Significance
Due to the heterogeneity and low efficiency of the primary cell reprogramming process, it has been challenging to identify and isolate the reprogramming initiating cells. To address this limitation, we engineered transgenic mice, from which many, if not all, cell types can be efficiently reprogrammed in vitro to produce induced pluripotent stem cells (iPSCs). We report the generation of two reprogrammable mouse lines whose cells reach pluripotency at an efficiency that highly exceeds that of previously described reprogramming systems. We incorporated two fluorescent reporters that enable partially and completely reprogrammed cells to be tracked and isolated. These reprogramming systems will facilitate the investigation of molecular events during somatic cell reprogramming and allow the future development of pluripotent cell therapeutic derivatives.
Keywords: somatic cell reprogramming, induced pluripotent stem cells, secondary reprogramming, reprogrammable mouse, transgenic mouse
Abstract
Revealing the molecular events associated with reprogramming different somatic cell types to pluripotency is critical for understanding the characteristics of induced pluripotent stem cell (iPSC) therapeutic derivatives. Inducible reprogramming factor transgenic cells or animals—designated as secondary (2°) reprogramming systems—not only provide excellent experimental tools for such studies but also offer a strategy to study the variances in cellular reprogramming outcomes due to different in vitro and in vivo environments. To make such studies less cumbersome, it is desirable to have a variety of efficient reprogrammable mouse systems to induce successful mass reprogramming in somatic cell types. Here, we report the development of two transgenic mouse lines from which 2° cells reprogram with unprecedented efficiency. These systems were derived by exposing primary reprogramming cells containing doxycycline-inducible Yamanaka factor expression to a transient interruption in transgene expression, resulting in selection for a subset of clones with robust transgene response. These systems also include reporter genes enabling easy readout of endogenous Oct4 activation (GFP), indicative of pluripotency, and reprogramming transgene expression (mCherry). Notably, somatic cells derived from various fetal and adult tissues from these 2° mouse lines gave rise to highly efficient and rapid reprogramming, with transgene-independent iPSC colonies emerging as early as 1 wk after induction. These mouse lines serve as a powerful tool to explore sources of variability in reprogramming and the mechanistic underpinnings of efficient reprogramming systems.
Reprogramming somatic cells to induced pluripotent stem cells (iPSCs) has promised to revolutionize cell-based therapeutic strategies by providing an unlimited source of cells to treat many degenerative diseases. Forced overexpression of the transcription factors, Oct4, Klf4, c-Myc, and Sox2 (OKMS), also known as the Yamanaka factors, reprograms differentiated cells to embryonic stem cell (ESC)–like iPSCs (1). When these reprogramming factors are stably introduced into a population of cells by randomly integrated transgenes, their expression levels differ from cell to cell due to the varying number and location of genomic insertion. The heterogeneity of OKMS expression not only contributes to the low efficiency of reprogramming but also confounds the interpretation of molecular events captured by existing omics datasets of the reprogramming process (2–5). Thus, investigations of the molecular processes of iPSC generation remain challenging, particularly for early events.
To overcome this limitation, 2° reprogramming systems have been developed. These systems produce a population of transgene integration–identical cells carrying one or more doxycycline (dox)-inducible transgene(s) encoding the reprogramming factors (6). Initially, 2° systems consisted of differentiated cells isolated from iPSC-derived chimeras (7–14). The introduction of OKMS factors by lentiviral vector–delivered transgenes produced a moderate increase in the reprogramming efficiency of these systems. A significant improvement in reprogramming yield (40% in chimera-derived mouse embryonic fibroblasts (MEFs)) was achieved when the transgenes were delivered by piggyBac (PB) transposons (11, 13). Several groups have since reported reprogrammable transgenic mice that carry OKMS as a single polycistronic transgene in a defined locus (13, 15–18). Although two of these mouse lines (13, 15) have been widely used, the reprogramming efficiency (number of iPSC colonies per input somatic cell) derived from most tissues in these lines is not much higher than 1%. An exception is the 40% efficiency rate reported for hematopoietic stem cells (HSCs) (13), which may be associated with the fact that these cells begin in a stem cell state prior to reprogramming. Thus, there is a need for 2° reprogramming systems that offer a high reprogramming efficiency across an array of starting somatic cell types.
2° reprogramming systems have opened the door to understanding the fate trajectories of reprogramming cells using both bulk (7, 12, 19–21) and single-cell reprogramming approaches (22–25). However, these systems vary in their reprogramming dynamics. For example, some give rise to stable alternative or intermediate states, while others do not (26). Furthermore, while 2° cells that carry the four Yamanaka factors in multiple insertion sites have offered a high reprogramming efficiency from a MEF state (11, 12) and have enabled comprehensive multiomics datasets of the reprogramming process (20), the presence of multiple transgene insertion sites makes the establishment of reprogrammable (transgenic) mouse lines practically impossible, and consequently, the study of adult somatic cells reprogramming has remained a challenge. While 2° cells carrying OKMS in a single locus circumvent this issue, they experience transgene expression levels influenced by transcriptional activity of the integration site, which varies across cellular lineages and can bottleneck reprogramming efficiency. Additionally, inopportune single insertion sites may be differentially silenced across cell types. While the efficiency of 2° reprogramming is higher than what has been reported for primary (1°) reprogramming, 2° systems have not eliminated the need for purification steps, such as fluorescence-activated cell sorting (FACS), to enrich reprogramming cells for analysis (19).
Given the ongoing impact of 2° cells for catalyzing mechanistic insights about the reprogramming process, there is a need to establish transgenic animals with inducible 2° reprogramming systems where a single-copy insertion of OKMS mediates sufficiently high transgene levels that are not or minimally affected by silencing. Such a system would address a critical gap in the capability of current 2° systems to offer highly efficient reprogramming across a wide array of somatic cell types. It would also open the door to a broad range of in vivo experiments.
