Significance
Arp2/3 complex is a protein assembly with seven subunits that forms actin filament branches on the sides of preexisting “mother” actin filaments. This process generates growing actin filaments for cellular locomotion and intracellular movements. Binding proteins called nucleation-promoting factors and to the side of a mother filament, activates intrinsically inactive Arp2/3 complex to initiate a daughter filament. We determined the structure of the branch junction with active Arp2/3 complex at 3.5 Å resolution by cryo-electron microscopy. Activation involves rotation of two blocks of structure to position the actin-related proteins Arp2 and Arp3 next to form the base for the actin filament branch and to allow interactions with five actin subunits in the mother filament.
Keywords: actin, Arp2/3 complex, cryo-electron microscopy, polymerization
Abstract
We reconstructed the structure of actin filament branch junctions formed by fission yeast Arp2/3 complex at 3.5 Å resolution from images collected by electron cryo-microscopy. During specimen preparation, all of the actin subunits and Arp3 hydrolyzed their bound adenosine triphosphate (ATP) and dissociated the γ-phosphate, but Arp2 retained the γ-phosphate. Binding tightly to the side of the mother filament and nucleating the daughter filament growing as a branch requires Arp2/3 complex to undergo a dramatic conformational change where two blocks of structure rotate relative to each other about 25° to align Arp2 and Arp3 as the first two subunits in the branch. During branch formation, Arp2/3 complex acquires more than 8,000 Å2 of new buried surface, accounting for the stability of the branch. Inactive Arp2/3 complex binds only transiently to the side of an actin filament, because its conformation allows only a subset of the interactions found in the branch junction.
Arp2/3 complex nucleates actin filaments as branches on the sides of preexisting actin filaments (1) to produce dendritic networks of growing filaments that drive cellular and organelle movements (2). Branch formation depends on interactions of Arp2/3 complex with both nucleation-promoting factors (NPFs), and a preexisting actin filament known as the mother filament (3). Members of the WASp family of NPFs share motifs that bind actin monomers (called either a V or W motif) and Arp2/3 complex (the C and A motifs) (4). NPF binding to Arp2/3 complex favors binding to the side of an actin filament, which promotes branch formation; the V motifs bring along actin subunits for the daughter filament (5). The SPIN90 family of proteins favors nucleation of a daughter filament without binding to the side of a mother filament (6).
The conformation of inactive Arp2/3 complex in high-resolution crystal structures (7) differs from lower-resolution structures of Arp2/3 complex in branch junctions obtained by electron microscopy (8). Inspection of the crystal structure of inactive Arp2/3 complex (7), molecular dynamics simulations (9), spectroscopic measurements (10), chemical crosslinking (11, 12), and a cryo-EM structure of fission yeast Arp2/3 complex bound to Dip1 on a short daughter filament (6) all provided evidence for a major conformational change in Arp2/3 complex during branch formation that positions Arp2 next to Arp3 at the pointed end of the daughter filament. We refer to this as the short-pitch conformation as found in actin filaments. However, understanding the pathway of branch formation depends on knowing the structure of the product of the reactions, the branch junction with Arp2/3 complex connecting the daughter filament to the mother filament, at higher resolution than prior studies.
Here we report a cryo-EM structure of the branch junction with six actin subunits in the mother filament and two actin subunits in the daughter filament assembled from Mg-ATP actin and Arp2/3 complex. The resolution of 3.5 Å is sufficient to build a model with virtually all of the 4,934 residues including their side chains and the nucleotides associated with the actin subunits and the two Arps. During specimen preparation, all of the actins and Arp3 hydrolyzed their bound ATP and dissociated the γ-phosphate, but Arp2 retained the γ-phosphate. The conformation of the mother filament is virtually identical to a bare adenosine diphosphate-actin filament, but Arp2/3 complex undergoes a massive conformational change to align Arp2 and Arp3 like two subunits of an actin filament to form the base for the daughter filament. Two blocks of subunits in Arp2/3 complex rotate about 25° around the C-terminal α-helices of the ARPC2 and ARPC4 subunits, as predicted by prior work cited above. We used changes in buried surface area as a rough measure of free energy changes (13) for clues about the thermodynamics of the reactions. Comparisons of structures of inactive Arp2/3 complex and the branch junction suggest that the free energy for this massive conformational change comes from surface area buried by interactions with the mother and daughter filaments and internal re-arrangements. Readers of this paper will be interested in an independently determined 3.9 Å resolution cryo-EM structure of the branch junction with mammalian Arp2/3 complex (14). We compare the two structures in the Discussion and SI Appendix, Supplemental Materials.
Results
Quality of Cryo-EM Structure.
A three dimensional (3D) construction of the branch junction from a sample of 79,000 branches selected from 131,000 hand-picked particles in 4,200 motion-corrected images had a resolution better than 3.5 Å and includes seven subunits of Arp2/3 complex, six actin subunits in the mother filament, and two actin subunits in the daughter filament. Table 1 and SI Appendix, Fig. S1–S3 have details of the structure determination.
Table 1.
Statistics for data collection, map reconstruction, and model refinement
| Actin filament branch (EMD- 27962, (49) PDB 8E9B (50)) | |
|---|---|
| Data collection and map reconstruction | |
| Magnification | 36,657 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 50.07 |
| Defocus range (µm) (set/measured) | [−2.5, −1.5]/[−2.9, −0.9] |
| Pixel size (Å) | 1.364 |
| Symmetry | C1 |
| Initial particle images (no.) | 131,393 |
| Final particle images (no.) | 79,467 |
| Map analysis in reconstructed/modeled regions | |
| Resolution (Å) at 0.143 threshold, Relion | 3.53/3.47 |
| Sharpening B factor (Å2), Relion | −101.52/−74.09 |
| Resolution (Å) at 0.143 threshold, 3DFSC | 3.38/3.35 |
| Sphericity (out of one), 3DFSC | 0.963/0.966 |
| Map resolution range (Å) | 3.2–5.6 |
| Model Refinement | |
| Initial model used (PDB ID) | 1K8K/6DJO/6W17/AlphaFold |
| Model-to-map fit correlation | 0.77 |
| Model resolution (Å) at 0.5 threshold | 3.6 |
| Model composition | |
| Heavy atoms | 39,160 |
| Protein residues | 4,933 |
| Ligands (ATP/ADP/Mg2+) | 1/9/10 |
| B factors (Å2) | |
| Protein | 94.59 |
| Ligands | 87.65 |
| RMS deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.711 |
| Geometry validation | |
| MolProbity score | 1.91 |
| Clashscore | 10.43 |
| rotamer/Cβ outliers (%) | 0.05/0.00 |
| Ramachandran plot | |
| Favored (%) | 94.89 |
| Allowed (%) | 5.11 |
| Disallowed (%) | 0.00 |
The map clearly shows the backbone and complete side chains of most of the 4,934 residues (except acidic residues and highly flexible side chains) as well as the nucleotides for all eight actin subunits and both Arps (Figs. 1 and 2). The only parts missing from the map are the 3 N-terminal residues of each actin subunit, the D-loop of Arp3 (also disordered in Arp2/3 complex in the inactive state and bound to Dip1), residues 298–313 in the loop-helix-loop insertion of ArpC1, and the N-termini of all the subunits of Arp2/3 complex except ARPC2 and ARPC4.
