Significance
Kinase inhibitor treatment in NRAS/BRAF mutant melanoma can sensitize tumors to immunotherapy, in part through an increase in average surface presentation of peptide MHC molecules. Here, we demonstrate that MEK inhibition selectively boosts epitope abundance of select tumor-associated antigens in vitro and in vivo, enhancing targeted immunotherapy efficacy against these treatment-modulated epitopes.
Keywords: MHC, immunopeptidomics, antigen presentation, melanoma, ligandomics
Abstract
Combining multiple therapeutic strategies in NRAS/BRAF mutant melanoma—namely MEK/BRAF kinase inhibitors, immune checkpoint inhibitors (ICIs), and targeted immunotherapies—may offer an improved survival benefit by overcoming limitations associated with any individual therapy. Still, optimal combination, order, and timing of administration remains under investigation. Here, we measure how MEK inhibition (MEKi) alters anti-tumor immunity by utilizing quantitative immunopeptidomics to profile changes in the peptide major histocompatibility molecules (pMHC) repertoire. These data reveal a collection of tumor antigens whose presentation levels are selectively augmented following therapy, including several epitopes present at over 1,000 copies per cell. We leveraged the tunable abundance of MEKi-modulated antigens by targeting four epitopes with pMHC-specific T cell engagers and antibody drug conjugates, enhancing cell killing in tumor cells following MEK inhibition. These results highlight drug treatment as a means to enhance immunotherapy efficacy by targeting specific upregulated pMHCs and provide a methodological framework for identifying, quantifying, and therapeutically targeting additional epitopes of interest.
In recent years, cancer treatment paradigms have increasingly incorporated information regarding a patient’s genetic profile to identify appropriate therapeutic modalities, otherwise known as “precision medicine.” Targeted therapies against aberrant activation of the mitogen-activated protein kinase (MAPK) signaling pathway, including BRAF and MEK inhibitors (BRAFi and MEKi), have transformed the standard of care for BRAF and NRAS mutant melanoma patients—representing ~50% and ~20% of melanomas, respectively (1, 2). Unfortunately, despite these targeted therapies showing some initial efficacy in extending progression free survival (PFS), either alone (MEKi, NRAS) or in combination (BRAF), a majority of patients acquire resistance and experience disease progression within 1 y (3–9). Immune checkpoint inhibitors (ICIs), which target cell surface receptors controlling the activation or inhibition of an immune response, have shown remarkable clinical success in melanoma (10, 11). However, only a subset of patients respond, and those who do frequently experience immune related adverse events and many develop resistance (12, 13).
It has been proposed that combining MAPK inhibitors and ICIs may increase efficacy, in part due to increasing evidence that MEK/BRAF inhibitors can sensitize tumors to immunotherapy through upregulation of class I major histocompatibility molecules (MHCs), as well as increased immune cell infiltration, T cell activation, antigen recognition, and more (14–22). NRAS-mutant melanoma trials have suggested that MEKi/ICI treatment may enhance PFS (9, 23). Additionally, several clinical trials evaluating a triple combination of MEKi, BRAFi, and ICIs have shown enhanced efficacy in BRAF-mutant melanoma, though at the expense of increased toxicity (24, 25). Therefore, despite promising initial results, there remains much to learn about how exposure to kinase inhibitors alters the immune system, and how these alterations can be leveraged with ICIs and/or targeted immunotherapies (26). While it has been shown that MAPK pathway inhibitors can increase gene and protein expression of a handful of well characterized melanoma antigens such as MART-1 (MLANA) and gp-100 (PMEL) (27–31), measuring how the antigen repertoire, referred to as the “immunopeptidome,” presented by class I MHCs (MHC-I) changes in response to therapy is central to understanding the relationship between drug treatment and immune response, as recent reports highlight the potential for dynamic repertoire shifts in both the identity and abundance of peptide MHCs (pMHCs) following perturbation (32–34). To better understand how to optimally combine targeted and immunotherapies in mutant melanoma and identify relevant pMHC therapeutic targets, a precise, global, molecular understanding of how changes in melanoma cell state translate to relative and absolute quantitative changes in pMHC presentation following treatment is required.
To this end, we used quantitative immunopeptidomics to measure the relative changes in presentation of pMHC repertoires in response to MEKi in vitro and in vivo. This analysis showed increased expression of both putative and well-characterized tumor associated antigens (TAA) following MEKi treatment. To interrogate the mechanisms underlying altered pMHC repertoires, we performed a quantitative multi-omics analysis and integrated the results with quantitative immunopeptidomic data to identify associations between intracellular response to MEKi and extracellular immune presentation. This analysis suggested the selective modulation of melanoma differentiation antigens and other TAAs by MEKi though a shared mechanism, highlighting potential antigen targets for targeted immunotherapy whose expression can be tuned with MEK inhibitor treatment.
Copies per cell estimations of 18 MEKi-modulated TAAs enabled the selection of four TAAs with high MEKi-induced expression as targets for pMHC-specific antibody-based therapies, which show enhanced ability to mediate T cell cytotoxicity with higher antigen expression levels (35–38). The pMHC-Abs were used to generate antibody–drug conjugates and T cell engagers, which reveal a strong relationship between epitope density, therapeutic modality, and cytotoxicity, and highlight MEKi as a means to enhance efficacy by increasing target antigen expression. This work provides a quantitative immunopeptidomics-driven methodological framework to discover and exploit highly expressed drug-induced pMHC complexes for new immunotherapies.
Results
MEK Inhibition Increases MHC-I Expression in Melanoma Cell Lines.
To evaluate how MEK inhibition alters pMHC expression in NRAS and BRAF mutant melanomas, we selected two NRAS and four BRAF mutant cell lines (V600E) which exhibited a range of sensitivities to binimetinib (SI Appendix, Fig. S1A). We measured MHC-I surface expression with flow cytometry and found 72 h of treatment resulted in a maximal increase in expression over a dimethyl sulfoxide (DMSO)-treated control without requiring cell passaging (SI Appendix, Fig. S1B). Hence, we selected 72 h as the timepoint for all subsequent experiments. All cell lines showed elevated surface MHC-I expression following low-dose (100 nM) or high-dose (1 µM) binimetinib treatment at 72 h, with high-dose treatment generally resulting in a larger increase (Fig. 1A and SI Appendix, Fig. S1C). Primary melanocytes treated with binimetinib did not show a strong change in surface human leukocyte antigen (HLA) expression, similar to previously reported results in trametinib-treated peripheral blood mononuclear cells (PBMCs) (15), suggesting this effect is specific to oncogenic cell phenotypes with amplified MAPK signaling (SI Appendix, Fig. S1D).
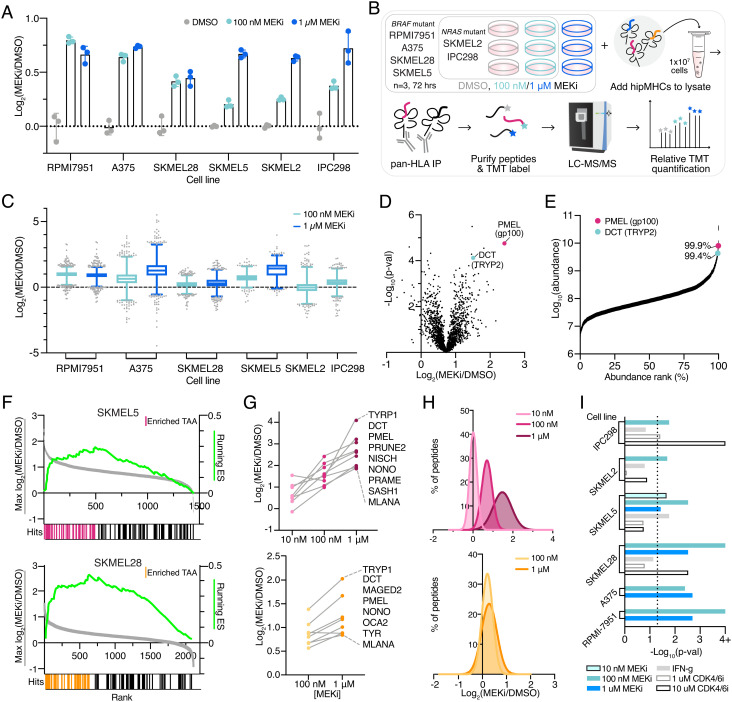
Fig. 1.