To address this need, our experience with the highly efficient 1B and 6C 2° MEF lines (11, 20), which carry multiple reprogramming factor insertion sites, encouraged us to generate mouse lines from which cells would reprogram with similar efficiencies but from a single insertion site that expresses robustly across somatic cell types. Here, we report the development of two transgenic mouse lines from which 2° cells reprogram with unprecedented efficiency from various somatic cells. These lines were instrumental in uncovering the exciting observation that heterogeneous subpopulations of MEFs engage in competitive interactions, leading a subset of clones to overtake the population (27). In addition to these lines, we present a strategy for deriving highly efficient reprogramming systems that is adaptable across the field. In deriving our reprogramming mice, we applied several key tools and techniques. First, we used a third-generation dox-inducible system (28) to activate the reprogramming transgenes at a higher level (29). This inducible promoter has reportedly increased the reprogramming efficiency by ninefold in MEFs, noting that this efficiency is below 10% (29). Second, we used a PB transposon approach to insert a polycistronic OKMS transgene cassette into the genome, leveraging the reduced epigenetic silencing that PB offers compared with viral insertions (30). Finally, we implemented a selection step to enrich for 1° iPSC clones capable of reinitiating the reprogramming process after early interruption of OKMS transgene expression. We then used these selected 1° iPSCs to generate reprogrammable mouse lines after germ line transmission. The two mouse lines generated using this strategy show unprecedented reprogramming efficiencies and rapid iPSC emergence. Importantly, these mice also carry two fluorescent reporters that facilitate the observation of important aspects of the reprogramming process in real time: the level of Yamanaka factor transgene expression (mCherry) and Oct4 promoter activity (GFP), making these systems highly practical and valuable for further research.
Results
Reprogramming with Short Interruption.
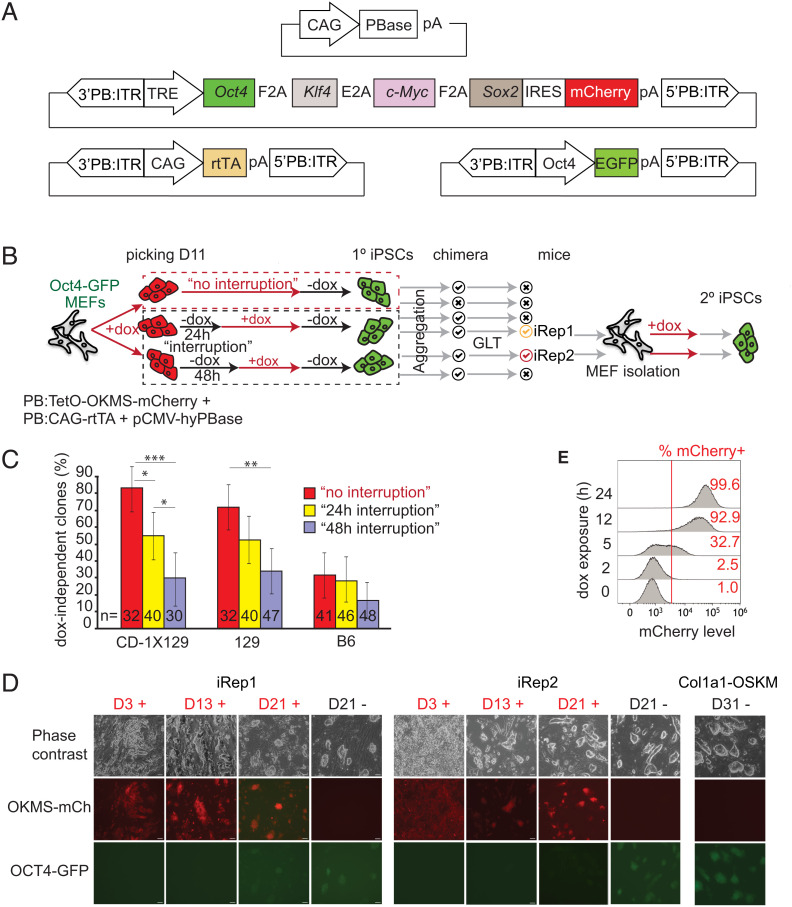
Previous studies demonstrated that when reprogramming factor expression is interrupted early during the reprogramming process, somatic cells revert to their original phenotype and typically become refractory to reinitiated reprogramming (31). We hypothesized that a short (24- to 48-h) interruption of an inducible OKMS expression during 1° reprogramming would select for clones that have an enhanced ability to return to the reprogramming process following dox reintroduction (i.e., clones that are not refractory to reprogramming), which results in sufficient transgene activation and high somatic cell reprogramming efficiency in 2° systems. To test this hypothesis, we transfected C57BL/6J (B6), 129S2/Sv (129), and CD-1x129/Sv MEFs carrying the previously characterized Oct4-GFP reporter transgene (32) with three PB transposon delivery constructs: 1) PB:TetO-OKMS-mCherry (30), a PB transposon containing a polycistronic OKMS linked to the mCherry reporter transgene with an internal ribosome entry site sequence, where the transgenes are under the control of an earlier-described (33) dox-inducible promoter variant (Fig. 1A); 2) PB:CAG-rtTA (11), a PB transposon delivering a CAG promoter (34)–driven rtTA transgene; and 3) pCMV-hyPBase (35), a PB hyperactive transient transposase expression plasmid to drive genomic insertion of aforementioned cassettes. Dox was added to the culture 24 h after the cotransfection, resulting in the reprogramming transgene activation. To select for clones that respond efficiently to dox activation, dox was interrupted at day 11 for a defined time interval (24 or 48 h) just prior to the acquisition of dox-independent iPSC stage (11). The reprogramming clones resulting from the dox interruption regimen were members of the “interruption” group, while clones that arose in cultures without transient dox removal were members of the no-interruption control group. The cells from both groups subsequently underwent dox removal at days 18 to 21 to assess for dox independence, self-renewal, and markers of pluripotency (Fig. 1B). As expected, a smaller fraction of the interruption clones successfully reached dox independence in all of the tested genetic backgrounds as compared with the no-interruption control group (36). Not surprisingly, 48 h of dox interruption led to a more stringent selection for cells that retained the ability to return to the reprogramming process (Fig. 1C).
Fig. 1.