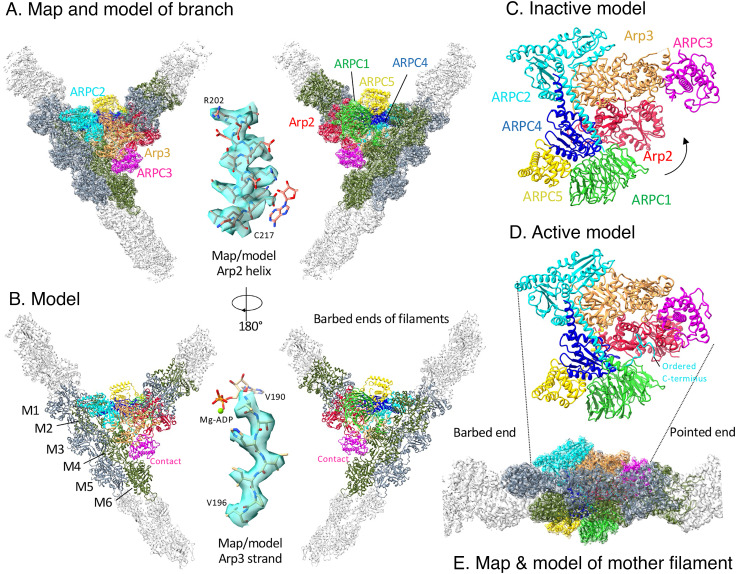
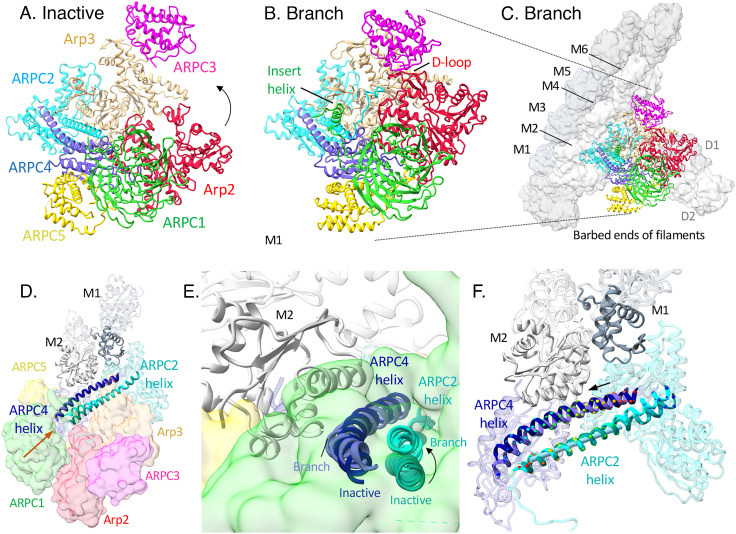
Fig. 1.
Map and model of the branch junction with color-coded subunits of Arp2/3 complex. (A) Two views of a semi-transparent rendering of the cryo-EM map around a ribbon diagram of the model. (B) Two views of ribbon diagrams of the model. In A and B, actin subunits close to Arp2/3 complex are colored slate gray on one strand and olive green on the other strand. Insets show higher magnification views of the map and model of an alpha helix from Arp2 and a beta strand from Arp3, both with stick figures of parts of the nucleotide. The stick rendering of the nucleotide shows the orientation. (C) Ribbon diagram of the classic view of inactive S. pombe Arp2/3 complex (cryo-EM structure; PDB: 6W18). (D) Ribbon diagram of the classic view of Arp2/3 complex in the branch junction. (E) Map and model of the branch junction, with Arp2/3 complex in the same orientation as panel D to show the mother filament relative to the classic view of Arp2/3 complex.
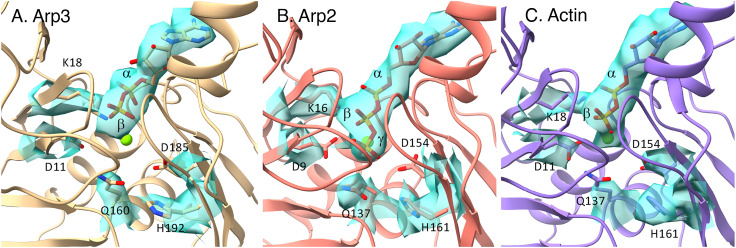
Fig. 2.
Maps and models of nucleotides in active sites of Arps and actin in the branch junction. (A) ADP in the active site of Arp3 without density for inorganic phosphate. The side chain of H192 is in an inactive conformation, similar to H161 in actin monomers. (B) ATP in the active site of Arp2 in spite of the catalytic residues Q137 and H161 being in active conformations. The distance from Q137 Cδ to ATP Pγ is 5.1 Å and that from H161 Nε2 to ATP Pγ is 6.2 Å; counterparts in the AMPPNP-actin filament are 4.7 Å and 6.7 Å. (C) ADP in the active site of mother filament actin subunit M1.
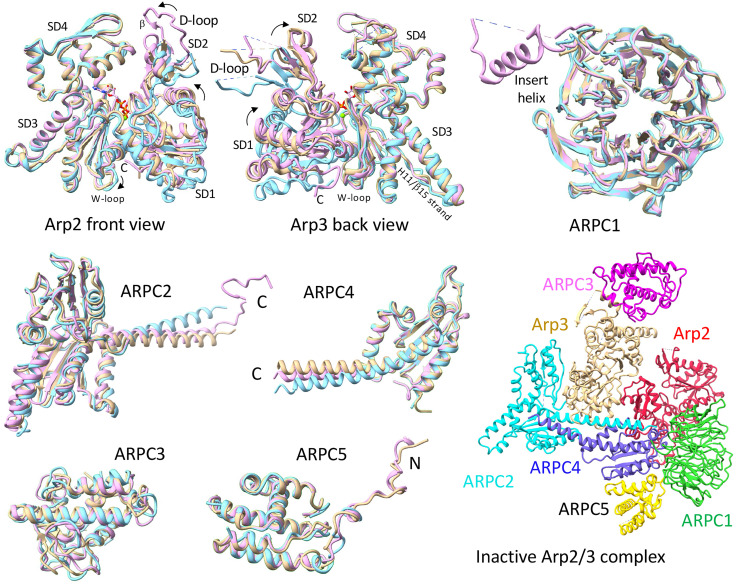
The quality of the map would have allowed building a model of the branch junction de novo, but we used prior knowledge to simplify the process. Our model of the actin filament (Chou Pollard, 2019) fits without modification of the backbone into the map for the six subunits of the mother filament and the two subunits of the daughter filament. To build a model of Arp2/3 complex in the branch junction, we took advantage of three sources of information: 1) the 2.0 Å resolution crystal structure of bovine Arp2/3 complex (7) and the 4.2 Å resolution cryo-EM structure of the free, inactive yeast Arp2/3 complex (6), which are very similar with an RMSD of 1.3 Å except for the presence of subdomains 1 and 2 of Arp2 in the electron microscope (EM) structure; 2) a 3.9 Å resolution cryo-EM structure of fission yeast Arp2/3 complex bound to Dip1 and a short daughter filament (6); and 3) models of the subunits generated by AlphaFold (15), which included information about side chains.
Comparisons with Previous Structures of the Branch Junction.
Our higher-resolution structure of branches formed by fission yeast Arp2/3 complex agrees overall with the structure of branches in mouse tissue culture cells (16), so the structure has been conserved over a billion years of divergent evolution. Both new structures share some features with the first three dimensional electron microscope (3D EM) structure of the branch junction (8), although that structure was limited to 25 Å resolution by negative staining and few particles (279) to image by electron tomography. In the Rouiller structure, the mother filament was distorted (perhaps by negative staining and drying), but is completely normal in our new structure. The Rouiller structure had the Arps positioned correctly as the base for the daughter filament, but the resolution was not good enough to place some other subunits correctly.
Nucleotides Bound to Arp2/3 Complex and Actin in the Branch Junction.
Actin.
All of the actin subunits in the mother and daughter filaments have adenosine diphosphate (ADP) bound in the active site (Fig. 2C), so they hydrolyzed ATP and dissociated the γ-phosphate during the 80 min of branch formation and preparation for freezing.
Arp2.
The map has a strong density for the γ-phosphate of ATP bound to Arp2 (Fig. 2B), which suggests at least 70% occupancy, and a clear density for Mg coordinated with the β- and γ-phosphates of the ATP. The conformations of the side chains of residues Q137 and H161 of Arp2 are identical to the catalytic residues in the active site of polymerized ADP-Pi actin (17). Nevertheless, Arp2 retained the γ-phosphate in the active site until the specimens were frozen. Dip1-activated Arp2/3 complex had ATP bound to Arp2 (6).
