptor, the development of drug against Nurr1 is difficult. Even though a few small molecules (13–14) have been shown capable of binding to Nurr1-LBD, their mechanism of action is not well defined. More interestingly, it is reported that a few agonists of RXRα can specifically activate Nurr1-RXRα heterodimer (22, 23, 40). It is imperative to understand the spatial arrangement and signaling pathway of Nurr1-RXRα heterodimer. Combining solution biophysical methods with molecular modeling, we obtained an integrated model of multi-domain Nurr1-RXRα heterodimer. The X-cross shape model we generated resembles the reported structure of RXRα-LXRβ on a DR4 motif (39). Comparing RXRα-LXRβ to our Nurr1-RXRα model shows that Nurr1 and LXRβ share very similar relative orientation between their DBD and LBD, albeit RXRα DBD location is distinctly different (SI Appendix, Fig. S9). And the two DBDs are closer in the Nurr1-RXRα model, which is most likely caused by the inversed binding motif in IR5. This is consistent with the hypothesis that the flexible hinge of RXRs helps nuclear receptor partners adapt to different DNA elements (33). Moreover, the X-cross shape could be a common structure feature for nuclear receptor heterodimers.
More importantly, the HDX-MS and molecular dynamics simulation results show that most of interfaces in the model remain relatively stable, except for the interface between Nurr1-DBD and RXRα-LBD, suggesting that Nurr1-DBD has a higher degree of flexibility in the dimer complex. On the other hand, a study from Piia Aarnisalo et.al showed that unliganded RXRα can suppress the constitutive activity of Nurr1 on NBRE (32). However, our study shows unliganded RXRα can slightly increase the activity of Nurr1 on IR5 (Fig. 3C). Previously, Sebastiaan H et al. proposed that DNA is a sequence-specific allosteric ligand of GR and tailors the activity of the receptor toward specific target genes (41). We speculate that, in the context of heterodimer, without RXRα agonist, similar to the monomeric Nurr1, different DNA can act as a sequence-specific allosteric trigger to modulate the conformational change of Nurr1, mostly relying on the orientation variability of Nurr1-DBD via the hinge region movement, to activate or inactivate the downstream gene activity.
In the study of Piia Aarnisalo et.al, RXRα agonist can rescue the repressive activity on Nurr1 (32). In our study, RXRα agonist Bexarotene can significantly increase the transcriptional activity of Nurr1-RXRα on IR5. We also performed XL-MS experiment for Nurr1258-RXRα98 in the absence of RXRα agonist and the data showed a significant difference from the data in the presence of Bexarotene (SI Appendix, Fig. S10 A and B). Previous studies showed that RXRα LBD exhibited very different conformation between apo and ligand-binding state (42) and the binding of its agonist can induce conformation change in its partner (43). We propose that agonist binding to RXRα will induce conformation allostery of Nurr1 LBD, mostly related to the Ω loop and Helix 10/11, and the active signaling will be transduced through the allostery from RXRα-DBD and Nurr1-DBD successively (Fig. 8B and SI Appendix, Fig. S7). And the allosteric effect caused by RXRα agonist would be stronger than that from DNA sequence. Interestingly, binding of RXRα agonist was reported to have different effects on various RXRα partners, affecting not only their binding to DNA but also their binding with their corresponding ligands (44). In this work, we have not investigated the effect of binding of RXRα agonist to potential Nurr1 ligand recognition, which warrants future study.
In summary, here we report a crystal structure of monomeric multi-domain Nurr1 binding to its response element NBRE, which contains two distinct conformations. The structure analysis reveals high flexibility for Nurr1 monomer. In the monomeric state, the transcriptional signal transduction mostly depends on the allostery triggered by the binding of DNA rather than the interaction between its DBD and LBD. Furthermore, we obtain an integrated model of multi-domain Nurr1-RXRα heterodimer binding with IR5, within which Nurr1 DBD is the most flexible domain. Nurr1 still has sufficient flexibility to modulate its own transcription activity in the absence of RXRα agonist. However, when the RXRα agonist binds to the Nurr1-RXRα heterodimer by interacting with RXRα LBD, it will greatly facilitate the conformation allostery of the whole complex to transmit the transcription signal to their target genes. These findings will provide important molecular mechanisms for therapeutic discovery targeting Nurr1 and Nurr1-RXRα.