Derivation and characterization of reprogrammable mouse lines. (A) Schematic of the elements of the PB transfection system used in this study. (B) Summary of methodology used to select 1° iPSC clones, generate chimeras, and characterize resulting 2° reprogramming and iPSC lines, iRep1 and iRep2. Red and black arrows represent incubation with and without dox, respectively. GLT, germ line transmission. (C) Selection pressure of dox removal for 24 or 48 h at D11 of reprogramming for obtaining dox-independent 1° iPSC clones in three different genetic backgrounds. The error bars indicate the 95% CI of efficiency measures, and statistical significance is represented as follows: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***). The experiment was repeated three times, and n is indicated in the bars. (D) Morphology and reporter gene expression during reprogramming of iRep1 and iRep2 2° MEFs compared with Col1a1-OSKM (R26rtTA*M2;Col1a14F2A) MEFs in the presence of dox (+) and dox-independent (−) ESC-like colonies at D21, D18, or D31. GFP reports for the endogenous OCT4, while the mCherry reports the reprogramming factor expression. (Scale bar, 20 µm.) Dox-independent cultures were captured at the earliest time a confluent culture was obtained (at the indicated times). The experiment was repeated three times. (E) Rapid transgene induction in iRep1 2° MEFs by dox incubation as indicated by mCherry fluorescence by flow cytometry analysis.
Investigating the Pluripotency and Reprogramming Efficiency of 2° MEFs Derived from Interruption 1° iPSCs.
We generated chimeric embryos to determine the 1° iPSC lines’ pluripotency-associated developmental potency and to explore the reprogramming efficiency of 2° MEFs obtained from the chimeras. Fourteen 1° iPSC lines (7 interruption and 7 no interruption) were used to generate chimeric embryos for MEF isolation (SI Appendix, Fig. S1 A and B). Four of the seven no-interruption iPSC lines gave rise to a total of 13 chimeric embryos and six of the seven interruption iPSCs gave rise to a total of 19 chimeric embryos. We then conducted reprogramming experiments with 2° MEFs derived from these chimeras to assess the reprogramming efficiency of these cells and the speed with which they could acquire the dox-independent secondary iPSC stage. We found that MEFs from chimeric embryos generated from the interruption iPSC lines had elevated reprogramming efficiency, depicted by higher alkaline phosphatase–positive (AP+) colony-forming ability from single cells, as compared with MEFs isolated from the control no-interruption group (SI Appendix, Fig. S1 C and D). They also reached the dox-independent iPSC stage faster (SI Appendix, Fig. S1E). We observed a positive correlation between the reprogramming factor expression level at 24 h following dox induction, indicated by the mCherry fluorescence intensity, and reprogramming efficiency measured by AP+ colony-forming ability (SI Appendix, Fig. S1F). This confirms an earlier observation (3) but at a higher reprogramming efficiency. In addition, mCherry intensity also correlates with a faster transition to a fully reprogrammed, dox-independent state (SI Appendix, Fig. S1G).
Generating 2° Reprogrammable Mouse Transgenic Lines.
Encouraged by these findings, chimeras from the interruption 1° iPSCs clones were allowed to develop to term (Fig. 1B). Highly chimeric mice (assessed by coat color) were born from almost all the tested lines. Chimeras, generated from two independent CD-1x129-derived iPSCs lines, transmitted the transgenes to their offspring and thereby generated two reprogrammable mouse lines designated as induced reprogrammable mouse (iRep) 1 and 2. iRep1 was established from a 1° iPSC line interrupted for 24 h, while the iRep2 originated from a 1° iPSC line interrupted for 48 h. We did not obtain germ line transmission from the no-interruption 1° iPSC clones.
The integration sites of the PB:TetO-OKMS-mCherry transgenes were determined via the splinkerette PCR (11). In both iRep1 and iRep2 lines, only one copy of the transgene is integrated into chromosomes 14 and 13 intergenic regions, respectively (SI Appendix, Fig. S2). To determine whether these integration sites may be influenced by reprogramming dynamics, we leveraged the dataset from our previous study in which we conducted a comprehensive analysis of gene expression and epigenetic changes throughout the reprogramming time course in 1B cells (20). Here, we focus our analysis from this dataset on a 10-kb genomic region centered around the transgene integration sites for both the iRep1 and iRep2 lines (SI Appendix, Fig. S3). For the iRep1, we found that no significant changes occurred during reprogramming in chromatin marks, DNA methylation, or RNA expression in the 10 kb vicinity of the insertion site (SI Appendix, Fig. S3). For iRep2, we found an increase in DNA methylation levels in iPSC/ESCs 4 kb away from integration compared with MEFs. This suggests that the area is more permissive for expression at a differentiated stage and neither during reprogramming nor in iPSCs. There was a small region with H3K4me3 marks (also at 4 kb away) in MEFs, which was rapidly decreased by reprogramming transgene expression. This indicated that this endogenous transcription might not influence the exogenous gene expression during the reprogramming process. Further support was obtained from our experiments, not observing significant transgene silencing during the reprogramming path. Both iRep1 and iRep2 consistently expressed mCherry (Fig. 1D).
To reduce the rtTA expression variability generated by the random integration of the PB:CAG-rtTA transgene, the PB:CAG-rtTA was replaced by breeding iRep1 and iRep2 mice to the ROSA26-rtTA mouse (37) in which a CAG-rtTA was inserted into the ubiquitous Rosa26 locus. The data presented herein were obtained from mice backcrossed to C57BL/6J between N3 and N7 generations.
Characterizing the iRep1 and iRep2 2° Mouse Lines.