Arp3.
The map of the active site has strong densities for the base, ribose, and α- and β-phosphates of ADP, low density for the coordinated Mg2+ but no density for the γ-phosphate (Fig. 2A). Thus, Arp3 hydrolyzed its bound ATP and dissociated the γ-phosphate during the preparation of branches. Residues Q160 and H192 are homologs of Q137 and H161 of actin and Arp2. The conformations of these side chains differ from Arp2. Instead they are similar to the crystal structures of bovine Arp2/3 complex with ADP (18) and catalytically inactive actin monomers (19). Arp3 in Dip1-activated Arp2/3 complex retained ATP (6).
Arp Phosphorylation.
The map has no extra density at Arp2 residues Y198, T233, and T234 where phosphorylation was reported in Arp2/3 complex (20) or Arp2 residue Y217 where mass spectrometry revealed phosphorylation of fission yeast Arp2/3 complex (21).
Absence of VCA Density.
The branches were assembled in the presence of 550 nM GCN4-VCA, which activates Arp2/3 complex, but the map of the branch junction had no density corresponding to any part of the CA motif bound to Arp2, Arp3, or ARPC1 as seen in an EM structure of inactive Arp2/3 complex (22). This is expected for two reasons. First, residues 46–49 in the D-loops of the daughter filament subunits D1 and D2 should displace the α-helical C-motifs from Arp3 and Arp2 (22). Second, residues W169–T180 in the loop after the hinge helix (Q160–S168) of Arp3 in our structure are in a conformation that blocks binding of the VCA C-helix. Similarly, the verprolin-connecting-acidic (VCA) V-motif must dissociate from the barbed end grooves of actin subunits D1 and D2 before actin subunits D3 and D4 can bind (19).
Structure of the Mother Filament.
The backbones and helical parameters of the actin subunits in the mother filament and nearly identical to the bare actin filament (17) (Figs. 1 and 3 and SI Appendix, Table S1). We use the Rouiller numbering scheme for the mother filament subunits with subunit M1 as the first subunit at the barbed end of the interface with Arp2/3 complex and subunit M6 at the pointed end of the interface (8) (Fig. 1); this is opposite to the numbering by Fäßler et al. (16). See below for a description of the interactions between Arp2/3 complex and subunits in the mother filament. The orientations of some side chains of the actin subunits in the mother filament differ from a bare actin filament due to steric clashes with Arp2/3 complex or other factors.
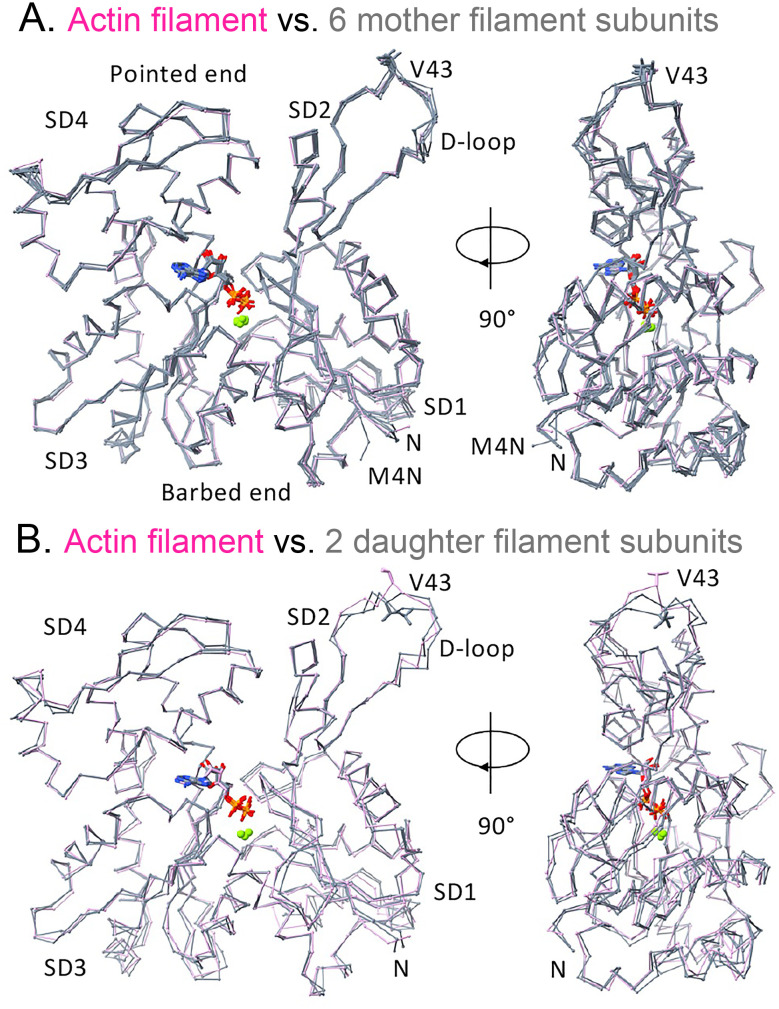
Fig. 3.
Backbone Cα-traces comparing the ADP-actin subunits of the mother and daughter filaments in the branch junction with the bare Mg-ADP-actin filament (PDB: 6DJO). All of these subunits are flattened like subunits in bare actin filaments. (A) Mother filament actin subunits M1–M6 (slate gray) superimposed on a subunit (plum) of bare Mg-ADP-actin filament. The side chain (stick) of V43 in the D-loop (SD2) faces the pointed end of the filament. The conformation of N terminus (rich in negative residues) of subunit M4 (M4N) differs from other actin subunits (N) due to interaction with the positively charged C-terminal loop of ARPC2, which is flexible in the absence of the mother filament. (B) Daughter filament subunits D1 and D2 (slate gray) superimposed on a subunit (plum) of the bare Mg-ADP-actin filament. The side chains (sticks) of V43 in both daughter subunits face the barbed end of the filament.
Structure of the Daughter Filament.
The map includes the first two actin subunits of the daughter filament (D1 and D2) (Figs. 1 and 2). Subunit D1 interacts with the barbed end of Arp3, much like subunits along the long-pitch helix of actin filaments, and with Arp2 and actin D2 like subunits along a short-pitch actin helix. Actin subunit D2 interacts longitudinally with Arp2 and laterally with actin D1. As in actin filaments, the D-loops of actins D1 and D2 make the major longitudinal contacts with Arp3 and Arp2. The conformations of the D-loops of both actin subunits are similar but different from those between subunits in actin polymers. For example, the side chain of V43 in both actin D-loops faces the daughter filament barbed end instead of the pointed end as in actin filaments (Fig. 3B).
Interactions of Daughter Filament Subunits with Arp2 and Arp3.
The longitudinal contact of actin D1 with Arp3 buries 1,395 Å2 of surface area, which is larger than between two subunits in an actin filament (1,180 Å2), because helix 11 is longer in Arp3 than actin. The 613 Å2 of surface area buried between actin D1 and Arp2 along the short-pitch helix is also larger than for two subunits in actin filaments (490 Å2), because the longer C-terminus of Arp2 (P386–T390) forms a loop that contacts actin D1 laterally. The D-loop of subunit D1 wraps around Y200 of Arp3, the homolog of actin Y169. The density of the side chain of D1 M44 is weaker than in actin filament structures (17), but as in actin filaments, M44 fits into a hydrophobic pocket formed by the W-loop (residues 196–203) and the hinge helix (residues 160–168) of Arp3.
The D-loop of actin subunit D2 binds to a surface of Arp2 homologous with actin but has a slightly different conformation (Fig. 3). The 978 Å2 of buried surface area between actin D2 and Arp2 (along the long-pitch helix) is slightly smaller than in actin filaments (1,180 Å2), because Arp2 residue V169 in middle of the D-loop of actin D2 is smaller than actin residue Y169. The side chain of actin D2 M44 fits into a hydrophobic pocket between the W-loop and the hinge helix of Arp2. Compared with actin, Arp2 has an insert (residues 322–332) after helix 11 (SI Appendix, Fig. S4) that interacts with the N-terminal residues 9–13 of ARPC5, burying 612 Å2 of surface area.