Materials and Methods
Expression, Purification, and Crystallization.
The Nurr1258 (258–598) was expressed in a modified pET28a vector in BL21(DE3) E. coli cells. Cells were induced with 0.1mM IPTG at 17°C overnight, harvested, and lysed in 20 mM Tris, pH 7.5, 500 mM NaCl, 20 mM imidazole. The protein was purified by Ni-sepharose, Heperin, and Superdex200 (GE Healthcare) gel filtration column successively, and the final elution buffer of Superdex 200 was 20 mM Tris, pH 7.5, 150 mM NaCl, and 2 mM DTT. The NBRE (5′-CCGAAAAGGTCATGCG-3′) and its complementary strand were synthesized by Sangon Biotech. Both strands were lysed with 50 mM Tris, 150 mM NaCl, pH 7.5 to 100 uM and then annealed by combining equimolar amounts of each strand and heating to 95°C followed by slow cooling to 20°C. Nurr1258 and NBRE were then combined in 1:1.2 molar ratio and then purified by Superdex 200. Finally, the complex was concentrated to 20 mg/mL and crystals were grown using the sitting drop method against a reservoir of 0.1M sodium malonate, pH5.0, 20% PEG3350, at 4°C. After optimization with a additive screen (Hampton research), the crystal we used for diffraction experiment was grown in 0.1M sodium malonate, pH5.0, 20% PEG3350,1.44% Myo-inositol.
The Nurr1258 and RXRα98 were co-cloned in pET-Duet vector for co-expression in BL21(DE3) E. coli cells. Cells were induced with 0.1 mM IPTG at 17°C overnight, harvested, and lysed in 20 mM Tris, pH 8.0, 500 mM NaCl, and 20 mM imidazole. The protein complex was purified by Ni-sepharose, Source Q and superdex200 (GE Healthcare) gel filtration column successively, and the final elution buffer of Superdex 200 was 20 mM HEPES, pH 7.5, 150 mM NaCl, and 1 mM TCEP.
IR5 (5′- AAAGGTCAAACTGTGACCTAT-3′) and its complementary strands were synthesized and annealed with the same method mentioned above.
Synthetic coactivator peptide (EKHKILHRLLQDSY) and the agonist Bexarotene of RXRα were added in 5× molar ratio to Nurr1258-RXRα98 complex, IR5 was added in 1× molar ratio to Nurr1258-RXRα98 complex, incubated at 4°C overnight. The whole complex was then purified by Superdex 200.
X-Ray Data Collection and Structure Determination.
Prior to data collection, crystals of Nurr1258-NBRE were soaked in mother liquor with the addition of 70% paraffin oil mixed with 30% paratone oil as cryoprotectant. Complete diffraction datasets for the crystals were collected at the Shanghai Synchrotron Radiation Facility at 100K. Diffraction patterns were integrated using DIALS (45). Then, datasets were scaled using Aimless from the CCP4 program suite (46). Five percent of the data were randomly selected for the R-free calculation. The initial structure solutions of Nurr1 bound with NBRE were obtained by Phaser-MR using the DBD structure of rat NGFI-B (PDB ID: 1CIT) and the LBD structure of human Nurr1 (PDB ID: 1OVL) as the search models (47). Then the model was refined by REFMAC5 and Coot (48–50). Structure figures were prepared using the program PyMOL (51).
Cross-Link Identifications via Mass Spectrometry.
Nurr1258-RXRα98-IR5 complex was prepared in cross-linking buffer (20 mM HEPES, pH 7.5, 150 mM NaCl). BS3 (Bis(Sulfosuccinimidyl) suberate) was freshly dissolved in cross-linking buffer at a final concentration of 2 mM. BS3 cross-linker was added to Nurr1258-RXRα98-IR5 complex at the ratio of 30:1 and incubated on ice for 2 h. Cross-link reactions were quenched for 15 min at room temperature with the addition of 50 mM TRIS pH 7.5. Control reactions were performed in parallel without the addition of cross-linker. Reaction produces were separated via SDS–PAGE.