To test the reprogramming ability and reporter gene readouts of our transgenic mouse systems, we derived 2° MEFs from iRep1 and iRep2 E13.5 embryos and cultured them in the presence of dox for up to 21 d. iRep1 and iRep2 cells rapidly expressed the OKMS-reporting mCherry (Fig. 1D). Over 90% of the iRep1 2° MEFs cultured in dox for 12 h activated the transgenes as seen by flow cytometry (mCherry fluorescence) (Fig. 1E). Rapid induction was confirmed by the time-lapse video microscopy, where mCherry expression was observed within a few hours of dox exposure (Movies S1 and S2). Colonies rapidly formed and at later stages of reprogramming, the cells became dox independent and expressed OCT4-GFP (Fig. 1D). After dox removal on day 21, the reprogramming iRep cells formed OCT4-GFP+, reprogramming transgene-independent, self-renewing 2° iPSCs (Fig. 1D) that could be cultured in ESC culture conditions for at least 10 passages. The emerging iPSC colonies resembled those from the previously characterized reprogrammable transgenic mouse line: R26rtTA*M2;Col1a1-OSKM4F2A (herein Col1a1-OSKM) (3, 15) that carried the Oct4-GFP transgene (31) (Fig. 1D). As expected (32), OCT4-GFP faithfully reported the expression of endogenous OCT4 as both staining correlated (Fig. 2A and SI Appendix, Fig. S4). These 2° iPSCs expressed endogenous OCT4, NANOG, SOX2, and KLF4 pluripotency factors as observed by immunofluorescence (Fig. 2A and SI Appendix, Fig. S4). Using quantitative reverse transcriptase PCR, we monitored the expression of the reprogramming transgenes, as well as their endogenous counterparts, during iRep1 and iRep2 MEF reprogramming. As expected (31), the Thy-1 fibroblast marker was down-regulated early, while Nanog was induced later in reprogramming. Consistent with previous reports, we observed earlier induction of Oct4 relative to Klf4 and Sox2 (20). Interestingly, Oct4,Klf4 and Sox2 but not c-Myc were induced more rapidly than the other reprogramming systems (Fig. 2B and SI Appendix, Fig. S5). The temporal expression of the transgenes across the reprogramming time course with iRep1 and iRep2 compared with the established 2° reprogramming MEFs, 1B (20) and Col1a1 (3, 15), also revealed that transgene expression was higher and better sustained in the iRep lines throughout the reprogramming time course, with relatively higher expression levels at days 2 to 12 (Fig. 2B and SI Appendix, Fig. S5).
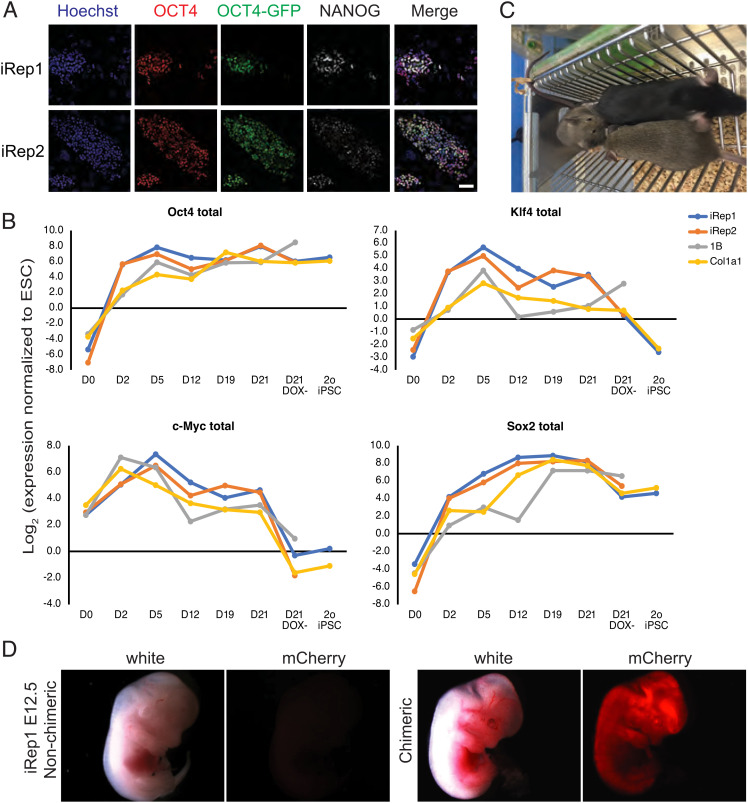
Fig. 2.
Characterization of pluripotency of reprogramming lines. (A) Representative fluorescence microscopy images of iRep1 dox-independent 2° iPSC colony stained with antibodies to OCT4 and NANOG and also observed for Oct4-GFP reporter expression. (Scale bar, 50 µm.) The experiment was repeated twice. (B) Relative gene expression levels of total Yamanaka factors during reprogramming of iRep1, iRep2, 1B, and Col1a1 reprogrammable 2° MEFs. Expression levels were normalized to the housekeeping gene mEEF2 and are shown relative to ROSA26-rtTA-ESCs. (C) iRep1 2° iPSC-derived mouse (100% chimera based on black coat color) and her 2 agouti germ line transmitted offspring. (D) Whole-mount white light and red fluorescence (mCherry) images of iRep1 E12.5 chimeric embryo (dark eye pigment, “pigment +”) and nonchimeric embryos (“pigment −”) from animals exposed to dox (drinking water, 1.5 µg/ml, and dox pellets) for 24 h prior to recovering the embryos. The experiment was repeated three times.
To further test the pluripotency of 2° iPSCs derived from iRep1 and iRep2 MEFs, we generated chimeras and observed, according to coat color, a high iPSC contribution. The germ line transmission of the iPSC genome to the offspring of the chimeras further confirmed the pluripotency of these 2° iPSC lines and is indicative of their appropriate epigenetic status (Fig. 2C) (38, 39). To test for in vivo induction of the OKMS-mCherry transgenes, these 2° iPSC-derived chimeric embryos were exposed to dox via drinking water and food pellets fed to the mothers for 24 h prior to dissection at E13.5. We found that OKMS-mCherry transgenes were successfully induced as determined by the whole-mount fluorescence microscopy (Fig. 2D) and by flow cytometry analysis of the disaggregated embryonic tissues (SI Appendix, Fig. S6A). Moreover, tissues dissected from adult transgenic iRep1 and iRep2 mice exposed to dox by water and food showed expression of the mCherry reporter (SI Appendix, Fig. S6B).
Quantifying Reprogramming Efficiency of iRep from Varied Somatic Cell Types.