MEKi enriches TAA presentation on pMHCs. (A) Fold change in median surface expression levels (over average DMSO control condition) of HLA-A/B/C in cell lines treated with vehicle control or binimetinib (MEKi) for 72 h. Error bars represent SD of n = 3 biological replicates. (B) Experimental setup for quantitative immunopeptidomics experiments. (C) Relative changes in pMHC expression +/− MEKi. Data are represented as a box and whiskers plot, with whiskers displaying the 1–99 percentiles. (D) Volcano plot of the average fold change in pMHC expression for SKMEL5 cells treated with 100 nM binimetinib for 72 h (n = 3 biological replicates for DMSO and MEKi treated cells) versus significance (mean-adjusted P value, unpaired two-sided t test). (E) pMHCs ranked by precursor ion area abundance. (F) TAA Enrichment plots of TAA enrichment in SKMEL5 +/− 100 nM MEKi (Top, pink) and SKMEL28 +/− 100 nM MEKi (Bottom, orange), displaying running enrichment scores (green, Right y-axis), and fold change in pMHC presentation (Left y-axis) versus rank (x-axis) for each peptide (gray). Hits denote TAA peptides, and colored hits represent enriched TAAs. SKMEL5 P = 0.001, SKMEL28 P = 0.001. (G) Selected enriched TAA peptides in SKMEL5 (Top) and SKMEL28 (Bottom) analyses. (H) Frequency distribution of pMHC fold change with MEK inhibition. SKMEL5 (Top): 10 nM: μ = 0.01, 100 nM: μ = 0.70, 1 μM: μ = 1.47. SKMEL28 (Bottom): 100 nM: μ = 0.21, 1μM: μ = 0.28. (I) Significance values for TAA pathway enrichment. Dotted line indicates and P < 0.05 and values ≥4 (Log10 adjusted) represent P < 0.0001.
We next investigated how the pMHC repertoires presented on these six cell lines were altered quantitatively in response to MEKi treatment. We employed our previously described framework for multiplexed, quantitative profiling of pMHC repertoires utilizing isobaric labeling (Tandem Mass Tags, “TMTs”) and heavy isotope-labeled peptide MHCs (hipMHC) standards for accurate relative quantitation of endogenous pMHCs (34). In triplicate, cells were treated with DMSO or binimetinib (100 nM NRAS mutant cells, 100 nM/1 µM BRAF mutant cells) for 72 h (Fig. 1B). Cells were lysed, and three hipMHC standards were spiked into the lysate mixture prior to immunoprecipitation (IP). Isolated endogenous and isotopically labeled peptides were subsequently labeled with TMT, combined and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) for quantitative immunopeptidomic profiling (Dataset S1). Peptides matched expected class I length distributions (SI Appendix, Fig. S2A), and a majority were predicted to be binders of each cell line’s HLA allelic profile (SI Appendix, Fig. S2B and Table S1).
Quantitative immunopeptidomics showed a median increase in pMHC expression levels following binimetinib treatment in most conditions, with similar average changes observed across peptides predicted to bind to HLA-A, HLA-B, and HLA-C (SI Appendix, Fig. S2C). However, in contrast to surface staining, measuring the average change in HLA expression, MS analysis showcased a wide distribution in presentation levels across peptides (Fig. 1C and SI Appendix, Fig. S3). For example, while the average fold change in HLA levels in A375 cells treated with 1 µM binimetinib was 2.45-fold, some peptides increased 16-fold or more in presentation while others decreased fourfold. In SKMEL2 cells, several peptides changed threefold to fourfold in presentation despite no change in surface HLA expression. These data illustrate the highly dynamic nature of the immunopeptidome, where individual pMHCs experience significant changes in presentation often not captured by surface staining alone.
TAA Are Selectively Enriched in Presentation with MEK Inhibition.
We investigated which peptides increased significantly relative to the median change in presentation to determine if any pMHCs were selectively enriched following MEK inhibition. We observed that two peptides in the SKMEL5 (low-dose MEKi) analysis, derived from known TAAs (dopachrome tautomerase [DCT or “TYRP2”] and premelanosome protein [PMEL or “gp100”]), had high changes in presentation, increasing 2.8- and 5.3-fold, respectively (Fig. 1D). These peptides were also highly abundant, ranking in the 99th percentile of precursor ion abundance (Fig. 1E).
To evaluate whether enriched presentation of DCT and PMEL peptides was indicative of increased expression of TAA-derived peptides following MEKi treatment broadly, we performed a nonparametric test to measure TAA enrichment significance. For this analysis, we compiled a custom TAA library derived from the literature and online databases, (SI Appendix, Table S2) (39–42) utilizing the peptide’s source proteins to generate a protein-based TAA library to accommodate peptides derived from all alleles (SI Appendix, Table S3). Peptide source proteins were rank-ordered by fold change in presentation with MEKi; in cases where multiple peptides were derived from the same source protein, the maximal/minimal fold change was selected to assess positive/negative enrichment.
In both SKMEL5 and SKMEL28 cells, TAAs were significantly positively enriched following low-dose MEKi (Fig. 1F). Beyond DCT, and PMEL, enriched TAAs included melanoma differentiation antigens from the MAGE family, MLANA (MART-1), and TYR, representing well-characterized antigens with demonstrated immunogenic potential (SI Appendix, Fig. S4A) (43, 44). Many TAAs showed a dose dependent increase in presentation, occurring regardless of whether mean HLA expression increases proportionally (Fig. 1 G and H). Even sub-cytotoxic doses of MEKi (10 nM) resulted in an increase in TAA presentation despite no change in average MHC surface expression (Fig. 1G and SI Appendix, Fig. S4B). Notably, RPMI-7951 cells exhibited a robust increase in HLA presentation and TAA enrichment despite having a limited cytotoxic response to binimetinib, suggesting MEKi may be able to modulate changes in antigen presentation even when the drug shows limited efficacy against cell killing.
TAA upregulation was not exclusive to binimetinib, as trametinib-treated SKMEL5 cells showed similar TAA enrichment (SI Appendix, Fig. S4C). Peptides rank-ordered by precursor ion abundance also reached significance, suggesting TAAs are both some of the most abundant peptides presented (SI Appendix, Fig. S4D). Applying the enrichment analysis framework to all cell lines and treatments revealed MEKi treatment significantly enriched (P < 0.05) TAA presentation in all cases, suggesting a mechanistic basis for this response (Fig. 1I). Other perturbations such as IFN-γ stimulation and CDK4/6 inhibitor treatment have also been shown to increase antigen presentation; however, the TAA enrichment analysis applied to these previously published datasets revealed that only a minority of cell lines/treatment conditions showed significant TAA enrichment (Fig. 1I) (34). Taken together, these data suggest MEK inhibition causes a distinct peptide repertoire shift from IFN-γ stimulation, robustly driving TAA upregulation distinct from other perturbations.
CLXs Show Enhanced TAA Presentation Following MEK and BRAF Inhibition In Vivo.
We next evaluated whether TAA enrichment following MEK inhibition translated in vivo at early timepoints. Four melanoma cell lines were inoculated subcutaneously in immunocompromised mice, and mice were treated with vehicle control or binimetinib for 1, 2, 3, or 5 d in triplicate prior to tumor harvesting (Fig. 2A). For BRAF mutant lines, three additional mice were treated for 3 d with encorafinib (BRAF inhibitor, BRAFi) or encorafinib and binimetinib as a combination therapy. Class-I pMHCs from tumors were isolated and subsequently profiled by quantitative multiplexed immunopeptidomics (Dataset S2 and SI Appendix, Fig. S5 A and B).
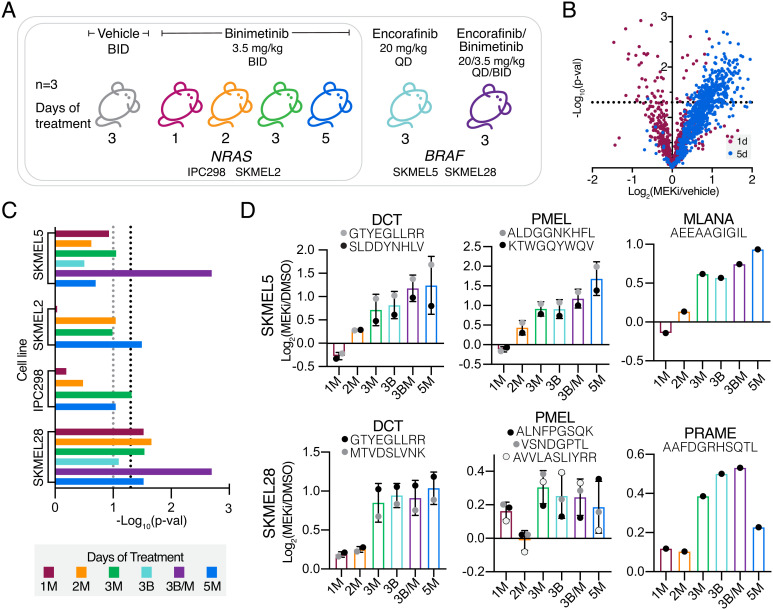
Fig. 2.