In-Gel Digestion.
The gel band was excised into small pieces and washed with ddH2O, 50% acetonitrile, and 100% acetonitrile sequentially. The sample was then reduced with 10 mM TCEP (in 100 mM NH4HCO3) for 30 min and alkylated with 55 mM iodoacetamide (in 100 mM NH4HCO3) in the dark for 30 min at room temperature. The gel pieces were washed with 100 mM NH4HCO3 and 100% acetonitrile and then dried out. 12.5 ng/μL Trypsin (in 100 mM NH4HCO3) was added to the tube for digestion at 37°C overnight. The tryptic digested peptides were extracted twice with 50% acetonitrile with 5% formic acid. And the peptide extraction was desalted using a MonoSpinTM C18 column for LC-MS/MS analysis.
LC/Tandem MS (MS/MS) Analysis and Data Analysis.
The desalted peptide mixture was loaded on a home-made 30-cm-long analytical column (75 μm I.D. packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch GmbH, Germany) by an Easy-nLC 1200 nano HPLC (Thermo Scientific, San Jose, CA). A 120-min HPLC gradient were used for peptide separation just as follows: 0–1 min, 6 to 10% B; 1–96 min, 10 to 36% B; 96–107 min, 36 to 60% B, 107–108 min, 60 to 100% B, 108–120 min, 100% B. The flow rate was set as 300 nL/min.MS analysis was performed with a Q Exactive mass spectrometer (Thermo Scientific, San Jose, CA) connected to the LC. Eluted peptides were electrosprayed into the mass spectrometer with a distal 2.5-kV spray voltage. Data-dependent acquisition (DDA) mode was applied for MS analysis. Briefly speaking, one full-scan MS spectrum (m/z 300-1800) was acquired followed by top 20 MS/MS events at a 28% normalized collision energy (NCE). MS resolution was set to 70,000 with automated gain control (AGC) target at 3e6, and MS/MS scan resolution was set to 17,500 with isolation window of 1.8 m/z and AGC target of 1e5. Maximum ion injection time was set at 50 ms and 100 ms, respectively. The dynamic exclusion settings were set at 5 s, 10 s, or 15 s. The pLink2 (pFind Team, Beijing, China) was used for data interpretation and cross-linking peptide identification as previously described (29, 30). Missed cleavages for trypsin was set at 3, and precursor and fragment tolerance were set at 20 ppm. Carbamidomethylation of cysteine and oxidation of methionine was set as fixed and variable modifications. The peptide level FDR was set at 1%.
Generation of Structural Model of Nurr1-RXRα on IR5.
Atomic models of Nurr1258-RXRα98 were generated from the crystal structures of their individual domains, Nurr1-DBD (PDB: 6L6Q), Nurr1-LBD (PDB: 1OVL), RXRα-DBD (PDB: 1BY4), and RXRα-LBD (PDB: 6A5Z) using DISVIS (52) and HADDOCK (53, 54), combined with the distance information provided by XL-MS. Firstly, DISVIS was used to analyze the information content of cross-linked restraints within macromolecular complex. It can identify the violated restraints and define the putative active and positive residues. Thirty-four medium confident cross-linked peptides (score > 5, calculated by pLINK2) were input and analyzed by DISVIS. We obtained 14 cross-linked peptides and sets of active residues on the surface between different domains. With these data, we used HADDOCK to dock the protein models and clusters of complex models were output. All high confident cross-linked peptides were used to rank the models with a satisfied score by setting the distance restraints between Cα atoms of the cross-linked residues to 30 Å. And finally, we used Paradock to perform rigid docking of IR5 onto the Nurr1-RXRα heterodimer model.
SAXS.