Interactions of Arp2/3 Complex Subunits with the Mother Filament.
Six subunits of Arp2/3 complex interact with five subunits of the mother filament (Fig. 1), burying a total of 3,381 Å2 of surface area (SI Appendix, Table S2). Our measurements agree in general with the contacts in the previous reconstructions of the branch junction (8, 16). However, the map of Fäßler et al. did not have a small contact of ARPC5 with the mother filament. Our structure does not have a direct contact of Arp2 with the mother filament, but residues 306–313 in the C-terminal extension of ARPC2 lie between the N-terminal residues 4–7 of M4 and a loop (residues 244–247) in SD4 of Arp2, which was interpreted as a direct contact at low resolution (8, 16).
In keeping with biochemical evidence that isolated ARPC2/ARPC4 can bind actin filaments with micromolar affinity (23), these two subunits contribute 60% of the total surface area buried on the mother filament. The ARPC1 insert helix (residues T318–N326; see Fig. 4C) contributes the next most buried surface.
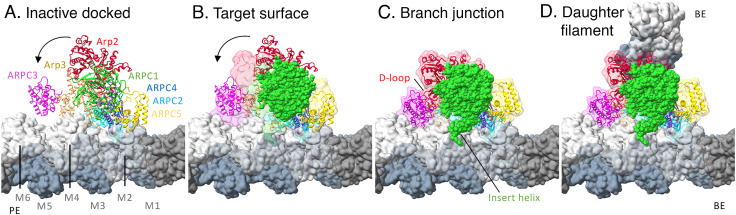
Fig. 4.
Docking inactive Arp2/3 complex on the mother actin filament followed by morphing to the active conformation in the branch junction. The space filling elements are surface representations of the model. (A) Inactive Arp2/3 complex docked on the mother actin filament. The two globular domains of ARPC2 and the outer domain of Arp3 in the branch junction were used to dock the inactive Arp2/3 complex on the mother filament. This buries 2,206 Å2 of surface area but precludes interactions of other subunits including Arp2, ARPC1, and ARPC3 with the mother filament. (B) Same as (A) but with a semi-transparent surface of Arp2/3 complex in the branch junction. (C) Active conformation of Arp2/3 complex in the branch junction with 3,337 Å2 of buried surface area against the mother filament. To reach this conformation a block of subunits consisting of Arp2 (red), ARPC1 (green), ARPC4 (slate blue), and ARPC5 (gold) rotates by about 25°. This rotation allows the D-loop of Arp2 to bind ARPC3, the insert helix of ARPC1 to bind mother filament subunit M4, ARPC3 to bind subunits M6, Arp3, ARPC2, and ARPC5 to increase their interactions with the mother filament. (D) Active Arp2/3 complex with the pointed ends (PE) of daughter filament subunits D1 and D2 bound to Arp3 and Arp2.
To explore why inactive and partially activated Arp2/3 complex binds slowly, weakly (24), and transiently to actin filaments (25), we used the branch structure to guide docking of ARPC2 and ARPC4 in the inactive Arp2/3 complex on an actin filament (Fig. 4A) as originally done by Goley et al. (26). The buried surface area is 1,100 Å2 less than in the branch junction (SI Appendix, Table S2), because the conformation of inactive Arp2/3 complex precludes many interactions found in the branch junction, contributing to the low affinity.
Conformational Changes in Arp2/3 Complex During Branch Junction Formation.
Comparing the cryo-EM structures of inactive fission yeast Arp2/3 complex (6) and Arp2/3 complex in the branch junction revealed that two blocks of structure rotate relative to each other during branch formation to position the Arps to initiate the daughter filament (Movie S1 and Figs. 4 and 5). The block with Arp2 includes ARPC1, ARPC4, and ARPC5, and the block with Arp3 includes ARPC2 and ARPC3. Morphing between the two structures illustrates the complicated motions dramatically (Movie S1). The following sections detail the conformational changes in each subunit.
Fig. 5.
Ribbon diagrams and space filling models of inactive Arp2/3 complex and Arp2/3 complex in the branch junction. (A) Inactive Arp2/3 complex. (B) Arp2/3 complex in the branch junction. (C) Arp2/3 complex in the branch junction including the mother and daughter filaments. (D and E) Changes during branch formation of the C-terminal helices of ARPC2 (inactive, dark cyan; branch, cyan) and ARPC4 (inactive, dark slate blue; branch, slate blue) shown as ribbon diagrams, semitransparent surfaces of other Arp2/3 complex subunits, and ribbon diagrams of mother filament subunits M1 (slate gray) and M2 (gray). The structures were aligned using the globular domains (1 and 2) of ARPC2 and subdomains 1 and 2 of Arp3. The orange arrow in (D) shows the view angle in (E). (F) During branch formation, bending and sliding rotate the C-terminal helices of ARPC2 and ARPC4. The two helices also move apart by 3.1 Å. Ribbon diagrams of ARPC2 and ARPC4 and mother filament subunits M1 and M2 in the branch junction with emphasis on the C-terminal helices of ARPC2 and ARPC4 (color code as in (E)). The structures were aligned on the long segment of ARPC2 C-terminal helices with a straight rod inside to show the bending of the ARPC2 C-terminal helix and sliding of ARPC4 C-terminal helix relative to ARPC2 C-terminal helix. Cα atoms in the two helices are rendered as colored spheres. The arrow indicates the direction of sliding.
Arp2.
Our map of the branch junction has clear density for the polypeptide from residue S3 to T389 including the D-loop of 15 residues (P36-K50), which is ordered except for residue G45. The D-loop is disordered in the cryo-EM structures of fission yeast Arp2/3 complex with or without Dip1 (6). The C-terminus of Arp2 in the branch (missing just T390) is more ordered than that in the structures of Arp2/3 complex alone (missing L385–T390) or with Dip1 and a daughter filament (missing R388–T390).
Conformational changes during branch formation move Arp2 next to Arp3, back-to-back with a half-subunit stagger, nearly identical to the short-pitch helix of an actin filament (SI Appendix, Table S1). This shift of about 25 Å eliminates the end-to-end contacts between subdomain (SD) four of Arp2 with subdomains 1 and 3 of Arp3, which preclude the addition of actin to Arp3 in inactive Arp2/3 complex. The net result is a modest increase of 220 Å2 (SI Appendix, Table S3) in the buried surface area between Arp2 and Arp3.
This shift allows the D-loop of Arp2 to circle around residues P113–P120 of ARPC3 and to form an anti-parallel β-sheet between Arp2 residues L38–R39 (Fig. 6, marked with β) and ARPC3 residues N59–E61. This intermolecular β-sheet is similar in concept to the intermolecular parallel β-sheet between the D-loop of actin and DNase-I (27). The D-loop of Arp2 does not have methionine residues corresponding to actin M44 or M47, which stabilize filaments.
Fig. 6.
Ribbon diagrams comparing the conformations of the subunits in fission yeast Arp2/3 complex in three cryo-EM structures: (sky blue) inactive structure; (tan) bound to Dip1; and (pink) branch junction. Curved arrows indicate domain rotations. The symbol β marks the part of the Arp2 D-loop that forms a β-sheet with ARPC3. The ribbon diagram at the lower right is the inactive Arp2/3 complex (PDB: 6W18) for orientation. The C-terminal helices of ARPC2 and ARPC4 bend more in the Dip1-activated Arp2/3 complex than in the branch junction.