In vivo analysis of TAA pMHC enrichment in CLXs. (A) Experimental setup for CLX studies of mice + binimetinib or encorafinib. X-axis describes days of therapy and drug treatment (M = MEKi, binimetinib and B = BRAFi, encorafinib). (B) Volcano plot, average fold change in pMHC expression with binimetinib treatment (n = 3 biological replicates for DMSO and MEKi treated cells) versus significance (mean-adjusted P value, unpaired two-sided t test) for IPC298 CLX. (C) TAA enrichment significance values for each analysis. Black dotted line represents P ≥ 0.05, gray = P ≥ 0.01. (D) Average changes in pMHC expression for select melanoma differentiation antigens (n = 3 mice per condition). Errors bars represent SD when >1 peptide from each source protein was identified.
Among cell line xenografts (CLXs), treatment with binimetinib for just 1 or 2 d minimally altered mean HLA presentation levels, with maximal changes in presentation observed after 3 or 5 d of treatment (Fig. 2B and SI Appendix, Fig. S5C). In BRAF mutant CLXs, combination therapy showed higher (SKMEL5) or similar (SKMEL28) changes in pMHC presentation compared to MEKi monotherapy, suggesting combination therapy in BRAF tumors may further improve antigenicity of tumors in some cases.
We next performed TAA enrichment analysis and observed significant enrichment in at least one treatment condition across CLXs, with SKMEL28 CLXs showing robust enrichment across timepoints (Fig. 2C). Melanoma differentiation antigens showed positive increases in presentation following MEK and BRAF inhibition across all cell lines, often above median fold-changes (Fig. 2D and SI Appendix, Fig. S5D). For example, the PMEL peptide “ALDGGNKHFL” had a nearly fourfold increase in presentation after 5 d of MEKi in SKMEL5 CLXs, far exceeding the median pMHC fold-change value of 1.15.
Finally, we performed TAA enrichment analysis with peptides rank ordered by peak area abundance for SKMEL5 and SKMEL28 CLX samples and found that both showed significant enrichment (P < 0.0001), as seen in the in vitro analyses (SI Appendix, Fig. S5E). High abundance TAAs mapped to peptides that had some of the highest changes in expression following MEKi (SI Appendix, Fig. S5F), further confirming our observations that TAAs are some the most abundant and differentially expressed pMHCs following MEK inhibition.
EMT-TF Switching Drives MITF and Melanoma Differentiation Antigen Expression.
To assess whether MEKi-induced, enriched TAA pMHC presentation could be predicted using other datatypes, we performed a multi-omics analysis and compared changes in protein and transcript expression, as well as changes in ubiquitination as a proxy for protein degradation, following MEKi treatment to changes in pMHC presentation (SI Appendix, Fig. S6A and Datasets S3–S5. Overall, there was no significant correlation between changes in pMHC expression and transcript/protein/ubiquitination after MEK inhibition, suggesting these datatypes cannot necessarily be used exclusively for predicting pMHC repertoire alterations (Fig. 3A and SI Appendix, Fig. S6B). For example, vimentin has increased pMHC presentation but decreased transcript and protein expression, suggesting the elevated pMHC expression is likely due to other post-translational processing such as enhanced degradation as measured by ubiquitination (Fig. 3B).
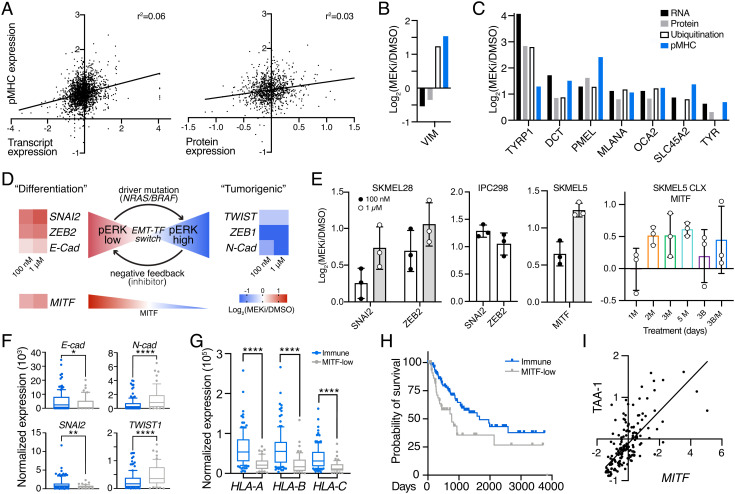
Fig. 3.
MEKi-induced EMT phenotype switching enhances TAA expression. (A) Correlation between pMHC expression and transcript expression (Left)/protein expression (Right) in SKMEL5 cells +/− 100 nM MEKi. Values represent Log2(MEKi/DMSO). (B), (C) Changes in RNA, protein, ubiquitination, and pMHC expression of selected targets. (D) Schematic of EMT-TF switching, with changes in transcription (SKMEL5 +/− 100 nM or 1 uM binimetinib) shown next to gene names. (E) Maximum average change in expression of pMHCs derived from EMT-related source proteins (n = 3). M = MEKi, B = BRAFi. Error bars = SD. (F–G) Normalized expression of select genes in NRAS/BRAF mutant immune (blue, n = 122) and MITF-low (gray, n = 54) classified tumors. *P < 0.05, **P < 0.01, ****P < 0.0001, unpaired 2-tailed t test. (H) Kaplan Meier curve of BRAF/NRAS immune and MITF-low melanomas. P = 0.012, log-rank test. (I) Correlation in expression of MITF and average TAA-1 gene set Z-scored expression for immune/MITF-low tumors, r = 0.723, P < 0.0001 (two-tail).
Despite the lack of general correlation, a clustering analysis of changes in pMHC, protein, and RNA expression following MEKi revealed a subset of source genes including DCT, PMEL, TYR, TRYP1, SLC45A2, and others, which showed increases across all datatypes (including ubiquitination when available) (Fig. 3C and SI Appendix, Fig. S6 C and D). This result demonstrates a clear connection between pMHC presentation and changes in transcription, translation, and degradation in the context of a subset of melanoma differentiation antigens.
To investigate the underlying biological mechanism responsible for select TAA enrichment, we performed gene set enrichment analysis against the cancer hallmarks pathway database and found significant negative enrichment of the epithelial to mesenchymal transition (EMT) pathway (SI Appendix, Fig. S6E) (45).
Transcript expression patterns in SKMEL5 cells provide evidence for a previously described EMT program in melanoma termed “EMT-transcription factor (TF) switching,” where SNAI2 and ZEB2 act as oncosuppressive proteins during melanocyte differentiation under MITF control (46, 47). In response to MAPK pathway activation, EMT-TFs ZEB1 and TWIST are upregulated to promote dedifferentiation and tumorigenesis, and the inhibition of this pathway (i.e., MEK or BRAF inhibitors, low phospho-ERK) can reverse the EMT-TF phenotype back to a “differentiation” state (high MITF). Here, cells showed increased differentiation and decreased “tumorigenic” marker transcript expression following MEK inhibition (Fig. 3D).
Previous studies have demonstrated that quantitative changes in pMHC repertoires reflect biological response to perturbation (34, 48). In line with this finding, we find pMHCs derived from differentiation-associated proteins like SNAI2, ZEB2, and MITF increasing in presentation following MEKi in in vitro and in vivo analyses, further connecting the intracellular response to treatment to extracellular immune presentation (Fig. 3E and SI Appendix, Fig. S7).
We next queried The Cancer Genome Atlas (TCGA) transcriptional data of 176 BRAF or NRAS mutant cutaneous melanoma patients to evaluate whether there was a relationship between melanoma differentiation antigen expression and EMT-TF phenotypes (49). Tumors were previously classified into subclasses by Akbani et al., including “immune” for tumors with high immune infiltration and “MITF-low” for low MITF and target gene expression, whose EMT-TF expression profiles matched the previously reported phenotypes (Fig. 3F) (46). MITF-low tumors had significantly lower HLA-A/B/C expression and a lower probability of survival, and MITF expression was significantly correlated with melanoma differentiation antigen expression (Fig. 3 G–I). These data suggest MITF-low BRAF/NRAS tumors may benefit from MEKi or other MAPK pathway inhibitors to induce a high-MITF, “differentiation” phenotype and suggest a common mechanism to augment TAA pMHC expression in melanoma.
Absolute Quantification of Treatment-Modulated TAA.