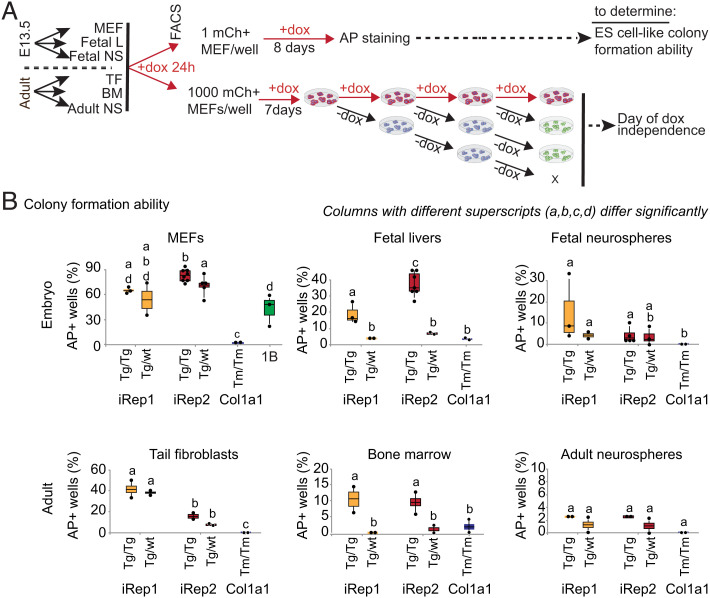
To address the effect of transgene copy number, cells from different tissues were isolated from mice that were homozygous for ROSA26-rtTA, either homozygous or hemizygous for the OKMS-mCherry transgene, and carried at least one copy of the Oct4-GFP transgene. Cells from the same tissues were isolated and tested from Col1a1-OSKM mice (3, 15) that were homozygous for R26rtTA*M2 and Col1a1-OSKM and that carried the Oct4-GFP transgene (32). Adult cell types (tail fibroblasts (TFs), CD45+ bone marrow cells (BMs), and neurospheres (NSs)) isolated from 4- to 7-wk-old mice and embryonic cell types isolated from E13.5 transgenic embryos (MEFs, CD45+ fetal liver cells (FLs), and fetal NSs) were single-cell sorted by flow cytometry into 96-well dishes and cultured in reprogramming medium (Fig. 3A). To characterize the reprogramming progress to pluripotency, we used two markers providing high-throughput readouts: AP expression and our previously characterized Oct4-GFP transgene (32). Each of the 48 wells plated with a single cell was assessed for colony-forming ability, determined by the presence of at least one AP+ ESC-like colony. Even though cells were single-cell plated, a very high efficiency of reprogramming was observed for iRep1 and iRep2. Indeed, for all cell types, the ability to form AP+ colonies was significantly higher in iRep1 and iRep2 than in Col1a1-OSKM (Fig. 3B). Of note, iPSCs were obtained from all tissues tested for iRep mice, which was not the case for the Col1a1-OSKM. Previous studies demonstrated that although AP is expressed in reprogramming cells and is used as an indicator of pluripotency, it is not a definitive biomarker for the most stringent pluripotency: the ability to support completely viable iPSC-derived animals (3). Mouse ESC studies (40) have shown that this ability is rare even among cell lines capable of germ line transmission and/or contributing high levels of chimerism in all tissues of the resulting embryo. Nevertheless, to test the correlation between AP expression and endogenous Oct4 activation in our system, we assessed dox independence, AP staining, and OCT4-GFP expression in parallel. We observed a very high agreement rate (>95%) between AP staining on day 8 and OCT4-GFP expression on day 21 (P = 2.2 × 10−16, chi-square test, and odds ratio of 57.5+/−1.47; Fig. 4A), suggesting that in our system, the expression of AP is a good indicator of reprogramming to pluripotency hallmarked by endogenous Oct4 activation. Overall, this assay, together with the AP versus OCT4-GFP proportional agreement, supports the high reprogramming yield of the cells isolated from the iRep1 and iRep2 mice.
Fig. 3.
Characterization of reprogramming efficiency of embryonic and adult tissues from iRep1 and iRep2 mouse lines. (A) Experimental layout for quantification of colony formation efficiency and the earliest time to exogenous reprogramming factor-independent pluripotency for embryonic and adult tissues from 1) iRep1 and iRep2 mice homozygous for ROSA26-rtTA targeted mutation (rtTAtm/tm), either homozygous (OKMStg/tg) or hemizygous for OKMS-mCherry (OKMStg/wt), and carried at least one Oct4-GFP transgene; and 2) Col1a1-OSKM homozygous for both R26rtTA*M2 and Col1a14F2A (Col1a1-OSKMTm/Tm;rtTATm/Tm) and carried at least one copy of Oct4-GFP transgene (32). MEFs and TFs were isolated from E13.5 embryos and adult mice, respectively, and used for fluorescence-activated cell sorting at passage 1. BMs and FLs were sorted and exposed to dox directly. NSs were grown for 2 passages before sorting and culturing in dox. (B) Quantification of fraction of single-cell sorted iRep1, iRep2, and Col1a1-OSKM cells that formed AP+ ESC-like colonies after 8 to 15 d in doxycycline: reprogramming cells were stained at Day 8 for MEFs and FLs, Day 11 for TFs, and Day 15 for BMs and adult NSs. The histograms show means +/− SEs of at least three independent experiments. Columns with different superscripts (a, b, c, and d) differ significantly (P < 0.05), while columns with the same superscripts do not differ significantly. Each dot on the plot represents the reprogramming efficiency of a single cell line (in %) obtained from a sample of n = 48 cells plated individually, one cell per well. Tg/Tg: homozygous OKMS-mCherry transgene; Tg/wt: heterozygous OKMS-mCherry transgene; and Tm/Tm: homozygous Col1a1-OSKM knock-in.
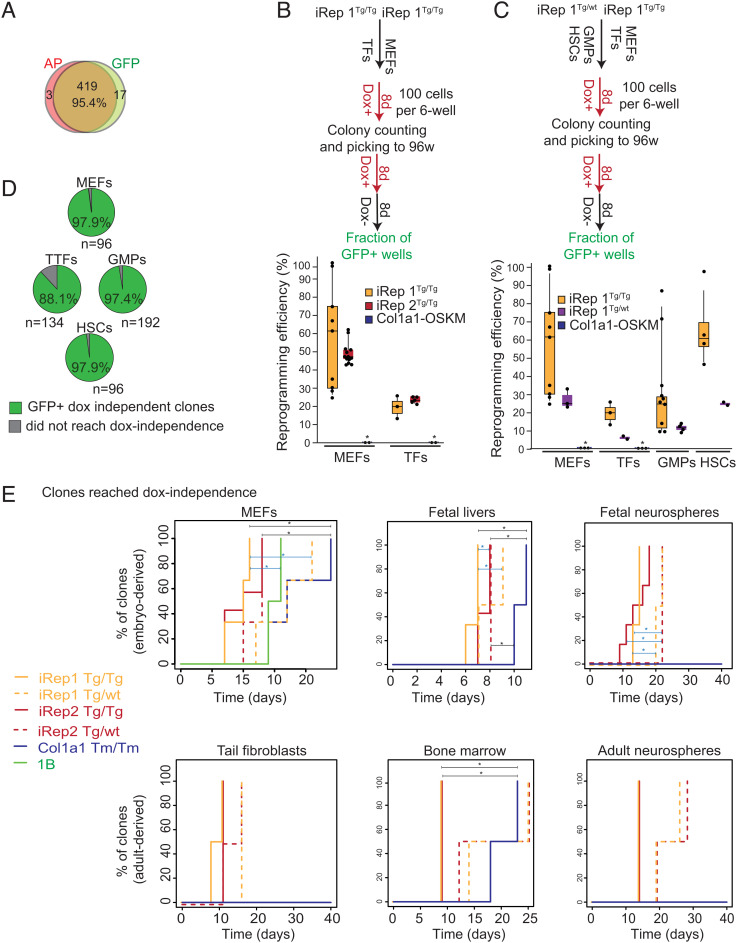
Fig. 4.