The conformation of Arp2 in the branch junction differs in two ways from inactive Arp2/3 complex (SI Appendix, Table S5). First, the nucleotide-binding cleft is closed more tightly around the bound ATP than in inactive or Dip1-activated Arp2/3 complex. The distances between T12 Cα and G158 Cα are 7.7 Å in the inactive state, 5.9 Å in the Dip1-activated state, and 5.1 Å in the branch junction. Second, Arp2 is flattened by rotation of subdomains 1 and 2 relative to subdomains 3 and 4. The changes are 11.3° (from −10.1° to 1.2°) in the branch junction and 8.6° (from −10.1° to −1.5°) in the Dip1-Arp2/3 complex structure with a daughter filament (Fig. 6). Flattening Arp2 opens the pocket between the hinge helix and W-loop for binding the side chain of actin D2 M44 (Fig. 6), but does not provide a pocket for binding actin residue V43, which faces the opposite direction as in actin filaments.
The changes in the backbone dihedral angles required to close the nucleotide-binding cleft and flatten Arp2 are relatively minor (SI Appendix, Table S4 and Fig. S4). Other changes in dihedral angles cause minor changes in the conformations of surface loops unrelated to the re-arrangement of the subunits (SI Appendix, Fig. S4). However, rotation of the Arp2 block of subunits during branch formation causes a steric clash with Arp3 that distorts the loop between Arp2 helices 5 and 6 (residues 199–207) (SI Appendix, Fig. S4).
Arp3.
The rotation of the two blocks of subunits during branch formation produces a steric clash between Arp2 and Arp3 accompanied by bending of Arp3 subdomains 3 and 4 by +13.7° to flatten Arp3 and close the nucleotide-binding cleft (Movie S1). Small changes of backbone dihedral angles of residues 368–374 (strand running parallel to helix 11) account for most of the movement of subdomains 3 and 4 (SI Appendix, Table S4) but are difficult to appreciate in ribbon diagrams (Fig. 6 and SI Appendix, Fig. S4). Compared with the branch junction, Arp3 in the Dip1 complex does not close the nucleotide-binding cleft as much and the dihedral angle between subdomains 1/2 and subdomains 3/4 changed only half as much (+7.3°) (SI Appendix, Table S5). Consequently, Arp3 is flatter in the branch junction than in the complex with Dip1, but not as flat as Arp2 in the branch junction or actin in the filament.
Flattening Arp3 causes multiple changes that favor addition of daughter subunit D1. 1) Flattening opens the hydrophobic pocket between the hinge helix and W-loop for binding the important side chain of M44 in the D-loop of actin (Fig. 6). 2) Flattening changes the conformation of a loop of residues (W169–T180) (SI Appendix, Fig. S4), so that it interferes with the VCA C-helix binding in the position observed in the inactive Arp2/3 complex (22), where the C-helix interferes with binding of the actin D-loop. The same is observed in Dip1-activated state (6). Dissociation of the C-helix should favor actin binding. 3) Flattening also changes the conformation of the Arp3 C-terminal residues A417–L427. Instead of interfering with binding of the D-loop of subunit D1 and inhibiting activation (11), residues F423–L427 are fully ordered and bent 120° between residues I422–F423 to allow actin binding and interactions with Arp2. These interactions include the backbone of Arp3 F423 with side chain of Arp2 R195; Arp3 N425 with H265 and L266 in the H-plug (residues F262–P272) of Arp2; and Arp3 L427 with Arp2 L223, H223, and Q263. Similarly, the C-terminus is less ordered (partially ordered from A417 to Y420) in the Dip1-activated state (6), allowing nucleation of the daughter filament.
New contacts of ARPC3 with Arp3 stabilize two loops in subdomain 4, residues 226–234 at the end of helix 5 and 261–265 at the end of helix 7 (SI Appendix, Table S4 and Fig S3) that lack density in prior EM (PDB: 6W18) and crystal (PDB: 3DWL) structures of inactive Arp2/3 complex. The ß12 and ß13 loop becomes ordered in the branch structure and contacts SD4 of M3.
ARPC1.
The conformation of the β-propeller domain is nearly identical in the inactive, Dip1 and branch structures, but differs for the loop-helix-loop of 49 residues (F293–N341) between blades 6 and 7. The whole loop-helix-loop is missing in the cryo-EM structures of fission yeast Arp2/3 complex with or without Dip1 (6), and only the helix is present in crystal structures of inactive Arp2/3 complex where it makes a crystal contact with Arp3 in an adjacent molecule (7).
The branch junction map has density for all the residues of the loop-helix-loop except S298–G313 of the first loop; these densities reveal new contacts not seen in inactive Arp2/3 complex. The second loop interacts with the C-terminal helices of both ARPC2 and ARPC4. The ARPC1 insert helix (residues T318–N326) interacts with the C-terminus of ARPC2, burying 229 Å2 of surface (SI Appendix, Table S3), as well as barbed end groove of the mother filament subunit M4, as modeled previously in lower-resolution maps (8, 16), burying 491 Å2 of surface (Fig. 4C and SI Appendix, Table S2). Helices in LifeAct (28) and the N-WASP V/WH2-motif (19) also bind to the barbed end groove, although the orientation of the V-helix is opposite.
ARPC2.
The conformations of the two homologous globular domains do not change during branch formation, but rotation round the long C-terminal helix (residues 255–297) moves the two blocks of structure. This helix is anti-parallel to the C-terminal helix of ARPC4. During branch formation, the two helices slide longitudinally and rotate laterally against each other reducing their contact area by 7.6% (176/2,309 Å2). In addition, the ARPC2 helix bends about 9° at residues 264–269 in the branch junction and about 12° in Dip1-activated structure (Figs. 5 and 6).
The branch structure has backbone densities for C-terminal residues D301–A317 of ARPC2 (Fig. 5) that are disordered in other structures. The side chain densities are weaker for the last residues. Positively charged residues in this random coil interact with the negatively charged N-terminal residues of mother filament subunit M4 (Fig. 1 A and B).
Fission yeast ARPC2 has a 17-residue insertion (residues 134–150) (Fig. 1C) not found in mammalian ARPC2. This insertion may enhance interactions with actin subunit M1.
ARPC3.
Only Arp3 contacts ARPC3 in inactive Arp2/3 complex, but rotation of the two blocks of structure during branch formation establishes new contacts that bury 931 Å2 of new surface, more than any other subunit (SI Appendix, Table S3). The overall conformation of ARPC3 is stable (Fig. 6) as the contacts with Arp3 increase and new contacts with the D-loop of Arp2 (Figs. 1D and 5B) and mother filament subunit M6 (Figs. 1B and 5C) form.
ARPC4.
The C-terminal α-helix (residues 128–165) of ARPC4 undergoes major changes in the branch structure: i) It bends about 8° at its origin in the globular domain near residues 141–147 (Figs. 5 and 6); ii) it twists about 25° relative to the ARPC2 helix (Fig. 5 D and E); iii) the ARPC4 and ARPC2 helices move apart by 3.1 Å to maintain interactions between their side chains (Fig. 5F). The changes are similar in the Dip1 structure except the ARPC4 helix bends about 12°. These changes allow the Arp2 block to rotate relative to the Arp3 block.
On the other hand, the conformation of the single globular domain (homologous to the globular domains of ARPC2) is similar in the inactive, Dip1, and branch structures (Fig. 6). Contacts with R95 in subdomain 1 of mother filament subunit M2 stabilize ARPC4 N-terminal residues M1 and S2 in the branch junction.
ARPC5.
The conformation of the globular domain is similar in all available structures, but the N-terminal residues (T2–E13) vary. They are absent in the EM structure of inactive Arp2/3 complex but contact Arp2 residues 322–332, ARPC1 and ARPC4 in the branch structure (SI Appendix, Table S3), and the Dip1 structure. Tension between the globular domain of ARPC5 and subdomain 3 of Arp2 may influence the order of the N-terminal residues of ARPC5. These two anchors are separated by 44.8 Å in the ordered branch junction structure and 42.4 Å in the less ordered structure of inactive bovine Arp2/3 complex (7).