MEK inhibitor-modulated TAAs present an attractive class of epitopes for targeted immunotherapy, as these antigens have high abundance relative to other epitopes and their expression can be further augmented in response to therapy. We hypothesized MEKi may enhance the anti-tumor immune response for immunotherapies targeted against MEKi-modulated antigens, although determining the appropriate immunotherapeutic strategy for each antigen requires knowledge of epitope abundance (37, 38).
To this end, we performed absolute quantification experiments to estimate copies per cell abundance of 18 HLA-A*02:01 epitope targets that increase in presentation following MEKi in SKMEL5 cells. We utilized a previously developed assay, “SureQuant Iso-MHC” (50), where a series of three peptide isotopologues with one, two, or three stable isotopically labeled (SIL) amino acids (1-3H) per target were loaded into MHC molecules (hipMHCs) and titrated into cell lysates across a 100-fold linear range as an embedded standard curve (Fig. 4A). A fourth isotopologue with four SIL-amino acids was added exogenously to leverage internal-standard triggered parallel reaction monitoring data acquisition (“SureQuant”) for sensitive and selective targeting of endogenous peptides and embedded peptide standards. We estimated copies per cell for our 18 TAA panel in A375 and RPMI-7951 cells treated with DMSO, low, or high dose MEKi for 72 h, and extended our previous analysis of SKMEL5 cells by measuring the target panel in SKMEL5 cells with high dose MEKi treatment and compared the data to DMSO and low dose measurements (50). Of note, these estimations represent total copies per cell, as this assay cannot differentiate intracellular from surface pMHC molecules.
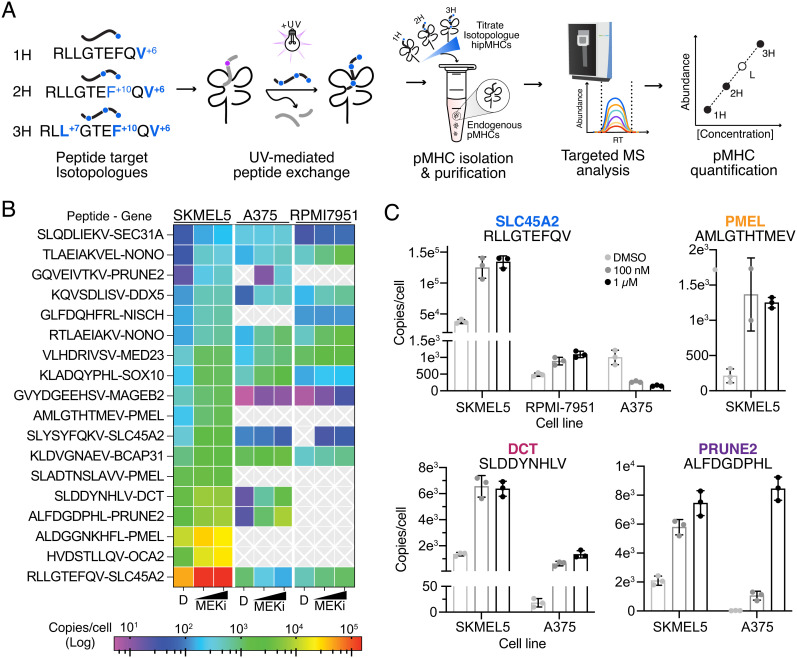
Fig. 4.
Absolute quantification of MEKi-inducible TAAs. (A) Schematic of SureQuant isoMHC workflow for multipoint absolute quantification of 18 TAAs. (B) Heatmap of copies/cell for each cell line treated with DMSO, 100 nM MEKi, or 1 μM MEK for n = 3 biological replicates. (C) Copies/cell for select epitopes across cell lines and treatment conditions.
All 18 peptides were quantifiable across high MEKi treated SKMEL5 cells, as expected since the panel was developed using pMHCs identified in SKMEL5 cells (Dataset S6). While we would not expect to detect the entire panel across A375 and RPMI-7951 cell lines (for example, A375 cells are PMEL−), 13 and 11 peptides were quantifiable within A375 and RPMI-7951 cells, respectively (Fig. 4B). Copies per cell estimations spanned five orders of magnitude across peptides, cell lines, and treatments, highlighting the wide range in epitope abundances presented by cells. Furthermore, these data showcase that even for epitopes derived from the same source protein and presented on the same HLA allele, pMHC presentation varies widely in both endogenous presentation levels, and the impact MEK inhibition has on changing presentation. For example, in SKMEL5 cells, three distinct A*02:01 PMEL-derived epitopes were measured at ~200, 1,300, and 12,000 copies per cell under basal conditions, and 1 uM MEKi treatment changed copies per cell estimations between twofold and sixfold (SI Appendix, Fig. S8). Quantitative immunopeptidomics measurements are uniquely suited to resolve epitope-specific dynamics, which bulk transcript/protein measurements cannot predict.
Generating pMHC-Specific Antibodies Against MEKi-Modulated TAAs.
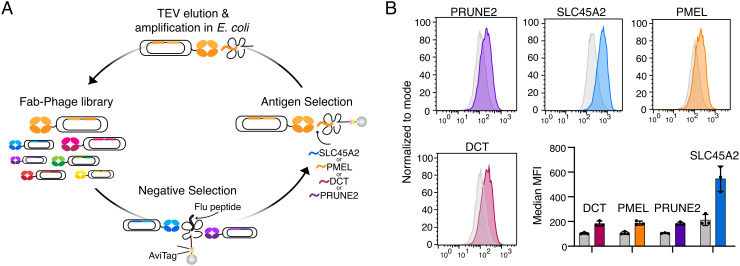
Antibody-based immunotherapies have shown the increasing promise of pMHC’s as therapeutic targets, in the context of both melanoma and cancer as a whole (35, 43, 51–53). MEKi induction of shared TAAs described here may present a therapeutic opportunity to use pMHC-targeted antibodies in combination with MEKi. We selected four HLA-A*02:01 associated TAAs with high epitope abundance in SKMEL5 cells as antigens for antibody generation. These peptides (derived from SLC45A2, PMEL, DCT, and PRUNE2) exhibited a range of basal and MEKi-induced presentation levels—three of which were also identified in at least one other cell line (Fig. 4C). To identify pMHC-specific antibodies, we performed a phage display campaign first clearing two Fab-phage libraries with an immobilized pMHC containing a decoy peptide (GILGFVFTL from influenza, “Flu peptide”). Remaining phage were incubated with pMHC’s of interest and bound phage were eluted via TEV protease and subsequently propagated to enrich for selective binders (Fig. 5A). After iterative rounds of selection, enzyme-linked immunoassay (ELISA) screening of individual clones identified 15 unique Fabs that showed strong specificity and high affinity (<20 nM) across our four pMHC targets (SI Appendix, Fig. S9). Flow cytometry using T2 lymphoblasts—an HLA-A*02:01+ cell line null for TAP which allows for exogenous peptide loading—revealed 1 Fab per pMHC that specifically recognized the pMHC on the surface of cells in a peptide-dependent manner (SI Appendix, Fig. S10). Upon conversion to IgG’s, these antibodies demonstrated selectivity in recognizing only peptide-specific target cells among cells pulsed with each of the four target peptides or the decoy flu peptide (SI Appendix, Fig. S11A), each with subnanomolar affinity (SI Appendix, Fig. S11B).
Fig. 5.
Generation of pMHC-specific antibodies. (A) Schematic of phage display selection. (B) Fluorescence intensity of SKMEL5 cells treated with DMSO (gray) or 1 μM MEKi (color) for 72 h stained with Alexa fluor 488-conjugated pMHC-specific antibodies. Data show a representative histogram and bar graph of median MFI. Error bars on bar graph display SD for n = 3 biological replicates per condition.
SKMEL5 cells treated with DMSO or high dose MEKi for 72 h displayed an increase in median fluorescence intensity in MEKi-treated cells compared to DMSO when stained with fluorophore-conjugated pMHC-specific IgG’s, in line with our immunopeptidomic analysis. (Fig. 5B). Due to the superior tumor-specific expression profiles in skin (SI Appendix, Fig. S12), as well poor biophysical properties of the antibody targeting the PRUNE2 pMHC, we selected SLC45A2, DCT, and PMEL-specific antibodies to evaluate for efficacy in vitro.
Therapeutic Modality, Antibody Properties, and Epitope Expression Influence Efficacy of pMHC-Specific Antibody-Based Therapies.