The SAXS data were collected at the BL19U2 beamline at National Facility for Protein Science Shanghai (NCPSS) and Shanghai Synchrotron Radiation Facility (SSRF). The wavelength was set as 1.033Å. For SEC-SAXS, 100 ug of ∼7 mg/ml Nurr1258-NBRE was injected on a size-exclusion chromatography Superdex 200 Increase 10/300 GL column equilibrated with 20 mM Tris-HCl, pH7.5, 150 mM NaCl. 2D scattering images were converted to 1D SAXS curves by the software package BioXTAS RAW (55). All preparations were analyzed by linearity in the Guinier region of the scattering data. Pair distribution functions of the particles P(r) and the maximum sizes D max were calculated by the program GNOM (56). Low-resolution shapes were determined from solution scattering data using DAMMIF, from the ATSAS program suite (57, 58). Twenty independent calculations were performed by DAMMIF programs for each dataset, using default parameters and no symmetry constraints. And the theoretical scattering patterns I(s) from the crystal structures was calculated by program CRYSOL (57).
Hydrogen/Deuterium Exchange (HDX) Mass Spectrometry Experiments.
Hydrogen/deuterium exchange (HDX) mass spectrometry experiments were performed as previously described (59). Briefly, protein complexes were processed automatically by a LEAP Technologies Hydrogen Deuterium Exchange PAL system (Carrboro, NC) combined with Thermo LTQ Elite MS. The timepoints for HDX were set at 0 s, 30 s, 90 s, 300 s, 900 s, 3600 s, and 7200 s at 4°C. At each time point, an aliquot of sample was transferred to a vial in a 0.5°C chamber for quenching by the addition of an equal volume of quench buffer (200 mM citric acid, 4 M guanidine-HCl, 500 mM TCEP in H2O, pH 2.3) for 0.5 min. All the steps, including pepsin digestion, desalting, and LC separation were then performed in the temperature-controlled compartment of the HDX PAL system at 4°C automatically. MS Data were acquired using a Thermo LTQ Orbitrap-Elite mass spectrometer (San Jose, CA). All the experiments were repeated three times at each timepoint. For peptide identification, a DDA mode was applied with one MS1 (60 k resolution at 400 m/z) at the m/z range of 300–1500 followed by ten CID MS2 scans with a ± 2.0 m/z isolation window. The deuterium uptake in HDX experiments was calculated by the intensity of the identified peptides. The MS data were interpretated and analyzed in PEAKS Studio X against the homemade database including all target proteins with a precursor mass tolerance at 20 ppm for FTMS mode and MS/MS fragment tolerance at 0.02 Da for IT mode. HDX uptake analysis was carried out using HDExaminer 2.0 (Sierra Analytics Inc., Modesto, CA). The uptake of deuterium was calculated using the software algorithm via matching the best theoretical isotope distribution pattern to the observed isotope distribution pattern. Deuterium uptake was also plotted according to the exchange time. Student’s t test at the 95% confidence level was used to confirm the consistency of the analytical results obtained based on the triplicate runs. Non-deuterated and fully deuterated samples were performed for the back-exchange correction.
Luciferase Assay.
Full-length Nurr1, RXRα, and their mutations were constructed in pCDNA 3.1 vector with a GFP-tag in the N terminal of the proteins, respectively, and their expressions were confirmed by western blot. 3 × IR5 was constructed in pGL3-promoter vector (Promega) which contains a SV40 promoter upstream of the luciferase gene. HEK293T cells were seeded in 96-well plate at 20,000 cells. Cells were then transfected using Lipofectamine 2,000 (Thermo Fisher Scientific) and Opti-MEM with protein plasmids, DNA plasmids, and Renilla as control, to a total of about 200 ng/well DNA. Bexarotene was added with different concentrations at 6 h before the cells were harvested for luciferase activity quantification. Dual-Glo luciferase assay system (Promega) was used to detect the luciferase signal.
BLI Kinetic Assay.
The binding affinities of IR5 with Nurr1258-RXRα98 heterodimer were evaluated by the Octet R8 (Sartorius) using the BLI approach. Purified Nurr1258-RXRα98 was diluted with Tris buffer (20 mM Tris pH8.0, 150 mM NaCl, 0.02% Tween-20) at different concentrations (i.e., 1,000 nM, 333 nM, 111 nM, 37 nM, 12 nM). Firstly, the streptavidin-coated biosensors were loaded with the biotinylated IR5 at 50 nM for 120 s. After washing with the Tris buffer for 120 s, the sensors were transferred to protein-containing solutions to allow probes and proteins to associate for180 s. Then the biosensors were transferred to Tris buffer to measure dissociation for 180 s. A sample without the protein (Tris buffer only) was used as a negative control. Results were analyzed by ForteBio Data Analysis software.