Characterization of reprogramming dynamics and derivation of fully reprogrammed clones from iRep1 and iRep2 mouse lines. (A) Observed proportional agreement between AP and GFP was assessed by the comparison between OCT4-GFP expression at day 21 and AP staining at day 8 from parallel 96-well plates (a total of 439 wells were compared). (B) Experimental design and the efficiency of colony formation as a percentage of input cells for embryonic and adult cell types homozygous for both OKMS-mCherry and ROSArtTA transgenes for iRep1 and iRep2 in comparison with Col1a1-OSKM homozygous for both R26rtTA*M2 and Col1a1-OSKM. Each dot on the plot represents the reprogramming efficiency of a single cell line (in %) obtained from a sample of 100 cells plated in a 35-mm well. The experiment was repeated twice. (C) Experimental design and the efficiency of colony formation for iRep1 homozygous for ROSA26-rtTA and either homozygous (Tg/Tg) or hemizygous (Tg/wt) for OKMS-mCherry transgene and Col1a1-OSKM (Tm/Tm) homozygous for both R26rtTA*M2 and Col1a1-OSKM. * = no reprogrammed colony was detected. Each dot on the plot represents the reprogramming efficiency of a single cell line (in %) obtained from a sample of 100 cells plated in a 35-mm well. The experiment was repeated twice. (D) Fraction of picked clones from iRep1 that generated OCT4-GFP+, dox-independent colonies. The rectangle height in Fig. 4D represents the interquartile range; the bar inside the rectangle represents the median and the length of the two whiskers, the 5% and 95% quantiles, respectively. (E). Comparison of the earliest day of dox-independent pluripotency for iRep1, iRep2, and Col1a1-OSKM reprogrammable tissues. For MEFs, n values were 3, 3, 7, 3, 3, and 2 for iRep1Tg/Tg, iRep1Tg/wt, iRep2Tg/Tg, iRep2Tg/wt, Col1a1-OSKM Tm/Tm, and 1B, respectively. For FLs and fetal neurospheres, n values were 3, 2, 7, 3, and 2 and 3, 2, 6, 2, and 2 for iRep1Tg/Tg, iRep1Tg/wt, iRep2Tg/Tg, iRep2Tg/wt, and Col1a1-OSKM Tm/Tm, respectively. For TFs, BMs, and adult NSs, n values were 2, 2, 2, 2, and 2 for iRep1Tg/Tg, iRep1Tg/wt, iRep2Tg/Tg, iRep2Tg/wt, and Col1a1-OSKM Tm/Tm, respectively. The experiment was repeated twice. An asterisk on top of the horizontal line indicates that the comparison between the 2 groups is significant (P < 0.05).
To further support the above conclusion, we determined the number of colonies that formed per total cells plated for MEFs, TFs, granulocyte–macrophage progenitor cells (GMPs), and HSCs sorted from the BMs of iRep1 and iRep2 mice (details in SI Appendix, Fig. S7). We also quantified the ability of the resulting individual reprogramming colonies to become dox independent. The colony formation efficiencies for MEFs, TFs, GMPs, and HSCs isolated from iRep1 Tg/Tg were 59.2%, 19%, 32%, and 65.3%, respectively (Fig. 4 B and C). Colonies were subsequently picked at day 8, disaggregated, and single-cell plated onto mitotically inactivated feeder MEFs in 96-well plates and assessed for 1) mCherry expression, 2) ESC-like colony morphology, 3) OCT4-GFP expression, and 4) the ability to form dox-independent colonies. Notably, over 88% of individually picked colonies for all cell types tested from the iRep1 line reached an OCT4-GFP+, dox-independent state with ESC-like morphology (Fig. 4D). In both assays, the efficiency of reprogramming for all cells tested was greatly increased compared with the reprogrammable mice reported earlier (i.e. for MEF: 50.3% and 63.8% in iRep1 and iRep2, respectively, vs. 1.5% (13) and <10% (15)). In contrast to a previous report (15), iPSCs could also be obtained from adult mice tissues carrying a single copy of the OKMS transgene (Figs. 3B and 4 C and E) and from tissues carrying a single copy of each ROSA26-rtTA and OKMS-mCherry transgene. Similar to previously reported studies (15), BM-derived HSCs had higher reprogramming efficiencies than GMPs. Furthermore, we observed greater colony-forming ability in embryo-derived cells as compared with adult counterparts, which is also consistent with previous findings (13, 41).
Determining the Commitment Point to iPSCs.
Given that the time to dox independence is a fundamental parameter defining the reprogramming efficiency, we proceeded to characterize the iRep systems by plating the cells, exposing to dox for 24 h, and subsequently sorting for mCherry+ cells, as described in Fig. 3A. at a density of 2500 cells/cm2 in reprogramming medium. For the bulk reprogramming experiments, passaging was done every 3 d, at which point the cultures were branched into dox-free or dox-containing medium at each passage (Fig. 3A). This dox independence test is stringent as cells are dissociated and passaged at the time of dox withdrawal, which requires ESC-like colony formation from disaggregated cells, in contrast to dox withdrawal from already-formed colonies. The day of dox independence was determined by the earliest day GFP+ ESC-like colonies were observed in dox-free media. Of the lines that became dox independent, both iRep1 and iRep2 showed faster reprogramming and over 10 d quicker time to dox independence as compared with the Col1a1-OSKM cells (Fig. 4E and SI Appendix, Fig. S8). Interestingly, dox independence was never achieved for Col1a1-OSKM adult and fetal NSs and TFs in the time period tested, while both fetal NSs and TFs from the iRep lines reached dox independence within 15 to 25 d. In addition, in all cases, reprogramming to dox independence coincided with endogenous Oct4 activation (SI Appendix, Fig. S8). No major difference between iRep1 and iRep2 cells was detected in terms of reprogramming speed and time to dox independence.