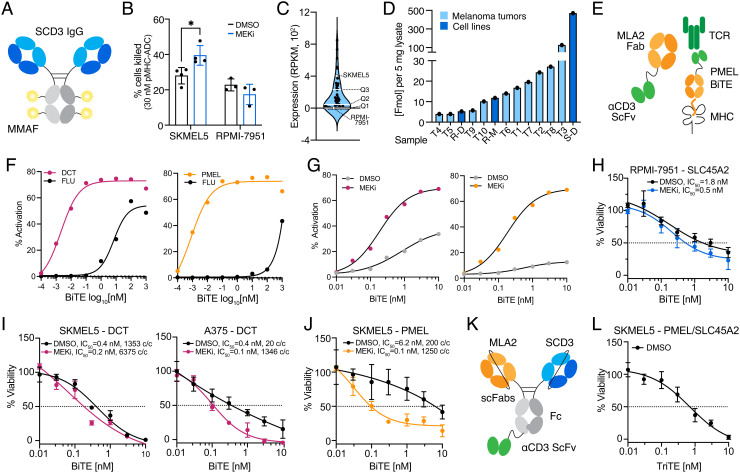
Previously reported data have demonstrated that ADCs targeting pMHCs require a high epitope density for efficacy, as only cells with expression levels generally above ~40,000 copies/cell showed an effect on viability greater than 20% (37). Here we hypothesized the high endogenous expression of the SLC45A2 “RLLGTEFQV” epitope in SKMEL5 cells may be effectively targeted by an ADC. To that end, we conjugated monomethyl auristatin F (MMAF), a tubulin polymerization inhibitor, to the anti-SLC45A2 pMHC IgG (Fig. 6A) and evaluated viability after 72 h in SKMEL5 and RPMI-7951 (low epitope density) cells pretreated with DMSO or 1 µM MEKi for 72 h to augment pMHC presentation of the target epitope. In SKMEL5 cells, MEKi pretreatment resulted in a superior therapeutic window following 72 h of ADC treatment, with a 40% reduction in viability achieved with MEKi compared to 28% with DMSO at 30 nM ADC (Fig. 6B and SI Appendix, Fig. S13A). In contrast, RPMI-7951 cells showed just an 18% reduction in viability in both conditions (<1,500 copies per cell), confirming that high epitope density is required for anti-pMHC ADC efficacy. Of note, DMSO or MEKi pretreated cells without ADC treatment showed comparable viability after 72 h, suggesting the effects in viability observed are specific to the ADC, not a loss of viability from MEKi pretreatment (SI Appendix, Fig. S13B).
Fig. 6.
MEKi enhances cytotoxicity of pMHC-specific antibody-based therapies. (A) Schematic of ADC targeting the SLC45A2 epitope. (B) Percent of cells killed with SLC45A2-ADC relative to the DMSO control. Error bars represent +/− SD for n = 4 (SKMEL5) and n = 3 (RPMI-7951) biological replicates. *Adjusted P value = 0.017, Sidak’s multiple comparisons test. (C) SLC45A2 transcript expression across n-57 BRAF/NRAS mutant melanoma cell lines. (D) RLLGTEFQV pMHC concentration across cell lines and n = 10 HLA-A*02:01+ melanoma tumors (50). S = SKMEL5, R = RPMI-7951, D = DMSO, M = 1 μM MEKi. (E) Schematic of PMEL-targeted BiTE. (F–G) Percent of GFP+ Jurkat cells following incubation with peptide-pulsed T2 cells (F) or SKMEL5 cells (G) and PMEL-BiTE. Lines represent a 4PL nonlinear fit, and error bars show +/− SEM for n = 3 biological replicates. (H–J), (K) Schematic of PMEL/SLC45A2-targeted TriTE. (L) Cell viability (percentage of untreated control) of target cells (pretreated for 72 h with DMSO) incubated with normal human T cells (effector:target 2:1) and a pMHC-specific BiTE/TriTE for 48 h.
Comparing SLC45A2 transcript expression across 57 BRAF/NRAS melanoma cell lines revealed that SKMEL5’s expression is in the upper quartile of abundances, and RLLGTEFQV epitope concentration in SKMEL5 cells relative to previously profiled melanoma tumors (50) suggests that the high SKMEL5 epitope abundance is unlikely to translate to a majority of patients (Fig. 6 C and D). Therefore, while a subset of patients may benefit modestly from an ADC approach, an alternative strategy with a lower threshold for presentation may be more efficacious. PMEL and DCT epitopes showed lower surface presentation levels compared to SLC45A2 in SKMEL5 cells, and thus we hypothesized bispecific T cell engagers (BiTEs) may be more potent against these epitopes, particularly in combination with MEKi (38, 54).
To this end, we generated BiTEs by fusing the PMEL, DCT, and SLC45A2 Fabs to the anti-CD3 single-chain variable fragment OKT3 (scFv, Fig. 6E). BiTE constructs showed selective T cell activation in a NFAT-GFP Jurkat reporter cell line when incubated with T2 lymphoblasts loaded with target peptide in comparison to the decoy Flu peptide (Fig. 6F and SI Appendix, Fig. S13C). We next tested Jurkat activation against SKMEL5 cells and saw that cells pretreated with 1 µM MEKi for 72 h showed superior activation across all three BiTES, suggesting that higher target expression leads to a higher proportion of activated effector cells (Fig. 6G and SI Appendix, Fig. S13D).
To assess cytotoxicity, we cocultured SKMEL5, RPMI-7951, or A375 cells pretreated with DMSO or high-dose MEKi for 72 h with primary human T cells isolated from healthy donor blood (effector to target ratio 2:1) in the presence of increasing concentrations of BiTE for 48 h. While RPMI-7951 cells were not responsive to the SLC45A2-ADC, the SLC45A2-BiTE did yield a cytotoxic response, with MEKi pretreated cells showing increased cell death with an IC50 of 0.5 nM with MEKi compared to 1.8 nM with DMSO (Fig. 6H and SI Appendix, Fig. S13E). By comparison, SKMEL5 cells showed a similar response regardless of MEKi treatment, likely due to the already high presentation levels at baseline (SI Appendix, Fig. S13F).
SKMEL5 cells showed a similar cytotoxic response to the DCT-BiTE regardless of treatment condition, possibly because both DMSO and MEKi-treated cells presented the target epitope at levels above 1,000 copies/cell (Fig. 6I). For A375 cells, the DCT-BiTE showed a superior reduction in cell viability with MEKi-pretreatment, where expression levels increased from 20 to 1,346 copies/cell. The PMEL-BiTE exhibited a similar trend in SKMEL5 cells, where a concentration of just 0.1 nM PMEL-BiTE was required to reduce SKMEL5 viability by 50% in MEKi pretreated cells, in contrast to 6.2 nM required in DMSO-treated cells (Fig. 6J). Similar to the ADC treatment, viability after 72 h of 0 nM BiTE treatment was comparable across cell lines regardless of MEKi pretreatment (SI Appendix, Fig. S13G). These data suggest that epitopes presented above ~1,000 copies/cell are most effectively targeted by BiTEs and that MEKi treatment can be used to augment presentation levels for increased efficacy when endogenous expression of the target epitope is low.
Utilizing a combination of BiTES that target patient epitopes may enhance cytotoxicity, particularly in cases where MEKi treatment may not be a viable strategy to augment presentation of target antigens (e.g., therapeutic resistance, non-BRAF/NRAS mutant melanoma) or in tumors with low endogenous levels of target antigens. Furthermore, two pMHC-specific antibodies can be combined to generate tri-specific T cell engager molecules (TriTEs), which may increase cytotoxic response and/or lower the concentration of therapy required for efficacy. Accordingly, we generated a TriTE against SLC45A2 and PMEL (Fig. 6K) and observed enhanced Jurkat activation when T2 lymphoblasts were pulsed with both peptides at TriTE concentrations below 1 nM compared to either peptide alone. In contrast, minimal Jurkat activation was observed below TriTE concentrations at 1 nM or below with the decoy FLU peptide (SI Appendix, Fig. S13H). In SKMEL5 cells (pretreated only with DMSO) co-cultured with human T cells and the SLC45A2/PMEL TriTE, we observed a greater cytotoxic response as compared to DMSO-treated cells incubated with the PMEL or SLC45A2 BiTEs alone (Fig. 6H and SI Appendix, Fig. S13E), reducing the IC50 to 0.7 nM. This result highlights pMHC-specific antibody combinations as an approach for further exploration for improved cytotoxic response for co-displayed antigen targets (Fig. 6L). Overall, we demonstrate T cell engagers against TAA pMHC’s can induce cytotoxicity in melanoma and in several cases, MEKi-treated melanoma lines can enhance this cytotoxic effect, thus potentially providing a therapeutic strategy using a combination of MEKi and targeted immunotherapies. Future work may focus on further characterizing the pMHC-Abs highlighted here and evaluating the potential for enhanced cytotoxicity in combination with MEKi treatment in an in vivo system across clinically relevant treatment timelines and dosing concentrations.