Molecular Dynamics Simulation.
All-atom MD simulations of models of the Nurr1 or Nurr1-RXRα heterodimer were conducted with AMBER 20 (University of California, http://ambermd.org) employing the ff14SB (60) force field for proteins, the TIP3P (61) water model with parameters for monovalent ions from Joung and Cheatham, and the Zinc AMBER force field (ZAFF) (62) for the four ZnCys4 sites in Nurr1. Missing Loops in the Nurr1 models were added using Rosetta. Then they were embedded in an octahedral TIP3P water box, and Na+ and Cl- ions were added to neutralize the system’s charge. SHAKE bond length constraints were applied to all bonds involving hydrogen. Nonbonded interactions were evaluated with a 10 Å cutoff, and electrostatic interactions were calculated with a particle-mesh Ewald summation method. The MD system was first minimized for 2,500 steps using steepest descent followed by 2,500 steps of conjugate gradient minimization, whereas other atoms were constrained with the harmonic force of 50 kcal mol−1 Å−2. Next, the harmonic force was set to 25, 5, and 0 kcal mol−1 Å−2 for the minimization. With protein restrained to their initial coordinates, the system was then heated to 310 K over 500 ps in the NVT ensemble using a step size of 1 fs and an Andersen thermostat with a heat bath coupling time constant of 1 ps. After switching to the NPT ensemble, positional restraints on protein were gradually removed over 6 ns, while Langevin dynamics with a collision frequency of 2.0 ps−1 was used for temperature regulation. Production MD was conducted for 1–2 μs using a step size of 2 fs, constant pressure periodic boundary conditions, isotropic pressure scaling, and Langevin dynamics. During MD simulations, the SHAKE method was applied to constrain the covalent bonds involving the hydrogen atoms. The Particle Mesh Ewald method was adopted to treat the long-range electrostatic interaction. The cutoff distances for the long-range electrostatic and van der Waals interactions were set to 8 Å. The analysis of MD trajectories with CPPTRAJ (version 18.0) (63) included the calculation of Cα atom root-mean-square deviations and root-mean-square fluctuations, measurement of DBD-LBD residue pair, and cross-link Cβ-Cβ distances, to assess the stability of the Nurr1-RXRα heterodimer docking model.
Dynamical network analysis was performed with the Network View plugin (34)in VMD. A node in the network was assigned to every amino acid in Nurr1-RXR model centered at Cα atom. Network edges were used in the analysis. Edges weights were derived from the pairwise residue correlation matrix calculated with the program Carma (64). Network communities were determined by applying the Girvan–Newman algorithm implemented in the program gncommunities as part of the Network View plugin.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Cong-Zhao Zhou for providing the modified expression vector and the staff from BL19U2 beamline at the National Facility for Protein Science in Shanghai (NFPS) and BL19U1 beamline at the Shanghai Synchrotron Radiation Facility, for assistance during the diffraction data collection. We are grateful for Prof. Xianyang Fang, Dr. Guangfeng Liu, and Linghui Xu for SAXS data analysis. We would also like to thank the support from the Guangzhou Branch of the Supercomputing Center of CAS. This work was supported by National Key Research and Development Program Grant (2022YFA1300015, 2017YFA0504104) (J. Liu), CAS Maintenance and Reconstruction Project DSS-WXGZ-2020-0001 (C.P.), the Youth Innovation Promotion Association of CAS 2021357 (T.X.), Guangdong Provincial Key Laboratory of Biocomputing (2016B030301007), and the National Basic Science Data Center “Database of Stem Cells and Metabolic Disease” (NO.NBSDC-DB-16). We thank the support from China HPC Environment Based Service Village for Biomedicine Application (http://biomed.cngrid.org) and some of the computational experiments were implemented in the “ORISE”HPC clusters.
Author contributions
C.P. and J. Liu designed research; M.Z., N.W., Y.G., J. Li, Y.Y., Y.D., J.Z., C.P., and T.X. performed research; M.Z., N.W., Y.G., T.X., and J. Liu analyzed data; and M.Z., N.W., T.X., and J. Liu wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Tingting Xu, Email: xu_tingting@gibh.ac.cn.