Discussion
Taken together, our studies demonstrate that a wide variety of adult and embryo tissues derived from iRep1 or iRep2 have ESC-like colony-forming ability and a time line to dox independence that surpasses that of previously characterized 2° systems reported to date (15, 16, 29). Recently, we used the iRep1 system to elucidate the competitive dynamics of elite reprogramming clones (27). In that study, we showed that the emergence of nonneutral clonal dominance leads a few reprogramming clones to overtake the population. Furthermore, by crossing the iRep2 reprogramming mouse with a Wnt1+ lineage-tracing mouse, we suggest that these dominating clones arise from the neural crest compartment and have an elite propensity to undergo reprogramming. The high efficiency of reprogramming achieved with this line was instrumental in enabling clonal tracking in this study, revealing that population reprogramming trajectory is not a sum of its clonal parts. Further applications of the highly efficient iRep1 and iRep2 lines include the possibility of Cre-conditional activation of reprogramming transgene expression using an established mouse line in which Cre-conditional rtTA is knocked into the Rosa26 locus to achieve cell type–specific reprogramming (37). For this purpose, the fact that iPSCs can be derived at high efficiency from adult mouse tissues carrying a single copy of the OKMS-mCherry and ROSA26-rtTA transgenes will save time and mice.
The iRep1 and iRep2 lines reported here were derived by interrupting the expression of transgenes in the early stages of reprogramming, leading to the derivation of clones that reprogram with unprecedented efficiencies. This builds on our previous work that demonstrated the elastic nature of early reprogramming, where dox removal leads to reacquisition of the somatic cell state (31). The mechanistic insight offered in our previous work suggested that an early and temporary interruption to OKMS expression can select for clones that are refractory to transgene silencing during exit and reentry to the somatic state, which is indicative of transgene insertion sites that offer robust OKMS expression that could be reactivated. This is reflected in our transcriptional analysis, which revealed the rapid and sustained expression of transgenes at higher levels than existing 2° reprogramming systems. Additionally, our observation that highly efficient reprogramming occurs across multiple somatic cell types, including adult cells, demonstrates the functional impact of our approach in selecting for iPSC clones with favorable transgene insertion sites.
The early interruption of reprogramming transgene expression has also been recently reported to lead to the derivation of a stable progenitor-like state, in which cells retain the ability to expand without attaining pluripotency (42). The expansion potential of these reprogramming interruption cells makes them amenable to genetic engineering while also opening the door to exploring mechanistic gaps in the reprogramming process by stabilizing cells in previously unexplored states along their trajectory. Thus, the approach of interrupting cells undergoing fate programming by premature switching off the transgene expression may provide avenues to understanding and better manipulating cell fate.
In conclusion, in addition to revealing a method to select for 1° iPSCs poised for 2° system generation, here we report two transgenic mouse lines carrying the four Yamanaka factors from which a variety of somatic cell types reprogram with unprecedented efficiency while harboring reporters for both transgene expression and the pluripotent state. We used single-copy insertion of the OKMS transgenes to derive iPSCs that are capable of germ line transmission, leveraging PB transposon insertion and a third-generation dox induction system to allow for stable and high transgene expression. These mouse lines have already proven to be a powerful tool for the study of multicellular reprogramming dynamics. They will continue to serve the iPSC research domain by enabling elucidation of reprogramming mechanisms from various cell types, isolated from individual tissues from live animals, or by conferring cell type specificity with restricted rtTA expression.
Notes: The iRep1 and iRep2 mouse lines are available at JAX as follows:
JAX#031011 iRep1 (or Oct4-GFP; ROSA26-rtTA(Δneo); OKMSCh250).
Strain Name: B6.Cg-Gt(ROSA)26Sor<tm1.1(rtTA,EGFP)Nagy> Tg(Pou5f1-EGFP)1Nagy TgTn(pb-tetO-Pou5f1,-Klf4,-Myc,-Sox2,-mCherry)250Nagy/J.
JAX#031009 iRep2 (or Oct4-GFP; ROSA26-rtTA(Δneo); OKMSCh72).
Strain Name: B6.Cg-Gt(ROSA)26Sor<tm1.1(rtTA,EGFP)Nagy> Tg(Pou5f1-EGFP)1Nagy TgTn(pb-tetO-Pou5f1,-Klf4,-Myc,-Sox2,-mCherry)72Nagy/J.
Supplementary Material
Appendix 01 (PDF)
iRep1 MEFs were seeded in 0.2% gelatin-coated 6-wells and doxinduced for live imaging. Brightfield images were collected every 10 min from the time of adding dox to the media, and fluorescence images were captured every hour for a total of 48-96h. mCherry was rapidly induced in both iRep transgenic mice and clusters of mCherry positive cells started to form within 48h in iRep1 and after 72h in iRep2.
iRep2 MEFs were seeded in 0.2% gelatin-coated 6-wells and doxinduced for live imaging. Brightfield images were collected every 10 min from the time of adding dox to the media, and fluorescence images were captured every hour for a total of 48-96h. mCherry was rapidly induced in both iRep transgenic mice and clusters of mCherry positive cells started to form within 48h in iRep1 and after 72h in iRep2.
Acknowledgments
We acknowledge the contribution of the Model Production Core at The Centre for Phenogenomics for technical support in generating chimeras. We are grateful to Malgosia Kownacka for the preparation of the mouse embryonic feeders, to Balazs Varga for his help with immunofluorescence, tissue culture, and scientific discussions, to Maria Shutova for support with bioinformatics, to Qin Liang and Kristina Nagy (K.N.) for their contribution to the site of insertion detection, to Claudio Monetti (C.M.) and Julia Nakanishi for artwork editing, K.N. and C.M. for comments on the manuscript, to Rebecca Cowling for help with mouse genotyping and microscopy, to Christina Dalrymple for her help with the mouse colony, and to Dr. Yitzhak Reizel for fruitful discussions regarding the epigenetic analysis in our study. This work was supported by an Ontario Research Fund Genome and Life Sciences (Round 2) Program grant from the Ontario Ministry of Research and Innovation (to A.N., GL2-01-028), the Canada Research Chairs Program (to A.N.), and a CIHR Foundation Program Grant (to A.N.).