Discussion
The structure of the branch junction at 3.5 Å resolution reveals in unprecedented detail the conformation of fully active Arp2/3 complex bound to five subunits of the structurally unperturbed mother filament and the base of the daughter filament (Fig. 1). The structure confirms the large-scale conformational change in Arp2/3 complex revealed by lower-resolution reconstructions from electron tomograms of negatively stained branches (8) and cryo-electron tomograms of the leading edge of motile mouse fibroblasts (16). During steered molecular dynamics simulations of Arp2/3 complex (9), the Arp2 block of structure including ARPC1, ARPC4, and ARPC5 rotated up to 25° relative to the Arp3 block of structure including ARPC2 and ARPC3. Fäßler et al. (16) confirmed the main features of the Dalhaimer mechanism. Rotation between the inactive and branch junction structures (Movie S1 and Figs. 4 and 5) aligns the two Arps back-to-back like two subunits along the short-pitch helix of an actin filament, so they can nucleate a daughter filament. The conformation of Arp2/3 complex in the branch junction is similar to that bound to Dip1 with a daughter filament (6), although the extent of many changes differs as noted throughout the Results.
Mechanics of the Activating Conformational Change of Arp2/3 Complex.
The anti-parallel C-terminal helices of ARPC2 and ARPC4 serve as the axis of rotation during the transition from inactive Arp2/3 complex to the short-pitch active conformation. The C-terminal helix of ARPC4 bends 8°, rotates 25°, and translocates 3.1 Å relative to C-terminal helix of ARPC2 (Figs. 5 and 6). This motion rotates of the two blocks of structure and positions the Arps next to each other. Steric interference between Arp2 and Arp3 during the rotation flattens both subunits and bends Arp3 subdomain 4 relative to subdomain 3. Subdomain 4 carries along ARPC3, which forms new contacts with both Arps that bury 931 Å2 of new surface, contributing a free energy change that stabilizes the active conformation of the complex ready to nucleate the daughter filament. The conformational changes extend to every subunit of Arp2/3 complex (Fig. 6).
Equilibrium Between Inactive and Active Arp2/3 Complex.
The structures of inactive Arp2/3 complex and the branch junction define two stable states of Arp2/3 complex. On its own, Arp2/3 complex is virtually all in the low energy inactive state. In the branch junction, Arp2/3 complex is in the high energy short-pitch conformation, but is also stable, because new contacts with the mother and daughter filament and internal changes bury a massive amount of new surface area (>8,000 Å2) and provide a large free energy change to stabilize the branch. We use buried surface changes to estimate free energy changes (13).
Crosslinking experiments provide strong evidence for an equilibrium between inactive and active Arp2/3 complex (11), where Arp2/3 complex spontaneously visits a conformation with the Arps close enough to the short-pitch helix conformation to be trapped irreversibly by chemical crosslinking of cysteine substitutions. However, the very slow pace of crosslinking between Arp2 and Arp3 indicates that little free Arp2/3 complex is in the short-pitch active state. The likely explanation is a substantial free energy barrier between the inactive and active states. Indeed, 11 of 12 steered molecular dynamics simulations of activation stalled with the Arps only part way toward the short-pitch conformation in spite of the applied external force (9). Morphing the structures of Arp2/3 complex from the inactive state to the branch junction (Movie S1) suggests that subdomains 3 and 4 of Arp3 are one barrier for the rotation of the Arp2 block. The barrier to the short-pitch conformation appears to be lower for budding yeast Arp2/3 complex, since it is spontaneously active (29) and crosslinks faster (11) than human Arp2/3 complex (22).
Binding two VCAs to free Arp2/3 complex shifts the equilibrium of conformational states toward the short-pitch conformation that can be trapped irreversibly by chemical crosslinking of cysteine substitutions (11). WDS proteins Dip1 (30) and SPIN90 (31) also increase the rate of crosslinking. VCA also favors both low and high affinity binding to the mother filament (24, 25). It is not known how bound VCA increases the probability of the high energy state.
However, even with bound VCA, the occupancy of the short-pitch conformation is low as indicated by the low probability of branching from Arp2/3 complex on an actin filament (25). Furthermore, no short-pitch conformations were detected in cryo-EM samples of human Arp2/3 complex saturated with two NPFs (22), and WDS proteins do not change the structure of inactive Arp2/3 complex (30) unless a daughter filament forms in the presence of VCA (6). VCA binding increases the efficiency of energy transfer modestly between Arp2 and Arp3, but less than binding to actin filaments (10).
Therefore, branch formation depends on interaction with a mother filament, which provides the free energy change to stabilize the short-pitch conformation (10). This differs from the proposal that NPFs favor a conformation stable enough to bind the mother filament and nucleate a branch without an equilibrium between the inactive and short-pitch conformations on the mother filament (25).
We suggest that full engagement of Arp2/3 complex with the mother filament provides the free energy change stabilizing the rotated state with extensive conformational changes throughout Arp2/3 complex, including flattening of Arp2 and Arp3. Narvaez-Ortiz and Nolen (32) argued against such a “concerted” mechanism, whereby VCA-actin and mother filaments act together to flatten and rotate the Arps into position. They proposed instead that VCA-actin drives twisted Arps into short-pitch dimer conformation followed by flattening of the Arps after binding to the mother filament.
Pathway of Branch Formation.
Arp2/3 complex appears to bind actin filaments in two stages: a weak, transient state that rarely converts to a stable state. Most single molecules of budding yeast Arp2/3 complex bind transiently to actin filaments with residence times less than 2 s without initiating a branch (25). We tested the hypothesis that the low affinity, transient species is inactive Arp2/3 complex by using the branch junction structure to dock ARPC2 and ARPC4 on a filament along with the rest of inactive Arp2/3 complex. The model with inactive Arp2/3 complex on the actin filament buries 2,206 Å2 of surface, including small contributions from Arp3 and ARPC5 (Fig. 4A and SI Appendix, Table S2). However, Arp2, ARPC1, and ARPC3 are physically separated from the filament surface, likely lowering the lifetime and the probability of rotating to the fully engaged active state. The Goley et al. model of inactive Arp2/3 complex on the mother filament also lacked these contacts (26).
On the other hand, stable binding of fission yeast Arp2/3 complex to actin filaments is extremely slow, k+ ~10−5 µM−1 s−1 (24), when measured by an increase in fluorescence of pyrene conjugated to a cysteine replacing alanine at the C-terminus of ARPC2 (33). This part of ARPC2 is disordered in inactive Arp2/3 complex and the Dip1 complex (Fig. 6) but becomes ordered during branch formation (Figs. 1 and 6), so the fluorescence change probably reflects an environmental change that likely depends on rotation toward the active state that can make a full set of interactions with the mother filament. NPFs favor this reaction by shifting the equilibrium from the inactive state toward active Arp2/3 complex as documented by chemical crosslinking (11, 12). Thus, we propose that strong binding of Arp2/3 complex to the mother filament requires a low probability, spontaneous conversion of Arp2/3 complex to the high energy short-pitch conformation on the mother filament.
Nucleation of the Daughter Filament.
Branch formation requires the alignment of the Arps in the short-pitch configuration and flattening to open critical sites for binding the D-loops of daughter subunits D1 and D2. Daughter filament subunit D1 interacts with Arp3, Arp2, and actin D2, and daughter filament subunit D2 interacts with Arp2 and actin subunit D1, all similar to subunits in an actin filament. The branch structure shows that flattening Arp3 is associated with two changes that open the binding site for the D-loop of actin D1. First, a change in the conformation of the C-terminal residues moves them out of binding site for the actin D-loop. Second, a change in the conformation of a loop (residues W169–T180) displaces the C-helix of VCA from Arp3. This change in the W169–T180 loop is also seen in the Dip1-activated state (6).
Flattening Arp2 opens the binding site for actin M44. Binding of the C-helix to Arp2 in inactive Arp2/3 complex would interfere with binding of actin D2 (22). Flattening Arp2 opens up the binding site for the C-helix and may favor its dissociation and the addition of actin D2.