Discussion
The emergence of drug resistance and/or toxicities to small molecule-targeted therapies and checkpoint immunotherapies remains a significant barrier to achieving complete remission in BRAF or NRAS mutant melanoma. To better understand how to optimally combine MEKi with immunotherapy, here we performed a comprehensive analysis characterizing pMHC repertoire response to MEK inhibition using relative and absolute quantitative immunopeptidomics. We identify significantly enriched TAA presentation as a common mechanism to MAPK pathway inhibition in vitro and in vivo in NRAS-mutant and BRAF-mutant melanomas. While elevated surface HLA presentation in response to MEKi has been previously reported, our data reveal that many of the enriched TAAs increased well beyond average changes in HLA surface expression, in some cases more than 10-fold. Enhanced TAA transcript expression following MEKi/BRAFi treatment in human melanoma tumors (similar to our in vitro findings) has been previously observed, suggesting repertoire-level alterations reported here are likely to translate in the clinic (29) Still, future work may focus on measuring changes in surface presentation on human tumors following MEKi, BRAFi, or combination therapy to better understand how our findings translate to changes in vivo at physiologically relevant concentrations of drug.
Elevated TAA presentation was observed across varying levels of sensitivities to MEKi, including at sub-cytotoxic doses and was common to NRAS-mutant and BRAF-mutant lines, suggesting that this response may be shared across many melanoma patients. A multi-omics analysis highlighted changes in cellular plasticity following MEKi as a likely mechanism for the upregulation of certain TAAs. This finding was supported by patient data from the TCGA and suggests that MAPK pathway inhibition may selectively enhance presentation of shared tumor antigens through enhanced MITF expression, particularly melanoma differentiation antigens, making them attractive therapeutic targets. Further exploration of this proposed mechanism though direct manipulation of MITF may provide additional evidence to elucidate the relationship between MITF and tumor antigen presentation and highlight opportunities for modulation outside the MAPK pathway.
One of the primary criticisms of utilizing shared tumor associated/tissue differentiation antigens (as opposed to tumor-specific antigens, i.e., “neoantigens”) as a therapeutic target for TCR-based therapies is that their low expression in non-tumor tissue can lead to off-target toxicity, likely attributed to the high sensitivity of T cells (55–57). We hypothesized these antigens with high basal or MEKi-induced expression could be intelligently leveraged using antibody-based therapies, which require higher thresholds of antigen presentation for efficacy than TCR-focused approaches, limiting off target toxicity in low-expressing, non-target tissue.
Here, four pMHC-specific antibodies were generated and incorporated into ADC and BiTE formats. Using these reagents, we demonstrated enhanced cell killing following MEKi treatment with either therapeutic modality. Cytotoxicity is observed using just the endogenous or MEKi-augmented antigen presentation levels, in contrast to engineered or overexpression cellular system, which may be less likely to represent physiologically relevant epitope densities (36). Importantly, this work connects targeted immunotherapy response to epitope abundance measurements made using embedded hipMHC multipoint calibrants for accurate quantitative estimations. This is distinct from studies employing exogenous peptide standards for absolute quantification, which underestimate copies per cell estimations due to significant losses occurring during sample processing, leading to inaccurate conclusions regarding the sensitivity profile of a given pMHC-targeted modality (35, 58).
Here, we confirm that high (>4e4 copies per cell) surface expression is required for ADC efficacy, though this high-level expression is rare and therefore not an optimal strategy for a majority of pMHC epitopes. In contrast, BiTES were effective at lower epitope densities, where the greatest difference in cytotoxic response was observed when cells had fewer than ~1,000 copies per cell. BiTES showed similar efficacy against targets present at 1,000 copies per cell or higher, though future studies exploring more pMHCs may further elucidate the relationship between antibody affinity and epitope density. While MEKi treatment did not augment HLA presentation levels in primary melanocytes, future studies may apply SureQuant-IsoMHC to estimate TAA epitope abundances in non-malignant cell lines or primary tissues to better understand the potential for off-target toxicity (59).
In this study, we primarily tested the cytotoxicity of a single pMHC-specific BiTE on tumor cells, yet BiTEs could be used in combination to enhance efficacy, or engineered as TriTEs against different epitopes for a single TAA or two different TAAs, offering a multitude of “off-the-shelf” targeted immunotherapy opportunities to target highly abundant, shared TAAs. Peptide MHC-specific antibodies and MEKi-induced expression could also be utilized for other antibody-based therapeutic strategies such as to initiate antibody-mediated cellular cytotoxicity (51), fabs conjugated to immunotoxins (43), or engineered as pMHC-specific chimeric antigen receptor T cells (60), where higher expression may also enhance efficacy and/or improve the therapeutic window. Furthermore, although the focus of the therapeutic modalities generated in this study was limited to HLA-A*2:01, the same strategy could be employed for other high-frequency alleles using MEKi-modulated TAAs identified within this study.
Though resistance to MEKi is inevitable for many melanoma patients, utilizing MEKi to boost TAA antigen presentation prior to or concurrently with ICI and antigen-specific immunotherapies like those described within this study or others (ex. vaccines, cell therapy) may improve therapeutic response. Beyond melanoma, a variety of different therapeutic modalities across cancer types have also been demonstrated to enhance HLA presentation (61–63). Employing quantitative immunopeptidomics in these settings may unlock additional treatment-modulated tumor antigens and provide critical insights as to how to appropriately leverage them for optimal therapeutic potential.
Methods
Human Cell Lines.
SKMEL5, SKMEL28, A375, RPMI-7951, and T2 cell lines were obtained from ATCC [ATCC HTB-70, ATCC HTB-72, CRL1619, HTB-66, and CRL-1992, respectively] and maintained in Dulbecco's Modified Eagle Medium (DMEM) (Corning). IPC298 and SKMEL2 cells were provided by Array Biopharma and maintained in Roswell Park Memorial Institute (RPMI) 1640 (Gibco) and Minimum Essential Medium Alpha (MEM-α, Gibco) mediums, respectively. Primary epidermal melanocytes (normal, human, adult) were obtained from ATCC (PCS-200-013) and maintained in dermal cell basal medium (ATCC PCS-200-030) supplemented with adult melanocyte growth kit (ATCC PCD-200-042). NFAT-GFP Jurkat cells were a generous gift from Arthur Weiss (Department of Medicine, UCSF) and were maintained in RPMI1640 + 2 mg/mL Geneticin (Gibco). All medium was supplemented with 10% Fetal Bovine Serum (FBS, Gibco) and 1% penicillin/streptomycin (p/s, Gibco) except for primary melanocytes (p/s only). Cells were routinely tested for mycoplasma contamination and maintained in 37°C, 5% CO2. All experiments were performed on passages 4–10.
CLXs.
SKMEL5, SKMEL28, SKMEL2, and IPC298 cell lines were used for CLX analyses in collaboration with Array Biopharma. 5 × 106 cells in 100 µL phosphate buffered saline (PBS) containing 50% Matrigel® were implanted via subcutaneous injection into NCr nu/nu mice on the right flank. Resultant tumors were randomized into study groups at a starting size of ~200–400 mg, dosed at 10 mL/kg for up to 5 d by oral gavage with vehicle, 3.5 mg/kg binimetinib (MEK162) or 20 mg/kg encorafenib (LGX818) prepared as suspensions in 1% critical micelle concentration (CMC)/0.5% Tween 80. Dosing continued for up to 5 d, and at the end of each time course tumors were harvested and flash frozen in liquid nitrogen. Animals were housed in groups of 3. Food, water, temperature, and humidity are according to Pharmacology Testing Facility performance standards (Standard Operating Protocols) which are in accordance with the 2011 Guide for the Care and Use of Laboratory Animals (National Research Council) and Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. Dosing schedules are listed in SI Appendix, Table S4.
Cell Viability Assays
Binimetinib dose response.
Half-maximal inhibitory concentrations (IC50) of binimetinib (Selleckchem, MEK162) were determined for each cell line using CellTiter-Glo (CTG) luminescent cell viability assay (Promega). Cells were seeded at density of 10,000 (SKMEL2, SKMEL28, IPC298) or 5,000 (SKMEL5, A375, RPMI-7951) cells/well in a 96-well plate and allowed to adhere overnight. Cells were then treated with binimetinib or DMSO as a vehicle control in fresh medium for 72h and assayed. All viability data was acquired using a Tecan plate reader Infinite 200 with Tecan icontrol version 1.7.1.12. IC50 values were calculated using a four-parameter logistic curve in Prism 9.0.0.
Antibody–drug conjugate cell killing assays.