Author contributions
J.E., M.C.P., P.W.Z., N.S., and A.N. designed research; J.E., M.C.P., M.F., K.W.A.B., L.N., B.M., M.G., and N.S. performed research; J.E., M.C.P., K.W.A.B., S.M.I.H., L.B., N.S., and A.N. analyzed data; M.F. contributed methodology; S.M.I.H. contributed epigenetic analysis; L.B. performed statistical analysis; and J.E., M.C.P., M.F., N.S., and A.N. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Some study data available (Mice strains used in this study are available from The Jackson Laboratory; details are provided in the SI Appendix).
Supporting Information
References
- 1.Takahashi K., Yamanaka S., Induction of Pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Papapetrou E. P., et al. , Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. U.S.A. 106, 12759–12764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey B. W., et al. , Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9, 588–598 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Tiemann U., et al. , Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry Part A 79A, 426–435 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zunder E. R., Lujan E., Goltsev Y., Wernig M., Nolan G. P., A continuous molecular roadmap to iPSC Reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell 16, 323–337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy A., Secondary cell reprogramming systems: As years go by. Curr. Opin. Genet. Dev. 23, 534–539 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen T. S., et al. , Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brambrink T., et al. , Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wernig M., et al. , A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26, 916–924 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna J., et al. , Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woltjen K., et al. , piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Malley J., et al. , High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 499, 1–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtfeld M., Maherali N., Borkent M., Hochedlinger K., A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Met. 7, 53–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin P., et al. , Optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells from Col1a1 4F2A-Oct4-GFP mice with high efficiency. Febs J. 285, 1667–1683 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Carey B. W., Markoulaki S., Beard C., Hanna J., Jaenisch R., Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods 7, 56–59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haenebalcke L., et al. , Efficient ROSA26-based conditional and/or inducible transgenesis using RMCE-compatible F1 hybrid mouse embryonic stem cells. Stem Cell Rev. Rep. 9, 774–785 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Abad M., et al. , Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Haenebalcke L., et al. , The ROSA26-iPSC mouse: A conditional, inducible, and exchangeable resource for studying cellular (De)differentiation. Cell Rep. 3, 335–341 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Polo J. M., et al. , A molecular roadmap of reprogramming somatic cells into iPS Cells. Cell 151, 1617–1632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussein S. M. I., et al. , Genome-wide characterization of the routes to pluripotency. Nature 516, 198–206 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Shakiba N., et al. , CD24 tracks divergent pluripotent states in mouse and human cells. Nat. Commun. 6, 7329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D. H., et al. , Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell 16, 88–101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao T., et al. , Single-Cell RNA-Seq reveals dynamic early embryonic-like programs during chemical reprogramming. Cell Stem Cell 23, 31–45.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Schiebinger G., et al. , Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran K. A., et al. , Defining reprogramming checkpoints from single-cell analyses of induced pluripotency. Cell Rep. 27, 1726–1741.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonge P. D., et al. , Divergent reprogramming routes lead to alternative stem-cell states. Nature 516, 192–197 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Shakiba N., et al. , Cell competition during reprogramming gives rise to dominant clones. Science 364, eaan0925-11 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Das A. T., et al. , Viral evolution as a tool to improve the tetracycline-regulated gene expression system. J. Biol. Chem. 279, 18776–18782 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Alaei S., et al. , An improved reprogrammable mouse model harbouring the reverse tetracycline-controlled transcriptional transactivator 3. Stem Cell Res. 17, 49–53 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Woltjen K., Kim S.-I., Nagy A., The piggyBac transposon as a platform technology for somatic cell reprogramming studies in mouse. Methods Mol. Biol. Clifton NJ 1357, 1–22 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Samavarchi-Tehrani P., et al. , Functional genomics reveals a bmp-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Viswanathan S., et al. , Supplementation-dependent differences in the rates of embryonic stem cell self-renewal, differentiation, and apoptosis. Biotechnol. Bioeng. 84, 505–517 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Agha-Mohammadi S., et al. , Second-generation tetracycline-regulatable promoter: Repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J. Gene. Med. 6, 817–828 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Hitoshi N., Ken-ichi Y., Jun-ichi M., Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Yusa K., Zhou L., Li M. A., Bradley A., Craig N. L., A hyperactive piggyBac transposase for mammalian applications. Proc. National. Acad. Sci. U.S.A. 108, 1531–1536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golipour A., et al. , A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11, 769–782 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Belteki G., et al. , Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33, e51 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messerschmidt D. M., Knowles B. B., Solter D., DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Gene. Dev. 28, 812–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hackett J. A., Zylicz J. J., Surani M. A., Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 28, 164–174 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C., Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markoulaki S., et al. , Transgenic mice with defined combinations of drug inducible reprogramming factors. Nat. Biotechnol. 27, 169–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo L., et al. , Interrupted reprogramming of alveolar type II cells induces progenitor-like cells that ameliorate pulmonary fibrosis. Npj Regen. Med. 3, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
iRep1 MEFs were seeded in 0.2% gelatin-coated 6-wells and doxinduced for live imaging. Brightfield images were collected every 10 min from the time of adding dox to the media, and fluorescence images were captured every hour for a total of 48-96h. mCherry was rapidly induced in both iRep transgenic mice and clusters of mCherry positive cells started to form within 48h in iRep1 and after 72h in iRep2.
iRep2 MEFs were seeded in 0.2% gelatin-coated 6-wells and doxinduced for live imaging. Brightfield images were collected every 10 min from the time of adding dox to the media, and fluorescence images were captured every hour for a total of 48-96h. mCherry was rapidly induced in both iRep transgenic mice and clusters of mCherry positive cells started to form within 48h in iRep1 and after 72h in iRep2.
Data Availability Statement
All study data are included in the article and/or SI Appendix. Some study data available (Mice strains used in this study are available from The Jackson Laboratory; details are provided in the SI Appendix).