The V motifs of NPFs are thought to deliver actin subunits to the Arps (5), but questions remain about the mechanism, since the C-motifs of the two NPFs must dissociate from Arp2 and Arp3 before daughter filament subunits D2 and D1 can bind to start the daughter filament. Perhaps the A motif alone can anchor VCA to Arp2/3 complex, allowing the V-motif to deliver actin subunits to the Arps.
Different mechanisms flatten the Arps in Arp2/3 complex and actin during polymerization (SI Appendix, Table S4 and Fig. S3). Rotation of the two blocks of structure produces a network of conformational changes that flatten Arp2 and Arp3, presumably before daughter filament subunits bind. On the other hand, in molecular dynamics simulations, actin subunits at the barbed end of a filament flattened progressively as they acquired neighboring subunits (34).
Nucleotide Hydrolysis by Schizosaccharomyces pombe Arp2/3 Complex.
The branch structure shows directly that Arp2 and Arp3 hydrolyze their bound ATP at substantially different rates. Like all seven actin subunits in the structure, Arp3 hydrolyzed its bound ATP and dissociated the γ-phosphate during the 80 min required for specimen preparation to leave just ADP in the active site. On the other hand, Arp2 retained most of the γ-phosphate, so hydrolysis and/or phosphate release was slower in spite of the catalytic residues in the active site being positioned to promote hydrolysis. ATP is bound to both Arps in the complex with Dip1 in spite of nucleating a daughter filament (6).
Our observation of the γ-phosphate in the active site of Arp2 in the branch junction is surprising given previous biochemical analysis and mutations of the active site. Dayel et al. (35) found that hydrolyzable ATP bound to Arp2/3 complex is required for branch nucleation, although the ATP analog may have influenced the observations. Both Dayel et al. (36) and Martin et al. (37) reported that Arp2 hydrolyzes its bound ATP coincident with actin filament nucleation. Martin et al. (38) found that mutations of residues in the active site of budding yeast Arp2 and Arp3 can compromise ATP binding and cause defects in clathrin-mediated endocytosis. However, mutations compromising ATP hydrolysis did not prevent budding yeast (38) or Drosophila (39) Arp2/3 complex from nucleating actin assembly but stabilized lamellipodial actin filament networks.
These differences from the nucleotides in the branch junction suggest that new work is needed on the order and rates of these events: formation of a short-pitch Arp2/Arp3 dimer; flattening of Arp2 and Arp3; delivery of actin subunits D1 and D2 from VCA to the Arps; dissociation of VCA; nucleotide hydrolysis; and phosphate release.
Comparison of the Structures of S. pombe and Bovine Arp2/3 Complex Branch Junctions.
In parallel with our work, Ding et al. determined a 3.9 Å resolution cryo-EM structure of the branch junction with bovine Arp2/3 complex (14). The two structures are nearly identical, although they differ in some details, illustrated in SI Appendix, Fig. S5 and enumerated in SI Appendix, Tables S6 and S7. For example, the 36 residue D-loop of Arp3 is mostly disordered in S. pombe branches, while the 22 residue D-loop is fully ordered in the bovine branch. On the other hand, the C-terminal 17 residues of ARPC2 are disordered in bovine but not S. pombe branches.
Inactive S. pombe Arp2/3 complex has several disordered regions in both crystal (40) and cryo-EM structures (6). Disordered residues 226–233, 261–265, and 271–279 in subdomain 4 of Arp3 in inactive S. pombe Arp2/3 complex fold during branch formation (SI Appendix, Fig. S4), while these residues are already folded in inactive bovine Arp2/3 complex (7). The C-terminus of S. pombe ARPC2 folds during branch formation establishing contacts with Arp2 and actin M4. Similarly, the N-terminus of S. pombe ARPC5 folds during branch formation establishing contact with Arp2. Folding of these disordered regions during branch formation contributes to the larger increase in internal buried surface area of S. pombe Arp2/3 complex compared with bovine Arp2/3 complex (SI Appendix, Tables S3 and S4). The two papers propose different pathways for branch formation.
Materials and Methods
Protein Purification.
We used published methods described in detail in SI Appendix, Supplemental Materials: chicken skeletal muscle actin (41); S. Arp2/3 complex (24); recombinant Saccharomyces cerevisiae GCN4 leucine zipper (residues 249–281) (UniProt ID: P03069) fused to bovine N-WASP VCA motif (residues 401–505) (UniProt ID: Q95107); and recombinant mouse capping protein CapZ α1/β2 heterodimer (42).
Assembly of Specimens with Short Branches.
The Ca2+ bound to actin monomers was exchanged for Mg2+ by incubating 50 µL of 22.5 µM Ca-ATP-actin monomers with 0.1 volumes of 10× ME buffer (0.5 mM MgCl2, 2 mM EGTA, and pH 7.5) at room temperature for 10 min. Actin monomers were polymerized into filaments by sequentially mixing 15 µL of 3.2 µM capping protein in 100 mM KCl, 10 mM HEPES, pH 7.0, 20 µL of 1× KMEH buffer (100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7.0, 1 mM DTT), and 35 µL of 20 µM Mg-ATP-actin monomer (10 µM actin monomers and 0.69 µM capping protein in 70 µL) and incubating at room temperature for 1 h. Separately, 1.0 µL of 25 µM Arp2/3 complex in QB buffer was pre-activated by mixing with 1.15 µL of 50 µM GCN4-VCA in KHT buffer and 1.0 µL of 10 mM ATP (pH 7.0 in H2O) and incubating at 4°C for 1 h before mixing with 70 µL of capped actin filaments from the previous step and incubating at room temperature for 5 min. Capped daughter filaments were grown in five cycles by adding sequentially to Arp2/3 complex with capped mother filaments 1.0 µL of 0.1 mM ATP, 1.0 µL of 3.2 µM capping protein, and 1.0 µL of 20 µM Mg-ATP-actin monomers. After each cycle, samples were incubated at room temperature for 5 min. The final sample contained in addition to the capped mother filaments 0.28 µM Arp2/3 complex, 1.15 µM actin, and 0.18 µM capping protein. The sample was directly used for vitrification in the next step without dilution. The density of branch junctions in electron micrographs of negatively stained specimens did not decrease overnight at 4°C, so the branches are stable for hours.
Sample Freezing and Image Acquisition.
As described in more detail in SI Appendix, Supplemental Materials, 3.0 µL of branched actin filaments were applied onto the carbon side of a holy carbon Quantifoil 1.2/1.3 300-mesh Au grids, attached to a Mark IV Vitrobot at 4°C and ~100% humidity and frozen by plunging into liquid ethane cooled to about −180°C. The ice thickness was estimated to be between 70 and 130 nm, thick enough to reduce the preferred orientation of the branches. Electron micrographs for image reconstruction were collected from two grids in a Titan Krios microscope equipped with a high brightness field emission gun (XPEG) at 300 kV, a nanoprobe, and a Gatan image filter (slit width: 20 eV). Movies were recorded at a series of defocus values between −2.5 µm and −1.2 µm on a K3 Summit camera in super-resolution mode, using the beam image shift strategy (nine holes each with a movie per stage movement) implemented in SerialEM. Each movie had 41 frames, and each frame time was 0.08 s. The dose rate at camera level was 28.4 counts/pixel/s, and physical pixel size was 1.364 Å.
EM Image Processing.
The dataset of images was processed mostly using Relion 4.0 (43), including the steps from motion correction and contrast transfer function (CTF) estimation to map post-processing, except for the particle picking step. Stand-alone Topaz (44) AI-based particle picking reliably identified branches with the branch lying approximately in the x-y plane of the grid but missed most branches oriented near the z-axis, which could only be picked by hand to assure a random sample of orientations around the filament axis. Manual picking was performed in Fesp (https://github.com/stevenzchou/Fesp), a program written in wxPython. A 3D reconstruction from a sample of 79,000 from a collection of 131,393 hand-picked branches gave a map with 3.5 Å resolution (Table 1).