SKMEL5 or A375 cells were pretreated for 72 h with DMSO or 1 µM binimetinib in 10-cm plates and subsequently seeded at a density of 5,000 cells/well in a 96-well plate. Cells were incubated antibody–drug conjugate with n = 4 technical replicates per treatment condition for an additional 72 h and similarly assayed with CTG.
T cell/target cell co-incubation cell killing assays.
Deidentified buffy coats from healthy human donors were obtained from Massachusetts General Hospital. PBMCs were isolated by density-based centrifugation using Ficoll (GE Healthcare). CD8+ T cells were isolated from PBMCs using a CD8+ T cell-negative selection kit (Stemcell). T cells were mixed with Human T-activator CD3/CD28 DynaBeads (Thermo Fisher Scientific) in a 1:1 ratio and maintained in R10 + IL-2 [RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% heat-inactivated FBS (Thermo Fisher Scientific), 1% HEPES (Corning), 1% L-glutamine (Thermo Fisher Scientific), 1% Pen/Strep (Corning) and 50 IU/mL of IL-2 (R&D Systems)] for 7 d prior to use in cell killing assays. DynaBeads were removed by magnetic separation prior to co-incubation of primary T cells with target cells. Target cells were treated with DMSO or 1 µM MEKi for 72 h and were subsequently seeded in a 96-well plate with primary T cells in R10 + IL-2 at an effector to target ratio of 2:1 and incubated with BiTEs for 48 h with n = 3 technical replicates per condition. Cells were assayed with CTG, and percent cytotoxicity was calculated by subtracting the average luminescence signal of the T cell only condition and normalizing to the no BiTE condition. ((X − [T cell only])/([average-no-BiTE] – [T cell only])) × 100.
Peptide MHC Isolation.
Cultured cells were seeded in 10-cm plates, allowed to adhere overnight, and treated for 72h with binimetinib or DMSO vehicle control. At the time of harvest, cells were washed with 1X PBS, and lifted using 0.05% Trypsin-Ethylenedamine tetraacetic acid (EDTA, Gibco). Cells were pelleted at 500 g for 5 min, washed twice more in 1X PBS, and pelleted again. Cells were resuspended in 1 mL lysis buffer [20 nM Tris-HCl pH 8.0, 150 mM NaCl, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1% CHAPS, and 1X Halt Protease/Phosphatase Inhibitor Cocktail (Thermo Scientific)], followed by brief sonication (3 × 10 s microtip sonicator pulses) to disrupt cell membranes. Lysate was cleared by centrifugation at 5,000 g for 5 min and quantified using bicinchoninic acid protein assay kit (Pierce). For in vitro analyses, 1 × 107 cells were used for each condition. Frozen CLX tumor samples were homogenized in lysis buffer, cleared by centrifugation, and quantified using a bicinchoninic acid (BCA) protein assay as described in the in vitro analyses. For each sample, 7 mg of lysate was used. For absolute quantification analyses, ~5 mg of lysate was used.
Peptide MHCs were isolated by IP and size exclusion filtration, as previously described (34). Briefly, 0.5 mg of pan-specific anti-human MHC Class I (HLA-A, HLA-B, HLA-C) antibody (clone W6/32, Bio X Cell [cat # BE0079]) was bound to 20 μL FastFlow Protein A Sepharose bead slurry (GE Healthcare) for 3 h rotating at 4°C. Beads were washed 2× with IP buffer (20 nM Tris-HCl pH 8.0, 150 mM NaCl) prior to lysate and hipMHC addition (in vitro analyses), and incubated rotating overnight at 4°C to isolate pMHCs. For TMT-labeled data-dependent acquisition (DDA) analyses, 30 fmol of the following hipMHC standards were added prior to IP for quantification correction: ALNEQIARL7, SLPEEIGHL7, and SVVESVKFL7. For absolute quantification analyses, 1, 10, or 100 fmol of 1-3H Iso18 hipMHCs standards were added to each IP. Beads were washed with 1X Tris buffered saline and water, and pMHCs were eluted in 10% formic acid for 20 min at room temperature (RT). Peptides were isolated from antibody and MHC molecules using a passivated 10K molecule weight cutoff filter (PALL Life Science), lyophilized, and stored at −80°C. Label-free MS analysis acquisition parameters and data analysis techniques are described in SI Appendix, Methods.
pMHC Labeling with TMTs and SP3 Cleanup.
For labeled analyses, 100 μg of prealiquoted TMT 6-plex, 10-plex, or TMT-pro was resuspended in 30 μL anhydrous acetonitrile, and lyophilized peptides were resuspended in 100 μL 150 mM triethylammonium bicarbonate, 50% ethanol. Both were gently vortexed, centrifuged at 13,400 g for 1 min, and combined. TMT/peptide mixtures were incubated on a shaker for 1 h at RT, followed by 15 min of vacuum centrifugation. After combining labeled samples, we washed tubes 2× with 25% acetonitrile (MeCN) in 0.1% acetic acid (AcOH) and added it to the labeled mixture, which was subsequently centrifuged to dryness.
Sample cleanup was performed using single-pot solid-phase-enhanced sample preparation (SP3) as previously described (64). Briefly, a 1:1 mix of hydrophobic/hydrophilic Sera-mag carboxylate-modified speed beads (GE Healthcare) was prepared at a final bead concentration of 10 μg μL−1. Labeled samples were resuspended in 30 μL of 100 mM ammonium bicarbonate (pH 7–8) and added to 500 μg of bead mix with 1 mL MeCN. Peptides were allowed to bind for 10 min at RT, washed 2× with MeCN, and eluted with 2% DMSO for 1 min of sonication in a bath sonicator. TMT-labeled peptides were transferred to a fresh microcentrifuge tube and centrifuged to dryness. Peptides were resuspended in 0.1% formic acid, 5% MeCN and analyzed by MS. MS acquisition parameters and data analysis techniques are described in SI Appendix, Methods.
Methods describing peptide synthesis, UV-mediated peptide exchange for hipMHCs, pMHC binding affinity, RNA sequencing, flow cytometry assays, cloning, protein expression/purification, fab-phage selection, phage ELISA, biolayer interferometry, IgG NHS-fluorophore conjugation, ADC conjugation, TCGA/gTEX/Cell line expression analyses, enrichment analyses, and sample preparation for mass spectrometry global protein expression profiling and ubiquitination analyses are described in SI Appendix, Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Acknowledgments
We thank Ryan Sullivan and Genevieve Boland for project guidance and feedback, Alex Jaegar and Connor Dobson for guidance on T-cell cytotoxicity assays, Iris Abrahantes Morales for assistance with T cell isolation, Susanna Elledge for assistance with bioconjugation of IgG’s, and Aaron Gajadhar, Bhavin Patel, and Sebastien Gallien from Thermo Fisher for project support on SureQuant-MHC. We also thank the MIT BioMicro Center (Stuart Levine) and the Swanson Biotechnology Center for technical support, specifically the flow cytometry (Glenn Paradis), biopolymers and proteomics (Richard Cook), and the Barbara K. Ostrom (1978) Bioinformatics (Charlie Whittaker) core facilities. This research was supported in part by NIH R35GM122451 and NCI R01CA248323 (J.A.W.), NIH U54 CA210180 and U01CA238720 (F.M.W.), as well as funding from the Melanoma Research Alliance (MRA Team Science Award 565436) and the MIT Center for Precision Cancer Medicine. J.A.W. was supported by generous funding from the Chan Zuckerberg Biohub Investigator Program, the Harry and Dianna Hind Professorship; N.J.R. was supported by the NSF Graduate Research Fellowship; L.E.S. was supported by an NIH training grant in Environmental Toxicology (T32-ES007020). Results shown here are in whole or part based upon data generated by the TCGA Research Network and The Genotype-Tissue Expression (GTEx) Project.