Jinsong Liu, Email: liu_jinsong@gibh.ac.cn.
Data, Materials, and Software Availability
X-ray crystallographic data and coordinates have been deposited in the Protein Data Bank (PDB) under accession code 7WNH, all other data that support the findings of this study are provided in the manuscript and the SI Appendix.
Supporting Information
References
- 1.Weikum E. R., Liu X., Ortlund E. A., The nuclear receptor superfamily: A structural perspective. Protein Sci. 27, 1876–1892 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zetterstrom R. H., Williams R., Perlmann T., Olson L., Cellular expression of the immediate early transcription factors Nurr1 and NGFI-B suggests a gene regulatory role in several brain regions including the nigrostriatal dopamine system. Brain Res. Mol. Brain Res. 41, 111–120 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Backman C., Perlmann T., Wallen A., Hoffer B. J., Morales M., A selective group of dopaminergic neurons express Nurr1 in the adult mouse brain. Brain Res. 851, 125–132 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Fan X., et al. , Nurr1 expression and its modulation in microglia. Neuroimmunomodulation 16, 162–170 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Li T., et al. , Alterations of NURR1 and cytokines in the peripheral blood mononuclear cells: Combined biomarkers for Parkinson’s disease. Front. Aging Neurosci. 10, 392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tippabathani J., Nellore J., Radhakrishnan V., Banik S., Kapoor S., Identification of NURR1 (Exon 4) and FOXA1 (Exon 3) haplotypes associated with mRNA expression levels in peripheral blood lymphocytes of Parkinson’s patients in small Indian population. Parkinsons Dis. 2017, 6025358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montarolo F., et al. , Effects of isoxazolo-pyridinone 7e, a potent activator of the Nurr1 signaling pathway, on experimental autoimmune encephalomyelitis in mice. PLoS One 9, e108791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojas P., Joodmardi E., Perlmann T., Ogren S. O., Rapid increase of Nurr1 mRNA expression in limbic and cortical brain structures related to coping with depression-like behavior in mice. J. Neurosci. Res. 88, 2284–2293 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Buervenich S., et al. , NURR1 mutations in cases of schizophrenia and manic-depressive disorder. Am. J. Med. Genet. 96, 808–813 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Rojas P., Joodmardi E., Hong Y., Perlmann T., Ogren S. O., Adult mice with reduced Nurr1 expression: An animal model for schizophrenia. Mol. Psychiatry 12, 756–766 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Wang Z., et al. , Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423, 555–560 (2003). [DOI] [PubMed] [Google Scholar]
- 12.de Vera I. M. S., et al. , Defining a canonical ligand-binding pocket in the orphan nuclear receptor nurr1. Structure 27, 66–77.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajan S., et al. , PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat. Chem. Biol. 16, 876–886 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruning J. M., et al. , Covalent modification and regulation of the nuclear receptor Nurr1 by a dopamine metabolite. Cell Chem. Biol. 26, 674–685.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C. H., et al. , Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 112, 8756–8761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson T. E., Paulsen R. E., Padgett K. A., Milbrandt J., Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science 256, 107–110 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Jiang L., et al. , Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements. J. Biol. Chem. 294, 19795–19803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlmann T., Jansson L., A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes. Dev. 9, 769–782 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Jolma A., et al. , DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Friling S., Bergsland M., Kjellander S., Activation of retinoid X receptor increases dopamine cell survival in models for Parkinson’s disease. BMC Neurosci. 10, 146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang Y. L., et al. , Docosahexaenoic acid promotes dopaminergic differentiation in induced pluripotent stem cells and inhibits teratoma formation in rats with Parkinson-like pathology. Cell Transplant. 21, 313–332 (2012). [DOI] [PubMed] [Google Scholar]
- 22.McFarland K., et al. , Low dose bexarotene treatment rescues dopamine neurons and restores behavioral function in models of Parkinson’s disease. ACS Chem. Neurosci. 4, 1430–1438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spathis A. D., et al. , Nurr1: RXRalpha heterodimer activation as monotherapy for Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, 3999–4004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinke G., Sigler P. B., DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat. Struct. Biol. 6, 471–477 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Merkley E. D., et al. , Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 23, 747–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z. L., et al. , A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 10, 3404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A windows PC-based system for small-angle scattering data analysis. J. App. Crystallogr. 36, 1277–1282 (2003). [Google Scholar]