Model Building and Refinement, and Structural Analysis and Visualization.
Our map allowed us to build models for most residues unambiguously in Coot (45). We also used a model of the Arp2/3 complex determined by cryo-EM (PDB: 6W18) and model of the individual subunits of Arp2/3 complex generated by AlphaFold (15) as references for cross-validation. We refined the modeled structure using PHENIX (46) in real space. Interdomain rotation angles were calculated with PyMOL. Rise (subunit translation) and twist (subunit translation), RMSDs, and buried surface areas were calculated using Chimera (47). Figures were generated using ChimeraX (48).
Supplementary Material
Appendix 01 (PDF)
Proposed pathway of branch formation. Step 1, inactive Arp2/3 complex is docked to the side of a mother filament using the interactions of the two globular domains of ARPC2 and the outer domain of Arp3 in the branch junction. Inactive Arp2/3 complex binds weakly and transiently (not shown). Step 2: Morphing from the structure of inactive Arp2/3 complex to its structure in the branch junctions shows the rotation of the two blocks of structure bringing Arp2 and Arp3 together in a short-pitch conformation and flattening both Arps. Step 3, the flattened Arps, aligned in the short-pitch conformation, nucleate the daughter filament.
Acknowledgments
Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01GM026132 and R01GM026338. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Ed Taylor for advice about free energy changes, Emily Guo for advice on sample freezing and for manually picking 60% of the particles, Shenping Wu, Jianfeng Lin, Marc Llaguno, Kaifeng Zhou, and Xinran Liu for managing the Yale Electron Cryo-Microscopy Resources, and the Yale Center for Research Computing for guidance and use of the research computing infrastructure.
Author contributions
S.Z.C., M.C., and T.D.P. designed research; S.Z.C., M.C., and T.D.P. performed research; S.Z.C. contributed new reagents/analytic tools; S.Z.C., M.C., and T.D.P. analyzed data; and S.Z.C., M.C., and T.D.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Electron microscopy structure maps and molecular models data have been deposited in Electron Microscopy Data Bank (EMDB) under accession number EMD-27962 (https://www.ebi.ac.uk/emdb/EMD-27962) (49) and Protein Data Bank (PDB) under accession number 8E9B (https://www.rcsb.org/structure/8E9B) (50).
Supporting Information
References
- 1.Mullins R. D., Heuser J. A., Pollard T. D., The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molinie N., Gautreau A., The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 98, 215–238 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Machesky L. M., et al. , Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 96, 3739–3744 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchand J. B., Kaiser D. A., Pollard T. D., Higgs H. N., Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76–82 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Gautreau A. M., Fregoso F. E., Simanov G., Dominguez R., Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol. 32, 421–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaaban M., Chowdhury S., Nolen B. J., Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson R. C., et al. , Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Rouiller I., et al. , The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalhaimer P., Pollard T. D., Molecular dynamics simulations of Arp2/3 complex activation. Biophys. J. 99, 2568–2576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinoza-Sanchez S., Metskas L. A., Chou S. Z., Rhoades E., Pollard T. D., Conformational changes in Arp2/3 complex induced by WASp-VCA and actin filaments. Proc. Nat. Acad. Sci. U.S.A. 115, E8642–E8651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodnick-Smith M., Liu S. L., Balzer C. J., Luan Q., Nolen B. J., Identification of an ATP controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat. Commun. 7, 12226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodnick-Smith M., Luan Q., Liu S. L., Nolen B. J., Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3834–E3843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Sawyer N., Regan L., Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 22, 510–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding B., et al. , Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 119, e2202723119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fäßler F., Dimchev G., Hodirnau V. V., Wan W., Schur F. K. M., Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nat. Commun. 11, 6437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Nat. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolen B. J., Pollard T. D., Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol. Cell 26, 449–457 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chereau D., et al. , Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeClaire L. L., Rana M., Baumgartner M., Barber D. L., The Nck-interacting kinase NIK increases Arp2/3 complex activity by phosphorylating the Arp2 subunit. J. Cell Biol. 208, 161–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein A. E., Espinoza-Sanchez S., Pollard T. D., Phosphorylation of Arp2 is not essential for Arp2/3 complex activity in fission yeast. Life Sci. All. 1, e201800202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmet A., et al. , Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci Adv. 6, eaaz7651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gournier H., Goley E. D., Niederstrasser H., Trinh T., Welch M. D., Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell 8, 1041–1052 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Ti S.-C., Jurgenson C., Nolen B. J., Pollard T. D., Structural and biochemical characterization of two binding sites for nucleation promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 108, E463–E471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith B. A., Daugherty-Clarke K., Goode B. L., Gelles J., Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 110, 1285–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goley E. D., et al. , An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc. Natl. Acad. Sci. U.S.A. 107, 8159–8164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C., Atomic structure of the actin: DNase I complex. Nature 347, 37–44 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Belyy A., Merino F., Sitsel O., Raunser S., Structure of the Lifeact-F-actin complex. PLoS Biol. 18, e3000925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen K. K., Rubenstein P. A., Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J. Biol. Chem. 280, 24168–24174 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Wagner A. R., Luan Q., Liu S. L., Nolen B. J., Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. 23, 1990–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luan Q., Liu S. L., Helgeson L. A., Nolen B. J., Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. EMBO J. 37, e100005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narvaez-Ortiz H. Y., Nolen B. J., Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks. Curr. Biol. 32, 975–987 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beltzner C. C., Pollard T. D., Pathway of actin filament branch formation by Arp2/3 complex. J. Biol. Chem. 283, 7135–7144 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Zsolnay V., Katkar H. H., Chou S. Z., Pollard T. D., Voth G. A., Structural basis for polarized elongation of actin filaments. Proc. Nat. Acad. Sci. U.S.A. 117, 30458–30464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dayel M. J., Holleran E. A., Mullins R. D., Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. U.S.A. 98, 14871–14876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayel M. J., Mullins R. D., Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2, E91 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin A. C., Welch M. D., Drubin D. G., Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8, 826–833 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Martin A. C., et al. , Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J. Cell Biol. 168, 315–328 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingerman E., Hsiao J. Y., Mullins R. D., Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol. 200, 619–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolen B., Pollard T. D., Crystal structure and characterization of fission yeast Arp2/3 complex lacking Arp2. J. Biol. Chem. 283, 26490–26498 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLean-Fletcher S., Pollard T. D., Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96, 18–27 (1980). [DOI] [PubMed] [Google Scholar]
- 42.Palmgren S., Ojala P. J., Wear M. A., Cooper J. A., Lappalainen P., Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155, 251–260 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimanius D., Dong L., Sharov G., Nakane T., Scheres S. H. W., New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bepler T., et al. , Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen E. F., et al. , UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou S.C., Chatterjee M., Pollard T. D., Cryo-EM maps and mask. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-27962. Deposited 26 August 2022.
- 50.Chou S.C., Chatterjee M., Pollard T. D., Coordinates. Protein Data Bank. https://www.ebi.ac.uk/emdb/EMD-27962. Deposited 26 August 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Proposed pathway of branch formation. Step 1, inactive Arp2/3 complex is docked to the side of a mother filament using the interactions of the two globular domains of ARPC2 and the outer domain of Arp3 in the branch junction. Inactive Arp2/3 complex binds weakly and transiently (not shown). Step 2: Morphing from the structure of inactive Arp2/3 complex to its structure in the branch junctions shows the rotation of the two blocks of structure bringing Arp2 and Arp3 together in a short-pitch conformation and flattening both Arps. Step 3, the flattened Arps, aligned in the short-pitch conformation, nucleate the daughter filament.
Data Availability Statement
Electron microscopy structure maps and molecular models data have been deposited in Electron Microscopy Data Bank (EMDB) under accession number EMD-27962 (https://www.ebi.ac.uk/emdb/EMD-27962) (49) and Protein Data Bank (PDB) under accession number 8E9B (https://www.rcsb.org/structure/8E9B) (50).