Author contributions
L.E.S., N.J.R., O.L., J.M.M., E.B., S.W., B.B., J.A.W., and F.M.W. designed research; L.E.S., N.J.R., O.L., J.M.M., and E.B. performed research; L.E.S. and N.J.R. contributed new reagents/analytic tools; L.E.S. and N.J.R. analyzed data; L.E.S., N.J.R., J.A.W., and F.M.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-sequencing data have been deposited into the NCBI Gene Expression Omnibus GSE218070. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029860 for DDA datasets and PXD029884 for targeted datasets. Previously published data were used for this work (PMID 34497125). All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Davies H., et al. , Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Muñoz-Couselo E., Adelantado E. Z., Ortiz C., García J. S., Perez-Garcia J., NRAS-mutant melanoma: Current challenges and future prospect. Onco. Targets. Ther. 10, 3941–3947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty K. T., et al. , Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Sullivan R. J., Lorusso P. M., Flaherty K. T., The intersection of immune-directed and molecularly targeted therapy in advanced melanoma: Where we have been, are, and will be. Clin. Cancer Res. 19, 5283–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim S. Y., Menzies A. M., Rizos H., Mechanisms and strategies to overcome resistance to molecularly targeted therapy for melanoma. Cancer 123, 2118–2129 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Long G. V., et al. , COMBI-d: A randomized, double-blinded, Phase III study comparing the combination of dabrafenib and trametinib to dabrafenib and trametinib placebo as first-line therapy in patients (pts) with unresectable or metastatic BRAF V600E/K mutation-positive cutaneous melanoma. J. Clin. Oncol. 32, 9011–9011 (2014). [Google Scholar]
- 7.Robert C., et al. , Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med. 381, 626–636 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Falchook G. S., et al. , Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: A phase 1 dose-escalation trial. Lancet Oncol. 13, 782–789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. Dummer, Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): A multicentre, open-label, randomised, phase 3 trial. www.thelancet.com/oncology (2017). [DOI] [PubMed]
- 10.Larkin J., et al. , Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodi F. S., et al. , Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol. 19, 1480–1492 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Martins F., et al. , Adverse effects of immune-checkpoint inhibitors: Epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 16, 563–580 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Weiss S. A., Wolchok J. D., Sznol M., Immunotherapy of melanoma: Facts and hopes. Clin. Cancer Res. 25, 5191–5201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapkota B., Hill C. E., Pollack B. P., Vemurafenib enhances MHC induction in BRAF(V600E) homozygous melanoma cells. Oncoimmunology 2, e22890 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brea E. J., et al. , Kinase regulation of human MHC class i molecule expression on cancer cells. Cancer Immunol. Res. 4, 936–947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley S. D., et al. , BRAFV600E co-opts a conserved MHC class I internalization pathway to diminish antigen presentation and CD8+ T-cell recognition of melanoma. Cancer Immunol. Res. 3, 602–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boni A., et al. , Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 70, 5213–5219 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Hu-Lieskovan S., et al. , Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAFV600E melanoma. Sci. Transl. Med. 7, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homet Moreno B., Mok S., Comin-Anduix B., Hu-Lieskovan S., Ribas A., Combined treatment with dabrafenib and trametinib with immune-stimulating antibodies for BRAF mutant melanoma. Oncoimmunology 5, e1052212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lelliott E. J., McArthur G. A., Oliaro J., Sheppard K. E., Immunomodulatory Effects of BRAF, MEK, and CDK4/6 Inhibitors: Implications for combining targeted therapy and immune checkpoint blockade for the treatment of melanoma. Front. Immunol., 1478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuske M., et al. , Immunomodulatory effects of BRAF and MEK inhibitors: Implications for melanoma therapy. Pharmacol. Res. 136, 151–159 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Liu L., et al. , The BRAF and MEK inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–51 (2015). 10.1158/1078-0432.CCR-14-2339. [DOI] [PubMed] [Google Scholar]
- 23.Kirchberger M. C., et al. , MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: Results of a retrospective multicentre analysis of 364 patients. Eur. J. Cancer 98, 10–16 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Ascierto P. A., et al. , Dabrafenib, trametinib and pembrolizumab or placebo in BRAF-mutant melanoma. Nat. Med. 25, 941–946 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ribas A., et al. , PD-L1 blockade in combination with inhibition of MAPK oncogenic signaling in patients with advanced melanoma. Nat. Commun. 111 (11), 1–10 (2020, 2020,). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vella L. J., et al. , The kinase inhibitors dabrafenib and trametinib affect isolated immune cell populations. Oncoimmunology 3, e946367 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kono M., et al. , Role of the mitogen-activated protein kinase signaling pathway in the regulation of human melanocytic antigen expression. Mol. Cancer Res. 4, 779–92 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Boni A., et al. , Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 70, 5213–5219 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Frederick D. T., et al. , BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin. Cancer Res. 19, 1225–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., et al. , The BRAF and MEK Inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–51 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Cooper Z. A., Reuben A., Austin-Breneman J., Wargo J. A., Does it MEK a difference? Understanding immune effects of targeted therapy. 21, 3102–3104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy J. P., et al. , Multiplexed relative quantitation with isobaric tagging mass spectrometry reveals class i major histocompatibility complex ligand dynamics in response to doxorubicin. Anal. Chem. 91, 5106–5115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaeger A. M., et al. , Rebalancing protein homeostasis enhances tumor antigen presentation. Clin. Cancer Res. 25, 6392–6405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stopfer L. E., Mesfin J. M., Joughin B. A., Lauffenburger D. A., White F. M., Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat. Commun. 11, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiue E. H. C., et al. , Targeting a neoantigen derived from a common TP53 mutation. Science 371, eabc8697, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai J., et al. , Elimination of melanoma by sortase A-generated TCR-like antibody-drug conjugates (TL-ADCs) targeting intracellular melanoma antigen MART-1. Biomaterials 178, 158–169 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Lowe D. B., et al. , TCR-like antibody drug conjugates mediate killing of tumor cells with low peptide/HLA targets. MAbs 9, 603–614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liddy N., et al. , Monoclonal TCR-redirected tumor cell killing. Nat. Med. 18, 980–987 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Gloger A., Ritz D., Fugmann T., Neri D., Mass spectrometric analysis of the HLA class I peptidome of melanoma cell lines as a promising tool for the identification of putative tumor-associated HLA epitopes Europe PMC Funders Group. Cancer Immunol. Immunother. 65, 1377–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen R. S., et al. , Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 72, 1642–50 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Van der Bruggen P., et al. , Tumor-specific shared antigenic peptides recognized by human T cells. Immunol. Rev. 188, 51–64 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Vita R., et al. , The Immune Epitope Database (IEDB): 2018 update. Nucleic. Acids. Res. 47, D339–D343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klechevsky E., et al. , Antitumor activity of immunotoxins with T-cell receptor-like specificity against human melanoma xenografts. Cancer Res. 68, 6360–6367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Middleton M. R., et al. , Tebentafusp, A TCR/Anti-CD3 bispecific fusion protein targeting gp100, potently activated antitumor immune responses in patients with metastatic melanoma. Clin. Cancer Res. 26, 5869–5878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caramel J., et al. , A switch in the expression of embryonic emt-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Tulchinsky E., Howard Pringle J., Caramel J., Ansieau S., Plasticity of melanoma and EMT-TF reprogramming. Oncotarget 5, 1–2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caron E., et al. , The MHC i immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol. Syst. Biol. 7, 533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akbani R., et al. , Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stopfer L. E., et al. , Absolute quantification of tumor antigens using embedded mhc-i isotopologue calibrants. Proc. Natl. Acad. Sci. U.S.A. 118, e2111173118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dao T., et al. , Targeting the intracellular WT1 oncogene product with a therapeutic human antibody. Sci. Transl. Med. 5, 176ra33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dao T., et al. , Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat. Biotechnol. 33, 1079–1086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Høydahl L. S., Frick R., Sandlie I., Løset G. Å., Targeting the MHC ligandome by use of TCR-like antibodies. Antibodies 8, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douglass J. “Bispecific antibodies targeting mutant RAS neoantigens” Sci Immunol. 2021 Mar 1 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson L. A., et al. , Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkhurst M. R., et al. , T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 19, 620–626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgan R. A., et al. , Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stopfer L. E., D’Souza A. D., White F. M., 1,2,3, MHC: A review of mass-spectrometry-based immunopeptidomics methods for relative and absolute quantification of pMHCs. Immuno-Oncology Technol. 11, 100042. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kubiniok P., et al. , Understanding the constitutive presentation of MHC class I immunopeptidomes in primary tissues. iScience 25, 103768 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yarmarkovich M., et al. , Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs. Nature 5997885 (599), 477–484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Chaganty B. K. R., et al. , Trastuzumab upregulates expression of HLA-ABC and T cell costimulatory molecules through engagement of natural killer cells and stimulation of IFNγ secretion. Oncoimmunology 5, e1100790. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goel S., et al. , CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwai T., et al. , Topoisomerase I inhibitor, irinotecan, depletes regulatory T cells and up-regulates MHC class I and PD-L1 expression, resulting in a supra-additive antitumor effect when combined with anti-PD-L1 antibodies. Oncotarget 9, 31411–31421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Browne C. M., et al. , A chemoproteomic strategy for direct and proteome-wide covalent inhibitor target-site identification. J. Am. Chem. Soc. 141, 191–203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Data Availability Statement
RNA-sequencing data have been deposited into the NCBI Gene Expression Omnibus GSE218070. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029860 for DDA datasets and PXD029884 for targeted datasets. Previously published data were used for this work (PMID 34497125). All other data are included in the manuscript and/or supporting information.