- 28.Svergun D., Barberato C., Koch M. H. J., CRYSOL a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995). [Google Scholar]
- 29.Lu S., et al. , Mapping native disulfide bonds at a proteome scale. Nat. Methods 12, 329–331 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Yang B., et al. , Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Li F., Song C., Zhang Y., Wu D., Structural overview and perspectives of the nuclear receptors, a major family as the direct targets for small-molecule drugs. Acta. Biochim. Biophys. Sin. (Shanghai) 54, 1–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aarnisalo P., Kim C. H., Lee J. W., Perlmann T., Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J. Biol. Chem. 277, 35118–35123 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Chandra V., et al. , The quaternary architecture of RARbeta-RXRalpha heterodimer facilitates domain-domain signal transmission. Nat. Commun. 8, 868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eargle J., Luthey-Schulten Z., NetworkView: 3D display and analysis of protein. RNA interaction networks. Bioinformatics 28, 3000–3001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q., et al. , Structural basis of RXR-DNA interactions. J. Mol. Biol. 296, 509–520 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Banitt I., Wolfson H. J., ParaDock: A flexible non-specific DNA–rigid protein docking algorithm. Nucleic. Acids Res. 39, e135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandra V., et al. , Multidomain integration in the structure of the HNF-4alpha nuclear receptor complex. Nature 495, 394–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandra V., et al. , Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 456, 350–356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lou X., et al. , Structure of the retinoid X receptor alpha-liver X receptor beta (RXRalpha-LXRbeta) heterodimer on DNA. Nat. Struct. Mol. Biol. 21, 277–281 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Morita K., et al. , Selective allosteric ligand activation of the retinoid X receptor heterodimers of NGFI-B and Nurr1. Biochem. Pharmacol. 71, 98–107 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Meijsing S. H., et al. , DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egea P. F., et al. , Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. EMBO J. 19, 2592–2601 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang N., Zou Q., Xu J., Zhang J., Liu J., Ligand binding and heterodimerization with retinoid X receptor alpha (RXRalpha) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding. J. Biol. Chem. 293, 18180–18191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez E. J., Allosteric pathways in nuclear receptors - potential targets for drug design. Pharmacol. Ther. 183, 152–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter G., et al. , DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 74, 85–97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project N, The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 47.McCoy A. J., Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger T. C., et al. , Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delano W. L., The PyMol molecular graphics system. Proteins Struct. Funct. Bioinf. 30, 442–454 (2002). [Google Scholar]
- 52.van Zundert G. C., Bonvin A. M., DisVis: Quantifying and visualizing accessible interaction space of distance-restrained biomolecular complexes. Bioinformatics 31, 3222–3224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dominguez C., Boelens R., Bonvin A. M., HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003). [DOI] [PubMed] [Google Scholar]
- 54.de Vries S. J., van Dijk M., Bonvin A. M., The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Nielsen S. S., et al. , BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. App. Crystallogr. 42, 959–964 (2009). [Google Scholar]
- 56.Svergu D. I., Determination of the regnlarization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992). [Google Scholar]
- 57.Petoukhov M. V., et al. , New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng H., et al. , Discovery of a cooperative mode of inhibiting RIPK1 kinase. Cell Discov. 7, 41 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maier J. A., et al. , ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horn H. W., et al. , Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 120, 9665–9678 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Peters M. B., et al. , Structural survey of zinc containing proteins and the development of the zinc AMBER force field (ZAFF). J. Chem. Theory Comput. 6, 2935–2947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roe D. R., Cheatham T. E. III, PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Glykos N. M., Software news and updates. Carma: A molecular dynamics analysis program. J. Comput. Chem. 27, 1765–1768 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
X-ray crystallographic data and coordinates have been deposited in the Protein Data Bank (PDB) under accession code 7WNH, all other data that support the findings of this study are provided in the manuscript and the SI Appendix.